- Bond Life Sciences Center, Division of Biological Sciences, University of Missouri, Columbia, MO, United States
Morphotypes of Brassica oleracea are the result of a dynamic interaction between genes that regulate the transition between vegetative and reproductive stages and those that regulate leaf morphology and plant architecture. In kales, ornate leaves, extended vegetative phase, and nutritional quality are some of the characters potentially selected by humans during domestication. We used a combination of developmental studies and transcriptomics to understand the vegetative domestication syndrome of kale. To identify candidate genes that are responsible for the evolution of domestic kale, we searched for transcriptome-wide differences among three vegetative B. oleracea morphotypes. RNA-seq experiments were used to understand the global pattern of expressed genes during a mixture of stages at one time in kale, cabbage, and the rapid cycling kale line TO1000. We identified gene expression patterns that differ among morphotypes and estimate the contribution of morphotype-specific gene expression that sets kale apart (3958 differentially expressed genes). Differentially expressed genes that regulate the vegetative to reproductive transition were abundant in all morphotypes. Genes involved in leaf morphology, plant architecture, defense, and nutrition were differentially expressed in kale. This allowed us to identify a set of candidate genes we suggest may be important in the kale domestication syndrome. Understanding candidate genes responsible for kale domestication is of importance to ultimately improve Cole crop production.
Introduction
Domestication is the process of selection used by humans to adapt wild plants to cultivation. During this process, a set of recurrent characters is acquired across a wide diversity of crops (known as the “domestication syndrome”) (Hammer, 1984; Doebley, 1992). Recurrent traits observed in domesticated plants include loss of seed shattering, changes in seed size, loss of photoperiod sensitivity, and changes in plant physiology and architecture (Doebley, 1992; Harlan, 1992; Allaby, 2014). Here, we explore the molecular basis behind the poorly defined vegetative domestication syndrome of kale, a Brassica oleracea morphotype, the leaves of which are consumed by humans and are also used as fodder. The domestication syndrome of kale includes apical dominance, ornate leaf patterns, the capacity to delay flower formation and maintain a vegetative state producing a higher yield of the edible portion, nutritional value of leaves, and the ability to defend against herbivores that also give the leaves a unique pungent taste.
The Cole crops (B. oleracea) are perennial plants native to Europe and the Mediterranean and include a range of domesticated and wild varieties (Snogerup, 1980; Tsunoda, 1980; Purugganan et al., 2000; Allender et al., 2007; Maggioni et al., 2010). Each crop type is distinguished by domestication traits not commonly found among the wild populations (Maggioni et al., 2010). They include dwarf plants with a main stem that has highly compressed internodes (cabbages), plants with an elongated main stem in which the lateral branches are highly compressed due to very short internodes (Brussels sprouts), plants with proliferation of floral meristems (broccoli) or proliferation of aborted floral meristems (cauliflower), and plants with swollen stems (kohlrabi, Marrow-stem kale) or ornate leaf patterns (kales) (Hodgkin, 1995; Lan and Paterson, 2000). It is not so easy to define which domestication traits are common to all of the above-mentioned types, as opposed to the wild species. At early stages in domestication, plants may have been selected for less bitter tasting, less fibrous, thicker stems, and more succulent storage organs (Maggioni et al., 2010).
The B. oleracea genome is derived from an ancient whole-genome triplication event and research has provided evidence that polyploidy and genetic redundancy contribute significantly to the phenotypic variation observed among these crops (Lukens et al., 2004; Paterson, 2005). The genetic basis underlying the phenotypic diversity present in Cole crops has been addressed for only some morphotypes. For example, the cauliflower phenotype of Arabidopsis is comparable to inflorescence development in the cauliflower B. oleracea morphotype, and it has been shown that the gene CAULIFLOWER is responsible for the phenotype observed in both species (Kempin et al., 1995; Carr and Irish, 1997; Purugganan et al., 2000; Smith and King, 2000). However, quantitative trait locus (QTL) mapping for the curd of cauliflower indicates that 86 QTLs are controlling eight curd-related traits, demonstrating this phenotype is under complex genetic control (Lan and Paterson, 2000). Other studies have found and confirmed BoAP-a1 to be involved in curd formation in cauliflower (Anthony et al., 1995; Carr and Irish, 1997; Gao et al., 2007) and have shown that morphologies in this cultivar are under complex genetic control (Sebastian et al., 2002). The molecular bases of broccoli have also been proposed as involving changes in three codons in the BoCAL and BoAP genes (Carr and Irish, 1997; Lowman and Purugganan, 1999). A total of five QTLs have been detected for leaf traits in Brussel sprouts, but no significant QTLs have been found for axillary buds (Sebastian et al., 2002). Head formation in cabbage has been suggested as additively inherited (Tanaka et al., 2009). In kale, a recent study suggest that the lobed-leaf trait is quantitatively inherited (Ren et al., 2019).
Definitions of domestication syndrome in kale include traits affecting cultivation, harvesting, and cooking—and its uses—including food, medicines, toxins, and fibers (Denham et al., 2020). Leafy kales are considered the earliest cultivated brassicas, originally used for both livestock feed and human consumption (Maggioni et al., 2010) due to leaves and floral buds with great nutritional value (Velasco et al., 2011). Apical dominance is manifest in kale varieties, involving selection for erect plants with fewer side branches, and more compact plants, which allows more plants to fit into each unit of cultivated soil (Denham et al., 2020).
In kale, leaves are green to purple and do not form a compact head like in cabbage. Crop varieties have surprising variation in leaf size, leaf type, and margin curliness. The over expression phenotype of knotted1-like homeobox (knox) transcription factors in Arabidopsis (Hay and Tsiantis, 2006, 2009; Blein et al., 2008; Kimura et al., 2008) is very reminiscent of the highly lobed margins phenotype of kale, while in other Brassicaceae, a different homeodomain protein is involved in evolutionary changes in leaf complexity at a later stage in leaf development (Vlad et al., 2014). Kale leaf margin characteristics including curliness, blade texture, and tissue overgrowth are some of the more variable morphological characters among varieties but remain to be studied at the molecular level.
Kale leaves also have antioxidative properties and have been shown to reduce cholesterol levels (Velasco et al., 2007; Soengas et al., 2012). Their high content of beta-carotene, vitamin K, vitamin C, and calcium make it one of the most nutritious vegetables available for human consumption (Stewarta and McDougalla, 2012). Kale is a source of two carotenoids: lutein and zeaxanthin (Velasco et al., 2007). Kale leaves also have mustard oils produced from glucosinolates including sulforaphane, a compound suggested to have anticancer properties (Velasco et al., 2007). These natural chemicals most likely contribute to plant defense against pests and diseases and impart a pungent flavor property characteristic of all cruciferous vegetables (Sønderby et al., 2010; Carmona et al., 2011; Hahn et al., 2016).
Here, we used a combination of developmental studies and transcriptomes to understand the vegetative domestication syndrome of kale. Comparisons of kale morphotypes with the “rapid-cycling” morphotype TO1000 and cabbage allow us to identify transcriptome-wide differences that set kale apart. RNA-seq of a mixture of leaf stages for the B. oleracea vegetative morphotypes cabbage, kale, and TO1000 reveal gene expression patterns unique to kale and associated developmental and metabolic processes. This allowed us to identify a set of candidate genes we suggest may be important in the kale domestication syndrome.
Materials and Methods
Plant Material Used for Microscopy Studies
Leaf shape development was recorded for different kale varieties: red winter kale with lobed leaf margins (B. oleraceae var. Red Winter) and Lacinato Nero Toscana kale (B. oleracea var. palmifolia) with crenulate leaf margins (Figures 1A,B). Structural features of the meristems and developing leaves were observed during 30 days using environmental scanning electron microscopy (ESEM) and light microscopy (Leica, Frankfurt, Germany). Images were processed with Adobe (San Jose, California, United States) Photoshop, version 7.0. Plants were visually observed and described in the green house until fully grown.

Figure 1. Comparison of mature leaf morphologies within kale varieties and TO1000 used for the developmental studies; red winter kale (Brassica oleraceae var. acephala Red Winter) was also used in transcriptomics experiments. (A) Red winter kale with lobed leaf margins, (B) Lacinato Nero Toscana kale (Brassica oleracea var. palmifolia) with crenulate leaf margins, (C) TO1000 (B. oleracea var. alboglabra) picture provided by Zachary Stansell. Scale bar = 10 cm.
Plant Material and Experimental Design Used for RNA-Seq Studies
Single accessions of cabbage PI 303629 South Africa (B. oleracea var. capitata), kale (B. oleraceae var. acephala Red Winter SI), and the rapid cycling doubled haploid kale-like type HRI TO1000 DH3 (B. oleracea var. alboglabra from Warwick) (Supplementary Figure 1; Parkin et al., 2014) were grown in an environmental chamber under uniform conditions: 10 h day (light, 7 AM–5 PM; dark, 5:01 PM–6:59 AM daily) and watered every other day. Morphotypes were planted in a completely randomized block design. Initially, two seeds per pot were planted; then, only one was picked (32 plants per morphotype total). A random number generator was used to move plants and flats around every other day.
Tissues were collected 24 days after seeds were planted (cabbage had 2–3 fully expanded leaves, kale had 3–4 fully expanded leaves, and TO1000 had 3–5 fully expanded leaves). The upper portion of the stems, including immature leaves, and apical and lateral meristems were flash frozen in liquid nitrogen. The assumption was made that genes involved in developing different morphologies should be expressed in younger leaves, shoots, and meristems. Three biological replicates per morphotype were collected, each replicate consisting of pooled tissue from eight plants.
RNA Extraction and cDNA Synthesis
Lysis and homogenization of fresh soft tissue were performed before RNA extraction; PureLink Micro-to-Midi Total RNA Purification System (Invitrogen, Cat. No 12183-018, Carlsbad, California, United States) was used. RNA extraction was done using the Purelink RNA Mini Kit (Ambion, Cat. No. 12183-018A, Carlsbad, California, United States). DNase was not used before proceeding with complementary DNA (cDNA) synthesis as recommended in the protocol. For cDNA synthesis, MINT (Evrogen, Cat. No SK001, Moscow, Russia) was used; this kit enzymes synthesize full-length-enriched double-stranded cDNA from a total polyA+ RNA. The cDNA was then amplified by 16–21 cycles of polymerase chain reaction and purified using a PCR Purification Kit (Invitrogen K3100-01 Carlsbad, California, United States).
Library Construction and Illumina Sequencing
Protocols previously published by Steele et al. (2012) were implemented here. End repair was performed on 400 μl with 3 μg of normalized ds-cDNA prior to ligating barcoding adapters for multiplexing, using NEB Prep kit E600L (New England Biolabs, Ipswich, Massachusetts, United States). Shearing was done using a Bioruptor® sonication device (Diagenode, Denville New Jersey, United States), at 4°C, continuously for 15 min total (7 mix) on high. Three replicates per morphotype were tagged with different adaptors and sequenced on a single lane of an Illumina GAIIx machine.
