
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 February 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.619158
This article is part of the Research TopicTowards the Identification of Useful Genes for Prunus BreedingView all 6 articles
 Jun Cheng†
Jun Cheng† Jingjing Ma†
Jingjing Ma† Xianbo Zheng†
Xianbo Zheng† Honglin Lv
Honglin Lv Mengmeng Zhang
Mengmeng Zhang Bin Tan
Bin Tan Xia Ye
Xia Ye Wei Wang
Wei Wang Langlang Zhang
Langlang Zhang Zhiqian Li
Zhiqian Li Jidong Li
Jidong Li Jiancan Feng*
Jiancan Feng*Peach (Prunus persica L. Batsch) trees grow vigorously and are subject to intense pruning during orchard cultivation. Reducing the levels of endogenous gibberellins (GAs) represents an effective method for controlling branch growth. Gibberellin 2-oxidases (GA2oxs) deactivate bioactive GAs, but little is known about the GA2ox gene family in peach. In this study, we identified seven PpGA2ox genes in the peach genome, which were clustered into three subgroups: C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I. Overexpressing representative genes from the three subgroups, PpGA2ox-1, PpGA2ox-5, and PpGA2ox-2, in tobacco resulted in dwarf plants with shorter stems and smaller leaves than the wild type. An analysis of the GA metabolic profiles of the transgenic plants showed that PpGA2ox-5 (a member of subgroup C19-GA2ox-II) is simultaneously active against both C19-GAs and C20-GAs,which implied that C19-GA2ox-II enzymes represent intermediates of C19-GA2oxs and C20-GA2oxs. Exogenous GA3 treatment of shoot tips activated the expression of all seven PpGA2ox genes, with different response times: the C19-GA2ox genes were transcriptionally activated more rapidly than the C20-GA2ox genes. GA metabolic profile analysis suggested that C20-GA2ox depletes GA levels more broadly than C19-GA2ox. These results suggest that the PpGA2ox gene family is responsible for fine-tuning endogenous GA levels in peach. Our findings provide a theoretical basis for appropriately controlling the vigorous growth of peach trees.
Peach (Prunus persica L.) is one of the most widely cultivated deciduous fruit trees worldwide. According to FAOSTAT data, the global cultivation area of peach and nectarine reached 1712.4 ha in 2018. Peach trees must be pruned to keep the tree size manageable and to facilitate harvesting, but most cultivated peach varieties grow vigorously, increasing the labor costs required for orchard management. One effective way to inhibit the rapid growth of peach branches is to treat the trees with paclobutrazol (PBZ), an inhibitor of gibberellin (GA) biosynthesis, indicating that endogenous GA is at least one of the phytohormones that promote the vigorous growth of peach branches.
The biosynthesis of GA has been studied in many plant species, and is catalyzed by seven enzymes: ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA 13-oxidase (GA13ox), GA 20-oxidase (GA20ox), and GA 3-oxidase (GA3ox) (Yamaguchi, 2008). Because GA homeostasis is crucial for proper plant growth and development, several mechanisms have evolved to reduce the levels of bioactive GAs in plants (Varbanova et al., 2007; Gao et al., 2016). The main mechanism involves catabolism through the 2β-hydroxylation of active GA, a process catalyzed by GA 2-oxidase (GA2ox) (Rieu et al., 2008).
GA2oxs belong to the 2OG-Fe (II) oxygenase superfamily (Rieu et al., 2008). GA2oxs are encoded by small gene families, as revealed in plants such as Arabidopsis thaliana (Schomburg et al., 2003; Rieu et al., 2008), rice (Lo et al., 2008), grapevine (Giacomelli et al., 2013), and tomato (Serrani et al., 2007). In Arabidopsis, GA2ox genes are expressed throughout plant development and in different tissues (Rieu et al., 2008). The role of GA2oxs in reducing the level of bioactive GAs has been demonstrated in many species, including rice (Lo et al., 2008), poplar (Gou et al., 2011), spinach (Lee and Zeevaart, 2005), and switchgrass (Wuddineh et al., 2015).
The expression levels of GA2ox genes vary in response to environmental changes and phytohormones. In Arabidopsis, bioactive GA is deactivated in the hypocotyls of light-grown seedlings via the activation of GA2ox1 transcription (Achard et al., 2007). Blue light-mediated hypocotyl elongation is promoted by repressing the transcription of GA2ox1 and GA2ox8 (Wang et al., 2011). Low temperature deactivates GA by activating the transcription of GA2ox genes in rice (Wang et al., 2018). GA2ox6 genes are upregulated under water-limiting conditions (Dubois et al., 2013). GA2ox7 is upregulated by high salinity, which results in lower levels of active GAs (Magome et al., 2008). Finally, GA2oxs are significantly upregulated in response to treatment with exogenous GA3 (Thomas et al., 1999; Tan et al., 2018). Therefore, regulating the expression of GA2ox genes is a vital way to control the levels of endogenous GAs in response to environmental changes.
The biosynthesis of GA produces intermediates (GA12, GA15, GA24, GA9, GA53, GA44, GA19, and GA20) in addition to bioactive GAs (GA1 and GA4). GAs are classified into different groups based on their number of carbon (C) atoms, including C20-GAs (such as GA12, GA15, GA24, GA53, GA44, and GA19) and C19-GAs (such as GA9, GA20, GA1, and GA4). GA2ox proteins are classified as C20-GA2ox or C19-GA2ox based on their preference for C20-GA or C19-GA substrates, respectively. Phylogenetic analysis of GA2ox proteins further divided the C19-GA2ox group into two subgroups (Lee and Zeevaart, 2005; Lo et al., 2008; Wuddineh et al., 2015), suggesting that members of these two C19-GA2ox subgroups do not have exactly the same functions.
In the current study, we identified seven PpGA2ox genes in the peach genome. Phylogenetic analysis clustered these genes into three subgroups: C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I. The overexpression of representative PpGA2ox genes from the three subgroups in tobacco resulted in a typical dwarf phenotype, with shorter stems and smaller leaves than the wild type. PpGA2ox5, a member of the C19-GA2ox-II subgroup, encodes an enzyme with activity against C20-GAs and C19-GAs. Our findings suggest that C19-GA2ox-II enzymes might be intermediate forms that arose during the divergence of the C20-GA2ox and C19-GA2ox lineages. Exogenous GA3 treatment of peach branches activated the expression of all seven PpGA2ox genes but with different time courses. PpGA2ox subgroup C19-GA2ox-I members were the earliest to be upregulated by this treatment, while PpGA2ox genes in subgroups C19-GA2ox-II and C20-GA2ox-I were activated later.
We identified seven putative GA2ox genes in the peach genome: PpGA2ox1, PpGA2ox2, PpGA2ox3, PpGA2ox4, PpGA2ox5, PpGA2ox6, and PpGA2ox7. These genes are distributed on Chromosomes 1, 3, and 4 (Figure 1). All PpGA2ox genes contain three exons and two introns, which is similar to the structure of GA2ox family members in Arabidopsis (Han and Zhu, 2011). PpGA2ox1 and PpGA2ox2 on Chromosome 1 share 26% sequence identity and have opposite gene orientations. PpGA2ox4, PpGA2ox5, PpGA2ox6, and PpGA2ox7 are located on Chromosome 4. The pairs PpGA2ox5-PpGA2ox6 (65% identity) and PpGA2ox4-PpGA2ox7 (52% identity) share the highest sequence identity among the GA2ox genes (Supplementary Table S1). PpGA2ox4, PpGA2ox6, and PpGA2ox7 are oriented in the same direction, while PpGA2ox5 is oriented in the opposite direction. These results suggest that intricate patterns of gene duplication occurred during the evolution of the peach GA2ox gene family.
