- 1Department of Biology, Syracuse University, Syracuse, NY, United States
- 2Department of Forest Resources and Environmental Conservation, Virginia Tech, Blacksburg, VA, United States
- 3Department of Plant Biology, University of Georgia, Athens, GA, United States
- 4Department of Genetics, University of Georgia, Athens, GA, United States
- 5Warnell School of Forestry and Natural Resources, University of Georgia, Athens, GA, United States
A major challenge for sustainable food, fuel, and fiber production is simultaneous genetic improvement of yield, biomass quality, and resilience to episodic environmental stress and climate change. For Populus and other forest trees, quality traits involve alterations in the secondary cell wall (SCW) of wood for traditional uses, as well as for a growing diversity of biofuels and bioproducts. Alterations in wood properties that are desirable for specific end uses can have negative effects on growth and stress tolerance. Understanding of the diverse roles of SCW genes is necessary for the genetic improvement of fast-growing, short-rotation trees that face perennial challenges in their growth and development. Here, we review recent progress into the synergies and antagonisms of SCW development and abiotic stress responses, particularly, the roles of transcription factors, SCW biogenesis genes, and paralog evolution.
Introduction
The plant secondary cell wall (SCW) plays important roles. In the stem, it participates in structure, form, and function, as a part of the water transport system. Throughout the plant, it is an important part of the defense system, as a barrier to attack, and in systematic response to biotic and abiotic stress. The cell wall acts as the first line of defense, and cell wall integrity sensing and maintenance are tightly integrated with biotic and abiotic stress signaling (Bacete et al., 2018). Cell wall plasticity is key to a plant’s capacity to adjust to environmental conditions, such as water and nutrient availability, and adapt to specific climates (Landi and Esposito, 2017; Lee et al., 2017). The stress response role of the cell wall has been previously reviewed, particularly with regards to cell wall integrity as a mechanism for sensing and responding to stress (Novakovic et al., 2018; Vaahtera et al., 2019; Anderson and Kieber, 2020). Here, we focus on trees, highlighting examples of regulatory and SCW metabolism genes that indicate both synergy and antagonism in achieving multiple goals of improved stress resilience, biomass yield, and biomass quality.
The SCW consists of a complex network of cellulose, hemicellulose, and lignin (reviewed by Kumar et al., 2016; Meents et al., 2018; Zhong et al., 2019). Prior to maturity, production of this network is influenced by external factors. These interactions and their effect on biomass yield and quality traits are especially complex in trees where harvested wood is the result of a multi-year history, from 2- to 3-year coppice cycles to decades of genotype × environment interactions. Optimum growth depends on developmental and physiological transitions being appropriately timed for the local climate (Cooke et al., 2012; Brunner et al., 2017). In temperate and boreal zones, this requires preparation for the cold and dehydration stresses of winter and subsequent reversal to allow resumption of growth with transitions occurring at seasonal times that avoid frost injury and optimally capture resources to support growth. Although photoperiod, prolonged chilling temperatures and accumulated exposure to warm temperature are primary signals for key phenological changes, other environmental factors modulate the timing and rate of these transitions (Cooke et al., 2012; Ding and Nilsson, 2016; Brunner et al., 2017; Maurya and Bhalerao, 2017). In temperate regions, water stress generally increases in mid to late summer, a trend thought to drive the transition from large vessel earlywood to dense, small vessel latewood (Plomion et al., 2001). In seasonally dry tropical climates, the intra-annual development of some tree taxa is characterized by distinct periods of rest and rapid shoot growth. Temperate taxa such as Populus, Betula, and many Salix species are free growing, a major factor for suitability as short rotation woody biomass crops. Their shoots have the capacity to continue to grow until a critical daylength threshold occurs; this capacity is limited by other factors, such as water and nutrient availability. Thus, achieving optimal woody biomass yield and quality requires increased understanding of the interplay among primary determinates of seasonal transitions and more episodic or site-specific stresses.
Not surprisingly, manipulation of SCW biosynthesis can result in widespread changes in both the metabolome and transcriptome that might lead to negative effects on growth and development (Xie et al., 2018b; Vanholme et al., 2019). Frequently, there are indications that altering SCW biosynthesis genes can have a direct or indirect impact on the plant’s response to abiotic stress. There is also evidence that sub-functionalization or neo-functionalization of cell wall biosynthesis gene duplicates can have an impact on the relationship between stress-resistance and cell wall structure. Here, we examine the interaction between the SCW and abiotic stress responses, highlighting examples of transcription factors (TFs) and SCW biogenesis genes that directly impact both biomass and stress response, as well as the sub-/neo-functionalization of SCW biosynthesis gene paralogs in the plant response to abiotic stress.
Transcriptional Regulation of SCW and Abiotic Stress
The intricate regulation of plant growth and stress response is directed in part by a large number of TFs, including MYB and NAC family members (Wilkins et al., 2009; Hu et al., 2010; Ye and Zhong, 2015; Chen et al., 2019). Dual roles of some TFs have been reported in both SCW formation and abiotic stress responses.
