
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 February 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.618984
This article is part of the Research Topic Genomics-enabled Triticeae Improvement View all 15 articles
The actin depolymerizing factor (ADF) gene family, which is conserved in eukaryotes, is important for plant development, growth, and stress responses. Cold stress restricts wheat growth, development, and distribution. However, genome-wide identification and functional analysis of the ADF family in wheat is limited. Further, because of the promising role of ADF genes in cold response, there is need for an understanding of the function of this family on wheat under cold stress. In this study, 25 ADF genes (TaADFs) were identified in the wheat genome and they are distributed on 15 chromosomes. The TaADF gene structures, duplication events, encoded conversed motifs, and cis-acting elements were investigated. Expression profiles derived from RNA-seq data and real-time quantitative PCR analysis revealed the tissue- and temporal-specific TaADF expression patterns. In addition, the expression levels of TaADF13/16/17/18/20/21/22 were significantly affected by cold acclimation or freezing conditions. Overexpression of TaADF16 increased the freezing tolerance of transgenic Arabidopsis, possibly because of enhanced ROS scavenging and changes to the osmotic regulation in cells. The expression levels of seven cold-responsive genes were up-regulated in the transgenic Arabidopsis plants, regardless of whether the plants were exposed to low temperature. These findings provide fundamental information about the wheat ADF genes and may help to elucidate the regulatory effects of the encoded proteins on plant development and responses to low-temperature stress.
Actin depolymerizing factor (ADF), which is a conserved protein family with a low molecular weight (15–22 kDa) (Staiger et al., 1997) in eukaryotic cells, is crucial for regulating the reorganization and rearrangement of the actin cytoskeleton (Maciver and Hussey, 2002). This protein was first identified in chick embryo brains (Bamburg et al., 1980), but it has since been detected in various plants (Gungabissoon et al., 1998; Smertenko et al., 1998; Dong et al., 2001; Wang et al., 2009; Fu et al., 2014). As a member of the actin-binding protein family, ADF remodels the actin cytoskeleton via the transition between F-actin and G-actin states (Du et al., 2016). The actin cytoskeleton influences cellular architecture as well as diverse processes, including the expression of cell polarity, cell expansion and division, intracellular transport, pathogen perception, abiotic stress responses, and signal transduction (Fowler and Quatrano, 1997; Drobak et al., 2004; Porter and Day, 2016; Allard et al., 2020).
Because of the various actin cytoskeleton functions affecting plant development and responses to stimuli, ADF proteins in higher plants are important for various cellular activities. For example, ADFs are involved in pollen development and pollen tube growth in Arabidopsis (Daher and Geitmann, 2012; Zheng et al., 2013), maize (Lopez et al., 1996), and tobacco (Chen et al., 2002). Previous studies revealed that a lack of AtADF9 results in early flowering (Burgos-Rivera et al., 2008) and knockout of AtADF4 increases the length of hypocotyls and epidermal cells (Henty et al., 2011). In cotton, GhADF1 is involved in fiber elongation and secondary cell wall formation (Wang et al., 2009). Additionally, the expression patterns of ADF genes appear to be tissue-specific. Earlier research indicated that OsADF2/4/5 are expressed in roots, stems, blades, sheaths, spikelets and seeds persistently, whereas OsADF9 expression is specific to spikelets at the heading stage in rice (Huang et al., 2012). The GhADF6/8 genes are preferentially expressed in petals, while GhADF7 is highly expressed in anthers of cotton (Li et al., 2010). Moreover, ADF proteins reportedly have a vital role in responses to abiotic and biotic stresses in Arabidopsis, wheat, rice, and other species (Ouellet et al., 2001; Huang et al., 2012; Fu et al., 2014; Sengupta et al., 2019). The overexpression of OsADF3 enhances the drought tolerance of transgenic Arabidopsis seedlings (Huang et al., 2012). The expression of wheat TaADF4 is induced by heat, but down-regulated by low temperatures or salt stress (Zhang et al., 2017). In contrast, TaADF3 expression is significantly up-regulated under cold conditions and water deficiency treatment, but is relatively un-affected by wounding or salt stress (Tang et al., 2016). Both TaADF4 and TaADF7 enhance the resistance of wheat plants to a Puccinia striiformis f. sp. tritici (Pst) infection, whereas TaADF3 has the opposite effect (Fu et al., 2014; Tang et al., 2016; Zhang et al., 2017).
Low-temperature stress is a key factor influencing the growth, yield, and distribution of wheat plants (Song et al., 2017). An exposure to low but nonfreezing temperatures (i.e., cold acclimation) is crucial for the freezing tolerance of over-wintering plants (Thomashow, 2001; Sung and Amasino, 2005). Although there are relatively few reports describing ADFs in wheat exposed to abiotic stress, two earlier investigations proved that ADF production is induced at low temperatures (Danyluk et al., 1996; Ouellet et al., 2001). During the cold acclimation process, the depolymerization of microtubules and actin filaments increases the fluidity of the plasma membrane (Danyluk et al., 1996). However, the molecular mechanism regulating the depolymerization of microtubules and actin filaments under cold conditions remains unknown. Thus, clarifying the relationship between ADF proteins and freezing tolerance response is warranted.
The development and application of genome sequencing technology has led to the identification of ADF genes in the genomes of several plant species, including rice, maize, tomato, and Arabidopsis (Feng et al., 2006; Khatun et al., 2016; Huang et al., 2020). The availability of a sequenced ‘Chinese Spring’ genome has helped to facilitate the genome-wide analysis of gene families in wheat (Hu et al., 2018; International Wheat Genome Sequencing Consortium (IWGSC), 2014; Liu et al., 2019; Zhou et al., 2019). However, to the best of our knowledge, there are no reports regarding the genome-wide identification and characterization of wheat ADF genes or the wheat ADF gene expression profiles in various tissues and in response to low-temperature stress. In this study, we identified 25 wheat ADF genes using the ‘Chinese Spring’ genome sequences (IWGSC, RefSeq V1.1), after which we analyzed the ADF gene structures and the encoded conserved motifs, determined the genomic locations and duplication events of the ADF genes, and predicted the putative cis-acting elements. Additionally, ADFs in Triticum dicoccoides, Hordeum vulgare, Triticum turgidum, Triticum urartu, and Aegilops tauschii, were identified and used along with the wheat ADFs to construct a phylogenetic tree. To further investigate the function of ADF in wheat, we analyzed the expression profiles of these genes in different tissues, as well as in response to cold stress. Furthermore, we examined the effects of TaADF16 overexpression on the freezing tolerance of Arabidopsis. The results of this study will enhance our understanding of the wheat ADF gene family and provide the basis for future investigations of ADFs in wheat.
The genome and protein sequences data of wheat were downloaded from Ensembl Plants database1. The actin-depolymerizing factor homology domain (ADF-H domain, with Pfam: PF00241) obtained from PFAM database2 was employed as a query for Hidden Markov Model (HMM) search using HMMER3.0 with a pre-defined threshold of E value ≤ 1e–10. The results obtained were used to construct a wheat-specific ADF HMM profile by hmm-build program, and the second HMM search was used to remove the redundant sequences among the identified ADF proteins with an E value ≤ 1e–10. After manual corrections applied as needed, the NCBI-CDD web server3, SMART database4 and Pfam database (see foot note 2) were used to further confirm the ADF_H domain in the putative ADF protein sequences. The biochemical parameters of TaADF proteins, such as isoelectric points (pI), molecular weights (MW), instability index (II), aliphatic index (AI) and calculated grand average of hydropathy index (GRAVY) of the putative ADF proteins were calculated using the ExPASy online protParam tool5 (Artimo et al., 2012). The prediction of subcellular location of the identified TaADFs were performed using Plant-mPLoc6 (Chou and Shen, 2010). Alignment analysis of TaADF proteins was performed using MEGA 7.0, and visualized by Jalview v2.11.1.3 (Waterhouse et al., 2009).
The phylogenetic tree was performed using the neighbor joining (NJ) method in MEGA 7.0, with 1,000 bootstrap replicates. Sequences of ADF proteins from select species were identified based on the corresponding genome (Supplementary Table 1) as described above. The accuracy of identified ADF was confirmed with Ensembl Plants1 and Uniport database7.
The exon/intron structures of TaADFs were constructed by gene structure display server (GSDS) program8 using the CDS and corresponding genomic sequences retrieved from the Ensemble plants database. Conserved motifs of TaADF discover were predicted using the Multiple Expectation Maximization for Motif Elication (MEME) 4.12.09, with the following parameters: maximum number of 20 motifs and optimum motif widths of 6-100 residues. The 1500 bp upstream of the transcription start site (−1) of all identified TaADF transcripts was extracted as promoter to predict cis-acting elements using Plant CARE10.
All the TaADF genes were mapped to wheat chromosomes based on physical location information from the database of wheat genome. The gene duplication in the wheat genome were analyzed with Multiple Collinearity Scan toolkit (MCScanX) (Wang et al., 2012) and visualized with Circos tool (Krzywinski et al., 2009). The calculation of ka and ks substitution of each duplicated TaADF genes were performed using KaKs_Calculator 2.0 (Wang et al., 2010). The syntenic maps between wheat and other species were constructed using the python version of MCScanX11.
The expression patterns of identified TaADF genes in different tissues and development stages were analyzed based on the publicly available wheat RNA-Seq datasets obtained from wheat eFP Browser12 (Ramírez-González et al., 2018). The expression levels were summarized as transcripts per million (TPM), and a heatmap of TaADF tissues-specific expression were conducted with R packages (Pheatmap and Stats).
