
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 April 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.652959
This article is part of the Research TopicRosaceae Fruit Development and QualityView all 11 articles
Loquat fruit accumulates lignin in its flesh when undergoing chilling injury during postharvest storage, making it a suitable model for the study of flesh lignification. Transcriptional regulation of lignin biosynthesis is principally controlled by the NAC-MYB transcriptional cascade in model plants. Previous research has demonstrated that EjMYB8 activates lignin biosynthesis through direct interaction with the promoter of Ej4CL1. However, the classic NAC-MYB gene regulation network has not been established. Here, the MADS-box gene EjAGL65 was discovered by screening a cDNA library using the EjMYB8 promoter as bait in yeast. A phylogenetic analysis and structural comparisons revealed that EjAGL65 belongs to the Mδ subgroup of the MADS-box family, whose members have not been reported as being involved in the regulation of lignin deposition. EjAGL65 transcription was downregulated at 0°C compared to 5°C, indicating a negative correlation with the change of lignin content. A dual-luciferase assay indicated that EjAGL65 is capable of inhibiting the promoter activity of EjMYB8 in vivo. These results showed that the Mδ MADS-box gene EjAGL65 transcriptionally regulates EjMYB8 during postharvest chilling induced flesh lignification, which differs from the classical regulation model of lignin biosynthesis that has been illustrated for developmental lignin accumulation.
Lignin is a pivotal component of vascular tissues that enabled plants to conquer dry land by reinforcing cell walls. In most cases, a lack of lignin may trigger severe defects in plant development. For example, disruption of lignin biosynthesis results in dwarfism in Arabidopsis (Im Kim et al., 2014) and crops, such as rice, suffer lodging due to low lignin deposition in stems (Peng et al., 2014). Although lignin is important for plant growth and development, it may negatively affect the quality of plant products. Many studies have attempted to alleviate the lignification progress in edible plant organs such as fruit (Dangcham et al., 2008; Cao et al., 2010; Suo et al., 2018). Loquat is a fruit common in China that is sensitive to low temperature. During postharvest storage and transportation, the flesh of loquat fruit readily lignifies if injured by chilling or mechanical force, which limits the distance the fruit can be transported. Lignified flesh becomes less juicy and hard to chew, resulting in the deterioration of fruit quality (Jin et al., 2014; Cao et al., 2018). Due to the negative impact on the postharvest storage of loquat, the molecular mechanism underlying flesh lignification must be determined.
Lignin deposition is a complicated process that includes monolignol synthesis and polymerization. Monolignols are synthesized from phenylalanine via the phenylpropanoid pathway in the cytoplasm before they are transported to the apoplast where they undergo oxidation before incorporation into the lignin polymer. The amino group of phenylalanine is first removed by phenylalanine ammonia-lyase (PAL), and p-coumaric acid is generated by adding a hydroxyl group to the C4 position of the aromatic ring by cinnamate 4-hydroxylase (C4H). The p-coumaric acid is then catalyzed sequentially by 4-coumarate: coenzyme A (CoA) ligase (4CL), p-hydroxycinnamoyl-CoA: quinate shikimate p-hydroxycinnamoyltransferase (HCT), p-coumarate 3-hydroxylase (C3H), caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), and ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), which results in methyl oxidation in the aromatic ring at the C3 (coniferaldehyde) site or both the C3 and C5 (sinapaldehyde) sites. The p-coumaraldehyde can only be converted by 4CL and CCR using p-coumaric acid as the substrate. Finally, the products of aldehydes are reduced to alcohols by cinnamyl alcohol dehydrogenase (CAD), generating p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol (Boerjan et al., 2003; Vanholme et al., 2019). These monomers are oxidized/radicalized by peroxidase (PRX) and laccase (LAC), followed by polymerization via oxidative coupling. Several genes encoding key lignin biosynthetic enzymes, such as Ej4CL1 (Li et al., 2017) and EjCAD5 (Xu et al., 2019b), have been identified and play a role in loquat flesh lignification; Ej4CL1 encodes 4CL, which catalyzes p-coumaric acid to p-coumaroyl-CoA. EjCAD5 encodes CAD, which converts p-coumaraldehyde, coniferaldehyde, and sinapaldehyde to their alcohol forms. Transcript abundance of these genes increases under chilling injury, which is in accordance with the higher lignin content and increased fruit firmness. Nevertheless, it is unknown how these enzymes are precisely regulated.
Flesh lignification is intensely regulated at the transcriptional level. Some evidence has been found indicating the involvement of various transcription factor families in the regulation of lignin biosynthesis. According to previous studies, ectopic expression of MYB (Xu et al., 2014b; Wang et al., 2016), NAC (NAM, ATAF1, ATAF2, and CUC2; Ge et al., 2017), APETALA2/Ethylene Response Factor (AP2/ERF; Zeng et al., 2015; Zhang et al., 2020), Heat Shock Factor (HSF; Zeng et al., 2016), and Homobox from Arabidopsis thaliana (HAT; Xu et al., 2019b) family members in loquat flesh resulted in changes in the expression of phenylpropanoid pathway genes. For example, the transcripts of Ej4CL1 can be induced by EjERF39 and EjHSF1 or repressed by EjAP2-1 and EjbHLH1, resulting in changes in the lignin content (Zeng et al., 2015, 2016; Xu et al., 2019b; Zhang et al., 2020). EjNAC3 and EjHAT1 can modify the expression of EjCAD-like and EjCAD5, respectively (Ge et al., 2017; Xu et al., 2019b). Although these transcription factors may be regulators of the phenylpropanoid pathway, most cannot physically interact with promoters of target genes on their own, indicating that mediators are required.
Recent publications have shown that MYB family proteins are mediators of most critical regulators of flesh lignification (Kamdee et al., 2014; Zhang et al., 2016). Through binding to the AC elements located in the promoter region, MYB family members provide other transcription factors with physical routes by which they can regulate the transcripts of Ej4CL1. For example, by protein interaction with EjMYB1, EjAP2-1 suppresses the EjMYB1-induced expression of Ej4CL1 (Zeng et al., 2015). Similarly, EjbHLH1 inhibits the transcription of Ej4CL1 by forming a protein complex with EjMYB2 (Xu et al., 2019a). In addition, the transcripts of Ej4CL1 that increased due to EjMYB8 could be further induced by the synergism between EjERF39 and EjMYB8 (Zhang et al., 2020). Thus, lignin biosynthesis can be regulated by protein-protein interactions with MYBs, which is a strategy frequently adopted by transcription factors that cannot directly bind to promoters of genes in the phenylpropanoid pathway.
Despite the numerous studies conducted on manipulating the biological functions of MYB members at the protein-protein level, limited research has been conducted on proteins that transcriptionally regulate MYB genes during flesh lignification. It is widely accepted that NAC domain proteins play a role in the development of the secondary cell wall by affecting transcript abundance of multiple MYB genes (Boerjan et al., 2003; Li et al., 2015; Yang et al., 2017). For instance, AtMYB58 and AtMYB63 directly control the monolignol biosynthesis gene At4CL1, and AtMYB58 and AtMYB63 are both regulated by the secondary cell wall master switch gene AtSND1, which represents a part of the complex NAC-MYB cascade (Zhou et al., 2009). Although several lignification-related NAC family members have been characterized in the loquat lignification process, interactions between NACs and MYBs have not yet been identified. EjNAC3 has been found to directly bind to EjCAD-like instead of forming a NAC-MYB cascade (Ge et al., 2017). The findings from studies on loquat support the hypothesis that the regulation of stress induced lignin deposition differs from that of developmental lignin (Cesarino, 2019). Though common regulators are employed, such as MYB and NAC genes, there may be novel regulators involved in regulating stress induced lignification. Therefore, further information is needed on partners that interact with EjMYBs to form a transcriptional cascade.
