
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 May 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.665014
Circular RNA (circRNA) is a novel class of endogenous long non-coding RNA (lncRNA) and participates in diverse physiological process in plants. From the dataset obtained by high-throughput RNA sequencing, we identified a circRNA encoded by the sense strand of the exon regions spanning two RuBisCO small subunit genes, RBCS2B and RBCS3B, in Arabidopsis thaliana. We further applied the single specific primer-polymerase chain reaction (PCR) and Sanger sequencing techniques to verify this circRNA and named it ag-circRBCS (antisense and across genic-circular RNA RBCS). Using quantitative real-time PCR (qRT-PCR), we found that ag-circRBCS shares a similar rhythmic expression pattern with other RBCS genes. The expression level of ag-circRBCS is 10–40 times lower than the expression levels of RBCS genes in the photosynthetic organs in Arabidopsis, whereas the Arabidopsis root lacked ag-circRBCS expression. Furthermore, we used the delaminated layered double hydroxide lactate nanosheets (LDH-lactate-NS) to deliver in vitro synthesized ag-circRBCS into Arabidopsis seedlings. Our results indicate that ag-circRBCS could significantly depress the expression of RBCS. Given that ag-circRBCS was expressed at low concentration in vivo, we suggest that ag-circRBCS may represent a fine-tuning mechanism to regulating the expression of RBCS genes and protein content in Arabidopsis.
Circular RNA (circRNA) is a class of covalently closed single-stranded RNA molecules that lack 5′ caps and 3′ poly(A) tails that generated by back-splicing events in eukaryotic cells (Wang et al., 2014). In 1976, Sanger et al. (1976) detected circRNAs in Solanum lycopersicum and Gynura as viroids by electron microcopy. With significant progress of high-throughput sequencing technologies and bioinformatic tools, scientists have verified that circRNAs are ubiquitous and abundant in eukaryotes, including in archaea (Danan et al., 2012), human (Jeck et al., 2013), Arabidopsis (Ye et al., 2015), and zebrafish (Shen et al., 2017). Several online databases have been constructed for deep analysis of circRNAs, including PlantcircBase (Chu et al., 2017), circBase ( GlaŽar et al., 2014), CIRCpedia (Zhang et al., 2016), circRNADb (Chen et al., 2016), and AtCircDB (Ye et al., 2017). Chu et al. (2020) analyzed these published databases and identified feature differences between plant circRNAs and animal circRNAs. GT/AG or CT/AC dinucleotides are general canonical splicing signals in animals and plants, but the proportions of circRNAs with non-canonical splicing signals were shown to be variable across 12 different plant species. Plant circRNAs show great variety, and based on genomic features, they can be classified into 10 unique types, namely, e-circRNA, ei-circRNA, i-circRNA, ie-circRNA, u-circRNA, ue-circRNA, ui-circRNA, ig-circRNA, igg-circRNA, and ag-circRNA (e, i, u, g, ig, and ag represent exon, intron, UTR region, genic region, intergenic region, and across-genic region, respectively) (Chu et al., 2020).
CircRNAs may regulate gene expression using different mechanisms at transcriptional, post-transcriptional, and translation levels. For example, circSEP3 and circSMARCA5 may form an R-loop with a host DNA locus and regulate gene expression or inhibit the DNA damage repairing (Conn et al., 2017; Xu et al., 2021). In this case, the circRNA, CDR1, regulates target gene expression by sequestering miR-7, and circPan3 stabilizes IL-13 mRNA subunits, leading to the maintenance of stem cells (Zhong et al., 2019; Zhu et al., 2019). Endogenous circRNA containing an internal ribosome entry site (IRES) can be translated in vivo (Wang and Wang, 2015; Zhang et al., 2020), and circRNA–protein complexes have been reported (Schneider et al., 2016). It is still unclear whether the plant and animal circRNAs share similar mechanisms for regulation of gene expression. For example, CDR1 contains 63 conserved miR-7 binding sites and sequesters miRNA. However, there is still no evidence in plant cells to support such a sequestration-based transcription inhibition mechanism. To date, no plant circRNA with an IRES has been reported to have peptide translation.
Additionally, plant circRNAs play important functional roles in various biological processes and in different environmental stresses (Zhang et al., 2020). In Arabidopsis, 1,583 and 36 circRNAs were shown to be differentially expressed under heat and drought stress, respectively. In rice (Oryza sativa), 27 exonic circRNAs (6 up-regulated and 21 down-regulated) were found to be differentially expressed between normal and phosphate-starvation condition (Ye et al., 2015). Moreover, various environmental stresses, including dehydration, low nitrogen, drought, cold, heat, copper, salt, calcium, low phosphorus, and pathogen invasion, can cause alteration of circRNA expression in different species (Zhang et al., 2020), such as rice (Ye et al., 2015), tomato (S. lycopersicum) (Wang et al., 2018), wheat (Triticum aestivum) (Ren et al., 2018), soybean (Glycine max) (Lv et al., 2020), and maize (Zea mays) (Ghorbani et al., 2018). Lately, the amount of circRNA datasets has sharply increased, suggesting that plant circRNAs may have important roles in different biological processes. However, their function roles and regulation mechanisms require more experimental investigation and verification. For example, the overexpression of circGORK (guard cell outward-rectifying K+-channel) confirmed its role in drought tolerance (Zhang et al., 2019), and the overexpression of lariat-derived circRNAs suggested that circRNAs altered developmental phenotypes (Cheng et al., 2018).