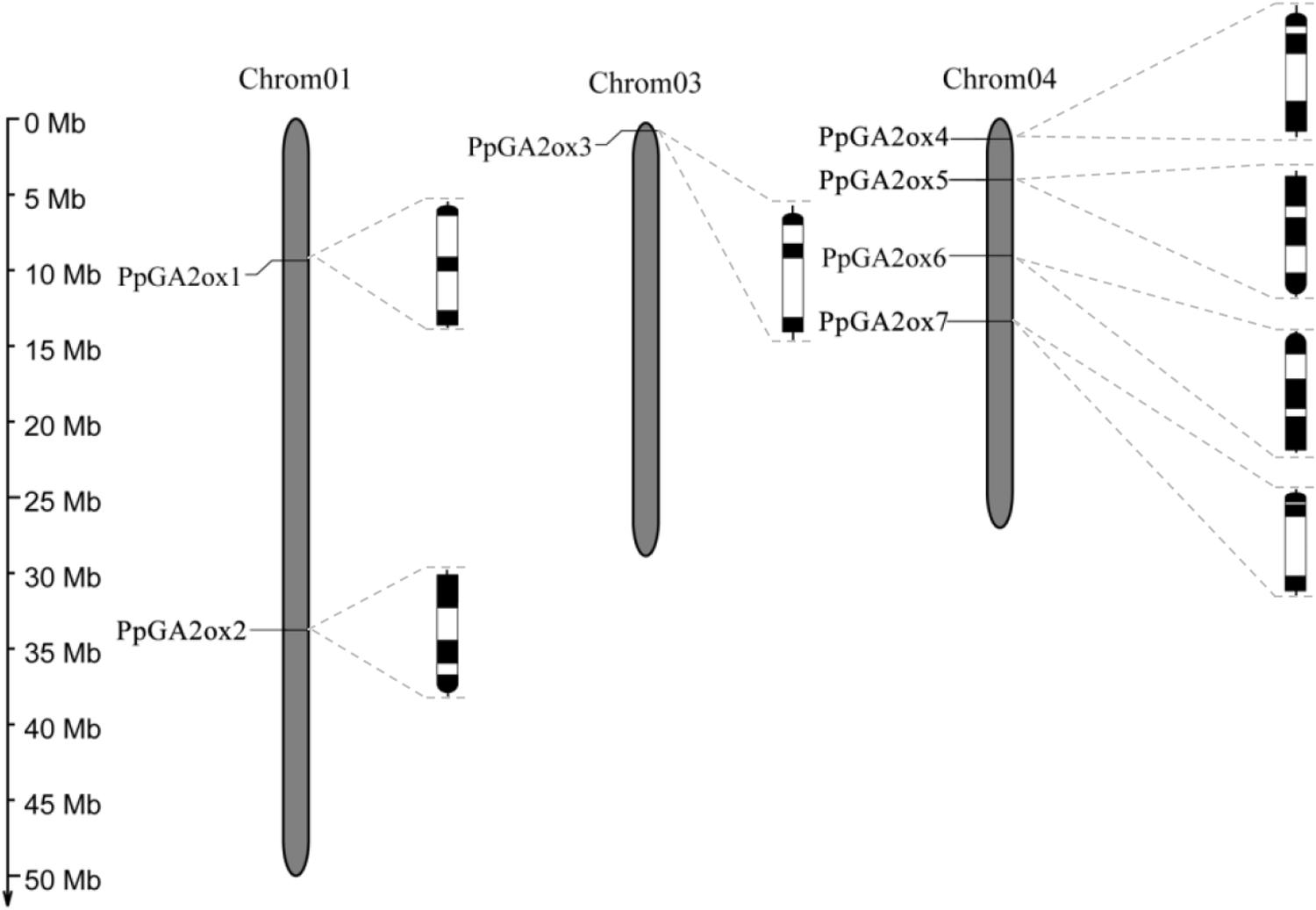
Figure 1. Chromosomal locations and genomic structures of PpGA2ox genes. Black boxes represent exons and white boxes represent introns. The semi-circle black boxes indicate the direction of the end of gene transcription.
Analysis of the conserved domains in the encoded enzymes revealed that the PpGA2oxs belong to the 2OG-Fe (II) oxygenase superfamily. All seven PpGA2oxs contain conserved amino acid residues that are thought to bind Fe2+ and 2-oxoglutarate (Supplementary Figure S1).
GA2oxs are usually active against C19-GAs (such as GA9, GA20, GA1, and GA4) or C20-GAs (such as GA12 and GA53), placing them into the C19-GA2ox or C20-GA2ox class (Rieu et al., 2008). To predict the functions of the PpGA2oxs, we collected reported GA2ox sequences from different plants and constructed a phylogenetic tree of the GA2ox family (Figure 2). The GA2ox family members fell into three groups: C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I. The reported members of C19-GA2ox-I and C19-GA2ox-II use C19-GAs as substrates, while the members in C20-GA2ox-I are mainly active against C20-GAs. PpGA2ox1 and PpGA2ox3 belong to subgroup C19-GA2ox-I; PpGA2ox5 and PpGA2ox6 belong to subgroup C19-GA2ox-II; and PpGA2ox2, PpGA2ox4, and PpGA2ox7 belong to subgroup C20-GA2ox-I. This clustering pattern predicts that PpGA2ox1, PpGA2ox3, PpGA2ox5, and PpGA2ox6 are active against C19-GAs, while PpGA2ox2, PpGA2ox4, and PpGA2ox7 are active against C20-GAs.
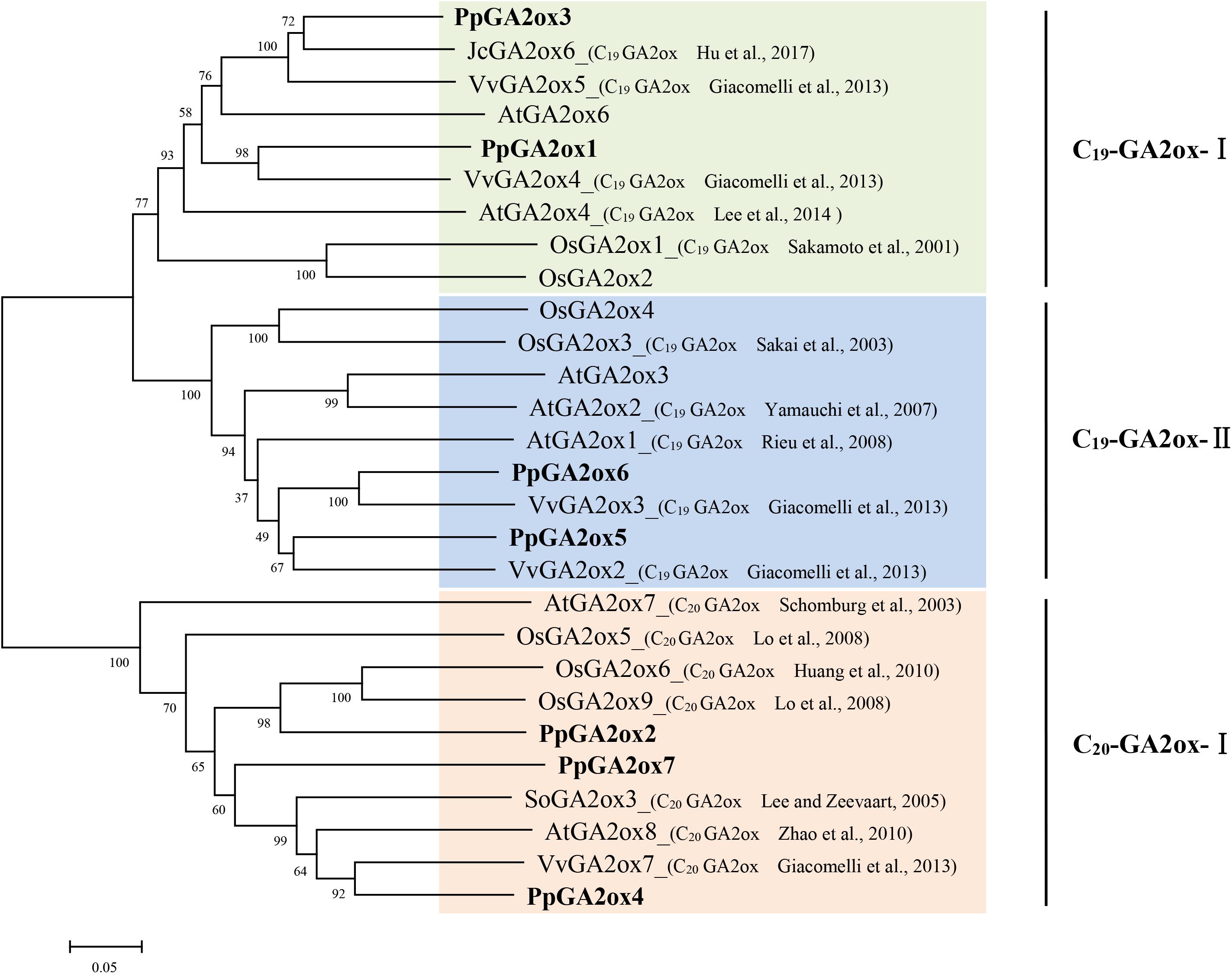
Figure 2. Phylogenetic tree of the predicted amino acid sequences of the GA2oxs from different species using MEGA5.1 program. The amino acid sequences formed three subgroup (C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I) and highlighted in different colors. Gene accession numbers of the sequences used in this tree are listed in Supplementary Table S2.
To determine whether the three subgroup division is conserved in other species, the reported GA2ox families from rice, grapevine, peach, tomato, and cucumber were selected and the number of GA2ox genes in each of the three subgroups was analyzed (Table 1). All species carry representatives of the C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I subgroups, suggesting that simultaneously possessing C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I genes is a highly conserved feature of both monocots and dicots.
The functional divergence of genes is a common occurrence during evolution. We constructed a phylogenetic tree based on GAox genes from liverwort (Marchantia polymorpha), the bryophyte Sphagnum fallax, and the lycophyte Selaginella moellendorffii and GA2ox genes from the flowering plants rice (Oryza sativa), peach (Prunus persica), and Arabidopsis thaliana (Supplementary Figure S2). The first three species represent basal clades, with Selaginella representing the earliest vascular plants. All GAoxs from M. polymorpha, Sph. fallax, and Sel. moellendorffii clustered with the C20-GA2oxs from monocots and dicots. This result implies that C20-GA2ox is a basal clade of the GA2ox family that possesses more ancient gene functions.