Oligonucleotides used as adapters were cabbage (1, ACGT; 2, GCTT; 3, TGCT), kale (1, TACT; 2, ATGT; 3, GTAT), and TO1000 (1, CTGT; 2, AGCT; 3, TCAT). To prepare adapters, equal volumes were combined of each adapter at 100 μmol/L floating them in a beaker of boiling water for 30 min and snap cooling them on ice. Ligation products were run on a 2% low-melt agarose gel and size selected for ∼300 bp using an x-tracta Disposable Gel Extraction Tool (Scientific, Ocala Florida, United States). Fragments were enriched using PCR in 50 μl volumes containing 3 μl of ligation product, 20 μl of ddH2O, 25 μl master mix (from NEB kit), and 1 μl each of a 25 μmol/L solution of each forward and reverse primer [forward 5’-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC∗ T-3’; reverse 5’-CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC∗-3’; both high-performance liquid chromatography (HPLC) purified]. Thermal cycle routine was as follows: 98°C for 30 s, followed by 15 cycles of 98°C for 10 s, 65°C for 30 s, and 72°C for 30 s, with a final extension step of 72°C for 5 min. Products were run on a 2% low-melt agarose gel, and the target product was excised and purified with the tools described above. DNA was purified at each step during the end repair, adapter ligation, size selection, and fragment enrichment with either a Gel Extraction kit (Qiagen, Germantown Maryland, United States) or a DNA QIAquick PCR Purification kit (Qiagen, Germantown Maryland, United States). The three replicates per morphotype were run on one-third of a lane with single-end 98-bp reads on an Illumina GAIIx Genome Analyzer using the Illumina Cluster Generation Kit v2-GA II, Cycle Sequencing Kit v3 (Illumina, San Diego, United States), and image analysis using Illumina RTA 1.4.15.0 (Illumina, San Diego, United States) at University of Missouri DNA Core.
Data Quality Control and Preprocessing, Alignment, and Differential Expression Analysis
Reads were first trimmed for primer and adapter sequences using Cutadapt v2.4 (Martin et al., 2010) and read quality assessed using FastQC (Andrews, 2010). These were then aligned against the B. oleracea TO1000 genome (Parkin et al., 2014) downloaded from EnsemblPlants 44 (Kersey et al., 2018) with two passes of STAR v2.7.1a (Dobin et al., 2013). In the first pass, reads for each sample were aligned for splice-junction discovery, in addition to those junctions provided in the TO1000 annotations. Splice junctions from all samples were combined and provided for a second alignment, after which read counts were calculated for each gene using STAR. Mapping statistics are provided in Supplementary Table 1. Raw sequencing data and unnormalized read counts are publicly available through the Gene Expression Omnibus accession GSE149483.
Read counts were imported into R v3.6.3 (R Core Development Team, 2008), and differential expression was determined using DESeq2 v1.26.0 (Love et al., 2014). Specifically, read counts were normalized, with the TO1000 samples as reference, and fitted to a parametric model. We looked for differentially expressed genes (DEGs) for each pairwise comparison (kale vs. TO1000, kale vs. cabbage, and cabbage vs. TO1000). Comparisons were done to ultimately separate unique kale-morphotype DEGs. Since each pairwise comparison increases the potential number of false positives, these were combined into a single table, and the false discovery rate (FDR) (Benjamini and Hochberg, 1995) was adjusted for the entire table. Genes with an FDR < 0.05 and log-fold change > 1 or < −1 were considered as DEGs. Plots were made using the R packages ggplot2 (Wickham, 2009), pheatmap (Kolde, 2019), and VennDiagram (Chen, 2018). Transcription-associated proteins (TAPs) were identified using the approach described for the construction of PlnTFDB v3.01 (Riaño-Pachón et al., 2007; Pérez-Rodríguez et al., 2010). Scripts for bioinformatic analysis and original figure pdfs and tables are available on GitHub2.
Subgenome Analysis
Assignment of each gene to their respective subgenome were obtained from Parkin et al. (2014). Genes assigned to a subgenome were previously determined by synteny to Arabidopsis thaliana. Unassigned genes were considered non-syntenic. Enrichment of DEGs for syntenic vs. non-syntenic and for each subgenome was determined using a Fisher’s exact test (Fisher, 1934) and considered significant with an FDR-corrected p < 0.05.
Gene Annotation, GO, and KEGG Analysis
Gene Ontology (GO) terms (Ashburner et al., 2000) are not currently available for the TO1000 assembly. In order to annotate TO1000 gene models with GO terms, TO1000 protein sequences were Blasted (blastp) against protein sequences from UniProtKB Swiss-Prot (release 2019_05) (UniProt Consortium, 2018) using Diamond (Buchfink et al., 2014), with an e-value cutoff of 1e–5 and a maximum of one hit per query. UniProt IDs were then used to extract GO terms and mapped to the corresponding TO1000 gene model. In total, 35,422 genes (out of 59,225 total, ∼59.8%) had a GO term assigned. GO term enrichment was performed using topGO v 2.38.1 (Alexa and Rahnenfuhrer, 2019) and GO.db v3.10.0 (Carlson, 2019) with the parent–child algorithm (Grossmann et al., 2007) and Fisher’s exact test. A minimum node size of 5 was required, i.e., each GO term required a minimum of five genes mapping to it in order to be considered. p-values were adjusted for FDR and considered significant with an adjusted p < 0.05.
KEGG annotations are available for TO1000 (see3), but the National Center for Biotechnology Information (NCBI) GeneID was used rather than the gene name in the Ensembl annotations. To address this, a reciprocal Blast (blastp) was performed using Diamond with protein sequences from the Ensemble TO1000 genome and translated coding sequences from NCBI TO1000 genome (accession GCA_000695525.1). NCBI GeneIDs were then assigned to the TO1000 Ensemble genome annotations based on the reciprocal best blast hit. Enrichment of KEGG pathways was then performed in R using clusterProfiler v3.18.0 (Yu et al., 2012), and pathways were considered enriched with an FDR-adjusted p < 0.05. Scripts, GO terms, and NCBI GeneID assignments for each gene are available on GitHub (see text footnote 2).
Results
Kale Leaf Development
Developmental transitions and morphological changes were observed in two kale varieties: red winter kale with lobed leaf margins (B. oleraceae var. Red Winter) and Lacinato Nero Toscana kale (B. oleracea var. palmifolia) with crenulate leaf margins and the rapid cycling TO1000 DH3 (B. oleracea var. alboglabra from Warwick) for 22 days (Figures 1, 2). Leaf development encompasses three continuous and overlapping phases. During leaf initiation, the leaf primordium emerges from the flanks of the shoot apical meristem (SAM) at positions determined by specific phyllotactic patterns (Figures 2M,N). In the second phase, the leaf expands laterally, and primary morphogenesis events occur from specific meristematic regions at the leaf margin (blastozones) (Figures 2A–F). In the third phase of secondary morphogenesis, extensive cell expansion and histogenesis occurs (Figures 2G–L,Q,R). A week after planting, all morphotypes had germinated; at first, only the cotyledons were observed. Lobed-kale grew faster than non-lobed kale and TO1000. Using environmental scanning electron microscopy (ESEM), tiny true leaves with lobed margins and abundant trichomes were observed for lobed-kale at 5 days (Figure 2N), while non-lobed kale and TO1000 had not developed lobes or leaves at this point (Figures 2M,O). In 8 day-old lobed kale, the first immature leaves were seen emerging from between the cotyledons and were covered by trichomes (Figure 2B). Ten days after germination, lobed-kale seedlings had abundant trichomes and highly lobed leaf margins; leaf shape was elliptic to obovate and leaves had red venation and red petioles (Figure 2E). In comparison to the kale varieties, TO1000 seedlings had taller hypocotyls and erect stems. Leaf shape was oblanceolate, and leaf margins were sinuate (Figure 2D). Leaf margins in the Lacinato Nero Toscana kale were more sinuate in comparison to TO1000 at this stage (Figure 2F). Twelve day-old lobed-kale produce ectopic meristems in the leaf margins and more sporadically in the leaf blade of mature leaves; microscopic scales surrounded these ectopic meristems (Figure 2Q). Ectopic meristems located at the tip of each lobe had a vascular bundle inserted to them and were always subtended by a trichome. Nineteen-day-old lobed kale and TO1000 had four leaves, the fourth one small; while non-lobed kale has three leaves and the third one small. Lobed-kale leaves at 20–22 day-old develop more lobes and less trichomes with time. TO1000 leaves were thinner with non-lobed margins. In summary, we observed that kale margins were more lobed than TO1000 (Figure 2).
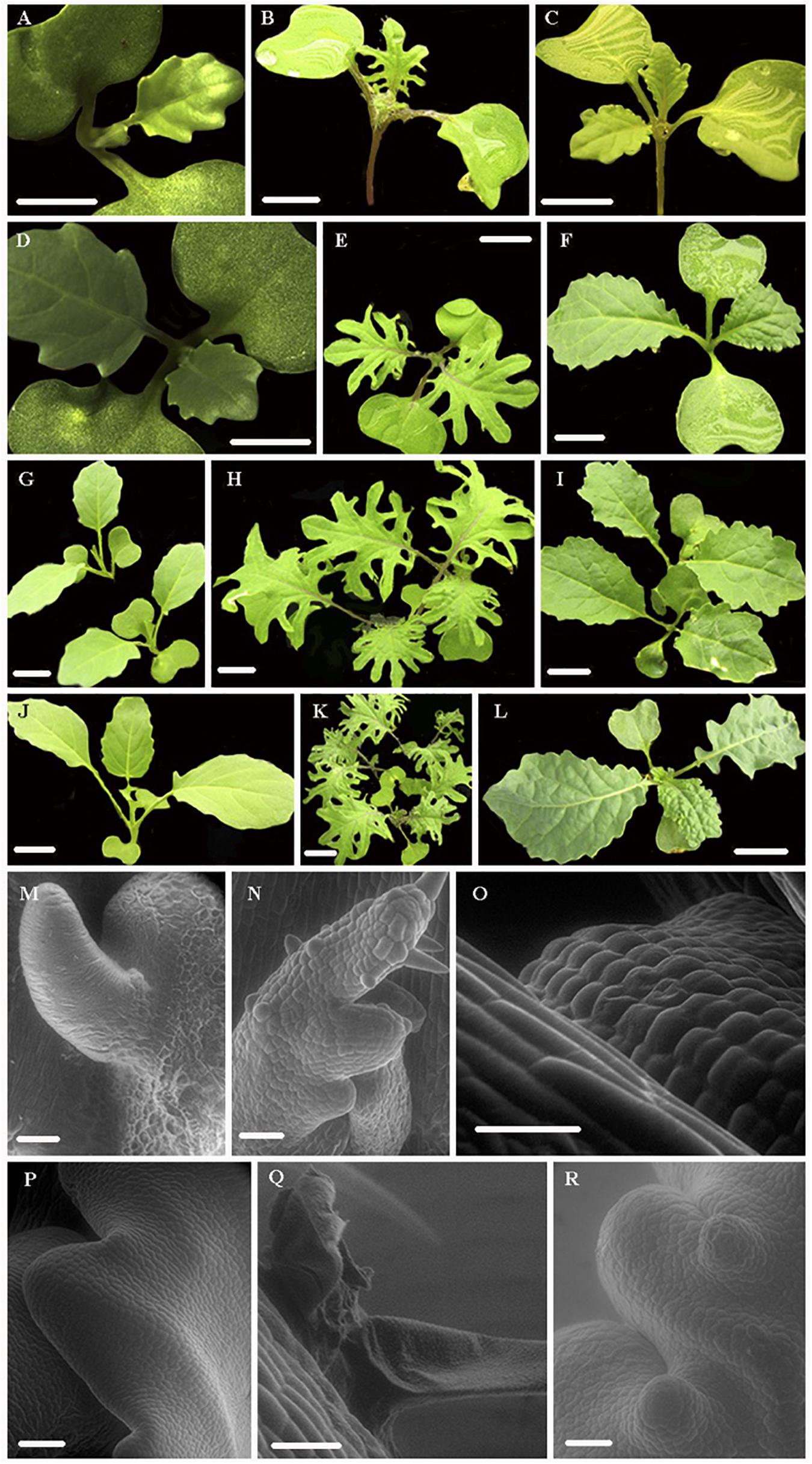
Figure 2. Leaf shape development in two Kale varieties, red winter kale (Brassica oleraceae var. acephala) and Lacinato Nero Toscana kale (B. oleracea var. palmifolia) and TO1000 (B. oleracea var. alboglabra). (A–L) Progressive development of the different kale varieties and the rapid cycling TO1000 control (stereoscope and regular pics). Scale bar = 1 cm. (M–R) Scanning electron microscopy (SEM) of apical meristem, primordial leaves, and details of lobes in different kale varieties and TO1000 control, scale bar = 5 μm. (A,D,G,J) Leaf development series for TO1000. (B,E,H,K) Leaf development series for kale with lobed leaf margins (red winter kale). (C,F,I,L) Leaf development series for Lacinato Nero Toscana kale with non-lobed leaf margins. (M,P) SEM of TO1000 leaf primordia and margin formation detail. (N) SEM of red winter kale leaf primordia. (O) SEM of Lacinato Nero Toscana kale leaf primordia. (Q) SEM of ectopic growth of leaf blade in mature leaves of red winter kale subtended by a trichome. (R) Margin formation detail in non-lobed kale (Lacinato Nero Toscana kale).
