
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 02 June 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.679140
This article is part of the Research Topic Plasmodesmata: Recent Progress and New Insights View all 12 articles
Plasmodesmata (PD) are cytoplasmic canals that facilitate intercellular communication and molecular exchange between adjacent plant cells. PD-associated proteins are considered as one of the foremost factors in regulating PD function that is critical for plant development and stress responses. Although its potential to be used for crop engineering is enormous, our understanding of PD biology was relatively limited to model plants, demanding further studies in crop systems. Recently developed genome editing techniques such as Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associate protein (CRISPR/Cas) might confer powerful approaches to dissect the molecular function of PD components and to engineer elite crops. Here, we assess several aspects of PD functioning to underline and highlight the potential applications of CRISPR/Cas that provide new insight into PD biology and crop improvement.
The discovery of plasmodesmata (PD) in 1885 by Edward Tangl has revolutionized the field of plant science. PD functions as one of the vital controllers in plant growth and development (Wu et al., 2018; Vu et al., 2020). Briefly, PD are symplasmic (cytoplasm-to-cytoplasm) nanochannels between adjacent cells, approximately 50-60 nm in size. Structurally, a plasmodesma (the singular form of PD) consists of a cytoplasmic sleeve and a desmotubule (Bell and Oparka, 2011). The space between plasmalemma connecting the cytosol of adjacent cells is the cytoplasmic sleeve. Tubes of appressed endoplasmic reticulum (ER) connecting two adjacent cells are termed desmotubules. As symplasmic tunnels, PD provide pathways for transport of a range of molecules from cell-to-cell, including sugars, ions, proteins, and other essential nutrients, as well as different types of RNA molecules (Wu and Gallagher, 2012; Sager and Lee, 2018; Li et al., 2020b). Cell-to-cell movement of molecules through PD is thought to be dependent on a PD-size exclusion limit (PD-SEL), which involves several aspects, such as PD permeability, PD morphology, PD-associated proteins, and their functions (Sager and Lee, 2018). SEL is determined by the size of the largest molecules that can diffuse through PD. PD-SEL regulates the effectiveness of intracellular communication, which is required for plants to fine-tune their biological and developmental processes under various environmental circumstances (Wu et al., 2018). PD permeability is highly dynamic. The up-and-down modes of PD permeability are controlled by callose, a polysaccharide formed by callose (or β-1,3-glucan) synthase (CalS) enzymes and degraded by glucanase (BG) proteins (Zavaliev et al., 2011; Wu et al., 2018). Callose degradation increases the PD-SEL, whereas callose deposits reduce the PD-SEL. Moreover, PD morphology is considered an essential factor in intercellular transport and can range from simple, twinned or funnel to more complex forms (Oparka et al., 1999; Roberts et al., 2001; Faulkner et al., 2008; Nicolas et al., 2017; Ross-Elliott et al., 2017; Sager and Lee, 2018; Dorokhov et al., 2019).
As plasma membrane (PM)-lined channels, PD-PMs are occupied by unique membrane domains named lipid rafts, sterols- and sphingolipid-enriched microdomains. Lipid rafts provide attractive places for PD-receptor-like proteins (PD-RLPs) and PD-receptor-like kinases (PD-RLKs) to perceive signaling molecules in response to prevailing environmental stimuli (Iswanto and Kim, 2017; Iswanto et al., 2020; Vu et al., 2020). As a gatekeeper of cell trafficking, dynamic PD structure permits the cell-to-cell movement of endogenous molecules and acts as a channel for spreading disease-causing factors. Genome sequencing and proteome analyses are expanding the database of putative or partially characterized PD-related proteins from different plant species (Fernandez-Calvino et al., 2011; Kraner et al., 2017; Leijon et al., 2018). Also, some genes encoding PD-related proteins are redundant in sequence and function. In this regard, to characterize the functions of redundant genes, recent techniques such as genome editing serve as an ideal tool for generating knockout mutants, inducing randomized mutagenesis of the targeted region, or modulating transcriptional regulation.
The most popular genome-editing tool is CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR associated) for engineering plants at the DNA and RNA levels (Shelake et al., 2019a; Pramanik et al., 2021). CRISPR/Cas technology has been widely optimized for various applications in several plant species. Such applications include knockout generation, DNA insertion, DNA deletion, gene replacement, chromosome rearrangement, nucleic acid imaging, precise nucleotide substitution, epigenetic modification, pathogen detection, transcription regulation, and more. In this article, we highlight the role of PD-SEL (including PD-associated proteins) in response to multiple external stimuli. We also summarize the characterization of viral/fungal/bacterial proteins targeted at PD, along with potential genome editing tools, strategies, and techniques to understand the basics and improve agronomic traits through PD-SEL engineering.
The characteristics of callose deposition in response to abiotic stresses (such as osmotic, drought, cold, heat, metal stress) have been reviewed in recent literature (Sager and Lee, 2014). Several reports have highlighted the factors that regulate callose accumulation, thereby conferring enhanced abiotic stress resistance. However, the mechanisms that connect callose-mediated cell-to-cell signaling to the perception of abiotic cues are elusive. This section highlights the PD-associated proteins that positively or negatively control PD-callose under abiotic stresses.
In Arabidopsis, callose deposition in response to salt (NaCl) stress was first characterized by Wrzaczek’s group. It has been reported that the receptor-like kinase (RLK), Cys-rich receptor-like kinase 2 (CRK2), can positively regulate the salt stress-dependent pathway in Arabidopsis (Hunter et al., 2019). CRK2-overexpressing lines showed an enhanced germination rate and root length under high salinity conditions. They also found that CRK2 relocalizes to PD after 15 min of mannitol treatment or 30 min of 150 mM NaCl treatment. Furthermore, CRK2 regulates callose deposition under salt-stress conditions by interacting with CalS1. They also highlighted that the CalS1 played an important role in PD permeability during salt stress. cals1 mutant plants showed impairment in callose accumulation and germination deficiency under high NaCl treatment, which indicated that the phenotype of cals1 was similar to the crk2 mutant. The exact mechanism of salt-stress tolerance mediated by CRK2 relocalization (from PM to PD) and callose deposition is not clear because the CRK2-overexpression plants showed enhanced PD callose deposition and reduced PD permeability even without salt stress. Therefore, it can be hypothesized that CRK2 is implicated not only in callose-dependent salt-stress tolerance but may also be involved in plant growth and development irrespective of salt-stress conditions. In the same year, another study demonstrated that Qian Shou kinase 1 (QSK1) and inflorescence meristem kinase 2 (IMK2), a different class of RLKs, relocalize from the PM to PD in response to salt stress (Grison et al., 2019). The mechanism of callose-mediated salt stress tolerance depends on QSK1 phosphorylation but not on sterol or sphingolipid membrane composition. Interestingly, QKS1 and IMK2 rapidly modulate its localization from PM to PD within 1–4 min post-treatment of 400 mM mannitol and 100 mM NaCl. QSK1 is involved in callose deposition, the PD transportation pathway, lateral root density control, and root development. QSK1 overexpression displays an increased lateral root number and a slightly delayed lateral root formation compared to wild-type and mutant. It was also suggested that the relationship between callose accumulation and tolerance phenotypes observed with QSK1 overexpression was unclear.
Some metals were reported to trigger PD-associated proteins. Calreticulin is a highly conserved Ca2+-sequestering protein that typically resides within the ER lumen, especially in maize and Medicago truncatula (Baluska et al., 1999; Sujkowska-Rybkowska and Znojek, 2018). Under Aluminum stress, calreticulin protein in M. truncatula mycorrhizal roots was induced and colocalized with Ca2+ at the interface of fungal structures and in the periphery of the infected cortex cells (Sujkowska-Rybkowska and Znojek, 2018). Microscopic observations suggested that this colocalization might be required for the calcium mobilization that controls fungal accommodation inside the cortical cells and arbuscular development under Al stress conditions. However, the interaction of calreticulin and Ca2+ at PD needs further characterization. Interestingly, calmodulin (CaML) proteins have been found to reside at PD during flg22 treatment in Arabidopsis, which raises the possibility of CaML and calreticulin involvement in stress response (Xu et al., 2017; Wu et al., 2018). Similarly, treatment with subtoxic levels of copper and iron can severely inhibit primary root growth and interfere with the cell-to-cell movement of green fluorescence protein (GFP) (O’Lexy et al., 2018). Iron and copper alter PD permeability in roots via the regulation of callose synthases (CalS5, CalS12) and β-1,3-glucanases (BG_ppap, β-1,3-glucanase-putative; BG6, β-1,3-glucanase 6), respectively.
Wound stress results in alteration of callose accumulation via CalS1 and CalS8 (Cui and Lee, 2016). Aniline blue staining and Drop-ANd-See assay revealed no accumulation of PD callose in mutant leaves lacking CalS1/8 compared with wild-type after wounding. Genetically, CalS8 regulates PD permeability independently with PD-located protein 5 (PDLP5) upon wounding-induced reactive oxygen species (ROS) stress, while CalS1 requires PDLP5 in salicylic-dependent plasmodesmal response. Notably, CalS1 and CalS8 are suggested to localize along with the PM and PD. It remains to deciphered how CalS1 and CalS8 overexpression control PD permeability to enhance plant defense during biotic stresses. Abiotic stresses like heat and light trigger multi-layer signaling pathways that produce systemic acquired acclimation in plants. For example, a recent study reported the involvement of PD proteins (PDLP1 and 5) in propagating systemic ROS-signal waves in response to high light stress in Arabidopsis by altering the PD pore size (Fichman et al., 2021). Further studies into the role of PD-associated proteins in regulating the relay of different systemic signals triggered by various stresses may uncover novel mechanisms of plant protection. The list of PD-associated proteins involved in abiotic stresses is summarized in Table 1. Overall, examining the dynamic relocalization of PD-associated proteins from PM-to-PD and their role in long-distance signaling during abiotic stresses will help to understand new dimensions of PD biology.