MADS-box genes constitute an important group of transcriptional regulators that have been characterized as regulators of plant morphology (Yu, et al., 2014), flower development (Pelaz et al., 2000), flowering time (Michaels and Amasino, 1999), fruit development, and ripening (Gimenez et al., 2010; Martel et al., 2011). Generally, MADS-box genes are rarely considered as regulators of lignin deposition. Although a thinner pericarp with altered lignin content was observed in tomato fruit with a silenced TAGL1 MADS-box gene (Gimenez et al., 2010), the detailed mechanism whereby MADS-box genes regulate components of the secondary cell wall remains unclear.
In the present study, the upstream regulators of a potent activator of lignin, EjMYB8, were investigated, with the aim of discovering the cascade regulation model. Using yeast one-hybrid screening, a MADS-box domain protein EjAGL65 was discovered to physically interact with the promoter region of EjMYB8. According to phylogenetic analyses, EjAGL65 was the most similar to AtAGL65, which belongs to the Mδ subgroup of the MADS-box family. Transcript abundance of EjAGL65 was monitored at 0 and 5°C, two treatments that promote and alleviate lignification, respectively. The expression pattern of EjAGL65 showed a negative correlation with lignin content and fruit firmness. A dual-luciferase assay confirmed that EjAGL65 inhibited the activity of both EjMYB8 and the Ej4CL1 promoter. These results revealed the involvement of MADS-box genes and the novel MADS-MYB transcriptional cascade in fruit flesh lignification.
Loquat fruits from Eriobotrya japonica Lindl. cultivar “LYQ” were collected in Luqiao, Zhejiang Province, China. The fruits and treatments were previously described in Xu et al. (2014b). Generally, the fruits were selected for uniformity and divided into two batches with approximately 150 fruits in each batch. One batch was kept at 5°C, while the other at 0°C, as the control. Fruits were sampled at 0, 1, 2, 4, and 6 days of storage. At each time point, 15 fruits with the pericarp and seeds removed were sampled; therefore, there were three mixed flesh groups containing five fruits each as three biological replicates. The collected flesh samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
Yeast one-hybrid screening was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech, Mountain View, CA, United States). The promoter of EjMYB8 was isolated using the GenomeWalker universal kit (Clontech) then inserted into the pAbAi vector. Primers used for genome walking and pAbAi construction are listed in Supplementary Tables S1, S2, respectively. The recombinant EjMYB8-pAbAi vector was linearized and transformed into a Y1HGold yeast strain. After testing for autoactivation, 100 ng/ml Aureobasidin A (AbA) was used for library screening (Supplementary Figure S1). The Y1HGold[EjMYB8/AbAi] yeast strain was transfected together with the cDNA library, which was previously constructed by Zhang et al. (2016). After 5 days of growth at 30°C, every individual yeast colony was picked for PCR analysis and then sequenced by Huajin Company (Shanghai, China). The obtained cDNA fragments were used for an in silico search for annotations using the Basic Local Alignment Search Tool.1 Five putative transcription factors were selected for further confirmation (listed in Supplementary Table S3). Primers designed for full-length CDS isolation and construction of pGADT7 vectors were listed in Supplementary Tables S4, S5, respectively. The obtained sequences were uploaded to the NCBI database (GenBank numbers are given in Supplementary Table S6).
To confirm true positive interactions, we separately transformed pGADT7 vectors with the genes listed in Supplementary Table S5 into the Y1HGold [EjMYB8/pAbAi] yeast strain. Empty pGADT7 plasmids were transformed as the negative control. Transformed yeast was grown on SD/-Leu medium containing 100 ng/ml of AbA.
Additional nine MADS-box genes were added to the previously reported 107 MADS-box genes by searching the TAIR database.2 A total of 116 Arabidopsis and two Populus MADS-box genes were used in the phylogenetic analysis (Supplementary Table S7). The analysis was completed by the ClustalX v1.81. The protein sequence of EjAGL65 was aligned with 116 MADS-box genes using the default settings of gap open and gap extension cost, and the Blosum 30 was selected as the protein weight matrix. A phylogenetic tree was constructed using the neighbor-joining (NJ) method, and the bootstrap value was set to 1,000. Results of the alignments were visualized using Figtree v1.4.4.3
The “LYQ” flesh samples collected at each time point were used for total RNA extraction following the method described by Shan et al. (2008). Potential genomic DNA contamination was eliminated using the TURBO DNA-free kit (Ambion, Austin, TX, United States). The quality and quantity of freshly extracted RNA were determined by gel electrophoresis and spectrophotometry, respectively (Implen, Westlake Village, CA, United States). A total of 1 μg DNA-free RNA were used to synthesize first-strand cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States). A 10-fold dilution of cDNA was made prior to the real-time PCR analysis.
Real-time PCR was carried out using the CFX96 Real-Time System (Bio-Rad) with SsoFast EvaGreen Supermix (Bio-Rad). The reaction mixture contained 10 μl SYBR PCR supermix, 6 μl diethylpyrocarbonate-treated H2O, 2 μl diluted cDNA template, and 1 μl of each primer (10 μM). Primers used for real-time PCR were designed using Primer3 (version 4.0.0) and are listed in Supplementary Table S8.4 The primers were tested to ensure their specificity for unique genes (Yin et al., 2010). The real-time PCR program was set as follows: a pre-denaturation step of 95°C for 30 s, followed by for 45 cycles of 95°C for 10 s and 60°C for 10 s. A melting curve analysis was also conducted. Three biological replicates were collected from the data from each time point.
Dual-luciferase assays were performed as described previously (Min et al., 2014). Full-length EjAGL65 genes were amplified with the primers listed in Supplementary Table S9 and integrated into the pGreen II 0029 62-SK vector (SK). The promoter of EjMYB1, EjMYB2, and EjMYB8 was individually inserted into the pGreen II 0800-LUC vector (LUC). The LUC vectors containing promoters of lignin biosynthesis genes in Arabidopsis were constructed by Xu et al. (2014b).
All of the recombinant SK and LUC vectors were transfected into Agrobacterium tumefaciens GV3101 and stored as glycerol stocks. Transfected Agrobacterium cultures were grown on LB plates containing 50 μg/ml kanamycin and 25 μg/ml gentamycin for 2 days, then restreaked on new LB plates and grown for 1 day. Agrobacterium cultures were suspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6) to optimal density (OD600 = 0.75), then 1 ml Agrobacterium culture containing transcription factors was mixed with 100 μl Agrobacterium containing promoters. Finally, the mixtures were injected into tobacco leaves with needleless syringes. Three days after infiltration, the fluorescence intensity of LUC and REN were measured using dual luciferase assay reagents (Promega, Madison, WI, United States). Five replicates were conducted for each transcription factor and promoter combination.
OriginPro 2020 software (Microcal Software Inc., Northampton, MA, United States) was used to perform statistical analyses and to draw figures. Student’s t-tests were conducted on data from luciferase assays; differences were considered significant at 5%. One-way ANOVA followed by means comparison using a Tukey’s test was applied for comparing data from real-time PCR at the 5% confidence level. The average value and SE of three replicates were calculated using Excel 2017 (Microsoft, Seattle, WA, United States).
