
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 01 March 2021
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.628366
This article is part of the Research Topic Nitrogen Use Efficiency and Sustainable Nitrogen Management in Crop Plants View all 23 articles
In agricultural cropping systems, relatively large amounts of nitrogen (N) are applied for plant growth and development, and to achieve high yields. However, with increasing N application, plant N use efficiency generally decreases, which results in losses of N into the environment and subsequently detrimental consequences for both ecosystems and human health. A strategy for reducing N input and environmental losses while maintaining or increasing plant performance is the development of crops that effectively obtain, distribute, and utilize the available N. Generally, N is acquired from the soil in the inorganic forms of nitrate or ammonium and assimilated in roots or leaves as amino acids. The amino acids may be used within the source organs, but they are also the principal N compounds transported from source to sink in support of metabolism and growth. N uptake, synthesis of amino acids, and their partitioning within sources and toward sinks, as well as N utilization within sinks represent potential bottlenecks in the effective use of N for vegetative and reproductive growth. This review addresses recent discoveries in N metabolism and transport and their relevance for improving N use efficiency under high and low N conditions.
Nitrogen (N) is essential for general plant functions as it is a component of amino acids, which are the elemental units of protein and enzymes. Amino acids are also precursors or N donors for many fundamental compounds including nucleic acids, hormones, chlorophyll, ureides, and other metabolites required for primary metabolism and specialized biological functions, respectively (Lam et al., 1996; Lea and Ireland, 1999; Epstein and Bloom, 2005; Zrenner et al., 2006; Züst and Agrawal, 2016). In addition, amino acids are the main long-distance transport forms of N in plants (Urquhart and Joy, 1982; Turgeon and Wolf, 2009; Patrick, 2013; Tegeder and Hammes, 2018). While amino acids, peptides, or even proteins may be taken up from the soil (Chapin et al., 1993; Näsholm et al., 2009; Tegeder and Rentsch, 2010), nitrate and ammonium are often preferred (Crawford and Glass, 1998; Loqué and von Wirén, 2004; Krapp et al., 2014). However, the inorganic N is only usable by plants when it is assimilated into amino acids, a process that primarily occurs in source organs such as the roots or leaves (Andrews et al., 1992; Lam et al., 1996; Miller et al., 2007; Xu et al., 2012; Krapp, 2015) but can also take place in sinks like seeds (Rochat and Boutin, 1992; Weber et al., 1998; Chen et al., 2020).
Plants that preferentially reduce N in roots move the newly produced amino acids through the xylem to source leaves (Miflin and Lea, 1977; Schobert and Komor, 1990). In addition, some xylem amino acids may be transferred to the phloem along the translocation pathway for immediate N supply to fast-growing sinks (Pate et al., 1975; van Bel, 1984; Zhang et al., 2010). However, in many plant species nitrate is allocated predominantly to source leaves as photosynthesis provides the carbon backbone and the energy for amino acid synthesis (Lam et al., 1996; Nunes-Nesi et al., 2010; Tegeder and Masclaux-Daubresse, 2018). In leaves, xylem-derived or leaf-synthesized amino acids may be used for metabolism, especially for photosynthesis (Wallsgrove et al., 1983; Schulze-Siebert et al., 1984), transiently stored as amino acids or proteins to be used for reproductive development (Millard, 1988; Staswick, 1994; Liu et al., 2005; Nunes-Nesi et al., 2010), or they are actively loaded into the phloem and transported to growing sinks (Urquhart and Joy, 1982; Atkins and Beevers, 1990; Tegeder and Masclaux-Daubresse, 2018). Once arrived in sink organs, the phloem unloading and movement of amino acids to sink cells takes place (Knoblauch et al., 2016; Milne et al., 2018; Tegeder and Hammes, 2018). Within the seed coat of seed sinks, amino acid catabolism, transamination, and re-assimilation events may occur (Atkins et al., 1975; Rainbird et al., 1984; Weber et al., 1995; Gallardo et al., 2007) followed by an efflux of amino acids into the seed apoplastic space and their subsequent uptake by the embryo for development, metabolism, and synthesis of seed storage compounds (Patrick, 1997; Offler et al., 2003; Ladwig et al., 2012; Tegeder et al., 2013).
Over the past decades, large amounts of N fertilizer have been applied to maximize crop yields while causing negative consequences for ecosystems and human health due to N leaching from the soil or degradation and subsequently releasing into the atmosphere (Matson et al., 1997; Fowler et al., 2013; Kopittke et al., 2019). In addition, crop plants are often not efficient in acquiring and using the supplied N and may take up less than 50% of the N fertilizer (Raun and Johnson, 1999; Zhu et al., 2016). Consequently, increasing N fertilization does not necessarily result in a proportional increase in yield (Ju et al., 2004; Mueller et al., 2014; Zhu et al., 2016). Therefore, improvements in yield and N use efficiency (NUE) in conjunction with a reduction in N application and N losses into the environment are pressing goals for a sustainable agriculture (Zhang et al., 2015a; Anas et al., 2020). However, progress in NUE is an ambitious target as crop yields are influenced by numerous factors, including genetic parameters and their variation among and within species (G), environmental effects such as location, soil conditions, and climate (E), and agronomic technologies and management practices (M; e.g., type, timing, amount, and place of N application or weed and pest control) (Hatfield and Walthall, 2015; Nguyen et al., 2017; Martinez-Feria et al., 2018; Nguyen and Kant, 2018; Gramma et al., 2020; Jensen et al., 2020; Lemaire and Ciampitti, 2020; Plett et al., 2020). Only the combination or interaction of these factors (G × E × M) will allow for sustainable and secure crop production (Swarbreck et al., 2019; Cooper et al., 2020; Hawkesford and Riche, 2020; Peng et al., 2020).
