- 1Key Laboratory of Saline-Alkali Vegetation Ecology Restoration, Ministry of Education, College of Life Sciences, Northeast Forestry University, Harbin, China
- 2State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 3Shanghai Center for Plant Stress Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, China
- 4National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 5Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Changchun, China
- 6School of Life Sciences and Agriculture and Forestry, Qiqihar University, Qiqihar, China
The plant cytoskeleton forms a stereoscopic network that regulates cell morphogenesis. The cytoskeleton also provides tracks for trafficking of vesicles to the target membrane. Fusion of vesicles with the target membrane is promoted by SNARE proteins, etc. The vesicle-SNARE, Sec22, regulates membrane trafficking between the ER and Golgi in yeast and mammals. Arabidopsis AtSEC22 might also regulate early secretion and is essential for gametophyte development. However, the role of AtSEC22 in plant development is unclear. To clarify the role of AtSEC22 in the regulation of plant development, we isolated an AtSEC22 knock-down mutant, atsec22-4, and found that cell morphogenesis and development were seriously disturbed. atsec22-4 exhibited shorter primary roots (PRs), dwarf plants, and partial abortion. More interestingly, the atsec22-4 mutant had less trichomes with altered morphology, irregular stomata, and pavement cells, suggesting that cell morphogenesis was perturbed. Further analyses revealed that in atsec22-4, vesicle trafficking was blocked, resulting in the trapping of proteins in the ER and collapse of structures of the ER and Golgi apparatus. Furthermore, AtSEC22 defects resulted in impaired organization and stability of the cytoskeleton in atsec22-4. Our findings revealed essential roles of AtSEC22 in membrane trafficking and cytoskeleton dynamics during plant development.
Introduction
Plant cell shape formation is crucial for development and morphogenesis, which are regulated by cytoskeletal dynamics (Fu et al., 2005; Panteris and Galatis, 2005; Kotzer and Wasteneys, 2006; Zhang et al., 2011; Szymanski, 2014; Armour et al., 2015; Chen et al., 2016; Christopher and Arun, 2018). The cytoskeleton, comprised of actin filaments (AFs) and microtubules (MTs), forms a three-dimensional intracellular network that provides tracks for cellular transport, including organelles, vesicles, protein complexes, and macromolecules (Wasteneys and Yang, 2004; Hussey et al., 2006; Akhmanova and Hammer, 2010). Cortical MTs control anisotropic cell expansion by directing the deposition pattern of cellulose microfibrils in the root and hypocotyl (Fu et al., 2005; Takatani et al., 2015). The cell shape is controlled by MT dynamics modulated by microtubule binding proteins (MAPs) and by the coordination between cortical MTs and AFs (Burk et al., 2001; Fu et al., 2005; Panteris and Galatis, 2005; Ambrose et al., 2007, 2011; Kirik et al., 2007; Wang et al., 2007; Zhang et al., 2013). Cortical AFs provide endomembrane guidance and structural support as well as a driving force by forming dynamic fringe structures at the leading edge during polarized cell growth (Vidali et al., 2009). The particular shape of a cell is important for its function and environmental adaptation (Robert, 2013). Trichomes, guard cells, and pavement cells are highly specialized cell types generated from the leaf epidermal layer (Vöfély et al., 2019). The Arabidopsis trichome is a unicellular structure with usually three branches; MTs are responsible for trichome initiation and branching, and AFs control the shape (Sambade et al., 2014; Chen et al., 2016). It has been shown in detail that AFs and MTs mediate vesicle trafficking in plants (Žárský et al., 2009; Mooren et al., 2012; Idilli et al., 2013; Kong et al., 2015).
In eukaryote, vesicle trafficking mediates the delivery of proteins to their destinations and maintains the endomembrane system. Most of the secreted proteins are synthesized in the rough ER and transported to the target compartments. Vesicle transport involves vesicle budding from the donor membrane, movement along the cytoskeleton, and fusion with the target membrane (Palade, 1975; Rothman, 2014). Cargo recruitment and vesicle formation in the ER are mediated by the GTPase SAR1, the guanine-nucleotide exchange factor (GEF) Sec12, and the coat protein II (COPII) complex (Tang et al., 2005; Gürkan et al., 2006). ER-derived COPII vesicles fuse to the cis-Golgi cisterna and release the cargoes (Kurokawa et al., 2014; Lee et al., 2016). Conversely, Golgi-derived COPI vesicles fuse to the ER membrane to retrieve the membrane component and ER-retention proteins for recycling (Yu et al., 2012; Brandizzi and Barlowe, 2013; Spang, 2013; Dodonova et al., 2015). Membrane fusion is mediated by conserved proteins including Rab GTPases, tethers, and soluble N-ethylmalemide-sensitive factor attachment protein receptors (SNAREs), which contain SNARE motifs (Söllner et al., 1993; Hanson et al., 1997; Bonifacino and Glick, 2004; Lipka et al., 2007; Saito and Ueda, 2009). Membrane fusion is conducted by the formation of a trans-SNARE complex composed of one vesicle membrane-SNARE (v-SNARE) and two or three target membrane-SNAREs (t-SNAREs) (Söllner et al., 1993; Li et al., 2016; Bruinsma et al., 2018). In yeast and mammalian cells, v-SNARE Sec22 mediates anterograde and retrograde transport between the ER and the Golgi complex (Newman et al., 1990; Hardwick and Pelham, 1992; Flanagan et al., 2015; Li et al., 2015; Zhao et al., 2015; Lee et al., 2016). In yeast, in anterograde transport, fusion of ER-derived COPII vesicles with Golgi cisterna requires Sec22p, which forms a SNARE complex with the cis-Golgi localized t-SNAREs, Sed5p, Bos1p, and Bet1p. Conversely, in retrograde transport, Sec22p is combined with ER-localized Ufe1p, Slt1p, and Sec20p (Liu and Barlowe, 2002; Burri et al., 2003; Morsomme et al., 2003; Brandizzi and Barlowe, 2013; Spang, 2013; Dodonova et al., 2015). COPI vesicles are implicated in the quick recycling of Sec22 from the Golgi to the ER under salt stress (Letourneur et al., 1994; Ossipov et al., 1999). In tobacco leaves, transiently overexpressed Arabidopsis SEC22 was localized on the ER and Golgi apparatus and induced collapse of Golgi membrane proteins and redistribution into the ER, suggesting that it functions in the early secretion pathway (Chatre et al., 2005). El-Kasmi et al. have reported that AtSEC22 is essential for gametophyte development and Golgi integrity. Loss of AtSEC22 has been shown to result in Golgi fragmentation and pollen lethality (El-Kasmi et al., 2011). Moreover, Sec22p/SEC22 specifically impacted Cs+ accumulation in yeast and plants. Sec22p has also been shown to enhance non-selective cation deposition (Draxl et al., 2013). Cooperating with Sar1 and Memb11, Sec22 facilitates ER export of Phyl1.1 (Phytolongin), which is important for vesicle formation and fusion in plants (de Marcos Lousa et al., 2016).
In order to further investigate SEC22 function in plant development, we isolated an AtSEC22 knockdown mutant, atsec22-4, which has developmental defects such as delayed germination, shorter primary roots (PRs), dwarf, and sterility. Interestingly, the morphology of trichomes, pavement cells, and stomata are altered in atsec22-4. Our investigation revealed that AtSEC22 interacts with the Golgi-resident syntaxin AtSYP32, to regulate vesicle transport between the ER and Golgi. Downregulation of AtSEC22 blocked ER export. Furthermore, we found that cytoskeleton organization and stability were disrupted in atsec22-4, which resulted in impaired cell morphogenesis, stomata movement, and delayed development. Our findings illustrate the essential role of AtSEC22 in cell morphogenesis during plant development.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as a wild-type plant. T-DNA-tagged lines were derived from Col-0. Homozygous plants were obtained by PCR screening using the insertion-specific primers shown in Supplementary Table 1. Arabidopsis seeds were surface-sterilized and sown either on soil or onto 0.8 or 1.2% agar with 1/2 Murashige and Skoog medium (PhytoTech) and 1% (w/v) sucrose. Plants were grown at 22°C under 16 h: 8 h light: dark cycles. atsec22-4 (SAIL_736_F03), atsyp32-1 (GABI-109A09), and atsyp32-2 (GABI_920F05) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. The fluorescence marker lines of ST-GFP (Boevink et al., 1998), GFP-HDEL (Matsushima et al., 2002), pUBQ:ABD2-GFP (Tian et al., 2015), p35S:MBD-GFP (Dyachok et al., 2014), p35S:TUA6-GFP (Eisinger W. et al., 2012), and pPIN1:PIN1-GFP (Benkova et al., 2003) were employed.
Plasmid Construction
An AtSEC22 cDNA fragment was amplified from Col-0 using the AtSEC22-specific primers SEC22TOPO-F and SEC22TOPO-R and ligated into pENTR/D-TOPO vectors (Invitrogen, Carlsbad, CA, United States). To generate TAP-tagged (containing 9xmyc myc) AtSEC22-expressing transgenic plants, the AtSEC22 cDNA fragment was transferred from the AtSEC22 entry clone to the destination vector pNTAPa (Rubio et al., 2005) by an LR reaction (Invitrogen). For generating AtSEC22 RNAi plants, a 390 bp fragment of AtSEC22 cDNA was amplified using the primers Sec22RNAi-F and Sec22RNAi-R, and cloned into the pENTR/D-TOPO vector and subsequently subcloned into the destination vector pK7GWIWG2 by the LR reaction. To generate pAtSEC22:gAtSEC22 complementation plants, the AtSEC22 genomic sequence was amplified using the primers Sec22comple-F and Sec22comple-R and cloned into the pDONR207 ENTRY vector (Invitrogen), and subsequently subcloned into the destination vector pMDC99 by the LR reaction. The primers used are listed in Supplementary Table 1.
Anti-AtSEC22 Antibody Generation
To prepare the antigen, the AtSEC22 cytosolic fragment corresponding to 1–195 amino acids was amplified using the primers SEC22TOPO-F and SEC22TOPO-R, ligated into pENTR/D-TOPO vectors, and subsequently introduced into the pET32a vector (Novagen). Recombinant protein was expressed in the Escherichia coli BL-21 strain, purified with a HiTrap chelating column, and entrusted to PhytoAB Inc., to generate polyclonal antibodies. Immunoblot detected the AtSEC22 band using anti-AtSEC22 antibody from total extracts of Col-0 and atsec22-4 seedlings.
Immunoblotting
Dodecyl sulfate, sodium salt-Polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses were performed as described previously (Li et al., 2006). Antibodies were diluted as follows: anti-AtSEC22, 1:500; anti-12S, 1:20,000; anti-2S3P, 1:5,000 (Li et al., 2006); anti-BiP (AS09 481, Agriser, Sweden), 1:2,500; anti-actin (AS13 2640, Agriser), 1:1,500; anti-tubulin A1 (At1g64740) (R0267-1a, Abiocode), 1:1,500; and MAP65-1 (0732B5, 9632B9, PhytoAB Inc.), 1:1,000, respectively. The secondary antibody was against rabbit IgG (ZB2301 and ZSGB-BIO), 1:5000. Immunoreactive signals were detected using an enhanced chemiluminescence detection system (LAS-4000 and FYJIFILM).