Previous research has demonstrated that NAC and MYB genes mediate lignification in loquat flesh (Xu et al., 2014b), but the classic NAC-MYB transcription cascade has not been previously revealed. Here, we chose to identify the upstream regulators of EjMYB8 because of its significance in cold triggered expression by yeast one-hybrid screening (Wang et al., 2016). The promoter region of EjMYB8 was integrated into the Y1HGold yeast strain, and we performed a screening under the stress of 100 ng/ml AbA to eliminate the background growth of yeast. Following the amplification of library fragments by yeast colony PCR, a total of 32 colonies were sequenced and annotated (listed in Supplementary Table S10). We chose five putative transcription factors for further confirmation. Consequently, a MADS-box domain-containing protein (GenBank No. MF942415) was identified as having a positive interaction with the promoter of EjMYB8; the other four proteins had false-positive results (Figure 1).
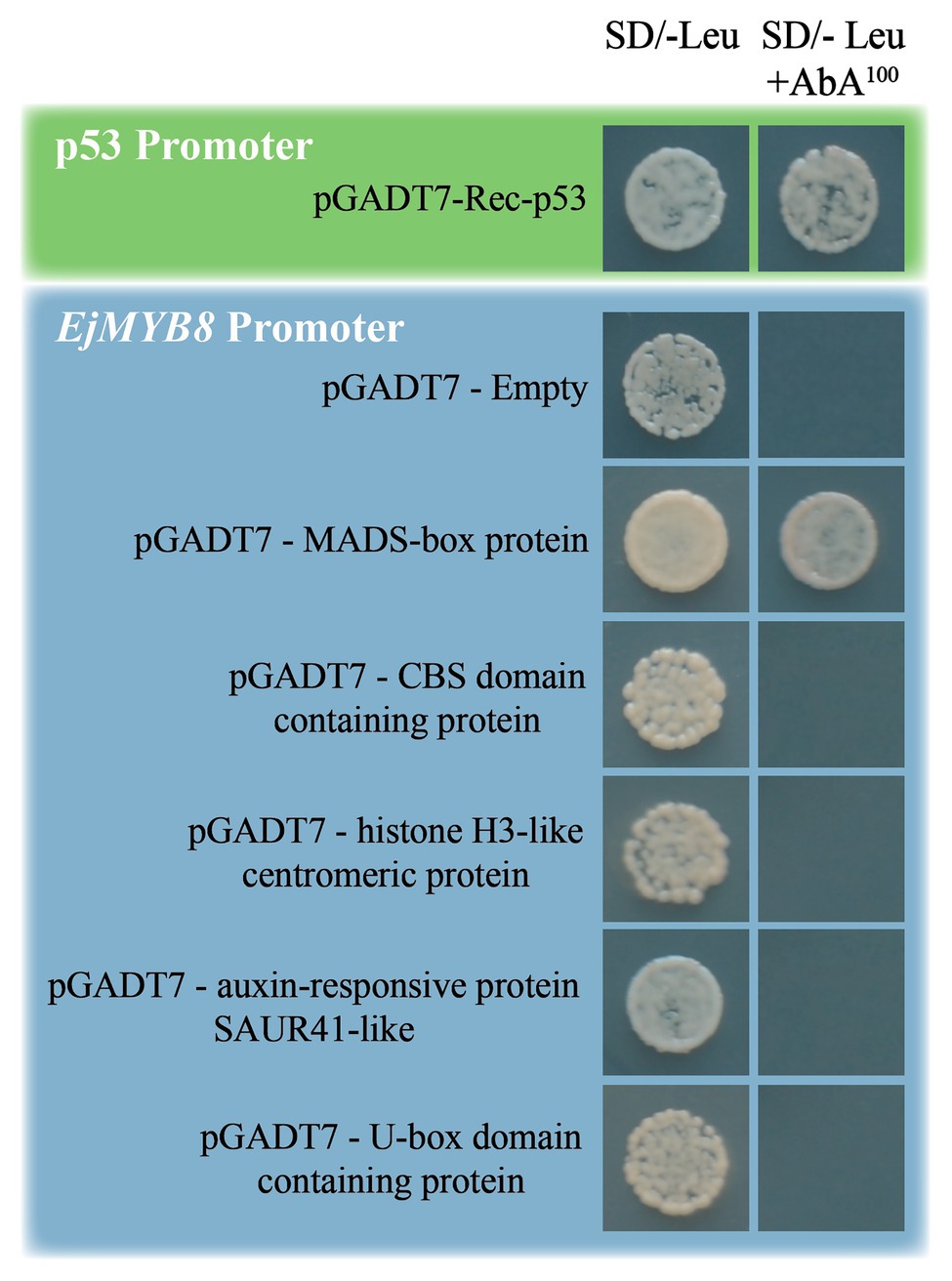
Figure 1. A yeast one-hybrid analysis was used to identify a MADS-box protein, which binds the EjMYB8 promoter. Sequences of five genes were inserted individually into the pGADT7 vector then transformed into Y1HGold[pro-EjMYB8] for an in vivo Protein-DNA binding assay based on the results of the yeast one-hybrid library screening. Growth of yeast on SD/-Leu plates with 100 ng/ml Aureobasidin A (AbA) indicates that the interaction is genuinely positive. The promoter of p53 was also transformed into yeast as the positive control. The negative control was an empty pGADT7 vector.
Previous studies have shown that MADS-box genes are further classified into five subgroups, named MIKC, Mα, Mβ, Mγ, and Mδ, based on four conserved domains in Arabidopsis (Parenicová et al., 2003), and each group has a unique biological function. To predict the potential function of the MADS-box domain gene, we conducted a phylogenetic analysis. By comparing the amino acid sequence with MADS-box isoforms in Arabidopsis, we found that the EjMYB8-associated loquat MADS-box protein clustered with AtAGL65, AtAGL30, and AtAGL94, which are in the Mδ subgroup (Figure 2). Therefore, the newly identified MADS-box gene was named EjAGL65. AtAGL65, which is the putative ortholog of EjAGL65, forms a heterodimer with AtAGL104 and regulates pollen activity (Adamczyk and Fernandez, 2009). Besides, the MIKC group gene AtAGL15 regulates the class III peroxidase PRX17 and the knockout line prx17 showed reduced lignin content in the stem and siliques. The expression level of PRX17 is downregulated in a 35S:AGL15 line, indicating the involvement of AtAGL15 in lignin metabolism (Cosio et al., 2017). However, EjAGL65 was not clustered with AtAGL15. These findings indicate that the EjAGL65 may not affect monolignol polymerization. The putative ortholog of EjAGL65 in Arabidopsis is not thought to be lignin-related.
