- 1Fruit Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 2Fujian Engineering and Technology Research Center for Deciduous Fruit Trees, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 3The New Zealand Institute for Plant and Food Research Limited, Mt Albert Research Centre, Auckland, New Zealand
Plum is one of the most important stone fruits in the world and anthocyanin-rich plums are increasingly popular due to their health-promoting potential. In this study, we investigated the mechanisms of anthocyanin accumulation in the flesh of the red-fleshed mutant of the yellow-fleshed plum ‘Sanyueli’. RNA-Seq and qRT-PCR showed that anthocyanin biosynthetic genes and the transcription factor PsMYB10.2 were upregulated in the flesh of the mutant. Functional testing in tobacco leaves indicated that PsMYB10.2 was an anthocyanin pathway activator and can activate the promoter of the anthocyanin biosynthetic genes PsUFGT and PsGST. The role of PsMYB10.2 in anthocyanin accumulation in the flesh of plum was further confirmed by virus-induced gene silencing. These results provide information for further elucidating the underlying mechanisms of anthocyanin accumulation in the flesh of plum and for the breeding of new red-fleshed plum cultivars.
Introduction
Plum is one of the most consumed stone fruits in the world and is a rich source of health-promoting compounds such as anthocyanins (Lee et al., 2009; Johnson et al., 2011; Valero et al., 2013; Fanning et al., 2014; Wang et al., 2020). Anthocyanins are responsible for the red pigmentation in plum skin and in the flesh of red-fleshed varieties (Cheng et al., 2016). It has been widely reported that the consumption of anthocyanin-rich foods promotes health (Santhakumar et al., 2015; Igwe et al., 2017; Jamar et al., 2017; Bakuradze et al., 2019). Increasing evidences have indicated that anthocyanins can delay the onset of diseases such as cancers, diabetes, cardiovascular and inflammatory diseases (He and Giusti, 2010). For example, Liu et al. (2017) reported that cyanidin, which is the major type of anthocyanin in plum fruits (Tomás-Barberán et al., 2001; Usenik et al., 2009; Wang R. et al., 2018), alleviates inflammation in vivo by blocking the binding of the cytokine interleukin-17A to the IL-17RA subunit, which suggested that cyanidin derived small molecules could be developed into drugs for the treatment of IL-17A–dependent inflammatory diseases and cancer.
China is the largest plum producer in the world (Milošević and Milošević, 2018), with a production >700 million tons in 20191. While red-fleshed fruits are rich in anthocyanin and favored by consumers (Espley et al., 2013; Silvestri et al., 2018), they represent only one-fifth of the plum accessions in China (Zhang and Zhou, 1998). Most of the cultivated cultivars are red-skinned but yellow-fleshed. Therefore, it is of great interest to elucidate the mechanisms controlling anthocyanin accumulation in flesh of plum and develop new red-fleshed cultivars.
The anthocyanin biosynthesis pathway has been widely investigated in plants. The structural genes, such as phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumaroyl:CoA-ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), flavonoid3′-hydroxylase (F3′H), dihydroflavonol-4-reductase (DFR), anthocyanin synthase (ANS), UDPglucose:flavonoid-3-O-glucosyltransferase (UFGT), and glutathione S-transferase (GST), that are involved in anthocyanin biosynthesis and accumulation are well documented (Tanaka et al., 2008; Jaakola, 2013). The expression of these genes are coordinately regulated by the MYB-bHLH-WD40 (MBW) complex (Allan et al., 2008; Rahim et al., 2014; Li et al., 2016) and other transcription factors, such as NACs, HD-Zips, bZIPs and WRKYs, which interact with the MBW complex or directly bind to the promoter of anthocyanin structural genes (Zhou H. et al., 2015; Jiang et al., 2017; An et al., 2018; Liu et al., 2018; Wang N. et al., 2018). Among them, the R2R3-MYBs have often been identified as the key positive or negative regulator (Espley et al., 2007; Feng et al., 2015; Tuan et al., 2015; Jin et al., 2016; Butelli et al., 2017; Plunkett et al., 2018; Zhang et al., 2018). In our previous study, we showed that a homolog of the Arabidopsis R2R3 MYB gene, AtMYB113, was upregulated during pigmentation of flesh in ‘Furongli’ plum (a red-fleshed cultivar) (Fang et al., 2016b). R2R3-MYB genes have been reported to be responsible for variation, loss or gain of anthocyanin pigmentation in several plants (Li et al., 2015; Kotepong et al., 2019; Wang et al., 2019; Jia et al., 2020; Xu et al., 2020). In addition, basic helix-loop-helix (bHLH) transcription factors also play an essential role in the regulation of anthocyanin biosynthesis in plant species such as Arabidopsis (Gonzalez et al., 2008), Petunia (Spelt et al., 2000), Medicago truncatula (Li et al., 2016), and grape (Hichri et al., 2010). Recently, it has been shown that abnormal MabHLH3 expression is responsible for loss of anthocyanin accumulation in fruits of ‘Baiyuwang’ mulberry (Li et al., 2020). It has been demonstrated that bHLHs are indispensable for the anthocyanin activating activity in Nicotiana of peach PpMYB10.2 (Zhou et al., 2018) and apple MdMYB10 (Espley et al., 2007) and significantly enhanced the anthocyanin activating activity of peach PpMYB10.1 (Zhou H. et al., 2015) and PpMYB10.4 (Zhou et al., 2014) and Chinese bayberry MrMYB1 (Niu et al., 2010). bHLH proteins are characterized by a conserved bHLH domain, which is responsible for DNA recognition and DNA-binding specificity (Feller et al., 2011; Hichri et al., 2011). Zhou et al. (2018) showed that substitution of Arg/Gly93 in bHLH-binding domain of peach PpMYB10.1 and PpMYB10.2 could significantly affect their anthocyanin-promoting activity by altering their binding affinity to PpbHLH3.
Previously, we identified a red-fleshed and late-ripening mutant (named ‘Fuhongli’) from the radiation-induced mutant population obtained from 60Co-γ ray treated buds of ‘Sanyueli’ plum (Fang et al., 2016a). Due to its largely similar genetic origins, the mutant provides opportunities to investigate the molecular mechanisms that modulate anthocyanin accumulation in flesh of plum. This study aims to clarify the mechanisms underlying the accumulation of anthocyanins in the flesh of ‘Fuhongli’. The results indicated that anthocyanin content in the flesh of ‘Fuhongli’ increased rapidly during fruit ripening, but no anthocyanin was accumulated in the flesh of ‘Sanyueli’. RNA-Seq, dual-luciferase assays and transient color assays in tobacco leaves and plum fruits identified PsMYB10.2, which activated the promoters of PsUFGT and PsGST, as a key transcription factor contributing to anthocyanin accumulation in the flesh of ‘Fuhongli’. The present study suggests that PsMYB10.2 is a weak anthocyanin activator and is responsible for anthocyanin biosynthesis in flesh of plum fruits.
Materials and Methods
Plant Materials
All fruits were harvested from 5-years old ‘Sanyueli’ (SY) plum (Prunus salicina Lindl.) tree and its red-fleshed mutant ‘Fuhongli’ (MT) grown in Fruit Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, Fujian Province, China. Fruit samples of ‘Sanyueli’ were collected at 95, 105, 115 days after flowering (DAF) and Fruit samples of red-fleshed mutant were collected at 95, 105, 115, and 125 DAF. Fruit maturity in ‘Sanyueli’ plums was reached 115 days after flowering (DAF), while ‘Fuhongli’ plums ripened 150 DAF. Fruits sampled from three different branches were used as biological replicates. Ten representative fruits sampled from each branch at each developmental stage were peeled and sliced into small pieces. The sliced fruit fleshes were pooled into three replicates and immediately frozen in liquid nitrogen and kept at −80°C for further experiments.
Determination of Total Sugar, Fructose, Glucose, Sucrose, and Organic Acid Contents
Concentrations of total sugar, fructose, glucose and sucrose were measured using total sugar (ZT-1-Y), fructose (GT-1-Y), glucose (PT-1-Y), sucrose (ZHT-1-Y) assay kits (Comin Biotechnology Co., Ltd., Suzhou, China) according to the manufacturer’s instructions. Titratable acid was measured as described by Chen and Zhu (2011). For the determination of malic acid content, 0.1 g powder was suspended with 1 ml ice-cold water and extracted by ultrasonic treatment for 1 h. The extracts were centrifugated at 8,000 × g for 10 min. The supernatant was filtrated ussing a 0.22 μm membrane filter and then analyzed with HPLC using an Agilent 1100 system (Agilent Technologies, Palo Alto, CA, United States). Separation was performed using a Kromasil C18 250 × 4.6mm, 5μm column (AkzoNobel, Sweden).
Determination of Anthocyanin and Carotenoid Content in Plum Flesh
For quantification of anthocyanin, approximately 2 g of fruit flesh was ground to fine powder in liquid nitrogen and extracted in 10 mL methanol with 0.05% HCl at 4°C for 24 h. Anthocyanin content was quantified by pH differential method as described previously (Fang et al., 2016b). Three measurements for each biological replicate sample were performed. The total carotenoid content was measured using the carotenoid assay kit (Comin Biotechnology Co., Ltd., Suzhou, China.) as previously described (Hou et al., 2020).
For HPLC analysis, approximately 2 g of the flesh of mature fruits of ‘Fuhongli’ was ground in liquid nitrogen and extracted in 20 ml methanol with 1% HCl at 4°C for 12 h in the dark. Samples were brought to room temperature then centrifuged for 10 min at 5,866 × g. HPLC analysis was performed using an Agilent 1120 system (Agilent Technologies, Palo Alto, CA). Separation was performed using a C-18 4.6 × 150 mm column (InfinityLab Poroshell 120 EC-C18, Agilent, United States). The mobile phase was 0.3% phosphoric acid in water (solvent A) and acetonitrile (solvent B) at a flow rate of 1 ml min–1. The linear gradient of phase B was as follows: 0–20 min, 5%; 20–30 min, 15%; 30–40 min, 25%; 40–40 min, 25%; 48–50 min, 5%. The injection volume was 10 μL. Anthocyanin components were detected at 510 nm and peaks identified by comparison of peaks to authentic standards.
RNA Preparation, Library Construction and RNA-Seq
Extraction of total RNA, library construction and RNA-Seq were performed by staff at Beijing BioMarker Technologies (Beijing, China) as described previously (Fang et al., 2016b). Sequencing of the cDNA library was carried out on the Illumina HiSeqTM X Ten sequencing platform. RNA-seq was performed in three replicates. The RNA-seq reads have been deposited in the NCBI Short Read Archive and are accessible under PRJNA690967.
Analysis of RNA-Seq Data
The RNA-seq raw reads were processed with in-house perl scripts to remove reads containing adapter, reads containing ploy-N and low-quality reads. All generated clean reads were mapped to the genome sequence of ‘Sanyueli’ (Fang et al., 2020) using HISAT2 (Kim et al., 2015). The expression level of genes was estimated by the fragments per kilobase of transcript per million fragments mapped (FPKM) values using the StringTie software package (Pertea et al., 2015). DESeq2 (Love et al., 2014) was employed to perform differential gene expression analysis. Genes with fold change ≥2 and a false discovery rate <0.01 were considered significantly differentially expressed. One sample of ‘Sanyueli’ at 105DAF (SY105d-2) which was not well correlated with other biological replicates at the same stage was excluded from further analysis. GO and KEGG pathway enrichment analysis was carried out as reported previously (Fang et al., 2020). The heat maps for differentially expressed genes were constructed using TBtools (Chen et al., 2020).
Prediction of Transcription Factors
All transcripts were subjected to the PlantTFDB v5.02 to predict transcription factors using the Transcription Factor Prediction module.