Yeast Two-Hybrid Assay
For the yeast two-hybrid assay, AtSYP81 plasmids were generated in a previous study (Li et al., 2006). The cytosolic regions of AtSEC22, AtSYP31, and AtSYP32 were amplified using corresponding specific primers, Sec22NdeI-F/BamHI-R, SYP31/32NdeI-F, and SYP31/32BamHI-R, respectively. Then these fragments were ligated into the pEASY-Blunt vector (TransGen, #CB101-01), respectively. After Sanger sequencing confirmation, the fragments were transferred into pGADT7 or pGBKT7 vectors, respectively. The paired constructs were introduced into Saccharomyces cerevisiae strain AH109 (Clontech) and selected on SD/-Leu/-Trp medium. The interactions were examined on SD/-Leu/-Trp/-His/-Ade medium.
RNA Extraction and RT-PCR Analysis
Total RNA was isolated from seedlings of Col-0 and atsec22-4 using the RNeasy kit (P4623, Tiangen, China). Total RNA (0.5–1 μg) was treated with DNase I (Invitrogen) to reduce genomic DNA contamination. Reverse transcription was performed using the All-in-One First Strand Synthesis MasterMix (NOVA). Semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed according to the manufacturer’s instructions. ACT2 was used as an endogenous control for RT-PCR and quantitative reverse transcription PCR (RT-qPCR). The specific primers are listed in Supplementary Table 1.
Pull-Down Assay
Pull-down assays were performed as described previously (Li et al., 2013) using an iMACS epitope tag protein isolation kit (anti-c-myc, Miltenyi Biotec1). Two grams of two-week-old seedlings were used for each sample. The beads were sent for Shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Shotgun LC-MS/MS Analysis
LC-MS/MS analysis was performed as described previously (Qin et al., 2017), with modifications. Briefly, interacting proteins were eluted and reduced from beads by SDS elution buffer at 95°C for 5 min. Proteins were alkylated and digested on a centrifugal filter unit (10 kDa MWCO) using the filter-aided sample preparation (FASP) method. Peptides were analyzed by nanoAcquity ultra performance LC (Waters, Milford, MA, United States) and Orbitrap Fusion mass spectrometry (Thermo Fisher Scientific, Waltham, MA, United States). MS survey scan was performed by Orbitrap at a resolution of 60,000 over a m/z range of 350–1,800, and the top 20 precursor ions were selected for MS/MS measurements by HCD scans. Dynamic exclusion was enabled for 60 s. MS/MS raw data were searched against the database of The Arabidopsis Information Resource (TAIR10) using Mascot Daemon 2.5 (Matrix Science, London, United Kingdom). Carbamidomethylation of Cys was designated as a fixed modification. Deamidation of Asn or Gln and oxidation of Met were considered as variable modifications. Peptide assignments were filtered by an ion score cut-off of 15, and false discovery rate (FDR) of peptides was set up to less than 1%. The original data are presented in Supplementary Table 2.
Chemical Treatments
For MT-depolymerization, 10 μM of oryzalin was applied to plant tissues for 10 min for hypocotyl and 20 min for leaf (Chen et al., 2016). For AF disruption, rosette leaves were incubated in 1 μM of LatB solution for 20 min (Konopka et al., 2008). Mocks were the same solvent without the reagents. For FM4-64 staining, plant tissues were incubated for 5 min in 4 μM of FM4-64 solution (Rigal et al., 2015).
Confocal Microscopy
Fluorescent images were obtained using a point scanning confocal microscope (Leica TCS SP8). Confocal imaging was preset for GFP (with Ex:488 nm, Em:500–550 nm) or for RFP and DsRED (with Ex:543 nm, Em: 580–640 nm).
Scanning Electron Microscope and Transmission Electron Microscopy Analysis
The 3rd or 4th rosette leaves from seven-week-old plants were collected and analyzed by Scanning Electron Microscope (SEM). Trichomes were observed using a JSM-6610LV Emission SEM (Hitachi). Transmission electron microscopy (TEM) observations were performed as described previously (Shimada et al., 2006) with leaves from two-week-old plants. The sections were observed and photographed using a transmission electron microscope H7700 (Hitachi).
Accession Numbers
Gene and protein sequence data regarding this study can be found in the TAIR2 databases under the following accession numbers: AtSEC22 (At1g11890), AtSYP32 (At3g243500), AtSYP31 (At5g05760), AtSYP81 (At1g51740), AtSec20 (At3g24315), and MAP65-1 (At5g55230).
Results
atsec22-4 Mutant Exhibits Serious Developmental Defects
To explore the mechanisms underlying AtSEC22-mediated regulation of plant development, we isolated the atsec22-4 mutant, in which T-DNA was inserted into the last exon of At1g11890 (Figure 1A). RT-PCR detection of the fragment before or spanning the T-DNA insertion site indicated that AtSEC22 expression was either very low in some regions (exons 2–4), or could not be detected in some other regions (exons 1–3, 3–6) (Figure 1B), suggesting that AtSEC22 expression is significantly downregulated in atsec22–4. We also generated AtSEC22 RNAi lines, while it was partially downregulated in RNAi lines (Figure 1C). We then generated an anti-AtSEC22 polyclonal antibody against 1–192 amino acids to detect protein levels. Immunoblot analysis indicated that very little AtSEC22 protein accumulated in atsec22-4, and protein levels were partially decreased in AtSEC22 RNAi lines (Figure 1D). atsec22-4 exhibited serious developmental defects, including delayed germination and lower germination rate (Supplementary Figures 1A,B), short PR, more adventitious roots (Figures 1E,F), and denser root hairs (Figures 1G,H). Moreover, atsec22-4 plants had dwarfism (Supplementary Figure 1C). To confirm the causal gene, we performed complementation experiments by generating pAtSEC22:gAtSEC22-expressing plants harboring a native promoter-driven AtSEC22 genomic fragment that was introduced into atsec22-4 by crossing. The complemented atsec22-4 plants recovered all phenotypes (Figure 1 and Supplementary Figure 2C). All these results show that AtSEC22 is the gene responsible for atsec22-4 phenotypes.
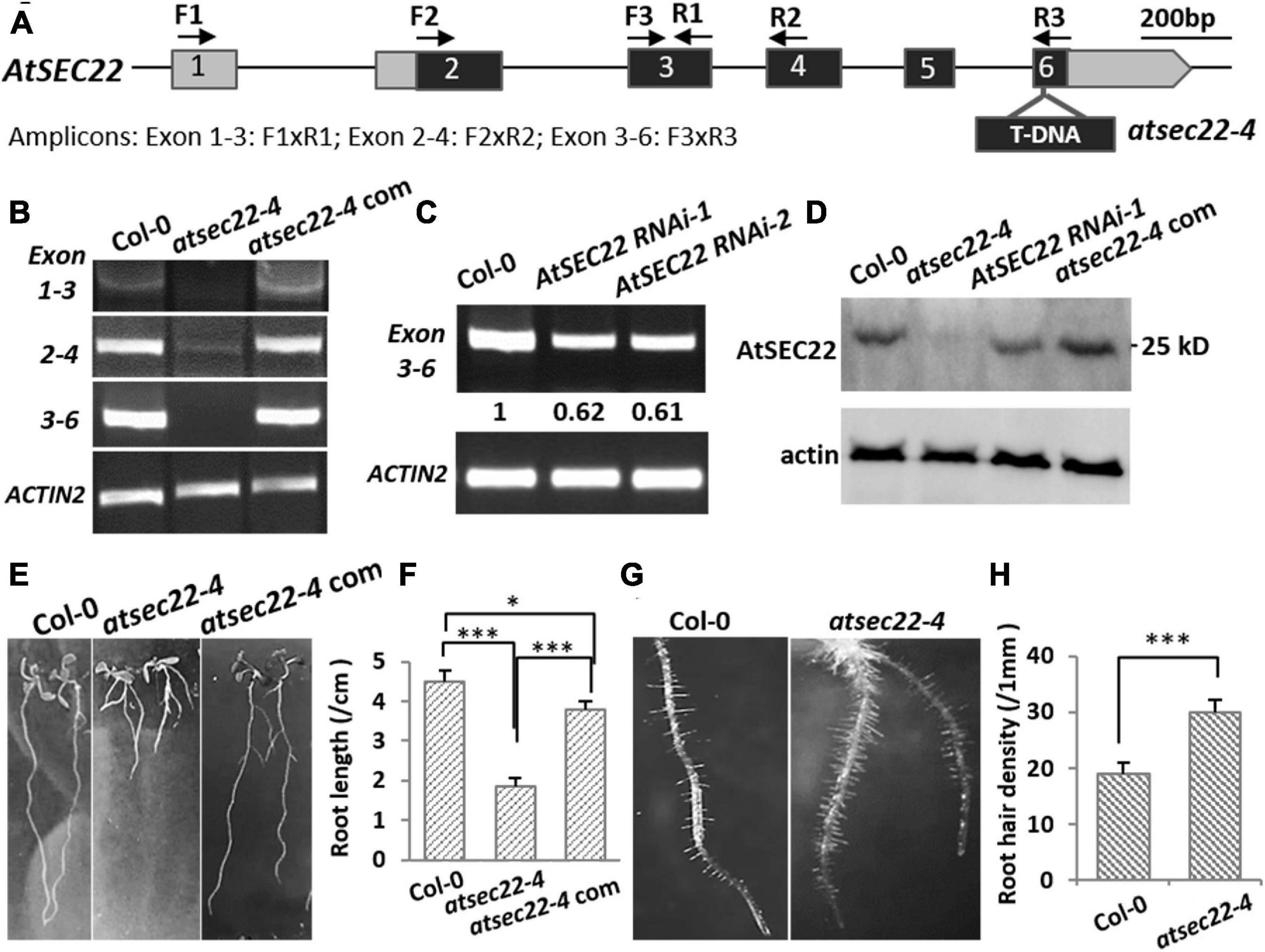
Figure 1. AtSEC22 gene and defective mutants. (A) The structure of AtSEC22 gene and the T-DNA insertion site in atsec22-4. Location of primers for RT-PCR analysis are indicated by arrows. Exons are indicated by boxes with numbers, and the solid lines indicate introns. (B,C) RT-PCR detection of expression level of AtSEC22 with different combinations of primers in Col-0, atsec22-4, and atsec22-4 complemented lines (atsec22-4 com). Actin2 was used as endogenous control. Exon 1-3: F1xR1; Exon 2-4: F2xR2; Exon 3-6: F3xR3. (D) Immunoblot was performed with ten-day-old seedlings of Col-0, atsec22-4, AtSEC22 RNAi, and atsec22-4 complemented lines using the anti-AtSEC22 antibody. (E,F) Ten-day-old seedlings of atsec22-4 exhibited short primary roots and more adventitious roots. Panel (F) shows statistics of panel (E). n ≥ 30, three independent experiments per sample. (G,H) Root hairs are denser in atsec22-4. Panel (H) shows statistics of panel (G). n ≥ 10 roots. ***P < 0.001. Significance was evaluated by Student’s t test.
