- 1CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 3Department of Biology, University of Nebraska at Kearney, Kearney, NE, United States
- 4South China Limestone Plants Center, College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
- 5Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 6Research Centre of Ecological Sciences, College of Agronomy, Jiangxi Agricultural University, Nanchang, China
Paralamium (Lamiaceae) is a monotypic genus within the subfamily Lamioideae and has a sporadic distribution in subtropical mountains of southeast Asia. Although recent studies have greatly improved our understanding of generic relationships within Lamioideae, the second most species-rich subfamily of Lamiaceae, the systematic position of Paralamium within the subfamily remains unclear. In this study, we investigate the phylogenetic placement of the genus using three datasets: (1) a 69,276 bp plastome alignment of Lamiaceae; (2) a five chloroplast DNA region dataset of tribe Pogostemoneae, and (3) a nuclear ribosomal internal transcribed spacer region dataset of Pogostemoneae. These analyses demonstrate that Paralamium is a member of Pogostemoneae and sister to the monotypic genus Craniotome. In addition, generic-level phylogenetic relationships within Pogostemoneae are also discussed, and a dichotomous key for genera within Pogostemoneae is provided.
Introduction
Lamiaceae, as currently defined, contains about 7000 species and is subdivided into 12 subfamilies (Li et al., 2016; Li and Olmstead, 2017; Zhao et al., 2021). Lamioideae, containing at least 1260 species and about 61 genera, is the second-largest subfamily (after Nepetoideae) within Lamiaceae in terms of both the number of species and genera (Harley et al., 2004). Although the subfamily has a subcosmopolitan distribution, it is most common in southwest Asia and the Mediterranean region, China, and sub-Saharan Africa. During the past two decades, relationships and circumscription of constituent genera of Lamioideae have largely been clarified through both morphological (Abu-Asab and Cantino, 1992, 1994; Cantino, 1992a, b; Cantino et al., 1992; Ryding, 1994a, b, c, 1995, 1998, 2003, 2008; Salmaki et al., 2008; Xiang et al., 2013a; Seyedi and Salmaki, 2015) and molecular phylogenetic studies at various taxonomic levels (Wink and Kaufmann, 1996; Lindqvist and Albert, 2002; Scheen and Albert, 2007, 2009; Scheen et al., 2008, 2010; Bendiksby et al., 2011, 2014; Salmaki et al., 2012, 2013; Xiang et al., 2013b; Chen et al., 2014; Roy and Lindqvist, 2015; Li et al., 2016; Yao et al., 2016; Siadati et al., 2018). In particular, the molecular phylogenetics analyses of Scheen et al. (2010), Bendiksby et al. (2011), and Zhao et al. (2021) have dramatically improved our understanding of both tribal classification and character evolution within Lamioideae. Systematic positions of several enigmatic genera which were previously unplaced within Lamioideae have been recently elucidated (Scheen et al., 2010; Bendiksby et al., 2011; Chen et al., 2014; Roy and Lindqvist, 2015; Olmstead, 2016; Zhao et al., 2021), while a few genera, namely the rare and monotypic Paralamium Dunn. and Metastachydium Airy Shaw ex C.Y. Wu & H.W. Li, and Roylea Wall. ex Benth., remain unclassified at the tribal level because of insufficient molecular data available to date. The aforementioned Paralamium and Metastachydium have not been included in any published molecular phylogenetic study.
The genus Paralamium was originally described by Dunn (1913) and reported to be endemic to southeast Asia with a sporadic distribution in humid regions of southwestern China (subtropical Yunnan), northern Vietnam, northern Burma, and eastern India (Assam) (Li and Hedge, 1994; Harley et al., 2004; Suddee and Paton, 2004). The genus is distinguished from other Lamioideae genera mostly based on calyx morphology. Paralamium has unequal calyx-lobes, with the posterior calyx tooth being the largest and having a truncate apex flanked by smaller triangular lateral lobes, and lanceolate-triangular anterior lobes (Figure 1). Harley et al. (2004) called this unique calyx morphology a 1/2/2 split, while Li and Hedge (1994) recognized this shape as a 1/4 split. In addition, this genus is characterized by possessing very small pollen grains with the polar length and/or equatorial width less than < 18 μm (Harley et al., 2004), which is an uncommon feature within Lamiaceae.
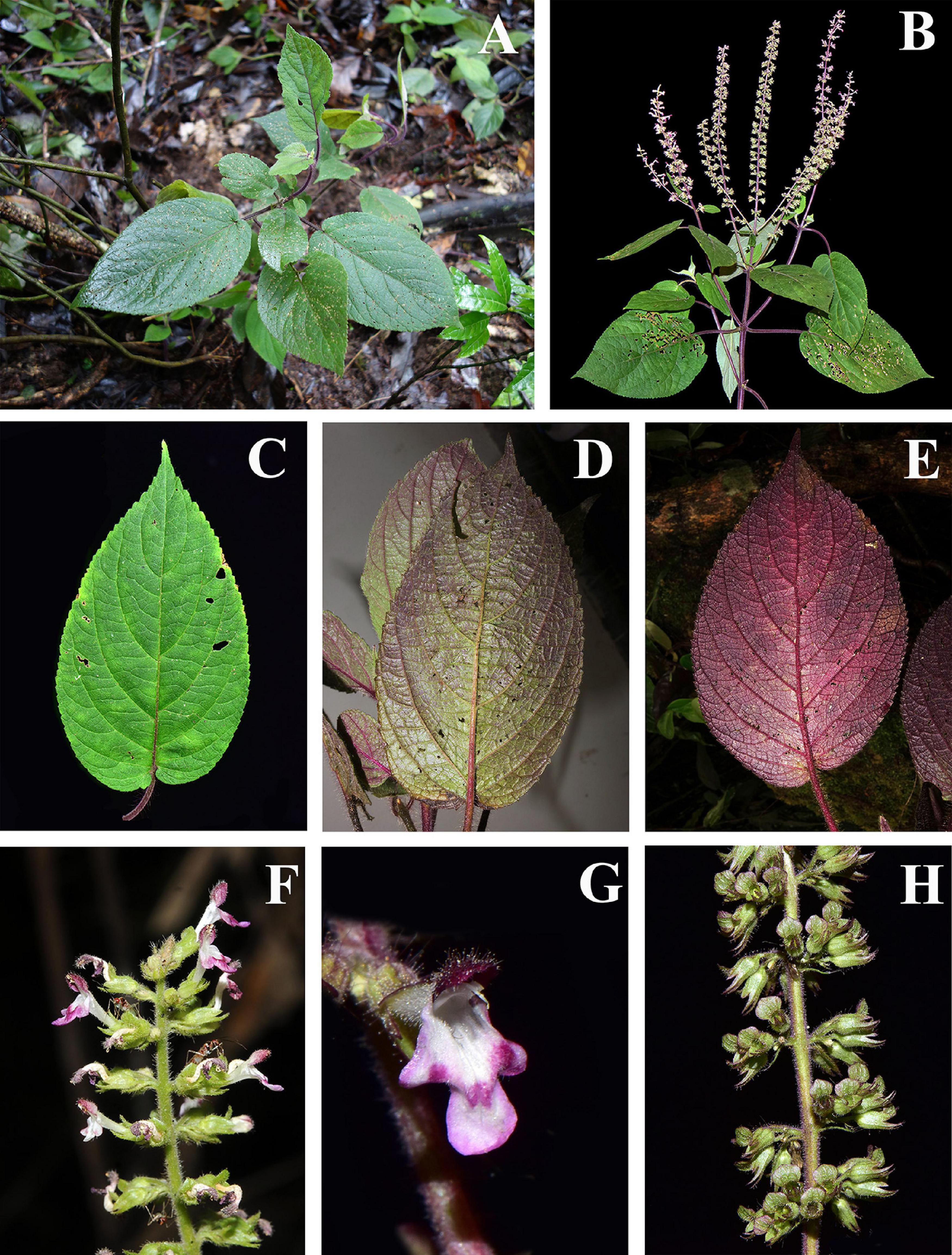
Figure 1. Morphology of Paralamium griffithii. (A) Habitat; (B) plant and inflorescence; (C–E) Leaf morphology, showing the color variation of leaf blades; (F) Lateral review of inflorescence; (G) Frontal view of flower; (H) Lateral of calyx. (Photo by XX Zhu).
Paralamium is monotypic, with the sole species, P. gracile Dunn (1913) described on the basis of a specimen collected from Yunnan, China (Henry 10636). However, before the description of this species, Hooker (1885) described Plectranthus griffithii Hook.f. based on a collection from Eastern Assam, India (Herb Griffith 4056). After careful examination of the type materials, Suddee and Paton (2004) suggested that Plectranthus griffithii Hook.f. and Paralamium gracile Dunn. were conspecific. Thus, they formerly transferred the former species to Paralamium and a new combination, Paralamium griffithii (Hook.f.) S. Suddee & A.J. Paton, was created, making the latter species (Paralamium gracile) as a synonym.
The systematic position of Paralamium has been enigmatic ever since its original description. When establishing the genus, Dunn (1913) noted that the calyx is the “most striking” character of Paralamium and similar to Orthosiphon Benth. (Nepetoideae), Coleus Lour. (Nepetoideae) and Teucrium L. (Ajugoideae) by virtue of the following calyx characters: a broad upper calyx tooth with recurved decurrent margins and a conspicuously veined calyx tube. However, in the protologue for Paralamium (Dunn, 1913), the genus was also considered to be closely related to Lamium L. (Lamioideae) based on nutlet and corolla characters, hence the name “Paralamium” which can be translated to mean “resembling Lamium.” Studies on the genus after its original description have been scarce. Li (1977) placed Paralamium within subtribe Lamiinae of tribe Lamieae in subfamily Lamioideae sensu Briquet (1895–1897) based on its morphology provided in the protologue (Dunn, 1913). Later, Cantino and Sanders (1986) considered Paralamium as an anomalous genus within Lamiaceae because of its morphology similar to various genera in different subfamilies, but discreetly suggested that it could probably be related to Lamium based on their similar tricolpate and two-celled pollens observed by Abu-Asab and Cantino (1994). Harley et al. (2004) also placed Paralamium within Lamioideae in their comprehensive classification of Lamiaceae. In the most recent classifications of Lamioideae based on molecular data (Scheen et al., 2010; Bendiksby et al., 2011), Paralamium was provisionally treated as incertae sedis within Lamioideae but additionally suggested to be a member of tribe Pogostemoneae based on nutlet morphology (e.g., small glossy nutlets) (Bendiksby et al., 2011). While in the updated online synoptical classification of Lamiales, Olmstead (2016) placed Paralamium within tribe Stachydeae of Lamioideae. However, Paralamium has never been included in a published molecular phylogenetic analysis, making the above empirical placement of Paralamium within Lamioideae untested.
The main reason that Paralamium has not be included in any molecular phylogenetic studies is a lack of suitable leaf tissue for DNA extraction. However, during collecting expeditions in the Yunnan province of China in 2018 and 2019, we discovered two populations of P. griffithii. These collections allowed us to investigate the phylogenetic position of this monotypic and enigmatic genus based on molecular data. Here, using both plastid and nuclear ribosomal DNA markers, we present molecular phylogenetic analyses using different sampling strategies to finally establish the tribal affinities of Paralamium within Lamioideae and provide an updated phylogeny of the tribe Pogostemoneae. Furthermore, we provide a dichotomous key for genera within Pogostemoneae.
Materials and Methods
Field Collections
Specimens from two populations of Paralamium griffithii were collected from Malipo County (Liu et al. 7859) and Jinping County (Z.Y. Cai and X.E. Ye czy-36) within the Yunnan Province of China. Fresh leaves were collected and dried with silica gel. Voucher specimens were deposited in the Herbarium of Kunming Institute of Botany (KUN), Chinese Academy of Sciences.
