- 1School of Biological Sciences, University of Bristol, Bristol, United Kingdom
- 2School of Mathematics, University of Bristol, Bristol, United Kingdom
The physical presence of roots and the compounds they release affect the cohesion between roots and their environment. However, the plant traits that are important for these interactions are unknown and most methods that quantify the contributions of these traits are time-intensive and require specialist equipment and complex substrates. Our lab developed an inexpensive, high-throughput phenotyping assay that quantifies root-substrate adhesion in Arabidopsis thaliana. We now report that this method has high sensitivity and versatility for identifying different types of traits affecting root-substrate adhesion including root hair morphology, vesicle trafficking pathways, and root exudate composition. We describe a practical protocol for conducting this assay and introduce its use in a forward genetic screen to identify novel genes affecting root-substrate interactions. This assay is a powerful tool for identifying and quantifying genetic contributions to cohesion between roots and their environment.
Introduction
Plants secrete compounds that help them adapt to their environment, sense and interact with other organisms and to improve water and nutrient uptake (reviewed in Bais et al., 2006; Sasse et al., 2018; Vives-Peris et al., 2020). These compounds (exudates) can vary in composition based on plant species, developmental stage, and environment (Naveed et al., 2017). Recent studies have shown that some compounds have bioadhesive properties that stick plants and soils together and aggregate soil particles, modifying the microenvironment around plant roots (Galloway et al., 2018). However, understanding how plant biology and physiology contribute to plant-soil interactions can be confounded by multiple factors, like other organisms living in the soil, which also secrete organic metabolites that can contribute to soil cohesion. Soil composition might affect plant exudate composition and vice versa. Also, plant root morphology, including root architecture and the presence of root hairs, can alter root-soil interactions (Bailey et al., 2002; De Baets et al., 2007; Stokes et al., 2009; Burylo et al., 2012; Ghestem et al., 2014). This complexity has made it difficult to characterize the relevant plant physiology.
We developed a high-throughput assay that identifies and quantifies the contribution of root hair cells to root-substrate cohesion in a less variable environment than the natural rhizosphere (De Baets et al., 2020). The assay uses a centrifuge to apply force to Arabidopsis thaliana seedlings grown on the surface of a sterile medium and measures the force required to detach seedlings from that surface, using a Cox proportional hazards regression (Prentice and Kalbfleisch, 2003) to quantify the adherence of candidate Arabidopsis lines relative to wild-type plants at increasing centrifugal force. Previous use of this assay assessed whether presence or absence of root hairs affected adhesion to the agar (De Baets et al., 2020) but did not address subtle aspects of root hair biology. We now wanted to test whether this assay is sensitive enough to identify other types of traits involved in root-substrate adhesion. To validate this extended use of the assay, we asked if it could detect the adhesive effects in (1) mutants with altered root hair shape, (2) vesicle trafficking mutants with no known root hair morphological defects, (3) exudate compositional mutants with no known root hair phenotypes, and (4) a forward genetic screen of a mutant population to identify novel genes that affect root-substrate adhesion.
Here, we show that this method has the sensitivity to identify how differences in root hair morphology and biological functions that do not present a visible root hair phenotype contribute to substrate cohesion. Using the genetic and molecular tools available in Arabidopsis and working in a sterile environment allows hypotheses about plant specific factors of root-substrate interactions to be tested. While plant-soil cohesion is a complex and dynamic interaction, our assay provides a way to efficiently probe root cellular functions that might be masked by confounding variables in a soil-based study system. Results from this assay provide a platform for more comprehensive studies of plant morphology and physiology that affect root-substrate cohesion.
Materials and Methods
Materials and Equipment
1. 20% (v/v) bleach solution
2. Sterile distilled water
3. Seed for candidate Arabidopsis lines
4. Half strength Murashige and Skoog basal medium
5. 1% (w/v) sucrose
6. 1% (w/v) agar
7. Laminar air hood
8. 90-mm triple vented Petri Plates
9. Parafilm
10. Growth room
11. Swing-out bucket centrifuge
12. Laboratory safety razor blade
13. Analytical balance
Plant Material and Growth Conditions
Candidate and wild-type lines of the Columbia-0 (Col-0) ecotype of A. thaliana single-gene mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC; Nottingham, United Kingdom) or were stocks maintained in the lab (Table 1). Homozygous lines were used in all experiments.
For all experiments, seeds were surface sterilized for 15 min in 20% (v/v) bleach and sterile water solution, followed by five washes in sterile water. All sterilized seeds were stratified in the dark at 4°C for 24–48 h in Eppendorf microtubes containing sterile water. All plants were reared in a growth room at 21–22°C with a continuous photoperiod (light intensity = 120–145 μmol m−2 s−1) at 60% humidity.
Half strength Murashige and Skoog basal medium was supplemented with 1% (w/v) sucrose, pH adjusted to 5.7 and solidified with 1% (w/v) agar. A 30 ml volume of sterilized media (at ~45–50°C) was pipetted into 90-mm triple vented Petri plates and left to set for ~30 min in a laminar air hood. Sterilized seeds were sown onto Petri plates containing the sterile medium and sealed with Parafilm. Plates were then placed in stacks of five at ~80° from the horizontal (lid side up) to encourage vertical growth along the gel surface. Plates were incubated in the growth chamber for 5–6 days before centrifugation. Plants for genomic DNA extractions were grown in compost (three parts Levington F3 compost and one-part J Arthur Bowers horticultural silver sand).
Centrifuge Assay Set-Up
To measure the adhesion of Arabidopsis seedling roots using a centrifuge, seedling roots must not penetrate the gel or touch each other, as both events could interfere with seedling detachment during centrifugation. Therefore, 10 seeds were sown 1-cm apart on each plate in two parallel rows (Figure 1Ai). The plates were then grown for 5–6 days (Figure 1Aii) and seedlings that were touching each other, had grown into the gel, or not uniform in size were omitted from the assay. The seedlings were then numbered so that plate positional effects could be included as a potential covariate in the final analysis (Figure 1Bi). Plates were inverted and placed into a swing-out bucket centrifuge (Beckman Allegra X-30R Centrifuge) that held four plates at a time and subjected to increments of increasing centrifugal force at 720, 1018, 1,247, 1,440, and 1,611 RPM for one minute each (Figure 1Bii). The proportion of seedlings that detached from the gel surface was recorded between each centrifugal speed; a detachment event was recorded when a seedling had partially or fully peeled away from the gel (Figure 1Biii). If the gel shattered during the assay, seedlings that had not detached from the gel were not recorded and seedlings that remained adhered to the gel after the maximum centrifugal speed were censored in the analysis. During method optimization, we encountered some common methodological challenges for which we provide troubleshooting options in Supplementary Table S1.