Cell Morphogenesis Was Perturbed in atsec22-4
In addition to the developmental defects, the more conspicuous phenotype in atsec22-4 was that atsec22-4 had fewer trichomes on the leaves (Figures 2A,B, and Supplementary Figure 2A) and fewer hairs on the stems (Supplementary Figure 2C). SEM and stereomicroscopic observation indicated that trichome morphology in atsec22-4 was abnormal. In the wild-type, most trichomes (83.3%) had three branches, but in atsec22-4, only 56.7% of trichomes had three branches, 26.7% had two branches, and the rest had four, five, or no branches (Figures 2C,D, and Supplementary Figure 2B). In the AtSEC22 RNAi lines, delayed development (Supplementary Figure 3A), decreased trichome number (Supplementary Figures 3B,D), and altered morphology (Supplementary Figures 3C,E) were also observed. Since the trichome is a specialized cell type originating from epidermal cells, we then examined epidermal cells. The pavement cells adopted jigsaw puzzle-like shapes with interlocking lobes and necks in the wild-type plant. However, in atsec22-4, pavement cells exhibited irregular shapes (Figures 2E, dotted lines, Supplementary Figure 2D). We measured the lobe length and neck width of the pavement cells, and found that the lobe length in atsec22-4 was significantly shorter, while the neck width was not significantly different (Figures 2F,F’). Moreover, an increased number of stomata with disordered distribution and uneven size were observed in atsec22-4 leaves (Figure 2E, dotted circles, Figure 2G, and Supplementary Figure 2D). These results indicated that cell morphogenesis was perturbed in atsec22-4.
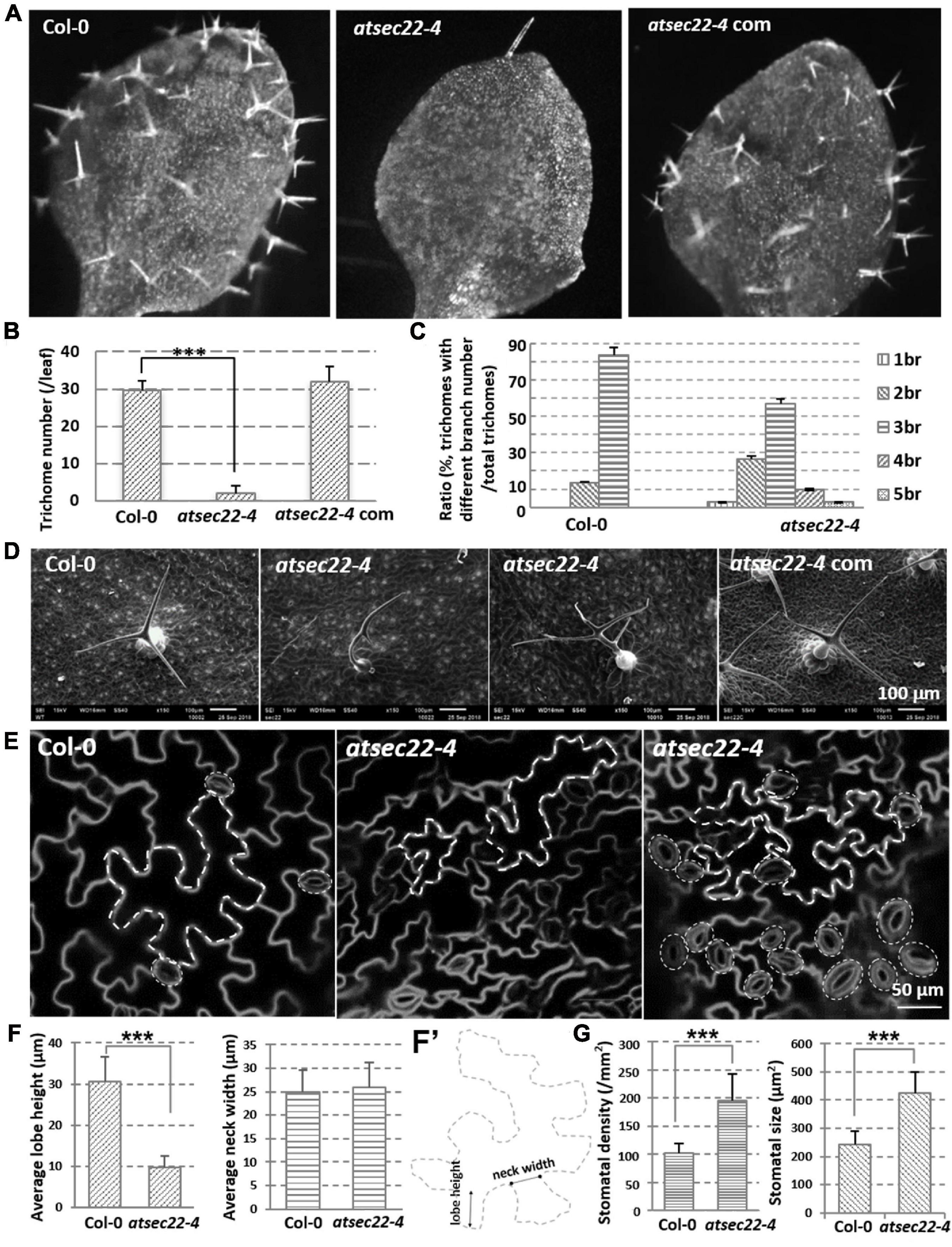
Figure 2. Morphology of epidermal cells was abnormal in atsec22-4. (A) Images of the first true leaves from 12-day-old seedlings. Images were the same scale. (B) Statistics of leaf trichome number in panel (A). nCol–0 = 12, natsec22–4 = 20 leaves. ***P < 0.001. Significance was evaluated by Student’s t test. (C) Statistics of leaf trichomes with different branch number from seven-week-old plants. nCol–0 = 66, natsec22–4 = 46 trichomes. br, branch. (D) SEM images of 3rd or 4th rosette leaves from seven-week-old plants. (E) FM4-64-labeled shape of pavement cells and stomata in the 4th leaves from 14-day-old seedlings. Scale bars are as shown. (F,F’) The measurement of lobe length and neck width of pavement cells in the middle part of leaves from 14-day-old seedlings. n ≥ 20. Panel (F’), the line with round dots indicates neck width; the line with arrows indicates lobe height. (G) Statistics of leaf stomata density and size in the middle part of the first pair true leaves from 12-day-old seedlings. For density statistics, 20 slices of an area of 0.02 mm2 were used; for size statistics, nCol–0 = 40, natsec22–4 = 50 stomata. ***P < 0.001. Significance was evaluated by Student’s t test.
Downregulation of AtSEC22 Affected Vesicle Trafficking and Integrity of Endomembrane System
To investigate the effects on vesicle transport in atsec22-4, we introduced the trans-Golgi marker ST-GFP (Boevink et al., 1998), the ER-retention marker GFP-HDEL (Matsushima et al., 2002), and the plasma membrane-targeting marker PIN1-GFP (Benkova et al., 2003), into atsec22-4 by crossing. Confocal images revealed that Golgi-resident ST-GFP proteins were collapsed and redistributed to the ER, and some ST-GFP aggregates accumulated in the ER tubular intersections in atsec22-4 (Supplementary Figure 4A), suggesting that ER export was blocked. The GFP-HDEL-visualized ER network exhibited a relatively looser structure with a larger space between the ER tubules in atsec22-4 (arrows), suggesting that the ER structure was disturbed in the mutant (Supplementary Figure 4B). Moreover, a large amount of intracellular-localized PIN1-GFP was observed in stele cells in atsec22-4 (Figure 3A, arrows), suggesting disturbed membrane trafficking. We then examined the accumulation of precursors of seed storage proteins (SSPs), which are considered as markers for ER-to-vacuolar transport (Li et al., 2006, 2013; Shimada et al., 2006; Zhao et al., 2018). As shown in Figure 3B, numerous SSP precursors accumulated in atsec22-4 seeds, indicating that the ER-to-vacuole pathway was blocked.
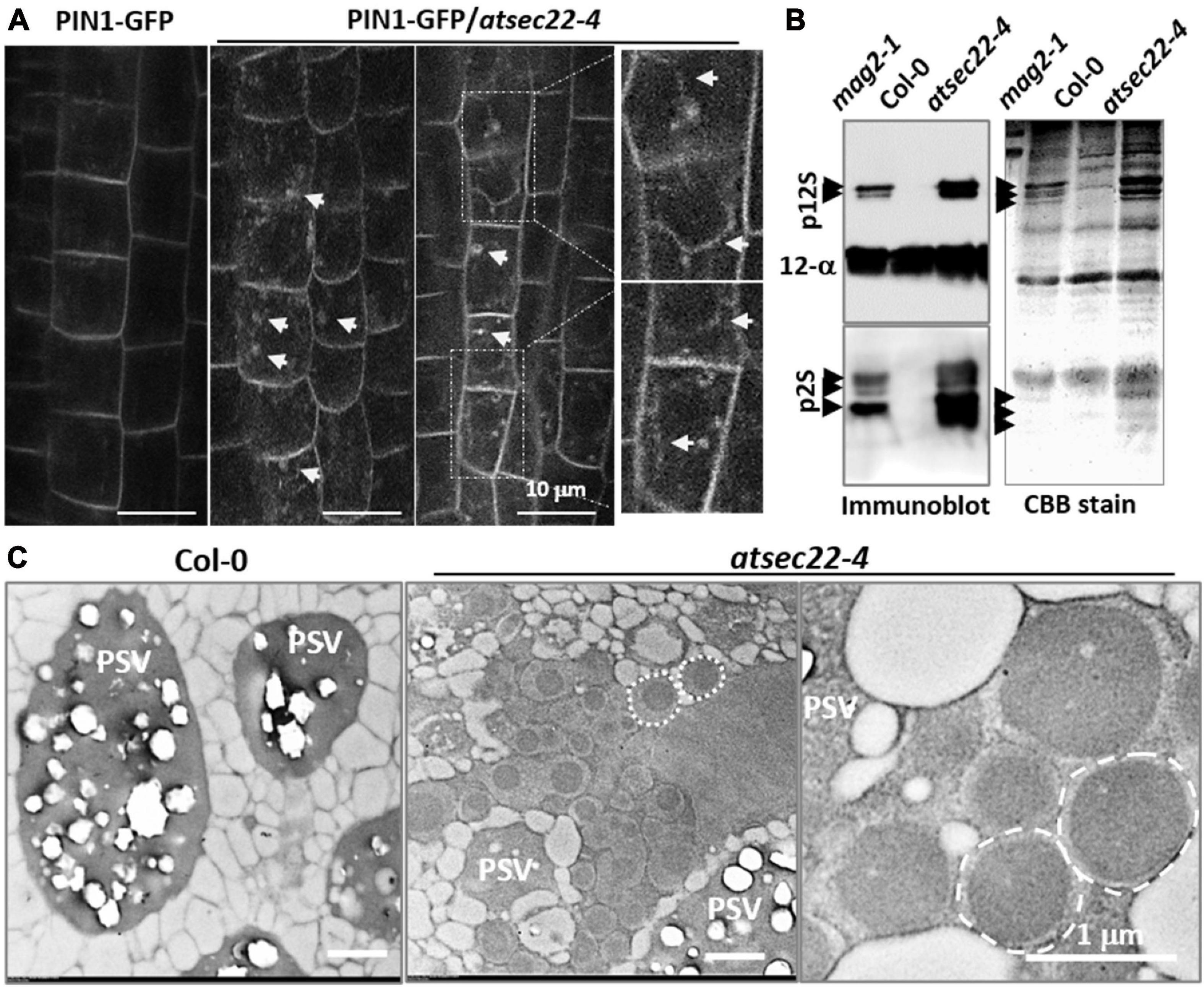
Figure 3. Vesicle trafficking was disturbed in atsec22-4. (A) PIN1-GFP were internalized in root cells in atsec22-4 (arrows). Magnified images emphasize multiple localization of PIN1-GFP (arrows). (B) Immunoblot of seed proteins with anti-12S globulin and anti-2S albumin antibodies (left), and total seed protein profiles (CBB-stained SDS-PAGE gel) (right). The atsec22-4 seeds accumulated numerous SSP precursors (arrows). mag2-1 was used as positive control, Col-0 was used as negative control. p12S, 12S globulin precursors; p2S, 2S albumin precursors; 12S-α, 12S globulin α subunits. Scale bars are as shown. (C) TEM images of dry seed cells. Unusual structures (dotted circles) with a high electron-dense core were developed in atsec22-4 seed cells. PSV, protein storage vacuole.