At crop level, N use efficiency is defined as seed yield relative to N availability (Moll et al., 1982). It is comprised of N uptake efficiency (NUpE) or the proportion of N in the shoot relative to the N supply, and N utilization efficiency (NUtE), which describes the amount of shoot N used for seed production. Many genetic factors contribute to plant biomass production, yield, and NUE including those responsible for plant structure and architecture (Gifford et al., 2008; Xu et al., 2012; Li et al., 2015; Xie et al., 2017; Luo et al., 2020), as well as N root uptake, (re)assimilation, remobilization, partitioning, utilization, and signaling processes (Coque and Gallais, 2006; Chardon et al., 2010; McAllister et al., 2012; Han et al., 2015; Zhao et al., 2018; Griffiths and York, 2020). Numerous genes related to these developmental and physiological processes have been identified and their importance for plant growth, function, and seed yield has been demonstrated. We would like to refer the readers to some recent reviews that provide comprehensive overviews on the discoveries (Miller et al., 2007, 2008; Xu et al., 2012; Han et al., 2015; Mandal et al., 2018; Tegeder and Masclaux-Daubresse, 2018; Wang et al., 2018a; Raghuram and Sharma, 2019; Chen et al., 2020; Fernie et al., 2020; Fichtner et al., 2020; Li et al., 2020; Vidal et al., 2020). The current work will focus on new findings in N metabolism and transport, along with their associated genes and proteins, and analyze their role in improving NUE under high and low N conditions.
Root uptake of inorganic N forms often dominates and requires the function of nitrate and ammonium transporters, respectively. Nitrate uptake is achieved by transporters of the NRT1 and NRT2 family, respectively (Williams and Miller, 2001; Tsay et al., 2007; Krapp et al., 2014; Fan et al., 2017). Studies on natural variation and genetic manipulation of NRT1 or NRT2 expression have resolved that NRT function in roots influences N acquisition, plant growth, and seed development (Huang et al., 1999; Cerezo et al., 2001; Remans et al., 2006; Li et al., 2007; Hu et al., 2015; Chen et al., 2016, 2017; Wang et al., 2020). To date, successes in improving NUE under both high and low N have mainly been achieved when NRT transporters were constitutively overexpressed (Fang et al., 2013; Fan et al., 2016a,b; Feng et al., 2017; c.f. Wang et al., 2018a) or expressed in both root and leaf tissues (Figures 1, 2; Hu et al., 2015; Chen et al., 2016, 2017; Wang et al., 2018b). While it remains to be determined if root-specific overexpression of NRTs may be sufficient for enhancing N uptake, research suggests that downstream nitrate transport processes may additionally or dominantly contribute to the observed increases in NUE. Indeed, recent work in Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and rice (Oryza sativa) showed that overexpression of the hyperactive chimeric nitrate transporter AtNC4N in the phloem of old leaves results in improved NUE under high and low N status (Chen et al., 2020). This study also supports the fact that during leaf senescence, besides amino acids and peptides (Winter et al., 1992; Feller and Fischer, 1994; Masclaux et al., 2000), nitrate recycling from the vacuole and subsequent source-to-sink partitioning contribute to sink N supply. In line with this, increased expression of the vacuolar OsNRT1.1A/OsNPF6.3 nitrate transporter either throughout rice plants, or in the epidermis and vascular tissues of roots and parenchyma cells of culms and leaf sheaths resulted in increased grain yield and NUE under high and low N supply, due to improved N utilization for grain development (Wang et al., 2018b). As nitrate transport, related N assimilation, and amino acid partitioning processes are highly intertwined (Tegeder and Rentsch, 2010; Tegeder, 2012, 2014; Tegeder and Masclaux-Daubresse, 2018), it would be interesting to examine if in AtNC4N and OsNRT1.1A overexpressors co-regulated by changes in amino acid pools and transport also add to the observed increases in seed yield, seed N content, and NUE (Wang et al., 2018b; Chen et al., 2020).
