
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 April 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.663309
This article is part of the Research Topic Lipids, Lipid-Related Biomolecules and Lipid-Protein Interactions Involvement in Photosynthesis View all 10 articles
Chlorophyll (Chl) is essential for photosynthesis and needs to be produced throughout the whole plant life, especially under changing light intensity and stress conditions which may result in the destruction and elimination of these pigments. All steps of the Mg-branch of tetrapyrrole biosynthesis leading to Chl formation are carried out by enzymes associated with plastid membranes. Still the significance of these protein-membrane and protein-lipid interactions in Chl synthesis and chloroplast differentiation are not very well-understood. In this review, we provide an overview on Chl biosynthesis in angiosperms with emphasis on its association with membranes and lipids. Moreover, the last steps of the pathway including the reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), the biosynthesis of the isoprenoid phytyl moiety and the esterification of Chlide are also summarized. The unique biochemical and photophysical properties of the light-dependent NADPH:protochlorophyllide oxidoreductase (LPOR) enzyme catalyzing Pchlide photoreduction and located to peculiar tubuloreticular prolamellar body (PLB) membranes of light-deprived tissues of angiosperms and to envelope membranes, as well as to thylakoids (especially grana margins) are also reviewed. Data about the factors influencing tubuloreticular membrane formation within cells, the spectroscopic properties and the in vitro reconstitution of the native LPOR enzyme complexes are also critically discussed.
Thylakoid membranes of photosynthetic organisms have a unique and highly conserved lipid composition: in addition to the phospholipid, phosphatidylglycerol (PG), they predominantly contain galactolipids (monogalactosyldiacylglycerol—MGDG, digalactosyldiacylglycerol—DGDG, and sulfoquinovosyldiacylglycerol—SQDG). Galactolipids are major components of plastid inner membranes that play an important role in chloroplast differentiation from proplastids or etioplasts, in chlorophyll (Chl) biosynthesis, in the accumulation of light-harvesting proteins (Fujii et al., 2019a,b), and have been also identified as structural components of several major protein complexes of the photosynthetic apparatus (PSII, PSI, LHCII, and cytochrome b6f) (Kobayashi, 2016). In this review, we provide an overview of the Mg-branch of tetrapyrrole biosynthesis, leading to Chl biosynthesis, that occurs in plastids and is associated to plastid membranes. We focus on the reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), and the role of lipids and plastid inner membranes in the process. Two distinct enzymes have been evolved to catalyze this reaction step: first a nitrogenase-like, oxygen-sensitive dark-operative NADPH:Pchlide oxidoreductase enzyme (DPOR), and later a light-dependent NADPH:protochlorophyllide oxidoreductase (LPOR), which requires light for its activity (Gabruk and Mysliwa-Kurdziel, 2015; Vedalankar and Tripathy, 2019). There is very low sequence homology between these two enzymes (Gabruk et al., 2012), and in most organisms they occur simultaneously. However, angiosperms lack LPOR and became unable to synthesize Chlide and chlorophylls (Chls) in the absence of light. Due to this special feature, Chl biosynthesis has been extensively studied in dark-germinated angiosperm seedlings which also have agricultural relevance as the seeds of many crops are sown deep into the soil and thus start to germinate in the dark. Due to space limitation our focus is on data available about the role of lipids and membranes on Chl biosynthesis in angiosperms.
Chlorophylls (i.e., Chl a and Chl b) are the main photosynthetic pigments in plants. Concerning their molecular structure, they belong to tetrapyrroles (Fiedor et al., 2019). Their biosynthesis takes place in plastids and shares some common steps with that of other tetrapyrroles. The tetrapyrrole biosynthesis route leading to Chl formation is often referred to as the Mg-branch (for review see Mochizuki et al., 2010; Tanaka et al., 2011; Tripathy and Pattanayak, 2012; Rebeiz, 2014; Willows, 2019; Bryant et al., 2020). Several new data indicate that galactolipids play crucial roles in Chl biosynthesis (Kobayashi et al., 2014; Fujii et al., 2019b) in addition to other factors regulating the Mg-branch at various levels and via different mechanisms as reviewed by Grimm (2010), Stenbaek and Jensen (2010), Zhang et al. (2011), Czarnecki and Grimm (2012), Richter and Grimm (2013), Brzezowski et al. (2015), Wang and Grimm (2015), Kobayashi and Masuda (2016), Yuan et al. (2017), and Herbst et al. (2019).
The incorporation of Mg2+ to protoporphyrin IX is the first reaction on the Mg-branch of tetrapyrrole biosynthesis and the protoporphyrin IX is the last common intermediate of both Chl and heme biosynthesis (Figure 1). This reaction is catalyzed by Mg-chelatase (EC 6.6.1.1) which is a large multisubunit complex composed of three subunits, CHLI, CHLD, and CHLH. Two of them, CHLD and CHLI, catalyze ATP hydrolysis, whereas the third one, CHLH, binds the substrate (protoporphyrin IX). During the two-steps reaction, first, an ATP- and Mg2+-dependent activation occurs, leading to the formation of the active Mg-chelatase complex, which is then followed by the ATP-dependent chelation step (reviewed by Masuda, 2008; Bryant et al., 2020). Recent detailed analyses unraveled the conformational changes and kinetic parameters of CHLH caused by the substrate binding (Adams et al., 2020). Two CHLI isoforms, CHLI1 and CHLI2, were found in A. thaliana, both having similar expression profiles. The dominant CHLI1 isoform is crucial for the chelatase activity, whereas CHLI2 plays a limited role in Chl biosynthesis but certainly contributes to the assembly of the Mg-chelatase complex (Kobayashi et al., 2008).
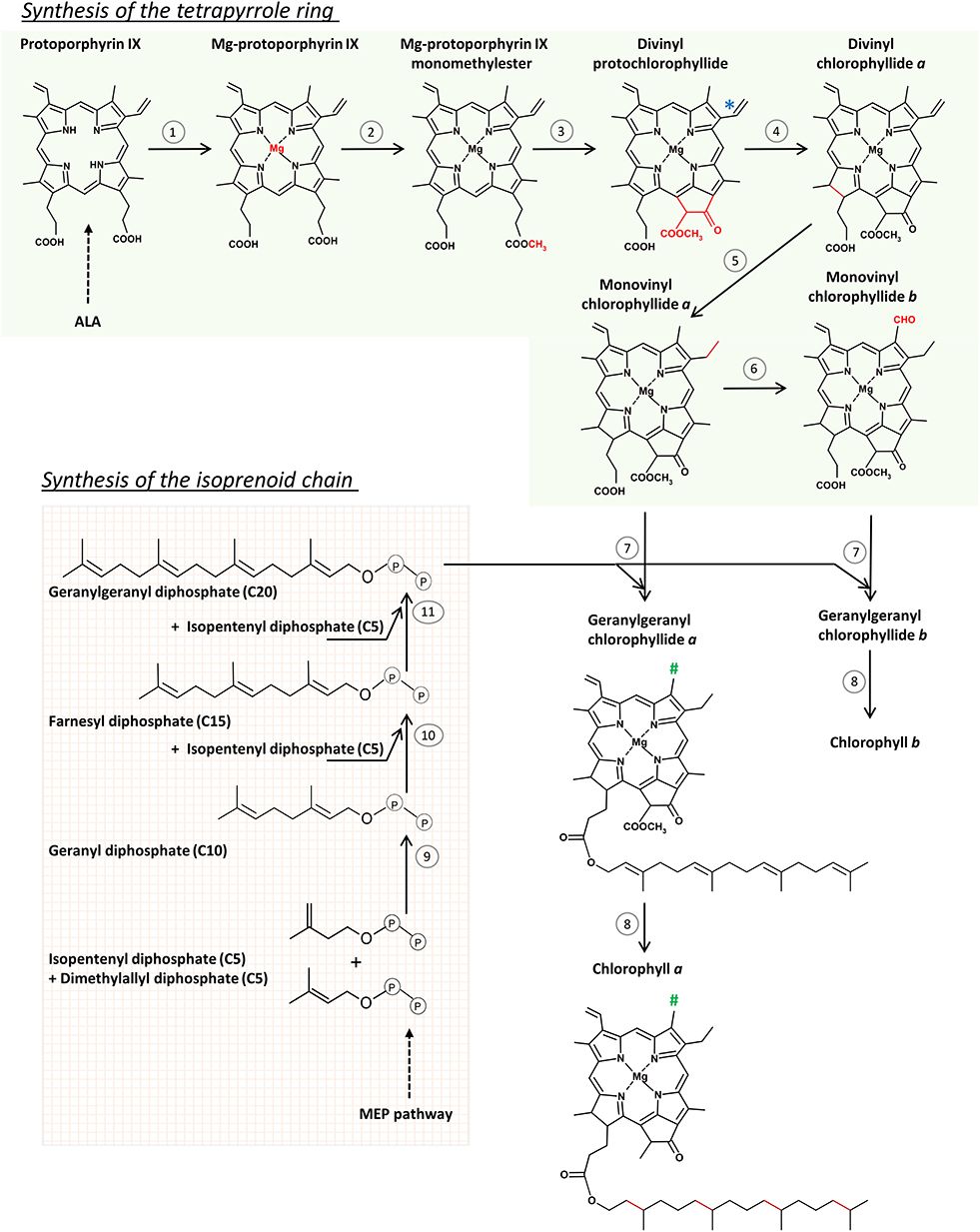
Figure 1. The scheme of chlorophyll (Chl) biosynthesis in angiosperms. Reactions of the tetrapyrrole ring formation route (1–6) are catalyzed by: (1) Mg-chelatase, (2) protoporphyrin IX methyltransferase, (3) Mg protoporphyrin IX monomethyl ester (oxidative) cyclase, (4) light-dependent NADPH:protochlorophyllide oxidoreductase, (5) divinyl reductase, (6) chlorophyllide oxygenase, (7) chlorophyll synthase, (8) geranylgeranyl reductase. (*) Divinyl reductase (5) can also convert the indicated divinyl group to a monovinyl group in protochlorophyllide molecule. (#) Methyl group in Chl a, and formyl group in Chl b. Reactions of the isoprenoid chain formation (9–11) are catalyzed by geranyl diphosphate and geranylgeranyl diphosphate synthases. See text for further explanations.
GUN4 is a positive regulator of Mg-chelatase and is involved in plastidic retrograde signaling (Larkin et al., 2003; Davison et al., 2005). Moreover, it is a key regulator of Chl biosynthesis acting on the posttranslational level (Peter and Grimm, 2009). Together with protoporphyrin IX, it was suggested to promote interactions between CHLH and chloroplast membranes (Adhikari et al., 2009). Changes in the activity of the Mg-branch under different light conditions is probably regulated by GUN4 phosphorylation (Richter et al., 2016).
Mg-protoporphyrin is converted to 3,8 vinyl-Pchlide (divinyl-Pchlide or DV-Pchlide) in two successive reactions. The first is the methylation of Mg-protoporphyrin IX catalyzed by Mg-protoporphyrin IX methyltransferase (EC 2.1.1.11). The second is the formation of the isocyclic ring E catalyzed by different cyclases in oxygenic and anoxygenic phototrophs, according to different mechanisms (Chen et al., 2017). In eukaryotic oxygenic phototrophs that include angiosperms, mostly discussed in this work, the formation of the E ring is catalyzed by Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase (EC 1.14.13.81). This is an oxygen-dependent enzyme, composed of multiple subunits (Chereskin et al., 1982; Wong and Castelfranco, 1984, 1985; Rzeznicka et al., 2005). To keep the enzymatic activity, Mg-protoporphyrin IX monomethylester cyclase (CHL27/CRD1) (Tottey et al., 2003) requires the presence of an auxiliary factor YCF54, which is the hypothetical chloroplast open reading frame 54 (Albus et al., 2012; Chen and Hunter, 2020). Among various factors, YCF54 interacts with ferredoxin-NADPH reductase (FNR1), which may deliver electrons from the photosynthetic electron transport chain to the cyclase (Herbst et al., 2018). However, the exact mechanism of the formation of the isocyclic E ring is still not fully elucidated.
A specific reductase, 8-vinyl reductase (EC 1.3.1.75 also called divinyl reductase, i.e., DVR), can reduce one of the two vinyl groups in DV-Pchlide converting DV-Pchlide to 3-vinyl Pchlide (monovinyl-Pchlide or MV-Pchlide), a reaction observed in etiolated seedlings (Belanger and Rebeiz, 1979, 1980; Shioi and Takamiya, 1992) but also during the night phases of photoperiodic growth (Carey and Rebeiz, 1985; Carey et al., 1985; Tripathy and Rebeiz, 1988). This observation contributed to the categorization of plants depending on whether they accumulate predominantly DV-Pchlide or MV-Pchlide during daytime or nights of photoperiodic growth (Ioannides et al., 1994; Mageed et al., 1997). In particular, Arabidopsis (DDV-LDV, i.e., a dark divinyl-light divinyl plant) accumulates mainly DV-Pchlide independently on light conditions whereas wheat and rice accumulate MV-Pchlide at night and mostly DV-Pchlide at daytime and are thus considered as DMV-LDV (i.e., dark monovinyl-light divinyl) plants. Further analysis of the accumulation of other MV- and DV- tetrapyrrole intermediates and their interconversion in various plant species led to a multibranched pathway, including parallel MV and DV paths of Chl biosynthesis interconnected by DVR (Tripathy and Rebeiz, 1986; Rebeiz et al., 1999; Wang et al., 2013). Plant DVR is a NADPH-dependent enzyme (Parham and Rebeiz, 1992; Whyte and Griffiths, 1993) and shows a broad substrate specificity (Wang et al., 2013), however, it is the most active for DV-Chlide as substrate (Parham and Rebeiz, 1992; Nagata et al., 2007; Wang et al., 2013). A putative transmembrane α helix has been identified near the C terminus of DVR (Nakanishi et al., 2005; Wang et al., 2010).
Both MV-Pchlide and DV-Pchlide are accepted as substrates by the enzymes catalyzing the subsequent reactions leading finally to the formation of the Chl a molecule (Knaust et al., 1993; Nagata et al., 2007). What is more, most literature data dealing with the last steps of Chl biosynthesis in angiosperms did not determine the exact nature of Pchlide (e.g., MV- or DV-Pchlide) in their samples. Therefore, and for simplicity, we decided to refer to the pigment below as Pchlide. Pchlide is a key intermediate of Chl biosynthesis in angiosperms. It is a porphyrin type compound, capable of light absorption. However, its Q absorption bands are strongly blue-shifted compared to that of Chls (Fiedor et al., 2019). The photophysical properties of Pchlide in various model systems and in plastids are thoroughly discussed in section LPOR—An Enzyme Operating in Lipid Environment.
DPOR (EC 1.3.7.7) is an ancestral but oxygen-sensitive enzyme catalyzing Pchlide reduction. According to a widely accepted long-held hypothesis, LPOR (EC 1.3.1.33) emerged as an evolutionary response to the increasing level of atmospheric oxygen (Fujita, 1996; Schoefs and Franck, 2003; Yamazaki et al., 2006; Reinbothe et al., 2010) caused by the emergence of oxygenic photosynthesis around 2.4 billion years ago (Suzuki and Bauer, 1995), and occurs only in oxygenic phototrophs. However, LPOR was discovered a few years ago in the aerobic anoxygenic phototrophic α-proteobacterium Dinoroseobacter shibae (Kaschner et al., 2014), and in other anoxygenic organisms (Chernomor et al., 2020), probably as a result of horizontal gene transfer from cyanobacteria. Further investigations are required to elucidate the evolutionary origin and distribution of LPOR. In angiosperms, the LPOR gene was duplicated several times, leading to the formation of LPOR isoforms in several species denoted as LPOR-A, LPOR-B, and LPOR-C (Gabruk and Mysliwa-Kurdziel, 2020) (discussed in detail in section Biochemical Characterization of LPOR).
In angiosperms, the reduction of Pchlide to Chlide is catalyzed by LPOR (for review see Scrutton et al., 2012; Gabruk and Mysliwa-Kurdziel, 2015). The photocatalytic reaction catalyzed by LPOR is the reduction of one of the conjugated double bonds in the porphyrin ring of Pchlide, thus converting it to a chlorine. The photophysical properties of the product, Chlide, are only slightly different from those of Chl (Fiedor et al., 2003, 2008). LPOR activity is induced by light and is fully inhibited in the dark. No chemical LPOR inhibitors are known. The biochemical characterization of LPOR and the reaction mechanism of the photoreduction are discussed in section LPOR—An Enzyme Operating in Lipid Environment.
The esterification of Chlide with an isoprenyl alcohol leads to the formation of Chl a molecule. Below, we'll first review the formation of the phytol chain, and then the esterification reaction itself.
The isoprenyl side chain of Chl is a phytyl moiety. Phytol is a 20-carbon (C20) alcohol, which is structurally related to geranylgeraniol, however, it is more saturated. The prenyl backbone of geranylgeraniol is synthesized in chloroplasts via the 2-methylerythritol-4-phosphate (MEP) pathway (Lichtenthaler et al., 1997), one of two pathways of isoprenoid biosynthesis present in plant cells. The characterization and detailed description of these pathways is beyond the scope of this paper and can be found in Rodriguez-Concepcion (2010), Vranová et al. (2013), Rodríguez-Concepción and Boronat (2015), and Gutbrod et al. (2019).
Geranylgeranyl diphosphate (C20) is formed from two prenyl diphosphate precursors: isopentenyl diphosphate (C5) and dimethylallyl diphosphate (C5), both produced in the MEP pathway, in sequential condensation reactions. Their condensation leads to geranyl diphosphate (C10). The condensation of geranyl diphosphate (C10) with an isopentenyl diphosphate (C5) molecule results in the formation of farnesyl diphosphate (C15), which can further condensate with another isopentenyl diphosphate (C5) molecule to form geranylgeranyl diphosphate (C20). These condensation reactions are catalyzed by the geranyl diphosphate and geranylgeranyl diphosphate synthases, EC 2.5.1.10 and EC 2.5.1.29, respectively, which are enzymes belonging to the family of isopentenyl-diphosphate synthases (prenyltransferases) (Gutbrod et al., 2019). Geranylgeranyl diphosphate (C20) is an important intermediate not only for Chl synthesis, but also for other biochemical pathways including the biosynthesis of carotenoids, prenyllipids and plant hormones (gibberellins). However, regulatory mechanisms of geranylgeranyl diphosphate consumption among these biosynthesis pathways are not yet fully understood.
Interestingly, isopentenyl diphosphate or farnesyl diphosphate formed in the cytosol by the alternative pathway of isoprenoid biosynthesis, the so-called mevalonate pathway (MVA), can contribute to the synthesis of chloroplast isoprenoids (Nagata et al., 2002; Bick and Lange, 2003; Opitz et al., 2014; Manzano et al., 2016; Pellaud and Saffrané, 2017). However, isoprenoids derived from the mevalonate pathway cannot substitute for the deficiency in the flux through the MEP pathway (Nagata et al., 2002). Geranylgeranyl diphosphate deficiency leads to overaccumulation of Chlide, which can be a source of photooxidative stress (Kim et al., 2013).
The plastid geranylgeranyl diphosphate synthase was first purified from Capsicum chromoplasts (Dogbo and Camara, 1987) and Sinapsis alba etioplasts (Laferrière and Beyer, 1991), and then cloned and further characterized (Kuntz et al., 1992). Multiple geranylgeranyl diphosphate synthase families were characterized in Arabidopsis (Beck et al., 2013) and some other plants (Wang and Dixon, 2009; Zhou et al., 2017; Wang et al., 2019). They show specific subcellular localizations and different expression patterns, which may be important for geranylgeranyl diphosphate synthesis depending on metabolic pathways, developmental stages, or specific tissues. Seven of 10 synthases found in Arabidopsis were localized to plastids. One of them (GGPPS11) mainly participates in the biosynthesis of plastid isoprenoids including Chls (Beck et al., 2013; Ruiz-Sola et al., 2016). GGPPS11 operates as a dimer with another geranylgeranyl diphosphate synthase, GGPPS12, which lacks motifs required for prenyl binding and is catalytically inactive (Beck et al., 2013; Ruiz-Sola et al., 2016). Binding of this accompanying protein regulates the enzyme specificity between the production of geranylgeranyl diphosphate and geranyl diphosphate. The accompanying protein in rice, which is called a recruiting molecule, controls the dimerization of geranylgeranyl diphosphate synthase and enhances its catalytic activity (Zhou et al., 2017). Moreover, it determines the allocation of the enzyme from the stroma to the thylakoid membranes, which is a way to control the flux of geranylgeranyl diphosphate toward Chl biosynthesis.
It is noteworthy to mention that independently from Chl biosynthesis, peculiar plastid inner membranes can be observed in plastids accumulating isoprenoids (Turner et al., 2000, 2012; reviewed e.g., in Solymosi and Schoefs, 2010) which indicates some direct or indirect interaction of these lipophilic molecules with the biomembranes.
Chlide is esterified with geranylgeranyl diphosphate to form geranylgeranyl Chlide (GG-Chlide). This reaction is catalyzed by chlorophyll synthase (EC 2.5.1.62) (Rüdiger et al., 1980). Pchlide is not accepted as the substrate for this enzyme (Griffiths, 1974; Rüdiger et al., 1980). However, in the two subsequent reactions, i.e., the photoreduction of Pchlide and the esterification of Chlide, the same part of the tetrapyrrole molecule is modified. Pchlide and Chlide are thus bound to the catalytic site of the respective enzymes, i.e., LPOR and chlorophyll synthase, in the same orientations (Helfrich et al., 1994, 1996; Rüdiger et al., 2005). Chlorophyll synthase binds the isoprenoid chain before binding of the second substrate, Chlide (Schmid et al., 2002).
