
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 February 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.621561
This article is part of the Research TopicMolecular Mechanisms of Flowering Plant ReproductionView all 19 articles
 Shiyu Sun1
Shiyu Sun1 Duoxiang Wang1
Duoxiang Wang1 Jingbin Li1
Jingbin Li1 Yaqi Lei1
Yaqi Lei1 Gang Li2
Gang Li2 WenGuo Cai1
WenGuo Cai1 Xiangxiang Zhao3
Xiangxiang Zhao3 Wanqi Liang1
Wanqi Liang1 Dabing Zhang1,2*
Dabing Zhang1,2*Environmental conditions, such as photoperiod and temperature, can affect male fertility in plants. While this feature is heavily exploited in rice to generate male-sterile lines for hybrid breeding, the underlying molecular mechanisms remain largely unknown. In this study, we use a transcriptomics approach to identify key genes and regulatory networks affecting pollen maturation in rice anthers in response to different day lengths. A total of 11,726 differentially expressed genes (DEGs) were revealed, of which 177 were differentially expressed at six time points over a 24-h period. GO enrichment analysis revealed that genes at all time points were enriched in transport, carbohydrate, and lipid metabolic processes, and signaling pathways, particularly phytohormone signaling. In addition, co-expression network analysis revealed four modules strongly correlated with photoperiod. Within these four modules, 496 hub genes were identified with a high degree of connectivity to other photoperiod-sensitive DEGs, including two previously reported photoperiod- and temperature-sensitive genes affecting male fertility, Carbon Starved Anther and UDP-glucose pyrophosphorylase, respectively. This work provides a new understanding on photoperiod-sensitive pollen development in rice, and our gene expression data will provide a new, comprehensive resource to identify new environmentally sensitive genes regulating male fertility for use in crop improvement.
Hybrid breeding has made a great contribution to the yield increase of rice (Oryza sativa) relative to inbred varieties, with improvements of up to 20% (Khush, 2001; Michel et al., 2001; Yuan, 2004; Cheng et al., 2007). Originally, after the discovery of a wild abortive cytoplasmic male sterile (CMS) line in the 1970s, a three-line system containing the male-sterile line, a maintainer line, and a restorer line was used (Yuan, 1984). Due to the time-consuming and germplasm-limited nature of using separate maintainer and restorer lines (Chen et al., 2010), a more modern two-line system was developed in 1973, based on the discovery and application of environmentally sensitive genic male sterile lines (EGMS) (Shi, 1981). Photoperiod-sensitive (PGMS) lines and thermosensitive (TGMS) lines have been successfully developed and applied widely in rice breeding (Mou, 2016).
Since photoperiod is more stable and predictable than temperature, breeders have focused on PGMS lines. Several genes, such as photoperiod-sensitive male sterility-1 (pms1) (Zhou et al., 2011; Fan et al., 2016), pms2 (Zhang et al., 1994), pms3 (Ding et al., 2012a,b; Zhou et al., 2012), Programmed Cell Death 5 (OsPDCD5) (Wang Y. et al., 2010), and Carbon Starved Anther (CSA) (Zhang et al., 2013), have been reported to control PGMS. Pms1 encodes a long non-coding RNA (lncRNA) that acts as a phased small-interfering RNA (phasiRNA)-generating locus to produce the PMS1T transcript (21-nt phasiRNA). PMS1T is highly expressed during pollen formation in wild-type plants under long-day (LD) conditions, and a mutation in PMS1T that causes an accumulation of phasiRNAs causes PGMS (Fan et al., 2016). A single-nucleotide polymorphism (SNP) in pms3, another lncRNA required for normal pollen development, reduces pms3 transcript levels under LD conditions, leading to PGMS in a japonica cultivar, NK58S (Ding et al., 2012a,b). Subsequently, Zhou et al. (2012) identified the same SNP in NK58S-derived cultivars and reported the role of this mutation in producing one mutated small RNA, leading to PGMS and TGMS in japonica and indica backgrounds, respectively (Zhou et al., 2012). OsPDCD5 is an ortholog of mammalian programmed cell death 5, whose decreased expression induces photoperiod-sensitive male sterility in rice under LD photoperiods (≥13.5 h sunlight) (Wang Y. et al., 2010). CSA encodes an R2R3 MYB transcription factor that regulates sugar accumulation in rice anther sink tissue; its knockout mutant accumulates insufficient sugar and starch levels in anthers under short-day (SD) conditions, resulting in male sterility (Zhang et al., 2010, 2013). A TGMS line currently used for hybrid seed production, thermosensitive genic male sterile 5 (tms5), has been shown to contain a mutation in a conserved ribonuclease, RNase Z (Zhou et al., 2014). These findings reveal the complexity of the interaction between genetic components and environmental signals in determining male fertility in plants (Kim and Zhang, 2018). However, the global understanding of male reproduction in plants in response to different photoperiods remains largely unknown.
To comprehensively analyze molecular mechanisms controlling plant male fertility in response to photoperiod, we performed a global RNA sequencing (RNA-seq) analysis on anthers grown in LD and SD conditions. Genes associated with carbohydrate transport and metabolism, and phytohormone signaling, were highly correlated with day length during late pollen developmental. This work provides insights into the molecular mechanisms of photoperiod-dependent male fertility, which can lead to identification of new PGMS genes for hybrid breeding.
Rice variety O. sativa L. ssp. japonica 9522 was grown in paddy fields around Shanghai (China) during the normal rice growing season (June–August). Photoperiod treatment was started after panicle initiation and continued until all samples were collected: LD was the natural daylight condition, with a photoperiod of ∼14 h; SD conditions were simulated by covering plants with black cloth for the last 2 h of natural daylight to create a 12-h photoperiod. Duplicate samples of anthers were collected at mitosis I (stage 11) based on anther length (about 2.2 mm), which was described by Zhang et al. (2011) at six time points (00:00, 04:00, 08:00, 12:00, 16:00, and 20:00; Supplementary Figure 1). All samples were frozen immediately in liquid nitrogen and stored at −80°C.
RNA isolation, library construction, and sequencing were performed by Novelbio Ltd (Shanghai, China). Briefly, TRIzol® reagents (Invitrogen, Carlsbad, CA, United States) were used to isolate RNA from each of two biological samples from each experimental condition. RNA quality was assessed by 1% agarose gel electrophoresis, Nanodrop (ThermoFisher), and Bioanalyzer (Agilent 2100), with A260/A280 = 1.8–2.2, and RNA integrity number (RIN) > 6.5. Libraries were synthesized using the standard Ion proton RNA-Seq Kit v2.0, according to the manufacturer’s instructions. Library sequencing was conducted by Ion ProtonTM (Life Technologies, Carlsbad, CA, United States).
The genome and annotation of the rice reference cultivar Nipponbare was downloaded from the MSU Rice Genome Annotation Project1 (Kawahara et al., 2013). FastQC (Version 0.11.1) was used to remove reads with 20% of the base quality lower than 13. Clean reads were mapped to the reference genome using Mapsplice v2.1.8 and then read counts were calculated (Wang K. et al., 2010), which were normalized to reads per kilobase per million (RPKM) to obtain relative levels of transcript expression. Counts and RPKM for each gene are given in Supplementary Data Sheet 1. A summary of sequencing data for each sample is given in Supplementary Table 1 and Supplementary Figure 2. The sequencing data were also uploaded in NCBI (GSE163030). Expression data for genes of interest in other vegetative and reproductive tissues (Supplementary Figure 5B) were sourced from the Rice Expression Profile Database (RiceXPro) (Sato et al., 2012).
cDNA was synthesized using the Fast RT Kit (Tiangen Biotech, Beijing), following the manufacturer’s instructions. qRT-PCR analysis was performed on the LightCycler 96 (Roche). The OsActin gene was used as internal control, and the relative expression level of target genes was calculated based on the 2–ΔΔCt method (Livak and Schmittgen, 2001). Primer sequences for qRT-PCR were found in qPrimerDB (Supplementary Data Sheet 9) (Lu et al., 2017).
Differentially expressed genes (DEGs) were identified by DEseq2, using | log2fold change| ≥ 1 and false discovery rate (FDR) < 0.05 (Audic and Claverie, 1997). The Venn diagram of all DEGs was plotted by Rpackage “venn” (Adrian, 2019), while heatmaps and dendrograms of gene expression were plotted with Rpackage “pheatmap” (Raivo Kolde, 2019). The cluster of all DEGs was also visualized by “pheatmap” based on k-means clustering algorithm. Gene ontology (GO) enrichment was performed by agriGo2, and significant enriched GO terms were defined with p value < 0.01 and q value < 0.05 (Tian et al., 2017). The visualization of GO enrichment results was performed by Rpackage “Clusterprofiler” (Yu et al., 2012). KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment was also performed using Rpackage “Clusterprofiler” (Kanehisa and Goto, 2000).
Weighted gene co-expression network analysis R package (version 3.6) (Langfelder and Horvath, 2008) was used to construct co-expression networks of all expressed genes with automatic, one-step network construction and module detection. First, the soft thresholding power β was chosen based on the lowest power for which the scale-free topology fit index reached a high value. The function “blockwiseModules” was then used to construct topological overlap matrix (TOM) and module detection. The associations between modules and trait were estimated using Pearson correlation coefficient between module eigengenes and phenotype where 0/1 were used to define SD/LD or light/dark conditions. The expression patterns of four trait-related modules were plotted by “ggplot2” (Wickham, 2016). Hub genes were identified by both gene significance and module membership. The module membership (MM) is the correlation of gene expression profile with module eigengenes, and gene significance (GS) is the association of individual genes with that trait. Hub genes were set to a threshold of GS and MM > 0.8 (Horvath and Dong, 2008).
Cytoscape (V3.7.2) (Shannon et al., 2003) was used to visualize co-expression networks. Input data from turquoise module were chosen based on the weight in terms of correlation value from the TOM (Topological Overlap Matrix Value), so that a higher value refers to a strong co-expression. The top 100 highest correlations with CSA and Ugp1 genes were chosen and highlighted the top 1% correlation value of genes.