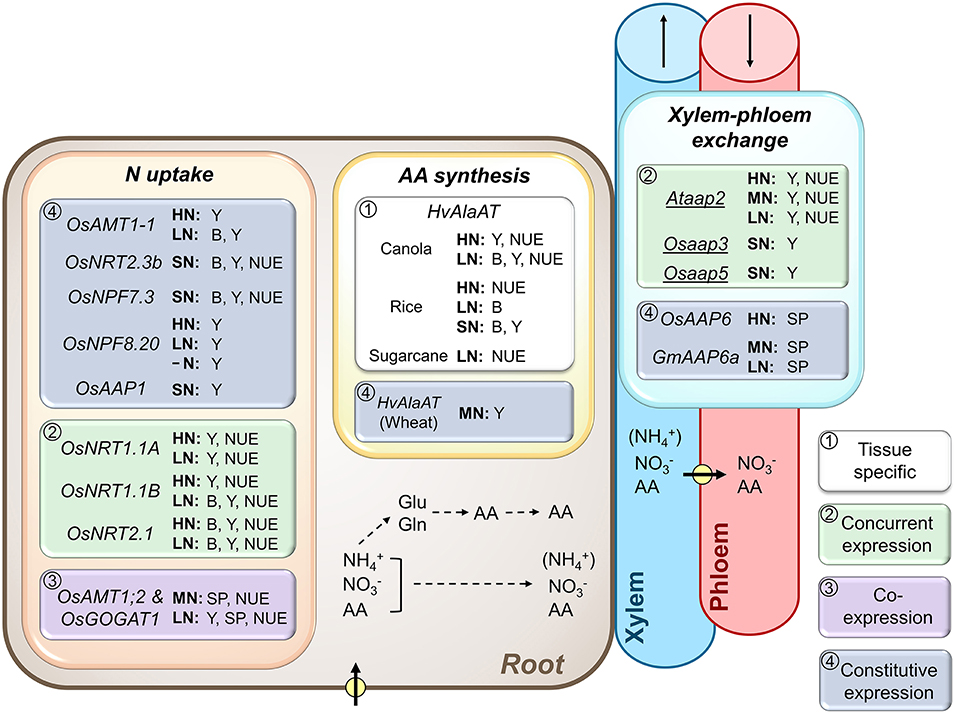
Figure 1. Importance of root N transport and metabolic processes, and xylem-phloem exchange along the transport pathway for plant biomass and yield production, and N use efficiency. Shown are ammonium (), nitrate (), and amino acid (AA) transporters, and AA synthesis genes that were successfully overexpressed (UPPERCASE) or repressed/knocked out (lowercase and underlined) and resulted in increased biomass (B), seed yield (Y), seed protein (SP), and/or plant N use efficiency (NUE) at varying N supply. As indicated by framed white, green, purple, or blue backgrounds, the transgenic strategies used either (1) tissue-specific gene manipulation, (2) concurrent expression of a particular gene in both root and shoot tissues (OsNRT1.1A, Wang et al., 2018b; OsNRT1.1B, Hu et al., 2015; OsNRT2.1, Chen et al., 2016), (3) co-expression of two genes in root and shoot tissues (OsAMT1;2 and OsGOGAT1, Lee et al., 2020a), or (4) constitutive gene expression throughout the plant. Positive results were achieved with (a) native expression of ammonium transporters (OsAMT1-1, Ranathunge et al., 2014; OsAMT1;2, Lee et al., 2020a), nitrate transporters (OsNRT1.1A, Wang et al., 2018b; OsNRT1.1B, Hu et al., 2015; OsNRT2.1, Chen et al., 2016; OsNRT2.3b, Fan et al., 2016b; OsNPF7.3, Fang et al., 2017; OsNPF8.20, Fang et al., 2013), and amino acid transporters (OsAAP1, Ji et al., 2020; OsAAP6, Peng et al., 2014; GmAAP6a, Liu et al., 2020) in rice (Os), Arabidopsis (At), and soybean (Gm), (b) expression of a barley alanine aminotransferase (HvAlaAT) in canola (Good et al., 2007), rice (Shrawat et al., 2008; Beatty et al., 2013), sugarcane (Snyman et al., 2015), and wheat (Peña et al., 2017), and (c) by knocking out native/endogenous genes (Ataap2, Perchlik and Tegeder, 2018; Osaap3, Lu et al., 2018; Osaap5, Wang et al., 2019). Plants were grown under high (HN), moderate (MN), sufficient (SN; N supply was not specified) and low N (LN), or without N (-N). Pathways of N uptake and xylem-phloem transfer (arrows with circle), as well as N assimilation into glutamate (Glu) and glutamine (Gln), and the subsequent synthesis of other AA and their loading into the xylem are indicated. may be transported in the xylem to the shoot, but in relatively low amounts (indicated by brackets). Arrows in xylem and phloem indicate the direction of translocation. For additional information see manuscript text.
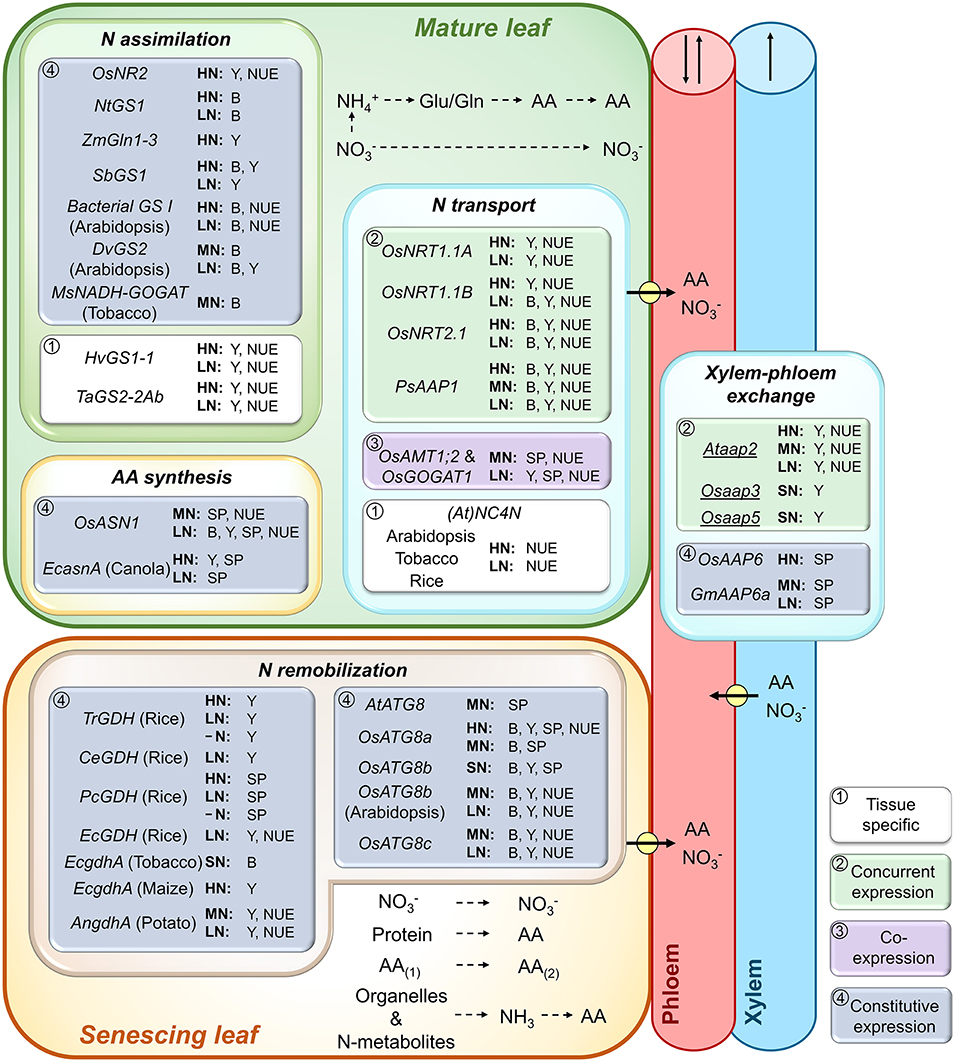
Figure 2. Role of leaf N (re)assimilation and remobilization, amino acid (AA) synthesis and N transport processes in plant biomass, yield production, and N use efficiency. Genes expressed in mature, photosynthetically active leaves, and senescing leaves, respectively, were analyzed. Shown are genes that were successfully overexpressed (UPPERCASE) or repressed/knocked out (lowercase and underlined) and resulted in increased biomass (B), seed yield (Y), seed protein (SP), and/or plant N use efficiency (NUE) at varying N supply. As indicated by framed white, green, purple, or blue backgrounds, the transgenic strategies either used (1) tissue-specific gene manipulation, (2) concurrent expression of a particular gene in different source or sink tissues, (3) co-expression of two genes in root and shoot tissues (OsAMT1;2 and OsGOGAT1, Lee et al., 2020a), or (4) constitutive gene expression throughout the plant. Plants were grown under high (HN), moderate (MN), sufficient (SN; N supply was not specified), or