
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 August 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.670369
This article is part of the Research Topic Hormonal Crosstalk on the Regulation of Stress Responses View all 11 articles
 Rajesh K. Singhal1†
Rajesh K. Singhal1† Debanjana Saha2
Debanjana Saha2 Milan Skalicky3
Milan Skalicky3 Udit N. Mishra4
Udit N. Mishra4 Jyoti Chauhan5
Jyoti Chauhan5 Laxmi P. Behera6
Laxmi P. Behera6 Devidutta Lenka7
Devidutta Lenka7 Subhash Chand1
Subhash Chand1 Vivek Kumar8
Vivek Kumar8 Prajjal Dey9
Prajjal Dey9 Indu1
Indu1 Saurabh Pandey10†
Saurabh Pandey10† Pavla Vachova3
Pavla Vachova3 Aayushi Gupta3
Aayushi Gupta3 Marian Brestic3,11
Marian Brestic3,11 Ayman El Sabagh12,13*
Ayman El Sabagh12,13*In the era of rapid climate change, abiotic stresses are the primary cause for yield gap in major agricultural crops. Among them, salinity is considered a calamitous stress due to its global distribution and consequences. Salinity affects plant processes and growth by imposing osmotic stress and destroys ionic and redox signaling. It also affects phytohormone homeostasis, which leads to oxidative stress and eventually imbalances metabolic activity. In this situation, signaling compound crosstalk such as gasotransmitters [nitric oxide (NO), hydrogen sulfide (H2S), hydrogen peroxide (H2O2), calcium (Ca), reactive oxygen species (ROS)] and plant growth regulators (auxin, ethylene, abscisic acid, and salicylic acid) have a decisive role in regulating plant stress signaling and administer unfavorable circumstances including salinity stress. Moreover, recent significant progress in omics techniques (transcriptomics, genomics, proteomics, and metabolomics) have helped to reinforce the deep understanding of molecular insight in multiple stress tolerance. Currently, there is very little information on gasotransmitters and plant growth regulator crosstalk and inadequacy of information regarding the integration of multi-omics technology during salinity stress. Therefore, there is an urgent need to understand the crucial cell signaling crosstalk mechanisms and integrative multi-omics techniques to provide a more direct approach for salinity stress tolerance. To address the above-mentioned words, this review covers the common mechanisms of signaling compounds and role of different signaling crosstalk under salinity stress tolerance. Thereafter, we mention the integration of different omics technology and compile recent information with respect to salinity stress tolerance.
Soil is an indispensable component of the environment and a fundamental prerequisite for nourishing optimistic plant growth and development. Along with its supporting role, soil provides essential nutrient and mineral elements for the vigorous and productive growth of plants. Despite this, the extensive use of chemical fertilizers, excess irrigation, farm mechanization, and other anthropogenic activities act as stress factors for soil natural properties and lead to soil salinity (Pessarakli and Szabolcs, 1999). Soil salinity is one of the global indispensable stress factors, affecting plant growth drastically in both irrigated and rain-fed areas (Hussain et al., 2019; Sabagh et al., 2019; Liu L. et al., 2020). According to the Land and Plant Nutrition Management Service report, approximately 6% (>45 Mha) of the world’s cultivated areas and about one-third of irrigated land on earth is affected by the salinity stress (Carillo et al., 2011; Deinlein et al., 2014; Parihar et al., 2015). Soil salinity situations emerge when glut salts leach and accrue in soil and at the same time, there is no room to flush out the accumulated salts to a well-managed drainage system (Qadir et al., 2008). At the same time, salt concentration swiftly reaches levels that are injurious to salt-sensitive species and beyond this, salt concentration levels can also affect salt-tolerant species. The initial stage of salinity stress is osmotic stress and perused by ion toxicity, which mainly targets uptake and transport of essential ions in plant roots (Serrano and Rodriguez-Navarro, 2001). Salinity stress induces considerable changes in physiological, biochemical, and molecular processes, depending on the extent and severity of the stress, types of genotypes, and crop stages which ultimately leads to huge yield penalty in important agricultural crops (Zeng et al., 2001; Thitisaksakul et al., 2015; Negrão et al., 2017). Osmotic stress culminates in loss of water absorption capacity of the root system, water potential of leaves, membrane damage, nutrients inequity, reduced photosynthetic and metabolic processes, and abatement of the antioxidant defense of plants (Amirjani, 2010; Yan et al., 2013; Parihar et al., 2015). Severe toxicity leads to alterations in crucial plant processes and destroys root functions via modifying redox potential, ion homeostasis, hormonal balance, transpiration, and generates a high amount of ROS [singlet oxygen, superoxide, hydroxyl radical, and hydrogen peroxide (H2O2)], which damages the cellular membrane integrity and macromolecular structure (carbohydrate, proteins, lipids, and DNA) (Läuchli and Grattan, 2007; Farkhondeh et al., 2012). The consequences of salinity on root and plant functions, fundamental processes, and at molecular levels are illustrated in Figure 1.
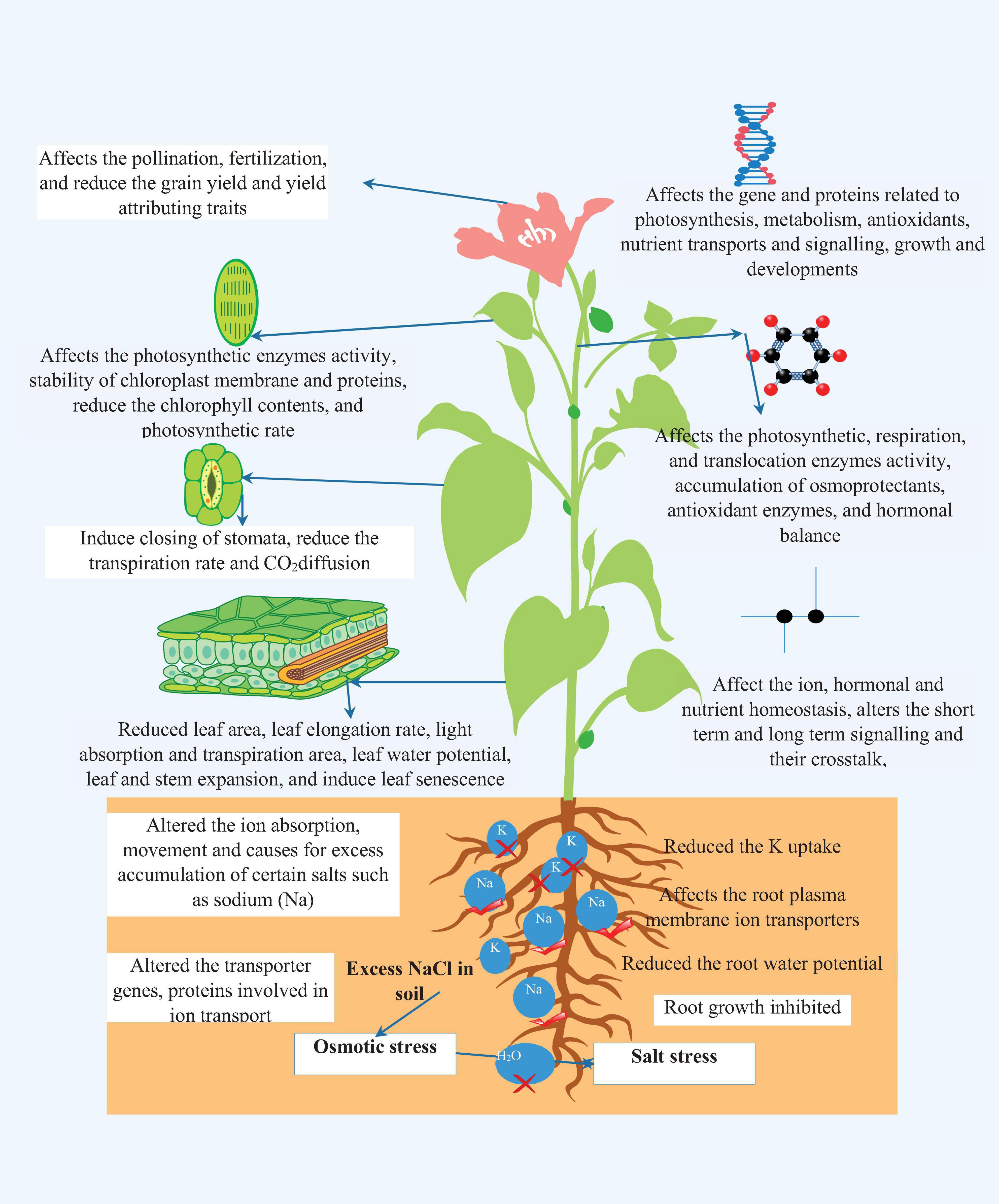
Figure 1. The effect of salinity stress on root growth, ionic homeostasis, physiological, biochemical, and molecular processes.
Hence, salt stress adaptivity or tolerance responses are very crucial to develop stress-tolerant varieties under unfavorable situations. As a consequence, plants activate some exclusive physiological, biochemical, and molecular mechanisms in order to survive under stress conditions, which involve the activation of antioxidant enzymes, compartmentalization, uptake and transport of ions, accumulation of osmoprotectants and compatible solutes, ion homeostasis, and secondary metabolites (Gupta and Huang, 2014; Singhal et al., 2017). Nevertheless, the natural capacity of plants enable them to achieve the desired food potential under these circumstances. Therefore, researchers are continuously working on finding strategies and mechanisms to boost their final potential. In this regard, identifying differentially expressed genes and gene products and transgenic approaches that are associated with stress tolerance are promising approaches to develop smart crops (Yang and Guo, 2018). Even if, due to the complex nature and effects of abiotic stresses on plant processes, these strategies are also completely efficient to achieve the goal of food security under climate change. Therefore, recently scientists have been working on finding and elucidating sophisticated signaling and molecular approaches to develop multiple stress-tolerant crops.
At this point, actuating the signal-transduction cascade for versatile climate plant responses includes various adjustments that are produced in an exceptionally well-coordinated way for exhibiting great opportunities to provide stress tolerance. The promoter-reporter approach has also been exceptionally helpful for identifying genes involved in osmotic stress (Ishitani et al., 1997), and has significantly improved salt-stress monitoring in higher plants. Ionic homeostasis under salt stress is mediated by the SOS (salt overlay sensitive) pathway in a Ca+2-dependent manner, which transduces the salt stress signal in a regulated pattern with the SOS3-SOS2 protein kinase complex at the cell membrane that adds an Na+ ion into the cell and balances ion homeostasis (Ji et al., 2013; Gupta and Huang, 2014). Interestingly, several other signaling compounds such as nitric oxide (NO), hydrogen sulfide (H2S), H2O2, Ca, ROS, and plant growth regulators salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and abscisic acid (ABA) have crucial roles during cell signaling and crosstalk as they provide tolerance to multiple stresses (Chauhan et al., 2017; Noctor et al., 2018; Pei et al., 2018). Moreover, germplasm resources and integrated “omics-assisted” approaches such as phenomics, ionomics, transcriptomics, proteomics, genomics, miRNAomics, lipidomics, and metabolomics are prominently used for developing salt tolerance in crop species (Ho et al., 2020). Correspondingly, epigenetics and next generation phenotyping also provide efficient platforms in context to the production of salt stress-tolerant species (Jha et al., 2019). All integrated omics-assisted approaches have contributed extraordinarily to understanding the outcomes of salinity stress and the alterations which plants adopt for survival and mitigation under unfavorable conditions (Mehta et al., 2019; Pan et al., 2020; Rasel et al., 2020). It is important to keep in mind that, in the present perspective, we point out the recent advances in the crosstalk of important signaling compounds and their role under salinity stress tolerance. Further, we address the recent advances in integrative multi-omics approaches, which are crucial to provide salinity tolerance and a future platform to develop promising salt-tolerant varieties for salt stress conditions.
Plants under stressed conditions such as abiotic (heat, cold, salinity, heavy metal) and biotic stresses must be acknowledged and the innate immune system must be activated for survival and better fitness. The survival of plants under stressful conditions depends on compact signaling networks and their crosstalk (Tena et al., 2011; Smékalová et al., 2014). Signaling pathway activities are activated after the sensation of a signal produced by a specified receptor that triggers the urging of secondary signals and protein phosphorylation cascades like MAPK signaling. Secondary messengers such as ROS, Ca2+, NO, H2S, H2O2, phospholipids, and PGRs act as secondary signals during signaling cascades (DeFalco et al., 2010; Suzuki et al., 2012). These signals are involved in signaling pathways through multiple ways and administer fundamental processes such as cell division and growth, differentiation, and programmed cell death under normal as well as abiotic and biotic interactions (Müller et al., 2010; Tena et al., 2011; Sasabe and Machida, 2012).
Salt stress changes to membrane structures induce metabolic stress, form ROS, and prevent photosynthesis leading to nutrient deficiency (Hasegawa et al., 2000; Tuteja, 2007). The growth responses to salinity include two distinct stages (Munns, 1993). Hormonal signals from the root’s delay development, and then switch off the signal when the plant is mature. Growth reduction can be attributed to a salt-specific effect, which often takes a while (varies between weeks to years) to create. The second step is a result of decreased water availability and accumulation of salt in transpiring leaves, adding to thresholds that surpass the capacity of a cell to sequester salts into vacuoles (Munns, 1993, 2005; Läuchli and Grattan, 2007). Na+ reaches cells quite quickly just after the occurrence of salt stress. The increased sodium in this water might influence its salinity adaptation. Therefore, the crucial mechanism of mitogen activates protein kinase and Ca signaling under stress conditions are discussed in the next section.
The function of MAPK relies on post translational phosphorylation signaling, established by a serine/threonine kinase, i.e., mitogen-activated protein kinase kinase kinase (MAPKKK or MAP3K) that reversibly phosphorylates MAPKK (a dual-specificity kinase), then that phosphorylates MAPKs (Keshet and Seger, 2010; Smékalová et al., 2014). MAPKs are involved in phosphorylation of transcription factors, cytoskeleton-associated protein, and protein kinase in plants (Nakagami et al., 2005). Recently, various researchers established that root growth initiation was due to the activity of Ca2+ channels and production of auxin which boost the accumulation of NO. This NO is culpable for the modulation in Ca2+ channel movement and MAPK cascade enzyme activities. ROS molecules, which form as metabolic by-products under stressed conditions, can also induce the activation of MAPKs (Nakagami et al., 2005).
Plant hormones act as an impressive signaling molecule under both normal and stress conditions. MAPK cascades also respond to various hormone signaling like auxin, SA, JA, brassinosteroids (BRs), strigolactones, ABA, and ET. These signaling molecules accomplish a distinct signaling network, which crosstalk to each other and respond under normal and stress conditions (Devoto and Turner, 2003; Depuydt and Hardtke, 2011; Chini et al., 2016). Treatment with natural and synthetic auxin triggers the prompt actuation of MAPKs in the roots of Arabidopsis (Mockaitis and Howell, 2000). ABA signaling has a massive role in the plant growth process that deals with the turgor and stomatal activity of plant cells. MPK4, MPK9, MPK12, and MPK15 proteins exist in guard cells (Zhao et al., 2008), which exhibit the decisive role in ABA signaling and are possibly associated with the activation of the ABA-dependent anion channel (Jammes et al., 2009). Under stress conditions, ABA induces the production of H2O2 and the expression of catalase (CAT) isoform CAT1. This expression is mediated by Arabidopsis MAP2K, in response to H2O2 (Xing et al., 2008). H2S is considered an endogenous gaseous transmitter that exhibits a specific role in the germination of seed, root growth, stomatal activity, photosynthesis, and abscission of plant organs under normal as well as stressed conditions (Corpas and Palma, 2020). H2S interacts with other signal molecules such as ABA, ethylene, auxin, Ca2+, CO, and NO, and controls post transitional modification of proteins (Hancock and Whiteman, 2016; Xuan et al., 2020). ABA induced H2S accumulation via activating SnRK2.6 activities at Cys131 and Cys137 by S-sulfhydration of SnRK2.6 that enhances the interaction of SnRK2.6 with ABA responsive element-binding factor ABF2 (Chen S. et al., 2020). NO is involved in ABA and ethylene crosstalk (Domingos et al., 2015). Indeed, NO was produced during the initial phase of seed germination and promoted seedling growth by inducing ABA 8′-hydroxylase gene expression and ethylene production. Ethylene protects the Brassicaceae seed from the inhibitory effect of ABA by stimulating weakening and rupturing seed testa and endosperm (Arc et al., 2013). NO donors inhibit ethylene biosynthesis and prevent the dormancy of seeds and stimulate germination in apples (Gniazdowska et al., 2007). Breaking of apple seed dormancy by NO encourages ROS production, which stimulates ethylene accumulation due to an increase in ACS and ACO activity (Gniazdowska et al., 2010). EREBPs, which are described as transcriptional factors induced by NO, and ethylene stimulate EREBP-3 just before the rupturing of endosperm during tobacco seed germination, which is inhibited by ABA (Leubner-Metzger et al., 1998). H2S effectively alleviated ethylene-mediated fruit softening in Kiwi fruits and enhanced the ascorbic acid, starch, sugar protein, and titratable acidity (Li T. T. et al., 2017). Combined treatment of H2S-ET inhibited ET synthesis and its related genes such as ACS6, ACO1, ACO4, ERF1, and ETR4, thus suppressed ET induced petiole abscission in tomatoes (Liu D. et al., 2020). Treatment with a higher level of NaHS inhibited primary root growth, initiated by ROS and NO accumulation and activation of the MPK6 gene (Zhang et al., 2017), which denoted that ROS-MPK6-NO cascading intermediates have repressive impacts of high concentration of H2S on root activity (Zhang et al., 2017). H2S strengthens the plant capacity to heat and aluminum (Al) tolerance by reducing oxidative damage after interaction with NO (Sun et al., 2016). Both H2S and NO interactions improved the survival rate of plants under heat stress conditions, due to a decrease in malondialdehyde (MDA) accumulation and enhanced antioxidant capacity in maize and strawberry (Uchida et al., 2002; Christou et al., 2014; Li J. et al., 2014; Li Z. G. et al., 2014).
Under salinity stress, plants depict two forms at the same time including osmotic and ionic stresses. “Cell apoptosis versus adaptation” is dependent on the timing of two cellular responses: the first prompted by Ca, and the second prompted by oxidative outburst in the apoplast. A delay in the formation and dissipation of a salinity-triggered Ca-dependent signal coupled with ROS activates JA signaling, leading to the death of the cells. In contrast to the same molecular signal, calcium will, when properly timed, activate various adaptive processes including sequestration and extrusion of sodium, and also through ABA signaling. With respect to the perception of external inputs, calcium transients, from a number of extracellular compartments, become cytosolic through transient Ca2+-dependent Ca2+ channels. The “Ca2+-signature/Ca2+-spiking” concept became common when it was defined by Webb et al. (1996). The pattern of calcium signaling is determined by the type and amplitude of the stimuli. The calcium level in the body has an enormous impact on the success level of life forms. Calcium-binding proteins, functioning as calcium receptors, relay the information to be conveyed from Ca signals. An unprecedented rate of sensitivity is accomplished by a group of calcium binding modules, that include ‘calmodulin’ (CaM), ‘calmodulin-like protein (CML) family,’ ‘Ca2+-dependent protein kinases’ (CDPK), Ca2+-binding proteins serving as “Ca2+ sensors,” ‘calcineurin B-like proteins (CBLs),’ “Ca2+-decoders,” and ‘CBL-interacting protein kinases (CIPKs)’ which all together transmit the information embedded within calcium signatures. CaM is conserved regardless of species, while CML, CDPK, and CBL are unique to plants and some prokaryotes (Day et al., 2002; Harper and Harmon, 2005; Batistič and Kudla, 2009). Single-cell systems, including pollen germination, provide an excellent model to unveil the coding mechanism and determinants of “Ca2+-signature.” Induction of calcium transients mainly occurs at the organ level via a single spike. The induction of “Ca2+-signature” is in accordance with (i) Ca2+ in various plasma membrane (PM) and endomembrane (EM) flux channels, (ii) cytosolic Ca2+ rallying (in and out) induced by Ca2+ influx and efflux transporters, respectively (McAinsh and Pittman, 2009; Kudla et al., 2010). Therefore, plant signaling is very complex in nature and numerous signaling compounds regulate the plant processes under normal and stress conditions.