Finally, the geranylgeranyl reductase (EC 1.3.1.83) reduces subsequently three double bonds in geranylgeranyl moiety of GG-Chlide and converts it to phytyl moiety, yielding this way Chl a. Observations that mainly GG-Chlide was detected shortly after illumination of etiolated seedlings (Schoch, 1978) whereas Chl a, having a phytyl moiety, was found in green barley seedlings (Soll et al., 1983) opened up a long-lasting discussion on the order of the esterification and reduction steps in plants. However, Schoefs and Bertrand (Schoefs and Bertrand, 2000) proved that the transformation of Chlide to Chl in developing seedlings is a four-steps process, which includes the successive formation of GG-Chlide, dihydrogeranylgeranyl Chlide and tetrahydrogeranylgeranyl Chlide as Chl a biosynthesis intermediates. The reduction of geranylgeranyl moiety of GG-Chlide was confirmed in tobacco cell cultures (Benz et al., 1984). Moreover, the preferential use of geranylgeranyl diphosphate than phytyl diphosphate by recombined Arabidopsis thaliana chlorophyll synthase was observed (Gaubier et al., 1995; Oster and Rüdiger, 1997). Chlides a and b were esterified at the same rate by recombinant chlorophyll synthase (Oster and Rüdiger, 1997).
In plastids, the same geranylgeranyl reductase may operate in different pathways of hydrogenation of geranylgeranyl moiety. The hydrogenation of GG-Chlide for Chl biosynthesis occurs in thylakoids whereas the reduction of geranylgeranyl diphosphate to phytyl diphosphate during the synthesis of tocopherols takes place in the envelopes (Soll et al., 1983; Keller et al., 1998). In etiolated seedlings the hydrogenation process is slowed down making it easier to observe than in green leaves. The activity of geranylgeranyl reductase is more affected by low temperature (273 K) than that of chlorophyll synthase (Schoefs and Bertrand, 2000). Cycloheximide is an inhibitor of the hydrogenation of geranylgeranyl to phytyl moiety in the irradiated etiolated seedlings, whereas it has no effect on Chlide esterification with geranylgeranyol (Rassadina et al., 2004).
Chl b is formed from Chlide in two reactions (Figure 1). The first is the two-step oxygenation of the side methyl group at the C7 in the ring B to a formyl group using molecular oxygen, which is catalyzed by chlorophyllide a oxygenase (CAO; EC 1.14.13.122) (Tanaka et al., 1998; for review see Rüdiger, 2002, 2006; Tanaka and Tanaka, 2007). Chlide a was shown to be the sole substrate for CAO activity, because neither Chl a nor Pchlide were accepted as the substrates of CAO in vitro (Oster et al., 2000; Rüdiger, 2002). The CAO is a membrane-bound protein, localized in thylakoid and envelope membranes (Eggink et al., 2004). The biosynthesis of Chl b plays an important regulatory role in the assembly and stabilization of light harvesting antennae (LHC) and as a consequence in granum stacking, which rely on the presence of this pigment (Espineda et al., 1999; Tanaka et al., 2001; Harper et al., 2004; Reinbothe et al., 2006). Judging from the low activity of the recombinant CAO in vitro it was suggested that additional accessory proteins might be required to reach the optimal catalytic oxygenase activity (Eggink et al., 2004), but these are yet unidentified.
Esterification of the D ring of Chlide, a reaction which is described in section Esterification of Chlide for Chl a formation, converts Chlide b into Chl b. The Chl a/b ratio in plants is regulated using the reversible conversion of Chlide a—Chl b—Chl(ide) a, which is known as “the chlorophyll cycle” (for a review see Rüdiger, 2002; Tanaka and Tanaka, 2019). Keeping the optimal level of Chl a/b ratio which correlates with the amount of the LHC complexes and supramolecular structure of photosynthetic complexes enables plant adaptation to changing environmental conditions (for a review see Tanaka and Tanaka, 2011; Voitsekhovskaja and Tyutereva, 2015).
The Mg-branch occurs at the plastid membranes and some enzymes are organized into macromolecular complexes. For example, LIL3, a protein belonging to LHC family, plays an essential role in the organization of complexes involved in Chl biosynthesis and in the delivery of Chls to photosynthetic complexes. By linking tetrapyrrole and terpenoid biosynthesis, LIL3 plays a critical role in the organization of later steps in Chl biosynthesis (Hey et al., 2017). Studies on the Arabidopsis mutant lacking LIL3 revealed the importance of this protein for Chl biosynthesis and in the stabilization of geranylgeranyl reductase (Tanaka et al., 2010). A direct interaction of geranylgeranyl reductase with LIL3 was demonstrated using a split ubiquitin assay, bimolecular fluorescence complementation as well as combined blue-native and SDS polyacrylamide gel electrophoresis (Tanaka et al., 2010). LIL3 was also shown to stabilize LPOR, and the direct LIL3-LPOR interaction was also confirmed using multiple analysis of protein-protein interactions (Hey et al., 2017). In addition to that, fluorescence in vitro analysis showed high binding affinity of LIL3 to Pchlide—the substrate of LPOR. However, no interactions with chlorophyll synthase were reported in this study. Similar complexes were detected in etio-chloroplasts and etioplasts of barley, using native gel electrophoresis with autofluorescence detection and mass spectrometry (Reisinger et al., 2008; Mork-Jansson et al., 2015). In these experiments, LIL3 formed complexes with LPOR, geranylgeranyl reductase and chlorophyll synthase. However, using split ubiquitin assay, the interaction between LIL3, LPOR and chlorophyll synthase was demonstrated, whereas no interaction with gerenylgeranyl reductase was proven (Mork-Jansson et al., 2015). Studies performed on thylakoids of rice (Zhou et al., 2017) revealed an additional protein called geranylgeranyl reductase recruiting protein, regulating the binding of geranylgeranyl reductase in the complexes clustered around LIL3.
Membrane complexes composed of Mg-protoporphyrin IX monomethylester cyclase, CHL27, LPOR (i.e., LPOR-B, LPOR-C), geranylgeranyl reductase and the FLU protein were identified in isolated thylakoids of Arabidopsis (Kauss et al., 2012). FLU is a negative regulator of Chl biosynthesis operating at the step of delta aminolevulinic acid (ALA) formation (Meskauskiene et al., 2001; Meskauskiene and Apel, 2002). In the absence of light, glutamyl-tRNA reductase is bound to FLU in these complexes and ALA formation is inhibited (Zhang et al., 2015). Formation of complexes of FLU with enzymes catalyzing the final steps of Chl biosynthesis was demonstrated using native electrophoresis, immunoprecipitation and mass spectrometry (Kauss et al., 2012).
It was also shown that, Mg-protoporphyrin IX monomethylester cyclase forms complexes with YCF54 and FNR1. However, YCF54 is probably a scaffold protein for a multi-subunit enzymatic complex, including other enzymes of late Chl biosynthesis, namely LPOR as well as the DVR and geranylgeranyl reductases (Kong et al., 2016; Herbst et al., 2018). Formation of such multi-subunit complexes favors the flow of intermediates during the Chl synthesis in light as well as the inhibition of the process in the dark. Taking into account that FNR1 provides the electrons for the cyclase activity, it is hypothesized that it might also deliver electrons for LPOR and DVR in the multi-enzymatic complex (Herbst et al., 2018, 2019). Nevertheless, the direct interaction of YCF54-FNR1 with LPOR and DVR has not yet been shown until now. Moreover, the question about the regulation of the electron flow in thylakoids remains open. Finally, it has not been elucidated how the late Chl synthesis is orchestrated at the beginning of angiosperm greening.
Another question which also remains open till date concerns the coexistence of CHL27-YCF54-FLU-LPOR-geranylgeranyl reductase complex with the LIL3-LPOR-geranylgeranyl reductase-geranylgeranyl diphosphate synthase-chlorophyll synthase complex in thylakoid membranes. LPOR and geranylgeranyl reductase were found in both type of complexes in thylakoid membranes. However, it remains unknown whether these complexes are somehow associated or stay separated. Recent analyses revealed that in Arabidopsis LPOR, curvature Thy1 (CURT1), the Mg2+-chelatase subunit 1 (CHLI) and Mg2+ protoporphyrin IX methyl transferase (CHLM) are also located to the granum margins (Wang et al., 2020). Further research is required to elucidate the exact localization, organization and regulation of the last steps of Chl biosynthesis. Answering these questions is important for the understanding of the regulatory mechanisms of the delivery of prenyl intermediates for Chl and tocopherol synthesis (Gutbrod et al., 2019).
Dual localization of enzymes involved in the Mg-branch in both chloroplast envelope and plastid inner membranes were documented based on biochemical studies (Joyard et al., 1990; Block et al., 2002; Tottey et al., 2003; Eggink et al., 2004; Tanaka and Tanaka, 2007), and later confirmed by proteomics (for review see Joyard et al., 2009; Bruley et al., 2012; Salvi et al., 2018). Dual localization was suggested to supply Chl to different Chl-binding proteins (PS core complexes and LHC) (Tottey et al., 2003). Additional research is required to elucidate the functional meaning of the dual localization and its relation to protein import mechanisms of plastids. Some yet unsolved problems that need to be mentioned: (i) interactions among enzymes and regulatory factors, (ii) structure and functioning of enzyme complexes, and (iii) their interaction with lipid membrane components, as well as (iv) the MV and DV heterogeneity of the biosynthesis route.
An interesting research area is the understanding of the regulatory networks connecting the biosynthesis of Chls, of protein components of photosynthetic complexes as well as lipids with the formation of the thylakoid membranes typical for chloroplasts. These processes need to be orchestrated to enable the proper assembly of the photosynthetic apparatus and to avoid the overaccumulation of unbound tetrapyrroles which may lead among others to photooxidative damage (Erdei et al., 2005; Hideg et al., 2010; Kim et al., 2013). It is nowadays known that the expression of Mg-chelatase and Mg-protoporphyrin IX monomethyl ester cyclase is linked to the synthesis of galactolipids (Fujii et al., 2014; Kobayashi et al., 2014; Kobayashi, 2016, 2018; Kobayashi and Masuda, 2016). Coordination of Chl and galactolipid biosyntheses goes through cytokinin and light signaling pathways (Kobayashi et al., 2014), however, further studies are needed to understand the molecular background of this regulation.
It has been shown that in etioplasts, the Mg-branch is sensitive to the membrane lipid environment, namely the MGDG and DGDG levels. Deficiency of any of these galactolipids in A. thaliana mutants resulted in overaccumulation of Mg-protoporphyrin IX pointing to the impairment of the following enzymes in the biosynthetic pathway (Figure 1) and leading to the accumulation of Pchlide intermediates and low Pchlide content (Fujii et al., 2017, 2018). On the contrary, the Mg-branch was reconstituted in E. coli cells from recombined Synechocystis proteins: Mg-chelatase, Mg-protoporphyrin IX methyltransferase, Mg-protoporphyrin IX monomethyl ester cyclase, LPOR, DVR, Chl synthase, and geranylgeranyl reductase without galactolipids (Chen et al., 2018). Thus, galactolipids rather enhance the activity of the enzymes, however, the regulatory mechanisms are yet unknown. Functioning of Mg-protoporphyrin IX methyltransferase, Mg-protoporphyrin IX monomethyl ester cyclase in barley etioplasts was also affected in the absence of carotenoids (La Rocca et al., 2007).
Among all reaction steps of Chl biosynthesis, the photoreduction of Pchlide is probably the most studied. This may be a result of several factors including (i) the unique catalytic activity of the enzyme involving an ultrafast light-activated photochemical reaction interesting from the biophysical and biochemical points of view, (ii) its important role in the regulation of tetrapyrrole biosynthesis, (iii) its association with arrested chloroplast differentiation and the formation of peculiar etioplast inner membranes, and (iv) the fact that dark-grown angiosperm seedlings represent a convenient plant material to study the last steps of the biosynthetic pathway in a synchronized way after illumination. Spectroscopy is also a useful tool to study this topic because there is a 30–40 nm difference in the spectral properties of the substrate, Pchlide (a porphyrin containing 11 double bonds in the tetrapyrrole ring) and the product, Chlide (a chlorine with 10 double bonds). This way and due to the high sensitivity of the delocalized electron system of the porphyrin ring to alterations in the molecular environment of the pigment, the reaction as well as the different populations of the pigments can be easily studied and characterized by relatively simple steady-state spectroscopic methods, which is summarized in the section Spectroscopic Properties of Pchlide in vivo. Pigments involved in similar molecular interactions and located in similar molecular micro-environments within the plastids exhibit similar spectral properties and thus represent a subpopulation of the entire pigment pool denoted as a pigment “form.” Pigment forms are often referred to by using the wavelength of their spectral maxima, but most of them have been also characterized from the biochemical and physiological points of view.
However, it is important to mention, that in green leaves containing significant amounts of Chls in the form of the various Chl-protein complexes of the photosynthetic apparatus, the fluorescence emission signal of Chl hinders the detection of its precursors such as e.g., Pchlide, which are present in much lower quantities (i.e., three order of magnitude lower amounts). Similarly, in green plants, the level of enzymes required for the steady-state Chl synthesis that produces Chl molecules to replace the pigments damaged during photosynthesis or stress conditions is very low and is hard to detect with classical biochemical or spectroscopic methods. Therefore, most studies related to the last steps of Chl biosynthesis have been performed on dark-germinated angiosperm seedlings in which the light-dependent LPOR enzyme is accumulating along with its substrate Pchlide, while Chl is absent.
When deprived from light during the early stages of their development, proplastids differentiate into a peculiar plastid type called etioplast in the photosynthetic tissues of dark-grown plants. The inner membranes of etioplasts consist of flat, sac-like membranes called prothylakoids, i.e., lipid bilayers encircling an inner aqueous phase called lumen, and a unique non-lamellar, but cubic phase inner membrane structure, the prolamellar body (PLB). Due to the inhibition of other light-signaling pathways [e.g., those involving phytochrome and cryptochrome (Sineshchekov and Belyaeva, 2019)] in darkness, the development of such dark-grown seedlings is also peculiar and is called skotomorphogenesis. When etiolated plants get illuminated, photomorphogenesis proceeds in them in parallel with the light-induced conversion of Pchlide to Chlide, resulting in the etioplast-to-chloroplast conversion which includes major reorganization of the plastid inner membranes, and active synthesis of the components of the photosynthetic apparatus (reviewed in Solymosi and Aronsson, 2013; Armarego-Marriott et al., 2020; Hernández-Verdeja et al., 2020).
Data about LPOR and its native structure and activity are reviewed below with emphasis on the lipid-protein interactions in them.
LPOR belongs to the short-chain dehydrogenase/reductase (SDR) protein superfamily (Yang and Cheng, 2004). It is a nuclear encoded protein which is then post-translationally imported to plastids (Aronsson et al., 2003b; Kim et al., 2005). Based on its amino acid sequence it is considered as a soluble and globular protein with surprisingly high contents of basic and hydrophobic amino acids (Schoefs and Franck, 2003; Masuda and Takamiya, 2004; Heyes and Hunter, 2005; Gabruk and Mysliwa-Kurdziel, 2015). Circular dichroism studies and bioinformatic tools predicted its secondary and tertiary structure (Schulz et al., 1989; Darrah et al., 1990; Birve et al., 1996; Dahlin et al., 1999; Townley et al., 2001; Buhr et al., 2008; Gabruk et al., 2012, 2015; Menon et al., 2016; Gholami et al., 2018), but hydropathy plots did not identify potential hydrophobic transmembrane segments in it (Benli et al., 1991; Spano et al., 1992). This is surprising as 98% of LPOR present in etioplasts has been localized to PLB membranes (Ryberg and Sundqvist, 1982a; Ikeuchi and Murakami, 1983), and it represents the major protein of isolated PLB fractions (Ryberg and Sundqvist, 1982a; Selstam and Sandelius, 1984; von Zychlinski et al., 2005; Blomqvist et al., 2008; Kanervo et al., 2008). Several in vitro and in vivo experiments involving washing and directed mutagenesis tried to identify the membrane association mechanism of LPOR. It has been shown that NADPH and ATP are required for its proper binding to plastid inner membranes such as PLBs, PTs and thylakoids (Dahlin et al., 1995; Engdahl et al., 2001). Washing experiments with various salts, detergents (Grevby et al., 1989; Selstam and Widell-Wigge, 1989) and proteases (Lütz and Tönissen, 1984; Dahlin et al., 1995; Engdahl et al., 2001) also revealed that LPOR binds more strongly to etioplast inner membranes (PLBs and PTs) than to thylakoid membranes. In chloroplasts, LPOR was predominantly found in the granum margins (Wang et al., 2020) which are, similarly to PLBs, also highly curved membranes with special lipid composition and special membrane organization. Based on its amino acid sequence LPOR is not an integral transmembrane protein, but because of its strong attachment to PLBs and PTs due to which it is difficult to solubilize it, it is probably not a peripheral membrane protein but an integral monotopic membrane protein which is permanently attached to one side of the plastid inner membrane. Using mutagenesis experiments, some amino acid residues (Cys) and the C-terminal have been shown to be involved in membrane association of LPOR (Dahlin et al., 1999; Aronsson et al., 2001), and recently a Chaperone-like Protein of POR1 (CPP1) has been identified which may be involved in anchoring LPOR to the PLBs (Lee et al., 2013b).
LPOR has a central β-sheet comprised of β-strands surrounded by α-helices, forming a typical dinucleotide binding Rossmann fold (Rossmann et al., 1974). The catalytic YxxxK and the NADPH-bounding G-rich (GxxxGxG) motifs are conservative (Wilks and Timko, 1995; Schoefs and Franck, 2003; Gabruk and Mysliwa-Kurdziel, 2020). LPOR crystal structure remained unknown for a long time; the structures of two cyanobacterial LPOR enzymes have been published only recently (Zhang et al., 2019; Dong et al., 2020). In general, both structures are compatible with the homology models, however, due to slightly different protocols there were some differences between these two works.
Most SDR proteins have tendency to form dimers and oligomers (Jörnvall et al., 1995), a property which was also found in LPOR macrocomplexes isolated from PLBs pre-treated with chemical cross-linkers (Wiktorsson et al., 1993), or isolated in fully photoactive native state from PLBs after mild solubilization and gel chromatography (Ouazzani Chahdi et al., 1998). Circular dichroism spectra (Mathis and Sauer, 1972; Böddi et al., 1989, 1990) and energy transfer studies (Kahn et al., 1970) also revealed that dimers or oligomers of the pigments are involved in the photoreduction of Pchlide. Cross-linking and subsequent mass spectrometric analysis of recombinant LPOR suggested that after substrate binding, structural changes occur in the LPOR oligomers which bring the catalytic motifs and the Pchlide molecules bound to the active site closer together (Gabruk et al., 2016). Similar observations were done in case of the analyses of cyanobacterial LPOR in which substrate binding induced oligomerization (Zhang et al., 2019) or monomers were observed in solution but homodimerization was observed during crystal formation (Dong et al., 2020). Experiments with recombinant pea LPOR also showed that it can form photoactive dimers in solution (Martin et al., 1997). A close distance between catalytic motifs brings the Pchlide molecules bound within the oligomers into close proximity, which enables energy transfer between them and also influences their spectral properties (Kahn et al., 1970). It has to be noted that the enzymatically active LPOR complexes have unique spectral and biochemical properties that are hardly reconstituted in vitro, especially in the absence of lipids (Gabruk et al., 2017). However, recent in vitro reconstitution studies successfully yielded crystal structures of oligomers: in case of cyanobacterial LPOR octamers were reported (Zhang et al., 2021) while in case of Arabidopsis LPOR helical structures associated with lipids were observed (Nguyen et al., 2021).
Data indicate the role of carotenoids [zeaxanthin and violaxanthin (Ouazzani Chahdi et al., 1998); neoxanthin and violaxanthin (Bykowski et al., 2020); the accumulation of poly-cis xanthophylls (Park et al., 2002; Cuttriss et al., 2007)] and lipids [MGDG (Aronsson et al., 2008; Fujii et al., 2017); and MGDG, PG, and SQDG (Gabruk et al., 2017; Nguyen et al., 2021)] in the formation of the photoactive enzyme complexes and the PLBs. Similarly, carotenoids (Denev et al., 2005) were suggested to be involved in the membrane association of LPOR. Lipid biosynthesis mutants had hindered LPOR activity, no LPOR oligomerization and abnormal PLB formation (Fujii et al., 2017, 2018, 2019b) which again outlines the strong relation between the LPOR oligomers, plastid lipids and inner membrane structures. Cryo electron microscopic investigations on in vitro reconstituted LPOR revealed that LPOR oligomers form helical filaments around lipid bilayer tubes and are inserted in the outer leaflet of the membranes (Nguyen et al., 2021).
LPOR gene probably appeared ~1.36 billion years ago, and since then it underwent several duplication events and mutations (Figure 2) (Gabruk and Mysliwa-Kurdziel, 2020). As a result, few organisms contain only one LPOR gene and thus one isoform [e.g., pea (Spano et al., 1992) and cucumber (Fusada et al., 2000)], but several organisms contain at least two different isoforms (termed in general or historically as LPOR-A and LPOR-B—barley, rice, tobacco, and wheat, see later for references) or even more than that [e.g., Arabidopsis also has LPOR-C (Oosawa et al., 2000; reviewed in Gabruk and Mysliwa-Kurdziel, 2020)]. The isoforms share ~75% sequence homology, but according to detailed functional analyses performed on a few species—barley (Holtorf et al., 1995; Garrone et al., 2015), Arabidopsis (Armstrong et al., 1995), tobacco (Masuda et al., 2002), wheat (Blomqvist et al., 2008), and rice (Kwon et al., 2017)—they have different expression patterns, substrate binding affinities, different plastid import mechanisms, different localization within the plastid membranes and different gene regulation patterns also depending on the developmental stage, temperature and light conditions (reviewed in Solymosi and Schoefs, 2008, 2010; Gabruk and Mysliwa-Kurdziel, 2020). The transport of the different isoforms into the plastids is peculiar as it shows a large substrate, cell, tissue, and organ specificity (Aronsson et al., 2003b; Kim et al., 2005), the detailed discussion of which is beyond the scope of this review.
