
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 February 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.634040
This article is part of the Research Topic Signaling Events in Regulating Leaf Senescence View all 11 articles
A correction has been applied to this article in:
Corrigendum: Transcription Factor NAC075 Delays Leaf Senescence by Deterring Reactive Oxygen Species Accumulation in Arabidopsis
Leaf senescence is a highly complex genetic process that is finely tuned by multiple layers of regulation. Among them, transcriptional regulation plays a critical role in controlling the initiation and progression of leaf senescence. Here, we found that the NAC transcription factor NAC075 functions as a novel negative regulator of leaf senescence. Loss of function of NAC075 promotes leaf senescence in an age-dependent manner, whereas constitutive overexpression of NAC075 delays senescence in Arabidopsis. Transcriptome analysis revealed that transcript levels of antioxidant enzymes such as catalase (CAT), ascorbate peroxidase (APX), and superoxide dismutase (SOD) are significantly suppressed in nac075 mutants compared with wild-type plants. Electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) analyses revealed that NAC075 directly binds the promoter of catalase 2 (CAT2). Moreover, genetic analysis showed that overexpression of CAT2 suppresses the overproduction of reactive oxygen species (ROS) and the early senescence phenotypes of nac075 mutants, suggesting that CAT2 acts downstream of NAC075 to delay leaf senescence by repressing ROS accumulation. Collectively, our findings provide a new regulatory module involving NAC075-CAT2-ROS in controlling leaf senescence in Arabidopsis.
Leaf senescence is a universal biological phenomenon in nature that contributes to the recycling of nutrients (Guo and Gan, 2005; Hughes and Reynolds, 2005). Senescence is the last stage of leaf development, accompanied by the hydrolysis of a series of macromolecules and the disassembly of chloroplasts and mitochondria, which ultimately leads to leaf death (Buchanan-Wollaston et al., 2005; Mao et al., 2017; Woo et al., 2019). Among them, yellowing of leaves from tip to base due to the loss of chlorophyll is the most striking marker of leaf senescence (Guo and Gan, 2005; Li et al., 2019; Woo et al., 2019). During leaf senescence, nutrients released by the catabolism of macromolecular substances such as proteins, lipids, and nucleic acids are transferred to active growing organs such as new buds and developing fruits and seeds, or stored for use in the next growing season (Guo and Gan, 2005; Lim et al., 2007; Woo et al., 2019). Efficient senescence is essential for maximizing viability in the next generation or season, while premature senescence induced by a variety of environmental factors declines crop plants’ yield and quality (Breeze et al., 2011). Thus, the appropriate onset and progression of leaf senescence are essential for plant fitness, suggesting that senescence evolves as a life history strategy. Significant advances in understanding the regulatory mechanisms of leaf senescence will provide valuable clues for the manipulation of traits of agronomical important plants.
Leaf senescence is a highly complex and orderly dynamic regulation process and is strictly controlled by multiple layers of regulation, including chromatin-mediated, transcriptional, post-transcriptional, translational, and post-translational regulation (Woo et al., 2013, 2019; Kim et al., 2016, 2018). Leaf senescence is not a passive but a highly coordinated process that is regulated by a number of senescence-associated genes (SAGs), whose transcripts increase as leaves age. The onset, development, and completion of leaf senescence involve extensive regulation of gene expression (Woo et al., 2013). Genome-wide transcriptional analysis revealed that the leaf senescence process is accompanied by differential expression of thousands of SAGs (Breeze et al., 2011; Woo et al., 2016). At the transcription level, transcription factors (TFs) act as core control elements to drive the drastic changes in SAGs expression during leaf senescence. The dynamic activation of TFs is triggered by internal signals such as plant hormones or environmental factors such as high salt (Guo and Gan, 2005; Guo, 2013; Li et al., 2018). Significant advances in dissecting the regulatory mechanisms underpinning leaf senescence have benefited from the identification and functional assessment of hundreds of SAGs and their corresponding mutants. Previous studies have identified numerous TFs that participate in the process of leaf senescence in Arabidopsis, including NAC (NAM, ATAF1, 2, and CUC2), WRKY, MYB, and bZIP families’ TFs, which play important roles in regulating leaf senescence (Lim et al., 2007; Balazadeh et al., 2008; Woo et al., 2019). As one of the largest TF families in plants, NAC TFs receive widespread attention due to their important role in regulating leaf senescence process in a variety of plant species (Kim et al., 2016). The regulatory roles of a number of NAC TFs in leaf senescence have been characterized in Arabidopsis. For instance, ORE1 (ANAC092), AtNAP (ANAC029), ATAF1 (ANAC002), JUB1 (ANAC042/ANAC2), VNI2 (ANAC083), and ANAC017/082/090 act as positive or negative regulators of leaf senescence (Guo and Gan, 2006; Balazadeh et al., 2010; Yang et al., 2011; Wu et al., 2012; Jensen et al., 2013; Garapati et al., 2015; Kim et al., 2018). Recent findings revealed that the molecular network of NAC TFs regulates leaf senescence by integrating internal developmental signals and numerous environmental signals (Kim et al., 2016). Although a growing body of evidence indicates that NAC TFs play important roles in leaf senescence, little is known regarding their importance and underlying regulatory mechanisms.
In this study, we found that NAC TF NAC075 functions as a negative regulator of leaf senescence. Mutation of NAC075 evidently hastens leaf senescence, whereas overexpression of NAC075 markedly prolongs leaf longevity. Biochemical and genetic evidence shows that NAC075 delays leaf senescence by directly upregulating CAT2 expression and suppressing the accumulation of reactive oxygen species (ROS) in Arabidopsis.
The Arabidopsis thaliana ecotype Columbia (Col-0) is the parent strain for all mutants and transgenic lines used in this study. The transfer DNA (T-DNA) insertional mutant nac075 (SALK_132120C) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). The nac075 CAT2ox was generated by genetic cross, and the homozygous plants were identified through PCR-based genotyping. Seeds were surface-sterilized and plated on Murashige and Skoog (MS) medium (4.3 g/L MS salts, 1% sucrose, pH 5.7–5.8, and 8 g/L agar). After stratifying at 4°C for 3 days to improve germination uniformity, the plates were transferred to an environmentally controlled growth room (PAR of 100–150 μE m–2 s–1) for 4 days. For plant leaf senescence phenotypic analysis, light-grown seedlings were transferred to soil and grown at 22°C under long-day conditions (16-h light/8-h dark).
To construct ProNAC075:GUS/Col-0, a 3-kb genomic promoter sequence was amplified and inserted into pCambia1391 vector (GenBank Accession-AF234308). To generate 35S:GFP-NAC075/Col-0, the full-length NAC075 CDS sequence was amplified and then inserted into pEGAD vector (Cutler et al., 2000). To generate inducible overexpressing lines, the full-length NAC075 CDS fused with 3xFLAG was into pER8 vector (Zuo et al., 2000). All constructs were transformed into Agrobacterium tumefaciens cells (strain GV3101), which was used to transform Col-0 plants by the floral dip method (Clough and Bent, 1998). Primers used for PCR are listed in Supplementary Table 1.
Total RNA was isolated by using plant RNA extraction kits (ER301; TransGen Biotech, China), and the complementary DNA was produced using TransScript All-in-One First-Strand cDNA Synthesis kit (AT341; TransGen Biotech). Transcript levels were detected with TransStart Green qPCR SuperMix (AQ111; TransGen Biotech) by using Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Carlsbad, California, United States). Ubiquitin-conjugating enzyme 21 (UBC21, AT5G25760) was used as an internal control to normalize the gene expression level. The primers used in this study are listed in Supplementary Table 1.
