
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 May 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.638788
The APETALA2 (AP2) and ethylene-responsive element-binding factor (ERF) gene family is one of the largest plant-specific transcription factor gene families, which plays a critical role in plant development and evolution, as well as response to various stresses. The TARGET OF EAT3 (TOE3) gene is derived from Halostachys caspica and belongs to the AP2 subfamily with two AP2 DNA-binding domains. Currently, AP2 family mainly plays crucial roles in plant growth and evolution, yet there are few reports about the role of AP2 in abiotic stress tolerance. Here, we report HcTOE3, a new cold-regulated transcription factor gene, which has an important contribution to freezing tolerance. The main results showed that the expression of HcTOE3 in the H. caspica assimilating branches was strongly induced by different abiotic stresses, including high salinity, drought, and extreme temperature (heat, chilling, and freezing), as well as abscisic acid and methyl viologen treatments. Overexpressing HcTOE3 gene (OE) induced transgenic Arabidopsis plant tolerance to freezing stress. Under freezing treatment, the OE lines showed lower content of malondialdehyde and electrolyte leakage and less accumulation of reactive oxygen species compared with the wild type. However, the survival rates, antioxidant enzyme activities, and contents of osmotic adjustment substance proline were enhanced in transgenic plants. Additionally, the OE lines increased freezing tolerance by up-regulating the transcription level of cold responsive genes (CBF1, CBF2, COR15, COR47, KIN1, and RD29A) and abscisic acid signal transduction pathway genes (ABI1, ABI2, ABI5, and RAB18). Our results suggested that HcTOE3 positively regulated freezing stress and has a great potential as a candidate gene to improve plant freezing tolerance.
Cold temperature is an important circumstance stress element, which restricts the plant growth, evolution, yield, and geographic dissemination, and it is a kind of natural disaster often encountered in agricultural production (Ding et al., 2019). Low-temperature stress impacted the plant cell membrane and the activities of reactive oxygen species (ROS)–scavenging enzymes, as well as protein stability (Orvar et al., 2000). These processes led to photoinhibition and impaired photosynthesis and caused severe membrane destruction (Siddiqui and Cavicchioli, 2006; Ruelland et al., 2009). According to the different degrees of cold temperatures, there are two major categories of plant damage: chilling injury (greater than 0°C) and freezing stress (less than 0°C) (Liu et al., 2018). Freezing stress not only caused chilling injury, but also generated more damage to affected plants, including cell and plant death (Pearce, 2001).
During long-history evolution process of plants, they have evolved different mechanisms for handling low-temperature stress, including activating ICE1–CBF–COR signaling pathway (Thomashow, 1999). The expression of CBF genes was quickly up-regulated by cold treatment, and further induced the overexpression of COR genes, thereby improving plant tolerance to cold stress (Gilmour et al., 1998). In addition, there is also an abscisic acid (ABA)–dependent cold signaling pathway. The expression of some cold temperature–related genes was regulated and induced by ABA (such as RAB18, RD29b) (Uno et al., 2000). Their promoters of these genes contained ABA response elements (ABREs), which initiated a series of plant responses to resist low temperature and enhanced plant cold tolerance (Agarwal and Jha, 2010). Under cold stress conditions, plants also synthesized certain cold-responsive proteins and small molecules, such as proline and soluble sugars for enhancing plant physiological response to cold stress (Kaplan and Guy, 2004; Kaplan et al., 2007). These cold-responsive proteins and small molecules were involved in regulation of cell membrane stability and osmotic potentials, as well as removing certain ROS during plant response to cold stress (Lee et al., 2002; Doherty et al., 2009).
Transcription factors (TFs) are an extensive class of proteins, which play important functions in regulating plant growth and development, as well as responding to various stresses (Xu et al., 2011; Mizoi et al., 2012). The APETALA2 (AP2) and ethylene-responsive element-binding factor (ERF) gene family is one of the largest plant-specific TF gene families. According to the number and binding sequence of AP2/ERF DNA-binding domain (BD), the AP2/ERF TF family was categorized into five subfamilies: AP2, ERF, DREB RAV, and Soloist (Nakano et al., 2006; Cao et al., 2020). Each subfamily contains different domain that can bind to different DNA sequence (Nakano et al., 2006; Cao et al., 2020).
There have been many reports on AP2/ERF genes conferring plant tolerance to abiotic stresses. BpERF13 was cloned from Betula platyphylla, and overexpressing of BpERF13 gene enhanced plant tolerance to cold stress in transgenic B. platyphylla (Lv et al., 2020). The DcDREB1A gene was isolated from carrots, and ectopic expression of DcDREB1 increased plant tolerance to drought in Arabidopsis (Li et al., 2020). Overexpressing of ERF1-V from Haynaldia villosa improved wheat resistance to fungal disease, and the transgenic wheat with ERF1-V also induced plant tolerance to salt and drought stresses (Xing et al., 2017). CmRAV1, a new RAV gene, was isolated from melon; the heterologous expression of CmRAV1 enhanced salt tolerance in transgenic Arabidopsis during seed germination and early developmental stages (Zhao et al., 2019). However, at present, members of the AP2 subfamily majorly play more functions in affecting floral meristem and determining the identity development of floral organs (Jofuku et al., 1994; Krogan et al., 2012). Additionally, AP2 also functions in maintaining the stem cell niche of bud meristem (Wurschum et al., 2006). Overexpression of AfAP2-2 gene from Aechmea fasciata reduced seed size and delayed flowering in transgenic Arabidopsis (Lei et al., 2019).
The halophyte Halostachys caspica belongs to Chenopodiaceae. It grows naturally in the semi-desert regions of Central Asia, where saline and alkaline are commonly observed. A previous study showed that this plant species had high tolerance to salt stress (Zeng et al., 2015). The TF TOE3 (TARGET OF EAT3) contains two AP2 domains and belongs to the AP2 subfamily. The AP2 subfamily mainly functions in plant growth and development; however, AP2 TF may be also involved in the regulation of abiotic stress tolerance. In our previous work, we isolated and cloned HcTOE3 gene from H. caspica. In this study, we comprehensively identified HcTOE3, a new cold stress–responsive AP2 gene, can improve freezing tolerance in Arabidopsis by ectopic expression. Our data demonstrated that HcTOE3 is a positive regulator in enhancing freezing tolerance of plants.
The gene and protein sequences of Arabidopsis AP2/ERF were downloaded from the PlantTFDB (Jin et al., 2017). PlantTFDB is a publicly available plant TF database. DNAMAN software was employed to construct a multiple amino-acid alignment of the AP2/ERF TFs, including the H. caspica TOE3 protein. MEGA7 was employed to construct a phylogenetic tree by the default neighbor-joining method with 1,000 bootstraps. NCBI and SMART1 online tools were employed to analyze the existence of the expected AP2 domains in amino acid (aa) sequences. The nuclear localization sequences of HcTOE3 were predicted by using online software2. All tested aa sequences can be found in Supplementary Table 1.
HcTOE3 gene was first isolated and cloned into pGBKT7 plasmid. The primers used in the cloning process can be found in the Supplementary Table 2. Then, the constructed pGBKT7-HcTOE3 vector was transformed into Y2H yeast cells. At the same time, empty pGBKT7 and pGBKT7-ScABI3 vectors (Zhang et al., 2018) were also transformed into Y2H yeast cells, which were served as negative and positive controls for transactivation assays. The transactivation activity of each protein was assessed by comparing cell growth on selective medium with SD/-Trp and SD/-Trp-His; the activity was also assessed by the activity of X-α-Gal.
pCAMBIA1304-HcTOE3-GFP vector was constructed by inserting the HcTOE3 open reading frame cDNA without the stop codon into the plant binary expression vector pCAMBIA1304. Both the constructed fusion vector (HcTOE3-GFP) and GFP vector (serving as negative control) were, respectively, introduced into Agrobacterium tumefaciens strain GV3101. Then, the transformed bacteria were used for transient expression analysis in onion epidermal cells (Xu et al., 2014). After 24 h of culture on MS medium, the cells were observed under Zeiss LSM800 laser scanning confocal microscope (Jena, Germany).