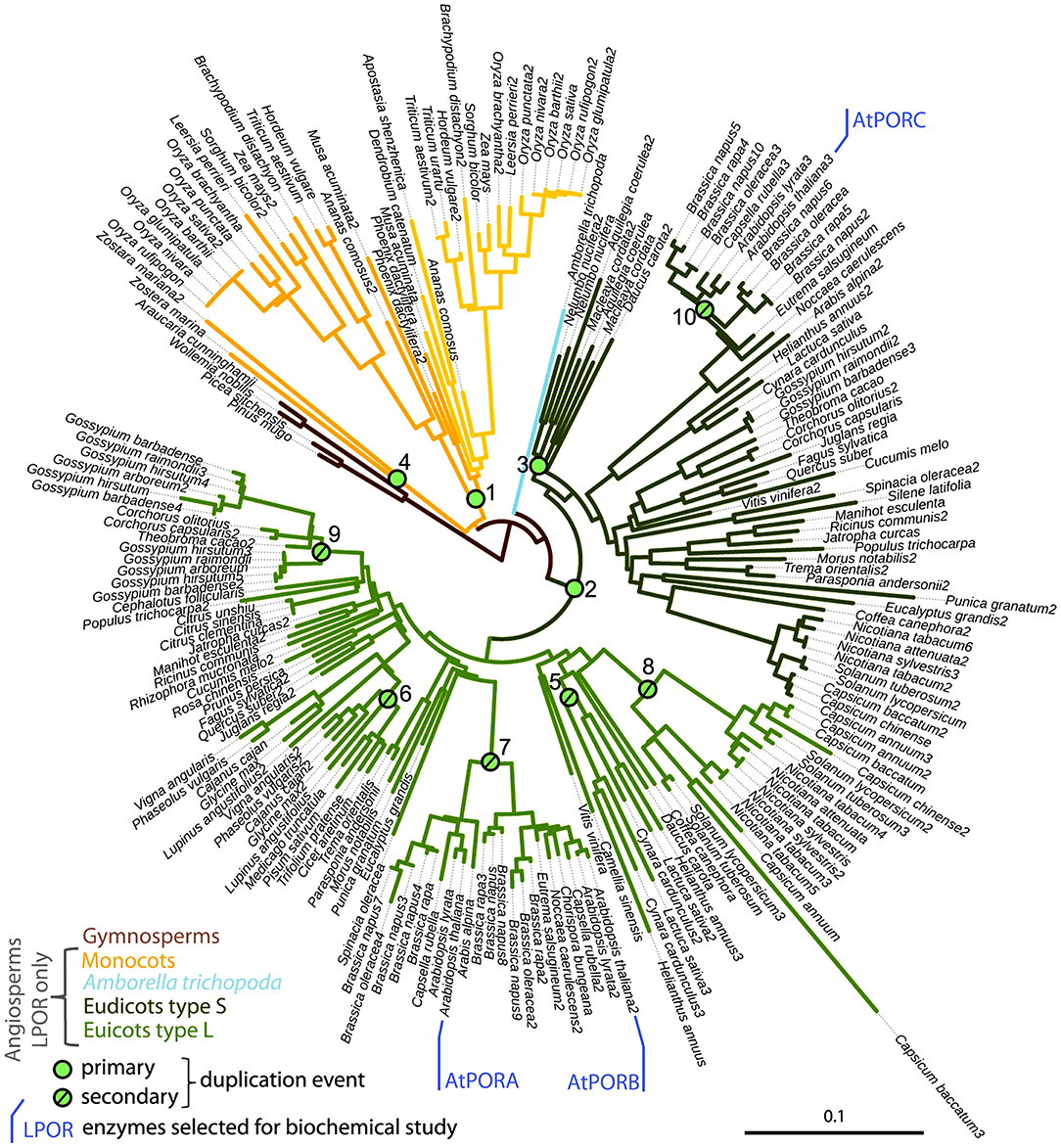
Figure 2. Subtree of LPOR sequences from seed plants, with Arabidopsis thaliana's isoforms AtPORA, AtPORB, and AtPORC being marked, and green dots indicating duplication events. Republished with permission of Portland Press Ltd., from The origin, evolution and diversification of multiple isoforms of light-dependent protochlorophyllide oxidoreductase (LPOR): Focus on angiosperms; Gabruk and Mysliwa-Kurdziel (2020), permission conveyed through Copyright Clearance Center, Inc.
LPOR-A and LPOR-B are present in etiolated material, while LPOR-C is expressed typically in green tissues (Oosawa et al., 2000; Aronsson et al., 2003a; Masuda et al., 2003; reviewed in Solymosi and Schoefs, 2010; Gabruk and Mysliwa-Kurdziel, 2015). LPOR-A is transiently expressed during early phases of development when large amounts of pigments need to be synthesized quickly, while LPOR-B and LPOR-C are thought to be responsible for the bulk Chl synthesis of adult or green plants (Paddock et al., 2010, 2012). Arabidopsis double mutants lacking LPOR-B and -C were shown to be unable to produce enough Chl under light conditions, indicating the importance of these isoforms in the biogenesis of the photosynthetic apparatus and also its membrane structures such as grana (Frick et al., 2003). Some ancestral LPOR genes (like that of Synechocystis) may even have different catalytic activity as they operate in lipid independent manner in contrast with other LPORs analyzed so far in angiosperms or gymnosperms (Gabruk and Mysliwa-Kurdziel, 2020). Taking into account the biochemical characteristics of LPOR isoforms, their interaction with lipids, as well as their phylogenetic relationships, Gabruk and Mysliwa-Kurdziel (Gabruk and Mysliwa-Kurdziel, 2020) proposed a new classification of LPOR family consisting of three LPOR types (Figure 2). The first one includes bacterial LPORs, termed “Z-type,” which are lipid-independent. Two other categories, termed L-type and S-type LPOR isoforms, are lipid-driven and present in angiosperms. L-type isoforms preferentially form complexes on the lipid membranes (like A. thaliana LPOR-A and LPOR-B), while the S-type ones (like A. thaliana LPOR-C) are active both with and without lipids. Further biochemical investigation is required to characterize the effect of lipids on LPOR isoforms from other angiosperm species.
Native spectral properties of Pchlide complexes in etiolated seedlings, and the spectral changes following the light-induced reduction of photoactive Pchlide were described already in the 50's (Figure 3) (Shibata, 1957). Photoreduction of Pchlide can take place at 203 K and reversible intermediates can be observed already at 77 K (Sironval and Brouers, 1970; Heyes et al., 2002, 2003; Belyaeva and Litvin, 2011, 2014), therefore, low temperature (typically 77 K) spectroscopic methods are needed to characterize the native state of the pigments. The absorption and fluorescence emission spectra of etiolated leaves contain two major spectral bands, each attributed to a specific pigment form also characterized at the biochemical level. Since low temperature absorption spectroscopy is less frequently used, we will refer to 77 K fluorescence properties of the pigments in this work.
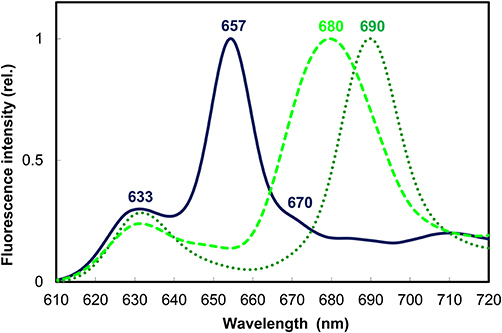
Figure 3. Normalized 77 K fluorescence emission spectra of 10-day-old dark-grown wheat (Triticum aestivum) leaves before (solid line) and after illumination with white light of 100 μmol s−1 m−2 photon flux density for 10 s (dotted line) and a subsequent 15 min dark incubation to reach the end stage of Shibata shift (broken line). Experimental conditions were as in Solymosi et al. (2002) and Smeller et al. (2003).
The short-wavelength band with fluorescence emission maximum at 633 nm represents a pool of monomeric Pchlide pigments bound either to the membrane surface or to a yet unidentified protein or to LPOR, but not in the active site of the enzyme or not to the active form of the enzyme (Figure 3). These pigments were primarily localized to the prothylakoid membranes of the etioplasts (Ryberg and Sundqvist, 1982a,b) and to the cytosolic side of the outer envelope (Joyard et al., 1990). They are not directly photoconvertible with a flash, and thus belong to the so-called “non-photoactive” Pchlide pool.
Another major band located at 655–657 nm belongs to the so-called “photoactive” Pchlide pool, i.e., Pchlide pigments bound to the active site of LPOR macrodomains, which correspond to oligomers of Pchlide:LPOR:NADPH ternary complexes and are strongly associated with the PLB membranes of the etioplasts (Figure 3) (Ryberg and Sundqvist, 1982a,b). Gaussian deconvolution and further spectral and biochemical analyses of isolated and fractionated etioplast inner membranes revealed the presence of other minor Pchlide forms, like for instance smaller oligomers (probably dimers) of Pchlide:LPOR:NADPH ternary complexes with emission maximum at 644 nm which are also photoactive and are suggested to be located to the edge of the PLB membranes (Böddi et al., 1990, 1991, 1992). In addition, a non-photoactive Pchlide molecular subpopulation hypothetically located to the central regions of PLBs and having fluorescence emission maximum at around 670 nm was also described (Figure 3) (Böddi et al., 1990; Bykowski et al., 2020). Upon short (already μs-long) illumination the fluorescence emission of Pchlide:LPOR:NADPH oligomers with emission maxima at 655 nm disappears, and that of the freshly produced Chlide:LPOR:NADP+ oligomers appears at 690 nm (Figure 3) (Böddi et al., 1990). After 10–15 min these oligomers and the pigments located in them undergo conformational changes and disaggregation as reflected by the blue shift of their emission maximum toward 680 nm, referred to as the so-called Shibata shift (Figure 3) (Shibata, 1957; Smeller et al., 2003; Solymosi et al., 2007a).
After the ultrafast transformation of the LPOR-bound, so-called “photoactive” Pchlide molecules into Chlide, and the subsequent slower conformational changes and reorganizations of LPOR oligomers including the dissociation of Chlide, the so-called “non-photoactive” Pchlide molecules can bind to the active site of LPOR and can be then also directly transformed into Chlide in the same ultrafast photochemical reaction step.
Depending on the oxidation state of NADPH in the ternary complexes, and also on the studied species and organs (e.g., leaves or stems) other spectral forms have been also characterized but discussion about them, as well as about developmental and other species-specific factors determining the ratios of the different forms is beyond the scope of this review, and can be found e.g., in Schoefs (2005), Belyaeva and Litvin (2007), and Solymosi and Schoefs (2008, 2010). Below we summarize knowledge about the molecular background of Pchlide photoreduction and the membrane association of LPOR.
The major hurdle in understanding LPOR structure and its exact molecular interaction with PLB membranes is the fact that the in vivo crystal structure of photoactive LPOR complexes is not available so far. In addition, photoactive dimeric and oligomeric complexes of LPOR are also hard to be isolated and fully purified from etiolated tissues because they often undergo disaggregation and dissociation during these processes. As stated earlier, it is even harder to detect or isolate them from fully green plant material. On the other hand, photoactive LPOR is also hard to reconstitute in vitro. Below we'll review some key spectral and structural features of Pchlide and LPOR, and also discuss how various in vitro experiments and reconstitution studies helped us to better understand the native structure of LPOR.
The key role of Pchlide in the light-triggered biosynthesis of Chl is related to its electronic properties. The Pchlide molecule captures sunlight and uses the absorbed energy to power the enzymatic reduction of its C17–C18 double bond. Data for understanding Pchlide photochemistry were collected from numerous studies of Pchlide in various model systems. It is now well-documented that Pchlide is an intrinsically reactive molecule. After light absorption, an electronically excited Pchlide molecule undergoes deexcitation using two parallel pathways (Dietzek et al., 2004, 2006b, 2009; Colindres-Rojas et al., 2011). The first one, called the reactive pathway, goes through an intramolecular charge-transfer state (SICT), which is non-fluorescent. The other, called the non-reactive pathway, goes through the lowest excited singlet state (S1), which then decays via fluorescence or by intersystem crossing to the long-lived Pchlide triplet state. Charge separation across Pchlide molecule in the SICT state depends on solvent polarity and is more stable in polar solvents (Dietzek et al., 2006a, 2010). A carbonyl group (C13) in the Pchlide isopentanone ring is of special importance for this charge separation (Sytina et al., 2010; Heyes et al., 2017), and the formation of the SICT state is important for Pchlide photocatalysis (Dietzek et al., 2006b, 2010; Heyes et al., 2015).
Fluorescence emission spectra of Pchlide in organic solvents have the maximum between 626 and 641.5 nm (Mysliwa-Kurdziel et al., 2004). The photophysical properties of Pchlide S1 state only weakly depend on non-specific solvation, as revealed from the small Stokes shifts (between 50 and 300 cm−1) observed in organic solvents (Mysliwa-Kurdziel et al., 2004). Specific solvation in protic (methanol, ethanol) and coordinating solvents (pyridine) enlarges the Stokes shift due to the lowering of the S1 state energy, and shortens the fluorescence lifetime. The site-specific solvation in Pchlide excited state via hydrogen bonding was confirmed experimentally (Mysliwa-Kurdziel et al., 2004; Sytina et al., 2010) and by theoretical calculation (Zhao and Han, 2008). DV-Pchlide has slightly red-shifted absorption and fluorescence maxima and differs in fluorescence lifetime when compared to MV-Pchlide (Kotzabasis et al., 1990; Kruk and Mysliwa-Kurdziel, 2004; Mysliwa-Kurdziel et al., 2008). The prenyl moiety, present in protochlorophyll molecule, only slightly changes the spectral and photophysical properties of the tetrapyrrole ring of Pchlide (Mysliwa-Kurdziel et al., 2004, 2008).
Pchlide in aqueous solutions forms aggregates even at low concentrations, which is manifested by a significant red shift of the absorption and emission maxima, and in strong fluorescence quenching (Mysliwa-Kurdziel et al., 2004, 2013b; Sytina et al., 2011a,b). Aggregation of the pigments was also observed in organic solvents at high Pchlide concentrations (Kotzabasis et al., 1990; Kruk and Mysliwa-Kurdziel, 2004; Mysliwa-Kurdziel et al., 2013b).
Liposomes composed of thylakoid lipids were used to model interactions of free Pchlide molecules with etioplast inner membranes (Mysliwa-Kurdziel et al., 2013a,b). Pchlide molecules were found at the interface area of the liposomes and/or the head-group area of the lipid bilayer. In the case of high Pchlide contents, aggregate formation was observed, which was facilitated in galactolipid liposomes. Pchlide aggregates had similar fluorescence emission to aggregated Pchlide:LPOR:NADPH complexes in vivo (i.e., at 656 nm), however, the excitation maximum was red-shifted to 480 nm. Experiments performed for Pchlide in reversed micelles from dioctyl sulfosuccinate sodium salt (AOT)/isooctane mixture showed that the molecular dynamics of water bound at the hydrophylic core of micelles is also important for Pchlide monomer-aggregate equilibrium (Mysliwa-Kurdziel et al., 2013b).
As observed in the experiments performed with in vitro enzyme-free model systems, the Pchlide chromophore is very sensitive to changes in its molecular environment. In the monomeric state its fluorescence emission varies between 626 and 641.5 nm, fluorescence emission maxima of Pchlide pigment forms above 641.5 nm can only be obtained when two or more Pchlide molecules are in close proximity, have overlapping delocalized electron systems and thus form aggregates in vitro (see above, and Mysliwa-Kurdziel et al., 2004, 2013a,b; Sytina et al., 2011a,b). A model summarizing the spectral properties and the molecular interactions of Pchlide in vitro is presented in Figure 4.
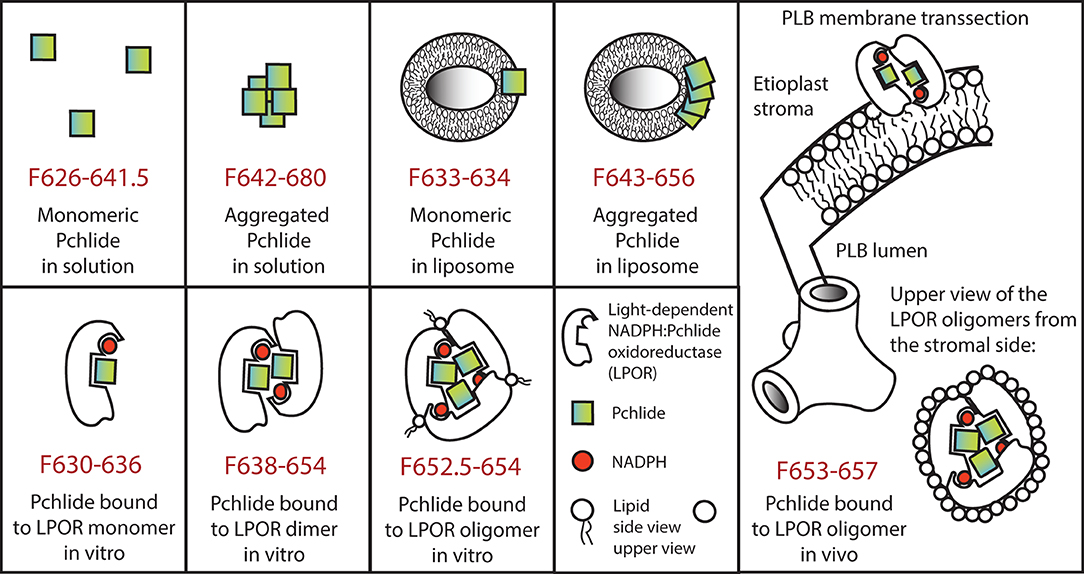
Figure 4. Model representing spectral properties and molecular interactions of protochlorophyllide in various in vitro model systems as well as in vivo in case of the major photoactive protochlorophyllide form located to the prolamellar bodies. In case of oligomers for simplicity trimers are represented, although larger aggregates (e.g., tetramers and octamers) have been also reported. For further details see the text.
Based on data obtained from in vitro model systems as well as other experimental evidence including CD spectroscopy of etioplast inner membrane fractions isolated from etiolated leaves (Böddi et al., 1989, 1990), it seems clear that the photoactive Pchlide complexes contain Pchlide pigments in at least dimeric or aggregated forms. This can be probably achieved by the distinct conformation of the LPOR dimers or oligomers in which the delocalized electron systems of neighboring Pchlide molecules overlap. Below we'll briefly review the latest in vitro reconstitution experiments of photoactive LPOR complexes. Again, we should mention that important pioneering observations have been done on isolated and partially purified photoactive complexes (in early works referred to as holochromes) from plants, but detailed discussion of these complexes and early data is provided elsewhere (e.g., Schoefs and Franck, 2003; Solymosi and Schoefs, 2010). Similarly, we only discuss data on in vitro reconstitution experiments of photoactive plant LPOR complexes, and not those about cyanobacterial LPOR (reviewed e.g., in Heyes and Hunter, 2005), because the peculiar membrane structures (prolamellar bodies) and direct LPOR-lipid and LPOR-membrane interactions have been only scarcely studied in such organisms (Schneidewind et al., 2019; Yamamoto et al., 2020).
Martin et al. (1997) successfully reconstituted photoactive complexes from recombinant pea LPOR with absorption maximum at around 630 nm, which were—based on their molecular mass—assigned to LPOR dimers. However, plant LPOR complexes with fluorescence emission maximum at 655 nm were reconstituted for the first time using a mixture of LPOR isoforms from barley (i.e., LPOR-A and LPOR-B), two different zinc derivatives of Pchlide as well as lipids extracted from isolated PLBs (Reinbothe et al., 1999). Later in vitro experiments using Pchlide and recombinant LPOR-A from Arabidopsis thaliana demonstrated that the formation of ternary LPOR complexes with fluorescence emission maximum at 655 nm requires the presence of MGDG and a negatively charged plant lipid (either PG or SQDG) (Figure 4) (Gabruk et al., 2017; Nguyen et al., 2021). Lipids did not only play a kind of structural role but also influenced the enzyme activity. Negatively charged lipids (PG and SQDG) did not influence the spectral properties of the complexes but affected their NADPH binding properties. When only PG was present, much lower concentrations of NADPH were required by the enzyme to form photoactive complexes, suggesting that these LPOR-A complexes are preferably associated to the lipid membranes, especially under low NADPH concentrations which are common in etiolated tissues (Gabruk et al., 2017).
Interestingly, the conical shaped, non-bilayer lipid, MGDG had a strong effect on the spectral properties of LPOR-A complexes in vitro. In the presence of MGDG, the fluorescence emission maximum of in vitro reconstituted LPOR complexes was shifted up to 652 nm, but the formation of complexes with native-like fluorescence properties (i.e., emission maximum at 655 nm) were only induced in the simultaneous presence of MGDG and PG (Figure 4) (Gabruk et al., 2017). Successful cryo electron microscopic analyses of in vitro assembled LPOR, NADPH, Pchlide in a mixture of lipids revealed that LPOR and Pchlide are inserted into the outer leaflet of the membranes, and LPOR forms oligomers arranged in helical filaments which are strongly associated with the membranes and have an important role in inducing and shaping their tubular organization (Nguyen et al., 2021).
As stated above, in etioplasts the photoactive LPOR proteins and their oligomers are mostly located to the PLBs and are only present in minor amounts in isolated prothylakoid fractions, in which monomeric, non-photoactive Pchlide complexes are dominating (Ryberg and Sundqvist, 1982b). LPOR (and especially LPOR-A) accounts for the vast majority of the proteins of the PLBs (Ryberg and Sundqvist, 1982a; Blomqvist et al., 2008).
