- 1Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 2VIB Center for Plant Systems Biology, Ghent, Belgium
Plant cells perceive and adapt to an ever-changing environment by modifying their plasma membrane (PM) proteome. Whereas secretion deposits new integral membrane proteins, internalization by endocytosis removes membrane proteins and associated ligands, largely with the aid of adaptor protein (AP) complexes and the scaffolding molecule clathrin. Two AP complexes function in clathrin-mediated endocytosis at the PM in plant cells, the heterotetrameric AP-2 complex and the hetero-octameric TPLATE complex (TPC). Whereas single subunit mutants in AP-2 develop into viable plants, genetic mutation of a single TPC subunit causes fully penetrant male sterility and silencing single subunits leads to seedling lethality. To address TPC function in somatic root cells, while minimizing indirect effects on plant growth, we employed nanobody-dependent delocalization of a functional, GFP-tagged TPC subunit, TML, in its respective homozygous genetic mutant background. In order to decrease the amount of functional TPC at the PM, we targeted our nanobody construct to the mitochondria and fused it to TagBFP2 to visualize it independently of its bait. We furthermore limited the effect of our delocalization to those tissues that are easily accessible for live-cell imaging by expressing it from the PIN2 promoter, which is active in root epidermal and cortex cells. With this approach, we successfully delocalized TML from the PM. Moreover, we also show co-recruitment of TML-GFP and AP2A1-TagRFP to the mitochondria, suggesting that our approach delocalized complexes, rather than individual adaptor complex subunits. In line with the specific expression domain, we only observed minor effects on root growth, yet realized a clear reduction of endocytic flux in epidermal root cells. Nanobody-dependent delocalization in plants, here exemplified using a TPC subunit, has the potential to be widely applicable to achieve specific loss-of-function analysis of otherwise lethal mutants.
Introduction
Cells are delineated by their plasma membrane (PM). The PM houses a plethora of proteins ranging from receptors and ion channels to structural membrane proteins. Many of these PM proteins, commonly termed cargo, are responsible for cellular communication with the outside world. In eukaryotes, endocytosis is the cellular process where cargoes, associated ligands as well as lipids are internalized from the PM. Endocytosis thereby provides a way to regulate the content and consequently modulate protein activity at the PM. A predominant and well-studied form of endocytosis is clathrin-mediated endocytosis (CME; Bitsikas et al., 2014). CME refers to the dependency of the scaffolding protein clathrin, which coats the developing and mature vesicles (Robinson, 2015). In plants, CME plays a role in hormone signaling (Irani et al., 2012; Martins et al., 2015; Zhang et al., 2017), nutrient availability (Wang et al., 2017; Dubeaux et al., 2018; Yoshinari et al., 2019), pathogen defense and susceptibility (Mbengue et al., 2016; Li and Pan, 2017), and other biotic and abiotic stresses (Li et al., 2011). Consequently, CME is essential for plant development.
Two early-arriving adaptor complexes, the heterotetrameric Adaptor Protein-2 complex (AP-2) and the hetero-octameric TPLATE complex (TPC) facilitate CME in plants (Gadeyne et al., 2014). In contrast to AP-2, TPC represents an evolutionary ancient protein complex, which is lost in yeast and mammalian cells (Hirst et al., 2014). The slime mold Dictyostelium discoideum contains a similar complex, named TSET. TSET, however, is a hexameric complex in contrast to TPC in Arabidopsis thaliana, which has two additional subunits. Also contrary to TPC, TSET is dispensable in D. discoideum (Hirst et al., 2014). The presence of a full or partial TSET complex in other eukaryotes was confirmed by additional homology searches, indicative of its ancient evolutionary origin (Hirst et al., 2014).
Adaptor Protein-2 complex and TPC have both common and distinct functions, possibly relating to cargo specificity and/or fate of the internalized cargo (Bashline et al., 2015; Sánchez-Rodríguez et al., 2018; Wang et al., 2019; Yoshinari et al., 2019). In addition, functional diversification of both complexes is reflected in their mutant phenotypes. Knockout plants in individual AP-2 subunits are affected at various stages of development but viable (Bashline et al., 2013; Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013). However, ap2 mutants show reduced internalization of the styryl dye N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide (FM4-64), which can be seen as proxy to a difference in cargo uptake (Jelínková et al., 2010), as well as known endocytic cargoes like the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1), the boron exporter BOR1, and auxin efflux carriers of the PIN-FORMED protein family (Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yoshinari et al., 2016, 2019).
The relatively mild phenotype of ap2 single subunit mutants in plants contrasts with the lethal phenotype of a single ap2 subunit knockout in mice (Mitsunari et al., 2005). Alternatively, the complex does not seem to be essential for yeast (Yeung et al., 2013). In Caenorhabditis elegans, AP-2 subunits are capable of assembling into hemicomplexes which partially retain their functionality (Gu et al., 2013). In plants, AP2M and AP2S are still recruited to the PM in ap2s and ap2m mutants, respectively (Wang et al., 2016), suggesting that AP-2 hemicomplexes might also confer partial functionality in plants.
In contrast to AP-2, single knockouts of TPC subunits result in fully penetrant male sterility with shriveled pollen and ectopic callose accumulation (Gadeyne et al., 2014). Similar pollen-lethal phenotypes are also reported for a mutant in DYNAMIN-RELATED PROTEIN 1C protein (drp1c; Backues et al., 2010), as well as a CLATHRIN LIGHT CHAIN mutant, clc1 (Wang et al., 2013), involved in vesicle fission and clathrin triskelion assembly, respectively.
So far, there is only one viable weak allele of one TPC subunit identified. This twd40-2-3 mutant (Bashline et al., 2015) is however likely merely a knockdown as twd40-2-1 and twd40-2-2 mutants are pollen lethal (Gadeyne et al., 2014). Knockdowns of TML and TPLATE resulted in seedling lethality with a reduced internalization of FM4-64, BRI1, RECEPTOR-LIKE PROTEIN 44 (RLP44), and the cellulose synthase subunit CESA6 (Irani et al., 2012; Gadeyne et al., 2014; Sánchez-Rodríguez et al., 2018; Gómez et al., 2019). Silencing works on the messenger level and phenotypes only become apparent following degradation of pre-made proteins. As AP complexes can be recycled following each round of internalization, approaches affecting these complexes at the protein level have a more direct effect. In animal cells, conditional delocalization using rapamycin to target AP-2 to mitochondria has been successfully applied to interfere with endocytosis (Robinson et al., 2010). Also, epidermal growth factor receptor substrate 15 (EPS15), a pioneer endocytic accessory protein, was successfully inactivated in HeLa cells by expressing an anti-EPS15 nanobody on endosomes or mitochondria (Traub, 2019).
Since their discovery, nanobodies, derived from camelid heavy chain-only antibodies (HCAb), have found their way into a wide variety of applications in biological fields. Nanobodies are similar to antibodies (Ab) in the sense that they can bind epitopes with high affinity in a highly selective manner (Ingram et al., 2018). Their applications range from drug discovery, crystallography, and imaging techniques to probing protein functions by degradation or delocalization (Caussinus et al., 2012; Früholz et al., 2018; Ingram et al., 2018). Nanobodies can be expressed as a single chain, compact and stable protein while still retaining high selectivity and affinity for its epitope (Muyldermans, 2013). This makes them more convenient to clone and to express compared to conventional Ab.
In plants, nanobodies have been used to selectively degrade proteins using the deGradFP method, originally developed in Drosophila melanogaster. DeGradFP links an anti-GFP nanobody to an F-box protein, thereby targeting it for ubiquitin-dependent degradation (Caussinus et al., 2012). This approach was shown to be functional in plants (Baudisch et al., 2018) and successfully used to deplete WUSCHEL-GFP in the Arabidopsis flowering meristem (Ma et al., 2019) and the centromeric Histon H3 of Arabidopsis in transgenic tobacco plants (Sorge et al., 2021). Nanobodies have also been used in Arabidopsis seedlings to lock down vacuolar sorting receptors (VSRs) in cellular compartments upstream of trans-Golgi network/early endosomes, allowing to determine their retrograde trafficking pathway (Früholz et al., 2018).
Here, we explore, similar to what has been done in animal cells (Robinson et al., 2010; Traub, 2019), the effects on CME caused by lockdown of the endocytic machinery. We use a GFP-tagged functional TML-GFP fusion protein in the homozygous tml-1(−/−) mutant background and delocalized it to the mitochondria using a nanobody directed against eGFP.