For expression analysis of TaADFs of different tissues, seeds of wheat ‘Chinese Spring’ were grown under 20°C with a 12/12 h photoperiod/dark in glass dish for 15 day (three-leaf stage). Roots and leaves were collected from five seedlings. For low temperature treatment, the wheat seedlings of ‘Jing 411’ were cultivated in the incubator at 20°C with a 12/12 h photoperiod/dark period for 15 days until three-leaf stage (TL), which was followed with different temperature treatments: 4°C for cold acclimation (CA), 20°C for un-cold acclimation (UCA). After 28 days, the CA and UCA seedlings were exposed to −5°C for 1 day (cold acclimation and freezing, CAF; un-cold acclimation and freezing, UCAF). All the samples were immediately frozen in liquid nitrogen and stored at −80°C. Three biological replications were performed.
The samples were subjected to total RNA extraction using a Trizol Reagent (Invitrogen, Carlsbad, CA, United States). The analysis of RNA-seq for low temperature treatment was based on our previous study (Zhao et al., 2019b), we re-analyzed the data based on reference genome of ‘Chinese Spring’ genome (IWGSC: RefSeq V1.1). Sampled crowns from TL, CA, UCA, CAF, and UCAF were used for RNA-seq analysis. Using the DESeq R package, the differently expressed genes (DEGs, | log2 (fold change)| > 1 and p < 0.05) were analyzed.
cDNA products were subjected to RT-PCR analysis, in which, TaGAPDH and TaTEF1-α were used as double internal reference genes for wheat. RT-PCR was performed with BCS Wiz SYBR Green RT-PCR Master Mix and the QuantStudio 5 Real-time PCR system (Applied Biosystems, Malham, MA, United States). The following amplification protocol was used: first step, 95°C for 30 s; second step, 40 cycles of 95°C for 5 s and 60°C for 30 s; final step, 95°C for 15 s, 60°C for 1 min, 95°C for 15 s, and 50°C for 30 s. The relative expression were calculated with 2–ΔΔCt method. Three biological replications were performed (each biological replication for three technical). Specific primers used in this study are shown in Supplementary Table 2.
The coding sequence (CDS) of TaADF11, TaADF14, TaADF15, and TaADF16 was cloned into the pBWA(V)HS-ccdb-GLosgfp vector containing the cauliflower mosaic virus 35S (CaMV 35S) promoter, respectively. Subsequently, the control plasmid and fusion plasmids were transiently expressed in A. thaliana protoplasts. Then, the transformed protoplasts were incubated for 24 h at 22°C darkness. Finally, green fluorescent protein (GFP) fluorescence signals were observed using Nikon C2-ER confocal laser scanning confocal microscope. The specific primers containing the restriction site are shown in Supplementary Table 2.
The CDS of TaADF16 (Gene id: TraesCS5A02G478500), was amplified by PCR with gene-specific primer (F: 5′-GG AGAGGACACGCTCGAGATGACTTTATCTCGCCGACATG-3′ and R 5′-TTAAAGCAGGACTCTAGATTAGGTGGTGTAGT CCTTGAGGAT-3′) and then cloned into the pART-CAM vector controlled by the CaMV 35S promoter. The 35S:TaADF16 plasmid were transformed into Agrobacterium tumefaciens GV3101 and then transformed into Arabidopsis. Seeds of T0 transgenic plants were selected on MS medium containing 100 μg/ml kanamycin and further confirmed by PCR. T3 homozygous of Arabidopsis transgenic plants was used for freezing tolerance assay.
For freezing tolerance analysis in WT and OE lines, all the seeds were planted in plastic pots filled nutrient soil for three weeks, with 14/10 h photoperiod and temperature at 22°C. Three weeks old seedlings were transfered to −5°C for 4 h for freezing stress and ion leakage was determined. For recovery treatment, the plants after freezing were placed in the dark at 4°C for 12 h followed by 4 day at 22°C and the survival rates was determined. Photos were taken to record the growth phenotype before treatment and after recovery. Leaves of plants after 4°C for 24 h were collected for analysis of POD and SOD activities, MDA and soluble sugar content as described by Li et al. (2000). Seven cold responsive genes were selected for RT-PCR assay. AtTUB2 and AtUBQ10 of Arabidopsis was used as double internal reference gene. Three biological replications were carried out for each sample. The primers of the genes for RT-PCR are listed in Supplementary Table 2.
Data were statistically processed by the SPSS 25.0 and Graphpad Prism 8 software. The mean value ± standard deviation (SD) of at least three replicates for each sample are presented. Statistical significance was assessed by Student’s t-test between control and treatment.
A total of 25, 18, 8, 5, 12, and 11 non-redundant putative ADF genes were identified in wheat (TaADF1–TaADF25), T. dicoccoides (TdADF1–TdADF18), Ae. tauschii (AetADF1–AetADF8), T. urartu (TuADF1–TuADF5), T. turgidum (TtADF1–TtADF 12), and barley (HvADF1–HvADF11), respectively (Table 1 and Supplementary Table 3). Of the encoded TaADF proteins, TaADF4/8 and TaADF22 were, respectively, revealed as the shortest (132 amino acids) and longest (235 amino acids). The molecular weights (MW) of the TaADF proteins ranged from 15.30 to 25.91 kDa and the isoelectric points (pI) ranged from 4.39 to 8.74. The GRAVY values (<0) reflected the hydrophilicity of the TaADF proteins. An analysis of the instability index suggested that 16 proteins (64%) may be unstable (instability index > 40) and nine proteins (36%) are probably stable (instability index ranging from 30.91 to 38.53). The aliphatic index, which ranged from 62.52 to 79.72, indicated the thermal stability of TaADF proteins.
The results of the chromosomal localization and collinear analysis revealed that the TaADF genes are unevenly distributed on 15 chromosomes, with the number of TaADF genes on each chromosome ranging from 1 (1A, 1B, 1D, 4A, 4B, 4D, 6A, 6B, and 6D) to 4 (5A and 5D) (Figure 1A). Nine, seven, and nine TaADF genes were detected in the A, B, and D sub-genomes, respectively, implying some of the TaADF genes in the B sub-genome may be lost during evolution. These lost ADF genes in 5B may have a redundant function with the ADF genes on 5A or 5D, with a low purifying selection, they were finally lost during evolution. We found 7 homologous gene groups with a copy on each of A, B and D sub-genomes, and 2 gene pairs had two homologous genes on A and D sub-genomes. In addition, the homologous TaADF genes shared high protein sequence similarity, with a range of 92.5% (TaADF16-5A, TaADF18-5B, TaADF22-5D) to 100% (TaADF15-5A and TaADF19-5D) (Supplementary Table 4). Gene duplication events affected the TaADF genes on 15 chromosomes. Thirty-one segmental duplications (Figure 1B) and one tandem duplication (Figure 1A) were detected. The rates of non-synonymous (Ka) and synonymous (Ks) nucleotide substitutions were calculated to evaluate the selection pressure on the TaADF gene duplication events (Supplementary Table 5). The Ka/Ks ratios for the 32 duplicated pairs were less than 1.00, implying the wheat ADF genes evolved under strong purifying selection.
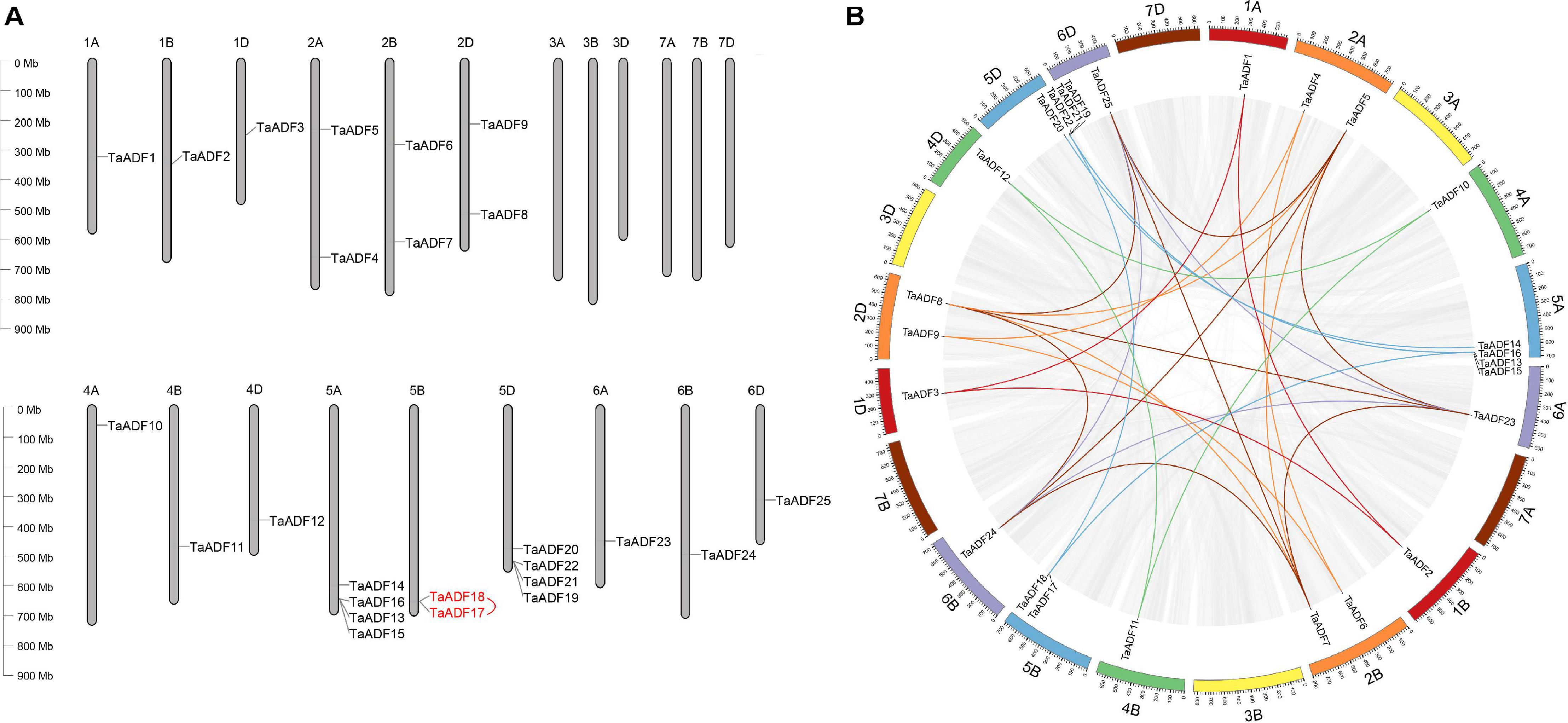
Figure 1. Chromosomal location and duplication events of ADF in wheat. (A) Chromosomal location of ADF genes in wheat. Red line between genes indicate tandem duplication of TaADF genes. (B) Segmental duplication in wheat genome. Gray lines indicate all synteny blocks in the wheat genome, and the different colors represent seven homeologous group of wheat chromosomes. Homologous genes of each group are linked by lines with corresponding color.