Salt tolerance is very complex in nature and affects various processes in plants. In this regard, signaling compounds such as NO, H2S, H2O2, ROS, and plant growth regulators crosstalk with each other and coordinate numerous plant functions and processes, which are associated with salinity tolerance. The crosstalk of various signaling compounds for salinity tolerance are discussed, followed, and represented in Table 1 and Figure 2.
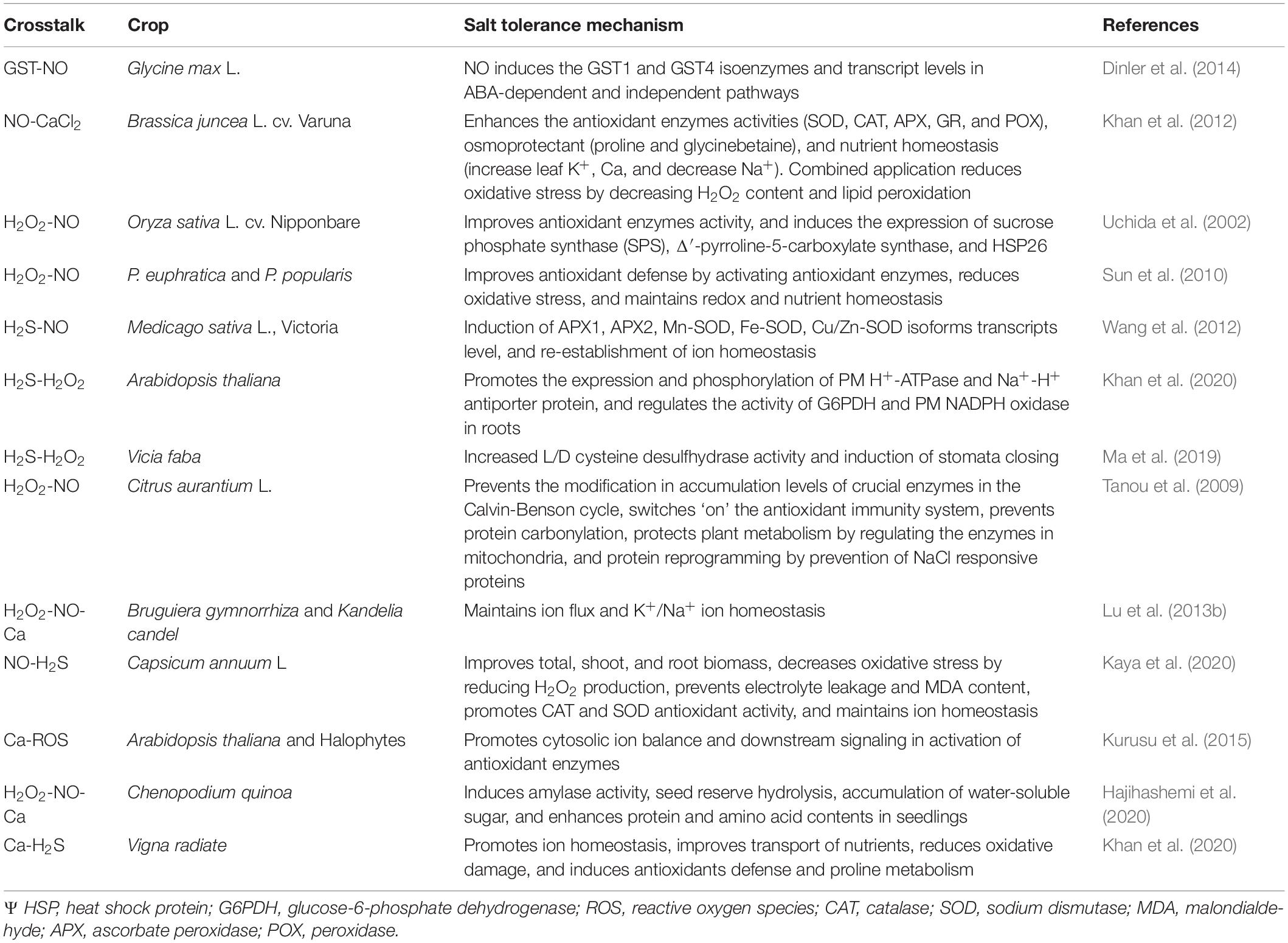
Table 1. The crosstalk of crucial signaling compounds under salinity stress and their salt tolerance mechanism in different crops.
Nitric oxide is the primary gasotransmitter, administering numerous physiological and signaling functions, and also promotes salinity tolerance in plants (Xie et al., 2008). In recent years, it was confirmed that NO crosstalk with other signaling compounds and phytohormone signaling pathways helps in alleviating salinity stress (Tanou et al., 2009; Poór and Tari, 2011; Zhao et al., 2018). Auxin (AUX), ET, and ABA are imperative plant hormones that move from salt-treated roots to leaves that induce synthesis of NO or are transported throughout the plant (Molassiotis et al., 2010). Further, an increase in antioxidant activity and a decrease in thiobarbituric acid, which is reactive material content, is associated with NO-induced alleviation of oxidative damage in saline areas (Xu J. et al., 2011). In cotton, supplying exogenously NO (using SNP sodium nitroprusside for the NO donor) reduces the salt-induced senescence in leaves through downregulating the manifestation of ABA biosynthesis genes such as NCED 9 (9-cis-epoxycarotenoid- dioxygenase) and NCED 2 (Kong et al., 2016). In Arabidopsis, a callus treated with 100 mM of NaCl stimulated NO accumulation that added to ET emission, and resulted in induction of H+-ATPase gene expression in the plasma membrane (PM) (Wang et al., 2009). However, an antagonistic relationship between NO and ET in a suspension culture of tomato cells treated with 100 and 200 mM of NaCl was reported and suggested that an increase in ET synthesis encourages ROS production that is associated with high dead cell ratio, whereas production of NO curtails the dead cell ratio (Poór and Tari, 2011). In the cell suspension culture and segments of apical root, both lack NO and ET-generated (Na+/K+) ionic imbalance, respectively, that leads to an increase in susceptibility toward salinity stress (Poór et al., 2011). Application of SA and SNP, in combination, reduces NaCl-induced toxicity by supplementing the accumulation of proline and stimulation of GPX (glutathione peroxidase), CAT (catalase), and APX (ascorbate peroxidase) in soybean seedlings (Simaei et al., 2011). SA interaction along with signaling flow of NO alters the photosynthetic capacity along with diminished accumulation of H2O2, which enhances the influx of H+-ATPase into PM. The collaborative effect of both SA and NO promotes the absorption of Ca2+/Mg2+ with decreased Na+ uptake in saline conditions (Dong et al., 2015).
Among metabolites, sulfur is the main constituent present in reduced glutathione (GSH), methionine, coenzyme A, cysteine (Cys), iron–sulfur (Fe–S), thioredoxin, and sulfo-lipid systems associated with regulating the physiological process in salt stress environments (Khan et al., 2013). Further, NO enhances S-assimilation which is linked with ET synthesis through cysteine production. Sulfur and NO interact to regulate ABA and ET level in the guard cell and regulate photosynthetic and stomatal activities under salt conditions (Fatma et al., 2016). NO acts as a crucial regulatory signal, which activates various biochemical activities and their interaction with the sulfhydryl and nitro class during nitration enhances tolerance against salinity (Leterrier et al., 2011). NO along with other signaling compounds like H2S helps in building tolerance toward salinity stress in plants. It is observed that exogenous application of NO under saline conditions alters proline (Pro) metabolism and enhances the ratio of free proline accumulation that maintains the turgor potential and protects cucumber seedlings from salinity (Fan et al., 2012). In mustard, CaCl2 and/or SNP application alleviates salt stress by influencing antioxidant enzyme activities along with promoting glycinebetaine (gb) and proline (pro) accumulation, which is associated with a decrease in H2O2, TBARS (thiobarbituric acid reactive substances), and electrolyte leakage (Khan et al., 2012). In Lactuca sativa, application of NaCl triggers osmotic, oxidative, and ionic stress that arise into hormonal imbalances and reduced growth of the plant. Exogenous NO application results in reduction of Na+ accumulation, balancing the concentration of mineral nutrient, which is associated with balanced photosynthetic rate along with the established growth (Campos et al., 2019). Through NO signaling, phytohormone balance leads to osmotic regulation and also activates the antioxidant system with subsequent increase in tolerance level against salinity. The corm of Crocus sativus treated with NO shows more growth under salt-stress and promotes biosynthesis of the secondary metabolites, deposition of compatible solutes, and accelerates antioxidative enzyme activity, whereas treatment with SA did not boost plant growth during salinity (Babaei et al., 2021). Under saline conditions, NO-releasing substances and melatonin application counteracted inhibition of NaCl-treated seedling growth in addition to redox and ion homeostasis which is proved by retardation of ROS overproduction, Na+/K+ ratio, and reduction in the synthesis of TBARS. Consequently, increased level of NO augments addition of melatonin in seedling roots under salt stress (Zhao et al., 2018). Therefore, NO acts as an important signaling network with different signaling factors in plant systems under salinity stress.
Hydrogen sulfide is a signaling molecule, which plays a major role in adventitious rooting, postharvest senescence, and seed germination (Deng et al., 2020), and provides a protective response toward multiple abiotic and biotic stresses (Corpas, 2019). Salt tolerance is enhanced through H2S by increased soluble protein content and chlorophyll under saline conditions but also inhibits ROS accumulation (Mostofa et al., 2015a). H2S donors which are identified/synthesized include CaS2, morpholin-4-ium 4-methoxyphenyl (morpholino) phosphinodithioate (GYY4137), sodium hydrosulfide (NaHS), NOSH-aspirin, dialkyldithiophosphate (ZDDP), AP39, and diallyl trisulfide (DATS). NOSH-aspirin releases two gasotransmitters simultaneously, H2S and NO (Kodela et al., 2012).
In plants, ROS levels are regulated by two ways, by scavenging excess ROS through antioxidant substances (like glutathione and ascorbate) and antioxidant enzymes linked with the AsA–GSH (ascorbate–glutathione) cycle. Accumulation of ROS is reduced by external application of H2S because H2S promotes antioxidant enzyme activities like SOD and CAT in Chinese cabbage (Zhang et al., 2015). From this we can assume that activity of antioxidant enzymes may be controlled by H2S through their protein expressions, thus decreasing accumulation of ROS due to Al toxicity. It can also maintain membrane integrity and ROS homeostasis by controlling the antioxidant mechanism (AsA-GSH cycle and enzymes), therefore enhancing the tolerance level in plants toward salinity stress. In recent studies, by maintaining Na+/K+ homeostasis, H2S helps in enhancing plant salt tolerance level. Under salinity conditions, the content of cellular Na+ mainly increased whereas K+ content reduced in rice, which shows a hike in Na+/K+ ratio in leaves and roots of rice. Therefore, exogenous H2S application maintains Na+/K+ homeostasis in saline conditions in rice (Mostofa et al., 2015b).
Along with NO as a molecule for signaling, H2S is also included in stress responses as they exist in a synergistic relationship (Hancock and Whiteman, 2016). The upstream and downstream relationship between H2S and NO focuses on two factors: NO involvement in the stress tolerance enhances H2S in plants and NO-induced stress tolerance where H2S acts as a downstream signal molecule. Nitrosothiol is a new compound produced when H2S reacts with NO and also results in a decrease of both the compound levels (Huang et al., 2020). In Arabidopsis roots, treatment with 100 mM of NaCl causes electrolyte discharge and also disturbs the Na+/K+ ratio but post treatment with NaHS promoted tolerance toward salt in roots (Li et al., 2016). Production of H2S in alfalfa plants boosted the tolerance level against salinity in the germination stage of seeds which may be induced through oxidative damage (Wang et al., 2012). H2S and NO modify the activity of many antioxidant enzymes like CAT, APX, and SOD to scavenge ROS which deceases their accumulation, thus helping in enhancing the tolerance level of plants toward higher salinity conditions (da-Silva et al., 2018).
Hydrogen sulfide interaction with various phytohormones such as ET, MT, ABA, and SA plays a fundamental role in response to abiotic stress (Huang et al., 2020). Gene expression related to ABA metabolism is increased by the exogenous H2S along with an upregulation of ABA receptor expression levels in roots of wheat during stress conditions, which indicates that the stress tolerance of wheat is promoted by H2S through the involvement of the ABA signaling pathway (Ma et al., 2016). When production of NO is restricted, ET is reduced which enhances the level of H2S. A study on Vicia faba L. showed that H2S biosynthesis inhibitors do not block the stomatal closure which is ET-induced with NO accumulation (Liu et al., 2016). Through the activation of endogenously synthesized H2S, SA enhances the tolerance level of plants toward several other abiotic stresses. Crosstalk of NO and H2S plays a crucial role in promoting the tolerance level against abiotic stresses. H2S not only regulates the uptake along with transport of metal ions and maintains Na+/K+ homeostasis during salinity but also shows an interaction with different signaling molecules such as Cys, phytohormones, and NO for enhancing the plant tolerance toward salinity stress.
Hydrogen peroxide is synthesized in a form of ROS and as a result of oxidative stress that cause damage due to excess accumulation of ROS under different stress conditions, which results in the death of cells (Fotopoulos et al., 2006). H2O2 production is induced in plants following exposure to a different range of environmental stimuli. Further, it was found that H2O2 acts as a signal to determine different physiological, biochemical, and molecular responses within plants and cells and their crosstalk in between other signaling pathways (Neill et al., 2002). NO and H2O2 signaling pathways are coordinated and tightly linked to different plant responses toward the environmental stimuli (Molassiotis and Fotopoulos, 2011). Alteration in production of both NO and H2O2 is considered under these plant responses toward salt stress (Zhang et al., 2006). Further, it was reported that expression of the AtNOA1 gene in Arabidopsis was suppressed by NaCl treatment which leads to a reduced NO level (Zhao M. G. et al., 2007). In contrast, expression of OsNOA1, the rice homolog of AtNOA1, was moderately enhanced due to salinity (Qiao et al., 2009). By pre-treating wheat seeds with H2O2, the salt tolerance level of the seedlings was improved (Wahid et al., 2007). Apart from the biosynthetic level, H2O2 and NO crosstalk are also included in protein activity and regulation of gene expression (Qiao et al., 2014). In Bermuda grass, NO and H2O2 regulate induction of CAT and SOD activity through ABA (Lu et al., 2009). From further studies, it has been observed that both ABA-dependent NO and H2O2 influenced the activity of antioxidant enzymes and transcription under salt tolerance (Zhang et al., 2009). Consequently, SA enhances endogenous H2O2 levels significantly through regulating activation of SOD. Therefore, SA and H2O2 work together in a self-amplifying process (Rao et al., 1997). The high concentration of H2O2 causes oxidative/nitrosative stress, while in low concentrations H2O2 acts as a signaling molecule to regulate stress responses.
Calcium is an important signaling molecule, and secondary messenger crosstalk with several other signaling compounds help in the mitigation of salinity stress. Several studies have shown that flavonoids and phenylalanine ammonia lyase (PAL) activity are elevated after adding calcium chloride or an ionophore to the nutrient medium of a plant species (Paranhos, 2014). Ca2+-mediated modulation for the development of the specialized metabolites is collectively induced by JA(Lee-Parsons and Ertürk, 2005), ABA (Vighi et al., 2019), and SA (Guo H. et al., 2015). The effects of MT and Ca, and how they affect the development of phenolic compounds (PCs), were investigated in the plant Dracaena kotschyi under stress conditions of salt water and compared with control. Vafadar et al. (2020b) reported that external NaCl application (mimicking salinity stress) reduces dry biomass of shoots but elevates H2O2 content, electrolyte leakage (EL) level, and 2,2-diphenyl-1-picrylhydrazyl (DPPH) scavenging ability, and upregulates gene expression of PAL, RAS, and TAL enzymes. Vafadar et al. (2020a) found that pre-treatment of D. kotschyi with a Mel biosynthetic pathway inhibitor has no effect on Ca2+-mediated production of PCs in salt-affected plants. On the contrary, pre-treatment of D. kotschyi with a plasma membrane channel blocker, Ca2+ chelator, and calmodulin (CaM) antagonist resulted in impairment of Mel effects under salinity stress. This suggests that biosynthetic triggering of phenolics is attributed to Mel application only when influx of Ca2+ (carry out Ca2+/CaM signaling) ions are there (Vafadar et al., 2020a,b,c).
Under stress conditions, H2O2 and NO interact with Ca2+ ions forming an intricate signaling web to withstand salinity (Hasanuzzaman et al., 2018). Under salt stress conditions, it was observed that pre-treated quinoa seeds with an NO donor (sodium nitroprusside; SNP), a reactive oxygen species (H2O2), and CaCl2 showed a significant positive linear correlation with germination rate (GR) and germination index (GI), whereas a reversed linear correlation occurred between them with mean germination time (MGT) (Hajihashemi et al., 2020). The pretreatment enabled seed germination and rapid seedling establishment in the salt-affected soil. Pre-treating with NaCl completely prevented the decrease in the activities of alpha amylase and beta amylase. Salinity stress decreases seed germination by inhibiting the main enzymes, α- and β-amylase, which hydrolyze starch during germination, and pretreatment lowered this negative effect of salinity on these enzymes. Studies indicate that exposure to molecules, such as H2O2 or CaCl2, may reduce the adverse effect of environmental stress on amylase activity and restore normal germination (Zheng et al., 2009; Li et al., 2013; Li Z. et al., 2017; Bouallègue et al., 2017). The most significant finding from the study conducted by Hajihashemi et al. (2020) was that the presence of NO, H2O2, and Ca2+ resulted in enhanced amylase activity. The rise in starch degradation increases germination, and more seeds sprout (Li et al., 2013). This connection could alleviate the negative impact of salt stress on quinoa germination.
Polyamines (PAs), like putrescine, spermidine, and spermine, are well regarded besides their substantial plant developmental processes and adaptation toward environmental cues (Pathak et al., 2014). It has been established that PAs play a vital role in a variety of cellular pathways including programmed cell death. Expression levels of PA biosynthesis-related genes were shown to be modulated under stress (Gupta et al., 2013; Shi and Chan, 2014). The modulatory response of arginine decarboxylase (ADC) (regulatory enzyme of PA biosynthesis) to salinity is a key regulator of the adaptive response in plants (Liu et al., 2006). Transcriptomics of PA biosynthetic genes in different varieties of rice revealed that the ADC gene is induced under salinity stress (Do et al., 2014). Studies indicated that perhaps the PA metabolic pathway is in intricate crosstalk with other signaling pathways including ABA, H2O2, and gamma-aminobutyrate (GABA) (Marco et al., 2011; Seifikalhor et al., 2019). Within that direction, Kalhor et al. (2018) have demonstrated that GABA helps increase salinity tolerance in lettuce. PA metabolism further induces NO output, which has been linked to several other stress intermediaries like Ca2+ ions and protein kinases. Under salinity conditions, PAs can engage ion channel proteins thereby affecting their conductivity (Zhao F. et al., 2007; Velarde-Buendía et al., 2012). Garufi et al. (2007) proposed that PAs perform through regulating the activity of multiple ion channels indirectly by enhancing interactions with “14–3–3 proteins” (a family of highly conserved regulatory molecules). In stressful conditions, it is possible that high levels of cytoplasmic Ca2+ can be deleterious to standard cellular metabolism. Since active Ca2+ efflux networks play a pivotal role in sustaining cell Ca2+ contents, PAs activate Ca2+ efflux mechanisms, such as the PM channel and the membrane Ca2+-ATPase, while maintaining steady plasma Ca2+ levels (Pottosin et al., 2012, 2014; Pottosin and Shabala, 2014). In this regard, PAs metabolism is related to Ca2+ signaling indirectly, even though underlying regulatory mechanisms remain unclear.
