
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 22 April 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.650894
This article is part of the Research TopicGenome-wide Analyses of Pectobacterium and Dickeya SpeciesView all 7 articles
 Collins Kipngetich Tanui1†
Collins Kipngetich Tanui1† Divine Yutefar Shyntum1†
Divine Yutefar Shyntum1† Precious K. Sedibane1,2
Precious K. Sedibane1,2 Daniel Bellieny-Rabelo1†
Daniel Bellieny-Rabelo1† Lucy N. Moleleki1,2*
Lucy N. Moleleki1,2*To adapt to changing environmental niches, bacteria require taxis, a movement toward or away from a stimulus (ligand). Chemotaxis has been studied in some members of the Soft Rot Pectobacteriaceae (SRP), particularly members of the genus Dickeya. On the contrary, there are fewer studies on this topic for the other genus in the SRP group, namely Pectobacterium. This study evaluated chemotactic responses in Pectobacterium brasiliense (Pb 1692) to various ligands. A total of 34 methyl-accepting chemotactic proteins (MCPs) were identified in the Pb 1692 genome and the domain architectures of these MCPs were determined. Four Pb 1692 MCPs previously shown to be differentially expressed during potato tuber infection were selected for further functional characterization. Toward this end, Pb 1692 mutant strains each lacking either AED-0001492, AED-0003671, AED-0000304, or AED-0000744 were generated. Two of these mutants (AED-0001492 and AED-0003671), were attenuated in their ability to grow and respond to citrate and are thus referred to as MCPcit2 and MCPcit1, respectively, while the other two, AED-0000304 (MCPxyl) and AED-0000744 (MCPasp), were affected in their ability to respond to xylose and aspartate, respectively. Trans-complementation of the mutant strains restored swimming motility in the presence of respective ligands. The four MCP mutants were not affected in virulence but were significantly attenuated in their ability to attach to potato leaves suggesting that ecological fitness is an important contribution of these MCPs toward Pb 1692 biology.
Bacteria employ a myriad of mechanisms to efficiently adapt to changing environmental conditions (Armitage, 1992; Blair, 1995). Such pieces of machinery include one-component systems (OCS), two-component systems (TCS), and chemoreceptor-based signaling, also known as chemotaxis (Matilla and Krell, 2017). Chemotaxis is an important process involving several host-pathogen interactions, and can ultimately determine the outcome of infection (Armitage, 1992; O’Toole et al., 1996; Antúnez-Lamas et al., 2009a). Chemotaxis and motility are critical during the early stages of infection when bacteria search for entry sites to penetrate the host apoplast (Stock and Baker, 2009; Reverchon and Nasser, 2013). The attracted bacteria swim toward a wounded site to which they attach and subsequently enter the host apoplastic environment (Antúnez-Lamas et al., 2009a).
In bacteria, methyl-accepting chemotaxis proteins (MCPs) or chemoreceptors are membrane-bound receptors that sense external stimuli and respond through a signal transduction pathway consisting of CheABRWYZ proteins (Charkowski et al., 2012). The molecular mechanism of taxis has been extensively studied in Escherichia coli and this bacterium has been shown to have five MCPs (Stock and Baker, 2009). However, unlike E. coli, many plant pathogens are predicted to encode more than 30 MCP receptors; with different Pectobacterium spp. encoding between 30 and 39 taxis receptors (Glasner et al., 2008).
Chemoreceptors respond to different ligands such as sugars, amino acids, and organic acids (Lacal et al., 2010). The ligand-binding domains (LBDs) of MCPs can either be located in the periplasmic or cytoplasmic space (Porter et al., 2011). The most abundant LBDs belong to three superfamilies, namely the 4-helix bundle (4HB), CACHE, and the PAS domain (the PAS domain is typically cytosolic in location) (Ortega et al., 2017). Following the perception of this signal, the LBD induces a conformational change that triggers the chemosensory signaling cascade leading ultimately to chemotaxis (Porter et al., 2011). This conformational change leads to alterations in CheA autokinase activity. CheA is a cytoplasmic histidine autokinase protein that interacts with MCPs through CheW, a coupling protein (Falke and Hazelbauer, 2001). The phosphorylated CheA then transfers its phosphoryl group to a diffusible cytoplasmic response regulator CheY (Falke and Hazelbauer, 2001). The phosphorylated CheY rotates the flagellar either clockwise or anti-clockwise by binding to the motor region of the flagellar, resulting in swimming motility away from repellents or toward favorable conditions (Falke and Hazelbauer, 2001). Pectobacterium brasiliense, a member of the soft rot Pectobacteriaceae (SRP), is a concern to potato growers worldwide (Duarte et al., 2004; van der Merwe et al., 2010; Panda et al., 2012; Onkendi et al., 2014). Unlike Dickeya dadantii, another member of the SRP, responses of P. brasiliense to various ligands are not well established (Rio-Alvarez et al., 2012). As a consequence, the roles of Pb 1692 chemoreceptors in response to various substrates during the infection process are still poorly understood. A previous report from our laboratory found several chemoreceptors differentially expressed in Pb 1692 during potato tuber infection (Bellieny-Rabelo et al., 2019). Based on these observations, four differentially expressed chemoreceptors were selected for further characterization. In this study, we evaluated the response of Pb 1692 wild-type and four MCP mutant strains to 20 sugars, amino, and organic acids. Furthermore, the four MCP mutant strains were assessed for their ability to confer fitness advantage during in vitro growth and during plant colonization relative to the wild-type strain.
Using the ASAP database1 we screened the Pb 1692 genome for the presence of methyl-accepting proteins (MCPs). The similarity-based approach (BLASTP) detected 34 taxis receptor proteins containing a methyl-accepting protein domain in the Pb 1692 genome (Supplementary Table 1). Domain architecture analyses indicated that all 34 chemoreceptor proteins harbor a cytosolic MCP domain and a few (5/34) have an additional HAMP (histidine kinases, adenylyl cyclases, MCPs, and phosphatases) linker domain. However, at this stage we cannot rule out the possibility that receptors identified here as being without a HAMP domain do indeed have one. The most abundant domains were those belonging to the 4HB and CACHE superfamilies. Other domain organizations include TarH, NIT, PAS, and the helical bimodular (HBM) domain (Figure 1 and Supplementary Table 1).