Taxon Sampling and Genetic Markers Selected
In order to better evaluate the systematic position of Paralamium and assess the phylogenetic relationships of this enigmatic genus and related genera, we experimented with three datasets. The first dataset included 79 plastid protein-coding genes within Lamiaceae (dataset CP79) aiming to confirm the subfamilial position of Paralamium. In total, 84 accessions from 84 species and 63 genera of Lamiaceae were included for this initial analysis, covering 11 of the 12 subfamilies recognized by Li et al. (2016) and Li and Olmstead (2017). The plastome of P. griffithii (Z.Y. Cai and X.E. Ye czy-36) was newly sequenced for this dataset. Outgroups of the dataset CP79 were selected from Mazaceae [Mazus pumilus (Burm. f.) Steenis)], Wightiaceae (Wightia speciosissima (D. Don) Merr.), Phrymaceae (Phryma leptostachya L. subsp. asiatica H. Hara), Paulowniaceae (Paulownia coreana Uyeki), and Orobanchaceae (Castilleja paramensis F. González et Pabón-Mora), according to recent Lamiales-wide phylogenies (Refulio-Rodriguez and Olmstead, 2014; Liu et al., 2020). GenBank accession numbers and the source publications for taxa in this dataset are provided in Supplementary Table 1. We used the phylogenetic results from this first set of analyses as a basis for a more focused second round of analyses.
Because the first set of analyses demonstrated that Paralamium has affinities with tribe Pogostemoneae of Lamioideae, we expanded the sampling of Pogostemoneae in a second round of analyses. These analyses focused on further exploring the placement of Paralamium within Pogostemoneae and explicating relationships among genera of the tribe. Chen et al. (2014) demonstrated that the monotypic genus Holocheila (Kudô) S. Chow is a member of Pogostemoneae, so we also included this genus for analysis. In total, for the first time, all 12 genera (including Paralamium) of Pogostemoneae were included as part of our Pogostemoneae-wide analyses. This comprehensive generic sampling offers the opportunity to clarify generic relationships of Pogostemoneae using five plastid regions (matK, rbcL, rps16, trnH-psbA, trnL-trnF; dataset CP5) and the nuclear ribosomal internal transcribed spacer (dataset nrITS). In total, 56 sequences were newly sequenced for 13 species in 8 genera, while others were taken from previous studies (Chen et al., 2014; Yao et al., 2016) or downloaded from GenBank (Table 1). Outgroups for the dataset CP5 and the dataset nrITS were sampled from tribe Gomphostemmateae (Chelonopsis souliei (Bonati) Merr., Gomphostemma lucidum Wall. ex Benth., and Gomphostemma sp.) according to Yao et al. (2016).
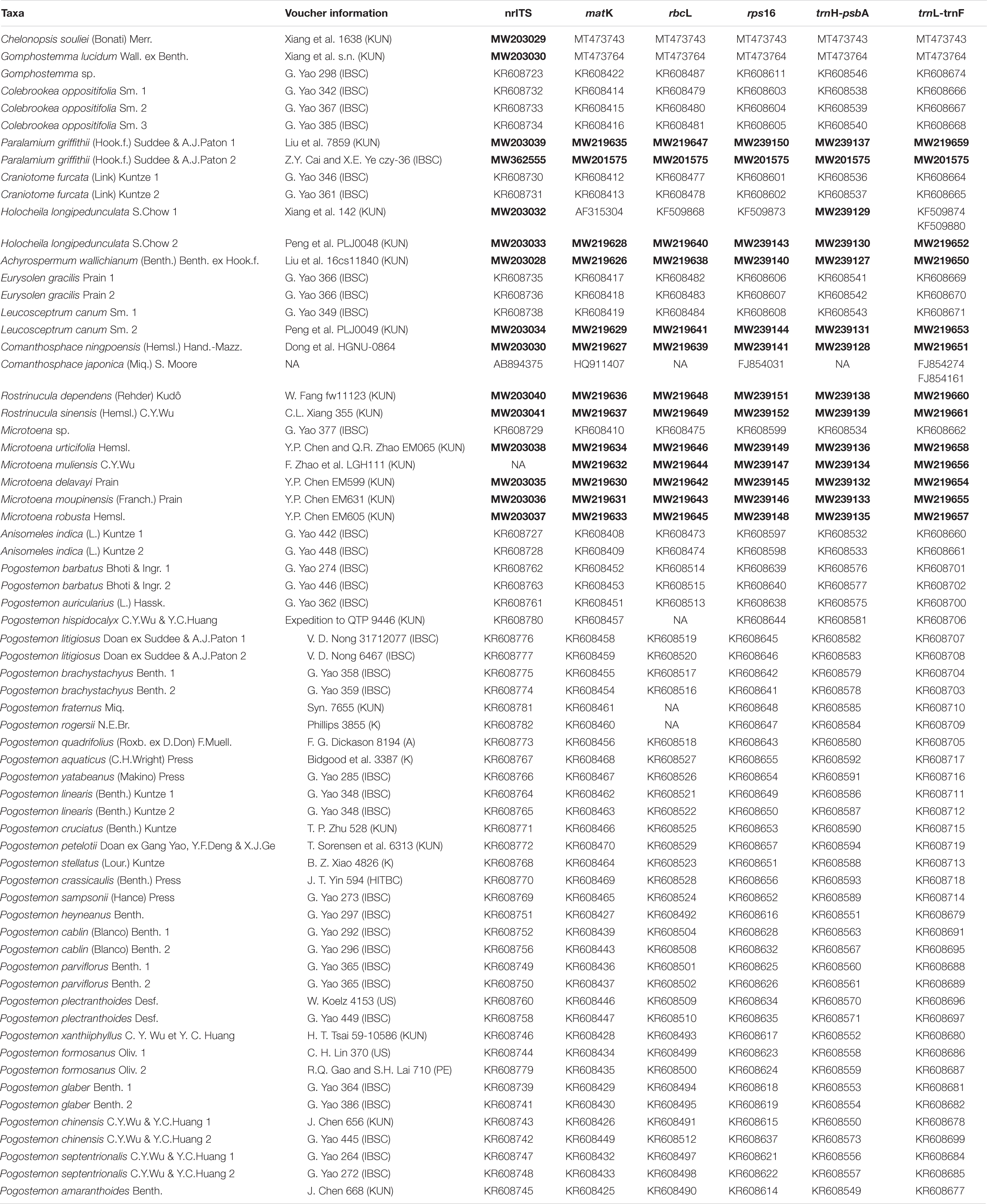
Table 1. The voucher information of the taxa from Tribe Pogostemoneae analysis in this study, the GenBank accession numbers of the new sequenced are shown in the bold, other sequence were from the previous studies (NA = not available).
DNA Extraction, Amplification, and Sequencing
Total genomic DNA was extracted from fresh or silica-gel-dried leaf fragments using the CTAB procedure of Doyle and Doyle (1987), then dissolved in double-distilled water and kept at −20°C for future polymerase chain reaction (PCR) amplification.
Primers and PCR thermal cycler settings for matK and rbcL followed Chen et al. (2014), and those for nrITS, trnL-trnF, rps16, and trnH-psbA were as described by Xiang et al. (2013b). Amplified PCR products were visualized on 1% TBE agarose gel, stained with ethidium bromide and then sequenced by an ABI-PRISM3730 sequencer after purification with a QIAquick PCR purification Kit (BioTek, Beijing, China). Voucher information for newly sequenced species and GenBank accession numbers for all sequences used in the current study are listed in the Table 1.
Plastome Sequencing, Assembly, Annotation, and Gene Region Extraction
The DNA concentration of Paralamium griffithii was at least 35 ng/μL as measured by a NanoDrop spectrophotometer 2000 (Thermo Scientific, Carlsbad, CA, United States). DNA integrity was detected and purified by 1% Agarose Gel Electrophoresis for 40 min at 150 V. Subsequently, the DNA samples were sheared into 300 bp fragments for paired-end library construction according to manufacturer’s instructions (Illumina, San Diego, CA, United States), details are provided in Zhao et al. (2020a).
Prior to genome assembly, adapter sequences and low-quality reads were removed using the ea-utils package1. Quality control of raw sequence reads was carried out using FastQC 0.11.8 (Andrews, 2018) with the parameter set as Q ≥ 25. We used the GetOrganelle pipeline (Jin et al., 2020) for the de novo assembling. The software Bandage v. 0.8.1 (Wick et al., 2015) was employed for contig visualization and editing. Lastly, in order to validate the assembly error, the raw reads were mapped to the assembled plastid genome sequences by the Bowtie2 (Langmead and Salzberg, 2012) plugin in Geneious v. 11.0.3 (Kearse et al., 2012). In addition to the newly sequenced plastome of Paralamium griffithii and downloaded plastomes of 54 species from GenBank (Supplementary Table 1), 32 data from the Sequences Read Archive (SRA) were included for reassembling.
The Initial annotations were implemented in the Plastid Genome Annotator (PGA) (Qu et al., 2019), and the published plastome of Phlomoides betonicoides (Diels) Kamelin & Makhm (MN617020; Zhao et al., 2020b) was set as a reference, then Geneious v.11.0.3 (Kearse et al., 2012), and tRNAscan-SE service (Lowe and Chan, 2016) were used adjusting of the putative starts, stops, intron positions, and tRNA boundaries as described by Zhao et al. (2021) and Xiang et al. (2020). Finally, the circular physical map of the plastome of Paralamium (Supplementary Figure 1) was drawn by the Organellar Genome DRAW tool (Lohse et al., 2013). The coding regions (CR) were extracted from the annotated complete plastome sequences for phylogenetic analyses.
Sequence Alignment and Phylogenetic Analyses
Sequences were assembled and edited using Geneious v.11.0.3 (Kearse et al., 2012). Sequences were aligned using MAFFT v.7.221 (Katoh and Standley, 2013), and then adjusted manually in PhyDE v.0.9971 (Müller et al., 2010) for minor corrections. All datasets were submitted to TreeBASE (study ID: S27475).
Since topological incongruence between the combined cpDNA and nrITS data was reported in Yao et al. (2016), the nrITS and cpDNA datasets were not combined for analyses here. However, because plastome regions typically have a shared genetic history, the five plastid DNA regions were combined for phylogenetic analyses. All datasets were analyzed using Maximum Likelihood (ML) and Bayesian Inference (BI) algorithms on the CIPRES Science Gateway (2 Miller et al., 2010). The ML analyses were implemented with RAxML v.8.2.9 (Stamatakis, 2014), bootstrap probabilities were generated by conducting 1000 bootstrap iterations, and details for parameter settings are described by Xiang et al. (2020). Bayesian inference analyses were performed using MrBayes v.3.2.2 (Ronquist et al., 2012). The best-fit nucleotide substitution models were selected under the Akaike Information Criterion (AIC) using jModelTest v.3.7 (Posada, 2008). The models used were the GTR+I+G for dataset CP79, TVM+I+G for dataset CP5, and for the nrITS dataset. In addition, a partitioned strategy for the dataset CP5 also used for Bayesian inference analyses (GTR + G for matK, GTR + I for rbcL, TVM + G for rps16, TPM1uf + G for trnH-psbA, GTR + I for trnL-trnF). Specific steps for analyses are described in detail in Chen et al. (2016) and references provided therein. Finally, we used FigTree v.1.4.2 (Rambaut, 2014) to visualize and edit all resulting trees. We defined branches with posterior probabilities (PP) ≥ 0.95 and bootstrap values (BS) ≥ 80% as strongly supported, PP = 0.90–0.95 and BS = 70–80% as moderately supported, while PP < 0.90 and BS < 70% were defined as weakly supported.