On the other hand, it has to be added that fluorescence emission typical for monomeric Pchlide has been described in the cytosolic side of the outer envelope membranes of chloroplasts in spinach (Joyard et al., 1990). On the long term (i.e., after 10 min) and in the presence of glycerol slow transformations of the Pchlide pigments was observed in such isolated envelope membrane fractions upon illumination. This indicates that Pchlide-Chlide transformation may take place in the envelope membrane when the LPOR protein conformation is influenced by glycerol (Joyard et al., 1990) which is a component that stabilizes and maybe preferentially favors oligomerization (Zhong et al., 1996; Klement et al., 2000; Solymosi et al., 2002, 2007a; Smeller et al., 2003).
Information about the transport of the LPOR protein from the envelope membranes where they were located (Joyard et al., 1990) toward the internal parts of the plastids is scarce. On the other hand, Dahlin et al. (1995) did not observe accumulation of LPOR in the stroma and the envelope membranes, but have shown that LPOR is a peripheral protein associated with the stromal side of the thylakoid membranes of chloroplasts where it is bound more loosely to the membranes than to PLBs or PTs. Other authors located them to grana margins (Wang et al., 2020). Concerning the import of LPOR from the envelope toward the membranes, it may use plastid vesicle trafficking pathways (Lindquist et al., 2016; Lindquist and Aronsson, 2018).
PLBs represent a highly peculiar membrane structure, in which the lipids do not form bilayers, but special cubic phase structures. Such special membranes termed tubular complexes or tubuloreticular inclusions were observed within various intracellular compartments of several organisms including animals, humans or plants. In the former, these are thought to represent the modification of the (rough) endoplasmic reticulum, are in general located inside its cisternae or in the perinuclear space (e.g., Boor et al., 1979) and are rich in acidic glycoproteins. Tubuloreticular complexes have been observed in the cytoplasm of various animal [e.g., dog (Krohn and Sandholm, 1975; Madewell and Munn, 1990), Rhesus monkey (Feldman et al., 1986), cynomolgus monkey (Geisbert et al., 1992), horse and mule (Madewell and Munn, 1989), chicken (Schaff et al., 1976), rat (Datsis, 1993)] and human cells [e.g., lymphoid cells, lymphocytes (Schaff et al., 1972, 1973; Grimley et al., 1973, 1985; Splinter et al., 1975; Kang et al., 1984), monocytes (Luu et al., 1989; Kostianovsky et al., 2016), brain endothelial cells (Lee et al., 1988), liver endothelial cells (Geisbert et al., 1992), heart endothelial cells (Boor et al., 1979), renal cells (Hurd et al., 1969; Grimley et al., 1973; Krohn and Sandholm, 1975; Lee et al., 1988, 2013a, 2017; Datsis, 1993; Elmaghrabi et al., 2017), fibroblasts (Boor et al., 1979; Feldman et al., 1986)] where they were often associated with various, first of all immunological disorders or viral diseases [lupus erythematosus (Hurd et al., 1969; Grimley et al., 1973; Schaff et al., 1973), AIDS (Maturi and Font, 1996), AIDS-associated Kaposi's sarcoma (Marquart, 2005), cytomegalovirus infection (Lee et al., 1988), Sjögren's syndrome (Daniels et al., 1974), SARS (Almsherqi et al., 2005), many other types of virus infections (Deng et al., 2010; reviewed in Grimley and Schaff, 1976; Luu et al., 1989; Almsherqi et al., 2006; Deng et al., 2010) or certain neoplasms [e.g., pituitary tumors (Landolt et al., 1976), connective tissue neoplasms (Madewell and Munn, 1989), plasmacytoma (Madewell and Munn, 1990), intracranial germinomas (Matsumura et al., 1984), lung carcinomas (Schaff et al., 1976), hepatomas (Schaff et al., 1976), Burkitt's type lymphoma (Popoff and Malinin, 1976)].
Tubuloreticular membrane organization has been also observed inside mitochondria and plastids (Almsherqi et al., 2009; Almsherqi, 2010). In the former, cubic membrane formation was induced by starvation and/or autophagy processes of the amoeba cells, during which they were slowing the degradation of the organelle and thus contributed to the survival of the cell upon stress conditions (Chong et al., 2017). In case of plastids, they occurred either in etioplasts or etio-chloroplasts in the form of PLBs and were thus strongly associated with intensive Chl biosynthesis and accumulation of LPOR (Figure 5) (Solymosi and Schoefs, 2008, 2010; Solymosi and Aronsson, 2013) or in secretory plastids having active isoprenoid biosynthesis involving the plastid located MEP-pathway or biosynthesis of hydrophobic molecules such as cutin or suberin (Solymosi and Schoefs, 2010; Böszörményi et al., 2020). Rarely, such tubuloreticular membrane organizations could be interpreted as vesicle clusters (Lindquist et al., 2016) or as structures that appeared under various stress conditions [e.g., UV irradiation (Kovács and Keresztes, 2002)].
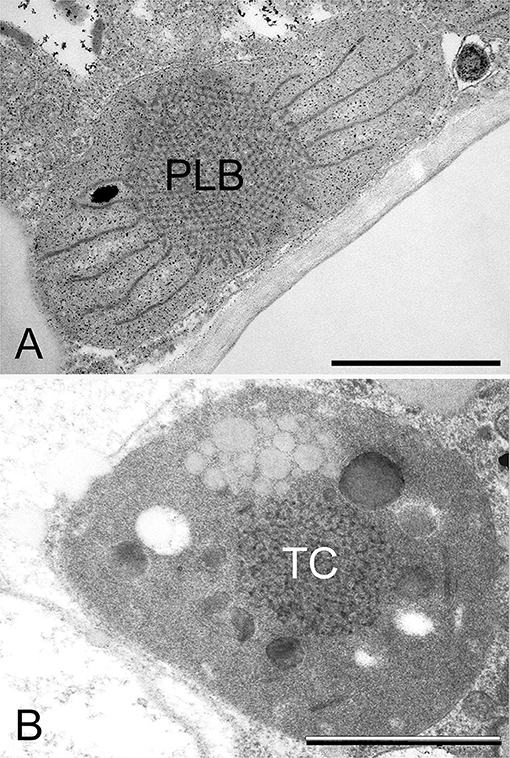
Figure 5. Transmission electron micrographs of the prolamellar body (PLB) present in the etioplast of the cotyledon of a 2-week-old dark-germinated rosemary (Rosmarinus officinalis) seedling (A), and tubular complex (TC) of a leucoplast of the neck cell of a peltate glandular hair on the surface of a light-grown adult rosemary plant (B). Scale bar: 1 μm. Sample preparation was as described in Böszörményi et al. (2020).
Secretory plastids containing tubuloreticular membranes were observed in various taxa and secretory tissues including extrafloral nectaries of Passiflora (Schnepf, 1961), plastids of various glandular hairs [e.g., Cannabis (Hammond and Mahlberg, 1978; Kim and Mahlberg, 1997; Solymosi and Köfalvi, 2017), Artemisia (Ascensao and Pais, 1982), Chrysanthemum (Vermeer and Peterson, 1979), Centrolobium (Matos and Paiva, 2012), Platanthera (Stpiczyńska et al., 2005)], Mentha piperita (Amelunxen, 1965; Turner et al., 2000, 2012), Perilla ocymoides (Kashina and Danilova, 1993) and Rosmarinus officinalis (Böszörményi et al., 2020). Some authors suggested that light may play a role in the formation of these peculiar membrane structures in secretory cells (Kashina and Danilova, 1993), while others observed identical structures in the neck cells of the peltate glandular hairs of both light-grown and dark-forced shoots (Böszörményi et al., 2020). Detailed comparative analyses about these peculiar non-bilayer membranes of Rosmarinus officinalis plastids have clearly demonstrated important structural differences. The PLBs of etioplasts represented a highly ordered and symmetrical membrane organization when compared with the loose and irregular tubuloreticular membranes of the secretory plastids (Figure 5) (Böszörményi et al., 2020).
The PLB should be thus considered as a highly regular subtype of tubular complexes, in which membrane tubules are arranged into tetrahedral (Figure 4) or hexapodel units, which are then joined into a paracrystalline 3D spatial network of membranes (Figure 5) (Gunning, 1965, 2001; Solymosi and Schoefs, 2008; Rudowska et al., 2012; Solymosi and Aronsson, 2013; Kowalewska et al., 2019; Bykowski et al., 2020). The different PLB types observed in different species and plastids, at different stages of development are beyond the scope of this review.
In spite of their widespread occurrence, it is still unclear whether tubuloreticular membranes have any specific function (e.g., in the production of secreted compounds of plastids or storage or accumulation of some metabolites) or they simply reflect some disturbance in the membrane homeostasis. The latter possibility may be outlined by the fact that such structures in animals or humans are mostly associated with stressful or diseased conditions [in case of AIDS tubuloreticular membranes have even been suggested to represent an ultrastructural pathological marker of the disease (Almsherqi et al., 2006)] and may be induced by exogenous or endogenous interferon (Grimley et al., 1985; Feldman et al., 1986; Orenstein et al., 1987), halogenated pyrimidines (Hulanicka et al., 1977) or even pyridoxine deficiency (Datsis, 1993). Stressful conditions such as starvation (Chong et al., 2017) or UV-irradiation (Kovács and Keresztes, 2002) also led to the formation of tubuloreticular complexes in amoeba mitochondria and apple plastids, respectively. On the other hand, some works suggest that such structures may be involved in regeneration processes of endothelial cells in wounded tissues (Eady and Odland, 1975), or in viral DNA or RNA uptake procedures (Almsherqi et al., 2006; Almsherqi, 2010).
In case of the PLBs, several data clearly indicate that due to their very high surface-to-volume ratio, these membranes represent a kind of membrane depot from which the photosynthetic membranes of the chloroplasts can be formed very fast upon illumination (reviewed in Solymosi and Schoefs, 2010; Solymosi and Aronsson, 2013). Thus, the etioplast-to-chloroplast transformation is much faster than the proplastid-to-chloroplast transformation pathway (Liebers et al., 2017), because during the latter important de novo synthesis of membrane lipids is required in parallel with protein and pigment biosynthesis. In addition, the PLBs and the proper organization and oligomerization of LPOR as well as the presence of carotenoids clearly play a role in photoprotection of the porphyrin pigments upon illumination, while in the absence of them, often photooxidative stress is induced (Sperling et al., 1997; Erdei et al., 2005; Hideg et al., 2010; reviewed by Solymosi and Schoefs, 2010). Similarly, cubic membranes efficiently prevented lipid peroxidation and RNA damage under oxidative stress conditions (Almsherqi, 2010).
There is no consensus on the factors inducing the formation of tubuloreticular membrane organization. Some authors suggested that they are the result of altered lipid (cholesterol) homeostasis, lipid and fatty acid composition induced by for instance viral infection (Almsherqi, 2010; Deng et al., 2010), others suggested that they may be associated with altered carotenoid composition (Park et al., 2002; Cuttriss et al., 2007), membrane symmetry (Larsson and Larsson, 2014), special protein-protein interactions or lipid-to-protein ratio (Almsherqi et al., 2009) or low divalent ion concentrations (Almsherqi et al., 2006; Brasnett et al., 2017). Such membrane organization was also observed in the aqueous lipid-protein film of lung surfactants (Larsson and Larsson, 2014). Therefore, it might be possible that a kind of specific water-lipid-protein composition is responsible for the formation of such structures. Similarly, we may speculate that the accumulation of lipophilic compounds (isoprenoids, terpenes, carotenoids, fatty acids, etc.) within the membranes could be involved in the formation of the PLBs. In the next section, we'll briefly discuss data about factors influencing PLB structure in etioplasts.
The lipid composition of the PLBs of etioplasts and chloroplast thylakoids is basically similar (Selstam and Sandelius, 1984; Selstam, 1998; Fujii et al., 2018, 2019a,b; Yu et al., 2020) with slightly lower amount of unsaturated (18:3) fatty acids being present in the PLB membranes than in the thylakoids (Selstam and Sandelius, 1984; Selstam, 1998). The ratio of the non-bilayer lipid MGDG to DGDG is slightly higher (1.6–1.8) in purified PLBs and in chloroplast thylakoids (1.7) than in prothylakoids (1.1–1.4) (Ryberg et al., 1983; Sandelius and Selstam, 1984; Selstam and Sandelius, 1984). Data on chloroplast structure of different lipid biosynthesis mutants revealed that minor alterations in the MGDG:DGDG ratio may have important effect on plastid structure (Mazur et al., 2019; Yu et al., 2020) and that dynamic local changes of the neutral/anionic lipid ratios may have important role in thylakoid arrangement (Kobayashi and Wada, 2016).
It is also evident from studies using lipid biosynthesis mutants (Fujii et al., 2017) and from in vitro reconstitution experiments (Gabruk et al., 2017) that some lipids (MGDG, DGDG and SQDG) are crucial for the proper assembly of PLBs and also for their transformation into thylakoids upon illumination (Fujii et al., 2018). Fujii et al. (2019b) have reviewed in detail the role of different plastid lipids on the Mg-branch of porphyrin biosynthesis, therefore, we prefer not to discuss these data in detail in this work.
In spite of the similar lipid composition, major differences are observed in the protein composition of chloroplast thylakoids and PLBs (Ryberg and Sundqvist, 1982a; Selstam and Sandelius, 1984; von Zychlinski et al., 2005; Blomqvist et al., 2008; Kanervo et al., 2008) with the latter containing much less proteins (and thus a relatively high lipid to protein ratio) and among them predominantly LPOR.
The lipid-dependent formation of the photoactive oligomers was proposed to be a mechanism inducing the formation of the PLBs (Gabruk et al., 2017), and correlations were found between PLB formation and proper lipid composition and accumulation, and proper amounts of LPOR in other works as well (reviewed in Fujii et al., 2019b). Similarly, a clear relationship was found between the formation and accumulation of LPOR oligomers (especially LPOR-A) and the occurrence and size of PLBs in etioplasts (Sperling et al., 1997; Franck et al., 2000; Frick et al., 2003; Paddock et al., 2010, 2012). Studies using Arabidopsis thaliana mutants showed that the amounts of LPOR-A and LPOR-B correlate with PLB size. Inhibition of LPOR-A expression led to reduced PLB size (Frick et al., 2003; Paddock et al., 2010, 2012), while its overexpression resulted in larger PLBs (Sperling et al., 1997; Franck et al., 2000; Paddock et al., 2010, 2012).
Similar data were observed in pea with overexpressed or antisense-LPOR (Seyedi et al., 2001). Furthermore, in organs with low LPOR levels, proplastids are present and PLBs are scarce or small [e.g., in non-leaf organs of pea (Böddi et al., 1994)]. Recent data on cyanobacterial LPOR indicated that it is present in dimerized form in vivo (Schneidewind et al., 2019) and its overexpression induced the formation of tubuloreticular membranes slightly resembling PLBs within the cells accumulating LPOR and Pchilde but being deficient in NADPH (Yamamoto et al., 2020).
Similarly, the disaggregation of the LPOR oligomers or the degradation of LPOR are also strongly associated with the disruption of the regular structure of the PLBs (e.g., Ryberg and Sundqvist, 1988) both under dark conditions (and induced by stress factors, e.g., Solymosi et al., 2006b) or during the light-induced greening of etiolated leaves, during which the PLB is fully reorganized and transformed into developing thylakoids (Kowalewska et al., 2016; reviewed in Solymosi and Schoefs, 2010; Solymosi and Aronsson, 2013; Kowalewska et al., 2019). These large-scale membrane reorganizations occur in parallel with important spectral changes (e.g., the Shibata shift—Figure 3, Shibata, 1957) which reflect the disaggregation of Chlide pigments and further outline the strong structural connection between LPOR macrodomains and PLB membranes. The Shibata shift is strongly inhibited by low temperature, high pressure as well as protein cross-linkers, or glycerol and sugars (Wiktorsson et al., 1993; Zhong et al., 1996; Solymosi et al., 2002, 2007a; Smeller et al., 2003), i.e., factors and conditions which stabilize the oligomeric structure, inhibit its conformational changes, disaggregation or the dissociation of peripheral proteins from membranes. However, under normal conditions, in parallel with the disruption of the PLB structure, the LPOR oligomers also undergo disaggregation and release Chlide from their active site. After this, the photoactive LPOR oligomers are again regenerated by binding Pchlide (Granick and Gassman, 1970; Amirjani and Sundqvist, 2004; Rassadina et al., 2004; Schoefs and Franck, 2008) from the pool of non-photoactive Pchlide molecules, and they again catalyze Pchlide photoreduction in them. Several authors reported that until Chl biosynthesis is still active in organs, because their final Chl content has not been reached, PLBs (Ikeda, 1970, 1971) and LPOR oligomers persist in their plastids and may even accumulate under special light conditions (e.g., Solymosi et al., 2006a,b; Schoefs and Franck, 2008; Solymosi et al., 2012). The plastids of such low-light-grown or partially etiolated or young tissues contain simultaneously developing grana and PLBs, thus an important spatial and structural heterogeneity can be clearly observed in the organization of the plastid inner membranes in these so-called etio-chloroplasts. Unfortunately, the PLBs of etio-chloroplasts seem to be versatile structures hard to isolate, therefore, we have not much information about the lipid and protein composition of these membranes.
Most data on Chl biosynthesis and especially about Pchlide photoreduction and its molecular details were obtained using etiolated seedlings. The use of etiolated systems to study these processes were often criticized because the major chloroplast differentiation pathway under natural light conditions is the proplastid-to-chloroplast pathway occurring in most seeds germinating on the soil surface or in new leaves produced by the shoot apical meristem (Charuvi et al., 2012; Yadav et al., 2019). However, several data indicate that etioplasts and accumulation of Pchlide and LPOR ternary complexes occur also under natural conditions in seedlings germinating in the soil (e.g., Vitányi et al., 2013; Kakuszi et al., 2017), in tissues partially covered by other organs (e.g., Solymosi et al., 2007b), in inner leaf primordia developing inside closed bud structures (e.g., Solymosi et al., 2004, 2006a, 2012), and in other systems (water plants, etc., reviewed in Solymosi and Aronsson, 2013; Armarego-Marriott et al., 2020). In such tissues or organs Chl biosynthesis and etioplast-to-chloroplast transformation may be similar to those described in completely etiolated seedlings, but the processes and their regulation still need to be elucidated in such naturally etiolated tissues.
In addition, photosynthesis requires a constant supply of Chls especially in plants under fluctuating or stressful conditions. Several data indicate that for instance after recovery from desiccation or drought stress (e.g., Solymosi et al., 2013; Liu et al., 2018, 2020), Chl synthesis genes are upregulated and an intensive biosynthesis occurs. So far only limited amount of information is available about LPOR catalytic activity and macromolecular organization in chloroplasts. Taken together, a better understanding of the Mg-branch of Chl biosynthesis, especially of LPOR activity, structure and location, and factors involved in its regulation in chloroplasts and etio-chloroplasts of various green (crop) plants may be also important for agriculture. The recent breakthroughs in molecular biology (e.g., next generation sequencing and tremendous developments in various omics techniques which enable the identification of different isoforms and their expression and translational patterns in various plant species and plastid subcompartments) and microscopic methods, as well as the use of various novel model systems (i.e., dark-forced tobacco shoots—Armarego-Marriott et al., 2019, duckweed—Monselise et al., 2015) will probably provide novel and interesting data on the exact molecular regulation of these processes and the role of lipids in them.
A large step into the understanding of LPOR-lipid interactions has been achieved by the in vitro reconstitution of photoactive LPOR macrodomains in the presence of lipids (Nguyen et al., 2021). These data demonstrate for the first time directly the role of LPOR and LPOR-lipid interactions in the formation of a special membrane phase and helical organization. In addition to analyses on different mutants, similar in vitro reconstitution experiments and membrane and lipid binding assays of various LPOR isoforms as well as other Chl biosynthesis enzymes of different species should be performed to increase our understanding of the membrane association and localization of Chl biosynthesis. It is also important to outline, that at the moment our data are mostly related to the 3 LPOR isoforms of Arabidopsis, and the 2 isoforms present in few crop species (rice, barley), but further experiments with other crops with less or more isoforms are necessary to understand LPOR activity and its universal or specific regulation in case of the various isoforms and species. The same applies to other enzymes of Chl biosynthesis. Due to its major and central role in autotrophic plant metabolism, we believe that an increased understanding of Chl biosynthesis may be crucial to breed plants with improved quality, e.g., higher yield or performance under adverse environmental conditions.
KS and BM-K equally contributed to the conceptualization, writing, and discussion about the manuscript's content. Both authors edited and approved the submitted version.
KS acknowledges the support from the Bolyai János Research Scholarship of the Hungarian Academy of Sciences and from the National Research, Development and Innovation Office (Grant OTKA FK 124748). BM-K acknowledges the support from the Polish National Science Centre (NCN; https://www.ncn.gov.pl) as a part of Grant UMO-2013/10/E/NZ3/00748.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
KS acknowledges ERASMUS+ Training Staff Mobility for financing her short trip to Krakow in June 2019.
Chl, chlorophyll; Chlide, chlorophyllide; DPOR, dark-operative NADPH:Pchlide oxidoreductase enzyme; DGDG, digalactosyldiacylglycerol; DV-Pchlide, divinyl-Pchlide (3;8 vinyl-Pchlide); DVR, divinyl reductase; FNR1, ferredoxin-NADPH reductase 1; GG-Chlide, geranylgeranyl chlorophyllide; LPOR, light-dependent NADPH:protochlorophyllide oxidoreductase; MEP, 2-methylerythritol-4-phosphate pathway; MGDG, monogalactosyldiacylglycerol; MV-Pchlide, protochlorophyllide; Pchlide, protochlorophyllide; PG, phosphatidylglycerol; PLB, prolamellar body; PTs, prothylakoids; SQDG, sulfoquinovosyldiacylglycerol.
