
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 24 March 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.608061
This article is part of the Research TopicAn Update on Brassinosteroids: Homeostasis, Crosstalk, and Adaptation to Environmental StressView all 7 articles
 Jaspreet Kour1
Jaspreet Kour1 Sukhmeen Kaur Kohli1
Sukhmeen Kaur Kohli1 Kanika Khanna1
Kanika Khanna1 Palak Bakshi1
Palak Bakshi1 Pooja Sharma1
Pooja Sharma1 Arun Dev Singh1
Arun Dev Singh1 Mohd Ibrahim1
Mohd Ibrahim1 Kamini Devi1
Kamini Devi1 Neerja Sharma1
Neerja Sharma1 Puja Ohri2
Puja Ohri2 Milan Skalicky3
Milan Skalicky3 Marian Brestic3,4
Marian Brestic3,4 Renu Bhardwaj1*
Renu Bhardwaj1* Marco Landi5*
Marco Landi5* Anket Sharma6*
Anket Sharma6*Brassinosteroids (BRs) are group of plant steroidal hormones that modulate developmental processes and also have pivotal role in stress management. Biosynthesis of BRs takes place through established early C-6 and late C-6 oxidation pathways and the C-22 hydroxylation pathway triggered by activation of the DWF4 gene that acts on multiple intermediates. BRs are recognized at the cell surface by the receptor kinases, BRI1 and BAK1, which relay signals to the nucleus through a phosphorylation cascade involving phosphorylation of BSU1 protein and proteasomal degradation of BIN2 proteins. Inactivation of BIN2 allows BES1/BZR1 to enter the nucleus and regulate the expression of target genes. In the whole cascade of signal recognition, transduction and regulation of target genes, BRs crosstalk with other phytohormones that play significant roles. In the current era, plants are continuously exposed to abiotic stresses and heavy metal stress is one of the major stresses. The present study reveals the mechanism of these events from biosynthesis, transport and crosstalk through receptor kinases and transcriptional networks under heavy metal stress.
Brassinosteroids (BRs) were discovered 40 years ago, and since then an enormous amount of work has been done to illuminate their role in plant physiology (Peres et al., 2019). BRs are poly-hydroxylated steroids that regulate developmental processes such as cell division, cell cycle, elongation, morphogenesis, reproduction, senescence and stress-protective responses (Clouse and Sasse, 1998). They function especially as master switches in triggering the metabolic response to noxious environmental conditions (Bajguz, 2010). BRs are considered as derivatives of 5α-cholesterol, but they vary in structure due to the carbon side-chains. They are present in all parts of plants but are mostly found in seeds and pollen (Bartwal and Arora, 2020). Caño-Delgado et al. (2004) suggested that as animal steroid hormones bind to nuclear receptors that modulate gene expression, BRs also bind to receptors on the cell surface initiating a signal cascade leading to alterations in gene expression. Recent progress in understanding the BR signaling pathways supports the idea that they do not follow a linear path but rather undergo crosstalk with other hormones to combat stress conditions (Nolan et al., 2017). BRs also provide tolerance to plants against abiotic stressors by modulating the activity of enzymatic and non-enzymatic antioxidants (Sharma A. et al., 2016; Sharma et al., 2017; Bartwal and Arora, 2020; Chi et al., 2020; Rattan et al., 2020; Chen et al., 2021). BRs are also reported to stimulate the formulation of phytochelatins as reported by Bajguz, 2002. Reports suggested that brassinolides along with the lead are responsible for the increase in the synthesis of phytochelatins in Chlorella vulgaris. To overcome the harmful effects of heavy metals, BRs interact with other hormones like auxin, cytokinin, abscisic acid (ABA), ethylene, and jasmonic acid (JA). In this review we have discussed in detail the molecular mechanism of BR biosynthesis, signaling, and the role of transcriptional networks in the response to heavy metal stress in plants, and the ways that BRs crosstalk with other phytohormones to prevent heavy metal damage.
Brassinosteroids share structural resemblance with animal steroids like cholestane, ergo- and stigmastane. The majority of BRs are 5α-cholestane derivatives and structural variation among them emerges because of C-substitutions within the side chains (Ali, 2019). They can be found in all plant parts, yet most of the synthesis occurs in the meristematic zones of seeds and pollen grains. These BR categories represent more than eighty plant species including angiosperms, pteridophytes, algae, and bryophytes (Yokota, 1999). Based on the number of carbon atoms and alkyl groups within side chains, BRs are divided into C-27, -28, and -29 steroids present freely or in conjugation with fatty acids and sugars (Fujioka and Yokota, 2003). Forty to fifty percent of BRs are C-28. The great diversity in the patterns of cyclic groups as well as side chains is mainly responsible for the conversion of BRs into active analogs such as 24-epibrassinolides and 28-homobrassinolides (Kang and Guo, 2011). BRs along with analogs like typhasterol, brassinolide, and castasterone are widely distributed within specialized plant structures like pollens, seeds, flowers, roots, shoots, leaves and stems. The greatest concentration of BRs (100 ng per g fresh weight) have been recorded within pollen grains and seeds compared to 0.01 and 0.1 ng per g fresh weight within shoots and leaves, respectively.
More than 69 different BRs and derivatives have been identified to date from various plants (Bajguz and Tretyn, 2003) including Castanea crenata and Catharanthus roseus. Along with various BRs there are five conjugates of BRs which are found together with 8 metabolites and 137 analogs of BRs (Liu et al., 2017). The distribution of BRs in various plant families from maximum to minimum runs from angiosperms to gymnosperms, pteridophytes, bryophytes, and algae (Bajguz and Tretyn, 2003). It is well documented that sterols within plants are transformed into brassinolide through teasterone, typhasterol as well as castasterone via the isoprenoid pathway in association with acetyl coA, isopentenyl pyrophosphate (IPP), geranyl pyrophosphate (GPP), mevalonate and farnesyl pyrophosphate (FPP) (Symons et al., 2008). Brassinolides are highly effective forms of BRs that are produced as the end product of BRs synthesis (Zhao and Li, 2012). According to a more limited description, among the metabolites produced during the biosynthesis of BR-lactones, only those that have been formed from 22α and 23α-dihydroxylation would be considered as true BRs, the rest being placed in the category of BR precursors (Zullo and Kohout, 2004). BRs containing 23-epoxy groups, 23-glycosidic groups, 23-ester groups, 3-oxygenated-5α-cholestane-23α diols along with alkyl derivatives were also considered natural BRs (Zullo, 2018). As reported, the bioactive potential of BRs lies with the side chains of rings (Zullo and Adam, 2002). Employment of the rice laminal inclination assay for ring structures showed that 22α and 23α dihydroxy BRs were active as 28-epibrassinolides (Watanabe et al., 2001). However, 23-dehydrogenation or conjugation of side chains may disrupt the biological activity (Watanabe et al., 2001). Further studies demonstrated that the biological activity of BRs was regulated by other active sites (Liu et al., 2017). A wide spectrum of structural variability was seen in ring A with fifteen types of structures starting from Δ2, 3-unsaturated to conjugated BRs. Even if analysis of the structural variation does not reveal the probable structures of this ring, it is assumed to belong to the category of BRs (Zullo, 2018). The biological functionality of BRs possessing rings increases in the order from 3β-hydroxy to 3α-dihydroxy rings (Liu et al., 2017). This variation in rings correlates with the biosynthetic pathway of BRs, suggesting that higher oxidation states are enhanced from the 6-deoxo and 6α-hydroxy forms toward the 6-oxo and 7-oxa lactone states (Vriet et al., 2013). It has been observed that there is decline in the functionality from 2α, 3α toward 2β, 3β and it is suggested that this decline is due to reason that α-oriented hydroxyl group that is present at C-2 is important for its activity (Zou et al., 2020). BRs have also been classified according to B-rings: 6-oxo-7-oxalactonic BRs (28-homobrassinolides, 2α,3β-dihydroxylated: 3-epibrassinolide, 2α,3α-dihydroxylated: brassinolide, 3β-hydroxylated: 3-epi-2-deoxybrassinolide, dolicholide), 6-oxo or 6-keto BRs (2α,3α-dihydroxylated: castasterone, 28-homocastasterone, 25-methylcastasterone, 2β,3α-dihydroxylated: 2-epicastasterone, 2α,3β-dihydroxylated: 3-epicastasterone, 2,3-diepi-25-methyl dolichosterone, 3β-monohydroxylated: teasterone, 28-homotyph asterol, 23-dehydro-2-epicastasterone, 1α,2α,3β-dihydroxylated: 1α-hydroxy-3-epicastasterone, 1α,2α,3β-dihydroxylated: 1α-hydroxy-3-epicastasterone, 23α-conjugates: 23-O-β-D-glucopy ranosyl-25-methyldolichosterone, 3-dehydro: 3-dehydrotea sterone), 6α-hydroxy BRs (6α-hydroxycastasterone), 6-deoxo BRs (2α,3α-dihydroxylated: 6-deoxocastasterone, 6-deoxo-28-homodolichosterone, 6-deoxo-24-epicastasterone, 3α-monohy droxylated: 6-deoxotyphasterol, 6-deoxo-28-homotyphasterol, 6-deoxo-28-norteasterone, 3-dehydro-6-deoxo-28-norteasterone), respectively (Zullo and Bajguz, 2019).
Genetic and biochemical studies have identified most of the enzymes associated with BR synthesis up to now, but the molecular mechanism of BR biosynthesis and release needs further work to provide a deeper understanding of the regulation of its synthesis, catalysis and conjugation. These activities were explained in a coordinated manner by Japanese scientists who systematically elucidated the biosynthetic pathways concerning BRs within plant cells (Fujioka and Sakurai, 1997). Their data illustrated that biosynthesis of brassinolide (28-epibrassinolide) was linked to two main pathways, the early and late C6 oxidation pathways (Fujioka et al., 1998). The plant steroid, campesterol, was thought to be the progenitor of brassinolides on account of the side chain as well as the bioactive potential. The molecular structure and concurrence of teasterone, typhasterol, and castasterone also suggested that brassinolides were synthesized from campesterol (Yokota et al., 1991). The BR-specific campesterol precursor is initially transformed into campestanol, after which the early as well as late C6-pathways of oxidation, also termed as campestanol-dependent pathways, are activated. The enzymes required for this biosynthetic process include DET2, DWF4, cytochrome monooxygenases (Cyp P450), ROT3, BAS1, BR oxidase 2(BRox2), constitutive photomorphogenesis and dwarfism (CPD), and BR oxidase 6(BR6ox1) (Bhanu, 2019). The synthesis of castasterone and brassinolide requires the enzyme Cyp450 (CYP85A2), that also contributes to the rate limiting step for the conversion of 6-deoxycatasterone into castasterone and finally into brassinolide (Kim et al., 2005). Thus, by acting on several intermediates upstream, DWF4 can bifurcate the pathway at campesterol to generate an initial C-22 hydroxylation pathway (Fujioka and Yokota, 2003).
The BR biosynthetic pathway is generally triterpenoid in nature; therefore, the triterpene squalane can also be a precursor for BR synthesis, where it gets cyclized to cycloartenol using specialized enzymatic machinery (Hartmann, 1998). It is mainly formed via condensation of two molecules of FPP using NADPH as reducing agent (Heldt and Piechulla, 2011). In addition, plant sterols containing different alkyl groups at C24 like cholesterol and sitisterol, as well as campesterol, can also act as precursors for different BRs (Fujioka and Yokota, 2003). The biosynthetic pathway of BRs is limited at various steps through regulatory feedback processes to maintain endogenous BR homeostasis by the expression of genes for BR degradation. For example, expression of the CPD, DET2, and DWARF4 genes is known to get modulated under such conditions. Also, those genes functioning in the C-22 hydroxylase pathway become active during the DWF4- or CPD-induced cascade depending on the availability of substrates. DWF4 can also bind large substrates like campesterol, 4-en-3-one, campestanol, and 6-oxo campesterol to enter the C-22 oxidation biosynthetic pathway. Likewise, CPD can metabolize campesterol and 22, 23-dihydroxy campesterol via parallel pathways. After campesterol hydroxylation by DWF4, the intermediates formed are altered and enter the 6-oxidation pathway. It was also assumed that CYP90A1/CPD encoding C23 hydroxylases converted teasterone into castasterone (Szekeres et al., 1996; Chung and Choe, 2013). CYP90A1/CPD was reported to be active in the C3 oxidation pathway as revealed through GC-MS analysis (Ohnishi et al., 2012). The transformation of 6-deoxo teasterone to 6-deoxo dehydroteasterone can also occur through CYP90A1/CPD activation. However, 22-hydroxycampesterol accumulation was observed along with reduced 6-deoxo 3-deoxyeasterone and 6-deoxo castasterone in CPD-knockout mutants. By this, it was inferred that the campestanol in-mediated pathway was principally involved in BR biosynthesis. CYP90C1/ROT3 and CYP90D1 were also important in C23 hydroxylase synthesis. This adds a new dimension to the BR synthesis pathway through conversion of 22-hydroxy-ergost-4-en-3-one and 22-hydroxy-5α-ergostan 3-one into C23 hydoxylated forms (Bishop, 2007). During the early stage of the C6 oxidation pathway, oxidation at C-6 occurs before the onset of DWF4-induced C22 hydroxylation. Studies were done with Arabidopsis CYP85A1 and CYP85A2 on the C-6 oxidation pathway that showed conversion of 6-deoxoteasterone and 6-deoxocastasterone into typhasterol and castasterone (Kim et al., 2005). CYP85A2 was also observed to induce the conversion of 6-deoxotyphasterol into brassinolides through castasterone by a downstream pathway. An investigation in CYP85A1 and CYP85A2 double mutants of endogenous BR levels suggested that increased levels of 6-oxocampesterol were present at the initial steps of BR-biosynthesis, revealing the activity of an unknown enzyme (Kwon et al., 2005). It is pertinent to mention here that CYP85A2-induced brassinolide synthesis only occurs in dicots. In rice, which is a monocot, brassinolides were not detected even in BRI1 mutants (Yamamuro et al., 2000). Also, monocots have only single copy CYP85A gene, while, dicots have duplicate genes (Kim et al., 2008). The evidence shows that castasterone is the product formed using these mechanistic pathways (Kim et al., 2008), but the additional genetic and biochemical strategies that enhance BR biosynthesis also need to be determined.
