
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 April 2021
Sec. Plant Proteomics and Protein Structural Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.645286
This article is part of the Research TopicPlant Glycobiology - A Sweet World of Glycans, Glycoproteins, Glycolipids, and Carbohydrate-Binding ProteinsView all 33 articles
 Hewot Allelign Ashagre1
Hewot Allelign Ashagre1 David Zaltzman1
David Zaltzman1 Anat Idan-Molakandov1
Anat Idan-Molakandov1 Hila Romano1
Hila Romano1 Oren Tzfadia2
Oren Tzfadia2 Smadar Harpaz-Saad1*
Smadar Harpaz-Saad1*The plasticity of root development represents a key trait that enables plants to adapt to diverse environmental cues. The pattern of cell wall deposition, alongside other parameters, affects the extent, and direction of root growth. In this study, we report that FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 18 (FLA18) plays a role during root elongation in Arabidopsis thaliana. Using root-specific co-expression analysis, we identified FLA18 to be co-expressed with a sub-set of genes required for root elongation. FLA18 encodes for a putative extra-cellular arabinogalactan protein from the FLA-gene family. Two independent T-DNA insertion lines, named fla18-1 and fla18-2, display short and swollen lateral roots (LRs) when grown on sensitizing condition of high-sucrose containing medium. Unlike fla4/salt overly sensitive 5 (sos5), previously shown to display short and swollen primary root (PR) and LRs under these conditions, the PR of the fla18 mutants is slightly longer compared to the wild-type. Overexpression of the FLA18 CDS complemented the fla18 root phenotype. Genetic interaction between either of the fla18 alleles and sos5 reveals a more severe perturbation of anisotropic growth in both PR and LRs, as compared to the single mutants and the wild-type under restrictive conditions of high sucrose or high-salt containing medium. Additionally, under salt-stress conditions, fla18sos5 had a small, chlorotic shoot phenotype, that was not observed in any of the single mutants or the wild type. As previously shown for sos5, the fla18-1 and fla18-1sos5 root-elongation phenotype is suppressed by abscisic acid (ABA) and display hypersensitivity to the ABA synthesis inhibitor, Fluridon. Last, similar to other cell wall mutants, fla18 root elongation is hypersensitive to the cellulose synthase inhibitor, Isoxaben. Altogether, the presented data assign a new role for FLA18 in the regulation of root elongation. Future studies of the unique vs. redundant roles of FLA proteins during root elongation is anticipated to shed a new light on the regulation of root architecture during plant adaptation to different growth conditions.
Developmental pliancy allows plants to respond to a wide range of environmental signals by altering the pattern of plant growth and development. This enables otherwise immobile plants to adjust to heterogeneous soil and ever-changing environmental conditions by continuous forage toward optimal conditions with respect to water, nutrients, and other resources. In practice, this is achieved by modification of root system architecture in response to different developmental and environmental cues. The adaptability and flexibility of the root system is one of the main determinants affecting the rate and extent of plant biomass production (Lynch, 1995; Malamy, 2005; Gruber et al., 2013; Tian et al., 2014; Slovak et al., 2016).
The plasticity of root development is obtained through embryonic and postembryonic organogenesis. The primary root (PR) apical meristem, which is responsible for PR growth, is formed as part of the developing embryo. In contrast, lateral roots (LRs) are repeatedly formed from the PR, or from previously formed LRs, and determine the shape of the plant root system dictating the efficiency of plant anchorage, water uptake and nutrient acquisition. It is noteworthy, that first-order LRs are formed from the PR, while second- and third-order LRs are formed from lower-order LRs. LR meristems are formed post-embryonically from the pericycle, a highly specified cell layer encircling the root vascular stele tissue. The intervals of LR formation vary according to developmental stage and growth conditions (Malamy and Benfey, 1997; Casimiro et al., 2001; De Smet et al., 2003; Lavenus et al., 2013; Vilches-Barro and Maizel, 2015; Slovak et al., 2016). While much is known about the mechanism employed in LR initiation and meristem formation, little is known about the mechanisms employed in PR vs. LR elongation and how it is modified in response to different environmental signals.
Cell expansion can occur through isotropic growth in which the cell expands to the same extent in all directions, or through anisotropic growth in which the cell expends in a non-uniform, directional manner. Root elongation is obtained through anisotropic cell expansion. While turgor pressure provides the driving force for this process, the pattern of cell wall deposition in the extra-cellular matrix is one of the parameters dictating growth extent and direction. The current model suggests that anisotropic cell expansion is obtained as a result of stiffer cell wall deposited perpendicular to the growth axis, yielding cell elongation driven by the cytosolic turgor pressure (Green, 1980; Taiz, 1984; Darley et al., 2001; Baskin, 2005). Primary cell wall is deposited in all plant cells and protect the protoplast as the cell grows, while secondary cell wall is synthesized in specific cell types as part of the process of cell differentiation which takes place after the cell reached its final size. The plant cell walls are composed primarily of three classes of polysaccharides: cellulose, hemicelluloses and pectins (Bacic et al., 1988; Carpita and Gibeaut, 1993; Cosgrove, 2005; Somerville, 2006). It was previously shown that cellulose synthesis, organization and cross-linking with other cell wall material is a key determinant dictating the extent of anisotropic cell expansion and subsequently, of root elongation (Arioli et al., 1998; Fagard et al., 2000; Schindelman et al., 2001). Cellulose is composed of β-(1,4)-linked glucan chains synthesized at the plasma membrane by the cellulose synthase complex (CSC; Mueller et al., 1976; Brown et al., 1996; Kimura et al., 1999). Each glucan chain is synthesized by CELLULOSE SYNTHASE A (CESA), a glycosyltransferase encoded by multi gene-families in different plant species (Pear et al., 1996; Delmer, 1999; Richmond and Somerville, 2000; McFarlane et al., 2014). In Arabidopsis, CESA1, CESA3, and CESA6 have been shown to be essential for cellulose synthesis as part of primary cell wall deposition during root elongation. Mutants in either of these three CESAs, display short roots with a swollen root tip (Hauser et al., 1995; Caño-Delgado et al., 2000; Fagard et al., 2000; Desprez et al., 2007). Similar results were obtained using chemical inhibitors of CESA (Heim et al., 1989; Desprez et al., 2002; Tateno et al., 2016). Additional components required for cell wall deposition and organization have been shown to play a role during root elongation, including KORRIGAN (KOR), COBRA (COB), FASCICLIN-LIKE 4/SALT-OVERLY SENSITIVE 5 (FLA4/SOS5), CHITINASE-LIKE 1 (CTL1), and others (Nicol, 1998; Schindelman et al., 2001; Shi et al., 2003; Hermans et al., 2010). However, so far the intricate mechanism regulating cell wall synthesis and remodeling in elongating roots under different growth conditions has not been fully established.
To identify new candidate-genes participating in cell wall deposition during root elongation, a tissue specific co-expression analysis was performed as part of the current study. The premise of this method is the well established phenomenon that genes functioning in the same pathway, or required for the same process, tend to express in a transcriptionally coordinated manner. Co-expression approaches have been used to assign function for genes involved in cellulose synthesis, lignin deposition, and other metabolic processes (Brown et al., 2005; Ruprecht et al., 2011; Yang et al., 2011, 2019; Wang et al., 2012; Ben-Tov et al., 2015; Endler et al., 2015; Ransbotyn et al., 2015; Voiniciuc et al., 2015a, b, 2018). In this work, ten genes, known to play a role in primary cell wall deposition during root elongation, were used as ‘baits’ to mine a spatio-temporal, high-resolution root gene expression dataset (Supplementary Table 1; Brady et al., 2007). The identified candidate genes were examined for perturbation of root elongation. Interestingly, two independent T-DNA insertion lines corresponding to different mutations in the FASCICLIN-LIKE ARABINOGALACTAN 18 (FLA18) gene (AT3G11700), which has not been assigned a function so far, showed modification of PR and LR elongation under different growth conditions. The presented data suggest that FLA18 plays a role in the regulation of root elongation.
A tissue specific co-expression analysis was performed using the Arabidopsis high-resolution spatio-temporal gene expression dataset generated for developing root (Brady et al., 2007). The ‘bait’ list for the co-expression analysis was generated based on genes known in the literature to be involved in primary cell wall deposition during root elongation (Supplementary Table 1). Pearson correlation identifies a tight correlation between FLA18 gene expression and that of CESA1, CESA3, and CESA6 involved in primary cell wall deposition during root elongation (Supplementary Figure 2B; Fagard et al., 2000; Desprez et al., 2007; Persson et al., 2007; Chen et al., 2016). A highly specific expression pattern was observed for representative ‘baits’ and FLA18 along the longitudinal root axis (Supplementary Figure 2A). This includes, low expression at the meristematic zone, high expression at the transition from the meristematic to the elongation zone and then again, reduced expression in the interface between the elongation and the differentiation zone (Supplementary Figure 2A). In contrast, CESA4, CESA7 or CESA8, involved in secondary cell wall deposition as part of root vascular system differentiation, display a very different expression pattern (Supplementary Figure 2A). No correlation in gene expression was observed between FLA18 and CESA4, CESA7, or CESA8 using the root-specific data-set (Supplementary Figure 2B; Turner and Somerville, 1997; Persson et al., 2005; Taylor-Teeples et al., 2015; Meents et al., 2018). In addition, the expression of FLA18 and other ‘baits’ used for the co-expression analysis (CESA1, CESA3, and CESA6 are presented as an example) is induced in the linage of cells destined to differentiate into root hairs, and to a lower extent in cell layers with non-root hair identity (Supplementary Figure 2A). Taken together, the co-expression relationship between FLA18 and established-players required for root elongation prompted us to further investigate the role of FLA18 in this developmental context.
To study FLA18 expression pattern throughout plant growth and development, we used Reverse Transcription PCR (RT-PCR) analysis and a GUS-reporter gene driven by the FLA18 promoter. The FLA18 transcript was detected in all organs, including leaves, stems, siliques, and flowers (Supplementary Figures 3A,B). GUS activity was detected in various developmental contexts, correlating with the RT-PCR results (Supplementary Figures 3C–F). The GUS staining identified FLA18 expression in stems, flowers, as well as elongating tissues like, root tips, and hypocotyls of dark-grown seedlings. In dark-grown seedlings, the induction of FLA18 gene expression was detected specifically in the upper part of the hypocotyl, which was previously shown to display high rate of cell elongation as compared to other parts of the hypocotyl (Crowell et al., 2011). Overall, FLA18 gene expression was confirmed in elongating roots and additional developmental contexts throughout plant development.
To investigate the role of FLA18 during root elongation, two independent T-DNA insertion lines were obtained, SALK_086944 named fla18-1 and SALK_039619 named fla18-2, both carrying a T-DNA insertion in the first exon of the FLA18 coding sequence (Figure 1A and Supplementary Figure 4A). Homozygous lines were identified by genotyping. The expression of FLA18 in both fla18-1 and fla18-2 was examined by RT-PCR analysis using FLA18-specific primers localized downstream (3′) of the T-DNA insertion site and TUBULIN as a reference gene. According to the RT-PCR analysis both fla18-1 and fla18-2 appeared to be knock-down alleles, as both exhibited very low expression compared to the wild-type, but neither completely abolished the FLA18 gene expression (Supplementary Figure 4B). The reduced expression level in fla18-1 was verified by nCounter NanoString analysis, using a FLA18-specific probe targeting the sequence localized downstream of the T-DNA insertion site (Figure 1B). An approximate 10-fold reduction in FLA18 gene expression was measured in the fla18-1 mutant background as compared to the wild type (Figure 1B).