Materials and Methods
Cloning
Gateway entry clones pDONR221-TagBFP2, pDONR221-MITOTagBFP2 and pDONRP2RP3-GFPNb were generated according to the manufacturer’s instructions (ThermoFisher Scientific BP clonase). pDONR221-TagBFP2 was amplified from pSN.5 mTagBFP2 (Pasin et al., 2014) with primers:
AttB1-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGTCATCTAAGGGTGAAGAGC TTATC AAAGAGAAT and AttB2-GGGGACCACTTTGTACAAGAAAGCTGGGTCACCTCCGCCACCTCCACCTCCCAGTCCTGCGTA.
pDONR221-MITOTagBFP2 was generated from pDONR221-TagBFP2 by including the import signal of the yeast mitochondrial outer membrane protein Tom70p as described before (Robinson et al., 2010). The following primers sequences were used:
AttB1-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGAAGAGCTTCATTACAAGGAACAAGACAGCCATTTTGGC AACCGTTGCTGCTACAGGTACTGCCATCGGTGCCTACTATTATTACAACCAATTGCAACAGGATCCACCGGTCGCCACCATGTCATCTAAGGGTGAAGAGCTT and AttB2-GGGGACCACTTTGTACAAGAAAGCTGGGTACGCTAAGTCTTCCTCTGAAATCAA.
pDONRP2RP3-GFPNb was generated from an anti-GFP Nanobody construct (Künzl et al., 2016) with primers attB2-GGGGACAGCT TTCTTGTACAAAG TGGGGATGTATCCTTA TGATGTTC and attB3r-GGGGACAACTT TGTATAATAAAGT TGTTTAAT GATGATGATGA TGATGAGAAGA including a HA-tag, a 3xHis-tag, and a stop codon.
The entry clones of the PIN2 promoter pDONRP4P1R_PIN2prom (Marquès-Bueno et al., 2016) or 35 s promoter, pDONR221-MITOTagBFP2 and pDONRP2RP3-GFPNb were used in a triple Gateway LR reaction, combining pB7m34GW (Karimi et al., 2005) to yield pB7m34GW_PIN2prom::MITOTagBFP2-GFPNb or pB7m34GW_p35sprom::MITOTagBFP2-GFPNb.
Nicotiana benthamiana Plant Growth and Transient Expression Assay
Nicotiana benthamiana plants were grown in a greenhouse under long-day conditions (6–22 h light, 100 PAR, 21°C) in soil (Saniflo Osmocote pro NPK: 16 − 11 − 10+ magnesium and trace elements). Transient expression was performed by leaf infiltration according to (Sparkes et al., 2006). The abaxial epidermis was imaged 48 h after infiltration.
Arabidopsis Plant Material and Transformation
Plants expressing pB7m34GW_PIN2prom::MITOTagBFP2-GFPNb were generated by floral dip (Clough and Bent, 1998). Constructs were dipped into Col-0 and tml-1(−/−) (At5g57460) mutant lines described previously (Gadeyne et al., 2014). Primary transformants (T1) were selected on BASTA containing ½ strength MS medium without sucrose and 0.6% Gelrite (Duchefa, Netherlands). PIN2prom::MITOTagBFP2-GFPNb expression was analyzed in the progeny of BASTA-resistant primary transformants (T2 seeds) by microscopy and T2 lines demonstrating strong expression were selected regardless of insert copy number. Next, T2 lines were crossed with the previously described TML-GFP complemented tml-1(−/−) mutant line expressing also RPS5Aprom::AP2A1-TagRFP (Gadeyne et al., 2014). Primary hybrids were analyzed via microscopy and best lines were selected on the basis of both PIN2prom::MITOTagBFP2-GFPNb and RPS5Aprom::AP2A1-TagRFP expression. For both Col-0 and tml-1(−/−) backgrounds, two independent lines (-Nb1 and -Nb2) were generated. Namely, Col-Nb1, Col-Nb2, TML-Nb1, and TML-Nb2. In order to synchronize the age and overall fitness of the seeds, all lines used in the root growth and carbon starvation study, including the Col-0 and tml-1(−/−) lines, were propagated together and collected at the same time.
Quantification of TML-GFP Endocytic Foci
Four to five days old seedlings of TML, TML-Nb1, and TML-Nb2 were grown on ½ strength MS medium without sucrose and 0.6% Gelrite (Duchefa, Netherlands). Acquired pictures were analyzed in Fiji/ImageJ (Schindelin et al., 2012; Schneider et al., 2012). In each analyzed root an ROI of constant dimensions was selected. In addition in TML-Nb1 and TML-Nb2, the ROI did not overlap with mitochondrial clusters. The median intensity in each root was recorded, followed by the counts of the endocytic foci using the Find Maxima function. Statistical differences in median intensity and foci count between the lines were analyzed with pairwise Wilcoxon tests. p-values were adjusted using the Bonferroni method.
Phenotypical Quantification of Root Growth
Arabidopsis seedlings were grown on ½ strength MS medium without sucrose and 0.6% Gelrite (Duchefa, Netherlands). Seeds of all lines were equally placed on the plates (3 seeds per line per plate, distributed over 14 plates). Plates were left for the stratification for 48 h at 4°C, and then placed at 21°C in continuous light. Two days after transfer to the light, seeds which did not germinate were marked and excluded in the further analysis. For root growth analysis, seedlings were grown in continuous light and 2 days after germination the root growth of every seedling was marked for 7 days, daily. For carbon starvation, seedlings were grown for 5 days, including the germination period, in continuous light after which the root growth of every seedling was marked. Subsequently, the plates were covered with aluminum foil and left for 7 days in dark after which root growth was marked again. Root growth and carbon starvation assays measurements were carried out with Fiji/ImageJ (Schindelin et al., 2012; Schneider et al., 2012). Seedlings, which stopped growing at early time points of the study, have grown into the medium or have grown in direct contact with the plate edge, were excluded from further analysis. Statistical differences for root growth assays were determined via a mixed model analysis. Mixed linear model analysis was applied to the root length of the lines Col-0, Col-Nb1, Col-Nb2, TML, TML-Nb1, and TML-Nb2 using the mixed procedure from SAS (SAS Studio 3.8 and SAS 9.4, SAS Institute Inc., Cary, NC). Fixed effects in the model were Line, Day, and the interaction term. An unstructured covariance structure was estimated to model the correlations between measurements done on the same plant. The degrees of freedom of the fixed effects were approximated with the Kenward-Rogers method. The hypotheses of interests were the differences between Col-0 and its respective nanobody lines, between TML and its respective nanobody lines and between the nanobody lines with the same background. These hypotheses were tested using the plm procedure. p-values were adjusted for multiple testing using the maxT procedure as implemented in the plm procedure. For the carbon starvation assay, statistical differences in root growth between the lines were analyzed with pairwise Wilcoxon tests. p-values were adjusted using the Bonferroni method.
FM-Uptake Quantification
Endocytic tracer FM4-64 stock solution was prepared prior to treatment (2 mM in DMSO, Thermo Fisher). Roots were stained with 2 μM FM4-64 by incubating the seedlings in FM-containing ½ strength MS medium without sucrose for 30 min. Treatment was followed by microscopy. Acquired pictures were analyzed in Fiji/ImageJ (Schindelin et al., 2012; Schneider et al., 2012). PM and cytosol of individual epidermal cells were outlined (using the Select Brush Tool and Freehand selections, respectively) and histograms of pixel intensities were generated. Pictures which contained more than 1% saturated pixels were excluded from the quantification. Cytoplasm/PM ratios were calculated from average intensities of the top 1% highest intensity pixels based on the histograms. Outliers were removed via interquartile range (IQR, data point ruled out if its value was either lower than first quartile Q1-1.5 × IQR, or higher than third quartile Q3-1.5 × IQR) in a single step. The normality assumption of the measurements was verified with the Shapiro-Wilk normality test. Due to violation of the normality assumption, statistical differences in intensity values between the lines were analyzed with pairwise Wilcoxon tests. p-values were adjusted using the Bonferroni method. All Shapiro-Wilk normality tests and Wilcoxon tests were performed using the R statistical software. (Rstudio Team, 2019, Version 1.2.5033, R Version 3.6.2).
