
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 07 June 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.650926
This article is part of the Research TopiclncRNA in Plants: Function, Mechanisms and ApplicationsView all 5 articles
Long non-coding RNAs (lncRNAs) play a vital role in a variety of biological functions in plant growth and development. In this study, we provided an overview of the molecular mechanisms of lncRNAs in interacting with other biomolecules with an emphasis on those lncRNAs validated only by low-throughput experiments. LncRNAs function through playing multiple roles, including sponger for sequestering RNA or DNA, guider or decoy for recruiting or hijacking transcription factors or peptides, and scaffold for binding with chromatin modification complexes, as well as precursor of microRNAs or small interfering RNAs. These regulatory roles have been validated in several plant species with a comprehensive list of 73 lncRNA–molecule interaction pairs in 16 plant species found so far, suggesting their commonality in the plant kingdom. Such initial findings of a small number of functional plant lncRNAs represent the beginning of what is to come as lncRNAs with unknown functions were found in orders of magnitude more than proteins.
Non-coding RNAs (ncRNAs), transcribed in over 90% of eukaryotic genomes of fungi, plants, and animals (Chekanova et al., 2007), were initially thought as “dark matter” in transcripts because their expressions are low, the sequences are poorly conserved, and the protein-coding potentials are absent (Palazzo and Koonin, 2020). With the advances in high-throughput sequencing and other experimental techniques, more ncRNAs begin to reveal their functional roles. In particular, they had been shown to play a key role in regulating the gene expressions at epigenetic, transcriptional, posttranscriptional, and posttranslational levels.
Long non-coding RNAs (lncRNAs) refer to those ncRNAs longer than 200 nucleotides (nt) in length (Rai et al., 2019; Palazzo and Koonin, 2020), as poorly conservative in sequence as other shorter ncRNAs (Chekanova, 2015). LncRNAs have been classified into three types according to their genomic locations: long intergenic ncRNAs (lincRNAs) in the intergenic regions, intronic ncRNAs (incRNAs) in the intronic regions, and natural antisense transcripts (NATs) from the antisense coding regions (Mattick and Rinn, 2015; Rai et al., 2019). Most lncRNAs are transcribed by RNA polymerase II (Pol II) or III (Pol III), and some could be produced by plant-specific RNA Pol IV and V (Wierzbicki et al., 2008). In this article, we summarized the molecular functions of plant lncRNAs discovered and validated by the low-throughput experiments with a specific focus on the relationship between lncRNAs and other biological macromolecules in regulating RNA activity, protein modification, and chromatin remodeling.
A large number of lncRNAs in plants were found by using transcriptome sequencing and bioinformatics analysis (Muers, 2011; Li et al., 2014, 2019; Hou et al., 2017; Yu T. et al., 2019). Several studies have summarized the bioinformatics tools and database resources for plant lncRNA identification and prediction (Jha et al., 2020; Waseem et al., 2020). However, until present, only a small number of these lncRNAs have been validated by the low-throughput experimental analysis. For example, Plant Long Non-Coding RNA Database version 2.0 (Jin et al., 2020) has 1,246,372 lncRNAs and predicted lncRNA targets in 80 species, and NONCODEV6 contains 94,697 lncRNAs in 23 plant species (Zhao et al., 2020). According to the experimentally validated functional lncRNA (EVLncRNA) predictions, nearly 30% of lncRNAs obtained by high-throughput sequencing have biological functions (Zhou et al., 2019). If lncRNAs are calculated on the order of millions, there are more than 200,000 predicted functional lncRNAs based on predictions (Zhou et al., 2019). By comparison, only 506 functional lncRNAs in 56 plant species are curated in the database of EVLncRNAs2.0 (Zhou et al., 2020). The majority of these EVLncRNAs belong to the model plant Arabidopsis thaliana (Figure 1). As shown in Figure 2, lncRNAs were found to regulate the downstream gene expression in cis or trans and play crucial roles in bud dormancy (Li et al., 2020), flowering time (Heo and Sung, 2011; Wang Z. W. et al., 2014; Kim et al., 2017), seedling photomorphogenesis (Wang Y. et al., 2014), root organogenesis (Ariel et al., 2014), sexual reproduction (Fan et al., 2016), gene silencing (Huang et al., 2011), and response to biotic and abiotic stress (Wunderlich et al., 2014; Kindgren et al., 2018; Zhao et al., 2018). In other words, the regulatory role of lncRNAs is extensive in eukaryotes (Long et al., 2017). The low-throughput experimental techniques include RNA fluorescence in situ hybridization (FISH), Northern blot, real-time quantitative polymerase chain reaction (RT-qPCR), overexpression, RNA interference (RNAi), clustered regularly interspaced short palindromic repeat-associated protein 9 (CRISPR/Cas9), RNA pull-down, RNA immunoprecipitation (RIP), and chromatin immunoprecipitation (ChIP) (Wu et al., 2020). Table 1 provides the most up-to-date (November 30, 2020) list of all validated 73 lncRNA–molecule interaction pairs in 16 plant species that offer a glimpse of the molecular interaction network of functional lncRNAs in plant development and stress response. These interactions according to the interaction partners of lncRNAs are presented in the following sections.
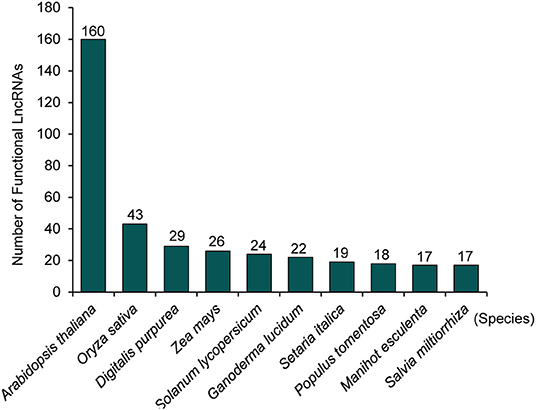
Figure 1. Statistics of the number of functional lncRNAs in top 10 plant species based on the experimentally validated functional lncRNAs in plants, according to EVLncRNA2.0 (Zhou et al., 2020).
