- Institute of Systems Biology, Universiti Kebangsaan Malaysia, Bangi, Malaysia
Hybridization is key to the evolution and diversity of plants in nature. Nepenthaceae comprises a family of diverse tropical carnivorous pitcher plant species with extensive hybridization. However, there is no study to date on the metabolite expression of hybrids in this family. We performed a non-targeted metabolomics analysis of the pitchers of two Nepenthes species with different dietary habits, namely, the semi-detritivorous N. ampullaria and carnivorous N. rafflesiana with their hybrid (N. × hookeriana) for a comparative study. The whole-pitcher samples were extracted in methanol:chloroform:water (3:1:1) via sonication-assisted extraction and analyzed using ultra-performance liquid chromatography time-of-flight mass spectrometry (UPLC-TOF-MS) followed by data analysis to profile chemical compositions. A total of 1,441 metabolite features were profiled from the three species in which 43.3% of features in the hybrid samples were not found in either of its parents. The partial least squares discriminant analysis (PLS-DA) found 324 metabolite features with variable in projection (VIP) values greater than one in which 55 features were statistically significant. This showed that the hybrid is closer to N. rafflesiana, which is consistent to the previous study on gene and protein expressions. A total of 105 metabolites were putatively identified with manual searches using public metabolite databases. Phenols were detected to be the most abundant secondary metabolites due to a high flavonoid content, especially in N. rafflesiana. The most abundant feature 476.3s:449.102 was found to be the most significant VIP for distinguishing between the three species as a chemical marker. This is the first study comparing metabolites in the carnivory organs of different Nepenthes species with comprehensive profiling and putative identification. The differential metabolite compositions in the pitchers of different species might have ecological implications with the hybrid showing intermediate phenotype between the parents as well as manifesting unique metabolites. However, there is no clear evidence of metabolites related to the differences in dietary habits between the hybrid and the two parent species.
Introduction
Nepenthes L. is the sole genus of paleotropical carnivorous pitcher plants from the monotypic Nepenthaceae family of Caryophyllales order that is highly distributed around the warm and humid equatorial countries, such as Indonesia, Philippines, and Malaysia (Adam, 1997). Nepenthes pitcher plants thrive on poor soils and acquire limiting nutrients mainly from insect prey via the unique fluid-filled pitcher organ developed at the leaf tip connected by a tendril, which allows prey trapping, digestion, and absorption (Clarke and Robinson, 2018). Fascinating pitcher shapes and sizes have been observed in this diverse family of carnivorous plants with over 150 extant species (Murphy et al., 2020), which are linked to the various ecological adaptations related to nutrient acquisition or dietary habits. For example, Nepenthes ampullaria Jack have evolved a semi-detritivorous habit with a vestigial pitcher lid bent away from the pitcher opening surrounded by a relatively flat peristome and growing on the forest floor to ease the trapping of fallen leaf litter, thus less dependent on catching prey (Moran et al., 2003; Pavlovič et al., 2011). In comparison, a typical carnivorous N. rafflesiana Jack produces larger pitchers with attractive coloration and a functional lid covering the pitcher opening, some of which secrete fragrance and sweet nectars to lure insect prey (Di Giusto et al., 2010). Additionally, the viscoelastic fluid (Bazile et al., 2015) and slippery epicuticular wax crystals of the inner pitcher wall with the anisotropic effect of lunate cells (Wang et al., 2018) also contribute to the success of prey capture and retention.
Hybridization is the formation of organisms by cross-fertilization between individuals of different species, which is key to the evolution and diversification of 30–70% flowering plants (Soltis and Soltis, 2009). Hybrids generally express phenotypes intermediate to those of parents, which sometimes result in heterosis or hybrid vigor such that the hybrids survive better than both parents, but the opposite is also possible (Orians, 2000; Goulet et al., 2017). Studies comparing the metabolite expression of hybrids with that of their parent species are limited, but existing evidence suggests that hybrids may express more diverse combinations of secondary metabolites or produce unique phytochemicals novel to both parental species (Cheng et al., 2011; Lätti et al., 2011). This leads to an interesting question on the expression of metabolites in the hybrids of Nepenthes species.
Hybridization in the species-rich Nepenthaceae is extensive, either naturally or artificially driven by high horticultural demand as ornamental plants. According to Harry James Veitch in 1906 (Veitch, 1979), the first Nepenthes artificial hybridization was initiated by John Dominy who produced N. × dominicana from N. rafflesiana with an unnamed species in 1862, followed by N. × hybrida from N. khasiana with an unnamed Bornean species in 1866. Despite the long history, studies on Nepenthes hybrids and their chemical compositions are generally limited. Studies of Nepenthes have largely focused on the proteins of pitcher fluids (Ravee et al., 2018). Recent studies have reported the effects of endogenous protein depletion on protein secretion in pitcher fluids of N. × ventrata (Wan Zakaria et al., 2019) and gene expression in pitchers of N. ampullaria with transcriptome analysis (Goh et al., 2020). The same proteomics informed by the transcriptomics approach was applied to explore the expression of genes and proteins in the pitchers and fluids, respectively, in N. ampullaria, N. rafflesiana, and their hybrid N. × hookeriana (Zulkapli et al., 2017; Zulkapli et al., 2021).
To date, information is only available on certain targeted metabolites that have been successfully isolated and characterized from some Nepenthes species. Advanced metabolite identification using high-performance liquid chromatography (HPLC), nuclear magnetic resonance (NMR), and gas chromatography–mass spectrometry (GC-MS) was performed on a few Nepenthes species in several studies, but the isolated and characterized compounds were targeted (Eilenberg et al., 2010; Kováčik et al., 2012). There is only one recent untargeted metabolite profiling study comparing the feeding effect on leaf blades and pitchers of N. × ventrata based on cheminformatics fingerprinting without compound identification (Dávila-Lara et al., 2020). The authors found little impact of feeding on the metabolite composition of pitchers with only small changes in the polar metabolites.
The current study aims to unveil the effects of hybridization in pitcher plants based on comparative metabolic profiling through an untargeted approach for identifying metabolites. Liquid chromatography–mass spectrometry (LC-MS) is used to classify and compare chemical profiles between N. × hookeriana, a natural hybrid with its parents, N. ampullaria and N. rafflesiana. The parentage of N. × hookeriana was previously discovered based on its morphological resemblance to N. ampullaria and N. rafflesiana (Danser, 1928), which was confirmed by molecular markers, including amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), and inter-simple sequence repeats (ISSR) (Kiew et al., 2003; Yulita and Mansur, 2012). Differences in the morphology and diet between N. ampullaria and N. rafflesiana may lead to differences in the metabolite compositions. Therefore, this study provides a comprehensive metabolite profiling of these three species to investigate how the parental molecular phenotypes are manifested in N. × hookeriana.
Materials and Methods
Pitcher Sampling
Pitcher tissues of closely related lowland Nepenthes species, N. ampullaria Jack and N. rafflesiana Jack (Bunawan et al., 2017) with their hybrid N. × hookeriana Lindl., all of which were originally transplanted from Endau-Rompin National Park, were sampled from an experimental terrace at Universiti Kebangsaan Malaysia (2°55′12.7″N, 101°46′59.7″E) (Supplementary Figure 1). The pitchers were monitored and measured throughout their development (Supplementary Figure 2) to capture the timing of pitcher opening. Pitchers after 7 days of lid opening were chosen as they are reported to be fully functional traps (Bauer et al., 2009). The sampling was performed in the morning between 09:00 and 11:00 h during October to December 2015 with at least four biological replicates from each species (Rosli et al., 2018). Whole pitchers were harvested without the tendril, emptied, and rinsed with deionized water before rapidly frozen in liquid nitrogen and stored at −80°C.
Phytochemical Extraction
The whole-pitcher samples excluding the tendril were ground until fine homogenous powders using pestle and mortar prechilled with liquid nitrogen, collected in a Falcon tube, and freeze-dried for 48 h. Dried powder samples (100 mg) were extracted using 200 μL of methanol:chloroform:water (3:1:1) via the sonication-assisted method (Shyur et al., 2013; Rosli et al., 2017). Samples were vortexed, sonicated in a water bath sonicator at room temperature for 15 min with maximum frequency, vortexed again, and then centrifuged at 10,000 g for 10 min. The extracts were filtered through a 0.22-μm PTFE membrane syringe filter before stored at −80°C.
Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis
The chromatographic separation was carried out using a Thermo Scientific C18 column (Acclaim^TM Polar Advantage II, 3 × 150 mm, 3 μm particle size) on UltiMate 3000 UHPLC (Dionex) system according to previous reports (Goh et al., 2016; Rosli et al., 2017). The gradient elution was performed for 22 min total run time, using (A) water containing 0.1% formic acid, and (B) 100% acetonitrile as the mobile phase with 0.4 mL/min flow rate at 40°C. The gradient was started at 5% solvent B for 3 min (0–3 min), then increased to 80% solvent B for 7 min (3–10 min) and maintained at 80% solvent B for 5 min (10–15 min). Finally, the gradient was returned to 5% solvent B in 7 min (15–22 min). The injection volume for each sample was 1 μL, and the samples were prepared by dissolving 10 μL of sample extracts in LC-grade methanol (990 μL) spiked with internal standards not present in the studied sample, namely, ribitol, riboflavin, and vanillin at a concentration of 50 ppm each with known basic characteristics, such as retention time (rt) and exact mass. Internal standards are important in metabolic profiling analysis serving as references for relative quantitative analysis and validation on the performance of chromatographic and MS systems (Stobiecki and Kachlicki, 2013). At least four independent biological replicates of pitcher extracts from each species were analyzed.