Motif analysis of GA2ox, GA20ox, and GA3ox proteins uncovered an amino acid sequence in the N-termini of these proteins that is the signature motif of GA2oxs (Han and Zhu, 2011), pointing to a close relationship between the signature motif and the functions of GA2oxs. To identify the possible cause of the functional divergence within the GA2ox family, we analyzed the signature motifs of the proteins in the C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I subgroups (Figure 3). The most highly conserved amino acids in C19-GA2ox-II proteins are the same as those in C19-GA2ox-I proteins, except for tryptophan (W; at position 16 in the wider multiple sequence alignment), which is highly conserved among C20-GA2ox-I proteins. This observation suggests that the C19-GA2ox-II subgroup is intermediate between the C19-GA2ox-I and C20-GA2ox-I subgroups.
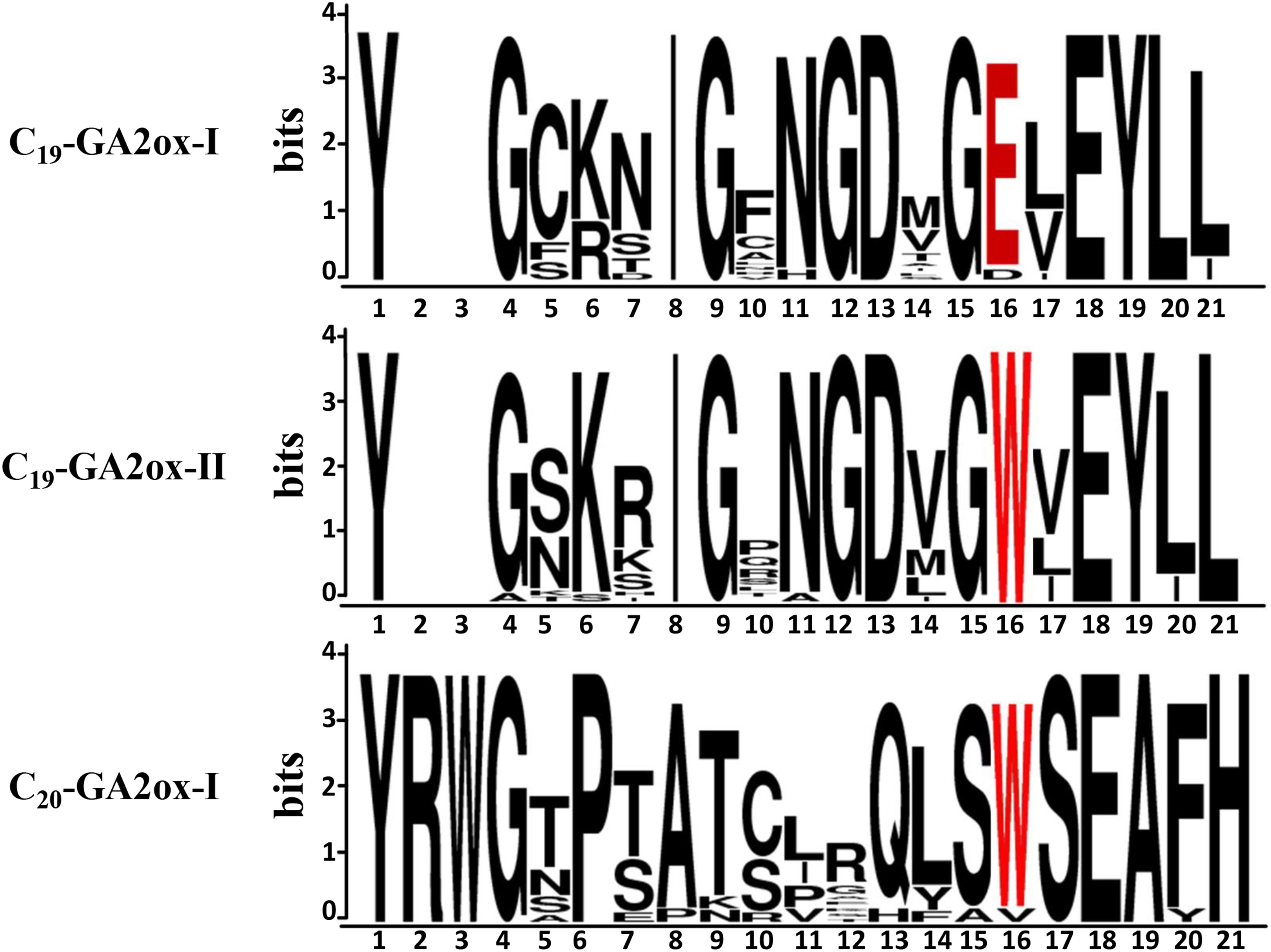
Figure 3. Graphical representation of the signature motif of GA 2-oxidases generated by WebLogo. Letter size is proportional to the degree of amino acid conservation. The 16th amino acid was highlighted in red font.
To investigate the expression profile of PpGA2ox genes in different tissues, we analyzed their transcript levels in six tissues (young fruit, flower bud, flower, young leaf, mature leaf, and shoot tip tissue) (Figure 4); PpTEF2 was used for normalization. PpGA2ox7 was expressed at the lowest level in all tissues analyzed compared to the six other PpGA2ox genes. PpGA2ox1 was expressed at higher levels in flowers and sepals than in other tissues. PpGA2ox2 and PpGA2ox4 were expressed at higher levels in sepals compared to other tissues. PpGA2ox1, PpGA2ox2, and PpGA2ox4 were expressed at relatively high levels in flower tissue. PpGA2ox3 and PpGA2ox5 were expressed at higher levels in young fruits compared to other tissues. PpGA2ox6 was expressed at lower levels in shoot tips compared to other tissues.
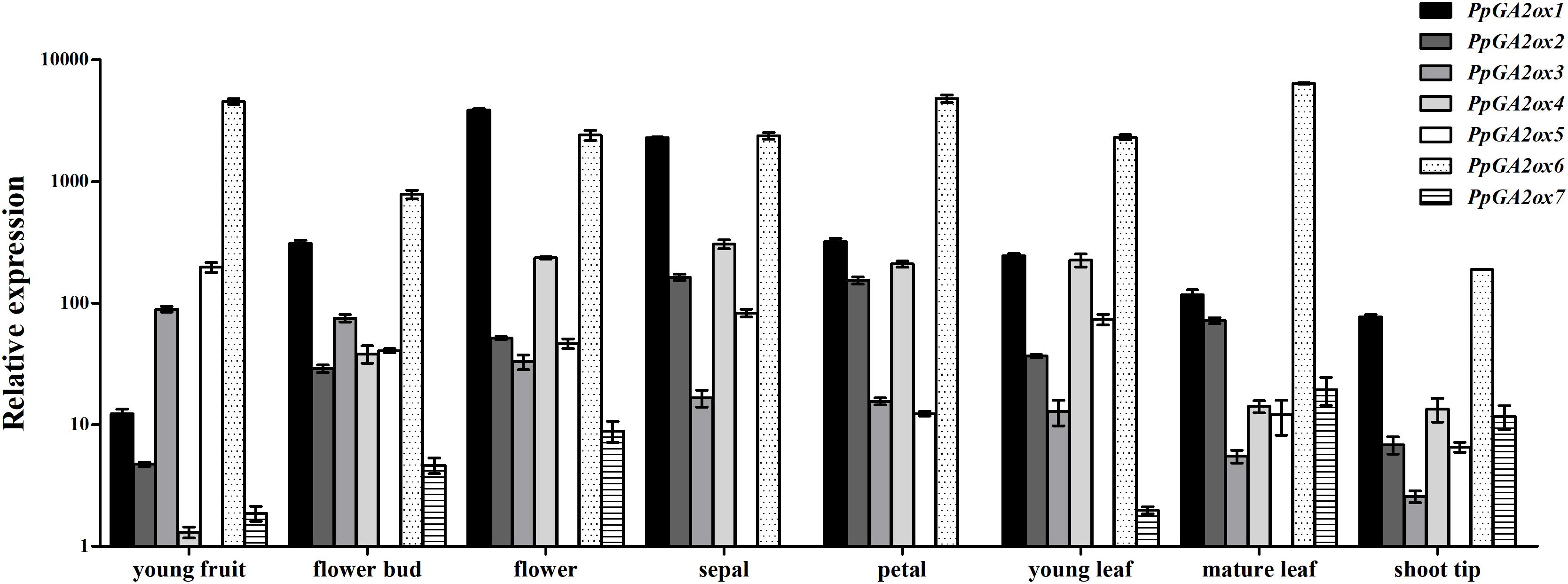
Figure 4. Expression of PpGA2ox genes in organs harvested from 3 year old “QMH” peach trees. Total RNA was isolated and analyzed by qRT-PCR, and the PpTEF2 was used as a reference gene. Results are plotted as the ratio to the lowest detected level (PpGA2ox7 in sepal). Note that the y-axis is on a logarithmic scale. All data are means of three replicates with error bars indicating standard deviation.
