
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 05 January 2022
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.644424
This article is part of the Research TopicRosaceae Fruit Development and QualityView all 11 articles
The function of floral organ identity genes, APETALA1/2/3, PISTILLATA, AGAMOUS, and SEPALLATA1/2/3, in flower development is highly conserved across angiosperms. Emerging evidence shows that these genes also play important roles in the development of the fruit that originates from floral organs following pollination and fertilization. However, their roles in fruit development may vary significantly between species depending on the floral organ types contributing to the fruit tissues. Fruits of the Rosaceae family develop from different floral organ types depending on the species, for example, peach fruit flesh develops from carpellary tissues, whereas apple and strawberry fruit flesh develop from extra-carpellary tissues, the hypanthium and receptacle, respectively. In this review, we summarize recent advances in understanding floral organ gene function in Rosaceae fruit development and analyze the similarities and diversities within this family as well as between Rosaceae and the model plant species Arabidopsis and tomato. We conclude by suggesting future research opportunities using genomics resources to rapidly dissect gene function in this family of perennial plants.
Floral organs are classified into four types, sepals, petals, stamens, and carpels. Their development is regulated by four different classes of floral organ identity genes according to the proposed ABCE model (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Krizek and Fletcher, 2005). In summary, for Arabidopsis sepal formation is specified by the class A genes APETALA1 (AP1) and AP2, petal formation is specified by the class A genes and two class B genes [APETALA3 (AP3) and PISTILLATA (PI)]; stamen formation requires AP3 and PI together with the class C gene AGAMOUS (AG); and carpel formation is controlled by the AG alone (Weigel and Meyerowitz, 1994). Proper development of all four whorls of floral organs also requires the class E SEPALLATA (SEP) genes. The consequence of mutating all four SEPALLATA genes (sep1, sep2, sep3, and sep4) simultaneously is a plant that produces flowers consisting of reiterating whorls of leaf-like organs (Krizek and Fletcher, 2005). All class A, B, C, and E genes encode MADS-box transcription factors, except for AP2, which encodes a transcription factor belonging to the super-family of AP2/ERF proteins (Okamuro et al., 1997). However, classification of a gene as being a member of the ABCE model is based on its function and not sequence similarity to other members of the ABCE model.
The diversity of fruit types seen between various flowering plant species is primarily a consequence of different carpellary and/or extra-carpellary floral tissues contributing to fruit formation (Spjut, 1994). Botanical fruit, also known as true fruit, is derived from carpellary tissues, whereas accessory fruit, also known as pseudo fruit, has fruit flesh derived from extra-carpellary tissues (Liu et al., 2020). With fruit being derived from floral organs, the genes that regulate the growth of floral organs also play important roles in the control of fruit development. This was clearly demonstrated by the functions of the Arabidopsis FRUITFULL (FUL), an AP1 homolog (Gu et al., 1998), tomato TM29, a SEPALLATA homolog (Ampomah-Dwamena et al., 2002), and poppy AP1/FUL homologs (Pabón-Mora et al., 2012).
Rosaceae species produce flowers with five sepals and petals, and a varied number of stamens and carpels. Their carpels show variations in shape, degrees of fusion, and relative position to other floral organs. These variations are the key contributing factors of fruit type diversity in Rosaceae (Liu et al., 2020). For example, peach develops a superior ovary as the single carpel is positioned above all other floral organs and goes on to develop a drupe. In contrast, apple has five fused carpels that are below the attachment point of all other floral organs, develops an inferior ovary that fuses with the surrounding hypanthium tissues formed from the bases of sepals, petals, and stamens and develops a pome (Liu et al., 2020).
In addition to floral structure variations, differential enlargement of specific floral tissues also contributes to fruit type diversity in Rosaceae. Strawberry and raspberry, for example, have a similar floral structure that consists of numerous carpels growing on a receptacle. Strawberry is an achenetum because its carpels are not enlarged after fertilization and become dry achenes, but the receptacle is enlarged and grows into the fleshy pseudo fruit. In contrast, raspberry is a drupetum because its carpels are enlarged after fertilization to form fleshy drupelets but the receptacle is not enlarged or fleshy (Liu et al., 2020).
Together with flower and fruit morphological differences, duplicated floral organ genes have the opportunity to play diversified roles in fruit development. In Rosaceae, whole-genome duplication (WGD) events have resulted in gene copies and their potential to have dosage effects, redundancy of function, or for copies to have undergone functional changes, such as neo-functionalization (one gene copy taking on a totally new function) or sub-functionalization (each gene copy retaining a subset of the original ancestral functions; Conant et al., 2014; Xiang et al., 2017). In the following sections, we summarize recent advances in understanding floral organ gene duplication, expression profiles, and function in relation to Rosaceae fruit development. We also analyze the similarities and diversities within this family predominantly using apple, peach, and strawberry as examples representing the diversity of fruit types in the family. The similarities and diversities between Rosaceae and other plants, in particular Arabidopsis and tomato, are also analyzed.
Prior to the assembly of reference genome sequences, sequence-based approaches using homology to the Arabidopsis genes were used to clone and identify floral organ MADS-box genes from Rosaceae species. In apple, several studies identified a total of two AP1-related genes, MdMADS5 (MdAP1; Yao et al., 1999; Kotoda et al., 2002) and MdMADS2 (Sung et al., 1999), one PI homolog, MdPI (Yao et al., 2001), two AP3 homologs, MdMADS13 (Van Der Linden et al., 2002) and MdTM6 (Kitahara et al., 2004), three AG-related genes, MdMADS10 (Yao et al., 1999), MdMADS14, and MdMADS15 (Van Der Linden et al., 2002), and five SEP-related genes, MdMADS4 (Sung et al., 2000), MdMADS6, 7, 8, and 9 (Yao et al., 1999). After the first apple reference genome was assembled (Velasco et al., 2010), three additional SEP-related genes were identified, MdMADS18, 104, and 118 (Ireland et al., 2013). All these genes, except for MdPI, have been annotated in the second apple reference genome generated using a haploid plant derived from the cultivar “Golden Delicious” (Daccord et al., 2017).
In this review, floral MADS-box genes are identified from the reference genome of peach (Verde et al., 2013), strawberry (Edger et al., 2017), and apple (Daccord et al., 2017). Their gene IDs are listed in Table 1 together with their class, based on gene lineage, and the relevant references. Their relationship is shown in a protein sequence-based phenology tree in Figure 1A. One or more genes from peach, strawberry, and apple are identified as homologs of each of the Arabidopsis floral MADS-box genes. In apple, 12 floral MADS-box genes are present as homolog pairs, most likely due to a recent apple-specific whole-genome duplication (Velasco et al., 2010).