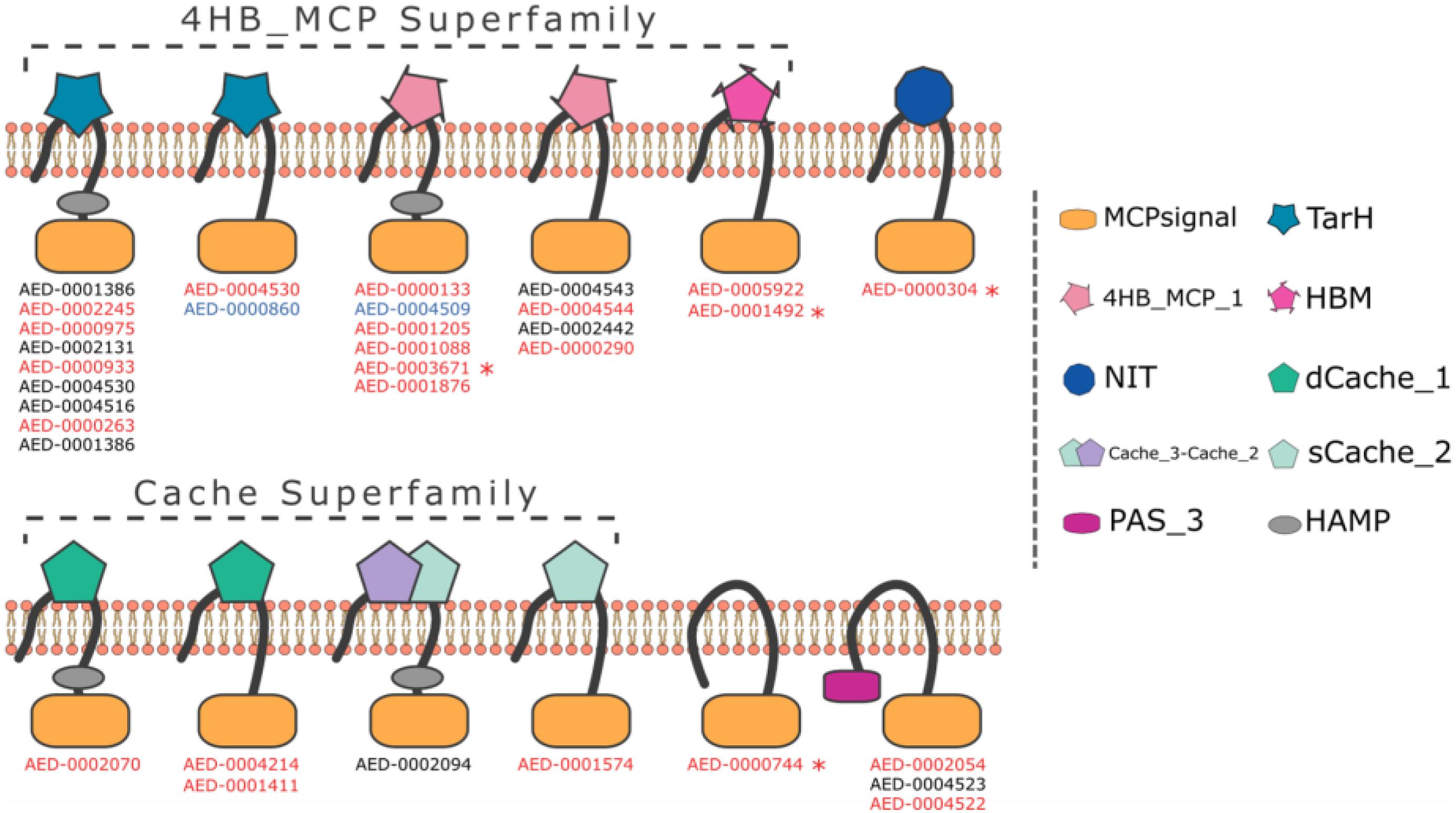
Figure 1. Representation of domain organization of all 34 MCP in Pb 1692 strain. The organization containing both MCP and HAMP domains is the most common in Pb 1692 strain. All predicted domain structures were made through HMMER software with the support of the Pfam-A database. Transmembrane regions were predicted by the Phobius algorithm. The genes highlighted: (1) black was not upregulated, (2) blue was down-regulated, and (3) red was up-regulated during in-planta infection. Furthermore, genes with “*” indicated those mutated and characterized.
Four MCPs (AED-0001492, AED-0003671, AED-0000304, and AED-0000744) representing different architectural classes were selected for further functional characterization. AED-0000304 has a NIT ligand-binding domain while AED-0000744 is cytosolic. Both AED-0001492 and AED-0003671 belong to the 4HB superfamily. An alignment of AED 00003671 and AED 0001492 showed a 61% similarity and both these proteins present transmembrane regions in similar locations (Supplementary Figures 1, 2). All four selected MCP genes were differentially expressed in Pb 1692 at 24 h post-infection (hpi) in potato tubers (Bellieny-Rabelo et al., 2019), suggesting they might play an important role at this stage of infection. To investigate the role of these four MCPs in Pb 1692 concerning ecological fitness as well as virulence, we generated isogenic mutant strains each lacking one of these genes (Supplementary Figure 3). The integrity of the mutant strains was confirmed by a Southern blot, PCR, and sequencing (Supplementary Figure 3, and results not shown). The mutant strains were successfully complemented by expressing the wild-type gene of interest extra-chromosomally (using a plasmid) in the mutated strains.
The Pb 1692 wild-type was screened for the ability to grow and respond to different chemo-attractants consisting of 20 amino acids (aa), sugars, and organic acids supplemented in M9 minimal media (Table 1). Responses were categorized into three groups namely; highly (2 cm and above), moderately (1.0–2.0 cm), and least (0.1–1.0 cm) responsive based on the Pb 1692 wild-type motility in M9 minimal soft-agar plates. The results indicated that Pb 1692 wild-type was highly responsive to some sugars and several amino acids listed in Table 1. We observed that Pb 1692 wild-type was moderately responsive to most amino acids as well as one sugar, sucrose. On the contrary, Pb 1692 wild-type was least responsive to, or in some cases unable to grow in the presence of the following; maleic acid, maltose, mannose, and cysteine (Table 1).