An ortholog of Arabidopsis MYB46 (Zhong et al., 2007) from Betula platyphylla was overexpressed and silenced in birch (Guo et al., 2017). Overexpression lines showed improved growth under both salt and osmotic stress, while silenced lines were reduced in growth including above and below ground biomass, as well as chlorophyll content. Overexpression lines had increased levels of proline and reactive oxygen species (ROS) scavenging, attributed to increased expression of Δ1-pyrroline-5-carboxylate synthetase (P5CS), superoxide dismutase (SOD), and peroxidase (POD) genes. The overexpressing lines had increased lignin and cellulose levels and thicker fiber cell walls, but decreased hemicellulose relative to WT; silenced lines showed the opposite pattern. This was attributed to alterations in expression of a suite of lignin biosynthesis genes as well as cellulose synthases (CesAs), with increased expression in the overexpression lines, but reduced expression in silenced lines. Hemicellulose-related genes displayed the opposite pattern. ChIP-PCR supported interactions between BpMYB46 and promoters of the above-mentioned genes involved in ROS, proline, and SCW biosynthesis.
AtMYB61 induces ectopic lignification and dark photomorphogenesis in Arabidopsis (Newman et al., 2004). AtMYB61 is also expressed in guard cells and its mis-expression has a direct impact on stomatal apertures, smaller in overexpressors and larger in knockout mutants relative to WT (Liang et al., 2005). The Populus ortholog PtoMYB170 positively regulates lignin biosynthetic genes, as evidenced by enhanced lignin deposition in PtoMYB170-overexpressing plants and reduced lignification in CRISPR-knockout (KO) lines (Xu et al., 2017). PtoMYB170 is specifically expressed in guard cells and confers enhanced drought tolerance when overexpressed in Arabidopsis (Xu et al., 2017). This suggests that the dual functionality in SCW biogenesis and abiotic stress responses is evolutionarily conserved.
A dual role has also been reported for NAC secondary wall thickening promoting factor/secondary wall-associated NAC domain protein (NST/SND) orthologs in the regulation of SCW formation and abiotic stress resistance in both Arabidopsis and birch. Arabidopsis snd1 mutants with impaired fiber SCW biogenesis (Zhong et al., 2006) were also shown to have reduced survival rate under salt stress (Jeong et al. 2018). Mutant lines had increased levels of ABA, which lends strength to the model that SND1 positively regulates MYB46 and lignin biosynthesis, and negatively regulates ABA signaling and biosynthesis (Jeong et al., 2018).
Hu et al. (2019) characterized the AtSND1 ortholog BpNAC012 in birch. BpNAC012 was expressed predominantly in stems and its expression in leaves increased in plants exposed to salt, osmotic, and drought stress. Silencing of BpNAC012 resulted in thinner fiber walls. While the cell wall thickness was unchanged in overexpression lines, these lines produced more biomass and were more tolerant to salt and osmotic stress, attributed to increased expression of P5CS1 and P5CS2 and increased SOD and POD activities. Multiple assays demonstrated interactions between BpNAC012 and the promoters of abiotic stress-responsive (SOD and POD) genes, as well as known lignin, cellulose, and hemicellulose biosynthesis genes and additional SCW TFs. The authors hypothesize a model in which BpNAC012 binds to the core sequence CGT[G/A] in regulation of genes associated with abiotic stress and binds to the SNBE site in regulation of genes involved in SCW biosynthesis (Hu et al., 2019).
Additional examples of transcriptional co-regulation of growth, defense, and lignification in herbaceous species are discussed in a recent review (Xie et al., 2018b). In trees, the dual role of TFs in regulation of SCW formation and abiotic stress resistance is likely to involve responses to seasonal signals and dormancy-growth transitions, as well as reactions to sporadic stress events.
Roles of SCW Biogenesis Genes in Abiotic Stress Responses
The effects of abiotic stresses on SCW biogenesis and wood formation have been covered in other reviews (Moura et al., 2010; Houston et al., 2016; Camargo et al., 2019; Eckert et al., 2019). Although few studies have investigated the role of SCW synthesis genes in stress responses in trees, many transcriptomic studies suggest roles for SCW biosynthesis in both seasonal adaptations and abiotic stress responses, with consequential effects on biomass utilization (Fox et al., 2017; Ployet et al., 2017; Wildhagen et al., 2017; Jokipii-Lukkari et al., 2018). For instance, drought-acclimated Populus nigra showed the expected reduction of cambial growth, with an unexpected increase of saccharification potential (Wildhagen et al., 2017). The increased sugar release was unrelated to lignin content but instead, was strongly associated with cell wall matrix polysaccharide biosynthesis and modification, based on gene coexpression network analysis (Wildhagen et al., 2017). Thus, besides well-documented effects of environmental stresses on lignin traits (Moura et al., 2010), the sensitivity of SCW polysaccharide biosynthesis to abiotic stresses also warrants attention.
A few studies show altered expression of stress-related genes in transgenics with modified wood characteristics. For instance, lignin-deficient poplars exhibited transcriptome reprogramming of genes associated with not only cell wall biogenesis and remodeling, but also ROS metabolism, detoxification, and response to various stimuli (Tsai et al., 2020). In particular, genes involved in the glutathione-ascorbate cycle, sulfate assimilation, and cadmium response were upregulated in lignin-reduced poplars. The patterns are in agreement with reported responses of cadmium-exposed plants, including poplars (Herbette et al., 2006; Van De Mortel et al., 2008; Elobeid et al., 2011; Ding et al., 2017), supporting a link between lignification and heavy metal-elicited oxidative stress responses.