High-resolution mass spectrometry analysis was carried out using the Bruker Daltonics MicrOTOF-Q III with the following settings: capillary voltage at 4,500 V, nebulizer pressure at 1.2 bar, drying gas flow at 8 L/min with the source temperature at 200°C, and m/z range from 50 to 1,000 Da. The mode of electrospray ionization (ESI) was compared preliminary before the metabolite profiling LC-MS analysis. Most of the metabolites identified through negative mode were present in the total ion chromatogram of positive mode (data not shown). Thus, the positive ESI mode was selected following a higher number of metabolites profiled in preliminary LC-MS analysis and other studies (Goh et al., 2016; Rosli et al., 2017; Dávila-Lara et al., 2020). Independent from the MS analysis of individual samples, the MS/MS analysis was performed for each species using the pooled replicates of all extracts at equal amounts using automated fragmentation settings (Auto-MS/MS) over a mass-to-charge precursor ion ranged between 500 and 1,000 Da (Vargas et al., 2016).
Mass Spectrometry Data Processing
Mass spectrometry data processing was done according to Krug et al. (2008) with some modifications. The mass spectrometry raw data was extracted from Bruker DataAnalysis (version 4.1) and aligned using Bruker Compass ProfileAnalysis (version 2.1). The advanced bucketing setting was selected with the following parameters: Δm/z = 20 mDa, Δrt = 10 s, threshold of signal-to-noise ratio = 5, and smoothing width = 4. The correlation coefficient was set at 0.7. The exact mass and predicted molecular formula obtained from SmartFormula, a feature in ProfileAnalysis (version 2.1), was selected based on the highest score with low molecular mass error tolerance range (Δppm = 5) and manually matched to accessible public databases, including METLIN1 developed for QTOF instruments (Guijas et al., 2018), MassBank2 the longest standing community database (Horai et al., 2010), and MetFrag3 (Ruttkies et al., 2016) for putative metabolite identification.
Putative metabolites were retrieved from the databases for having molecular weights within the molecular mass error tolerance range of Δppm < 20 to the query m/z values via positive mode adduct. Full-scan LC-MS data were acquired for statistical analysis to shortlist molecular ions with significant differences between samples, followed by independent precursor-ion (PI) scans to acquire MS/MS data of the PIs. These MS/MS data of precursor m/z and retention time were used to derive the structural information with confidence level 1 (CL1), while metabolite features without MS/MS data were putatively identified at confidence level 2 (CL2) according to the Metabolomics Standards Initiative (Blaženović et al., 2018). Erroneous metabolite identifications were removed through manual inspection based on chemotaxonomy information with reference to the KNApSAcK species–metabolite relationship database (Afendi et al., 2012).
Statistical Analysis
XCMS version 3.7.14 was used for a preliminary inspection of feature detection using the mzXML files and default parameters (ID#6675) optimized for “UPLC/Bruker Q-TOF pos” to generate the statistics of identified features and mirror plots of pairwise comparisons between the samples (Huan et al., 2017). For further statistical analysis, the data matrix generated from ProfileAnalysis (version 2.1) comprising peak intensity of each retention time (rt) and m/z value was normalized to the intensity of the most consistent internal standard (vanillin) using MetaboAnalyst 3.05, a free online tool for statistical and pathway analyses (Xia and Wishart, 2016). The normalized peak intensity data from MetaboAnalyst was exported into SIMCA-P+ (version 14.1, Umetrics, Umeå, Sweden) for multivariate analysis (MVA). The data was preprocessed with Pareto (Par) scaling which was applied to the normalized data to reduce the effects of background noise interference on sample clustering. MVA based on non-supervised principal component analysis (PCA) and supervised partial least squares discriminant analysis (PLS-DA) were applied to cluster samples according to the profiled metabolite features and to identify distinct metabolites that differentiate between the three species. One-way analysis of variance (ANOVA) with a significance of P < 0.05 was conducted on the normalized peak intensity data to differentiate the statistically significant metabolites among the three species. The variable importance in the projection (VIP) ranks the overall contribution of each variable to the PLS-DA model, and those variables with VIP > 1.0 were considered relevant for group discrimination (Xie et al., 2008). The list of important VIPs was based on PLS-DA loading plots with VIP scores > 1. Venn diagrams were generated using a web tool6.
Results
Morphological Changes During Pitcher Development
For the sampling of pitchers, daily observations of morphological changes were performed in situ (Supplementary Figure 1) throughout pitcher development (Figure 1). The three species showed different pitcher longevity with N. ampullaria pitchers being the most long-lived (>6 months) compared to the short-lived (≤2 months) N. rafflesiana pitchers with the hybrid pitchers showing intermediate longevity (2–3 months). It took around 1 month (22–33 days) from pitcher initiation at the tendril tip to pitcher opening for all three species. Pitcher development continues after opening and maturity within 7 days with a stable morphology, namely, the positioning of the lid, peristome structure, coloration, and pitcher size (Figure 1 and Supplementary Figure 2). Therefore, 7-day-old mature pitchers after lid opening were chosen to standardize sampling for metabolomics analysis.
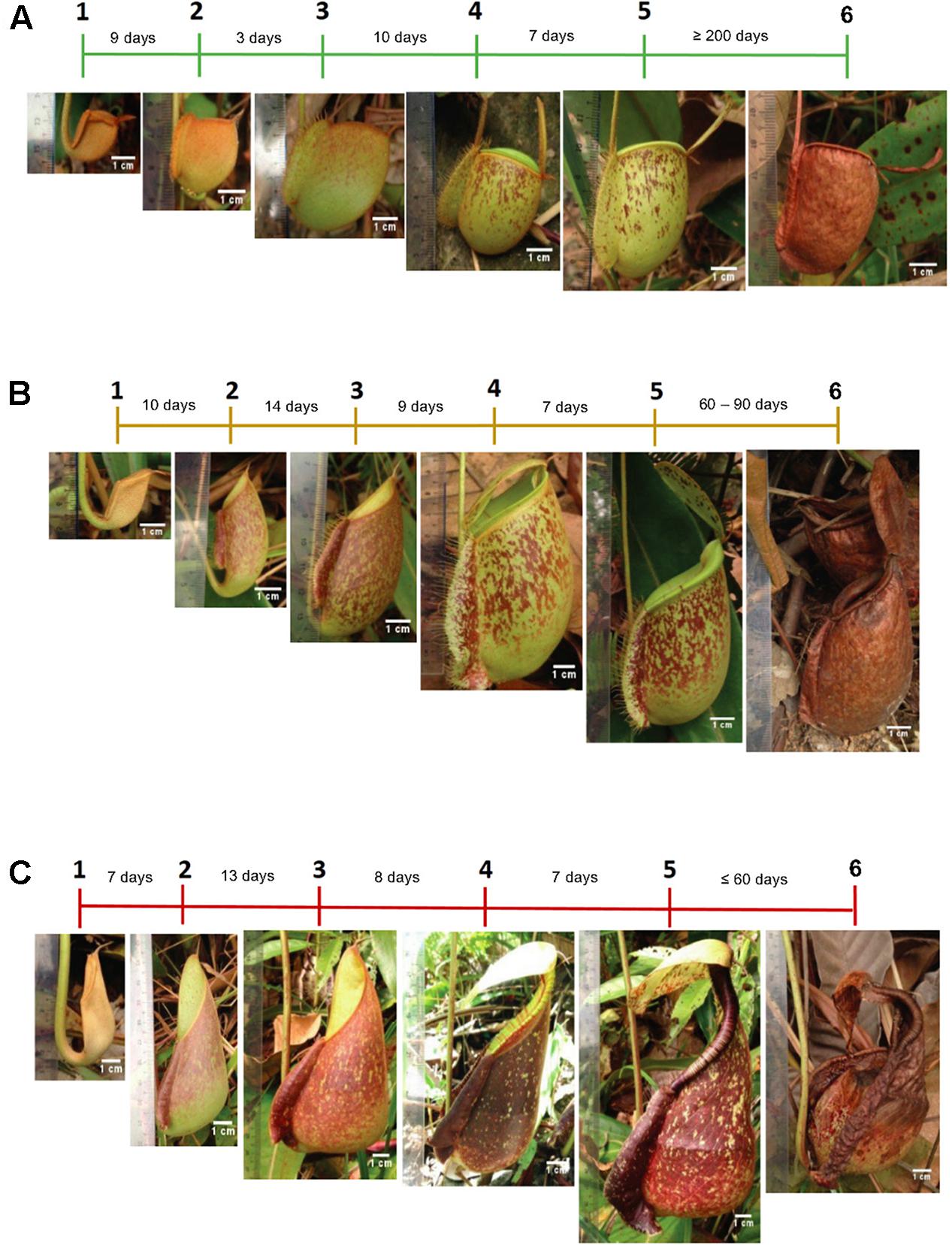
Figure 1. The timeline of pitcher morphological changes of (A) N. ampullaria, (B) N. × hookeriana, and (C) N. rafflesiana. Different phases and durations of pitcher development are indicated in the timeline.
Metabolite Profiling
The chromatographic separation of the three Nepenthes samples and internal standards can be visualized on a base peak chromatogram (BPC), which shows the peak intensity based on the m/z value and retention time (rt) with extracted ion chromatogram (EIC). All three internal standards were successfully identified in the chromatogram, including ribitol at rt 1.9 min, riboflavin at rt 8.4 min, and vanillin at rt 9.5 min (Figure 2). Based on a comparison of all the chromatograms, all three internal standards were present and separated at similar rt in each sample. Samples from the three species showed metabolic separation patterns with different numbers of metabolite features. There were metabolite features uniquely present in one species or all species with different intensities. For instance, a metabolite feature with the highest intensity after internal standards at rt 8 min exhibited the highest intensity in N. rafflesiana, moderate in the hybrid, N. × hookeriana, and the lowest in N. ampullaria.