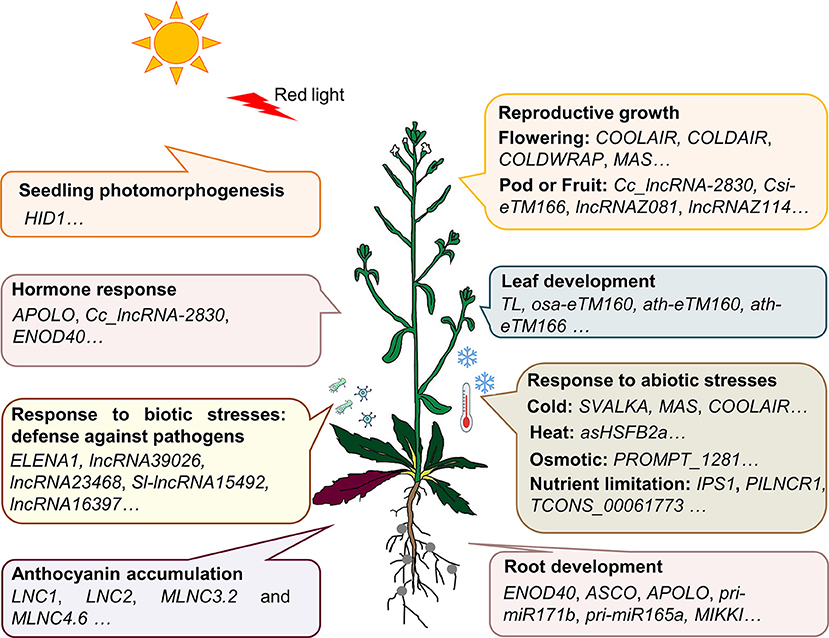
Figure 2. LncRNAs play extensive roles in most aspects of plant growth and development. During plant growth and development, multiple lncRNAs are validated to play crucial roles. HID1 plays an active role in seedling photomorphogenesis under continuous red light; APOLO, Cc_lncRNA-2830, and ENOD40 were demonstrated to respond to the auxin treatment; ELENA1, lncRNA39026, and lncRNA23468 enhance resistance to pathogen attacks; COOLAIR, COLDAIR, COLDWRAP, and MAS respond to cold, and involve in regulating the flowering time in vernalization; TL, osa-eTM160, ath-eTM160, and ath-eTM166 affect the leaf shape; SVALKA regulates cold signal transduction; asHSFB2a is induced by heat stress and affects grain yield; IPS1 and PILNCR1 are induced by phosphate deficiency; TCONS_00061773 respond to nitrate signal; LNC1, LNC2, MLNC3.2, and MLNC4.6 are involved in anthocyanin accumulation; ENOD40, APOLO, pri-miR171b, pri-miR165a, and MIKKI are involved in root nodule organogenesis or lateral root development.
The association between lncRNAs and small RNAs is perhaps the most reported relationship in plants. MicroRNAs (miRNAs) that are small ncRNAs (21–23 nt in length) play vital roles in diverse biological processes such as root development (Bazin et al., 2012), vegetative-to-reproductive transition (Yang et al., 2013), formation of phytohormones, and biotic/abiotic stress response (Yamamuro et al., 2016; Song X. et al., 2019). In principle, lncRNA could work as a “sponger” to adsorb a complementary miRNA to indirectly regulate the target genes of the miRNA. These lncRNAs are known as competitive endogenous RNAs (ceRNAs) or “endogenous target mimic (eTM).” In Arabidopsis, the lncRNA IPS1 (INDUCED BY PHOSPHATE STARVATION1) is complementary to miR399, but it contains a mismatched loop to interrupt the pairing at the miRNA cleavage site (Franco-Zorrilla et al., 2007). As illustrated in Figure 3A, under the phosphate (Pi) starvation condition, IPS1 is induced to sequester miR399. Sequestration of miR399 increases the transcriptional level of miR399 target PHO2 (encoding an E2 ubiquitin conjugase-related enzyme), which subsequently reduces the Pi content of the shoot (Franco-Zorrilla et al., 2007). The family members of IPS1 have been identified in several species with the same functional mechanism as the eTM of miR399s, including At4, At4-1, AT4-2, and AT4-3 in A. thaliana (Shin et al., 2006), TPSI1 in Solanum lycopersicum (Liu et al., 1997), Mt4 in barrel clover (Burleigh and Harrison, 1999), PILNCR1 in maize (Du et al., 2018), PDIL1 in Medicago truncatula (Wang et al., 2017), and HvIPS1 and HvIPS2 in Hordeum vulgare (Huang et al., 2011) (Table 1).
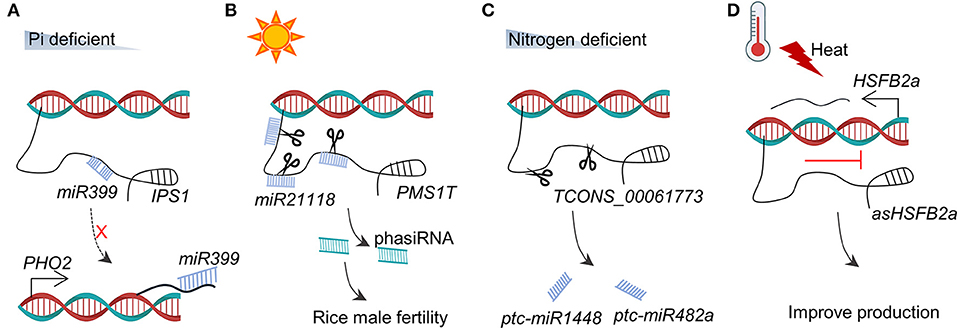
Figure 3. The relationship between lncRNAs and RNAs. (A) Under Pi deficiency, IPS1 serves as an eTM to bind with miR399, and prevents the interaction between miR399 and PHO2 mRNA. (B) PMS1T is sensitive to photoperiod and is targeted by miR2118 to produce phasiRNA. (C) TCONS_00061773 acts as the precursor of ptc-miR1448 and ptc-miR482a in response to nitrogen deficiency. (D) Heat stress induces asHSFB2a and then represses HSFB2a.
The above-mentioned eTM regulatory mechanism that occurs between other lncRNAs and miRNAs is conserved in different plant species and in biological pathway. As shown in Table 1, lncRNAs act as ceRNAs to sequestrate the silencing of miRNA for target genes, such as regulating leaf shapes in Arabidopsis (Wu et al., 2013), altering anthocyanin contents in Hippophae rhamnoides (Zhang et al., 2018), affecting the citrus fruit development (Ke et al., 2019), responding to auxin signal (Das et al., 2019), promoting jasmonic acid (JA) and methyl jasmonate (MeJA) biosynthesis and signal transduction pathways (Zhu et al., 2019), regulating rice root development (Cho and Paszkowski, 2017), and enhancing resistance to disease (Jiang et al., 2019; Hou et al., 2020).
The long non-coding RNAs that are binding with the complementary miRNA may lead to their cleavage to yield phased small-interfering RNAs (phasiRNAs). In eukaryote, lncRNAs or other RNAs are complemented and cleaved by miRNA to generate single-stranded RNAs first, and then to form double-stranded RNAs from the lysed RNA via RNA-dependent RNA polymerase, which are further processed into phasiRNA of 21–24 nt. Finally, phasiRNAs were loaded into AGONAUTE proteins for silencing RNA (Komiya, 2017). As shown in Figure 3B, the lncRNA PMS1T, encoded by photoperiod-sensitive genic male sterility 1 (Pms1) locus in rice, is cleaved by miR2118 to generate 21-nt phasiRNAs. The abundance of phasiRNAs is associated with the photoperiod-sensitive male sterile (Fan et al., 2016; Komiya, 2017). Two other anther-specifically expressed lncRNAs, Male Sterility-related PhasiRNA Precursor LincRNAs1 (MSPPL1) and MSPPL2, are also cleaved by miR2118 to yield phasiRNAs that play a role in the posttranscriptional regulation during meiosis in rice (Zhang et al., 2020). This mechanism is well-known in different plant development processes (Table 1); among these, Cs1g09600 and Cs1g09635 are cleaved by miR3954 to yield phasiRNAs, playing a role in the regulation of flowering in Citrus sinensis (Liu et al., 2017), MuLnc1 is cleaved by miR3954 to produce phasiRNAs, disrupting the expression of the calmodulin-like protein gene CML27 in Morus multicaulis (Gai et al., 2018), and Sl-lncRNA15492-miR482a-phasiRNAs affect the resistance to Phytophthora infestans in tomato (Jiang et al., 2020).