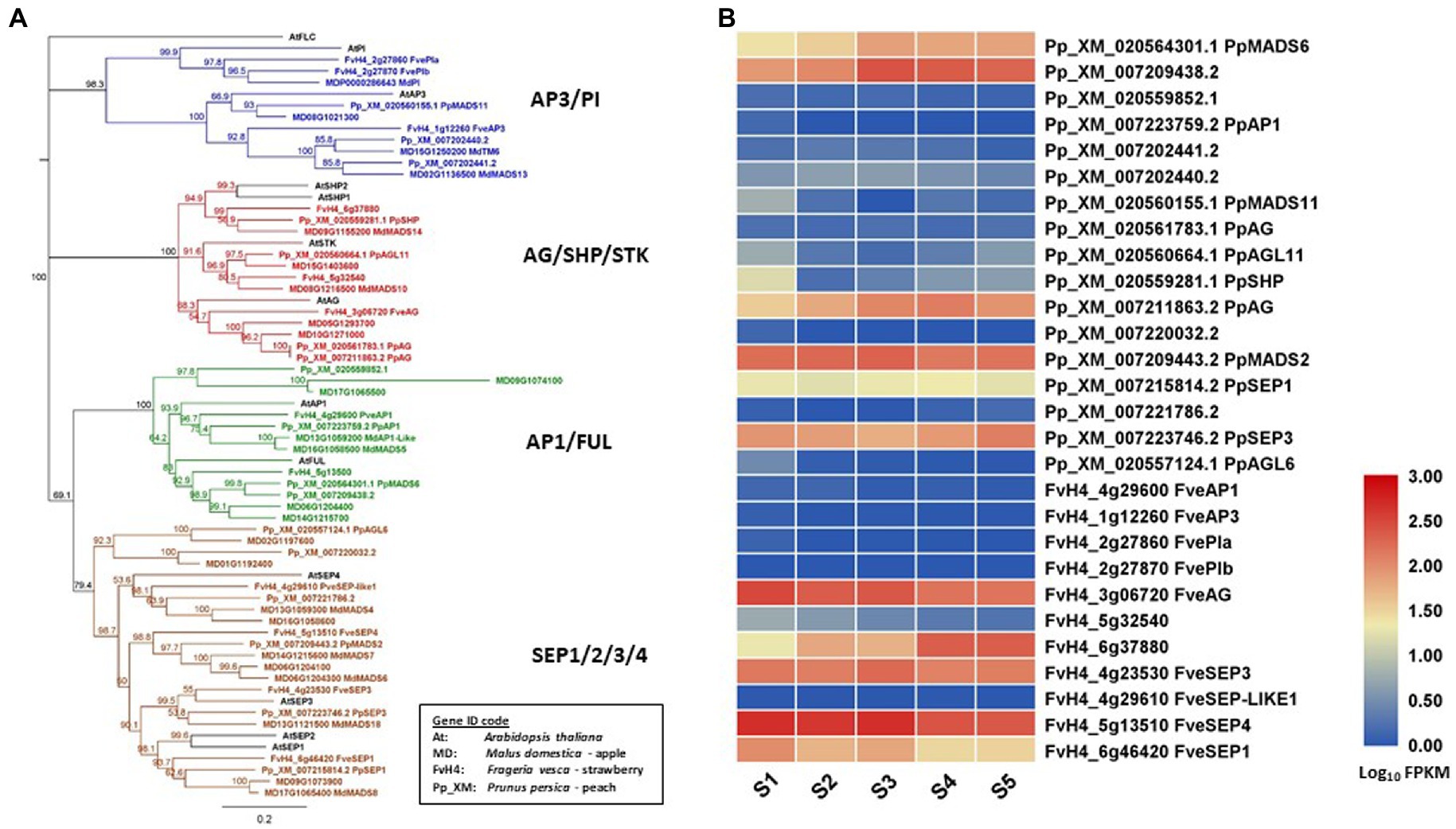
Figure 1. Phylogeny tree of floral MADS-box proteins and their gene expression in developing fruit tissues. (A) a Neighbor-Joining consensus tree was constructed using Geneious software (v10.0.9) with 1,000 bootstrap replicates after protein sequences were aligned using Geneious Alignment software (v10.0.9). MADS-box protein sequences downloaded from the reference genome of apple (Daccord et al., 2017), peach (Verde et al., 2013), strawberry (Edger et al., 2017), and Arabidopsis. The Arabidopsis protein IDs are AT1G69120.1 (AtAP1), AT5G60910.1 (AtFUL), AT5G20240.1(AtPI), AT3G54340.1 (AtAP3), AT4G18960.1 (AtAG), AT3G58780.1 (AtSHP1), AT2G42830.1 (AtSHP2), AT4G09960.1 (AtSTK), AT5G15800.1 (AtSEP1), AT3G02310.1 (AtSEP2), AT1G24260.1 (AtSEP3), AT2G03710.1 (AtSEP4), and AT5G10140.1 (AtFLC). (B) a heatmap shows the expression patterns of floral MADS-box genes in fruit tissues of peach and strawberry at five developmental stages (S1-S5) based on previously published RNA-seq data (Pei et al., 2020). Rosaceae Gene IDs are coded as: MD for apple (Malus domestica), Pp_XM for peach (based on Prunus_persica_NCBIv2) and FvH4 for strawberry (based on Frageria vesca H4). S1 to S5: 21, 42, 56, 84, and 105 days after full bloom for peach, and 11, 27, 31, 43, and 47 DAFB for strawberry.
Expression patterns of SEP-like genes in apple fruit have been described previously, such as the AtSEP1/2-like MADS1/MADS8, and MADS9 showing high expression at two to seven days after pollination, the PhFBP9/23-like MADS3/MADS7 and MADS6 showing high expression at four to eight weeks after pollination, and the AtSEP4-like MADS4 showing strong expression in young fruits (Sung and An, 1997; Yao et al., 1999; Sung et al., 2000). In addition, Yao et al. (1999) demonstrated the expression of class AP1- and AG-related genes in the fruit flesh of apple. Detection of their expression in fruit indicates that AP1-, AG-, and SEP-related MADS-box genes have a potential role in apple fruit development.
Transcriptome analyses of whole fruit tissues without seeds collected from five developmental stages revealed that two FUL homologs (PpMADS6 and XM_007209438), an AG homolog (PpAG), and three SEP homologs (PpMADS2, PpSEP1, and PpSEP3) are expressed in peach fruit. The same analyses showed that an AG homolog (FveAG), a SHP homolog (FvH6g37880), and three SEP homologs (FveSEP1/3/4) are expressed in strawberry (Fragaria x ananassa) fruit (Pei et al., 2020; Figure 1B). In addition, two AP3-related genes (XM007202440 and PpMADS11) are also expressed in peach fruit during development although their expression levels are relatively low (Figure 1B). The expression of AP3-related genes in peach fruit is interesting because this expression is not detected in apple or strawberry fruit. In contrast, expression of AP1/FUL homologs is at a high level in apple and peach but not detected in strawberry. It would be worthwhile determining the functions of theAP1/FUL and AP3/PI homologs in regulating fruit development of Rosaceae.
AP2 homologous genes have been identified from the reference genomes of apple (Daccord et al., 2017), peach (Verde et al., 2013), and strawberry (Edger et al., 2017; Figure 2A). In apple, there are eight euAP2 (Kim et al., 2006) homologs that contain a miR172 target site in the coding sequences and encode proteins containing two AP2 domains (Yao et al., 2015). In peach and strawberry, five and four such AP2 homologs have been identified, respectively. The AP2 homologs of these species are classified into four distinct clades. Each clade contains two apple and one strawberry homolog. One clade contains two peach homologs and of other three clades each contains one peach homolog (Figure 2A). The specific duplication of AP2 homologs in apple provides new potential of dosage effect or functional changes. Among the euAP2 homologs, one in each species (MD15G1286400, FvH_1g16350, and Pp_XM_007207942) is expressed predominantly in the developing fruit tissues (Yao et al., 2015; Pei et al., 2020; Figures 2A,B). These three homologs are in the same clade as Arabidopsis AP2 (AtAP2) in the phylogenetic tree (Figure 2A). Further insights into the roles of these floral MADS-box and euAP2-like genes in fruit development may be obtained by analyzing their expression patterns using transcriptome data sets generated in Rosaceae species (Kang et al., 2013; Hawkins et al., 2017; Hu et al., 2018; Pei et al., 2019; Shahan et al., 2019; Tan et al., 2019; Ying et al., 2019; Shang et al., 2020).
