- 1Center for Plant Molecular Biology (ZMBP), University of Tübingen, Tübingen, Germany
- 2Institute of Biology, University of Hohenheim, Stuttgart, Germany
The nuclear lamina (NL) is a complex network of nuclear lamins and lamina-associated nuclear membrane proteins, which scaffold the nucleus to maintain structural integrity. In animals, type V intermediate filaments are the main constituents of NL. Plant genomes do not encode any homologs of these intermediate filaments, yet plant nuclei contain lamina-like structures that are present in their nuclei. In Arabidopsis thaliana, CROWDED NUCLEI (CRWN), which are required for maintaining structural integrity of the nucleus and specific perinuclear chromatin anchoring, are strong candidates for plant lamin proteins. Recent studies revealed additional roles of Arabidopsis Nuclear Matrix Constituent Proteins (NMCPs) in modulating plants’ response to pathogen and abiotic stresses. However, detailed analyses of Arabidopsis NMCP activities are challenging due to the presence of multiple homologs and their functional redundancy. In this study, we investigated the sole NMCP gene in the liverwort Marchantia polymorpha (MpNMCP). We found that MpNMCP proteins preferentially were localized to the nuclear periphery. Using CRISPR/Cas9 techniques, we generated an MpNMCP loss-of-function mutant, which displayed reduced growth rate and curly thallus lobes. At an organelle level, MpNMCP mutants did not show any alteration in nuclear morphology. Transcriptome analyses indicated that MpNMCP was involved in regulating biotic and abiotic stress responses. Additionally, a highly repetitive genomic region on the male sex chromosome, which was preferentially tethered at the nuclear periphery in wild-type thalli, decondensed in the MpNMCP mutants and located in the nuclear interior. This perinuclear chromatin anchoring, however, was not directly controlled by MpNMCP. Altogether, our results unveiled that NMCP in plants have conserved functions in modulating stress responses.
Introduction
The nuclear lamina (NL) is a fibrillar network that is composed of intermediate filaments and membrane-associated proteins, proving mechanical support to the nucleus (Dechat et al., 2008; Prokocimer et al., 2009). In addition, the NL also functions at the level of gene expression. Reports show that the NL serves as an attachment surface for chromosomes where it provides a repressive environment for gene silencing (Shevelyov and Nurminsky, 2012; van Steensel and Belmont, 2017). Accordingly, transcriptional activation of genes interacting with the NL correlates with local chromatin rewiring and NL detachment (Brueckner et al., 2020). For instance, in mammals and nematodes, X chromosome inactivation is mediated partially by tethering to the NL (Crane et al., 2015), which is dependent on the Lamin B receptor in mammals (Chen et al., 2016; van Steensel and Belmont, 2017). NL–chromatin interactions also contribute to genome organization. In eukaryotic nuclei, active chromatin and inactive chromatin occupy physically separated nuclear compartments (Zheng et al., 2018; Ulianov et al., 2019; Chang et al., 2020). Recent reports with various animal model systems revealed that deficiency in nuclear lamina proteins gave rise to increased chromatin interactions between active and inactive compartments (Zheng et al., 2018; Ulianov et al., 2019; Chang et al., 2020).
Animal nuclear lamins are type V intermediate filaments (Herrmann et al., 2009; Dittmer and Misteli, 2011). Plants do not have lamin-like analogs; however, there exists a lamina-like structure that helps to maintain nuclear morphology in the plant nucleus (McNulty and Saunders, 1992; Minguez and Moreno Diaz de la Espina, 1993; Masuda et al., 1997). Key components of this structure are a group of proteins known as nuclear matrix constituent proteins (NMCPs), which are plant-specific and highly conserved in the plant kingdom (Ciska et al., 2013). NMCPs are diverged into two phylogenetic groups, NMCP1 and NMCP2 (Ciska et al., 2019). In Arabidopsis thaliana, CROWDED NUCLEI (CRWN) proteins are the four NMCPs annotated as lamin candidates (Wang et al., 2013; Mikulski et al., 2019), among which CRWN1 to 3 and CRWN4 are classified as NMCP1 and NMCP2 type, respectively (Tamura et al., 2015). Functional analyses of CRWNs indicate their conserved function in regulating nuclear morphology as animal lamins (Zwerger et al., 2011; Tamura et al., 2015; Meier et al., 2016). At the cellular level, single crwn1 and crwn4 mutants show reduced nuclear size, and this phenotype is more pronounced in higher-order crwn mutants (Sakamoto and Takagi, 2013; Wang et al., 2013; Goto et al., 2014). Although individual single crwn growth similarly as wild-type (WT) plants, a low but non-trivial level of CRWN activities is required to complete the plant life cycle, as mutants lacking all the four CRWN genes are inviable (Wang et al., 2013).
Arabidopsis CRWN genes, especially CRWN1, are so far the most intensively investigated plant NMCP. In addition to regulating nuclear size and morphology, CRWN1 has also been found to be involved in protecting DNA against oxidative damage (Wang et al., 2019), pathogen response (Guo et al., 2017; Choi et al., 2019; Jarad et al., 2019), promoting copper tolerance (Sakamoto et al., 2020), ABA signaling pathway (Zhao et al., 2016), and chromatin activities. CRWN1 and CRWN4 proteins are localized at the nuclear periphery, where they are both involved in regulating specific perinuclear chromatin anchoring (Sakamoto and Takagi, 2013; Poulet et al., 2017b; Hu et al., 2019). CRWN1 was shown recently to tether repressed chromatin domains to the NL by binding to inaccessible chromatin (Hu et al., 2019). Furthermore, CRWN1 and CRWN4 play a role in regulating heterochromatin integrity and condensation (Wang et al., 2013; Poulet et al., 2017a). Additionally, CRWN is also able to mediate active gene expression. A recent report by Sakamoto and colleagues shows that CRWN1 participates in positioning copper tolerance genes at the NL and mediating their transcriptional upregulation under excess copper conditions (Sakamoto et al., 2020). With the use of proteomic approaches, CRWN1 has been shown to interact with transcriptional regulators, such as Polycomb Repressive Complex 2-associated factor PWO1 (Mikulski et al., 2019) and NAC transcription factor NTL9 (Guo et al., 2017). A CRWN homolog in rice, OsNMCP1, has been found to interact with a subunit belonging to SWITCH/SUCROSE NON-FERMENTING (SWI/SNF) chromatin remodeling complex to modulate drought stress response (Yang et al., 2020).
Studies of multiple Arabidopsis CRWN genes also unveiled their functional redundancy in suppressing autoimmunity. RNA sequencing (RNA-seq) experiments of several crwn single and double mutants indicated that the gene expression profile of these mutants was concomitant with the activation of pathways related to plant–pathogen interactions, for example, in the activation of PR1 (PATHOGENESIS-RELATED1) and PR2 and PR5 expression (Guo et al., 2017; Choi et al., 2019). With ectopically activated biotic stress response pathways, crwn1 crwn2 and crwn1 crwn4 double mutants showed greater resistance to infection with virulent bacterial pathogens (Guo et al., 2017; Choi et al., 2019).
The liverwort Marchantia polymorpha is a dioecious non-vascular plant with a mostly haploid life cycle and a highly streamlined genome: small size (ca. 280 Mbp) with low genetic redundancy (Tanurdzic and Banks, 2004). Early phylogenetic analysis indicates that the Marchantia genome possesses only one NMCP homolog (hereafter referred to as “MpNMCP”) (Ciska et al., 2019). It is not known if this sole NMCP gene in Marchantia shares conserved functions as those discovered in higher plants. In this study, we report that MpNMCP is a lamina-like nuclear periphery protein. We found that while the growth of Mpnmcp mutants was suppressed, loss of NMCP did not block vegetative growth of Marchantia, and Mpnmcp mutant nuclei did not show altered nuclear size or shape. We also found that a heterochromatic region from the Marchantia male sex chromosome displayed preferential perinuclear localization, but it detached from the NL in Mpnmcp nuclei and decondensed. However, MpNMCP proteins did not show direct interactions with chromatin at the nuclear periphery. Nevertheless, transcriptome experiments indicated that MpNMCP was involved in suppressing ectopic stress responses. Overall, the Marchantia NMCP has remarkable differences in functional conservation compared with NMCPs in higher vascular plants, except the sphere of plant stress responses, where it retains a similar function.
Results
MpNMCP Promotes Basic Growth of Marchantia
The Marchantia genome assembly has been recently upgraded from a scaffold (v3.1) to chromosomal scale version (v5.1) containing slightly more annotated genes (Montgomery et al., 2020). Thus, we performed a homology search to check if there were additional Marchantia NMCP homologs missed in the earlier phylogenetic analyses using the older genome assembly. The protein sequences of Arabidopsis CRWN1 and another two NMCPs from Physcomitrella patens were used to search for Marchantia NMCP homologs with Basic Local Alignment Search Tool for protein sequences (BLASTp). For each of the inquiry sequences, only one hit was found, confirming that the Marchantia genome encodes only one putative NMCP (Supplementary Figure 1; Ciska et al., 2019).
Next, to clarify the function of MpNMCP, we generated Mpnmcp mutants using the CRISPR/Cas9 system, by which two guide RNAs were designed to delete most of MpNMCP’s coding region (Figure 1A). All T1 mutant lines carrying a deletion allele displayed severely delayed thallus growth; among them, we chose two lines in the Takaragaike-1 (Tak-1) background, namely, Mpnmcp-1 and Mpnmcp-2, for further phenotype characterization. Sequencing of these mutants revealed that both of them had a ∼3-kb deletion between the two single-guide RNAs (sgRNAs) (Figures 1A,B). Under a standard greenhouse condition, Mpnmcp mutant thalli showed dwarf phenotypes and curly branches as compared with the WT Tak-1 plants (Figure 1D). Asexual reproduction of Mpnmcp mutants was largely unaffected: they could produce gemma cups similarly to WT Tak-1 thalli (Figure 1E). Smaller gemmae, however, were found within these gemmae cups (Figure 1F).