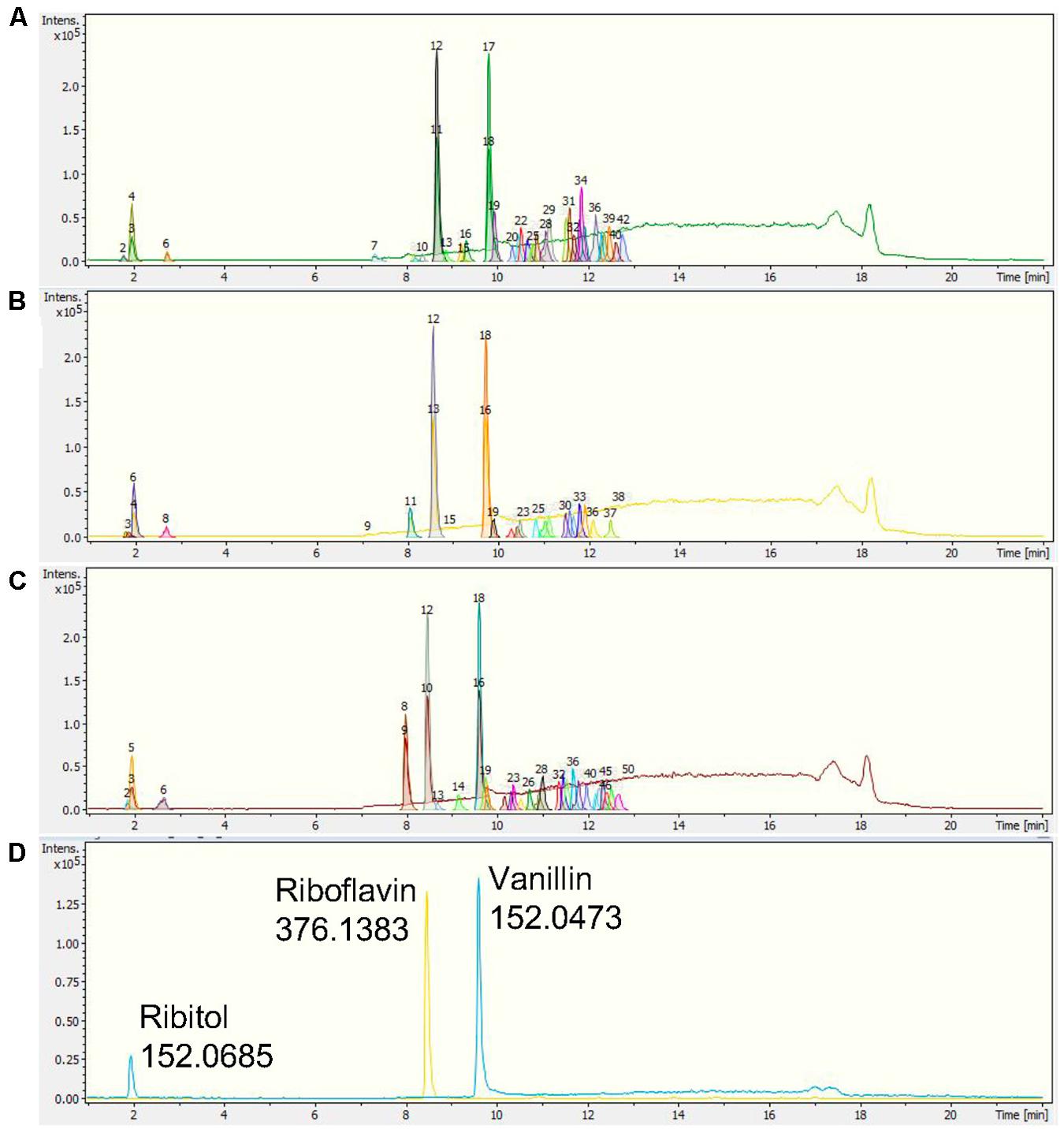
Figure 2. Representative base peak chromatograms (BPCs) with extracted ion chromatograms (EICs) of (A) N. ampullaria, (B) N. × hookeriana, (C) N. rafflesiana, and (D) a blank with internal standards, namely, ribitol (1.9 min), riboflavin (8.4 min), and vanillin (9.5 min).
A statistical analysis was performed using XCMS using the raw data from all the samples for pairwise comparisons (Supplementary Figure 3) and feature detection (Supplementary Figure 4). Comparison between the two parent species showed the biggest difference with 22 significant differentially accumulated features (DAFs), followed by the hybrid and N. ampullaria (9 DAFs), and lastly between the hybrid and N. rafflesiana (2 DAFs) (Supplementary Figure 3).
After filtering and bucketing using Bruker Compass ProfileAnalysis, we successfully profiled a total of 1,441 metabolite features in pitcher samples from all three species (Supplementary File 1). The distribution of pitcher metabolite features for each species is shown in a Venn diagram (Figure 3A). The pitcher extracts with the highest number of profiled metabolite features were found in N. ampullaria with 841 features, followed by the hybrid species, N. × hookeriana (632) and N. rafflesiana (577). Only 221 metabolite features were present in all three species with many features unique to individual species. It is noteworthy that there were 274 (43.3%) features found in N. × hookeriana which were not found in any of the parent species. Similar numbers of metabolite features were shared by the hybrid with N. ampullaria (292, 46.2%) and N. rafflesiana (287, 45.4%). However, there were many more unique metabolite features in N. ampullaria (549, 65.3%) that were absent in the hybrid, compared to N. rafflesiana which shared 49.7% of its total metabolite features with the hybrid. The mass-to-charge ratio (m/z) of metabolite features were rather evenly distributed compared to the retention time (rt) (Figure 3B). Most of the unique features found in N. ampullaria were detected during early rt (25 s), while the majority of the shared features were found in the middle of rt (400–800 s).
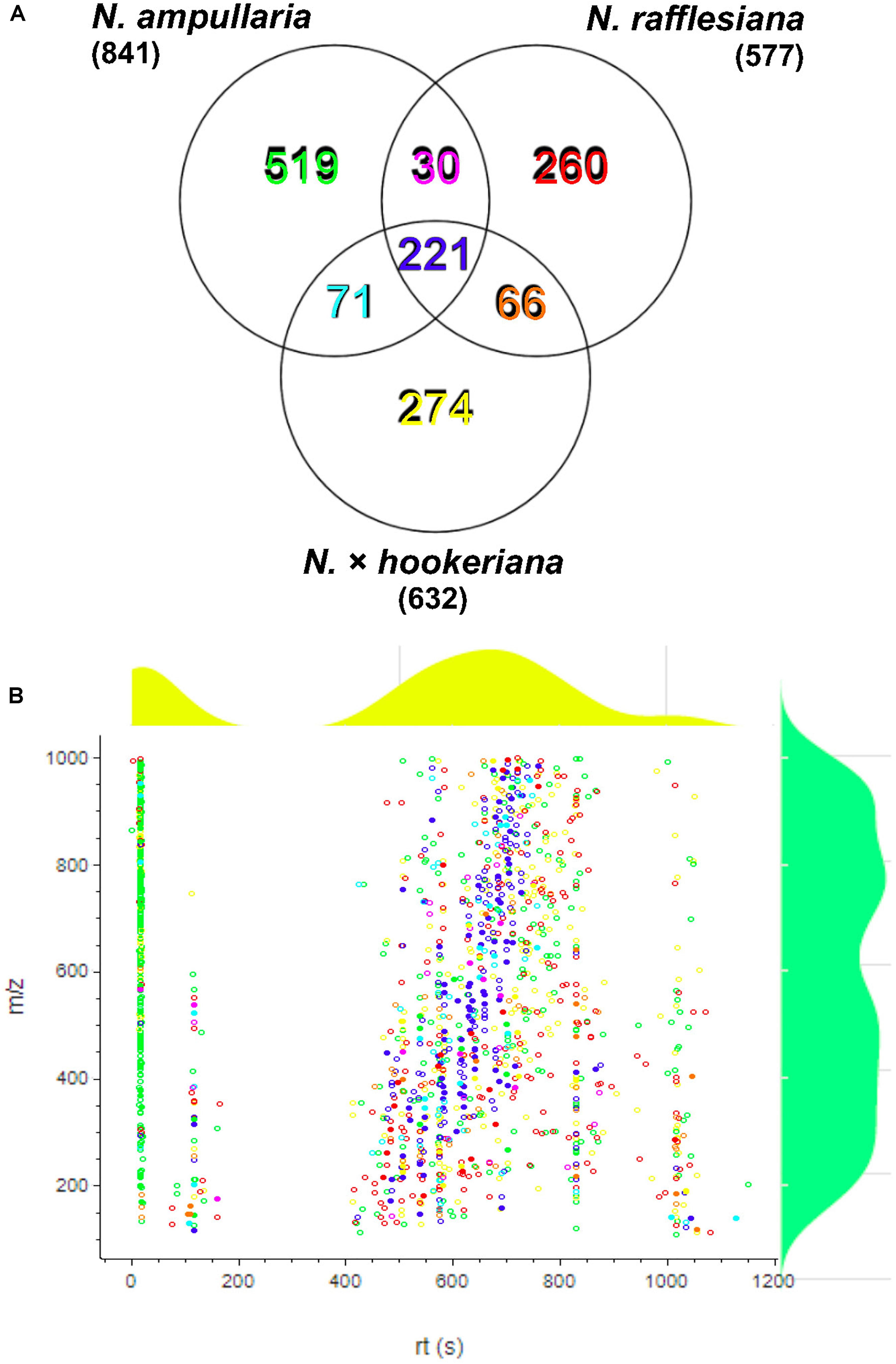
Figure 3. Metabolite profiling. (A) Venn analysis of metabolite features identified from LCMS analysis of all three species. Numbers in parentheses show the total number for each species. (B) Scatter plot of all metabolite features with overlaying relative density plots for retention time (rt) along the X-axis and mass over charge (m/z) along the Y-axis. Symbol colors correspond to the font colors in the Venn diagram. Filled symbols represent features with putative identification.