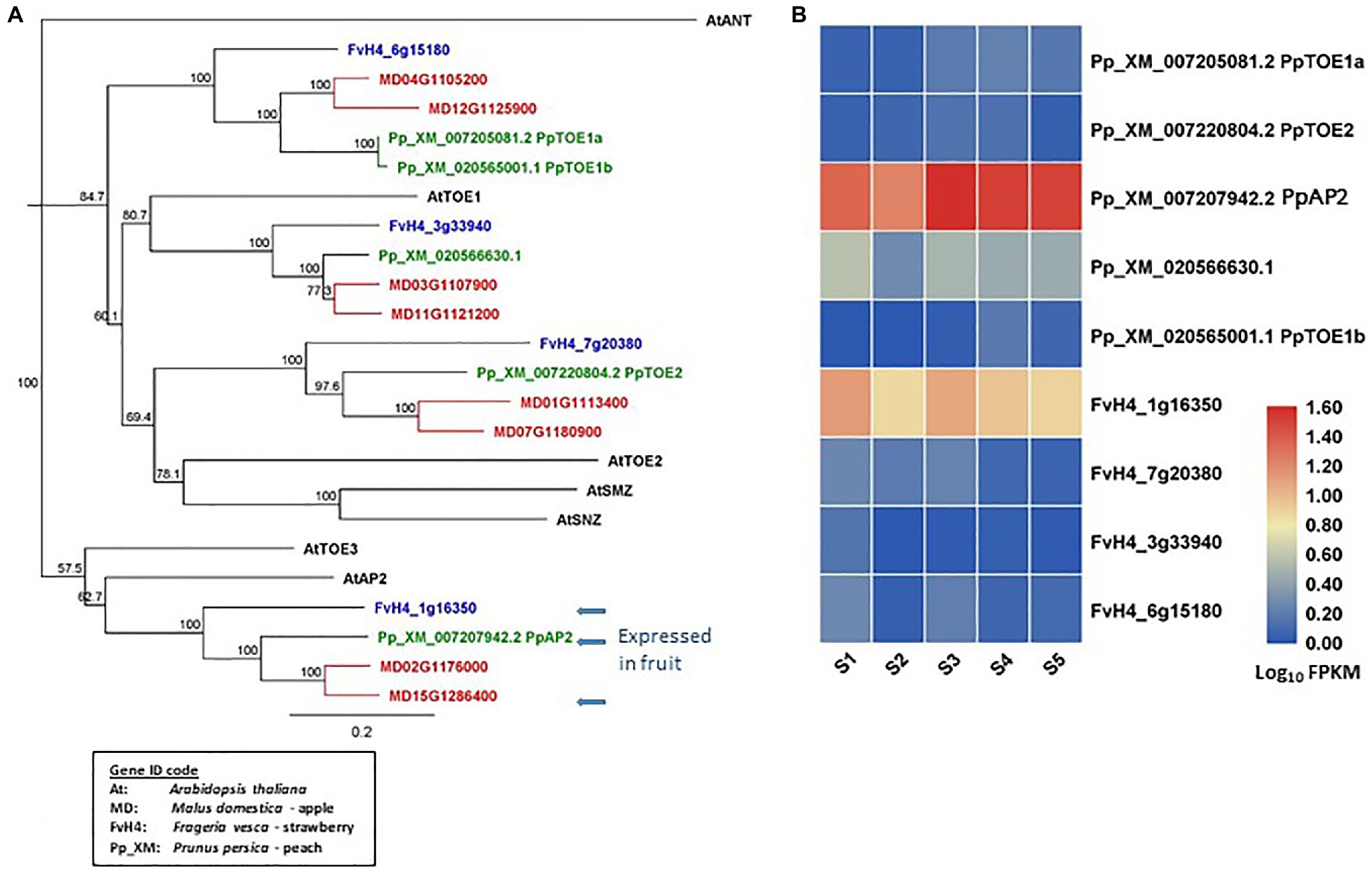
Figure 2. Phylogeny tree of AP2-like proteins and their gene expression in developing fruit tissues. (A) a Neighbor-Joining consensus tree was constructed using Geneious software (v10.0.9) with 1,000 bootstrap replicates after protein sequences were aligned using Geneious Alignment software (v10.0.9). AP2-like protein sequences downloaded from the reference genome of apple, peach, strawberry, and Arabidopsis. The Arabidopsis protein IDs are AT4G36920.1 (AtAP2), AT2G28550.3 (AtTOE1), AT5G60120.2 (AtTOE1), AT5G67180.1 (AtTOE3), AT3G54990.1 (AtSMZ), AT2G39250.1 (AtSNZ), and AT4G37750.1 (AtANT). (B) a heatmap shows the expression patterns of AP2-like genes in fruit tissues of peach and strawberry at five developmental stages (S1-S5) based on previously published RNA-seq data (Pei et al., 2020). Rosaceae Gene IDs are coded as: MDs for apple (Malus domestica), Pp_XM for peach (based on Prunus_persica_NCBIv2), and FvH4 for strawberry (based on Frageria vesca H4). S1 to S5: 21, 42, 56, 84, and 105 days after full bloom for peach, and 11, 27, 31, 43, and 47 DAFB for strawberry. The arrows indicate AP2 genes predominantly expressed in fruits.
From the above analyses of gene sequences and expression patterns, it is clear that these three species in the Rosaceae family contain similar genes in the floral MADS-box and AP2 clades, but the gene number varies among the three species (Figure 2A). Expression of these genes generally shows a degree of similarity across the three species described, although specific temporal and spatial expression patterns are present (Figure 2B; Kang et al., 2013; Hawkins et al., 2017; Hu et al., 2018; Pei et al., 2019; Shahan et al., 2019; Tan et al., 2019; Ying et al., 2019; Shang et al., 2020). Evidence to date would indicate that there are conserved and diversified functions for these genes in different species, although the research to understanding these functions is very much in its infancy.
In plants, microRNA172 (miR172) is highly conserved and targets a subfamily of AP2-like genes to repress their expression, by initiating degradation and/or inhibiting translation of the target mRNA (Aukerman and Sakai, 2003; Chen, 2004; Zhu et al., 2009; Zhu and Helliwell, 2011). As members of the euAP2 subfamily have different spatial and temporal patterns of expression (Okamuro et al., 1997; Tang et al., 2007; Yao et al., 2015) and interact with different proteins (Teotia and Tang, 2015), the MIR172 gene family has the potential to affect a number of aspects of plant development.
In annual plant species, early flowering is a phenotypic change commonly observed for over-expression of miR172 (Aukerman and Sakai, 2003; Chen, 2004; Mlotshwa et al., 2006; Zhu and Helliwell, 2011; Teotia and Tang, 2015), and in some instances abnormalities of the floral organs have been observed (Mlotshwa et al., 2006; Zhu et al., 2009). As AP2 governs floral organ size (Jofuku et al., 2005) and floral organ development (Yant et al., 2010), then miR172 has the potential to influence growth of fruit.
In Arabidopsis, miR172 positively regulates Arabidopsis silique growth (Ripoll et al., 2015). The silique is derived from carpel tissues and its growth is negatively regulated by AP2 (Ripoll et al., 2011). The expression of AP2 is repressed in the silique valve by miR172 (Ripoll et al., 2015). Therefore, suppression of the function of miR172, through the use of microRNA mimicry technology, results in a reduction in silique size. Similarly, a reduction in the size of the silique occurs in transgenic plants expressing a modified AP2 gene without a functional miR172-target sequence (Ripoll et al., 2015).
Apple fruit flesh, is derived largely from the hypanthium, hypothesized to consist of the fused bases of the stamens, petals, and sepals (Pratt, 1988) and fruit core is derived from the carpellary tissues. Further molecular studies have indicated that sepal bases make the greatest contribution to apple flesh formation (Yao et al., 2001). Negative regulation of apple fruit growth by miR172 was demonstrated in transgenic plants over-expressing miR172 and also observed in natural variants of apple. Over-expression of miR172 in apple converted segments of sepals to petal-like tissues, indicating that AP2-like transcription factors are required for sepal development and furthermore the miR172 over-expression reduced fruit weight more than 60-fold (Figure 3; Yao et al., 2015). A transposable element (TE) insertional allele of the apple MIR172p gene is associated with reduction of MIR172 expression and co-located with one of four fruit size QTLs (Yao et al., 2015). In the progeny of a controlled cross, the TE insertional allele is associated with large fruit and underlies the evolution of fruit size from the small wild crabapples to large cultivated apples. Although the correlation between fruit size and the level of AP2 expression during fruit development could be interpreted as a direct effect of AP2 on fruit growth, one cannot discount that this is an indirect effect because AP2 represses the function of class C genes (e.g., AG) in the first whorl, according to the ABCE model (Wollmann et al., 2010). Therefore, miRNA-mediated reduction of AP2, resulting in elevated expression of class C genes in the first whorl during flower development, could lead to a conversion of sepals to carpel-like structures. As sepals contribute to the apple fruit flesh we postulate that this conversion of floral organs could account for the reduction in fruit size. However, a sepal to carpel conversion in apple over-expressing miR172 was not reported (Yao et al., 2015).