RuBisCO is the most abundant enzyme on the Earth, and it is also solely responsible for all carbon fixation via photosynthetic assimilation of atmospheric CO2 (Bracher et al., 2017; Hayer-Hartl and Hartl, 2020). Therefore, it is the crucial enzyme for both feeding humanity and controlling the climate on Earth (Caetano-Anollés, 2017; Baluška and Mancuso, 2020; von Caemmerer, 2020). In higher plants and green algae, RuBisCO is composed of eight small subunits (RBCS) encoded by RBCS multigene family in the nuclear genome and eight large subunits (RbcL) encoded by a single RbcL gene in the chloroplast genome (Huang et al., 2020), denoted as RbcL8S8 (Bracher et al., 2017). For synthesis of the RuBisCO holoenzyme, both genes need to be expressed coordinately (Makino and Suzuki, 2012). RBCS subunits have been shown to influence the catalytic efficiency, CO2 specificity, assembly, activity, and stability of RuBisCO (Yamada et al., 2019), whereas the large subunits contain the active sites for catalytic activity. Therefore, the precise regulation of genes of the expression of large and small subunits is critical in the maintaining and regulation of RuBisCO activities.
In our previous study, we generated a circRNA database for etiolated and de-etiolation Arabidopsis seedlings (accession number: SUB3747127) (Liu et al., 2019). Among this dataset, a small group of circRNAs encoded by RBCS attracted our attention. Our RNA sequencing results suggested that these circRNAs were encoded by the sense strand of DNA, with complement sequence to the RBCS3B and RBCS2B mRNAs. Therefore, we verified these circRNA sequences and analyzed their spatial and temporal expression. We also used the delaminated layered double hydroxide lactate nanosheets (LDH-lactate-NS), a proven nanotransporter for intact plant cells (Bao et al., 2017; Song et al., 2019), to deliver the in vitro synthesized circRNA into Arabidopsis seedlings to investigate their biological functions.
From our previously reported strand-specific RNA sequencing database, we identified a small group of antisense circRNAs encoded by the sense strand of exon regions across RuBisCO small subunit genes in Arabidopsis thaliana, which we named ath_circ_362-366 (Supplementary Figure 1). We designed pairs of convergent and divergent primers for polymerase chain reaction (PCR) to confirm the existence of these circRNAs. The divergent primers were designed across back-splice junctions, whereas the convergent primers were used as controls to validate linear sequences (Figure 1A). In the PCR reactions carried out with gDNA templates, only the convergent primers led to a positive band. Meanwhile, the divergent primers clearly indicated the existence of circRNAs using cDNA templates (Figure 1B). We tried to confirm the existence of ath_circ_362-366 (data not shown), but only the primer pairs for ath_circ_364 led to positive results (Supplementary Table 1).
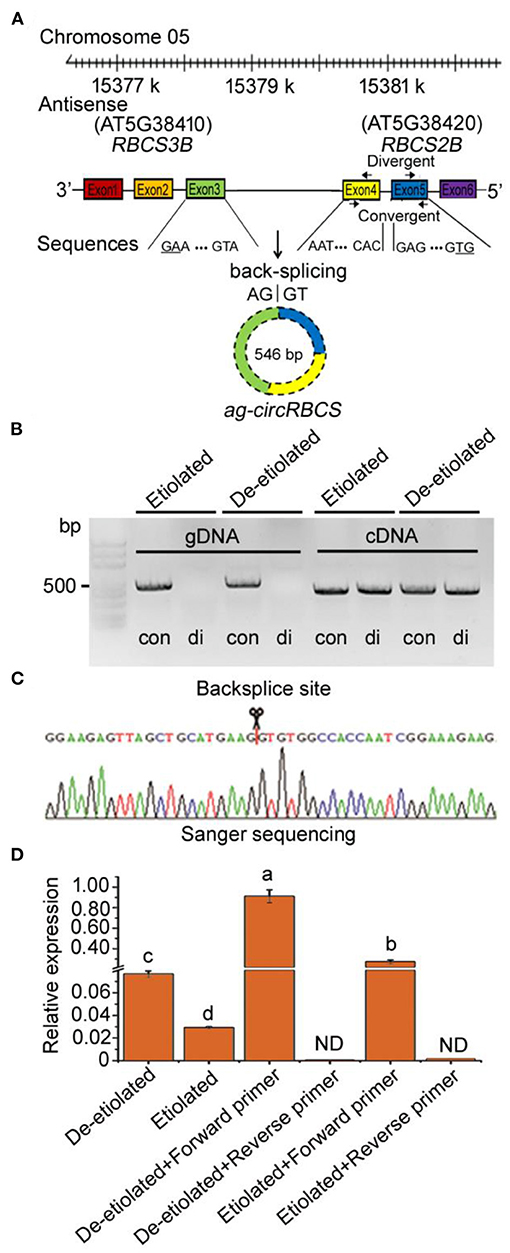
Figure 1. Identification of circular RNA from Arabidopsis seedlings. (A) ag-circRBCS is encoded by three exonic regions, exon3 of RBCS3B and exon4, 5 of RBCS2B. (B) Divergent primers successfully amplified circular RNAs in cDNA but failed in genomic DNA. Convergent primers worked on both cDNA and genomic DNA. con, convergent primers; di, divergent primers. (C) Sanger sequencing further confirmed head-to-tail back-splicing. (D) The circular RNA was confirmed as an antisense RNA via the single specific primer-PCR. All the data were analyzed for significant differences using ANOVA with Duncan's test. Different lowercase letters represent statistical significances of p < 0.05 (n = 3). ND, not detected.
Sanger sequencing based on these PCR products indicated that the ath_circ_364 reflected a gene sequence encoded by three exonic regions, exon3 of RBCS3B and exon4, 5 of RBCS2B, meaning that this was a cross genic circRNA. Therefore, we named it ag-circRBCS (antisense and across genic-circular RNA RBCS) (Figure 1A). ag-circRBCS had a canonical site where the splice site was flanked by GT/AG and was formed by joining a splice donor to an upstream splice acceptor (Figure 1C).