To clarify the function of a representative peach GA2ox gene from each of the three subgroups (C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I), we generated the 35S:PpGA2ox1, 35S:PpGA2ox5, and 35S:PpGA2ox2 constructs and introduced them into tobacco by Agrobacterium-mediated transformation. For each construct, more than three independent transgenic tobacco lines were obtained, which were confirmed by PCR and qRT-PCR (Supplementary Figure S3). All transgenic plants exhibited dwarf phenotypes compared to the non-transgenic tobacco control (WT) (Figure 5A). The heights of the transgenic plants were no more than 30% that of the WT (Figure 5B). In addition, the mature leaves of the transgenic plants were crinkled and smaller than those of the WT (Figures 5C,D). These results suggest that PpGA2ox1, PpGA2ox5, and PpGA2ox2 negatively regulate plant height and strongly affect leaf development.
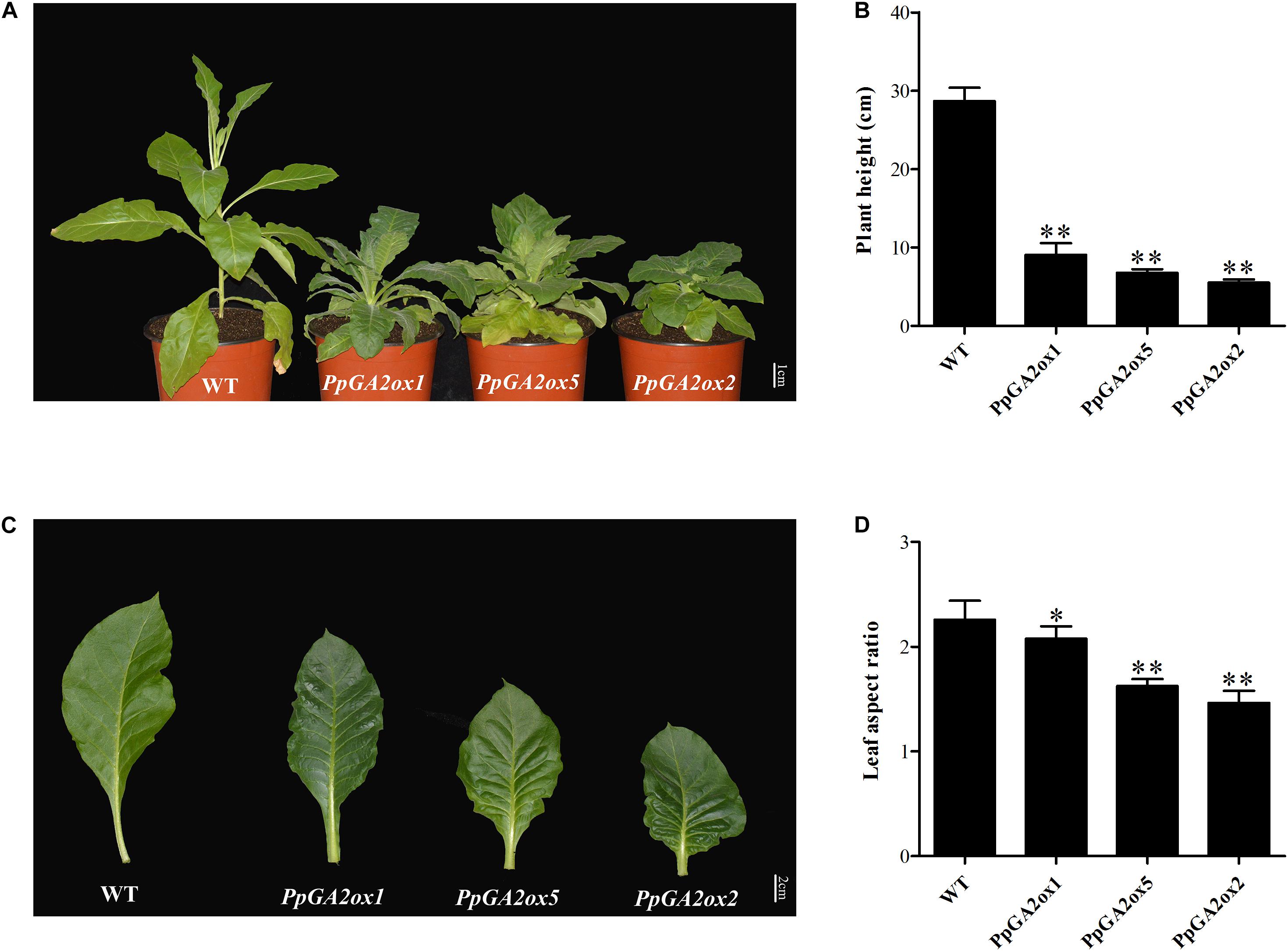
Figure 5. Ectopic expression of PpGA2ox1, PpGA2ox5, and PpGA2ox2 in tobacco induces dwarfism and smaller leaves. (A) Transgenic tobacco plants were grown in a climate chamber for 3 months. (B) Heights of the WT and transgenic tobacco plants. (C) Mature leaves of WT and transgenic tobacco. (D) Leaf aspect ratios of the WT and transgenic tobacco plants. The values are presented as the means ± standard deviation. ∗Significantly different from the wild type at P < 0.05. ∗∗Significantly different from the wild type at P < 0.01.
Based on our phylogenetic analysis (Figure 2), we predicted that PpGA2ox1 and PpGA2ox5 would be active against C19-GAs, while PpGA2ox2 would be active against C20-GAs. To elucidate the different metabolic functions of these enzymes, we analyzed the responses of the PpGA2ox1-OE, PpGA2ox5-OE, and PpGA2ox2-OE tobacco lines to exogenous GA1, a C19-GA (Figures 6A,B). GA3, which is resistant to enzyme-catalyzed degradation by GA2ox due to the presence of a double bond between C-2 and C-3 (Kanno et al., 2016), was used as a positive control. The wild type and all transgenic plants showed rapid growth after GA3 treatment. By contrast, after GA1 treatment, wild-type plants and plants overexpressing PpGA2ox2 showed rapid growth, whereas plants overexpressing PpGA2ox1 showed no significant difference in growth from the negative control (PpGA2ox1-OE with 0 mg/L GA1 treatment). These results strongly suggest that PpGA2ox1 is a C19-GA2ox and that PpGA2ox2 is a C20-GA2ox. Interestingly, plants overexpressing PpGA2ox5 showed an intermediate phenotype compared to the PpGA2ox1-OE and PpGA2ox2-OE lines. These results suggest that PpGA2ox5 is active against GA1. However, PpGA2ox5 is less efficient than PpGA2ox1, suggesting that PpGA2ox5 might not be a classical type of C19-GA2ox.
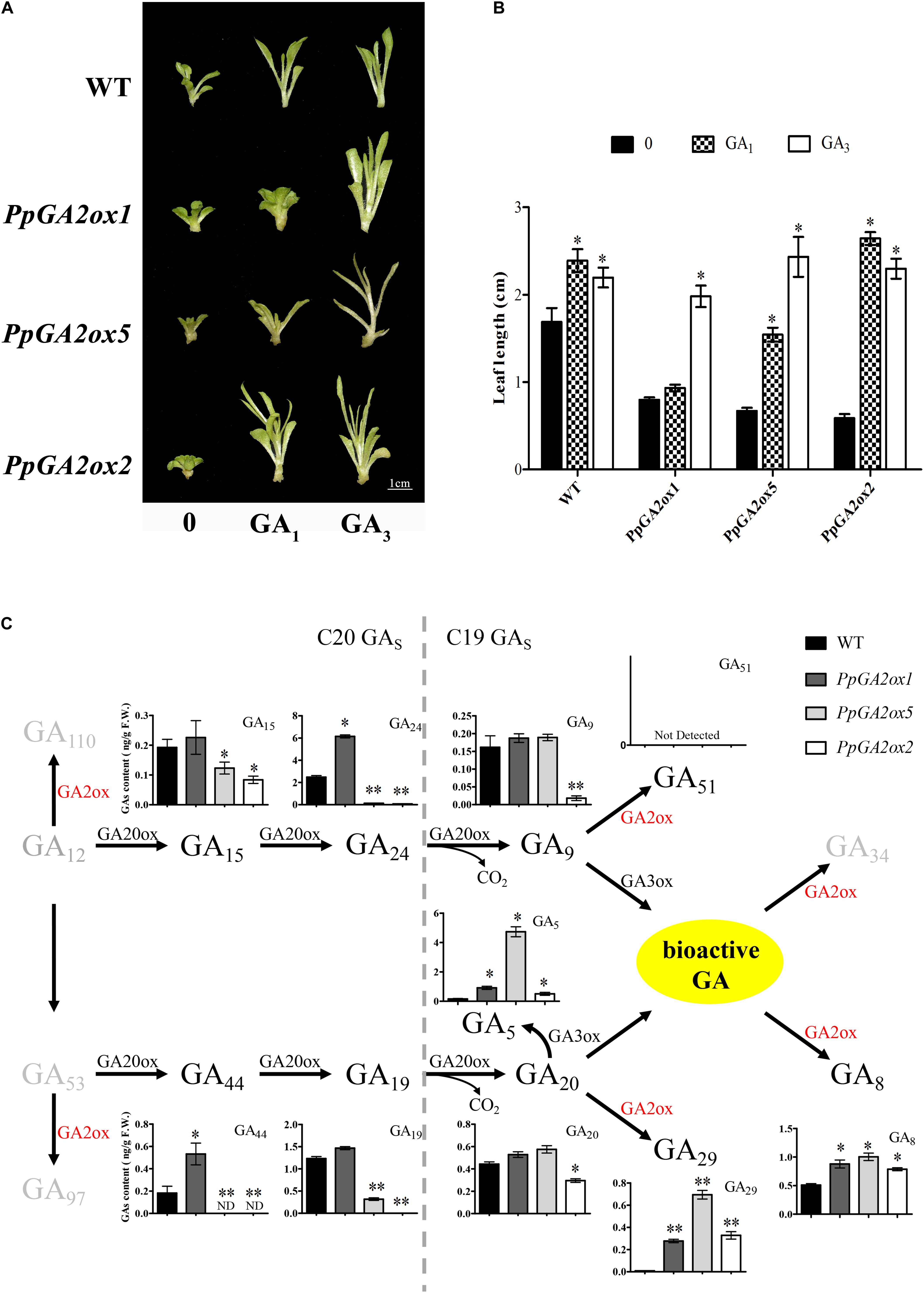
Figure 6. Analysis the function of PpGA2ox1, PpGA2ox5, and PpGA2ox2. (A,B) Effects of GA1 and GA3 on phenotypes of transgenic plants. Shoots cultivated in MS medium containing 0.4 or 0.0 mg/L GA1 or GA3. WT represents the non-transgenic plant. Bar represents 1 cm. (C) Effects of PpGA2ox1, PpGA2ox5, and PpGA2ox2 on the biosynthesis pathway of GA in tobacco measured using HPLC-MS/MS. The values are presented as the means ± standard error. ∗Significantly different from the wild type or untreated plant (P < 0.05). ∗∗Significantly different from the wild type (P < 0.01).
