- The Milner Centre for Evolution, Department of Biology and Biochemistry, University of Bath, Bath, United Kingdom
Chromosomes are dynamic entities in the eukaryotic nucleus. During cell development and in response to biotic and abiotic change, individual sections as well as entire chromosomes re-organise and reposition within the nuclear space. A focal point for these processes is the nuclear envelope (NE) providing both barrier and anchor for chromosomal movement. In plants, positioning of chromosome regions and individual genes at the nuclear envelope has been shown to be associated with distinct transcriptional patterns. Here, we will review recent findings on the interplay between transcriptional activity and gene positioning at the nuclear periphery (NP). We will discuss potential mechanisms of transcriptional regulation at the nuclear envelope and outline future perspectives in this research area.
Introduction
The eukaryotic nucleus is defined by the presence of the nuclear envelope (NE) acts as a physical barrier dividing the nuclear content from the cytoplasm (Hetzer, 2010). The NE creates an intranuclear environment that shields the genome, enabling coordinated processes, including DNA duplication and transcription (Lammerding and Wolf, 2016). Within the nucleus, the spatial organisation of eukaryotic genomes and association with the NE has been firmly associated with transcriptional regulation (Fedorova and Zink, 2008; van Steensel and Belmont, 2017).
Here, we will discuss the nuclear envelope in plants and its association with different patterns of gene expression. We will compare structure and function of the plant NE with its counterparts in other eukaryotes. We will focus on our current understanding of the link between gene positioning at the NE and transcriptional activity and outline future directions in the field.
Organisational Principles and Functions of the Nuclear Envelope
The NE is organised into two lipid bilayers, the outer nuclear membrane (ONM) and the inner nuclear membrane (INM), divided by a luminal perinuclear space and the nuclear pore complex (NPC; Goodchild et al., 2015; Meier et al., 2017). The ONM is classified as an extension of the endoplasmic reticulum (ER), and ribosomes are attached to its cytoplasmic surface. Protein content of the ONM is largely shared with the ER (Foresti et al., 2014; Meier et al., 2017). The INM encloses the nucleoplasm and shows a more distinct protein composition (Foresti et al., 2014; Meier et al., 2017). The ONM and INM are connected at the NPCs. These protein channels consist of multiple proteins called nucleoporins (Tamura et al., 2010; Hoelz et al., 2011).
The main role of the INM and ONM is to ensure a spatial continuum between the NE, the cytoplasm, and the nucleoplasm (De Magistris and Antonin, 2018). NPCs act as a bidirectional transport system for macromolecules, which relocate between the nucleus and cytoplasm (Tamura et al., 2015; Meier et al., 2017; De Magistris and Antonin, 2018). Furthermore, NPCs have been implicated in chromatin organisation, transcriptional and post-transcriptional regulation of gene expression, and the progression of the cell cycle (Lince-Faria et al., 2009; Tan-Wong et al., 2009; Ahmed et al., 2010; Kylberg et al., 2010; Strambio-De-Castillia et al., 2010; Vaquerizas et al., 2010; Smith et al., 2015; Tamura et al., 2017). The perinuclear space serves as ion storage and linkage site for interacting proteins from ONM and INM. As such, it functions in nuclear signalling and force transmission between cytoplasm and nucleoplasm (Razafsky and Hodzic, 2009; Starr, 2009; Capoen et al., 2011; Mauger, 2012). It is the site of interaction between linker of nucleoskeleton and cytoskeleton (LINC) complexes that directly link components of the cytoplasm and nucleoplasm (Rothballer and Kutay, 2013).
In metazoan, a dense layer of protein filaments called the nuclear lamina structurally supports the NE underneath the INM (de Leeuw et al., 2018; Alvarado-Kristensson and Rossello, 2019; Tenga and Medalia, 2020). Nuclear lamina composition varies across the eukaryotes. For example, lamin proteins which represent the major constituents of the animal lamina have not been identified in plants (Meier et al., 2017). Instead, three types of plant-specific proteins, CROWDED NUCLEI (CRWN) or nuclear matrix constituent protein (NMCP), KAKU4 and nuclear envelope-associated proteins (NEAPs) have been suggested to function as lamin-like proteins in plants (Masuda et al., 1997; Ciska and de la Espina, 2014; Goto et al., 2014; Pawar et al., 2016; McKenna et al., 2021). Beyond its scaffolding role, the nuclear lamina is of functional importance in chromatin folding, DNA repair, and gene transcription (Burke and Stewart, 2013).
All elements of the NE exhibit high structural variability and may re-organise during cellular differentiation and division. Loss of function of NE components may cause severe diseases in humans and may lead to developmental and signalling defects in plants (Hatch and Hetzer, 2014; Meier et al., 2017; Tang et al., 2020).
Chromatin Organisation at the NE
Components of the NE control nuclear shape and are directly linked to the nucleoplasmic chromatin. As such, it has a dominant role in defining the organisation of chromosomes in the nuclear space and takes in an important position in the transcriptional regulation of gene expression.