Adams, N. B. P., Bisson, C., Brindley, A. A., Farmer, D. A., Davison, P. A., Reid, J. D., et al. (2020). The active site of magnesium chelatase. Nat. Plants 6, 1491–1502. doi: 10.1038/s41477-020-00806-9
Adhikari, N. D., Orler, R., Chory, J., Froehlich, J. E., and Larkin, R. M. (2009). Prophyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J. Biol. Chem. 284, 24783–24796. doi: 10.1074/jbc.M109.025205
Albus, C. A., Salinas, A., Czarnecki, O., Kahlau, S., Rothbart, M., Thiele, W., et al. (2012). LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 160, 1923–1939. doi: 10.1104/pp.112.206045
Almsherqi, Z. A. (2010). The Missing Dimension of Cell Membrane Organization. Study on Cubic Membrane Structure and Function. Available online at: https://scholarbank.nus.edu.sg/handle/10635/22981.
Almsherqi, Z. A., Kohlwein, S. D., and Deng, Y. (2006). Cubic membranes: a legend beyond the flatland of cell membrane organization. J. Cell Biol. 173, 839–844. doi: 10.1083/jcb.200603055
Almsherqi, Z. A., Landh, T., Kohlwein, S. D., and Deng, Y. (2009). Chapter 6 cubic membranes. The missing dimension of cell membrane organization. Int. Rev. Cell Mol. Biol. 274, 275–342. doi: 10.1016/S1937-6448(08)02006-6
Almsherqi, Z. A., McLachlan, C. S., Mossop, P., Knoops, K., and Deng, Y. (2005). Direct template matching reveals a host subcellular membrane gyroid cubic structure that is associated with SARS virus. Redox Rep. 10, 167–171. doi: 10.1179/135100005X57373
Amelunxen, F. (1965). Elektronenmikroskopische Untersuchungen an den Drüsenshuppen von Mentha piperita L. Planta Med. 13, 457–453. doi: 10.1055/s-0028-1100141
Amirjani, M. R., and Sundqvist, C. (2004). Regeneration of protochlorophyllide in green and greening leaves of plants with varying proportions of protochlorophyllide forms in darkness. Physiol. Plant. 121, 377–390. doi: 10.1111/j.1399-3054.2004.00340.x
Armarego-Marriott, T., Kowalewska, Ł., Burgos, A., Fischer, A., Thiele, W., Erban, A., et al. (2019). Highly resolved systems biology to dissect the etioplast-to-chloroplast transition in tobacco leaves. Plant Physiol. 180, 654–681. doi: 10.1104/pp.18.01432
Armarego-Marriott, T., Sandoval-Ibañez, O., and Kowalewska, Ł. (2020). Beyond the darkness: recent lessons from etiolation and de-etiolation studies. J. Exp. Bot. 71, 1215–1225. doi: 10.1093/jxb/erz496
Armstrong, G. A., Runge, S., Frick, G., Sperling, U., and Apel, K. (1995). Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 108, 1505–1517. doi: 10.1104/pp.108.4.1505
Aronsson, H., Schöttler, M. A, Kelly, A. A, Sundqvist, C., Dörmann, P., Karim, S., et al. (2008). Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol. 148, 580–592. doi: 10.1104/pp.108.123372
Aronsson, H., Sundqvist, C., and Dahlin, C. (2003a). POR - import and membrane association of a key element in chloroplast development. Physiol. Plant. 118, 1–9. doi: 10.1034/j.1399-3054.2003.00088.x
Aronsson, H., Sundqvist, C., and Dahlin, C. (2003b). POR hits the road: import and assembly of a plastid protein. Plant Mol. Biol. 51, 1–7. doi: 10.1023/A:1020795415631
Aronsson, H., Sundqvist, C., Timko, M. P., and Dahlin, C. (2001). The importance of the C-terminal region and Cys residues for the membrane association of the NADPH:protochlorophyllide oxidoreductase in pea. FEBS Lett. 502, 11–15. doi: 10.1016/S0014-5793(01)02595-9
Ascensao, L., and Pais, M. S. S. (1982). Secretory trichomes from Artemisia crithmifolia: some ultrastructural aspects. Bull. la Société Bot. Fr. 129, 83–87. doi: 10.1080/01811789.1982.10826552
Beck, G., Coman, D., Herren, E., Ruiz-Sola, M. Á., Rodríguez-Concepción, M., Gruissem, W., et al. (2013). Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 82, 393–416. doi: 10.1007/s11103-013-0070-z
Belanger, F. C., and Rebeiz, C. A. (1979). Chloroplast biogenesis XXVII detection of novel chlorophyll and chlorophyll precursors in higher plants. Biochem. Biophys. Res. Commun. 88, 365–372. doi: 10.1016/0006-291X(79)92057-6
Belanger, F. C., and Rebeiz, C. A. (1980). Chloroplast biogenesis. Detection of divinyl protochlorophyllide in higher plants. J. Biol. Chem. 255, 1266–1272. doi: 10.1016/S0021-9258(19)86024-6
Belyaeva, O. B., and Litvin, F. F. (2007). Photoactive pigment-enzyme complexes of chlorophyll precursor in plant leaves. Biochemistry 72, 1458–1477. doi: 10.1134/S0006297907130044
Belyaeva, O. B., and Litvin, F. F. (2011). Advances in understanding of the primary reactions of protochlorophyll(ide) photoreduction in cells and model systems. J. Biophys. Chem. 02, 1–9. doi: 10.4236/jbpc.2011.21001
Belyaeva, O. B., and Litvin, F. F. (2014). Mechanisms of phototransformation of protochlorophyllide into chlorophyllide. Biochem. Biokhimiia 79, 337–348. doi: 10.1134/S0006297914040038
Benli, M., Schulz, R., and Apel, K. (1991). Effect of light on the NADPH-protochlorophyllide oxidoreductase of Arabidopsis thaliana. Plant Mol. Biol. 16, 615–625. doi: 10.1007/BF00023426
Benz, J., Lempert, U., and Rüdiger, W. (1984). Incorporation of phytol precursors into chlorophylls of tobacco cell cultures. Planta 162, 215–219. doi: 10.1007/BF00397442
Bick, J. A., and Lange, B. M. (2003). Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 415, 146–154. doi: 10.1016/S0003-9861(03)00233-9
Birve, S. J., Selstam, E., and Johansson, L. B. (1996). Secondary structure of NADPH: protochlorophyllide oxidoreductase examined by circular dichroism and prediction methods. Biochem. J. 317, 549–555. doi: 10.1042/bj3170549
Block, M. A., Tewari, A. K., Albrieux, C., Maréchal, E., and Joyard, J. (2002). The plant S-adenosyl-L-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur. J. Biochem. 269, 240–248. doi: 10.1046/j.0014-2956.2001.02643.x
Blomqvist, L. A., Ryberg, M., and Sundqvist, C. (2008). Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth. Res. 96, 37–50. doi: 10.1007/s11120-007-9281-y
Böddi, B., Lindsten, A., Ryberg, M., and Sundqvist, C. (1989). On the aggregational states of protochlorophyllide and its protein complexes in wheat etioplasts. Physiol. Plant. 76, 135–143. doi: 10.1111/j.1399-3054.1989.tb05622.x
Böddi, B., Lindsten, A., Ryberg, M., and Sundqvist, C. (1990). Phototransformation of aggregated forms of protochlorophyllide in isolated etioplast inner membranes. Photochem. Photobiol. 52, 83–87. doi: 10.1111/j.1751-1097.1990.tb01759.x
Böddi, B., McEwen, B., Ryberg, M., and Sundqvist, C. (1994). Protochlorophyllide forms in non-greening epicotyls of dark-grown pea (Pisum sativum). Physiol. Plant. 92, 160–170. doi: 10.1111/j.1399-3054.1994.tb06667.x
Böddi, B., Ryberg, M., and Sundqvist, C. (1991). The formation of a short-wavelength chlorophyllide form at partial phototransformation of protochlorophyllide in etioplast inner membranes. Photochem. Photobiol. 53, 667–673. doi: 10.1111/j.1751-1097.1991.tb08495.x
Böddi, B., Ryberg, M., and Sundqvist, C. (1992). Identification of four universal protochlorophyllide forms in dark-grown leaves by analyses of the 77 K fluorescence emission spectra. J. Photochem. Photobiol. B Biol. 12, 389–401. doi: 10.1016/1011-1344(92)85043-T
Boor, P. J., Ferrans, V. J., Jones, M., Kawanami, O., Thiedemann, K. U., Herman, E. H., et al. (1979). Tubuloreticular structures in myocardium: an ultrastructural study. J. Mol. Cell. Cardiol. 11, 977–979. doi: 10.1016/0022-2828(79)90388-2
Böszörményi, A., Dobi, A., Skribanek, A., Pávai, M., and Solymosi, K. (2020). The effect of light on plastid differentiation, chlorophyll biosynthesis, and essential oil composition in rosemary (Rosmarinus officinalis) leaves and cotyledons. Front. Plant Sci. 11:196. doi: 10.3389/fpls.2020.00196
Brasnett, C., Longstaff, G., Compton, L., and Seddon, A. (2017). Effects of cations on the behaviour of lipid cubic phases. Sci. Rep. 7:8229. doi: 10.1038/s41598-017-08438-4
Bruley, C., Dupierris, V., Salvi, D., Rolland, N., and Ferro, M. (2012). AT_CHLORO: a chloroplast protein database dedicated to sub-plastidial localization. Front. Plant Sci. 3:205. doi: 10.3389/fpls.2012.00205
Bryant, D. A., Hunter, C. N., and Warren, M. J. (2020). Biosynthesis of the modified tetrapyrroles—the pigments of life. J. Biol. Chem. 295, 6888–6925. doi: 10.1074/jbc.REV120.006194
Brzezowski, P., Richter, A. S., and Grimm, B. (2015). Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985. doi: 10.1016/j.bbabio.2015.05.007
Buhr, F., El Bakkouri, M., Valdez, O., Pollmann, S., Lebedev, N., Reinbothe, S., et al. (2008). Photoprotective role of NADPH:protochlorophyllide oxidoreductase A. Proc. Natl. Acad. Sci. U.S.A. 105, 12629–12634. doi: 10.1073/pnas.0803950105
Bykowski, M., Mazur, R., Buszewicz, D., Szach, J., Mostowska, A., and Kowalewska, Ł. (2020). Spatial nano-morphology of the prolamellar body in Etiolated Arabidopsis thaliana plants with disturbed pigment and polyprenol composition. Front. Cell Dev. Biol. 8:586628. doi: 10.3389/fcell.2020.586628
Carey, E., and Rebeiz, C. (1985). Chloroplast biogenesis 49: differences among angiosperms in the biosynthesis and accumulation of monovinyl and divinyl protochlorophyllide during photoperiodic greening. Plant Physiol. 79, 1–6. doi: 10.1104/pp.79.1.1
Carey, E. E., Tripathy, B. C., and Rebeiz, C. A. (1985). Chloroplast biogenesis 51. Plant Physiol. 79, 1059–1063. doi: 10.1104/pp.79.4.1059
Charuvi, D., Kiss, V., Nevo, R., Shimoni, E., Adam, Z., and Reich, Z. (2012). Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of arabidopsis. Plant Cell 24, 1143–1157. doi: 10.1105/tpc.111.094458
Chen, G. E., Canniffe, D. P., Barnett, S. F. H., Hollingshead, S., Brindley, A. A., Vasilev, C., et al. (2018). Complete enzyme set for chlorophyll biosynthesis in Escherichia coli. Sci. Adv. 4:eaaq1407. doi: 10.1126/sciadv.aaq1407
Chen, G. E., Canniffe, D. P., and Neil Hunter, C. (2017). Three classes of oxygen-dependent cyclase involved in chlorophyll and bacteriochlorophyll biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 114, 6280–6285. doi: 10.1073/pnas.1701687114
Chen, G. E., and Hunter, C. N. (2020). Protochlorophyllide synthesis by recombinant cyclases from eukaryotic oxygenic phototrophs and the dependence on Ycf54. Biochem. J. 477, 2313–2325. doi: 10.1042/BCJ20200221
Chereskin, B. M., Wong, Y. S., and Castelfranco, P. A. (1982). In vitro synthesis of the chlorophyll isocyclic ring: transformation of magnesium-protoporphyrin IX and magnesium-protoporphyrin IX monomethyl ester into magnesium-2,4-divinyl pheoporphyrin A(5). Plant Physiol. 70, 987–993. doi: 10.1104/pp.70.4.987
Chernomor, O., Peters, L., Schneidewind, J., Loeschcke, A., Knieps-Grünhagen, E., Schmitz, F., et al. (2020). Complex evolution of light-dependent protochlorophyllide oxidoreductases in aerobic anoxygenic phototrophs: origin, phylogeny, and function. Mol. Biol. Evol. 38, 819–837. doi: 10.1093/molbev/msaa234
Chong, K., Almsherqi, Z. A., Shen, H., and Deng, Y. (2017). Cubic membrane formation supports cell survival of amoeba Chaos under starvation-induced stress. Protoplasma 255, 517–525. doi: 10.1007/s00709-017-1169-x
Colindres-Rojas, M., Wolf, M. M. N., Grob, R., Seidel, S., Dietzek, B., Schmitt, M., et al. (2011). Excited-state dynamics of protochlorophyllide revealed by subpicosecond infrared spectroscopy. Biophys. J. 100, 260–267. doi: 10.1016/j.bpj.2010.11.054
Cuttriss, A. J., Chubb, A. C., Alawady, A., Grimm, B., and Pogson, B. J. (2007). Regulation of lutein biosynthesis and prolamellar body formation in Arabidopsis. Funct. Plant Biol. 34, 663–672. doi: 10.1071/FP07034
Czarnecki, O., and Grimm, B. (2012). Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63, 1675–1687. doi: 10.1093/jxb/err437
Dahlin, C., Aronsson, H., Wilks, H. M., Lebedev, N., Sundqvist, C., and Timko, M. P. (1999). The role of protein surface charge in catalytic activity and chloroplast membrane association of the pea NADPH: protochlorophyllide oxidoreductase (POR) as revealed by alanine scanning mutagenesis. Plant Mol. Biol. 39, 309–323. doi: 10.1023/A:1006135100760
Dahlin, C., Sundqvist, C., and Timko, M. P. (1995). The in vitro assembly of the NADPH-protochlorophyllide oxidoreductase in pea chloroplasts. Plant Mol. Biol. 29, 317–330. doi: 10.1007/BF00043655
Daniels, T. E., Sylvester, R. A., Silverman, S., Polando, V., and Talal, N. (1974). Tubuloreticular structures within labial salivary glands in sjögren's syndrome. Arthritis Rheum. 17, 593–597. doi: 10.1002/art.1780170514
Darrah, P. M., Kay, S. A., Teakle, G. R., and Griffiths, W. T. (1990). Cloning and sequencing of protochlorophyllide reductase. Biochem. J. 265, 789–798. doi: 10.1042/bj2650789
Datsis, A. G. S. (1993). Tubuloreticular structures in pyridoxine deficiency. Exp. Toxicol. Pathol. 45, 55–59. doi: 10.1016/S0940-2993(11)80458-6
Davison, P. A., Schubert, H. L., Reid, J. D., Iorg, C. D., Heroux, A., Hill, C. P., et al. (2005). Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44, 7603–7612. doi: 10.1021/bi050240x
Denev, I. D., Yahubyan, G. T., Minkov, I. N., and Sundqvist, C. (2005). Organization of protochlorophyllide oxidoreductase in prolamellar bodies isolated from etiolated carotenoid-deficient wheat leaves as revealed by fluorescence probes. Biochim. Biophys. Acta Biomembr. 1716, 97–103. doi: 10.1016/j.bbamem.2005.09.001
Deng, Y., Almsherqi, Z. A., Ng, M. M. L., and Kohlwein, S. D. (2010). Do viruses subvert cholesterol homeostasis to induce host cubic membranes? Trends Cell Biol. 20, 371–379. doi: 10.1016/j.tcb.2010.04.001
Dietzek, B., Kiefer, W., Hermann, G., Popp, J., and Schmitt, M. (2006a). Solvent effects on the excited-state processes of protochlorophyllide: a femtosecond time-resolved absorption study. J. Phys. Chem. B 110, 4399–4406. doi: 10.1021/jp0556456
Dietzek, B., Kiefer, W., Yartsev, A., Sundström, V., Schellenberg, P., Grigaravicius, P., et al. (2006b). The excited-state chemistry of protochlorophyllide a: a time-resolved fluorescence study. ChemPhysChem 7, 1727–1733. doi: 10.1002/cphc.200600172
Dietzek, B., Maksimenka, R., Siebert, T., Birckner, E., Kiefer, W., Popp, J., et al. (2004). Excited-state processes in protochlorophyllide a - a femtosecond time-resolved absorption study. Chem. Phys. Lett. 397, 110–115. doi: 10.1016/j.cplett.2004.08.075
Dietzek, B., Tschierlei, S., Hanf, R., Seidel, S., Yartsev, A., Schmitt, M., et al. (2010). Dynamics of charge separation in the excited-state chemistry of protochlorophyllide. Chem. Phys. Lett. 492, 157–163. doi: 10.1016/j.cplett.2010.04.027
Dietzek, B., Tschierlei, S., Hermann, G., Yartsev, A., Pascher, T., Sundström, V., et al. (2009). Protochlorophyllide a: a comprehensive photophysical picture. ChemPhysChem 10, 144–150. doi: 10.1002/cphc.200800536
Dogbo, O., and Camara, B. (1987). Purification of isopentenyl pyrophosphate isomerase and geranylgeranyl pyrophosphate synthase from Capsicum chromoplasts by affinity chromatography. Biochim. Biophys. Acta Lipids Lipid Metab. 920, 140–148. doi: 10.1016/0005-2760(87)90253-0
Dong, C. S., Zhang, W. L., Wang, Q., Li, Y. S., Wang, X., Zhang, M., et al. (2020). Crystal structures of cyanobacterial light-dependent protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 117, 8455–8461. doi: 10.1073/pnas.1920244117
Eady, R. A. J., and Odland, G. F. (1975). Intraendothelial tubular aggregates in experimental wounds. Br. J. Dermatol. 93, 165–173. doi: 10.1111/j.1365-2133.1975.tb06736.x
Eggink, L. L., LoBrutto, R., Brune, D. C., Brusslan, J., Yamasato, A., Tanaka, A., et al. (2004). Synthesis of chlorophyll b: localization of chlorophyllide a oxygenase and discovery of a stable radical in the catalytic subunit. BMC Plant Biol. 4:5. doi: 10.1186/1471-2229-4-5
Elmaghrabi, A., Brown, E., Khin, E., Hassler, J., and Hendricks, A. R. (2017). Tubuloreticular inclusions in the absence of systemic lupus erythematosus and HIV infection: a report of three pediatric cases. Case Rep. Nephrol. Dial. 7, 91–101. doi: 10.1159/000477661
Engdahl, S., Aronsson, H., Sundqvist, C., Timko, M. P., and Dahlin, C. (2001). Association of the NADPH:protochlorophyllide oxidoreductase (POR) with isolated etioplast inner membranes from wheat. Plant J. 27, 297–304. doi: 10.1046/j.1365-313x.2001.01094.x
Erdei, N., Barta, C., Hideg, É., and Böddi, B. (2005). Light-induced wilting and its molecular mechanism in epicotyls of dark-germinated pea (Pisum sativum L.) seedlings. Plant Cell Physiol. 46, 185–191. doi: 10.1093/pcp/pci012
Espineda, C. E., Linford, A. S., Devine, D., and Brusslan, J. A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 10507–10511. doi: 10.1073/pnas.96.18.10507
Feldman, D., Hoar, R. M., Niemann, W. H., Valentine, T., Cukierski, M., and Hendrickx, A. G. (1986). Tubuloreticular inclusions in placental chorionic villi of rhesus monkeys after maternal treatment with interferon. Am. J. Obstet. Gynecol. 155, 413–424. doi: 10.1016/0002-9378(86)90844-6
Fiedor, L., Kania, A., Mysliwa-Kurdziel, B., Orzeł, Ł., and Stochel, G. (2008). Understanding chlorophylls: central magnesium ion and phytyl as structural determinants. Biochim. Biophys. Acta Bioenerg. 1777, 1491–1500. doi: 10.1016/j.bbabio.2008.09.005
Fiedor, L., Stasiek, M., Mysliwa-Kurdziel, B., and Strzałka, K. (2003). Phytol as one of the determinants of chlorophyll interactions in solution. Photosynth. Res. 78, 47–57. doi: 10.1023/A:1026042005536
Fiedor, L., Zbyradowski, M., and Pilch, M. (2019). “Tetrapyrrole pigments of photosynthetic antennae and reaction centers of higher plants: structures, biophysics, functions, biochemistry, mechanisms of regulation, applications,” in Advances in Botanical Research, Vol. 90, ed B. Grimm (London: Academic Press), 1–33. doi: 10.1016/bs.abr.2019.04.001
Franck, F., Sperling, U., Frick, G., Pochert, B., Van Cleve, B., Apel, K., et al. (2000). Regulation of etioplast pigment-protein complexes, inner membrane architecture and protochlorophyllide a chemical heterogeneity by light-dependent I NADPH:protochlorophyllide oxidoreductases A and B. Plant Physiol. 124, 1678–1696. doi: 10.1104/pp.124.4.1678
Frick, G., Su, Q., Apel, K., and Armstrong, G. A. (2003). An Arabidopsis porB porC double mutant lacking light-dependent NADPH:protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 35, 141–153. doi: 10.1046/j.1365-313X.2003.01798.x
Fujii, S., Kobayashi, K., Nagata, N., Masuda, T., and Wada, H. (2017). Monogalactosyldiacylglycerol facilitates synthesis of photoactive protochlorophyllide in etioplasts. Plant Physiol. 174, 2183–2198. doi: 10.1104/pp.17.00304
Fujii, S., Kobayashi, K., Nagata, N., Masuda, T., and Wada, H. (2018). Digalactosyldiacylglycerol is essential for organization of the membrane structure in etioplasts. Plant Physiol. 177, 1487–1497. doi: 10.1104/pp.18.00227
Fujii, S., Kobayashi, K., Nakamura, Y., and Wada, H. (2014). Inducible knockdown of monogalactosyldiacylglycerol synthase1 reveals roles of galactolipids in organelle differentiation in arabidopsis cotyledons. Plant Physiol. 166, 1436–1449. doi: 10.1104/pp.114.250050
Fujii, S., Nagata, N., Masuda, T., Wada, H., and Kobayashi, K. (2019a). Galactolipids are essential for internal membrane transformation during etioplast-to-chloroplast differentiation. Plant Cell Physiol. 60, 1224–1238. doi: 10.1093/pcp/pcz041
Fujii, S., Wada, H., and Kobayashi, K. (2019b). Role of galactolipids in plastid differentiation before and after light exposure. Plants 8, 1–17. doi: 10.3390/plants8100357
Fujita, Y. (1996). Protochlorophyllide reduction: a key step in the greening of plants. Plant Cell Physiol. 37, 411–421. doi: 10.1093/oxfordjournals.pcp.a028962
Fusada, N., Masuda, T., Kuroda, H., Shiraishi, T., Shimada, H., Ohta, H., et al. (2000). NADPH-protochlorophyllide oxidoreductase in cucumber is encoded by a single gene and its expression is transcriptionally enhanced by illumination. Photosynth. Res. 64, 147–154. doi: 10.1023/A:1006418608647
Gabruk, M., Grzyb, J., Kruk, J., and Mysliwa-Kurdziel, B. (2012). Light-dependent and light-independent protochlorophyllide oxidoreductases share similar sequence motifs -in silico studies. Photosynthetica 50, 529–540. doi: 10.1007/s11099-012-0057-z
Gabruk, M., and Mysliwa-Kurdziel, B. (2015). Light-dependent protochlorophyllide oxidoreductase: phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262. doi: 10.1021/acs.biochem.5b00704
Gabruk, M., and Mysliwa-Kurdziel, B. (2020). The origin, evolution and diversification of multiple isoforms of light-dependent protochlorophyllide oxidoreductase (LPOR): focus on angiosperms. Biochem. J. 477, 2221–2236. doi: 10.1042/BCJ20200323
Gabruk, M., Mysliwa-Kurdziel, B., and Kruk, J. (2017). MGDG, PG and SQDG regulate the activity of lightdependent protochlorophyllide oxidoreductase. Biochem. J. 474, 1307–1320. doi: 10.1042/BCJ20170047
Gabruk, M., Nowakowska, Z., Skupien-Rabian, B., Kȩdracka-Krok, S., Mysliwa-Kurdziel, B., and Kruk, J. (2016). Insight into the oligomeric structure of PORA from A. thaliana. Biochim. Biophys. Acta Proteins Proteomics 1864, 1757–1764. doi: 10.1016/j.bbapap.2016.09.015
Gabruk, M., Stecka, A., Strzałka, W., Kruk, J., Strzałka, K., and Mysliwa-Kurdziel, B. (2015). Photoactive protochlorophyllide-enzyme complexes reconstituted with PORA, PORB and PORC proteins of A. thaliana: fluorescence and catalytic properties. PLoS ONE 10:e0116990. doi: 10.1371/journal.pone.0116990
Garrone, A., Archipowa, N., Zipfel, P. F., Hermann, G., and Dietzek, B. (2015). Plant protochlorophyllide oxidoreductases A and B: catalytic efficiency and initial reaction steps. J. Biol. Chem. 290, 28530–28539. doi: 10.1074/jbc.M115.663161
Gaubier, P., Wu, H. J., Laudié, M., Delseny, M., and Grellet, F. (1995). A chlorophyll synthetase gene from Arabidopsis thaliana. MGG Mol. Gen. Genet. 249, 58–64. doi: 10.1007/BF00290236
Geisbert, T. W., Jahrling, P. B., Hanes, M. A., and Zack, P. M. (1992). Association of Ebola-related Reston virus particles and antigen with tissue lesions of monkeys imported to the United States. J. Comp. Pathol. 106, 137–152. doi: 10.1016/0021-9975(92)90043-T
Gholami, S., Nenov, A., Rivalta, I., Bocola, M., Bordbar, A. K., Schwaneberg, U., et al. (2018). Theoretical model of the protochlorophyllide oxidoreductase from a hierarchy of protocols. J. Phys. Chem. B 122, 7668–7681. doi: 10.1021/acs.jpcb.8b04231
Granick, S., and Gassman, M. (1970). Rapid regeneration of protochlorophyllide650. Plant Physiol. 45, 201–205. doi: 10.1104/pp.45.2.201
Grevby, C., Engdahl, S., Ryberg, M., and Sundqvist, C. (1989). Binding properties of NADPH-protochlorophyllide oxidoreductase as revealed by detergent and ion treatments of isolated and immobilized prolamellar bodies. Physiol. Plant. 77, 493–503. doi: 10.1111/j.1399-3054.1989.tb05382.x
Griffiths, W. T. (1974). Protochlorophyll and protochlorophyllide as precursors for chlorophyll synthesis in vitro. FEBS Lett. 49, 196–200. doi: 10.1016/0014-5793(74)80510-7
Grimley, P., Davis, G., Kang, Y., Dooley, J., Strohmaier, J., and Hoofnagle, J. (1985). Tubuloreticular inclusions in peripheral blood mononuclear cells related to systemic therapy with alpha-interferon. Lab Invest. 52, 638–649.