Several living organisms, specifically fungi, bacteria, yeast, nematodes, insects, arachnids, and weeds, interact with plants. These plant interactions with other species could be beneficial (mutualism), useful to another partner only (commensalism), or harmful to a partner (parasitism). When viruses, fungi, or bacteria attack the plants, it often causes disease due to their virulence activities. In many cases, invasion by pathogens causes plant growth retardation and significant losses in crop quality and productivity. To protect from pathogens, host plants have evolved diverse barricades and remarkable immune machinery for pathogen recognition and the activation of defense signaling modes (Jones and Dangl, 2006; Nguyen et al., 2021). However, some viral, fungal, and bacterial pathogens target PD to mediate intercellular spread in host plant cells.
Viruses are neither “living” nor “non-living” and depend on host organisms to replicate and propagate, such as animals, bacteria, fungi, and plants. When viruses invade host plants, they form three major types of proteins, replication proteins (RPs), structural proteins (SPs), and movement proteins (MPs) which are classified based on their functions. RP is crucial for nucleic acid production; SP forms the outer protein shell and other units in the virions, whereas MP is employed to facilitate virus spread between host plant cells (Lefeuvre et al., 2019). The first study on plant viruses began in the 1890s; an infectious virus causing leaf spots in tobacco was characterized, Tobacco mosaic virus (TMV). TMV was the first virus of any host ever to be identified. So far, hundreds of plant viruses have been identified, almost all of which are infectious viruses of crop plants (Roossinck, 2010). Ten important plants viruses were ranked based on the scientific and economic importance, including TMV, Tomato spotted wilt virus (TSWV), Tomato yellow leaf curl virus (TYLCF), Cucumber mosaic virus (CMV), Potato virus y (PVY), Cauliflower mosaic virus (CaMV), African cassava mosaic virus (ACMV), Plum pox virus (PPV), Brome mosaic virus (BMV), and Potato virus x (PVX) (Scholthof et al., 2011). Plant viruses are transmitted from one plant to another by different modes such as seeds or pollen, vectors, grafting, or mechanical wounds (Hipper et al., 2013). Upon entry into the plant cell, viral components replicate and move from cell to cell through PD or are transported to long-distant organs through the vascular system. Plant viruses have evolved mechanisms of cell-to-cell movement, which involves the MP to facilitate intercellular trafficking of the plant viruses to and through the PD (Heinlein, 2015). Strikingly, some plant viruses encode multiple MPs, epitomized by triple gene block (TGB) proteins. Each TGB protein is involved in different stages of virus replication and cell-to-cell movement. In addition to MPs, some viral movement machinery requires additional virus-encoded proteins to deliver the viral genome. For instance, PVX also requires capsid protein (CP), whereas some potyviruses which do not encode MP require cylindrical inclusion protein for their cell-to-cell and long-distance dissemination (Carrington et al., 1998; Tilsner et al., 2013; Kumar et al., 2015). Many MP and other virus-encoded proteins are targeted to be localized at intercellular host regions such as the chloroplast, vesicles, ER, Golgi apparatus, nucleus, PM, and PD apertures. The list of plant virus-encoded proteins-targeted PD is summarized in Table 2.
Since many plant virus-encoded proteins localize to PD, it has been assumed that these symplasmic channels play a pivotal role in the viral spread. Plant viruses have evolved in several ways to achieve virulence and pathogenicity. However, PD-SEL is considered one of the main factors limiting the spread of virus infection (Kumar et al., 2015). The PD-SEL is highly linked to the callose accumulation at the edges of PD; therefore, the regulation of CalSs or BGs are depicted as the central signaling pathways to maintain intercellular trafficking via PD (Wu et al., 2018). It has been reported that increased callose accumulation at PD through the suppression of class I BG (GLU I, β-1,3-glucanase) inhibits intercellular movement of TMV, PVX, CMV in the tobacco plants. In contrast, increased PD flux by class III BG (GLU III) overexpression dilates the spread of potato virus YNTN (PVYNTN) in the potato plants (Iglesias et al., 2000; Bucher et al., 2001; Dobnik et al., 2013), see Table 2. The alteration of callose-mediated PD-SEL upon virus infection also involves the physical interaction between PD-associated proteins and virus-encoded proteins. A cytoplasmic receptor ankyrin repeat-containing protein 1 (ANK1) from Nicotiana tabacum recruited and interacted directly with TMV MP at PD, resulting in callose attenuation, subsequently enhancing the cell-to-cell movement of TMV MP (Ueki et al., 2010). In addition to PD-associated proteins, PDLP1 interacts with 2B MP from Grapevine fanleaf virus (GFLV) at PD, and a pdlp1/2/3 triple mutant leads to reduced intercellular movement of GFLV (Amari et al., 2010). Besides PDLP1, PDLP5 may also be essential for the movement of other viral proteins. It has been reported that PDLP5 regulates PD permeability in a callose-dependent manner, and reduced callose accumulation in the pdlp5 mutant exhibits increased cell-to-cell movement of TMV MP30 (Cui and Lee, 2016). However, it remains unknown whether PDLP1 regulates the cell-to-cell movement of GFLV through a callose-dependent manner, and it has not yet been explicitly verified whether PDLP5 physically interacts with TMV MP30.
In addition to PD-associated proteins, a plant-specific lipid microdomain and PD protein, Solanum tuberosum Remorin 1.3 (StREM1.3), physically interacts with PVX TGB1 protein (Raffaele et al., 2009). The overexpression of StREM1.3 significantly inhibits the cell-to-cell movement of PVX TGB1, TMV MP30 as well as PVY Hc-Pro (Raffaele et al., 2009; Perraki et al., 2014). Another study on plant REM has shown that Nicotiana benthamiana REM1 (NbREM1) is a negative regulator of the intercellular movement of Rice stripe virus (RSV) through the S-acylation suppression process (Fu et al., 2018). Although a PVX TGB2 protein interacts indirectly with a BG protein (Fridborg et al., 2003) and grain setting defect 1 (GSD1) (Gui et al., 2015), a REM protein identified from Oryza sativa interacts directly with OsACT1 at PD in controlling PD permeability (Gui et al., 2014). Most recent studies on plant REM indicate that the restriction of PVX spread occurs in a REM-induced callose accumulation-dependent manner and may involve the activation of salicylic acid (SA) signaling (Perraki et al., 2018; Huang et al., 2019). Overall, REM proteins from different plant species were reported to be implicated in callose deposition at PD, a key mechanism in plant development and stress responses. Therefore, REM interaction with viral components could be targeted by genome editing or transgenic technology for imparting viral-stress tolerance depending on the negative or positive effect on the viral spread, respectively.
In structure, PD represents membrane-lined canals that provide a suitable compartment for plant receptors to perceive diverse environment-related stimuli. Some of the plant receptors are predominantly localized or recruited at PD in response to abiotic and biotic stresses (Vu et al., 2020). In the case of viral infection, host plants have evolved an antiviral defense mechanism, namely RNA interference (RNAi) mediated by small interfering RNA (siRNA) (Borges and Martienssen, 2015). This RNAi moves from cell to cell through PD to overcome virus infectivity (Smith et al., 2007). However, viruses also develop viral suppressors of RNA silencing (VSR) to target multiple parts of the RNAi machinery (Csorba et al., 2015). In the recent study of virus-related PD-RLKs, BARELY ANY MERISTEM 1 and 2 (BAM1 and BAM2) are essential for the cell-to-cell movement of RNAi whereby they interact with C4 protein from TYLCV (Rosas-Diaz et al., 2018) and the viral silencing suppressor P19 from Tomato bushy stunt virus (TBSV) at PD (Garnelo Gomicronmez et al., 2021). However, the role of BAM1 and BAM2 in callose-mediated PD closure is still elusive. In addition to PD-PM protein, SYNAPTOTAGMIN A (SYTA)- an ER-PM contact site protein- can be recruited at PD to facilitate the cell-to-cell movement of Turnip vein-clearing virus (TVCV) MP (Levy et al., 2015). SYTA also interacts with the TMV MP and PD localization signal (PLS) of TMV MP and other virus-encoded proteins from Cabbage leaf curl virus (CaLCuV). The suppression of SYTA leads to reduced cell-to-cell movement of TMV MP, inhibited the systemic spread of CaLCuV, Turnip mosaic virus (TuMV), and TVCV, and disrupted PD targeting of TMV PLS (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014; Yuan et al., 2016; Yuan et al., 2018). However, it remains unknown whether SYTA-mediated viral movement occurs in a callose-mediated PD closure-dependent manner or not. A recent study highlighted the importance of phosphorylatable amino acid residues of CMV MP in symptom development and PD localization (Sáray et al., 2021). Investigating such new aspects will shed light on virus-plant host interactions in detail and provide potential clues toward designing novel crop protection strategies in the future.
