
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 July 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.655127
 Yi Wang
Yi Wang Wanting Yu
Wanting Yu Lingfang Ran
Lingfang Ran Zhong Chen
Zhong Chen Chuannan Wang
Chuannan Wang Yang Dou
Yang Dou Yuanyuan Qin
Yuanyuan Qin Qingwei Suo
Qingwei Suo Yaohua Li
Yaohua Li Jianyan Zeng
Jianyan Zeng Aimin Liang
Aimin Liang Yonglu Dai
Yonglu Dai Yiping Wu
Yiping Wu Xufen Ouyang
Xufen Ouyang Yuehua Xiao*
Yuehua Xiao*Gibberellins (GAs) promote secondary cell wall (SCW) development in plants, but the underlying molecular mechanism is still to be elucidated. Here, we employed a new system, the first internode of cotton, and the virus-induced gene silencing method to address this problem. We found that knocking down major DELLA genes via VIGS phenocopied GA treatment and significantly enhanced SCW formation in the xylem and phloem of cotton stems. Cotton DELLA proteins were found to interact with a wide range of SCW-related NAC proteins, and virus-induced gene silencing of these NAC genes inhibited SCW development with downregulated biosynthesis and deposition of lignin. The findings indicated a framework for the GA regulation of SCW formation; that is, the interactions between DELLA and NAC proteins mediated GA signaling to regulate SCW formation in cotton stems.
Plant secondary cell walls (SCWs) are particularly important in adopting erect growth and land environment because they provide mechanical support and water transportation tunnels (Kumar et al., 2016). On the other hand, SCWs, comprising most of the plant biomass, are central to generate woods, biofuel, and other industrial uses. In general, SCWs are deposited inside the primary cell wall (PCW) after the cessation of cell expansion. The transition from PCM to SCW deposition encompasses a series of cell and metabolic changes, such as microtubule reorganization, biosynthesis of SCW-specific polysaccharides and lignin, and even activation of SCW-specific cellulose synthase (CESA) genes for cellulose synthesis (Zhong and Ye, 2015; Zhao, 2016; Meents et al., 2018).
In the last decades, a wealth of efforts has been taken to unveil molecular bases that control SCW differentiation and deposition. A conserved pyramid-shaped regulatory hierarchy has been identified to regulate gene expressions required for SCW formation in plants, especially in vascular tissues (Zhong et al., 2008; Nakano et al., 2015; Kumar et al., 2016; Zhao, 2016). This regulatory network contains basically three layers of transcription factors (TFs). In Arabidopsis, a group of NAC (NAM, ATAF1/2 and CUC2) family TFs, such as NAC secondary wall thickening (NST) promoting factors 1–3 and vascular-related NAC domains (VNDs) 1–7 (Zhong et al., 2006; Mitsuda et al., 2007; Yamaguchi et al., 2008; Zhou et al., 2014), comprise tier 3 and function as top master switches, which control the transcription of tier 2 TFs, such as MYB proteins (MYBs 46, 55, 61, 83, and 103) and NAC proteins (SND3 and XND1) (Zhong et al., 2010). Tier 2 TFs regulate a number of tier 1 TFs, which directly bind to promoters of structural genes (Hussey et al., 2011; Huang et al., 2015; Kumar et al., 2016). In addition, E2Fc can regulate the transcription of tier 3 TFs and is designated as top-level TF for SCW regulation (Taylor-Teeples et al., 2015). Furthermore, E2FC and tier 3 TFs can also bind to promoters of structural genes and thus promote their expressions as well, and some tier 1 TFs can negatively regulate the expression of tier 3 TFs (Hussey et al., 2011; Huang et al., 2015; Kumar et al., 2016; Zhang et al., 2018).
Gibberellins (GAs) are key plant hormones for many processes of plant development, such as seed germination, stem elongation, leaf expansion, and floral transition (Achard and Genschik, 2009; Daviere and Achard, 2013). DELLA proteins are the major repressor of GA signaling, and GAs generally promote DELLA-repressed downstream activities by mediating their degradation (Daviere and Achard, 2013; Van De Velde et al., 2017). It has been recognized for a long time that GAs promote SCW development, especially the formation of wood (xylem with highly thickened SCW) (Wareing, 1958; Digby and Wareing, 1966; Wang et al., 2013, 2017; Bai et al., 2014; Yuan et al., 2019). The upregulation of GA biosynthesis enzyme genes significantly enhanced SCW (wood) formation in stems of several plants (Israelsson et al., 2003; Biemelt et al., 2004; Park et al., 2015; Jeon et al., 2016). In the process of wood formation, the main roles of GAs are thought to regulate the early stages of wood differentiation, such as cell elongation and expansion (Israelsson et al., 2005; Ye and Zhong, 2015), and xylem differentiation from the cambium (Mauriat and Moritz, 2009). Recently, DELLAs were found to physically interact with a class-I KNOX transcription factor (KNAT1/ BREVIPEDICELLUS) and impair its function in inducing xylary fibers (Felipo-Benavent et al., 2018). In addition, GAs were found to regulate more processes in SCW formation, e.g., cellulose biosynthesis. Huang et al. revealed that a conserved GA-mediated DELLA-NAC signaling cascade regulates SCW synthesis, in which GAs promote cellulose synthesis by releasing SCW master switches NAC29/31 from DELLA and promoting the expression of CESA activator MYB61 in rice (Huang et al., 2015). Another CESA activator, OsMYB103L, could directly interact with DELLA, and GA-mediated DELLA degradation may promote cellulose synthesis and SCW thickening by releasing OsMYB103L (Ye et al., 2015). These results suggested that DELLAs may interact with multiple SCW regulators and form a complex regulatory network for GA signaling in SCW development.
In this study, we employed a new system, the first internode of cotton, to analyze the role of DELLA proteins (GhGAIs) in the GA regulation of SCW development. Yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays indicated that the cotton DELLA protein interacted with the SCW-related NAC TFs of different tiers, whose knockdown resulted in reduced SCW development in cotton stems. The results suggested that the interactions between DELLA and SCW-related TFs, such as NACs, KNAT1, and MYBs, may form a multiple-knotted regulatory network and mediate GA signaling to promote SCW formation in plant stems.
Upland cotton (Gossypium hirsutum cv Jimian No. 14) plants were used throughout this study and grown in a growth chamber at 25°C under 16 h light (40,000 lux)/8 h dark cycle with 50% humidity conditions.
Gibberellin (GA4, Sangon Biotech, Shanghai, China) and paclobutrazol (PAC, Sangon Biotech, Shanghai, China) were dissolved in dimethyl sulfoxide (DMSO) and diluted into water to 100 and 50 μM, respectively. Ten-day-old cotton seedlings were sprayed with GA and/or PAC. The first internode length was measured every 2 days, and the treated plants were collected for phenotype observation and gene expression analyses 8 days later.
To investigate DELLA protein level by immunoblotting, the primary antibody anti-GhGAI1 used in this study was customized at Shanghai Youke Biotechnology Co., Ltd (Shanghai, China). The antibody was generated in rabbits by immunization with 246 amino acids at the N terminal of GhGAI1D. The first internodes from the seedlings were ground in liquid nitrogen and homogenized in three volumes of an extraction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerin, 1% Triton X-100, 1 mM DTT, 1× protease inhibitor cocktail). Extracts were centrifuged three times for 10 min at 15,000 × g and 4°C, and the supernatant was quantified by BCA assay. Total soluble proteins were boiled in an SDS sample buffer and further subjected to immunoblotting. The western blots were incubated with a 1:1,000 diluted anti-GhGAI1 antibody.