Image Acquisition
Nicotiana benthamiana infiltration assay imaging (Figure 1A) was performed on a PerkinElmer UltraView spinning-disk system, attached to a Nikon Ti inverted microscope and operated using the Volocity software package. Images were acquired on an ImagEMccd camera (Hamamatsu C9100-13) using frame-sequential imaging with a 60x water immersion objective (NA = 1.20). Specific excitation and emission was performed using a 405 nm laser excitation combined with a single band pass filter (454–496 nm) for TagBFP2 and 561 nm laser excitation combined with a dual band pass filter (500–530 and 570–625 nm) for RFP. Images shown are Z-stack projections. Z-stacks were acquired in sequential frame mode with a 1 μm interval using the UltraView (Piezo) focus drive module.
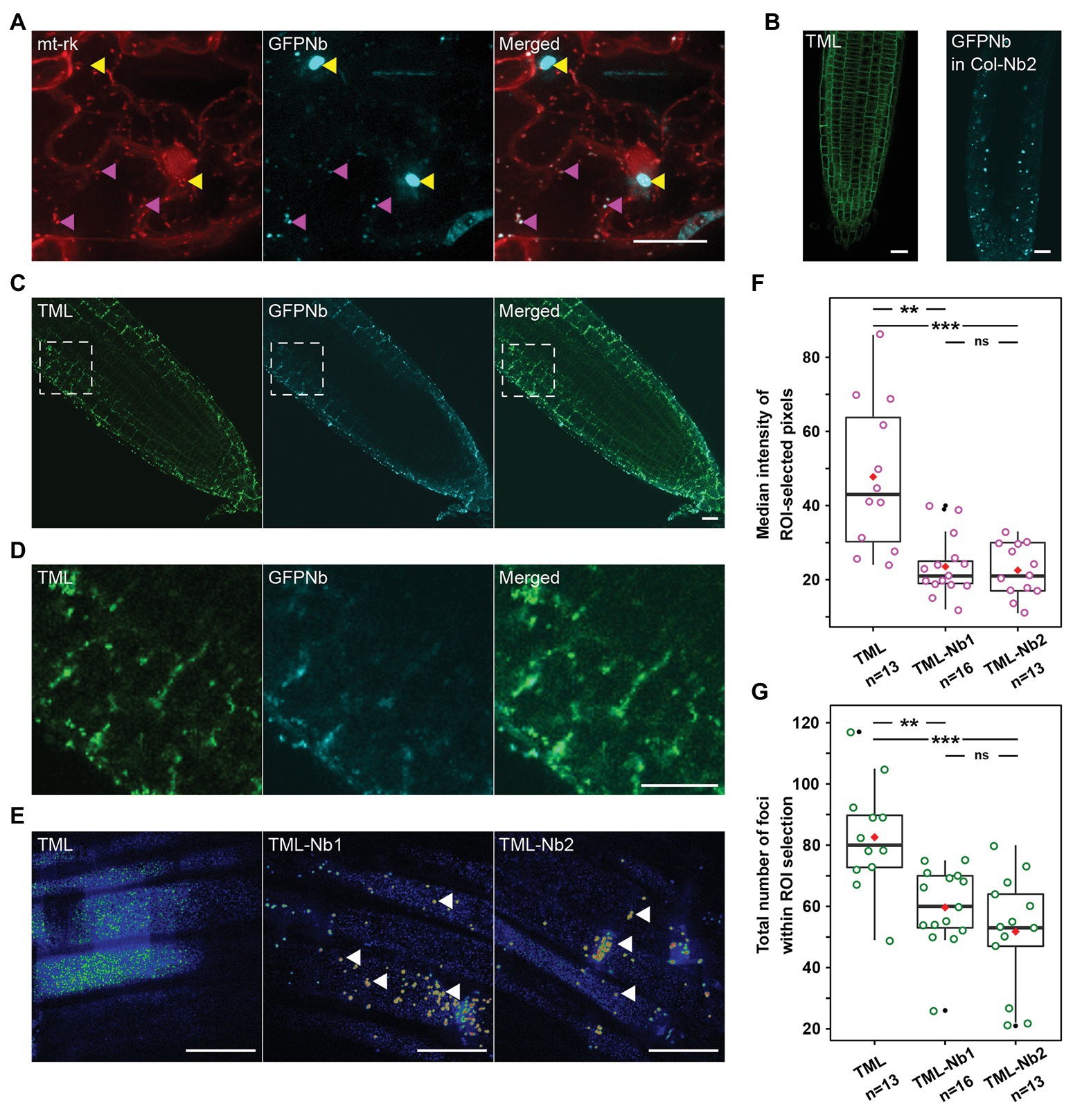
Figure 1. Expression of a mitochondrial-targeted nanobody against GFP allows delocalization of TML-GFP. (A) Representative Z-stack projection of epidermal Nicotiana benthamiana cells transiently expressing the mitochondrial marker mt-rk (red) and the 35S::MITO-TagBFP2-GFP-Nanobody (GFPNb, cyan). Both fusion proteins colocalize at small cytosolic punctae (magenta arrowheads), but not at big aggregates, likely representing dysfunctional clustered mitochondria (yellow arrowheads). (B) Representative Arabidopsis root image of tml-1(−/−) complemented with TML-GFP showing that the functional TML fusion is predominantly targeted to the PM (left) as well as localization of GFPNb in cytosolic punctae in the WT (Col-0) background (right). (C,D) Representative overview images and respective zooms of the outlined region of Arabidopsis roots where TML-GFP in tml-1(−/−) was combined with MITOTagBFP2-GFPNb expression, leading to its delocalization from the PM. (E) Representative, rainbow intensity colored, grazing sections through the PM, showing the recruitment of TML to endocytic foci without (left) and with partial delocalization of TML-Nb1 and TML-Nb2 (middle, right, white arrowheads). Scale bars equal 20 μm. (F,G) Box plots showing the median intensity (F) or total number (G) of endosomal TML-GFP positive foci in the Arabidopsis roots. In the roots expressing GFPNb, both the intensity of the foci as well as their number are significantly reduced compared to TML-GFP (Wilcoxon pairwise comparison tests with Bonferroni adjusted p-values. p < 0.001 are represented as ***, <0.01 as **, and non-significant values as “ns”). The black lines represent the median and the red diamonds represent the mean of the analyzed values. Each magenta or green dot represents an individual cell. Black dot refers to outliers. n refers to the total number of analyzed cells.
Confocal Arabidopsis images were taken using Leica SP8X confocal microscope equipped with a White Light Laser and using the LASX software (Figures 1B-D, 2, 4). Images were acquired on Hybrid (HyD, gating 0.3–10.08 ns) and Photomultiplier (PMT) Detectors using bidirectional line-sequential imaging with a 40x water objective (NA = 1.10) and frame or line signal averaging. Specific excitation and emission were used: 405 nm laser and filter range 410–470 nm for TagBFP2, 488 nm laser and filter range 500–550 nm for GFP, 488 nm laser, and filter range 600–740 nm for FM4-64, 555nm laser and filter range 560–670 for TagRFP. Focal planes of PMs (Figure 1E) were acquired with the PerkinElmer UltraView spinning-disk system using a 100x oil immersion objective (NA = 1.45). Specific excitation and emission was performed using a 488 nm laser combined with a single band pass filter (500–550 nm) for GFP. Images shown are single-slice.