Our RNA sequencing database suggested that ag-circRBCS had a higher expression in de-etiolated plants than in etiolated Arabidopsis, which was confirmed by quantitative real-time PCR (qRT-PCR) (Figure 1D). We further used the single specific primer-PCR (SSP-PCR) to define this strand-specific sequence. After multiple rounds of SSP-PCR, the cDNA contents of target sequences amplified with a complementary primer should be higher than before amplification. Therefore, a qRT-PCR assay followed SSP-PCR to indicate strand-specific cDNA sequences. In this study, we designed a forward primer that was complementary to the antisense sequence and the reversed primer as the control (Supplementary Table 2). After 35 rounds of SSP-PCR, the content of PCR products using the forward primer was much higher than those using the reverse primer, and in fact, the latter was almost undetectable. This result confirmed that ag-circRBCS reflected the antisense sequence of template DNAs. In addition, the de-etiolated seedlings had a much higher concentration of this than the etiolated seedlings (Figure 1D).
To investigate the relationship of the transcription level of ag-circRBCS and its parental gene, RBCS2B and RBCS3B, we used the qRT-PCR to quantify their spatial and temporal expression profile in Arabidopsis. Divergent primers were again used to quantify the expression of ag-circRBCS, and convergent primers were applied to understand the expression level of parental genes. Since sequences of RBCS2B and RBCS3B were very similar, the PCR products generated using convergent primers did not allow us the distinction between RBCS2B and RBCS3B. Therefore, we termed these genes RBCS. In underground tissue, the expression of both RBCS and ag-circRBCS was undetectable. In aerial tissues, RBCS in stems and leaves were not significantly differentially expressed, whereas the expression level of RBCS in pods and flowers was significantly lower than that in stems and leaves. Among all these tissues, the ag-circRBCS expressions level was the highest in photosynthetic tissue, namely, in the leaves. The expression of ag-circRBCS gradually decreased in stems, pods, and flowers. Moreover, the expression of RBCS was much higher than the expression of ag-circRBCS. In pods, flowers, stems, and leaves, the RBCS expression was 24.8, 40.2, 25.9, and 10.3 times higher than ag-circRBCS, respectively (Figure 2A). Meanwhile, the ratio of ag-circRBCS to RBCS was the largest in the leaves. This ratio was gradually decreased in pods, stems, and flowers (Figure 2B).
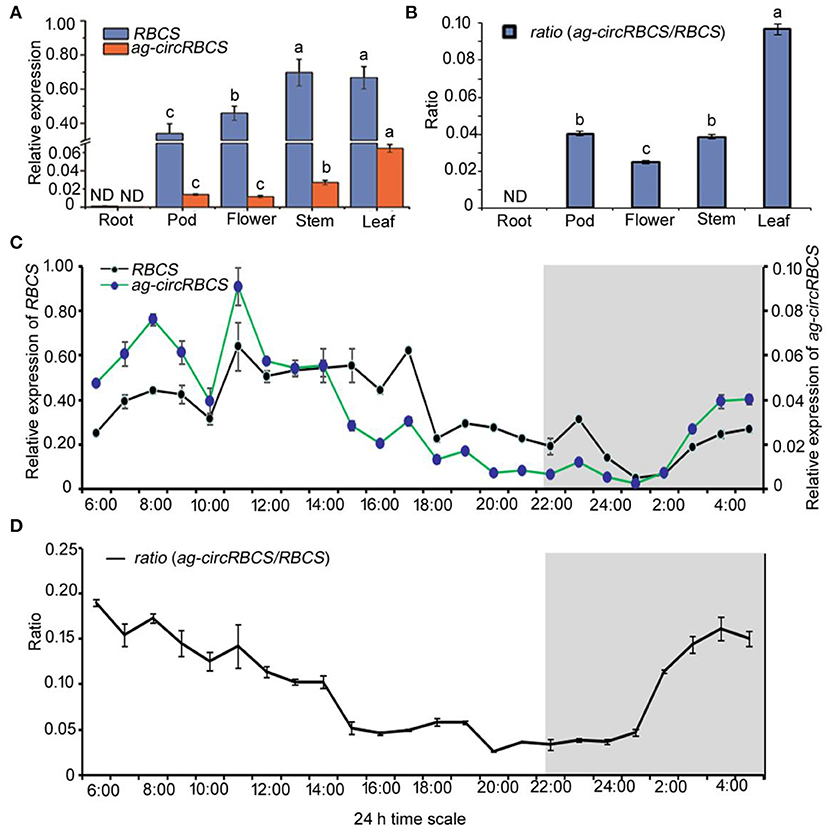
Figure 2. The expression profile of RBCS and ag-circRBCS. (A) The relative expression levels of RBCS and ag-circRBCS in different tissues. (B) The expression ratio of ag-circRBCS to RBCS in different tissues. Actin was used as an internal control. (C) The expression levels of RBCS and ag-circRBCS in different time points. (D) The expression ratio of ag-circRBCS to RBCS in different time points. Actin was used as an internal control. Data are expressed as mean ± standard deviation from three independent experiments. All the data were analyzed for significant differences using ANOVA with Duncan's test. Different lowercase letters represent statistical significances of p < 0.05 (n = 3) of RBCS, ag-circRBCS, and ratio, respectively. ND, not detected.
We further investigated the 24 h rhythmic expression pattern of ag-circRBCS in Arabidopsis seedlings. Arabidopsis seedlings were cultured under a routine long day illumination, e.g., 16 h in light and 8 h in dark, with illumination from 6:00 to 22:00. The profile of 24-h expression of both RBCS and ag-circRBCS showed similar typical periodic pattern (Figure 2C). The expression peak of ag-circRBCS occurred at 7:00 and 11:00, and the expression level of ag-circRBCS significantly decreased after 14:00. However, the expression level of RBCS remained relatively high between 11:00 and 17:00 and decreased after 18:00. Additionally, the peak of the ratio between the relative expression level of ag-circRBCS to RBCS occurred at 6:00, whereas the ratio of the off-peak occurred at 15:00–1:00. This ratio decreased slowly during the daytime and increased rapidly at night (Figure 2D).