To investigate the effects of protein accumulation on subcellular structures in the mutant, dry seeds and leaves were examined using TEM analyses. In atsec22-4 seed cells, numerous unusual structures with a high electron-dense core were developed (dotted circles) (Figure 3C). The unusual structures resembled those in mag2, mag4, mag5, and mip1/2/3 mutants defective in ER export. The structures have been shown to be composed of precursors of 2S albumins and 12S globulins (Li et al., 2006, 2013; Takahashi et al., 2010; Takagi et al., 2013), and the ER chaperons BiP and PDI, implying their ER lumen-localization (Li et al., 2006). These results suggested that ER export was blocked in atsec22-4. TEM observations of leaf cells revealed that in atsec22-4, the ER network lost most of the tubular structures and produced expanded and fragmented ER sheets (Supplementary Figure 4C, black arrows). Furthermore, the Golgi stacks became smaller with fewer cisternae, indicating that the endomembrane system was collapsed due to a block in vesicle trafficking. The trap of proteins inside the ER usually induces ER stress. Determination of the expression levels of the ER stress-specific marker BiP3 indicated that its transcriptional expression was substantially increased in atsec22-4 (Supplementary Figure 5A), suggesting serious ER stress. ER stress usually induces the unfolded protein response (UPR) pathway (Sundaram et al., 2018). We examined the expression levels of UPR pathway markers, IRE1A, IRE1B, and bZIP60, and found that bZIP60 expression was significantly upregulated (Supplementary Figure 5B), suggesting that the UPR pathway was affected in atsec22-4. Taken together, these results indicated that vesicle trafficking and integrity of the endomembrane network were affected in atsec22-4.
AtSEC22 Interacted With SNARE Protein AtSYP32
In yeast and mammalian cells, Sec22 mediates anterograde and retrograde transport between the ER and Golgi complex (Flanagan et al., 2015; Li et al., 2015; Zhao et al., 2015). To investigate which transport pathway is regulated by AtSEC22, we performed yeast two-hybrid (Y2H) analysis. Y2H analysis detected an interaction between AtSEC22 and the Golgi-resident syntaxin AtSYP32 (At3g243500), but no interaction of AtSEC22 with AtSYP31 or the ER-localized syntaxin AtSYP81 (Figure 4A). We further performed pull-down analysis using TAP (myc)-tagged AtSEC22-overexpressing plants followed by shotgun LC-MS/MS analysis, and identified AtSYP32. To confirm this interaction, we generated myc-tagged AtSYP32-over-expressing plants and performed pull-down analysis followed by shotgun LC-MS/MS analysis. As expected, AtSEC22 was identified (Figure 4B), suggesting that AtSEC22 interacts with AtSYP32 in plant cells and might regulate anterograde transport. We then isolated the atsyp32-1 and atsyp32-2 mutants and found that they were homozygously lethal. This sterile phenotype was similar to the homozygous mutant, atsec22-1, which has pollen lethality (El-Kasmi et al., 2011), suggesting that AtSEC22 and AtSYP32 are essential for reproductive development. The heterozygotes, atsyp32-1± and atsyp32-2±, had shorter siliques and were partially abortive; these phenotypes were similar to those of atsec22-4, the downregulation mutant (Figures 4C–E).
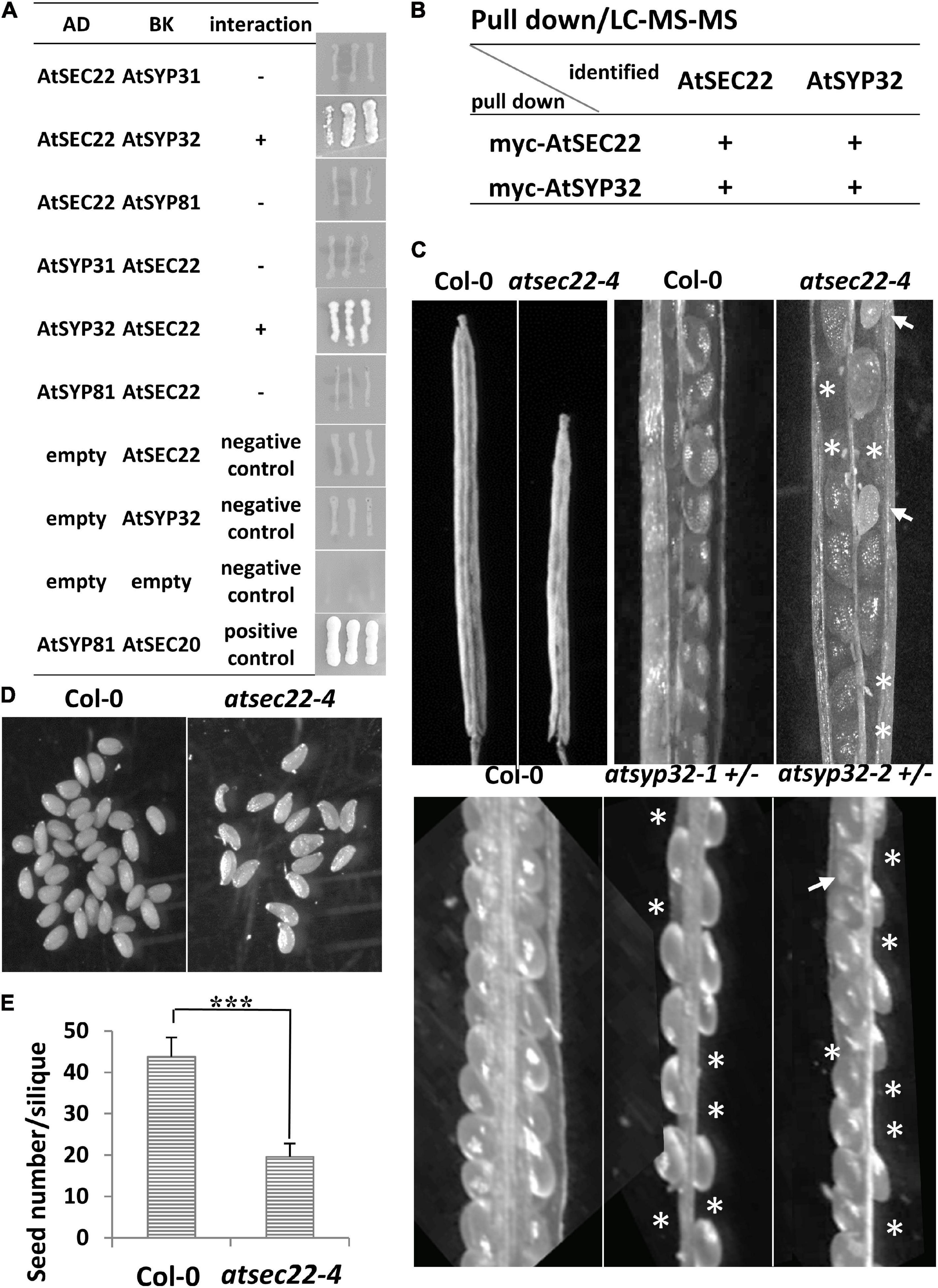
Figure 4. AtSEC22 interacts with Golgi-localized syntaxin AtSYP32. (A) AtSEC22 interacts with AtSYP32 rather than with AtSYP31 and AtSYP81. Plasmid combinations are shown. Yeast strain AH109 was transformed with the paired constructs. Transformants were streaked onto SD/_Leu/_Trp/_His/_Ade medium. +, interaction; –, no interaction; AD, pGADT7; BK, pGBKT7. (B) Pull down assay using myc-AtSEC22-overexpressing plants followed by shotgun LC-MS/MS analysis. As a results, AtSYP32 was identified. Inverse experiments identified AtSEC22. (C) atsec22-4, atsyp32-1±, and atsyp32-2 ± had smaller siliques and were partially abortive. Asterisks, positions of abortive seeds; arrows, poorly developed seeds. (D) Siliques in atsec22-4 had fewer seeds than those in wild-type. The seeds were from one silique. (E) Statistics of panel (D). n ≥ 15. ***P < 0.001. Significance was evaluated by Student’s t test.
Cytoskeleton Organization Is Disturbed in atsec22-4
It has been reported that the cytoskeleton controls trichome morphogenesis, MTs determine trichome formation and branch number, and AFs control trichome morphology (Kim et al., 2002; Mathur, 2005; Szymanski, 2005; Sambade et al., 2014). Therefore, we introduced the MT markers, MBD-GFP and TUA6-GFP, and the AF marker ABD2-GFP into atsec22-4 by crossing. Confocal microscope analysis revealed that the organization of cortical MTs and AFs was altered in atsec22-4. In hypocotyl from five-day-old seedlings grown in the dark, MBD-GFP-visualized cortical MTs in atsec22-4 were thinner and denser than that in the wild-type, in the top, middle, and base parts of hypocotyl (Figures 5A,B). Furthermore, the MT array orientation was also altered (Figure 5C). ABD2-GFP-visualized AFs maintained transverse or oblique alignment in most epidermal cells in wild-type hypocotyl. However, in atsec22-4 hypocotyl, AF arrays became more extensive (Figures 5D,E), and longitudinally aligned AFs and thicker actin bundles were more frequently observed (Figure 5F). All these observations suggested that cytoskeleton organization was disturbed in atsec22-4.