Like pathogenic viruses, plant pathogenic fungi and bacteria cause different diseases that hinder crop quality and productivity. The following plant pathogenic fungi and bacteria have been listed based on their scientific and economic importance. The list of pathogenic fungi includes Magnaporthe oryzae, Botrytis cinerea, Puccinia spp., Fusarium graminearum, Fusarium oxysporum, Blumeria graminis, Mycosphaerella graminicola, Colletotrichum spp., Ustilago maydis, and Melampsora lini (Dean et al., 2012). The list of pathogenic bacteria includes Pseudomonas syringae pathovars, Ralstonia solanacearum, Agrobacterium tumefaciens, Xanthomonas oryzae pv. oryzae, Xanthomonas campestris pathovars, Xanthomonas axonopodis pathovars, Erwinia amylovora, Xylella fastidiosa, Dickeya (dadantii and solani), Pectobacterium carotovorum, and Pectobacterium atrosepticum (Mansfield et al., 2012). Like viruses, pathogenic fungi and bacteria have also evolved sophisticated machinery to invade their host plants. The most common approach for invasion among pathogenic fungi and bacteria is to deploy various effector proteins that can target and modulate PD channels, thus activating various processes in host plants (Lee and Lu, 2011). A hemibiotrophic rice blast fungus M. oryzae utilizes invasive hyphae to exploit PD channels (Kankanala et al., 2007) and spread to neighboring cells through PD to expand its vicinity, possibly by delivering an effector Pathogenicity toward Weeping Lovegrass (PWL2) protein (Khang et al., 2010). In addition to M. oryzae, the effectiveness of fungal growth from one cell to a neighboring cell is mainly controlled by the attenuation of callose deposition at PD in which a single fungal mitogen-activated protein kinase (MAPK), PmK1, is involved (Sakulkoo et al., 2018). Melampsora larici-populina causes rust disease and severe problems in the genus Populus plants and other family Salicaceae plants. M. larici-populina is grouped into biographic plant-parasites that secrete an assortment of effectors to determine host cell colonization. A recent study indicates that one of the effectors from M. larici-populina, MLP37347, is located at PD (Germain et al., 2018). Even though MLP37347 is targeted to PD, there is no unequivocal evidence showing that MLP37347 effector regulates PD function during M. larici-populina infection. It will be interesting to explore the role of the MLP37347 effector in correlation with PD biology. Other effectors from F. oxysporum, Avr2 and Six5, have been reported to interact at PD. This interaction is required to manipulate PD apertures, allowing Avr2 to move from one cell to neighboring cells. The presence of Six5 is required for Avr2 cell-to-cell movement through PD, whereas without Avr2, the Six5 effector alone is not sufficient to alter PD permeability. This experiment indicates that to trigger PD opening upon F. oxysporum infection, the interaction between Avr2 and Six5 effectors in host cells is required (Cao et al., 2018).
To manipulate the immunity and physiology of host plants, pathogenic fungi and bacteria not only secrete effectors but also target them into PD aperture or other host interiors. Like viruses, it can be assumed that some fungi or bacteria effectors target PD and interact directly with PD-associated proteins to regulate symplasmic continuity. Recently, two pathogen effectors, RxLR3 from Phytophthora brassicae and HopO1-1 from P. syringae, were reported to localize at PD and interact with PD proteins (Aung et al., 2020; Tomczynska et al., 2020). RxLR3 targets CalS1, CalS2, and CalS3 to control symplasmic trafficking through callose turnover at PD (Tomczynska et al., 2020). Unlike the RxLR3 effector, HopO1-1 physically associates with other PD proteins, such as PDLP5 and PDLP7, to hamper their stability (Table 3). The destabilization of PDLP5 and PDLP7 proteins upon HopO1-1 infection leads to enhanced symplasmic conductivity (Aung et al., 2020). It has been shown that PDLP5 is involved in the immune response upon bacterial infection through maintaining callose accumulation at PD (Lee et al., 2011; Cui and Lee, 2016). Furthermore, the mechanism of HopO1-1-enhanced PD permeability seems to be PDLP5/PDLP7-regulated callose accumulation-dependent. In addition to HopO1-1, recent studies reported that several effectors from P. syringae not only localized at PD, but they also moved symplastically between the cells through these channels (Kang et al., 2021; Li et al., 2021). It was also suggested that the intercellular movement of effectors is PD permeability dependent manner (Li et al., 2021). However, the molecular linkage between the intercellular movement of effectors and PD regulation is still poorly understood.
Recent advancements in genome engineering tools based on CRISPR/Cas systems have opened new doors to fine-tune the plant genome at all layers of the central dogma (Pramanik et al., 2021). Another major advantage of CRISPR-based tools is the ability to customize a strategy to precisely edit the redundant genes or simultaneously edit multiple homologs (Wang et al., 2019; Hong et al., 2020). CRISPR-based tools have been employed in the editing of PD-related genes in recent times (Rosas-Diaz et al., 2018), but their real potential is yet to be explored for manipulating PD biology. In the following sections, we present and discuss the CRISPR/Cas tools and their future applications to investigate fundamental aspects of PD biology or PD engineering for stress management strategies.
The first report demonstrating the potential of CRISPR/Cas components for genome editing was published in 2012 (Jinek et al., 2012). Since then, tremendous progress has been made in developing novel CRISPR-based tools (Figure 1). The primary CRISPR/Cas tool consists of two components comprising a nuclease enzyme (Cas) and a programmable RNA guide (gRNA) complementary to the target DNA. Cas enzyme bound with scaffold-fused gRNA (sgRNA) recognizes the target site followed by a short recognition motif called protospacer adjacent motif (PAM). The protein-RNA-DNA complex formation leads to the generation of DNA double-strand breaks (DSBs) at desired sites in the complex genome, and endogenous DNA repair pathways make precise or error-prone DNA modifications. The Cas9 and Cas12a (Cpf1) are the most commonly applied Cas enzymes for mutant creation in different organisms (Shelake et al., 2019b). Various Cas variants and orthologs have been characterized to maximize the editing scope and different PAM specificities. Simultaneous targeting of multiple loci in the genome is a significant advantage of CRISPR-based tools compared to other genome engineering methods.
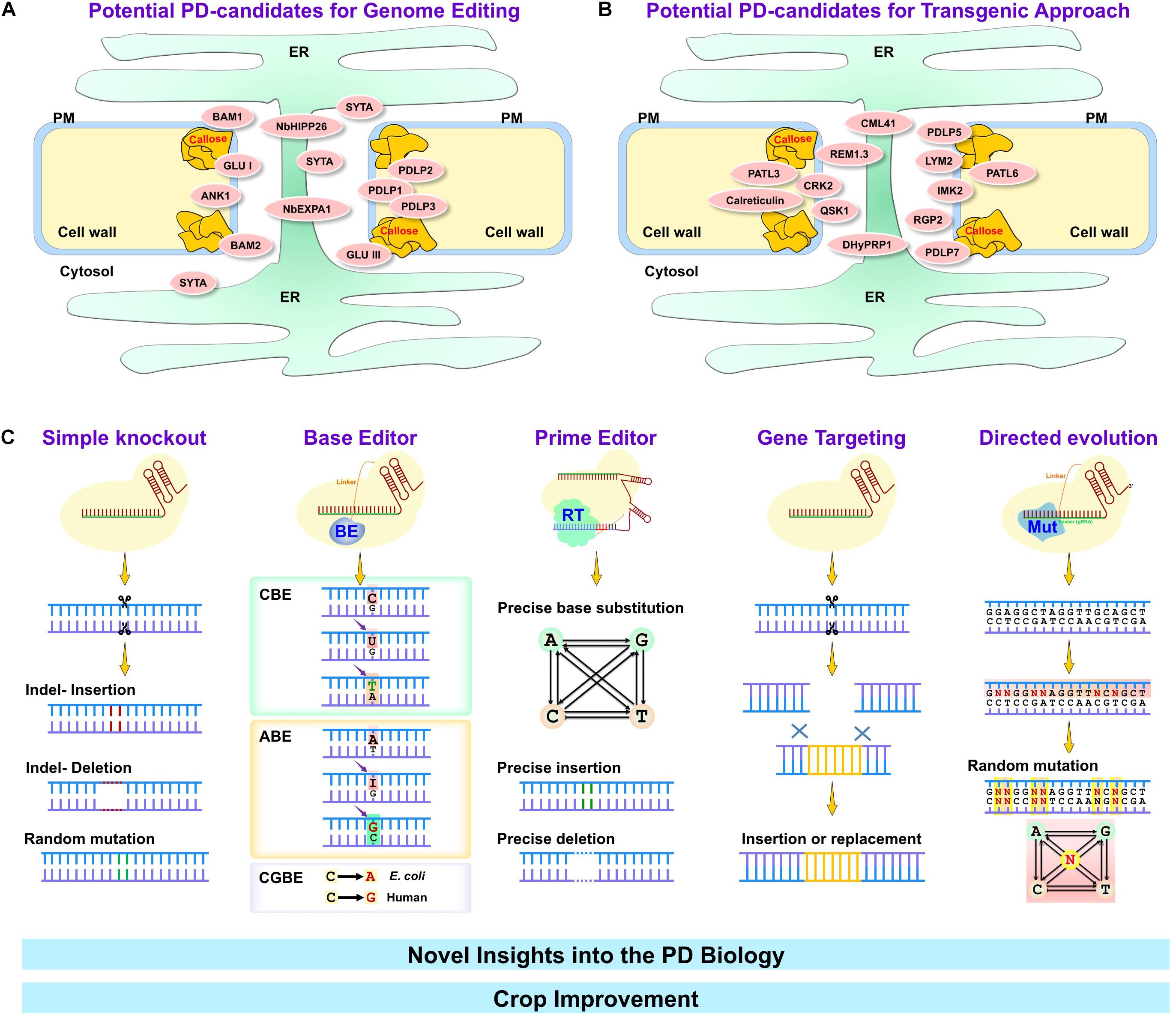
Figure 1. Major genome editing techniques for crop improvement through plasmodesmal engineering. Many PD-associated proteins are involved in a variety of environmental stresses (abiotic and biotic stresses). Depending on the impact on stress mechanism, particular PD-associated proteins can be classified as negative (A) or positive (B) regulators. PD-associated proteins that negatively regulate stress tolerance in plants (summarized in panel A) can be targeted using genome editing tools such as simple knockout by CRISPR/Cas, base editor, prime editor, gene targeting and directed evolution tools (C). On the other hand, PD-associated proteins that positively regulate stress tolerance in plants (summarized in panel B) may provide the easiest way to overexpress them by transgenic approach. Targeting PD-associated proteins by genome editing or transgenically possess potential avenues to improve crop quality and productivity. ER, Endoplasmic reticulum; PM, plasma membrane; BAM1 and 2, barely any meristem 1 and 2; GLU I and III, β-1,3-glucanase class I and III; ANK1, ankyrin repeat-containing protein; SYTA, Synaptotagmin A; NbHIPP26, Nicotiana benthamiana heavy metal-associated isoprenylated plant protein; NbEXPA1, N. benthamiana α-expansin 1; PDLP1, 2, 3, 5, and 7, Plasmodesmata-located protein 1, 2, 3, 5, and 7; PATL3 and 6, patellin 3 and 6; CML41, calmodulin-like protein 41; REM1.3, Remorin 1.3; CRK2, Cys-rich receptor-like kinase 2; QSK1, Qian Shou kinase 1; DHyPRP1, double hybrid proline-rich protein 1; LYM2, lysin motif domain-containing glycosylphosphatidylinositol-anchored protein 2; IMK2, inflorescence meristem kinase 2; RGP2, reversibly glycosylated polypeptide 2.