Chlorophyll contents were measured in the third and fourth rosette leaves of Arabidopsis using a chlorophyll meter Konica Minolta SPAD502 Plus (Sakura-machi, Hino-shi Tokyo, Japan), and three biological replicates were performed. Maximal photochemical efficiencies of Photosystem II (PSII, Fv/Fm) were measured by using a MultiSpeQ instrument (East Lansing, MI, United States).
GUS (β-Glucuronidase) staining was performed as described previously (Jefferson, 1989). Plant tissues were incubated with GUS staining solution (100 mM Na3PO4, pH 7.0, 1 mM EDTA, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1% Triton X-100, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-glucuronide) for 8 to 12 h at 37°C in the dark, followed by decolonization using 95% ethanol.
The third and fourth leaves (18-day-old) were vacuum-infiltrated with diaminobenzidine tetrahydrochloride (DAB) solution (1 mg/ml 3,3’-diaminobenzidine-4HCl, pH 3.8) and NBT (nitroblue-tetrazole) solution (0.5 mg/ml NBT, 10 mM potassium phosphate, pH 7.8, and 10 mM sodium azide) to detect hydrogen peroxide and superoxide, respectively, incubated in the dark for 8–10 h, and decolorized in 95% ethanol. The intensity of brown and blue coloration indicates H2O2 and O2– contents, respectively.
Trypan blue staining was performed as described previously with minor modifications (Kim et al., 2009). Briefly, the third and fourth rosette leaves were soaked in trypan blue staining solution (10 g phenol, 10 ml glycerol, 10 ml lactic acid, 10 ml ddH2O2, and 0.02 g trypan blue) and stained in a boiling water bath for 3–5 min. Three leaves were completely submerged in trypan blue staining solution. After leaving overnight at room temperature, the leaves were carefully clamped into the decolorizing solution (2.5 g/ml chloral hydrate).
The third and fourth rosette leaves of 24-day-old Col-0 and nac075 mutant plants were collected and ground into a powder in liquid nitrogen. Total RNA was extracted using an RNeasy Plant kit (Qiagen), and the quality and quantity of RNA were detected using an IMPLEN NanoPhotometer (GmbH). RNA-seq data were generated with an Illumina HiSeq 2000 system at Biomarker Ltd. (Beijing, China). Raw reads (fastq format) were trimmed and filtered through in-house perl scripts (Biomarker Ltd. China). The reads were then mapped to Arabidopsis reference genome using Hisat2 algorithm. DEGs were filtered using the following criteria: | Log2 (fold change)| > 1.0, P < 0.05. Gene ontology (GO) enrichment analysis was performed by using the GO database1. Default parameters were used for all bioinformatics software. Raw RNA-seq reads are available at the National Center for Biotechnology Information2.
The full-length coding region of NAC075 was produced by quantitative RT-PCR (qRT-PCR) and used for developing the DNA constructs to pET32a for the expression of recombinant proteins in Escherichia coli BL21 (DE3). Purification of NAC075 protein was conducted according to the protocol included with the His-Trap HP pre-packed minicolumns (GE Healthcare Life Sciences, Uppsala, Sweden). EMSA was performed according to the user guide from the LightShift Chemiluminescent EMSA Kit (Lot#20148, Thermo Scientific). Briefly, the binding reaction was performed in a total volume of 20 μl by incubation of an appropriate amount of NAC075 proteins with 20 fm of biotin-labeled probe DNA and 1 μg of poly (dI–dC) in a reaction buffer (25 mM HEPES-KOH, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 10% [v/v] glycerol, and 1 mM DTT) at room temperature for 30 min. The binding reaction products were resolved on a 6% polyacrylamide gel run in 0.5 × TBE buffer. 5’-biotin-labeled oligonucleotide of CAT2 was synthesized and used as probes in EMSA (Supplementary Table 1).
ChIP experiments were performed as described previously with minor modifications (Saleh et al., 2008). Briefly, 5 g of 4-week-old 35S:GFP-NAC075 leaves was collected into 50-ml Falcon tubes with 37 ml of cross-linked buffer (10 mM Tris–HCl, pH 8.0, 0.4 M sucrose, 1 mM EDTA, 1 mM PMSF, and 1% formaldehyde). Next, 2 M glycine was added for 5 min to quench the cross-linking reaction. The leaves were then washed three times with sterile deionized water, frozen in liquid nitrogen, and quickly ground into a powder. Next, the ground powder was added into 25 ml of nuclear separation buffer and vortexed to isolate chromatin DNA. The sonicated chromatin supernatant (300 μl) was diluted and 50 μl of salmon sperm DNA/protein A agarose beads (Upstate) was added for pre-clearing at 4°C for 1 h with gentle rotation (12 rpm). The solutions were then transferred into two new tubes. Add 10 μl of anti-GFP monoclonal antibody with a dilution of 1:150 (v/v) to one tube, but not the other (as a negative control). After incubation at 4°C overnight, beads were washed with low-salt wash buffer, high-salt wash buffer, and Tris–EDTA (TE) buffer, followed by followed by Proteinase K (10 mg/ml; Sigma-Aldrich) treatment and reverse cross-linking with 5 M NaCl. DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1), and then ethanol precipitated with 2 μl of 20 mg/ml glycogen. The purified DNA was resuspended in 20 μl of distilled water and stored at -20°C. All oligonucleotide sequences used here are listed in Supplementary Table 1.
The third and fourth leaves (20-day-old) detached from Col-0, nac075, and NAC075ox plants were treated with 5 mM MES (Mock), 10 μM ACC, 50 mM MeJA, 1 mM SA, 50 μM ABA, 10 mM H2O2, or 100 mM NaCl in dark conditions for 3 days, respectively.
The 28-day-old pER8-FLAG-NAC075 transgenic plants were sprayed with 50 μM β-estradiol. After treatment for 0.5, 1, and 4 h, the third and fourth rosette leaves were detached and used for RNA extraction and qRT-PCR analysis.
Previous studies have shown that NAC TF NAC075 is related to leaf senescence (Woo et al., 2016; Li et al., 2020), but the underlying regulatory mechanism remains unclear. Toward this end, we firstly performed Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) to examine the transcript levels of NAC075 in Arabidopsis leaves at young, mature, early, and late stage of senescence (Figure 1A). Time-course analysis of mRNA level monitored by qRT-PCR revealed that the transcript level of NAC075 gradually increased during leaf development and senescence (Figure 1A). SAG12, a widely used marker gene of leaf senescence (Noh and Amasino, 1999; Pontier et al., 1999), was specifically expressed in the senescing leaves (Supplementary Figure 1A). We also measured the transcript levels of AtNAP and ORE1, two well-known positive regulators of leaf senescence (Guo and Gan, 2006; Kim et al., 2009), and found that their expressions also increased as leaves age (Supplementary Figures 1B,C). To further verify the age-dependent regulation of NAC075 mRNA in planta, we generated transgenic Arabidopsis expressing GUS (β-glucuronidase gene) driven by NAC075 promoter containing a 3-kb fragment upstream promoter of the start codon (ProNAC075:GUS/Col-0). Histochemical staining assay of rosette leaves of 30-day-old ProNAC075:GUS/Col-0 plants revealed that the old yellowing leaves displayed higher GUS activity than that in young green leaves (Figure 1B), indicative of an increase in NAC075 expression level during the leaf senescence process.
