
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Plant Sci., 01 March 2021
Sec. Plant Systematics and Evolution
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.652483
This article is a correction to:
New Insights Into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae)
A Corrigendum on
New Insights Into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae)
by Oyebanji, O., Zhang, R., Chen, S.-Y., and Yi, T.-S. (2020). Front. Plant Sci. 11:151. doi: 10.3389/fpls.2020.00151
In the original article, there was a mistake in the legend for Table 1 as published. The use of “C. gracilis” in the h legend is incorrect. The correct legend appears below.
“hDuplicated in the IR of all species except D. araripensis, L. domingensis, O. pinnata, P. violacea, X. stuhlmannii, I. linifolia and tinctoria.”
In the original article, there was a mistake in Figure 1 and 4 as published. The mistakes were: Cochlianthus gracilis (Phaseoleae), Craspedolobium schochii (Millettieae), and Shuteria vestita (Desmodieae). The corrected taxonomic names appear below.
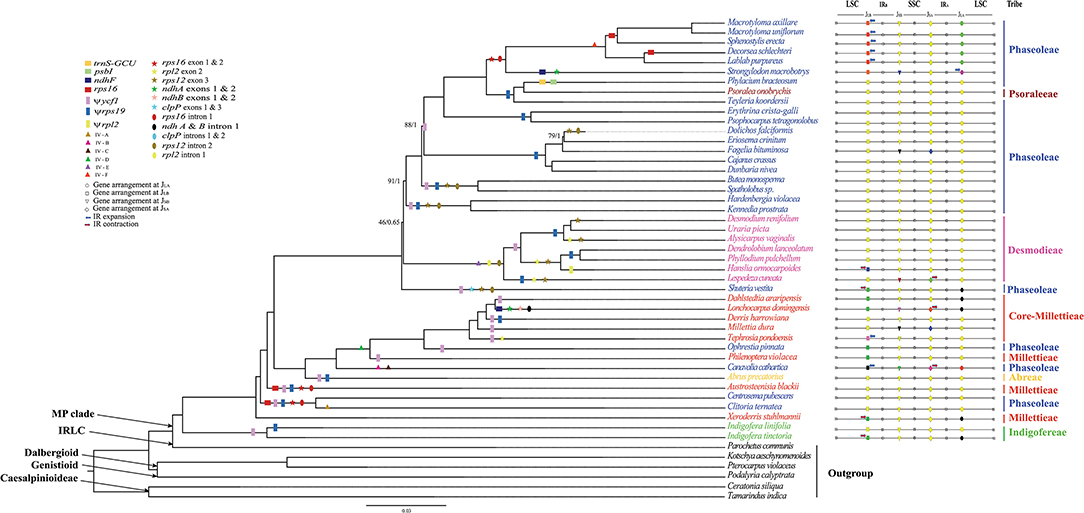
Figure 1. The ML tree of the MP clade reconstructed based on the CP and the variation of IR/SC junctions. Numbers at nodes correspond to ML bootstrap percentages (only values <100% are shown) and Bayesian inference (BI) posterior probabilities (only probabilities <1.0 are shown). Genes loss, pseudogenes, inversions (IV), exon and intron loss, in the plastome, are indicated on the branches using coloured squares, rectangles, triangles, stars and oval shapes, respectively. The IR expansion and contraction are shown by blue and red arrow, respectively.
Inserted CORRECTED names: Philenoptera violacea (Millettieae), Spatholobus sp. (Phaseoleae), and Shuteria vestita (Phaseoleae), respectively.
In the original article, there was a mistake in Supplementary Figure S1 as published. The mistakes were: Cochlianthus gracilis and Craspedolobium schochii.
Inserted CORRECTED names: Philenoptera violacea and Spatholobus sp. respectively.
In the original article, there was a mistake in Supplementary Figure S2 as published. The mistakes were: Cochlianthus gracilis and Craspedolobium schochii.
Inserted CORRECTED names: Philenoptera violacea and Spatholobus sp., respectively.
In the original article, there was a mistake in Supplementary Figure S3 as published. The mistakes were: Cochlianthus gracilis and Craspedolobium schochii.
Inserted CORRECTED names: Philenoptera violacea and Spatholobus sp., respectively.
In the original article, there was a mistake in Supplementary Table S1 as published. The mistakes were: Cochlianthus gracilis and Craspedolobium schochii.
