- 1Centre of Agricultural Biochemistry and Biotechnology (CABB), University of Agriculture, Faisalabad, Pakistan
- 2UWA School of Agriculture and Environment, The University of Western Australia, Perth, WA, Australia
- 3College of Allied Health Professional, Faculty of Medical Sciences, Government College University Faisalabad, Faisalabad, Pakistan
- 4Mountain Research Center for Field Crops, Khudwani, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, Srinagar, India
Climate change is a threat to global food security due to the reduction of crop productivity around the globe. Food security is a matter of concern for stakeholders and policymakers as the global population is predicted to bypass 10 billion in the coming years. Crop improvement via modern breeding techniques along with efficient agronomic practices innovations in microbiome applications, and exploiting the natural variations in underutilized crops is an excellent way forward to fulfill future food requirements. In this review, we describe the next-generation breeding tools that can be used to increase crop production by developing climate-resilient superior genotypes to cope with the future challenges of global food security. Recent innovations in genomic-assisted breeding (GAB) strategies allow the construction of highly annotated crop pan-genomes to give a snapshot of the full landscape of genetic diversity (GD) and recapture the lost gene repertoire of a species. Pan-genomes provide new platforms to exploit these unique genes or genetic variation for optimizing breeding programs. The advent of next-generation clustered regularly interspaced short palindromic repeat/CRISPR-associated (CRISPR/Cas) systems, such as prime editing, base editing, and de nova domestication, has institutionalized the idea that genome editing is revamped for crop improvement. Also, the availability of versatile Cas orthologs, including Cas9, Cas12, Cas13, and Cas14, improved the editing efficiency. Now, the CRISPR/Cas systems have numerous applications in crop research and successfully edit the major crop to develop resistance against abiotic and biotic stress. By adopting high-throughput phenotyping approaches and big data analytics tools like artificial intelligence (AI) and machine learning (ML), agriculture is heading toward automation or digitalization. The integration of speed breeding with genomic and phenomic tools can allow rapid gene identifications and ultimately accelerate crop improvement programs. In addition, the integration of next-generation multidisciplinary breeding platforms can open exciting avenues to develop climate-ready crops toward global food security.
Global Scenario of Climate Changes and Food Security
Food security is the biggest challenge in feeding the continuous up-surging population. The dream of a world without hunger is only possible if agricultural productivity increases in a sustainable manner (Tilman et al., 2011). About two billion people are facing extreme micronutrient deficiencies, and over 815 million are suffering from chronic hunger. Recent evidence has revealed the increased number of undernourished people (Figure 1A) in the developing countries of Western Asia and Africa since 2014 (FAO, IFAD, UNICEF, WFP, and WHO, 2018). The situation is more deteriorated by the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which severely affects the food supply chain, and its impact on global food security is yet to be understood. It is forecasted that this food crisis will upsurge people with acute hunger from 135 to 265 million by 2020 (Henry, 2020). The availability of food largely relies on economic growth, which is still crucial in dealing with both hidden and chronic hunger and poverty in underdeveloped and developing countries (Gödecke et al., 2018). Still, it might not be adequate to lessen malnutrition and hunger as there are many factors affecting global food security.
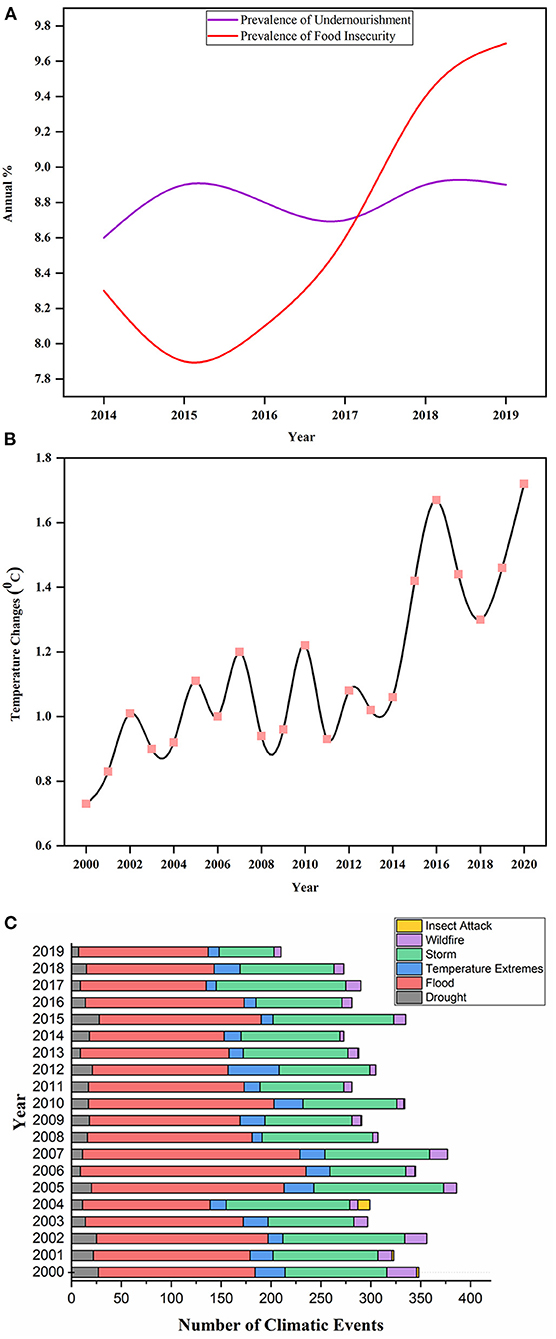
Figure 1. Displaying the annual prevalence of undernourishment and food insecurity percentage during 2014–2019 in (A). Source: Food and Agriculture Organization (FAO) (http://www.fao.org/faostat/en/#data/FS/visualize). Illustration of changing trends in the world's temperature annually from 2000 to 2020 in (B). Source: FAO (http://www.fao.org/home/en/). (C) depicted the total number of climatic events that occurred from 2000 to 2020 around the world. The climatic events include drought, extreme temperature, flood, storm, wildfire, and insect attack. Source: Emergency Disaster Database2.
The two big challenging factors to food security are climate changes and population growth. The world's population is growing rapidly, which increases the food demand and exerts more pressure on agricultural land and other resources (Abberton et al., 2016). It is projected that the population will cross eight billion mark at the end of 2030 and is anticipated to exceed 9.7 billion before the end of 2050 (Ray et al., 2013). Agricultural productivity needs to boost to meet 49% more food requirements by 2050 to avoid extreme hunger fears1. An annual increase of 1.1–1.3% of the major cereal crops from the current pace is required to tackle the hunger and severe food shortage by 2050 (Fischer et al., 2014). The failure to enhance crop yield will badly affect the developing countries and can lead to famines and social discomfort. Also, there are many factors, like economic, agronomic, societal, and climatic factors, which adversely hampered crop productivity. It is a huge task for plant breeders and policymakers to cope with this massive assignment of global food safety and security (Ray et al., 2013).
Climate change is a leading aspect threatening agricultural yield worldwide. However, it is impossible to envisage the exact costs of damages caused by climate changes, in a broader perspective the crop yields will be greatly reduced. Climate change elevates the global temperature (Figure 1B) that changes geographical orders of rainfall and instigates greater concerns to agricultural production. These changes cause global warming, and an increased CO2 level is anticipated to affect the nutritive quality of numerous cultivars while many varieties may become unsafe because of chemical alterations in the cells. The extreme events of climate change increase the loss of agricultural land and accelerate biotic and abiotic stresses (Raza et al., 2019). The future projection and impacts of climate changes are uncertain (Allen et al., 2019), which lead to crop adaptability under diverse range of climate stress, i.e., a tough breeding goal. The dilemma becomes even bigger because the fluctuations in annual rainfall and temperature negatively affect crop growth and encourage the attacks of crop pathogens (Heeb et al., 2019). It was reported that 10–25% yield of major staple crops, including wheat, maize and rice, was reduced due to per degree rise in temperature (Deutsch et al., 2018). This is due to an increase in temperature accelerating the metabolic activities of insects and enhancing insect's food consumption rate. Likewise, the elevated level of CO2 makes soybean more susceptible to insect pathogens (Zavala et al., 2008). The major stress events that occurred due to climate change in the last two decades are illustrated in Figure 1C.
The reported rise in productivity of major staple crops, such as wheat, rice, maize, and soy, sharing two-thirds of total calories consumed by people is very less, although there has been a significant increase in the crop yield in recent years (Figure 2; FAO, 2017; Kim et al., 2019a). Meanwhile, the actual increase in population is much quicker than forecasted before, with the updated figure being over 9.9 billion by 2050 (Change, 2020). These current climate and population growth trends are supposed to further hamper the crop yield, thus extending the gap between food demand and food production. These challenges will greatly decide the future scenario of global food security.
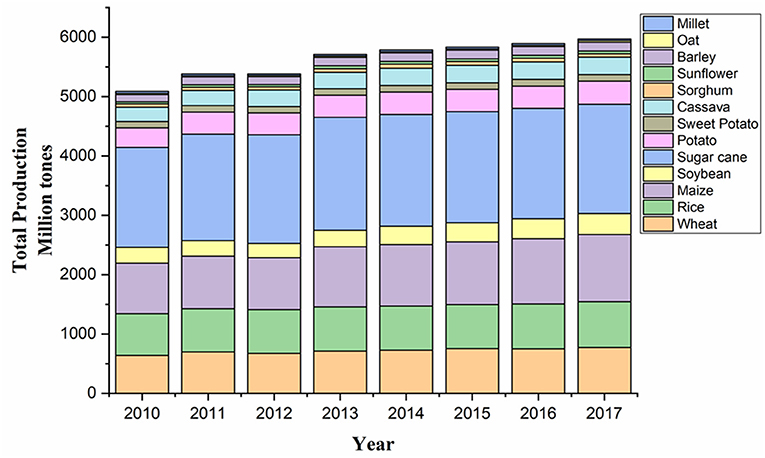
Figure 2. Graphical representation of the total production of major crops in the world (2010–2017). Source: Data retrieved from FAOSTAT1.
A novel agricultural model, including the integrated systems of modern breeding, different agronomic practices, and plant microbiome analysis, is needed. Microbiomes or plant-associated microbes can offer crucial ecosystem facilities, which can boost crop growth and help to mitigate abiotic stresses and pathogen attacks (Arif et al., 2020). Climate-smart agriculture is gaining interest to develop climate-resilient crop varieties by adopting the next-generation breeding approaches that can withstand multidimensional stresses, including salinity, waterlogging, heat, cold, drought, and insect-pests attack.
In the present review, we argue that climate change and the unchecked growing population may reverse the progress achieved at present toward the sustainability goal of zero hunger. The current pace of crop production is insufficient to meet future demands. We highlight significant breakthrough in the plant breeding history. We advocate the next-generation breeding as a reasonably practical way forward to mitigate the impacts of climate changes and develop climate-ready crops for better resilience and improved yield. Indeed, there is a need for a comprehensive strategy, including an integrated multidisciplinary strategy (seed production, pathology, agronomy, post-harvest approaches, agriculture extension, and different breeding practices); still breeding is an obvious point for moving ahead. Hence, we discuss the fascinating technologies of genomic-assisted breeding (GAB) with a focus on advances in crop pan-genome assemblies and their application for crop improvement. We pay specific attention to cutting-edge genome editing tools like clustered regularly interspaced short palindromic repeat/CRISPR-associated (CRISPR/Cas) systems and spotlight their expanding toolbox. Furthermore, we discuss plant phenomics and their major bottlenecks that need to be overcome to bridge the gap between genomics and phenomics and focus on high-throughput phenotyping for crop improvement. We also highlight the next-generation phenomics approaches, including artificial intelligence (AI) and machine learning (ML), that can revolutionize the digital agriculture. Finally, we describe speed breeding and propose the integration of next-generation breeding technologies to expedite the crop production for food security.
Plant Breeding the Savior
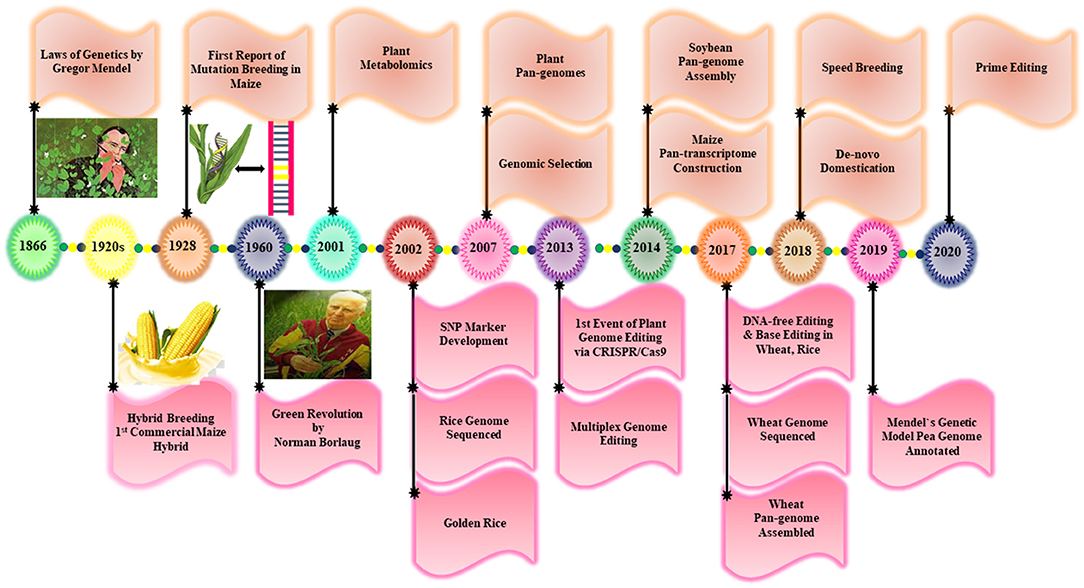
Approximately 10,000 years ago, plant breeding emerged as a central approach for plant domestication by exploiting wild relatives to select the desired traits through a continuous selection process over several generations for crop improvement (Purugganan and Fuller, 2009). Many important crops are cultivated extensively all around the world, which have been developed through a breeding process. Some of the major milestones achieved in the plant breeding history are spotlighted in Figure 3. For example, in the pre-genetic era, many agronomic traits were incorporated blindly into different crop species (Purugganan and Fuller, 2009). The discovery of Mendel laws of inheritance and the continuous detection of genetic elements provide new insight into plant breeding. After 100 years of constant research, the scientists are allowed to identify the genomic regions, which are named later as genes that regulate the agronomic traits in plants (McCouch et al., 2013). In the 1960's, the Green Revolution remarkably increased the yield potential of some major crops, including rice and wheat, to meet the growing food demands (Pingali, 2012). Although it has brought an enormous benefit for agriculture and humanity but also a lot of negative environmental consequences because of the unchecked application of synthetic fertilizers and pesticides. Additionally, the Green Revolution encouraged intensive breeding that resulted in the reduction of genetic diversity (GD) and the loss of several unique genes. This led to an increased attack of various insect-pests pathogens and the reduction of plant vigor to withstand extreme heat, drought salinity, and flooding conditions (Tilman et al., 2002). However, plant breeding has been under immense pressure after this period to maintain constant agricultural yield with limited resources of water, land, and fertilizers. To tackle these problems, plant scientists need to elucidate the unique genetic resources to produce superior cultivars having better stress resilience and increased grain yield.
Generally, in classical breeding, elite crop varieties have been selected via hybridization and a continuous screening process (Purugganan and Fuller, 2009). The fundamental features of basic plant breeding lead to the incorporation of GD through intercrossing of the plants with novel agronomic features with wild relatives or crop landraces and select the best genotypes having outstanding characters (Lavarenne et al., 2018). Investigations of wild populations can offer greater GD to introduce the desired traits to develop new crop varieties, which provide an excellent roadmap for crop improvement. For example, the introduction of diverse genetic recombination through hybridization among species may offer an exceptional chance to combat climate stresses (Becker et al., 2013). However, conventional breeding strategies for crop improvement are of limited use because of genetic drag, genetic erosion, hybridization bottlenecks, and laborious selection process (Abberton et al., 2016). It could take 10–20 years to develop a crop variety with desired traits, which make it a complicated and time-consuming endeavor (Fischer et al., 2014). On the other hand, modern breeding approaches like genetic engineering to produce genetically modified (GM) crops gained a significant progress in the last three decades to overcome the hurdles posed by conventional breeding techniques. For example, in 1994, “Flavr Savr” transgenic tomato having an improved shelf life has been approved by Food and Drug Administration (FDA) in the USA. After that, a series of GM crops like glyphosate-resistant soybeans, Bt cotton, Bt maize, and Bt Potato, etc., have been approved for commercial use in the USA (James, 1997). A total of 529 transgenic events reported in 33 crops have been commercialized (ISAAA database, 2021). About 91% of the total area coverage under GM crops are offered by USA, Canada, Brazil, Argentina, and India. USA is the largest grower of GM crops in the world and cultivates different improved varieties of maize (43), soybean (25), potato (43) cotton (32), and canola (22). Asian countries, such as China (canola, cotton, maize, and soybean), India (soybean and cotton), Pakistan (maize and cotton), and Bangladesh (Bt Brinjal), grow only a considerable number of GM crops on a limited area (ISAAA database, 2021). In spite of all these achievements, there are numerous serious concerns associated with GM crops, such as biosafety for human consumption, resistance breakdown in several pathogens, hazardous impacts on nontarget organisms, and the cost of commercialization. Also, all the European countries banned all kinds of GM organisms in their country, which make this a controversial technique for crop improvement. Owing to public safety concerns and unwillingness to accept GM crops, we need alternative next-generation breeding technologies to increase the genetic gains in plant breeding and develop intelligent models to bridge the gap between genotype and phenotype for the next-generation crop improvement.
