- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Populations, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 3School of Life Sciences, Yunnan University, Kunming, China
- 4School of Integrative Plant Science, Section of Plant Biology and the L.H. Bailey Hortorium, Cornell University, Ithaca, NY, United States
- 5School of Life Sciences, Yunnan Normal University, Kunming, China
Accurate species delimitation and identification, which is a challenging task in traditional morphology-based taxonomy, is crucial to species conservation. Ottelia acuminata (Hydrocharitaceae) is a severely threatened submerged macrophyte endemic to southwestern China. The taxonomy of O. acuminata, which has long been in dispute, remains unresolved, impeding effective conservation and management practices. Here, we aim to address the long-standing issues concerning species boundary and intraspecific subdivision of O. acuminata using complete plastome sequences as super-barcodes. The taxonomic delimitation of O. acuminata was explored using phylogenetic inference and two independent sequence-based species delimitation schemes: automatic barcode gap discovery (ABGD) and multi-rate Poisson tree processes (mPTP). The reciprocally reinforcing results support the reduction of the closely related congeneric species, O. balansae and O. guanyangensis, as two conspecific varieties of O. acuminata. Within the newly defined O. acuminata, accurate varietal identification can be achieved using plastome super-barcodes. These findings will help inform future decisions regarding conservation, management and restoration of O. acuminata. This case study suggests that the use of plastome super-barcodes can provide a solution for species delimitation and identification in taxonomically difficult plant taxa, thus providing great potential to lessen the challenges of inventorying biodiversity, as well as biologically monitoring and assessing threatened species.
Introduction
Species are a fundamental unit of biodiversity (Claridge et al., 1997). Estimating how many species are under threat is an essential step in setting conservation priorities (May, 1988; May and Beverton, 1990; Margules and Pressey, 2000; Dirzo and Raven, 2003; Mace et al., 2003). Given that prioritizing species for natural conservation heavily depends on reliable species identification, often requiring prior species delimitation (Mace, 2004), accurate delineation of species boundaries is crucial for species conservation (Rojas, 1992; Sites and Crandall, 1997; Prance et al., 2000; Mace, 2004). Nevertheless, species delimitation is a challenging task in numerous plant taxa due to a lack of taxonomically robust morphologies, especially given that the majority of plant species are recognized based on morphological differences alone which can fail to discriminate cryptic species (Duminil and Di Michele, 2009). Recent general acceptance suggests that species should be recognized as evolutionarily distinct entities possessing significant morphological and evolutionary distinctiveness, or niche differentiation (de Queiroz, 1998; Wiley and Mayden, 2000; Sites and Marshall, 2003). This acceptance drove the development of a multidisciplinary approach that utilizes morphological, genetic, ecological, and even metabolomic data in discriminating species (Sites and Marshall, 2003; Duminil and Di Michele, 2009; Su et al., 2015; Eisenring et al., 2016; Cheng et al., 2020).
Analysis of DNA sequence variation can provide useful genetic information to develop robust species delimitation for the purpose of conservation and utilization (Hebert et al., 2003; Duminil et al., 2012; Puillandre et al., 2012b). DNA barcoding, a technique that involves the standardized use of one or a few DNA regions (DNA barcodes) for identification and discrimination of species (Hebert et al., 2003; Kress et al., 2005; Hollingsworth, 2011; Hollingsworth et al., 2011, 2016), has proven useful in facilitating species delimitation (e.g., Duminil et al., 2012; Puillandre et al., 2012a; Kekkonen and Hebert, 2014; Mutanen et al., 2015; Hausmann et al., 2016). Nevertheless, the efficacy of standard DNA barcodes (i.e., rbcL, matK, trnH-psbA, and ITS) in either identification or delimitation of plant species remains problematic, especially in recently diverged or rapidly radiating taxa (Hollingsworth et al., 2009, 2011, 2016; Hollingsworth, 2011; Coissac et al., 2016). Benefiting from the development and advancement of next-generation DNA sequencing (NGS) technologies, genome-wide sequence data are increasingly used as extended DNA barcodes for species identification and delimitation, providing a possible solution for credibly delineating species boundaries in plants, especially in taxonomically perplexing taxa (Coissac et al., 2016; Hollingsworth et al., 2016).
The complete plastid genomes (plastomes) possess more variable loci by orders of magnitude than standard DNA barcodes and thus have great potential to improve resolution in species discrimination (Nock et al., 2011; Kane et al., 2012; Ruhsam et al., 2015). Additionally, they are highly repetitive genome components in each plant cell, making plastome assembly feasible via a relatively shallow sequencing depth (Straub et al., 2012). Due to these advantages, complete plastome DNA sequences have been recommended for consideration as “super-barcodes” for plant species discrimination and delimitation (Dodsworth, 2015; Li et al., 2015). Several recent studies attempted to use plastome super-barcodes to decipher species boundaries in a wide spectrum of plant lineages (e.g., Firetti et al., 2017; Ji et al., 2019, 2020; Zhu et al., 2019; Li L. et al., 2020; Ślipiko et al., 2020). However, most of the studies inferred tree topology solely under the premise of reciprocal monophyly to explore species boundaries, thus likely producing biased delimitation schemes. In view of this, empirical studies that employ multiple delimitation methods are urgently needed to evaluate the usefulness of plastome super-barcodes in species delimitation especially for conservation and management purposes.
Ottelia Persoon (Hydrocharitaceae) is a pantropic genus with ∼21 species of submerged macrophytes primarily distributed in tropic Africa and southeast Asia, according to the most comprehensive taxonomic revision of the genus (Cook et al., 1983). However, recent studies based on DNA sequence data reveal that two widespread species within the genus, namely Ottelia alismoides (Ito et al., 2019) and O. ulvifolia (Li et al., 2020b), contain cryptic species, suggesting that the alpha taxonomy of Ottelia remains ambiguous. Given that the majority of species within the genus are threatened with local or global extinction (Phillips et al., 2016; Zhang et al., 2017), a credible taxonomy is necessary for conservation and management of extant Ottelia species.
Ottelia acuminata, a severely threatened submerged macrophyte, consisting of six phenotypic varieties, is an endemic species occurring in freshwater lakes, ponds and rivers in Southwest China (Jiang et al., 2005; Wang et al., 2010). This species is categorized as vulnerable (VU) under the criteria “A2c” in the China Species Red List (Qin et al., 2017), and many wild populations have deteriorated or even perished during the past 30 years due to habitat degradation, anthropogenic disturbances, and introduction of herbivorous fish (Li, 1985, 1988; Godo et al., 2003; Liang and Li, 2007; Jiang et al., 2010; Yang et al., 2012). Remarkably, the taxonomic delimitation of O. acuminata as well as its varieties (O. acuminata var. acuminata, O. acuminata var. crispa, O. acuminata var. jingxiensis, O. acuminata var. lunanensis, O. acuminata var. songmingensis, and O. acuminata var. tonghaiensis) remains controversial. For instance, O. acuminata var. crispa recognized by Li (1981) was treated as a separate species, O. crispa, by Dandy (1935) and Wang (1986). Ottelia acuminata var. lunanensis was accepted as a variety by Li (1981) and Wang et al. (2010), while Cook et al. (1983) reduced it to a synonym of O. acuminata var. acuminata. Despite O. acuminata var. tonghaiensis being described as a variety by Li (1981), Wang et al. (2010) combined it with O. acuminata var. acuminta. Additionally, genus-level phylogenetic analyses fail to resolve O. acuminata as a monophyletic unit (Li et al., 2020c). Therefore, the taxonomic delimitation of O. acuminata and its phenotypic varieties needs to be re-evaluated.
In this study, we aim to clarify the long-standing controversies in species delimitation and intraspecific subdivision of O. acuminata using plastome super-barcodes. We employed a genome skimming approach (Straub et al., 2012) to generate complete plastome DNA sequences as well as sampling multiple accessions of each varieties within the species. Under a phylogenetic framework, we first examined whether O. acuminata and the phenotypic varieties are monophyletic entities. Next, we used two independent molecular species delimitation methods to explore the species boundaries of O. acuminata and closely related species. The results suggest that the species boundary of O. acuminata should be expanded to accommodate O. balansae and O. guanyangensis as two conspecific varieties. The newly circumscribed O. acuminata, including the conspecific varieties, can be distinguished with plastome super-barcodes. The findings will help inform future decisions regarding conservation, management and restoration of O. acuminata. Inferred from this case study, we discuss the perspectives on the use of plastome super-barcodes in plant species conservation.