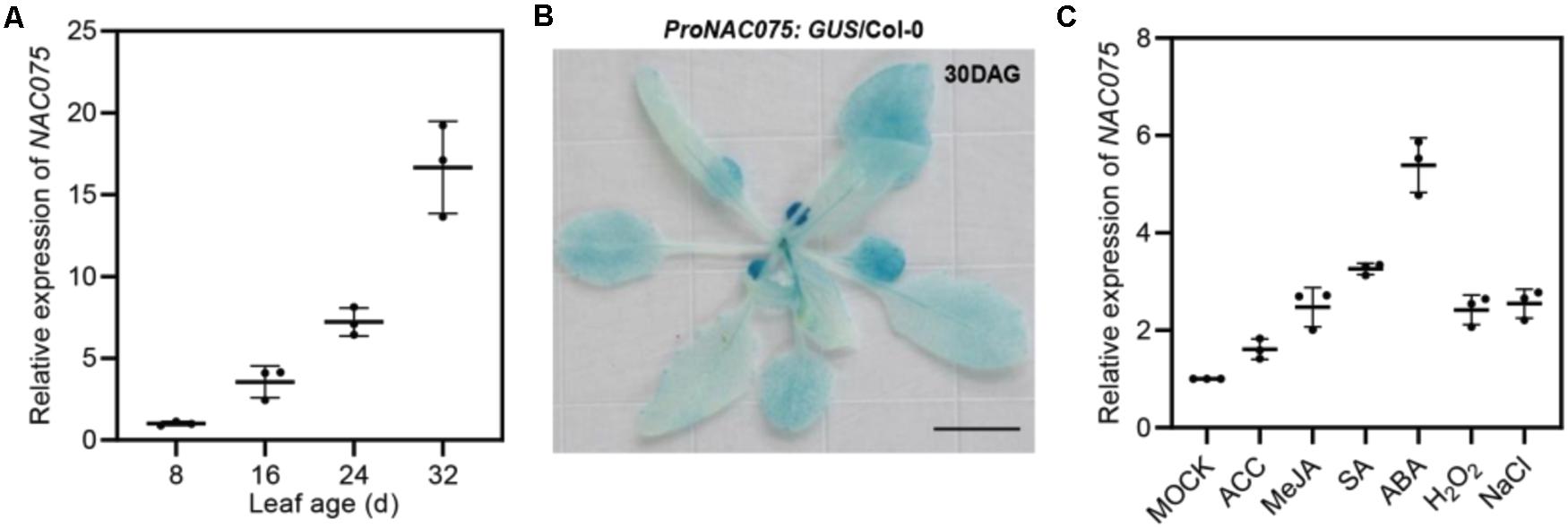
Figure 1. NAC075 transcription factor is a senescence-associated gene that is induced by age, plant hormones, ROS, and salinity. (A) qRT-PCR analyses of NAC075 expression in the third or fourth rosette leaves at the indicated leaf age. Data are represented as means ± SD (n = 3). (B) GUS staining of rosette leaves of 30-day-old plants harboring the GUS transgene driven by the promoter of NAC075 (ProNAC075:GUS/Col-0). Scale bar, 1 cm. (C) qRT-PCR analyses of NAC075 expression in the leaves of 20-day-old Col-0 plants upon treatment with 5 mM MES (Mock), 10 μM ACC, 50 mM MeJA, 1 mM SA, 50 μM ABA, 10 mM H2O2, or 100 mM NaCl for 6 h. Data are represented as means ± SD (n = 3).
We next performed qRT-PCR to investigate the influences of other senescence-regulating signals such as plant hormones, ROS, and salt stress on the transcription of NAC075. We found that treatment with the ethylene precursor ACC (1-aminocyclopropane-1-carboxylate), methyl jasmonate (MeJA), salicylic acid (SA), abscisic acid (ABA), H2O2, and salt evidently increased the expression levels of NAC075 compared to the mock-treated plants (Figure 1C). Among them, treatment with ABA greatly enhanced NAC075 transcription (Figure 1C), suggesting that NAC075 may be involved in ABA-induced leaf senescence process.
To investigate the function of NAC075 in leaf senescence, we examined the senescence-associated phenotypes of nac075 knockout mutant carrying a T-DNA insert in the third intron of NAC075 (Supplementary Figure 2A) and the transgenic plants overexpressing NAC075 (NAC075ox) (Figure 2A). Genotyping analysis demonstrated that nac075 mutant is a null allele (Supplementary Figure 2B), which was confirmed further by gene expression analysis (Supplementary Figure 2C). We performed qRT-PCR to detect the NAC075 transcription in three NAC075ox lines (#1, #2, and #3), and selected NAC075ox #2 with the highest expression level for subsequent experiments (Supplementary Figure 3). We found that nac075 mutant exhibited an early-senescence phenotype in comparison to Col-0 plants under long-day conditions (Figure 2A), while NAC075ox plant displayed a delayed senescence phenotype (Figure 2A), suggesting that NAC075 is a negative regulator of leaf senescence. Interestingly, the nac075 mutants also displayed early silique senescence (Supplementary Figure 4), suggesting that NAC075 is also involved in silique development. Next, we examined the senescence characteristics of single leaf at different ages. Leaf yellowing occurred in the third or fourth rosette leaves of nac075 mutant plants at 24 days after emergence (DAE), whereas the leaves of Col-0 and NAC075ox plants remained green. At 28 DAE, the third and fourth rosette leaves of nac075 mutants were completely yellowed, which was not observed in Col-0 and NAC075ox plants (Figure 2B). At 32 DAE, the leaves of Col-0 plant began to turn yellow, while the leaves of NAC075ox plants remained green. Leaf yellowing caused by chloroplast decomposition and chlorophyll loss are typical characteristics of leaf senescence (Woo et al., 2001). We also monitored the chlorophyll contents, and photochemical efficiency of photosystem II (PSII; Fv/Fm) decreased more quickly and evidently in nac075 mutant than in Col-0 (Figures 2C–E). The NAC075ox plants displayed delayed leaf senescence phenotypes, with elevated chlorophyll content and Fv/Fm compared with Col-0, demonstrating further that NAC075 is a negative regulator of leaf senescence. We also found that aging-induced cell death was accelerated in the nac075 mutants, which was delayed in NAC075ox plants, as shown by the earlier emergence of trypan blue-stained cells in 16 and 24-day-old leaves (Figure 2F). Thus, NAC075 is a negative regulator of aging-induced cell death and senescence in Arabidopsis leaves.