Real-Time Quantitative RT-PCR (qRT-PCR) Analysis
The expression of 15 differentially expressed genes were validated using qRT-PCR. The extraction of total RNA from fruit flesh was carried out using a modified CTAB method (Xuan et al., 2015). All primers used for qRT-PCR were provided in Supplementary Table 1. The synthesis of cDNA was carried out using the PrimeScript® RT reagent kit with gDNA Eraser (Takara, Dalian, China). qRT-PCR was performed using the Eppendorf Realplex4 real-time PCR system (Hamburg, Germany) as described previously (Fang et al., 2016b). qRT-PCR conditions were 5 min at 95°C, followed by 40 cycles of 5 s at 95°C, 15 s at 60°C, and 30 s at 72°C, followed by 60–95°C melting curve detection. Actin (PsSY0026636) gene was used as the reference. The expression levels were calculated as described previously (Fang et al., 2016b). Three biological and three technical replications were performed.
Phylogenetic Analysis and Multiple Sequence Alignment
Phylogenetic and evolutionary analyses were performed using the neighbor-joining method with 1000 bootstrap replicates by MEGA-X. Additional sequences include Arabidopsis thaliana AtMYB4 (At4G38620), AtMYB11 (AT3G62610), AtMYB12 (AT2G47460), AtMYB75 (AT1G56650), AtMYB90(AT1G66390), AtMYB111 (AT5G49330), AtMYB113 (AT1G66370), AtMYB114 (AT1G66380), AtMYB123 (AT5G35550); Actinidia chinensis AcMYB10 (PSS35990) and AcMYB110 (AHY00342); Fragaria × ananassa FaMYB1 (AAK84064.1), FaMYB9(JQ989281), FaMYB10 (ABX79947.1); Gossypium hirsutum GhMYB36(AF336284); Lotus japonicus LjMYB12 (AB334529); Myrciaria cauliflora McMYB (MH383068); Malus × domestica MdMYB9 (DQ267900), MdMYB10 (EU518249), MdMYB11 (DQ074463); Medicago truncatula MtLAP1 (ACN79541.1) and MtMYB14 (XP_003594801.1); Prunus cerasifera PcMYB10.1 (KP772281) and PcMYB10.2 (KP772282); Petunia hybrida PhMYB27 (AHX24372), PhAN2 (AB982128), PhDEEP PURPLE (ADQ00393.1), PhPURPLE HAZE (ADQ00388.1), and PhMYB4 (ADX33331.1); Prunus persica PpMYB10.1(XM_007216468), PpMYB10.2(XM_007216161), and PpMYB18 (KT159234); Prunus salicina PsMYB10.1(MK105923) and PsMYB18 (MK284223); Pyrus pyrifolia PyMYB10(GU253310); Solanum lycopersicum SlMYB12 (ACB46530.1); Trifolium arvense TaMYB14 (AFJ53053.1); Trifolium repens TrMYB4 AMB27079), TrMYB133 (AMB27081) and TrMYB134 (AMB27082); Vaccinium corymbosum VcMYBA (MH105054); Vaccinium uliginosum VuMYBC2; Vitis vinifera VvMYBA1 (BAD18977), VvMYBA2(BAD18978), VvMYBF1 (ACV81697), VvMYBC2-L1 (AFX64995.1), VvMYB-L2 (ACX50288.2), VvMYBPA1 (CAJ90831.1), and VvMYBPA2 (ACK56131.1). Multiple sequence alignment of R2R3 MYB amino acid sequences from plum, sweet cherry and peach was performed by DANMAN7.
Vector Construction
cDNA was generated from red pigmented flesh of ‘Fuhongli’ using a RevertAid First-Strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA). PsMYB10.2 cDNA and PsbHLH3 cDNA were isolated using 2 × Phanta Max Master Mix (Vazyme, Nanjing, China) and inserted into pSAK277. Genomic DNA was isolated from young plum leaves using a Plant Genomic DNA kit (Tiangen, China) and used as a template for promoter amplification. Promoter sequences of PsUFGT (2523bp) and PsGST (2339bp) were amplified and inserted into pGreenII 0800–LUC vector using ClonExpress Ultra One Step Cloning Kit (Vazyme, Nanjing, China). A fragment of PsMYB10.2 (296–645 bp) was amplified using specific primers (TRV2-PsMYB10.2F and pTRV2-PsMYB10.2R) and inserted into pTRV2 to generate pTRV2-PsMYB10.2. Primer sequences used for the vector construction are listed in Supplementary Table 2.
Transient Color Assay in Tobacco Leaf and Quantification of Anthocyanins
Transient overexpression assays were performed in young leaves of 2-week-old seedlings of Nicotiana tabacum grown in the greenhouse as reported by Zhou et al. (2019) using Agrobacterium tumefaciens strain GV3101. A. tumefaciens GV3101 carrying constructs were incubated at 28°C for 2 days and resuspended in infiltration buffer containing 10 mM MgCl2, 100 μM acetosyringone (pH = 5.7) to an OD600 of 0.5. Separate strains containing PsMYB10.2 and PsbHLH3 fused to the 35S promoter in the pSAK277 vector and empty pSAK277 vector were infiltrated or co-infiltrated into the abaxial leaf surface. Peach PpMYB10.1 (Zhou H. et al., 2015) co-infiltrated with PsbHLH3 was used as a positive control. Photographs were taken 7 days after infiltration.
For quantification of anthocyanins, 10 mg of freeze-dried tobacco leaves from the infiltrated area was mixed in 1 mL of methanol: water: formic acid (80: 19.5: 0.5, v/v/v) and shaken for 4 h at room temperature. The tube containing the mixture was centrifuged at 10,000 g for 15 min. The supernatants were filtered through a 0.45 μm PTFE syringe filter and submitted for high performance liquid chromatography (HPLC) analysis according to a method reported by Andre et al. (2012) with a few modifications. Briefly, quantification of the anthocyanins was performed using a Dionex Ultimate 3000 system (Sunnyvale, CA) equipped with a diode array detector (DAD). A 5 μL aliquot was injected onto a Thermo C18 Acclaim PolarAdvantage II column (150 × 2.1 mm i.d.; 3 μm particle size) (Waltham, MA). The mobile phases were (A) H2O with 5% formic acid and (B) MeCN with 0.1% formic acid. The flow rate was 0.35 mL min–1, and the column temperature was 35°C. The 28 min gradient was as follows: 0-5 min, 7% B constant; 5-10 min, 7-12% B; 10-20 min, 12-25% B; 20-21 min, 25-100% B; 21-24 min, 100% B constant; 24 min, 7% B; 24-28 min, 7% B re-equilibration time. Monitoring was set at 520 nm for quantification. Anthocyanins were identified by their spectral data and were quantified as cyanidin-3-glucoside using five-point calibration curves. A validation standard was injected after every 10th injection.
Dual-Luciferase Assay
Dual-luciferase assays were conducted in N. benthamiana leaves as reported previously (Lin-Wang et al., 2010). Promoter constructs were individually transformed into A. tumefaciens GV3101 with the pSoup plasmid. Agrobacteria cultivation was performed as described above for transient color assay. For dual-luciferase assays, Agrobacteria strain GV3101 were transformed with the constructs and incubated at 28°C for 2 days and resuspended in infiltration buffer containing 10 mM MgCl2, 100 μM acetosyringone (pH = 5.7) to an OD600 of 0.5. Two Agrobacteria suspensions carrying the overexpression vectors (empty vector, PsMYB10.2 and PsbHLH3) were mixed equally and then combined with one of the dual-luciferase promoter construct (pPsUFGT:LUC and pPsGST: LUC) suspensions in a 5:1 ratio. The mixture of Agrobacteria suspension carrying an empty vector control construct and the dual-luciferase promoter construct, again suspensded in a 5:1 ratio, was infiltrated as control. The final mixture was infiltrated into the abaxial leaf surface of N. benthamiana using a needleless 1 ml syringe. Four days after infiltration, 3-mm diameter leaf punches from infiltrated patches were harvested and subjected to dual-luciferase assay.
RNAi Transient Assay of Plum Fruit Using the Virus-Induced Gene Silencing (VIGS) System
pTRV1, pTRV2, and pTRV2-PsMYB10.2 were individually transformed into Agrobacterium strain GV3101. These Agrobacterium strains were suspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, 150 μM acetosyringone) and kept at room temperature for 3 h. Fruit infiltration was performed using maturing fruit of ‘Fuhongli’ as previously described (Zhou H. et al., 2015). Flesh tissues around the infiltration site were harvested and used for quantification of anthocyanin concentration and qRT-PCR analysis. Quantification of anthocyanin content was performed as described previously (Fang et al., 2016b).
Results
Accumulation of Anthocyanin in ‘Fuhongli’
The dynamic changes in content of total soluble sugar, fructose, sucrose, glucose, total acid and malic acid were analyzed in the ‘Sanyueli’ and ‘Fuhongli’ during ripening. The accumulation pattern of total soluble sugar, fructose, sucrose and glucose were similar in ‘Sanyueli’ and ‘Fuhongli’ (Figures 1A–D). The content of total acid and malic acid decreased during the stages of fruit ripening in both ‘Sanyueli’ and ‘Fuhongli’ (Figures 1E,F). The flesh of unripe ‘Sanyueli’ fruit was green and changed into yellow during the late stages of ripening (Figure 2A). The flesh color of ‘Fuhongli’ was similar to that of ‘Sanyueli’ during the early stages of fruit development. However, the flesh of ‘Fuhongli’ showed weak red pigmentation at 105DAF and rapidly turned red during the late stages of ripening (Figure 2A). Analysis of fruit flesh anthocyanin contents demonstrated that no anthocyanin was accumulated in the flesh of ‘Sanyueli’ (Figure 2B). In contrast, a small amount of anthocyanins was detected in the flesh of ‘Fuhongli’ at 105DAF, when the fruit skin was still green, and the content of anthocyanin increased significantly during the late stages of ripening (Figure 2B). The carotenoid content increased in the flesh of ‘Sanyueli’, but slightly decreased in the flesh of ‘Fuhongli’ (Figure 2C). According to HPLC analysis, the anthocyanins accumulated in the flesh of mature fruits of ‘Fuhongli’ were cyanidin-3-rutinoside and cyanidin-3-glucoside (Figure 2D).
