- 1National Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 2Biology and Food Engineering School, Fuyang Normal University, Fuyang, China
- 3Shandong Lufeng Group Co., Ltd., Anqiu, China
Abscisic acid (ABA) is an important phytohormone regulating plant growth, development and stress responses. A multitude of key factors implicated in ABA signaling have been identified; however, the regulation network of these factors needs for further information. AtS40.4, a plant-specific DUF584 domain-containing protein, was identified previously as a senescence regulator in Arabidopsis. In this study, our finding showed that AtS40.4 was negatively involved in ABA signaling during seed germination and early seedling growth. AtS40.4 was highly expressed in seeds and seedlings, and the expression level was promoted by ABA. AtS40.4 was localized both in the nucleus and the cytoplasm. Moreover, the subcellular localization pattern of AtS40.4 was affected by ABA. The knockdown mutants of AtS40.4 exhibited an increased sensitivity to ABA, whereas the overexpression of AtS40.4 decreased the ABA response during seed germination and seedling growth of Arabidopsis. Furthermore, AtS40.4 was involved in ABRE-dependent ABA signaling and influenced the expression levels of ABA INSENTIVE (ABI)1-5 and SnRK2.6. Further genetic evidence demonstrated that AtS40.4 functioned upstream of ABI4. These findings support the notion that AtS40.4 is a novel negative regulator of the ABA response network during seed germination and early seedling growth.
Introduction
Abscisic acid (ABA) is a key phytohormone, which plays versatile functions in developmental regulation and various environmental stress response. The ABA plays roles in spatiotemporal and dose-dependent ways, which is connected with its diverse functions (Liu et al., 2019; Lu et al., 2019; Chandrasekaran et al., 2020; Xie et al., 2020). ABA is well-known for its stress-related properties, but it also has been proven to regulate plant growth and development such as seed germination, root growth and flowering transition (Huang et al., 2017; Belda-Palazon et al., 2018; Xiong et al., 2019). ABA content in seeds rapidly decreases after imbibition, which facilitates seed germination; whereas the exposure of seeds to ABA during germination rapidly arrests seed growth (Zhu et al., 2011; Ye et al., 2012). ABA maintains or represses the primary root elongation and the lateral root formation during plants response to different environmental stimuli such as water deficit, osmotic and salt stress (Spollen et al., 2000; Zhang et al., 2010; Duan et al., 2013; Jiang et al., 2013). Recent researches reveal that ABA also regulate plant flowering time. Several core components of ABA signaling modulate expression levels of key genes in flowering transition (Shu et al., 2018; Conti, 2019; Xiong et al., 2019).
Many signaling details of ABA have been well-elucidated. ABA signaling network is tightly regulated and modulated by various factors at transcription and post-transcription levels in response to endogenous and exogenous cues (Yu et al., 2019; Ali et al., 2020; Gong et al., 2020). A subset of key genes involved in ABA signaling has been identified. Among these genes, ABA INSENTIVE (ABI) 1, ABI2, ABI3, ABI4, and ABI5, encode five key negative or active regulators of ABA response (Leung et al., 1997; Skubacz et al., 2016; Chandrasekaran et al., 2020). ABI1 and ABI2 encode type 2C protein phosphatases (PP2Cs) that play roles as negative regulators of ABA signaling. PP2Cs interact with and inactivate ABA-activated sucrose non-fermenting 1-related protein kinase 2 (SnRK2) members, a core kinase in ABA signaling (Leung et al., 1997). ABI3, ABI4, and ABI5 are transcription factors (TFs) of the B3, APETALA2, and basic leucine zipper families, respectively, and share overlapping functions in ABA signaling as positive regulators (Finkelstein et al., 2002; Cheng et al., 2014; Liu et al., 2017; Chandrasekaran et al., 2020).
In addition to these TFs as trans-acting factors, there are several important cis-acting elements have been identified and well-characterized in ABA signaling such as ABA-responsive element (ABRE), coupling element and dehydration-responsive element (DRE) (Narusaka et al., 2003; Min et al., 2020). ABREs are major cis-acting regulators in ABA-dependent gene expression (Kim et al., 2004; Wang et al., 2019; Chen et al., 2020). ABI3, ABI4, and ABI5 bind directly to the ABREs of the promoter regions of a subset of genes, thereby controlling their transcription levels (Carles et al., 2002; Ezcurra et al., 2000; Mizoi et al., 2012; Shu et al., 2013; Skubacz et al., 2016; Wang et al., 2019; Chandrasekaran et al., 2020; Collin et al., 2020). Notably, ABI3, ABI4, and ABI5 either activate or repress gene expression in diverse processes depending on DNA binding. For instance, ABI4 directly or indirectly induces expression of genes involved in seed maturation; whereas represses transcription of genes functioning in photosynthesis and pigment metabolism, or ABA catabolic pathways (Wind et al., 2013; Baek et al., 2020; Khan et al., 2020). Considering that the core position of these ABIs in ABA signaling, it is very important to understand the expression regulation of ABIs genes.
The plant-specific senescence 40 (S40) family proteins that have a molecular weight of 12–25 kD and contain a plant-specific domain of unknown function 548 (DUF548) are widely present in plants (Fischer-Kilbienski et al., 2010; Jehanzeb et al., 2017; Zheng et al., 2019). DUF548 is an intriguing C-terminal domain sharing the sequence GRXLKGR(D/E) (L/M)XXXR(D/N/T)X(I/V)XXXXG(F/I) which is highly conserved in plant species (Jehanzeb et al., 2017). A subset of genes encoding DUF548 proteins have been identified in barley, Arabidopsis and rice (Krupinska et al., 2002; Fischer-Kilbienski et al., 2010; Zheng et al., 2019). Notably, multiple members of S40 genes exist in one plant species. For example, there are 15 S40s (AtS40s) in Arabidopsis and 16 S40s (OsS40s) in rice (Fischer-Kilbienski et al., 2010; Jehanzeb et al., 2017). AtS40s, OsS40s, and barley S40 (HvS40) participate in regulation of natural and stress-reduced leaf senescence (Krupinska et al., 2002; Fischer-Kilbienski et al., 2010; Janack et al., 2016; Jehanzeb et al., 2017; Zheng et al., 2019). AtS40.3, HvS40, OsS40-1, OsS40-13, and OsS40-14 are partly or completely localized in the nucleus. Moreover, AtS40.3 and HvS40 are assigned as DNA binding proteins (Krupinska et al., 2002; Fischer-Kilbienski et al., 2010). Interestingly, AtS40.3 also affects the sex ratio of cyst nematodes in Arabidopsis (Anwer et al., 2018). Until now, the regulation of expression of these S40 genes, the subcellular localization of these DUF548 proteins and their molecular mechanisms remain largely uncovered.
AtS40.4 (At2G28400) encodes a cytoplasmic S40 protein and is associated with natural and darkness-induced senescence (Fischer-Kilbienski et al., 2010). AtS40.4 expression is regulated by dark-induced senescence, salicylic acid and ABA treatments, and pathogen infections in rosette leaves of 4 weeks old plants. In this study, we showed that AtS40.4 functions in seed germination and early seedling growth as a negative regulator in the ABA signaling pathway. AtS40.4 was localized both in the nucleus and the cytoplasm, and the localization pattern of AtS40.4 was affected by ABA. Moreover, AtS40.4 affects the expression of ABIs and ABRE-containing genes. These findings provide a new insight to understand the molecular features of these plant-specific DUF584 proteins in modulating ABA signaling.
Materials and Methods
Plant Materials and Growth Conditions
The material used in the experiment was Arabidopsis thaliana (Columbia, Col-0). Seeds of T-DNA insertion lines, ats40.4-1 (GABI_561G08) and ats40.4-2 (GABI_863C05), were obtained from the Arabidopsis Biological Resources Center (ABRC). Seeds were surface-sterilized, and placed on 0.5 × MS solid medium with or without ABA (Shi et al., 2018). After stratification for 3 days at 4°C in the dark to break dormancy, the plates were incubated at 20–22°C with a 16/8 h light/dark cycle. The genotyping was identified by using PCR with primers listed in Supplementary Table 1.
Generation of Transgenic Plants and Double Mutant
To generate 35S:AtS40.4 construct, the full-length coding sequence (CDS) of AtS40.4 was amplified using the specific primers (Supplementary Table 2) and was subcloned into the pMD18-T vector (Takara). Subsequently, the fragment was ligated into the binary vector pBI121 by restriction enzymes XbaI and KpnI, resulting in the 35S:AtS40.4 construct. For proAtS40.4:GUS construct, a 1,403 bp genomic fragment upstream of the ATG start codon of AtS40.4 was cloned into the Gateway Entry vector (pCR8/GW/TOPO vector, Invitrogen) via gateway technology, and subsequently was introduced into the pMDC163 vector using the LR Clonase II Plus enzyme (Invitrogen) as described by Xiong et al. (2019). A promoter region (1,403 bp upstream of the start codon) with gene region of AtS40.4 (489 bp with mutation of termination codon TAA to TAT) was subcloned into the pMDC107 vector via gateway technology (Invitrogen) to generate the proAtS40.4:AtS40.4-GFP construct. These constructs were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into wild-type (proAtS40.4:GUS and 35S:AtS40.4) or ats40.4-1 mutant plants (proAtS40.4:AtS40.4-GFP) using the floral dip method (Clough and Bent, 1998). The transgenic seedlings were selected on 0.5 × MS medium containing 25 mg/L hygromycin or kanamycin and then identified by RCR using the primers (Supplementary Table 2). We used T3 seeds for further study.