A total of 11,726 unique DEGs were identified between SD and LD conditions (Supplementary Data Sheet 2). Six genes from the RNA-seq data were selected for confirmation by quantitative reverse-transcription PCR (qRT-PCR), and patterns of expression with the two methods were found to be consistent (Supplementary Figure 3).
The highest numbers of DEGs were expressed at 12:00, with 4552 genes up-regulated and 4662 genes down-regulated in SD anthers compared with LD anthers (Figure 1A and Supplementary Figure 4). Interestingly, the lowest number of DEGs was observed 4 h later at 16:00, with only 824 up-regulated and 546 down-regulated genes in SD anthers. All 11,726 DEGs assembled into just seven clusters, confirming that the largest differences in expression between LD and SD appeared at 12:00, followed by 08:00 and 04:00 (Figure 1B). Additionally, the 12:00 time point saw expression of the largest number of unique DEGs (4502), whereas the least numbers of unique DEGs occurred at 20:00 (55) and 00:00 (64) (Figure 1C). These results suggest that photoperiod dramatically affects gene expression patterns in anthers, with the largest changes occurring in the morning (04:00, 08:00, and 12:00), even before dawn.
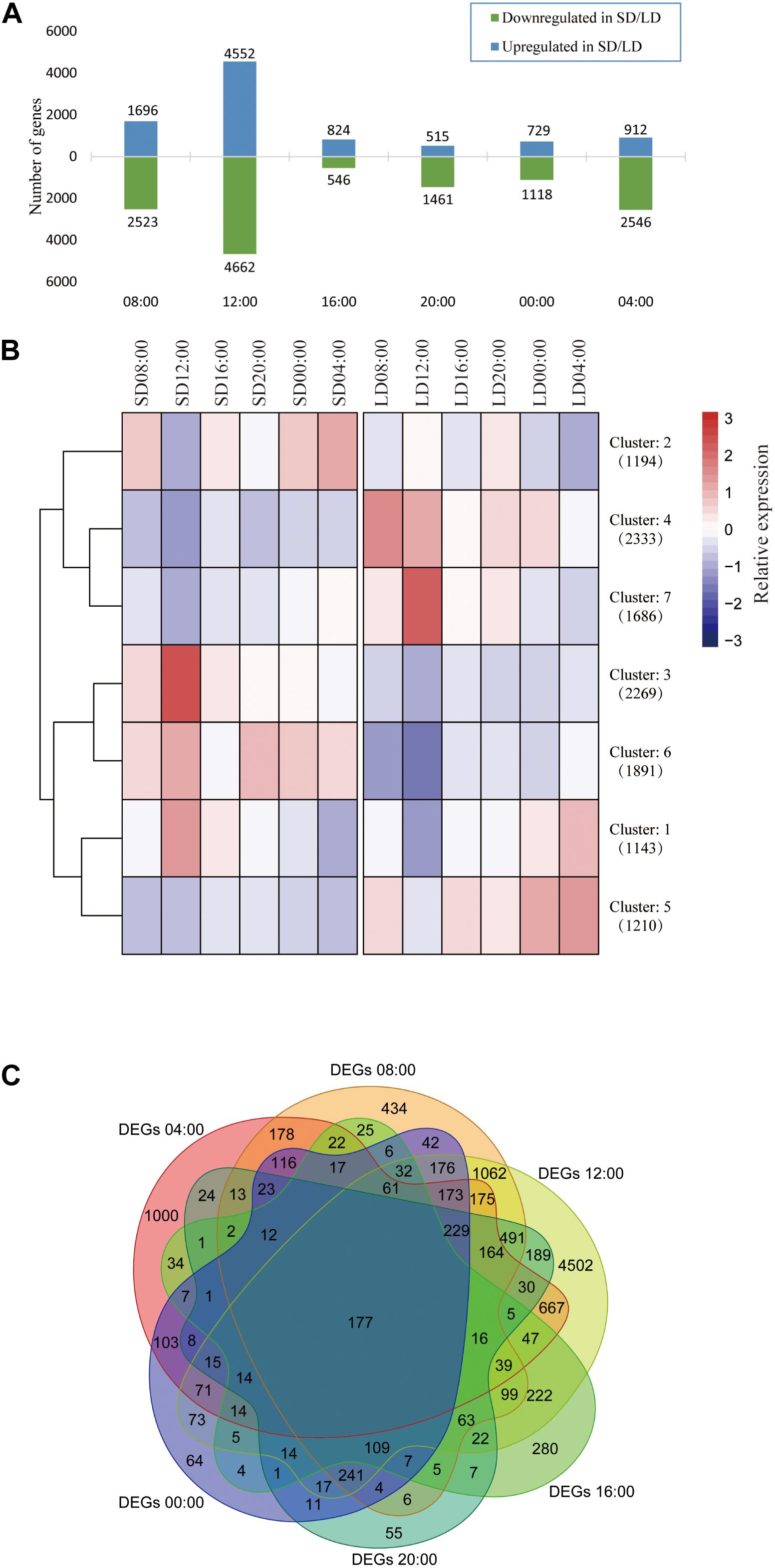
Figure 1. Expression of 11,726 DEGs in response to photoperiod conditions. SD, short day; LD, long day. (A) Number of up- and down-regulated DEGs at each time point. (B) Heatmap and dendrogram of all DEGs in all samples (k = 7). Gene numbers in each cluster given in brackets. DEGs assembled to seven clusters: three clusters up-regulated in SD (2, 3, and 6); three clusters up-regulated in LD (4, 5, and 7); and Cluster 1, with higher daytime expression under SD conditions, and nighttime expression under LD conditions. The value of center point of seven clusters represents relative expression. (C) Venn diagram showing all DEGs at six time points. A total of 177 genes showed differential expression between SD and LD at all time points.
A total of 177 genes were differentially expressed at all time points in SD and LD anthers (Figure 1C). Most of these genes (138) were up-regulated in LD anthers, with low or undetectable expression in SD anthers (Supplementary Figure 5A). A large proportion of these DEGs (51 genes, 29%) exhibit significantly higher expression in the anther than in other rice tissues (Supplementary Figure 5B), including previously reported anther-related genes such as ATP Binding Cassette G15 (OsABCG15) (Wu et al., 2014), LESS ADHESIVE POLLEN 6 (OsLAP6) (Zou et al., 2017), and HOTHEAD-Like1 (HTH1) (Liu et al., 2016). These 177 DEGs were found in GO categories primarily associated with cell wall synthesis and metabolism, including hydrolytic enzyme expression and regulation, indicating that the anther cell wall structure and composition may be photoperiod-sensitive (Supplementary Figure 5C). These genes may play key roles in anther development and pollen formation, and can be used as markers to distinguish SD and LD conditions.
Gene ontology and kyoto encyclopedia of genes and genomes functional analysis of anther DEGs indicated that genes involved in transport, lipid and carbohydrate metabolism, and metabolic regulation were most affected by photoperiod (Figure 2 and Supplementary Figure 6). GO enrichment analysis revealed that DEGs at all time points were enriched in GO “biological process” categories of “transport,” “localization,” “establishment of localization” (transport is a sub-item of these two terms), “carbohydrate metabolic process,” and “lipid metabolic process” (Figure 2A and Supplementary Data Sheet 3). DEGs involved in regulatory processes were generally enriched at 12:00 and 04:00, while metabolic processes were enriched at 08:00 and 12:00 (Figure 2A). In “molecular function” categories, the GO terms of “transport activity” and “enzyme regular activity” were enriched at most time points except 16:00 and 04:00, respectively, while “catalytic activity” was enriched at 08:00 and 00:00 (Figure 2B). For the “cell component” GO category, DEGs associated with “membrane” and “cell wall” were seen at all time points, while specific cellular components were most affected at 12:00 and 16:00 (Figure 2C).
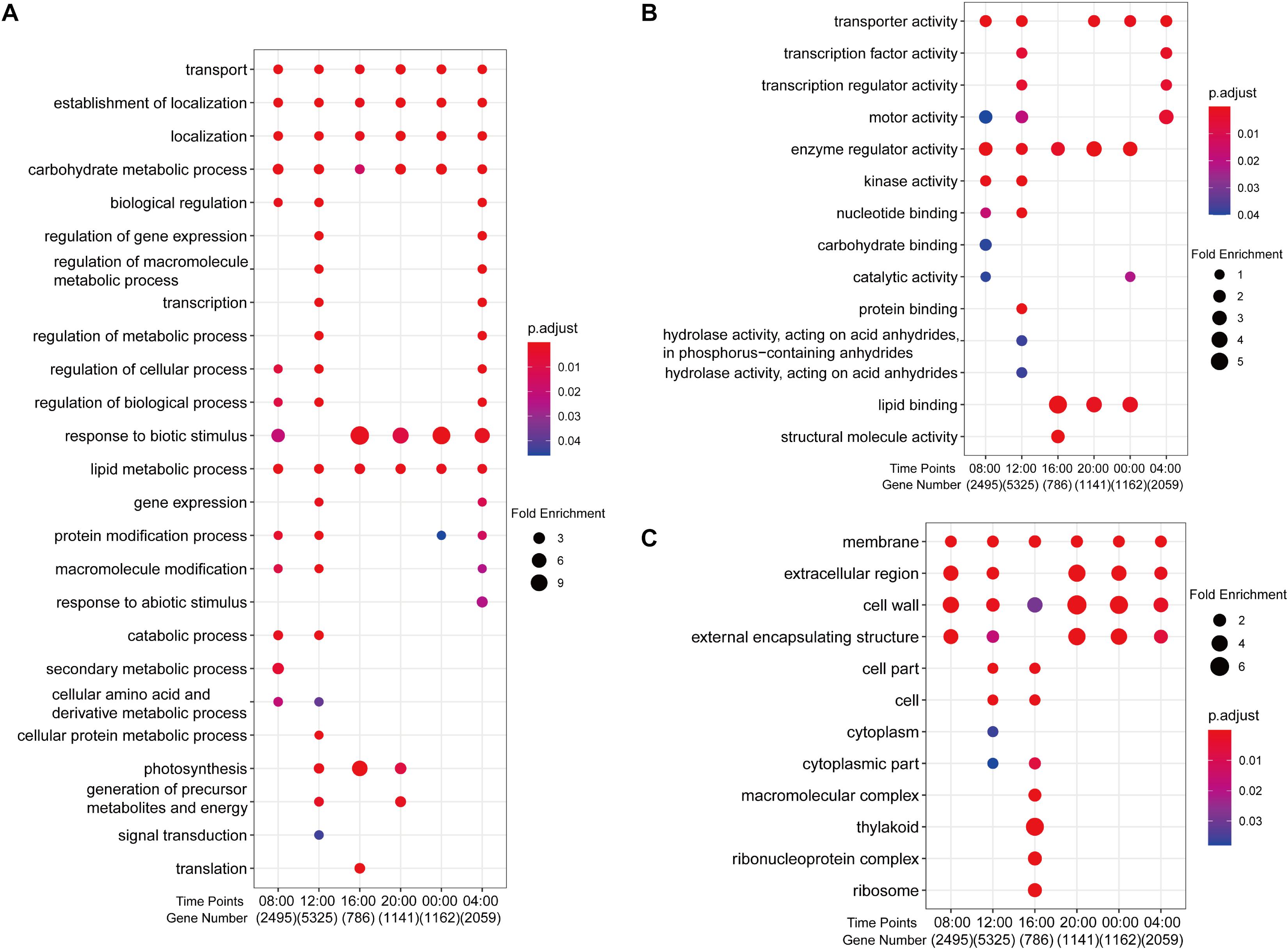
Figure 2. Enriched GO terms of 11,726 DEGs between SD and LD rice anther samples. (A) Biological processes. (B) Molecular function. (C) Cell component. Fold enrichment, depicted by circle size, which calculated by GeneRatio/BgRatio; p.adjust, shown by color, indicates significance level of enrichment results from blue to red.