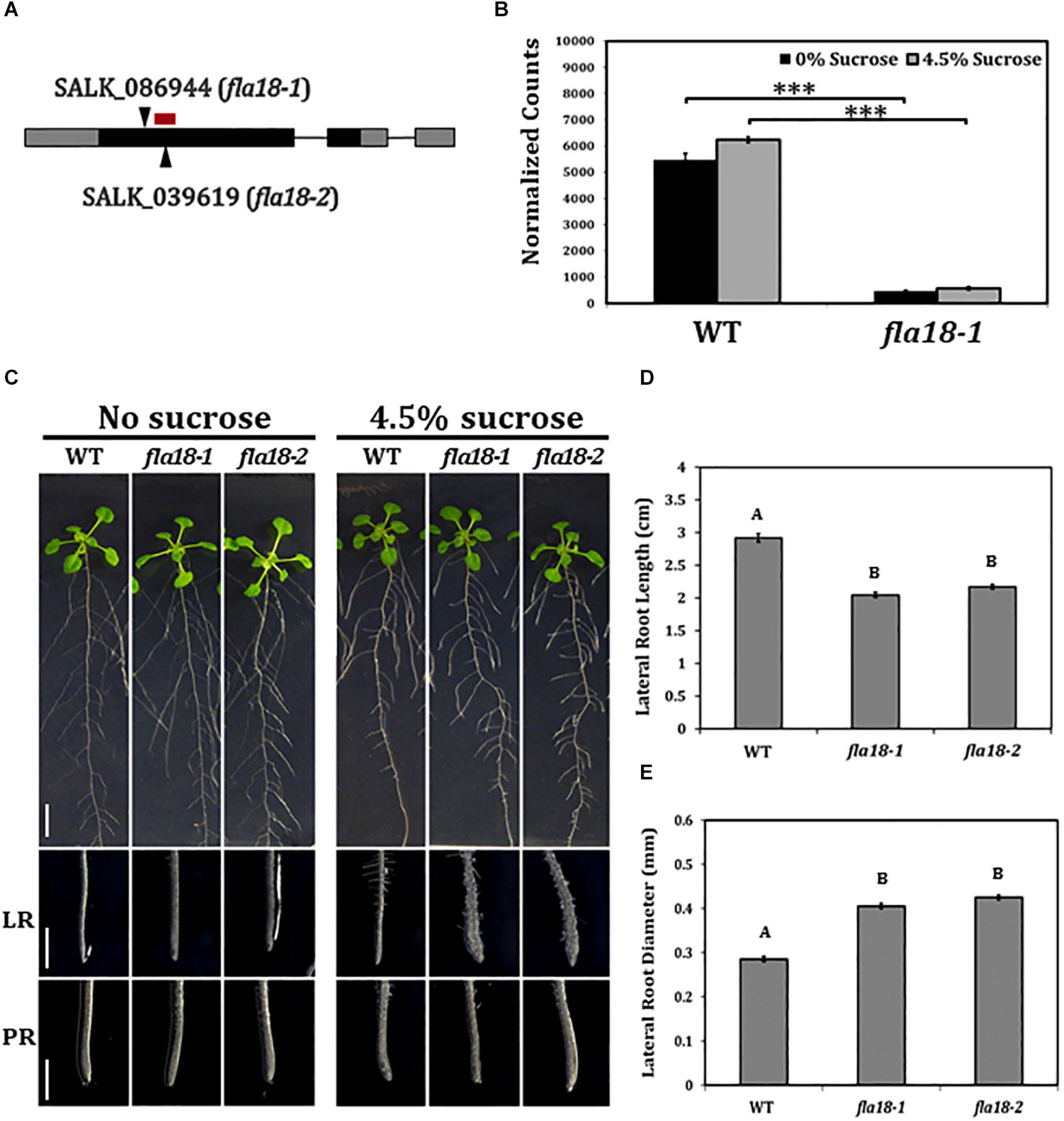
Figure 1. fla18 mutants display conditional perturbation of root elongation. (A) Schematic presentation of the FLA18 gene as annotated by The Arabidopsis Information Resource, pinpointing the location of the T-DNA insertion in both alleles examined, i.e., fla18-1 (SALK_086944) and fla18-2 (SALK_039619). The red bar represents the location of the nCounter NanoString probe. (B) FLA18 gene expression at the primary root of the indicated genotypes. Seedlings were germinated on medium containing 1% sucrose, for 4 d, and then transferred to either sucrose-free or 4.5% sucrose-containing medium for 6 h. Gene expression was measured using the nCounter NanoString technology, in four biological replicates. Expression level is indicated in normalized counts, as calculated by the nSolver platform using GAPC, UBQ10, EF1a, F-BOX, and AP2 as reference genes. (C) Seedlings of the indicated genotypes were grown on sucrose-free MS medium for 4 days and then, transferred to either new sucrose-free MS medium or MS medium supplemented with 4.5% sucrose, for an additional 10 days. Whole seedlings (upper panel), lateral root tips (LR, middle panel) and primary root tips (PR, lower panel) were documented. Total lateral root length (D) and diameter (E) were measured 7 days after the transfer to restrictive conditions (MS plus 4.5% sucrose). Error bars represent SE. The results were analyzed through JMPpro13 for statistical analysis, applying Tukey’s HSD test (B; ***p < 0.0001) or a non-parametric comparison for all pairs using the Steel-Dwass Method (D,E; p < 0.0001). D and E, n = 10. Scale bars = 1 cm (C; upper panel) and 1 mm (C; middle and lower panel).
To study the possible role of FLA18 in the context of root elongation, the mutant lines were grown under either ambient conditions [Murashige and Skoog (MS) medium with no sucrose added] or restrictive conditions (MS supplemented with 4.5% sucrose). These restrictive conditions have been previously shown to act as sensitizing conditions that may serve to uncover perturbations in cell wall deposition during root elongation (Hauser et al., 1995; Schindelman et al., 2001; Xu et al., 2008; Basu et al., 2016). When grown under ambient conditions, the mutant lines resembled the wild type (Figure 1C), except from a slight increase in PR elongation rate and final length (Supplementary Figures 2, 5). When grown under restrictive conditions, the LRs of both fla18-1 and fla18-2 displayed an approximate 30% reduction in length compared to the wild type (Figure 1D and Supplementary Figure 6). Furthermore, a significant increase of about 30% in LR width was also detected (Figure 1E). It is important to note that the fla18 LR phenotype can be modified by different MS or agar types used in the media. Different batches (lot numbers) of the same product, MS or gelling agent, showed different LR phenotype in fla18 background, ranging from short and swollen to indistinguishable from the wild-type (data not shown). No specific difference in nutrient composition could be associated with this effect on the fla18 root phenotype. Nonetheless, under the experimental conditions mentioned above, this phenotype was observed in multiple independent experiments. Transgenic lines (T3) expressing the FLA18 coding sequence driven by the 35S promoter, complemented the LR phenotype of the fla18-1 allele when grown on high-sucrose containing medium (Supplementary Figure 7). Note that complementation line #2, which is a strong suppressor of the fla18 root phenotype, seem to display reduced LR density as compared to the wild-type. It is noteworthy, that despite the fact that FLA18 gene expression was detected in other developmental contexts additional to the root, no other phenotypes were detected for either fla18-1 or fla18-2. In summary, these observations identify a new role for FLA18 in the regulation of root elongation and suggest that it may differentially affect root elongation in PR vs. LR under different growth conditions.
The finding that both fla18 and fla4/sos5, herein referred to as sos5, display perturbation of LR elongation suggests a non-redundant role in this developmental context under restrictive conditions of high-sucrose containing medium. Taking into account the phylogenetic distance between these two FLA-encoding genes this might be expected (Supplementary Figure 1). We therefore generated the fla18-1sos5 and fla18-2sos5 double mutants to study the relationship between these two FLA-encoding genes in the context of root elongation. The results with both fla18 alleles were the same (Figure 2 and Supplementary Figure 9, data not shown) and hence quantification is presented only for fla18-1sos5. The severity of the fla18sos5 phenotype was modified in response to different growth conditions. Under ambient conditions (MS media with no sucrose), no root swelling was observed, but slight differences in the rate of root elongation were detected (Figure 2). As previously noted (Figure 1), the rate of fla18-1 PR elongation was slightly higher as compared to the wild type (Figure 2C) and the final root length was slightly longer (Supplementary Figure 8). At the same time, sos5 and to a greater extent fla18-1sos5, displayed a reduced PR elongation rate, as compared to the wild type (Figure 2C and Supplementary Figure 8). When grown on MS media supplemented with 1% sucrose, the fla18sos5 LRs and PRs displayed swollen root tips, unlike any of the single mutants or the wild type (Supplementary Figure 9). Last, under restrictive conditions (MS supplemented with 4.5% sucrose), both wild type and fla18-1 displayed normal root growth, while sos5 displayed a short and swollen root phenotype in both LR and PR, consistent with previous findings (Figure 2B; Shi et al., 2003; Xu et al., 2008; Basu et al., 2016). Interestingly, under these conditions, the fla18sos5 double mutants displayed a more severe perturbation of anisotropic root growth, as indicated by the very short and swollen root phenotype observed in both PR and LRs (Figure 2B). Quantification of PR elongation rate, following the transfer to restrictive conditions, demonstrated that the fla18-1sos5 roots display a much more severe response to the restrictive conditions, compared to all other genotypes examined in this experiment (Figures 2C,D and Supplementary Figure 8). Altogether, the fla18sos5 double mutant displays a severe effect on both PR and LR elongation, as compared to either of the single mutants or the wild-type, when grown on mild- or high-sucrose containing media.