The abi4 ats40.4 double mutant was created by genetic crossing. The abi4 (SALK_080095) mutant was described previously (Shu et al., 2013). We created the 6xABRE_A:erGFP ats40.4-1 plants by crossing 6xABRE_A:erGFP into ats40.4-1 background to mark the expression of ABRE-containing genes (Wu et al., 2018).
Phenotypic Analysis
Phenotypic analysis was performed as described previously (Li Z. et al., 2016; Shi et al., 2018). Sterilized seeds of wild type, ats40.4 mutants and AtS40.4-overexpressing lines were plated on ABA (Sigma-Aldrich) medium. For root elongation assays, the seedlings grown on 0.5 × MS medium for 3 days were transferred to new 0.5 × MS medium or medium containing ABA for further growth on vertical plates. All the results were calculated based on at least three replicate experiments, and more than 60 seeds or seedlings for each line were used for the quantification.
Gene Expression Analysis
Gene expression analysis was performed by the reverse transcription- polymerase chain reaction (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR) with the specific primers (Supplementary Tables 3, 4). Total RNA was isolated from samples with RNAprep pure Plant Kit (TIANGEN, DP441) and was used for reverse transcription using PrimeScriptTM RT reagent Kit (TAKARA). TUBULIN2 and GAPDH were used as the control genes for RT-PCR and qRT-PCR, respectively. All experiments were performed from three biological replicates.
For analysis of expression levels of genes after ABA treatment, 5-day-old seedlings grown on 0.5 × MS liquid medium were used to exogenous ABA treatment. The seedlings were placed into 0.5 × MS liquid medium with ABA (10 μM, in ethanol), and the same amount of ethanol as a negative control. Only the roots of seedlings were immersed in ABA medium and whole seedlings were used for extraction of total RNA (Xiong et al., 2019). The expression levels of genes selected in ABA signaling were detected as descripted by Mei et al. (2014).
GUS Histochemical Staining
Six independent lines expressing the proAtS40.4:GUS construct in Col-0 background were analyzed for GUS histochemical staining. GUS staining was performed as described by Xiong et al. (2019). Samples were fixed using 90% acetone for 20 min and then were immersed in the staining solution were incubated in staining solution (50 mM NaPO4, pH 7.2, 2 mM X-gluc, 0.5 mM K3Fe (CN)6, and 0.5 mM K4Fe (CN)6, 0.1% Triton X-100) at 37°C overnight after vacuum infiltration for 10 min on ice. Then stained materials were washed with 30, 50, 75, 85, 95, 100% ethanol for 1 h each to remove the chlorophyll.
To analyze the effect of ABA on AtS40.4 expression, 3-day-old seedings transformed proAtS40.4:GUS were transferred into 0.5 × MS liquid medium with or without 10 μM ABA for 6 h before GUS staining.
Fluorescence Imaging and Quantification
To explore the subcellular localization of AtS40.4, six independent lines transformed proAtS40.4:AtS40.4-GFP construct in ats40.4-1 plants were selected. The green fluorescence in root tip cells of 7-day-old seedlings were detected. To test the effects of ABA on the subcellular localization of AtS40.4, the roots of 7-day-old seedlings were treated with 0.5 × MS liquid medium containing 100 μM ABA for 2 or 4 h (Xu et al., 2019). PEG-induced moderate severity (-0.75 MPa) was used to simulate drought stress to roots (Verslues et al., 2006; Chong et al., 2019).
The fluorescence signals of green fluorescent protein (GFP) and 4′,6-diamidino-2-phenylindole (DAPI) were captured using an LSM 880 laser scanning confocal microscope (Carl Zeiss) with the 488/505–550 and 358/460 nm excitation/emission wavelengths, respectively. Quantification of fluorescence intensity was performed using more than 30 plants in each sample. ImageJ was used to quantify the GFP fluorescence intensity.
Results
Ats40.4 Mutations Increased Sensitivity to ABA During Seed Germination and Early Seedling Growth
To understand the roles of these plant-specific genes which encode DUF-containing proteins in ABA signaling, we screened the Arabidopsis T-DNA insertional population with different responses to exogenous ABA from the wild type. Among these mutants, GABI_561G08 and GABI_863C05 (named as ats40.4-1 and ats40.4-2), two mutants of AtS40.4 (At2G28400), showed increased sensitivity to ABA during seed germination and seedling growth. The T-DNA insertion in the AtS40.4 5’ UTR in ats40.4-1 and ats40.4-2 mutants led to a significantly reduced expression of AtS40.4 (Figures 1A–C).
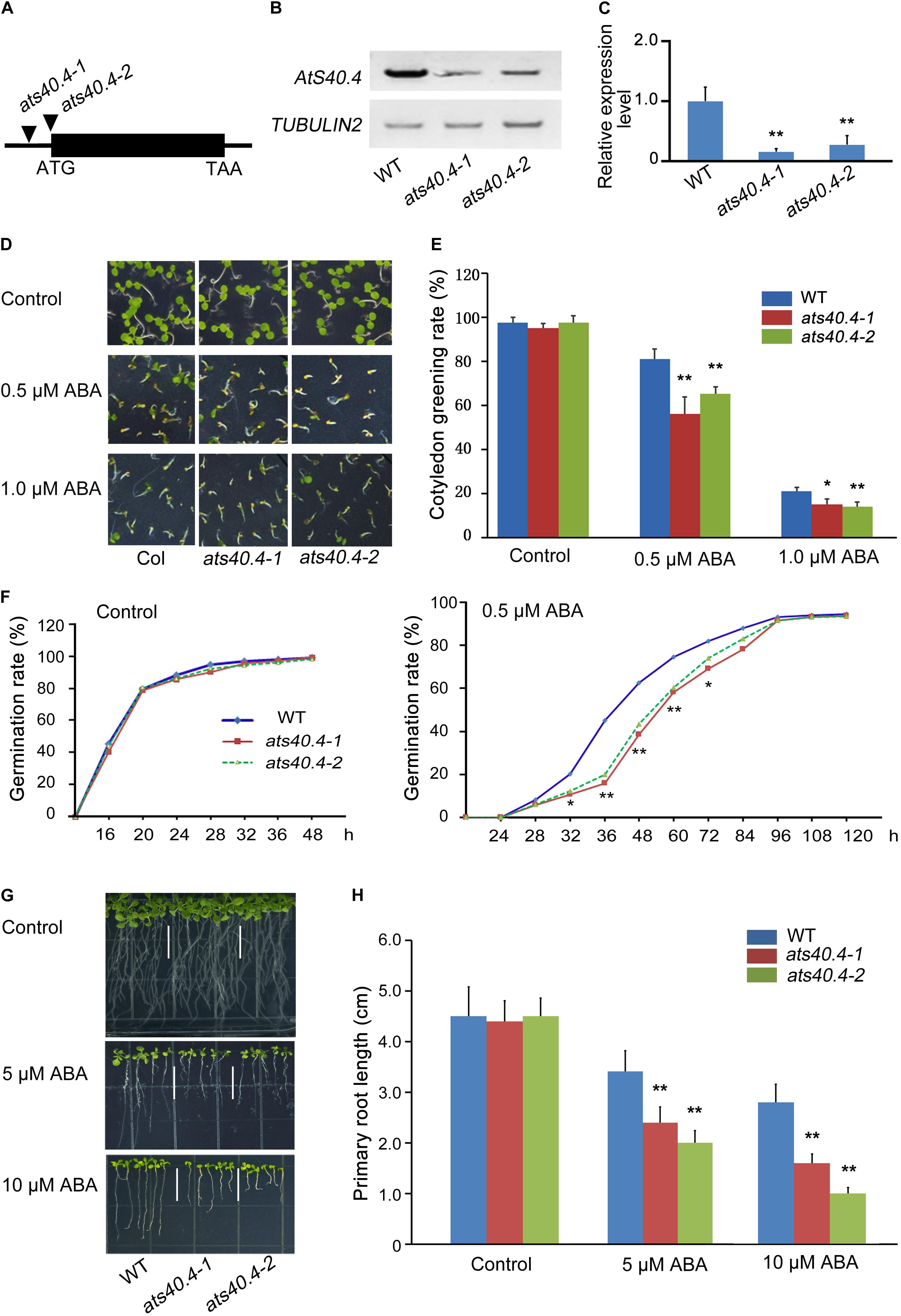
Figure 1. AtS40.4 negatively regulates ABA signaling during seed germination and early seedling growth. (A) Genomic structure of AtS40.4 (At2g28400) and the positions of T-DNA insertions (triangles) of ats40.4-1 (GABI_561G08) and ats40.4-2 (GABI_863C05). (B,C) RT-PCR and qRT-PCR analysis of AtS40.4 expression in the seedlings of Col-0 wild-type (WT) and ats40.4 mutants. TUBULIN2 and GAPDH were used as internal controls for RT-PCR (B) and qRT-PCR (C), respectively. Significant differences are denoted with double asterisks (C) (Student’s t-test, **P < 0.01). (D) Seed germination and seedling growth of the WT and ats40.4 mutants grown on 0.5 × MS medium containing 0, 0.5, and 1.0 μM ABA. (E) Cotyledon greening rates of seedlings grown on 0.5 × MS medium containing 0, 0.5, and 1.0 μM ABA. Error bars indicate the SEs of three replicated experiments. Single and double asterisks indicate a significant difference (Student’s t-test; *P < 0.05; **P < 0.01). (F) Seed germination rates of WT and ats40.4 mutants. Seeds were grown on 0.5 × MS medium containing 0 (left) and 0.5 μM ABA (right). Asterisks indicate a significant difference from the control value (*P < 0.05; **P < 0.01). (G,H) Comparison of root elongation of WT with ats40.4 mutants. The 3-day-old seedlings grown on 0.5 × MS medium were transferred onto the medium containing 0, 5 and 10 μM ABA for further growth 14 days (G). Data (H) are means ± SEs based on three biological replicates. Double asterisks indicate statistically significant differences between the mutants and WT calculated by Student’s t-test (**P < 0.01).