Materials and Methods
Taxon Sampling
Among the six phenotypic varieties of O. acuminata, O. acuminata var. tonghaiensis was not found after many field explorations within the areas of known distribution and is most likely extinct. For the remaining varieties, our field collections approximately cover all known populations. According to the phylogeny recovered by Li et al. (2020c), we also included O. alismoides, O. balansae, and O. guanyangensis in phylogenetic and species delimitation analyses to investigate their relationships to O. acuminata, and to explore taxonomic boundaries among these taxa. In total 60 accessions (Figure 1 and Table 1), representing O. acuminata var. acuminata (18 accessions from 3 populations), O. acuminata var. crispa (8 accessions from 1 populations), O. acuminata var. jingxiensis (15 accessions from 3 populations), O. acuminata var. lunanensis (3 accessions from 1 populations), O. acuminata var. songmingensis (2 accessions from 1 population), O. alismoides (2 accessions from 1 populations), O. balansae (9 accessions from 4 populations), and O. guanyangensis (3 accessions from 1 populations) were sampled from wild populations. The sampling size of these taxa was determined according to their distribution range and population size. For those taxa possessing narrowly restricted distribution and extremely small population size, such as O. acuminata var. lunanensis, O. acuminata var. songmingensis, and O. guanyangensis, we tried to sample at least two individuals per taxon. The extensive sampling strategy adopted in this study allows a robust test for the current taxonomic designation of the target taxa.
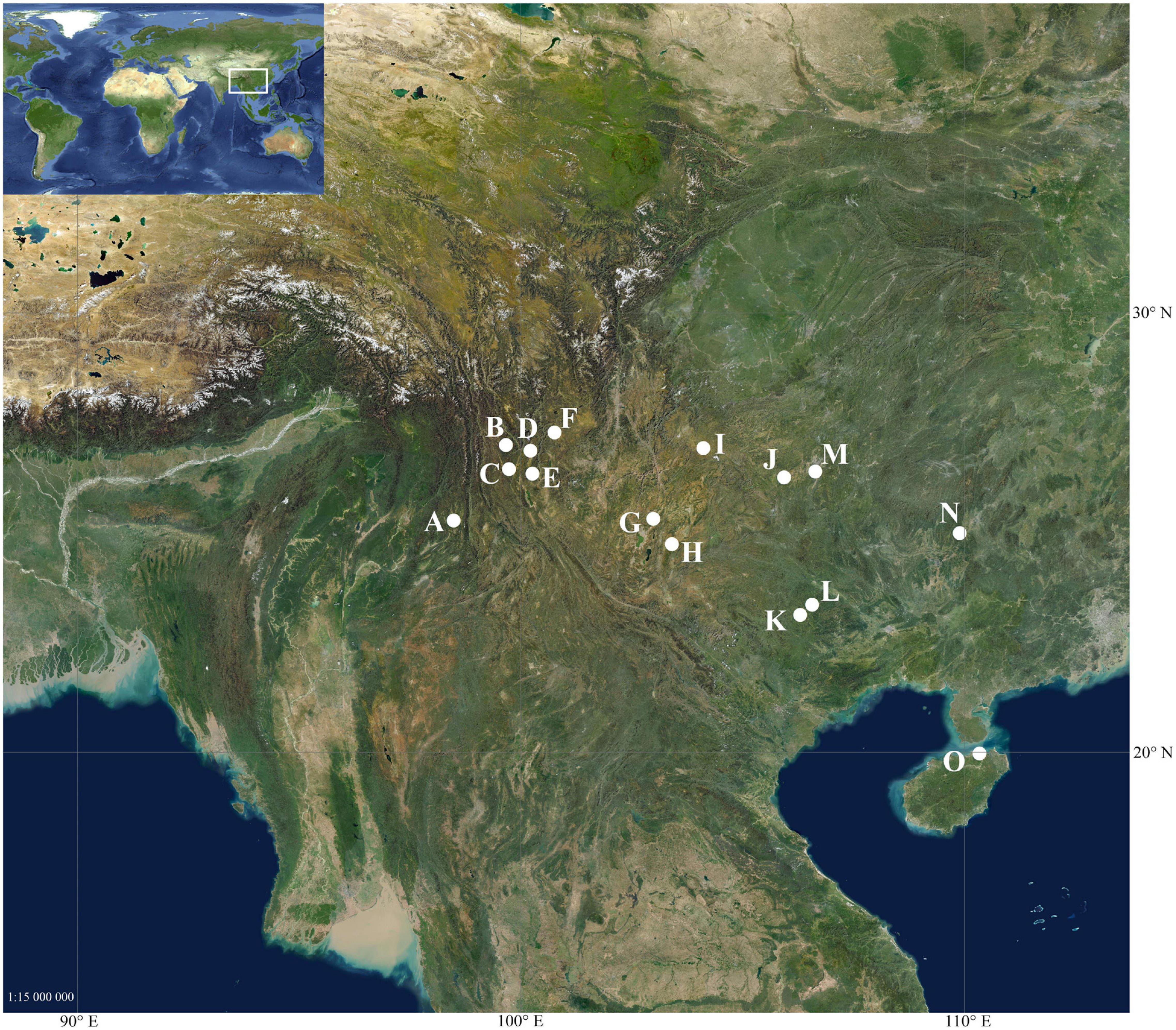
Figure 1. Geographic distribution of sampled populations. (A) Beihai Lake, Tengchong, Yunnan, China (25.276° N, 98.494° E); (B) Qixian Lake, Yulong, Yunnan China (26.979° N, 99.671° E); (C) Jian Lake, Jianchuan, Yunnan, China (26.436° N, 99.743° E); (D) O. Lashihai Lake, Lijiang, Yunnan, China (26.855° N, 100.226° E); (E) Heilongtan, Heqing, Yunnan, China (26.334° N, 100.270° E); (F) Lugu Lake, Ninglang, Yunnan, China (27.263° N, 100.770° E); (G) Heilongtan, Songming, Yunnan, China (25.314° N, 102.998° E); (H) Changhu Lake, Shilin, Yunnan, China (24.744° N, 103.417° E); (I) Caohai Lake, Weining, Guizhou, China (26.910° N, 104.129° E); (J) Anshun, Guizhou, China (26.253° N, 105.948° E); (K) Equan River, Jingxi, Guangxi; China (23.146° N, 106.311° E); (L) Longtan River, Debao, Guangxi, China (23.379° N, 106.583° E); (M) Huaxi, Guiyang, Guizhou, China (26.384° N, 106.654° E); (N) Baishou River, Yongfu, Guangxi, China (24.993° N, 109.907° E); (O) Haikou, Hainan, China (20.018° N, 110.349° E).