The seeds of H. caspica were collected from the field, located on the edge of Gurbantunggut Desert. The field is extremely saline-alkaline and also considered as a semidesert area in Xinjiang, China. The healthy seeds were cultivated in flowerpots with substrate (perlite: vermiculite: flower soil = 1: 1: 3) and cultured under natural light and temperature (25–28°C). After 8 weeks of growth, H. caspica plants were used for profiling the expression of HcTOE3 gene under 600 mM NaCl, 1,000 mM mannitol, 45°C high temperature, 100 μM methyl viologen (MV), 4°C chilling injury, 0°C freezing point, −2°C freezing stress, and 300 μM ABA treatments, and those seedlings were only irrigated with water as the controls. For each treatment, the assimilating branches were harvested at 0, 3, and 24 h of treatments, respectively. The samples were instantly frozen in liquid nitrogen. Finally, the samples were stored at −80°C until RNA extraction and gene expression profiling.
RNAs were isolated from 100 mg sample using Plant RNA Kit (Omega BioTek, United States) by following the instruction provided in the kit. After treatment with DNase I, the RNAs were reversely transcribed into cDNA using SuperScriptTM II reverse transcriptase (Takara, Japan) and a poly(dT)18 primer. An ABI Prism 7500 Fast Sequence Detection System (Applied Biosystems, United States) was employed to detect the changes in gene expression after stress treatments. A Power SYBR® Green polymerase chain reaction (PCR) Master Mix (Applied Biosystems, United States) was used in the reaction. HcUBC10 and Atactin2 were used as the reference genes in these experiments (Shi et al., 2012; Zeng et al., 2015). The relative gene expression level was measured by using the 2–ΔΔCT comparative method (Livak and Schmittgen, 2001). The information of all genes in the quantitative reverse transcription (qRT)–PCR experiments was listed in the Supplementary Table 2.
Floral dip method (Clough and Bent, 1998) was employed to transfer HcTOE3 gene into Arabidopsis wild-type (WT) Colombia-0. After harvesting the seeds, all seeds were grown on MS medium containing kanamycin to select the transgenic plants. The selected transgenic plants were verified by the integration and expression analysis of HcTOE3 gene using genomic PCR and qRT-PCR.
Arabidopsis were grown in a greenhouse according to a previous report (Hu et al., 2012). For freezing treatment, plants grown for 4 weeks in the greenhouse were put in a light incubator at −2°C for 60 h and then removed to the greenhouse for continuous growth. At the same time, the phenotypes of each plant were carefully monitored at each treatment stage. The survival rates of each transgenic line and the WT were counted with 46 biological replicates. Among them, the Arabidopsis leaf samples were collected for further physiological experiments, histochemical staining, and gene expression at 24 h after freezing treatment.
Membrane permeability was measured according to a previous report (Chen and Wang, 2006). Each repeated collection of 0.1 g leaves was dipped into distilled water for 24 h and then boiled. The conductivity meter (DSS-307, Shanghai, China) was used to measure the initial and postboiling conductivity to calculate the relative conductivity. The contents of malondialdehyde (MDA) and proline were detected. The activities of peroxidase (POD), superoxide dismutase (SOD), or catalase (CAT) were measured by following the manufacturer’s instructions (Beijing Solarbio Technology Co., Ltd., China). Briefly, 0.1 g of tissues was collected and grounded for each sample. The contents of superoxide radical (O) and hydrogen peroxide (H2O2) were detected using NBT and DAB staining method (Shi et al., 2010). The cell damage degree was detected using Evans blue and trypan blue staining. DAB (1 mg/mL, pH 3.8), NBT (1 mg/mL), Evans blue, or trypan blue solution was used to treat each selected leaf from both treatment and the controls at 25°C in darkness for 6 h, respectively. Ten plant leaves (biological replicates) were run for each treatment. Then, the dyed leaves were treated in 95% ethanol with boiled temperature to remove chlorophyll and placed on a light box to take photographs, as described by a previous study (Fryer et al., 2002).
Two-way analysis of variance was used to analyze the significant differences between the treatments and the controls. During the gene expression analysis, three biological replicates were run for each treatment; for avoiding the potential pipetting errors, three technical replicates were run for each gene. For physiological traits, each treatment was repeated for at least three times. Post hoc Duncan multiple-comparisons test was performed with significant levels of ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. All data were represented as means ± SD.
In our previous work, TOE3 was cloned from H. caspica by homologous cloning and RACE (Zhang et al., 2016). Based on the sequence analysis, there are a total of 1,329 bp in the HcTOE3 ORF. HcTOE3 gene encodes a 48.8-kDa protein containing 443 aa. By comparing with Arabidopsis TOE proteins, two AP2 DNA-BDs were found in the HcTOE3 protein, which are located at 130–186 and 222–279 aa, respectively. HcTOE3 protein also contains a nuclear localization signal (NLS) sequence that is located at 118–125 aa (Figure 1A). A phylogenetic tree containing both HcTOE3 and 150 Arabidopsis AP2/ERF TFs showed four distinct clades among these proteins. These four clades were AP2, RAV, ERF, and DREB clades comprising of 21, 7, 65, and 56 proteins, respectively. HcTOE3 was the closest to AtTOE3 protein; HcTOE3 was clustered into AP2 subfamily (Figure 1B).
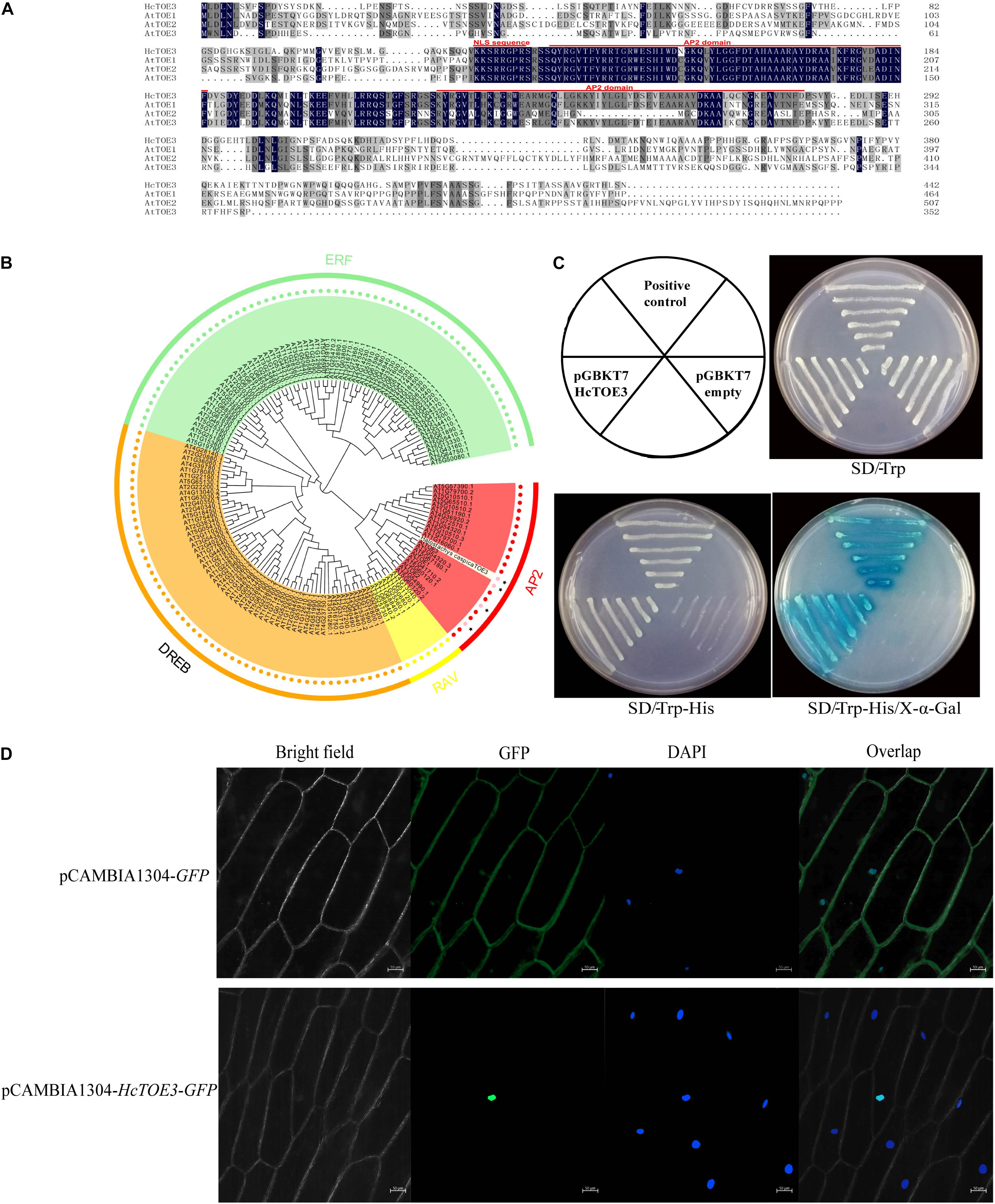
Figure 1. The analysis of the conserved domains (A), phylogenetic relationship (B), transcriptional activation of HcTOE3 (C), and subcellular localization of HcTOE3 (D).
