
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 31 May 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.656696
Wheat is one of the most important food crops worldwide. In recent decades, fertilizers, especially nitrogen (N), have been increasingly utilized to maximize wheat productivity. However, a large proportion of N is not used by plants and is in fact lost into the environment and causes serious environmental pollution. Therefore, achieving a low N optimum via efficient physiological and biochemical processes in wheat grown under low-N conditions is highly important for agricultural sustainability. Although N stress-related N capture in wheat has become a heavily researched subject, how this plant adapts and responds to N starvation has not been fully elucidated. This review summarizes the current knowledge on the signaling mechanisms activated in wheat plants in response to N starvation. Furthermore, we filled the putative gaps on this subject with findings obtained in other plants, primarily rice, maize, and Arabidopsis. Phytohormones have been determined to play essential roles in sensing environmental N starvation and transducing this signal into an adjustment of N transporters and phenotypic adaptation. The critical roles played by protein kinases and critical kinases and phosphatases, such as MAPK and PP2C, as well as the multifaceted functions of transcription factors, such as NF-Y, MYB, DOF, and WRKY, in regulating the expression levels of their target genes (proteins) for low-N tolerance are also discussed. Optimization of root system architecture (RSA) via root branching and thinning, improvement of N acquisition and assimilation, and fine-tuned autophagy are pivotal strategies by which plants respond to N starvation. In light of these findings, we attempted to construct regulatory networks for RSA modification and N uptake, transport, assimilation, and remobilization.
Nitrogen (N) is an essential macronutrient in plant growth and development, acting as the cellular constituent for a large number of molecules, including amino acids, nucleic acids, chlorophyll, and phytohormones. In recent decades, increasing the input of N fertilizers has contributed considerably to improving crop productivity. However, it is alarming to note that major cereal crops, such as wheat (Triticum aestivum L.), rice (Oryza sativa L.), and maize (Zea mays L.), only utilize 30–40% of the applied N. Apart from the unnecessary expense of wasted fertilizer, excessive input of N has also caused serious environmental problems and some negative effects on plant growth in certain cases. Therefore, improving the nitrogen use efficiency (NUE) of crops under suboptimal N conditions has emerged as an effective strategy to promote agricultural sustainability worldwide (Liu et al., 2018).
Plants can immediately sense nutrient availability, generate signal messengers, and translocate to the cell nucleus to induce genetic and downstream responses and thereby maintain nutrient homeostasis (Paul et al., 2015). To date, increasing numbers of investigations have been performed that have focused on understanding the physiological and biochemical mechanisms underlying the plant response to N starvation. Upon N stress, plants induce specific N signaling and construct signal transduction pathways, including biosynthesis of signal molecules, initiation of transcriptional modulation, and induction of the expression of regulatory and functional genes encoding N transporters and the main enzymes involved in N assimilation (Remans et al., 2006; Gojon et al., 2011; Wang et al., 2012, 2019b). These systematic signaling networks play important roles in the response to low-N stress (Wang et al., 2019b). Although a large number of investigations have been carried out, the sensing and perception of N starvation signaling and tolerance to low-N stress in plants have not been fully elucidated, especially in cereal crops (Jung et al., 2018; Liu et al., 2018; Qiao et al., 2018). Thus, first, we explored the signaling regulatory pathways that may be associated with morphological adaptation, N uptake and assimilation, and the corresponding biochemical processes. A better understanding of signal transduction for high NUE may establish a foundation for the molecular breeding of N starvation-tolerant crop cultivars.
Wheat is one of three most important cereal crops worldwide. This crop exhibits a dominant production of 772 Mt year–1, which is lower than 1135 Mt year–1 for maize but slightly higher than the 770 Mt year–1 for rice (FAO Stats 2017). These three crops represent approximately 90% of all cereal production worldwide (Ladha et al., 2016). Wheat production requires a large amount of N for both grain yield and quality (Zörb et al., 2018). Thus, timing and precise N application or using genetic means to improve the ability of wheat plants to grow well under suboptimal N are required for sustainable agriculture. Although most N-responsive genes are regulated by hormones and other signals, how wheat plants establish networks by which wheat tolerates or efficiently uses limited N has not been fully elucidated at the molecular, posttranscriptional, biochemical, and developmental levels.
The most important strategies for responding to N starvation include a number of pathways, such as signal generation, regulation of gene expression and metabolism, and morphological adaptation. In this review, we summarize the significant progress made in our understanding of environmental N sensing and signaling in recent years and discuss how low N signaling is integrated with other regulatory elements and regulates the expression of genes involved in the N uptake and assimilation pathways in wheat. Although many N sensing and regulatory components are borrowed from other species, such as Arabidopsis thaliana, rice, and maize, the components identified to date tend to have conserved functions and may be characterized by their orthologs in wheat. This review may help to elucidate the mechanism underlying which wheat responds to N starvation and benefits genetically engineered crop cultivars with improved NUE under low-N cultivation conditions.
Calcium (Ca2+) is a ubiquitous secondary messenger and is widely involved in signal transduction pathways that regulate eukaryote responses to biotic and abiotic stresses. Ca2+ signaling is mediated by three sensor protein families, calmodulins (CaMs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinases (CDPKs), and its downstream signaling cascade includes kinases, such as CIPK8 and CIPK23, and TFs, such as ANR1, nodule inception-like protein (NLP) 7, and SPL9.
In rice, N starvation increases the leaf Ca2+ contents, especially in line with high tolerance to low-N stress (Tang et al., 2018). In particular, Ca2+ binding is an essential way to regulate the expression of genes in wheat roots that are associated with N stress responses (Zhang et al., 2016). In wheat, CaM, which can bind four calcium ions, is upregulated by low-N treatment and is involved in the promotion of primary root growth (Xu et al., 2019a). The CaM-related calcium sensor proteins, OsCML22 (predicted target gene of miR164) and Ca2+-transporting ATPases (predicted target gene of miR1318), are significantly upregulated in rice (Nischal et al., 2012), and transcripts of CaM, CaM-binding protein, and Ca2+-binding EF-hand protein are upregulated in maize roots under N starvation (Mascia et al., 2019).
CBLs can interact and activate CIPKs, forming the CBL-CIPK network as a component of the Ca2+ signaling pathway. CBL7 is upregulated and positively regulates the NO3–-dependent induction of NRT2.4 and NRT2.5 gene expression under NO3– deprivation (Ma et al., 2015; Sharma et al., 2017). CIPK8 acts as a positive regulator in nitrate low-affinity responses under low-N stress and is involved in long-term nitrate-modulated primary root growth (Hu et al., 2009). CIPK23, which is activated by CBL1 and CBL9 and is dephosphorylated by ABI2 (a member of the PP2C family), phosphorylates NRT1.1 under low-NO3– conditions, thereby converting it from a low- to high-affinity transporter (Sharma et al., 2017). In wheat, both TaCIPK8 and TaCIPK23 are upregulated under low-N conditions (Mahmoud et al., 2020). Overexpressing OsCIPK2 increases Ca2+ uptake, with plants exhibiting higher NO3– uptake and root and shoot growth in rice under low N stress (Khan et al., 2019). Taken together, these findings suggest that Ca2+ signaling pathways may be involved in a regulatory cascade for metabolic processes, including modification of root architecture, N uptake, and transport under N starvation, mainly via reversible protein phosphorylation.
An increasing number of TFs have been identified to be involved in the N-starvation response via transcriptional modulation of numerous N-responsive genes. TFs may be important signaling components and can modify root plasticity, increase N uptake, and promote N metabolism (Table 1). NF-Y TFs are induced or upregulated in the roots and shoots of wheat seedlings by low-N stress, increasing N uptake and grain yield (Qu et al., 2015; Curci et al., 2017). TaNFYA1-6B is a low nutrient-inducible TF on chromosome 6B that encodes one of the subunits of NF-Ys. TaNAC2-5A is a cereal-specific NAC (no apical meristem protein, Arabidopsis transcription activation factor, and CUp-shaped cotyledon) TF and can directly bind to the promoter regions of the genes TaNRT2.1-B1, TaNPF7.1-D1, and TaGS2-2A. Overexpression of TaNFYA1-6B and TaNAC2-5A TFs significantly stimulates lateral branching, enhances the expression of NRT1 and NRT2 families, and positively regulates TaGS2 expression and root growth, thereby promoting N uptake, N assimilation, tiller numbers, spikelet number, and grain yield in wheat under low-N conditions (He et al., 2015; Qu et al., 2015; Figure 1).
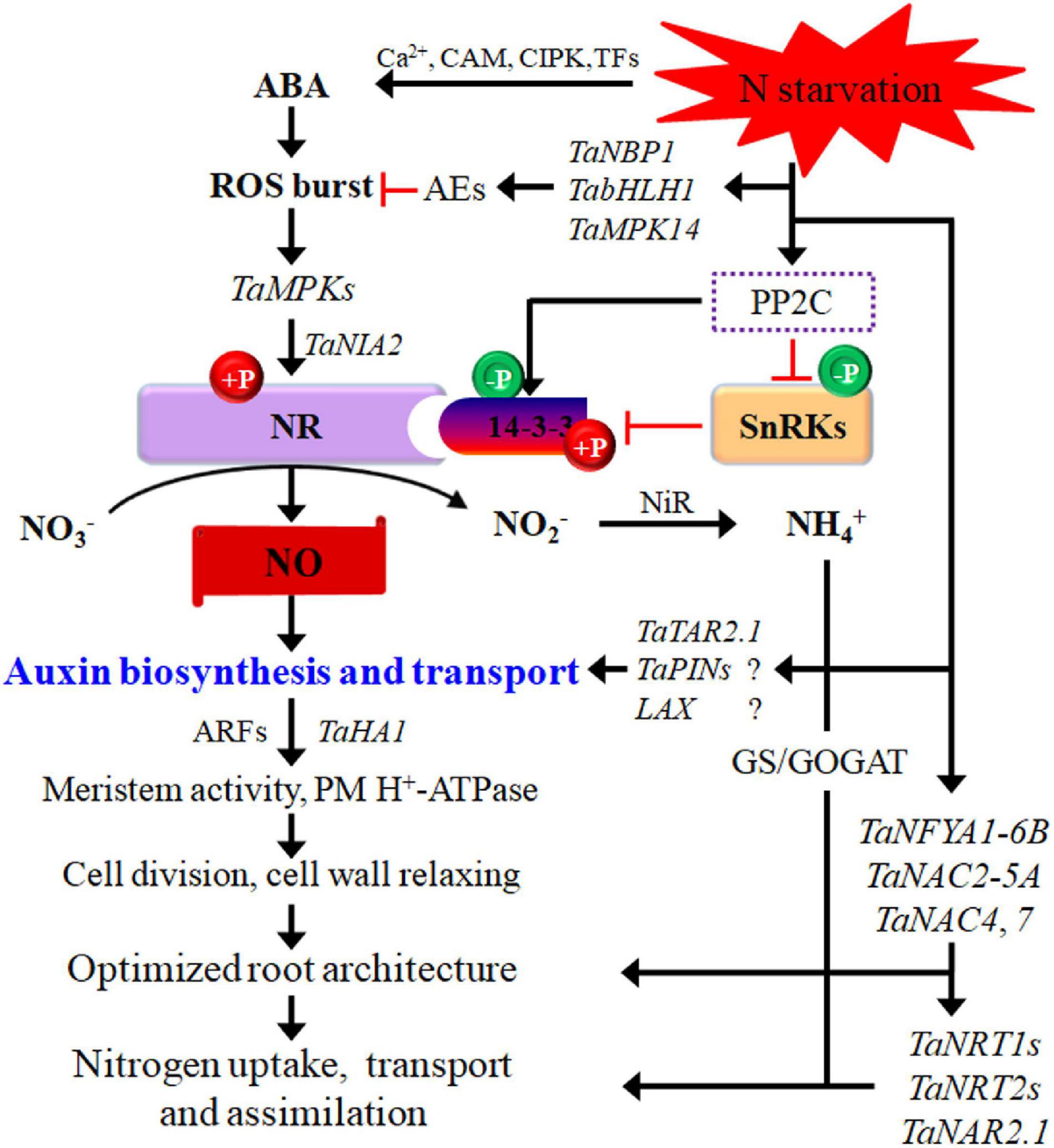
Figure 1. Potential pathways regulating wheat responses to low-N stress. N starvation interacts with phytohormones signaling. The interactions between them regulate root cell activity via ARFs and thus modify the root system architecture. Black arrows indicate positive regulation of N transport or assimilatory pathways, and red lines with short vertical columns indicate negative signaling steps. AEs, antioxidant enzymes; ARF, auxin responsive factors; bHLH, basic helix–loop–helix; GOGAT, glutamate synthase; GS, glutamine synthetase; MPK, mitogen-activated protein kinase; NBP, nucleotide-binding protein; NFYA, nuclear factor Y subunit A; NR, nitrate reductase; SnRKs, SNF1-related protein kinases; TAR, tryptophan aminotransferase-related. Question marks in the figure indicate that the important components in low-N signaling networks remain to be elucidated in wheat.