To investigate the evolutionary relationships and characteristics of the ADF genes, 117 ADF proteins from wheat and other species were used to construct a phylogenetic tree (Figure 2 and Supplementary Table 3). The evolutionary relationships between wheat and eight other species were determined (Figure 2A). The phylogenetic tree revealed that the ADF genes can be classified into four main groups, with each clade consisting of 15–56 members (Figure 2B and Supplementary Table 6). More specifically, 3, 5, 6, and 11 TaADF are clustered in Groups I, II, III, and IV, respectively.
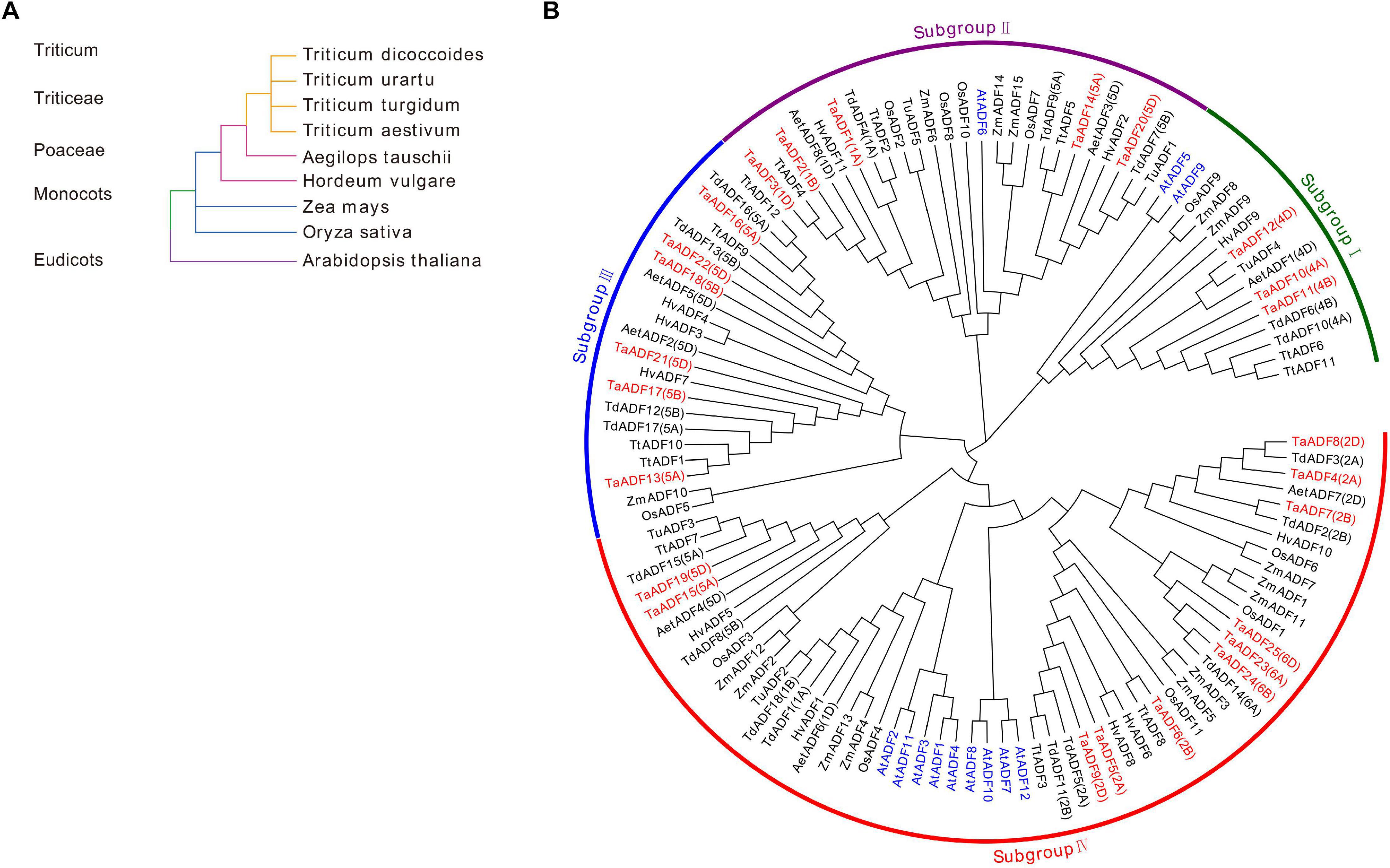
Figure 2. Systematic evolutionary relationships of ADF in wheat and eight other species. (A) Evolution relationship among the nine species; (B) Phylogenetic tree of ADF family members in wheat and eight other species. ADF proteins in Triticum aestivum. L., Oryza sativa, Arabidopsis thaliana, Zea mays, Hordeum vulgare, Triticum turgidum, Triticum urartu, Triticum dicoccoides and Aegilops tauschii are prefixed by Ta, Os, At, Zm, Hv, Tt, Tu, Td, and Aet, respectively. ADF proteins in wheat and Arabidopsis are marked in red and blue, respectively. The chromosome locations of ADF in Triticum aestivum. L, Triticum turgidum, and Aegilops tauschii were provided in the bracket followed with each gene name.
To more thoroughly determine the phylogenetic mechanisms of TaADF genes, we examined the synteny between wheat and other four gramineous species, including Ae. tauschii, T. dicoccoides, barley, and rice. A total of 16, 26, 18, and 22 orthologous gene pairs between hexaploid wheat (T. aestivum) and other species (Ae. tauschii, T. dicoccoides, barley, and rice) were identified (Figure 3 and Supplementary Table 7). Some collinear pairs (with eight TaADF genes) were identified in all of the four syntenic maps, suggesting that these orthologous pairs were relatively well conserved during the evolution of gramineous species. Each of ADF genes in 2A, 2B, 4A, 4B of tetraploid wheat showed synteny to several ones in hexaploid wheat. However, some orthologous gene pairs were only identified between chromosome 5A (or 5B) of T. dicoccoides and 5A of hexaploid wheat, but not 5B of hexaploid wheat, which may be due either to the quality of the genome assembly or the gene deletion or chromosomal recombination during evolution and polyploidization.
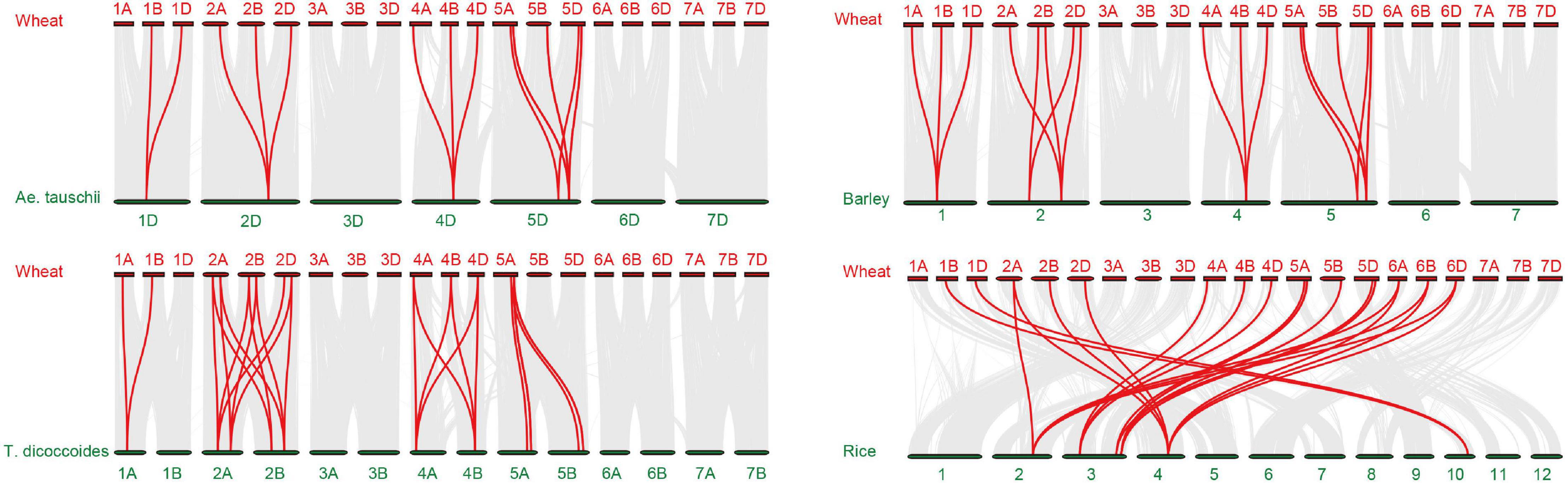
Figure 3. Synteny analysis of ADF genes between wheat and other four species. Gray lines in the background and red lines between different species indicate the collinear blocks and syntenic ADF pairs between wheat and other species, respectively.