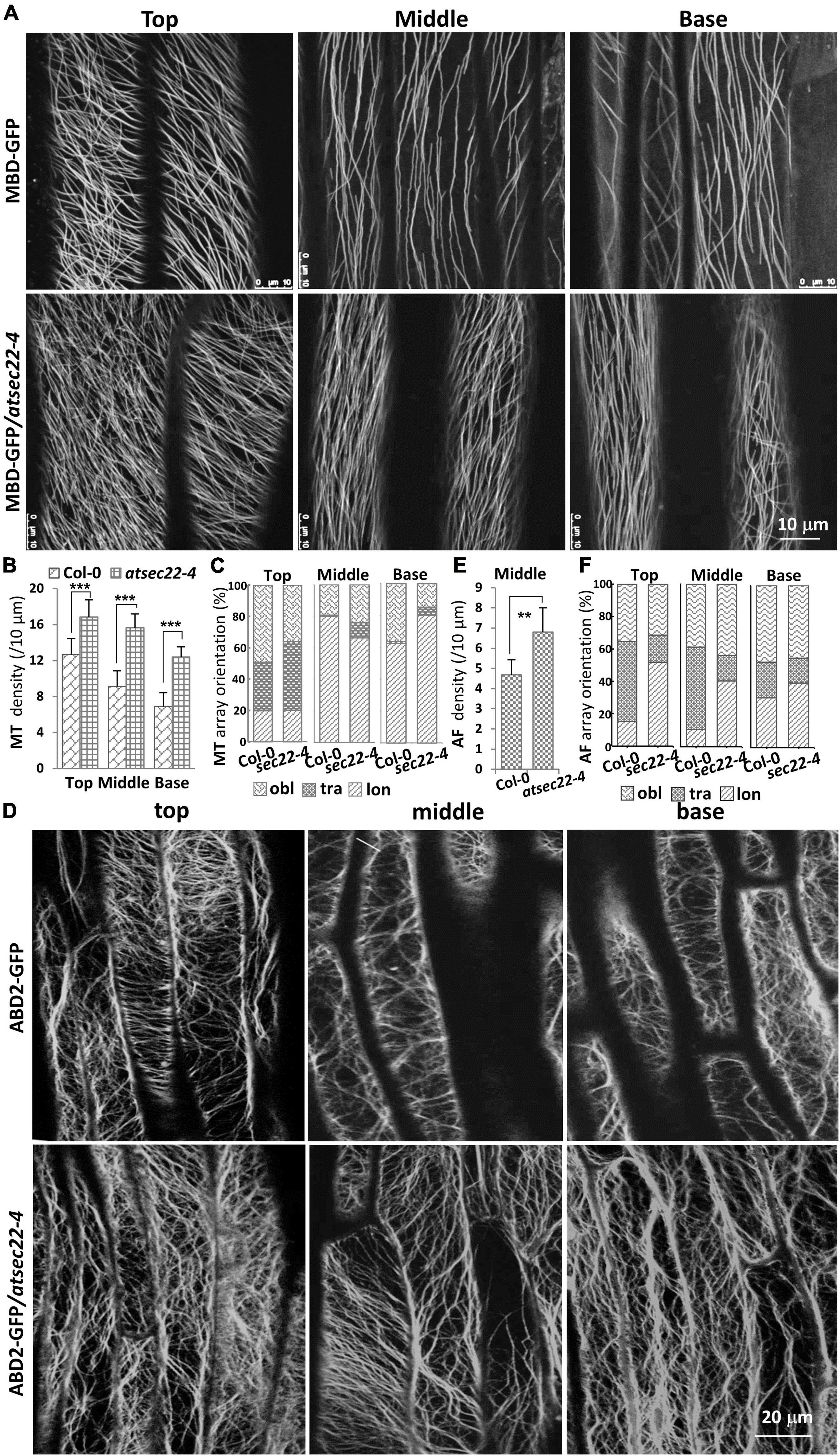
Figure 5. Cytoskeleton organization was disturbed in atsec22-4. (A) Confocal images of MBD-GFP-visualized cortical MTs in the top, middle, and basal regions of hypocotyls from five-day-old seedlings grown in the dark. (B) Statistics of cortical MT density in panel (A). (C) Statistics of cortical MT array orientations in (A). (D) Confocal images of ABD2-GFP-labeled AFs in the top, middle, and basal regions of hypocotyls. (E) Statistics of AF density in middle region of hypocotyls (D). (F) Statistics of AF array orientations in panel (D). lon, longitudinal; tra, transverse; obl, oblique. n ≥ 10 cells. Scale bars are as shown. **P < 0.01, ***P < 0.001. Significance was evaluated by Student’s t test.
AtSEC22 Modulates Cytoskeleton Stability
We further examined the stability of MTs and AFs using the microtubule-disrupting drug, oryzalin, and the actin polymerization inhibitor, LatB, respectively. After 10 min of treatment with 10 μM of oryzalin to hypocotyls and 20 min to the leaves, TUA6-GFP-visualized cortical MTs in wild-type leaf cells (Figure 6A) and MBD-GFP-visualized MTs (Supplementary Figure 6A) in wild-type hypocotyl cells just began to be depolymerized, appearing as GFP-labeled dots. However, in most of the atsec22-4 leaf and hypocotyl cells, cortical MTs were almost completely depolymerized. After 2 h of washing, the alignment of cortical MTs in the wild-type was recovered, whereas that in atsec22-4 was significantly delayed. A large number of dots remained, but they were organized along the MT arrays (Figure 6A and Supplementary Figure 6A). As for AFs, after 20 min treatment with 1 μM of LatB, most of the ABD2-GFP-visualized AFs in wild-type leaf cells lost their linear alignment, whereas AFs in atsec22-4 showed significant delay (Figure 6B). These results suggest that AtSEC22 is vital for cytoskeleton stability.
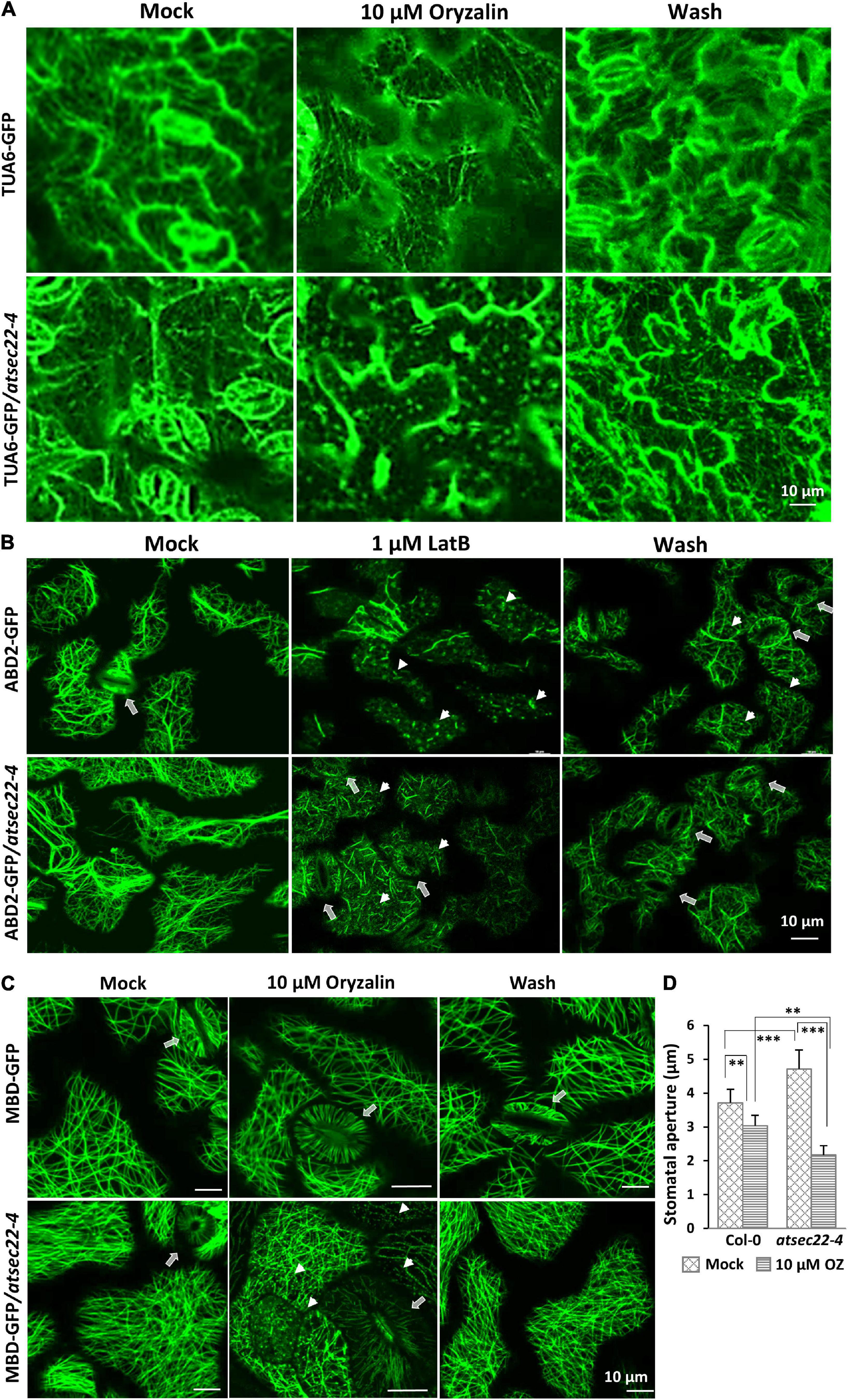
Figure 6. Cytoskeleton stability was perturbed in atsec22-4. (A) Cortical MTs in atsec22-4 were sensitive to oryzalin treatment. TUA6-GFP-visualized cortical MTs in leaves from 12-day-old seedlings were treated with 10 μM of oryzalin for 20 min, and followed by a 2 h wash. (B) Cortical AFs in atsec22-4 were resistant to LatB treatment. ABD2-GFP-visualized cortical AFs in leaves from 12-day-old seedlings were treated with 1 μM of LatB for 20 min, and followed by a 2 h wash. Arrows, stomata; arrow heads, depolymerized AFs. (C) Stomata movement was abnormal in atsec22-4. MBD-labeled cortical MTs in leaves from 12-day-old seedlings were treated with 10 μM of oryzalin for 20 min, and followed by a 2 h wash. Arrows, stomata; arrow heads, depolymerized MTs. (D) Statistics of stomata aperture in panel (C). n ≥ 30. Scale bars are as shown. **P < 0.01, ***P < 0.001. Significance was evaluated by Student’s t test.
Stomatal movement is closely correlated with AF and MT dynamics (Lemichez et al., 2001; Zhang et al., 2007; Zhao et al., 2011; Eisinger W. R. et al., 2012). In wild-type, stomata are usually open with proper apertures. However, in atsec22-4, the stomata seemed open with a relatively wider and uneven aperture (Figure 2E and Supplementary Figure 2D). Following treatment with 10 μM of oryzalin for 20 min, stomata in atsec22-4 closed with a larger range than those in the wild-type (Figures 6C,D). The sensitivity of stomata to oryzalin further indicated altered cytoskeleton stability in atsec22-4.
To determine the protein levels of tubulin and actin in atsec22-4, we performed immunoblot analysis. The results indicated no significant change in tubulin and actin levels in atsec22-4 (Figure 7A), indicating that the abnormalities of the cytoskeleton were not due to the cytoskeletal component proteins. To determine the reasons for defects in cytoskeleton dynamics, we re-analyzed the pull down-LC-MS/MS results and identified some cytoskeleton-associated proteins (Figure 7B). We found that the expression levels of AtMAP65-1, AtMAP65-5, and ADF11 were significantly altered (Figure 7C). We further examined the protein level of MAP65-1, an MT-associated protein required for MT depolymerization and reorganization (Smertenko et al., 2004; Gaillard et al., 2008; Lucas et al., 2011; Zhou et al., 2017), using two antibodies against two different parts of the MAP65-1 protein. The results indicated that the protein levels of MAP65-1 were also dramatically decreased in atsec22-4 (Figure 7D), suggesting that the stability of the cytoskeleton regulators was affected.