Most MYB genes are differentially (up- or downregulated) expressed in roots and leaves/stems of durum wheat under N starvation (Curci et al., 2017). In wheat, the expression of genes for MYB78 in leaves is significantly downregulated, while MYB33 in roots is significantly upregulated under low-N stress (Wang et al., 2019b). The newly identified miRNA ttu-novel-106 is immediately and strongly downregulated in roots after N stress and is negatively correlated with the expression of its putative target gene MYB-A (Zuluaga et al., 2018). These results suggest that the ttu-novel-106/TtMYB-A coupling in response to low-N stress may contribute to the adaptation or resistance of plants to low-N conditions. In the roots of OsMYB305-overexpressing rice lines, the expression of OsNRT2.1, OsNRT2.2, OsNAR2.1, and OsNiR2 is upregulated, and 15NO3– influx, tiller number, shoot dry weight, and total N content are significantly increased under low-N conditions (Wang et al., 2020a). MYB TFs are associated with cell development and the cell cycle in wheat under low-N stress (Zhang et al., 2012).
The MYB-related TFs bZIP77 and bZIP53 encoded by MRP2 are upregulated by low-N stress and may play a positive regulatory role in maize root growth under low-N stress (Ma et al., 2020). TabZIP60 is a wheat ABRE (ABA-responsive elements)-binding factor (ABF)-like leucine zipper TF; knockdown of TabZIP60 through RNAi promotes lateral root branching, increases N uptake, NADH-dependent glutamate synthase (NADH-GOGAT) activity, and spike number and improves grain yield under field conditions, while overexpression of TabZIP60-6D has shown the opposite effects (Yang et al., 2019a), indicating that this TF acts as a negative regulator of the NO3– response. Recently, TabZIP1 (an ortholog of AtbZIP16) and TaPIMP1 TFs have been found to be upregulated in both roots and shoots and identified as new players in the low-N response in wheat (Mahmoud et al., 2020).
TaZFP593;l is a C2H2-type gene encoding ZFP in wheat that bears a conserved C2H2 motif and targets the nucleus after sorting from the ER. TaZFP593;l plays a pivotal role in mediating RSA establishment, N acquisition, and cellular ROS homeostasis by transcriptionally regulating the genes associated with N starvation adaptation in wheat (Chen et al., 2017). Similarly, the transcription of TaZFP15 is induced in wheat roots by N deficiency, suggesting that TaZFP15 is involved in mediating low-N signal transduction (Sun et al., 2012).
WRKY genes have been reported to be signaling genes that respond to abiotic and biotic stresses, including low-N stress. The expression levels of WRKY TF genes are significantly upregulated in the roots of wheat under low-N stress (Wang et al., 2019b). However, most identified members of the WRKY family are downregulated in the roots of durum wheat in response to N chronic stress (Curci et al., 2017). Although there is still no direct evidence linking them to other components in regulatory networks, WRKY proteins may contribute to the establishment of complex signaling networks and thereby increase the N stress tolerance of plants (Curci et al., 2017).
DOF (DNA-binding with one finger) TFs are involved in various biological processes, including hormone signaling (Noguero et al., 2013). The DOF TF-encoding genes, particularly DOF1, act as crucial mediators of N starvation tolerance by regulating the external nitrate response and internal N assimilation (Yanagisawa et al., 2004). Expression of the DOF1.3 gene is upregulated in the roots of durum wheat plants when exposed to N starvation, and C2C2-Dof TFs are also upregulated in roots and shoot tissues (Curci et al., 2017).
In rice, the OsDOF18 gene can induce ammonium transporter family members (AMT1, AMT2, and AMT3), thereby promoting NH4+ uptake in roots (Wu et al., 2017). Transgenic expression of the ZmDOF1 gene increases N assimilation and plant growth under low-N conditions (Kurai et al., 2011). Furthermore, the expression of EcDof1 is accompanied by the expression of NR, GS and GOGAT in finger millet (Eleusine coracana L.), indicating that Dof1 probably regulates the expression of these genes (Gupta et al., 2009). On the other hand, overexpressing phosphoenolpyruvate carboxylase (PEPC) confers low-N tolerance and a higher grain yield by increasing carbon levels under low-N conditions (Tang et al., 2018). Overexpression of ZmDof1 in Arabidopsis increases the expression of PEPC and several genes involved in the tricarboxylic acid cycle, thereby producing more carbon skeletons for the assimilation of N (Yanagisawa et al., 2004). This view is strongly supported by the finding in rice that TFs increase the net photosynthesis rate and carbon flow toward N assimilation and thus improve N assimilation and plant growth under low-N conditions (Kurai et al., 2011).
RWP-RKs represent a small family of TFs that are unique to plants and function particularly under N starvation conditions. NLPs (NIN-like proteins, one subfamily of RWP-RKs) have been confirmed to regulate the tissue-specific expression of genes involved in NUE (Konishi and Yanagisawa, 2013; Chardin et al., 2014; Kumar et al., 2018). The expression of two TaNLP genes (TaNLP1 and TaNLP2) is induced by low-N conditions in mature leaves and is implicated in metabolic and regulatory processes associated with NUE (Konishi and Yanagisawa, 2013). The NLP7 gene (Kumar et al., 2018) and TaNLP4 (Mahmoud et al., 2020) are significantly upregulated in wheat under N-limited conditions. The TaNLP7 gene has already been characterized in Arabidopsis and wheat and regulates the expression of NRT1.1 and auxin signaling F-box (AFB3) genes (Castaings et al., 2009; Yu et al., 2016; Kumar et al., 2018). In wheat, NLPs may be involved in the nitrate signal transduction pathway, probably by functioning through their N-terminal regions that can bind specifically to nitrate-responsive elements (Kumar et al., 2018). In maize, TF ZmNLP6 is involved in modulating the early N response and root architecture through the mechanism of alternative splicing, rather than altering transcriptional abundance (Wang et al., 2020d).
Plant miRNAs are highly conserved non-coding small RNAs that are involved in regulating cell signaling mechanisms, plant growth, and development by depressing gene expression at the posttranscriptional level or via translational inhibition. The expression of miRNAs can be induced or suppressed by N deprivation (Paul et al., 2015). To date, a large number of miRNA families have been identified in the transcriptional response to N starvation in wheat (Zhao et al., 2013; Sinha et al., 2015; Gao et al., 2016). The interactions between low N-responsive miRNAs and their target genes may establish distinct miRNA-mediated pathways for regulating the plant response to N deprivation (Zhao et al., 2015; Figure 2 and Appendix Table 1). miRNA-guided regulatory networks should provide new tools for the genetic improvement of wheat NUE. However, the mechanisms underlying the miRNA-modulated adaptation to N starvation remain to be further characterized due to the complexity of modeling the miRNA-mediated stress response.
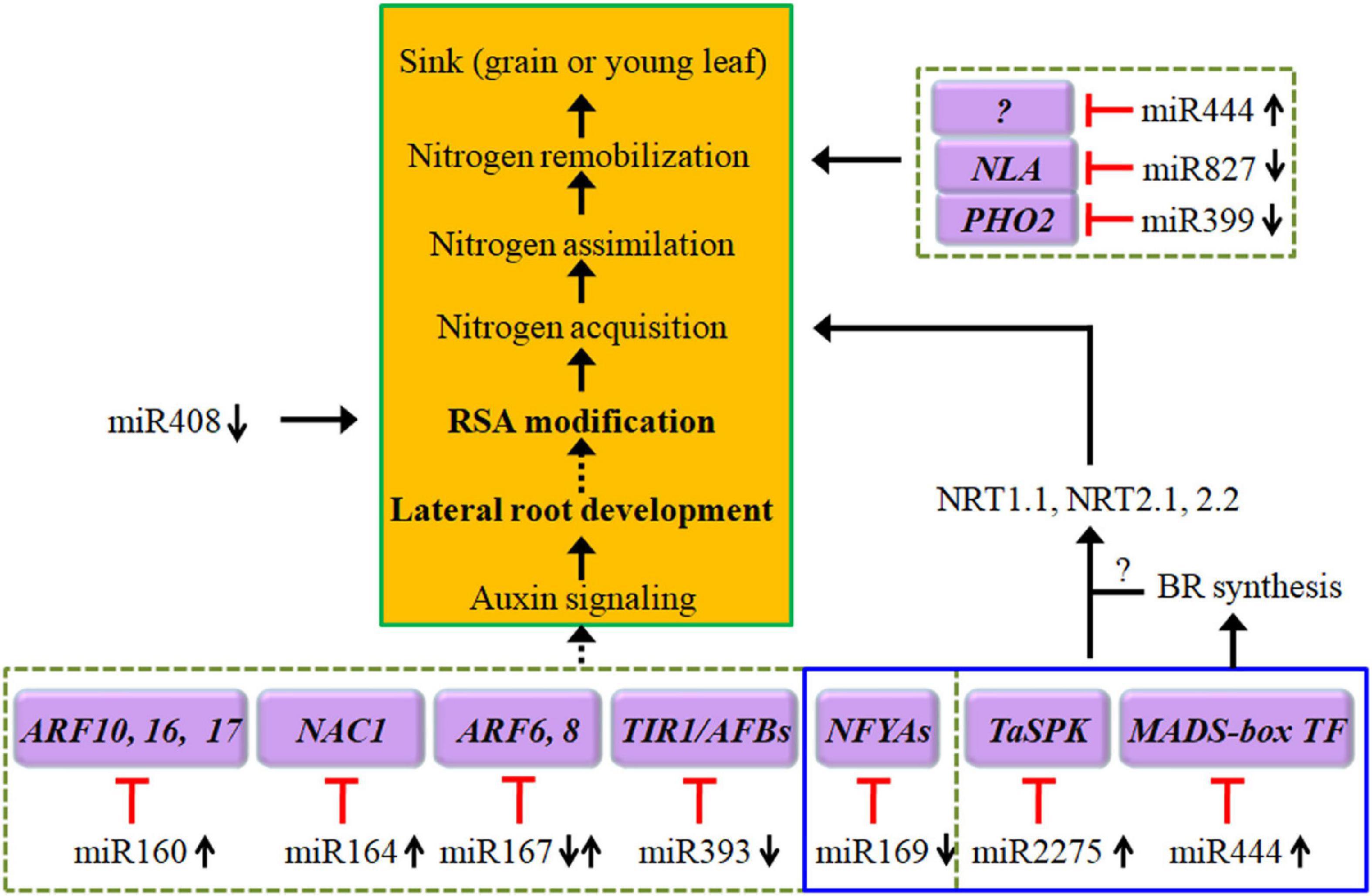
Figure 2. Diagram of low-N stress-induced alteration in the expression of a set of miRNAs that in turn regulate the expression of their targeted genes and then construct several regulatory pathways, including auxin signaling for root architecture modifications, expression of NRTs for N uptake and transport, and expression of NLA (nitrogen limitation adaptation) and PHO2 for N remobilization. The black arrows indicate positive signaling steps, the red lines with short vertical columns indicate negative signaling steps, and the dashed lines with solid arrowheads indicate putative regulation. See the text for further explanation. NFYA, nuclear factor Y, subunit A.
In the roots of wheat, miR1118 is N-responsive and plays a role in RSA development under N starvation (Sinha et al., 2015; Zhao et al., 2015). In addition to TamiR1118, TamiR1129 and TamiR1136 are upregulated, whereas TamiR1133 is downregulated in roots under N-deprived conditions (Zhao et al., 2015). The target genes of these miRNAs are listed in Appendix Table 1. Notably, some circRNAs are involved in the regulation of low-N-promoted root growth, and TamiR1118 and TamiR1133 can interact with circRNA2473 and thus regulate root elongation (Xu et al., 2019b).
miR159 is depressed in the grains of wheat under low N conditions (Hou et al., 2020). In root tissues, miR159a and miR159b are differentially expressed in response to N availability (Sinha et al., 2015). The N-responsive expression of miR159 has also been observed in the roots of maize (Zhao et al., 2012). Li et al. (2016) suggested that miR159 may function as a regulator of MYB and Teosinte branched1/cycloidea/proliferating cell factor (TCP) TFs and is associated with GA signaling by regulating the target GAMYB genes, thus influencing plant development.
