
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 10 March 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.634393
This article is part of the Research Topic Photoperiod Control of Growth and Reproduction View all 15 articles
The length of the day (photoperiod) is a robust seasonal signal originated by earth orbital and translational movements, a resilient external cue to the global climate change, and a predictable hint to initiate or complete different developmental programs. In eukaryotic algae, the gene expression network that controls the cellular response to photoperiod also regulates other basic physiological functions such as starch synthesis or redox homeostasis. Land plants, evolving in a novel and demanding environment, imbued these external signals within the regulatory networks controlling organogenesis and developmental programs. Unlike algae that largely have to deal with cellular physical cues, within the course of evolution land plants had to transfer this external information from the receiving organs to the target tissues, and mobile signals such as hormones were recruited and incorporated in the regulomes. Control of senescence by photoperiod, as suggested in this perspective, would be an accurate way to feed seasonal information into a newly developed function (senescence) using an ancient route (photoperiodic signaling). This way, the plant would assure that two coordinated aspects of development such as flowering and organ senescence were sequentially controlled. As in the case of senescence, there is growing evidence to support the idea that harnessing the reliability of photoperiod regulation over other, more labile signaling pathways could be used as a robust breeding tool to enhance plants against the harmful effects of climate change.
Due to their particular static nature, plants have adapted a high number of interconnected pathways that respond to external and internal stimuli to execute their development programs (Pajoro et al., 2014; Jing and Lin, 2020). Inherently, plants must mature in a plastic way that ensures that their development programs are closely coordinated with the seasonal changes in their environment. In this way, they can optimize all physiological decisions by synchronizing them with the correct time of the year and growth stage (Casal et al., 2004). Each plant species has thus optimized their developmental plans for their particular habitats to maximize growth and the production of offspring. Therefore, to understand and predict plant behavior at any particular physiological stage and organ, we need to interconnect all this information. This could be crucial to protect existing plants or design new varieties capable of coping with the unpredictable weather conditions promoted by global climate change (GCC; Nicotra et al., 2010).
Arabidopsis thaliana as an annual model plant has provided a wealth of developmental information, which can be applied to other species, including crops (Ferrier et al., 2011), so this review will focus on annual plants. The advent of the genomic era and the generation of a massive amount of data on plant development from Systems Biology experiments in recent years have increased the need for using computer-aided approaches to handle the accumulated Terabytes of information (Kinoshita and Richter, 2020). However, as already mentioned, this complexity reflects the complex developmental responses of plants to internal and environmental changes. That is why our ability to interconnect different pathways becomes increasingly important to understand the behavior of plants (Franks and Hoffmann, 2012; Majeed et al., 2020; Zhang et al., 2020).
The correct response to external physical stimuli such as light or temperature is critical for the survival of any organism, and early plants developed a complex gene network to respond successfully to them (Serrano-Bueno et al., 2017; Cheng et al., 2019). With the increasing signaling complexity of the new aerial habitats and the production of new organs (Bowman et al., 2017; de Vries and Archibald, 2018), land plants developed new forms of regulation that included transportable signals such as florigens, tuberigens, signal peptides, and hormones, among other mobile effectors (Thomas et al., 2009; Wang et al., 2015; Briones-Moreno et al., 2017; Figure 1). This may explain why evolutionarily modern and complex developmental programs, such as flower formation or senescence, are deeply intertwined with hormonal signals (Thomas et al., 2009), whereas ancient physiological responses, such as photosynthesis modulation or photoperiodic signaling in the leaf or algae, often respond to more physical stimuli such as changes in light or temperature (Serrano-Bueno et al., 2017). For example, during the flowering process in Arabidopsis, there is a relatively low abundance of hormonal regulation in early photoperiodic responses in the leaf, whereas hormones play a more important role in the shoot apical meristem (SAM) and in the stages later in flower development (Lee et al., 2019; Sang et al., 2020).
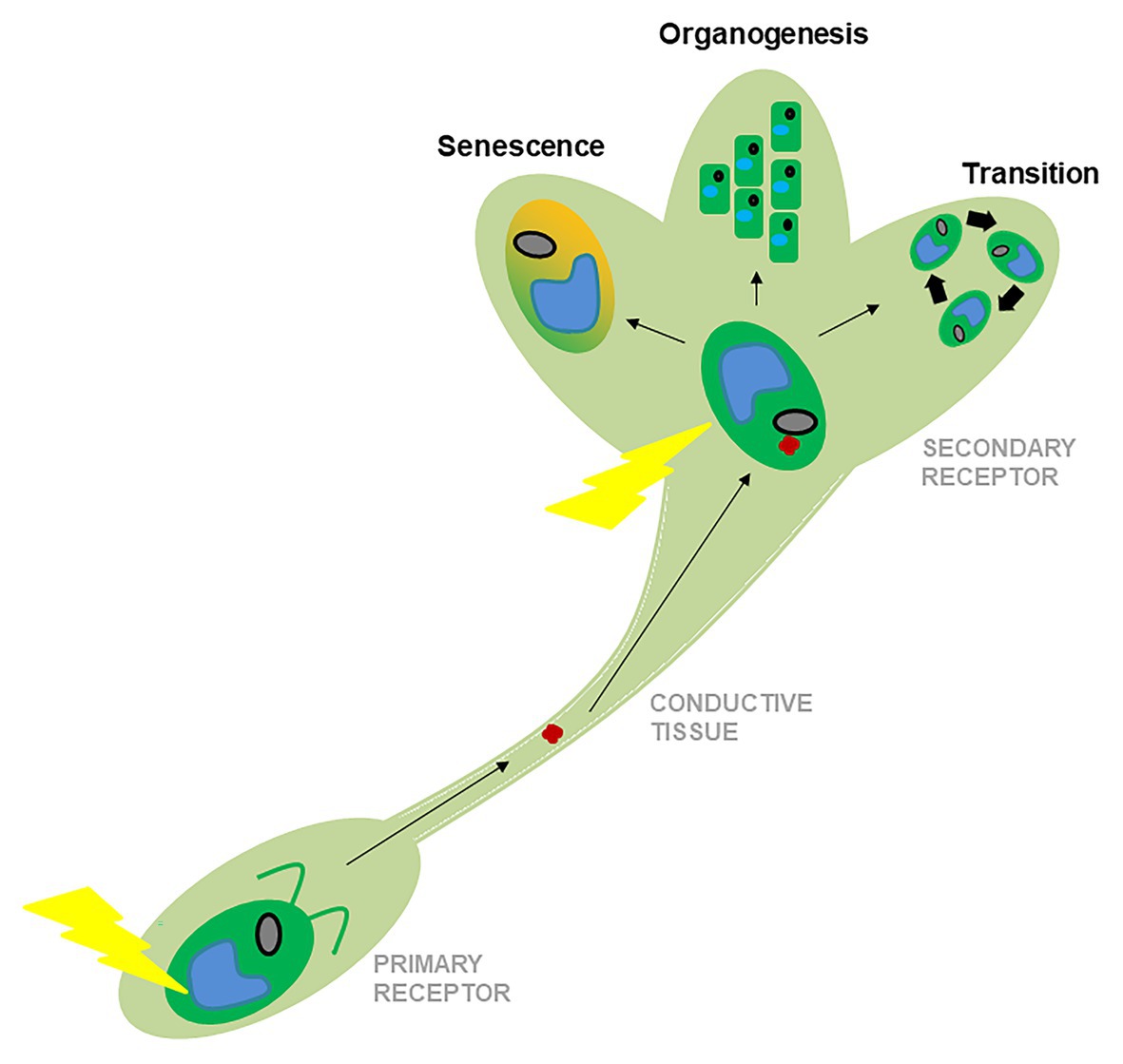
Figure 1. Schematic representation of the evolution of the responses to environmental stimuli from algae to land plants. The primary receptor, like a leave (represented by an algal cartoon, below), responds primarily to physical stimuli (yellow ray). The secondary receptor, like the SAM (represented by a shoot cartoon, above) receives mobile signals that move along the conductive tissues (red dot). After receiving reinforcement from physical stimuli, target organs make different developmental decisions represented here by phase transitions, organogenesis, or senescence.
In this review, we propose a connection between two processes generally considered independently, such as the photoperiodic response and the senescence program (Michelson et al., 2018). Recent results show a seasonal input in the maturation and senescent programs (Körner and Basler, 2010; Kim et al., 2016), which allow us to propose a model through which photoperiod and senescence would be coordinated to ensure a correct developmental program in the plant life cycle.
Senescence is a naturally-occurring phenomenon that involves a gradual decline of functional cells and tissues (Van Deursen, 2014). In many plants, senescence is the final stage in their developmental programs, eventually leading to the death of the organism. However, despite its apparently deteriorating character, it is often a tightly controlled process whose main objective is to allow recycling, remobilization, and reassignment of nutrients from decaying tissues to developing organs (Wen et al., 2020). In plants with short life cycles, this recycling takes place in seeds or fruits, whereas in perennial plants, it mainly happens in storage organs such as stems or roots (Gan and Amasino, 1997; Lim et al., 2007). In annual species, this process provides enough resources for the initiation, progression, and culmination of its reproductive stage, while in perennial species it often implies the beginning of a resting vegetative stage (Woo et al., 2019).
Plant senescence is the result of massive physiological, biochemical, and metabolic changes that take place in all organs, but which have been well described in leaves and flowers. Although essentially dependent on age, senescence occurs when multiple internal and external signals are integrated into age-related information through different regulatory pathways (Buchanan-Wollaston et al., 2003; Lim et al., 2007; Majeed et al., 2020). Considering their given spatial and temporal niches, plants can fine-tune the onset, progression rate, and nature of senescence to ensure successful offspring production and survival. Therefore, senescence is not only a precisely, fine-tuned process for the controlled degradation of macromolecules, but it is also considered a refined evolutionary strategy that plants have acquired to ensure reproduction and survival (Thomas and Stoddart, 1980; Buchanan-Wollaston et al., 2003; Lim et al., 2007; Thomas et al., 2009; Zhang et al., 2020).