Calculating Seedling Detachment Force
To calculate the centrifugal force acting on a seedling, the aerial tissue mass for each seedling was separated from the root by making an incision at the hypocotyl-root junction using a laboratory safety razor blade and immediately measured using an analytical balance (Figures 1Ci,ii). The aerial tissue weight of each seedling was measured because a plant with heavier aerial tissue experiences more force than a lighter one subjected to the same centrifugal speed. Centrifugal force, Fc (mN), acting on a seedling was calculated as previously reported in De Baets et al. (2020). Aerial tissue weight (kg), the angular velocity (ω), and the distance between the seedling and the axis of rotation on the centrifuge (radius = 0.07 m) were used to give the following equation:
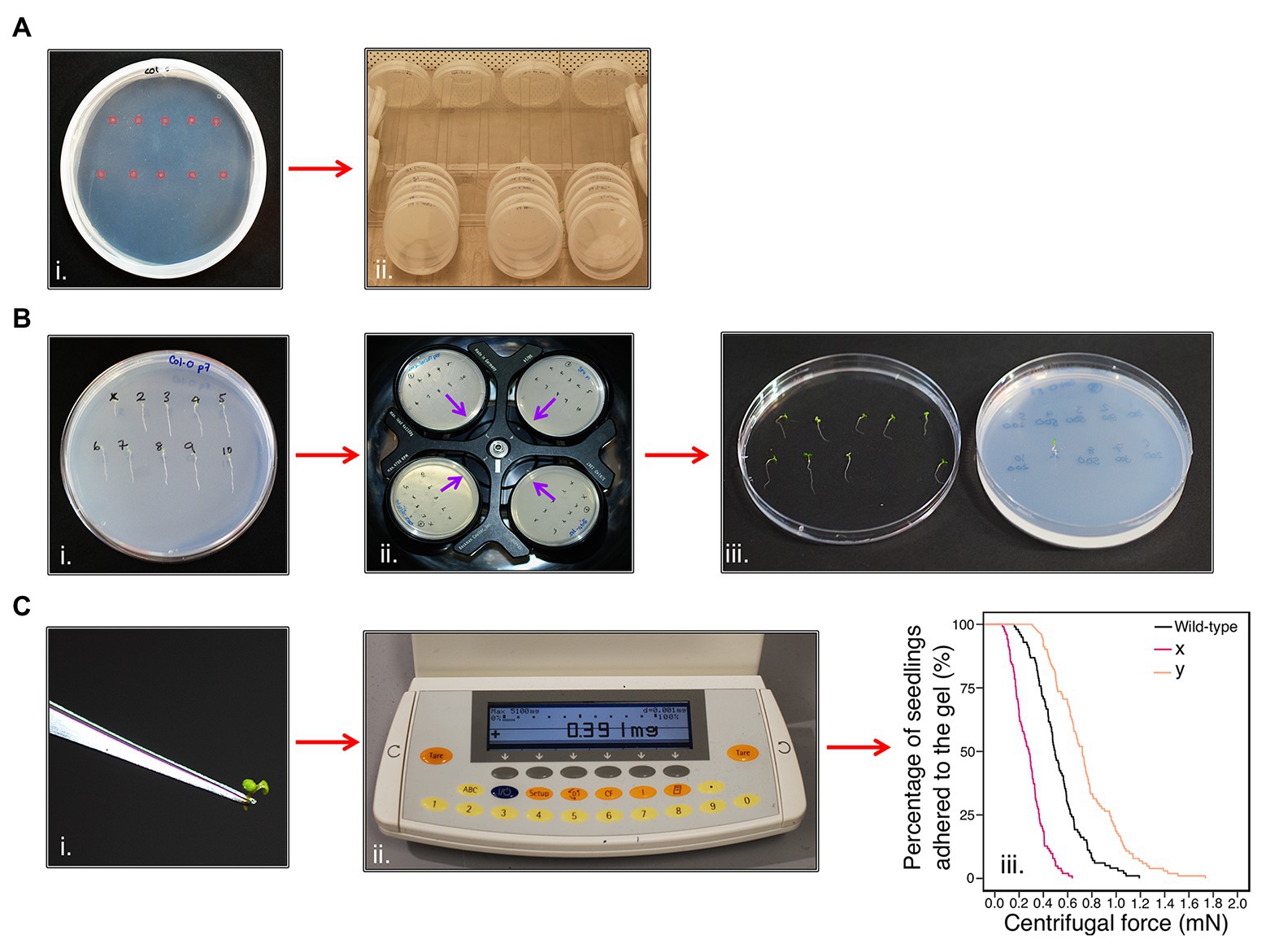
Figure 1. Centrifuge assay workflow. (Ai) Ten seeds (highlighted in pink) are sown onto the surface of sterile, solid gel growth medium in a single 90 mm Petri plate in two horizontal rows. (Aii) The plates are stacked in groups of five and orientated vertically at approximately 80° to encourage the roots to grow down the surface of the gel medium. Plates are grown in a growth chamber with constant light (120–145 μmol m−2 s−1) conditions at 22°C and 60% relative humidity. (B) After 5–6 days, (Bi) seedlings are visually analyzed and numbered. (Bii) Petri plates are placed into a swing-out-bucket centrifuge in an inverted orientation with their roots pointing inward (indicated by the purple arrows). (Biii) After seedlings have been subjected to each 1-min pulse of increasing centrifugal speed, seedling detachment is recorded. (Ci) The aerial tissue mass of each seedling is determined using (Cii) an analytical scale to (Ciii) determine the root-gel adhesion properties of candidate lines are compared to wild-type to assess whether they have increased (e.g., line y) or decreased (e.g., line x) adhesion to the sterile gel.
Identification of abcg43-1 Mutant T-DNA Insert
Genomic DNA was extracted from a pool of vegetative tissue taken from 10 3-week old plants using an adapted protocol (Healey et al., 2014) for Illumina Next-Generation two x paired-end sequencing conducted at the Bristol Genomics Facility (University of Bristol, United Kingdom). The Illumina TruSeq Nano LT gDNA kit (Illumina Inc.) was used to generate a genomic DNA sequencing library following the manufacturer’s protocol. The final library was diluted to a loading concentration of 1.4 pM for cluster generation and 2 × 150 bp paired-end sequencing on the Illumina NextSeq500 system (Illumina Inc.) alongside a 5% PhiX spike-in control library. Read summary statistics were generated using the RTA 2.4.6 Primary Analysis Software (Illumina Inc.). The read summaries were analyzed in the Sequencing Analysis Viewer (Illumina Inc.). Filtered paired reads were subject to paired-end alignment using the Bowtie2 2.3.4.2 aligner (Langmead and Salzberg, 2012). A bespoke reference genome was produced that combined the TAIR10 Arabidopsis genome (Lamesch et al., 2012) and the pROK2 vector sequence (Baulcombe et al., 1986). Alignments were viewed in the Integrative Genomics Viewer IGV 2.3 (Robinson et al., 2011).
For Sanger sequencing, DNA was extracted from a pool of vegetative tissue taken from 2-week-old plants using a modified Edwards prep (Edwards et al., 1991). High fidelity PCR was conducted on the abcg43-1 line using the Q5 High-Fidelity 2X Master Mix (NEB). PCR products were purified and extracted from a 1% agarose gel using the QIAquick Gel Extraction Kit (QIAGEN), following the manufacturer’s instructions. Purified PCR product plus the abcg43-1 forward, reverse or border genotyping primer (Supplementary Table S2) was used for Sanger sequencing (using the Mix2Seq overnight sequencing kit, Eurofins Genomics). Chromatograms and FASTA files were obtained from Eurofin Genomics and following manual low-quality end trimming, the final sequences were aligned to the pROK2 vector sequence and ABCG43 gene sequence using the MUSCLE alignment tool (Madeira et al., 2019).
Genotyping
To genotype the abcg43 mutant alleles, genomic DNA was extracted from the vegetative tissue of 2-week-old plants using a modified Edwards prep (Edwards et al., 1991). T-DNA border and gene-specific primers were used in PCR analyses to confirm the genotype (Supplementary Table S2).
Morphological Root Trait Analyses
Five-day-old Arabidopsis seedlings were imaged with a Leica MZ FLIII microscope (Leica) with dark-field lighting. Root hair length and density were measured using microscope images of 5-day-old Arabidopsis seedlings and Fiji version 1.0 (Schindelin et al., 2012) and the Bio-Formats Importer plugin. At least two experimental repeats were conducted; in each experiment, we imaged 8–10 individual plants from each line and measured at least 30 root hairs per plant.
Statistical Analyses
All statistical analyses were conducted using RStudio, version 1.1453 (R Core Team, 2014), and all graphs were generated using the R package ggplot2 (Wickham, 2016).
Centrifuge Assay
Survival analysis was used to study when seedlings detach from the sterile gel medium (i.e., seedling detachment is the event) at increasing force (i.e., force is modeled rather than time). We used the Cox proportional hazards (PH) regression function, coxph, within the survival package in R to statistically test for differences between the detachment of experimental seedlings relative to the wild-type control (Col-0), using centrifugal force as the predictor variable (Figure 1Ciii). We incorporated seedling position and individual plate number as covariates within our Cox PH regression models to account for any environmental heterogeneity such as minor inconsistencies in the water content or gelling of the medium. When these covariates had no significant effect, they were removed from the model. Seedlings that remained adhered to the gel after the maximum centrifugal speed were censored in our Cox PH regression model. Comparisons of different candidate lines relative to the wild-type line were analyzed a priori; therefore, a series of contrasts were set up using the R function contr.treatment rather than using post-hoc testing methods.
For each Cox PH regression model run, we report the value of p of the Wald Statistic (z-score) and the hazard ratio with the upper and lower bound confidence intervals. Hazard ratios are exponentiated coefficients that give the instantaneous risk of root-gel detachment (Cox and Oakes, 1984; Bradburn et al., 2003; Spruance et al., 2004). For each Cox PH regression model conducted, we used the hazard ratio as a measure of effect size to assess differences between the root-gel adhesion of candidate lines and wild type (Cox and Oakes, 1984). Wild type was used as the baseline group for all models, with a hazard ratio of one. A candidate line with the higher risk of detachment from gel than wild type will have a hazard ratio above one. Conversely, lines with a lower risk of detachment than wild type will have a hazard ratio below one.
For all Cox PH regression models, the assumption of proportionality was satisfied and an alpha level of 0.01 was used. The root-gel detachment of at least 70 plants from each line was assessed within a single experiment.
Root Hair Analyses
Two sample t-tests were conducted using the t.test function in R to test for differences in the mean root hair density or mean root hair length (of 8–10 individual plants from each line) between candidate lines relative to wild type (Col-0). In all cases, the assumptions of normality and homoscedasticity were satisfied. Because comparisons of different mutant lines relative to the wild-type line were a priori, a series of contrasts were conducted using the contr.treatment function in R. To prevent type I errors from multiple testing, we applied the Bonferroni method and adjusted the alpha level to 0.025 (0.05/2).