Nutlets Morphology
Mature nutlets were collected from both wild-collected or herbarium plant specimens from the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, for light microscope (LM) and scanning electron microscope (SEM) observation. With the exception of the monotypic Holocheila, nutlet morphology was examined for all genera of Pogostemoneae. Based on previous studies (Yao et al., 2016; Wang, 2018) and our phylogenetic analyses, three species representing both major clades of Pogostemon were selected, and four species from Microtoena were sampled. For each species, at least five mature nutlet samples were examined. In total, nutlets of 17 species representing 11 out of 12 genera of Pogostemoneae were included for morphological investigation. Measurements and LM investigation were done with the Keyence VHX-6000 digital microscope. For SEM examination, mericarps were directly affixed to stubs with double-sided tape and the sputter-coated with gold-palladium. Observations were conducted using ZEISS EVO LS10 scanning electron microscope (Carl ZEISS NTS, Germany) with 10 kV voltage (Kunming Institute of Botany, Yunnan, China). Nutlets terminology followed Moon et al. (2009).
Results
Genome Assembly, Features and Gene Content of Paralamium griffithii
The newly sequenced and annotated plastome was submitted to the National Center for Biotechnology Information (NCBI) database with the accession number MW201575. Illumina paired-end sequencing generated 20,321,882 clean reads, with coverage of 179 × for P. griffithii. The plastome size was 152,664 bp and displayed the typical quadripartite structure consisting of a pair of IR regions (25,617 bp) separated by the large single copy (LSC; 83,788 bp) and small single copy (SSC; 17,642 bp) regions (Supplementary Figure 1). In total, 114 unique genes (80 protein-coding genes, 30 tRNAs, and 4 rRNAs; Supplementary Table 2) were identified (duplicated genes in IR regions were counted only once). We used 79 common protein-coding genes for phylogenetic analyses based on Zhao et al. (2021) with the exclusion of the ycf15 gene because it could not be extracted from most plastome reassembed from SRA database.
Sequence Characterization
Properties for different datasets are summarized in Table 2. The aligned length of the combined 79 protein coding regions (CP79) was 70,100 bp. Removal of ambiguous sites and single taxon insertions resulted in an aligned length of 69,276 bp, of which 47,566 sites were constant (68.66%). The aligned regions and the excluded ambiguous sites of the individual loci are listed in Supplementary Table 3.
In the second set of analyses, the combined cpDNA dataset was 3,439 bp (832 bp for matK, 574 bp for rbcL, 880 bp for trnL-trnF, 861 bp for rps16, and 292 bp for trnH-psbA) after excluding ambiguously aligned characters. The nrITS matrix contained 656 aligned positions (Table 2).
Phylogenetic Analysis
For each combined dataset (CP79, CP5, and nrITS), ML and BI analyses yielded identical topologies, respectively (Figures 2–4; Supplementary Figures 2–8). Therefore, only the trees resulting from maximum likelihood analysis of each dataset are presented, with posterior probability values from BI analyses indicated.
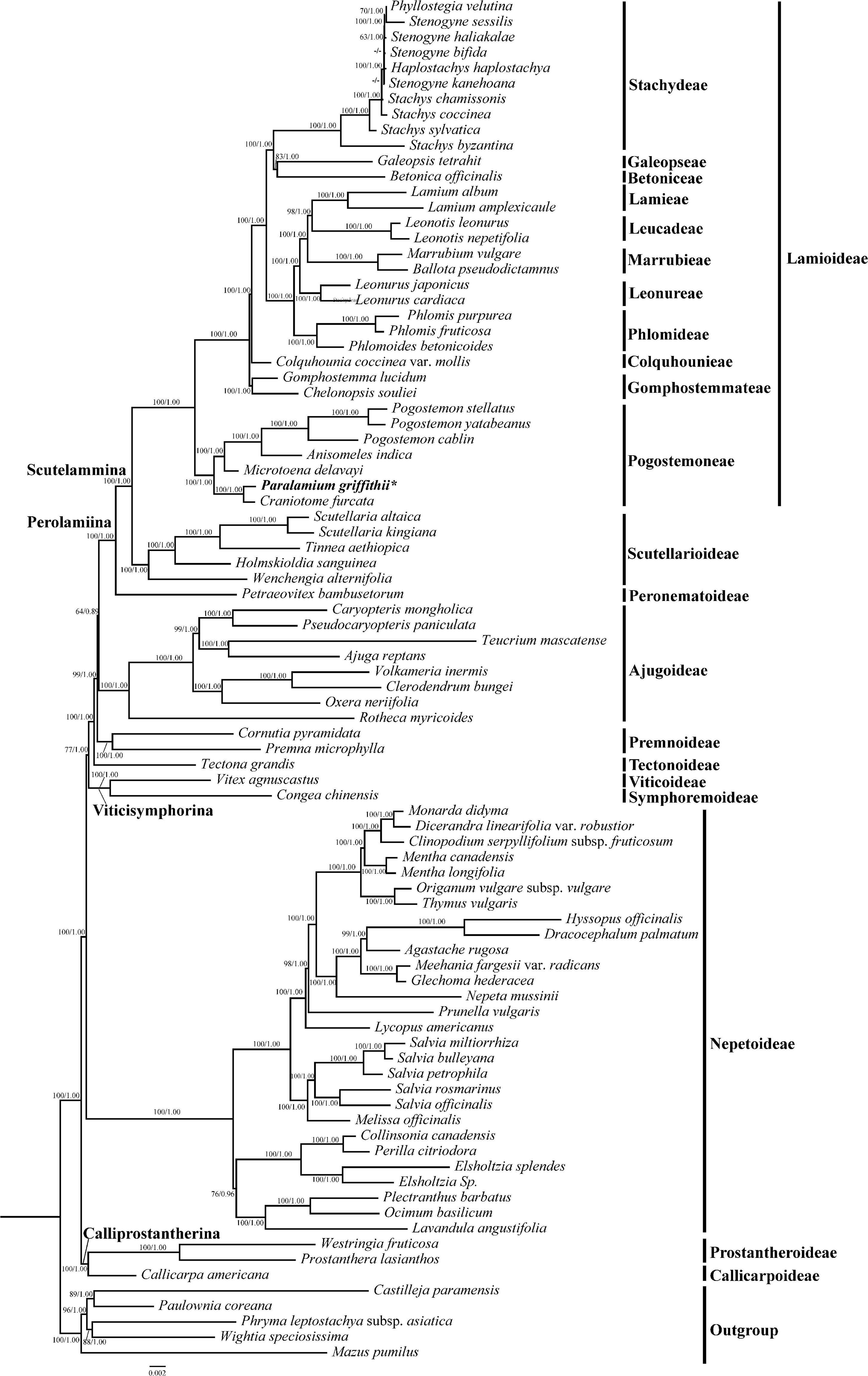
Figure 2. Phylogeny of Lamiaceae inferenced by maximum likelihood (ML) based on 79 coding regions (dataset CP79), with ambiguously aligned sites excluded from analysis. Bootstrap values ≥ 50% in ML and posterior probability values ≥ 0.90 in BI analyses displayed on the branch follow the order MLBS/BIPP (“-” indicates a support value BS < 50% or PP values < 0.9). Subfamilial classification of Lamiaceae is based on Li et al. (2016) and Li and Olmstead (2017).
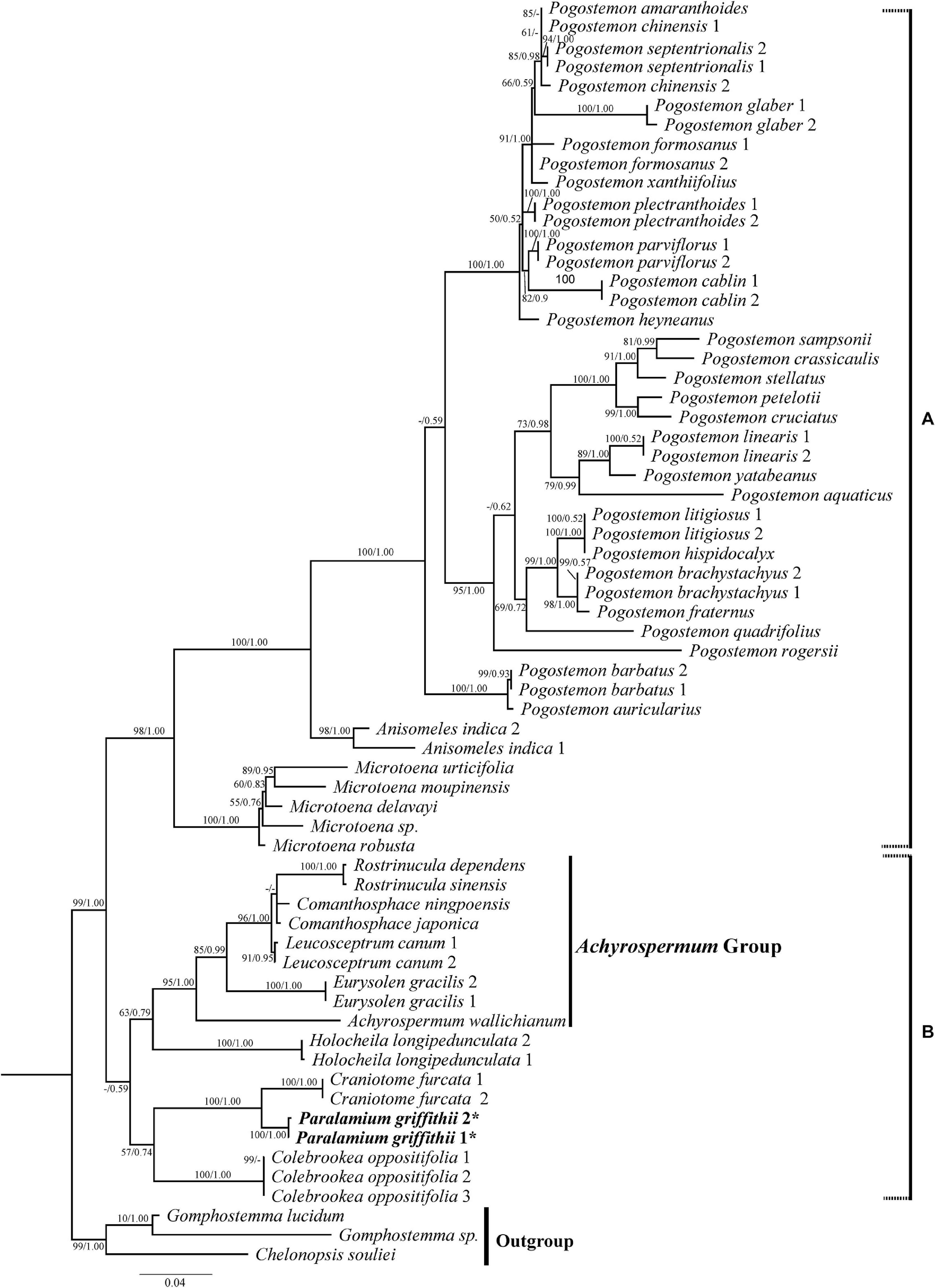
Figure 3. Phylogenetic relationships of Pogostemoneae based on the nrITS dataset. The support values (BS/PP) are indicated above branches. BS values < 50% and PP support < 90% indicated by -. The outgroup and other major groups are labeled at the right.