In plants, the biogenesis of miRNAs is precisely controlled (Yu Y. et al., 2019). The production of miRNA undergoes a complex process in which MIR genes mainly located in the intergenic regions are first transcribed into primary miRNAs, then modified into the precursor miRNAs, forming miRNA duplexes, and finally forming mature miRNA to guide the posttranscriptional gene silencing (Sanei and Chen, 2015). Some lncRNAs act as the precursors of mature miRNAs (Liu et al., 2015; Yu Y. et al., 2019). In Arabidopsis, two lncRNAs, npc83, and npc521, are found to be the miRNA precursors for MIR86lncRNA9A and MIR160C, respectively (Ben Amor et al., 2009). The same phenomena are found in other plant species. TCONS_00061773 are the precursors of ptc-miR1448 and ptc-miR482a, which involve in the defense mechanism to prevent nitrogen deficiency in S. lycopersicum (Chen et al., 2016) (Figure 3C). In tomato fruit, lncRNAZ081 is the precursor of miRNA6027, and lncRNAZ114 is the precursor of miRNA1919b and miRNA1919c (Wang et al., 2018a). In this study, we described only miRNA precursors for those lncRNAs that were validated by the low-throughput experiments.
Some lncRNAs can be produced as antisense transcripts in the coding regions, and cis-regulate mRNA transcript through indirect physical interaction. As shown in Figure 3D, asHSFB2a in A. thaliana is a natural long non-coding antisense RNA of HSFB2a induced by heat stress (Wunderlich et al., 2014). The overexpressed asHSFB2a leads to an interrupt of HSFB2a, which subsequently improves the production in vegetative development (Wunderlich et al., 2014). Antisense lncRNAs were also found in S. lycopersicum and Oryza sativa. The antisense transcript of SlGRX22, lncRNA16397, upregulates the SlGRX22 and SlGRX21 expression and enhances the disease resistance of P. infestans by tomato (Cui et al., 2017). TWISTED LEAF (TL) in rice is an antisense transcript of R2R3-MYB transcription factor gene, namely, MYB60 locus. The RNA interference and the overexpression experiments indicate that TL cis-suppresses the MYB60 expression in regulating leaf blade flattening in rice by altering the chromatin structure (Liu et al., 2018).
Dynamic chromatin topology is closely associated with the gene expression patterns. In Arabidopsis, the lncRNA AUXIN-REGULATED PROMOTER LOOP (APOLO) was shown essential for the lateral root development by regulating the transcription level of auxin-responsive genes. APOLO controls these auxin-responsive genes in cis or trans by modulating chromatin loops (Mas and Huarte, 2020; Figure 4A). APOLO is activated by AUXIN RESPONSE FACTORS7 (ARF7) after treating with auxin and cis-activates the neighboring gene PID through formatting a chromatin loop with the promoter of PID by reducing H3K27me3 in the loop (Ariel et al., 2014). The opened loop is closed after APOLO generated from RNA Pol II further recruits polycomb repressive complex1 (PRC1) to deposit the DNA methylation on the genomic region of APOLO-PID (Ariel et al., 2014). APOLO transcripts can also regulate several distant target genes such as WAG2 and AZG2 through forming R-loops in trans (Ariel et al., 2020). APOLO recognizes its distant target genes by base complementarity through the formation of DNA–RNA duplexes. Two complementary TTCTTC boxes in APOLO RNA are confirmed to be essential for the formation of APOLO-target DNA loops (Ariel et al., 2020).
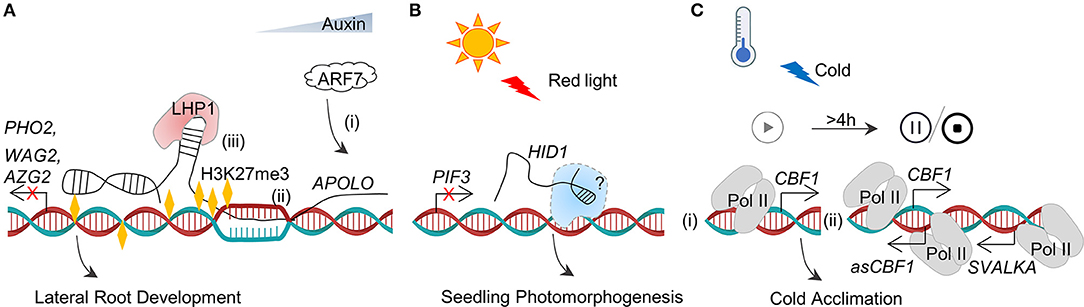
Figure 4. LncRNA–DNA interactions. (A) (i) Upregulation of APOLO by ARF7 after auxin treatment, (ii) direct binding between APOLO and auxin-responsive genes chromatin through base complementarity to form R-loop in cis and trans. Expressions of these auxin-responsive genes contribute to the lateral development, and (iii) recruitment of LHP1 by APOLO to deposit H3K27me3 for closing the loop and then for silencing the APOLO target. (B) HID1 binds to the proximal promoter of PIF3 and represses its expression to modulate seedling photomorphogenesis. HID1 may function by combining with proteins although the detail is still unknown. (C) (i) Sense CBF1 is initiated by cold exposure, and the expression peaks after 4 h. (ii) Contemporaneously, SVALKA is induced in the antisense region of CBF1. RNA Pol II read-through transcription of SVALKA leads to the expression of asCBF1 and an increase of RNA Pol II occupancy on strands. The collision of RNA Pol II stalls CBF1 sense transcription.
The long non-coding RNAs can interact with DNA to regulate gene transcriptions. The knockdown of HIDDEN TREASURE1 (HID1) (Wang Y. et al., 2014) in Arabidopsis displays elongated hypocotyls in continuous red light. ChIP-qPCR experiments revealed that HID1 directly binds to the chromatin of the first intron of PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) to repress its transcription in trans (Wang Y. et al., 2014). HID1 is detected in nuclear. However, its protein partner is still unknown (Figure 4B). It should be noted that the sequence and secondary structure of HID1 are highly conserved from moss to Arabidopsis, with a similar function in seedling photomorphogenesis (Wang Y. et al., 2014).
In plants, a novel transcriptional regulation mechanism of coding gene depends on the transcription of the adjacent lncRNA (Kindgren et al., 2018). The lncRNA SVALKA, found in the cold-sensitive region of the Arabidopsis genome, is the antisense transcript between C-repeat/dehydration-responsive element binding factor3 (CBF3) and CBF1. RNA Pol II read-through transcription of SVALKA leads to the expression of antisense CBF1 (asCBF1), and the cascade of SVALKA-asCBF1 represses CBF1 transcribing through RNA Pol II collision stemming, leading to maximize the low-temperature tolerance of plants with minimal adaptation changes (Kindgren et al., 2018) (Figure 4C).
