
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 March 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.658679
Mechanisms governing plant–microbe interaction in the rhizosphere attracted a lot of investigative attention in the last decade. The rhizosphere is not simply a source of nutrients and support for the plants; it is rather an ecosystem teeming with diverse flora and fauna including different groups of microbes that are useful as well as harmful for the plants. Plant–microbe interaction occurs via a highly complex communication network that involves sophisticated machinery for the recognition of friend and foe at both sides. On the other hand, nitric oxide (NO) is a key, signaling molecule involved in plant development and defense. Studies on legume–rhizobia symbiosis suggest the involvement of NO during recognition, root hair curling, development of infection threads, nodule development, and nodule senescence. A similar role of NO is also suggested in the case of plant interaction with the mycorrhizal fungi. Another, insight into the plant–microbe interaction in the rhizosphere comes from the recognition of pathogen-associated molecular patterns (PAMPs)/microbe-associated molecular patterns (MAMPs) by the host plant and thereby NO-mediated activation of the defense signaling cascade. Thus, NO plays a major role in mediating the communication between plants and microbes in the rhizosphere. Interestingly, reports suggesting the role of silicon in increasing the number of nodules, enhancing nitrogen fixation, and also the combined effect of silicon and NO may indicate a possibility of their interaction in mediating microbial communication underground. However, the exact role of NO in mediating plant–microbe interaction remains elusive. Therefore, understanding the role of NO in underground plant physiology is very important, especially in relation to the plant’s interaction with the rhizospheric microbiome. This will help devise new strategies for protection against phytopathogens and enhancing plant productivity by promoting symbiotic interaction. This review focuses on the role of NO in plant–microbe communication underground.
Plants develop a close association with the microorganisms inhabiting the areas in and around the roots which are influenced by the chemicals (and/or enzymes) released from the plant roots. The collective term for microbial communities in the rhizosphere is the root microbiome. The rhizosphere harbors the largest reservoir of microbial diversity on earth. Several beneficial microbes are not only able to suppress diseases in host plants by inhibiting the growth of soil-borne phytopathogens, but they also stimulate growth and defense responses in host plants by the production of growth regulators, phytohormones, and other substances like antibiotics, antifungal metabolites, and defense enzymes for pathogen control and trigger induced systemic resistance (ISR) via methyl jasmonate and methyl salicylate in plants, displaying a sophisticated signaling network through which they communicate. Signaling generally involves signal perception through the cell wall or cell membrane receptors, ion channels and sensors, signal transduction through secondary messengers including cyclic nucleotides, lipid messengers, ions, free radicals, and gases like nitric oxide (NO). These secondary messengers then activate target-responsive genes and stimulate metabolic and physiological responses (Yamaguchi-Shinozaki and Shinozaki, 2006; Pérez-Clemente et al., 2013). Particularly, in the rhizosphere, there exists a multitude of microbial processes where NO is involved. In plants, two isoforms of the nitrate reductase (NR) enzyme catalyze the production of NO from nitrate, though multiple other pathways for NO production also exist. However, being highly reactive NO may prove toxic to the plants. It, therefore, needs to be converted to a non-toxic and ubiquitous form. This is accomplished by the covalent attachment of NO group (–NO) to exposed cysteine thiols to form S-nitrosothiols (SNOs), a process termed S-nitrosation which is a highly ubiquitous post-translational modification of proteins with diverse regulatory roles across the different kingdoms of life. These SNOs serve as mobile reservoirs, providing a sustained supply of NO in vivo, playing key roles in plant physiology under basal as well as stress conditions. At the cellular level, NO reacts with glutathione (GSH) to form S-nitrosoglutathione (GSNO) a major cellular reservoir of NO, capable of transferring NO bioactivity to other proteins. The Cellular GSNO levels are controlled by an evolutionarily conserved cytosolic enzyme GSNO reductase 1 (GSNOR1).
At the cellular level, biotic and abiotic stress responses are characterized by immediate redox bursts potentiated by reactive oxygen and reactive nitrogen species. Decades of research reveal an intricate balance between various ROS and RNS to maintain a physiological equilibrium under stress. The magnitude, intensity, and other characteristics of RNS-mediated redox bursts vary depending upon the type, magnitude, and intensity of the stress. The role of NO in plant–pathogen interaction has been studied. However, a lot is still unknown, about the different roles NO plays during plant–microbe interactions. Information about the role of NO during the plant’s interaction with useful microorganisms in the rhizosphere such as the rhizobacteria, mycorrhizae, free-living plant growth-promoting rhizobacteria and fungi, and others is specifically scarce. In this review, we discuss the role of NO in plant–microbe interaction in the rhizosphere.
The role of NO in morphological growth and developmental pattern of root and other developmental processes like photomorphogenesis, senescence, and leaf expansion in many plant has already been elucidated (Beligni and Lamattina, 2001; Kwon et al., 2012; Baudouin and Hancock, 2014). Also, reports of its role are suggested in rapid physiological responses like stomatal closure (Desikan et al., 2004) and the cytokinin signaling pathway (Tun et al., 2001). However, its role in influencing root growth and developmental processes via auxin-regulated signaling cascade is considered as one of the most important functions in plant biology. The role of NO in root development has been thoroughly reviewed by Molina-Favero et al. (2007). NO influences auxin signaling through S-nitrosation of the auxin receptor TIR1 at cys140 (Terrile et al., 2012). The view that NO is involved in root development also seems to have some connections with the root microbiome. Therefore, we aimed to review the studies that could suggest links between NO role in root development and its links with the root microbiome.
Several studies suggest the involvement of auxin-induced NO-dependent pathway during adventitious root formation (Pagnussat et al., 2004; Lanteri et al., 2006). Pagnussat et al. (2002) reported the first study suggesting that auxin induces adventitious root formation through an increase of the NO concentration at the base of cucumber hypocotyls. Soon after this study, the same group demonstrated that auxin and NO trigger both cGMP-dependent and cGMP-independent pathways leading to adventitious root formation. Moreover, the role of NO in promoting root elongation has also been reported in maize plants (Gouvea et al., 1997). Other studies also support the involvement of NO in phosphorous deficiency-induced lateral root development and growth in Lupinus albus (Wang et al., 2010; Meng et al., 2012), NO-induced NADPH oxidase in adventitious root growth in Panax ginseng (Tewari et al., 2008; Tewari and Paek, 2014), Azospirillum brasilense-induced NO in lateral root formation in tomato (Creus et al., 2005), and NO-induced modulation in adventitious root growth in Vigna radiata (Sharma et al., 2019) and rice (Kushwaha et al., 2019). Furthermore, a recent study also suggests that NO is involved in the brassinolide-induced adventitious root development in cucumber (Li et al., 2020). Root hair initiation and elongation in lettuce also involves NO (Lombardo et al., 2006) and is critical in determining root hair differentiation and elongation, mediating an auxin-triggered signaling cascade. Similar regulation by NO and auxin has also been reported in Arabidopsis (Lombardo et al., 2006). Interestingly, bacterial quorum-sensing molecules are also reported to have a role in plant defense responses and root development. For instance, molecules like N-3-oxo-decanoyl-L-homoserine-lactone can induce adventitious root formation through the activation of cGMP signaling in a hydrogen peroxide and NO-dependent manner in mung bean (Bai et al., 2012). Another study has also found enhanced plant growth and root development accelerated by arbuscular mycorrhizal fungi in trifoliate orange (Tian et al., 2017). Furthermore, the role of NO in root architecture has been recently reviewed by Mukherjee and Corpas (2020).