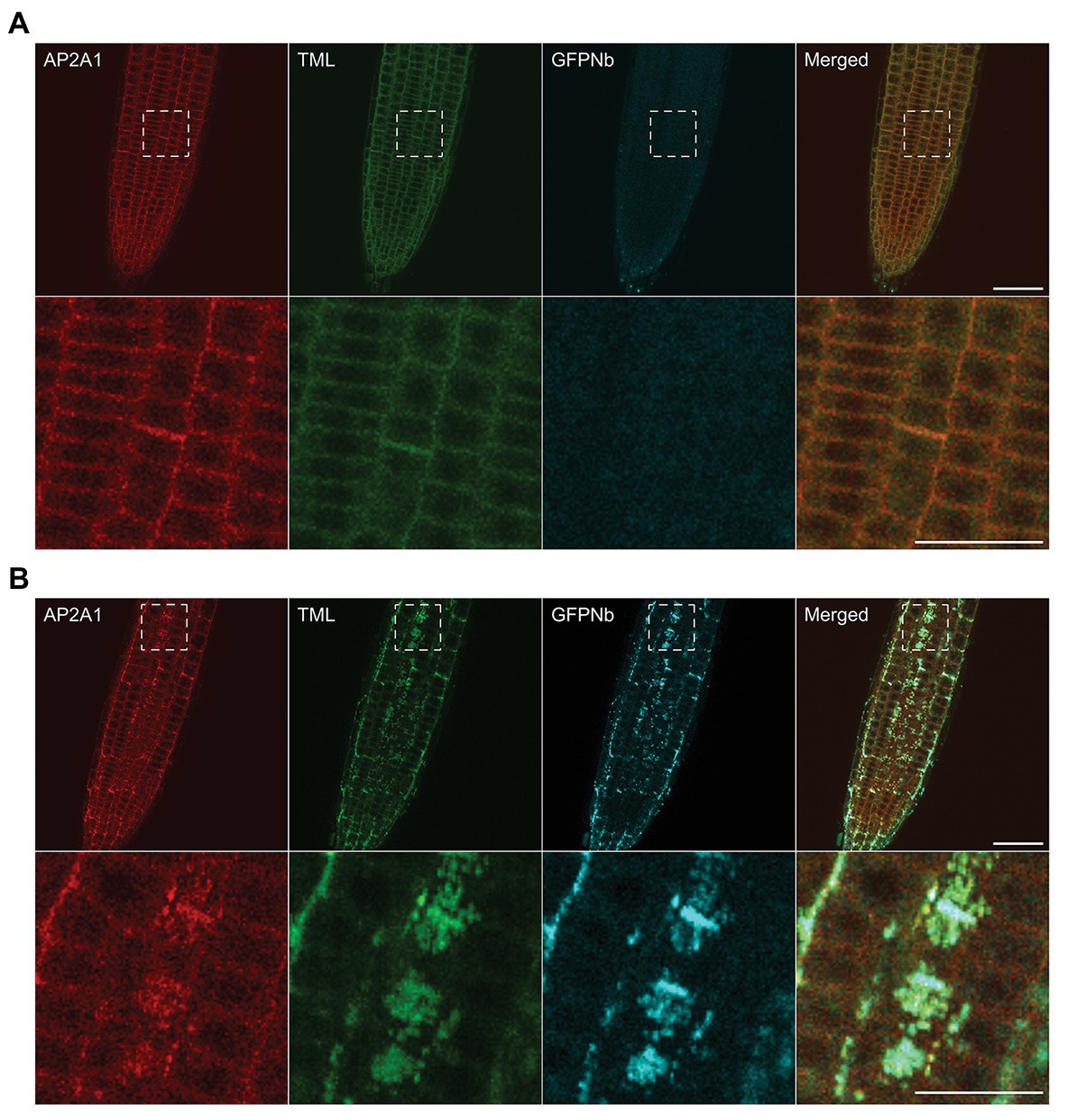
Figure 2. Delocalization of TML also affects the targeting of other endocytic players. (A,B) Representative images and blow-ups of the outlined regions of Arabidopsis roots expressing TML-GFP and AP2A1-TagRFP without (A) and with (B) MITOTagBFP2-GFPNb expression. GFPNb expression causes delocalization of both TML and AP2A1. Scale bars equal 20 μm (overview pictures) or 10 μm (blow-up pictures).
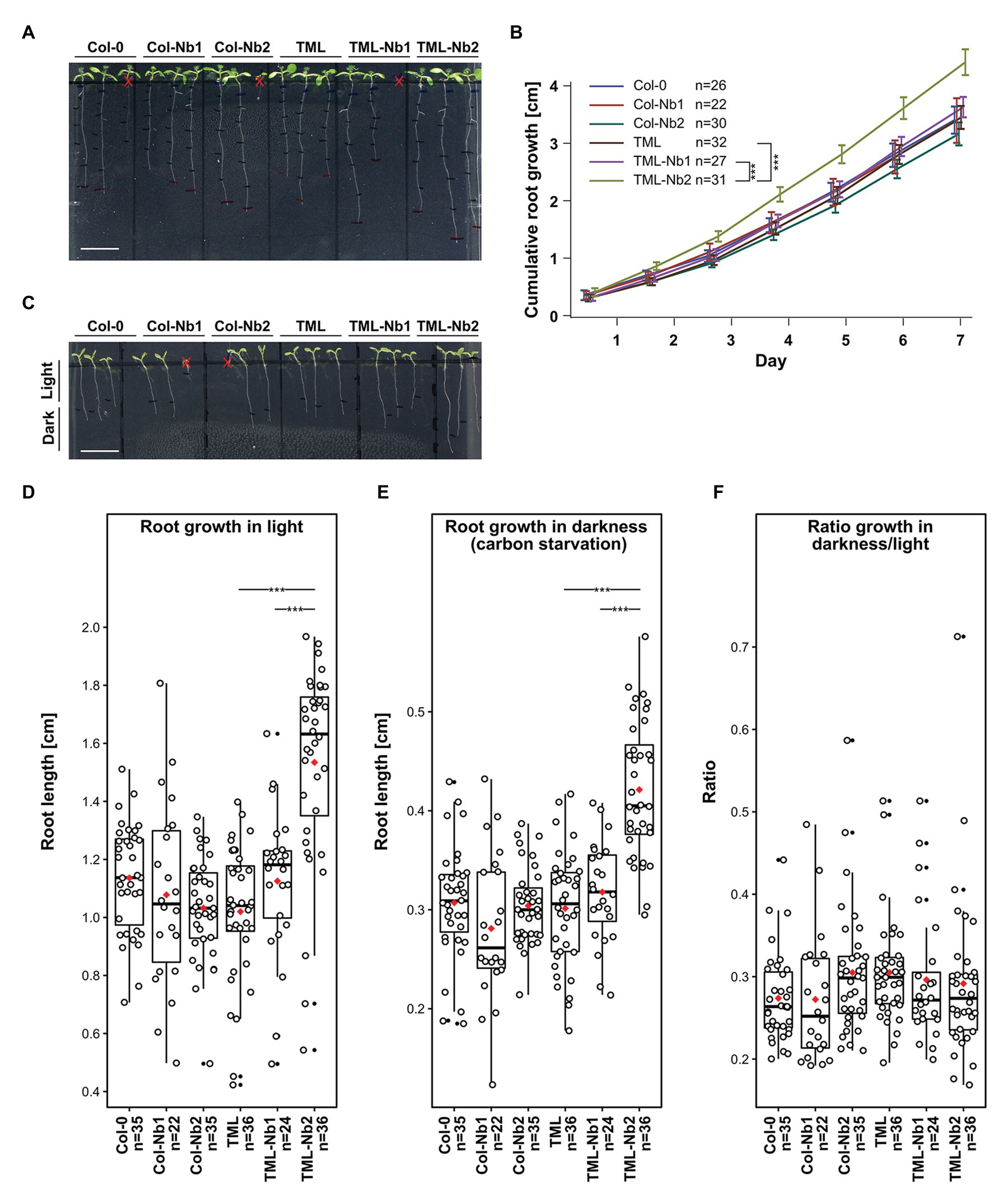
Figure 3. Delocalizing TML-GFP in root epidermal and cortical cells does not adversely affect root growth. Comparison of wild type seedlings (Col-0), wild type seedlings expressing MITOTagBFP2-GFPNb (Col-Nb1 and Col-Nb2) complemented tml-1(−/−) mutants expressing TML-GFP (TML) and complemented tml-1(−/−) mutants expressing TML-GFP and MITOTagBFP2-GFPNb (TML-Nb1 and TML-Nb2) in different light conditions. (A,B) Representative images of seedlings and quantification of root growth in continuous light. There are no statistical differences between the lines except TML-Nb2 which showed enhanced cumulative root growth. (C-F) Representative images of seedlings grown for 5 days in continuous light and subsequently for 7 days in continuous dark. Consistent with the cumulative root growth assay, the TML-Nb2 grew bigger compared to the TML-GFP and TML-Nb1 under both growth conditions (D,E). However, there were no significant differences in the analysis of the ratio of darkness/light growth (F). Scale bars in A and C equal 1 cm. Red crosses in A and C are marking seedlings that were excluded from the analysis due to delayed germination or due to roots growing in the agar. The measurements of growth in light, dark, as well as the respective dark/light ratio, are represented as jitter box plots. The black lines represent the median and the red diamonds represent the mean of the analyzed values. Each dot represents an individual cell and black dost refer to outliers. n refers to the total number of analyzed cells. Significant statistical differences in growth between the lines, based on Wilcoxon pairwise comparisons tests are indicated. p < 0.001 are represented as *** (Bonferroni adjusted p-values).