In Arabidopsis, overexpression of miR160 promotes lateral root development, resulting in more lateral roots under N-limited conditions (Gifford et al., 2008; Liang et al., 2012). The miR160/ARF17 module may function in RSA formation through the auxin-related regulatory pathway (Li et al., 2016; Figure 2). In durum wheat, ttu-miR160 tends to be upregulated, while ARF22 (the ortholog of a T. aestivum gene similar to the Oryza sativa ARF22) is significantly downregulated in roots (Zuluaga et al., 2018). These results imply that N deficiency-induced expression of miR160, in combination with decreased ARFs, promotes lateral root production to access more N (Guo et al., 2005; Gifford et al., 2008; Liang et al., 2012). However, the expression of miR160 is only upregulated in roots in response to transient low-N conditions in maize (Xu et al., 2011) but is repressed by chronic N stress in wheat roots (Sinha et al., 2015). Moreover, several members of miR160 are downregulated in roots and shoots in rice (Shin et al., 2018) under both short-term and long-term low-N stress conditions. Therefore, the distinct roles of miR160/ARFs in response to low-N stress have not been clarified. In any case, some evidence has shown that overexpression of miR160 enhances lateral root development under N-limited conditions, which is believed to improve or boost the capability of plants to maximize the uptake of limited N availability (Gifford et al., 2008).
miR164 targets and cleaves the transcripts of NAC1 (a NAC domain-containing gene); this miR164/NAC module may function in the production of more lateral roots through the auxin-related regulatory pathway because NAC1 transduces auxin signals for lateral root emergence (Guo et al., 2005; Li et al., 2016; Figure 2). In the currently available literature, reduced expression of miR164 has been observed in wheat under chronic N stress (Sinha et al., 2015) and in durum wheat in short-term N stress, where ttu-miR164d is repressed corresponded to an upregulation of NAC7 (an ortholog of T. aestivum NAC7) gene (Zuluaga et al., 2018), which may be consistent with the findings in Arabidopsis that plants with downregulated expression of miR164 produce more lateral roots due to its role in regulating the expression of NAC TFs (Guo et al., 2005). miR164 can interact with another predicted target of ubiquitin family proteins that is involved in protein degradation and remobilization, thus conferring plants with high low-N tolerance (Nischal et al., 2012).
miR164 and miR167 are coordinately responsive to N deficiency in wheat, Arabidopsis, maize and rice. Decreased expressions of both miR164s and miR167s are closely associated with low-N tolerance in rice (Nischal et al., 2012). In Arabidopsis, miR164 is upregulated (Guo et al., 2005), but miR167 is downregulated (Liang et al., 2012; Paul et al., 2015) under N starvation conditions. Similarly, Xu et al. (2011) found that under long-term low-N conditions, miR164 is upregulated in leaves, while miR167 is downregulated in the roots of maize. Once again, in roots of durum wheat with a higher NUE, miR164d is strongly upregulated; conversely, miR167h is significantly downregulated under N stress (Zuluaga et al., 2017). These results suggest that a spatial miR164/miR167 complex may be responsible for specific RSA adaptation to low-N stress (Zuluaga et al., 2017). The downregulation of miR167 by N starvation positively regulates adventitious root growth via a higher expression of its target genes ARF6 and ARF8 in roots (Xu et al., 2011; Liang et al., 2012); the latter is induced in pericycle and lateral root cap cells under N-limiting conditions as the regulator of lateral roots (Gifford et al., 2008). However, the opposite results have also been observed; N starvation induces the expression of miR167a and represses the expression of ARF8, which positively mediates lateral root initiation in Arabidopsis, indicating that ARF6 and ARF8 are associated with the repression of lateral root development during NO3– limitation (Gifford et al., 2008; Figure 2). In the shoots and roots of maize, the expression of miR167s has also been reported to be upregulated by N deficiency (Zhao et al., 2012).
Relative to other miRNAs, such as miR160, miR164, and miR167, the expression of miR169 is consistently repressed in the grain of wheat (Hou et al., 2020) and in the roots and/or shoots of Arabidopsis, rice, and maize upon N starvation (Xu et al., 2011; Liang et al., 2012; Trevisan et al., 2012b; Zhao et al., 2012; Qu et al., 2015; Yang et al., 2019b). The transcriptional repression of miR169s upon NO3– starvation is a crucial step in integrating nitrate signals into metabolic, physiological, and morphological adaptations in maize roots (Trevisan et al., 2012b). Importantly, miR169 may function as a potential long-distance signal to regulate N starvation responses because a sharp decrease in miR169 in phloem sap has been observed in oilseed rape (Brassica napus) under N-limiting conditions (Pant et al., 2009). This long distance signaling occurring through the vasculature may play a fundamental role in maintaining nutrient homeostasis across the whole plant.
In wheat and rice under low-N stress, the miR169 family is downregulated, while nuclear factor Y subunit A (NFYA), the target of miR169, is upregulated (Zhao et al., 2011; Qu et al., 2015; Shin et al., 2018). In contrast, the constitutive expression of miR169a in transgenic Arabidopsis suppresses the accumulation of NFYA family members and decreases the expression of AtNRT1.1 and AtNRT2.1, and transgenic plants accumulate less N and are more sensitive to N stress than wild-type plants, which may be attributed to impaired N uptake systems (Zhao et al., 2011; Figure 2). Overexpressing TaNFYA-B1, a low-N-inducible NFYA TF, significantly increases N uptake and grain yield, likely by upregulating auxin biosynthetic genes and the expression of N transporters in the roots and by stimulating root development (Qu et al., 2015). Furthermore, determination of the biological function of these targets using gene ontology (GO) analysis revealed that miR169/targets may also be involved in the regulation of N metabolism (Yang et al., 2019b). These results suggest that miR169 and NFYA family members may control a complex regulatory network.
Target genes of wheat TamiR2275 encode proteins involved in various biological processes, including signal transduction, transcriptional regulation, and stress response. miR2275 is gradually upregulated upon N starvation and thus positively regulates plant tolerance to low-N stress through transcriptional regulation of target genes, conferring plants improved N acquisition, better phenotype, increased biomass, and enhanced photosynthetic function (Zuluaga et al., 2017; Qiao et al., 2018; Figure 2).
miR393 targets transcripts encoding the bHLH TF (bHLH77), and the auxin receptors transport TIR1 (inhibitor response protein 1), AFB1, AFB2, and AFB3 (Jones-Rhoades and Bartel, 2004; Vidal et al., 2010; Chen et al., 2011). In durum wheat, miR393c is downregulated, and the inverse correlation for miRNA393/AFB2 has been observed in roots, leaves, and stems (Zuluaga et al., 2017). Another module, miR393/AFB3, also contributes to plant developmental plasticity in response to N stress via lateral root initiation and primary root elongation (Vidal et al., 2010; Tang et al., 2018; Figure 2).
The expression of miR399s is downregulated in the roots and/or grains of wheat (Sinha et al., 2015; Hou et al., 2020), durum wheat (Zuluaga et al., 2017), and Arabidopsis (Liang et al., 2012) upon N starvation. However, in the roots of rice, miR399i is upregulated by N stress and has been identified as a regulator of plant nutrient homeostasis (Cai et al., 2012). In maize, miR399 is repressed in response to chronic low NO3– conditions, while it is upregulated in response to transient low N stress (Xu et al., 2011).
Under N stress, the downregulated miR399 shows an inverse correlation with its target PHO2 gene (Zuluaga et al., 2017), which encodes a ubiquitin-conjugating enzyme and is involved in the protein degradation pathway (Cai et al., 2012). This miRNA/target interaction may enhance proteasome-mediated N remobilization of putative targets, such as Rubisco, and play an important role in the adaptation of durum wheat to a low-N environment (Zuluaga et al., 2017).
In root tissues of wheat, miR408 is differentially expressed in response to N availability (Sinha et al., 2015). Upon N starvation, the expression of miR408 is repressed in the roots/shoots of maize (Xu et al., 2011; Yang et al., 2019b) and Arabidopsis (Liang et al., 2012; Zhao et al., 2012). GO analysis revealed that miR408 may be involved in the regulation of N metabolism; in addition, decreased expression of this microRNA is also involved in ROS scavenging through its target gene encoding SOD1A (Yang et al., 2019b). In maize, miR408/b is preferentially expressed in root tips, epidermal cells of primary roots, and lateral root primordia (Trevisan et al., 2012a,b). These authors proposed that the repression of the transcription of miR408/b is crucial for integrating nitrate signals into developmental regulation and the adjustment of root architecture; during this process, ROS signaling may mediate this transduction pathway in response to an N shortage (Figure 2).
miR444 is a critical regulator mediating plant tolerance to low-N stress. In wheat roots and/or leaves, miR444 is upregulated, whereas the transcription of their target genes, such as the MIKC-type MADS-box TFs WM32B and WM30, shows the opposite expression patterns in the above tissues (Zhao et al., 2015; Gao et al., 2016; Figure 2). In particular, TamiR444a transcriptionally regulates a large number of genes, including NRT1.1-s, NET1.1-t, NRT2.1, and antioxidant enzyme-encoding genes, including CAT1;1, POD1;3, and POD4, thus establishing a complicated gene network, which is associated with signaling perception and transduction, N acquisition through the modulation of NRT genes, cellular ROS homeostasis, phytohormone response, and plant growth (Gao et al., 2016; Figure 2).
However, in durum wheat, the upregulation of miR444 in response to low N is only observed in roots of one of two experimental varieties (Zuluaga et al., 2017). In rice, miR444 is only slightly upregulated by N starvation, but the levels of its three targets (MADS23, MADS27a, and MADS57) are decreased significantly (Yan et al., 2014). These authors suggested that miR444 modulates N homeostasis by mediating ANR1-like MADS-box FTs; however, they then found that upregulation of miR444a reduced the adaptability of rice plants to N-limiting conditions by reducing N remobilization from old leaves to young leaves (Yan et al., 2014; Figure 2). Moreover, the root-specific expression of osa-miR444a.4-3p is downregulated in response to N starvation, and OsMADS25 (a novel target gene) has been shown to be targeted by osa-miR444a.4-3p; therefore, the miR444a/MADS25 module may be involved in N acquisition and homeostasis in roots (Shin et al., 2018). Considering these conflicting results, we have to mention the most recent findings in rice roots in which NH4+ promotes the biosynthesis of brassinosteroid (BR) through miR444 to regulate rice root growth. During this process, miR444 positively regulates BR biosynthesis through its MADS-box targets, resulting in decreased root elongation (Jiao et al., 2020).
Generally, miR827 is downregulated in roots under N starvation in durum wheat (ttu-miR827a), Arabidopsis, and maize (Liang et al., 2012; Zhao et al., 2012; Zuluaga et al., 2017; Yang et al., 2019b). In contrast to miR827 under low-N conditions, the expression of its target NLA gene increases in Arabidopsis (Pant et al., 2009; Liang et al., 2012) and maize (Yang et al., 2019b). GO analysis revealed that miR827 may be involved in the regulation of N metabolism by regulating the expression of the NLA gene, which encodes a RING-type ubiquitin ligase and has been identified as an essential component in adaptive responses to N limitation, as confirmed using an NLA mutation (Peng et al., 2007; Yang et al., 2019b; Figure 2).
Conversely, in the shoots or roots of oilseed rape, the expression of miR827 is upregulated by N limitations, opposite to the expression of NLA1s (Zhang et al., 2018). Further study showed that the miR827-NLA1-NRT1.7 regulatory circuit functions as a pivotal pathway involving the adaptive responses of plants to N limitations (Zhang et al., 2018). NLA negatively regulates NRT1.7 through the protein ubiquitination pathway to meet the N requirements of plants (Liu et al., 2017). In the leaves of maize, miR827 is downregulated by short-term N deficiency but upregulated under long-term N deficiency (Xu et al., 2011). The expression of miR827 is upregulated at 12 h after low-N treatment, followed by a trend to decrease; thus, the predicted target gene CLP (ATP-dependent Clp protease ATP-binding subunit) shows an increase in expression at 48 h (Zuluaga et al., 2018).
In addition, a large number of miRNAs that mediate N metabolism pathways in other plants, such as miR854 (regulating ethylene-responsive transcription factor), miR2630, and miR1074 (both LRR receptor-like serine/threonine-protein kinase), have not been investigated thus far in wheat.
Plants have two types of nitrate uptake systems: a low-affinity transport system (LATS) when NO3– availability is > 0.5 mM and a high-affinity transport system (HATS) when external NO3– availability is < 0.5 mM (Wang et al., 2012). Under low-N conditions, plants trigger constitutive and inducible expression of HATS to promote N uptake (Wang et al., 2012). In wheat, a large number of NRT and AMT (encoding ammonium transporter) genes are orthologous to those of Arabidopsis, barley, maize, and rice (Bajgain et al., 2018). TaNRT1, TaNRT2 (Guo et al., 2014), and TaAMTs (Li et al., 2017) are closely associated with N starvation tolerance. TaNRT2.1 plays a major role in NO3– uptake (Taulemesse et al., 2015). The expression of the TaNRT2.1 gene is invariably upregulated at the early stage (14 d) of NO3– starvation; however, it is subsequently reduced (Sinha et al., 2020). Similarly, transcription of the TaNRT2.2 gene is rapidly induced by low-N stress (Melino et al., 2015), but its level rapidly declines (Melino et al., 2018). Low N induces more accumulation of NRT2.5 and NAR2.1 (a partner protein of NRT2 proteins), which enhance N uptake, transport, and remobilization and are also involved in root growth in wheat (Guo et al., 2014; Xu et al., 2019a; Wang et al., 2020c). TaNRT1.1, TaNRT2.1, TaNRT2.2, and TaNAR2.1 are upregulated under low NO3– conditions to increase NO3– uptake and are closely associated with varietal differences in NUE (Jiang et al., 2017; Figure 1). In maize under N deficiency stress, the expressions of ZmNRT2.1, ZmNRT2.2, and ZmNRT2.5 are significantly increased, and ZmAMT1.3 and ZmAMT3.3 do not respond to N starvation in roots and/or shoots (Dechorgnat et al., 2019). NRT2.2, AMT2, GLNs, NR1, and NR2 are upregulated to increase N uptake and facilitate N assimilation, thereby stimulating plant growth (Ma et al., 2020). BdNRT2 gene expression is also governed by both internal and external N status (Wang et al., 2019a). BdNRT2/3 genes are potentially involved in HATS for root NO3– uptake in Brachypodium distachyon (David et al., 2019). In the bdnrt2.1 mutant, HATS have been shown to be reduced by 30% (Wang et al., 2019a). NRT2 is more evolutionarily conserved, and a conserved expression pattern of NRT2 genes between species has been observed, suggesting that functional conservation may exist between species (Dechorgnat et al., 2019; Wang et al., 2019c).