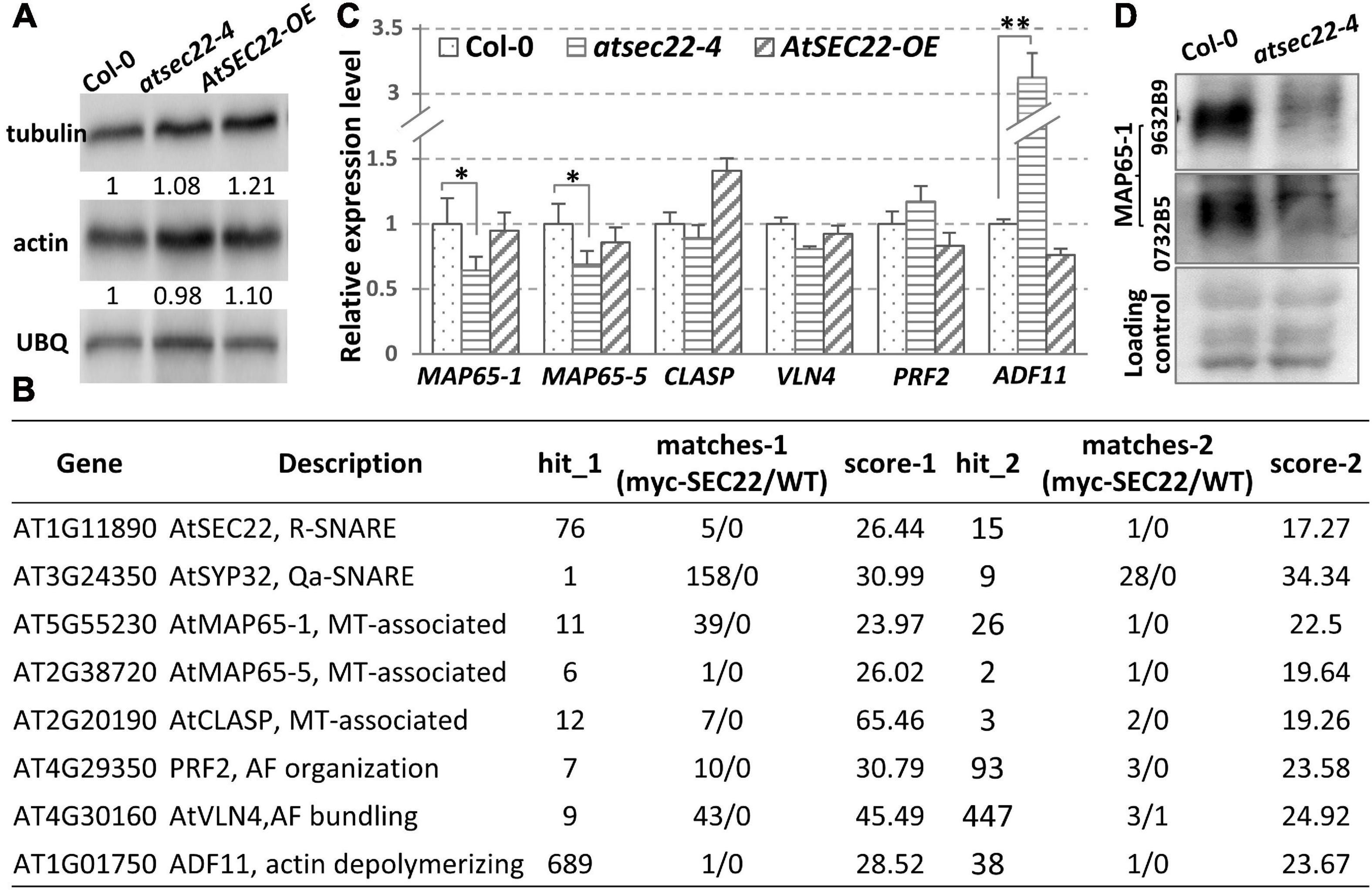
Figure 7. Transcription levels of cytoskeleton-associated proteins were affected in atsec22-4. (A) Immunoblot analysis using ten-day-old seedlings with anti-actin and anti-tubulin antibodies. Ubiquitin (UBQ) was used as endogenous control. (B) LC-MS/MS identified cytoskeleton-regulators from pull down assay using myc-AtSEC22-overexpressing plants. Hit_1, peptide_hit_number_1; score-1, peptide score-1; WT, Col-0. (C) Transcriptional levels of cytoskeleton-regulator genes. Two independent experiments for each sample, three replicates for each experiment. *P < 0.05, **P < 0.01. Significance was evaluated by Student’s t test. (D) Protein levels of MAP65-1 were determined with two anti-MAP65-1 antibodies against two different peptides of MAP65-1 using ten-day-old seedlings.
Discussion
AtSEC22 Regulates Secretion Pathway
SNARE proteins contribute to efficient membrane fusion. In yeast and mammals, Sec22 localizes to the ER and Golgi and regulates anterograde and retrograde transport (Flanagan et al., 2015; Li et al., 2015; Zhao et al., 2015), and Sec22 is recruited to the COPII vesicles by Sec23/24 (Mancias and Goldberg, 2007). Plant SEC22 has been reported to function in the early secretory pathway and is essential for the integrity of the ER network and Golgi complex (Chatre et al., 2005; El-Kasmi et al., 2011). In this study, we found that AtSEC22 interacts with the Golgi-syntaxin AtSYP32. Since AtSYP32 is predicted as a Qa-SNARE localized in cis-Golgi, it is quite possible that AtSEC22 cooperates with AtSYP32 to regulate ER-Golgi transport. However, even if AtSYP32 is usually used as a Golgi marker, its biological function has not been reported so far. Interestingly, interactome analysis of Qa-SNARE proteins revealed that cytoskeleton components were enriched in the SYP3 (SYP31 and SYP32) group (Fujiwara et al., 2014), suggesting potential relations between the cytoskeleton and AtSYP32.
A large amount of intracellular-localized PIN1-GFP was observed in stele cells in atsec22-4. That it resulted from the blocked ER-Golgi trafficking or another transport pathway affected in atsec22-4 needs further investigation to clarify. Because the ER and Golgi morphology were disturbed seriously in atsec22-4, combined with the fact that the Golgi apparatus is the hub for intracellular vesicle trafficking, it is possible that the secretion pathway and potentially also the recycling pathway were defective in atsec22-4. Further investigation is needed to clarify these points.
AtSEC22 Is Involved in Cell Morphogenesis and Plant Development
AtSEC22 has previously been reported to regulate gametophyte development (El-Kasmi et al., 2011), our study found that atsec22-4 exhibited serious developmental and reproduction defects. The impaired morphogenesis in atsec22-4 were due to disrupted organization and stability of the MT and AF cytoskeleton. And the reproduction defects could have resulted from dysfunction of the spindle, which is composed of MTs. These abnormalities might be due to dis-homeostasis of the MT- and AF-associated proteins. These regulators might be synthesized in cytoplasm, however, some of them need modification for activation. Such as the MAP65-1 phosphorylation form which is required for MT depolymerization and reorganization. Its phosphorylation is regulated by Aurora-, cyclin dependent kinases (CDK)-, and MAPK-dependent pathways for different functions (Smertenko et al., 2004, 2006; Sasabe and Machida, 2012; Smékalová et al., 2014; Boruc et al., 2017; Zhou et al., 2017; Vavrdová et al., 2019). These enzymes are usually synthesized in the ER then delivered to the Golgi apparatus. It is quite possible that the export of them from the ER was affected and subsequently disturbed cytoskeleton regulators’ modification in atsec22-4. Reduction of MAP65-1 might be one of the triggers for MT disorganization and/or depolymerization. It has been reported that the morphology of leaf pavement cells in AtMAP65-1-overexpressing plants was significantly altered, and the lobe length was obviously decreased resulting in a smooth cell shape. Whereas the atmap65-1 mutant had denser root hairs, and some of them seemed swollen. Furthermore, the alignment of MTs in both the mutant and overexpression lines was altered (Chen, 2009). MAP65-1 was recently reported to inhibit katanin to bind to MT bundles, thus protecting them from severing (Burkart and Dixit, 2019), suggesting that MAP65-1 is essential for MTs stability. Moreover, MAP65-1 and its homolog MAP65-5 have spatiotemporal colocalization, they are concentrated at the midzones of the spindle during anaphase B and the phragmoplast (Smertenko et al., 2008), and they also colocalize with cortical MTs (van Damme et al., 2004; Lucas et al., 2011), suggesting their functional redundancy. Taken together, AtSEC22 regulates plant morphogenesis and development by controlling cytoskeleton dynamics.
Largely, plant development relies on the pattern of cell development. It is well known that cytoskeleton dynamics are vital for vesicle trafficking. Our findings provide evidence that cytoskeleton dynamics are essential for ER-Golgi trafficking. The impaired alignment of the cytoskeleton in atsec22-4 strongly suggested the dependence of AtSEC22-mediated vesicle trafficking on the cortical MTs and AFs. On the other hand, the cytoskeleton organization and stability are dependent on vesicle trafficking which probably control homeostasis of MT-/AF-associated regulators. Extensive in-depth research is expected to clarify the crosstalk. In conclusion, vesicle trafficking and cytoskeleton are closely interdependent.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LL and XL conceived the project. LL, LG, and XL designed the experiments. LG and SY generated all material used in this study (cloning, transformations, transgenic plants, and crosses). LG, SL, YL, YY, YQL, and TW were involved in the confocal observation, trichome statistics, immunoblot, plant handling, and post-acquisition image analysis. GQ performed the LC-MS/MS. HW generated the TSM images of seeds. J-KZ, YW, XF, and ZW contributed the reagents, materials, and the analytical platform. LL and LG drafted the manuscript. All authors commented on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31570246 and 31570291) and Fundamental Research Funds for the Central Universities (2572019CT03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Ikuko Hara-Nishimura (Kyoto University) for the gifts of NaTAP and pK7GWIWG2 vectors, and GFP-HDEL and ST-GFP seeds; Prof. Shuang Wu (Fujian Agriculture and Forestry University) for providing 35S:MBD-GFP and 35S:TUA6-GFP seeds, and Prof. Zhaosheng Kong (Chinese Academy of Sciences) for pUBQ:ABD2-GFP seeds. We are grateful to Dr. Yimin She, Dr. Li Tan, and Dr. Hong Zhu (Shanghai Center for Plant Stress Biology, CAS Center of Excellence in Molecular Plant Sciences, CAS) for support in mass spectrometry and cell biological analysis. We would like to thank Editage (www.editage.cn) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.635732/full#supplementary-material
Supplementary Figure 1 | Development of atsec22-4 was delayed. (A) Germination of atsec22-4 was delayed. Germination ratio was quantified using three-day-old seedlings. (B) Statistics of panel (A). n ≥ 30. Three independent experiments per sample. ∗∗P < 0.01. (C) atsec22-4 plants were dwarf.
Supplementary Figure 2 | Morphogenesis was disturbed in atsec22-4. (A) There were less leaf trichomes in 40-day-old plants in atsec22-4. (B) Morphology of trichomes in atsec22-4 were altered. Trichomes were from the 3rd/4th rosette leaves in seven-week-old plants. (C) There were less hairs on stems in atsec22-4. (D) Morphology of pavement cells and stomata were altered in atsec22-4.