In plants, NO is produced through enzymatic and non-enzymatic reactions. One of the enzymatic reactions involves L-arginine as a primary substrate for NO synthase (NOS) leading to the production of NO through oxidative pathway (Gupta et al., 2011). The first report of putative NOS activity in plants was from the roots and nodules of L. albus via indirect detection of NOS antagonist N(G)-monomethyl-L-arginine, inhibiting the production of L-citrulline (Cueto et al., 1996). A unique AtNOS1 gene was isolated from Arabidopsis thaliana and its mutant (Atnos1) showed lesser NO content as compared to the wild type (Guo et al., 2003). Later on, it was renamed NO-associated protein 1 (AtNOA1) due to its inability to bind and oxidize arginine (Moreau et al., 2008). In 2010, a study described the presence of NOS enzyme in Ostreococcus tauri, a single-celled alga with mostly those characteristics that are present in animal NOS, thus suggesting the presence of NOS in plants (Foresi et al., 2010). However, until now the production of NO via a canonical NOS enzyme remains controversial (Hancock, 2020). NO production through enzymatic reaction also occurs through NR which catalyzes the conversion of nitrate and nitrite to NO (Tejada-Jimenez et al., 2019). This process is well established for NO production in several plant species including maize, cucumber, spinach, and sunflower (reviewed in detail by Cohen et al., 2009).
In the case of bacteria, the reduction of nitrite to NO is catalyzed by nitrite reductase which acts as a key enzyme in the dissimilative denitrification chain. Dissimilative nitrite reductase is therefore considered the major known source of NO in bacteria. Two distinct classes of dissimilatory nitrite reductases may contain either copper or heme as a cofactor, out of which heme-containing enzyme is more frequent. The biological relevance of this redundancy is only speculated to be associated with the availability of copper and iron in different microenvironments because enzymes containing copper and heme never coexist within the same bacterial species (Cutruzzolà, 1999). Moreover, bacterial NOS (bNOS) activity was first reported in the genus Nocardia (Chen and Rosazza, 1994). Later, it was reported in many plant-associated and free-living bacteria including Sinorhizobium, Mycobacterium, Nocardia, Rhodococcus, Streptomyces, Bacillus, Geobacillus, and Paenibacillus by Cohen et al. (2009). Since bacteria and plants share several similar features of NO-producing pathways, NO is suggested to act as a signaling molecule in plant–microbe interaction.
Common NO-mediated signaling links in plants and microbes have also been reported in a few studies. An important role of NO in plants is its influence on cellular processes through protein S-nitrosylation. Interestingly, endogenous S-nitrosylation is also reported to occur in Escherichia coli as a prominent feature of anerobic respiration which is regulated by OxyR, a regulon that controls endogenous S-nitrosylation in E. coli (Seth et al., 2012). Studies have also suggested the involvement of OxyR in influencing virulence of plant pathogens (Flores-Cruz and Allen, 2011; Ishiga and Ichinose, 2016).
Several plant associate microbes are pathogens that impair growth and reproduction. As such, there would hardly be any defense without the pathogen. Hence, the co-evolution of plant–microbe interaction shapes the defense system of both plants and microbes over time. Jones and Dangl (2006) proposed that plants possess a two-branched innate immune system that responds to infection. The first branch which is recognized by a set of receptors, also known as pattern-recognition receptors (PRRs), recognizes conserved microbe-associated molecular patterns (MAMPs). Typical examples of these receptors are LRR receptor-like kinases including Arabidopsis FLAGELLIN-SENSING 2 (FLS2) which recognize a peptide sequence (22 amino acids) present in bacterial flagellin and EF-Tu receptor (EFR), recognizing active epitope of bacterial elongation factor. Therefore, these receptors and the microbial molecules recognized by them constitute an essential part of defense related to innate immunity. On the other hand, the second branch comprises large families of intracellular immune receptors that encode NBS-LRR (nucleotide-binding site leucine-rich repeat) proteins. Meyers et al. (2003) reported the presence of 140 NBS-LLR encoding proteins from the Arabidopsis genome, whereas the japonica rice genome encodes 500 NBS-LRR proteins (Zhou et al., 2004). However, PRRs activate their specific immune response through pathogen-triggered immunity (PTI), and the NBS-LRRs activate it through effector-triggered immunity. The activation of downstream innate immune response through both these branches initiates with changes in the cytoplasmic Ca2+ levels, production of reactive oxygen species, and post translational activation of mitogen-activated protein kinase (MAPK) cascade (Jonak et al., 2002). Furthermore, NO is produced when plants are treated with MAMPs or during fungal infection which indicates an important role of NO in activating the innate defense response (Clarke et al., 2000). In fact, literature reports the production of NO during the establishment of interaction between nominally compatible plants and bacteria (Conrath et al., 2004; Mur et al., 2005), and also when plants are treated with lipopolysaccharide (LPS), NO mediates the activation of innate defense genes (Zeidler et al., 2004). Moreover, NO also mediates the regulation of NADPH-generating enzymes through post-translational modifications particularly, tyrosine nitration and S-nitrosation. NADPH is a versatile coenzyme involved in cellular growth, proliferation, detoxification processes, and maintenance of cellular redox status (Corpas and Barroso, 2014).
The important pathways which confer plant immune response are either salicylic acid or jasmonic acid/ethylene pathways. While salicylic acid plays a key role in plant immunity against biotrophic pathogens, it has antagonistic function against the jasmonic acid pathway which is essential role player in providing immunity against necrotrophic pathogens and also during physical wounding (Glazebrook, 2005). Along each signaling pathway, NO can either induce or suppress signaling. It initiates salicylic acid biosynthesis and enables nitrosylation of key cysteines on TGA-class transcription factors to initiate salicylic acid-dependent gene expression. During pathogenic attack, salicylic acid concentration increases in response to the changes in the cellular redox state. This leads to the reduction of disulfide bonds of NPR1 oligomer releasing monomers which translocate to the nucleus activating pathogenesis-related (PR) genes. However, oligomerization of NPR1 occurs under higher levels of SNO via S-nitrosylation. This was shown in Arabidopsis mutant atgsnor1-3 where NPR1 oligomer accumulates due to increased GSNO levels which enabled S-nitrosylation at cysteine (Cys)-156, while reducing monomerization by salicylic acid thus resulting in delayed activation of PR genes and enhanced pathogen susceptibility (Tada et al., 2008). Furthermore, Mur et al. (2013) have reviewed the salicylic acid and jasmonic acid/ethylene pathways mediated by NO during plant defense response in much detail. However, investigation is required to understand how this signaling network is mediated and controlled during plant–pathogen interaction in the rhizosphere.