Typically, transcriptionally inactive heterochromatic regions of the genome are located adjacent to the NE. In animals, such ‘lamina associated domains’ (LADs) possess distinct properties and are specifically localised towards the NE. LADs, usually 10 Kb–10 Mb in size, comprise a significant part of the genome and incorporate around a third of the human and mice genomes. LADs are rich in repressive histone marks and gene sparse (Guelen et al., 2008; Kind et al., 2015; van Steensel and Belmont, 2017). LADs may associate stably as well as transiently with the nuclear lamina. Constitutive LADs (cLADs) interact with the nuclear lamina across cell types while facultative LADs (fLADs) are localised at the nuclear lamina in distinct cell types (Peric-Hupkes et al., 2010; Meuleman et al., 2013). cLADs are enriched in repetitive elements and depleted in gene content compared to fLADs (Meuleman et al., 2013).
Despite the absence of nuclear lamin proteins in plants, recent studies have shown that the genome of Arabidopsis thaliana is organised non-randomly at the nuclear periphery (NP), in a similar manner to that of LADs in animals. Regions which are associated with the NP in plants have been termed plantLADs (pLADS; Bi et al., 2017; Hu et al., 2019). Heterochromatic domains of centromeric and pericentromeric regions, also called chromocenters, as well as 10–20% of chromosomal arms have been reported to associate with the nuclear periphery in A. thaliana (Fransz et al., 2002; Bi et al., 2017; Hu et al., 2019). Like LADs, pLADs are characterised by an enrichment for heterochromatin and transposable elements. However, unlike LADs, pLADs identified in chromosomal arms of A. thaliana are neither gene poor nor A/T rich (Bi et al., 2017; Hu et al., 2019). pLAD composition remains largely stable across different plant organs (Bi et al., 2017).
In animals, chromatin is anchored at the NE by lamins and INM associated proteins. B-type lamins, for example, interact with lamin B receptors. These receptors bind to the heterochromatic histone marks H3K9me3 and H4K20me2 via their heterochromatin binding and tudor domains (Hirano et al., 2012; Gruenbaum and Foisner, 2015). A-type lamins interact with proteins of the INM and the nucleoplasm. Furthermore, A-type lamins have been shown to associate with GAGA DNA sequence motifs along chromosomes (Zullo et al., 2012; Gruenbaum and Foisner, 2015). Lamins themselves can directly bind to DNA and histones (Taniura et al., 1995; Stierle et al., 2003; Mattout et al., 2007).
In plants, CRWN proteins have been suggested as mediators of pLAD recruitment to the NE (Hu et al., 2019; Sakamoto, 2020). crwn mutants display large scale changes in chromatin organisation. In crwn mutants, chromatin positioning at the nuclear periphery is significantly altered (Hu et al., 2019). Additionally, crwn mutants show an increased chromosomal compaction and disruption of chromocenter organisation (Wang et al., 2013; Grob et al., 2014). By chromatin immunoprecipitation, an interaction of CRWN1 proteins with chromatin has been shown (Hu et al., 2019). Furthermore, CRWN1 proteins have been shown to bind the transcription factor (TF) NTL9, a transcription factor involved in the regulation of plant immunity (Guo et al., 2017). Another candidate for chromosome anchoring at the plant NE is the transcription factor bZIP18, which has been suggested to interact with NEAP1 proteins (Pawar et al., 2016).
Transcriptional Pattern at the NE
The association of chromatin to the NE has profound implications on transcriptional activity of genes. LADs are largely transcriptionally inactive (Guelen et al., 2008). Introduction of reporter genes into LADs is accompanied by reduced transcriptional activity compared to reporter activity in non-LAD locations (Akhtar et al., 2013). Artificial targeting of genes towards the NE has been shown to cause transcriptional downregulation (Finlan et al., 2008; Reddy et al., 2008). In accordance, release from the nuclear lamina may lead to de-repression of genes (Shevelyov et al., 2009; Kohwi et al., 2013; Chen et al., 2014). Like LADs, pLADs are also characterised by low transcriptional activity (Bi et al., 2017).
Peripheral location of genes at the NE, however, may also lead to increased gene expression. For example, the NPC has been described as a site of high transcriptional activity (Strambio-De-Castillia et al., 2010; Vaquerizas et al., 2010). It is suggested that this trait is based on an enhanced efficiency of mRNA export from the nucleoplasm to the cytoplasm. Nucleoporins of the NPC have been implicated in the recruitment of chromosomal regions to the NPC across eukaryotes. In yeasts, binding of nucleoporins to chromatin modifying complexes with positive function in transcriptional activation is shown to be involved in the upregulation of the NE associated GAL gene cluster (Cabal et al., 2006). In A. thaliana, fusion of the LacI DNA binding protein to the nucleoporin Seh1 resulted in peripheral localisation of the LacO-Luciferase reporter construct and increased reporter activity (Smith et al., 2015).
Furthermore, up to 10% of genes encoded in LADs show high expression levels (Guelen et al., 2008; Peric-Hupkes et al., 2010; Wu and Yao, 2017). It is suggested that these genes are driven by promoters that are more resistant to the repressive chromatin environment at the NE and bound by pioneering transcription factors. They may also be preferentially localised at LAD sites with weak NE association (Leemans et al., 2019). Increased transcriptional activity has also been linked to NE association via binding to CRWN proteins. A cluster of coordinately expressed copper-associated (CA) genes is located towards the nuclear periphery upon transcriptional induction (Figure 1). Chromatin integration labelling (ChIL) confirmed association of the CA genes to CRWN1 proteins. Indeed, loss of CRWN1 and CRWN4 results in lower transcription levels of CA genes and reduced NE association (Sakamoto et al., 2020).