ag-circRBCS was encoded by sense strand from the exonic regions of RBCS2B and RBCS3B genes. Obviously, the overexpression, knock-out, or knock-down of this circRNA would be expected to change its parental gene sequences, located on the antisense strand. Thus, we synthesized linear ag-circRBCS and circularized it in vitro. Next, LDH-lactate-NS was used to transport this synthesized circRNA into the leaf cells of Arabidopsis (Figure 3A). The linear antisense sequence of exon 3–5 of RBCS3B and RBCS2B was expressed in the T4 RNA ligase 1 system, and the successful synthesis and circulation of this RNA was validated by PCR with convergent and divergent primers (Figure 3B). By electrophoresis analysis, the PCR products with convergent primers could be detected in linear and cyclized RNA products, whereas the PCR products with divergent primers could only be found using cyclized RNA products, confirming the successful synthesis of ag-circRBCS (Figure 3B). The synthesized ag-circRBCS was then absorbed using LDH-lactate-NS in vitro. The efficiency of ag-circRBCS adsorption via LDH-lactate-NS was further investigated (Figure 3C). When the LDH was added to the synthesized ag-circRBCS at a weight ratio of 1:3, no ag-circRBCS repelled down. When natural air was bubbled into the LDH-ag-circRBCS solution for 1 h, the ag-circRBCS was replaced by CO2, and the obvious target band of ag-circRBCS appeared again (Figure 3C).
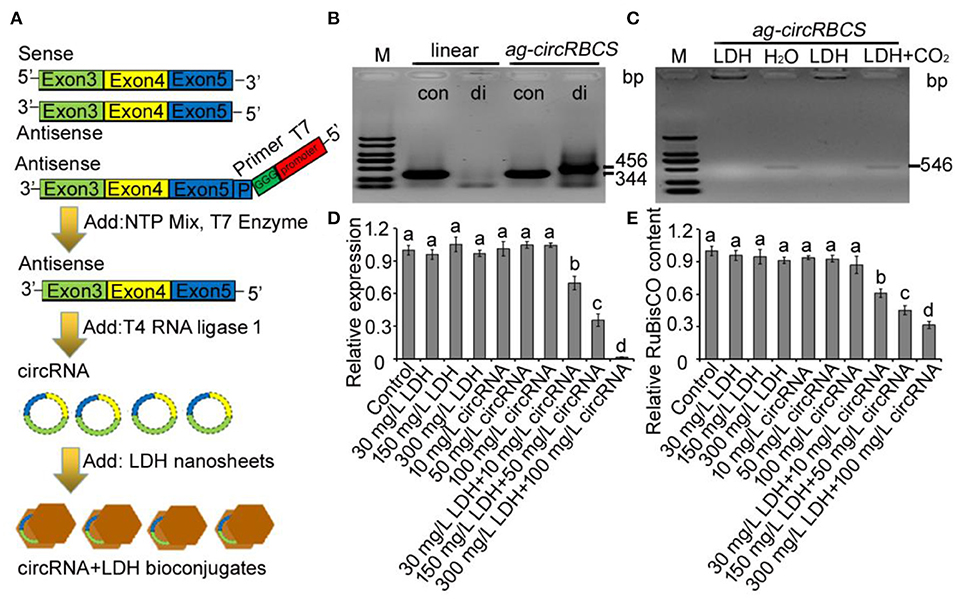
Figure 3. ag-circRBCS regulates RBCS gene expression. (A) Flowchart for artificial synthesis of ag-circRBCS and transported via layered double hydroxide lactate nanosheets (LDH-lactate-NS) in vitro. (B) The identification of synthesis ag-circRBCS via PCR. The convergent (con) and divergent (di) primers were used for analysis. (C) Analysis of the adsorption efficiency of LDH-lactate-NS in vitro. (D) Relative expression level of RBCS genes under different treatments. Numbers in (B,C) indicate the expected RNA length. (E) Analysis of the RBCS protein content via ELISA. M, marker; linear, synthesized single strand RNA before circulation; LDH, layered double hydroxide lactate nanosheets. Actin was used as an internal control. Data are expressed as mean ± standard deviation from three independent experiments. All the data were analyzed for significant differences using ANOVA with Duncan's test. Different lowercase letters represent statistical significance of p < 0.05 (n = 3).
To investigate the effect of ag-circRBCS on the expression of parental or target genes, we used qRT-PCR to quantify the expression of RBCS genes and used enzyme-linked immunosorbent assay (ELISA) to quantify the RuBisCO protein contents with an RbcL antibody. When Arabidopsis seedlings were immersed in distilled water with LDH-lactate-NS or ag-cricRBCS separately, the expression level of RBCS was not affected, except in the solution with high LDH content. When the Arabidopsis seedlings were immersed in the solution with premixed LDH-lactate-NS and ag-cricRBCS, the expression of RBCS was significantly down-regulated (Figure 3D). Moreover, ELISA assay showed that the RuBisCO protein contents were down-regulated (Figure 3E).
In Arabidopsis, the RbcS gene family is further divided into two groups. Gene duplication and loss events of RBCS occurred during the evolution of Arabidopsis (Schwarte and Tiedemann, 2011). Therefore, three members of RBCS group B genes are homologous and tandemly distribute to chromosome 5. The expression of individual members of RbcS gene family has been shown to be separately regulated. For example, the RBCS1A is the only member distributed in root (Sawchuk et al., 2008), and the expression level of RBCS1A, RBCS2B, and RBCS3B is regulated by light signals, except RBCS1b (Dedonder et al., 1993). The tissue-specific expression and rhythmic expression of ag-circRBCS and its parental genes provide further clues for understanding the functional role of ag-circRBCS. ag-circRBCS was co-expressed with its parental genes, whereas both had their highest expression in photosynthetic tissues, coinciding with previous analysis of RBCS mRNA levels (Suzuki et al., 2009a,b). The rhythmic expression of ag-circRBCS and RBCS had similar profiles, too. The expression of RBCS had a circadian rhythm, such as maximally accumulated during subjective day and dropping to the lowest levels in the early evening (Choudhary et al., 2016). Our results agreed with this previous study and further suggested that the ratio of ag-circRBCS/RBCS expression reached a peak value in midnight (2:00). The rhythmic changes of the ag-circRBCS/RBCS expression ratio suggested that the ag-circRBCS/RBCS is not a simple by-product of RBCS expression.
ag-circRBCS has a unique and novel structure according to known antisense RNAs. It is encoded by the exon region from the sense DNA strand of two neighboring genes. This RNA should be synthesized from an antisense pre-RNA with cross-genic sequences, and the splicing and back-splicing processes occur exactly as in sense pre-RNA (Figure 4). Thus, the expression of both antisense and sense strands was regulated by similar but unknown mechanisms. ag-circRBCS is not a by-product of RBCS expression, and its function needs to be further investigated.