To ascertain whether HcTOE3 functions as a TF, transactivation assay was performed in yeast. As shown in Figure 1C, the positive control and yeast containing pGBKT7-HcTOE3 gene divided quickly and formed the big biomass with blue color; however, no significant growth was observed in the negative control, and the yeast did not show the blue color. This suggests that HcTOE3 is activated and functions well in the yeast cells.
Protein sequence alignment showed that HcTOE3 contained a conserved NLS domain. To localize where the HcTOE3 protein functions, the HcTOE3 gene was linked to a GFP report gene and then transformed into onion epidermal cells. The green fluorescence was only observed in the nucleus (Figure 1D). This suggests that HcTOE3 is a nuclear-localized protein; after it is translated, it will be translocated into the nucleus.
Gene expression analysis showed that HcTOE3 was strongly induced by NaCl, mannitol (simulated drought stress), heat (45°C), MV, chilling injury (4°C), freezing point (0°C), freezing stress (−2°C), and ABA (Figure 2). These results suggested that HcTOE3 participated in abiotic stress responses and ABA-mediated exogenous hormone signal transduction pathways, and HcTOE3 may be a rapid response gene in the early stage of stress response.
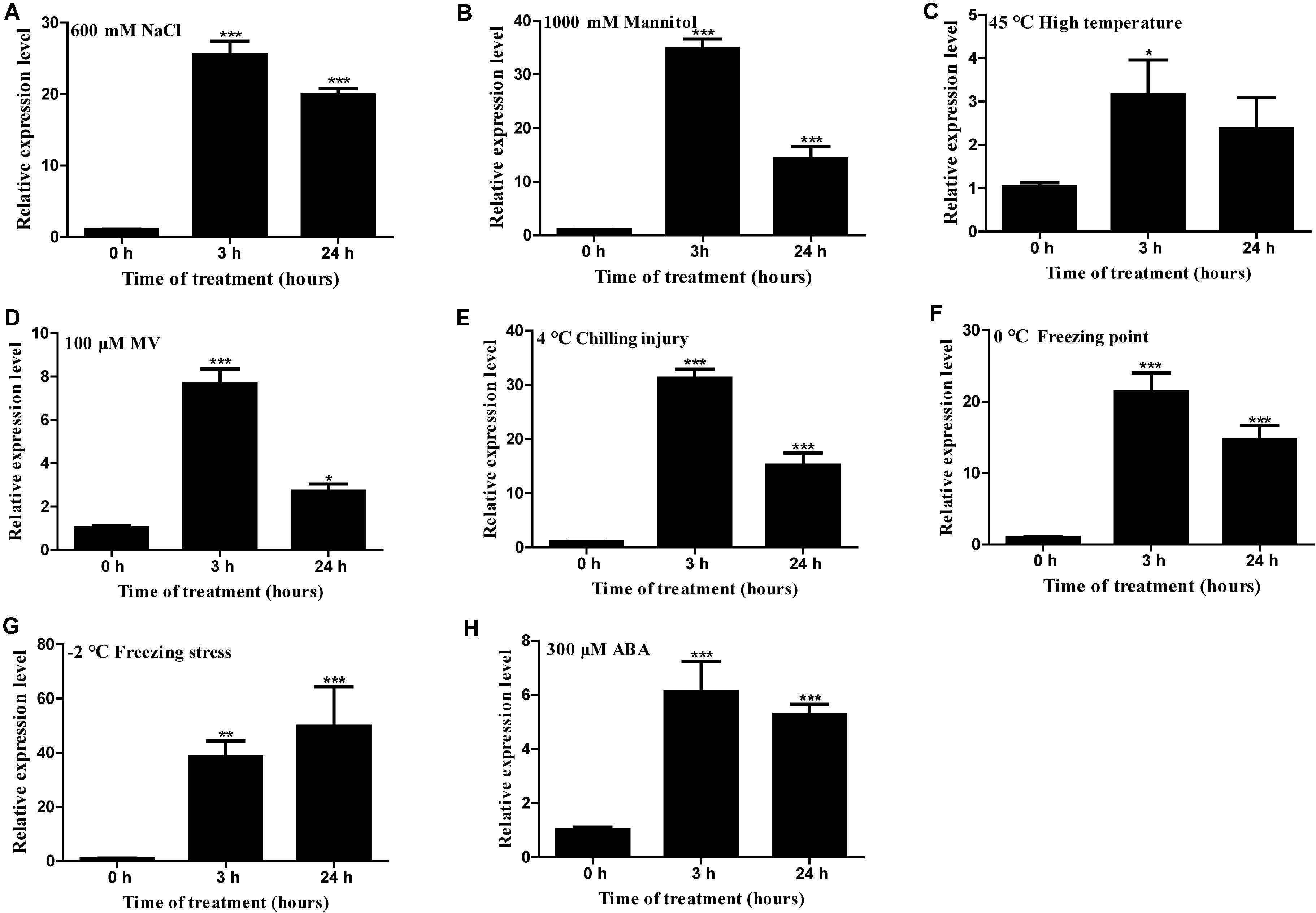
Figure 2. Expression profile of H. caspica assimilating branch TOE3 gene under different abiotic stress and abscisic acid treatments: 600 mM NaCl (A), 1,000 mM mannitol (B), 45°C high temperature (C), 100 μM MV (D), 4°C chilling injury (E), 0°C freezing point (F), −2°C freezing stress (G), 300 μM abscisic acid (ABA) (H). HcUBC10 served as reference gene, and the expression of HcTOE3 gene in the control (0 h) was adjusted to 1. Data represented the average of three biological replicates. Significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001.
To elucidate the functions of HcTOE3 gene, HcTOE3 was transferred into Arabidopsis by Agrobacteria-mediated floral dip transformation with CaMV 35S promoter. Four independent transgenic overexpression lines (OE, T2) were obtained, as confirmed by genomic DNA PCR and qRT-PCR analysis (Figure 3). Two independent homozygous T2 lines (OE2 and OE3) with the highly expressed HcTOE3 were selected for testing plant tolerance to freezing stress.