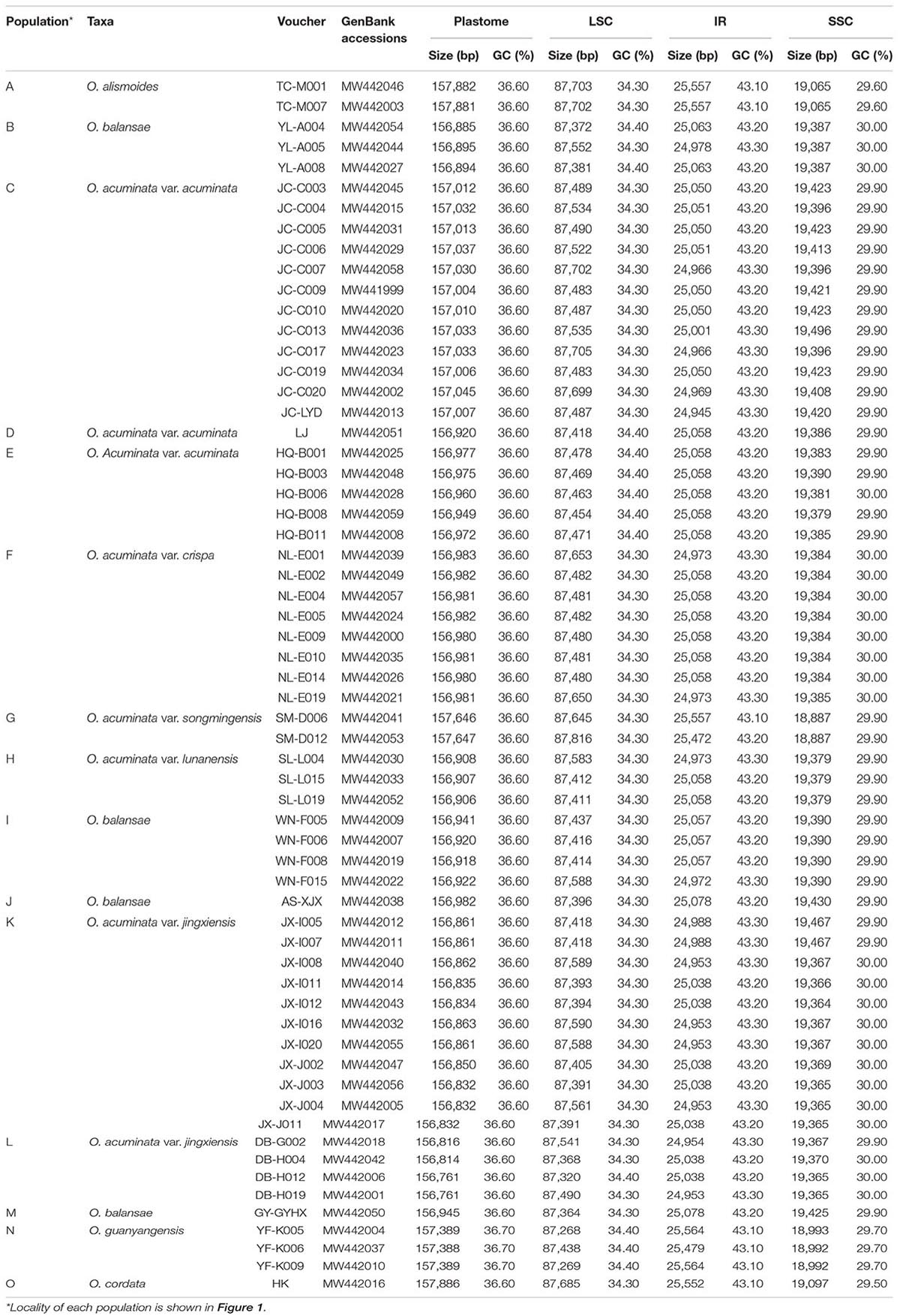
Table 1. Samples used in this study with population, voucher, GenBank accessions, and plastome features.
Shotgun Sequencing, Plastome Assembly and Annotation
Total genomic DNA for each accession was isolated from ∼20 mg silica gel dried leaf tissues using the cetyltrimethylammonium bromide method of Doyle and Doyle (1987). Approximately 5 μg of purified genomic DNA was used to construct paired-end libraries with a TruSeq DNA Sample Prep Kit (Illumina, Inc., San Diego, CA, United States) following the manufacturer’s instructions. Shotgun sequencing was performed on the Illumina HiSeq 2500 system with 2 × 150 reads. Raw reads were subjected to the NGS QC Toolkit (Patel and Jain, 2012) to remove adaptors and low-quality reads with the default parameters.
Using the filtered reads, de novo assembly of complete plastome was performed by NOVOPlasty v2.7.0 (Dierckxsens et al., 2017) with k–mer of 31, and using the large subunit of RuBisCO gene of O. acuminata (HM257638) as the seed for iterative extension of contigs to recover the whole plastome of each accession. The newly assembled plastomes were annotated with the Dual Organellar Genome Annotator database (Wyman et al., 2004). The annotation of protein-coding genes was further confirmed with a BLAST search against the NCBI protein database. Genes putatively annotated as transfer RNA (tRNA) were further verified by tRNAscan-SE 1.21 (Schattner et al., 2005) with default parameters. The boundary of the large-single copy (LSC), small-single copy (SSC), and inverted-repeat (IR) regions for each plastome were visually examined and manually adjusted with Geneious V10.2.3 (Kearse et al., 2012).
Phylogenetic Analyses
Based on inferred phylogenetic trees, we tested whether O. acuminata and allied species (O. alismoides, O. balansae, and O. guanyangensis) are evolutionarily distinctive entities. The complete plastome sequences were aligned using MAFFT v7.450 (Katoh and Standley, 2013) with manual adjustment where necessary. Phylogenetic trees were reconstructed using both maximum likelihood (ML) and Bayesian inference (BI) methods. Ottelia cordata, the closest relative of the ingroup (Li et al., 2020c), was used to root the phylogenetic tree. The best-fit sequence substitution model for complete plastomes (GTR + G) was selected using MODELTEST v3.7 (Posada and Crandall, 1998) with the Akaike information criterion (Posada and Buckley, 2004).
Maximum likelihood analyses were performed using RAxML-HPC BlackBox v8.1.24 (Stamatakis, 2006). The best-scoring ML tree was generated with 1,000 bootstrap (BS) replicates to obtain branch support. BI reconstructions were conducted using MrBayes v3.2 (Ronquist and Huelsenbeck, 2003). Two independent Markov Chain Monte Carlo runs were performed with 1,000,000 generations, sampling every 100 generations. An initial 25% of the sampled trees were discarded as burn-in. Posterior probability (PP) values were computed based on the remaining trees. Stationarity was considered to be reached when the average standard deviation of the split frequencies was <0.01.
Sequence-Based Species Delimitation
There has been an explosion in molecular-based species delimitation approaches over the past 15 years (e.g., Yang and Rannala, 2010; Ence and Carstens, 2011; Masters et al., 2011; Puillandre et al., 2012b; Rannala and Yang, 2013; Zhang et al., 2013). As revealed by previous studies, using different approaches simultaneously to delineate species boundary allows the methods to compensate for each other’s weaknesses (e.g., Hebert et al., 2003; Kekkonen and Hebert, 2014; Mutanen et al., 2015). Based on the alignments of complete plastomes sequences, we used two sequence-based species delimitation tools to estimate the number of species-like units among O. acuminata and related taxa. The first was a distance-based method, automatic barcode gap discovery (ABGD), which statistically infers the barcode gap from the sequence data and clusters sequences into putative species based on the pairwise distances among group of individuals (Puillandre et al., 2012a). The ABGD analyses were conducted on the online server1 using three different distance models (JC69, K2P, and P-distances) with default settings (Pmin = 0.001, Pmax = 0.1, Steps = 10, X = 1.5, and Nb bins = 20). All assignments for intraspecific divergence (P) values between 0.0001 and 0.0100 were recorded. Next, we used the tree-based methods: multi-rate Poisson tree processes model (mPTP; Kapli et al., 2017) to explore putative species boundaries of the target species. Based on the phylogenetic species concept, this method uses nucleotide substitutions along the branches of the phylogenetic tree to determine putative species boundaries under the assumption that the number of intraspecific substitutions is smaller than that of interspecific substitutions, with both substitutions following a distinct Poisson distribution (Zhang et al., 2013). The mPTP v0.2.3 algorithm (Kapli et al., 2017), an improvement to PTP (Zhang et al., 2013), was run on the web server2 with standard default settings, using the ML trees of complete plastome as inputs, because the branch lengths of ML tree represent number of sequence mutations.
Results
Shotgun Sequencing and Plastome Assembly
Based on cleaned shotgun reads, de novo assembly generated the complete plastome of all samples. These newly sequenced plastomes possess a typical quadripartite structure, with the sequence length varying from 156,761–157,886 bp, containing a pair of inverted repeats (IRs; 24,945–25,564 bp) separated by the LSC (87,268–87, 816 bp) and SSC (18,887–19,496 bp) regions (Figure 2 and Table 1). The gene content of each plastome includes 114 genes, including 79 protein-coding genes, 30 tRNA genes, and four plastid rRNA genes (Supplementary Table 1).