To determine how PpGA2ox5 may differ from PpGA2ox1 and PpGA2ox2, we analyzed the GA metabolic profiles in shoot tips with young leaves from wild-type tobacco plants and transgenic lines overexpressing PpGA2ox1, PpGA2ox2, or PpGA2ox5 (Figure 6C). We measured the contents of four C20-GAs (GA15, GA24, GA44, and GA19) and seven C19-GAs (GA5, GA8, GA9, GA20, GA29, GA34, and GA51) using HPLC-MS/MS. The levels of the four C20-GAs (GA15, GA24, GA4, and GA19) were higher in the PpGA2ox1-OE line and lower in the PpGA2ox5-OE and PpGA2ox2-OE lines vs. the wild type. GA24 and GA44 levels were significantly higher in the PpGA2ox1-OE line, whereas the levels of all four C20-GAs were significantly lower in the PpGA2ox5-OE and PpGA2ox2-OE lines compared to the wild type. Only PpGA2ox2-OE plants showed significantly lower levels of both C19-GAs (GA9 and GA20) compared to the wild type. GA8 and GA29 levels were significantly higher in all transgenic plants compared to the wild type. GA5 levels were significantly higher in all three transgenic lines vs. the wild type as well. Moreover, GA5 contents were 30.3-fold higher in PpGA2ox5-OE than in the wild type. GA12, GA110, GA97, GA51, and GA34 were not detected in any of the plants. These results suggest that PpGA2ox5 is active not only against C19-GAs, but also against C20-GAs.
Surprisingly, there were no differences in GA8 content among PpGA2ox1-OE, PpGA2ox5-OE, and PpGA2ox2-OE plants, but PpGA2ox1-OE plants showed higher activity against GA1 compared to PpGA2ox5-OE and PpGA2ox2-OE. The main bioactive GA in tobacco is GA4, whereas GA1 levels are low in this plant (Xiao et al., 2016). This paradoxical result could be due to the different concentrations of GA1 (endogenous vs. exogenous GA1) in the plants.
Increased levels of bioactive GA significantly activate the transcription of GA2oxs in Arabidopsis (Rieu et al., 2008). In the current study, overexpressing a C20-GA2ox reduced the levels of GA15, GA24, GA44, GA19, GA9, and GA20 in tobacco, while overexpressing a C19-GA2ox promoted the accumulation of GA24 and GA44 (Figure 6C). These findings suggest that C20-GA2oxs and C19-GA2oxs play different roles in regulating the levels of bioactive GAs. We therefore investigated the responses of C20-GA2ox and C19-GA2ox genes in shoot tips to treatment with GA3 for 0.5, 2, and 4 h (Figure 7). The seven PpGA2ox genes were divided into three types based on their transcriptional responses. The type 1 response, which was observed only for PpGA2ox3, was the most rapid response to GA3 treatment, with peak expression at 0.5 h followed by a reduction at 2 and 4 h after GA3 treatment. The type 2 response, which was observed for PpGA2ox1, PpGA2ox6, and PpGA2ox7, was characterized by peak transcript levels at 2 h after GA3 treatment that remained high at 4 h after treatment. The type 3 response to GA3, as observed for PpGA2ox2, PpGA2ox4, and PpGA2ox5, was slower, with gradually increasing transcript levels at the first two time points and a peak at 4 h. These results suggest that the transcription of the C19-GA2ox genes is activated more rapidly than the transcription of C20-GA2ox genes after GA3 treatment.
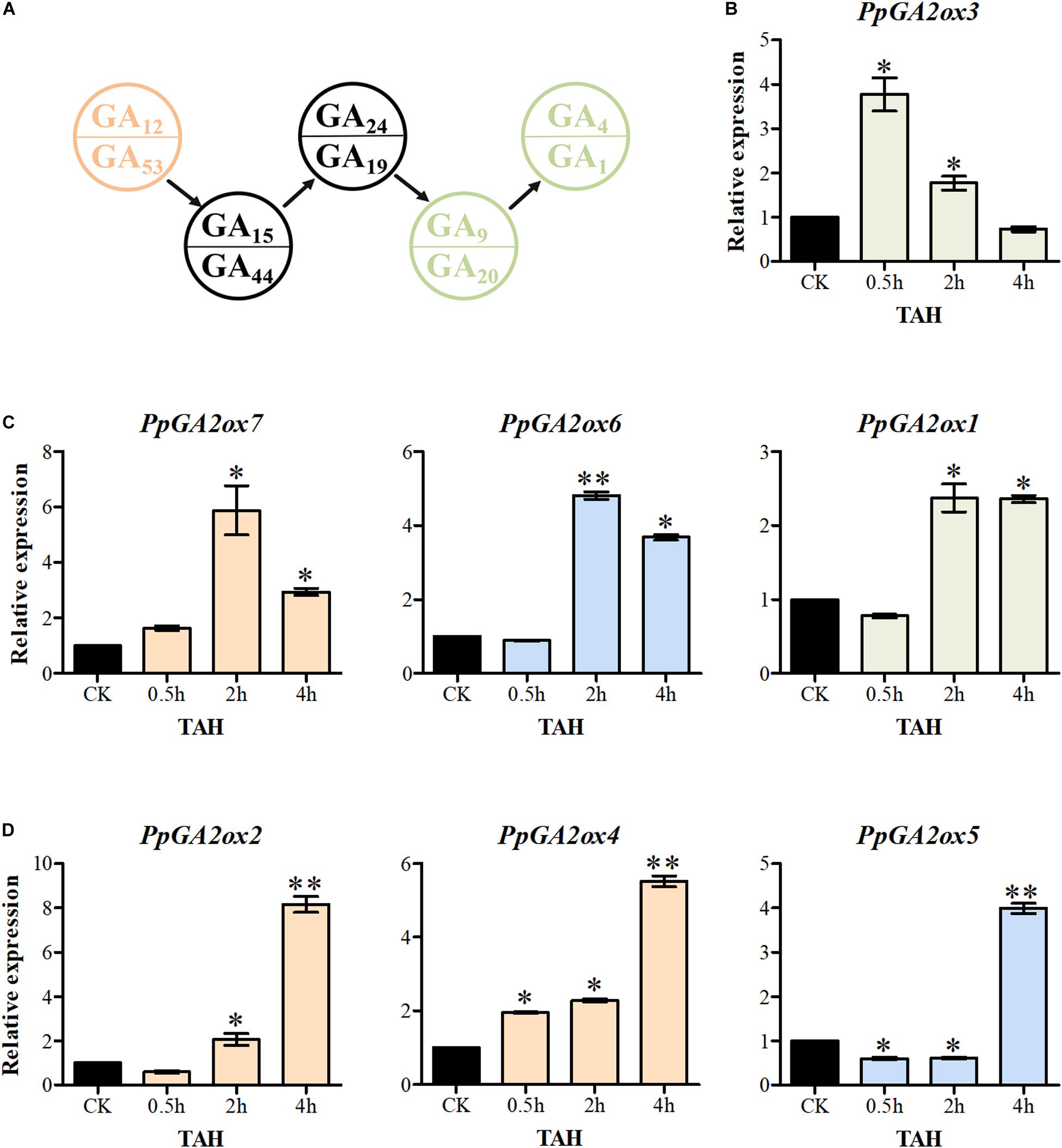
Figure 7. The transcription of PpGA2ox genes after GA3 treatment. (A) Part of GA biosynthesis way. Pink and green font represent the substrates for C20-GA2ox and C19-GA2ox, respectively. Seven PpGA2ox genes were divided into three type based on the response time to GA3 treatment. (B) The type 1 showed the most rapid response to GA3 treatment. (C) The type 2 reached to the peak transcript level at 2 h. (D) The type 3 reached to the peak transcript level at 4 h. Green, blue and pink columns represent the genes from the C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I subfamilies, respectively. TAH, treatment after hours. CK was the control treatment sprayed with distilled water. The values are presented as the means ± standard deviation. ∗Significantly different from the control (P < 0.05). ∗∗Significantly different from the control (P < 0.01).