The long non-coding RNAs regulate the expression of genes by interacting with RNA-binding proteins. As mentioned earlier, after binding to the complementary DNA to form an R-loop, APOLO acts as a guide to recruit PRC1 component LIKE HETEROCHROMATIC PROTEIN1 (LHP1) to deposit the repressive histone marks (histone H3 Lys 27 trimethylation, H3K27me3) at the target chromatin and to repress the auxin-responsive gene transcription (Ariel et al., 2014; Figure 4A). The polycomb complex is recruited by lncRNAs during the process of vernalization-triggered flowering (Questa et al., 2016). The vernalization also induces three lncRNAs, such as COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs (COOLAIR) (Baurle and Dean, 2006), COLDWRAP (Kim and Sung, 2017), and COLD ASSISTED INTRONIC NON-CODING RNA (COLDAIR) (Heo and Sung, 2011) which are derived from the antisense strand, the repressed promoter, and the first intron of FLOWERING LOCUS C (FLC), respectively (Figure 5A). These three lncRNAs involve in the deposition of H3K27me3 at FLC chromatin to repress FLC transcription (Kim and Sung, 2017). COLDAIR and COLDWRAP directly interact with PRC2, which catalyzes H3K27me3 (Heo and Sung, 2011; Kim and Sung, 2017; Kim et al., 2017), whereas COOLAIR binds to the protein FCA, which interacts with PRC2 subunit CURLY LEAF (CLF) to enrich H3K27me3 at FLC chromatin (Figure 5A). The protein phosphatase SSU72 physically interacts with FCA to antagonize the FCA binding with COOLAIR (Tian et al., 2019). In particular, an antisense lncRNA that derived by the same promoter of COOLAIR is independent on the vernalization in early-flowering Arabidopsis ecotypes. ASL1 transcript physically associates with nuclear exosome component AtRRP6L1 (Callahan and Butler, 2008), maintaining H3K27me3 levels for silencing FLC (Shin and Chekanova, 2014).
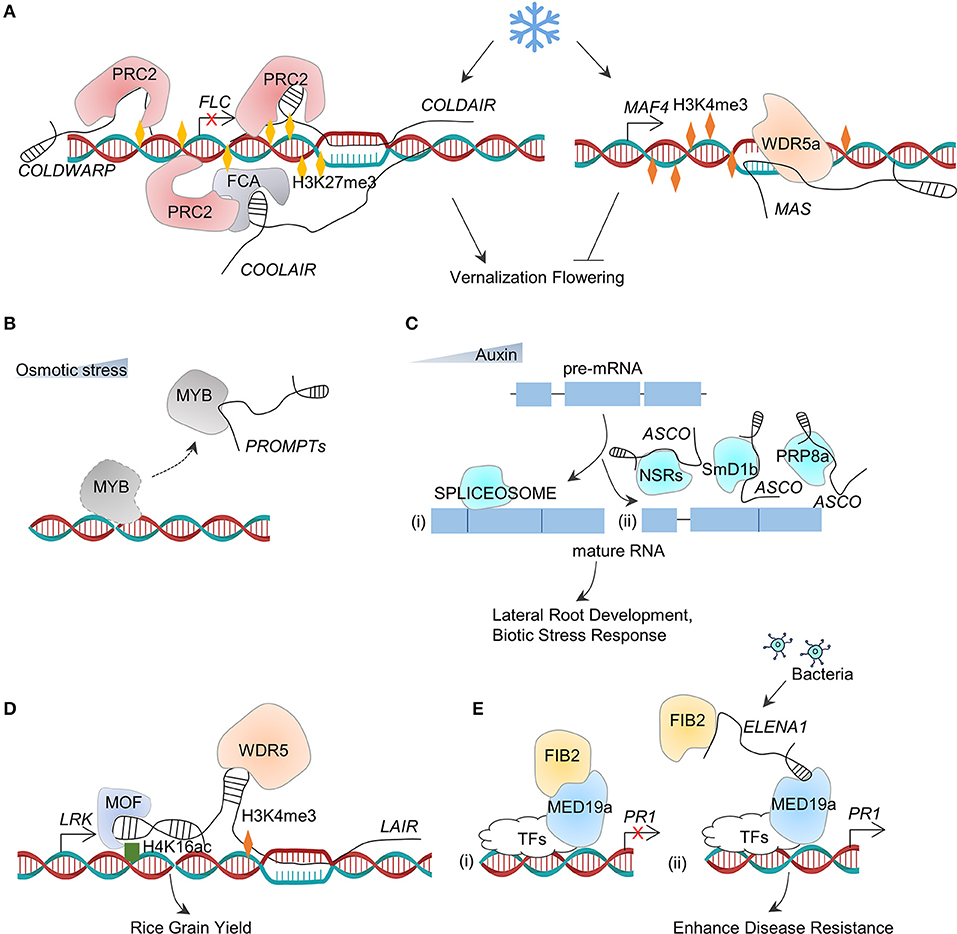
Figure 5. LncRNA–protein interactions. (A) During vernalization, cold temperature induces COLDAIR, COLDWARP, and COOLAIR. Then, COLDAIR and COLDWARP recruit PRC2 to enrich H3K27me3 in the regulation region of FLC to repress its expression, whereas COOLAIR directly binds with FCA to recruit PCR2 and deposit H3K27me3 in the chromatin of FLC. At the same time, cold signal induces the expression of MAS and then recruits WDR5a to deposit H3K4me3 in the chromatin of MAF4 to increase its expression. Downregulation of FLC and upregulation of MAF4 ensure the accurate flowering time during vernalization. (B) Under osmotic stress, PROMPTs act as decoys to hijack the MYB transcription factors (TFs) for preventing their regular functions as TFs in regulating gene transcription. (C) During alternative splicing of pre-mRNA, multiple splicing factors including NSRs, SmD1b, and PRP8a binds (ii) or not (i) to ASCO, together with pre-mRNA to modulate transcriptome diversity dynamically. (D) LAIR, derived from the antisense transcript of LRK, directly interacts with the genomic region of LRK and then acts as a scaffold to recruit MOF and WDR5 to deposit H4K16ac and H3K4me3, respectively. This leads to the upregulation of LRK expression and increase of grain yield. (E) (i) In the absence of pathogen (bacteria) treatment, FIB2 directly interacts with MED19a, and this complex represses PR1 expression; (ii) after the pathogen treatment, the transcription level of lncRNA ELENA1 increases and then binds to FIB2 and MED19a. After FIB2 dissociates, MED19a continues to bind on the promoter of PR1 and activate its expression.
Another cold-induced lncRNA is MAS that plays a regulatory role in the flowering time (Zhao et al., 2018). MAS is an NAT of the MADS AFFECTING FLOWERING4 (MAF4) gene discovered in Arabidopsis, and it activates MAF4 transcription involved in H3K4me3 deposition. Further assays showed that MAS binds to the locus of MAF4 directly and recruits the COMPASS-like complexes (component WD repeat domain 5a, WDR5a) to enhance H3K4me3. The cold-induced formation of the MAS–WAR5a complex suppresses the premature flowering in vernalization (Zhao et al., 2018; Figure 5A). The interaction between lncRNAs and chromatin proteins modulates the chromatin 3D conformations of the neighboring or distant locus and affects the transcriptional activity in cis or trans (Figures 4A, 5A).
PROMPT_1281 is a promoter upstream transcript (PROMPT) lncRNAs, which is sensitive to osmotic stress in Populus simonii, through binding with MYBs transcription factors to interrupt them from interacting with DNA and lead to an increase in the expression of target genes in cis and trans (Song Y. et al., 2019; Figure 5B).