Results
The Targeting Sequence of Yeast Tom70p Targets the Nanobody to the Mitochondria
We expressed a nanobody directed against eGFP (GFPNb; Künzl et al., 2016), which we visualized by fusing it to TagBFP2. We targeted the fusion protein to the mitochondria using the import signal of the yeast mitochondrial outer membrane protein Tom70p as described before (Robinson et al., 2010). This targeting signal is functional in plants as constructs containing this signal colocalized with MitoTracker in N. benthamiana leaf epidermal cells (Winkler et al., 2021). To verify that the GFP nanobody did not interfere with the mitochondrial targeting, we expressed it under the control of the 35Sprom together with the red mitochondrial mt-rk marker (Nelson et al., 2007) in N. benthamiana abaxial leaf epidermal cells and visualized both fusion proteins using spinning disk confocal microscopy. Both constructs colocalized at discrete punctae, but not exclusively. We observed that the mt-rk marker did not localize to big aggregates labeled with the GFPNb (Figure 1A). The mt-rk marker line consists of yeast cytochrome c oxidase IV (Nelson et al., 2007). It has been shown that release of cytochrome c oxidase into the cytosol is associated with changes of mitochondrial integrity (Kadenbach et al., 2004). It is, therefore, possible that the observed signal from aggregates originates from mitochondrial clustering similarly to what we observed with our knocksideways in plants system (Winkler et al., 2021). The absence of the mt-rk fusion protein from those clusters might therefore be a consequence of altered fitness of the clustered mitochondria.
A Mitochondrially Targeted Nanobody Can Delocalize TML
TPLATE complex is a robust multi-subunit complex functioning at the PM and can be affinity purified using any of its subunits as bait (Gadeyne et al., 2014). In order to delocalize, and thereby inactivate TPC, we took advantage of the functionally complemented homozygous tml-1(−/−) mutant expressing TMLprom::TML-GFP (Gadeyne et al., 2014). In complemented tml-1(−/−) Arabidopsis roots, TML-GFP is recruited predominantly at the PM (Figure 1B, left). We introduced our MITO-TagBFP2-GFPNb nanobody into this background and used PIN2prom to drive expression of the construct. PIN2prom expresses in epidermis and cortex root cell files, which, with respect to future experiments, would allow us easily to perform confocal microscopy. Two independent lines, TML-Nb1 and TML-Nb2, were selected. MITO-TagBFP2-GFPNb (GFPNb) analogous to N. benthamiana leaves, localized to discrete punctae also in Arabidopsis wild type roots (Figure 1B, right).
Co-expression with GFPNb changed the uniform PM labeling of TML to a denser staining of discrete punctae in epidermis and cortex. Most of those were still near the PM and colocalized with the fluorescent signal from the nanobody, indicating effective delocalization of TML-GFP (Figure 1C and enhanced in Figure 1D). This delocalization was not apparent in the deeper layers of the root, where TML remained uniformly recruited to the PM (Figure 1C). Detailed analysis using spinning disk confocal microscopy confirmed the strong recruitment of TML to mitochondria that were present in the focal plane of the PM (Figure 1E, arrowheads). Next to the mitochondria, however, TML remained recruited to endocytic foci at the PM in root epidermal cells. The density of endocytic foci in epidermal root cells is very high (Dejonghe et al., 2016, 2019; Sánchez-Rodríguez et al., 2018). The density and intensity of the endocytic foci, marked by TML-GFP, appeared higher in epidermal cells in the complemented mutant (control) compared to the two independent lines expressing the GFPNb. Quantification showed a marked decrease in median signal intensity at the PM in the GFPNb expressing lines in regions devoid of mitochondria. The lower intensity of the signal also led to a statistically reduced number of foci (maxima) that could be detected. The reduced median intensity and lower amount of foci detected are in agreement with a substantial amount of TML-GFP accumulating at the mitochondria (Figures 1F,G).
Nanobody-Dependent Delocalization of TML Also Affects Other Endocytic Players
In plants, the heterotetrameric AP-2 complex and the octameric TPC are presumed to function largely, but not exclusively, together to execute CME (Gadeyne et al., 2014; Bashline et al., 2015; Wang et al., 2016; Adamowski et al., 2018). Both TPC and AP-2 have been shown to be involved in the internalization of cellulose synthase (CESA) complexes or the brassinosteroid receptor BRI1 for example (Bashline et al., 2013, 2015; Di Rubbo et al., 2013; Gadeyne et al., 2014; Sánchez-Rodríguez et al., 2018).
Moreover, a joint function is also suggested from proteomics analyses, which could identify subunits of both complexes when the AtEH1/Pan1 TPC subunit was used as bait in tandem-affinity purification assays (Gadeyne et al., 2014). To investigate whether our tool, aimed at delocalizing TPC, would also interfere with AP-2 recruitment at the PM, we tested the localization of AP-2 when TML was targeted to the mitochondria. To do so, we crossed our TML-GFP line, in tml-1(−/−) and expressing PIN2prom::MITOTagBFP2-GFPNb with the homozygous complemented tml-1(−/−) line, expressing TML-GFP as well as one of the large AP-2 subunits, AP2A1, fused to TagRFP (Gadeyne et al., 2014). Offspring plants that did not inherit the nanobody construct showed PM and cell plate recruitment of TML and AP2A1, and only background fluorescence in the TagBFP2 channel (Figure 2A). In the offspring plants that inherited the nanobody construct, however, the localization of the adaptor complex subunits changed. Both TML and AP2A1 accumulated at punctae, which clearly colocalized with the TagBFP2-fused nanobody construct (Figure 2B). The observed delocalization of AP2A1 to the mitochondria, together with TML strongly suggests that our approach has the capacity to delocalize TPC and AP-2 rather than TML alone, given that TPC and AP-2 are presumed to be linked via the AtEH1/Pan1 subunit (Gadeyne et al., 2014).
Mistargeting Adaptor Complexes in Epidermis and Cortex Affects Root Endocytic Uptake With Only Minor Effects on Root Growth
In contrast to AP-2, genetic interference with TPC subunits causes fully penetrant male sterility (Van Damme et al., 2006; Di Rubbo et al., 2013; Fan et al., 2013; Kim et al., 2013; Yamaoka et al., 2013; Gadeyne et al., 2014). TPC functionality, therefore, requires all subunits, and constitutive homozygous loss-of-function backgrounds are therefore non-existing. Abolishing endocytosis in plants, by silencing TPC subunits (Gadeyne et al., 2014) or overexpression of the uncaging proteins AUXILLIN-LIKE 1 or 2 (Adamowski et al., 2018) severely affects seedling development. The effect of silencing TPC subunits only indirectly affects protein levels and targeting clathrin might interfere with trafficking at endosomes besides the PM. As TPC and AP-2 only function at the PM, inactivating their function should not directly interfere with more downstream aspects of endosomal trafficking. Furthermore, by restricting the expression domain where the adaptor complex function is tuned down to the two outermost layers in the root should allow to study internalization from the PM, independently of possible indirect effects caused by the severe developmental alterations.
We evaluated the growth of several different lines expressing either GFPNb alone: Col-Nb1 and Col-Nb2, or GFPNb combined with TML-GFP in the complemented tml-1(−/−) mutant background: TML-Nb1 (−/−) and TML-Nb2 (−/−). At the seedling level, we did not observe any major adverse developmental effects (Figure 3A). Root length measurements of light grown seedlings revealed enhanced growth in the TML-Nb2 line compared to TML and TML-Nb1 (Figure 3B). The observed variability of this line, compared to all other lines that behaved similarly, probably results from a positional effect of the insertion. Nevertheless, our results show that nanobody expression in the PIN2prom domain and partial delocalization of TML has no negative effect on seedling development under normal growth conditions.