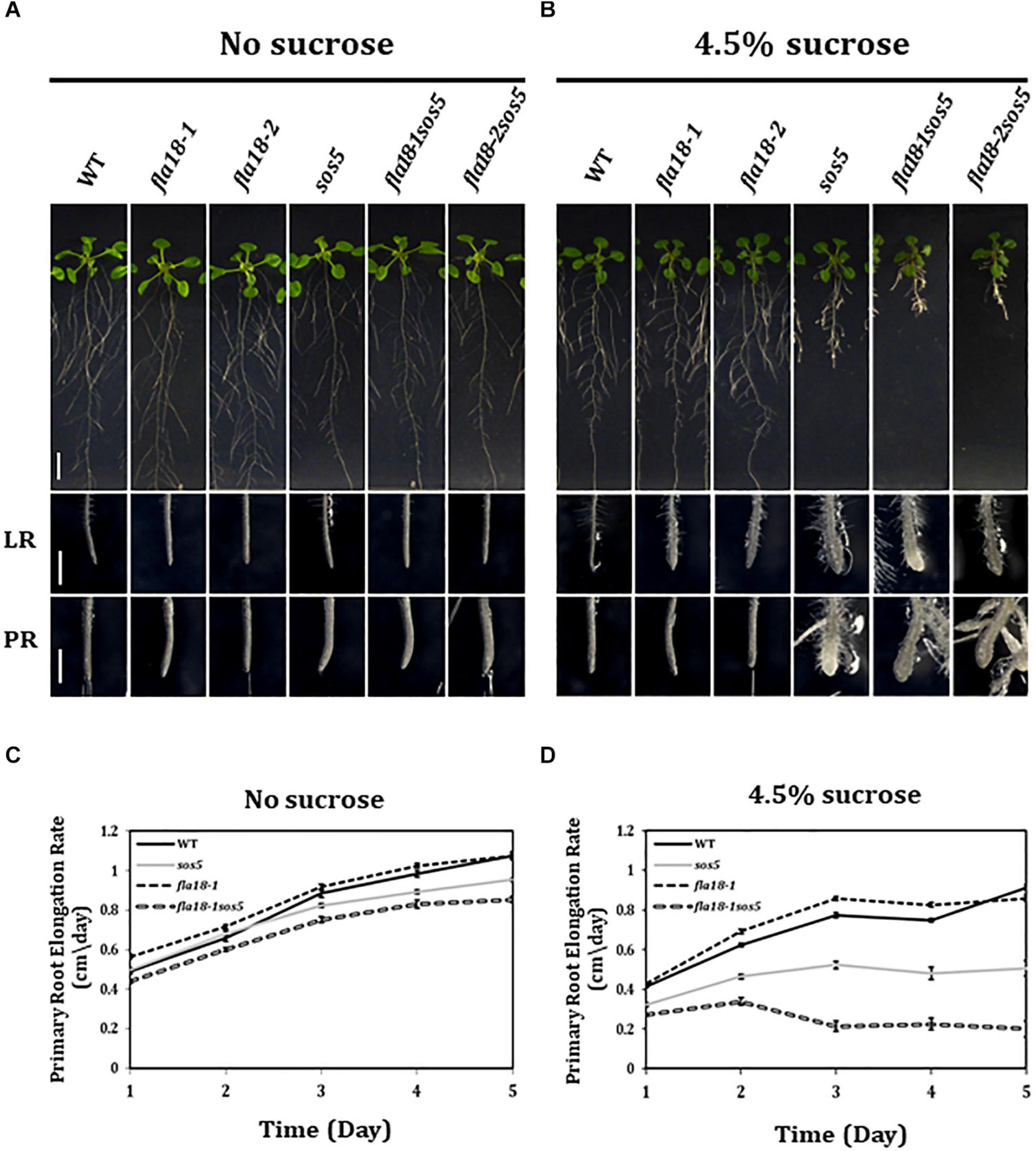
Figure 2. fla18sos5 double mutants impart a synergistic effect on root elongation. (A,B) Seedlings of the indicated genotypes were grown on sucrose-free MS medium for 4 days and then transferred to either fresh sucrose-free medium (A; permissive conditions) or medium containing 4.5% sucrose (B; restrictive conditions), for an additional 7 days. Whole seedlings (upper panel), lateral root tips (LR, middle panel) and primary root tips (PR, lower panel) were documented. Note, the synergistic effect of the double mutant on both PR and LR elongation and swelling. (C,D) Seedlings of the indicated genotypes were grown on MS medium for 4 days and then transferred to sucrose-free MS medium (C) or MS medium plus 4.5% sucrose (D). Root length was measured daily and elongation rate was calculated; n = 10 (fla18-1; n = 5). Error bars represent SE. Scale bars = 1 cm (A,B; upper panel) and 1 mm (A,B; middle and lower panel).
The effect of the different genotypes on root elongation was examined also under salt stress conditions (Figure 3). As mentioned above, sos5 is a conditional mutant that displays root growth arrest and root tip swelling when grown on high-sucrose or high-salt containing media (Shi et al., 2003; Xu et al., 2008; Xue and Seifert, 2015; Basu et al., 2016). In the current study, when grown on MS supplemented with 100 nM NaCl, roots of the fla18-1 allele looked similar to the wild type, yet a slight but significant increase in PR length could be detected (Figures 3A,B). Unlike sos5, no phenotype was detected in fla18 LRs (Figure 3A). Short and swollen LRs in the fla18 mutants were observed only on high-sucrose and not on high-salt containing media (Figures 1–3). However, the most prominent phenotype was that of the fla18-1sos5 double mutant. When root elongation of the fla18-1sos5 seedlings was examined under salt-stress conditions, a significant reduction in PR length was observed of about 84% compared to either fla18-1 or the wild-type and about 52% compared to the sos5 single mutant (Figure 3B). Interestingly, under these conditions, fla18-1sos5 displayed also a small and chlorotic shoot phenotype, that was not detected in any of the other genotypes examined (Figure 3A).
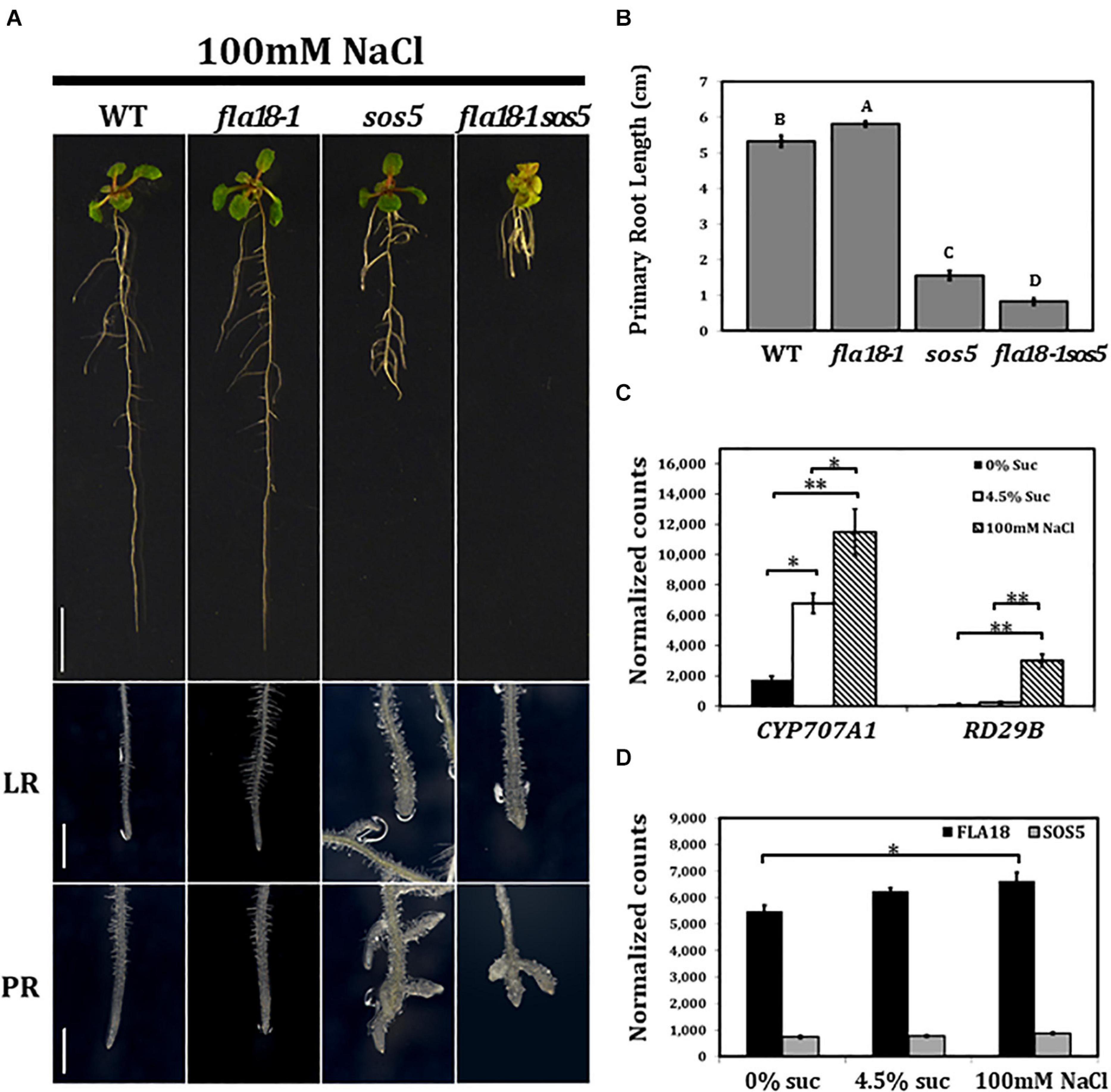
Figure 3. fla18sos5 double mutants impart a synergistic effect on root elongation under salt stress conditions. Seedlings of the indicated genotypes were grown on MS medium containing 1% sucrose, for 4 days, and then transferred to medium containing 1% sucrose and 100 mM NaCl for an additional 7 days. (A) Whole seedlings (upper panel), lateral root tips (LR, middle panel) and primary root tips (PR, lower panel). (B) Measurements of primary root length were conducted 7 d after seedlings were transferred to restrictive conditions (MS supplemented with 1% sucrose and 100 mM NaCl). n = 6. (C,D) Gene expression of the indicated genes was examined in the PR of wild type seedlings germinated on medium containing 1% sucrose, for 4 days, and then transferred to medium containing either no sucrose, 4.5% sucrose or 1% sucrose + 100 mM NaCl, for 6 h. Gene expression was measured using nCounter NanoString technology, in four biological replicates. Expression level is indicated by normalized counts, as calculated by the nSolver platform using GAPC, UBQ10, EF1a, F-BOX, and AP2 as reference genes. The results were analyzed using JMPpro13 for statistical analysis and analyzed using Tukey’s HSD (honestly significant difference) test (*p < 0.05**p < 0.001). Scale bars = 1 cm (A; upper panel) and 1 mm (A; middle and lower panel).
To investigate the effect of the different growth conditions on FLA gene expression in roots, total RNA was extracted from roots of 4-days old seedlings, 6 h after the transfer from ambient to either of the examined conditions, and subjected to nCounter NanoString analysis. After 6 h no phenotype could be observed, yet stress response could already be detected, as indicated by the induced expression of RESPONSIVE TO DESICCATION 29B (RD29B) and CYTOCHROME P450, FAMILY 707, SUBFAMILY A, POLYPEPTIDE 1 (CYP7071A; Ingram and Bartels, 1996; Xiong et al., 2002; Nambara and Marion-Poll, 2005; Seifert et al., 2014). At this time point, the expression of RD29B in wild type was 20-fold higher under salt-stress conditions compared to ambient conditions (Figure 3C), but was not significantly altered in roots transffered to high-sucrose containing medium (Figure 3C). In contrast, CYP7071A, which is involved in ABA catabolism, showed an approximate fourfold increase in roots grown on high sucrose-containing medium and an approximate sixfold increase in roots grown on high salt-containing medium, as compared to ambient conditions (Figure 3C). Neither FLA18 nor SOS5 gene expression was significantly modified by the abiotic, high-sucrose or high-salt, stress conditions (Figure 3D). To learn more more about the interaction between FLA18 and SOS5, we followed the expression of each of these genes in the different mutant backgrounds (Supplementary Figure 10). As expected, FLA18 gene expression was significantly reduced in fla18-1 and fla18-1sos5 mutant background, compared to the wild-type. Interestingly, a mild but significant increase in FLA18 gene expression could be detected in sos5 mutant background when grown under either permissive (MS with no-sucrose added) or restrictive conditions (MS supplemented with 4.5% sucrose; Supplementary Figure 10). For SOS5, a significant twofold decrease was detected in sos5 and fla18-1sos5 mutant background, as compared to the wild-type. This was somewhat surprising as the sos5-2 allele used in this study is a T-DNA insertion line that was previously described as a null allele (Xu et al., 2008; Harpaz-Saad et al., 2011; Griffiths et al., 2014; Seifert et al., 2014; Xue and Seifert, 2015; Basu et al., 2016; Xue et al., 2017). One explanation could be that the probe recognition site in this case is localized upstream of the T-DNA insertion site. Nonetheless, a minor increase in SOS5 gene expression could be detcetd in fla18-1 mutant background, which was statistically significant under restrictive conditions (Supplementary Figure 10). Altogether, these results pinpoint to the different mechanisms induced under different stress conditions and suggest that FLA18 and FLA4 each plays a unique vs. partially overlapping roles during plant development under different growth conditions.