Fast-Forward Gab
Modern plant breeding approaches have revolutionized plant breeding and are emerged as a powerful alternative to conventional breeding. Plant genomics is extremely vital to accelerate breeding programs and crucial to improve crop performance, including trait identification and the discovery of genetic variations within the crop genome, that regulate crop performance and increase stress resilience (Bevan et al., 2017). Plant genomics is an important player of omics science that deals with the whole plant genomes for determining the structure and assessing the function of genomes. It is important to draw the information inside the genome by defining a specific arrangement of DNA sequences, which help to probe the genomic evolution and interpret molecular phylogenetic relationships (Varshney, 2016). It also assists in elucidating the function of genes and their interaction that govern the plant growth and functions under diverse agroclimatic conditions (Unamba et al., 2015).
Fast-forward genotyping and phenotyping platforms have facilitated multitrait association studies via genome-wide association studies (GWAS) to accurately explore the genetic make-up of crop traits. Quantitative trait loci (QTL) analysis through mapping agronomically important traits enabled to dissect the relationship between genotype and phenotype. Gene cloning and characterization, haplotype-based breeding, allele mining for stress tolerance, and tapping natural variations have open new avenues toward GAB (Leng et al., 2017). In the coming year, large data sets of genomics can offer wealth of information about the plant genomes, and GAB would become an instrumental practice to accelerate breeding to design future crops (Varshney et al., 2021).
Exclusive understanding of plant genomes is essential for next-generation sequencing- (NGS-) based trait mapping that has allowed rapid gene identification. Current advances in genome sequencing techniques have transformed plant breeding by providing access to huge plant genomes and open a new era of genomics (Bassi et al., 2016). The advanced NGS techniques enable to explore the diverse and multidimensional spectrum of genetic variations that can be linked to elucidate complex phenotypes (Unamba et al., 2015).
There are numerous genomic markers that can be utilized for the identification of plant genes through positional cloning (Bassi et al., 2016). For example, single-nucleotide polymorphism (SNP) cost-efficient, accurate, and ubiquitous in crop genomes and has been used on a large scale to screen thousands of crop germplasm (Voss-Fels and Snowdon, 2016). Although SNPs are the chief molecular markers to study the phenotypic and GD among different crop breeds, but they can also be very crucial for probing a wide range of variations linked with some important stress tolerance and agronomic characters (Saxena et al., 2014). Sequencing techniques can capture SNP accurately but are unable to generate long reads sufficient to represent the diversity. To overcome these issues, optical mapping can be executed for generating long read maps having a greater variation and complex regions (Golicz et al., 2016a). Crops like wheat and maize have larger genomes, genotyping by sequencing (GBS) coupled with NGS have been widely used for multiplexed sample sequencing (Arthur and Bennetzen, 2018). Fixed SNP genotyping arrays can be used instead of NGS-based techniques because they are economical and precise, and require less data analysis. Recently, some genotyping platforms and crop breeding chips have been complied, which could be used in GAB (Rasheed et al., 2017).
A comprehensive knowledge of genes and their regulating pathways that control the dynamic traits, such as quality, yield, and stress tolerance, would increase our knowledge to develop next-generation crop varieties. However, the less availability of genomic information and lack of significant understanding are not completely accredited to the inadequate genomic innovations but also depend on the environment–genotype interactions and phenotypic pitfalls (van Bezouw et al., 2019). In the last two decades, a significant development has been achieved in the genomic technologies that accelerate breeding programs (Varshney et al., 2017). Advanced multidisciplinary breeding platforms are required to mitigate the climate changes and accelerate the genetic gains for achieving the target demands of crop yield (Bevan et al., 2017). Upon the integration of fast-forward genomic technologies, such as genomic selection (GS), NGS, SNP-mediated marker-assisted selection, and GWAS, together with holistic phenotyping and more sophisticated bioinformatics, data analysis, and decision support tools (Varshney et al., 2018) can drive the next-generation breeding.
The Pan-Genomes and In-Depth Exploration of Natural Variation
Crop domestication and natural evolution have severe impacts on crop genomes resulting in the impairment of major loci regulating important agronomic traits and ultimately reduce GD (Warschefsky et al., 2014). Also, the selection of superior lines for abiotic/biotic stress tolerance to improve crop yield has worsened the condition, directing to the elimination of novel stress-tolerant genes that were abundantly present in the gene pool of crop wild relatives (CWRs; Brozynska et al., 2016). Due to the reduction in GD, the crops are now more vulnerable to adverse climatic stresses, and there is an urgency to transfer all the intentions toward CWRs that contain a rich-gene pool of stress-responsive genes (McCouch et al., 2013).
Although the principal focus on studying GD is carried out by detecting the structural variations (SVs) within the genome by employing SNPs, the SVs has been considered as a principal source of GD and may include presence/absence variants (PAVs), copy number variants (CNVs), and some other diverse variations such as chromosomal translocation, inversion, and transversions (Wang et al., 2015). CNVs occur in the distinctive form of copies among individuals while PAVs occur in one genome and are absent in another (Saxena et al., 2014). PAVs and CNVs are commonly present within species in the plant genomes and play a key part to understand plant genetics (Hirsch et al., 2014). A few studies showed that the genetic variations that are mainly studied via SNPs/InDels are insufficient to depict the whole genomic pool of a species (Saxena et al., 2014). Furthermore, the re-sequencing technologies are centered only on a single-reference genome that has inadequate capability to detect the entire spectrum of large SVs, like CNVs and PAVs, and thus providing insufficient information for crop GD (Tao et al., 2019).
Hence, there is a need to develop multireference genomes to investigate the genome composition of all individuals, including cultivated, landraces, or wild progenitors. With the advancement in NGS technologies, a huge wave of pan-genomic studies have been launched, which open a new roadmap to determine crop evolution and adaptation across the genus, and study deep insights into genome functions with the possible application for crop breeding (Hirsch et al., 2014; Zhao et al., 2018a). Pan-genomics provides an excellent platform to study the GD, compares multiple genomes simultaneously, and recaptures the whole genetic repertoire of a species (Zhao et al., 2018a).
Crop Pan-Genomes
The term pan-genome was firstly opted to describe the bacterial genome, and it depicts the whole gene stock of different individuals present in a species. Pan-genomes are comprised of two regions, including the core genome that represents the core genes present in all individuals and dispensable genome consisting of accessory or variable genes, particularly shared by few group members and missing in remaining individuals (Tettelin et al., 2005). The core genes tend to be conserved and perform important functions while the dispensable genome provides a greater GD and is supposed to carry the stress-responsive genes in a species. In addition, the dispensable genome is a key component to regulate the phenotypic variation related to agronomic traits, and is crucial for increasing agricultural productivity. It is also a major contributor to the domestication and adaptive evolution of species (Tranchant-Dubreuil et al., 2018; Jayakodi et al., 2021). In recent years, pan-genomes concepts have moved toward crop plant research, and pan-genomes of some major crops have been constructed (Hurgobin et al., 2018).
The resources of crop pan-genomes rather than single-reference genomes will accelerate molecular breeding and provide a multidimensional understanding of genomic variations (Golicz et al., 2016a,b). SVs and PAVs are vital to understanding the variable genes, which have been revealed to control the genes related to stress, like biotic stress, tolerance in soybean (McHale et al., 2012), Brassica oleracea (Bayer et al., 2019), muskmelon (González et al., 2013), and phosphor uptake capacity of rice (Schatz et al., 2014). In general, pan-genomes explore new trends to study GD, which will play a key part in probing genomic variations and offer a speedy approach to scanning complex gene sets for crop improvement (Tao et al., 2019).
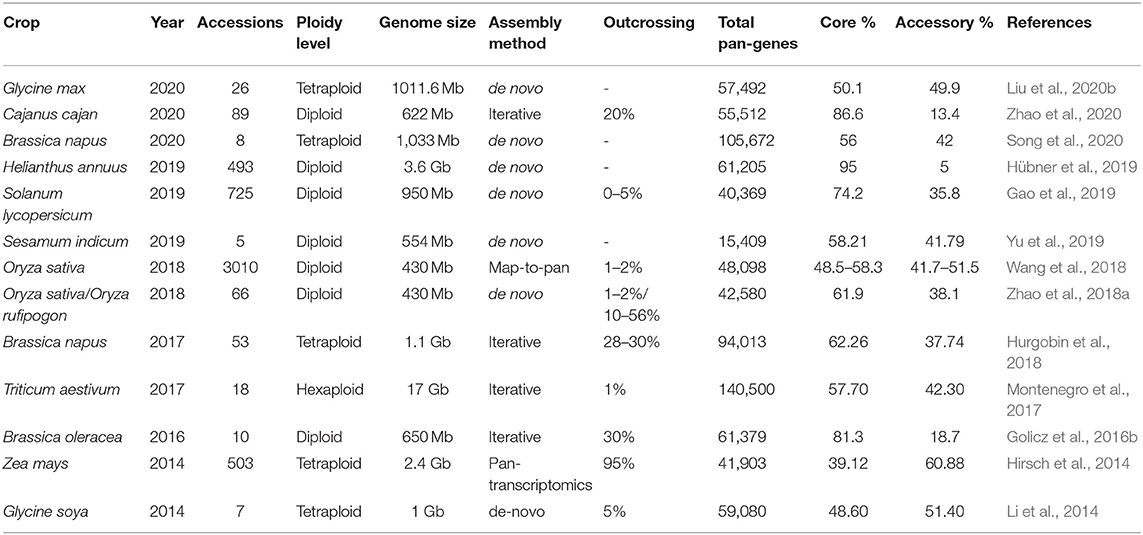
Pan-genomes of rice (Schatz et al., 2014; Zhao et al., 2018a), tomato (Gao et al., 2019), soybean (Li et al., 2014; Liu et al., 2020b), sunflower (Hübner et al., 2019), maize (Hirsch et al., 2014), Brassica rapa (Lin et al., 2014), Brassica napus (Hurgobin et al., 2018; Song et al., 2020), B. oleracea (Golicz et al., 2016b), Sesamum indicum (Yu et al., 2019), wheat (Montenegro et al., 2017), Brachypodium distachyon (Gordon et al., 2017), and Cajanus cajan (Zhao et al., 2020) have been mapped to open new horizons for the crop improvement studies. A detailed summary of the currently reported pan-genomes is summarized in Table 1.
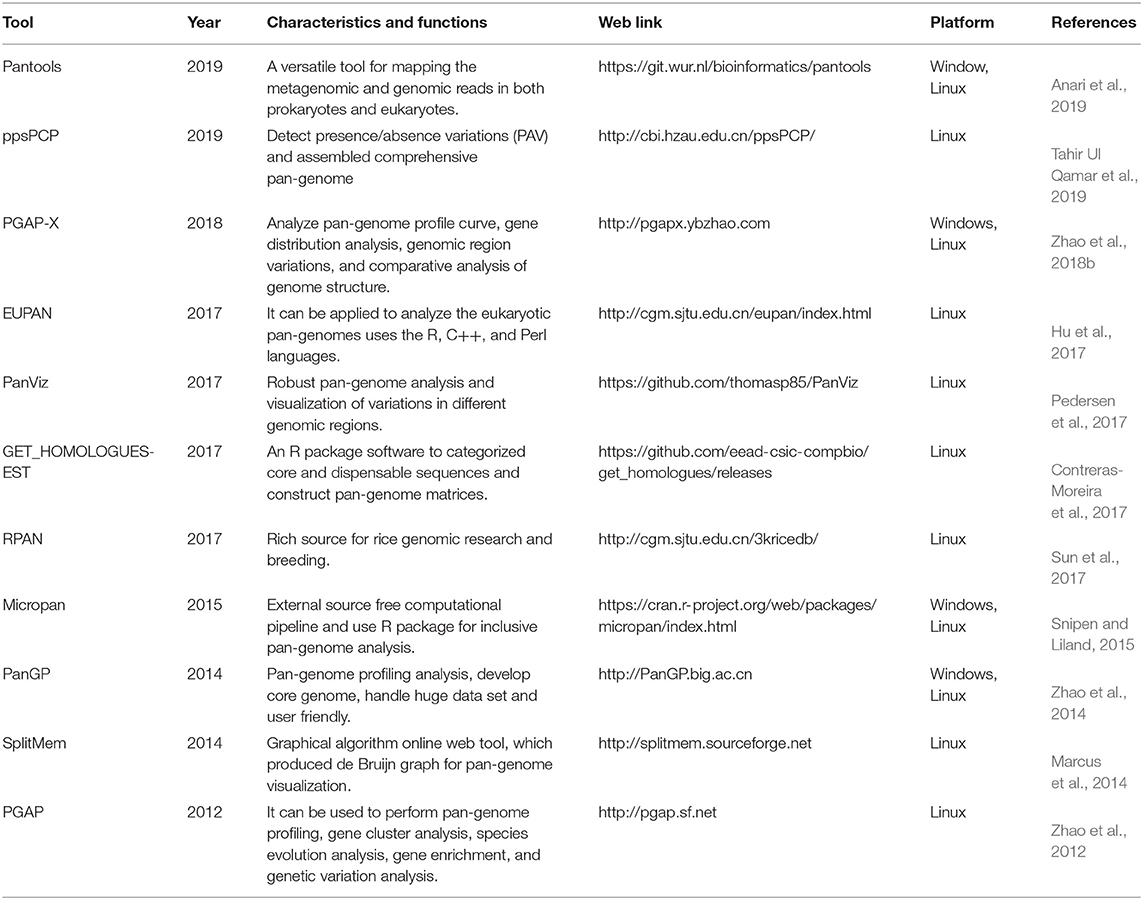
In recent years, several visualization platforms have been designed for crop pan-genomes analyses like GET_HOMO LOGUES (Contreras-Moreira et al., 2017), PanViz (Pedersen et al., 2017), SplitMem (Marcus et al., 2014), RPAN (Sun et al., 2017), Pantools (Anari et al., 2019), ppsPCP (Tahir Ul Qamar et al., 2019), seq-seq-pan, ITEP (Jandrasits et al., 2018), EUPAN (Hu et al., 2017), PGAP-X (Zhao et al., 2018b), PanGP (Zhao et al., 2014), Micropan (Snipen and Liland, 2015), and PGAP (Zhao et al., 2012). These visualization tools permit analyzing the pan-genomes for retrieving the genes in databank, enabling access to PAV, gene annotation, gene expression, and genomic sequence information (Danilevicz et al., 2020). The design of advanced visualization tools is needed to allow a robust integrated examination of pan-genomes for future applications in crop improvement. A more detailed description of these analytical platforms is summarized in Table 2.
Pan-Genomes and Crop Improvement
The crop pan-genomes studies enable us to catch up with the genes that are lost in reference genomes during crop domestication. Availability of the crop pan-genome that comprises all its CWRs, landraces, and cultivated varieties gives a well-defined system to gather all the information about genotypic and phenotypic variations and permit the identification of missing genes within the reference genomes (Danilevicz et al., 2020). Superior knowledge about accessory genome would help to screen the elite cultivars for abiotic and biotic stresses harboring stress-responsive genes (Bayer et al., 2019). Pan-genomes offer a great opportunity to understand the role of GD in genomic-based crop improvement. Fully annotated hexaploid wheat pangenome of 18 elite lines showed 140,500 ± 102 genes and 36 million intervariatel SNPs. The functional analysis of dispensable genes revealed their association with important agronomic traits (Montenegro et al., 2017). Similarly, several SNPs controlling the nine agronomic traits and variable genes for disease resistance have been identified in the pigeon pea pangenome that can be very useful for crop improvement (Zhao et al., 2020).
For example, SVs of 25 maize inbred lines have been studied and found a similar order among heterotic set and PAVs showing that SVs can be crucial for the identification of parentage for hybrid development (Darracq et al., 2018). The yield-related traits have been subjected to a rigorous selection during breeding, and the desired alleles for improved productivity have been identified in major crops like soybean (Concibido et al., 2003), rice (Thalapati et al., 2012), and wheat (Huang et al., 2003). Díaz et al. (2012) studied the genes controlling the flowering time in wheat and discovered that the genes show CNV. Similarly, FLC gene family regulates flowering in B. oleracea having variations in the CNV and found four core genes and two variable genes (Golicz et al., 2016b). The Sub1A gene regulating the submergence tolerance has been identified in rice under submergence (Schatz et al., 2014).
Several biotic stress-responsive genes have revealed the PAVs in a large number of species (Cook et al., 2012). Recently, Dolatabadian et al. (2020) analyzed the pan-genome of 50 B. napus genotypes to characterize the disease-resistant genes. The pan-genome analysis unveiled 1,749 resistance genes, from which 753 are dispensable and 996 are core gene while 368 genes are not detected in the reference genome. Furthermore, SNPs studies revealed 15,318 unique hotspots within 1,030 resistance gene orthologs and identified 106 putative QTLs related to blackleg resistance (Dolatabadian et al., 2020). Sunflower pangenome identified 61,205 genes across a diverse range of wild and cultivated species. Functional annotation of biotic stress-related genes showed an allelic variation for disease resistance including downy mildew (Hübner et al., 2019). The resistance gene orthologs vary among different cultivars, and capture 59 candidate genes that are linked with the Fusarium wilt, clubroot, and Sclerotinia-resistant QTLs in B. oleracea pan-genome (Bayer et al., 2019). The progressive information gathered from these studies can be used to develop improved crop cultivars. The workflow of pan-genome assembly and its exploitation for crop improvement is illustrated in Figure 4.