To determine cell length, the epidermis in the middle area of the first internodes were removed with forceps and observed with an optical microscope (Olympus IX81, Olympus, Japan). Cell length was measured in the collected images using ImageJ (http://imagej.net/Fiji). To localize the lignified cell wall, hand-cut transverse sections from the first internodes were incubated in a 1% phloroglucinol solution (in 75% ethanol) for 5 min, treated with concentrated hydrochloric acid for 1 min, and photographed using a stereomicroscope (Zeiss Discovery V20, Carl Zeiss AG, Germany). The autofluorescence of lignin was visualized using an optical microscope (Olympus IX81, Olympus, Japan) under UV conditions.
To knockdown the expression of GhGAIs, GhSND2s, GhFSNs, and GhVND4s genes by VIGS, 150–200 bp fragments of GhGAI1D, GhGAI2D, GhGAI3D, GhGAI4D, GhSND2-1A, GhSND2-2A, GhSND2-3A, GhFSN1A, GhFSN2A, GhVND4-1A, GhVND4-2A, GhVND4-3A, GhVND4-4A, and GhVND4-5A were amplified from the cDNA of developing cotton stems using the VIGS primers (Supplementary Table 1). Asymmetric overlap extension polymerase chain reaction (PCR) methods were employed to fuse related fragments (GhGAI1D-GhGAI2D, GhGAI3D-GhGAI4D, GhSND2-1A-GhSND2-2A-GhSND2-3A, GhFSN1A-GhFSN2A, and GhVND4-1A-GhVND4-2A-GhVND4-3A-GhVND4-4A-GhVND4-5A) (Xiao et al., 2007), which were subsequently inserted into the pTRV2 vector with EcoRI and KpnI sites to generate VIGS vectors pTRV2-GhGAI1 and GhGAI2, pTRV2-GhGAI3 and GhGAI4, pTRV2-GhSND2s, pTRV2-GhFSNs, and pTRV2-GhVND4s, respectively. The pTRV1, pTRV2, and derived pTRV2 VIGS vectors were transformed into the Agrobacterium strain GV3101 by electroporation (MicroPulser, Bio-Rad, California, United States). The VIGS assays were carried out as described previously (Ma et al., 2016).
Total RNAs were extracted from various cotton tissues using a rapid plant RNA extraction kit (Aidlab, Beijing, China). First-strand cDNAs were generated from 1 μg total RNA using a PrimeScript™ RT reagent kit (TaKaRa, Dalian, China) with gDNA eraser to degrade trace genomic DNAs according to the instructions of the manufacturer. Quantitative PCRs were performed with SYBR-Green PCR Mastermix (Vazyme, Nanjing, China). Amplification was monitored on a real-time basis using a CFX96 real-time PCR system (Bio-Rad, California, United States). The thermocycling parameters were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 5 s, 57°C for 20 s, and a standard melting curve program was added to monitor PCR specificity. GhACT was used as an internal control (Artico et al., 2010). The qRT-PCR results were analyzed using Bio-Rad CFX Manager 2.0 provided by the manufacturer (Bio-Rad, California, United States). The primers are listed in Supplementary Table 1.
The first internodes were dried and crushed into powder and passed through an 80-mesh sieve before component analysis. Lignin content was quantified by a modified protocol (Syros et al., 2004). In brief, a 50-mg sample was weighed, placed in a test tube, and treated with 5 ml 80% (v/v) ethanol in a water bath at 80°C for 2 h. The extracts were centrifuged at 10,000 × g for 10 min, and the supernatants were discarded. Extraction was repeated twice. The pellet was suspended in 4 ml chloroform and incubated for 1 h at 62°C. After centrifuging at 10,000 × g for 10 min, the precipitate was dried for 2 days at 50°C and dissolved in a 2.6 ml acetyl bromide/acetic acid solution (1:3, v/v). After incubation in a 70°C water bath for 1 h, 580 μl of a mixed solution of 17.24% (v/v) 2 N sodium hydroxide and 82.76% (v/v) acetic acid was added, and 100 μl of each sample was transferred to a fresh tube. Finally, 20 μl of 7.5 M hydroxylamine hydrochloride was added to the tube to terminate the reaction, and the volume was fixed to 2 ml with acetic acid. The absorbance at 280 nm was determined using a spectrophotometer (Varioskan LUX multimode microplate reader, Thermo Fisher Scientific, Massachusetts, United States).
Cellulose content was measured using a modified Updegraff method (Updegraff, 1969). Briefly, a 50-mg sample was first treated with 5 ml 80% (v/v) ethanol in an 80°C water bath for 2 h. The mixture was centrifuged at 10,000 × g for 10 min, and the supernatant was discarded. These steps were repeated three times. The precipitate was further digested with a 5 ml acetic/nitric reagent (mixing 150 ml 80% acetic acid and 15 ml concentrated nitric acid) in a boiling water bath for 30 min. After cooling at room temperature and centrifuging at 10,000 × g for 10 min, the supernatant was discarded. The digestion steps were repeated twice until the insoluble pellet became completely white. The pellet was washed twice with 10 ml distilled water and twice with 5 ml acetone, and left to air dry overnight. The dried pellet was re-suspended in 5 ml 72% (v/v) H2SO4, fully mixed, and let to stand at room temperature for 1 h or more until the pellet was completely hydrolyzed. After centrifuging at 10,000 × g for 10 min, 1 ml supernatant was diluted to 100 ml with distilled water. Then 2 ml dilution were transferred to a fresh test tube, and mixed with 0.5 ml freshly prepared 2% anthrone in ethyl acetate and 5 ml concentrated H2SO4. After incubation in boiling water for 5 min, the absorbance at 620 nm was measured using a spectrophotometer (Varioskan LUX multimode microplate reader, Thermo Fisher Scientific, Massachusetts, United States). Cellulose concentration was measured against a standard curve.
Total RNAs were extracted from the first internode of seedlings using a plant RNA extraction kit (AidLab, Beijing, China). RNA detection, sequencing, and routine data analysis were performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd (www.majorbio.com). Raw data were deposited in GenBank (SRA accession No.PRJNA703439). Paired-end clean reads of 150 nt in size (over 6.29 Gb) were aligned to the revised genome assemble of G. hirsutum (https://phytozome.jgi.doe.gov/pz/portal.html#!bulk?org=Org_Ghirsutum_er) using hisat2 v2.1.0. RSEM v1.3.1 was used to count the reads mapped to each gene, and then the number of fragments per kilobase of transcript sequence per million base pairs (FPKM) of each gene was calculated as the normalized expression level (Anders and Huber, 2010). Differential expression analyses were performed using the DEGSeq R package v1.38.0 (http://bioconductor.org/packages/release/bioc/html/DEGseq.html). Genes with more than two fold change and P ≤ 0.001 were defined as differentially expressed genes (DEGs). Pathway analysis was mainly based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2008).
For the Y2H assay, full length coding sequences of GhVND1-4D, GhVND4-3A, GhVND4-5D, GhFSN1A, GhFSN2D, GhSND2-1A, GhSND2-1D, GhSND2-2A, GhSND2-2D, and GhSND2-3A were inserted into a pGBKT7 bait vector, and the C-terminal fragments of GhGAI1A and GhGAI1D (GRAS domain) were cloned into a pGADT7 prey vector. Final bait and prey constructs were co-transformed into yeast strain AH109 according to the manual of the manufacturer (Clontech, Dalian, China). Transformed yeast cells were grown on a SD/-Trp/-Leu medium for selection. Protein–protein interactions were tested on a SD/-Trp/-Leu/-His/-Ade medium.