Previous studies demonstrated that FLA4/SOS5 functions as a positive regulator of cell wall deposition by modulating ABA signaling (Seifert et al., 2014). To examine the potential interaction between FLA18 function and ABA, we used exogenous ABA treatment and Fluridon, an inhibitor of the carotenoid pathway leading to ABA synthesis (Nakamura and Asami, 2014). Seedlings were grown for 5 days under ambient conditions and then transferred to medium containing either 4.5% sucrose (DMSO that served as the Fluridon solvent was added as a control), or 4.5% sucrose with the addition of ABA or Fluridon, for two additional days (Figure 4). As previously described, on MS supplemented with high-sucrose concentration, the PR of the wild type and fla18-1 remained unswollen, while the PR of sos5 and fla18-1sos5 was short and swollen (Figure 4A). The same can be seen for the PR of procuste1 (prc1), a mutant in CESA6 encoding for the cellulose synthase catalytic subunit that served as a positive control for this set of experiments (Figure 4A). When exposed to ABA, the root length of the wild-type was shorter, as compared to control with no-ABA added. Interestingly, PR length of all fla-mutants was significantly longer than the wild type (Figure 4B). In addition, the swollen root phenotype of sos5, fla18-1sos5 and prc1 was suppressed (Figure 4C). On the other hand, Fluridon treatment, which also results in shorter wild-type PR as compared to control conditions, led to a reduction in PR length in all examined genotypes, as compared to the wild-type (Figure 4). fla18sos5 and prc1 displayed the most severe perturbation in root elongation in the presence of Fluridon (Figure 4B). This was accompanied by significant PR-swelling in all genotypes apart from the wild type (Figure 4C). These findings were further supported by the transgenic lines (T2) expressing the coding sequence of FLA18, driven by 35S promoter, in fla18 mutant background. FLA18 overexpression reversed the Fluridon hypersensitivity of the fla18 mutants (Supplementary Figure 11). Complementation of the fla18 phenotype was evident in both PR length and diameter and was documented for three independent lines (Supplementary Figure 11). To further examine the relationship between ABA and the FLA proteins during root development we followed the expression of CYP707A1, encoding an ABA 8’-hydroxylase employed in ABA catabolism and C-TERMINAL DOMAIN PHOSPHATASE LOCI 1 (CPL1), encoding a negative regulator of ABA-induced genes. The expression of CPL1 remained un-altered in all fla mutants examined, as compared to the wild type (Supplementary Figure 12). However, the level of CYP707A1 expression was slightly but significantly induced in all fla-mutants examined, under permissive conditions, as compared to the wild-type. Under restrictive conditions, the levels of CYP707A1 expression was slightly reduced in both single mutants and slightly increased in the fla18-1sos5 double mutant background, as compared to the wild type (Supplementary Figure 12). To summarize, these observations support a role for FLA18 in both LR and PR elongation. Moreover it suggests that, similar to FLA4/SOS5, FLA18 may also display cross-talk with ABA through a yet to be identified mechanism.
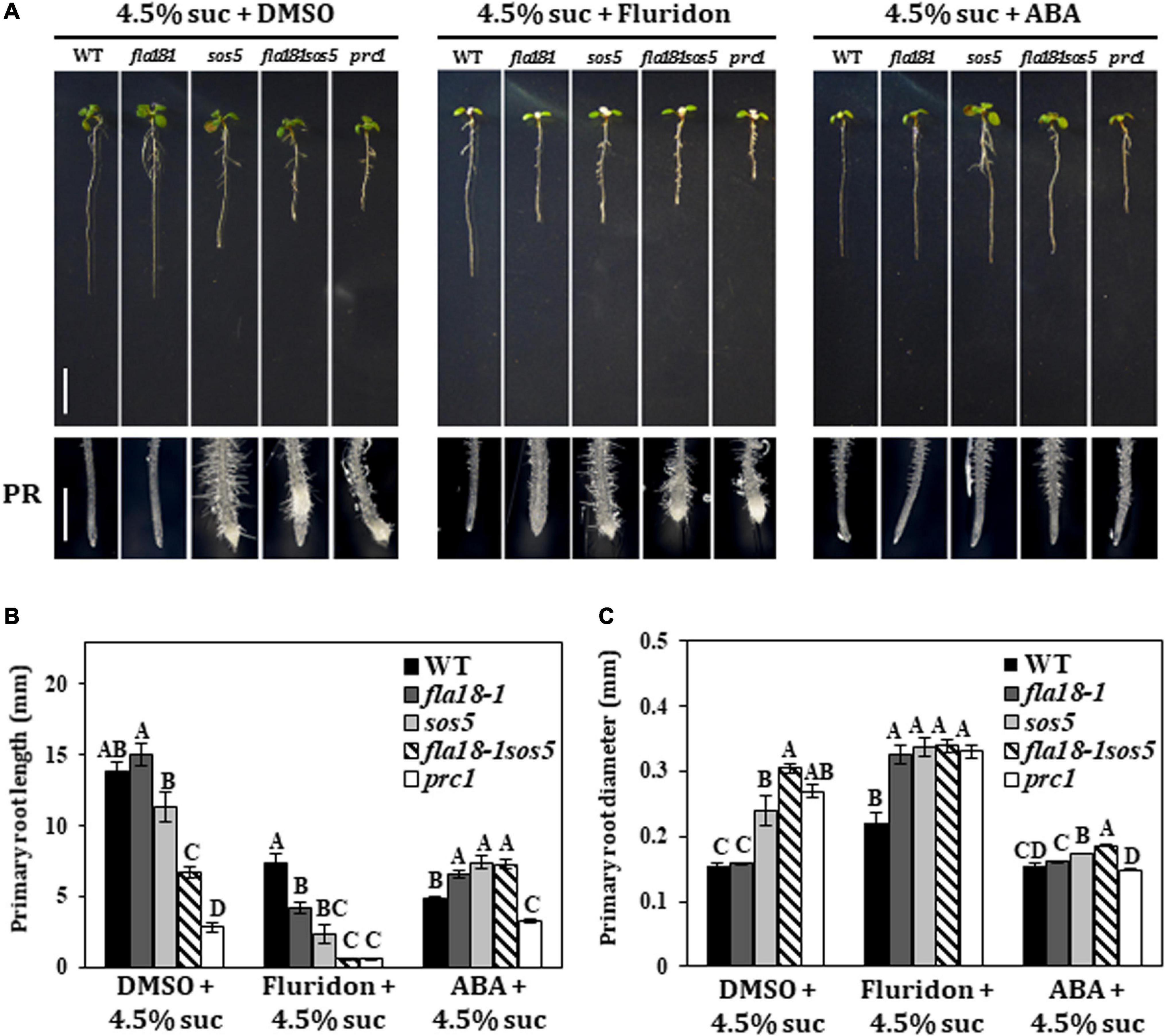
Figure 4. Root elongation in fla18 and fla18sos5 is suppressed by ABA and hypersensitive to the ABA-synthesis inhibitor, Fluridon. (A) Seedlings of the indicated genotypes were grown on MS medium with 1% sucrose, for 5 days, and then transferred to medium containing 4.5% sucrose and either 5 mM ABA, 5 mM Fluridon or similar volume of DMSO, the ABA/Fluridon solvent. (B,C) Measurements of primary root length (B) and diameter (C) were conducted 48 h after seedlings were transferred to new media. The results were analyzed using JMPpro13 for statistical analysis and run through Tukey’s HSD test (p < 0.05); n = 6. Error bars represent SE. Scale bars = 1 cm (A; upper panel) and 1 mm (A; lower panel).
Various cell wall mutants, including sos5 and prc1, have been shown to be hypersensitive to the cellulose synthase inhibitor, Isoxaben (Desprez et al., 2002; Fridman et al., 2014; Basu et al., 2016). To investigate the effect of the fla18 mutation on cell wall properties, we examined the sensitivity of root elongation to Isoxaben in fla18-1 and fla18-1sos5 mutant background. Seedlings were grown for 4 days under ambient conditions and then transferred to MS medium supplemented with 1% sucrose and increasing Isoxaben concentrations for an additional 48 h. sos5 and prc1, served as positive controls for this assay. The fla18 PR is hypersensitive to Isoxaben as reflected by the reduction in PR length as well as the increase in PR tip width compared to the wild type. The fla18sos5 double mutant, proved even more hypersensitive to isoxaben, as compared to wild-type and the single mutants (Figure 5). This was evident by the shorter root length and increased radial swelling observed at lower concentrations of Isoxaben. The effect of isoxaben on fla18sos5 PR length was already observed at 0.5 nM, a lower concentration compared to both wild type and the single mutants (Figure 5B) while the effect on the root diameter was only observed at 1 nM isoxaben (Figure 5C).