The senescence process involves many morphological, cytological, physiological, and molecular changes that are regulated and carried out following a specific order (Wojciechowska et al., 2018). In this section, we will briefly describe how senescence initiation and progression are regulated at organ-specific level.
Leaf senescence is a degenerative process that culminates in the death of leaf cells, and during which they undergo well-defined cell structure and metabolic changes, as well as modifications in gene expression (Lim et al., 2007). Progression of leaf senescence is characterized by a change from assimilation to remobilization of nutrients and the involvement of degenerative events in cellular structures (Masclaux et al., 2000). The earliest cell structural change involves the progressive loss of functionality and breaking down of chloroplasts, where up to 70% of the leaf protein is contained. Concomitantly, a drastic metabolic shift in the chloroplast from anabolism to catabolism takes place, and chlorophyll is massively degraded together with other macromolecules such as RNAs, structural lipids, and proteins. This issue leads to the green-to-yellow color change of leaves that is visible during grain ripening and maturation in crops and during autumn in trees and other perennial plants (Thomas and Stoddart, 1980; Gan and Amasino, 1997; Buchanan-Wollaston et al., 2003; Lim et al., 2007). Unlike chloroplasts, the nucleus and mitochondria remain intact from the onset of senescence until their last stages (Lim et al., 2007). In Arabidopsis, mitochondrial integrity and energy production via respiration are maintained along the senescence process, although their numbers diminish significantly (Chrobok et al., 2016).
Flower senescence is the terminal phase of its development and includes flower wilting, blossoms fading, and the shedding of floral sub-structures (Tripathi and Tuteja, 2007). Regulation of flower lifespan is not only essential to ensure that its maintenance is energetically cost-effective for the plant, but also to avoid flowers being misused after fulfilling their role (Ashman and Schoen, 1994; Rogers, 2006). Petals constitute relatively simple organs with similar characteristics to leaves that can be used as a useful model to study the regulation of senescence. The senescence of the petal is the final stage of its development and constitute a tightly regulated programmed cell death process (PCD; Rogers, 2006, 2013; Van Doorn and Woltering, 2008). Although common physiological and biochemical changes are shared between petal and leaf senescence, both processes differ in terms of reversibility, nutrient remobilization purposes, and speed of progression (Ma et al., 2018). Furthermore, flower or petal senescence patterns exhibit a wide variation across species, being flower wilting or withering followed by abscission the most prominent and visibly shown events (Van Doorn and Woltering, 2008; Shahri and Tahir, 2011).
As plant development progresses, many of the physiological stages are regulated by hormones, often coordinating a complex response. Senescence is not an exception, and many hormones play an important role in the process (Figure 2). With the aim of comparing photoperiod and senescence, this section briefly describes the hormonal control of senescence.
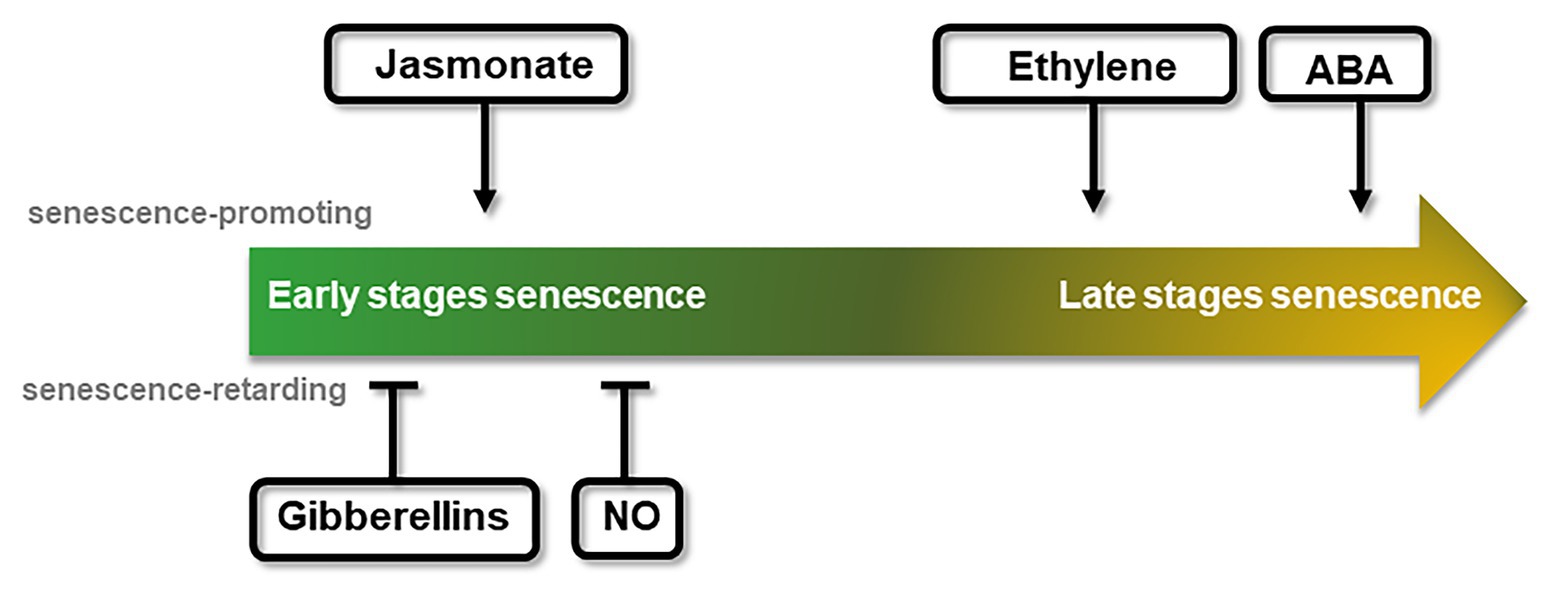
Figure 2. Schematic representation of the chronological effect of different phytohormones on senescence progression. Promotion or delay of senescence can take place at different stages throughout the course of the senescence process, reflected by the central arrow stream. The effect of each hormone or signaling molecule is indicated on the approximate time of their effect. Arrows indicate positive effects over senescence (senescence promoting, above), while bars indicate negative effects (senescence retarding, below).
Jasmonate (JA), ethylene, and abscisic acid (ABA) are regarded as senescence-inducing hormones (Jing et al., 2005; Jibran et al., 2013; Hu et al., 2017; Ma et al., 2018). An external addition of methyl jasmonates (MeJA) led to a rapid loss of chlorophyll and photochemical efficiency, as well as to an increased expression of developmental senescence-associated genes (SAGs; Xiao et al., 2004; Jung et al., 2007), whereas the expression of photosynthesis-associated genes was reduced (Jibran et al., 2013). JA content was reported to increase during the progress of senescence (He et al., 2002). Consistent with increasing JA levels during leaf and flower senescence, genes involved in JA synthesis and signaling pathways showed an increased expression during organ senescence (Porat et al., 1993, 1995; He et al., 2002; Van Der Graaff et al., 2006; Breeze et al., 2011). Also, a raise in transcript abundance of JA biosynthetic genes has been found previous to any visible signs of chlorophyll loss, suggesting a JA role from early stages of leaf senescence (Figure 2; Jibran et al., 2013). Curiously, the JA-insensitive mutant coi1-1 from CORONATINE INSENSITIVE 1 (COI1) does not show signs of JA-induced leaf senescence (He et al., 2002), although the repression of Rubisco activase (RCA) observed in coi1-1 has been described as a mechanism by which increased JA content can promote senescence (Shan et al., 2011).
In a similar way to JA, ethylene application accelerates leaf and flower senescence, while inhibition of its synthesis or signaling promotes senescence delay (Buchanan-Wollaston et al., 2005; Jing et al., 2005; Kim et al., 2014). Similarly, a reduced expression of the enzyme involved in the ethylene biosynthesis, 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO), delayed flower senescence and flower abscission in some cultivars of petunia, torenia, and carnation (Savin et al., 1995; Aida et al., 1998; Huang et al., 2007; Tan et al., 2014). Mutant plants in ethylene signaling (ethylene-insensitive2, ein2) also displayed an arrest in developmental senescence (Buchanan-Wollaston et al., 2005). Another central factor of ethylene signaling, ETHYLENE-INSENSITIVE3 (EIN3), was shown to activate two senescence-promoting transcription factors (TFs), ORE1 and AtNAP, that positively regulate leaf senescence (Kim et al., 2014). Increase of transcript abundance of ethylene synthesis and signaling genes has been found to occur in the same timeframe in which a decline of chlorophyll concentration and transcripts of photosynthetic genes is observed, which suggests that ethylene promotes the latter stages of leaf senescence (Figure 2; Breeze et al., 2011).
As in the case of JA, endogenous ABA levels increase in leaf tissues as they mature, which is accompanied by the upregulation of genes associated with biosynthesis and signaling of ABA (Philosoph-Hadas et al., 1993; Buchanan-Wollaston et al., 2005; He et al., 2005; Van Der Graaff et al., 2006; Breeze et al., 2011). Exogenous application of ABA promotes senescence and abscission (Figure 2; Nooden, 1988; Borochov and Woodson, 1989; Becker and Apel, 1993; Panavas et al., 1998; Yang et al., 2002) and plants under environmental stresses showing leaf senescence have an increased ABA content in their leaves (Lim et al., 2007; Sah et al., 2016). Moreover, ABA regulates the expression of SAGs (Zhang and Gan, 2012; Gao et al., 2016; Zhao et al., 2016; Asad et al., 2019). Regarding the phase in which they function, different studies have pointed out to an effect of ABA on leaf senescence that depends on age, concomitant with rising of ABA levels in later stages of flower development. This suggests that ABA may play a role in the enhancement of senescence rather than in its onset (Figure 2; Hunter et al., 2004; Lee et al., 2011; Arrom and Munné-Bosch, 2012; Zhang et al., 2012; Gao et al., 2016).