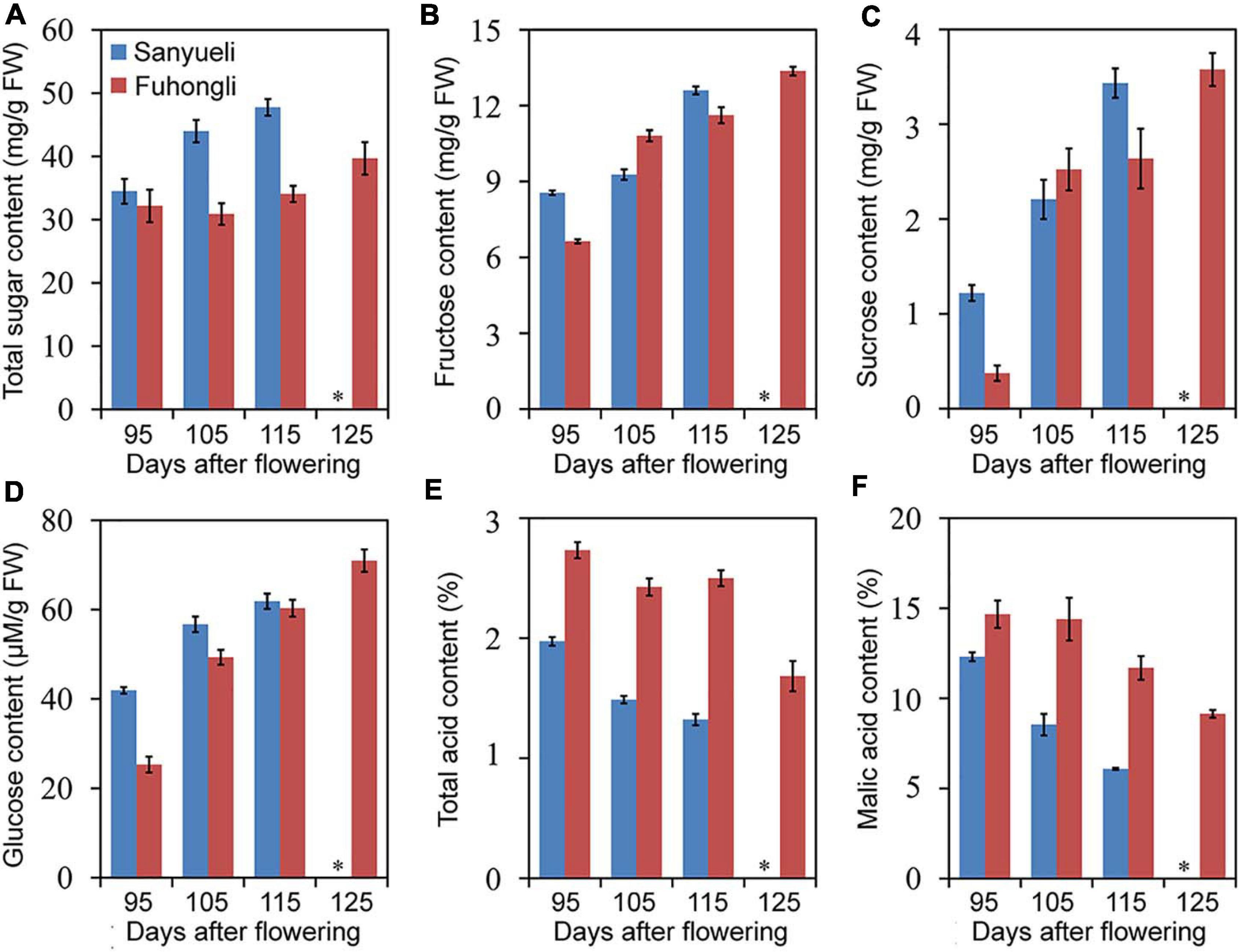
Figure 1. Trends in total soluble sugar (A), fructose (B), sucrose (C), glucose (D), total acid (E), and malic acid (F) content in the flesh of ‘Sanyueli’ and ‘Fuhongli’ plum during fruit ripening. ∗ indicates there is no value for ‘Sanyueli’ 125 DAF as the fruits ripened earlier. Error bars represent standard error of three replicates.
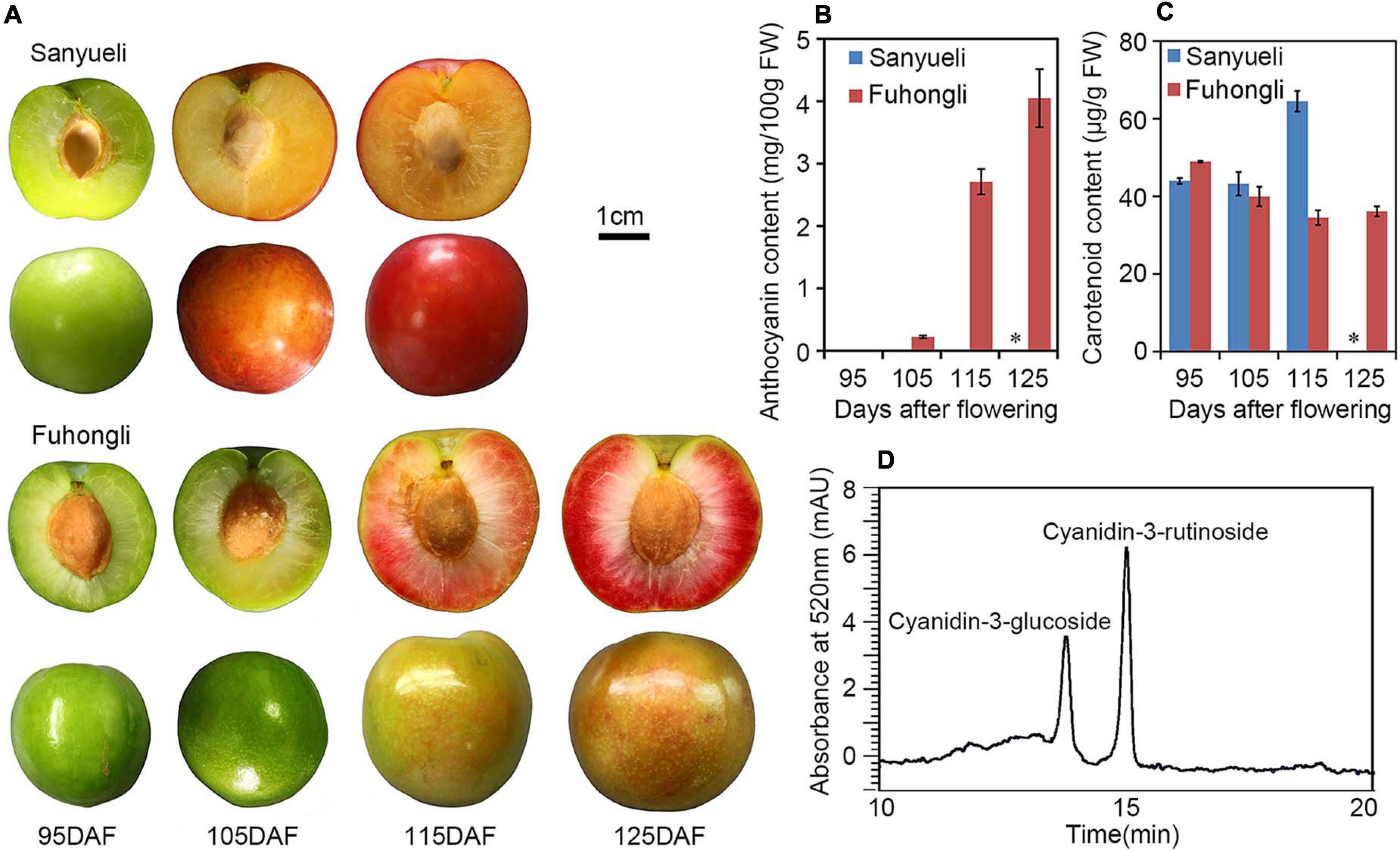
Figure 2. Pigmentation changes during fruit ripening of ‘Sanyueli’ and ‘Fuhongli’ plum. (A) Fruits of ‘Sanyueli’ plum and ‘Fuhongli’ at different stages. (B) Comparison of anthocyanin content in different ripening stages of ‘Sanyueli’ and ‘Fuhongli’. (C) Carotenoid content in different ripening stages of ‘Sanyueli’ and ‘Fuhongli’. (D) HPLC chromatogram of anthocyanins extracted from flesh of ‘Fuhongli’. ∗ indicates there is no value for ‘Sanyueli’ 125 DAF as the fruits ripened earlier. Error bars represent standard error of three replicates.
RNA-Seq and Identification of Differentially Expressed Genes(DEGs) During Fruit Ripening of ‘Sanyueli’ and ‘Fuhongli’
Flesh of ‘Sanyueli’ and ‘Fuhongli’ at different stages were subjected to RNA-seq analysis. A total of 163.32Gb clean data were obtained from these samples after discarding low-quality reads and adaptor sequences (Supplementary Table 3). The clean reads were mapped to the genome of ‘Sanyueli’ (Fang et al., 2020). The mapping ratio to the reference genome ranged between 81.29% and 85.35% (Supplementary Table 3). Correlation analysis showed that the biological replicates were highly correlated within each stage (Supplementary Figure 1). A total of 2,091 putative new genes that have not appeared in the genome of ‘Sanyueli’ were identified and 1,641 of them was annotated by databases including NCBI non-redundant protein database (NR), Swiss-Prot protein database (Swiss-Prot), Gene Ontology (GO), Clusters of Orthologous Groups database (COG), euKaryotic Ortholog Groups of proteins database (KOG), protein family database (Pfam) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Supplementary Table 4).
A total of 9,594 genes were identified to be differentially expressed by pairwise comparison (Supplementary Figure 2). Among them, 3,073 and 3,000 genes were differentially expressed during ripening of ‘Sanyueli’ and ‘Fuhongli’, respectively. The comparison of transcriptome between ‘Sanyueli’ and ‘Fuhongli’ at different stages enabled the identification of 8477 DEGs (Supplementary Figure 2).
Identification of Differentially Expressed Structural Genes Involved in Anthocyanin Accumulation
To investigate in which step of anthocyanin synthesis was activated in the red flesh of ‘Fuhongli’, we identified differentially expressed structural genes of anthocyanin biosynthesis according to the KEGG and GO annotation results. In total, 249 genes were assigned to phenylpropanoid biosynthesis and flavonoid biosynthesis pathway according to KEGG enrichment analysis (Supplementary Table 5). Ten of them, including PAL (PsSY0020810), C4H (PsSY0029112), 4CL (PsSY0028380), CHS (PsSY0019243 and PsSY0011918), CHI (PsSY0003309), F3H (PsSY0015860), F3′H (PsSY0012653), DFR (PsSY0000799), and ANS (PsSY0019761), were identified as DEGs (Figure 3). PsSY0019615 was annotated as CHI and encodes a protein with molecular function designated as chalcone isomerase activity (GO:0045430) and involved in anthocyanin-containing compound biosynthetic process (GO:0009718). PsSY0020267 was annotated as F3′H and predicted to have flavonoid 3′,5′-hydroxylase activity (GO:0033772). PsSY0022880 was annotated as anthocyanidin 3-O-glucosyltransferase having anthocyanidin 3-O-glucosyltransferase activity (GO:0047213). In addition, PsSY0017871 was predicted to encode a GST which was shown to be upregulated in the flesh of red-fleshed cultivar ‘Furongli’ plum during fruit ripening in our previous study (Fang et al., 2016b). All of these anthocyanin-related genes, except 4CL, were upregulated during fruit ripening of ‘Fuhongli’ (Figure 3 and Supplementary Table 5). There was no significant change in the expression of PAL (PsSY0020810), C4H (PsSY0029112), CHS (PsSY0003309), F3H (PsSY0015860), F3′H (PsSY0012653), and ANS (PsSY0019761) in the flesh of ‘Sanyueli’ during fruit ripening, while that of CHS (PsSY0019243 and PsSY0011918), CHI (PsSY0019615), and DFR (PsSY0000799) was significantly downregulated (Figure 3 and Supplementary Table 5). It is noteworthy that the expression of C4H (PsSY0029112), CHS (PsSY0003309) and F3′H (PsSY0012653) in the flesh of ‘Sanyueli’ was comparable to the highest level in the flesh of ‘Fuhongli’, but UFGT (PsSY0022880) and GST (PsSY0017871) were only expressed at extremely low levels in the flesh of ‘Sanyueli’ (Figure 3 and Supplementary Table 5). The expression profiles of 11 differentially expressed structural genes were validated using qRT-PCR. The results showed that the expression patterns detected by qRT-PCR experiments of the selected genes were consistent with the expression patterns investigated by RNA-seq analysis (Figure 4).