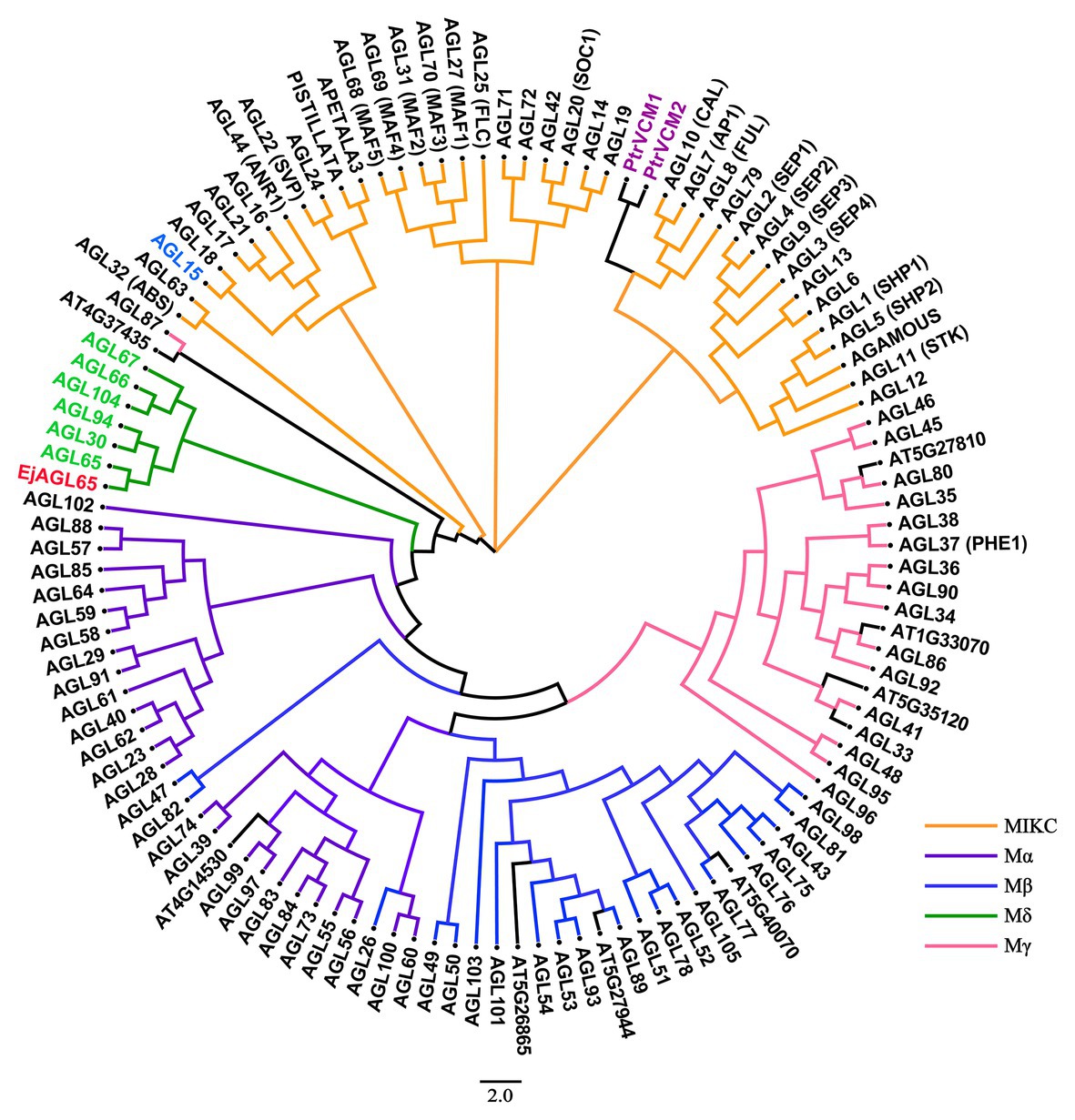
Figure 2. Phylogenetic analysis of EjAGL65 with 116 Arabidopsis MADS-box proteins and two Populus MADS-box proteins. The previously defined Mδ group genes are shown in green. EjAGL65 was structurally closest to Arabidopsis AGL65 (At1g18750) and is shown in red. AGL15 (At5g13790), which was expressed in the lignified tissue of Arabidopsis, is shown in blue. PtrVCM1 and PtrVCM2, which affect vascular cambium proliferation activity, are in purple. The color of clades represents MIKC (orange), Mα (violet), Mβ (blue), Mδ (green), and Mγ (pink) subgroups of Arabidopsis MADS-box genes. Genes without annotations are labeled with an ID from the TAIR database.
The expression of EjAGL65 was investigated to determine whether it responds to chilling injury, which is critical evidence in support of it regulating chilling-induced lignification. Treatments at 0 and 5°C were routinely applied when characterizing candidate genes that relate to chilling injury of loquat fruits because 0°C induces lignification, while 5°C alleviates it (Cai et al., 2006). As shown in Figure 3, the average transcripts of EjAGL65 decreased at 0°C on the first and second days, though the difference was not significant according to Turkey’s test. The transcription of EjAGL65 did not decrease at 5°C during the first 2 days of treatment, followed by significant increases later. Based on its expression profile, EjAGL65 was more sensitive to treatment at 0°C than 5°C. Xu et al. (2014b) tested fruit firmness and lignin content using the same batch of materials. The firmness and lignin content increased rapidly at 0 and 5°C during the first 2 days, which was opposite from the observed change of EjAGL65 transcripts. This information indicates that EjAGL65 is negatively correlated with changes in fruit firmness and lignin content. In addition, the decrease in EjAGL65 transcripts stopped after 4 days of storage at 0°C, indicating an unknown mechanism preventing EjAGL65 transcripts from continuously decreasing. EjAGL65 is a candidate negative regulator of the cold-induced lignification process in loquat fruit flesh.
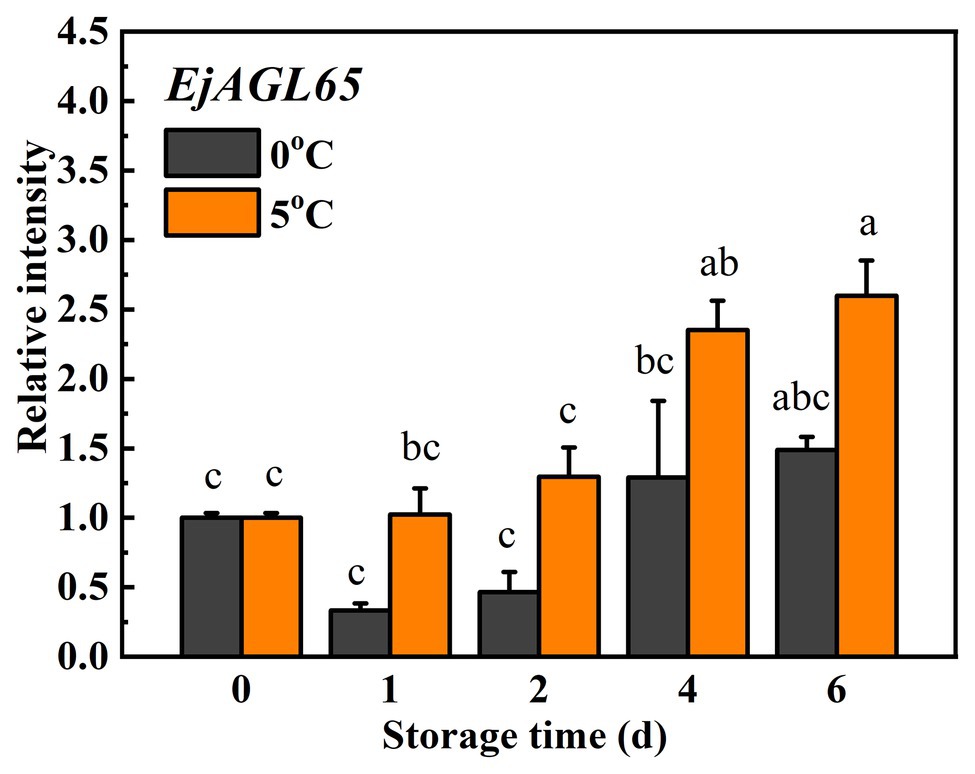
Figure 3. Expression of EjAGL65 in response to temperature treatments at 0°C (control) and 5°C in “LYQ” loquat. Transcript levels of EjAGL65 before the temperature treatment were set to 1. Error bars indicate the SE of three biological replicates. One-way ANOVA followed by Tukey’s comparison was performed at p = 0.05.