On the contrary, the phytohormone gibberellin (GA) and the gaseous signaling molecule nitric oxide (NO) have been reported as senescence-retarding effectors whose content declines during the progression of developmental senescence (Figure 2; Schippers et al., 2007; Procházková and Wilhelmová, 2011; Bruand and Meilhoc, 2019). Different studies in GA biosynthesis or GA signaling deficient mutants further support GA role as a negative player in regulating senescence (Van Der Graaff et al., 2006; Chen et al., 2014; Lü et al., 2014). In Arabidopsis, expression of the GA deactivating enzyme GA 2-oxidase 2 was reported to be increased during senescence (Van Der Graaff et al., 2006), while silencing of the GA biosynthetic gene GA 20-oxidase resulted in accelerating petal senescence in cut rose (Lü et al., 2014). Leaf senescence in Arabidopsis was retarded in the GA biosynthesis mutant ga1-3, in which negative regulators of GA signaling pathways abnormally accumulate. Regarding the gaseous signaling NO, exogenous application of NO or NO-donor compounds extended fruits and vegetables post-harvesting life and arrested the senescence of flowers (Leshem et al., 1998). Although NO has been linked to other molecules involved in senescence, no mechanism of NO-preventing effect over leaf senescence has been described yet. Different studies using NO-deficient Arabidopsis plants have demonstrated that NO regulates expression of photosynthetic genes and SAGs (Mishina et al., 2007; Liu and Guo, 2013). The recent identification of TFs that respond to NO levels in Arabidopsis (Imran et al., 2018) can pave the way to further understand how NO contributes to the regulation of senescence.
In any living organisms, changes in developmental processes throughout the year often define their living strategy. This is particularly true of annual plants, such as Arabidopsis, as well many of the crops that feed humanity (rice, corn, and wheat), as they have to precisely plan for germination, growth, reproduction, and senescence to complete a life cycle in 1 year (Preston and Fjellheim, 2020). The way they respond to each seasonal change is also a tactical decision, for example, to coincide with pollinators, outsmart potential opponents, or conversely, modify flowering time to avoid competition (Franks and Hoffmann, 2012). These decisions are closely related to how they respond to fluctuating seasonal changes in environmental conditions and have evolutionarily shaped how their developmental programs respond today.
For a plant, a particularly reliable seasonal change is day length, since its constant change throughout the year establishes the succession of seasons and indicates the duration and intensity of energy availability. Therefore, day length has been used since early in plant evolution as a reliable source of information for making crucial developmental decisions (Serrano-Bueno et al., 2017). In the green alga Chlamydomonas, day length (photoperiodic) decisions regulate starch accumulation, reproductive behavior, cell division program (Serrano et al., 2009), photosynthesis protection (Tokutsu et al., 2019), or the retrograde signal from the chloroplast to the nucleus (Gabilly et al., 2019). This evolutionarily conserved mechanism also regulates flowering time and starch synthesis in higher plants, but involves a much larger number of genes, reflecting how evolution often responds to increasingly demanding complexity by amplifying the gene network associated with it (Ortiz-Marchena et al., 2014). But it also indicates that a seasonal detection system based on photoperiod signals was established very early in evolution and still governs many physiological responses in plants (Romero-Campero et al., 2013).
One of the best studied photoperiodic responses in higher plants is the floral transition (Andrés and Coupland, 2012; Kinoshita and Richter, 2020). It is becoming increasingly clear that a conserved central module in the developmental processes of plants is designed to receive, process, and transfer signals coming from changes in day length in the leaves to decide the precise moment of the floral transition (Song et al., 2018). This central floral module dates back to gymnosperms, evolved from an ancestral algal system to regulate photoperiodic signaling (Serrano-Bueno et al., 2017), and is conserved in monocots and dicots (Shrestha et al., 2014). The core gene module consists of genes that encode a family of B-Box proteins called BBX or more specifically CONSTANS-like (COL) that can transfer light and time information (from the circadian clock) to the developmental regulatory program (Valverde, 2011; Shim et al., 2017). The presence of these central TFs of which CONSTANS (CO or BBX1) in Arabidopsis was the first to be identified (Putterill et al., 1995), must be strictly controlled to assure a perfectly synchronized floral transition (Suárez-López et al., 2001; Valverde et al., 2004). In this way, CO expression is controlled at the transcriptional level by a set of TEMPRANILLO (TEM), BHLH (FBHs), and DOF (CDFs) TFs (Castillejo and Pelaz, 2008; Fornara et al., 2009; Ito et al., 2012) whose expression is simultaneously controlled by microRNAs, photoreceptors (PHYs and CRYs), clock genes (GIGANTEA, GI), and LOV-containing ubiquitin ligases (Mizoguchi et al., 2005; Sawa et al., 2007; Kubota et al., 2017), a bryophyte design that is capable of detecting light and sending proteins for degradation through the proteasome (members of the ADAGIO family of E3 ligases such as ZLP or FKF; Song et al., 2014). These set of proteins ensures that CO expression is high in the leaves during the day in Arabidopsis only in long days (LD) but not in short days (SD; Suárez-López et al., 2001; Figure 3A).
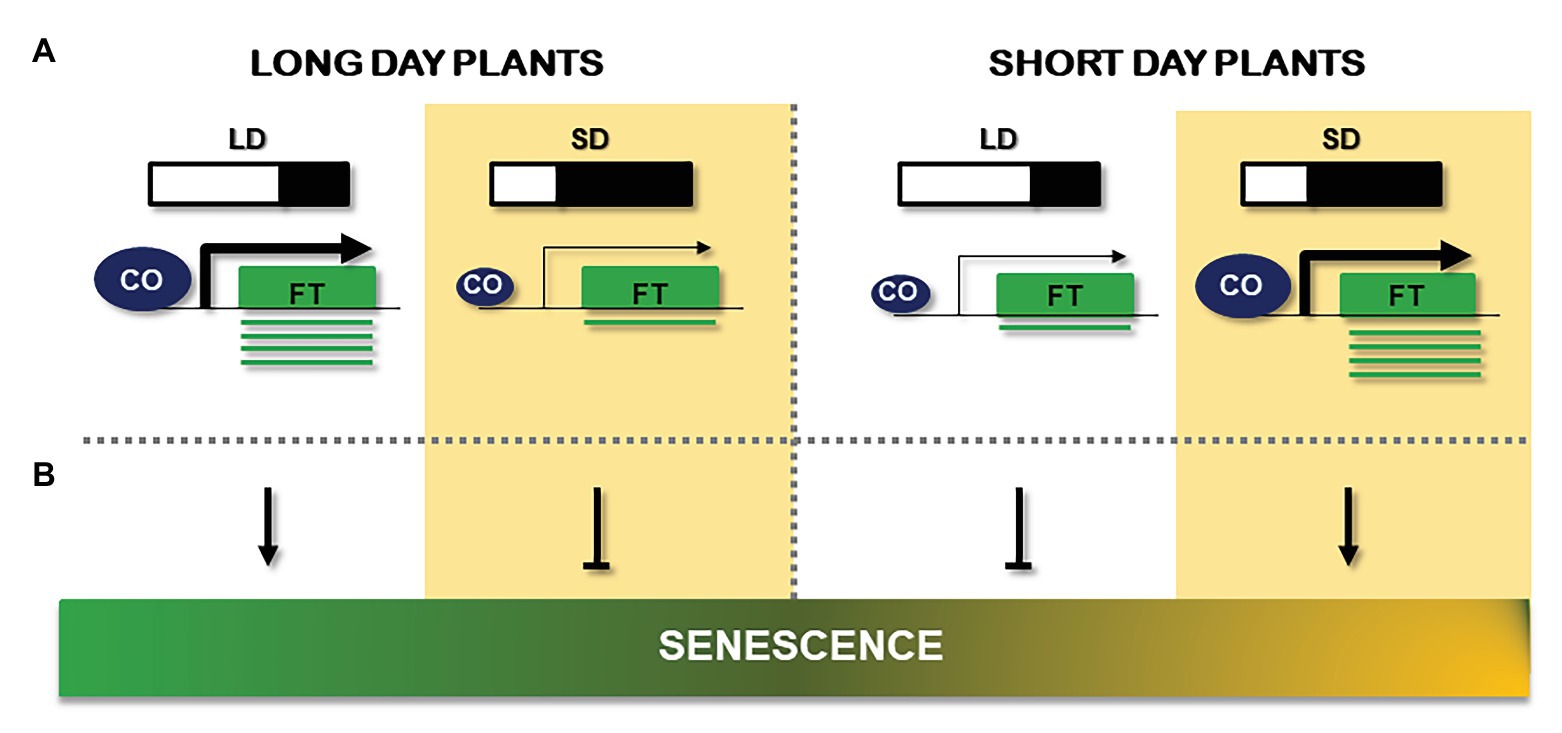
Figure 3. Comparison of the effect of long days (LD) and short days (SD) signals on flowering and senescence. (A) Schematic description of day length effect on flowering and its capcity to activate FT gene (green box) expression in LD plants (left) and SD plants (right). CONSTANS (CO) abundance is represented by the size of the blue circle, while the arrow size reflects its capacity to activate FT mRNA (green line) production. (B) Observed results of LD or SD on senescence. Length of the day is represented by a light/black diagram and a white background (LD) or yellow background (SD). Arrows indicate positive effects; bars indicate repressive effects.