Several studies indicate the control of NO metabolism during salt tolerance (Wimalasekera et al., 2011; Ahmad et al., 2016). As a major NO production pathway, NO synthase (NOS) enzyme leads to most of the NO production in animals (Santolini et al., 2017) and plants (Moreau et al., 2008). When realizing that NO is a molecule linked to PAs via the common precursor of 1-arginine, it could be probable that PAs like spermidine and spermine produce NO in plants. The role of NO in signaling may be influenced by mobilization of intracellular calcium or interaction with calcium channels which ultimately elicit Ca2+ signaling (Courtois et al., 2008). These underlying mechanisms of Ca2+ and NO signaling affect each other. NO synthesis mediated by NOS operates via Ca2+ and CaM signaling (Corpas et al., 2004, 2006). Lamotte et al. (2006) found that NO plays a massive part in the activation of plasma membrane channels and the subsequent release of Ca2+ under salt stress recovery.
The ‘salt overly sensitive’ (SOS) pathway could potentially play an important role in the membrane conductance of the root epidermal cell of plants to the extracellular acidic environment, thereby helping to detoxify this ion in the root epidermal cells and thereby extruding this ion form the root epidermal cells (Quintero et al., 2002; Martínez-Atienza et al., 2007). The “salt overly sensitive” (SOS) pathway forms a critical pillar for preserving ion homeostasis when exposed to salinity stress (Ji et al., 2013). Nevertheless, during salinity, sustained accumulation and scavenging of ROS serve mostly as distress indicators, whereas redox homeostasis as well as antioxidant signaling at the cellular levels are engaged in stress sensing and tolerance. However, high levels of ROS accumulation can impair essential plant metabolic processes and development (Gill and Tuteja, 2010; Bose et al., 2014; Jajic et al., 2015). Electrons released by oxidants serve as a signaling cue in the cell to alert the plant to stress adaptation (Mittler et al., 2011). Salinity stress also causes ROS-mediated damage to lipids and proteins, and contributes to programmed cell death (Poór et al., 2012). Yet, ROS and calcium (Ca2+) are considered to be effective intracellular signals (Gilroy et al., 2014). In response to a high salinity environment, cytosolic calcium increases, which stimulates calcium sensors and calcium signaling pathways (Boudsocq and Sheen, 2009). However, plants also employ ion transport control (e.g., sodium and potassium accumulation), compatible solute aggregation, and expression of genes under salt stress (Kurusu et al., 2015).
Reactive oxygen species are continually formed in plant organelles as inevitable byproducts of metabolic activities (Apel and Hirt, 2004; Abogadallah, 2010). Although, the increased oxidative stress that would lead to cell death through apoptosis is preventable and can be counteracted with antioxidants, it would also be a point of differentiation. It is clearly right, as many plants often undergo necroptosis, including those that do not typically reside in a stressful climate (Coll et al., 2011). In comparison, singlet oxygen is utilized as a substrate of lipoxygenases activating a metabolic cascade that will produce a further essential stress signal, JA (Farmer and Mueller, 2013). Furthermore, ABA synthesis is also triggered by ROS (Xiong and Zhu, 2003). Plant species under salinity or drought stress will close their stomata and thus reduce moisture loss and their CO2 influx (Hsu and Kao, 2003). Subsequently, carbon reduction and photosynthetic NADPH utilization by the Calvin cycle decreases, resulting in the development of electron holes in photosystem I that leads to electron leakage to O2 (Türkan and Demiral, 2009).
Peroxisomal glycolate oxidase is a major source of ROS that is involved both in basal conditions and stress conditions (Mittler, 2002). ROS quenching can also be accomplished by other signaling molecules such as NO. NO nullifies Fenton-type oxidative stress by scrounging superoxide thus inhibiting the development of oxidizing agents that leads to recovery of redox homeostasis (Lamattina et al., 2003). As an alternate regulator of NO and GSH, H2S, recently identified as a signaling molecule in crops, enhances GSH concentrations, affects enzymatic activity, and communicates with NO (Paul and Snyder, 2012; Lisjak et al., 2013). The NO formed by the phytohormone will serve as a crosstalk intermediary between the two signaling pathways. This signal attenuation is comprehensive, since even ROS derived from NADPH oxidase activity in the plasma membrane are vital to trigger ABA signaling (Kwak et al., 2003).
Reactive oxygen species aggregation throughout stress conditions often depends on the capabilities of ROS recycling through the scavenging system. The buildup of different reactive oxygen forms gradually contributes to the adaptability to stress factors and culminates in apoptosis. SA administered through the root system resulted in a raised H2O2 concentration in young leaf and root tissue which caused plant death (Gémes et al., 2011). Szepesi et al. (2009) observed that even under salinity, plants pre-treated with SA contained even more Na+ than controls. The leaves did not exhibit any signs of salt damage, and rather, their photo- and membrane-integrity remained intact. As intracellular ROS is hugely involved in salt responses and active oxygen radical production within the cell is sometimes correlated with abiotic stress, it was of concern whether intracellular ROS and NO could make a significant contribution to the loss of vitality of distressed leaf protoplasts.
Reactive oxygen species-activated calcium-permeable channels including “respiratory burst oxidase homolog” (Rboh) are being hypothesized to entail an optimistic feedback loop that activates calcium ion and active radical signals in root cells (Takeda et al., 2008). RbohC/RHD2 has an affect on the production of ROS and controls Arabidopsis root hair elongation in a Ca2+-dependent fashion (Takeda et al., 2008; Monshausen et al., 2009). Salinity-induced (Ca2+) cyt is enhanced through hyperpolarization- and depolarization-activated PM Ca2+-permeable channels (Tracy et al., 2008). Salinity-induced escalation of cytosolic Ca2+ entry plays an important role in ROS signaling and saline tolerance (Shabala and Newman, 2000; Kader and Lindberg, 2010). Polyamines induced by salinity can produce oxygen-derived free radicals as substrates for apoplastic ROS enzymes (Kärkönen and Kuchitsu, 2015). Both OH and polyamines may significantly change the cationic and anionic conducting pathways and affect Ca2+ signaling in plants (Pottosin et al., 2014). It can unwrap a novel feature of ROS development during salinity adaptation and/or the acclimation process. A notable research paper demonstrates that perhaps the recruitment of Ca2+/CaM-like proteins (CDPKs) are far more pronounced in halophytes compared to glycophytes which experiences salt stress (Xu P. et al., 2013). Such calcium-binding proteins can play a pivotal role as critical amplifiers of initial calcium influx under salt stress. Ca2+ signaling is also very important in seawater stress’s impact on signaling mechanisms. It is suggested that TPC1 is engaged in the dissemination of salt stress cues and seems also to participate in the plant defense response (Choi et al., 2014). Inflammation triggered by Ca2+-ROS pulses concerning TPC1 can be one of the plant stresses tolerating mechanisms (Choi et al., 2014; Gilroy et al., 2014). Furthermore, NO has been demonstrated to have potent antioxidant activity; preventing and minimizing lipid peroxidation and protein oxidation (Fancy et al., 2017). Saline conditions enhance NO output that minimizes salinity-induced damages (Fatma et al., 2016; Da Silva et al., 2017). At about the same time, numerous sources claim a preventive action for NO in salt-stress tolerance due to upregulating antioxidant activity in various native plants (Zheng et al., 2009; Sheokand et al., 2010).
The productivity of agricultural crops has continuously and adversely been affected by soil salinity. A plant’s ability to tolerate salinity stress can be augmented by application of signaling molecules such as H2S, NO, and H2O2. For example, exogenous application of NO decreases the accumulation of Na+ and stabilizes the concentration of mineral nutrients, and thereby results in a balanced photosynthetic rate and re-establishment of vegetative growth in lettuce (Lactuca sativa). Osmotic regulation, activation of the antioxidant system, and balanced phytohormones help to increase plant tolerance against salinity stress (Campos et al., 2019). Salinity stress adversely affects plant growth through upregulation of proline, MDA, and ABA content while downregulating K+/Na+ ratio and electrolyte leakage. Exogenous application of sodium nitroprusside (SNP) and 24-epibrassinolide (EBL) in combined form increases the endogenous level of ABA in Indian mustard (Brassica juncea var, Varuna) through proline, nitrogen, and ABA metabolism (Gupta et al., 2017). Soil salinity has a detrimental effect on agricultural crops through water deficiency and modified K+/Na+. This process leads to altered cellular redox pathways by producing reactive oxygen species such as H2O2, superoxide (O2–), and hydroxyl radicles (•OH). These free radicals pose a severe toxic impact on a molecular, biochemical, physiological, and cellular level via the lipid peroxidation pathway and lead to protein and nucleic acid destruction, and thereby negatively affect several vital pathways such as gaseous exchange, plant growth and development, and proline and nitrogen metabolism (Siddiqui et al., 2012; Gupta et al., 2017). The crosstalk of PGRs with other signaling compounds under salinity stress tolerance are illustrated in Table 2. NO metabolism regulates several biochemical pathways of ABA homeostasis in plants such as seed germination, dormancy, leaf senescence, stomatal movement, and fruit ripening in normal and stress conditions. The signaling molecule NO induces post translational modifications such as tyrosine nitration and sulfur-nitrosylation of proteins which regulate ABA signaling pathways. NO modulates antioxidant systems such as SODs and the catalase and ascorbate GSH cycle, and also affect ABA-induced reactive oxygen species production (Prakash et al., 2019). Phytohormones play a critical role in plants to adapt them to an unfavorable environment such as salinity via modulating physiological responses. Both phytohormone-ethylene and GAs play crucial roles to mitigate salinity stress by activating defense regulatory genes or increasing plant growth. However, both ethylene and GA are interconnected with each other; GA is well known to increase ethylene synthesis while its signaling is also dependent on ethylene (Iqbal et al., 2012). Transcript-based meta-analysis studies have illustrated that both ethylene and GA metabolism-related genes are expressed in plants under salinity stress. The precursor ACC may be synthesized unanimously for ethylene and GA synthesis. Exogenous application of ethephon and GA3 reduces the adverse effect of salinity on seed germination of Amaranthus caudatus (Bialecka and Kepczynski, 2009). Ethephon showed a more stimulatory effect on seed germination than GA3 under soil salinity. Foo et al. (2006) studied the interaction effect of ethylene and GA synthesis in pea and revealed that ethylene synthesis was negatively controlled by phytohormones and also tended to reduce GA production. Similarly, ethylene and GA have a positive effect on hypocotyl elongation in Arabidopsis (De Grauwe et al., 2007). It has been reported that GA alone is ineffective but acts synergistically with ethylene and promotes the number of penetrating roots and growth rate of emerged roots (Steffens et al., 2006). Their effect is not additive in nature, but both are synergistic with each other.
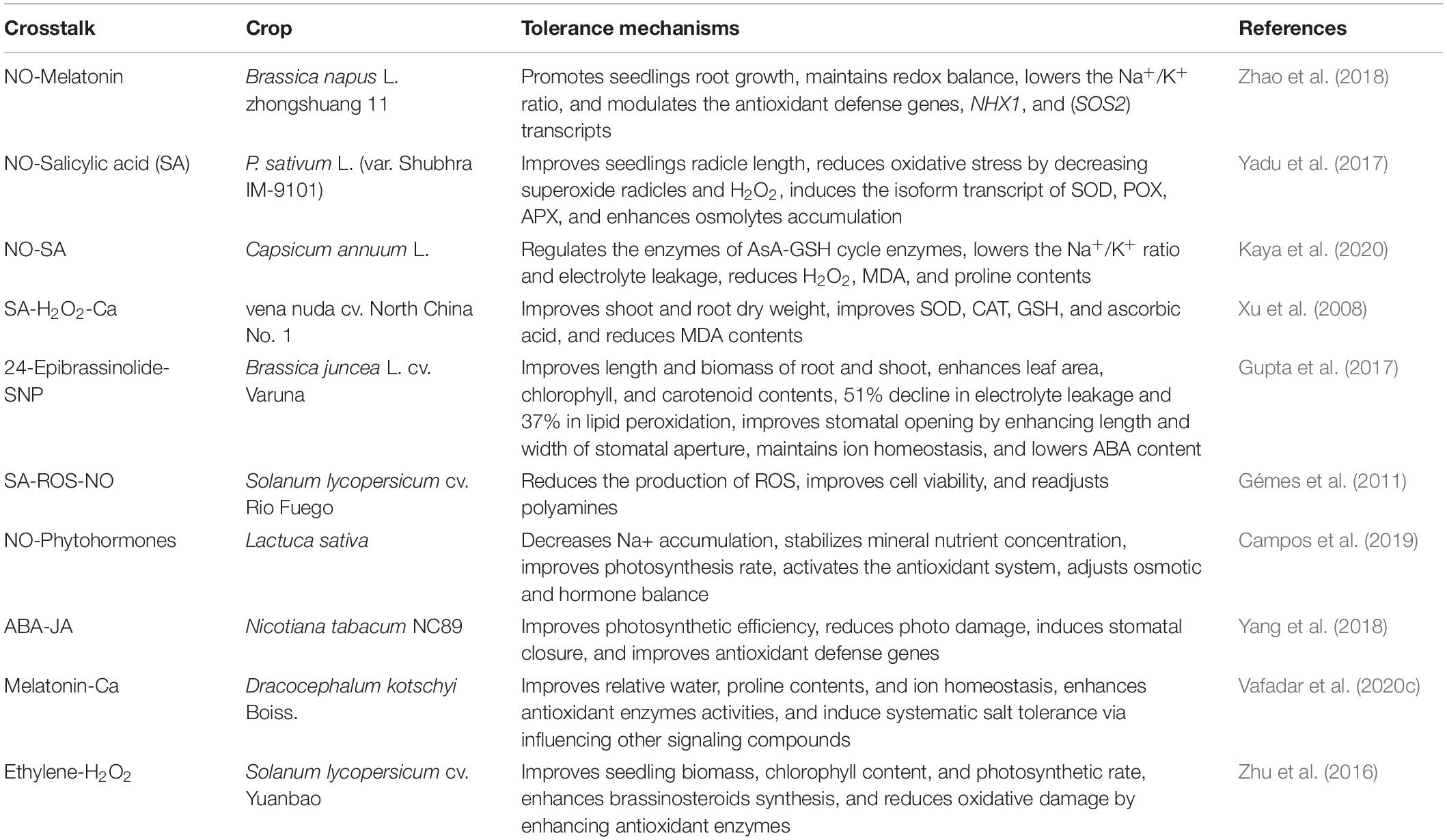
Table 2. The crosstalk of plant growth regulators with crucial signaling compounds under salinity stress and their salt tolerance mechanism in different crops.
Seed germination is affected by both ethylene and NO under salinity in plants. Exogenous application of ACC (a precursor of ethylene biosynthesis) or SNP (an NO donor) inhibits the negative impact of salinity on seed germination of Arabidopsis (Lin et al., 2013). However, the stimulatory effect of both ACC and SNP was reduced by the inhibitor of ethylene biosynthesis, i.e., aminoisobutyric acid (AIB) or NO scavenger compound, i.e., 2-phenyl-4,4,5,5-tetramethyl-imidazoline-1-oxyl-3-oxide (cPTIO) and indicated the interaction effect of both NO and ethylene on seed germination under salinity. Moreover, NO production was increased by ACC and overexpression of the ACS2 gene was noticed by SNP which is directly involved in ethylene biosynthesis. Thus, this indicates the importance of both in their production under salinity stress. Interestingly, exogenous application of ACC increased seed germination under oxidative stress induced by H2O2. However, NO-treated wild-type Arabidopsis plants were less affected and no effect was observed on ethylene-insensitive mutant seeds.
The major influences on plant growth limitation in salt-affected soil are due to osmotic stress and ion toxicity (Munns and Tester, 2008; Bargaz et al., 2016). Plants adapt distinct strategies (molecular, biochemical, and physiological adjustments) to modulate metabolic pathways and at the same time, to combat cellular salt levels via regulating water and ion homeostasis. In this regard, the modern integrative “omics” approach in plant biology has taken momentum over the last two decades in the research area powered by advances in platforms for nucleic acid sequence, peptide sequencing platforms, mass spectrometry (MS), efficient computational skills, and data analysis methodologies. This integrated “omics” system offers a snapshot of cells, tissues or organisms’ developments, functions, and relationships by characterizing and quantifying all their biomolecules using a high-performance approach (Soda et al., 2015; Mosa et al., 2017; Parida et al., 2018). In recent years, omics technologies have shown promising results and been tested in numerous crops, which are highlighted in Table 3. Among them the application of some omics approaches under salinity stress tolerance are represented in Figure 3 and discussed below.
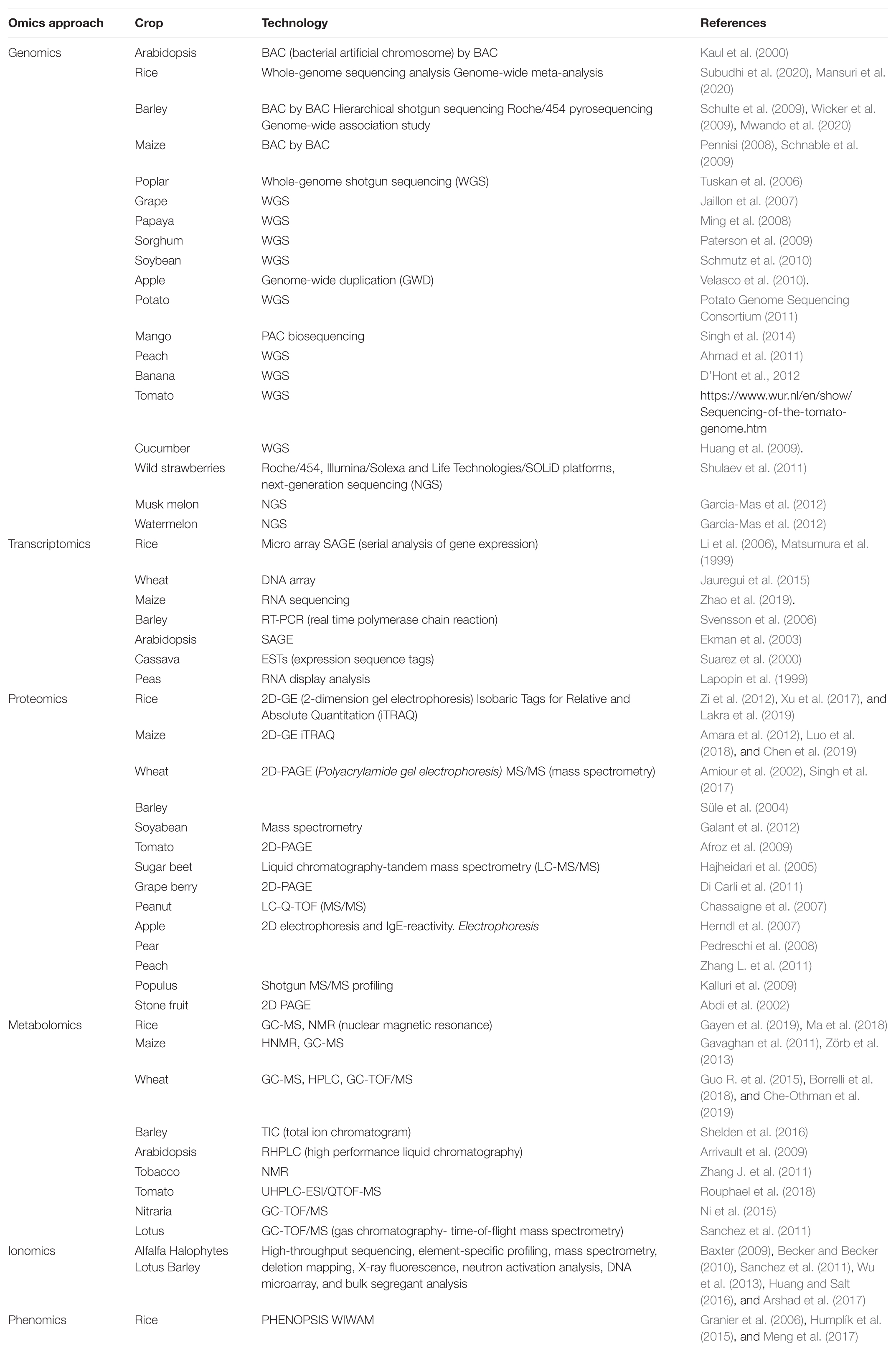
Table 3. The crucial genomics, transcriptomics, metabolomics, and proteomics approaches used in different crops.