The TaADF amino acid sequences were aligned and a phylogenetic tree was constructed using the MEGA 7.0 program (Figure 4A). A structural analysis of the TaADF genes indicated that 19 TaADF genes have three exons, whereas the remaining genes have two exons (Figure 4B). Additionally, 19 TaADF genes consist of a 150-bp exon at the C-terminus and a second exon comprising 247–289 bp. Thus, the exon lengths and positions appear to be highly conserved in wheat TaADF genes. A total of 14 conversed motifs were identified (motifs 1–14) (Figures 4C,D). Motifs 1, 2, and 5 are three conserved regions that form the ADF-H domain in all TaADF proteins, whereas motif 3 is present in the N-terminus of 23 TaADF proteins (Figure 4C). Alignment of the predicted TaADFs revealed that the ADF-H domain position and actin binding sites were conserved in all of the ADFs (Supplementary Figure 1). Additionally, our analysis indicated that closely related homologous TaADF genes in the A, B, or D sub-genomes are usually similar regarding their structures and encoded motifs, suggesting wheat TaADF genes were conserved during evolution.
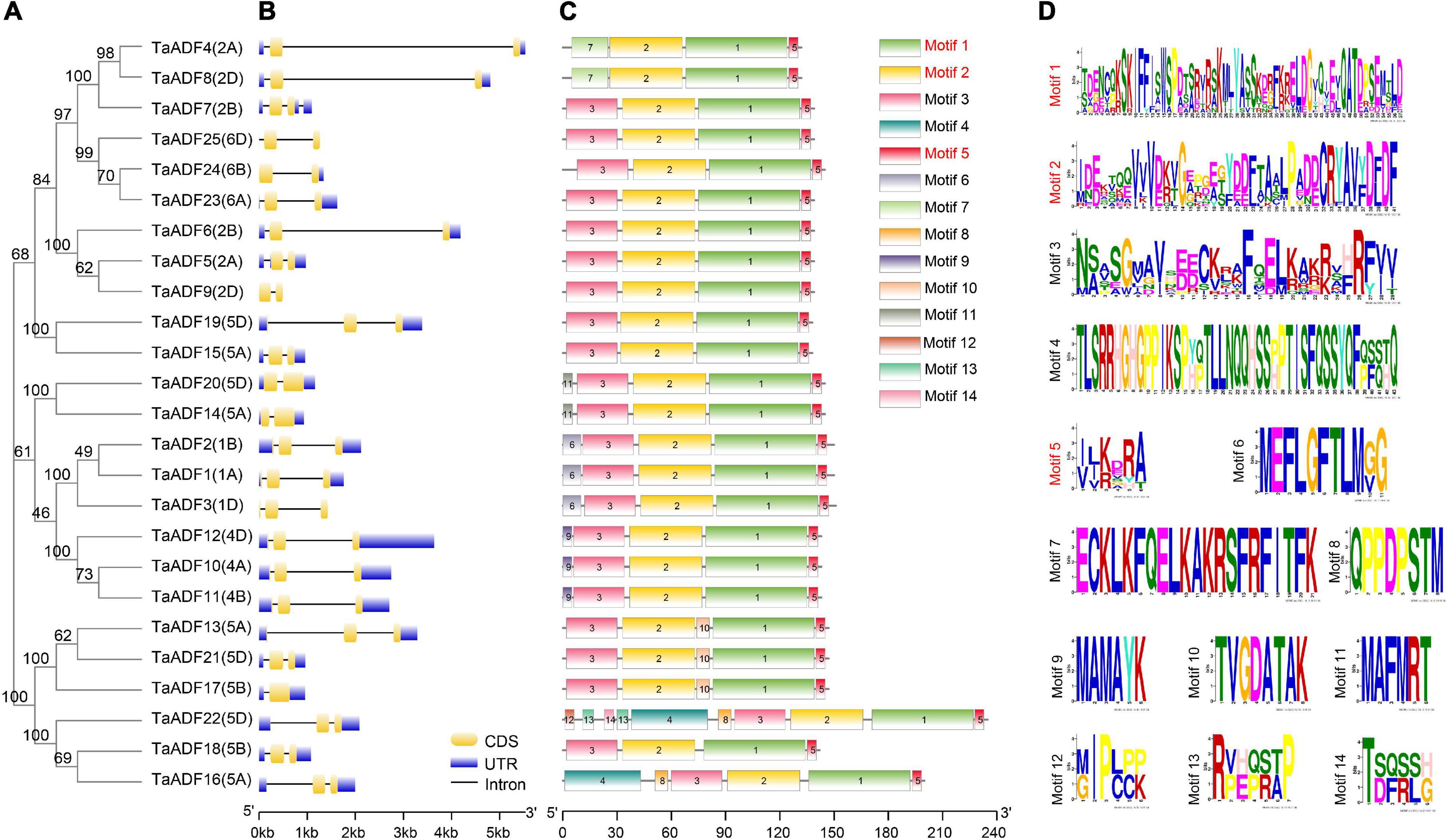
Figure 4. Phylogenetic relationship, exon-intron structures and motif composition of TaADFs. (A) Phylogenetic relationship of TaADFs. (B) Exon-intron structures of TaADF genes. Exons-intron are indicated by wide color bar and black line, respectively; (C) Distribution of conserved motifs of TaADFs predicted by MEME tool. (D) Conserved motifs in TaADFs. The motifs, numbered 1–14, are displayed in different colored boxes. Motifs correspond to the ADF-H domain region are shown in red.
The cis-acting elements in the 1,500-bp upstream promoter region of the identified TaADF genes were predicted using the PlantCARE online program. Fifty-seven cis-acting elements related to cell cycle regulation, plant development, hormone responses, stress responses, and transcription are presented in Supplementary Figure 2. Many cis-acting elements were associated with responses to various hormones, including abscisic acid (ABRE), methyl jasmonate (CGTCA-motif and TGACG-motif), auxin (TGA-element and AuxRR-core), and gibberellin (P-box and GARE-motif). Some of the identified cis-acting elements may regulate the development of specific tissues such as the endosperm (GCN4-motif), seed (RY-element), and meristem (CAT-box). The promoters of 16 and 13 TaADF genes included cis-acting elements responsive to drought (MBS) and low temperature (LTR), respectively. The presence of multiple cis-acting elements in the TaADF promoters may be indicative of the diversity in the biological functions of the encoded proteins.
To gain insights into the TaADF expression patterns in diverse wheat tissues, the available RNA-seq data for various wheat tissues across different developmental stages were obtained from the Wheat eFP database (Figure 5A). The TaADF expression levels varied among tissues at the same growth stage. At the flag leaf stage, the TPM of TaADF15 was 113.68, 248.09, and 302.91 in the leaf blade, root, and shoot axis, respectively. Nine genes (TaADF4/5/6/7/8/9/23/24/25), located on chromosome 2 or 6, were highly expressed in the anther (TPM: 239.41–823.54), but were expressed at low or undetectable levels (TPM < 1) in the leaf blade, root apical meristem, root, shoot axis, and grain. Additionally, the expression of most TaADF genes in specific tissues varied substantially at different growth stages. For example, TaADF15/16/19/18/22 expression levels in the root apical meristem and root were higher at the tillering stage than at the three-leaf stage. During the four examined grain developmental stages, the expression levels of these genes were highest and lowest at the milk grain stage and ripening stage, respectively. The tissue- and temporal-specific expression of TaADFs may help to clarify the complex functions of TaADF in various cellular processes.