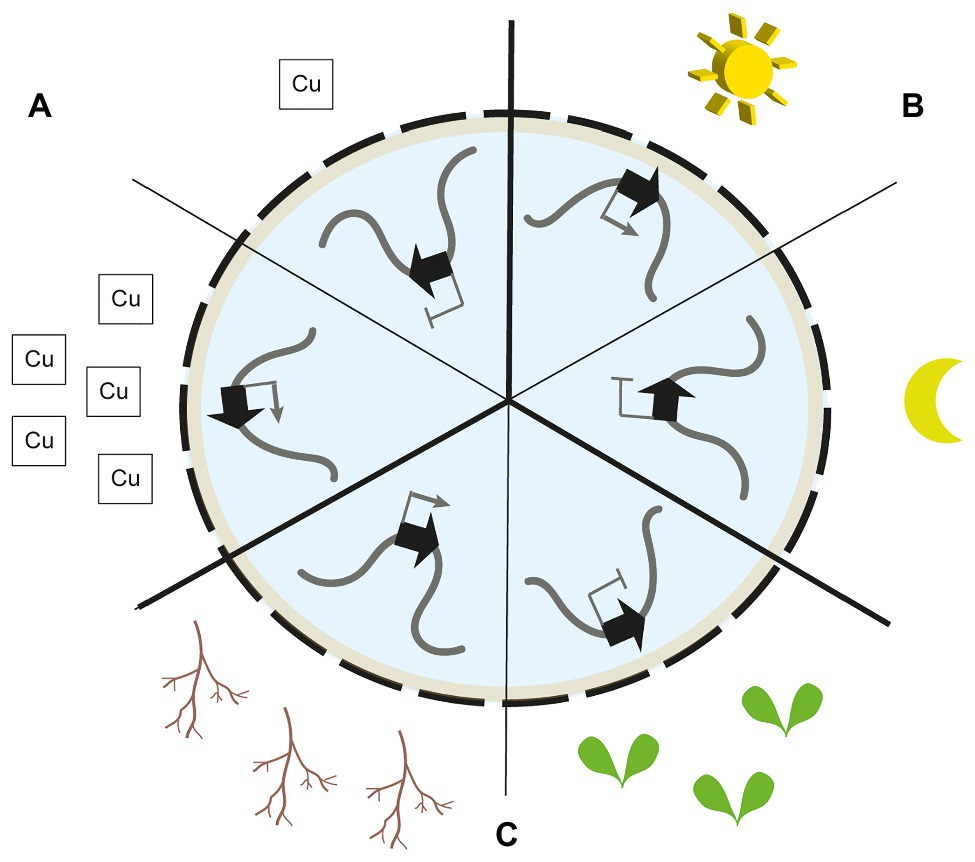
Figure 1. Nuclear gene re-positioning in response to environmental and developmental signals. (A) The copper-associated (CA) gene cluster is located at the nuclear envelope (NE) upon plant exposure to copper. Peripheral localisation is accompanied by transcriptional activation. In the absence of copper, the cluster is located away from the NE and silenced (Sakamoto et al., 2020). (B) The light-induced CAB gene cluster is located at the NE upon plant exposure to light. In absence of light, the cluster is located away from the NE and silenced (Feng et al., 2014). (C) The silenced thalianol biosynthetic gene cluster is located at the nuclear periphery (NP) in leaf cells. In roots, when transcriptionally active, the cluster is positioned away from the nuclear periphery (Nützmann et al., 2020).
Such transient re-localisation towards the NE has also been observed for light-responsive genes in A. thaliana (Figure 1). The CAB gene cluster encoding chlorophyll a/b-binding proteins as well as the RBCS1A, GUN5, and PC genes are re-positioned from the interior of the nucleus towards the periphery upon light exposure to seedlings (Feng et al., 2014). The peripheral localisation of these genes is accompanied by an increased transcriptional activity. Interestingly, the CAB gene cluster is re-localised towards the nuclear periphery before the highest transcription levels are reached, thus, showing that chromatin repositioning precedes full transcriptional activation. In photoreceptor mutants, the dynamic re-positioning of the CAB gene cluster is lost (Feng et al., 2014).
Biosynthetic gene clusters in A. thaliana show the opposite direction in nuclear re-positioning associated with their transcriptional activity (Nützmann et al., 2020; Figure 1). Silenced biosynthetic gene clusters are located at the nuclear periphery while active clusters are located away from the periphery. Such clusters encode biosynthesis genes of specialised metabolic pathways and are exclusively transcribed in root tissues and silenced in the aerial organs of the plant. Chromosome conformation capture and 3D DNA FISH analyses showed that silenced biosynthetic gene clusters are associated with the chromocentric sections of chromosomes in close proximity to the NE. Interestingly, silenced clusters are delineated by H3K27me3 histone marks, and loss of PRC2 activity leads to reduced chromocentric localisation of the loci (Yu et al., 2016; Nützmann et al., 2020).
Discussion
The link between NE association of genes and transcriptional activity is firmly established. However, how positioning of genes at the NE leads to distinct transcriptional outputs in plant nuclei remains largely unknown (Parry, 2015; Santos et al., 2020).