The expression of NRT2/NAR genes in wheat under limited NO3– supply is regulated by ABA signaling, contributing to the optimization of NO3– uptake (Wang et al., 2020c). TaNBP1, a guanine nucleotide-binding protein subunit beta gene, improves RSA establishment, N acquisition, and cellular reactive oxygen species (ROS) homeostasis through transcriptional regulation of NtNRT2.2 and distinctive antioxidant enzyme genes, such as NtSOD1, NtSOD2, and NtCAT1, resulting in a higher N-starvation tolerance for wheat (Liu et al., 2018). Basic helix–loop–helix (bHLH) TFs comprise a large TF family, act as crucial regulators in various biological processes in plants, and are involved in stress signaling. TabHLH1, a wheat bHLH TF member, is upregulated after N and phosphorus deprivation in roots, leaves, and/or shoots and plays an important role in transcriptionally regulating genes encoding NRTs and antioxidant enzymes that mediate cellular ROS homeostasis under nutrient stresses (Yang et al., 2016). BIM2 (encoding a bHLH type) is highly upregulated under N stress, and alternate splicing in this gene may play an important role in N uptake through the modulation of root architecture (Subudhi et al., 2020).
NPFs (nitrate transporter 1/peptide transporters) are also transporters that are mainly involved in NO3– uptake. In Arabidopsis, AtNPF6.3 acts as a low-affinity transporter when the NO3– level > 1 mM switches to a high-affinity mode when the nitrate level < 1 mM due to the function of CBL-interacting protein kinase (CIPK) 23 to phosphorylate intracellular threonine (Liu and Tsay, 2003). In wheat, TaNPF6.1, TaNPF6.2, and TaNPF6.3 are identified as co-orthologous to Arabidopsis AtNPF6.3 (Buchner and Hawkesford, 2014). Recently, alternative splicing has been considered a pattern that increases proteins and enriches the function of a gene. In rice, two variants, OsNPF7.7-1 and OsNPF7.7-1, have been identified, and the expression of OsNPF7.7s was relatively higher in the roots or axillary buds upon low-N treatment than upon high-N treatment; altered expression of each variant could regulate shoot branching and nitrogen utilization efficiency (NUtE) and improve NO3– or NH4+ in roots (Huang et al., 2018). However, ZmNPF6.6 expression is downregulated, and ZmNPF6.4, ZmAMT1.3, and ZmAMT3.3 show no response to N starvation (Dechorgnat et al., 2019).
Recently, a large number of NRT/NPF genes have been identified and are heterogeneous in terms of their gene structures and mRNA abundance (Wang et al., 2019c); in addition to NO3– and peptides, NPFs are also involved in plant hormone transport (auxin, ABA, jasmonates, and GAs) (Wang et al., 2020b), indicating that NPF and hormone signals may act as a component of regulatory networks in response to low-N stress. Therefore, the systematic identification of gene composition, chromosomal locations, evolutionary relationships, and expression profiles contributes to a better understanding of the roles of NPF genes in wheat (Wang et al., 2020b).
Amino acid transporters (AATs) play an important role in amino acid uptake and remobilization from vegetative tissues to the grain. AATs are highly expressed in roots and in senescing leaves and stems, suggesting that AATs are good candidates for high NUE (Wan et al., 2017). However, the role of AATs in the wheat plant response to N starvation has not been elucidated.
As discussed above, under low-NO3– conditions, PM H+-ATPase activity increases in wheat roots due to the upregulated expression of TaHA1 (Jiang et al., 2017; Lv et al., 2021; Figure 1). In addition to the acid promotion of root growth, low-N stress has been shown to induce a greater PM H+-ATPase transcript abundance and increase the energy supply for N uptake (Jiang et al., 2017).
GS assimilates ammonia and glutamate to form glutamine, and ferredoxin-dependent GOGAT catalyzes the synthesis of glutamate from glutamine and 2-oxoglutarate through transamidation reactions. In wheat, low N induces the activities of both enzymes in roots and leaves, not only to promote ammonia assimilation but also to achieve osmotic homeostasis via glutamate-based synthesis of proline (Kichey et al., 2005; Yousuf et al., 2017; Lv et al., 2021). TaNADH-GOGAT and its interaction with TabZIP60 play important roles in mediating N use and wheat growth (Yang et al., 2019a). The increased activities of N assimilation enzymes may be a signal for the promotion of N uptake by plant roots (Jiang et al., 2017). As discussed above, in wheat and rice, nitric oxide (NO) plays a pivotal signaling role not only in improving the N acquisition capacity and N transport by promoting lateral root initiation (Sun et al., 2015) but also in N assimilation by transcriptional and posttranscriptional regulation of the activity of N assimilation enzymes (NR, GS) under low N conditions (Balotf et al., 2018; Tang et al., 2018).
Autophagy is an evolutionarily conserved biological process in all eukaryotes for the degradation of intracellular components for nutrient (mainly N) recycling, thereby playing an important role in N remobilization and seed filling (Guiboileau et al., 2012).
In wheat, TaATG4a (an autophagy-related gene) shows higher transcript abundance under N deficiency (Pei et al., 2014). N starvation has been shown to enhance the expression of most of the autophagy genes tested and is involved in the regulation of N metabolism (Bedu et al., 2020), suggesting that autophagy is regulated at the transcriptomic level by N status signals. In maize, N deficiency-induced autophagy is critical during N stress and severely impacts productivity, while atg12 mutants show reduced grain yield because of impaired N recycling (Li et al., 2015a). In foxtail millet, SiATG8a is localized in the membrane and cytoplasm, and its transcriptional level is dramatically induced by N starvation (Li et al., 2015b). SiATG8a transgenic plants have shown larger root and leaf areas and accumulated more total N than wild-type plants under N starvation conditions (Li et al., 2015b). The expression of OsATG8a, OsATG8b, and OsATG8c is significantly increased under N-deficient conditions and therefore enhances the activity of autophagy, N uptake, NUE, N uptake efficiency, and yield in rice and Arabidopsis (Xia et al., 2012; Yu et al., 2019; Zhen et al., 2019a,b; Fan et al., 2020). Increasing the expression of the ATG8, AtAtg8f, and MdATG8i genes in Arabidopsis increases autophagosome number; promotes autophagy activity, N remobilization efficiency, and grain filling; and confers tolerance to N limitation (Slavikova et al., 2008; Wang et al., 2016; Chen et al., 2019). Based on these findings, ATG8 is considered a great candidate gene to increase NUE and grain yield in cereals (Yu et al., 2019).
In addition, overexpression of MdATG18a upregulates NRT2.1/2.4/2.5 and NIA2 genes and NO3– uptake and assimilation and therefore enhances N deficiency tolerance and plant growth in both Arabidopsis and apple (Sun et al., 2018c). To date, no published study has reported that ATG8s are directly related to low-N tolerance in wheat; however, Yue et al. (2018) observed that the TaAtg8 subfamily plays a crucial role in wheat tissue autophagy and stress defense, indicating that TaAtg8 may possibly function in response to low-N stress in wheat. Regardless, it has been demonstrated in wheat that autophagy is involved in the regulation of low-N-induced promotion of root growth (Xu et al., 2019a), likely due to the regulation of oxidative stress (Masclaux-Daubresse, 2016) and N remobilization from shoots to roots.
Hormone signaling may be involved in N starvation-induced autophagy (Masclaux-Daubresse, 2016). TaATG promoter regions contain putative IAA and ARF elements and abiotic stress-related transcription elements, implying cross talk between autophagy and hormonal signaling, and the interactions between stresses and hormone signaling may coordinately regulate TaAtg gene expression in plants upon stress response (Yue et al., 2018; Liao and Bassham, 2020). In tomato (Solanum lycopersicum), the transcription of ATGs and the formation of autophagosomes are triggered by enhanced levels of BRs and involve BR-activated TF brassinazole-resistant1 (BZR1), which in turn enhances the degradation of denatured and unfolded proteins by autophagy, suggesting that BZR1-dependent BR signaling upregulates the expression of ATGs and autophagosome formation, which plays a critical role in the plant response to N starvation (Wang et al., 2019d).
Plant roots are the first organ to sense the external nutrient status and therefore can immediately respond to N starvation signaling, developing an optimized RSA to enhance the acquisition of limited N nutrients (Sinha et al., 2015). The total root size consistently increased in all wheat genotypes that were examined due to nitrate (NO3–) starvation, although the shoot fresh weight decreased (Sinha et al., 2015, 2020). In wheat, the number of lateral roots, total root area, and root fresh mass significantly increased, root diameter significantly decreased, and the shoot fresh weight showed no difference at 48 h of exposure to low-N treatment (Lv et al., 2021). Interestingly, low NO3– had a greater influence on RSA development than low NH4+ (Lv et al., 2021). In maize under long-term N stress (40 d), the length and surface area of the primary root and the average length of the seminal root-derived lateral root increased, although the whole root fresh weight decreased (Ma et al., 2020). In Arabidopsis, induction of lateral root growth by N deprivation is a “foraging” mechanism used by the plant to capture more N (Gruber et al., 2013). Therefore, optimized RSA is undoubtedly considered a morphological response of plants to N deficiency; however, to date, the detailed mechanisms by which plants perceive and transduce N stress signaling and form morphological adaptations remain largely unknown in wheat (Xu et al., 2019a).
A number of studies on wheat, rice, and maize have revealed that N starvation rapidly induces an ROS burst, which in turn functions as a component of plant signal transduction cascades in response to N limitation to induce antioxidant defense systems and adjust root architecture, thus facilitating the exploration of new regions of the soil to find additional nutrient elements (Lian et al., 2006; Trevisan et al., 2012a; Hsieh et al., 2018; Singh et al., 2018; Figure 1). However, with increasing time of N-starvation stress, the ROS content in plant tissues is negatively associated with tolerance to low-N stress (Tang et al., 2018), and the ROS burst triggers the expression of numerous genes to construct a transduction pathway, including increases in the gene expression of antioxidant enzymes, such as superoxide dismutase (SOD), peroxidase (POD), and CAT (Shin et al., 2005; Wang et al., 2019b; Yang et al., 2019b). Interestingly, miR398 and miR408, which both target [Cu-Zn]4AP/SOD, are downregulated after N stress, implying that the ROS signal and homeostasis due to the regulation of antioxidant enzyme expression might adapt to N stress (Trevisan et al., 2012a; Yang et al., 2019b). Similarly, in wheat, miR398 is downregulated under K deficiency and regulates SOD activity (Zhao et al., 2020). Additionally, in wheat, biosynthesis inhibition of miR398 in roots increases SOD activity in response to oxidative toxicity (Li et al., 2020). Increased antioxidants, such as glutathione and ascorbic acid, contribute to ROS homeostasis to maintain root development in barley (Hordeum vulgare); however, ROS are elevated in shoots, leading to decreased shoot dry biomass (Kováčik et al., 2014).
Protein kinases and phosphatases are well-known regulatory proteins involved in various signal transduction pathways. Many genes for protein kinases and phosphatases are upregulated or downregulated by low-N stress (Lian et al., 2006). In particular, the MAPK cascade is an evolutionarily conserved signal transduction module in plants. The MAPK signaling pathway is involved in the wheat plant response to low-N stress (Wen et al., 2015; Sultana et al., 2020; Figure 1). TaMPK14, a MAPK family gene in wheat, is characterized for its role in mediating the N starvation response. Compared with the wild type, the TaMPK14-overexpressing lines display higher antioxidant enzyme activities, improved cellular ROS homeostasis, and N accumulation under N deficiency, indicating the crucial roles of the MAPK gene in mediating the N starvation response (Shi et al., 2020; Figure 1). In durum wheat, a large number of protein kinases are differentially regulated by N starvation during grain filling, mainly including kinases functioning in MAPK cascades, tyrosine kinase-like kinases and those carrying leucine-rich repeat (LRR) domains, PKA–PKG–PKC, calcium- and calmodulin-regulated kinases, and CMGC (cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinase, and cyclin-dependent kinases) (Curci et al., 2017). Consequently, critical kinases and phosphatases that are involved in the hormone signaling pathway and regulate RSA plasticity and the expression of NRTs, AMTs, N assimilation, and autophagy need to be identified.
