
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 14 July 2021
Sec. Crop and Product Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.659307
Seed vigor is a complex trait, including the seed germination, seedling emergence, and growth, as well as seed storability and stress tolerance, which is important for direct seeding in rice. Seed vigor is established during seed development, and its level is decreased during seed storage. Seed vigor is influenced by genetic and environmental factors during seed development, storage, and germination stages. A lot of factors, such as nutrient reserves, seed dying, seed dormancy, seed deterioration, stress conditions, and seed treatments, will influence seed vigor during seed development to germination stages. This review highlights the current advances on the identification of quantitative trait loci (QTLs) and regulatory genes involved in seed vigor at seed development, storage, and germination stages in rice. These identified QTLs and regulatory genes will contribute to the improvement of seed vigor by breeding, biotechnological, and treatment approaches.
Rice (Oryza sativa L.) is one of the most popular crops in the world. Direct seeding of rice at present is popular in many countries because of its low cost and convenience compared to the conventional transplantation (Farooq et al., 2011). Direct seeding can be classified into three types, including wet direct seeding, dry direct seeding, and water direct seeding, in rice (Mahender et al., 2015). However, the low seed germination, seedling establishment, and the poor seedling growth in field are the major constraints in the production of direct seeding. Seed vigor is an agronomical trait that determines the rapid and uniform germination, the final seedling emergence, and the seed storability and stress tolerance (Sun et al., 2007). Thus, rice varieties with high seed vigor are required for direct seeding. Seed vigor is regulated by a lot of genetic and environmental factors during seed development, storage, and germination stages. Here, the current advances on the identification of quantitative trait loci (QTLs) and regulatory genes involved in seed vigor at seed development, storage, and germination stages were mainly summarized in rice. Identification of the QTLs and genes involved in seed vigor will contribute to develop high-vigor varieties for direct seeding in rice.
Seed vigor is the sum of those properties that determine the germination activity and performance of seed lots in a wide range of environments (ISTA, 2015). Seed vigor is thus a complex trait, including the seed germination, seedling emergence and growth, and seed storability and stress tolerance (Sun et al., 2007; Finch-Savage and Bassel, 2016), in which the germination speed and uniformity are usually conducted using the indexes of germination potential, time to 50% germination (T50), and germination index in rice. The final germination percentage (GP) is used to test the seed viability, and the indexes of seedling percentage, vigor index, and seedling weight are conducted to evaluate seedling growth (Wang et al., 2010; He et al.,2019a,b). The above traits are usually used to evaluate seed vigor by traditional germination methods under normal, low-temperature, salinity, and flooding stress conditions (Wang et al.,2011a,b; Chamara et al., 2018). However, the conventional methods for seed vigor testing have several limitations as they involve visual assessment and are destructive. Thus, the non-contact- and non-destructive-type imaging techniques, such as visible imaging, hyperspectral imaging, and X-ray imaging, for conducting seed vigor might be an attractive alternative to the existing vigor assessment methods for rice (Mahajan et al., 2018; Zhang T. et al., 2020).
Seed vigor is influenced by a lot of factors, such as reserves accumulation and seed dormancy during development, seed deterioration during storage, and stress conditions and seed treatments during germination, in rice. Seed vigor progressively increases with physiological maturity (Bewley et al., 2013) and begins to decline before and after harvest (Finch-Savage and Bassel, 2016). Seed dormancy is established during seed maturation, which helps to prevent preharvest sprouting (PHS) at harvest stage (Sugimoto et al., 2010). However, deep dormancy will inhibit seed germination when sowing (Gubler et al., 2005). Thus, seed dormancy is established during development that has both advantages and disadvantages for seed vigor in terms of cultivation and utilization (Sugimoto et al., 2010). Seed vigor will ultimately decline during storage because of deterioration in rice (Sasaki et al., 2015; Finch-Savage and Bassel, 2016). Thus, seed longevity, the maintenance of viability during dry storage, is a crucial factor to preserve seed vigor (Pellizzaro et al., 2020). Additionally, seed vigor will be reduced by various stresses, such as salt, low temperature, and flooding stresses, during seed germination in rice (Wang et al.,2011a,b; Chamara et al., 2018), but it can be enhanced by seed priming treatment, an invigoration technique of low-vigor seeds (Rajjou et al., 2012). In most studies, the influencing factors of rice seed vigor is mainly considered at seed germination stage, whereas other stages are limitedly considered. To accurately characterize seed vigor of rice, the traits that influence seed vigor deserve to be studied systematically during seed development to germination stages. It will greatly help us in exploring the elite varieties for direct seeding, as well as unraveling its regulatory mechanisms and promoting variety breeding for high seed vigor in rice.
Grain size is one of the key agronomic traits that determine grain yield in rice. Therefore, uncovering the genetic and molecular mechanisms of grain size is mainly conducted during seed development in rice, but seed vigor is less considered. More than 500 QTLs associated with grain size have been identified in rice, of which approximately 20 genes have been map-based cloned (Li et al., 2018). For example, GS3 (Mao et al., 2010), qGL3/GL3.1 (Qi et al., 2012; Zhang et al., 2012), TGW6 (Ishimaru et al., 2013), GLW7 (Si et al., 2016), OsLG3 (Yu et al., 2017), GL4 (Wu et al., 2017), qLGY3/OsLG3b (Liu et al., 2018; Yu et al., 2018a), qTGW3/GL3.3 (Hu et al., 2018; Xia et al., 2018), and GL6 (Wang et al., 2019) for gain length, and GW2 (Song et al., 2007), qSW5/GW5/GSE5 (Shomura et al., 2008; Weng et al., 2008; Duan et al., 2017), GS5 (Li et al., 2011), GW8 (Wang S. et al., 2012), and TGW2 (Ruan et al., 2020) for gain width, as well as GL7/GW7/SLG7 (Wang S. et al., 2015; Wang Y. et al., 2015; Zhou et al., 2015), GW6a (Song et al., 2015), and GS9 (Zhao et al., 2018) for both gain length and width, have been cloned in rice. Several reports have shown that large seeds have the advantages of producing more vigorous seedlings (Douglas et al., 1994; Bockous and Shroyer, 1996), but the lack of a significant positive relation between seed weight and seedling dry weight has also been reported in several studies (Carleton and Cooper, 1972; Johnson and Wax, 1978). One rice population of recombinant inbred lines (RILs) was used to determine the genetic characteristics of the establishment of seed vigor at 4 (early), 5 (middle), and 6 weeks (late) after heading during seed development in 2 years in our previous study (Liu et al., 2014). We observed that the QTLs for seed vigor were rarely colocalized among the different maturity stages; more QTLs were expressed at the early maturity stage followed by the late and middle stages. The stably expressed QTLs for seed vigor are likely to coincide with QTL for grain size, low-temperature germinability, and seed dormancy (Liu et al., 2014). It is interesting to reveal that whether the above-described genes associated with grain size are also involved in controlling seed vigor in rice.
Seed dormancy is affected by multiple genes and environmental factors. Many QTLs have been identified for seed dormancy in rice. For example, two dormancy QTLs qSdn-1 and qSdn-5 have been obtained from the segregation analysis of the advanced backcross populations derived from the cross between cultivars N22 (strong dormancy) and Nanjing35 (weak dormancy) (Lu et al., 2011). Three putative QTLs, Sdr6, Sdr9, and Sdr10, for seed dormancy have been identified using chromosome segment substitution lines (CSSLs) derived from a cross between Nona Bokra (strong dormancy) and Koshihikari (weak dormancy) (Marzougui et al., 2012). A total of four, one, and three additive QTLs for seed dormancy have been identified at the early, middle, and late development stages, respectively, using one RIL population (Wang et al., 2014). A total of 12 and 27 QTLs for seed dormancy have been detected using CSSL and backcross inbred line (BIL) population, both derived from the same parents Nipponbare, a japonica cultivar with seed dormancy, and 9311, an indica cultivar lacking seed dormancy (Zhang C. et al., 2020). A genome-wide association study (GWAS) based on seed GP in freshly harvested seeds (FHSs) and after-ripened seeds (ARS) in 350 worldwide accessions has revealed that 16 loci significantly associated with GP in FHS and 38 in ARS (Magwa et al., 2016). Meanwhile, a GWAS has detected nine significant single-nucleotide polymorphisms (SNPs) associated with seed dormancy using an indica-only population consisting of 453 accessions, and a total of 212 candidate genes have been predicted (Lu et al., 2018). However, only few QTLs, such as Sdr4 (Sugimoto et al., 2010) and qSD1-2 (Ye et al., 2015), have been map-based cloned.
Seed storability, defined as the ability to remain alive during storage, is an important agronomic and physiological characteristic. Several studies have detected some QTLs controlling seed storability in rice. For example, three putative QTLs for seed longevity, qLG-2, qLG-4, and qLG-9, have been detected using 98 BILs derived from a cross between a japonica variety Nipponbare and an indica variety Kasalath (Miura et al., 2002). A total of five and three QTLs associated with seed germination capability have been identified in various storage periods using two sets of RILs, derived from crosses between Milyang 23 and Tong 88-7 and between Dasanbyeo and TR22183, respectively (Jiang et al., 2011). A total of seven QTLs have been identified for seed storability using two segregating populations with N22 (indica) as a common parent, viz. a set of 122 BILs derived from the backcross Nanjing35 (japonica)/N22//Nanjing35 and another population comprising 189 RILs from the cross of USSR5 (japonica) and N22 (Lin et al., 2015). Seven putative QTLs have been identified for seed storability under natural storage using the BILs that was derived from a cross of a japonica cultivar, Nipponbare, and an indica cultivar, 9311 (Yuan et al., 2019). A GWAS has shown that eight major loci are associated with seed longevity parameters using 299 indica accessions, and the candidate genes conferring high seed longevity might be related to DNA repair and transcription, sugar metabolism, reactive oxygen species (ROS) scavenging, and embryonic/root development (Lee et al., 2019). These identified QTLs will be further studied to elucidate the mechanisms controlling seed longevity/storability; however, few QTLs, such as qLG-9, for seed longevity have been fine-mapped (Sasaki et al., 2015).
A total of 10 QTLs have been detected to control rice seed vigor, including germination rate, final GP, and germination index, using one RIL population derived from a cross between japonica Daguandao and indica IR28 under normal conditions (Wang et al., 2010). Most of the QTLs are likely to coincide with QTLs for seed weight, seed size, or seed dormancy, suggesting that the rice seed vigor might be correlated with seed weight, seed size, and seed dormancy (Wang et al., 2010). A total of 27 marker-trait associations with seed vigor have been detected using 540 rice cultivars (419 from China and 121 from Vietnam) under normal condition (Dang et al., 2014). Conditional QTL analysis identified that qSV-1, qSV-5b, qSV-6a, qSV-6b, and qSV-11 influenced seedling establishment and that qSV-5a, qSV-5c, and qSV-8 influenced only germination using an RIL population from a cross between indica cultivars ZS97 and MH63 (Xie et al., 2014). Recently, 43 QTLs have been identified governing the early germination and seedling vigor traits related to weed competitive ability in rice using a total of 167 BC1F5 selective introgression lines developed from a backcross population involving Weed Tolerant Rice-1 as the recipient parent and Y-134 as the donor parent (Dimaano et al., 2020).