RNA-Seq and Differential Gene Expression
To identify genes that might be associated with domestication traits including leaf development in lobed kale, RNA-seq was used to assess transcriptional differences between lobed kale, cabbage, and TO1000. Three biological replicates each were sequenced; however, one of the cabbage replicates was not used for assembly because very few reads of low quality were obtained after sequencing. Between 9 and 25 million trimmed reads were obtained per sample, with ∼78–89% reads mapping uniquely to the TO1000 B. oleracea genome (Supplementary Table 1). After normalization with DESeq2, hierarchical clustering and principal component analysis showed that samples were grouped by morphotype, with greater variance between morphotypes than between replicates (Supplementary Figure 2).
Differentially expressed genes (DEGs) were called for all pair-wise comparisons between morphotypes (kale vs. TO1000, kale vs. cabbage, cabbage vs. TO1000), with the aim of identifying genes that were differentially expressed in kale compared to other morphotypes. Genes were considered differentially expressed if they had a twofold change in expression and a p < 0.05 after correction for multiple testing. In total, 8854 DEGs were identified between kale and TO1000 (4599 higher expressed in kale, 4255 lower expressed in kale), 8037 DEGs between kale and cabbage (4704 higher expressed in kale, 3333 lower expressed in kale), and 8189 DEGs between cabbage and TO1000 (3406 higher expressed in cabbage, 4783 lower expressed in cabbage) (Figure 3A). There was a total of 3958 DEGs shared between the kale–TO1000 and kale–cabbage comparisons, 2960 found only in the kale comparisons and 998 found in all three comparisons (Figure 3B).
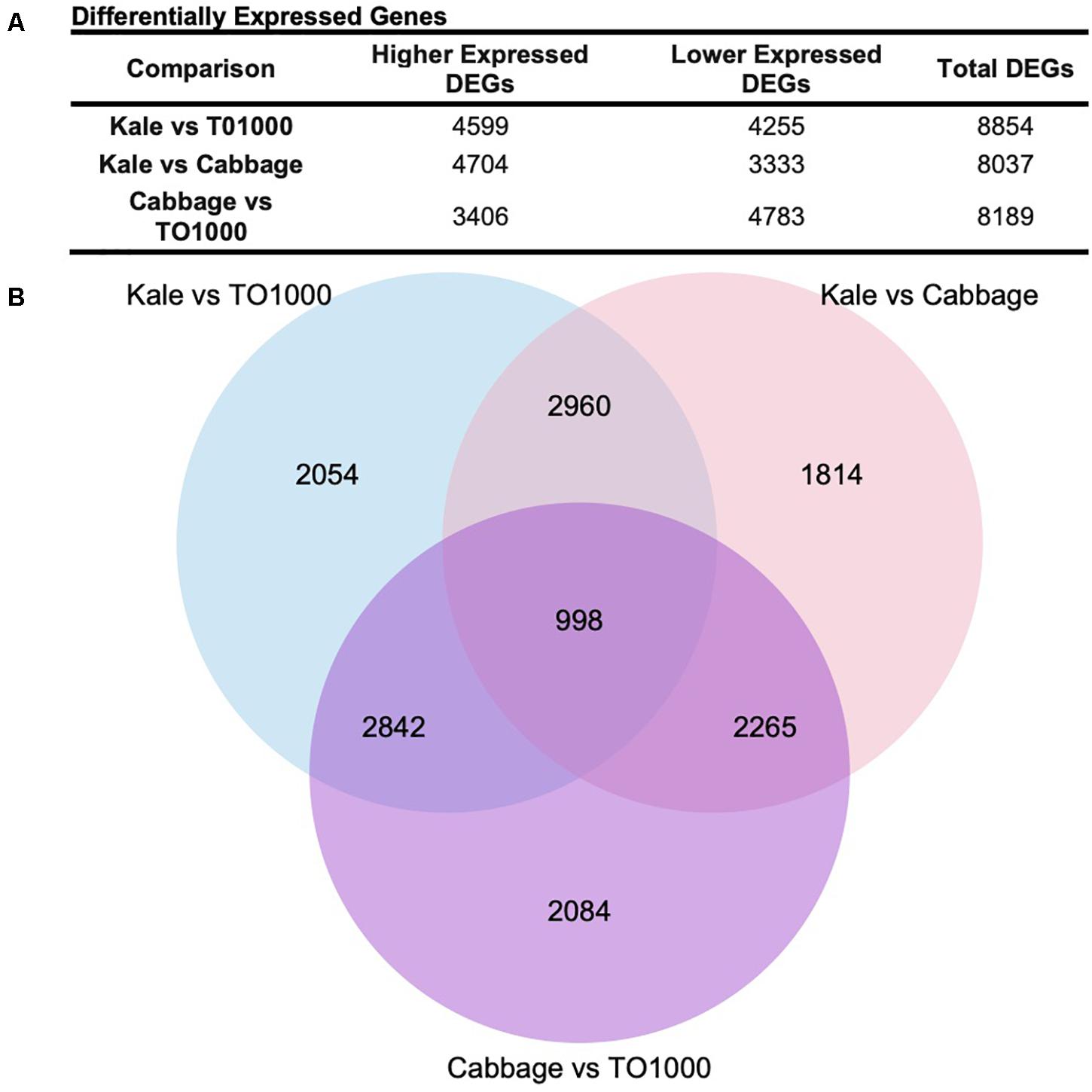
Figure 3. Number of genes differentially expressed in this study after comparing three different Brassica oleracea morphotypes (cabbage, kale, and TO1000). (A) Number of genes differentially expressed. (B) Venn Diagram showing shared genes after doing all possible comparisons between three different B. oleracea morphotypes.
Polyploidy and Its Contribution to the Kale Phenotype
It has been previously shown that the whole-genome triplication (WGT) in B. oleracea has contributed to its phenotypic diversity (Lukens et al., 2004; Paterson, 2005). As a result, the B. oleracea genome is composed of three subgenomes. Previous studies have used conservation of gene order (synteny) with A. thaliana to assign ∼41% of annotated genes to one of the three subgenomes, which have been named the least-fractionated (LF) subgenome, more-fractionated 1 (MF1) subgenome, and more-fractionated 2 (MF2) subgenome (Parkin et al., 2014). Using these gene lists, we explored how this ancient WGT contributed to differential gene expression between morphotypes. Syntenic genes, which includes genes assigned to all three subgenomes, were significantly overrepresented, while non-syntenic genes were underrepresented among DEGs in the kale–cabbage and cabbage–TO1000 comparisons but not the kale–TO1000 comparisons (Figure 4A). We further tested the enrichment of individual subgenomes among DEGs (Figures 4C,D). The LF subgenome was underrepresented in kale–TO1000 comparison (odds ratio = 0.9162346. FDR-corrected p-value = 4.116097e-03) and overrepresented in the cabbage–TO1000 comparison (odds ratio = 1.0816227. FDR-corrected p-value = 9.948104e-03). The MF1 subgenome was significantly overrepresented for DEGs in all three comparisons (kale–TO1000: odds ratio = 1.0912796, FDR-corrected p-value < 1.103789e-02; kale–cabbage: odds ratio = 1.2091591, FDR-corrected p-value < 5.909977e-08; cabbage–TO1000: odds ratio = 1.1776233, FDR-corrected p-value = 2.913654e-06). The MF2 subgenome was overrepresented in the cabbage–TO1000 comparisons (odds ratio = 1.1118552. FDR-corrected p-value = 4.931485e-03).