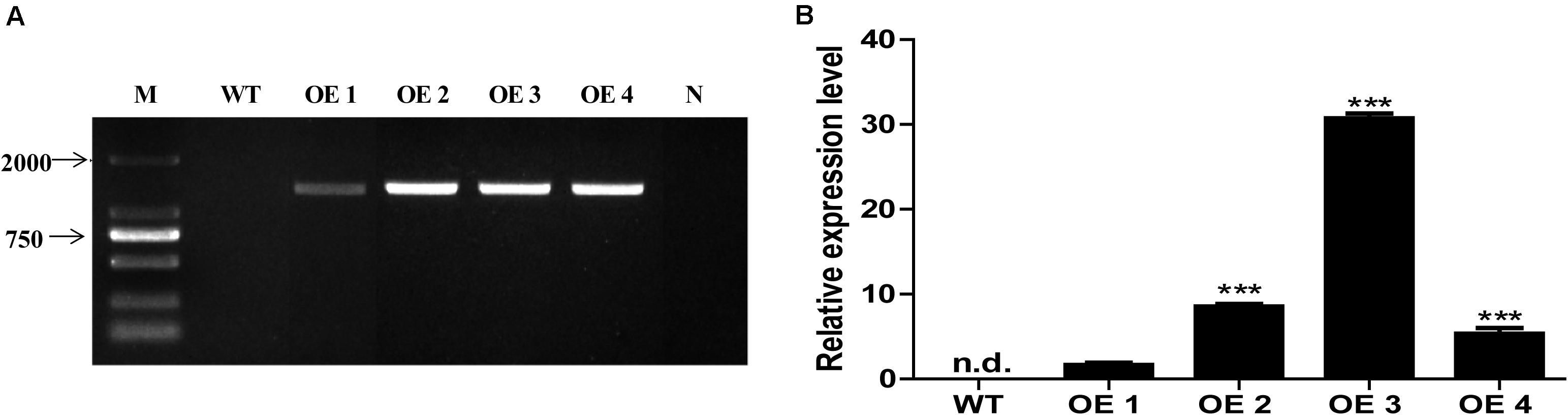
Figure 3. The identification and selection of overexpression Arabidopsis. (A) Genomic DNA PCR analysis of HcTOE3 gene from WT and four transgenic lines. M, DNA marker; N, negative control using water as template. (B) Relative expression of HcTOE3 in WT and four transgenic lines with AtActin2 as an internal control using qRT-PCR. Data represented the average of three biological replicates. Significant differences were at ***p < 0.001.
To examine the effects of HcTOE3 transgenic lines on freezing tolerance condition, a freezing stress treatment was carried out with the T2 generation of homozygous transgenic Arabidopsis (OE2 and OE3) and the WT plants growing in pots under normal conditions for 4 weeks. As shown in Figure 4A, transgenic lines showed significantly enhanced freezing stressed tolerance than the controls. Under freezing treatment, the survival rate of HcTOE3 transgenic lines was all greater (60%) compared to 20% for the WT (Figure 4B). Electrolyte leakage (EL) and MDA were inversely correlated with cell membrane stability and used as indicators to assess cell membrane damage (Bajji et al., 2002; Ashrafi et al., 2015). Here, all HcTOE3-overexpressing plants showed clearly lower EL values and MDA content than WT after freezing treatment (Figures 4C,D). These results indicated that expressing HcTOE3 increase plant tolerance to freezing treatment.
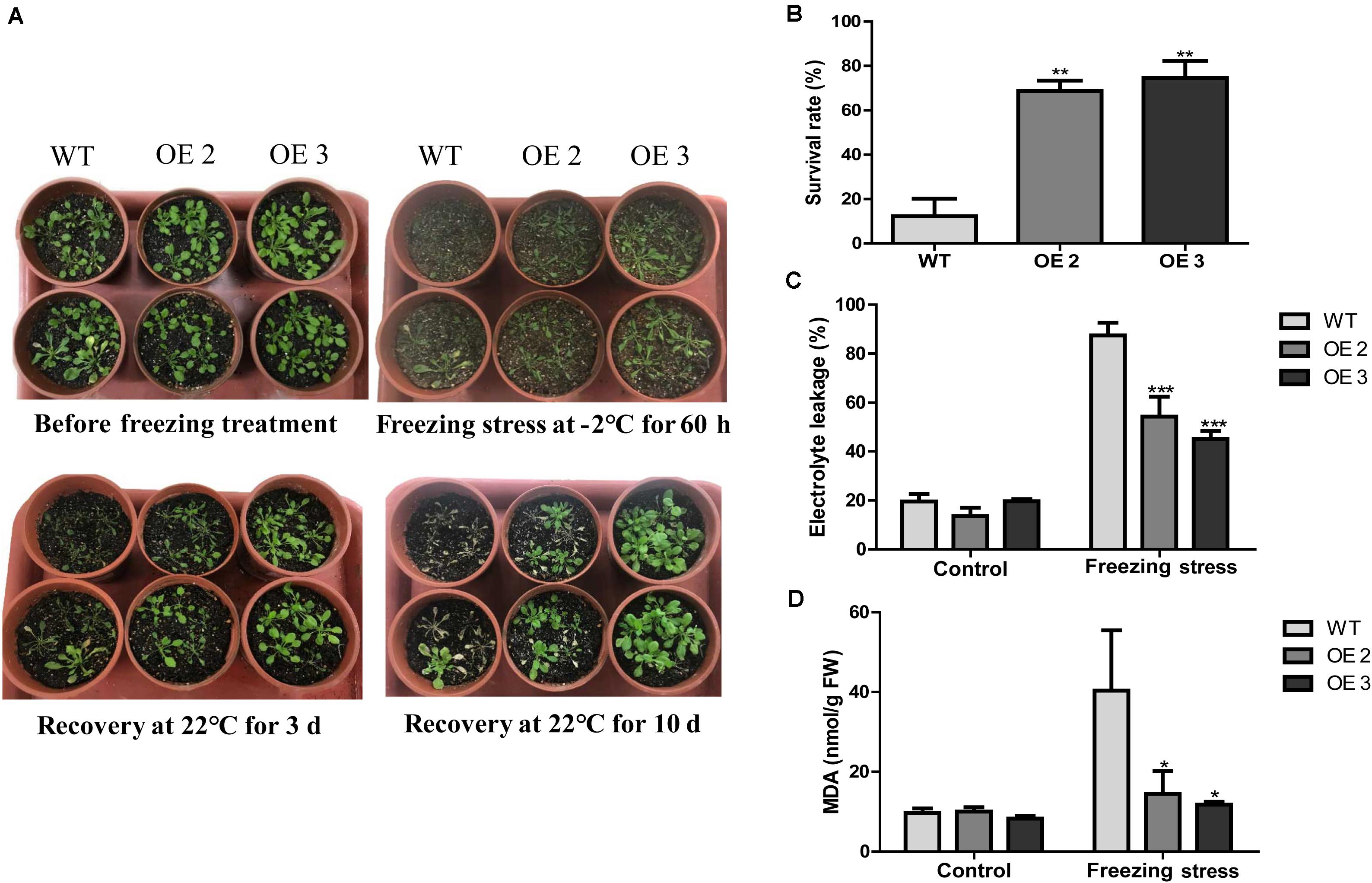
Figure 4. Overexpression of HcTOE3 confers freezing tolerance in Arabidopsis. (A) Phenotype of the HcTOE3-overexpressing plants and the non-transgenic controls under freezing stress. (B) Survival rates were counted from 46 biological replicates. (C) Electrolyte leakage. (D) MDA content. Data represented the average of three biological replicates. Significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001.
It is well-known that unfavorable stress induces ROS accumulation (Dinakar et al., 2012). DAB and NBT staining were used to detect H2O2 and O2– accumulation levels; trypan and Evans blue staining were used to examine the cell death in leaves from HcTOE3 transgenic lines and WT under freezing stress. As shown in Figures 5A–D, less intensively brown and blue staining can be observed in the transgenic plants than those in the WT after freezing treatment, suggesting that HcTOE3 transgenic plants had less accumulation of H2O2 and O2– and less severe cell damage after suffering freezing stress.