Some mechanism of recognition or exchange of molecular signals between plants and microbes must be present which determines the specificity of host–microbe interactions. Certainly, for plant–microbe interaction to occur, there must be exchange of some signaling molecules, which is necessary for interaction among them. The initiation of plant–microbe interaction generally involves signal exchange through specific molecules produced by host or microbe or both during recognition which leads to the biochemical, physiological, and molecular responses that affects development of their interaction. Signals initiating, maintaining, and controlling the interaction determine the process of successful interactions. Only the compatible microbes can alter the host defense response out of the millions of microbes present in the rhizosphere and their communication affects plant health and growth. Therefore, the focus of recent research is to determine the signals and regulatory networks which counteract immune detection and recognize beneficial/compatible rhizospheric microbes. Here, we are focusing on the plant growth-promoting microbes and the various suggested mechanisms through which they interact with the plants. Plant growth-promoting microbes are the beneficial soil microbes (bacteria or fungi) colonizing the plant roots and useful in promoting plant growth and health (Spaepen et al., 2009). They are well known for their role and application in agriculture (Reddy et al., 2014).
The plant–microbe interaction occurs in a highly sophisticated manner, and several specialized metabolites and exudates control the process. As a result, gene expression is altered in either plant or microbe or both the interacting partners leading to either “plant growth, inhibition of soil pathogens, nutrient availability, biofilm development, or accumulation of soil microbes” (Li et al., 2013; Mommer et al., 2016; Sasse et al., 2018). The communication may be inter-/intra-species (microbial quorum sensing (QS) or signaling volatile organic compounds) or inter-kingdom communication (root exudates, exchange of symbiotic signals, bacterial QS signals, and exchange of antimicrobial and volatile organic compounds) (Bukhat et al., 2020). Out of the two, our main focus for this review is the inter-kingdom signaling in the rhizosphere which is widespread, common, and diverse. Microbial association with plants, particularly, symbiotic ones is established through the secretion and emission of beneficial molecules which includes growth regulators like auxins, cytokinins, gibberellins, abscisic acid, salicylic acid, and jasmonic acid and volatile-organic compounds like 2-heptanol, 2-endecanone, and pentadecane (Fahad et al., 2015). These molecules activate the plant transcriptome for specific changes or adjustments.
Some chemical signals are also transmitted from the plant’s side which are sensed by specific microbes for association. Cucumber root exudates are reported to attract Bacillus amyloliquefaciens SQR9, while fumaric acid from banana root attracts Bacillus subtilis N11; as a result, biofilm formation is reported in both the cases (Zhang et al., 2014). Other root exudates involved in plant–microbe interaction are flavonoids (derivatives of 2-phenyl-1,4-benzopyrone) which trigger nod genes in bacteria to produce lipochitooligosaccharides (LCOs) for initiating nodule formation in the roots of leguminous plants. These LCOs are also reported to participate in the interaction between arbuscular mycorrhizal fungi and plants, mimicking bacterial QS molecules, and playing role in influencing bacterial metabolism (Hassan and Mathesius, 2012). Moreover, plant exudates like aminocyclopropane-1-carboxylic acid (ACC) (Haichar et al., 2012) and tryptophan (Haichar et al., 2014) are also used by rhizobacteria as precursors of growth regulators ethylene and auxin, respectively. A recent study by Ding et al. (2020) suggests that there is alteration in the transcriptome of the plant pathogen, Fusarium graminearum which triggers NO production during sensing the presence of host before establishing physical contact with host’s roots. Further, virulence in F. graminearum and NO production during host sensing requires FgANK1, a protein containing an ankyrin-repeat domain. FgANK1, which normally resides in the cytoplasm, is translocated to the nucleus in response to the host signal where it interacts with a zinc finger transcription factor (FgZC1) which is also required for the specific binding to the NR promoter, NO production, and virulence in F. graminearum. Hence, NO is produced in F. graminearum after host is recognized and FgANK1 and FgZC1 regulate it, but still the exact pathway of NO biosynthesis and production in F. graminearum or other fungi is still unknown (Ding et al., 2020). Furthermore, one of the studies from our group has also reported the growth promotory effects of Bacillus aryabhattai strain SRB02 in soybean which is modulated by the production of phytohormones (Park et al., 2017). Although legume–rhizobia interaction provides much evidence in support of the involvement of NO during plant–microbe interaction, studies need to be conducted in order to understand how NO promotes their interaction.
Rhizosphere is home to numerous microbes that may or may not affect plant growth, health and productivity directly. However, microbes that do not affect plants directly have several indirect roles to play. The uncertainty about their roles comes from the limited knowledge of the impact that these plant-associated microbes may have on the plants. In-depth studies on rhizospheric symbiotic rhizobia and mycorrhizal fungi has enabled a greater understanding of their association with plants. Moreover, as the review focuses on the role of nitric oxide as an important signaling molecule in mediating the mutual association, our focus is mainly on plant-rhizobium signaling because it is the best understood association where nitric oxide is involved at several steps. Further, this review may draw attention of the researchers involved in studying particular plant-microbe interactions, towards studying the speculated yet unknown role of nitric oxide signaling network involved in mediating the plant-microbe interaction.
Recent research studies have demonstrated some important roles of NO during plant–bacterial interactions (Table 1) (Vaishnav et al., 2018). It has been known that the association of hemoglobins (Hbs) and NO is an important factor between symbiotic interactions. In addition, NO contents in plant and symbiont cells are intimately regulated by Hbs and S-nitrosoglutathione (GSNO) reductase to minimize toxic effects. For pathogen interactions, the expression level of Hb is maintained at low level to perpetuate NO production and promote defense response. In the event of symbiotic interactions, the expression of Hb is rapidly induced and it causes decrease of NO contents and suppression of plant defense system (Hichri et al., 2016). Plant phytoglobins also regulate NO accumulation by functioning as NO dioxygenases, metabolizing NO to nitrate. Studies also suggest its role during plant–microbe interaction in controlling NO bioactivity (Shimoda et al., 2009; Bai et al., 2016). PHYTOGB1 is reported as one of the early activated during root–mycorrhizal interaction (Hogekamp and Küster, 2013). Furthermore, PHYTOGB1, is also demonstrated to regulate NO levels in tomato roots during arbuscular mycorrhizal symbiosis with R. irregularis in order to promote and control the interaction (Martínez-Medina et al., 2019). Overall, the role(s) of NO in the arbuscular mycorrhizal fungi still remains elusive and its regulation is yet unclear. In arbuscular mycorrhizae interaction, another NO detoxification gene VfLb29 in Vicia faba is also upregulated to repress defense response. Therefore, the best example of the NO as a signaling molecule in plant–microbe interaction is during the process of biological nitrogen fixation. The process of nitrogen fixation is symbiotic association between legume (leguminous plants) and rhizobia (a small group of bacteria) which increases the level of nitrogen in the soil. These rhizobia may be free living or may be associated with epiphytes and endophytes. In some cases, they can also establish endocellular symbiosis with legumes (Masson-Boivin et al., 2009). Thus, the role of NO as a potential signaling molecule during plant–microbe interaction is highlighted in several studies.