Another study investigated tubulin genes encoding components of cortical microtubules that have long been thought to direct cellulose microfibril deposition during cell wall biogenesis (Baskin, 2001). Consistent with this role, several tubulin genes are among the most abundant transcripts in SCW-rich xylem (Hu et al., 2016). However, manipulation of tubulin genes can be lethal or result in abnormal development (Anthony et al., 1999; Burk et al., 2006; Ishida et al., 2007). In poplar, constitutive expression of xylem-biased tubulins led to abnormal vascular development, and plant regeneration was achieved only with post-translational modification mimics of tubulins (Swamy et al., 2015). Those plants showed tissue-dependent tubulin transgene expression, much higher in leaves than xylem, opposite to the expression of endogenous tubulins. No differences in major SCW constituents were detected in transgenic wood; however, extractability of lignin-bound pectin and xylan polysaccharides was increased, as was expression of genes encoding cell wall-modifying enzymes (Swamy et al., 2015). The authors suggest an association between pectin, xylan, and lignin during early stage of SCW biogenesis that is sensitive to subtle tubulin perturbation. In transgenic leaves with elevated expression of tubulin transgenes, pectin levels increased, while expression and activity of pectin methylesterase were reduced (Harding et al., 2018). Transgenic leaves also exhibited altered stomatal behavior, with delayed opening in response to light and delayed closure in response to drought (Swamy et al., 2015), consistent with microtubule involvement in guard cell dynamics. These studies add to the functional multiplicity of tubulins and microtubules in different phases of cell wall biogenesis, associated with both cellulosic and non-cellulosic polysaccharide assembly, and impacting both wood formation and stress responses.
Manipulation of SCW genes can cause tradeoffs between stress resistance and growth. Xyloglucan endotransglycosylase/hydrolase (XTH) acts in the relaxation of the cell wall, which is key in cell expansion during normal growth, and in cell wall remodeling during stress. XTH has been shown to allow or restrict cell wall expansion (Takeda et al., 2002), and to respond to drought stress (Iurlaro et al., 2016). In poplar, PtoXTH27 and PtoXTH34 were indicated to play a role in osmotic stress responses (Jiang et al., 2020).
Given the role of xylem in water transport (Rodriguez-Zaccaro and Groover, 2019), it is not surprising that alterations in wood composition often result in reduced water transport (Kitin et al., 2010). In Arabidopsis, this can be mitigated by restoring expression of SCW-related genes in vessels (De Meester et al., 2018). Hence, it will be interesting to test whether similar strategies can be an effective in improving wood quality and yield without negative effects on abiotic stress resilience.
Sub-Functionalization of Duplicated Genes
A largely unexplored area is the contribution of gene duplication and evolution to the integration or separation of genetic pathways involved in growth, stress resilience, and wood development. Members of the Salicaceae share a relatively recent WGD estimated to have occurred ~60 million years ago, and Populus retains ~8,000 Salicoid duplicate gene pairs (Tuskan et al., 2006; Rodgers-Melnick et al., 2012; Dai et al., 2014). Evidence of paralog regulatory divergence can be inferred from the growing wealth of RNA-seq datasets. Spatially-detailed expression profiling of the poplar secondary stem showed that Salicoid duplicates with peak expression during SCW deposition tended to exhibit highly-similar profiles, suggesting that many SCW-associated paralogs have functionally redundant roles (Sundell et al., 2017). However, increasing evidence supports a role for WGD in plant environmental adaptation (Wu et al., 2020); thus, we compared published transcriptomic studies for evidence that paralogs showing highly-correlated expression in the Populus woody stem (Sundell et al., 2017) exhibit regulatory divergence in response to different abiotic stresses (Luo et al., 2015; Swamy et al., 2015; Xue et al., 2016; Lu et al., 2019; Yao et al., 2020), as well as to SCW modification (Tsai et al., 2020). Strikingly, in all these stress-tissue combinations, only one member of these paralog pairs was differentially expressed in response to the stress more often than both paralogs (Figure 1). Examples of functional diversification of gene duplicates with regards to SCW synthesis are provided below. With increasing efforts in functional characterization of gene duplicates, the expectation is that neo-/sub-functionalization of wood-expressed paralogs will continue to be identified as one of the mechanisms for tree adaptation to varying environment.