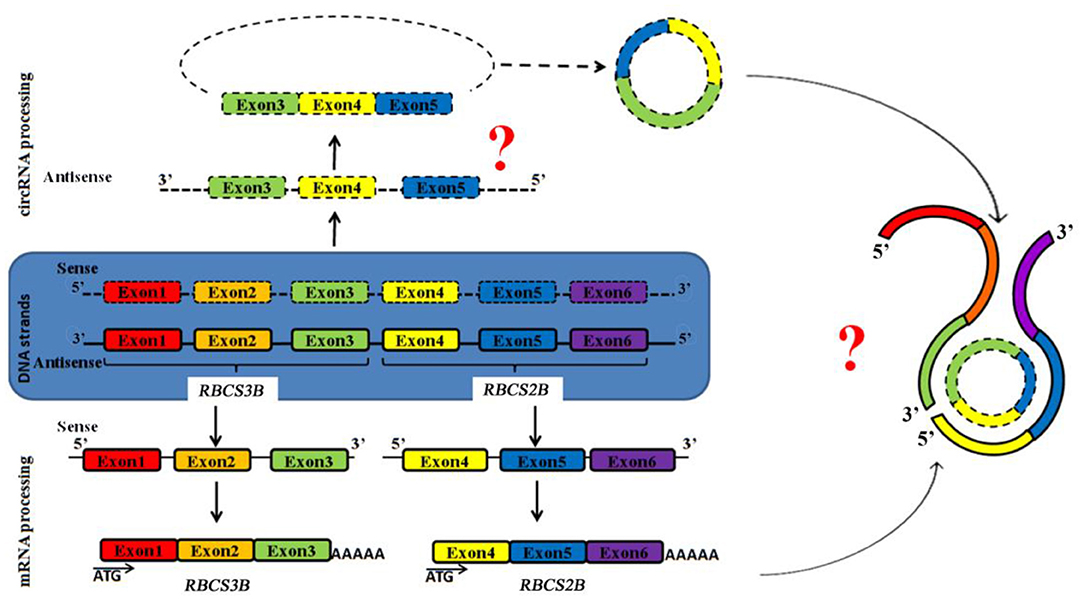
Figure 4. Schematic model to show the expression of both strands of genomic DNA of RBCS3B and RBCS2B genes. Square frames exhibit the exon regions. Two question marks indicate two critical steps: edition of antisense RNA and function of this antisense circRNA.
Antisense RNA is an efficient means of regulating the expression of endogenous and foreign genes in eukaryotes (Xu et al., 2018). However, only a few functional antisense RNAs in plants have been reported. For example, a natural antisense RNA was shown to be involved in the regulation of MADS AFFECTING FLOWERING4 (MAF4) in Arabidopsis (Zhao et al., 2018). The overexpression of an antisense RNA of the maize gene ZmRLK7 in Arabidopsis resulted in regulating plant architecture and organ size formation (He et al., 2020). An antisense RNA may regulate target gene expression using different mechanisms. For example, antisense long non-coding RNAs (lncRNAs) may form L-loops with genomic DNA (Tan-Wong et al., 2019) or act as competing endogenous RNAs (ecRNA) (Tay et al., 2014). In this study, we did not investigate these mechanisms, but rather used a proven nanotransporter, LDH-lactate-NS, to deliver in vitro synthesized ag-circRBCS into plant cells. Our results indicate that a high concentration of ag-circRBCS can significantly depress the expression of its target genes, RBCS2B and RBCS3B, and even alter the expression of the RuBisCO protein itself. Given that ag-circRBCS has only 1/10–1/40 the relative expression level to RBCS, we suggest that it may provide fine-tuning to the regulation of its parental genes, the RBCS2B and RBCS3B. Moreover, an antisense circRNA provides an RNA formation with high stability during its life cycle, giving it resistance to RNase R. This conclusion coincides with an early study on rice, where antisense RBCS down-regulate the expression level of RuBisCO (Makino et al., 1997).
In conclusion, we report here that a novel circRNA, ag-circRBCS, may have roles in the precise regulation of RuBisCO expression. A possible model for its expression and formation is presented in Figure 4. We believe that this unique structure of an RNA sequence will provide new avenues for a better understanding of non-coding RNAs in plants.
The wild-type A. thaliana ecotype Col-0 (Columbia-0) was used for experiments. Single seed was chosen for further experiment within three generations. Seeds were placed on 1/2 MS medium (half-strength Murashige and Skoog-containing) with 0.4% Phytagels. Afterward, the plates were incubated in light chambers at 22°C with a 16-/8-h light cycle at 120 μmol photons m−2 s−1. The 10-day-old Arabidopsis seedlings were used for rhythmic transcriptional level assessments of RBCS and circRNAs. Arabidopsis seedling leaves were sampled every 1 h for 24 h. For tissue-specific expression, plants were cultivated in mixed soil (vermiculite/nutrition soil = 2:1, v/v). The Arabidopsis seedlings were germinated and grown in a 16-h light/8-h dark photoperiod at 22°C. The 30-day-old Arabidopsis seedlings were used to assessing the specific transcriptional level of RBCS and circRNAs.