The involvement of NO in legume–rhizobia symbiosis has been thoroughly studied in the last decade. NO is involved at almost every step of the interaction between the legume host and rhizobia (Figure 1). Legume–rhizobium symbiosis is established following the recognition of rhizobia by the plants and the formation of root nodules that host the bacteria for nitrogen fixation. Nodule prevents the inhibition of nitrogenase activity by providing low oxygen environment. The process of nodulation begins with the production of bacterial nodulation factors (Nod-factors) in response to the chemical signals released by the root hairs. These Nod-factors trigger division in the roots and other responses downstream (Geurts and Bisseling, 2002). The growth of rhizobia induces root hair curling and the formation of infection thread which is a tubular and intracellular structure. Plant hormones cytokinin and auxin induce the formation of infection thread (Roy et al., 2020). The reproduction of rhizobia allows the continuous growth of rhizobia from where it reaches to the nodule primordium after reaching the base of the root hair cells (Geurts and Bisseling, 2002). Thereafter, the bacterial cells are released into the parenchyma cells of the nodule and the differentiation of bacterial cells to bacteroids begins. At this stage, morphological and physiological changes are required for this transformation. In the entire nodulation process, the involvement of NO is well documented. NO was reported to be induced at early stages (4 h post inoculation) but not at 10 and 24 h of plant–rhizobia interactions in Lotus japonicus–Mesorhizobium loti and Medicago sativa–Ensifer meliloti suggesting its role in signal recognition (Nagata et al., 2008). Similar results about NO induction and involvement during symbiosis establishment were observed in Medicago truncatula–E. meliloti symbiosis by another group (Del Giudice et al., 2011). Additionally, they have also reported NO detection during curling of root hair and formation of infection thread 2 and 6 days post-infection, respectively. Furthermore, scavenging the endogenously produced NO in the infection thread delays the nodule development.
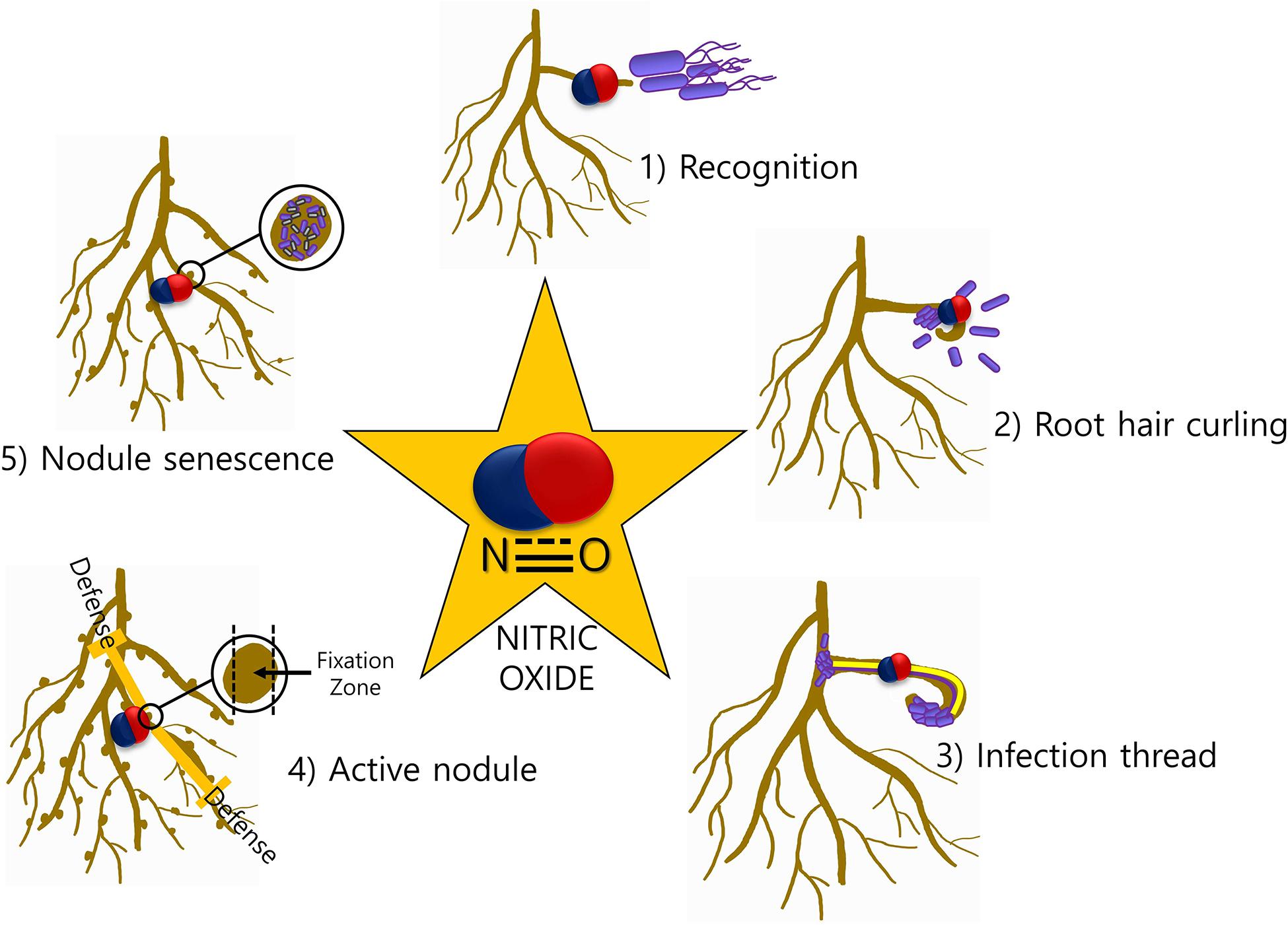
Figure 1. NO-mediated signaling during legume–rhizobia symbiosis. The presence of NO at various stages (1–5) of symbiosis is indicative of a major role of NO in mediating the communication between plants and microbes.
Significant quantity of silicon present in the soil makes it the second most abundant element on the earth’s crust, and hence studies were carried out to understand its role in the rhizosphere. It was discovered that silicon could not only increase the biomass production but also had role in enhancing tolerance to biotic and abiotic stresses, thus supporting plants with stability and protection (Luyckx et al., 2017; López-Pérez et al., 2018). Interestingly, a study also discovered silicon uptake transporters from some legumes like soybean which were similar to the silicon transporters in hyper accumulating grasses (Deshmukh et al., 2013). The abundance of silicon in the rhizosphere and its role in plants lead to the investigations on the role of silicon in root nodulation and nitrogen fixation which started about 21 years back. The first study in Vigna unguiculate reported that silicon significantly affected root nodulation and nitrogen fixation depending upon the concentration of silicon, its forms (metasilicic acid or silicic acid), and growing substrate (liquid culture or sand culture) (Nelwamondo and Dakora, 1999). Later on, the same group reported that other than affecting nodule numbers and nitrogen fixation, silicon also affected the internal structure of the nodule. An increase in the size of peribacteroid space and cell wall thickness at high concentrations was reported along with an increase in the number of bacteroids and symbiosomes (Nelwamondo et al., 2001). A characteristic feature of silicon integration in most of the high silicon accumulating plants is an increase in cell wall thickness (Kumar et al., 2017). Interestingly, reports of the presence of silicon in the root nodules were also observed in Cajanus cajan (pigeonpea) genotypes (Garg and Singh, 2018). Nodule activity was promoted in two different genotypes of pigeonpea through the production of leghemoglobin. Another study in M. sativa also demonstrated the positive impact of silicon on root nodulation and also found it to be involved in biosynthesis of foliar amino acids (Johnson et al., 2017). Overall, it is clear that silicon employs beneficial impact on the symbiotic traits.