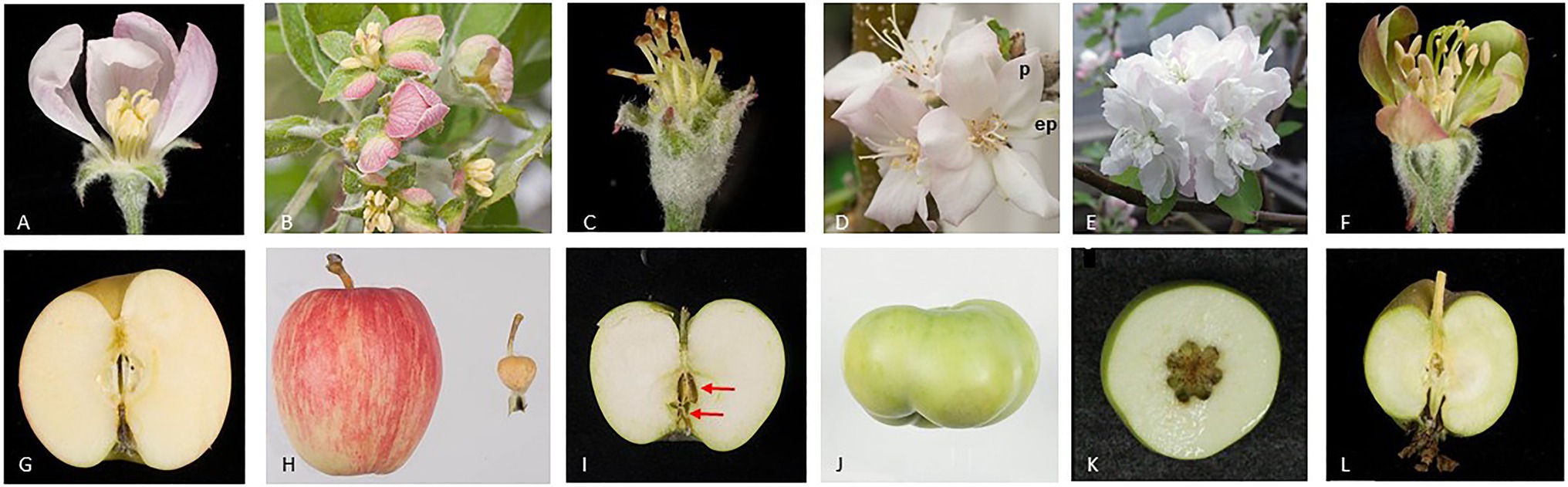
Figure 3. Apple flower and fruit phenotypes after alteration of the expression of floral organ genes. The images show open flowers (A–F) and mature fruit (G–L) of wild-type (A,G) and transgenic apple plants (B–F, H–L). miR172 is known to inhibit the expression of the AP2- and AP2-like genes. miR172 over-expression converts sepal segments to petal-like tissues (B) outer petals are removed to show sepal-petal conversion and dramatically reduces fruit size (H), wild-type fruit on left and transgenic fruit on right (Yao et al., 2015). Repressing expression of the MdPI gene converts petals to sepals, and stamens to carpels (C), and results in parthenocarpic (seedless) fruit with two whorls of carpels marked using two red arrows (I). However, over-expressing MdPI converts sepals to petals (p, petal; ep, ectopic petal) (D) and suppresses fruit tissue growth resulting in altered fruit shape (J) (Yao et al., 2001, 2018). Repressing expression of two AG homologs, MdMADS15/22, converts stamens to petals (E) and increases carpel number and therefore fruit locule number (K) (Klocko et al., 2016). Repressing expression of two SEP homologs, MdMADS8/9, partially converts petals to greenish sepal-like organs (F) and suppresses fruit flesh growth (L) (Ireland et al., 2013).
In tomato, over-expression of miR172 produced flowers consisting entirely of ovary tissues that develop into parthenocarpic fruit, with a fruit-in-fruit phenotype observed in some extreme cases (Yao et al., 2016). As in apple, the size of the tomato fruit was reduced significantly compared with wild-type control fruit, although the tomato fruit size reduction (4-5-fold) was not as dramatic as that observed in apple. Growth of tomato fruit flesh tissue is dependent largely on division and expansion of cells occurring post-fertilization through the stimulation by hormones released from the seeds. Although over-expression of miR172 may promote fruit development in the absence of fertilization, the parthenocarpic fruit is smaller than wild-type fruit. This “smaller than wild-type” size of plants over-expressing miR172 indicates that the direct or indirect influence of the miR172 transgene on promoting tomato fruit growth in these parthenocarpic fruit does not fully compensate for the growth stimulation of seed-derived hormones in wild-type fruit.
The reports described above have shown miR172/AP2 playing different roles in the regulation of the development of different fruit types represented by apple, Arabidopsis, and tomato. This suggests that miR172/AP2 has the potential for a diversified function in regulating the development of different fruits of the Rosaceae family, although to date, their function has not been reported in any Rosaceae species other than apple. It is interesting to note that the extreme phenotype of apple transgenic plants over-expressing miR172 displays abnormal flowers without sepals, petals, or stamens, but with separate carpels rather than the fused the carpels of normal apples, and no hypanthium surrounding the ovary (Figure 4; Yao et al., 2015). If these flowers were to develop into fruit, the fruit would resemble peach rather than apple, indicating miR172/AP2 could be involved in the diversification of fruit types in Rosaceae.

Figure 4. Flowers of apple transgenic plants over-expressing miR172. (A) Floral inflorescences show abnormal flowers consisting of leaf-like carpel tissues (lct), style, and stigma, but no sepals, petals, or stamens. (B) A florescence of (A) with leaf-like carpels removed reveals separate carpels consisting of ovary, style, and stigma. (C) A single carpel removed from the inflorescence in (B) reveals two ovules after cutting the ovary open (This figure is re-produced from Yao et al., 2015).
The Arabidopsis PI and AP3 are single-copy class B genes, expressed specifically in petals and stamens during their development (Goto and Meyerowitz, 1994; Jack et al., 1992). In PI or AP3 loss of function mutants, the flower phenotype is double the number of carpels, two whorls of sepals, and the absence of stamens or petals (Goto and Meyerowitz, 1994; Jack et al., 1992). Three apple varieties, Rae Ime, Spencer Seedless, and Wellington Bloomless, produce abnormal flowers that resemble Arabidopsis pi and ap3 mutants. Interestingly, the flowers of these three apple varieties develop parthenocarpic fruit, and their single-copy MdPI gene is not expressed owing to the presence of a long-terminal-repeat TE (Yao et al., 2001). Consistently, a similar flower phenotype and parthenocarpic fruit development are achieved with MdPI cosuppression in apple transgenic plants (Yao et al., 2018). The parthenocarpic trait of apple in MdPI loss of function mutants and cosuppression transgenic plants may be explained by the conversion of petals to sepals that are the key floral organs contributing to apple flesh tissue growth (Yao et al., 2001). An additional or alternative explanation for this parthenocarpic trait is that the loss of stamens, as a consequence of suppression of the class B MdPI, results in male sterility. Male sterility has been associated with parthenocarpic fruit production in multiple plant species, including tomato (Ingrosso et al., 2011; Medina et al., 2013; Takei et al., 2019), citrus (Vardi et al., 2008), and grape (Royo et al., 2016). In contrast, ectopic MdPI expression coverts sepals to petals and inhibits apple fruit growth toward the base of the fruit resulting in short and flat fruit (Figure 3). Histological analyses showed that suppression of cell expansion was likely to be the key reason for the reduced tissue growth, although the possibility of inhibition of cell division could not be ruled out (Yao et al., 2018). These two studies clearly indicated MdPI plays an inhibitory role on fruit growth.