Results
Root Hair Morphological Effects on Root-Substrate Adhesion
We screened mutant lines with known root hair morphology defects to ask if the centrifuge assay could determine if root hair shape affects root-substrate adhesion. The mostly bald mutant csld3, which encodes the CESA-like 3D protein (Favery et al., 2001; Wang et al., 2001; Yang et al., 2020) was included as a root hairless example. For lines that produce short or malformed root hairs, we included the gdi1-2 Rho GTPase GDP dissociation inhibitor mutant, rol1-2 rhamnose biosynthesis 1 mutant allele, can of worms 1 (cow1-3) phosphatidylinositol transferase protein mutant, and lrx1-4 leucine rich extensin 1 mutant (Grierson et al., 1997; Baumberger et al., 2001; Böhme et al., 2004; Carol et al., 2005; Diet et al., 2006). The rol1-2 and gdi1-2 mutants have short root hairs and the root hairs on gdi1-2 plants can also branch (Parker et al., 2000; Diet et al., 2006; Ringli et al., 2008; Kang et al., 2017); cow1-3 plants have short, wide root hairs that can branch; and lrx1-4 plants can develop root hairs that abort, swell, branch, or collapse in a growth condition-dependent manner (Grierson et al., 1997; Parker et al., 2000; Baumberger et al., 2001; Böhme et al., 2004). Assay results for ectopic and long root hairs were reported previously (De Baets et al., 2020). Under our growth conditions, we confirmed the reported phenotypes for each mutant line (Figure 2A). Wild type was used as the baseline group for all centrifuge experiments, with a hazard ratio of one. Therefore, a candidate line with a higher risk of detachment from the gel than wild type will have a hazard ratio above one, while lines with a lower risk of detachment than wild type will have a hazard ratio below one.
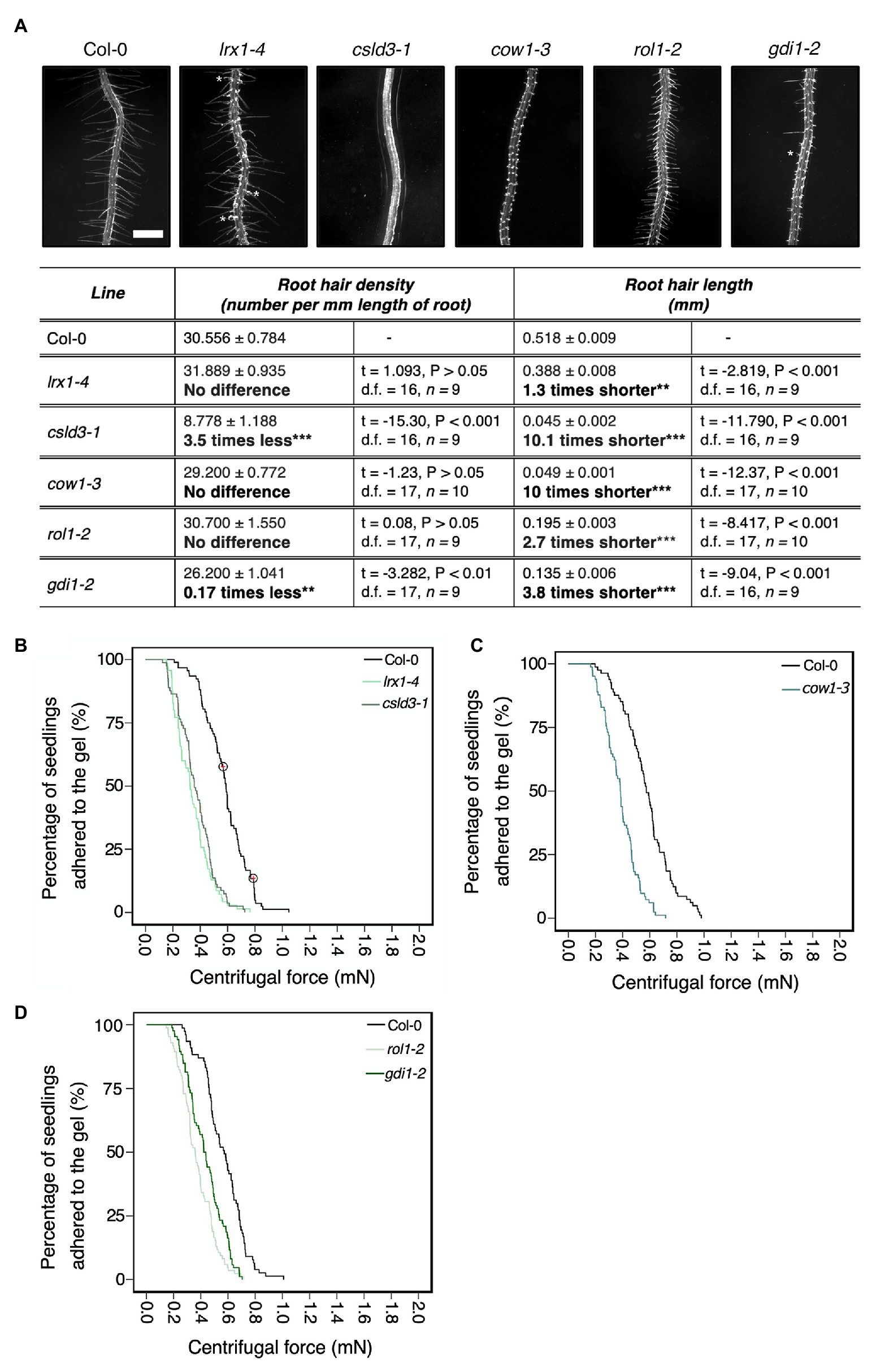
Figure 2. Physical root hair properties contribute to root-substrate adhesion in Arabidopsis thaliana. (A) Root hair phenotypes of 5-day-old wild-type (Col-0), lrx 1-4, csld3-1, cow1-3, rol1-2, and gdi1-2 seedlings grown on gel medium and statistical comparisons of mean root hair density (number per mm length of root) and mean root hair length (mm) for each mutant line relative to wild type. White asterisks on the root hair images indicate the characteristic root hair bulging phenotype in the lrx1-4 mutant and the short, branching root hair phenotype in gdi1-2. Scale bar = 0.5 mm. In the table, the mean ± standard error is given for root hair density and root hair length of wild type and each mutant as well as a mean comparison to wild type, which is listed in bold. “No difference” is stated when there was no statistically significant difference between wild type and a mutant line. The statistical output of each univariate linear model is given. Alpha = 0.025, statistical significance codes: “***” = <0.001 and “**” = <0.01. The results shown are from a representative experiment for at least two independent experiments. Survival curves displaying the proportion of seedlings that adhered to the gel at increasing centrifugal force for (B) 92 wild type (Col-0 – black); 81 lrx1-4 (light green); 70 csld3-1 (green); (C) 82 wild type (Col-0 – black); 83 cow1-3 (green); and (D) 77 wild type (Col-0‐ black); 85 rol1-2 (light green); and 86 gdi1-2 (dark green). Circled red crosses on the survival curves represent seedlings that remained adhered to the gel after the maximum centrifugal speed (1,611 RPM). Each graph shows a representative experiment for at least two independent experiments showing a statistically significant difference in adhesion between mutant lines relative to wild type (Cox PH regression; alpha = 0.01). A single experiment included >70 biological replicates for each candidate line.
Consistent with previous results (De Baets et al., 2020), bald (csld3) seedlings detached at lower centrifugal forces relative to wild-type seedlings, with 4.6 times the risk of detachment than wild-type plants (p < 0.001; Table 2; Figure 2B). Mutants with deformed root hair phenotypes including gdi1-2, rol1-2, cow1-3, and lrx1-4 had risks of detachment that were 2.5, 4, 3.8, and 3.9 times that of wild-type plants, respectively (for all lines p < 0.001, Table 2; Figures 2B–D). These results indicated that the centrifuge assay can quantify the effects of root hair shape on root-substrate cohesion.