KEGG analysis showed that genes encoding proteins involved in “pentose and glucuronate interconversions” in carbohydrate metabolism were enriched at 08:00, 12:00, 20:00, and 00:00 (Supplementary Data Sheet 4 and Supplementary Figure 6). UDP-glucose pyrophosphorylase 2 (OsUgp2), a reported TGMS gene required for starch accumulation in pollen (Mu et al., 2009), was present in this pathway. In addition, genes associated with “starch and sucrose metabolism” and “carbon metabolism” were specifically enriched at 12:00, indicating a major difference in carbohydrate metabolism between SD and LD anthers at this time. Genes encoding “photosynthesis-antenna proteins” were enriched at all time points except 08:00. Rice chlorophyll a/b binding protein gene, OsCAB1R, a circadian clock-controlled gene (Sugiyama et al., 2001), is part of this pathway, indicating that rhythm genes may play photoperiod-sensitive roles in the anther. Calmodulin-related genes were mainly enriched in “plant–pathogen interaction” at 08:00, 20:00, and 04:00, such as calcium-dependent protein kinase 9 (OsCPK9), OsCPK21, and OsCPK25/26. OsCPK9 overexpression has been shown to improve pollen fertility under drought stress and shows sensitivity to ABA (Wei et al., 2014). Specific enrichment of mitogen-activated protein kinase (MAPK) pathway genes involved in defense against biotic stress was observed at 04:00.
Genes involved in transport were differentially expressed in anthers at all time points under the two photoperiod conditions, including 189 genes encoding proteins involved in ion transport, 82 genes associated with protein transport, and 54 genes associated with lipid transport (Figure 3A and Supplementary Data Sheet 5). These genes could be grouped in two main clusters that were generally up-regulated in either LD or SD conditions. Genes up-regulated in SD anthers were generally most highly expressed at 12:00, but genes up-regulated in LD anthers had high expression at the 08:00 time point as well. For ion transport, DEGs predicted to encode potassium transporters, including high-affinity K+ transporter (HKT) (Jabnoune et al., 2009) and HAK family (Bañuelos et al., 2002) proteins, as well as sodium, calcium, hydrogen (proton), iron, boron, and zinc transporters were represented at all time points. For protein transport, genes encoding 15 rat sarcoma-related transporter and 17 mitochondrial translocase proteins were differentially expressed. A small GTPase protein, OsRacD, which has been shown to participate in light signal transduction and the fertility switch of NK58S (Ye et al., 2004), was more highly expressed under SD conditions at 08:00 and 12:00. For lipid transport, DEGs encoding 44 lipid transfer protein (LTP) family members were detected. Among these was Photoperiod-sensitive dwarf 1 (Psd1), a non-specific LTP gene whose mutation can cause dwarfism under LD and low-temperature conditions (Deng W. et al., 2020). Psd1 was up-regulated under SD conditions at 12:00. Other transport-associated DEGs included sugar transporters, such as monosaccharide transporters (MSTs) and sucrose transporters (SUTs). For example, MST7 was up-regulated under SD conditions at all time points, while MST5 and SUT1 were up-regulated under LD conditions at all times except 16:00.
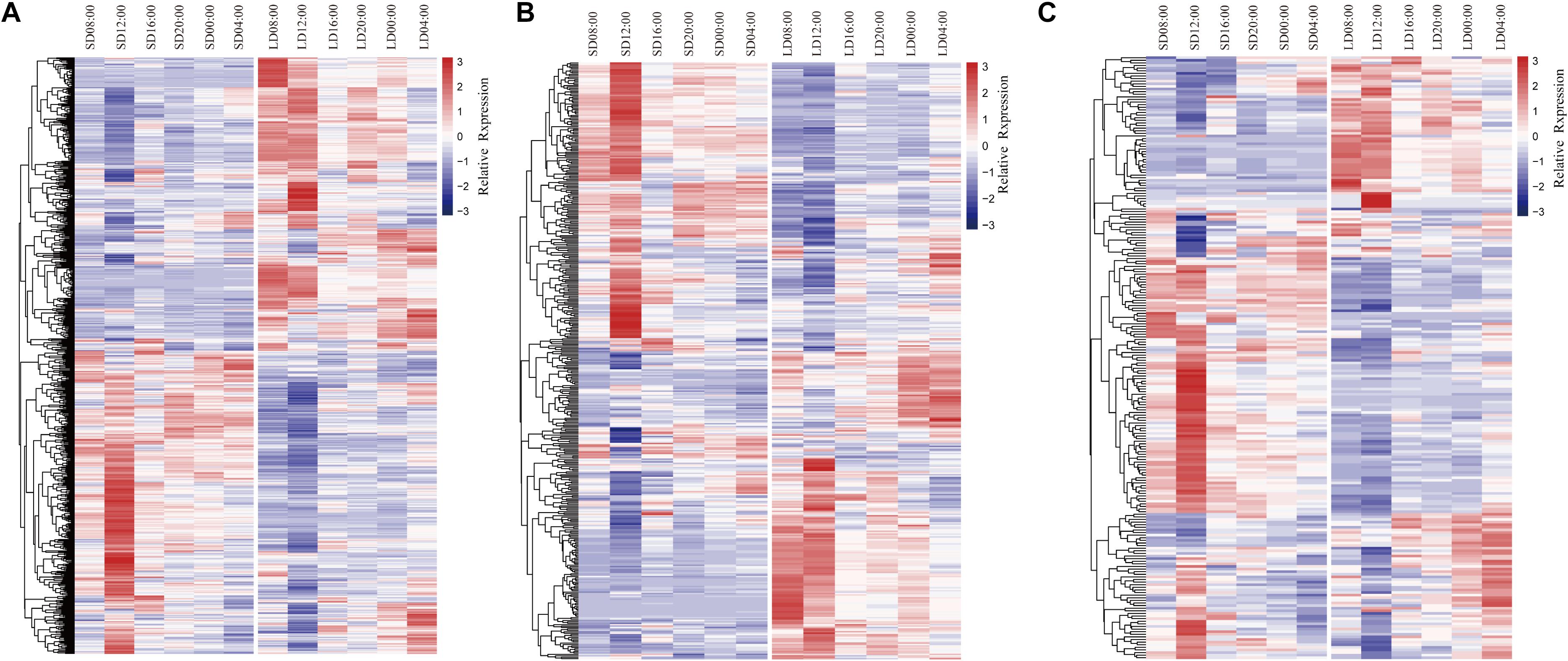
Figure 3. Heatmaps and dendrograms of DEGs encoding proteins involved in (A) transport; (B) carbohydrate metabolism; (C) lipid metabolism. Blue color indicates down-regulation in anthers; red color indicates up-regulation in anthers.
In the GO category “carbohydrate metabolism,” DEGs encoding glycosyl hydrolase and transferase proteins were highly represented, including 250 hydrolases and 118 transferases (Figure 3B and Supplementary Data Sheet 5). These genes again clustered into two groups, each up-regulated under one of the two photoperiod conditions, and genes up-regulated under SD conditions were again most highly expressed at 12:00 (Figure 3B). DEGs up-regulated under LD conditions exhibited two patterns of expression: a larger group most highly expressed at 08:00 and 12:00 pm, while a second, smaller group exhibited peak expression at 00:00 and 04:00 (Figure 3B). Hexokinase (HXK) family genes including HXK5 (Cho et al., 2009), HXK7 (Kim et al., 2016), and HXK10 (Xu et al., 2008) as well as four invertases (INV) (Oliver et al., 2005; Deng X. et al., 2020) were identified, and these were spread across all three major patterns of expression.
DEGs encoding proteins involved in lipid metabolism related genes also exhibited three distinct patterns of expression (Figure 3C and Supplementary Data Sheet 5). Genes generally up-regulated in SD anthers had maximum expression at 12:00; genes up-regulated in LD anthers generally peaked at 08:00 and 12:00, while a third group of genes exhibited maximum expression in SD anthers at 12:00 and in LD anthers at 04:00. Two genes associated with male sterility, Wax-deficient anther 1 (Wda1) and HUMIDITY-SENSITIVE GENIC MALE STERILITY 1 (HMS1) (Jung et al., 2006; Chen et al., 2020), were among the genes up-regulated in SD anthers. Wda1 is associated with biosynthesis of very-long-chain fatty acids for the formation of anther epicuticular wax crystals, whose mutation causes complete male sterility (Jung et al., 2006). HMS1 encodes a β-ketoacyl-CoA synthase, and its mutant is humidity-sensitive genic male sterile (Chen et al., 2020).