The use of CRISPR/Cas in homologous recombination-based gene targeting (HR-GT) has demonstrated the potential to improve gene-targeting efficiency through precise DSB induction if a donor template is provided to promote the homology-directed repair (HDR) pathway (Capdeville et al., 2021). The HDR-based GT mostly occurs in dividing cells and desired HR-GT products are often mixed with additional indels (insertions/deletions) due to preferred non-homologous end joining (NHEJ). To address this issue, partial catalyzed (nickase, D10A, or H840A) or fully deactivated (dead D10A together with H840A) Cas9 nuclease is engineered for delivering the effector molecules to the target locus for many applications beyond simple DSB-mediated knockout generation (Adli, 2018). Primary tools based on the fusion of effector molecules with nCas9 or dCas9 include base editors that introduce base substitutions without the need for HDR, DSBs, or donor templates (Komor et al., 2016; Gaudelli et al., 2017). A recent addition to the CRISPR toolbox is the prime editor, which needs a template (Anzalone et al., 2019). Although the prime editor tool can introduce customized changes (small insertions or deletions, all 12 base substitutions) at the targeted genomic locus, optimization for plant use is desirable in the near future. Several in silico and in vivo protocols are being devised for target site selection, validation of gRNAs, appropriate choice of CRISPR tool for the desired application, and suitable delivery strategies depending on the species (Huang et al., 2021). Overall, several features like simple design, high precision, efficiency, lower cost, choice of versatile tools, and a broad range of targeting in the genome have enabled the wider adoption of CRISPR/Cas technology for various purposes in plants.
Despite the discovery of PD in the nineteenth century, precise knowledge about PD structure and function is still elusive. Although PD operating mode remains challenging to understand, applications of contemporary techniques are revealing their novel facets. Broadly, CRISPR-based technologies can be applied in PD research with bidirectional aims. The first direction is the understanding of PD biology, and secondly, targeting PD-related proteins for the development of stress-tolerant crops. The choice of the CRISPR tool predominantly depends on the possible outcome. As discussed earlier, CRISPR tools may produce a variety of genetic modifications- for example, simple knockout, base substitution, precise insertion/deletion/replacement, strong/weak allele generation, epigenetic modulation, transcriptional regulation, and chromosomal rearrangements (Shelake et al., 2019a). We describe the potential of CRISPR tools for exploiting plasmodesmal biology in two parts: understanding the basics of PD functioning and their modulation for stress management.
The PD interactome can be roughly divided into three parts depending on their direct or indirect role in PD formation and functioning. Group 1 consists of the actual players that form the PD structure itself; the second group involves the molecules that regulate the PD SEL. The third group contains the molecules trafficking through PD. The interplay between the molecules from these three groups is crucial not only to plant physiology and development but also to plant stress responses and environmental signals (Azim and Burch-Smith, 2020). On the one hand, long-distance trafficking of soluble molecules and defense signals occurs through PD. On the other hand, pathogens also hijack the PD cell-to-cell movement machinery to spread from infected to non-infected plant parts. Therefore, the PD-PM interface is at the forefront of the battle between pathogens and plant defense molecules.
Considerably, several studies have uncovered different facets of PD-mediated spread of viruses and plant defense signaling molecules such as siRNAs. For example, the C4 protein of TYLCV primarily interacts with proteins implicated in plant defense, ubiquitination, and translation from host tomato plants (Kim et al., 2016). Recent reports showed that the RLK homologs (BAM1 and BAM2) act as a positive regulator of siRNA spread through PD (Rosas-Diaz et al., 2018). CRISPR-mediated double knockout mutants (bam1 bam2) were generated, confirming the redundant role of BAM1 and BAM2 in promoting the cell-to-cell spread of RNAi. Also, this study suggested the C4 interaction halts the BAM1/2 function and eventually the spread of RNA silencing. In the follow-up study, another viral protein, P19 from TBSV, was demonstrated to interact with BAM1/2 in a similar fashion like C4, indicating that BAM1 and BAM2 are good candidates for CRISPR targeting of C4/P19-interacting domains to develop geminiviral-resistant plants (Garnelo Gomicronmez et al., 2021). Overall, CRISPR-mediated genome editing of PD-related genes is valuable to explore their function and provides attractive potential candidates from the PD interactome to edit and develop biotic and abiotic stress tolerance (Table 4).
The symbiotic interaction between host plant-PD and nitrogen-fixing microbes is another research area that needs to be explored. Recent work has shed some light on the molecular dialog between the host plant and associated microbes confirming that PD regulation is a key early event for establishing the symbiotic legume plant-microbe association (Gaudioso-Pedraza et al., 2018). The PD-localized β-1,3-glucanase from Medicago truncatula MtBG2 promoted the symplasmic connectivity, thereby facilitating the nodule formation. The increased PD permeability (Complainville et al., 2003) or higher number of PD pores (Schubert et al., 2013) substantially increased nodule number in M. truncatula and Casuarina glauca, respectively. Also, some tetraspanin proteins like TET3 from Arabidopsis (Fernandez-Calvino et al., 2011), PvTET3, and PvTET6 from the common bean were reported to be localized at the PD-PM interface during nodule formation with rhizobia (Jimenez-Jimenez et al., 2019), suggesting their direct role in symplasmic interaction through PD regulation and cellular trafficking. Thus, the use of CRISPR tools in altering PD may help to promote the positive interaction of symbiotic association of nitrogen-fixing microbes and host plants.
The new set of plant breeding techniques, collectively known as new plant breeding technologies (NPBT), includes the concept of grafting wild-type onto genetically modified (GM) rootstock (Langner et al., 2018). The proper combination of scion and rootstock is advantageous to develop improved crop traits. The bi-directional interaction between rootstock and scion involves exchanging all three major macromolecules (DNA, RNA, and protein) through the PD. A recent report showed that even the genomes could transfer horizontally via organelle travel during the remodeling of PD and vascular connection at the root-scion junction (Hertle et al., 2021). Previous reports have successfully used transgenic rootstocks to transfer transgene-mediated traits to the wild-type scion parts- for example, the development of CMV resistance in tomato (Bai et al., 2016), Pierce’s disease resistance in grape (Dandekar et al., 2019), PPV resistance in plum (Sidorova et al., 2021), and increased nitrogen levels in walnut overexpressing an ammonium transporter gene (Liu et al., 2021). In such studies, engineering of PD trafficking and the use of transgene-free CRISPR techniques to attain desired traits would be highly desirable because the non-transgenic genome editing approach may easily avoid the GM issues and related-regulatory hurdles.
PD-mediated symplasmic transport permits cell-to-cell communication in multicellular plants, regulating the harmonized physiological growth and development during environmental stresses. Even though the dynamic nature of PD allows surprisingly high intercellular transport of molecules, PD plasticity makes it challenging to establish the regulatory mechanisms of PD functioning. In this regard, advanced techniques like genome editing and high-resolution microscopy are promising to solve the mysteries around PD structure and function. The primary goal of crop improvement is to design climate-resilient varieties with superior traits. The crucial role of the PD interactome in plant defense is now well-known. The use of genome editing in PD engineering has a vast potential to improve molecule transport for higher nutrition quality for human health, to protect plants against biotic and abiotic stresses, to design improved symbiotic interactions for plant nutrition, and to enhance grafting-based strategies for crop improvement.