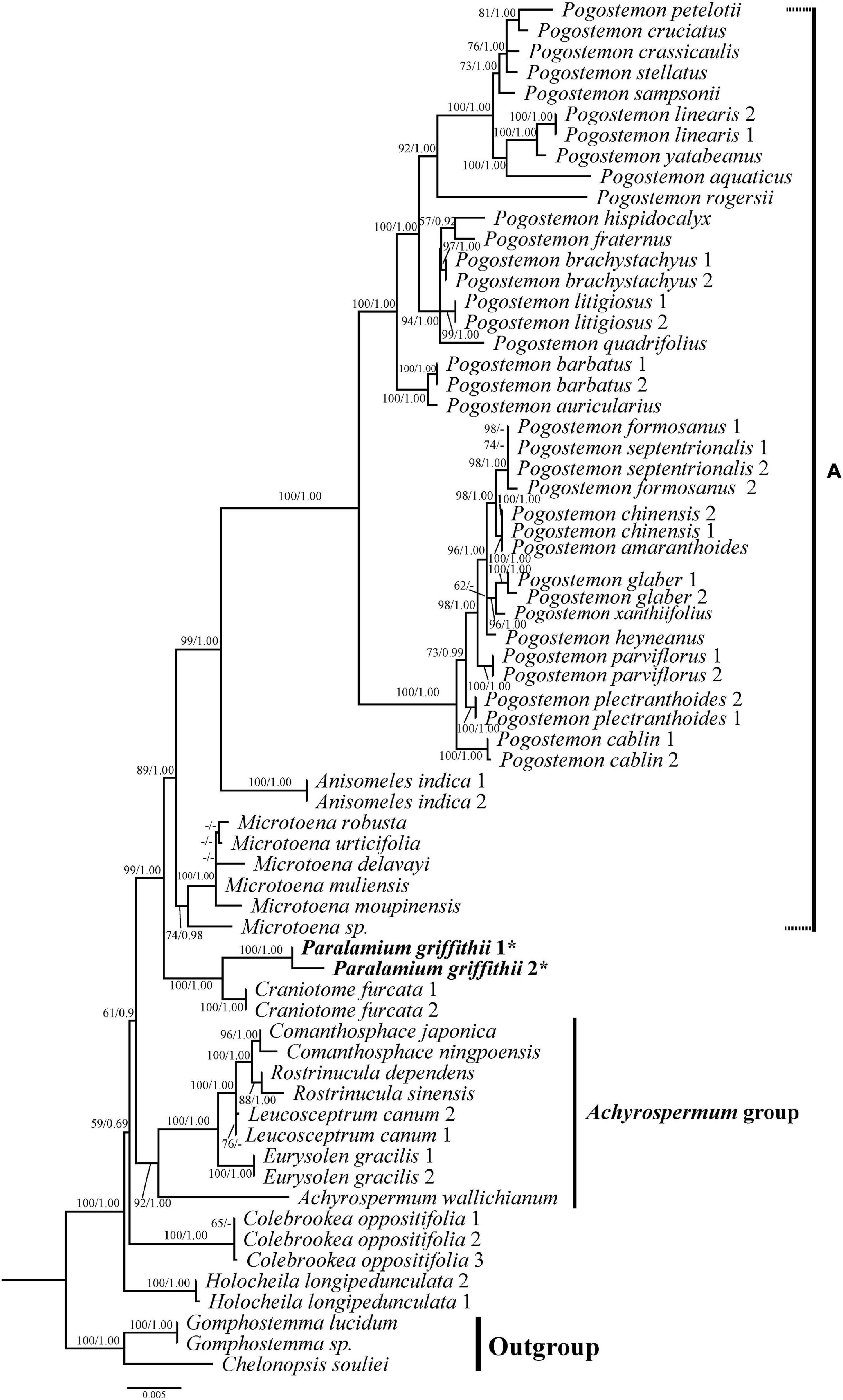
Figure 4. Phylogenetic relationships of Pogostemoneae based on the dataset CP5. The support values (BS/PP) are indicated above branches. BS values < 50% and PP support < 90% indicated by -. The outgroup and other major groups are labeled at the right.
In our phylogenetic analyses based on 79 coding plastome sequences (CP79), Lamiaceae are supported as monophyletic (Figure 2; ML-BS = 100%/BI-PP = 1.00; all support values follow this order hereafter) and subfamilial relationships are identical to those recovered by Zhao et al. (2021), and 11 tribes were recovered within Lamioideae (Figure 2). In all analyses, the focal species Paralamium griffithii was sister to Craniotome furcata (Link) Kuntze (100%, 1.00) within tribe Pogostemoneae of subfamily Lamioideae.
This recognition guided the second set of analyses, which aimed to further clarify the position of Paralamium, reassess generic relationships within Pogostemoneae, and update the phylogeny of Pogostemoneae by including as comprehensive taxon sampling as possible using both nrITS and cpDNA data. In all analyses, Pogostemoneae is robustly supported as monophyletic (Figures 3, 4), but the topologies differed between the nrITS and cpDNA phylogenetic trees. In the nrITS phylogeny, Pogostemoneae was found to have two major clades (labelled A and B in Figure 3). Clade A, or the Pogostemon group, includes Pogostemon Desf., Anisomeles R. Br., and Microtoena Prain, in which the former two genera formed a clade (100%, 1.00) sister to Microtoena (98%, 1.00). Clade B is poorly supported (59%, -) and includes nine genera. Clade B in turn is comprised of two subclades: one containing Colebrookea Sm., Paralamium + Craniotome Rchb., weakly supported (57%, -); and another subclade composed of Holocheila and the “Achyrospermum group” (i.e., Achyrospermum Blume, Eurysolen Prain, Leucosceptrum Sm., Comanthosphace S. Moore, and Rostrinucula Kudô), also poorly supported (63%, -).
All analyses based on the combined cpDNA dataset (CP5) also strongly supported the monophyly of Pogostemoneae (Figure 4; 100%, 1.00). At this point in the Pogostemoneae topology, the two samples of Holocheila formed a well-supported clade (100%, 1.00) and were recovered as sister to the remaining Pogostemoneae, which formed a weakly supported clade (59%, -). This “remaining Pogostemoneae” clade included Colebrookea (100%, 1.00), the Achyrospermum group (92%, 1.00), Paralamium + Craniotome (100%, 1.00), and Clade A (i.e., the Pogostemon group, 89%, 1.00).
Nutlets Morphology
The nutlets of the genera in clade A (Figure 3) are glossy and smooth (Figures 5, 6A–P) compared with those of genera in clade B (Figure 3). As reported previously (Ryding, 1994a; Bongcheewin et al., 2017), the nutlets of Pogostemon (Figures 5A–P) and Anisomeles indica (L.) Kuntze (Figures 5Q–T) are orbicular to subglobose, dark-brown to black, and the surface is very smooth (P. chinensis C.Y. Wu et Y.C. Huang, Figures 5A–D; P. glaber Benth., Figures 5E–H) or finely striato-reticulate (P. brachystachyus Benth., Figures 5I–L; P. amaranthoides Benth.; Figures 5M–P). In Microtoena (Figures 6A–P), nutlets are ovoid or subglobose, brown to black, glossy, and the surface is relatively smooth (M. delavayi Prain, Figures 6A–D; M. prainiana Diels, Figures 6E–H; M. stenocalyx C.Y. Wu et S.J. Hsuan, Figures 6M–P), or finely granulated (M. esquirolii H. Lév.; Figures 6I–L).
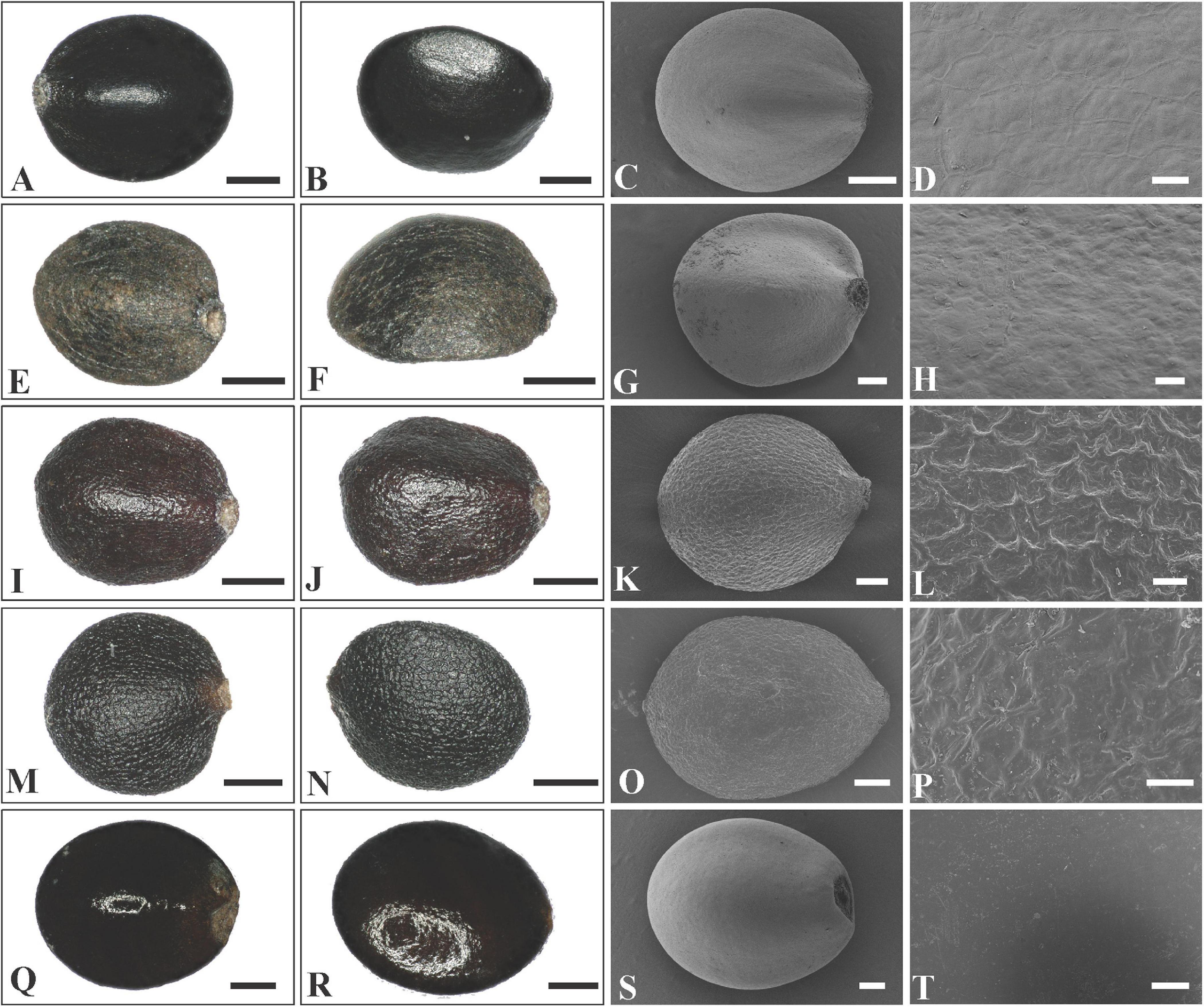
Figure 5. LM and SEM micrographs of mericarps of Pogostemon and Anisomeles. (A–D) P. chinensis (Guo et al., 12CS5297), (E–H) P. glaber (Li Yongliang, LiYL1637), (I–L) P. brachystachyus (Sun Xingxu, SunXX154), (M–P) P. amaranthoides (Sun Xingxu, SunXX148), (Q–T) Anisomeles indica (Liu et al., SCSB-B-0000020).— Scale bars: (A–C,E–F,I–J,M–N,S) 200 μm; (D,H,L,P) 20 μm; (G,K,O) 100 μm; (Q,R) 400 μm; (T) 40 μm.