It is very crucial to understand the plant genomic response toward environmental stress. Plants are acutely complex and consist of a large number of genes in the reaction to salinity. It has been difficult to absolutely understand how plants react to salinity because of their multi-genetic nature. Genomics has made considerable strides over the past decade and has played a critical role in delivering the information needed to promote crop production. Genomics is a branch of “omics” that studies a certain genome and discloses useful knowledge on the organism’s biology (Gilliham et al., 2017). By genomics, the genes involved in salinity stress response have been identified and characterized; signaling pathways have been mapped and certainly this information can be used for salinity tolerance of existing plants. It is significant to mention that genomics as a tool primarily improves and does not replace current technologies. Several high-throughput approaches, such as forward genetics, candidate gene approach, serial analysis of gene expression (SAGE), expression sequence tag (EST), next generation sequencing (NGS), high resolution melting (HRM), targeting-induced local lesion in genomes (TILLING), RNA interference (RNAi), and genome wide association study (GWAS), etc. have enabled us to not only understand salinity tolerance in plants but have also opened the path for developing plants under salinity stress. Using forward genetics, fundamental genes (SOS1, SOS2, SOS3) of the salt tolerance pathway have been identified (Zhu et al., 1998) and this knowledge of candidate gene approach has been pursued to efficiently identify the SOS gene orthologs in rice (Martínez-Atienza et al., 2007), Poplar (Tang et al., 2010), and tomato (Olías et al., 2009). Eleven single nucleotide polymorphisms (SNPs) were identified in the coding region of five salt-tolerant rice genotypes by exploring the advance method of TILLING, i.e., Eco-TILLING (Negrão et al., 2013). Because of its ability to boost the resolution of quantitative trait loci (QTL) detection without spending additional efforts in population development, GWAS receives greater attention. Recently, three novel QTLs were identified on chromosomes 4, 6, and 7, which are associated with salt tolerance in rice, through the use of molecular breeding approach GWAS (Kumar et al., 2015). Salinity tolerance may also be correlated with six genomic regions in soybean confirmed through SoySNP50K BeadChip iSelect (Huang et al., 2018). Consequently, nine SNP-rich regions were identified in 215 accessions of Asian cotton using the same GWAS approach as those related to plant parameters in salt stress conditions (Dilnur et al., 2019).
The use of RNAi technology in salt tolerances shows the positive control of tocopherol cyclase (Ouyang et al., 2011). New technologies of genomics like zinc finger nuclease (ZFN), transcription activator-like effector nucleases (TALENs), CRISPR-Cas9 (Pennisi, 2013), and Speed Breeding (Li et al., 2018) provides the opportunity to generate precisely engineered crops for salinity tolerance. The latest study of genotyping-by-sequencing in cowpea has been used to discover the effects of salinity tolerance on seed germination and seedling growth (Ravelombola et al., 2018). Nine haplotypes, two salt-tolerant and seven salt-sensitive, were addressed by a new genome sequencing experiment of 31 landraces and 22 wild soybeans (Guan et al., 2014).
Soil salinization is recognized as a major problem for agricultural production and sustainability at a global level. The mechanisms of salinity tolerance are well known to be complicated and governed by polygenic traits (Munns and Tester, 2008). Therefore, improvements in understanding other “omics” beyond genomics have helped assign functional roles to candidate gene(s)/QTL(s) that relate to multiple abiotic stresses including salinity stress in crop plants (Salt et al., 2008). Another branch of “omics” technology is transcriptomics which deals with the RNA expression profile of organisms at temporal and spatial bases (Duque et al., 2013; El-Metwally et al., 2014; Shen et al., 2019). Unlike genomics, the transcriptome is highly complex and dynamic, and changes depending on diverse factors (El-Metwally et al., 2014). Recent shifting of RNA sequencing (RNA-seq) high-performance technology from the microarray accelerated the response of the candidate gene to stress (Liu et al., 2014; Vu et al., 2015; Conesa et al., 2016). The recently developed transcriptomics measure the abundance of transcripts of thousands of genes in parallel. RNA profiling is currently being carried out by means of RNA sequencing, microarray platforms, digital gene expression profiling, and SAGE (Molina et al., 2011; Raney, 2012; Duque et al., 2013; Xu Y. et al., 2013; Leisner et al., 2017; Li P. et al., 2017; Kreszies et al., 2019). This technology improves the ability, in salt stress, to identify transcripts/genes that are essential in controlling transcription and translation machinery (Sahi et al., 2006; Jamil et al., 2011).
To understand the transcriptomic changes during salt stress, RNA sequencing (RNA-Seq) has become the most used method for identification of novel genes and their expression pathways (Hrdlickova et al., 2017). Transcriptome profiling has been extensively and successfully used to analyze salt stress response mechanisms of plants. It is an effective method to find common sets of genes that are differentially expressed between stress-tolerant and sensitive genotypes with diverse genetic backgrounds (Peng et al., 2014). Comparing the difference in transcriptional levels between tolerant and sensitive genotypes under stress conditions, the genes related to stress tolerance can be isolated. A transcriptomics study also revealed different up- and downregulated transcriptional factors such as MYB, MYB-related, AP2-EREBP, NAC, and WRKY (Chen F. et al., 2020). This transcriptional profiling study gives a better insight into the understanding of the key components in the plant salt tolerance network which is important for developing more salt-tolerant plants. NGS and SAGE techniques were employed together by Molina et al. (2008, 2011) to classify the entire chickpea salt transcriptome. Likewise, for Arabidopsis thaliana responses to salt stress, Rasmussen et al. (2013) used large-scale microarray analysis. The comprehensive genome-wide study of common beans was used to recover a total of 155 bHLH (helix loop helix) genes related to salt stress response (Kavas et al., 2016). A research was performed with Solexa/Illumina to investigate the transcriptome expression profiles for Poplar (Populus simonii X Populus nigra) under salinity stress (Chen et al., 2012). Similarly, differential expression levels were checked in salinity stress for WRKY-TF genes (Garg and Singla, 2016). Most currently, miR156 working in reaction to salinity stress in alfalfa was shown by Arshad et al. (2017).
Salt stress is predicted to cause salinization of arable lands within the next 25 years, which may result in 30% land loss and up to 50% by the year 2050. Salinity stress causes different genes to be expressed and the result is reflected in the protein profiles. It could thus be essential to collect whole proteins created by various abiotic stresses, including salt stress, to enhance our knowledge of protein networks associated with salt-interacting pathways (Ji et al., 2016). Recently developed “omics” technologies are being designed in plant sciences to determine key proteins or metabolites that are novel, covering metabolomics, proteomics, and genomics responsible for plants stress tolerance and also biomolecules that regulate the genes. These omics studies give us a better insight into the agents affecting plant growth and development. Proteomics deals unshakably with the identification of proteins, expression profile, post-translational modifications (PTMs), and protein–protein interactions underneath stress and non-stress conditions. Proteomics studies offer a new replacement approach to obtain proteins and pathways related to crop physiological and stress responses. Thus, determining plants at proteomic levels might facilitate the finding of pathways concerned in stress tolerance.
Plant responses to salt stress through the proteomics approach have been studied in both glycophytes and halophytes. Plant scientists have worked with model plants under saline stress at proteomic levels, Razavizadeh et al. (2009) in Nicotiana tabacum, Chen et al. (2012) in Populus cathayana, Chattopadhyay et al. (2011) in grass pea, and Xu C. et al. (2011) in Agrostis stolonifera. Moreover, agricultural plants have also been examined under saline stress in different analyses, e.g., durum wheat (Peng et al., 2009; Jacoby et al., 2010), canola (Bandehagh et al., 2011), sugarbeet (Wakeel et al., 2011), soybean (Sobhanian et al., 2010), peanut (Jain et al., 2006), S. bicolor (Swami et al., 2011; Ngara et al., 2012), maize (Zörb et al., 2010), tomato (Chen et al., 2009; Manaa et al., 2011), potato (Aghaei et al., 2008), and cucumber (Du et al., 2010), etc.
Plant roots exhibit the foremost negative symptoms of salt stress because the few genes that are responsive to salinity stress are induced more in roots than in shoots as evident from the findings of different workers in soybean, rice, wheat, maize, and potato (Hasanuzzaman et al., 2013). A proteomics study in soybean was carried out under salt stress with the use of different tissues. They recognized that 50S ribosome protein was downregulated in leaves and that it was thought to participate in the biosynthesis of soybean protein and cause a decrease in plant growth. A phosphoproteome study of the roots of rice on exposure to NaCl (150 mM) for a few hours by using Pro-Q Diamond stain revealed that 20 proteins were upregulated and 18 downregulated. They positively identified 17 of the 20 upregulated proteins and 11 of the 18 downregulated ones. Proteins related to GST, Hsp70, and mannose binding rice lectin were upregulated, while protein kinase, ATP synthase beta-chain, GALP hydrogenase were downregulated. They believed that phosphorylated proteins could be identified using Pro-Q Diamond stain under saline conditions. Of all proteins, 17 overexpressed proteins were responsive to salinity, however, some other proteins identified did not express in any of the proteomic reports on rice on exposure to salinity (Chitteti and Peng, 2007).
Higher plants have an excellent capacity to synthesize a broad variety of different molecules and play an important role in chemical defenses against biotic and abiotic stress. The synthesis and accumulation of all small molecule’s metabolites (less than 1.5 kDa) is an evolved, conserved, and ubiquitous process that shows immense variety in chemical structure and function known as metabolome, analogous to transcriptome, and proteome. Metabolism is an effective plant physiology method that is closer to phenotype than genes and proteins in response to abiotic stresses and metabolites, quite accurately representing the overall effects of genetic expressions and complex regulatory procedures (Scherling et al., 2010; Arbona et al., 2013; Ramalingam et al., 2015). Metabolites include a number of organic acids, hormones, amino acids, ketones, vitamins, and steroids. In this regard, metabolomics (i.e., the study of the metabolome, or the set of metabolites found in a given plant tissue or organ) play an essential role in procuring metabolic fingerprints or metabolic profiles based on the physiochemical properties of each metabolite using different test instruments and separation technologies (Jogaiah et al., 2013).
Relative to proteome and transcriptomics, this method generates more reliable information (Dos Santos et al., 2017). The illustration of stress tolerance mechanisms and metabolite profiling in plants has been improved by progress in mass spectrometry liquid chromatography or gas chromatography (LC-MS and GC-MS), high performance liquid chromatography nuclear magnetic resonance (NMR), direct injection mass spectrometry (DIMS), and other metabolomics techniques (Wolfender et al., 2013; Parida et al., 2018). Different researchers have noticed that variations in metabolites involving carbohydrate metabolism, tri carboxylic and glycolytic acid, amino acid biosynthesis, and at other protective antioxidant levels under stress are easily analyzed using metabolomics (Kumari et al., 2015; Jiao et al., 2018). A study of Lu et al. (2013a) revealed that a comparative study between soybean wild-type W05 and cultivated soybean C08 indicated abundance at a metabolic level of several compounds in a wild form, such as disaccharides, sugar alcohols, and acetylated amino acids. The increase of tocopherol in maize shoots and the sharp decrease in ascorbic acid levels after salt stress were reported by AbdElgawad et al. (2016). Wang et al. (2015) also stated that in Kosteletzkya virginica seedlings, proline levels increased when introduced to high salinity. In a study by Shen al. (2016), a decrease in the levels of glycolysis pathway-associated sugars occurred in barley in response to salt stress. A review of metabolomics comparison reported by Jiao et al. (2018) in common wild-type soybean W1 and W2 salinity-tolerant wild-type soybean revealed increased accumulation of various organic acids, TCA cycle metabolites, and various amino acids, which in turn gave W2 greater tolerance to salinity than W1. The study of metabolome data from foxtail millet roots showed in this research that 17 associated genes of flavonoid biosynthesis were significantly raised 2- to 11-fold under salinity in Yugu 2 (Pan et al., 2020). Salinity stress-specific metabolites could therefore serve as biomarkers to evaluate a salt-tolerant and sensitive genotype.
The “ionome” is said to be the mineral and elemental collection of an organism (Salt et al., 2008). A landmark in ionomics research was identified in A. thaliana (Hirschi, 2003; Rea, 2003) where more than a thousand plants were analyzed for ionomic mutants. Ionomic circuits in crops are orchestrated and require critical reviews for high efficiency elemental profiling (Salt et al., 2008; Baxter, 2009). High-throughput sequencing, element-specific profiling, mass spectrometry, deletion mapping, X-ray fluorescence, neutron activation analysis, DNA microarray, bulk segregant analysis, and various reverse genetic tools confirmed the involvement of multiple regulators that regulate the ionome (Baxter, 2009; Becker and Becker, 2010; Sanchez et al., 2011; Wu et al., 2013; Huang and Salt, 2016). This has provided another possible avenue for exploration for plant-based genetic engineering for stress tolerance. Ionomics could lead to better management of root mineral nutrients status in plants (Shelden and Roessner, 2013). Ionomics has been deciphering the key elucidation toward ion homeostasis and ion detoxification in response to salinity stress in crops (Sanchez et al., 2011; Wu et al., 2013). The research findings indicate altered expression of calcium, magnesium, manganese, iron, and zinc within plants manifested with salinity stress. Phytogeographically plants reacted differently with increasing salinity. Research showed that a higher expression of salinity-responsive miR156 in alfalfa plants results in the accumulation of lower levels of Na+ (Arshad et al., 2017).
To adjust high salinity, plants both manage an ion uptake and distribution system (Sanchez et al., 2008). Since normal plant cells require high amounts of K1 and Na1, this ratio should be high. Low retention of K1 results in high K1 levels in the cytosol at higher Na1 concentrations that get depolarized through membrane currents and causes K1 efflux through outward-rectifying potassium (KOR) channels (Shabala and Mackay, 2011; Bose et al., 2014). Entry of Na1 into the cytosol occurs either via selective transporters or via cation channels, in a saltier environment (Sanchez et al., 2011). During salt stress, Ca2+ alters Na+ influx through extracellular channels and then Na1, K1, and Ca1 remain in balance through SOS pathways (Mahajan and Tuteja, 2005). Membrane transporters maintain appropriate levels of ions such as sodium (Na), potassium (K), hydrogen (H), and others (Shi et al., 2002; Maathuis, 2006; Sanadhya et al., 2015). To expel Na1 from the cell, SOS1 controls SOS3 along with a Ca21 sensor pathway (Khan, 2011). Through Ca21 messages, SOS3 experiences the extracellular salt environment. SOS2 activates the SOS system.
Na1 transport proteins keep Na1 concentrations low in cell cytosol. In soil, HKT1 contributes to the import of anions into plant roots, and the uptake of cations. The electrochemical potential results in accumulation of Na+ ions in the leaves in hyper-salinity environments (Su et al., 2003). The H1-ATPase complex consists of 11 heteromeric subunits (Shabala et al., 2014). By moving protons across the tonoplast of the endoplasmic reticulum (ER), V-type H1-ATPase produces the proton motive force which allows invisible influx of Na+ into the vesicle and thus lowers Na+ toxicity in the cell’s cytoplasm. Therefore, Na+ accumulation inside the vesicle of the cell emerged as an effective framework for osmotic pressure regulation in plant cells (Du et al., 2010). Halophytes have developed salt glands that remove surplus salts from metabolically active tissues (Agarie et al., 2007; Flowers and Colmer, 2008; Shabala et al., 2014).
With the increasing output of genomics data, phenotyping ability does not offer a crucial benefit in regard to the understanding of phenotypically affected genetic variants which are significantly affected by the environment (Furbank and Tester, 2011). Multi-laboratory automated phenotyping systems are in development. Of the next-generation techniques, phenotyping techniques provided multiple advantages over traditional tests including non-destructive testing, automating data, and spectral imaging (Berger et al., 2012; Campbell et al., 2015; Al-Tamimi et al., 2016; Negrão et al., 2017). The technique for capturing molecular phenotypes at “high levels of detail, at unprecedented times and spatial scales” is gaining attention (Negrão et al., 2017). Some findings have been reported on the color of leaves, as well as stomatal responses to a variety of stresses applied to growing progeny (Berger et al., 2012; Hairmansis et al., 2014; Campbell et al., 2015; Awlia et al., 2016). Relative growth rate dynamics of chickpea plants under high salinity stress are notable and worth working on (Atieno et al., 2017). High-throughput phenotyping would be useful in assessing the impact of salt stress on photosynthesis, transpiration, ionic relationships, plant senescence, and on yield. Automatic and digital imaging of plants can further increase understanding of diversity of response of genotype to salinity. Besides, active vision cell technology, deep learning, and other modern phenotyping techniques may be used for defining, quantifying, and predicting plant salinity response with enhanced precision.
Salinity stress can affect seedling growth and seed germination. During vegetative growth, it decreases germination percentage, leaf area, total chlorophyll content, total biomass, and root and shoot length. Visible imaging is used for research, while RGB is used to classify the chlorophyll content of various samples (Mishra et al., 2016a, b). Next-generation phenotyping assays are used to determine salt tolerance, for example, “PHENOPSIS” (Granier et al., 2006) and “WIWAM”1 in rice and other field crops (Humplík et al., 2015; Meng et al., 2017).
The regulation of plant growth and development processes under salinity stress is very complex. Its effect varies with the type of crop species, their growth habit, growth stages, and with environmental conditions. It affects germination to vegetative stage and up to maturity stage from very low to very high levels depending upon the mechanisms adapted or acclimatized by plants. At present, research on salinity stress tolerance is mainly based on the physio-morphological, biochemical, and molecular levels. The most promising ways to enhance salinity stress tolerance will be (1) screening of stress-tolerant genotypes, (2) a deep understanding of the effects and mechanisms of salinity stress in plants, and (3) identifying new genes, proteins, alleles, and transcription factors in respect to higher salinity tolerance. This study aims to understand the signaling mechanisms under salinity stress and we focused on the important signaling components MAPK and Ca under stress conditions. Thereafter, we addressed the crosstalk of important signaling compounds and plant growth regulators and cleared some ideas regarding their function and regulations under salinity stress. Then we discussed the role of recent advanced technology “omics” (genomics, proteomics, transcriptomics, and metabolomics) and how these technologies have helped in recent developments in salinity stress tolerance.
To address the challenges identified by existing research and studies, we came to following conclusions, which can be considered for future research in salinity stress tolerance.
(1) Plant stress tolerance is very complex, therefore existing strategies such as physiological, biochemical, soil, agronomical, and molecular approaches should be integrated to achieve salinity stress tolerance.
(2) Gasotransmitters and plant growth regulators have a crucial role in cell signaling, which needs to be focused on more to help understand the complexity of signaling pathways.
(3) “Omics” technologies are very promising to develop smart crops under environmental fluctuations. Integration of omics technology is a good choice for stress crop improvement programs.
Therefore, this study comprised recent progress in signaling mechanisms, crosstalk mechanisms of signaling compounds, and omics technology for salinity stress tolerance. These developments give a novel insight into understanding the signaling mechanisms and crosstalk under salinity stress tolerance and development of salinity stress tolerance genotypes by applying omics approaches.