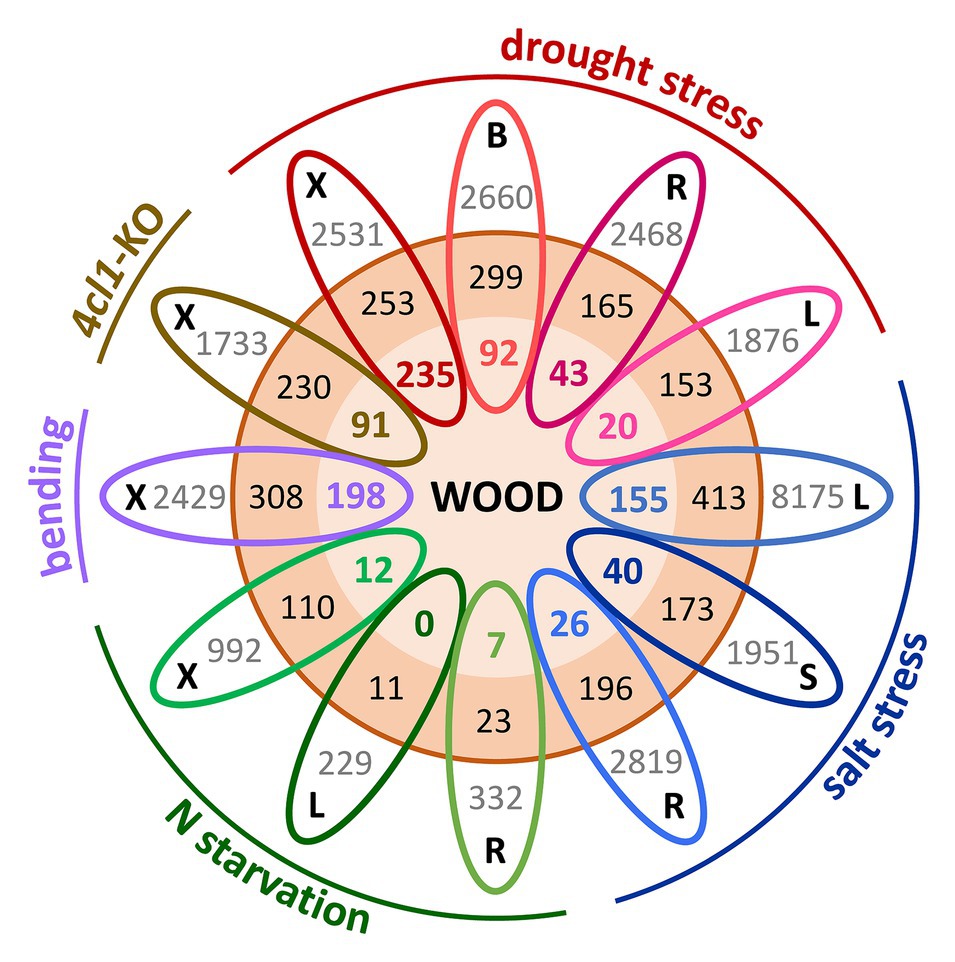
Figure 1. Gene expression divergence in response to abiotic stress among Populus wood-expressed paralogs. Salicoid duplicates (3,428 genes or 1,714 paralogs pairs, orange circle) showing expression correlations of ≥0.75 across the secondary stem (Sundell et al., 2017) were interrogated for their responsiveness to different perturbations, including nitrogen (N) starvation in roots, leaves (Luo et al., 2015), and xylem (Lu et al., 2019), tension wood induction by bending (Swamy et al., 2015), drought-stressed xylem, bark, roots, and leaves (Xue et al., 2016), xylem of lignin-deficient trees (4CL1-KO; Tsai et al., 2020), and salt-stressed leaves, stem, and roots (Yao et al., 2020). Each oval represents one tissue (B, bark; L, leaf; R, root; S, stem; and X, xylem) from a given experiment, color-coded by perturbation type. Gray values indicate the total number of differentially expressed genes for the specific tissue-stress combination as reported in the source paper. Boldfaced color values (inner orange circle) indicate wood-expressed paralogous gene pairs where both members of the pair were differentially expressed in response to the indicated stress, whereas black values (outer orange circle) denote the number of cases where only one paralog of a pair showed differential expression. For clarity of illustration, each stress dataset was compared with the wood paralogs individually without considering overlapping gene responses across tissues or stress conditions.
One example is the poplar 5-enolpyruvylshikimiate 3-phosphate synthase duplicate (Xie et al., 2018a); one encoding a classic EPSP synthase (PtrEPSP-SY) of the shikimate pathway, and the other harboring an extended N terminus with a helix-turn-helix DNA-binding motif (PtrEPSP-TF) with xylem-biased expression. Using linkage-disequilibrium based associate mapping, PtrEPSP-TF was found to exhibit associations with lignin content and syringyl-to-guaiacyl (S/G) ratio (Xie et al., 2018a). PtrEPSP-TF overexpression induces ectopic lignin and flavonoid biosynthesis through transcriptional repression of a hAT transposase PtrhAT. PtrhAT represses PtrMYB021, a MYB46 ortholog that regulates biosynthesis of SCW components, including lignin, cellulose, and xylan (Xie et al., 2018a). Neo-functionalization of a primary (shikimate) biosynthetic pathway gene with an additional role in transcriptional regulation of downstream secondary (phenylpropanoid) pathways represents an example of protein moonlighting conferring enhanced fitness of complex organisms (Copley, 2014).
A second example is the poplar paralogs of AtMYB61 involved in regulation of lignin biosynthesis and stomatal aperture noted above. While PtoMYB170 exhibits conserved dual functionality, guard cell expression was not detected for its Salicoid duplicate PtoMYB216 (Xu et al., 2017). In this case, sub-functionalization might have resulted in more specific involvement of the poplar MYB216 in SCW biogenesis (Tian et al., 2013; Wei et al., 2019).