ABBI and RMS designed the manuscript structure. ABBI, RMS, and MHV wrote the manuscript. J-YK and SHK designed the manuscript structure and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Research Foundation of Korea (the Bio & Medical Technology Development Program 2020M3A9I4038352 and Priority Research Centers Program 2020R1A6A1A03044344), and a grant from the New breeding technologies development Program (Project No. PJ01483601), Rural Development Administration, South Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACLSV, Apple chlorotic leaf spot virus; ACMV, African cassava mosaic virus; ADV, Alfalfa dwarf virus; ANK1, Ankyrin 1; BAM, BARELY ANY MERISTEM; BBWV-2, broad bean wilt virus 2; BDMV, Bean dwarf mosaic virus; BG, β -1,3-glucanase; BMV, Brome mosaic virus; BNYVV, Beet necrotic yellow vein virus; BSMV, Barley stripe mosaic virus; BYV, Beet yellows virus; CaCV, Capsicum chlorosis virus; CaLCuV, Cabbage leaf curl virus; CalS, callose synthase; CaML, calmodulin; CaMV, Cauliflower mosaic virus; CCYV, Cucurbit chlorotic yellows virus; CMV, Cucumber mosaic virus; CP, capsid protein; CPMV, Cowpea mosaic virus; CPsV, Citrus psorosis virus; CRISPR, clustered regularly interspaced short palindromic repeats; CRK2, cys-rich receptor-like kinase 2; CTV, Citrus tristeza virus; CWMV, Chinese wheat mosaic virus; DHyPRP1, double hybrid proline-rich protein; DSB, double-strand break; ER, endoplasmic reticulum; EXPA1, expansin 1; GFLV, Grapevine fanleaf virus; GM, genetically modified; GSD1, grain setting defect 1; GVA, Grapevine virus A; GVB, Grapevine virus B; HDR, homology-directed repair; HIPP26, heavy metal-associated isoprenylated plant protein 26; HR-GT, homologous recombination-based gene targeting; IMK2, inflorescence meristem kinase 2; LIYV, Lettuce infectious yellows virus; LNYV, Lettuce necrotic yellows virus; LYM2, Lysin motif domain-containing glycosylphosphatidylinositol-anchored protein 2; MAPK, mitogen-activated protein kinase; MiLBVV, Mirafiori lettuce big-vein virus; MNSV, Melon necrotic spot virus; MP, movement protein; MSV, Maize streak virus; NHEJ, non-homologous end joining; NPBT, new plant breeding technologies; OLV-2, Olive latent virus 2; PAM, protospacer adjacent motif; PATL, Patellin; PD, plasmodesmata; PDLP, plasmodesmata-located protein; PeVYV, Pepper vein yellows virus; PLRV, Potato leafroll virus; PLS, PD localization signal; PM, plasma membrane; PMTV, Potato mop-top pomovirus; PPV, Plum pox virus; PepRSV, Pepper ringspot virus; PVX, Potato virus x; PVY, Potato virus y; PWL, Pathogenicity toward Weeping Lovegrass; QSK1, Qian Shou Kinase 1; RBSDV, Rice black-streaked dwarf virus; RCMV, Red clover mottle virus; REM, Remorin; RGP2, reversibly glycosylated polypeptide 2; RGSV, Rice grassy stunt virus; RLBV, Raspberry leaf blotch emaravirus; RLK, receptor-like kinase; RLP, receptor-like protein; RNAi, RNA interference; ROS, reactive oxygen species; RP, replication protein; RSV, Rice stripe virus; RTYV, Rice transitory yellowing virus; SA, salicylic acid; SEL, size exclusion limit; sgRNA, scaffold-fused gRNA; siRNA, small interfering RNA; SP, structural protein; SYTA, SYNAPTOTAGMIN A; TBSV, Tomato bushy stunt virus; TGB, triple gene block; TMV, Tobacco mosaic virus; TSWV, Tomato spotted wilt virus; TuMV, Turnip mosaic virus; TVCV, Turnip vein-clearing virus; TYLCF, Tomato yellow leaf curl virus; TYLCV, Tomato yellow leaf curl virus; VSR, viral suppressors of RNA.
Amari, K., Boutant, E., Hofmann, C., Schmitt-Keichinger, C., Fernandez-Calvino, L., Didier, P., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog 6:e1001119. doi: 10.1371/journal.ppat.1001119
Andika, I. B., Zheng, S., Tan, Z., Sun, L., Kondo, H., Zhou, X., et al. (2013). Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology 435, 493–503. doi: 10.1016/j.virol.2012.10.024
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Aung, K., Kim, P., Li, Z., Joe, A., Kvitko, B., Alfano, J. R., et al. (2020). Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues. Plant Cell 32, 595–611. doi: 10.1105/tpc.19.00707
Avisar, D., Prokhnevsky, A. I., and Dolja, V. V. (2008). Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J. Virol. 82, 2836–2843. doi: 10.1128/jvi.02246-07
Azim, M. F., and Burch-Smith, T. M. (2020). Organelles-nucleus-plasmodesmata signaling (ONPS): an update on its roles in plant physiology, metabolism and stress responses. Curr. Opin. Plant Biol. 58, 48–59. doi: 10.1016/j.pbi.2020.09.005
Bai, M., Chen, W.-T., Xie, B.-Y., and Yang, G.-S. (2016). A novel strategy to enhance resistance to Cucumber mosaic virus in tomato by grafting to transgenic rootstocks. J. Int. Agriculture 15, 2040–2048. doi: 10.1016/s2095-3119(16)61330-8
Baluska, F., Samaj, J., Napier, R., and Volkmann, D. (1999). Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. 19, 481–488. doi: 10.1046/j.1365-313x.1999.00530.x
Bell, K., and Oparka, K. (2011). Imaging plasmodesmata. Protoplasma 248, 9–25. doi: 10.1007/s00709-010-0233-6
Blackman, L. M., Boevink, P., Cruz, S. S., Palukaitis, P., and Oparka, K. J. (1998). The movement protein of cucumber mosaic virus traffics into sieve elements in minor veins of nicotiana clevelandii. Plant Cell 10, 525–538. doi: 10.2307/3870730
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Bucher, G. L., Tarina, C., Heinlein, M., Di Serio, F., and Meins, F. Jr., et al. (2001). Local expression of enzymatically active class I beta-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28, 361–369. doi: 10.1046/j.1365-313x.2001.01181.x
Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., and Takken, F. L. W. (2018). The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 11, 691–705. doi: 10.1016/j.molp.2018.02.011
Capdeville, N., Merker, L., Schindele, P., and Puchta, H. (2021). Sophisticated CRISPR/Cas tools for fine-tuning plant performance. J. Plant Physiol. 257:153332. doi: 10.1016/j.jplph.2020.153332
Carrington, J. C., Jensen, P. E., and Schaad, M. C. (1998). Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14, 393–400. doi: 10.1046/j.1365-313x.1998.00120.x
Chai, M., Wu, X., Liu, J., Fang, Y., Luan, Y., Cui, X., et al. (2020). P3N-PIPO interacts with P3 via the shared N-Terminal domain to recruit viral replication vesicles for cell-to-cell movement. J. Virol. 94:e01898-19.
Chen, M. H., Tian, G. W., Gafni, Y., and Citovsky, V. (2005). Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 138, 1866–1876. doi: 10.1104/pp.105.064386
Complainville, A., Brocard, L., Roberts, I., Dax, E., Sever, N., Sauer, N., et al. (2003). Nodule initiation involves the creation of a new symplasmic field in specific root cells of medicago species. Plant Cell 15, 2778–2791. doi: 10.1105/tpc.017020
Cowan, G. H., Roberts, A. G., Jones, S., Kumar, P., Kalyandurg, P. B., Gil, J. F., et al. (2018). Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiol. 176, 2052–2070. doi: 10.1104/pp.17.01698
Csorba, T., Kontra, L., and Burgyan, J. (2015). viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 47, 85–103. doi: 10.1016/j.virol.2015.02.028
Cui, W., and Lee, J. Y. (2016). Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2:16034.