Based on our finding that OsCAB1R, a circadian clock-controlled gene, was a DEG in the “photosynthesis-antenna proteins” GO class (Supplementary Data Sheet 4), we analyzed, and found, differential expression of other genes known to be affected by circadian rhythm (Supplementary Figure 7 and Supplementary Data Sheet 5). Rice CIRCADIAN CLOCK ASSOCIATED 1 (OsCCA1), homologous to Arabidopsis AtCCA1 (Izawa et al., 2002), showed a higher expression in the early morning, with almost twofold higher expression under LD compared with SD conditions at 04:00; ZEITLUPE 2 (ZTL2) had a similar pattern but peaked under LD at 08:00. At 12:00, OsCCA1 was expressed nearly seven times higher in SD compared with LD anthers. Rice PSEUDO-RESPONSE REGULATOR (OsPRR), OsPRR95, homologous to Arabidopsis circadian oscillator AtPRR5 and AtPRR9 (Murakami et al., 2005), showed fivefold expression higher at 12:00 under SD compared with that of LD conditions. OsPRR37 shows up-regulation at night under LD (20:00, 00:00, and 04:00), four times higher at 00:00 and 04:00, while OsPRR73, Early flowering 3 (OsELF3), and ZTL1 showed little difference between SD and LD conditions. Rice LUX ARRHYTHMO (OsLUX), homologous to a nighttime repressor of circadian gene expression AtLUX (Murakami et al., 2007), was also up-regulated at 16:00 in SD conditions. Other rice circadian clock or flowering genes FLAVIN BINDING, KELCH REPEAT, F-BOX1 (OsFKF1), TIMING OF CAB EXPRESSION 1 (OsTOC1), and Gigantea (OsGI) (Hayama et al., 2003; Murakami et al., 2003) were expressed more highly at 16:00 in LD anthers than in SD anthers, although by less than a twofold difference. These results indicated that rhythm genes in the anther are sensitive to photoperiod, particularly under SD conditions.
To examine the regulatory networks of genes under SD and LD conditions, 39,966 genes (99.8%) were categorized into 34 modules based on scale-free topology model (β = 7; Supplementary Figure 8). The remaining 78 genes that did not assemble into existing modules were grouped into a 35th “module” (gray). Standard modules ranged in size from 177 to 5852 genes, e.g., black contained 1496 genes that generally expressed highly at LD 12:00; dark red had 555 genes that generally expressed highly at SD 16:00 (Supplementary Table 2 and Supplementary Figure 9B).
Variation in gene expression was higher between SD and LD conditions than between dark/light conditions (Figure 4), suggesting that photoperiod has a significant effect on anther development. Expression of 36% of genes correlated with SD or LD conditions, suggesting that a large proportion of genes responded directly or indirectly to day length. Gene expression in four modules in particular was observed to correlate significantly (p < 0.05) with photoperiod: genes in turquoise (5852 genes, r = 0.76) and magenta (1132 genes, r = 0.71) modules were up-regulated in LD, while genes in yellow (4929 genes, r = −0.71), and blue (2,698 genes, r = −0.71) modules were up-regulated in SD (Supplementary Table 2, Supplementary Figure 9, and Supplementary Data Sheet 6).

Figure 4. Module-trait associations. Correlation coefficients between different modules and traits show in matrix. Each cell contains correlation coefficient with p value in bracket. Photoperiod, short-day, and long-day condition; LD acts as positive control, in which red color shows positive correlation with LD; SD is the opposite; Light and Dark: daytime vs. night conditions, daytime is defined as positive blank including samples at 08:00, 12:00, and 16:00; night includes 20:00, 00:00, and 04:00.
Gene ontology analysis revealed enrichment of different functional categories of genes among modules (Figure 5). The turquoise, magenta, and blue modules were enriched for biological processes including “transport,” “localization,” “establishment of localization,” and “carbohydrate metabolic process,” and contained genes encoding various potassium (HKT and HAK family) and sugar (MST, SUT) transporters, and carbohydrate metabolic proteins such as sucrose synthase (SUS) and invertases (Supplementary Data Sheet 7).
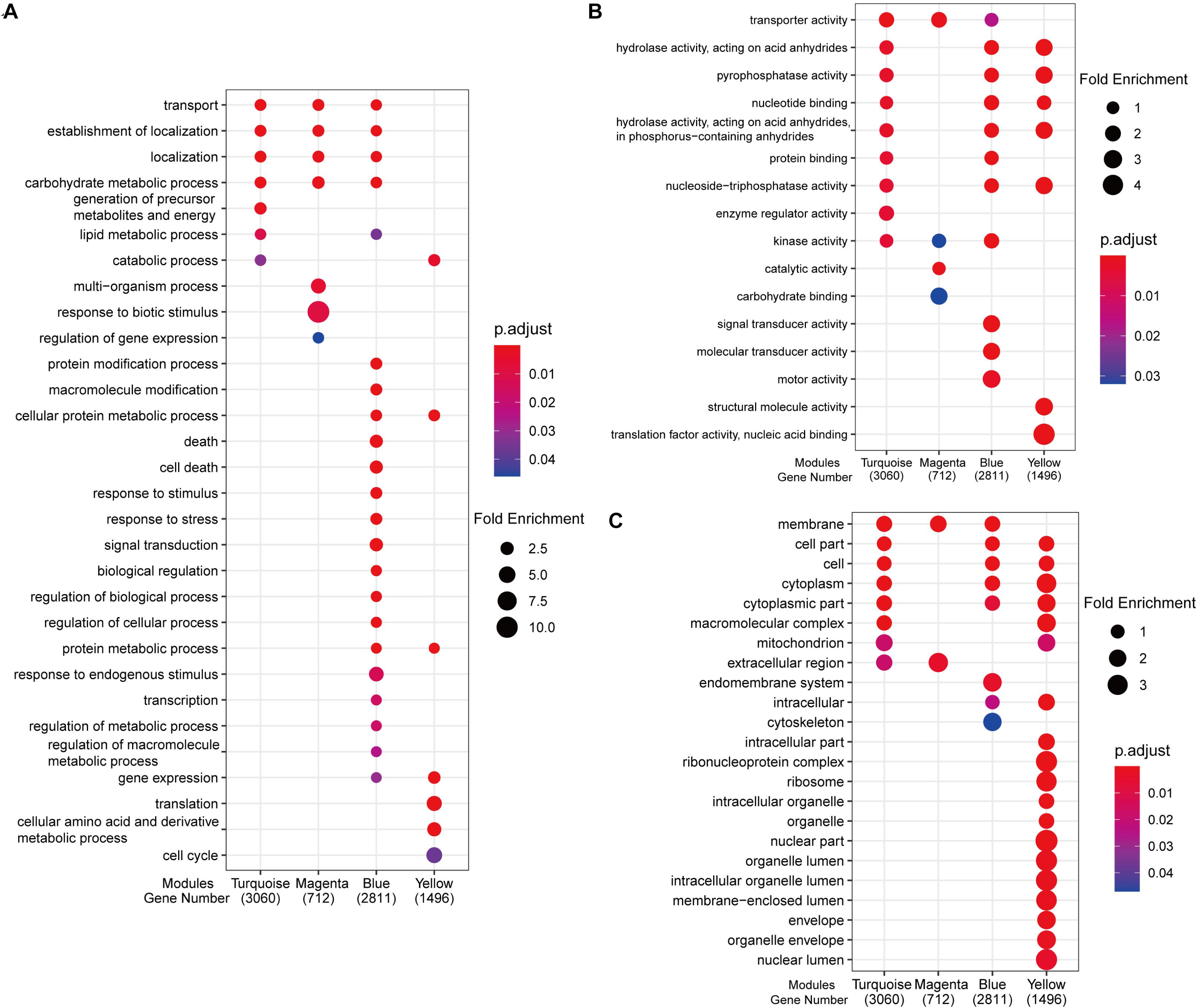
Figure 5. Enriched GO terms of genes in four co-expression modules. (A) Biological processes. (B) Molecular function. (C) Cell component. Fold enrichment, depicted by circle size, which is calculated by GeneRatio/BgRatio; p.adjust, shown by color, indicates significance level of enrichment results from blue to red.
Genes associated with “lipid metabolic process” were enriched in turquoise and blue modules (Figure 5A), which is also consistent with the GO enrichment of DEGs (Figure 2A). The blue module was enriched in more GO categories than other modules, including “protein metabolic process” and “macromolecule modification” (Figure 5A). In the “protein metabolic process” category were TGMS genes, such as Temperature-sensitive Male Sterile 10 (TMS10) and TMS10-Like gene (TMS10L) (Yu et al., 2017) and ubiquitin fusion ribosomal protein L40 (UbL404), which acts downstream of TMS5 (Zhou et al., 2014), as well as genes encoding light-responsive proteins, such as photoreceptors Phytochrome B(PHYB) and PHYC, and Heading date 6 (Hd6) (Takahashi et al., 2001; Takano et al., 2005) (Supplementary Data Sheet 7).
KEGG enrichment analysis revealed genes associated with “oxidative phosphorylation,” the “TCA (tricarboxylic acid) cycle,” and “carbon metabolism pathway” in the turquoise module (Supplementary Figure 10). The magenta module was enriched in diterpenoid biosynthesis, while yellow module was enriched in categories associated with DNA replication, RNA transport, and ribosome biosynthesis. These results indicate that protein metabolism and transcription/translation processes are also affected by day length.