The accumulation of BZR1 (brassinazole-resistant 1) and BRI1 EMS suppressor BES1/BZR2 reduced the expression of BR biosynthetic genes (Bartwal and Arora, 2020).BAS1, which encodes PhyB-activated tagged suppressor 1, is the foremost BR-inactivating gene in plants for maintaining optimal hormone levels. Mutations in the pathway for BR synthesis have also been implicated in male sterility, leaf curl, and dwarfing and BR-induced signal transduction is essential in plant metabolism (Bartwal and Arora, 2020). Earlier studies on mutants showed that exogenous application of BRs in wild type plants restored mutant BR-biosynthetic genes, while this did not occur in strains where BR signaling was hindered. Later studies were conducted on mutants of Arabidopsis, to elucidate the BR-signaling and transcriptional regulatory process. It was reported that leucine-rich repeated receptors and protein kinases (LRR-RLK) and BRI1 were the primary receptors for BRs. The resulting signal cascade led to trans-phosphorylation of BRI1 and BAK1, and further downstream signal transduction events (Cheng et al., 2017). Figure 1 represents the detailed biosynthetic pathway of BRs showing the early C-6 oxidation pathway, late C-6 oxidation pathway and early C-22 oxidation pathway.
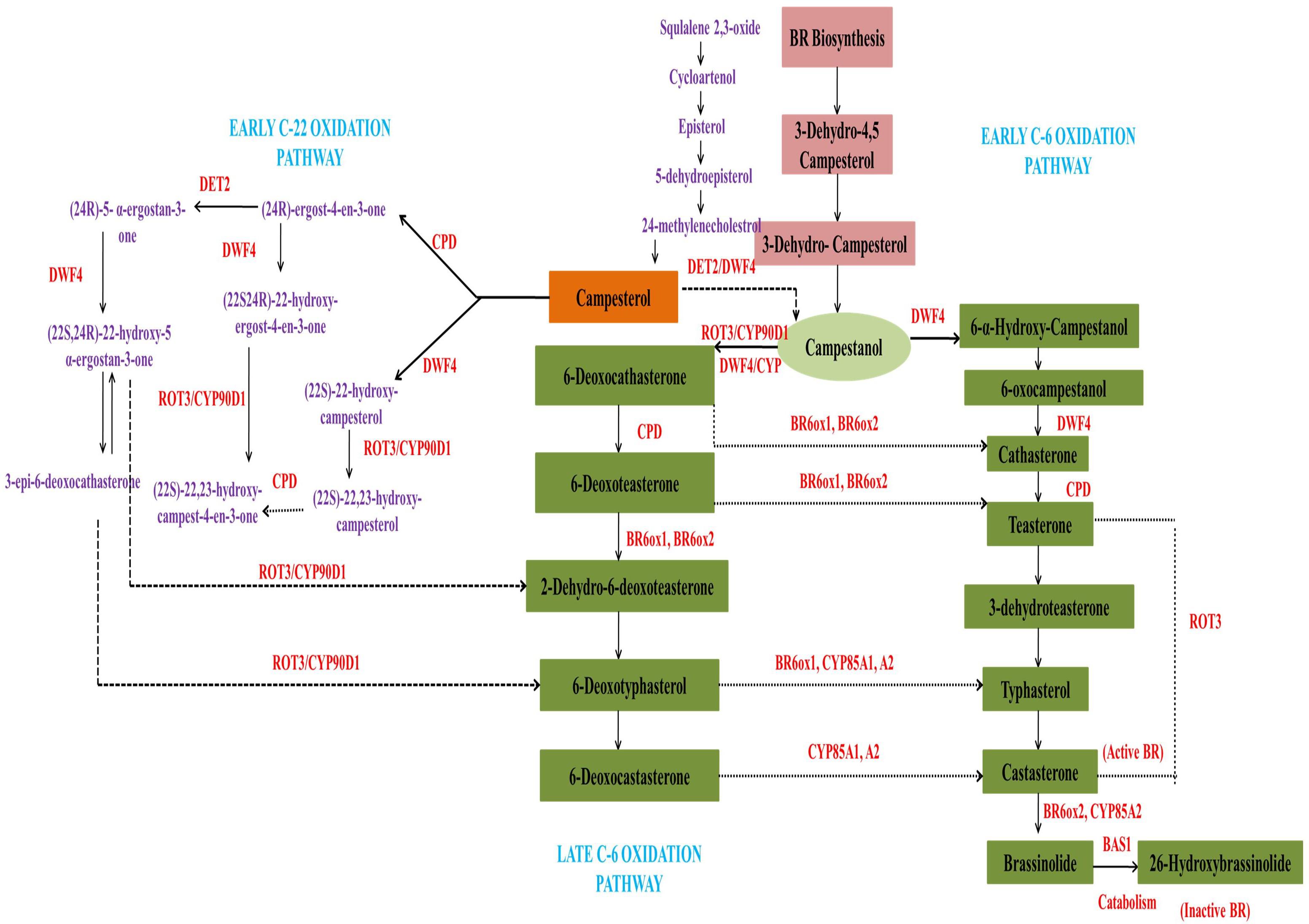
Figure 1. Schematic representation of the BR biosynthetic pathway, showing the early C-6 oxidation pathway, the late C-6 oxidation pathway and the early C-22 oxidation pathway.
With evolution, plants have developed strategies for optimal growth with minimal energy input. When BRs levels are adequate, the regulatory mechanism of positive and negative feedback loops are activated to down-regulate the endogenous BR levels. The endogenous BR level is either regulated by the down-regulation of BR biosynthetic genes or by the inactivation of already available bioactive BRs. Exogenous BL application down-regulates the BR biosynthetic genes, DWF-4, CPD, BR6ox1, and ROT-3 (rotundifolia 3), but up-regulates Brz, an inhibitor of BR synthesis. Exogenous BL also up-regulates BAS-1, another inhibitor of BR synthesis and down-regulates BES/BZR, a key transcription factor in the BR biosynthesis and signaling network (Yu et al., 2011).
DWF-4 genes expressed in bacterial cells were found to not be allosterically regulated. Exogenous application of epi-BR or Brz inhibitors of BR biosynthesis altered the functioning of DWF-4 genes and the transcription of DWF-4 exceeded the threshold levels, even in BR signaling mutants. The accumulation of DWF-4 mRNA was observed after Brz application, but BL reduced DWF-4 mRNA. Some other genes like CPD, BR6ox1, and ROT-3 were transcriptionally regulated by BL and Brz, but DET-2 was sensitive to Brz only (Tanaka et al., 2005). The sterol methyltransferase 2 (SMT2) and Dwarf1(DWF1) genes related to sterol biosynthesis are sensitive to both BL and Brz, but the sterol C-14 reductase, FACKEL (FK), is responsive to Brz only. In Bri1 mutants exposure to BL and Brz can trigger the expression of DWF-4 but the other genes required the normal BRI1 for transcriptional regulation (Tanaka et al., 2005).
Wang et al. (2002) reported that the two downstream factors BES and BZR controlled the whole BR regulation pathway. BZR1 is involved in transcriptional regulation of multiple BR-biosynthesis genes necessary for triggering specific growth processes and in down-regulating these genes by negative feed-back mechanism (He et al., 2005). At low BL levels, BIN2 phosphorylates BZR1, which inactivates it and blocks initiation of the BR signaling cascade. Normal BL levels initiate the BR signaling cascade, which either activates or inhibits the expression of intermediate genes. Thus, starting from campesterol as the primary precursor for BL formation, and proceeding through the activation or inactivation of BR-related genes, there exists a regulatory feed-back mechanism for controlling BR biosynthesis (Figure 2).

Figure 2. Outline of the transcriptional regulation of BR biosynthetic genes. BZR1 controls the expression of these genes through a negative feed-back loop (shown by black arrows). BES1, another downstream transcription factor, positively regulates the expression of BR-biosynthetic genes (shown as green and brown arrows) (It is a conclusion diagram based on reports in corresponding section “Transcriptional Regulation”).
In the brxs mutant of Arabidopsis, expression of the CPD gene is arrested, which results in defective cell division and altered root growth (Mouchel et al., 2006). Normal BRX is required for CPD expression, as evidenced by exogenous BL application or by over-expression of CPD genes that convert the mutant root trait into the wild type one (Poppenberger et al., 2011). CESTA (CES), a basic helix-loop helix (bHLH) transcription factor, is a well-known trigger of CPD gene expression in Arabidopsis. CES binds to the CPD promoter region at the G-Box sequence elements and modulates its transcription.
Data from ChIP (chromatin immunoprecipitation) assays has shown that the expression of DWF4 can be regulated by TCP1 via direct or indirect binding to the DWF-4 promotor. TCP1 expression is up-regulated by BL treatment but down-regulated by the application of Brz (Guo et al., 2010).
RAVL-1 and BRI-1 exhibit similar activity in loss of function and gain of function BR synthesis mutants. In-vivo and in-vitro experiments revealed that RAVL-1 bound to the B-box motif of the BR biosynthetic genes, D2, D11, BRD-1, and OsBRI1, and controlled their expression (Je et al., 2010).
In Arabidopsis, auxin can induce the expression of DWF4. Treatment with the auxin, indole acetic acid (IAA), or its synthetic analog, 2,4-D, up-regulates the expression of DWF4 and other intermediates such as 22-OHCR and 22-OH-3, and typically enhances endogenous BR levels in roots (Chung et al., 2011; Yoshimitsu et al., 2011). Thus, auxin signaling is required for independent DWF4 regulation with respect to BR biosynthesis. BZR1 binds strongly to the BRRE motif on the DWF-4 promotor with or without BL treatment. The DWF4 promotor was found to have multiple regulatory sequences including one Aux/IAA response element (Aux/IAA-RE, TGTCTC) (Ulmasov et al., 1997), three BRRE motifs (Chung et al., 2011) and one Aux/IAA-RE-like element (TGTGCTC) (Hagen and Guilfoyle, 2002). The auxin response factor, ARF-7, also directly bound to the DWF4 promotor. At high auxin concentrations, the inhibitor of Aux/IAA was degraded with the release of ARF-7, which then bound to the DWF-4 promotor and initiated the expression of BR response and BR biosynthetic genes. It was also found that BZR1 left its binding site to accommodate the binding of ARF-7. CHX treatment masked the expression of DWF-4, which suggests that some other transcription factor must be required to mediate auxin-dependent DWF-4 regulation. One study conducted at a higher temperature (29°C) supported the hypothesis that auxin is involved in the expression of DWF-4 (Maharjan and Choe, 2011). Seedlings of a DWF-4 proGUS line showed increased DWF-4 gene expression at the higher ambient temperature. Hypocotyl elongation is stimulated by auxin, and higher temperature increases the synthesis and transport of auxins (Gray et al., 1998). Hypocotyl elongation was arrested in BR-biosynthetic mutants, and it was concluded that high BR levels were required for hypocotyl elongation under temperature stress (Nemhauser et al., 2004).
Brassinosteroids are recognized by the receptor kinase, BRASSINOSTEROID-INSENSITIVE 1 (BRI1), and the subsequent downstream signal cascade activates the brassinazole-resistant 1 (BZR1) family of transcription factors, which plays a vital role in brassinosteroid-regulated gene expression (Guo R. et al., 2013). When the levels of BRs are low, BZR1 is phosphorylated by the BRASSINOSTEROID-INSENSITIVE 2 (BIN2) GSK3-like kinase. As a result, its DNA-binding activity is lost and it remains in the cytoplasm. When BR levels are high, BZR1 is dephosphorylated by protein phosphatase 2A (PP2A). Activation of BRI1 results in a sequence of phosphorylation reactions which activates brassinosteroid-signaling kinases (BSKs), cytoplasmic kinase (CDG1) and members of the BRI1 Suppressor 1 (BSU1) family. This phosphatase activity leads to the inactivation of glycogen synthase kinase 3 (GSK3) through dephosphorylation of tyrosine. Dephosphorylated BZR1 is translocated into the nucleus where it regulates various target genes (Bai et al., 2012). It was documented in Arabidopsis that BRZ1 accumulated in the nucleus at elevated temperatures where it induced the expression of growth-promoting genes. BZR1 induced the expression of PIF4 by binding to its promoter and acting as a temperature-dependent positive growth regulator (Ibañez et al., 2018). A recent study revealed that organ size in Arabidopsis is regulated by BZR1. Enhanced expression of Zea mays BZR1 resulted in phenotypes of enlarged cotyledons, and increased size of floral organs, rosette leaves and seeds in transgenic Arabidopsis (Zhang et al., 2020).
BZR1 and BES1 have been documented as positive regulators of freezing tolerance. Gain-of-function mutants (bzr1-1D and bes1-D) of the transcription factors, BZR1 and BES1, showed increased tolerance to freezing, and the accumulation of dephosphorylated BZR1 upon cold treatment (Li et al., 2017). Chen et al., 2019 revealed that BZR1 was essential for locule development in Arabidopsis. Disruption of all BZRs resulted in generation of a hextuple mutant (bzr-h) which showed vegetative growth phenotypes that were similar to those of the BR receptor null mutant. Yin et al. (2018) demonstrated the role of BRZ1 in heat stress tolerance in tomato by regulating the feronia (FER) homologs. Mutations in BZR1 impeded the respiratory burst oxidase homolog 1 (RBOH1) generation of hydrogen peroxide (H2O2) in the apoplast and heat tolerance. Addition of exogenous H2O2 restored the heat tolerance of the tomato bzr1 mutant, and enhanced expression of BZR1 increased the generation of apoplastic H2O2 and improved heat stress tolerance.