Possible regulatory actions of EjAGL65 were tested with the promoter of EjMYB8 because of the direct binding confirmed in the yeast one-hybrid assay. The dual-luciferase assay showed that the activity of the EjMYB8 promoter was significantly repressed after transient overexpression of EjAGL65. We also tested the promoter activity of EjMYB1 and EjMYB2, another two EjMYB members related to lignin. However, the transient overexpression of EjAGL65 did not significantly change the promoter activity of EjMYB1 and EjMYB2 (Figure 4).
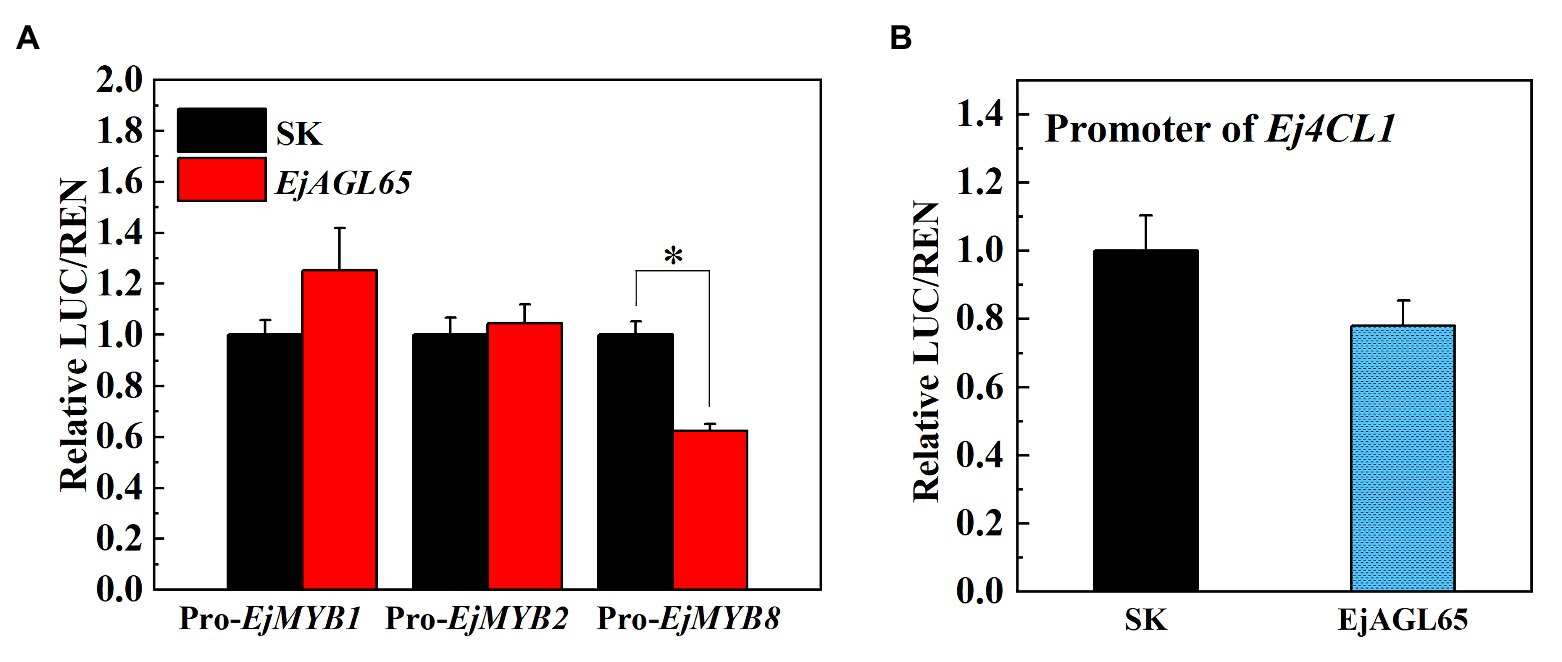
Figure 4. Luciferase assay between EjAGL65 and lignin biosynthesis-related genes. The SK vector contains the coding sequence of EjAGL65; the LUC vectors with the insertion of the EjMYB1, EjMYB2, EjMYB8 (A), and Ej4CL1 (B) promoter were pairwise transformed into the Agrobacterium strain GV1301 followed by leaf infection. The LUC/REN value represents the activation (above 1) or inhibition (below 1) effect generated by EjAGL65. A combination of the Empty SK vector and LUC vectors with different promoters was used as a calibration (set as 1). Error bars represent five replicates. The asterisk indicates a significant difference according to the Student’s t-test at a significance level of 0.05 (*p < 0.05).
Ej4CL1 is a key gene for loquat fruit lignification and the direct target of EjMYB8. The interaction between EjAGL65 and the promoter of Ej4CL1 was also tested to further characterize the effect of EjAGL65 on the Ej4CL1 promoter without EjMYB8. Protein-DNA interaction was analyzed using a yeast one-hybrid assay and dual-luciferase assay. Consequently, the activity of the Ej4CL1 promoter was slightly but not significantly reduced due to the transient overexpression of EjAGL65 (Figure 4). In addition, EjAGL65 cannot bind the promoter of Ej4CL1 in vivo as shown by the yeast one-hybrid assay (Supplementary Figure S2), suggesting that EjAGL65 alone is not sufficient to affect the activity of the Ej4CL1 promoter.
In addition, we chose the well-defined lignin biosynthesis-related genes from Arabidopsis to discover potential regulatory targets of EjAGL65. Consequently, most of the promoter activity of selected genes could not be regulated by EjAGL65. It is unclear whether the statistically significant activity against AtC4H and AtCCR1 promoters (marked with asterisks in Figure 5) is sufficient to alter the biological function. Overall, the target preference of EjAGL65 suggests it might selectively affect transcription factors, such as MYB family members, rather than structural genes of lignin biosynthesis.
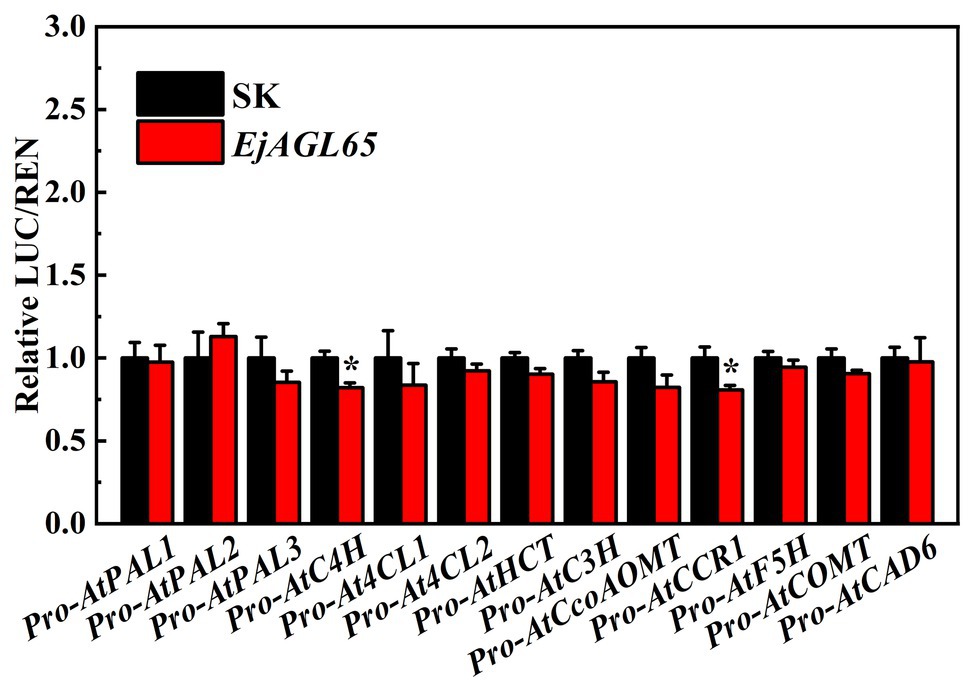
Figure 5. Interactions between EjAGL65 and Arabidopsis genes involved in lignin biosynthesis. A combination of the empty SK vector and LUC vectors with different promoters was used as a calibration (set as 1). Error bars resulted from five replicates. The asterisk indicates that the relative LUC/REN value was significantly different between SK and EjAGL65 based on the Student’s t-test at a significance level of 0.05.
