- 1College of Life Science, Taizhou University, Taizhou, China
- 2National Key Laboratory of Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
Mediator complex is a multiprotein complex that regulates RNA polymerase II-mediated transcription. Moreover, it functions in several signaling pathways, including those involved in response to biotic and abiotic stresses. We used virus-induced gene silencing (VIGS) to study the functions of two genes, namely OsMED16 and OsMED25 in response to biotic and abiotic stresses in rice. Both genes were differentially induced by Magnaporthe grisea (M. grisea), the causative agent of blast disease, hormone treatment, and abiotic stress. We found that both BMV: OsMED16- and BMV: OsMED25-infiltrated seedlings reduced the resistance to M. grisea by regulating the accumulation of H2O2 and expression of defense-related genes. Furthermore, BMV: OsMED16-infiltrated seedlings decreased the tolerance to cold by increasing the malondialdehyde (MDA) content and reducing the expression of cold-responsive genes.
Introduction
Mediator complex, a multiprotein complex, was first discovered in yeast as a cofactor of RNA polymerase II that participates in RNA polymerase II-mediated transcription (Conaway and Conaway, 2011). Bioinformatics analysis and wet experiments revealed it to be a conserved multiprotein complex containing approximately 20 to 30 subunits. Although the number of these subunits vary in different organisms, they all form a four-module complex consisting of the head domain, middle domain, tail domain, and a cyclin-dependent kinase domain (Bourbon, 2008). Interestingly, the mediator complex is not stationary and uses isomerism to transfer numerous signals (Conaway and Conaway, 2011). Mediator complex was first purified from plants in 2007 (Backstrom et al., 2007). The sequences of its subunits in plants are considerably different from those in other eukaryotic organisms; however, their secondary structure is similar. For example, out of the 27 subunits present in the Arabidopsis mediator complex, 21 are conserved across eukaryotic organisms, whereas the remaining six are specific to plants (Backstrom et al., 2007; Kidd et al., 2011; Mathur et al., 2011). Recently, 28 conserved MEDs were identified in Arabidopsis including HAC1 and HAC5 (Guo J. et al., 2020). The mediator complex functions in several signaling pathways involved in development, response to biotic and abiotic stresses, and cellular movement (Samanta and Thakur, 2015; Malik et al., 2017; Chong et al., 2020a,b; Crawford et al., 2020).
MED8, MED12, MED13, MED14, MED15, MED16, MED17, MED18, MED19, MED20, MED21, MED25, CDK8, and MED37 have been implicated in the development (Autran et al., 2002; Lalanne et al., 2004; Wang and Chen, 2004; Gillmor et al., 2010; Ito et al., 2011; Xu and Li, 2011; Canet et al., 2012; Zheng et al., 2013; Lai et al., 2014; Hasan et al., 2020; Sun et al., 2020; Zhang and Guo, 2020; Agrawal et al., 2021). A study reported that a mutation in MED18 resulted in dwarfism, delayed formation of floral organs, late flowering, and reduced fertility (Kim et al., 2011). The underlying mechanisms of the function of MEDs in the development are being studied. MED16 regulates iron homeostasis by modulating iron uptake and gene expression and by associating with EIN3/EIL1 through the MED25 subunit (Yang et al., 2014; Zhang et al., 2014). MED8 and MED25 affect the size of plant organs by regulating cell proliferation (Lalanne et al., 2004; Xu and Li, 2011). MED18 affects flowering time and the formation of floral organs by regulating the expression of Flowering Locus C (FLC) and AGAMOUS (AG) (Zheng et al., 2013; Lai et al., 2014).
Mediator complex subunits CDK8, MED7, MED14, MED16, MED17, MED25, and MED36a are involved in response to abiotic stresses (Knight et al., 2009; Hemsley et al., 2014; Zhang et al., 2017; Ohama et al., 2021). CDK8 regulates abscisic acid (ABA) signaling and drought response in Arabidopsis by associating with RAP2.6, and SnRK2.6 (Zhu et al., 2020). MED16 regulates the response to cold stress, MED25 regulates the response to salt and drought stress (Boyce et al., 2003; Elfving et al., 2011). Furthermore, subunits MED8, MED14, MED15, MED16, MED18, MED19a, MED20, MED21, MED25, and MED36a are involved in response to biotic stress. The med8 mutants, med18 mutants, and MED21-silencing plants reduced the resistance to both Alternaria brassicicola (A. brassicicola) and Botrytis cinerea (B. cinerea) in Arabidopsis (Dhawan et al., 2009; Kidd et al., 2009; Lai et al., 2014). In addition, Arabidopsis med8 mutants increased the resistance to Fusarium oxysporum (F. oxysporum) and Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). Arabidopsis med18 mutants increased the resistance to RNA and DNA viral infections (Kidd et al., 2009; Hussein et al., 2020). The Arabidopsis mutant med25 decreased the resistance to A. brassicicola and B. cinerea, whereas it increased the resistance to F. oxysporum (Kidd et al., 2009). The mutants med14 and med15 decreased the resistance to Pst DC3000 in Arabidopsis (Canet et al., 2012; Zhang et al., 2013). In wheat, the MED15 mutant suppressed the resistance to stem rust (Hiebert et al., 2020). However, in Arabidopsis, med16 mutants displayed decreased resistance to Pst DC3000/avrRpt2, Pst DC3000, A. brassicicola, B. cinerea, and Sclerotinia sclerotiorum (S. sclerotiorum) (Wathugala et al., 2012; Zhang et al., 2012; Wang et al., 2015). MED19a positively regulate the resistance to Hyaloperonospora arabidopsidis (Caillaud et al., 2013).
Our team focus on the functions of MEDs in plants such as tomato and rice. MEDs were found to have functions in the resistance to Botrytis cinerea in tomato, including MED16 and MED25. The silencing of MED16 or MED25 strongly decreased the resistance to Botrytis cinerea in tomato. However, MED16 or MED25 do not have functions on the tolerance to cold and drought stresses in tomato. Whether MED16 or MED25 have functions in responses to biotic and abiotic stresses in rice were studied in this study. In this study, we studied the functions of two genes, OsMED16 (Os10 g35560) and OsMED25 (Os09 g13610) in rice. The expression of OsMED16 and OsMED25 was induced by pathogen inoculation, hormone treatment, and stresses. The higher susceptibility of BMV:OsMED16- and BMV:OsMED25-infiltrated plants to M. grisea, when compared with the control, could be attributed to regulation of H2O2 accumulation and expression of defense-related genes. BMV: OsMED16-infiltrated plants decreased the tolerance to cold stress by regulating the MDA content and the expression of cold-responsive genes.
Materials and Methods
Characterization of OsMED16 and OsMED25
Rice genome database was searched using BlastP program with Arabidopsis AtMED16 and AtMED25 as queries, and the predicted nucleotide and amino acid sequences for OsMED16 and OsMED25 were downloaded. Phylogenetic trees for tomato, Arabidopsis, rice, and other MEDs were constructed using the Neighbor-joining method by MEGA6 program with the p-distance, complete deletion, and 1000 bootstraps.
Plant Growth Conditions and Different Treatments
Yuanfengzao, a pair of isogenic lines (H8R and H8S), and IR64 were used for different analyses. For hormone treatment, 1.5 mM SA (salicylic acid, pH 6.5), 100 μM JA (jasmonic acid), 100 μM ACC (1-amino cyclopropane-1-carboxylic acid), or 100 μM ABA was sprayed on the leaves of 4-week-old Yuanfengzao seedlings. The same volume of water or 0.1% ethanol was used as control. Three-week-old plants were placed at 42°C for expression analysis of response to heat and at 4°C for cold. For drought treatment, the hydroponic 3-week-old plants were placed on the floor of the frame after the water was dried by filter paper. For NaCl stress, the hydroponic 3-week-old plants were treated with 200 mM NaCl solution. Samples were collected at specific time points for gene expression. Roots, flowers, stems, coats, leaves, sheaths, and ligules were collected for gene expression analysis in different tissues.
To infect the plants with M. grisea (strain 85–14B1, race ZB1), H8R and H8S lines were sprayed with spore solution as described earlier (Luo et al., 2005). IR64 was used for virus-induced gene silencing (VIGS) assays. A month later, silenced plants were used for different analyses. In every experiment, the samples were collected at indicated time points and stored at −80°C until use. All plants were kept in a room at 26°C, with a cycle of 14 h light (>3000 lux)/10 h dark, except those for VIGS assay, which were kept at 24°C, with a cycle of 14 h light (>3000 lux)/10 h dark.
Vector Construction and VIGS Infiltration
PCR fragments (200–400) (sequences were showed in Supplementary Material) were digested by AvrII and NcoI, then ligated to modified BMV vector. The recombinant plasmids confirmed by sequencing were electroporated into Agrobacterium tumefaciens strain C58C1; the agrobacteria were used for VIGS infiltration as described earlier (Sun et al., 2013). Briefly, agrobacteria carrying recombinant plasmids were cultured in liquid YEP medium with antibiotics at 28°C in a shaker incubator overnight. The cells were collected by centrifugation and resuspended in an induction buffer. Next, the solution was kept at room temperature for 5 h, centrifuged, resuspended in infiltration solution, and kept at room temperature. Infiltration was stopped until the OD value reached 2.0. It was subsequently mixed with the same volume of Agrobacterium harboring pC13/F1 + 2 before vacuum infiltration. The aerial parts of 10-day-old IR64 seedlings were mixed with Agrobacterium suspension for 7 min at 20 Kpa. The obtained plants were recorded as BMV:OsMED16-infiltrated plants and BMV:OsMED25-infiltrated plants. A group of rice seedlings were infiltrated with agrobacteria harboring a construct of BMV:OsPDS (Phytoene desaturase) and used as positive controls for silencing evaluation of the VIGS procedure (data showed in Supplementary Material). Plants transfected with empty vector were used as controls, which were recorded as BMV:00-infiltrated plants.
qRT-PCR Analysis of Gene Expression
RNA was extracted with TRIzol (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized by AMV reverse transcriptase (TaKaRa, Dalian, China). The qPCR was performed in a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States) with a reaction volume of 25 μL containing 12.5 μL 2 × SYBR Premix Ex TaqTM (TaKaRa, Dalian, China), 0.1 μg of cDNA, and 7.5 pmol of each gene-specific primer (see Table 1).