Grimley, P., and Schaff, Z. (1976). Significance of tubuloreticular inclusions in the pathobiology of human diseases. Pathobiol Annu. 6, 221–257.
Grimley, P. M., Decker, J. L., Michelitch, H. J., and Frantz, M. M. (1973). Abnormal structures in circulating lymphocytes from patients with systemic lupus erythematosus and related diseases. Arthritis Rheum. 16, 313–323. doi: 10.1002/art.1780160305
Grimm, B. (2010). “Chapter 3: Control of the metabolic flow in tetrapyrrole biosynthesis: regulation of expression and activity of enzymes in the Mg branch of tetrapyrrole biosynthesis,” in The Chloroplast, Advances in Photosynthesis and Respiration, Vol. 31, eds C. A. Rebeiz, C. Benning, H. Bohnert, H. Daniell, J. K. Hoober, H. K. Lichtenthaler, et al. (Dordrecht: Springer), 39–54.
Gunning, B. (2001). Membrane geometry of “open” prolamellar bodies. Protoplasma 215, 4–15. doi: 10.1007/BF01280299
Gunning, B. E. S. (1965). The greening process in plastids - 1. The structure of the prolamellar body. Protoplasma 60, 111–130. doi: 10.1007/BF01248133
Gutbrod, K., Romer, J., and Dörmann, P. (2019). Phytol metabolism in plants. Prog. Lipid Res. 74, 1–17. doi: 10.1016/j.plipres.2019.01.002
Hammond, C. T., and Mahlberg, P. G. (1978). Ultrastructural development of capitate glandular hairs of Cannabis sativa L. (Cannabaceae). Am. J. Bot. 65, 140–151. doi: 10.1002/j.1537-2197.1978.tb06051.x
Harper, A. L., Von Gesjen, S. E., Linford, A. S., Peterson, M. P., Faircloth, R. S., Thissen, M. M., et al. (2004). Chlorophyllide a oxygenase mRNA and protein levels correlate with the chlorophyll a/b ratio in Arabidopsis thaliana. Photosynth. Res. 79, 149–159. doi: 10.1023/B:PRES.0000015375.40167.76
Helfrich, M., Schoch, S., Schäfer, W., Ryberg, M., and Rüdiger, W. (1996). Absolute configuration of protochlorophyllide a and substrate specificity of NADPH-protochlorophyllide oxidoreductase. J. Am. Chem. Soc. 118, 2606–2611. doi: 10.1021/ja953440c
Helfrich, M., Schoch, S., Ulrika, L., Cmiiel, E., and Rudiger, W. (1994). Chlorophyll synthetase cannot synthesize chlorophyll a′. Eur. J. Biochem. 219, 267–275. doi: 10.1111/j.1432-1033.1994.tb19938.x
Herbst, J., Girke, A., Hajirezaei, M. R., Hanke, G., and Grimm, B. (2018). Potential roles of YCF54 and ferredoxin-NADPH reductase for magnesium protoporphyrin monomethylester cyclase. Plant J. 94, 485–496. doi: 10.1111/tpj.13869
Herbst, J., Hey, D., and Grimm, B. (2019). “Posttranslational control of tetrapyrrole biosynthesis: interacting proteins, chaperones, auxiliary factors,” in Advances in Botanical Research, Vol. 91, ed B. Grimm (London: Academic Press), 163–194. doi: 10.1016/bs.abr.2019.01.001
Hernández-Verdeja, T., Vuorijoki, L., and Strand, Å. (2020). Emerging from the darkness: interplay between light and plastid signaling during chloroplast biogenesis. Physiol. Plant. 169, 397–406. doi: 10.1111/ppl.13100
Hey, D., Rothbart, M., Herbst, J., Wang, P., Müller, J., Wittmann, D., et al. (2017). LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiol. 174, 1037–1050. doi: 10.1104/pp.17.00505
Heyes, D. J., Hardman, S. J. O., Hedison, T. M., Hoeven, R., Greetham, G. M., Towrie, M., et al. (2015). Excited-state charge separation in the photochemical mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Angew. Chemie Int. Ed. 54, 1512–1515. doi: 10.1002/anie.201409881
Heyes, D. J., Hardman, S. J. O., Mansell, D., Ní Cheallaigh, A., Gardiner, J. M., Johannissen, L. O., et al. (2017). Excited-state properties of protochlorophyllide analogues and implications for light-driven synthesis of chlorophyll. J. Phys. Chem. B 121, 1312–1320. doi: 10.1021/acs.jpcb.7b00528
Heyes, D. J., and Hunter, C. N. (2005). Making light work of enzyme catalysis: protochlorophyllide oxidoreductase. Trends Biochem. Sci. 30, 642–649. doi: 10.1016/j.tibs.2005.09.001
Heyes, D. J., Ruban, A. V., and Hunter, C. N. (2003). Protochlorophyllide oxidoreductase: “dark” reactions of a light-driven enzyme. Biochemistry 42, 523–528. doi: 10.1021/bi0268448
Heyes, D. J., Ruban, A. V., Wilks, H. M., and Hunter, C. N. (2002). Enzymology below 200 K: the kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 99, 11145–11150. doi: 10.1073/pnas.182274199
Hideg, É., Vitányi, B., Kósa, A., Solymosi, K., Bóka, K., Won, S., et al. (2010). Reactive oxygen species from type-I photosensitized reactions contribute to the light-induced wilting of dark-grown pea (Pisum sativum) epicotyls. Physiol. Plant. 138, 485–492. doi: 10.1111/j.1399-3054.2009.01329.x
Holtorf, H., Reinbothe, S., Reinbothe, C., Bereza, B., and Apel, K. (1995). Two routes of chlorophyllide synthesis that are differentially regulated by light in barley (Hordeum vulgare L.). Proc. Natl. Acad. Sci. U.S.A. 92, 3254–3258. doi: 10.1073/pnas.92.8.3254
Hulanicka, B., Barry, D. W., and Grimley, P. M. (1977). Induction of tubuloreticular inclusions in human lymphoma cells (Raji line) related to S-phase treatment with halogenated pyrimidines. Cancer Res. 37, 2105–2113.
Hurd, E., Eigenbrodt, E., Ziff, M., and Strunk, S. (1969). Cytoplasmic tubular structures in kidney biopsies in systemic lupus erythematosus. Arthritis Rheum. 12, 541–542. doi: 10.1002/art.1780120517
Ikeda, T. (1970). Changes in Fine Structure of prolamellar body in relation to the formation of the chloroplast. Bot. Mag. Tokyo 83, 1–9. doi: 10.15281/jplantres1887.83.1
Ikeda, T. (1971). Prolamellar body formation under different light and temperature conditions. Shokubutsugaku Zasshi 84, 363–375. doi: 10.15281/jplantres1887.84.363
Ikeuchi, M., and Murakami, S. (1983). Separation and characterization of prolamellar bodies and prothylakoids from squash etioplasts. Plant Cell Physiol. 24, 71–80. doi: 10.1093/oxfordjournals.pcp.a076515
Ioannides, I. M., Fasoula, D. A., Robertson, K. R., and Rebeiz, C. A. (1994). An evolutionary study of chlorophyll biosynthetic heterogeneity in green plants. Biochem. Syst. Ecol. 22, 211–220. doi: 10.1016/0305-1978(94)90010-8
Jörnvall, H., Persson, B., Krook, M., Atrian, S., Gonzàlez-Duarte, R., Jeffery, J., et al. (1995). Short-chain dehydrogenases/reductases(SDR). Biochemistry 34, 6003–6013. doi: 10.1021/bi00018a001
Joyard, J., Block, M., Pineau, B., Albrieux, C., and Douce, R. (1990). Envelope membranes from mature spinach chloroplasts contain a NADPH:protochlorophyllide reductase on the cytosolic side of the outer membrane. J. Biol. Chem. 265, 21820–21827. doi: 10.1016/S0021-9258(18)45813-9
Joyard, J., Ferro, M., Masselon, C., Seigneurin-Berny, D., Salvi, D., Garin, J., et al. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2, 1154–1180. doi: 10.1093/mp/ssp088
Kahn, A., Boardman, N. K., and Thorne, S. W. (1970). Energy transfer between protochlorophyllide molecules: evidence for multiple chromophores in the photoactive protochlorophyllide-protein complex in vivo and in vitro. J. Mol. Biol. 48, 85–101. doi: 10.1016/0022-2836(70)90220-2
Kakuszi, A., Solymosi, K., and Böddi, B. (2017). Transformation of plastids in soil-shaded lowermost hypocotyl segments of bean (Phaseolus vulgaris) during a 60-day cultivation period. Physiol. Plant. 159, 483–491. doi: 10.1111/ppl.12519
Kanervo, E., Singh, M., Suorsa, M., Paakkarinen, V., Aro, E., Battchikova, N., et al. (2008). Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol. 49, 396–410. doi: 10.1093/pcp/pcn016
Kang, Y. H., Yaffe, L., and Grimley, P. M. (1984). Immunoelectron microscopic application of monoclonal antibodies for identification of lymphocyte subsets bearing tubuloreticular inclusions or parallel tubular arrays. J. Exp. Pathol. 1, 157–173.
Kaschner, M., Loeschcke, A., Krause, J., Minh, B. Q., Heck, A., Endres, S., et al. (2014). Discovery of the first light-dependent protochlorophyllide oxidoreductase in anoxygenic phototrophic bacteria. Mol. Microbiol. 93, 1066–1078. doi: 10.1111/mmi.12719
Kashina, T. K., and Danilova, M. F. (1993). Ultrastructure of glandular hair plastids and nictophylness of Perilla ocymoides L. Russ. J. Plant Physiol. 40, 785–790.
Kauss, D., Bischof, S., Steiner, S., Apel, K., and Meskauskiene, R. (2012). FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg ++ -branch of this pathway. FEBS Lett. 586, 211–216. doi: 10.1016/j.febslet.2011.12.029
Keller, Y., Bouvier, F., D'Harlingue, A., and Camara, B. (1998). Metabolic compartmentation of plastid prenyllipid biosynthesis - evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251, 413–417. doi: 10.1046/j.1432-1327.1998.2510413.x
Kim, C., Ham, H., and Apel, K. (2005). Multiplicity of different cell- and organ-specific import routes for the NADPH-protochlorophyllide oxidoreductases A and B in plastids of Arabidopsis seedlings. Plant J. 42, 329–340. doi: 10.1111/j.1365-313X.2005.02374.x
Kim, E.-S., and Mahlberg, P. G. (1997). Plastid development in disc cells of glandular trichomes of Cannabis (Cannabaceae). Mol. Cells 7, 352–359.
Kim, S., Schlicke, H., Van Ree, K., Karvonen, K., Subramaniam, A., Richter, A., et al. (2013). Arabidopsis chlorophyll biosynthesis: an essential balance between the methylerythritol phosphate and tetrapyrrole pathways. Plant Cell 25, 4984–4993. doi: 10.1105/tpc.113.119172
Klement, H., Oster, U., and Rüdiger, W. (2000). The influence of glycerol and chloroplast lipids on the spectral shifts of pigments associated with NADPH:protochlorophyllide oxidoreductase from Avena sativa L. FEBS Lett. 480, 306–310. doi: 10.1016/S0014-5793(00)01887-1
Knaust, R., Seyfried, B., Schmidt, L., Schulz, R., and Senger, H. (1993). Phototransformation of monovinyl and divinyl protochlorophyllide by NADPH:protochlorophyllide oxidoreductase of barley expressed in Escherichia coli. J. Photochem. Photobiol. B Biol. 20, 161–166. doi: 10.1016/1011-1344(93)80146-Z
Kobayashi, K. (2016). Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 129, 565–580. doi: 10.1007/s10265-016-0827-y
Kobayashi, K. (2018). Correction to: role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development (Journal of Plant Research, (2016), 129, 4, (565-580), 10.1007/s10265-016-0827-y). J. Plant Res. 131:563. doi: 10.1007/s10265-018-1011-3
Kobayashi, K., Fujii, S., Sasaki, D., Baba, S., Ohta, H., Masuda, T., et al. (2014). Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front. Plant Sci. 5:272. doi: 10.3389/fpls.2014.00272
Kobayashi, K., and Masuda, T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7:1811. doi: 10.3389/fpls.2016.01811
Kobayashi, K., Mochizuki, N., Yoshimura, N., Motohashi, K., Hisabori, T., and Masuda, T. (2008). Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 7, 1188–1195. doi: 10.1039/b802604c
Kobayashi, K., and Wada, H. (2016). “Role of lipids in chloroplast biogenesis,” in Lipids in Plant and Algae Development. Subcellular Biochemistry, Vol. 86, eds Y. Nakamura, and Y. Li-Beisson (Cham: Springer). doi: 10.1007/978-3-319-25979-6_5
Kong, W., Yu, X., Chen, H., Liu, L., Xiao, Y., Wang, Y., et al. (2016). The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol. Biol. 92, 177–191. doi: 10.1007/s11103-016-0513-4
Kostianovsky, M., Kang, Y. H., and Grimley, P. M. (2016). Disseminated tubuloreticular inclusions in acquired immunodeficiency syndrome (AIDS). Ultrastruct. Pathol. 4, 331–336. doi: 10.3109/01913128309140585
Kotzabasis, K., Senge, M., Seyfried, B., and Senger, H. (1990). Aggregation of monovinyl- and divinyl-protochlorophyllide in organic solvents. Photochem. Photobiol. 52, 95–101. doi: 10.1111/j.1751-1097.1990.tb01761.x
Kovács, E., and Keresztes, A. (2002). Effect of gamma and UV-B/C radiation on plant cells. Micron 33, 199–210. doi: 10.1016/S0968-4328(01)00012-9
Kowalewska, Ł., Bykowski, M., and Mostowska, A. (2019). Spatial organization of thylakoid network in higher plants. Bot. Lett. 166, 326–343. doi: 10.1080/23818107.2019.1619195
Kowalewska, L., Mazur, R., Suski, S., Garstka, M., and Mostowska, A. (2016). Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell. 28, 875–891. doi: 10.1105/tpc.15.01053
Krohn, K., and Sandholm, M. (1975). Myxovirus-like structures in the glomerular endothelial cell cytoplasm in canine nephritis. Acta Pathol Microbiol. Scand. A 83, 355–359. doi: 10.1111/j.1699-0463.1975.tb01883.x
Kruk, J., and Mysliwa-Kurdziel, B. (2004). Separation of monovinyl and divinyl protochlorophyllides using C-30 reverse phase high performance liquid chromatography column: analytical and preparative applications. Chromatographia 60, 117–123. doi: 10.1365/s10337-004-0320-y
Kuntz, M., Römer, S., Suire, C., Hugueney, P., Weil, J. H., Schantz, R., et al. (1992). Identification of a cDNA for the plastid-located geranylgeranyl pyrophosphate synthase from Capsicum annuum: correlative increase in enzyme activity and transcript level during fruit ripening. Plant J. 2, 25–34. doi: 10.1046/j.1365-313X.1992.t01-47-00999.x
Kwon, C. T., Kim, S. H., Song, G., Kim, D., and Paek, N. C. (2017). Two NADPH: protochlorophyllide oxidoreductase (POR) isoforms play distinct roles in environmental adaptation in rice. Rice 10, 1–14. doi: 10.1186/s12284-016-0141-2
La Rocca, N., Rascio, N., Oster, U., and Rüdiger, W. (2007). Inhibition of lycopene cyclase results in accumulation of chlorophyll precursors. Planta 225, 1019–1029. doi: 10.1007/s00425-006-0409-7
Laferrière, A., and Beyer, P. (1991). Purification of geranylgeranyl diphosphate synthase from Sinapis alba etioplasts. Biochim. Biophys. Acta Protein Struct. Mol. 1077, 167–172. doi: 10.1016/0167-4838(91)90054-4
Landolt, A., Ryffel, U., Hosbach, H., and Wyler, R. (1976). Ultrastructure of tubular inclusions in endothelial cells of pituitary tumors associated with acromegaly. Virchows Arch. A Path. Anat. Histol. 370, 129–140. doi: 10.1007/BF00430809
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Larsson, M., and Larsson, K. (2014). Periodic minimal surface organizations of the lipid bilayer at the lung surface and in cubic cytomembrane assemblies. Adv. Colloid Interface Sci. 205, 68–73. doi: 10.1016/j.cis.2013.07.003
Lee, C., Suh, K., and Kim, K. (2013a). The clinicopathologic significance of endothelial tubuloreticular inclusions in glomerular diseases. Ultrastruct. Pathol. 37, 386–394. doi: 10.3109/01913123.2013.814738
Lee, J. Y., Lee, H. S., Song, J. Y., Jung, Y. J., Reinbothe, S., Park, Y., et al. (2013b). Cell growth defect factor1/CHAPERONE-LIKE PROTEIN OF POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis. Plant Cell 25, 3944–3960. doi: 10.1105/tpc.113.111096
Lee, J. Y., Song, S. H., Kim, Y. S., Lim, B. J., Il Kim, S., Kim, M. S., et al. (2017). Tubuloreticular inclusions in peritubular capillaries of renal allografts. Pathol. Res. Pract. 213, 1185–1190. doi: 10.1016/j.prp.2017.06.009
Lee, S., Harris, C., Hirschfeld, A., and Dickson, D. W. (1988). Cytomembranous inclusions in the brain of a patient with the acquired immunodeficiency syndrome. Acta Neuropathol. 76, 101–106. doi: 10.1007/BF00687686
Lichtenthaler, H. K., Schwender, J., Disch, A., and Rohmer, M. (1997). Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 400, 271–274. doi: 10.1016/S0014-5793(96)01404-4
Liebers, M., Grübler, B., Chevalier, F., Lerbs-Mache, S., Merendino, L., Blanvillain, R., et al. (2017). Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front. Plant Sci. 8:23. doi: 10.3389/fpls.2017.00023
Lindquist, E., and Aronsson, H. (2018). Chloroplast vesicle transport. Photosynth. Res. 138, 361–371. doi: 10.1007/s11120-018-0566-0
Lindquist, E., Solymosi, K., and Aronsson, H. (2016). Vesicles are persistent features of different plastids. Traffic 17, 1125–1138. doi: 10.1111/tra.12427
Liu, X., Li, L., Li, M., Su, L., Lian, S., Zhang, B., et al. (2018). AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Sci. Rep. 8:2250. doi: 10.1038/s41598-018-20542-7
Liu, X., Li, L., Zhang, B., Zeng, L., and Li, L. (2020). AhHDA1-mediated AhGLK1 promoted chlorophyll synthesis and photosynthesis regulates recovery growth of peanut leaves after water stress. Plant Sci. 294:110461. doi: 10.1016/j.plantsci.2020.110461
Lütz, C., and Tönissen, H. (1984). Effects of enzymatic cleavage on prolamellar bodies and prothylakoids prepared from oat ehoplasts. Isr. J. Bot. 33, 195–209.