Rice originated from tropical or subtropical areas is a low-temperature–sensitive crop. A large number of QTLs for seed germination have been detected under low-temperature conditions compared to that of normal conditions in rice. For example, four QTLs for low-temperature vigor of germination have been identified using F2:3 population, which included 200 individuals and lines derived from a cross of indica Milyang 23 and japonica Jileng 1 (Han et al., 2006). Three putative QTLs involved in low-temperature germination have been detected using an RIL population derived from a cross of japonica USSR5 and indica N22, and qLTG-9 has been fine-mapped (Li et al., 2013). Using a mini core collection of 174 Chinese rice accessions, 22 QTLs for cold tolerance have been detected at the germination stage (Pan et al., 2015). A GWAS of low-temperature germinability in 187 rice natural accessions has revealed that 53 QTLs are associated with low-temperature germinability, and Stress-Associated Protein 16 (OsSAP16), coding for a zinc-finger domain protein, is a causal gene for one of the major QTLs (Wang X. et al., 2018). Recently, a total of 27 main-effect QTLs have been detected for germination and early seedling growth traits under low-temperature conditions using 230 introgression lines (ILs) in BC1F7 generation derived from the Weed Tolerant Rice-1 and Haoannong (Najeeb et al., 2020). A total of 20 QTLs for cold tolerance and cold recovery during germination have been identified in the subset of japonica accessions, whereas nine QTLs have been identified in the subset of indica accessions using a diversity panel of 257 rice accessions by GWAS approach (Thapa et al., 2020). Six and five QTLs for low-temperature tolerance have been detected during the germination and bud stages using RILs derived from indica rice H335 (tolerant) and indica rice CHA-1 (sensitive), and loci 3 has been detected during both the germination and bud stages (Yang et al., 2020). However, few QTLs, such as qLTG3-1, for low-temperature germinability have been cloned in rice (Fujino et al., 2008).
Salt tolerance of rice at the seed germination stage is one of the major determinants for the stable stand establishment in salinity soil. Several QTLs for seed germination have been detected under salt conditions in rice. For example, a total of 16 QTLs for seed imbibition and GP have been identified under control (water) and salt stress (100 mM NaCl) using one population of RILs, derived from a cross between a japonica rice landrace tolerant to salt stress and a sensitive indica rice variety (Wang et al., 2011b). A GWAS has revealed that 11 loci are associated with salt tolerance using 478 diverse rice accessions (Shi et al., 2017). A total of six QTLs for salt tolerance have been detected at the germination stage using 208 rice mini-core accessions by GWAS approach (Naveed et al., 2018). A total of 12 associated peaks have been detected for salt tolerance during seed germination by GWAS approach in rice, and the characterized 17 genes may contribute to salt tolerance (Yu et al., 2018b). Recently, a total of 13 QTLs have been identified for seed germination, i.e., 10 QTLs under H2O conditions and nine QTLs under salt conditions, using a BC1F2 population derived from the crossing Wujiaozhan (salt tolerant) and Nipponbare (salt sensitive), and LOC_Os06g10650, encoding tyrosine phosphatase family protein, might be the causal candidate gene for qGR6.2 (Zeng et al., 2021).
A few QTLs for seed germination under flooding conditions have been detected in rice. For example, five QTLs for anaerobic germination have been detected on chromosomes 1 (qAG-1-2), 3 (qAG-3-1), 7 (qAG-7-2), and 9 (qAG-9-1 and qAG-9-2) using a backcross population (Angaji et al., 2010). Using GWAS approach, 20 significant genes have been identified contributing to anaerobic germination in rice (Rohilla et al., 2020). Of them, two most relevant genes are OsXDH1, involved in purine catabolism pathway and acting as a scavenger of ROS in plants, and SSXT, which is a GRF1-interacting factor 3. Rapid elongation of the coleoptile is a perfect response to flooding during germination in rice. Nearly 20 SNP markers associated with the length of flooded coleoptiles have been detected using a pool of 432 indica varieties (Zhang et al., 2017). A total of 11 significant marker-trait associations for coleoptile length have been discovered by GWAS using 273 japonica accessions under submergence, and some candidate genes, such as auxin transporter AUX1, have been identified (Nghi et al., 2019). A novel QTL qACE3.1 likely affecting the expression of genes involved in fermentative metabolism has been identified for coleoptile elongation under submergence using CSSLs (Nishimura et al., 2020). Recently, a total of 26 loci have been detected for coleoptile length and coleoptile diameter under submergence using 209 natural rice populations by GWAS approach, and four reliable candidate genes have been identified related to anaerobic germination (Su et al., 2021).
Seed vigor has not been selected as an important breeding trait in traditional breeding programs because of its quantitative inherence in rice. As described above, these detected QTLs for seed germination under various conditions in rice, and these candidate genes represent valuable resources for molecular breeding and genetic improvement of seed vigor during germination. However, only few QTLs have been map-based cloned. It is necessary to fine-map these regions and to find functional markers for marker-assisted selection in rice breeding programs for seed vigor. Usually, the indica rice presents stronger seed vigor during the germination stage than japonica rice under normal conditions (Wang et al., 2010). Typically, indica rice is more tolerant to salt stress than the japonica rice at seedling stage (Lee et al., 2003), whereas japonica rice exhibits better cold tolerance than indica rice (Zhang et al., 2005; Andaya and Tai, 2007; Cheng et al., 2007). However, this is not observed in salt and cold tolerance at the germination stage in rice (Wang et al.,2011a,b). It is interesting to reveal the mechanisms of seed vigor differences during seed germination between rice subspecies at various conditions.
Most seeds contain large quantities of nutrient reserves, mainly carbohydrates, proteins, and lipids, biosynthesized and deposited during seed development. Only several genes regulating reserves accumulation have been reported to be involved in seed vigor in rice. For example, the mutation of rice PFP1 (Pyrophosphate-fructose 6-phosphate 1-phosphotransferase) exhibits remarkably low grain weight and starch content but significantly increases protein and lipid content and then causes a significantly low germination rate (Figure 1; Chen et al., 2020). Imprinted polycomb group (PcG) complex binds to and alters chromatin condensation, resulting in the down-regulation of homeotic target gene expression. The PcG gene Fertilization Independent Endosperm (FIE) plays important roles in controlling seed development (Luo et al., 1999). The complex PcG-OsFIE1 of rice has been reported to regulate the development of embryo and endosperm and influences seed storage proteins and amino acids (Ile, Leu, and Val), which will affect the following seed germination (Huang et al., 2016). Downregulated expression of a lipid transporter OsLTPL36 in plants will lead to decreasing seed setting rate and 1,000-grain weight, causing chalky endosperm, reducing fat acid content, and impeding seed germination in rice (Figure 1; Wang X. et al., 2015). These results indicate that the genes regulating nutrient reserves are vital components for seed vigor during seed development in rice. Starch is the most predominant storage substance, accounting for approximately 90% of the dry seed weight, in rice. A large number of regulatory genes involved in starch biosynthesis have been detected in rice. For example, the mRNA cap-binding protein gene DU3 with pre-mRNA splicing, RNA nuclear export, and non-sense-mediated decay functions (Masayuki et al., 2008), rice starch regulator1 RSR1 (Fu and Xue, 2010), basic leucine zipper transcription factor OsZIP58 (Wang et al., 2013), substandard starch grain SSG4 (Matsushima et al., 2014), SSG6 (Matsushima et al., 2016), floury endosperm FLO7 (Zhang et al., 2016), and floury shrunken endosperm1 FSE1 (Long et al., 2018) have been demonstrated to affect starch biosynthesis and endosperm development in rice (Figure 1). However, whether these genes are involved in the regulation of seed vigor is still unclear in rice. Additionally, the relationships between the seed storage proteins, including glutelins, prolamins, and globulin, with seed vigor are understood limitedly in rice, although several genes involved in the accumulation of storage proteins have been reported (Lee et al., 2015).
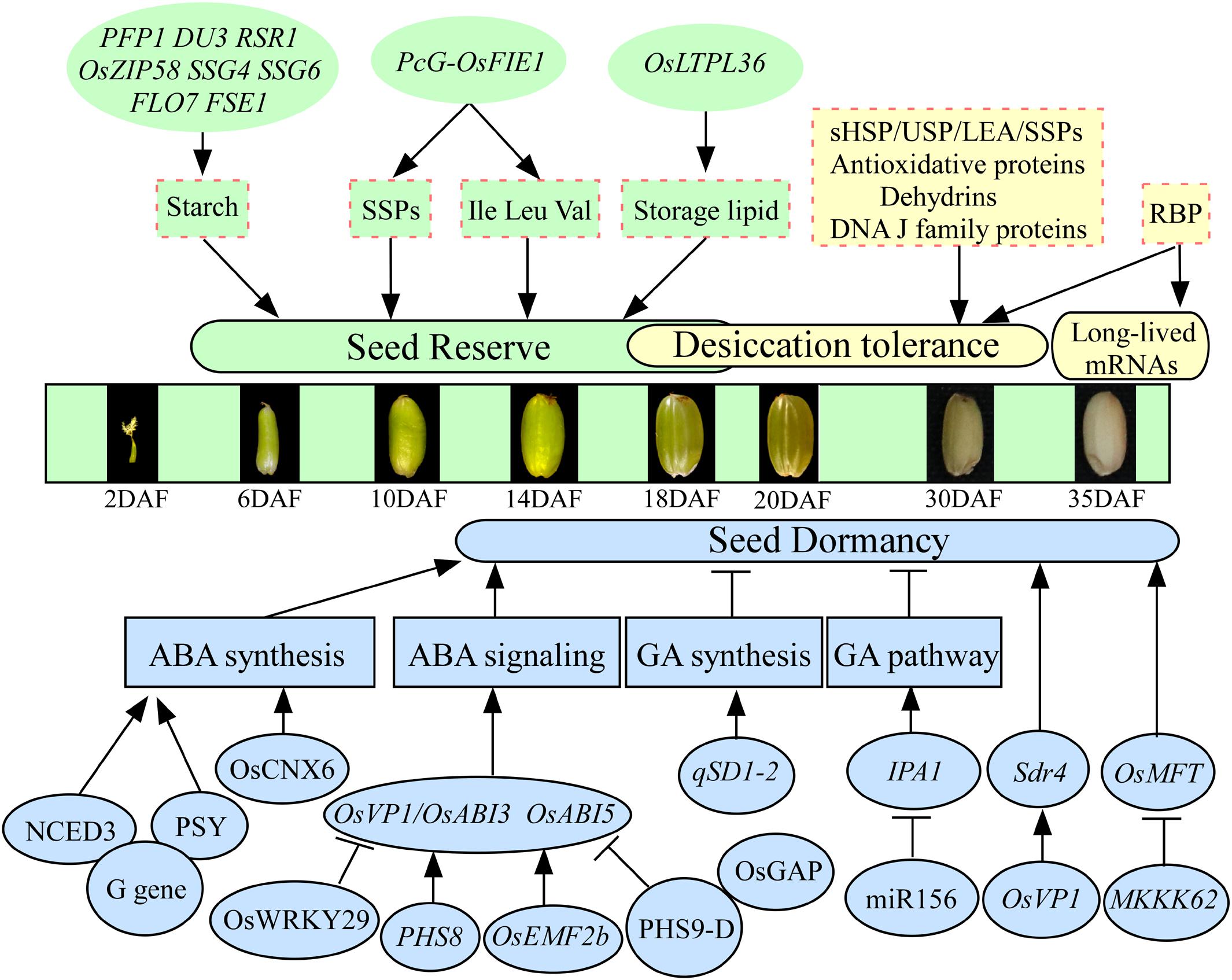
Figure 1. The regulatory genes involved in seed vigor during seed development in rice. The states of seed development according to Xu et al. (2008). Only several genes, such as PFP1, PcG-OsFIE1, and OsLTPL36, regulating reserves accumulation have been reported to be involved in seed vigor. The LEA, sHSP, USP, antioxidative proteins, dehydrins, and DNA J family are involved in the acquisition of desiccation tolerance. The RBP plays important roles in the stability of long-lived mRNAs. The genes regulate seed dormancy mainly through ABA and GA pathways in rice. The G, PHS8, OsEMF2b, PHS9-D, OsCNX6, and OsWRKY29 regulate seed dormancy involved in ABA pathway, and qSD1-2 and MIR156 regulate seed dormancy involved in GA pathway. Arrows and lines with slanted dashes indicate positive and negative effects, respectively. ABA, abscisic acid; GA, gibberellin; SSPs, seed storage proteins; LEA, late embryonic abundant; RBP, RNA-binding protein; PFP1, pyrophosphate-fructose 6-phosphate 1-phosphotransferase; DU3, mRNA cap-binding protein gene; RSR1, rice starch regulator1; OsZIP58, basic leucine zipper transcription factor; SSG4/SSG6, substandard starch grain; FLO7, floury endosperm; FSE1, floury shrunken endosperm 1; PcG, polycomb group; OsFIE1, fertilization-independent endosperm 1; OsLTPL36, lipid transporter; Sdr4, seed dormancy gene; PHS8/9-D, preharvest sprouting; OsABI3/5, ABA-responsive transcription factor; OsVP1, a homolog of the ABA-responsive transcription factor OsABI3; OsGAP, an interaction partner of the ABA receptor OsRCAR1; OsEMF2b, polycomb gene; G gene, stay-green G gene; NCED3, 9-cis-epoxycarotenoid dioxygenase; PSY, phytoene synthase; miR156, microRNA; IPA1, ideal plant architecture 1; MKKK62, protein kinases of MAPK cascade system; OsMFT, a homolog gene controlling wheat seed dormancy.