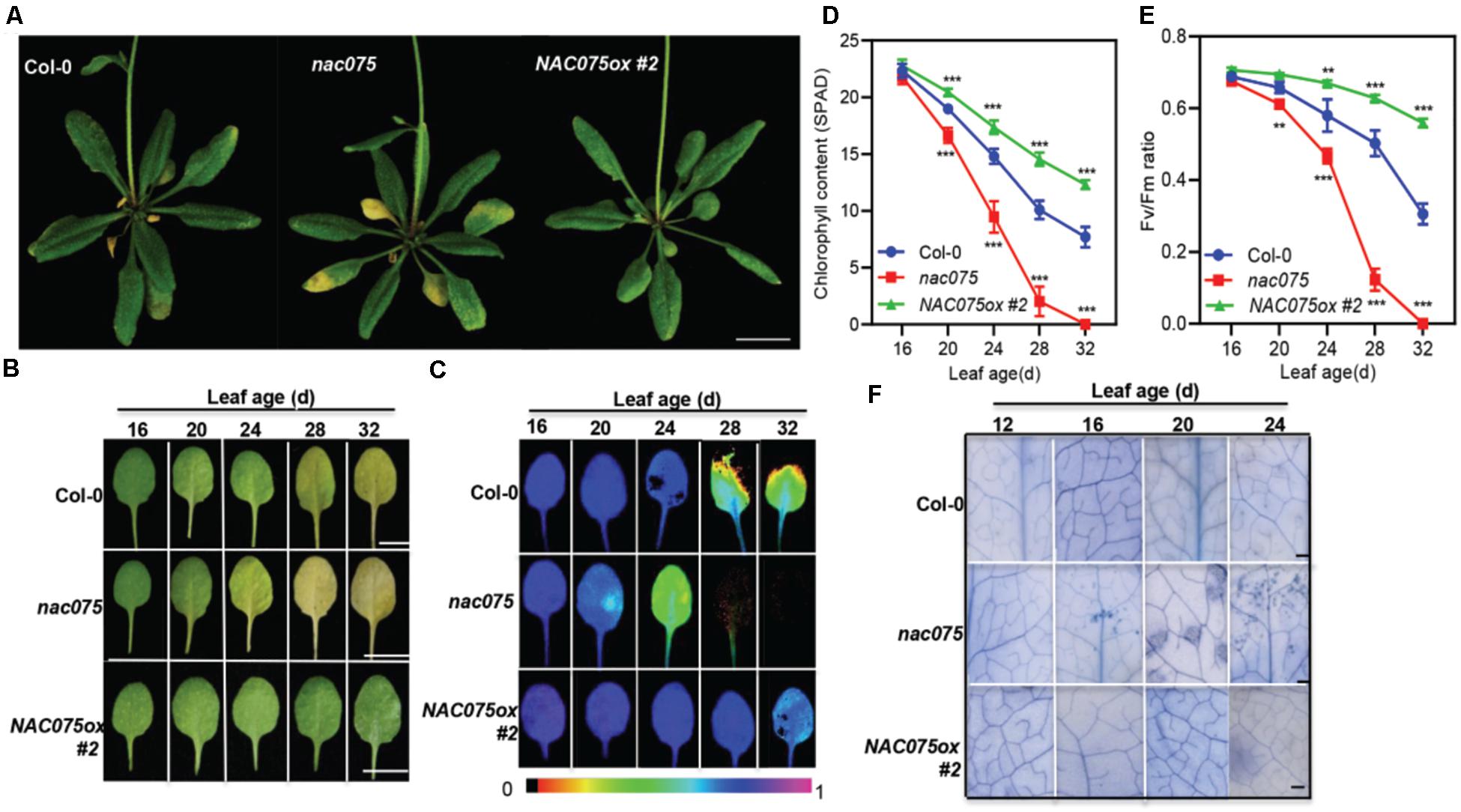
Figure 2. Age-dependent senescence symptoms in the nac075 mutants and NAC075ox plants. (A) The senescence phenotypes of 30-day-old Col-0, nac075 mutants, and NAC075ox plants. Early onset of leaf senescence in nac075 mutant was observed compared with Col-0 plants grown under long-day condition. Scale bar, 1 cm. (B) The age-dependent leaf senescence phenotype of Col-0, nac075 mutants, and NAC075ox plants. Photographs show the third or fourth rosette leaves at the indicated days after emergence (DAE). Scale bar, 1 cm. (C) Analysis of Fv/Fm in Col-0, nac075 mutants, and NAC075ox plants as leaves age. Image generation was performed by IMAGING-PAM. Image processing was performed by Imaging Win software. (D,E) Chlorophyll content (D) and Fv/Fm (E) in Col-0, nac075 mutants and NAC075ox plants as leaves age. Error bars indicate SD (n = 3). Student’s t test, ∗∗P < 0.01, ∗∗∗P < 0.001. (F) Trypan blue staining of leaves at the indicated leaf age. In each plant leaf, dead or dying leaf areas formed blue-colored patches of cells by trypan blue staining. Bar = 500 μm.
Given that NAC075 transcription was induced by multiple plant hormones and stresses (Figure 1C), we examined whether NAC075 is involved in the leaf senescence process triggered by these factors. To this end, the third or fourth rosette leaves of 20-day-old Col-0, nac075 mutants, and NAC075ox plants were treated with darkness, plant hormones, ROS, and salt. We found that the leaves of NAC075ox plants exhibited the delayed senescence phenotypes upon treatment with these factors (Supplementary Figure 5), suggesting that NAC075 delays the leaf senescence process caused by numerous factors. In contrast, the senescence phenotype of nac075 leaves was not evidently different from that of the wild-type Col-0, indicative of the existence of the functional redundancy among NAC TFs (Supplementary Figure 5).
To elucidate the underlying mechanisms of NAC075 in the regulation of leaf senescence, we performed genome-wide mRNA expression analysis of wild-type and mutant leaves at the presenescent stage (24-day-old) to identify the candidate target genes. We compared the transcriptomes of wild-type leaves with those of nac075 mutants and identified 2225, 2241, and 2156 differentially expressed genes (DEGs) in three biological replicates, respectively (Figure 3A). Out of them, 1721 genes (491 up-regulated genes and 1211 down-regulated genes) exhibited overlapping differential expression in three biological replicate samples (Figure 3B), suggesting substantial regulation by NAC075.
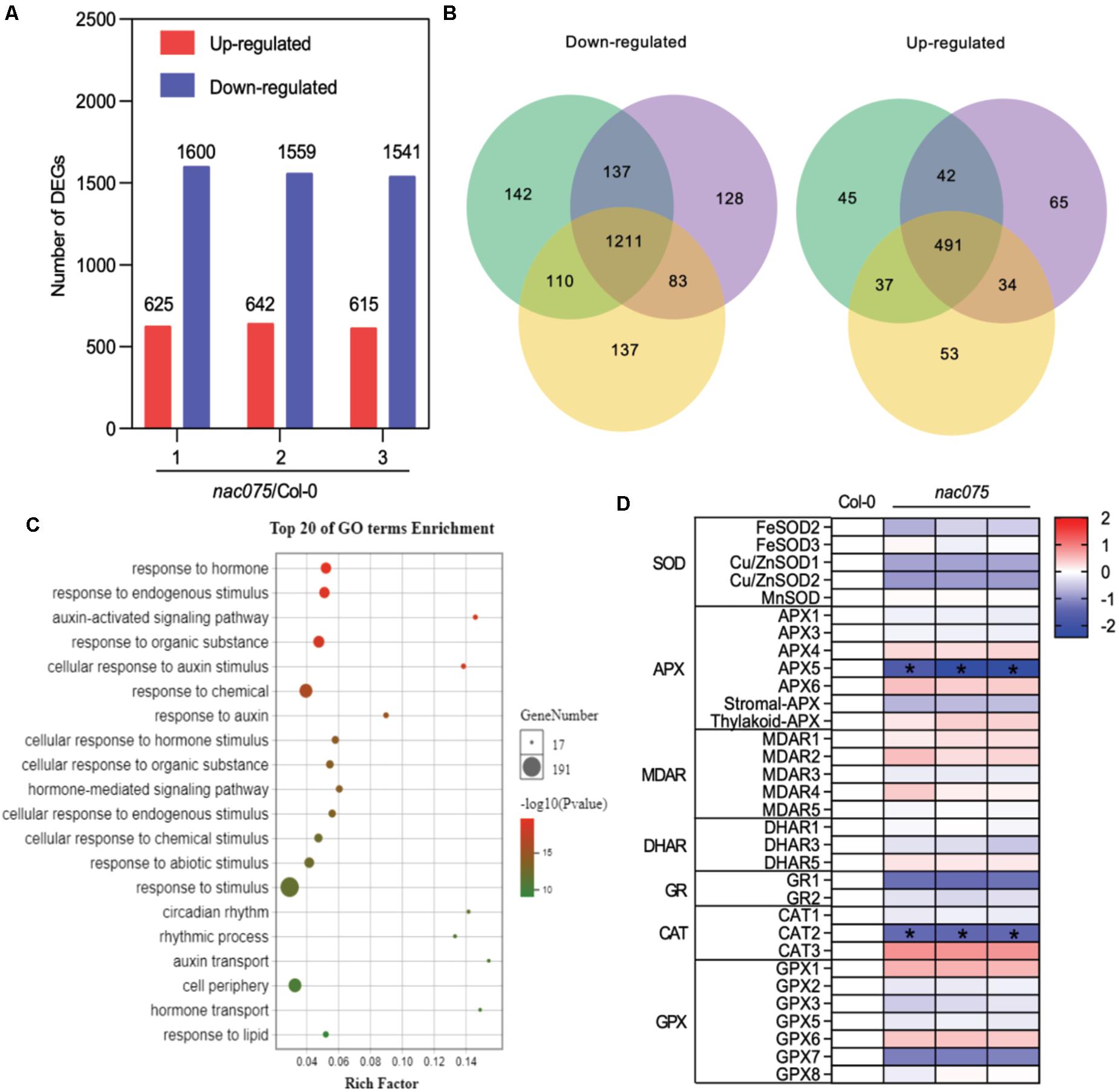
Figure 3. Genome-wide transcriptome analysis of nac075 mutant plants. (A) The number of up-regulated and down-regulated genes in three biological replicates of nac075 mutant plant rosette leaves vs. Col-0 with RNA-seq. Red represents up-regulated genes, and blue represents down-regulated genes. (B) Schematic of the Venn diagram analysis of common down-regulated and up-regulated genes [DEG; | Log2 (FC)| > 1, FDR < 0.05] in three nac075 mutant plant leaves relative to the Col-0 control. (C) GO bubble diagram of down-regulated genes among the DEGs in three nac075 rosette leaf samples. The bubble size represents the number of DEGs, and the bubble color represents the P-value. The rich factor equals the number of DEGs in a certain signaling pathway. (D) Heat map showing ROS-related genes in nac075 mutant plant rosette leaves compared to Col-0. Means of three experiments are shown. The log2 fold change scale is indicated on the right side of the heat map. SOD, superoxide dismutase; APX, ascorbate peroxidase; MDAR, monodehydroascorbate reductase; DHAR, dehydroascorbate reductase; GR, glutathione reductase; CAT, catalase; GPX, glutathione peroxidate.