Luu, J., Bockus, D., Remington, F., Bean, M. A., and Hammar, S. P. (1989). Tubuloreticular structures and cylindrical confronting cisternae: a review. Hum. Pathol. 20, 617–627. doi: 10.1016/0046-8177(89)90148-2
Madewell, B., and Munn, R. (1989). Tubuloreticular inclusions in equine connective tissue neoplasms. J. Comp. Pathol. 100, 449–452. doi: 10.1016/0021-9975(89)90011-X
Madewell, B. R., and Munn, R. J. (1990). Canine lymphoproliferative disorders. J. Vet. Intern. Med. 4, 63–70. doi: 10.1111/j.1939-1676.1990.tb03105.x
Mageed, H. A. A., El, El Sahhar, K. F., Robertson, K. R., Parham, R., and Rebeiz, C. A. (1997). Chloroplast biogenesis 77: two novel monovinyl and divinyl light-dark greening groups of plants and their relationship to the chlorophyll a biosynthetic heterogeneity of green plants. Photochem. Photobiol. 66, 89–96. doi: 10.1111/j.1751-1097.1997.tb03143.x
Manzano, D., Andrade, P., Caudepón, D., Altabella, T., Arró, M., and Ferrer, A. (2016). Suppressing farnesyl diphosphate synthase alters chloroplast development and triggers sterol-dependent induction of jasmonate- and Fe-related responses. Plant Physiol. 172, 93–117. doi: 10.1104/pp.16.00431
Marquart, K. H. (2005). Occurrence of tubuloreticular structures and intracisternal paracrystalline inclusions in endothelial cells of tissue from different epidemiological types of Kaposi's sarcoma. Ultrastruct. Pathol. 29, 85–93. doi: 10.1080/01913120590912205
Martin, G. E., Timko, M. P., and Wilks, H. M. (1997). Purification and kinetic analysis of pea (Pisum sativum L.) NADPH:protochlorophyllide oxidoreductase expressed as a fusion with maltose-binding protein in Escherichia coli. Biochem. J. 325, 139–45. doi: 10.1042/bj3250139
Masuda, T. (2008). Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 96, 121–143. doi: 10.1007/s11120-008-9291-4
Masuda, T., Fusada, N., Oosawa, N., Takamatsu, K., Yamamoto, Y. Y., Ohto, M., et al. (2003). Functional analysis of isoforms of NADPH: protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol. 44, 963–974. doi: 10.1093/pcp/pcg128
Masuda, T., Fusada, N., Shiraishi, T., Kuroda, H., Awai, K., Shimada, H., et al. (2002). Identification of two differentially regulated isoforms of protochlorophyllide oxidoreductase (POR) from tobacco revealed a wide variety of light- and development-dependent regulations of POR gene expression among angiosperms. Photosynth. Res. 74, 165–172. doi: 10.1023/A:1020951409135
Masuda, T., and Takamiya, K. I. (2004). Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth. Res. 81, 1–29. doi: 10.1023/B:PRES.0000028392.80354.7c
Mathis, P., and Sauer, K. (1972). Circular dichroism studies on the structure and the photochemistry of protochlorophyllide and chlorophyllide holochrome. Biochim. Biophys. Acta Bioenerg. 267, 498–511. doi: 10.1016/0005-2728(72)90178-8
Matos, E., and Paiva, E. (2012). Structure, function and secretory products of the peltate glands of Centrolobium tomentosum (Fabaceae, Faboideae). Aust. J. Bot. 60, 301–309. doi: 10.1071/BT12009
Matsumura, H., Setoguti, T., Mori, K., Ross, E. R., and Koto, A. (1984). Endothelial tubuloreticular structures in intracranial germinomas. Pathol. Int. 34, 1–9. doi: 10.1111/j.1440-1827.1984.tb02176.x
Maturi, R. K., and Font, R. L. (1996). Ultrastructural features and prevalence of tubuloreticular structures in the ocular vasculature of patients with AIDS: a study of 23 cases. Br. J. Ophthalmol. 80, 252–255. doi: 10.1136/bjo.80.3.252
Mazur, R., Mostowska, A., Szach, J., Gieczewska, K., Wójtowicz, J., Bednarska, K., et al. (2019). Galactolipid deficiency disturbs spatial arrangement of the thylakoid network in Arabidopsis thaliana plants. J. Exp. Bot. 70, 4689–4703. doi: 10.1093/jxb/erz219
Menon, B. R. K., Hardman, S. J. O., Scrutton, N. S., and Heyes, D. J. (2016). Multiple active site residues are important for photochemical efficiency in the light-activated enzyme protochlorophyllide oxidoreductase (POR). J. Photochem. Photobiol. B Biol. 161, 236–243. doi: 10.1016/j.jphotobiol.2016.05.029
Meskauskiene, R., and Apel, K. (2002). Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett. 532, 27–30. doi: 10.1016/S0014-5793(02)03617-7
Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. (2001). FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831. doi: 10.1073/pnas.221252798
Mochizuki, N., Tanaka, R., Grimm, B., Masuda, T., Moulin, M., Smith, A. G., et al. (2010). The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498. doi: 10.1016/j.tplants.2010.05.012
Monselise, E. B. I., Levkovitz, A., and Kost, D. (2015). Ultraviolet radiation induces stress in etiolated Landoltia punctata, as evidenced by the presence of alanine, a universal stress signal: a 15N NMR study. Plant Biol. 1, 101–107. doi: 10.1111/plb.12198
Mork-Jansson, A., Bue, A. K., Gargano, D., Furnes, C., Reisinger, V., Arnold, J., et al. (2015). Lil3 assembles with proteins regulating chlorophyll synthesis in barley. PLoS ONE 10:e133145. doi: 10.1371/journal.pone.0133145
Mysliwa-Kurdziel, B., Kruk, J., and Strzałka, K. (2004). Fluorescence lifetimes and spectral properties of protochlorophyllide in organic solvents in relation to the respective parameters in vivo. Photochem. Photobiol. 79, 62–67. doi: 10.1562/0031-8655(2004)79<62:FLASPO>2.0.CO;2
Mysliwa-Kurdziel, B., Kruk, J., and Strzałka, K. (2013a). Protochlorophyllide and protochlorophyll in model membranes - an influence of hydrophobic side chain moiety. Biochim. Biophys. Acta 1828, 1075–1082. doi: 10.1016/j.bbamem.2012.12.007
Mysliwa-Kurdziel, B., Kruk, J., and Strzałka, K. (2013b). Protochlorophyllide in model systems - an approach to in vivo conditions. Biophys. Chem. 175–176, 28–38. doi: 10.1016/j.bpc.2013.02.002
Mysliwa-Kurdziel, B., Solymosi, K., Kruk, J., Böddi, B., and Strzałka, K. (2008). Solvent effects on fluorescence properties of protochlorophyll and its derivatives with various porphyrin side chains. Eur. Biophys. J. 37, 1185–1193. doi: 10.1007/s00249-008-0288-x
Nagata, N., Suzuki, M., Yoshida, S., and Muranaka, T. (2002). Mevalonic acid partially restores chloroplast and etioplast development in Arabidopsis lacking the non-mevalonate pathway. Planta 216, 345–350. doi: 10.1007/s00425-002-0871-9
Nagata, N., Tanaka, R., and Tanaka, A. (2007). The major route for chlorophyll synthesis includes [3,8-divinyl]- chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol. 48, 1803–1808. doi: 10.1093/pcp/pcm153
Nakanishi, H., Nozue, H., Suzuki, K., Kaneko, Y., Taguchi, G., and Hayashida, N. (2005). Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol. 46, 467–473. doi: 10.1093/pcp/pci053
Nguyen, H. C., Melo, A. A., Kruk, J., Frost, A., and Gabruk, M. (2021). Photocatalytic LPOR forms helical lattices that shape membranes for chlorophyll synthesis. Nat. Plants. doi: 10.1038/s41477-021-00885-2
Oosawa, N., Masuda, T., Awai, K., Fusada, N., Shimada, H., Ohta, H., et al. (2000). Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett. 474, 133–136. doi: 10.1016/S0014-5793(00)01568-4
Opitz, S., Nes, W. D., and Gershenzon, J. (2014). Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry 98, 110–119. doi: 10.1016/j.phytochem.2013.11.010
Orenstein, J. M., Olivia, T., Kind, P., and Washington, G. (1987). The relationship of serum alpha-interferon and ultrastructural markers in HIV-seropositive individuals. Ultrastruct. Pathol. 11, 673–679. doi: 10.3109/01913128709048453
Oster, U., and Rüdiger, W. (1997). The G4 gene of Arabidopsis thaliana encodes a chlorophyll synthase of etiolated plants. Bot. Acta 110, 420–423. doi: 10.1111/j.1438-8677.1997.tb00658.x
Oster, U., Tanaka, R., Tanaka, A., and Rüdiger, W. (2000). Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 21, 305–310. doi: 10.1046/j.1365-313x.2000.00672.x
Ouazzani Chahdi, M. A., Schoefs, B., and Franck, F. (1998). Isolation and characterization of photoactive complexes of NADPH:protochlorophyllide oxidoreductase from wheat. Planta 206, 673–680. doi: 10.1007/s004250050446
Paddock, T., Lima, D., Mason, M. E., Apel, K., and Armstrong, G. A. (2012). Arabidopsis light-dependent NADPH: protochlorophyllide oxidoreductase A (PORA) is essential for normal plant growth and development: an addendum. Plant Mol. Biol. 80, 237–240. doi: 10.1007/s11103-012-9944-8
Paddock, T. N., Mason, M. E., Lima, D. F., and Armstrong, G. A. (2010). Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 72, 445–457. doi: 10.1007/s11103-009-9582-y
Parham, R., and Rebeiz, C. (1992). Chloroplast biogenesis: [4-vinyl] chlorophyllide a reductase is a divinyl chlorophyllide a-specific, NADPH-dependent enzyme. Biochemistry 31, 8460–8464. doi: 10.1021/bi00151a011
Park, H., Kreunen, S. S., Cuttriss, A. J., DellaPenna, D., and Pogson, B. J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14, 321–332. doi: 10.1105/tpc.010302
Pellaud, S., and Saffrané, L. M. (2017). Metabolic origins and transport of vitamin E biosynthetic precursors. Front. Plant Sci. 8:1959. doi: 10.3389/fpls.2017.01959
Peter, E., and Grimm, B. (2009). GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210. doi: 10.1093/mp/ssp072
Popoff, A., and Malinin, T. (1976). Cytoplasmic tubular arrays in cells of American Burkitt's type lymphoma. Cancer 37, 275–284. doi: 10.1002/1097-0142(197601)37:1<275::AID-CNCR2820370138>3.0.CO;2-V
Rassadina, V., Domanskii, V., Averina, N. G., Schoch, S., and Rüdiger, W. (2004). Correlation between chlorophyllide esterification, Shibata shift and regeneration of protochlorophyllide650 in flash-irradiated etiolated barley leaves. Physiol. Plant. 121, 556–567. doi: 10.1111/j.1399-3054.2004.00362.x
Rebeiz, C. A. (2014). “The Chl a carboxylic biosynthetic routes: reactions between Mg-Protoporphyrin IX and protochlorophyllide a,” in Chlorophyll Biosynthesis and Technological Applications (Berlin: Springer), 197–214. doi: 10.1007/978-94-007-7134-5_7
Rebeiz, C. A., Ioannides, I. M., Kolossov, V., and Kopetz, K. J. (1999). Chloroplast biogenesis 80. Proposal of a unified multibranched chlorophyll a/b biosynthetic pathway. Photosynthetica 36, 117–128. doi: 10.1023/A:1007027005903
Reinbothe, C., Buhr, F., Bartsch, S., Desvignes, C., Quigley, F., Pesey, H., et al. (2006). In vitro-mutagenesis of NADPH:protochlorophyllide oxidoreductase B: two distinctive protochlorophyllide binding sites participate in enzyme catalysis and assembly. Mol. Genet. Genomics 275, 540–552. doi: 10.1007/s00438-006-0109-9
Reinbothe, C., El Bakkouri, M., Buhr, F., Muraki, N., Nomata, J., Kurisu, G., et al. (2010). Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 15, 614–624. doi: 10.1016/j.tplants.2010.07.002
Reinbothe, C., Lebedev, N., and Reinbothe, S. (1999). A protochlorophyllide light-harvesting complex involved in de-etiolation of higher plants. Nature 397, 80–84. doi: 10.1038/16283
Reisinger, V., Plöscher, M., and Eichacker, L. A. (2008). Lil3 assembles as chlorophyll-binding protein complex during deetiolation. FEBS Lett. 582, 1547–1551. doi: 10.1016/j.febslet.2008.03.042
Richter, A. S., and Grimm, B. (2013). Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway in plants. Front. Plant Sci. 4:371. doi: 10.3389/fpls.2013.00371
Richter, A. S., Hochheuser, C., Fufezan, C., Heinze, L., Kuhnert, F., and Grimm, B. (2016). Phosphorylation of GENOMES UNCOUPLED 4 alters stimulation of Mg chelatase activity in angiosperms. Plant Physiol. 172, 1578–1595. doi: 10.1104/pp.16.01036
Rodriguez-Concepcion, M. (2010). Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 504, 118–122. doi: 10.1016/j.abb.2010.06.016
Rodríguez-Concepción, M., and Boronat, A. (2015). Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 25, 17–22. doi: 10.1016/j.pbi.2015.04.001
Rossmann, M. G., Moras, D., and Olsen, K. W. (1974). Chemical and biological evolution of a nucleotide-binding protein. Nature 250, 194–199. doi: 10.1038/250194a0
Rüdiger, W. (2002). Biosynthesis of chlorophyll b and the chlorophyll cycle. Photosynth. Res. 74, 187–193. doi: 10.1023/A:1020959610952
Rüdiger, W. (2006). “Biosynthesis of chlorophylls a and b: the last steps,” in Chlorophylls and Bacteriochlorophylls. Advances in Photosynthesis and Respiration, Vol. 25, eds B. Grimm, R. J. Porra, W. Rüdiger, and H. Scheer (Dordrecht: Springer), 189–200. doi: 10.1007/1-4020-4516-6_14
Rüdiger, W., Benz, J., and Guthoff, C. (1980). Detection and partial characterization of activity of chlorophyll synthetase in etioplast membranes. Eur. J. Biochem. 109, 193−200. doi: 10.1111/j.1432-1033.1980.tb04784.x
Rüdiger, W., Böhm, S., Helfrich, M., Schulz, S., and Schoch, S. (2005). Enzymes of the last steps of chlorophyll biosynthesis: modification of the substrate structure helps to understand the topology of the active centers. Biochemistry 44, 10864–10872. doi: 10.1021/bi0504198
Rudowska, Ł., Gieczewska, K., Mazur, R., Garstka, M., and Mostowska, A. (2012). Chloroplast biogenesis - Correlation between structure and function. Biochim. Biophys. Acta Bioenerg. 1817, 1380–1387. doi: 10.1016/j.bbabio.2012.03.013
Ruiz-Sola, M. Á., Coman, D., Beck, G., Barja, M. V., Colinas, M., Graf, A., et al. (2016). Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 209, 252–264. doi: 10.1111/nph.13580
Ryberg, M., Sandelius, A. S., and Selstam, E. (1983). Lipid composition of prolamellar bodies and prothylakoids of wheat etioplasts. Physiol. Plant. 57, 555–560. doi: 10.1111/j.1399-3054.1983.tb02785.x
Ryberg, M., and Sundqvist, C. (1982a). Characterization of prolamellar bodies and prothylakoids fractionated from wheat etioplasts. Physiol. Plant. 56, 125–132. doi: 10.1111/j.1399-3054.1982.tb00313.x
Ryberg, M., and Sundqvist, C. (1982b). Spectral forms of protochlorophyllide in prolamellar bodies and prothylakoids fractionated from wheat etioplasts. Physiol. Plant. 56, 133–138. doi: 10.1111/j.1399-3054.1982.tb00314.x
Ryberg, M., and Sundqvist, C. (1988). The regular ultrastructure of isolated prolamellar bodies depends on the presence of membrane- bound NADPH-protochlorophyllide oxidoreductase. Physiol. Plant. 73, 218–226. doi: 10.1111/j.1399-3054.1988.tb00589.x
Rzeznicka, K., Walker, C. J., Westergren, T., Kannangara, C. G., Von Wettstein, D., Merchant, S., et al. (2005). Xantha-I encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 5886–5891. doi: 10.1073/pnas.0501784102
Salvi, D., Bournais, S., Moyet, L., Bouchnak, I., Kuntz, M., Bruley, C., et al. (2018). AT_CHLORO: the first step when looking for information about subplastidial localization of proteins. Methods Mol. Biol. 1829, 395–406. doi: 10.1007/978-1-4939-8654-5_26
Sandelius, A. S., and Selstam, E. (1984). Localization of galactolipid biosynthesis in etioplasts isolated from dark-grown wheat (Triticum aestivum L.). Plant Physiol. 76, 1041–1046. doi: 10.1104/pp.76.4.1041
Schaff, Z., Barry, D. W., and Grimley, P. M. (1973). Cytochemistry of tubuloreticular structures in lymphocytes from patients with systemic lupus erythematosus and in cultured human lymphoid cells. Comparison to a paramyxovirus. Lab. Investig. 29, 577–586.
Schaff, Z., Heine, U., and Dalton, A. J. (1972). Ultramorphological and ultracytochemical studies on tubuloreticular structures in lymphoid cells. Cancer Res. 32, 2696–2706.