Desiccation tolerance ensures seed survival through natural maturation or artificial drying, which is important for the establishment of seed vigor. Rice seeds are orthodox seeds, which tolerate water losses of up to 7% of their water content and can be stored at low temperature. In rice, dehydration begins 10–20 days after flowering (DAF), and desiccation phase occurs in the period from 20 to 40 DAF (Figure 1; Sano et al., 2013a). Proteomic analysis has revealed that late embryogenesis abundant (LEA) proteins, small heat shock proteins (sHSPs), universal stress proteins (USPs), antioxidative proteins, dehydrins, and DNA J family accumulate at the beginning of dehydration and remain at a high level in the desiccation phase, suggesting that these proteins are involved in the acquisition of desiccation tolerance in rice (Sano et al., 2013a; He and Yang, 2013; Wang W. Q. et al., 2015). Interestingly, it has been reported that more than 17,000 different mRNA species are stored in mature dry seeds of rice (Howell et al., 2009), which are designated “long-lived mRNAs” because they remain active and translatable even if seeds undergo severe desiccation. Why can these mRNAs live long after seed desiccation or artificially drying? Previous study has shown that the RNA-binding protein (RBP) may have important roles in the stability of long-lived mRNAs in rice (Figure 1; Sano et al., 2013b). After seed imbibition, a population of stored mRNAs is selectively loaded into polysomes, and the mRNAs, involved in processes such as redox, glycolysis, and protein synthesis, are actively translated for germination (Sano et al., 2020). Additionally, the selective oxidation of stored mRNAs has been reported to be involved in dormancy release during after-ripening, and non-selective oxidation and degradation of stored mRNAs have been reported to be involved in seed deterioration during long-term storage (Sano et al., 2012, 2020). However, which specific long-lived mRNAs are sufficient for the seed germination, dormancy, and storability is still limitedly understood.
Seed dormancy has both advantages and disadvantages for seed vigor. In rice, most of the dormancy-related genes are involved in the synthesis or signal transduction of gibberellin (GA) and abscisic acid (ABA; Figure 1). For example, the classical stay-green G gene has conserved functions in controlling seed dormancy in soybean, rice, and Arabidopsis. Stay green G gene affects seed dormancy via the interactions with 9-cis-epoxycarotenoid dioxygenase NCED3 and phytoene synthase PSY and in turn modulates ABA synthesis (Wang M. et al., 2018). The expression of the first rice seed dormancy gene Seed dormancy 4 (Sdr4), a key gene controlling PHS, is controlled by the ABA-responsive transcription factor OsVP1 (Sugimoto et al., 2010). A rice mutant, phs8, encodes a starch debranching enzyme named isoamylase1, and it exhibits PHS phenotype by affecting ABA signaling. Mutation in PHS8 results in the decreased expression of OsABI3 and OsABI5, as well as reduced sensitivity to ABA (Du et al., 2018). Polycomb gene OsEMF2b could directly bind to the promoter of OsVP1 and could affect H3K27me3 enrichments of OsVP1 in seedling, which will influence seed dormancy (Chen et al., 2017). Rice PHS9-D could interact with OsGAP, a partner of ABA receptor OsRCAR1, and PHS9 and OsGAP play important roles in the regulation of rice PHS through the integration of ROS and ABA signaling (Xu et al., 2019). Additionally, mutation of qSD1-2, which controls GA synthesis, leads to decreasing GA levels and enhancing seed dormancy (Ye et al., 2015). The grain yield modulator miR156 regulates seed dormancy involved in GA pathway through de-repression of the miR156 target gene ideal plant architecture 1 (IPA1; Miao et al., 2019). The transcription factor IPA1 reduces unproductive tillers and increases grains per panicle, which results in improved rice yield (Jiao et al., 2010; Miura et al., 2010; Lu et al., 2013). Protein kinase MKKK62 has been reported to negatively control seed dormancy in rice, and it regulates seed dormancy through altering OsMFT transcription (Figure 1; Mao et al., 2019). Molybdenum cofactor (MoCo) is required for ABA biosynthesis in plants. Recently, map-based cloning has shown that OsCNX6 encoding homologs of MoaE participate in MoCo biosynthesis and is essential for rice development, especially for seed dormancy and germination (Liu et al., 2019). A new ABA signaling repressor OsWRKY29 represses seed dormancy by directly down-regulating the expression of OsABF1 and OsVP1 (Zhou et al., 2020). As mentioned previously, the genes regulate seed dormancy mainly through ABA and GA pathways in rice. However, the regulations of other hormones (ethylene, brassinosteroids, salicylic acid, cytokinin, auxin, and jasmonic acid) and radicals (reactive oxygen and nitrogen species), as well as their crosstalk networks, on seed dormancy need to be further investigated. Additionally, proteomic studies showed that dormancy release is involved in metabolism, energy production, protein synthesis and destination, storage protein, cell growth and division, signal transduction, cell defense, and rescue in rice (Xu et al., 2016). These identified genes need to be further confirmed. Because of the selection for the rapid and uniform germination during rice domestication and breeding activities, many modern cultivars have lost part or all of their dormancy at development stage (Gu et al., 2008). Therefore, seeds of rice require a well-balanced level of seed dormancy; data on how to use the above genes to develop the cultivars with deep dormancy during development stage but non-deep dormancy when seed sowing are urgent (Cheng et al., 2014).
Seed deterioration occurs always during seed storage in rice, resulting in the gradually loss of seed vigor. Seed deterioration has shown to be regulated by a number of genetic factors in rice. Of them, lipid peroxidation is a major factor influencing seed vigor (Figure 2). Lipid peroxidation is catalyzed by the enzyme lipoxygenase 3 (LOX3), and the suppression of LOX3 expression increases grain storability in rice (Xu et al., 2015). Similarly, OsLOX2 acts in the opposite directions between seed germination and seed longevity. Appropriate repression of the OsLOX2 gene may delay the aging process during seed storage (Huang et al., 2014). Aldehyde dehydrogenases (ALDHs) catalyze the irreversible oxidation of a wide range of reactive aldehydes to their corresponding carboxylic acids. Rice OsALDH7 plays an important role in maintaining seed viability by detoxifying the aldehydes generated with lipid peroxidation (Shin et al., 2009). Lipid peroxidation mediates the reactive carbonyl compounds (RCCs) and non-enzymatic modifications of proteins through Maillard and Amadori products to reduce seed vigor. Rice aldo-ketoreductase (AKR1) enzyme detoxifies cytotoxic compounds, which results in lower levels of cytotoxic compounds and glycation products, to improve seed vigor (Nisarga et al., 2017). Lipid degradation caused by phospholipase D (PLD) activity is known to be responsible for seed deterioration (Wang S. et al., 2012). The transcript levels of OsPLDα1/α3 will be increased during seed aging in rice, indicating that OsPLD plays an important role in seed deterioration (Figure 2; Nisarga et al., 2017). Overall, the lipid peroxidation causes the loss of membrane integrity, reduced energy metabolism, impairment of RNA and protein synthesis, and formation of malondialdehyde and RCCs, which will lead to seed deterioration during storage in rice (Figure 2; Kapoor et al., 2011).