All authors have prepared the draft of the manuscript. And also contributed during writing the manuscript, advised scientific suggestion as well as revised/edited the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AbdElgawad, H., Zinta, G., Hegab, M. M., Pandey, R., Asard, H., and Abuelsoud, W. (2016). High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 7:276. doi: 10.3389/fpls.2016.00276
Abdi, N., Holford, P., and Mcglasson, B. (2002). Application of two-dimensional gel electrophoresis to detect proteins associated with harvest maturity in stonefruit. Postharvest Biol. Technol. 26:197. doi: 10.1016/S0925-5214(01)00197-1
Abogadallah, G. M. (2010). Insights into the significance of antioxidative defense under salt stress. Plant Signal. Behav. 5, 369–374. doi: 10.4161/psb.5.4.10873
Afroz, A., Khan, M. R., Ahsan, N., and Komatsu, S. (2009). Comparative proteomic analysis of bacterial wilt susceptible and resistant tomato cultivars. Peptides 30, 1600–1607. doi: 10.1016/j.peptides.2009.06.005
Agarie, S., Shimoda, T., Shimizu, Y., Baumann, K., Sunagawa, H., Kondo, A., et al. (2007). Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J. Exp. Bot. 58, 1957–1967. doi: 10.1093/jxb/erm057
Aghaei, K., Ehsanpour, A. A., and Komatsu, S. (2008). Proteome analysis of potato under salt stress. J. Proteome Res. 7, 4858–4868. doi: 10.1021/pr800460y
Ahmad, P., Abdel Latef, A. A., Hashem, A., Abd-Allah, E. F., Gucel, S., and Tran, L. S. P. (2016). Nitric oxide mitigates salt stress by regulating levels of osmolytes and antioxidant enzymes in chickpea. Front. Plant Sci. 7:347. doi: 10.3389/fpls.2016.00347
Ahmad, R., Parfitt, D. E., Fass, J., Ogundiwin, E., Dhingra, A., Gradziel, T. M., et al. (2011). Whole genome sequencing of peach (Prunus persica L.) for SNP identification and selection. BMC Genome 12:569. doi: 10.1186/1471-2164-12-569
Al-Tamimi, N., Brien, C., Oakey, H., Berger, B., Saade, S., Ho, Y. S., et al. (2016). Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Communi. 7:3342. doi: 10.1038/ncomms13342
Amara, I., Odena, A., Oliveira, E., Moreno, A., Masmoudi, K., Pages, M., et al. (2012). Insights into Maize LEA proteins: from proteomics to functional approaches. Plant Cell Physiol. 53, 312–329. doi: 10.1093/pcp/pcr183
Amiour, N., Merlino, M., Leroy, P., and Branlard, G. (2002). Proteomic analysis of amphiphilic proteins of hexaploid wheat kernels. Proteomics 2, 632–641. doi: 10.1002/1615-9861(200206)2:6<632::aid-prot632>3.0.co;2-m
Amirjani, M. R. (2010). Effect of salinity stress on growth, mineral composition, proline content, antioxidant enzymes of soybean. Am. J. Plant Physiol. 5, 350–360. doi: 10.3923/ajpp.2010.350.360
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arbona, V., Manzi, M., Ollas, D. C., and Gómez-Cadenas, A. (2013). Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 14, 4885–4911. doi: 10.3390/ijms14034885
Arc, E., Sechet, J., Corbineau, F., Rajjou, L., and Marion-Poll, A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63. doi: 10.3389/fpls.2013.00063
Arrivault, S., Guenther, M., Ivakov, A., Feil, R., Vosloh, D., Van Dongen, J. T., et al. (2009). Use of reverse−phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59, 826–839. doi: 10.1111/j.1365-313X.2009.03902.x
Arshad, M., Gruber, M. Y., Wall, K., and Hannoufa, A. (2017). An insight into microRNA156 role in salinity stress responses of alfalfa. Front. Plant Sci. 8:356. doi: 10.3389/fpls.2017.00356
Atieno, J., Li, Y., Langridge, P., Dowling, K., Brien, C., Berger, B., et al. (2017). Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-01211-7
Awlia, M., Nigro, A., Fajkus, J., Schmoeckel, S. M., Negrão, S., Santelia, D., et al. (2016). High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front. Plant Sci. 7:1414. doi: 10.3389/fpls.2016.01414
Babaei, S., Niknam, V., and Behmanesh, M. (2021). Comparative effects of nitric oxide and salicylic acid on salinity tolerance in saffron (Crocus sativus). Plant Biosys. 155, 73–82. doi: 10.1080/11263504.2020.1727975
Bandehagh, A., Salekdeh, G. H., Toorchi, M., Mohammadi, A., and Komatsu, S. (2011). Comparative proteomic analysis of canola leaves under salinity stress. Proteomics 11, 1965–1975. doi: 10.1002/pmic.201000564
Bargaz, A., Nassar, R. M. A., Rady, M. M., Gaballah, M. S., Thompson, S. M., Brestic, M., et al. (2016). Improved Salinity Tolerance by Phosphorus Fertilizer in Two Phaseolus vulgaris Recombinant Inbred Lines Contrasting in Their P-Efficiency. J. Agron. Crop Sci. 202, 497–507. doi: 10.1111/jac.12181
Batistič, O., and Kudla, J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. BBA 1793, 985–992. doi: 10.1016/j.bbamcr.2008.10.006
Baxter, I. (2009). Ionomics: studying the social network of mineral nutrients. Curr. Opin. Plant Biol. 12, 381–386. doi: 10.1016/j.pbi.2009.05.002
Becker, J. S., and Becker, J. S. (2010). Imaging of metals, metalloids, and non-metals by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) in biological tissues. Totowa, NJ: Humana Press, 51–82.
Berger, B., De Regt, B., and Tester, M. (2012). Trait dissection of salinity tolerance with plant phenomics. Totowa, NJ: Humana Press, 399–413.
Bialecka, B., and Kepczynski, J. (2009). Effect of ethephon and gibberellin A3 on Amaranthus caudatus seed germination and alpha-and beta-amylase activity under salinity stress. Acta Biol. Cracov. Ser. Bot. 2, 119–125.
Borrelli, G. M., Fragasso, M., Nigro, F., Platani, C., Papa, R., Beleggia, R., et al. (2018). Analysis of metabolic and mineral changes in response to salt stress in durum wheat (Triticum turgidum ssp. durum) genotypes, which differ in salinity tolerance. Plant Physiol. Biochem. 133, 57–70. doi: 10.1016/j.plaphy.2018.10.025
Bose, J., Rodrigo-Moreno, A., and Shabala, S. (2014). ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257. doi: 10.1093/jxb/ert430
Bouallègue, A., Souissi, F., Nouairi, I., Souibgui, M., Abbes, Z., and Mhadhbi, H. (2017). Salicylic acid and hydrogen peroxide pretreatments alleviate salt stress in faba bean (Vicia faba) seeds during germination. Seed Sci. Tech. 45, 675–690. doi: 10.15258/sst.2017.45.3.07
Boudsocq, M., and Sheen, J. (2009). Stress signaling II: calcium sensing and signaling. In Abiotic stress adaptation in plants. Dordrecht: Springer, 75–90.
Campbell, M. T., Knecht, A. C., Berger, B., Brien, C. J., Wang, D., and Walia, H. (2015). Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 168, 1476–1489. doi: 10.1104/pp.15.00450
Campos, F. V., Oliveira, J. A., Pereira, M. G., and Farnese, F. S. (2019). Nitric oxide and phytohormone interactions in the response of Lactuca sativa to salinity stress. Planta 250, 1475–1489. doi: 10.1007/s00425-019-03236-w
Carillo, P., Annunziata, M. G., Pontecorvo, G., Fuggi, A., and Woodrow, P. (2011). Salinity stress and salt tolerance. Abiotic Stress Plants Mechan. Adapt. 1, 21–38.
Chassaigne, H., Nørgaard, J. V., and Van Hengel, A. J. (2007). Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS). J. Agric. Food Chem. 55, 4461–4473. doi: 10.1021/jf063630e
Chattopadhyay, A., Subba, P., Pandey, A., Bhushan, D., Kumar, R., and Datta, et al. (2011). Analysis of the grasspea proteome and identification of stress-responsive proteins upon exposure to high salinity, low temperature, and abscisic acid treatment. Phytochem 72, 1293–1307. doi: 10.1016/j.phytochem.2011.01.024
Chauhan, J., Singhal, R. K., Chaudhary, S., and Sodani, R. (2017). Calmodulin in Plant Responses to Abiotic Stresses and Signalling. Int. J. Pure App. Biosci. 5, 1122–1131. doi: 10.18782/2320-7051.5235
Chen, F., Fang, P., Peng, Y., Zeng, W., Zhao, X., Ding, Y., et al. (2019). Comparative proteomics of salt-tolerant and salt-sensitive maize inbred lines to reveal the molecular mechanism of salt tolerance. Int. J. Mol. Sci. 20:4725. doi: 10.3390/ijms20194725
Chen, F., Fang, P., Zeng, W., Ding, Y., Zhuang, Z., and Peng, Y. (2020). Comparing transcriptome expression profiles to reveal the mechanisms of salt tolerance and exogenous glycine betaine mitigation in maize seedlings. PLoS One 15:e0233616. doi: 10.1371/journal.pone.0233616
Chen, S., Gollop, N., and Heuer, B. (2009). Proteomic analysis of salt-stressed tomato (Solanumly copersicum) seedlings: effect of genotype and exogenous application of glycinebetaine. J. Exp. Bot. 60, 2005–2019. doi: 10.1093/jxb/erp075
Chen, S., Jia, H., Wang, X., Shi, C., Wang, X., Ma, P., et al. (2020). Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2. 6 in guard cells. Mol. Plant 13, 732–744. doi: 10.1016/j.molp.2020.01.004
Chen, S., Jiang, J., Li, H., and Liu, G. (2012). The salt-responsive transcriptome of Populus simonii× Populus nigra via DGE. Gene 504, 203–212. doi: 10.1016/j.gene.2012.05.023
Che-Othman, M. H., Jacoby, R. P., Millar, A. H., and Taylor, N. L. (2019). Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 225, 1166–1180. doi: 10.1111/nph.15713
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opinion Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Chitteti, B. R., and Peng, Z. (2007). Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 6, 1718–1727. doi: 10.1021/pr060678z
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., and Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Nat. Acad. Sci. U. S. A. 111, 6497–6502. doi: 10.1073/pnas.1319955111
Christou, A., Filippou, P., Manganaris, G. A., and Fotopoulos, V. (2014). Sodium hydrosulfide induces systemic thermotolerance to strawberry plants through transcriptional regulation of heat shock proteins and aquaporin. BMC Plant Biol. 14:42. doi: 10.1186/1471-2229-14-42
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Diff. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Conesa, A., Madrigal, P., Tarazona, S., Gomez-Cabrero, D., Cervera, A., McPherson, A., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 1–19. doi: 10.1186/s13059-016-0881-8
Corpas, F. J. (2019). Hydrogen sulfide: a new warrior against abiotic stress. Trends Plant Sci. 24, 983–988. doi: 10.1016/j.tplants.2019.08.003
Corpas, F. J., and Palma, J. M. (2020). H2S signaling in plants and applications in agriculture. J. Adv. Res. 24, 131–137. doi: 10.1016/j.jare.2020.03.011
Corpas, F. J., Barroso, J. B., Carreras, A., Quirós, M., León, A. M., Romero-Puertas, M. C., et al. (2004). Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant physiol. 136, 2722–2733. doi: 10.1104/pp.104.042812
Corpas, F. J., Barroso, J. B., Carreras, A., Valderrama, R., Palma, J. M., León, A. M., et al. (2006). Constitutive arginine-dependent nitric oxide synthase activity in different organs of pea seedlings during plant development. Planta 224, 246–254. doi: 10.1007/s00425-005-0205-9
Courtois, C., Besson, A., Dahan, J., Bourque, S., Dobrowolska, G., Pugin, A., et al. (2008). Nitric oxide signalling in plants: interplays with Ca2+ and protein kinases. J. Exp. Bot. 59, 155–163. doi: 10.1093/jxb/erm197
Da Silva, C. J., Fontes, E. P. B., and Modolo, L. V. (2017). Salinity-induced accumulation of endogenous H2S and NO is associated with modulation of the antioxidant and redox defense systems in Nicotiana tabacum L. cv. Havana. Plant Sci. 256, 148–159. doi: 10.1016/j.plantsci.2016.12.011
da-Silva, C. J., Mollica, D. C., Vicente, M. H., Peres, L. E., and Modolo, L. V. (2018). NO, hydrogen sulfide does not come first during tomato response to high salinity. Nitric Oxide 76, 164–173. doi: 10.1016/j.niox.2017.09.008
Day, I. S., Reddy, V. S., Ali, G. S., and Reddy, A. S. N. (2002). Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3, 1–24. doi: 10.1186/gb-2002-3-10-research0056
De Grauwe, L., Vriezen, W. H., Bertrand, S., Phillips, A., Vidal, A. M., Hedden, P., et al. (2007). Reciprocal influence of ethylene and gibberellins on response-gene expression in Arabidopsis thaliana. Planta 226, 485–498. doi: 10.1007/s00425-007-0499-x
DeFalco, T. A., Bender, K. W., and Snedden, W. A. (2010). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425, 27–40. doi: 10.1042/BJ20091147
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Deng, G., Zhou, L., Wang, Y., Zhang, G., and Chen, X. (2020). Hydrogen sulfide acts downstream of jasmonic acid to inhibit stomatal development in Arabidopsis. Planta 251, 1–13. doi: 10.1007/s00425-019-03334-9
Depuydt, S., and Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, 365–373. doi: 10.1016/j.cub.2011.03.013
Devoto, A., and Turner, J. G. (2003). Regulation of jasmonate−mediated plant responses in Arabidopsis. Ann. Bot. 92, 329–337. doi: 10.1093/aob/mcg151
D’Hont, A., Denoeud, F., Aury, J. M., Baurens, F. C., Carreel, F., Garsmeur, O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. doi: 10.1038/nature11241
Di Carli, M., Zamboni, A., Pé, M. E., Pezzotti, M., Lilley, K. S., Benvenuto, E., et al. (2011). Two-dimensional differential in gel electrophoresis (2D-DIGE) analysis of grape berry proteome during postharvest withering. J. Proteome Res. 10, 429–446. doi: 10.1021/pr1005313
Dilnur, T., Peng, Z., Pan, Z., Palanga, K. K., Jia, Y., Gong, W., et al. (2019). Association analysis of salt tolerance in Asiatic cotton (Gossypium arboretum) with SNP markers. Int. J. Mol. Sci. 20:2168. doi: 10.3390/ijms20092168
Dinler, B. S., Antoniou, C., and Fotopoulos, V. (2014). Interplay between GST and nitric oxide in the early response of soybean (Glycine max L.) plants to salinity stress. J. Plant Physiol. 171, 1740–1747. doi: 10.1016/j.jplph.2014.07.026
Do, P. T., Drechsel, O., Heyer, A. G., Hincha, D. K., and Zuther, E. (2014). Changes in free polyamine levels, expression of polyamine biosynthesis genes, and performance of rice cultivars under salt stress: a comparison with responses to drought. Front. Plant Sci 5:182. doi: 10.3389/fpls.2014.00182
Domingos, P., Prado, A. M., Wong, A., Gehring, C., and Feijo, J. A. (2015). Nitric oxide: a multitasked signaling gas in plants. Mol. Plant 8, 506–520. doi: 10.1016/j.molp.2014.12.010
Dong, F., Simon, J., Rienks, M., Lindermayr, C., and Rennenberg, H. (2015). Effects of rhizopheric nitric oxide (NO) on N uptake in Fagus sylvatica seedlings depend on soil CO2 concentration, soil N availability and N source. Tree Physiol. 35, 910–920. doi: 10.1093/treephys/tpv051
Dos Santos, V. S., Macedo, F. A., do Vale, J. S., Silva, D. B., and Carollo, C. A. (2017). Metabolomics as a tool for understanding the evolution of Tabebuia sensu lato. Metabolomics 13:72. doi: 10.1007/s11306-017-1209-8
Du, C. X., Fan, H. F., Guo, S. R., Tezuka, T., and Li, J. (2010). Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71, 1450–1459. doi: 10.1016/j.phytochem.2010.05.020
Duque, A. S., de Almeida, A. M., da Silva, A. B., da Silva, J. M., Farinha, A. P., Santos, D., et al. (2013). Abiotic stress responses in plants: unraveling the complexity of genes and networks to survive. Abiot. Stress Plant Resp. Appl. Agricult. 2013, 49–101.
Ekman, D. R., Lorenz, W. W., Przybyla, A. E., Wolfe, N. L., and Dean, J. F. (2003). SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2, 4, 6-trinitrotoluene. Plant Physiol. 133, 1397–1406. doi: 10.1104/pp.103.028019
El-Metwally, S., Ouda, O. M., and Helmy, M. (2014). First-and next-generations sequencing methods. In Next Generation Sequencing Technologies and Challenges in Sequence Assembly. New York, NY: Springer, 29–36.
Fan, H. F., Du, C. X., and Guo, S. R. (2012). Effect of nitric oxide on proline metabolism in cucumber seedlings under salinity stress. J. Am. Soc. Hort. Sci. 137, 127–133. doi: 10.21273/JASHS.137.3.127
Fancy, N. N., Bahlmann, A. K., and Loake, G. J. (2017). Nitric oxide function in plant abiotic stress. Plant Cell Environ. 40, 462–472. doi: 10.1111/pce.12707
Farkhondeh, R., Nabizadeh, E., and Jalilnezhad, N. (2012). Effect of salinity stress on proline content, membrane stability and water relations in two sugar beet cultivars. Int. J. Agri. Sci. 2, 385–392.
Farmer, E. E., and Mueller, M. J. (2013). ROS-mediated lipid peroxidation and RES-activated signaling. Ann. Rev. Plant Biol. 64, 429–450. doi: 10.1146/annurev-arplant-050312-120132
Fatma, M., Masood, A., Per, T. S., and Khan, N. A. (2016). Nitric oxide alleviates salt stress inhibited photosynthetic performance by interacting with sulfur assimilation in mustard. Front. Plant Sci. 7:521. doi: 10.3389/fpls.2016.00521
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 2008, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Foo, E., Ross, J. J., Davies, N. W., Reid, J. B., and Weller, J. L. (2006). A role for ethylene in the phytochrome−mediated control of vegetative development. Plant J. 46, 911–921. doi: 10.1111/j.1365-313X.2006.02754.x
Fotopoulos, V., Sanmartin, M., and Kanellis, A. K. (2006). Effect of ascorbate oxidase over-expression on ascorbate recycling gene expression in response to agents imposing oxidative stress. J. Exp.Bot. 57, 3933–3943. doi: 10.1093/jxb/erl147
Furbank, R. T., and Tester, M. (2011). Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 16, 635–644. doi: 10.1016/j.tplants.2011.09.005
Galant, A., Koester, R. P., Ainsworth, E. A., Hicks, L. M., and Jez, J. M. (2012). From climate change to molecular response: redox proteomics of ozone-induced responses in soybean. New Phytol. 194, 220–229. doi: 10.1111/j.1469-8137.2011.04037.x
Garcia-Mas, J., Benjak, A., Sanseverino, W., Bourgeois, M., Mir, G., González, V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Nat. Acad. Sci. U. S. A. 109, 11872–11877. doi: 10.1073/pnas.1205415109
Garg, N., and Singla, P. (2016). Stimulation of nitrogen fixation and trehalose biosynthesis by naringenin (Nar) and arbuscular mycorrhiza (AM) in chickpea under salinity stress. Plant Growth Regul. 80, 5–22. doi: 10.1007/s10725-016-0146-2
Garufi, A., Visconti, S., Camoni, L., and Aducci, P. (2007). Polyamines as physiological regulators of 14-3-3 interaction with the plant plasma membrane H+-ATPase. Plant Cell Physiol. 48, 434–440. doi: 10.1093/pcp/pcm010
Gavaghan, C. L., Li, J. V., Hadfield, S. T., Hole, S., Nicholson, J. K., Wilson, I. D., et al. (2011). Application of NMR−based metabolomics to the investigation of salt stress in maize (Zea mays). Phytochem. Analysis 22, 214–224. doi: 10.1002/pca.1268
Gayen, D., Barua, P., Lande, N. V., Varshney, S., Sengupta, S., Chakraborty, S., et al. (2019). Dehydration-responsive alterations in the chloroplast proteome and cell metabolomic profile of rice reveals key stress adaptation responses. Environ. Exp. Bot. 160, 12–24. doi: 10.1016/j.envexpbot.2019.01.003
Gémes, K., Poór, P., Horváth, E., Kolbert, Z., Szopkó, D., and Szepesi, Á, et al. (2011). Cross−talk between salicylic acid and NaCl−generated reactive oxygen species and nitric oxide in tomato during acclimation to high salinity. Physiol. Plant 142, 179–192. doi: 10.1111/j.1399-3054.2011.01461.x
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gilliham, M., Able, J. A., and Roy, S. J. (2017). Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 90, 898–917. doi: 10.1111/tpj.13456
Gilroy, S., Suzuki, N., Miller, G., Choi, W. G., Toyota, M., Devireddy, A. R., et al. (2014). A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19, 623–630. doi: 10.1016/j.tplants.2014.06.013
Gniazdowska, A., Dobrzyńska, U., Babańczyk, T., and Bogatek, R. (2007). Breaking the apple embryo dormancy by nitric oxide involves the stimulation of ethylene production. Planta 225, 1051–1057. doi: 10.1007/s00425-006-0384-z
Gniazdowska, A., Krasuska, U., and Bogatek, R. (2010). Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 232, 1397–1407. doi: 10.1007/s00425-010-1262-2
Granier, C., Aguirrezabal, L., Chenu, K., Cookson, S. J., Dauzat, M., Hamard, P., et al. (2006). PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 169, 623–635. doi: 10.1111/j.1469-8137.2005.01609.x
Guan, R., Qu, Y., Guo, Y., Yu, L., Liu, Y., Jiang, J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in G m SALT 3. Plant J. 80, 937–950. doi: 10.1111/tpj.12695
Guo, H., Zhu, N., Deyholos, M. K., Liu, J., Zhang, X., and Dong, J. (2015). Calcium mobilization in salicylic acid-induced Salvia miltiorrhiza cell cultures and its effect on the accumulation of rosmarinic acid. Appl. Biochem. Biotech. 175, 2689–2702. doi: 10.1007/s12010-014-1459-0
Guo, R., Yang, Z., Li, F., Yan, C., Zhong, X., Liu, Q., et al. (2015). Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 15:170. doi: 10.1186/s12870-015-0546-x
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014:701596. doi: 10.1155/2014/701596
Gupta, K., Dey, A., and Gupta, B. (2013). Plant polyamines in abiotic stress responses. Acta Physiol. Planta 35, 2015–2036. doi: 10.1007/s11738-013-1239-4
Gupta, P., Srivastava, S., and Seth, C. S. (2017). 24-Epibrassinolide and sodium nitroprusside alleviate the salinity stress in Brassica juncea L. cv. Varuna through cross talk among proline, nitrogen metabolism and abscisic acid. Plant Soil 411, 483–498. doi: 10.1007/s11104-016-3043-6
Hairmansis, A., Berger, B., Tester, M., and Roy, S. J. (2014). Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice 7, 1–10. doi: 10.1186/s12284-014-0016-3
Hajheidari, M., Abdollahian-Noghabi, M., Askari, H., Heidari, M., Sadeghian, S. Y., Ober, E. S., et al. (2005). Proteome analysis of sugar beet leaves under drought stress. Proteomics 5, 950–960. doi: 10.1002/pmic.200401101
Hajihashemi, S., Skalicky, M., Brestic, M., and Pavla, V. (2020). Cross-talk between nitric oxide, hydrogen peroxide and calcium in salt-stressed Chenopodium quinoa Willd. At seed germination stage. Plant Physiol. Biochem. 154, 657–664. doi: 10.1016/j.plaphy.2020.07.022
Hancock, J. T., and Whiteman, M. (2016). Hydrogen sulfide signaling: interactions with nitric oxide and reactive oxygen species. Ann. New York Acad. Sci. 1365, 5–14. doi: 10.1111/nyas.12733
Harper, J. F., and Harmon, A. (2005). Plants, symbiosis and parasites: a calcium signalling connection. Nat. Rev. Mol. Cell Biol. 6, 555–566. doi: 10.1038/nrm1679
Hasanuzzaman, M., Nahar, K., and Fujita, M. (2013). “Plant response to salt stress and role of exogenous protectants to mitigate salt-induced damages,” in Ecophysiology and Responses of Plants Under Salt Stress, eds P. Ahmed, M. M. Azooz, and M. N. V. Prasad (New York, NY: Springer), 25–87. doi: 10.1007/978-1-4614-4747-4_2
Hasanuzzaman, M., Oku, H., Nahar, K., Bhuyan, M. B., Al Mahmud, J., Baluska, F., et al. (2018). Nitric oxide-induced salt stress tolerance in plants: ROS metabolism, signaling, and molecular interactions. Plant Biotech. Rep. 12, 77–92. doi: 10.1007/s11816-018-0480-0
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Ann. Rev. Plant Biol. 51, 463–499.