To determine the specific ligands associated with each of the four Pb 1692 MCP mutant strains, we screened for chemotactic responses of the four Pb 1692 MCP mutants, wild-type and complemented mutant strains toward the 20 chemo-attractants (Table 1 and Figures 2A–L). The results demonstrated that AED-0000304 and AED-0000744 mutants were least attracted to xylose and aspartate, respectively (Figures 2E,F) hence these were denoted Pb1692ΔMCPxyl and Pb1692ΔMCPasp, respectively. On the other hand, both Pb 1692 AED-0001492 and AED-0003671 mutants were least attracted to citrate compared to other Pb strains (Figures 2G,H) hence these were denoted Pb1692ΔMCPcit2 and Pb1692ΔMCPcit1, respectively. Trans-expression of (MCPxyl), (MCPasp), and MCPcit2 and MCPcit1 genes in the corresponding mutant strains restored chemotaxis of the complemented strains to wild-type levels (Figures 2I–L). Given that MCPs have been associated with the ability of bacteria to optimally utilize different amino acids and carbon sources, we compared the growth rate of Pb 1692 wild-type and its respective MCP mutant strains in different amino acids and carbon sources. Our findings were in agreement with the results obtained from carbon utilization assays where utilization of sole carbon source was determined (Table 1). The response toward glucose, fructose, sucrose, mannose, ribose, glutamate, valine, alanine, serine, methionine, and asparagine as sole sources of carbon was also found to be similar between the different Pb 1692 MCP mutant strains and the wild-type (Table 1). It was also observable that, Pb 1692 wild-type and the mutant strains generated in this study were least attracted to or unable to grow in the presence of mannose, maltose, maleic acid, and cysteine (Table 1).
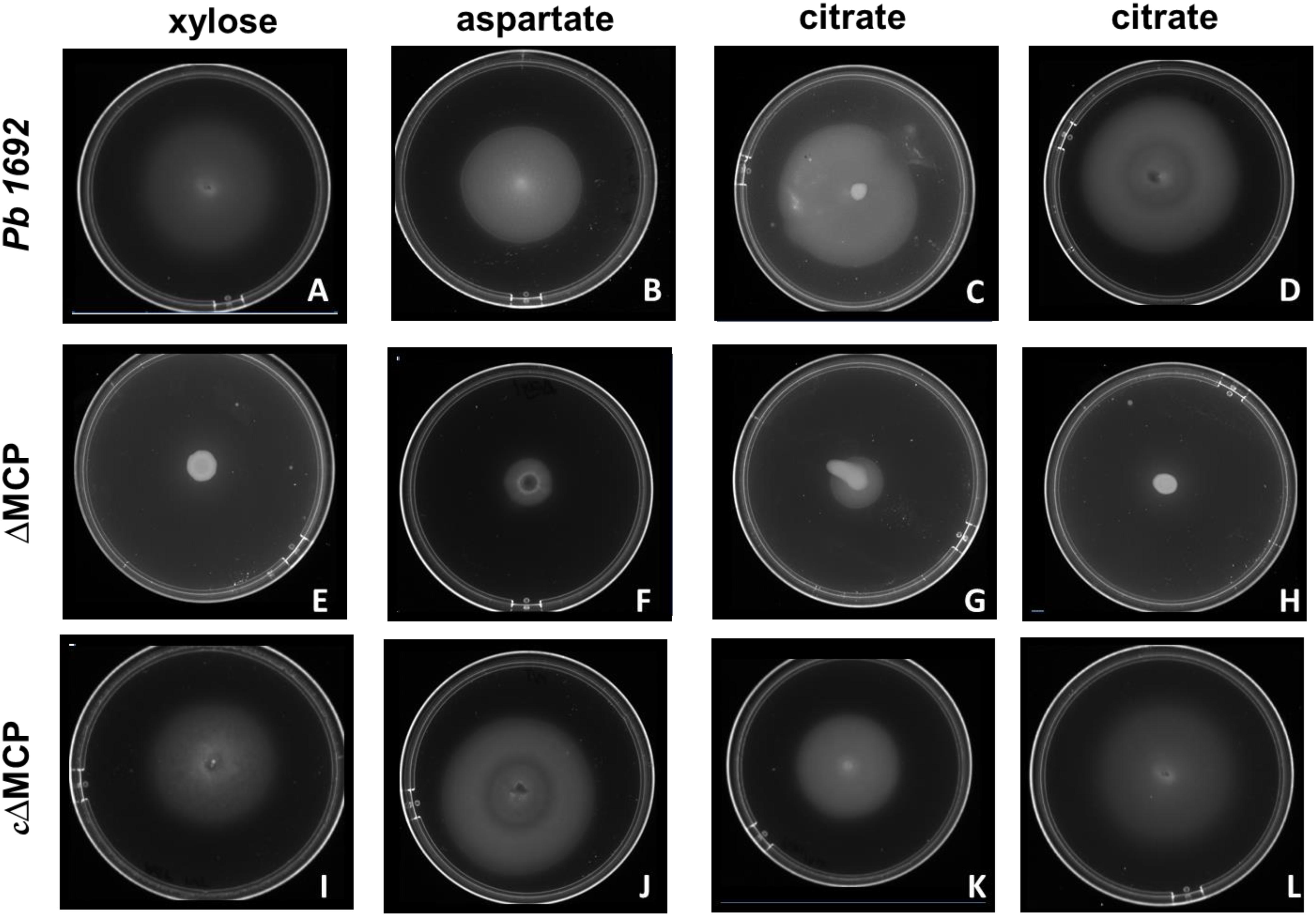
Figure 2. Swimming motility assay. Pb 1692 wild-type was spotted on minimal media supplemented with 1 mM of xylose (A), aspartate (B), and citrate (C,D). Similarly, mutant strains lacking AED-0000304 (Pb1692ΔMCPxyl), AED-0000744 (Pb1692ΔMCPasp), AED-0003671 (Pb1692ΔMCPcit1), and AED-0001492 (Pb1692ΔMCPcit2) were spotted on minimal media supplemented with 1 mM of xylose (E), aspartate (F) and citrate (G,H), respectively. Complemented strains Pb1692ΔMCPxyl–pxyl Pb1692ΔMCPasp–pasp, Pb1692ΔMCPcit1–pcit1, and Pb1692ΔMCPcit2–pcit2, were spotted on minimal media supplemented with 1 mM of aspartate (I), xylose (J) citrate (K,L), respectively.