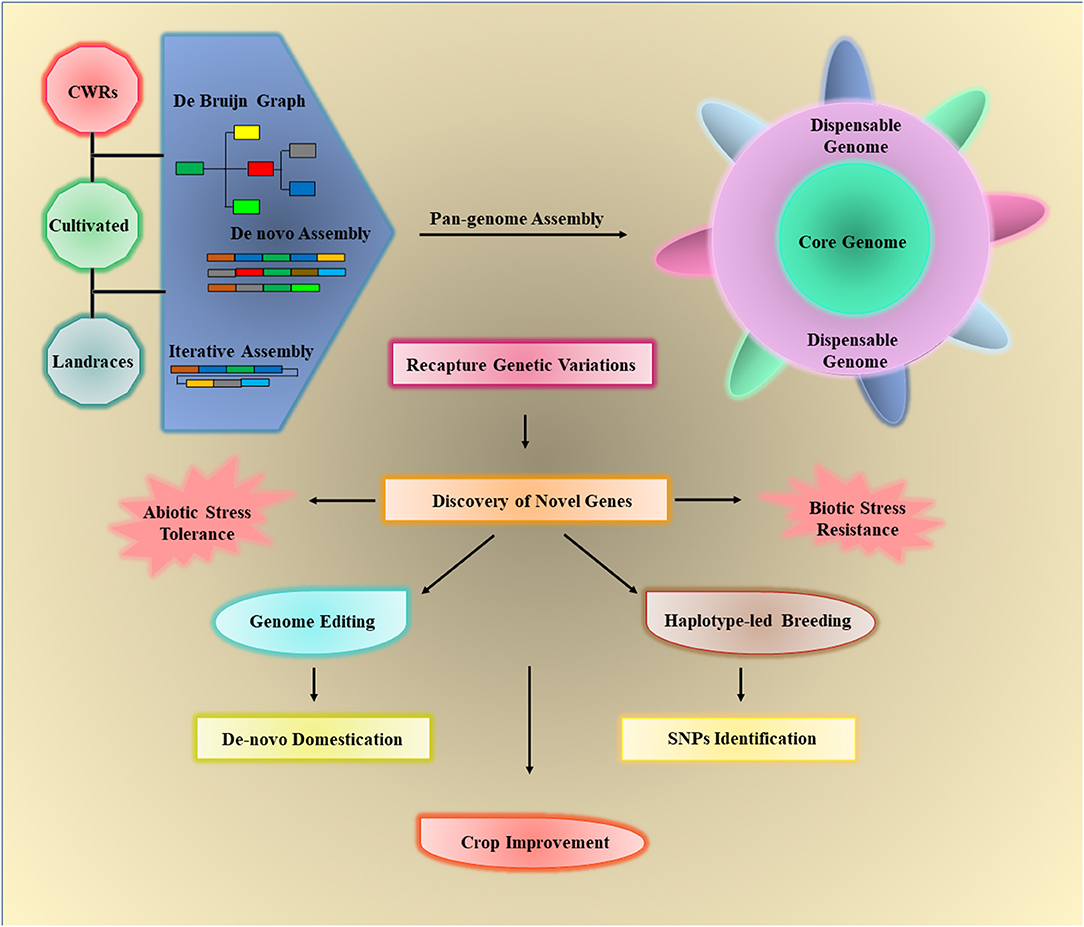
Figure 4. The crop wild relatives (CWRs), landraces, and cultivated varieties of crops can be used to assemble the crop pan-genomes via three approaches such as de novo assembly, de Bruijn graph, and iterative assembly. The core genome includes all the genes of individuals while the dispensable or assessor genome contains all remaining genes, which are not necessary to present in all individuals. Pan-genomes can be used to identify different structural variations (SVs) in any individual and detect novel genes that are lost in cultivated varieties during the breeding process. The elucidation of desired traits/genes can be used for crop improvement by providing biotic/abiotic stress tolerance through haplotype-based breeding and de novo domestication.
A Breakthrough Is Still Needed
Numerous pitfalls remain to be dealt with in achieving economical, robust, and perfectly annotated pan-genome assembly. First, the main prerequisite for pan-genomic analysis is the accessibility to a well-annotated reference genome. However, the major drawback in transferring the adjoining sequences is the short genomic reads that cause hindrance in repetitive region assembly. Novel techniques like single-molecule sequencing can offer longer reads but have low accuracy. With the advent of highly sophisticated algorithms tools, the assembly of long reads has become easy and robust, which result in the assembly of high-quality genomes. The incorporation of long reads can help in resolving the highly complex polypoid genomes and repetitive motifs that are quite challenging to read short sequences, and this strategy can be useful for the assembly of pan-genomes. Another crucial aspect for the pan-genome analysis is the useful association among phenotypes and functional data sets. Only accurate functional data will create a linkage map between trait diversity and pan-genome.
Second, the size and complexity of several genomes pose a big hurdle for developing the visualization tools for pan-genomic interpretations. To tackle this, scientists have come up with cloud-based solutions to assist the availability of pan-genome visualization tools. The cloud-based system could harness the multi-omics approaches and established a huge platform for data sharing, pan-genomic analysis, and developing the standardized protocols for the pan-genome studies. Third, the presentation and storage of large data sets of pan-genome results are also challenging tasks. Databases can store a large number of pan-genomic data, including transposable elements, noncoding RNAs, indels, and SNPs. Developing a coordinate system like SuperGenome requires to be upgraded to address the challenges related to presentation and storage. Fourth, the information collected from the pan-genome needs GWAS/QTL analysis and re-sequencing to unravel the candidate genes for crop improvement. Thus, overcoming these hurdles to develop inclusive genomic resources would beneficial to expand the pan-genomic assemblies to other crop species. Furthermore, the applications of ML and deep learning (DL) in pan-genomic research may facilitate to cross the barriers of pan-genome construction and visualization as ML tools can autonomously detect the sequence reads in huge data sets. Also, a complete information flow reflecting gene-transcriptome-metabolome-epigenome-phenome can provide an excellent platform to study a phenotypic variation within different microenvironments. As the omics tools continue to attain more precision and sensitivity for analytical investigation, the era of crop “pan-omics” is not far away. The upcoming future is expecting to see the data science explosion, where “Crop Pan-omes” studies will be one of the key players in giving a new direction to future agriculture.
The Era of Plant Genome Editing
To date, classical breeding is speedier in comparison to 50 years ago but still inadequate to accelerate the agricultural production with respect to the global demands (Breseghello and Coelho, 2013; Voss-Fels et al., 2019). In addition, mutation breeding and transgenic technology can be applied to introduce novel genes for crop improvement, but GMOs are banned in many countries because of public health safety and regulatory concerns. It normally takes 10–12 years to design a crop cultivar through conventional, mutational, and transgenic breeding (Razzaq et al., 2019b).
The rise of first-generation genome engineering nulceases, like transcriptional activator-like effector nucleases (TALENs) and zinc finger nucleases (ZFNs), brought a revolution in plant research and accelerated the plant research (Lloyd et al., 2005; Cermak et al., 2011). Genome editing has been applied to generate insertion/deletion (indels), substitution, replacement, and overcome all the concerns of nonspecific, cross species, and unstable integration of foreign DNA into the host cell (Kim and Kim, 2014). Although, ZFNs and TALENs have been extensively applied for site-specific plant genome editing in the past but certain drawbacks, like cumbersome cloning and vector construction protocols, large size, ineffective delivery methods, repetitive nature, less specificity, and large off-target, limit their use today (Puchta, 2017).
Broadening the CRISPR/Cas Toolbox
Clustered regularly interspaced short palindromic repeat/CRISPR-associated system is the most fascinating and ground-breaking technology for genome editing (Jinek et al., 2012). Continuous efforts are in progress to minimize the drawbacks of the CRISPR/Cas systems in plants and to develop next-generation genome editing tools.
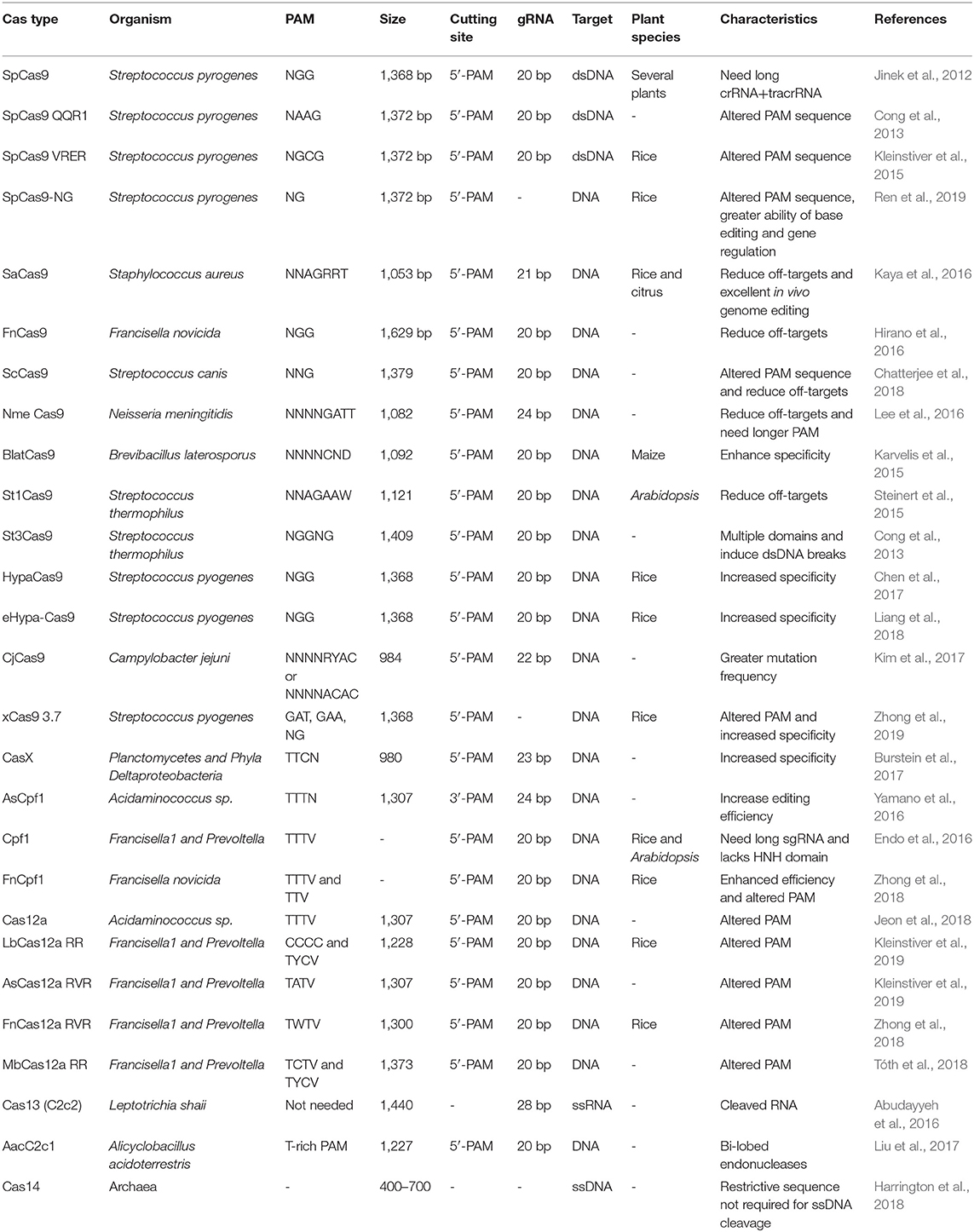
The present classification of a CRISPR toolkit is incomplete as new classes of variants are discovered continuously (Koonin et al., 2017). A CRISPR/Cas system is still evolving and has yet to fulfill its potential. Currently, there are two major classes (classes 1 and 2), six types, and more than 30 subtypes of the CRISPR system according to their respective signature protein. The class 1 includes multiple Cas effector proteins to perform many tasks and comprises type I, III, and IV with the corresponding proteins like Cas3, Cas10, and Csf1 (Makarova et al., 2015). Whereas the class 2 system with only single signature protein is the most extensively adopted genome editing system and consists of type II (Cas9), V [Cas12a (Cpf1), Cas12b, Cas12c, Cas12d (CasY), Cas12e (CasX), Cas12g, Cas12h, Cas12i, Cas14a, Cas14b, Cas14c], and VI [Cas13a (C2c2), Cas13b (C2c6), Cas13c-d] systems (Koonin et al., 2017). Several Cas orthologs as depicted in Table 3 have been discovered to overcome the bottlenecks in the CRISPR/Cas system.
Naturally, Cas9 is found in Streptococcus pyogenes (SpCas9) and contains three subunits: a Cas9 protein, CRISPR RNA (crRNA), and trans-activating crRNA (tracrRNA) (Jinek et al., 2012; Mali et al., 2013). Cas9 comprises two lobe-like structures: a recognition domain (REC) linked with a nuclear domain (NUC). The NUC lobe consists of two catalytic sites like HNH and RuvC that target the protospacer adjacent motif (PAM) present at 3 bp upstream of the desired DNA region. The mechanism of CRISPR/Cas9 editing initiates by designing 20-bp guide RNA (gRNA) to form gRNA/Cas9 assembly and recognize PAM site to produce double-standard breaks (DSBs) at a specific site (Cong et al., 2013). CRISPR/Cas12a is considered as another important Cas ortholog, which is also called as Cpf1. It is an RNA-based editing tool and presents some inimitable characters in contrast to a CRISPR/Cas9 system. Cpf1 needs a T-rich spacer region at 5'-end having 42-nt crRNA and create DSBs with staggered ends (Zetsche et al., 2015). For example, Cpf1-based genome editing was performed in allotetraploid cotton, and the results indicated zero off-target cleavage with 87% editing efficiency (Li et al., 2019a). CRISPR/Cas9 and CRISPR/Lsh Cas13a have been applied against RNA potyvirus to make disease-resistant plants and can be applied against many other invading viruses (Aman et al., 2018). CRISPR/Cas14a is a highly ideal tool to target the plant single-standard DNA viruses like Nanoviridae and Geminiviridae families. It has the ability to develop viral-resistant crops by allowing unrestricted cleavage without any dependency on the sequence (Khan et al., 2019).
Engineering Crops for Improved Stress Resilience
A tremendous progress in plant genome engineering has been achieved by exploiting the CRISPR/Cas system for crop improvement (Li et al., 2013; Shan et al., 2013). CRISPR/Cas technology is revolutionizing the plant breeding due to its immense application to develop climate-resilient crops (Puchta, 2017). There are numerous studies reported for improved agronomic traits and stress tolerance to abiotic/biotic stresses (Table 4). For example, drought is the most damageable abiotic stress causing severe loss to crop production. A CRISPR/Cas9 system was used to produce the knockout mutants of SlLBD40 in tomato subjected to drought stress. The mutant lines showed improved water-holding capacity as compared to the SlLBD40 overexpressing line. The results demonstrated that the SlLBD40 gene negatively regulates drought stress in tomato (Liu et al., 2020a). CRISPR/Cas9 was employed to study the effect of SlNPR1 mutants under drought stress and revealed that the knockout tomato lines were highly susceptible to drought stress (Li et al., 2019b). Ogata et al. (2020) applied CRISPR/Cas9-mediated frame shift mutations to develop the rice mutant lines for the OsERA1 gene under drought stress. The results showed an increase in tolerance to drought and positively induce primary root development under normal conditions. Ramírez Gonzales et al. (2021) provided the evidence about the StCDF1–StFLORE locus that regulates the water homeostasis and vegetative growth in potato by disrupting the StFLORE using a CRISPR/Cas9 system. Loss of function of this gene resulted in enhanced drought tolerance. Pan et al. (2020) reported that the gene ZmSRL5 is very vital for a cuticular wax structure, which protects the maize plant from different stresses. Loss of functional mutant progenies of maize showed that the ZmSRL5 gene involved in drought response by keeping the structure of cuticle wax intact.
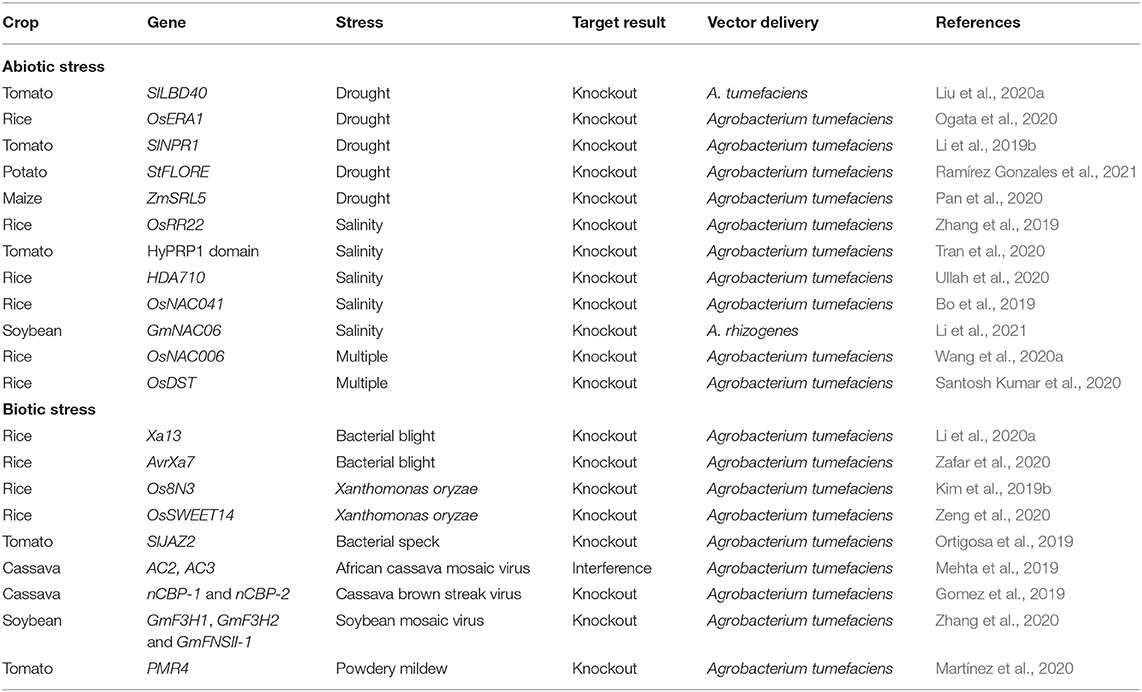
Table 4. Applications of clustered regularly interspaced short palindromic repeat/CRISPR-associated (CRISPR/Cas) system to engineered abiotic/biotic stress tolerance.