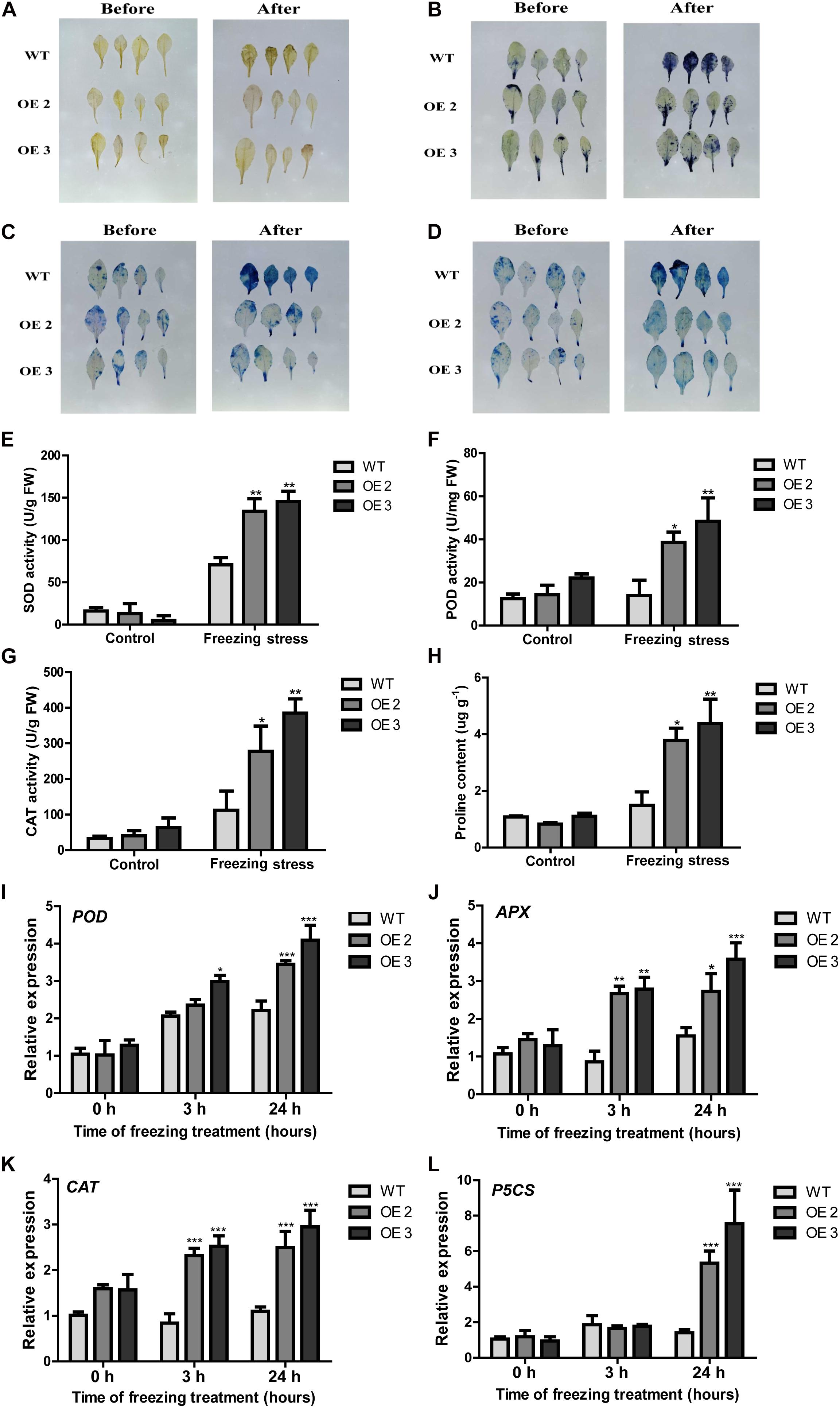
Figure 5. The performance of HcTOE3 transgenic Arabidopsis under freezing treatment. (A) H2O2 detection in plant leaves by DAB staining. (B) O2– detection in plant leaves by NBT staining. (C, D) Dead cell detection in plant leaves by Evans blue and trypan blue staining. (E) SOD activity. (F) POD activity. (G) CAT activity. (H) Proline content. (I–K) Expression levels of antioxidant enzyme genes, including POD, APX, and CAT. (L) Expression level of osmotic regulation gene P5CS. Data represented the average of three biological replicates. Significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001.
To further explore the function of HcTOE3 in enhancing plant tolerance to freezing stress, we examined physiological indicators in connection with ROS scavenging and osmotic adjustment substance contents in the plants during freezing stress treatment and the non-treated controls. For the controls, there were no difference on the activities of the antioxidant enzymes (SOD, CAT, and POD) between the transgenic plants and the WT controls. However, although freezing treatment increased the activities of all tested antioxidant enzymes in both transgenic plants and the controls, the activities were induced more in the transgenic plants than those in the WT controls (Figures 5E–G). Additionally, the accumulation of proline content was higher in the transgenic Arabidopsis plants after freezing stress (Figure 5H). To further confirm the above physiological data, we also examined the expression of the osmotic-related genes P5CS and the ROS detoxification-associated genes (POD, CAT, and APX). The qRT-PCR results demonstrated that freezing treatment induced the expression of all four tested genes more in the Arabidopsis OE plants than that in the non-transgenic controls (Figures 5I–L). All these evidence suggested that overexpressing HcTOE3 enhances transgenic plant tolerance to freezing stress by increasing the contents of osmotic solutes and antioxidant enzyme activities.
HcTOE3 is a TF controlling the expression of other genes. To answer the question whether or not HcTOE3 induced the expression of low temperature–responsive genes and how they are related to each other. We detected the expression of several cold-associated genes, such as CBF1 and CBF2 and their associated genes, including COR15A, COR47, KIN1, and RD29A. During normal growth condition, there was no significant difference on the expression of six tested genes between the transgenic plants and the non-transgenic controls. However, under freezing treatment, the expression levels of the six tested genes were significantly higher in HcTOE3-overexpressing plants than those in the non-transgenic controls (Figures 6A–F). Additionally, we also examined the expression of genes associated with ABA signaling pathway, which included ABI1, ABI2, ABI5, and RAB18. The results indicated that freezing stress induced the expression of four ABA-responsive genes (Figures 6G–J). All of the above results suggested that HcTOE3 plays a critical role in enhancing plant tolerance to freezing stress potentially by regulating cold and ABA-responsive genes under freezing stress.