Speculations about the joint effects of silicon and NO have been suggested recently. Combination of silicon and NO application in maize plants under cadmium stress is reported to increase plant growth and reduce cadmium uptake, accumulation, translocation, and bioaccumulation factors (Liu et al., 2020). However, the combined effect of silicon and NO in promoting root–microbiome interaction remains to be elucidated (Figure 2).
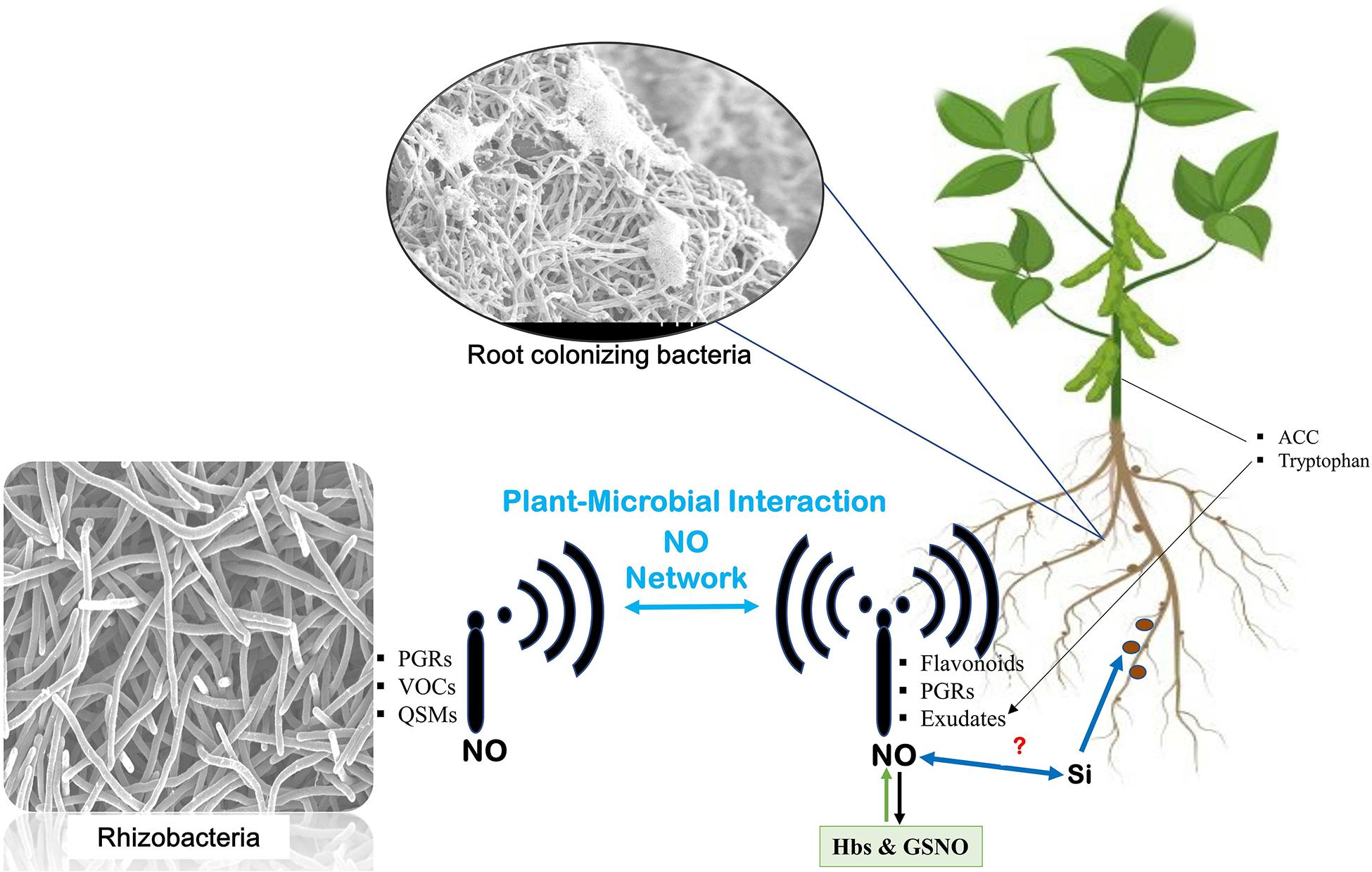
Figure 2. Nitric oxide network underground. Communication is established between plants and microbes through signals transmitted by microbes [such as plant growth regulators (PGRs), volatile organic compounds (VOCs), and quorum sensing molecules (QSMs) from microbes and similar signal transmission from host plants]. The signals from the host include flavonoids, PGRs, and exudates from the roots and aerial parts of the plant like 1-aminocyclopropane-1-carboxylic acid (ACC) and tryptophan. At every step, nitric oxide mediates the signaling and helps establish their interaction. Role of silicon in enhancing nitrogen fixation also leaves speculations about its relation with NO in mediating the signaling events.
Rhizosphere is the area around a plant root inhabited by a diverse population of microorganisms; however, only the unique population can influence or can be influenced by the chemical signals released by either of them. Recent research has shown that NO participates in early basal signaling during plant–bacterial interactions. Bacteria and plants share several features of NO-producing pathways which suggests that NO acts as a signaling molecule during plant–microbe association.
Nitric oxide has been thoroughly studied as a signaling molecule in the last decade. NO may be produced endogenously by plant cells or may be generated as a result of activity of soil microorganisms. Soils are an important source of NO, and the presence of inorganic fertilizers can affect NO emissions into the environment. Since NO is a direct intermediate of nitrification and denitrification processes carried out by microorganisms during nitrogen fixation (Payne, 1981; Skiba and Smith, 1993; Feelisch and Martin, 1995), it suggests the possible contribution of roots and microbes in NO production and influencing the synthesis of NO on each other during symbiotic association (Molina-Favero et al., 2007). Furthermore, research needs to be conducted in understanding the role of silicon in plant–microbe signaling along with its combined effect with NO in mediating the microbial communication underground.
AP and B-GM have contributed equally in conceiving the idea and structure of the manuscript, and writing the first draft of the manuscript. D-SL illustrated the figures. G-ML contributed to the writing of the Section “NO in Plants and Bacteria.” MK contributed to the Section “Plant–Microbe Interaction: NO and the Plant Innate Immune System.” AH revised and improved the manuscript and figures. B-WY revised the whole manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant No. 2020R1I1A3073247).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alen’kina, S. A., Bogatyrev, V. A., Matora, L. Y., Sokolova, M. K., Chernyshova, M. P., Trutneva, K. A., et al. (2014). Signal effects of the lectin from the associative nitrogen-fixing bacterium Azospirillum brasilense Sp7 in bacterial–plant root interactions. Plant Soil 381, 337–349. doi: 10.1007/s11104-014-2125-6
Asai, S., and Yoshioka, H. (2009). Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana. Mol. Plant Microbe Interact. 22, 619–629. doi: 10.1094/mpmi-22-6-0619
Bai, X., Long, J., He, X., Yan, J., Chen, X., Tan, Y., et al. (2016). Overexpression of spinach non-symbiotic hemoglobin in Arabidopsis resulted in decreased NO content and lowered nitrate and other abiotic stresses tolerance. Sci. Rep. 6:26400.