In animals, specific binding and increased local concentrations of chromatin modifying enzymes (CME) at the NE have been suggested as key mechanistic factors in gene silencing at the nuclear periphery (Harr et al., 2016; van Steensel and Belmont, 2017). The animal INM protein emerin, for example, directly interacts with the histone deacetylase HDAC3 and modulates its activity (Demmerle et al., 2012). As such, a transient contact of a gene with the NE may result in changes to its histone modifications. Higher abundance of histone lysine 9 methyltransferases at the nuclear periphery may promote heterochromatin formation of NE associated chromosomal segments (Towbin et al., 2012; Gonzalez-Sandoval et al., 2015). Similarly, chromatin modifiers that act positively on transcription, e.g., histone acetylation complexes, are suggested to be actively retained in euchromatic areas of the nucleus and are as such excluded from the NE (Cabianca et al., 2019). Localisation within heterochromatin at the nuclear periphery may also prevent access of activating transcription factors to its target loci or recruitment of genes to transcription factories in the interior of the nucleus (Yao et al., 2011; van Steensel and Belmont, 2017; Figure 2). Furthermore, it has been suggested that gene positioning within LADs may prevent access of gene promoters to enhancers located outside of LADs (Yaffe and Tanay, 2011; Amendola and van Steensel, 2014). Active retention of transcription factors at the nuclear periphery and away from target chromatin may represent another mechanism of NE associated gene regulation (Mirza et al., 2019).
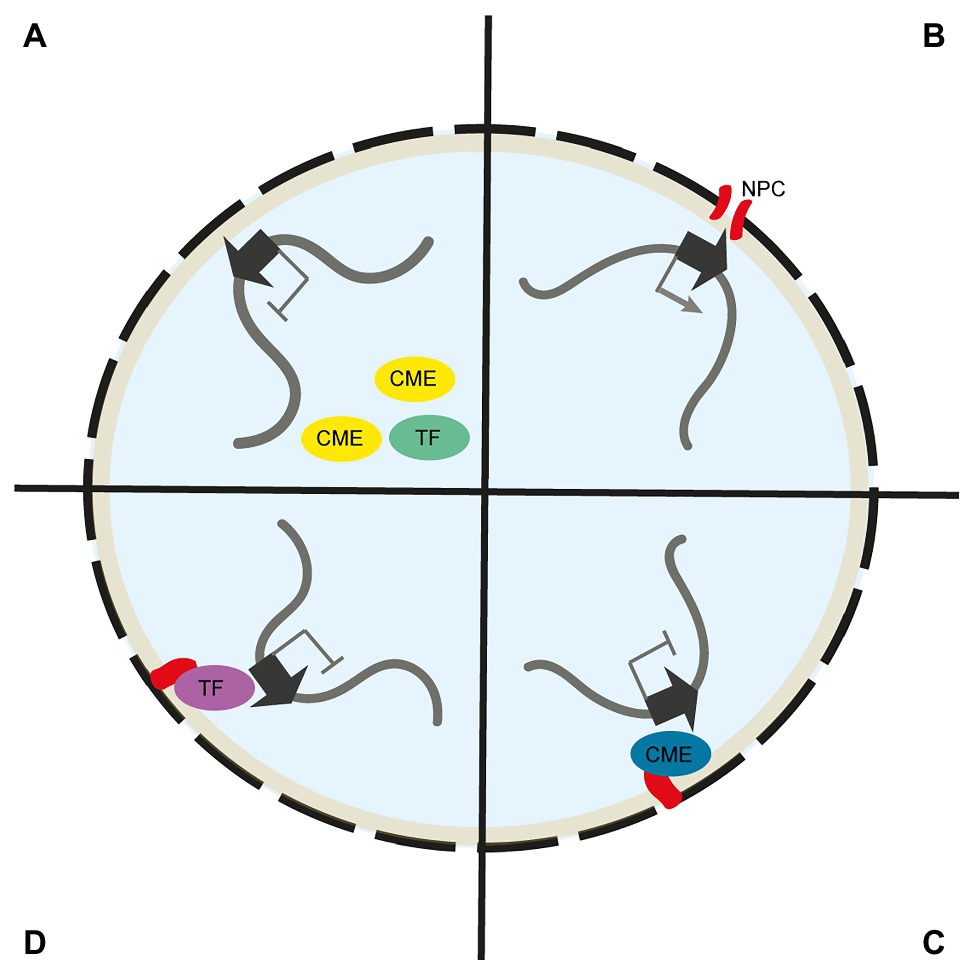
Figure 2. Mechanisms of gene regulation at the nuclear envelope. (A) Chromatin modifying enzymes (CMEs) and transcription factors (TFs) with gene activating function are excluded from the NE. As such, peripheral chromosome regions are devoid of active chromatin marks and activating TFs. (B) Chromosome regions are recruited to the nuclear pore complex (NPC) via nucleoporin interaction. At the NPC, mRNA export is streamlined and loci may interact with NPC associated CMEs. (C) Chromosome regions at the NE are exposed to CMEs with gene silencing activity, such as H3K9 methyltransferases, that are directly linked to the NE. (D) Chromosome regions at the NE are exposed to repressive TFs that are directly linked to the NE.