Fungal Culture and Disease Assay
Magnaporthe grisea was cultivated on oatmeal agar for 15 days until spores covered the whole surface. Next, the spores were washed to prepare a solution. For detached leaf analysis, the completely expanded leaves were obtained from 4-week-old plants and placed on a wet filter paper. Next, 5 μL of spore suspension was dropped on the surface of leaves. For whole-plant analysis, the plants were sprayed with the spore solution. Afterward, the tray was covered with a preservative film and kept in the room used for silencing plant growth. The images were acquired 7 days later, the lesion size was recorded, and the fungal growth was measured.
Cold Stress Assay and Physiological and Biochemical Measurements
For cold stress assay, 4-week-old BMV:OsMED16-infiltrated plants and BMV:00-infiltrated plants (control) were grown in the same pot at 4°C for 1 day, then transferred to normal conditions for recovery. Plants with more than 20% green leaves were considered to have survived, and the others were considered dead. The survival rate was calculated as the percentage of survival among total plants. The relative electrolyte leakage, chlorophyll content, malondialdehyde (MDA) content, H2O2 content, SOD activity, and CAT activity were measured as described previously (Lichtenthaler, 1987; Mittova et al., 2000; Alexieva et al., 2011; Song et al., 2011). In situ detection of H2O2 in leaf tissues was performed by DAB staining (Thordal-Christensen et al., 1997).
Results
Characterization of OsMED16 and OsMED25 in Rice
By BlastP searches against the rice genome database using the characterized Arabidopsis AtMED16 and AtMED25 as queries, OsMED16 and OsMED25 were obtained. Phylogenetic tree analysis revealed that OsMED16 and OsMED25 showed more than 40% of sequence identity to Arabidopsis AtMED16 and AtMED25 which were already reported to have functions in response to biotic and abiotic stresses (Figure 1).
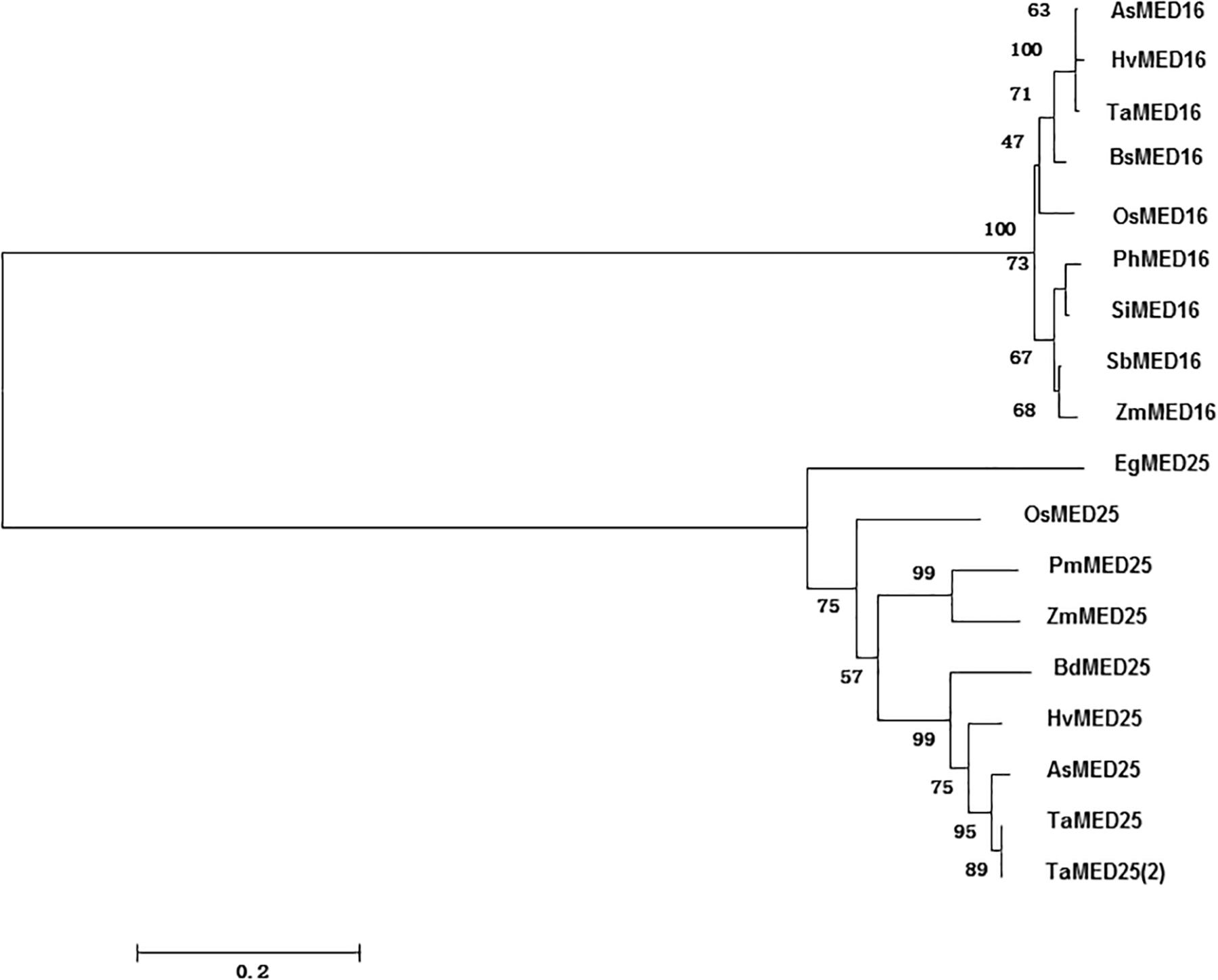
Figure 1. Phylogenetic tree of OsMED16 and OsMED25. Phylogenetic tree was constructed by neighbor-joining method using MEGA program version 6.0.
The Expression of OsMED16 and OsMED25 Was Induced by Different Stimuli
To study if OsMED16 or OsMED25 contributed to disease resistance, we first analyzed the responsiveness of these two genes in rice after pathogen infection and treatment with defense-related signal molecules. As shown in Figure 2A, the expression of OsMED16 and OsMED25 increased significantly in incompatible interaction but was different from each other. The expression of OsMED16 was the highest in the incompatible interaction after 24 h, whereas that of OsMED25 was highest in the incompatible interaction after 48 h. In the treatment of ACC, JA, and SA, OsMED16 and OsMED25 were induced significantly by all three signal molecules (Figure 2B).
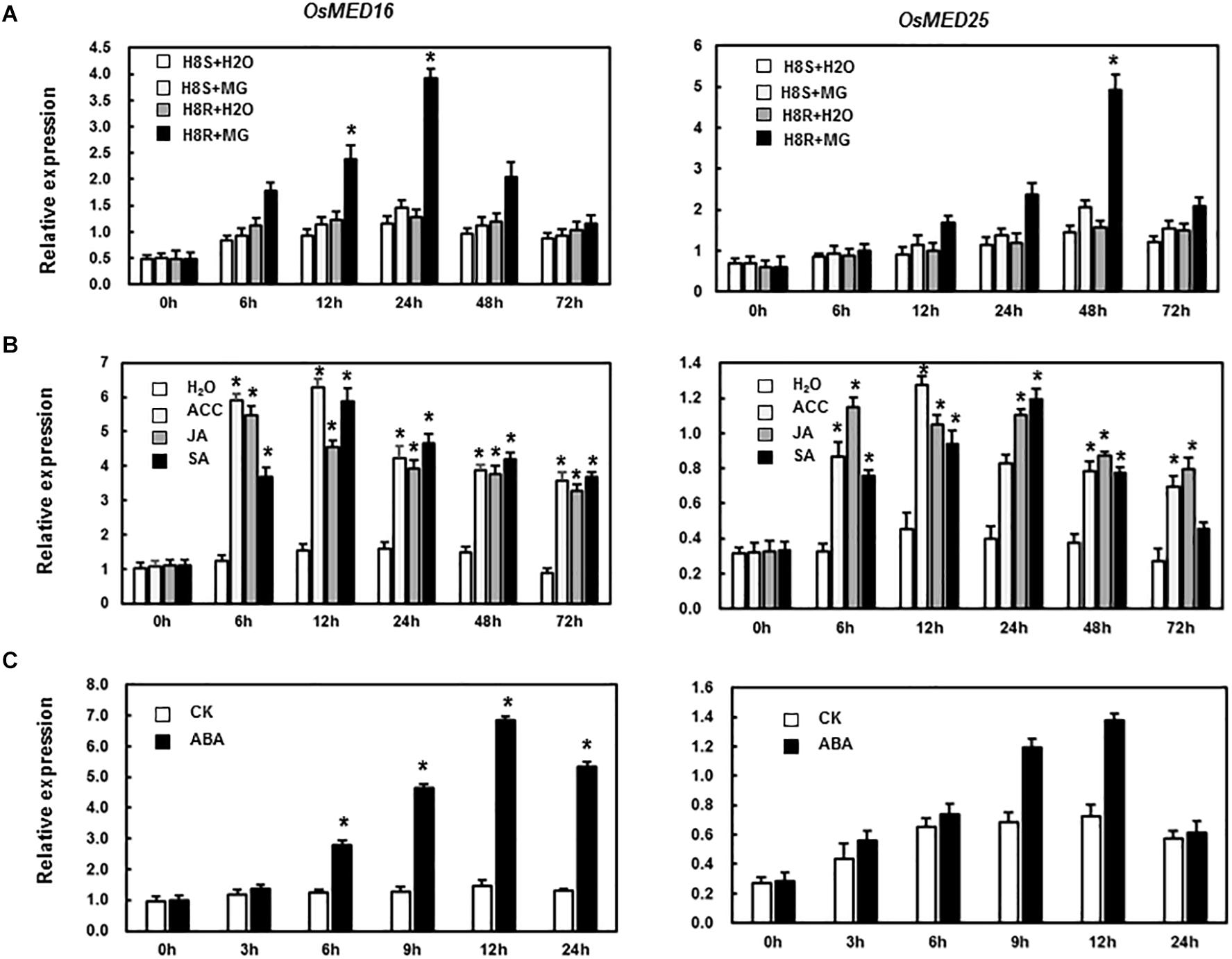
Figure 2. Expression patterns of OsMED16 and OsMED25 genes in response to Magnaporthe grisea infection and hormone treatment. (A) The expression patterns of OsMED16 and OsMED25 following inoculation with M. grisea. (B) The expression patterns of OsMED16 and OsMED25 following treatment with 1.5 mM SA, 100 μM JA, and 100 μM ACC. (C) The expression patterns of OsMED16 and OsMED25 following treatment with 100 μM ABA. The data were normalized against OsActin gene, and the relative expression was shown as fold expression of OsActin. Data are presented as means ± SD from three independent experiments and * above the columns indicate significant differences at p < 0.05 between the pathogen-inoculated or hormone-treated plants and the mock-inoculated/treated plants.