Salinity tolerance in important crop species can also be attained by exploiting CRISPR/Cas9-mediated genome editing. CRISPR/Cas9 technology was executed to generate OsRR22 mutant lines of rice, which exhibit enhanced salt tolerance at the seedling stage (Zhang et al., 2019). Regulatory domain editing of multidomain proteins is another important aspect that can be used to engineer the negatively regulated domains associated with abiotic stresses. Tomato hybrid proline-rich protein 1 (HyPRP1) domain was targeted successfully by using multiplex genome editing via CRISPR/Cas9, which is a negative regulator for salinity stress. The results revealed that the elimination of this domain produces tomato with high salinity tolerance (Tran et al., 2020). Ullah et al. (2020) disrupted the HDA710 gene through CRISPR/Cas9, and the mutant lines displayed reduced abscisic acid sensitivity and increased salinity tolerance in rice. A large number of transcription factors are involved in the salinity tolerance mechanism including NAC transcription factors. A CRISPR/Cas9 system was used to develop the knockout mutants of the OsNAC041 gene in rice. The study indicated a direct function of the OsNAC041 gene under salt stress and can offer an excellent potential to target other NAC genes for rice-resistant breeding (Bo et al., 2019). Similarly, another NAC gene GmNAC06 was targeted by using CRISPR/Cas9-based gene editing and overexpression technology in soybean, and the results demonstrated that the GmNAC06 gene improved the salinity tolerance by alleviating ROS effects, accumulating glycine betaine and proline, and maintaining ionic homeostasis (Li et al., 2021).
Wang et al. (2020a) executed CRISPR/Cas9-mediated genome editing to mutate the OsNAC006 gene in rice and the mutant lines showing a high susceptibility to heat and drought stress. Likewise, CRISPR/Cas9 was applied to develop the mutant alleles for the OsDST gene, and the result exhibited a reduction in stomatal density while increasing tolerance against drought, salinity, and osmotic stress without damaging the rice grain yield (Santosh Kumar et al., 2020).
Clustered regularly interspaced short palindromic repeat/CRISPR-associated gene editing also provide a great opportunity to combat against plant pathogens and invading organisms efficiently. Such a resistance to bacterial blight disease was improved in rice by editing the promoter region of Xa13 gene. The knockdown progenies were transgene-free edited plants and showed an increase in resistance to bacterial blight (Li et al., 2020a). In a similar report, the AvrXa7 gene was the target for bacterial blight resistance in basmati rice (Zafar et al., 2020). A CRISPR/Cas9 system was developed by two independent groups to manipulate the Os8N3 and OsSWEET14 gene in rice. The results showed that the mutations were transferred in successive progenies, and homozygous knockouts demonstrated an increased resistance to Xanthomonas oryzae pv. oryzae without any yield in plenty (Kim et al., 2019b; Zeng et al., 2020). Ortigosa et al. (2019) designed a CRISPR/Cas9 system to develop bacterial speck-resistant tomatoes. In this experiment, SlJAZ2 functional ortholog, which was found in stomata, was edited, which provide resistance to bacterial speck-causing agent Pseudomonas syringae pv.
Clustered regularly interspaced short palindromic repeat/CRISPR-associated genome editing can also be applied to cope with the different plant viruses. Mehta et al. (2019) developed a CRISPR/Cas9 interference system to target AC2 and AC3 genes and to have an engineered resistance against African cassava mosaic virus. This system is very useful to cope with the Geminiviruses attack in plants. The resistance against soyabean mosaic virus has been achieved by targeting the genes involved in metabolic pathways of isoflavone, including GmF3H1, GmF3H2, and GmFNSII-1 using the multiplex CRISPR/Cas9 system (Zhang et al., 2020).
Martínez et al. (2020) examined the role of the tomato PMR4 gene against powdery mildew by mutating it via a CRISPR/Cas9 tool. The mutant lines exhibited a higher susceptibility to fungal infection as compared to the normal plants. This evidence can be useful to characterize and analyze S-genes under different fungal pathogens. CRISPR/Cas9 was used to produce the knockouts for nCBP-1 and nCBP-2 genes to study the resistant mechanism against cassava brown streak virus. The knockouts lines displayed little symptoms and reduced disease severity in contrast to control lines (Gomez et al., 2019).
Recent Innovations in CRISPR/Cas System
Without any doubt, the next-generation CRISPR/Cas technology continuously opens up new avenues in plant breeding research. Some breakthrough strategies were reported for the CRISPR/Cas system that removes certain constraints prevailing in genome editing as shown in Figure 5. Recent advancement in the CRISPR/Cas toolbox results in the improvement of several novel features, like target specificity, broader target range, minimizing off-targets, precise nuclease activity, various PAM sites, and efficient delivery methods (Koonin et al., 2017). It is the preferred editing system for carrying out other genetic modifications such as probing mutation patterns (Jia et al., 2018), the introduction of exogenous genes (Collonnier et al., 2017), gene regulation (Qi et al., 2013), and cell imaging (Xue and Acar, 2018).
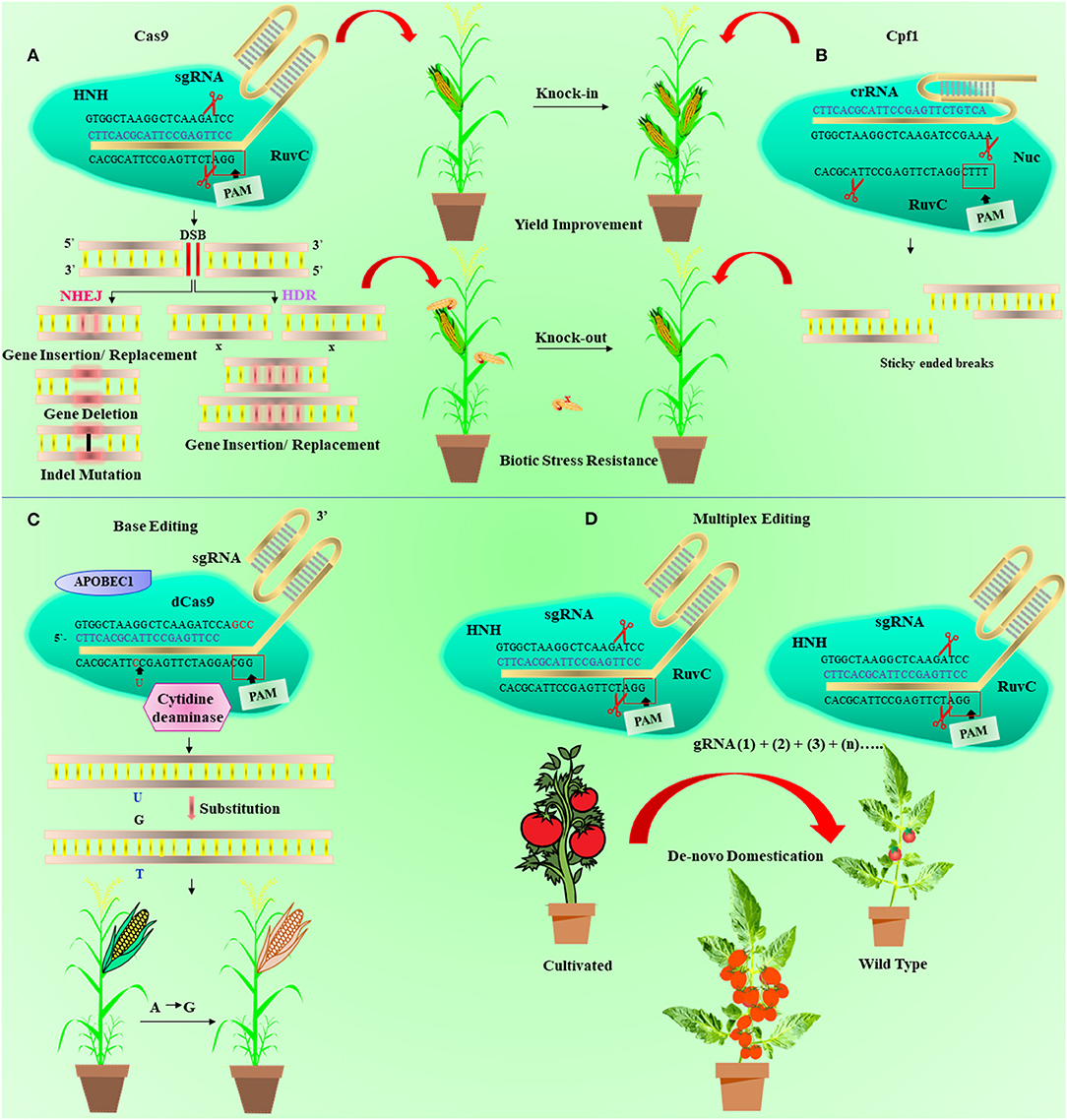
Figure 5. Diagrammatic illustration of the base editing, clustered regularly interspaced short palindromic repeat/CRISPR-associated 9 (CRISPR/Cas9) and Cpf1 mechanism, and de novo domestication. (A) In the CRISPR/Cas9 mechanism, Cas9 protein is guided and activated with the help of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA), respectively, to produce double-standard breaks (DSBs) in DNA. The single-guide RNA (sgRNA) (blue) is developed with the grouping of tracrRNA and crRNA and identifies the 20-nucleotide (orange) target sequence. This makes a complex of Cas9-sgRNA, which moves along the target site and cuts double-standard DNA 3 bases upstream of protospacer adjacent motif (PAM) through HNH and RuvC domains. The DSBs are reconstructed via nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathway. (B) Shows the Cpf1 mechanism that recognize the 24-nucleotide target sequence (blue) of crRNA and cleaves five nucleotides opposite to T-rich (TTTN) spacer at 5′ end. (C) Representing the base editing in which dead Cas9 (dCas9) is associated with cytidine deaminase (brown). It is directed by sgRNA (blue) for base substitute at target sequence (orange) distal to PAM site at 3' end. (D) depicted the de novo domestication process in wild plant using multiplex genome editing. Multiple guide RNA (gRNA) can be used to edit more than one gene simutanelously linked to some agronomic traits.
For example, Maher et al. (2020) reported a protocol to remove all the barriers caused by the laborious and time-consuming protocols of tissue culture. They have developed de novo meristem induction by transferring all the editing machinery into the somatic cells to produce the shoots having targeted manipulations (Maher et al., 2020). Ren et al. (2019) developed a didirectional promoter (BiP) system to express gRNA and Cas9 cassettes in the opposite direction to enhance the editing effectiveness of about 75.9–93.3% in rice. Decaestecker et al. (2019) constructed a CRISPR-TSKO system to produce tissue-specific knockout mutants to overcome the pleiotropic effect of a mutated gene. This will open up new possibilities for crop improvement to target a tissue-specific gene (Decaestecker et al., 2019). The editing efficiency of Cas nucleases is greatly influenced by the preferred PAM sites as it has a limited range to target the GC-rich region. Recently, Ren et al. (2021) designed a PAM-free editing tool by developing the CRISPR-SpRY toolbox in rice. It can resolve the PAM sites' limitations to target a wide range of specific sequences in the DNA molecule. Uranga et al. (2021) engineered potato virus X to construct a vector delivery system for multiple gRNAs and achieved 80% indels mutation rate in targeting the Nicotiana benthamiana genes. Also, the virus-free lines can be screened from the progeny developed via infected seeds or tissues that demonstrate greater biallelic mutations.
Base Editing
Production of the precise point mutations in plant genomic base editing is an emerging strategy to disrupt a single base using CRISPR/nCas9 (Cas9 nickase) attached with cytidine deaminase. An efficient system called base editor 3 (BE3) has been developed for cotton to create the targeted base substitutions with the mutation rate of 26.67–57.78% (Qin et al., 2020). A novel adenine base editor (ABE) was designed by Li et al. (2018a) to produce herbicide-resistant wheat and rice plants. ABE was enabled G to C and A to T point mutations with 59.1% successful rate in regenerated lines (Li et al., 2018a). Sretenovic et al. (2020) engineered the iSpyMacCas9 tool for the targeted mutation at A-rich PAM sites that substitute A to G and C to A base. The constructed vector system is well-suited for gateway cloning and also very compatible for all types of gRNA. This platform can be used to improve other editing systems like CRISRP activation, CRISPR interference, homology-directed repair (HDR) system, and prime editing. Plant virus can also be used for a fast and an efficient delivery of CRISPR/Cas machinery for virus-induced editing. While the APOBEC3-Cas9 fusion-induced deletion was designed to cleave 5′-deaminated C bases in rice and wheat protoplasts (Wang et al., 2020b).
Prime Editing
Recently, prime editing is emerged as an ideal toolbox that can remove all the previous hurdles and produces insertion, deletions, and base substitutions without generating DSBs (Marzec et al., 2020). Xu group developed the first prime-editing system in rice by designing the plant prime editors (pPE2) toolkit to induce mutations at different genomic regions with a frequency of 0–31.3% (Xu et al., 2020a). After this study, a huge wave of genome editing using prime editing have been conducted in different crops (Hua et al., 2020). For example, a prime editor system was optimized by promoter and codon to introduce insertion, deletions, and point mutations in wheat and rice protoplast (Lin et al., 2020), generate single and multiple base edits in rice (Xu et al., 2020b) having the mutation rate of 21.8 and 26%, respectively. In later experiment, Lin et al. (2021) designed two prime-editing gRNAs, which led to an increase in editing efficiencies from 2.9-fold to 17.4-fold in rice. Furthermore, the prime-editing system was executed to target both endogenous and exogenous genes to produce homozygous and heterozygous mutated lines with minimum off-targets and developed herbicide-resistant lines of rice through base substitutions (Butt et al., 2020; Li et al., 2020b).
De novo Domestication
De novo domestication is another important breeding pipeline benefited by the powerful technology of CRISPR/Cas. De novo domestication can be used to exploit the GD and has a great potential to introduce desriable traits in CWRs (Razzaq et al., 2021). De novo domestication of four wild cultivars of tomato was performed by mutating the four genes SlWUS, SlCLV3, SP5G, and SP through CRISPR/Cas9-mediated multiplex genome editing to develop improved tomato fruits (Li et al., 2018b). Similarly, Zsögön et al. (2018) executed de novo domestication of wild tomato by disrupting the six genes related to useful agronomic traits using multiplex editing, which controls the nutritional and yield-related traits. The engineered wild progenies showed 500% increase in the accumulation of lycopene compared to the cultivated species. Also, they exhibited a 3-fold greater fruit size and 10-fold extra fruit number as compared to their wild parents (Zsögön et al., 2018). Recently, Yu et al. (2021) established an efficient transformation system in allotetraploid rice. The de novo domestication strategy was used for improving the six agronomically important traits in allotetraploid rice. De novo domestication using next-generation genome editing tools can provide an alternative strategy to explore the GD of CWRs for crop improvement.
Beyond Editing
Apart from genome editing, the CRISPR/Cas toolbox can be applied to regulate gene expression and epigenome editing. A newly fine-tuning gene expression regulation system has created to control the translation process in plants. A CRISPR/Cas9 system with the modified protocol is used to edit the upstream open reading frames to produce transgene-free mutated plants. This approach can be utilized to study the functions of several genes and facilitate rapid crop improvement programs (Si et al., 2020). Another genome editing system named as Cas12b has been designed for site-specific genome manipulation in plants. It is considered as the third most exceptional CRISPR tool after Cas12a and Cas9 system. It also has an excellent potential for gene regulation through transcriptional activation and repression mechanism in plants (Ming et al., 2020). Nuclease-dead Cas9 (dCas9) allows programmable regulation of multiple genes via transcriptional effectors without damaging the target site. CRISPR/dCas9 has vast applications, such as DNA-free genome engineering, live-cell chromatin imaging, chromatin topology, epigenome editing, and gene regulation, in plants (Moradpour and Abdulah, 2020).
Technological Barriers and Public Concerns for CRISPR Technology
The major drawback in the applications of CRISPR-mediated engineering for plant breeding is not experimental or technical but consumer acceptance, public concerns, and strict regulatory affairs for the approval of edited crops. Technological improvement in genome editing tools would develop the similar traits just like the traits produced during conventional breeding in nature. However, CRISPR/Cas systems could be applied to incorporate foreign genes into the host genome, but this can easily be captured. Therefore, genome-edited crops developed through the CRISPR/Cas system should not be regarded as GM crops. Also, the use of CRISPR/Cas technology is still under strict constraints due to the ban imposed by European countries and cumbersome regulatory protocols adopted by USA, Australia, and Canada to ensure the biosafety of genome-edited crops. All these regulatory barriers will overcome only with a strong political determination and consensus among all stakeholders of different countries to consider the CRISPR-edited plants as non-GMOs.
There are still remaining certain bottlenecks that need to be fixed to exploit the full potential of the CRISPR/Cas system (Figure 6). For example, SpCas9 has a larger size, greater off-target mutations, and can only detect the NGG PAM sequence that limits its use. The emergence of other Cas orthologs like Cpf1, Cas13a, and Cas14 (a,b) can solve this problem due to their small size, broader PAM sites, and reduced off-target effects. The establishment of tissue culture-free, transient CRISPR system like prime editing is required to make this technology in a more robust and simpler way. The use of nonhomologous end-joining (NHEJ) inhibitors or HDR boosters can improve the HDR efficiency but yet to be reported in plants. In future, the next-generation CRISPR systems can offer sustainable agriculture production by overcoming the technical and regulatory barriers.