But a simultaneous posttranslational regulatory level is needed to fully confer the day length information to the core photoperiodic floral regulome of the leaf (Valverde et al., 2004; Shim et al., 2017). In this way, CO is controlled at the protein level by a specific association with a set of ring-finger E3 ligases (SPA1, COP1, and HOS1) that are activated through the interaction of CRYs and PHYs, thus transmitting a second light information level to the photoperiod pathway (Jang et al., 2008; Liu et al., 2008; Lazaro et al., 2012). In Arabidopsis leaves, stable and active CO protein in the evening of a LD is able to associate with NF-YB, NF-YC TFs, substituting NF-YA from the trimeric conformation (Wenkel et al., 2006). The CO/NF-YB/NF-YC trimeric complex is capable of interacting with DNA and specifying transcriptional activation at CO responsive element (CORE) sites of target promoters, such as the florigen FT (Tiwari et al., 2010; Shen et al., 2020) or the starch synthase GBSSI (Ortiz-Marchena et al., 2014). In fact, this trimeric conformation is observed in Chlamydomonas (Tokutsu et al., 2019) and possibly in other proteins of the CONSTANS, CONTANS-LIKE, TOC1 (CCT) family (Shen et al., 2020). This so-called external coincidence model of CO protein explains why Arabidopsis and other long day plants will flower earlier in LD than in SD (Andrés and Coupland, 2012; Yang et al., 2014). But in other short day plants, CO protein has an almost opposite role, functioning as a repressor of FT expression in LD and activating FT expression in SD as in rice or Pharbitys species (Hayama et al., 2003, 2007).
In long day plants, therefore, CO will activate FT expression in the leaves during LD and will function as a repressor in SD (Samach et al., 2000; Luccioni et al., 2019), while in short day plants, CO will function in a repressing complex in LD and activate transcription in SD (Hayama et al., 2003; Figure 3A). How CO is able to differentiate both stages and function as a repressor or activator is not fully understood, but it could be at the core of making a plant long day or short day flowering. The so-called neutral plants, which are not able to respond to changes in day length, such as tomato, often present a defective photoperiod response or have lost some of the regulatory components that would respond to light signals (Cao et al., 2015; Gaudinier and Blackman, 2019).
The activation of CO in the leaves is a physical phenomenon that depends upon light density, quality, and exposure length, a complex regulatory mechanism originated from a relatively simple algal toolkit (Serrano-Bueno et al., 2017). When life on land evolved into aerial structures that allowed reproduction independently from water, floral structures, and seeds were created (Pires and Dolan, 2012; Morris et al., 2018), but then, the external information had to be transported from the photosynthetic tissues where it was originated, to the meristems where the reproductive structures were produced. Therefore, different long-distance effectors were designed to transfer developmental and physiological signals from receiving organs to target tissues such as the tuberigen StSP6A (Navarro et al., 2011), the metabolic signal HY5 (Chen et al., 2016), or the clock signal ELF4 (Chen et al., 2020a). In the case of the floral transition, the main florigenic signal is the production of the protein FT in the leaves and its controlled transport to the SAM (Corbesier et al., 2007; Liu et al., 2013). Briefly, the transformation of the vegetative apical meristem into a floral meristem starts with the import of FT into the apical cells via the phloem (Abe et al., 2019). Once in the first layers of the SAM, FT can interact with a 14-3-3 chaperon that allows the binding of the TF FLOWERING LOCUS D (FD), and this so-called florigen complex (FC) is then able to activate the expression of other TFs like SUPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), LEAFY (LFY), or APETALA1 (AP1) that eventually activate the cascade of MADS box TFs producing the different floral whorls (Abe et al., 2005; Wigge et al., 2005; Collani et al., 2019). In fact, QTL analyses have shown that senescence is influenced by functional alleles of the FT repressor, MADS box TF FLOWERING LOCUS C (FLC), and its positive regulator FRIGIDA (FRI), whose expression levels negatively correlated with those from senescence-induced genes as well as the floral promoters FT and SOC1 (Wingler, 2011). These studies provide a link between flowering and senescence in Arabidopsis independent of photoperiodic signaling.
However, several experiments indicate that CO regulation and function in development maybe more complex than above described, such as participating in an interplay between CO and GA signaling (Andrés et al., 2014), having an active role on stomata opening (Ando et al., 2013) and promoting a link with the circadian clock by the interaction with PRR proteins (Hayama et al., 2017), among others (Kinoshita and Richter, 2020). Recently, a protective role for chloroplast photo redox defense and retrograde signaling has been reported in algae (Gabilly et al., 2019) that could also be conserved in plants, as well as an active role in sugar mobilization from starch (Ortiz-Marchena et al., 2014) and a regulation by phosphorylation (Sarid-Krebs et al., 2015; Chen et al., 2020b). The complex aspects of CO regulation, the different roles it is playing and its presence in different organs suggest that seasonal information is not only controlling floral transition but also other important physiological processes (Valverde, 2011). Here, we present some evidences that suggest that CO may also be involved in senescence by providing a seasonal input to this important developmental process.
Many physiological processes in plants are affected by photoperiodic signaling, and particularly important for this perspective review, they include flowering and senescence (Nooden et al., 1996; Valverde, 2011). In general terms, Arabidopsis developmental processes are accelerated under LD. In this sense, Kim et al. (2016) compared the expression of the senescence marker SENESCENCE 4 (SEN4) in leaves of Arabidopsis plants grown under LD and SD conditions. SEN4 expression increased under both conditions; however, the increase was higher under LD than under SD, suggesting a possible senescence dependence on photoperiod. This effect was also observed in the long day plant Pea (Pisum sativum L.). A pea early flowering genetic line named G2, showed early apical senescence under LD, while in SD, it extended the reproductive phase and showed delayed apical senescence (Proebsting et al., 1976, 1978). Parrott et al. (2012) showed an acceleration of leaf senescence associated to LDs in barley (Hordeum vulgare L.). Under LD, they observed the beginning of leaf senescence at day 77 after sawing, while under SD treatment, the first symptoms of leaf senescence showed up at day 105. Another point of connection between photoperiod and senescence is mediated by the FBHs TFs. In Arabidopsis, overexpression of FBH4 promoted a high increase in CO levels and led to an early flowering phenotype, while CO expression was reduced in the fbh1-4 mutant (Ito et al., 2012). In Petunia flowers (Petunia hybrida), PhFBH4 levels were significantly increased during senescence, indicating a possible connection with photoperiodic signaling (Yin et al., 2015). PhFBH4 overexpression line showed early flower senescence, whereas phfbh4 antisense silencing lines extended flower longevity. In addition, the expression of senescence associated genes (SAG12 and SAG29) was drastically altered in Petunia PhFBH4ox flowers.
On the contrary, in short day plants, the effect of day length over senescence is the opposite to what was observed in LD. This way, post flowering SD treatment promoted leaf senescence while LD delayed aging in the short day plant Soybean. Han et al. (2006) proposed that the photoperiodic control of development is active from germination through maturation, and the photoperiodic signals are likewise mediated by phytochromes throughout plant development. In rice, the CO like gene, Ghd2 (Grain number, plant height, and heading date2) is involved in the regulation of leaf senescence and drought resistance. The accelerated senescence and the increase of many SAGs transcripts in Ghd2ox rice plants grown under drought stress revealed the implication of Ghd2 in drought-induced senescence (Liu et al., 2016).
From the above referred data, it seems that in annual plants, the day length effect over senescence seems to be opposite in long day and short day plants: while LD condition accelerates senescence in long day plants, in short day plants, this process is delayed. On the contrary, SD treatment seems to induce aging in short day plants, whereas reduces senescence in LD plants. Therefore, a correlation between flowering phenotype/CO activity and senescence can be deduced, in both LD and SD plants and this is reflected in Figure 3B. Although an early study in the Arabidopsis early flowering accession Ler showed that leaf senescence was unaffected in the co-2 mutant grown under continuous illumination (Hensel et al., 1993), it has been argued that such light regime could cause the uncoupling of flowering from the senescence process (Wingler, 2011). However, what can be deduced from experiments in the literature run in different light regimes, is that early flowering phenotype and high CO activity are associated with accelerated plant senescence, while late flowering phenotype and low CO activity correlate well with a delay in this effect. These facts reveal that the relationship between photoperiod and senescence may be due in part to CO function.
Phytohormones play an important role in plant senescence as discussed above. This signal also affects flowering time/CO activity in Arabidopsis through the photoperiod pathway (Davis, 2009), particularly in late developmental stages (Figure 4). A short description of the effect of phytohormones in photoperiodic flowering follows and will help to understand the relationship between both processes.
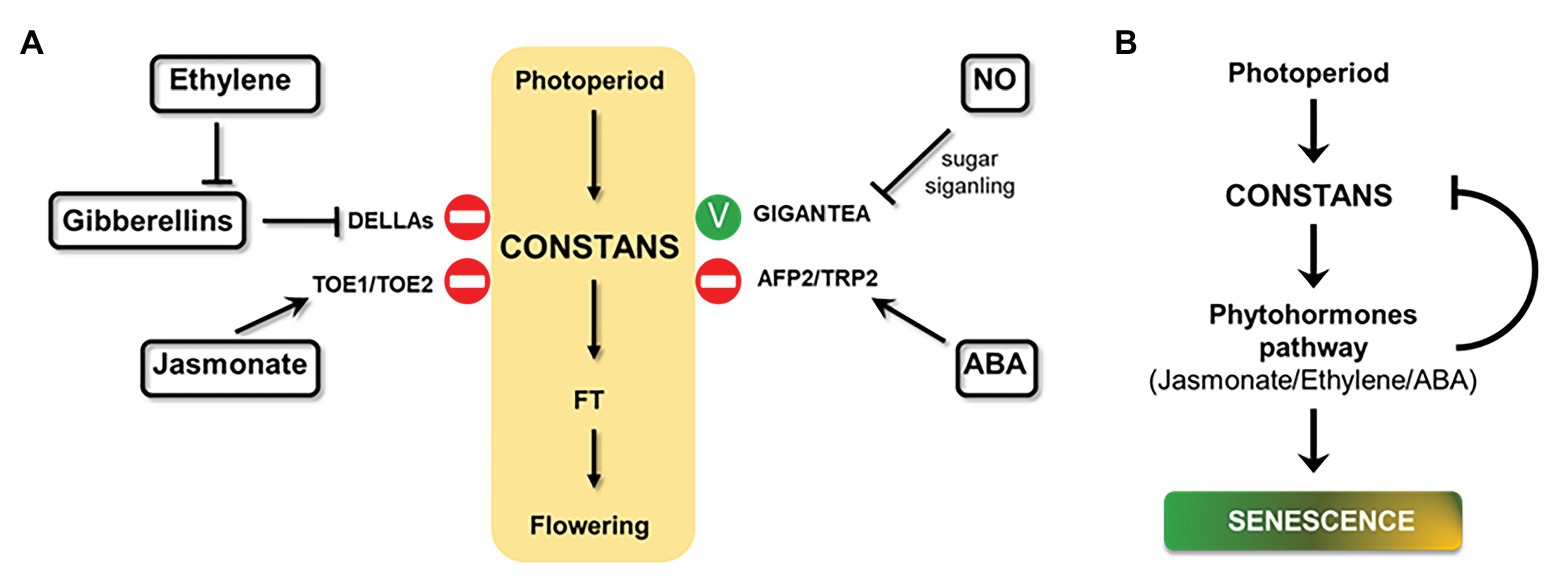
Figure 4. Effect of hormones on photoperiodic signaling and a proposed model for photoperiod and senescence connection. (A) Schematic overview of phytohormones effects on central photoperiod module (yellow box) through different effectors (in bold) in Arabidopsis plants under LDs (positive effect: green stick and negative effect: stop symbol). (B) A model of how photoperiod, phytohormones, and senescence could be related. Arrows indicate positive effects; bars indicate repressive effects.