The inhibitory role of the class B genes in fruit growth is also supported by studies in other plant species. For example, ectopic expression of VvPI, caused by an insertion of a miniature inverted-repeat TE, inhibits the growth of fruit flesh tissue and results in small, fleshless berries in grape (Vitis vinifera; Fernandez et al., 2013). In tomato, DEFICIENS (an AP3 homolog) expression is temporarily upregulated before and throughout anthesis in the wild-type but not in the parthenocarpic fruit (pat) mutant (Mazzucato et al., 2008), and silencing of TM6 (an AP3 homolog) expression using RNA interference produced parthenocarpic fruit (De Martino et al., 2006). These results suggest that the AP3 and PI homologs in other fruit crops also may suppress ovary and fruit development.
In Arabidopsis, PI and AP3 function together through forming protein heterodimers in petals and stamens (Goto and Meyerowitz, 1994). As apple and grape have several copies of AP3 paralogues, some have acquired new expression patterns, like the sepal and fruit tissue expression observed in apple (Van Der Linden et al., 2002; Kitahara et al., 2004) and grape (Poupin et al., 2007). Ectopic expression of VvPI in grape or MdPI in apple inhibits fruit flesh tissue growth (Fernandez et al., 2013; Yao et al., 2018). A possible explanation for this phenotype in apple and grape is that the ectopic expression of PI and the new expression patterns of AP3 homologues results in the concomitant presence of AP3 and PI in fruit flesh tissue allowing the formation of functional AP3/PI heterodimers which repress fruit flesh tissue growth.
In cultivated strawberry (Fragaria x ananassa), two AP3 homologues (FaAP3 and FaTM6) exhibited an expression pattern equivalent to that of AP3 in Arabidopsis. CRISPR/Cas9-mediated genome editing of strawberry to knockout FaTM6 inhibited anther development although normal fruit development occurred after pollination with wild-type pollen (Martin-Pizarro et al., 2019), indicating FaTM6 plays an important role in strawberry anther development. Comparison of the phenotypes between MdPI mutants of apple and FaTM6 knockout of strawberry indicates the function of AP3/PI homologs is conserved in regulating floral organ development, but different in promoting fruit growth.
Apple has one SHATTERPROOF (SHP), two AG, and two SEEDSTICK (STK) homologs (Table 1; Figure 1A). When two AG homologs (MdMADS15 and MdMADS22) are knocked out together the transgenic plants produce flowers with an increased number of petals, but these flowers still have stamens and carpels and can be pollinated to produce normal sized fruit with increased number of locules (Figure 3; Klocko et al., 2016). However, the simultaneous knockout of three genes, MdMADS15/22and MdMADS14 (a SHP homolog), results in the flowers of these transgenic plants having increased numbers of petals and no stamens or carpels. This phenotype resembles the floral phenotype of Arabidopsis agamous mutants. The absence of the carpel in these apple mutant flowers means that they cannot be pollinated; however, they can be induced to produce pseudo fruit without any seed or fruit core by treatments with a combination of plant growth regulators including auxin, cytokinin, and gibberellin (Schaffer et al., 2017; Ireland et al., 2021). This is the first study to show pseudo fruit development without the development of carpel tissue. It is not fruit development in the true sense but extra-carpellary tissue continuing its development when carpels are not formed.
In peach, expression of PrpPLENA, a homolog of SHP, increases during fruit ripening. Ectopic expression of PrpPLENA in tomato plants results in the conversion of sepals into carpel-like structures that, like real fruits, become fleshy and ripen. These transgenic tomatoes also show accelerated ripening (Tadiello et al., 2009). This phenotype is the same as that of tomato over-expressing a SHP homolog TAGL1 (Vrebalov et al., 2009), indicating that PrpPLENA is likely the functional homologue of tomato’s TAGL1. Another class C-related gene of peach, PrpSHP, is expressed from the full anthesis until the fruit harvest stages. PrpSHP expression levels differ between those peach cultivars sensitive to split-pit and those resistant to split-pit (Tani et al., 2007). These findings suggest that temporal regulation of PrpSHP expression may influence the split-pit process of peach, which is a process similar to the silique dehiscence in Arabidopsis regulated by SHP1 and SHP2 (Liljegren et al., 2000).
The AG/SHP homologs of apple and peach have similar functions in regulating the growth of carpellary tissues during fruit development. The difference is that the carpellary tissues contribute to the growth of the whole fruit in peach, but only to the fruit core of apple. As demonstrated in the MADS15/15/22 knockout transgenic plants coreless apple pseudo fruit can be induced to develop from the extra-carpellary hypanthium when the function of all AG/SHP homologs is lost. In contrast, complete loss of function of AG/SHP homologs in peach would likely result in no fruit being produced, although such experiments have yet to be reported.
The class E SEP genes show partial redundancy in Arabidopsis with single SEP mutants showing only subtle phenotypes, in contrast to the triple sep1/2/3 mutant which displays indeterminate flowers and whorls of sepals (Pelaz et al., 2000) and the quadruple sep1/2/3/4 mutant which has whorls of leaf-like organs but no flower-like structures (Ditta et al., 2004). The role of AtSEP genes as floral meristem identity genes has been highlighted by their over-expression. Plants with a SEP4 transgene under the control of the CaMV35S promoter exhibit replacement of the inflorescence meristem with fused terminal flowers (Ditta et al., 2004), while plants with a SEP3 transgene expressed from the CaMV35S promoter are early flowering with solitary flowers subtended by curled cauline leaves (Pelaz et al., 2001).
The functions of SEP homologs in regulating fruit development have been shown in tomato. Suppression of expression of either the AtSEP1/2-like gene TM29 or AtSEP3-like gene TM5 promotes parthenocarpic fruit development in transgenic tomato (Pnueli et al., 1994; Ampomah-Dwamena et al., 2002). This suggests that SEP genes repress tomato fruit development. Vrebalov et al. (2002) demonstrated that the AtSEP4-like LeMADS-RIN (RIN) gene is a “master regulator” of fruit ripening in tomato. RIN is proposed to act upstream of, and independently to, ethylene-mediated regulation of ripening. Rin mutant plants show inhibition of all ripening-related traits, including autocatalytic ethylene production, carotenoid accumulation, softening, and volatile production. The rin mutant fails to ripen even under ethylene treatment (Vrebalov et al., 2002). Furthermore, RIN can bind and activate the promoter of ethylene responsive genes, such as LeACS2 (Ito et al., 2008) and in the rin mutant, the expression of LeACS2 is suppressed (Barry et al., 2000).
In strawberry (Fragaria x ananassa Duch.) downregulation of FaMADS9 (an AtSEP1/2-like gene) results in increased green coloration of petals toward a sepaloid form (Seymour et al., 2011), similar to the sepaloid petals seen in Arabidopsis sep3 single mutants (Pelaz et al., 2001). Seymour et al. (2011) also showed that although fruit development appears normal when FaMADS9 is downregulated, ripening is delayed with respect to the accumulation of anthocyanin, degreening of achenes, and fruit softening, akin to the SEP4-like rin mutant of tomato. Furthermore, complete suppression of FaMADS9 leads to severe inhibition of fruit development such that the receptacle develops only to an immature stage (Seymour et al., 2011). Another study showed that over-expression of FaMADS1α, also belonging to the SEP1/2 clade, reduced the expression of anthocyanin-related genes and delayed fruit ripening (Lu et al., 2018). Point mutations of FveSEP3 (FvH4_4g23530) generated by EMS treatment and CRISPR/Cas9-mediated gene editing convert petals, stamens, and carpels to sepaloid organs, promote parthenocarpic fruit growth, and delay fruit ripening in woodland strawberry (Pi et al., 2021).
In apple (Malus x domestica), suppression of MdMADS8/9, a member of the SEP1/2 clade, partially converts petals to sepals and reduces fruit flesh growth, resulting in small fruit (Figure 3). At fruit maturity, the MADS8/9-suppressed apples produce no ethylene and fail to ripen, showing no starch degradation or ethylene-modulated ripening traits (Ireland et al., 2013). Unlike the tomato rin mutant, the MADS8/9-suppressed apples ripen when supplied exogenous ethylene, suggesting that in this case the SEP-like genes are not acting as a master regulator controlling the competency of the fruit to ripen, but instead control the developmental regulation of distinct facets of apple fruit ripening, including the initiation of ethylene production.