low N (LN). For native/endogenous gene expression only gene names are provided, while the expression of non-native genes is indicated by the gene name and the common name of the transgenic plant species. Positive results were achieved for genes of a nitrate reductase (OsNR2, Gao et al., 2019b), glutamine synthetases (NtGS1, Oliveira et al., 2002; ZmGln1-3, Martin et al., 2006; SbGS1, Urriola and Rathore, 2015; bacterial GS I, Zhu et al., 2015; DvGS2, Zhu et al., 2014), glutamate synthases (MsNADH-GOGAT, Chichkova et al., 2001; OsGOGAT1, Lee et al., 2020a), asparagine synthetases (OsASN1, Lee et al., 2020b; E. coli EcasnA, Seiffert et al., 2004), nitrate (OsNRT1.1A, Wang et al., 2018b; (At)NC4N, a synthetic gene construct based on Arabidopsis gene sequences, Chen et al., 2020), and amino acid transporters (PsAAP1, Perchlik and Tegeder, 2017) as well as for glutamate dehydrogenases (Trichurus sp. TrGDH, Du et al., 2019; C. ehrenbergii CeGDH, Zhou et al., 2015b; P. cystidiosus PcGDH, Zhou et al., 2014; E. cheralieri EcGDH, Tang et al., 2018; E. coli EcgdhA, Ameziane et al., 2000; Lightfoot et al., 2007; Aspergillus nidulans AngdhA, Egami et al., 2012) and autophagy-related proteins (AtATG8, Chen et al., 2019; OsATG8a, Yu et al., 2019; OsATG8b, Zhen et al., 2019a, Fan et al., 2020; OsATG8c, Zhen et al., 2019b). Pathways of N (re)assimilation, AA synthesis and conversion (AA(1) to AA(2)), organelle and N metabolite degradation, as well as N phloem loading and xylem-phloem exchange (arrows with circle) are shown. Arrows in xylem and phloem indicate possible directions of translocation. For additional information see manuscript text.
Uptake of ammonium is regulated by ammonium transporters (AMTs) as well as aquaporins or cation channels (Glass et al., 2002; Sonoda et al., 2003; Jahn et al., 2004; Loqué et al., 2005; Guo et al., 2007; Bárzana et al., 2014). Although overexpression of OsAMT1;1 in rice under the control of a ubiquitin promoter resulted in increased seed yield under suboptimal and optimal N conditions (Figure 1; Ranathunge et al., 2014), numerous studies on genetic manipulation of ammonium uptake had rather limited success (Meister et al., 2014; Li et al., 2017; Zhang et al., 2020a). For example, approaches using CaMV-35S promoter-OsAMT1;1 or OsAMT1;3 constructs in rice resulted in increased ammonium acquisition but had no effect on plant performance (Kumar et al., 2006) or even showed a decrease in biomass and seed yield (Bao et al., 2015). Generally, the use of ammonium transporters as targets for improving N uptake and utilization may be problematic since changes in cellular ammonium pools or excess ammonium can be toxic for the plant cell (Britto and Kronzucker, 2002; Bittsánszky et al., 2015). Nevertheless, detrimental effects of ammonium can potentially be avoided if downstream N assimilation keeps up with its increased uptake. This is supported by a study in rice using activation tagging lines, where OsAMT1;2 and OsGOGAT1 genes were simultaneously overexpressed. The modified plants showed an increased grain yield under N-limited growth conditions and improved grain N as well as protein content under both limiting and sufficient N supply (Figures 1, 2; Lee et al., 2020a). Overall, the activation tagging lines were more efficient in N uptake, remobilization, and utilization for sink development and/or N nutrition, clearly presenting an encouraging strategy for improving NUE in rice and most probably other plant species.