In M. truncatula, an lncRNA named as MtENOD40 has a short open reading frame (sORF) and is important in root nodule organogenesis (Campalans et al., 2004). The functional analysis indicates that MtENOD40 can interact with M. truncatula RNA-binding protein 1 (MtRBP1, also named as M. truncatula Nuclear Speckles RNA-binding protein 1, MtNSR1). When MtENOD40 expressed in root tissues, the ENOD40-MtRBP1 complex would relocate from the nucleus to the cytoplasm (Campalans et al., 2004). Interestingly, the Arabidopsis orthologs of MtRBP1 (i.e., AtNSRa and AtNSRb), when co-expressed with MtENOD40, also lead to the relocation of AtNSRs from the nucleus to the cytoplasm, indicating the conserved relocation activity of MtENOD40 (Bardou et al., 2014). The above-mentioned AtNSR proteins participate in alternative splicing (AS) of pre-mRNA in Arabidopsis (Bardou et al., 2014). Lnc351, also called as AS competitor long-non-coding RNA (ASCO-lncRNA, ASCO), is induced by auxin. This lncRNA would competitively bind AtNSRs with AS targets in vitro and in vivo, regulating transcriptome diversity in the formation of lateral root development (Bardou et al., 2014). Additionally, ASCO binds with multiple splicing factors including PRP8a and SmD1b and modulates the spliced genes in response to bacterial flagellin (Rigo et al., 2020) (Figure 5C). ASCO acts as an integrator through interacting with the spliceosome to dynamically modulate the transcriptome diversity.
The long non-coding RNAs can act as scaffolds to link multiple proteins together for forming the functional ribonucleoprotein complexes. The lncRNA LAIR (LRK Antisense Intergenic RNA) is derived from the antisense strand of neighboring gene LRK (leucine-rich repeat receptor kinase) cluster. Its overexpression leads to the upregulated expression of LRK genes and increased rice grain yields (Wang et al., 2018b). It was found that LAIR directly binds to the genomic regions of LRK1 and functions as a scaffold to link histone modification proteins such as Males-absent-on-the-first (OsMOF) and OsWDR5 in vivo, which in turn gives a rise in the enrichment of H3K4me3 and H4K16 acetylation (H4K16ac) at chromatin to active LRK1 (Wang et al., 2018b; Figure 5D).
Similarly, as a part of plant immune response, the lncRNA ELF18-INDUCED LONG NON-CODING RNA 1 (ELENA1) activates PATHOGENESIS-RELATED GENE1 (PR1) by directly binding with Mediator subunit 19a (MED19a) and negative transcription regulator FIBRILLARIN2 (FIB2) to enrich MED19a on PR1 promoter (Seo et al., 2017). More specifically, an association of ELENA1 with FIB2 (Seo et al., 2019) hinders the inhibitory effect of FIB2 on PR1 and allows the accumulation of more MED19a on PR1 promoter to activate the gene expression (Figure 5E).
In addition to interacting with RNA-binding proteins, MtENOD40 was found to bind and repress two small nodulins, namely, Small Nodulin Acidic RNA-Binding Protein1 (MtSNARP1) and MtSNARP2 (Laporte et al., 2010). SNARPs are acidic peptides and are involved in the nitrogen-fixing symbiosis between M. truncatula and Sinorhizobium meliloti (Figure 6A). However, the interaction between MtSNARPs and MtENOD40 in cytoplasm, as well as the functional role of MtRBP1 in cytoplasm require further studies.
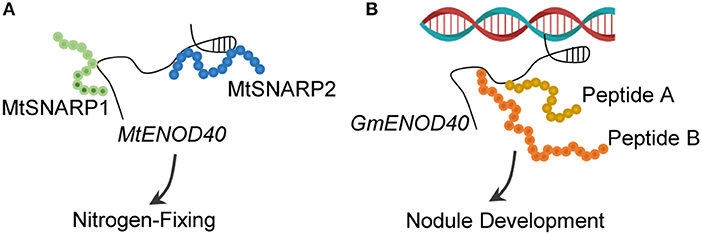
Figure 6. LncRNA–peptide relations. (A) MtENOD40 acts as a decoy to titrate two small peptides, namely, MtSNARP1 and MtSNARP2. Both peptides play a key role in establishing nitrogen-fixing symbiosis between Medicago truncatula and Sinorhizobium meliloti. (B) GmENOD40 contains sORFs that encode two small peptides (Peptide A and Peptide B) of 12 and 24 amino acids each. Both peptides involve in the development of nitrogen-fixing nodules.
The long non-coding RNAs typically lack the protein-coding potential. However, some lncRNAs were found to code peptides. Soybean ENOD40 could produce two small peptides of 12 and 24 amino acids long, both of which bind to a subunit of sucrose synthase nodulin 100 for modulating sucrose use in nitrogen-fixing nodules (Rohrig et al., 2002; Figure 6B). Another example of lncRNA-coding peptides is pri-miRNAs, which are primary transcripts of miRNAs. The pri-miR171b of M. truncatula and the pri-miR165a of Arabidopsis contain sORFs that code the peptides miPEP171b and miPEP165a, respectively (Lauressergues et al., 2015). Increasing the expression levels of miPEP171b and miPEP165a alters the accumulation of miR171b and miR165a, reduces the lateral root development, and promotes the primary root growth (Lauressergues et al., 2015). A similar mechanism has been reported in other life processes, such as those involved in miPEP858a encoded by pri-miR858a to modulate flavonoid biosynthesis in Arabidopsis (Sharma et al., 2020) and miPEP171d1 encoded by pri-miR171d to accumulate miR171d and regulate adventitious root formation in Vitis vinifera (Chen et al., 2020), indicating the universality of this mechanism in plants (Prasad et al., 2020).
The finding of plant lncRNAs and their functional mechanisms is still in its early phase, and all the experimentally validated interaction pairs in plants are shown in Table 1. Although a large number of plant lncRNAs with unknown functions have been suggested by high-throughput sequencing, not all of them are functional. So far, only a small number of lncRNAs have been analyzed by the low-throughput biochemical and molecular biology functional assays. In fact, only 506 functional lncRNAs and 41 interactions of lncRNAs biomolecules in 56 plant species have been recorded in the recent update of the database for EVLncRNAs (https://www.sdklab-biophysics-dzu.net/EVLncRNAs2/) (Zhou et al., 2020). These data are only a tiny fraction of what can be possibly found. More experiments are needed to expand our knowledge in plant lncRNAs. Many questions that are to be addressed are as follows:
• What are the patterns in the secondary structure of lncRNAs in recognizing interacting molecules?
• What are the key sequence or structural factors that affect lncRNA functions?
• What is the degradation mechanism of lncRNAs in organisms?
These questions highlight the importance of RNA structures in providing functional clues. Recent advances in deep learning of protein structure prediction (Senior et al., 2020) and RNA secondary structure (Singh et al., 2019) highlight the importance of huge data in structure and function prediction. However, unlike proteins, the RNA structural and functional data remain scarce. As a result, the power of deep learning is quite limited until more data are available.
As illustrated in Figure 2, functional lncRNAs identified so far respond to the plant development signals, which act as regulators in plant growth and development in association with other macromolecules. LncRNAs in plants can respond to a variety of signals, including light (Wang Y. et al., 2014), phytohormones (Ariel et al., 2020), salt (Qin et al., 2017), temperature (Wunderlich et al., 2014; Kindgren et al., 2018), deficiency of nutritions such as phosphorus (Franco-Zorrilla et al., 2007; Zhang et al., 2019), and biotic stresses such as pathogen infection (Hou et al., 2020).