Multivariate Analysis
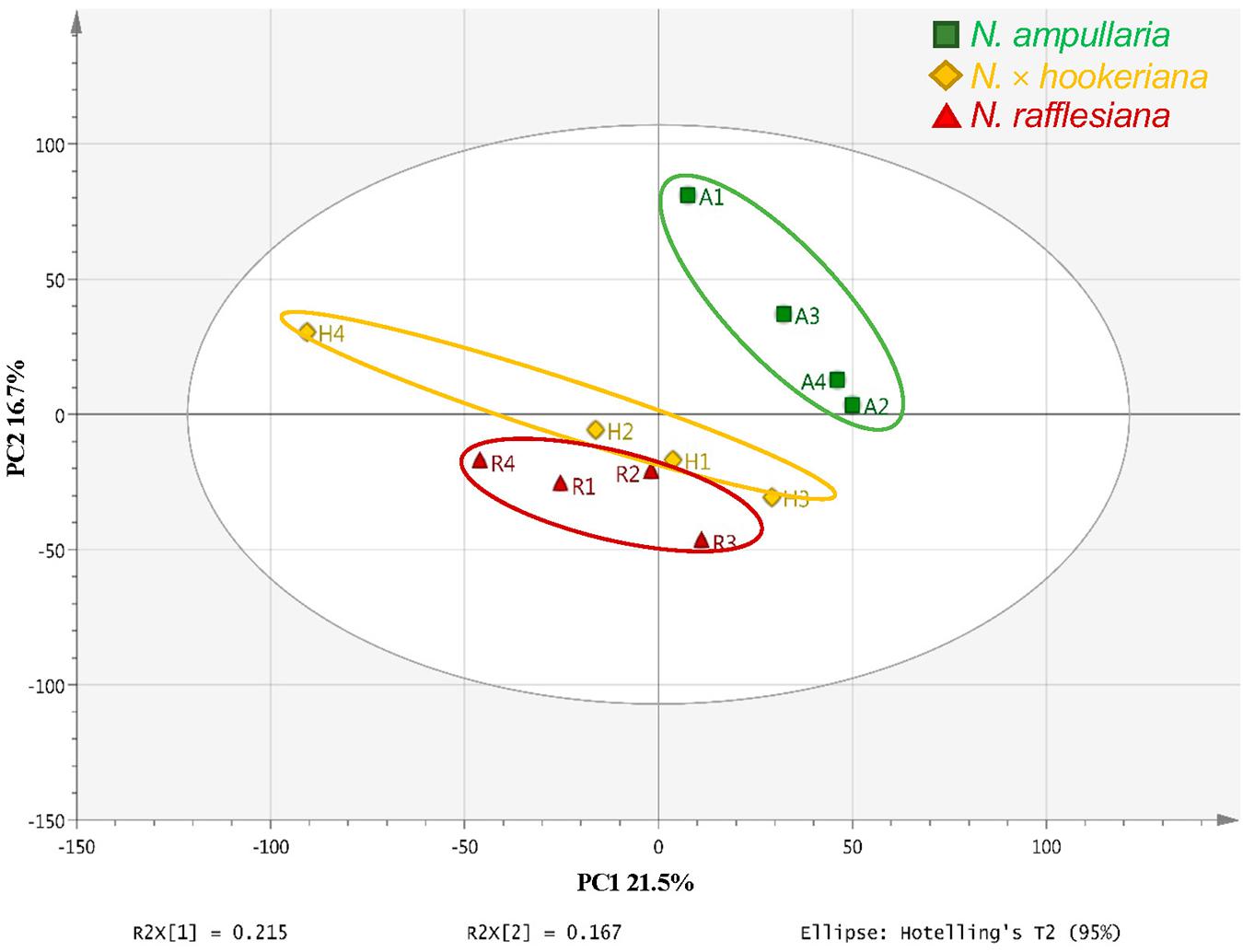
An unsupervised principal component analysis (PCA) analysis shows the projections of each sample to other samples in a multidimensional space with each point representing an individual sample. The sample dispersions are related to the differences in metabolite compositions such that samples with higher similarities are closer together while samples with bigger differences are further apart. The parent clusters of N. ampullaria and N. rafflesiana were separated from each other, but the hybrid cluster was dispersed and closer to N. rafflesiana (Figure 4). This separation only accounted for 21.5% of variation on the first principal component, and the cumulative values of R2 and Q2 were lower than 0.5 despite having no outlier. This indicates high variations between biological replicates, which could be due to environmental factors since sampling was performed on-site in the field.
A supervised partial least squares-discriminant analysis (PLS-DA) was then performed for sample clustering with species information (Figure 5A). N. ampullaria samples were closely clustered and separated from N. rafflesiana along the first component that accounted for 19.1% variations. N. × hookeriana samples were more dispersed and closer to N. rafflesiana on the first component with separation mainly on the second component that explained 10.8% variations. These two components appeared to be a good fit model with a predictive value of confidence prediction (Q2) at 35% lower than the value of R2Y at 88%, which suggests no overfitting as supported by a permutation test (Supplementary Figure 5).
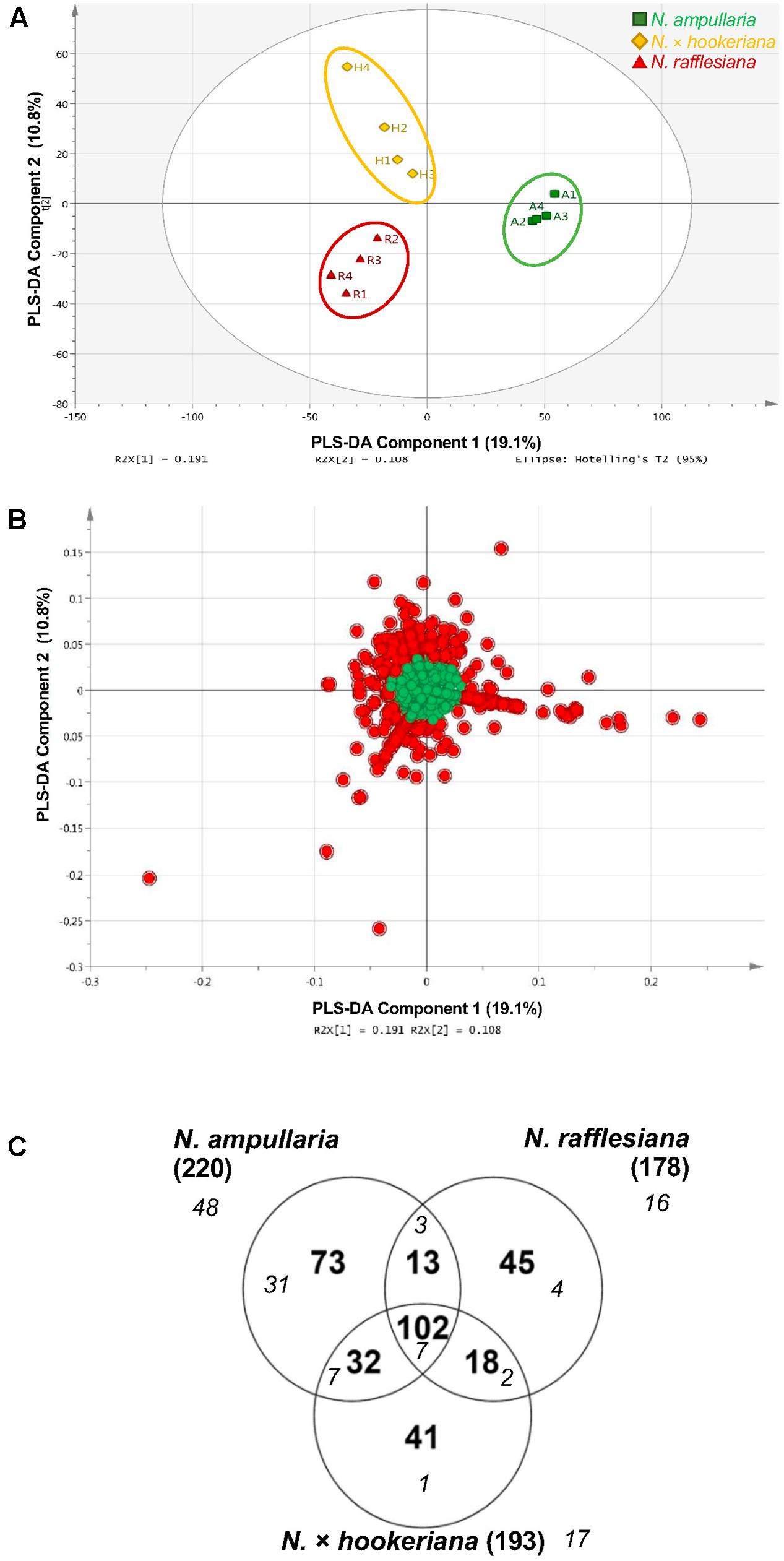
Figure 5. PLS-DA analysis of pitcher extracts from different Nepenthes species. (A) PLS-DA score plot. (B) PLS-DA loading plot with red dots showing the VIP features. (C) Venn analysis of VIP features in three Nepenthes species. Numbers in parentheses show the total number for each species. Italic numbers in smaller font size represent the numbers of significant (P < 0.05) VIP.