Dandekar, A. M., Jacobson, A., Ibanez, A. M., Gouran, H., Dolan, D. L., Aguero, C. B., et al. (2019). Trans-Graft protection against pierce’s disease mediated by transgenic grapevine rootstocks. Front. Plant Sci. 10:84. doi: 10.3389/fpls.2019.00084
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dickinson, V. J., Halder, J., and Woolston, C. J. (1996). The product of maize streak virus ORF V1 is associated with secondary plasmodesmata and is first detected with the onset of viral lesions. Virology 220, 51–59. doi: 10.1006/viro.1996.0285
Dobnik, D., Baebler, S., Kogovsek, P., Pompe-Novak, M., Stebih, D., Panter, G., et al. (2013). beta-1,3-glucanase class III promotes spread of PVY(NTN) and improves in planta protein production. Plant Biotechnol. Rep. 7, 547–555. doi: 10.1007/s11816-013-0300-5
Dorokhov, Y. L., Ershova, N. M., Sheshukova, E. V., and Komarova, T. V. (2019). Plasmodesmata conductivity regulation: a mechanistic model. Plants (Basel) 8:595. doi: 10.3390/plants8120595
Erhardt, M., Morant, M., Ritzenthaler, C., Stussi-Garaud, C., Guilley, H., Richards, K., et al. (2000). P42 movement protein of beet necrotic yellow vein virus is targeted by the movement proteins P13 and P15 to punctate bodies associated with plasmodesmata. Mol. Plant Microbe Interact. 13, 520–528. doi: 10.1094/mpmi.2000.13.5.520
Faulkner, C., Akman, O. E., Bell, K., Jeffree, C., and Oparka, K. (2008). Peeking into pit fields: a multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20, 1504–1518. doi: 10.1105/tpc.107.056903
Faulkner, C., Petutschnig, E., Benitez-Alfonso, Y., Beck, M., Robatzek, S., Lipka, V., et al. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. U S A. 110, 9166–9170. doi: 10.1073/pnas.1203458110
Fernandez-Calvino, L., Faulkner, C., Walshaw, J., Saalbach, G., Bayer, E., Benitez-Alfonso, Y., et al. (2011). Arabidopsis plasmodesmal proteome. PLoS One 6:e18880. doi: 10.1371/journal.pone.0018880
Fichman, Y., Myers, R. J., Grant, D. G., and Mittler, R. (2021). Plasmodesmata-localized proteins and ROS orchestrate light-induced rapid systemic signaling in Arabidopsis. Sci. Signal. 14:eabf0322. doi: 10.1126/scisignal.abf0322
Fridborg, I., Grainger, J., Page, A., Coleman, M., Findlay, K., and Angell, S. (2003). TIP, a novel host factor linking callose degradation with the cell-to-cell movement of Potato virus X. Mol. Plant Microbe Interact. 16, 132–140. doi: 10.1094/mpmi.2003.16.2.132
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., and Zhou, X. (2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Garnelo Gomicronmez, B., Rosas-Diaz, T., Shi, C., Fan, P., Zhang, D., Rufian, J. S., et al. (2021). The viral silencing suppressor P19 interacts with the receptor-like kinases BAM1 and BAM2 and suppresses the cell-to-cell movement of RNA silencing independently of its ability to bind sRNA. New Phytol. 229, 1840–1843. doi: 10.1111/nph.16981
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A⋅T to G⋅C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Gaudioso-Pedraza, R., Beck, M., Frances, L., Kirk, P., Ripodas, C., Niebel, A., et al. (2018). Callose-Regulated symplastic communication coordinates symbiotic root nodule development. Curr. Biol. 28:e3566.
Genoves, A., Pallas, V., and Navarro, J. A. (2011). Contribution of topology determinants of a viral movement protein to its membrane association, intracellular traffic, and viral cell-to-cell movement. J. Virol. 85, 7797–7809. doi: 10.1128/jvi.02465-10
Germain, H., Joly, D. L., Mireault, C., Plourde, M. B., Letanneur, C., Stewart, D., et al. (2018). Infection assays in Arabidopsis reveal candidate effectors from the poplar rust fungus that promote susceptibility to bacteria and oomycete pathogens. Mol. Plant Pathol. 19, 191–200. doi: 10.1111/mpp.12514
Grangeon, R., Jiang, J., Wan, J., Agbeci, M., Zheng, H., and Laliberte, J. F. (2013). 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 4:351. doi: 10.3389/fmicb.2013.00351
Grieco, F., Castellano, M. A., Di Sansebastiano, G. P., Maggipinto, G., Neuhaus, J. M., and Martelli, G. P. (1999). Subcellular localization and in vivo identification of the putative movement protein of olive latent virus 2. J. Gen. Virol. 80(Pt 5), 1103–1109. doi: 10.1099/0022-1317-80-5-1103
Grison, M. S., Kirk, P., Brault, M. L., Wu, X. N., Schulze, W. X., Benitez-Alfonso, Y., et al. (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol. 181, 142–160. doi: 10.1104/pp.19.00473
Gui, J., Liu, C., Shen, J., and Li, L. (2014). Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 166, 1463–1478. doi: 10.1104/pp.114.246769
Gui, J., Zheng, S., Shen, J., and Li, L. (2015). Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 6:804. doi: 10.3389/fpls.2015.00804
Haviv, S., Moskovitz, Y., and Mawassi, M. (2012). The ORF3-encoded proteins of vitiviruses GVA and GVB induce tubule-like and punctate structures during virus infection and localize to the plasmodesmata. Virus Res. 163, 291–301. doi: 10.1016/j.virusres.2011.10.015
Heinlein, M. (2015). Plant virus replication and movement. Virology 479-480, 657–671. doi: 10.1016/j.virol.2015.01.025
Hertle, A. P., Haberl, B., and Bock, R. (2021). Horizontal genome transfer by cell-to-cell travel of whole organelles. Sci. Adv. 7:eabd8215. doi: 10.1126/sciadv.abd8215
Hipper, C., Brault, V., Ziegler-Graff, V., and Revers, F. (2013). Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 4:154. doi: 10.3389/fpls.2013.00154
Hiraguri, A., Hibino, H., Hayashi, T., Netsu, O., Shimizu, T., Uehara-Ichiki, T., et al. (2012). The movement protein encoded by gene 3 of rice transitory yellowing virus is associated with virus particles. J. Gen. Virol. 93, 2290–2298. doi: 10.1099/vir.0.044420-0
Hiraguri, A., Netsu, O., Shimizu, T., Uehara-Ichiki, T., Omura, T., Sasaki, N., et al. (2011). The nonstructural protein pC6 of rice grassy stunt virus trans-complements the cell-to-cell spread of a movement-defective tomato mosaic virus. Arch. Virol. 156, 911–916. doi: 10.1007/s00705-011-0939-6
Hong, W. J., Kim, Y. J., Kim, E. J., Kumar Nalini, Chandran, A., Moon, S., et al. (2020). CAFRI-Rice: CRISPR applicable functional redundancy inspector to accelerate functional genomics in rice. Plant J. 104, 532–545. doi: 10.1111/tpj.14926
Huang, D., Sun, Y., Ma, Z., Ke, M., Cui, Y., Chen, Z., et al. (2019). Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U S A. 116, 21274–21284. doi: 10.1073/pnas.1911892116
Huang, T. P., Newby, G. A., and Liu, D. R. (2021). Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 16, 1089–1128. doi: 10.1038/s41596-020-00450-9
Hunter, K., Kimura, S., Rokka, A., Tran, H. C., Toyota, M., Kukkonen, J. P., et al. (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDalpha1. Plant Physiol. 180, 2004–2021. doi: 10.1104/pp.19.00560
Iglesias, V. A., Meins, F., and Jr. (2000). Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 21, 157–166. doi: 10.1046/j.1365-313x.2000.00658.x
Iswanto, A. B., and Kim, J. Y. (2017). Lipid raft, regulator of plasmodesmal callose homeostasis. Plants (Basel) 6:15. doi: 10.3390/plants6020015
Iswanto, A. B. B., Shon, J. C., Liu, K. H., Vu, M. H., Kumar, R., and Kim, J. Y. (2020). Sphingolipids modulate secretion of glycosylphosphatidylinositol-anchored plasmodesmata proteins and callose deposition. Plant Physiol. 184, 407–420. doi: 10.1104/pp.20.00401
Jiang, Z., Zhang, K., Li, Z., Li, Z., Yang, M., Jin, X., et al. (2020). The barley stripe mosaic virus gammab protein promotes viral cell-to-cell movement by enhancing ATPase-mediated assembly of ribonucleoprotein movement complexes. PLoS Pathog 16:e1008709. doi: 10.1371/journal.ppat.1008709
Jimenez-Jimenez, S., Santana, O., Lara-Rojas, F., Arthikala, M. K., Armada, E., Hashimoto, K., et al. (2019). Differential tetraspanin genes expression and subcellular localization during mutualistic interactions in Phaseolus vulgaris. PLoS One 14:e0219765. doi: 10.1371/journal.pone.0219765
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kaido, M., Inoue, Y., Takeda, Y., Sugiyama, K., Takeda, A., Mori, M., et al. (2007). Downregulation of the NbNACa1 gene encoding a movement-protein-interacting protein reduces cell-to-cell movement of Brome mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Interact. 20, 671–681. doi: 10.1094/mpmi-20-6-0671
Kang, H., Nguyen, Q.-M., Iswanto, A. B. B., Hong, J. C., Bhattacharjee, S., Gassmann, W., et al. (2021). Nuclear localization of HopA1Pss61 is required for effector-triggered immunity. Plants 10:888. doi: 10.3390/plants10050888
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Khang, C. H., Berruyer, R., Giraldo, M. C., Kankanala, P., Park, S. Y., Czymmek, K., et al. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22, 1388–1403. doi: 10.1105/tpc.109.069666
Kim, N., Kim, J., Bang, B., Kim, I., Lee, H. H., Park, J., et al. (2016). Comparative analyses of tomato yellow leaf curl virus C4 Protein-Interacting host proteins in healthy and infected tomato tissues. Plant Pathol. J. 32, 377–387. doi: 10.5423/ppj.ft.08.2016.0165
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Kraner, M. E., Muller, C., and Sonnewald, U. (2017). Comparative proteomic profiling of the choline transporter-like1 (CHER1) mutant provides insights into plasmodesmata composition of fully developed Arabidopsis thaliana leaves. Plant J. 92, 696–709. doi: 10.1111/tpj.13702
Kumar, D., Kumar, R., Hyun, T. K., and Kim, J. Y. (2015). Cell-to-cell movement of viruses via plasmodesmata. J. Plant Res. 128, 37–47. doi: 10.1007/s10265-014-0683-6
Langner, T., Kamoun, S., and Belhaj, K. (2018). CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi: 10.1146/annurev-phyto-080417-050158
Lee, J. Y., and Lu, H. (2011). Plasmodesmata: the battleground against intruders. Trends Plant Sci. 16, 201–210. doi: 10.1016/j.tplants.2011.01.004
Lee, J. Y., Wang, X., Cui, W., Sager, R., Modla, S., Czymmek, K., et al. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23, 3353–3373. doi: 10.1105/tpc.111.087742
Lefeuvre, P., Martin, D. P., Elena, S. F., Shepherd, D. N., Roumagnac, P., and Varsani, A. (2019). Evolution and ecology of plant viruses. Nat. Rev. Microbiol. 17, 632–644.