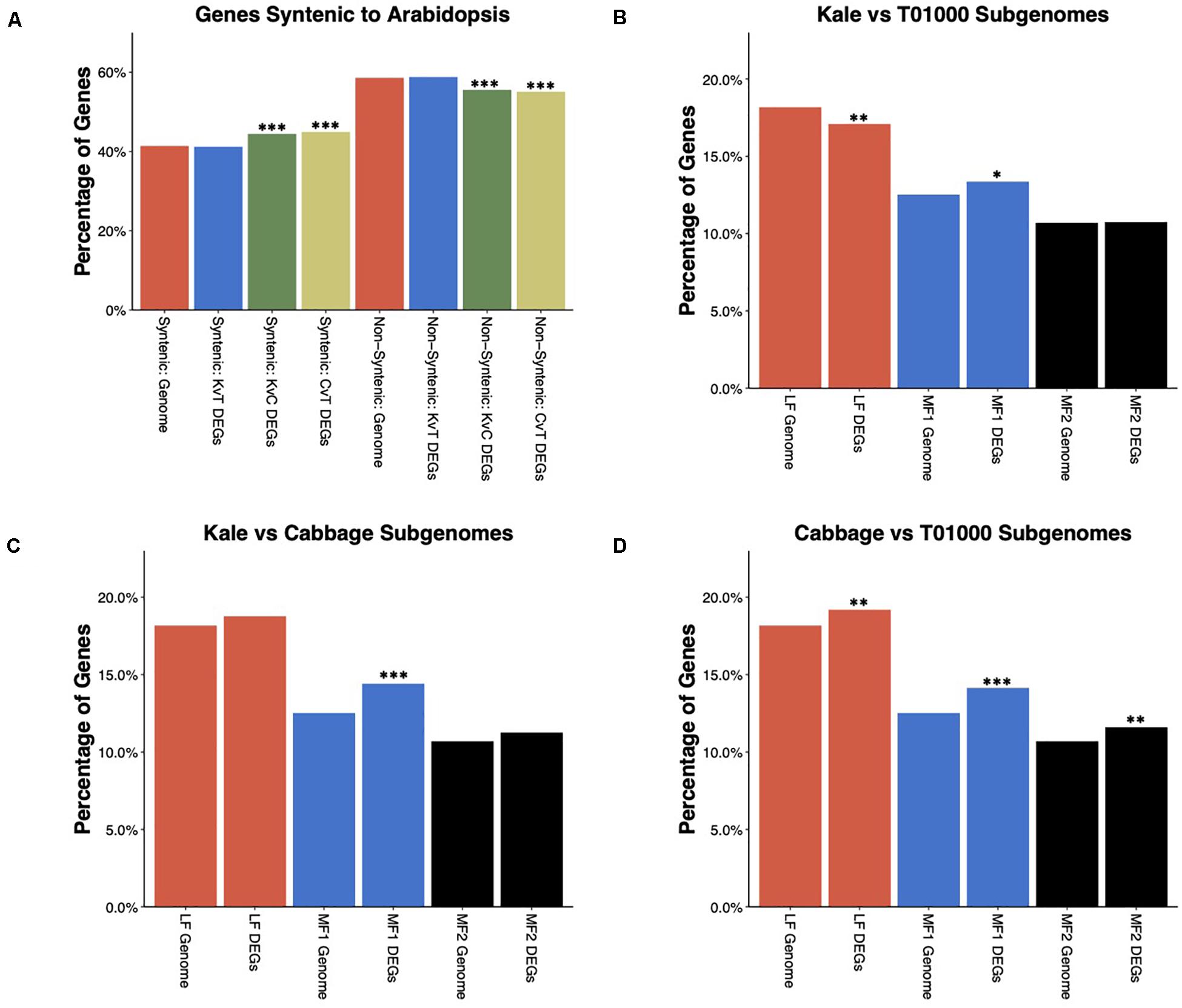
Figure 4. (A) Enrichment/depletion of differentially expressed genes (DEGs) amongst B. oleracea genes syntenic or non-syntenic to Arabidopsis thaliana. DEGs in the Kale vs TO1000 comparison show no enrichment or depletion, while the DEGs in the Kale vs Cabbage and Cabbage vs TO1000 comparisons are enriched in syntenic genes and depleted in non-syntenic genes. (B–D) Enrichment/depletion of DEGs for each of the three subgenomes of B. oleracea. (B) Kale vs TO1000 DEGs are depleted for LF subgenome genes, enriched for MF1 subgenome genes, and show no difference for MF2 sub genome genes. (C) Kale vs Cabbage DEGs are enriched for MF1 subgenome genes and show no difference for LF or MF2 subgenome genes. (D) Cabbage vs TO1000 DEGs show enrichment in genes from all three subgenomes. ∗significant at FDR corrected p-value < 0.05, ∗∗significant at FDR corrected p-value < 0.01, ∗∗∗significant at FDR corrected p-value < 0.001.
Gene Ontology and KEGG Pathway Enrichment Analysis
DEGs for each pair-wise comparison and the list of DEGs shared between the two kale comparisons (3958 genes) were separated into lists of higher- and lower-expressed genes and then tested for enrichment of Gene Ontology (GO) terms (Supplementary Tables 5–12) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Supplementary Tables 13–20). We focus here specifically on those kale comparisons to better understand the kale domestication syndrome, in particular GO terms in the biological process (BP) category, as this includes developmental terms. GO terms and KEGG pathways related to the cabbage vs. TO1000 comparison are provided as supplement (Supplementary Figure 2).
Kale vs. TO1000
There were 91 (64 BP) and 17 (1 BP) GO terms enriched in the higher (Supplementary Table 5) and lower (Supplementary Table 6) expressed DEGs, respectively. Higher expressed genes were characterized by a number of metabolic processes: “lipid metabolism,” “nitrile metabolism,” “sulfur compound metabolism,” “benzene-containing compound metabolism,” “organic hydroxy compound biosynthesis,” and “carbohydrate derivative catabolism.” There were also multiple GO terms related to photosynthesis and plastid organization and environmental responses to both biotic and abiotic stresses. The only developmental terms enriched were related to senescence (Supplementary Figure 3A). Lower expressed genes showed little enrichment, except for genes involved in “cellular nitrogen compound metabolism” (Supplementary Figure 3B). Six KEGG pathways were enriched in higher expressed genes (Supplementary Table 13) and largely correspond to metabolic pathways identified in the GO term analysis. These include processes related to “photosynthesis-antenna proteins,” “fatty acid elongation,” “flavonoid biosynthesis,” “steroid biosynthesis,” “glyoxylate and dicarboxylate metabolism,” “other glycan degradation,” and “cyanoamino acid metabolism.” Lower expressed genes were enriched in “spliceosome” and “circadian rhythm” pathways (Supplementary Table 14).
Kale vs. Cabbage
Higher expressed genes had 21 (15 BP) enriched GO terms (Supplementary Table 7), while lower expressed genes (Supplementary Table 8) had 26 (11 BP). Similar to the kale vs. TO1000 comparison, there was enrichment for processes involved in “fatty acid derivative metabolism,” “nitrile metabolism,” and “photosynthesis” (Supplementary Figure 3C). There was also enrichment for environmental responses, specifically responses to herbivores. Additionally, there was enrichment for processes in “carbon fixation,” “benzoate metabolism,” “endosome organization,” and the “negative regulation of ion transport.” Lower expressed genes (Supplementary Figure 3D) include “glycosinolate metabolism,” “response to chitin,” and “gene expression.” “Sulfur compound metabolism” was enriched in both comparisons but was enriched among lower-expressed genes in the kale–cabbage comparison while being enriched in higher-expressed genes in the kale–TO1000 comparison. These are again reflected in KEGG pathways where higher expressed genes were enriched for “other glycan degradation” KEGG pathways (Supplementary Table 15). Lower expressed genes were enriched for “glucosinolate biosynthesis” and “glutathione metabolism” KEGG pathways (Supplementary Table 16).
Shared Genes
Higher expressed genes were enriched for 32 (22 BP) GO terms (Supplementary Table 11 and Supplementary Figure 3G). These again include multiple metabolic terms, especially related to “fatty acid derivative metabolism,” “nitrile metabolism,” “carbohydrate derivative catabolism,” and “benzoate metabolism.” Also similar to the individual comparisons, “photosynthesis” and terms related to abiotic and biotic stresses, especially herbivores, were enriched. Unique to this subgroup was an enrichment for protein phosphorylation terms. Only four GO terms were enriched in the lower expressed genes, and these were all in the cellular component GO term class (Supplementary Table 12). Higher expressed DEGs were enriched for “other glycan degradation” and “Sesquiterpenoid and triterpenoid biosynthesis” KEGG pathways, the former being also enriched in both of the individual comparisons. No KEGG pathways were enriched in the shared lower expressed DEGs (Supplementary Tables 19, 20).
Candidate Genes Potentially Involved in the Domestication of Kale
A set of candidate genes were identified based on their expression pattern (Figure 5) and functions related to the suite of kale domestication syndrome characteristics. We further identified transcription factor (TF) families and found a total of 141 TFs in a total of 3010 differentially expressed genes (Figure 3B). Some of the most abundant included bHLH, ERF, and MYB TFs (Supplementary Table 21).
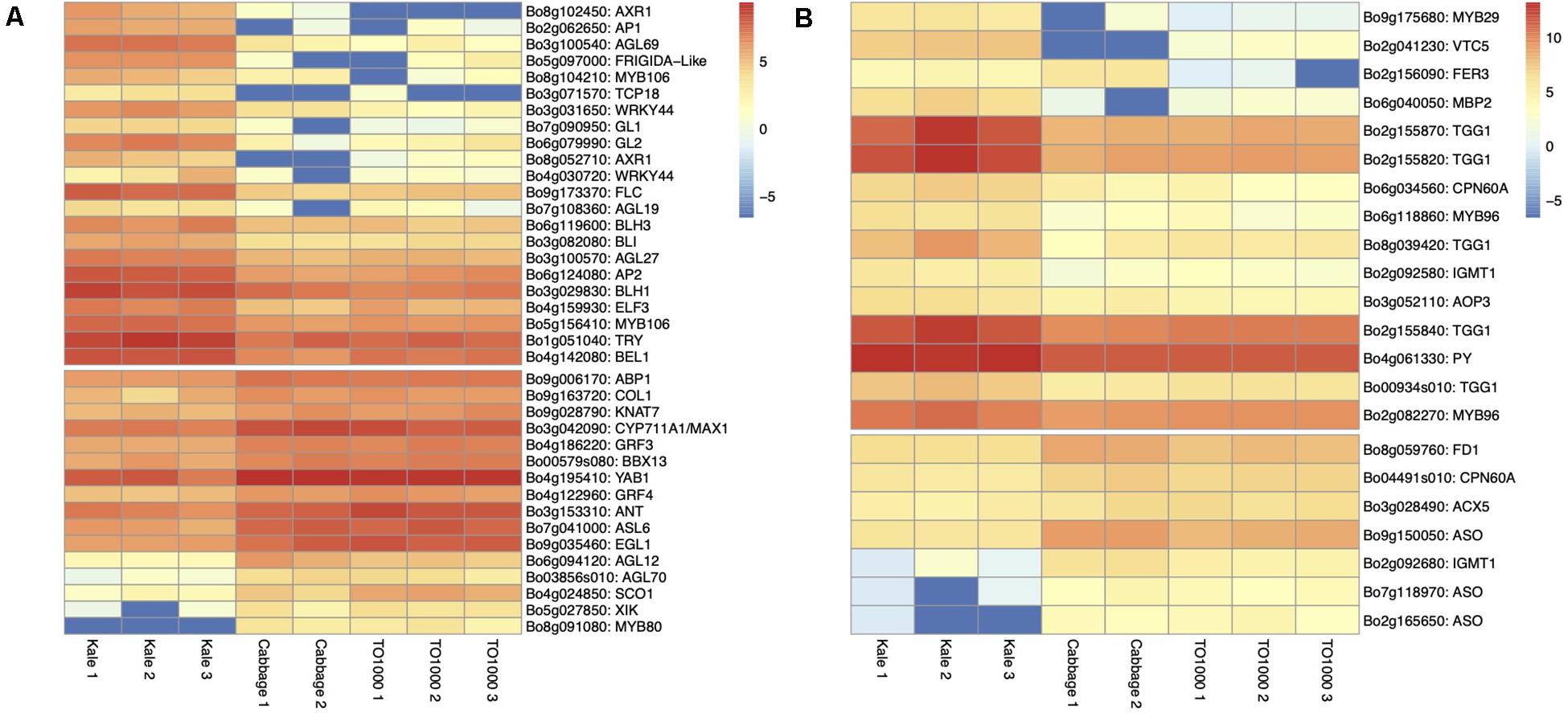
Figure 5. Heatmap of differentially expressed genes for lobed kale (red winter kale). Up- and downregulated genes involved in (A) plant and leaf development and flowering (B) plant defense responses and taste and nutrition.