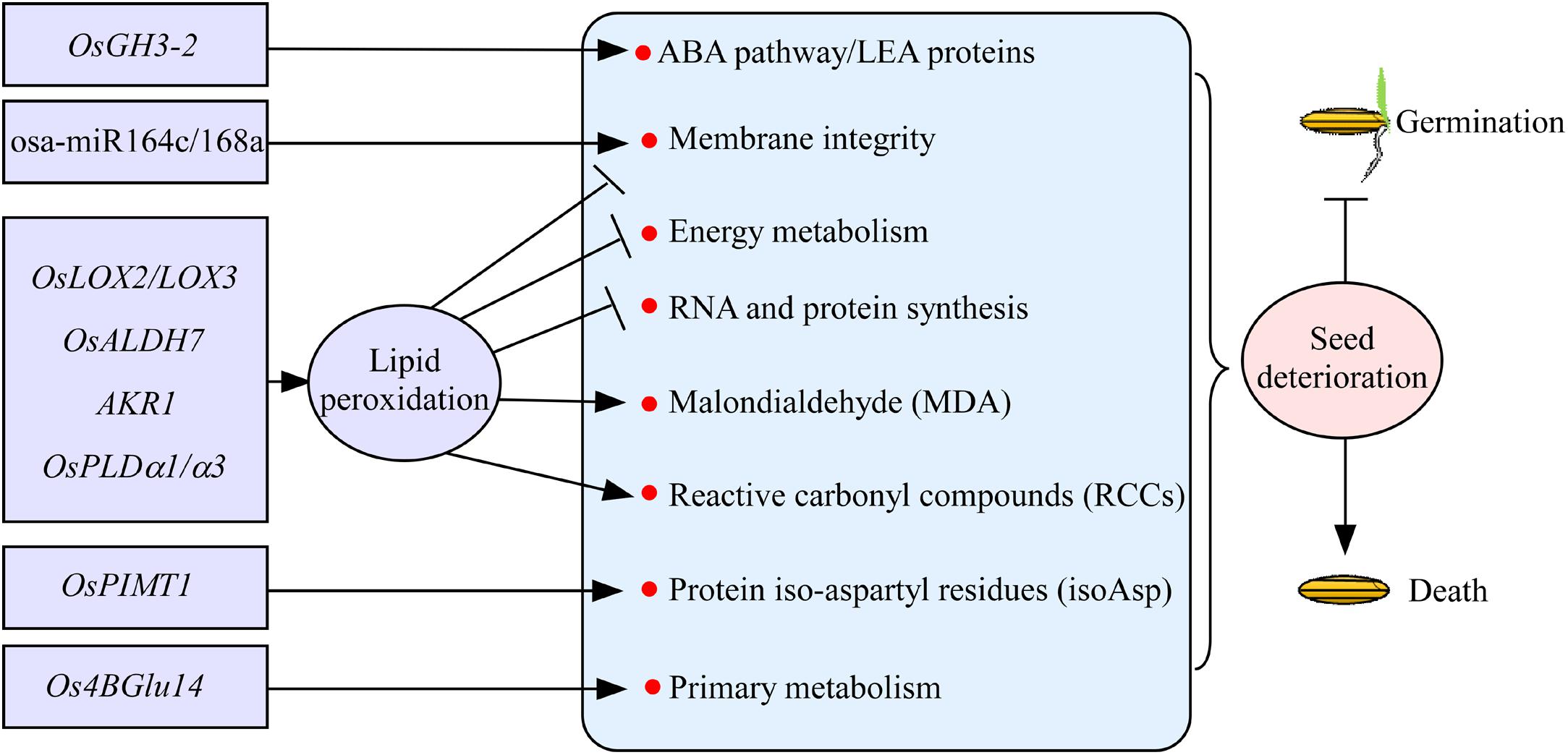
Figure 2. The regulatory genes involved in seed deterioration during seed storage in rice. Most of the genes, such as OsLOX2/LOX3, OsALDH7, AKR1, and OsPLDα1/α3, regulate seed deterioration mainly through lipid peroxidation. Rice OsPIMT1, osa-miR164c/168a, Os4BGlu14, and OsGH3-2 regulate seed deterioration involved in the information of protein iso-aspartyl residues, membrane integrity, primary metabolism, and ABA pathway, respectively. Arrows and lines with slanted dashes indicate positive and negative effects, respectively. osa-miR164c/168a, microRNAs; OsLOX2/LOX3, lipoxygenase 2/3; OsALDH7, aldehyde dehydrogenases; AKR1, aldo-ketoreductase; OsPLDα1/α3, phospholipase D; OsPIMT1, protein–lisoaspartyl methyltransferase; Os4BGlu14, a monolignol β-glucosidase; OsGH3-2, indole-3-acetic acid (IAA)–amido synthetase gene GRETCHEN HAGEN3-2.
Damaged proteins containing abnormal isoaspartyl (isoAsp) accumulate as seeds age, and the abnormality is thought to undermine seed vigor. Protein–lisoaspartyl methyltransferase (PIMT) involved in both seed longevity and germination vigor is first reported in Arabidopsis (Ogé et al., 2008). In rice, the improvement of seed longevity by OsPIMT1 is probably due to the repair of detrimental isoAsp containing proteins (Figure 2; Wei et al., 2015). MicroRNAs (miRNAs) are the key regulators of gene expression in many important biological processes of plants. Recently, miRNAs osa-miR164c and osa-miR168a have been shown to regulate seed vigor under aging condition by influencing the membrane integrity of seeds in rice (Figure 2; Zhou et al., 2020). Rice Os4BGlu14, a monolignol β-glucosidase, plays a negative role in seed longevity by affecting primary metabolism during seed development and aging (Ren et al., 2020). Recently, indole-3-acetic acid (IAA)–amido synthetase gene GRETCHEN HAGEN3-2 (OsGH3-2) has been detected controlling seed storability, and it acts as a negative regulator of seed storability by modulating many genes related to the ABA pathway and probably subsequently LEA proteins at the transcription level (Yuan et al., 2021). Proteomic study has shown that several redox regulation proteins, mainly glutathione-related proteins, and some disease/defense proteins, including DNA-damage-repair/toleration proteins, might be correlated with seed storability (Gao et al., 2016). Metabolomic study has shown that raffinose has potential roles in seed storability (Yan et al., 2018). Altogether, only few factors involved in seed deterioration have been verified in rice (Figure 2). The underlying genetic basis of seed vigor during seed storage needs to be further studied in rice.
Seed germination begins with the imbibition of water and ends with the protrusion of the coleoptile and/or radicle (Bewley, 1997). The mobilization of stored starch upon imbibition is important to supply the energy and nutrients for seed germination and seedling growth. In rice, most α-amylases (RAmy1, RAmy2, and RAmy3) are exclusively expressed in the germinating seeds, which are critical to motivate the stored starch to nourish the developing seedling (Figure 3; Damaris et al., 2019). The expression of αAmy genes is negatively regulated by sugar in embryo during seed germination in rice (Chen et al., 2019). The on/off switch of αAmy expression is regulated by two MYB transcription factors competing for the same promoter element. MYBS1 promotes αAmy expression under sugar starvation, whereas MYBS2 represses it (Chen et al., 2019). Meanwhile, αAmy expression is positively regulated by GA in endosperm (Damaris et al., 2019). However, few studies have reported on the roles of storage lipid mobilization for seed germination in rice. Recently, the lipidome analysis has revealed the distinct patterns of molecular species distribution in individual lipid classes and displayed the metabolic connections between lipid mobilization and rice seedling growth (Dolui and Vijayaraj, 2020). The roles of lipid homeostasis for seed germination are still limitedly understood in rice. Seedling establishment begins with the appearance of the radicle and terminates when the seedling has exhausted the seed’s energy reserves and starts to carry out photosynthesis. The trait of seed reserve utilization, e.g., the conversion efficiency of utilized seed reserve into seedling tissue, plays important roles in rice vigorous seedling growth (Cheng et al., 2013). However, only three αAmy genes (OsAmy3B, OsAmy3C, and OsAmy3E) and sucrose synthase (OsSus2, OsSus3, and OsSus4) have been reported to be involved in seed reserve utilization in rice (Cheng et al., 2015). The genetic characteristics of seed reserve utilization are still unclear in rice.
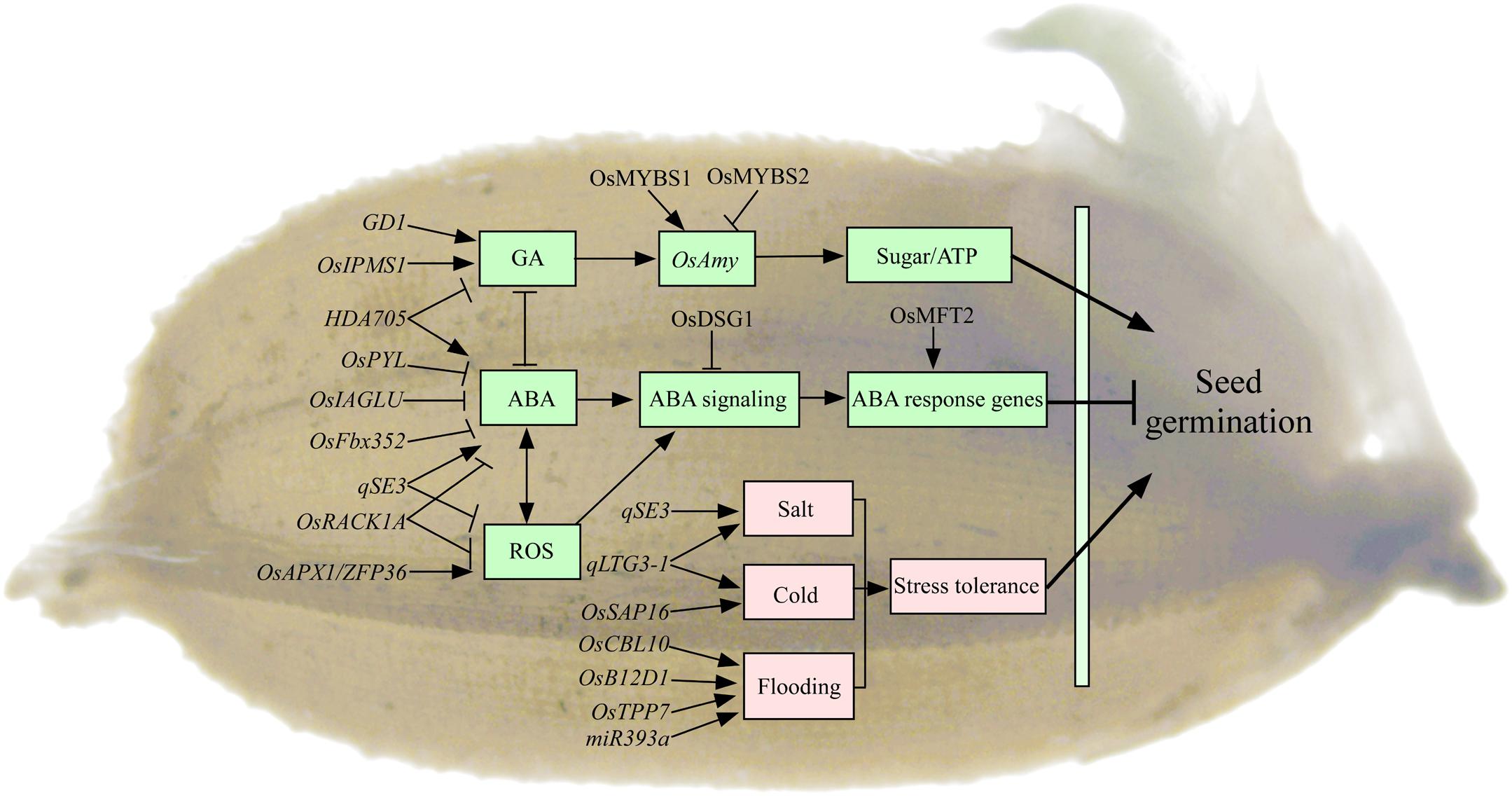
Figure 3. The regulatory genes involved in seed germination in rice. Most of the genes regulate seed germination mainly through GA, ABA, and ROS pathways in rice. Rice GD1, OsIPMS1, and MYBS1/MYBS2 regulate seed germination by influencing αAmy expression through GA pathway. Rice OsPYL, OsDSG1, OsIAGLU, OsFbx352, and OsMFT2 regulate seed germination involved in ABA pathway. Rice HDA705 regulates seed germination through GA and ABA pathways. Rice qSE3, OsAPX1, and OsRACK1A regulate seed germination involved in the crosstalk between ABA and ROS pathways. A number of genes have been reported to be involved in seed germination under stress conditions. Rice qLTG3-1 and OsSAP16 regulate low-temperature germinability, and qSE3 regulates seed germination under salt condition, and OsB12D1, TPP7, miR393a, and OsCBL10 regulate seed germination under flooding condition. Arrows and lines with slanted dashes indicate positive and negative effects, respectively. ABA, abscisic acid; GA, gibberellin; ROS, reactive oxygen species; ATP, adenosine-triphosphate; LTG, low-temperature germinability; GD1, germination-defective 1; OsIPMS1, isopropylmalate synthase; OsAmy, alpha-amylases; HDA705, RPD3/HDA1-type histone deacetylase; OsFbx352, F-box protein; OsPYL, pyrabactin resistance 1 like; OsDSG1, delayed seed germination 1; OsMFT2, mother of FT and TFL 1; OsIAGLU, indole-3-acetate betaglucosyltransferase; qSE3, QTL for seedling establishment under salinity stress; OsRACK1A, rice receptor for activated C kinase 1; OsAPX1, ascorbate peroxidase; ZFP36, zinc finger transcription factor; qLTG3-1, QTL for seed germination under low temperature; OsSAP16, stress-associated protein 16; OsB12D1, an unknown subgroup protein of the barley aleurone and embryo; miR393a, MicroRNAs; TPP7, trehalose 6 phosphate phosphatase 7; OsCBL10, calcineurin B–like protein 10.