Although the contribution of organic vs. inorganic N acquisition to total root N uptake has not been resolved (Näsholm et al., 2009), it has been demonstrated that plants are highly capable in acquiring organic N, and especially amino acids, via root-localized transporters (Komarova et al., 2008; Paungfoo-Lonhienne et al., 2008; Tegeder and Masclaux-Daubresse, 2018). Such uptake systems are probably of special relevance when amino acid levels are relatively high as observed in soils of organic farming or other cropping systems that rely on manure or compost for N nutrition (Jones et al., 2005a,b; Moran-Zuloaga et al., 2015; Enggrob et al., 2019) or when conservation agriculture is used (Li et al., 2018; Alam et al., 2020). Indeed, it has been suggested that a high proportion of fertilized organic matter compared to mineral N fertilizer results in higher crop N uptake, grain yields, and NUE, with the additional benefit of reduced N losses into the environment (Xia et al., 2017).
Root importers for amino acids include amino acid permeases AAP1 and AAP5, proline transporter ProT2, and lysine-histidine-type transporters LHT1 and LHT6 (Grallath et al., 2005; Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2007, 2008, 2011; Lehmann et al., 2011; Perchlik et al., 2014; Ganeteg et al., 2017; Wang et al., 2019; Guo et al., 2020a,b), and their importance for plant growth has been demonstrated (Tegeder and Masclaux-Daubresse, 2018; Guo et al., 2020a). In rice, constitutive overexpression of OsAAP1 led to increased N uptake and reallocation, positively affecting tiller number and final grain yield (Figure 1; Ji et al., 2020). Nevertheless, it remains to be examined if and how root-specific overexpression of OsAAP1 (or other amino acid transporters) influences N uptake and NUE, including under varying soil amino acid concentrations.
For root-to-shoot translocation, amino acids taken up from the soil or synthesized in roots are exported from root endodermal cells, the pericycle, or vascular parenchyma into the apoplast (Tegeder, 2014) via usually multiple acids move in and out transporters (UmamiTs; Ladwig et al., 2012; Müller et al., 2015; Besnard et al., 2016) and subsequently loaded into the xylem. Most of the xylem amino acids are then moved to photosynthetically active, transpiring leaves (Miflin and Lea, 1977; Schobert and Komor, 1990) followed by their import into the mesophyll cells, which in Arabidopsis is mediated by LHT1 (Hirner et al., 2006; Liu et al., 2010; Svennerstam et al., 2011). It has not yet been analyzed if and how transporter function in root amino acid export/xylem loading and import into leaf cells affects N uptake and allocation to the shoot, and NUE.
Not all amino acids that are transported out of the root are moved to leaves. Especially during the vegetative phase, up to 21% of the organic N may be retrieved from the transpiration stream along the pathway (van Bel, 1984) for metabolism (Bailey and Leegood, 2016), establishment of N storage pools (Streeter, 1979; Millard, 1988), or xylem-to-phloem transfer to directly supply growing sinks with N (Pate et al., 1975; van Bel, 1984, 1990; Dickson et al., 1985). In Arabidopsis, transporters of the AAP family have been shown to be involved in the removal of amino acids from the xylem (i.e., AtAAP6; Hunt et al., 2010) and their loading into the phloem (i.e., AtAAP2, AtAAP3, and potentially AtAAP5; Okumoto et al., 2004; Zhang et al., 2010; Tegeder and Ward, 2012; Perchlik and Tegeder, 2018). GmAAP6a and OsAAP6 have been overexpressed in soybean and rice, respectively, using the CaMV-35S promoter (Figures 1, 2; Peng et al., 2014; Liu et al., 2020). In the transgenic soybean plants, total seed N as well as protein content were improved under sufficient and low N conditions, respectively. Similarly, the rice overexpressors showed improvement in grain protein content under relatively high N supply (Peng et al., 2014). When knocking out AtAAP2, which functions in the xylem-phloem exchange of amino acids in Arabidopsis, more N was allocated to leaves resulting in increased photosynthetic N use efficiency [pNUE, i.e., the ratio of CO2 fixation to organic N per leaf area (Poorter and Evans, 1998)], carbon fixation, and sucrose transport from source-to-sink independent of the N supply (Perchlik and Tegeder, 2018). The Ataap2 mutants displayed increased growth, seed yield, and seed carbon storage pools under high, limiting, and highly deficient N supply (Figures 1, 2). NUE efficiency was improved due to increased NUpE, while NUtE was not changed leading to a decrease in N content in individual seeds. In these Ataap2 plants, increased amounts of N were trapped in the straw/stubble tissue, indicating that remobilization was not adjusted and presented a bottleneck for seed storage protein accumulation (Perchlik and Tegeder, 2018). Similar to the Arabidopsis AAPs, both rice OsAAP3 and OsAAP5 seem to be, at least to some extent, expressed in the vasculature and may function in amino acid exchange between the xylem and phloem (Lu et al., 2018; Wang et al., 2019). Indeed, rice OsAAP3 and OsAAP5 RNAi lines also showed significant improvements under tested sufficient N conditions, including increases in tiller number and grain yield (Figures 1, 2).