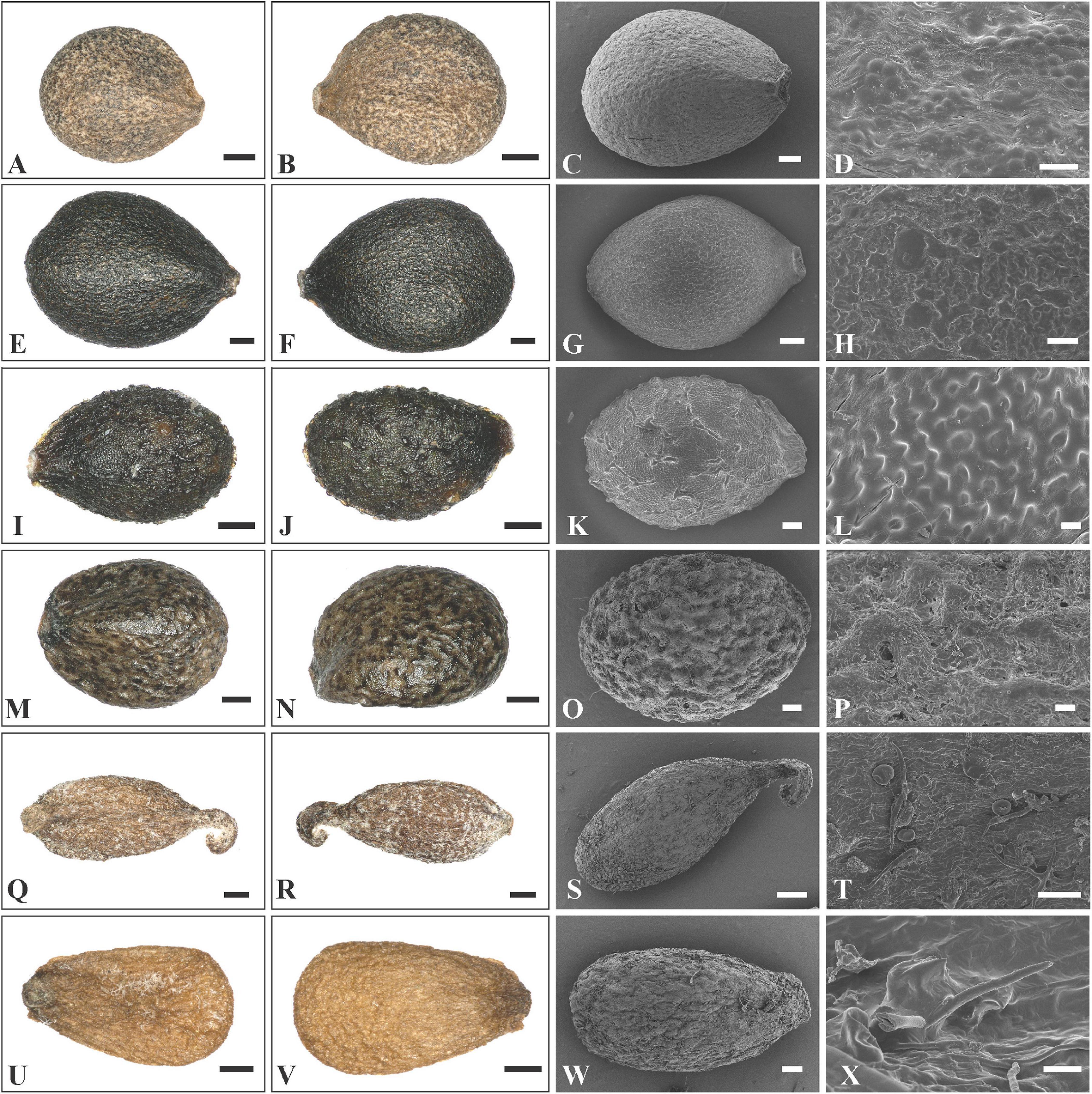
Figure 6. LM and SEM micrographs of mericarps of Microtoena, Rostrinucula, and Comanthosphace. (A–D) M. delavayi (Wang et al., SCSB-TBG-147), (E–H) Microtoena praineana (Yi Sirong and Tan Qiuping, YISR432), (I–L) M. esquirolii (Cai Jie and Zhang Ting, 12CS5726), (M–P) M. stenocalyx (Li et al., LiYL1849), (Q–T) Rostrinucula sinensis (Zhang Daigui, 3756), (U–X) Comanthosphace ningpoensis (Zhu et al., ZhuXX121). — Scale bars: (A–B,Q–S,U–V) 400 μm; (C,E–F,I–J,M–N,T,W) 200 μm; (D): 40 μm, (G,K,O) 100 μm; (H,L,P,X) 20 μm.
In Rostrinucula, the nutlets are narrowly ellipsoid with curved hook-like apices, brown, pubescent outside with glands and eglandular trichomes (R. sinensis (Hemsl.) C.Y. Wu, Figures 6Q–T). Nutlets of Comanthosphace are obovate, light brown, and the surface is rough and has subsessile and eglandular trichomes (C. ningpoensis (Hemsl.) Hand.-Mazz., Figures 6U–X). In Leucosceptrum canum Sm. the nutlets are oblong, brown, with sharp edges or ribs apically, and a surface more or less smooth but with sparse subsessile glands (Figures 7A–D). Nutlets of Eurysolen are also obovate, dark brown, dull, and densely glandular along the ventral side (Figures 7E–H). Only one species of Achyrospermum, A. wallichianum (Benth.) Benth. ex Hook. f., was included for this study. Achyrospermum wallichianum has somewhat elliptic light brown nutlets that are hairy at apex and reticulate on the surface (Figures 7I–L). Nutlets of Craniotome (Figures 7M–P) and Paralamium (Figures 7Q–T) are subspheric, brown and black respectively, and slightly reticulate outside. Nutlets of Colebrookea (Figures 7U–X) are obovoid to oblong, light brown, with apices and fruit navels densely covered with glands, and a surface that is smooth and sometimes with subsessile glands.
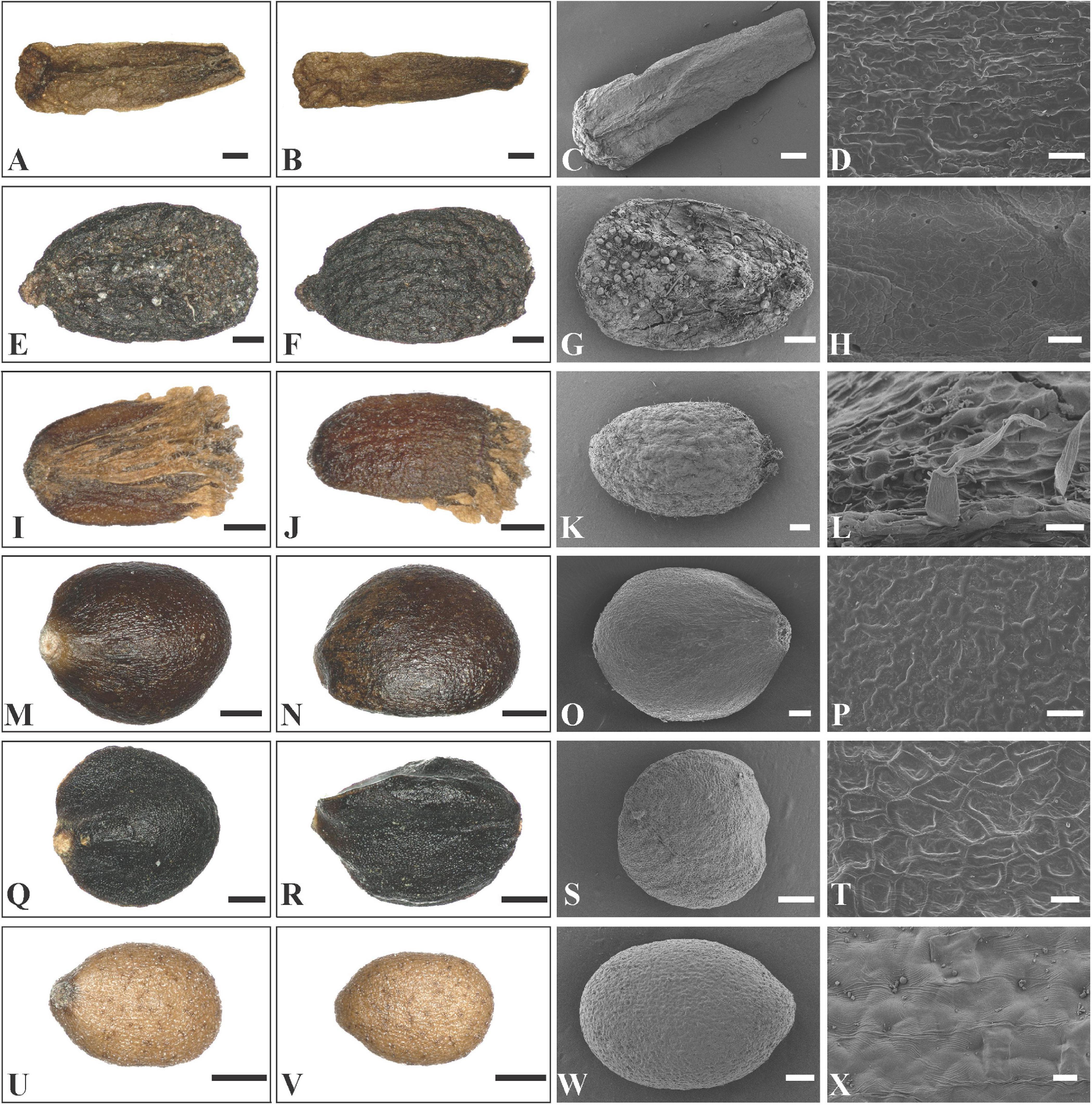
Figure 7. LM and SEM micrographs of mericarps of Leucosceptrum, Eurysolen, Achyrospermum, Craniotome, Paralamium and Colebrookea. (A–D) Leucosceptrum canum (Li Yongliang, YDDXS00058), (E–H) Eurysolen gracilis (Chen Yaping and Jiang Lei, EM1436), (I–L) Achyrospermum wallichianum (Liu et al., 16CS11840), (M–P) Craniotome furcata (Liu et al., 16CS11942), (Q–T) Paralamium griffithii (Liu et al., 7859), (U–X) Colebrookea oppositifolia (Li Yongliang, YDDXS1001). — Scale bars: (A–C,I,J) 400 μm; (D,H,L,P) 20 μm; (E–G,K,M,N,Q,R,U,V) 200 μm; (O,S,W) 100 μm; (T,X) 10 μm.
Discussion
Paralamium as a Member of Pogostemoneae in Subfamily Lamioideae
The resulting topologies of Lamiaceae from the dataset CP79 are consistent with that of previous studies (Li et al., 2016) based on five cpDNA regions and relationships among these subfamilies are well resolved. Moreover, all tribes of Lamioideae are strongly supported as monophyletic (Figure 2), which is in concordance with previous studies (Scheen et al., 2010; Bendiksby et al., 2011; Roy and Lindqvist, 2015; Zhao et al., 2021).
Cantino and Sanders (1986) considered Paralamium as an anomalous genus within Lamiaceae because of its morphological similarities to genera from various subfamilies (i.e. Orthosiphon and Coleus of Nepetoideae, Ajuga of Ajugoideae, and Lamium of Lamioideae). However, the presence of tricolpate and two-celled pollens in Paralamium suggested its placement within Lamioideae (Cantino and Sanders, 1986). The genus was suggested to be closely related to Pogostemoneae by Bendiksby et al. (2011) based on nutlet morphology, but they explicitly treated it as incertae sedis within Lamioideae due to the lack of molecular phylogenetic data. Here, both the plastid and nuclear DNA data (Figures 2–4) support that Paralamium is a member of tribe Pogostemoneae, and is sister to the monotypic genus Craniotome.