It has been reported that several genes regulate seed germination involved in GA pathway in rice (Figure 3). For example, rice GD1 (germination-defective1) regulates seed germination through directly or indirectly regulating GA4 level and carbohydrate homeostasis (Guo et al., 2013). Disruption of isopropylmalate synthase OsIPMS1 results in low seed vigor, which might be tightly associated with the reduction of amino acids and GA biosynthesis, as well as energy level, in germinating seeds (He et al., 2019a). Hormone ABA is an essential repressor of seed germination (Figure 3). Transgenic rice plants expressing OsPYL/RCAR5 (pyrabactin resistance 1 like/regulatory components of ABA receptors) are hypersensitive to ABA during seed germination and early seedling growth (Kim et al., 2012). Rice Delayed Seed Germination 1 (OsDSG1) could bind to OsABI3 that is a major regulator of ABA signaling in germinating seeds (Park et al., 2010). Rice F-box protein OsFbx352 plays a regulatory role in the glucose-induced suppression of seed germination by targeting ABA metabolism, including ABA synthesis (OsNCED2 and OsNCED3) and ABA catabolism (OsABA-ox2 and OsABA-ox3), in the presence of glucose (Song et al., 2012). Rice mother of FT and TFL 1 (OsMFT2) negatively regulates seed germination, and it positively regulates ABA response genes through interacting with three bZIP transcription factors OsbZIP23/66/72 (Song et al., 2020). Several genes regulate seed vigor involved in the crosstalk between hormone and/or ROS pathways in rice (Figure 3). For example, the overexpression of rice HDA705, a RPD3/HDA1-type histone deacetylase (HDAC) gene, in plants delays seed germination because of the down-regulated expression of GA biosynthetic genes and up-regulation of ABA biosynthetic genes (Zhao et al., 2016). Disruption of indole-3-acetate betaglucosyltransferase OsIAGLU gene results in low seed vigor in rice. The regulation of seed vigor by OsIAGLU is through modulating auxin (IAA) and ABA levels to alert OsABIs expression in germinating seeds of rice (He et al., 2020b). Rice OsAPX1 coding an ascorbate peroxidase (APX) has the most affinity for H2O2 (substrate; a type of ROS) that is a direct target of zinc finger transcription factor ZFP36. The LEA protein OsLEA5 interacts with ZFP36 to coregulate OsAPX1 in seed germination in rice (Huang et al., 2018). Rice receptor for activated C kinase 1 (OsRACK1A), one member of the most important WD repeat–containing family of proteins, positively regulates seed germination by controlling endogenous levels of ABA and H2O2 and their interactions (Zhang et al., 2014). RNA sequencing approach has been used to understand the signaling responses in the initial imbibition (8 h) of rice seed germination. The majority of signaling response-related genes are hormone- and transcription factor–related genes, and the largest number of genes belongs to the AP2-domain–containing regulators (He et al., 2020a). Similar with above seed dormancy, the genes regulate seed germination mainly through ABA and GA pathways in rice. However, the regulations of other hormones, radicals, and transcription factors, as well as their crosstalk networks, on seed germination also need to be further investigated.
Seed germination of rice is also influenced by a series of abiotic factors, such as low temperature, salt, and flooding stress. A number of genes have been reported to be involved in rice seed germination under stress conditions (Figure 3). For example, the low-temperature germinability qLTG3-1 is preferentially expressed in the coleorhiza and epiblast of rice, and it increases seed germination through the cell vacuolation and tissue relaxation (Fujino et al., 2004, 2008). Stress-Associated Protein 16 (OsSAP16), coding for a zinc-finger domain protein, is a causal gene for the major low-temperature germinability QTL in rice. The variation in expression of the OsSAP16 gene contributes to low-temperature germinability variation in rice (Wang X. et al., 2018). One major QTL qSE3, which encodes a K+ transporter gene OsHAK21, positively regulates seed germination and seedling establishment by increasing ABA biosynthesis, activating ABA signaling responses, and decreasing H2O2 level in germinating seeds under salinity stress in rice (He et al., 2019b). Rice OsB12D1 belongs to a function unknown subgroup of the Balem (Barley aleurone and embryo) protein that positively regulates flooding tolerance during seed germination and early seedling growth. This tolerance regulated by OsB12D1 is possibly through enhancing electron transport to mediate Fe and oxygen availability under flooded conditions (He et al., 2014). The trehalose 6 phosphate phosphatase 7 (TPP7) gene plays a key role in the mobilization of starch under water in terms of coleoptile elongation and embryo development (Kretzschmar et al., 2015). The miR393a/target plays a role in coleoptile elongation and stomata development via the modulation of auxin signaling during seed germination and seedling establishment under submergence (Guo et al., 2016). Natural variation in the promoter of rice Calcineurin B–like Protein10 (OsCBL10) has been identified affecting flooding tolerance during seed germination among subspecies (Ye et al., 2018). The adaptation to flooding stress during rice germination is associated with two different OsCBL10 promoters, which in turn affects OsCBL10 expression in different cultivars and negatively affects calcineurin B–like protein-interacting protein kinase (OsCIPK15) protein accumulation and its downstream cascade (Ye et al., 2018). Altogether, these above identified genes might be promising genes to enhance seedling establishment for direct seeding under various field conditions in rice. Seed germination is a complex trait that is influenced by many genetic and environmental factors, but the key event(s) associated with seed germination are still poorly understood. It is necessary to integrate the available genomic, transcriptomic, and metabolomic data in order to comprehensively understand the process of germination in rice (He and Yang, 2013; Yang et al., 2019).
Seed priming techniques limit the availability of water to the seed, in which there is sufficient water to initiate germination processes while preventing radicle emergence (Heydecker et al., 1973; Bewley et al., 2013; Cheng et al., 2017). Priming is widely used to enhance seed vigor in terms of the traits of germination potential and stress tolerance (Sheteiwy et al., 2016; Singh et al., 2020). On-farm seed priming with water in maize, rice, and chickpea in India has been chosen as a low-cost, low-risk intervention appropriate to the farmers’ needs (Harris et al., 1999). The activation of DNA repair and antioxidant mechanisms, the de novo synthesis of nucleic acids and proteins, and the production of ATP when seed rehydration are all generally agreed benefits of seed priming for improving seed vigor (Balestrazzi et al., 2010; Paparella et al., 2015). However, few genes have been reported to be involved in seed priming to date in rice. Proteomics have identified that a series of proteins, such as those related to metabolism, storage, protein synthesis, ROS scavenging, and signaling, as well as stress response, might be involved in seed priming in rice (He and Yang, 2013). Recently, our data have demonstrated that the expression of OsIPMS1 contributes to seed priming due to ATP production (He et al., 2019a). Seed priming involves a dehydration step after the controlled imbibition back to the original moisture content of the treated seeds. Thus, the choice of correct stop time-point for seed priming is a critical step, but it is still difficult to monitor in rice because of the variation of priming effects among varieties and seed batches. The phenotypic, physiological, and biochemical markers are difficultly used in the control of priming process because these traits are affected by environmental factors (Cheng et al., 2017). To unravel molecular markers of seed priming in rice, it is possible to characterize the genes as ideal candidate biomarkers for rice seed priming (Cheng et al., 2017; He et al., 2019a). Seed priming with various priming agents has been reported to improve seed vigor under different stress conditions such as salinity, cold, and drought in rice (Salah et al., 2015; Hussain et al., 2016; Samota et al., 2017; Du et al., 2019; Subramanyam et al., 2019). However, the mechanisms of seed priming contributing to seed vigor under stress conditions are limitedly understood in rice. The molecular markers to determine the best stop time-point of priming also need to be investigated further in the future.
In this review, we have summarized findings about the QTLs and genes associated with seed vigor in rice. Seed vigor is a complex trait that is determined by a lot of factors during seed development, storage, and germination stages in rice. Further developing a systems approach to accurately evaluate seed vigor will greatly help in unraveling its molecular mechanisms. Nowadays, the studies of seed vigor are mainly focused on seed dormancy and germination, whereas there are few studies on seed longevity and seed priming associated with seed vigor in rice. Seed vigor is regulated by a very complicated network of signaling and gene expression in rice, and its molecular mechanisms are still largely unknown. Postgenomics studies at the three levels of transcripts, proteins, and metabolites have provided important new information about the mechanisms underlying seed vigor in rice. However, only limited factors have been verified. Characterizing the more seed vigor-related genes will provide us a better understanding of the regulatory mechanisms of seed vigor in rice, which may allow us to enhance seed vigor by breeding and seed treatment approaches.
ZW designed the manuscript. ZW, YH, JZ, and SH wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31971995, 31901601, and 31771889), the Key-Area Research and Development Program of Guangdong Province (Grant Nos. 2018B020202012 and 2020B020219004), the Natural Science Foundation of Guangdong Province (Grant No. 2020A1515011053), the Postdoctoral Science Foundation of China (Grant No. 2020M672654), the Science Foundation of Huzhou (2018GZ07), and the startup funding from South China Agricultural University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank reviewers for the careful reading of the manuscript and constructive comments.
Andaya, V. C., and Tai, T. H. (2007). Fine mapping of the qCTS4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol. Breeding 20, 349–358. doi: 10.1007/s11032-007-9096-8
Angaji, S. A., Septiningsih, E. M., Mackill, D. J., and Ismail, A. M. (2010). QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172, 159–168. doi: 10.1007/s10681-009-0014-5
Balestrazzi, A., Confalonieri, M., Macovei, A., and Carbonera, D. (2010). Seed imbibition in Medicago truncatula Gaertn.: expression profiles of DNA repair genes in relation to PEG-mediated stress. J. Plant Physiol. 168, 706–713. doi: 10.1016/j.jplph.2010.10.008
Bewley, J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. doi: 10.1105/tpc.9.7.1055
Bewley, J. D., Bradford, K. J., Hilhorst, H. W. M., and Nonogaki, H. (2013). Seeds: Physiology of Development, Germination and Dormancy. New York, NY: Springer.