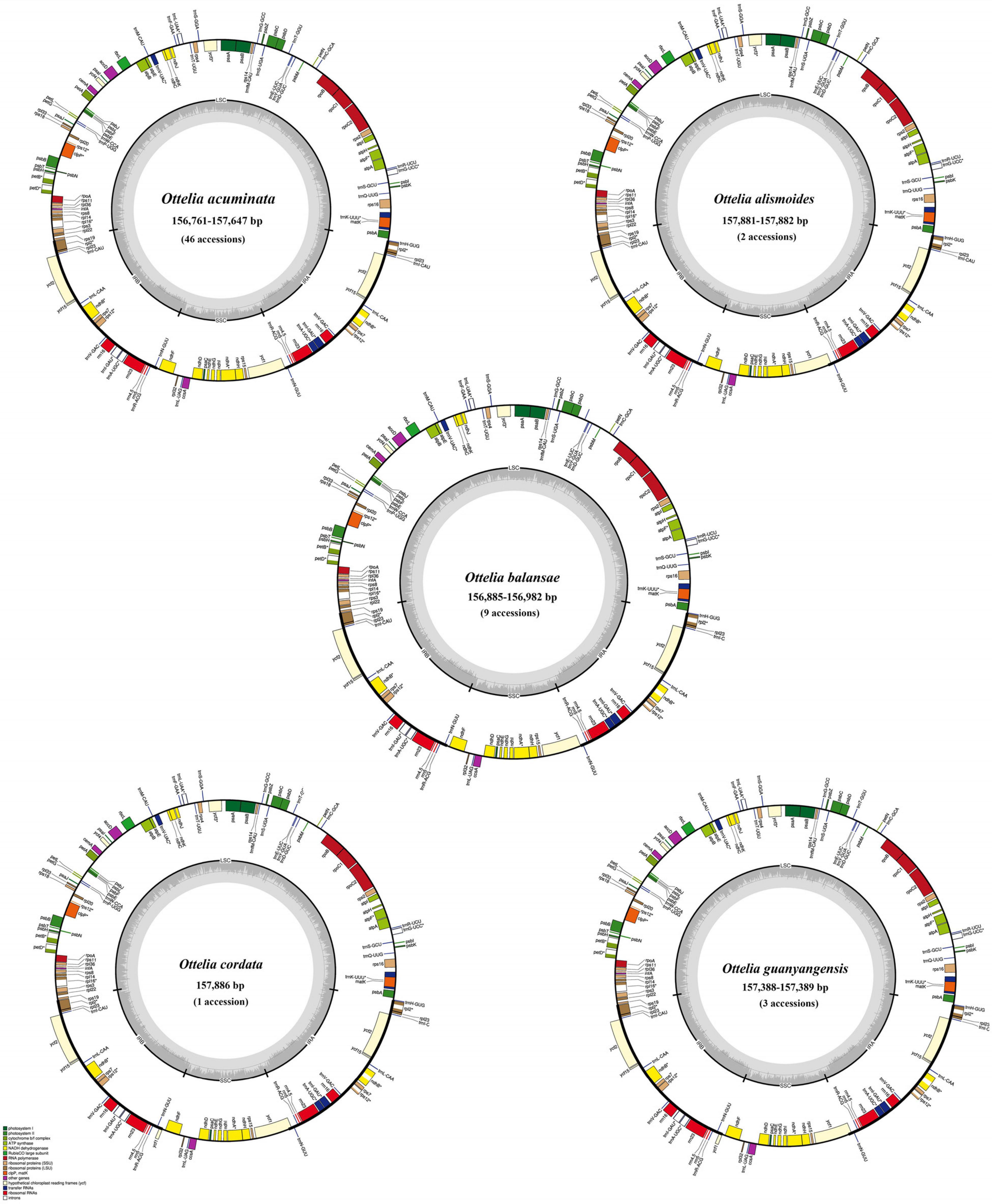
Figure 2. Map of Ottelia plastomes. Genes shown outside the circle are transcribed clockwise, and those inside are transcribed counterclockwise.
Phylogenetic Reconstruction
Alignment of the plastome sequences yielded a matrix of 159,782 positions, in which 1,818 variable sites (1.14%) were identified and 1,745 (1.09%) were parsimoniously informative. ML and BI analyses of complete plastomes produced identical tree topologies (Figure 3). Overall, all five varieties of O. acuminata (O. acuminata var. acuminata, O. acuminata var. crispa, O. acuminata var. jingxiensis, O. acuminata var. lunanensis, and O. acuminata var. songmingensis), as well as O. alismoides, O. balansae, and O. guanyangensis were resolved as well-supported monophyletic entities. The monophyly of O. acuminata as a single species, however, was not supported by either ML or BI phylogeny. Among the taxa, O. alismoides (Clade I) was sister to the clade that included the remaining taxa (Clade II). Within Clade II, O. acuminata var. songmingensis and O. guanyangensis formed the earliest diverging branch (BS = 100%, PP = 1.00), and the remaining taxa grouped into two well-supported subclades. The first one comprised accessions of O. acuminata var. acuminata and O. acuminata var. crispa. Within the second one, O. acuminata var. jingxiensis was sister to O. acuminata var. lunanensis, and these two taxa, in turn, were sister to O. balansae.
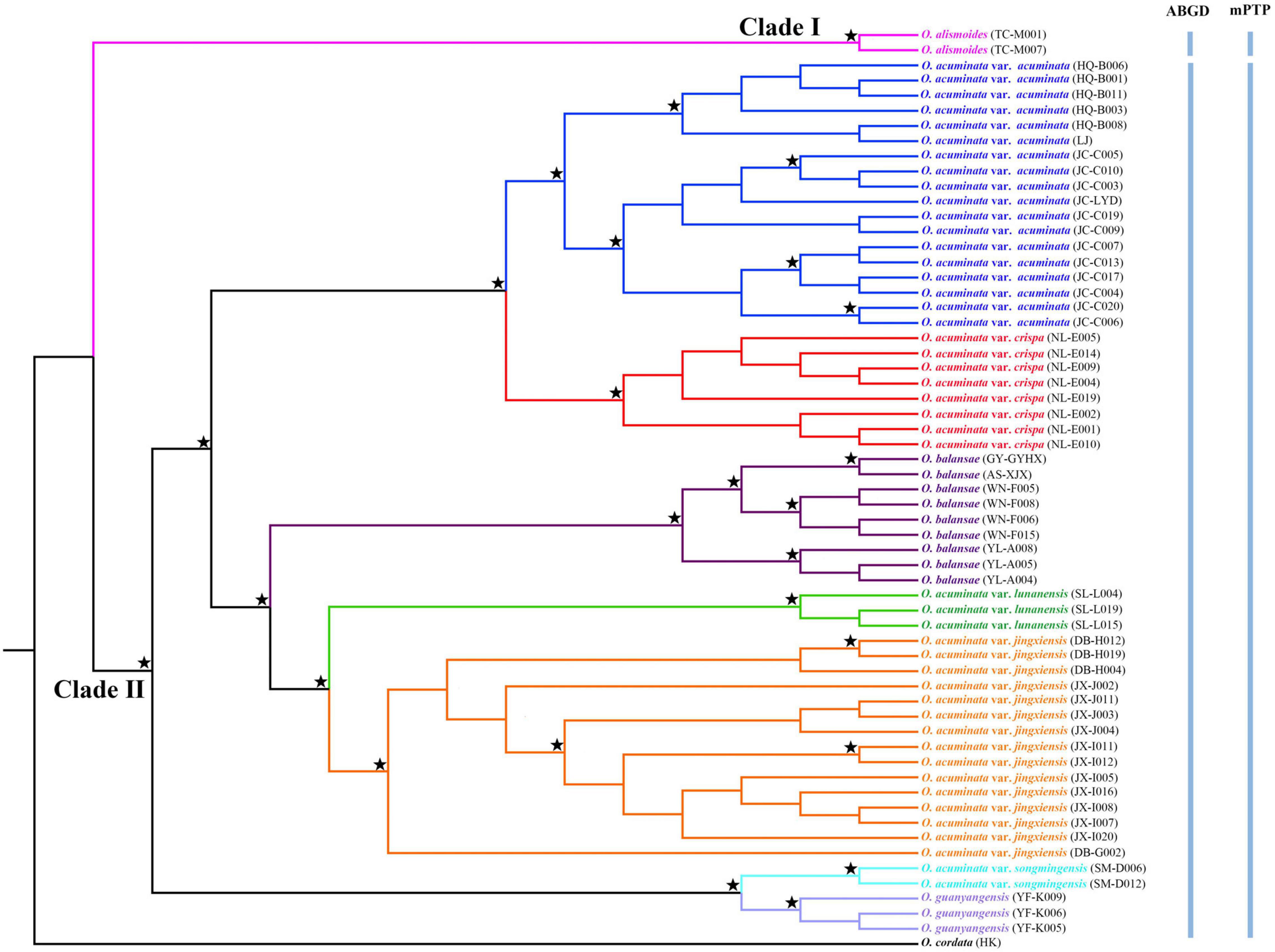
Figure 3. Phylogenetic relationships between Ottelia acuminata and closely related species based on Maximum likelihood (ML) and Bayesian inference (BI) of complete plastomes. Well-supported nodes (BS > 90% and PP > 0.95) are indicated with a star. Species delimitation schemes proposed by automatic barcode gap discovery (ABGD) and multi-rate Poisson tree processes model (mPTP) are reflected on the tree topology.