It has been described that a mutant of the JA signal receptor CORONATINE INSENSITIVE 1 (COI1) showed early flowering phenotype, while overexpression of the JA signal repressor JASMONATE-ZIM-DOMAIN PROTEIN 1 (JAZ1) delayed this process (Robson et al., 2010; Zhai et al., 2015). While the loss-of-function coi1-1 mutant showed early flowering phenotype and high FT expression, the molecular mechanism behind the phenotype is unknown (Zhai et al., 2015). Zhang et al. (2015) described that JAZ1 controls the activity of TARGET OF EAT1 (TOE1) and TOE2, which repress FT transcription. In the morning, TOE1 and TOE2 can form a complex with CO, while in the afternoon, both proteins interact with CO stabilizer FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1) to suppress CO activity in both cases (Song et al., 2012; Zhang et al., 2015). Regarding the effect of photoperiod over JA pathway, Cagnola et al. (2018) analyzed the transcriptome of Arabidopsis plants grown under SD and then transferred to LD. The study revealed that LD enhanced JA response to increase plant defense; however, this effect was independent of the hormone levels.
Among phytohormones, the role of GAs in Arabidopsis flowering is probably the best understood (Davis, 2009). Exogenous application of GAs as well as overexpression of the biosynthetic gene GA5 promoted flowering (Huang et al., 1998; Coles et al., 1999). Diverse genetic studies also suggested that GA signaling promoted flowering under both SD and LD (Galvão et al., 2012; Porri et al., 2012; Yu et al., 2012). Two studies published in 2016 revealed that GAs induce FT expression by a CO-dependent pathway. Moreover, DELLA proteins, the main repressors of GA signaling, can directly interact with CO and inhibit CO/FT-mediated flowering in LDs (Wang et al., 2016; Xu et al., 2016). These facts evidence an integration of GA pathway and photoperiodic signaling to modulate flowering under LDs. Regarding flowering induction by GAs in SD, a recent study stablished that MYC3, a bHLH TF, is stabilized by DELLA proteins in the GA pathway to suppress FT expression by opposing CO activation (Bao et al., 2019). MYC3 regulates flowering under SD through FT suppression. This TF competes with CO to regulate FT transcription. Therefore, GAs promote flowering in SD through DELLA proteins interaction with MYC3. This interaction promotes GA-mediated degradation of MYC3, releasing CO/FT-mediated flowering in SD (Bao et al., 2019).
Ethylene production results in a delayed floral transition. Arabidopsis plants grown in the presence of ethylene or a precursor, showed late flowering phenotype (Achard et al., 2006). Similarly, the Arabidopsis mutant ctr1, a main negative regulator of ethylene signaling, showed the same flowering phenotype under LD and SD photoperiodic conditions (Achard et al., 2007). This delay of flowering can be partially rescued by mutation of genes encoding DELLAs. This finding indicates that the effect of ethylene on flowering may be in part due to modulating the activity of DELLA proteins. Also, activated ethylene signaling enhanced the accumulation of DELLAs by reducing bioactive GA levels (Achard et al., 2007). It has been demonstrated that the effect of ethylene on flowering depends on EIN3, so that ethylene stabilizes EIN3 and EIN3-like proteins by inhibiting the activity of their proteases Cullin1-based E3 complexes EBF1/EBF2 (Binder et al., 2007). Actually, GAs application partially restored flowering time in ebf1ebf2 double mutant, indicating that ethylene effect over flowering cannot be exclusively due to the inhibition of GA signaling. This fact also reveals the existence of an unknown ethylene control of flowering independent of DELLAs (Achard et al., 2007). Regarding the effect of photoperiod over ethylene pathway, an early target gene of CO activity is involved in ethylene biosynthesis (Samach et al., 2000). ACS10, which encodes a putative synthase involved in ethylene biosynthesis, was differentially expressed in the CO-activity inducible plant 35S:CO:GR in response to the inducer DEXAMETASONE (DEX). Treatment of 35S:CO:GR plants with DEX increased the abundance of ACS10 mRNA (Samach et al., 2000).
Abscisic acid signal delays flowering by upregulating FLC and downregulating CO through ABI5-BINDING PROTEIN 2 (AFP2). The bZIP TF ABSCISIC ACID-INSENSITIVE MUTANT 5 (ABI5) can bind to the FLC promoter, activate FLC expression and delay flowering (Wang et al., 2013). Furthermore, flowering time was significantly delayed, and CO expression was reduced, in an Arabidopsis AFP2ox line under LD conditions, while, in the loss-of-function afp2 mutant, flowering time was markedly accelerated and CO expression was increased. This study showed that AFP2 interacts with CO and the transcriptional corepressor TOPLESS-related protein2 (TPR2) to form the CO-AFP2-TPR2 complex that mediates CO degradation during the night (Chang et al., 2019). These studies reveal a role for AFP2 in photoperiodic flowering by modulating CO levels. Regarding the effect of photoperiod over ABA pathway, Zeevaart (1971) analyzed ABA content in the LD plant spinach (Spinacia oleracea L.) transferred from SD to LD, and the ABA content increased 2 to 3-fold.
It has been described that the gaseous signaling molecule NO repressed flowering in Arabidopsis (He et al., 2004). Plants treated with a NO-promoting agent, as well as a mutant overproducing NO (nox1), showed late flowering phenotype, while the Arabidopsis NO deficient mutant (nos1) flowered early. nox1 mutant showed upregulation of the FLC transcript; however, the molecular mechanism still awaits further investigation. Late flowering phenotype associated to NO levels cannot be exclusively assigned to the interaction with the vernalization pathway, since expression levels of the photoperiodic genes CO and GI were reduced in the nox1 mutant (He et al., 2004). Thus, NO was proposed to interact with the photoperiod pathway to regulate CO expression through a GI-dependent pathway. A recent study revealed that this interaction depends on sugar signaling (Zhang et al., 2019). Sucrose supplementation reversed the effects of NO treatment over CO and GI transcripts. While NO induced S-nitrosation modification on CO and GI, sucrose reduced the levels of this modification in both proteins (Figure 4A).
The influence of phytohormones on the central regulatory module of photoperiodic signaling often involves the central hub CONSTANS, as shown in Figure 4A. Interestingly, many senescence-inducing phytohormones, such as ethylene, jasmonato, and ABA, are also involved in the photoperiodic-dependent flowering signal. In these cases, phytohormones delay flowering time by activating CO-repressors, DELLAs, TOE1/TOE2, and AFP2/TRP2 proteins. Although there are many works that analyze the effects of phytohormones over the photoperiodic pathway, few of them describe the effect of photoperiodic signaling on phytohormones and their responses in Arabidopsis (Samach et al., 2000; Cagnola et al., 2018), or in other species (Zeevaart, 1971). It would be of particular interest to further investigate this relationship in order to establish a complete overview of photoperiod, phytohormones, and senescence cycle in plants.
Most of the studies we have referred to in this review have been conducted on the annual model plant Arabidopsis. It is not clear whether photoperiodic regulation of senescence could also occur in perennials. It has been reported that branches of the perennial Arabis alpina could behave as annuals, since leaves from flower-containing branches senesced earlier than those from flower-devoid branches (Wingler, 2011). Another major question concerns the directionality of the connection between flowering and senescence in perennials, since flower promotion does not always result in early senescence. Miryeganeh et al. (2018) showed that senescence is synchronized in Arabidopsis regardless of flowering initiation; however, a strong synchronization of flowering termination and whole-plant senescence was observed. Senescence-related genes were upregulated before flowering termination, pointing out that nutrient remobilization preceded reproduction termination (Miryeganeh et al., 2018). Further studies will be needed to explore whether in perennials flowering and senescence are connected in a similar manner to annuals.
Based on all the data collected in this perspective article, we propose a model on how photoperiod and senescence could be related, where CO regulation could be the central axis (Figure 4B). This model includes the implication of phytohormones, such as JA, ethylene, and ABA, on this relationship. In this model, we suggest that senescence regulation by photoperiod is due to CO activation of phytohormone responses. This scenario also includes a negative feedback loop, where phytohormones, in turn, inhibit CO activity.
In conclusion, this perspective review tries to shed new light on the increasingly complex regulation of plant development by integrating two independent, but chronologically interconnected programs, such as photoperiodic signaling and senescence. Early physiological responses (light and temperature) would be transmitted through physical signaling systems of archaic origin, while more complex regulatory pathways of modern origin would involve mobile signals and/or hormonal actions. Developing genetic strategies to modulate robust and constant photoperiodic signals to control plant development could have the added value of balancing the deleterious effects that other less consistent signals such as temperature, drought, or salinity will have on plant growth in the future GCC scenario.