Supplementary Figure 3 | Plant development and morphogenesis were disturbed in AtSEC22 RNAi lines. (A) Development of AtSEC22 RNAi plant was delayed. (B) Trichome number was decreased in AtSEC22 RNAi leaves. (C) Trichome morphology was altered in AtSEC22 RNAi lines. (D) Statistics of trichome number of panel (B). n ≥ 10 leaves. ∗∗P < 0.01. (E) Statistics of leaf trichomes with different branch number. nCol–0 = 20, nRNAi = 30.
Supplementary Figure 4 | Vesicle trafficking was disturbed in atsec22-4. in atsec22-4. (A) Confocal images of ST-GFP-visualized Golgi apparatus in root cells. ST-GFP was collapsed from the Golgi and redistributed in the ER in atsec22-4 (arrows). (B) Confocal images of GFP-HDEL-visualized ER network in pavement cells. The ER exhibited relatively looser structure with larger space between the ER tubules in atsec22-4 (arrows). (C) TEM images of leaf cells from 21-day-old plants. In atsec22-4, the ER tubules became expanded and fragmented (black arrows), and the Golgi stacks became smaller in width and consisted of less cisternae.
Supplementary Figure 5 | ER stress was induced in atsec22-4. (A) Expression level of BiP3 was detected by RT-PCR. (B) Expression levels of IRE1A, IRE1B and bZIP60 were detected by RT-qPCR.
Supplementary Figure 6 | Microtubule stability was perturbed in atsec22-4. Confocal images of MBD-GFP-visualized cortical MTs in hypocotyls treated with 10 μM of oryzalin for 10 min (A), and followed by a 2 h wash (B). Arrows in panel (A), depolymerized MTs; arrows in panel (B), MTs started to line up.
Supplementary Table 1 | List of primers used in this study.
Supplementary Table 2 | Original data of LC-MS/MS.
Footnotes
References
Akhmanova, A., and Hammer, J. A. III (2010). Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22, 479–487. doi: 10.1016/j.ceb.2010.04.008
Ambrose, C., Allard, J. F., Cytrynbaum, E. N., and Wasteneys, G. O. (2011). A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2:430.
Ambrose, J. C., Shoji, T., Kotzer, A. M., Pighin, J. A., and Wasteneys, G. O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19, 2763–2775. doi: 10.1105/tpc.107.053777
Armour, W. J., Barton, D. A., Law, A. M. K., and Overall, R. L. (2015). Differential growth in periclinal and anticlinal walls during lobe formation in Arabidopsis cotyledon pavement cells. Plant Cell 27, 2484–2500. doi: 10.1105/tpc.114.126664
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/s0092-8674(03)00924-3
Boevink, P., Oparka, K., Cruz, S. S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. doi: 10.1046/j.1365-313x.1998.00208.x
Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. doi: 10.1016/s0092-8674(03)01079-1
Boruc, J., Weimer, A. K., Stoppin-Mellet, V., Mylle, E., Kosetsu, K., Cedeño, C., et al. (2017). Phosphorylation of MAP65-1 by Arabidopsis Aurora kinases is required for efficient cell cycle progression. Plant Physiol. 173, 582–599. doi: 10.1104/pp.16.01602
Brandizzi, F., and Barlowe, C. (2013). Organization of the ER–Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392. doi: 10.1038/nrm3588
Bruinsma, S., James, D. J., Quintana Serrano, M., Esquibel, J., Woo, S. S., Kielar-Grevstad, E., et al. (2018). Small molecules that inhibit the late stage of Munc13-4-dependent secretory granule exocytosis in mast cells. J. Biol. Chem. 293, 8217–8229. doi: 10.1074/jbc.ra117.001547
Burk, D. H., Liu, B., Zhong, R., Morrison, W. H., and Ye, Z. H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–828. doi: 10.2307/3871342
Burkart, G., and Dixit, R. (2019). Microtubule bundling by MAP65-1 protects against severing by inhibiting the binding of katanin. Mol. Biol. Cell 30, 1587–1597. doi: 10.1091/mbc.e18-12-0776
Burri, L., Varlamov, O., Doege, C. A., Hofmann, K., Beilharz, T., Rothman, J. E., et al. (2003). A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 100, 9873–9877. doi: 10.1073/pnas.1734000100
Chatre, L., Brandizzi, F., Hocquellet, A., Hawes, C., and Moreau, P. (2005). Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 139, 1244–1254. doi: 10.1104/pp.105.067447
Chen, J. (2009). The Role of Two Microtubule-Associated Proteins in Cell Morphogenesis. Ph.D.thesis. Beijing: Capital Normal University.
Chen, L., Peng, Y., Tian, J., Wang, X., Kong, Z., Mao, T., et al. (2016). TCS1, a Microtubule-binding protein, Interacts with KCBP/ZWICHEL to regulate trichome cell shape in Arabidopsis thaliana. PLoS Genet. 12:e1006266. doi: 10.1371/journal.pgen.1006266
Christopher, E. R., and Arun, S. (2018). Getting into shape: the mechanics behind plant morphogenesis. Curr. Opin. Plant Biol. 46, 25–31. doi: 10.1016/j.pbi.2018.07.002
de Marcos Lousa, C., Soubeyrand, E., Bolognese, P., Wattelet-Boyer, V., Bouyssou, G., Marais, C., et al. (2016). Subcellular localization and trafficking of phytolongins (non-SNARE longins) in the plant secretory pathway. J. Exp. Bot. 67, 2627–2639. doi: 10.1093/jxb/erw094
Dodonova, S. O., Diestelkoetter-Bachert, P., von Appen, A., Hagen, W. J. H., Beck, R., Beck, M., et al. (2015). A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198. doi: 10.1126/science.aab1121
Draxl, S., Muller, J., Li, W. B., Michalke, B., Scherb, H., Hense, B. A., et al. (2013). Caesium accumulation in yeast and plants is selectively repressed by loss of the SNARE Sec22p/SEC22. Nat. Commun. 4:2092.
Dyachok, J., Sparks, J. A., Liao, F., Wang, Y. S., and Blancaflor, E. B. (2014). Fluorescent protein-based reporters of the actin cytoskeleton in living plant cells: fluorophore variant, actin binding domain, and promoter considerations. Cell Motil. Cytoskeleton 71, 311–327. doi: 10.1002/cm.21174
Eisinger, W., Ehrhardt, D., and Briggs, W. (2012). Microtubules are essential for guard-cell function in Vicia and Arabidopsis. Mol. Plant 5, 601–610. doi: 10.1093/mp/sss002
Eisinger, W. R., Kirik, V., Lewis, C., Ehrhardt, D. W., and Briggs, W. R. (2012). Quantitative changes in microtubule distribution correlate with guard cell function in Arabidopsis. Mol. Plant 5, 716–725. doi: 10.1093/mp/sss033
El-Kasmi, F., Pacher, T., Strompen, G., Stierhof, Y. D., Muller, L. M., Koncz, C., et al. (2011). Arabidopsis SNARE protein SEC22 is essential for gametophyte development and maintenance of Golgi-stack integrity. Plant J. 66, 268–279. doi: 10.1111/j.1365-313x.2011.04487.x
Flanagan, J. J., Mukherjee, I., and Barlowe, C. (2015). Examination of Sec22 homodimer formation and role in SNARE-dependent membrane fusion. J. Biol. Chem. 290, 10657–10666. doi: 10.1074/jbc.m114.626911
Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687–700. doi: 10.1016/j.cell.2004.12.026
Fujiwara, M., Uemura, T., Ebine, K., Nishimori, Y., Ueda, T., Nakano, A., et al. (2014). Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 55, 781–789. doi: 10.1093/pcp/pcu038
Gaillard, J., Neumann, E., Van Damme, D., Stoppin-Mellet, V., Ebel, C., Barbier, E., et al. (2008). Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell 19, 4534–4544. doi: 10.1091/mbc.e08-04-0341
Gürkan, C., Stagg, S. M., LaPointe, P., and Balch, W. E. (2006). The COPII cage: unifying principles of vesicle coat assembly. Nat. Rev. Mol. Cell Biol. 7, 727–738. doi: 10.1038/nrm2025
Hanson, P. I., Heuser, J. E., and Jahn, R. (1997). Neurotransmitter release - four years of SNARE complexes. Curr. Opin. Neurobiol. 7, 310–315. doi: 10.1016/s0959-4388(97)80057-8
Hardwick, K. G., and Pelham, H. R. (1992). SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J. Cell Biol. 119, 513–521. doi: 10.1083/jcb.119.3.513
Hussey, P. J., Ketelaar, T., and Deeks, M. J. (2006). Control of the Actin Cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125. doi: 10.1146/annurev.arplant.57.032905.105206
Idilli, A. I., Morandini, P., Onelli, E., Rodighiero, S., Caccianiga, M., and Moscatelli, A. (2013). Microtubule depolymerization affects endocytosis and exocytosis in the tip and influences endosome movement in tobacco pollen tubes. Mol. Plant 6, 1109–1130. doi: 10.1093/mp/sst099
Kim, G. T., Shoda, K., Tsuge, T., Cho, K. H., Uchimiya, H., Vokoyama, R., et al. (2002). The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 21, 1267–1279. doi: 10.1093/emboj/21.6.1267
Kirik, V., Herrmann, U., Parupalli, C., Sedbrook, J. C., Ehrhardt, D. W., and Hulskamp, M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120, 4416–4425. doi: 10.1242/jcs.024950
Kong, Z., Ioki, M., Braybrook, S., Li, S., Ye, Z. H., Julie Lee, Y. R., et al. (2015). Kinesin-4 functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant 8, 1011–1023. doi: 10.1016/j.molp.2015.01.004
Konopka, C. A., Backues, S. K., and Bednarek, S. Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1c and a clathrin light chain at the plasma membrane. Plant Cell Online 20, 1363–1380. doi: 10.1105/tpc.108.059428
Kotzer, A. M., and Wasteneys, G. (2006). Mechanisms behind the puzzle: microtubule-microfilament cross-talk in pavement cell formation. Can. J. Bot. 84, 594–603. doi: 10.1139/b06-023
Kurokawa, K., Okamoto, M., and Nakano, A. (2014). Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 5:3653.
Lee, H., Noh, H., Mun, J., Gu, C., Sever, S., and Park, S. (2016). Anks1a regulates COPII-mediated anterograde transport of receptor tyrosine kinases critical for tumorigenesis. Nat. Commun. 7:12799.