For the BiFC assay, full-length coding sequences of GhVND1-4D, GhVND4-3A, GhVND4-5D, GhFSN1A, GhFSN2D, GhSND2-1A, GhSND2-1D, GhSND2-2A, GhSND2-2D, GhSND2-3A, and GhGAI1D were transferred into pEaelyGate201-YN or pEaelyGate202-YC vectors (BioVector, Beijing, China) to generate GhVND1-4D-nYFP, GhVND4-3A-nYFP, GhVND4-5D-nYFP, GhFSN1A-nYFP, GhFSN2D-nYFP, GhSND2-1A-nYFP, GhSND2-1D-nYFP, GhSND2-2A-nYFP, GhSND2-2D-nYFP, GhSND2-3A-nYFP, and GhGAI1D-cYFP. Agrobacterium strains (GV3101) containing the above plasmids were co-infiltrated into leaves of Nicotiana benthamiana. A yellow fluorescent protein (YFP) fluorescence signal was observed 2 days after infiltration with a confocal microscope (Olympus FV1000, Olympus, Japan). To indicate the nuclei, 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/ml, Sigma-Aldrich, United States) was infiltrated into leaves 30 min before observation.
All statistical analyses were performed by Student's t-test (GraphPad Prism 8.0). The quantitative differences between the two groups of data for comparison with P < 0.05 or <0.01) were shown to be statistically significant or extremely significant, respectively.
To investigate the roles of the gibberellin signaling pathway during cotton stem development, we treated the cotton seedlings with gibberellin (GA) and/or its biosynthesis inhibitor, PAC. As previously reported (Achard and Genschik, 2009), the GA treatment significantly promoted plant growth and stem elongation, while the PAC treatment showed opposite effects, which can be restored by GA feeding (Figures 1A,B). Next, phloroglucinol staining was employed to detect lignification in the newly developed first internode. It was observed that the GA and PAC treatments significantly increased and decreased the staining area and intensity in xylem and phloem, respectively, compared with the control, indicating that GA enhanced lignification and SCW development in cotton stems (Figure 1C). Furthermore, the immunoblot analysis revealed that the DELLA protein level in the cotton stems was significantly lowered by GA and elevated upon PAC treatment (Figure 1D), indicating that the common GA signaling process, GA-induced DELLA degradation, functioned in the cotton stems. Collectively, these results suggested that GA promoted stem elongation and SCW development via the mediation of the degradation of DELLA proteins in the cotton seedlings.
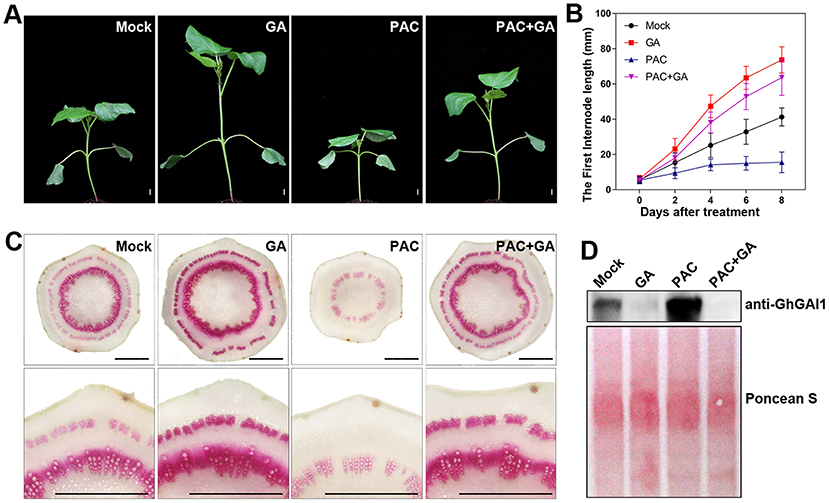
Figure 1. Effects of exogenous GA and PAC on stem elongation and lignification, and DELLA protein level in cotton. Ten-day-old seedlings sprayed with water (Mock), 100 μM GA (GA), 50 μM PAC (PAC), and mixed GA and PAC (GA + PAC) were grown at 25°C under 16 h/8 h (light/dark) conditions for 8 days. (A) The phenotype of cotton plants. Bar = 10 mm. (B) Time courses of first internode elongation. (C) Phloroglucinol staining of cross-sections of the first internode, aligned with the enlarged view at the lower panel. Bar = 1 mm. (D) Cotton DELLA protein level detected with anti-GhGAI1 in the first internodes. The amounts of total proteins transferred onto membrane are indicated by Ponceau S staining.
To genetically analyze the GA function in stem development, we tried to employ the VIGS method to downregulate DELLA genes in the cotton stems. First, we performed a genome-wide analysis of the DELLA gene family in cotton using Arabidopsis DELLA protein GAI as a query probe. A total of eight and four DELLA genes were identified in the assembled G. hirsutum and extant diploid ancestor species (G. raimondii and G. arboretum) genomes (Supplementary Table 2), respectively. According to their sequence similarity, cotton DELLA proteins could be divided into four homeologous groups (GoGAI1~4), among which GoGAI1 and GoGAI2 belonged to clade I (DELLA) protein and GoGAI3 and GoGAI 4 belonged to clade II (DGLLA) protein (Supplementary Table 2 and Supplementary Figure 1) (Hu et al., 2011). These genes were named as species abbreviations plus homeologous groups plus subgenome (A or D) in tetraploid (Supplementary Table 2). Based on normalized transcriptomic data in the Cotton FGD collections (https://cottonfgd.org/), we found that clade I DELLA protein genes, especially GhGAI1A/D, predominantly expressed in all the tissues investigated, although the expression of clade II DELLA protein genes was also detectable (Supplementary Figure 2).
Next, we ask whether the VIGS of GhGAIs may reproduce the effects of the GA treatment on cotton stem development. As shown in Figures 2A,B, TRV:GhGAI1 and GhGAI2 and TRV:GhGAI3 and GhGAI 4, targeting clade I and clade II DELLA genes, respectively, could silence the target genes specifically in the cotton stems. Both GhGAI1 and GhGAI2- and GhGAI3 and GhGAI4-silenced cotton displayed faster plant growth and stem elongation (Figures 2C,D), similar to the effects of the exogenous application of GA. These effects were additive to some extent (Figure 2D), but GhGAI1&2-silencing could well-reproduce the effects of the GA treatment, such as lowered DELLA levels, enhanced elongation, and lignification (comparing Figures 2, 3, and Supplementary Figure 3 with Figure 1). Comparing the GhGAI1&2-silenced cotton with the control, the diameters of the internodes were significantly increased (Figure 3A) with enhanced SCW development in the xylem and phloem (Supplementary Figure 4). Lignification was significantly enhanced in both vascular bundles and interfascicular regions (Figures 3B,C). It was confirmed that the lignin and cellulose contents in the first internode were significantly elevated in the GhGAI1&2-silenced cotton 15 and/or 20 days post infiltration (Figures 3D,E). Collectively, the VIGS of GhGAIs (especially GhGAI1 and GhGAI2) could phenocopy the GA treatment to promote stem elongation and SCW formation in the xylem and phloem of the cotton seedlings.