FV and GS-B designed the manuscript and figures. All authors contributed to the article and approved the submitted version.
The authors gratefully acknowledge the help of the Spanish Ministry of Science and Education in the project BIO2017-83629-R and the European Union for the “LONGFLOW” MSCFA 838317 project from the H2020 Program. We also acknowledge the support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank José M. Pardo, José M. Romero, Jose R. Pérez-Castiñeira, and Miguel A. Blázquez for critical reading of parts of the manuscript.
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Abe, M., Kosaka, S., Shibuta, M., Nagata, K., Uemura, T., Nakano, A., et al. (2019). Transient activity of the florigen complex during the floral transition in Arabidopsis thaliana. Development 146:dev171504. doi: 10.1242/dev.171504
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U. S. A. 104, 6484–6489. doi: 10.1073/pnas.0610717104
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Aida, R., Yoshida, T., Ichimura, K., Goto, R., and Shibata, M. (1998). Extension of flower longevity in transgenic torenia plants incorporating ACC oxidase transgene. Plant Sci. 138, 91–101. doi: 10.1016/S0168-9452(98)00139-3
Ando, E., Ohnishi, M., Wang, Y., Matsushita, T., Watanabe, A., Hayashi, Y., et al. (2013). TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis thaliana. Plant Physiol. 162, 1529–1538. doi: 10.1104/pp.113.217984
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Andrés, F., Porri, A., Torti, S., Mateos, J., Romera-Branchat, M., García-Martínez, J. L., et al. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. U. S. A. 111, E2760–E2769. doi: 10.1073/pnas.1409567111
Arrom, L., and Munné-Bosch, S. (2012). Hormonal changes during flower development in floral tissues of Lilium. Planta 236, 343–354. doi: 10.1007/s00425-012-1615-0
Asad, M. A. U., Zakari, S. A., Zhao, Q., Zhou, L., Ye, Y., and Cheng, F. (2019). Abiotic stresses intervene with aba signaling to induce destructive metabolic pathways leading to death: premature leaf senescence in plants. Int. J. Mol. Sci. 20:256. doi: 10.3390/ijms20020256
Ashman, T. L., and Schoen, D. J. (1994). How long should flowers live? Nature 371, 788–791. doi: 10.1038/371788a0
Bao, S., Hua, C., Huang, G., Cheng, P., Gong, X., Shen, L., et al. (2019). Molecular basis of natural variation in photoperiodic flowering responses. Dev. Cell 50, 90–101. doi: 10.1016/j.devcel.2019.05.018
Becker, W., and Apel, K. (1993). Differences in gene expression between natural and artificially induced leaf senescence. J. Optim. Theory Appl. 189, 74–79.
Binder, B. M., Walker, J. M., Gagne, J. M., Emborg, T. J., Hemmann, G., Bleecker, A. B., et al. (2007). The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19, 509–523. doi: 10.1105/tpc.106.048140
Borochov, A., and Woodson, W. R. (1989). Physiology and biochemistry of flower petal senescence. Hortic. Rev. 11, 15–43.
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304. doi: 10.1016/j.cell.2017.09.030
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Briones-Moreno, A., Hernández-García, J., Vargas-Chávez, C., Romero-Campero, F. J., Romero, J. M., Valverde, F., et al. (2017). Evolutionary analysis of DELLA-associated transcriptional networks. Front. Plant Sci. 8:626. doi: 10.3389/fpls.2017.00626
Bruand, C., and Meilhoc, E. (2019). Nitric oxide in plants: pro- or anti-senescence. J. Exp. Bot. 70, 4419–4427. doi: 10.1093/jxb/erz117
Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., et al. (2003). The molecular analysis of leaf senescence - a genomics approach. Plant Biotechnol. J. 1, 3–22. doi: 10.1046/j.1467-7652.2003.00004.x
Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P. O., Nam, H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585. doi: 10.1111/j.1365-313X.2005.02399.x
Cagnola, J. I., Cerdán, P. D., Pacín, M., Andrade, A., Rodríguez, V., Zurbriggen, M. D., et al. (2018). Long-day photoperiod enhances jasmonic acid-related plant defense. Plant Physiol. 178, 163–173. doi: 10.1104/pp.18.00443
Cao, K., Cui, L., Zhou, X., Ye, L., Zou, Z., and Deng, S. (2015). Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front. Plant Sci. 6:1213. doi: 10.3389/fpls.2015.01213
Casal, J. J., Fankhauser, C., Coupland, G., and Blázquez, M. A. (2004). Signalling for developmental plasticity. Trends Plant Sci. 9, 309–314. doi: 10.1016/j.tplants.2004.04.007
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Chang, G., Yang, W., Zhang, Q., Huang, J., Yang, Y., and Hu, X. (2019). ABI5-BINDING PROTEIN2 coordinates CONSTANS to delay flowering by recruiting the transcriptional corepressor TPR2. Plant Physiol. 179, 477–490. doi: 10.1104/pp.18.00865
Chen, M., Maodzeka, A., Zhou, L., Ali, E., Wang, Z., and Jiang, L. (2014). Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci. 219–220, 26–34. doi: 10.1016/j.plantsci.2013.11.016
Chen, Y., Song, S., Gan, Y., Jiang, L., Yu, H., and Shen, L. (2020b). SHAGGY-like kinase 12 regulates flowering through mediating CONSTANS stability in Arabidopsis. Sci. Adv. 6:eaaw0413. doi: 10.1126/sciadv.aaw0413
Chen, W. W., Takahashi, N., Hirata, Y., Ronald, J., Porco, S., Davis, S. J., et al. (2020a). A mobile ELF4 delivers circadian temperature information from shoots to rrots. Nat. Plants 6, 416–426. doi: 10.1038/s41477-020-0634-2
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., and Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Cheng, S., Xian, W., Fu, Y., Marin, B., Keller, J., Wu, T., et al. (2019). Genomes of subaerial Zynematophyceae provide insights into land plant evolution. Cell 179, 1057–1067. doi: 10.1016/j.cell.2019.10.019
Chrobok, D., Law, S. R., Brouwer, B., Lindén, P., Ziolkowska, A., Liebsch, D., et al. (2016). Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol. 172, 2132–2153. doi: 10.1104/pp.16.01463
Coles, J. P., Phillips, A. L., Croker, S. J., García-Lepe, R., Lewis, M. J., and Hedden, P. (1999). Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 17, 547–556. doi: 10.1046/j.1365-313x.1999.00410.x
Collani, S., Neumann, M., Yant, L., and Schmid, M. (2019). FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol. 180, 367–380. doi: 10.1104/pp.18.01505
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Davis, S. J. (2009). Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 32, 1201–1210. doi: 10.1111/j.1365-3040.2009.01968.x
de Vries, J., and Archibald, M. J. (2018). Plant evolution: landmarks on the path to terrestrial life. New Phytol. 217, 1428–1434. doi: 10.1111/nph.14975
Ferrier, T., Matus, J. T., Jin, J., and Riechmann, J. L. (2011). Arabidopsis paves the way: genomic and network analyses in crops. Curr. Opin. Biotechnol. 22, 260–270. doi: 10.1016/j.copbio.2010.11.010
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Franks, S. J., and Hoffmann, A. A. (2012). Genetics of climate change adaptation. Annu. Rev. Genet. 46, 185–208. doi: 10.1146/annurev-genet-110711-155511
Gabilly, S. T., Baker, C. R., Wakao, S., Crisanto, T., Guan, K., Bi, K., et al. (2019). Regulation of photoprotection gene expression in Chlamydomonas by a putative E3 ubiquitin ligase complex and a homolog of CONSTANS. Proc. Natl. Acad. Sci. U. S. A. 116, 17556–17562. doi: 10.1073/pnas.1821689116
Galvão, V. C., Horrer, D., Küttner, F., and Schmid, M. (2012). Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139, 4072–4082. doi: 10.1242/dev.080879
Gan, S., and Amasino, R. M. (1997). Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 113, 313–319. doi: 10.1104/pp.113.2.313
Gao, S., Gao, J., Zhu, X., Song, Y., Li, Z., Ren, G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9, 1272–1285. doi: 10.1016/j.molp.2016.06.006
Gaudinier, A., and Blackman, B. K. (2019). Evolutionary processes from the perspective of flowering time diversity. New Phytol. 225, 1883–1898. doi: 10.1111/nph.16205
Han, T., Wu, C., Tong, Z., Mentreddy, R. S., Tan, K., and Gai, J. (2006). Postflowering photoperiod regulates vegetative growth and reproductive development of soybean. Environ. Exp. Bot. 55, 120–129. doi: 10.1016/j.envexpbot.2004.10.006
Hayama, R., Agashe, B., Luley, E., King, R., and Coupland, G. (2007). A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell 19, 2988–3000. doi: 10.1105/tpc.107.052480
Hayama, R., Sarid-Krebs, L., Richter, R., Fernández, V., Jang, S., and Coupland, G. (2017). PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 36, 904–918. doi: 10.15252/embj.201693907
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
He, Y., Fukushige, H., Hildebrand, D. F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
He, P., Osaki, M., Takebe, M., Shinano, T., and Wasaki, J. (2005). Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J. Exp. Bot. 56, 1117–1128. doi: 10.1093/jxb/eri103
He, Y., Tang, R. H., Hao, Y., Stevens, R. D., Cook, C. W., Ahn, S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. doi: 10.1126/science.1098837
Hensel, L. L., Grbić, V., Baumgarten, D. A., and Bleecker, A. B. (1993). Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5, 553–564. doi: 10.1105/tpc.5.5.553
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx0004
Huang, L. C., Lai, U. L., Yang, S. F., Chu, M. J., Kuo, C. I., Tsai, M. F., et al. (2007). Delayed flower senescence of Petunia hybrida plants transformed with antisense broccoli ACC synthase and ACC oxidase genes. Postharvest Biol. Technol. 46, 47–53. doi: 10.1016/j.postharvbio.2007.03.015
Huang, S., Raman, A. S., Ream, J. E., Fujiwara, H., Cerny, R. E., and Brown, S. M. (1998). Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 118, 773–781. doi: 10.1104/pp.118.3.773
Hunter, D. A., Ferrante, A., Vernieri, P., and Reid, M. S. (2004). Role of abscisic acid in perianth senescence of daffodil (Narcissus pseudonarcissus “Dutch Master”). Physiol. Plant. 121, 313–321. doi: 10.1111/j.0031-9317.2004.0311.x
Imran, Q. M., Hussain, A., Lee, S. U., Mun, B. G., Falak, N., Loake, G. J., et al. (2018). Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 8:771. doi: 10.1038/s41598-017-18850-5