Schaff, Z., Lapis, K., and Grimley, P. (1976). Undulating membraneous structures associated with the endoplasmic reticulum in tumour cells. Int. J. Cancer 18, 697–702. doi: 10.1002/ijc.2910180519
Schmid, H. C., Rassadina, V., Oster, U., Schoch, S., and Rüdiger, W. (2002). Pre-loading of chlorophyll synthase with tetraprenyl diphosphate is an obligatory step in chlorophyll biosynthesis. Biol. Chem. 383, 1769−1778. doi: 10.1515/BC.2002.198
Schneidewind, J., Krause, F., Bocola, M., Stadler, A. M., Davari, M. D., Schwaneberg, U., et al. (2019). Consensus model of a cyanobacterial light-dependent protochlorophyllide oxidoreductase in its pigment-free apo-form and photoactive ternary complex. Commun. Biol. 2:351. doi: 10.1038/s42003-019-0590-4
Schnepf, E. (1961). Piastidenstrukturen bei passiflora. Protoplasma 54, 310–313. doi: 10.1007/BF01260360
Schoch, S. (1978). The esterification of chlorophyllide a in greening bean leaves. Zeitschrift für Naturforsch. C 33, 712–714. doi: 10.1515/znc-1978-9-1018
Schoefs, B. (2005). Protochlorophyllide reduction - what is new in 2005? Photosynthetica 43, 329–343. doi: 10.1007/s11099-005-0056-4
Schoefs, B., and Bertrand, M. (2000). The formation of chlorophyll from chlorophyllide in leaves containing proplastids is a four-step process. FEBS Lett. 486, 243–246. doi: 10.1016/S0014-5793(00)02309-7
Schoefs, B., and Franck, F. (2003). Protochlorophyllide reduction: mechanisms and evolutions. Photochem. Photobiol. 78, 543–557. doi: 10.1562/0031-8655(2003)078<0543:PRMAE>2.0.CO;2
Schoefs, B., and Franck, F. (2008). The photoenzymatic cycle of NADPH: protochlorophyllide oxidoreductase in primary bean leaves (Phaseolus vulgaris) during the first days of photoperiodic growth. Photosynth. Res. 96, 15–26. doi: 10.1007/s11120-007-9274-x
Schulz, R., Steinmüller, K., Klaas, M., Forreiter, C., Rasmussen, S., Hiller, C., et al. (1989). Nucleotide sequence of a cDNA coding for the NADPH-protochlorophyllide oxidoreductase (PCR) of barley (Hordeum vulgare L.) and its expression in Escherichia coli. MGG Mol. Gen. Genet. 217, 355–361. doi: 10.1007/BF02464904
Scrutton, N. S., Louise Groot, M., and Heyes, D. J. (2012). Excited state dynamics and catalytic mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Phys. Chem. Chem. Phys. 14, 8818–8824. doi: 10.1039/c2cp23789j
Selstam, E. (1998). “Development of thylakoid membranes with respect to lipids,” in Lipids in Photosynthesis: Structure, Function and Genetics. Advances in Photosynthesis and Respiration, eds S. Paul-André and M. Norio (Dordrecht: Springer), 209–224.
Selstam, E., and Sandelius, A. S. (1984). A comparison between prolamellar bodies and prothylakoid membranes of etioplasts of dark-grown wheat concerning lipid and polypeptide composition. Plant Physiol. 76, 1036–1040. doi: 10.1104/pp.76.4.1036
Selstam, E., and Widell-Wigge, A. (1989). Hydrophobicity of protochlorophyllide oxidoreductase, characterized by means of Triton X-114 partitioning of isolated etioplast membrane fractions. Physiol. Plant. 77, 401–406. doi: 10.1111/j.1399-3054.1989.tb05660.x
Seyedi, M., Timko, M. P., and Sundqvist, C. (2001). The distribution of protochlorophyllide and chlorophyll within seedlings of the lip1 mutant of Pea. Plant Cell Physiol. 42, 931–941. doi: 10.1093/pcp/pce118
Shibata, K. (1957). Spectroscopic studies on chlorophyll formation in intact leaves. J. Biochem. 44, 147–173. doi: 10.1093/oxfordjournals.jbchem.a126741
Shioi, Y., and Takamiya, K. (1992). Monovinyl and divinyl protochlorophyllide pools in etiolated tissues of higher plants. Plant Physiol. 100, 1291–1295. doi: 10.1104/pp.100.3.1291
Sineshchekov, V. A., and Belyaeva, O. B. (2019). Regulation of chlorophyll biogenesis by phytochrome A. Biochemistry 84, 491–508. doi: 10.1134/S0006297919050043
Sironval, C., and Brouers, M. (1970). The reduction of protochlorophyllide into chlorophyllide. II. The temperature dependence of the P657-647-P688-676 phototransformation. Photosynthetica 4, 38–47.
Smeller, L., Solymosi, K., Fidy, J., and Böddi, B. (2003). Activation parameters of the blue shift (Shibata shift) subsequent to protochlorophyllide phototransformation. Biochim. Biophys. Acta Proteins Proteomics 1651, 130–138. doi: 10.1016/S1570-9639(03)00261-9
Soll, J., Schultz, G., Rüdiger, W., and Benz, J. (1983). Hydrogenation of geranylgeraniol. Two pathways exist in spinach chloroplasts. Plant Physiol. 71, 849–854. doi: 10.1104/pp.71.4.849
Solymosi, K., and Aronsson, H. (2013). “Etioplasts and their significance in chloroplast biogenesis,” in Plastid Development in Leaves During Growth and Senescence: Advances in Photosynthesis and Respiration, eds B. Biswal, K. Krupińska, and A. K. Biswal (Dordrecht: Springer), 39–71.
Solymosi, K., Bóka, K., and Böddi, B. (2006a). Transient etiolation: protochlorophyll(ide) and chlorophyll forms in differentiating plastids of closed and breaking leaf buds of horse chestnut (Aesculus hippocastanum). Tree Physiol. 26, 1087–1096. doi: 10.1093/treephys/26.8.1087
Solymosi, K., and Köfalvi, A. (2017). Cannabis: a treasure trove or Pandora's box? Mini Rev. Med. Chem. 17, 1223–1291. doi: 10.2174/1389557516666161004162133
Solymosi, K., Martinez, K., Kristóf, Z., Sundqvist, C., and Böddi, B. (2004). Plastid differentiation and chlorophyll biosynthesis in different leaf layers of white cabbage (Brassica oleracea cv. capitata). Physiol. Plant. 121, 520–529. doi: 10.1111/j.0031-9317.2004.00349.x
Solymosi, K., Morandi, D., Bóka, K., Böddi, B., and Schoefs, B. (2012). High biological variability of plastids, photosynthetic pigments and pigment forms of leaf primordia in buds. Planta 235, 1035–1049. doi: 10.1007/s00425-011-1559-9
Solymosi, K., Mysliwa-Kurdziel, B., Bóka, K., Strzałka, K., and Böddi, B. (2006b). Disintegration of the prolamellar body structure at high concentrations of Hg2+. Plant Biol. 8, 627–635. doi: 10.1055/s-2006-924110
Solymosi, K., and Schoefs, B. (2008). “Prolamellar body: a unique plastid compartment, which does not only occur in dark-grown leaves,” in Plant Cell Compartments - Selected Topics, ed B. Schoefs (Trivandrum: Research Signpost), 151–202.
Solymosi, K., and Schoefs, B. (2010). Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105, 143–166. doi: 10.1007/s11120-010-9568-2
Solymosi, K., Smeller, L., Böddi, B., and Fidy, J. (2002). Activation volumes of processes linked to the phototransformation of protochlorophyllide determined by fluorescence spectroscopy at high pressure. Biochim. Biophys. Acta Bioenerg. 1554, 1–4. doi: 10.1016/S0005-2728(02)00209-8
Solymosi, K., Smeller, L., Ryberg, M., Sundqvist, C., Fidy, J., and Böddi, B. (2007a). Molecular rearrangement in POR macrodomains as a reason for the blue shift of chlorophyllide fluorescence observed after phototransformation. Biochim. Biophys. Acta Biomembr. 1768, 1650–1658. doi: 10.1016/j.bbamem.2007.02.022
Solymosi, K., Tuba, Z., and Böddi, B. (2013). Desiccoplast-etioplast-chloroplast transformation under rehydration of desiccated poikilochlorophyllous Xerophyta humilis leaves in the dark and upon subsequent illumination. J. Plant Physiol. 170, 583–590. doi: 10.1016/j.jplph.2012.11.022
Solymosi, K., Vitányi, B., Hideg, É., and Böddi, B. (2007b). Etiolation symptoms in sunflower (Helianthus annuus) cotyledons partially covered by the pericarp of the achene. Ann. Bot. 99, 857–867. doi: 10.1093/aob/mcm034
Spano, A. J., He, Z., Michel, H., Hunt, D. F., and Timko, M. P. (1992). Molecular cloning, nuclear gene structure, and developmental expression of NADPH:protochlorophyllide oxidoreductase in pea (Pisum sativum L.). Plant Mol. Biol. 18, 967–972. doi: 10.1007/BF00019210
Sperling, U., van Cleve, B., Frick, G., Apel, K., and Armstrong, G. A. (1997). Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 12, 649–658. doi: 10.1046/j.1365-313X.1997.d01-11.x
Splinter, T., Helder, A., Lucas, C., and Feltkamp-Vroom, T. (1975). Spontaneous occurrence of TRS The available lymphoid cell lines were It is very remarkable that all 4 cell patients are negative and all 5 cell lines. Br. J. Exp. Path. 56, 124–132.
Stenbaek, A., and Jensen, P. E. (2010). Redox regulation of chlorophyll biosynthesis. Phytochemistry 71, 853–859. doi: 10.1016/j.phytochem.2010.03.022
Stpiczyńska, M., Milanesi, C., Faleri, C., and Cresti, M. (2005). Ultrastructure of the nectary spur of Platanthera chlorantha (Custer) Rchb. (Orchidaceae) during successive stages of nectar secretion. Acta Biol. Cracoviensia Ser. Bot. 47, 111–119.
Suzuki, J. Y., and Bauer, C. E. (1995). A prokaryotic origin for light-dependent chlorophyll biosynthesis of plants. Proc. Natl. Acad. Sci. U.S.A. 92, 3749–3753. doi: 10.1073/pnas.92.9.3749
Sytina, O. A., Novoderezhkin, V. I., Van Grondelle, R., and Groot, M. L. (2011a). Modeling of multi-exciton transient absorption spectra of protochlorophyllide aggregates in aqueous solution. J. Phys. Chem. A 115, 11944–11951. doi: 10.1021/jp204395z
Sytina, O. A., van Stokkum, I. H. M., Heyes, D. J., Hunter, C. N., van Grondelle, R., and Groot, M. L. (2010). Protochlorophyllide excited-state dynamics in organic solvents studied by time-resolved visible and mid-infrared spectroscopy. J. Phys. Chem. B 114, 4335–4344. doi: 10.1021/jp9089326
Sytina, O. A., Van Stokkum, I. H. M., Van Grondelle, R., and Groot, M. L. (2011b). Single and multi-exciton dynamics in aqueous protochlorophyllide aggregates. J. Phys. Chem. A 115, 3936–3946. doi: 10.1021/jp108317u
Tanaka, A., Ito, H., Tanaka, R., Tanaka, N. K., Yoshida, K., and Okada, K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. U.S.A. 95, 12719–12723. doi: 10.1073/pnas.95.21.12719
Tanaka, A., and Tanaka, R. (2019). “Chapter Six - The biochemistry, physiology, and evolution of the chlorophyll cycle,” in Metabolism, Structure and Function of Plant Tetrapyrroles: Introduction, Microbial and Eukaryotic Chlorophyll Synthesis and Catabolism, Vol. 90, ed B. Grimm (London: Academic Press), 183–212. doi: 10.1016/bs.abr.2019.03.005
Tanaka, R., Kobayashi, K., and Masuda, T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. Arab. B. 9, e0145. doi: 10.1199/tab.0145
Tanaka, R., Koshino, Y., Sawa, S., Ishiguro, S., Okada, K., and Tanaka, A. (2001). Overexpression of chlorophyllide a oxygenase (CAO) enlarges the antenna size of photosystem II in Arabidopsis thaliana. Plant J. 26, 365–373. doi: 10.1046/j.1365-313X.2001.2641034.x
Tanaka, R., Rothbart, M., Oka, S., Takabayashi, A., Takahashi, K., Shibata, M., et al. (2010). LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 16721–16725. doi: 10.1073/pnas.1004699107
Tanaka, R., and Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448
Tanaka, R., and Tanaka, A. (2011). Biochimica et biophysica acta chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta Bioenerg. 1807, 968–976. doi: 10.1016/j.bbabio.2011.01.002
Tottey, S., Block, M. A., Allen, M., Westergren, T., Albrieux, C., Scheller, H. V., et al. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. U.S.A. 100, 16119–16124. doi: 10.1073/pnas.2136793100
Townley, H. E., Sessions, R. B., Clarke, A. R., Dafforn, T. R., and Trevor Griffiths, W. (2001). Protochlorophyllide oxidoreductase: a homology model examined by site-directed mutagenesis. Proteins Struct. Funct. Genet. 44, 329–335. doi: 10.1002/prot.1098
Tripathy, B., and Pattanayak, G. (2012). “Chlorophyll biosynthesis in higher plants,” in Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation, Advances in Photosynthesis and Respiration, Vol. 34, eds J. J. Eaton-Rye, B. C. Tripathy, and T. D. Sharkey (Springer Science+Business Media B.V.), 63–94.
Tripathy, B. C., and Rebeiz, C. A. (1986). Chloroplast biogenesis. Demonstration of the monovinyl and divinyl monocarboxylic routes of chlorophyll biosynthesis in higher plants. J. Biol. Chem. 261, 13556–13564. doi: 10.1016/S0021-9258(18)67055-3
Tripathy, B. C., and Rebeiz, C. A. (1988). Chloroplast biogenesis 60. Plant Physiol. 87, 89–94. doi: 10.1104/pp.87.1.89
Turner, G. W., Davis, E. M., and Croteau, R. B. (2012). Immunocytochemical localization of short-chain family reductases involved in menthol biosynthesis in peppermint. Planta 235, 1186–1195. doi: 10.1007/s00425-011-1567-9
Turner, G. W., Gershenzon, J., and Croteau, R. B. (2000). Development of peltate glandular trichomes of peppermint. Plant Physiol. 124, 665–680. doi: 10.1104/pp.124.2.665
Vedalankar, P., and Tripathy, B. C. (2019). Evolution of light-independent protochlorophyllide oxidoreductase. Protoplasma 256, 293–312. doi: 10.1007/s00709-018-1317-y
Vermeer, J., and Peterson, R. L. (1979). Glandular trichomes on the inflorescence of Chrysanthemum morifolium cv dramatic (Compositae). 1. Development and morphology. Can. J. Bot. 57, 705–713. doi: 10.1139/b79-090
Vitányi, B., Kósa, A., Solymosi, K., and Böddi, B. (2013). Etioplasts with protochlorophyll and protochlorophyllide forms in the under-soil epicotyl segments of pea (Pisum sativum) seedlings grown under natural light conditions. Physiol. Plant. 148, 307–315. doi: 10.1111/j.1399-3054.2012.01714.x
Voitsekhovskaja, O. V., and Tyutereva, E. V. (2015). Chlorophyll b in angiosperms: functions in photosynthesis, signaling and ontogenetic regulation. J. Plant Physiol. 189, 51–64. doi: 10.1016/j.jplph.2015.09.013
von Zychlinski, A., Kleffmann, T., Krishnamurthy, N., Sjölander, K., Baginsky, S., and Gruissem, W. (2005). Proteome analysis of the rice etioplast: metabolic and regulatory networks and novel protein functions. Mol. Cell. Proteomics 4, 1072–1084. doi: 10.1074/mcp.M500018-MCP200
Vranová, E., Coman, D., and Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi: 10.1146/annurev-arplant-050312-120116
Wang, G., and Dixon, R. A. (2009). Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 106, 9914–9919. doi: 10.1073/pnas.0904069106
Wang, J., Lin, H. X., Su, P., Chen, T., Guo, J., Gao, W., et al. (2019). Molecular cloning and functional characterization of multiple geranylgeranyl pyrophosphate synthases (ApGGPPS) from Andrographis paniculata. Plant Cell Rep. 38, 117–128. doi: 10.1007/s00299-018-2353-y
Wang, L., Leister, D., Guan, L., Zheng, Y., Schneider, K., Lehmann, M., et al. (2020). The Arabidopsis SAFEGUARD1 suppresses singlet oxygen-induced stress responses by protecting grana margins. Proc. Natl. Acad. Sci. U.S.A. 117, 6918–6927. doi: 10.1073/pnas.1918640117
Wang, P., Gao, J., Wan, C., Zhang, F., Xu, Z., Huang, X., et al. (2010). Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol. 153, 994–1003. doi: 10.1104/pp.110.158477
Wang, P., and Grimm, B. (2015). Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth. Res. 126, 189–202. doi: 10.1007/s11120-015-0154-5
Wang, P., Wan, C., Xu, Z., Wang, P., Wang, W., Sun, C., et al. (2013). One divinyl reductase reduces the 8-vinyl groups in various intermediates of chlorophyll biosynthesis in a given higher plant species, but the isozyme differs between species. Plant Physiol. 161, 521–534. doi: 10.1104/pp.112.208421
Whyte, B. J., and Griffiths, W. T. (1993). 8-Vinyl reduction and chlorophyll a biosynthesis in higher plants. Biochem. J. 291, 939–944. doi: 10.1042/bj2910939
Wiktorsson, B., Engdahl, S., Zhong, L. B., Boddi, B., Ryberg, M., and Sundqvist, C. (1993). The effect of cross-linking of the subunits of nadph- protochlorophyllide oxidoreductase on the aggregational state of protochlorophyllide. Photosynthetica 29, 205–218.
Wilks, H. M., and Timko, M. P. (1995). A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc. Natl. Acad. Sci. U.S.A. 92, 724–728. doi: 10.1073/pnas.92.3.724
Willows, R. D. (2019). “The Mg branch of chlorophyll synthesis: biosynthesis of chlorophyll a from protoporphyrin IX,” in Advances in Botanical Research, Vol. 90, ed B. Grimm (London: Academic Press Inc.), 141–182. doi: 10.1016/bs.abr.2019.03.003
Wong, Y.-S., and Castelfranco, P. A. (1984). Resolution and reconstitution of Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase, the enzyme system responsible for the formation of the chlorophyll isocyclic ring. Plant Physiol. 75, 658–661. doi: 10.1104/pp.75.3.658
Wong, Y.-S., and Castelfranco, P. A. (1985). Properties of the Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase system. Plant Physiol. 79, 730–733. doi: 10.1104/pp.79.3.730
Yadav, D., Zemach, H., Belausov, E., and Charuvi, D. (2019). Initial proplastid-to-chloroplast differentiation in the developing vegetative shoot apical meristem of Arabidopsis. Biochem. Biophys. Res. Commun. 519, 391–395. doi: 10.1016/j.bbrc.2019.09.019
Yamamoto, H., Kojima-Ando, H., Ohki, K., and Fujita, Y. (2020). Formation of prolamellar-body-like ultrastructures in etiolated cyanobacterial cells overexpressing light-dependent protochlorophyllide oxidoreductase in Leptolyngbya boryana. J. Gen. Appl. Microbiol. 66, 129–139. doi: 10.2323/jgam.2020.01.009
Yamazaki, S., Nomata, J., and Fujita, Y. (2006). Differential operation of dual protochlorophyllide reductases for chlorophyll biosynthesis in response to environmental oxygen levels in the cyanobacterium Leptolyngbya boryana. Plant Physiol. 142, 911–922. doi: 10.1104/pp.106.086090
Yang, J., and Cheng, Q. (2004). Origin and evolution of the light-dependent protochlorophyllide oxidoreductase (LPOR) genes. Plant Biol. 6, 537–544. doi: 10.1055/s-2004-821270
Yu, C.-W., Lin, Y.-T., and Li, H. (2020). Increased ratio of galactolipid MGDG: DGDG induces jasmonic acid overproduction and changes chloroplast shape. New Phytol. 228, 1327–1335. doi: 10.1111/nph.16766
Yuan, M., Zhao, Y. Q., Zhang, Z. W., Chen, Y. E., Ding, C. B., and Yuan, S. (2017). Light regulates transcription of chlorophyll biosynthetic genes during chloroplast biogenesis. CRC. Crit. Rev. Plant Sci. 36, 35–54. doi: 10.1080/07352689.2017.1327764
Zhang, M., Zhang, F., Fang, Y., Chen, X., Chen, Y., Zhang, W., et al. (2015). The non-canonical tetratricopeptide repeat (TPR) domain of fluorescent (FLU) mediates complex formation with glutamyl-tRNA reductase. J. Biol. Chem. 290, 17559–17565. doi: 10.1074/jbc.M115.662981
Zhang, S., Godwin, A. R. F., Taylor, A., Hardman, S. J. O., Jowitt, T. A., Johannissen, L. O., et al. (2021). Dual role of the active site ‘lid’ regions of protochlorophyllide oxidoreductase in photocatalysis and plant development. FEBS J. 288, 175–189. doi: 10.1111/febs.15542
Zhang, S., Heyes, D. J., Feng, L., Sun, W., Johannissen, L. O., Liu, H., et al. (2019). Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574, 722–725. doi: 10.1038/s41586-019-1685-2
Zhang, Z. W., Yuan, S., Feng, H., Xu, F., Cheng, J., Shang, J., et al. (2011). Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs - new evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 168, 714–721. doi: 10.1016/j.jplph.2010.10.016
Zhao, G.-J., and Han, K.-L. (2008). Site-specific solvation of the photoexcited protochlorophyllide a in methanol: formation of the hydrogen-bonded intermediate state induced by hydrogen-bond strengthening. Biophys. J. 94, 38–46. doi: 10.1529/biophysj.107.113738
Zhong, L. B., Wiktorsson, B., Ryberg, M., and Sundqvist, C. (1996). The Shibata shift; effects of in vitro conditions on the spectral blue-shift of chlorophyllide in irradiated isolated prolamellar bodies. J. Photochem. Photobiol. B Biol. 36, 263–270. doi: 10.1016/S1011-1344(96)07394-0
Keywords: chlorophyll biosynthesis, chloroplast, etioplast, NADPH:protochlorophyllide oxidoreductase, phytol, prolamellar body, protochlorophyllide, tubular complex
Citation: Solymosi K and Mysliwa-Kurdziel B (2021) The Role of Membranes and Lipid-Protein Interactions in the Mg-Branch of Tetrapyrrole Biosynthesis. Front. Plant Sci. 12:663309. doi: 10.3389/fpls.2021.663309
Received: 02 February 2021; Accepted: 22 March 2021;
Published: 28 April 2021.
Edited by:
Yoshitaka Nishiyama, Saitama University, JapanReviewed by:
Ayumi Tanaka, Hokkaido University, JapanCopyright © 2021 Solymosi and Mysliwa-Kurdziel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beata Mysliwa-Kurdziel, Yi5teXNsaXdhLWt1cmR6aWVsQHVqLmVkdS5wbA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.