AtS40.4 encodes a DUF584 protein involved in the leaf senescence (Fischer-Kilbienski et al., 2010). To investigate the effect of AtS40.4 on the ABA responses during seed germination and seedling growth, we analyzed the changes of seed germination and seedling growth of ats40.4 mutants. In the absence of exogenous ABA, ats40.4-1 and ats40.4-2 mutants did not exhibit apparent difference during seed germination and seedling growth (Figure 1D). However, these ats40.4 mutants displayed a significantly increased sensitivity of seed germination and cotyledon greening after ABA application (Figures 1E,F). Moreover, the root growth of the ats40.4-1 and ats40.4-2 mutants was more inhibited than that of wild-type seedlings by ABA (Figures 1G,H). The ABA hypersensitivity of ats40.4-1 mutant was significantly decreased by expressing the AtS40.4 CDS under the control of its native promoter (proAtS40.4:AtS40.4-GFP) (Supplementary Figures 1A–D), indicating that the disruption of the AtS40.4 causes the ABA-hypersensitive phenotype.
To further confirm that AtS40.4 functions in the ABA response during seed germination and seedling growth, we then overexpressed AtS40.4 CDS in the wild-type Arabidopsis under the control of CaMV 35S promoter (35S:AtS40.4). The transcription level of AtS40.4 in the transgenic lines (referred to as OE-1, OE-2, and OE-3) was 3–7-fold higher than that in the wild-type seedlings (Supplementary Figure 2A). The overexpression of AtS40.4 decreased the sensitivity of seed germination and cotyledon greening to exogenous ABA (Supplementary Figures 2B–D). The primary roots of the AtS40.4 overexpressing lines were significantly longer than those of the wild type (Supplementary Figures 2E,F).
Abiotic stresses stimulate the accumulation of endogenous ABA and multiple proteins are involved in the response of plants to abiotic stress through ABA-dependent way (Quesada et al., 2000; González-Guzmán et al., 2002; Lu et al., 2019). To investigate whether or not AtS40.4 plays a role in abiotic stress-induced ABA signaling, we tested the seed germination of ats40.4 mutants and AtS40.4-overexpressing lines on 0.5 × MS medium containing 175 mM NaCl. The germination rate was decreased in the ats40.4 mutants, while increased in the AtS40.4-overexpressing lines under NaCl stress (Supplementary Figures 3A,B), indicating that AtS40.4 might function in slat-induced ABA effects on seed germination. Altogether, these results indicate that AtS40.4 participates in the ABA signaling pathway during seed germination and early seedling growth as a negative regulator.
The ABRE is a major cis-element for ABA-responsive gene expression (Nakashima and Yamaguchi-Shinozaki, 2013; Gong et al., 2020). The 6 × ABRE_A:erGFP is an ABA responsive reporter that can mark the transcriptional response of tissues to ABA (Wu et al., 2018). To further investigate the AtS40.4 functions in the ABA-mediated transcriptional regulation and the sites of action, we introduced 6 × ABRE_A:erGFP into the ats40.4-1 plants and investigated the GFP fluorescence intensity and patterns. ABA application significantly enhanced the GFP-fluorescence intensity in the lateral root cap, columella root cap, epidermis and quiescent center cells in root tips of the seedlings (Figure 2), consisting with the previous report by Wu et al. (2018). Notably, the fluorescence intensity of GFP was significantly increased in the root cap cells of ats40.4-1 background seedlings compared to those in the wild-type Arabidopsis after 10 μM ABA treatment for 12, or 24 h, indicating that AtS40.4 might function in the root cap to sense and response the soil conditions in ABA signaling during seedling growth. This supports that AtS40.4 is involved in ABRE-dependent ABA signaling.
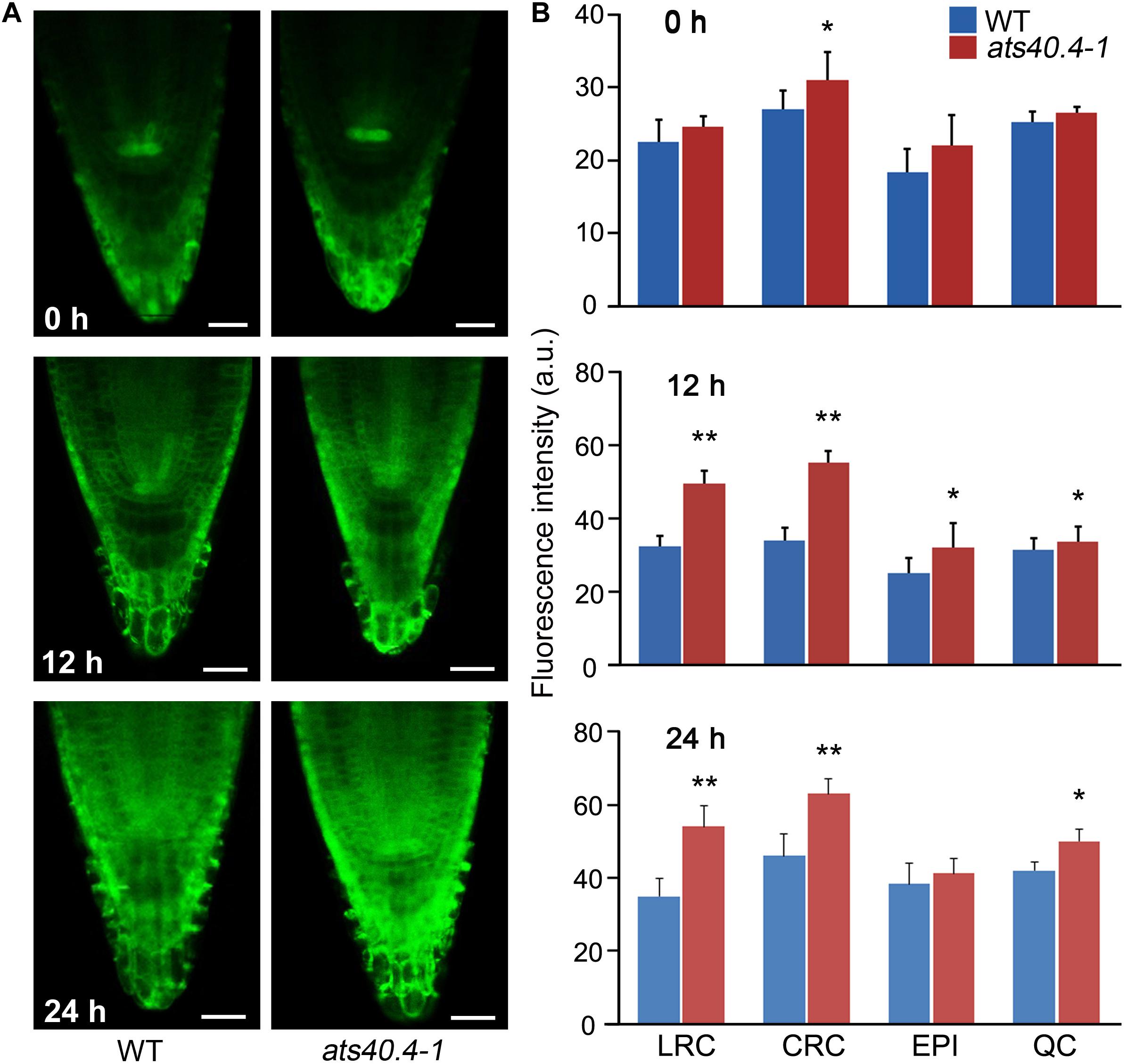
Figure 2. AtS40.4 is involved in the ABRE-dependent ABA signaling. (A) Expression pattern of 6xABRE_A:erGFP in root tips of wild type and ats40.4 background plants under 10 μM ABA applications for 0, 12 and 24 hours. (B) Quantified fluorescence of 6xABRE_A:erGFP in lateral root cap (LRC), columella root cap (CRC), epidermis (EPI) and quiescent center (QC) cells of root tip. Scale bars, 20 μm.