The metabolite features responsible for the discrimination between the three species can be observed in the PLS-DA loading plot (Figure 5B) with a higher contribution by the metabolites that are projected further from the center. A total of 324 (22.5%) metabolite features with variables important in projection (VIP) values of more than 1 were identified in the PLS-DA loading plot (Figure 5B), in which 102 VIPs were shared among the three species with N. ampullaria having the highest number of VIPs (Figure 5C). Apart from VIPs, univariate statistical analysis can identify statistically significant metabolites by comparing intensity differences of metabolite features in each sample, whereby only statistically significant features are considered important and maybe biological or chemical markers. Only 55 (17%) out of 324 VIPs were statistically significant (P < 0.05) based on the one-way variance analysis (ANOVA) in which 31 were unique to N. ampullaria. PCA performed based on these 55 significant VIPs can separate the three species into three distinct clusters and explained 63.2% of variation on the first principal component (PC1) and 14.2% for PC2 (Supplementary Figure 6). This supports that these metabolite features contribute significantly to the distinction of each species and account for their differences in metabolism.
Some of these significant metabolite features were plotted in a heat map with normalized intensity values (Figure 6). A total of 27 out of the 36 metabolite features showed higher abundance in N. ampullaria compared to six metabolites in N. rafflesiana and only three in the hybrid. This indicates that N. ampullaria displayed a more distinct metabolite distribution compared to N. rafflesiana and N. × hookeriana, which is consistent with the results from the multivariate analysis.
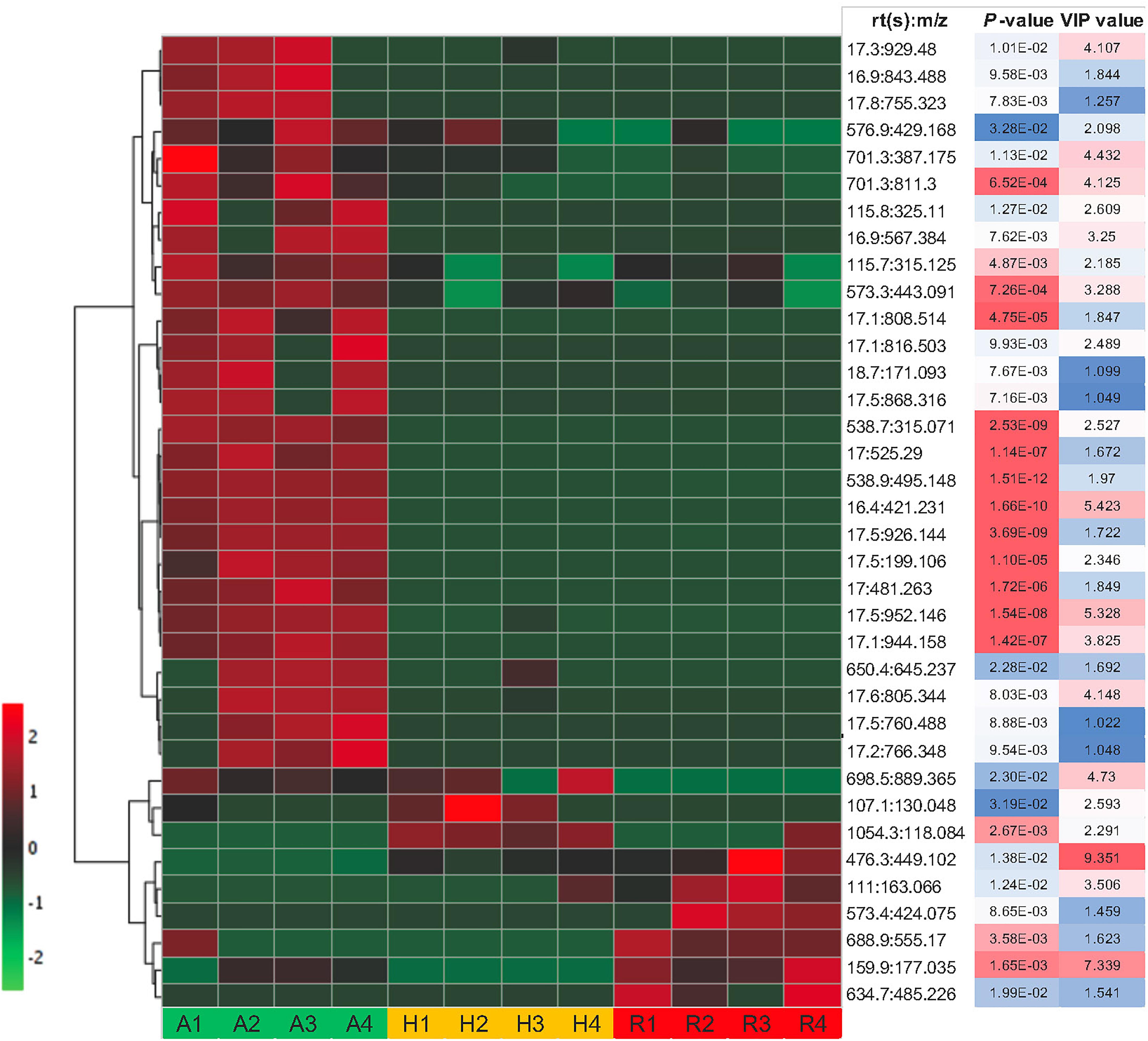
Figure 6. Heat map of statistically significant features in all three species. The color temperature represents the relative peak intensity with hotter red color showing higher intensity. Putative identification of metabolite features can refer to Supplementary File 1.
Metabolite Identification
Due to the labor-intensive metabolite identification, we mainly focused on VIP and metabolite features with high abundance for putative identification of metabolites by manually matching the accurate masses, fragmentation patterns, and MS/MS spectral data with reference to public metabolite databases, including METLIN, MassBank, and MetFrag (Supplementary File 1). For every annotated metabolite, we manually checked against the KNApSAcK database to make sure the metabolite is produced in plants. A total of 95 (29.3%) of 324 VIPs were among the 105 (7.3%) putatively identified metabolites from 1,441 metabolite features based on our manual approach.
Discussion
Untargeted Metabolite Profiling of Nepenthes Pitcher Extracts
This untargeted metabolite profiling study is the first comparative metabolomics analysis of Nepenthes pitchers from different species. For non-model organisms, LC-MS analysis is the preferred method for metabolic profiling with good selection and sensitivity to analyze various types of metabolites with large mass range differences and different physicochemical properties, such as volatility, heat loss, and polarity (Stobiecki and Kachlicki, 2013). The chromatographic separation can reduce the sample complexity and increase the MS detection sensitivity with less background noise. Furthermore, the electrospray ionization (ESI) mode of soft ionization can produce large amounts of ions through the exchange of charges in solution and often form intact molecular ions for initial identification of metabolites (Zhou et al., 2012). Time-of-flight (ToF) technology applied in this study further improves the mass accuracy to facilitate the identification and relative quantification of metabolites.
To compare the three Nepenthes species with four biological replicates of pitcher extracts from each species, their metabolite profile data were processed and normalized based on vanillin because it exhibited the most consistent peak intensities in all samples with the lowest coefficient of variation (CV) compared to ribitol and riboflavin (Supplementary File 1). Overall, multivariate analysis (MVA) showed that the metabolite composition of the hybrid is more like N. rafflesiana than N. ampullaria. This corroborates the recent report that the hybrid showed more similar transcriptome and secretome to N. rafflesiana than N. ampullaria (Zulkapli et al., 2021). Previously, it was proposed that the variation of colors and motifs were determined by male Nepenthes based on a genetic study (Yulita and Mansur, 2012). N. × hookeriana “spotted” as a hybrid between male N. rafflesiana and female N. ampullaria “green” was more like N. rafflesiana while N. × hookeriana “green” was hypothesized to be a hybrid between male N. ampullaria “green” and female N. rafflesiana. Hence, this explains the molecular compositions of samples in this study with N. × hookeriana “spotted” (Figure 1).
A recent untargeted metabolite profiling study of N. × ventrata pitchers was based on cheminformatics fingerprinting to infer metabolite classification without compound identification, which hindered functional interpretation (Dávila-Lara et al., 2020). At present, metabolite identification for untargeted metabolomics analysis largely depends on mass-based search using the m/z values of molecular ions of interest against databases (Xiao et al., 2012; Blaženović et al., 2018). In this study, manual searches against different public databases putatively identified a total of 105 metabolites with 95 VIPs in which only four metabolites were identified with MS/MS data. These putative metabolites are only suggestive because of limited information on Nepenthes metabolites in the public databases and the low hit of auto MS/MS spectra for most of the compounds. To our knowledge, this is the most comprehensive study of metabolite profiling ever reported in Nepenthes studies.
Due to the complexity and diversity of the chemical composition of metabolites, metabolite identification is still a big challenge in metabolomics analysis (Aretz and Meierhofer, 2016). Commonly, there are less than 20% identified compounds in untargeted analysis, such as the Metabolomics Workbench (Sud et al., 2016) or the European metabolomics repository MetaboLights (Haug et al., 2013) even for the model organisms. A comprehensive spectral database for LC-MS is almost impossible with various instruments, methods, and acquisition of metabolic data, such as collision, fragmentation, and various settings resulting in varied retention time (rt) and molecular fragments (Aretz and Meierhofer, 2016).
Functional Implications of Putatively Identified Metabolites in Nepenthes Pitchers
The most significant VIP metabolite feature (476.3s:449.102) was the most abundant metabolite in the pitchers with its peak clearly detectable at the eighth minute with different intensities in each species showing the highest abundance in N. rafflesiana, intermediate in the hybrid, and the lowest in N. ampullaria (Figure 2). VIP refers to metabolites that contribute to the discrimination between samples, and metabolites with high VIP values can serve as biological or chemical markers (Beatriz et al., 2014). A MS/MS spectrum match search using the precursor m/z with MS/MS peaks (Supplementary Figure 7) found 476.3s:449.102 matches to kaempferol 3-glucoside (astragalin) and cyanidin 3-O-glucoside (chrysenthemin), both with a METLIN score of 100 and Δppm = 11. However, further fragment similarity search only found hit to astragalin at a fragment peak of m/z 287.054 with Δppm = 9 (Supplementary Figure 7D). Nonetheless, we cannot exclude the possibility that 476.3s:449.102 could be other isomers, kaempferol O-glucosides, or luteolin O-glucosides without further validation and structural elucidation using reference compounds and NMR. Astragalin is a flavonol involved in the biosynthesis of flavones and flavonols. Astragalin is a type of yellow pigment that could protect plants from UV radiation (Nakabayashi, 1955). Therefore, this compound may contribute to the pitcher color and confer protection from UV radiation (Lavola et al., 2003). Astragalin is associated with various pharmacological and biological activities, including anti-inflammatory, antioxidant activity, anti-atopic dermatitis activity, TNF-a, IL-1b, and IL-6 production inhibiting activity, and inhibiting histamine release in human blood cells (Riaz et al., 2018).