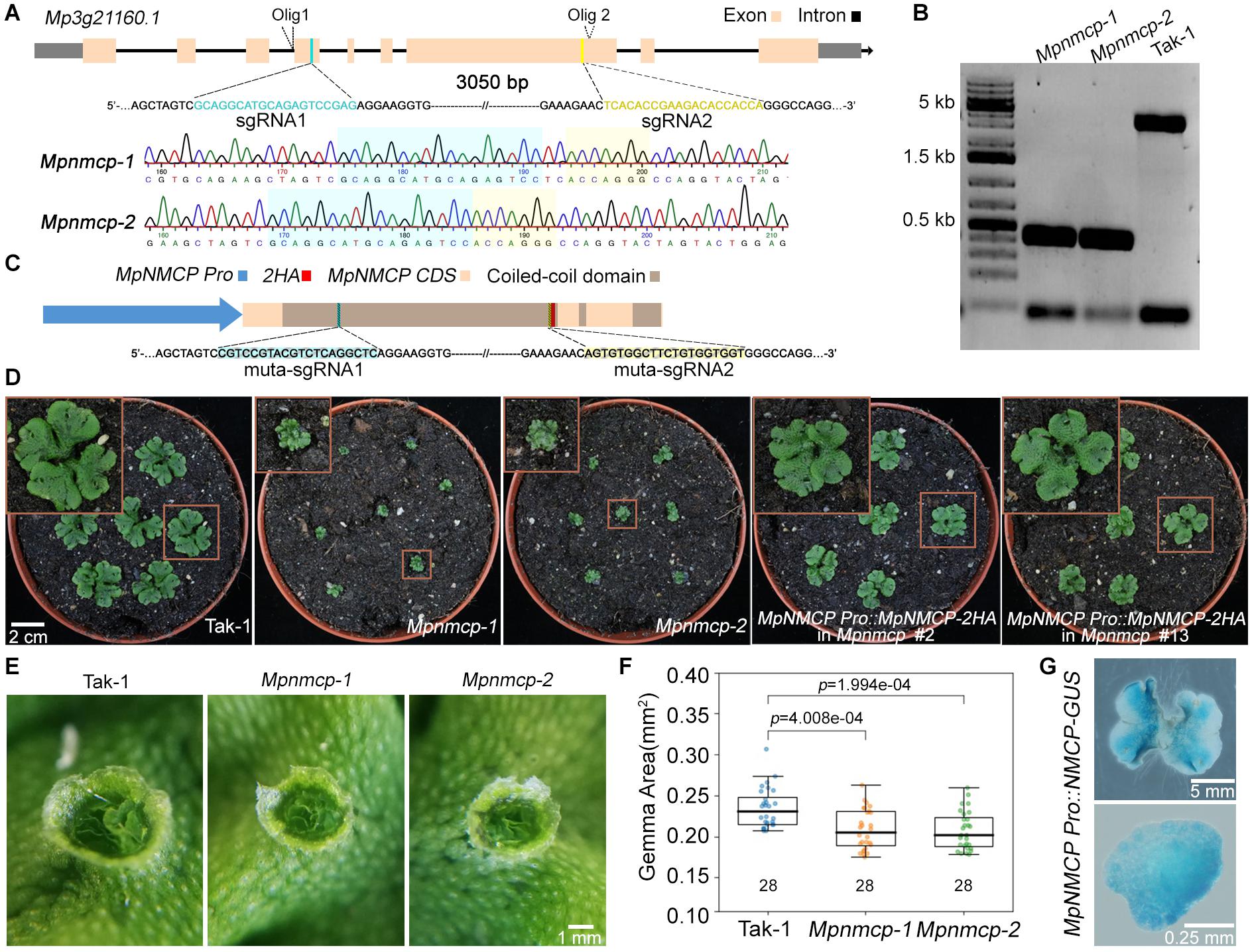
Figure 1. Mpnmcp mutants show dwarf phenotype. (A) Schematic depiction of the MpNMCP locus and single-guide RNAs (sgRNAs) aiming for site-directed mutagenesis (top). Representative sequencing tracks of using Olig1 (Supplementary Table 2) showing the deletion of a ∼3-kb region flanked by the two sgRNAs (middle and bottom). (B) Genotyping PCR results of individual T3 Mpnmcp mutants. Genotyping was carried out by using genotype specific primers: Olig1 (forward primer) and Olig2 (reverse primer) (primer sequences are given in Supplementary Table 2). (C) Schematic depiction of the MpNMCP CDS with MpNMCP promoter (2-kb upstream of the gene model). HA, hemagglutinin tag. muta-sgRNA1 and muta-sgRNA2 indicate the two regions to which silent mutations are introduced to avoid recognition by the sgRNAs used for site-directed mutagenesis of the endogenous MpNMCP locus. (D) Phenotype of 3-week-old wild-type (WT) Tak-1, Mpnmcp mutants, and two independent MpNMCP Pro:MpNMCP-2HA/Mpnmcp transgenic lines. The MpNMCP Pro:MpNMCP-2HA in the Mpnmcp mutant displayed normal development. (E) Close-up of one gemma cup from 20-day-old WT Tak-1 or Mpnmcp mutants, which have mature gemmae inside. (F) Boxplot shows the size of gemmae collected from gemma cups of Mpnmcp mutants and Tak-1 plants. n = 28. p values were determined by Mann–Whitney U test. (G) Glucuronidase (GUS) staining of MpNMCP Pro:GUS plants. Pictures show the representative distribution pattern of GUS staining in 2-week-old thallus (top panel, scale bar = 5 mm) and gemmae (bottom panel, scale bar = 0.25 mm).
To confirm that the mentioned Mpnmcp phenotypes were causal to the lack of functional MpNMCP, we generated complementation lines by double transformation, in which the WT MpNMCP coding sequence fused with a tandem hemagglutinin (HA) tag driven by the native MpNMCP promoter and introduced it into null Mpnmcp mutant background (Figure 1C). The growth defects exhibited by the Mpnmcp mutants were largely rescued in the complementation lines (Figure 1D and Supplementary Figure 10). Altogether, phenotypic characterization of Mpnmcp mutants revealed that the absence of MpNMCP leads to general growth repression.
To characterize MpNMCP spatial–temporal expression patterns in greater detail, we generated reporter lines in which the same MpNMCP promoter region (Figure 1C) was placed in front of the beta-glucuronidase (GUS) reporter coding sequence. We analyzed GUS staining patterns in more than 30 independent transgenic MpNMCP Pro:GUS lines, all of which showed GUS activity throughout thalli except for older parts at thallus margins (Figure 1G). In gemmae, the GUS staining was visible across the entire sample (Figure 1G).
MpNMCP Suppresses Ectopic Onset of Stress Response
To gain a better understanding of how MpNMCP regulates thallus growth, we performed RNA-seq to compare the gene expression profile of 2-week-old Mpnmcp mutant and WT thallus tissues. In total, 571 and 472 genes were annotated as up- and downregulated genes shared between two independent mutant lines, respectively (Supplementary Figure 2 and Supplementary Table 1). As the function of NMCP genes in both Arabidopsis and rice has been linked to stress response (Guo et al., 2017; Choi et al., 2019; Jarad et al., 2019; Yang et al., 2020), we postulated that the Mpnmcp mutant transcriptome would show correlation to those stress conditions. To this end, we compared differentially expressed genes (DEGs) in Mpnmcp mutants and several published stress-related Marchantia transcriptomes. To capture pathogen response, we used a transcriptome dataset of Marchantia infected by hemi-biotrophic oomycete Phytophthora palmivora (Carella et al., 2019). Among the 571 upregulated genes in Mpnmcp, 100 (p = 1.02e–24, Fisher’s exact test) and 143 (p = 6.03e–46, Fisher’s exact test) were upregulated in 3-dpi (days post inoculation) and 4-dpi samples, respectively (Figure 2A). A similar extent of overlap was found for downregulated genes, where 85 (p = 3.20e–44, Fisher’s exact test) and 152 (p = 1.19e–96, Fisher’s exact test) downregulated genes in Mpnmcp showed suppressed expression in 3- and 4-dpi samples, respectively (Figure 2A). The jasmonate (JA) pathway has recently been shown as a suppressor of defense in Marchantia (Monte et al., 2019; Matsui et al., 2020). Interestingly, for the Mpjaz mutant that exhibits constitutive activation of the JA pathway, its transcriptome also resembled that of the Mpnmcp mutant (Figure 2B). However, only one of the three JA maker genes MpAOS2 (Mp5g16260), MpAOC (Mp7g06220), and MpCHL (Mp8g10460), which were highly upregulated in Mpjaz (Monte et al., 2019), showed noticeable upregulation in Mpnmcp, suggesting a partially activated JA pathway in our mutants (Supplementary Figure 3). We also compared Mpnmcp transcriptome with a salt-stress transcriptome dataset (Tanaka et al., 2018). Due to a lower sequencing coverage of this salt-stress transcriptome dataset and our filter settings of DEG calling, there were only 109 genes annotated as salt-stress responsive. Nevertheless, more than half of them (p = 2.787e–62, Fisher’s exact test) were upregulated in the Mpnmcp mutant (Figure 2C). Altogether, the noticeable overlap of DEGs between the Mpnmcp mutant and different stress-related transcriptome suggests that the role of plant NMCP genes in regulating stress response is conserved in Marchantia.
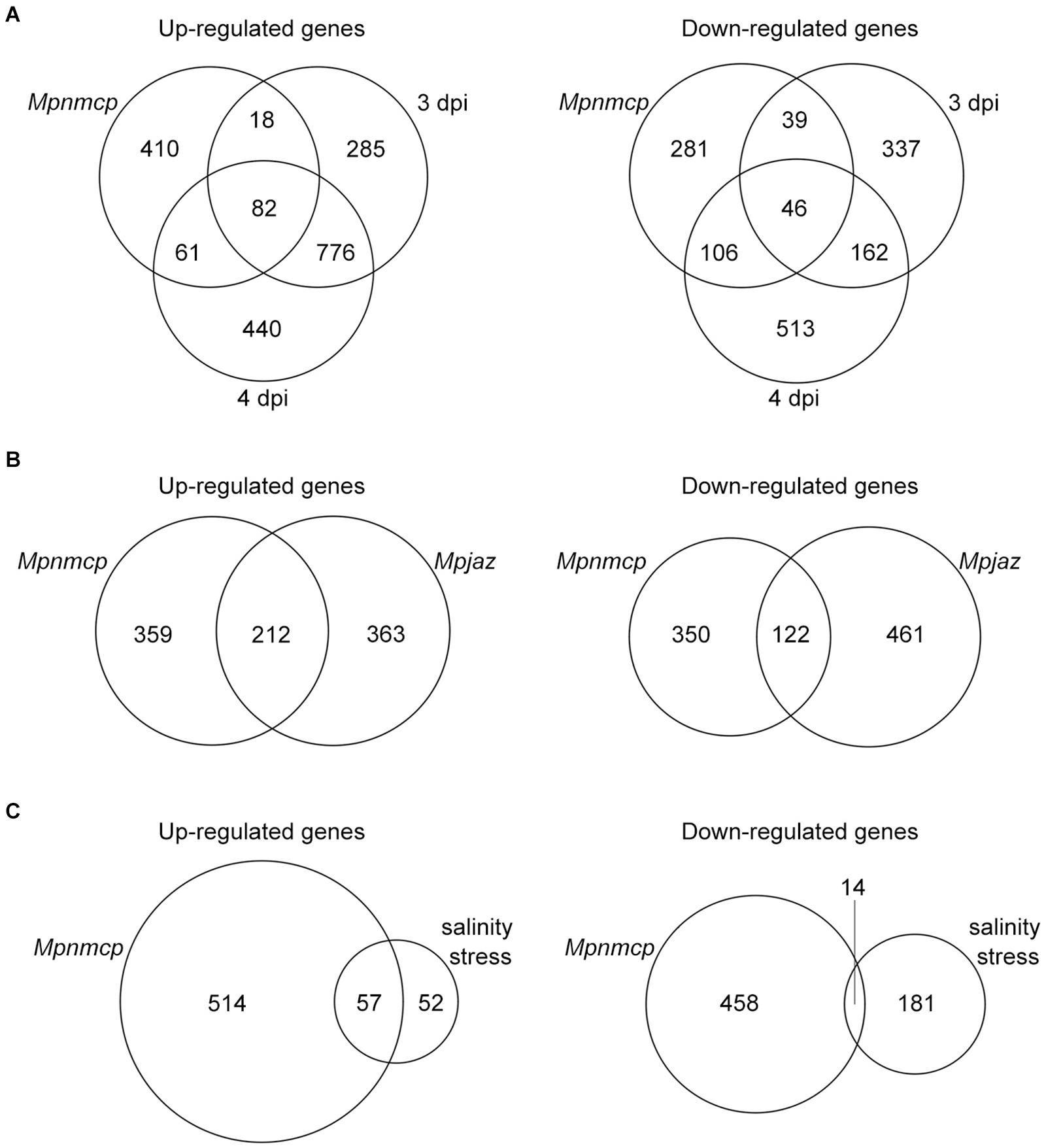
Figure 2. Characteristics of differentially expressed genes (DEGs). (A) Venn diagram showing the overlap of DEGs among Mpnmcp, 3 dpi (days post inoculation with Phytophthora palmivora) and 4 dpi. (B) Venn diagram showing the overlap of DEGs between Mpnmcp and Mpjaz. (C) Venn diagram showing the overlap of DEGs between Mpnmcp and Marchantia under salinity stress.