MED16 and MED25 were reported to differentially regulate ABA signaling in Arabidopsis (Guo P. C. et al., 2020). So we tested whether the expression levels of OsMED16 or OsMED25 were induced by ABA treatment in rice. As shown in Figure 2C, after treatment with ABA, the expression of OsMED16 increased dramatically, whereas that of OsMED25 showed no significant difference as compared with the control. To study whether OsMED16 or OsMED25 responded to abiotic stresses, we analyzed the responsiveness of these two genes in rice under abiotic stresses. In the drought and heat stresses, the expression of OsMED16 and OsMED25 showed no significant difference as compared with the control (Figures 3A,B). In cold stress, the expression of OsMED16 was induced dramatically, whereas that of OsMED25 showed no significant difference as compared with the control (Figure 3C). In salt stress, the expression of OsMED25 decreased slightly, whereas OsMED16 showed no significant difference when compared with the control (Figure 3D).
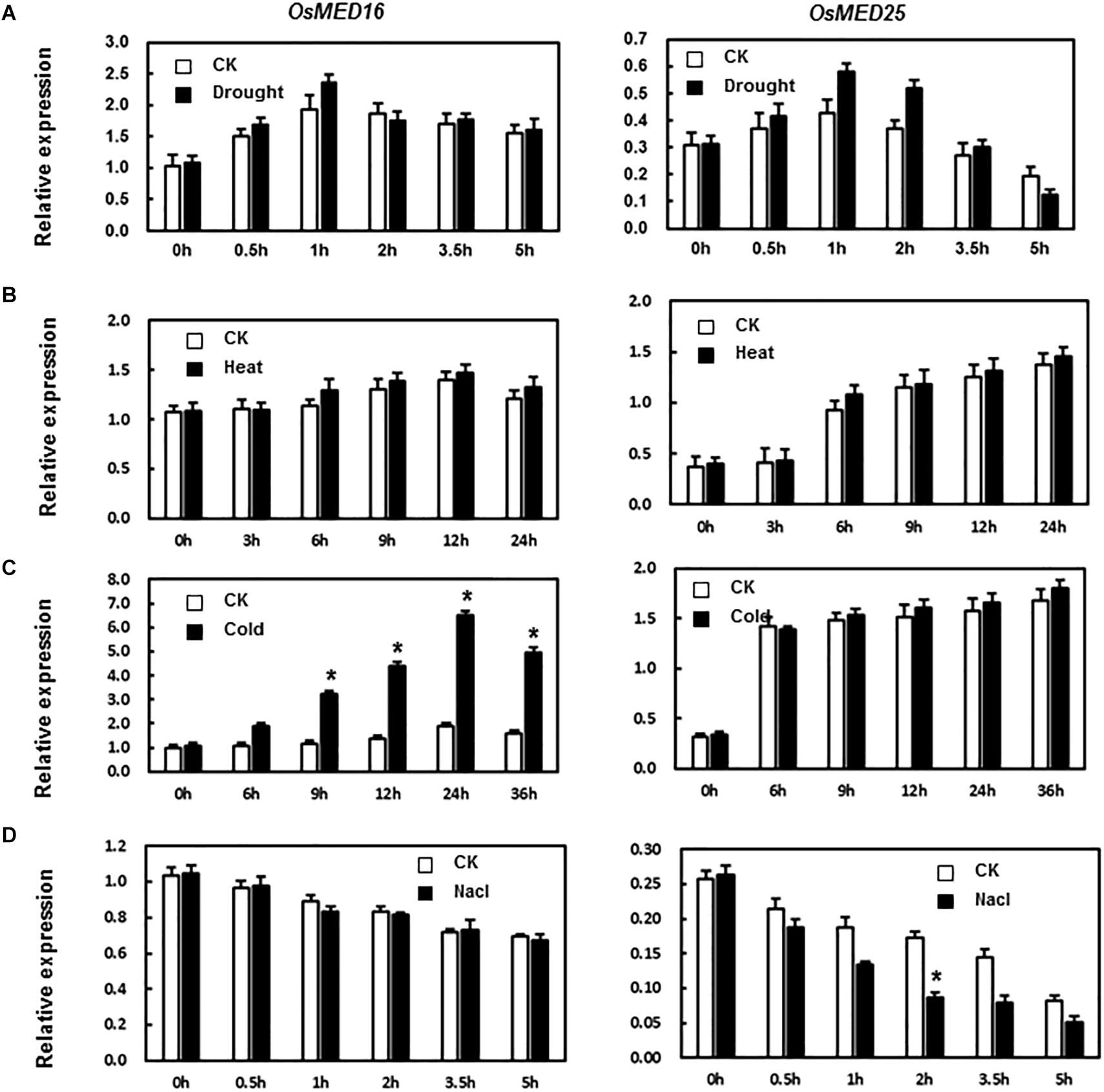
Figure 3. The expression patterns of OsMED16 and OsMED25 genes in response to drought, heat, cold, and NaCl stress. (A) The expression patterns of OsMED16 and OsMED25 under drought stress. (B) The expression patterns of OsMED16 and OsMED25 under heat stress. (C) The expression patterns of OsMED16 and OsMED25 under cold stress. (D) The expression patterns of OsMED16 and OsMED25 under NaCl stress. The samples were collected at different time points for analysis of gene expression by qRT-PCR. Expression data were normalized against OsActin gene as the reference, and the relative expression is shown as fold expression of OsActin. Data are presented as means ± SD from three independent experiments, and * above the columns indicate significant differences at p < 0.05 between the treated plants and the control.
In addition, we analyzed the expression patterns of these genes in various tissues of rice seedlings. Both genes were expressed in all tissues assessed to varying levels in different tissues. OsMED16 was highly expressed in roots, stems, leaves, and ligules; however, the expression was low in flowers, sheaths, and coats (Figure 4A). Moreover, OsMED25 was highly expressed in the stems, leaves, sheaths, and ligules; however, the expression was low in roots, flowers, and coats (Figure 4B).
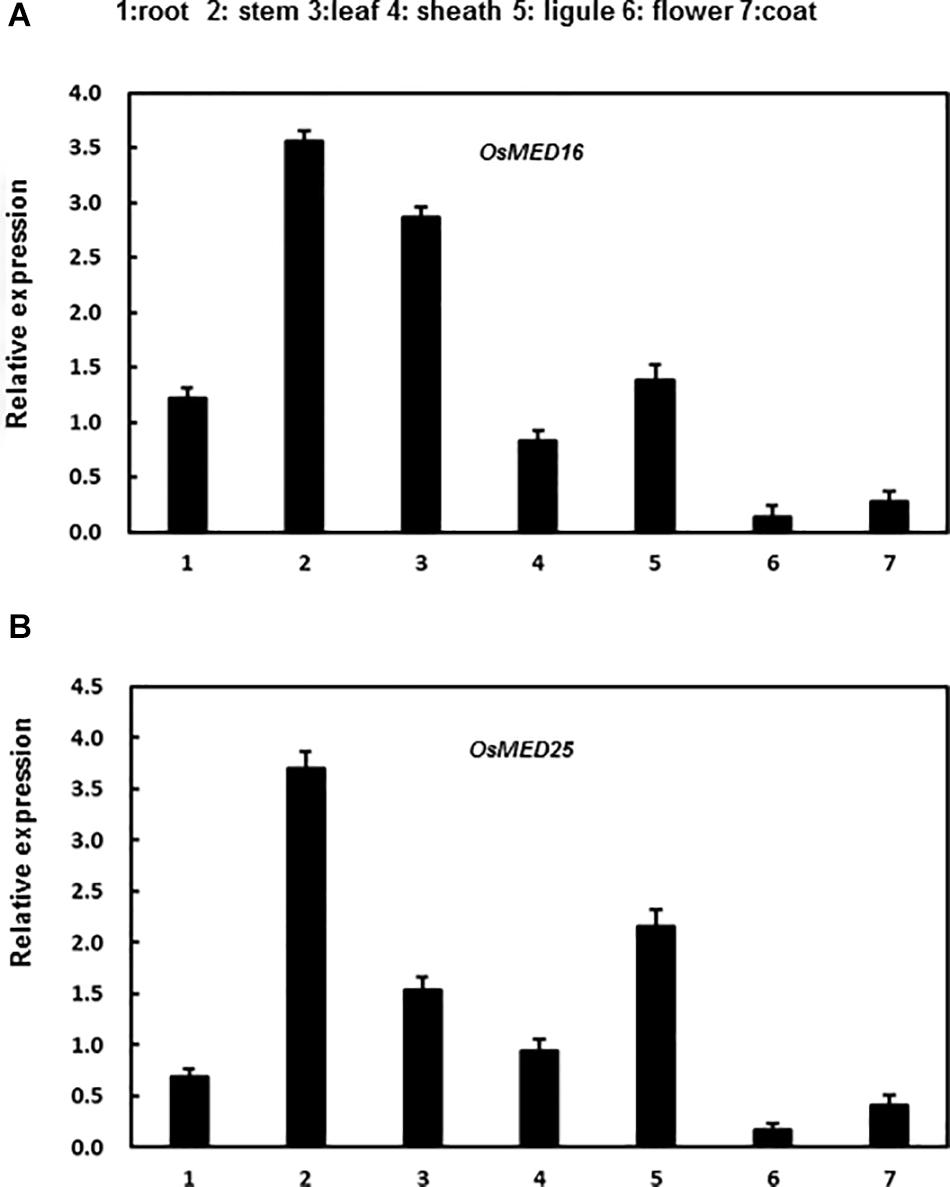
Figure 4. The expression patterns of OsMED16 and OsMED25 genes in different tissues. (A) The expression patter of OsMED16 in different tissues. (B) The expression patter of OsMED25 in different tissues. Roots, flowers, stems, coats, leaves, sheaths, and ligules were collected to analyze the gene expression by qRT-PCR. Expression data were normalized against OsActin gene, and the relative expression is shown as fold expression of OsActin. Data are presented as means ± SD from three independent experiments.