This finding agrees with Kováčik et al. (2012) that found phenols constituted the most abundant metabolites in Nepenthes. The results of several other phytochemical studies also emphasized phenolic compounds in Nepenthes roots and leaves, particularly naphthoquinones (Cannon et al., 1980; Aung et al., 2002; Van Thanh et al., 2015). Flavonoids were found to be the dominant phenols in the pitcher extracts, apart from quinones, quinic acids, monolignols, lignans, and phenylpropanoids (Supplementary File 1). These flavonoids have antioxidant properties that protect plants from environmental stresses, especially oxidative stress and ultraviolet (UV) light (Pollastri and Tattini, 2011). Flavonoids involved in UV light protection include flavones and flavonols that also act as antioxidants in protecting plants from oxidative stress caused by temperatures higher than suboptimal temperatures (Herrmann, 1976; Jaakola and Hohtola, 2010). Flavan is a product of a double reduction of flavanone. Most of the flavans are soluble in lipids, making these compounds often found on the fruit peel and the cutin layer of the leaf surface (Swanson, 2003). According to Mierziak et al. (2014), the strongest antifungal activity is shown by unmodified flavones and flavanones. Therefore, these compounds are commonly used in plants as phytoalexins to fight fungi and insects (Swanson, 2003). Chalcones are usually intermediates in flavonoid biosynthesis and generally have low compositions in most plants but display a variety of biological activities as traditional medicine (Rozmer and Perjési, 2016).
Anthocyanins are responsible for color in plants. Cyanidin and delphinidin are parts of anthocyanins that contribute to a variety of colors, including orange, red, and purple, depending on the types, levels, and structural modifications of anthocyanins, the presence of co-pigments such as flavones or flavonol, and pH variations (Yu and McGonigle, 2005). Anthocyanidins were present in pitchers of all species but in relatively low abundance (Supplementary File 1). These compounds may be involved in the pitcher pigmentation and coloration, such as spots on the pitchers.
The presence of flavonols and flavones, which were highly abundant in N. rafflesiana, may also contribute to the red coloration of pitchers, although the distribution of anthocyanidins in the species is low (Supplementary File 1). A high temperature can inhibit anthocyanin biosynthesis caused by chemical degradation from the accumulation of polyphenol oxidase and peroxide enzymes (Vaknin et al., 2005; Mori et al., 2007). This partly explains the effect of latitude on plant flavonoid biosynthesis (Jaakola and Hohtola, 2010). For example, a tropical herb plant Polygonum minus growing in the lowland and exposed to higher temperatures contained higher amounts of flavones and flavonols but lower anthocyanidin content compared to those from highland and exposed to lower temperatures (Goh et al., 2016). However, whether or not N. rafflesiana plants growing at lowland and highland exhibit different compositions of anthocyanidins remains to be investigated.
In previous reports, phenolic compound naphthoquinones such as plumbagin with numerous bioactivities are known to be highly abundant in Nepenthes (Likhitwitayawuid et al., 1998; Gwee et al., 2014). Plumbagin has been suggested with a possible role in attracting their insect prey with distinct volatile preferences toward the sticky trap of another insectivorous plant, Drosera auriculata (El-Sayed et al., 2016; Widhalm and Rhodes, 2016). According to Shin et al. (2007), some naphthoquinones with antifungal activities can be found in the roots of N. rafflesiana, N. thorelii, N. gracilis, and N. insignis. Studies have also shown that naphthoquinones can be found in the leaves of N. khasiana and N. gracilis (Aung et al., 2002; Eilenberg et al., 2010; Raj et al., 2011). Naphthoquinones might have several functions in Nepenthes, including photosynthesis, pigmentation and coloration, and protection against UV light, desiccation, and insects (Widhalm and Rhodes, 2016). Raj et al. (2011) speculated toxic or anesthetic effects of naphthoquinones on insect prey in N. khasiana pitchers. Furthermore, derivatives of naphthoquinones, droserone, and 5-O-methyl droserone could protect and maintain the sterility of the pitcher fluids against microbes from the trapped prey and together with plumbagin may act as molecular triggers in prey capture and digestion (Raj et al., 2011).
In this study, there were a few naphthoquinones putatively identified (Supplementary File 1), which have never been reported. There might be several factors that contribute to the detection of naphthoquinones, including the differences in the extraction using different solvents or analytical methods such as GC-MS (Raj et al., 2011). Besides that, Eilenberg et al. (2010) showed that naphthoquinones in N. khasiana pitcher fluids only present after chitin induction, which suggests the induced production of naphthoquinones.
As mentioned above, this study is limited by the uncertain metabolite identification for a detailed discussion of metabolites in Nepenthes pitchers. Furthermore, this also hindered further discussion on whether the different metabolite compositions in the pitchers were accountable by the different dietary habits of the parent and hybrid species. Nevertheless, this study is meaningful in finding potential chemical markers and bioactive compounds. Various reports described the curative effects of extracts from Nepenthes on diseases, for example, jaundice, hepatitis, gastric ulcers, ureteral stones, diarrhea, diabetes, cough, fever, hypertension, urinary system infections (Thao et al., 2016), malaria (Likhitwitayawuid et al., 1998), and Staphylococcus infection (Wiart et al., 2004), and recently on different kinds of oral cancer cells (Tang et al., 2019). Thus, further works on the validation of metabolites from Nepenthes as well as the analysis of their putative pharmaceutical uses are promising to explore new compounds and therapeutics. Subsequently, compounds with bioactivities of interest can be targeted for isolation, structural elucidation, and functional characterization.
Conclusion
This study provides a comprehensive profiling and putative identification of metabolites from Nepenthes pitchers through the untargeted LC-MS approach with a total of 1,441 metabolite features and 105 putatively identified metabolites. MVA performed on profiled metabolites showed that the chemical compositions of hybrid pitchers were more similar to that of N. rafflesiana than N. ampullaria. Astragalin, which is a pharmacologically useful flavonol, was putatively identified as the most significant compound in distinguishing between the three Nepenthes species with potential as a chemical marker. However, the influence of dietary habit on the differences in the metabolite compositions between the three taxa remains unclear.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MR and H-HG conceived the study and experiments. MR performed the experiments and analyzed data. MR, AM, KA, SB, and H-HG discussed the data and wrote the manuscript. H-HG acquired the funding for this work. All authors read and agreed to the present version of the manuscript.
Funding
This work was funded by FRGS/2/2014/SG05/UKM/02/4 from the Malaysian Ministry of High Education and UKM Research University Grant DIP-2014-008 awarded to H-HG. The research group is currently supported by DIP-2020-005 and FRGS/1/2019/STG05/UKM/02/10.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Dr. Jumaat Haji Adam for providing access to the experimental terrace and contributing the pitcher samples. We are also grateful to both reviewers for their constructive comments in improving this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.655004/full#supplementary-material
Supplementary Figure 1 | Nepenthes pitcher plants in this study. (A–C) Nepenthes ampullaria. (D,E) Nepenthes × hookeriana. (F–H,J,K) Nepenthes rafflesiana (A,D,F) Whole pitcher plants. (B) A cluster of N. ampullaria pitchers on the ground. (C,E,H) A single pitcher. (G) An aerial pitcher of N. rafflesiana. (I) Experimental terrace for pitcher sampling of all three species. (J) Characteristics of a typical Nepenthes pitcher. (K) External and internal structures of a N. rafflesiana pitcher.
Supplementary Figure 2 | (A) Measurement of pitcher length. (B) Pitcher length at each growth phase. Error bars showing standard errors of six biological replicates.
Supplementary Figure 3 | Mirror cloud plots of differentially accumulated features (DAFs) in pairwise comparisons of (A) N. ampullaria and N. rafflesiana, (B) N. ampullaria and N. × hookeriana, and (C) N. rafflesiana and N. × hookeriana. The numbers of significant (P < 0.001, rt > 1) DAFs are shown in parentheses and listed on the side with details in ascending P-value. DAFs are shown for lower (top) or higher (bottom) accumulation in the first species of comparison. Circle size depicts the average fold change. Y-axis indicates the m/z value of the feature; the further away from the origin, the higher the m/z. The superimposed, raw base-peak chromatograms (BPC) of all runs are shown in the background, colored according to different samples. Insets show peak intensity boxplots of selected DAFs with m/z values.
Supplementary Figure 4 | Venn analysis of identified features in XCMS pairwise comparisons of all three species. A: N. ampullaria; H: N. × hookeriana; R: N. rafflesiana.
Supplementary Figure 5 | Permutation test of PLS-DA validation model.
Supplementary Figure 6 | PCA score plot analysis based on significant features. Each symbol represents different species of Nepenthes, green square: N. ampullaria, yellow diamond: N. × hookeriana, and red triangle: N. rafflesiana.