Brassinazole (Brz) was reported as the first brassinosteroid biosynthesis inhibitor. Arabidopsis thaliana treated with Brz showed the phenotype of a brassinosteroid mutant but application of brassinolide restored wild type activity (Nagata et al., 2000). The triazole compound, propiconazol (Pcz), is a potent BR metabolism inhibitor in plants. Arabidopsis seedlings treated with Pcz showed various BR-deficient phenotypes like reduced primary root growth, reduced size of cotyledons, and epinastic growth of cotyledons. However, seedlings co-treated with Pcz and 24-epibrassinolide showed improved root length compared to controls (Hartwig et al., 2012). YCZ-18 is another BR inhibitor that targets C23-hydroxylation in the BR biosynthetic pathway. Wild-type Arabidopsis plants grown in medium supplemented with YCZ-18 had short opened cotyledons. A decline in endogenous BR levels was also observed in Arabidopsis treated with YCZ-18 (Oh et al., 2015a). A pyrimidine-type fungicide, fenarimol, was also reported to inhibit BR biosynthesis in Arabidopsis and cause loss of etiolation and dwarfism in the dark (Oh et al., 2015b). An imidazole fungicide, Imazalil used as a post-harvest antifungal agent binds to CYP51 and inhibits fungal biosynthesis of ergosterol. In A. thaliana, hypocotyl shortening was observed with Imazalil treatment, which was reversed by application of 24-epibrassinolide (Rozhon et al., 2019). It has been reported by Bajguz, 2019 that brassinozole (BRz) decreases the level of BRs in the leaves of Barley and this effect is reversed by the application of BRs exogenously. Voriconazole, fenpropimorph, and fluconazole are also some of the BR inhibitors that inhibit the synthesis of cycloeucalenol-obtusifoliol isomers that is responsible for decreasing the synthesis of BRs (Rozhon et al., 2019).
The metabolic pathways associated with BRs are related to successive signaling networks. Therefore, an inclusive understanding of BRs homeostasis in plants is imperative for establishing a comprehensive view of BR signaling cascades (Chung and Choe, 2013). The BR-linked pathways have been elucidated over the past decades and the studies revealed a complex BR signaling network with a crucial role in proper growth and development of plants (Anwar et al., 2018). The signaling cascade initiated by BR was categorized into three steps: (i) BR recognition and early activation of BRI1 receptor kinases, (ii) inactivation of BIN2 inhibitors, phosphatases, and kinases, and (iii) regulation of transcriptional factors such as BES1 and BZR1 (Clouse, 2011).
The BRs are initially recognized by brassinosteroid-insensitive 1 (BRI1) receptor kinase on the surface of the cells (Chakraborty et al., 2015). BRI1 is comprized of a leucine-enriched repeat receptor kinase with an extra-cellular domain that binds BRs and trans-phosphorylates signals. BRI1 is associated with a co-receptor, somatic embryogenesis receptor kinase (SERK), and belonging to a smaller LLRK family. BRI1 and SERK in combination form an active intricate complex that stimulates a downstream signal network involving activation of a wide array of kinases and phosphatases. Consequently, many transcription factors are activated which in turn alter the expression of specific genes (Belkhadir et al., 2012). Exogenously supplemented BRs bind to BRI1 and induce an association with BRI1-associated receptor kinase 1 (BAK1) and a disassociation of BRI1 kinase inhibitor 1 (BKI1). BKI1 is a membrane-bound negative regulator of BR signals, which combines with BRI1 and is responsible for prevention of co-receptor interference. It has been widely reported in Oryza sativa and A. thaliana plants (Jiang et al., 2015). Another protein, OsREM4.1, has similar functions to BRI1. It was identified in O. sativa and has corroborated that disruption of the BRI1/SERK complex suppresses downstream BR signaling. The expression of OsREM4.1 was positively modulated by ABA and elevated in response to increased ABA levels, and showed significant participation of the bZIP transcription factor (Gui et al., 2016). Various studies support the hypothesis that BRI1 is a multi-function kinase with the ability to phosphorylate Ser, Thr, and Tyr residues and it was affirmed that phosphorylation of Tyr residue is vital for specific aspects of BR signaling in plants (Oh et al., 2009). BRI1 was localized to the plasma membrane as a homodimer. It is ligand-independent and responsible for stabilization and initiation of BR binding and signaling (Albrecht et al., 2008). BAK1 (also termed SERK3) has been associated with phosphorylation of BRI1 and BAK1 both in-vivo and in-vitro (Wang et al., 2008). SERK4, also termed as BAK1-like (BKK1), has been shown to interact with BRI1 in-vivo in a BR-dependent manner (He et al., 2007). Another LRR RLK, FLS2, interacts with BAK1 and augments its function in plant defense (Bar et al., 2010). Karlova et al. (2006) observed a significant role for SERK1 in embryogenesis where it heterodimerizes with BRI1 and augments BR signaling.
In order to expound the entire mechanism of phosphorylation and oligomerization of BRI1 and BAK1 in response to exogenously supplemented BR, Wang et al. (2008) expressed various combinations of kinase-inactive and wild-type Arabidopsis BRI1 and BAK1 in the same transgenic plant. It was concluded that an active BRI1 was responsible for BR-dependent association of the pair, and not BAK1 kinase. Additionally, it was observed that when BAK1-green fluorescent protein (GFP) was highly expressed in the bri1-1 null mutant background, the phosphorylation was drastically lowered in BAK1-GFP. In-vitro studies indicated that BAK1 positively triggered BRI1 activity and that they can transphosphorylate each other at specific residues (Wang et al., 2008). The above studies with LC-MS/MS assays, biochemical analysis and functional characterization have aided in development of a new sequential transphosphorylation model of BRI1/BAK1 interaction. These studies further suggested that plant receptor kinases have certain properties similar to those of TGF-b and RTKs receptor kinases present in mammals, although they have plant-specific characteristics (Wang et al., 2008). Similar to BRI1, Tyr residue are also autophosphorylated and mutation studies revealed that Tyr phosphorylation regulated the expression of a BAKI subset in-vivo (Oh et al., 2010).
Increased BR signaling activity results in de-activation of brassinosteroidinsensitive-2(BIN2) kinase, which is considered the principal effect of BR signaling (Chung and Choe, 2013). Sequence analysis of BIN2 revealed significant similarity with mammalian glycogensynthasekinase3 (GSK3). There was also similarity to a member of a small family of ten related genes identified in Arabidopsis that have an essential role in BR signal transduction (Rozhon et al., 2010). The inactivation of BR results in activation of two related transcription factors, BRI-EMS suppressor-1(BES1) (Yin et al., 2005) and brassinazole-resistant 1 (BZR1) (He et al., 2005). BES1 is also known as BZR2 (Wang et al., 2002). More recent studies of downstream BR signaling in Arabidopsis have elucidated the association between BRI1 and BAK1 activation (Kim and Wang, 2010). Another observation by Wang and Chory (2006) using a yeast two-hybrid screen for BRI1 integrators suggested that BRI1 kinase inhibitor-1 (BKI1) acted as a negative modulator of BR signaling. In the absence of BAK1 it bound to BRI1 and inactivated its function and prevented further binding to BAK1.
Exogenous BR treatment results in dissociation of BKI1 from the outer cell surface and further inhibits the BR signaling network. Association of BRI1 and BKI1 was demonstrated in-vivo and in-vitro, which resulted in BKI1 phosphorylation and interaction with recombinant BRI1-CD in vitro. More recent observations showed that BKI1 interacted with BRI1 through a C-terminal 20-residue conserved segment (Jaillais et al., 2011). They additionally indicated that a peptide surrounding this binding site suppressed the association of BRI1 with BAK1. A Lys-Arg-enriched domain present within the BKI1 directed the protein toward the plasma membrane. This localization consequently phosphorylated Tyr-211 within the motif and released BKI1 from the plasma membrane in a BRI1- and BR-dependent manner. Over-expression of a BKI1-Y211F mutant construct in transgenic Arabidopsis consequently resulted in extremely dwarfed plants. This protein was reportedly membrane bound and found at the same location even after BR supplementation. This observation suggests that upon recognition, BR phosphorylates BRI1 on Tyr-211, resulting in its disassociation from the membrane. This permits BRI1 to freely associate with BAK1 and initiate BR signaling (Lemmon and Schlessinger, 2010; Lim and Pawson, 2010).
A proteomic screening of BR-modulated proteins identified a plethora of members of receptor-like cytoplasmic kinase families such as RLCK-XII, specifically BR-signaling kinases (BSKs). BSKs are a direct substrate of BRI1 and up-regulate BR signaling (Tang et al., 2008). BSKs have a presumed trans-membrane sequence with N-myristylation sites that are probably responsible for directing their membrane localization. Additionally, BSK1 and BSK3 have been observed to interact with BRI1 in-situ in the absence of other ligands. BRI1 phosphorylated BSK1, most likely on Ser-230, resulting in activation and its release from the receptor complex (Tang et al., 2008). This phosphorylated BSK1 interacted with BRI suppressor-1 (BSU1) phosphatase which then negatively modulated BIN2 (Kim et al., 2009). Ample biochemical and genetic evidence affirmed that BSK1 binding to BSU1 activated BSU1-mediated dephosphorylation of phosphoTyr-200 in BIN2, resulting in suppression of the BR signaling cascade (Kim et al., 2009).
Brassinazole-resistant 1(BZR1) and BRI 1-EMS suppressor-1 (BES1) are the chief transcriptional factors activated when BR binds to BRI1. BZR1 and BES1 (also known as BZR2) regulate a number of genes involved in various physiological processes (Peres et al., 2019). BIN2 (brassinosteroid insensitive-2), a GSK3/serine-threonine protein kinase, plays a key role in regulation of BZR1 AND BES1. In the BR-mediated signaling cascade, BIN2 acts as negative regulator by repressing the activity of BZR1 and BES1. In the absence of BR signaling, cytoplasmic CDG1 and BSKs interact with inactive BRI1 to prevent binding of BAK1 to the receptor. BES1 and BZR1 are phosphorylated by BIN2, and the phosphorylated transcription factors are expelled from the nucleus and retained in the cytoplasm by 14-3-3 proteins (Chung and Choe, 2013). The dephosphorylated BIN2 may be involved in degradation of BZR1 and BES1. Concomitantly, in the presence of BR, BRI1 recognized the signal followed by association with BAK1 and disassociation from BKI1. Auto- or trans-phosphorylation of BRI1 and BAK1 activates the main receptor, which is followed by phosphorylation of BSKs and CDG1. Activated BSKs and CDG1 stimulate BSU1 phosphatase, which dephosphorylates the major repressor protein, BIN2. Inactivation of BIN2 positively affects BZR1 and BES1 by terminating their degradation. In the absence of BIN2, phosphatase 2A dephosphorylates BZR1 and BES2 in the cytoplasm. The dephosphorylated transcription factors are than translocated to the nucleus where they affect expression of various BR-mediated genes. The mechanism of BRs signaling is illustrated in the Figure 3.
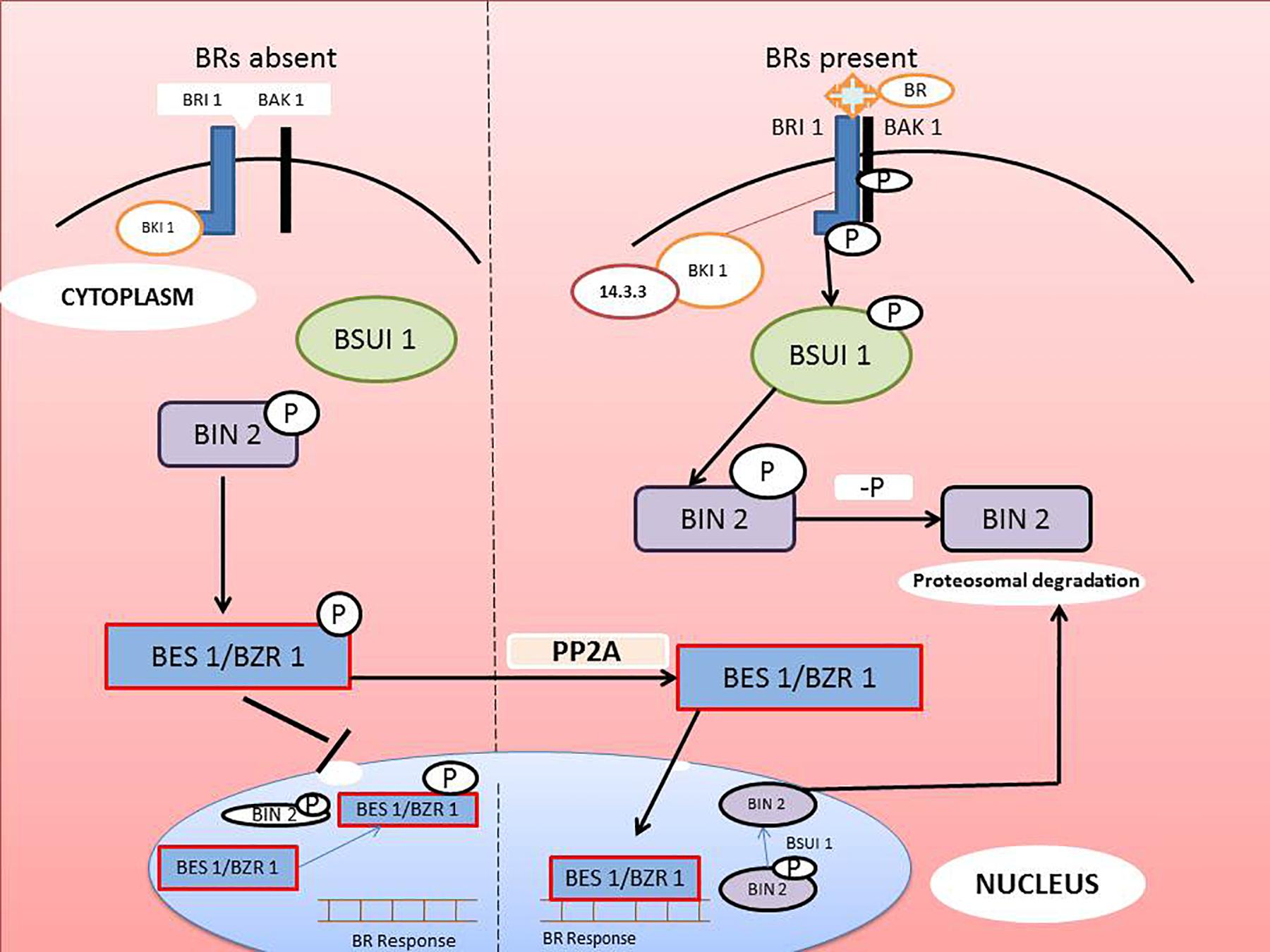
Figure 3. Molecular mechanism of BRs signaling from recognition at the cell surface to transcriptional activation of specific genes in the nucleus (modified from Chung and Choe, 2013).