OsMED16 and OsMED25 Silencing Did Not Affect Growth and Development
To better understand the biological functions of OsMED16 and OsMED25, we generated BMV:OsMED16- and BMV:OsMED25-infiltrated seedlings by VIGS. The silencing efficiency of the target gene was assessed by qRT-PCR. As shown in Figure 5B, the silencing efficiency for these two genes was approximately 60%. The silenced seedlings were selected for further experiments. The BMV:OsMED16- and BMV:OsMED25-infiltrated seedlings grew and developed normally during our observation as compared with BMV:00-infiltrated plants (Figures 5A,C). We believe that OsMED16 or OsMED25 silencing did not affect the growth and development of rice seedlings.
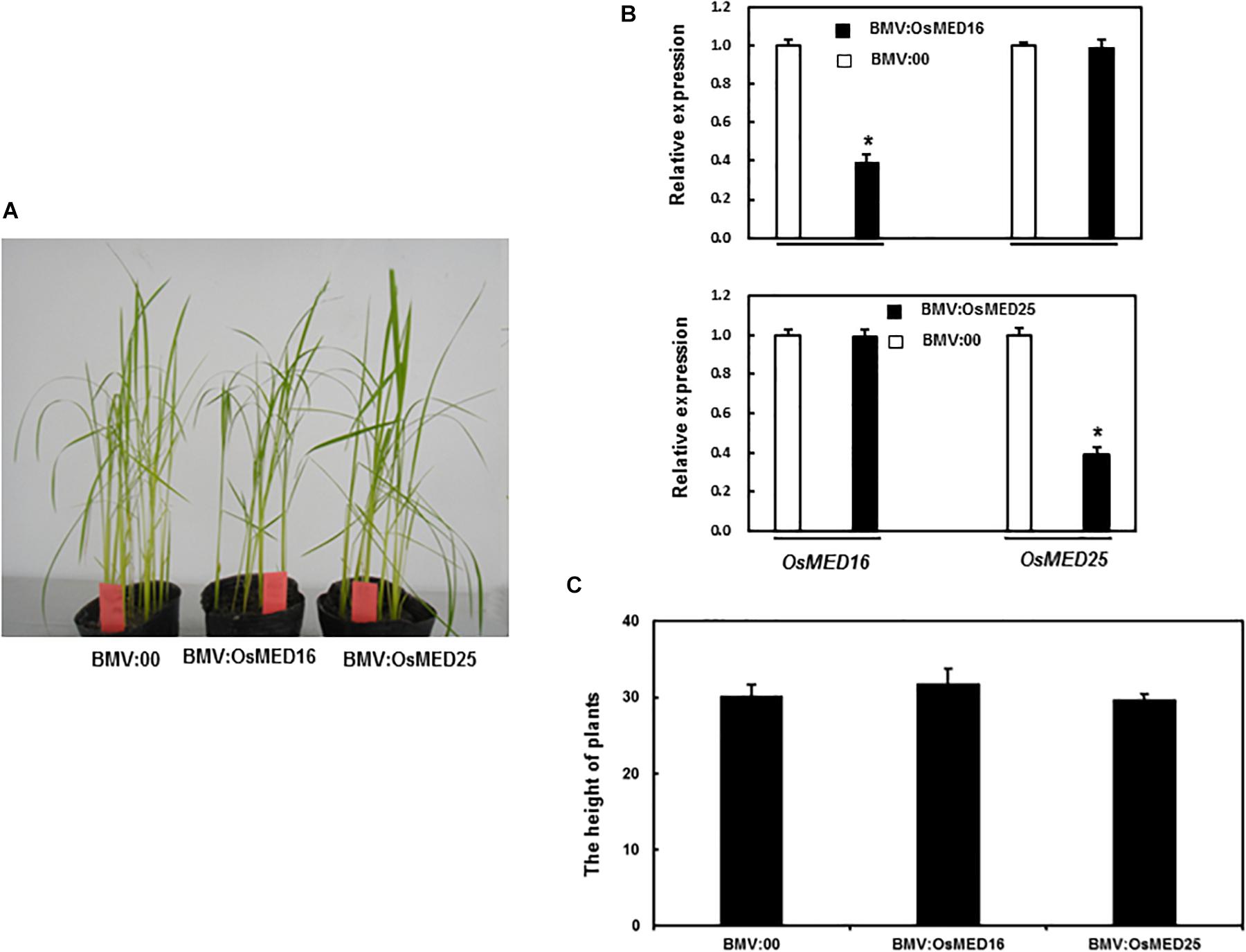
Figure 5. The growth condition of BMV:OsMED16- and BMV:OsMED25- infiltrated plants. (A) Growth conditions of BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants. (B) The silencing specificity of OsMED16 and OsMED25 in BMV:OsMED16- or BMV:OsMED25-infiltrated plants. 10-day-old IR64 plants were infiltrated with agrobacteria carrying BMV: OsMED16-, BMV:OsMED25 or BMV:00 constructs and leaf samples were collected at 4 weeks after agroinfiltration. Transcript levels for OsMED16 and OsMED25 were analyzed by qRT-PCR using a rice OsActin gene as an internal control. (C) Heights of BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants. *Above the columns indicate significant differences at p < 0.05 between the silencing plants and the control.
OsMED16 and OsMED25 Silencing Decreased the Resistance to M. grisea
Magnaporthe grisea was inoculated into the detached leaves and whole plants following the above-described method. The detached leaf assay revealed that the lesions on the leaves of BMV:OsMED16- and BMV:OsMED25-infiltrated plants were considerably larger than those on BMV:00-infiltrated plants (Figures 6A,B). The whole plant analysis showed similar results. The disease phenotype of BMV:OsMED16- or BMV:OsMED25-infiltrated plants was severe than that of BMV:00-infiltrated plants (Figure 6C) with more fungal growth (Figure 6D). This implied that compared with BMV:00-infiltrated plants, BMV:OsMED16- and BMV:OsMED25-infiltrated plants increased the susceptibility to M. grisea.