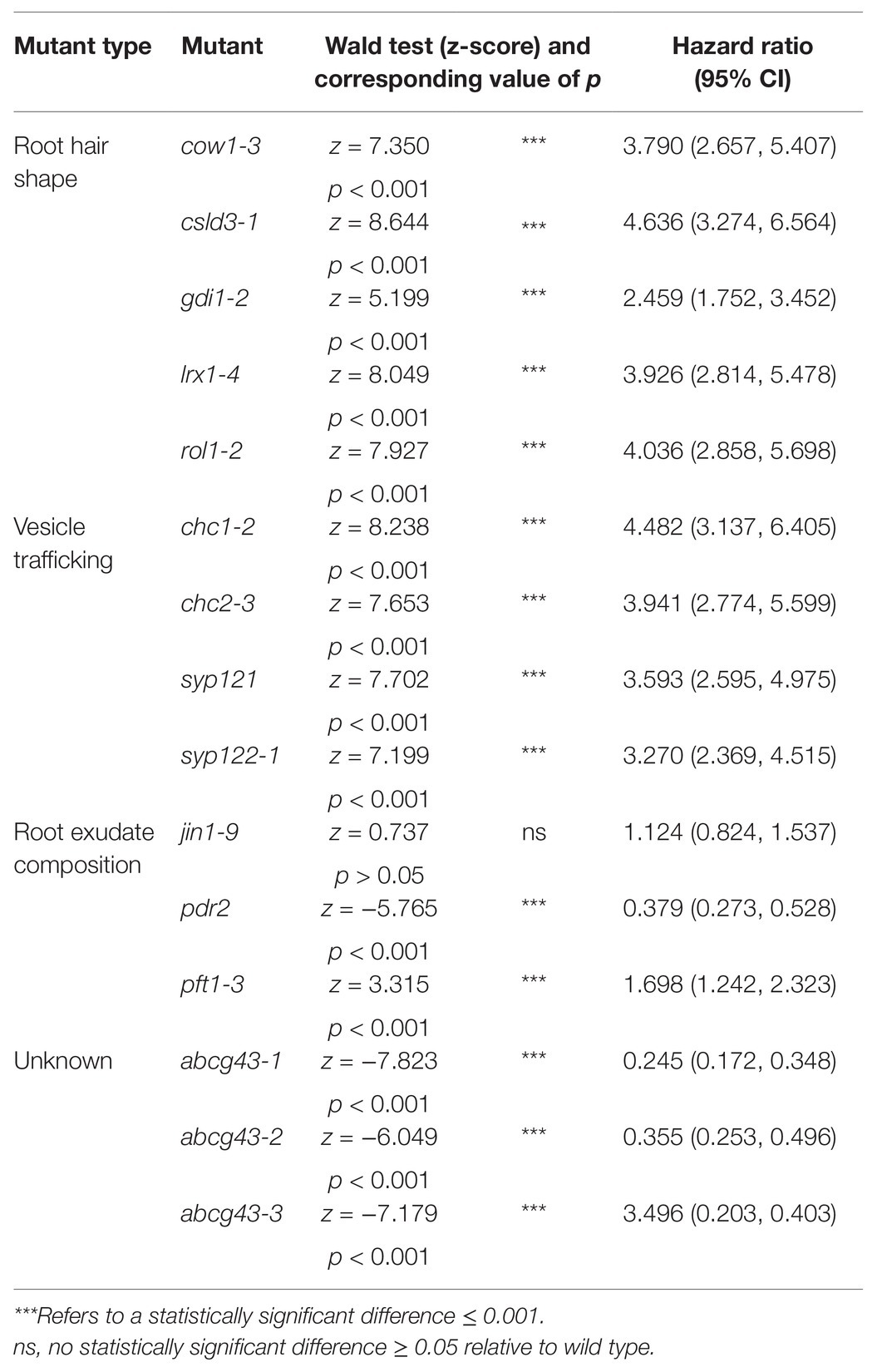
Table 2. Output of the Cox PH regression models comparing the root-gel detachment of root hair shape, vesicle trafficking, and root exudate composition mutants relative to wild type.
Vesicle Trafficking Mutations Affect Root-Substrate Adhesion
We hypothesized that Arabidopsis mutants with defective vesicle trafficking pathways might alter cell wall and apoplast characteristics that contribute to root-substrate interactions and adhesion. We chose the secretory Soluble NSF (N-ethylmaleimide sensitive fusion protein) Attachment proteins (SNAP) REceptor (SNARE) mutants syp121 and syp122-1, and the endocytic mutants, chc1-2 and chc2-3 as candidates for our assay. SYP121 is the primary secretory SNARE found on the plasma membrane (Geelen et al., 2002; Assaad et al., 2004) and has characterized aboveground phenotypes, including small rosettes and stomatal mobility defects (Eisenach et al., 2012; Larson et al., 2017); however, the mutant does not have a reported root hair phenotype. Although the related SNARE SYP122 is thought to share partial functionality with SYP121, the syp122-1 mutant does not share these phenotypes with syp121 and a recent proteomic analysis reported differences in their vesicle cargoes, which could contribute to their functional independence (Waghmare et al., 2018). The chc1-2 and chc2-3 mutants are defective in the heavy chain subunits of the clathrin coat complex, which is required for vesicle traffic at the plasma membrane (Kitakura et al., 2011; Mbengue et al., 2016). Endo‐ and exo-cytic rates are impaired in both mutants (Larson et al., 2017), but no root hair phenotypes have been reported. For the centrifuge assay, it was important to select mutants that do not have root hair phenotypes given the effect root hair morphology has on root-gel adhesion. Therefore, we evaluated the root hairs of these vesicle trafficking mutants and found that the root hairs of these lines did not significantly differ from wild-type plants (Figure 3A).
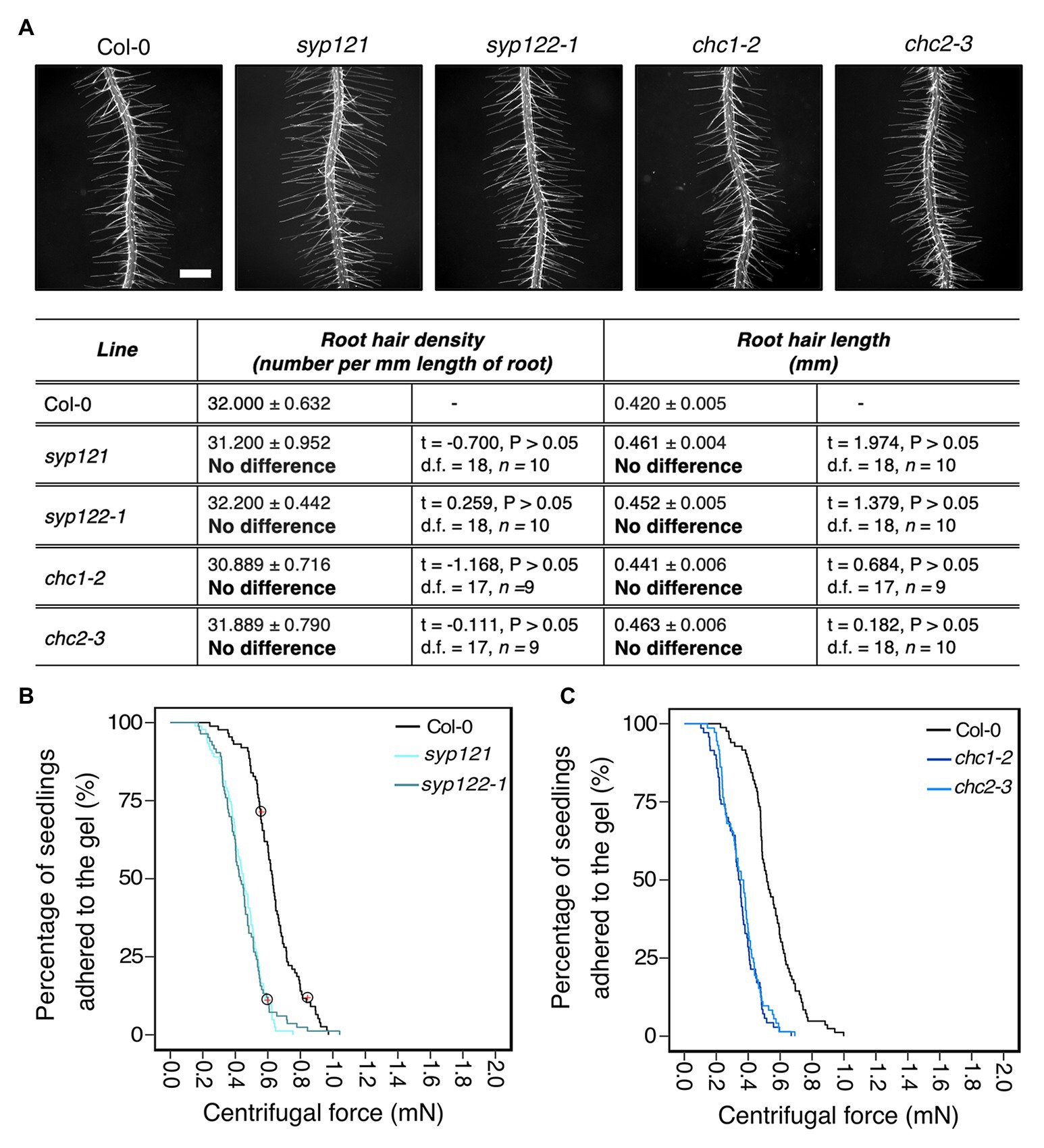
Figure 3. Vesicle trafficking mechanisms that contribute to root-substrate adhesion in Arabidopsis thaliana. (A) Root hair phenotypes of 5-day-old wild-type (Col-0), syp121, syp122-1, chc1-2, and chc2-3 seedlings grown on a gel medium and statistical comparisons of mean root hair density (number per mm length of root) and mean root hair length (mm) for each mutant line relative to wild type. Scale bar = 0.5 mm. In the table, the mean ± standard error is given for the root hair density and root hair length of wild type and each mutant as well as a mean comparison to wild type, which is listed in bold. “No difference” is stated when there was no statistically significant difference between wild type and a mutant line. The statistical output of each univariate linear model is given. Alpha = 0.025. The results shown are from a representative experiment for at least two independent experiments. Survival curves displaying the proportion of seedlings that adhered to the gel at increasing centrifugal force for (B) 87 wild type (Col-0 – black); 91 syp121 (turquoise); 83 syp122-1 (blue/grey); and (C) 83 wild type (Col-0 – black); 70 chc1-2 (dark blue); and 72 chc2-3 (medium blue). Red crosses circled on the survival curves represent seedlings that remained adhered to the gel after the maximum centrifugal speed (1,611 RPM). Each graph shows a representative experiment for at least two independent experiments showing a statistically significant difference in adhesion between all mutant lines relative to wild type (Cox PH regression; alpha = 0.01). A single experiment included >70 biological replicates for each candidate line.