Currently, plant lncRNAs were shown to function through the formation of eTM with miRNA to regulate the expression of miRNA target genes indirectly (Figure 3A), acting as decoys to hijack nucleic acids or proteins to prevent their functions (Figures 3A, 5B), the recruitment of modified proteins to remodel chromatin status at the epigenetic level (Figures 4A, 5A), or the linkage of macromolecules to form functional complexes (Figures 5D,E). Similar mechanisms of interaction between plant lncRNA and other macromolecules can be found in eukaryotes such as humans, mammals, insects, and even fungi (Quinn and Chang, 2016; Golicz et al., 2018; Kazimierczyk et al., 2020), further suggesting the conservation of biological evolution on the earth. It should be noted that lncRNA is a single-stranded RNA that can recognize the complementary RNA or DNA sequences. Although some tools have been developed to predict the potential complementary nucleic acid molecules of lncRNA, the low abundance of lncRNAs makes it necessary to be certain about their biological effects by conducting the low-throughput experiments.
Taken together, understanding the functions of plant lncRNAs is just at the beginning stage. Model plants such as A. thaliana and O. sativa are the most studied plants (Figure 1). Significantly more studies are needed to cover the ocean of lncRNAs with unknown structures and functions. These fundamental studies are practically important because these lncRNAs are implicated in plant yield and quality and disease resistance. According to the current experimental identification technology, such as CRISPR/Cas9, RNAi, and loss and gain functions, the rate of functional lncRNA verified by the biological experiments is ~600 per year (Zhou et al., 2020). It will be a long journey to fully understand the role of the massive amount of potential functional lncRNAs in the living organisms. Moreover, the growth of new high-throughput lncRNAs is accelerating. Thus, it is time to shed more light on the lncRNA “dark matter” to discover new “stars.”
QC conceived the topic of the article. QC, KL, RY, PH, and ZC wrote the manuscript. BZ, YZ, and JW reviewed and revised the manuscript. All authors have directly contributed to the work and also read and approved the final published version.
We acknowledge the support from the National Natural Science Foundation of China (32000199, 62071085, 61671107, and 61801081); the Natural Science Foundation of Shandong of China (ZR2020QC034); Youth Innovation Team Lead-education Project of Shandong Educational Committee; and Talent Introduction Project of Dezhou University of China (2019xjrc322, 2019xjrc325, and 2019xjrc326). The support of Shenzhen Science and Technology Program (Grant No. KQTD20170330155106581) and the Major Program of Shenzhen Bay Laboratory S201101001 is also acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ariel, F., Jegu, T., Latrasse, D., Romero-Barrios, N., Christ, A., Benhamed, M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55, 383–396. doi: 10.1016/j.molcel.2014.06.011
Ariel, F., Lucero, L., Christ, A., Mammarella, M. F., Jegu, T., Veluchamy, A., et al. (2020). R-Loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell 77, 1055–1065.e1054. doi: 10.1016/j.molcel.2019.12.015
Bardou, F., Ariel, F., Simpson, C. G., Romero-Barrios, N., Laporte, P., Balzergue, S., et al. (2014). Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30, 166–176. doi: 10.1016/j.devcel.2014.06.017
Baurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125, 655–664. doi: 10.1016/j.cell.2006.05.005
Bazin, J., Bustos-Sanmamed, P., Hartmann, C., Lelandais-Briere, C., and Crespi, M. (2012). Complexity of miRNA-dependent regulation in root symbiosis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 1570–1579. doi: 10.1098/rstb.2011.0228
Ben Amor, B., Wirth, S., Merchan, F., Laporte, P., d'Aubenton-Carafa, Y., Hirsch, J., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19, 57–69. doi: 10.1101/gr.080275.108
Burleigh, S. H., and Harrison, M. J. (1999). The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 119, 241–248. doi: 10.1104/pp.119.1.241
Callahan, K. P., and Butler, J. S. (2008). Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 36, 6645–6655. doi: 10.1093/nar/gkn743
Campalans, A., Kondorosi, A., and Crespi, M. (2004). Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16, 1047–1059. doi: 10.1105/tpc.019406
Chekanova, J. A. (2015). Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 27, 207–216. doi: 10.1016/j.pbi.2015.08.003
Chekanova, J. A., Gregory, B. D., Reverdatto, S. V., Chen, H., Kumar, R., Hooker, T., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353. doi: 10.1016/j.cell.2007.10.056
Chen, M., Wang, C., Bao, H., Chen, H., and Wang, Y. (2016). Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genomics 291, 1663–1680. doi: 10.1007/s00438-016-1210-3
Chen, Q. J., Deng, B. H., Gao, J., Zhao, Z. Y., Chen, Z. L., Song, S. R., et al. (2020). A miRNA-encoded small peptide, vvi-miPEP171d1, regulates adventitious root formation. Plant Physiol. 183, 656–670. doi: 10.1104/pp.20.00197
Cho, J., and Paszkowski, J. (2017). Regulation of rice root development by a retrotransposon acting as a microRNA sponge. Elife 6:e30038. doi: 10.7554/eLife.30038
Cui, J., Luan, Y., Jiang, N., Bao, H., and Meng, J. (2017). Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 89, 577–589. doi: 10.1111/tpj.13408
Das, A., Nigam, D., Junaid, A., Tribhuvan, K. U., Kumar, K., Durgesh, K., et al. (2019). Expressivity of the key genes associated with seed and pod development is highly regulated via lncRNAs and miRNAs in Pigeonpea. Sci. Rep. 9:18191. doi: 10.1038/s41598-019-54340-6
Du, Q., Wang, K., Zou, C., Xu, C., and Li, W. X. (2018). The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 177, 1743–1753. doi: 10.1104/pp.18.00034
Fan, Y., Yang, J., Mathioni, S. M., Yu, J., Shen, J., Yang, X., et al. (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. U.S.A. 113, 15144–15149. doi: 10.1073/pnas.1619159114
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gai, Y. P., Yuan, S. S., Zhao, Y. N., Zhao, H. N., Zhang, H. L., and Ji, X. L. (2018). A novel LncRNA, MuLnc1, associated with environmental stress in mulberry (Morus multicaulis). Front. Plant Sci. 9: 669. doi: 10.3389/fpls.2018.00669
Golicz, A. A., Bhalla, P. L., and Singh, M. B. (2018). lncRNAs in plant and animal sexual reproduction. Trends Plant Sci. 23, 195–205. doi: 10.1016/j.tplants.2017.12.009