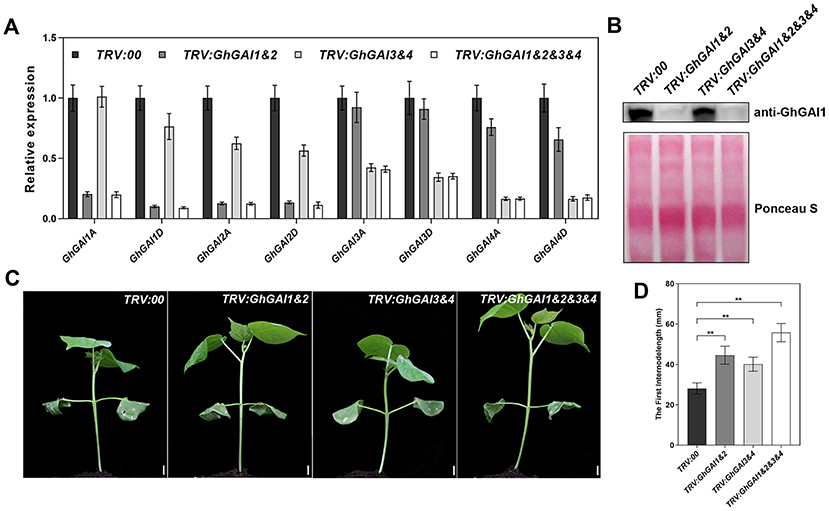
Figure 2. Functional characterization of GhGAIs by VIGS analysis. Agrobacterium cultures carrying pTRV2-GhGAI1&2 (TRV:GhGAI1&2), pTRV2-GhGAI3&4 (TRV:GhGAI3&4) or both (TRV:GhGAI1&2&3&4), mixed with pTRV1 Agrobacterium, were infiltrated into two fully expanded cotyledons of 7-day-old cotton seedlings. Empty vector pTRV2-00 (TRV:00) was used as control. (A) Transcript levels of 8 GhGAIs in the first internode were detected 15 days post infiltration (DPI) by qRT-PCR. (B) DELLA protein levels detected with anti-GhGAI1 in immunoblot analysis. The amounts of total proteins transferred onto the membrane are indicated by Ponceau S staining. (C) Fifteen DPI seedlings. Bar = 10mm. (D) The first internode length measured 15 DPI. ** indicates a significant difference compared with control (t-test, P < 0.01, n ≥ 10). Error bars indicate SD.
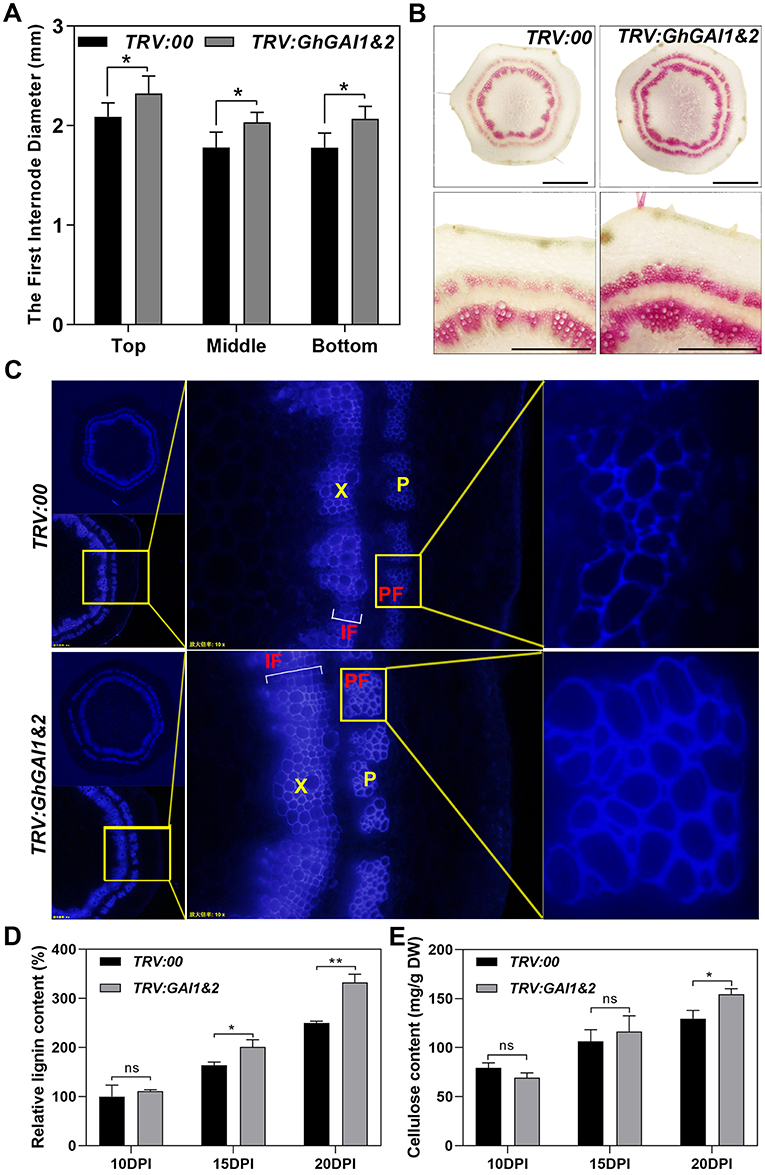
Figure 3. Silencing of GhGAI1 and GhGAI 2 enhances SCW development in the first internodes. Agrobacterium cultures carrying empty vector pTRV2-00 (TRV:00) or pTRV2-GhGAI1&2 (TRV:GAI1&2), mixed with pTRV1 Agrobacterium, were infiltrated into two fully expanded cotyledons of 7-day-old cotton seedlings. (A) The first internode diameters at the top, middle and bottom parts measured 20 DPI. * indicates a significant difference compared with control (t-test, P < 0.05, n ≥ 10). Error bars indicate SD. (B) Phloroglucinol staining of the cross-sections of 20 DPI first internode in the middle region. Bar = 1 mm or 500 μm (amplification). (C) Microscopic observation of cross-sections of 20 DPI first internode in the middle region under UV conditions. (D) Lignin and (E) cellulose contents in the first internodes, respectively. The internodes were sampled 10, 15, and 20 DPI. Lignin content is shown relative to the control level 10 days post infiltration. * and ** indicate a significant difference compared with the control, with P-values of 0.05 and 0.01, respectively (t-test, n = 3). Error bars indicate SD. P, phloem; PF, phloem fibers; X, xylem; and IF, interfascicular region.
To clarify the mechanism of DELLA proteins that regulate secondary cell walls, we carried out RNA-seq analyses using the first internode of a wild type treated with mock, GA, or PAC for 8 days, as well as TRV:00 and GhGAI1&2-silenced plants 15 and 20 DPI. A total of 148 differentially expressed genes (DEGs), which responded to GA and PAC in the cotton stems, were identified in GhGAI1&2-silenced plants 15 DPI (fold change > 2), including 79 upregulated and 69 downregulated DEGs. Under the same filter conditions, 102 DEGs (47 upregulated and 55 downregulated DEGs) were identified in GhGAI1&2-silenced plants 20 DPI (Supplementary Figure 5A). Out of these DEGs, we performed union computation, which resulted in the identification of 97 upregulated and 94 downregulated DEGs as GA signal-responsive genes in the cotton stems, and these DEGs were subjected to subsequent analyses (Supplementary Figure 5B). The KEGG pathway analysis showed that the upregulated genes were mainly involved in phenylpropanoid biosynthesis, starch and sucrose metabolism, galactose metabolism, and zeatin biosynthesis (Supplementary Figure 5C). In contrast, the downregulated genes were enriched in glutathione metabolism and plant hormone signal transduction (Supplementary Figure 5D). Notably, gibberellin 2-oxidase (GA2ox) was upregulated, while gibberellin 3-oxidase (GA3ox) and gibberellin receptor GID1 were downregulated in GhGAI1&2-silenced plants, indicating feedback regulation of the gibberellin signal in developing cotton stems.