In the BR-mediated signaling cascade, BES1 and BZR1 interact with the target genome product and play important roles in regulating gene expression. BES1/ BZR1 can act as an inducer as well as a repressor in the BR signaling pathway. BES1 binds to the CANNTG sequences (E-box) to stimulate gene expression, whereas BZR1 interacts with the CGTGT/CG sequences (BRRE) to suppress gene expression (He et al., 2005). MYB30, a BES1 target interacts with BES1 to strengthen BR induced signals. MYBL2 also functions as an important regulator in BR signaling. It is phosphorylated and stabilized by BIN2 kinase, thus playing an effective role in the transcriptional process of BR (Ye et al., 2012). Studies revealed that in the BR-repressed condition, there were more BRRE expressed, whereas E-boxes showed enhanced expression in BR-induced genes (Yu et al., 2011). The E-box interacts with BES1 and different transcription factors and cofactors. Interactions between BES1, PIL6, GLK1, and GLK2 have been associated with the transcriptional network pathway that triggers BR-induced gene expression. BZR1 interacted with PIF4 (a target gene product) to form a heterodimer that recognized a promoter element, CACGTG (G-box) (Oh et al., 2012). Interaction of BZR1 and PIFs are very important for normal growth of hypocotyls. Evidence has been obtained by studying mutants of PIF4 and the homologs (pif1 pif3 pif4 pif5), which showed reduced plant growth in darkness and inhibition of BR-induced growth. Plants have up to 2,000 target genes common for both BZR1 and PIF4, which also have various PREs involved in cell elongation (Guo H. et al., 2013). In the GA signaling pathway, in the absence of GA, the DELLA protein is stably localized to the nucleus and binds to PIFs and BZR, impeding the genome binding activity. An active GA signal leads to degradation of the DELLA protein thereby activating PIF and BZR, which target specific genes (Liu et al., 2018). Pioneer studies have revealed that in inhibition of chloroplast BES1 plays a role through GLK1 and GLK2. Inhibition of chloroplasts causes enlarged plastoglobules and other alterations in the structure of the organelle (Yu et al., 2011).
Plants survival is threatened in several ways but heavy metal stress is one of the most important concerns in agricultural research. The effects of metal contamination on plants is primarily ameliorated by hormones such as the BRs, that scavenge reactive oxygen species (ROS) and activate the antioxidant defense enzymes, superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), ascorbate peroxidase (APOX), glutathione reductase (GR), guaiacol peroxidase (GPOX), and glutathione-S-transferase (GST) (Bücker-Neto et al., 2017). BRs help the plant to become metal-tolerant, thereby increasing crop yield and quality (Vriet et al., 2012). The bioactive BRs, 28-homobrassinolide (HBL) and 24-epibrassinolide (EBL), are part of a system called assisted phytoremediation, which helps the plant to eliminate toxic metals (Barbafieri and Tassi, 2011). BRs reduce the uptake of heavy metals by altering cell membrane permeability and also induce a group of defensive enzymes. They stimulate the production of stress-proteins through actions on anti-stress genes due to increased expression of ATPase (Madhan et al., 2014). External application of BRs induces transient H2O2 formation, which activates MAPK, leading to the production of NADPH oxidase (Jiang et al., 2012) and upregulation of stress-proteins and defensive enzymes that curtail the metal stress (Yin et al., 2016). Jiang et al. (2012) suggested that low doses of EBR (0.1–0.15 μM) suppressed photosynthetic efficiency. According to Xia et al. (2014), 0.1 μM EBR stimulated stomatal opening while 1.0 μM EBR facilitated the closing of stomata (Figure 4).
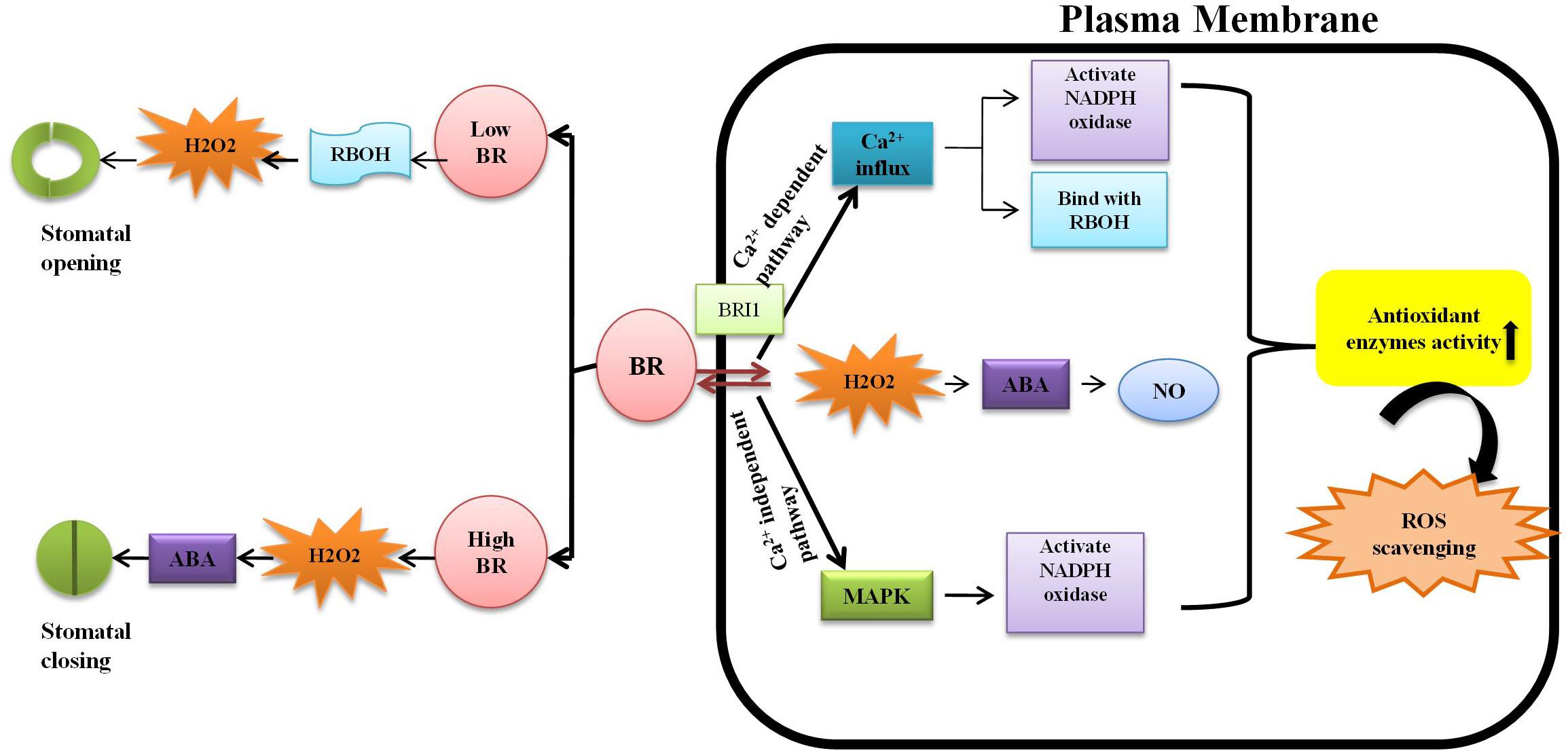
Figure 4. Different doses of Brassinosteroid regulates the opening and closing of stomata: at low concentration of BR activate respiratory burst oxidase homolog (RBOH) which further increases the level of H2O2 resulting in opening of stomata whereas at high concentration, it causes the accumulation of H2O2 and also reducing the uptake of K+ ions in guard cells lead to the activation of abscisic acid (ABA) cause closure of stomata (Daszkowska-Golec and Szarejko, 2013). Brassinosteroid hormone received by a specific receptor, brassinosteroid insensitive1 (BR1), on plasma membrane. Furthermore, there is increase in Ca2+ infux that activates the NADPH oxidase and then bind with EF-motif of RBOH enhancing the production of ROS (Xia et al., 2015); another pathway is upregulation of mitogen-activated protein kinase (MAPK) which further scavenge the reactive oxygen species (ROS); BR also stimulates the production of nitric oxide (NO) resulting the enhancement in antioxidative defense system (Zhang et al., 2011).
The mechanism of regulation of heavy metal toxicity via BRs involves: (a) stimulating H2O2 production, (b) scavenging ROS through boosting the defensive antioxidant system, (c) up-regulating MAPK expression, and (d) alleviating metal toxicity by increasing the concentration of potassium and sodium ions, proline, antioxidants, and osmolytes (Rajewska et al., 2016). Some reports indicated that NADPH oxidase was an important apoplastic source of H2O2 from conversion of O2– by superoxide dismutase in the plasma membrane of plant cells, which further increased H++ ATPase activity via upregulation of the CsHA gene (Jakubowska et al., 2015). Thus, while uptake of toxic metals can negatively affect plant cell membranes by ROS peroxidation of lipids and oxidation of proteins, phytohormones like BRs boost the level of antioxidants and defensive enzymes to ameliorate this toxicity and restore normal osmoregulation (Shahzad et al., 2018). The BR-specific inhibitor, brassinazole (Brz), and the bioactive brassinosteroid, 24-brassinolide (EBL), were applied for alleviating the toxicity of metals, and restoring the photosynthetic machinery and defense system of A. thaliana (Wu et al., 2019). BRs are vital for plant cellular homeostasis. They restore CO2 absorption and enhance antioxidant capacity, thus overcoming the toxic effects of heavy metals (Ahammed et al., 2020). BRI1 is a BR receptor on the plasma membrane (Nolan et al., 2020) that activates the signaling cascade up-regulating the expression of transcription factors, which enhance the transcription of brassinosteroid genes (Tong and Chu, 2018; Planas-Riverola et al., 2019). Increased expression of these genes enhances the endogenous level of BRs that help in mitigating metal stress (Xia et al., 2018), but the mechanism remains unclear. Table 1 summarizes the role of brassinosteroids in regulation of physiological and biochemical responses of plants growing under metal toxicity.
Brassinosteroids play a diverse and vital role in regulating plant metabolism because of their synergy with other plant hormones such as auxin, cytokinins (CK), ethylene, polyamines (PA), gibberellins (GA), salicylic acid (SA), JA, and ABA (Ohri et al., 2019). The bioactive BRs (HBL, EBL) can protect the plant from toxic metals by assisted phytoremediation (Barbafieri and Tassi, 2011). BRs reduce the uptake of toxic metals by altering cell permeability and reduce damage by activating defensive enzymes. The mechanism of BRs signaling and its interplay with other hormones at the molecular level (Peres et al., 2019) is illustrated in Figure 5.