Over-expression of the peach (Prunus persica) PrpMADS5, an AtSEP3-like gene, in Arabidopsis results in an early flowering phenotype and over-expression of the peach PrpMADS7, an AtSEP1/2-like gene causes extremely early flowering, with bolting after the cotyledon stage (Xu et al., 2008). The PrpSEP1 gene is closely related to apple MdMADS8 and MdMADS9 and is expressed in peach flesh. Its expression pattern during the storage of melting flesh peach is consistent with that of ethylene- and ripening-related genes. Suppression of PrpSEP1 expression by virus-induced gene silencing (VIGS) delays fruit ripening and softening in melting flesh peach. PrpSEP1 can interact with the promoter of a ripening-related gene encoding a cell wall softening enzyme polygalacturonase (PG; Li et al., 2017).
From the studies discussed above, it is clear that SEP homologs play similar function in regulating fruit ripening among apple, peach, and strawberry. When the expression level of SEP homologs is reduced, fruit ripening is delayed in all three species. However, SEP homologs show opposite functions in regulating fruit flesh growth between apple and strawberry, i.e., promoting flesh tissue growth in apple (Ireland et al., 2013) but inhibiting flesh tissue growth function in strawberry (Pi et al., 2021). This difference may be related the difference of floral organs contributing to development of fruit flesh between apple and strawberry. Apple fruit flesh develops from a hypanthium containing the bases of sepals, petals, and stamens, whereas strawberry flesh develops from a receptacle known as a stem tip. However, further research is required to understand the mechanisms of this differential regulation.
The studies on apple floral MADS-box genes started in the mid-1990s. Recent advancements of reference genome assembly for multiple Rosaceae species have facilitated the genome-wide identification of floral MADS-box and AP2 genes, and their expression patterns in developing fruit tissue have been analyzed using transcriptome data. These genome and transcriptome resources have allowed identification of promising candidate genes for analyzing their function in the regulation of fruit development and quality. So far, functional analyses of these genes have been predominantly in apple and strawberry because these two species are relatively easy to transform, and strawberry has a relatively short generation time. The function of a few genes, such as MdPI and miR172, has also been shown using natural mutants. The functional analyses of floral organ genes in regulating apple fruit development are summarized in Figure 5.
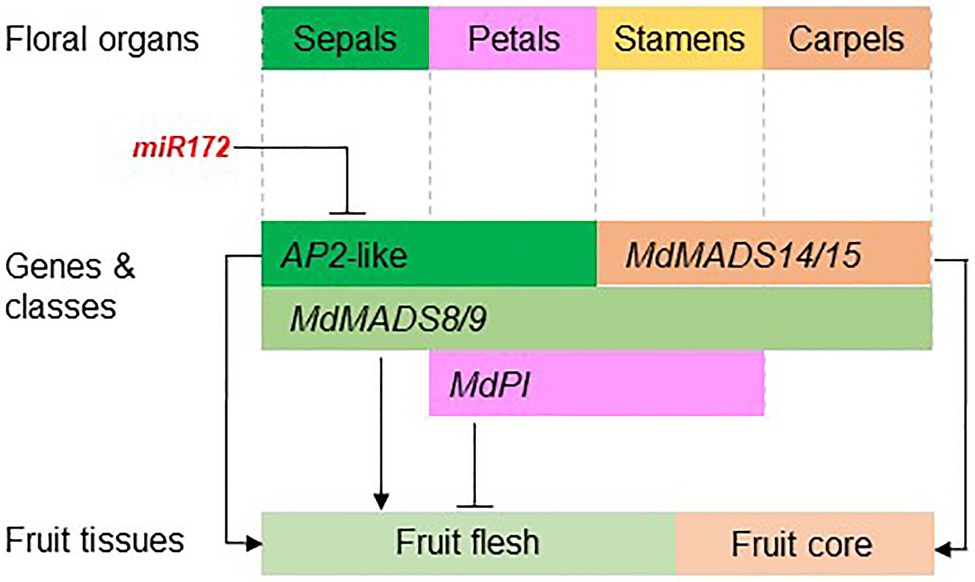
Figure 5. The apple floral organ genes regulate fruit development. The diagram shows the apple floral organ genes that have been functionally studied in apple transgenic plants. miR172 inhibits the expression of AP2-like genes that promote apple fruit flesh growth (Yao et al., 2015). Ectopic expression of MdPI inhibits fruit flesh growth (Yao et al., 2018) while knockout of MdPI promotes parthenocarpic fruit development (Yao et al., 2001). The AG/SHP homologs MADS14/15 are required for fruit core growth and coreless fruit can be produced by knocking out MADS14/15 (Schaffer et al., 2017; Ireland et al., 2021). The class E genes MdMADS8/9 are required for fruit flesh growth and fruit ripening (Ireland et al., 2013).
Future studies should be directed to functionally analyzing the roles of floral organ genes in a wider range of Rosaceae species for a greater understanding of the molecular mechanisms regulating fruit type diversity and fruit development in the family. Gene editing, using CRISPR-Cas9 or similar, offers the opportunity to disrupt the function of specific gene family members to elucidate their precise role, the degree of redundancy within the gene family and any neo- and sub-functionalization that has evolved. However, as most Rosaceae plants are woody perennials, the challenges of elucidating a gene’s role in the regulation of fruit development and fruit quality are the relatively long time to the plants first flowering and fruiting and their recalcitrance to transformation. These challenges may be overcome by using transgenes to promote early flowering, as has been achieved using the Betula pendula (silver birch) BpMADS4 gene to accelerate apple flowering (Weigl et al., 2015), and transgenes to enhance the efficiency of plant transformation and regeneration, as has been achieved using the Baby boom gene to improve maize and apple transformation (Lowe et al., 2016; Chen et al., 2021). In addition, recent accumulation of genome sequence data from a large number of cultivars and germplasm accessions of some of the Rosaceae family, along with transcriptomics data, is proving useful for the identification of alleles associated with key fruit development traits. With the ongoing expansion of genomics data, more refined spatio-temporal transcriptomics, new gene editing, and transformation technologies and their application across a wider range of Rosaceae family members, we would expect that significantly faster progress will be made in the next decade to understand how floral MADS-box and AP2 genes regulate fruit development and fruit quality in Rosaceae, a family that produces such a wide array of fruit types.
All authors contributed to the design of the review and approved the manuscript. J-LY completed the first draft. J-LY, CK, and AG revised the manuscript.