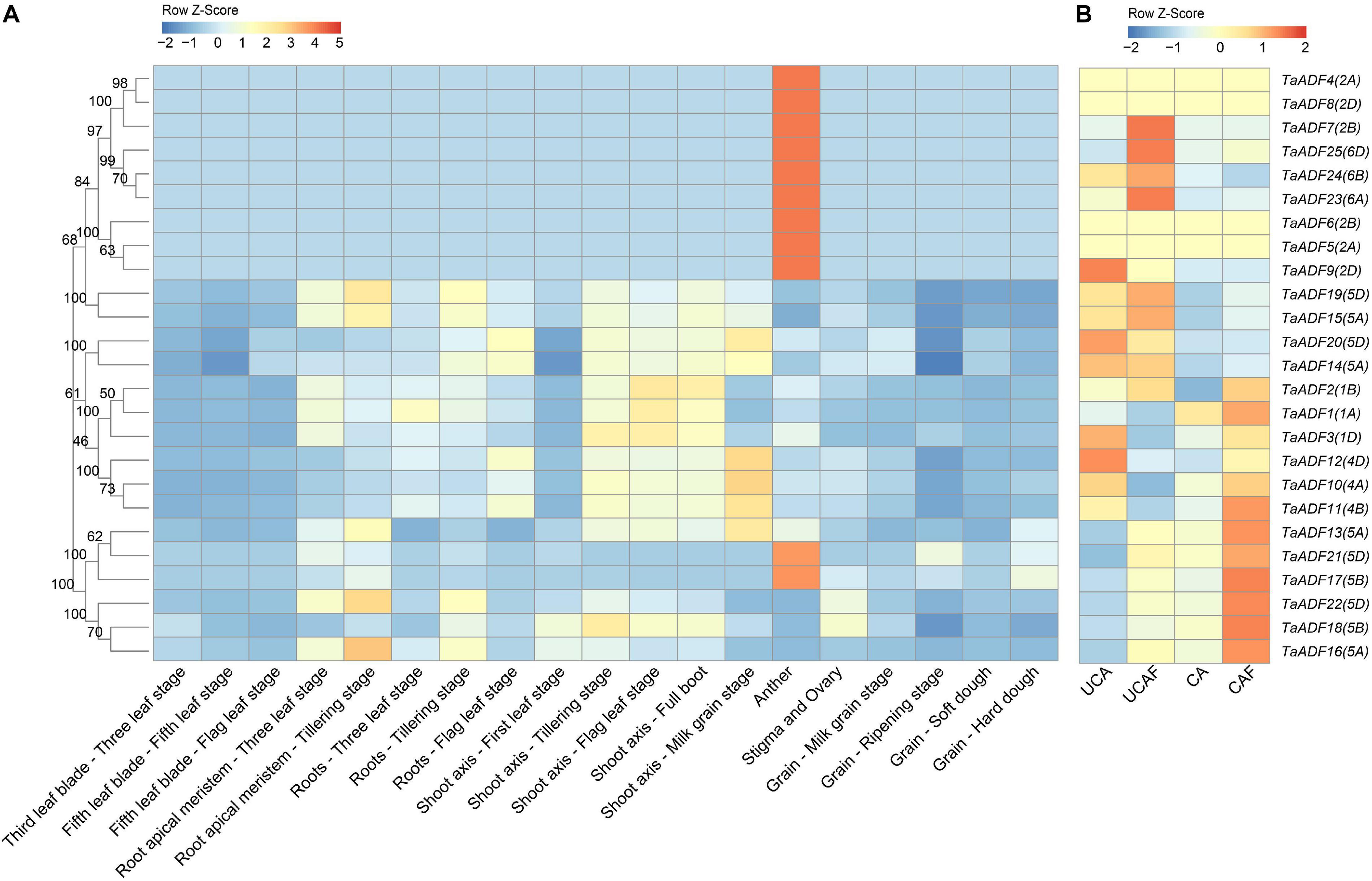
Figure 5. Expression patterns of 25 TaADFs in wheat. (A) Expression patterns of TaADFs in different tissues and growth stages. (B) Expression patterns of TaADFs under low temperature. Expression is scaled across genes (z score). CA: cold acclimation at 4°C for 28 days; UCA: un-cold acclimation at 20°C for 28 days; UCAF: UCA followed with −5°C for 1 day; CAF: CA followed with −5°C for 1 day.
The expression pattern of TaADFs were examined by RT-PCR in leaf and root at three-leaf stage (Supplementary Figure 3). The expression of homologous genes TaADF15-5A and TaADF19-5D were extremely high in both leaf and root, while the homologous genes TaADF10-4A, TaADF11-4B, and TaADF12-4D, showed significantly lower expression abundances. These results indicated that homologous TaADF genes in closely related clades appear to be expressed in different tissues similarity, implying they may be functionally similar.
To further evaluate the potential functions of ADFs in response to low temperatures, the ADF gene expression patterns under cold acclimation and freezing conditions were analyzed based on the fragment per kilobase of transcript per million reads mapped (FPKM). Results showed that TaADFs had differential expression under different temperature treatment. The expression of TaADF13/16/17/18/21/22 was induced by the cold acclimation and freezing treatments, while the expression of TaADF14/20 were decreased (Figure 5B). In total, seven DEGs were identified in three comparison (CA vs. UCA, CAF vs. CA and UCAF vs. UCA) (Supplementary Table 8). With the exception of TaADF20, the expression levels of all DEGs were up-regulated under the cold acclimation or freezing conditions. Significant changes to TaADF13/20 expression were detected only in the CA vs. UCA. Up-regulated TaADF17 expression was detected only in the CAF vs. CA. The TaADF16/18 expression levels were up-regulated after the CA and UCA samples when exposed to freezing stress (CAF vs. CA and UCAF vs. UCA), with a greater change in expression in the samples that did not undergo the cold acclimation process. These results suggest that TaADF16/17/18 contribute to the freezing tolerance of wheat plants acclimated to the cold.
To verify the TaADF expression patterns induced by the cold acclimation and freezing conditions, the expression levels of 10 TaADF genes were analyzed by RT-PCR. The consistency between the RT-PCR data and the RNA-seq data was reflected by the calculated correlation coefficient (R2 = 0.83) (Supplementary Figure 4). These results confirmed the accuracy of the RNA-seq results.
Based on the predicted subcellular localizations, all the TaADFs are cytoplasmic proteins, whereas TaADF22 is present in the cytoplasm and nucleus (Table 1). To further confirm the prediction of subcellular localization of TaADFs, we constructed a fusion vector, which was transformed into A. thaliana protoplasts and observed by laser scanning confocal microscope (Figure 6). All the four proteins (TaADF11, TaADF14, TaADF15, and TaADF16) have strong fluorescence signal in transformed A. thaliana protoplasts, the four protein are localized in the cytoplasm and nucleus.
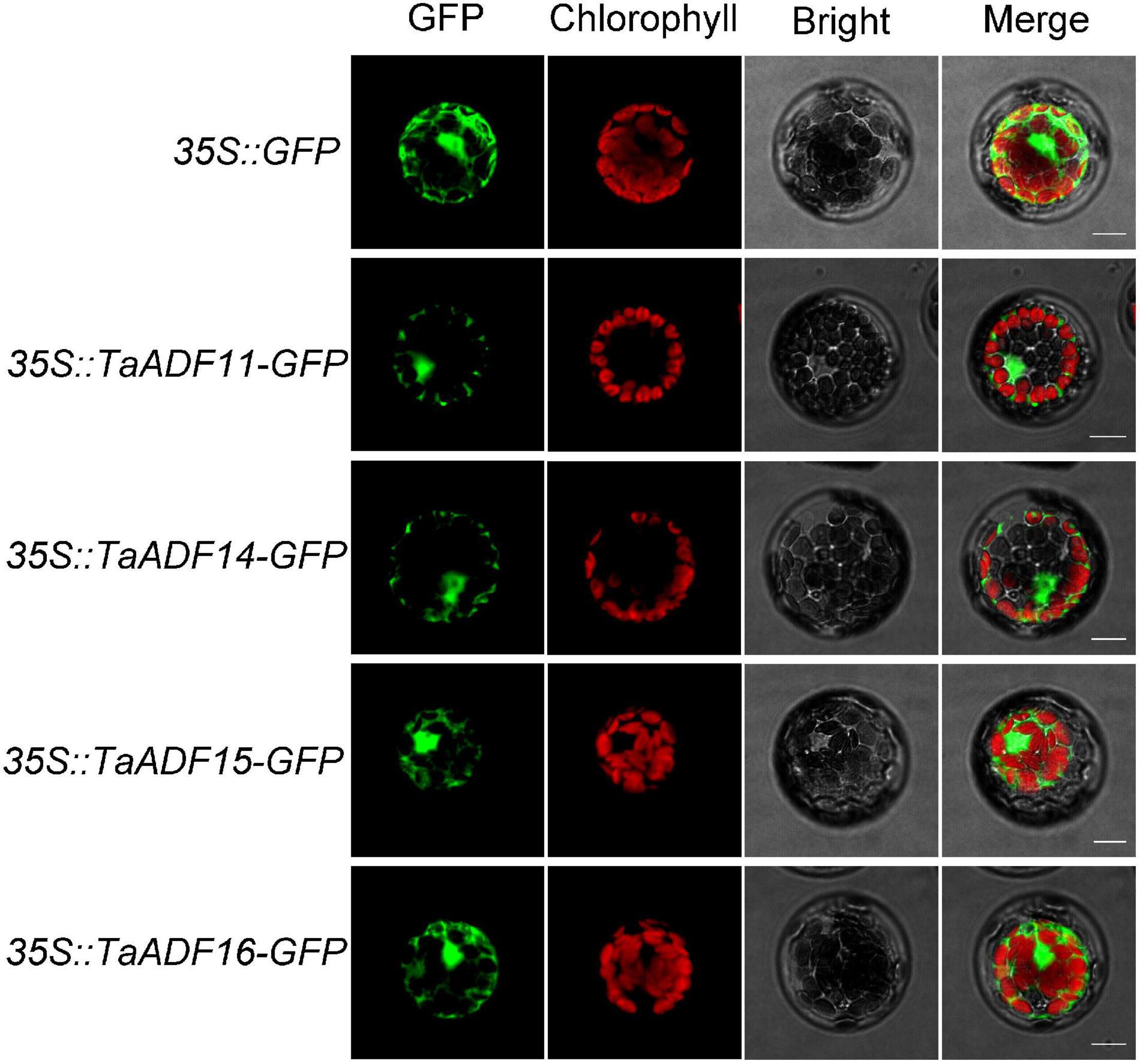
Figure 6. Subcellular location of TaADF11, TaADF14, TaADF15, and TaADF16 in A. thaliana protoplasts. Scale bars = 10 μm. 35S::GFP was used as a negative control.