Some bacteria use different MCPs to sense and move toward different plant signals, for example, jasmonic acid produced by wounded plants (Río-Álvarez et al., 2015). Here, we wanted to determine the role played by these four Pb 1692 MCPs in the attachment to potato leaves. The CFU/ml count indicated that mutant strains were significantly reduced in their ability to attach and colonize potato leaves when compared to Pb 1692 wild-type strain (Figure 3). Conversely, in vitro growth and virulence in potato tubers indicated that the mutant strains and Pb 1692 wild-type strain had no significant difference (Supplementary Figures 4A,B). Together, these findings demonstrate that in planta and in vitro growth as well as virulence on potato tubers are not affected by deletion of these selected MCP-encoding genes in Pb 1692.
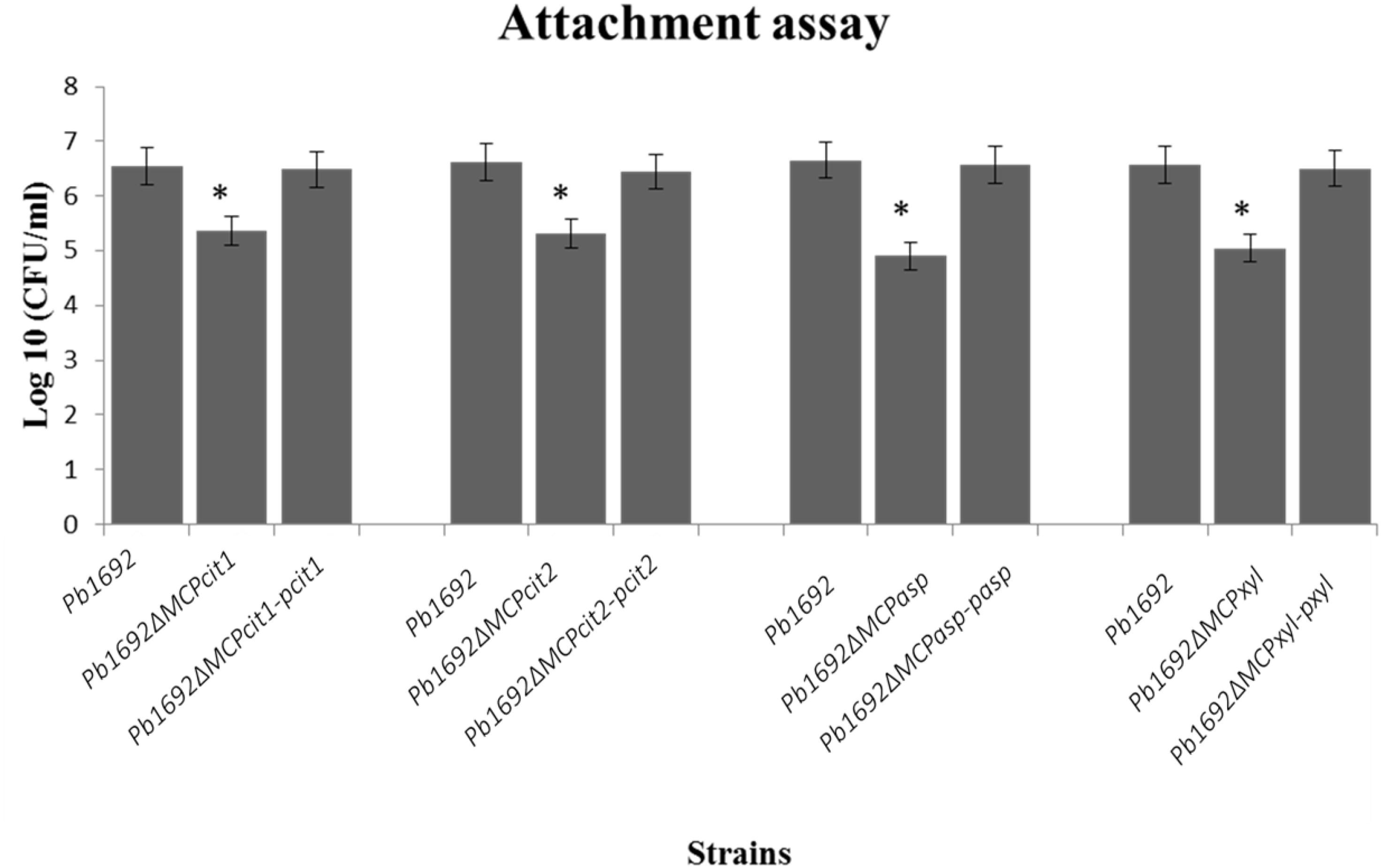
Figure 3. MCP mutants were attenuated in their ability to attach to potato leaves. Determination of CFU/ml from potato leaves after 2 h incubation with 10 μl of each mutant strain (OD600 equivalent to 1.0) in M9 media. Three independent experiments were performed each with three technical replicates with a corresponding standard error of the mean. Statistically significant differences (p < 0.05) between mutants, wild-type, and complemented strains are shown by an asterisk.