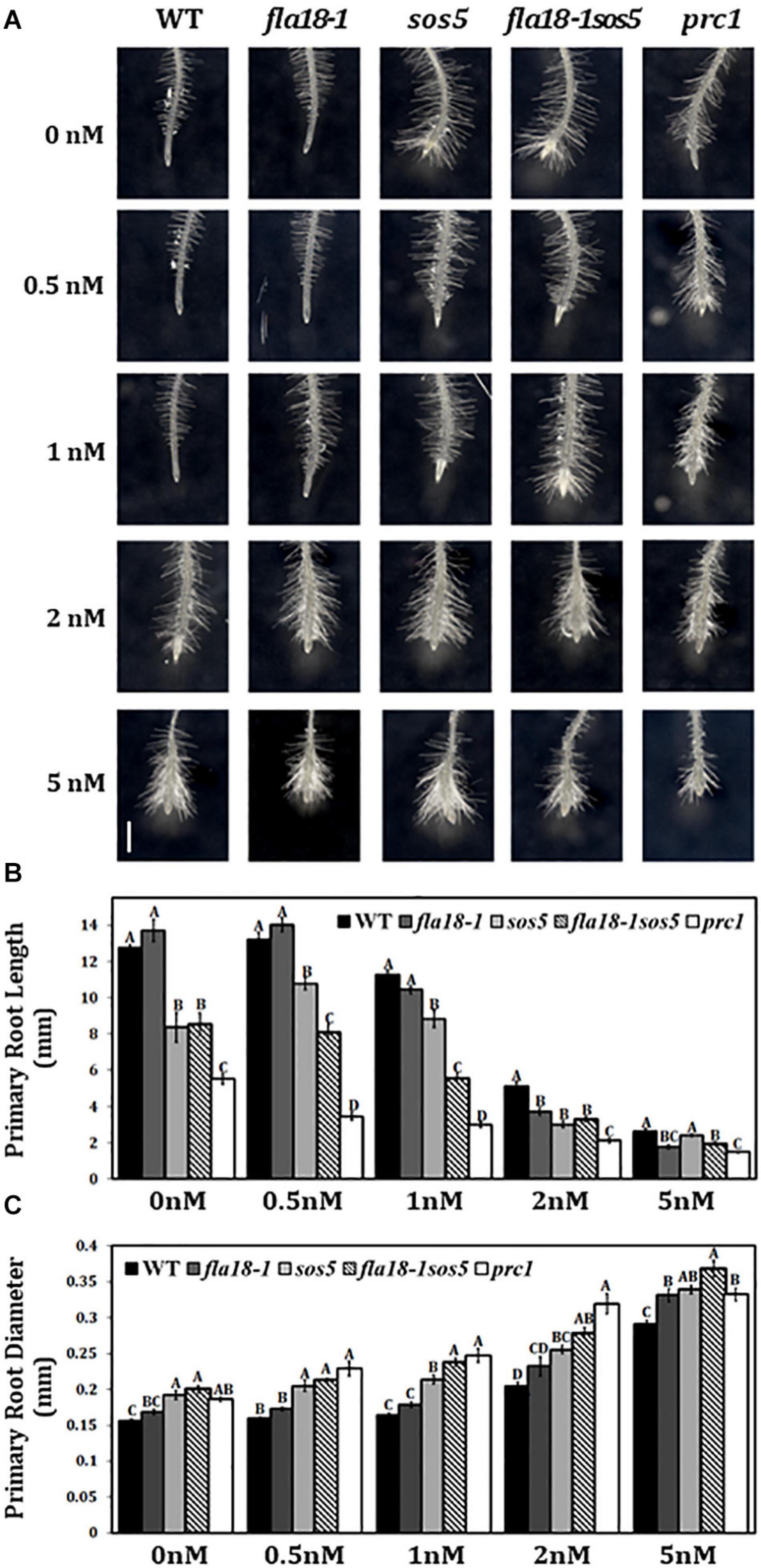
Figure 5. fla18 and fla18sos5 are hypersensitive to the cellulose synthase inhibitor, Isoxaben. (A) Primary root tips of seedlings of the indicated genotypes were grown on MS medium with 1% sucrose for 4 days and then transferred to medium containing 1% sucrose and the indicated concentration of isoxaben, for 48 h. (B,C) Measurements of PR length (B) and diameter (C) at the end of the experiment. The results were analyzed using JMPpro13 for statistical analysis and run through Tukey’s HSD test (p < 0.05). Error bars represent SE. PR length: n = 6 (prc1; n = 5). PR diameter: n = 4. Scale bar = 1 mm (A).
Previous studies done on various cell wall mutants have been able to show that in some cases, perturbation of cell wall deposition can lead to ectopic lignin deposition. Phloroglucinol is a histochemical stain that specifically detects 4-O-linked hydroxy-cinnamyl aldehydes, which form part of the lignin polymer (Supplementary Figure 13; Pomar et al., 2002). The mutants prc1 and sos5 served as positive controls as they were previously shown to display ectopic lignin deposition stained by phloroglucinol (Desprez et al., 2002; Caño-Delgado et al., 2003; Denness et al., 2011; Basu et al., 2016). Phloroglucinol stain demonstrated ectopic lignin deposition also in fla18sos5 (Supplementary Figure 13). Altogether, these results suggest that FLA18 may affect root elongation through modification of cell wall deposition. Further research will be required to test this hypothesis.
In plants, organ growth occurs as a result of highly synchronized cell expansion of numerous cells simultaneously. To enable growth without protoplast rupture, cell wall loosening and reconstruction must occur in a tightly regulated manner. In the current study, we used tissue specific co-expression analysis to uncover new gene-products involved in the regulation of root elongation. This led to the identification of FLA18, a fasciclin-like arabinogalactan protein, as a new regulator of PR and LR elongation in Arabidopsis. The expression of FLA18 suggests various roles in different developmental contexts along the course of plant development. Yet, the most prominent phenotype detected was the modification of root architecture by differential regulation of LR vs. PR growth in response to different growth conditions.
The FLA proteins are a sub-family of extra-cellular arabinogalactan-proteins (AGPs) characterized by the presence of one or two fasciclin (FAS) domains (Schultz et al., 2000, 2002; Johnson et al., 2003; Seifert and Roberts, 2007; Showalter et al., 2010). The FAS domains are 110 to 150 amino acids long, they share low overall sequence similarity, but possess two highly conserved regions of approximately 15 amino acids each, and a conserved central YH motif (Johnson et al., 2003; Seifert, 2018). FLA proteins were identified in all kingdoms including animals, yeast, bacteria, algae, lichens, and seed plants (Elkins et al., 1990; Huber and Sumper, 1994; Kawamoto et al., 1998; Gaspar et al., 2001; Paulsrud and Lindblad, 2002; Johnson et al., 2003; Novakoviæ et al., 2018; Yeats et al., 2018; Shafee et al., 2020). They were shown to be highly glycosylated and frequently predicted to have a glycosylphosphatidylinositol (GPI)-anchor that attaches them to the outer leaflet of the plasma membrane, facing the extra-cellular matrix (Schultz et al., 2002; Johnson et al., 2003; Seifert and Roberts, 2007; Ellis et al., 2010; Basu et al., 2016; Xue et al., 2017; Yeats et al., 2018). Proteins containing FAS domains, from a broad spectrum of organisms, have been shown to function as adhesion molecules (Elkins et al., 1990; Huber and Sumper, 1994; Kawamoto et al., 1998; Schultz et al., 2000).
The FLA gene-family in Arabidopsis is composed of 21 genes (Supplementary Figure 1; Schultz et al., 2000; Johnson et al., 2003; MacMillan et al., 2010; Costa et al., 2019; Shafee et al., 2020). He et al. (2019) revised this number and suggested there are only 20 FLA genes in Arabidopsis. The biological function of most FLA-encoded proteins remained largely unknown. Partly, because of high levels of functional redundancy and the fact that mutant analysis uncovered very few phenotypes so far. Of the limited number of FLA genes with established function, FLA4, also known as SOS5, was also shown to affect cell wall deposition. The sos5 mutant displays short and swollen roots under restrictive conditions of high-sucrose or high-salt containing media. This phenotype was associated with perturbation of the middle lamella required for cell-cell adhesion (Shi et al., 2003; Xu et al., 2008; Xue and Seifert, 2015; Basu et al., 2016; Xue et al., 2017). The sos5 mutant displays also a mild-swelling of the hypocotyls of dark-grown seedlings and perturbation of seed mucilage organization (Xu et al., 2008; Harpaz-Saad et al., 2011; Griffiths et al., 2014, 2016; Basu et al., 2016). FLA11 and FLA12, which are expressed during xylem cell differentiation, were assigned a function in secondary cell wall deposition. The fla11fla12 double mutant displays reduced cellulose content, alteration in cellulose microfibril angle, reduced tensile strength and reduced tensile modulus of elasticity. Similar results were obtained also for FLA11 and FLA12 orthologs in other species like cotton, poplar, eucalypt and hemp (Huang et al., 2008; MacMillan et al., 2015; Bygdell et al., 2017; Guerriero et al., 2017). Recent study, identified FLA16 as an additional player required for secondary cell wall deposition during stem elongation in Arabidopsis. The presented data, demonstrate that he fla16 mutant displays reduced stem length, alteration in stem biomechanical properties and reduced cellulose levels suggesting a role in secondary cell wall deposition (Liu et al., 2020). FLA3 was assigned a function in microspore development. FLA3 RNA interference lines displayed reduced intine cell wall synthesis and reduced calcofluor stain for glycans in aborted pollen grains (Li et al., 2010). Hence multiple FLA proteins were assigned function in cell wall deposition or organization. In addition to the FLA proteins mentioned above, mutant analysis and gene expression studies have suggested that FLA1 plays a role in early events of shoot regeneration in tissue culture (Johnson et al., 2011). The phenotypic analysis of mutants in FLA9 suggest it is essential to prevent seed abortion (Cagnola et al., 2018). The function of the other FLA-encoding genes, fourteen in Arabidopsis, is yet to be identified.
The Arabidopsis FLA genes were classified into four groups based on the number of FAS domains (one or two), the number of AGP domains (one or two), and whether they contain a predicted GPI-anchor modification site (Johnson et al., 2003; MacMillan et al., 2010). Phylogenetic studies suggest that these four groups are maintained throughout the evolution of land plants (Costa et al., 2019; He et al., 2019; Shafee et al., 2020). FLA18 was classified as part of group B which contains FLA15, FLA16, FLA17 and FLA18. Group B FLAs have two FAS1 domains separated by an AGP-region and are not predicted to contain a GPI-anchor. Recent study by Liu et al. (2020), assigned function to the first group B-FLA, FLA16. The fla16 mutant displays short stems, reduced stem diameter, reduced stem pith cell number, reduced stem cellulose levels, and modification of stem biomechanical properties. These suggest that FLA16 plays a role as part of the mechanism regulating secondary cell wall deposition and cell wall integrity during stem growth. This was further supported by the localization of FLA16-fusion protein, driven by the endogenous promoter, to the cell wall and plasma membrane-wall interface of cells producing secondary cell wall like fiber and xylem cells. Interestingly, etiolated seedlings of the fla16 mutant display reduced hypocotyl length as compared to the wild type and hypersensitivity to the cellulose synthase inhibitor, Isoxaben. Given that secondary cell wall cellulose synthesis is not affected by isoxaben (Watanabe et al., 2018) this suggests a possible role also as part of the mechanism required for primary cell wall deposition in the context of dark-grown seedlings (Liu et al., 2016). In the current study, we identified FLA18 through tissue-specific co-expression analysis using established players involved in primary cell wall deposition required for PR elongation. Given that genes involved in the same metabolic process tend to express in a transcriptionally coordinated manner (Persson et al., 2005; Ruprecht and Persson, 2012; Ben-Tov et al., 2015; Voiniciuc et al., 2015a, b, 2018) this suggests that FLA18 may play a role in this process. Further support for this hypothesis arise from the short and swollen LR phenotype of the fla18 mutants, which is a characteristic feature of various cell wall mutants, like: CELLULOSE SYNTHASE A 6 (Fagard et al., 2000), CELLULOSE SYNTHASE A 3 (Caño-Delgado et al., 2003), COBRA (Schindelman et al., 2001), and others. Moreover, the double mutants fla18-1sos5 and fla18-2sos5 display a more severe perturbation of both LR and PR anisotropy, compared to each of the single mutants. Additionally, both fla18 and fla18sos5 are hypersensitive to the cellulose synthase inhibitor, Isoxaben. Altogether, these results support a role for FLA18, a group B-FLA, as part of the complex mechanism required for primary cell wall deposition during LR and PR elongation. Further studies will be required to prove whether FLA18 is indeed part of the mechanism employed in cell wall deposition.