Authors AG and J-LY were employed by company The New Zealand Institute for Plant and Food Research Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B., and Yao, J. L. (2002). Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130, 605–617. doi: 10.1104/pp.005223
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Barry, C. S., Llop-Tous, M. I., and Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123, 979–986. doi: 10.1104/pp.123.3.979
Chen, X. M. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chen, J., Tomes, S., Gleave, A., Hall, W., Luo, Z., Xu, J., et al. (2021). Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby Boom transcription factor Horticulture Research. doi: 10.1093/hr/uhab014 (in press)
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Conant, G. C., Birchler, J. A., and Pires, J. C. (2014). Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 19, 91–98. doi: 10.1016/j.pbi.2014.05.008
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106. doi: 10.1038/ng.3886
De Martino, G., Pan, I., Emmanuel, E., Levy, A., and Irish, V. F. (2006). Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18, 1833–1845. doi: 10.1105/tpc.106.042978
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Edger, P. P., Vanburen, R., Colle, M., Poorten, T. J., Wai, C. M., Niederhuth, C. E., et al. (2017). Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7, 1–7. doi: 10.1093/gigascience/gix124
Fernandez, L., Chaib, J., Martinez-Zapater, J. M., Thomas, M. R., and Torregrosa, L. (2013). Mis-expression of a PISTILLATA-like MADS box gene prevents fruit development in grapevine. Plant J. 73, 918–928. doi: 10.1111/tpj.12083
Gattolin, S., Cirilli, M., Pacheco, I., Ciacciulli, A., Da Silva Linge, C., Mauroux, J. B., et al. (2018). Deletion of the miR172 target site in a TOE-type gene is a strong candidate variant for dominant double-flower trait in Rosaceae. Plant J. 96, 358–371. doi: 10.1111/tpj.14036
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene pistillata. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Gu, Q., Ferrandiz, C., Yanofsky, M. F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. doi: 10.1242/dev.125.8.1509
Hawkins, C., Caruana, J., Li, J. M., Zawora, C., Darwish, O., Wu, J., et al. (2017). An eFP browser for visualizing strawberry fruit and flower transcriptomes. Horticulture Res. 4:17029. doi: 10.1038/hortres.2017.29
Hollender, C. A., Kang, C., Darwish, O., Geretz, A., Matthews, B. F., Slovin, J., et al. (2014). Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol. 165, 1062–1075. doi: 10.1104/pp.114.237529
Hu, P. P., Li, G., Zhao, X., Zhao, F. L., Li, L. J., and Zhou, H. C. (2018). Transcriptome profiling by RNA-Seq reveals differentially expressed genes related to fruit development and ripening characteristics in strawberries (Fragaria x ananassa). Peerj 6:e4976. doi: 10.7717/peerj.4976
Ingrosso, I., Bonsegna, S., De Domenico, S., Laddomada, B., Blando, F., Santino, A., et al. (2011). Over-expression of a grape stilbene synthase gene in tomato induces parthenocarpy and causes abnormal pollen development. Plant Physiol. Biochem. 49, 1092–1099. doi: 10.1016/j.plaphy.2011.07.012
Ireland, H. S., Tomes, S., Hallett, I. C., Karunairetnam, S., David, K. M., Yao, J.-L., et al. (2021). Coreless apples generated by the suppression of carpel genes and hormone-induced fruit set. Fruit Res. 1, 1–9. doi: 10.48130/FruRes-2021-0002
Ireland, H. S., Yao, J.-L., Tomes, S., Sutherland, P. W., Nieuwenhuizen, N., Gunaseelan, K., et al. (2013). Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 73, 1044–1056. doi: 10.1111/tpj.12094
Ito, Y., Kitagawa, M., Ihashi, N., Yabe, K., Kimbara, J., Yasuda, J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223. doi: 10.1111/j.1365-313X.2008.03491.x
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U. S. A. 102, 3117–3122. doi: 10.1073/pnas.0409893102
Kang, C. Y., Darwish, O., Geretz, A., Shahan, R., Alkharouf, N., and Liu, Z. C. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25, 1960–1978. doi: 10.1105/tpc.113.111732
Kim, S., Soltis, P. S., Wall, K., and Soltis, D. E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23, 107–120. doi: 10.1093/molbev/msj014
Kitahara, K., Ohtsubo, T., Soejima, J., and Matsumoto, S. (2004). Cloning and characterization of apple class MADS-box genes including a novel AP3 homologue MdTM6. J. Japanese Soci. Horticultural Sci. 73, 208–215. doi: 10.2503/jjshs.73.208
Klocko, A. L., Borejsza-Wysocka, E., Brunner, A. M., Shevchenko, O., Aldwinckle, H., and Strauss, S. H. (2016). Transgenic suppression of AGAMOUS genes in apple reduces fertility and increases floral attractiveness. PLoS One 11:e0159421. doi: 10.1371/journal.pone.0159421
Kotoda, N., Wada, M., Kusaba, S., Kano-Murakami, Y., Masuda, T., and Soejima, J. (2002). Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci. 162, 679–687. doi: 10.1016/S0168-9452(02)00024-9
Krizek, B. A., and Fletcher, J. C. (2005). Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Li, J. J., Fang, L., Qian, M., Han, M. Y., Liu, H. K., Zhang, D., et al. (2017). Characteristics and regulatory pathway of the PrupeSEP1 SEPALLATA gene during ripening and softening in peach fruits. Plant Sci. 257, 63–73. doi: 10.1016/j.plantsci.2017.01.004
Liljegren, S. J., Ditta, G. S., Eshed, H. Y., Savidge, B., Bowman, J. L., and Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liu, Z., Ma, H., Jung, S., Main, D., and Guo, L. (2020). Developmental mechanisms of fleshy fruit diversity in Rosaceae. Annu. Rev. Plant Biol. 71, 547–573. doi: 10.1146/annurev-arplant-111119-021700
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Lu, W. J., Chen, J. X., Ren, X. C., Yuan, J. J., Han, X. Y., Mao, L. C., et al. (2018). One novel strawberry MADS-box transcription factor FaIVIADS1 alpha acts as a negative regulator in fruit ripening. Sci. Hortic. 227, 124–131. doi: 10.1016/j.scienta.2017.09.042
Martin-Pizarro, C., Trivino, J. C., and Pose, D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 885–895. doi: 10.1093/jxb/ery400
Mazzucato, A., Olimpieri, I., Siligato, F., Picarella, M. E., and Soressi, G. P. (2008). Characterization of genes controlling stamen identity and development in a parthenocarpic tomato mutant indicates a role for the DEFICIENS ortholog in the control of fruit set. Physiol. Plant. 132, 526–537. doi: 10.1111/j.1399-3054.2007.01035.x
Medina, M., Roque, E., Pineda, B., Cañas, L., Rodriguez-Concepción, M., Beltrán, J. P., et al. (2013). Early anther ablation triggers parthenocarpic fruit development in tomato. Plant Biotechnol. J. 11, 770–779. doi: 10.1111/pbi.12069
Mlotshwa, S., Yang, Z., Kim, Y., and Chen, X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61, 781–793. doi: 10.1007/s11103-006-0049-0
Okamuro, J. K., Caster, B., Villarroel, R., Vanmontagu, M., and Jofuku, K. D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 94, 7076–7081. doi: 10.1073/pnas.94.13.7076
Pabón-Mora, N., Ambrose, B. A., and Litt, A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158, 1685–1704. doi: 10.1104/pp.111.192104
Pei, M. S., Cao, S. H., Wu, L., Wang, G. M., Xie, Z. H., Gu, C., et al. (2020). Comparative transcriptome analyses of fruit development among pears, peaches, and strawberries provide new insights into single sigmoid patterns. BMC Plant Biol. 20:108. doi: 10.1186/s12870-020-2317-6
Pei, M. S., Gu, C., and Zhang, S. L. (2019). Genome-wide identification and expression analysis of genes associated with peach (Prunus persica) fruit ripening. Sci. Hortic. 246, 317–327. doi: 10.1016/j.scienta.2018.10.065
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L., and Yanofsky, M. F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. doi: 10.1046/j.1365-313X.2001.2641042.x
Pi, M., Hu, S., Cheng, L., Zhong, R., Cai, Z., Liu, Z., et al. (2021). The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Horticulture. Demogr. Res. 8, 1–5. doi: 10.1038/s41438-021-00673-1
Pnueli, L., Hareven, D., Broday, L., Hurwitz, C., and Lifschitz, E. (1994). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6, 175–186. doi: 10.2307/3869637
Poupin, M. J., Federici, F., Medina, C., Matus, J. T., Timmermann, T., and Arce-Johnson, P. (2007). Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTALLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404, 10–24. doi: 10.1016/j.gene.2007.08.005
Pratt, C. (1988). Apple flower and fruit: morphology and anatomy. Hortic. Rev. 10, 273–308. doi: 10.1002/9781118060834.ch8
Ripoll, J. J., Bailey, L. J., Mai, Q.-A., Wu, S. L., Hon, C. T., Chapman, E. J., et al. (2015). microRNA regulation of fruit growth. Nature Plants 1:15036. doi: 10.1038/nplants.2015.36
Ripoll, J. J., Roeder, A. H., Ditta, G. S., and Yanofsky, M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138, 5167–5176. doi: 10.1242/dev.073031
Royo, C., Carbonell-Bejerano, P., Torres-Pérez, R., Nebish, A., Martínez, Ó., Rey, M., et al. (2016). Developmental, transcriptome, and genetic alterations associated with parthenocarpy in the grapevine seedless somatic variant Corinto bianco. J. Exp. Bot. 67, 259–273. doi: 10.1093/jxb/erv452
Schaffer, R. J., Ireland, H. S., and Yao, J.-L. (2017). Methods and Materials for Producing Coreless Fruit. US Patent App. 15/329,229.