Another example concerns the poplar 4-coumarate:CoA ligase (4CL) duplicates. In poplar, 4CL1 normally comprises ~90% of xylem 4CL transcripts and encodes the predominant isoform involved in lignin biosynthesis (Hu et al., 1999; Voelker et al., 2010). 4CL1-knockout led to ~20% lignin reduction and uniform wood discoloration (Zhou et al., 2015). Knockout of its Salicoid duplicate 4CL5, the only other xylem-expressed 4CL gene, has no effect on lignin accrual, suggesting a conditional role (Tsai et al., 2020). Nonetheless, the 4cl1 mutants maintain ~80% WT lignin levels, which must be sustained by 4CL5. 4CL5 expression was not significantly changed in the 4cl1 mutants; however, caffeoylshikimate esterase1 (CSE1) involved in caffeate biosynthesis and caffeoyl-CoA O-methyltransferase1 (CCoAOMT1) that acts downstream of 4CL product caffeoyl-CoA were upregulated (Tsai et al., 2020). In contrast, the S lignin-specific ferulate/coniferaldehyde 5-hydroxylases were downregulated. These, along with elevated levels of caffeic acid in the mutant xylem hint at a novel mechanism for in vivo enhancement of 4CL5 function to sustain G lignin biosynthesis at the expense of S lignin (Tsai et al., 2020).
The preferential reductions of S lignin in the 4cl1 poplars (Zhou et al., 2015) contrasts with maize, sorghum, Arabidopsis, and switchgrass mutants where 4CL-knockout led to strong G lignin reductions (Saballos et al., 2008; Van Acker et al., 2013; Park et al., 2017; Xiong et al., 2019). The molecular responses also differ between poplar and Arabidopsis mutants, with the latter showing upregulation of early pathway genes phenylalanine ammonia-lyase2, cinnamate 4-hydroxylase, and 4-coumaroylshikimate 3'-hydroxylase (Vanholme et al., 2012). Gene coexpression network modeling revealed distinct associations between Salicoid paralogs of 4CL1/4CL5, CSE1/CSE2, and CCoAOMT1/CCoAOMT2 duplicates, with 4CL5, CSE1 and CCoAOMT1 belonging to the same coexpression module in the 4cl1 mutant network (Tsai et al., 2020). The data provide evidence for coordinated subfunctionalization of multiple gene duplicates in the lignin pathway with conditional roles that may be key for lineage-specific adaptation.
Conclusion
To meet the grand challenge for sustainable food, fuel, and fiber under changing climate requires a holistic understanding of diverse roles of SCW genes during plant growth, development, and interactions with the environment. Expanding functional characterization efforts promise to provide additional insights into many of the hidden/conditional roles. This is especially important for woody perennials with a rich repertoire of gene duplicates, many of which likely have evolved via sub-/neo-functionalization. The specificity of CRISPR genome editing allows the dissection of functional redundancy vs. specificity of gene duplicates, and the targeted selection of genes and gene duplicates to better understand connections between SCW formation and abiotic stress resistance.
The necessity of exploring the largely unexamined, but clear intersectional implications of SCW development and abiotic stress responses, particularly in the face of changing climate, is clear. Trees present the challenge of integrating multi-year growth accumulation with recurring seasonal phenology and episodic stress events. Reflecting this diversity of environmental interactions, poplar transgenics with altered expression of lignin biosynthesis genes have shown phenotypic differences between greenhouse and field studies (reviewed in Chanoca et al., 2019). Although transgenic tree responses to stress and seasonal cues in controlled conditions provides insight, more field studies are needed to delineate gene functions in trees, and to advance genetic engineering for simultaneous improvement of wood yield, quality, and resilience to environmental stress and climate change.
Author Contributions
All authors contributed equally to writing and editing of this manuscript.
Funding
This work was supported by the USDA National Institute of Food and Agriculture, McIntire Stennis project 1025004 (AB), the Department of Energy, Office of Biological and Environmental Research grants DE-SC0005140 (C-JT) and DE-SC0008470 (C-JT), and the Center for Bioenergy Innovation (C-JT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anderson, C. T., and Kieber, J. J. (2020). Dynamic construction, perception, and remodeling of plant cell walls. Annu. Rev. Plant Biol. 71, 39–69. doi: 10.1146/annurev-arplant-081519-035846
Anthony, R. G., Reichelt, S., and Hussey, P. J. (1999). Dinitroaniline herbicide-resistant transgenic tobacco plants generated by co-overexpression of a mutant alpha-tubulin and a beta-tubulin. Nat. Biotechnol. 17, 712–716. doi: 10.1038/10931
Bacete, L., Mélida, H., Miedes, E., and Molina, A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. doi: 10.1111/tpj.13807
Baskin, T. I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215, 150–171. doi: 10.1007/BF01280311
Brunner, A. M., Varkonyi-Gasic, E., and Jones, R. C. (2017). “Phase change and phenology in trees” in Comparative and evolutionary genomics of angiosperm trees. eds. A. Groover and Q. Cronk (Cham, Switzerland: Springer), 227–274.