Chemotaxis plays an important role in phytopathogenic bacteria host colonization, infection, and disease development. In our previous work (Bellieny-Rabelo et al., 2019), we observed that the master regulator for flagellar biogenesis (FlhDC) together with other genes encoding flagellar biosynthesis (Flg and Fli), chemotactic signaling cascade (CheADVWYZ), and various MCPs were mostly upregulated at 24 hpi during Pb 1692 infection of potato tubers. This indicated that the bacteria are potentially responding to some host-derived environmental cues or ligands leading to effective host colonization. However, to date, responses of Pb 1692 to different types of ligands have not yet been documented. Furthermore, unlike Dickeya, chemotaxis is less studied for most Pectobacterium spp. (Antúnez-Lamas et al.,2009a,b; Río-Álvarez et al., 2015; Velando et al., 2020). Toward this end, our first aim was to determine Pb 1692 responses to various ligands. Our results show that citrate, xylose, glucose, fructose, jasmonic acid, and a number of amino acids are amongst those ligands to which Pb 1692 is highly attracted to. This can be expected since potato tubers have high concentrations of sugars such as fructose and glucose, organic acids such as malic and citric acid as well as various amino acids, albeit in varying concentrations per cultivar (Dobson et al., 2008; Uri et al., 2014). Also, xylose is a monomer of xylan released by plant cells upon damage (Beliën et al., 2006). Interestingly, the gene that drives the strong response to xylose (MCPxyl) was recently reported as part of a SlyA regulatory network in Pb 1692 (Bellieny-Rabelo et al., 2020). More specifically, the SlyA regulon at 12 hpi on potato tubers is enriched with genes associated with carbohydrate metabolism, and it also includes the MCPxyl gene. Hence, the results presented here are consistent with these findings and may point to a yet larger carbohydrate-responsive network under the regulation of SlyA than previously thought, one that comprises xylose as one of its triggers.
Given that D. dadantii has been shown to swim toward jasmonic acid, it is also not surprising that jasmonic acid is one of those metabolites that Pb 1692 was highly attracted to Hugouvieux-Cotte-Pattat and Charaoui-Boukerzaza (2009). These results are also generally reflected in our previously reported transcriptome data (Bellieny-Rabelo et al., 2019) where genes involved in glucose, D-galacturonate, citrate, valine, methionine, and aspartate uptake and metabolism were up-regulated in Pb 1692 potato tuber infection (Bellieny-Rabelo et al., 2019). Malic acid is another major metabolite of plant cells, typically represented in root exudates as well as potato tuber extracts (Dobson et al., 2008; Uri et al., 2014; Feng et al., 2018, 2019). Yet, under these specific experimental conditions, we found that Pb 1692 was unable to grow in malic acid and least responsive to sugars such as maltose. It must be noted that the concentration of a ligand can affect whether binding to the receptor occurs or not (Uri et al., 2014; Feng et al., 2018, 2019). Furthermore, in vitro concentrations and conditions may differ from those in vivo, thus it stands to reason that the outcome of our assays will have been greatly influenced by our specific experimental conditions.
The MCP mutant strains retained growth rates similar to those of the wild-type strain and were not affected in virulence. Therefore, we can conclude that these four MCP proteins do not contribute to the maceration of potato tubers. On the contrary, attachment assays showed that all the MCP mutants were significantly reduced in their ability to attach to potato leaf-surfaces compared to Pb 1692 wild-type. This would therefore suggest that MCPs play an important role in sensing chemo-attractants and subsequently initiating flagella-mediated swimming motility toward the host. Therefore, it can be argued that the ability of the Pb 1692 strain to sense and attach to potato leaves is a measure of its ecological fitness. SRP are ubiquitous by nature and can be found in different ecological niches such as soil, water and the host (Charkowski et al., 2012). Nonetheless, it has to be noted that survival outside the host for the SRP is only for short periods of time (Toth et al., 2021). Hence, the ability (conferred by MCPs as demonstrated here) to sense and move toward the host enables them to move from unfavorable niches to the host environment where survival is more likely (Van Gijsegem et al., 2021). As demonstrated in our results, these MCPs respond to xylose, aspartate, and citrate which are major metabolites found in various plant tissue types including potato tubers. However, it is important to take cognisance of the fact that a single chemoreceptor can bind several different ligands (Falke and Hazelbauer, 2001; Porter et al., 2011; Matilla and Krell, 2017). Thus, while these were the ligands that these mutant strains were least responsive to, it is important to note that our screening did not exhaust all potential ligands. It is therefore likely that these MCPs could bind many other ligands not tested in this study. For instance, in this study we identified a chemoreceptor that responds to Asp. A recent study by Cerna-Vargas et al. (2019) elegantly demonstrated that a Pseudomonas syringae tomato chemoreceptor, PsPto-PscA, responds to L-Asp, Glu and D-Asp (Cerna-Vargas et al., 2019). It would therefore be interesting to determine whether Pb 1692 (MCPasp) also responds to other amino acids similar to those identified for PsPto-PscA.
Interestingly, two of the MCP mutant strains appear to be affected in their response to citrate. Protein alignment of these two proteins indicated that these have a 61 % similarity. Thus these two proteins appear to be paralogs and could be dependent on each other in sensing citrate ligands in Pb 1692 strain. Possibly, effective chemotactic action toward citrate by Pb 1692 could be synergistically performed by these two MCP paralogs or they could extend the response range of Pb 1692 to citrate. This phenomenon was observed in Pseudomonas putida KT2440 where McpQ and McpS are paralogous chemoreceptors both involved in sensing citrate ligands (Martín-Mora et al., 2016). It is noteworthy that architectures of MCPcit2 and MCPcit1 present transmembrane regions in similar locations (Supplementary Figures 1, 2); furthermore, while the first protein remains with both HAMP-MCP domains, the latter conserves HMB-MCP, suggesting that the ancestor-gene architecture could be HMB-HAMP-MCP since there is no sequence-overlapping among the copies’ domains (Supplementary Figure 2). In addition, similar to Salmonella typhimurium TCP, the LBD of the two Pb MCPcit is 4HB while the P. petuda McpQ forms a 4HMB fold (Martín-Mora et al., 2016). This demonstrates how diverse bacterial receptors that recognize citrate are and thus underscores the importance of citrate as a major component in plants.
In conclusion, this study has cataloged some of the chemo-attractants that Pb 1692 responds to. Furthermore, mutation of four Pb 1692 MCPs resulted in attenuated ability to adhere to their host implying that intact chemotaxis machinery in the Pb 1692 strain is required for effective colonization during the early stages of infections. Our findings also show that the four MCPs of Pb 1692 could be associated with chemo-attractants such as xylose, aspartate, and citrate.
All bacterial strains and plasmids used in this study are indicated in Table 2. The Pectobacterium brasiliense 1692 strains were regularly cultured in liquid Luria-Bertani (LB) broth or M9 media at 37°C with constant shaking (370 rpm) (Sambrook et al., 1989). Growth medium was supplemented with either 50 μg/ml kanamycin (Sigma-Aldrich) or 100 μg/ml ampicillin (Sigma-Aldrich) for mutant and complementation strains, respectively. When required, growth media was supplemented with amino acids and other carbon sources to a final concentration of 1 mM.