In plants, pioneering work by Mikulski et al. (2019) showed that CRWN1 associated with the PROLINE-TRYPTOPHANE-TRYPTOPHANE-PROLINE (PWWP) INTERACTOR OF POLYCOMBS1 (PWO1) protein, a component of the Polycomb Repressive Complex 2 (PRC2). As such, it is suggested that the localisation of PWO1 links PRC2 to the NE and thus promotes the formation of facultative H3K27me3-marked chromatin at the NE (Mikulski et al., 2019). Indeed, chromosome simulations support the important role of H3K27me3 in chromosome organisation in A. thaliana (Di Stefano et al., 2021). Furthermore, different DNA methylation pathways are suggested to target transposable elements at the periphery and interior of the A. thaliana nucleus (Bi et al., 2017). This may indicate a mechanistic role for distinct DNA methylation pathways in the silencing of transposable elements at the NE in plants.
A largely unexplored aspect of NE-related influence on gene expression is how mechanical stress put on the exterior of the cell and mediated by the NE affects transcription. LINC complexes link the cytoskeleton with the nucleoskeleton (Rothballer and Kutay, 2013). Indeed, mechanical stress applied to mammalian cells has been shown to spread through the actin cytoskeleton and onto lamin proteins (Ramdas and Shivashankar, 2015; Tajik et al., 2016). These forces change chromatin conformation by stretching it and lead to increased expression levels in affected chromosome regions. In plants, both abiotic and biotic stressors may change the mechanical force applied on a cell. For example, appressoria of fungal plant pathogens induce strong mechanical forces on cells (Bastmeyer et al., 2002). These may be transduced via the NE into the nucleus and lead to changes in chromosome organisation and gene expression.
Future research investigating the interplay between the NE and gene transcription in plants may expand its focus from A. thaliana to other species and move from whole-plant and organ-level to cell type and single cell level. A. thaliana is characterised by its relatively small and gene-dense genome. Plants with larger genomes and lower gene density are likely to exhibit different pattern of NE associated chromatin and gene expression. This would follow the significant differences in chromosome organisation identified in species, such as wheat, maize, and tomato compared to A. thaliana (Dong et al., 2017; Concia et al., 2020). Furthermore, elucidating the NE proteome and the NE associated chromatin on cell-type specific level and in response to biotic and abiotic challenges may lead to the identification of functionally important NE associated chromatin anchors and chromatin modifying complexes. These may become targets for manipulation of gene expression in plants. Future studies may further resolve which locus-specific features cause some genes to be upregulated upon transfer to the nuclear periphery and some genes to be downregulated. These may be accompanied by investigations aiming to shed light on the molecular mechanisms involved in the movement of loci towards and away from the periphery and how flanking genes are affected by gene-positioning at the NE. In human cell lines, for example, about 50–100 Kb of flanking chromatin is detached from the nuclear lamina upon activation of single genes (Brueckner et al., 2020). It remains to be seen whether plant genomes with diverse sizes and gene densities exhibit a similar pattern of chromatin detachment and how this affects the transcription of neighbouring genes.
Author Contributions
All authors wrote the manuscript. HWN developed the concept and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the University of Bath (FV, JB, HS, and HWN) and the following Royal Society awards: University Research Fellowship UF160138 (HWN), Research Fellows Enhancement Award RGF\EA\201054 (FV and HWN), and Research Grants for Research Fellows RGF\R1\181018 (HS and HWN). We thank for support by the EU COST Action INDEPTH (CA16212).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmed, S., Brickner, D. G., Light, W. H., Cajigas, I., Mcdonough, M., Froyshteter, A. B., et al. (2010). DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12, 111–118. doi: 10.1038/ncb2011
Akhtar, W., de Jong, J., Pindyurin, A. V., Pagie, L., Meuleman, W., De Ridder, J., et al. (2013). Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154, 914–927. doi: 10.1016/j.cell.2013.07.018
Alvarado-Kristensson, M., and Rossello, C. A. (2019). The biology of the nuclear envelope and its implications in cancer biology. Int. J. Mol. Sci. 20:2586. doi: 10.3390/ijms20102586
Amendola, M., and van Steensel, B. (2014). Mechanisms and dynamics of nuclear lamina-genome interactions. Curr. Opin. Cell Biol. 28, 61–68. doi: 10.1016/j.ceb.2014.03.003
Bastmeyer, M., Deising, H. B., and Bechinger, C. (2002). Force exertion in fungal infection. Annu. Rev. Biophys. Biomol. Struct. 31, 321–341. doi: 10.1146/annurev.biophys.31.091701.170951