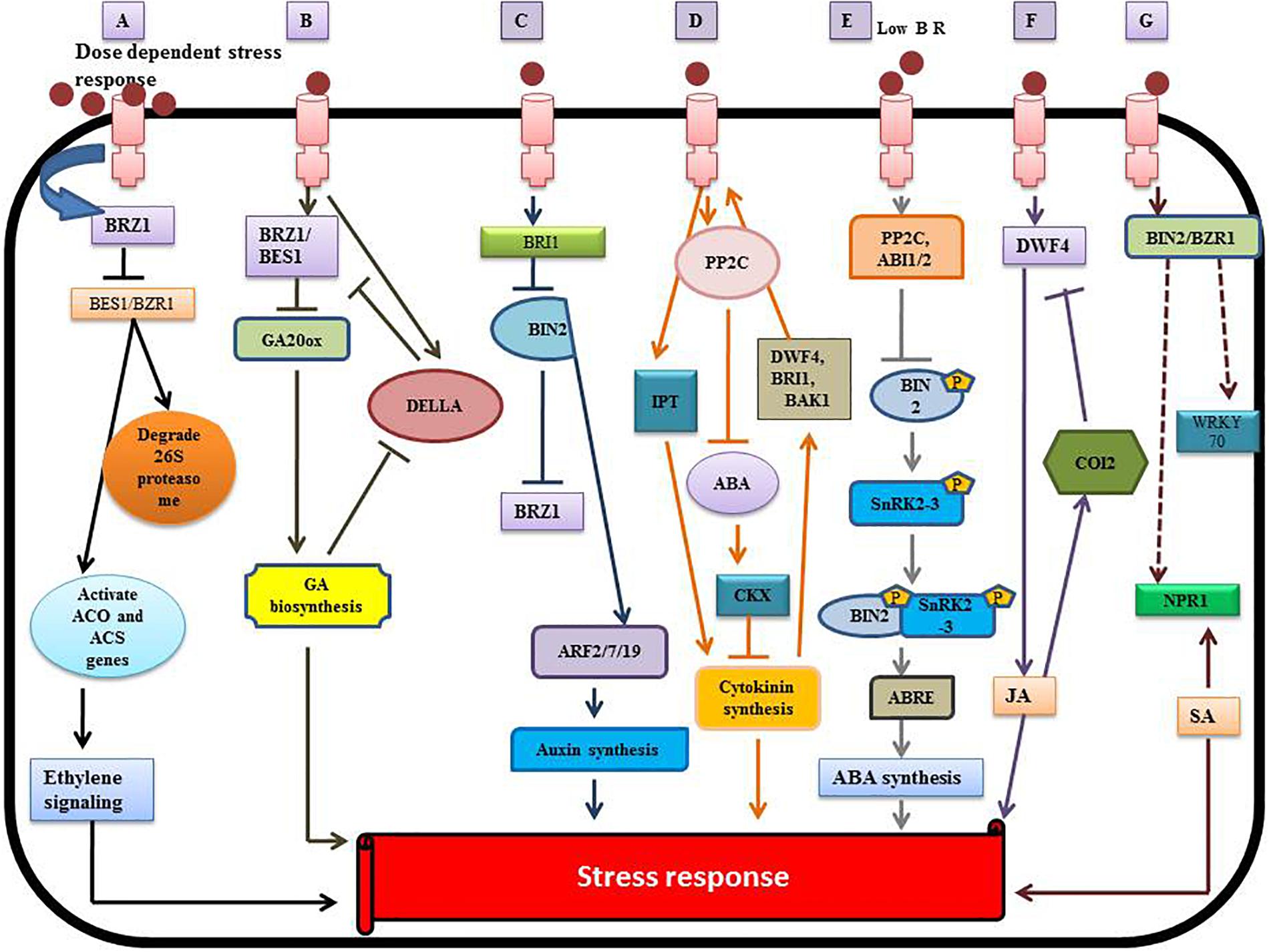
Figure 5. Crosstalk between BRs and other phytohormones. (A) Interplay between BR and ethylene. (B) Interaction between BR and gibberellin (GA). (C) Brassinosteroid – auxin crosstalk. (D) Brassinoster oid-cytokinin interaction. (E) Brassinosteroid-Abscisic acid (ABA) interplay. (F) Brassinosteroid-jasmonic acid (JA) crosstalk. (G) Brassinosteroid-salicylic acid (SA) [modified from Saini et al. (2015), Peres et al. (2019), and Ohri et al. (2019)], involving the regulation of various transcription factors.
Brassinosteroids stimulate the synthesis of ethylene by activating the expression of ACO (1-aminocyclopropane-1-carboxylic acid oxidase) and ACS (1-aminocyclopropane-1-carboxylic acid (ACC)-synthase enzyme) genes (Figure 5). BR regulates ethylene synthesis at the transcriptional and post-transcriptional level by increasing the ACS5 protein half-life (Hansen et al., 2009). BR regulates ethylene biosynthesis negatively or positively via a dose-dependent pathway (Lv et al., 2018). Some reports showed that exogenous application of BR enhanced the ripening of banana (Musa acuminata L.) by elevating the expression of MaACS1, MaACO13, and MaACO14 genes and regulating ethylene production (Guo et al., 2019). BR acts at the post-transcriptional level by increasing the expression of ACS2 and ACS4 in Solanum lycopersicum (Zhu et al., 2015). BR also stabilizes other ACS proteins like ACS6, ACS9 and ACS5 by degrading the 26S proteasome (Hansen et al., 2009). BR indirectly regulates the hyponastic growth by regulating expression of the ROT3/CYP90C1 gene to increase ethylene synthesis (Polko et al., 2013). The overproduction of BR genes (BRI1 and DWARF) enhances the biosynthesis of ethylene in tomato (Li and He, 2016; Nie et al., 2017).
The interplay between BRs (BZR1/BES1) and gibberellin (DELLA) genes is complex (Sun et al., 2010; Sun, 2011; Tong et al., 2014; Hu et al., 2017), reflecting interaction with both proteins and DNA (Li Q.F. et al., 2018; Li W. et al., 2018). The exogenous application of BR activates BZR1, which upregulates the expression of GA20ox genes and enhances GA production (Stewart Lilley et al., 2013). The DELLA gene has an inhibitory effect on the transcriptional activity of BZR1, but GA induces the degradation of DELLA (Bai et al., 2012). Therefore, both GA and BR affect expression of the target gene, BRZ1 (Figure 5). Thus, BES1/BRZ1 promotes production of GA, resulting in greater degradation of DELLA (Ross and Quittenden, 2016). There is direct interaction between DELLA and BZR1 that inhibits the binding of BZR1 to DNA. This suppresses the signal that is required for maintaining the etiolation of seedlings and elongation of cells (Li and He, 2013). The level of ROS-scavenging enzymes increases because of the interaction between DELLA protein and BRZ1 under heavy metal stress (Achard et al., 2008).
Both BRs and auxins are considered “master regulators” with synergistic effects on plant growth and development (Chaiwanon and Wang, 2015). BIN2-mediated phosphorylation reduces the inhibitory effect of auxin response factors (ARF2) (Vert et al., 2008) that leads to the enhancement of ARF promoter activities and stimulates the expression of BR-regulated genes promoting auxin synthesis. The indole-3-acetic acid/auxin (IAA/AUX) genes are also involved in BR-regulated auxin production (Li Q.F. et al., 2018; Li W. et al., 2018). The expression of genes associated with auxin transport such as PIN3, PIN4, PIN7, and LAX are suppressed by the BR signaling pathway (Nemhauser et al., 2004). This crosstalk suggests an interplay between auxin and BR in heavy metal tolerance through the auxin transport genes. Wang et al., 2015 reported that the movement and local concentration of auxin was regulated by the expression of CYP79B2, the ABCB family, Yucca (YUC), the PIN family, the Gretchen Hagen (GH3) genes, and phosphoribosyl anthranilate transferase 1 (PAT1), in response to heavy metals (Hacham et al., 2012). The auxin secretion transporters, PIN7 and PIN4, that regulate the movement and distribution of auxin, are managed by BES1 (Paponov et al., 2005). In the root elongation zone, BIN2 stimulates the post-transcriptional modification of ARF, and BZR1 transcriptionally activates ARF expression and auxin-responsive genes (Tian et al., 2018). Both ARF and auxin-related genes were suppressed by BZR1 in the quiescent region of the root (Chaiwanon and Wang, 2015). It has been reported in A. thaliana that auxin treatment increases DWF4 expression, which stimulates production of BR through the auxin-induced Bravis radix (BRX) gene (Chung et al., 2011); there is feedback inhibition of DWF4 by BR. Several reports discussed the adversary aspect of auxin and BR in controlling BR synthesis genes and the DWF4 gene (Maharjan et al., 2011).
Cytokinins (CKs) are adenine-derived compounds, which regulate the plant growth processes under abiotic stress (Perilli et al., 2010). CKs stimulate the defense mechanism that mitigates heavy metal toxicity and restores the photosynthetic apparatus. Piotrowska-Niczyporuk et al., 2012 reported the alleviation of Cd toxicity by enhancing photosynthesis efficiency and the level of primary metabolites. Two key enzymes regulate the biosynthesis of CKs: isopentenyltransferases (IPTs) which promote synthesis of CKs and CK oxidase/dehydrogenases (CKXs) which suppress genes involved in CK synthesis. Both enzymes target the responses mediated by BR (Werner and Schmülling, 2009). Exogenous application of BR leads to overexpression of CKX3 and ectopic expression of BRI1, which increases the leaf and root length. This suggests crosstalk between BR and CKs, which may be involved in enhancing crop yield (Yuldashev et al., 2012). Some reports showed that interaction between BR and CKs resulted in the accumulation of anthocyanins (Yuan et al., 2014). ABA is also involved in the interaction between BR and CKs, as it suppresses BR synthesis during metal toxicity (Zhang et al., 2009). Therefore, there are three hormones interacting in a complex way that remains unclear.
Abscisic acid is known to be a “sensing element” for abiotic stress that protects the plant from various kinds of stress (Fujii and Zhu, 2009). The BR and ABA interplay depend upon the regulation of gene expression and modulation of protein activity. The complex formed by the merging of histone deacetylase19 (HDAC19), topless (TPL/TPR) and BRI1-EMS suppressor1 (BES1) affects the E-box promoter causing suppression of ABA insensitive 3 (ABI3) gene expression in the presence of BR. There is also repression of ABI3 and ABI5 gene expression through the interaction of the BZR1 transcription factor with the ABI5 G-Box promoter sequences. This reduces the stress response by down regulating the ABA-regulated gene expression. BIN2 promotes activation of SnRK2.3 that stimulates the stress response at low levels of BR. ABA-related gene expression is upregulated through phosphorylation of the ABI5 transcription factor by BIN2 (Wang et al., 2018). Some reports also showed auto-stimulation of downstream expression of ABA-related SnRK2s genes and kinase activity (Belin et al., 2006; Yunta et al., 2011).
CORONATINE INSENSITIVE 1 (COI1) is an F-box protein responsible for JA signaling and responses in A. thaliana (Yan et al., 2009; Peng et al., 2011) reported that blocking JA signaling caused an accumulation of anthocyanins in Arabidopsis under the influence of brassinazole. Upregulation and downregulation of JA transcript factors as well as signaling genes depended upon the relative concentration of BR in plants (Peng et al., 2011). The exogenous application of JA decreased expression of the BR signaling gene, OSBRI1 and the BR biosynthesis gene, OsDWF4, revealing the interplay between JA and BR in O. sativa (Nahar et al., 2013).
Activation of systemic acquired resistance (SAR) is a putative role of SA during abiotic and biotic stress (Roychoudhury et al., 2016). Divi et al., 2010, studied the induction of BR-mediated stress tolerance by regulating BIN2 and BZR1. During the stress response, the OsWRKY45 transcription factor was expressed in the presence of SA (Huangfu et al., 2016). Some reports revealed that the metabolite composition was affected by the cumulative action of both SA and BR (24-epibrassinolide) leading to a decline in lead toxicity (Kohli et al., 2018). The interplay between BR and SA may be due to the NPR1 gene (non-expressor of pathogenesis-related genes 1) which stimulates expression of the SA-related genes involved in plant defense (Ohri et al., 2015). NPR1 also regulates the BR signaling genes, BIN2 and BZRI, which induces stress tolerance in plants (Divi et al., 2010).
Polyamines are associated with disease and the aging process. Previous studies reported that PAs did not affect the biosynthesis and signaling of BR (Anwar et al., 2015), butco-treatment with BR and PAs improved tolerance against copper stress. Choudhary et al. (2012b), reported enhanced Cu tolerance in radish through exogenous application of EBR and spermidine (Spd), which decreased the uptake of Cu and upregulated the expression of RsCOPT2 and RsCOPT1 genes. BR controls the spermidine level in plants, which further raises the concentration of putrescine as required for stress tolerance as well as diminishing cadaverine levels to counterbalance the oxidative burst (Takahashi and Kakehi, 2010). Pál et al., 2017, showed that in cadmium toxicity, Pas caused phytochelatin production in O. sativa. Recent reports suggest that the crosstalk between BR and PAs requires modulation of the expression of various enzymes associated with PA synthesis and its interplay with other plant hormones. As summarized in recent findings, this crosstalk between BRs and other phytohormones as well as their interaction showed both positive and antagonistic effects in response to stress.
In this review, we have discussed the molecular mechanisms involved in the biosynthesis of the BRs, the roles of various transcriptional factors on gene expression, the interactions with auxin and other molecules, and the modulating effects of BES1/BZR1 in the biosynthesis of BRs. The molecular mechanism of BR signaling from its recognition on the plasma membrane to the transcriptional activation of specific genes in the nucleus has also been reviewed in detail. As a leading example of the importance of the BRs, we examined their role in heavy metal stress and their crosstalk with other phytohormones under stress conditions. But more work needs to be done for a detailed understanding of how BRs are transported out of cells and how they protect the plant during oxidative stress. There is a knowledge gap that needs attention to unravel the interactions between these phytohormones and various metabolites and transcription factors, which will add a new direction to the study of stress responses in plants.
AS, ML, and RB designed the outline and revised the initial draft. All authors were equally involved in writing of current version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ABI1/2, abscisic acid-insensitive 1 and 2; ABRE, abscisic acid response elements; ACO, 1-aminocyclopropane-1-carboxylic acid (ACC)-oxidase; ACS, 1-aminocyclopropane-1-carboxylic acid (ACC)-synthase enzyme; APOX, ascorbate peroxidase; BIN2, brassinosteroid-insensitive 2; BZR1, brassinazole-resistant 1; BZR1/BES1, brassinazole-resistant 1/BRI1-EMS suppressor1; CAT, catalase; Cd, cadmium; CKX, CK oxidases/dehydrogenases; Cr, chromium; Cu, copper; DHAR, dehydroascorbate reductase; EBL, 24-epibrassinolide; GPOX, guaiacol peroxidase; GR, glutathione reductase; GSH, reduced glutathione; GST, glutathione-S-transferase; H2O2, hydrogen peroxide; HBL, 28-homobrassinolide; Hg, mercury; IPT, isopentenyltransferases; MDA, malondialdehyde; MDHAR, monodehydroascorbate reductase; Mn, manganese; Ni, nickel; NPR1, non-expressor of pathogenesis-related genes 1; Pb, lead; POD, peroxidase; PP2C, protein phosphatase 2C; PPO, polyphenol oxidase; ROS, reactive oxygen species; Sb, antimony; SOD, superoxide dismutase; Zn, zinc.