Species Delimitation
Automatic barcode gap discovery and mPTP analyses of complete plastome sequences produced highly congruent results that are reflected in the inferred phylogenetic tree (Figure 3). The ABGD analyses (Table 2) resulted in a consistent count of species division (n = 2) with a range of prior intraspecific values (P = 0.0046–0.00836) using JC69, K2P, and P-distances with initial and recursive partitions. One putative species includes individuals of O. alismoides, while the other comprises individuals of O. balansae, O. guanyangensis and O. acuminata (Figure 3). The mPTP analyses yielded the same delimitation scheme as ABGD did: all individuals were grouped into two species-like units that coincide with the two putative species proposed by ABGD analyses (Figure 3), with both of them receiving high posterior support (PP = 1.00).
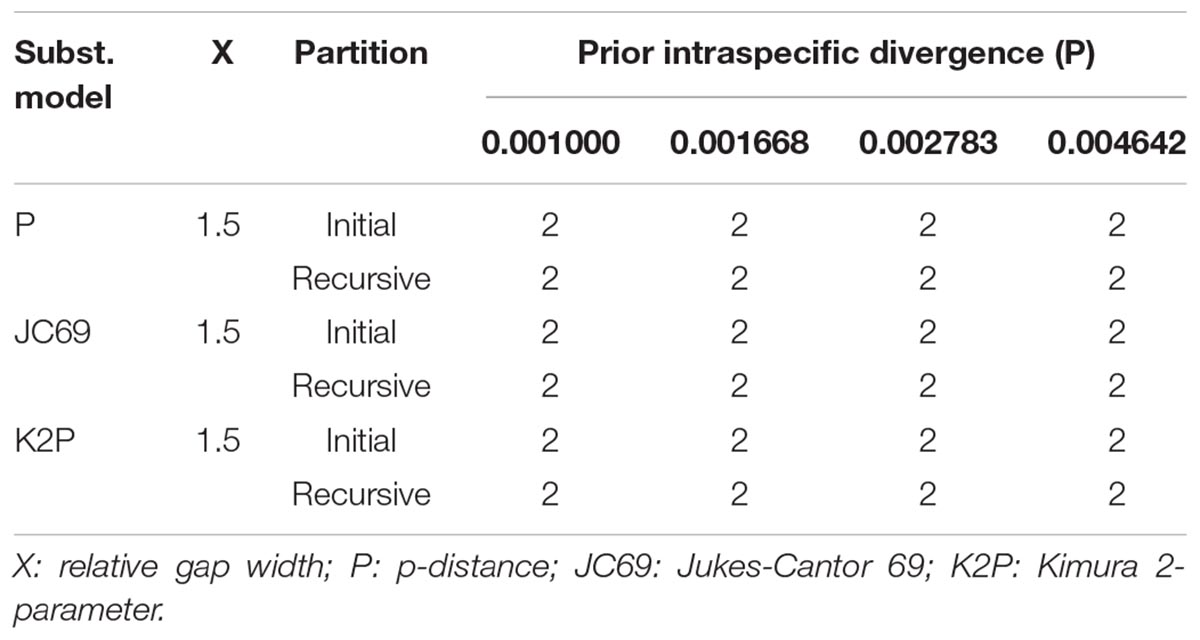
Table 2. The number of putative species recognized by Automatic Barcode Gap Discovery (ABGD) analyses of complete plastome sequences using three distance metrics.
Discussion
Taxonomic Delimitation of Ottelia acuminata and Conspecific Varieties
In this study, a sampling strategy that included multiple individuals within a species (or variety) representing different localities (if any) was employed to test for the evolutionary distinctiveness of each taxon. Additionally, two independent delimitation methods (mPTP and ABGD) were used to explore species delimitation, which allows the methods to compensate for weaknesses of the other method to develop a robust taxonomic delimitation framework (e.g., Hebert et al., 2003; Kekkonen and Hebert, 2014; Mutanen et al., 2015). Similar to an earlier study that did not include multiple individuals per taxon in a phylogenetic context (Li et al., 2020c), our plastome-based phylogeny failed to resolve O. acuminata as a monophyletic unit given two congeneric relatives (O. balansae and O. guanyangensis) were embedded within O. acuminata in the tree topology. This implies that the three species may have been incompletely separated from each other, thus recognizing them as distinct species may not hold (de Queiroz, 1998, 2007). This inference is justified by the results of the species delimitation analyses. Specifically, the ABGD analyses partitioned all the samples into two species-level entities corresponding to the two clades recovered by phylogenetic analyses, suggesting that Clade I (O. alismoides) and Clade II (O. acuminata + O. balansae + O. guanyangensis) represent two distinct species with significant genetic gaps between them (Puillandre et al., 2012b). Moreover, the mPTP analyses grouped all accessions into two putative species with high delimitation posterior probability, coinciding with the results found in the ABGD analyses. These reciprocal reinforcing results suggest that only two species-level taxonomic units that, respectively, correspond to the O. alismoides clade and the O. acuminata + O. balansae + O. guanyangensis clade can be recognized.
A previous study (Li et al., 2020c) suggests that the divergence between O. alismoides and the O. acuminata + O. balansae + O. guanyangensis clade occurred at ∼6.01 million years ago (Mya). The long-term differentiation developed a high degree evolutionary independence between them. As shown by phylogenetic analysis, they are two completely separated lineages in the tree topologies. Given that they reflect the unity of morphological uniqueness, genetic distinctiveness, and evolutionary independence, it is reasonable to recognized them as distinct species under the unified species concept (de Queiroz, 1998, 2007). Comparatively, it is indicated that the stem age of the O. acuminata + O. balansae + O. guanyangensis clade are more recent at ∼3.88 Mya (Li et al., 2020c). This implies that these taxa may have incompletely separated from each other due to their relatively short evolutionary histories. The speculation is justified by our phylogenetic analyses and sequence-based species delimitation schemes, which consistently indicate that O. balansae and O. guanyangensis are neither evolutionarily nor genetically distinct from O. acuminata. Under the updated concept of subspecies that recognizes incompletely separated lineages within a more inclusive lineage as subspecific taxa (de Queiroz, 2020), it is reasonable to reduce O. balansae and O. guanyangensis as conspecific varieties of O. acuminata.
On the other hand, O. balansae and O. guanyangensis share high levels of similarity in leaf, spathe, sepal, petal, and fruit morphologies with O. acuminata, but differ from the latter species in having bisexual flowers (versus unisexual flowers in O. acuminata) (Wang et al., 2010; Liu et al., 2018). However, it has been reported that there are scattered individuals with bisexual flowers in natural O. acuminata populations (Li, 1981; Jiang et al., 2010), and our field observations found that most individuals of O. balansae and O. guanyangensis produce unisexual flowers and only few individuals have bisexual flowers in wild populations. This suggests that the diagnostic character (bisexual versus unisexual flowers) used to distinguish O. balansae and O. guanyangensis from O. acuminata can be a plastic trait. Therefore, prior morphology-based taxonomic studies overemphasized intraspecific morphological differences to establish species, thus leading to taxonomic over-splitting of species. As a result, it is reasonable to merge O. balansae and O. guanyangensis into O. acuminata.
The lack of continuous water systems among lakes, ponds, and rivers in southwest China, which severely restricted pollen and seed dispersal among fragmented populations, may have led to significant isolation events in O. acuminata (Zhang et al., 2009; Long et al., 2010; Chen et al., 2017; Guo et al., 2019). The limited gene flow among isolated populations would result in significant genetic differentiations in O. acuminata, and thus triggered the formation of diverse conspecific varieties (Chen et al., 2017). Although there is no significant morphological difference between these phenotypic varieties (Li et al., 2020c), they were identically resolved as well-supported monophyletic units by our complete plastome-based phylogenies. This suggests the genetic boundaries among these taxa are large enough to ensure accurate varietal identification using complete plastome DNA sequences as super-barcodes. Nevertheless, previous studies revealed that these varieties identically harbor low genetic diversity and weak population genetic differentiation (Zhang et al., 2009; Long et al., 2010; Chen et al., 2017; Zhai et al., 2018; Guo et al., 2019). Moreover, except for O. acuminata var. acuminata, O. acuminata var. jingxiensis, and O. balansae, the remaining taxa occur in only a single population and possess extremely small population size (Li, 1985; Jiang et al., 2005, 2010; Wang et al., 2010). From this perspective, they are more likely to represent genetically differentiated and geographically isolated intraspecific populations than separately evolving metapopulation lineages. Taken together, these findings provide good support to our taxonomic proposal that recognizes O. acuminata var. acuminata, O. acuminata var. crispa, O. acuminata var. jingxiensis, O. acuminata var. lunanensis, and O. acuminata var. songmingensis as distinct varieties, and reduces O. balansae and O. guanyangensis as conspecific varieties of O. acuminata.