Overall, the risk of detachment for syp121, syp122-1, chc1-2, and chc2-3 plants was 3.6, 3.3, 4.5, and 3.9 times that of wild type, respectively (for all lines p < 0.001; Table 2; Figures 3B,C). These results indicated that the centrifuge assay can be used to probe for intracellular molecular machinery that affects root-substrate interactions.
Exudate Composition Changes Root Detachment Rates
Over 20% of plant assimilated carbon is released as root exudates (Huang et al., 2016). These are complex mixtures of diverse molecular forms that can modify the external environment in response to abiotic and biotic stimuli. Some exudate components contribute to soil adhesion (Akhtar et al., 2018; Galloway et al., 2018). Root epidermal and hair cells secrete compounds that contribute to root exudate profiles that can be plant species specific (Naveed et al., 2017). Because mutants in both root hair development and vesicle trafficking pathways showed root-substrate adhesion phenotypes (Figures 2, 3), we asked if the centrifuge assay could identify effects of exudate composition on root-substrate adhesion. We selected Arabidopsis mutants reported to have altered exudate composition compared to wild type, including pft1-3, jin1-9, and pdr2 (Berger et al., 1996; Badri et al., 2009; Kidd et al., 2009; Carvalhais et al., 2015). PFT1 (MED25) encodes the MEDIATOR25 subunit of the Mediator nuclear protein, and JIN1 (MYC2) encodes a basic helix-loop-helix leucine zipper transcription factor; both proteins are involved in the jasmonate signaling pathway (Lorenzo et al., 2004; Kidd et al., 2009). pft1-3 and jin1-9 mutants are reported to have altered root exudate composition, including lower amounts of the amino acids asparagine, ornithine, and tryptophan than wild-type plants (Carvalhais et al., 2015). PDR2 (ABCG30) encodes a pleiotropic drug resistance (PDR) full-length ABC transporter that is involved in ABA transport and the exudation of secondary metabolites (Badri et al., 2008; Kang et al., 2015). We did not observe root hair growth phenotypes in these mutant lines compared to wild type (Figure 4A).
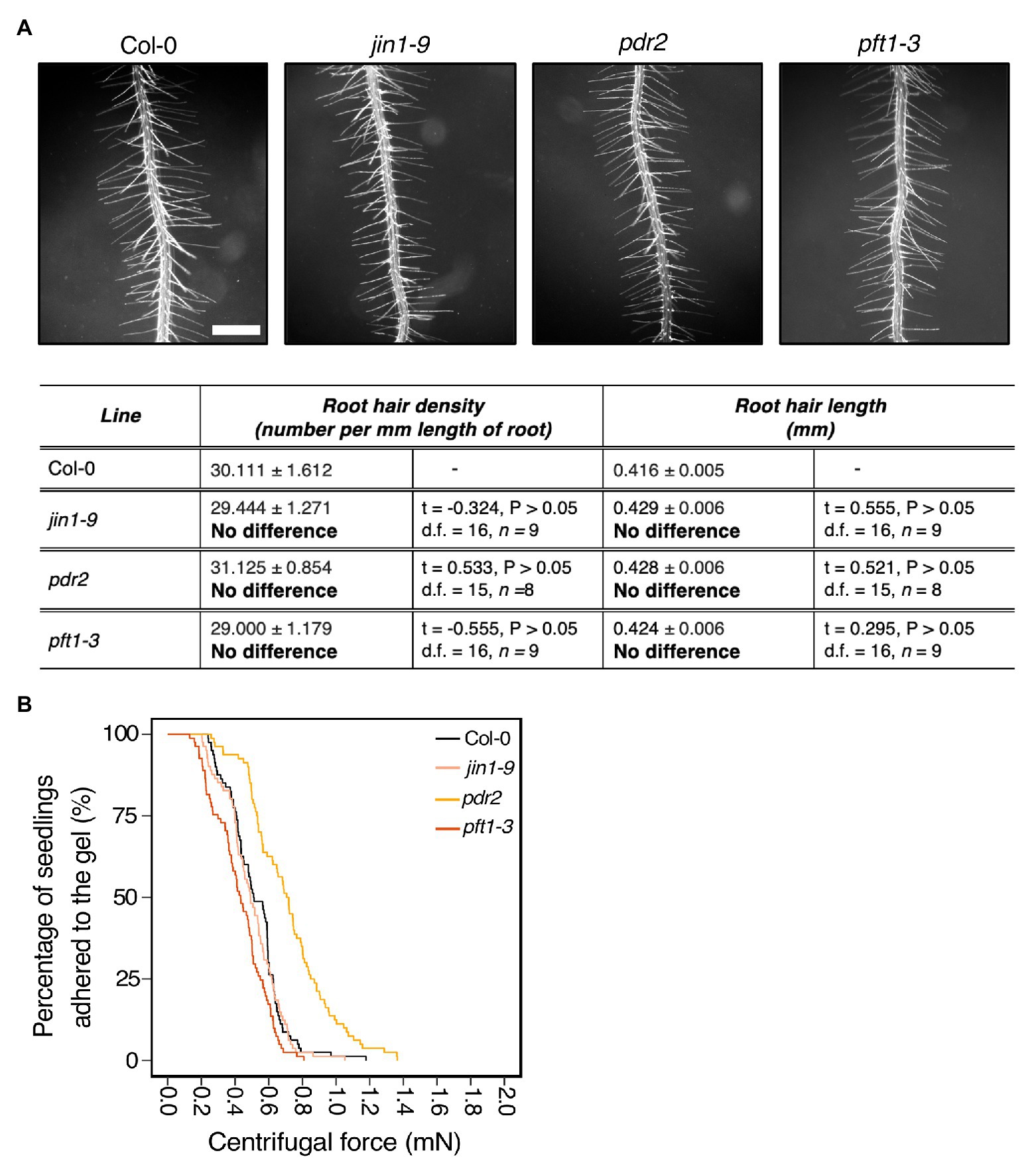
Figure 4. Exudate composition changes root-substrate adhesion properties in Arabidopsis thaliana. (A) Root hair phenotypes of 5-day-old wild-type (Col-0), jin1-9, pdr2, and pft1-3 seedlings grown on a gel medium and statistical comparisons of mean root hair density (number per mm length of root) and mean root hair length (mm) for each mutant line relative to wild type. Scale bar = 0.5 mm. In the table, the mean ± standard error is given for the root hair density and root hair length of wild type and each mutant as well as a mean comparison to wild type, which is listed in bold. “No difference” is stated when there was no statistically significant difference between wild-type and a mutant line. The statistical output of each univariate linear model is given. Alpha = 0.025. The results shown are from a representative experiment for at least two independent experiments. (B) Survival curves displaying the proportion of seedlings that adhered to the gel at increasing centrifugal force for 80 wild type (Col-0 – black), 81 jin1-9 (light pink), 80 pdr2 (light orange), and 81 pft1-3 (dark orange). The results shown are from a representative experiment for at least two independent experiments showing a statistically significant difference in adhesion between mutant lines relative to wild type (Cox PH regression; alpha = 0.01), except for jin1-9. A single experiment included >70 biological replicates for each candidate line.
Compared to wild-type seedlings, pdr2 seedlings resisted detachment, with 0.38 times the risk of detaching from the gel (p < 0.001; Table 2; Figure 4B). Conversely, pft1-3 seedlings had an increased risk of detachment 1.7 times that of wild-type plants (p < 0.001) and there was no difference in gel adhesion between jin1-9 and wild-type plants (p > 0.05; Table 2; Figure 4B). These results show that exudate composition can alter root-substrate interactions, although this may not be the case for all exudate compositional changes, which may lay outside the sensitivity range of the assay.