NO is a small redox signaling molecule in plant cells that regulates plant responses to biotic and abiotic stresses, even though the complete scenario of NO generation in plants is still elusive. High homogeneity is observed during changes in ROS and NO; namely, both are simultaneously elevated in shoots, and both show no changes in the roots of barley (Kováčik et al., 2014). In Arabidopsis, NO production is promoted by ROS via the enhanced accumulation of mitogen-activated protein kinase (MPK) 6 during lateral root development, and NIA2 (encoding one nitrate reductase (NR) isoform) is required for the MAPK6-mediated production of NO (Wang et al., 2010). In this process, NIA2 interacts directly with MPK6 and can be phosphorylated by MPK6, thus leading to an increase in NR activity and NO production and thereby regulating root development (Wang et al., 2010). Furthermore, in rice, N deficiency-induced NO production involves an NIA2-dependent NR pathway (Sun et al., 2015, 2016). Considering that TaNIA2 is also upregulated under low-N stress (Mahmoud et al., 2020), and TaMPK6 (the homologous gene of the Arabidopsis MPK6 gene in wheat) is involved in the NR-catalyzed NO generation in wheat roots (Zhang et al., 2013), we reasonably speculate that NIA2 may play a vital role in MPK6 regulated NO generation under low-N stress (Figure 1).
On the other hand, PP2C9, a protein phosphatase, enhances NR activation by downregulating SnRK1 and 14-3-3 proteins, influencing N uptake and assimilation in rice under low-N stress; overexpression of PP2C9 promotes NO biosynthesis by NR, which is activated via dephosphorylation of 14-3-3 and SnRK (Waqas et al., 2018). In wheat, WPK4, a SnRK, has been found to inactivate NR by assembling the NR and 14-3-3 complex through its phosphorylation specificity (Ikeda et al., 2000), suggesting that WPKs may be responsible for controlling N metabolism via the NO signaling pathway. As a consequence, NO generated via the NR pathway plays a pivotal role in increasing lateral root initiation, root branching, and the N uptake rate, thus improving the N acquisition capacity in wheat and rice under low-N stress (Sun et al., 2015; Balotf et al., 2018; Waqas et al., 2018; Figure 1). Considering that both NO and low NO3– are involved in root growth modulation and are metabolically connected, it is reasonable to postulate that NO may be an important signal participating in low-N signaling and mediating plant adaptation.
A study demonstrated that low-N-induced NO in root tips are involved in cross talk with strigolactones (SLs), leading to the elongation of seminal roots via induction of meristem cell activity in rice (Sun et al., 2016). A subsequent study revealed that auxin acts as a downstream regulator of NO and SL signals to induce meristem activity in root tips in rice under low N, and OsPIN1b, encoding an auxin efflux protein, is involved in auxin transport (Sun et al., 2018a; Figure 1).
Plant hormones are important components of signal transduction for N stress (Sultana et al., 2020). A large body of studies has demonstrated the existence of interactions between N nutrition and auxin signaling and revealed the roles of auxin polar transport and signal transduction in the regulation of RSA modifications in response to N availability. Low-N conditions induce the biosynthesis of auxin in the roots of Arabidopsis seedlings (Kiba et al., 2011) and wheat (Lv et al., 2021). Pyridoxal phosphate-dependent transferase, an important coenzyme for catalyzing transamination of L-tryptophan for auxin biosynthesis, is upregulated in roots of durum wheat (T. turgidum ssp. Durum) under N starvation; accordingly, auxin-responsive factors (ARF2 and ARF18) are also upregulated in the same tissues (Curci et al., 2017). In Arabidopsis, the tryptophan aminotransferase-related gene (TAR2) is induced by low N conditions, and this gene functions in the tryptophan-dependent pathway of auxin biosynthesis and is expressed in the pericycle and the vasculature of the mature root zone near the root tip (Ma et al., 2014). In wheat, TaTAR2.1 is upregulated by low-N availability and expressed mainly in roots. Overexpressing TaTAR2.1-3A enhances the auxin content in the root tip and promotes lateral root branching and shoot N accumulation (Shao et al., 2017). These results suggest that the interaction between TAR2 and auxin, as well as enhanced auxin biosynthesis, is required for modifying RSA in response to low-N conditions.
In maize and Arabidopsis, low-N availability enhances shoot-to-root auxin transport and auxin accumulation, which induces lateral root growth and RSA development via auxin-dependent acid growth and the auxin-regulated target of the rapamycin pathway (Tian et al., 2008; Asim et al., 2020; Sun et al., 2020b). OsPIN1b, an auxin efflux carrier gene, is significantly downregulated in rice root under low-N stress (Sun et al., 2018a). In maize, ZmPINY is also downregulated, and the ZmLAX4 auxin influx carrier gene is induced in roots by N starvation; accordingly, a large proportion of the downstream auxin response genes are upregulated, implying that auxin polar transport and flux may play an important role in the reprogramming of the root architecture of plants under N deficiency (Ma et al., 2020). In wheat, expression of TaPIN1, TaPIN2, TaPIN3, and TaPIN4 is reduced in seedlings under low-Pi availability, thus stimulating lateral root initiation (Talboys et al., 2014). Assay using a non-invasive micro-test technology showed that the rate of IAA influx at the root surface significantly increases at early hours of low-N stress; as a consequence, the enhanced root IAA concentration stimulates the root branching and elongation (Lv et al., 2021). However, the roles of TaPINs and LAX genes in wheat response to low-N stress remain to be elucidated.
In contrast, Maghiaoui et al. (2020) proposed that low-NO3– stress represses LAX3 and TAR2 gene expression through NRT1.1, which acts as a negative regulator of the auxin biosynthetic gene expression, thus preventing the growth of lateral root primordia. However, researches are needed to understand how N affects the underlying auxin signaling processes in governing RSA, especially in wheat.
Under low-N conditions, significantly increased lateral root growth is accompanied by enhanced expression of plasma membrane (PM) H+-ATPase AHA2 (Młodzińska et al., 2015). In this scenario, H+-ATPase activity is probably regulated by endogenous auxin transport because treatment with 2,3,5-triiodobenzoic acid, an inhibitor of the polar transport of auxin, significantly decreases the H+-ATPase activity in the root cells and inhibits low-N-induced lateral root initiation and growth (Sun et al., 2020b; Lv et al., 2021), suggesting that the increased auxin level may enhance root elongation by acidifying the root cell wall and enhancing cell elongation in response to the low-N supply, which is crucial for the development of the root architecture and the plant to explore a greater space for N resources.
NRT1.1, a nitrate transporter, has a role in signaling and promotes auxin transport from the lateral root tip toward the shoot to prevent auxin accumulation in lateral root primordia and young lateral roots under low NO3– levels (Zhang et al., 2007; Gojon et al., 2011), thus reducing lateral root development (Krouk et al., 2010). In contrast, NRT2.1, a high-affinity nitrate transporter, facilitates NO3– transport across the plasma membrane and acts as a positive regulator of lateral root initiation under low-N conditions in Arabidopsis (Remans et al., 2006). Whether this process is related to auxin polar transport is still unknown. Additionally, Loqué et al. (2006) observed that the ammonia transporter gene AMT1.1 and AMT1;3 play a critical role in restructuring lateral root architecture in Arabidopsis under N starvation.
A low NO3– (< 1.0 mM) supply stimulates lateral root elongation, while a high level of NO3– inhibits root development, where ABA plays an important role in mediating the effects of NO3– on lateral root formation (Signora et al., 2001). Wheat zeaxanthin epoxidase, an important ABA synthesis-related enzyme, is significantly upregulated, and the ABA contents are significantly enhanced in the roots and leaves of wheat under low-N stress, suggesting a role of ABA in the tolerance of plants to N deficiency (Kang et al., 2019; Mahmoud et al., 2020). Kang et al. (2019) proposed a putative ABA-dependent schematic model in higher plants under N deficiency. Plant cells can perceive external N deficiency in the cell membrane, and this signaling is subsequently transduced by Ca2+ sensors, kinases, or other signaling components, which then induce translational changes in many functional proteins related to ABA. ABA and other phytohormones then regulate downstream posttranscriptional responses, followed by morphological and physiological adaptive changes that assist survival under N-starvation stress (Kang et al., 2019; Figure 1). However, Ma et al. (2020) reported that all ABA-related genes are downregulated in maize under low-N stress. ABA receptor PYL2 is also downregulated by low-N treatment in roots, which is accompanied by longer primary roots (Xu et al., 2019a).
In rice, RPN10 (26S proteasome regulatory particle non-ATPase subunit 10) regulates plant sensitivity to both auxin and ABA, which is repressed by low-N treatment (Ding et al., 2011), whereas the expression of key TF genes for ABA and auxin is rapidly induced by N starvation (Hsieh et al., 2018). These results suggest that the interactions between phytohormone signals may be involved in regulating the responses to low-N stress. A persuasive argument has been proposed that the interactions between auxin and ABA induce cell division and elongation and thereby promote primary root growth (Hsieh et al., 2018; Figure 1).
Cytokinins are also closely associated with N deficiency signaling, act as both local and systemic signals, and participate in the regulation of RSA modifications and N acquisition under N-deficient conditions (Kiba et al., 2011; Lv et al., 2021). Two genes encoding cytokinin dehydrogenase are significantly downregulated by N deficiency as a strategic response to N deficiency stress (Shao et al., 2020). Adenosine kinase plays an important role in the conversion of cytokinins toward nucleotides (Kwade et al., 2005). In response to low-N stress, adenosine kinase is downregulated, suggesting that adenosine kinase may be involved in cytokinin signaling to regulate low-N tolerance (Ding et al., 2011).
Recently, Xiong et al. (2019) observed that the mutation of gibberellin (GA)-responsive protein genes results in resistance to low-N stress in wheat. The level of GA3 is significantly increased under low-N stress (Lv et al., 2021). Most notably, low-N nutrition promotes the expression of the AtNPF3.1 gene, which is involved in GA transport in plants under low-nitrate conditions (David et al., 2016). Eight genes related to GA synthesis are differentially expressed under N starvation (Ma et al., 2020). In addition to these hormones, JA and SA are also implicated in the response to low-N conditions (Lv et al., 2021; Sun et al., 2020a). JA signaling may function by regulating sugar biosynthesis, amino acid biosynthesis, and absorption of NH4+ (Sun et al., 2020a).
Taken together, most hormones, such as GAs, ABA, and cytokinins, may be involved in the regulation of root growth and development, likely through crosstalk with auxin (Ma et al., 2020).
First, it is imperative to identify the cell membrane and intracellular receptors that perceive N starvation and downstream signals and to investigate how these signals are transduced to gene expression and protein translation and then integrated with pathways to regulate root growth and development. At the transcriptional level, the exact functions of the differentially expressed genes and the timing and tissue specificity of the transcriptional responses to low-N stress are most important for cereal crops with high NUE under suboptimal N conditions. Thus, employing a combination of multiomics-assisted analyses, such as SWATH-MS-based proteomics, proteogenomics, and de novo peptide sequencing, is useful for deciphering low-N signaling, screen downstream signaling components, and generate metabolic pathways and networks to better understand the regulatory mechanisms at developmental, molecular, and physiological levels. The identification of central members, such as the regulatory circuit miR827-NLA-NRTs, in the network will facilitate our understanding of the molecular mechanisms underlying the plant response to external N signaling mediated by miRNA/target pathways. The plant signaling system is very important for plant breeders and agronomists and enables them to improve NUE.
The generation of transgenic lines based on valuable gene resources, such as TAR2.1, NBP1, bZIP60, and NADH-GOGAT, is critical for the development of large RSAs, improving N uptake and assimilation, and shows potential for genetic engineering to increase the grain yield of wheat under N-saving cultivation conditions. Transcription factors may be a potential gene for molecular breeding and cultivation of wheat to improve N remobilization, utilization, and yield in crops simultaneously. Genetic programing through temperature- or photoperiod-controlled timing expression of genes, such as ATGs, would promote N remobilization during grain filling and increase NUE.
In recent years, genome editing with engineered nucleases has been used in plant breeding. Further identification of the genes that negatively control NUE and agronomically important traits will facilitate the use of genome editing to increase NUE and yield. These large-scale datasets provide valuable information for the generation of new wheat cultivars with higher NUE or greater resistance to N starvation in future breeding programs.
In conclusion, many links between signal perception, transduction, and regulation of gene functions at the transcriptional, posttranscriptional, and translational levels are still missing. Although we have filled the putative gaps with knowledge translated from other plants, a genetics-based approach is needed to find molecular targets in the field of N starvation, which is an effective way to screen signaling components and to construct regulatory networks in response to N starvation.