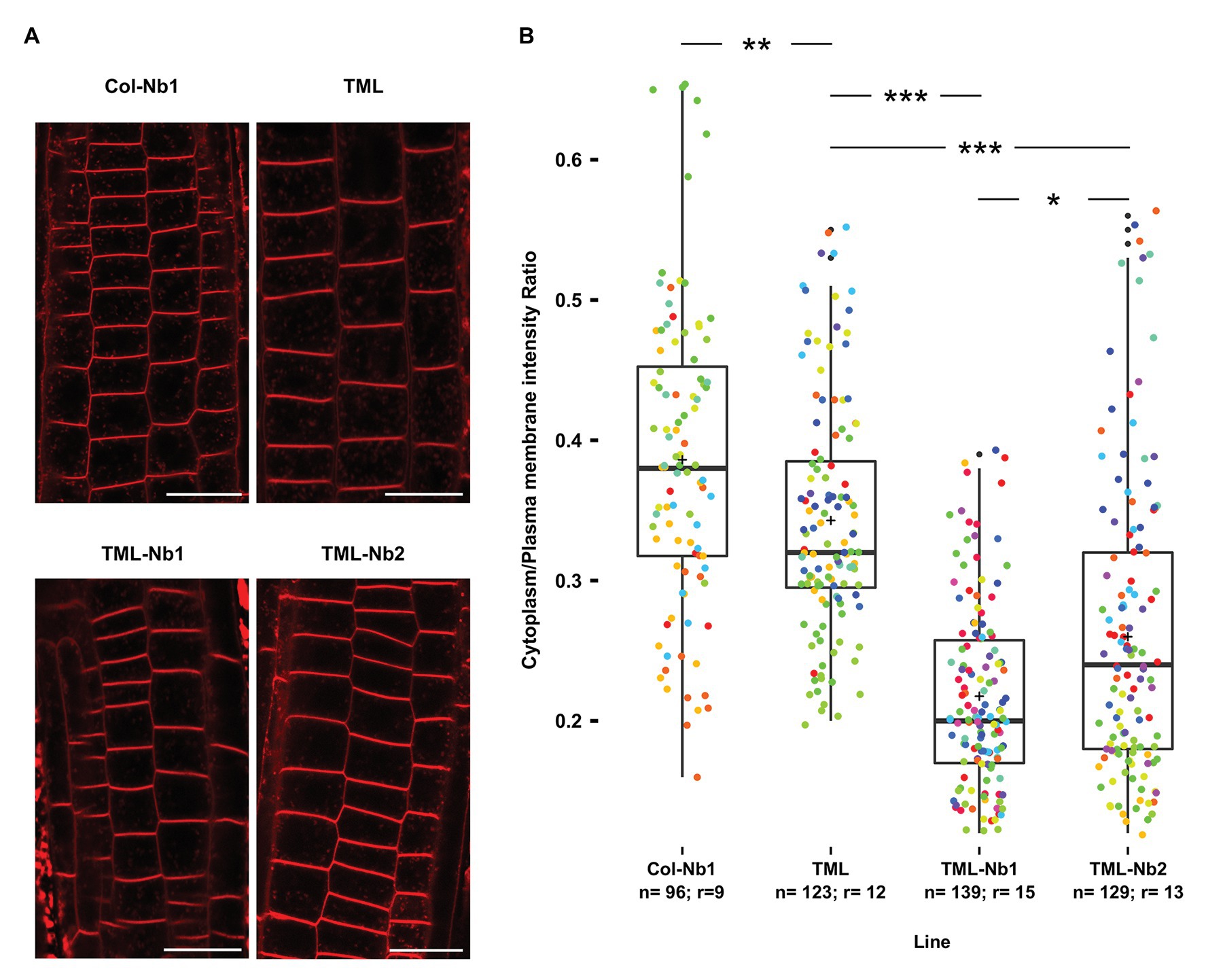
Figure 4. Nanobody-dependent delocalization reduces endocytic flux. (A) Representative single confocal slices of FM4-64 stained root cells of the different lines for which endocytic flux was quantified. FM4-64 uptake was compared between wild type Arabidopsis expressing MITOTagBFP2-GFPNb (Col-Nb1), the TML-GFP expressing complemented tml-1(−/−) mutant (TML), and two independent lines of the TML-GFP expressing complemented tml-1(−/−) mutant expressing MITOTagBFP2-GFPNb (TML-Nb1 and TML-Nb2). Scale bars equal 20 μm. (B) Jittered box plot representation of the quantification of the cytoplasm/plasma membrane intensity of FM4-64 as proxy for endocytic flux. The black lines represent the median and the crosses represent the mean values. The dots represent individual measurements of cells. The rainbow-colored indication of the dots groups the cells from the different roots that were analyzed. The number of cells (n) and the number of individual roots (r) are indicated in the graph. The indicated p-values were calculated using pairwise Wilcoxon tests and corrected using the Bonferroni method. Significant statistical differences between the lines based on Wilcoxon pairwise comparisons tests are indicated. p < 0.001 are represented as ***, < 0.01 are represented as ** and < 0.1 are represented as * (Bonferroni adjusted p-values).
The AtEH/Pan1 TPC subunits were recently implicated in growth under nutrient-depleted conditions as downregulation of AtEH1/Pan1 expression rendered plants hyper-susceptible to carbon starvation (Wang et al., 2019). We, therefore, assessed if delocalizing TML-GFP, as well as other endocytic players, would also render these plants susceptibility to nutrient stress. To do so, we measured root lengths of seedlings grown for 5 days in continuous light and afterwards we placed them in the dark for an additional 7 days. Measurements of root growth in the dark under carbon stress conditions did not show any differences between WT and Col-Nb lines. However, also here, the TML-Nb2 line exhibited increased root growth compared to TML and TML-Nb1 in both light and dark conditions (Figures 3C-E). We calculated the ratio of root growth in dark over root growth in light to avoid overestimation of the results due to the extraordinary growth of TML-Nb2 line. The ratios revealed that sequestering TML in TML-Nb lines did not cause any negative effect as the ratios were similar between all lines tested (Figure 3F). Overall, the effects of TML relocalization did not reveal any defects on seedling development, even under nutrient-stress conditions.
The subtle differences observed by comparing the effect of delocalization of TML on plant growth are likely a consequence of the restricted expression domain of GFPNb. We, therefore, monitored the effects of delocalizing TML more directly by visualizing the internalization of the styryl dye FM4-64, which in plants is commonly used as proxy for endocytic flux (Rigal et al., 2015; Jelínková et al., 2019). To rule out indirect effects of targeting GFPNb to the mitochondria, we compared endocytic flux between Col-Nb1, TML-GFP in tml-1(−/−), TML-Nb1 (−/−), and TML-Nb2 (−/−). We observed a slight decrease in endocytic flux when comparing wild type seedlings with the complemented tml-1(−/−) line and a strong reduction in endocytic flux between the complemented mutant and both complemented mutant lines where TML was partially delocalized (Figures 4A,B). Direct visualization of endocytic flux, therefore, allowed us to conclude that expression of the PIN2prom::MITOTagBFP2-GFPNb has the capacity to interfere with endocytosis in Arabidopsis root epidermal cells and that this tool certainly has the capacity to generate knockdown, and maybe even knockout lines at the protein level.
Discussion
Analyzing how impaired TPC function directly affects endocytosis is hampered by the male sterility and/or seedling lethal mutant phenotypes following genetic interference of individual subunits (Gadeyne et al., 2014). Here, we explored to impair TPC function at the protein level by delocalizing a functional and essential subunit in its respective complemented mutant background. We were inspired by previous work in animal cells. However, instead of using rapamycin-dependent rerouting of one of the large AP-2 subunits, combined with silencing the endogenous subunit (Robinson et al., 2010), we took advantage of the complemented tml-1(−/−) mutant line expressing TML-GFP (Gadeyne et al., 2014) in combination with targeting a nanobody directed against GFP (GFPNb; Künzl et al., 2016) to the mitochondria. We expressed the GFPNb in epidermis, cortex and lateral root cap as we expected ubiquitous constitutive expression to be lethal for the plant. Moreover, the epidermis and cortex cell files are easily accessible for imaging purposes. Proteins fused to this mitochondrial targeting signal colocalized with MitoTracker in transient N. benthamiana experiments (Winkler et al., 2021) and also here, we found our GFPNb to colocalize with the mt-rk mitochondrial marker in small punctae (Nelson et al., 2007). We also observed larger aggregates of signal, which we assume to be clustered dysfunctional mitochondria, similar to what we observed with our knocksideways strategy (Winkler et al., 2021). Constitutively decorating mitochondria with a GFPNb construct in the root epidermis and cortex cell files, therefore, might affect mitochondrial functionality without however causing a severe penalty on overall plant growth. The GFPNb system was capable of delocalizing TML-GFP and this caused the appearance of strongly fluorescent GFP-positive aggregations. Detailed inspection revealed however that our approach was insufficient to remove all TML from the PM. Compared to the control cells, sequestration of TML-GFP led to an overall reduction in signal intensity at the endocytic foci, as well as a reduction in overall density of endocytic foci when this was calculated as the amount of maxima that could be identified within a region of interest. The observed reduction of TML at the PM correlated with a significant reduction in endocytic tracer uptake, a proxy for reduced endocytosis.