Herndl, A., Marzban, G., Kolarich, D., Hahn, R., Boscia, D., Hemmer, W., et al. (2007). Mapping of Malus domestica allergens by 2-D electrophoresis and IgE-reactivity. Electrophoresis 28, 437–448. doi: 10.1002/elps.200600342
Hirschi, K. D. (2003). Strike while the ionome is hot: making the most of plant genomic advances. Trends Biotech. 21, 520–521. doi: 10.1016/j.tibtech.2003.09.013
Ho, W. W. H., Hill, C. B., Doblin, M. S., Shelden, M. C., van de Meene, A., Rupasinghe, T., et al. (2020). Integrative multi-omics analyses of barley rootzones under salinity stress reveal two distinctive salt tolerance mechanisms. Plant Communi. 1:100031. doi: 10.1016/j.xplc.2020.100031
Hrdlickova, R., Toloue, M., and Tian, B. (2017). RNA−Seq methods for transcriptome analysis. Wiley Interdisc. Rev. RNA 8:e1364. doi: 10.1002/wrna.1364
Hsu, S. Y., and Kao, C. H. (2003). Differential effect of sorbitol and polyethylene glycol on antioxidant enzymes in rice leaves. Plant Growth Regul. 39, 83–90.
Huang, D., Huo, J., and Liao, W. (2020). Hydrogen Sulfide: Roles in Plant Abiotic Stress Response and Crosstalk with Other Signals. Plant Sci. 2020:110733. doi: 10.1016/j.plantsci.2020.110733
Huang, L., Zeng, A., Chen, P., Wu, C., Wang, D., and Wen, Z. (2018). Genomewide association analysis of salt tolerance in soybean [Glycine max (L.) Merr.]. Plant Breed. 137, 714–720. doi: 10.1111/pbr.12623
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Gene. 41, 1275–1281.
Huang, X. Y., and Salt, D. E. (2016). Plant ionomics: from elemental profiling to environmental adaptation. Mol. Plant 9, 787–797. doi: 10.1016/j.molp.2016.05.003
Humplík, J. F., Lazár, D., Husičková, A., and Spíchal, L. (2015). Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses–a review. Plant Met. 11, 1–10.
Hussain, S., Shaukat, M., Ashraf, M., Zhu, C., Jin, Q., and Zhang, J. (2019). Salinity stress in arid and semi-arid climates: Effects and management in field crops. Climate Change Agricult. 72, 123–145.
Iqbal, N., Masood, A., and Khan, N. A. (2012). Phytohormones in salinity tolerance: ethylene and gibberellins cross talk. In Phytohormones and Abiotic Stress Tolerance in Plants. Heidelberg: Springer, 77–98. doi: 10.1007/978-3-642-25829-9_3
Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9, 1935–1949. doi: 10.1105/tpc.9.11.1935
Jacoby, R. P., Millar, A. H., and Taylor, N. L. (2010). Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. J. Proteome Res. 9, 6595–6604. doi: 10.1021/pr1007834
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463. doi: 10.1038/nature06148
Jain, S., Srivastava, S., Sarin, N. B., and Kav, N. N. (2006). Proteomics reveals elevated levels of PR 10 proteins in saline-tolerant peanut (Arachis hypogaea) calli. Plant Physiol. Biochem. 44, 253–259. doi: 10.1016/j.plaphy.2006.04.006
Jajic, I., Sarna, T., and Strzalka, K. (2015). Senescence, stress, and reactive oxygen species. Plants 4, 393–411. doi: 10.3390/plants4030393
Jamil, A., Riaz, S., Ashraf, M., and Foolad, M. R. (2011). Gene expression profiling of plants under salt stress. Critical Rev. Plant Sci. 30, 435–458. doi: 10.1080/07352689.2011.605739
Jammes, F., Song, C., Shin, D., Munemasa, S., Takeda, K., and Gu, et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proceed. Nat. Acad. Sci. U. S. A. 106, 20520–20525. doi: 10.1073/pnas.0907205106
Jauregui, I., Aroca, R., Garnica, M., Zamarreño, ÁM., García−Mina, J. M., Serret, M. D., et al. (2015). Nitrogen assimilation and transpiration: key processes conditioning responsiveness of wheat to elevated [CO2] and temperature. Physiol. Plant. 155, 338–354. doi: 10.1111/ppl.12345
Jha, U. C., Bohra, A., Jha, R., and Parida, S. K. (2019). Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 38, 255–277. doi: 10.1007/s00299-019-02374-5
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Ji, W., Cong, R., Li, S., Li, R., Qin, Z., Li, Y., et al. (2016). Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front. Plant Sci. 7:573. doi: 10.3389/fpls.2016.00573
Jiao, Y., Bai, Z., Xu, J., Zhao, M., Khan, Y., Hu, Y., et al. (2018). Metabolomics and its physiological regulation process reveal the salt-tolerant mechanism in Glycine soja seedling roots. Plant Physiol. Biochem. 126, 187–196. doi: 10.1016/j.plaphy.2018.03.002
Jogaiah, S., Govind, S. R., and Tran, L. S. P. (2013). Systems biology-based approaches toward understanding drought tolerance in food crops. Critical Rev. Biotech. 33, 23–39. doi: 10.3109/07388551.2012.659174
Kader, M. A., and Lindberg, S. (2010). Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 5, 233–238. doi: 10.4161/psb.5.3.10740
Kalhor, M. S., Aliniaeifard, S., Seif, M., Asayesh, E. J., Bernard, F., Hassani, B., et al. (2018). Enhanced salt tolerance and photosynthetic performance: Implication of γ-amino butyric acid application in salt-exposed lettuce (Lactuca sativa L.) plants. Plant Physiol. Biochem. 130, 157–172. doi: 10.1016/j.plaphy.2018.07.003
Kalluri, U. C., Hurst, G. B., Lankford, P. K., Ranjan, P., and Pelletier, D. A. (2009). Shotgun proteome profile of Populus developing xylem. Proteomics 9, 4871–4880. doi: 10.1002/pmic.200800854
Kärkönen, A., and Kuchitsu, K. (2015). Reactive oxygen species in cell wall metabolism and development in plants. Phytochem 112, 22–32. doi: 10.1016/j.phytochem.2014.09.016
Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T., Tallon, L. J., et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Kavas, M., Baloğlu, M. C., Atabay, E. S., Ziplar, U. T., Das̨gan, H. Y., and Ünver, T. (2016). Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Gen. Genom. 291, 129–143. doi: 10.1007/s00438-015-1095-6
Kaya, C., Higgs, D., Ashraf, M., Alyemeni, M. N., and Ahmad, P. (2020). Integrative roles of nitric oxide and hydrogen sulfide in melatonin−induced tolerance of pepper (Capsicum annuum L.) plants to iron deficiency and salt stress alone or in combination. Physiol. Plant. 168, 256–277. doi: 10.1111/ppl.12976
Keshet, Y., and Seger, R. (2010). The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. MAP Kinase Sign. Prot. 2, 3–38. doi: 10.1007/978-1-60761-795-2_1
Khan, A., Ahmad, I., Shah, A., Ahmad, F., Ghani, A., Nawaz, M., et al. (2013). Amelioration of salinity stress in wheat (Triticum aestivum L) by foliar application of phosphorus. Phyton 82:281. doi: 10.32604/phyton.2013.82.281
Khan, M. N., Siddiqui, M. H., Mohammad, F., and Naeem, M. (2012). Interactive role of nitric oxide and calcium chloride in enhancing tolerance to salt stress. Nitric Oxide 27, 210–218. doi: 10.1016/j.niox.2012.07.005
Khan, M. N., Siddiqui, M. H., Mukherjee, S., Alamri, S., Al-Amri, A. A., Alsubaie, Q. D., et al. (2020). Calcium-hydrogen sulfide crosstalk during K+-deficient NaCl stress operates through regulation of Na+/H+ antiport and antioxidative defense system in mung bean roots. Plant Physiol. Biochem. 159, 211–225. doi: 10.1016/j.plaphy.2020.11.055
Khan, M. S. (2011). Role of sodium and hydrogen (Na+/H+) antiporters in salt tolerance of plants: present and future challenges. Afr. J. Biotech. 10, 13693–13704. doi: 10.5897/ajb11.1630
Kodela, R., Chattopadhyay, M., and Kashfi, K. (2012). NOSH-aspirin: a novel nitric oxide–hydrogen sulfide-releasing hybrid: a new class of anti-inflammatory pharmaceuticals. ACS Med. Chem. Lett. 3, 257–262. doi: 10.1021/ml300002m
Kong, X., Wang, T., Li, W., Tang, W., Zhang, D., and Dong, H. (2016). Exogenous nitric oxide delays salt-induced leaf senescence in cotton (Gossypium hirsutum L.). Acta Physiol. Plant. 38:79 doi: 10.1007/s11738-016-2079-9
Kreszies, T., Shellakkutti, N., Osthoff, A., Yu, P., Baldauf, J. A., and Zeisler−Diehl, V. V. (2019). Osmotic stress enhances suberization of apoplastic barriers in barley seminal roots: analysis of chemical, transcriptomic and physiological responses. New Phytol. 221, 180–194. doi: 10.1111/nph.15351
Kudla, J., Batistič, O., and Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22, 541–563. doi: 10.1105/tpc.109.072686
Kumar, V., Singh, A., Mithra, S. A., Krishnamurthy, S. L., Parida, S. K., Jain, S., et al. (2015). Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 22, 133–145. doi: 10.1093/dnares/dsu046
Kumari, A., Das, P., Parida, A. K., and Agarwal, P. K. (2015). Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front. Plant Sci. 6:537. doi: 10.3389/fpls.2015.00537
Kurusu, T., Kuchitsu, K., and Tada, Y. (2015). Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 6:427. doi: 10.3389/fpls.2015.00427
Kwak, J. M., Mori, I. C., Pei, Z. M., Leonhardt, N., Torres, M. A., Dangl, J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS−dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Lakra, N., Kaur, C., Singla-Pareek, S. L., and Pareek, A. (2019). Mapping the ‘early salinity response’triggered proteome adaptation in contrasting rice genotypes using iTRAQ approach. Rice 12, 1–22. doi: 10.1007/s10142-015-0460-1
Lamattina, L., García-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Ann. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Lamotte, O., Courtois, C., Dobrowolska, G., Besson, A., Pugin, A., and Wendehenne, D. (2006). Mechanisms of nitric-oxide-induced increase of free cytosolic Ca2+ concentration in Nicotiana plumbaginifolia cells. Free Radi. Biol. Med. 40, 1369–1376. doi: 10.1016/j.freeradbiomed.2005.12.006
Lapopin, L., Gianinazzi-Pearson, V., and Franken, P. (1999). Comparative differential RNA display analysis of arbuscular mycorrhiza in Pisum sativum wild type and a mutant defective in late stage development. Plant Mol. Biol. 41, 669–677.
Läuchli, A., and Grattan, S. R. (2007). Plant growth and development under salinity stress. In Advances in molecular breeding toward drought and salt tolerant crops. Dordrecht: Springer, 1–32.
Lee-Parsons, C. W., and Ertürk, S. (2005). Ajmalicine production in methyl jasmonate-induced Catharanthus roseus cell cultures depends on Ca 2+ level. Plant Cell Rep. 24, 677–682. doi: 10.1007/s00299-005-0026-0
Leisner, C. P., Yendrek, C. R., and Ainsworth, E. A. (2017). Physiological and transcriptomic responses in the seed coat of field-grown soybean (Glycine max L. Merr.) to abiotic stress. BMC Plant Biol. 17:1188. doi: 10.1186/s12870-017-1188-y
Leterrier, M., Chaki, M., Airaki, M., Valderrama, R., Palma, J. M., Barroso, J. B., et al. (2011). Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 6, 789–793. doi: 10.4161/psb.6.6.15161
Leubner-Metzger, G., Petruzzelli, L., Waldvogel, R., Vögeli-Lange, R., and Meins, F. (1998). Ethylene-responsive element binding protein (EREBP) expression and the transcriptional regulation of class I β-1, 3-glucanase during tobacco seed germination. Plant Mol. Biol. 38, 785–795. doi: 10.1023/A:1006040425383
Li, H., Rasheed, A., Hickey, L. T., and He, Z. (2018). Fast-forwarding genetic gain. Trends Plant Sci. 23, 184–186. doi: 10.1016/j.tplants.2018.01.007
Li, J., Jia, H., Wang, J., Cao, Q., and Wen, Z. (2014). Hydrogen sulfide is involved in maintaining ion homeostasis via regulating plasma membrane Na+/H+ antiporter system in the hydrogen peroxide-dependent manner in salt-stress Arabidopsis thaliana root. Protoplasma 251, 899–912. doi: 10.1007/s00709-013-0592-x
Li, L., Wang, X., Stolc, V., Li, X., Zhang, D., Su, N., et al. (2006). Genome-wide transcription analyses in rice using tiling microarrays. Nat. Gen. 38, 124–129. doi: 10.1038/ng1704
Li, P., Cao, W., Fang, H., Xu, S., Yin, S., Zhang, Y., et al. (2017). Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 8:290. doi: 10.3389/fpls.2017.00290
Li, T. T., Li, Z. R., Hu, K. D., Hu, L. Y., Chen, X. Y., and Li, et al. (2017). Hydrogen sulfide alleviates kiwifruit ripening and senescence by antagonizing effect of ethylene. HortSci 52, 1556–1562. doi: 10.21273/HORTSCI12261-17
Li, X., Jiang, H., Liu, F., Cai, J., Dai, T., Cao, W., et al. (2013). Induction of chilling tolerance in wheat during germination by pre-soaking seed with nitric oxide and gibberellin. Plant Growth Regul. 71, 31–40. doi: 10.1007/s10725-013-9805-8
Li, Z. G., Min, X., and Zhou, Z. H. (2016). Hydrogen sulfide: a signal molecule in plant cross-adaptation. Front. Plant Sci. 7:1621. doi: 10.3389/fpls.2016.01621
Li, Z. G., Yi, X. Y., and Li, Y. T. (2014). Effect of pretreatment with hydrogen sulfide donor sodium hydrosulfide on heat tolerance in relation to antioxidant system in maize (Zea mays) seedlings. Biologia 69, 1001–1009. doi: 10.2478/s11756-014-0396-2
Li, Z., Xu, J., Gao, Y., Wang, C., Guo, G., Luo, Y., et al. (2017). The synergistic priming effect of exogenous salicylic acid and H2O2 on chilling tolerance enhancement during maize (Zea mays L.) seed germination. Front. Plant Sci. 8:1153. doi: 10.3389/fpls.2017.01153
Lin, Y., Yang, L., Paul, M., Zu, Y., and Tang, Z. (2013). Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol. Biochem. 73, 211–218. doi: 10.1016/j.plaphy.2013.10.003
Lisjak, M., Teklic, T., Wilson, I. D., Whiteman, M., and Hancock, J. T. (2013). Hydrogen sulfide: environmental factor or signalling molecule? Plant Cell Environ. 36, 1607–1616. doi: 10.1111/pce.12073
Liu, D., Li, J., Li, Z., and Pei, Y. (2020). Hydrogen sulfide inhibits ethylene-induced petiole abscission in tomato (Solanum lycopersicum L.). Hort. Res. 7, 1–11. doi: 10.1038/s41438-019-0237-0
Liu, J. H., Nada, K., Honda, C., Kitashiba, H., Wen, X. P., Pang, X. M., et al. (2006). Polyamine biosynthesis of apple callus under salt stress: importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 57, 2589–2599. doi: 10.1093/jxb/erl018
Liu, L., Nakamura, Y., Taliman, N. A., Sabagh, A. E., Moghaieb, R. E., and Saneoka, H. (2020). Differences in the growth and physiological responses of the leaves of Peucedanum japonicum and Hordeum vulgare exposed to salinity. Agric 10:317. doi: 10.3390/agriculture10080317
Liu, X., Chen, J., Wang, G. H., Wang, W. H., Shen, Z. J., Luo, M. R., et al. (2016). Hydrogen sulfide alleviates zinc toxicity by reducing zinc uptake and regulating genes expression of antioxidative enzymes and metallothioneins in roots of the cadmium/zinc hyperaccumulator Solanum nigrum L. Plant Soil 400, 177–192. doi: 10.1007/s11104-015-2719-7
Liu, Y., Zhou, J., and White, K. P. (2014). RNA-seq differential expression studies: more sequence or more replication? Bioinform 30, 301–304. doi: 10.1093/bioinformatics/btt688
Lu, S., Su, W., Li, H., and Guo, Z. (2009). Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2-and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47, 132–138. doi: 10.1016/j.plaphy.2008.10.006
Lu, Y., Lam, H., Pi, E., Zhan, Q., Tsai, S., Wang, C., et al. (2013a). Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J. Agric. Food Chem. 61, 8711–8721. doi: 10.1021/jf402043m
Lu, Y., Li, N., Sun, J., Hou, P., Jing, X., Zhu, H., et al. (2013b). Exogenous hydrogen peroxide, nitric oxide and calcium mediate root ion fluxes in two non-secretor mangrove species subjected to NaCl stress. Tree Physiol. 33, 81–95. doi: 10.1093/treephys/tps119
Luo, M., Zhao, Y., Wang, Y., Shi, Z., Zhang, P., Zhang, Y., et al. (2018). Comparative proteomics of contrasting maize genotypes provides insights into salt-stress tolerance mechanisms. J. Prote. Res. 17, 141–153. doi: 10.1021/acs.jproteome.7b00455
Ma, D., Ding, H., Wang, C., Qin, H., Han, Q., Hou, J., et al. (2016). Alleviation of drought stress by hydrogen sulfide is partially related to the abscisic acid signaling pathway in wheat. PLoS one 11:e163082 doi: 10.1371/journal.pone.0163082
Ma, N. L., Lah, W. A. C., Kadir, N. A., Mustaqim, M., Rahmat, Z., Ahmad, A., et al. (2018). Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections. PLoS One 2018:e0192732. doi: 10.1371/journal.pone.0192732
Ma, Y., Zhang, W., Niu, J., Ren, Y., and Zhang, F. (2019). Hydrogen sulfide may function downstream of hydrogen peroxide in salt stress-induced stomatal closure in Vicia faba. Funct. Plant Biol. 46, 136–145. doi: 10.1071/FP18096
Maathuis, F. J. (2006). The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 57, 1137–1147. doi: 10.1093/jxb/erj001
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophy. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Manaa, A., Ben Ahmed, H., Valot, B., Bouchet, J. P., Aschi-Smiti, S., Causse, M., et al. (2011). Salt and genotype impact on plant physiology and root proteome variations in tomato. J. Exp. Bot. 62, 2797–2813. doi: 10.1093/jxb/erq460
Mansuri, R. M., Shobbar, Z. S., Jelodar, N. B., Ghaffari, M., Mohammadi, S. M., and Daryani, P. (2020). Salt tolerance involved candidate genes in rice: an integrative meta-analysis approach. BMC Plant Biol. 20:114.