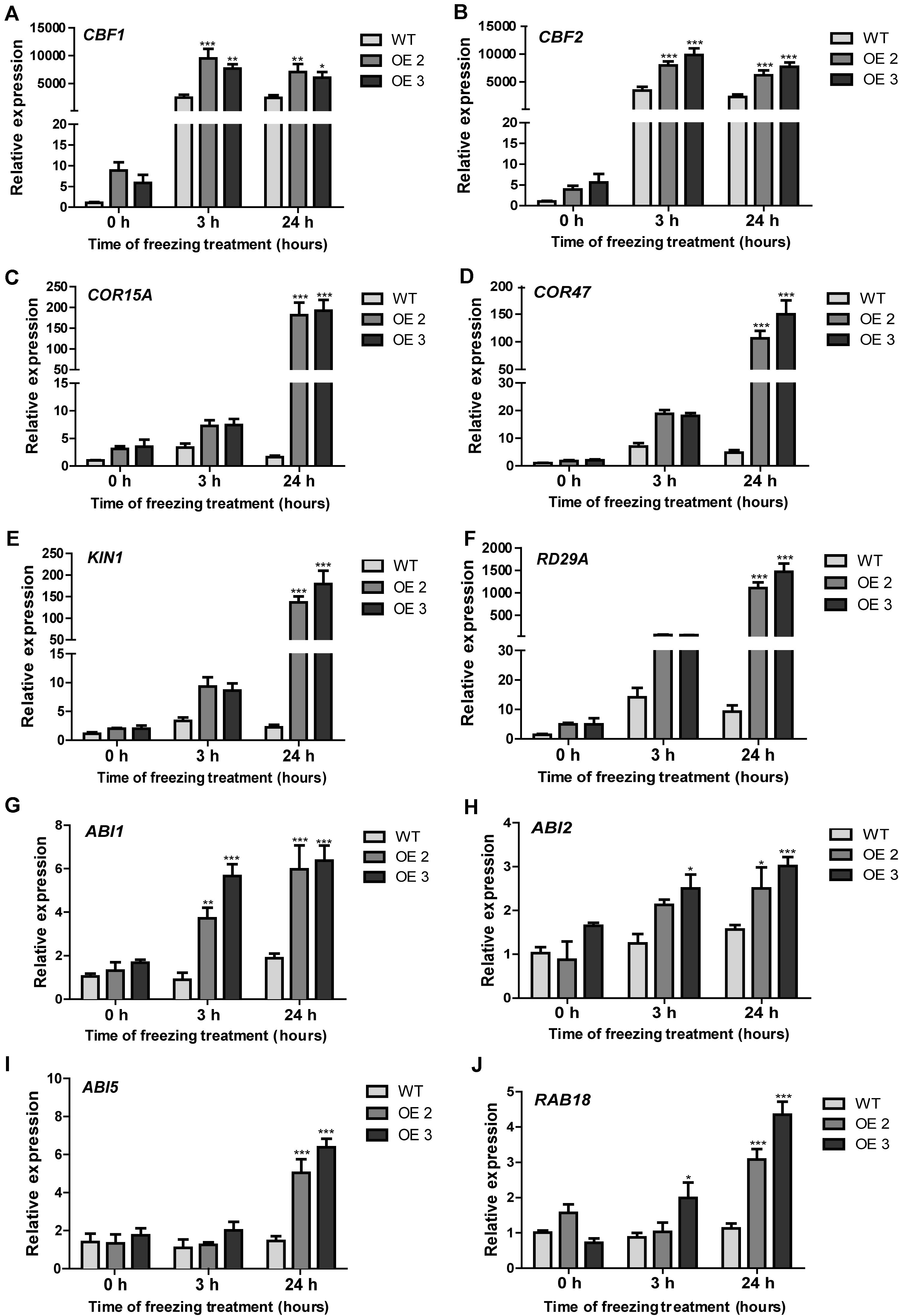
Figure 6. Expression patterns of cold-responsive and ABA-associated genes in the transgenic Arabidopsis and the non-transgenic plants under freezing stress treatment. (A–F) Expression profiles of cold responsive genes, including CBF1, CBF2, COR15A, COR47, KIN1, and RD29A. (G–J) Expression profiles of ABA-related genes, including ABI1, ABI2, ABI5, and RAB18. Data represented the average of three biological replicates. Significant differences at *p < 0.05, **p < 0.01, and ***p < 0.001.
AP2/ERF TFs are one of the major TF gene families with a larger number of gene members. According to their gene structure and evolution relationship, AP2/ERF family can be divided into different subfamilies. Although different member of AP2/ERF may play different roles in plant growth and development, some members, such as ERF, DREB, and RAV, play an important function in plant response to various abiotic stresses (Xing et al., 2017; Zhao et al., 2019; Li et al., 2020; Lv et al., 2020). TOEs are AP2 TF family members, which inhibit CONSTANS and regulate flowering time by transmitting photoperiodic signal (Zhang et al., 2015). However, there is no report on the AP2 TFs in H. caspica. This study is the first report on the freezing tolerance identification of a novel TF gene HcTOE3 from H. caspica. The HcTOE3 protein contains two typical AP2 domains belonging to AP2 subfamily and has a close relationship with AtTOE3 (Figures 1A,B). HcTOE3 exhibited transcriptional activation activity and localized in the nucleus (Figures 1C,D). In Lily, the expression of LlDREB2B was rapidly increased under cold, salt, mannitol, and heat treatment (Wu et al., 2018). Similarly in H. caspica, HcTOE3 gene was strongly induced by salinity, mannitol (simulated drought), heat, MV, chilling injury, freezing point, freezing stress, and ABA treatments (Figure 2). These results indicated that HcTOE3 may play significant roles in enhancing plant tolerance to abiotic stresses.
Under abiotic stresses, plants usually produce different ROS with a high level. As we know, ROS is a class of oxidative chemicals that damage plant cell membrane and cell structure, cause cell EL, and further inhibit plant growth and development (Caverzan et al., 2019). It was reported that low temperature significantly induced AP2/ERF gene expression in Brassica napus (Du et al., 2016). Overexpression of AtERF102 and AtERF103 demonstrated the important role of AP2/ERF gene in plant tolerance to cold stress potentially due to maintenance of electrolyte balance between inside and outside of the cells (Illgen et al., 2020). To study the function of HcTOE3, we obtained HcTOE3-overexpressing Arabidopsis (Figure 3), and the freezing tolerance of mature plants was examined; we found that overexpressing HcTOE3 reduced EL in the transgenic plants; we also found MDA content was also decreased in the transgenic plants than that in the WT (Figures 4C,D). Our results agree with the previous results reported by Illgen et al. (2020). Overexpressing HcTOE3 had less accumulation of O2– and H2O2 in transgenic Arabidopsis plants than those in the WT by DAB and NBT staining (Figure 5). All the evidence demonstrated that overexpressing HcTOE3 enhanced plant tolerance to freezing stress.
Freezing stress destroys water potential dispensation in plant cells, which further causes cell dehydration and even death (Pearce, 2001). Accumulation of proline and soluble proteins was associated with enhanced tolerance to freezing stress in plants (Kaplan and Guy, 2004; Kaplan et al., 2007). In our experiments, compared with WT plants, HcTOE3 overexpressed plants showed higher proline content under freezing-stressed treatment (Figure 5H). P5CS is a crucial gene in the proline glutamate biosynthesis pathway, which increased proline accumulation and enhanced cold tolerance in several plants (Liu et al., 2020). After freezing treatment, the expression of P5CS was higher in transgenic plants than that in non-transgenic controls, which was consistent with proline content (Figure 5L). These results suggest that enhanced osmolytes may increase freezing tolerance in the HcTOE3 OE plants.
Plants have evolved antioxidative systems to sustain ROS at low homeostasis levels. Plant enhanced their tolerance to various abiotic stresses by scavenging excessive ROS through antioxidant enzyme system (Chen et al., 2014; Ma et al., 2018). Here, we found that the activities of all tested antioxidant enzymes were increased in both transgenic and non-transgenic plants under freezing stress; however, the increased levels were higher in the transgenic plants than those in the non-transgenic plants (Figures 5E–G). AtCAT, AtPOD, and AtAPX encode CAT, POD, and ascorbate peroxidase (APX), respectively. They are thought to catalyze a lot of oxidative reactions by using H2O2 as an electron acceptor (Orvar and Ellis, 1995; Pasqualini et al., 2007). The transcriptional abundance of antioxidant genes was induced by freezing treatment in plants with overexpression of HcTOE3 gene (Figures 5I–K). These results suggest that the freezing tolerance of HcTOE3 transgenic Arabidopsis may be related to the enhanced antioxidative system.
There are several known pathways to mediate plant response to cold, of which the most classic one is the CBF pathway, which is associated with transcription regulators CBF1–3. Some targeted downstream genes of CBF have been identified, including COR15A and COR47 (Wang et al., 2017). Our gene expression analysis showed that freezing treatment induced the expression of many low temperature–responsive genes, such as CBF1, CBF2, COR15A, COR47, KIN1, and RD29A, which were tested in this study. Freezing stress also induced the expression of the genes associated with ABA signaling pathway, which included ABI1, ABI2, ABI5, and RAB18. These results indicated that HcTOE3 may depend on the CBF signaling pathway to control plant response to freezing stress. The ABA-dependent cold signaling pathway has long been considered to be mostly independent of the CBF pathway (Bolt et al., 2017). However, certain downstream genes, such as the TF MYB96, may integrate and activate both ABA and CBF pathways (Lee and Seo, 2015). Future work may be needed to investigate how HcTOE3 proteins are integrated into the widespread transcriptional networks that regulate plant response to cold stress. More research may also need to test the function of ABA regulation of HcTOE3 in the integrated freezing response pathway.