Conservation Implications
Aquatic macrophytes are non-negligible targets for conservation management since many species have become locally or even globally extinct during the past decades due to water pollution, eutrophication, changes in hydrological regime, and biological invasion (Sand-Jensen et al., 2000; Phillips et al., 2016; Zhang et al., 2017). Credible delineation of species boundaries is an essential step in species conservation (Hopkins and Freckleton, 2002; Sites and Marshall, 2003; Mace, 2004). It is generally accepted that well delimited species are fundamental to allow adequate conservation and biodiversity management (Mace, 2004). Contrarily, poor delineation of species boundaries usually makes it difficult to properly address conservation issues. For instance, species delimitations that are too broad will result in the underestimation of true species richness and improper assignation of conservation priorities because some threatened species are overlooked (Rojas, 1992; Sites and Crandall, 1997; Prance et al., 2000; Mace, 2004), while taxonomic over-splitting of species may lead to the misallocation of limited conservation resource to widespread species that are not at risk of extinction (Agapow et al., 2004; Mace, 2004; Joppa et al., 2011). In this study, analyses of plastome super-barcodes develop a clear-cut taxonomic delimitation of O. acuminata and its conspecific varieties, which will help to better inform future decisions regarding the conservation and management of this severely threatened submerged macrophyte.
Based on our data, several strategies can be proposed for the conservation and restoration of O. acuminata. Although the species as a whole is not at a high risk of extinction due to its relatively wide distribution range and large population size, it is highly relevant for conservation since the conspecific varieties possess high level of evolutionary and genetic distinctiveness with mostly small distributions. The preservation of genetic diversity and evolutionary potential is a primary goal for the conservation of threatened species (Milligan et al., 1994; Margules and Pressey, 2000). The conservation strategy for O. acuminata should be aimed at preserving all its conspecific varieties because they represent an indispensable evolutionary legacy. Among them, O. acuminata var. crispa, O. acuminata var. lunanensis, and O. acuminata var. songmingensis are recommended here as the prioritized taxa for conservation, because they possess a particularly narrow distribution and occur in only one lake or pond (Li, 1985; Jiang et al., 2005, 2010; Wang et al., 2010) thus are more vulnerable to environmental changes and anthropogenic disturbances (Mckinney, 1997; Henle et al., 2004).
Extant populations of O. acuminata are facing severe threats, such as habitat degradation, anthropogenic disturbances, and introduction of herbivorous fish (Li, 1985, 1988; Godo et al., 2003; Liang and Li, 2007; Jiang et al., 2010; Yang et al., 2012). In addition to in situ conservation, it is necessary to establish full-scale germplasm collections for ex situ conservation (Heywood and Iriondo, 2003). Due to the significant evolutionary and genetic distinctiveness between O. acuminata varieties, their germplasms need to be separately collected and propagated to prevent mixing divergent gene pools. Moreover, for restoration of natural O. acuminata populations, the reciprocal introduction and reintroduction between different varieties should be avoided, as this action may lead to outbreeding that may change the gene pool of locally adapted populations (Fischer and Lindenmayer, 2000; Edmands, 2007). Therefore, there is considerable need to identify O. acuminata varieties and to characterize their germplasms for conservation purposes. Our data show that the use of plastome super-barcodes meet this requirement, thus serving as a useful tool for proper conservation, management, and restoration of O. acuminata.
Perspectives on the use of Plastome Super-Barcodes in Plant Species Conservation
Taxonomy and biodiversity conservation are interdependent practices (Mace, 2004). On global and regional scales, a taxonomic diversity inventory and estimates of the number of species that are under threat are essential for managing and conserving biodiversity (May, 1988; May and Beverton, 1990; Margules and Pressey, 2000; Mace et al., 2003). Recently, DNA barcoding has been widely used as a rapid and cost-effective tool for biodiversity inventory and for monitoring and assessment of threatened species (reviewed by Kress et al., 2015; Hollingsworth et al., 2016; Wilson et al., 2016). Nevertheless, the use of standard DNA barcodes may result in biased estimates of species diversity and ambiguous species identification due to insufficient performance in discriminating plant species, especially in lineages that have experienced rapid radiations or complicated evolutionary histories (Hollingsworth et al., 2009, 2011, 2016; Hollingsworth, 2011; Coissac et al., 2016). As revealed by previous studies (e.g., Kane et al., 2012; Ruhsam et al., 2015; Firetti et al., 2017; Ji et al., 2019, 2020; Zhu et al., 2019; Li et al., 2020; Ślipiko et al., 2020) and our data, the use of plastome super-barcodes performs well in species identification and delimitation as well as in discovery of cryptic or overlooked diversity. This tool has great potential to lessen the challenges of biodiversity inventory and setting conservation priorities for threatened species.
Currently, complete plastomes of most plants can be easily obtained through a relatively low coverage shotgun sequencing of genomic DNA (Straub et al., 2012). Compared with Restriction-site Associated DNA sequencing (Baird et al., 2008), another recommended technique for plant barcoding 2.0 (Hollingsworth et al., 2016), a promising advantage of using plastome super-barcodes for species identification and delimitation is its universality (Kane et al., 2012). Additionally, with the advances of NGS technology, it has become much easier to generate complete plastome sequences even with using trace and highly degraded genomic DNA to prepare shotgun libraries (Zeng et al., 2018), making it feasible to catalog species diversity and to monitor threatened plants with herbarium specimens and other plant products. With the plastomes of a wide spectrum of plant lineages increasingly available in public database (e.g., NCBI GenBank), the plastome super-barcode reference libraries of known taxa are constantly enriched. We are optimistic that the plastome super-barcoding approach will likely to product more information of conservation values.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
YJ and YZ conceived the research. YJ, JY, SW, and ZY collected and analyzed the data. YJ wrote the manuscript. JL discussed the results and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was granted by Science and Technology Basic Resources Investigation Program of China (Survey and Germplasm Conservation of Plant Species with Extremely Small Populations in South-west China, No. 2017FY100100), and a grant of the Large-scale Scientific Facilities of the Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.681270/full#supplementary-material
Footnotes
References
Agapow, P. M., Bininda-Emonds, O. R., Crandall, K. A., Gittleman, J. L., Mace, G. M., Marshall, J. C., et al. (2004). The impact of species concept on biodiversity studies. Q. Rev. Biol. 79, 161–179. doi: 10.1086/383542
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3:e3376. doi: 10.1371/journal.pone.0003376
Chen, J. M., Du, Z. Y., Long, Z. C., Gichira, A. W., and Wang, Q. F. (2017). Molecular divergence among varieties of Ottelia acuminata (Hydrocharitaceae) in the Yunnan-Guizhou Plateau. Aquat. Bot. 140, 62–68. doi: 10.1016/j.aquabot.2017.03.001
Cheng, Z., Shu, H., Zhang, S., Luo, B., Gu, R., Zhang, R., et al. (2020). From folk taxonomy to species confirmation of Acorus (Acoraceae): evidences based on phylogenetic and metabolomic analyses. Front. Plant Sci. 11:965. doi: 10.3389/fpls.2020.00965
Claridge, M. F., Dawah, H. A., and Wilson, M. R. (1997). Species: The Units of Biodiversity. Q. Rev. Biol. 73:529. doi: 10.1086/420496
Coissac, E., Hollingsworth, P. M., Lavergne, S., and Taberlet, P. (2016). From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 25, 1423–1428. doi: 10.1111/mec.13549
Cook, C. D. K., Symoens, J. J., and Urmi-König, K. (1983). A revision of the genus Ottelia (Hydrocharitaceae). I. Generic considerations. Aquat. Bot. 18, 263–274. doi: 10.1016/0304-3770(84)90068-8
de Queiroz, K. (1998). “The general lineage concept of species, species criteria, and the process of speciation,” in Endless Forms: Species and Speciation, eds D. J. Howard and S. H. Berlocher (Oxford: Oxford University Press), 57–75.
de Queiroz, K. (2020). An updated concept of subspecies resolves a dispute about the taxonomy of incompletely separated lineages. Herpetol. Rev. 51, 459–461.
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955
Dirzo, R., and Raven, P. H. (2003). Global state of biodiversity and loss. Annu. Rev. Environ. Resour. 28, 137–167.