Apical Dominance and Plant Architecture. Several genes known to be involved in axillary shoot growth inhibition were differentially expressed in kale tissues and upregulated; these include an auxin-resistance AXR1 (Bo8g102450, Bo8g052710) (Stirnberg et al., 1999), TCP18 (Teosinte Branched 1-Like 1) (Bo3g071570), two copies of REVOLUTA (Bo9g130730, Bo7g021280) (Poza-Carrión et al., 2007), and the beta-carotene isomerase D27 chloroplastic (protein DWARF-27 homolog) (Bo9g035190) (Waters et al., 2012). The more axillary branches gene MAX1 (Bo3g042090) (Bennett et al., 2006) was also differentially expressed and downregulated in kale (Table 1 and Figure 5A).
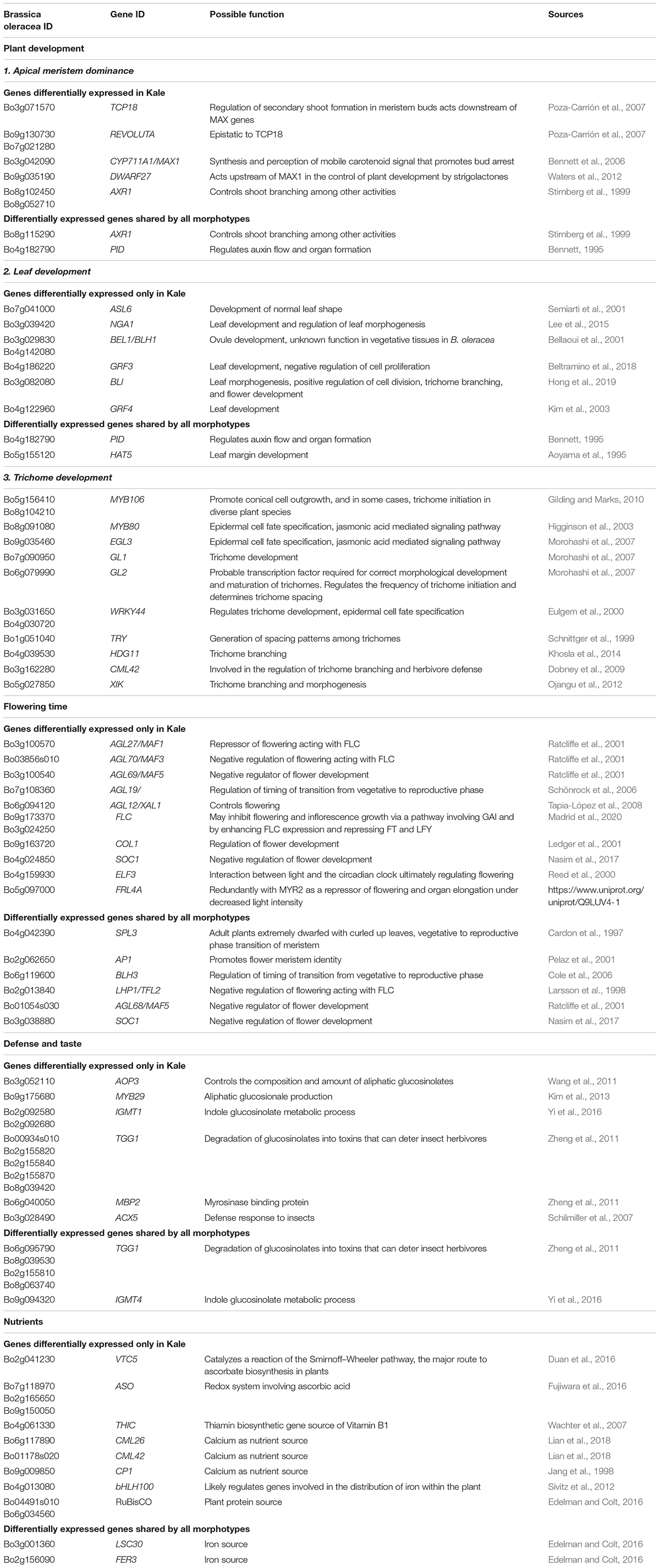
Table 1. Candidate genes that are differentially expressed in lobed kale (red winter kale) or the three morphotypes.
Leaf Development. Differentially expressed genes in kale include two upregulated copies of AXR1 (Bo8g052710; Bo8g102450). The lateral organ boundaries (LOB) domain-containing protein ASYMMETRIC LEAVES 2-like protein 6 (ASL6) (Bo7g041000) was downregulated (Semiarti et al., 2001). These genes could have a major role in the highly lobed margin kale phenotype (Figure 5A). Genes differentially expressed in all three pair-wise comparisons that were potentially involved in the kale domestication syndrome (Figure 3) include PID (Bo4g182790), which regulates auxin flower and organ formation (Bennett, 1995), and HAT5 (Bo5g155120), which is involved in leaf formation and has been characterized as involved in leaf margin development (Aoyama et al., 1995).
Identified transcription factors included the AP2-like ethylene-responsive ANT (Bo3g153310) (Figure 5A). This gene is a positive regulator of cell proliferation, expansion, and regulation of ectopic meristems (Shigyo et al., 2006), similar to what was observed in the kale phenotypes (Figure 2Q). The growth-regulating factors 3 and 4 (GRF3 and GRF4; Bo4g186220 and Bo4g122960) involved in the regulation of cell expansion in leaf and cell proliferation and the basic helix–loop–helix protein 64 AtbHLH64 (Bo3g094060), an atypical bHLH transcription factor, were also downregulated in kale (Figure 5A and Supplementary Table 21).
A significant number of trichomes in kale leaves were observed during development (Figures 2N,H). More than 10 genes related to trichome formation (Table 1 and Figure 5A) were differentially expressed in kale; some of those included two upregulated copies of an Myb-related proteins MYB106 (Bo5g156410 and Bo8g104210). These genes can act as both positive and negative regulators of cellular outgrowth and promote trichome morphogenesis and expansion (Gilding and Marks, 2010). A Homeobox-leucine zipper protein GLABRA 2-like GL2 (Bo6g079990) was upregulated in kale. This gene is required for correct morphological development and maturation of trichomes, regulates the frequency of trichome initiation, and determines trichome spacing in Arabidopsis (Morohashi et al., 2007). Two expressed copies of the WRKY transcription factor 44 (Bo3g031650 and Bo4g030720), and the transcription factor TRIPTYCHON TRY (Bo1g051040) involved in trichome branching, were also differentially expressed in kale (Table 1 and Figure 5A).
Flowering Time. Kale phenotypes display a significant delaying in flowering time in comparison to other B. oleracea morphotypes, while TO1000 has been bred to be rapid cycling. We hypothesized that this maintenance in a vegetative stage was an important character during domestication due mainly to the harvest of fresh leaves for consumption. Our mature kale phenotypes did not display flowering during the course of our experiments (24 days) (Figure 1). A series of Agamous-like MADS-box proteins were exclusively expressed in kale (Figure 3B) including two downregulated genes AGL12 (Bo6g094120) and AGL70 (Bo03856s010) and an upregulated AGL27 (Bo3g100570). MADS-box proteins such a SOC1 (Bo4g024850) was downregulated, and two copies of FLOWERING LOCUS C (Bo9g173370, Bo3g024250) were upregulated (Table 1 and Figure 5A), both involved in the negative regulation of flowering (Supplementary Table 21). Other genes differentially expressed in all three pair-wise comparisons that were potentially involved in the negative regulation of flowering (Figure 3) included SUPPRESSOR OF OVEREXPRESSION OF CO 1 SCO1 (Bo4g024850, Bo3g038880), SPL3 (Bo4g042390), AP1 (Bo2g062650), BLH3 (Bo6g119600), and AGL68/MAF5 (Bo3g100540) (Table 1 and Figure 5A).
Plant Defense. Differentially expressed genes involved in plant defense exclusively expressed in lobed kale leaves (Figure 3B) include two copies of the upregulated Myb-related protein MYB96 (Bo2g082270) involved in the activation of cuticular wax biosynthesis that could potentially deter herbivores. Five upregulated copies of a glucosinolate synthesis and regulation enzyme Myrosinase TGG1 (Bo00934s010, Bo2g155820, Bo2g155840, Bo2g155870, Bo8g039420) are involved in the degradation of glucosinolates to produce toxic degradation products that can deter insect herbivores (Zheng et al., 2011). Lastly, two copies of the indole glucosinolate O-methyltransferase, one was upregulated IGMT1 (Bo2g092580) and the second one downregulated IGMT1 (Bo2g092680) (Table 1 and Figure 5B), and MYB29 (Bo9g175680) were all involved in plant defense against fungi (Supplementary Table 21). Refer to Table 1 and Figure 5B for additional candidate genes potentially involved in plant taste and defense.
Nutritional Value of Leaves. Kale is known to have great nutritional value including nutrients like Vitamin C, calcium, and iron (Edelman and Colt, 2016). Most notable were three copies of the L-ascorbate oxidase (AAO) (Ascorbase) (EC 1.10.3.3) (Bo2g165650, Bo9g150050, Bo7g118970) involved in Vitamin C synthesis, all three of which were downregulated in kale (Table 1 and Figure 5B) and the enzyme ferritin-1 LSC30 (Bo3g001360) proposed as a source of iron. Refer to Table 1 and Figure 5B for other genes potentially involved in nutritional value of kale leaves.
Discussion
The vegetative domestication syndrome of kale is characterized by apical dominance, leaf morphology, maintenance of a vegetative stage for high leaf production, taste/defense, and nutritional value. Differentially expressed genes (DEGs) were identified in all pair-wise comparisons. In the cabbage comparisons, DEG were enriched among syntenic genes while being depleted in non-syntenic genes (Figure 4). This aligns with recent findings that genes derived from the ancient Brassica polyploidy are also more likely involved in domestication in B. rapa (Qi et al., 2019). However, in the kale–TO1000 comparison, no such enrichment was found. As the subgenomes of B. oleracea have been previously reconstructed, we further explored the relative contribution of each. The LF subgenome contains the most genes and, unsurprisingly, also had the most DEGs. The LF subgenome was overrepresented in the cabbage–TO1000 comparison; however, it was underrepresented in the kale–TO1000 comparison. In contrast, the MF1 subgenome was consistently overrepresented. The more-fractionated 2 subgenome was overrepresented in the cabbage–TO1000 comparison. Given the limited number of morphotypes in this study, we cannot conclude a general pattern regarding the relative importance of syntenic vs. non-syntenic genes and each subgenome; however, the overrepresentation of the MF1 subgenome in each comparison should be explored further.