Mpnmcp Is Localized at the Nuclear Periphery but Does Not Control Nuclear Morphology
We next examined nuclear localization patterns of MpNMCP proteins by immunohistostaining, in which functional MpNMCP:2HA fusion proteins were visualized with fluorophore-conjugated anti-HA antibodies. As shown by representative images in Figure 3A, MpNMCP is preferentially localized at the nuclear periphery in vivo, which is similar to many NMCP proteins from various plants species (Ciska et al., 2013; Sakamoto and Takagi, 2013; Yang et al., 2020; McKenna et al., 2021). Besides, we found that MpNMCP proteins were not evenly distributed at the nuclear periphery but exhibited a punctate-like pattern, which was similar to that from onion (Ciska et al., 2013).
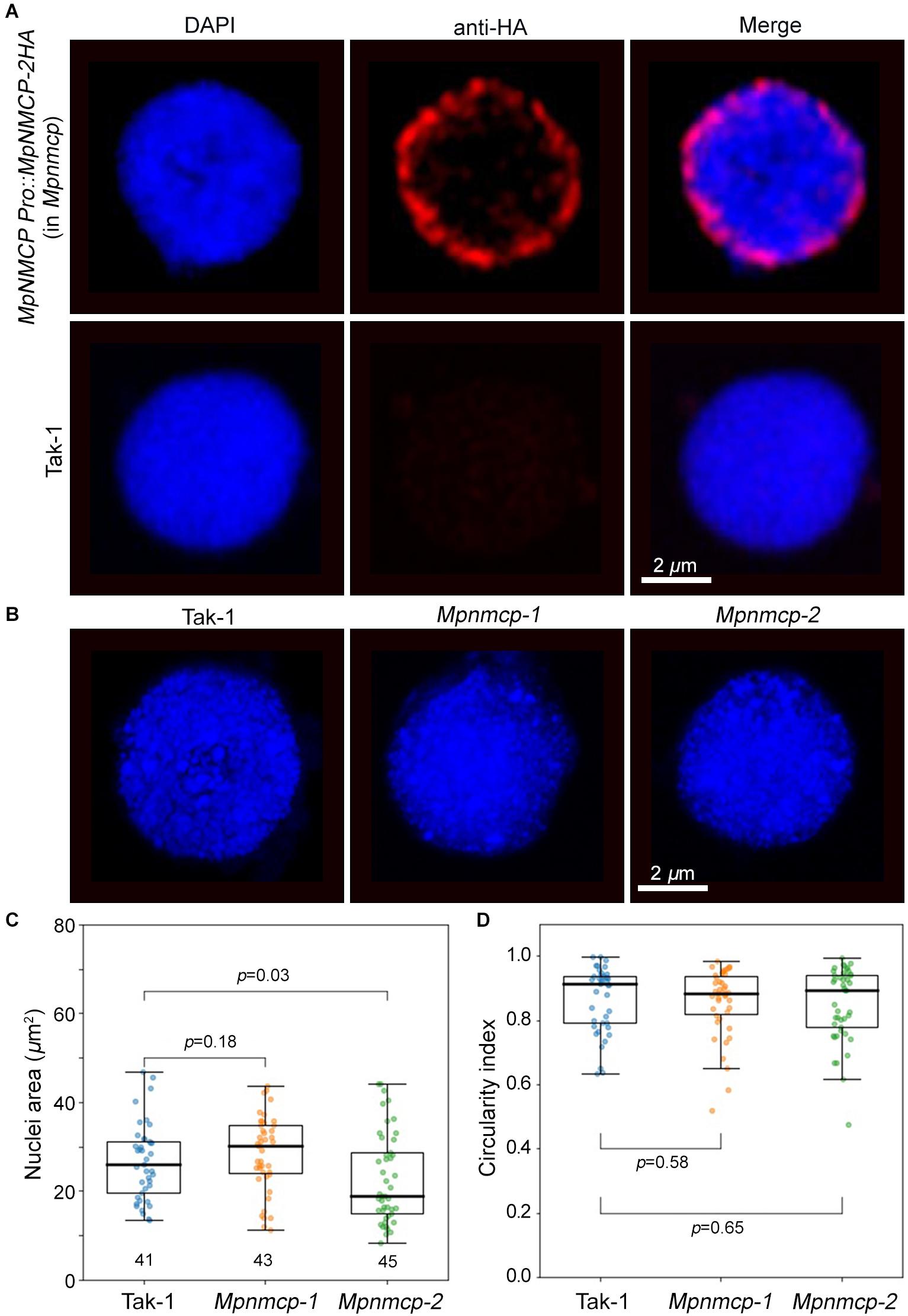
Figure 3. Mpnmcp mutants do not show observable defects in nuclear morphology. (A) Immunohistostaining shows perinuclear localization of MpNMCP:2HA proteins (red, top panel). The image shown in the bottom panels serves as a negative control. 4′,6-Diamidino-2-phenylindole (DAPI) staining (blue) was used for visualizing nuclei. Scale bar shows 2 μm. (B) Representative Takaragaike-1 (Tak-1) (left) and Mpnmcp (middle and right-hand panels) nuclei stained with DAPI. (C,D) Comparison of nuclear morphology. For each genotype, measurements were taken from the maximum z-projected images of around 45 nuclei collected from gemmae to compute their nuclear size (C) and circularity index (D). p values were determined using Mann–Whitney U test.
At the nuclear periphery, Arabidopsis NMCPs CRWN1 and CRWN4 are required to maintain nuclear size and shape; mutant plants losing either CRWN1 or CRWN4 show smaller and round nuclei (Wang et al., 2013). Additionally, overexpression of Arabidopsis CRWN1 can partially deform the nuclear envelope by forming invaginations (Goto et al., 2014). Similar effects with the maize NMCP homolog, NCH1, have also been reported recently (McKenna et al., 2021). The conserved localization of Marchantia NMCP at the NL prompted us to ask if knocking out MpCRWN would result in similar defects in nuclear morphology and whether these defects would underlie the suppressed growth phenotype seen in the Mpnmcp mutants. After quantifying nuclear size and shape, we were not able to detect any differences between Mpnmcp-1 and WT Tak-1 (Figures 3C,D). However, we found that Mpnmcp-2 mutant had smaller nuclei (p = 0.03, Mann–Whitney U test) (Figure 3C). As the two mutant lines did not display consistent changes in nuclear size, we conclude that losing MpNMCP does not cause an effect on nuclear size.
To further examine if MpNMCP proteins possess abilities in shaping nuclei in higher plants, we exogenously over-expressed MpNMCP in the Arabidopsis crwn1 crwn2 double mutant that shows nuclear morphology changes. Loss-of-function of both CRWN1 and CRWN2 had synergistic effects that give rise to much smaller nuclei than crwn1 or crwn2 single mutants (Wang et al., 2013). We examined over 20 independent lines of crwn1 crwn2 35S:MpNMCP:2HA plants that had confirmed MpNMCP:2HA transgene expression (Supplementary Figure 11). We found that all of them developed nuclei with similar morphology compared with crwn1 crwn2, indicating that MpNMCP could not regulate nuclear shape or size in Arabidopsis (Figures 4A–C). In addition, crwn1 crwn2 35S:MpNMCP:2HA seedlings phenocopied crwn1 crwn2 double mutants that showed growth retardation and dwarfism (Figure 4D). This growth phenotype has been related to ectopic activation of biotic stress response (Guo et al., 2017; Choi et al., 2019) part of which involves a hyperactive salicylic acid pathway.