It is interesting to note, that in most cell wall mutants, the perturbation of root anisotropy can be observed in both PR and LRs. This includes mutants in genes required for cell wall synthesis and deposition, including: CELLULOSE SYNTHASE A 6 (Fagard et al., 2000), CELLULOSE SYNTHASE A 3 (Caño-Delgado et al., 2003), CELLULOSE SYNTHASE A 1 (Williamson et al., 2001) COBRA (Schindelman et al., 2001), CHITINASE LIKE 1 (Hermans et al., 2010), CELLULOSE SYNTHASE INTERACTIVE PROTEIN 1 (Hauser et al., 1995), and others. Additionally, mutants like fei1fei2 and sos5, affecting cell wall organization through a yet to be identified mechanism, also demonstrate perturbation of root growth anisotropy in both PR and LRs, under restrictive conditions (Shi et al., 2003; Xu et al., 2008; Basu et al., 2016; Xue et al., 2017). Altogether, this accumulating data, led to the hypothesis that the same set of core components is required for root elongation in both PR and LRs. To our surprise, when we examined the fla18 mutants, perturbation of root anisotropy was observed only in LRs, when examined under restrictive conditions of high-sucrose containing medium. While the PR displays slight but consistent increase in elongation rate, under permissive conditions, as compared to the wild type. This suggests a divergence in the mechanism involved in the regulation of PR and LR elongation. Various studies have shown that PRs and LRs may display distinct growth dynamics in response to different developmental or environmental cues. For example, mild nitrate deficiency significantly enhances LR growth, but does not affect the growth of the PR (Zhang, 1998; Zhang et al., 1999; López-Bucio et al., 2003; Gruber et al., 2013). In another example, low phosphate levels result in modification of root architecture by reducing the rate of both PR and LR elongation but each responds to different phosphate concentrations (Williamson et al., 2001; López-Bucio et al., 2002; Pérez-Torres et al., 2008; Gruber et al., 2013). Similarly, accumulating data demonstrate that the plant hormone ABA has a much stronger inhibitory effect on LR growth as compared to the effect on PR growth (Signora et al., 2002; De Smet et al., 2003; Duan et al., 2013). Altogether, these results suggest that root morphogenesis can be altered by differential regulation of PR vs. LR elongation in response to different developmental and environmental signals. Moreover it suggests that FLA18 is not a core component of cell wall deposition but rather can affect PR and LR elongation in opposite ways yielding modification of root architecture.
Phenotypic analysis of fla18, sos5, and the fla18sos5 double mutant suggests that a sub-set of FLA proteins is required to maintain root growth under different growth conditions. The genetic interaction between fla18 and sos5 demonstrate that in the fla18 background the phenotype of sos5 becomes much more severe, as compared to each of the single mutants and the wild type. Despite the phylogenetic distance between these two FLA proteins (Supplementary Figure 1), the genetic interaction between these two mutants suggests some levels of functional redundancy. This is supported by the following observations: (i) the fla18sos5 mutant displays reduced root growth under permissive conditions in which the sos5 root resembles the wild-type and fla18 display slightly longer PR compared to the wild type; (ii) The double mutant displays a synergistic effect on both PR and LR elongation leading to shorter and more swollen roots, as compared to each of the single mutants and the wild type, under restrictive conditions of either high-sucrose or high-salt containing media; and (iii) a small and chlorotic shoot phenotype that can be detected when seedlings are grown on high-salt containing medium, a phenotype that cannot be detected in any of the single mutants. These results indicate that FLA18, FLA4/SOS5, and potentially additional FLA proteins (like FLA17, the closest homolog of FLA18) each plays a unique role and yet they are partially functionally redundant in various developmental contexts. Previous studies already present evidence for functional redundancy within the FLA-gene family. For example, MacMillan et al. (2010), found that while single-mutants in fla11 or fla12 exhibited a very mild phenotype, the fla11fla12 double mutant displayed a more pronounced phenotype compared to the wild-type and either of the single mutants. Recent study, demonstrated that FLA16 is an additional player affecting secondary cell wall deposition during stem growth (Liu et al., 2020). Moreover, gene expression studies suggested that other group B FLAs like FLA15 and FLA18 are also highly expressed in elongating stems, suggesting additional FLAs may play a role in this context (Liu et al., 2020). It will be fascinating to see whether FLA18 functions in both primary and secondary cell wall deposition. Future research will be required in order to identify the sub-set of FLAs that function in each developmental context and the way specific vs. overlapping roles are determined.
The Columbia (Col-0) ecotype of Arabidopsis thaliana was used in this study. The fla18 alleles (fla18-1, SALK_086944; fla18-2, SALK_039619), sos5 [SALK_125874; (Xu et al., 2008; Harpaz-Saad et al., 2011; Seifert et al., 2014; Xue et al., 2017; Basu et al., 2016; Griffiths et al., 2016)], and prc1 (Fagard et al., 2000) mutants were obtained from the Arabidopsis Biological Resource Center (Alonso, 2003). Homozygous plants of each line were identified via PCR-based genotyping, using gene and insert-specific primers (detailed in Supplementary Table 2).
For growth in soil, plants were grown at 22°C in 75 μE, under long day conditions, with a light regime of 16/8 h. For growth in vitro, seeds were surface-sterilized, sown on bacteriological square petri dishes (120 mm x 120 mm x 17 mm, Greiner Bio-One, 688102) containing1x Murashige and Skoog (MS) medium (Tivan Biotech, MSP01-50LT) with pH adapted to 5.8 using KOH and 0.6% Phytagel (Sigma-Aldrich, P8169). Following cold treatment at 4°C, for 4 d in the dark, plants were grown for 4–7 days in an upright position, at 22°C in 75 μE, under long-day conditions, with a light regime of 16/8 h. For induction of restrictive conditions in vitro, plants were transferred after 4 d in the light to plates maintained in an upright position, with MS medium containing 0.6% phytagel and one of the following additions: 4.5% sucrose, 100 mM NaCl plus 1% sucrose or the indicated concentrations of Isoxaben (Sigma-Aldrich, 36138) or Fluridon (Sigma-Aldrich, 45511). Plants transferred to MS medium containing 0.6% phytagel and the corresponding solvent served as control (ambient conditions). The root images were documented using a stereo-microscope (SMZ1270, Nikon) equipped with a camera (NIKON, DS- Ri2).
Root length was measured using Image J (FIJI) software (Schindelin et al., 2012) and the results were analyzed using the JMP pro13 or JMP pro 15 software for statistical analysis (Statistical DiscoveryTM, SAS). For the measurement of root elongation rate, the root tip of each seedling was marked daily for 5 consecutive days after their transfer to the indicated medium. Measurements of PR and LR length were conducted 7 d after the transfer to the indicated conditions. Due to large variability in LR length, in each plant, 2 of the longest roots, located in the upper third part of the root system, were selected and used for LR length measurements (Supplementary Figure 6). PR and LR diameter was measured at the widest point of the root tip, approximately 1–4 mm from the root tip.
Total RNA was isolated from the indicated tissues using a plant/fungi total RNA extraction kit (Norgen Biotek, 25800), followed by Turbo-DNase treatment (Invitrogen, AM1907). The first strand of complementary DNA (cDNA) was synthesized from 1 μg of the total RNA using SuperScript II reverse transcriptase (Invitrogen, 18064022), according to the manufacturer’s instructions. PCR products from the cDNA of the indicated genotypes (Wild type, fla18-1, fla18-2, and fla4/sos5) were amplified, 28 cycles for each of the reactions, using FLA18 or TUBULIN-specific primers. TUBULIN served as a reference gene (Supplementary Table 2).
Wild type genomic fragments comprising the promoter region of FLA18 (2260 bp) or coding sequence region (1390 bp) were amplified by PCR (Supplementary Table 2) using the Phusion Taq polymerase (Thermo Fisher Scientific, F-549S), as described by the manufacturer. The fragments were cloned into pENTR-TOPO-D (Invitrogen, K240020). The resultant entry plasmid was used for the LR-clonase reaction (as described by the manufacturer; Invitrogen, 11791020), introducing: i) the FLA18-promoter sequence into the binary vector pGWB3 for the expression of a FLA18-promoter driven β-GUS protein; or, ii) the FLA18-coding sequence into the pGWB5 binary vector (Nakagawa et al., 2007). The FLA18-promoter-pGWB3 vector was transformed to wild-type Columbia background while the FLA18-CDS-pGWB5 was transformed to fla18-1 for complementation. Transgenic plants were selected on hygromycin and independent lines were identified.
Plants from three independent transgenic lines harboring the FLA18 promoter driving GUS expression, were used for histochemical activity assay performed using 5-bromo-4-chloro-3-indolyl β-D-glucuronide sodium salt, as previously described (Twell et al., 1990). Briefly, samples were immersed in staining solution composed of 100 mM sodium phosphate buffer (pH 7.0) with 10 mM EDTA (pH 8.0), 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 1 mM 5-bromo-4-chloro-3-indolyl-b-glucuronic acid, and 0.1% Triton X-100. The tissue was stained either for 1 h or overnight at 37°C, as indicated. Chlorophyll was removed using 95% ethanol. T2 plants from 8 independent transgenic lines were analyzed at different developmental stages, and 3 representative lines were documented using stereo-microscope (SMZ1270, Nikon).
Phloroglucinol staining (Sigma-Aldrich, P3502) was performed according to a previously described procedure (Caño-Delgado et al., 2003). Seedlings were grown for 4 d on plates with sucrose-free MS and then transferred to restrictive conditions for 7 d growing period in the light. The seedlings were then stained for approximately 5 min with a 2% phloroglucinol-HCl solution.
nCounter NanoString is a hybridization-based platform used to follow gene expression (Geiss et al., 2008; Amit et al., 2009). To monitor gene expression during root elongation, seedlings were grown for 4 d on sucrose-free MS and then transferred to fresh sucrose-free MS, MS supplemented with 4.5% sucrose or supplemented with 100 mM NaCl plus 1% sucrose - for 6 h. A total of 20–25 root tips were harvested and stored in −80°C. Total RNA was extracted using the Single Cell RNA Purification Kit (Norgen Biotek, 51800). The NanoString probes were designed and synthesized by NanoString Technologies (detailed in Supplementary Table 3)1. As reference genes we used GAPC (AT3G04120), UBQ10 (AT5G53300), EF1a (AT5G60390) and F-BOX FAMILY PROTEIN (AT5G15710), AP2 (AT5G46630). Counts were normalized using the nSolver, according to the nCounter Gene Expression Assay Manual2.
GenBank accession numbers: FLA18 (AT3G11700), FLA4 (AT3G46550), CYP707A1 (AT4G19230), RD29B (AT5G52300), GAPC (AT3G04120), UBQ10 (AT5G53300), EF1a (AT5G60390), F-BOX FAMILY PROTEIN (AT5G15710), and AP2 (AT5G46630).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HA performed the experiments, analyzed and interpreted the data, and wrote the article with under the supervision of SH-S and assistance of the other authors. HR assisted with experimental procedures. AI-M assisted with RNA extraction and nCounter NanoString analysis. DZ did the bioinformatic analysis under the supervision of OT. All authors contributed to the article and approved the submitted version.