GAs are important phytohormones that control diverse aspects of plant growth and development, including seed germination, stem elongation, leaf expansion, and flower and seed development (Yamaguchi, 2008). The homeostasis of bioactive GA levels is crucial for plant growth and development. GA2ox enzymes catalyze the deactivation of GA, thereby helping to maintain GA homeostasis. In the current study, we identified seven PpGA2ox genes in the peach genome. Our findings suggest that C19-GA2ox-II enzymes might be evolutionary intermediates between C20-GA2oxs and C19-GA2oxs and that the PpGA2ox family is responsible for maintaining the delicate balance of GA homeostasis.
The GA2 oxidase family is divided into three groups: C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I. These groups are highly conserved in monocots and dicots (Table 1), suggesting there are likely functional differences among the groups. Here, we demonstrated that PpGA2ox5 belongs to subfamily C19-GA2ox-II and is simultaneously active against both C20-GAs and C19-GAs. Tobacco plants overexpressing PpGA2ox5 contained lower levels of four C20-GAs (GA15, GA24, GA44, and GA19) than the wild type, which was similar to the results for PpGA2ox2-OE (a predicted C20-GA2ox) but opposite to those for of PpGA2ox1-OE (a predicted C19-GA2ox). These changes in C20-GA levels in the PpGA2ox5-OE lines were consistent with those in Arabidopsis c19-ga2ox and c20-ga2ox mutants (Magome et al., 2008; Rieu et al., 2008). In addition, PpGA2ox5-OE contained significantly higher levels of GA5 and GA29 than the wild type. These results suggest that PpGA2ox5-OE is active against C19-GAs.
PpGA2ox1-OE showed higher levels of GA15, GA24, GA44, and GA19 than the WT. Perhaps these plants exhibited increased GA 20-oxidase activity resulting from feedback regulation due to reduced levels of bioactive GAs. PpGA2ox2-OE also contained significantly higher levels of GA5 and GA29 than the wild type, suggesting that PpGA2ox2 also has activity against C19-GAs. While PpGA2ox2-OE plants had a dwarf phenotype, they grew rapidly after GA1 treatment. These results suggest that PpGA2ox2-OE might also have two activities against C20-GAs and C19-GAs, although its major activity is against C20-GAs. VvGA2ox7, a member of C20-GA2ox-I in grapevine, is also active against C19-GAs (GA9, GA20, GA4, and GA1) (Giacomelli et al., 2013).
Understanding the evolutionary relationships among gene subfamilies can uncover information about the course of functional divergence of genes. C19-GA2oxs, C20-GA2oxs, and GA biosynthetic oxidases (GA20ox and GA3ox) are all members of the 2OG-Fe (II) oxygenase superfamily (Han and Zhu, 2011). Although the evolutionary relationships among the C19-GA2ox, C20-GA2ox, GA20ox, and GA3ox subfamilies is unclear, the four subfamilies likely share a common ancestor (Giacomelli et al., 2013; Huang et al., 2015). Our results suggest that C20-GA2ox-I is an older clade of the GA2ox family compared to C19-GA2ox. Indeed, several studies have suggested that C19-GA2ox genes were derived from C20-GA2ox genes (Han and Zhu, 2011; Giacomelli et al., 2013; Huang et al., 2015). In an addition, only ent-kaurenoic acid, a gibberellin precursor, was detected in Physcomitrella patens, and ent-kaurenoic acid oxidase (KAO), GA20ox, and GA3ox were absent in this moss (Hirano et al., 2007; Miyazaki et al., 2018). ent-kaurenoic acid contains 20 carbons and can be converted into the bioactive GAs by KAO, GA20ox, and GA3ox. This observation suggested that C20-GA arose before C19-GA during evolution, implying that enzymes active against C20-GAs evolved earlier than enzymes active against C19-GAs.
Since PpGA2ox2 showed activity against C19-GAs, perhaps the ancient GA2ox was able to catalyze the 2β-hydroxylation of both C19-GAs and C20-GAs but was mainly active against C20-GAs and was only weakly active against C19-GAs. Perhaps once the ancient GA2ox evolved into two clades, one of the clades kept the original activity and formed the C20-GA2ox-I subfamily, while the other clade gradually gained increased activity against C19-GAs, resulting in the formation of the C19-GA2ox clade. The C19-GA2ox-II clade, containing PpGA2ox5, might have retained the ancient function of catalyzing the 2β-hydroxylation of C20-GAs (Figure 8).
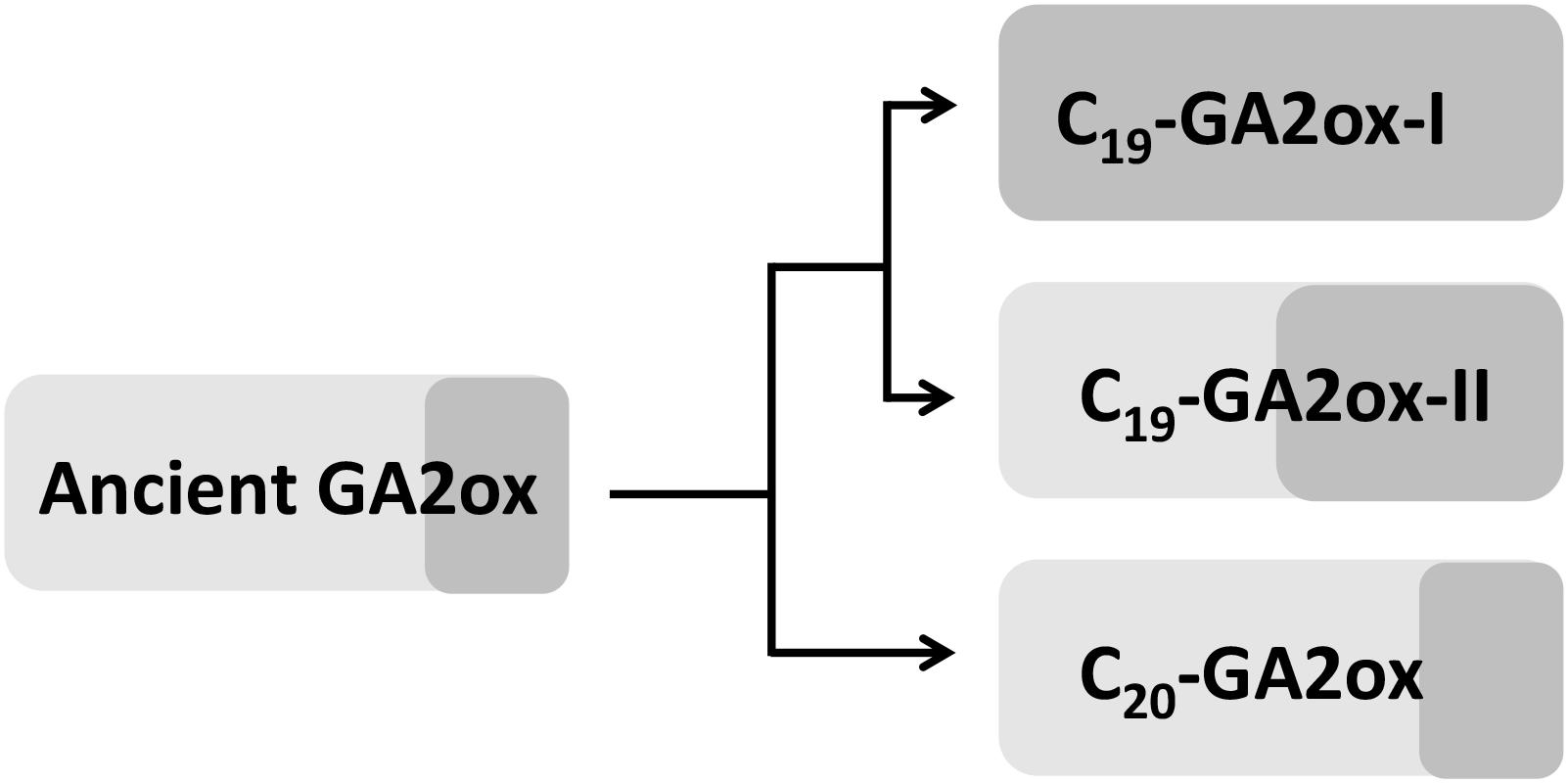
Figure 8. Hypothesis for the evolution of GA2 oxidases in higher plants. Light and dark gray box represents the activity of GA2ox against C20-GAs and C19-GAs, respectively. The area of box is proportional to the degree of enzyme activity.