Lemichez, E., Wu, Y., Sanchez, J. P., Mettouchi, A., Mathur, J., and Chua, N. H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15, 1808–1816. doi: 10.1101/gad.900401
Letourneur, F. O., Gaynor, E. C., Hennecke, S., Démollière, C., Duden, R., Emr, S. D., et al. (1994). Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79, 1199–1207. doi: 10.1016/0092-8674(94)90011-6
Li, F., Tiwari, N., Rothman, J. E., and Pincet, F. (2016). Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc. Natl. Acad. Sci. U.S.A. 113, 10536–10541. doi: 10.1073/pnas.1604000113
Li, L., Shimada, T., Takahashi, H., Koumoto, Y., Shirakawa, M., Takagi, J., et al. (2013). MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J. 76, 781–791. doi: 10.1111/tpj.12347
Li, L., Shimada, T., Takahashi, H., Ueda, H., Fukao, Y., Kondo, M., et al. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant cell 18, 3535–3547. doi: 10.1105/tpc.106.046151
Li, X., Wu, Y., Shen, C., Belenkaya, T. Y., Ray, L., and Lin, X. (2015). Drosophila p24 and Sec22 regulate Wingless trafficking in the early secretory pathway. Biochem. Biophys. Res. Commun. 463, 483–489. doi: 10.1016/j.bbrc.2015.04.151
Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu.Rev.Cell Dev.Biol. 23, 147–174. doi: 10.1146/annurev.cellbio.23.090506.123529
Liu, Y., and Barlowe, C. (2002). Analysis of Sec22p in endoplasmic Reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13, 3314–3324. doi: 10.1091/mbc.e02-04-0204
Lucas, J. R., Courtney, S., Hassfurder, M., Dhingra, S., Bryant, A., and Shaw, S. L. (2011). Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23, 1889–1903. doi: 10.1105/tpc.111.084970
Mancias, J. D., and Goldberg, J. (2007). The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 26, 403–414. doi: 10.1016/j.molcel.2007.03.017
Mathur, J. (2005). The ARP2/3 complex: giving plant cells a leading edge. Bioessays 27, 377–387. doi: 10.1002/bies.20206
Matsushima, R., Hayashi, Y., Kondo, M., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130, 1807–1814. doi: 10.1104/pp.009464
Mooren, O. L., Galletta, B. J., and Cooper, J. A. (2012). Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81, 661–686. doi: 10.1146/annurev-biochem-060910-094416
Morsomme, P., Prescianotto-Baschong, C., and Riezman, H. (2003). The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J. Cell Biol. 162, 403–412. doi: 10.1083/jcb.200212101
Newman, A. P., Shim, J., and Ferronovick, S. (1990). BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol. Cell. Biol. 10:3405. doi: 10.1128/mcb.10.7.3405
Ossipov, D., Schroderkohne, S., and Schmitt, H. D. (1999). Yeast ER-Golgi v-SNAREs Bos1p and Bet1p differ in steady-state localization and targeting. J. Cell Sci. 112, 4135–4142.
Palade, G. (1975). Intracellular aspects of the process of protein synthesis. Science 189, 347–358. doi: 10.1126/science.1096303
Panteris, E., and Galatis, B. (2005). The morphogenesis of lobed plant cells in the mesophyll and epidermis: organization and distinct roles of cortical microtubules and actin filaments. New Phytol. 167, 721–732. doi: 10.1111/j.1469-8137.2005.01464.x
Qin, G., Ma, J., Chen, X., Chu, Z., and She, Y.-M. (2017). Methylated-antibody affinity purification to improve proteomic identification of plant RNA polymerase Pol V complex and the interacting proteins. Sci. Rep. 7:42943.
Rigal, A., Doyle, S. M., and Robert, S. (2015). Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods Mol. Biol. 1242, 93–103. doi: 10.1007/978-1-4939-1902-4_9
Robert, M. (2013). Understanding of leaf development-the science of complexity. Plants (Basel) 2, 396–415. doi: 10.3390/plants2030396
Rothman, E. J. (2014). The principle of membrane fusion in the cell (Nobel Lecture). Angew. Chem. 53, 12676–12694. doi: 10.1002/anie.201402380
Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S. P., et al. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778. doi: 10.1111/j.1365-313x.2004.02328.x
Saito, C., and Ueda, T. (2009). Chapter 4: functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 274, 183–233. doi: 10.1016/s1937-6448(08)02004-2
Sambade, A., Findlay, K., Schaeffner, A. R., Lloyd, C. W., and Buschmann, H. (2014). Actin-dependent and -independent functions of cortical microtubules in the differentiation of Arabidopsis leaf trichomes. Plant Cell 26, 1629–1644. doi: 10.1105/tpc.113.118273
Sasabe, M., and Machida, Y. (2012). Regulation of organization and function of microtubules by the mitogen-activated protein kinase cascade during plant cytokinesis. Cytoskeleton 69, 913–918. doi: 10.1002/cm.21072
Shimada, T., Koumoto, Y., Li, L., Yamazaki, M., Kondo, M., Nishimura, M., et al. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47, 1187–1194. doi: 10.1093/pcp/pcj103
Smékalová, V., Luptovčiak, I., Komis, G., Šamajová, O., Ovečka, M., Doskočilová, A., et al. (2014). Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203, 1175–1193. doi: 10.1111/nph.12880
Smertenko, A. P., Chang, H. Y., Sonobe, S., Fenyk, S. I., Weingartner, M., Bögre, L., et al. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119, 3227–3237. doi: 10.1242/jcs.03051
Smertenko, A. P., Chang, H.-Y., Wagner, V., Kaloriti, D., Fenyk, S., Sonobe, S., et al. (2004). The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16, 2035–2047. doi: 10.1105/tpc.104.023937
Smertenko, A. P., Kaloriti, D., Chang, H. Y., Fiserova, J., Opatrn, Z., and Husseya, P. J. (2008). The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20, 3346–3358. doi: 10.1105/tpc.108.063362
Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., et al. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. doi: 10.1038/362318a0
Spang, A. (2013). Retrograde traffic from the Golgi to the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013391–a013391.
Sundaram, A., Appathurai, S., Plumb, R., and Mariappan, M. (2018). Dynamic changes in complexes of IRE1α, PERK, and ATF6α during endoplasmic reticulum stress. Mol. Biol. Cell 29, 1376–1388. doi: 10.1091/mbc.e17-10-0594
Szymanski, D. B. (2005). Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8, 103–112. doi: 10.1016/j.pbi.2004.11.004
Szymanski, D. B. (2014). The kinematics and mechanics of leaf expansion: new pieces to the Arabidopsis puzzle. Curr. Opin. Plant Biol. 22, 141–148. doi: 10.1016/j.pbi.2014.11.005
Takagi, J., Renna, L., Takahashi, H., Koumoto, Y., Tamura, K., Stefano, G., et al. (2013). MAIGO5 functions in protein export from golgi-associated endoplasmic reticulum exit sites in Arabidopsis. Plant Cell 25, 4658–4675. doi: 10.1105/tpc.113.118158
Takahashi, H., Tamura, K., Takagi, J., Koumoto, Y., Hara-Nishimura, I., and Shimada, T. (2010). MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant Cell Physiol. 51, 1777–1787. doi: 10.1093/pcp/pcq137
Takatani, S., Hirayama, T., Hashimoto, T., Takahashi, T., and Motose, H. (2015). Abscisic acid induces ectopic outgrowth in epidermal cells through cortical microtubule reorganization in Arabidopsis thaliana. Sci. Rep. 5:11364.
Tang, B. L., Wang, Y., Yan, S. O., and Hong, W. (2005). COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta 1744, 293–303.
Tian, J., Han, L., Feng, Z., Wang, G., Liu, W., Ma, Y., et al. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. Elife 19:e09351.
van Damme, D., van Poucke, K., Boutant, E., Ritzenthaler, C., Inzé, D., and Geelen, D. (2004). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136, 3956–3967. doi: 10.1104/pp.104.051623
Vavrdová, T., Šamaj, J., and Komis, G. (2019). Phosphorylation of plant microtubule-associated proteins during cell division. Front. Plant Sci. 10:238. doi: 10.3389/fpls.2019.00238
Vidali, L., Rounds, C. M., Hepler, P. K., and Bezanilla, M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS One 4:e5744. doi: 10.1371/journal.pone.0005744
Vöfély, R. V., Gallagher, J., Pisano, G. D., Bartlett, M., and Braybrook, S. A. (2019). Of puzzles and pavements: a quantitative exploration of leaf epidermal cell shape. New phytol. 221, 540–552. doi: 10.1111/nph.15461
Wang, X., Zhu, L., Liu, B., Wang, C., Jin, L., Zhao, Q., et al. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19, 877–889. doi: 10.1105/tpc.106.048579
Wasteneys, G. O., and Yang, Z. (2004). New views on the plant cytoskeleton. Plant Physiol. 136, 3884–3891. doi: 10.1104/pp.104.900133
Yu, X., Breitman, M., and Goldberg, J. (2012). A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell 148, 530–542. doi: 10.1016/j.cell.2012.01.015
Žárský, V., Cvrčkov, F., Potocký, M., and Hála, M. (2009). Exocytosis and cell polarity in plants-exocyst and recycling domains. New Phytol. 183, 255–272.
Zhang, C., Halsey, L. E., and Szymanski, D. B. (2011). The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol. 11:27. doi: 10.1186/1471-2229-11-27
Zhang, Q., Fishel, E., Bertroche, T., and Dixit, R. (2013). Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23, 2191–2195. doi: 10.1016/j.cub.2013.09.018
Zhang, W., Fan, L.-M., and Wu, W.-H. (2007). Osmo-sensitive and stretch-activated calcium-permeable channels in Vicia faba guard cells are regulated by actin dynamics. Plant Physiology 143, 1140–1151. doi: 10.1104/pp.106.091405
Zhao, X., Guo, X., Tang, X., Zhang, H., Wang, M., Kong, Y., et al. (2018). Misregulation of ER-Golgi vesicle transport induces ER stress and affects seed vigor and stress response. Front. Plant Sci. 9:658. doi: 10.3389/fpls.2018.00658
Zhao, X., Yang, H., Liu, W., Duan, X., and Tong, C. (2015). Sec22 regulates endoplasmic reticulum morphology but not autophagy and is required for eye development in drosophila. J. Biol. Chem. 290, 7943–7951. doi: 10.1074/jbc.m115.640920
Zhao, Y., Zhao, S., Mao, T., Qu, X., Cao, W., Zhang, L., et al. (2011). The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 23, 2314–2330. doi: 10.1105/tpc.111.086546
Keywords: AtSEC22, membrane trafficking, cytoskeleton dynamics, cell morphogenesis, plant growth and development
Citation: Guan L, Yang S, Li S, Liu Y, Liu Y, Yang Y, Qin G, Wang H, Wu T, Wang Z, Feng X, Wu Y, Zhu J-K, Li X and Li L (2021) AtSEC22 Regulates Cell Morphogenesis via Affecting Cytoskeleton Organization and Stabilities. Front. Plant Sci. 12:635732. doi: 10.3389/fpls.2021.635732
Received: 30 November 2020; Accepted: 01 April 2021;
Published: 04 June 2021.
Edited by:
Csaba Mathe, University of Debrecen, HungaryReviewed by:
Luciana Renna, Michigan State University, United StatesCaiji Gao, South China Normal University, China
Copyright © 2021 Guan, Yang, Li, Liu, Liu, Yang, Qin, Wang, Wu, Wang, Feng, Wu, Zhu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixin Li, bGl4aW5saTA1MTVAbmVmdS5lZHUuY24=
 Li Guan
Li Guan Shurui Yang2
Shurui Yang2 Yi Yang
Yi Yang Guochen Qin
Guochen Qin Tao Wu
Tao Wu Zhigang Wang
Zhigang Wang Yongrui Wu
Yongrui Wu Xugang Li
Xugang Li Lixin Li
Lixin Li