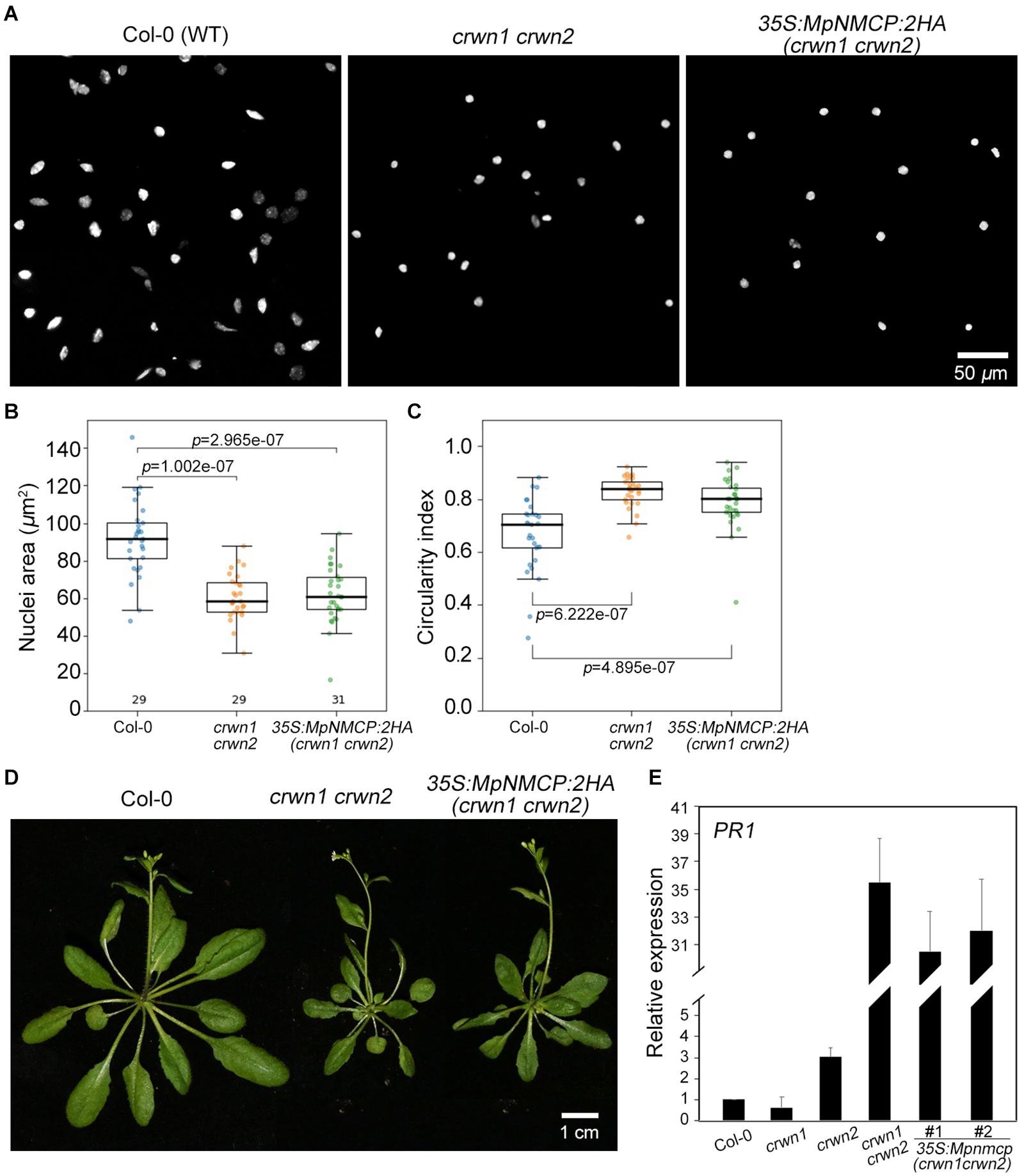
Figure 4. MpNMCP could not complement Arabidopsis crwn mutants. (A) Comparison of 8C nuclei morphology. The nuclei were isolated from the first true leaf of 2-week-old seedlings. 4′,6-Diamidino-2-phenylindole (DAPI) staining (blue) was used for visualization of nuclei. Scale bar, 50 μm. (B,C) Comparison of nuclear morphology. Quantifications of nuclear size (B) and nuclear circularity index (C) in Col-0, crwn1 crwn2, and crwn1 crwn2 35S:MpNMCP-2HA. (D) Phenotypes of transgenic Arabidopsis plants over-expressing MpNMCP:2HA. Scale bar, 1 cm. (E) The relative expression level of PR1 in different mutants detected by real-time PCR. Values were normalized to the expression of ubiquitin 5 (UBQ5). Error bars display ± SD. PR1 expression in individual biological replicates is set as 1.
As our transcriptome analyses revealed a connection between MpNMCP and biotic stress-signaling pathway in Marchantia (Figure 2), we checked if MpNMCP could partially suppress salicylic acid pathway as its Arabidopsis homologs via analyzing the PR1 gene expression. Loss-of-function of CRWN1 and CRWN2 produces a synergistic effect in activating PR1. In 10-day-old crwn1 crwn2 double-mutant seedlings, PR1 was expressed approximately 35 times as high as in WT plants (Figure 4E). Compared with crwn1 crwn2, in crwn1 crwn2 35S:NMCP:2HA, only a subtle decrease in PR1 expression was observed (Figure 4E). Together, these data indicate that MpNMCP could not functionally complement Arabidopsis NMCPs.
V Chromosome Repeats Lost Perinuclear Localization and Decondense in Mpnmcp
In addition to regulating nuclear morphology, Arabidopsis NMCPs also participates in perinuclear chromatin anchoring and heterochromatin organization (Wang et al., 2013; Poulet et al., 2017a; Hu et al., 2019). Our recent study on Arabidopsis CRWN1 revealed that it tethers repressed chromatin domains to the nuclear periphery via direct protein–chromatin interactions (Hu et al., 2019). We hypothesized that if MpNMCP proteins had such a function, Marchantia chromatin regions with preferential localization at the nuclear periphery would show detachment from the NL in Mpnmcp mutants. In most eukaryotic cells, heterochromatin regions are enriched at the nuclear periphery (Buchwalter et al., 2019). Across the Marchantia genome, the V chromosome stands out as a good candidate with potential perinuclear localization patterns, because it carries a much higher level of heterochromatic marks (e.g., H3K9me1 and H3K27me1) and repeats than autosomes (Montgomery et al., 2020). Thus, we used a probe derived from previously identified V chromosome-specific repeats, which had multiple copies located in the 6.4- to 7.5-Mb interval, to examine the nuclear location of the corresponding genomic regions (Okada et al., 2001). The specificity of the probe could be confirmed, as hybridization only occurred in the V chromosome (Figure 5A). With this probe, our fluorescence in situ hybridization (FISH) experiment on Tak-1 thallus nuclei showed preferential localization of the hybridization signal at the nuclear periphery (Figure 5B and Supplementary Figures 4A,B). At times, more than one signal spot was observed, probably reflecting the presence of multiple copies of genomic regions bearing probe sequences (Supplementary Figure 4B).
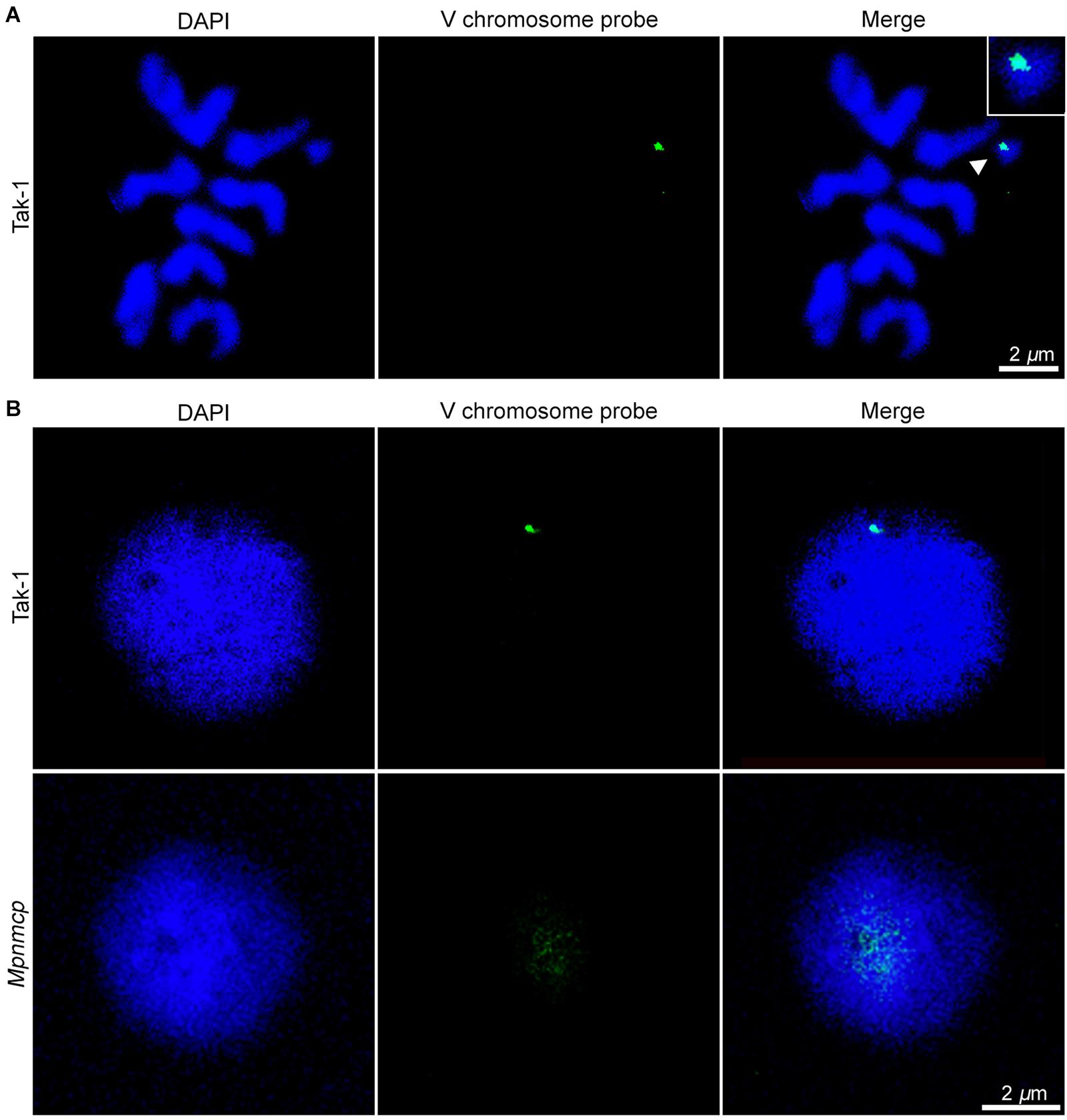
Figure 5. Fluorescence in situ hybridization (FISH) analysis of repeats from the V chromosome. (A) Chromosome spread confirms the specificity of FISH probes to the V chromosome (pointed by an arrowhead). The scale bar represents 2 μm. (B) Distribution of the probed genomic region from V chromosome in Tak-1 (top row) and Mpnmcp mutant (bottom row) nuclei. The nuclei were isolated from vegetative thalli fixed with 0.1% formaldehyde. The scale bar represents 2 μm. Nuclei are counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue).
Surprisingly, we could not detect any hybridization signal in Mpnmcp mutant nuclei with our default FISH protocol (Supplementary Figure 4, third row). We ruled out the possibility that the target genome regions were lost in the Mpnmcp mutant, as the probe could produce positive signals when subjected to chromosome spread FISH (data not shown). Instead, the hybridization of probe to Mpnmcp nuclei appeared to be sensitive to sample preparation, in that a much lower concentration of formaldehyde had been used for nuclear fixation (Figure 5B). With a revised sample fixation procedure, we found that the target genomic regions in Mpnmcp adopted a decondensed configuration and were not positioned at the nuclear periphery, suggesting that Marchantia NMCP functions in perinuclear chromatin tethering (Figure 5B).