We focused our search on DEGs that were among comparisons with kale, reasoning that these were more likely to underly the unique characteristics of kale. We tested for enrichment in GO terms and KEGG pathways to gain a global view of the differences. Many more terms and pathways were enriched among higher expressed genes, despite several thousand DEGs being identified in both groups. This may suggest more coordination in the regulation of kale higher-expressed genes or simply better annotation of genes in this list. Enriched terms and pathways in all categories were typically involved in metabolism or defense. Changes in these functions would affect the taste, texture, and nutrition of leaves and thus likely have been affected by human selection. There was a relative absence of developmentally related GO terms, with the exception of senescence-related terms in the kale–TO1000 comparison. This overall lack of enrichment for developmentally related GO terms could result from either insufficient annotation of developmental genes or due to more subtle expression differences that would require tissue-specific or single-cell approaches. Alternatively, the large developmental changes observed could be caused by differential expression of only a small number of key genes. To address this, we examined sets of genes, whose function, based on homology and previous work, suggests an involvement in the developmental traits examined in this study. This analysis found many differentially expressed candidate genes shared among comparisons with kale, supporting their potential role in the kale domestication syndrome.
We identified potential candidate genes for the morphological and chemical differences among Brassica morphotypes. First, we found differentially expressed genes that set kale apart in terms of its morphology. Apical dominance has been proposed as a main character involved in crop domestication (Doebley et al., 1997). Suppression of axillary branches aiming to concentrate resources in the main stem is a salient phenotypic character in most Brassica crops. Apical dominance during domestication in kale can be explained by the indirect theory of apical dominance, as auxin-induced stem growth inhibits bud outgrowth by diverting sugars away from buds since growing stems are a strong sink for sugars (Kebrom, 2017). Here, genes involved in apical dominance and branching suppression were differentially expressed (Table 1 and Figure 5).
Kale displays morphological diversity in leaves, including leaf shape, size and margins, trichome development, colors among others, which is likely the result of genetic variation selected by plant breeders. ESEM showed that many of the characteristic leaf traits of kale are established early in leaf development. We collected tissues during kale leaf initiation and primary morphogenesis (the upper portion of the stem including immature leaves and apical and lateral meristems) to capture candidate genes related to the establishment of the basic leaf structure such as lamina initiation, specification of lamina domains, and the formation of marginal structures such as lobes or serrations.
Three types of differentially expressed genes related to the kale leaf morphogenesis phenotype were found in our samples (Table 1 and Figures 3, 5): (1) involved in margin development; (2) involved in cell proliferation, expansion, and the development of ectopic meristems; and (3) involved in trichome development. The LOB domain-containing protein 4 (asymmetric leaves 2-like protein 6) ASL6 (Bo7g041000) was identified as a potential important gene responsible of the lobed kale leaf phenotype (Semiarti et al., 2001; Figures 1, 2E,H,K). Loss-of-function mutations in the AS2 homolog in Arabidopsis have ectopic expression of class 1 KNOX genes in the leaves resulting in asymmetric leaf serration, with generation of leaflet-like structures from petioles and malformed veins (Matsumura et al., 2009). ASL6 was differentially expressed and upregulated in kale. A second set of candidates involved two copies of the BEL1-like homeodomain protein 1 BEL1/BHL1 (Bo3g029830, Bo4g142080) that were found to be overexpressed in kale. We have also found several downregulated growth-regulating factors (GRF3 and GRF4) that have been shown to interact with KNOX genes (Kuijt et al., 2014). A third gene involved in cell proliferation, expansion, and the development of ectopic meristems included the NGATHA NGA1 (Bo3g039420) involved in cell proliferation (Lee et al., 2015); however, it has been reported as downregulated for cell overproliferation and was upregulated in kale (Table 1).
In some species with simple leaves, KNOXI overexpression can lead to variable phenotypes, which include knot-like structures on the leaves, curled or lobed leaves, and ectopic meristems on leaves (Tsiantis and Hay, 2003). Our differential expression analysis did not show KNOXI overexpression; however, the differential expression of BELL genes, another class of TALE HD proteins found in plants, was upregulated (Bo3g029830). Yeast two-hybrid studies have indicated that KNOX proteins interact with BEL1-like (BELL) proteins. The messenger RNA (mRNA) expression patterns of KNOX and BELL proteins overlap in meristems, suggesting that they may potentially interact in vivo (Smith et al., 2002). Analogous BLH–KNOX interactions have also been reported in other plants, such as potato (Solanum tuberosum) and barley (Hordeum vulgare), indicating that these interactions are evolutionarily conserved and that the interaction is probably required for their biological function (Müller et al., 2001; Chen et al., 2003). A matter of debate is whether KNOX and BLH can exert some of their functions independently of each other or whether the formation of KNOX/BLH heterodimers is mandatory for TALEs to work. We did not find differential expression of KNOX genes in this analysis perhaps for several reasons: (1) KNOX do not participate in lobe formation in B. oleracea; (2) we did not target the correct point in development to capture KNOX expression in relation to leaf lobation; (3) a change in BELL expression is sufficient to affect KNOX and BELL interactions and alter leaf morphology without a corresponding change in KNOX expression. Moreover, some species, such as legumes, with dissected leaves do not express KNOX genes; on the contrary KNOX expression has been observed in leaf primordia of species with unlobed leaves, such as Lepidium oleraceum (Piazza et al., 2010; Vlad et al., 2014).
Epidermal cells are stimulated to differentiate into trichomes when a regulatory complex including transcription factors are triggered (Yang and Ye, 2013). ESEM results showed hairiness, and individual trichomes are seen in kale leaf blades (Figures 2N,Q). Differentially expressed genes involved in trichome formation included two MYB transcription factors (Higginson et al., 2003; Gilding and Marks, 2010; Pattanaik et al., 2014), the enhancer of GL3 EGL3 (Bo9g035460), the Homeobox-leucine zipper proteins GL1 (Bo7g090950) and GL2 (Bo6g079990) (Morohashi et al., 2007), the transcription factor TRY (Bo1g051040) (Schnittger et al., 1999), and homeodomain GLABROUS 11, HDG11 (Bo4g039530) (Khosla et a., 2014). These five genes either interact or act redundantly to promote trichome differentiation and were upregulated in kale (except HDG11) (Table 1 and Figure 5A). Trichome regulatory genes have been studied in Brassica villosa, a wild C genome relative B. oleracea that is densely covered by trichomes. Nayidu et al. (2014) found that TRY was upregulated in trichomes of B. villosa in contrast to Arabidospis and other Brassica species where this gene has been proposed as a negative regulator. Here, we found that TRY was more highly upregulated in leaves and meristems of kale in comparison to ELG3 (Table 1), GL1 and GL2 (Figure 5A).
Another set of candidate genes for the kale domestication syndrome we looked for are floral transition genes. Floral transition is a major development switch controlled by regulatory pathways, integrating endogenous and environmental cues (Koornneef et al., 2004). We suggest that during kale domestication, delaying of flowering time was necessary to maintain leaf production as the main consumable. Four main regulatory pathways directly or indirectly involved in flowering time have been described in A. thaliana: photoperiodic, autonomous, vernalization, and gibberellin (Okazaki et al., 2007). The central integrator of flowering signals is the flowering locus C (FLC) that encodes a transcription factor of the MADS box family. While A. thaliana contains only one FLC gene, five copies have been described for B. oleracea (Okazaki et al., 2007). We found two copies of this gene being differentially expressed in kale. One was upregulated (Bo9g173370) and, together with Agamous-like MADS-box protein AGL27/MAF1 (Bo3g100570), has been proposed as floral repressors (Ratcliffe et al., 2001; Table 1 and Figure 5A). A second copy of the FLC (Bo3g024250) gene was found to be shared by the three morphotypes kale, cabbage, and TO1000 (Table 1).
Pathways to domestication in kale may involve selection of plants with alterations of phytochemical concentrations (Hahn et al., 2016). Cole crops herbivore defense involves secondary metabolites (e.g., glucosinolates) and physical plant traits (e.g., waxes) that influence the feeding patterns of arthropod herbivores (Carmona et al., 2011). Glucosinolates are the main class of secondary metabolites found in cruciferous crops. They generally appear to be constitutively synthesized at rather low concentrations, but their synthesis is induced by herbivores through jasmonate and other signaling pathways. Cole crops have a two-part defense system involving glucosinolate compounds and a myrosinase protein complex. A series of copies of the Myrosinase enzyme TGG1 were differentially expressed and upregulated in kale (Bo00934s010, Bo2g155820, Bo2g155840, Bo2g155870, Bo8g039420) (Zheng et al., 2011). This enzyme breaks down glucosinolates into toxins (isothiocyanates, thiocynates, nitriles, and epithionitriles) upon leaf tissue damage. Therefore, glucosinolates may act as a potent feeding deterrent for generalist insect species, as their toxicity causes developmental and fitness damage (Santolamazza-Carbone et al., 2014; Yi et al., 2015). Gene Ontology terms related to glucosinolates and response to herbivores were also found to be enriched in our analysis specifically in the comparison between kale vs. cabbage terms. We proposed that persistence of pungency and bitterness would have reduced competition from mammals as well as loss to pests, during cultivation and storage.
Several genes were identified to be related to the nutritional content in kale, for example, precursors and enzymes related to vitamin C; GDP-L-galactose phosphorylase 2 VTC5 was upregulated in kale (Bo2g041230) (Duan et al., 2016), and several copies of L-ascorbate oxidase (ASO) (Ascorbase) (EC 1.10.3.3) and AAO (Bo2g165650, Bo9g150050, Bo7g118970) (Fujiwara et al., 2016) were downregulated. In addition, DEG was the precursor of the vitamin B1 phosphomethylpyrimidine synthase chloroplastic (EC 4.1.99.17) THIC (Bo4g061330) (Wachter et al., 2007). The RuBisCO large subunit-binding protein subunit alpha chloroplastic (60 kDa chaperonin subunit alpha (Bo04491s010) and large subunit-binding protein subunit beta chloroplastic (60 kDa chaperonin subunit beta) (Bo6g034560) were upregulated in kale (Edelman and Colt, 2016). Differentially expressed genes found in the three morphotypes included Ferredoxin-1 chloroplastic (AtFd1) LSC30 (Bo8g059760) (Table 1 and Figure 5B).
Conclusion
This study provided leaf developmental analysis and transcriptomics for B. oleracea morphotypes kale, cabbage, and TO1000, three economically important Cole crops with distinct plant morphologies and domestication syndromes. RNA-seq experiments allowed the discovery of novel candidate genes proposed to be involved in domestication syndromes. We identified DEG in B. oleracea that are different for each vegetative morphotypes cabbage and kale, compared to the “rapid-cycling” morphotype TO1000, and estimated the contribution of morphotype-specific gene expression patterns that set kale apart. We propose that during kale domestication, farmers could have been selecting for different vernalization times, different taste, resistance to herbivores, or flower at different times during development. The DEG we identified provides candidates for further testing the molecular basis for all of these traits.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TA co-developed questions and framework, obtained funding, made collections, performed microscopy analysis, growth chamber experiments, and lab work including library sequencing preparation, performed analyses, and wrote manuscript. CN performed analyses, performed lab work, and wrote and edited manuscript. PM co-developed questions and framework, provided funding, and mentored students. JP co-developed questions and framework, provided funding, and mentored students. All authors contributed to the article and approved the submitted version.