Achard, P., Renou, J. P., Berthomé, R., Harberd, N. P., and Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18, 656–660. doi: 10.1016/j.cub.2008.04.034
Ahammed, G. J., Li, X., Liu, A., and Chen, S. (2020). Brassinosteroids in Plant Tolerance to Abiotic Stress. J. Plant Growth Regul. 2020, 1–14.
Albrecht, C., Russinova, E., Kemmerling, B., Kwaaitaal, M., and de Vries, S. C. (2008). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and-independent signaling pathways. Plant Physiol. 148, 611–619. doi: 10.1104/pp.108.123216
Ali, B. (2019). Brassinosteroids: The Promising Plant Growth Regulators in Horticulture. In Brassinosteroids: Plant Growth and Development. Singapore: Springer, 349–365.
Anwar, A., Liu, Y., Dong, R., Bai, L., Yu, X., and Li, Y. (2018). The physiological and molecular mechanism of brassinosteroid in response to stress: a review. Biol. Res. 51:46.
Anwar, R., Mattoo, A. K., and Handa, A. K. (2015). Polyamine interactions with plant hormones: crosstalk at several levels. In Polyamines. Tokyo: Springer, 267–302.
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Bajguz, A. (2010). An enhancing effect of exogenous brassinolide on the growth and antioxidant activity in Chlorella vulgaris cultures under heavy metals stress. Environ. Exp. Bot. 68, 175–179. doi: 10.1016/j.envexpbot.2009.11.003
Bajguz, A., and Tretyn, A. (2003). The chemical characteristic and distribution of brassinosteroids in plants. Phytochem 62, 1027–1046. doi: 10.1016/s0031-9422(02)00656-8
Bajguz, A. (2019). Brassinosteroids in microalgae: application for growth improvement and protection against abiotic stresses. In Brassinosteroids: Plant Growth and Development. Singapore: Springer, 45–58.
Bajguz, A. (2002). Brassinosteroids and lead as stimulators of phytochelatins synthesis in Chlorella vulgaris. J. Plant Physiol. 159, 321–324. doi: 10.1078/0176-1617-00654
Bar, M., Sharfman, M., Ron, M., and Avni, A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63, 791–800. doi: 10.1111/j.1365-313x.2010.04282.x
Barbafieri, M., and Tassi, E. (2011). Brassinosteroids for phytoremediation application. In Brassinosteroids: a class of plant hormone. Dordrecht: Springer, 403–437.
Bartwal, A., and Arora, S. (2020). Brassinosteroids: Molecules with Myriad Roles. Co-Evol. Sec. Metabol. 2020, 869–895. doi: 10.1007/978-3-319-96397-6_18
Belin, C., de Franco, P. O., Bourbousse, C., Chaignepain, S., Schmitter, J. M., and Vavasseur, A. (2006). Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 141, 1316–1327. doi: 10.1104/pp.106.079327
Belkhadir, Y., Jaillais, Y., Epple, P., Balsemão-Pires, E., Dangl, J. L., and Chory, J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. 109, 297–302. doi: 10.1073/pnas.1112840108
Bhanu, A. N. (2019). Brassinosteroids: Relevance in Biological Activities of Plants and Agriculture. J. Plant Sci. Res. 35, 1–15. doi: 10.32381/jpsr.2019.35.01.1
Bishop, G. J. (2007). Refining the plant steroid hormone biosynthesis pathway. Trends Plant Sci. 12, 377–380. doi: 10.1016/j.tplants.2007.07.001
Bücker-Neto, L., Paiva, A. L. S., Machado, R. D., Arenhart, R. A., and Margis-Pinheiro, M. (2017). Interactions between plant hormones and heavy metals responses. Genet. Mol. Bio. 40, 373–386. doi: 10.1590/1678-4685-gmb-2016-0087
Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. doi: 10.1242/dev.01403
Chaiwanon, J., and Wang, Z. Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25, 1031–1042. doi: 10.1016/j.cub.2015.02.046
Chakraborty, N., Sharma, P., Kanyuka, K., Pathak, R. R., Choudhury, D., Hooley, R., et al. (2015). G-protein α-subunit (GPA1) regulates stress, nitrate and phosphate response, flavonoid biosynthesis, fruit/seed development and substantially shares GCR1 regulation in A. thaliana. Plant Mol. Biol. 89, 559–576. doi: 10.1007/s11103-015-0374-2
Chen, J., Fei, K., Zhang, W., Wang, Z., Zhang, J., and Yang, J. (2021). Brassinosteroids mediate the effect of high temperature during anthesis on the pistil activity of photo-thermosensitive genetic male-sterile rice lines. Crop J. 9, 109–119.
Chen, L. G., Gao, Z., Zhao, Z., Liu, X., Li, Y., Zhang, Y., et al. (2019). BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mole. Plant 12, 1408–1415. doi: 10.1016/j.molp.2019.06.006
Cheng, X., Gou, X., Yin, H., Mysore, K. S., Li, J., and Wen, J. (2017). Functional characterisation of brassinosteroid receptor MtBRI1 in Medicago truncatula. Sci. Rep. 7, 1–12.
Chi, C., Li, X., Fang, P., Xia, X., Shi, K., Zhou, Y., et al. (2020). Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 71, 1092–1106. doi: 10.1093/jxb/erz466
Choudhary, S. P., Kanwar, M., Bhardwaj, R., Yu, J. Q., and Tran, L. S. P. (2012a). Chromium stress mitigation by polyamine-brassinosteroid application involves phytohormonal and physiological strategies in Raphanus sativus L. PLoS One 7:e33210. doi: 10.1371/journal.pone.0033210
Choudhary, S. P., Oral, H. V., Bhardwaj, R., Yu, J. Q., and Tran, L. S. P. (2012b). Interaction of brassinosteroids and polyamines enhances copper stress tolerance in Raphanus sativus. J. Exp. Bot. 63, 5659–5675. doi: 10.1093/jxb/ers219
Chung, Y., and Choe, S. (2013). The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit. Rev. Plant Sci. 32, 396–410. doi: 10.1080/07352689.2013.797856
Chung, Y., Maharjan, P. M., Lee, O., Fujioka, S., Jang, S., Kim, B., et al. (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 66, 564–578. doi: 10.1111/j.1365-313x.2011.04513.x
Clouse, S. D. (2011). Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 23, 1219–1230. doi: 10.1105/tpc.111.084475
Clouse, S. D., and Sasse, J. M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
Dalyan, E., Yüzbaşıoğlu, E., and Akpınar, I. (2018). Effect of 24-epibrassinolide on antioxidative defence system against lead-induced oxidative stress in the roots of Brassica juncea L. seedlings. Russian J. Plant Physiol. 65, 570–578. doi: 10.1134/s1021443718040118
Daszkowska-Golec, A., and Szarejko, I. (2013). Open or close the gate–stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 4:138.
Divi, U. K., Rahman, T., and Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Bio. 10:151. doi: 10.1186/1471-2229-10-151
dos Santos, L. R., da Silva, B. R. S., Pedron, T., Batista, B. L., and da Silva, and Lobato, A. K. (2020). 24-Epibrassinolide improves root anatomy and antioxidant enzymes in soybean plants subjected to zinc stress. J. Soil Sci. Plant Nutri. 20, 105–124.
Fariduddin, Q., Ahmed, M., Mir, B. A., Yusuf, M., and Khan, T. A. (2015). 24-Epibrassinolide mitigates the adverse effects of manganese induced toxicity through improved antioxidant system and photosynthetic attributes in Brassica juncea. Env. Sci. and Pollut. Res. 22, 11349–11359. doi: 10.1007/s11356-015-4339-4
Filová, A., Sytar, O., and Krivosudská, E. (2013). Effects of brassinosteroid on the induction of physiological changes in Helianthus annuus L. under copper stress. Acta Uni. Agri. et Silvi. Mend. Brun. 61, 623–629. doi: 10.11118/actaun201361030623
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fujioka, S., and Sakurai, A. (1997). Brassinosteroids. Nat. Prod. Rep. 14, 1–10. doi: 10.1007/978-94-017-0948-4_1
Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Ann. Rev. Plant Biol. 54, 137–164.
Fujioka, S., Noguchi, T., Yokota, T., Takatsuto, S., and Yoshida, S. (1998). Brassinosteroids in Arabidopsis thaliana. Phytochem. 48, 595–599. doi: 10.1016/s0031-9422(98)00065-x
Gray, W. M., Östin, A., Sandberg, G., Romano, C. P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. 95, 7197–7202. doi: 10.1073/pnas.95.12.7197
Gui, J., Zheng, S., Liu, C., Shen, J., Li, J., and Li, L. (2016). OsREM4. 1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell. 38, 201–213. doi: 10.1016/j.devcel.2016.06.011
Guo, H., Li, L., Aluru, M., Aluru, S., and Yin, Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Bio. 16, 545–553. doi: 10.1016/j.pbi.2013.08.002
Guo, R., Qian, H., Shen, W., Liu, L., Zhang, M., Cai, C., et al. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64, 2401–2412. doi: 10.1093/jxb/ert094
Guo, Y. F., Shan, W., Liang, S. M., Wu, C. J., Wei, W., Chen, J. Y., et al. (2019). MaBZR1/2 act as transcriptional repressors of ethylene biosynthetic genes in banana fruit. Physio. Planta. 165, 555–568. doi: 10.1111/ppl.12750
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., and Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 22, 1161–1173. doi: 10.1105/tpc.109.069203
Hacham, Y., Sela, A., Friedlander, L., and Savaldi-Goldstein, S. (2012). BRI1 activity in the root meristem involves post-transcriptional regulation of PIN auxin efflux carriers. Plant Sign. Behav. 7, 68–70. doi: 10.4161/psb.7.1.18657
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Bio. 49, 373–385. doi: 10.1007/978-94-010-0377-3_9
Hansen, M., Chae, H. S., and Kieber, J. J. (2009). Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 57, 606–614. doi: 10.1111/j.1365-313x.2008.03711.x
Hartmann, M. A. (1998). Plant sterols and the membrane environment. Trends Plant Sci. 3, 170–175. doi: 10.1016/s1360-1385(98)01233-3
Hartwig, T., Corvalan, C., Best, N. B., Budka, J. S., Zhu, J.-Y., et al. (2012). Propiconazole Is a Specific and Accessible Brassinosteroid (BR) Biosynthesis Inhibitor for Arabidopsis and Maize. PLoS One. 7:e36625. doi: 10.1371/journal.pone.0036625
Hasan, S. A., Hayat, S., and Ahmad, A. (2011). Brassinosteroids protect photosynthetic machinery against the cadmium induced oxidative stress in two tomato cultivars. Chemosphere 84, 1446–1451. doi: 10.1016/j.chemosphere.2011.04.047
Hayat, S., Alyemeni, M. N., and Hasan, S. A. (2012). Foliar spray of brassinosteroid enhances yield and quality of Solanum lycopersicum under cadmium stress. Saudi J. Bio. Sci. 19, 325–335. doi: 10.1016/j.sjbs.2012.03.005
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., et al. (2007). BAK1 and BKK1 regulate brassinosteroid dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115. doi: 10.1016/j.cub.2007.05.036
Heldt, H. W., and Piechulla, B. (2011). In the photorespiratory pathway phosphoglycolate formed by the oxygenase activity of Rubisco is recycled. Plant Biochem. London: Academic Press, 193–209.
Hu, S., Wang, C., Sanchez, D. L., Lipka, A. E., Liu, P., Yin, Y., et al. (2017). Gibberellins promote brassinosteroids action and both increase heterosis for plant height in maize (Zea mays L.). Front. Plant Sci. 8:1039.
Huangfu, J., Li, J., Li, R., Ye, M., Kuai, P., Zhang, T., et al. (2016). The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 17:697. doi: 10.3390/ijms17060697
Hussain, A., Nazir, F., and Fariduddin, Q. (2019). 24-epibrassinolide and spermidine alleviate Mn stress via the modulation of root morphology, stomatal behavior, photosynthetic attributes and antioxidant defense in Brassica juncea. Physio. Mol. Bio. Plants. 25, 905–919. doi: 10.1007/s12298-019-00672-6
Ibañez, C., Delker, C., Martinez, C., Bürstenbinder, K., Janitza, P., Lippmann, R., et al. (2018). Brassinosteroids dominate hormonal regulation of plant thermo morphogenesis via BZR1. Curr. Biol. 28, 303–310. doi: 10.1016/j.cub.2017.11.077
Jaillais, Y., Hothorn, M., Belkhadir, Y., Dabi, T., Nimchuk, Z. L., Meyerowitz, E. M., et al. (2011). Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–237. doi: 10.1101/gad.2001911
Jakubowska, D., and Janicka, M. (2017). The role of brassinosteroids in the regulation of the plasma membrane H+-ATPase and NADPH oxidase under cadmium stress. Plant Sci. 264, 37–47. doi: 10.1016/j.plantsci.2017.08.007
Jakubowska, D., Janicka-Russak, M., Kabała, K., Migocka, M., and Reda, M. (2015). Modification of plasma membrane NADPH oxidase activity in cucumber seedling roots in response to cadmium stress. Plant Sci. 234, 50–59. doi: 10.1016/j.plantsci.2015.02.005
Jan, S., Alyemeni, M. N., Wijaya, L., Alam, P., Siddique, K. H., and Ahmad, P. (2018). Interactive effect of 24-epibrassinolide and silicon alleviates cadmium stress via the modulation of antioxidant defense and glyoxalase systems and macronutrient content in Pisum sativum L. seedlings. BMC Plant Biol. 18, 1–18. doi: 10.1186/s12870-018-1359-5
Je, B. I., Piao, H. L., Park, S. J., Park, S. H., Kim, C. M., Xuan, Y. H., et al. (2010). RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell. 22, 1777–1791. doi: 10.1105/tpc.109.069575
Jiang, J., Zhang, C., and Wang, X. (2015). A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell. 27, 361–374. doi: 10.1105/tpc.114.133678
Jiang, Y. P., Cheng, F., Zhou, Y. H., Xia, X. J., Mao, W. H., Shi, K., et al. (2012). Cellular glutathione redox homeostasis plays an important role in the brassinosteroid−induced increase in CO2 assimilation in Cucumis sativus. New Phytol. 194, 932–943. doi: 10.1111/j.1469-8137.2012.04111.x
Kang, Y. Y., and Guo, S. R. (2011). Role of brassinosteroids on horticultural crops. In Brassinosteroids: A class of plant hormone. Dordrecht: Springer, 269–288.