Next, in order to determine the cellular processes associated with the DEGs, we carried out enrichment analysis of GO biological processes (GOBPs) by subjecting the sequences to GO annotations (Young et al., 2010). Interestingly, the GOBP-association analysis revealed that responses to stimulus or chemical (such as salt stresses) and responses to oxygen-containing compounds (such as oxidative/ROS) are the top senescence-promoting processes regulated by NAC075 among all processes (Figure 3C). Overproduction of ROS caused by various stresses has been demonstrated as potentially critical for induction and maintenance of senescence in animals and plants (Woo et al., 2013). Therefore, we performed GO analysis on seven types of ROS-clearance genes in DEGs. The heat map showed that several ROS-clearance genes, such as APX5 and CAT2, were down-regulated in mutants (Figure 3D), which is consistent with the early-senescence phenotype of nac075 mutants (Figure 2). Collectively, these data suggest that NAC075 delays leaf senescence process through negatively regulating senescence-promoting processes such as responses to oxidative/ROS stress.
The above data pushed us to explore whether NAC075 directly regulates expressions of genes related to ROS clearance. Interestingly, transcripts of CATALASE2 (CAT2), an important ROS scavenging enzyme, were significantly decreased in the leaves of nac075 mutants in comparison with Col-0 (Figure 4A), which is consistent with the transcriptome data (Supplementary Dataset 1). To examine whether CAT2 is a direct target of NAC075, we firstly identified the NAC075 binding sites (NBSs) in the promoter regions of CAT2. Based on a previous study (Lindemose et al., 2014), two NBSs (TG/ACGT) were identified and then used for ChIP assay using 35-day-old Pro35S:NAC075-GFP/Col-0 (NAC075ox) transgenic plants. ChIP-qPCR analysis showed that NAC075 is significantly enriched in TACGT regions, indicating that NAC075 binds to these regions in vivo (Figure 4B). We next performed EMSAs to examine the in vitro binding activity of NAC075. The results revealed that NAC075 protein tagged with His (NAC075-His) was capable of binding probes containing P2, while it was unable to bind probes containing P1 (Figure 4C). Using unlabeled probes as competitors, competitive binding assays were carried out to confirm the binding specificity by adding an excess of unlabeled competitor DNA fragments (Figure 4C), suggesting further that NAC075 directly binds the promoter of CAT2. To further investigate the regulatory roles of NAC075 on CAT2, we generated inducible overexpressing plants pER8-FLAG-NAC075. Upon treatment with β-estradiol, NAC075 transcripts increased (Figure 4D). As expected, expression levels of CAT2 also increased (Figure 4D). Collectively, the above data reveal that NAC075 can directly bind the promoter of CAT2 and regulate its expression.