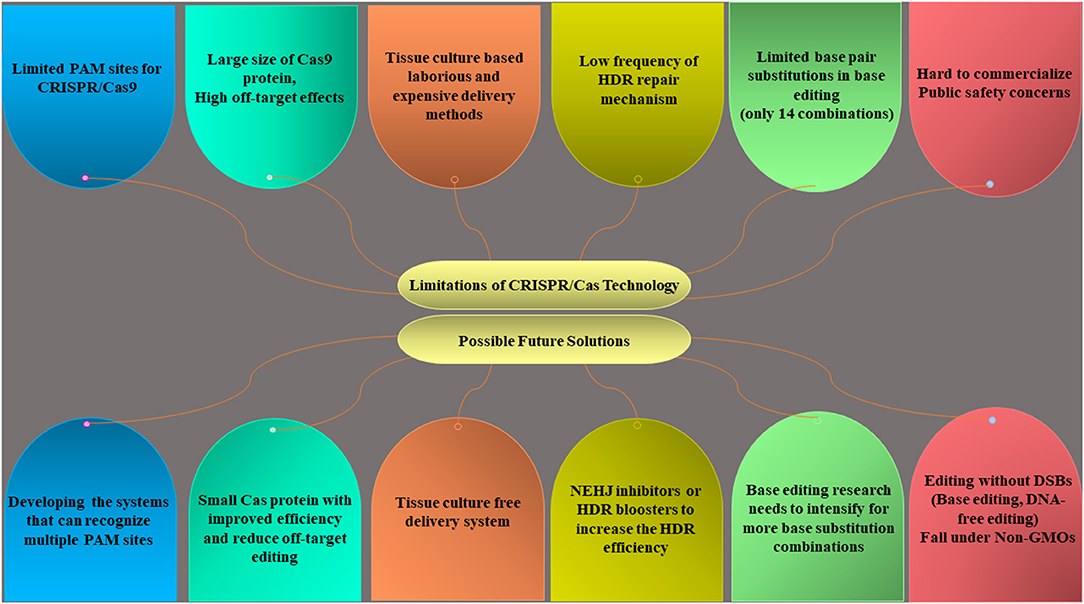
Figure 6. Limitations of the CRISPR/Cas system and a future way forward to develop ideal editing systems.
Next-Generation Plant Phenotyping Platforms
Plant trait phenotyping has been vital for successful crop domestication since thousands of years. The word “phenome” implies the whole phenotypic profile of any plant, and a phenotype is the blend of evident genomic expression with respect to its environment (Houle et al., 2010). Plant phenomics has unfolded within an evolving niche to a blooming investigative platform. It can be defined as the multidimensional application of advanced tools and procedures applied for capturing the detailed data on plant growth, function, structure, and behavior in a given environment. It deals with the acquisition, organization, and evaluation of huge phenotypic data sets, and the design of intelligent models for the prediction of plant growth in multiple scenarios (Houle et al., 2010). Plant phenotyping is a crucial approach to study the relationship of plants with their environment, and can conduct at various grades of resolution from genome to the whole plant under diverse climatic conditions, from field to systematic controlled environments. However, for every level spotlight on specific traits, the final objective is to connect the information from bottom up to develop elite crop varieties (Walter et al., 2015; Araus et al., 2018). So, the plant phenomics tools are indispensable in modern breeding and provide an excellent way forward to develop next-generation crops.
High-Throughput Phenotyping: A Step Toward Digital Agriculture
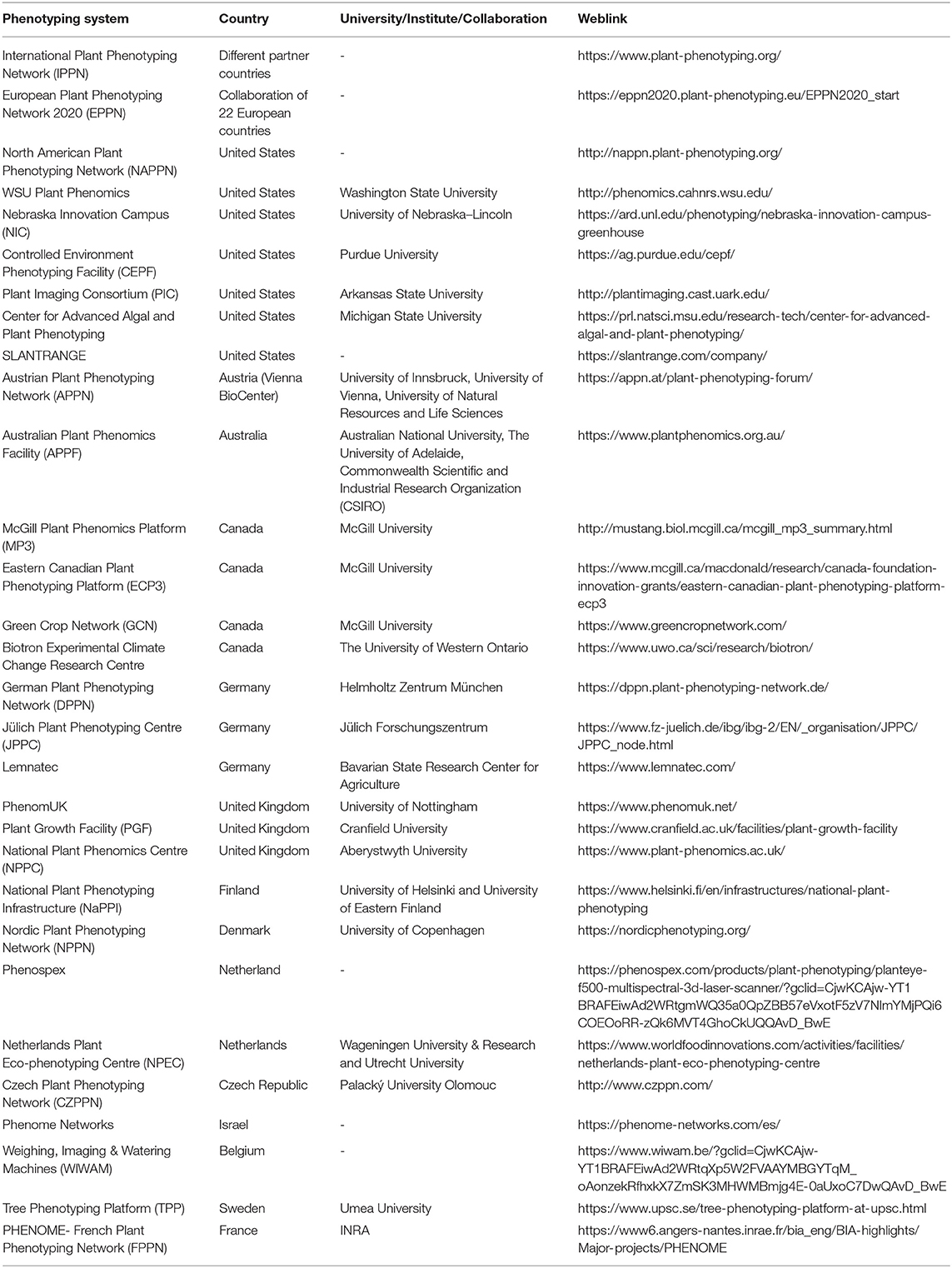
In recent years, state-of-the-art advanced phenotyping has emerged as a joint venture of multidisciplinary research groups to facilitate the launching of high-throughput phenotyping to expedite the next-generation breeding programs to ensure food security (Fasoula et al., 2020). High-throughput phenotyping allows high resolution imaging of thousands of plants for a better understanding of the insights of plant phenomics and genetics (Roitsch et al., 2019). It is the most promising technology that successfully incorporates plant science, engineering, math, information science, and computation with highly sophisticated tools of AI and ML to uncover diverse and intractable phenotypes of larger genotypes that are important to develop the best crop cultivars (Furbank et al., 2019). At present, several state-of-the-art phenotyping centers have been established in Europe, Australia, and USA as described in Table 5.
High-throughput plant phenomics operates within three approaches; firstly, the identification of a target trait regulating a unique stress response for precise, reproducible, accurate, and rapid data acquisition. The second and the third step based on a cutting-edge computer vision system enable the estimation and imaging examination of data and the computational analysis to forecast a biological response, respectively (Esposito et al., 2020). These hi-tech computer systems can lead current agriculture to digital agriculture, which uses AI and ML approaches to determine several factors like crop diseases, weeds identification, irrigation requirements, pesticide control, crop yield, and quality prediction (Zhang et al., 2017). There are numerous vital phenotypes that need to be investigated in order to dig out mysterious plant functions, or candidate phenotypes related to abiotic/biotic stresses and several agronomic traits. These multigenic traits must be spilt into constituent traits that can be observed, assessed, and evaluated via high-throughput advanced tools (Roitsch et al., 2019).
Tools like imaging, robotics, sensors, and software platforms have revolutionized the crop phenomics. The most common phenotyping platforms are unmanned aerial vehicles (UAVs), such as satellites, drones, phenotyping towers, environmental sensor networks, phenomobiles, autonomous ground vehicles, air crafts, helicopters, zeppelins, and field scanning platforms, which are currently used on a large scale (Liebisch et al., 2015). Non-invasive sensor-based phenotyping tools included; laser triangulation or red-green-blue (RGB) imaging system to determine the phenotypes of size, color, morphology, structure, texture, and growth of plant canopies. The hyperspectral and multispectral sensors can be used to measure moisture content, pigments composition, nitrogen content, and biophysical parameters. The thermal sensors were used for canopy temperature measurements to understand root physiology phenotypes. Light detection and ranging (LIDAR) provide the three-dimensional (3D) data for plant structural phenotypes (Araus et al., 2018).
Similarly, the automated image-based high-throughput phenotyping deals with the remote sensing and quantification of a large number of plant traits by capturing and analyzing the images at regular intervals with great accuracy (Jimenez-Berni et al., 2018). Image-based phenotyping is mainly non-destructive, enabling the desire traits to be assessed regularly during plant‘s life (Bao et al., 2019). At present, 3D imaging technology is getting attention in modern phenomics, including laser-based scanning and image-based approaches, which can produce 3D models to extract volumetric and spatial plant traits simultaneously (Paulus et al., 2014). For example, multiview stereo (MVS) is an excellent and a cost-effective 3D phenotyping platform for multiview imaging of plants at organ level (Hui et al., 2018). Nguyen et al. (2015) designed a 3D imaging system mounted with 10 digital high-resolution cameras supported by an illumination source to increase the surface imaging of plants. In another study, a 3D stereo-imaging unit was developed to visualize the traits related to plant height and rape seedling leaf area by installing two RGB cameras in an imaging chamber having a bright illuminous system (Xiong et al., 2017).
The emergence of field-based phenotyping provides a way forward to grab the full leverage of genetic gain and overcome the hurdles in breeding programs as it is the end-phenotypic expression of any genetic factor in relation to its environment (Singh et al., 2016; Araus et al., 2018; Bao et al., 2019). Field-based phenotyping platforms generally encompass ground wheeled, UAV, robotic-assisted system, tractor-driven, and cable-suspended phenotyping units, connected with high-performance sensors and cameras (Roitsch et al., 2019). These tools have been employed to estimate crop adaptability in natural conditions and can determine the canopy photosynthesis rate, leaf area index, plant height, performance, biomass, and disease symptoms (Jimenez-Berni et al., 2018). Recently, a cost-effective field-based high-throughput system MVS-Pheno is designed to monitor the shoot size of maize and offers exceptional ability to study large plant populations under diverse ecological zones (Wu et al., 2020). Similarly, a robotic field-based phenotyping system, which provide side-view stereo imaging, was developed to capture the plant height of sorghum at regular intervals of time (Bao et al., 2019).
Pitfalls in Plant Phenomics
In modern breeding, genotype-to-phenotype is a major drawback, which hinders the advanced breeding programs mediated by high-throughput genomic and phenomic tools (Harfouche et al., 2019). The connection between genome-environment-phenotype offers an excellent understanding to study the high-throughput data indicating that the plant stress mechanism is far more challenging due to multidimensional impacts of environmental changes on phenotypic plasticity and eventually widening of the genotype–phenotype gap (Gosa et al., 2019). Currently, modern crop improvement platforms heavily rely on advanced GAB and meticulous assessment of plant traits to define experimental lines and map the desired genes (Yang et al., 2020). The integration of genomic data with high-throughput phenotypic data to extract biologically fruitful information is a key to success in modern breeding (Harfouche et al., 2019).
However, despite a recent progress in genomic tools, the phenotyping technology did not grow at a competing rate to integrate the phenotypic data with genomics. The failure to capture phenotypic data effectively has a major pitfall, which hampers crop improvement programs (Harfouche et al., 2019; Wu et al., 2020; Yang et al., 2020). Due to the lack of phenomics data, our ability to measure phenotypic traits lags behind the existing capability to draw genomic data. Hence, the bottleneck is moving from genomics to phenomics (Großkinsky et al., 2018). There are some critical bottlenecks in plant phenotyping like the less efficient assessment of captured trait data that could result in poor identification of candidate genes and to capture the allelic variations for traits. Phenotyping of germplasm required a control range of conditions in replicated trails, which was costly and labor intensive (Junker et al., 2015). Accurate phenotyping in natural environment is also a big pitfall in many crop breeding schemes due to the highly heterogeneous natural conditions. Additionally, traditional phenotyping technologies are laborious, subjective, tedious, often causing damage to plants, and keeping record end-point phenotype (Naik et al., 2017). Therefore, an ample advancement in plant phenomics is needed for crop improvement in the long run, which will improve screening ability, fast-tracking of the genetic gains, accurate scanning of plant health status, and filling the hole between genotypic and phenotypic variations (Wu et al., 2020).
AI for Agriculture
In this digital world, AI is the most expeditiously rising technology in computer science and deals with the building of intelligent machines that mimic the intelligence of human mind (Harfouche et al., 2019). AI comprises ML algorithm models like deep neural network (DNN), artificial neural network (ANN), random forest (RF), support vector machine (SVM), and advanced hi-tech technology such as internet of things (IoT). AI is a mesmerizing hi-tech system with infinite applications in agriculture and opens up new horizons for digital agriculture (Montesinos-López et al., 2018). Systems are being designed to help the agricultural scientists for a better understanding of the plant behavior under diverse climatic conditions (Jeong et al., 2016). Recently, Summit, the world's most powerful supercomputer has been launched, which has the capacity to store 27,000 graphical processing units (GPUs) and unfolds an exciting way forward. AI can be a game changer and is pivotal for the next-generation crop revolution in the near future (Streich et al., 2020).
Recently, next-generation AI has gained significant attention in plant breeding to solve the problems related to abiotic/biotic stresses, herbicide resistance, crop yield, and soil fitness by developing the intelligent predictive models (Muraya et al., 2017). The applications of AI in agricultural production can be enormous because making AI-assisted spatial feature mining approaches can offer an exceptional prospect to incorporate the multi-omics results with high-throughput data sets captured via modern phenomics tools (Bolger et al., 2019). Al needs efficient and intelligent data mining that can assist breeders to accurately predict the agronomic factors and can also forecast crop performance under different conditions such as temperature, humidity, and soil type (Großkinsky et al., 2018; Harfouche et al., 2019).
Jiang et al. (2004) have developed more precise yield predicting models and projected wheat yield using multiple linear regression (MLR) and ANN models with satellite-assisted climate and vegetation indices in North China. An UAV-mediated phenotyping system aided by multispectral imaging and AI was performed to estimate the phenotypic features on field crops (Ampatzidis and Partel, 2019). Hemming et al. (2019) carried out AI to successfully control a greenhouse as compared to manually controlled greenhouse for vegetable production.
ML and Big Data Analytics
Machine learning is an emerging and a promising application of AI, which can be defined as the state-of-the-art computer-based systems that make the machine more intelligent to learn automatically and improve its ability without being stringently computed (Singh et al., 2016). Advanced ML algorithms have revealed a great potential in making highly precise and efficient pipelines for data analysis to enhance the breeding performance and ultimately crop productivity (Singh et al., 2018). For an accurate trait detection, several ML approaches have been employed, which can be divided into supervised/unsupervised and generative/discriminative learning model (Ghosal et al., 2018). In addition, many easily available programming languages or packages like MATLAB, ImageJ, and Python have been made to support or execute computer-based image pre-processing (Schindelin et al., 2012). ML tools can be applied to breakdown the multimodel data and identify the plant stresses and examine plant-pathogen association and interaction of other stresses with plants (Singh et al., 2018). One of the main benefits of ML tools for plant biologists is to get a chance to discover data sets in order to explore the patterns by analyzing the multitrait simultaneously (Shakoor et al., 2017).
Recently, an economical and a high resolving power image-based phenotyping system coupled with ML was designed to capture the root images of hundreds of plants to study the root system architecture traits in soybean (Falk et al., 2020). Likewise, root phenotyping of mature plants was performed via ML algorithms (RFs and SVM) to screen the most distinguishing root traits (Zhao et al., 2016). Additionally, sensor tools with ML algorithms can be applied to forecast crop yield under field conditions (Pantazi et al., 2016) and the plant growth trends for future predictions (Lee et al., 2018). An accurate ML model was developed to predict the photosynthesis activity in crops, and the result showed that the spectra-based phenotyping technique has the ability to improve photosynthesis capacity (Heckmann et al., 2017). ML has been employed for the early detection of various plant diseases. For example, ANNs and RFs tools were used to envisage the risk assessment of wheat Stagonospora nodorum blotch (Mehra et al., 2016). A realtime pipeline for phenotyping using ML was established to estimate the severity of abiotic and biotic stress in soybean. This system can be assisted to enhance the genetic gains by allowing the automatic stress trait identification and stress scouting applications (Naik et al., 2017).
ML-Assisted GS
Genomic selection enables the quick screening of elite germplasm and expedites the crop breeding cycle (Crossa et al., 2017). Currently, GS depends on innovations in ML tools and retrieval of large genotyping data sets related to agronomically important phenotypic traits for genomic prediction (Tong and Nikoloski, 2021). So, GS models are the core elements of ML that intend to design and examine the model performance through an array of training data. In GS models, the genotyping data such as SNPs are the input, and the predicted phenotypic trait is the final output. The leverage of exploiting ML, especially DL to analyze the GS is that it may obtain highly complex interactions and deliver greater predictability (González-Camacho et al., 2012). There are several examples of DL application for GS in crop improvement programs because CNNs are very accurate in predicting phenotypic traits (Pérez-Enciso and Zingaretti, 2019).