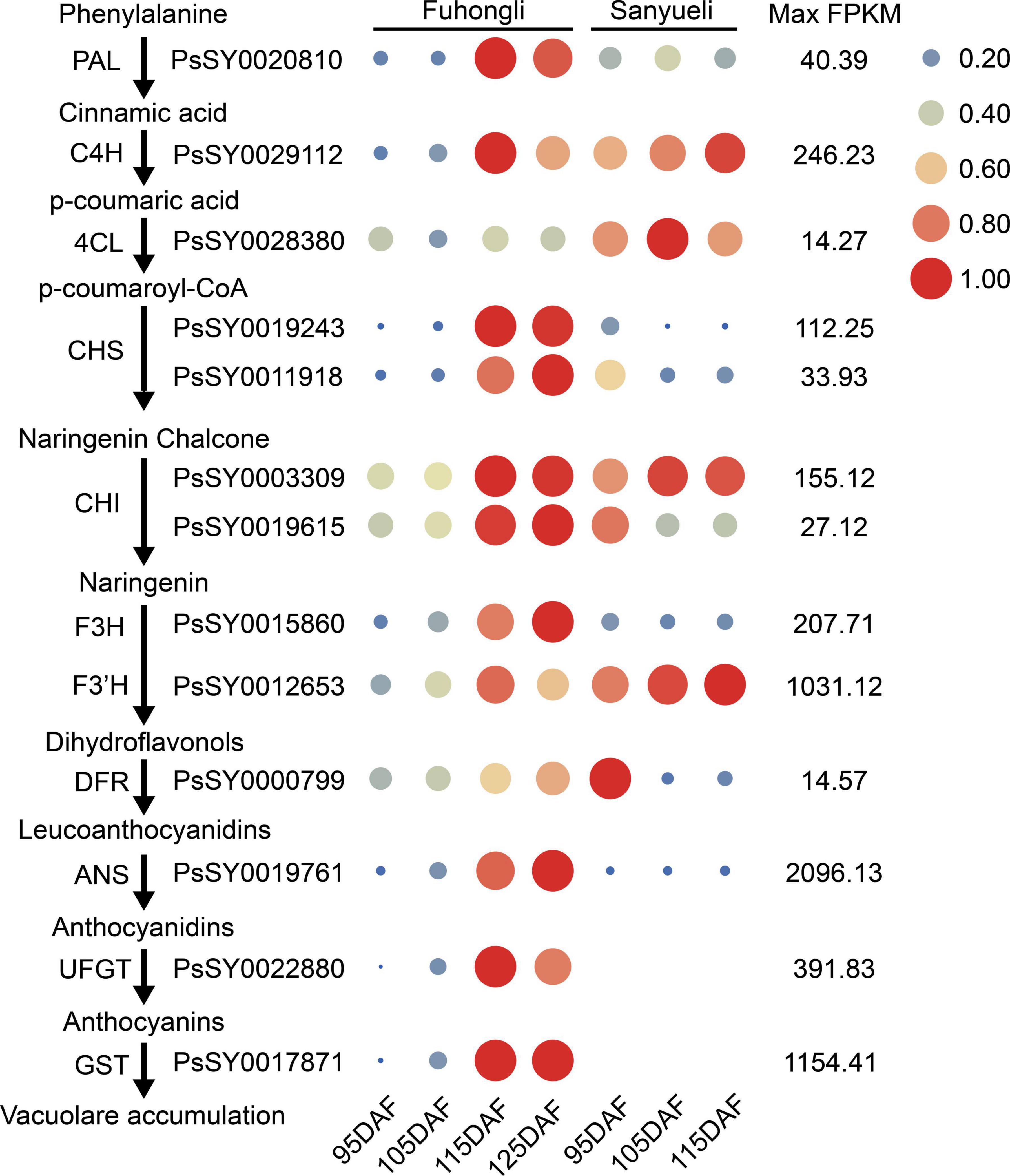
Figure 3. Expression of structural genes involved in the phenylpropanoid and anthocyanin biosynthesis, as evaluated by RNA-sequencing in the plum flesh of ‘Sanyueli’ and ‘Fuhongli’. FPKM values of all genes were normalized with maximum FPKM values of each gene. The expression level of genes was indicated using filled circle with different size and color. The larger the circle, the higher the expression. High expression was indicated in red, while low expression was indicated in blue. The values on the right indicates the highest FPKM value of each gene. Abbreviations for pathway genes as described in the Introduction.
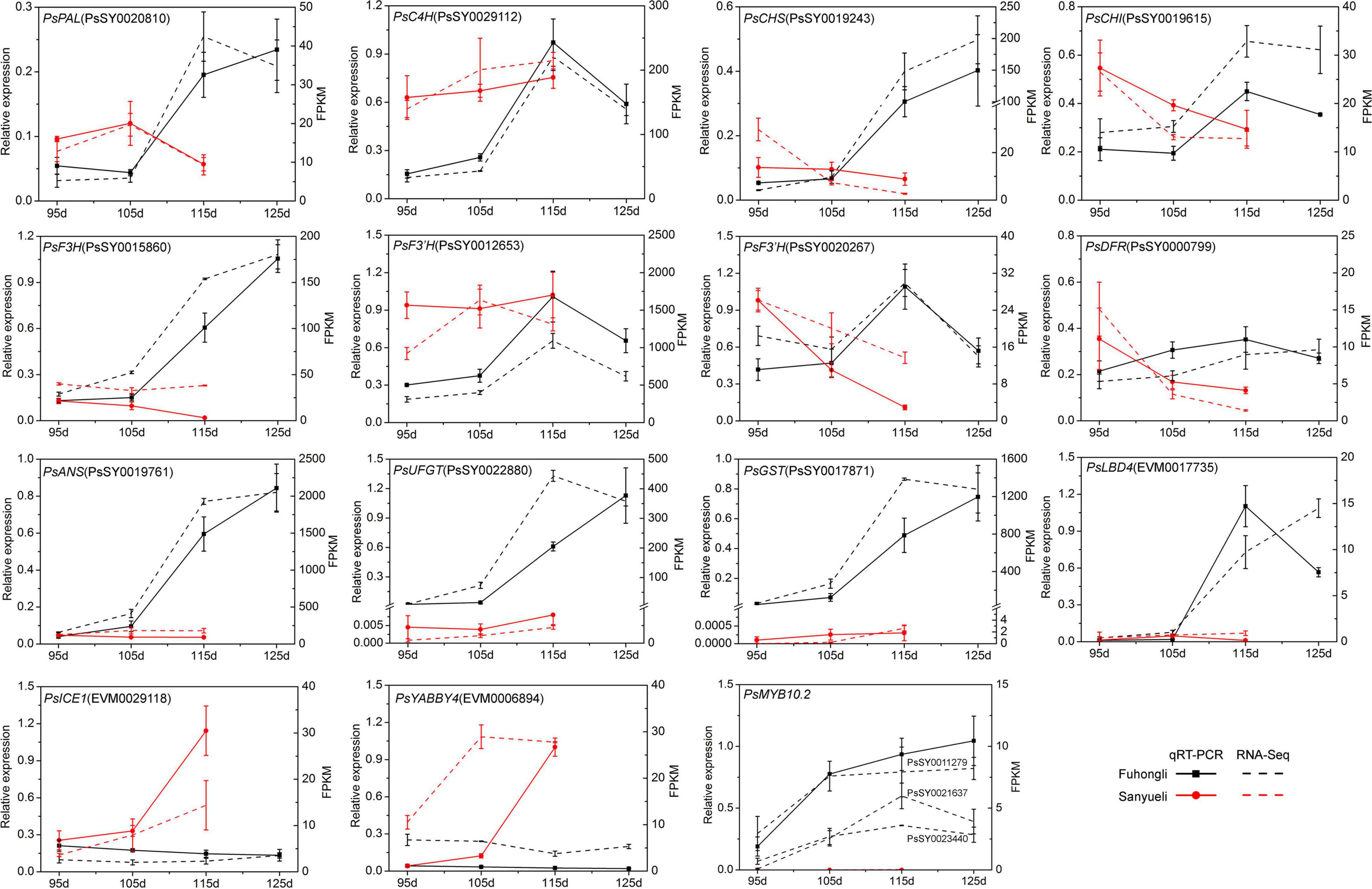
Figure 4. Expression profiles of selected differentially expressed transcription factors in the flesh of ‘Sanyueli’ and ‘Fuhongli’. FPKM values of all genes were normalized with maximum FPKM values of each gene. The expression level of genes was indicated using filled circle with different size and color. The larger the circle, the higher the expression. The values on the right indicates the highest FPKM value of each gene.
Identification of Transcription Factors
Since the structural genes involved in anthocyanin accumulation are transcriptionally regulated, we examined identify transcription factor potentially related to anthocyanin accumulation in the flesh of ‘Fuhongli’. From this, 1687 genes were predicted to encode transcription factors and 536 of them were differentially expressed (Supplementary Table 6). A total of 29 transcription factor genes, including 19 genes (Cluster A) that were increased in the flesh of ‘Fuhongli’ but expressed at low level or decreased in the flesh of ‘Sanyueli’ and 10 genes (Cluster B) that were expressed at low level in the flesh of ‘Fuhongli’ but increased or expressed in higher level in the flesh of ‘Sanyueli’, were identified (Figure 5 and Supplementary Table 6). These include five MYBs and the best hit in Arabidopsis to four of them is AtMYB113 (Figure 5 and Supplementary Table 6). PsSY0018232, which was decreased in both ‘Sanyueli’ and ‘Fuhongli’ but more abundant in ‘Fuhongli’ during fruit ripening, has high sequence similarity with MYB repressors such as PpMYB6. Intriguingly, the expression of a gene annotated as LOB domain-containing protein 4 (LBD4, PsSY0017735) was significantly enhanced in the flesh of ‘Fuhongli’ during pigmentation but only expressed at a low level in the flesh of ‘Sanyueli’ (Figures 4, 5 and Supplementary Table 6). In addition, many other TF types including NACs, Dof, AP2, G2-like, FAR1, and MADS were also included in the differentially expressed transcription factor sets (Figure 5 and Supplementary Table 6). The expression pattern of PsICE1 and PsYABBY4, which are lowly expressed in the flesh of ‘Sanyueli’, was also confirmed by qRT-PCR (Figure 4).
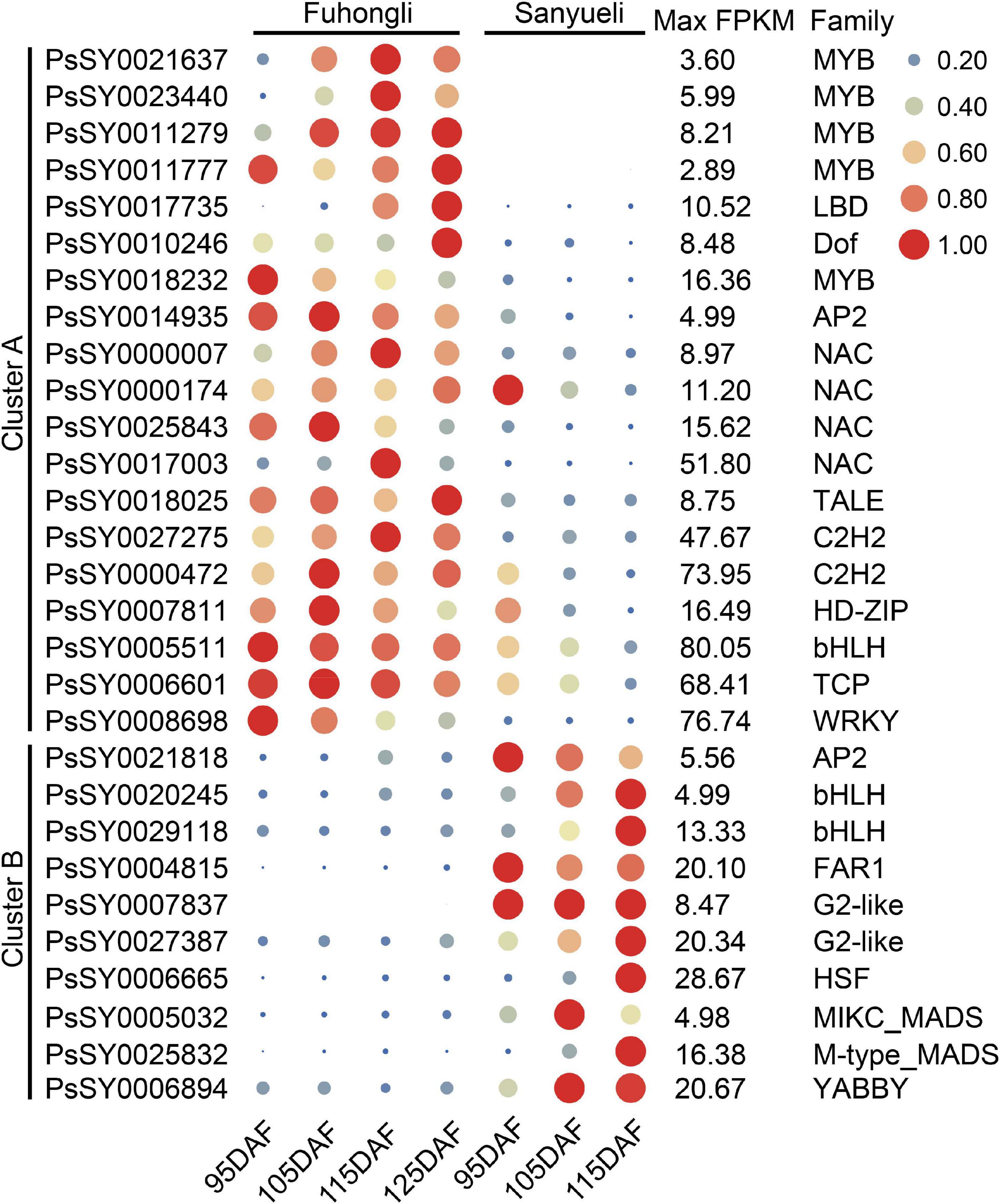
Figure 5. qRT-PCR validation of DEGs in flesh of ‘Sanyueli’ and ‘Fuhongli’ during fruit ripening. Actin was used as the reference gene. The error bars represent the standard error of three biological replicates. The left y-axis shows the relative gene expression levels obtained by qRT-PCR (solid lines). The right y-axis indicates the corresponding results of RNA-seq (dashed lines).