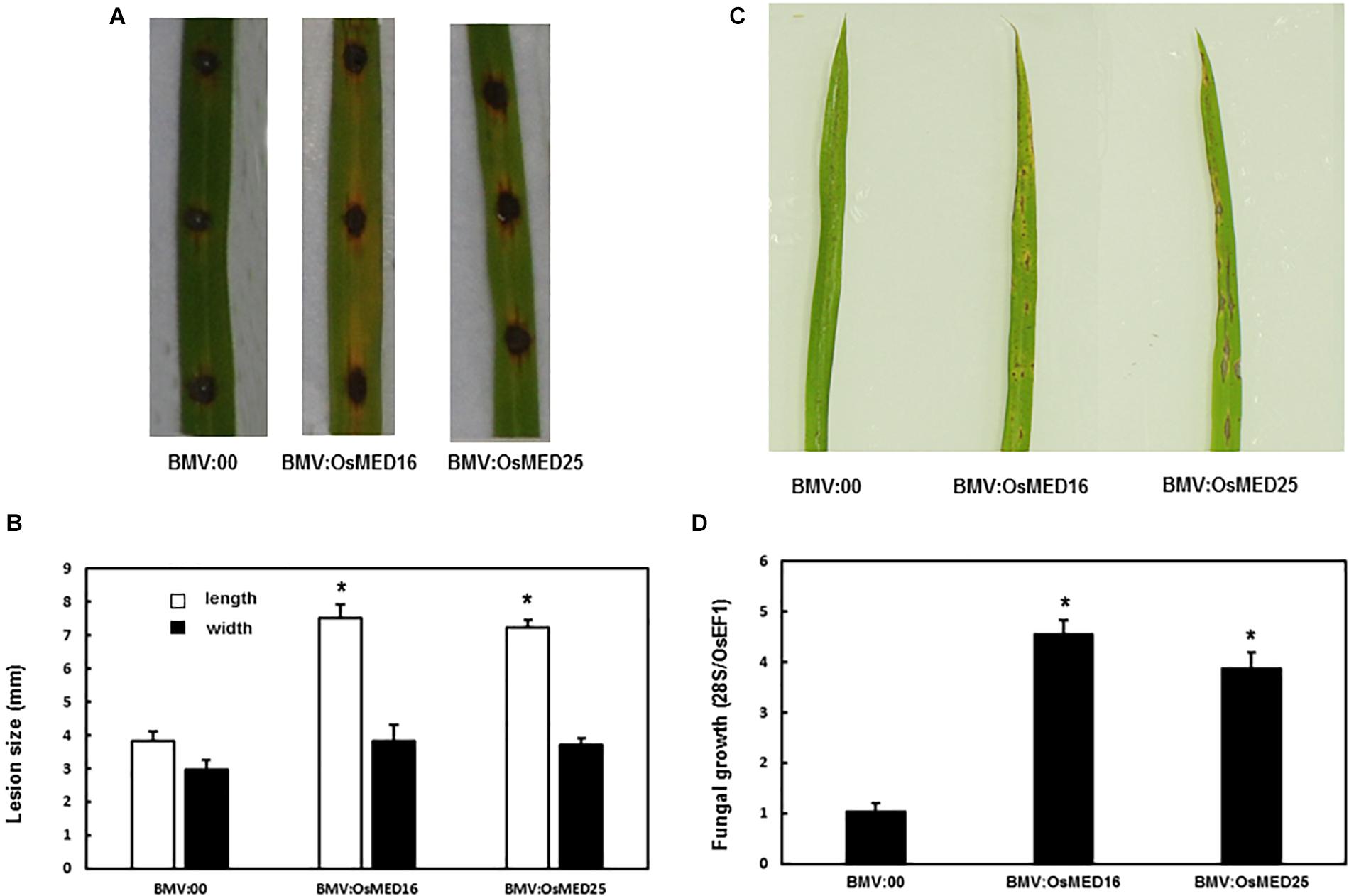
Figure 6. BMV:OsMED16- and BMV:OsMED25-infiltrated plants displayed reduced resistance to Magnaporthe grisea when compared with BMV:00-infiltrated plants. (A) The phenotype of BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants following M. grisea inoculation by detached leaf analysis. (B) The lesion size was measured using the detached leaf analysis. (C) The phenotype of BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants following M. grisea inoculation by whole plant analysis. (D) Fungal growth in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants following M. grisea inoculation. *Above the columns indicate significant differences at p < 0.05 between the silencing plants and the control.
To explore the mechanism for reduced resistance to M. grisea following OsMED16 and OsMED25 silencing, H2O2 content and the expression patterns of defense-related genes were analyzed. First, H2O2 condition was analyzed. DAB staining results showed that BMV:OsMED16- and BMV:OsMED25-infiltrated plants accumulated more H2O2 than BMV:00-infiltrated plants following the inoculation with M. grisea (Figure 7A). However, there were no significant differences in H2O2 accumulation in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants without the inoculation of M. grisea. This finding was further confirmed by quantification of H2O2 concentrations (Figure 7B). The SOD activity in BMV:OsMED16- and BMV:OsMED25-infiltrated plants was higher than that in BMV:00-infiltrated plants after the inoculation of M. grisea, whereas the CAT activity was lower (Figures 7C,D).
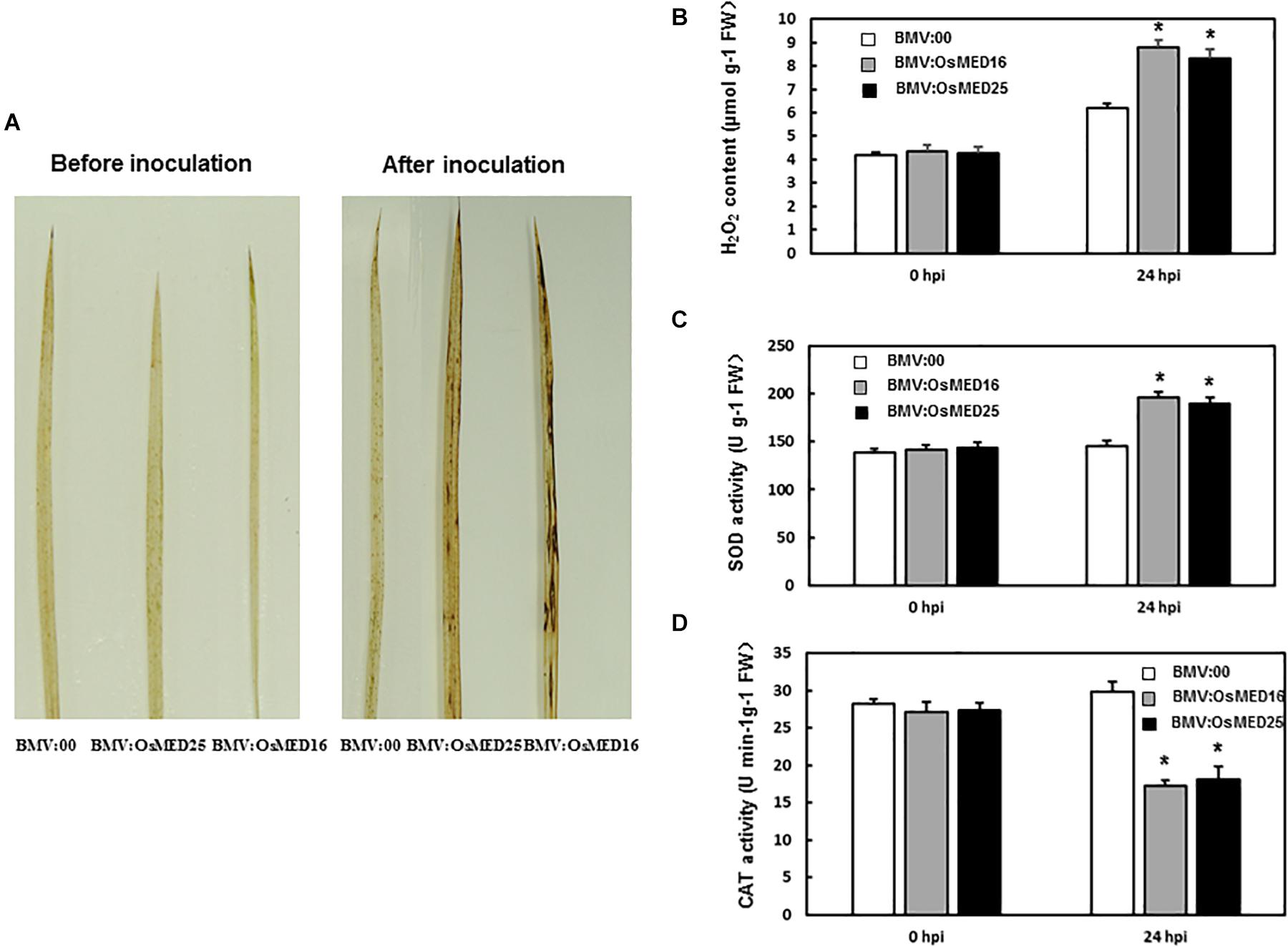
Figure 7. The H2O2 condition in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after Magnaporthe grisea inoculation. (A) The DAB staining of leaves from BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after M. grisea inoculation. (B) The H2O2 content in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after M. grisea inoculation. (C) The SOD activity in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after M. grisea inoculation. (D) The CAT activity in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after M. grisea inoculation. *Above the columns indicate significant differences at p < 0.05 between the silencing plants and the control.
Next, the expression of defense-related genes was analyzed. There existed no significant difference in the expression of OsLOX1, OsNH1, OsPR1a, OsPR3, and OsWRKY45 between BMV:target genes- and BMV:00-infiltrated plants before the inoculation of M. grisea. Three days after the inoculation of M. grisea, the expression levels of OsNH1 and OsPR1a in BMV:OsMED16-infiltrated plants increased significantly, whereas that of OsLOX1, OsPR3, and OsWRKY45 decreased when compared BMV:00-infiltrated plants (Figure 8A). After the inoculation of M. grisea, the expression of OsLOX1, OsPR3, and OsWRKY45 decreased in BMV:OsMED25-infiltrated plants, whereas that of OsNH1 and OsPR1a remained unchanged when compared with that in BMV:00-infiltrated plants (Figure 8B).