In recent years, knowledge on fruit flesh lignification has advanced partially due to experiments on protein-DNA interactions (Xu et al., 2015; Wang et al., 2016) that helped to unravel key aspects of the underlying regulatory network. Previous studies have demonstrated that EjAP2-1 works in coordination with EjMYB1 and EjMYB2 for transcriptional regulation of Ej4CL1 (Zeng et al., 2015). Zhang et al. (2020) recently found that the EjERF39 and EjMYB8 protein complex activates Ej4CL1 by protein-DNA interaction with the promoter of Ej4CL1. Furthermore, utilization of the loquat transcriptome has allowed for the discovery of more lignification-related transcription factors (Lin et al., 2018; Liu et al., 2019). However, MADS-box genes are not frequently studied. In the presented study, we found the MADS-box domain gene EjAGL65 using a yeast one-hybrid library screening assay. Data from a luciferase assay suggests that transient overexpression of EjAGL65 represses the transcription of EjMYB8, which is considered as a key activator of Ej4CL1 (Wang et al., 2016), indicating the involvement of EjAGL65 in lignin biosynthesis. Interestingly, there is limited evidence of MADS-box genes being associated with lignin biosynthesis. Instead, MADS-box genes play vital roles in plant morphological development (Yu et al., 2014) and precise control of flowering and fruit development (Michaels and Amasino, 1999). AtAGL65, AtAGL30, and AtAGL94, which were determined to be the closest homologs of EjAGL65, control the maturation of pollen and pollen tube growth (Adamczyk and Fernandez, 2009). Tomato MADS-RIN is a primary ripening regulator that is highly expressed in fruit tissue (Vrebalov et al., 2002). The cell wall components were altered in the rin mutant, resulting in firmer texture, but the influence of RIN on lignin deposition was not clear (Li et al., 2018). Although the repression of another tomato MADS-box gene, SlTAGL1, results in lignin accumulation, the regulatory mechanism is unknown. Recently, a link between the MADS-box gene and lignin was established. AtAGL15, a MIKC type MADS-box gene, has been found to directly interact with the class III peroxidase PRX17, which causes ectopic lignin distribution in Arabidopsis stems if silenced (Cosio et al., 2017). Meanwhile, ectopic expression of VCM1 and VCM2, two Populus MADS-box genes, leads to abnormal secondary growth by the regulation of the auxin transporter PIN5 (Zheng et al., 2020). By comparing amino acid sequences with Arabidopsis MADS-box genes, we found that AtAGL15, VCM1, and VCM2 belong to the MIKC group members. However, EjAGL65 lacks the K-box domain and is located in the Mδ subgroup (Supplementary Figure S3), indicating different mechanisms of lignin regulation between EjAGL65 and the other reported secondary cell wall related MADS-box genes. The Mδ subgroup consists of only six members in Arabidopsis and is crucial for pollen maturation (Verelst et al., 2007). The function of the Mδ genes is conserved in the land plants studied thus far (Kwantes et al., 2012), indicating their importance during plant evolution. The discovery of EjAGL65, which can alter lignin biosynthesis-related transcription factors, might suggest a novel function of the Mδ subgroup that is distinct from pollen development.
Several studies have been conducted on the expression of the MADS-box gene transcripts during different developmental stages (Qin et al., 2012; Niu et al., 2016). However, in fruit, the dynamic modulations of MADS-box genes in response to environmental stimuli, such as chilling, have rarely been studied. Here, transcript levels of EjAGL65 were measured during the cold storage of loquat fruits. EjAGL65 mRNA levels rapidly decreased during the first 48 h, but gradually recovered to their initial level, representing a response to chilling at days 1 and 2. Considering that the lignin content of loquat fruits also correlates with the pattern of EjAGL65 transcripts, there may be specific mechanisms preventing the EjAGL65 transcripts from continuing to decrease. Generally, genes responsible for cold perception have various means of preventing overexpression or insufficient expression at the transcriptional or translational level, such as SIZ1 and HOS1 proteins, which antagonistically determine the degradation of the ICE1 protein (Miura et al., 2007). Although, we cannot yet conclude whether EjAGL65 is located in the cold signal transduction network, revealing such a feedback mechanism for precisely controlling the expression of EjAGL65 would provide a better understanding of chilling induced flesh lignification.
The relationship between MYB genes and lignin biosynthesis, as well as NAC master switches have been extensively studied in model plants, trees, and grasses. Lignin-related MYB genes are primarily regulated by NAC domain proteins at the transcription level in vascular tissue (Ohtani and Demura, 2019). The NAC-MYB based regulatory network is relatively conserved in the plant kingdom; it even exists in Physcomitrella patens, which is a moss that does not accumulate lignin (Xu et al., 2014a). Nevertheless, information about genes other than the NAC family that affect lignin-related MYB genes is still limited. Only the hAT transposase family gene PtrhAT was reported as the regulator of PtrMYB021 (Xie et al., 2018). Several NAC genes have been characterized as activators of the lignification process in loquat flesh. However, no evidence indicates that NAC genes target lignin-related MYB genes, such as EjMYB1, whose homologs in Arabidopsis (AtMYB58/63) are regulated by the NAC-MYB transcription cascade (McCarthy et al., 2009; Zhou et al., 2009). On the contrary, the reported loquat NAC genes were confirmed as neither having a biological effect on MYB genes nor directly binding their promoter (Xu et al., 2015; Ge et al., 2017). These results suggest that the loquat may recruit new players for regulating stress induced lignification in flesh tissue. In this study, we performed a library-scale screening to determine the upstream regulators of EjMYB8. Nevertheless, no NAC genes were found in the library pool after in vivo screening in yeast. Interestingly, a MADS-box domain protein, EjAGL65, physically binds to the promoter of EjMYB8, forming the MADS-MYB cascade. The results provide additional information about how to unravel a new hub in the complicated regulatory network of fruit flesh lignin biosynthesis.
Research on transcriptional regulation in the lignification of loquat flesh began with identifying two MYB family genes, EjMYB1 and EjMYB2 (Xu et al., 2014b). Subsequently, EjAP2-1 was found to be involved in the regulatory network by showing protein-protein interactions with either EjMYB1 or EjMYB2 (Zeng et al., 2015). With the further screening of MYB genes, EjMYB8 was proposed as an effective activator of lignin biosynthesis (Wang et al., 2016). The activation effect of EjMYB8 could be further enhanced by forming a protein complex with EjERF39 (Zhang et al., 2020), but there was limited information about negative regulators of EjMYB8. These MYB genes share the same target, located in the promoter region of Ej4CL1, a gene that plays an essential role in the biosynthesis of lignin monomers (Li et al., 2017), which complicates the mechanism by which Ej4CL1 transcripts are adjusted.