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hou, X., Cui, J., Liu, W., Jiang, N., Zhou, X., Qi, H., et al. (2020). LncRNA39026 enhances tomato resistance to phytophthora infestans by decoying miR168a and inducing PR gene expression. Phytopathology 110, 873–880. doi: 10.1094/PHYTO-12-19-0445-R
Hou, X., Du, Y., Liu, X., Zhang, H., Liu, Y., Yan, N., et al. (2017). Genome-Wide analysis of long non-coding RNAs in potato and their potential role in tuber sprouting process. Int. J. Mol. Sci. 19:101. doi: 10.3390/ijms19010101
Huang, C. Y., Shirley, N., Genc, Y., Shi, B., and Langridge, P. (2011). Phosphate utilization efficiency correlates with expression of low-affinity phosphate transporters and noncoding RNA, IPS1, in barley. Plant Physiol. 156, 1217–1229. doi: 10.1104/pp.111.178459
Jha, U. C., Nayyar, H., Jha, R., Khurshid, M., Zhou, M., Mantri, N., et al. (2020). Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 20:466. doi: 10.1186/s12870-020-02595-x
Jiang, N., Cui, J., Hou, X., Yang, G., Xiao, Y., Han, L., et al. (2020). Sl-lncRNA15492 interacts with Sl-miR482a and affects Solanum lycopersicum immunity against Phytophthora infestans. Plant J. 103, 1561–1574. doi: 10.1111/tpj.14847
Jiang, N., Cui, J., Shi, Y., Yang, G., Zhou, X., Hou, X., et al. (2019). Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-phytophthora infestans interaction. Hortic. Res. 6:28. doi: 10.1038/s41438-018-0096-0
Jin, J., Lu, P., Xu, Y., Li, Z., Yu, S., Liu, J., et al. (2020). PLncDB V2.0: a comprehensive encyclopedia of plant long noncoding RNAs. Nucleic Acids Res. 49, D1489–D1495. doi: 10.1093/nar/gkaa910
Kazimierczyk, M., Kasprowicz, M. K., Kasprzyk, M. E., and Wrzesinski, J. (2020). Human long noncoding RNA interactome: detection, characterization and function. Int. J. Mol. Sci. 21:1027. doi: 10.3390/ijms21031027
Ke, L., Zhou, Z., Xu, X. W., Wang, X., Liu, Y., Xu, Y., et al. (2019). Evolutionary dynamics of lincRNA transcription in nine citrus species. Plant J. 98, 912–927. doi: 10.1111/tpj.14279
Kim, D. H., and Sung, S. (2017). Vernalization-Triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40, 302–312.e304. doi: 10.1016/j.devcel.2016.12.021
Kim, D. H., Xi, Y., and Sung, S. (2017). Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 13:e1006939. doi: 10.1371/journal.pgen.1006939
Kindgren, P., Ard, R., Ivanov, M., and Marquardt, S. (2018). Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun. 9:4561. doi: 10.1038/s41467-018-07010-6
Komiya, R. (2017). Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and dicer-processing. J. Plant Res. 130, 17–23. doi: 10.1007/s10265-016-0878-0
Laporte, P., Satiat-Jeunemaitre, B., Velasco, I., Csorba, T., Van de Velde, W., Campalans, A., et al. (2010). A novel RNA-binding peptide regulates the establishment of the MEDICAGO truncatula-sinorhizobium meliloti nitrogen-fixing symbiosis. Plant J. 62, 24–38. doi: 10.1111/j.1365-313X.2009.04121.x
Lauressergues, D., Couzigou, J. M., Clemente, H. S., Martinez, Y., Dunand, C., Becard, G., et al. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93. doi: 10.1038/nature14346
Li, J., Hull, J. J., Liang, S., Wang, Q., Chen, L., Zhang, Q., et al. (2019). Genome-Wide analysis of cotton miRNAs during whitefly infestation offers new insights into plant-herbivore interaction. Int. J. Mol. Sci. 20:5357. doi: 10.3390/ijms20215357
Li, L., Eichten, S. R., Shimizu, R., Petsch, K., Yeh, C. T., Wu, W., et al. (2014). Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 15:R40. doi: 10.1186/gb-2014-15-2-r40
Li, L., Liu, J., Liang, Q., Zhang, Y., Kang, K., Wang, W., et al. (2020). Genome-wide analysis of long noncoding RNAs affecting floral bud dormancy in pears in response to cold stress. Tree Physiol. 39, 1821–1837. doi: 10.1093/treephys/tpaa147
Liu, C., Muchhal, U. S., and Raghothama, K. G. (1997). Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol. Biol. 33, 867–874. doi: 10.1023/a:1005729309569
Liu, J., Wang, H., and Chua, N. H. (2015). Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 13, 319–328. doi: 10.1111/pbi.12336
Liu, X., Li, D., Zhang, D., Yin, D., Zhao, Y., Ji, C., et al. (2018). A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 218, 774–788. doi: 10.1111/nph.15023
Liu, Y., Ke, L., Wu, G., Xu, Y., Wu, X., Xia, R., et al. (2017). miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J. 92, 263–275. doi: 10.1111/tpj.13650
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3:eaao2110. doi: 10.1126/sciadv.aao2110
Mas, A. M., and Huarte, M. (2020). lncRNA-DNA hybrids regulate distant genes. EMBO Rep. 21:e50107. doi: 10.15252/embr.202050107
Mattick, J. S., and Rinn, J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7. doi: 10.1038/nsmb.2942
Muers, M. (2011). RNA: genome-wide views of long non-coding RNAs. Nat. Rev. Genet. 12, 742. doi: 10.1038/nrg3088
Palazzo, A. F., and Koonin, E. V. (2020). Functional long non-coding RNAs evolve from junk transcripts. Cell 183, 1151–1161. doi: 10.1016/j.cell.2020.09.047
Prasad, A., Sharma, N., and Prasad, M. (2020). Noncoding but CODING: Pri-miRNA into the action. Trends Plant Sci. 26, 204–206. doi: 10.1016/j.tplants.2020.12.004
Qin, T., Zhao, H., Cui, P., Albesher, N., and Xiong, L. (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 175, 1321–1336. doi: 10.1104/pp.17.00574
Questa, J. I., Song, J., Geraldo, N., An, H., and Dean, C. (2016). Arabidopsis transcriptional repressor VAL1 triggers polycomb silencing at FLC during vernalization. Science 353, 485–488. doi: 10.1126/science.aaf7354
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Rai, M. I., Alam, M., Lightfoot, D. A., Gurha, P., and Afzal, A. J. (2019). Classification and experimental identification of plant long non-coding RNAs. Genomics 111, 997–1005. doi: 10.1016/j.ygeno.2018.04.014
Rigo, R., Bazin, J., Romero-Barrios, N., Moison, M., Lucero, L., Christ, A., et al. (2020). The Arabidopsis lncRNA ASCO modulates the transcriptome through interaction with splicing factors. EMBO Rep. 21:e48977. doi: 10.15252/embr.201948977
Rohrig, H., Schmidt, J., Miklashevichs, E., Schell, J., and John, M. (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. U.S.A. 99, 1915–1920. doi: 10.1073/pnas.022664799
Sanei, M., and Chen, X. (2015). Mechanisms of microRNA turnover. Curr. Opin. Plant Biol. 27, 199–206. doi: 10.1016/j.pbi.2015.07.008
Senior, A. W., Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., Green, T., et al. (2020). Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710. doi: 10.1038/s41586-019-1923-7