Following uptake, nitrate is reduced in the cytosol to nitrite which is transported into the plastid for further reduction to ammonium (Lam et al., 1996; Tegeder and Rentsch, 2010; Liu et al., 2015). The two-step reduction is achieved by nitrate reductase (NR) and nitrite reductase (NiR), respectively. NR catalyzes the main regulatory step in the N assimilation process and its activity is highly regulated by factors such as nitrate, light, or water availability (Beevers and Hageman, 1969; Djennane et al., 2002; Chamizo-Ampudia et al., 2017; Mauceri et al., 2020). In addition, amino acids (e.g., glutamine) present signals regulating uptake and assimilation by affecting both the activity of nitrate transporters (Rawat et al., 1999; Vidmar et al., 2000; Thornton, 2004; Miller et al., 2008) and NR (Dzuibany et al., 1998; Migge et al., 2000a; Fan et al., 2006). Overexpression of NR or NiR has often resulted in increased N uptake, however improvements in seed yield, seed protein level, or NUE have generally not been achieved, potentially due to negative feedback of N-containing metabolites and related transcriptional or post-transcriptional regulation (Crawford, 1995; Crété et al., 1997; Good et al., 2004; Li et al., 2020). Nevertheless, recent research in japonica rice showed that constitutive expression of indica OsNR2, which encodes a NADH/NADPH-dependent NR, confers increased tiller number, grain yield, and NUE at high N supply (Figure 2; Gao et al., 2019a). These effects were further enhanced when indica OsNR2 and OsNRT1.1B were concurrently expressed, indicating that OsNR2 positively regulates OsNRT1.1B and thereby regulates nitrate uptake (Gao et al., 2019a).
Ammonium is assimilated through the coordinated activity of glutamine synthetase (GS) and glutamate synthase (GOGAT) into glutamine and glutamate, respectively, which serve as N donors for the biosynthesis of other amino acids and amides (Chichkova et al., 2001; Brauer et al., 2011; Lea and Miflin, 2011). Two isoforms exist for GS, the cytosolic GS1 and plastidic GS2, which have distinct functions in N assimilation and are encoded by multigenic or single gene families (Miflin and Habash, 2002; Chardon et al., 2012). GS2 is involved in primary N assimilation (i.e., de novo amino acid synthesis) as well as re-assimilation of ammonium produced during photorespiration (Cren and Hirel, 1999; Ferreira et al., 2019). GS1 isoforms are mainly involved in N re-assimilation and remobilization, with some exceptions such as AtGLN1;2 in Arabidopsis, which is important for N assimilation under high nitrate supply (Lothier et al., 2011), and OsGS1;1 involved in ammonium assimilation in rice shoots and roots (Kusano et al., 2011). Due to their fundamental role in N metabolism and to improve plant growth and seed yield, GS1 and GS2 genes have been overexpressed in numerous species, including in tobacco (Migge et al., 2000b; Fuentes et al., 2001; Oliveira et al., 2002; Wang et al., 2013a), Arabidopsis (Zhu et al., 2014; Hu et al., 2015), Sorghum bicolor (sorghum; Urriola and Rathore, 2015), Zea mays (maize; Martin et al., 2006), Triticum aestivum (wheat; Habash et al., 2001; Hu et al., 2018), Hordeum vulgare (barley; Gao et al., 2019b), and rice (Cai et al., 2009; Brauer et al., 2011; Bao et al., 2014). While in some cases no or negative effects on plant performance have been reported (Migge et al., 2000b; Cai et al., 2009; Brauer et al., 2011; Bao et al., 2014; c.f. Thomsen et al., 2014; Garnett et al., 2015; Sweetlove et al., 2017), other studies have shown improvements in seed yield, NUE, NUpE, or NUtE for both GS1 and GS2, and repeatedly at high and low N supply (Figure 2; Oliveira et al., 2002; Martin et al., 2006; Brauer et al., 2011; Zhu et al., 2014; Hu et al., 2015, 2018; Urriola and Rathore, 2015; Gao et al., 2019b). For example, increased expression of HvGS1-1 under the control of its own promoter in barley led to improved grain yields and NUE under high and low N conditions (Gao et al., 2019b). Zhu et al. (2015) constitutively expressed bacterial GS I genes from Klebsiella sp. D1-5 and Lactococcus sp. Zjy3, respectively, in Arabidopsis and showed, for both genes, improved plant growth and NUE at low and sufficient N. Similarly, overexpression of TaGS2-2Ab in wheat using its endogenous promoter resulted in higher seed yield and NUE when N supply was high and low, due to increased N uptake and remobilization (Hu et al., 2018).