Inserted CORRECTED names: Philenoptera violacea and Spatholobus sp. have been inserted to replace the initial names respectively.
In the original article, there was a mistake in Supplementary Table S2 as published. The mistakes were: Cochlianthus gracilis (Phaseoleae), Craspedolobium schochii (Millettieae), and Shuteria vestita (Desmodieae).
Inserted CORRECTED names: Philenoptera violacea (Millettieae), Spatholobus sp. (Phaseoleae), and Shuteria vestita (Phaseoleae), respectively.
In the original article, there was an error: The mean plastome coverage ranged between 162.0 × (Cochlianthus gracilis Benth., Phaseoleae) and 1,536.4 × [Cajanus crassus (Prain ex King) Maesen, Phaseoleae]. A correction has been made to Section: Results, Sub-section- Plastome Organization and Size.
Inserted CORRECTED paragraph: The mean plastome coverage ranged between 162.0 × (Philenoptera violacea (Klotzsch) Schrire, Millettieae) and 1,536.4 × [Cajanus crassus (Prain ex King) Maesen, Phaseoleae].
In the original article, there was an error: C. gracilis. A correction has been made to Section: Results, Sub-section Plastome Structural Variations in the MP Clade.
Inserted CORRECTED paragraph: P. violacea.
In the original article, there was an error: However, the lineage consisting of Butea monosperma (Lam.) Kuntze and Craspedolobium schochii Harms has different phylogenetic position in trees of CP and NCDs, and that of CDs, but both relationships were weakly supported. Also, the tribe Desmodieae was weakly supported to be monophyletic in CDs, but being weakly supported to be paraphyletic in CP and NCDs. The tribe Indigofereae was strongly supported as sister to the remainder of the MP clade (BS = 100%, and PP = 1.0). Based on the current sampling, it is not sure if the tribe Desmodieae is monophyletic, while the tribes Millettieae and Phaseoleae appear non-monophyletic. correction has been made to Section: Results, Sub-section Phylogenetic Relationships of the MP Clade.
Inserted CORRECTED paragraph: However, the lineage consisting of Butea monosperma (Lam.) Kuntze and Spatholobus Hassk sp. has different phylogenetic position in trees of CP and NCDs, and that of CDs, but both relationships were weakly supported. Also, the tribe Desmodieae was weakly supported to be monophyletic in CP and NCDs data matrices whereas strongly supported by CDs data. The tribe Indigofereae was strongly supported as sister to the remainder of the MP clade (BS = 100%, and PP = 1.0). Based on the current sampling, it is not sure if the tribe Desmodieae is monophyletic, while the tribes Millettieae and Phaseoleae appear non-monophyletic.
In the original article, there was an error: According to this study, with the exception of the loss of the clpP introns 1 and 2 in a single species of S. vestita (Desmodieae) and the loss of ndh A and ndh B intron 1 in a single species of L. domingensis (Millettieae), two other introns (rps16 and rps12) have experienced multiple independent loss during the plastome evolution of the species from the MP clade. This finding agrees with the previous studies on the independent loss of rpsl2, rps16, and clpP introns in the MP clade (Guo et al., 2007; Schwarz et al., 2015; Kaila et al., 2016). A correction has been made to Section: Discussion, Sub-section Evolutionary Pattern of PSV in the MP Clade.
Inserted CORRECTED paragraph: According to this study, with the exception of the loss of the clpP introns 1 and 2 in a single species of S. vestita (Phaseoleae) and the loss of ndh A and ndh B intron 1 in a single species of L. domingensis (Millettieae), two other introns (rps16 and rps12) have experienced multiple independent loss during the plastome evolution of the species from the MP clade. This finding agrees with the previous studies on the independent loss of rpsl2, rps16, and clpP introns in the MP clade (Guo et al.,2007; Schwarz et al., 2015; Kaila et al., 2016).
In the original article, there was an error: Desmodieae was supported as monophyletic group in previous studies (Bruneau et al., 1994; Doyle et al., 1997; Kajita et al., 2001; Stefanovic et al., 2009; Cardoso et al., 2013; de Queiroz et al., 2015; Egan et al., 2016), however this tribe was weakly supported as monophyletic by CDs but paraphyletic by CP and NCDs (Figure 4). A correction has been made to Section: Discussion, Sub-section Phylogenetic Relationships in the MP Clade.