Leijon, F., Melzer, M., Zhou, Q., Srivastava, V., and Bulone, V. (2018). Proteomic analysis of plasmodesmata from populus cell suspension cultures in relation with callose biosynthesis. Front. Plant Sci. 9:1681. doi: 10.3389/fpls.2018.01681
Levy, A., Zheng, J. Y., and Lazarowitz, S. G. (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 25, 2018–2025. doi: 10.1016/j.cub.2015.06.015
Lewis, J. D., and Lazarowitz, S. G. (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. U S A. 107, 2491–2496. doi: 10.1073/pnas.0909080107
Li, B. C., Zhang, C., Chai, Q. X., Han, Y. Y., Wang, X. Y., Liu, M. X., et al. (2014). Plasmalemma localisation of DOUBLE HYBRID PROLINE-RICH PROTEIN 1 and its function in systemic acquired resistance of Arabidopsis thaliana. Funct. Plant Biol. 41, 768–779. doi: 10.1071/fp13314
Li, S., Su, X., Luo, X., Zhang, Y., Zhang, D., Du, J., et al. (2020a). First evidence showing that Pepper vein yellows virus P4 protein is a movement protein. BMC Microbiol. 20:72. doi: 10.1186/s12866-020-01758-y
Li, Z. P., Paterlini, A., Glavier, M., and Bayer, E. M. (2020b). Intercellular trafficking via plasmodesmata: molecular layers of complexity. Cell Mol. Life Sci. 78, 799–816. doi: 10.1007/s00018-020-03622-8
Li, Z., Variz, H., Chen, Y., Liu, S.-L., and Aung, K. (2021). Plasmodesmata-dependent intercellular movement of bacterial effectors. Front. Plant Sci. 12:640277. doi: 10.3389/fpls.2021.640277
Lim, G. H., Shine, M. B., De Lorenzo, L., Yu, K., Cui, W., Navarre, D., et al. (2016). Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19, 541–549. doi: 10.1016/j.chom.2016.03.006
Liu, C., Ye, L., Lang, G., Zhang, C., Hong, J., and Zhou, X. (2011). The VP37 protein of Broad bean wilt virus 2 induces tubule-like structures in both plant and insect cells. Virus Res. 155, 42–47. doi: 10.1016/j.virusres.2010.08.013
Liu, H.-J., Zhang, J.-Q., Hu, H.-K., Huang, Y.-J., Xv, C.-M., Hu, Y.-Y., et al. (2021). Overexpression of JrAMT2 in walnut (Juglans regia L.) rootstock enhances nitrogen level in grafted wild-type walnut scions. Sci. Horticulturae 280:109928. doi: 10.1016/j.scienta.2021.109928
Mann, K. S., Bejerman, N., Johnson, K. N., and Dietzgen, R. G. (2016). Cytorhabdovirus P3 genes encode 30K-like cell-to-cell movement proteins. Virology 489, 20–33. doi: 10.1016/j.virol.2015.11.028
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
McGavin, W. J., Mitchell, C., Cock, P. J. A., Wright, K. M., and Macfarlane, S. A. (2012). Raspberry leaf blotch virus, a putative new member of the genus Emaravirus, encodes a novel genomic RNA. J. Gen. Virol. 93, 430–437. doi: 10.1099/vir.0.037937-0
Navarro, J. A., and Pallas, V. (2017). An update on the intracellular and intercellular trafficking of carmoviruses. Front. Plant Sci. 8:1801. doi: 10.3389/fpls.2017.01801
Nguyen, Q.-M., Iswanto, A. B. B., Son, G. H., and Kim, S. H. (2021). Recent advances in effector-triggered immunity in plants: new pieces in the puzzle create a different paradigm. Int. J. Mol. Sci. 22:4709. doi: 10.3390/ijms22094709
Nicolas, W. J., Grison, M. S., Trepout, S., Gaston, A., Fouche, M., Cordelieres, F. P., et al. (2017). Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 3:17082.
O’Lexy, R., Kasai, K., Clark, N., Fujiwara, T., Sozzani, R., and Gallagher, K. L. (2018). Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J. Exp. Bot. 69, 3715–3728. doi: 10.1093/jxb/ery171
Oparka, K. J., Roberts, A. G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K. S., et al. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. doi: 10.1016/s0092-8674(00)80786-2
Park, S. H., Li, F., Renaud, J., Shen, W., Li, Y., Guo, L., et al. (2017). NbEXPA1, an alpha-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 92, 846–861. doi: 10.1111/tpj.13723
Peiro, A., Izquierdo-Garcia, A. C., Sanchez-Navarro, J. A., Pallas, V., Mulet, J. M., and Aparicio, F. (2014). Patellins 3 and 6, two members of the plant patellin family, interact with the movement protein of Alfalfa mosaic virus and interfere with viral movement. Mol. Plant Pathol. 15, 881–891. doi: 10.1111/mpp.12146
Perraki, A., Binaghi, M., Mecchia, M. A., Gronnier, J., German-Retana, S., Mongrand, S., et al. (2014). StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett. 588, 1699–1705. doi: 10.1016/j.febslet.2014.03.014
Perraki, A., Gronnier, J., Gouguet, P., Boudsocq, M., Deroubaix, A. F., Simon, V., et al. (2018). REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14:e1007378. doi: 10.1371/journal.ppat.1007378
Pramanik, D., Shelake, R. M., Kim, M. J., and Kim, J. Y. (2021). CRISPR-Mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 14, 127–150. doi: 10.1016/j.molp.2020.11.002
Raffaele, S., Bayer, E., Lafarge, D., Cluzet, S., German Retana, S., Boubekeur, T., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555. doi: 10.1105/tpc.108.064279
Roberts, I. M., Boevink, P., Roberts, A. G., Sauer, N., Reichel, C., and Oparka, K. J. (2001). Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218, 31–44. doi: 10.1007/bf01288358
Robles Luna, G., Pena, E. J., Borniego, M. B., Heinlein, M., and Garcia, M. L. (2013). Ophioviruses CPsV and MiLBVV movement protein is encoded in RNA 2 and interacts with the coat protein. Virology 441, 152–161. doi: 10.1016/j.virol.2013.03.019
Rodrigues, K. B., Orilio, A. F., Blawid, R., Melo, F. L., and Nagata, T. (2015). Subcellular localization of p29, a putative movement protein of pepper ringspot virus. Arch. Virol. 160, 359–364. doi: 10.1007/s00705-014-2237-6
Rodriguez, A., Angel, C. A., Lutz, L., Leisner, S. M., Nelson, R. S., and Schoelz, J. E. (2014). Association of the P6 protein of Cauliflower mosaic virus with plasmodesmata and plasmodesmal proteins. Plant Physiol. 166, 1345–1358. doi: 10.1104/pp.114.249250
Roossinck, M. J. (2010). Lifestyles of plant viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1899–1905. doi: 10.1098/rstb.2010.0057
Rosas-Diaz, T., Zhang, D., Fan, P., Wang, L., Ding, X., Jiang, Y., et al. (2018). A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. U S A. 115, 1388–1393. doi: 10.1073/pnas.1715556115
Ross-Elliott, T. J., Jensen, K. H., Haaning, K. S., Wager, B. M., Knoblauch, J., Howell, A. H., et al. (2017). Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLife 6:e24125.