Funding
The authors acknowledge the following research grants and granting institutions: National Science Foundation (DEB 1146603 and DEB 1209137), Ministerio de Ciencia y Tecnologia de Colombia (64777), and The Newton Fund, UK-Colombia mobility grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank G. Conant, J. Birchler, M. Liscum, C. Henriquez, M. Tang, and A. Reyes and the reviewers for valuable comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.637115/full#supplementary-material
Footnotes
- ^ http://plntfdb.bio.uni-potsdam.de
- ^ https://github.com/niederhuth/The-molecular-basis-of-Kale-domestication
- ^ https://www.genome.jp/kegg/catalog/org_list.html
References
Alexa, A., and Rahnenfuhrer, J. (2019). topGO: Enrichment Analysis for Gene Ontology. R Package. Available online at: https://www.bioconductor.org/packages/release/bioc/html/topGO.html
Allaby, R. G. (2014). “Domestication syndrome in plants,” in Encyclopedia of Global Archaeology, ed. C. Smith (New York, NY: Springer), 2182–2184.
Allender, C. J., Allainguillaume, J., Lynn, J., and King, G. J. (2007). Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n = 9) wild relatives. Theoret. Appl. Genet. 114, 609–618.
Andrews, S. (2010). FastQC a Quality Control Tool for High Throughput Sequence Data version 0.11.8. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Anthony, R. G., James, P. E., and Jordan, B. R. (1995). The cDNA sequence of a cauliflower apetala-1/squamosa homolog. Plant Physiol. 108, 441–442. doi: 10.1104/pp.108.1.441
Aoyama, T., Dong, C. H., Wu, Y., Carabelli, M., Sessa, G., Ruberti, I., et al. (1995). Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 7, 1773–1785. doi: 10.2307/3870186
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology: the gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bellaoui, M., Pidkowich, M. S., Samach, A., Kushalappa, K., Kohalmi, S. E., Modrusan, Z., et al. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470. doi: 10.1105/tpc.13.11.2455
Beltramino, M., Ercoli, M. F., Debernardi, J. M., Goldy, C., Rojas, A. M., Nota, F., et al. (2018). Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 8, 1–13.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bennett, G. J. (1995). United States Patent and Trademark Office. U.S. Patent No. 5,384,526. Washington, DC: U.S. Patent and Trademark Office.
Bennett, T., Sieberer, T., Willett, B., Booker, J., Luschnig, C., and Leyser, O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16, 553–563. doi: 10.1016/j.cub.2006.01.058
Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., An, L., et al. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835–1839. doi: 10.1126/science.1166168
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12:59. doi: 10.1038/nmeth.3176
Cardon, G. H., Höhmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 12, 367–377. doi: 10.1046/j.1365-313x.1997.12020367.x
Carlson, M. (2019). GO.db: A Set of Annotation Maps Describing the Entire Gene Ontology. R Package. Available online at: https://www.bioconductor.org/packages/release/data/annotation/html/GO.db.html
Carmona, D., Lajeunesse, M. J., and Johnson, M. T. (2011). Plant traits that predict resistance to herbivores. Funct. Ecol. 25, 358–367. doi: 10.1111/j.1365-2435.2010.01794.x
Carr, S. M., and Irish, V. F. (1997). Floral homeotic gene expression defines developmental arrest stages in Brassica oleracea L. vars. botrytis and italica. Planta 201, 179–188. doi: 10.1007/bf01007702
Chen, H. (2018). VennDiagram: Generate High-Resolution Venn and Euler Plots. R package version 1.6.20. Available online at: https://CRAN.R-project.org/package=VennDiagram
Chen, H., Rosin, F. M., Prat, S., and Hannapel, D. J. (2003). Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 132, 1391–1404. doi: 10.1104/pp.103.022434
Cole, M., Nolte, C., and And Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34, 1281–1292. doi: 10.1093/nar/gkl016
Denham, T., Barton, H., Castillo, C., Crowther, A., Dotte-Sarout, E., Florin, S. A., et al. (2020). The domestication syndrome in vegetatively propagated field crops. Ann. Bot. 125, 581–597. doi: 10.1093/aob/mcz212
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dobney, S., Chiasson, D., Lam, P., Smith, S. P., and Snedden, W. A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284, 31647–31657. doi: 10.1074/jbc.m109.056770
Doebley, J. (1992). “Molecular systematics and crop evolution,” in Molecular Systematics of Plants, eds P. S. Soltis, D. E. Soltis, and J. J. Doyle (Boston, MA: Springer), 202–222. doi: 10.1007/978-1-4615-3276-7_9
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Duan, W., Ren, J., Li, Y., Liu, T., Song, X., Chen, Z., et al. (2016). Conservation and expression patterns divergence of Ascorbic Acid d-mannose/l-galactose pathway genes in Brassica rapa. Front. Plant Sci. 7:778. doi: 10.3389/fpls.2016.00778
Edelman, M., and Colt, M. (2016). Nutrient value of leaf vs. seed. Front. Chem. 4:32. doi: 10.3389/fchem.2016.00032
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Fujiwara, A., Togawa, S., Hikawa, T., Matsuura, H., Masuta, C., and Inukai, T. (2016). Ascorbic acid accumulates as a defense response to Turnip mosaic virus in resistant Brassica rapa cultivars. J. Exper. Bot. 67, 4391–4402. doi: 10.1093/jxb/erw223
Gao, M., Li, G., Yang, B., Qiu, D., Farnham, M., and Quiros, C. (2007). High-density Brassica oleracea linkage map: identification of useful new linkages. Theoret. Appl. Genet. 115, 277–287. doi: 10.1007/s00122-007-0568-3
Gilding, E. K., and Marks, M. D. (2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64, 304–317. doi: 10.1111/j.1365-313x.2010.04329.x
Grossmann, S., Bauer, S., Robinson, P. N., and Vingron, M. (2007). Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics 23, 3024–3031. doi: 10.1093/bioinformatics/btm440
Hahn, C., Müller, A., Kuhnert, N., and Albach, D. (2016). Diversity of kale (Brassica oleracea var. sabellica), glucosinolate content and phylogenetic relationships. J. Agric. Food Chem. 64, 3215–3225. doi: 10.1021/acs.jafc.6b01000
Hay, A., and Tsiantis, M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38, 942–947. doi: 10.1038/ng1835
Hay, A., and Tsiantis, M. (2009). A KNOX family TALE. Curr. Opin. Plant Biol. 12, 593–598. doi: 10.1016/j.pbi.2009.06.006
Higginson, T., Li, S. F., and Parish, R. W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35, 177–192. doi: 10.1046/j.1365-313x.2003.01791.x
Hodgkin, T. (1995). “Cabbages, kales, etc. Brassica oleracea (Cruciferae),” in Evolution of Crop Plants, 2nd Edn, eds J. Small and N. W. Simmonds (Harlow: Longman group), 76–82.
Hong, Z. H., Qing, T., Schubert, D., Kleinmanns, J. A., and Liu, J. X. (2019). BLISTER-regulated vegetative growth is dependent on the protein kinase domain of ER stress modulator IRE1A in Arabidopsis thaliana. PLoS Genet. 15:e1008563. doi: 10.1371/journal.pgen.1008563
Jang, H. J., Pih, K. T., Kang, S. G., Lim, J. H., Jin, J. B., Piao, H. L., et al. (1998). Molecular cloning of a novel Ca 2+-binding protein that is induced by NaCl stress. Plant Mol. Biol. 37, 839–847.
Kebrom, T. H. (2017). A growing stem inhibits bud outgrowth-the overlooked theory of apical dominance. Front. Plant Sci. 8:1874. doi: 10.3389/fpls.2017.01874
Kempin, S. A., Savidge, B., and Yanofsky, M. F. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267, 522–525. doi: 10.1126/science.7824951
Kersey, P. J., Julian, P., Allen, J. E., Allot, A., Barba, M., Boddu, S., et al. (2018). Ensembl Genomes 2018: an integrated Omics infrastructure for non-vertebrate species. Nucleic Acids Res. 46, D802–D808.
Khosla, A., Boehler, A. P., Bradley, A. M., Neumann, T. R., and Schrick, K. (2014). HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26, 2184–2200. doi: 10.1105/tpc.113.120360
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313x.2003.01862.x
Kim, Y. B., Li, X., Kim, S. J., Kim, H. H., Lee, J., Kim, H., et al. (2013). MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 18, 8682–8695. doi: 10.3390/molecules18078682
Kimura, S., Koenig, D., Kang, J., Yoong, F. Y., and Sinha, N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18, 672–677. doi: 10.1016/j.cub.2008.04.008
Kolde, R. (2019). Pheatmap: Pretty Heatmaps. R Package version 1.0.12. Available online at: https://CRAN.R-project.org/package=pheatmap
Koornneef, M., Feusier, J., Corwin, J., Rubin, M., Lin, C., Muok, A., et al. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55, 141–172. doi: 10.1146/annurev.arplant.55.031903.141605
Kuijt, S. J., Greco, R., Agalou, A., Shao, J., Cj‘t Hoen, C., Övernäs, E., et al. (2014). Interaction between the growth-regulating factor and knotted1-like homeobox families of transcription factors [W]. Plant Physiol. 164, 1952–1966. doi: 10.1104/pp.113.222836
Lan, T. H., and Paterson, A. H. (2000). Comparative mapping of quantitative trait loci sculpting the curd of Brassica oleracea. Genetics 155, 1927–1954.
Larsson, A. S., Landberg, K., and Meeks-Wagner, D. R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149, 597–605.
Ledger, S., Strayer, C., Ashton, F., Kay, S. A., and Putterill, J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22. doi: 10.1046/j.1365-313x.2001.01003.x
Lee, B. H., Kwon, S. H., Lee, S. J., Park, S. K., Song, J. T., Lee, S., et al. (2015). The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol. Biol. 89, 529–538. doi: 10.1007/s11103-015-0386-y
Lian, X. P., Zeng, J., Zhang, H. C., Yang, X. H., Zhao, L., and Zhu, L. Q. (2018). Molecular cloning and expression analysis of CML27 in Brassica oleracea pollination process. Acta Biol. Cracovien. Ser. Botan. 60, 15–23.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Lowman, A. C., and Purugganan, M. D. (1999). Duplication of the Brassica oleracea APETALA1 floral homeotic gene and the evolution of domesticated cauliflower. J. Hered. 90, 514–520. doi: 10.1093/jhered/90.5.514
Lukens, L. N., Quijada, P. A., Udall, J., Pires, J. C., Schranz, M. E., and Osborn, T. C. (2004). Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linnean Soc. 82, 665–674. doi: 10.1111/j.1095-8312.2004.00352.x
Madrid, E., Chandler, J. W., and Coupland, G. (2020). Gene regulatory networks controlled by FLOWERING LOCUS C that confer variation in seasonal flowering and life history. J. Exper. Bot. 72:eraa216.