Compared to GS, relatively few studies have looked at altering the expression of genes encoding for plastid-localized ferredoxin-dependent (Fd-GOGAT) or NADH-dependent (NADH-GOGAT) GOGAT (Good et al., 2004; Xu et al., 2012). While Fd-GOGAT is primarily involved in the assimilation of ammonium that originates either from nitrate reduction or leaf photorespiration, NADH-GOGAT is highly expressed in non-photosynthesizing cells and has a distinct function in non-photorespiratory ammonium reduction (Somerville and Ogren, 1980; Lancien et al., 2002; Lee et al., 2020). Using Arabidopsis, barley, rice, or Medicago sativa (alfalfa) mutants, essential roles in plant growth and seed development have been demonstrated for both Fd-GOGAT and NADH-GOGAT (Somerville and Ogren, 1980; Kendall et al., 1986; Schoenbeck et al., 2000; Lancien et al., 2002; Potel et al., 2009; Tamura et al., 2010; Yang et al., 2016; Zeng et al., 2017). However, relatively few studies manipulated GOGAT expression to enhance seed yield and NUE, and those focused on NADH-GOGAT with rather moderate outcomes. For instance, OsNADH-GOGAT overexpression in rice using its endogenous promoter only resulted in increased grain weight at low N fertilization (Yamaya et al., 2002). Further, constitutive expression of MsNADH-GOGAT showed higher biomass production in tobacco when N was sufficient (Figure 2; Chichkova et al., 2001), whereas in maize, overexpression of ZmNADH-GOGAT resulted in a lower shoot biomass and no changes in kernel yield (Cañas et al., 2020). Interestingly, and as pointed out above, combined overexpression of OsAMT1;2 and OsNADH-GOGAT1 in rice improved NUE at both sufficient and low N conditions (Figure 2). At sufficient N, the transgenic lines achieved higher seed protein levels without altering seed yield, while at low N both seed protein and yield were improved (Lee et al., 2020a). The results suggest that by increasing both N uptake and N assimilation, negative intrinsic effects that might happen when only AMT (see above) or GOGAT is overexpressed (e.g., through unbalanced pools of N metabolites) may be avoided (Lee et al., 2020a).
Relatively few studies have described improvements in NUE by altering the expression of genes related to amino acid synthesis, with the exception of asparagine synthetase (ASN) and alanine aminotransferase (AlaAT) (Lea and Azevedo, 2007; McAllister et al., 2012; Xu et al., 2012). Both asparagine and alanine are major long-distance transport forms of N in certain plants (Atkins et al., 1975; Winter et al., 1992; Miflin and Habash, 2002; Lea et al., 2007). Asparagine is synthesized through ASN, which functions in both N assimilation and remobilization (Sieciechowicz et al., 1988; Lam et al., 1995, 1996; Gaufichon et al., 2013, 2017; Moison et al., 2018; Lee et al., 2020b). Constitutive overexpression of OsASN1 was somewhat successful in rice and showed increased seed N content and NUE under limiting and sufficient N treatments, as well as additional improvements in shoot biomass and seed yield at low N (Figure 2; Lee et al., 2020b). On the other hand, expression of Escherichia coli EcasnA in Brassica napus (canola) seems to only be successful at high N status (Seiffert et al., 2004). AlaAT is involved in alanine synthesis and catabolism, and the constitutive or root-specific expression of barley HvAlaAT resulted in improved seed yields in wheat (Peña et al., 2017), or in canola (Good et al., 2007) and rice (Shrawat et al., 2008) (Figure 1). In addition, NUE was increased in rice, canola, and Saccharum officinarum (sugarcane) when HvAlaAT expression was targeted to roots and engineered plants were exposed to high N (Beatty et al., 2013) and/or low N (Good et al., 2007; Snyman et al., 2015). How AlaAT function in roots affects seed yield and NUE is unclear, as it seems to not be vital for plant N metabolism and source-to-sink partitioning (Lam et al., 1996; Miyashita et al., 2007; Beatty et al., 2013; McAllister and Good, 2015).
A relatively high portion of N that is delivered to seeds or other storage sinks may derive from the recycling or remobilization of N in source leaves, rather than directly from root uptake (Cliquet et al., 1990; Patrick and Offler, 2001; Masclaux-Daubresse et al., 2010; Masclaux-Daubresse and Chardon, 2011). This includes nitrates, amino acids, and ureides that are temporarily stored in vacuoles during the vegetative stage (Staswick, 1994; Diaz et al., 2008; Masclaux-Daubresse and Chardon, 2011), as well as N from proteins and organelles, especially from chloroplast degradation (Havé et al., 2017). Overall, the degree of N remobilization in source tissues and its allocation to sinks is assumed to have a significant impact on grain filling, yield, and NUE, while decreasing the residual N in vegetative tissues at harvest (Masclaux-Daubresse and Chardon, 2011). N remobilization and reassimilation occurs especially during leaf senescence and mainly involves the activity of the cytosolic GS1, which has been shown to affect NUE (see above; Masclaux-Daubresse et al., 2010; Avila-Ospina et al., 2014; Havé et al., 2017). Glutamate dehydrogenase (GDH) may also have some role in N assimilation (Lam et al., 1996; Melo-Oliveira et al., 1996; Good et al., 2004; Lea and Miflin, 2011), but it seems to predominantly function in glutamate deamination and ammonium supply for GS1 (Havé et al., 2017; Moison et al., 2018) during N remobilization or when C is limited (Robinson et al., 1991; Masclaux-Daubresse et al., 2006; Fontaine et al., 2012; Grzechowiak et al., 2020). Efforts to enhance plant performance and NUE by overexpressing GDH have been inconclusive. Studies have been unsuccessful when using constitutive overexpression of tomato Slgdh-NAD;B1 in tobacco (Purnell et al., 2005), Nicotiana plumbaginifolia GDHA and GDHB in N. tabacum (Tercé-Laforgue et al., 2013), and GDH from fungi Sclerotinia sclerotiorum and Magnaporthe grisea in rice (Du et al., 2014; Zhou et al., 2015a). On the other hand, there is good evidence that constitutive overexpression of GDH can also lead to increased growth, seed yield, and/or NUE at different N regimes (Figure 2). Specifically, expression of EcgdhA from E. coli in tobacco and maize resulted in increased biomass under sufficient N conditions (Ameziane et al., 2000) and increased seed yield under high N supply (Lightfoot et al., 2007), respectively. Further, expression of AngdhA from Aspergillus nidulans in potato (Solanum tuberosum) caused increased photosynthesis rates, tuber numbers and tuber dry weight, as well as improved NUE at moderate and low N levels (Egami et al., 2012). In rice, increases in NUE and/or grain yield were achieved at low N when fungal GDH from Cylindrocarpon ehrenbergii (CeGDH) or Eurotium cheralieri (EcGDH) were expressed (Zhou et al., 2015b; Tang et al., 2018). In addition, improvements in rice grain yield and weight, or in grain storage protein levels were obtained at low and high N as well as without N fertilization by using fungal GDHs from Trichurus sp. (TrGDH, Du et al., 2019) and Pleurotus cystidiosus (PcGDH, Zhou et al., 2014), respectively. Why constitutive overexpression of GDH was successful in some but not all studies is unclear, but it could potentially result from the genes and plant species that were analyzed. Also, a leaf-specific engineering approach may help to more reliably promote source N remobilization through GDH and enhance source-to-sink N allocation.