Ito, S., Song, Y. H., Josephson-Day, A. R., Miller, R. J., Breton, G., Olmstead, R. G., et al. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 3582–3587. doi: 10.1073/pnas.1118876109
Jang, S., Marchal, V., Panigrahi, K. C. S., Wenkel, S., Soppe, W., Deng, X. W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288. doi: 10.1038/emboj.2008.68
Jibran, R., Hunter, D. A., and Dijkwel, P. P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 82, 547–561. doi: 10.1007/s11103-013-0043-2
Jing, Y., and Lin, R. (2020). Transcriptional regulatory network of the light signaling pathways. New Phytol. 227, 683–697. doi: 10.1111/nph.16602
Jing, H. C., Schippers, J. H. M., Hille, J., and Dijkwel, P. P. (2005). Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 56, 2915–2923. doi: 10.1093/jxb/eri287
Jung, C., Lyou, S. H., Yeu, S. Y., Kim, M. A., Rhee, S., Kim, M., et al. (2007). Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep. 26, 1053–1063. doi: 10.1007/s00299-007-0311-1
Kim, H., Kim, Y., Yeom, M., Lim, J., and Nam, H. G. (2016). Age-associated circadian period changes in Arabidopsis leaves. J. Exp. Bot. 67, 2665–2673. doi: 10.1093/jxb/erw097
Kim, H. J., Hong, S. H., Kim, Y. W., Lee, I. H., Jun, J. H., Phee, B. K., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. doi: 10.1093/jxb/eru112
Kinoshita, A., and Richter, R. (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71, 2490–2504. doi: 10.1093/jxb/eraa057
Körner, C., and Basler, D. (2010). Phenology under global warming. Science 327, 1461–1462. doi: 10.1126/science.1186473
Kubota, A., Ito, S., Shim, J. S., Johnson, R. S., Song, Y. H., Breton, G., et al. (2017). TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 13:e1006856. doi: 10.1371/journal.pgen.1006856
Lazaro, A., Valverde, F., Piñeiro, M., and Jarillo, J. A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24, 982–999. doi: 10.1105/tpc.110.081885
Lee, I. C., Hong, S. W., Whang, S. S., Lim, P. O., Nam, H. G., and Koo, J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52, 651–662. doi: 10.1093/pcp/pcr026
Lee, Z. H., Hirakawa, T., Yamaguchi, N., and Ito, T. (2019). The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 20:4065. doi: 10.3390/ijms20164065
Leshem, Y. Y., Wills, R. B. H., and Ku, V. V. V. (1998). Evidence for the function of the free radical gas - nitric oxide (NO) - as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol. Biochem. 36, 825–833. doi: 10.1016/S0981-9428(99)80020-5
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu, F., and Guo, F. Q. (2013). Nitric oxide deficiency accelerates chlorophyll breakdown and stability loss of thylakoid membranes during dark-induced leaf senescence in Arabidopsis. PLoS One 8:e56345, doi: 10.1371/journal.pone.0085660
Liu, J., Shen, J., Xu, Y., Li, X., Xiao, J., and Xiong, L. (2016). Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 67, 5785–5798. doi: 10.1093/jxb/erw344
Liu, L., Zhu, Y., Shen, L., and Yu, H. (2013). Emerging insights into florigen transport. Curr. Opin. Plant Biol. 16, 607–613. doi: 10.1016/j.pbi.2013.06.001
Liu, L. J., Zhang, Y. C., Li, Q. H., Sang, Y., Mao, J., Lian, H. L., et al. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. doi: 10.1105/tpc.107.057281
Lü, P., Zhang, C., Liu, J., Liu, X., Jiang, G., Jiang, X., et al. (2014). RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J. 78, 578–590. doi: 10.1111/tpj.12494
Luccioni, L., Krzymuski, M., Sánchez-Lamas, M., Karayekov, E., Cerdán, P. D., and Casal, J. (2019). CONSTANS delays Arabidopsis flowering under short days. Plant J. 97, 923–932. doi: 10.1111/tpj.14171
Ma, N., Ma, C., Liu, Y., Shahid, M. O., Wang, C., and Gao, J. (2018). Petal senescence: a hormone view. J. Exp. Bot. 69, 719–732. doi: 10.1093/jxb/ery009
Majeed, N., Panigrahi, K. C. S., Sukla, L. B., John, R., and Panigrahy, M. (2020). Regulatory mechanisms across networks of the circadian clock and senescence pathways. J. Plant Biochem. Biotechnol. 29, 665–674. doi: 10.1007/s13562-020-00612-6
Masclaux, C., Valadier, M. H., Brugière, N., Morot-Gaundry, J. H., and Hirel, B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. doi: 10.1007/s004250000310
Michelson, I. H., Ingvarsson, P. K., Robinson, K. M., Edlund, E., Eriksson, M. E., Nilsson, O., et al. (2018). Autumn senescence in Aspen is not triggered by day length. Physiol. Plant. 162, 123–134. doi: 10.1111/ppl.12593
Miryeganeh, M., Yamaguchi, M., and Kudoh, H. (2018). Synchronisation of Arabidopsis flowering time and whole-plant senescence in seasonal environments. Sci. Rep. 8:10282. doi: 10.1038/s41598-018-28580-x
Mishina, T. E., Lamb, C., and Zeier, J. (2007). Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ. 30, 39–52. doi: 10.1111/j.1365-3040.2006.01604.x
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U. S. A. 115, 2274–2283. doi: 10.1073/pnas.1719588115
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi: 10.1038/nature10431
Nicotra, A. B., Atkin, O. K., Bonser, S. P., Davidson, A. M., Finnegan, E. J., Mathesius, U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. doi: 10.1016/j.tplants.2010.09.008
Nooden, L. D. (1988). “The phenomena of senescence and ageing” in Senescence and aging in plants. eds. L. D. Nooden and A. C. Leopold (San Diego, California, USA: Academic Press), 1–50.
Nooden, L. D., Hillsberg, J. W., and Schneider, M. J. (1996). Induction of leaf senescence in Arabidopsis thaliana by long days through a light-dosage effect. Physiol. Plant. 96, 491–495. doi: 10.1111/j.1399-3054.1996.tb00463.x
Ortiz-Marchena, M. I., Albi, T., Lucas-Reina, E., Said, F. E., Romero-Campero, F. J., Cano, B., et al. (2014). Photoperiodic control of carbon distribution during the floral transition in Arabidopsis. Plant Cell 26, 565–584. doi: 10.1105/tpc.114.122721
Pajoro, A., Biewers, S., Dougali, E., Valentim, F. L., Mendes, M. A., Porri, A., et al. (2014). The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: a two-decade history. J. Exp. Bot. 65, 4731–4745. doi: 10.1093/jxb/eru233
Panavas, T., Walker, E. L., and Rubinstein, B. (1998). Possible involvement of abscisic acid in senescence of daylily petals. J. Exp. Bot. 49, 1987–1997. doi: 10.1093/jxb/49.329.1987
Parrott, D. L., Downs, E. P., and Fischer, A. M. (2012). Control of barley (Hordeum vulgare L.) development and senescence by the interaction between a chromosome six grain protein content locus, day length, and vernalization. J. Exp. Bot. 63, 1329–1339. doi: 10.1093/jxb/err360
Philosoph-Hadas, S., Hadas, E., and Aharoni, N. (1993). Characterization and use in ELISA of a new monoclonal antibody for quantitation of abscisic acid in senescing rice leaves. Plant Growth Regul. 12, 71–78. doi: 10.1007/BF00144585
Pires, N. D., and Dolan, L. (2012). Morphological evolution in land plants: new designs with old genes. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 367, 508–518. doi: 10.1098/rstb.2011.0252
Porat, R., Borochov, A., and Halevy, A. H. (1993). Enhancement of petunia and dendrobium flower senescence by jasmonic acid methyl ester is via the promotion of ethylene production. Plant Growth Regul. 13, 297–301. doi: 10.1007/BF00024851
Porat, R., Halevy, A. H., Serek, M., and Borochov, A. (1995). An increase in ethylene sensitivity following pollination is the initial event triggering an increase in ethylene production and enhanced senescence of Phalaenopsis orchid flowers. Physiol. Plant. 93, 778–784. doi: 10.1111/j.1399-3054.1995.tb05131.x
Porri, A., Torti, S., Romera-Branchat, M., and Coupland, G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209. doi: 10.1242/dev.077164
Preston, J. C., and Fjellheim, S. (2020). Understanding past, and predicting future, niche transitions based on grass flowering time variation. Plant Physiol. 183, 822–839. doi: 10.1104/pp.20.00100
Procházková, D., and Wilhelmová, N. (2011). Nitric oxide, reactive nitrogen species and associated enzymes during plant senescence. Nitric Oxide 24, 61–65. doi: 10.1016/j.niox.2011.01.005
Proebsting, W. M., Davies, P. J., and Marx, G. A. (1976). Photoperiodic control of apical senescence in a genetic line of peas. Plant Physiol. 58, 800–802. doi: 10.1104/pp.58.6.800
Proebsting, W. M., Davies, P. J., and Marx, G. A. (1978). Photoperiod-induced changes in gibberellin metabolism in relation to apical growth and senescence in genetic lines of peas (Pisum sativum L.). Planta 141, 231–238. doi: 10.1007/BF00388337
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Robson, F., Okamoto, H., Patrick, E., Harris, S. R., Wasternack, C., Breaerly, C., et al. (2010). Jasmonate and phytochrome a signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160. doi: 10.1105/tpc.109.067728
Rogers, H. J. (2006). Programmed cell death in floral organs: how and why do flowers die? Ann. Bot. 97, 309–315. doi: 10.1093/aob/mcj051
Rogers, H. J. (2013). From models to ornamentals: how is flower senescence regulated? Plant Mol. Biol. 82, 563–574. doi: 10.1007/s11103-012-9968-0
Romero-Campero, F. J., Lucas-reina, E., Said, F. E., Romero, J. M., and Valverde, F. (2013). A contribution to the study of plant development evolution based on gene co-expression networks. Front. Plant Sci. 4:291. doi: 10.3389/fpls.2013.00291
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7, 1–26. doi: 10.3389/fpls.2016.00571
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Sang, Q., Pajoro, A., Sun, H., Song, B., Yang, X., Stolze, S. C., et al. (2020). Mutagenesis of a quintuple mutant impaired in environmental responses reveals roles for CHROMATIN REMODELING4 in the Arabidopsis floral transition. Plant Cell 32, 1479–1500. doi: 10.1105/tpc.19.00992
Sarid-Krebs, L., Panigrahi, K. C. S., Fornara, F., Takahashi, Y., Hayama, R., Jang, S., et al. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84, 451–463. doi: 10.1111/tpj.13022
Savin, K. W., Baudinette, S. C., Graham, M. W., Michael, M. Z., Nugent, G. D., Lu, C. Y., et al. (1995). Antisense ACC oxidase RNA delays carnation petal senescence. Hort. Sci. 30, 970–972.