Supplementary Figure 7 | Putative identification of metabolite feature 476.3s:449.102 based on tandem MS data. (A) MS/MS spectra of 476.3s:449.102 with the list of peaks (m/z and intensity). (B) Chemical structure of astragalin and chrysenthemin. Comparison for putative metabolite identification based on (C) METLIN MS/MS spectrum and (D) Fragment Similarity Search. The blue square in (A) shows the precursor ion. For (C), the asterisk indicates the highest peak of precursor ion, while the black and cyan lines represent the MS/MS spectrum of the putative metabolite and the search fragments, respectively.
Supplementary File 1 | Normalized peak intensity values of metabolite features with putative metabolite identification.
Footnotes
- ^ https://metlin.scripps.edu/
- ^ http://www.massbank.jp/Search
- ^ https://msbi.ipb-halle.de/MetFrag/
- ^ https://xcmsonline.scripps.edu/
- ^ http://www.metaboanalyst.ca
- ^ http://bioinformatics.psb.ugent.be/webtools/Venn/
References
Adam, J. H. (1997). Prey spectra of bomean Nepenthes species (Nepenthaceae) in relation to their habitat. Pertanika J. Trop. Agric. Sci. 20, 121–134.
Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Nakamura, K., et al. (2012). KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53:e1. doi: 10.1093/pcp/pcr165
Aretz, I., and Meierhofer, D. (2016). Advantages and pitfalls of mass spectrometry based metabolome profiling in systems biology. Int. J. Mol. Sci. 17:632. doi: 10.3390/ijms17050632
Aung, H., Chia, L., Goh, N., Chia, T., Ahmed, A., Pare, P., et al. (2002). Phenolic constituents from the leaves of the carnivorous plant Nepenthes gracilis. Fitoterapia 73, 445–447. doi: 10.1016/s0367-326x(02)00113-2
Bauer, U., Willmes, C., and Federle, W. (2009). Effect of pitcher age on trapping efficiency and natural prey capture in carnivorous Nepenthes rafflesiana plants. Ann. Bot. 103, 1219–1226. doi: 10.1093/aob/mcp065
Bazile, V., Moguédec, G. L., Marshall, D. J., and Gaume, L. (2015). Fluid physico-chemical properties influence capture and diet in Nepenthes pitcher plants. Ann. Bot. 115, 705–716. doi: 10.1093/aob/mcu266
Beatriz, G.-P., Lennart, E., and Johan, T. (2014). Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chemom. 28, 623–632. doi: 10.1002/cem.2627
Blaženović, I., Kind, T., Ji, J., and Fiehn, O. (2018). Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 8:31. doi: 10.3390/metabo8020031
Bunawan, H., Yen, C. C., Yaakop, S., and Noor, N. M. (2017). Phylogenetic inferences of Nepenthes species in peninsular Malaysia revealed by chloroplast (trnL intron) and nuclear (ITS) DNA sequences. BMC Res. Notes 10:67. doi: 10.1186/s13104-017-2379-1
Cannon, J., Lojanapiwatna, V., Raston, C., Sinchai, W., and White, A. (1980). The quinones of Nepenthes rafflesiana. the crystal structure of 2, 5-dihydroxy-3, 8-dimethoxy-7-methylnaphtho-1, 4-quinone (nepenthone-E) and a synthesis of 2, 5-dihydroxy-3-methoxy-7-methylnaphtho-1, 4-quinone (nepenthone-C). Aust. J. Chem. 33, 1073–1093. doi: 10.1071/ch9801073
Cheng, D., Vrieling, K., and Klinkhamer, P. G. L. (2011). The effect of hybridization on secondary metabolites and herbivore resistance: implications for the evolution of chemical diversity in plants. Phytochem. Rev. 10, 107–117. doi: 10.1007/s11101-010-9194-9
Clarke, S. J., and Robinson, A. S. (2018). “Species of carnivorous plants,” in Carnivorous Plants: Physiology, Ecology, and Evolution, eds A. Ellison and L. Adamec (Oxford: Oxford University Press), 411–413.
Danser, B. H. (1928). “The Nepenthaceae of the Netherlands Indies,” in Bulletin du Jardin Botanique de Buitenzorg, Série III, Vol. 9. Research Triangle Park, NC: Natural History Publications, 249–438.
Dávila-Lara, A., Rodríguez-López, C. E., O’Connor, S. E., and Mithöfer, A. (2020). Metabolomics analysis reveals tissue-specific metabolite compositions in leaf blade and traps of carnivorous Nepenthes plants. Int. J. Mol. Sci. 21:4376. doi: 10.3390/ijms21124376
Di Giusto, B., Bessière, J. M., Guéroult, M., Lim, L. B., Marshall, D. J., Hossaert-McKey, M., et al. (2010). Flower-scent mimicry masks a deadly trap in the carnivorous plant Nepenthes rafflesiana. J. Ecol. 98, 845–856. doi: 10.1111/j.1365-2745.2010.01665.x
Eilenberg, H., Pnini-Cohen, S., Rahamim, Y., Sionov, E., Segal, E., Carmeli, S., et al. (2010). Induced production of antifungal naphthoquinones in the pitchers of the carnivorous plant Nepenthes khasiana. J. Exp. Bot. 61, 911–922. doi: 10.1093/jxb/erp359
El-Sayed, A. M., Byers, J. A., and Suckling, D. M. (2016). Pollinator-prey conflicts in carnivorous plants: when flower and trap properties mean life or death. Sci. Rep. 6:21065.
Goh, H. H., Baharin, A., Mohd Salleh, F. I., Ravee, R., Wan Zakaria, W. N. A., and Mohd Noor, N. (2020). Transcriptome-wide shift from photosynthesis and energy metabolism upon endogenous fluid protein depletion in young Nepenthes ampullaria pitchers. Sci. Rep. 10:6575.
Goh, H. H., Khairudin, K., Sukiran, N. A., Normah, M. N., and Baharum, S. N. (2016). Metabolite profiling reveals temperature effects on the VOCs and flavonoids of different plant populations. Plant Biol. 18, 130–139. doi: 10.1111/plb.12403
Goulet, B. E., Roda, F., and Hopkins, R. (2017). Hybridization in plants: old ideas, new techniques. Plant Physiol. 173, 65–78. doi: 10.1104/pp.16.01340
Guijas, C., Montenegro-Burke, J. R., Domingo-Almenara, X., Palermo, A., Warth, B., Hermann, G., et al. (2018). METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 90, 3156–3164. doi: 10.1021/acs.analchem.7b04424
Gwee, P. S., Khoo, K. S., Ong, H. C., and Sit, N. W. (2014). Bioactivity-guided isolation and structural characterization of the antifungal compound, plumbagin, from Nepenthes gracilis. Pharm. Biol. 52, 1526–1531. doi: 10.3109/13880209.2014.902083
Haug, K., Salek, R. M., Conesa, P., Hastings, J., De Matos, P., Rijnbeek, M., et al. (2013). MetaboLights - An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–D786.
Herrmann, K. (1976). Flavonols and flavones in food plants: a review. Int. J. Food Sci. 11, 433–448. doi: 10.1111/j.1365-2621.1976.tb00743.x
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714. doi: 10.1002/jms.1777
Huan, T., Forsberg, E. M., Rinehart, D., Johnson, C. H., Ivanisevic, J., Benton, H. P., et al. (2017). Systems biology guided by XCMS Online metabolomics. Nat. Methods 14, 461–462. doi: 10.1038/nmeth.4260
Jaakola, L., and Hohtola, A. (2010). Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 33, 1239–1247.
Kiew, R., Teo, L., and Gan, Y. (2003). Assessment of the hybrid status of some malesian plants using amplified fragment length polymorphism. Telopea 10, 225–233. doi: 10.7751/telopea20035617
Kováčik, J., Klejdus, B., and Repčáková, K. (2012). Phenolic metabolites in carnivorous plants: inter-specific comparison and physiological studies. Plant Physiol. Biochem. 52, 21–27. doi: 10.1016/j.plaphy.2011.11.007
Krug, D., Zurek, G., Schneider, B., Garcia, R., and Müller, R. (2008). Efficient mining of myxobacterial metabolite profiles enabled by liquid chromatography-electrospray ionisation-time-of-flight mass spectrometry and compound-based principal component analysis. Anal. Chim. Acta 624, 97–106. doi: 10.1016/j.aca.2008.06.036
Lätti, A. K., Riihinen, K. R., and Jaakola, L. (2011). Phenolic compounds in berries and flowers of a natural hybrid between bilberry and lingonberry (Vaccinium × intermedium Ruthe). Phytochemistry 72, 810–815. doi: 10.1016/j.phytochem.2011.02.015
Lavola, A., Aphalo, P. J., Lahti, M., and Julkunen-Tiitto, R. (2003). Nutrient availability and the effect of increasing UV-B radiation on secondary plant compounds in Scots pine. Environ. Exp. Bot. 49, 49–60. doi: 10.1016/s0098-8472(02)00057-6
Likhitwitayawuid, K., Kaewamatawong, R., Ruangrungsi, N., and Krungkrai, J. (1998). Antimalarial naphthoquinones from Nepenthes thorelii. Planta Med. 64, 237–241. doi: 10.1055/s-2006-957417
Mierziak, J., Kostyn, K., and Kulma, A. (2014). Flavonoids as important molecules of plant interactions with the environment. Molecules 19, 16240–16265. doi: 10.3390/molecules191016240
Moran, J. A., Clarke, C. M., and Hawkins, B. J. (2003). From carnivore to detritivore? Isotopic evidence for leaf litter utilization by the tropical pitcher plant Nepenthes ampullaria. Int. J. Plant Sci. 164, 635–639. doi: 10.1086/375422
Mori, K., Goto-Yamamoto, N., Kitayama, M., and Hashizume, K. (2007). Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 58, 1935–1945. doi: 10.1093/jxb/erm055
Murphy, B., Forest, F., Barraclough, T., Rosindell, J., Bellot, S., Cowan, R., et al. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Mol. Phylogenet. Evol. 144:106668. doi: 10.1016/j.ympev.2019.106668
Nakabayashi, T. (1955). Isolation of astragalin and isoquercitrin from bracken, Pteridium aquilinum. J. Agric. Chem. Soc. Jpn. 19, 104–109. doi: 10.1271/bbb1924.19.104
Orians, C. M. (2000). The effects of hybridization in plants on secondary chemistry: implications for the ecology and evolution of plant - Herbivore interactions. Am. J. Bot. 87, 1749–1756. doi: 10.2307/2656824
Pavlovič, A., Slováková, L., and Šantrůček, J. (2011). Nutritional benefit from leaf litter utilization in the pitcher plant Nepenthes ampullaria. Plant Cell Environ. 34, 1865–1873. doi: 10.1111/j.1365-3040.2011.02382.x
Pollastri, S., and Tattini, M. (2011). Flavonols: old compounds for old roles. Ann. Bot. 108, 1225–1233. doi: 10.1093/aob/mcr234
Raj, G., Kurup, R., Hussain, A. A., and Baby, S. (2011). Distribution of naphthoquinones, plumbagin, droserone, and 5-O-methyl droserone in chitin-induced and uninduced Nepenthes khasiana: molecular events in prey capture. J. Exp. Bot. 62, 5429–5436. doi: 10.1093/jxb/err219
Ravee, R., Salleh, F. M., and Goh, H. H. (2018). Discovery of digestive enzymes in carnivorous plants with focus on proteases. Peer J. 2018:e4914. doi: 10.7717/peerj.4914
Riaz, A., Rasul, A., Hussain, G., Zahoor, M. K., Jabeen, F., Subhani, Z., et al. (2018). Astragalin: a bioactive phytochemical with potential therapeutic activities. Adv. Pharmacol. Sci. 2018:9794625.