Burk, D. H., Zhong, R. Q., Morrison, W. H., and Ye, Z. H. (2006). Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged alpha-tubulin 6 causes a marked reduction in cell wall synthesis. J. Integr. Plant Biol. 48, 85–98. doi: 10.1111/j.1744-7909.2006.00202.x
Camargo, E. L. O., Ployet, R., Cassan-Wang, H., Mounet, F., and Grima-Pettenati, J. (2019). “Chapter seven - digging in wood: new insights in the regulation of wood formation in tree species” in Advances in botanical research. ed. F. M. Cánovas (Cambridge, MA, USA: Academic Press), 201–233.
Chanoca, A., de Vries, L., and Boerjan, W. (2019). Lignin engineering in forest trees. Front. Plant Sci. 10:912. doi: 10.3389/fpls.2019.00912
Chen, H., Wang, J. P., Liu, H., Li, H., Lin, Y.-C. J., Shi, R., et al. (2019). Hierarchical transcription factor and chromatin binding network for wood formation in Populus trichocarpa. Plant Cell 31, 602–626. doi: 10.1105/tpc.18.00620
Cooke, J. E. K., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Copley, S. D. (2014). An evolutionary perspective on protein moonlighting. Biochem. Soc. Trans. 42, 1684–1691. doi: 10.1042/BST20140245
Dai, X., Hu, Q., Cai, Q., Feng, K., Ye, N., Tuskan, G. A., et al. (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24, 1274–1277. doi: 10.1038/cr.2014.83
De Meester, B., De Vries, L., Ozparpucu, M., Gierlinger, N., Corneillie, S., Pallidis, A., et al. (2018). Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176, 611–633. doi: 10.1104/pp.17.01462
Ding, S., Ma, C., Shi, W., Liu, W., Lu, Y., Liu, Q., et al. (2017). Exogenous glutathione enhances cadmium accumulation and alleviates its toxicity in Populus × canescens. Tree Physiol. 37, 1697–1712. doi: 10.1093/treephys/tpx132
Ding, J. H., and Nilsson, O. (2016). Molecular regulation of phenology in trees - because the seasons they are a-changin’. Curr. Opin. Plant Biol. 29, 73–79. doi: 10.1016/j.pbi.2015.11.007
Eckert, C., Sharmin, S., Kogel, A., Yu, D., Kins, L., Strijkstra, G.-J., et al. (2019). What makes the wood? Exploring the molecular mechanisms of xylem acclimation in hardwoods to an ever-changing environment. Forests 10:358. doi: 10.3390/f10040358
Elobeid, M., Göbel, C., Feussner, I., and Polle, A. (2011). Cadmium interferes with auxin physiology and lignification in poplar. J. Exp. Bot. 63, 1413–1421. doi: 10.1093/jxb/err384
Fox, H., Doron-Faigenboim, A., Kelly, G., Bourstein, R., Attia, Z., Zhou, J., et al. (2017). Transcriptome analysis of Pinus halepensis under drought stress and during recovery. Tree Physiol. 38, 423–441. doi: 10.1093/treephys/tpx137
Guo, H., Wang, Y., Wang, L., Hu, P., Wang, Y., Jia, Y., et al. (2017). Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 15, 107–121. doi: 10.1111/pbi.12595
Harding, S. A., Hu, H., Nyamdari, B., Xue, L.-J., Naran, R., and Tsai, C.-J. (2018). Tubulins, rhythms and cell walls in poplar leaves: it’s all in the timing. Tree Physiol. 38, 397–408. doi: 10.1093/treephys/tpx104
Herbette, S., Taconnat, L., Hugouvieux, V., Piette, L., Magniette, M. L., Cuine, S., et al. (2006). Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88, 1751–1765. doi: 10.1016/j.biochi.2006.04.018
Houston, K., Tucker, M. R., Chowdhury, J., Shirley, N., and Little, A. (2016). The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7:984. doi: 10.3389/fpls.2016.00984
Hu, H., Gu, X., Xue, L.-J., Swamy, P. S., Harding, S. A., and Tsai, C.-J. (2016). Tubulin C-terminal post-translational modifications do not occur in wood forming tissue of Populus. Front. Plant Sci. 7:1493. doi: 10.3389/fpls.2016.01493
Hu, W. J., Harding, S. A., Lung, J., Popko, J. L., Ralph, J., Stokke, D. D., et al. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17, 808–812. doi: 10.1038/11758
Hu, R., Qi, G., Kong, Y., Kong, D., Gao, Q., and Zhou, G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10:145. doi: 10.1186/1471-2229-10-145
Hu, P., Zhang, K., and Yang, C. (2019). BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol. 179, 700–717. doi: 10.1104/pp.18.01167
Ishida, T., Kaneko, Y., Iwano, M., and Hashimoto, T. (2007). Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104, 8544–8549. doi: 10.1073/pnas.0701224104
Iurlaro, A., De Caroli, M., Sabella, E., De Pascali, M., Rampino, P., De Bellis, L., et al. (2016). Drought and heat differentially affect XTH expression and XET activity and action in 3-day-old seedlings of durum wheat cultivars with different stress susceptibility. Front. Plant Sci. 7:1686. doi: 10.3389/fpls.2016.01686