The absence of major developmental defects, observed when TML-GFP was delocalized in the GFPNb lines can be explained by the fact that not all complex was delocalized as well as by the limited-expression domain of the PIN2prom. The increased root growth observed for the TML-Nb2 line is likely not linked to the delocalization of TML-GFP as we did not observe this in the TML-Nb1 line. An alternative explanation could be that the growth-promoting effect might possibly be a consequence of a positional effect of the insert. Inducible overexpression of AUXILIN-LIKE1/2 results in complete seedling growth arrest with drastic effects on cell morphology (Adamowski et al., 2018). The same holds true for inducible expression of dominant-negative clathrin HUB and DRP1A (Kitakura et al., 2011; Yoshinari et al., 2016). Furthermore, estradiol-inducible TPLATE and TML knockdown lines are noticeably shorter and show bulging cells (Gadeyne et al., 2014). As we did not observe cellular effects in epidermal or cortical cell files, we conclude that our approach lacked the required strength to block endocytosis, but only reduced it.
Recent results suggest that plant cells very likely contain a feedback loop controlling TPC expression, as carbon starved plants contained roughly the same amount of full-length TPLATE-GFP, next to an extensive amount of TPLATE-GFP degradation products (Wang et al., 2019). In case plant cells make more TPC upon depleting the complex at the PM, DeGradFP could provide a viable solution to this problem (Baudisch et al., 2018; Ma et al., 2019; Sorge et al., 2021). By applying this method in GFP-complemented tml-1(−/−) mutants, newly synthesized TML-GFP would be broken down immediately, preventing to achieve functional levels of TPC at the PM. Stronger or inducible promotors and/or the use of a different targeting location might also increase the delocalization capacity. To avoid lethality due to ubiquitous sequestration, engineered anti-GFP nanobodies, whose affinity can be controlled by small molecules, could also be used (Farrants et al., 2020).
Untangling the function of TPC and AP-2 in CME at the PM requires tools that allow interfering specifically with the functionality of both complexes. Our nanobody-dependent approach targeting TPC via TML resulted in the co-delocalization of one of the large subunits of AP-2, indicating that we likely are not only targeting TPC, but also AP-2 function. Whether a complementary approach, by delocalizing AP-2, using AP2S or AP2M in their respective complemented mutant backgrounds, would also delocalize TPC is something that would be worth trying. Furthermore, as AP2S and AP2M subunits are still recruited in ap2m and ap2s single mutant backgrounds (Wang et al., 2016), AP-2 in plants might also function as hemicomplexes similar to what is reported in C. elegans (Gu et al., 2013). Single mutants therefore might not reflect functional null ap2 mutants and a similar approach as performed here might also provide tools to inactivate AP-2 as a whole, which can be highly complementary to working with the single subunit mutants.
In conclusion, the data presented here is a first step toward the development of specific tools, which are required to help us understand the functions of AP-2 and TPC. In the long-term, this will generate insight into endocytosis at the mechanistic level and this will bring us closer to being able to modulate CME-dependent processes, and thereby modulating plant development, nutrient uptake as well as defense responses to our benefit.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
JW, ADM, EM, and PG designed and performed experiments. DVD designed experiments and wrote the initial draft together with ADM and JW. VS performed root growth assay statistical analysis. All authors contributed to the article and approved the submitted version.
Funding
Research in the Van Damme lab is supported by the European Research Council (T-Rex project number 682436 to DVD, JW, and ADM) and by the Research Foundation Flanders (FWO postdoctoral fellowship grant 1226420N to PG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the ENPER members for forming a vibrant and open research community for more than 20 years already. We would also like to thank Steffen Vanneste (PSB, VIB/UGent, Belgium) for providing research tools. This manuscript has been released as a pre-print at BioRxiv (Winkler et al., 2021).
References
Adamowski, M., Narasimhan, M., Kania, U., Glanc, M., De Jaeger, G., and Friml, J. (2018). A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30, 700–716. doi: 10.1105/tpc.17.00785
Backues, S. K., Korasick, D. A., Heese, A., and Bednarek, S. Y. (2010). The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22, 3218–3231. doi: 10.1105/tpc.110.077727
Bashline, L., Li, S., Anderson, C. T., Lei, L., and Gu, Y. (2013). The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a Clathrin-mediated endocytosis Adaptin. Plant Physiol. 163, 150–160. doi: 10.1104/pp.113.221234
Bashline, L., Li, S., Zhu, X., and Gu, Y. (2015). The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. 112, 12870–12875. doi: 10.1073/pnas.1509292112
Baudisch, B., Pfort, I., Sorge, E., and Conrad, U. (2018). Nanobody-directed specific degradation of proteins by the 26S-proteasome in plants. Front. Plant Sci. 9:130. doi: 10.3389/fpls.2018.00130
Bitsikas, V., Corrêa, I. R., and Nichols, B. J. (2014). Clathrin-independent pathways do not contribute significantly to endocytic flux. elife 3:e03970. doi: 10.7554/eLife.03970
Caussinus, E., Kanca, O., and Affolter, M. (2012). Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–122. doi: 10.1038/nsmb.2180
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dejonghe, W., Kuenen, S., Mylle, E., Vasileva, M., Keech, O., Viotti, C., et al. (2016). Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat. Commun. 7:11710. doi: 10.1038/ncomms11710
Dejonghe, W., Sharma, I., Denoo, B., De Munck, S., Lu, Q., Mishev, K., et al. (2019). Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 15, 641–649. doi: 10.1038/s41589-019-0262-1
Di Rubbo, S., Irani, N. G., Kim, S. Y., Xu, Z. -Y., Gadeyne, A., Dejonghe, W., et al. (2013). The Clathrin adaptor complex AP-2 mediates endocytosis of BRASSINOSTEROID INSENSITIVE1 in Arabidopsis. Plant Cell 25, 2986–2997. doi: 10.1105/tpc.113.114058
Dubeaux, G., Neveu, J., Zelazny, E., and Vert, G. (2018). Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69, 953.e5–964.e5. doi: 10.1016/j.molcel.2018.02.009
Fan, L., Hao, H., Xue, Y., Zhang, L., Song, K., Ding, Z., et al. (2013). Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140, 3826–3837. doi: 10.1242/dev.095711
Farrants, H., Tarnawski, M., Müller, T. G., Otsuka, S., Hiblot, J., Koch, B., et al. (2020). Chemogenetic control of nanobodies. Nat. Methods 17, 279–282. doi: 10.1038/s41592-020-0746-7
Früholz, S., Fäßler, F., Kolukisaoglu, Ü., and Pimpl, P. (2018). Nanobody-triggered lockdown of VSRs reveals ligand reloading in the Golgi. Nat. Commun. 9:643. doi: 10.1038/s41467-018-02909-6
Gadeyne, A., Sánchez-Rodríguez, C., Vanneste, S., Di Rubbo, S., Zauber, H., Vanneste, K., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704. doi: 10.1016/j.cell.2014.01.039
Gómez, B. G., Lozano-Durán, R., and Wolf, S. (2019). Phosphorylation-dependent routing of RLP44 towards brassinosteroid or phytosulfokine signalling. BioRxiv [Preprint]. doi: 10.1101/527754
Gu, M., Liu, Q., Watanabe, S., Sun, L., Hollopeter, G., Grant, B. D., et al. (2013). AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. elife 2:e00190. doi: 10.7554/eLife.00190
Hirst, J., Schlacht, A., Norcott, J. P., Traynor, D., Bloomfield, G., Antrobus, R., et al. (2014). Characterization of TSET, an ancient and widespread membrane trafficking complex. elife 3:e02866. doi: 10.7554/eLife.02866