Marco, F., Alcázar, R., Tiburcio, A. F., and Carrasco, P. (2011). Interactions between polyamines and abiotic stress pathway responses unraveled by transcriptome analysis of polyamine overproducers. OMICS 15, 775–781. doi: 10.1089/omi.2011.0084
Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Matsumura, H., Nirasawa, S., and Terauchi, R. (1999). Transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J. 20, 719–726. doi: 10.1046/j.1365-313x.1999.00640.x
McAinsh, M. R., and Pittman, J. K. (2009). Shaping the calcium signature. New Phytol. 181, 275–294. doi: 10.1111/j.1469-8137.2008.02682.x
Mehta, S., James, D., and Reddy, M. K. (2019). Omics technologies for abiotic stress tolerance in plants: current status and prospects. In Recent Approaches in Omics for Plant Resilience to Climate Change. Cham: Springer, 1–34. doi: 10.1007/978-3-030-21687-0_1
Meng, R., Saade, S., Kurtek, S., Berger, B., Brien, C., Pillen, K., et al. (2017). Growth curve registration for evaluating salinity tolerance in barley. Plant Met. 13, 1–9.
Ming, R., Hou, S., Feng, Y., Yu, Q., Dionne-Laporte, A., Saw, J. H., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya L.). Nature 452, 991–996.
Mishra, S., Singh, B., Misra, P., Rai, V., and Singh, N. K. (2016a). Haplotype distribution and association of candidate genes with salt tolerance in Indian wild rice germplasm. Plant Cell Rep. 35, 2295–2308. doi: 10.1007/s00299-016-2035-6
Mishra, S., Singh, B., Panda, K., Singh, B. P., Singh, N., Misra, P., et al. (2016b). Association of SNP haplotypes of HKT family genes with salt tolerance in Indian wild rice germplasm. Rice 9, 1–13.
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G. A. D., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Mockaitis, K., and Howell, S. H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. doi: 10.1111/j.1365-313X.2000.00921.x
Molassiotis, A., and Fotopoulos, V. (2011). Oxidative and nitrosative signaling in plants: two branches in the same tree? Plant Signal. Behav. 6, 210–214. doi: 10.4161/psb.6.2.14878
Molassiotis, A., Tanou, G., and Diamantidis, G. (2010). NO says more than ‘YES’to salt tolerance: salt priming and systemic nitric oxide signaling in plants. Plant Signal. Behav. 5, 209–212. doi: 10.4161/psb.5.3.10738
Molina, C., Rotter, B., Horres, R., Udupa, S. M., Besser, B., Bellarmino, L., et al. (2008). SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genom. 9:553. doi: 10.1186/1471-2164-9-553
Molina, C., Zaman-Allah, M., Khan, F., Fatnassi, N., Horres, R., Rotter, B., et al. (2011). The salt-responsive transcriptome of chickpea roots and nodules via deep SuperSAGE. BMC Plant Biol. 11:31. doi: 10.1186/1471-2229-11-31
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H., and Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356. doi: 10.1105/tpc.109.068395
Moreau, M., Lee, G. I., Wang, Y., Crane, B. R., and Klessig, D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. doi: 10.1074/jbc.M804838200
Mosa, K. A., Ismail, A., and Helmy, M. (2017). Omics and system biology approaches in plant stress research. In Plant stress tolerance. Cham: Springer, 21–34. doi: 10.1007/978-3-319-59379-1_2
Mostofa, M. G., Rahman, A., Ansary, M. M. U., Watanabe, A., Fujita, M., and Tran, L. S. P. (2015a). Hydrogen sulfide modulates cadmium-induced physiological and biochemical responses to alleviate cadmium toxicity in rice. Sci. Rep. 5, 1–17. doi: 10.1038/srep14078
Mostofa, M. G., Saegusa, D., Fujita, M., and Tran, L. S. P. (2015b). Hydrogen sulfide regulates salt tolerance in rice by maintaining Na+/K+ balance, mineral homeostasis and oxidative metabolism under excessive salt stress. Front. Plant Sci 6:1055. doi: 10.3389/fpls.2015.01055
Müller, J., Beck, M., Mettbach, U., Komis, G., Hause, G., Menzel, D., et al. (2010). Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre−prophase band, phragmoplast, trans−Golgi network and plasma membrane. Plant J. 61, 234–248. doi: 10.1111/j.1365-313X.2009.04046.x
Munns, R. (1993). Physiological processes limiting plant growth in saline soils: some dogmas and hypotheses. Plant Cell Environ. 16, 15–24. doi: 10.1111/j.1365-3040.1993.tb00840.x
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Mwando, E., Han, Y., Angessa, T. T., Zhou, G., Hill, C. B., Zhang, X. Q., et al. (2020). Genome-wide association study of salinity tolerance during germination in barley (Hordeum vulgare L.). Front. Plant Sci. 11:118. doi: 10.3389/fpls.2020.00118
Nakagami, H., Pitzschke, A., and Hirt, H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10, 339–346. doi: 10.1016/j.tplants.2005.05.009
Negrão, S., Cecília Almadanim, M., Pires, I. S., Abreu, I. A., Maroco, J., Courtois, B., et al. (2013). New allelic variants found in key rice salt−tolerance genes: an association study. Plant Biotech. J. 11, 87–100. doi: 10.1111/pbi.12010
Negrão, S., Schmöckel, S. M., and Tester, M. (2017). Evaluating physiological responses of plants to salinity stress. Ann. Bot. 119, 1–11. doi: 10.1093/aob/mcw191
Neill, S. J., Desikan, R., Clarke, A., Hurst, R. D., and Hancock, J. T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53, 1237–1247. doi: 10.1093/jexbot/53.372.1237
Ngara, R., Ndimba, R., Borch-Jensen, J., Jensen, O. N., and Ndimba, B. (2012). Identification and profiling of salinity stress-responsive proteins in Sorghum bicolor seedlings. J. Proteomics 75, 4139–4150. doi: 10.1016/j.jprot.2012.05.038
Ni, J., Yang, X., Zhu, J., Liu, Z., Ni, Y., Wu, H., et al. (2015). Salinity-induced metabolic profile changes in Nitraria tangutorum Bobr. suspension cells. PCTOC 122, 239–248. doi: 10.1007/s11240-015-0744-0
Noctor, G., Reichheld, J. P., and Foyer, C. H. (2018). ROS-related redox regulation and signaling in plants. In Seminars in Cell & Developmental Biology, Vol. 80. New York: Academic Press, 3–12.
Olías, R., Eljakaoui, Z., Pardo, J. M., and Belver, A. (2009). The Na+/H+ exchanger SOS1 controls extrusion and distribution of Na+ in tomato plants under salinity conditions. Plant Signal. Behav. 4, 973–976. doi: 10.4161/psb.4.10.9679
Ouyang, S., He, S., Liu, P., Zhang, W., Zhang, J., and Chen, S. (2011). The role of tocopherol cyclase in salt stress tolerance of rice (Oryza sativa). Sci. China Life Sci. 54, 181–188. doi: 10.1007/s11427-011-4138-1
Pan, J., Li, Z., Dai, S., Ding, H., Wang, Q., Li, X., et al. (2020). Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-70520-1
Paranhos, A. (2014). Interplay of calcium, cAMP and PKA in flavonoid accumulation by cell cultures of Hypericum androsaemum L. Planta Medica 80:53. doi: 10.1055/s-0034-1395053
Parida, A. K., Panda, A., and Rangani, J. (2018). Metabolomics-guided elucidation of abiotic stress tolerance mechanisms in plants. In: Plant metabolites and regulation under environmental stress. San Diego, CA: Academic, 89–131. doi: 10.1016/B978-0-12-812689-9.00005-4
Parihar, P., Singh, S., Singh, R., Singh, V. P., and Prasad, S. M. (2015). Effect of salinity stress on plants and its tolerance strategies: a review. Env. Sci. Pol. Res. 22, 4056–4075. doi: 10.1007/s11356-014-3739-1
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556.
Pathak, M. R., Teixeira, da Silva, J. A., and Wani, S. H. (2014). Polyamines in response to abiotic stress tolerance through transgenic approaches. GM Crops Food 5, 87–96. doi: 10.4161/gmcr.28774
Paul, B. D., and Snyder, S. H. (2012). H 2 S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507. doi: 10.1038/nrm3391
Pedreschi, R., Hertog, M., Robben, J., Noben, J. P., and Nicolai, B. (2008). Physiological implications of controlled atmosphere storage of ‘Conference’ pears (Pyrus communis L.): a proteomic approach. Postharvest Biol. Technol. 50, 110–116. doi: 10.1016/j.postharvbio.2008.04.004
Pei, Y., Jin, Z., Liu, Z., Fang, H., Zhang, L., Hao, X., et al. (2018). Gasotransmitters in Plants. Gasotransmitters 12:235. doi: 10.1039/9781788013000-00235
Peng, Z., He, S., Gong, W., Sun, J., Pan, Z., Xu, F., et al. (2014). Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genom. 15:760. doi: 10.1186/1471-2164-15-760
Peng, Z., Wang, M., Li, F., Lv, H., Li, C., and Xia, G. (2009). A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol. Cellu. Proteom. 8, 2676–2686. doi: 10.1074/mcp.M900052-MCP200
Pennisi, E. (2008). Corn genomics pops wide open. Science 319:1333. doi: 10.1126/science.319.5868.1333
Pessarakli, M., and Szabolcs, I. (1999). Soil salinity and sodicity as particular plant/crop stress factors. Handb. Plant Crop Stress 1998:19.
Poór, P., and Tari, I. (2011). Ethylene-regulated reactive oxygen species and nitric oxide under salt stress in tomato cell suspension culture. Acta Biol. Szeged. 55, 143–146.
Poór, P., Gémes, K., Horváth, F., Szepesi, A., Simon, M. L., and Tari, I. (2011). Salicylic acid treatment via the rooting medium interferes with stomatal response, CO2 fixation rate and carbohydrate metabolism in tomato, and decreases harmful effects of subsequent salt stress. Plant Biol. 13, 105–114. doi: 10.1111/j.1438-8677.2010.00344.x
Poór, P., Szopkó, D., and Tari, I. (2012). Ionic homeostasis disturbance is involved in tomato cell death induced by NaCl and salicylic acid. Vitro Cell. Devel. Biol. Plant 48, 377–382. doi: 10.1007/s11627-011-9419-7
Potato Genome Sequencing Consortium. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475:189. doi: 10.1038/nature10158
Pottosin, I., and Shabala, S. (2014). Polyamines control of cation transport across plant membranes: implications for ion homeostasis and abiotic stress signaling. Front. Plant Sci 5:154. doi: 10.3389/fpls.2014.00154
Pottosin, I., Velarde-Buendía, A. M., Bose, J., Zepeda-Jazo, I., Shabala, S., and Dobrovinskaya, O. (2014). Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. J. Exp. Bot. 65, 1271–1283. doi: 10.1093/jxb/ert423
Pottosin, I., Velarde-Buendía, A. M., Zepeda-Jazo, I., Dobrovinskaya, O., and Shabala, S. (2012). Synergism between polyamines and ROS in the induction of Ca2+ and K+ fluxes in roots. Plant Signal. Behav. 7, 1084–1087. doi: 10.4161/psb.21185
Prakash, V., Singh, V. P., Tripathi, D. K., Sharma, S., and Corpas, F. J. (2019). Crosstalk between nitric oxide (NO) and abscisic acid (ABA) signalling molecules in higher plants. Environ. Exp. Bot. 161, 41–49. doi: 10.1016/j.envexpbot.2018.10.033
Qadir, M., Qureshi, A. S., and Cheraghi, S. A. M. (2008). Extent and characterisation of salt−affected soils in Iran and strategies for their amelioration and management. Land Degrad. Develop. 19, 214–227. doi: 10.1002/ldr.818
Qiao, W., Li, C., and Fan, L. M. (2014). Cross-talk between nitric oxide and hydrogen peroxide in plant responses to abiotic stresses. Environ. Exp. Bot. 100, 84–93. doi: 10.1016/j.envexpbot.2013.12.014
Qiao, W., Xiao, S., Yu, L., and Fan, L. M. (2009). Expression of a rice gene OsNOA1 re-establishes nitric oxide synthesis and stress-related gene expression for salt tolerance in Arabidopsis nitric oxide-associated 1 mutant Atnoa1. Environ. Exp. Bot. 65, 90–98. doi: 10.1016/j.envexpbot.2008.06.002
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Nat. Acad. Sci. U. S. A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Ramalingam, A., Kudapa, H., Pazhamala, L. T., Weckwerth, W., and Varshney, R. K. (2015). Proteomics and metabolomics: two emerging areas for legume improvement. Front. Plant Sci. 6:1116. doi: 10.3389/fpls.2015.01116
Raney, J. A. (2012). Transcriptome analysis of drought induced stress in Chenopodium quinoa. Am. J. Plant Sci. 5, 338–357. doi: 10.4236/ajps.2014.53047
Rao, M. V., Paliyath, G., Ormrod, D. P., Murr, D. P., and Watkins, C. B. (1997). Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes (salicylic acid-mediated oxidative damage requires H2O2). Plant Physiol. 115, 137–149. doi: 10.1104/pp.115.1.137
Rasel, M., Tahjib-Ul-Arif, M., and Hossian, M. A. (2020). Screening of Salt-Tolerant Rice Landraces by Seedling Stage Phenotyping and Dissecting Biochemical Determinants of Tolerance Mechanism multidimensional roles in salt-stressed plants. J. Plant Growth Regul. 20:235. doi: 10.1007/s0034 4-020-10235 -9
Rasmussen, S., Barah, P., Suarez-Rodriguez, M. C., Bressendorff, S., Friis, P., Costantino, P., et al. (2013). Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 161, 1783–1794. doi: 10.1104/pp.112.210773
Ravelombola, W., Shi, A., Weng, Y., Mou, B., Motes, D., Clark, J., et al. (2018). Association analysis of salt tolerance in cowpea (Vigna unguiculata (L.) Walp) at germination and seedling stages. Theoret. Appl. Genet. 131, 79–91. doi: 10.1007/s00122-017-2987-0
Razavizadeh, R., Ehsanpour, A. A., Ahsan, N., and Komatsu, S. (2009). Proteome analysis of tobacco leaves under salt stress. Peptides 30, 1651–1659. doi: 10.1016/j.peptides.2009.06.023
Rouphael, Y., Raimondi, G., Lucini, L., Carillo, P., Kyriacou, M. C., Colla, G., et al. (2018). Physiological and metabolic responses triggered by omeprazole improve tomato plant tolerance to NaCl stress. Front. Plant Sci. 9:249.
Sabagh, A. E., Hossain, A., Islam, S., Barutcular, C., Hussain, S., Hasanuzzaman, M., et al. (2019). Drought and salinity stresses in barley: consequences and mitigation strategies. Aus. J. Crop Sci. 13, 810–820. doi: 10.21475/ajcs.19.13.06.p1286
Sahi, C., Singh, A., Kumar, K., Blumwald, E., and Grover, A. (2006). Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct. Integ. Genomics 6, 263–284. doi: 10.1007/s10142-006-0032-5
Salt, D. E., Baxter, I., and Lahner, B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733. doi: 10.1146/annurev.arplant.59.032607.092942
Sanadhya, P., Agarwal, P., and Agarwal, P. K. (2015). Ion homeostasis in a salt-secreting halophytic grass. AoB Plants 7:1. doi: 10.1201/b19862-2
Sanchez, D. H., Pieckenstain, F. L., Escaray, F., Erban, A., Kraemer, U. T. E., Udvardi, M. K., et al. (2011). Comparative ionomics and metabolomics in extremophile and glycophytic Lotus species under salt stress challenge the metabolic pre−adaptation hypothesis. Plant Cell Environ. 34, 605–617. doi: 10.1111/j.1365-3040.2010.02266.x
Sanchez, D. H., Siahpoosh, M. R., Roessner, U., Udvardi, M., and Kopka, J. (2008). Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol. Plant. 132, 209–219.
Santolini, J., André, F., Jeandroz, S., and Wendehenne, D. (2017). Nitric oxide synthase in plants: where do we stand? Nitric Oxide 63, 30–38. doi: 10.1016/j.niox.2016.09.005
Sasabe, M., and Machida, Y. (2012). Regulation of organization and function of microtubules by the mitogen−activated protein kinase cascade during plant cytokinesis. Cytoskeleton 69, 913–918. doi: 10.1002/cm.21072
Scherling, C., Roscher, C., Giavalisco, P., Schulze, E. D., and Weckwerth, W. (2010). Metabolomics unravel contrasting effects of biodiversity on the performance of individual plant species. PLoS One 5:e12569. doi: 10.1371/journal.pone.0012569
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183.
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115.
Schulte, D., Close, T. J., Graner, A., Langridge, P., Matsumoto, T., Muehlbauer, G., et al. (2009). The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiol. 149, 142–147. doi: 10.1104/pp.108.128967
Seifikalhor, M., Aliniaeifard, S., Hassani, B., Niknam, V., and Lastochkina, O. (2019). Diverse role of γ-aminobutyric acid in dynamic plant cell responses. Plant Cell Rep. 19, 1–21. doi: 10.1007/s00299-019-02396-z
Serrano, R., and Rodriguez-Navarro, A. (2001). Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13, 399–404. doi: 10.1016/S0955-0674(00)00227-1
Shabala, S., and Mackay, A. (2011). Ion transport in halophytes. Adv. Bot. Res. 57, 151–199. doi: 10.1016/b978-0-12-387692-8.00005-9
Shabala, S., and Newman, I. (2000). Salinity effects on the activity of plasma membrane H+ and Ca2+ transporters in bean leaf mesophyll: masking role of the cell wall. Ann. Bot. 85, 681–686. doi: 10.1006/anbo.2000.1131
Shabala, S., Shabala, L., Barcelo, J., and Poschenrieder, C. (2014). Membrane transporters mediating root signalling and adaptive responses to oxygen deprivation and soil flooding. Plant Cell Environ. 37, 2216–2233.
Shelden, M. C., and Roessner, U. (2013). Advances in functional genomics for investigating salinity stress tolerance mechanisms in cereals. Front. Plant Sci. 4:123.