Total RNA was extracted, and DNA contamination was removed by using the E.Z.N.A. Total RNA Kit (OMEGA, GA, USA). DNase I was added to the total RNA extraction for 10 min at 37°C to digest DNA and then kept at 65°C for 10 min to inactivate the DNase I. The quality and concentration of total RNA samples were assessed by 1.5% agarose gel electrophoresis and NanoDrop 2000 spectrophotometer. The purified RNA served as a template for synthesizing first-strand cDNA using the TransScript One-Step gDNA Removal kit and cDNA Synthesis Super Mix (TransGen, Beijing, China).
qRT-PCR was performed using the SYBR® Premix Ex Taq™ II (Perfect Real Time) (TaKaRa, Dalian, China) in a typical 20 μl PCR mixture including 10 μl of SYBR® Premix Ex Taq™ II, 1 μl of template cDNA, and 0.2 μM of each PCR primer, with double distilled water up to 20 μl. Samples were mixed gently and centrifuged briefly to collect droplets. The cycling conditions were 95°C for 5 min, followed by 40 cycles at 95°C for 20 s, 53°C for 20 s, and 72°C for 30 s, and samples were run on a Real-Time PCR Detection System CFX96 (Bio-Rad, CA, USA). All of the relative gene expression levels were calculated using the 2−ΔΔCt method, with A. thaliana actin (AT3G18780) used as an internal control. The primers used for qRT-PCR are listed in Supplementary Table 2.
For SSP-PCR, cDNA of de-etiolated or etiolated Arabidopsis seedlings was used as template, and divergent forward primer and divergent reverse primer were added for first round of PCR, respectively. These fragments were amplified by first round of PCR using the Pyrobest DNA polymerase (TaKaRa, Dalian, China) for 35 cycles in a 50 μl reaction with these primers (Supplementary Table 2). The reactions were amplified using the qRT-PCR mix using the “Quantitative real-time PCR” program on a Biorad thermocycler.
For RuBisCO quantification, Arabidopsis seedlings were suspended in phosphate buffered solution (PBS, pH 7.4, 0.15 M) on ice. Then, the RuBisCO in the supernatant was quantified using a plant RuBisCO ELISA kit (Jiangsu Meimian Industrial Co., Ltd., Jiangsu, China) according to the manufacturer's instructions. Briefly, the supernatant was added to a 96-well plate and combined with horseradish peroxidase (HRP)-labeled RbcL enzyme antibody at 37°C for 1 h. After washing 5 times, 3,3′,5,5-tetramethylbenzidine (TMB) was quickly added to the reaction, and then sulfuric acid solution was quickly added to terminate the reaction. The absorbance of reaction mixtures was then measured at 450 nm with the ELx800 microplate reader (Bio Tek, EV, UK). The concentration in each well was then calculated based on a standard curve. The amounts of RBCS were calculated from the RuBisCO holoenzyme and the ratio of molecular mass between RBCS and RbcL (Makino and Suzuki, 2012; Suganami et al., 2020).
Large quantities of antisense linear circRNA were synthesized in Dongxuan jiyin (Jiangsu dongxuan jiyin Jiangsu Technology Co., Ltd., Jiangsu, China) by T7 RNA polymerase using the TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, NY, USA). The sequences are listed in Supplementary Table 1. After DNase I treatment for 10 min at 37°C, RNA was purified by LiCl precipitation and then treated with T4 polynucleotide kinase (New England Biolabs, MA, USA) in the presence of ATP. Finally, the linear RNA was circularized with T4 RNA ligase 1 (New England Biolabs, MA, USA) for 16 h at 16°C. Any residual linear RNA was eliminated by RNase R for 10 min at 37°C. After electrophoresis, RNA in bonds with a size of about 500 bp was collected by E.Z.N.A. ploy gel RNA extraction kit (OMEGA, GA, USA). Finally, the collected circRNA was precipitated and concentrated via equal volume ethanol.
Divergent and convergent PCR primers were designed for circRNA validation (Supplementary Table 2). First-strand cDNA was synthesized using divergent primers or convergent primers. The PrimeSTAR® Max DNA Polymerase (Takara, Dalian, China) was used for cDNA amplification with PCR to detect circRNA templates. The PCR mixture included 25 μl PrimeSTAR Max Premix (2×), 1 μl of template cDNA, and 1 μl of each PCR primer, with double distilled water up to 50 μl. The cycling conditions used were 95°C for 5 min, followed by 40 cycles at 95°C for 20 s, 53°C for 20 s, and 72°C for 30 s on a Biometra T1 Thermocycler. Then, Sanger sequencing was performed on all PCR products to validate their sequence. Before performing Sanger sequencing, the PCR products were electrophoresed on 1% agarose gel and collected by GeneJET gel extraction kit (Thermo Scientific, Lithuania).
Mixtures of circRNAs and LDH with weight ratios of 1:3 were incubated at 25°C for 1 h in airtight tubes. After that, the mixtures were transferred to DNase-, RNase-, and DNA-free 6-well tissue culture plates (Thomas Scientific, NJ, USA) and exposed in air at 25°C for 1 h. The adsorption effect was detected via 1% agarose gel electrophoresis with Tris/Borate/EDTA buffer at 5 V/cm.
For function analysis, 10-day-old seedlings were infiltrated at 6-well tissue culture plates with 3 ml of different circRNA:LDH solutions, namely, 300 mg/L LDH + 100 mg/L circRNA, 150 mg/L LDH + 50 mg/L circRNA, and 30 mg/L LDH + 10 mg/L circRNA. Then, 100 mg/L circRNA, 50 mg/L circRNA, 10 mg/L circRNA, 300 mg/L LDH, 150 mg/L LDH, and 30 mg/L LDH were used as negative controls, respectively. Afterward, seedlings were transferred to 22°C conditions with a 16-h light/8-h dark photoperiod for 3 days. Then, the seedlings' leaves were stored at −80°C for future use.