PsMYB10.2 Is a Positive Regulator of Anthocyanin Biosynthesis
Nucleotide sequence alignment results demonstrated that the coding region of the four PsMYB genes (PsSY0021637, PsSY0023440, PsSY0011279, and PsSY0011777) that were upregulated in the flesh of ‘Fuhongli’ were nearly identical (Supplementary Figure 3). The genomic DNA sequence of PsMYB genes was also nearly identical. However, a 7,045 nt insertion was detected in the second intron of PsSY0011777 (Supplementary Figure 4). The PsSY0021637, PsSY0023440, and PsSY0011279 encode a 732 bp coding sequence and predicted protein with 243 amino acid residues while PsSY0011777 encode a 741 bp coding sequence and predicted protein with 247 amino acid residues. Phylogenetic analysis showed that all of them fell in the anthocyanin clade (SG6) and were clustered into a subgroup with previously reported anthocyanin-activating MYB proteins from other Rosacea species (Figure 6A). The two proteins most closely related to the PsMYBs were PcMYB10.2 and PpMYB10.2 from cherry plum and peach (Figure 6A). Multiple sequence alignment showed the PsMYBs contain conserved R2R3 domain and ‘ANDV’ and SG6 motif for anthocyanin-promoting MYBs. They are highly homologous to PcMYB10.2 and PpMYB10.2, but PpMYB10.2 had an 18-amino acid deletion and PsMYBs (PsSY0021637 and PsSY0011279) had a 3-amino acid deletion in the C-terminal region (Figure 6B). These results suggest that the PsMYBs are likely activators of anthocyanin biosynthesis.
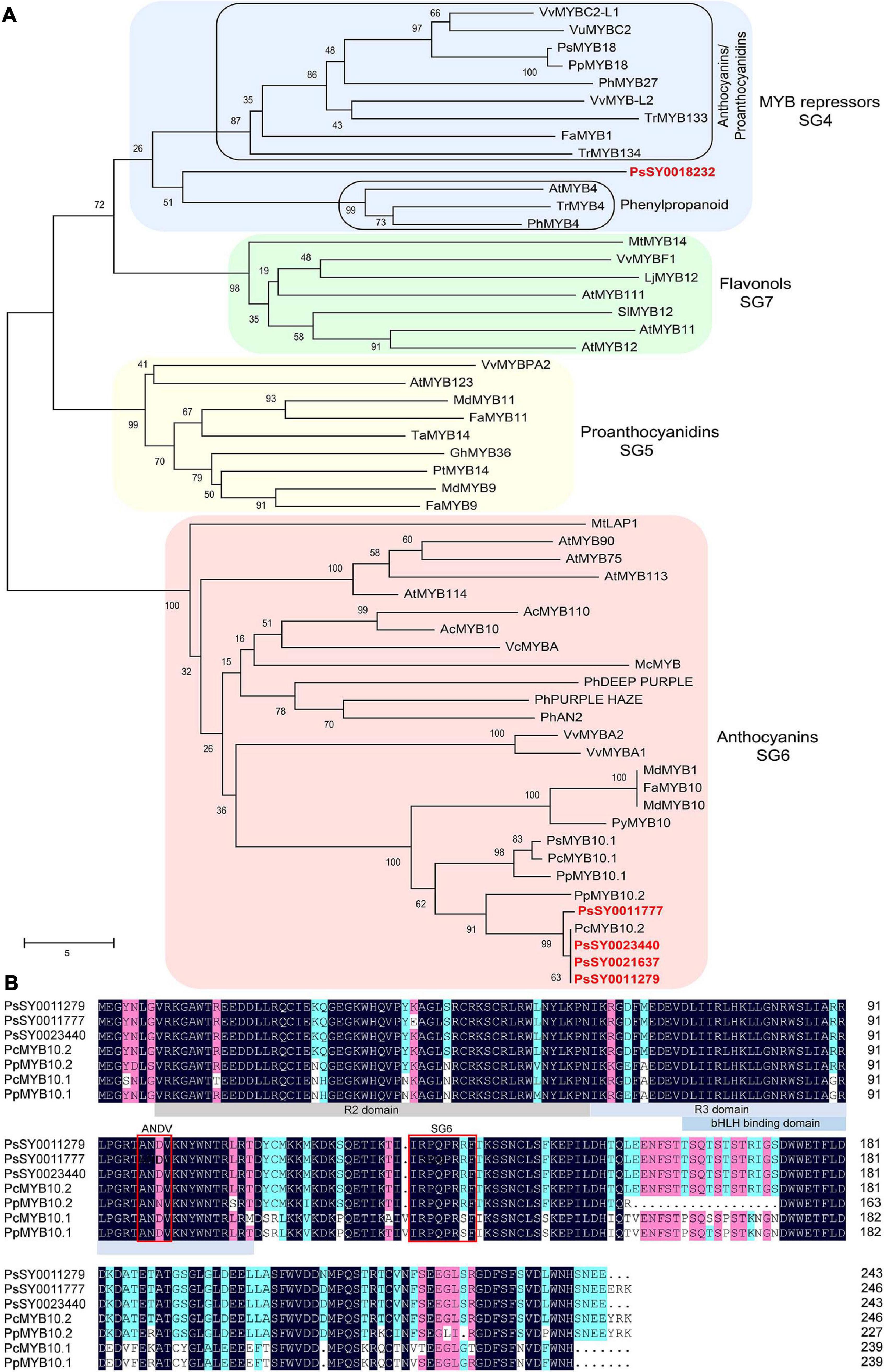
Figure 6. Sequence analysis of selected R2R3 MYBs. (A) Phylogenetic analysis of PsMYBs and R2R3 MYBs from other plant species. Plum MYBs are indicated in red font. (B) Multiple sequence alignment of PsMYBs and R2R3 MYBs from peach and cherry plum. R2, R3, and bHLH binding domain are highlighted in gray and blue colors, conserved ‘ANDV’ and SG6 motif for anthocyanin-promoting MYBs are indicated in red boxes.
The cDNA sequence of PsSY0011279 was cloned from the red flesh of ‘Fuhongli’ and designated as PsMYB10.2 (GenBank accession No. MK340932). The coding sequence of PsMYB10.2 is identical to the differentially expressed R2R3 MYB gene identified from red-fleshed ‘Furongli’ plum in our previous study (Fang et al., 2016b). PsMYB10.2 was nearly undetectable in the flesh of ‘Sanyueli’ but its expressed increased in the flesh of ‘Fuhongli’ during pigmentation (Figure 4). Tobacco leaf transient expression assays were carried out to test the function of PsMYB10.2. Coinfiltration of PsMYB10.2 and PsbHLH3 resulted in faint red coloration at infiltration sites 7 days after transformation, but no pigmentation was found when PsMYB10.2 or PsbHLH3 were infiltrated alone (Figure 7A). Quantification of anthocyanins in leaves of N. tabacum indicated that no anthocyanin was detected in the infiltrated areas transformed with empty vector, PsMYB10.2 or PsbHLH3 (Figure 7B). Anthocyanin was detected in tobacco leaves infiltrated with PsMYB10.2 and PsbHLH3, but the content was significantly lower than that in tobacco leaves infiltrated with PpMYB10.1 and PsbHLH3 (Figure 7B). In addition, promoter structure analysis revealed there are multiple potential MYB binding sites in the promoter of PsUFGT and PsGST (Figure 7C). The promoters of PsUFGT and PsGST were isolated and fused to a luciferase reporter and dual luciferase assays were employed to investigate the interaction of PsMYB10.2 with the promoter sequences of structural genes involved in anthocyanin accumulation. Infiltration of PsMYB10.2 was able to activate the promoters of both PsUFGT and PsGST, and co-infiltration with PsbHLH3 enhanced the activity of PsMYB10.2 (Figure 7D). These results confirmed the role of PsMYB10.2 as a positive regulator of anthocyanin accumulation.
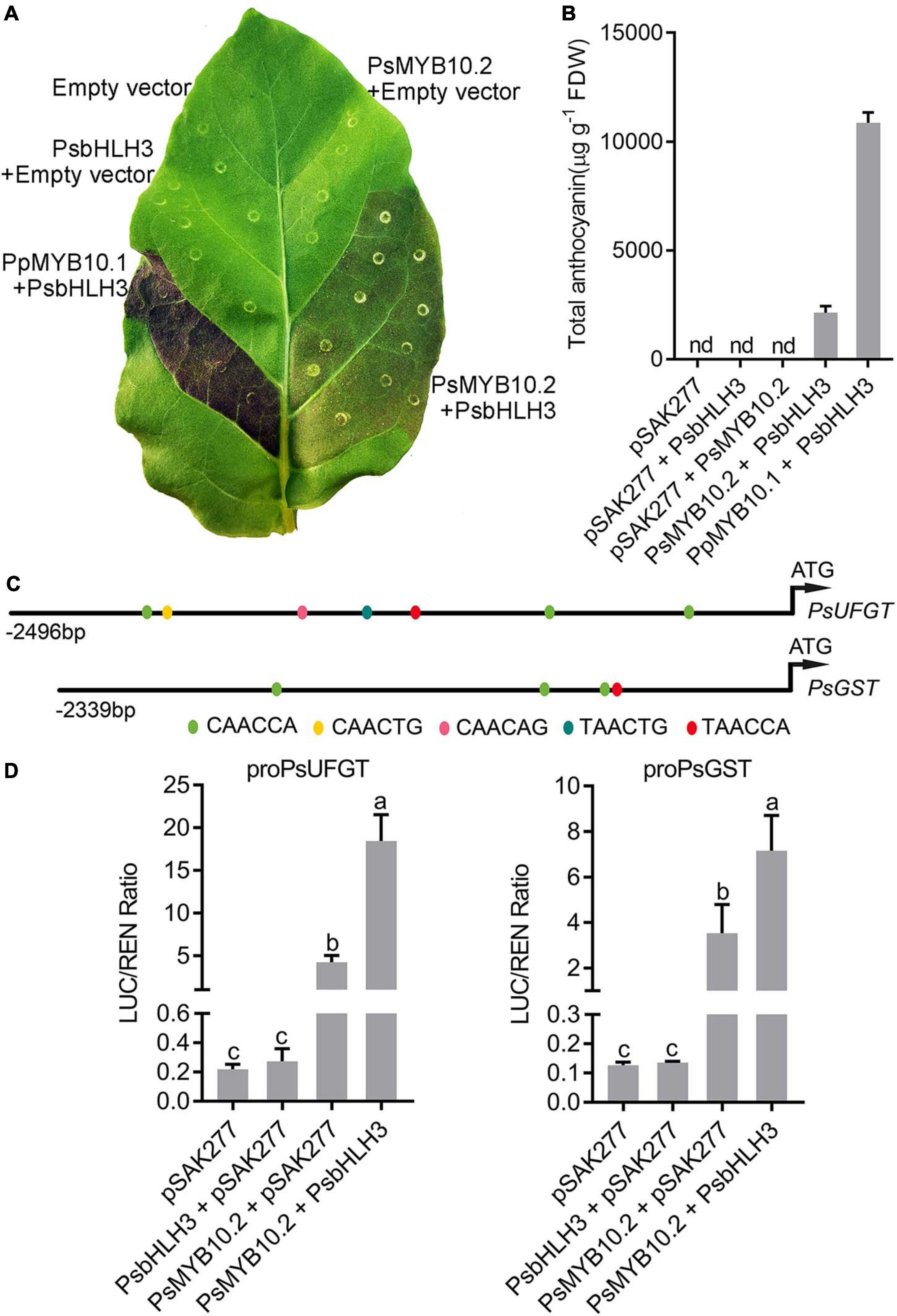
Figure 7. Functional analysis of the PsMYB10.2 gene using transient assay in tobacco. (A) Transient expression of PsMYB10.2 in tobacco leaf. The photo was taken 7 days after infiltration. Peach PpMYB10.1/PsbHLH3 was used as positive control. (B) Anthocyanin content in transformed leaves of tobacco. Each value represents the mean of three biological replicates. FDW, freeze dry weight. nd, non-detected. (C) Schematic diagram of the PsUFGT and PsGST promoter. The prediction of the cis-acting elements in the promoter of PsUFGT and PsGST was performed using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database. Color ellipses represent predicted MYB binding sites. (D) Analysis of the interaction of the PsMYB10.2 gene with the promoters of PsUFGT and PsGST genes using dual-luciferase reporter assay Nicotiana benthamiana leaves. Error bars represent the standard errors for three replicates. Different lowercase letters indicate significant differences among treatments according to one-way ANOVA test (P < 0.05). Means with the same letter are not significantly different at the 0.05 level.