Next, we asked if the V chromosome as a whole had preferential localization at the nuclear periphery. As tiling bacterial artificial chromosome (BAC) clones covering the V chromosome were not available, we approximated V chromosome FISH by mixing labeled Tak-1 genomic DNA with excessive unlabeled Tak-2 genomic DNA so that the latter could suppress autosome DNA hybridizing with the probes. In the absence Tak-2 DNA, FISH signals were found throughout Tak-1 nuclei (Supplementary Figure 5A). By contrast, the addition of Tak-2 DNA resulted in specific signals occupying a subdomain in the Tak-1 nucleus, which presumably corresponded to the V chromosome territory (Supplementary Figure 5B). Overall, most of the V chromosome FISH signals did not show contacts with the nuclear periphery (Supplementary Figure 5B). In summary, perinuclear chromatin anchoring was limited to parts of the V chromosome.
Mpnmcp Genome Does Not Show Drastic Conformational Changes
Given conformational changes (decondensation) in the abovementioned repeat region at the V chromosome in Mpnmcp mutants, we performed Hi-C experiment to compare their genome-wide chromatin organization patterns with those of WT Tak-1 (Supplementary Figure 6). Because the repeat region (6.5–7.5 Mb) at the V chromosome had poor read mapping quality, it was masked during Hi-C map normalization. Therefore, we could not examine how the V chromosome chromatin packing pattern was or how it interacted with other genomic regions. The rest part of the V chromosome, based on manual inspection, did not show drastic conformational changes, including topologically associated domains (TADs) that we reported in an earlier study (Figure 6; Montgomery et al., 2020). We further performed quantitative analyses to assess possible chromatin packing changes. We reasoned that if the V chromosome in Mpnmcp became decondensed, it would gain more contacts with autosomes and show weaker long-range intra-chromosomal interactions. Compared with WT Tak-1, we found that genomic regions along the V chromosome in Mpnmcp did not exhibit an altered level of trans-interactions (Figure 6B). Additionally, we found that the V chromosome did not show decreased long-range intra-chromosomal interactions, which were approximated from compactness values (sum of cis-contacts up to 200 kb) from Hi-C map normalized at 10-kb resolution (Figure 6C). These results did not support our hypothesis that other parts at the V chromosome adapted a decondensed state in Mpnmcp mutants.
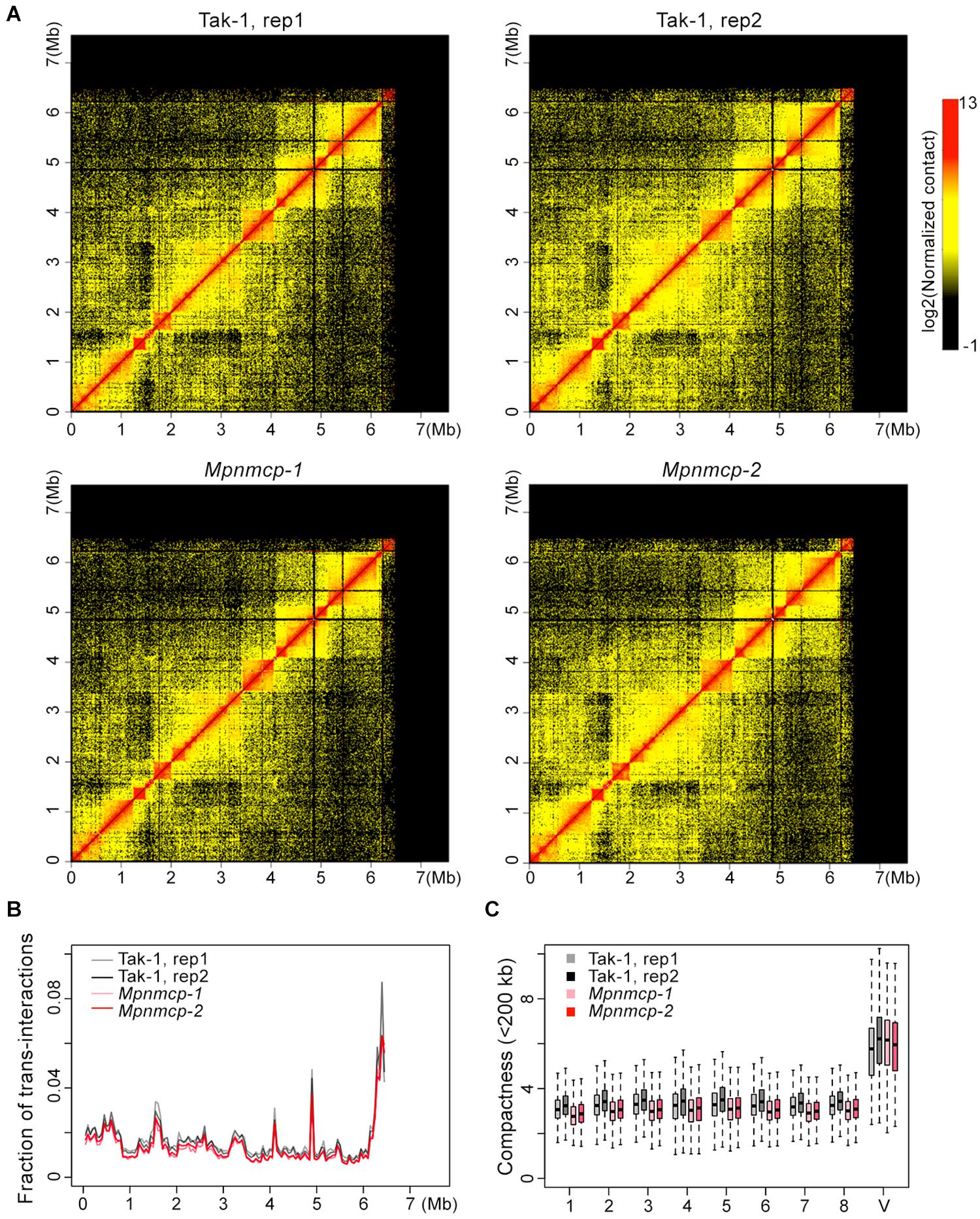
Figure 6. Comparison of the V chromosome organization patterns in wild-type (WT) Tak-1 and Mpnmcp. (A) Hi-C maps of the V chromosome normalized at 10-kb resolution. (B) Inter-chromosomal interaction patterns across the V chromosome. (C) Comparison of chromatin compactness of individual chromosomes derived from Hi-C maps.
However, individual autosomes in Mpnmcp showed a mild drop in their compactness values, suggesting that they became slightly decondensed in the mutants (Figure 6C). We further calculated interaction decay exponents of the autosomes (up to 2.5 Mb), which was a measure of how fast chromatin cis-contacts drop with increasing genomic distance. Consistent with their compactness values, autosomes in Mpnmcp showed a slightly faster decay of chromatin contact strength as genomic distance increased (Supplementary Figure 7A). Nevertheless, such nuanced chromatin decondensation was not accompanied with elevated inter-chromosomal interactions at autosomes (Supplementary Figure 7B). Altogether, we conclude that in spite of having chromatin decondensation at the end of the V chromosome (approximately 6.5 to 7.5 Mb), lost-of-function of MpNMCP did not result in drastic alteration in genomic organization.
Discussion
MpNMCP Is Not Involved in Regulating Nuclear Morphology
In this study, we functionally characterized the sole NMCP homolog encoded by the Marchantia genome and compared it with AtCRWN1. While the MpNMCP proteins have similar domain architecture and identical nuclear localization patterns to AtCRWN1 (N-terminal conserved coiled-coil domains), it functions differently in liverwort.
Early functional characterization of AtCRWN1 unveiled its role in regulating nuclear morphology (Dittmer et al., 2007). Functional studies of other NMCPs have been conducted on multiple plants, including rice (Yang et al., 2020), maize (McKenna et al., 2021), and onion (Ciska et al., 2013); yet it is unknown whether the corresponding NMCP genes in these plants play a structural role in the nucleus. In this study, we showed that although MpNMCP proteins were located at the nuclear periphery (Figure 3A), the Mpnmcp mutant does not show changes in nuclear size or shape (Figures 3B–D). While measuring nuclear sizes, we found noticeable variation of data values (Figure 3C), and in our opinion, this was due to sampling of different cell types. At the moment, we do not have Marchantia cell-lineage maker lines that allow us to analyze nuclear morphology of a specific cell type. However, according to studies in Arabidopsis, the effect of losing CRWNs (i.e., smaller nuclei) can be found consistently in multiple cell types (Poulet et al., 2017a). Therefore, we expected a shift in nuclear size, regardless of cell type in Mpnmcp mutants if the Marchantia NMCP homolog is required for nuclear size regulation. As a statistically significant difference was only observed in one of the Mpnmcp mutant line (Figure 3C), we conclude that losing MpNMCP does not influence the nuclear size.
Furthermore, MpNMCP could not rescue the Arabidopsis crwn1 crwn2 mutant phenotype (Figure 4A). It should be noted that the absence of functional complementation of crwn1 crwn2 by MpNMCP might be attributed to other technical issues; for example, Arabidopsis plant cannot generate post-transcriptional modifications or homologous proteins that are important for MpNMCP to be fully functional. Nevertheless, together with nuclear morphology analysis on Mpnmcp mutants, these results suggest that the nuclear morphology in Marchantia is not controlled by MpNMCP but by other structural proteins. In higher plants, two other types of putative nuclear lamina-like genes that regulate nuclear size are KAKU4 (Goto et al., 2014) and Nuclear Envelope-Associated Proteins (NEAPs) (Pawar et al., 2016). Our sequence analysis revealed that the Marchantia genome encodes no KAKU4 homologs but one putative NEAP (Mp4g20150). It is therefore tempting to speculate a potential structural role of MpNEAP concerning nuclear morphology regulation.
Conserved Function of MpNMCP in Stress Signaling
Both Arabidopsis and rice NMCP proteins have been found to be involved in stress signaling. AtCRWNs have also been shown to regulate transcriptional levels of stress and defense response genes (Guo et al., 2017; Choi et al., 2019; Choi and Richards, 2020). crwn1 crwn2 and crwn1 crwn4 mutants showed greater resistance to infection with virulent bacterial pathogens and increased Pathogenesis-Related1 (PR1), PR2, and PR5 transcript levels (Guo et al., 2017; Choi et al., 2019). PR1 expression is inhibited by the NAC transcription factor-like 9 (NTL9). NTL9 interacts with AtCRWN1 and Suppressor of NPR1 inducible 1 (SNI1), possibly forming a protein complex to suppress PR1 expression and inhibit the activity of NPR1, a master regulator of pathogen resistance in Arabidopsis (Kim et al., 2012; Guo et al., 2017). OsNMCP1 has also been associated with drought tolerance. Overexpression of OsNMCP1 improves rice drought resistance and results in deeper and thicker root systems, which are of pivotal importance in alleviating water stress (Yang et al., 2020).