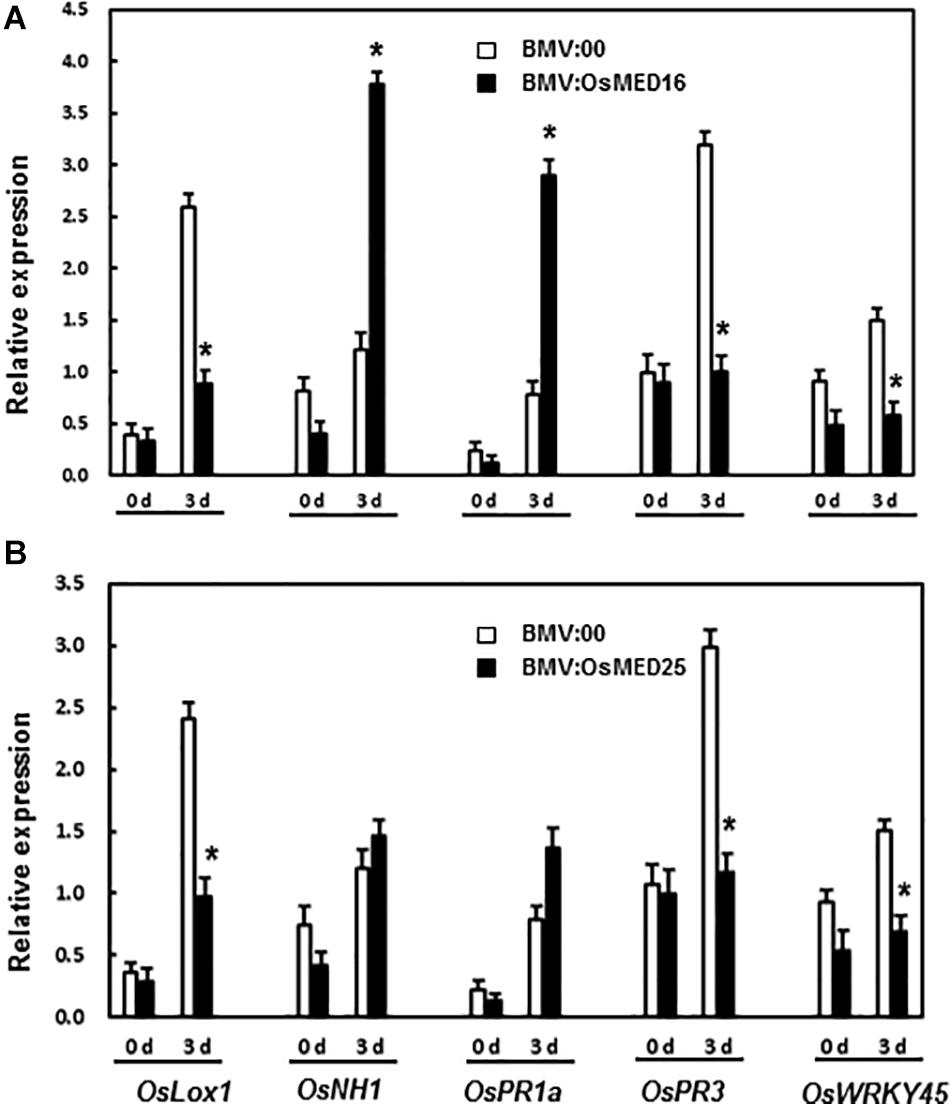
Figure 8. The expression pattern of defense-related genes in BMV:OsMED16- (A) and BMV:OsMED25-infiltrated plants (B) before and after Magnaporthe grisea inoculation. The expression of defense related genes was analyzed by qRT-PCR. Expression data were normalized against OsActin gene, and relative expression is shown as fold expression of OsActin. Data are presented as means ± SD from three independent experiments and * above the columns indicate significant differences at p < 0.05 between the silenced plants and the control.
BMV:OsMED16-Infiltrated Plants Showed Decreased Tolerance to Cold
To study whether OsMED16 or OsMED25 contributed to the response to cold stress, the tolerance of BMV:OsMED16- and BMV:OsMED25-infiltrated plants to cold was assessed. The 4-week-old BMV:target genes- and BMV:00-infiltrated plants in the same pot were placed in the incubator at 4°C with a 14 h/10 h light/dark cycle. After 24 h, the plants were transferred to normal growth conditions for recovery. As shown in Figure 9A, less number of BMV:OsMED16-infiltrated plants were recovered from cold stress when compared with BMV:00-infiltrated plants. The survival rate of BMV:OsMED16-infiltrated plants was approximately 25% of BMV:00-infiltrated plants (Figure 9B).
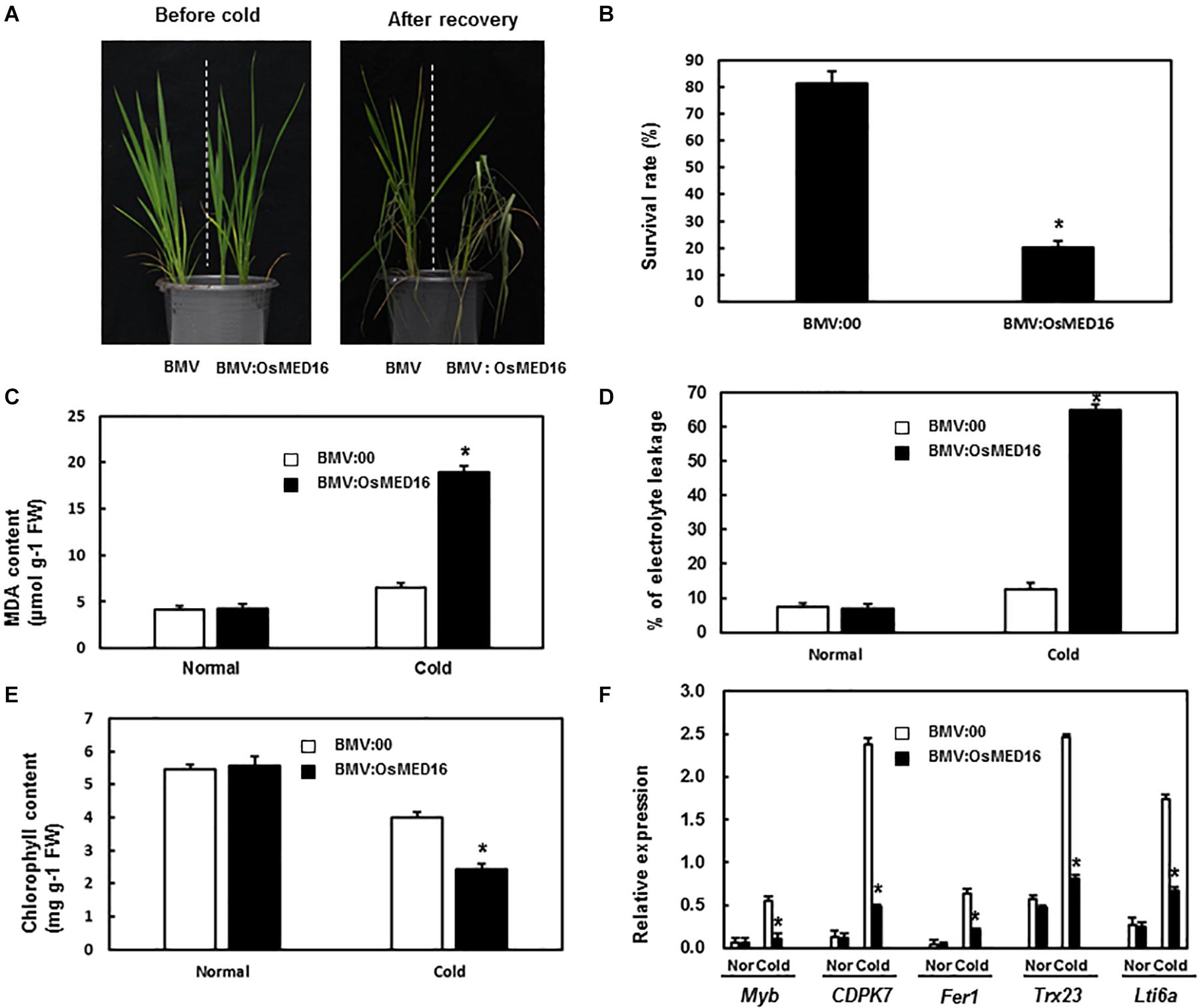
Figure 9. BMV:OsMED16-infiltrated plants showed decreased tolerance to cold stress when compared with BMV:00-infiltrated plants. (A) The phenotype of BMV:OsMED16- and BMV:00-infiltrated plants under cold stress. (B) The survival rate of BMV:OsMED16- and BMV:00-infiltrated plants after cold stress. (C) The MDA content in BMV:OsMED16- and BMV:00-infiltrated plants with and without cold stress. (D) The relative electrolyte leakage in BMV:OsMED16- and BMV:00-infiltrated plants with and without cold stress. (E) The chlorophyll content in BMV:OsMED16- and BMV:00-infiltrated plants with and without cold stress. (F) The expression of cold-responsive genes in BMV:OsMED16- and BMV:00-infiltrated plants before and after cold stress. *Above the columns indicate significant differences at p < 0.05 between the silencing plants and the control.
To find out the possible mechanism of OsMED16 silencing resulting in a decreased tolerance to cold, the MDA content, relative electrolyte leakage, chlorophyll content, and the expression of cold-responsive genes in BMV:OsMED16- and BMV:00-infiltrated plants under unstressed or cold stress were analyzed. The MDA content and relative electrolyte leakage in BMV:OsMED16-infiltrated plants increased dramatically after cold stress when compared with BMV:00-infiltrated plants, whereas there existed no significant difference under unstressed conditions (Figures 9C,D). The chlorophyll content in BMV:OsMED16-infiltrated plants decreased dramatically after cold stress when compared with BMV:00-infiltrated plants (Figure 9E). Finally, the expression of cold-responsive genes was measured in BMV:OsMED16- and BMV:00-infiltrated plants. As shown in Figure 9F, the expression of Myb, CDPK7, Fer1, Trx23, and Lti6a decreased dramatically in BMV:OsMED16-infiltrated plants when compared with BMV:00-infiltrated plants under cold stress.
Discussion
The plant mediator complex was first purified from Arabidopsis in 2007 (Backstrom et al., 2007). Since then, several studies have been conducted on its functions in plants. The complex has been implicated in several processes, such as development, abiotic and biotic stress responses, and many other cellular activities, including on-encoding RNA functions, regulation of DNA and protein stability, and secondary metabolism (Kidd et al., 2011; Kim and Chen, 2011; Kim et al., 2011; Bonawitz et al., 2014; Gillmor et al., 2014; Hemsley et al., 2014; Raya-González et al., 2014; Yang et al., 2014; Zhang et al., 2014; Li et al., 2015). The majority of the work has been performed in Arabidopsis, with only a few studies in rice. In our present study, the functions of OsMED16 and OsMED25 were studied in rice.
The expression of the mediator complex is induced by pathogen inoculation and exposure to stresses (Hussein et al., 2020; Nath et al., 2020). For instance, the transcription of ScMED7 is induced by heavy metals (CdCl2), low temperatures (4°C), and hormones (SA and MeJA), whereas it is inhibited by osmotic stresses by NaCl and PEG (Zhang et al., 2017). The expression of MED16 is induced by Pseudomonas syringae, ultraviolet-C (UV-C) irradiation, SA, and JA (Wathugala et al., 2012). In our study, the expression of OsMED16 and OsMED25 was induced by the inoculation of rice plants with M. grisea, hormone treatment, and abiotic stresses (Figures 2, 3). The results showed different expression patterns implying varying functions in response to biotic and abiotic stresses. The expression of OsMED14 was high in roots, leaves, anthers, and seeds at younger stages (Malik et al., 2020). Although ScMED7 is constitutively expressed, the expression is significantly higher in bud tissues. Furthermore, in our study, OsMED16 and OsMED25 were constitutively expressed in all tissues we studied, however, with different patterns (Figure 4).