Bai, X., Todd, C. D., Desikan, R., Yang, Y., and Hu, X. (2012). N-3-oxo-decanoyl-L-homoserine-lactone activates auxin-induced adventitious root formation via hydrogen peroxide-and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol. 158, 725–736. doi: 10.1104/pp.111.185769
Baudouin, E., and Hancock, J. (2014). Nitric oxide signaling in plants. Front. Plant Sci. 4:553. doi: 10.3389/fpls.2013.00553
Beligni, M. V., and Lamattina, L. (2001). Nitric oxide in plants: the history is just beginning. Plant Cell Environ. 24, 267–278. doi: 10.1046/j.1365-3040.2001.00672.x
Berger, A., Boscari, A., Horta Araújo, N., Maucourt, M., Hanchi, M., Bernillon, S., et al. (2020). Plant nitrate reductases regulate nitric oxide production and nitrogen-fixing metabolism during the Medicago truncatula–Sinorhizobium meliloti symbiosis. Front. Plant Sci. 11:1313. doi: 10.3389/fpls.2020.01313
Bukhat, S., Imran, A., Javaid, S., Shahid, M., Majeed, A., and Naqqash, T. (2020). Communication of plants with microbial world: exploring the regulatory networks for PGPR mediated defense signaling. Microbiol. Res. 238:126486. doi: 10.1016/j.micres.2020.126486
Chen, Y. J., and Rosazza, J. P. (1994). A bacterial, nitric oxide synthase from a Nocardia species. Biochem. Biophys. Res. Commun. 203, 1251–1258. doi: 10.1006/bbrc.1994.2317
Clarke, A., Desikan, R., Hurst, R. D., Hancock, J. T., and Neill, S. J. (2000). NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24, 667–677. doi: 10.1046/j.1365-313x.2000.00911.x
Cohen, M. F., Lamattina, L., and Yamasaki, H. (2009). “Nitric oxide signaling by plant-associated bacteria”, in Nitric Oxide Plant Physiol. eds S. Hayat, M. Mori, J. Pichtel, and A. Ahmad (Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA) 161–172. doi: 10.1002/9783527629138.ch11
Conrath, U., Amoroso, G., Köhle, H., and Sültemeyer, D. F. (2004). Non-invasive online detection of nitric oxide from plants and some other organisms by mass spectrometry. Plant J. 38, 1015–1022. doi: 10.1111/j.1365-313x.2004.02096.x
Corpas, F. J., and Barroso, J. B. (2014). NADPH-generating dehydrogenases: their role in the mechanism of protection against nitro-oxidative stress induced by adverse environmental conditions. Front. Environ. Sci. 2:55. doi: 10.3389/fenvs.2014.00055
Creus, C. M., Graziano, M., Casanovas, E. M., Pereyra, M. A., Simontacchi, M., Puntarulo, S., et al. (2005). Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221, 297–303. doi: 10.1007/s00425-005-1523-7
Cueto, M., Hernández-Perera, O., Martín, R., Bentura, M. L., Rodrigo, J., Lamas, S., et al. (1996). Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett. 398, 159–164. doi: 10.1016/s0014-5793(96)01232-x
Cutruzzolà, F. (1999). Bacterial nitric oxide synthesis. Biochim. Biophys. Acta (BBA) Bioenerg. 1411, 231–249.
Del Giudice, J., Cam, Y., Damiani, I., Fung-Chat, F., Meilhoc, E., Bruand, C., et al. (2011). Nitric oxide is required for an optimal establishment of the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 191, 405–417.
Deshmukh, R. K., Vivancos, J., Guérin, V., Sonah, H., Labbé, C., Belzile, F., et al. (2013). Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 83, 303–315. doi: 10.1007/s11103-013-0087-3
Desikan, R., Cheung, M. K., Bright, J., Henson, D., Hancock, J. T., and Neill, S. J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55, 205–212. doi: 10.1093/jxb/erh033
Ding, Y., Gardiner, D. M., Xiao, D., and Kazan, K. (2020). Regulators of nitric oxide signaling triggered by host perception in a plant pathogen. Proc. Natl. Acad. Sci. U.S.A. 117, 11147–11157. doi: 10.1073/pnas.1918977117
Fahad, S., Hussain, S., Bano, A., Saud, S., Hassan, S., Shan, D., et al. (2015). Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: consequences for changing environment. Environ. Sci. Pollut. Res. 22, 4907–4921. doi: 10.1007/s11356-014-3754-2
Feelisch, M., and Martin, J. F. (1995). The early role of nitric oxide in evolution. Trends Ecol. Evol. 10, 496–499. doi: 10.1016/s0169-5347(00)89206-x
Flores-Cruz, Z., and Allen, C. (2011). Necessity of OxyR for the hydrogen peroxide stress response and full virulence in Ralstonia solanacearum. Appl. Environ. Microbiol. 77, 6426–6432. doi: 10.1128/aem.05813-11
Foresi, N., Correa-Aragunde, N., Parisi, G., Caló, G., Salerno, G., and Lamattina, L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22, 3816–3830. doi: 10.1105/tpc.109.073510
Garg, N., and Singh, S. (2018). Arbuscular mycorrhiza Rhizophagus irregularis and silicon modulate growth, proline biosynthesis and yield in Cajanus cajan L. Millsp.(pigeonpea) genotypes under cadmium and zinc stress. J. Plant Growth Regul. 37, 46–63. doi: 10.1007/s00344-017-9708-4
Geurts, R., and Bisseling, T. (2002). Rhizobium Nod factor perception and signalling. Plant Cell 14(Suppl. 1), S239–S249.
Ghiglione, J. F., Gourbiere, F., Potier, P., Philippot, L., and Lensi, R. (2000). Role of respiratory nitrate reductase in ability of Pseudomonas fluorescens YT101 To Colonize the Rhizosphere of Maize. Appl. Environ. Microbiol. 66, 4012–4016. doi: 10.1128/aem.66.9.4012-4016.2000
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gouvea, C. M. C. P., Souza, J. F., Magalhaes, A. C. N., and Martins, I. S. (1997). NOcdot–releasing substances that induce growth elongation in maize root segments. Plant Growth Regul. 21, 183–187.
Guo, F. Q., Okamoto, M., and Crawford, N. M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103. doi: 10.1126/science.1086770
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and van Dongen, J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168.
Haichar, F. E. Z., Roncato, M. A., and Achouak, W. (2012). Stable isotope probing of bacterial community structure and gene expression in the rhizosphere of Arabidopsis thaliana. FEMS Microbiol. Ecol. 81, 291–302. doi: 10.1111/j.1574-6941.2012.01345.x
Haichar, F. E. Z., Santaella, C., Heulin, T., and Achouak, W. (2014). Root exudates mediated interactions belowground. Soil Biol. Biochem. 77, 69–80. doi: 10.1016/j.soilbio.2014.06.017
Hassan, S., and Mathesius, U. (2012). The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Hichri, I., Meilhoc, E., Boscari, A., Bruand, C., Frendo, P., and Brouquisse, R. (2016). “Nitric Oxide: jack-of-all-trades of the nitrogen-fixing symbiosis?,” in Advances in Botanical Research, Vol. 77, ed. J. A. Callow (Cambridge, MA: Academic Press), 193–218.