Ingram, J. R., Schmidt, F. I., and Ploegh, H. L. (2018). Exploiting nanobodies’ singular traits. Annu. Rev. Immunol. 36, 695–715. doi: 10.1146/annurev-immunol-042617-053327
Irani, N. G., Di Rubbo, S., Mylle, E., Van Den Begin, J., Schneider-Pizoń, J., Hniliková, J., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589. doi: 10.1038/nchembio.958
Jelínková, A., Malínská, K., and Petrášek, J. (2019). Using FM dyes to study endomembranes and their dynamics in plants and cell suspensions. Methods Mol. Biol. 1992, 173–187. doi: 10.1007/978-1-4939-9469-4_11
Jelínková, A., Malínská, K., Simon, S., Kleine-Vehn, J., Pařezová, M., Pejchar, P., et al. (2010). Probing plant membranes with FM dyes: tracking, dragging or blocking? Plant J. 61, 883–892. doi: 10.1111/j.1365-313X.2009.04102.x
Kadenbach, B., Arnold, S., Lee, I., and Hüttemann, M. (2004). The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochim. Biophys. Acta 1655, 400–408. doi: 10.1016/j.bbabio.2003.06.005
Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10, 103–105. doi: 10.1016/j.tplants.2005.01.008
Kim, S. Y., Xu, Z. -Y., Song, K., Kim, D. H., Kang, H., Reichardt, I., et al. (2013). Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25, 2970–2985. doi: 10.1105/tpc.113.114264
Kitakura, S., Vanneste, S., Robert, S., Löfke, C., Teichmann, T., Tanaka, H., et al. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931. doi: 10.1105/tpc.111.083030
Künzl, F., Früholz, S., Fäßler, F., Li, B., and Pimpl, P. (2016). Receptor-mediated sorting of soluble vacuolar proteins ends at the trans-Golgi network/early endosome. Nat. Plants 2:16017. doi: 10.1038/nplants.2016.17
Li, X., and Pan, S. Q. (2017). Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci. Adv. 3:e1601528. doi: 10.1126/sciadv.1601528
Li, X., Wang, X., Yang, Y., Li, R., He, Q., Fang, X., et al. (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23, 3780–3797. doi: 10.1105/tpc.111.091454
Ma, Y., Miotk, A., Šutiković, Z., Ermakova, O., Wenzl, C., Medzihradszky, A., et al. (2019). WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 10:5093. doi: 10.1038/s41467-019-13074-9
Marquès-Bueno, M. M., Morao, A. K., Cayrel, A., Platre, M. P., Barberon, M., Caillieux, E., et al. (2016). A versatile multisite gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 85, 320–333. doi: 10.1111/tpj.13099
Martins, S., Dohmann, E. M. N., Cayrel, A., Johnson, A., Fischer, W., Pojer, F., et al. (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6:6151. doi: 10.1038/ncomms7151
Mbengue, M., Bourdais, G., Gervasi, F., Beck, M., Zhou, J., Spallek, T., et al. (2016). Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. 113, 11034–11039. doi: 10.1073/pnas.1606004113
Mitsunari, T., Nakatsu, F., Shioda, N., Love, P. E., Grinberg, A., Bonifacino, J. S., et al. (2005). Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25, 9318–9323. doi: 10.1128/MCB.25.21.9318-9323.2005
Muyldermans, S. (2013). Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797. doi: 10.1146/annurev-biochem-063011-092449
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Pasin, F., Kulasekaran, S., Natale, P., Simón-Mateo, C., and García, J. A. (2014). Rapid fluorescent reporter quantification by leaf disc analysis and its application in plant-virus studies. Plant Methods 10:22. doi: 10.1186/1746-4811-10-22
Rigal, A., Doyle, S. M., and Robert, S. (2015). Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods Mol. Biol. 1242, 93–103. doi: 10.1007/978-1-4939-1902-4_9
Robinson, M. S. (2015). Forty years of Clathrin-coated vesicles. Traffic 16, 1210–1238. doi: 10.1111/tra.12335
Robinson, M. S., Sahlender, D. A., and Foster, S. D. (2010). Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell 18, 324–331. doi: 10.1016/j.devcel.2009.12.015
Sánchez-Rodríguez, C., Shi, Y., Kesten, C., Zhang, D., Sancho-Andrés, G., Ivakov, A., et al. (2018). The cellulose synthases are cargo of the TPLATE adaptor complex. Mol. Plant 11, 346–349. doi: 10.1016/j.molp.2017.11.012
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sorge, E., Demidov, D., Lermontova, I., Houben, A., and Conrad, U. (2021). Engineered degradation of EYFP-tagged CENH3 via the 26S proteasome pathway in plants. PLoS One 16:e0247015. doi: 10.1371/journal.pone.0247015
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Traub, L. M. (2019). A nanobody-based molecular toolkit provides new mechanistic insight into clathrin-coat initiation. elife 8:e41768. doi: 10.7554/eLife.41768
Van Damme, D., Coutuer, S., De Rycke, R., Bouget, F. -Y., Inze, D., and Geelen, D. (2006). Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18, 3502–3518. doi: 10.1105/tpc.106.040923
Wang, C., Hu, T., Yan, X., Meng, T., Wang, Y., Wang, Q., et al. (2016). Differential regulation of Clathrin and its adaptor proteins during membrane recruitment for endocytosis. Plant Physiol. 171, 215–229. doi: 10.1104/pp.15.01716
Wang, P., Pleskot, R., Zang, J., Winkler, J., Wang, J., Yperman, K., et al. (2019). Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 10:5132. doi: 10.1038/s41467-019-12782-6
Wang, C., Yan, X., Chen, Q., Jiang, N., Fu, W., Ma, B., et al. (2013). Clathrin light chains regulate Clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25, 499–516. doi: 10.1105/tpc.112.108373
Wang, S., Yoshinari, A., Shimada, T., Hara-Nishimura, I., Mitani-Ueno, N., Feng Ma, J., et al. (2017). Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. Plant Cell 29, 824–842. doi: 10.1105/tpc.16.00825
Winkler, J., Mylle, E., De Meyer, A., Pavie, B., Merchie, J., Grones, P., et al. (2021). Visualizing protein–protein interactions in plants by rapamycin-dependent delocalization. Plant Cell doi: 10.1093/plcell/koab004
Yamaoka, S., Shimono, Y., Shirakawa, M., Fukao, Y., Kawase, T., Hatsugai, N., et al. (2013). Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25, 2958–2969. doi: 10.1105/tpc.113.114082
Yeung, B. G., Phan, H. L., and Payne, G. S. (2013). Adaptor complex-independent Clathrin function in yeast. Mol. Biol. Cell 10, 3643–3659. doi: 10.1091/mbc.10.11.3643
Yoshinari, A., Fujimoto, M., Ueda, T., Inada, N., Naito, S., and Takano, J. (2016). DRP1-dependent endocytosis is essential for polar localization and boron-induced degradation of the borate transporter BOR1 in Arabidopsis thaliana. Plant Cell Physiol. 57, 1985–2000. doi: 10.1093/pcp/pcw121
Yoshinari, A., Hosokawa, T., Amano, T., Beier, M. P., Kunieda, T., Shimada, T., et al. (2019). Polar localization of the borate exporter BOR1 requires AP2-dependent endocytosis. Plant Physiol. 179, 1569–1580. doi: 10.1104/pp.18.01017
Keywords: nanobody, endocytosis, Arabidopsis, protein delocalization, fluorescence microscopy, TPLATE complex (TPC)
Citation: Winkler J, De Meyer A, Mylle E, Storme V, Grones P and Van Damme D (2021) Nanobody-Dependent Delocalization of Endocytic Machinery in Arabidopsis Root Cells Dampens Their Internalization Capacity. Front. Plant Sci. 12:538580. doi: 10.3389/fpls.2021.538580
Edited by:
Erika Isono, University of Konstanz, GermanyReviewed by:
Takashi Ueda, Graduate University for Advanced Studies (Sokendai), JapanMatthieu Platre, Salk Institute for Biological Studies, United States
Copyright © 2021 Winkler, De Meyer, Mylle, Storme, Grones and Van Damme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniël Van Damme, ZGFuaWVsLnZhbmRhbW1lQHBzYi52aWItdWdlbnQuYmU=; ZGFkYW1AcHNiLnZpYi11Z2VudC5iZQ==
†These authors have contributed equally to this work
 Joanna Winkler
Joanna Winkler Andreas De Meyer
Andreas De Meyer Evelien Mylle
Evelien Mylle Veronique Storme
Veronique Storme Peter Grones
Peter Grones Daniël Van Damme
Daniël Van Damme