We further employed the qRT-PCR method to verify the expression levels of SCW formation-related genes. It was shown that SCW-specific cellulose biosynthesis (CesA4, CesA7, and CesA8), hemicellulose biosynthesis (IRX9 and IRX14), lignin biosynthesis (PAL, 4CL, CCoAOMT, and F5H), and polymerization genes (LAC4, PER4, and PER21) were upregulated in GhGAI1&2-silenced plants (Figure 4). The expression levels of these genes were also induced by the GA treatment and reduced by the PAC treatment (Figure 4). These results suggested that DELLA proteins mediated the GA signal to regulate SCW formation by regulating the genes related to cellulose, hemicellulose and lignin biosynthesis, and polymerization.
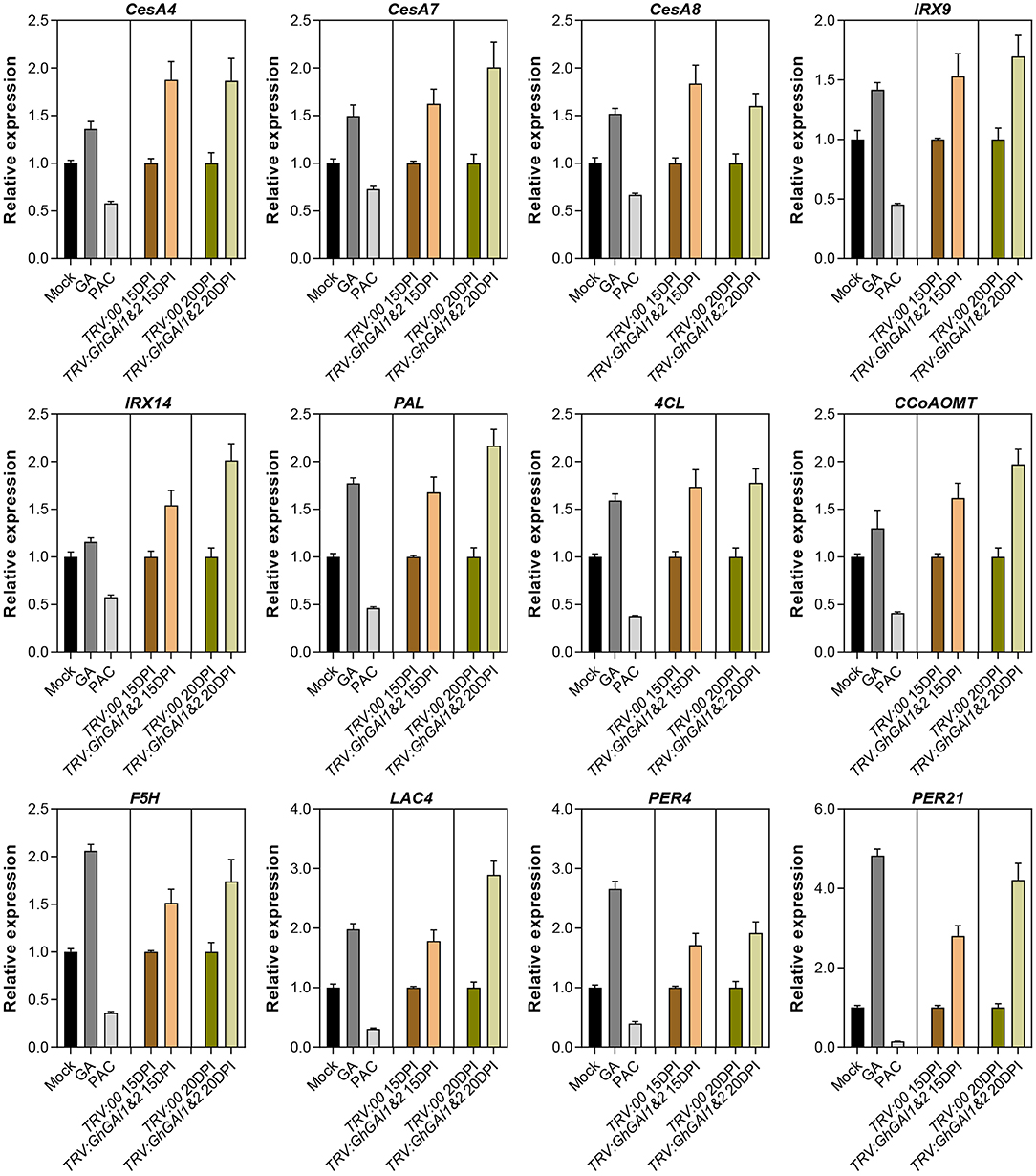
Figure 4. Quantitative RT-PCR analysis of genes related to SCW formation. Total RNAs were isolated from the first internodes. Mock, GA, and PAC represent 10-day-old seedlings treated with water, 100 μM GA, and 50 μM PAC for 8 days. VIGS-treated materials are indicated with pTRV2 vectors (TRV:00 and TRV:GhGAI1&2) plus sampling time (15 or 20 DPI). The relative expression levels in GA/PAC-treated and GhGAI1&2-silenced seedlings are then normalized to Mock and TRV:00, respectively. Gene accession numbers in cotton FGD: CesA4 (Gh_D08G0509); CesA7 (Gh_D05G0079); CesA8 (Gh_D10G0333); IRX9 (Gh_A09G1418); IRX14 (Gh_D11G1966); PAL (Gh_D10G2528); 4CL (Gh_D10G0473); CCoAOMT (Gh_A04G1032); F5H (Gh_Sca004990G01); LAC4 (Gh_A11G2936); PER4 (Gh_A03G0199); and PER21 (Gh_A09G1415).
Next, we attempted to identify any mediator between the GA-DELLA signal and SCW formation in the cotton stems. To this end, we aimed to test the possible interaction between the GhGAI1D and cotton homologs of SCW-related NAC proteins. On the basis of the previous report (Zhang et al., 2018), a total of 62 SCW-related NAC genes were identified in the upland cotton genome (Supplementary Figure 6 and Supplementary Table 3). These genes included 34 homologs of tier 3 NAC TFs (VND1–7, NST1–3) and six SND2/3 homologs, and 18 were homologous to SCW negative regulators (XND1 and VNI2) (Zhao et al., 2007; Yamaguchi et al., 2010). As shown in Table 1 and Figure 5, BiFC analyses showed significant interactions in nuclei between DELLA protein GhGAI1D and all the investigated tier 3 TFs and SND2/3 homologs (each of five NAC proteins), while in the Y2H assay, interactions between GhGAI1D and seven NACs were confirmed, and false-negative results were observed for the rest of the three NACs (Table 1 and Supplementary Figure 7). These results demonstrated that predominant DELLA proteins in the cotton stems (GhGA1D) may have interacted with SCW-related NAC switches of different tiers, implying that these NACs function as mediators of the GA-DELLA signal to regulate SCW formation in cotton stems.