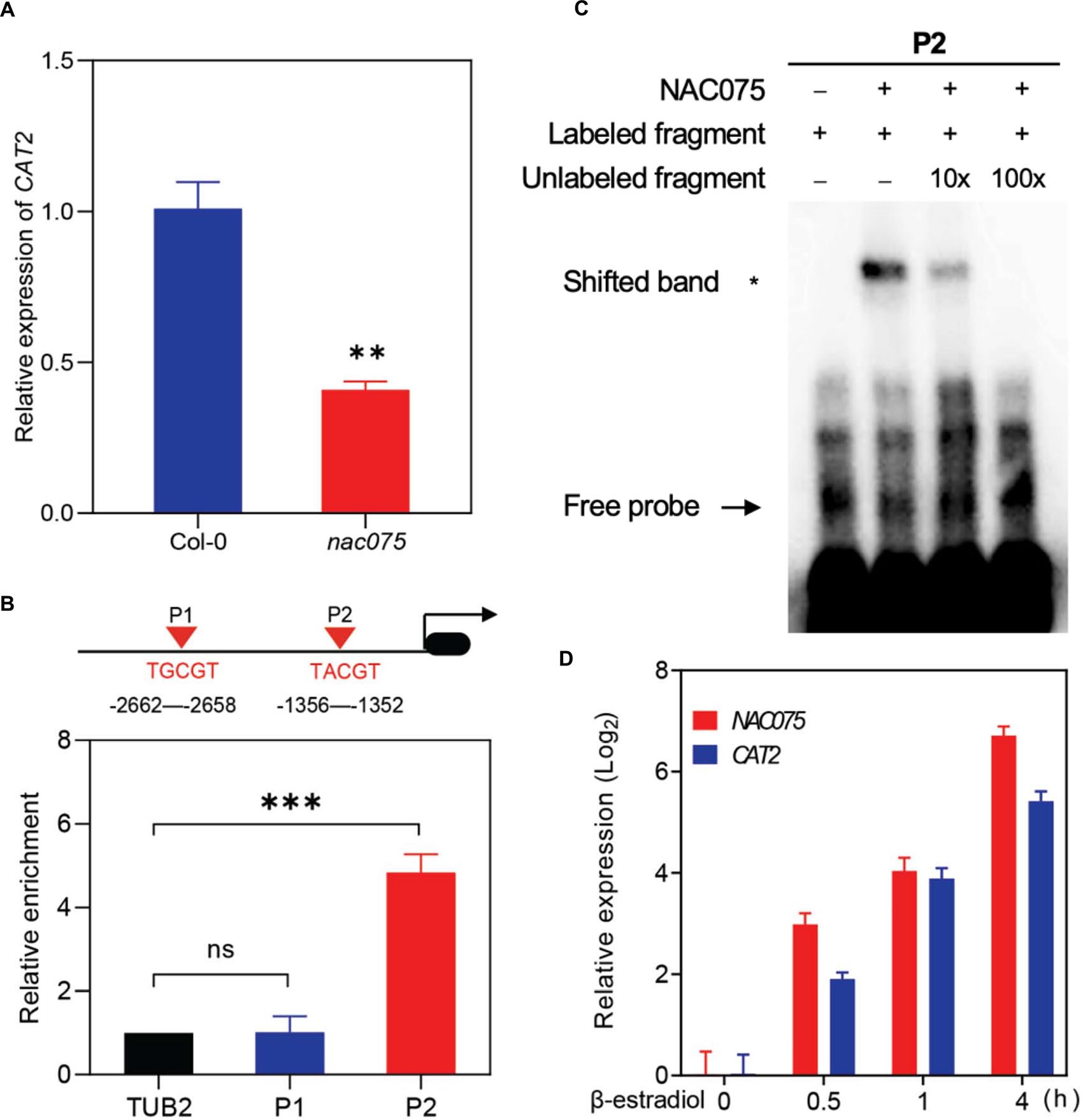
Figure 4. CAT2 is a direct target of NAC075. (A) Relative expression level of CAT2 in the third and fourth rosette leaves of 25-day-old Col-0 and nac075 mutant plants. Data are means ± SD, n = 3. (B) NAC075 directly binds to the promoter of CAT2. Schematic diagram indicates the locations of two putative NAC075 binding sites (P1 and P2) in the CAT2 promoter (top). ChIP-qPCR was performed to examine the relative NAC075 binding to the CAT2 promoter (bottom). An anti-GFP monoclonal antibody was used for DNA immunoprecipitation from 28-day-old Pro35S:NAC075-GFP/Col-0 transgenic plants. The relative enrichment of NAC075 binding to CAT2 promoter was normalized to TUBULIN2 (TUB2). Student’s t-test, ∗∗P < 0 .01, ∗∗∗P < 0.001, ns, not significant. Data are means ± SD of three independent biological replicates. (C) EMSA assay of the binding of NAC075 to the CAT2 promoter in vitro. Biotin-labeled probe was used to EMSA experiment and non-labeled fragments were used as competitors. The + and − symbols represent the presence and absence of components. Three biological replicates were performed with similar results. (D) qRT-PCR analysis of NAC075 and CAT2 expressions in the pER8-FLAG-NAC075 plants treated with 50 mM β-estradiol for the indicated time points. Data are represented as means ± SD, n = 3.
To further explore the genetic regulatory relationship between NAC075 and CAT2 in leaf senescence, we generated nac075 CAT2ox plants by crossing the CAT2ox (Guo et al., 2017) transgenic plants to nac075 mutant. Under long-day conditions, plants with the combined nac075 CAT2ox genotype showed an obvious delayed senescence phenotype compared with nac075 mutant plants, indicating that the accelerated leaf senescence phenotype of nac075 mutant plants was effectively suppressed by CAT2 (Figure 5A). Moreover, DAB staining showed that H2O2 levels in the nac075 CAT2ox and CAT2ox plants was significantly lower than that of nac075 and Col-0 plants (Figure 5B). This indicates that overexpression of CAT2 suppresses the elevated H2O2 levels in nac075 mutant plants, which is in line with the observation that CAT2 is the downstream target gene of NAC075. Similarly, higher chlorophyll content and Fv/Fm further confirmed the delayed senescence phenotype of nac075 CAT2ox plants (Figures 5C,D). Taken together, these results reveal that overexpression of CAT2 suppresses the early senescence phenotype in nac075 mutant plants by reducing H2O2 accumulation. Therefore, a regulatory module is proposed, which is the NAC075-CAT2 pathway modulates leaf senescence by regulating ROS levels (Figure 5E).
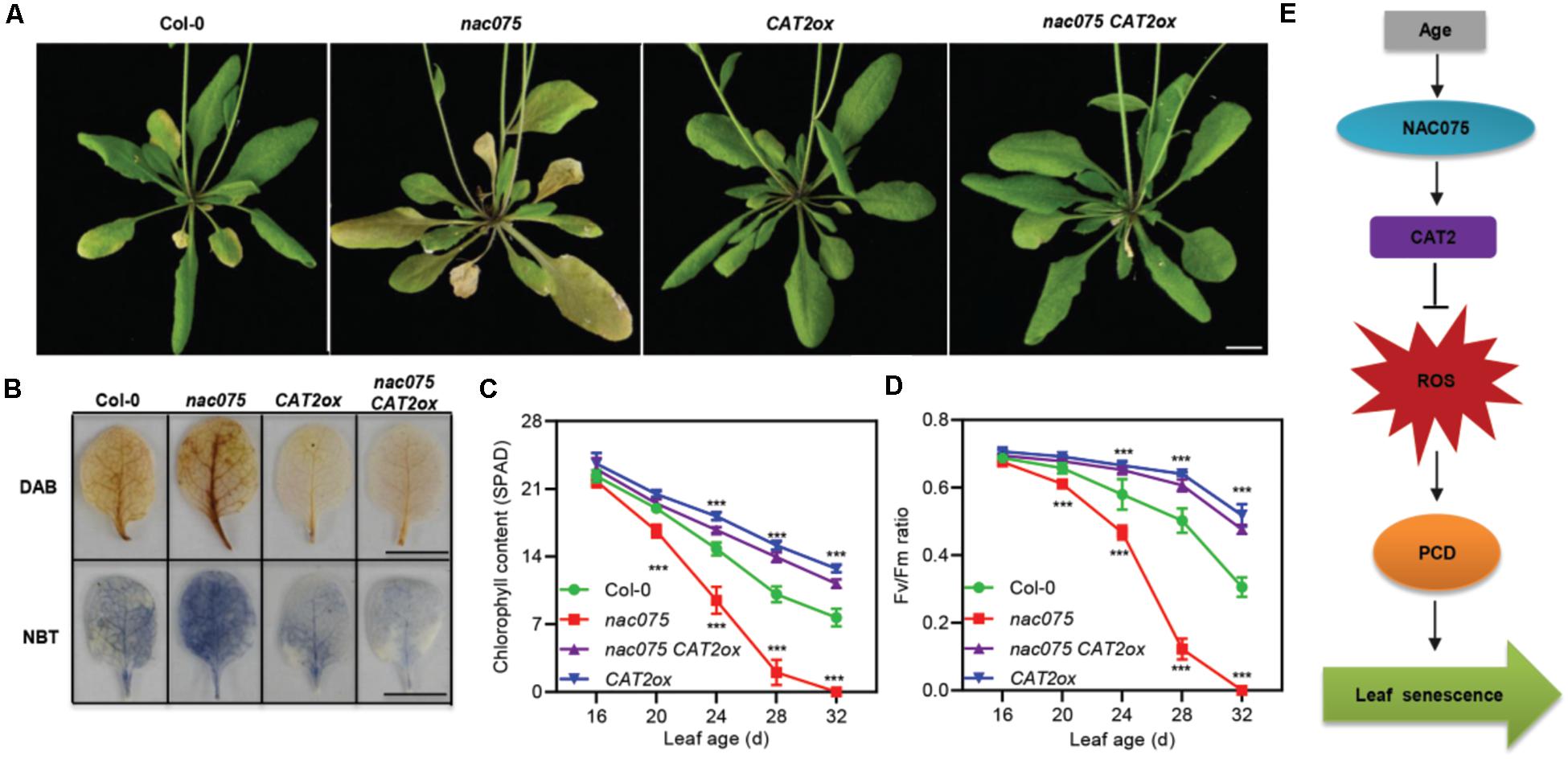
Figure 5. Overexpression of CAT2 suppresses the early senescence phenotype of nac075 mutant plants. (A) Leaf senescence phenotype of 32-day-old Col-0, nac075, nac075 CAT2ox, and CAT2ox plants. The scale bar represents 1 cm. (B) DAB and NBT staining were used to detect H2O2 and O−2 accumulation, respectively, in the third or fourth leaves of Col-0, nac075, nac075 CAT2ox, and CAT2ox plants. The brown and blue color represent H2O2 and O−2 accumulation, respectively. Scale bar, 1 cm. (C,D) Measurements of chlorophyll contents (C) and photochemical efficiency (Fv/Fm) (D) in Col-0, nac075, nac075 CAT2ox, and CAT2ox plants as leaves age. Data are represented as means ± SD, n = 3. The experiment was performed three times with similar results. Student’s t-test, ***P < 0.001. (E) A proposed model illustrates the transcription factor NAC075 that delays leaf senescence by deterring reactive oxygen species accumulation in Arabidopsis. NAC075 promotes CAT2 transcription by directly binding to its promoter, which is able to suppress ROS overproduction. Decreased ROS levels are capable of reducing programmed cell death, thereby delaying leaf senescence.