Kanwar, M. K., Bhardwaj, R., Arora, P., Chowdhary, S. P., Sharma, P., and Kumar, S. (2012). Plant steroid hormones produced under Ni stress are involved in the regulation of metal uptake and oxidative stress in Brassica juncea L. Chemosphere 86, 41–49. doi: 10.1016/j.chemosphere.2011.08.048
Kapoor, D., Kaur, S., and Bhardwaj, R. (2014). Physiological and biochemical changes in Brassica juncea plants under Cd-induced stress. BioMed Res. Int. 2:2014.
Karlova, R., Boeren, S., Russinova, E., Aker, J., Vervoort, J., and de Vries, S. (2006). The Arabidopsis Somatic Embryogenesis Receptor-Like Kinase1 Protein Complex Includes Brassinosteroid- Insensitive1. Plant Cell. 18, 626–638. doi: 10.1105/tpc.105.039412
Kim, B. K., Fujioka, S., Takatsuto, S., Tsujimoto, M., and Choe, S. (2008). Castasterone is a likely end product of brassinosteroid biosynthetic pathway in rice. Biochem. Biophysio. Res. Commun. 374, 614–619. doi: 10.1016/j.bbrc.2008.07.073
Kim, T. W., and Wang, Z. Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681–704. doi: 10.1146/annurev.arplant.043008.092057
Kim, T. W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J. X., et al. (2009). Brassinosteroid signaltransduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260. doi: 10.1038/ncb1970
Kim, T. W., Hwang, J. Y., Kim, Y. S., Joo, S. H., Chang, S. C., Lee, J. S., et al. (2005). Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell. 17, 2397–2412. doi: 10.1105/tpc.105.033738
Kohli, S. K., Handa, N., Sharma, A., Gautam, V., Arora, S., Bhardwaj, R., et al. (2018). Combined effect of 24-epibrassinolide and salicylic acid mitigates lead (Pb) toxicity by modulating various metabolites in Brassica juncea L. seedlings. Protoplasma 255, 11–24. doi: 10.1007/s00709-017-1124-x
Kwon, M., Fujioka, S., Jeon, J. H., Kim, H. B., Takatsuto, S., Yoshida, S., et al. (2005). A double mutant for theCYP85A1 andCYP85A2 Genes of Arabidopsis exhibits a Brassinosteroid dwarf phenotype. J. Plant Biol. 48, 237–244. doi: 10.1007/bf03030413
Lemmon, M. A., and Schlessinger, J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134. doi: 10.1016/j.cell.2010.06.011
Li, Q.-F., and He, J.-X. (2013). Mechanisms of signaling crosstalk between brassinosteroids and gibberellins. Plant Signal Behav. 8:e24686. doi: 10.4161/psb.24686
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., and Yang, S. (2017). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant. 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Li, Q. F., and He, J. X. (2016). BZR1 interacts with HY5 to mediate brassinosteroid-and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant. 9, 113–125. doi: 10.1016/j.molp.2015.08.014
Li, Q. F., Lu, J., Yu, J. W., Zhang, C. Q., He, J. X., and Liu, Q. Q. (2018). The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta 1861, 561–571. doi: 10.1016/j.bbagrm.2018.04.003
Li, W., Nishiyama, R., Watanabe, Y., Van Ha, C., Kojima, M., An, P., et al. (2018). Effects of overproduced ethylene on the contents of other phytohormones and expression of their key biosynthetic genes. Plant Physio. Biochem. 128, 170–177. doi: 10.1016/j.plaphy.2018.05.013
Lim, W. A., and Pawson, T. (2010). Phosphotyrosine signaling: Evolving a new cellular communication system. Cell 142, 661–667. doi: 10.1016/j.cell.2010.08.023
Lima, M. D. R., Junior, U. D. O. B., Batista, B. L., and da Silva, and Lobato, A. K. (2018). Brassinosteroids mitigate iron deficiency improving nutritional status and photochemical efficiency in Eucalyptus urophylla plants. Trees 32, 1681–1694. doi: 10.1007/s00468-018-1743-7
Liu, F., Wang, P., Zhang, X., Li, X., Yan, X., Fu, D., et al. (2018). The genetic and molecular basis of crop height based on a rice model. Planta 247, 1–26. doi: 10.1007/s00425-017-2798-1
Liu, J., Zhang, D., Sun, X., Ding, T., Lei, B., and Zhang, C. (2017). Structure-activity relationship of brassinosteroids and their agricultural practical usages. Steroids 124, 1–17. doi: 10.1016/j.steroids.2017.05.005
Lv, B., Tian, H., Zhang, F., Liu, J., Lu, S., Bai, M., et al. (2018). Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genetics 14:e1007144. doi: 10.1371/journal.pgen.1007144
Madhan, M., Mahesh, K., and Rao, S. S. (2014). Effect of 24-epibrassinolide on aluminium stress induced inhibition of seed germination and seedling growth of Cajanus cajan (L.) Millsp. Int. J. Multidiscipl. Curr. Res. 2, 286–290.
Maharjan, P. M., and Choe, S. (2011). High temperature stimulates DWARF4 (DWF4) expression to increase hypocotyl elongation in Arabidopsis. J. Plant Bio. 54:425. doi: 10.1007/s12374-011-9183-6
Maharjan, P. M., Schulz, B., and Choe, S. (2011). BIN2/DWF12 antagonistically transduces brassinosteroid and auxin signals in the roots of Arabidopsis. J. Plant Bio. 54, 126–134. doi: 10.1007/s12374-010-9138-3
Mir, B. A., Khan, T. A., and Fariduddin, Q. (2015). 24-epibrassinolide and spermidine modulate photosynthesis and antioxidant systems in Vigna radiata under salt and zinc stress. Int. J. Adv. Res. 3, 592–608.
Mouchel, C. F., Osmont, K. S., and Hardtke, C. S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461. doi: 10.1038/nature05130
Nagata, N., Min, Y. K., Nakano, T., Asami, T., and Yoshida, S. (2000). Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta 211, 781–790. doi: 10.1007/s004250000351
Nahar, K., Kyndt, T., Hause, B., Höfte, M., and Gheysen, G. (2013). Brassinosteroids suppress rice defense against root-knot nematodes through antagonism with the jasmonate pathway. Mol. Plant-Microbe Inter. 26, 106–115. doi: 10.1094/mpmi-05-12-0108-fi
Nemhauser, J. L., Mockler, T. C., and Chory, J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2:e258. doi: 10.1371/journal.pbio.0020258
Nie, S., Huang, S., Wang, S., Cheng, D., Liu, J., Lv, S., et al. (2017). Enhancing brassinosteroid signaling via overexpression of tomato (Solanum lycopersicum) SlBRI1 improves major agronomic traits. Front. Plant Sci. 8:1386.
Nolan, T. M., Vukašinoviæ, N., Liu, D., Russinova, E., and Yin, Y. (2020). Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 32, 295–318. doi: 10.1105/tpc.19.00335
Nolan, T., Chen, J., and Yin, Y. (2017). Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474, 2641–2661. doi: 10.1042/bcj20160633
Oh, E., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Oh, K., Matsumoto, T., Yamagami, A., Hoshi, T., Nakano, T., and Yoshizawa, Y. (2015b). Fenarimol, a pyrimidine-type fungicide, inhibits brassinosteroid biosynthesis. Int. J. Mol. Sci. 16, 17273–17288. doi: 10.3390/ijms160817273
Oh, K., Matsumoto, T., Yamagami, A., Ogawa, A., Yamada, K., Suzuki, R., et al. (2015a). YCZ-18 is a new brassinosteroid biosynthesis inhibitor. PLoS One. 10:e0120812. doi: 10.1371/journal.pone.0120812
Oh, M. H., Wang, X., Kota, U., Goshe, M. B., Clouse, S. D., and Huber, S. C. (2009). Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. 106, 658–663. doi: 10.1073/pnas.0810249106
Oh, M. H., Wang, X., Wu, X., Zhao, Y., Clouse, S. D., and Huber, S. C. (2010). Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. U. S. A. 107, 17827–17832. doi: 10.1073/pnas.0915064107
Ohnishi, T., Godza, B., Watanabe, B., Fujioka, S., Hategan, L., Ide, K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287, 31551–31560. doi: 10.1074/jbc.m112.392720
Ohri, P., Bhardwaj, R., Bali, S., Kaur, R., Jasrotia, S., Khajuria, A., et al. (2015). The common molecular players in plant hormone crosstalk and signaling. Curr. Prot. Pep. Sci. 16, 369–388. doi: 10.2174/1389203716666150330141922
Ohri, P., Bhardwaj, R., Kaur, R., Jasrotia, S., Parihar, R. D., Khajuria, A., et al. (2019). Emerging Trends on Crosstalk of BRS with Other Phytohormones. In Brassinosteroids: Plant Growth and Development. Singapore: Springer, 425–441.
Pál, M., Csávás, G., Szalai, G., Oláh, T., Khalil, R., Yordanova, R., et al. (2017). Polyamines may influence phytochelatin synthesis during Cd stress in rice. J. Hazardous Mater. 340, 272–280. doi: 10.1016/j.jhazmat.2017.07.016
Paponov, I. A., Teale, W. D., Trebar, M., Blilou, I., and Palme, K. (2005). The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10, 170–177. doi: 10.1016/j.tplants.2005.02.009
Planas-Riverola, A., Gupta, A., Betegón-Putze, I., Bosch, N., Ibañes, M., and Caño-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146:dev151894. doi: 10.1242/dev.151894
Peng, Z., Han, C., Yuan, L., Zhang, K., Huang, H., and Ren, C. (2011). Brassinosteroid enhances jasmonate−induced anthocyanin accumulation in Arabidopsis seedlings. J. Integr. Plant Bio. 53, 632–640. doi: 10.1111/j.1744-7909.2011.01042.x
Peres, A. L. G., Soares, J. S., Tavares, R. G., Righetto, G., Zullo, M. A., Mandava, N. B., et al. (2019). Brassinosteroids, the sixth class of phytohormones: a molecular view from the discovery to hormonal interactions in plant development and stress adaptation. Int. J. Mol. Sci. 20:331. doi: 10.3390/ijms20020331
Perilli, S., Moubayidin, L., and Sabatini, S. (2010). The molecular basis of cytokinin function. Curr. Opin. Plant Bio. 13, 21–26. doi: 10.1016/j.pbi.2009.09.018
Piotrowska-Niczyporuk, A., Bajguz, A., Zambrzycka, E., and Godlewska-Żyłkiewicz, B. (2012). Phytohormones as regulators of heavy metal biosorption and toxicity in green alga Chlorella vulgaris (Chlorophyceae). Plant Phys. Biochem. 52, 52–65. doi: 10.1016/j.plaphy.2011.11.009
Polko, J. K., Pierik, R., van Zanten, M., Tarkowská, D., Strnad, M., Voesenek, L. A., et al. (2013). Ethylene promotes hyponastic growth through interaction with ROTUNDIFOLIA3/CYP90C1 in Arabidopsis. J. Exp. Bot. 64, 613–624. doi: 10.1093/jxb/ers356
Poppenberger, B., Rozhon, W., Khan, M., Husar, S., Adam, G., Luschnig, C., et al. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30, 1149–1161. doi: 10.1038/emboj.2011.35
Poonam, R. K., Bali, S., Singh, R., Pati, P. K., and Bhardwaj, R. (2014). Treatment of 24-EBL to Brassica juncea plants under Cu-metal stress lowers oxidative burst by activity antioxidative enzymes. J. Stress Physiol. Biochem. 10, 315–327.
Rattan, A., Kapoor, D., Kapoor, N., Bhardwaj, R., and Sharma, A. (2020). Brassinosteroids Regulate Functional Components of Antioxidative Defense System in Salt Stressed Maize Seedlings. J. Plant Growth Reg. 2020, 1–11.
Rady, M. M. (2011). Effect of 24-epibrassinolide on growth, yield, antioxidant system and cadmium content of bean (Phaseolus vulgaris L.) plants under salinity and cadmium stress. Sci. Horticult. 129, 232–237. doi: 10.1016/j.scienta.2011.03.035
Rady, M. M., and Osman, A. S. (2012). Response of growth and antioxidant system of heavy metal-contaminated tomato plants to 24-epibrassinolide. Afr. J. Agric. Res. 7, 3249–3254.
Rajewska, I., Talarek, M., and Bajguz, A. (2016). Brassinosteroids and response of plants to heavy metals action. Front. Plant Sci. 7:629. doi: 10.3389/fpls.2016.00629
Ramakrishna, B., and Rao, S. S. R. (2015). Foliar application of brassinosteroids alleviates adverse effects of zinc toxicity in radish (Raphanus sativus L.) plants. Protoplasma 252, 665–677. doi: 10.1007/s00709-014-0714-0
Ross, J. J., and Quittenden, L. J. (2016). Interactions between brassinosteroids and gibberellins: synthesis or signaling? Plant Cell. 28, 829–832. doi: 10.1105/tpc.15.00917
Roychoudhury, A., Ghosh, S., Paul, S., Mazumdar, S., Das, G., and Das, S. (2016). Pre-treatment of seeds with salicylic acid attenuates cadmium chloride-induced oxidative damages in the seedlings of mungbean (Vigna radiata L. Wilczek). Acta Physiol. Planta. 38:11.