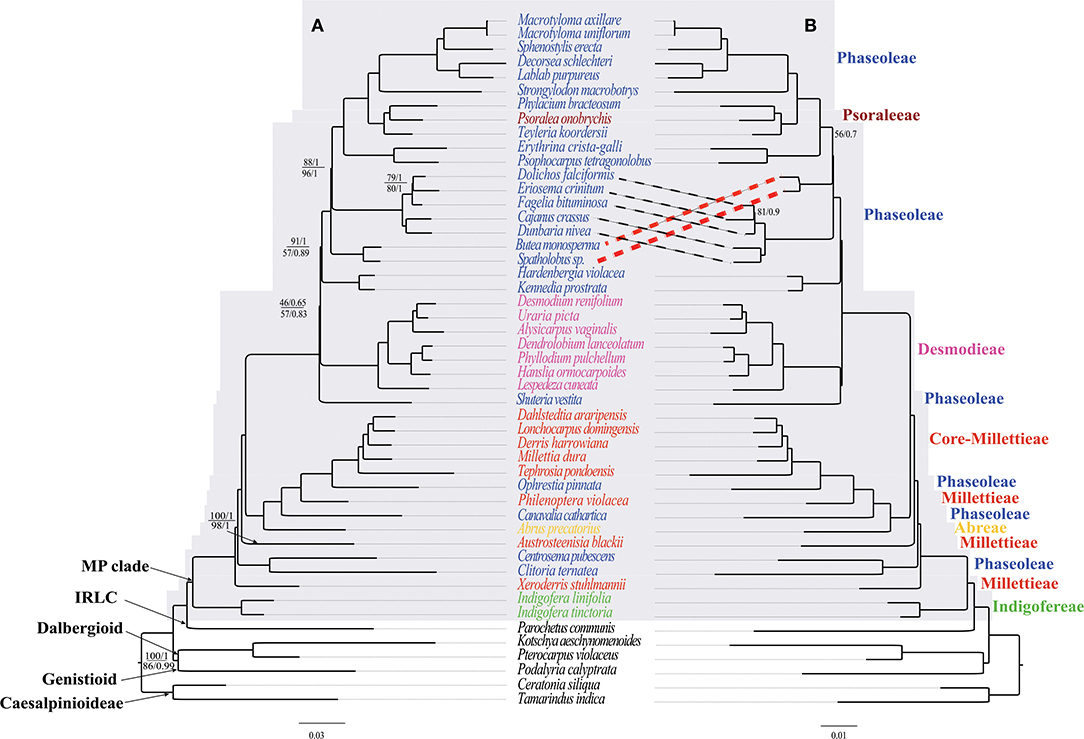
Figure 4. The ML and BI phylogenetic relationships reconstructed for the MP clade. (A) CP and NCDs, and (B) CDs. Numbers at nodes correspond to ML bootstrap percentages (only values <100% are shown) and Bayesian inference (BI) posterior probabilities (only probabilities <1.0 are shown). For (A), the values above and below the line represents support values for the CP and NCDs, respectively. The thick dotted lines indicate topology differences. The scale bar represents the mean nucleotide substitutions per site along the branch.
Inserted CORRECTED paragraph: Desmodieae was supported as monophyletic group in previous studies (Bruneau et al., 1994; Doyle et al., 1997; Kajita et al., 2001; Stefanovic et al., 2009; Cardoso et al., 2013; de Queiroz et al., 2015; Egan et al., 2016), however this tribe was strongly supported as monophyletic by CDs but weakly supported by CP and NCDs (Figure 4).
In the original article, there was an error: Notably, our multi-locus plastome data strongly supported (BS = 100%, PP = 1) the evolutionary position of S. vestita within the tribe Desmodieae, in contrast with previous placement in the tribe Phaseoleae (Lackey et al., 1981; de Queiroz et al., 2015). Formerly, the genus Shuteria was included in the tribe Phaseoleae based on flower structures shared with core Phaseoleae species (e.g., Amphicarpaea Elliott ex Nutt., Cologania Kunth, and Dumasia DC., Lackey et al., 1981). It is noteworthy that a similar phylogenetic placement in the MP clade has been shown from analysis based on the single plastid region matK (de Queiroz et al., 2015). Therefore, our phylogeny supports the placement of S. vestita within the tribe Desmodieae. A correction has been made to Section: Discussion, Sub-section Phylogenetic Relationships in the MP Clade.