Leaf senescence is a process of programmed cell death (PCD), which not only is affected by various internal and external factors but also involves highly complex regulatory processes with the coordinated actions of multiple pathways (Lim et al., 2007; Woo et al., 2013, 2019). Deep dissection of the molecular mechanism underlying the leaf senescence may provide a theoretical basis for crop genetic breeding. As leaf senescence involves extensive transcriptional reprogramming, the dynamic activation of transcription factors is considered as a key mechanism that controls the age-dependent expression of thousands of SAGs (Woo et al., 2013). Transcriptome profiling has revealed that a number of NAC genes showed enhanced expression during leaf senescence in Arabidopsis, indicating that they play important roles in the senescence process (Kim et al., 2016). Genetic analyses reveal that a number of NAC TFs function as positive (ANAC016, ANAC029/AtNAP, ANAC046, ANAC059/ORS1, and ANAC092/ORE1) (Guo and Gan, 2006; Balazadeh et al., 2010, 2011; Kim et al., 2013; Oda-Yamamizo et al., 2016) or negative (ANAC042/JUB1, ATAF1/ANAC002 and ANAC083/VNI2) regulators of leaf senescence (Yang et al., 2011; Wu et al., 2012; Garapati et al., 2015). Recently, ANAC017, ANAC082, and ANAC090, referred to as a “NAC troika,” are responsible for governing the positive-to-negative regulatory shift and function as negative regulators of leaf senescence in Arabidopsis (Kim et al., 2018). Here, our study revealed that NAC TF NAC075 acts as a novel negative regulator in the age-dependent leaf senescence.
Our data demonstrated that NAC075 is a functional SAG whose transcription level increases with age (Figure 1A). To this end, we screened knockout lines and generated overexpression transgenic plants to investigate its function in leaf senescence. Loss of function of NAC075 significantly accelerated leaf senescence, whereas overexpression of NAC075 delayed leaf senescence (Figure 2A), further supporting its negative function in regulating leaf senescence. In addition, overexpression of NAC075 also evidently suppressed numerous plant hormones or stress-induced leaf senescence. We also found that NAC75 and other NAC TFs may have functional redundancy in regulating leaf senescence and will construct multiple mutants to verify this possibility in the future. RNA-seq profiling analysis revealed that most of the DEGs are enriched in response to stimulus, and a large portion of ROS-clearance genes were significantly downregulated in nac075 mutant plants (Figures 3C,D). Consequently, we observed that the ROS content in nac075 mutant plants was increased compared with the wild type (Figure 5B). This indicates that NAC075 functions during leaf senescence likely by regulating the expression of ROS-clearance genes. Accordingly, we found that ROS scavenging enzyme CAT2 is one of the putative target genes of NAC075. H2O2 is a well-defined inducers of leaf senescence and CAT2 is a key gene responsible for removing H2O2 (Vandenabeele et al., 2004; Hieno et al., 2019). Our ChIP-qPCR and EMSA experiments demonstrated that NAC075 bound directly to the CAT2 promoter, indicating that CAT2 is a direct target of NAC075 (Figures 4C,D). In addition, CAT2 overexpression suppresses the early senescence phenotype of nac075 mutant plants (Figure 5B), providing genetic evidence for the importance of CAT2 transcription promotion by NAC075 to leaf senescence and ROS accumulation. Based on these results, we conclude that NAC075 suppresses ROS production and leaf senescence by inducing CAT2 expression. Currently, the upstream TFs regulating age-dependent NAC075 transcription are unclear.
It is reported that increased ROS levels due to decreased antioxidant capacity is highly correlated with leaf senescence (Rogers and Munne-Bosch, 2016). A number of studies have previously reported that NAC TFs regulate leaf senescence by modulating ROS levels, such as JUB1 (ANAC042) (Wu et al., 2012), ATAF1 (ANAC002) (Garapati et al., 2015), ORS1 (ANAC059) (Balazadeh et al., 2011), NTL4 (NAC53) (Lee et al., 2012), ANAC017 (Kim et al., 2018), and ANAC032 (Mahmood et al., 2016). Previous studies reveal that the expressions of JUB1 and ORS1 are induced by H2O2 (Balazadeh et al., 2011; Wu et al., 2012), while our work found that NAC075 is not response to ROS, suggesting that NAC075 acts as an upstream negative regulator of ROS accumulation but is not induced by ROS.
Based on our data, we proposed a NAC075-CAT2-ROS model to clarify how NAC075 is responsible for delaying leaf senescence (Figure 5E). In this model, NAC075 transcription is induced by age. Elevated ROS levels lead to PCD and accelerate the senescence process of leaves (Lee et al., 2012; Wu et al., 2012; Rogers and Munne-Bosch, 2016), whereas NAC075 is able to deter the accumulation of ROS by promoting CAT2 transcription and thereby delay leaf senescence. It is reported that NAC075 is involved in the secondary cell wall formation and the regulation of flowering (Sumire and Nobutaka, 2016). Transcriptome analysis also shows that NAC075 is involved in an array of biotic and abiotic stresses as well as the signal transduction process of plant hormones. Given that the downstream regulatory networks dictated by NAC075 in these processes are still unclear, our finding of the regulatory role of NAC075 in ROS scavenging in leaf senescence offers a potential mechanism for these processes as well.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/, PRJNA689040.