Ats40.4 Is a Ubiquitously Expressed Gene and Highly Expressed in the Seeds and Seedlings
The participation of AtS40.4 in ABA response during seed germination and seedling growth suggested that this gene may play roles at the early stages of Arabidopsis development besides the senescence leaves as described previously (Fischer-Kilbienski et al., 2010). We prepared the total RNA from the seedlings and roots, stems, rosette and cauline leaves, flowers, old-yellowing siliques, and mature seeds of Arabidopsis to examine the expression levels of AtS40.4. The qRT-PCR result showed that AtS40.4 was expressed highly in the seedlings, stems, roots, siliques, and seeds (Figure 3A).
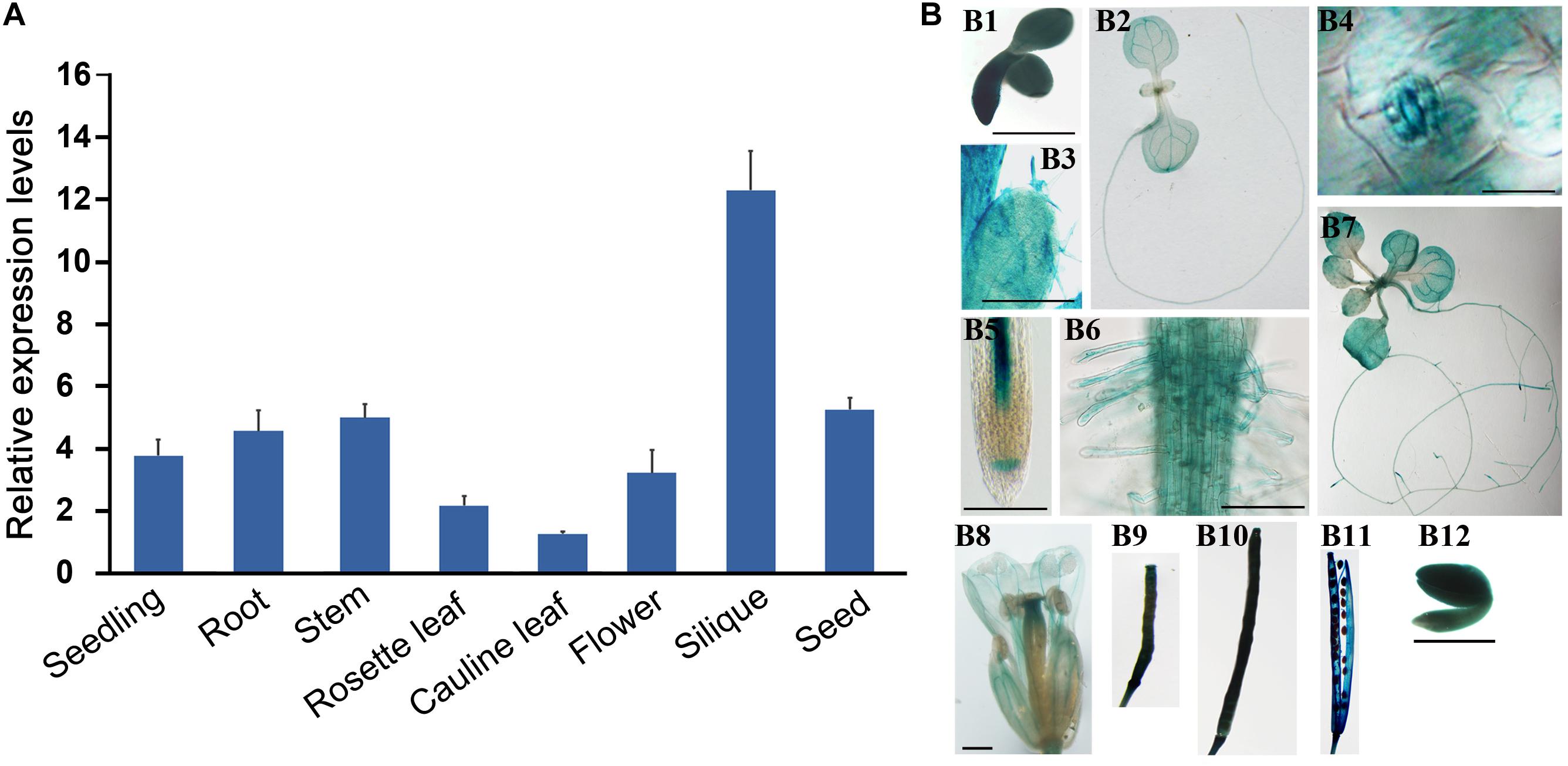
Figure 3. The expression patterns of AtS40.4 in Arabidopsis. (A) Expression level of AtS40.4 in Arabidopsis including seedlings, roots, stems, rosette leaves, cauline leaves, flowers, siliques, and developing seeds detected by qRT-PCR using the specific primers for AtS40.4. GAPDH was used as an internal control. Error bars indicate the SEs of three experimental replicates. (B) GUS histochemical staining shows the expression patterns of AtS40.4 in Arabidopsis expressing the proAtS40.4:GUS construct. The strong GUS signal was observed in the germinating seed (B1), 2-day-old seedling (B2), guard cell (B3) and trichomes (B4) in leaf, root tip (B5), and root hairs (B6) of 2-week old seedling (B7), flower (B8), young (1 and 2 weeks after pollination, B9 and B10) and old-yellowing siliques (B11), and cotyledons of mature seeds (B12). The experiment was repeated twice with a similar result. Scale bars, 100 μm.
To further investigate the temporal and spatial expression patterns of AtS40.4 in Arabidopsis, we generated a proAtS40.4:GUS construct, and transformed this construct into the Col-0 Arabidopsis. Four independent T3 transgenic lines were used to investigate the AtS40.4 expression patterns during Arabidopsis development. The high GUS activity was detected in the cotyledons of 24 h imbibed seeds, 7-day- and 2-week-old seedlings; rosette and cauline leaves, stem, inflorescences, young and old-yellowing siliques, and cotyledons of mature seeds (Figure 3B). The trichomes and guard cells of the leaves, root tips and root hairs, and old-yellowing siliques exhibited strong GUS staining signals. These results demonstrated that AtS40.4 is globally expressed in Arabidopsis at the vegetative growth and reproductive stages.
The Gene Expression and Subcellular Localization of Ats40.4 Are Regulated by ABA
Considering that AtS40.4 functions in ABA signal during early seedling development, we investigated whether the expression of AtS40.4 was regulated by ABA by using qRT-PCR. Five-day-old wild-type seedlings were placed into 0.5 × MS liquid medium with ABA (10 μM, in ethanol), and the ethanol-treatment as a negative control. As shown in Figure 4A, the expression levels of AtS40.4 increased to 12- and 36-fold after ABA treatment for 1 and 6 h, respectively. The transgenic lines expressing proAtS40.4:GUS were next used to explore the response of AtS40.4 to exogenous ABA. In the 3-day-old seedlings of the transgenic line, the GUS signal was detected in the cotyledon, hypocotyl, and root. After 10 μM ABA treatment for 6 h, the GUS reaction in the root of the seedling evidently enhanced (Figure 4B). These results indicate that AtS40.4 is an ABA-responsive gene.