Previously identified MCP gene homologs from D. dandantii (Rio-Alvarez et al., 2012) were used as bait to identify homologous genes in Pb 1692, using (BLASTN) in the ASAP database2. The search for the conserved domain in Pb 1692 MCPs was performed using the Pfam identifier. Using HMMER v 3.0, the alignment of MCPs proteins was used to construct hidden Markov models (HMMs) to search and identify conserved domains. Amino acid alignment of two MCPs was performed using BLASTP (Altschul et al., 1990) and visual processing by Jalview (Waterhouse et al., 2009). Recognition of conserved domains in protein sequences was made by using HMMER3 package, and transmembrane regions prediction by Phobius; graphical representation of these results was made by IBS software.
The four methyl-accepting chemotaxis genes (AED-0001492, AED-0003671, AED-0000304, and AED-0000744) in the Pb 1692 wild-type strain were inactivated using the lambda red recombinase one-step PCR inactivation method as previously described by Datsenko and colleagues (Datsenko and Wanner, 2000; Tanui et al., 2017; Supplementary Figure 3). Briefly, the open reading frames (ORF) of the targeted genes were replaced by a kanamycin resistance gene. Using kanamycin specific primers with upstream and downstream extended nucleotides, the kanamycin resistance gene was PCR-amplified from pKD4. Subsequently, upstream and downstream of the target gene were PCR amplified, excised, and purified using gel extraction kits following manufactures instruction (Zymo PCR purification kit). The three PCR products were fused using 5′ forward and 3′ reverse primers denoted with an asterisk∗ (Table 3). The fused products were gel purified and electroporated into electro-competent Pb 1692 cells carrying pKD20 plasmid. All primers used in this study are listed in Table 3. Each PCR reaction contained 1 μl of DNA template, 12.5 μl of HiFi HotStart PCR Kit (KAPA Biosystems), 10 μl of nuclease-free H2O, and 0.75 μl (0.5 μM) of each forward and reverse primer making a reaction volume of 25 μl. The PCR conditions were 96°C for 3 min, 30 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by 72°C for 2 min. Detection of kanamycin insert was confirmed using test primers which bind up and downstream of the gene thus amplify the upstream, kanamycin, and downstream sequences of the gene (Table 3). The transformants were selected on an LB agar plate supplemented with 50 μg/ml–1 kanamycin (Table 2). The number of kanamycin insertion sites on Pb 1692 was determined using Southern blot hybridization.
The complemented strains were generated by amplifying individual methyl-accepting genes AED-0001492, AED-0003671, AED-0000304, and AED-0000744 with their cognate promoter region in Pb1692 using primers CFcit2/CRcit2, CFcit1F/CRcit1R, CFxyl/CRxyl, and CFasp/CFasp set of primers, respectively (Table 3). The DNA fragments were cloned into the bacterial cloning vector pJET1.2 (Thermo-Fisher) to generate pJetcit2, pJetcit1, pJetxyl, and pJetasp, which were individually electroporated into the electrocompetent corresponding mutant strains to generate the complemented mutant strains Pb1692ΔMCPcit2-pcit2, Pb1692ΔMCPcit1-pcit1, Pb1692ΔMCPxyl-pxyl, and Pb1692ΔMCPasp-pasp. The transformants were selected on LB agar plate amended with 100 μg/ml–1Ampicilin (Table 2). All generated complements were further confirmed using PCR and DNA sequencing.
To determine what effect deletion of the different Pb 1692 MCP genes has on the growth of the mutant strains when compared to the Pb 1692 wild-type strain, a growth rate curve was performed. Pb 1692 wild-type and the corresponding MCP mutant strains generated in this study were grown in LB broth at 37°C for 16 h with continuous agitation at 370 rpm. The optical density (OD) of the cultures were adjusted to an OD600 equivalent to 0.4 (OD600 = 0.4). Two milliliters of each Pb 1692 strain was inoculated into 100 ml of LB broth and incubated at 37°C with constant agitation at 370 rpm. The optical density (OD600) of each culture was recorded every 1 h for 18 h. The experiment was performed in triplicates, and the mean values were used to calculate statistically significant differences between the wild-type and the MCP mutant strains using a one-way ANOVA. P-values less than 0.05 (p < 0.05) were considered to represent a statistically significant difference.
Chemotaxis and motility tests were investigated on semi-solid 0.3 % agar minimal media medium. Each plate was individually supplemented with different chemo-attractant at the same concentration (1 mM). Optical densities of all strains were standardized (OD600 = 1). A 5 μl of bacterial suspension aliquot was pipetted at the center of each plate then incubated at 37°C for 24 h. Motility toward attractant between Pb 1692 wild-type and the mutant strains was compared 24 h after incubation. Chemotaxis assays were generally performed no less than three independent occasions with at least three plates per assay.
Attachment and entry of the Pb 1692 wild-type, Pb1692ΔMCPcit1, Pb1692ΔMCPcit2, Pb1692ΔMCPasp, and Pb1692ΔMCPxyl into potato leaves was investigated. To this end, Pb 1692 cultures were prepared in M9 minimal media as previously described. Thereafter, potato leaves with a diameter of 3 cm were dipped into 10 ml of M9 minimal media inoculated with 10 μl of each strain equivalent to a density of 107 CFU/ml. Minimal media was supplemented with different ligands to a final concentration of 1mM. Inoculated leaves were incubated at 28°C for 2 h without shaking. Thereafter, the leaves were removed washed with double distilled water and crushed by grinding in 1 ml of 10 mM MgSO4. Viable cell counts were determined by serial dilution and plating on LB medium. Unwounded potato leaves were used as the control experiment. Each experiment consisted of three biological and three technical repeats.