Dodsworth, S. (2015). Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 20, 525–527. doi: 10.1016/j.tplants.2015.06.012
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Duminil, J., and Di Michele, M. (2009). Plant species delimitation: a comparison of morphological and molecular markers. Plant Biosyst. 143, 528–542. doi: 10.1080/11263500902722964
Duminil, J., Kenfack, D., Viscosi, V., Grumiau, L., and Hardy, O. J. (2012). Testing species delimitation in sympatric species complexes: the case of an African tropical tree, Carapa spp. (Meliaceae). Mol. Phylogenet. Evol. 62, 275–285. doi: 10.1016/j.ympev.2011.09.020
Edmands, S. (2007). Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16, 463–475. doi: 10.1111/j.1365-294X.2006.03148.x
Eisenring, M., Altermatt, F., Westram, A. M., and Jokela, J. (2016). Habitat requirements and ecological niche of two cryptic amphipod species at landscape and local scales. Ecosphere 7:e01319. doi: 10.1002/ecs2.1319
Ence, D. D., and Carstens, B. C. (2011). SpedeSTEM: a rapid and accurate method for species delimitation. Mol. Ecol. Res. 11, 473–480. doi: 10.1111/j.1755-0998.2010.02947.x
Firetti, F., Zuntini, A. R., Gaiarsa, J. W., Oliveira, R. S., Lohmann, L. G., and Van Sluys, M. A. (2017). Complete chloroplast genome sequences contribute to plant species delimitation: A case study of the Anemopaegma species complex. Am. J. Bot. 104, 1493–1509. doi: 10.3732/ajb.1700302
Fischer, J., and Lindenmayer, D. B. (2000). An assessment of the published results of animal relocations. Biol. Conserv. 96, 1–11. doi: 10.1016/S0006-3207(00)00048-3
Godo, T., Lu, Y., and Guan, K. (2003). Present state of Ottelia acuminata (Gagnep.) Dandy (Hydrocharitaceae) in Yunnan Province, China. Bull. Bot. Gard. Toyama 8, 49–54.
Guo, J. L., Yu, Y. H., Zhang, J. W., Li, Z. M., Zhang, Y. H., and Volis, S. (2019). Conservation strategy for aquatic plants: endangered Ottelia acuminata (Hydrocharitaceae) as a case study. Biodivers. Conserv. 28, 1533–1548. doi: 10.1007/s10531-019-01740-9
Hausmann, A., Miller, S. E., Holloway, J. D., Dewaard, J. R., Pollock, D., Prosser, S. W. J., et al. (2016). Calibrating the taxonomy of a megadiverse insect family: 3000 DNA barcodes from geometrid type specimens (Lepidoptera, Geometridae). Genome 59, 671–684. doi: 10.1139/gen-2015-0197
Hebert, P. D. N., Cywinska, A., Ball, S. L., and De-Waard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Henle, K., Davies, K. F., Kleyer, M., Margules, C., and Settele, J. (2004). Predictors of species sensitivity to fragmentation. Biodivers. Conserv. 13, 207–251. doi: 10.1023/B:BIOC.0000004319.91643.9e
Heywood, V. H., and Iriondo, J. M. (2003). Plant conservation: old problems, new perspectives. Biol. Conserv. 113, 321–335.
Hollingsworth, P. M. (2011). Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19451–19452. doi: 10.1073/pnas.1116812108
Hollingsworth, P. M., Forrest, L. L., Spouge, J. L., Hajibabaei, M., Ratnasingham, S., and van der Bank, M. (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6:e19254. doi: 10.1371/journal.pone.0019254
Hollingsworth, P. M., Li, D. Z., Michelle, V. D. B., and Twyford, A. D. (2016). Telling plant species apart with DNA: from barcodes to genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150338. doi: 10.1098/rstb.2015.0338
Hopkins, G. W., and Freckleton, R. P. (2002). Declines in the numbers of amateur and professional taxonomists: implications for conservation. Anim. Conserv. 5, 245–249. doi: 10.1017/s1367943002002299
Ito, Y., Tanaka, N., Barfod, A. S., Bogner, J., Li, J., Yano, O., et al. (2019). Molecular phylogenetic species delimitation in the aquatic genus Ottelia (Hydrocharitaceae) reveals cryptic diversity within a widespread species. J. Plant Res. 132, 335–344. doi: 10.1007/s10265-01109-7
Ji, Y. H., Liu, C. K., Yang, J., Jin, L., Yang, Z. Y., and Yang, J. B. (2020). Ultra-barcoding discovers a cryptic species in Paris yunnanensis (Melanthiaceae), a medicinally important plant. Front. Plant Sci. 11:411. doi: 10.3389/fpls.2020.00411
Ji, Y. H., Liu, C. K., Yang, Z. Y., Yang, L. F., He, Z. S., Wang, H. C., et al. (2019). Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol. Ecol. Resour. 19, 1333–1345. doi: 10.1111/1755-0998.13050
Jiang, Z., Li, H., and Dao, Z. (2005). Ottelia acuminata var. songmingensis, a new variety of the Hydrocharitaceae from Yunnan, China. Guihaia 25, 424–425. doi: 10.1360/aps040037
Jiang, Z. T., Li, H., Dao, Z. L., and Long, C. L. (2010). Ethnobotanical study on Ottelia acuminata, an aquatic edible plant occurring in Yunnan. J. Inner Mong. Norm. Univ. 39, 163–168.
Joppa, L. N., Roberts, D. L., and Pimm, S. L. (2011). How many species of flowering plants are there? Proc. Biol. Sci. 278, 554–559. doi: 10.1098/rspb.2010.1004
Kane, N. S., Sveinsson, S., Dempewolf, H., Yang, J. Y., Zhang, D., Engels, J. M. M., et al. (2012). Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 99, 320–329. doi: 10.3732/ajb.1100570
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., et al. (2017). Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33, 1630–1638. doi: 10.1093/bioinformatics/btx025
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kekkonen, M., and Hebert, P. D. N. (2014). DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Mol. Ecol. Resour. 14, 706–715. doi: 10.1111/1755-0998.12233
Kress, W. J., García-Robledo, C., Uriarte, M., and Erickson, D. L. (2015). DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 30, 25–35. doi: 10.1016/j.tree.2014.10.008
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., and Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Li, H. (1981). Classification, distribution and phylogeny of the genus Ottelia. J. Syst. Evol. 19, 29–42.
Li, H. (1985). The flourishing and declining of Ottelia acuminata in the lake Dian Chi. J. Yunnan Univ. 7, 138–142.