Jeong, C. Y., Lee, W. J., Truong, H. A., Trinh, C. S., Jin, J. Y., Kim, S., et al. (2018). Dual role of SND1 facilitates efficient communication between abiotic stress signalling and normal growth in Arabidopsis. Sci. Rep. 8:10114. doi: 10.1038/s41598-018-28413-x
Jiang, Y., Li, Y., Lu, C., Tang, Y., Jiang, X., and Gai, Y. (2020). Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 155, 1277–1287. doi: 10.1016/j.ijbiomac.2019.11.099
Jokipii-Lukkari, S., Delhomme, N., Schiffthaler, B., Mannapperuma, C., Prestele, J., Nilsson, O., et al. (2018). Transcriptional roadmap to seasonal variation in wood formation of Norway spruce. Plant Physiol. 176, 2851–2870. doi: 10.1104/pp.17.01590
Kitin, P., Voelker, S. L., Meinzer, F. C., Beeckman, H., Strauss, S. H., and Lachenbruch, B. (2010). Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiol. 154, 887–898. doi: 10.1104/pp.110.156224
Kumar, M., Campbell, L., and Turner, S. (2016). Secondary cell walls: biosynthesis and manipulation. J. Exp. Bot. 67, 515–531. doi: 10.1093/jxb/erv533
Landi, S., and Esposito, S. (2017). Nitrate uptake affects cell wall synthesis and modeling. Front. Plant Sci. 8:1376. doi: 10.3389/fpls.2017.01376
Lee, Y., Karunakaran, C., Lahlali, R., Liu, X., Tanino, K. K., and Olsen, J. E. (2017). Photoperiodic regulation of growth-dormancy cycling through induction of multiple bud-shoot barriers preventing water transport into the winter buds of Norway spruce. Front. Plant Sci. 8:2109. doi: 10.3389/fpls.2017.02109
Liang, Y. K., Dubos, C., Dodd, I. C., Holroyd, G. H., Hetherington, A. M., and Campbell, M. M. (2005). AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 15, 1201–1206. doi: 10.1016/j.cub.2005.06.041
Lu, Y., Deng, S., Li, Z., Wu, J., Liu, Q., Liu, W., et al. (2019). Competing endogenous rna networks underlying anatomical and physiological characteristics of poplar wood in acclimation to low nitrogen availability. Plant Cell Physiol. 60, 2478–2495. doi: 10.1093/pcp/pcz146
Luo, J., Zhou, J., Li, H., Shi, W., Polle, A., Lu, M., et al. (2015). Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 35, 1283–1302. doi: 10.1093/treephys/tpv091
Maurya, J. P., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Meents, M. J., Watanabe, Y., and Samuels, A. L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121, 1107–1125. doi: 10.1093/aob/mcy005
Moura, J. C. M. S., Bonine, C. A. V., De Oliveira Fernandes Viana, J., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Newman, L. J., Perazza, D. E., Juda, L., and Campbell, M. M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37, 239–250. doi: 10.1046/j.1365-313X.2003.01953.x
Novakovic, L., Guo, T., Bacic, A., Sampathkumar, A., and Johnson, K. L. (2018). Hitting the wall-sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plan. Theory 7:89. doi: 10.3390/plants7040089
Park, J.-J., Yoo, C. G., Flanagan, A., Pu, Y., Debnath, S., Ge, Y., et al. (2017). Defined tetra-allelic gene disruption of the 4-coumarate:coenzyme a ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol. Biofuels 10:284. doi: 10.1186/s13068-017-0972-0
Plomion, C., Leprovost, G., and Stokes, A. (2001). Wood formation in trees. Plant Physiol. 127, 1513–1523. doi: 10.1104/pp.010816
Ployet, R., Soler, M., Carocha, V., Ladouce, N., Alves, A., Rodrigues, J.-C., et al. (2017). Long cold exposure induces transcriptional and biochemical remodelling of xylem secondary cell wall in Eucalyptus. Tree Physiol. 38, 409–422. doi: 10.1093/treephys/tpx062
Rodgers-Melnick, E., Mane, S. P., Dharmawardhana, P., Slavov, G. T., Crasta, O. R., Strauss, S. H., et al. (2012). Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 22, 95–105. doi: 10.1101/gr.125146.111
Rodriguez-Zaccaro, F. D., and Groover, A. (2019). Wood and water: how trees modify wood development to cope with drought. Plants, People, Planet 1, 346–355. doi: 10.1002/ppp3.29
Saballos, A., Vermerris, W., Rivera, L., and Ejeta, G. (2008). Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenergy Res. 1, 193–204. doi: 10.1007/s12155-008-9025-7
Sundell, D., Street, N. R., Kumar, M., Mellerowicz, E. J., Kucukoglu, M., Johnsson, C., et al. (2017). AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604. doi: 10.1105/tpc.17.00153
Swamy, P. S., Hu, H., Pattathil, S., Maloney, V. J., Xiao, H., Xue, L.-J., et al. (2015). Tubulin perturbation leads to unexpected cell wall modifications and affects stomatal behaviour in Populus. J. Exp. Bot. 66, 6507–6518. doi: 10.1093/jxb/erv383