Virulence assays were performed as described previously (Moleleki et al., 2017). Briefly, fresh and healthy susceptible potato tubers (cv Mondial) were socked in 10% sodium hypochlorite for 10 min, rinsed twice with double-distilled H2O, and air-dried. The optical density of all Pb 1692 strains was adjusted to 1 (OD600 = 1). A 1 cm hole was made in sterilized potato tubers using sterilized tips and 10 μl of each strain was inoculated onto each hole. All Pb 1692 strains (wild-type, mutants, and complemented strains) were inoculated in a single potato tuber, including 10 mM MgSO4 which served as a control. Inoculated potato tubers were then sealed with petroleum jelly and incubated in moist sterilized plastic boxes at 25°C for 72 h. Thereafter, the macerated tissue from the inoculated site was scooped and quantified. Three biological experiments with three technical repeats were performed.
To investigate the mutants’ multiplication ability during in planta infection, Pb 1692 strains were inoculated into potato tubers as described in the virulence assays. For bacterial CFU/ml enumeration, 0.2 g of macerated tissue was resuspended in 1 ml of double-distilled H2O. Plant tissue was removed by centrifuging at 11904.464 g for 3 min, and the supernatant which contains bacteria were collected and serially diluted on plates containing appropriate antibiotics. The plates were incubated at 37°C for 24 h for CFU counts. Three biological experiments with three technical repeats were performed.
In this study, experiments were performed in triplicate and three independent times. Where applicable, a one-way Analysis of Variance (ANOVA) was performed to determine statistical significance and a p-value less than 0.05 (p < 0.05) was considered to be a statistically significant difference.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
CKT and LNM contributed to the conception and design of the study. CKT and PKS performed the experiment, organized, and analyzed the data. CKT, PKS, and DB-R performed the bioinformatics and statistical analyses. CKT wrote the first draft of the manuscript. LNM provided resources and supervised this study. DYS generated all mutant strains and complementation plasmids. All authors contributed to the manuscript reviewed, edited, and approved the submitted version.
This research study was funded by the National Research Foundation (NRF), South Africa, through Competitive Funding for Rated Researchers (CFRR 98993) and NRF Bioinformatics and Functional Genomics (BFG 93685). DYS received an NRF and BFG postdoctoral fellowship. DB-R received a University of Pretoria postdoctoral fellowship and CKT received a University of Pretoria Ph.D. studentship. Any opinion, findings, conclusions, or recommendations expressed in this material are those of the authors, and the NRF does not accept any liability in this regard.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.650894/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antúnez-Lamas, M., Cabrera-Ordóñez, E., López-Solanilla, E., Raposo, R., Trelles-Salazar, O., Rodríguez-Moreno, A., et al. (2009a). Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 155, 434–442. doi: 10.1099/mic.0.022244-0
Antúnez-Lamas, M., Cabrera, E., Lopez-Solanilla, E., Solano, R., González-Melendi, P., Chico, J. M., et al. (2009b). Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol. Microbiol. 74, 662–671. doi: 10.1111/j.1365-2958.2009.06888.x
Beliën, T., Van Campenhout, S., Robben, J., and Volckaert, G. (2006). Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant Microbe Interact. 19, 1072–1081. doi: 10.1094/MPMI-19-1072
Bellieny-Rabelo, D., Nkomo, N. P., Shyntum, D. Y., and Moleleki, L. N. (2020). Horizontally acquired quorum-sensing regulators recruited by the PhoP regulatory network expand the host adaptation repertoire in the Phytopathogen Pectobacterium brasiliense. mSystems 5:e00650-19. doi: 10.1128/msystems.00650-19
Bellieny-Rabelo, D., Tanui, C. K., Miguel, N., Kwenda, S., Shyntum, D. Y., and Moleleki, L. N. (2019). Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and Interbacterial Competition in Pectobacterium and Dickeya spp. Appl. Environ. Microbiol. 85:e02050–18. doi: 10.1128/AEM.02050-18
Blair, D. F. (1995). How bacteria sense and swim. Annu. Rev. Microbiol. 49, 489–520. doi: 10.1146/annurev.mi.49.100195.002421
Cerna-Vargas, J. P., Santamaría-Hernando, S., Matill, M. A., Rodríguez-Herva, J. J., Daddaou, A., Rodríguez-Palenzuela, P., et al. (2019). Chemoperception of specific amino acids controls phytopathogenicity in Pseudomonas syringae pv. tomato. MBio 10:e01868–19. doi: 10.1128/mBio.01868-19
Charkowski, A., Blanco, C., Condemine, G., Expert, D., Franza, T., Hayes, C., et al. (2012). The role of secretion systems and small molecules in soft-rot enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. doi: 10.1146/annurev-phyto-081211-173013
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dobson, G., Shepherd, T., Verrall, S. R., Conner, S., Mcnicol, J. W., Ramsay, G., et al. (2008). Phytochemical diversity in tubers of potato cultivars and landraces using a gc-ms metabolomics approach. J. Agric. Food Chem. 56, 10280–10291. doi: 10.1021/jf801370b
Duarte, V., De Boer, S. H., Ward, L. J., and De Oliveira, A. M. R. (2004). Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 96, 535–545. doi: 10.1111/j.1365-2672.2004.02173.x
Falke, J. J., and Hazelbauer, G. L. (2001). Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265. doi: 10.1016/S0968-0004(00)01770-9
Feng, H., Zhang, N., Du, W., Zhang, H., Liu, Y., Fu, R., et al. (2018). Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria bacillus amyloliquefaciens SQR9. Mol. Plant-Microbe Interact. 31, 995–1005. doi: 10.1094/MPMI-01-18-0003-R