Silencing of the PsMYB10.2 Reduces Anthocyanin Accumulation in Flesh of ‘Fuhongli’
To further validate the role of the PsMYB10.2, the VIGS system was used to transiently suppress the expression of PsMYB10.2 in the flesh of ‘Fuhongli’. Coinjection with pTRV1 and pTRV2-PsMYB10.2 reduced pigmentation in infiltration sites (Figure 8A). The anthocyanin content in the flesh around the injection site of pTRV1/pTRV2-PsMYB10.2 was significantly lower than the corresponding opposite non-injected site (Figure 8B). Anthocyanin content in the flesh tissues around the infiltration site of pTRV1/pTRV2 was reduced, but significantly higher than that of pTRV1/pTRV2-PsMYB10.2. In addition, the expression level of PsMYB10.2 and PsUFGT at the injection site of pTRV1/pTRV2-PsMYB10.2decreased by approximately 57 and 66%, respectively, compared with the injection site of pTRV1/pTRV2 (Figure 8C). These results further confirm that PsMYB10.2 is involved in the regulation of anthocyanin accumulation in flesh of ‘Fuhongli’.
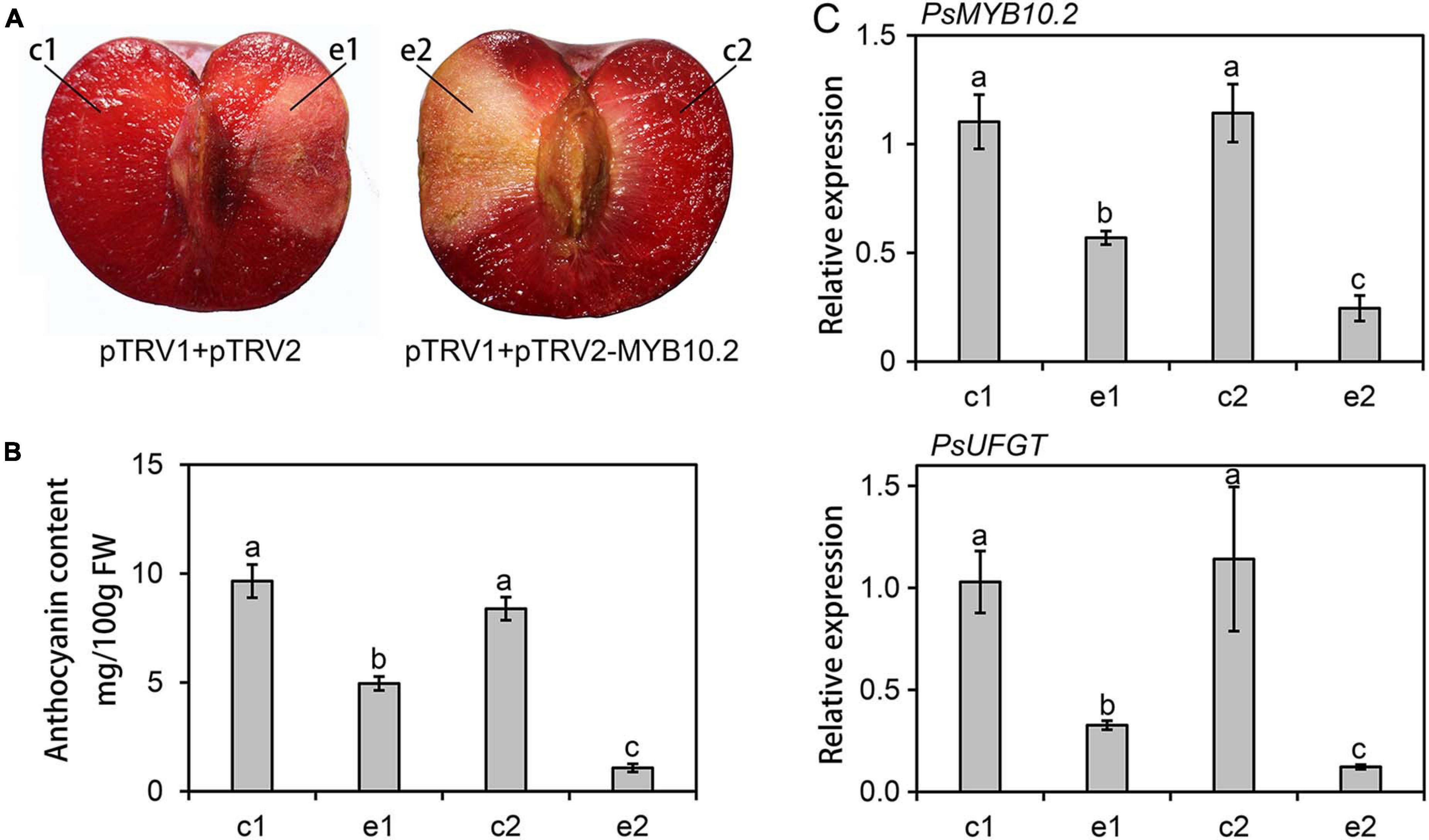
Figure 8. Functional analysis of the PsMYB10.2 gene by TRV-based virus-induced gene silencing. (A) Silencing of the PsMYB10.2 gene in ‘Fuhongli’ plum fruit at ripening stages. (B) Anthocyanin content in the flesh tissues around the infiltration sites (e1 and e2) and the opposite non-infiltrated sites (c1 and c2). FW, fresh weight. (C) Expression levels of the PsMYB10.2 and PsUFGT in the flesh tissues. The flesh tissues were collected 7 days after infiltration. Error bars represent the standard errors for three replicates. Different lowercase letters indicate significant differences among treatments according to one-way ANOVA test (P < 0.05).
Discussion
The association of anthocyanins with fruit quality and potential health benefits, has driven efforts to produce anthocyanin-rich plums through breeding and postharvest treatments in recent years (Cevallos-Casals et al., 2006; Fanning et al., 2014; Santhakumar et al., 2015; Fang et al., 2016a; Wang et al., 2020). To our knowledge, ‘Fuhongli’ is the first red-fleshed mutant induced by γ-radiation. In this study, the mechanism involved in anthocyanin accumulation in the flesh of ‘Fuhongli’ was investigated using HPLC, RNA-Seq, dual-luciferase assays and transient color assays in tobacco leaves and plum fruits.
‘Fuhongli’ is a red-fleshed and late-ripening mutant of ‘Sanyueli’ plum (Fang et al., 2016a). A delayed sugar accumulation was observed in the flesh of ‘Fuhongli’, together with delayed reduction of organic acid concentration through ripening (Figure 1), corroborating its late-ripening trait. Our results indicated also that anthocyanin content increased rapidly in fruit flesh of ‘Fuhongli’ from 115 DAF. The presence of cyanidin-3-rutinoside and cyanidin-3-glucoside, which have been identified as the main anthocyanins in Chinese plum (Fanning et al., 2014), was confirmed in ‘Fuhongli’, while no anthocyanin was detected in flesh of ‘Sanyueli’. Our previous study indicated that the structural genes involved in anthocyanin accumulation were upregulated during fruit ripening (Fang et al., 2016b). It is possible that ‘Fuhongli’ has gained the ability to accumulate anthocyanins due to the activation of anthocyanin biosynthetic genes. In this study, comparison of ‘Sanyueli’ and ‘Fuhongli’ fruit flesh transcriptomes indicated that most genes involved in biosynthesis and accumulation of anthocyanin were also upregulated in flesh of ‘Fuhongli’. On the other hand, most biosynthetic enzymes were lowly expressed or decreased in flesh of ‘Sanyueli’ flesh during ripening. The transcripts of PsUFGT and PsGST were barely in ‘Sanyueli’ flesh. This suggests a direct link to the lack of anthocyanin accumulation in flesh of ‘Sanyueli’.
The upregulation of structural genes suggested that anthocyanin accumulation in ‘Fuhongli’ flesh might be transcriptionally regulated. Several studies have suggested R2R3 MYBs as key genes for anthocyanin accumulation in Rosaceae species (Lin-Wang et al., 2010; Rahim et al., 2014; Gu et al., 2015). Our previous study showed that PsMYB10.2 was likely the key gene involved in anthocyanin biosynthesis in flesh of ‘Furongli’ plum (Fang et al., 2016b). Results from the RNA-seq and qRT-PCR analyses performed in this study further indicated that transcripts encoding PsMYB10.2 were significantly upregulated in flesh of ‘Fuhongli’ but not expressed in flesh of ‘Sanyueli’. Sequence analysis indicated that PsMYB10.2 belongs to the anthocyanin activator clade, similar to peach PpMYB10.2 (Zhou et al., 2016, 2018) and shows a high amino acid sequence homology to the cherry plum activator PcMYB10.2. Ectopic expression of PsMYB10.2 and PsbHLH3 resulted in anthocyanin accumulation and red pigmentation in tobacco leaves, but no pigmentation was observed when PsMYB10.2 was infiltrated alone. PsMYB10.2 silencing by VIGS reduced the expression of PsUFGT and the concentration of anthocyanins in the flesh of ‘Fuhongli’. These results suggested that PsMYB10.2 is a key anthocyanin activator responsible for red pigmentation in the flesh of ‘Fuhongli’.
The upregulation of PsMYB10.2 in flesh of ‘Fuhongli’ suggests mutations are responsible for its transcriptional activation. For example, changes in the upstream regulators of PsMYB10.2 or a loss of repression. Consistent with this, several transcription factors that were more abundant in the flesh ‘Fuhongli’ or ‘Sanyueli’ were identified. These include the transcription factors that have been suggested to regulate the expression of anthocyanin MYB activators such as NAC (Zhou H. et al., 2015) and MADS (Jaakola et al., 2010). Interestingly, our results demonstrated that the expression level of PsLBD4 was upregulated and more abundant in the flesh of ‘Fuhongli’. LBD genes were first identified for their role in lateral organ development in Arabidopsis (Shuai et al., 2002) and have been reported to be involved in regulation of anthocyanin biosynthesis but they act as repressors. Rubin et al. (2009) demonstrated that AtLBD37, AtLBD38, and AtLBD39 are anthocyanin repressors. They showed that overexpression of these genes inhibited nitrogen deficiency induced anthocyanin via suppressing the expression of anthocyanin activators PAP1 and PAP2, while loss of function enhanced anthocyanin accumulation. Li et al. (2017) reported a nitrate-induced LBD gene MdLBD13, a homolog of Arabidopsis LBD37/38, repressed anthocyanin biosynthesis by reducing expression of the flavonoid pathway genes in apple. However, Zhang et al. (2019) found that CsLOB_3 and CsLBD36_2 can bind to and activate the promoter of structural genes involved in flavonoid biosynthesis. However, it remains to be investigated as to whether these transcription factors are involved in regulating the expression of PsMYB10.2 and anthocyanin biosynthesis in the flesh of ‘Fuhongli’.