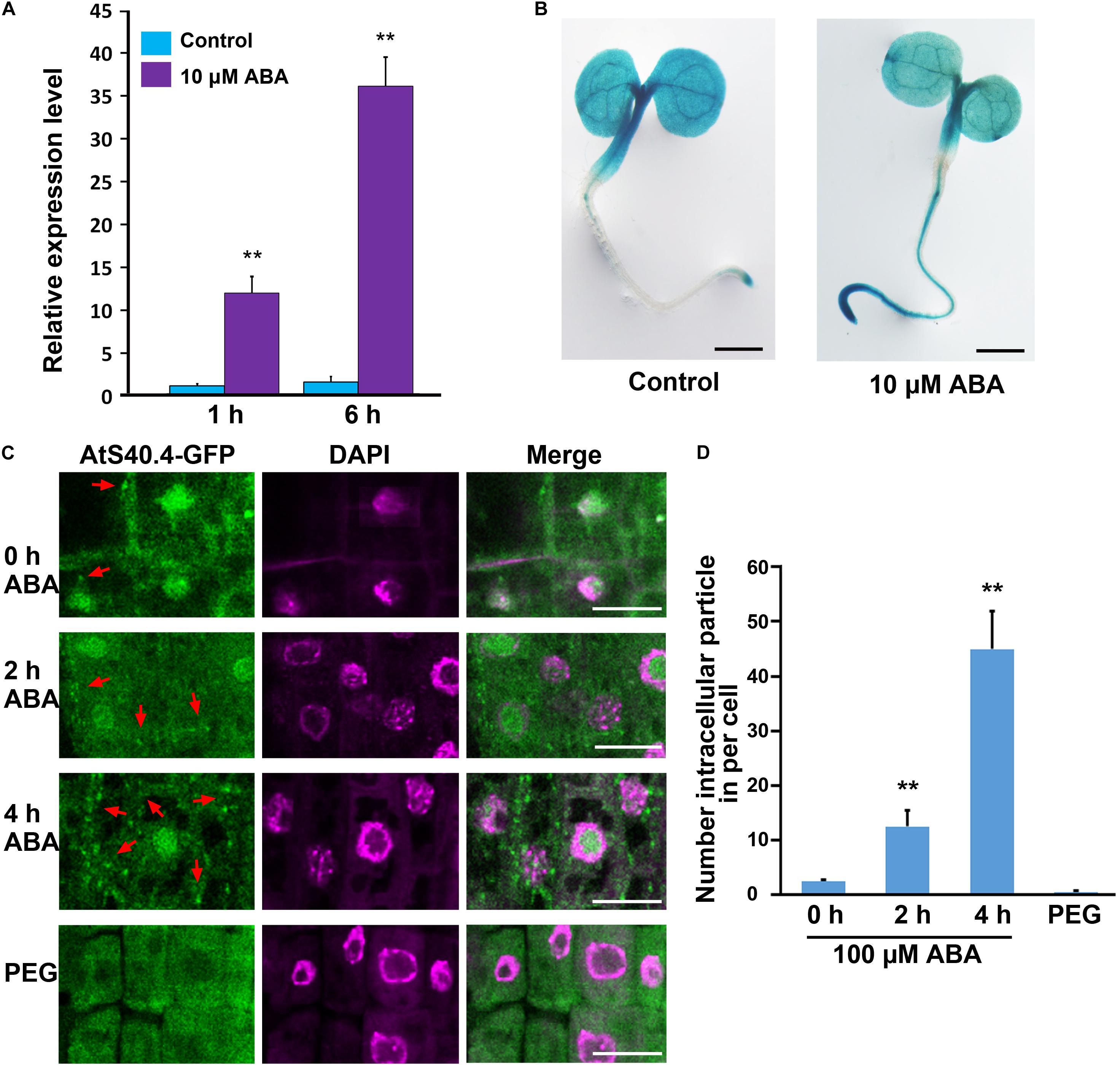
Figure 4. ABA affects AtS40.4 expression and protein localization. (A) qRT-PCR analysis of AtS40.4 mRNA level in seedlings. The 5-day-old seedlings grown on 0.5 × MS medium were cultured with free and 10 μM ABA treatment for 1 and 6 h. GAPDH was used as an internal control. Data are means ± SEs of three biological replicates. Double asterisks indicate significant differences (Student’s t-test; **P < 0.01). (B) GUS activity of transgenic proAtS40.4:GUS seedlings with or without ABA treatment. The 3-day-old seedlings grown on 0.5 × MS medium were treated with 10 μM ABA or mock treatment solution for 6 h before GUS staining. This experiment was repeated three times with similar results. Scale bars, 200 μm. (C) Subcellular localization of AtS40.4 in root tip cells of ats40.4 seedlings expressing proAtS40.4:AtS40.4-GFP. Seedling roots were immersed into 100 μM ABA for 0, 2, or 4 h before observation or were transferred onto 24% PEG-6000 (−0.75 Mpa) medium for further growth 1 day as moderate drought stress. Nuclei were labeled with DAPI (shown in a pseudo color representation). Scale bars, 20 μm. (D) Numbers of AtS40.4-GFP particles in a root tip cell of seedlings with free or 100 μM ABA, and 24% PEG-6000 applications. Data are means ± SEs based on more than 10 roots each treatment. Double asterisks denote statistical differences with Student’s t-test at P < 0.01.
To elucidate how AtS40.4 plays roles at the cellular level, we investigated the subcellular localization of AtS40.4 in the ats40.4-1 background plants expressing proAtS40.4:AtS40.4-GFP construct. The green fluorescent signals of GFP were localized in the cytoplasm and the nucleus of the root tip cells (Figure 4C and Supplementary Figure 4A). These results indicate that the AtS40.4 protein is a cytoplasm-nucleus dual-localized protein.
The dual cytosolic/nuclear localization and involvement in ABA signaling of AtS40.4 prompted us to examine whether the distribution of AtS40.4 between the cytoplasm and the nucleus was regulated by ABA as recent reports which the distribution of nucleocytoplasmic trafficking of WD40 protein 1 and C2-domain ABA-related proteins is regulated by ABA (Qin et al., 2019; Xu et al., 2019). Surprisingly, after 100 μM ABA application to the seedling roots, numerous intracellular fluorescent particles appeared in the root tip cells (Figure 4C and Supplementary Figures 4B,C). Further statistical analysis showed that the GFP-particles in the root tip cells under 100 μM ABA treatment for 2 or 4 h were 6- and 20-fold higher than those in the control (Figure 4D), indicating that the formation of these particles containing AtS40.4 was time dependent.
To investigated whether or not the localization changes of AtS40.4 is caused by the stress response from ABA application, we used PEG-6000 to treatment the seedling root as described previously (Chong et al., 2019). The fluorescent of GFP-particles was rarely detected in the root cells after PEG-induced moderate drought severity (-0.75 MPa, 24% PEG-6000) for 1 day (Figures 4C,D and Supplementary Figure 4D). This indicates that the ABA-induced alteration of AtS40.4 localization patterns is different from the stress imposed by drought. Taken together, these results suggest that AtS40.4 is a novel ABA-responsive member.
Ats40.4 Regulates the Expression of ABIs and Functions Upstream of ABI4
Because AtS40.4 was partly localized in the nucleus, we then explored whether AtS40.4 has an impact on gene expression. ABIs are core genes in ABA signaling (Leung et al., 1997; Skubacz et al., 2016; Chandrasekaran et al., 2020). Therefore, we tested the transcription levels of these genes in the ats40.4 mutants and AtS40.4-overexpressing lines with and without ABA treatment. The expression levels of ABI1 and ABI2 were downregulated in the ats40.4 seedlings but upregulated in the AtS40.4-overexpressing lines after 10 μM ABA application in roots for 5 h (Figure 5). By contrast, the expression levels of ABI3-5 were enhanced in the ats40.4 mutants but decreased in the overexpressing lines. In addition, the expression level of SnRK2.6, another core gene in ABA signaling, was increased in ats40.4 mutants while decreased in AtS40.4-overexpressing lines after ABA treatment (Supplementary Figure 5). We also tested the expression of RD29A, an important gene response to ABA and abiotic stress such as drought and salt. The promoter region of RD29A contains two cis-acting regulatory elements, an ABRE and two DREs (Narusaka et al., 2003). The transcription level of RD29A was promoted by ABA in the AtS40.4-overexpressing line, whereas no significant change was detected in the ats40.4 mutant (Supplementary Figure 5).
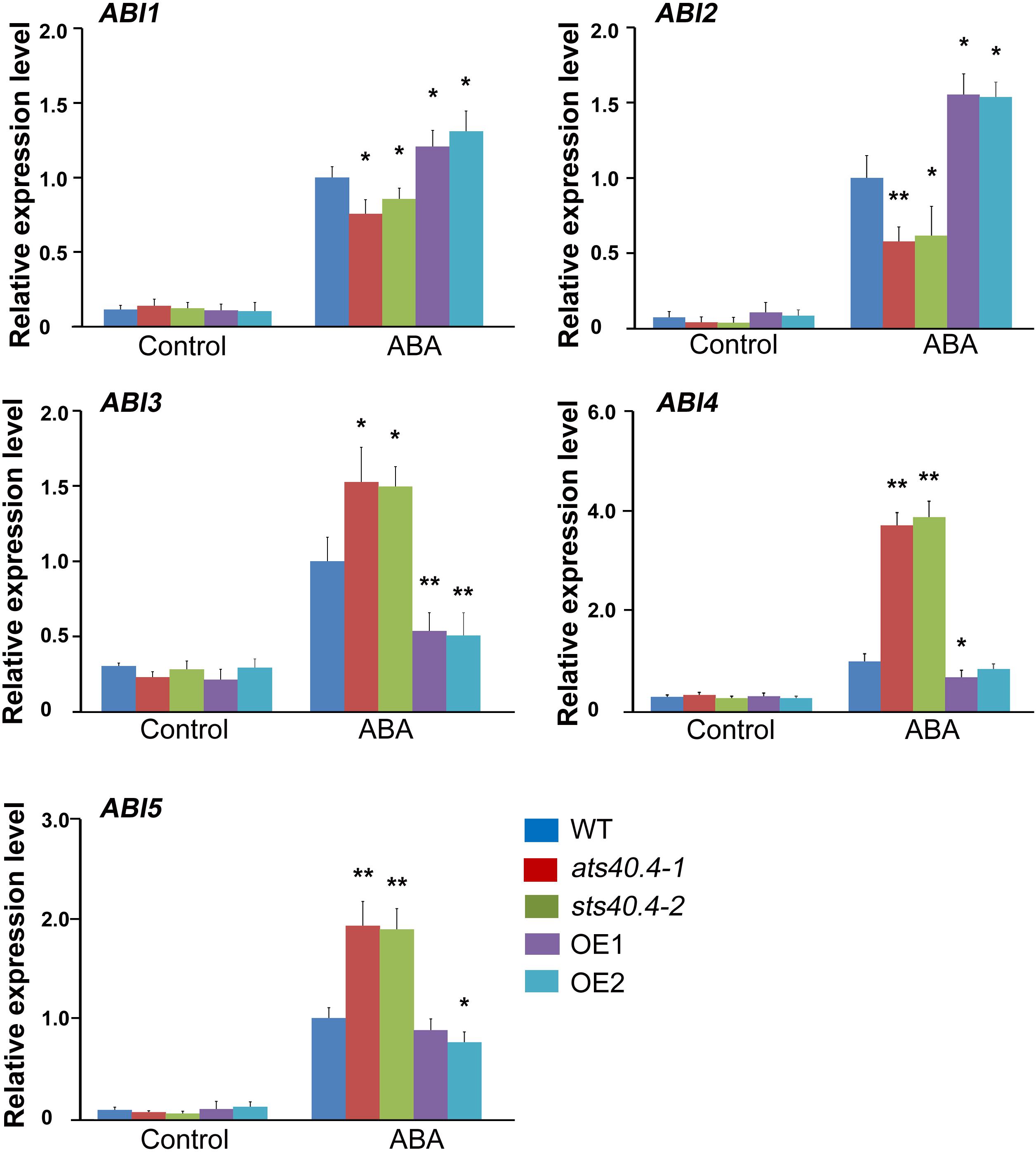
Figure 5. AtS40.4 affects expression of ABI1-5. qR-PCR analysis of the expression levels of ABI1-5. The 7-day-old seedlings of WT, ats40.4 mutants and AtS40.4-overexpressing lines (OE-1 and OE-2) were treatment with 0 or 10 μM ABA for 5 h. GAPDH was used as the internal control. Each value is the mean ± SE of three biological determinations. Significant differences are denoted with an asterisk (P < 0.05) or double asterisks (P < 0.01).