SH-S is a recipient of an I-CORE grant number 757/12 and personal grant number 474/17 from The Israel Science Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Harpaz-Saad lab members for fruitful discussions, friendship, and always being there to lend a hand and help in any way required.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.645286/full#supplementary-material
Supplementary Figure 1 | Evolutionary relationships between FASCICLIN-LIKE proteins in Arabidopsis.
Supplementary Figure 2 | FLA18 is co-expressed with genes involved in primary cell wall deposition during root elongation.
Supplementary Figure 3 | FLA18 expression pattern in Arabidopsis plants.
Supplementary Figure 4 | FLA18 gene expression in T-DNA insertion mutants, as compared to wild type.
Supplementary Figure 5 | Primary root elongation in fla18 mutants.
Supplementary Figure 6 | Lateral root length measurements for quantification.
Supplementary Figure 7 | Expression of FLA18 suppresses the short and swollen lateral root phenotype of fla18-1.
Supplementary Figure 8 | Primary root length of fla18-1sos5.
Supplementary Figure 9 | The fla18sos5 double mutant phenotype on media containing 1% sucrose.
Supplementary Figure 10 | FLA18 and SOS5 gene expression in fla18-1, sos5 and the double mutant fla18-1sos5.
Supplementary Figure 11 | Expression of FLA18 suppresses the fla18-1 primary root hyper-sensitivity to the ABA-synthesis inhibitor, Fluridon.
Supplementary Figure 12 | Expression of genes involved in ABA signaling and metabolism in fla-mutant background.
Supplementary Figure 13 | Histochemical Staining of the fla18-1sos5 double mutant with phloroglucinol.
Supplementary Table 1 | The genes used as ‘baits’ for the tissue-specific co-expression analysis.
Supplementary Table 2 | Primers used in this study.
Supplementary Table 3 | Probes used in the nCounter NanoString analysis for gene expression.
Alonso, J. M. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Amit, I., Garber, M., Chevrier, N., Leite, A. P., Donner, Y., Eisenhaure, T., et al. (2009). Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263. doi: 10.1126/science.1179050
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. doi: 10.1126/science.279.5351.717
Bacic, A., Harris, P. J., and Stone, B. A. (1988). “Structure and function of plant cell walls,” in The Biochemistry of Plants, ed. J. Priess (New York, NY: Academic Press), 297–371.
Baskin, T. I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21, 203–222. doi: 10.1146/annurev.cellbio.20.082503.103053
Bygdell, J., Srivastava, V., Obudulu, O., Srivastava, M. K., Nilsson, R., Sundberg, B., et al. (2017). Protein expression in tension wood formation monitored at high tissue resolution in Populus. J. Exp. Bot. 68, 3405–3417. doi: 10.1093/jxb/erx186
Basu, D., Tian, L., Debrosse, T., Poirier, E., Emch, K., Herock, H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. doi: 10.1371/journal.pone.0145092
Ben-Tov, D., Abraham, Y., Stav, S., Thompson, K., Loraine, A., Elbaum, R., et al. (2015). COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol. 167, 711–724. doi: 10.1104/pp.114.240671
Brady, S. M., Orlando, D. A., Lee, J. Y. L., Wang, J. Y., Koch, J., Dinneny, J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. doi: 10.1126/science.1146265
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542.major
Brown, M. R. Jr., Saxena, I., and Kudlicka, K. (1996). Cellulose biosynthesis in higher plants. Trends Plant Sci. 1, 149–156. doi: 10.1016/S1360-1385(96)80050-1
Cagnola, J. I., Dumont, de Chassart, G. J., Ibarra, S. E., Chimenti, C., Ricardi, M. M., et al. (2018). Reduced expression of selected FASCICLIN-LIKE ARABINOGALACTAN PROTEIN genes associates with the abortion of kernels in field crops of Zea mays (maize) and of Arabidopsis seeds. Plant Cell Environ. 41, 661–674. doi: 10.1111/pce.13136
Caño-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34, 351–362. doi: 10.1046/j.1365-313X.2003.01729.x
Caño-Delgado, A. I., Metzlaff, K., and Bevan, M. W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405.
Carpita, N. C., and Gibeaut, D. M. (1993). Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x
Casimiro, I., Marchant, A., Bhalerao, R. P., Beeckman, T., Dhooge, S., Swarup, R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. doi: 10.1105/tpc.13.4.843
Chen, S., Jia, H., Zhao, H., Liu, D., Liu, Y., Liu, B., et al. (2016). Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiol. 171, 242–250. doi: 10.1104/pp.15.01874
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Costa, M., Pereira, A. M., Pinto, S. C., Silva, J., Pereira, L. G., and Coimbra, S. (2019). In silico and expression analyses of fasciclin-like arabinogalactan proteins reveal functional conservation during embryo and seed development. Plant Reprod. 32, 353–370. doi: 10.1007/s00497-019-00376-7
Crowell, E. F., Timpano, H., Desprez, T., Franssen-Verheijen, T., Emons, A.-M., Hofte, H., et al. (2011). Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis Hypocotyl. Plant Cell 23, 2592–2605. doi: 10.1105/tpc.111.087338
Darley, C. P., Forrester, A. M., and McQueen-Mason, S. J. (2001). The molecular basis of plant cell wall extension. Plant Mol. Biol. 47, 179–195. doi: 10.1023/A:1010687600670
De Smet, I., Signora, L., Beeckman, T., Inzé, D., Foyer, C. H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555. doi: 10.1046/j.1365-313X.2003.01652.x
Delmer, D. P. (1999). Cellulose biosynthesis: exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. doi: 10.1146/annurev.arplant.50.1.245
Denness, L., McKenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Desprez, T., Juraniec, M., Crowell, E. F., Jouy, H., Pochylova, Z., Parcy, F., et al. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15572–15577. doi: 10.1073/pnas.0706569104
Desprez, T., Vernhettes, S., Fagard, M., Refrégier, G., Desnos, T., Aletti, E., et al. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128, 482–490. doi: 10.1104/pp.010822
Duan, L., Dietrich, D., Ng, C. H., Chan, P. M. Y., Bhalerao, R., Bennett, M. J., et al. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25, 324–341. doi: 10.1105/tpc.112.107227
Elkins, T., Hortsch, M., Bieber, A. J., Snow, P. M., and Goodman, C. S. (1990). Drosophila fasciclin I is a novel homophilic adhesion molecule that along with fasciclin III can mediate cell sorting. J. Cell Biol. 110, 1825–1832. doi: 10.1083/jcb.110.5.1825
Ellis, M., Egelund, J., Schultz, C. J., and Bacic, A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419. doi: 10.1104/pp.110.156000
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., et al. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell Online 12, 2409–2424. doi: 10.1105/tpc.12.12.2409
Fridman, Y., Elkouby, L., Holland, N., Vragoviæ, K., Elbaum, R., and Savaldi-Goldstein, S. (2014). Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev. 28, 912–920. doi: 10.1101/gad.239335.114
Gaspar, Y., Johnson, K. L., McKenna, J. A., Bacic, A., and Schultz, C. J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176.
Geiss, G. K., Bumgarner, R. E., Birditt, B., Dahl, T., Dowidar, N., Dunaway, D. L., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325. doi: 10.1038/nbt1385
Green, P. B. (1980). Organogenesis-a biophysical view. Annu. Rev. Plant Physiol. 31, 51–82. doi: 10.1146/annurev.pp.31.060180.000411
Griffiths, J. S., Crepeau, M.-J., Ralet, M.-C., Seifert, G. J., and North, H. M. (2016). Dissecting seed mucilage adherence mediated by FEI2 and SOS5. Front. Plant Sci. 7, 1–13. doi: 10.3389/fpls.2016.01073
Griffiths, J. S., Tsai, A. Y. L., Xue, H., Voiniciuc, C., oola, K., Seifert, G. J., et al. (2014). SALT-OVERLY SENSITIVE5 mediates arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol. 165, 991–1004. doi: 10.1104/pp.114.239400
Gruber, B. D., Giehl, R. F. H., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Guerriero, G., Mangeot-Peter, L., Legay, S., Behr, M., Lutts, S., Siddiqui, K. S., et al. (2017). Identification of fasciclin-like arabinogalactan proteins in textile hemp (Cannabis sativa L.): in silico analyses and gene expression patterns in different tissues. BMC Genomics 18, 1–13. doi: 10.1186/s12864-017-3970-5Google Scholar
Harpaz-Saad, S., McFarlane, H. E., Xu, S., Divi, U. K., Forward, B., Western, T. L., et al. (2011). Cellulose synthesis via the FEI2 RLK/SOS5 pathway and CELLULOSE SYNTHASE 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J. 68, 941–953. doi: 10.1111/j.1365-313X.2011.04760.x
Hauser, M. T., Morikami, A., and Benfey, P. N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252.
He, J., Zhao, H., Cheng, Z., Ke, Y., Liu, J., and Ma, H. (2019). Evolution analysis of the fasciclin-like arabinogalactan proteins in plants shows variable fasciclin-AGP domain constitutions. Int. J. Mol. Sci. 20, 1–17. doi: 10.3390/ijms20081945
Heim, D. R., Roberts, J. L., Pike, P. D., and Larrinua, I. M. (1989). Mutation of a locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiol. 90, 146–150. doi: 10.1104/pp.90.1.146
Hermans, C., Porco, S., Verbruggen, N., and Bush, D. R. (2010). Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 152, 904–917. doi: 10.1104/pp.109.149849
Huang, G. Q., Xu, W. L., Gong, S. Y., Li, B., Wang, X. L., Xu, D., et al. (2008). Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol. Plant. 134, 348–359. doi: 10.1111/j.1399-3054.2008.01139.x
Huber, O., and Sumper, M. (1994). Algal-CAMs: isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to Drosophila fasciclin I. Embo J. 13, 4212–4222.
Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. doi: 10.1146/annurev.arplant.47.1.377
Johnson, K. L., Jones, B. J., Bacic, A., and Schultz, C. J. (2003). The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules 1. Plant Physiol. 133, 1911–1925. doi: 10.1104/pp.103.031237.kinases
Johnson, K. L., Kibble, N. A. J., Bacic, A., and Schultz, C. J. (2011). A Fasciclin-Like Arabinogalactan-Protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS One 6:e25154. doi: 10.1371/journal.pone.0025154
Kawamoto, T., Noshiro, M., Shen, M., Nakamasu, K., Hashimoto, K., Kawashima-Ohya, Y., et al. (1998). Structural and phylogenetic analyses of RGD-CAP/βig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim. Biophys. Acta Gene Struct. Exp. 1395, 288–292. doi: 10.1016/S0167-4781(97)00172-3
Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, C. R., and Brown, R. M. (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11, 2075–2085. doi: 10.1105/tpc.11.11.2075
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lavenus, J., Goh, T., Roberts, I., Guyomarc, S., Smet, I., and De, et al. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. doi: 10.1016/j.tplants.2013.04.006
Li, J., Yu, M., Geng, L.-L., and Zhao, J. (2010). The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 64, 482–497. doi: 10.1111/j.1365-313X.2010.04344.x
Liu, E., MacMillan, C. P., Shafee, T., Ma, Y., Ratcliffe, J., Van de Meene, E., et al. (2020). Fasciclin-Like Arabinogalactan-Protein 16 (FLA16) is required for stem development in Arabidopsis. Front. Plant Sci. 11:615392. doi: 10.3389/fpls.2020.615392
López-Bucio, J., Cruz-Ramírez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. doi: 10.1016/S1369-5266(03)00035-9
López-Bucio, J., Hernández-Abreu, E., Sánchez-Calderón, L., Nieto-Jacobo, M. F., Simpson, J., Herrera-estrella, L., et al. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. doi: 10.1104/pp.010934
Lynch, J. (1995). Root architecture and plant productivity. Plant Physiol. 109, 7–13. doi: 10.1104/pp.109.1.7
MacMillan, C. P., Mansfield, S. D., Stachurski, Z. H., Evans, R., and Southerton, S. G. (2010). Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 62, 689–703. doi: 10.1111/j.1365-313X.2010.04181.x
MacMillan, C. P., Taylor, L., Bi, Y., Southerton, S. G., Evans, R., and Spokevicius, A. (2015). The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 206, 1314–1327. doi: 10.1111/nph.13320
Malamy, J. E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77. doi: 10.1111/j.1365-3040.2005.01306.x
Malamy, J. E., and Benfey, P. N. (1997). Down and out in arabidopsis: the formation of lateral roots. Trends Plant Sci. 2, 390–396. doi: 10.1016/S1360-1385(97)01118-7
McFarlane, H. E., Döring, A., and Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Meents, M. J., Watanabe, Y., and Samuels, A. L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121, 1107–1125. doi: 10.1093/aob/mcy005
Mueller, S. C., Brown, R. M., and Scott, T. K. (1976). Cellulosic microfibrils: nascent stages of synthesis in a higher plant cell. Science 194, 949–951. doi: 10.1126/science.194.4268.949
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nakamura, H., and Asami, T. (2014). Target sites for chemical regulation of strigolactone signaling. Front. Plant Sci. 5:623. doi: 10.3389/fpls.2014.00623
Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Nicol, F. (1998). A plasma membrane-bound putative endo-1,4-beta -D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. Embo J. 17, 5563–5576. doi: 10.1093/emboj/17.19.5563
Novakoviæ, L., Guo, T., Bacic, A., Sampathkumar, A., and Johnson, K. (2018). Hitting the wall—sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 7:89. doi: 10.3390/plants7040089
Paulsrud, P., and Lindblad, P. (2002). Fasciclin domain proteins are present in nostoc symbionts of lichens. Appl. Environ. Microbiol. 68, 2036–2039. doi: 10.1128/AEM.68.4.2036-2039.2002
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., and Stalker, D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 12637–12642. doi: 10.1073/pnas.93.22.12637
Pérez-Torres, C.-A., López-Bucio, J., Cruz-Ramírez, A., Ibarra-Laclette, E., Dharmasiri, S., Estelle, M., et al. (2008). Phosphate availability alters lateral root development in arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20, 3258–3272. doi: 10.1105/tpc.108.058719
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Persson, S., Wei, H., Milne, J., Page, G. P., and Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638. doi: 10.1073/pnas.0503392102
Pomar, F., Merino, F., and Barceló, A. R. (2002). O -4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220, 17–28. doi: 10.1007/s00709-002-0030-y
Ransbotyn, V., Yeger-Lotem, E., Basha, O., Acuna, T., Verduyn, C., Gordon, M., et al. (2015). A combination of gene expression ranking and co-expression network analysis increases discovery rate in large-scale mutant screens for novel Arabidopsis thaliana abiotic stress genes. Plant Biotechnol. J. 13, 501–513. doi: 10.1111/pbi.12274
Richmond, T. A., and Somerville, C. R. (2000). The cellulose synthase superfamily. Plant Physiol. 124, 495–498. doi: 10.1104/pp.124.2.495
Ruprecht, C., and Persson, S. (2012). Co-expression of cell wall-related genes: New tools and insights. Front. Plant Sci. 3, 1–7. doi: 10.3389/fpls.2012.00083
Ruprecht, C., Mutwil, M., Saxe, F., Eder, M., Nikoloski, Z., and Persson, S. (2011). Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Front. Plant Sci. 2:23. doi: 10.3389/fpls.2011.00023
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schindelman, G., Morikami, A., Jung, J., Baskin, T. I., Carpita, N. C., Derbyshire, P., et al. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. doi: 10.1101/gad.879101
Schultz, C. J., Johnson, K. L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1767. doi: 10.1105/tpc.12.9.1751
Schultz, C. J., Rumsewicz, M. P., Johnson, K. L., Jones, B. J., Gaspar, Y. M., and Bacic, A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. doi: 10.1104/pp.003459
Seifert, G. (2018). Fascinating fasciclins: a surprisingly widespread family of proteins that mediate interactions between the cell exterior and the cell surface. Int. J. Mol. Sci. 19:1628. doi: 10.3390/ijms19061628
Seifert, G. J., and Roberts, K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161. doi: 10.1146/annurev.arplant.58.032806.103801
Seifert, G. J., Xue, H., and Acet, T. (2014). The Arabidopsis thaliana fasciclin like arabinogalactan protein 4 gene acts synergistically with abscisic acid signalling to control root growth. Ann. Bot. 114, 1125–1133. doi: 10.1093/aob/mcu010
Shafee, T., Bacic, A., and Johnson, K. (2020). Evolution of sequence-diverse disordered regions in a protein family: order within the chaos. Mol. Biol. Evol. 37, 2155–2172. doi: 10.1093/molbev/msaa096
Shi, H., Kim, Y., and Guo, Y. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. doi: 10.1105/tpc.007872.)
Showalter, A. M., Keppler, B., Lichtenberg, J., Gu, D., and Welch, L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513. doi: 10.1104/pp.110.156554
Signora, L., De Smet, I., Foyer, C. H., and Zhang, H. (2002). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28, 655–662. doi: 10.1046/j.1365-313x.2001.01185.x
Slovak, R., Ogura, T., Satbhai, S. B., Ristova, D., and Busch, W. (2016). Genetic control of root growth: from genes to networks. Ann. Bot. 117, 9–24. doi: 10.1093/aob/mcv160
Somerville, C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22, 53–78. doi: 10.1146/annurev.cellbio.22.022206.160206
Taiz, L. (1984). Plant cell expansion: regulation of cell wall mechanical properties. Annu. Rev. Plant Physiol. 35, 585–657. doi: 10.1146/annurev.pp.35.060184.003101
Tateno, M., Brabham, C., and Debolt, S. (2016). Cellulose biosynthesis inhibitors - a multifunctional toolbox. J. Exp. Bot. 67, 533–542. doi: 10.1093/jxb/erv489
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tian, H., Smet, I., and De, and Ding, Z. (2014). Shaping a root system?: regulating lateral versus primary root growth. Trends Plant Sci. 19, 426–431. doi: 10.1016/j.tplants.2014.01.007
Turner, S. R., and Somerville, C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. doi: 10.1105/tpc.9.5.689
Twell, D., Yamaguchi, J., and Mccormick, S. (1990). Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–713.
Vilches-Barro, A., and Maizel, A. (2015). Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23, 31–38. doi: 10.1016/j.pbi.2014.10.005
Voiniciuc, C., Engle, K. A., Günl, M., Dieluweit, S., Schmidt, M. H.-W., Yang, J., et al. (2018). Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 178, 1045–1064. doi: 10.1104/pp.18.00584
Voiniciuc, C., Guenl, M., Schmidt, M. H.-W., and Usadel, B. (2015a). Highly branched xylan made by IRX14 and MUCI21 links mucilage to Arabidopsis seeds. Plant Physiol. 169, 2481–2495. doi: 10.1104/pp.15.01441
Voiniciuc, C., Schmidt, M. H.-W., Berger, A., Yang, B., Ebert, B., Scheller, H. V., et al. (2015b). MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 169, 403–420. doi: 10.1104/pp.15.00851
Wang, S., Yin, Y., Ma, Q., Tang, X., Hao, D., and Xu, Y. (2012). Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol. 12:138. doi: 10.1186/1471-2229-12-138
Watanabe, Y., Schneider, R., Barkwill, S., Gonzales-Vigil, E., Hill, J. L. Jr., Samuels, A. L., et al. (2018). Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E6366–E6374.
Williamson, L. C., Ribrioux, S. P. C. P., Fitter, A. H., and Leyser, H. M. O. (2001). Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 126, 875–882. doi: 10.1104/pp.126.2.875
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” Browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Xiong, L., Lee, H., Ishitani, M., and Zhu, J.-K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596. doi: 10.1074/jbc.M109275200
Xu, S.-L., Rahman, A., Baskin, T. I., and Kieber, J. J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell Online 20, 3065–3079. doi: 10.1105/tpc.108.063354
Xue, H., and Seifert, G. J. (2015). Fasciclin like arabinogalactan protein 4 and respiratory burst oxidase homolog D and F independently modulate abscisic acid signaling. Plant Signal. Behav. 10:e989064. doi: 10.4161/15592324.2014.989064
Xue, H., Veit, C., Abas, L., Tryfona, T., Maresch, D., Ricardi, M. M., et al. (2017). Arabidopsis thaliana FLA4 functions as a glycan-stabilized soluble factor via its carboxy-proximal Fasciclin 1 domain. Plant J. 91, 613–630. doi: 10.1111/tpj.13591
Yang, B., Voiniciuc, C., Fu, L., Dieluweit, S., Klose, H., and Usadel, B. (2019). TRM4 is essential for cellulose deposition in Arabidopsis seed mucilage by maintaining cortical microtubule organization and interacting with CESA3. New Phytol. 221, 881–895. doi: 10.1111/nph.15442
Yang, X., Ye, C.-Y., Bisaria, A., Tuskan, G. A., and Kalluri, U. C. (2011). Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci. 181, 675–687. doi: 10.1016/j.plantsci.2011.01.020
Yeats, T. H., Bacic, A., and Johnson, K. L. (2018). Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J. Integr. Plant Biol. 60, 649–669. doi: 10.1111/jipb.12659
Zhang, H. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. doi: 10.1126/science.279.5349.407
Zhang, H., Jennings, A., Barlow, P. W., and Forde, B. G. (1999). Dual pathways for regulation of root branching by nitrate. Plant Biol. 96, 6529–6534. doi: 10.1073/pnas.96.11.6529
Keywords: fasciclin-like, arabinogalactan protein, cell wall, root elongation, ABA, abiotic stress
Citation: Allelign Ashagre H, Zaltzman D, Idan-Molakandov A, Romano H, Tzfadia O and Harpaz-Saad S (2021) FASCICLIN-LIKE 18 Is a New Player Regulating Root Elongation in Arabidopsis thaliana. Front. Plant Sci. 12:645286. doi: 10.3389/fpls.2021.645286
Received: 22 December 2020; Accepted: 19 February 2021;
Published: 07 April 2021.
Edited by:
Georg J. Seifert, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Catherine Rayon, University of Picardie Jules Verne, FranceCopyright © 2021 Allelign Ashagre, Zaltzman, Idan-Molakandov, Romano, Tzfadia and Harpaz-Saad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Smadar Harpaz-Saad, c21hZGFyLmhhcnBhekBtYWlsLmh1amkuYWMuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.