Rozhon, W., Mayerhofer, J., Petutschnig, E., Fujioka, S., and Jonak, C. (2010). ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62, 215–223. doi: 10.1111/j.1365-313x.2010.04145.x
Rozhon, W., Akter, S., Fernandez, A., and Poppenberger, B. (2019). Inhibitors of brassinosteroid biosynthesis and signal transduction. Molecules 24:4372. doi: 10.3390/molecules24234372
Saini, S., Sharma, I., and Pati, P. K. (2015). Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 6:950. doi: 10.3389/fpls.2015.00950
Sharma, I., Pati, P. K., and Bhardwaj, R. (2011). Effect of 24-epibrassinolide on oxidative stress markers induced by nickel-ion in Raphanus sativus L. Acta Physiol. Plant 33, 1723–1735. doi: 10.1007/s11738-010-0709-1
Sharma, A., Thakur, S., Kumar, V., Kanwar, M. K., Kesavan, A. K., Thukral, A. K., et al. (2016). Pre-sowing seed treatment with 24-epibrassinolide ameliorates pesticide stress in Brassica juncea L. through the modulation of stress markers. Front. Plant Sci. 7:1569.
Sharma, A., Thakur, S., Kumar, V., Kesavan, A. K., Thukral, A. K., and Bhardwaj, R. (2017). 24-epibrassinolide stimulates imidacloprid detoxification by modulating the gene expression of Brassica juncea L. BMC Plant Bio. 17:56.
Shahzad, B., Tanveer, M., Che, Z., Rehman, A., Cheema, S. A., Sharma, A., et al. (2018). Role of 24-epibrassinolide (EBL) in mediating heavy metal and pesticide induced oxidative stress in plants: A review. Ecotoxico. Environ. Safety. 147, 935–944. doi: 10.1016/j.ecoenv.2017.09.066
Sharma, N., Hundal, G. S., Sharma, I., and Bhardwaj, R. (2014). 28-Homobrassinolide alters protein content and activities of glutathione-S-transferase and polyphenol oxidase in Raphanus sativus L. plants under heavy metal stress. Toxico. Int. 21:44.
Sharma, P., Kumar, A., and Bhardwaj, R. (2016). Plant steroidal hormone epibrassinolide regulate–Heavy metal stress tolerance in Oryza sativa L. by modulating antioxidant defense expression. Environ. Exp. Bot. 122, 1–9. doi: 10.1016/j.envexpbot.2015.08.005
Singh, S., and Prasad, S. M. (2017). Effects of 28-homobrassinoloid on key physiological attributes of Solanum lycopersicum seedlings under cadmium stress: photosynthesis and nitrogen metabolism. Plant Growth Regul. 82, 161–173. doi: 10.1007/s10725-017-0248-5
Soares, C., de Sousa, A., Pinto, A., Azenha, M., Teixeira, J., Azevedo, R. A., et al. (2016). Effect of 24-epibrassinolide on ROS content, antioxidant system, lipid peroxidation and Ni uptake in Solanum nigrum L. under Ni stress. Environ. Exp. Bot. 122, 115–125. doi: 10.1016/j.envexpbot.2015.09.010
Stewart Lilley, J. L., Gan, Y., Graham, I. A., and Nemhauser, J. L. (2013). The effects of DELLA s on growth change with developmental stage and brassinosteroid levels. Plant J. 76, 165–173.
Sun, T. P. (2011). The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Bio. 21, R338–R345.
Sun, Y., Fan, X. Y., Cao, D. M., Tang, W., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Symons, G. M., Ross, J. J., Jager, C. E., and Reid, J. B. (2008). Brassinosteroid transport. J. Exp. Bot. 59, 17–24. doi: 10.1093/jxb/erm098
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/s0092-8674(00)81094-6
Tadayon, M. S., and Moafpourian, G. (2019). Effects of Exogenous epi-brassinolid, zinc and boron foliar nutrition on fruit development and ripening of grape (Vitis vinifera L. clv.‘Khalili’). Scient. Horticult. 244, 94–101. doi: 10.1016/j.scienta.2018.09.036
Takahashi, T., and Kakehi, J. I. (2010). Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Anna. Bot. 105, 1–6. doi: 10.1093/aob/mcp259
Tanaka, K., Asami, T., Yoshida, S., Nakamura, Y., Matsuo, T., and Okamoto, S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138, 1117–1125. doi: 10.1104/pp.104.058040
Tang, W., Kim, T. W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560. doi: 10.1126/science.1156973
Tian, H., Lv, B., Ding, T., Bai, M., and Ding, Z. (2018). Auxin-BR interaction regulates plant growth and development. Front. Plant Sci. 8: 2256.
Tong, H., and Chu, C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23, 1016–1028. doi: 10.1016/j.tplants.2018.08.007
Tong, H., Xiao, Y., Liu, D., Gao, S., Liu, L., Yin, Y., et al. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell. 26, 4376–4393. doi: 10.1105/tpc.114.132092
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 9, 1963–1971. doi: 10.2307/3870557
Vert, G., Walcher, C. L., Chory, J., and Nemhauser, J. L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. 105, 9829–9834. doi: 10.1073/pnas.0803996105
Vriet, C., Russinova, E., and Reuzeau, C. (2012). Boosting crop yields with plant steroids. Plant Cell. 24, 842–857. doi: 10.1105/tpc.111.094912
Vriet, C., Russinova, E., and Reuzeau, C. (2013). From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant. 6, 1738–1757. doi: 10.1093/mp/sst096
Wang, H., Tang, J., Liu, J., Hu, J., Liu, J., Chen, Y., et al. (2018). Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant. 11, 315–325. doi: 10.1016/j.molp.2017.12.013
Wang, R., Wang, J., Zhao, L., Yang, S., and Song, Y. (2015). Impact of heavy metal stresses on the growth and auxin homeostasis of Arabidopsis seedlings. Biometals 28, 123–132. doi: 10.1007/s10534-014-9808-6
Wang, X., and Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122. doi: 10.1126/science.1127593
Wang, X., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M. B., et al. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell. 15, 220–235. doi: 10.1016/j.devcel.2008.06.011
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2, 505–513. doi: 10.1016/s1534-5807(02)00153-3
Watanabe, T., Noguchi, T., Yokota, T., Shibata, K., Koshino, H., Seto, H., et al. (2001). Synthesis and biological activity of 26-norbrassinolide, 26-norcastasterone and 26-nor-6-deoxocastasterone. Phytochem 58, 343–349.
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Bio. 12, 527–538.
Wu, C., Li, F., Xu, H., Zeng, W., Yu, R., Wu, X., et al. (2019). The potential role of brassinosteroids (BRs) in alleviating antimony (Sb) stress in Arabidopsis thaliana. Plant Physio Biochem. 141, 51–59.
Xia, X. J., Fang, P. P., Guo, X., Qian, X. J., Zhou, J., Shi, K., et al. (2018). Brassinosteroid−mediated apoplastic H2O2−glutaredoxin 12/14 cascade regulates antioxidant capacity in response to chilling in tomato. Plant. Cell ENV. 41, 1052–1064.
Xia, X. J., Gao, C. J., Song, L. X., Zhou, Y. H., Shi, K. A. I., and Yu, J. Q. (2014). Role of H2O2 dynamics in brassinosteroid−induced stomatal closure and opening in Solanum lycopersicum. Plant. Cell ENV. 37, 2036–2050.
Xia, X. J., Zhou, Y. H., Shi, K., Zhou, J., Foyer, C. H., and Yu, J. Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66, 2839–2856.
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 12, 1591–1605.
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Ye, H., Li, L., Guo, H., and Yin, Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. 109, 20142–20147.
Yin, Y. L., Zhou, Y., Zhou, Y. H., Shi, K., Zhou, J., Yu, Y., et al. (2016). Interplay between mitogen-activated protein kinase and nitric oxide in brassinosteroid-induced pesticide metabolism in Solanum lycopersicum. J. Hazard. Mater. 316, 221–231.
Yin, Y., Qin, K., Song, X., Zhang, Q., Zhou, Y., Xia, X., et al. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physio. 59, 2239–2254.
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid regulated gene expression in Arabidopsis. Cell 120, 249–259.
Yokota, T. (1999). The history of brassinosteroids: discovery to isolation of biosynthesis and signal transduction mutants. Brassinosteroids 1999, 1–20.
Yokota, T., Ogino, Y., Suzuki, H., Takahashi, N., Saimoto, H., Fujioka, S., et al. (1991). Metabolism and biosynthesis of brassinosteroids. In: Brassinosteroids: Chemistry. Bioact. Appl. 1991, 86–96.
Yoshimitsu, Y., Tanaka, K., Fukuda, W., Asami, T., Yoshida, S., Hayashi, K. I., et al. (2011). Transcription of DWARF4 plays a crucial role in auxin-regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PLoS One. 6:e23851.
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome−wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646.
Yuan, L. B., Peng, Z. H., Zhi, T. T., Zho, Z., Liu, Y., Zhu, Q., et al. (2014). Brassinosteroid enhances cytokinin-induced anthocyanin biosynthesis in Arabidopsis seedlings. Biol. Plant. 59, 99–105.
Yuldashev, R., Avalbaev, A., Bezrukova, M., Vysotskaya, L., Khripach, V., and Shakirova, F. (2012). Cytokinin oxidase is involved in the regulation of cytokinin content by 24-epibrassinolide in wheat seedlings. Plant Physio Biochem. 55, 1–6.
Yunta, C., Martínez-Ripoll, M., Zhu, J. K., and Albert, A. (2011). The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress. J. Mol. Bio. 414, 135–144.
Yusuf, M., Fariduddin, Q., and Ahmad, A. (2012). 24-Epibrassinolide modulates growth, nodulation, antioxidant system, and osmolyte in tolerant and sensitive varieties of Vigna radiata under different levels of nickel: a shotgun approach. Plant Physio Biochem. 57, 143–153.
Yusuf, M., Fariduddin, Q., Hayat, S., Hasan, S. A., and Ahmad, A. (2011). Protective response of 28-homobrassinolide in cultivars of Triticum aestivum with different levels of nickel. Arch. Environ. Cont. Toxico. 60, 68–76.
Zhang, A., Zhang, J., Zhang, J., Ye, N., Zhang, H., Tan, M., et al. (2011). Nitric oxide mediates brassinosteroid-induced ABA biosynthesis involved in oxidative stress tolerance in maize leaves. Plant Cell Physio. 52, 181–192.
Zhang, S., Cai, Z., and Wang, X. (2009). The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. 106, 4543–4548.
Zhang, X., Guo, W., Du, D., Pu, L., and Zhang, C. (2020). Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 292: 110378.
Zhao, B., and Li, J. (2012). Regulation of brassinosteroid biosynthesis and inactivation F. J. Integr. Plant Biol. 54, 746–759.
Zhou, Y. L., Huo, S. F., Wang, L. T., Meng, J. F., Zhang, Z. W., and Xi, Z. M. (2018). Exogenous 24-Epibrassinolide alleviates oxidative damage from copper stress in grape (Vitis vinifera L.) cuttings. Plant Physio. Biochem. 130, 555–565.
Zhu, T., Tan, W. R., Deng, X. G., Zheng, T., Zhang, D. W., and Lin, H. H. (2015). Effects of brassinosteroids on quality attributes and ethylene synthesis in postharvest tomato fruit. Postharvest Bio Techno. 100, 196–204.
Zou, L., Qu, M., Zeng, L., and Xiong, G. (2020). The molecular basis of the interaction between Brassinosteroid induced and phosphorous deficiency induced leaf inclination in rice. Plant Growth Regul. 2020, 1–14.
Zullo, M. A. T. (2018). Brassinosteroids and related compounds. New York: LAP LAMBERT Academic Publishing.
Zullo, M. A. T., and Adam, G. (2002). Brassinosteroid phytohormones: structure, bioactivity and applications. Braz. J. Plant Physiol. 14, 143–181.
Zullo, M. A. T., and Bajguz, A. (2019). The brassinosteroids family–structural diversity of natural compounds and their precursors. In Brassinosteroids: Plant Growth and Dev. Singapore: Springer, 1–44.
Keywords: BR biosynthetic pathway, BR signaling, transcription, heavy metal, stress, hormone crosstalk
Citation: Kour J, Kohli SK, Khanna K, Bakshi P, Sharma P, Singh AD, Ibrahim M, Devi K, Sharma N, Ohri P, Skalicky M, Brestic M, Bhardwaj R, Landi M and Sharma A (2021) Brassinosteroid Signaling, Crosstalk and, Physiological Functions in Plants Under Heavy Metal Stress. Front. Plant Sci. 12:608061. doi: 10.3389/fpls.2021.608061
Received: 18 September 2020; Accepted: 27 January 2021;
Published: 24 March 2021.
Edited by:
Shamsul Hayat, Aligarh Muslim University, IndiaReviewed by:
Norbert Mehlmer, Technical University of Munich, GermanyCopyright © 2021 Kour, Kohli, Khanna, Bakshi, Sharma, Singh, Ibrahim, Devi, Sharma, Ohri, Skalicky, Brestic, Bhardwaj, Landi and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anket Sharma, YW5rZXRzaGFybWFAZ21haWwuY29t; Marco Landi, bWFyY28ubGFuZGlAdW5pcGkuaXQ=; Renu Bhardwaj, cmVudWJoYXJkd2FqODJAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.