Our study reveals a connection between MpNMCP and stress response in Marchantia. First, by comparing DEGs in Mpnmcp mutants and several published stress-related Marchantia transcriptome datasets, we found a noticeable overlap between Mpnmcp DEGs and stress-related DEGs (Figure 2A). Second, we found transcriptome resemblance between Mpnmcp and Mpjaz mutants (Figure 2B). The expression of MpCHL, which is the precursor of JA biosynthesis (12-oxo-phytodienoic acid, OPDA)-marker gene, was strongly promoted (ca. 50-fold) in Mpnmcp (Supplementary Figure 3). Such gene upregulation, however, is considerably lower than that in the Mpjaz mutants (around 4,000-fold) (Monte et al., 2019). Besides, compared with those in Mpjaz, changes of the relative expression of another two JA marker genes, MpAOS2 and MpAOC, were much attenuated in Mpnmcp (Supplementary Figure 3). These data suggest a partially activated JA pathway in the Mpnmcp mutants. Interestingly, loss-of-function of AtCRWN1 also results in activated JA pathway in Arabidopsis (Jarad et al., 2019). JAs are lipid-derived hormones released as chemical signals required for wounding response, defense against pathogens, and development (Huang et al., 2017). Recent studies show that Marchantia utilizes a conserved “JA” signaling module that functions through OPDA rather than JA (Wasternack and Strnad, 2016; Monte et al., 2018). Our results imply that, in the absence of MpNMCP, the OPDA may step in to help Marchantia defend itself against pathogens. Overall, it appears that the MpNMCP have largely retained their function in the sphere of modulating stress response. Alternatively, the Mpnmcp mutant transcriptome might largely reflect secondary defects downstream of primary effects from losing MpNMCP. To obtain better insights into how this is linked to stress response, one can use an inducible system, for example, estradiol-mediated gene knock-down (Flores-Sandoval et al., 2016), to study immediate downstream events in the nucleus after losing MpNMCP activities.
MpNMCP Indirectly Regulates Perinuclear Location of Chromatin
We also used our functional MpNMCP:2HA tagging lines to perform ChIP-seq (chromatin immunoprecipitation coupled to sequencing) to assess if MpNMCP proteins directly participated in perinuclear chromatin anchoring. We speculated that the 6.5- to 7.5-Mb interval region at the V chromosome, which showed preferential localization at the nuclear periphery (Figure 5), would be a good candidate. However, analysis of ChIP-seq libraries did not reveal such enrichment in this genomic interval. Moreover, inconsistent ChIP signals between individual MpNMCP:2HA tagging lines were found across the autosomes (data not shown). The ChIP experiments were performed in accordance with our working protocol that we used for MpTCP1 proteins (Karaaslan et al., 2020b), and we verified successful immunoprecipitation of MpNMCP:2HA proteins with Western blot (Supplementary Figure 8). Thus, our data suggest that MpNMCP proteins do not directly interact with chromatin at the nuclear periphery. Still, we cannot fully rule out technical issues generating such negative ChIP results. For instance, MpNMCP proteins might change their conformation or interact with other proteins upon binding to chromatin, leading to epitope masking that prevents immunoprecipitation. Nonetheless, in our view, the negative ChIP-seq results are in favor of a hypothesis that MpNMCP is not involved in organizing genome folding at the nuclear periphery like the Arabidopsis CRWN1. This notion is supported by our Hi-C data that there was no increase of inter-chromosomal contacts in Mpnmcp mutant nuclei (Figure 6B and Supplementary Figure 6), which were otherwise observed in mutants having deficiency in perinuclear chromatin tethering (Hu et al., 2019; Chang et al., 2020). Nevertheless, it is clear that MpNMCP is required for proper chromatin organization, at least for the repeat region at the V chromosome, since it detached from the nuclear periphery and became decondensed in Mpnmcp mutants (Figure 5). Such regulation at limited genomic regions is likely to be indirect. The conformational changes of this V chromosome region might be a secondary consequence of losing MpNMCP, for example, due to the thallus growth retardation and the alteration of transcriptome in Mpnmcp mutants.
Materials and Methods
Plant Material and Plasmid Construction
Marchantia polymorpha Tak-1 (male accession) was used as WT (Ishizaki et al., 2008). Plants were cultivated on half-strength Gamborg’s B5 medium containing 1% agar. For growth analysis, gemmae were grown in a controlled environment room at 23°C in long-day conditions (8-h dark and 16-h light) under white light unless specified otherwise.
CRISPR/Cas9 mutations were generated as previously described in Sugano et al. (2014) using pMpGE_En03, pMpGE010 for gRNA1, and pMpGE011 for gRNA2 (Addgene entry nos. 71535, 71536, and 71537). A double CRISPR/Cas9 approach was carried out to delete a 3-kb genomic fragment at the MpNMCP locus, which included most of its protein-coding region (Figure 1). In this regard, two guide RNAs (sgRNA1, 5′-GCAGG CATGCAGAGTCCGAGAGG-3′, and sgRNA2, 5′-TCACACCG AAGACACCACCAGGG-3′; Supplementary Table 2) were generated. Transformation of spores with assembled vector containing the CRISPR/Cas9 and the two sgRNA expression cassettes was performed by following the protocol described in Kubota et al. (2013). T0 plants were selected on plates containing 100 μg/ml of cefotaxime, 10 μg/ml of hygromycin, and 0.5 μM of chlorsulfuron; individual candidate plants with deletion at the MpNMCP locus were identified by genotyping PCR.
To generate the MpNMCP:2HA complementation construct, the MpNMCP CDS was amplified using Tak-1 cDNA as a template with oligos 5′-TGTACAAAAAAGCAGGCTTTACT GCTAAGTGCTGTGAGAGAA-3′ and 5′-CTTTGTACAAGA AAGCTGGGTTACGTGATTAAGAAGTCCCAG-3′; its 2-kb upstream region was amplified by using Tak-1 genomic DNA with oligos 5′-TGTACAAAAAAGCAGGCTTTAGACCT GAATGTAAACCACCG-3′ and 5′-GTGCCACTTCTCTCAC AGCACTTAGCAGTCTTTCTTTCTA-3′. Next, the amplified MpNMCP coding sequence was mutagenized with the following two pairs of oligos: first, 5′-GAGCCTGAGACGTACGGACG GACTAGCTTCTGCACGGATAGCTTG-3′ and 5′-CGTCCGT ACGTCTCAGGCTCAGGAAGGTGCAGGAGGCGGA-3′, and second, 5′-AGGGTATCCAGCATAATCTGGTACGTCGTA TGGGTATCCTTCTTTCCCAGAATCTCTCAC-3′ and 5′-GA TTATGCTGGATACCCTTACGACGTACCAGATTACGCTCAG TGTGGCTTCTGTGGTGGTGGGCCAGGTACTAGTACTG-3′. Each pair of these mutagenesis oligos introduced silent mutations to the MpNMCP coding region to prevent it from being recognized by sgRNAs. The second pair of oligos also inserted a 2HA tag to create the MpNMCP:2HA fusion protein. Finally, the resulting expression cassette was further subcloned into pMpGWB301 (Ishizaki et al., 2015) and was used to rescue Mpnmcp mutants.
To generate MpNMCP Pro:GUS construct, MpNMCP promoter (2-kb upstream of gene model) was amplified using Tak-1 genomic DNA with oligos 5′-TGTAC AAAA AAGCA GGCTT TAGA CCTGA ATGT AAAC CACCG-3′ and 5′-GTGCCAC TTCTC TCACAG CACTT AG CAGT CTTTC TTT CTA-3′; the resulting PCR product was subcloned into pMpGWB224 (Ishizaki et al., 2015).
Glucuronidase Staining
GUS assays of MpNMCP Pro:GUS transgenic thalli were performed as described previously (Althoff et al., 2014). Plants were fixed with 90% ice-cold acetone for 20 min. Fixed plants were immersed in GUS staining solution (100 mM of potassium ferrocyanide, 100 mM of potassium ferricyanide, and 1 mM of X-Gluc), placed under vacuum for 5 min, incubated for 3 h at 37°C, and cleared with ethanol.
Fluorescence in situ Hybridization
The respective repeat of V chromosome (Okada et al., 2001) was amplified from genomic DNA of Tak-1, with oligos 5′-GGATCCGGATGCCGAATATAT-3′ and 5′-GGATCCGCGAAACTTCTGGG-3′ (Supplementary Table 2). To label probes, PCR amplification was carried out using DNA polymerase with 50% of dTTP replaced with digoxigenin-11-dUTP (Product Number 3359247910, Roche, United States) according to the manufacturer’s instructions. The test of probe male-specificity was done by chromosome FISH and was detected with Alexa Fluor 488 (Green) (Abcam, Cat# ab150077).
Preparation of chromosome spreads from young thalli of Marchantia and FISH were performed following a standard procedure with minor modifications (Nakayama et al., 2001; Okada et al., 2001; Yamato et al., 2007). In brief, for chromosome preparation, 2-week-old thalli were fixed by a fixative (ethanol:acetic acid 3:1) at 4°C for 30 min. Then, the fixative was replaced with 70% ethanol and stored at 4°C overnight. Fixed thalli were washed with de-ionized water, followed by the enzyme maceration at 37°C for 1 h. After being rinsed, each piece of thallus was laid onto a clean glass slide, and cells were spread by an easy “SteamDrop” method (Kirov et al., 2014). The next FISH steps were performed exactly as described in the method part of Okada et al. (2000). Briefly, 25 ng digoxigenin (DIG)-labeled V chromosome targeting probe was further hybridized with preparations in 20 ml of a hybridization mixture (Bi et al., 2017). The mixture was denatured at 85°C for 10 min and transferred to preparations. Then, the preparations were heated at 85°C for 30 min and hybridized in a hybridizer (Agilent, United States) at 37°C overnight. For FISH probe detection, Monoclonal Anti-Digoxin (Sigma, Catalog # D8156) and Alexa Fluor 488-conjugated goat anti-Mouse IgG (Invitrogen, Catalog # A-11017) were used as the primary and secondary antibodies, respectively.
DNA FISH in Marchantia nuclei was adapted from Bi et al. (2017). Around 5,000 nuclei were sorted with a Beckman Coulter MofFlo XDP as described in Karaaslan et al. (2020a). The sorted nuclei were centrifuged for 3,000 × g at 4°C for 7 min. The supernatant was discarded after centrifugation, and the pellet was resuspended in 30 μL of 1 × phosphate-buffered saline (PBS). The nuclei were incubated at 65°C for 30 min and mixed with 5 μL of 0.1 mg/ml RNase A. A circle (∼1 cm2) was drawn on the Superfrost Ultra Plus Adhesion Slide (Thermo Fisher Scientific) using a DAKO pen to avoid deforming the specimen once mounted with a coverslip. All the suspensions were transferred onto the slide and incubated in a moist chamber at 37°C. At the end of RNase A treatment, the nuclei became attached to the glass slide. Next, the slide was rinsed in 2 × SSC and dehydrated in a graded series of alcohol solutions at room temperature (RT). Twenty microliters of the probe mixture (same contents as in chromosome FISH) was denatured at 95°C for 10 min and transferred immediately to ice. Twenty microliters of denatured probe was added to the slide and hybridized in a moist chamber at 37°C overnight. All subsequent steps, including washing and detection, were strictly followed (Bi et al., 2017).