CORRECTED paragraph: Notably, our multi-locus plastome data suggested (BS = 100%, PP = 1) the evolutionary position of S. vestita as sister to the tribe Desmodieae, in contrast with previous placement close to the subtribe Kennediinae of the tribe Phaseoleae (e.g., de Queiroz et al., 2015). Formerly, the genus Shuteria was included in the tribe Phaseoleae based on flower structures shared with core Phaseoleae species (e.g., Amphicarpaea Elliott ex Nutt., Cologania Kunth, and Dumasia DC., Lackey et al., 1981). It is noteworthy that a similar phylogenetic placement in the MP clade has been shown from analysis based on the single plastid region matK (de Queiroz et al., 2015). Therefore, our phylogeny supports the placement of S. vestita as sister to the tribe Desmodieae. Nevertheless, we expect that future phylogenetic studies would improve the understanding of the phylogenetic relationships of the genus Shuteria within the clade.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way.
The original article has been updated.
Bruneau, A., Doyle, J. J., and Doyle, J. A. (1994). “Phylogenetic relationships in Phaseoleae: evidence from chloroplast DNA restriction site characters,” in Advances in legume systematics, Part 7. Eds. Crisp, M., Doyle, J. J. (Richmond, Surrey, UK: Royal Botanic Gardens, Kew), 309–330.
Cardoso, D., Pennington, R. T., de Queiroz, L. P., Boatwright, J. S., Van Wyk, B. E., Wojciechowski, M. F., et al. (2013). Reconstructing the deep-branching relationships of the papilionoid legumes. S. Afr. J. Bot. 89, 58–75. doi: 10.1016/j.sajb.2013.05.001
de Queiroz, L. P., Pastore, J. F., Cardoso, D., Snak, C., de C Lima, A. L., Gagnon, E., et al. (2015). A multilocus phylogenetic analysis reveals the monophyly of a recircumscribed papilionoid legume tribe diocleae with well-supported generic relationships. Mol. Phylogenet. Evol. 90, 1–19. doi: 10.1016/j.ympev.2015.04.016
Doyle, J. J., Doyle, J. L., Ballenge, J. A., Dickson, E. E., Kajita, T., and Ohashi, H. (1997). A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am. J. Bot. 84, 541–554. doi: 10.2307/2446030
Egan, A. N., Vatanparast, M., and Cagle, W. (2016). Parsing polyphyletic Pueraria: delimiting distinct evolutionary lineages through phylogeny. Mol. Phylogenet. Evol. 104, 44–59. doi: 10.1016/j.ympev.2016.08.001
Guo, X., Castillo-Ramírez, S., González, V., Bustos, P., Fernández-Vázquez, J. L., Santamaría, R. I., et al. (2007). Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genom. 8, 228. doi: 10.1186/1471-2164-8-228
Kaila, T., Chaduvla, P. K., Saxena, S., Bahadur, K., Gahukar, S. J., Chaudhury, A., et al. (2016). Chloroplast Genome Sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome organization and comparison with other legumes. Front. Plant Sci. 7, 1847. doi: 10.3389/fpls.2016.01847
Kajita, T., Ohashi, H., Tateishi, Y., Bailey, C. D., and Doyle, J. J. (2001). rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst. Bot. 26, 15–536. doi: 10.1043/0363-6445-26.3.515
Lackey, J. A., Polhill, R. M., and Raven, P. H. (1981). “Phaseoleae,” in Advances in Legume Systematics, part 1 (UK: Royal Botanic Gardens, Kew), 301–327.
Schwarz, E. N., Ruhlman, T. A., Sabir, J. S. M., Hajrah, N. H., Alharbi, N. S., Al-Malki, A. L., et al. (2015). Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 53, 458–468. doi: 10.1111/jse.12179
Keywords: evolutionary relationships, inversion, IR expansion/contraction, Leguminosae, Plastome, the Millettioid/Phaseoloid clade
Citation: Oyebanji O, Zhang R, Chen S-Y and Yi T-S (2021) Corrigendum: New Insights Into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae). Front. Plant Sci. 12:652483. doi: 10.3389/fpls.2021.652483
Received: 12 January 2021; Accepted: 18 January 2021;
Published: 01 March 2021.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2021 Oyebanji, Zhang, Chen and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting-Shuang Yi, dGluZ3NodWFuZ3lpQG1haWwua2liLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.