The mediator complex was reported to be involved in development. MED25 exerts effects on organ size, root hair differentiation, and root system architecture (Xu and Li, 2011; Sundaravelpandian et al., 2013a,b; Raya-González et al., 2014). However, no significant difference was found between the root of BMV:OsMED25-infiltrated plants and those of BMV:00-infiltrated plants (data not shown). The mutation in MED17, MED18, or MED20 resulted in dwarfism, delayed flowering, and reduced fertility (Kim et al., 2011). However, we did not observe any significant difference in growth and development between BMV:target gene- and BMV:00-infiltrated plants (Figure 5). We speculated that OsMED16 and OsMED25 did not function in the vegetative stage. Whether OsMED16 and OsMED25 have a function in the reproductive stage needs to be further studied.
The mediator complex also functions in plant immunity (Canet et al., 2012; Cevik et al., 2012; Chen et al., 2012; Caillaud et al., 2013; Zhang et al., 2013). For example, MED16 mutations in Arabidopsis decreased the resistance to Pst DC3000/avrRpt2, Pst DC3000, A. brassicicola, and B. cinerea. MED16 could be a positive regulator of SAR, regulating the responsive capability of SA and the basal resistance (Wathugala et al., 2012; Zhang et al., 2012). The med25 mutants decreased the resistance to A. brassicicola and B. cinerea and increased the resistance to F. oxysporum (Kidd et al., 2009). OsMED16 and OsMED25 showed sequence similarity to AtMED16 and AtMED25, respectively (Figure 1). Therefore, we studied whether OsMED16 and OsMED25 functioned in plant immunity in rice and our results indicated that the silencing of OsMED16 and OsMED25 reduced the resistance to M. grisea, produced larger lesions, and increased fungal growth (Figure 6).
The relationship between plant immunity and ROS generation is well described. The ROS accumulation is beneficial for pathogen infection. The function of H2O2, a kind of ROS, in disease resistance, has gained extensive attention. H2O2 accumulation was found in several plant-pathogen systems. We first analyzed H2O2 conditions to explore the mechanism of reduced resistance caused by the silencing of OsMED16 and OsMED25. As shown in Figure 7A, BMV:OsMED16- and BMV:OsMED25-infiltrated plants accumulated more H2O2 than BMV:00-infiltrated plants following M. grisea inoculation, which was confirmed by H2O2 quantification (Figure 7B). The SOD and CAT activities were analyzed to explain the reason for elevated H2O2 levels. SOD catalyzes superoxide anions to H2O2 and O2, and CAT catalyzes H2O2 to H2O and O2. Higher SOD activity in BMV:OsMED16- and BMV:OsMED25-infiltrated plants implied more H2O2 production, whereas a lower CAT activity in BMV:OsMED16- and BMV:OsMED25-infiltrated plants meant less H2O2 consumption (Figures 7C,D). This could explain the high H2O2 content in BMV:OsMED16- and BMV:OsMED25-infiltrated plants.
Next, the expression of defense-related genes in BMV: OsMED16-, BMV: OsMED25-, and BMV:00-infiltrated plants before and after M. grisea inoculation was analyzed to explore the mechanism of decreased resistance caused by the silencing of OsMED16 and OsMED25. The expression of both OsNH1 and OsPR1a increased dramatically in BMV:OsMED16-infiltrated plants after the infection of M. grisea when compared with BMV:00-infiltrated plants, whereas the expression of OsLOX1, OsPR3, and OsWRKY45 decreased (Figure 8A). This result indicated that the expression of JA-responsive genes decreased, whereas SA-responsive genes increased in BMV:OsMED16-infiltrated plants after the infection of M. grisea. And OswWRKY45 is a positive regulator of resistance to fungal pathogens. As reported previously, MED25 is a positive regulator of the expression of JA-responsive genes. The expression of several defense genes, especially JA-responsive genes, was decreased dramatically in med25 mutants, whereas that of SA-responsive genes showed no dramatic difference. The mutants showed no difference in the resistance to PstDC3000 and SAR induced by organisms compared with the control (Kidd et al., 2009). In our study, the expression of JA-responsive genes (such as OsLOX1 and OsPR3) decreased, whereas that of SA-responsive genes (such as OsNH1 and OsPR1a) remained unchanged in BMV:OsMED25-infiltrated plants after the infection of M. grisea when compared with BMV:00-infiltrated plants (Figure 8B).
MED25 that interacts with DREB2A, ZFHD1, and Myb transcription factor regulates the response to abiotic stresses. Its mutants were more sensitive to salt stress with reduced resistance to drought (Elfving et al., 2011). However, BMV:OsMED25-infiltrated seedlings showed no difference from BMV:00-infiltrated seedlings in response to abiotic stresses. MED16, along with MED2 and MED14, regulates cold stress and its mutants decreased the resistance to cold and osmotic stress (Boyce et al., 2003; Knight et al., 2009; Hemsley et al., 2014). Moreover, we observed that the BMV:OsMED16-infiltrated plants displayed reduced resistance to cold when compared with the control (Figures 9A,B).
Under abiotic stresses, plants produce excessive radicals causing peroxidation injury, metabolic disorders, and antioxidant imbalance. Plants have evolved enzymatic and non-enzymatic systems to scavenge ROS and maintain the balance between ROS generation and ROS scavenging ability. Reduced activity of scavenging enzymes increases ROS species, including peroxides and MDA. MDA content is often considered as a physiological index for the degree of cell membrane damage and lipid peroxidation. High levels of MDA are toxic to plant cells (You and Chan, 2015). Furthermore, relative electrolyte leakage is an important index to judge the extent of damage after abiotic stresses. The extent of damage to the plants increases with the severity of plant stresses. Chlorophyll content partly implies the growth condition of plants. In contrast, the expression of cold-responsive genes was one of the contributing factors to cold tolerance. Therefore, the MDA content, relative electrolyte leakage, chlorophyll content, and the expression of cold-responsive genes were analyzed to explore the mechanism of decreased tolerance to cold stress caused by OsMED16 silencing. In our study, the MDA content and relative electrolyte leakage in BMV:OsMED16-infiltrated seedlings increased notably when compared with the BMV:00-infiltrated seedlings, whereas the chlorophyll content decreased (Figures 9C–E). The expression of cold-responsive genes downregulated in BMV:OsMED16-infiltrated seedlings when compared with BMV:00-infiltrated seedlings (Figure 9F). These results showed that BMV:OsMED16-infiltrated seedlings reduced the tolerance to cold stress through increased MDA content and decreased expression of cold-responsive genes.
Altogether, OsMED16 and OsMED25 contribute to the response to biotic stresses, probably by regulating the H2O2 content and the expression of defense-related genes. BMV:OsMED16-infiltrated plants displayed decreased tolerance to cold stress, probably by regulating MAD content and the expression of cold-responsive genes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
HZ, DZ, LY, MJ, and FS carried out most of the experiments. MJ and HZ designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Taizhou Municipal Science and Technology Project (20ny18), Science Foundation for Distinguished Young Scholars of Taizhou University (2019JQ001), and Zhejiang Provincial Natural Science Foundation of China (LY19C150004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.652453/full#supplementary-material
References
Agrawal, R., Jiri, F., and Thakur, J. K. (2021). The kinase module of the Mediator complex: an important signalling processor for the development and survival of plants. J. Exp. Bot. 72, 224–240. doi: 10.1093/jxb/eraa439
Alexieva, V., Sergiev, I., Mapelli, S., and Karanov, E. (2011). The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 24, 1337–1344. doi: 10.1046/j.1365-3040.2001.00778.x
Autran, D., Jonak, C., Belcram, K., Beemster, G. T., Kronenberger, J., Grandjean, O., et al. (2002). Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049.
Backstrom, S., Elfving, N., Nilsson, R., Wingsle, G., and Bjorklund, S. (2007). Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729. doi: 10.1016/j.molcel.2007.05.007
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Bourbon, H. M. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008. doi: 10.1093/nar/gkn349
Boyce, J. M., Knight, H., Deyholos, M., Openshaw, M. R., Galbraith, D. W., Warren, G., et al. (2003). The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34, 395–406.
Caillaud, M. C., Asai, S., Rallapalli, G., Piquerez, S., Fabro, G., and Jones, J. D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11:e1001732. doi: 10.1371/journal.pbio.1001732
Canet, J. V., Dobon, A., and Tornero, P. (2012). Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24, 4220–4235.
Cevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916.