The MYB responsible for anthocyanin biosynthesis in peach flesh is PpMYB10.1 (Zhou H. et al., 2015), while PpMYB10.2 is involved in peach flower coloration (Zhou et al., 2016). Tuan et al. (2015) showed that PpMYB10.2 was expressed in peach leaves that do not contain anthocyanin. Gu et al. (2015) suggested that PcMYB10.2 is involved in purple coloration in sepals of cv. Ziyeli. They showed that PcMYB10.2 was expressed in green-colored flesh of cherry plum, but it was undetectable in purple-colored flesh (Gu et al., 2015). High expression of PcMYB10.2 was also observed in both purple and green fruit skin, but its expression was much higher in green fruit skin than in purple fruit skin (Gu et al., 2015). These results suggest divergence of the function of MYB10.2 gene in Rosaceous species.
Our results showed that PsMYB10.2 was closely related to peach PpMYB10.2, the anthocyanin-promoting ability of which is much weaker than PpMYB10.1. Zhou et al. (2018) showed that PpMYB10.2 only induced pale red coloration in tobacco leaves, when infiltrated alone no pigmentation was observed. Similarly, transient expression of PsMYB10.2 and PsbHLH3 only induced faint anthocyanin production in tobacco leaves. It is noteworthy that the red coloration induced by PsMYB10.2 and PsbHLH3 was observed one a week after infiltration and was stable, while PpMYB10.2 and PpbHLH3 induced faint coloration in tobacco leaves 2 weeks after infiltration and the faint pigmentation was only observed about one-third of the replicates (Zhou et al., 2018). Our results demonstrated that PsMYB10.2 was weaker than PpMYB10.1 and possibly stronger than PpMYB10.2. Sequence alignment indicated that there was an 18-amino acids deletion in C-terminal of PpMYB10.2. Whether the 18-amino acids deletion in PpMYB10.2 is responsible for the difference of anthocyanin-promoting activity between PsMYB10.2 and PpMYB10.2 need further investigation. In most cases, the anthocyanin content is low (often <20 mg per 100 g fresh weight) in the flesh of red-fleshed plums and is much lower than that in the peel of red or black skin plums (Fanning et al., 2014; Zhou D. et al., 2015). Similarly low anthocyanin content was also observed in the flesh of ‘Furongli’(Fang et al., 2016b) and ‘Fuhongli’ in this study. As PsMYB10.2 was responsible for anthocyanin accumulation in the flesh of plum (Fang et al., 2016b), the weak anthocyanin-promoting ability of PsMYB10.2 may explain the low anthocyanin concentration in flesh of red-fleshed plums. An anthocyanin-rich plum cultivar Queen Garnet, which was shown to contain 107 mg per 100 g fresh weight in flesh, was developed in Queensland (Netzel et al., 2012; Fanning et al., 2014). It would be interesting to identify anthocyanin activators and elucidate underlying mechanisms responsible for anthocyanin biosynthesis in the flesh of anthocyanin-rich plum cultivars such as Queen Garnet.
Our study shows that the direct mechanism for the presence of red color in the flesh of ‘Fuhongli’ plum fruit is via the upregulation of the anthocyanin biosynthetic and transport pathway genes. This is mediated by the transcriptional activator PsMYB10.2. Further work will be required to establish the causation of PsMYB10.2 upregulation in the mutagenized ‘Fuhongli’ plum. Since this is unlikely to be due to actual changes in the coding sequences of the activating MYBs, then it may be due to changes in upstream regulators of the MYB or some loss of repression. Future studies should focus on investigating mutations that are responsible for activating the expression of PsMYB10.2 in flesh of ‘Fuhongli’. Taken together, the results of our study provide useful information for the development of new red-fleshed plums.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA690967.
Author Contributions
Z-ZF and X-FY supervised the project. Z-ZF designed and performed the whole experiments and wrote the manuscript. D-RZ, Y-JL, and C-CJ participate in the experiments and data analysis. KL-W and RE provided scientific suggestion and revised the manuscript. SL-P prepared and handled plum samples. CA performed quantification of anthocyanins in tobacco leaves and revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the Natural Science Foundation of Fujian Province (2017J01043), the National Natural Science Foundation of China (31801916), the Basic Scientific Research Funds of Public Welfare Scientific Research Institutes of Fujian Province (2018R1013-1), the China Scholarship Council (201909350001), and Projects of Fujian Academy of Agricultural Sciences (YC2015-9 and AGY2018-3).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This was a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.680469/full#supplementary-material
Supplementary Figure 1 | Heat map of correlation coefficients between samples.
Supplementary Figure 2 | Differentially expressed genes (DEG) identified by RNA-seq analysis in the flesh of ‘Sanyueli’ and the mutant. (A) DEGs of different comparison groups. (B) Venn diagram showing the number of differentially expressed genes. SY indicates DEGs during ripening of ‘Sanyueli’, MT indicates DEGs during ripening of the red-fleshed mutant, SY vs. MT indicates DEGs between ‘Sanyueli’ and its red-fleshed mutant.
Supplementary Figure 3 | Nucleotide alignment of the coding sequence of PsSY0021637, PsSY0023440, PsSY0011279, and PsSY0011777.
Supplementary Figure 4 | Gene structure of PsSY0021637, PsSY0023440, PsSY0011279, and PsSY0011777. (A) Schematic overview of introns and exons in PsSY0021637, PsSY0023440, PsSY0011279, and PsSY0011777. (B) Structural alignments of PsSY0011279 and PsSY0011777. Boxes and lines represent exons and introns, respectively.
Supplementary Table 1 | Primer sequences for qRT-PCR analysis.
Supplementary Table 2 | Sequences of primers used for vector construction.
Supplementary Table 3 | Summary of sequencing and mapping results.
Supplementary Table 4 | Summary of functional annotations for putative new genes.
Supplementary Table 5 | Genes assigned to phenylpropanoid biosynthesis and flavonoid biosynthesis pathway.
Supplementary Table 6 | Differentially expressed transcription factor genes.
Abbreviations
4CL, 4-coumaroyl:CoA-ligase; ANS, anthocyanin synthase; C4H, cinnamate-4-hydroxylase; CHI, chalcone isomerase; CHS, chalcone synthase; DAF, days after flowering; DFR, dihydroflavonol-4-reductase; DEG, differentially expressed genes; F3H, flavanone-3-hydroxylase; F3′H, flavonoid3′-hydroxylase; FDW, freeze dry weight; FPKM, fragments per kilobase of transcript per million fragments mapped; GO, gene ontology; GST, glutathione S-transferase; HPLC, high performance liquid chromatography; KEGG, Kyoto Encyclopedia of Genes and Genomes; MBW, MYB-bHLH-WD40; NR, non-redundant protein sequence database; PAL, phenylalanine ammonia-lyase; qRT-PCR, real-time quantitative RT-PCR; UFGT, UDPglucose:flavonoid-3-O-glucosyltransferase; VIGS, virus-induced gene silencing; Swiss-Prot, Swiss-Prot protein database; COG, Clusters of Orthologous Groups database; KOG, euKaryotic Ortholog Groups of proteins database; Pfam, protein families database; LBD, LOB domain-containing protein.
Footnotes
References
Allan, A. C., Hellens, R. P., and Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., and Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates ABA-promoted anthocyanin accumulation. Plant Cell Environ. 41, 2678–2692. doi: 10.1111/pce.13393
Andre, C. M., Greenwood, J. M., Walker, E. G., Rassam, M., Sullivan, M., Evers, D., et al. (2012). Anti-inflammatory procyanidins and triterpenes in 109 apple varieties. J. Agric. Food Chem. 60, 10546–10554. doi: 10.1021/jf302809k
Bakuradze, T., Tausend, A., Galan, J., Groh, I. A. M., Berry, D., Tur, J. A., et al. (2019). Antioxidative activity and health benefits of anthocyanin-rich fruit juice in healthy volunteers. Free Radic. Res. 53, 1045–1055. doi: 10.1080/10715762.2019.1618851
Butelli, E., Garcia-Lor, A., Licciardello, C., Las Casas, G., Hill, L., Reforgiato Recupero, G., et al. (2017). Changes in anthocyanin production during domestication of Citrus. Plant Physiol. 173, 2225–2242. doi: 10.1104/pp.16.01701
Cevallos-Casals, B. A., Byrne, D., Okie, W. R., and Cisneros-Zevallos, L. (2006). Selecting new peach and plum genotypes rich in phenolic compounds and enhanced functional properties. Food Chem. 96, 273–280.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202.
Chen, Z., and Zhu, C. (2011). Combined effects of aqueous chlorine dioxide and ultrasonic treatments on postharvest storage quality of plum fruit (Prunus salicina L.). Postharvest Biol. Technol. 61, 117–123. doi: 10.1016/j.postharvbio.2011.03.006
Cheng, Y., Liu, L., Yuan, C., and Guan, J. (2016). Molecular characterization of ethylene-regulated anthocyanin biosynthesis in plums during fruit ripening. Plant Mol. Biol. Rep. 34, 777–785. doi: 10.1007/s11105-015-0963-x
Espley, R. V., Bovy, A., Bava, C., Jaeger, S. R., Tomes, S., Norling, C., et al. (2013). Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 11, 408–419. doi: 10.1111/pbi.12017
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fang, Z.-Z., Lin-Wang, K., Dai, H., Zhou, D.-R., Jiang, C.-C., Espley, R. V., et al. (2020). The genome of low-chill Chinese plum ‘Sanyueli’ (Prunus salicina Lindl.) provides insights into the regulation of the chilling requirement of flower buds. bioRxiv [Preprint]. doi: 10.1101/2020.07.31.193243
Fang, Z. Z., Ye, X., Zhou, D., Pan, S., and Jiang, C. (2016a). ISSR analysis on red-flesh and late-ripening mutant of Sanyue Plum (Prunus salicina Lindl.) induced by irradiation. J. Nucl. Agric. Sci. 30, 209–215.
Fang, Z. Z., Zhou, D. R., Ye, X. F., Jiang, C. C., and Pan, S. L. (2016b). Identification of candidate anthocyanin-related genes by transcriptomic analysis of ‘Furongli’ plum (Prunus salicina Lindl.) during fruit ripening using RNA-Seq. Front. Plant Sci. 7:1338. doi: 10.3389/fpls.2016.01338
Fanning, K. J., Topp, B., Russell, D., Stanley, R., and Netzel, M. (2014). Japanese plums (Prunus salicina Lindl.) and phytochemicals – breeding, horticultural practice, postharvest storage, processing and bioactivity. J. Sci. Food Agric. 94, 2137–2147. doi: 10.1002/jsfa.6591
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, S., Sun, S., Chen, X., Wu, S., Wang, D., and Chen, X. (2015). PyMYB10 and PyMYB10.1 interact with bHLH to enhance anthocyanin accumulation in pears. PLoS One 10:e0142112. doi: 10.1371/journal.pone.0142112
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gu, C., Liao, L., Zhou, H., Wang, L., Deng, X., and Han, Y. (2015). Constitutive activation of an anthocyanin regulatory gene PcMYB10.6 is related to red coloration in purple-foliage plum. PLoS One 10:e0135159. doi: 10.1371/journal.pone.0135159
He, J., and Giusti, M. M. (2010). Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 1, 163–187. doi: 10.1146/annurev.food.080708.100754
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 3, 509–523.