Bi, X. L., Cheng, Y. J., Hu, B., Ma, X. L., Wu, R., Wang, J. W., et al. (2017). Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 27, 1162–1173. doi: 10.1101/gr.215186.116
Brueckner, L., Zhao, P. A., van Schaik, T., Leemans, C., Sima, J., Peric-Hupkes, D., et al. (2020). Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 39:e103159. doi: 10.15252/embj.2019103159
Burke, B., and Stewart, C. L. (2013). The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 14, 13–24. doi: 10.1038/nrm3488
Cabal, G. G., Genovesio, A., Rodriguez-Navarro, S., Zimmer, C., Gadal, O., Lesne, A., et al. (2006). SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441, 770–773. doi: 10.1038/nature04752
Cabianca, D. S., Munoz-Jimenez, C., Kalck, V., Gaidatzis, D., Padeken, J., Seeber, A., et al. (2019). Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 569, 734–739. doi: 10.1038/s41586-019-1243-y
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U. S. A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Chen, H. Y., Zheng, X. B., and Zheng, Y. X. (2014). Age-associated loss of Lamin-B leads to systemic inflammation and gut hyperplasia. Cell 159, 829–843. doi: 10.1016/j.cell.2014.10.028
Ciska, M., and De La Espina, S. M. D. (2014). The intriguing plant nuclear lamina. Front. Plant Sci. 5:166. doi: 10.3389/fpls.2014.00166
Concia, L., Veluchamy, A., Ramirez-Prado, J. S., Martin-Ramirez, A., Huang, Y., Perez, M., et al. (2020). Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biol. 21:104. doi: 10.1186/s13059-020-01998-1
de Leeuw, R., Gruenbaum, Y., and Medalia, O. (2018). Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 28, 34–45. doi: 10.1016/j.tcb.2017.08.004
De Magistris, P., and Antonin, W. (2018). The dynamic nature of the nuclear envelope. Curr. Biol. 28, R487–R497. doi: 10.1016/j.cub.2018.01.073
Demmerle, J., Koch, A. J., and Holaska, J. M. (2012). The nuclear envelope protein Emerin binds directly to histone Deacetylase 3 (HDAC3) and activates HDAC3 activity. J. Biol. Chem. 287, 22080–22088. doi: 10.1074/jbc.M111.325308
Di Stefano, M., Nützmann, H. W., Marti-Renom, M. A., and Jost, D. (2021). Polymer modelling unveils the roles of heterochromatin and nucleolar organizing regions in shaping 3D genome organization in Arabidopsis thaliana. Nucleic Acids Res. 49, 1840–1858. doi: 10.1093/nar/gkaa1275
Dong, P. F., Tu, X. Y., Chu, P. Y., Lu, P. T., Zhu, N., Grierson, D., et al. (2017). 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10, 1497–1509. doi: 10.1016/j.molp.2017.11.005
Fedorova, E., and Zink, D. (2008). Nuclear architecture and gene regulation. Biochim. Biophys. Acta 1783, 2174–2184. doi: 10.1016/j.bbamcr.2008.07.018
Feng, C. M., Qiu, Y. J., Van Buskirk, E. K., Yang, E. J., and Chen, M. (2014). Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 5:3027. doi: 10.1038/ncomms4027
Finlan, L. E., Sproul, D., Thomson, I., Boyle, S., Kerr, E., Perry, P., et al. (2008). Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4:e1000039. doi: 10.1371/journal.pgen.1000039
Foresti, O., Rodriguez-Vaello, V., Funaya, C., and Carvalho, P. (2014). Quality control of inner nuclear membrane proteins by the Asi complex. Science 346, 751–755. doi: 10.1126/science.1255638
Fransz, P., de Jong, J. H., Lysak, M., Castiglione, M. R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. U. S. A. 99, 14584–14589. doi: 10.1073/pnas.212325299
Gonzalez-Sandoval, A., Towbin, B. D., Kalck, V., Cabianca, D. S., Gaidatzis, D., Hauer, M. H., et al. (2015). Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163, 1333–1347. doi: 10.1016/j.cell.2015.10.066
Goodchild, R. E., Buchwalter, A. L., Naismith, T. V., Holbrook, K., Billion, K., Dauer, W. T., et al. (2015). Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J. Cell Sci. 128, 2854–2865. doi: 10.1242/jcs.167452
Goto, C., Tamura, K., Fukao, Y., Shimada, T., and Hara-Nishimura, I. (2014). The novel nuclear envelope protein KAKU4 modulates nuclear morphology in Arabidopsis. Plant Cell 26, 2143–2155. doi: 10.1105/tpc.113.122168
Grob, S., Schmid, M. W., and Grossniklaus, U. (2014). Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55, 678–693. doi: 10.1016/j.molcel.2014.07.009
Gruenbaum, Y., and Foisner, R. (2015). Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 84, 131–164. doi: 10.1146/annurev-biochem-060614-034115
Guelen, L., Pagie, L., Brasset, E., Meuleman, W., Faza, M. B., Talhout, W., et al. (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951. doi: 10.1038/nature06947
Guo, T. T., Mao, X. G., Zhang, H., Zhang, Y., Fu, M. D., Sun, Z. F., et al. (2017). Lamin-like proteins negatively regulate plant immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol. Plant 10, 1334–1348. doi: 10.1016/j.molp.2017.09.008
Harr, J. C., Gonzalez-Sandoval, A., and Gasser, S. M. (2016). Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep. 17, 139–155. doi: 10.15252/embr.201541809
Hatch, E., and Hetzer, M. (2014). Breaching the nuclear, envelope in development and disease. J. Cell Biol. 205, 133–141. doi: 10.1083/jcb.201402003