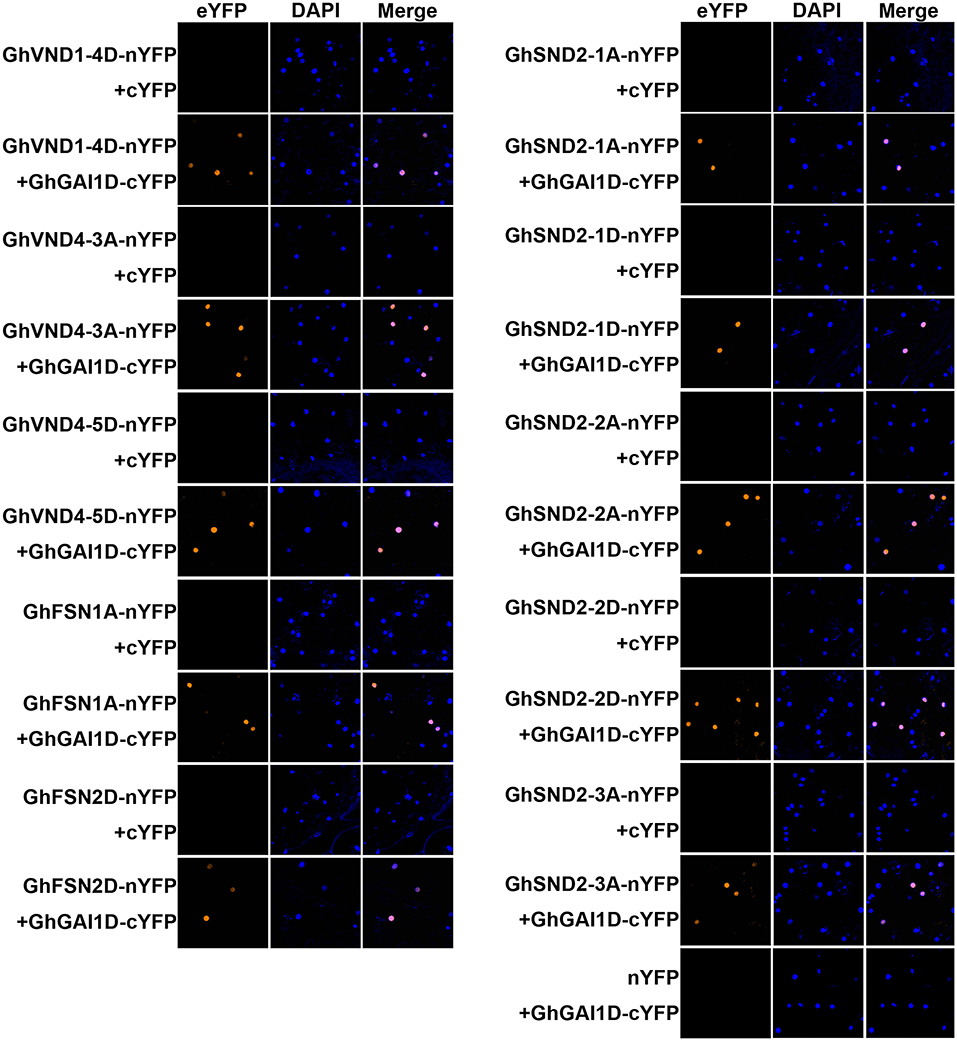
Figure 5. BiFC analyses of interactions between GhGAI1D and SCW-related NACs. NACs fused to N-terminal fragment of YFP (NAC-nYFP) were co-infiltrated with GhGAI1D fused to C-terminal fragment of YFP (GhGAI1D-cYFP) or unfused cYFP into N. benthamiana leaves. Unfused cYFP and nYFP were used as negative controls. The nuclei were indicated by DAPI staining.
To further validate the function of these DELLA-interacting NACs in cotton stem development, we employed the VIGS method to downregulate their expressions. The effects of TRV:GhSND2s treatment is shown in Figure 6. The expression levels of GhSND2-1A, GhSND2-1D, GhSND2-2A, GhSND2-2D, GhSND2-3A, and GhSND2-3D were all downregulated in the first internode of TRV:GhSND2s plants 15 DPI (Figure 6A). Meanwhile, lighter phloroglucinol staining intensity in the xylem area and phloem fibers was observed in the first internode of ChSND2s-silenced plants compared with TRV:00 plants (Figure 6B), indicating the knockdown of GhSND2s resulted in reduced lignification and SCW development in cotton stems. We further examined the expression levels of SCW-related genes and found that the knockdown of GhSND2s suppressed the expression of genes associated with cellulose (CesA8) and hemicellulose biosynthesis (IRX14), and lignin polymerization (LAC4, PER4, and PER21). Monolignol biosynthetic genes (such as PAL, 4CL, CCoAOMT, COMT, and F5H), however, had no differential expression levels between TRV:GhSND2s and TRV:00 cotton plants (Figure 6C), which was consistent with the report on Arabidopsis (Hussey et al., 2011). The VIGS of the other groups of DELLA-interacting NACs (GhFSNs and GhVNDs) also resulted in reduced SCW formation in cotton stems (Supplementary Figure 8). These results demonstrated that these DELLA-interacting NACs positively regulate the SCW formation in cotton stems.
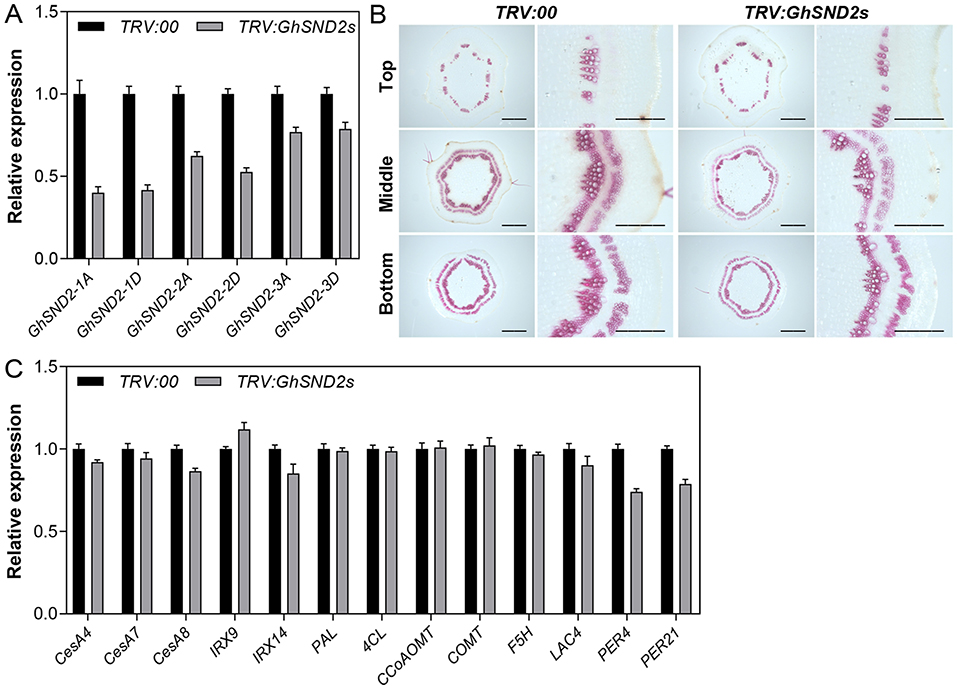
Figure 6. VIGS analysis of GhSND2s. Agrobacterium cultures that carry pTRV1 and pTRV2-GhSND2s (TRV:GhSND2s) were infiltrated into two fully expanded cotyledons of 7-day-old cotton seedlings. Empty vector pTRV2-00 (TRV:00) was used as control. (A) Transcript levels of six GhSND2 genes in the first internodes 15 days post infiltration (DPI) by qRT-PCR analysis. (B) Phloroglucinol staining of the cross-sections of 15 DPI first internodes. Bar = 1 mm or 500 μm (amplification). (C) Quantitative RT-PCR analysis of the genes related to SCW formation in the first internodes.