ZL and HG conceived the project and designed the experiments. YS and XX designed part of the experiments. CK carried out most of the experiments. YZ conducted ChIP and EMSA assays. H-LW analyzed the RNA-seq data. ZL and CK wrote the manuscript with input from all co-authors. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31970196 to ZL, 32011540381 to ZL, 31570286 to HG, 31900173 to HW, and 31770649 to XX), the National Key Research and Development Program of China (No. 2019YFA0903904 to HG), Shenzhen Science and Technology Program (KQTD20190929173906742 to HG), Chinese Postdoctoral Science Foundation (2019M650514 to YZ and 2019M650516 to HW), and the startup funding for plant aging research from “Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University.”
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the lab members for their helpful suggestions and discussions. T-DNA mutant lines were purchased from the NASC.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.634040/full#supplementary-material
Supplementary Figure 1 | Transcript level of SAG12, AtNAP and ORE1 increases as leaf ages.
Supplementary Figure 2 | PCR genotyping of the nac075 mutants.
Supplementary Figure 3 | Overexpression of NAC075 delayed leaf senescence.
Supplementary Figure 4 | The age-dependent pods senescence phenotype of 30-d-old Col-0 and nac075 mutants.
Supplementary Figure 5 | Senescence phenotypes of Col-0, nac075 mutants and NAC075ox plants.
Supplementary Table 1 | Primers used in this study.
Supplementary Dataset 1 | The differential expressed genes in nac075 mutant.
Balazadeh, S., Kwasniewski, M., Caldana, C., Mehrnia, M., Zanor, M. I., Xue, G. P., et al. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4, 346–360. doi: 10.1093/mp/ssq080
Balazadeh, S., Riano-Pachon, D. M., and Mueller-Roeber, B. (2008). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 10(Suppl. 1), 63–75. doi: 10.1111/j.1438-8677.2008.00088.x
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana-Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313x.2010.04151.x
Breeze, E., Harrison, E., Mchattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P. O., Nam, H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585. doi: 10.1111/j.1365-313x.2005.02399.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP: : cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U. S. A. 97, 3718–3723. doi: 10.1073/pnas.97.7.3718
Garapati, P., Xue, G. P., Munne-Bosch, S., and Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. doi: 10.1104/pp.15.00567
Guo, P., Li, Z., Huang, P., Li, B., Fang, S., Chu, J., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29, 2854–2870. doi: 10.1105/tpc.17.00438
Guo, Y. (2013). Towards systems biological understanding of leaf senescence. Plant Mol. Biol. 82, 519–528. doi: 10.1007/s11103-012-9974-2
Guo, Y., and Gan, S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71, 83–112. doi: 10.1016/s0070-2153(05)71003-6
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313x.2006.02723.x
Hieno, A., Naznin, H. A., Inaba-Hasegawa, K., Yokogawa, T., Hayami, N., Nomoto, M., et al. (2019). Transcriptome analysis and identification of a transcriptional regulatory network in the response to H2O2. Plant Physiol. 180, 1629–1646. doi: 10.1104/pp.18.01426
Hughes, K. A., and Reynolds, R. M. (2005). Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 50, 421–445. doi: 10.1146/annurev.ento.50.071803.130409
Jensen, M. K., Lindemose, S., De Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3, 321–327. doi: 10.1016/j.fob.2013.07.006
Kim, H. J., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, H. J., Park, J. H., Kim, J., Kim, J. J., Hong, S., Kim, J., et al. (2018). Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, E4930–E4939.
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Kim, Y. S., Sakuraba, Y., Han, S. H., Yoo, S. C., and Paek, N. C. (2013). Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54, 1660–1672. doi: 10.1093/pcp/pct113
Lee, S., Seo, P. J., Lee, H. J., and Park, C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70, 831–844. doi: 10.1111/j.1365-313x.2012.04932.x
Li, W., Li, X., Chao, J., Zhang, Z., Wang, W., and Guo, Y. (2018). NAC family transcription factors in Tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 9:1900. doi: 10.3389/fpls.2018.01900
Li, X., Ahmad, S., Ali, A., Guo, C., Li, H., Yu, J., et al. (2019). Characterization of somatic embryogenesis receptor-like kinase 4 as a negative regulator of leaf senescence in Arabidopsis. Cells 8:50. doi: 10.3390/cells8010050
Li, Z., Zhang, Y., Zou, D., Zhao, Y., Wang, H. L., Zhang, Y., et al. (2020). LSD 3.0: a comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 48, D1069–D1075.
Lindemose, S., Jensen, M. K., Van De Velde, J., O’shea, C., Heyndrickx, K. S., Workman, C. T., et al. (2014). A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 42, 7681–7693. doi: 10.1093/nar/gku502
Mahmood, K., El-Kereamy, A., Kim, S. H., Nambara, E., and Rothstein, S. J. (2016). ANAC032 positively regulates age-dependent and stress-induced senescence in Arabidopsis thaliana. Plant Cell Physiol. 57, 2029–2046. doi: 10.1093/pcp/pcw120
Mao, C., Lu, S., Lv, B., Zhang, B., Shen, J., He, J., et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763. doi: 10.1104/pp.17.00542
Noh, Y. S., and Amasino, R. M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41, 181–194.
Oda-Yamamizo, C., Mitsuda, N., Sakamoto, S., Ogawa, D., Ohme-Takagi, M., and Ohmiya, A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 6:23609.
Pontier, D., Gan, S., Amasino, R. M., Roby, D., and Lam, E. (1999). Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39, 1243–1255.
Rogers, H., and Munne-Bosch, S. (2016). Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: similar but different. Plant Physiol. 171, 1560–1568. doi: 10.1104/pp.16.00163
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Sumire, F., and Nobutaka, M. (2016). ANAC075, a putative regulator of vascular-related NAC-DOMAIN7, is a repressor of flowering. Plant Biotechnol. 33, 255–265. doi: 10.5511/plantbiotechnology.16.0215b
Vandenabeele, S., Vanderauwera, S., Vuylsteke, M., Rombauts, S., Langebartels, C., Seidlitz, H. K., et al. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58. doi: 10.1111/j.1365-313x.2004.02105.x
Woo, H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. doi: 10.1105/tpc.13.8.1779
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Woo, H. R., Kim, H. J., Nam, H. G., and Lim, P. O. (2013). Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell. Sci. 126, 4823–4833. doi: 10.1242/jcs.109116
Woo, H. R., Koo, H. J., Kim, J., Jeong, H., Yang, J. O., Lee, I. H., et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171, 452–467. doi: 10.1104/pp.15.01929
Wu, A., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506. doi: 10.1105/tpc.111.090894
Yang, S.-D., Seo, P. J., Yoon, H.-K., and Park, C.-M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23, 2155–2168. doi: 10.1105/tpc.111.084913
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14.
Keywords: leaf senescence, NAC transcription factor, reactive oxygen species, catalase, Arabidopsis thaliana
Citation: Kan C, Zhang Y, Wang H-L, Shen Y, Xia X, Guo H and Li Z (2021) Transcription Factor NAC075 Delays Leaf Senescence by Deterring Reactive Oxygen Species Accumulation in Arabidopsis. Front. Plant Sci. 12:634040. doi: 10.3389/fpls.2021.634040
Received: 26 November 2020; Accepted: 22 January 2021;
Published: 24 February 2021.
Edited by:
Yongfeng Guo, Tobacco Research Institute (CAAS), ChinaReviewed by:
Pyung Ok Lim, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaCopyright © 2021 Kan, Zhang, Wang, Shen, Xia, Guo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Guo, Z3VvaHdAc3VzdGVjaC5lZHUuY24=; Zhonghai Li, bGl6aG9uZ2hhaUBiamZ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.