Previous studies based on cpDNA sequences showed that Craniotome grouped with Microtoena, Anisomeles, and Pogostemon (Scheen et al., 2010; Bendiksby et al., 2011; Chen et al., 2014). Using low-copy nuclear pentatricopeptide repeat (PPR) data, Roy and Lindqvist (2015) also recovered a close relationship among Craniotome, Anisomeles, and Pogostemon (Microtoena not sampled). In our analyses, however, Craniotome consistently grouped with Paralamium with high support values (Figures 2–4). Some morphological characters support the close relationship between Paralamium and Craniotome. For example, the size of pollen grains is very similar in Paralamium (16.3 × 15.0 μm) and Craniotome (16.6 × 14.9 μm), and are smaller than other lamioid genera (Abu-Asab and Cantino, 1994). Additionally, nutlet morphology supports the sister relationship between Paralamium and Craniotome. Nutlets in both genera are obovoid and glossy (Figures 7O,S) and have reticulate ornamentation on the surface (Figure 7P, × 750; Figure 7T, × 1200), while in other related genera in clade B (Figure 3), nutlets are oblong (Leucosceptrum, Figures 7A–C), hooked (Rostrinucula, Figures 6Q–S) or hairy (Achyrospermum, Figures 7I–J) at apex, or has eglaudular (Comanthosphace, Figure 6X) or glandular (Eurysolen, Figure 7G; Colebrookea, Figure 7X) trichomes. At the same time, Paralamium has some unique morphological characters, especially its unequal calyx lobs (i.e., 1/2/2 split), can differentiate it from other genera (calyx lobs 3/2 or 1/4 split, or (sub)equal) within Pogostemoneae.
Circumscription and Relationships Within Pogostemoneae
The monophyly of Pogostemoneae was supported by most studies (Scheen et al., 2010; Bendiksby et al., 2011; Chen et al., 2014) based on cpDNA sequences, but not by Roy and Lindqvist (2015) using PPR data, who revealed that genera of Pogostemoneae were included in two separate clades. The first clade was referred as the Achyrospermum group (i.e., subclade A in Figure 2 sensu Roy and Lindqvist, 2015), forming the first-diverging clade within Lamioideae. The second clade consist of Pogostemon, Anisomeles, and Craniotome (i.e., subclade B in Figure 2 sensu Roy and Lindqvist, 2015), forming the second diverging clade sister to remainder of Lamioideae.
In our analyses, the cpDNA datasets strongly support the monophyly of Pogostemoneae (Figure 2), and the monophyly of this tribe was recovered based on nrITS dataset, although only two genera were selected as outgroup (Figure 3). Based on the results from present as well as previous studies (Scheen et al., 2010; Bendiksby et al., 2011; Chen et al., 2014; Roy and Lindqvist, 2015), Pogostemoneae comprises 12 genera: Pogostemon, Anisomeles, Microtoena, Rostrinucula, Comanthosphace, Leucosceptrum, Eurysolen, Achyrospermum, Holocheila, Craniotome, Paralamium, and Colebrookea. Most genera are monotypic or oligotypic, excepting Pogostemon (80 spp.), Achyrospermum (ca. 25 spp.), and Microtoena (19 spp.). Morphologically, Pogostemoneae is a very heterogeneous group, and synapomorphies for the tribe are still unclear. However, some morphological and anatomical characters can be used to distinguish Pogostemoneae from other lamioid members. Most genera of Pogostemoneae possess small and relatively glossy nutlets with pericarps often lacking a sclerenchyma region (Ryding, 1994a, 1995), generally long-exserted stamens with bearded filaments, weakly 2-lipped corollas, and broad bracts (Scheen et al., 2010). Additionally, pollen grains of Pogostemoneae are typically smaller (less than 28 × 27 μm) than that of most genera of Lamioideae (Abu-Asab and Cantino, 1994).
In addition to the confirmation of the systematic position of Paralamium and sister relationship between Paralamium and Craniotome, some other well supported groups within Pogostemoneae are also recovered in this study, which enables us to further discuss the relationships within the tribe. Based on nrITS phylogeny (Figure 3), two subclades (i.e., clade A and clade B) can be recognized. Clade A is strongly supported and composed of three genera (Pogostemon, Anisomeles, and Microtoena), while clade B is composed of the remaining genera of Pogostemoneae. Although clade B is weakly supported (0.59, -), this split is supported by nutlet morphology. In the present study, nutlets of 17 species representing 11 out of 12 genera (except Holocheila) of Pogostemoneae were included for analyses. Based on our LM and SEM observations, we found that nutlets of genera in clade A (Figure 3; Pogostemon, Anisomeles, and Microtoena) are glossy and relatively glabrous (Figures 5, 6A–P), and the sclerenchyma region is very distinctive (Bendiksby et al., 2011), while genera in clade B (Rostrinucula, Comanthosphace, Leucosceptrum, Eurysolen, Achyrospermum, Paralamium, Craniotome) have dull and glandular nutlets (Figures 7Q–X), and the sclerenchyma region is often absent or indistinct (Ryding, 1994a, 1995).
Within clade A, Anisomeles is sister to Pogostemon, with Microtoena sister to the Anisomeles-Pogostemon clade (Figures 3, 4). The three genera form a clade referred as clade A, which was supported by previous molecular phylogenetic studies (Scheen et al., 2010; Bendiksby et al., 2011). Cantino (1990, 1992a, 1992b)) suggested a close relationship between Anisomeles and Pogostemon based on their bearded staminal filaments and lustrous pericarps, as well as the presence of minute glands with unicellular caps on the leaf epidermis. Later, Abu-Asab and Cantino (1994) found that the two genera have very similar pollen grains with regular polygonal lumina and large perforations (see also Bean, 2015).
The close relationship between Microtoena and the Pogostemon-Anisomeles clade has been reported in previous studies (Scheen et al., 2010; Bendiksby et al., 2011; Chen et al., 2014; Roy and Lindqvist, 2015). The three genera are similar in terms of calyx morphology, with the calyx splitting the upper two and bottom three lobes up to ca. 1/2 of its length. Furthermore, linear bracts are present in Anisomeles and most species of Microtoena, while lanceolate or ovate bracts can be found in some species of Microtoena and Pogostemon (Wang, 2018). Geographically, most of the species of clade A are distributed in tropical East Asia (Scheen et al., 2010), although some species occur on islands within the Pacific and West Indian Oceans (Anisomeles), Africa (Pogostemon), and the Himalayas (Craniotome and Pogostemon glaber).
Microtoena was shown to be polyphyletic in some studies (Bendiksby et al., 2011; Roy and Lindqvist, 2015) based on cpDNA regions, but our results recover it as monophyletic with convincing support (Figure 3). A possible reason for this discrepancy may be that only two species and three cpDNA markers (matK, trnL-trnF, rps16 intron) were used in previous studies. Wang (2018) included 11 species for the phylogenetic reconstruction of Microtoena. Though his study was based only on two cpDNA regions (matK, trnL-trnF), the monophyly of Microtoena was well supported, as in our present study using nrITS (Figure 3, 100%, 1.00) and additional cpDNA markers (Figure 4, 74%, 0.98). Microtoena is a poorly understood genus and was previously placed within Stachydeae (Prain, 1889; Briquet (1895–1897)). Although recent molecular phylogenetic studies (Bendiksby et al., 2011; Yao et al., 2016; Zhao et al., 2021) confirmed its placement within Pogostemoneae, corroborating the taxonomic treatment of Harley et al. (2004), species relationships within Microtoena remain unresolved.
Another subclade (i.e., Achyrospermum group) composed of Achyrospermum, Eurysolen, Leucosceptrum, Rostrinucula, and Comanthosphace is also strongly supported in both the nrITS (Figure 3) and cpDNA trees (Figure 4), among which Rostrinucula and Comanthosphace are consistently resolved as sister genera (Figures 3, 4). The Achyrospermum group was first reported by Bendiksby et al. (2011) using cpDNA markers and subsequently recovered by Roy and Lindqvist (2015) based on the PPR region, but neither of them sampled Leucosceptrum. Species of the Achyrospermum group are distributed mainly in tropical East Asia and share several morphological characters. For example, the sclerenchyma region in the fruit pericarp is present in most lamioid members (Ryding, 1995; Bendiksby et al., 2011), but is obsolete, indistinct, or absent in the Achyrospermum group (Ryding, 1994b, 1995). Moreover, genera in this subclade have dull and glandular nutlets (Figures 6Q–X, 7A–L), while other genera within Pogostemoneae have glossy and glabrous nutlets (Bendiksby et al., 2011). Stamens long-exserted from the corolla are rare in Lamioideae, and are restricted to Comanthosphace, Rostrinucula, and Leucosceptrum in the Achyrospermum group, as well as a few species of Pogostemon in clade A. As suggested by Scheen et al. (2010), this character may be a synapomorphy for the small clade consisting of Comanthosphace, Rostrinucula, and Leucosceptrum. Molecular phylogenetic and morphological studies based on a broader sampling and more DNA sequences may further help to elucidate relationships within Pogostemoneae and identify morphological synapomorphies for the tribe.
Incongruence Between Nuclear and Plastid Phylogenies
In this study we provide the first comprehensive molecular phylogenetic study of Pogostemoneae. Though the intergeneric relationships within this tribe are generally well resolved, the placement of four monotypic genera (Colebrookea, Holocheila, Paralamium and Craniotome) is still uncertain due to incongruent topologies between nrITS and cpDNA trees. In the nrITS phylogeny (Figure 3), the Paralamium-Craniotome clade is sister to Colebrookea but weakly supported (57%, -). The Paralamium-Craniotome-Colebrookea clade is then sister to a clade including Holocheila and the Achyrospermum group, which is also weakly supported (-, 0.90) again. In the cpDNA tree (Figure 4), however, Holocheila is the first diverging clade, followed by Colebrookea, the Achyrospermum group, and then Paralamium-Craniotome + clade A, which is largely consistent with the topology of Chen et al. (2014). Most genera in clade B (excepting Achyrospermum, 25 spp.), all other genera are monotypic (Colebrookea, Craniotome, Eurysolen, Holocheila, Leucosceptrum, Paralamium, Rostrinucula) or oligotypic (Comanthosphace, 4 spp.) and mainly distributed in East Asia.
Incongruence between genomes have been noted within several genera in Lamiaceae, and ancient hybridization and chloroplast capture has often been posited to have contributed to the discordance (e.g., Albaladejo et al., 2005; Drew and Sytsma, 2013; Drew et al., 2014; Deng et al., 2015; Walker et al., 2015; Hu et al., 2018). Roy and Lindqvist (2015) suggested ancient reticulation events are likely to be responsible for the discordance between the plastid and PPR topologies of Pogostemoneae. They also demonstrated that ancestors of Pogostemoneae may have undergone rapid diversification during the middle Miocene in East Asia, which may have been triggered by climatic changes resulting from the uplift of the Qinghai-Tibetan Plateau (QTP) (Roy and Lindqvist, 2015). Considering that incomplete lineage sorting (ILS) among taxa is often associated with rapid radiations (Enard and Paabo, 2004; Pollard et al., 2006), ILS may also be a cause of the incongruences between the nuclear and plastid trees of Pogostemoneae. In the present study, two clades (clade A and clade B) are recognized based on nrITS phylogeny, but clade B is weakly supported by nrITS data and not recovered using cpDNA data. Although nutlet morphology supported the division of these two clades, futures studies involving next-generation sequencing and increased taxon sampling are need to provide insights into the complex evolutionary history of this group.