Rosli, M. A. F., Azizan, K. A., Baharum, S. N., and Goh, H.-H. (2017). Mass spectrometry data of metabolomics analysis of Nepenthes pitchers. Data Brief. 14, 295–297. doi: 10.1016/j.dib.2017.07.068
Rosli, M. A. F., Azizan, K. A., and Goh, H.-H. (2018). Antioxidant activity of pitcher extracts from three Nepenthes species. Sains Malays. 47, 3069–3075. doi: 10.17576/jsm-2018-4712-17
Rozmer, Z., and Perjési, P. (2016). Naturally occurring chalcones and their biological activities. Phytochem. Rev. 15, 87–120. doi: 10.1007/s11101-014-9387-8
Ruttkies, C., Schymanski, E. L., Wolf, S., Hollender, J., and Neumann, S. (2016). MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 8:3.
Shin, K.-S., Lee, S.-K., and Cha, B.-J. (2007). Antifungal activity of plumbagin purified from leaves of Nepenthes ventricosa x maxima against phytopathogenic fungi. Plant Pathol. J. 23, 113–115. doi: 10.5423/ppj.2007.23.2.113
Shyur, L. F., Liu, C. P., and Chien, S. C. (2013). “Metabolomics in herbal medicine research,” in The Handbook of Plant Metabolomics, eds W. Weckwerth and G. Kahl (Hoboken, NJ: Wiley), 155–174. doi: 10.1002/9783527669882.ch8
Soltis, P. S., and Soltis, D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. doi: 10.1146/annurev.arplant.043008.092039
Stobiecki, M., and Kachlicki, P. (2013). “Liquid chromatographic-mass spectrometric analysis of flavonoids,” in The Handbook of Plant Metabolomics, eds W. Weckwerth and G. Kahl (Hoboken, NJ: Wiley), 197–213. doi: 10.1002/9783527669882.ch10
Sud, M., Fahy, E., Cotter, D., Azam, K., Vadivelu, I., Burant, C., et al. (2016). Metabolomics workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470.
Swanson, B. G. (2003). “Tannins and polyphenols,” in Encyclopedia of Food Sciences and Nutrition, 2nd Edn, ed. B. Caballero (Oxford: Academic Press), 5729–5733. doi: 10.1016/b0-12-227055-x/01178-0
Tang, J. Y., Peng, S. Y., Cheng, Y. B., Wang, C. L., Farooqi, A. A., Yu, T. J., et al. (2019). Ethyl acetate extract of Nepenthes adrianii x clipeata induces antiproliferation, apoptosis, and DNA damage against oral cancer cells through oxidative stress. Environ. Toxicol. 34, 891–901. doi: 10.1002/tox.22748
Thao, N. P., Luyen, B. T. T., Koo, J. E., Kim, S., Koh, Y. S., Van Thanh, N., et al. (2016). In vitro anti-inflammatory components isolated from the carnivorous plant Nepenthes mirabilis (Lour.) Rafarin. Pharm. Biol. 54, 588–594. doi: 10.3109/13880209.2015.1067234
Vaknin, H., Bar-Akiva, A., Ovadia, R., Nissim-Levi, A., Forer, I., Weiss, D., et al. (2005). Active anthocyanin degradation in Brunfelsia calycina (yesterday-today-tomorrow) flowers. Planta 222, 19–26. doi: 10.1007/s00425-005-1509-5
Van Thanh, N., Thao, N. P., Huong, P. T. T., Lee, S. H., Jang, H. D., Cuong, N. X., et al. (2015). Naphthoquinone and flavonoid constituents from the carnivorous plant Nepenthes mirabilis and their anti-osteoporotic and antioxidant activities. Phytochem. Lett. 11, 254–259. doi: 10.1016/j.phytol.2015.01.009
Vargas, L. H. G., Neto, J. C. R., de Aquino Ribeiro, J. A., Ricci-Silva, M. E., Souza, M. T., Rodrigues, C. M., et al. (2016). Metabolomics analysis of oil palm (Elaeis guineensis) leaf: evaluation of sample preparation steps using UHPLC-MS/MS. Metabolomics 12:153.
Wan Zakaria, W. N. A., Aizat, W. M., Goh, H. H., and Mohd Noor, N. (2019). Protein replenishment in pitcher fluids of Nepenthes × ventrata revealed by quantitative proteomics (SWATH-MS) informed by transcriptomics. J. Plant Res. 132, 681–694. doi: 10.1007/s10265-019-01130-w
Wang, L., Tao, D., Dong, S., Li, S., and Tian, Y. (2018). Contributions of lunate cells and wax crystals to the surface anisotropy of Nepenthes slippery zone. R. Soc. Open Sci. 5:180766. doi: 10.1098/rsos.180766
Wiart, C., Mogana, S., Khalifah, S., Mahan, M., Ismail, S., Buckle, M., et al. (2004). Antimicrobial screening of plants used for traditional medicine in the state of Perak, Peninsular Malaysia. Fitoterapia 75, 68–73. doi: 10.1016/j.fitote.2003.07.013
Widhalm, J. R., and Rhodes, D. (2016). Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic. Res. 3:16046.
Xia, J., and Wishart, D. S. (2016). Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 14.10.11–14.10.91.
Xiao, J. F., Zhou, B., and Ressom, H. W. (2012). Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TRAC Trend Anal. Chem. 32, 1–14. doi: 10.1016/j.trac.2011.08.009
Xie, G. X., Ni, Y., Su, M. M., Zhang, Y. Y., Zhao, A. H., Gao, X. F., et al. (2008). Application of ultra-performance LC-TOF MS metabolite profiling techniques to the analysis of medicinal Panax herbs. Metabolomics 4, 248–260. doi: 10.1007/s11306-008-0115-5
Yu, O., and McGonigle, B. (2005). Metabolic engineering of isoflavone biosynthesis. Adv. Agron. 86, 147–190. doi: 10.1016/s0065-2113(05)86003-1
Yulita, K. S., and Mansur, M. (2012). The occurrence of hybrid in Nepenthes hookeriana Lindl. from Central Kalimantan can be detected by RAPD and ISSR markers. Hayati J. Biosci. 19:18. doi: 10.4308/hjb.19.1.18
Zhou, B., Xiao, J. F., Tuli, L., and Ressom, H. W. (2012). LC-MS-based metabolomics. Mol. Biosyst. 8, 470–481.
Zulkapli, M. M., Ab Ghani, N. S., Ting, T. Y., Aizat, W. M., and Goh, H.-H. (2021). Transcriptomic and proteomic analyses of Nepenthes ampullaria and Nepenthes rafflesiana reveal parental molecular expression in the pitchers of their hybrid, Nepenthes × hookeriana. Front. Plant Sci. 11:625507. doi: 10.3389/fpls.2020.625507
Keywords: carnivorous plant, flavonoid, hybrid, LC-MS, metabolomics, Nepenthes, pitcher
Citation: Rosli MAF, Mediani A, Azizan KA, Baharum SN and Goh H-H (2021) UPLC-TOF-MS/MS-Based Metabolomics Analysis Reveals Species-Specific Metabolite Compositions in Pitchers of Nepenthes ampullaria, Nepenthes rafflesiana, and Their Hybrid Nepenthes × hookeriana. Front. Plant Sci. 12:655004. doi: 10.3389/fpls.2021.655004
Received: 18 January 2021; Accepted: 15 March 2021;
Published: 21 April 2021.
Edited by:
Jeremy D. Rentsch, Francis Marion University, United StatesReviewed by:
Axel Mithöfer, Max Planck Institute for Chemical Ecology, GermanyWeng Ngai Lam, National University of Singapore, Singapore
Copyright © 2021 Rosli, Mediani, Azizan, Baharum and Goh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hoe-Han Goh, Z29oaGhAdWttLmVkdS5teQ==
 Muhammad Aqil Fitri Rosli
Muhammad Aqil Fitri Rosli Ahmed Mediani
Ahmed Mediani Kamalrul Azlan Azizan
Kamalrul Azlan Azizan Syarul Nataqain Baharum
Syarul Nataqain Baharum Hoe-Han Goh
Hoe-Han Goh