LK drafted the manuscript. YZ and WD searched the literature and provided suggestions for writing. HX and SF modified the manuscript. BZ conceived the project, constructed figures, and gave suggestions on the revision of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Shandong Province (ZR2020MC087), the National Key R&D Program of China (2016YFD0300403 and 2017YFD03002), the National Natural Science Foundation of China (31801282), and the Shandong Modern Agricultural Technology and Industry System (SDAIT-01-06).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Asim, M., Ullah, Z., Oluwaseun, A., Wang, Q., and Liu, H. (2020). Signalling overlaps between nitrate and auxin in regulation of the root system architecture: insights from the Arabidopsis thaliana. Int. J. Mol. Sci. 21:2880. doi: 10.3390/ijms21082880
Bajgain, P., Russell, B., and Mohammadi, M. (2018). Phylogenetic analyses and in-seedling expression of ammonium and nitrate transporters in wheat. Sci. Rep. 8:7082. doi: 10.1038/s41598-018-25430-8
Balotf, S., Islam, S., Kavoosi, G., Kholdebarin, B., Juhasz, A., and Ma, W. (2018). How exogenous nitric oxide regulates nitrogen assimilation in wheat seedlings under different nitrogen sources and levels. PLoS One 13:e0190269. doi: 10.1371/journal.pone.0190269
Bedu, M., Marmagne, A., Masclaux-Daubresse, C., and Chardon, F. (2020). Transcriptional plasticity of autophagy-related genes correlates with the genetic response to nitrate starvation in Arabidopsis thaliana. Cells 9:1021. doi: 10.3390/cells9041021
Buchner, P., and Hawkesford, M. J. (2014). Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J. Exp. Bot. 65, 5697–5710. doi: 10.1093/jxb/eru231
Cai, H., Lu, Y., Xie, W., Zhu, T., and Lian, X. (2012). Transcriptome response to nitrogen starvation in rice. J. Biosci. 37, 731–747. doi: 10.1007/s12038-012-9242-2
Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435. doi: 10.1111/j.1365-313X.2008.03695.x
Chardin, C., Girin, T., Roudier, F., Meyer, C., and Krapp, A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587. doi: 10.1093/jxb/eru261
Chen, Q., Soulay, F., Saudemont, B., Elmayan, T., Marmagne, A., and Masclaux-Daubresse, C. (2019). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60, 343–352. doi: 10.1093/pcp/pcy214
Chen, Y., Yang, M., Ding, W., Zhao, Y., Li, X., and Xiao, K. (2017). Wheat ZFP gene TaZFP593;l mediates the N-starvation adaptation of plants through regulating N acquisition and the ROS metabolism. Plant Cell Tiss. Org. 129, 271–288. doi: 10.1007/s11240-017-1176-9
Chen, Z. H., Bao, M. L., Sun, Y. Z., Yang, Y. J., Xu, X. H., Wang, J. H., et al. (2011). Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 77, 619–629. doi: 10.1007/s11103-011-9838-1
Curci, P. L., Cigliano, R. A., Zuluaga, D. L., Janni, M., Sanseverino, W., and Sonnante, G. (2017). Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 7:1176. doi: 10.1038/s41598-017-01377-0
David, L. C., Berquin, P., Kanno, Y., Seo, M., Daniel-Vedele, F., and Ferrario-Mery, S. (2016). N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 244, 1315–1328. doi: 10.1007/s00425-016-2588-1
David, L. C., Girin, T., Fleurisson, E., Phommabouth, E., Mahfoudhi, A., Citerne, S., et al. (2019). Developmental and physiological responses of Brachypodium distachyon to fluctuating nitrogen availability. Sci. Rep. 9:3824. doi: 10.1038/s41598-019-40569-8
Dechorgnat, J., Francis, K. L., Dhugga, K. S., Rafalski, J. A., Tyerman, S. D., and Kaiser, B. N. (2019). Tissue and nitrogen-linked expression profiles of ammonium and nitrate transporters in maize. BMC Plant Biol. 9:206. doi: 10.1186/s12870-019-1768-0
Ding, C., You, J., Liu, Z., Rehmani, M. I., Wang, S., Li, G., et al. (2011). Proteomic analysis of low nitrogen stress-responsive proteins in roots of rice. Plant Mol. Biol. Rep. 29, 618–625. doi: 10.1007/s11105-010-0268-z
Fan, T., Yang, W., Zeng, X., Xu, X., Xu, Y., Fan, X., et al. (2020). A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 11:588. doi: 10.3389/fpls.2020.00588
Gao, S., Guo, C., Zhang, Y., Zhang, F., Du, X., Gu, J., et al. (2016). Wheat microRNA member TamiR444a is nitrogen deprivation-responsive and involves plant adaptation to the nitrogen-starvation stress. Plant Mol. Biol. Rep. 34, 931–946. doi: 10.1007/s11105-016-0973-3
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M., and Birnbaum, K. D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U S A. 105, 803–808. doi: 10.1073/pnas.0709559105
Gojon, A., Krouk, G., Perrine-Walker, F., and Laugier, E. (2011). Nitrate transceptor(s) in plants. J. Exp. Bot. 62, 2299–2308. doi: 10.1093/jxb/erq419
Gruber, B. D., Giehl, R. F. H., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U S A. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataillé, M. P., Avice, J. C., and Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Guo, H. S., Xie, Q., Fei, J. F., and Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Guo, T. C., Xuan, H. M., Yang, Y. Y., Wang, L. N., Wei, L. T., Wang, Y. H., et al. (2014). Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 33, 837–848. doi: 10.1007/s00344-014-9435-z
Gupta, A. K., Gaur, V. S., Gupta, S., and Kumar, A. (2009). Nitrate signals determine the sensing of nitrogen through differential expression of genes involved in nitrogen uptake and assimilation in finger millet. Mol. Biol. Rep. 36, 2209–2220. doi: 10.1007/s10142-013-0311-x
He, X., Qu, B., Li, W., Zhao, X., Teng, W., Ma, W., et al. (2015). The nitrate inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169, 1991–2005. doi: 10.1104/pp.15.00568
Hou, G., Du, C., Gao, H., Liu, S., Sun, W., Lu, H., et al. (2020). Identification of microRNAs in developing wheat grain that are potentially involved in regulating grain characteristics and the response to nitrogen levels. BMC Plant Biol. 20:87. doi: 10.1186/s12870-020-2296-7
Hsieh, P. H., Kan, C. C., Wu, H. Y., Yang, H. C., and Hsieh, M. H. (2018). Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci. Rep. 8:12207. doi: 10.1038/s41598-018-30632-1
Hu, H. C., Wang, Y. Y., and Tsay, Y. F. (2009). AtCIPK8, CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57, 264–278. doi: 10.1111/j.1365-313X.2008.03685.x
Huang, W., Bai, G., Wang, J., Zhu, W., Zeng, Q., Lu, K., et al. (2018). Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front. Plant Sci. 9:300. doi: 10.3389/fpls.2018.00300
Ikeda, Y., Koizumi, N., Kusano, T., and Sano, H. (2000). Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J. Biol. Chem. 275, 31695–31700. doi: 10.1074/jbc.M004892200
Jiang, S., Sun, J., Tian, Z., Hu, H., Michel, E. J., Gao, J., et al. (2017). Root extension and nitrate transporter up-regulation induced by nitrogen deficiency improves nitrogen status and plant growth at the seedling stage of winter wheat (Triticum aestivum L.). Environ. Exp. Bot. 141, 28–40. doi: 10.1016/j.envexpbot.2017.06.006
Jiao, X., Wang, H., Yan, J., Kong, X., Liu, Y., Chu, J., et al. (2020). Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth. Plant Physiol. 182, 1454–1466. doi: 10.1104/pp.19.00190
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cells 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Jung, J. Y., Ahn, J. H., and Schachtman, D. P. (2018). CC-type glutaredoxins mediate plant response and signaling under nitrate starvation in Arabidopsis. BMC Plant Biol. 8:281. doi: 10.1186/s12870-018-1512-1
Kang, G., Wu, Y., Li, G., Wang, P., Han, Q., Wang, Y., et al. (2019). Proteomics combined with BSMV-VIGS methods identified some N deficiency-responsive protein species and ABA role in wheat seedling. Plant Soil 444, 177–191. doi: 10.1007/s11104-019-04260-1
Khan, M. U., Li, P., Amjad, H., Khan, A. Q., Arafat, Y., Waqas, M., et al. (2019). Exploring the potential of overexpressed OsCIPK2 rice as a nitrogen utilization efficient crop and analysis of its associated rhizo-compartmental microbial communities. Int. J. Mol. Sci. 20:3636. doi: 10.3390/ijms20153636
Kiba, T., Kudo, T., Kojima, M., and Sakakibara, H. (2011). Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409. doi: 10.1093/jxb/erq410
Kichey, T., Le Gouis, J., Sangwan, B., Hirel, B., and Dubois, F. (2005). Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant Cell Physiol. 46, 964–974. doi: 10.1093/pcp/pci105
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4:1617. doi: 10.1038/ncomms2621
Kováčik, J., Klejdus, B., Babula, P., and Jarošová, M. (2014). Variation of antioxidants and secondary metabolites in nitrogen-deficient barley plants. J. Plant Physiol. 171, 260–268. doi: 10.1016/j.jplph.2013.08.004
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Kumar, A., Batra, R., Gahlaut, V., Gautam, T., Kumar, S., Sharma, M., et al. (2018). Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS One 13:e0208409. doi: 10.1371/journal.pone.0208409
Kurai, T., Wakayama, M., Abiko, T., Yanagisawa, S., Aoki, N., and Ohsugi, R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9, 826–837. doi: 10.1111/j.1467-7652.2011.00592.x
Kwade, Z., Swiatek, A., Azmi, A., Goossens, A., InzéD, Onckelen, H. V., et al. (2005). Identification of four adenosine kinase isoforms in tobacco By-2 cells and their putative role in the cell cycle-regulated cytokinin metabolism. J. Biol. Chem. 280, 17512–17519. doi: 10.1074/jbc.M411428200
Ladha, J. K., Tirol-Padre, A., Reddy, C. K., Cassman, K. G., Verma, S., Powlson, D. S., et al. (2016). Global nitrogen budgets in cereals: a 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 6:19355. doi: 10.1038/srep19355
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015a). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, W., Chen, M., Zhong, L., Liu, J., Xu, Z., Li, L., et al. (2015b). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.). confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Bioph. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Li, H., Hu, B., Wang, W., Zhang, Z. H., Liang, Y., Gao, X. K., et al. (2016). Identification of microRNAs in rice root in response to nitrate and ammonium. J. Genet. Genomics 43, 651–661. doi: 10.1016/j.jgg.2015.12.002
Li, J., Shen, Y., Zhu, J., Liu, S., Zeng, N., and Zhan, X. (2020). miR398 is involved in the relief of phenanthrene-induced oxidative toxicity in wheat roots. Environ. Pollut. 258:113701. doi: 10.1016/j.envpol.2019.113701
Li, T., Liao, K., Xu, X., Gao, Y., Wang, Z., Zhu, X., et al. (2017). Wheat ammonium transporter (AMT) gene family: diversity and possible role in host-pathogen interaction with stem rust. Front. Plant Sci. 8:1637. doi: 10.3389/fpls.2017.01637
Lian, X., Wang, S., Zhang, J., Feng, Q., Zhang, L., Fan, D., et al. (2006). Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol. Biol. 60, 617–631. doi: 10.1007/s11103-005-5441-7
Liang, G., He, H., and Yu, D. (2012). Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7:e48951. doi: 10.1371/journal.pone.0048951
Liao, C. Y., and Bassham, D. C. (2020). Combating stress: the interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 71, 1723–1733. doi: 10.1093/jxb/erz515