HcTOE3 is an AP2/ERF TF and belongs to the AP2 subfamily. AP2 subfamily, including TOE3, mainly plays a crucial role in plant growth and evolution. However, under abiotic stress, the function of HcTOE3 remains poorly understood. Transcription of HcTOE3 in H. caspica was enhanced by environmental stresses, as well as exogenous plant hormone ABA treatments. Overexpression of HcTOE3 resulted in elevated freezing tolerance of transgenic Arabidopsis. HcTOE3 transgenic Arabidopsis reduced membrane lipid peroxidation and cell electrolyte extravasation. HcTOE3 transgenic Arabidopsis plants also enhanced plant tolerance to freezing stress through increasing antioxidant enzyme activities and the contents of osmoregulation substances, such as proline. HcTOE3 mediates cold and ABA signal transduction by increasing the transcription level of stress-responsive genes, thereby improving plant tolerance to freezing stress in transgenic Arabidopsis. Our findings put forward a model to propose how HcTOE3 plays a significant role in enhancing freezing tolerance and provide valuable insight into the plant response to cold stress (Figure 7).
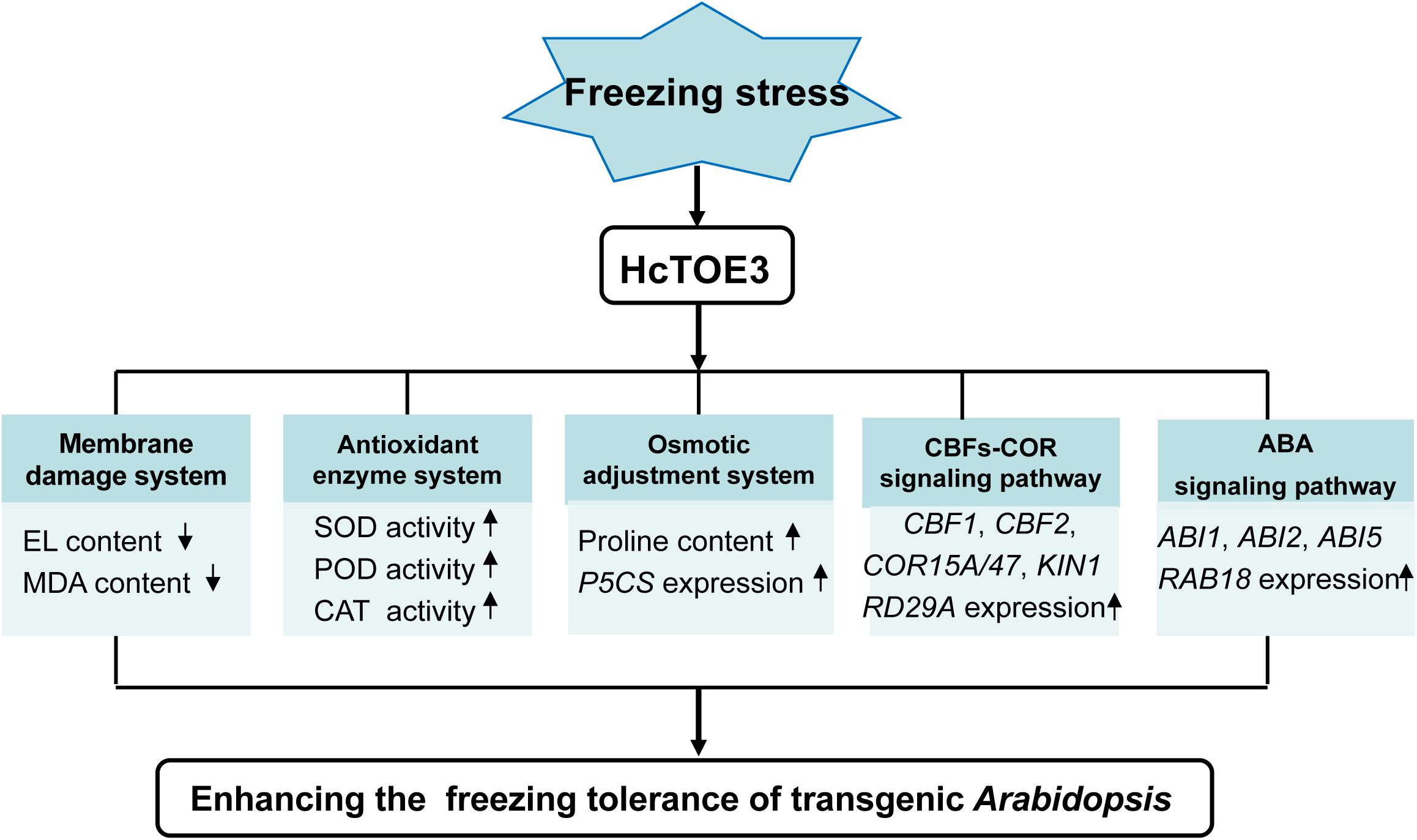
Figure 7. Model of HcTOE3 regulation of the freezing stress response in transgenic Arabidopsis. The model indicated that overexpression of HcTOE3 up-regulates genes taking part in CBFs-COR signaling, ABA signaling, proline biosynthesis, and ROS scavenging. These genes lead to an increase in SOD, POD, CAT activities and proline content; a reduction ROS content and cell membrane peroxidation; and ultimately enhancing the freezing tolerance of Arabidopsis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
FY and YoZ designed the experiments, analyzed the data, and wrote the manuscript text. FY, JJ, PW, YuZ, and WL performed the experiments. All authors reviewed the manuscript.
This study was supported by the National Natural Science Foundation of China (No. 31760071), the Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2015211C274), and the Graduate Scientific Research Innovation Project of Xinjiang Uygur Autonomous Region (No. XJ2020G047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.638788/full#supplementary-material
Supplementary Table 1 | The summary of the amino acid sequences from Arabidopsis AP2/ERF transcription factor members and HcTOE3 used to generate phylogenetic tree.
Supplementary Table 2 | Primer sequences used in the present study.
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 54, 201–212. doi: 10.1007/s10535-010-0038-7
Ashrafi, E., Razmjoo, J., Zahedi, M., and Pessarakli, M. (2015). Screening Alfalfa for salt tolerance based on lipid peroxidation and antioxidant enzymes. Agron. J. 107, 167–173. doi: 10.2134/agronj14.0248
Bajji, M., Kinet, J. M., and Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70.
Bolt, S., Zuther, E., Zintl, S., Hincha, D. K., and Schmuelling, T. (2017). ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 40, 108–120. doi: 10.1111/pce.12838
Cao, S. L., Wang, Y., Li, X. T., Gao, F., Feng, J. C., and Zhou, Y. J. (2020). Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants (Basel) 9:455. doi: 10.3390/plants9040455
Caverzan, A., Piasecki, C., Chavarria, G., Stewart, C. N., and Vargas, L. (2019). Defenses against ROS in crops and weeds: the effects of interference and herbicides. Int. J. Mol. Sci. 20:1086. doi: 10.3390/ijms20051086
Chen, J. X., and Wang, X. F. (2006). Experimental Guidance of Plant Physiology. Guangzhou: South China University of Technology Press, 115–116.