Shelden, M. C., Dias, D. A., Jayasinghe, N. S., Bacic, A., and Roessner, U. (2016). Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 67, 3731–3745. doi: 10.1093/jxb/erw059
Shen, Q., Fu, L., Dai, F., Jiang, L., Zhang, G., and Wu, D. (2016). Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC genomics, 17:889. doi: 10.1186/s12864-016-3242-9
Shen, W., Li, H., Teng, R., Wang, Y., Wang, W., and Zhuang, J. (2019). Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Genomics 111, 1142–1151. doi: 10.1016/j.ygeno.2018.07.009
Sheokand, S., Bhankar, V., and Sawhney, V. (2010). Ameliorative effect of exogenous nitric oxide on oxidative metabolism in NaCl treated chickpea plants. Brazil. J. Plant Physiol. 22, 81–90. doi: 10.1590/S1677-04202010000200002
Shi, H., and Chan, Z. (2014). Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J. Integ. Plant Biol. 56, 114–121. doi: 10.1111/jipb.12128
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Shulaev, V., Sargent, D. J., Crowhurst, R. N., Mockler, T. C., Folkerts, O., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Gen. 43, 109–116.
Siddiqui, M. H., Mohammad, F., Khan, M. M. A., and Al-Whaibi, M. H. (2012). Cumulative effect of nitrogen and sulphur on Brassica juncea L. genotypes under NaCl stress. Protoplasma 249, 139–153. doi: 10.1007/s00709-011-0273-6
Simaei, M., Khavarinejad, R. A., Saadatmand, S., Bernard, F., and Fahimi, H. (2011). Interactive effects of salicylic acid and nitric oxide on soybean plants under NaCl salinity. Russ. J. Plant Physiol. 58, 783–790. doi: 10.1134/S1021443711050220
Singh, N. K., Mahato, A. K., Sharma, N., Gaikwad, K., Srivastava, M., Tiwari, K., et al. (2014). “A draft genome of the king of fruit, mango (Mangifera indica L.),” in Plant and Animal Genome XXII Conference. (New York: Acadamic Press).
Singh, R. P., Runthala, A., Khan, S., and Jha, P. N. (2017). Quantitative proteomics analysis reveals the tolerance of wheat to salt stress in response to Enterobacter cloacae SBP-8. PLoS One 12:e0183513. doi: 10.1371/journal.pone.0183513
Singhal, R. K., Sodani, R., Chauhan, J., Sharma, M. K., and Yashu, B. R. (2017). Physiological Adaptation and Tolerance Mechanism of Rice (Oryza sativa L.) in Multiple Abiotic Stresses. Int. J. Pure App. Biosci 5, 459–466. doi: 10.18782/2320-7051.5036
Smékalová, V., Doskočilová, A., Komis, G., and Šamaj, J. (2014). Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotech. Adv. 32, 2–11. doi: 10.1016/j.biotechadv.2013.07.009
Sobhanian, H., Razavizadeh, R., Nanjo, Y., Ehsanpour, A. A., Jazii, F. R., Motamed, N., et al. (2010). Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 8, 1–15. doi: 10.1186/1477-5956-8-19
Soda, N., Wallace, S., and Karan, R. (2015). Omics study for abiotic stress responses in plants. Adv. Plants Agri. Res. 2, 00037.
Steffens, B., Wang, J., and Sauter, M. (2006). Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta 223, 604–612. doi: 10.1007/s00425-005-0111-1
Su, H., Balderas, E., Vera-Estrella, R., Golldack, D., Quigley, F., Zhao, C., et al. (2003). Expression of the cation transporter McHKT1 in a halophyte. Plant Mol. Biol. 52, 967–980.
Suarez, M. C., Bernal, A., Gutierrez, J., Tohme, J., and Fregene, M. (2000). Developing expressed sequence tags (ESTs) from polymorphic transcript-derived fragments (TDFs) in cassava (Manihot esculenta Crantz). Genome 43, 62–67. doi: 10.1139/g99-073
Subudhi, P. K., Shankar, R., and Jain, M. (2020). Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 10, 1–13.
Süle, A., Vanrobaeys, F., Hajós, G., Van Beeumen, J., and Devreese, B. (2004). Proteomic analysis of small heat shock protein isoforms in barley shoots. Phytochem 65, 1853–1863. doi: 10.1016/j.phytochem.2004.03.030
Sun, C., Lu, L., Yu, Y., Liu, L., Hu, Y., and Ye, et al. (2016). Decreasing methylation of pectin caused by nitric oxide leads to higher aluminium binding in cell walls and greater aluminium sensitivity of wheat roots. J. Exp. Bot. 67, 979–989. doi: 10.1093/jxb/erv514
Sun, J., Li, L., Liu, M., Wang, M., Ding, M., Deng, S., et al. (2010). Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl-stressed callus cells of two contrasting poplars. Plant Cell Tiss. Organ Cult. 103, 205–215. doi: 10.1007/s11240-010-9768-7
Suzuki, N., Koussevitzky, S. H. A. I., Mittler, R. O. N., and Miller, G. A. D. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Env. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Svensson, J. T., Crosatti, C., Campoli, C., Bassi, R., Stanca, A. M., Close, T. J., et al. (2006). Transcriptome analysis of cold acclimation in barley Albina and Xantha mutants. Plant Physiol. 141, 257–270. doi: 10.1104/pp.105.072645
Swami, A. K., Alam, S. I., Sengupta, N., and Sarin, R. (2011). Differential proteomic analysis of salt stress response in Sorghum bicolor leaves. Environ. Exp. Bot. 71, 321–328. doi: 10.1016/j.envexpbot.2010.12.017
Szepesi, Á, Csiszár, J., Gémes, K., Horváth, E., Horváth, F., Simon, M. L., et al. (2009). Salicylic acid improves acclimation to salt stress by stimulating abscisic aldehyde oxidase activity and abscisic acid accumulation, and increases Na+ content in leaves without toxicity symptoms in Solanum lycopersicum L. J. Plant Physiol. 166, 914–925. doi: 10.1016/j.jplph.2008.11.012
Takeda, S., Gapper, C., Kaya, H., Bell, E., Kuchitsu, K., and Dolan, L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244. doi: 10.1126/science.1152505
Tang, R. J., Liu, H., Bao, Y., Lv, Q. D., Yang, L., and Zhang, H. X. (2010). The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 74, 367–380. doi: 10.1007/s11103-010-9680-x
Tanou, G., Job, C., Rajjou, L., Arc, E., Belghazi, M., Diamantidis, G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804. doi: 10.1111/j.1365-313x.2009.04000.x
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opinion Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Thitisaksakul, M., Tananuwong, K., Shoemaker, C. F., Chun, A., Tanadul, O. U. M., Labavitch, J. M., et al. (2015). Effects of timing and severity of salinity stress on rice (Oryza sativa L.) yield, grain composition, and starch functionality. J. Agri. Food Chem. 63, 2296–2304. doi: 10.1021/jf503948p
Tracy, F. E., Gilliham, M., Dodd, A. N., Webb, A. A., and Tester, M. (2008). NaCl−induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 31, 1063–1073. doi: 10.1111/j.1365-3040.2008.01817.x
Türkan, I., and Demiral, T. (2009). Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67, 2–9. doi: 10.1016/j.envexpbot.2009.05.008
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604.
Tuteja, N. (2007). Mechanisms of high salinity tolerance in plants. Met. Enzymol. 428, 419–438. doi: 10.1016/S0076-6879(07)28024-3
Uchida, A., Jagendorf, A. T., Hibino, T., Takabe, T., and Takabe, T. (2002). Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 163, 515–523. doi: 10.1016/S0168-9452(02)00159-0
Vafadar, F., Amooaghaie, R., Ehsanzadeh, P., and Ghanadian, M. (2020a). Salinity stress alters ion homeostasis, antioxidant activities and the production of rosmarinic acid, luteolin and apigenin in Dracocephalum kotschyi Boiss. Biologia 75, 2147–2158. doi: 10.2478/s11756-020-00562-3
Vafadar, F., Amooaghaie, R., Ehsanzadeh, P., Ghanadian, M., Talebi, M., and Ghanati, F. (2020c). Melatonin and calcium modulate the production of rosmarinic acid, luteolin, and apigenin in Dracocephalum kotschyi under salinity stress. Phytochemistry 177:112422. doi: 10.1016/j.phytochem.2020.112422
Vafadar, F., Amooaghaie, R., Ehsanzadeh, P., Ghanati, F., and Sajedi, R. H. (2020b). Crosstalk between melatonin and Ca2+/CaM evokes systemic salt tolerance in Dracocephalum kotschyi. J. Plant Physiol. 252:153237. doi: 10.1016/j.jplph.2020.153237
Velarde-Buendía, A. M., Shabala, S., Cvikrova, M., Dobrovinskaya, O., and Pottosin, I. (2012). Salt-sensitive and salt-tolerant barley varieties differ in the extent of potentiation of the ROS-induced K+ efflux by polyamines. Plant Physiol. Biochem. 61, 18–23. doi: 10.1016/j.plaphy.2012.09.002
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus× domestica Borkh.). Nat. Gen. 42, 833–839.
Vighi, I. L., Crizel, R. L., Perin, E. C., Rombaldi, C. V., and Galli, V. (2019). Crosstalk during fruit ripening and stress response among abscisic acid, calcium-dependent protein kinase and phenylpropanoid. Critical Rev. Plant Sci. 38, 99–116. doi: 10.1080/07352689.2019.1602959
Vu, W. T., Chang, P. L., Moriuchi, K. S., and Friesen, M. L. (2015). Genetic variation of transgenerational plasticity of offspring germination in response to salinity stress and the seed transcriptome of Medicago truncatula. BMC Evoluti. Biol. 15:322. doi: 10.1186/s12862-015-0322-4
Wahid, A., Perveen, M., Gelani, S., and Basra, S. M. (2007). Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J. Plant physiol. 164, 283–294. doi: 10.1016/j.jplph.2006.01.005
Wakeel, A., Asif, A. R., Pitann, B., and Schubert, S. (2011). Proteome analysis of sugar beet (Beta vulgaris L.) elucidates constitutive adaptation during the first phase of salt stress. J. Plant Physiol. 168, 519–526. doi: 10.1016/j.jplph.2010.08.016
Wang, H., Liang, X., Wan, Q., Wang, X., and Bi, Y. (2009). Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta 230, 293–307. doi: 10.1007/s00425-009-0946-y
Wang, H., Tang, X., Wang, H., and Shao, H. B. (2015). Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 6:792. doi: 10.3389/fpls.2015.00792
Wang, Y., Li, L., Cui, W., Xu, S., Shen, W., and Wang, R. (2012). Hydrogen sulfide enhances alfalfa (Medicago sativa) tolerance against salinity during seed germination by nitric oxide pathway. Plant Soil 351, 107–119. doi: 10.1007/s11104-011-0936-2
Webb, A. A., McAinsh, M. R., Taylor, J. E., and Hetherington, A. M. (1996). Calcium ions as intracellular second messengers in higher plants. Adv. Bot. Res. 22, 45–96. doi: 10.1016/S0065-2296(08)60056-7
Wicker, T., Taudien, S., Houben, A., Keller, B., Graner, A., Platzer, M., et al. (2009). A whole−genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 59, 712–722. doi: 10.1111/j.1365-313x.2009.03911.x
Wimalasekera, R., Tebartz, F., and Scherer, G. F. (2011). Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181, 593–603. doi: 10.1016/j.plantsci.2011.04.002
Wolfender, J. L., Rudaz, S., Hae Choi, Y., and Kyong Kim, H. (2013). Plant metabolomics: from holistic data to relevant biomarkers. Curr. Med. Chem. 20, 1056–1090. doi: 10.2174/092986713805288932
Wu, D., Shen, Q., Cai, S., Chen, Z. H., Dai, F., and Zhang, G. (2013). Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 54, 1976–1988. doi: 10.1093/pcp/pct134
Xie, Y., Ling, T., Han, Y., Liu, K., Zheng, Q., Huang, L., et al. (2008). Carbon monoxide enhances salt tolerance by nitric oxide−mediated maintenance of ion homeostasis and up−regulation of antioxidant defence in wheat seedling roots. Plant Cell Environ. 31, 1864–1881. doi: 10.1111/j.1365-3040.2008.01888.x
Xing, Y., Jia, W., and Zhang, J. (2008). AtMKK1 mediates ABA−induced CAT1 expression and H2O2 production via AtMPK6−coupled signaling in Arabidopsis. Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x
Xiong, L., and Zhu, J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36. doi: 10.1104/pp.103.025395
Xu, C., Jiang, Z., and Huang, B. (2011). Nitrogen deficiency-induced protein changes in immature and mature leaves of creeping bentgrass. J. Am. Soc. Hort. Sci. 136, 399–407. doi: 10.21273/JASHS.136.6.399
Xu, E., Chen, M., He, H., Zhan, C., Cheng, Y., Zhang, H., et al. (2017). Proteomic analysis reveals proteins involved in seed imbibition under salt stress in rice. Front. Plant Sci. 7:2006.
Xu, J., Wang, W., Sun, J., Zhang, Y., Ge, Q., Du, L., et al. (2011). Involvement of auxin and nitric oxide in plant Cd-stress responses. Plant Soil 346, 107–119. doi: 10.1007/s11104-011-0800-4
Xu, P., Liu, Z., Fan, X., Gao, J., Zhang, X., Zhang, X., et al. (2013). De novo transcriptome sequencing and comparative analysis of differentially expressed genes in Gossypium aridum under salt stress. Gene 525, 26–34. doi: 10.1016/j.gene.2013.04.066
Xu, Q., Xu, X., Zhao, Y., Jiao, K., Herbert, S. J., and Hao, L. (2008). Salicylic acid, hydrogen peroxide and calcium-induced saline tolerance associated with endogenous hydrogen peroxide homeostasis in naked oat seedlings. Plant Growth Regul. 54, 249–259. doi: 10.1007/s10725-007-9247-2
Xu, Y., Gao, S., Yang, Y., Huang, M., Cheng, L., Wei, Q., et al. (2013). Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics 14:662. doi: 10.1186/1471-2164-14-662
Xuan, L., Li, J., Wang, X., and Wang, C. (2020). Crosstalk between hydrogen sulfide and other signal molecules regulates plant growth and development. Int. J. Mol. Sci. 21:4593. doi: 10.3390/ijms21134593
Yadu, S., Dewangan, T. L., Chandrakar, V., and Keshavkant, S. (2017). Imperative roles of salicylic acid and nitric oxide in improving salinity tolerance in Pisum sativum L. Physiol. Mol. Biol. Plants 23, 43–58. doi: 10.1007/s12298-016-0394-7
Yan, K., Shao, H. B., Shao, C. H., Chen, P., Zhao, S., Brestic, M., et al. (2013). Physiological adaptive mechanisms of plant grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Planta 325, 2867–2878. doi: 10.1007/s11738-013-1325-7
Yang, T., Lv, R., Li, J., Lin, H., and Xi, D. (2018). Phytochrome A and B negatively regulate salt stress tolerance of Nicotiana tobacum via ABA–jasmonic acid synergistic cross-talk. Plant Cell Physiol. 59, 2381–2393. doi: 10.1093/pcp/pcy164
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt−stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Zeng, L., Shannon, M. C., and Lesch, S. M. (2001). Timing of salinity stress affects rice growth and yield components. Agri. Water Manag. 48, 191–206. doi: 10.1016/S0378-3774(00)00146-3
Zhang, J., Zhang, Y., Du, Y., Chen, S., and Tang, H. (2011). Dynamic metabolomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 10, 1904–1914. doi: 10.1021/pr101140n
Zhang, L., Pei, Y., Wang, H., Jin, Z., Liu, Z., Qiao, Z., et al. (2015). Hydrogen sulfide alleviates cadmium-induced cell death through restraining ROS accumulation in roots of Brassica rapa L. ssp. pekinensis. Oxidat. Med. Cell. Long. 2015:804603. doi: 10.1155/2015/804603
Zhang, L., Yu, Z., Jiang, L., Jiang, J., Luo, H., and Fu, L. (2011). Effect of post-harvest heat treatment on proteome change of peach fruit during ripening. J. Proteomics 74, 1135–1149. doi: 10.1016/j.jprot.2011.04.012
Zhang, P., Luo, Q., Wang, R., and Xu, J. (2017). Hydrogen sulfide toxicity inhibits primary root growth through the ROS-NO pathway. Scientific Rep. 7, 1–11. doi: 10.1038/s41598-017-01046-2
Zhang, Y., Tan, J., Guo, Z., Lu, S., He, S., Shu, W., et al. (2009). Increased abscisic acid levels in transgenic tobacco over−expressing 9 cis−epoxycarotenoid dioxygenase influence H2O2 and NO production and antioxidant defences. Plant Cell Environ. 32, 509–519. doi: 10.1111/j.1365-3040.2009.01945.x
Zhang, Y., Wang, L., Liu, Y., Zhang, Q., Wei, Q., and Zhang, W. (2006). Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 224, 545–555. doi: 10.1007/s00425-006-0242-z
Zhao, F., Song, C. P., He, J., and Zhu, H. (2007). Polyamines improve K+/Na+ homeostasis in barley seedlings by regulating root ion channel activities. Plant Physiol. 145, 1061–1072. doi: 10.1104/pp.107.105882
Zhao, G., Zhao, Y., Yu, X., Kiprotich, F., Han, H., Guan, R., et al. (2018). Nitric oxide is required for melatonin-enhanced tolerance against salinity stress in rapeseed (Brassica napus L.) seedlings. Int. J. Mol. Sci. 19:912. doi: 10.3390/ijms19071912
Zhao, M. G., Tian, Q. Y., and Zhang, W. H. (2007). Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 144, 206–217. doi: 10.1104/pp.107.096842
Zhao, Y., Hu, F., Zhang, X., Wei, Q., Dong, J., Bo, C., et al. (2019). Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC Plant Biol. 19:273. doi: 10.1186/s12870-019-1878-8
Zhao, Z., Zhang, W., Stanley, B. A., and Assmann, S. M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20, 3210–3226. doi: 10.1105/tpc.108.063263
Zheng, C., Jiang, D., Liu, F., Dai, T., Liu, W., Jing, Q., et al. (2009). Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environ. Exp. Bot. 67, 222–227. doi: 10.1016/j.envexpbot.2009.05.002
Zhu, J. K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10, 1181–1191. doi: 10.1105/tpc.10.7.1181
Zhu, T., Deng, X., Zhou, X., Zhu, L., Zou, L., Li, P., et al. (2016). Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Scientific Rep. 6:392. doi: 10.1038/srep35392
Zi, J., Zhang, J., Wang, Q., Lin, L., Tong, W., Bai, X., et al. (2012). Proteomics study of rice embryogenesis: discovery of the embryogenesis-dependent globulins. Electrophoresis 33, 1129–1138. doi: 10.1002/elps.201100398
Zörb, C., Geilfus, C. M., Mühling, K. H., and Ludwig-Müller, J. (2013). The influence of salt stress on ABA and auxin concentrations in two maize cultivars differing in salt resistance. J. Plant Physiol. 170, 220–224. doi: 10.1016/j.jplph.2012.09.012
Keywords: antioxidant defense, crosstalk, homeostasis, omics approaches, signaling network, plant growth regulators, salinity stress tolerance
Citation: Singhal RK, Saha D, Skalicky M, Mishra UN, Chauhan J, Behera LP, Lenka D, Chand S, Kumar V, Dey P, Indu, Pandey S, Vachova P, Gupta A, Brestic M and El Sabagh A (2021) Crucial Cell Signaling Compounds Crosstalk and Integrative Multi-Omics Techniques for Salinity Stress Tolerance in Plants. Front. Plant Sci. 12:670369. doi: 10.3389/fpls.2021.670369
Received: 21 February 2021; Accepted: 28 May 2021;
Published: 13 August 2021.
Edited by:
Bok-Rye Lee, Chonnam National University, South KoreaReviewed by:
Bhaskar Gupta, Government General Degree College, Singur, IndiaCopyright © 2021 Singhal, Saha, Skalicky, Mishra, Chauhan, Behera, Lenka, Chand, Kumar, Dey, Indu, Pandey, Vachova, Gupta, Brestic and El Sabagh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ayman El Sabagh, YXltYW4uZWxzYWJhZ2hAYWdyLmtmcy5lZHUuZWc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.