Among the TaADF genes, TaADF16 was the most highly expressed (FPKM: CA 684.5, CAF 1837.33, UCAF 932.94) and up-regulated gene (CA vs. UCA 3.72-folds, CAF vs. CA, 1.56-folds, UCAF vs. UCA, 4.12-folds) under cold acclimation or freezing treatment (Figure 5B and Supplementary Table 8). Therefore, it was functionally characterized using transgenic Arabidopsis plants. Three transgenic lines with high TaADF16 expression levels were selected for further analyses (Figures 7A,B). Before the freezing treatment, there were no obvious differences between the wild-type (WT) and TaADF16-overexpressing (OE) plants. Morphological changes consistent with freezing damage were detected in the WT and OE lines, but the damage was more severe in the WT plants (Figure 7C). The survival rate (%) of WT was 16.67%, whereas the survival rate of OE8, OE9, and OE11 lines were obviously higher (75.83%, 71.67, and 74.87%, respectively) after recovery (Figure 7D). The electrolyte leakage after the freezing treatment was significantly greater in the WT (70.65%) plants than in the OE lines (OE8 43.78%, OE9 42.28%, and OE11 44.87%) (Figure 7E), suggesting the cell membranes were more severely damaged in the WT plants than in the transgenic lines.
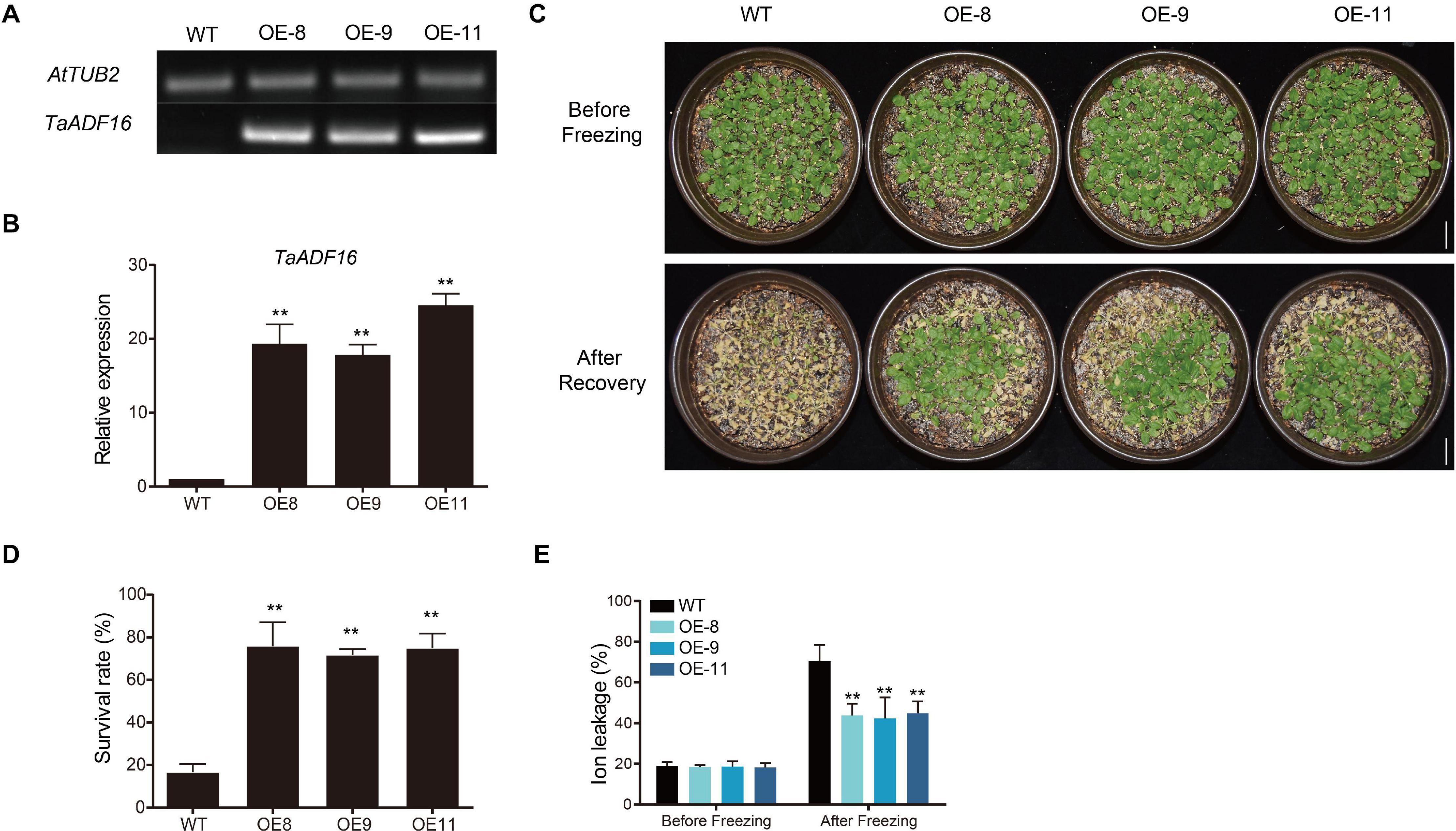
Figure 7. Overexpression of TaADF16 enhances freezing tolerance in Arabidopsis. Detection of TaADF16 mRNA in transgenic Arabidopsis using (A) Semi-quantitative RT-PCR and (B) RT-PCR; (C) Freezing tolerance of the transgenic Arabidopsis. Line bar: 2 cm. (D) Survival rate (%) and (E) Ion leakage (%) of WT and OE lines. Before freezing: three-week-old Arabidopsis seedlings under control; after freezing: three-week-old Arabidopsis seedlings under −5°C for 4 h and recovery at 22 °C for 4 days. Error bar indicated SD among at least three independent replicates. **P < 0.01 (Student’s t-test).
The activity of POD and SOD, as well as the soluble sugar content of the OE lines were similar to those in the WT before cold treatment, but the OE lines had higher POD and SOD activities and accumulated more soluble sugar than the WT plants after a 24-h incubation at 4°C (Supplementary Figures 5A–C). Following the cold treatment, the MDA content increased in both WT and OE lines, but MDA was significantly less abundant in the OE lines (OE8 3.38 μmol/g, OE9 2.80 μmol/g, and OE11 2.83 μmol/g) than in the WT plants (4.40 μmol/g) (Supplementary Figure 5D). Therefore, the overexpression of TaADF16 appeared to enhance the freezing tolerance of Arabidopsis by modulating the scavenging of ROS and by altering the osmotic regulation.
To further investigate the regulatory effects of TaADF16 in response to low-temperature stress, seven cold-responsive genes (CBF1, CBF2, CBF3, COR15A, COR15B, COR47, and RD22) were selected for a RT-PCR analysis. The basal and cold-induced expression levels of these genes were higher in the OE lines than in the WT plants (Supplementary Figure 6). These results suggest that TaADF16 overexpression induces the expression of cold-responsive genes, thus enhancing the cold resistance of transgenic plants.
The ADF gene family is relatively small in higher plants, with only 12, 12, 13, 11, and 14 members in rice, Arabidopsis, maize, tomato and poplar, respectively (Feng et al., 2006; Roy-Zokan et al., 2015; Khatun et al., 2016; Huang et al., 2020). In this study, we identified 25, 18, 12, 11, 8, and 5 ADF genes in the wheat, T. dicoccoides, T. turgidum, barley, Ae. Tauschii, and T. urartu genomes. The fact that more ADF genes were detected in wheat than in the other species may be attributed to the two rounds of polyploidization that occurred during wheat evolution (Marcussen et al., 2014). The presence of only a few ADF genes in the genomes of wheat relatives indicates that TaADF genes evolved after naturally occurring genomic hybridizations and fusions. The ADF gene family is believed to be structurally and functionally conserved in plants (McCurdy et al., 2001). Analyses of the phylogenetic relationships, gene structures, and encoded motifs indicated that closely related TaADF homologous in the A, B, or D sub-genomes have similar exon–intron structures and encode the same conserved motifs, which is consistent with the results of an earlier study (Feng et al., 2006). Our phylogenetic tree revealed that the ADF genes in the examined monocots are clustered in four main groups, with the ADF genes in the eudicot Arabidopsis distributed in subgroups I, II, and V. The phylogenetic relationships among the ADF genes from selected species were consistent with those described in published reports (Khatun et al., 2016; Huang et al., 2020). Accordingly, the ADF genes in the analyzed flowering plants likely evolved from a common ancestor.
Tandem and segmental duplications are considered to be the major driving force of gene family expansions during evolution (Cannon et al., 2004). Segmental duplications were revealed in this study as the main events responsible for the evolution of the TaADF gene family (Figure 1B). Similar events likely occurred in maize and tomato (Khatun et al., 2016; Huang et al., 2020). Therefore, we speculate that the ADF gene families in higher plants expanded primarily because of segmental duplications. The homologous ADF genes at the branch ends of each clade of sub-genome A, B, or D are likely the putative homoalleles of genes that evolved in Ae. tauschii, T. dicoccoides, and bread wheat (Figure 2B).
Tissue- and temporal-specific expression patterns of genes in growing plants usually reflect the differences in the biological functions of gene family members as well as the cross-talk among the associated pathways (Hu et al., 2018; Zhao et al., 2019a). Nine TaADF genes (TaADF4/5/6/7/8/9/23/24/25) were highly expressed in the anther, stigma, and ovary, but were expressed at low levels in the other examined tissues (Figure 5A). Similar results were reported for PhADF1/2 in petunia (Mun et al., 2000), SlADF1/2/10/11 in tomato (Khatun et al., 2016), and ZmADF1/2/7/12/13 in maize (Huang et al., 2020). The actin genes in the Arabidopsis genome have been divided into the vegetative class (expressed predominantly in the leaves, roots, stems, petals, and sepals) and the reproductive class (highly expressed in pollen) (Meagher et al., 1999). Because ADF proteins interact with actin, researchers also classified the ADF genes into vegetative and pollen-specific groups (Mun et al., 2000). We predict that the TaADF4/5/6/7/8/9/23/24/25 genes clustered in subgroup IV belong to the reproductive class, whereas the other TaADF genes are grouped in the vegetative class. The TaADF14/15/19/20 genes were more highly expressed than the other TaADF genes in the vegetative class, implying these four TaADF genes are important for wheat growth.