Hetzer, M. W. (2010). The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000539. doi: 10.1101/cshperspect.a000539
Hirano, Y., Hizume, K., Kimura, H., Takeyasu, K., Haraguchi, T., and Hiraoka, Y. (2012). Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J. Biol. Chem. 287, 42654–42663. doi: 10.1074/jbc.M112.397950
Hoelz, A., Debler, E. W., and Blobel, G. (2011). The structure of the nuclear pore complex. Annu. Rev. Biochem. 80, 613–643. doi: 10.1146/annurev-biochem-060109-151030
Hu, B., Wang, N., Bi, X. L., Karaaslan, E. S., Weber, A. L., Zhu, W. S., et al. (2019). Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 20:87. doi: 10.1186/s13059-019-1694-3
Kind, J., Pagie, L., de Vries, S. S., Nahidiazar, L., Dey, S. S., Bienko, M., et al. (2015). Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163, 134–147. doi: 10.1016/j.cell.2015.08.040
Kohwi, M., Lupton, J. R., Lai, S. L., Miller, M. R., and Doe, C. Q. (2013). Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152, 97–108. doi: 10.1016/j.cell.2012.11.049
Kylberg, K., Bjork, P., Fomproix, N., Ivarsson, B., Wieslander, L., and Daneholt, B. (2010). Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp. Cell Res. 316, 1028–1038. doi: 10.1016/j.yexcr.2009.10.016
Lammerding, J., and Wolf, K. (2016). Nuclear envelope rupture: actin fibers are putting the squeeze on the nucleus. J. Cell Biol. 215, 5–8. doi: 10.1083/jcb.201609102
Leemans, C., van Der Zwalm, M. C. H., Brueckner, L., Comoglio, F., van Schaik, T., Pagie, L., et al. (2019). Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell 177, 852–864. doi: 10.1016/j.cell.2019.03.009
Lince-Faria, M., Maffini, S., Orr, B., Ding, Y., Florindo, C., Sunkel, C. E., et al. (2009). Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184, 647–657. doi: 10.1083/jcb.200811012
Masuda, K., Xu, Z. J., Takahashi, S., Ito, A., Ono, M., Nomura, K., et al. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp. Cell Res. 232, 173–181. doi: 10.1006/excr.1997.3531
Mattout, A., Goldberg, M., Tzur, Y., Margalit, A., and Gruenbaum, Y. (2007). Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J. Cell Sci. 120, 77–85. doi: 10.1242/jcs.03325
Mauger, J. P. (2012). Role of the nuclear envelope in calcium signalling. Biol. Cell. 104, 70–83. doi: 10.1111/boc.201100103
McKenna, J. F., Gumber, H. K., Turpin, Z. M., Jalovec, A. M., Kartick, A. C., Graumann, K., et al. (2021). Maize (Zea mays L.) nucleoskeletal proteins regulate nuclear envelope remodeling and function in stomatal complex development and pollen viability. Front. Plant Sci. 12:645218. doi: 10.3389/fpls.2021.645218
Meier, I., Richards, E. J., and Evans, D. E. (2017). Cell biology of the plant nucleus. Annu. Rev. Plant Biol. 68, 139–172. doi: 10.1146/annurev-arplant-042916-041115
Meuleman, W., Peric-Hupkes, D., Kind, J., Beaudry, J. B., Pagie, L., Kellis, M., et al. (2013). Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23, 270–280. doi: 10.1101/gr.141028.112
Mikulski, P., Hohenstatt, M. L., Farrona, S., Smaczniak, C., Stahl, Y., Kalyanikrishna,, et al. (2019). The chromatin-associated protein PWO1 interacts with plant nuclear lamin-like components to regulate nuclear size. Plant Cell 31, 1141–1154. doi: 10.1105/tpc.18.00663
Mirza, A. N., Mckellar, S. A., Urman, N. M., Brown, A. S., Hollmig, T., Aasi, S. Z., et al. (2019). LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell 176, 198–212. doi: 10.1016/j.cell.2018.10.054
Nützmann, H. W., Doerr, D., Ramirez-Colmenero, A., Sotelo-Fonseca, J. E., Wegel, E., Di Stefano, M., et al. (2020). Active and repressed biosynthetic gene clusters have distinct chromosome states. Proc. Natl. Acad. Sci. U. S. A. 117, 13800–13809. doi: 10.1073/pnas.1920474117
Parry, G. (2015). The plant nuclear envelope and regulation of gene expression. J. Exp. Bot. 66, 1673–1685. doi: 10.1093/jxb/erv023
Pawar, V., Poulet, A., Detourne, G., Tatout, C., Vanrobays, E., Evans, D. E., et al. (2016). A novel family of plant nuclear envelope-associated proteins. J. Exp. Bot. 67, 5699–5710. doi: 10.1093/jxb/erw332
Peric-Hupkes, D., Meuleman, W., Pagie, L., Bruggeman, S. W. M., Solovei, I., Brugman, W., et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions duringdifferentiation. Mol. Cell 38, 603–613. doi: 10.1016/j.molcel.2010.03.016
Ramdas, N. M., and Shivashankar, G. V. (2015). Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol. 427, 695–706. doi: 10.1016/j.jmb.2014.09.008
Razafsky, D., and Hodzic, D. (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186, 461–472. doi: 10.1083/jcb.200906068
Reddy, K. L., Zullo, J. M., Bertolino, E., and Singh, H. (2008). Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247. doi: 10.1038/nature06727
Rothballer, A., and Kutay, U. (2013). The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122, 415–429. doi: 10.1007/s00412-013-0417-x
Sakamoto, Y. (2020). Nuclear lamina CRWN proteins regulate chromatin organization, gene expression, and nuclear body formation in plants. J. Plant Res. 133, 457–462. doi: 10.1007/s10265-020-01184-1