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schippers, J. H. M., Jing, H. C., Hille, J., and Dijkwel, P. P. (2007). Developmental and hormonal control of leaf senescence. Annu. Plant Rev. 26, 145–170. doi: 10.1002/9781119312994.apr0268
Serrano, G., Herrera-Palau, R., Romero, J. M., Serrano, A., Coupland, G., and Valverde, F. (2009). Chlamydomonas CONSTANS and the evolution of plant photoperiodic signalling. Curr. Biol. 19, 359–368. doi: 10.1016/j.cub.2009.01.044
Serrano-Bueno, G., Romero-Campero, F. J., Lucas-Reina, E., Romero, J. M., and Valverde, F. (2017). Evolution of photoperiod sensing in plants and algae. Curr. Opin. Plant Biol. 37, 10–17. doi: 10.1016/j.pbi.2017.03.007
Shahri, W., and Tahir, I. (2011). Flower senescence-strategies and some associated events. Bot. Rev. 77, 152–184. doi: 10.1007/s12229-011-9063-2
Shan, X., Wang, J., Chua, L., Jiang, D., Peng, W., and Xie, D. (2011). The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155, 751–764. doi: 10.1104/pp.110.166595
Shen, C., Liu, H., Guan, Z., Yan, J., Zheng, T., Yan, W., et al. (2020). Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell 32, 3469–3484. doi: 10.1105/tpc.20.00067
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Shrestha, R., Gómez-Ariza, J., Brambilla, V., and Fornara, F. (2014). Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann. Bot. 114, 1445–1458. doi: 10.1093/aob/mcu032
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., MacCoss, M. J., et al. (2014). Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U. S. A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Song, Y. H., Kubota, A., Kwon, M. S., Covington, M. F., Lee, N., Taagen, E. R., et al. (2018). Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants 4, 824–835. doi: 10.1038/s41477-018-0253-3
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Tan, Y., Liu, J., Huang, F., Guan, J., Zhong, S., Tang, N., et al. (2014). PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Mol. Plant 7, 1384–1387. doi: 10.1093/mp/ssu024
Thomas, H., Huang, L., Young, M., and Ougham, H. (2009). Evolution of plant senescence. BMC Evol. Biol. 9:163. doi: 10.1186/1471-2148-9-163
Thomas, H., and Stoddart, J. L. (1980). Leaf senescence. Annu. Rev. Plant Physiol. 31, 83–111. doi: 10.1146/annurev.pp.31.060180.000503
Tiwari, S. B., Shen, Y., Chang, H. C., Hou, Y., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Tokutsu, R., Fujimura-Kamada, K., Matsuo, T., Yamasaki, T., and Minagawa, J. (2019). The CONSTANS flowering complex controls the protective response of photosynthesis in the green alga Chlamydomonas. Nat. Commun. 10:4099. doi: 10.1038/s41467-019-11989-x
Tripathi, S. K., and Tuteja, N. (2007). Integrated signaling in flower senescence: an overview. Plant Signal. Behav. 2, 437–445. doi: 10.4161/psb.2.6.4991
Valverde, F. (2011). CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 62, 2453–2463. doi: 10.1093/jxb/erq449
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Van Der Graaff, E., Schwacke, R., Schneider, A., Desimone, M., Flügge, U. I., and Kunze, R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. doi: 10.1104/pp.106.079293
Van Deursen, J. M. (2014). The role of senescent cells in ageing. Nature 509, 439–446. doi: 10.1038/nature13193
Van Doorn, W. G., and Woltering, E. J. (2008). Physiology and molecular biology of petal senescence. J. Exp. Bot. 59, 453–480. doi: 10.1093/jxb/erm356
Wang, C., Liu, Y., Li, S. S., and Han, G. Z. (2015). Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886. doi: 10.1104/pp.114.247403
Wang, H., Pan, J., Li, Y., Lou, D., Hu, Y., and Yu, D. (2016). The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 172, 479–488. doi: 10.1104/pp.16.00891
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Wen, B., Xiao, W., Mu, Q., Li, D., Chen, X., Wu, H., et al. (2020). How does nitrate regulate plant senescence? Plant Physiol. Biochem. 157, 60–69. doi: 10.1016/j.plaphy.2020.08.041
Wenkel, S., Turck, F., Singer, K., Gissot, L., Gourrierec, J. L., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Wingler, A. (2011). Interactions between flowering and senescence regulation and the influence of low temperature in Arabidopsis and crop plants. Ann. Appl. Biol. 159, 320–338. doi: 10.1111/j.1744-7348.2011.00497.x
Wojciechowska, N., Sobieszczuk-Nowicka, E., and Bagniewska-Zadworna, A. (2018). Plant organ senescence – regulation by manifold pathways. Plant Biol. 20, 167–181. doi: 10.1111/plb.12672
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Xiao, S., Dai, L., Liu, F., Wang, Z., Peng, W., and Xie, D. (2004). COS1: an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16, 1132–1142. doi: 10.1105/tpc.020370
Xu, F., Li, T., Xu, P. B., Li, L., Du, S. S., Lian, H. L., et al. (2016). DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590, 541–549. doi: 10.1002/1873-3468.12076
Yang, S., Weers, B. D., Morishige, D. T., and Mullet, J. E. (2014). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14:148. doi: 10.1186/1471-2229-14-148
Yang, J., Zhang, J., Wang, Z., Zhu, Q., and Liu, L. (2002). Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta 215, 645–652. doi: 10.1007/s00425-002-0789-2
Yin, J., Chang, X., Kasuga, T., Bui, M., Reid, M. S., and Jiang, C. Z. (2015). A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2:15059. doi: 10.1038/hortres.2015.59
Yu, S., Galvão, V. C., Zhang, Y. C., Horrer, D., Zhang, T. Q., Hao, Y. H., et al. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24, 3320–3332. doi: 10.1105/tpc.112.101014
Zeevaart, J. A. D. (1971). (+)-abscisic acid content of spinach in relation to photoperiod and water stress. Plant Physiol. 48, 86–90. doi: 10.1104/pp.48.1.86
Zhai, Q., Zhang, X., Wu, F., Henf, H., Deng, L., Xu, L., et al. (2015). Transcriptional mechanism of Jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27, 2814–2828. doi: 10.1105/tpc.15.00619
Zhang, Z. W., Fu, Y. F., Zhou, Y. H., Wang, C. Q., Lan, T., Chen, G. D., et al. (2019). Nitrogen and nitric oxide regulate Arabidopsis flowering differently. Plant Sci. 284, 177–184. doi: 10.1016/j.plantsci.2019.04.015
Zhang, K., and Gan, S. S. (2012). An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 158, 961–969. doi: 10.1104/pp.111.190876
Zhang, Y., Wang, H. L., Li, Z., and Guo, H. (2020). Genetic network between leaf senescence and plant immunity: crucial regulatory nodes and new insights. Plan. Theory 9:495. doi: 10.3390/plants9040495
Zhang, B., Wang, L., Zeng, L., Zhang, C., and Ma, H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29, 975–987. doi: 10.1101/gad.251520.114
Zhang, K., Xia, X., Zhang, Y., and Gan, S. S. (2012). An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 69, 667–678. doi: 10.1111/j.1365-313X.2011.04821.x
Keywords: plant development, photoperiod, senescence, flowering, evolution, phytohormones
Citation: Serrano-Bueno G, Sánchez de Medina Hernández V and Valverde F (2021) Photoperiodic Signaling and Senescence, an Ancient Solution to a Modern Problem? Front. Plant Sci. 12:634393. doi: 10.3389/fpls.2021.634393
Received: 27 November 2020; Accepted: 12 February 2021;
Published: 10 March 2021.
Edited by:
Manuel Pineiro, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainReviewed by:
Lu Liu, Shanghai Jiao Tong University, ChinaCopyright © 2021 Serrano-Bueno, Sánchez de Medina Hernández and Valverde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federico Valverde, ZmVkZXJpY28udmFsdmVyZGVAaWJ2Zi5jc2ljLmVz; Gloria Serrano-Bueno, Z2xvcmlhLnNlcnJhbm9AaWJ2Zi5jc2ljLmVz
†Present address: Víctor Sánchez de Medina Hernández, Gregor Mendel Institute (GMI), Austrian Academy of Sciences, Vienna BioCenter (VBC), Vienna, Austria
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.