Takeda, T., Furuta, Y., Awano, T., Mizuno, K., Mitsuishi, Y., and Hayashi, T. (2002). Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc. Natl. Acad. Sci. U. S. A. 99, 9055–9060. doi: 10.1073/pnas.132080299
Tian, Q., Wang, X., Li, C., Lu, W., Yang, L., Jiang, Y., et al. (2013). Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS One 8:e76369. doi: 10.1371/journal.pone.0076369
Tsai, C. J., Xu, P., Xue, L. J., Hu, H., Nyamdari, B., Naran, R., et al. (2020). Compensatory guaiacyl lignin biosynthesis at the expense of syringyl lignin in 4CL1-knockout poplar. Plant Physiol. 183, 123–136. doi: 10.1104/pp.19.01550
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Vaahtera, L., Schulz, J., and Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932. doi: 10.1038/s41477-019-0502-0
Van Acker, R., Vanholme, R., Storme, V., Mortimer, J., Dupree, P., and Boerjan, W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 6:46. doi: 10.1186/1754-6834-6-46
Van De Mortel, J. E., Schat, H., Moerland, P. D., Van Themaat, E. V. L., Van Der Ent, S., Blankestijn, H., et al. (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 31, 301–324. doi: 10.1111/j.1365-3040.2007.01764.x
Vanholme, R., De Meester, B., Ralph, J., and Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Vanholme, R., Storme, V., Vanholme, B., Sundin, L., Christensen, J. H., Goeminne, G., et al. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24, 3506–3529. doi: 10.1105/tpc.112.102574
Voelker, S. L., Lachenbruch, B., Meinzer, F. C., Jourdes, M., Ki, C., Patten, A. M., et al. (2010). Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol. 154, 874–886. doi: 10.1104/pp.110.159269
Wei, K., Zhao, Y., Zhou, H., Jiang, C., Zhang, B., Zhou, Y., et al. (2019). PagMYB216 is involved in the regulation of cellulose synthesis in Populus. Mol. Breed. 39:65. doi: 10.1007/s11032-019-0970-y
Wildhagen, H., Paul, S., Allwright, M., Smith, H. K., Malinowska, M., Schnabel, S. K., et al. (2017). Genes and gene clusters related to genotype and drought-induced variation in saccharification potential, lignin content and wood anatomical traits in Populus nigra. Tree Physiol. 38, 320–339. doi: 10.1093/treephys/tpx054
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Wu, S. D., Han, B. C., and Jiao, Y. N. (2020). Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13, 59–71. doi: 10.1016/j.molp.2019.10.012
Xie, M., Muchero, W., Bryan, A. C., Yee, K., Guo, H. B., Zhang, J., et al. (2018a). A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus. Plant Cell 30, 1645–1660. doi: 10.1105/tpc.18.00168
Xie, M., Zhang, J., Tschaplinski, T. J., Tuskan, G. A., Chen, J.-G., and Muchero, W. (2018b). Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 9:1427. doi: 10.3389/fpls.2018.01427
Xiong, W., Wu, Z., Liu, Y., Li, Y., Su, K., Bai, Z., et al. (2019). Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 12:82. doi: 10.1186/s13068-019-1421-z
Xu, C., Fu, X., Liu, R., Guo, L., Ran, L., Li, C., et al. (2017). PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 37, 1713–1726. doi: 10.1093/treephys/tpx093
Xue, L.-J., Frost, C. J., Tsai, C.-J., and Harding, S. A. (2016). Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression. Sci. Rep. 6:33655. doi: 10.1038/srep33655
Yao, W., Li, C., Lin, S., Wang, J., Zhou, B., and Jiang, T. (2020). Transcriptome analysis of salt-responsive and wood-associated NACs in Populus simonii × Populus nigra. BMC Plant Biol. 20:317. doi: 10.1186/s12870-020-02507-z
Ye, Z.-H., and Zhong, R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. doi: 10.1093/jxb/erv081
Zhong, R., Cui, D., and Ye, Z.-H. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Richardson, E. A., and Ye, Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. doi: 10.1105/tpc.107.053678
Keywords: abiotic stress, secondary cell wall, Populus, drought, nutrient stress, gene duplication
Citation: Coleman HD, Brunner AM and Tsai C-J (2021) Synergies and Entanglement in Secondary Cell Wall Development and Abiotic Stress Response in Trees. Front. Plant Sci. 12:639769. doi: 10.3389/fpls.2021.639769
Edited by:
Paula Casati, Centro de Estudios Fotosintéticos y Bioquímicos (CEFOBI), ArgentinaReviewed by:
Qingzhang Du, Beijing Forestry University, ChinaJin-Gui Chen, Oak Ridge National Laboratory (DOE), United States
Copyright © 2021 Coleman, Brunner and Tsai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heather D. Coleman, aGNvbGVtYW5Ac3lyLmVkdQ==
 Heather D. Coleman
Heather D. Coleman Amy M. Brunner
Amy M. Brunner Chung-Jui Tsai
Chung-Jui Tsai