Mean and standard deviation (SD) are displayed as representative values for data in the figures. Analysis of variance (ANOVA) with Duncan's test was used to assess statistical significance, which was done with SPSS v10, unless otherwise noted. p < 0.05 was regarded as statistically significant.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HZ and SL did most of the experimental works and wrote the manuscript. SL did the database analysis. XL and HW synthesized the LDH. LY did the 24 h analysis. FB helped in the manuscript writing and supported this project. YW supervised this project. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31671489) and the Hainan Provincial Natural Science Foundation of China (2019RCI55).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.665014/full#supplementary-material
Supplementary Figure 1. Library preparation for circRNA sequencing.
Supplementary Table 1. Sequences of circular RNAs encoding the antisense RBCS genes.
Supplementary Table 2. Primers used in this study.
Baluška, F., and Mancuso, S. (2020). Plants, climate and humans: plant intelligence changes everything. EMBO Rep. 21:e50109. doi: 10.15252/embr.202050109
Bao, W. L., Wan, Y. L., and Baluska, F. (2017). Nanosheets for delivery of biomolecules into plant cells. Trends Plant Sci. 22, 445–447. doi: 10.1016/j.tplants.2017.03.014
Bracher, A., Whitney, S. M., Hartl, F. U., and Hayer-Hartl, M. (2017). Biogenesis and metabolic maintenance of Rubisco. Annu. Rev. Plant Biol. 68, 29–60. doi: 10.1146/annurev-arplant-043015-111633
Caetano-Anollés, G. (2017). RubisCO and the search for biomolecular culprits of planetary change. BioEssays 39:1700174. doi: 10.1002/bies.201700174
Chen, L., Yu, Y. Y., Zhang, X. C., Chen, L., Ye, C. Y., and Fan, L. J. (2016). PcircRNA_finder: a software for circRNA prediction in plants. Bioinformatics 32, 3528–3529. doi: 10.1093/bioinformatics/btw496
Cheng, J. P., Zhang, Y., Li, Z. W., Wang, T. Y., Zhang, X. T., and Zheng, B. L. (2018). A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci. China Life Sci. 61, 204–213. doi: 10.1007/s11427-017-9182-3
Choudhary, M. K., Nomura, Y., Shi, H., Nakagami, H., and Somers, D. E. (2016). Circadian profiling of the Arabidopsis proteome using 2D-DIGE. Front. Plant Sci. 7:1007. doi: 10.3389/fpls.2016.01007
Chu, Q. J., Bai, P. P., Zhu, X. T., Zhang, X. C., Mao, L. F., Zhu, Q. H., et al. (2020). Characteristics of plant circular RNAs. Brief. Bioinform. 21, 135–143. doi: 10.1093/bib/bby111
Chu, Q. J., Zhang, X. C., Zhu, X. T., Mao, L. F., Ye, C. Y., Zhu, Q. H., et al. (2017). PlantcircBase:a database for plant circular RNAs. Mol. Plant 10, 1126–1128. doi: 10.1016/j.molp.2017.03.003
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3:17053. doi: 10.1038/nplants.2017.53
Danan, M., Schwartz, S., Edelheit, S., and Sorek, R. (2012). Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40, 3131–3142. doi: 10.1093/nar/gkr1009
Dedonder, A., Rethy, R., Fredericq, H., Van Montagu, M., and Krebbers, E. (1993). Arabidopsis rbcS gene are differentially regulated by light. Plant Physiol. 101, 801–808. doi: 10.1104/pp.101.3.801
Ghorbani, A., Izadpansh, K., Peters, J. R., Dietzgen, R. G., and Mitter, N. (2018). Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci. 274, 402–409. doi: 10.1016/j.plantsci.2018.06.016
GlaŽar, P., Papavasileiou, P., and Rajewsky, N. (2014). circBase: a database for circular RNAs. RNA 20, 1666–1670. doi: 10.1261/rna.043687.113
Hayer-Hartl, M., and Hartl, F. U. (2020). Chaperone machineries of Rubisco-the most abundat enzyme. Trends Biochem. Sci. 45, 748–763. doi: 10.1016/j.tibs.2020.05.001
He, C., Wang, J., Dong, R., Guan, H., Liu, T., Liu, C., et al. (2020). Overexpression of an antisense RNA of maize receptor-like kinase gene ZmRLK7 enlarges the organ and seed size of transgenic Arabidopsis plants. Front. Plant Sci. 11:579102. doi: 10.3389/fpls.2020.579120
Huang, F., Kong, W. W., Sun, Y. Q., Chen, T. Y., Dykes, G. F., Jiang, Y. L., et al. (2020). Rubisco accumulation factor 1(Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biolgenesis. Pro. Natl. Acad. Sci. U. S. A. 117, 17418–17428. doi: 10.1073/pnas.2007990117
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Liu, S., Wang, Q. J., Li, X. Y., Wang, G. B., and Wan, Y. L. (2019). Detecting of chloroplast circular RNAs in Arabidopsis thaliana. Plant Signal. Behav. 14:1621088. doi: 10.1080/15592324.2019.1621088
Lv, L., Yu, K., Lü, H., Zhang, X., Liu, X., Sun, C., et al. (2020). Transcriptome-wide identification of novel circular RNAs in soybean in response to low-phosphorus stress. PLoS ONE 15:e0227243. doi: 10.1371/journal.pone.0227243
Makino, A., Shimada, T., Takumi, S., Kaneko, K., Matsuoka, M., Shimamoto, K., et al. (1997). Does decrease in Ribulose-1,5-Bisphosphate Carboxylase by antisense RbcS lead to a higher N-use efficiency of photosynthesis under conditions of saturating CO2 and light in rice plants? Plant Physiol. 114, 483–491. doi: 10.1104/pp.114.2.483
Makino, A., and Suzuki, Y. (2012). Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assenbly of its holoenzyme in rice. Plant Physiol. 160, 533–540. doi: 10.1104/pp.112.201459