Hub genes are those with a high degree of connectivity and co-relationships with genes in modules of interest for a given phenotype (Langfelder and Horvath, 2008). Based on the module membership and gene significance, 239, 62, 139, and 56 hub genes were identified in turquoise, magenta, blue, and yellow modules, respectively (Supplementary Data Sheet 6). As identified in our earlier GO and KEGG analyses, these genes were involved a broad range of processes, including macromolecule, carbohydrate, and lipid metabolisim and transport. Of particular interest, six hub genes have previously been reported to be involved in male sterlity or low fertility, including LOC_Os01g16810 (CSA) (Zhang et al., 2010, 2013), LOC_Os09g38030 (UDP-glucose pyrophosphorylase gene 1, OsUgp1) (Chen et al., 2007), LOC_Os01g54620 (cellulose synthase catalytic subunit genes 4, OsCesA4) (Tanaka et al., 2003), LOC_Os02g04840 (collapsed abnormal pollen 1, CAP1) (Ueda et al., 2013), LOC_Os04g39980 (dioxygenase for auxin oxidation gene, DAO) (Zhao et al., 2013), and LOC_Os06g04090 (secondary wall NAC 1, OsSWN1) (Chai et al., 2015). The discovery of known key regulators of pollen sterility indicates the robustness of our co-expression and hub gene construction method to mine for key anther regulator genes. CSA encodes a key R2R3 MYB transcription factor that controls male fertility in a photoperiod-dependent manner (Zhang et al., 2013). OsUgp1 is required for callose depostion in pollen mother cells in meiosis; knockdown of OsUgp1 causes a thermosensitive male sterile defect (Chen et al., 2007). OsCesA4 is required for cellulose synthesis, and its mutant shows brittle culm and low fertility (Tanaka et al., 2003). CAP1 encodes an arabinokinase-like protein required for nucleus, cytoplasm, and intine cell wall development in rice pollen. DAO encodes a 2-oxoglutarate-dependent-Fe (II) dioxygenase essential for anther formation and dehiscence, pollen fertility, and seed initiation (Zhao et al., 2013). OsSWN1 is a NAC transcription factor gene involved in secondary cell wall biosynthesis, whose down-regulation causes a decrease in lignin and an increase in polysaccharide content in cell walls, resulting in anther dehiscence defects (Chai et al., 2015). The identification of these genes in our analyses suggests that these previously characterized pollen development regulators could also have a day length-dependent role.
An additional 30 hub genes encoding transcription factors were detected, including 5 previously unreported MYB proteins and 10 zinc-finger proteins (Table 1). The function of these unreported genes in regulating anther development needs to be further characterized, but it seems likely that they will be key regulators of rice anther development particularly affected by photoperiod.
Two hub genes, CSA and Ugp1, have been reported to be involved in rice photoperiod-sensitive and thermosensitive male fertility, respectively. We visualized genes with high correlation to CSA and Ugp1 (Figure 6 and Supplementary Data Sheet 8). Notably, CSA and Ugp1 shared 74 overlapped correlation genes, such as MATRILINEAL (OsMATL) and OsHXK5 (Cho et al., 2009; Yao et al., 2018), which were up-regulated in LD anthers, unlike that of CSA and Ugp1—as undirected network relationships. Four co-expressed genes were also selected for qRT-PCR analysis and confirmed this prediction outcome (Supplementary Figure 11). OsMATL encodes a pollen-specific phospholipase that is involved in haploid induction, whose knockout mutant displays reduced seed set (Yao et al., 2018). OsHXK5 encodes a hexokinase that functions as a glucose sensor. Plants overexpressing OsHXK5 showed a hypersensitive and slow growth phenotype (Cho et al., 2009). Six TFs such as MYB, Zinc Finger, and MADS-Box proteins were in network; 5 lipid metabolic-associated genes, 11 carbohydrate metabolic genes, and 11 transport genes were also detected. In addition, there were 38 hub genes highly co-expressing with CSA and Ugp1. These results indicate that CSA and Ugp1 are likely to share regulatory pathways in controlling environmentally mediated male development in rice.
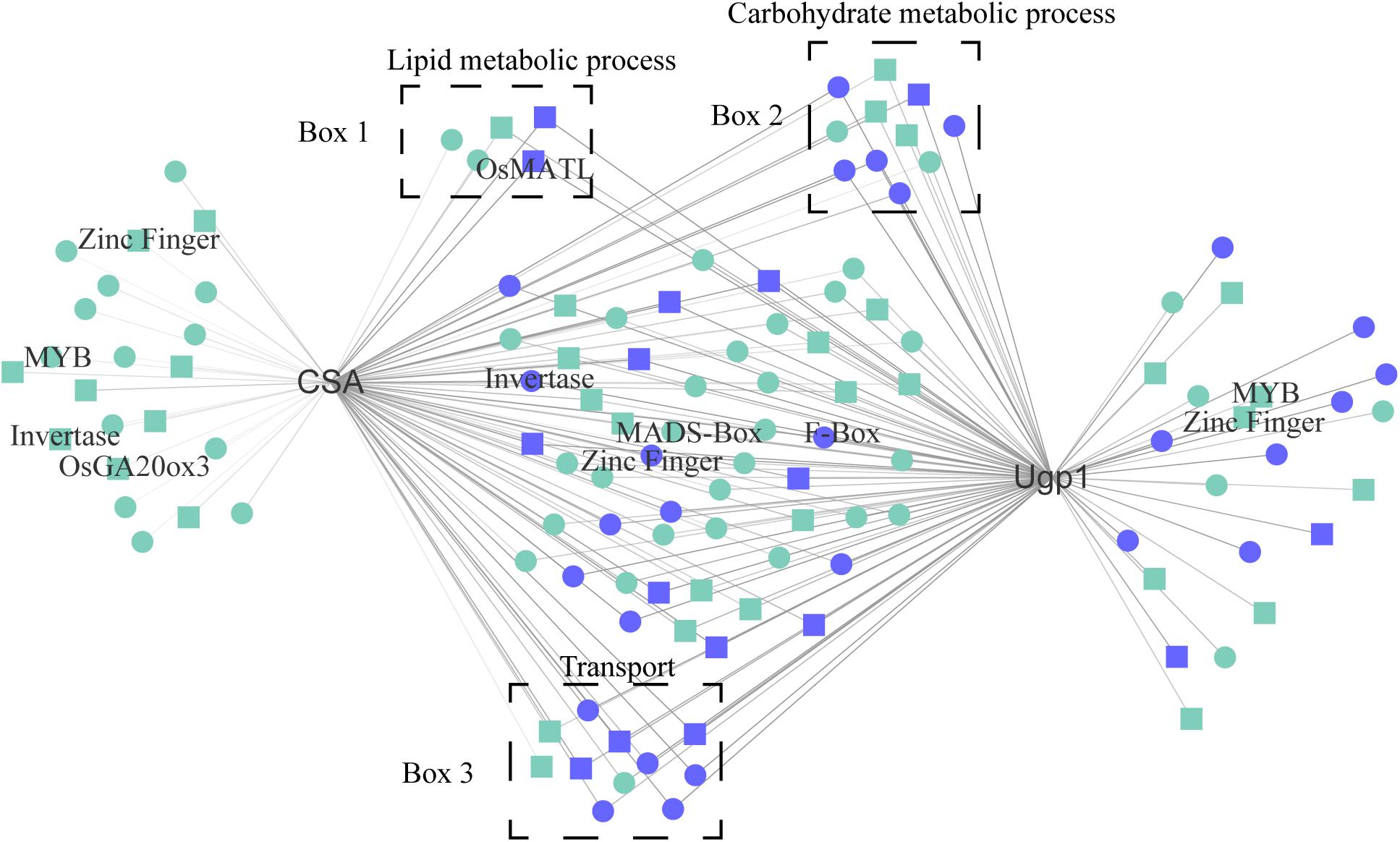
Figure 6. High co-expressed genes of CSA and Ugp1. Blue indicates the top 1% TOM value genes; rectangle represents hub genes. OsGA20ox3 (gibberellin 20 oxidase 3, LOC_Os07g07420); OsMATL (MATRILINEAL, LOC_Os03g27610); CSA (Carbon Starved Anther, LOC_Os01g16810); Ugp1 (UDP-glucose pyrophosphorylase, LOC_Os09g38030); Invertase (LOC_Os12g37480, LOC_Os02g01310); MYB-TFs (LOC_Os01g51260, LOC_Os06g46560); MADS-box genes (LOC_Os06g11970); Zinc Finger genes (LOC_Os02g44120, LOC_Os01g68900, and LOC_Os12g08070); F-box (LOC_Os04g19800); Box 1, lipid metabolic process-related genes; Box 2, carbohydrate metabolic process-related genes; Box 3, Transport-related genes.
Transcript profiling of anthers grown under different photoperiod conditions provides information to help understand the molecular mechanisms that affect male fertility in response to day length and thus improve our ability to create new PGMS lines for rice breeding. In this study, we have conducted the first genome-wide transcriptomic and gene network analysis of anthers grown under short and long day lengths and revealed functional and regulatory genes that respond to photoperiod.
Approximately one-third of expressed genes (11,726 out of ∼30,000 in different samples) were differentially expressed in anthers grown under SD and LD conditions, with gene expression generally down-regulated under SD conditions (Figure 1A, Supplementary Table 1, and Supplementary Data Sheet 2). A total of 177 genes were differentially expressed at all time points (Figure 1C), of which 51 were specifically expressed in the anther and other reproductive tissues (Supplementary Figure 5B and Supplementary Data Sheet 2). Several genes whose mutation is known to cause male sterility were found in this set of 51 genes. OsABCG15 plays a role in the formation of the anther cuticle and pollen exine and is involved in the export of lipid precursors from the tapetum to anther locules (Wu et al., 2014; Zhao et al., 2015). OsLAP6 functions in sporopollenin metabolism and affects bacula elongation to regulate pollen exine formation (Zou et al., 2017). HOTHEAD 1 (HTH1) encodes a glucose–methanol–choline (GMC) oxidoreductase; its RNAi line and mutant both display defective anther walls and aborted pollen with decreased amounts of long-chain fatty acids and cutin (Xu et al., 2017). Other genes in this set of 177 DEGs, especially genes that specifically expressed in anthers, may also affect male fertility, particularly in a photoperiod-sensitive manner.