Bockous, W. W., and Shroyer, J. P. (1996). Effect of seed size on seedling vigor and forage production of winter wheat. Can. J. Plant Sci. 76, 101–105. doi: 10.4141/cjps96-015
Carleton, A. E., and Cooper, C. S. (1972). Seed size effects upon seedling vigor of three forage legumes. Crop Sci. 12, 183–186. doi: 10.2135/cropsci1972.0011183x001200020008x
Chamara, B. S., Marambe, B., Kumar, V., Ismail, A. M., Septiningsih, E. M., and Chauhan, B. S. (2018). Optimizing sowing and flooding depth for anaerobic germination-tolerant genotypes to enhance crop establishment, early growth, and weed management in dry-seeded rice (Oryza sativa L.). Front. Plant Sci. 9:1654. doi: 10.3389/fpls.2018.01654
Chen, C., He, B., Liu, X., Ma, X., Liu, Y., Yao, H. Y., et al. (2020). Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa L.). Plant Biotechnol. J. 18, 83–95. doi: 10.1111/pbi.13173
Chen, M., Xie, S., Ouyang, Y., and Yao, J. (2017). Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1. Plant Sci. 260, 80–89. doi: 10.1016/j.plantsci.2017.04.005
Chen, Y. S., Ho, T. D., Liu, L., Lee, D. H., Lee, C. H., Chen, Y. R., et al. (2019). Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth stress tolerance and grain weight in rice. Proc. Natl. Acad. Sci. U.S.A. 116, 21925–21935. doi: 10.1073/pnas.1904818116
Cheng, C., Yun, K. Y., Ressom, H. W., Mohanty, B., Bajic, V. B., Jia, Y., et al. (2007). An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics 8:175. doi: 10.1186/1471-2164-8-175
Cheng, J., Cheng, X., Wang, L., He, Y., An, C., Wang, Z., et al. (2015). Physiological characteristics of seed reserve utilization during the early seedling growth in rice. Braz. J. Bot. 38, 1–9. doi: 10.1007/s40415-015-0190-6
Cheng, J., Wang, L., Du, W., Lai, Y., Huang, X., Wang, Z., et al. (2014). Dynamic quantitative trait locus analysis of seed dormancy at three development stages in rice. Mol. Breeding 34, 501–510. doi: 10.1007/s11032-014-0053-z
Cheng, J., Wang, L., Zeng, P., He, Y., Zhou, R., Zhang, H., et al. (2017). Identification of genes involved in rice seed priming in the early imbibition stage. Plant Biol. (Stuttg) 19, 61–69. doi: 10.1111/plb.12438
Cheng, X., Cheng, J., Huang, X., Lai, Y., Wang, L., Du, W., et al. (2013). Dynamic quantitative trait loci analysis of seed reserve utilization during three germination stages in rice. PLoS One 8:e80002. doi: 10.1371/journal.pone.0080002
Damaris, R. N., Lin, Z., Yang, P., and He, D. (2019). The rice alpha-amylase, conserved regulator of seed maturation and germination. Int. J. Mol. Sci. 20:450. doi: 10.3390/ijms20020450
Dang, X., Thi, T. G., Dong, G., Wang, H., Edzesi, W. M., and Hong, D. (2014). Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 239, 1309–1319. doi: 10.1007/s00425-014-2060-z
Dimaano, N. G. B., Ali, J., Mahender, A., Sta Cruz, P. C., Baltazar, A. M., Diaz, M. G. Q., et al. (2020). Identification of quantitative trait loci governing early germination and seedling vigor traits related to weed competitive ability in rice. Euphytica 216:159. doi: 10.1007/s10681-020-02694-8
Dolui, A. K., and Vijayaraj, P. (2020). Functional omics identifies serine hydrolases that mobilize storage lipids during rice seed germination. Plant Physiol. 184, 693–708. doi: 10.1104/pp.20.00268
Douglas, C. L., Wilkins, D. E., and Churchill, D. B. (1994). Tillage, seed size and seed density effects on performance of soft white winter wheat. Agron. J. 86, 707–711. doi: 10.2134/agronj1994.00021962008600040023x
Du, B., Luo, H., He, L., Zhang, L., Liu, Y., Mo, Z., et al. (2019). Rice seed priming with sodium selenate: effects on germination, seedling growth, and biochemical attributes. Sci. Rep. 9:4311. doi: 10.1038/s41598-019-40849-3
Du, L., Xu, F., Fang, J., Gao, S., Tang, J., Fang, S., et al. (2018). Endosperm sugar accumulation caused by mutation of PHS8/ISA1 leads to pre-harvest sprouting in rice. Plant J. 95, 545–556. doi: 10.1111/tpj.13970
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Farooq, M., Siddique, K. H. M., Rehman, H., Aziz, T., Lee, D. J., and Wahid, A. (2011). Rice direct seeding: experiences, challenges and opportunities. Soil Tillage Res. 111, 87–98. doi: 10.1016/j.still.2010.10.008
Finch-Savage, W. E., and Bassel, G. W. (2016). Seed vigour and crop establishment: extending performance beyond adaptation. J. Exp. Bot. 67, 567–591. doi: 10.1093/jxb/erv490
Fu, F. F., and Xue, H. W. (2010). Coexpression analysis identifies rice starch regulator1, a Rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. doi: 10.1104/pp.110.159517
Fujino, K., Sekiguchi, H., and Matsuda, Y. (2008). Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 12623–12628. doi: 10.1073/pnas.0805303105
Fujino, K., Sekiguchi, H., Sato, T., Kiuchi, H., Nonoue, Y., Takeuchi, Y., et al. (2004). Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 108, 794–799. doi: 10.1007/s00122-003-1509-4
Gao, J., Fu, H., Zhou, X., Chen, Z., Luo, Y., Cui, B., et al. (2016). Comparative proteomic analysis of seed embryo proteins associated with seed storability in rice (Oryza sativa L) during natural aging. Plant Physiol. Biochem. 103, 31–44. doi: 10.1016/j.plaphy.2016.02.026
Gu, X. Y., Turnipseed, B. E., and Foley, M. E. (2008). The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics 179, 2263–2273. doi: 10.1534/genetics.108.092007
Gubler, F., Millar, A. A., and Jacobsen, J. V. (2005). Dormancy release, ABA and preharvest sprouting. Curr. Opin. Plant Biol. 8, 183–187. doi: 10.1016/j.pbi.2005.01.011
Guo, F., Han, N., Xie, Y., Fang, K., Yang, Y., Zhu, M., et al. (2016). The miR393a/target module regulates seed germination and seedling establishment under submergence in rice (Oryza sativa L.). Plant Cell Environ. 39, 2288–2302. doi: 10.1111/pce.12781
Guo, X., Hou, X., Fang, J., Wei, P., Xu, B., Chen, M., et al. (2013). The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 75, 403–416. doi: 10.1111/tpj.12209
Han, L. Z., Zhang, Y. Y., Qiao, Y. L., Cao, G. L., Zhang, S. Y., Kim, J. H., et al. (2006). Genetic and QTL analysis for low-temperature vigor of germination in rice. Acta Genet. Sin. 33, 998–1006. doi: 10.1016/S0379-4172(06)60135-2
Harris, B. D., Joshi, A., Khan, P. A., Gothkar, P., and Sodhi, P. S. (1999). On-farm seed priming in semi-arid agriculture: development and evaluation in maize, rice and chickpea in India using participatory methods. Exp. Agric. 35, 15–29. doi: 10.1017/S0014479799001027
He, D., and Yang, P. (2013). Proteomics of rice seed germination. Front. Plant Sci. 4:246. doi: 10.3389/fpls.2013.00246
He, D., Zhang, H., and Yang, P. (2014). The mitochondrion-located protein OsB12D1 enhances flooding tolerance during seed germination and early seedling growth in rice. Int. J. Mol. Sci. 15, 13461–13481. doi: 10.3390/ijms150813461
He, Y., Cheng, J., He, Y., Yang, B., Cheng, Y., Yang, C., et al. (2019a). Influence of isopropylmalate synthase OsIPMS1 on seed vigor associated with amino acid and energy metabolism in rice. Plant Biotechnol. J. 17, 322–337. doi: 10.1111/pbi.12979
He, Y., Yang, B., He, Y., Zhan, C., Cheng, Y., Zhang, J., et al. (2019b). A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 97, 1089–1104. doi: 10.1111/tpj.14181
He, Y., Zhao, J., Feng, D., Huang, Z., Liang, J., Zheng, Y., et al. (2020a). RNA-seq study reveals AP2-domain-containing signalling regulators involved in the initial imbibition of seed germination in rice. Rice Sci. 27, 302–314. doi: 10.1016/j.rsci.2020.05.006
He, Y., Zhao, J., Yang, B., Sun, S., Peng, L., and Wang, Z. (2020b). Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol. J. 18, 1933–1945. doi: 10.1111/pbi.13353
Heydecker, W., Higgins, J., and Gulliver, R. L. (1973). Accelerated germination by osmotic seed treatment. Nature 246, 42–44. doi: 10.1038/246042a0
Howell, K. A., Narsai, R., Carroll, A., Ivanova, A., Lohse, M., Usadel, B., et al. (2009). Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 149, 961–980. doi: 10.1104/pp.108.129874
Hu, Z., Lu, S., Wang, M., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, J., Cai, M., Long, Q., Liu, L., Lin, Q., Jiang, L., et al. (2014). OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 23, 643–655. doi: 10.1007/s11248-014-9803-2
Huang, L., Jia, J., Zhao, X., Zhang, M. Y., Huang, X., Ji, E., et al. (2018). The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem. Biophys. Res. Commun. 495, 339–345. doi: 10.1016/j.bbrc.2017.10.128
Huang, X., Lu, Z., Wang, X., Ouyang, Y., Chen, W., Xie, K., et al. (2016). Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 87, 305–317. doi: 10.1111/tpj.13202
Hussain, S., Khan, F., Hussain, H. A., and Nie, L. (2016). Physiological and biochemical mechanisms of seed priming-induced chilling tolerance in rice cultivars. Front. Plant Sci. 7:116. doi: 10.3389/fpls.2016.00116
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi: 10.1038/ng.2612
ISTA (2015). International Rules for Seed Testing. Bassersdorf: The International Seed Testing Association.