In this study, we detected dynamic changes to TaADF expression levels during various plant growth and developmental stages. In tomato, SlADF1/5/7/9 are highly expressed in immature fruit, whereas SlADF3/11 expression levels peak in the mature fruit stage (Khatun et al., 2016). In the current study, in developing wheat grains, the expression of most TaADF genes peaked in the milk grain stage, markedly decreased in the ripening stage, and then increased in the soft dough and hard dough stages. These findings suggest the encoded TaADF proteins have similar regulatory effects on actin filaments in wheat plants. However, there were no obvious expression-level trends common to all TaADF genes during the development of other tissues (e.g., leaf, root apical meristem, root, and shoot axis). These observations have compelled us to investigate the complex ADF regulatory mechanisms underlying wheat growth.
Previous studies confirmed that the expression of ADF gene is induced in plants exposed to low temperatures (Kerr and Carter, 1990; Danyluk et al., 1996; Ouellet et al., 2001; Tang et al., 2016). The reorganization regulated by ADFs may influence various cytoskeletal-associated cell processes. In response to low-temperature stress, microtubules are more easily depolymerized in cold-acclimated rye root tip cells than in non-acclimated cells, and this depolymerization enhances the freezing tolerance of the root tips (Kerr and Carter, 1990). We identified six TaADF genes (TaADF13/16/17/18/21/22) with significantly up-regulated expression under cold acclimation or freezing conditions, which is consistent with the cold-induced changes in SlADF2/11 expression in tomato (Khatun et al., 2016). However, TaADF20 expression was down-regulated by cold stress. This phenomenon might be explained by the antagonistic relationships among ADFs. For example, in Arabidopsis, AtADF9 adversely affects AtADF1 by regulating its ability to depolymerize actin, whereas the opposite effect is observed when AtADF9 and AtADF1 are ectopically expressed in tobacco cells (Tholl et al., 2011). Notably, TaADF16/18/22 expression levels were low during all examined wheat growth stages, but were highly up-regulated by low-temperature stress. Accordingly, to cope with cold conditions, TaADF gene expression is induced in wheat plants, thereby increasing the remodeling of actin. These findings reveal the complexity of the TaADF regulatory mechanism under cold conditions. Osmotic and ROS homoeostasis is essential for plant cold tolerance (Zuo et al., 2019). In our study, the overexpression of TaADF16 increased the freezing tolerance of Arabidopsis plants, likely because of the positive effects on ROS scavenging and osmotic regulation (Supplementary Figure 5). Furthermore, the expression of cold-responsive genes was induced in the transgenic Arabidopsis (Supplementary Figure 6), suggesting that TaADF16 may regulate cold tolerance by interacting with ICE-CBF-related genes. However, to more comprehensively characterize the relationship between the remodeling of the actin cytoskeleton and wheat responses to cold stress, additional ADF genes may need to be identified and functionally annotated.
In this study, we identified 25 ADF genes in wheat. Based on the protein sequence alignment, 117 ADFs from wheat and the other analyzed species were clustered into four main groups. Segmental duplications during evolution were important for the expansion of the TaADF gene family. Analyses of the phylogenetic relationships, gene structures, and encoded motifs suggested that TaADF were conserved during evolution. The tissue- and temporal-specific expression patterns of TaADF genes were revealed in this study. Nine genes preferentially expressed in the anther (TaADF4/5/6/7/8/9/23/24/25) are likely associated with pollen development. Additionally, we identified seven differentially expressed TaADF genes after low-temperature treatments. Specifically, the expression of homologous genes TaADF16/18/22 were considerably induced by cold stress, implying these genes are critical for the freezing tolerance of wheat. Overexpression of TaADF16 substantially increased the tolerance of transgenic plants to freezing stress because of the associated effects on the cell membrane and ROS homeostasis, as well as the CBF/DREB pathway genes. These results provide new insights into the regulatory functions of TaADF proteins related to wheat responses to low temperature, and provides candidate gene resources for breeding new wheat varieties with enhanced freezing tolerance.
Publicly available datasets were analyzed in this study. This data can be found here: NCBI GEO database:GSE135474 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135474).
YZ and XY conceived the project, set the scientific objectives, and revised the manuscript. KX, SiZ, HL, WW, and ShZ conducted the experiments and analyzed the data. KX wrote the manuscript. All authors discussed the results as well as read and approved the final manuscript for publication.
This work was funded by the National Key Research and Development Program of China (2017YFD0101000), The Technology Innovation of Winter Wheat of Science and Technology Planning Project of Hebei Province (16226320D), and The Innovation Fund Project for Graduate Student of Hebei Province (CXZZBS2019101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.618984/full#supplementary-material
Supplementary Figure 1 | Multiple alignments of TaADFs. The solid arrow and open arrow indicate the binding site of G-actin (ADFs shown in red) and F-actin (all the 25 ADFs), respectively. The red solid line indicates the ADF-H domain position.
Supplementary Figure 2 | Analysis of cis-acting elements in TaADFs.
Supplementary Figure 3 | Analysis of expression profiles of TaADF genes in leaf (A) and root (B) by RT-PCR. Error bar indicated SD among at least three independent replicates, and the values differed significantly when P < 0.05. Varied letters meant a significant difference.
Supplementary Figure 4 | Confirmation of RNA-seq results by RT-PCR. (A) Expression patterns of TaADFs by RNA-seq and RT-PCR. (B) Correlation between RNA-seq and RT-PCR for selected TaADF genes. The relative expression of TaADFs in crown of three leaf stage seedlings (TL) was set as the control. Three biological replicates for each sample were performed and bars represent the SD. CA: cold acclimation at 4°C for 28 days; UCA: un-cold acclimation at 20°C for 28 days; UCAF: UCA followed with −5°C for 1d; CAF: CA followed with −5°C for 1d.
Supplementary Figure 5 | Analysis of SOD activity (A), POD activity (B), MDA content (C), and soluble sugar content (D) in WT and OE lines of Arabidopsis. Before cold: three-week-old Arabidopsis seedlings under control (22°C); after cold: three-week-old Arabidopsis seedlings under 4°C for 24 h. Error bar indicates SD among at least three independent replicates. ∗∗P < 0.01 (Student’s t-test).
Supplementary Figure 6 | Expression levels of seven cold response genes in WT and OE lines before or after cold stress. Before cold: three-week-old Arabidopsis seedlings under control; after cold: three-week-old Arabidopsis seedlings under 4°C for 24 h. Error bar indicates SD among at least three independent replicates. ∗∗P < 0.01 (Student’s t-test).
Supplementary Table 1 | The data of genome sequences used in this study.
Supplementary Table 2 | Specific primers used in this study.
Supplementary Table 3 | The information of ADF protein of different plant species in this study.
Supplementary Table 4 | Sequence similarities among the TaADF proteins.
Supplementary Table 5 | Ka/Ks ratios of collinearity TaADF genes in wheat.
Supplementary Table 6 | The number of ADF proteins of different subgroups in phylogenetic tree.
Supplementary Table 7 | ADF collinear gene pairs between wheat and other selected species.
Supplementary Table 8 | The log2 (fold change) of differently expressed genes (DEGs) under cold stresses.