Deep learning techniques have been used in a multitrait situation, for example, wheat population exhibited that the prediction of multitrait DL is the same as a single-trait model, but somewhat superior when analyzing in relation to univariate DL models (Montesinos-López et al., 2019a,b). González-Camacho et al. (2016) demonstrated that the probabilistic neural network (PNN) is a promising strategy for GS in crop breeding by testing the wheat and maize population to predict the genotypes related to good or bad groups. The results showed that PNN models are more efficient than multilayer perceptron (MLP) models. Liu et al. (2019) study soybean to predict five traits employing CNN models and revealed better performance. A CNN-based DeepGS method was developed to study the grain-related yield in wheat population (Ma et al., 2018).
Speed Breeding
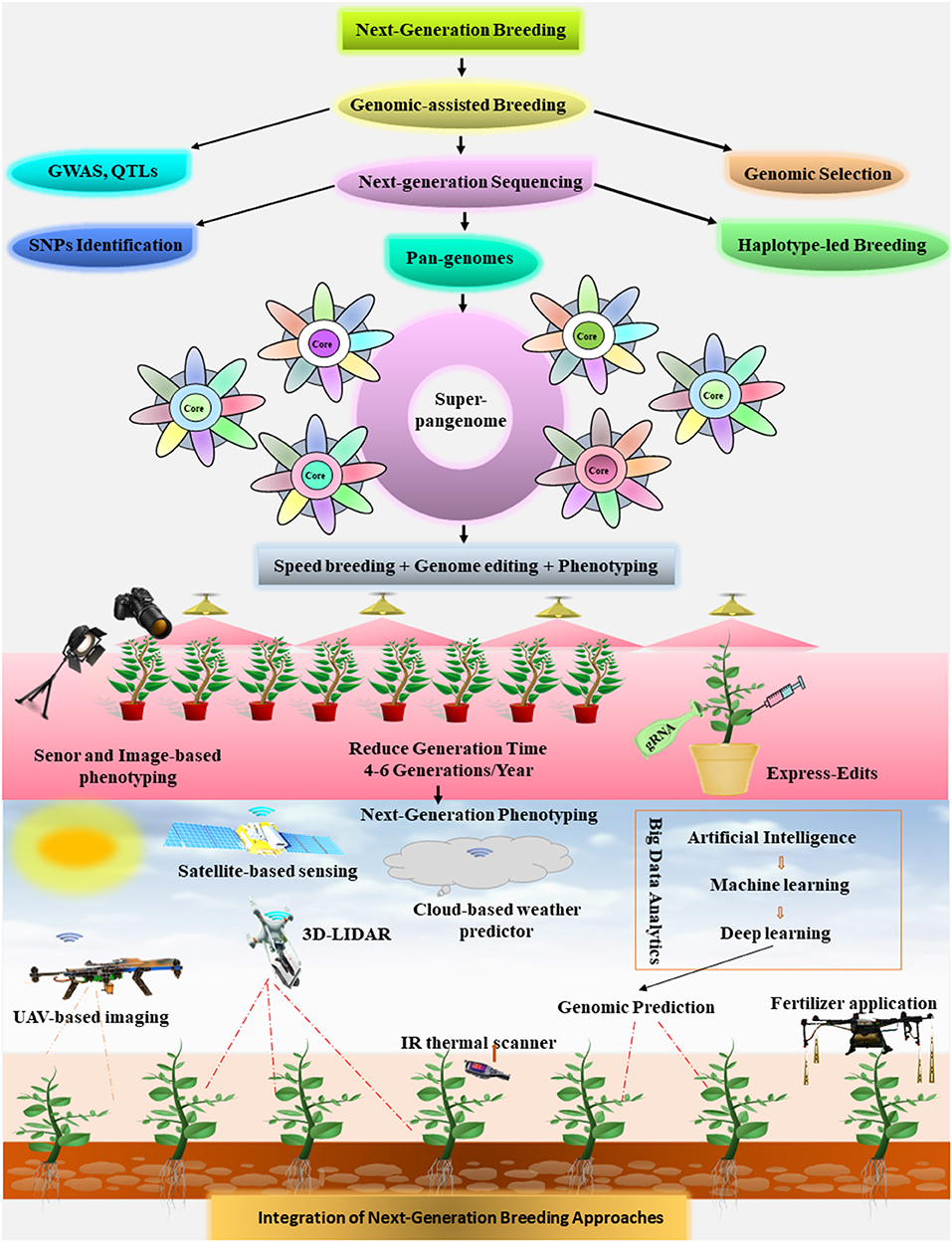
The world is attracted by the most fascinating technology of speed breeding. The scientist from the University of Queensland inspired by the NASA to grow the wheat plants in space under artificial lights. Watson et al. (2018) successfully developed the protocols for different plant species under a speed breeding system. Speed breeding is a powerful strategy to shorten the crop generation time and expedite the breeding programs for crop improvement (Watson et al., 2018). Speed breeding mimics daily dawn and dusk, and plants are subjected to an extended photoperiod of about 22 h by using a combination of different light sources. It provides an extended day length with optimal light intensity coupled with controlled temperature to increase the photosynthesis activity, which results in quick flowering and early seed development to reduce generation time (Ghosh et al., 2018).
Speed breeding is revolutionizing the agriculture and can be executed to accelerate the crop breeding activities such as crossing, back crossing, rapid gene identification, mapping population, pyramiding of traits, and developing transgenic pipelines (Hickey et al., 2019). In conventional breeding, only 1–2 generations per season of any crop can be achieved but in speed breeding up to four generations of B. napus and six generations of Hordeum vulgare, Triticum aestivum, Pisum sativum, Cicer arietinum, and B. distachyon (Watson et al., 2018). Furthermore, it can provide a robust, an efficient, and an economical platform to carry out the crop improvement project in an integrated way from genomics to phenomics. It may include the candidate genes discovery through GAB approaches such as pan-genome assembly, GS, and GBS to multiplex gene editing, or metabolic pathway editing for desired traits followed by high-throughput phenotyping to visualize the results. The integration of speed breeding with next-generation metabolomic tools can also be used for a rapid risk assessment of gene-edited crops in multiple generations in a robust manner (Razzaq et al., 2019a). Hence, speed breeding technology will offer an exciting way forward for crop improvement by integrating it with next-generation OMICS tools to accelerate the crop breeding programs.
Outlook
Human population explosion and adverse climatic changes are posing extreme challenges for promising global food security. Next-generation breeding technologies offer a robust platform to develop high yielding and climate-resilient crops. GAB has made a significant impact on plant research, and ground-breaking pan-genome techniques permit to capture the full landscape of genetic variations. Hence, we are expecting a genomic data explosion as third-generation sequencing combined with the pan-genome concept will possibly construct large gene repertoires of landraces, cultivated plants, and CWR. The development of super-pangenomes, a combination of different pan-genomes, depicts the whole genetic profile of any specific genus and will help for pangenome-led haplotype breeding for crop improvement. The assembly of plant pan-omes will allow to study the genetic variation in single cell level and will provide huge data sets from genome to phenome for a quick and an accurate phenotypic identification. The integration of pan-omes with speed breeding and genome editing will accelerate the trait-specific breeding. Applications of AI such as DL in super pangenomes construction will help to retrieve huge data sets. The potential of DL in combining the sequencing data will enable to construct the models for GS and trait prediction.
Next-generation CRISPR systems, like de novo domestication, tissue-specific editing, prime editing, and fine tuning of gene expression, are now giving plant scientists an unprecedented prospect to manipulate the plant genomes with more accuracy and precision. The potential of an CRISPR/Cas system to edit the plant organelles like chloroplast and mitochondria is yet to be explored because of the unavailability of delivery vectors that can enter into these organelles. In the near future, chloroplast and mitochondria can be targeted by transferring multiple gRNAs to achieve cell-based editing. Also, some breakthroughs like the delivery of CRISPR/Cas machinery being the biggest drawback in genome editing are still needed. The RNP or carbon nanotubes- (CNTs-) based delivery system is transferred directly into inflorescence tissues, pollen grains, and apical meristems, or it is sprayed to already grown plant to get express edits in the field or under speed breeding chambers (Hickey et al., 2019). Performing the genome editing under the speed breeding condition will provide a controlled system to speed up the crop breeding cycles with a minimum cost and will be a technological breakthrough to get non-transgenic plants without tissue culturing. It will tag as non-GMOs with more public acceptance and avoid the strict regulatory affairs.
The exciting phenotyping platforms inspired by AI, like robotics, 3D imaging sensors, and their integration with other OMICS date, are taking plant science to new heights. With multiscale, multidimensional, and multidomain phenomics knowledge, we promptly require the next-generation approaches of swarm intelligence, hybrid intelligence, AI, data fusion, and DL to establish big data handling methods.
To conclude, a powerful unified strategy is needed to tackle the current challenges by integrating the multidisciplinary next-generation strategies and scientists to work under one umbrella and carry out mega projects to boost up the crop production in a rapid way as proposed in Figure 7.
The multidisciplinary tools from genomics to phenomics should be carried out in a well-integrated manner to overcome the current hurdles in plant research. By employing the integrated next-generation approaches, future crop breeding can attain irreversible success to ensure food security and will fulfill the food demands of rapidly increasing population in decades to come.
Author Contributions
AR conceived the idea. AR and FS led the writing of manuscript. NA helped in revising the manuscript. PK and SW reviewed, edited, and improved the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the researchers whose contribution have been cited in this review paper. AR was thankful to Professor Robert Henry for his valuable comments and suggestions while preparing the initial draft. PK was supported by the University of Western Australia with additional computational resources and support from the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia.
Footnotes
1. ^FAOSTAT (2020). Available: http://www.fao.org/faostat/en/#data (accessed July 30, 2020).
2. ^EM-DAT (2020). Available: https://www.emdat.be/ (accessed March 29, 2020).
References
Abberton, M., Batley, J., Bentley, A., Bryant, J., Cai, H., Cockram, J., et al. (2016). Global agricultural intensification during climate change: a role for genomics. Plant Biotechnol. J. 14, 1095–1098. doi: 10.1111/pbi.12467
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Allen, M., Antwi-Agyei, P., Aragon-Durand, F., Babiker, M., Bertoldi, P., Bind, M., et al. (2019). Technical Summary: Global Warming of 1.5°C. An IPCC Special Report on the Impacts of Global Warming of 1.5°C Above Pre-Industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty.
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19, 1–9. doi: 10.1186/s13059-017-1381-1
Ampatzidis, Y., and Partel, V. (2019). “UAV-based high throughput phenotyping in specialty crops utilizing artificial intelligence,” in 2019 ASABE Annual International Meeting. St Joseph, MI: American Society of Agricultural and Biological Engineers, 1. doi: 10.13031/aim.201900293
Anari, S. S., de Ridder, D., Schranz, M. E., and Smit, S. (2019). Pangenomic read mapping. bioRxiv 813634. doi: 10.1101/813634
Araus, J. L., Kefauver, S. C., Zaman-Allah, M., Olsen, M. S., and Cairns, J. E. (2018). Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 23, 451–466. doi: 10.1016/j.tplants.2018.02.001
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Arthur, R. A., and Bennetzen, J. L. (2018). Discovery of lineage-specific genome change in rice through analysis of resequencing data. Genetics 209, 617–626. doi: 10.1534/genetics.118.300848
Bao, Y., Tang, L., Breitzman, M. W., Salas Fernandez, M. G., and Schnable, P. S. (2019). Field-based robotic phenotyping of sorghum plant architecture using stereo vision. J. Field Robot. 36, 397–415. doi: 10.1002/rob.21830
Bassi, F. M., Bentley, A. R., Charmet, G., Ortiz, R., and Crossa, J. (2016). Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 242, 23–36. doi: 10.1016/j.plantsci.2015.08.021
Bayer, P. E., Golicz, A. A., Tirnaz, S., Chan, C. K. K., Edwards, D., and Batley, J. (2019). Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 17, 789–800. doi: 10.1111/pbi.13015
Becker, M., Gruenheit, N., Steel, M., Voelckel, C., Deusch, O., Heenan, P. B., et al. (2013). Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nat. Climate Change 3, 1039–1043. doi: 10.1038/nclimate2027
Bevan, M. W., Uauy, C., Wulff, B. B., Zhou, J., Krasileva, K., and Clark, M. D. (2017). Genomic innovation for crop improvement. Nature 543, 346–354. doi: 10.1038/nature22011
Bo, W., Zhaohui, Z., Huanhuan, Z., Xia, W., Binglin, L., Lijia, Y., et al. (2019). Targeted mutagenesis of NAC transcription factor gene, OsNAC041, leading to salt sensitivity in rice. Rice Sci. 26, 98–108. doi: 10.1016/j.rsci.2018.12.005
Bolger, A. M., Poorter, H., Dumschott, K., Bolger, M. E., Arend, D., Osorio, S., et al. (2019). Computational aspects underlying genome to phenome analysis in plants. Plant J. 97, 182–198. doi: 10.1111/tpj.14179
Breseghello, F., and Coelho, A. S. G. (2013). Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J. Agri. Food Chem. 61, 8277–8286. doi: 10.1021/jf305531j
Brozynska, M., Furtado, A., and Henry, R. J. (2016). Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotechnol. J. 14, 1070–1085. doi: 10.1111/pbi.12454
Burstein, D., Harrington, L. B., Strutt, S. C., Probst, A. J., Anantharaman, K., Thomas, B. C., et al. (2017). New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241. doi: 10.1038/nature21059
Butt, H., Rao, G. S., Sedeek, K., Aman, R., Kamel, R., and Mahfouz, M. (2020). Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020:13399. doi: 10.1111/pbi.13399
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucl. Res. 39:e82. doi: 10.1093/nar/gkr739
Change, G. E. (2020). Meeting the food security challenge for nine billion people in 2050: what impact on forests? Glob. Environ. Change 62:102056. doi: 10.1016/j.gloenvcha.2020.102056
Chatterjee, P., Jakimo, N., and Jacobson, J. M. (2018). Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 4:eaau0766. doi: 10.1126/sciadv.aau0766
Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., et al. (2017). Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410. doi: 10.1038/nature24268
Collonnier, C., Guyon-Debast, A., Maclot, F., Mara, K., Charlot, F., and Nogué, F. (2017). Towards mastering CRISPR-induced gene knock-in in plants: survey of key features and focus on the model Physcomitrella patens. Methods 121, 103–117. doi: 10.1016/j.ymeth.2017.04.024
Concibido, V., La Vallee, B., Mclaird, P., Pineda, N., Meyer, J., Hummel, L., et al. (2003). Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theoret. Appl. Genet. 106, 575–582. doi: 10.1007/s00122-002-1071-5
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Contreras-Moreira, B., Cantalapiedra, C. P., García-Pereira, M. J., Gordon, S. P., Vogel, J. P., Igartua, E., et al. (2017). Analysis of plant pan-genomes and transcriptomes with GET_HOMOLOGUES-EST, a clustering solution for sequences of the same species. Fronti. Plant Sci. 8:184. doi: 10.3389/fpls.2017.00184
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., de los Campos, G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Danilevicz, M. F., Fernandez, C. G. T., Marsh, J. I., Bayer, P. E., and Edwards, D. (2020). Plant pangenomics: approaches, applications and advancements. Curr. Opin. Plant Biol. 54, 18–25. doi: 10.1016/j.pbi.2019.12.005
Darracq, A., Vitte, C., Nicolas, S., Duarte, J., Pichon, J.-P., Mary-Huard, T., et al. (2018). Sequence analysis of European maize inbred line F2 provides new insights into molecular and chromosomal characteristics of presence/absence variants. BMC Genom. 19, 1–20. doi: 10.1186/s12864-018-4490-7
Decaestecker, W., Buono, R. A., Pfeiffer, M. L., Vangheluwe, N., Jourquin, J., Karimi, M., et al. (2019). CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887. doi: 10.1105/tpc.19.00454
Deutsch, C. A., Tewksbury, J. J., Tigchelaar, M., Battisti, D. S., Merrill, S. C., Huey, R. B., et al. (2018). Increase in crop losses to insect pests in a warming climate. Science 361, 916–919. doi: 10.1126/science.aat3466
Díaz, A., Zikhali, M., Turner, A. S., Isaac, P., and Laurie, D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 7:e33234. doi: 10.1371/journal.pone.0033234
Dolatabadian, A., Bayer, P. E., Tirnaz, S., Hurgobin, B., Edwards, D., and Batley, J. (2020). Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 18, 969–982. doi: 10.1111/pbi.13262
Endo, A., Masafumi, M., Kaya, H., and Toki, S. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 6:38169. doi: 10.1038/srep38169
Esposito, S., Carputo, D., Cardi, T., and Tripodi, P. (2020). Applications and trends of machine learning in genomics and phenomics for next-generation breeding. Plants 9:34. doi: 10.3390/plants9010034
Falk, K. G., Jubery, T. Z., Mirnezami, S. V., Parmley, K. A., Sarkar, S., Singh, A., et al. (2020). Computer vision and machine learning enabled soybean root phenotyping pipeline. Plant Methods 16:5. doi: 10.1186/s,13007-019-0550-5
FAO (2017). Benefits of Farm Level Disaster Risk Reduction Practices in Agriculture. United Nations, 33, 661. Available online at: http://www.fao.org/3/i7319e/i7319e.pdf (accessed May 15, 2021).
FAO IFAD, UNICEF, WFP, and WHO. (2018). The State of Food Security and Nutrition in the World 2018: Building Climate Resilience for Food Security and Nutrition. Rome: Food & Agriculture Org.
Fasoula, D. A., Ioannides, I. M., and Omirou, M. (2020). Phenotyping and plant breeding: overcoming the barriers. Front. Plant Sci. 10:1713. doi: 10.3389/fpls.2019.01713
Fischer, R., Byerlee, D., and Edmeades, G. (2014). Crop Yields and Global Food Security. Canberra, ACT: ACIAR, 8–11.