Centrifuge Assay as a Method for Forward Genetic Screening
Having demonstrated that our centrifugation technique can identify different types of traits that affect root-substrate adhesion, we used it in a forward genetic screen to find novel genes involved in root-substrate adhesion. To conduct this screen, the root-gel adhesion properties of individual plants from a pooled SALK T-DNA insertion mutant collection were analyzed (Alonso et al., 2003). Individual plants that detached at the lowest centrifugal speed or remained attached to the gel after being spun at the highest centrifugal speed were recovered from the screen and self-fertilized to obtain progeny and identify the putative mutations. The centrifuge assay was used to rescreen the progeny (using a sample size of ≥70 n) to confirm whether candidate lines had significantly increased or decreased root-gel adhesion relative to wild-type plants. From this screen, we identified a line with significantly enhanced root-gel adhesion and conducted genomic Next-Generation sequencing followed by Sanger sequencing of T-DNA flanking PCR products to confirm a T-DNA insertion in the ABC transporter gene, ABCG43, whose function is unknown. We named this as T-DNA insertion line abcg43-1 and identified two additional mutant alleles, abcg43-2 (SALK_201207c) and abcg43-3 (SALKseq_30713) (Figure 5A).
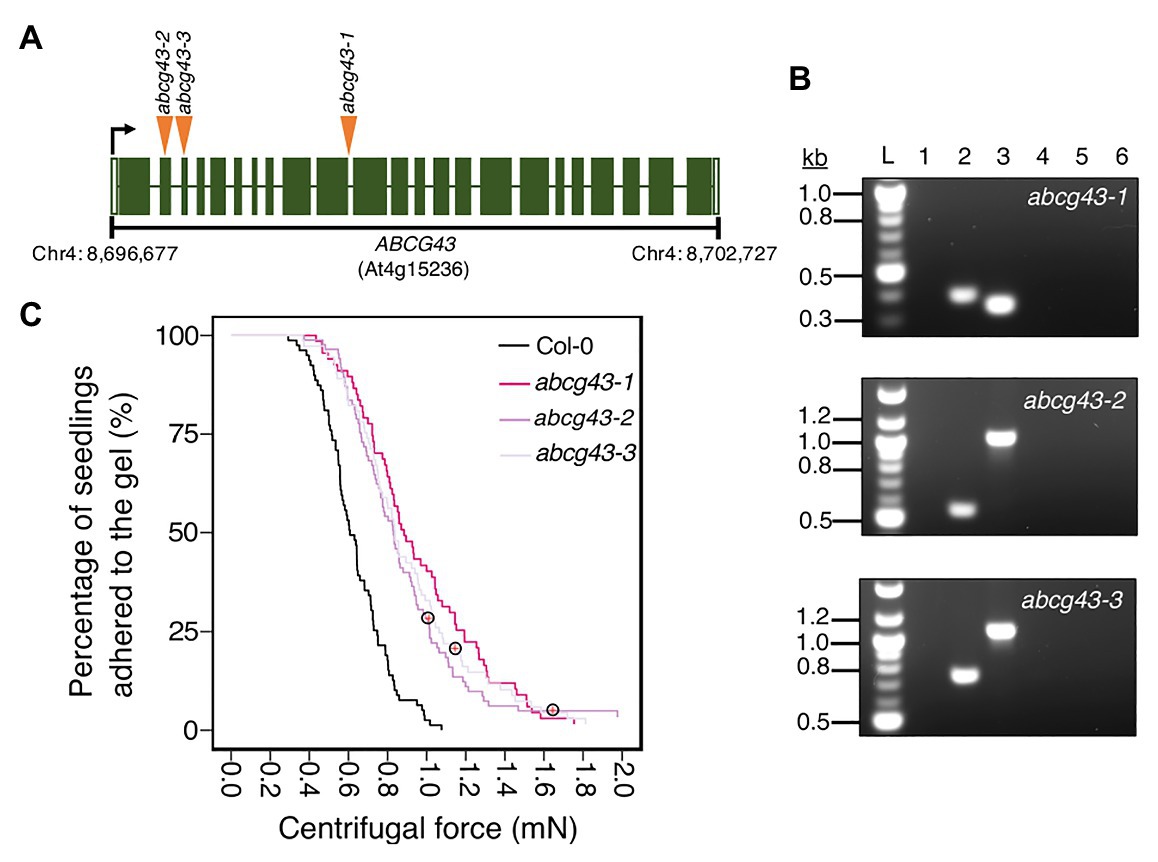
Figure 5. Using the centrifuge assay in a forward genetic screen to identify Arabidopsis thaliana root-substrate adhesion mutants. (A) T-DNA insert locations in ABCG43 for each abcg43 mutant allele, with insertions indicated by orange arrowheads. The ABCG43 gene is located on chromosome four at position Chr4: 8,696,677–8,702,727 and contains 23 exons (dark green) and 22 introns (white gaps). abcg43-1 has a T-DNA insert in the exon10/intron10 boundary, abcg43-2 (SALK_201207) has a T-DNA insert in exon 2 and abcg43-3 (SALKseq_30713) has a T-DNA insert in exon 3. (B) Genomic PCR confirming the homozygosity of each abcg43 mutant for the respective T-DNA inserts. Lanes 1 and 2 were loaded with the gene-specific and T-DNA-border PCR products from abcg43 mutant lines. Lanes 3 and 4 were loaded with the gene-specific and T-DNA-border PCR products from a wild type (Col-0) genomic DNA template. Lanes 5 and 6 were loaded with the water controls for the gene-specific and T-DNA-border PCR reactions, respectively. “L” indicates the 100-bp ladder. The expected product sizes for the gene-specific PCRs were ~350, ~1,025, and ~1,090 nt; the T-DNA-border PCR product sizes were ~400, ~550, and ~750 nt for abcg43-1, abcg43-2, and abcg43-3 alleles, respectively. (C) Survival curves displaying the proportion of seedlings that adhered to the gel at increasing centrifugal force for 79 wild type (Col-0 – black), 70 abcg43-1 (dark pink), 85 abcg43-2 (purple), and 73 abcg43-3 (light purple). Red crosses circled on the survival curves represent seedlings that remained adhered to the gel after the maximum centrifugal speed (1,611 RPM). The results are from a representative experiment for at least two independent experiments showing a statistically significant difference in adhesion between the mutant lines relative to wild type (Cox PH regression; alpha = 0.01). A single experiment included >70 biological replicates for each candidate line.
We analyzed the root-gel adhesion of the homozygous insertional ABCG43 mutant alleles and found abcg43-1, abcg43-2, and abcg43-3 had detachment risks 0.25, 0.36, and 0.35 times that of wild-type plants, respectively (for all lines p < 0.001; Table 2; Figures 5B,C). These results illustrate the use of the centrifuge assay as a powerful screening method for identifying novel genes involved in root-substrate adhesion, as well as a tool for evaluating the effects of known gene function on root-substrate interactions.
Discussion
To date, most published methods that quantify the contribution of particular traits to plant-soil interactions are time-intensive and expensive (Bailey et al., 2002; Toukura et al., 2006; De Baets and Poesen, 2010; De Baets et al., 2020). In contrast, our assay can produce data within a week, is available to any laboratory with access to a bench top centrifuge and does not require speciality consumables. This method is a high-throughput and quantitative way to test the effects of root morphology and cell function on root-substrate adhesion and to screen for new biological and molecular factors that can alter plant-substrate interactions. Using a defined model system like Arabidopsis and sterile medium conditions permits the identification of plant-specific characteristics that can then be probed in other substrate‐ or soil-based conditions.
Using the Centrifuge Assay to Investigate the Role of Root Hairs in Root-Substrate Interactions
Results presented in this paper show that the centrifuge assay can quantify effects of root hair morphology on plant-substrate adhesion. Previous work has shown that the presence of root hairs can significantly enhance root-substrate adhesion compared to the absence of root hairs (Haling et al., 2013; Choi and Cho, 2019; De Baets et al., 2020). We now show that our centrifuge assay can distinguish the adherence strength of seedlings with altered root hair shapes and sizes (Figure 2) and can quantify the contribution these morphological traits make to plant-substrate interactions. Some mutants with defective root hairs have mutations in cell wall biosynthesis and modification proteins, so it is possible that an altered root-substrate interface compromises adhesion in these mutants not just root hair shape. Using our method to rapidly phenotype mutants with root hair growth defects could direct additional experiments to characterize biochemical and morphological properties of genes of interest. Indeed, current evidence for the importance of root hairs for the deposition of complex polysaccharides with soil-binding properties (Galloway et al., 2020) supports the need for assays such as ours to expedite the identification of genes and mechanisms that improve soil-remediation, top soil maintenance, and erosion tolerance of at-risk land.