Feng, H., Zhang, N., Fu, R., Liu, Y., Krell, T., Du, W., et al. (2019). Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Glasner, J. D., Marquez-Villavicencio, M., Kim, H. S., Jahn, C. E., Ma, B., Biehl, B. S., et al. (2008). Niche-specificity and the variable fraction of the Pectobacterium pan-genome. Mol. Plant-Microbe Interact. 21, 1549–1560. doi: 10.1094/MPMI-21-12-1549
Hugouvieux-Cotte-Pattat, N., and Charaoui-Boukerzaza, S. (2009). Catabolism of raffinose, sucrose, and melibiose in Erwinia chrysanthemi 3937. J. Bacteriol. 191, 6960–6967. doi: 10.1128/JB.00594-09
Lacal, J., Alfonso, C., Liu, X., Parales, R. E., Morel, B., Conejero-Lara, F., et al. (2010). Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J. Biol. Chem. 285, 23126–23136. doi: 10.1074/jbc.M110.110403
Martín-Mora, D., Reyes-Darias, J. A., Ortega, Á, Corral-Lugo, A., Matilla, M. A., and Krell, T. (2016). McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 18, 3284–3295. doi: 10.1111/1462-2920.13030
Matilla, M. A., and Krell, T. (2017). Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 45, 8–14. doi: 10.1016/j.copbio.2016.11.021
Moleleki, L. N., Pretorius, R. G., Tanui, C. K., Mosina, G., and Theron, J. (2017). A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol. Plant Pathol. 18, 32–44. doi: 10.1111/mpp.12372
O’Toole, R., Milton, D. L., and Wolf-Watz, H. (1996). Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19, 625–637. doi: 10.1046/j.1365-2958.1996.412927.x
Onkendi, E. M., Maluleke, L. N., and Moleleki, L. N. (2014). First report of Pectobacterium carotovorum subsp. brasiliense causing soft rot and blackleg of potatoes in Kenya. Plant Dis. 98:684. doi: 10.1094/PDIS-09-13-0988-PDN
Ortega, Á, Zhulin, I. B., and Krell, T. (2017). Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 81:e00033–17. doi: 10.1128/mmbr.00033-17
Panda, P., Fiers, M. A. W. J., Armstrong, K., and Pitman, A. R. (2012). First report of blackleg and soft rot of potato caused by Pectobacterium carotovorum subsp. brasiliensis in New Zealand. New Dis. Rep. 26:15. doi: 10.5197/j.2044-0588.2012.026.015
Porter, S. L., Wadhams, G. H., and Armitage, J. P. (2011). Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9, 153–165. doi: 10.1038/nrmicro2505
Reverchon, S., and Nasser, W. (2013). Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 5, 622–636. doi: 10.1111/1758-2229.12073
Río-Álvarez, I., Muñoz-Gómez, C., Navas-Vásquez, M., Martínez-García, P. M., Antúnez-Lamas, M., Rodríguez-Palenzuela, P., et al. (2015). Role of Dickeya dadantii 3937 chemoreceptors in the entry to Arabidopsis leaves through wounds. Mol. Plant Pathol. 16, 685–698. doi: 10.1111/mpp.12227
Rio-Alvarez, I., Rodríguez-Herva, J. J., Cuartas-Lanza, R., Toth, I., Pritchard, L., Rodríguez-Palenzuela, P., et al. (2012). Genome-wide analysis of the response of Dickeya dadantii 3937 to plant antimicrobial peptides. Mol. Plant Microbe Interact. 25, 523–533. doi: 10.1094/MPMI-09-11-0247
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Laboratory. Available online at: https://ci.nii.ac.jp/naid/10003540621/ (accessed November 23, 2020).
Stock, J. B., and Baker, M. D. (2009). “Chemotaxis,” in Encyclopedia of Microbiology, ed. T. M. Schmidt (Amsterdam: Elsevier Inc), 71–78. doi: 10.1016/B978-012373944-5.00068-7
Tanui, C. K., Shyntum, D. Y., Priem, S. L., Theron, J., and Moleleki, L. N. (2017). Influence of the ferric uptake regulator (Fur) protein on pathogenicity in Pectobacterium carotovorum subsp. brasiliense. PLoS One 12:e0177647. doi: 10.1371/journal.pone.0177647
Toth, I. K., Barny, M., Brurberg, M. B., Condemine, G., Czajkowski, R., Elphinstone, J. G., et al. (2021). “Pectobacterium and Dickeya: environment to disease development,” in Plant Diseases Caused by Dickeya and Pectobacterium Species, eds F. Van Gijsegem, I. K. Toth, and J. M. van der Wolf (New York, NY: Springer International Publishing), 39–84. doi: 10.1007/978-3-030-61459-1_3
Uri, C., Juhász, Z., Polgár, Z., and Bánfalvi, Z. (2014). A GC-MS-based metabolomics study on the tubers of commercial potato cultivars upon storage. Food Chem. 159, 287–292. doi: 10.1016/j.foodchem.2014.03.010
Velando, F., Gavira, J. A., Rico-Jiménez, M., Matilla, M. A., and Krell, T. (2020). Evidence for Pentapeptide-Dependent and Independent CheB Methylesterases. Int. J. Mol. Sci. 21:8459. doi: 10.3390/ijms21228459
van der Merwe, J. J., Coutinho, T. A., Korsten, L., and van der Waals, J. E. (2010). Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 126, 175–185. doi: 10.1007/s10658-009-9531-2
Van Gijsegem, F., Hugouvieux-Cotte-Pattat, N., Kraepiel, Y., Lojkowska, E., Moleleki, L. N., Gorshkov, V., et al. (2021). “Molecular interactions of pectobacterium and dickeya with plants,” in Plant Diseases Caused by Dickeya and Pectobacterium Species, eds F. Van Gijsegem, I. K. Toth, and J. M. van der Wolf (New York, NY: Springer International Publishing), 85–147. doi: 10.1007/978-3-030-61459-1_4
Keywords: chemotaxis, soft rot, blackleg disease, Pectobacterium brasiliense, ecological fitness, virulence, methyl accepting chemotaxis proteins
Citation: Tanui CK, Shyntum DY, Sedibane PK, Bellieny-Rabelo D and Moleleki LN (2021) Pectobacterium brasiliense 1692 Chemotactic Responses and the Role of Methyl-Accepting Chemotactic Proteins in Ecological Fitness. Front. Plant Sci. 12:650894. doi: 10.3389/fpls.2021.650894
Received: 08 January 2021; Accepted: 24 March 2021;
Published: 22 April 2021.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Tino Krell, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 Tanui, Shyntum, Sedibane, Bellieny-Rabelo and Moleleki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucy N. Moleleki, bHVjeS5tb2xlbGVraUB1cC5hYy56YQ==
†Present address: Collins Kipngetich Tanui, Department of Nutrition and Food Science, University of Maryland, College Park, MD, United States; Divine Yutefar Shyntum, Biotechnology Center, Silesian University of Technology, Gliwice, Poland; Daniel Bellieny-Rabelo, Department of Ecology and Environmental Science (EMG), Umeå University, Umeå, Sweden
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.