Gene ontology enrichment analysis of DEGs at different time points revealed a high proportion of genes associated with transport and metabolism, particularly of ions, carbohydrates, and lipids (Figure 2). Generally, these genes were up-regulated at 12:00 under SD and at 08:00 and 12:00 under LD conditions, indicating that these pathways may be associated with the growth and physiological adaption of plants in response to photoperiod.
In stage 11 anthers, mitosis and accumulation of nutrients are the main developmental processes, consistent with our GO enrichment results. Correct ion homeostasis is essential for normal plant growth; accordingly, many genes encoding ion transporters were detected (Supplementary Data Sheet 3). Several potassium transporter family genes were enriched, like HKT, HAK, and AKT family genes (Rodríguez-Navarro and Rubio, 2006). HKT1;4 can also exclude Na+ in productive stage of rice (Suzuki et al., 2016). Cation/H+ exchanger 14 (OsCHX14), involved in K+ homeostasis, may play an important role during rice flowering stages (Chen et al., 2016). OsBOR1 and OsBOR4 DEGs encode boron transporters; pollen from osbor4 mutants have defects in pollen tube elongation (Nakagawa et al., 2007; Tanaka et al., 2013). OsFRDL1 is a MATE (multidrug and toxic compound extrusion) family gene, a citrate transporter of iron, whose mutant exhibits a decrease in pollen viability and grain fertility (Yokosho et al., 2009, 2016). SULTR-like phosphorus distribution transporter (SPDT) encodes a plasma-membrane-localized phosphorus transporter that preferentially allocates phosphorus to grains (Yamaji et al., 2017). Psd1 encodes a non-specific lipid transfer protein (nsLTP) that regulates cell division and elongation; psd1 mutants show photoperiod-thermosensitive dwarfism (Deng W. et al., 2020).
Many DEGs were also found to encode sugar transporters and catabolic proteins, such as monosaccharide transporters (MST1 to MST8), sucrose transporters (SUT1, SUT2, SUT3, and SUT5), and invertases (INV1, INV2, INV3, and INV4). Proteins from these three family genes cover sucrose transport from source to sink. Sucrose is loaded into the phloem by SUTs and unloaded in sink tissues, like anthers or seeds, where sucrose is converted by invertases into hexoses and imported into cells by MSTs (Lim et al., 2006). OsMST4 has been shown to play a role in monosaccharide supply during grain filling (Wang et al., 2007), and OsMST5 is expressed in panicles before pollination and may be involved in pollen development (Ngampanya et al., 2003). MST8 is predicted to transport monosaccharides from anther wall layers to the microspore, affecting the unloading process of the source–sink tissue during male reproduction (Zhang et al., 2010). SUT1 is predicted to play a role in the uptake of sucrose from the apoplast, and its lack during anther development suppresses pollen germination (Hirose et al., 2010). INV4 is expressed specifically in the anther, down-regulated by cold stress, resulting in abnormal accumulation of sucrose (Oliver et al., 2005).
Other DEGs associated with carbohydrate metabolism could also affect anther viability. Glucan synthase-like 5 (GSL5) encodes a callose synthase, and its mutant showed decreased fertility due to defects in the pollen callose wall (Shi et al., 2015). Oryza sativa glucanase 1 (Osg1) encodes a plant β-1,3-glucanase essential for callose degradation during tetrad dissolution; RNAi plants exhibited delayed release of young microspores into anther locules, causing male sterility (Wan et al., 2011). Hexokinase (HXK) family proteins are involved in sugar sensing and signaling (Cho et al., 2006), and genes encoding HXK5, HXK7, and HXK10 were among DEGs detected in our analysis. HXK10 can also phosphorylate hexose sugars; RNAi knockdown lines showed non-dehiscent anthers (Xu et al., 2008). Genes encoding disproportionating enzymes DPE1 and DPE2 were also among our DEGs; these proteins are involved in starch metabolism and maturation of starch granules (Akdogan et al., 2011; Hwang et al., 2016).
Differentially expressed genes encoding transport and metabolic genes clustered into two, similarly sized groups that were up-regulated in either SD or LD anthers, suggesting that these biological processes may occur, or be regulated by, different pathways depending on photoperiod to maintain nutrient balances essential for normal development (Figures 3A–C). While previously reported genes affecting male sterility were found among our photoperiod-sensitive DEGs, their mutations generally caused total male sterility rather than PGMS (i.e., Osg1, OsFRDL1, and OsBOR4). Discovery of new potential PGMS regulators will require careful analysis of differential expression of candidate genes under SD and LD conditions.
We analyzed the expression pattern of genes associated with the control of circadian rhythm and found that OsLUX, OsFKF1, OsGI, and OsTOC1 show higher expression in LD over SD anthers at16:00, OsPRR37 was up-regulated under night at LD, while OsPRR95 has higher expression at 12:00. OsCCA1 was up-regulated at 12:00 and 16:00 in SD compared with LD anthers (Supplementary Figure 7). These genes and proteins interact with each other in complex ways. In Arabidopsis, CCA1 positively regulates PRR7 and PRR9 expression, which in turn affect feedback regulation of CCA1 (Farré et al., 2005). CCA1 can also interact with TOC1 to bind the GI promoter (Hsu and Harmer, 2014). LUX is a MYB transcript factor required for normal circadian rhythm that can repress PRR9 expression (Chow et al., 2012). FKF1 is an F-box protein that forms a complex with GI to regulate the transition to flowering (Sawa et al., 2007). In rice, overexpression of CCA1 or GI delays flowering (Hayama et al., 2003). The presence of these important circadian genes in our DEGs suggests that they may also play a critical role in anther development in response to photoperiod and warrant further investigation as potential candidates for PGMS line creation.
Weighted gene co-expression network analysis (WGCNA), which evaluates potential interaction between all expressed genes, revealed large gene clusters and regulatory networks accociated with photoperiod length and confirmed that gene expression differs significantly between SD and LD anthers. Four modules were identified with a high correlation to photoperiod: genes in turquoise (5852 genes) and magenta (1132 genes) modules were up-regulated in LD anthers, while genes in blue (4929 genes) and yellow (2698 genes) modules were up-regulated in SD anthers (Figure 4 and Supplementary Figure 8).
These modules were generally enriched with the same GO terms as DEGs (Figures 2, 5), but the blue module was also enriched in genes from new categories, i.e., protein-related and macromolecule-related processes, which contained TMS10 (thermosensitive male sterility 10), TMS10L (thermosensitive male sterility 10-like), and UbL404 (ubiquitin fusion ribosomal protein L40) genes. TMS10 and TMS10L are two thermosensitive genes that redundantly regulate post-meiotic tapetal development and pollen development in low and high temperatures (Yu et al., 2017). UbL404 is a target of TMS5, which encodes a conserved RNase Z protein. UbL404 is degraded by RNase Z in wild-type plants, but accumulates in tms5 plants at high temperatures, causing temperature-sensitive male sterility (Zhou et al., 2014). A subsequent mutation in GATA10, which decreases UbL404 levels, can restore fertility at high temperatures (Jin et al., 2019). Our modules thus contain known PGMS and TGMS effectors and are likely to be a good source to uncover as-yet-unknown genes affecting PGMS.
The “protein process” GO category also contained genes associated with light response and anther-specific genes (Figure 5 and Supplementary Data Sheet 7). Light response genes included PHYB, PHYC, and Hd6. PHYB and PHYC are photoreceptors in rice, single mutants of which induce early flowering; a triple mutant with PHYA causes incomplete sterility (Takano et al., 2005). Hd6 encode a casein kinase II that delays heading time in LD conditions (Takahashi et al., 2001). Anther-specific genes included Pollen Mitosis Relative (PMR), essential for correct pollen mitosis (Liu et al., 2017), and DWARF AND RUNTISH SPIKELET (DRUS1) and DRUS2, which regulate anther development and pollen maturation redundantly through sugar utilization or conversion (Pu et al., 2017). Genes involved in the MAPK (mitogen-activated protein kinase) and Ca2+-related signaling pathway were also found, such as Calcium-dependent protein kinase 2 (OsCDPK2), which is repressed by light and disrupts seed development (Frattini et al., 1999; Morello et al., 2000). Genes associated with phytohormone signaling included brassinosteroid-signaling kinase (BSK) family members and Ethylene Receptor 2 (ETR2). Thus, while mitosis and sugar transport are key biological processes occurring in stage 11 anthers, hormone, and other signaling pathways are also very active during anther late development.
Our WGCNA analysis revealed 496 hub genes in these four modules (Supplementary Data Sheet 6). A known PGMS gene, CSA, and TGMS gene, Ugp1, were among these hub genes, both known to affect male sterility through carbohydrate metabolism (Chen et al., 2007; Zhang et al., 2013). Other genes affecting carbohydrate metabolism were also among the hub genes, including CAP1, DPE2, and OsSWN1. These results provide further evidence that carbohydrate metabolism is a key characteristic of late anther development affected by photoperiod. Multiple phytohormone-related genes were also present in the hubs: ABA 8’-Hydroxylase gene 3 (OsABA8ox3) is a key gene in abscisic acid (ABA) catabolism (Zhu et al., 2009); OsNAP can be induced by ABA and, in turn, affects ABA biosynthesis (Liang et al., 2014); Gibberellin 20-oxidase 3 (OsGA20ox3) and Gibberellin 2-oxidase 4 (OsGA2ox4) affect gibberellic acid biosynthesis, GA20ox3, in a temperature-dependent manner (Lo et al., 2008; Sakata et al., 2014); Dioxygenase of auxin oxidation (DAO) affects the auxin pathway (Zhao et al., 2013); and SK3/SHAGGY-like kinase (GSK2) acts as a negative regulator of brassinosteroid signaling (Tong et al., 2012). Phytohormones thus appear to be key regulators mediating the adaption of anther development to changing day length.
Given the presence of CSA and Ugp1 as hub genes, we searched for, and found, other hub genes regulating environment-sensitive male sterility (Table 1 and Supplementary Data Sheet 6). TMS5 was clustered in the red module, which did not show significant correlation with photoperiod. OsPDCD5, Ugp2, and Osgata10 clustered in the turquoise module, while TMS10 clustered in the blue module. Both OsPDCD5 and TMS10 are involved in PCD, which, in rice anthers, begins before stage 11; had younger anthers been used in our analysis, WGCNA significance scores for these genes may have been higher.