Hou, F., Xie, B., Zhen, Q., Li, A., Dong, S., Zhang, H., et al. (2020). Sweetpotato leaf curl virus decreased storage root yield and quality of edible sweetpotato (Ipomoea batatas (L.) Lam) in China. Agron. J. 112, 3948–3962. doi: 10.1002/agj2.20025
Igwe, E. O., Charlton, K. E., Roodenrys, S., Kent, K., Fanning, K., and Netzel, M. E. (2017). Anthocyanin-rich plum juice reduces ambulatory blood pressure but not acute cognitive function in younger and older adults: a pilot crossover dose-timing study. Nutr. Res. 47, 28–43.
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jaakola, L., Poole, M., Jones, M. O., Kämäräinen-Karppinen, T., Koskimäki, J. J., Hohtola, A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. doi: 10.1104/pp.110.158279
Jamar, G., Estadella, D., and Pisani, L. P. (2017). Contribution of anthocyanin-rich foods in obesity control through gut microbiota interactions. Biofactors 43, 507–516. doi: 10.1002/biof.1365
Jia, D., Li, Z., Dang, Q., Shang, L., Shen, J., Leng, X., et al. (2020). Anthocyanin biosynthesis and methylation of the MdMYB10 promoter are associated with the red blushed-skin mutant in the red striped-skin ‘Changfu 2’ apple. J. Agric. Food Chem. 68, 4292–4304. doi: 10.1021/acs.jafc.9b07098
Jiang, Y., Liu, C., Yan, D., Wen, X., Liu, Y., Wang, H., et al. (2017). MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 68, 1055–1069. doi: 10.1093/jxb/erx029
Jin, W., Wang, H., Li, M., Wang, J., Yang, Y., Zhang, X., et al. (2016). The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. doi: 10.1111/pbi.12568
Johnson, C. D., Lucas, E. A., Hooshmand, S., Campbell, S., Akhter, M. P., and Arjmandi, B. H. (2011). Addition of fructooligosaccharides and dried plum to soy-based diets reverses bone loss in the ovariectomized rat. Evid. Based Complement. Alternat. Med. 2011:836267.
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kotepong, P., Paull, R. E., and Ketsa, S. (2019). Anthocyanin accumulation and differential gene expression in wild-type and mutant Syzygium malaccense fruits during their growth and ripening. Biol. Plant. 63, 710–720.
Lee, S. H., Lillehoj, H. S., Cho, S. M., Chun, H. K., Park, H. J., Lim, C. I., et al. (2009). Immunostimulatory effects of oriental plum (Prunus salicina Lindl.). Comp. Immunol. Microbiol. Infect. Dis. 32, 407–417. doi: 10.1016/j.cimid.2007.12.001
Li, H., Yang, Z., Zeng, Q., Wang, S., Luo, Y., Huang, Y., et al. (2020). Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 7:83. doi: 10.1038/s41438-020-0302-8
Li, H.-H., Liu, X., An, J.-P., Hao, Y.-J., Wang, X.-F., You, C.-X. J. P. C., et al. (2017). Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. 130, 47–59. doi: 10.1007/s11240-017-1203-x
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921. doi: 10.1111/nph.13816
Li, Z., Wang, J., Zhang, X., and Xu, L. (2015). Comparative transcriptome analysis of Anthurium ‘Albama’ and its anthocyanin-loss mutant. PLoS One 10:e0119027. doi: 10.1371/journal.pone.0119027
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Liu, C., Zhu, L., Fukuda, K., Ouyang, S., Chen, X., Wang, C., et al. (2017). The flavonoid cyanidin blocks binding of the cytokine interleukin-17A to the IL-17RA subunit to alleviate inflammation in vivo. Sci. Signal. 10:eaaf8823. doi: 10.1126/scisignal.aaf8823
Liu, C.-C., Chi, C., Jin, L.-J., Zhu, J., Yu, J.-Q., and Zhou, Y.-H. (2018). The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41, 1762–1775. doi: 10.1111/pce.13171
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Milošević, T., and Milošević, N. (2018). ‘Plum (Prunus spp.) breeding,’ in Advances in Plant Breeding Strategies: Fruits, Vol. 3, eds J. M. Al-Khayri, S. M. Jain, and D. V. Johnson (Cham: Springer), 165–215.
Netzel, M., Fanning, K., Netzel, G., Zabaras, D., Karagianis, G., Treloar, T., et al. (2012). Urinary excretion of antioxidants in healthy humans following queen garnet plum juice ingestion: a new plum variety rich in antioxidant compounds. J. Food Biochem. 36, 159–170. doi: 10.1111/j.1745-4514.2010.00522.x
Niu, S., Xu, C., Zhang, W., Zhang, B., Li, X., Lin-Wang, K., et al. (2010). Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 231, 887–899. doi: 10.1007/s00425-009-1095-z
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Plunkett, B. J., Espley, R. V., Dare, A. P., Warren, B. A. W., Grierson, E. R. P., Cordiner, S., et al. (2018). MYBA from blueberry (Vaccinium Section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 9:1300. doi: 10.3389/fpls.2018.01300
Rahim, M. A., Busatto, N., and Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. doi: 10.1007/s00425-014-2078-2
Rubin, G., Tohge, T., Matsuda, F., Saito, K., and Scheible, W.-R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. doi: 10.1105/tpc.109.067041
Santhakumar, A. B., Kundur, A. R., Fanning, K., Netzel, M., Stanley, R., and Singh, I. (2015). Consumption of anthocyanin-rich Queen Garnet plum juice reduces platelet activation related thrombogenesis in healthy volunteers. J. Funct. Foods 12, 11–22. doi: 10.1016/j.jff.2014.10.026
Shuai, B., Reynaga-Peña, C. G., and Springer, P. S. (2002). The Lateral Organ Boundaries gene defines a novel, plant-specific gene damily. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Silvestri, C., Cirilli, M., Zecchini, M., Muleo, R., and Ruggieri, A. (2018). Consumer acceptance of the new red-fleshed apple variety. J. Food Prod. Mark. 24, 1–21. doi: 10.1080/10454446.2016.1244023
Spelt, C., Quattrocchio, F., Mol, J. N. M., and Koes, R. (2000). anthocyanin1 of Petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1632. doi: 10.1105/tpc.12.9.1619
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Tomás-Barberán, F. A., Gil, M. I., Cremin, P., Waterhouse, A. L., Hess-Pierce, B., and Kader, A. A. (2001). HPLC-DAD-ESIMS analysis of phenolic compounds in nectarines, peaches, and plums. J. Agric. Food Chem. 49, 4748–4760. doi: 10.1021/jf0104681
Tuan, P. A., Bai, S., Yaegaki, H., Tamura, T., Hihara, S., Moriguchi, T., et al. (2015). The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 15:280. doi: 10.1186/s12870-015-0664-5
Usenik, V., Štampar, F., and Veberič, R. (2009). Anthocyanins and fruit colour in plums (Prunus domestica L.) during ripening. Food Chem. 114, 529–534. doi: 10.1016/j.foodchem.2008.09.083
Valero, D., Díaz-Mula, H. M., Zapata, P. J., Guillén, F., Martínez-Romero, D., Castillo, S., et al. (2013). Effects of alginate edible coating on preserving fruit quality in four plum cultivars during postharvest storage. Postharvest Biol. Technol. 77, 1–6. doi: 10.1016/j.postharvbio.2012.10.011
Wang, H., Wang, X., Song, W., Bao, Y., Jin, Y., Jiang, C., et al. (2019). PdMYB118, isolated from a red leaf mutant of Populus deltoids, is a new transcription factor regulating anthocyanin biosynthesis in poplar. Plant Cell Rep. 38, 927–936. doi: 10.1007/s00299-019-02413-1
Wang, L., Sang, W., Xu, R., and Cao, J. (2020). Alteration of flesh color and enhancement of bioactive substances via the stimulation of anthocyanin biosynthesis in ‘Friar’ plum fruit by low temperature and the removal. Food Chem. 310:125862.
Wang, N., Liu, W., Zhang, T., Jiang, S., Xu, H., Wang, Y., et al. (2018). Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 66, 7076–7086. doi: 10.1021/acs.jafc.8b01273
Wang, R., Wang, L., Yuan, S., Li, Q., Pan, H., Cao, J., et al. (2018). Compositional modifications of bioactive compounds and changes in the edible quality and antioxidant activity of ‘Friar’ plum fruit during flesh reddening at intermediate temperatures. Food Chem. 254, 26–35.
Xu, Z.-S., Yang, Q. Q., Feng, K., Yu, X., and Xiong, A. S. (2020). DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 18, 1585–1597. doi: 10.1111/pbi.13325
Xuan, J., Jia, Z., Qain, M., Zhang, J., Wang, G., and Guo, Z. (2015). Comparative of methods for RNA extraction from plum (Prunus salicina Lindl.) fruit flesh. North. Hort. 22, 110–113.
Zhang, J., and Zhou, E. (1998). China Fruit-Plant Monograph Chinese Plum. Beijing: China Forestry Press.
Zhang, X., He, Y., He, W., Su, H., Wang, Y., Hong, G., et al. (2019). Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 9:15651. doi: 10.1038/s41598-019-52027-6
Zhang, Y., Fang, Z., Ye, X., and Pan, S. (2018). Identification of candidate genes involved in anthocyanin accumulation in the peel of jaboticaba (Myrciaria cauliflora) fruits by transcriptomic analysis. Gene 676, 202–213.
Zhou, D., Ye, X., Fang, Z., Pan, S., Jiang, C., Liao, R., et al. (2015). Research on relativity of color with contents of anthocyanin, flavonoids and carotenoid in plum. Fujian J. Agric. Sci. 30, 33–37.
Zhou, H., Liao, L., Xu, S., Ren, F., Zhao, J., Ogutu, C., et al. (2018). Two amino acid changes in the R3 repeat cause functional divergence of two clustered MYB10 genes in peach. Plant Mol. Biol. 98, 169–183. doi: 10.1007/s11103-018-0773-2
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Zhou, H., Lin-Wang, K., Wang, H., Gu, C., Dare, A. P., Espley, R. V., et al. (2015). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. doi: 10.1111/tpj.12792
Zhou, H., Peng, Q., Zhao, J., Owiti, A., Ren, F., Liao, L., et al. (2016). Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 7:1557. doi: 10.3389/fpls.2016.01557
Zhou, Y., Zhou, H., Lin-Wang, K., Vimolmangkang, S., Espley, R. V., Wang, L., et al. (2014). Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 14:388. doi: 10.1186/s12870-014-0388-y
Keywords: transcriptome, red-fleshed mutant, anthocyanin biosynthesis, PsMYB10.2, plum
Citation: Fang Z-Z, Lin-Wang K, Zhou D-R, Lin Y-J, Jiang C-C, Pan S-L, Espley RV, Andre CM and Ye X-F (2021) Activation of PsMYB10.2 Transcription Causes Anthocyanin Accumulation in Flesh of the Red-Fleshed Mutant of ‘Sanyueli’ (Prunus salicina Lindl.). Front. Plant Sci. 12:680469. doi: 10.3389/fpls.2021.680469
Received: 14 March 2021; Accepted: 21 May 2021;
Published: 22 June 2021.
Edited by:
Zeng-Yu Wang, Qingdao Agricultural University, ChinaReviewed by:
Ulhas S. Kadam, Gyeongsang National University, South KoreaDang Fengfeng, South China Agricultural University, China
Copyright © 2021 Fang, Lin-Wang, Zhou, Lin, Jiang, Pan, Espley, Andre and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Zhen Fang, ZnpoemgyMDA4QDE2My5jb20=; ZmFuZ3poaXpoZW5AZmFhcy5jbg==; Xin-Fu Ye, c3BlY2lhbGZydWl0QDEyNi5jb20=; eWV4aW5mdUAxMjYuY29t
 Zhi-Zhen Fang
Zhi-Zhen Fang Kui Lin-Wang
Kui Lin-Wang Dan-Rong Zhou1,2
Dan-Rong Zhou1,2 Richard V. Espley
Richard V. Espley Christelle M. Andre
Christelle M. Andre