The present study showed that the MADS-box gene EjAGL65 affects loquat flesh lignin accumulation via repressing the transcription of the characterized lignin activator EjMYB8 (Figure 6). EjAGL65 transcripts showed a temperature-dependent pattern similar to its Arabidopsis homologs, providing a new hub for characterizing a chilling induced regulation network of flesh lignification. The system contains three MYB genes, two members of the AP2/ERF gene family, and a MADS-box gene, forming three regulatory pathways. Interestingly, only two of the abovementioned genes negatively regulate lignin biosynthesis at a transcription level, while the others either have identical activation effects or function through protein-protein interaction. Identifying additional factors and understanding the synergistic or antagonistic effects among the four pathways of the network will be the main focus of future research.
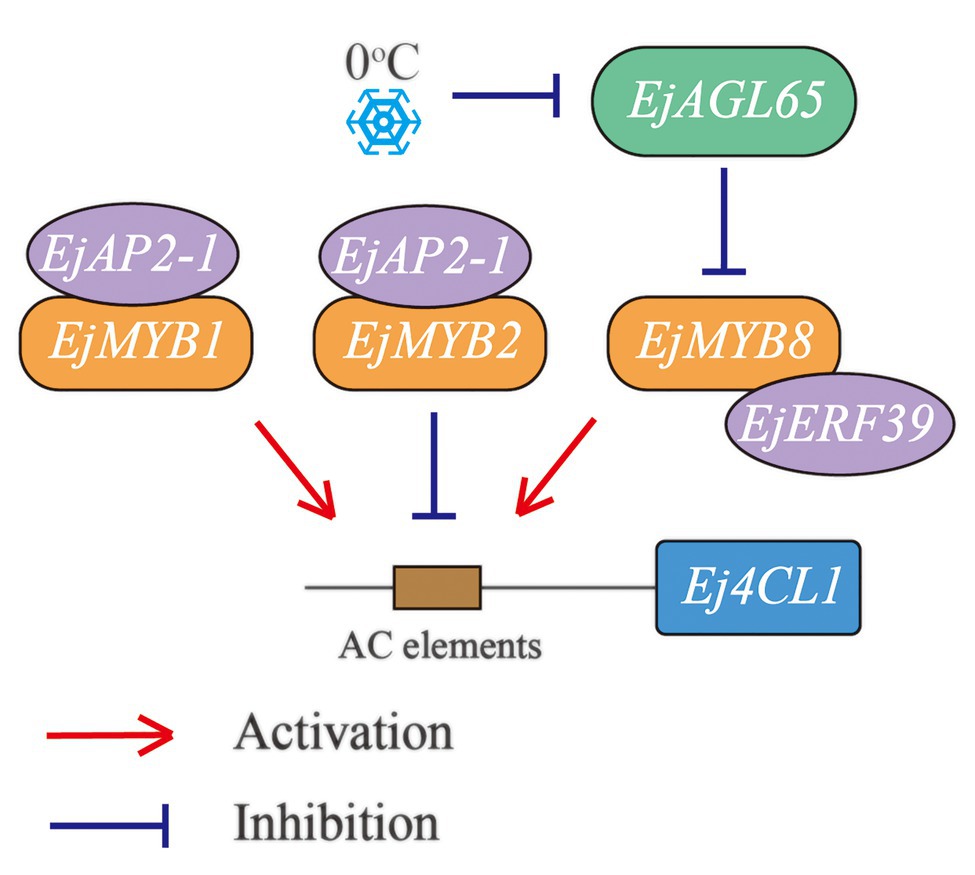
Figure 6. Regulatory routes identified based on Ej4CL1. Three MYB genes are physically associated with Ej4CL1, acting as mediators for binding with other transcription factors. Both EjMYB1 and EjMYB2 can recruit EjAP2-1 to repress their transcriptional activities. EjMYB8 is controlled at both protein level (by EjERF39) and mRNA level (by EjAGL65). Lines with an arrow or a dashed terminal indicate activation or repression, respectively. Two genes shown in a sequence indicate a confirmed protein-protein interaction with each other.
The present study discovered a chilling-repressed MADS-box gene named EjAGL65. Results of dual luciferase and yeast one-hybrid assays indicated that EjAGL65 transcriptionally inhibits the promoter of EjMYB8, forming a MADS-MYB transcriptional cascade. The homologs of EjAGL65 were not known to affect the lignin biosynthesis pathway in model plants, revealing a novel route for the regulation of stress induced lignin.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
KC conceived the experiments. HG and MZ conducted the study and processed the data. HG and YS wrote the manuscript. KC, XL, and XY revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (31630067 and 31901740) and the 111 Project (B17039).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.652959/full#supplementary-material
1. https://www.ncbi.nlm.nih.gov/
2. https://www.arabidopsis.org/
Adamczyk, B. J., and Fernandez, D. E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149, 1713–1723. doi: 10.1104/pp.109.135806
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Cai, C., Xu, C. J., Shan, L. L., Li, X., Zhou, C. H., Zhang, W. S., et al. (2006). Low temperature conditioning reduces postharvest chilling injury in loquat fruit. Postharvest Biol. Technol. 41, 252–259. doi: 10.1016/j.postharvbio.2006.04.015
Cao, S., Shao, J., Shi, L., Xu, L., Shen, Z., Chen, W., et al. (2018). Melatonin increases chilling tolerance in postharvest peach fruit by alleviating oxidative damage. Sci. Rep. 8:806. doi: 10.1038/s41598-018-19363-5
Cao, S., Zheng, Y., Wang, K., Rui, H., and Tang, S. (2010). Effect of methyl jasmonate on cell wall modification of loquat fruit in relation to chilling injury after harvest. Food Chemistry 118, 641–647. doi: 10.1016/j.foodchem.2009.05.047
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012
Cosio, C., Ranocha, P., Francoz, E., Burlat, V., Zheng, Y., Perry, S. E., et al. (2017). The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 213, 250–263. doi: 10.1111/nph.14127
Dangcham, S., Bowen, J., Ferguson, I. B., and Ketsa, S. (2008). Effect of temperature and low oxygen on pericarp hardening of mangosteen fruit stored at low temperature. Postharvest Biol. Technol. 50, 37–44. doi: 10.1016/j.postharvbio.2008.02.005
Ge, H., Zhang, J., Zhang, Y. J., Li, X., Yin, X. R., Grierson, D., et al. (2017). EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. J. Exp. Bot. 68, 5129–5136. doi: 10.1093/jxb/erx330
Gimenez, E., Pineda, B., Capel, J., Anton, M. T., Atares, A., Perez-Martin, F., et al. (2010). Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS One 5:e14427. doi: 10.1371/journal.pone.0014427
Im Kim, J., Ciesielski, P. N., Donohoe, B. S., Chapple, C., and Li, X. (2014). Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol. 164, 584–595. doi: 10.1104/pp.113.229393
Jin, P., Duan, Y. F., Wang, L., Wang, J., and Zheng, Y. H. (2014). Reducing chilling injury of loquat fruit by combined treatment with hot air and methyl jasmonate. Food Bioprocess Technol. 7, 2259–2266. doi: 10.1007/s11947-013-1232-3
Kamdee, C., Imsabai, W., Kirk, R., Allan, A. C., Ferguson, I. B., and Ketsa, S. (2014). Regulation of lignin biosynthesis in fruit pericarp hardening of mangosteen (Garcinia mangostana L.) after impact. Postharvest Biol. Technol. 97, 68–76. doi: 10.1016/j.postharvbio.2014.06.004
Kwantes, M., Liebsch, D., and Verelst, W. (2012). How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302. doi: 10.1093/molbev/msr200
Li, C. F., Wang, X. Q., Ran, L. Y., Tian, Q. Y., Fan, D., and Luo, K. M. (2015). PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol. 56, 2436–2446. doi: 10.1093/pcp/pcv157
Li, S., Xu, H., Ju, Z., Cao, D., Zhu, H., Fu, D., et al. (2018). The RIN-MC fusion of MADS-box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol. 176, 891–909. doi: 10.1104/pp.17.01449
Li, X., Zang, C., Ge, H., Zhang, J., Grierson, D., Yin, X. R., et al. (2017). Involvement of PAL, C4H, and 4CL in chilling injury-induced flesh lignification of loquat fruit. HortScience 52, 127–131. doi: 10.21273/HORTSCI11304-16
Lin, S., Wu, T., Lin, H., Zhang, Y., Xu, S., Wang, J., et al. (2018). De novo analysis reveals transcriptomic responses in Eriobotrya japonica fruits during postharvest cold storage. Genes 9:639. doi: 10.3390/genes9120639