Key to All Genera of Pogostemoneae
The following circumscription of Pogostemoneae is based on this as well as previous studies (Scheen et al., 2010; Bendiksby et al., 2011). We provide a key to the 12 genera of Pogostemoneae below.
1 Creeping herb; corolla with two entire lips (1/1)…………………………………… Holocheila
1 Shrub, subshrubs or erect herb; corolla 2-lipped, 4-lobed (1/3, 1/3 or 2/3)…………………………………… 2
2 Calyx 5-lobed, lobes unequal (1/2/2), posterior lip very broad ………………………………… Paralamium
2 Calyx 5-lobed unequal (3/2 or 1/4) or (sub)equal …… 3
3 Flowers dioecious with dimorphic male and female flowers ………………………………………… Colebrookea
3 Flowers monoecious ……………………………… 4
4 Corolla-tube longer than 1.5 cm …………… Microtoena
4 Corolla-tube less than 1 cm long …………………… 5
5 Nutlet narrowly ellipsoid, hooked at apex … Rostrinucula
5 Nutlet not narrowly ellipsoid, unhooked at apex ……… 6
6 Filaments usually bearded along center with moniliform hairs …………………………………… Pogostemon
6 Filaments without moniliform hairs ………………… 7
7 Stamens long-exserted from corolla ………………… 8
7 Stamens not or shortly exserted from corolla ………… 9
8 Shrub or small tree; nutlets cylindrical-oblong ……………………………………… Leucosceptrum
8 Rhizomatous perennial herbs; nutlets obovate …………………………………… Comanthosphace
9 Nutlets scaly at apex ………………… Achyrospermum
9 Nutlets never scaly at apex ………………………… 10
10 Anthers 1-celled; corolla tube saccate in front … Eurysolen
10 Anthers 2-celled; corolla tube not saccate in front …… 11
11 Verticillasters in dense or interrupted, long, terminal spikes ………………………………………… Anisomeles
11 Cymes pedunculate, helicoid or sometimes dichotomous, in axillary or terminal panicles ………………………………… Craniotome
Conclusion
This study confirms the systematic placement of Paralamium for the first time inferred from chloroplast and nuclear DNA data. Paralamim is a member of the tribe Pogostemoneae within Lamioideae and is sister to Craniotome. As currently defined, the tribe Pogostemoneae is composed of 12 genera, and the monophyly of Pogostemoneae is supported in all analyses. Phylogenetically, Pogostemoneae are the first diverging tribe and sister to the remaining Lamioideae. Morphologically, Pogostemoneae are a remarkably diverse group and lack clear synapomorphies. Although some well-supported groups were identified within Pogostemoneae, relationships of some monotypic genera (e.g., Holocheila, Colebrookea, Paralamium and Craniotome) remain unclear. Thus, studies using broad sampling of low-copy and/or single-copy intrageneric phylogenies and detailed comparative morphological investigation are needed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FZ, C-LX, BD, and BL conceived the idea and designed the research. Y-WW, FZ, and Y-PC conducted experiments. E-DL and JC conducted specimen and seed collection. FZ, Y-WW, Y-PC, GY, BD, JC, E-DL, BL, and C-LX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was jointly supported by the “Ten Thousand Talents Program of Yunnan” (Grant No. YNWR-QNBJ-2018-279), Yunnan Fundamental Research Projects (Grant No. 2019FI009), the CAS “Light of West China” program), and the Large-scale Scientific Facilities of the Chinese Academy of Sciences (2017-LSFGBOWS-02) granted to C-LX, and NSF-DEB grant DEB-1655611 granted to BD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Xing-Er Ye of South China Botanical Garden, Chinese Academy of Sciences, Dr. Hong-Jin Dong of Huanggang Normal University, Hubei, for their help in field collection, and Dr. Xin-Xin Zhu of Xinyang Normal University for providing photos. We gratefully thank Mr. Lian-Yi Li and Mr. Zhi-Jia Gu for their technical assistance in LM and SEM. Thanks are also extended to the staff Germplasm Bank of Wild Species in Southwest China for their valuable help in research facilities and providing seed materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.646133/full#supplementary-material
Supplementary Figure 1 | Gene map of the complete chloroplast genome of Paralamium griffithii. Genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional categories are color-coded.
Supplementary Figure 2 | Maximum likelihood phylogeny of Lamiaceae based on 79 chloroplast DNA regions (dataset CP79) of coding regions dataset, ambiguous sites were excluded for analyses. Maximum likelihood bootstrap support (MLBS) are near the branches. A “–” indicates MLBS values < 50%.
Supplementary Figure 3 | Phylograms from Bayesian Inference (BI) analyses of Lamiaceae based on concatenated nucleotide sequences of 79 protein-coding genes (dataset CP79), ambiguous sites were excluded for analyses. Bayesian inference posterior probability (BIPP) are near the branches. A “–” indicates BIPP values < 0.8.
Supplementary Figure 4 | Maximum likelihood phylogeny of Pogostemoneae based on the nrITS region, ambiguous sites were excluded for analyses. Maximum likelihood bootstrap support (MLBS) are near the branches. A “–” indicates MLBS values < 50%.
Supplementary Figure 5 | Phylograms from Bayesian Inference (BI) analyses of Pogostemoneae based on the nrITS region, ambiguous sites were excluded for analyses. Bayesian inference posterior probability (BIPP) are near the branches. A “–” indicates BIPP values < 0.8.
Supplementary Figure 6 | Maximum likelihood phylogeny of Pogostemoneae based on the combined dataset of five cpDNA regions (matK, rbcL, rps16, trnH-psbA, and trnL-trnF), ambiguously aligned sites were excluded from analyses. Maximum likelihood bootstrap support (MLBS) are near the branches. A “–” indicates MLBS values < 50%.
Supplementary Figure 7 | Phylograms from Bayesian Inference (BI) analyses of Pogostemoneae based on the combined dataset of five cpDNA regions(matK, rbcL, rps16, trnH-psbA, and trnL-trnF), ambiguously aligned sites were excluded from analyses. Bayesian inference posterior probability (BIPP) are near the branches. A “–” indicates BIPP values < 0.8.
Supplementary Figure 8 | Phylograms from Bayesian Inference (BI) analyses of Pogostemoneae based on the combined dataset of five cpDNA regions (matK, rbcL, rps16, trnH-psbA, and trnL-trnF), under a partitioned strategy model, ambiguously aligned sites were excluded from analyses. Bayesian inference posterior probability (BIPP) are near the branches. A “–” indicates BIPP values < 0.8.
Supplementary Table 1 | List of taxa sampled with information related to taxonomy, GenBank accession numbers and references.
Supplementary Table 2 | Features of newly sequenced plastome of Paralamium griffithii.
Supplementary Table 3 | Excluded ambiguous sites for each dataset.
Footnotes
References
Abu-Asab, M. S., and Cantino, P. D. (1992). “Pollen morphology in subfamily Lamioideae (Labiatae) and its phylogenetic implications,” in Advances in Labiatae Science, eds R. M. Hartley and T. Reynolds (London: Royal Botanic Gardens, Kew), 361–379.
Abu-Asab, M. S., and Cantino, P. D. (1994). Systematic implications of pollen morphology in subfamilies Lamioideae and Pogostemonoideae (Labiatae). Ann. Mo. Bot. Gard. 81, 653–686. doi: 10.2307/2399915
Albaladejo, R. G., Aguilar, J. F., Aparicio, A., and Feliner, G. N. (2005). Contrasting nuclear-plastidial phylogenetic patterns in the recently diverged Iberian Phlomis crinita and P. lychnitis lineages (Lamiaceae). Taxon 54, 987–998. doi: 10.2307/25065483
Andrews, S. (2018). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed October 18, 2020)
Bendiksby, M., Salmaki, Y., Brauchler, C., and Ryding, O. (2014). The generic position of Stachys tibetica Vatke and amalgamation of the genera Eriophyton and Stachyopsis (Lamiaceae subfam. Lamioideae). Plant. Syst. Evol. 300, 961–971. doi: 10.1007/s00606-013-0935-2
Bendiksby, M., Thorbek, L., Scheen, A. C., Lindqvist, C., and Ryding, O. (2011). An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon 60, 471–484. doi: 10.1002/tax.602015
Bongcheewin, B., Pramali, K., Traiperm, P., Chantaranothai, P., and Paton, A. (2017). Pogostemon nudus sp. nov. (Lamiaceae) from Thailand. Nord. J. Bot. 35, 289–299. doi: 10.1111/njb.01439
Briquet, J. (1895–1897). “Labiatate,” in Die natürlichen Pflanzenfamilien, eds A. Engler and K. Prantl (Leipzig: Wilhelm Engelmann), 183–357.
Cantino, P. D. (1990). The phylogenetic significance of stomata and Trichomes in the Labiatae and verbenaceae. J. Arnold. Arboretum. 71, 323–370.
Cantino, P. D. (1992a). Evidence for a polyphyletic origin of the labiatae. Ann. Mo. Bot. Gard. 79, 361–379. doi: 10.2307/2399774
Cantino, P. D. (1992b). “Toward a phylogenetic classification of the Labiatae,” in Advances in Labiatae Science, eds R. M. Hartley and T. Reynolds (Richmond: Royal Botanic Gardens, Kew), 27–37.
Cantino, P. D., Harley, R. M., and Wagstaff, S. J. (1992). “Genera of Labiatae: status and classification,” in Advances in Labiatae Science, eds R. M. Hartley and T. Reynolds (London: Royal Botanic Gardens, Kew), 511–522.
Cantino, P. D., and Sanders, R. W. (1986). Subfamilial classification of labiatae. Syst. Bot. 11, 163–185. doi: 10.2307/2418955
Chen, Y. P., Drew, B. T., Li, B., Soltis, D. E., Soltis, P. S., and Xiang, C. L. (2016). Resolving the phylogenetic position of Ombrocharis (Lamiaceae), with reference to the molecular phylogeny of tribe Elsholtzieae. Taxon 65, 123–136. doi: 10.12705/651.8
Chen, Y. P., Li, B., Olmstead, R. G., Cantino, P. D., Liu, E. D., and Xiang, C. L. (2014). Phylogenetic placement of the enigmatic genus Holocheila (Lamiaceae) inferred from plastid DNA sequences. Taxon 63, 355–366. doi: 10.12705/632.8
Deng, T., Nie, Z. L., Drew, B. T., Volis, S., Kim, C., Xiang, C. L., et al. (2015). Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of Northern Hemisphere herbaceous plants? The case of Meehania (Lamiaceae). PLoS One 10:e0117171. doi: 10.1371/journal.pone.0117171
Doyle, J., and Doyle, J. (1987). Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem. Bull. Bot. Soc. Amer. 19, 11–15.
Drew, B. T., Cacho, N. I., and Sytsma, K. J. (2014). The transfer of two rare monotypic genera, Neoeplingia and Chaunostoma, to Lepechinia (Lamiaceae), and notes on their conservation. Taxon 63, 831–842. doi: 10.12705/634.6
Drew, B. T., and Sytsma, K. J. (2013). The South American radiation of Lepechinia (Lamiaceae): phylogenetics, divergence times and evolution of dioecy. Bot. J. Linn. Soc. 171, 171–190. doi: 10.1111/j.1095-8339.2012.01325.x
Enard, W., and Paabo, S. (2004). Comparative primate genomics. Annu Rev. Genomics Hum Genet. 5, 351–378. doi: 10.1146/annurev.genom.5.061903.180040
Harley, R. M., Atkins, S., Budantsev, A. L., Cantino, P. D., Conn, B. J., Grayer, R., et al. (2004). “Labiatae,” in The Families and Genera of Vascular Plants, eds K. Kubitzki and J. W. Kadereit (Berlin: Springer), 167–275.