AtS40.4 mutations led to a large increment of ABI4 transcription, indicating that ABI4 is an important target gene in the AtS40.4 response to ABA signaling. To further verify the genetic relationship between AtS40.4 and ABI4, we introduced AtS40.4 mutation into the abi4 mutant (SALK_080095) and generated the ats40.4-1 abi4 double mutant. Then, the ABA sensitivity of these different genotypes was investigated. No significant difference was found among the ats40.4-1 abi4 double mutant, ats40.4-1 and abi4 single mutants, and wild type during seed germination and root elongation on 0.5 × MS medium (Figures 6A,B). The ats40.4-1 abi4 double mutant showed less sensitivity to ABA similar to that of the abi4 mutant when seeds were germinated on ABA medium (Figures 6A,B). Moreover, the seedlings of the ats40.4-1 abi4 double mutant displayed an ABA-hyposensitive phenotype as abi4 seedlings in the presence of ABA (Figures 6C,D). These results indicate that the disruption of ABI4 abolishes the negative role of AtS40.4 in ABA signaling, supporting that ABI4 is genetically epistatic to AtS40.4 in the ABA-regulated seed germination and early seedling growth.
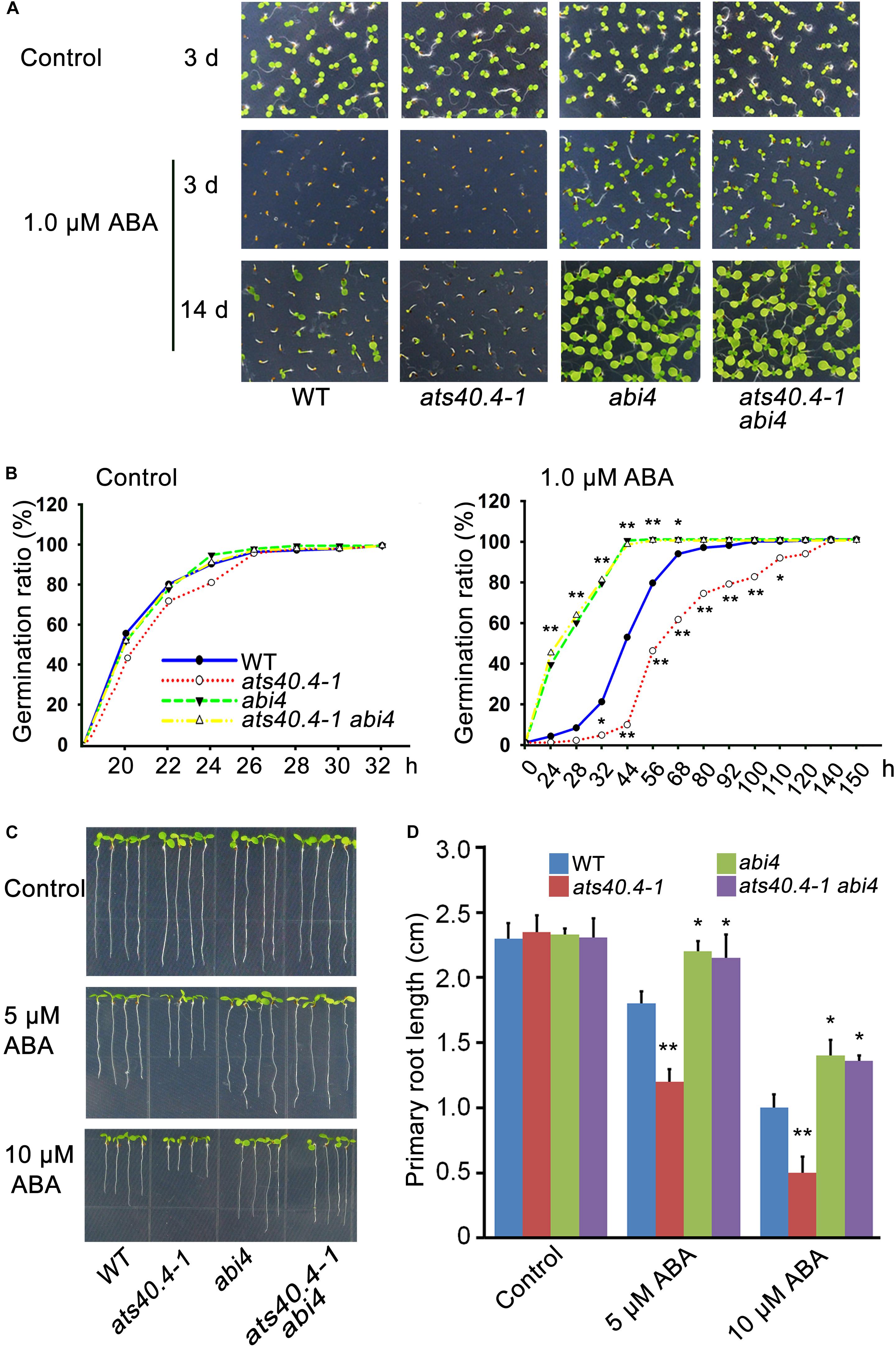
Figure 6. AtS40.4 functions upstream of ABI4 in ABA signaling. (A) Seed germination and seedling growth of wild type, ats40.4-1 and abi4 single mutants, and ats40.4-1 abi4 double mutant. Seeds were geminated and grown on 0.5 × MS medium containing 0, and 1.0 μM ABA. (B) Seed germination ratios of wild type, ats40.4-1, abi4 and ats40.4-1 abi4 mutants. Seeds were grown on 0.5 × MS medium containing 0 (left) and 1.0 μM ABA (right). The asterisks indicate a significant difference of mutants from the wild-type Arabidopsis (Student’s t-test; *P < 0.05; **P < 0.01). (C,D) Root elongation of seedlings of wild-type, ats40.4-1, abi4, and ats40.4-1 abi4 mutants. The 3-day-old seedlings grown on 0.5 × MS medium were transferred onto the 0, 5, and 10 μM ABA medium for further growth 7 days. (C). Values are means ± SE of three independent biological replicates. Significant differences (D) are denoted with an asterisk (P < 0.05) or double asterisks (P < 0.01).
Discussion
The role of AtS40.4 in leaf senescence has been previously characterized in Arabidopsis (Fischer-Kilbienski et al., 2010). In this study, we found a novel function of AtS40.4 which acts as a negative regulator in ABA signaling during seed germination and early seedling growth. AtS40.4 is globally expressed in Arabidopsis including the seedlings, both developing and germinated seeds. Moreover, expression of AtS40.4 is dramatically promoted by ABA in early seedlings, especially in the root cells.