Immunohistostaining
Around 5,000 sorted nuclei from 1% formaldehyde fixed Tak-1 or MpNMCP Pro:MpNMCP-2HA seedlings were used. After the nuclei were treated with RNase A at 37°C for 1 h, the nuclei were washed three times with PBS. Then, an antigen retrieval step was performed with Universal HIER antigen retrieval reagent (Abcam) by following manufacturer’s instructions. The nuclei were then incubated in blocking solution [5% bovine serum albumin (BSA) in 0.2% Tween 20 4 × SSC] and then mounted unto glass microscopy slides. Slides were incubated with HA Tag Alexa Fluor 647 conjugate (Thermo Fisher Scientific) at 1:500 diluted with blocking solution at 37°C for 1 h. After three washes with PBS, slides were mounted with SlowFade Diamond Antifade Mountant, to which 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, United States) was added to visualize nuclei (Sigma-Aldrich, St. Louis, United States).
Fluorescence Microscopy and Image Processing
All confocal image acquisition of nuclei were performed on an LSM 880 confocal laser scanning microscope (Zeiss) with Airyscan system equipped with a 63 × /1.4 numerical aperture (NA) water objective. For the processing of acquired images, ImageJ (NIH) was used to pseudocolor and merge.
In situ Hi-C and Data Processing
The in situ Hi-C library preparation was strictly followed as described (Karaaslan et al., 2020b). In total, two replicates of 2-week-old Tak-1 and Mpnmcp mutant thallus Hi-C libraries were made, and for each replicate, around 0.5 g of fixed sample was homogenized for nuclei isolation. The libraries were sequenced on an Illumina HiSeq 3000 instrument with 2 × 150 bp reads. Hi-C reads mapping, filtering, and Hi-C map normalization were done according to our recent Hi-C study (Karaaslan et al., 2020b). The bin size setting for genome-wide and individual chromosome Hi-C maps was 50 and 10 kb, respectively.
RNA-seq Library Preparation and Analysis
RNA-seq was performed with three biological replicates per sample. Total RNA was extracted from 23-day-old Mpnmcp mutants or Tak-1 thalli using an RNeasy Plant Mini Kit (Qiagen). After treatment with DNase I (Thermo Scientific, Waltham, MA, United States), the RNA was mixed with first-strand buffer (Invitrogen), heated at 80°C for 2 min followed by 94°C for 1.5 min. Then the Poly A+ RNA was purified from the total RNA and processed strictly according to manufacturer’s recommendations. After performing end repair, the free end of the newly synthesized double-stranded cDNA was ligated to the reverse adaptor, and final enrichment PCR was performed to generate libraries for sequencing.
RNA-seq reads were aligned against the Tak-1 v5 genome using TopHat 2 (v2.1.1). DEGs were identified with the DESeq2 package in R. The criteria for calling DEGs was set as false discovery rate smaller than 0.01 and expression fold change more than 3. Details of the reads count table and DEGs can be found in Supplementary Table 1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA692743. Large datasets including Hi-C matrices (10 kb bin size for individual chromosomes, 50 kb bin size for the whole genome) are accessible with the following link: https://figshare.com/articles/dataset/Characterization_of_a_plant_lamin-like_protein_in_Marchantia/13568276. The following public RNA-seq datasets were downloaded and re-analyzed as for those generated in this study: Phytophthora palmivora infection (PRJNA397637, https://www.ncbi.nlm.nih.gov/sra), and salt stress (PRJDB6783, https://ddbj.nig.ac.jp). DEGs from in the Mpjaz mutants, which were based on a microarray experiment, were from Monte et al. (2019); we converted their gene annotation to the Tak1 v5 format with an online tool at https://marchantia.info/utils/id_converter/.
Author Contributions
CL conceived this study. CL and NW designed the experiments. NW performed most of the experiments. NW, EK, and NF established and characterized transgenic lines. KB performed nuclei sorting. CL and NW prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Deutsche Forschungsgemein- schaft (LI 2862/4) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 757600).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank members of the COST Action CA1612 INDEPTH network for fruitful discussion. We would also like to thank computing support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant no. INST 37/935-1 FUGG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.670306/full#supplementary-material
Supplementary Figure 1 | Sequence alignment of NMCP homologs.
Supplementary Figure 2 | RNA-seq analysis of Mpnmcp mutant lines.
Supplementary Figure 3 | Relative expression of selected jasmonate marker genes.
Supplementary Figure 4 | Perinuclear localization of a V chromosome region.
Supplementary Figure 5 | FISH experiment of the entire V-chromosome.
Supplementary Figure 6 | Genome-wide Hi-C maps of WT Tak-1 and Mpnmcp mutant thalli.
Supplementary Figure 7 | Comparison of chromatin organization patterns of autosomes in WT Tak-1 and Mpnmcp mutants.
Supplementary Figure 8 | Western blot confirms MpNMCP-2HA protein IP.
Supplementary Figure 9 | Fluorescence-activated cell sorting (FACS) of 8C nuclei.
Supplementary Figure 10 | Thalli area measurements of WT Tak-1, Mpnmcp mutants and Mpnmcp complementation lines.
Supplementary Figure 11 | Semi-quantitative RT-PCR showing the expression of MpNMCP in the crwn1 crwn2 35S:MpNMCP-2HA plants.
Supplementary Table 1 | Read count and DEGs tables.
Supplementary Table 2 | Sequences of primer pairs used in this study.
Supplementary Data Set 1 | The NMCP sequences used to reconstruct Supplementary Figure 1.
References
Althoff, F., Kopischke, S., Zobell, O., Ide, K., Ishizaki, K., Kohchi, T., et al. (2014). Comparison of the MpEF1alpha and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic. Res. 23, 235–244. doi: 10.1007/s11248-013-9746-z
Bi, X., Cheng, Y. J., Hu, B., Ma, X., Wu, R., Wang, J. W., et al. (2017). Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 27, 1162–1173. doi: 10.1101/gr.215186.116
Brueckner, L., Zhao, P. A., van Schaik, T., Leemans, C., Sima, J., Peric-Hupkes, D., et al. (2020). Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 39:e103159. doi: 10.15252/embj.2019103159
Buchwalter, A., Kaneshiro, J. M., and Hetzer, M. W. (2019). Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 20, 39–50. doi: 10.1038/s41576-018-0063-5
Carella, P., Gogleva, A., Hoey, D. J., Bridgen, A. J., Stolze, S. C., Nakagami, H., et al. (2019). Conserved Biochemical Defenses Underpin Host Responses to Oomycete Infection in an Early-Divergent Land Plant Lineage. Curr. Biol. 29, 2282–2294.e2285. doi: 10.1016/j.cub.2019.05.078
Chang, L., Li, M., Shao, S., Li, C., Ai, S., Xue, B., et al. (2020). Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell 2020:8. doi: 10.1007/s13238-020-00794-8
Chen, C. K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. doi: 10.1126/science.aae0047
Choi, J., and Richards, E. J. (2020). The role of CRWN nuclear proteins in chromatin-based regulation of stress response genes. Plant Signal. Behav. 15:1694224. doi: 10.1080/15592324.2019.1694224
Choi, J., Strickler, S. R., and Richards, E. J. (2019). Loss of CRWN Nuclear Proteins Induces Cell Death and Salicylic Acid Defense Signaling. Plant Physiol. 179, 1315–1329. doi: 10.1104/pp.18.01020
Ciska, M., Hikida, R., Masuda, K., and Moreno Diaz de la Espina, S. (2019). Evolutionary history and structure of nuclear matrix constituent proteins, the plant analogues of lamins. J. Exp. Bot. 70, 2651–2664. doi: 10.1093/jxb/erz102
Ciska, M., Masuda, K., and Moreno Diaz de la Espina, S. (2013). Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J. Exp. Bot. 64, 1553–1564. doi: 10.1093/jxb/ert020
Crane, E., Bian, Q., McCord, R. P., Lajoie, B. R., Wheeler, B. S., Ralston, E. J., et al. (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244. doi: 10.1038/nature14450
Dechat, T., Pfleghaar, K., Sengupta, K., Shimi, T., Shumaker, D. K., Solimando, L., et al. (2008). Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853. doi: 10.1101/gad.1652708
Dittmer, T. A., and Misteli, T. (2011). The lamin protein family. Genome Biol. 12:222. doi: 10.1186/gb-2011-12-5-222
Dittmer, T. A., Stacey, N. J., Sugimoto-Shirasu, K., and Richards, E. J. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19, 2793–2803. doi: 10.1105/tpc.107.053231
Flores-Sandoval, E., Dierschke, T., Fisher, T. J., and Bowman, J. L. (2016). Efficient and Inducible Use of Artificial MicroRNAs in Marchantia polymorpha. Plant Cell Physiol. 57, 281–290. doi: 10.1093/pcp/pcv068
Goto, C., Tamura, K., Fukao, Y., Shimada, T., and Hara-Nishimura, I. (2014). The Novel Nuclear Envelope Protein KAKU4 Modulates Nuclear Morphology in Arabidopsis. Plant Cell 26, 2143–2155. doi: 10.1105/tpc.113.122168
Guo, T., Mao, X., Zhang, H., Zhang, Y., Fu, M., Sun, Z., et al. (2017). Lamin-like Proteins Negatively Regulate Plant Immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol. Plant 10, 1334–1348. doi: 10.1016/j.molp.2017.09.008
Herrmann, H., Strelkov, S. V., Burkhard, P., and Aebi, U. (2009). Intermediate filaments: primary determinants of cell architecture and plasticity. J. Clin. Invest. 119, 1772–1783. doi: 10.1172/JCI38214
Hu, B., Wang, N., Bi, X., Karaaslan, E. S., Weber, A. L., Zhu, W., et al. (2019). Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 20:87. doi: 10.1186/s13059-019-1694-3
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Ishizaki, K., Chiyoda, S., Yamato, K. T., and Kohchi, T. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. doi: 10.1093/pcp/pcn085
Ishizaki, K., Nishihama, R., Ueda, M., Inoue, K., Ishida, S., Nishimura, Y., et al. (2015). Development of Gateway Binary Vector Series with Four Different Selection Markers for the Liverwort Marchantia polymorpha. PLoS One 10:e0138876. doi: 10.1371/journal.pone.0138876
Jarad, M., Mariappan, K., Almeida-Trapp, M., Mette, M. F., Mithofer, A., Rayapuram, N., et al. (2019). The Lamin-Like LITTLE NUCLEI 1 (LINC1) Regulates Pattern-Triggered Immunity and Jasmonic Acid Signaling. Front. Plant Sci. 10:1639. doi: 10.3389/fpls.2019.01639
Karaaslan, E. S., Faiss, N., Liu, C., and Berendzen, K. W. (2020a). Isolation of Lineage Specific Nuclei Based on Distinct Endoreduplication Levels and Tissue-Specific Markers to Study Chromatin Accessibility Landscapes. Plants 9:91114678. doi: 10.3390/plants9111478
Karaaslan, E. S., Wang, N., Faiss, N., Liang, Y., Montgomery, S. A., Laubinger, S., et al. (2020b). Marchantia TCP transcription factor activity correlates with three-dimensional chromatin structure. Nat. Plants 6, 1250–1261. doi: 10.1038/s41477-020-00766-0
Kim, H. S., Park, H. C., Kim, K. E., Jung, M. S., Han, H. J., Kim, S. H., et al. (2012). A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 40, 9182–9192. doi: 10.1093/nar/gks683
Kirov, I., Divashuk, M., Van Laere, K., Soloviev, A., and Khrustaleva, L. (2014). An easy “SteamDrop” method for high quality plant chromosome preparation. Mol. Cytogenet. 7:21. doi: 10.1186/1755-8166-7-21
Kubota, A., Ishizaki, K., Hosaka, M., and Kohchi, T. (2013). Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Biochem. 77, 167–172. doi: 10.1271/bbb.120700
Masuda, K., Xu, Z. J., Takahashi, S., Ito, A., Ono, M., Nomura, K., et al. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp. Cell Res. 232, 173–181. doi: 10.1006/excr.1997.3531
Matsui, H., Iwakawa, H., Hyon, G. S., Yotsui, I., Katou, S., Monte, I., et al. (2020). Isolation of Natural Fungal Pathogens from Marchantia polymorpha Reveals Antagonism between Salicylic Acid and Jasmonate during Liverwort-Fungus Interactions. Plant Cell Physiol. 61:442. doi: 10.1093/pcp/pcz214
McKenna, J. F., Gumber, H. K., Turpin, Z. M., Jalovec, A. M., Kartick, A. C., Graumann, K., et al. (2021). Maize (Zea mays L.) Nucleoskeletal Proteins Regulate Nuclear Envelope Remodeling and Function in Stomatal Complex Development and Pollen Viability. Front. Plant Sci. 12:645218. doi: 10.3389/fpls.2021.645218
McNulty, A. K., and Saunders, M. J. (1992). Purification and immunological detection of pea nuclear intermediate filaments: evidence for plant nuclear lamins. J. Cell Sci. 103(Pt 2), 407–414.