Jiang, W., Lee, J., Jin, Y. M., Qiao, Y., Piao, R., Jang, S. M., et al. (2011). Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice. Mol. Cells 31, 385–392. doi: 10.1007/s10059-011-0049-z
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Johnson, D. R., and Wax, L. M. (1978). Relationships of soybean germination and vigor tests to field performance. Agron. J. 70, 273–278. doi: 10.2134/agronj1978.00021962007000020014x
Kapoor, N., Arya, A., Siddiqui, M. A., Kumar, H., and Amir, A. (2011). Physiological and biochemical changes during seed deterioration in aged seeds of rice (Oryza sativa L.). Am. J. Plant Physiol. 6, 28–35. doi: 10.3923/ajpp.2011.28.35
Kim, H., Hwang, H., Hong, J. W., Lee, Y. N., Ahn, I. P., Yoon, I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi: 10.1093/jxb/err338
Kretzschmar, T., Pelayo, M. A. F., Trijatmiko, K. R., Gabunada, L. F. M., Alam, R., Jimenez, R., et al. (2015). A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 1:15124. doi: 10.1038/nplants.2015.124
Lee, H. J., Jo, Y. M., Lee, J. Y., Lim, S. H., and Kim, Y. M. (2015). Lack of globulin synthesis during seed development alters accumulation of seed storage proteins in rice. Int. J. Mol. Sci. 16, 14717–14736. doi: 10.3390/ijms160714717
Lee, J. S., Velasco-Punzalan, M., Pacleb, M., Valdez, R., Kretzschmar, T., McNally, K. L., et al. (2019). Variation in seed longevity among diverse Indica rice varieties. Ann. Bot. 124, 447–460. doi: 10.1093/aob/mcz093
Lee, K. S., Choi, W. Y., Ko, J. C., Kim, T. S., and Gregorio, G. B. (2003). Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216, 1043–1046. doi: 10.1007/s00425-002-0958-3
Li, L., Liu, X., Xie, K., Wang, Y., Liu, F., Lin, Q., et al. (2013). qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor. Appl. Genet. 126, 2313–2322. doi: 10.1007/s00122-013-2137-2
Li, N., Xu, R., Duan, P., and Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 237–251. doi: 10.1007/s00497-018-0333-6
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Lin, Q., Wang, W., Ren, Y., Jiang, Y., Sun, A., Qian, Y., et al. (2015). Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22. Breed. Sci. 65, 411–419. doi: 10.1270/jsbbs.65.411
Liu, L., Lai, Y., Cheng, J., Wang, L., Du, W., Wang, Z., et al. (2014). Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS One 9:e115732. doi: 10.1371/journal.pone.0115732
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-protein βγ subunits determine grain size through interaction with 85 MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Liu, X., Wang, J., Yu, Y., Kong, L., Liu, Y., Liu, Z., et al. (2019). Identification and characterization of the rice pre-harvest sprouting mutants involved in molybdenum cofactor biosynthesis. New Phytol. 222, 275–285. doi: 10.1111/nph.15607
Long, W., Wang, Y., Zhu, S., Jing, W., Wang, Y., Ren, Y., et al. (2018). FLOURY SHRUNKEN ENDOSPERM1 connects phospholipid metabolism and amyloplast development in rice. Plant Physiol. 177, 698–712. doi: 10.1104/pp.17.01826
Lu, B., Xie, K., Yang, C., Wang, S., Liu, X., Zhang, L., et al. (2011). Mapping two major effect grain dormancy QTL in rice. Mol. Breeding 28, 453–462. doi: 10.1007/s11032-010-9495-0
Lu, Q., Niu, X., Zhang, M., Wang, C., Xu, Q., Feng, Y., et al. (2018). Genome-wide association study of seed dormancy and the genomic consequences of improvement footprints in rice (Oryza sativa L.). Front. Plant Sci. 8:2213. doi: 10.3389/fpls.2017.02213
Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759. doi: 10.1105/tpc.113.113639
Luo, M., Bilodeau, P., Koltunow, A., Dennis, E. S., Peacock, W. J., and Chaudhury, A. M. (1999). Genes controlling fertilization−independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 296–301. doi: 10.1073/pnas.96.1.296
Magwa, R. A., Zhao, H., and Xing, Y. (2016). Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet. 17:28. doi: 10.1186/s12863-016-0340-2
Mahajan, S., Mittal, S. K., and Das, A. J. (2018). Machine vision based alternative testing approach for physical purity, viability and vigour testing of soybean seeds (Glycine max). Food Sci. Technol. 55, 3949–3959. doi: 10.1007/s13197-018-3320-x
Mahender, A., Anandan, A., and Pradhan, S. K. (2015). Early seedling vigour, an imperative trait for direct-seeded rice: an overview on physio-morphological parameters and molecular markers. Planta 241, 1027–1050. doi: 10.1007/s00425-015-2273-9
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. doi: 10.1073/pnas.1014419107
Mao, X., Zhang, J., Liu, W., Yan, S., Liu, Q., Fu, H., et al. (2019). The MKKK62-MKK3-MAPK7/14 module negatively regulates seed dormancy in rice. Rice 12:2. doi: 10.1186/s12284-018-0260-z
Marzougui, S., Sugimoto, K., Yamanouchi, U., Shimono, M., Hoshino, T., Hori, K., et al. (2012). Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice. Theor. Appl. Genet. 124, 893–902. doi: 10.1007/s00122-011-1753-y
Masayuki, I., Yasuyuki, M., Aya, T., Ling, W. H., Hikaru, S., and Ko, S. (2008). Du3, a mRNA cap-binding protein gene, regulates amylose content in japonica rice seeds. Plant Tissue Cult. Lett. 25, 483–487. doi: 10.1016/j.fm.2004.01.015
Matsushima, R., Maekawa, M., Kusano, M., Kondo, H., Fujita, N., Kawagoe, Y., et al. (2014). Amyloplast-localized substandard starch GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 164, 623–636. doi: 10.1104/pp.113.229591
Matsushima, R., Maekawa, M., Kusano, M., Tomita, K., Kondo, H., Nishimura, H., et al. (2016). Amyloplast membrane protein substandard starch GRAIN6 controls starch grain size in rice endosperm. Plant Physiol. 170, 1445–1459. doi: 10.1104/pp.15.01811
Miao, C., Wang, Z., Zhang, L., Yao, J., Hua, K., Liu, X., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10:3822. doi: 10.1038/s41467-019-11830-5
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Miura, K., Lin, Y., Yano, M., and Nagamine, T. (2002). Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 104, 981–986. doi: 10.1007/s00122-002-0872-x
Najeeb, S., Ali, J., Mahender, A., Pang, Y. L., Zilhas, J., Murugaiyan, V., et al. (2020). Identification of main-effect quantitative trait loci (QTLs) for low-temperature stress tolerance germination- and early seedling vigor-related traits in rice (Oryza sativa L.). Mol. Breeding 40:10. doi: 10.1007/s11032-019-1090-4
Naveed, S. A., Zhang, F., Zhang, J., Zheng, T. Q., Meng, L. J., Pang, Y. L., et al. (2018). Identification of QTN and candidate genes for salinity tolerance at the germination and seedling stages in rice by genome-wide association analyses. Sci. Rep. 8:6505. doi: 10.1038/s41598-018-24946-3
Nghi, K. N., Tondelli, A., Valè, G., Tagliani, A., Marè, C., Perata, P., et al. (2019). Dissection of coleoptile elongation in japonica rice under submergence through integrated genome-wide association mapping and transcriptional analyses. Plant Cell Environ. 42, 1832–1846. doi: 10.1111/pce.13540
Nisarga, K. N., Vemanna, R. S., Chandrashekar, B. K., Rao, H., Vennapusa, A. R., Narasimaha, A., et al. (2017). Aldo-ketoreductase 1 (AKR1) improves seed longevity in tobacco and rice by detoxifying reactive cytotoxic compounds generated during ageing. Rice 10:11. doi: 10.1186/s12284-017-0148-3
Nishimura, T., Sasaki, K., Yamaguchi, T., Takahashi, H., Junko Yamagishi, J., and Kato, Y. (2020) Detection and characterization of quantitative trait loci for coleoptile elongation under anaerobic conditions in rice. Plant Prod. Sci. 23, 374–383. doi: 10.1080/1343943X.2020.1740600
Ogé, L., Bourdais, G., Bove, J., Collet, B., Godin, B., Granier, F., et al. (2008). Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20, 3022–3037. doi: 10.1105/tpc.108.058479
Pan, Y., Zhang, H., Zhang, D., Li, J., Xiong, H., Yu, J., et al. (2015). Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS One 10:e0120590. doi: 10.1371/journal.pone.0120590
Paparella, S., Araujo, S. S., Rossi, G., Wijayasinghe, M., Carbonera, D., and Balestrazzi, A. (2015). Seed priming: state of the art and new perspectives. Plant Cell Rep. 34, 1281–1293. doi: 10.1007/s00299-015-1784-y
Park, G. G., Park, J. J., Yoon, J., Yu, S. N., and An, G. (2010). A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 74, 467–478. doi: 10.1007/s11103-010-9687-3
Pellizzaro, A., Neveu, M., Lalanne, D., Ly Vu, B., Kanno, Y., Seo, M., et al. (2020). A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 225, 284–296. doi: 10.1111/nph.16150
Qi, P., Lin, Y. S., Song, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi: 10.1146/annurev-arplant-042811-105550
Ren, R. J., Wang, P., Wang, L. N., Su, J. P., Sun, L. J., Sun, Y., et al. (2020). Os4BGlu14, a monolignol beta-Glucosidase, negatively affects seed longevity by influencing primary metabolism in rice. Plant Mol. Biol. 104, 513–527. doi: 10.1007/s11103-020-01056-1
Rohilla, M., Singh, N., Mazumder, A., Sen, P., Roy, P., Chowdhury, D., et al. (2020). Genome-wide association studies using 50 K rice genic SNP chip unveil genetic architecture for anaerobic germination of deep-water rice population of Assam. India Mol. Genet. Genomics 295, 1211–1226. doi: 10.1007/s00438-020-01690-w
Ruan, B., Shang, L., Zhang, B., Hu, J., Wang, Y., Lin, H., et al. (2020). Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 227, 629–640. doi: 10.1111/nph.16540
Salah, S. M., Yajing, G., Dongdong, C., Jie, L., Aamir, N., Qijuan, H., et al. (2015). Seed priming with polyethylene glycol regulating the physiological and molecular mechanism in rice (Oryza sativa L.) under nano-ZnO stress. Sci. Rep. 5:14278. doi: 10.1038/srep14278
Samota, M. K., Sasi, M., Awana, M., Yadav, O. P., Amitha Mithra, S. V., Tyagi, A., et al. (2017). Elicitor-induced biochemical and molecular manifestations to improve drought tolerance in rice (Oryza sativa L.) through seed-priming. Front. Plant Sci. 8:934. doi: 10.3389/fpls.2017.00934
Sano, N., Masaki, S., Tanabata, T., Yamada, T., Hirasawa, T., and Kanekatsu, M. (2013a). Proteomic analysis of stress-related proteins in rice seeds during the desiccation phase of grain filling. Plant Biotechnol. 30, 147–156. doi: 10.5511/plantbiotechnology.13.0207a
Sano, N., Masaki, S., Tanabata, T., Yamada, T., Hirasawa, T., Kashiwagi, M., et al. (2013b). RNA-binding proteins associated with desiccation during seed development in rice. Biotechnol. Lett. 35, 1945–1952. doi: 10.1007/s10529-013-1283-6
Sano, N., Permana, H., Kumada, R., Shinozaki, Y., Tanabata, T., Yamada, T., et al. (2012). Proteomic analysis of embryonic proteins synthesized from long-lived mRNAs during germination of rice seeds. Plant Cell Physiol. 53, 687–698. doi: 10.1093/pcp/pcs024
Sano, N., Rajjou, L., and North, H. M. (2020). Lost in translation: physiological roles of stored mRNAs in seed germination. Plants (Basel) 9:347. doi: 10.3390/plants9030347
Sasaki, K., Takeuchi, Y., Miura, K., Yamaguchi, T., Ando, T., Ebitani, T., et al. (2015). Fine mapping of a major quantitative trait locus, qLG-9, that controls seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 769–778. doi: 10.1007/s00122-015-2471-7
Sheteiwy, M. S., Fu, Y., Hu, Q., Nawaz, A., Guan, Y., Li, Z., et al. (2016). Seed priming with polyethylene glycol induces antioxidative defense and metabolic regulation of rice under nano-ZnO stress. Environ. Sci. Pollut. Res. Int. 23, 19989–20002. doi: 10.1007/s11356-016-7170-7