Furbank, R. T., Jimenez-Berni, J. A., George-Jaeggli, B., Potgieter, A. B., and Deery, D. M. (2019). Field crop phenomics: enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. 223, 1714–1727. doi: 10.1111/nph.15817
Gao, L., Gonda, I., Sun, H., Ma, Q., Bao, K., Tieman, D. M., et al. (2019). The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 51, 1044–1051. doi: 10.1038/s41588-019-0410-2
Ghosal, S., Blystone, D., Singh, A. K., Ganapathysubramanian, B., Singh, A., and Sarkar, S. (2018). An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. U. S. A. 115, 4613–4618. doi: 10.1073/pnas.1716999115
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protocols 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
Gödecke, T., Stein, A. J., and Qaim, M. (2018). The global burden of chronic and hidden hunger: trends and determinants. Glob. Food Security 17, 21–29. doi: 10.1016/j.gfs.2018.03.004
Golicz, A. A., Batley, J., and Edwards, D. (2016a). Towards plant pangenomics. Plant Biotechnol. J. 14, 1099–1105. doi: 10.1111/pbi.12499
Golicz, A. A., Bayer, P. E., Barker, G. C., Edger, P. P., Kim, H., Martinez, P. A., et al. (2016b). The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7, 1–8. doi: 10.1038/ncomms13390
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
González, V. M., Aventín, N., Centeno, E., and Puigdomènech, P. (2013). High presence/absence gene variability in defense-related gene clusters of Cucumis melo. BMC Genom. 14:782. doi: 10.1186/1471-2164-14-782
González-Camacho, J. M., Crossa, J., Pérez-Rodríguez, P., Ornella, L., and Gianola, D. (2016). Genome-enabled prediction using probabilistic neural network classifiers. BMC Genom. 17, 1–16. doi: 10.1186/s12864-016-2553-1
González-Camacho, J. M., de Los Campos, G., Pérez, P., Gianola, D., Cairns, J. E., Mahuku, G., et al. (2012). Genome-enabled prediction of genetic values using radial basis function neural networks. Theoret. Appl. Genet. 125, 759–771. doi: 10.1007/s00122-012-1868-9
Gordon, S. P., Contreras-Moreira, B., Woods, D. P., Des Marais, D. L., Burgess, D., Shu, S., et al. (2017). Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8, 1–13. doi: 10.1038/s41467-017-02292-8
Gosa, S. C., Lupo, Y., and Moshelion, M. (2019). Quantitative and comparative analysis of whole-plant performance for functional physiological traits phenotyping: new tools to support pre-breeding and plant stress physiology studies. Plant Sci. 282, 49–59. doi: 10.1016/j.plantsci.2018.05.008
Großkinsky, D. K., Syaifullah, S. J., and Roitsch, T. (2018). Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 69, 825–844. doi: 10.1093/jxb/erx333
Harfouche, A. L., Jacobson, D. A., Kainer, D., Romero, J. C., Harfouche, A. H., Mugnozza, G. S., et al. (2019). Accelerating climate resilient plant breeding by applying next-generation artificial intelligence. Trends Biotechnol. 37, 1217–1235. doi: 10.1016/j.tibtech.2019.05.007
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., et al. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842. doi: 10.1126/science.aav4294
Heckmann, D., Schlüter, U., and Weber, A. P. (2017). Machine learning techniques for predicting crop photosynthetic capacity from leaf reflectance spectra. Mol. Plant 10, 878–890. doi: 10.1016/j.molp.2017.04.009
Heeb, L., Jenner, E., and Cock, M. J. (2019). Climate-smart pest management: building resilience of farms and landscapes to changing pest threats. J. Pest Sci. 92, 951–969. doi: 10.1007/s10340-019-01083-y
Hemming, S., de Zwart, F., Elings, A., Righini, I., and Petropoulou, A. (2019). Remote control of greenhouse vegetable production with artificial intelligence—greenhouse climate, irrigation, and crop production. Sensors 19:1807. doi: 10.3390/s19081807
Henry, R. (2020). Innovations in agriculture and food supply in response to the COVID-19 pandemic. Mol. Plant 13, 1095–1097. doi: 10.1016/j.molp.2020.07.011
Hickey, L. T., Hafeez, A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hirano, H., Gootenberg, J. S., Horii, T., Abudayyeh, O. O., Kimura, M., Hsu, P. D., et al. (2016). Structure and engineering of Francisella novicida Cas9. Cell 164, 950–961. doi: 10.1016/j.cell.2016.01.039
Hirsch, C. N., Foerster, J. M., Johnson, J. M., Sekhon, R. S., Muttoni, G., Vaillancourt, B., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26, 121–135. doi: 10.1105/tpc.113.119982
Houle, D., Govindaraju, D. R., and Omholt, S. (2010). Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866. doi: 10.1038/nrg2897
Hu, Z., Sun, C., Lu, K.-c., Chu, X., Zhao, Y., Lu, J., et al. (2017). EUPAN enables pan-genome studies of a large number of eukaryotic genomes. Bioinformatics 33, 2408–2409. doi: 10.1093/bioinformatics/btx170
Hua, K., Jiang, Y., Tao, X., and Zhu, J. K. (2020). Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020, 1–3. doi: 10.1111/pbi.13395
Huang, X., Cöster, H., Ganal, M., and Röder, M. (2003). Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theoret. Appl. Genet. 106, 1379–1389. doi: 10.1007/s00122-002-1179-7
Hübner, S., Bercovich, N., Todesco, M., Mandel, J. R., Odenheimer, J., Ziegler, E., et al. (2019). Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 5, 54–62. doi: 10.1038/s41477-018-0329-0
Hui, F., Zhu, J., Hu, P., Meng, L., Zhu, B., Guo, Y., et al. (2018). Image-based dynamic quantification and high-accuracy 3D evaluation of canopy structure of plant populations. Ann. Bot. 121, 1079–1088. doi: 10.1093/aob/mcy016
Hurgobin, B., Golicz, A. A., Bayer, P. E., Chan, C. K. K., Tirnaz, S., Dolatabadian, A., et al. (2018). Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 16, 1265–1274. doi: 10.1111/pbi.12867
ISAAA database (2021). GM Approval Database. Available online at: https://www.isaaa.org/gmapprovaldatabase/default.asp (accessed May 22, 2021).
James, C. (1997). Global Status of Transgenic Crops in 1997. ISAAA Brief No. 5. Ithaca, NY: ISAAA, 31. Available online at: https://www.isaaa.org/purchasepublications/itemdescription.asp?ItemType=BRIEFS&Control=IB005–1997 (accessed May 16, 2021).
Jandrasits, C., Dabrowski, P. W., Fuchs, S., and Renard, B. Y. (2018). seq-seq-pan: building a computational pan-genome data structure on whole genome alignment. BMC Genom. 19:47. doi: 10.1186/s12864-017-4401-3
Jayakodi, M., Schreiber, M., Stein, N., and Mascher, M. (2021). Building pan-genome infrastructures for crop plants and their use in association genetics. DNA Res. 28:dsaa030. doi: 10.1093/dnares/dsaa030
Jeon, Y., Choi, Y. H., Jang, Y., Yu, J., Goo, J., Lee, G., et al. (2018). Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-05245-x
Jeong, J. H., Resop, J. P., Mueller, N. D., Fleisher, D. H., Yun, K., Butler, E. E., et al. (2016). Random forests for global and regional crop yield predictions. PLoS ONE 11:e0156571. doi: 10.1371/journal.pone.0156571
Jia, C., Huai, C., Ding, J., Hu, L., Su, B., Chen, H., et al. (2018). New applications of CRISPR/Cas9 system on mutant DNA detection. Gene 641, 55–62. doi: 10.1016/j.gene.2017.10.023
Jiang, D., Yang, X., Clinton, N., and Wang, N. (2004). An artificial neural network model for estimating crop yields using remotely sensed information. Int. J. Remote Sens. 25, 1723–1732. doi: 10.1080/0143116031000150068
Jimenez-Berni, J. A., Deery, D. M., Rozas-Larraondo, P., Condon, A. T. G., Rebetzke, G. J., James, R. A., et al. (2018). High throughput determination of plant height, ground cover, and above-ground biomass in wheat with LiDAR. Front. Plant Sci. 9:237. doi: 10.3389/fpls.2018.00237
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Junker, A., Muraya, M. M., Weigelt-Fischer, K., Arana-Ceballos, F., Klukas, C., Melchinger, A. E., et al. (2015). Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Front. Plant Sci. 5:770. doi: 10.3389/fpls.2014.00770
Karvelis, T., Gasiunas, G., Young, J., Bigelyte, G., Silanskas, A., Cigan, M., et al. (2015). Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 16, 1–13. doi: 10.1186/s13059-015-0818-7
Kaya, H., Mikami, M., Endo, A., Endo, M., and Toki, S. (2016). Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6:26871. doi: 10.1038/srep26871
Khan, M. Z., Haider, S., Mansoor, S., and Amin, I. (2019). Targeting plant ssDNA viruses with engineered miniature CRISPR-Cas14a. Trends Biotechnol. 37, 800–804. doi: 10.1016/j.tibtech.2019.03.015
Kim, E., Koo, T., Park, S. W., Kim, D., Kim, K., Cho, H.-Y., et al. (2017). In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8:14500. doi: 10.1038/ncomms14500
Kim, H., and Kim, J.-S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334. doi: 10.1038/nrg3686
Kim, W., Iizumi, T., and Nishimori, M. (2019a). Global patterns of crop production losses associated with droughts from 1983 to 2009. J. Appl. Meteorol. Climatol. 58, 1233–1244. doi: 10.1175/JAMC-D-18-0174.1
Kim, Y. A., Moon, H., and Park, C. J. (2019b). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 12, 1–13. doi: 10.1186/s12284-019-0331-9
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. doi: 10.1038/nature14592
Kleinstiver, B. P., Sousa, A. A., Walton, R. T., Tak, Y. E., Hsu, J. Y., Clement, K., et al. (2019). Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282. doi: 10.1038/s41587-018-0011-0
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Lavarenne, J., Guyomarc'h, S., Sallaud, C., Gantet, P., and Lucas, M. (2018). The spring of systems biology-driven breeding. Trends Plant Sci. 23, 706–720. doi: 10.1016/j.tplants.2018.04.005
Lee, C. M., Cradick, T. J., and Bao, G. (2016). The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Therapy 24, 645–654. doi: 10.1038/mt.2016.8
Lee, U., Chang, S., Putra, G. A., Kim, H., and Kim, D. H. (2018). An automated, high-throughput plant phenotyping system using machine learning-based plant segmentation and image analysis. PLoS ONE 13:e0196615. doi: 10.1371/journal.pone.0196615
Leng, P. F., Lübberstedt, T., and Xu, M. L. (2017). Genomics-assisted breeding–a revolutionary strategy for crop improvement. J. Integr. Agri. 16, 2674–2685. doi: 10.1016/S2095-3119(17)61813-6
Li, B., Rui, H., Li, Y., Wang, Q., Alariqi, M., Qin, L., et al. (2019a). Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol. J. 17:1862. doi: 10.1111/pbi.13147
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., and Lin, Y. (2020a). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315. doi: 10.1111/pbi.13217
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018a). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19:59. doi: 10.1186/s13059-018-1443-z
Li, H., Li, J., Chen, J., Yan, L., and Xia, L. (2020b). Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13, 671–674. doi: 10.1016/j.molp.2020.03.011
Li, J.-F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, M., Chen, R., Jiang, Q., Sun, X., Zhang, H., and Hu, Z. (2021). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105, 333–345. doi: 10.1007/s11103-020-01091-y
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Yu, W., et al. (2019b). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19, 1–13. doi: 10.1186/s12870-018-1627-4
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018b). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi: 10.1038/nbt.4273
Li, Y.-h., Zhou, G., Ma, J., Jiang, W., Jin, L.-g., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi: 10.1038/nbt.2979
Liang, Z., Chen, K., Yan, Y., Zhang, Y., and Gao, C. (2018). Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol. J. 16, 2053–2062. doi: 10.1111/pbi.12938
Liebisch, F., Kirchgessner, N., Schneider, D., Walter, A., and Hund, A. (2015). Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 11:9. doi: 10.1186/s13007-015-0048-8
Lin, K., Zhang, N., Severing, E. I., Nijveen, H., Cheng, F., Visser, R. G., et al. (2014). Beyond genomic variation-comparison and functional annotation of three Brassica rapa genomes: a turnip, a rapid cycling and a Chinese cabbage. BMC Genom. 15:250. doi: 10.1186/1471-2164-15-250
Lin, Q., Jin, S., Zong, Y., Yu, H., Zhu, Z., Liu, G., et al. (2021). High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 1–5. doi: 10.1038/s41587-021-00868-w
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, L., Chen, P., Wang, M., Li, X., Wang, J., Yin, M., et al. (2017). C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell 65, 310–322. doi: 10.1016/j.molcel.2016.11.040
Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., et al. (2020a). CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 301:110683. doi: 10.1016/j.plantsci.2020.110683
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020b). Pan-genome of wild and cultivated soybeans. Cell 182, 162–176. doi: 10.1016/j.cell.2020.05.023
Liu, Y., Wang, D., He, F., Wang, J., Joshi, T., and Xu, D. (2019). Phenotype prediction and genome-wide association study using deep convolutional neural network of soybean. Front. Genet. 10:1091. doi: 10.3389/fgene.2019.01091
Lloyd, A., Plaisier, C. L., Carroll, D., and Drews, G. N. (2005). Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102, 2232–2237. doi: 10.1073/pnas.0409339102
Ma, W., Qiu, Z., Song, J., Li, J., Cheng, Q., Zhai, J., et al. (2018). A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 248, 1307–1318. doi: 10.1007/s00425-018-2976-9
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Marcus, S., Lee, H., and Schatz, M. C. (2014). SplitMEM: a graphical algorithm for pan-genome analysis with suffix skips. Bioinformatics 30, 3476–3483. doi: 10.1093/bioinformatics/btu756
Martínez, M. I. S., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 20, 1–13. doi: 10.1186/s12870-020-02497-y
Marzec, M., Braszewska-Zalewska, A., and Hensel, G. (2020). Prime editing: a new way for genome editing. Trends Cell Biol. 30, 257–259. doi: 10.1016/j.tcb.2020.01.004
McCouch, S., Baute, G. J., Bradeen, J., Bramel, P., Bretting, P. K., Buckler, E., et al. (2013). Feeding the future. Nature 499, 23–24. doi: 10.1038/499023a
McHale, L. K., Haun, W. J., Xu, W. W., Bhaskar, P. B., Anderson, J. E., Hyten, D. L., et al. (2012). Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159, 1295–1308. doi: 10.1104/pp.112.194605
Mehra, L. K., Cowger, C., Gross, K., and Ojiambo, P. S. (2016). Predicting pre-planting risk of Stagonospora nodorum blotch in winter wheat using machine learning models. Front. Plant Sci. 7:390. doi: 10.3389/fpls.2016.00390
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S. S. E. A., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20, 1–10. doi: 10.1186/s13059-019-1678-3
Ming, M., Ren, Q., Pan, C., He, Y., Zhang, Y., Liu, S., et al. (2020). CRISPR–Cas12b enables efficient plant genome engineering. Nat. Plants 6, 202–208. doi: 10.1038/s41477-020-0614-6
Montenegro, J. D., Golicz, A. A., Bayer, P. E., Hurgobin, B., Lee, H., Chan, C. K. K., et al. (2017). The pangenome of hexaploid bread wheat. Plant J. 90, 1007–1013. doi: 10.1111/tpj.13515
Montesinos-López, O. A., Martín-Vallejo, J., Crossa, J., Gianola, D., Hernández-Suárez, C. M., Montesinos-López, A., et al. (2019a). New deep learning genomic-based prediction model for multiple traits with binary, ordinal, and continuous phenotypes. Genes Genomes Genet. 9, 1545–1556. doi: 10.1534/g3.119.300585
Montesinos-López, O. A., Montesinos-López, A., Crossa, J., Gianola, D., Hernández-Suárez, C. M., and Martín-Vallejo, J. (2018). Multi-trait, multi-environment deep learning modeling for genomic-enabled prediction of plant traits. Genes Genomes Genet. 8, 3829–3840. doi: 10.1534/g3.118.200728
Montesinos-López, O. A., Montesinos-López, A., Tuberosa, R., Maccaferri, M., Sciara, G., Ammar, K., et al. (2019b). Multi-trait, multi-environment genomic prediction of durum wheat with genomic best linear unbiased predictor and deep learning methods. Front. Plant Sci. 10:1311. doi: 10.3389/fpls.2019.01311
Moradpour, M., and Abdulah, S. N. A. (2020). CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol. J. 18, 32–44. doi: 10.1111/pbi.13232
Muraya, M. M., Chu, J., Zhao, Y., Junker, A., Klukas, C., Reif, J. C., et al. (2017). Genetic variation of growth dynamics in maize (Zea mays L.) revealed through automated non-invasive phenotyping. Plant J. 89, 366–380. doi: 10.1111/tpj.13390
Naik, H. S., Zhang, J., Lofquist, A., Assefa, T., Sarkar, S., Ackerman, D., et al. (2017). A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 13:23. doi: 10.1186/s13007-017-0173-7
Nguyen, T. T., Slaughter, D. C., Max, N., Maloof, J. N., and Sinha, N. (2015). Structured light-based 3D reconstruction system for plants. Sensors 15, 18587–18612. doi: 10.3390/s150818587