Chen, Y., Jiang, J., Chang, Q., Gu, C., Song, A., Chen, S., et al. (2014). Cold acclimation induces freezing tolerance via antioxidative enzymes, proline metabolism and gene expression changes in two chrysanthemum species. Mol. Biol. Rep. 41, 815–822. doi: 10.1007/s11033-013-2921-8
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dinakar, C., Djilianov, D., and Bartels, D. (2012). Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 182, 29–41. doi: 10.1016/j.plantsci.2011.01.018
Ding, Y. L., Shi, Y. T., and Yang, S. H. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Du, C. F., Hu, K. N., Xian, S. S., Liu, C. Q., Fan, J. C., Tu, J. X., et al. (2016). Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus L.). Mol. Genet. Genom. 291, 1053–1067. doi: 10.1007/s00438-015-1161-0
Fryer, M. J., Oxborough, K., Mullineaux, P. M., and Baker, N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. doi: 10.1093/jxb/53.372.1249
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Hu, Y. Z., Zeng, Y. L., Guan, B., and Zhang, F. C. (2012). Overexpression of a vacuolar H+-pyrophosphatase and a B subunit of H+-ATPase cloned from the halophyte Halostachys caspica improves salt tolerance in Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 108, 63–71. doi: 10.1007/s11240-011-0013-9
Illgen, S., Zintl, S., Zuther, E., Hincha, D. K., and Schmuelling, T. (2020). Characterisation of the ERF102 to ERF105 genes of Arabidopsis thaliana and their role in the response to cold stress. Plant Mol. Biol. 103, 303–320. doi: 10.1007/s11103-020-00993-1
Jin, J. P., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J. C., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Jofuku, K. D., Denboer, B. G., Vanmontagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Kaplan, F., and Guy, C. L. (2004). Beta-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 135, 1674–1684. doi: 10.1104/pp.104.040808
Kaplan, F., Kopka, J., Sung, D. Y., Zhao, W., Popp, M., Porat, R., et al. (2007). Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50, 967–981. doi: 10.1111/j.1365-313X.2007.03100.x
Krogan, N. T., Hogan, K., and Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. doi: 10.1242/dev.085407
Lee, B. H., Lee, H. J., Xiong, L. M., and Zhu, J. K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14, 1235–1251. doi: 10.1105/tpc.010433
Lee, H. G., and Seo, P. J. (2015). The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 82, 962–977. doi: 10.1111/tpj.12866
Lei, M., Li, Z. Y., Wang, J. B., Fu, Y. L., and Xu, L. (2019). Ectopic expression of the Aechmea fasciata APETALA2 gene AfAP2-2 reduces seed size and delays flowering in Arabidopsis. Plant Physiol. Biochem. 139, 642–650. doi: 10.1016/j.plaphy.2019.03.034
Li, T., Huang, Y., Khadr, A., Wang, Y. H., Xu, Z. S., and Xiong, A. S. (2020). DcDREB1A, a DREB-binding transcription factor from Daucus carota, enhances drought tolerance in transgenic Arabidopsis thaliana and modulates lignin levels by regulating lignin-biosynthesis-related genes. Environ. Exp. Bot. 169:103896. doi: 10.1016/j.envexpbot.2019.103896
Liu, G. S., Zhang, Y. X., Yun, Z., Hu, M. J., Liu, J. L., Jiang, Y. M., et al. (2020). Melatonin enhances cold tolerance by regulating energy and proline metabolism in Litchi Fruit. Foods 9:454. doi: 10.3390/foods9040454
Liu, J. Y., Shi, Y. T., and Yang, S. H. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. doi: 10.1111/jipb.12657
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, K. W., Li, J., Zhao, K., Chen, S., Nie, J., Zhang, W. L., et al. (2020). Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 292:110375. doi: 10.1016/j.plantsci.2019.110375
Ma, N. L., Lah, C. W. A., Kadir, N. A., Mustaqim, M., Rahmat, Z., Ahmad, A., et al. (2018). Susceptibility and tolerance of rice crop to salt threat: physiological and metabolic inspections. PLoS One 13:e0192732. doi: 10.1371/journal.pone.0192732
Mizoi, J., Shinozaki, K., and Shinozaki, Y. K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Orvar, B. L., and Ellis, B. E. (1995). Isolation of a cDNA encoding cytosolic ascorbate peroxidase in tobacco. Plant Physiol. 108, 839–840. doi: 10.1104/pp.108.2.839
Orvar, B. L., Sangwan, V., Omann, F., and Dhindsa, R. S. (2000). Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 23, 785–794. doi: 10.1046/j.1365-313x.2000.0084.x
Pasqualini, S., Paolocci, F., Borgogni, A., Morettini, R., and Ederli, L. (2007). The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants. Plant Cell Environ. 30, 1545–1556. doi: 10.1111/j.1365-3040.2007.01730.x
Ruelland, E., Vaultier, M. N., Zachowski, A., and Hurry, V. (2009). Cold signalling and cold acclimation in plants. Adv. Bot. Res. 49, 35–150. doi: 10.1016/s0065-2296(08)00602-2
Shi, J., Fu, X. Z., Peng, T., Huang, X. S., Fan, Q. J., and Liu, J. H. (2010). Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 30, 914–922. doi: 10.1093/treephys/tpq030
Shi, Y. T., Tian, S. W., Hou, L. Y., Huang, X. Z., Zhang, X. Y., Guo, H. W., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Ann. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Shinozaki, Y. K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U. S. A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Wang, D. Z., Jin, Y. N., Ding, X. H., Wang, W. J., Zhai, S. S., Bai, L. P., et al. (2017). Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry (Mosc.) 82, 1103–1117. doi: 10.1134/s0006297917100030
Wu, Z., Liang, J. H., Zhang, S., Zhang, B., Zhao, Q. C., Li, G. Q., et al. (2018). A canonical DREB2-type transcription factor in Lily is post-translationally regulated and mediates heat stress response. Front. Plant Sci. 9:243. doi: 10.3389/fpls.2018.00243
Wurschum, T., Gross-Hardt, R., and Laux, T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18, 295–307. doi: 10.1105/tpc.105.038398
Xing, L. P., Di, Z. C., Yang, W. W., Liu, J. Q., Li, M. N., Wang, X. J., et al. (2017). Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8:1948. doi: 10.3389/fpls.2017.01948
Xu, K. D., Huang, X. H., Wu, M. M., Wang, Y., Chang, Y. X., Liu, K., et al. (2014). A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS One 9:e83556. doi: 10.1371/journal.pone.0083556
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Zeng, Y. L., Li, L., Yang, R. R., Yi, X. Y., and Zhang, B. H. (2015). Contribution and distribution of inorganic ions and organic compounds to the osmotic adjustment in Halostachys caspica response to salt stress. Sci. Rep. 5:13639. doi: 10.1038/srep15867
Zhang, B. L., Wang, L., Zeng, L. P., Zhang, C., and Ma, H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29, 975–987. doi: 10.1101/gad.251520.114
Zhang, Y. F., Yang, R. R., and Zeng, Y. L. (2016). Cloning, expression and correlated analysis of HcmiR172e with predicted target gene HcTOE3 in Halostachys caspica under salt stress. Plant Physiol. J. 52, 505–513. doi: 10.13592/j.cnki.ppj.2015.0642
Zhang, Y. G., Liu, X. J., Zhang, K. C., Zhang, D. Y., and Guan, K. Y. (2018). An Abscisic Acid Insensitive3-like gene from the desert moss Syntrichia caninervis confers abiotic stress tolerance and reduces ABA sensitivity. Plant Cell Tiss. Org. Cult. 133, 417–435. doi: 10.1007/s11240-018-1394-9
Keywords: Halostachys caspica, HcTOE3, AP2/ERF transcription factor, transgenic Arabidopsis, freezing tolerance
Citation: Yin F, Zeng Y, Ji J, Wang P, Zhang Y and Li W (2021) The Halophyte Halostachys caspica AP2/ERF Transcription Factor HcTOE3 Positively Regulates Freezing Tolerance in Arabidopsis. Front. Plant Sci. 12:638788. doi: 10.3389/fpls.2021.638788
Received: 07 December 2020; Accepted: 26 March 2021;
Published: 13 May 2021.
Edited by:
Heng Zhang, Shanghai Center for Plant Stress Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Yong Hwa Cheong, Sunchon National University, South KoreaCopyright © 2021 Yin, Zeng, Ji, Wang, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youling Zeng, emVuZ195bHhqdUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.