Shi, Y., Gao, L., Wu, Z., Zhang, X., Wang, M., Zhang, C., et al. (2017). Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 7:92. doi: 10.1186/s12870-017-1044-0
Shin, J. H., Kim, S. R., and An, G. (2009). Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol. 149, 905–915. doi: 10.1104/pp.108.130716
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Singh, A., Banerjee, A., and Roychoudhury, A. (2020). Seed priming with calcium compounds abrogate fluoride-induced oxidative stress by upregulating defence pathways in an indica rice variety. Protoplasma 257, 767–782. doi: 10.1007/s00709-019-01460-5
Song, S., Dai, X., and Zhang, W. H. (2012). A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice. J. Exp. Bot. 63, 5559–5568. doi: 10.1093/jxb/ers206
Song, S., Wang, G., Wu, H., Fan, X., Liang, L., Zhao, H., et al. (2020). OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 103, 532–546. doi: 10.1111/tpj.14748
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Song, X. J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 76–81. doi: 10.1073/pnas.1421127112
Su, L., Yang, J., Li, D., Peng, Z., Xia, A., Yang, M., et al. (2021). Dynamic genome-wide association analysis and identification of candidate genes involved in anaerobic germination tolerance in rice. Rice 14:1. doi: 10.1186/s12284-020-00444-x
Subramanyam, K., Du Laing, G., and Van Damme, E. J. M. (2019). Sodium selenate treatment using a combination of seed priming and foliar spray alleviates salinity stress in rice. Front. Plant Sci. 10:116. doi: 10.3389/fpls.2019.00116
Sugimoto, K., Takeuchi, Y., Ebana, K., Miyao, A., Hirochika, H., Hara, N., et al. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. U.S.A. 107, 5792–5797. doi: 10.1073/pnas.0911965107
Sun, Q., Wang, J. H., and Sun, B. Q. (2007). Advances on seed vigor physiological and genetic mechanisms. Agric. Sci. China 6, 1060–1066. doi: 10.1016/S1671-2927(07)60147-3
Thapa, R., Tabien, R. E., Thomson, M. J., and Septiningsih, E. M. (2020). Genome-wide association mapping to identify genetic loci for cold tolerance and cold recovery during germination in rice. Front. Genet. 11:22. doi: 10.3389/fgene.2020.00022
Wang, A., Hou, Q., Si, L., Huang, X., Luo, J., Lu, D., et al. (2019). The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 180, 2077–2090. doi: 10.1104/pp.18.01574
Wang, F., Wang, R., Jing, W., and Zhang, W. (2012). Quantitative dissection of lipid degradation in rice seeds during accelerated aging. Plant Growth Regul. 66, 49–58. doi: 10.1007/s10725-011-9628-4
Wang, J. C., Xu, H., Zhu, Y., Liu, Q. Q., and Cai, X. L. (2013). OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466. doi: 10.1093/jxb/ert187
Wang, L., Cheng, J., Lai, Y., Du, W., Huang, X., Wang, Z., et al. (2014). Identification of QTLs with additive, epistatic and QTL development interaction effects for seed dormancy in rice. Planta 239, 411–420. doi: 10.1007/s00425-013-1991-0
Wang, M., Li, W., Fang, C., Xu, F., Liu, Y., Wang, Z., et al. (2018). Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat. Genet. 50, 1435–1441. doi: 10.1038/s41588-018-0229-2
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, W. Q., Liu, S. J., Song, S. Q., and Møller, I. M. (2015). Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 86, 1–15. doi: 10.1016/j.plaphy.2014.11.003
Wang, X., Zhou, W., Lu, Z., Ouyang, Y., O, C. S., and Yao, J. (2015). A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 239, 200–208. doi: 10.1016/j.plantsci.2015.07.016
Wang, X., Zou, B., Shao, Q., Cui, Y., Lu, S., Zhang, Y., et al. (2018). Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 69, 413–421. doi: 10.1093/jxb/erx413
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wang, Z. F., Wang, F. H., Zhou, R., Wang, J. F., and Zhang, H. S. (2011a). Identification of quantitative trait loci for cold tolerance during the germination and seedling stages in rice (Oryza sativa L.). Euphytica 181, 405–413. doi: 10.1007/s10681-011-0469-z
Wang, Z. F., Wang, J. F., Bao, Y. M., Wang, F. H., and Zhang, H. S. (2010). Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B 11, 958–964. doi: 10.1631/jzus.B1000238
Wang, Z. F., Wang, J. F., Bao, Y. M., Wu, Y., and Zhang, H. S. (2011b). Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 178, 297–307. doi: 10.1007/s10681-010-0287-8
Wei, Y., Xu, H., Diao, L., Zhu, Y., Xie, H., Cai, Q., et al. (2015). Protein repair L-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol. Biol. 89, 475–492. doi: 10.1007/s11103-015-0383-1
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Wu, W., Liu, X., Wang, M., Meyer, R. S., Luo, X., Ndjiondjop, M. N., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3:17064. doi: 10.1038/nplants.2017.64
Xia, D., Zhou, H., Liu, R., Dan, W., Li, P., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to form extra-long grains in rice. Mol. Plant 11, 754–756. doi: 10.1016/j.molp.2018.03.006
Xie, L., Tan, Z., Zhou, Y., Xu, R., Feng, L., Xing, Y., et al. (2014). Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J. Integr. Plant Biol. 56, 749–759. doi: 10.1111/jipb.12190
Xu, F., Tang, J., Gao, S., Cheng, X., Du, L., and Chu, C. (2019). Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 100, 1036–1051. doi: 10.1111/tpj.14501
Xu, H. H., Liu, S. J., Song, S. H., Wang, R. X., Wang, W. Q., and Song, S. Q. (2016). Proteomics analysis reveals distinct involvement of embryo and endosperm proteins during seed germination in dormant and non-dormant rice seeds. Plant Physiol. Biochem. 103, 219–242. doi: 10.1016/j.plaphy.2016.03.00
Xu, H., Wei, Y., Zhu, Y., Lian, L., Xie, H., Cai, Q., et al. (2015). Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotechnol J. 13, 526–539. doi: 10.1111/pbi.12277
Xu, S. B., Li, T., Deng, Z. Y., Chong, K., Xue, Y., and Wang, T. (2008). Dynamic proteomic analysis reveals a switch between central carbon metabolism and alcoholic fermentation in rice filling grains. Plant Physiol. 148, 908–925. doi: 10.1104/pp.108.125633
Yan, S., Huang, W., Gao, J., Fu, H., and Liu, J. (2018). Comparative metabolomic analysis of seed metabolites associated with seed storability in rice (Oryza sativa L.) during natural aging. Plant Physiol. Biochem. 127, 590–598. doi: 10.1016/j.plaphy.2018.04.020
Yang, J., Li, D., Liu, H., Liu, Y., Huang, M., Wang, H., et al. (2020). Identification of QTLs involved in cold tolerance during the germination and bud stages of rice (Oryza sativa L.) via a high-density genetic map. Breed. Sci. 70, 292–302. doi: 10.1270/jsbbs.19127
Yang, M., Yang, J., Su, L., Sun, K., Li, D., Liu, Y., et al. (2019). Metabolic profile analysis and identification of key metabolites during rice seed germination under low-temperature stress. Plant Sci. 289:110282. doi: 10.1016/j.plantsci.2019.110282
Ye, H., Feng, J., Zhang, L., Zhang, J., Mispan, M. S., Cao, Z., et al. (2015). Map-based cloning of Seed Dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169, 2152–2165. doi: 10.1104/pp.15.01202
Ye, N. H., Wang, F. Z., Shi, L., Chen, M. X., Cao, Y. Y., Zhu, F. Y., et al. (2018). Natural variation in the promoter of rice calcineurin B-like protein10 (OsCBL10) affects flooding tolerance during seed germination among rice subspecies. Plant J. 94, 612–625. doi: 10.1111/tpj.13881
Yu, J., Miao, J., Zhang, Z., Xiong, H., Zhu, X., Sun, X., et al. (2018a). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. doi: 10.1111/pbi.12903
Yu, J., Xiong, H., Zhu, X., Zhang, H., Li, H., Miao, J., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 15:28. doi: 10.1186/s12915-017-0365-7
Yu, J., Zhao, W., Tong, W., He, Q., Yoon, M. Y., Li, F. P., et al. (2018b). A genome-wide association study reveals candidate genes related to salt tolerance in rice (Oryza sativa) at the germination stage. Int. J. Mol. Sci. 19:3145. doi: 10.3390/ijms19103145
Yuan, Z., Fan, K., Wang, Y., Tian, L., Zhang, C., Sun, W., et al. (2021). OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances. Plant Physiol. kiab059. doi: 10.1093/plphys/kiab059 [Epub ahead of print].
Yuan, Z., Fan, K., Xia, L., Ding, X., Tian, L., Sun, W., et al. (2019). Genetic dissection of seed storability and validation of candidate gene associated with antioxidant capability in rice (Oryza sativa L.). Int. J. Mol. Sci. 20:4442. doi: 10.3390/ijms20184442
Zeng, P., Zhu, P., Qian, L., Qian, X., Mi, Y., Lin, Z., et al. (2021). Identification and fine mapping of qGR6.2, a novel locus controlling rice seed germination under salt stress. BMC Plant Biol. 21:36. doi: 10.1186/s12870-020-02820-7
Zhang, C., Yuan, Z., Wang, Y., Sun, W., Tang, X., Sun, Y., et al. (2020). Genetic dissection of seed dormancy in rice (Oryza sativa L.) by using two mapping populations derived from common parents. Rice 13:52. doi: 10.1186/s12284-020-00413-4
Zhang, D., Chen, L., Li, D., Lv, B., Chen, Y., Chen, J., et al. (2014). OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa, L.). PLoS One 9:e97120. doi: 10.1371/journal.pone.0097120
Zhang, L., Ren, Y., Lu, B., Yang, C., Feng, Z., Liu, Z., et al. (2016). FLOURY ENDOSPERM 7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J. Exp. Bot. 67, 633–647. doi: 10.1093/jxb/erv469
Zhang, M., Lu, Q., Wu, W., Niu, X., Wang, C., Feng, Y., et al. (2017). Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in Indica rice. Front. Plant Sci. 8:678. doi: 10.3389/fpls.2017.00678
Zhang, T., Fan, S., Xiang, Y., Zhang, S., Wang, J., and Sun, Q. (2020). Non-destructive analysis of germination percentage, germination energy and simple vigour index on wheat seeds during storage by Vis/NIR and SWIR hyperspectral imaging. Spectrochim. Acta A Mol. Biomol. Spectrosc. 239:118488. doi: 10.1016/j.saa.2020.118488
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, Z. H., Li, S., Wei, L., Wei, C., and Zhu, Y. G. (2005). A major QTL conferring cold tolerance at the early seedling stage using recombinant inbred lines of rice (Oryza sativa L.). Plant Sci. 168, 527–534. doi: 10.1016/j.plantsci.2004.09.021
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhao, J., Li, M., Gu, D., Liu, X., Zhang, J., Wu, K., et al. (2016). Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 470, 439–444. doi: 10.1016/j.bbrc.2016.01.016
Zhou, Y., Miao, J., Gu, H., Peng, X., Leburu, M., Yuan, F., et al. (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201, 1591–1599. doi: 10.1534/genetics.115.181115
Keywords: rice, seed development, seed dormancy, seed deterioration, seed germination
Citation: Zhao J, He Y, Huang S and Wang Z (2021) Advances in the Identification of Quantitative Trait Loci and Genes Involved in Seed Vigor in Rice. Front. Plant Sci. 12:659307. doi: 10.3389/fpls.2021.659307
Received: 27 January 2021; Accepted: 22 March 2021;
Published: 14 July 2021.
Edited by:
María Serrano, Miguel Hernández University of Elche, SpainReviewed by:
Wenguan Zhou, Centre for Research in Agricultural Genomics (CRAG), SpainCopyright © 2021 Zhao, He, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhoufei Wang, d2FuZ3pmQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.