Our analysis showed that the conserved tryptophan (W) in the signature motif of GA2ox is highly conserved in both C19-GA2ox-II and C20-GA2ox proteins, while a glutamic acid (E) is highly conserved in C19-GA2ox-I proteins. This tryptophan is also highly conserved in four clades of the 2OG-Fe (II) oxygenase superfamily (GA20ox, GA3ox, GAox-A, and GAox-C) (Huang et al., 2015). We speculate that this tryptophan is important for the ancient function of catalyzing the 2β-hydroxylation of the C20-GAs. More work is needed to verify this hypothesis.
Plants must cope with variable environmental conditions and are subjected to daily variations in light and temperature along with various biotic and abiotic stresses. Endogenous phytohormones help regulate plant growth and development and function in plant responses to the variable environment. GAs generally stimulate organ growth by enhancing cell elongation and cell division (Hedden and Thomas, 2012). The regulation of endogenous GA levels is important for regulating plant growth and development. The deactivation of GAs by GA2 oxidases is an important mechanism that influences GA responses. For example, bioactive GAs accumulate in the hypocotyls of dark-grown Arabidopsis seedlings and are deactivated in the hypocotyls of light-grown seedlings via the activated transcription of GA2ox1 (Achard et al., 2007). Low temperature (15°C) promotes GA deactivation by activating the transcription of GA2ox genes in rice (Wang et al., 2018). High salinity upregulates GA2ox7 in leaves, hypocotyls, and roots, resulting in reduced levels of active GAs (Magome et al., 2008). GA2ox7 is also upregulated by cold stress (Zhou et al., 2017).
In the current study, we treated tobacco shoot tips with a high concentration of GA3, which appeared to be responsible for increasing the expression of GA2ox genes over time. The different response times of different PpGA2ox genes coupled with the different GA targets of the subgroups within the GA2ox family (C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I) illustrate how these enzymes fine-tune GA levels. Three PpGA2ox genes, one each from the C19-GA2ox-I, C19-GA2ox-II, and C20-GA2ox-I subgroups, were significantly upregulated after GA3 treatment at similar time points, whereas one gene from C19-GA2ox-II and one from C20-GA2ox-I were significantly upregulated more slowly after GA3 treatment. After GA3 treatment, the C19-GA2oxs were activated first, followed by the C20-GA2oxs. Six C20-GAs and four C19-GAs are intermediates in the GA biosynthesis pathway. The C20-GA2oxs significantly depleted the C20-GAs and some of the C19-GAs, while the C19-GA2oxs significantly depleted the C19-GAs but increased the levels of some of the C20-GAs (Figure 6). Ultimately, the C20-GA2oxs were activated, thereby blocking GA biosynthesis. These findings indicate that the plants employed an active response to cope with high GA3 treatment. Together, these results suggest that the PpGA2ox family contributes to the fine-tuning of endogenous GA levels via the different response times and different enzymatic activities of the individual PpGA2 oxidases.
The peach cultivar used in this study, “QiuMiHong” (QMH), was provided by the Fruit Tree Germplasm Repository of Henan Agricultural University (Henan Province, China). For RNA extraction, young fruits, flower buds, flowers, sepals, petals, young leaves, mature leaves, and shoot tips were collected from 3 year old “QMH” trees in the spring, immediately frozen in liquid nitrogen, and stored at −80°C for subsequent experiments. Tobacco variety “K326” seedlings were grown in vitro in a temperature-controlled growth chamber at 24 ± 2°C under a 16 h light/8 h dark cycle. To calculate the leaf aspect ratio, a digital Vernier caliper was used to measure the distance from the bottom to the top of the leaf (length) and the maximum width of the leaf (width).
To identify GA2ox genes in the peach genome, a BlastP with default parameters of the Phytozome database1 was performed based on the conserved domain of genes in the 2-oxoglutarate-dependent oxygenase family using the amino acid sequences of seven Arabidopsis GA2oxs as queries. The search results were aligned by ClustalW algorithm with default parameters to reduce duplicate and redundant sequences (Larkin et al., 2007). Gene information from the JGI database was then used to predict the genomic positions of the PpGA2ox genes. The PpGA2ox genes were renamed, plotted on the peach chromosomes, and visualized with MapGene2 Chrom2 (Jiangtao et al., 2015).
The amino acid sequences of all GA oxidases used in this study were downloaded from the Phytozome and NCBI databases3. Sequence logos were generated using the online WebLogo platform4 (Crooks et al., 2004). A phylogenetic tree was constructed using the neighbor-joining method (NJ) method in MEGA (version 5.1) (Tamura et al., 2011). The main parameters were set as follows: the model was set to “p-distance,” the GAPs was set to “pairwise deletion,” and bootstrapping was performed with 1000 replicates. Default parameters were used for all bioinformatics analyses, except for those specifically mentioned.
The full-length sequences of the PpGA2ox genes were cloned from RNA obtained from peach shoot tips using specific primers and ligated into the pEASY®-Blunt Vector (TransGen Biotech). PCR products that produced a single band on the gel were purified and sent to Sangon Biotech (Shanghai5) for sequencing. All primers used in this study are listed in Supplementary Table S3.
The full-length sequences of the PpGA2oxs were amplified from the pEASY® -Blunt Vector (TransGen Biotech) using specific primers containing restriction sites and inserted into the pSAK277 vector containing the CaMV35S promoter. The plant overexpression vectors 35S:PpGA2ox1, 35S:PpGA2ox5, and 35S:PpGA2ox2 were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform tobacco leaf discs as described previously (Horsch et al., 1985; Lee and Zeevaart, 2005). Briefly, the leaves of aseptic tobacco seedlings were cut into small pieces (∼1 cm × 1 cm), pre-cultured in Murashige and Skoog (MS) medium for 2 days, incubated with A. tumefaciens (OD600 = 0.6) harboring the chosen construct for 5 min, and transferred to MS medium. After 2 days of co-culture in the dark, the leaf discs were transferred to selection medium (MS, 1 mg/L 6-benzylaminopurine, 0.1 mg/L NAA, 50 mg/L kanamycin, and 400 mg/L cefotaxime). When shoots appeared, individual shoots were cut and transferred to root-inducing medium (MS, 0.1 mg/L NAA, 50 mg/L kanamycin, and 400 mg/L cefotaxime). After rooting, the plantlets were grown in pots in a greenhouse and used for phenotypic analysis and further experiments. A pSAK277-GFP vector was transformed into tobacco in our lab and the transgenic lines showed no dwarf phenotype and smaller leaves.
Total RNA was extracted from the samples using a Column-type Plant Total RNA Extraction and Purification Kit (Sangon Biotech. Shanghai, China). Purified total RNA (0.5–2 μg) was reverse transcribed into first-strand cDNA using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa Biotechnology, Beijing, China). The cDNA product was used for semiquantitative RT-PCR and qRT-PCR analysis. qRT-PCR analysis was conducted using SYBR Select Master Mix (Applied Biosystems, United States) according to the manufacturer’s protocol on an ABI PRISM 7500 FAST Sequence Detection System (Applied Biosystems, Madrid, CA, United States). All experiments were conducted in triplicate. To quantify relative transcript level, TEF2 (Peach EST database accession number: TC3544) and β-tubulin (accession number: U91564) were used for data normalization (Tong et al., 2009; Schmidt and Delaney, 2010).
One year old branches of “QMH” peach trees were sprayed with GA3 (100 mg/L) or with distilled water (as a control). Three biological replicates were performed for all treatments. Shoot tips were collected at 0.5, 2, and 4 h after treatment for RNA extraction to analyze GA2ox transcript levels.
To measure endogenous phytohormone contents, approximately 1.0 g of young leaf tissue was collected from transgenic and wild-type tobacco plants and frozen in liquid nitrogen. The concentrations of different GAs were measured by high-performance liquid chromatography (HPLC) with tandem mass spectrometry (MS/MS) as described previously (Cheng et al., 2019). Three biological replicates were used to measure the level of each hormone.
The shoots of transgenic tobacco were cultivated in MS medium containing 0.4 mg/L GA1 or 0.4 mg/L GA3. Changes in growth were observed 10 days after treatment. The lengths of 8–10 leaves were measured and averaged. The experiments were performed three times.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JF, XZ, and JC conceived and designed the experiments. JM, HL, and MZ performed the experiments. JF and JC wrote the manuscript. BT, XY, and WW performed the GA metabolic analysis. LZ, ZL, and JL revised the manuscript. All authors contributed to the article and approved the submitted version.
The work was conducted at the Henan Provincial Key Laboratory of Fruit and Cucurbit Biology and supported by the National Key Research and Development Program of China (2019YFD1000104), the Joint Funds of the National Natural Science Foundation of China (U1804114), and Innovation team project of Henan University (19IRTSTHN009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Anita K. Snyder for critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.619158/full#supplementary-material
Supplementary Figure 1 | Amino acid residues presumed to bind Fe at the active site (∗). Putative 2-oxoglutarate binding sites (▲) and the signature motif of GA2ox (Black overline). Two conserved domains DIOX_N (red overline) and 2OG-FeII_Oxy (green overline) were detected in all GA2ox proteins. Black shading indicates identity, magenta and cyan shading indicate that similar residues are greater than 75 and 50%, respectively.