Analysis of interaction networks of key hub genes revealed further interactions between PGMS gene CSA and TGMS gene Ugp1 that two genes share large part of co-expressed genes. Although these genes were up- and down-regulated in an opposite pattern CSA and Ugp1, these genes are likely to be common targets of Ugp1 and CSA regulation and have similar roles in co-regulating photoperiod- or temperature-sensitive anther development. This common list included genes encoding zinc finger, MADS-Box, and MYB transcript factors, which may play a key relational role in mediating photoperiod-associated male reproduction. Moreover, carbohydrate metabolic genes and transport genes like invertase were also detected, and these genes may be associated with the sugar transport and metabolism under SD and LD. Further analysis on 38 hub genes highly co-expressed with CSA and Ugp1 may reveal key genes regulating photoperiod-dependent male fertility.
In summary, we provided a transcriptomic view in anther development at different photoperiods and narrowed down candidates not only for future discovery of new PGMS but also for genes that interacted with reported EGMS, CSA and Ugp1, based on DEG analysis and WGCNA. Further investigations to confirm the biological function of these genes will reveal new genetic and molecular controls of PGMS in rice and possible other crops.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE163030.
DZ designed the study. DW, JL, YL, and SS performed experiments. SS analyzed the RNA-seq data and drafted the manuscript. WC, GL, XZ, and DZ revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (grant nos. 2016YFD0100804 and 2016YFD0100903), the National Natural Science Foundation of China (grant nos. 31861163002 and 31700276), and the Innovative Research Team, Ministry of Education, and 111 Project (grant no. B14016).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Mr. Zhijing Luo and Ms. Mingjiao Chen for their assistance in rice growth, and Dr. Natalie Betts for helpful comments on the article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.621561/full#supplementary-material
Adrian, D. (2019). venn: Draw Venn Diagrams. R Package Version 1.8. Available online at: https://Cran.R-project.org/package=venn (Accessed May 25, 2019).
Akdogan, G., Kubota, J., Kubo, A., Takaha, T., and Kitamura, S. (2011). Expression and characterization of rice disproportionating enzymes. J. Appl. Glycosci. 58, 99–105. doi: 10.5458/jag.jag.JAG-2010_026
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Bañuelos, M. A. A., Garciadeblas, B., Cubero, B., and Rodrıìguez-Navarro, A. (2002). Inventory and functional characterization of the HAK potassium transporters of Rice. Plant Physiol. 130, 784–795. doi: 10.1104/pp.007781
Chai, M., Bellizzi, M., Wan, C., Cui, Z., Li, Y., and Wang, G.-L. (2015). The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 58, 44–51. doi: 10.1007/s12374-014-0400-y
Chen, H., Zhang, Z., Ni, E., Lin, J., Peng, G., Huang, J., et al. (2020). HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 225, 2077–2093. doi: 10.1111/nph.16288
Chen, L.-Y., Xiao, Y.-H., and Lei, D.-Y. (2010). Mechanism of sterility and breeding strategies for photoperiod/thermo-sensitive genic male sterile rice. Rice Sci. 17, 161–167. doi: 10.1016/s1672-6308(09)60012-3
Chen, R., Zhao, X., Shao, Z., Wei, Z., Wang, Y., Zhu, L., et al. (2007). Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19, 847–861. doi: 10.1105/tpc.106.044123
Chen, Y., Ma, J., Miller, A. J., Luo, B., Wang, M., Zhu, Z., et al. (2016). OsCHX14 is involved in the K+ homeostasis in rice (Oryza sativa) flowers. Plant Cell Physiol. 57, 1530–1543. doi: 10.1093/pcp/pcw088
Cheng, S. H., Zhuang, J. Y., Fan, Y. Y., Du, J. H., and Cao, L. Y. (2007). Progress in research and development on hybrid rice: a super-domesticate in China. Ann. Bot. 100, 959–966. doi: 10.1093/aob/mcm121
Cho, J. I., Ryoo, N., Eom, J. S., Lee, D. W., Kim, H. B., Jeong, S. W., et al. (2009). Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759. doi: 10.1104/pp.108.131227
Cho, J.-I., Ryoo, N., Ko, S., Lee, S.-K., Lee, J., Jung, K.-H., et al. (2006). Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224, 598–611. doi: 10.1007/s00425-006-0251-y
Chow, B. Y., Helfer, A., Nusinow, D. A., and Kay, S. A. (2012). ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signaling Behav. 7, 170–173. doi: 10.4161/psb.18766
Deng, W., Li, R., Xu, Y., Mao, R., Chen, S., Chen, L., et al. (2020). A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 71, 1294–1305. doi: 10.1093/jxb/erz500
Deng, X., Han, X., Yu, S., Liu, Z., Guo, D., He, Y., et al. (2020). OsINV3 and its homolog, OsINV2, control grain size in rice. Int J Mol Sci 21:2199. doi: 10.3390/ijms21062199
Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., et al. (2012a). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 109, 2654–2659.
Ding, J., Shen, J., Mao, H., Xie, W., Li, X., and Zhang, Q. (2012b). RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5, 1210–1216. doi: 10.1093/mp/sss095
Fan, Y., Yang, J., Mathioni, S. M., Yu, J., Shen, J., Yang, X., et al. (2016). PMS1T, producing phased small-interfering RNAs regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. U.S.A. 113, 15144–15149.
Farré, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., and Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/j.cub.2004.12.067
Frattini, M., Morello, L., and Breviario, D. (1999). Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development. Plant Mol. Biol. 41, 753–764. doi: 10.1023/A:1006316422400
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hirose, T., Zhang, Z., Miyao, A., Hirochika, H., Ohsugi, R., and Terao, T. (2010). Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J. Exp. Bot. 61, 3639–3646. doi: 10.1093/jxb/erq175
Horvath, S., and Dong, J. (2008). Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 4:e1000117. doi: 10.1371/journal.pcbi.1000117
Hsu, P. Y., and Harmer, S. L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19, 240–249. doi: 10.1016/j.tplants.2013.11.007
Hwang, S.-K., Koper, K., Satoh, H., and Okita, T. W. (2016). Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 291, 19994–20007.
Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16, 2006–2020.
Jabnoune, M., Espeout, S., Mieulet, D., Fizames, C., Verdeil, J.-L., Conéjéro, G., et al. (2009). Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 150, 1955–1971. doi: 10.1104/pp.109.138008
Jin, J., Gui, S., Li, Q., Wang, Y., Zhang, H., Zhu, Z., et al. (2019). The transcription factor GATA10 regulates fertility conversion of a two-line hybrid tms5 mutant rice via the modulation of UbL40 expression. J. Integr. Plant Biol. 62, 1034–1056. doi: 10.1111/jipb.12871
Jung, K. H., Han, M. J., Lee, D. Y., Lee, Y. S., Schreiber, L., Franke, R., et al. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18, 3015–3032. doi: 10.1105/tpc.106.042044
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Khush, G. S. (2001). Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822. doi: 10.1038/35093585
Kim, H.-B., Cho, J.-I., Ryoo, N., Shin, D.-H., Park, Y.-I., Hwang, Y.-S., et al. (2016). Role of rice cytosolic hexokinase OsHXK7 in sugar signaling and metabolism. J. Integr. Plant Biol. 58, 127–135. doi: 10.1111/jipb.12366
Kim, Y.-J., and Zhang, D. (2018). Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9:559. doi: 10.1186/1471-2105-9-559
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Lim, J. D., Cho, J.-I., Park, Y.-I., Hahn, T.-R., Choi, S.-B., and Jeon, J.-S. (2006). Sucrose transport from source to sink seeds in rice. Physiol. Plant. 126, 572–584. doi: 10.1111/j.1399-3054.2006.00654.x
Liu, L., Zheng, C., Kuang, B., Wei, L., Yan, L., and Wang, T. (2016). Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet. 12:e1006085. doi: 10.1371/journal.pgen.1006085
Liu, Y., Xu, Y., Ling, S., Liu, S., and Yao, J. (2017). Anther-preferential expressing gene PMR is essential for the mitosis of pollen development in rice. Plant Cell Rep. 36, 919–931. doi: 10.1007/s00299-017-2123-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, S.-F., Yang, S.-Y., Chen, K.-T., Hsing, Y.-I., Zeevaart, J. A. D., Chen, L.-J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618. doi: 10.1105/tpc.108.060913
Lu, K., Li, T., He, J., Chang, W., Zhang, R., Liu, M., et al. (2017). qPrimerDB: a thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 46, D1229–D1236. doi: 10.1093/nar/gkx725
Michel, D., Jérôme, S., Richard, C., Christophe, S., Farid, R., Pierre, L., et al. (2001). Rice genomics: present and future. Plant Physiol. Biochem. 39, 323–334.
Morello, L., Frattini, M., Gianì, S., Christou, P., and Breviario, D. (2000). Overexpression of The calcium-dependent protein kinase OsCDPK2 in transgenic rice is repressed by light in leaves and disrupts seed development. Transgenic Res. 9, 453–462. doi: 10.1023/A:1026555021606
Mou, T. (2016). The research progress and prospects of two-line hybrid rice in China. Chin. Sci. Bull. 61, 3761–3769. doi: 10.1360/n972016-01045
Mu, H., Ke, J., Liu, W., Zhuang, C., and Yip, W. (2009). UDP-glucose pyrophosphorylase2 (OsUgp2), a pollen-preferential gene in rice, plays a critical role in starch accumulation during pollen maturation. Sci. Bull. 54, 234–243. doi: 10.1007/s11434-008-0568-y
Murakami, M., Ashikari, M., Miura, K., Yamashino, T., and Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44, 1229–1236. doi: 10.1093/pcp/pcg135
Murakami, M., Matsushika, A., Ashikari, M., Yamashino, T., and Mizuno, T. (2005). Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci. Biotechnol. Biochem. 69, 410–414. doi: 10.1271/bbb.69.410
Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 110–121. doi: 10.1093/pcp/pcl043
Nakagawa, Y., Hanaoka, H., Kobayashi, M., Miyoshi, K., Miwa, K., and Fujiwara, T. (2007). Cell-type specificity of the expression of Os BOR1, a rice efflux boron transporter gene, is regulated in response to boron availability for efficient boron uptake and xylem loading. Plant Cell 19, 2624–2635. doi: 10.1105/tpc.106.049015
Ngampanya, B., Sobolewska, A., Takeda, T., Toyofuku, K., Narangajavana, J., Ikeda, A., et al. (2003). Characterization of rice functional monosaccharide transporter, OsMST5. Biosci. Biotechnol. Biochem. 67, 556–562. doi: 10.1271/bbb.67.556
Oliver, S. N., Van Dongen, J. T., Alfred, S. C., Mamun, E. A., Zhao, X., Saini, H. S., et al. (2005). Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. PlantCell Environ. 28, 1534–1551.