Hooker, J. D. (1885). “Labiatae,” in Flora of British India, ed. J. D. Hooker (London: Reeve), 604–705.
Hu, G. X., Takano, A., Drew, B. T., Liu, E. D., Soltis, D. E., Soltis, P. S., et al. (2018). Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot. 122, 649–668. doi: 10.1093/aob/mcy104
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., Cantino, P. D., Olmstead, R. G., Bramley, G. L., Xiang, C. L., Ma, Z. H., et al. (2016). A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep. 6:34343. doi: 10.1038/srep34343
Li, B., and Olmstead, R. G. (2017). Two new subfamilies in Lamiaceae. Phytotaxa 313, 222–226. doi: 10.11646/phytotaxa.313.2.9
Li, H. W. (1977). “Paralamium Hand.-Mazz,” in Flora Reipublicae Popularis Sinicae, Vol. 65, ed. C. Y. Wu (Beijing: Science Press), 544–545.
Li, X. W., and Hedge, I. (1994). “Lamiaceae,” in Flora of China, Vol. 17, eds Z. Y. Wu and P. H. Raven (Beijing: St. Louis: Science Press;Missouri Botanical Garden Press), 196–224.
Lindqvist, C., and Albert, V. A. (2002). Origin of the Hawaiian endemic mints within North American Stachys (Lamiaceae). Am. J. Bot. 89, 1709–1724. doi: 10.3732/ajb.89.10.1709
Liu, B., Tan, Y. H., Liu, S., Olmstead, R. G., Min, D. Z., Chen, Z. D., et al. (2020). Phylogenetic relationships of Cyrtandromoea and Wightia revisited: a new tribe in Phrymaceae and a new family in Lamiales. J. Syst. Evol. 58, 1–17. doi: 10.1111/jse.12513
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. doi: 10.1093/nar/gkt289
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1007/978-1-4939-9173-0_1
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), (New York, NY: IEEE), 1–8.
Moon, H. K., Hong, S. P., Smets, E., and Huysmans, S. (2009). Micromorphology and character evolution of nutlets in tribe Mentheae (Nepetoideae, Lamiaceae). Syst. Bot. 34, 760–776. doi: 10.1600/036364409790139592
Müller, K., Müller, J., and Quandt, D. (2010). PhyDE: Phylogenetic Data Editor, Version 0.9971. Available online at: http://www.phyde.de/index.html (accessed November 14, 2020)
Olmstead, R. (2016). A Synoptical Classification of the Lamiales, Version 2.6.2. Available online at: http://depts.washington.edu/phylo/Classification.pdf (accessed 25 Feb 2020)
Pollard, D. A., Iyer, V. N., Moses, A. M., and Eisen, M. B. (2006). Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2:e173. doi: 10.1371/journal.pgen.0020173
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Prain, D. (1889). “Microtoena cymosa,” in Hooker’s Icones Plantarum, ed. J. D. Hooker (London: Longman), 1827.
Qu, X. J., Moore, M. J., Li, D. Z., and Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15:50. doi: 10.1186/s13007-019-0435-7
Rambaut, A. (2014). FigTree, v.1.4.2. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed October 13, 2018)
Refulio-Rodriguez, N. F., and Olmstead, R. G. (2014). Phylogeny of Lamiidae. Am. J. Bot. 101, 287–299. doi: 10.3732/ajb.1300394
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Roy, T., and Lindqvist, C. (2015). New insights into evolutionary relationships within the subfamily Lamioideae (Lamiaceae) based on pentatricopeptide repeat (PPR) nuclear DNA sequences. Am. J. Bot. 102, 1721–1735. doi: 10.3732/ajb.1500233
Ryding, O. (1994a). Pericarp structure and phylogeny of Lamiaceae subfamily Pogostemonoideae. Nord. J. Bot. 14, 59–63. doi: 10.1111/j.1756-1051.1994.tb00572.x
Ryding, O. (1994b). The pericarp structure in subtribe Melittidinae (Lamiaceae-Lamioideae) and its systematic implications. Bot. Jahrb. Syst. 115, 547–555.
Ryding, O. (1994c). Pericarp structure in the tribe Prasieae (Lamiaceae-Lamioideae) and its systematic implications. Bot. Jahrb. Syst. 116, 391–399.
Ryding, O. (1995). Pericarp structure and phylogeny of the Lamiaceae-Verbenaceae-complex. Plant Syst. Evol. 198, 101–141. doi: 10.1007/bf00985109
Ryding, O. (1998). Phylogeny of the Leucas Group (Lamiaceae). Syst. Bot. 23, 235–247. doi: 10.2307/2419591
Ryding, O. (2003). Reconsideration of Wiedemannia and notes on the circumscription of Lamium (Lamiaceae). Bot. Jahrb. Syst. 124, 325–335. doi: 10.1127/0006-8152/2003/0124-0325
Ryding, O. (2008). Pericarp structure and phylogeny of the Phlomis group (Lamiaceae subfam. Lamioideae). Bot. Jahrb. Syst. 127, 299–316. doi: 10.1127/0006-8152/2008/0127-0002
Salmaki, Y., Zarre, S., and Jamzad, Z. (2008). Nutlet micromorphology and its systematic implication in Stachys L. (Lamiaceae) in Iran. Feddes Rep. 119, 607–621. doi: 10.1002/fedr.200811187
Salmaki, Y., Zarre, S., Ryding, O., Lindqvist, C., Brauchler, C., Heubl, G., et al. (2013). Molecular phylogeny of tribe Stachydeae (Lamiaceae subfamily Lamioideae). Mol. Phylogenet. Evol. 69, 535–551. doi: 10.1016/j.ympev.2013.07.024
Salmaki, Y., Zarre, S., Ryding, O., Lindqvist, C., Scheunert, A., Brauchler, C., et al. (2012). Phylogeny of the tribe Phlomideae (Lamioideae: Lamiaceae) with special focus on Eremostachys and Phlomoides: new insights from nuclear and chloroplast sequences. Taxon 61, 161–179. doi: 10.1002/tax.611012
Scheen, A. C., and Albert, V. A. (2007). Nomenclatural and taxonomic changes within the Leucas clade (Lamioideae; Lamiaceae). Syst. Geogr. Pl. 77, 229–238.
Scheen, A. C., and Albert, V. A. (2009). Molecular phylogenetics of the leucas group (Lamioideae; Lamiaceae). Syst. Bot. 34, 173–181. doi: 10.1600/036364409787602366
Scheen, A. C., Bendiksby, M., Ryding, O., Mathiesen, C., Albert, V. A., and Lindqvist, C. (2010). Molecular phylogenetics, character evolution, and suprageneric classification of Lamioideae (Lamiaceae). Ann. Mo. Bot. Gard. 97, 191–217. doi: 10.3417/2007174
Scheen, A. C., Lindqvist, C., Fossdal, C. G., and Albert, V. A. (2008). Molecular phylogenetics of tribe Synandreae, a North American lineage of lamioid mints (Lamiaceae). Cladistics 24, 299–314. doi: 10.1111/j.1096-0031.2007.00180.x
Seyedi, Z., and Salmaki, Y. (2015). Trichome morphology and its significance in the systematics of Phlomoides (Lamiaceae; Lamioideae; Phlomideae). Flora 213, 40–48. doi: 10.1016/j.flora.2015.04.003
Siadati, S., Salmaki, Y., Mehrvarz, S. S., Heubl, G., and Weigend, M. (2018). Untangling the generic boundaries in tribe Marrubieae (Lamiaceae: Lamioideae) using nuclear and plastid DNA sequences. Taxon 67, 770–783. doi: 10.12705/674.6
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suddee, S., and Paton, A. (2004). Some nomenclatural changes in South East Asian Lamiaceae. Kew Bull. 59, 315–318. doi: 10.2307/4115869
Walker, J. B., Drew, B. T., and Sytsma, K. J. (2015). Unravelling species relationships and diversification within the iconic California Floristic Province Sages (Salvia subgenus Audibertia, Lamiaceae). Syst. Bot. 40, 826–844. doi: 10.1600/036364415x689285
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wink, M., and Kaufmann, M. (1996). Phylogenetic relationships between some members of the Subfamily Lamioideae (Family Labiatae) Inferred from nucleotide sequences of the rbcL Gene. Bot. Acta 109, 139–148. doi: 10.1111/j.1438-8677.1996.tb00554.x
Xiang, C. L., Dong, H. J., Landrein, S., Zhao, F., Yu, W. B., Soltis, D. E., et al. (2020). Revisiting the phylogeny of Dipsacales: new insights from phylogenomic analyses of complete plastomic sequences. J. Syst. Evol. 58, 103–117. doi: 10.1111/jse.12526
Xiang, C. L., Funamoto, T., Evangelista, E. V., Zhang, Q., and Peng, H. (2013a). Pollen morphology of the East Asiatic genus Chelonopsis (Lamioideae: Lamiaceae) and allied genera, with reference to taxonomic implications and potential pollination ecology. Plant Biosyst. 147, 620–628. doi: 10.1080/11263504.2012.748099
Xiang, C. L., Zhang, Q., Scheen, A. C., Cantino, P. D., Funamoto, T., and Peng, H. (2013b). Molecular phylogenetics of Chelonopsis (Lamiaceae: Gomphostemmateae) as inferred from nuclear and plastid DNA and morphology. Taxon 62, 375–386. doi: 10.12705/622.11
Yao, G., Drew, B. T., Yi, T. S., Yan, H. F., Yuan, Y. M., and Ge, X. J. (2016). Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol. Phylogenet. Evol. 98, 184–200. doi: 10.1016/j.ympev.2016.01.020
Zhao, F., Chen, Y. P., Salmaki, Y., Drew, B. T., Wilson, T. C., Scheen, A. C., et al. (2021). An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 19:2. doi: 10.1186/s12915-020-00931-z
Zhao, F., Li, B., Drew, B. T., Chen, Y. P., Wang, Q., Yu, W. B., et al. (2020a). Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae). PLoS One 15:e0232602. doi: 10.1371/journal.pone.0232602
Keywords: Lamioideae, molecular phylogenetics, nutlet morphology, plastome phylogenomics, Paralamium, Pogostemoneae
Citation: Zhao F, Wu Y-W, Drew BT, Yao G, Chen Y-P, Cai J, Liu E-D, Li B and Xiang C-L (2021) Systematic Placement of the Enigmatic Southeast Asian Genus Paralamium and an Updated Phylogeny of Tribe Pogostemoneae (Lamiaceae Subfamily Lamioideae). Front. Plant Sci. 12:646133. doi: 10.3389/fpls.2021.646133
Received: 25 December 2020; Accepted: 22 March 2021;
Published: 16 April 2021.
Edited by:
Petr Koutecký, University of South Bohemia, CzechiaReviewed by:
Tilottama Roy, Missouri Western State University, United StatesMario Fernández-Mazuecos, Complutense University of Madrid, Spain
Copyright © 2021 Zhao, Wu, Drew, Yao, Chen, Cai, Liu, Li and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun-Lei Xiang, eGlhbmdjaHVubGVpQG1haWwua2liLmFjLmNu; Bo Li, aGFuYm9saWp4QDE2My5jb20=
†These authors have contributed equally to this work
 Fei Zhao
Fei Zhao Yi-Wen Wu2†
Yi-Wen Wu2† Bryan T. Drew
Bryan T. Drew Gang Yao
Gang Yao Bo Li
Bo Li Chun-Lei Xiang
Chun-Lei Xiang