Liu, K. H., and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Liu, W., Sun, Q., Wang, K., Du, Q., and Li, W. X. (2017). Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol. 214, 734–744. doi: 10.1111/nph.14396
Liu, Z., Zhao, Y., Wang, X., Yang, M., Guo, C., and Xiao, K. (2018). TaNBP1, a guanine nucleotide-binding subunit gene of wheat, is essential in the regulation of N starvation adaptation via modulating N acquisition and ROS homeostasis. BMC Plant Biol. 18:167. doi: 10.1186/s12870-018-1374-6
Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48, 522–534. doi: 10.1111/j.1365-313X.2006.02887.x
Lv, X., Zhang, Y., Hu, L., Zhang, Y., Zhang, B., Xia, H., et al. (2021). Low-nitrogen stress stimulates lateral root initiation and nitrogen assimilation in wheat: roles of phytohormone signaling. J. Plant Growth Reg. 40, 436–450. doi: 10.1007/s00344-020-10112-5
Ma, N., Dong, L., Lü, W., Lü, J., Meng, Q., and Liu, P. (2020). Transcriptome analysis of maize seedling roots in response to nitrogen-, phosphorus-, and potassium deficiency. Plant Soil 447, 637–658. doi: 10.1007/s11104-019-04385-3
Ma, Q., Tang, R. J., Zheng, X. J., Wang, S. M., and Luan, S. (2015). The calcium sensor CBL7 modulates plant responses to low nitrate in Arabidopsis. Biochem. Bioph. Res. Commun. 468, 59–65. doi: 10.1016/j.bbrc.2015.10.164
Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79. doi: 10.1111/tpj.12448
Maghiaoui, A., Bouguyon, E., Moliner, C. C., Perrine-Walker, F., Alcon, C., Krouk, G., et al. (2020). The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport for regulating root branching in response to nitrate. J. Expe. Bot. 71, 4480–4494. doi: 10.1093/jxb/eraa242
Mahmoud, D., Pandey, R., Sathee, L., Dalal, M., Singh, M. P., and Chinnusamy, V. (2020). Regulation of expression of genes associated with nitrate response by osmotic stress and combined osmotic and nitrogen deficiency stress in bread wheat (Triticum aestivum L.). Plant Physiol. Rep. 25, 200–215. doi: 10.1007/s40502-020-00503-x
Mascia, M., Sega, D., Zamboni, A., and Varanini, Z. (2019). Nitrogen starvation differentially influences transcriptional and uptake rate profiles in roots of two maize inbred lines with different NUE. Int. J. Mol. Sci. 20:4856. doi: 10.3390/ijms20194856
Masclaux-Daubresse, C. (2016). Autophagy controls carbon, nitrogen, and redox homeostasis in plants. Autophagy 12, 896–897. doi: 10.4161/auto.36261
Melino, V. J., Casartelli, A., George, J., Rupasinghe, T., Roessner, U., Okamoto, M., et al. (2018). RNA catabolites contribute to the nitrogen pool and support growth recovery of wheat. Front. Plant Sci. 9:1539. doi: 10.3389/fpls.2018.01539
Melino, V. J., Fiene, G., Enju, A., Cai, J., Buchner, P., and Heuer, S. (2015). Genetic diversity for root plasticity and nitrogen uptake in wheat seedling. Funct. Plant Biol. 42, 942–956. doi: 10.1071/FP15041
Młodzińska, E., Kłobus, G., Christensen, M. D., and Fuglsang, A. T. (2015). The plasma membrane H+−ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plantarum 154, 270–282. doi: 10.1111/ppl.12305
Nischal, L., Mohsin, M., Khan, I., Kardam, H., Wadhwa, A., Abrol, Y. P., et al. (2012). Identification and comparative analysis of microRNAs associated with low-N tolerance in rice genotypes. PLoS One 7:e50261. doi: 10.1371/journal.pone.0050261
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF). transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Pant, B. D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J., et al. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555. doi: 10.1104/pp.109.139139
Paul, S., Datta, S. K., and Datta, K. (2015). miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 6:232. doi: 10.3389/fpls.2015.00232
Pei, D., Zhang, W., Sun, H., Wei, X., Yue, J., and Wang, H. (2014). Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33, 1697–1710. doi: 10.1007/s00299-014-1648-x
Peng, M., Hannam, C., Gu, H., Bi, Y. M., and Rothstein, S. J. (2007). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50, 320–337. doi: 10.1111/j.1365-313X.2007.03050.x
Qiao, Q., Wang, X., Yang, M., Zhao, Y., Gu, J., and Xiao, K. (2018). Wheat miRNA member TamiR2275 involves plant nitrogen starvation adaptation via enhancement of the N acquisition-associated process. Acta Physiol. Plantarum 40:183. doi: 10.1007/s11738-018-2758-9
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Remans, T., Nacry, P., Pervent, M., Girin, T., Tillard, P., Lepetit, M., et al. (2006). A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140, 909–921. doi: 10.1104/pp.105.075721
Shao, A., Ma, W., Zhao, X., Hu, M., He, X., Teng, W., et al. (2017). The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2. 1-3A increases grain yield of wheat. Plant Physiol. 174, 2274–2288. doi: 10.1104/pp.17.00094
Shao, C. H., Qiu, C. F., Qian, Y. F., and Liu, G. R. (2020). Nitrate deficiency decreased photosynthesis and oxidation-reduction processes, but increased cellular transport, lignin biosynthesis and flavonoid metabolism revealed by RNA-Seq in Oryza sativa leaves. PLoS One 15:e0235975. doi: 10.1371/journal.pone.0235975
Sharma, A., Shankhdhar, D., and Shankhdhar, S. C. (2017). “The role of calcium in plant signal transduction under macronutrient deficiency stress,” in Plant Macronutrient Use Efficiency, eds M. A. Hossain, T. Kamiya, D. J. Burrit, L. S. P. Tran, and T. Fujiwara, (Cambridge, MA: Academic Press), 181–196.
Shi, M., Wang, Z., Ma, Z., Song, W., Lu, W., and Xiao, K. (2020). Characterization on TaMPK14, an MAPK family gene of wheat, in modulating N-starvation response through regulating N uptake and ROS homeostasis. Plant Cell Rep. 39, 1285–1299. doi: 10.1007/s00299-020-02564-6
Shin, R., Berg, R. H., and Schachtman, D. P. (2005). Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 46, 1350–1357. doi: 10.1093/pcp/pci145
Shin, S. Y., Jeong, J. S., Lim, J. Y., Kim, T., Park, J. H., Kim, J. K., et al. (2018). Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics 19:532. doi: 10.1186/s12864-018-4897-1
Signora, L., De Smet, I., Foyer, C. H., and Zhang, H. (2001). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28, 655–662. doi: 10.1046/j.1365-313x.2001.01185.x
Singh, P., Jaiswal, S., Sheokand, S., and Duhan, S. (2018). Morpho-physiological and oxidative responses of nitrogen and phosphorus deficiency in wheat (Triticum aestivum L.). Indian J. Agr. Res. 52, 40–45. doi: 10.18805/IJARe.A-4905
Sinha, S. K., Kumar, A., Tyagi, A., Venakatesh, K., Paul, D., Singh, N. K., et al. (2020). Root architecture traits variation and nitrate-influx responses in diverse wheat genotypes under different external nitrogen concentrations. Plant Physiol. Biochem. 148, 246–259. doi: 10.1016/j.plaphy.2020.01.018
Sinha, S. K., Rani, M., Bansal, N., Gayatri, B., Venkatesh, K., and Mandal, P. K. (2015). Nitrate starvation induced changes in root system architecture, carbon:nitrogen metabolism, and miRNA expression in nitrogen-responsive wheat genotypes. App. Biochem. Biotechnol. 177, 1299–1312. doi: 10.1007/s12010-015-1815-8
Slavikova, S., Ufaz, S., Avinwittenberg, T., Levanony, H., and Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. doi: 10.1093/jxb/ern244
Subudhi, P. K., Garcia, R. S., Coronejo, S., and Tapia, R. (2020). Comparative transcriptomics of rice genotypes with contrasting responses to nitrogen stress reveals genes influencing nitrogen uptake through the regulation of root architecture. Int J. Mol, Sci. 21:5759. doi: 10.3390/ijms21165759
Sultana, N., Islam, S., Juhasz, A., Yang, R., She, M., Alhabbar, Z., et al. (2020). Transcriptomic study for identification of major nitrogen stress responsive genes in Australian bread wheat cultivars. Front. Genetics 11:583785. doi: 10.3389/fgene.2020.583785
Sun, H., Bi, Y., Tao, J., Huang, S., Hou, M., Xue, R., et al. (2016). Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ. 39, 1473–1484. doi: 10.1111/pce.12709
Sun, H., Li, J., Song, W., Tao, J., Huang, S., Chen, S., et al. (2015). Nitric oxide generated by nitrate reductase increases nitrogen uptake capacity by inducing lateral root formation and inorganic nitrogen uptake under partial nitrate nutrition in rice. J. Exp. Bot. 66, 2449–2459. doi: 10.1093/jxb/erv030
Sun, H., Tao, J., Bi, Y., Hou, M., Lou, J., Chen, X., et al. (2018a). OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to low nitrogen and phosphate. Sci. Rep. 8:13014. doi: 10.1038/s41598-018-29784-x
Sun, Q., Liu, X., Yang, J., Liu, W., Du, Q., Wang, H., et al. (2018b). MicroRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant 11, 806–814. doi: 10.1016/j.molp.2018.03.013
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018c). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Sun, H., Tao, J., Liu, S., Huang, S., Chen, S., Xie, X., et al. (2014). Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746. doi: 10.1093/jxb/eru029
Sun, L., Di, D. W., Li, G., Li, Y., Kronzucker, H. J., and Shi, W. (2020a). Transcriptome analysis of rice (Oryza sativa L.) in response to ammonium resupply reveals the involvement of phytohormone signaling and the transcription factor OsJAZ9 in reprogramming of nitrogen uptake and metabolism. J. Plant Physiol. 246:153137. doi: 10.1016/j.jplph.2020.153137
Sun, X., Chen, H., Wang, P., Chen, F., Yuan, L., and Mi, G. (2020b). Low nitrogen induces root elongation via auxin-induced acid growth and auxin-regulated target of rapamycin (TOR) pathway in maize. J. Plant Physiol. 254:153281. doi: 10.1016/j.jplph.2020.153281
Sun, Z., Ding, C., Li, X., and Xiao, K. (2012). Molecular characterization and expression analysis of TaZFP15, a C2H2-type zinc finger transcription factor gene in wheat (Triticum aestivum L.). J. Integr. Agr. 11, 31–42. doi: 10.1016/s1671-2927(12)60780-9
Talboys, P. J., Healey, J. R., Withers, P. J., and Jones, D. L. (2014). Phosphate depletion modulates auxin transport in Triticum aestivum leading to altered root branching. J. Exp. Bot. 65, 5023–5032. doi: 10.1093/jxb/eru284
Tang, Y., Li, X., Lu, W., Wei, X., Zhang, Q., Lv, C., et al. (2018). Enhanced photorespiration in transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase gene contributes to alleviating low nitrogen stress. Plant Physiol. Biochem. 130, 577–588. doi: 10.1016/j.plaphy.2018.08.013
Taulemesse, F., Le Gouis, J., Gouache, D., Gibon, Y., and Allard, V. (2015). Post-floweringnitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT2.1. PLoS One 10:e0120291. doi: 10.1371/journal.pone.0120291
Tian, Q., Chen, F., Liu, J., Zhang, F., and Mi, G. (2008). Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 165, 942–951. doi: 10.1016/j.jplph.2007.02.011
Trevisan, S., Begheldo, M., Nonis, A., and Quaggiotti, S. (2012a). The miRNA-mediated post-transcriptional regulation of maize response to nitrate. Plant Signal. Behav. 7, 822–826. doi: 10.4161/psb.20462
Trevisan, S., Nonis, A., Begheldo, M., Manoli, A., Palme, K., Caporale, G., et al. (2012b). Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 35, 1137–1155. doi: 10.1111/j.1365-3040.2011.02478.x