Supplementary Figure 2 | Analysis the evolutionary relationship of C19-GA2ox-I, C19-GA2ox-II and C20-GA2ox-I subgroups. A phylogenetic tree of GA2ox and GAox. All GA2oxs were searched through the Phytozome database (https://phytozome.jgi.doe.gov). Gene accession numbers of the sequences used in this tree are listed in Supplementary Table S2. ▲ represents the GA oxidases from liverwort (Marchantia polymorpha), Sphagnum fallax, Selaginella moellendorffii; ● represents the GA2 oxidases from rice, peach and Arabidopsis thaliana.
Supplementary Figure 3 | qRT-PCR analysis of PpGA2ox-overexpressing transgenic and non-transgenic (WT) lines using RNA isolated from the tobacco young leaves. The expression levels of each gene were relative to Nt β-Tubulin. Bars represent mean values of three biological repeats indicated standard deviation. ∗∗∗Significantly different from the wild type (P < 0.001).
Supplementary Table 1 | Comparison of the deduced amino acid sequences between PpGA2ox proteins.
Supplementary Table 2 | The accession numbers of GAoxs used in this study.
Supplementary Table 3 | Primer used in this study.
Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. doi: 10.1104/pp.106.092254
Chen, S., Wang, X., Zhang, L., Lin, S., Liu, D., Wang, Q., et al. (2016). Identification and characterization of tomato gibberellin 2-oxidases (GA2oxs) and effects of fruit-specific SlGA2ox1 overexpression on fruit and seed growth and development. Hortic. Res. 3:16059. doi: 10.1038/hortres.2016.59
Cheng, J., Zhang, M., Tan, B., Jiang, Y., Zheng, X., Ye, X., et al. (2019). A single nucleotide mutation in GID1c disrupts its interaction with DELLA1 and causes a GA-insensitive dwarf phenotype in peach. Plant Biotechnol. J. 17, 1723–1735. doi: 10.1111/pbi.13094
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Gao, S., Fang, J., Xu, F., Wang, W., and Chu, C. (2016). Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 28, 680–695. doi: 10.1105/tpc.15.01021
Giacomelli, L., Rota-Stabelli, O., Masuero, D., Acheampong, A. K., Moretto, M., Caputi, L., et al. (2013). Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J. Exp. Bot. 64, 4403–4419. doi: 10.1093/jxb/ert251
Gou, J., Ma, C., Kadmiel, M., Gai, Y., Strauss, S., Jiang, X., et al. (2011). Tissue-specific expression of Populus C19 GA 2-oxidases differentially regulate above- and below-ground biomass growth through control of bioactive GA concentrations. New Phytol. 192, 626–639. doi: 10.1111/j.1469-8137.2011.03837.x
Han, F., and Zhu, B. (2011). Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene 473, 23–35. doi: 10.1016/j.gene.2010.10.010
Hedden, P., and Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hirano, K., Nakajima, M., Asano, K., Nishiyama, T., Sakakibara, H., Kojima, M., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19, 3058–3079.
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hu, Y. X., Tao, Y. B., and Xu, Z. F. (2017). Overexpression of Jatropha Gibberellin 2-oxidase 6 (JcGA2ox6) induces dwarfism and smaller leaves, flowers and fruits in Arabidopsis and Jatropha. Front. Plant Sci. 8:2103. doi: 10.3389/fpls.2017.02103
Huang, J., Tang, D., Shen, Y., Qin, B., Hong, L., You, A., et al. (2010). Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genomics 37, 23–36. doi: 10.1016/S1673-8527(09)60022-9
Huang, Y., Wang, X., Ge, S., and Rao, G. Y. (2015). Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evol. Biol. 15:207. doi: 10.1186/s12862-015-0490-2
Jiangtao, C., Yingzhen, K., Qian, W., Yuhe, S., Daping, G., Jing, L., et al. (2015). MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 37, 91–97. doi: 10.16288/j.yczz.2015.01.013
Kanno, Y., Oikawa, T., Chiba, Y., Ishimaru, Y., Shimizu, T., Sano, N., et al. (2016). AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 7:13245. doi: 10.1038/ncomms13245
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, D. H., Lee, I. C., Kim, K. J., Kim, D. S., Na, H. J., Lee, I. J., et al. (2014). Expression of gibberellin 2-oxidase 4 from Arabidopsis under the control of a senescence-associated promoter results in a dominant semi-dwarf plant with normal flowering. J. Plant Biol. 57, 106–116. doi: 10.1007/s12374-013-0528-1
Lee, D. J., and Zeevaart, J. A. (2005). Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 138, 243–254. doi: 10.1104/pp.104.056499
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618. doi: 10.1105/tpc.108.060913
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. Cell Mol. Biol. 56, 613–626. doi: 10.1111/j.1365-313X.2008.03627.x
Miyazaki, S., Hara, M., Ito, S., Tanaka, K., Asami, T., Hayashi, K., et al. (2018). An ancestral gibberellin in a moss physcomitrella patens. Mol. Plant 11, 1097–1100. doi: 10.1016/j.molp.2018.03.010
Pimenta Lange, M. J., Liebrandt, A., Arnold, L., Chmielewska, S. M., Felsberger, A., Freier, E., et al. (2013). Functional characterization of gibberellin oxidases from cucumber, Cucumis sativus L. Phytochemistry 90, 62–69. doi: 10.1016/j.phytochem.2013.02.006
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20, 2420–2436. doi: 10.1105/tpc.108.058818
Sakai, M., Sakamoto, T., Saito, T., Matsuoka, M., Tanaka, H., and Kobayashi, M. (2003). Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J. Plant Res. 116, 161–164. doi: 10.1007/s10265-003-0080-z
Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., et al. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125, 1508–1516. doi: 10.1104/pp.125.3.1508
Schmidt, G. W., and Delaney, S. K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics MGG 283, 233–241. doi: 10.1007/s00438-010-0511-1
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A., and Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. doi: 10.1105/tpc.005975
Serrani, J. C., Sanjuán, R., Ruiz-Rivero, O., Fos, M., and García-Martínez, J. L. (2007). Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 145, 246–257. doi: 10.1104/pp.107.098335
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, P. H., Zhang, L., Yin, S. X., and Teng, K. (2018). Heterologous expression of a novel poa pratensis gibberellin 2-oxidase gene, PpGA2ox, caused dwarfism, late flowering, and increased chlorophyll accumulation in Arabidopsis. Biol. Plant 62, 462–470. doi: 10.1007/s10535-018-0788-1
Thomas, S. G., Phillips, A. L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. U.S.A. 96, 4698–4703. doi: 10.1073/pnas.96.8.4698
Tong, Z., Gao, Z., Wang, F., Zhou, J., and Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:71. doi: 10.1186/1471-2199-10-71
Varbanova, M., Yamaguchi, S., Yang, Y., McKelvey, K., Hanada, A., Borochov, R., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45. doi: 10.1105/tpc.106.044602
Wang, Q., Zeng, J., Deng, K., Tu, X., Zhao, X., Tang, D., et al. (2011). DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 233, 13–23. doi: 10.1007/s00425-010-1274-y
Wang, Y., Cui, Y., Hu, G., Wang, X., Chen, H., Shi, Q., et al. (2018). Reduced bioactive gibberellin content in rice seeds under low temperature leads to decreased sugar consumption and low seed germination rates. Plant Physiol. Biochem. PPB 133, 1–10. doi: 10.1016/j.plaphy.2018.10.020
Wuddineh, W. A., Mazarei, M., Zhang, J., Poovaiah, C. R., Mann, D. G., Ziebell, A., et al. (2015). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 13, 636–647. doi: 10.1111/pbi.12287
Xiao, Z., Fu, R., Li, J., Fan, Z., and Yin, H. (2016). Overexpression of the gibberellin 2-oxidase gene from Camellia lipoensis induces dwarfism and smaller flowers in Nicotiana tabacum. Plant Mol. Biol. Reporter 34, 182–191. doi: 10.1007/s11105-015-0917-3
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48, 555–561. doi: 10.1093/pcp/pcm023
Zhao, X. Y., Zhu, D. F., Zhou, B., Peng, W. S., Lin, J. Z., Huang, X. Q., et al. (2010). Over-expression of the AtGA2ox8 gene decreases the biomass accumulation and lignification in rapeseed (Brassica napus L.). J. Zhejiang Univ. Sci. B 11, 471–481. doi: 10.1631/jzus.B1000161
Keywords: Prunus persica, peach, GA2ox, functional divergence, GA homeostasis
Citation: Cheng J, Ma J, Zheng X, Lv H, Zhang M, Tan B, Ye X, Wang W, Zhang L, Li Z, Li J and Feng J (2021) Functional Analysis of the Gibberellin 2-oxidase Gene Family in Peach. Front. Plant Sci. 12:619158. doi: 10.3389/fpls.2021.619158
Received: 20 October 2020; Accepted: 28 January 2021;
Published: 17 February 2021.
Edited by:
Carlos Romero, Polytechnic University of Valencia, SpainReviewed by:
Jonathan Elias Maldonado, Pontificia Universidad Católica de Chile, ChileCopyright © 2021 Cheng, Ma, Zheng, Lv, Zhang, Tan, Ye, Wang, Zhang, Li, Li and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiancan Feng, amNmZW5nQGhlbmF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.