Ogata, T., Ishizaki, T., Fujita, M., and Fujita, Y. (2020). CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 15:e0243376. doi: 10.1371/journal.pone.0243376
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
Pan, Z., Liu, M., Zhao, H., Tan, Z., Liang, K., Sun, Q., et al. (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 62, 1895–1909. doi: 10.1111/jipb.12982
Pantazi, X. E., Moshou, D., Alexandridis, T., Whetton, R. L., and Mouazen, A. M. (2016). Wheat yield prediction using machine learning and advanced sensing techniques. Comput. Electron. Agri. 121, 57–65. doi: 10.1016/j.compag.2015.11.018
Paulus, S., Schumann, H., Kuhlmann, H., and Léon, J. (2014). High-precision laser scanning system for capturing 3D plant architecture and analysing growth of cereal plants. Biosyst. Eng. 121, 1–11. doi: 10.1016/j.biosystemseng.2014.01.010
Pedersen, T. L., Nookaew, I., Wayne Ussery, D., and Månsson, M. (2017). PanViz: interactive visualization of the structure of functionally annotated pangenomes. Bioinformatics 33, 1081–1082. doi: 10.1093/bioinformatics/btw761
Pérez-Enciso, M., and Zingaretti, L. M. (2019). A guide on deep learning for complex trait genomic prediction. Genes 10:553. doi: 10.3390/genes10070553
Pingali, P. L. (2012). Green revolution: impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. U. S. A. 109, 12302–12308. doi: 10.1073/pnas.0912953109
Puchta, H. (2017). Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36, 1–8. doi: 10.1016/j.pbi.2016.11.011
Purugganan, M. D., and Fuller, D. Q. (2009). The nature of selection during plant domestication. Nature 457, 843–848. doi: 10.1038/nature07895
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Qin, L., Li, J., Wang, Q., Xu, Z., Sun, L., Alariqi, M., et al. (2020). High-efficient and precise base editing of C∙ G to T∙ A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 18, 45–56. doi: 10.1111/pbi.13168
Ramírez Gonzales, L., Shi, L., Bergonzi, S. B., Oortwijn, M., Franco-Zorrilla, J. M., Solano-Tavira, R., et al. (2021). Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 105, 855–869. doi: 10.1111/tpj.15093
Rasheed, A., Hao, Y., Xia, X., Khan, A., Xu, Y., Varshney, R. K., et al. (2017). Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant 10, 1047–1064. doi: 10.1016/j.molp.2017.06.008
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8:e66428. doi: 10.1371/journal.pone.0066428
Raza, A., Razzaq, A., Mehmood, S. S., Zou, X., Zhang, X., Lv, Y., et al. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8:34. doi: 10.3390/plants8020034
Razzaq, A., Sadia, B., Raza, A., Khalid Hameed, M., and Saleem, F. (2019a). Metabolomics: a way forward for crop improvement. Metabolites 9:303. doi: 10.3390/metabo9120303
Razzaq, A., Saleem, F., Kanwal, M., Mustafa, G., Yousaf, S., Imran Arshad, H. M., et al. (2019b). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci. 20:4045. doi: 10.3390/ijms20164045
Razzaq, A., Wani, S. H., Saleem, F., Yu, M., Zhou, M., and Shabala, S. (2021). Rewilding crops for climate resilience: economic analysis and de novo domestication strategies. J. Exp. Bot. erab276. doi: 10.1093/jxb/erab276
Ren, Q., Sretenovic, S., Liu, S., Tang, X., Huang, L., He, Y., et al. (2021). PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat. Plants 7, 25–33. doi: 10.1038/s41477-020-00827-4
Ren, Q., Zhong, Z., Wang, Y., You, Q., Li, Q., Yuan, M., et al. (2019). Bidirectional promoter-based CRISPR-Cas9 systems for plant genome editing. Front. Plant Sci. 10:1173. doi: 10.3389/fpls.2019.01173
Roitsch, T., Cabrera-Bosquet, L., Fournier, A., Ghamkhar, K., Jiménez-Berni, J., Pinto, F., et al. (2019). New sensors and data-driven approaches—a path to next generation phenomics. Plant Sci. 282, 2–10. doi: 10.1016/j.plantsci.2019.01.011
Santosh Kumar, V. V., Verma, R. K., Yadav, S. K., Yadav, P., Watts, A., Rao, M. V., et al. (2020). CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 26, 1099–1110. doi: 10.1007/s12298-020-00819-w
Saxena, R. K., Edwards, D., and Varshney, R. K. (2014). Structural variations in plant genomes. Brief. Funct. Genom. 13, 296–307. doi: 10.1093/bfgp/elu016
Schatz, M. C., Maron, L. G., Stein, J. C., Wences, A. H., Gurtowski, J., Biggers, E., et al. (2014). Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 15:506. doi: 10.1186/PREACCEPT-2784872521277375
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shakoor, N., Lee, S., and Mockler, T. C. (2017). High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 38, 184–192. doi: 10.1016/j.pbi.2017.05.006
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Si, X., Zhang, H., Wang, Y., Chen, K., and Gao, C. (2020). Manipulating gene translation in plants by CRISPR–Cas9-mediated genome editing of upstream open reading frames. Nat. Protocols 15, 338–363. doi: 10.1038/s41596-019-0238-3
Singh, A., Ganapathysubramanian, B., Singh, A. K., and Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124. doi: 10.1016/j.tplants.2015.10.015
Singh, A. K., Ganapathysubramanian, B., Sarkar, S., and Singh, A. (2018). Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 23, 883–898. doi: 10.1016/j.tplants.2018.07.004
Snipen, L., and Liland, K. H. (2015). micropan: an R-package for microbial pan-genomics. BMC Bioinformat. 16:79. doi: 10.1186/s12859-015-0517-0
Song, J.-M., Guan, Z., Hu, J., Guo, C., Yang, Z., Wang, S., et al. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Sretenovic, S., Yin, D., Levav, A., Selengut, J. D., Mount, S. M., and Qi, Y. (2020). Expanding plant genome editing scope by an engineered iSpyMacCas9 system targeting the a-rich PAM Sequences. Plant Commun. 2:100101. doi: 10.1016/j.xplc.2020.100101
Steinert, J., Schiml, S., Fauser, F., and Puchta, H. (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84, 1295–1305. doi: 10.1111/tpj.13078
Streich, J., Romero, J., Gazolla, J. G. F. M., Kainer, D., Cliff, A., Prates, E. T., et al. (2020). Can exascale computing and explainable artificial intelligence applied to plant biology deliver on the United Nations sustainable development goals? Curr. Opin. Biotechnol. 61, 217–225. doi: 10.1016/j.copbio.2020.01.010
Sun, C., Hu, Z., Zheng, T., Lu, K., Zhao, Y., Wang, W., et al. (2017). RPAN: rice pan-genome browser for~ 3000 rice genomes. Nucl. Acids Res. 45, 597–605. doi: 10.1093/nar/gkw958
Tahir Ul Qamar, M., Zhu, X., Xing, F., and Chen, L. L. (2019). ppsPCP: a plant presence/absence variants scanner and pan-genome construction pipeline. Bioinformatics 35, 4156–4158. doi: 10.1093/bioinformatics/btz168
Tao, Y., Zhao, X., Mace, E., Henry, R., and Jordan, D. (2019). Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 12, 156–169. doi: 10.1016/j.molp.2018.12.016
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Thalapati, S., Batchu, A. K., Neelamraju, S., and Ramanan, R. (2012). Os11Gsk gene from a wild rice, Oryza rufipogon improves yield in rice. Funct. Integr. Genom. 12, 277–289. doi: 10.1007/s10142-012-0265-4
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U. S. A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. doi: 10.1038/nature01014
Tong, H., and Nikoloski, Z. (2021). Machine learning approaches for crop improvement: leveraging phenotypic and genotypic big data. J. Plant Physiol. 257:153354. doi: 10.1016/j.jplph.2020.153354
Tóth, E., Czene, B. C., Kulcsár, P. I., Krausz, S. L., Tálas, A., Nyeste, A., et al. (2018). Mb-and FnCpf1 nucleases are active in mammalian cells: activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucl. Acids Res. 46, 10272–10285. doi: 10.1093/nar/gky815
Tran, M. T., Doan, D. T. H., Kim, J., Song, Y. J., Sung, Y. W., Das, S., et al. (2020). CRISPR/Cas9-based precise excision of SlHyPRP1 domain (s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2020, 1–13. doi: 10.1007/s00299-020-02622-z
Tranchant-Dubreuil, C., Rouard, M., and Sabot, F. (2018). Plant pangenome: impacts on phenotypes and evolution. Ann. Plant Rev. Online 2, 453–478. doi: 10.1002/9781119312994.apr0664
Ullah, F., Xu, Q., Zhao, Y., and Zhou, D. X. (2020). Histone deacetylase HDA710 controls salt tolerance by regulating ABA signaling in rice. J. Integr. Plant Biol. 63, 451–467. doi: 10.1111/jipb.13042
Unamba, C. I., Nag, A., and Sharma, R. K. (2015). Next generation sequencing technologies: the doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 6:1074. doi: 10.3389/fpls.2015.01074
Uranga, M., Aragonés, V., Selma, S., Vázquez-Vilar, M., Orzáez, D., and Daròs, J. A. (2021). Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 106, 555–565. doi: 10.1111/tpj.15164
van Bezouw, R. F., Keurentjes, J. J., Harbinson, J., and Aarts, M. G. (2019). Converging phenomics and genomics to study natural variation in plant photosynthetic efficiency. Plant J. 97, 112–133. doi: 10.1111/tpj.14190
Varshney, R. K. (2016). Exciting journey of 10 years from genomes to fields and markets: some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci. 242, 98–107. doi: 10.1016/j.plantsci.2015.09.009
Varshney, R. K., Bohra, A., Yu, J., Graner, A., Zhang, Q., and Sorrells, M. E. (2021). Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 26, 631–649. doi: 10.1016/j.tplants.2021.03.010
Varshney, R. K., Saxena, R. K., Upadhyaya, H. D., Khan, A. W., Yu, Y., Kim, C., et al. (2017). Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 49:1082. doi: 10.1038/ng.3872
Varshney, R. K., Singh, V. K., Kumar, A., Powell, W., and Sorrells, M. E. (2018). Can genomics deliver climate-change ready crops? Curr. Opin. Plant Biol. 45, 205–211. doi: 10.1016/j.pbi.2018.03.007
Voss-Fels, K., and Snowdon, R. J. (2016). Understanding and utilizing crop genome diversity via high-resolution genotyping. Plant Biotechnol. J. 14, 1086–1094. doi: 10.1111/pbi.12456
Voss-Fels, K. P., Cooper, M., and Hayes, B. J. (2019). Accelerating crop genetic gains with genomic selection. Theoret. Appl. Genet. 132, 669–686. doi: 10.1007/s00122-018-3270-8
Walter, A., Liebisch, F., and Hund, A. (2015). Plant phenotyping: from bean weighing to image analysis. Plant Methods 11, 1–11. doi: 10.1186/s13007-015-0056-8
Wang, B., Zhong, Z., Wang, X., Han, X., Yu, D., Wang, C., et al. (2020a). Knockout of the OsNAC006 transcription factor causes drought and heat sensitivity in rice. Int. J. Mol. Sci. 21:2288. doi: 10.3390/ijms21072288
Wang, S., Zong, Y., Lin, Q., Zhang, H., Chai, Z., Zhang, D., et al. (2020b). Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC–Cas9. Nat. Biotechnol. 4, 1–6. doi: 10.1038/s41587-020-0566-4
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Warschefsky, E., Penmetsa, R. V., Cook, D. R., and Von Wettberg, E. J. (2014). Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 101, 1791–1800. doi: 10.3732/ajb.1400116
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M.-D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Wu, S., Wen, W., Wang, Y., Fan, J., Wang, C., Gou, W., et al. (2020). MVS-Pheno: a portable and low-cost phenotyping platform for maize shoots using multiview stereo 3D reconstruction. Plant Phenom. 2020, 1–17. doi: 10.34133/2020/1848437
Xiong, X., Yu, L., Yang, W., Liu, M., Jiang, N., Wu, D., et al. (2017). A high-throughput stereo-imaging system for quantifying rape leaf traits during the seedling stage. Plant Methods 13:7. doi: 10.1186/s13007-017-0157-7
Xu, R., Li, J., Liu, X., Shan, T., Qin, R., and Wei, P. (2020a). Development of plant prime-editing systems for precise genome editing. Plant Commun. 1:43. doi: 10.1016/j.xplc.2020.100043
Xu, W., Zhang, C., Yang, Y., Zhao, S., Kang, G., He, X., et al. (2020b). Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 13, 675–678. doi: 10.1016/j.molp.2020.03.012
Xue, Y., and Acar, M. (2018). Live-cell imaging of chromatin condensation dynamics by CRISPR. Iscience 4, 216–235. doi: 10.1016/j.isci.2018.06.001
Yamano, T., Nishimasu, H., Zetsche, B., Hirano, H., Slaymaker, I. M., Li, Y., et al. (2016). Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962. doi: 10.1016/j.cell.2016.04.003
Yang, W., Feng, H., Zhang, X., Zhang, J., Doonan, J. H., Batchelor, W. D., et al. (2020). Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol. Plant 13, 187–214. doi: 10.1016/j.molp.2020.01.008
Yu, H., Lin, T., Meng, X., Du, H., Zhang, J., Liu, G., et al. (2021). A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156–1170. doi: 10.1016/j.cell.2021.01.013
Yu, J., Golicz, A. A., Lu, K., Dossa, K., Zhang, Y., Chen, J., et al. (2019). Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol. J. 17, 881–892. doi: 10.1111/pbi.13022
Zafar, K., Khan, M. Z., Amin, I., Mukhtar, Z., Yasmin, S., Arif, M., et al. (2020). Precise CRISPR-Cas9 mediated genome editing in super basmati rice for resistance against bacterial blight by targeting the major susceptibility gene. Front. Plant Sci. 11:575. doi: 10.3389/fpls.2020.00575
Zavala, J. A., Casteel, C. L., DeLucia, E. H., and Berenbaum, M. R. (2008). Anthropogenic increase in carbon dioxide compromises plant defense against invasive insects. Proc. Natl. Acad. Sci. U. S. A. 105, 5129–5133. doi: 10.1073/pnas.0800568105
Zeng, X., Luo, Y., Vu, N. T. Q., Shen, S., Xia, K., and Zhang, M. (2020). CRISPR/Cas9-mediated mutation of OsSWEET14 in rice cv. Zhonghua11 confers resistance to Xanthomonas oryzae pv. oryzae without yield penalty. BMC Plant Biol. 20, 1–11. doi: 10.1186/s12870-020-02524-y
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breeding 39, 1–10. doi: 10.1007/s11032-019-0954-y
Zhang, J., Naik, H. S., Assefa, T., Sarkar, S., Reddy, R. C., Singh, A., et al. (2017). Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 7, 1–11. doi: 10.1038/srep44048
Zhang, P., Du, H., Wang, J., Pu, Y., Yang, C., Yan, R., et al. (2020). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. doi: 10.1111/pbi.13302
Zhao, J., Bayer, P. E., Ruperao, P., Saxena, R. K., Khan, A. W., Golicz, A. A., et al. (2020). Trait associations in the pangenome of pigeon pea (Cajanus cajan). Plant Biotechnol. J. 18, 1946–1954. doi: 10.1111/pbi.13354
Zhao, J., Bodner, G., and Rewald, B. (2016). Phenotyping: using machine learning for improved pairwise genotype classification based on root traits. Front. Plant Sci. 7:1864. doi: 10.3389/fpls.2016.01864
Zhao, Q., Feng, Q., Lu, H., Li, Y., Wang, A., Tian, Q., et al. (2018a). Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284. doi: 10.1038/s41588-018-0041-z
Zhao, Y., Jia, X., Yang, J., Ling, Y., Zhang, Z., Yu, J., et al. (2014). PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30, 1297–1299. doi: 10.1093/bioinformatics/btu017
Zhao, Y., Sun, C., Zhao, D., Zhang, Y., You, Y., Jia, X., et al. (2018b). PGAP-X: extension on pan-genome analysis pipeline. BMC Genom. 19, 115–124. doi: 10.1186/s12864-017-4337-7
Zhao, Y., Wu, J., Yang, J., Sun, S., Xiao, J., and Yu, J. (2012). PGAP: pan-genomes analysis pipeline. Bioinformatics 28, 416–418. doi: 10.1093/bioinformatics/btr655
Zhong, Z., Sretenovic, S., Ren, Q., Yang, L., Bao, Y., Qi, C., et al. (2019). Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 12, 1027–1036. doi: 10.1016/j.molp.2019.03.011
Zhong, Z., Zhang, Y., You, Q., Tang, X., Ren, Q., Liu, S., et al. (2018). Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol. Plant 11, 999–1002. doi: 10.1016/j.molp.2018.03.008
Keywords: food security, climate change, next-generation breeding, genomics, genome editing, CRISPR/Cas, abiotic stress, crop improvement
Citation: Razzaq A, Kaur P, Akhter N, Wani SH and Saleem F (2021) Next-Generation Breeding Strategies for Climate-Ready Crops. Front. Plant Sci. 12:620420. doi: 10.3389/fpls.2021.620420
Received: 22 October 2020; Accepted: 14 June 2021;
Published: 21 July 2021.
Edited by:
James Lloyd, Stellenbosch University, South AfricaReviewed by:
Atanas Ivanov Atanassov, Joint Genomic Center, BulgariaMehanathan Muthamilarasan, University of Hyderabad, India
Mahendar Thudi, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Manish Kumar Pandey, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Copyright © 2021 Razzaq, Kaur, Akhter, Wani and Saleem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fozia Saleem, fozia@ualberta.ca
†ORCID: Ali Razzaq orcid.org/0000-0002-9122-3714
Parwinder Kaur orcid.org/0000-0003-0201-0766
Shabir Hussain Wani orcid.org/0000-0002-7456-4090
Fozia Saleem orcid.org/0000-0002-9725-1329
 Ali Razzaq
Ali Razzaq Parwinder Kaur
Parwinder Kaur Naheed Akhter
Naheed Akhter Shabir Hussain Wani
Shabir Hussain Wani Fozia Saleem
Fozia Saleem