Meier, I., Griffis, A. H., Groves, N. R., and Wagner, A. (2016). Regulation of nuclear shape and size in plants. Curr. Opin. Cell Biol. 40, 114–123. doi: 10.1016/j.ceb.2016.03.005
Mikulski, P., Hohenstatt, M. L., Farrona, S., Smaczniak, C., Stahl, Y., Kalyanikrishna, et al. (2019). The Chromatin-Associated Protein PWO1 Interacts with Plant Nuclear Lamin-like Components to Regulate Nuclear Size. Plant Cell 31, 1141–1154. doi: 10.1105/tpc.18.00663
Minguez, A., and Moreno Diaz de la Espina, S. (1993). Immunological characterization of lamins in the nuclear matrix of onion cells. J. Cell Sci. 106(Pt 1), 431–439.
Monte, I., Franco-Zorrilla, J. M., Garcia-Casado, G., Zamarreno, A. M., Garcia-Mina, J. M., Nishihama, R., et al. (2019). A Single JAZ Repressor Controls the Jasmonate Pathway in Marchantia polymorpha. Mol. Plant 12, 185–198. doi: 10.1016/j.molp.2018.12.017
Monte, I., Ishida, S., Zamarreno, A. M., Hamberg, M., Franco-Zorrilla, J. M., Garcia-Casado, G., et al. (2018). Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 14, 480–488. doi: 10.1038/s41589-018-0033-4
Montgomery, S. A., Tanizawa, Y., Galik, B., Wang, N., Ito, T., Mochizuki, T., et al. (2020). Chromatin Organization in Early Land Plants Reveals an Ancestral Association between H3K27me3. Transposons, and Constitutive Heterochromatin. Curr. Biol. 30:e577. doi: 10.1016/j.cub.2019.12.015
Nakayama, S., Fujishita, M., Sone, T., and Ohyama, K. (2001). Additional locus of rDNA sequence specific to the X chromosome of the liverwort, Marchantia polymorpha. Chromosome Res. 9, 469–473. doi: 10.1023/a:1011676328165
Okada, S., Fujisawa, M., Sone, T., Nakayama, S., Nishiyama, R., Takenaka, M., et al. (2000). Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J. 24, 421–428. doi: 10.1046/j.1365-313x.2000.00882.x
Okada, S., Sone, T., Fujisawa, M., Nakayama, S., Takenaka, M., Ishizaki, K., et al. (2001). The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. U S A. 98, 9454–9459. doi: 10.1073/pnas.171304798
Pawar, V., Poulet, A., Detourne, G., Tatout, C., Vanrobays, E., Evans, D. E., et al. (2016). A novel family of plant nuclear envelope-associated proteins. J. Exp. Bot. 67, 5699–5710. doi: 10.1093/jxb/erw332
Poulet, A., Duc, C., Voisin, M., Desset, S., Tutois, S., Vanrobays, E., et al. (2017a). The LINC complex contributes to heterochromatin organisation and transcriptional gene silencing in plants. J. Cell Sci. 130, 590–601. doi: 10.1242/jcs.194712
Poulet, A., Probst, A. V., Graumann, K., Tatout, C., and Evans, D. (2017b). Exploring the evolution of the proteins of the plant nuclear envelope. Nucleus 8, 46–59. doi: 10.1080/19491034.2016.1236166
Prokocimer, M., Davidovich, M., Nissim-Rafinia, M., Wiesel-Motiuk, N., Bar, D. Z., Barkan, R., et al. (2009). Nuclear lamins: key regulators of nuclear structure and activities. J. Cell Mol. Med. 13, 1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x
Sakamoto, Y., Sato, M., Sato, Y., Harada, A., Suzuki, T., Goto, C., et al. (2020). Subnuclear gene positioning through lamina association affects copper tolerance. Nat. Commun. 11:5914. doi: 10.1038/s41467-020-19621-z
Sakamoto, Y., and Takagi, S. (2013). LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 54, 622–633. doi: 10.1093/pcp/pct031
Shevelyov, Y. Y., and Nurminsky, D. I. (2012). The nuclear lamina as a gene-silencing hub. Curr. Issues Mol. Biol. 14, 27–38.
Sugano, S. S., Shirakawa, M., Takagi, J., Matsuda, Y., Shimada, T., Hara-Nishimura, I., et al. (2014). CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55, 475–481. doi: 10.1093/pcp/pcu014
Tamura, K., Goto, C., and Hara-Nishimura, I. (2015). Recent advances in understanding plant nuclear envelope proteins involved in nuclear morphology. J. Exp. Bot. 66, 1641–1647. doi: 10.1093/jxb/erv036
Tanaka, H., Suzuki, R., Okabe, N., Suzuki, T., and Kodama, Y. (2018). Salinity stress-responsive transcription factors in the liverwort Marchantia polymorpha. Plant Biotechnol. 35, 281–284. doi: 10.5511/plantbiotechnology.18.0501a
Tanurdzic, M., and Banks, J. A. (2004). Sex-determining mechanisms in land plants. Plant Cell 16(Suppl.), S61–S71. doi: 10.1105/tpc.016667
Ulianov, S. V., Doronin, S. A., Khrameeva, E. E., Kos, P. I., Luzhin, A. V., Starikov, S. S., et al. (2019). Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat Commun. 10, 1176. doi: 10.1038/s41467-019-09185-y
van Steensel, B., and Belmont, A. S. (2017). Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 169, 780–791. doi: 10.1016/j.cell.2017.04.022
Wang, H., Dittmer, T. A., and Richards, E. J. (2013). Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13:200. doi: 10.1186/1471-2229-13-200
Wang, Q., Liu, S., Lu, C., La, Y., Dai, J., Ma, H., et al. (2019). Roles of CRWN-family proteins in protecting genomic DNA against oxidative damage. J. Plant Physiol. 233, 20–30. doi: 10.1016/j.jplph.2018.12.005
Wasternack, C., and Strnad, M. (2016). Jasmonate signaling in plant stress responses and development - active and inactive compounds. N. Biotechnol. 33, 604–613. doi: 10.1016/j.nbt.2015.11.001
Yamato, K. T., Ishizaki, K., Fujisawa, M., Okada, S., Nakayama, S., Fujishita, M., et al. (2007). Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl. Acad. Sci. U S A. 104, 6472–6477. doi: 10.1073/pnas.0609054104
Yang, J., Chang, Y., Qin, Y., Chen, D., Zhu, T., Peng, K., et al. (2020). A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. N. Phytol. 227, 65–83. doi: 10.1111/nph.16518
Zhao, W., Guan, C., Feng, J., Liang, Y., Zhan, N., Zuo, J., et al. (2016). The Arabidopsis CROWDED NUCLEI genes regulate seed germination by modulating degradation of ABI5 protein. J. Integr. Plant Biol. 58, 669–678. doi: 10.1111/jipb.12448
Zheng, X., Hu, J., Yue, S., Kristiani, L., Kim, M., Sauria, M., et al. (2018). Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 71:e807. doi: 10.1016/j.molcel.2018.05.017
Keywords: Marchantia, NMCP, nuclear periphery, nuclear lamina, nuclear envelope
Citation: Wang N, Karaaslan ES, Faiss N, Berendzen KW and Liu C (2021) Characterization of a Plant Nuclear Matrix Constituent Protein in Liverwort. Front. Plant Sci. 12:670306. doi: 10.3389/fpls.2021.670306
Received: 20 February 2021; Accepted: 01 April 2021;
Published: 07 May 2021.
Edited by:
Yangnan Gu, University of California, Berkeley, United StatesReviewed by:
Eric J. Richards, Boyce Thompson Institute, United StatesKentaro Tamura, University of Shizuoka, Japan
Copyright © 2021 Wang, Karaaslan, Faiss, Berendzen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Liu, Y2hhbmcubGl1QHVuaS1ob2hlbmhlaW0uZGU=
 Nan Wang
Nan Wang Ezgi Süheyla Karaaslan1
Ezgi Süheyla Karaaslan1 Chang Liu
Chang Liu