SCW formation in stems is essential to plant growth and has a wealth of industrial applications (Kumar et al., 2016; Hofte and Voxeur, 2017; Zhong et al., 2019). This research represented an effort to study this important process with a rapid genetic method (VIGS) in a new system (the first internode of cotton). In this system, the effects of gene knockdown could be observed in a short period (<1 month), as exemplified in the silencing of DELLA and NAC genes. Cotton has a stem structure typical of woody plants. Cultivar cotton exhibits the characteristics of herbaceous plants, such as annual and shrub-like plants, because of artificial selection (Mcgarry et al., 2016). Considering their accessibility and readiness, the first internode of growing cotton may be an alternative model, other than Arabidopsis and poplar (Kumar et al., 2016; Zhong et al., 2019), for biological research on SCW (wood) formation.
Most dicots have DELLA proteins that belong to two clades (Van De Velde et al., 2017), such as cotton (Supplementary Figure 1). DELLAs in clade II were characterized by a conserved DGLLA motif instead of the DELLA motif in clade I. Rice DGLLA proteins SLRL1 and SLRL2 may suppress plant growth like DELLA protein (Itoh et al., 2005). In cotton stems, clade I DELLAs have dominant expression compared with clade II DELLAs (Supplementary Figure 2). Silencing of both clades of DELLA genes resulted in increased stem growth. The clade II DELLA genes showed lesser effects than those of clade I, and simultaneous silencing of these DELLA proteins had additive effects on promoting cotton growth (Figures 2C,D), indicating that the two clades of DELLA may have redundant functions in regulating the development of cotton stems. Although western blot only detected GhGAI1 degradation induced by GAs (Figure 1), we envisioned that both clade I and II DELLAs shared a similar mechanism to affect stem development in cotton.
DELLA proteins are the major repressor of GA signaling, and mediate GA response by interacting with a wealth of downstream regulators (Daviere and Achard, 2013). In SCW regulation, KNAT1 was reported to interact with DELLA protein and mediated GA promotion of SCW differentiation in Arabidopsis. DELLA proteins also repress the functions of NAC29/31 and MYB103L in SCW formation in rice. Here, we indicated that DELLA protein interacted with at least 10 NAC proteins, which were homologous to most switches that promote SCW formation. It can be predicted that there existed other SCW regulators that have not yet been identified, and DELLA proteins and their interactors constituted a transcriptional regulatory network mediating GA regulation of secondary cell walls in plants.
In addition to GAs, other phytohormones are involved in the formation of wood. Auxin plays a key role in regulating wood formation by affecting cambium activity and xylem development (Savidge, 1996; Uggla et al., 1996, 1998; Tuominen et al., 1997). Brassinosteroids are thought to be produced in procambial cells and trigger xylem precursor cells to induce xylem differentiation and regulate secondary cell wall biosynthesis (Milhinhos and Miguel, 2013; Yuan et al., 2019). Ethylene synthesis occurs in the apoplast of xylem elements; and ethylene participates, in a paracrine manner, in the control of the cambial stem cell pool size during secondary xylem formation (Pesquet and Tuominen, 2011). More and more studies have revealed a comprehensive view of DELLAs by identifying interactions with several types of TFs, which are considered to be central command systems that integrate various signals (Daviere and Achard, 2013). A previous study on Arabidopsis has revealed that DELLA interacts with ARFs, BZR1, and EIN3 (An et al., 2012; Bai et al., 2012; Oh et al., 2014) to mediate crosstalk between gibberellin and auxin, brassinosteroids, and ethylene, respectively. Therefore, DELLA may be a key component that integrates GA with other phytohormones in wood formation regulation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA703439.
YWa and YX designed and wrote the manuscript. YWa, WY, LR, YDo, YQ, YDa, YWu, and XO performed experiments. YWa, WY, ZC, CW, YDa, YWu, and XO analyzed data. QS, JZ, YL, and AL discussed and corrected the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by National Natural Science Foundation of China (31571582, 31871539, and U2003209 to XY) and by Chongqing Science and Technology Bureau (cstc2018jscx-msybX0382 to XY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.655127/full#supplementary-material
Achard, P., and Genschik, P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. doi: 10.1093/jxb/ern301
An, F., Zhang, X., Zhu, Z., Ji, Y., He, W., Jiang, Z., et al. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22, 915–927. doi: 10.1038/cr.2012.29
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Artico, S., Nardeli, S. M., Brilhante, O., Grossi-de-Sa, M., and Alves-Ferreira, M. (2010). Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 10:71. doi: 10.1186/1471-2229-10-49
Bai, M.-Y., Shang, J.-X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Bai, W. Q., Xiao, Y. H., Zhao, J., Song, S. Q., Hu, L., Zeng, J. Y., et al. (2014). Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS ONE 9:e96537. doi: 10.1371/journal.pone.0096537
Biemelt, S., Tschiersch, H., and Sonnewald, U. (2004). Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol. 135, 254–265. doi: 10.1104/pp.103.036988
Daviere, J. M., and Achard, P. (2013). Gibberellin signaling in plants. Development 140, 1147–1151. doi: 10.1242/dev.087650
Digby, J., and Wareing, P. F. (1966). The effect of applied growth hormones on cambial division and the differentiation of the cambial derivatives. Ann. Botany 30, 539–548. doi: 10.1093/oxfordjournals.aob.a084095
Felipo-Benavent, A., Urbez, C., Blanco-Tourinan, N., Serrano-Mislata, A., Baumberger, N., Achard, P., et al. (2018). Regulation of xylem fiber differentiation by gibberellins through DELLA-KNAT1 interaction. Development 145:164962. doi: 10.1242/dev.164962
Hofte, H., and Voxeur, A. (2017). Plant cell walls. Curr. Biol. 27, R865–R870. doi: 10.1016/j.cub.2017.05.025
Hu, M.-Y., Luo, M., Xiao, Y.-H., Li, X.-B., Tan, K.-L., Hou, L., et al. (2011). Brassinosteroids and Auxin Down-Regulate DELLA Genes in Fiber Initiation and Elongation of Cotton. Agricul. Sci. China 10, 1168–1176. doi: 10.1016/S1671-2927(11)60107-7
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27:15. doi: 10.1105/tpc.15.00015
Hussey, S. G., Mizrachi, E., Spokevicius, A. V., Bossinger, G., Berger, D. K., and Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11:173. doi: 10.1186/1471-2229-11-173
Israelsson, M., Eriksson, M. E., Hertzberg, M., Aspeborg, H., Nilsson, P., and Moritz, T. (2003). Changes in gene expression in the woodforming tissue of transgenic hybrid aspen with increased secondary growth. Plant Mol. Biol. 52, 893–903. doi: 10.1023/A:1025097410445
Israelsson, M., Sundberg, B., and Moritz, T. (2005). Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in woodforming tissues in aspen. Plant J. 44, 494–504. doi: 10.1111/j.1365-313X.2005.02547.x