Hogekamp, C., and Küster, H. (2013). A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14:306. doi: 10.1186/1471-2164-14-306
Horchani, F., Prévot, M., Boscari, A., Evangelisti, E., Meilhoc, E., Bruand, C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. doi: 10.1104/pp.110.166140
Ishiga, Y., and Ichinose, Y. (2016). Pseudomonas syringae pv. tomato OxyR is required for virulence in tomato and Arabidopsis. Mol. Plant Microbe Interact. 29, 119–131. doi: 10.1094/mpmi-09-15-0204-r
Johnson, S. N., Hartley, S. E., Ryalls, J. M., Frew, A., DeGabriel, J. L., Duncan, M., et al. (2017). Silicon-induced root nodulation and synthesis of essential amino acids in a legume is associated with higher herbivore abundance. Funct. Ecol. 31, 1903–1909. doi: 10.1111/1365-2435.12893
Jonak, C., Ökrész, L., Bögre, L., and Hirt, H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5, 415–424. doi: 10.1016/s1369-5266(02)00285-6
Kumar, S., Soukup, M., and Elbaum, R. (2017). Silicification in grasses: variation between different cell types. Front. Plant Sci. 8:438. doi: 10.3389/fpls.2017.00438
Kushwaha, B. K., Singh, S., Tripathi, D. K., Sharma, S., Prasad, S. M., Chauhan, D. K., et al. (2019). New adventitious root formation and primary root biomass accumulation are regulated by nitric oxide and reactive oxygen species in rice seedlings under arsenate stress. J. Hazard. Mater. 361, 134–140. doi: 10.1016/j.jhazmat.2018.08.035
Kwon, E., Feechan, A., Yun, B. W., Hwang, B. H., Pallas, J. A., Kang, J. G., et al. (2012). AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236, 887–900. doi: 10.1007/s00425-012-1697-8
Lanteri, M. L., Pagnussat, G. C., and Lamattina, L. (2006). Calcium and calcium-dependent protein kinases are involved in nitric oxide-and auxin-induced adventitious root formation in cucumber. J. Exp. Bot. 57, 1341–1351. doi: 10.1093/jxb/erj109
Leach, J., Keyster, M., Du Plessis, M., and Ludidi, N. (2010). Nitric oxide synthase activity is required for development of functional nodules in soybean. J. Plant Physiol. 167, 1584–1591. doi: 10.1016/j.jplph.2010.06.019
Li, X. G., Zhang, T. L., Wang, X. X., Hua, K., Zhao, L., and Han, Z. M. (2013). The composition of root exudates from two different resistant peanut cultivars and their effects on the growth of soil-borne pathogen. Int. J. Biol. Sci. 9:164. doi: 10.7150/ijbs.5579
Li, Y., Wu, Y., Liao, W., Hu, L., Dawuda, M. M., Jin, X., et al. (2020). Nitric oxide is involved in the brassinolide-induced adventitious root development in cucumber. BMC Plant Biol. 20:102. doi: 10.1186/s12870-020-2320-y
Liu, X., Yin, L., Deng, X., Gong, D., Du, S., Wang, S., et al. (2020). Combined application of silicon and nitric oxide jointly alleviated cadmium accumulation and toxicity in maize. J. Hazard. Mater. 395:122679. doi: 10.1016/j.jhazmat.2020.122679
Lombardo, M. C., Graziano, M., Polacco, J. C., and Lamattina, L. (2006). Nitric oxide functions as a positive regulator of root hair development. Plant Signal. Behav. 1, 28–33. doi: 10.4161/psb.1.1.2398
López-Pérez, M. C., Pérez-Labrada, F., Ramírez-Pérez, L. J., Juárez-Maldonado, A., Morales-Díaz, A. B., González-Morales, S., et al. (2018). Dynamic modeling of silicon bioavailability, uptake, transport, and accumulation: applicability in improving the nutritional quality of tomato. Front. Plant Sci. 9:647. doi: 10.3389/fpls.2018.00647
Luyckx, M., Hausman, J. F., Lutts, S., and Guerriero, G. (2017). Impact of silicon in plant biomass production: focus on bast fibres, hypotheses, and perspectives. Plants 6:37. doi: 10.3390/plants6030037
Martínez-Medina, A., Pescador, L., Fernández, I., Rodríguez-Serrano, M., García, J. M., Romero-Puertas, M. C., et al. (2019). Nitric oxide and phytoglobin PHYTOGB1 are regulatory elements in the Solanum lycopersicum-Rhizophagus irregularis mycorrhizal symbiosis. New Phytol. 223, 1560–1574. doi: 10.1111/nph.15898
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Meng, Z. B., Chen, L. Q., Suo, D., Li, G. X., Tang, C. X., and Zheng, S. J. (2012). Nitric oxide is the shared signalling molecule in phosphorus-and iron-deficiency-induced formation of cluster roots in white lupin (Lupinus albus). Ann. Bot. 109, 1055–1064. doi: 10.1093/aob/mcs024
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Modolo, L. V., Augusto, O., Almeida, I. M., Magalhaes, J. R., and Salgado, I. (2005). Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 579, 3814–3820. doi: 10.1016/j.febslet.2005.05.078
Molina-Favero, C., Creus, C. M., Lanteri, M. L., Correa-Aragunde, N., Lombardo, M. C., Barassi, C. A., et al. (2007). Nitric oxide and plant growth promoting rhizobacteria: common features influencing root growth and development. Adv. Bot. Res. 46, 1–33. doi: 10.1016/s0065-2296(07)46001-3
Mommer, L., Kirkegaard, J., and van Ruijven, J. (2016). Root–root interactions: towards a rhizosphere framework. Trends Plant Sci. 21, 209–217. doi: 10.1016/j.tplants.2016.01.009
Moreau, M., Lee, G. I., Wang, Y., Crane, B. R., and Klessig, D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. doi: 10.1074/jbc.m804838200
Mukherjee, S., and Corpas, F. J. (2020). Crosstalk among hydrogen sulfide (H2S), nitric oxide and carbon monoxide (CO) in root-system development and its rhizosphere interactions: a gaseous interactome. Plant Physiol. Biochem. 155, 800–814. doi: 10.1016/j.plaphy.2020.08.020
Mur, L. A., Kenton, P., and Draper, J. (2005). In planta measurements of oxidative bursts elicited by avirulent and virulent bacterial pathogens suggests that H2O2 is insufficient to elicit cell death in tobacco. Plant Cell Environ. 28, 548–561. doi: 10.1111/j.1365-3040.2005.01301.x
Mur, L. A., Prats, E., Pierre, S., Hall, M. A., and Hebelstrup, K. H. (2013). Integrating nitric oxide into salicylic acid and jasmonic acid/ethylene plant defense pathways. Front. Plant Sci. 4:215. doi: 10.3389/fpls.2013.00215
Nagata, M., Murakami, E. I., Shimoda, Y., Shimoda-Sasakura, F., Kucho, K. I., Suzuki, A., et al. (2008). Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant Microbe Interact. 21, 1175–1183. doi: 10.1094/mpmi-21-9-1175
Nelwamondo, A., and Dakora, F. D. (1999). Silicon promotes nodule formation and nodule function in symbiotic cowpea (Vigna unguiculata). New Phytol. 142, 463–467. doi: 10.1046/j.1469-8137.1999.00409.x
Nelwamondo, A., Jaffer, M. A., and Dakora, F. D. (2001). Subcellular organization of N 2-fixing nodules of cowpea (Vigna unguiculata) supplied with silicon. Protoplasma 216:94. doi: 10.1007/bf02680136
Pagnussat, G. C., Lanteri, M. L., Lombardo, M. C., and Lamattina, L. (2004). Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 135, 279–286. doi: 10.1104/pp.103.038554
Pagnussat, G. C., Simontacchi, M., Puntarulo, S., and Lamattina, L. (2002). Nitric oxide is required for root organogenesis. Plant Physiol. 129, 954–956. doi: 10.1104/pp.004036
Park, Y. G., Mun, B. G., Kang, S. M., Hussain, A., Shahzad, R., Seo, C. W., et al. (2017). Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS One 12:e0173203. doi: 10.1371/journal.pone.0173203
Perchepied, L., Balagué, C., Riou, C., Claudel-Renard, C., Rivière, N., Grezes-Besset, B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant Microbe Interact. 23, 846–860. doi: 10.1094/mpmi-23-7-0846
Pérez-Clemente, R. M., Vives, V., Zandalinas, S. I., López-Climent, M. F., Muñoz, V., and Gómez-Cadenas, A. (2013). Biotechnological approaches to study plant responses to stress. BioMed Res. Int. 2013:654120.