Sakamoto, Y., Sato, M., Sato, Y., Harada, A., Suzuki, T., Goto, C., et al. (2020). Subnuclear gene positioning through lamina association affects copper tolerance. Nat. Commun. 11:5914. doi: 10.1038/s41467-020-19621-z
Santos, A. P., Gaudin, V., Mozgova, I., Pontvianne, F., Schubert, D., Tek, A. L., et al. (2020). Tidying-up the plant nuclear space: domains, functions, and dynamics. J. Exp. Bot. 71, 5160–5178. doi: 10.1093/jxb/eraa282
Shevelyov, Y. Y., Lavrov, S. A., Mikhaylova, L. M., Nurminsky, I. D., Kulathinal, R. J., Egorova, K. S., et al. (2009). The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. U. S. A. 106, 3282–3287. doi: 10.1073/pnas.0811933106
Smith, S., Galinha, C., Desset, S., Tolmie, F., Evans, D., Tatout, C., et al. (2015). Marker gene tethering by nucleoporins affects gene expression in plants. Nucleus 6, 471–478. doi: 10.1080/19491034.2015.1126028
Starr, D. A. (2009). A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122, 577–586. doi: 10.1242/jcs.037622
Stierle, V. N., Couprie, J. L., Ostlund, C., Krimm, I., Zinn-Justin, S., Hossenlopp, P., et al. (2003). The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry 42, 4819–4828. doi: 10.1021/bi020704g
Strambio-De-Castillia, C., Niepel, M., and Rout, M. P. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, 490–501. doi: 10.1038/nrm2928
Tajik, A., Zhang, Y. J., Wei, F. X., Sun, J., Jia, Q., Zhou, W. W., et al. (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15, 1287–1296. doi: 10.1038/nmat4729
Tamura, K., Fukao, Y., Hatsugai, N., Katagiri, F., and Hara-Nishimura, I. (2017). Nup82 functions redundantly with Nup136 in a salicylic acid-dependent defense response of Arabidopsis thaliana. Nucleus 8, 301–311. doi: 10.1080/19491034.2017.1279774
Tamura, K., Fukao, Y., Iwamoto, M., Haraguchi, T., and Hara-Nishimura, I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084–4097. doi: 10.1105/tpc.110.079947
Tamura, K., Goto, C., and Hara-Nishimura, I. (2015). Recent advances in understanding plant nuclear envelope proteins involved in nuclear morphology. J. Exp. Bot. 66, 1641–1647. doi: 10.1093/jxb/erv036
Tang, Y., Huang, A. B., and Gu, Y. N. (2020). Global profiling of plant nuclear membrane proteome in Arabidopsis. Nat. Plants 6, 838–847. doi: 10.1038/s41477-020-0700-9
Taniura, H., Glass, C., and Gerace, L. (1995). A chromatin binding-site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 131, 33–44. doi: 10.1083/jcb.131.1.33
Tan-Wong, S. M., Wijayatilake, H. D., and Proudfoot, N. J. (2009). Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23, 2610–2624. doi: 10.1101/gad.1823209
Tenga, R., and Medalia, O. (2020). Structure and unique mechanical aspects of nuclear lamin filaments. Curr. Opin. Struct. Biol. 64, 152–159. doi: 10.1016/j.sbi.2020.06.017
Towbin, B. D., Gonzalez-Aguilera, C., Sack, R., Gaidatzis, D., Kalck, V., Meister, P., et al. (2012). Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947. doi: 10.1016/j.cell.2012.06.051
van Steensel, B., and Belmont, A. S. (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791. doi: 10.1016/j.cell.2017.04.022
Vaquerizas, J. M., Suyama, R., Kind, J., Miura, K., Luscombe, N. M., and Akhtar, A. (2010). Nuclear pore proteins Nup153 and Megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6:e1000846. doi: 10.1371/journal.pgen.1000846
Wang, H. Y., Dittmer, T. A., and Richards, E. J. (2013). Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13:200. doi: 10.1186/1471-2229-13-200
Wu, F. N., and Yao, J. (2017). Identifying novel transcriptional and epigenetic features of nuclear lamina-associated genes. Sci. Rep. 7:100. doi: 10.1038/s41598-017-00176-x
Yaffe, E., and Tanay, A. (2011). Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065. doi: 10.1038/ng.947
Yao, J., Fetter, R. D., Hu, P., Betzig, E., and Tjian, R. (2011). Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 25, 569–580. doi: 10.1101/gad.2021411
Yu, N., Nützmann, H. W., Macdonald, J. T., Moore, B., Field, B., Berriri, S., et al. (2016). Delineation of metabolic gene clusters in plant genomes by chromatin signatures. Nucleic Acids Res. 44, 2255–2265. doi: 10.1093/nar/gkw100
Keywords: nuclear envelope, plants, transcription, epigenetics, chromosome organisation
Citation: Bishop J, Swan H, Valente F and Nützmann H-W (2021) The Plant Nuclear Envelope and Its Role in Gene Transcription. Front. Plant Sci. 12:674209. doi: 10.3389/fpls.2021.674209
Edited by:
Kentaro Tamura, University of Shizuoka, JapanReviewed by:
Yuki Sakamoto, Osaka University, JapanEmmanuel Vanrobays, Université Clermont Auvergne, France
Copyright © 2021 Bishop, Swan, Valente and Nützmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hans-Wilhelm Nützmann, aC5udWV0em1hbm5AYmF0aC5hYy51aw==
†These authors have contributed equally to this work and share first authorship
 Jade Bishop†
Jade Bishop† Francesco Valente
Francesco Valente Hans-Wilhelm Nützmann
Hans-Wilhelm Nützmann