Allard, A., Bouzid, M., Betz, T., Simon, C., Abou-Ghali, M., Lemiere, J., et al. (2020). Actin modulates shape and mechanics of tubular membranes. Sci. Adv. 6:eaaz3050. doi: 10.1126/sciadv.aaz3050
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, 597–603. doi: 10.1093/nar/gks400
Bamburg, J. R., Harris, H. E., and Weeds, A. G. (1980). Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 121, 178–182. doi: 10.1016/0014-5793(80)81292-0
Burgos-Rivera, B., Ruzicka, D. R., Deal, R. B., McKinney, E. C., King-Reid, L., and Meagher, R. B. (2008). ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68, 619–632. doi: 10.1007/s11103-008-9398-1
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, C. Y., Wong, E. I., Vidali, L., Estavillo, A., Hepler, P. K., Wu, H. M., et al. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14, 2175–2190. doi: 10.1105/tpc.003038
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. doi: 10.1371/journal.pone.0011335
Daher, F. B., and Geitmann, A. (2012). Actin depolymerizing factors ADF7 and ADF10 play distinct roles during pollen development and pollen tube growth. Plant Signal. Behav. 7, 879–881. doi: 10.4161/psb.20436
Danyluk, J., Carpentier, E., and Sarhan, F. (1996). Identification and characterization of a low temperature regulated gene encoding an actin-binding protein from wheat. FEBS Lett. 389, 324–327. doi: 10.1016/0014-5793(96)00599-6
Dong, C. H., Xia, G. X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N. H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. doi: 10.1105/tpc.13.6.1333
Drobak, B. K., Franklin-Tong, V. E., and Staiger, C. J. (2004). The role of the actin cytoskeleton in plant cell signaling. New Phytol. 163, 13–30. doi: 10.1111/j.1469-8137.2004.01076.x
Du, J., Wang, X., Dong, C. H., Yang, J. M., and Yao, X. J. (2016). Computational study of the binding mechanism of actin-depolymerizing factor 1 with actin in Arabidopsis thaliana. PLoS One 11:e0159053. doi: 10.1371/journal.pone.0159053
Feng, Y., Liu, Q., and Xue, Q. (2006). Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. J. Plant Physiol. 163, 69–79. doi: 10.1016/j.jplph.2005.01.015
Fowler, J. E., and Quatrano, R. S. (1997). Plant cell morphogenesis: plasma membrane interactions with the cytoskeleton and cell wall. Annu. Rev. Cell Dev. Biol. 13, 697–743. doi: 10.1146/annurev.cellbio.13.1.697
Fu, Y., Duan, X., Tang, C., Li, X., Voegele, R. T., Wang, X., et al. (2014). TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 78, 16–30. doi: 10.1111/tpj.12457
Gungabissoon, R. A., Jiang, C. J., Drobak, B. K., Maciver, S. K., and Hussey, P. J. (1998). Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J. 16, 689–696. doi: 10.1046/j.1365-313x.1998.00339.x
Henty, J. L., Bledsoe, S. W., Khurana, P., Meagher, R. B., Day, B., Blanchoin, L., et al. (2011). Arabidopsis actin depolymerizing factor 4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23, 3711–3726. doi: 10.1105/tpc.111.090670
Hu, C. H., Wei, X. Y., Yuan, B., Yao, L. B., Ma, T. T., Zhang, P. P., et al. (2018). Genome-wide identification and functional analysis of NADPH oxidase family genes in wheat during development andenvironmental stress responses. Front. Plant Sci. 9:906. doi: 10.3389/fpls.2018.00906
Huang, J., Sun, W., Ren, J., Yang, R., Fan, J., Li, Y., et al. (2020). Genome-wide identification and characterization of actin-depolymerizing factor (ADF) family genes and expression analysis of responses to various stresses in Zea Mays L. Int. J. Mol. Sci. 21:1751. doi: 10.3390/ijms21051751
Huang, Y. C., Huang, W. L., Hong, C. Y., Lur, H. S., and Chang, M. C. (2012). Comprehensive analysis of differentially expressed rice actin depolymerizing factor gene family and heterologous overexpression of OsADF3 confers Arabidopsis Thaliana drought tolerance. Rice 5:33. doi: 10.1186/1939-8433-5-33
International Wheat Genome Sequencing Consortium (IWGSC) (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
Kerr, G. P., and Carter, J. V. (1990). Relationship between freezing tolerance of root-tip cells and cold stability of microtubules in Rye (Secale cereale L. cv Puma). Plant Physiol. 93, 77–82. doi: 10.1104/pp.93.1.77
Khatun, K., Robin, A. H., Park, J. I., Kim, C. K., Lim, K. B., Kim, M. B., et al. (2016). Genome-wide identification, characterization and expression profiling of ADF family genes in Solanum lycopersicum L. Genes 7:79. doi: 10.3390/genes7100079
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Li, H. S., Sun, Q., Zhao, S. J., Zhang, W. H., Chen, C. L., and Hong, Y. Z. (2000). The Experiment Principle and Technique on Plant Physiology and Biochemistry. Beijing: Higher Education Press.
Li, X. B., Xu, D., Wang, X. L., Huang, G. Q., Luo, J., Li, D. D., et al. (2010). Three cotton genes preferentially expressed in flower tissues encode actin-depolymerizing factors which are involved in F-actin dynamics in cells. J. Exp. Bot. 61, 41–53. doi: 10.1093/jxb/erp280
Liu, D., Sun, J., Zhu, D., Lyu, G., Zhang, C., Liu, J., et al. (2019). Genome-wide identification and expression profiles of late embryogenesis-abundant (LEA) genes during grain maturation in wheat (Triticum aestivum L.). Genes 10:696. doi: 10.3390/genes10090696
Lopez, I., Anthony, R. G., Maciver, S. K., Jiang, C. J., Khan, S., Weeds, A. G., et al. (1996). Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 7415–7420. doi: 10.1073/pnas.93.14.7415
Maciver, S. K., and Hussey, P. J. (2002). The ADF/cofilin family: actin-remodeling proteins. Genome Biol 3, reviews3007.1–reviews3007.12. doi: 10.1186/gb-2002-3-5-reviews3007
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092. doi: 10.1126/science.1250092
McCurdy, D. W., Kovar, D. R., and Staiger, C. J. (2001). Actin and actin-binding proteins in higher plants. Protoplasma 215, 89–104. doi: 10.1007/Bf01280306
Meagher, R. B., McKinney, E. C., and Vitale, A. V. (1999). The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet. 15, 278–284. doi: 10.1016/s0168-9525(99)01759-x
Mun, J. H., Yu, H. J., Lee, H. S., Kwon, Y. M., Lee, J. S., Lee, I., et al. (2000). Two closely related cDNAs encoding actin-depolymerizing factors of petunia are mainly expressed in vegetative tissues. Gene 257, 167–176. doi: 10.1016/S0378-1119(00)00412-1
Ouellet, F., Carpentier, E., Cope, M. J., Monroy, A. F., and Sarhan, F. (2001). Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiol. 125, 360–368. doi: 10.1104/pp.125.1.360
Porter, K., and Day, B. (2016). From filaments to function: the role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J. Integr. Plant Biol. 58, 299–311. doi: 10.1111/jipb.12445
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:eaar6089. doi: 10.1126/science.aar6089
Roy-Zokan, E. M., Dyer, K. A., and Meagher, R. B. (2015). Phylogenetic patterns of codon evolution in the ACTIN-DEPOLYMERIZING FACTOR/COFILIN (ADF/CFL) gene family. PLoS One 10:e0145917. doi: 10.1371/journal.pone.0145917
Sengupta, S., Mangu, V., Sanchez, L., Bedre, R., Joshi, R., Rajasekaran, K., et al. (2019). An actin-depolymerizing factor from the halophyte smooth cordgrass, Spartina alterniflora (SaADF2), is superior to its rice homolog (OsADF2) in conferring drought and salt tolerance when constitutively overexpressed in rice. Plant Biotechnol. J. 17, 188–205. doi: 10.1111/pbi.12957
Smertenko, A. P., Jiang, C. J., Simmons, N. J., Weeds, A. G., Davies, D. R., and Hussey, P. J. (1998). Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. 14, 187–193. doi: 10.1046/j.1365-313x.1998.00107.x
Song, G., Zhang, R., Zhang, S., Li, Y., Gao, J., Han, X., et al. (2017). Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics 18:212. doi: 10.1186/s12864-017-3556-2
Staiger, C. J., Gibbon, B. C., Kovar, D. R., and Zonia, L. E. (1997). Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends Plant Sci. 2, 275–281. doi: 10.1016/S1360-1385(97)86350-9
Sung, S., and Amasino, R. M. (2005). Remembering winter: toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56, 491–508. doi: 10.1146/annurev.arplant.56.032604.144307
Tang, C. L., Deng, L., Chang, D., Chen, S. T., Wang, X. J., and Kang, Z. S. (2016). TaADF3, an actin-depolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 6:1214. doi: 10.3389/Fpls.2015.01214
Tholl, S., Moreau, F., Hoffmann, C., Arumugam, K., Dieterle, M., Moes, D., et al. (2011). Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585, 1821–1827. doi: 10.1016/j.febslet.2011.05.019
Thomashow, M. F. (2001). So what’s new in the field of plant cold acclimation? Lots! Plant Physiol. 125, 89–93. doi: 10.1104/pp.125.1.89
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, H. Y., Wang, J., Gao, P., Jiao, G. L., Zhao, P. M., Li, Y., et al. (2009). Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 7, 13–23. doi: 10.1111/j.1467-7652.2008.00367.x
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Zhang, B., Hua, Y., Wang, J., Huo, Y., Shimono, M., Day, B., et al. (2017). TaADF4, an actin-depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. Plant J. 89, 1210–1224. doi: 10.1111/tpj.13459
Zhao, Y., Ma, R. Y., Xu, D. L., Bi, H. H., Xia, Z. L., and Peng, H. R. (2019a). Genome-wide identification and analysis of the AP2 transcription factor gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 10:1286. doi: 10.3389/Fpls.2019.01286
Zhao, Y., Zhou, M., Xu, K., Li, J., Li, S., Zhang, S., et al. (2019b). Integrated transcriptomics and metabolomics analyses provide insights into cold stress response in wheat. Crop J. 7, 857–866. doi: 10.1016/j.cj.2019.09.002
Zheng, Y., Xie, Y., Jiang, Y., Qu, X., and Huang, S. (2013). Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25, 3405–3423. doi: 10.1105/tpc.113.117820
Zhou, M., Zheng, S., Liu, R., Lu, J., Lu, L., Zhang, C., et al. (2019). Genome-wide identification, phylogenetic and expression analysis of the heat shock transcription factor family in bread wheat (Triticum aestivum L.). BMC Genomics 20:505. doi: 10.1186/s12864-019-5876-x
Keywords: actin depolymerizing factor, wheat (Triticum aestivum L.), low temperature, genome-wide identification, transgenic Arabidopsis
Citation: Xu K, Zhao Y, Zhao S, Liu H, Wang W, Zhang S and Yang X (2021) Genome-Wide Identification and Low Temperature Responsive Pattern of Actin Depolymerizing Factor (ADF) Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 12:618984. doi: 10.3389/fpls.2021.618984
Received: 09 November 2020; Accepted: 05 February 2021;
Published: 24 February 2021.
Edited by:
Xianchun Xia, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Mingming Xin, China Agricultural University, ChinaCopyright © 2021 Xu, Zhao, Zhao, Liu, Wang, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhao, emhhb3lvbmdfMDQyM0AxNjMuY29t; Xueju Yang, c2hteXhqQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.