Ren, Y. Z., Yue, H. F., Li, L., Xu, Y. H., Wang, Z. Q., Xin, Z. Y., et al. (2018). Identification and characterization of circRNAs involved in the regulation of low nitrogen-promoted root growth in hexaploid wheat. Biol. Res. 51:43. doi: 10.1186/s40659-018-0194-3
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschnidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Sawchuk, M. G., Donner, T. J., Head, P., and Scarpella, E. (2008). Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol. 148, 1908–1924. doi: 10.1104/pp.108.126946
Schneider, T., Hung, L. H., Schreiner, S., Starke, S., Eckhof, H., Rossbach, O., et al. (2016). CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci. Rep. 6:31313. doi: 10.1038/srep31313
Schwarte, S., and Tiedemann, R. (2011). A gene duplication/loss event in the Ribulose-1,5-bisphosphate-carboxylase/oxygenase (Rubisco) small subunit gene family among accessions of Arabidopsis thaliana. Mol. Biol. Evol. 28, 1861–1876. doi: 10.1093/molbev/msr008
Shen, Y., Guo, X., and Wang, W. (2017). Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 591, 213–220. doi: 10.1002/1873-3468.12500
Song, Y. P., Xuan, A. R., Bu, C. H., Ci, D., Tian, M., and Zhang, D. Q. (2019). Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. J. 17, 164–177. doi: 10.1111/pbi.12955
Suganami, M., Suzuki, Y., Kondo, E., Nishida, S., Konno, S., and Makino, A. (2020). Effects of overproduction of Rubisco activase on Rubisco content in transgenic rice grown at different N levels. Int. J. Mol. Sci. 21:1626. doi: 10.3390/ijms21051626
Suzuki, Y., Miyamoto, T., Yoshizawa, R., Mae, T., and Makino, A. (2009a). Rubisco content and photosynthesis of leaves at different positions in transgenic rice with an overexpression of RBCS. Plant Cell Environ. 32, 417–427. doi: 10.1111/j.1365-3040.2009.01937.x
Suzuki, Y., Nakabayashi, K., Yoshizawa, R., Mae, T., and Makino, A. (2009b). Differences in expression of RBCS multigene family and Rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol. 50, 1851–1855. doi: 10.1093/pcp/pcp120
Tan-Wong, S. M., Dhir, S., and Proudfoot, N. J. (2019). R-loops promote antisense transcription across the mammalian genome. Mol. Cell 76, 600–616. doi: 10.1016/j.molcel.2019.10.002
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. doi: 10.1038/nature12986
von Caemmerer, S. (2020). Rubisco carboxylase/oxygenase: from the enzyme to the globe: a gas, exchange perspective. J. Plant Physiol. 252:153240. doi: 10.1016/j.jplph.2020.153240
Wang, J. Y., Yang, Y. W., Jin, L. M., Ling, X. T., Liu, T. L., Chen, T. Z., et al. (2018). Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 18:104. doi: 10.1186/s12870-018-1332-3
Wang, P. L., Bao, Y., Yee, M. C., Barrett, S. P., Hogan, G. J., Olsen, M. N., et al. (2014). Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9:e90859. doi: 10.1371/journal.pone.0090859
Wang, Y., and Wang, Z. (2015). Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179. doi: 10.1261/rna.048272.114
Xu, J. Z., Zhang, J. L., and Zhang, W. G. (2018). Antisense RNA: the new favorite in genetic research. J. Zhejiang Univ. Sci. B 19, 739–749. doi: 10.1631/jzus.B1700594
Xu, Y., Leng, K., Yao, Y., Kang, P., Liao, G., Han, Y., et al. (2021). A Circular RNA, cholangiocarcinoma-associated Circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology 73, 1419–1435. doi: 10.1002/hep.31493
Yamada, K., Davydov, I. I., Besnard, G., and Salamin, N. (2019). Duplication history and molecular evolution of the rbcS multigene family in angiosperms. J. Exp. Bot. 70, 6127–6139. doi: 10.1093/jxb/erz363
Ye, C. Y., Chen, L., Liu, C., Zhu, Q. H., and Fan, L. J. (2015). Widespread noncoding circular RNAs in plants. New Phytol. 208, 88–95. doi: 10.1111/nph.13585
Ye, C. Y., Zhang, X., Chu, Q., Liu, C., Yu, Y., Jiang, W., et al. (2017). Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 14, 1055–1063. doi: 10.1080/15476286.2016.1245268
Zhang, P., Fan, Y., Sun, X., Chen, L., Terzaghi, W., Bucher, E., et al. (2019). A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 98, 697–713. doi: 10.1111/tpj.14267
Zhang, P. J., Li, S. D., and Chen, M. (2020). Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 7:91. doi: 10.3389/fmolb.2020.00091
Zhang, X. O., Dong, R., Zhang, Y., Zhang, J. L., Luo, Z., Zhang, J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26, 1277–1287. doi: 10.1101/gr.202895.115
Zhao, X. Y., Li, J. R., Lian, B., Gu, H. Q., Li, Y., and Qi, Y. J. (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 9:5056. doi: 10.1038/s41467-018-07500-7
Zhong, Q., Huang, J., Wei, J., and Wu, R. (2019). Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer. Cell Int. 19:252. doi: 10.1186/s12935-019-0959-y
Keywords: antisense RNA, across-genic RNA, circular RNA, RBCS, Arabidopsis thaliana, expression regulation
Citation: Zhang H, Liu S, Li X, Yao L, Wu H, Baluška F and Wan Y (2021) An Antisense Circular RNA Regulates Expression of RuBisCO Small Subunit Genes in Arabidopsis. Front. Plant Sci. 12:665014. doi: 10.3389/fpls.2021.665014
Received: 13 February 2021; Accepted: 06 April 2021;
Published: 24 May 2021.
Edited by:
Alistair McCormick, University of Edinburgh, United KingdomReviewed by:
Hussein Gherli, University of Essex, United KingdomCopyright © 2021 Zhang, Liu, Li, Yao, Wu, Baluška and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglang Wan, eWx3YW5AaGFpbmFudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.