Chong, L., Guo, P. C., and Zhu, Y. F. (2020a). Mediator complex: a pivotal regulator of ABA signaling pathway and abiotic stress response in plants. Int. J. Mol. Sci. 21:7755. doi: 10.3390/ijms21207755
Chong, L., Shi, X. N., and Zhu, Y. F. (2020b). Signal integration by cyclin-dependent kinase 8 (CDK8) module and other mediator subunits in biotic and abiotic stress responses. Int. J. Mol. Sci. 22:354. doi: 10.3390/ijms22010354
Conaway, R. C., and Conaway, J. W. (2011). Function and regulation of the mediator complex. Curr. Opin. Genet. Dev. 21, 225–230. doi: 10.1016/j.gde.2011.01.013
Crawford, T., Karamat, F., Lehotai, N., Rentoft, M., Blomberg, J., Strand, A., et al. (2020). Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 10:5073. doi: 10.1038/s41598-020-61758-w
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H. N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex andregulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.2307/40537446
Elfving, N., Davoine, C., Benlloch, R., Blomberg, J., Brannstrom, K., Muller, D., et al. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. U.S.A. 108, 8245–8250. doi: 10.1073/pnas.1002981108
Gillmor, C. S., Park, M. Y., Smith, M. R., Pepitone, R., Kerstetter, R. A., and Poethig, R. S. (2010). The MED12-MED13 module of mediator regulates the timing of embryo patterning in Arabidopsis. Development 137, 113–122. doi: 10.1242/dev.043174
Gillmor, C. S., Silva-Ortega, C. O., Willmann, M. R., Buendía-Monreal, M., and Poethig, R. S. (2014). The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development 141, 4580–4589. doi: 10.1242/dev.111229
Guo, J., Wei, L., Chen, S. S., Cai, X. W., Su, Y. N., Li, L., et al. (2020). The CBP/p300 histone acetyltransferases function as plant-specific MEDIATOR subunits in Arabidopsis. J. Integr. Plant Biol. doi: 10.1111/jipb.13052 Online ahead of print.
Guo, P. C., Chong, L., Wu, F. M., Hsu, C. C., Li, C. Y., Zhu, J. K., et al. (2020). Mediator tail module subunits MED16 and MED25 differentially regulate abscisic acid signaling in Arabidopsis. J. Integr. Plant Biol. doi: 10.1111/jipb.13062 Online ahead of print.
Hasan, A., Schoor, J., Hecht, V., and Weller, L. W. (2020). The CYCLIN-DEPENDENT KINASE module of the mediator complex promotes flowering and reproductive development in pea. Plant Physiol. 182, 1375–1386. doi: 10.1104/pp.19.01173
Hemsley, P. A., Hurst, C. H., Kaliyadasa, E., Lamb, R., Knight, M. R., De Cothi, E. A., et al. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26, 465–484. doi: 10.1105/tpc.113.117796
Hiebert, C. W., Moscou, M. J., Hewitt, T., Steuernagel, B., Hernández-Pinzón, I., Green, P., et al. (2020). Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat. Commun. 11:1123. doi: 10.1038/s41467-020-14937-2
Hussein, N. K., Sabr, L. J., Lobo, E., Booth, J., Ariens, E., Detchanamurthy, S., et al. (2020). Suppression of Arabidopsis Mediator subunit-encoding MED18 confers broad resistance against DNA and RNA viruses while MED25 is required for virus defense. Front. Plant Sci. 11:162.
Ito, J., Sono, T., Tasaka, M., and Furutani, M. (2011). MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesisin Arabidopsis. Plant Cell Physiol. 52, 539–552. doi: 10.1093/pcp/pcr013
Kidd, B. N., Cahill, D. M., Manners, J. M., Schenk, P. M., and Kazan, K. (2011). Diverse roles of the mediator complex in plants. Semin. Cell Dev. Biol. 22, 741–748. doi: 10.1016/j.semcdb.2011.07.012
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Kim, Y. J., and Chen, X. (2011). The plant Mediator and its role in noncoding RNA production. Front. Biol. 6:125. doi: 10.1007/s11515-011-1133-7
Kim, Y. J., Zheng, B., Yu, Y., Won, S. Y., Mo, B., and Chen, X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–822. doi: 10.1038/emboj.2011.3
Knight, H., Mugford, S. G., Ulker, B., Gao, D., Thorlby, G., and Knight, M. R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58, 97–108.
Lai, Z., Schluttenhofer, C. M., Bhide, K., Shreve, J., Thimmapuram, J., Lee, S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5:3064. doi: 10.1038/ncomms4064
Lalanne, E., Michaelidis, C., Moore, J. M., Gagliano, W., Johnson, A., Patel, R., et al. (2004). Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167, 1975–1986. doi: 10.1534/genetics.104.030270
Li, W., Yoshida, A., Takahashi, M., Maekawa, M., Kojima, M., Sakakibara, H., et al. (2015). SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 81, 282–291. doi: 10.1111/tpj.12725
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol. 18, 350–382. doi: 10.1016/0076-6879(87)48036-1
Luo, H. L., Song, F. M., and Zheng, Z. (2005). Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J. Exp. Bot. 56, 2673–2682. doi: 10.1093/jxb/eri260
Malik, N., Agarwal, P., and Tyagi, A. (2017). Emerging functions of multi-protein complex Mediator with special emphasis on plants. Crit. Rev. Biochem. Mol. Biol. 52, 475–502.
Malik, N., Ranjan, R., Parida, S. K., Agarwal, P., and Tyagi, A. K. (2020). Mediator subunit OsMED14-1 plays an important role in rice development. Plant J. 101, 1411–1429. doi: 10.1111/tpj.14605
Mathur, S., Vyas, S., Kapoor, S., and Tyagi, A. K. (2011). The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157, 1609–1627. doi: 10.1104/pp.111.188300
Mittova, V., Volokita, M., Guy, M., and Tal, M. (2000). Activities of SOD and the ascorbateglutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Plant Physiol. 110, 42–51.
Nath, V. S., Shrestha, A., Awasthi, P., Mishra, A. K., Kocabek, T., Matousek, J., et al. (2020). Mapping the gene expression spectrum of mediator subunits in response to viroid infection in plants. Int. J. Mol. Sci. 21:2498. doi: 10.3390/ijms21072498
Ohama, N., Moo, T. L., and Chua, N. H. (2021). Differential requirement of MED14/17 recruitment for activation of heat inducible genes. New Phytol. 229, 3360–3376. doi: 10.1111/nph.17119
Raya-González, J., Ortiz-Castro, R., Ruíz-Herrera, L. F., Kazan, K., and López-Bucio, J. (2014). PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 165, 880–894. doi: 10.1104/pp.114.239806
Samanta, S., and Thakur, J. K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6:757. doi: 10.3389/fpls.2015.00757
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345. doi: 10.1007/s00425-011-1403-2
Sun, L. J., Zhang, H. J., Li, D. Y., Huang, H., Hong, H. Y., Ding, X. S., et al. (2013). Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 81, 41–56. doi: 10.1186/s12870-016-0897-y
Sun, W. J., Han, H. Y., Deng, L., Sun, C. L., Xu, Y. R., Lin, L. H., et al. (2020). Mediator subunit MED25 physically interacts with PHYTOCHROME INTERACTING FACTOR4 to regulate shade-induced hypocotyl elongation in tomato. Plant Physiol. 184, 1549–1562. doi: 10.1104/pp.20.00587
Sundaravelpandian, K., Chandrika, N. N., and Schmidt, W. (2013a). PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol. 197, 151–161. doi: 10.1111/nph.12000
Sundaravelpandian, K., Chandrika, N., Tsai, Y. H., and Schmidt, W. (2013b). PFT1-controlled ROS balance is critical for multiple stages of root hair development in Arabidopsis. Plant Signal. Behav. 8:e24066.
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Wang, C. G., Yao, J., Du, X. Z., Zhang, Y. P., Sun, Y. J., Rollins, J. A., et al. (2015). The Arabidopsis Mediator complex Subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol. 169, 856–872.
Wang, W., and Chen, X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131, 3147–3156. doi: 10.1104/pp.15.00351
Wathugala, D. L., Hemsley, P. A., Moffat, C. S., Cremelie, P., Knight, M. R., and Knight, H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195, 217–230. doi: 10.1111/j.1469-8137.2012.04138.x
Xu, R., and Li, Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138, 4545–4554. doi: 10.1242/dev.071423
Yang, Y., Ou, B., Zhang, J., Si, W., Gu, H., Qin, G., et al. (2014). Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77, 838–851. doi: 10.1111/tpj.12440
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Zhang, L. J., and Guo, C. K. (2020). The important function of mediator complex in controlling the developmental transitions in plants. Int. J. Mol. Sci. 21:2733. doi: 10.3390/ijms21082733
Zhang, X., Wang, C., Zhang, Y., Sun, Y., and Mou, Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24, 4294–4309. doi: 10.1105/tpc.112.103317
Zhang, X., Yang, Y. T., Zou, J. K., Chen, Y., Wu, Q. B., Guo, J. L., et al. (2017). ScMED7, a sugarcane mediator subunit gene, acts as a regulator of plant immunity and is responsive to diverse stress and hormone treatments. Mol. Genet. Genomics 292, 1363–1375. doi: 10.1007/s00438-017-1352-y
Zhang, X., Yao, J., Zhang, Y., Sun, Y., and Mou, Z. (2013). The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75, 484–497. doi: 10.1111/tpj.12216
Zhang, Y., Wu, H., Wang, N., Fan, H., Chen, C., Cui, Y., et al. (2014). Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 203, 770–783. doi: 10.1111/nph.12860
Zheng, Z., Guan, H., Leal, F., Grey, P. H., and Oppenheimer, D. G. (2013). Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS One 8:e53924. doi: 10.1371/journal.pone.0053924
Keywords: MED, resistance, M. grisea, cold, VIGS
Citation: Zhang H, Zheng D, Yin L, Song F and Jiang M (2021) Functional Analysis of OsMED16 and OsMED25 in Response to Biotic and Abiotic Stresses in Rice. Front. Plant Sci. 12:652453. doi: 10.3389/fpls.2021.652453
Received: 12 January 2021; Accepted: 08 March 2021;
Published: 31 March 2021.
Edited by:
Aardra Kachroo, University of Kentucky, United StatesReviewed by:
Yingfang Zhu, Henan University, ChinaAjay Singh, Indian Council of Agricultural Research (ICAR), India
Copyright © 2021 Zhang, Zheng, Yin, Song and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Jiang, amlhbmdtaW5nMTk3M0AxMzkuY29t
 Huijuan Zhang
Huijuan Zhang Dewei Zheng1
Dewei Zheng1 Fengming Song
Fengming Song Ming Jiang
Ming Jiang