Seo, J. S., Diloknawarit, P., Park, B. S., and Chua, N. H. (2019). ELF18-induced long noncoding RNA 1 evicts fibrillarin from mediator subunit to enhance pathogenesis-related gene 1 (PR1) expression. New Phytol. 221, 2067–2079. doi: 10.1111/nph.15530
Seo, J. S., Sun, H. X., Park, B. S., Huang, C. H., Yeh, S. D., Jung, C., et al. (2017). ELF18-induced long-noncoding RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29, 1024–1038. doi: 10.1105/tpc.16.00886
Sharma, A., Badola, P. K., Bhatia, C., Sharma, D., and Trivedi, P. K. (2020). Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 6, 1262–1274. doi: 10.1038/s41477-020-00769-x
Shin, H., Shin, H. S., Chen, R., and Harrison, M. J. (2006). Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45, 712–726. doi: 10.1111/j.1365-313X.2005.02629.x
Shin, J. H., and Chekanova, J. A. (2014). Arabidopsis RRP6L1 and RRP6L2 function in flowering locus C silencing via regulation of antisense RNA synthesis. PLoS Genet. 10:e1004612. doi: 10.1371/journal.pgen.1004612
Singh, J., Hanson, J., Paliwal, K., and Zhou, Y. (2019). RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat. Commun. 10:5407. doi: 10.1038/s41467-019-13395-9
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Song, Y., Xuan, A., Bu, C., Ci, D., Tian, M., and Zhang, D. (2019). Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol J. 17, 164–177. doi: 10.1111/pbi.12955
Tian, Y., Zheng, H., Zhang, F., Wang, S., Ji, X., Xu, C., et al. (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5:eaau7246. doi: 10.1126/sciadv.aau7246
Wang, T., Zhao, M., Zhang, X., Liu, M., Yang, C., Chen, Y., et al. (2017). Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 68, 5937–5948. doi: 10.1093/jxb/erx384
Wang, Y., Fan, X., Lin, F., He, G., Terzaghi, W., Zhu, D., et al. (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U.S.A. 111, 10359–10364. doi: 10.1073/pnas.1409457111
Wang, Y., Gao, L., Li, J., Zhu, B., Zhu, H., Luo, Y., et al. (2018a). Analysis of long-non-coding RNAs associated with ethylene in tomato. Gene 674, 151–160. doi: 10.1016/j.gene.2018.06.089
Wang, Y., Luo, X., Sun, F., Hu, J., Zha, X., Su, W., et al. (2018b). Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat Commun 9, 3516. doi: 10.1038/s41467-018-05829-7
Wang, Z. W., Wu, Z., Raitskin, O., Sun, Q., and Dean, C. (2014). Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. U.S.A. 111, 7468–7473. doi: 10.1073/pnas.1406635111
Waseem, M., Liu, Y., and Xia, R. (2020). Long non-coding RNAs, the dark matter: an emerging regulatory component in plants. Int. J. Mol. Sci. 2286. doi: 10.3390/ijms22010086
Wierzbicki, A. T., Haag, J. R., and Pikaard, C. S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. doi: 10.1016/j.cell.2008.09.035
Wu, H. J., Wang, Z. M., Wang, M., and Wang, X. J. (2013). Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161, 1875–1884. doi: 10.1104/pp.113.215962
Wu, L., Liu, S., Qi, H., Cai, H., and Xu, M. (2020). Research progress on plant long non-coding RNA. Plants 9:408. doi: 10.3390/plants9040408
Wunderlich, M., Gross-Hardt, R., and Schoffl, F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85, 541–550. doi: 10.1007/s11103-014-0202-0
Yamamuro, C., Zhu, J. K., and Yang, Z. (2016). Epigenetic modifications and plant hormone action. Mol. Plant 9, 57–70. doi: 10.1016/j.molp.2015.10.008
Yang, L., Xu, M., Koo, Y., He, J., and Poethig, R. S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2:e00260. doi: 10.7554/eLife.00260
Yang, T., Ma, H., Zhang, J., Wu, T., Song, T., Tian, J., et al. (2019). Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit. Plant J. 100, 572–590. doi: 10.1111/tpj.14470
Yu, T., Tzeng, D. T. W., Li, R., Chen, J., Zhong, S., Fu, D., et al. (2019). Genome-wide identification of long non-coding RNA targets of the tomato MADS box transcription factor RIN and function analysis. Ann. Bot. 123, 469–482. doi: 10.1093/aob/mcy178
Yu, Y., Zhang, Y., Chen, X., and Chen, Y. (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 35, 407–431. doi: 10.1146/annurev-cellbio-100818-125218
Zhang, G., Chen, D., Zhang, T., Duan, A., Zhang, J., and He, C. (2018). Transcriptomic and functional analyses unveil the role of long non-coding RNAs in anthocyanin biosynthesis during sea buckthorn fruit ripening. DNA Res. 25, 465–476. doi: 10.1093/dnares/dsy017
Zhang, Y. C., Lei, M. Q., Zhou, Y. F., Yang, Y. W., Lian, J. P., Yu, Y., et al. (2020). Reproductive phasiRNAs regulate reprogramming of gene expression and meiotic progression in rice. Nat. Commun. 11:6031. doi: 10.1038/s41467-020-19922-3
Zhang, Z., Zheng, Y., Ham, B. K., Zhang, S., Fei, Z., and Lucas, W. J. (2019). Plant lncRNAs are enriched in and move systemically through the phloem in response to phosphate deficiency. J. Integr. Plant Biol. 61, 492–508. doi: 10.1111/jipb.12715
Zhao, L., Wang, J., Li, Y., Song, T., Wu, Y., Fang, S., et al. (2020). NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171. doi: 10.1093/nar/gkaa1046
Zhao, X., Li, J., Lian, B., Gu, H., Li, Y., and Qi, Y. (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 9:5056. doi: 10.1038/s41467-018-07500-7
Zhou, B., Ji, B., Liu, K., Hu, G., Wang, F., Chen, Q., et al. (2020). EVLncRNAs 2.0: an updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 46, D86–D91. doi: 10.1093/nar/gkaa1076
Zhou, B., Yang, Y., Zhan, J., Dou, X., Wang, J., and Zhou, Y. (2019). Predicting functional long non-coding RNAs validated by low throughput experiments. RNA Biol. 16, 1555–1564. doi: 10.1080/15476286.2019.1644590
Zhu, C., Zhang, S., Fu, H., Zhou, C., Chen, L., Li, X., et al. (2019). Transcriptome and phytochemical analyses provide new insights into long non-coding rnas modulating characteristic secondary metabolites of oolong tea (Camellia sinensis) in solar-withering. Front. Plant Sci. 10:1638. doi: 10.3389/fpls.2019.01638
Keywords: plants, long non-coding RNAs, interaction, functional molecular, gene expression
Citation: Chen Q, Liu K, Yu R, Zhou B, Huang P, Cao Z, Zhou Y and Wang J (2021) From “Dark Matter” to “Star”: Insight Into the Regulation Mechanisms of Plant Functional Long Non-Coding RNAs. Front. Plant Sci. 12:650926. doi: 10.3389/fpls.2021.650926
Received: 08 January 2021; Accepted: 05 May 2021;
Published: 07 June 2021.
Edited by:
Jean Molinier, UPR2357 Institut de Biologie Moléculaire des Plantes (IBMP), FranceReviewed by:
Thomas Blein, UMR9213 Institut des Sciences des Plantes de Paris Saclay (IPS2), FranceCopyright © 2021 Chen, Liu, Yu, Zhou, Huang, Cao, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jihua Wang, amh3MjUzMzZAMTI2LmNvbQ==; Yaoqi Zhou, emhvdXlxQHN6YmwuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.