Pu, C.-X., Han, Y.-F., Zhu, S., Song, F.-Y., Zhao, Y., Wang, C.-Y., et al. (2017). The rice receptor-like kinases dwarf and runtish spikelet1 and 2 repress cell death and affect sugar utilization during reproductive development. Plant Cell 29, 70–89. doi: 10.1105/tpc.16.00218
Raivo Kolde (2019). pheatmap: Pretty Heatmaps. R package Version 1.0.12. Available online at: https://CRAN.R-project.org/package=pheatmap (accessed January 04, 2019).
Rodríguez-Navarro, A., and Rubio, F. (2006). High-affinity potassium and sodium transport systems in plants. J. Exp. Bot. 57, 1149–1160. doi: 10.1093/jxb/erj068
Sakata, T., Oda, S., Tsunaga, Y., Shomura, H., Kawagishi-Kobayashi, M., Aya, K., et al. (2014). Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164, 2011–2019. doi: 10.1104/pp.113.234401
Sato, Y., Takehisa, H., Kamatsuki, K., Minami, H., Namiki, N., Ikawa, H., et al. (2012). RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, D1206–D1213. doi: 10.1093/nar/gks1125
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261. doi: 10.1126/science.1146994
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504.
Shi, M. S. (1981). Breeding and using of a two-usage natural mutant in late japonica rice (in Chinese). Hubei. Agric. Sci. 7, 1–3.
Shi, X., Sun, X., Zhang, Z., Feng, D., Zhang, Q., Han, L., et al. (2015). Glucan synthase-like 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 56, 497–509. doi: 10.1093/pcp/pcu193
Sugiyama, N., Izawa, T., Oikawa, T., and Shimamoto, K. (2001). Light regulation of circadian clock-controlled gene expression in rice. Plant J. 26, 607–615. doi: 10.1046/j.1365-313x.2001.01063.x
Suzuki, K., Yamaji, N., Costa, A., Okuma, E., Kobayashi, N. I., Kashiwagi, T., et al. (2016). OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 16:22. doi: 10.1186/s12870-016-0709-4
Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. U.S.A. 98, 7922–7927. doi: 10.1073/pnas.111136798
Takano, M., Inagaki, N., Xie, X., Yuzurihara, N., Hihara, F., Ishizuka, T., et al. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17, 3311–3325. doi: 10.1105/tpc.105.035899
Tanaka, K., Murata, K., Yamazaki, M., Onosato, K., Miyao, A., and Hirochika, H. (2003). Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133, 73–83. doi: 10.1104/pp.103.022442
Tanaka, N., Uraguchi, S., Saito, A., Kajikawa, M., Kasai, K., Sato, Y., et al. (2013). Roles of pollen-specific boron efflux transporter, OsBOR4, in the rice fertilization process. Plant Cell Physiol. 54, 2011–2019. doi: 10.1093/pcp/pct136
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). Dwarf and low-tillering acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24, 2562–2577. doi: 10.1105/tpc.112.097394
Ueda, K., Yoshimura, F., Miyao, A., Hirochika, H., Nonomura, K., and Wabiko, H. (2013). Collapsed abnormal pollen1 gene encoding the Arabinokinase-like protein is involved in pollen development in rice. Plant Physiol. 162, 858–871. doi: 10.1104/pp.113.216523
Wan, L., Zha, W., Cheng, X., Liu, C., Lv, L., Liu, C., et al. (2011). A rice beta-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233, 309–323. doi: 10.1007/s00425-010-1301-z
Wang, Y., Zha, X., Zhang, S., Qian, X., Dong, X., Sun, F., et al. (2010). Down-regulation of the OsPDCD5 gene induced photoperiod-sensitive male sterility in rice. Plant Sci. 178, 221–228. doi: 10.1016/j.plantsci.2009.12.001
Wang, K., Singh, D., Zeng, Z., Coleman, S. J., Huang, Y., Savich, G. L., et al. (2010). MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 38:e178. doi: 10.1093/nar/gkq622
Wang, Y., Xu, H., Wei, X., Chai, C., Xiao, Y., Zhang, Y., et al. (2007). Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant Mol. Biol. 65, 439–451. doi: 10.1007/s11103-007-9228-x
Wei, S., Hu, W., Deng, X., Zhang, Y., Liu, X., Zhao, X., et al. (2014). A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 14:133. doi: 10.1186/1471-2229-14-133
Wu, L., Guan, Y., Wu, Z., Yang, K., Lv, J., Converse, R., et al. (2014). OsABCG15 encodes a membrane protein that plays an important role in anther cuticle and pollen exine formation in rice. Plant Cell Rep. 33, 1881–1899. doi: 10.1007/s00299-014-1666-8
Xu, F. Q., Li, X. R., and Ruan, Y. L. (2008). RNAi-mediated suppression of hexokinase gene OsHXK10 in rice leads to non-dehiscent anther and reduction of pollen germination. Plant Sci. 175, 674–684. doi: 10.1016/j.plantsci.2008.07.002
Xu, Y., Liu, S., Liu, Y., Ling, S., Chen, C., and Yao, J. (2017). Hothead-like HTH1 is involved in anther cutin biosynthesis and is required for pollen fertility in rice. Plant Cell Physiol. 58, 1238–1248. doi: 10.1093/pcp/pcx063
Yamaji, N., Takemoto, Y., Miyaji, T., Mitani-Ueno, N., Yoshida, K. T., and Ma, J. F. (2017). Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541, 92–95. doi: 10.1038/nature20610
Yao, L., Zhang, Y., Liu, C., Liu, Y., Wang, Y., Liang, D., et al. (2018). OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 4, 530–533. doi: 10.1038/s41477-018-0193-y
Ye, J., Huang, M., Zhao, S., and Wu, N. (2004). The relationship between the expression of osRACD and the fertility changes of photoperiod-sensitivegenic male sterile rice (in Chinese). Prog. Nat. Sci 14, 166–172.
Yokosho, K., Yamaji, N., and Ma, J. F. (2016). OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J. Exp. Bot. 67, 5485–5494. doi: 10.1093/jxb/erw314
Yokosho, K., Yamaji, N., Ueno, D., Mitani, N., and Ma, J. F. (2009). OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149, 297–305. doi: 10.1104/pp.108.128132
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Yu, J., Han, J., Kim, Y. J., Song, M., Yang, Z., He, Y., et al. (2017). Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc. Natl. Acad. Sci.U.S.A. 114, 12327–12332. doi: 10.1073/pnas.1705189114
Zhang, D., Luo, X., and Zhu, L. (2011). Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390. doi: 10.1016/j.jgg.2011.08.001
Zhang, H., Liang, W., Yang, X., Luo, X., Jiang, N., Ma, H., et al. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22, 672–689. doi: 10.1105/tpc.109.073668
Zhang, H., Xu, C., He, Y., Zong, J., Yang, X., Si, H., et al. (2013). Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl. Acad. Sci. U.S.A. 110, 76–81.
Zhang, Q., Shen, B. Z., Dai, X. K., Mei, M. H., Saghai Maroof, M. A., and Li, Z. B. (1994). Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc. Natl. Acad. Sci. U.S.A. 91, 8675–8679. doi: 10.1073/pnas.91.18.8675
Zhao, G., Shi, J., Liang, W., Xue, F., Luo, Q., Zhu, L., et al. (2015). Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 169, 2064–2079. doi: 10.1104/pp.15.00262
Zhao, Z., Zhang, Y., Liu, X., Zhang, X., Liu, S., Yu, X., et al. (2013). A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 27, 113–122. doi: 10.1016/j.devcel.2013.09.005
Zhou, H., Liu, Q., Li, J., Jiang, D., Zhou, L., Wu, P., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22, 649–660. doi: 10.1038/cr.2012.28
Zhou, H., Zhou, M., Yang, Y., Li, J., Zhu, L., Jiang, D., et al. (2014). RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 5:4884. doi: 10.1038/ncomms5884
Zhou, Y. F., Zhang, X. Y., and Xue, Q. Z. (2011). Fine mapping and candidate gene prediction of the photoperiod and thermo-sensitive genic male sterile gene pms1(t) in rice. J. Zhejiang Univ. Sci. B 12, 436–447. doi: 10.1631/jzus.B1000306
Zhu, G., Ye, N., and Zhang, J. (2009). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 50, 644–651. doi: 10.1093/pcp/pcp022
Keywords: transcriptome, rice anther, photoperiod, WGCNA, carbohydrate, transport, phytohormone, PGMS
Citation: Sun SY, Wang DX, Li JB, Lei YQ, Li G, Cai WG, Zhao X, Liang WQ and Zhang DB (2021) Transcriptome Analysis Reveals Photoperiod-Associated Genes Expressed in Rice Anthers. Front. Plant Sci. 12:621561. doi: 10.3389/fpls.2021.621561
Received: 26 October 2020; Accepted: 13 January 2021;
Published: 26 February 2021.
Edited by:
David Smyth, Monash University, AustraliaReviewed by:
Shiyong Song, Zhejiang University, ChinaCopyright © 2021 Sun, Wang, Li, Lei, Li, Cai, Zhao, Liang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dabing Zhang, emhhbmdkYkBzanR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.