Li, L., Jiang, Y., Niu, Z., Xue, Q., Liu, W., and Ding, X. (2020). The large single-copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Acta Pharm. Sin. B 10, 1989–2001. doi: 10.1016/j.apsb.2020.01.012
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Li, Z. Z., Lehtonen, S., Martins, K., Gichira, A. W., Wu, S., Li, W., et al. (2020b). Phylogenomics of the aquatic plant genus Ottelia (Hydrocharitaceae): implications for historical biogeography. Mol. Phylogenet. Evol. 152:106939. doi: 10.1016/j.ympev.2020.106939
Li, Z. Z., Ngarega, B. K., Lehtonen, S., Gichira, A. W., Karichu, M. J., Wang, Q. F., et al. (2020c). Cryptic diversity within the African aquatic plant Ottelia ulvifolia (Hydrocharitaceae) revealed by population genetic and phylogenetic analyses. J. Plant Res. 133, 373–381. doi: 10.1007/s10265-020-01175-2
Liu, Y., Wang, Q. F., Chen, J. M., Li, Z. Z., Liao, K., Zou, C. Y., et al. (2018). Ottelia guanyangensis (Hydrocharitaceae), a new species from southwestern China. Phytotaxa 361, 294–300. doi: 10.11646/phytotaxa.361.3.5
Long, C. L., Jiang, Z. T., and Dao, Z. L. (2010). Genetic diversity of Ottelia acuminata (Hydrocharitaceae) from the Eastern Himalayas, revealed by ISSR markers. Bot. Orientalis 7, 56–63. doi: 10.3126/botor.v7i0.4374
Mace, G. M. (2004). The role of taxonomy in species conservation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 711–719. doi: 10.1098/rstb.2003.1454
Mace, G. M., Gittleman, J. L., and Purvis, A. (2003). Preserving the tree of life. Science 300, 1707–1709. doi: 10.1126/science.1085510
Margules, C. R., and Pressey, R. L. (2000). Systematic conservation planning. Nature 405, 243–253. doi: 10.1111/fme.12236
Masters, B. C., Fan, V., and Ross, H. A. (2011). Species delimitation – a geneious plugin for the exploration of species boundaries. Mol. Ecol. Res. 11, 154–157. doi: 10.1111/j.1755-0998.2010.02896.x
May, R. M. (1988). How many species are there on earth? Science 241, 1441–1449. doi: 10.1126/science.241.4872.1441
May, R. M., and Beverton, R. (1990). How many species? Philos. Trans. R. Soc. Lond. B Biol. Sci. 330, 293–304. doi: 10.1098/rstb.1990.0200
Mckinney, M. L. (1997). Extinction vulnerability and selectivity: combining ecological and paleontological views. Annu. Rev. Ecol. Syst. 28, 495–516. doi: 10.1146/annurev.ecolsys.28.1.495
Milligan, B. G., Leebens-Mack, J., and Strand, A. E. (1994). Conservation genetics: beyond the maintenance of marker diversity. Mol. Ecol. 3, 423–435. doi: 10.1111/j.1365-294X.1994.tb00082.x
Mutanen, M., Kekkonen, M., Prosser, S. W. J., Hebert, D. N., and Kaila, L. (2015). One species in eight: DNA barcodes from type specimens resolve a taxonomic quagmire. Mol. Ecol. Resour. 15, 967–984. doi: 10.1111/1755-0998.12361
Nock, C. J., Waters, D. L. E., Edwards, M. A., Bowen, S. G., Rice, N., Cordeiro, G. M., et al. (2011). Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 9, 328–333. doi: 10.1111/j.1467-7652.2010.00558.x
Patel, R. K., and Jain, M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Phillips, G., Willby, N., and Moss, B. (2016). Submerged macrophyte decline in shallow lakes: what have we learnt in the last forty years? Aquat. Bot. 135, 37–45. doi: 10.1016/j.aquabot.2016.04.004
Posada, D., and Buckley, T. R. (2004). Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808. doi: 10.1080/10635150490522304
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Prance, G. T., Beentje, H., Dransfield, J., and Johns, R. (2000). The tropical flora remains undercollected. Ann. Mo. Bot. Gard. 87, 67–71. doi: 10.2307/2666209
Puillandre, N., Lambert, A., Brouillet, S., and Achaz, G. (2012a). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Puillandre, N., Modica, M. V., Zhang, Y., Sirovich, L., Boisselier, M. C., Cruaud, C., et al. (2012b). Large-scale species delimitation method for hyperdiverse groups. Mol. Ecol. 21, 2671–2691. doi: 10.1111/j.1365-294X.2012.05559.x
Qin, H. N., Yang, Y., Dong, S. Y., He, Q., Jia, Y., Zhao, L. N., et al. (2017). Threatened species list of China’s higher plants. Biodivers. Sci. 25, 696–744. doi: 10.17520/biods.2017144
Rannala, B., and Yang, Z. H. (2013). Improved reversible jump algorithms for Bayesian species delimitation. Genetics 194, 245–253. doi: 10.1534/genetics.112.149039
Rojas, M. (1992). The species problem and conservation: what are we protecting? Conserv. Biol. 6, 170–178. doi: 10.1046/j.1523-1739.1992.620170.x
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ruhsam, M., Rai, H. S., Mathews, S., Ross, G., Graham, S. W., Raubeson, L. A., et al. (2015). Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour. 15, 1067–1078. doi: 10.1111/1755-0998.12375
Sand-Jensen, K., Riis, T., Vestergaard, O., and Larsen, S. E. (2000). Macrophyte decline in Danish lakes and streams over the past 100 years. J. Ecol. 88, 1030–1040. doi: 10.1046/j.1365-2745.2000.00519.x
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, 686–689. doi: 10.1093/nar/gki366
Sites, J. W., and Crandall, K. A. (1997). Testing species boundaries in biodiversity studies. Conserv. Biol. 11, 1289–1297. doi: 10.1046/j.1523-1739.1997.96254.x
Sites, J. W., and Marshall, J. C. (2003). Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 18, 462–470. doi: 10.1016/S0169-5347(03)00184-8
Ślipiko, M., Myszczyński, K., Bączkowska, K., Bączkiewicz, A., Szczecińska, M., and Sawicki, J. (2020). Molecular delimitation of European leafy liverworts of the genus Calypogeia based on plastid super-barcodes. BMC Plant Biol. 20:243. doi: 10.1186/s12870-020-02435-y
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Straub, S. C. K., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., and Liston, A. (2012). Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 99, 349–364. doi: 10.3732/ajb.1100335
Su, X., Wu, G., Li, L., and Liu, J. (2015). Species delimitation in plants using the Qinghai-Tibet plateau endemic Orinus (Poaceae: Tridentinae) as an example. Ann. Bot. 116, 35–48. doi: 10.1093/aob/mcv062
Wang, N. Z. (1986). Chromosome numbers and Karyotype analysis of 9 species in Hydrocharitaceae. J. Syst. Evol. 24, 370–375.
Wang, Q. F., Guo, Y. H., Haynes, R. R., and Hellquist, C. B. (2010). “Hydrocharitaceae,” in Flora of China, Vol. 23, eds Z. Y. Wu and H. R. Peter (Beijing: Science Press), 91–102.
Wiley, E. O., and Mayden, R. (2000). “The evolutionary species concept,” in Species Concepts and Phylogenetic Theory: A Debate, eds Q. D. Wheeler and R. Meier (New York, NY: Columbia University Press), 70–89.
Wilson, J. J., Sing, K. W., Lee, P. S., and Wee, A. K. S. (2016). Application of DNA barcodes in wildlife conservation in Tropical East Asia. Conserv. Biol. 30, 982–989. doi: 10.1111/cobi.12787
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Yang, J., Li, J., Li, J. W., Li, M., and Zhang, Y. (2012). Summarize on the research of the hydro-bios and aquatic environment in Fuxian lake and Xingyun lake. Yunnan Geogr. Environ. Res. 24, 98–102.
Yang, Z. H., and Rannala, B. (2010). Bayesian species delimitation using multilocusmsequence data. Proc. Natl. Acad. Sci. U.S.A. 107, 9264–9269. doi: 10.1073/pnas.0913022107
Zeng, C. X., Hollingsworth, P. M., Yang, J., He, Z. S., Zhang, Z. R., Li, D. Z., et al. (2018). Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14:43. doi: 10.1186/s13007-018-0300-0
Zhai, S. H., Yin, G. S., and Yang, X. H. (2018). Population genetics of the endangered and wild edible plant Ottelia acuminata in southwestern China using novel SSR markers. Biochem. Genet. 56, 235–254. doi: 10.1007/s10528-018-9840-2
Zhang, H. Y., Tian, K., Yu, Y., Li, L. Y., and Yang, Y. M. (2009). Genetic diversity among natural populations of Ottelia acuminata (Gaghep.) Dandy revealed by ISSR. Afr. J. Biotechnol. 8, 6089–6093. doi: 10.5897/AJB09.680
Zhang, J. J., Kapli, P., Pavlidis, P., and Stamatakis, A. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876. doi: 10.1093/bioinformatics/btt499
Zhang, Y., Jeppesen, E., Liu, X., Qin, B., Shi, K., Zhou, Y., et al. (2017). Global loss of aquatic vegetation in lakes. Earth Sci. Rev. 173, 259–265. doi: 10.1016/j.earscirev.2017.08.013
Keywords: species boundary, molecular identification, DNA barcoding, taxonomy, threatened species, aquatic plant
Citation: Ji Y, Yang J, Landis JB, Wang S, Yang Z and Zhang Y (2021) Deciphering the Taxonomic Delimitation of Ottelia acuminata (Hydrocharitaceae) Using Complete Plastomes as Super-Barcodes. Front. Plant Sci. 12:681270. doi: 10.3389/fpls.2021.681270
Received: 16 March 2021; Accepted: 10 June 2021;
Published: 15 July 2021.
Edited by:
Silvia Pressel, Natural History Museum (United Kingdom), United KingdomReviewed by:
Muthama Muasya, University of Cape Town, South AfricaRafael Lira, National Autonomous University of Mexico, Mexico
Copyright © 2021 Ji, Yang, Landis, Wang, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunheng Ji, aml5aEBtYWlsLmtpYi5hYy5jbg==
†These authors have contributed equally to this work
 Yunheng Ji
Yunheng Ji Jin Yang1,3
Jin Yang1,3 Jacob B. Landis
Jacob B. Landis Zhenyan Yang
Zhenyan Yang Yonghong Zhang
Yonghong Zhang