Identifying New Genes Involved in Root-Substrate Adhesion
Mutations in cellular functions that do not alter root hair growth but do affect substrate adhesion can also be investigated using this assay. Vesicle trafficking pathways are required for cell wall deposition and maintenance (De Caroli et al., 2011; Larson et al., 2014; Rodriguez-Furlán et al., 2016); therefore, we hypothesized that mutants in vesicle trafficking pathways, particularly those at the plasma membrane, could affect root-substrate adhesion. Similarly, the composition and deposition of plant exudates help plants to modify and optimize their growth conditions (Carvalhais et al., 2015; Naveed et al., 2017), suggesting that modification to plant exudate composition could affect root-substrate interactions. Consistent with these hypotheses, mutants with vesicle trafficking defects and altered exudate composition had root-substrate adhesion properties that differed from wild type, indicating that this method can identify aspects of plant cell biology important for root-substrate interactions that do not affect root or root hair morphology (Figures 3, 4). Our results further indicate that while root hairs provide significant adhesive and cohesive support both on solid gel and soil media (De Baets et al., 2020), there are additional cellular factors contributing to plant-substrate interactions that are not directly observable without the use of expensive reagents, development of time-intensive transgenic lines, and microscopy equipment. Our centrifuge assay provides an affordable and quick alternative or complement to these current methodologies.
The compounds and interactions that contribute to root-substrate adhesion are complex, and the direct and indirect factors that participate in these interactions still remain to be characterized in detail. Many root cell wall or secreted compounds might directly affect adhesive characteristics of the root surface such as arabinogalactan proteins involved in the adhesion of English ivy (Huang et al., 2016). Other complex polysaccharides have also been shown to enhance soil binding in both Arabidopsis and cereal root systems (Galloway et al., 2018, 2020). Indeed, mutants in genes associated with the production of these compounds would be potentially excellent candidates for the application of this assay, which would quantify the contribution these compounds make to root-substrate adhesion.
We cannot expect all exudate components to present adhesive phenotypes because not all will affect root-substrate adhesion, and some will affect adhesion in a way that is not detected by this assay. For example, the jin1-9 mutant tested in our exudate cohort was not significantly more or less adhesive compared to wild-type roots (Figure 4) despite its altered exudate composition (Anderson et al., 2004). Some exudate components will play no role in adhesion, while others may require additional co-factors present in soil or non-sterile media to participate in root-substrate interactions. Although these are examples of the limitations of the assay, this assay does allow for the identification of direct contributions that plants make to root-substrate cohesion and its robust statistical power and high-throughput aspects will significantly improve genetic screening timelines and enhance large-scale experimental design.
Forward genetic screening relies on distinctive phenotypes that distinguish differences within large, genetically diverse populations. High throughput, creative techniques for identifying phenotypes within morphologically similar populations, such as thermal imaging (Plessis et al., 2011) and confocal imaging platforms (Robert et al., 2008; Drakakaki et al., 2011), require specific equipment that not all laboratories have at their disposal. In contrast, the centrifuge assay can be used as a screen to identify new genes affecting plant-substrate interactions without specialist equipment, making it an accessible option for research groups that may not otherwise be able to efficiently conduct genetic screens. Moreover, the centrifuge assay does not require morphological or conditional differences for candidate selection. As an example, we report the identification in a forward genetics screen of a mutant population in which abcg43 mutant seedlings were identified and shown to be more adhesive to gel than wild type (Figure 5). The function of ABCG43 in Arabidopsis is uncharacterized, indicating that this assay can identify novel candidate genes that have no previously known association with root morphological defects, adhesion, or exudate composition for future study.
Conclusion
This assay has applications in plant cell biology and genetics to investigate how diverse gene activities, cell functions, and root structure affect plant-soil adhesion and identify the molecular pathways involved. Potential applications not tested in this report include the measurement of the effects environmental conditions have on plant-substrate adhesion and how other plant species or ecotypes respond to detachment forces. Given the recent discovery that root hair presence is important for Arabidopsis cohesion between roots and gel and compost or soil (De Baets et al., 2020), this assay can broadly contribute to plant and soil sciences through the identification of plant traits that increase root-substrate cohesion and potentially stabilize slopes or reduce soil erosion.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://doi.org/10.5523/bris.21loiw3fpw372g99l93meaja1.
Author Contributions
CG designed the initial project, conducted proof-of-concept pilot experiments, obtained funding, and recruited and advised the team. TL and IC developed the theoretical parameters for the centrifugal assay and the bounds on the errors from other mechanical forces. BE and EL planned and conducted all experiments apart from the initial genetic screen, which was carried out by LW and KS, wrote the initial manuscript, and analyzed all the data presented herein. AS and BE optimized the centrifuge assay data collection and data analysis methods. BE, EL, and CG edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Leverhulme Trust project grant RPG-2013-260 to CG and TL. BE was supported by a BBSRC SWBio PhD studentship (grant BB/M009122/1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Don Grierson FRS and Enrico Coen FRS who independently suggested the centrifuge assay concept, Timothy Quine for providing insightful guidance during method development, and Thomas Denbigh for providing technical advice. We are grateful to Jill Harrison, Victoria Spencer, Zoe Nemec Venza, Sophie Carpenter, and Nicholas H. Fair for their helpful comments during revision of the manuscript. This manuscript has been released as a preprint at http://doi.org/10.1101/2020.05.14.095448 (Eldridge et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.602486/full#supplementary-material
References
Akhtar, J., Galloway, A. F., Nikolopoulos, G., Field, K. J., and Knox, P. (2018). A quantitative method for the high throughput screening for the soil adhesion properties of plant and microbial polysaccharides and exudates. Plant Soil 428, 57–65. doi: 10.1007/s11104-018-3670-1
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Assaad, F. F., Qiu, J. -L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129. doi: 10.1091/mbc.e04-02-0140
Badri, D. V., Loyola-Vargas, V. M., Broeckling, C. D., De-la-Peña, C., Jasinski, M., Santelia, D., et al. (2008). Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol. 146, 762–771. doi: 10.1104/pp.107.109587
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Bailey, P. H. J., Currey, J. D., and Fitter, A. H. (2002). The role of root system architecture and root hairs in promoting anchorage against uprooting forces in Allium cepa and root mutants of Arabidopsis thaliana. J. Exp. Bot. 53, 333–340. doi: 10.1093/jexbot/53.367.333
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Baulcombe, D. C., Saunders, G. R., Bevan, M. W., Mayo, M. A., and Harrison, B. D. (1986). Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature 321, 446–449. doi: 10.1038/321446a0
Baumberger, N., Ringli, C., and Keller, B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15, 1128–1139. doi: 10.1101/gad.200201
Berger, S., Bell, E., and Mullet, J. E. (1996). Two methyl jasmonate-insensitive mutants Show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. doi: 10.1104/pp.111.2.525
Böhme, K., Li, Y., Charlot, F., Grierson, C., Marrocco, K., Okada, K., et al. (2004). The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40, 686–698. doi: 10.1111/j.1365-313X.2004.02245.x
Bradburn, M. J., Clark, T. G., Love, S. B., and Altman, D. G. (2003). Survival analysis part II: multivariate data analysis – an introduction to concepts and methods. Br. J. Cancer 89, 431–436. doi: 10.1038/sj.bjc.6601119
Burylo, M., Rey, F., Mathys, N., and Dutoit, T. (2012). Plant root traits affecting the resistance of soils to concentrated flow erosion. Earth Surf. Process. Landf. 37, 1463–1470. doi: 10.1002/esp.3248
Carol, R. J., Takeda, S., Linstead, P., Durrant, M. C., Kakesova, H., Derbyshire, P., et al. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438, 1013–1016. doi: 10.1038/nature04198
Carvalhais, L. C., Dennis, P. G., Badri, D. V., Kidd, B. N., Vivanco, J. M., and Schenk, P. M. (2015). Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant-Microbe Interact. 28, 1049–1058. doi: 10.1094/MPMI-01-15-0016-R