Ruiz-Ruiz, S., Spano, R., Navarro, L., Moreno, P., Pena, L., and Flores, R. (2018). Citrus tristeza virus co-opts glyceraldehyde 3-phosphate dehydrogenase for its infectious cycle by interacting with the viral-encoded protein p23. Plant Mol. Biol. 98, 363–373. doi: 10.1007/s11103-018-0783-0
Sager, R., and Lee, J. Y. (2014). Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J. Exp. Bot. 65, 6337–6358. doi: 10.1093/jxb/eru365
Sakulkoo, W., Oses-Ruiz, M., Oliveira Garcia, E., Soanes, D. M., Littlejohn, G. R., Hacker, C., et al. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359, 1399–1403. doi: 10.1126/science.aaq0892
Sáray, R., Fábián, A., Palkovics, L., and Salánki, K. (2021). The 28 Ser amino acid of Cucumber mosaic virus movement protein has a role in symptom formation and plasmodesmata localization. Viruses 13:222. doi: 10.3390/v13020222
Schmitz, J., Stussi-Garaud, C., Tacke, E., Prufer, D., Rohde, W., and Rohfritsch, O. (1997). In situ localization of the putative movement protein (pr17) from potato leafroll luteovirus (PLRV) in infected and transgenic potato plants. Virology 235, 311–322. doi: 10.1006/viro.1997.8679
Scholthof, K. B., Adkins, S., Czosnek, H., Palukaitis, P., Jacquot, E., Hohn, T., et al. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. doi: 10.1111/j.1364-3703.2011.00752.x
Schubert, M., Koteyeva, N. K., Zdyb, A., Santos, P., Voitsekhovskaja, O. V., Demchenko, K. N., et al. (2013). Lignification of cell walls of infected cells in Casuarina glauca nodules that depend on symplastic sugar supply is accompanied by reduction of plasmodesmata number and narrowing of plasmodesmata. Physiol. Plant. 147, 524–540. doi: 10.1111/j.1399-3054.2012.01685.x
Shanks, M., Tomenius, K., Clapham, D., Huskisson, N. S., Barker, P. J., Wilson, I. G., et al. (1989). Identification and subcellular localization of a putative cell-to-cell transport protein from red clover mottle virus. Virology 173, 400–407. doi: 10.1016/0042-6822(89)90552-7
Shelake, R. M., Pramanik, D., and Kim, J.-Y. (2019a). Evolution of plant mutagenesis tools: a shifting paradigm from random to targeted genome editing. Plant Biotechnol. Rep. 13, 423–445. doi: 10.1007/s11816-019-00562-z
Shelake, R. M., Pramanik, D., and Kim, J. Y. (2019b). Exploration of plant-microbe interactions for sustainable agriculture in CRISPR Era. Microorganisms 7:269. doi: 10.3390/microorganisms7080269
Sidorova, T., Miroshnichenko, D., Kirov, I., Pushin, A., and Dolgov, S. (2021). Effect of grafting on viral resistance of non-transgenic plum scion combined with transgenic PPV-Resistant rootstock. Front. Plant Sci. 12:621954. doi: 10.3389/fpls.2021.621954
Smith, L. M., Pontes, O., Searle, I., Yelina, N., Yousafzai, F. K., Herr, A. J., et al. (2007). An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19, 1507–1521. doi: 10.1105/tpc.107.051540
Stewart, L. R., Medina, V., Sudarshana, M. R., and Falk, B. W. (2009). Lettuce infectious yellows virus-encoded P26 induces plasmalemma deposit cytopathology. Virology 388, 212–220. doi: 10.1016/j.virol.2009.03.016
Sui, X., Liu, X., Lin, W., Wu, Z., and Yang, L. (2018). Targeting of rice grassy stunt virus pc6 protein to plasmodesmata requires the ER-to-Golgi secretory pathway and an actin-myosin VIII motility system. Arch. Virol. 163, 1317–1323. doi: 10.1007/s00705-018-3726-9
Sujkowska-Rybkowska, M., and Znojek, E. (2018). Localization of calreticulin and calcium ions in mycorrhizal roots of Medicago truncatula in response to aluminum stress. J. Plant Physiol. 229, 22–31. doi: 10.1016/j.jplph.2018.05.014
Sun, Z., Zhang, S., Xie, L., Zhu, Q., Tan, Z., Bian, J., et al. (2013). The secretory pathway and the actomyosin motility system are required for plasmodesmatal localization of the P7-1 of rice black-streaked dwarf virus. Arch. Virol. 158, 1055–1064. doi: 10.1007/s00705-012-1585-3
Tilsner, J., Cowan, G. H., Roberts, A. G., Chapman, S. N., Ziegler, A., Savenkov, E., et al. (2010). Plasmodesmal targeting and intercellular movement of potato mop-top pomovirus is mediated by a membrane anchored tyrosine-based motif on the lumenal side of the endoplasmic reticulum and the C-terminal transmembrane domain in the TGB3 movement protein. Virology 402, 41–51. doi: 10.1016/j.virol.2010.03.008
Tilsner, J., Linnik, O., Louveaux, M., Roberts, I. M., Chapman, S. N., and Oparka, K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. doi: 10.1083/jcb.201304003
Tomczynska, I., Stumpe, M., Doan, T. G., and Mauch, F. (2020). A Phytophthora effector protein promotes symplastic cell-to-cell trafficking by physical interaction with plasmodesmata-localised callose synthases. New Phytol. 227, 1467–1478. doi: 10.1111/nph.16653
Uchiyama, A., Shimada-Beltran, H., Levy, A., Zheng, J. Y., Javia, P. A., and Lazarowitz, S. G. (2014). The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 5:584. doi: 10.3389/fpls.2014.00584
Ueki, S., Spektor, R., Natale, D. M., and Citovsky, V. (2010). ANK, a host cytoplasmic receptor for the Tobacco mosaic virus cell-to-cell movement protein, facilitates intercellular transport through plasmodesmata. PLoS Pathog 6:e1001201. doi: 10.1371/journal.ppat.1001201
Vu, M. H., Iswanto, A. B. B., Lee, J., and Kim, J. Y. (2020). The role of plasmodesmata-associated receptor in plant development and environmental response. Plants (Basel) 9:216.
Wang, W., Pan, Q., Tian, B., He, F., Chen, Y., Bai, G., et al. (2019). Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 100, 251–264. doi: 10.1111/tpj.14440
Wei, T., Zhang, C., Hong, J., Xiong, R., Kasschau, K. D., Zhou, X., et al. (2010). Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6:e1000962. doi: 10.1371/journal.ppat.1000962
Wei, Y., Shi, Y., Han, X., Chen, S., Li, H., Chen, L., et al. (2019). Identification of cucurbit chlorotic yellows virus P4.9 as a possible movement protein. Virol. J. 16:82.
Wellink, J., Van Lent, J. W., Verver, J., Sijen, T., Goldbach, R. W., and Van Kammen, A. (1993). The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J. Virol. 67, 3660–3664. doi: 10.1128/jvi.67.6.3660-3664.1993
Widana Gamage, S. M. K., and Dietzgen, R. G. (2017). Intracellular localization, interactions and functions of capsicum chlorosis virus proteins. Front. Microbiol. 8:612. doi: 10.3389/fmicb.2017.00612
Wolf, S., Deom, C. M., Beachy, R. N., and Lucas, W. J. (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379. doi: 10.1126/science.246.4928.377
Wu, S., and Gallagher, K. L. (2012). Transcription factors on the move. Curr. Opin. Plant Biol. 15, 645–651. doi: 10.1016/j.pbi.2012.09.010
Wu, S. W., Kumar, R., Iswanto, A. B. B., and Kim, J. Y. (2018). Callose balancing at plasmodesmata. J. Exp. Bot. 69, 5325–5339.
Xu, B., Cheval, C., Laohavisit, A., Hocking, B., Chiasson, D., Olsson, T. S. G., et al. (2017). A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 215, 77–84. doi: 10.1111/nph.14599
Xu, Y., and Zhou, X. (2012). Role of rice stripe virus NSvc4 in cell-to-cell movement and symptom development in Nicotiana benthamiana. Front. Plant Sci. 3:269. doi: 10.3389/fpls.2012.00269
Yoshikawa, N., Oogake, S., Terada, M., Miyabayashi, S., Ikeda, Y., Takahashi, T., et al. (1999). Apple chlorotic leaf spot virus 50 kDa protein is targeted to plasmodesmata and accumulates in sieve elements in transgenic plant leaves. Arch. Virol. 144, 2475–2483. doi: 10.1007/s007050050660
Yu, C., Karlin, D. G., Lu, Y., Wright, K., Chen, J., and Macfarlane, S. (2013). Experimental and bioinformatic evidence that raspberry leaf blotch emaravirus P4 is a movement protein of the 30K superfamily. J. Gen. Virol. 94, 2117–2128. doi: 10.1099/vir.0.053256-0
Yuan, C., Lazarowitz, S. G., and Citovsky, V. (2016). Identification of a functional plasmodesmal localization signal in a plant viral Cell-To-Cell-Movement protein. mBio 7:e02052-15.
Yuan, C., Lazarowitz, S. G., and Citovsky, V. (2018). The plasmodesmal localization signal of TMV MP is recognized by plant synaptotagmin SYTA. mBio 9:e01314-18.
Yuan, Z., Chen, H., Chen, Q., Omura, T., Xie, L., Wu, Z., et al. (2011). The early secretory pathway and an actin-myosin VIII motility system are required for plasmodesmatal localization of the NSvc4 protein of Rice stripe virus. Virus Res. 159, 62–68. doi: 10.1016/j.virusres.2011.04.023
Zavaliev, R., Sagi, G., Gera, A., and Epel, B. L. (2010). The constitutive expression of Arabidopsis plasmodesmal-associated class 1 reversibly glycosylated polypeptide impairs plant development and virus spread. J. Exp. Bot. 61, 131–142. doi: 10.1093/jxb/erp301
Zavaliev, R., Ueki, S., Epel, B. L., and Citovsky, V. (2011). Biology of callose (beta-1,3-glucan) turnover at plasmodesmata. Protoplasma 248, 117–130. doi: 10.1007/s00709-010-0247-0
Keywords: plasmodesmata, CRISPR/Cas, genome editing, plant stress, crop engineering
Citation: Iswanto ABB, Shelake RM, Vu MH, Kim J-Y and Kim SH (2021) Genome Editing for Plasmodesmal Biology. Front. Plant Sci. 12:679140. doi: 10.3389/fpls.2021.679140
Received: 11 March 2021; Accepted: 10 May 2021;
Published: 02 June 2021.
Edited by:
Tessa Maureen Burch-Smith, The University of Tennessee, Knoxville, United StatesReviewed by:
Kuan-Ju Lu, National Chung Hsing University, TaiwanCopyright © 2021 Iswanto, Shelake, Vu, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Hee Kim, c2FuZ2hlZWtpbUBnbnUuYWMua3I=; Jae-Yean Kim, a2ltanlAZ251LmFjLmty
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.