AtS40.4 functions in ABRE-dependent ABA signaling and affects transcription of ABIs, SnRK2.6 and RD29A. Interestingly, AtS40.4 enhanced the expression ABI1 and ABI2, two negative regulators of ABA signaling; whereas inhibited the transcription levels of ABI3-5 and SnRK2.6. These results support that AtS40.4 acts as an upstream regulator of ABA signaling and indirectly affects expression of the key genes in ABA signaling. The core negative components ABI1 and ABI2, and positive factors ABI3-5 play diverse roles in the ABA- regulated plant growth and development (Leung et al., 1997; Wind et al., 2013; Cheng et al., 2014; Li P. C. et al., 2016; Gong et al., 2020; Xu et al., 2020). Especially, ABI4, as a transcription factor, either activates or represses multigene expression in ABA signaling (Baek et al., 2020; Chandrasekaran et al., 2020). Moreover, ABI4 acts as a node of integration for different signals, including environmental stimuli, sugar sense, and various phytohormones (Wind et al., 2013; Chandrasekaran et al., 2020). Therefore, the expression regulation of ABI4 is important for plant growth, development, and response to environmental cues. Several regulators of ABI4, such as WRKY and MYB transcription factors, promote or inhibit ABI4 expression during seed maturation and early seedling development (Cui et al., 2013; Yan et al., 2014; Huang et al., 2016; Phukan et al., 2016; Mu et al., 2017; Ma et al., 2019; Zhao et al., 2019). Our findings reveal that AtS40.4 is a novel upstream regulator of ABI4 expression.
AtS40.4 is partly localized in the nucleus, consisting with its functions in effecting expression of ABIs. AtS40.3 and HvS40 exhibiting nuclear localization have been assigned as DNA binding proteins (Krupinska et al., 2002; Fischer-Kilbienski et al., 2010). Among the nuclear S40 proteins, the probability of AtS40.4 acting as a DNA-binding protein (76%) is lower than that of AtS40.3 (99.5%), HvS40 (95.7%), OsS40-1 (97.4%), and OsS40-14 (88.4%), but higher than OsS40-13 (33.2%) (Supplementary Figures 6, 7) (DNA Binding Protein Prediction)1. However, the effect of AtS40.4 on the expression between negative regulators of ABA signaling, ABI1-2, and positive factors of ABA signaling, ABI3-5 and SnRK2.6, is opposite. This indicates that AtS40.4 does not regulated directly gene transcription although it has a role of binding to DNA. This study, together with the previous findings, is not sufficient to explain functions of S40 proteins in the nucleus. Therefore, our work has just laid the groundwork for further exploration of precise biological roles and mechanism of S40 members in the nucleus.
The localization and partitioning of AtS40.4 are regulated by ABA. Similar result has been previously reported on Arabidopsis RopGEF1, a guanine nucleotide exchange factor protein, which plays an important role in the ABA-mediated growth regulation of root. RopGEF1 is rapidly sequestered to intracellular particles after exogenous ABA application, which results in ABA-mediated degradation (Li P. C. et al., 2016). Recent reports have indicated that the nucleocytoplasmic distribution of XPO1-interacting WD40 protein 1 (XIW1) and C2-domain ABA-related (CAR) proteins is also regulated by ABA (Qin et al., 2019; Xu et al., 2019). XIW1 interacts with ABI5 in the nucleus and positively regulates the ABA response in Arabidopsis. CAR proteins play roles in ABA response via recruiting ABA receptors to PM to facilitate ABA signaling (Diaz et al., 2016; Qin et al., 2019). The dynamic localization and stability of CAR proteins are regulated by LOWER TEMPERATURE 1 (LOT1) (Qin et al., 2019). ABA promotes XIW1 accumulation in the nucleus, leading to a decreased distribution of XIW1 in the cytoplasm (Xu et al., 2019); whereas ABA reduces the CAR9–LOT1 interaction in the nucleus, resulting in the increased localization of CAR9 to the plasma membrane (Qin et al., 2019). Therefore, the regulatory ways of spatial distribution of the ABA-response proteins are diverse. The small protein AtS40.4 is partitioned into intracellular particles after ABA application, implying that AtS40.4 distribution is rapidly responsive to the ABA. Moreover, the different localization changes of AtS40.4 after ABA and PEG-6000 treatments reveal that AtS40.4 response to ABA is distinct from that of PEG-induced drought stress. Further exploration of the regulation mechanisms of AtS40.4 intro-cellular distribution will shed new light on the molecular characteristics of these DUF584 proteins.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
X-LW, X-PS, and J-JR conceived and designed the research. X-PS and J-JR performed the experiments with the help from D-FL, H-DQ, YL, and Y-YW. X-LW and X-PS wrote the manuscript with contributions of L-JK. All authors reviewed and approved the final version.
Funding
This work was funded by the National Natural Science Foundation of China (31970330) and Major Basic Research Projects of Shandong Natural Science Foundation (ZR2018ZC08N3).
Conflict of Interest
D-FL was employed by the company Shandong Lufeng Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ying-Gao Liu (National Key Laboratory of Crop Biology, Shandong Agricultural University) for sharing the abi4 seeds, and GABI-Kat for providing the T-DNA insertion lines.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.622201/full#supplementary-material
Footnotes
References
Ali, A., Pardo, J. M., and Yun, D. J. (2020). Desensitization of ABA-signaling: the swing from activation to degradation. Front. Plant Sci. 11:379. doi: 10.3389/fpls.2020.00379
Anwer, M. A., Anjam, M. S., Shah, S. J., Hasan, M. S., Naz, A. A., Grundler, F., et al. (2018). Genome-wide association study uncovers a novel QTL allele of AtS40-3 that affects the sex ratio of cyst nematodes in Arabidopsis. J. Exp. Bot. 69, 1805–1814. doi: 10.1093/jxb/ery019
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., and Yun, D. J. (2020). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11:143. doi: 10.3389/fpls.2020.00143
Belda-Palazon, B., Gonzalez-Garcia, M. P., Lozano-Juste, J., Coego, A., Antoni, R., Julian, J., et al. (2018). PYL8 mediates ABA perception in the root through non-cell-autonomous and ligand-stabilization-based mechanisms. Proc. Natl. Acad. Sci. U.S.A. 115, E11857–E11863. doi: 10.1073/pnas.1815410115
Carles, C., Bies-Etheve, N., Aspart, L., Léon-Kloosterziel, K. M., Koornneef, M., Echeverria, M., et al. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30, 373–383. doi: 10.1046/j.1365-313X.2002.01295.x
Chandrasekaran, U., Luo, X., and Shu, K. (2020). Multifaceted signaling networks mediated by abscisic acid insensitive 4 (ABI4). Plant Commun. 2020:ery019.
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., and Zhao, Y. (2020). Abscisic acid dynamics signaling and functions in plants. Plant Biol. 62, 25–54. doi: 10.1111/jipb.12899
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Chong, G. L., Foo, M. H., Lin, W. D., Wong, M. M., and Verslues, P. E. (2019). Highly ABA-Induced 1 (HAI1)-Interacting protein HIN1 and drought acclimation-enhanced splicing efficiency at intron retention sites. Proc. Natl. Acad. Sci. U.S.A. 116, 22376–22385. doi: 10.1073/pnas.1906244116
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Collin, A., Daszkowska-Golec, A., Kurowska, M., and Szarejko, I. (2020). Barley ABI5 (Abscisic Acid INSENSITIVE 5) is involved in abscisic acid-dependent drought response. Front. Plant Sci. 11:1138. doi: 10.3389/fpls.2020.01138
Conti, L. (2019). The A-B-A of floral transition: the to do list for perfect escape. Mol. Plant 12, 289–291. doi: 10.1016/j.molp.2019.02.002
Cui, M. H., Yoo, K. S., Hyoung, S., Nguyen, H. T., Kim, Y. Y., Kim, H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor AtMYB20 negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587, 1773–1778. doi: 10.1016/j.febslet.2013.04.028
Diaz, M., Sanchez-Barrena, M. J., Gonzalez-Rubio, J. M., Rodriguez, L., Fernandez, D., Antoni, R., et al. (2016). Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 113, E396–E405. doi: 10.1073/pnas.1512779113
Duan, L., Dietrich, D., Ng, C. H., Chan, P. M., Bhalerao, R., Bennett, M. J., et al. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25, 324–341. doi: 10.1105/tpc.112.107227
Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerström, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. doi: 10.1046/j.1365-313x.2000.00857.x
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. doi: 10.1105/tpc.010441
Fischer-Kilbienski, I., Miao, Y., Roitsch, T., Zschiesche, W., Humbeck, K., and Krupinska, K. (2010). Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 73, 379–390. doi: 10.1007/s11103-010-9618-3
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
González-Guzmán, M., Apostolova, N., Bellés, J. M., Barrero, J. M., Piqueras, P., Ponce, M. R., et al. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. doi: 10.1105/tpc.002477