Seymour, G. B., Ryder, C. D., Cevik, V., Hammond, J. P., Popovich, A., King, G. J., et al. (2011). A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 62, 1179–1188. doi: 10.1093/jxb/erq360
Shahan, R., Li, D. D., and Liu, Z. C. (2019). Identification of genes preferentially expressed in wild strawberry receptacle fruit and demonstration of their promoter activities. Horticulture Res. 6:50. doi: 10.1038/s41438-019-0134-6
Shang, X. L., Zhang, J. P., Ma, Y. H., and Wang, L. R. (2020). Preliminary identification of candidate genes associated with the peach fruit sorbitol content based on comparative transcriptome analysis. Sci. Hortic. 263:109151. doi: 10.1016/j.scienta.2019.109151
Spjut, R.W. (1994). A Systematic Treatment of Fruit Types. Bronx, New York: New York Botanical Garden.
Sung, S. K., and An, G. H. (1997). Molecular cloning and characterization of a MADS-box cDNA clone of the Fuji apple. Plant Cell Physiol. 38, 484–489. doi: 10.1093/oxfordjournals.pcp.a029193
Sung, S. K., Yu, G. H., and An, G. H. (1999). Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiol. 120, 969–978. doi: 10.1104/pp.120.4.969
Sung, S. K., Yu, G. H., Nam, J., Jeong, D. H., and An, G. (2000). Developmentally regulated expression of two MADS-box genes, MdMADS3 and MdMADS4, in the morphogenesis of flower buds and fruits in apple. Planta 210, 519–528. doi: 10.1007/s004250050040
Tadiello, A., Pavanello, A., Zanin, D., Caporali, E., Colombo, L., Rotino, G. L., et al. (2009). A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. J. Exp. Bot. 60, 651–661. doi: 10.1093/jxb/ern313
Takei, H., Shinozaki, Y., Yano, R., Kashojiya, S., Hernould, M., Chevalier, C., et al. (2019). Loss-of-function of a tomato receptor-Like kinase impairs male fertility and induces Parthenocarpic fruit set. Front. Plant Sci. 10:403. doi: 10.3389/fpls.2019.00403
Tan, B., Lian, X. D., Cheng, J., Zeng, W. F., Zheng, X. B., Wang, W., et al. (2019). Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties. BMC Genomics 20:892. doi: 10.1186/s12864-019-6258-0
Tang, M., Li, G., and Chen, M. (2007). The phylogeny and expression pattern of APETALA2-like genes in rice. J. Gene. Genom. 34, 930–938. doi: 10.1016/S1673-8527(07)60104-0
Tani, E., Polidoros, A. N., Flemetakis, E., Stedel, C., Kalloniati, C., Demetriou, K., et al. (2009). Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol. Biochem. 47, 690–700. doi: 10.1016/j.plaphy.2009.03.013
Tani, E., Polidoros, A. N., and Tsaftaris, A. S. (2007). Characterization and expression analysis of FRUITFULL- and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiol. 27, 649–659. doi: 10.1093/treephys/27.5.649
Teotia, S., and Tang, G. (2015). To bloom or not to bloom: role of MicroRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Van Der Linden, C. G., Vosman, B., and Smulders, M. J. M. (2002). Cloning and characterization of four apple MADS box genes isolated from vegetative tissue. J. Exp. Bot. 53, 1025–1036. doi: 10.1093/jexbot/53.371.1025
Vardi, A., Levin, I., and Carmi, N. (2008). Induction of seedlessness in citrus: from classical techniques to emerging biotechnological approaches. J. Am. Soc. Hortic. Sci. 133, 117–126. doi: 10.21273/JASHS.133.1.117
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S. Q., Marroni, F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, U487–U447. doi: 10.1038/ng.2586
Vrebalov, J., Pan, I. L., Arroyo, A. J. M., Mcquinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Weigl, K., Wenzel, S., Flachowsky, H., Peil, A., and Hanke, M. V. (2015). Integration of BpMADS4 on various linkage groups improves the utilization of the rapid cycle breeding system in apple. Plant Biotechnol. J. 13, 246–258. doi: 10.1111/pbi.12267
Wollmann, H., Mica, E., Todesco, M., Long, J. A., and Weigel, D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633–3642. doi: 10.1242/dev.036673
Xiang, Y. Z., Huang, C. H., Hu, Y., Wen, J., Li, S. S., Yi, T. S., et al. (2017). Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34:msw242. doi: 10.1093/molbev/msw242
Xu, Z. D., Zhang, Q. X., Sun, L. D., Du, D. L., Cheng, T. R., Pan, H. T., et al. (2014). Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Mol. Gen. Genomics. 289, 903–920. doi: 10.1007/s00438-014-0863-z
Xu, Y., Zhang, L., Xie, H., Zhang, Y.-Q., Oliveira, M. M., and Ma, R.-C. (2008). Expression analysis and genetic mapping of three SEPALLATA-like genes from peach (Prunus persica (L.) Batsch). Tree Genet. Genomes 4, 693–703. doi: 10.1007/s11295-008-0143-3
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. doi: 10.1105/tpc.110.075606
Yao, J.-L., Dong, Y.-H., Kvarnheden, A., and Morris, B. (1999). Seven MADS-box genes in apple are expressed in different parts of the fruit. J. Am. Soc. Hortic. Sci. 124, 8–13. doi: 10.21273/JASHS.124.1.8
Yao, J.-L., Dong, Y.-H., and Morris, B. A. (2001). Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc. Natl. Acad. Sci. 98, 1306–1311. doi: 10.1073/pnas.98.3.1306
Yao, J. L., Tomes, S., Xu, J., and Gleave, A. P. (2016). How microRNA172 affects fruit growth in different species is dependent on fruit type. Plant Signal. Behav. 11:e1156833. doi: 10.1080/15592324.2016.1156833
Yao, J.-L., Xu, J., Cornille, A., Tomes, S., Karunairetnam, S., Luo, Z., et al. (2015). A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 84, 417–427. doi: 10.1111/tpj.13021
Yao, J. L., Xu, J., Tomes, S., Cui, W., Luo, Z. W., Deng, C., et al. (2018). Ectopic expression of the PISTILLATA homologous MdPI inhibits fruit tissue growth and changes fruit shape in apple. Plant Direct 2:e00051. doi: 10.1002/pld3.51
Ying, H., Shi, J., Zhang, S. S., Pingcuo, G. S., Wang, S., Zhao, F., et al. (2019). Transcriptomic and metabolomic profiling provide novel insights into fruit development and flesh coloration in Prunus mira Koehne, a special wild peach species. BMC Plant Biol. 19:463. doi: 10.1186/s12870-019-2074-6
Zhang, L., Xu, Y., and Ma, R. (2008). Molecular cloning, identification, and chromosomal localization of two MADS box genes in peach (Prunus persica). J. Gene. genom. 35, 365–372. doi: 10.1016/S1673-8527(08)60053-3
Zhu, Q.-H., and Helliwell, C. A. (2011). Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 62, 487–495. doi: 10.1093/jxb/erq295
Keywords: AP2, MADS-box, miR172, fruit development, apple, peach, strawberry
Citation: Yao J-L, Kang C, Gu C and Gleave AP (2022) The Roles of Floral Organ Genes in Regulating Rosaceae Fruit Development. Front. Plant Sci. 12:644424. doi: 10.3389/fpls.2021.644424
Received: 21 December 2020; Accepted: 22 November 2021;
Published: 05 January 2022.
Edited by:
Barbara Ambrose, New York Botanical Garden, United StatesReviewed by:
Vicente Balanzà, Polytechnic University of Valencia, SpainCopyright © 2022 Yao, Kang, Gu and Gleave. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew Peter Gleave, YW5kcmV3LmdsZWF2ZUBwbGFudGFuZGZvb2QuY28ubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.