Rasul, S., Dubreuil-Maurizi, C., Lamotte, O., Koen, E., Poinssot, B., Alcaraz, G., et al. (2012). Nitric oxide production mediates oligogalacturonide-triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana. Plant Cell Environ. 35, 1483–1499. doi: 10.1111/j.1365-3040.2012.02505.x
Reddy, M. S., Ilao, R. I., and Faylon, P. S. (2014). Recent Advances in Biofertilizers and Biofungicides (PGPR) for Sustainable Agriculture. Newcastle upon Tyne: Cambridge Scholars Publishing.
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Seth, D., Hausladen, A., Wang, Y. J., and Stamler, J. S. (2012). Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science 336, 470–473. doi: 10.1126/science.1215643
Sharma, S., Singh, H. P., Batish, D. R., and Kohli, R. K. (2019). Nitric oxide induced modulations in adventitious root growth, lignin content and lignin synthesizing enzymes in the hypocotyls of Vigna radiata. Plant Physiol. Biochem. 141, 225–230. doi: 10.1016/j.plaphy.2019.05.028
Shimoda, Y., Shimoda-Sasakura, F., Kucho, K. I., Kanamori, N., Nagata, M., Suzuki, A., et al. (2009). Overexpression of class 1 plant hemoglobin genes enhances symbiotic nitrogen fixation activity between Mesorhizobium loti and Lotus japonicus. Plant J. 57, 254–263. doi: 10.1111/j.1365-313x.2008.03689.x
Skiba, U., and Smith, K. A. (1993). Nitrification and denitrification as sources of nitric oxide and nitrous oxide in a sandy loam soil. Soil Biol. Biochem. 25, 1527–1536. doi: 10.1016/0038-0717(93)90007-x
Spaepen, S., Vanderleyden, J., and Okon, Y. (2009). Plant growth-promoting actions of rhizobacteria. Adv. Bot. Res. 51, 283–320. doi: 10.1016/s0065-2296(09)51007-5
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tejada-Jimenez, M., Llamas, A., Galván, A., and Fernández, E. (2019). Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plants 8:56. doi: 10.3390/plants8030056
Terrile, M. C., París, R., Calderón-Villalobos, L. I., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313x.2011.04885.x
Tewari, R. K., Hahn, E. J., and Paek, K. Y. (2008). Function of nitric oxide and superoxide anion in the adventitious root development and antioxidant defence in Panax ginseng. Plant Cell Rep. 27, 563–573. doi: 10.1007/s00299-007-0448-y
Tewari, R. K., and Paek, K. Y. (2014). “Role of Nitric Oxide in adventitious root development,” in Production of Biomass and Bioactive Compounds Using Bioreactor Technology, eds H. N. Murthy, J.-J. Zhong, and K.-Y. Paek (Dordrecht: Springer), 429–443. doi: 10.1007/978-94-017-9223-3_18
Tian, L., Nasrullah, X. Y. H., and Wu, Q. S. (2017). Nitric oxide accelerates mycorrhizal effects on plant growth and root development of trifoliate orange. Sains Malays. 46, 1687–1691. doi: 10.17576/jsm-2017-4610-03
Tun, N. N., Holk, A., and Scherer, G. F. (2001). Rapid increase of NO release in plant cell cultures induced by cytokinin. FEBS Lett. 509, 174–176. doi: 10.1016/s0014-5793(01)03164-7
Vaishnav, A., Sharma, S. K., Choudhary, D. K., Sharma, K. P., Ahmad, E., Sharma, M. P., et al. (2018). “Nitric oxide as a signaling molecule in plant-bacterial interactions,” in Plant Microbiome: Stress Response, eds D. Egamberdieva and P. Ahmad (Singapore: Springer), 183–199. doi: 10.1007/978-981-10-5514-0_8
Wang, B. L., Tang, X. Y., Cheng, L. Y., Zhang, A. Z., Zhang, W. H., Zhang, F. S., et al. (2010). Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol. 187, 1112–1123. doi: 10.1111/j.1469-8137.2010.03323.x
Wang, Y., Yang, Q., Tosa, Y., Nakayashiki, H., and Mayama, S. (2005). Nitric oxide-overproducing transformants of Pseudomonas fluorescens with enhanced biocontrol of tomato bacterial wilt. J. Gen. Plant Pathol. 71, 33–38. doi: 10.1007/s10327-004-0157-0
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Zeidler, D., Zähringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816. doi: 10.1073/pnas.0404536101
Zhang, N., Wang, D., Liu, Y., Li, S., Shen, Q., and Zhang, R. (2014). Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374, 689–700. doi: 10.1007/s11104-013-1915-6
Keywords: rhizosphere, microbes, nitrogen fixation, silicon, signaling, nitric oxide
Citation: Pande A, Mun B-G, Lee D-S, Khan M, Lee G-M, Hussain A and Yun B-W (2021) NO Network for Plant–Microbe Communication Underground: A Review. Front. Plant Sci. 12:658679. doi: 10.3389/fpls.2021.658679
Received: 26 January 2021; Accepted: 24 February 2021;
Published: 17 March 2021.
Edited by:
Andrea Chini, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Manda Yu, University of Wisconsin–Milwaukee, United StatesCopyright © 2021 Pande, Mun, Lee, Khan, Lee, Hussain and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adil Hussain, YWRpbGh1c3NhaW5AYXdrdW0uZWR1LnBr; Byung-Wook Yun, Ynd5dW5Aa251LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.