Huang, Y., Feng, C. Z., Ye, Q., Wu, W. H., and Chen, Y. F. (2016). Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 12:e1005833. doi: 10.1371/journal.pgen.1005833
Huang, Y., Sun, M. M., Ye, Q., Wu, X. Q., Wu, W. H., and Chen, Y. F. (2017). Abscisic Acid modulates seed germination via ABA INSENSITIVE5-mediated PHOSPHATE1. Plant Physiol. 175, 1661–1668. doi: 10.1104/pp.17.00164
Janack, B., Sosoi, P., Krupinska, K., and Humbeck, K. (2016). Knockdown of WHIRLY1 affects drought stress-induced leaf senescence and histone modifications of the senescence-associated gene HvS40. Plants 5:37. doi: 10.3390/plants5030037
Jehanzeb, M., Zheng, X., and Miao, Y. (2017). The of role the S40 gene family in leaf senescence. Int. J. Mol. Sci. 18:2152. doi: 10.3390/ijms18102152
Jiang, S., Zhang, D., Wang, L., Pan, J., Liu, Y., Kong, X., et al. (2013). A maize calcium-dependent protein kinase gene ZmCPK4 positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 71, 112–120. doi: 10.1016/j.plaphy.2013.07.004
Khan, I. U., Ali, A., Khan, H. A., Baek, D., Park, J., Lim, C. J., et al. (2020). PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in arabidopsis. Front. Plant Sci. 11:623. doi: 10.3389/fpls.2020.00623
Kim, S., Kang, J. Y., Cho, D. I., Park, J. H., and Kim, S. Y. (2004). ABF2 an ABRE-binding bZIP factor is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313X.2004.02192.x
Krupinska, K., Haussühl, K., Schäfer, A., van der Kooij, T. A., Leckband, G., Lörz, H., et al. (2002). A novel nucleus-targeted protein is expressed in barley leaves during senescence and pathogen infection. Plant Physiol. 130, 1172–1180. doi: 10.1104/pp.008565
Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. doi: 10.1105/tpc.9.5.759
Li, Z., Waadt, R., and Schroeder, J. I. (2016). Release of GTP exchange factor mediated down-regulation of abscisic acid signal transduction through ABA-induced rapid degradation of RopGEFs. PLoS Biol. 14:e1002461. doi: 10.1371/journal.pbio.1002461
Li, P. C., Huang, J. G., Yu, S. W., Li, Y. Y., Sun, P., Wu, C. A., et al. (2016). Arabidopsis YL1/BPG2 is involved in seedling shoot response to salt stress through ABI4. Sci. Rep. 6:30163. doi: 10.1038/srep30163
Liu, K., Zou, W., Gao, X., Wang, X., Yu, Q., and Ge, L. (2019). Young seedlings adapt to stress by retaining starch and retarding growth through ABA-dependent and -independent pathways in Arabidopsis. Biochem. Biophys. Res. Commun. 515, 699–705. doi: 10.1016/j.bbrc.2019.06.023
Liu, X. J., Liu, X., An, X. H., Han, P. L., You, C. X., and Hao, Y. J. (2017). An apple protein kinase MdSnRK1.1 interacts with MdCAIP1 to regulate ABA sensitivity. Plant Cell Physiol. 58, 1631–1641. doi: 10.1093/pcp/pcx096
Lu, C., Chen, M. X., Liu, R., Zhang, L., Ho, X., Liu, S., et al. (2019). Abscisic acid regulates auxin distribution to mediate maize lateral root development under salt stress. Front. Plant Sci. 10:716. doi: 10.3389/fpls.2019.00716
Ma, Q., Xia, Z., Cai, Z., Li, L., Cheng, Y., Liu, J., et al. (2019). GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. Front. Plant Sci. 9:1979. doi: 10.1186/s12870-018-1397-z
Mei, C., Jiang, S. C., Lu, Y. F., Wu, F. Q., Yu, Y. T., Liang, S., et al. (2014). Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signaling. J. Exp. Bot. 65, 5317–5330. doi: 10.1093/jxb/eru293
Min, M. K., Kim, R., Moon, S. J., Lee, Y., Han, S., Lee, S., et al. (2020). Selection and functional identification of a synthetic partial ABA agonist, S7. Sci Rep. 10:4. doi: 10.1038/s41598-019-56343-9
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mu, Y., Zou, M., Sun, X., He, B., Xu, X., Liu, Y., et al. (2017). Basic pentacysteine proteins repress abscisic acid insensitive4 expression via direct recruitment of the polycomb-repressive complex 2 in Arabidopsis root development. Plant Cell Physiol. 58, 607–621. doi: 10.1093/pcp/pcx006
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313x.2003.01708.x
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qin, T., Tian, Q., Wang, G., and Xiong, L. (2019). LOWER TEMPRERATUE 1 enhances ABA responses and plant drought tolerance by modulating the stability and localization of C2-Domain ABA-related proteins in Arabidopsis. Mol. Plant 12, 1243–1258. doi: 10.1016/j.molp.2019.05.002
Quesada, V., Ponce, M. R., and Micol, J. L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436.
Shi, X. P., Ren, J. J., Yu, Q., Zhou, S. M., Ren, Q. P., Kong, L. J., et al. (2018). Overexpression of SDH confers tolerance to salt and osmotic stress but decreases ABA sensitivity in Arabidopsis. Plant Biol. 20, 327–337. doi: 10.1111/plb.12664
Shu, K., Chen, F., Zhou, W., Luo, X., Dai, Y., Shuai, H., et al. (2018). ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol Biol Rep. 45, 2727–2731. doi: 10.1007/s11033-018-4290-9
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Spollen, W. G., LeNoble, M. E., Samuels, T. D., Bernstein, N., and Sharp, R. E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122, 967–976. doi: 10.1104/pp.122.3.967
Verslues, P. E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J. K. (2006). Methods and concepts in quantifying resistance to drought salt and freezing abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Wang, X., Guo, C., Peng, J., Li, C., Wan, F., Zhang, S., et al. (2019). ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 221, 341–355. doi: 10.1111/nph.15345
Wind, J. J., Peviani, A., Snel, B., Hanson, J., and Smeekens, S. C. (2013). ABI4: versatile activator and repressor. Trends Plant Sci. 18, 125–132. doi: 10.1016/j.tplants.2012.10.004
Wu, R., Duan, L., Pruneda-Paz, J. L., Oh, D. H., Pound, M., Kay, S., et al. (2018). The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional Regulation. Plant Physiol. 177, 1650–1665. doi: 10.1104/pp.18.00401
Xie, Q., Essemine, J., Pang, X., Chen, H., and Cai, W. (2020). Exogenous application of abscisic acid to shoots promotes primary root cell division and elongation. Plant Sci. 292:110385. doi: 10.1016/j.plantsci.2019.110385
Xiong, F., Ren, J. J., Yu, Q., Wang, Y. Y., Lu, C. C., Kong, L. J., et al. (2019). AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 223, 277–292. doi: 10.1111/nph.15756
Xu, D., Wu, D., Li, X. H., Jiang, Y., Tian, T., Chen, Q., et al. (2020). Light and abscisic acid coordinately regulate greening of seedlings. Plant Physiol. 83, 1281–1294. doi: 10.1104/pp.20.00503
Xu, X., Wan, W., Jiang, G., Xi, Y., Huang, H., Cai, J., et al. (2019). Nucleocytoplasmic trafficking of the Arabidopsis WD40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Mol. Plant 12, 1598–1611. doi: 10.1016/j.molp.2019.07.001
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Ye, N., Zhu, G., Liu, Y., Zhang, A., Li, Y., Liu, R., et al. (2012). Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by Abscisic acid in rice seeds. J. Exp. Bot. 63, 1809–1822. doi: 10.1093/jxb/err336
Yu, Z., Zhang, D., Xu, Y., Jin, S., Zhang, L., Zhang, S., et al. (2019). CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 70, 5457–5469. doi: 10.1093/jxb/erz302
Zhang, H., Han, W., De Smet, I., Talboys, P., Loya, R., Hassan, A., et al. (2010). ABA promotes quiescence of the quiescent center and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64, 764–774. doi: 10.1111/j.1365-313X.2010.04367.x
Zhao, X. Y., Qi, C. H., Jiang, H., You, C. X., Guan, Q. M., Ma, F. W., et al. (2019). The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. 6:66. doi: 10.1038/s41438-019-0147-1
Zheng, X., Jehanzeb, M., Habiba, O., Zhang, Y., Li, L., and Miao, Y. (2019). Characterization of S40-like proteins and their roles in response to environmental cues and leaf senescence in rice. BMC Plant Biol. 19:174. doi: 10.1186/s12870-019-1767-1
Keywords: Arabidopsis thaliana, AtS40.4, DUF584, abscisic acid signaling, ABI4, seed germination, early seedling growth
Citation: Shi X-P, Ren J-J, Qi H-D, Lin Y, Wang Y-Y, Li D-F, Kong L-J and Wang X-L (2021) Plant-Specific AtS40.4 Acts as a Negative Regulator in Abscisic Acid Signaling During Seed Germination and Seedling Growth in Arabidopsis. Front. Plant Sci. 12:622201. doi: 10.3389/fpls.2021.622201
Received: 28 October 2020; Accepted: 15 January 2021;
Published: 04 February 2021.
Edited by:
Eric Ruelland, UMR 7618 Institut d’écologie et des Sciences de l’environnement de Paris (IEES), FranceReviewed by:
Miguel Gonzalez-Guzman, University of Jaume I, SpainOksana Iakovenko, Institute of Bioorganic Chemistry and Petrochemistry (NAN Ukraine), Ukraine
Copyright © 2021 Shi, Ren, Qi, Lin, Wang, Li, Kong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiu-Ling Wang, eGx3YW5nQHNkYXUuZWR1LmNu
†These authors have contributed equally to this work
 Xiao-Pu Shi
Xiao-Pu Shi Jing-Jing Ren1†
Jing-Jing Ren1† Yi Lin
Yi Lin Yu-Yi Wang
Yu-Yi Wang Xiu-Ling Wang
Xiu-Ling Wang