Liu, W. L., Zhang, J., Jiao, C., Yin, X. R., Fei, Z. J., Wu, Q. B., et al. (2019). Transcriptome analysis provides insights into the regulation of metabolic processes during postharvest cold storage of loquat (Eriobotrya japonica) fruit. Hortic. Res. 6, 1–11. doi: 10.1038/s41438-019-0131-9
Martel, C., Vrebalov, J., Tafelmeyer, P., and Giovannoni, J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579. doi: 10.1104/pp.111.181107
McCarthy, R. L., Zhong, R. Q., and Ye, Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Min, T., Fang, F., Ge, H., Shi, Y. N., Luo, Z. R., Yao, Y. C., et al. (2014). Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLoS One 9:e97043. doi: 10.1371/journal.pone.0097043
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Niu, Q., Li, J., Cai, D., Qian, M., Jia, H., Bai, S., et al. (2016). Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 67, 239–257. doi: 10.1093/jxb/erv454
Ohtani, M., and Demura, T. (2019). The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 56, 82–87. doi: 10.1016/j.copbio.2018.10.002
Parenicová, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Peng, D. L., Chen, X. G., Yin, Y. P., Lu, K. L., Yang, W. B., Tang, Y. H., et al. (2014). Lodging resistance of winter wheat (Triticum aestivum L.): lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid. Field Crop Res. 157, 1–7. doi: 10.1016/j.fcr.2013.11.015
Qin, G., Wang, Y., Cao, B., Wang, W., and Tian, S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70, 243–255. doi: 10.1111/j.1365-313X.2011.04861.x
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., De Sun, C., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. doi: 10.1007/s00425-008-0696-2
Suo, J. T., Li, H., Ban, Q. Y., Han, Y., Meng, K., Jin, M. J., et al. (2018). Characteristics of chilling injury-induced lignification in kiwifruit with different sensitivities to low temperatures. Postharvest Biol. Technol. 135, 8–18. doi: 10.1016/j.postharvbio.2017.08.020
Vanholme, R., De Meester, B., Ralph, J., and Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotech. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Verelst, W., Twell, D., de Folter, S., Immink, R., Saedler, H., and Munster, T. (2007). MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 8:R249. doi: 10.1186/gb-2007-8-11-r249
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato Ripening-Inhibitor (Rin) locus. Science 296:343. doi: 10.1126/science.1068181
Wang, W. Q., Zhang, J., Ge, H., Li, S. J., Li, X., Yin, X. R., et al. (2016). EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS One 11:e0154399. doi: 10.1371/journal.pone.0154399
Xie, M., Muchero, W., Bryan, A. C., Yee, K., Guo, H. B., Zhang, J., et al. (2018). A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus. Plant Cell 30, 1645–1660. doi: 10.1105/tpc.18.00168
Xu, M., Li, S. J., Liu, X. F., Yin, X. R., Grierson, D., and Chen, K. S. (2019a). Ternary complex EjbHLH1-EjMYB2-EjAP2-1 retards low temperature-induced flesh lignification in loquat fruit. Plant Physiol. Biochem. 139, 731–737. doi: 10.1016/j.plaphy.2019.04.032
Xu, B., Ohtani, M., Yamaguchi, M., Toyooka, K., Wakazaki, M., Sato, M., et al. (2014a). Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508. doi: 10.1126/science.1248417
Xu, Q., Wang, W. Q., Zeng, J. K., Zhang, J., Grierson, D., Li, X., et al. (2015). A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 102, 25–31. doi: 10.1016/j.postharvbio.2015.02.002
Xu, Q., Yin, X. R., Zeng, J. K., Ge, H., Song, M., Xu, C. J., et al. (2014b). Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 65, 4349–4359. doi: 10.1093/jxb/eru208
Xu, M., Zhang, M. X., Shi, Y. N., Liu, X. F., Li, X., Grierson, D., et al. (2019b). EjHAT1 participates in heat alleviation of loquat fruit lignification by suppressing the promoter activity of key lignin monomer synthesis gene EjCAD5. J. Agric. Food Chem. 67, 5204–5211. doi: 10.1021/acs.jafc.9b00641
Yang, L., Zhao, X., Ran, L. Y., Li, C. F., Fan, D., and Luo, K. M. (2017). PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 7, 1–14. doi: 10.1038/srep41209
Yin, X. R., Allan, A. C., Chen, K. S., and Ferguson, I. B. (2010). Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 153, 1280–1292. doi: 10.1104/pp.110.157081
Yu, L. H., Miao, Z. Q., Qi, G. F., Wu, J., Cai, X. T., Mao, J. L., et al. (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zeng, J. K., Li, X., Zhang, J., Ge, H., Yin, X. R., and Chen, K. S. (2016). Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 39, 1780–1789. doi: 10.1111/pce.12741
Zhang, J., Ge, H., Zang, C., Li, X., Grierson, D., Chen, K. S., et al. (2016). EjODO1, a MYB transcription factor, regulating lignin biosynthesis in developing loquat (Eriobotrya japonica) fruit. Front. Plant Sci. 7:1360. doi: 10.3389/fpls.2016.01360
Zhang, J., Yin, X.-R., Li, H., Xu, M., Zhang, M.-X., Li, S.-J., et al. (2020). ETHYLENE RESPONSE FACTOR EjERF39-EjMYB8 complex activates cold-induced lignification of loquat fruit, via the biosynthetic gene Ej4CL1. J. Exp. Bot. 71, 3172–3184. doi: 10.1093/jxb/eraa085
Zheng, S., He, J., Lin, Z., Zhu, Y., Sun, J., and Li, L. (2020). Two MADS-box genes regulate vascular cambium activity and secondary growth by modulating auxin homeostasis in Populus. Plant Commun. 100134. doi: 10.1016/j.xplc.2020.100134 (in press).
Keywords: loquat, MADS, MYB, chilling injury, lignin biosynthesis, Mδ subgroup
Citation: Ge H, Shi Y-n, Zhang M-x, Li X, Yin X-r and Chen K-s (2021) The MADS-Box Transcription Factor EjAGL65 Controls Loquat Flesh Lignification via Direct Transcriptional Inhibition of EjMYB8. Front. Plant Sci. 12:652959. doi: 10.3389/fpls.2021.652959
Received: 26 January 2021; Accepted: 19 March 2021;
Published: 07 April 2021.
Edited by:
Jia-Long Yao, The New Zealand Institute for Plant and Food Research Ltd, New ZealandReviewed by:
Yongping Cai, Anhui Agricultural University, ChinaCopyright © 2021 Ge, Shi, Zhang, Li, Yin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun-song Chen, YWt1bkB6anUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.