Choi, H. -S., and Cho, H. -T. (2019). Root hairs enhance Arabidopsis seedling survival upon soil disruption. Sci. Rep. 9:11181. doi: 10.1038/s41598-019-47733-0
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. -L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. doi: 10.1038/nature02076
De Baets, S., Denbigh, T. D., Smyth, K. M., Eldridge, B. M., Weldon, L. M., Higgins, B. W., et al. (2020). Micro-scale interactions between Arabidopsis root hairs and soil particles influence soil erosion. Commun. Biol. 3:164. doi: 10.1038/s42003-020-0886-4
De Baets, S., and Poesen, J. (2010). Empirical models for predicting the erosion-reducing effects of plant roots during concentrated flow erosion. Geomorphology 118, 425–432. doi: 10.1016/j.geomorph.2010.02.011
De Baets, S., Poesen, J., Knapen, A., and Galindo, P. (2007). Impact of root architecture on the erosion-reducing potential of roots during concentrated flow. Earth Surf. Process. Landf. 32, 1323–1345. doi: 10.1002/esp.1470
De Caroli, M., Lenucci, M. S., Di Sansebastiano, G. -P., Dalessandro, G., De Lorenzo, G., and Piro, G. (2011). Dynamic protein trafficking to the cell wall. Plant Signal. Behav. 6, 1012–1015. doi: 10.4161/psb.6.7.15550
Diet, A., Link, B., Seifert, G. J., Schellenberg, B., Wagner, U., Pauly, M., et al. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18, 1630–1641. doi: 10.1105/tpc.105.038653
Drakakaki, G., Robert, S., Szatmari, A. -M., Brown, M. Q., Nagawa, S., Van Damme, D., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. 108, 17850–17855. doi: 10.1073/pnas.1108581108
Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349. doi: 10.1093/nar/19.6.1349
Eisenach, C., Chen, Z. -H., Grefen, C., and Blatt, M. R. (2012). The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J. 69, 241–251. doi: 10.1111/j.1365-313X.2011.04786.x
Eldridge, B. M., Larson, E. R., Weldon, L., Smyth, K. M., Sellin, A. N., Chenchiah, I. V., et al. (2020). A centrifuge-based method for identifying novel genetic traits that affect root-substrate adhesion in Arabidopsis thaliana. BioRxiv [Preprint]. doi: 10.1101/2020.05.14.095448
Favery, B., Ryan, E., Foreman, J., Linstead, P., Boudonck, K., Steer, M., et al. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89. doi: 10.1101/gad.188801
Galloway, A. F., Akhtar, J., Marcus, S. E., Fletcher, N., Field, K., and Knox, P. (2020). Cereal root exudates contain highly structurally complex polysaccharides with soil-binding properties. Plant J. 103, 1666–1678. doi: 10.1111/tpj.14852
Galloway, A. F., Pedersen, M. J., Merry, B., Marcus, S. E., Blacker, J., Benning, L. G., et al. (2018). Xyloglucan is released by plants and promotes soil particle aggregation. New Phytol. 217, 1128–1136. doi: 10.1111/nph.14897
Geelen, D., Leyman, B., Batoko, H., Di Sansebastiano, G. -P., Moore, I., Blatt, M. R., et al. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14, 387–406. doi: 10.1105/tpc.010328
Ghestem, M., Cao, K., Ma, W., Rowe, N., Leclerc, R., Gadenne, C., et al. (2014). A framework for identifying plant species to be used as “ecological engineers” for fixing soil on unstable slopes. PLoS One 9:e95876. doi: 10.1371/journal.pone.0095876
Grierson, C. S., Roberts, K., Feldmann, K. A., and Dolan, L. (1997). The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 115, 981–990. doi: 10.1104/pp.115.3.981
Haling, R. E., Brown, L. K., Bengough, A. G., Young, I. M., Hallett, P. D., White, P. J., et al. (2013). Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength. J. Exp. Bot. 64, 3711–3721. doi: 10.1093/jxb/ert200
Healey, A., Furtado, A., Cooper, T., and Henry, R. J. (2014). Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:21. doi: 10.1186/1746-4811-10-21
Huang, Y., Wang, Y., Tan, L., Sun, L., Petrosino, J., Cui, M. -Z., et al. (2016). Nanospherical arabinogalactan proteins are a key component of the high-strength adhesive secreted by English ivy. Proc. Natl. Acad. Sci. U. S. A. 113, E3193–E3202. doi: 10.1073/pnas.1600406113
Kang, E., Zheng, M., Zhang, Y., Yuan, M., Yalovsky, S., Zhu, L., et al. (2017). The microtubule-associated protein map18 affects ROP2 GTPase activity during root hair growth1[OPEN]. Plant Physiol. 174, 202–222. doi: 10.1104/pp.16.01243
Kang, J., Yim, S., Choi, H., Kim, A., Lee, K. P., Lopez-Molina, L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6:8113. doi: 10.1038/ncomms9113
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Kitakura, S., Vanneste, S., Robert, S., Löfke, C., Teichmann, T., Tanaka, H., et al. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931. doi: 10.1105/tpc.111.083030
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Larson, E. R., Domozych, D. S., and Tierney, M. L. (2014). SNARE VTI13 plays a unique role in endosomal trafficking pathways associated with the vacuole and is essential for cell wall organization and root hair growth in Arabidopsis. Ann. Bot. 114, 1147–1159. doi: 10.1093/aob/mcu041
Larson, E. R., Van Zelm, E., Roux, C., Marion-Poll, A., and Blatt, M. R. (2017). Clathrin heavy chain subunits coordinate endo‐ and exocytic traffic and affect stomatal movement. Plant Physiol. 175, 708–720. doi: 10.1104/pp.17.00970
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mbengue, M., Bourdais, G., Gervasi, F., Beck, M., Zhou, J., Spallek, T., et al. (2016). Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 113, 11034–11039. doi: 10.1073/pnas.1606004113
Naveed, M., Brown, L. K., Raffan, A. C., George, T. S., Bengough, A. G., Roose, T., et al. (2017). Plant exudates may stabilize or weaken soil depending on species, origin and time. Eur. J. Soil Sci. 68, 806–816. doi: 10.1111/ejss.12487
Parker, J. S., Cavell, A. C., Dolan, L., Roberts, K., and Grierson, C. S. (2000). Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12, 1961–1974. doi: 10.1105/tpc.12.10.1961
Plessis, A., Cournol, R., Effroy, D., Silva Pérez, V., Botran, L., Kraepiel, Y., et al. (2011). New ABA-hypersensitive Arabidopsis mutants are affected in loci mediating responses to water deficit and Dickeya dadantii infection. PLoS One 6:e20243. doi: 10.1371/journal.pone.0020243
Prentice, R. L., and Kalbfleisch, J. D. (2003). Mixed discrete and continuous Cox regression model. Lifetime Data Anal. 9, 195–210. doi: 10.1023/A:1022935019768
R Core Team (2014). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Ringli, C., Bigler, L., Kuhn, B. M., Leiber, R. -M., Diet, A., Santelia, D., et al. (2008). The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20, 1470–1481. doi: 10.1105/tpc.107.053249
Robert, S., Chary, S. N., Drakakaki, G., Li, S., Yang, Z., Raikhel, N. V., et al. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. PNAS 105, 8464–8469. doi: 10.1073/pnas.0711650105
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Rodriguez-Furlán, C., Salinas-Grenet, H., Sandoval, O., Recabarren, C., Arraño-Salinas, P., Soto-Alvear, S., et al. (2016). The root hair specific SYP123 regulates the localization of cell wall components and contributes to rizhobacterial priming of induced systemic resistance. Front. Plant Sci. 7:1081. doi: 10.3389/fpls.2016.01081
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Spruance, S. L., Reid, J. E., Grace, M., and Samore, M. (2004). Hazard ratio in clinical trials. Antimicrob. Agents Chemother. 48, 2787–2792. doi: 10.1128/AAC.48.8.2787-2792.2004
Stokes, A., Atger, C., Bengough, A. G., Fourcard, T., and Sidle, R. C. (2009). Desirable plant root traits for protecting natural and engineered slopes against landslides. Plant Soil 324, 1–30. doi: 10.1007/s11104-009-0159-y
Toukura, Y., Devee, E., and Hongo, A. (2006). Uprooting and shearing resistances in the seedlings of four weedy species. Weed Biol. Manag. 6, 35–43. doi: 10.1111/j.1445-6664.2006.00192.x
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Waghmare, S., Lileikyte, E., Karnik, R., Goodman, J. K., Blatt, M. R., and Jones, A. M. E. (2018). SNAREs SYP121 and SYP122 mediate the secretion of distinct cargo subsets. Plant Physiol. 178, 1679–1688. doi: 10.1104/pp.18.00832
Wang, X., Cnops, G., Vanderhaeghen, R., De Block, S., Van Montagu, M., and Van Lijsebettens, M. (2001). AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 126, 575–586. doi: 10.1104/pp.126.2.575
Keywords: Arabidopsis, centrifuge assay, forward genetics, root exudates, root hairs, root-substrate adhesion, survival analysis, vesicle trafficking, secretion
Citation: Eldridge BM, Larson ER, Weldon L, Smyth KM, Sellin AN, Chenchiah IV, Liverpool TB and Grierson CS (2021) A Centrifuge-Based Method for Identifying Novel Genetic Traits That Affect Root-Substrate Adhesion in Arabidopsis thaliana. Front. Plant Sci. 12:602486. doi: 10.3389/fpls.2021.602486
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Paul Knox, University of Leeds, United KingdomShenghua Fan, University of Kentucky, United States
Copyright © 2021 Eldridge, Larson, Weldon, Smyth, Sellin, Chenchiah, Liverpool and Grierson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claire S. Grierson, bGFjc2dAYnJpc3RvbC5hYy51aw==
§ORCID:
Bethany M. Eldridge
Emily R. Larson
Laura Weldon
Kevin M. Smyth
Isaac V. Chenchiah
Tanniemola B. Liverpool
Claire S. Grierson
‡Present address: Laura Weldon, Wildfowl and Wetlands Trust (WWT), Slimbridge, United Kingdom
†These authors have contributed equally to this work
 Bethany M. Eldridge
Bethany M. Eldridge Emily R. Larson
Emily R. Larson Laura Weldon
Laura Weldon Kevin M. Smyth1§
Kevin M. Smyth1§ Annabelle N. Sellin
Annabelle N. Sellin Isaac V. Chenchiah
Isaac V. Chenchiah Claire S. Grierson
Claire S. Grierson