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Wakayama, M., Abiko, T., Yanagisawa, S., Aoki, N., and Ohsugi, R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9, 826–837. doi: 10.1111/j.1467-7652.2011.00592.x
Wan, Y., King, R., Mitchell, R. A. C., Hassani-Pak, K., and Hawkesford, M. J. (2017). Spatiotemporal expression patterns of wheat amino acid transporters reveal their putative roles in nitrogen transport and responses to abiotic stress. Sci. Rep. 7:5461. doi: 10.1038/s41598-017-04473-4473
Wang, D., Xu, T., Yin, Z., Wu, W., Geng, H., Li, L., et al. (2020a). Overexpression of OsMYB305 in rice enhances the nitrogen uptake under low-nitrogen condition. Front. Plant Sci. 11:369. doi: 10.3389/fpls.2020.00369
Wang, H., Wan, Y., Buchner, P., King, R., Ma, H., and Hawkesford, M. J. (2020b). Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Triticum aestivum. J. Exp. Bot. 71, 4531–4546. doi: 10.1093/jxb/eraa210
Wang, M., Zhang, P., Liu, Q., Li, G., Di, D., Xia, G., et al. (2020c). TaANR1-TaBG1 and TaWabi5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol. 182, 1440–1453. doi: 10.1104/pp.19.01482
Wang, Y., Xu, J., Ge, M., Ning, L., Hu, M., and Zhao, H. (2020d). High-resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize. BMC Genomics 21:353. doi: 10.1186/s12864-020-6769-8
Wang, J., Hüner, N., and Tian, L. (2019a). Identification and molecular characterization of the Brachypodium distachyon NRT2 family, with a major role of BdNRT2.1. Physiol. Plant. 165, 498–510. doi: 10.1111/ppl.12716
Wang, J., Song, K., Sun, L., Qin, Q., Sun, Y., Pan, J., et al. (2019b). Morphological and transcriptome analysis of wheat seedlings response to low nitrogen stress. Plants 8:98. doi: 10.3390/plants8040098
Wang, J., Li, Y., Zhu, F., Ming, R., and Chen, L. Q. (2019c). Genome-wide analysis of nitrate transporter (NRT/NPF) family in Sugarcane Saccharum spontaneum L. Trop. Plant Biol. 12, 133–149. doi: 10.1007/s12042-019-09220-8
Wang, Y., Cao, J. J., Wang, K. X., Xia, X. J., Shi, K., Zhou, Y. H., et al. (2019d). BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 179, 671–685. doi: 10.1104/pp.18.01028
Wang, P., Du, Y., Li, Y., Ren, D., and Song, C. P. (2010). Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22, 2981–2998. doi: 10.1105/tpc.109.072959
Wang, P., Sun, X., Jia, X., Wang, N., Gong, X., and Ma, F. (2016). Characterization of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 7:720. doi: 10.3389/fpls.2016.00720
Wang, Y. Y., Hsu, P. K., and Tsay, Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. doi: 10.1016/j.tplants.2012.04.006
Waqas, M., Feng, S., Amjad, H., Letuma, P., Zhan, W., Li, Z., et al. (2018). Protein phosphatase (PP2C9) induces protein expression differentially to mediate nitrogen utilization efficiency in rice under nitrogen-deficient condition. Int. J. Mol. Sci. 19:2827. doi: 10.3390/ijms19092827
Wen, Y., Li, X., Guo, C., Ma, C., Duan, W., Lu, W., et al. (2015). Characterization and expression analysis of mitogen-activated protein kinase cascade genes in wheat subjected to phosphorus and nitrogen deprivation, high salinity, and drought. J. Plant Biochem. Biotechnol. 24, 184–196. doi: 10.1007/s13562-014-0256-8
Wu, Y., Yang, W., Wei, J., Yoon, H., and An, G. (2017). Transcription factor OsDOF18 controls ammonium uptake by inducing ammonium transporters in rice roots. Mol. Cells 40, 178–185. doi: 10.14348/molcells.2017.2261
Xia, T., Xiao, D., Liu, D., Chai, W., Gong, Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS One 7:e37217. doi: 10.1371/journal.pone.0037217
Xing, Y., Guo, S., Chen, X., Du, D., Liu, M., Xiao, Y., et al. (2018). Nitrogen metabolism is affected in the nitrogen-deficient rice mutant esl4 with a calcium-dependent protein kinase gene mutation. Plant Cell Physiol. 59, 2512–2525. doi: 10.1093/pcp/pcy169
Xiong, H., Guo, H., Zhou, C., Guo, X., Xie, Y., Zhao, L., et al. (2019). A combined association mapping and t-test analysis of SNP loci and candidate genes involving in resistance to low nitrogen traits by a wheat mutant population. PLoS One 14:e0211492. doi: 10.1371/journal.pone.0211492
Xu, Y., Ren, Y., Li, J., Li, L., Chen, S., Wang, Z., et al. (2019a). Comparative proteomic analysis provides new insights into low nitrogen-promoted primary root growth in hexaploid wheat. Front. Plant Sci. 10:151. doi: 10.3389/fpls.2019.00151
Xu, Y., Ren, Y., Lin, T., and Cui, D. (2019b). Identification and characterization of CircRNAs involved in the regulation of wheat root length. Biol. Res. 52:19. doi: 10.1186/s40659-019-0228-5
Xu, Z., Zhong, S., Li, X., Li, W., Rothstein, S. J., Zhang, S., et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS One 6:e28009. doi: 10.1371/journal.pone.0028009
Yan, Y., Wang, H., Hamera, S., Chen, X., and Fang, R. (2014). MiR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 78, 44–55. doi: 10.1111/tpj.12446
Yanagisawa, S., Akiyama, A., Kisaka, H., Uchimiya, H., and Miwa, T. (2004). Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. U S A. 101, 7833–7838. doi: 10.1073/pnas.0402267101
Yang, J., Wang, M., Li, W., He, X., Teng, W., Ma, W., et al. (2019a). Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotechnol. J. 17, 1823–1833. doi: 10.1111/pbi.13103
Yang, Z., Wang, Z., Yang, C., Yang, Z., Li, H., and Wu, Y. (2019b). Physiological responses and small RNAs changes in maize under nitrogen deficiency and resupply. Genes Genomics 41, 1183–1194. doi: 10.1007/s13258-019-00848-0
Yang, S. Y., Hao, D. L., Song, Z. Z., Yang, G. Z., Wang, L., and Su, Y. H. (2015). RNA-Seq analysis of differentially expressed genes in rice under varied nitrogen supplies. Gene 555, 305–317. doi: 10.1016/j.gene.2014.11.021
Yang, T., Hao, L., Yao, S., Zhao, Y., Lu, W., and Xiao, K. (2016). TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiol. Biochem. 104, 99–113. doi: 10.1016/j.plaphy.2016.03.023
Yousuf, P. Y., Abd_Allah, E. F., Nauman, M., Asif, A., Hashem, A., Alqarawi, A. A., et al. (2017). Responsive proteins in wheat cultivars with contrasting nitrogen efficiencies under the combined stress of high temperature and low nitrogen. Genes 8:356. doi: 10.3390/genes8120356
Yu, J., Zhen, X., Li, X., Li, N., and Xu, F. (2019). Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 10:584. doi: 10.3389/fpls.2019.00584
Yu, L., Wu, J., Tang, H., Yuan, Y., Wang, S., Wang, Y., et al. (2016). Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6:27795. doi: 10.1038/srep27795
Yue, W., Nie, X., Cui, L., Zhi, Y., Zhang, T., Du, X., et al. (2018). Genome-wide sequence and expressional analysis of autophagy gene family in bread wheat (Triticum aestivum L.). J. Plant Physiol. 229, 7–21. doi: 10.1016/j.jplph.2018.06.012
Zhang, H., Rong, H., and Pilbeam, D. (2007). Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J. Exp. Bot. 58, 2329–2338. doi: 10.1093/jxb/erm114
Zhang, L., Zhang, L., Xia, C., Zhao, G., Liu, J., Jia, J., et al. (2015). A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plantarum 153, 538–554. doi: 10.1111/ppl.12261
Zhang, L., Zhao, G., Jia, J., Liu, X., and Kong, X. (2012). Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 63, 203–214. doi: 10.1093/jxb/err264
Zhang, S. C., Li, M. J., Guo, J. K., Shi, Z. L., Fu, X. Y., Di, R. Y., et al. (2016). Comparative transcriptome analysis of Triticum aestivum in response to nitrogen stress. Russ. J. Plant Physiol. 63, 365–374. doi: 10.1134/S1021443716020175
Zhang, Y., Liu, H., Yin, H., Wang, W., Zhao, X., and Du, Y. (2013). Nitric oxide mediates alginate oligosaccharides-induced root development in wheat (Triticum aestivum L.). Plant Physiol. Biochem. 71, 49–56. doi: 10.1016/j.plaphy.2013.06.023
Zhang, Z. H., Zhou, T., Liao, Q., Yao, J. Y., Liang, G. H., Song, H. X., et al. (2018). Integrated physiologic, genomic and transcriptomic strategies involving the adaptation of allotetraploid rapeseed to nitrogen limitation. BMC Plant Biol. 18:322. doi: 10.1186/s12870-018-1507-y
Zhao, M., Ding, H., Zhu, J. K., Zhang, F., and Li, W. X. (2011). Involvement of miR169 in the nitrogen starvation responses in Arabidopsis. New Phytol. 190, 906–915. doi: 10.1111/j.1469-8137.2011.03647.x
Zhao, M., Tai, H., Sun, S., Zhang, F., Xu, Y., and Li, W. X. (2012). Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7:e29669. doi: 10.1371/journal.pone.0029669
Zhao, X., Liu, X., Guo, C., Gu, J., and Xiao, K. (2013). Identification and characterization of microRNAs from wheat (Triticum aestivum L.). under phosphorus deprivation. J. Plant Biochem. Biotechnol. 22, 113–123. doi: 10.1007/s13562-012-0117-2
Zhao, Y., Guo, L., LuW, Li, X., Chen, H., Guo, C., et al. (2015). Expression pattern analysis of microRNAs in root tissue of wheat (Triticum aestivum L.) under normal nitrogen and low nitrogen conditions. J. Plant Biochem. Biotechnol. 24, 143–153. doi: 10.1007/s13562-013-0246-2
Zhao, Y., Xu, K., Liu, G., Li, S., Zhao, S., Liu, X., et al. (2020). Global identification and characterization of miRNA family members responsive to potassium deprivation in wheat (Triticum aestivum L.). Sci. Rep. 10:15812. doi: 10.1038/s41598-020-72642-y
Zhen, X., Li, X., Yu, J., and Xu, F. (2019a). OsATG8c-mediated increased autophagy regulates the yield and nitrogen use efficiency in rice. Int. J. Mol. Sci. 20:4956. doi: 10.3390/ijms20194956
Zhen, X., Xu, F., Zhang, W., Li, N., and Li, X. (2019b). Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS One 14:e0223011. doi: 10.1371/journal.pone.0223011
Zörb, C., Ludewig, U., and Hawkesford, M. J. (2018). Perspective on wheat yield and quality with reduced nitrogen supply. Trends Plant Sci. 23, 1029–1037. doi: 10.1016/j.tplants.2018.08.012
Zuluaga, D. L., De Paola, D., Janni, M., Curci, P. L., and Sonnante, G. (2017). Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS One 12:e0183253. doi: 10.1371/journal.pone.0183253
Zuluaga, D. L., Liuzzi, V., Curci, P. L., and Sonnante, G. (2018). MicroRNAs in durum wheat seedlings under chronic and short-term nitrogen stress. Funct. Integr. Genomics 18, 645–657. doi: 10.1007/s10142-018-0619-7
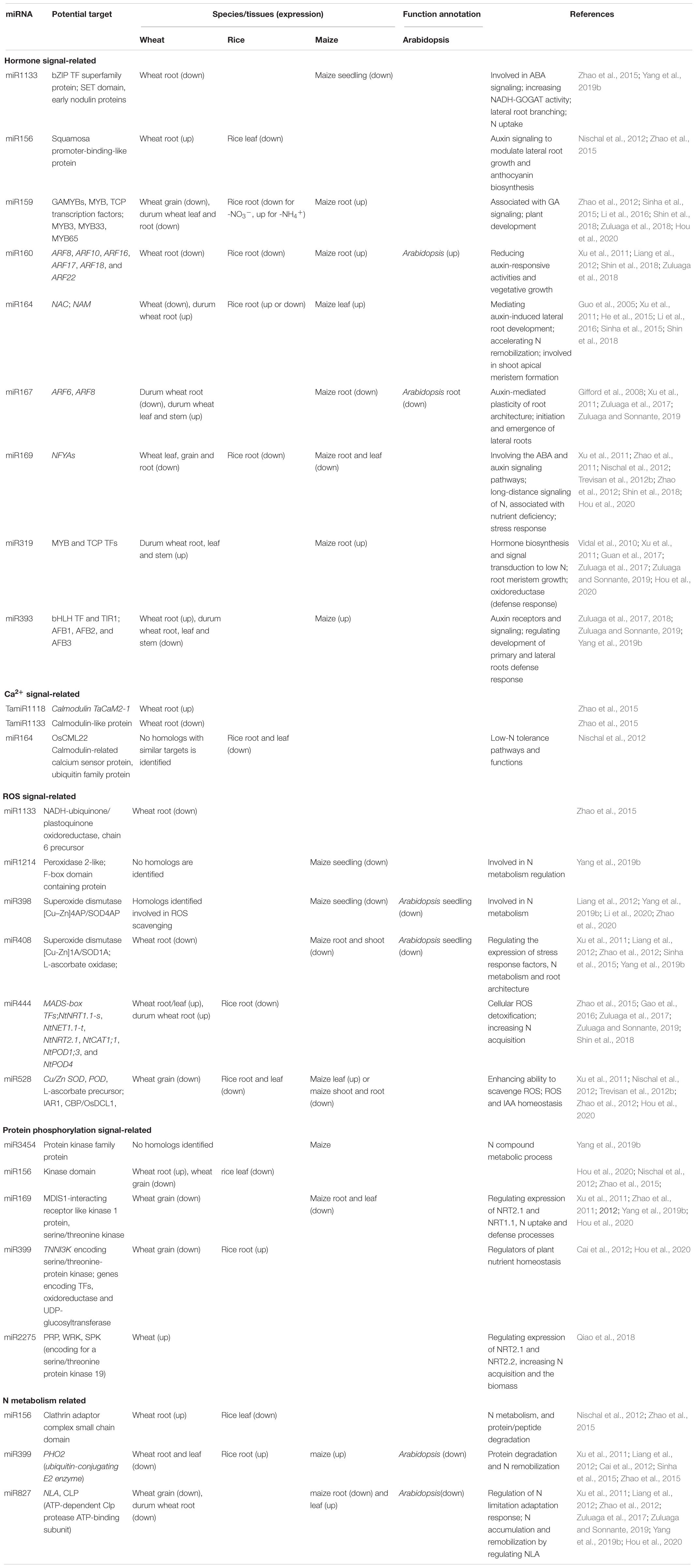
Appendix Table 1. Signaling or functions of differentially expressed miRNA families in response to low-N stress.
Keywords: microRNA, nitrogen starvation, phytohormone, root system architecture, signal, wheat
Citation: Kong L, Zhang Y, Du W, Xia H, Fan S and Zhang B (2021) Signaling Responses to N Starvation: Focusing on Wheat and Filling the Putative Gaps With Findings Obtained in Other Plants. A Review. Front. Plant Sci. 12:656696. doi: 10.3389/fpls.2021.656696
Received: 21 January 2021; Accepted: 08 April 2021;
Published: 31 May 2021.
Edited by:
Shi Xiao, Sun Yat-sen University, ChinaReviewed by:
Fabien Chardon, INRA UMR 1318 Institut Jean-Pierre Bourgin, FranceCopyright © 2021 Kong, Zhang, Du, Xia, Fan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Zhang, dmI1MzM3QDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.