Maggioni, L., von Bothmer, R., Poulsen, G., and Branca, F. (2010). Origin and domestication of cole crops (Brassica oleracea L.), Linguistic and literary considerations. Econ. Bot. 64, 109–123. doi: 10.1007/s12231-010-9115-2
Martin, J., Bruno, V. M., Fang, Z., Meng, X., Blow, M., Zhang, T., et al. (2010). Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genom. 11:663. doi: 10.1186/1471-2164-11-663
Matsumura, Y., Iwakawa, H., Machida, Y., and Machida, C. (2009). Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58, 525–537. doi: 10.1111/j.1365-313x.2009.03797.x
Morohashi, K., Zhao, M., Yang, M., Read, B., Lloyd, A., Lamb, R., et al. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746. doi: 10.1104/pp.107.104521
Müller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27, 13–23. doi: 10.1046/j.1365-313x.2001.01064.x
Nasim, Z., Fahim, M., and Ahn, J. H. (2017). Possible role of MADS AFFECTING FLOWERING 3 and B-BOX DOMAIN PROTEIN 19 in flowering time regulation of Arabidopsis mutants with defects in nonsense-mediated mRNA decay. Front. Plant Sci. 8:191. doi: 10.3389/fpls.2017.00191
Nayidu, N. K., Tan, Y., Taheri, A., Li, X., Bjorndahl, T. C., Nowak, J., et al. (2014). Brassica villosa, a system for studying non-glandular trichomes and genes in the Brassicas. Plant Mol. Biol. 85, 519–539. doi: 10.1007/s11103-014-0201-1
Ojangu, E. L., Tanner, K., Pata, P., Järve, K., Holweg, C. L., Truve, E., et al. (2012). Myosins XI-K, XI-1, and XI-2 are required for development of pavement cells, trichomes, and stigmatic papillae in Arabidopsis. BMC Plant Biol. 12:81. doi: 10.1186/1471-2229-12-81
Okazaki, K., Sakamoto, K., Kikuchi, R., Saito, A., Togashi, E., Kuginuki, Y., et al. (2007). Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theoret. Appl. Genet. 114, 595–608. doi: 10.1007/s00122-006-0460-6
Parkin, I. A., Koh, C., Tang, H., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 15:R77.
Paterson, A. H. (2005). Polyploidy, evolutionary opportunity, and crop adaptation. Genetica 123, 191–196. doi: 10.1007/1-4020-3836-4_17
Pattanaik, S., Patra, B., Singh, S. K., and Yuan, L. (2014). An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front. Plant Sci. 5:259. doi: 10.3389/fpls.2014.00259
Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L., and Yanofsky, M. F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. doi: 10.1046/j.1365-313x.2001.2641042.x
Pérez-Rodríguez, P., Riano-Pachon, D. M., Corrêa, L. G. G., Rensing, S. A., Kersten, B., and Mueller-Roeber, B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38(Suppl._1), D822–D827.
Piazza, P., Bailey, C. D., Cartolano, M., Krieger, J., Cao, J., Ossowski, S., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20, 2223–2228. doi: 10.1016/j.cub.2010.11.037
Poza-Carrión, C., Aguilar-Martínez, J. A., and Cubas, P. (2007). Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis. Plant Signal. Behav. 2, 551–552. doi: 10.4161/psb.2.6.4811
Purugganan, M. D., Boyles, A. L., and Suddith, J. I. (2000). Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea. Genetics 155, 855–862.
Qi, X., An, H., Hall, T. E., Di, C., Blischak, P. D., Pires, J. C., et al. (2019). Genes derived from ancient polyploidy have higher genetic diversity and are associated with domestication in Brassica rapa. bioRxiv [Preprint], doi: 10.1101/842351v1
R Core Development Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L., and Riechmann, J. L. (2001). Regulation of flowering in Arabidopsis by an FLCHomologue. Plant Physiol. 126, 122–132. doi: 10.1104/pp.126.1.122
Reed, J. W., Nagpal, P., Bastow, R. M., Solomon, K. S., Dowson-Day, M. J., Elumalai, R. P., et al. (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122, 1149–1160. doi: 10.1104/pp.122.4.1149
Ren, J., Liu, Z., Du, J., Fu, W., Hou, A., and Feng, H. (2019). Fine-mapping of a gene for the lobed leaf, BoLl, in ornamental kale (Brassica oleracea L. var. acephala). Mol. Breed. 39:40.
Riaño-Pachón, D. M., Ruzicic, S., Dreyer, I., and Mueller-Roeber, B. (2007). PlnTFDB: an integrative plant transcription factor database. BMC Bioinform. 8:42. doi: 10.1186/1471-2105-8-42
Santolamazza-Carbone, S., Velasco, P., Soengas, P., and Cartea, M. E. (2014). Bottom-up and top-down herbivore regulation mediated by glucosinolates in Brassica oleracea var. acephala. Oecologia 174, 893–907. doi: 10.1007/s00442-013-2817-2
Schilmiller, A. L., Koo, A. J., and Howe, G. A. (2007). Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 143, 812–824. doi: 10.1104/pp.106.092916
Schnittger, A., Folkers, U., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11, 1105–1116. doi: 10.2307/3870802
Schönrock, N., Bouveret, R., Leroy, O., Borghi, L., Köhler, C., Gruissem, W., et al. (2006). Polycomb-group proteins repressthe floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678. doi: 10.1101/gad.377206
Sebastian, R. L., Velasco, P., Soengas, P., and Cartea, M. E. (2002). Identification of quantitative trait loci controlling developmental characteristics of Brassica oleracea L. Theoret. Appl. Genet. 104, 601–609. doi: 10.1007/s001220100743
Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783.
Shigyo, M., Hasebe, M., and Ito, M. (2006). Molecular evolution of the AP2 subfamily. Gene 366, 256–265. doi: 10.1016/j.gene.2005.08.009
Sivitz, A. B., Hermand, V., Curie, C., and Vert, G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS One 7:e44843. doi: 10.1371/journal.pone.0044843
Smith, H. M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 99, 9579–9584. doi: 10.1073/pnas.092271599
Smith, L. B., and King, G. J. (2000). The distribution of BoCAL-a alleles in Brassica oleracea is consistent with a genetic model for curd development and domestication of the cauliflower. Mol. Breed. 6, 603–613.
Snogerup, S. (1980). “The wild forms of the Brassica oleraceae group (2n=18) and their possible relations to the cultivated ones,” in Brassica Crops and Wild Allies, Biology and Breeding, eds S. Tsunoda, K. Hinata, and C. Gomez-Campo (Tokyo: Scientific Societies Press), 121–132.
Soengas, P., Cartea, M. E., Francisco, M., Sotelo, T., and Velasco, P. (2012). New insights into antioxidant activity of Brassica crops. Food Chem. 134, 725–733. doi: 10.1016/j.foodchem.2012.02.169
Sønderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Steele, R. P., Geu-Flores, F., and Halkier, B. A. (2012). Quality and quantity of data recovered from massively parallel sequencing: examples in Asparagales and Poaceae. Am. J. Bot. 99, 330–348. doi: 10.3732/ajb.1100491
Stewarta, D., and McDougalla, G. (2012). The Brassicas - An Undervalued Nutritional and Health Beneficial Plant Family. Edinburgh: The James Hutton Institute.
Stirnberg, P., Chatfield, S. P., and Leyser, H. O. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121, 839–847. doi: 10.1104/pp.121.3.839
Tanaka, N., Niikura, S., and Takeda, K. (2009). Inheritance of cabbage head formation in crosses of cabbage x ornamental cabbage and cabbage x kale. Plant Breed. 128, 471–477. doi: 10.1111/j.1439-0523.2008.01572.x
Tapia-López, R., García-Ponce, B., Dubrovsky, J. G., Garay-Arroyo, A., Pérez-Ruíz, R. V., Kim, S. H., et al. (2008). An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182–1192. doi: 10.1104/pp.107.108647
Tsiantis, M., and Hay, A. (2003). Comparative plant development: the time of the leaf? Nat. Rev. Genet. 4, 169–180. doi: 10.1038/nrg1002
Tsunoda, S. (1980). “Eco-physiology of wild and cultivated forms in Brassica and allied genera,” in Brassica Crops and Wild Allies, Biology and Breeding, eds S. Tsunoda, K. Hinata, and C. Gomez-Campo (Tokyo: Japan Scientific Societies Press), 109–116.
UniProt Consortium (2018). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515.
Velasco, P., Cartea, M. E., González, C., Vilar, M., and Ordás, A. (2007). Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group). J. Agric. Food Chem. 55, 955–962.
Velasco, P., Francisco, M., Moreno, D. A., Ferreres, F., García-Viguera, C., and Cartea, M. E. (2011). Phytochemical fingerprinting of vegetable Brassica oleracea and Brassica napus by simultaneous identification of glucosinolates and phenolics. Phytochem. Analys. 22, 144–152. doi: 10.1002/pca.1259
Vlad, D., Kierzkowski, D., Rast, M. I., Vuolo, F., Ioio, R. D., Galinha, C. P., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780–783. doi: 10.1126/science.1248384
Wachter, A., Tunc-Ozdemir, M., Grove, B. C., Green, P. J., Shintani, D. K., and Breaker, R. R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3’ end processing of mRNAs. Plant Cell 19, 3437–3450. doi: 10.1105/tpc.107.053645
Wang, H., Wu, J., Sun, S., Liu, B., Cheng, F., Sun, R., et al. (2011). Glucosinolate biosynthetic genes in Brassica rapa. Gene 487, 135–142. doi: 10.1016/j.gene.2011.07.021
Waters, M. T., Brewer, P. B., Bussell, J. D., Smith, S. M., and Beveridge, C. A. (2012). The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159, 1073–1085. doi: 10.1104/pp.112.196253
Yang, C., and Ye, Z. (2013). Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Yi, G. E., Robin, A. H. K., Yang, K., Park, J. I., Hwang, B. H., and Nou, I. S. (2016). Exogenous methyl jasmonate and salicylic acid induce subspecies-specific patterns of glucosinolate accumulation and gene expression in Brassica oleracea L. Molecules 21:1417. doi: 10.3390/molecules21101417
Yi, G.-E., Robin, A. H. K., Yang, K., Park, J. I., Kang, J. G., Yang, T. J., et al. (2015). Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies. Molecules 20, 13089–13111. doi: 10.3390/molecules200713089
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Keywords: Brassica oleracea, domestication, kale, mustards, plant development, RNA-seq, transcriptomics
Citation: Arias T, Niederhuth CE, McSteen P and Pires JC (2021) The Molecular Basis of Kale Domestication: Transcriptional Profiling of Developing Leaves Provides New Insights Into the Evolution of a Brassica oleracea Vegetative Morphotype. Front. Plant Sci. 12:637115. doi: 10.3389/fpls.2021.637115
Received: 02 December 2020; Accepted: 18 January 2021;
Published: 05 March 2021.
Edited by:
Ive De Smet, Flanders Institute for Biotechnology, BelgiumReviewed by:
Polina Yu. Novikova, Max Planck Institute for Plant Breeding Research, GermanyDirk Carl Albach, University of Oldenburg, Germany
Copyright © 2021 Arias, Niederhuth, McSteen and Pires. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatiana Arias, dGF0aWFuYS50YXRpYW5hYXJpYXNAZ21haWwuY29t; dGF0aWFuYS5hcmlhczQ4QHRkZWEuZWR1LmNv
†Present address: Tatiana Arias, Tecnológico de Antioquia, Medellín, Colombia Chad E. Niederhuth, Department of Plant Biology, Michigan State University, East Lansing, MI, United States
‡These authors have contributed equally to this work
 Tatiana Arias
Tatiana Arias Chad E. Niederhuth
Chad E. Niederhuth Paula McSteen
Paula McSteen J. Chris Pires
J. Chris Pires