Itoh, H., Shimada, A., Ueguchi-Tanaka, M., Kamiya, N., Hasegawa, Y., Ashikari, M., et al. (2005). Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 44, 669–679. doi: 10.1111/j.1365-313X.2005.02562.x
Jeon, H. W., Cho, J. S., Park, E. J., Han, K. H., Choi, Y. I., and Ko, J. H. (2016). Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 14, 1161–1170. doi: 10.1111/pbi.12484
Kanehisa, M., Araki, M., Goto, S., Hattori, M., and Itoh, M. (2008). KEGG for linking genomes to life and the environment. Nuclc Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kumar, M., Campbell, L., and Turner, S. (2016). Secondary cell walls: biosynthesis and manipulation. J. Exp. Bot. 67, 515–531. doi: 10.1093/jxb/erv533
Ma, D., Hu, Y., Yang, C., Liu, B., Fang, L., Wan, Q., et al. (2016). Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7:10456. doi: 10.1038/ncomms10456
Mauriat, M., and Moritz, T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. The Plant Journal 58, 989–1003. doi: 10.1111/j.1365-313X.2009.03836.x
Mcgarry, R. C., Prewitt, S. F., Culpepper, S., Eshed, Y., Lifschitz, E., and Ayre, B. G. (2016). Monopodial and sympodial branching architecture in cotton is differentially regulated by theGossypium hirsutum SINGLE FLOWER TRUSSandSELF-PRUNINGorthologs. New Phytol. 212, 244–258. doi: 10.1111/nph.14037
Meents, M. J., Watanabe, Y., and Samuels, A. L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121, 1107–1125. doi: 10.1093/aob/mcy005
Milhinhos, A., and Miguel, C. M. (2013). Hormone interactions in xylem development: a matter of signals. Plant Cell Rep. 32, 867–883. doi: 10.1007/s00299-013-1420-7
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC Transcription Factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Oh, E., Zhu, J.-Y., Bai, M.-Y., Arenhart, R. A., Sun, Y., and Wang, Z.-Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3:25. doi: 10.7554/eLife.03031.025
Park, E. J., Kim, H. T., Choi, Y. I., Lee, C., Nguyen, V. P., Jeon, H. W., et al. (2015). Overexpression of gibberellin 20-oxidase1 from Pinus densiflora results in enhanced wood formation with gelatinous fiber development in a transgenic hybrid poplar. Tree Physiol. 35, 1264–1277. doi: 10.1093/treephys/tpv099
Pesquet, E., and Tuominen, H. (2011). Ethylene stimulates tracheary element differentiation in Zinnia elegans cell cultures. New Phytol. 190, 138–149. doi: 10.1111/j.1469-8137.2010.03600.x
Savidge, R. A. (1996). Xylogenesis, Genetic and Environmental Regulation-A Review. Iawa J. 17, 269–310. doi: 10.1163/22941932-90001580
Syros, T., Yupsanis, T., Zafiriadis, H., and Economou, A. (2004). Activity and isoforms of peroxidases, lignin and anatomy, during adventitious rooting in cuttings of Ebenus cretica L. J. Plant Physiol. 161, 69–77. doi: 10.1078/0176-1617-00938
Taylor-Teeples, M., Lin, L., De Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tuominen, H., Puech, L., Fink, S., and Sundberg, B. (1997). A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 115, 577–585. doi: 10.1104/pp.115.2.577
Uggla, C., Mellerowicz, E. J., and Sundberg, B. R. (1998). Indole-3-acetic acid controls cambial growth in scots pine by positional signaling. Plant Physiol. 117, 113–121. doi: 10.1104/pp.117.1.113
Uggla, C., Moritz, T., Sandberg, G., and Sundberg, B. (1996). Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. U.S.A. 93, 9282–9286. doi: 10.1073/pnas.93.17.9282
Updegraff, D. M. (1969). Semimicro determination of cellulose in biological materials. Analyt. Biochem. 32, 420–424. doi: 10.1016/S0003-2697(69)80009-6
Van De Velde, K., Ruelens, P., Geuten, K., Rohde, A., and Van Der Straeten, D. (2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22, 880–893. doi: 10.1016/j.tplants.2017.07.010
Wang, G. L., Que, F., Xu, Z. S., Wang, F., and Xiong, A. S. (2017). Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 254, 839–848. doi: 10.1007/s00709-016-0995-6
Wang, Z. G., Chai, G. H., Wang, Z. Y., Tang, X. F., Sun, C. J., Zhou, G. K., et al. (2013). Molecular mechanism of AtGA3OX1 and AtGA3OX2 genes affecting secondary wall thickening in stems in Arabidopsis. Yi Chuan 35, 655–665. doi: 10.3724/SP.J.1005.2013.00655
Wareing, P. F. (1958). Interaction between indole-acetic and gibberellic acid in cambial activity. Nature 181, 1744–1745. doi: 10.1038/1811744a0
Xiao, Y.-H., Yin, M.-H., Hou, L., Luo, M., and Pei, Y. (2007). Asymmetric overlap extension PCR method bypassing intermediate purification and the amplification of wild-type template in site-directed mutagenesis. Biotechnol. Lett. 29, 925–930. doi: 10.1007/s10529-007-9327-4
Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. doi: 10.1111/j.1365-313X.2008.03533.x
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Ye, Y., Liu, B., Zhao, M., Wu, K., Cheng, W., Chen, X., et al. (2015). CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol. Biol. 89, 385–401. doi: 10.1007/s11103-015-0376-0
Ye, Z. H., and Zhong, R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. doi: 10.1093/jxb/erv081
Yuan, H., Zhao, L., Guo, W., Yu, Y., Tao, L., Zhang, L., et al. (2019). Exogenous application of phytohormones promotes growth and regulates expression of wood formation-related genes in populus simonii × P. nigra. Int. J. Mol. Sci. 20:792. doi: 10.3390/ijms20030792
Zhang, J., Huang, G.-Q., Zou, D., Yan, J.-Q., Li, Y., Hu, S., et al. (2018). The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 217, 625–640. doi: 10.1111/nph.14864
Zhao, C., Avci, U., Grant, E. H., Haigler, C. H., and Beers, E. P. (2007). XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 53, 425–436. doi: 10.1111/j.1365-313X.2007.03350.x
Zhao, Q. (2016). Lignification: flexibility, biosynthesis and regulation. Trends Plant Sci. 21,713–721. doi: 10.1016/j.tplants.2016.04.006
Zhong, R., Cui, D., and Ye, Z. H. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., and Ye, Z.-H. (2010). Global analysis of direct targets of secondary wall NAC master switches in arabidopsis. Mol. Plant 3, 1087–1103. doi: 10.1093/mp/ssq062
Zhong, R., Lee, C., Zhou, J., Mccarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., and Ye, Z.-H. (2015). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Phys. 56, 195–214. doi: 10.1093/pcp/pcu140
Keywords: gibberellin, DELLA proteins, secondary cell wall formation, NAC proteins, cotton stem
Citation: Wang Y, Yu W, Ran L, Chen Z, Wang C, Dou Y, Qin Y, Suo Q, Li Y, Zeng J, Liang A, Dai Y, Wu Y, Ouyang X and Xiao Y (2021) DELLA-NAC Interactions Mediate GA Signaling to Promote Secondary Cell Wall Formation in Cotton Stem. Front. Plant Sci. 12:655127. doi: 10.3389/fpls.2021.655127
Received: 18 January 2021; Accepted: 18 May 2021;
Published: 09 July 2021.
Edited by:
David William McCurdy, The University of Newcastle, AustraliaReviewed by:
Anindya Ganguly, University of Virginia, United StatesCopyright © 2021 Wang, Yu, Ran, Chen, Wang, Dou, Qin, Suo, Li, Zeng, Liang, Dai, Wu, Ouyang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuehua Xiao, eGlhb3l1ZWh1YUBzd3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.