Autophagy, an essential mechanism for the degradation of proteins, organelles, and cytosolic macromolecules, is enhanced during leaf senescence and strongly contributes to N remobilization and source-to-sink transport of N (Suzuki and Ohsumi, 2007; Avila-Ospina et al., 2014; Chen et al., 2019). Altering expression of autophagy-related proteins (ATGs) has been a successful strategy to enhance N remobilization in Arabidopsis and rice (Figure 2; Chen et al., 2019; Yu et al., 2019; Zhen et al., 2019a,b; Fan et al., 2020). Depending on the study, the outcome was increased plant growth, seed yield, or seed N and protein levels (Figure 2). In Arabidopsis, constitutive overexpression of AtATG8 resulted in higher seed protein levels when transgenic plants were grown with moderate N supply (Chen et al., 2019), while OsATG8b expression led to increased biomass, seed yield, and NUE at moderate and low N (Zhen et al., 2019a). In rice, shoot biomass, seed yield, and protein levels, as well as NUE were improved when OsATG8a was overexpressed and plants were exposed to high N nutrition, while at moderate N, positive effects were only observed for biomass and seed protein (Yu et al., 2019). Similarly, constitutive expression of OsATG8b in rice resulted in more biomass as well as higher grain yield and protein under sufficient N (Fan et al., 2020), and OsATG8c overexpression triggered improvements in shoot biomass, grain yield, and NUE under moderate and low N fertilization (Zhen et al., 2019b). To date, studies on autophagy-related proteins have focused on Arabidopsis and rice, and work is now needed to address their importance for N remobilization and subsequently yield and NUE improvements in other crop species.
De novo synthesis of most amino acids occurs in plastids (Rolland et al., 2012). Once produced, amino acids are used for plastid function or they are exported from the plastids in support of metabolism in diverse cellular compartments, for transient storage in the vacuole, or for translocation in the phloem to developing sinks (Tegeder and Rentsch, 2010; Taniguchi and Miyake, 2012; Tan et al., 2019). Very little is known about the transport proteins that move amino acids across the chloroplast envelope, as well as between subcellular compartments (Tegeder and Rentsch, 2010; Widhalm et al., 2015; Dinkeloo et al., 2018; Tegeder and Masclaux-Daubresse, 2018; Bouchnak et al., 2019). So far, only two plastidic amino acid transporters have been identified in plants, Arabidopsis malate/glutamate translocators (AtDiT2, Renné et al., 2003; Eisenhut et al., 2015) and Petunia hybrida cationic amino-acid transporter (PhpCAT, Widhalm et al., 2015). In addition, a few amino acid transporters have been localized to the tonoplast, specifically Arabidopsis AtCAT2, AtCAT4, and vacuolar amino acid transporter AtAVT3 as well as tomato (Solanum lycopersicum) SlCAT9 (Yang et al., 2014; Snowden et al., 2015; Fujiki et al., 2017; Dinkeloo et al., 2018). Further, AtBAC1 and AtBAC2 have been shown to transport basic amino acids across the mitochondrial membrane (Hoyos et al., 2003; Palmieri et al., 2006; Toka et al., 2010). Although the function of these intracellular transporters in NUE has not yet been addressed, their regulatory role is most probably fundamental to the effective use of N, for example through, (i) regulating N levels in the chloroplasts and thereby chloroplast function and photosynthetic N use efficiency, (ii) balancing pools of specific amino acids that may otherwise negatively affect plant metabolism and performance through end-product inhibition or substrate limitations of the respective amino acid synthesis pathways, (iii) making amino acids available for phloem loading and source-to-sink allocation throughout the plant's life cycle, and (iv) regulating transient storage pools of amino acids and their remobilization for reproductive growth.
Phloem loading of amino acids represents a bottleneck in N allocation from source leaves to sink (Tegeder, 2012, 2014) and influences not only sink development but also source physiology including carbon fixation, assimilation, and partitioning to sinks (Tan et al., 2010; Zhang et al., 2015b; Santiago and Tegeder, 2016, 2017). For example, increased expression of the endogenous amino acid permease 1 (PsAAP1) in the pea (Pisum sativum) phloem (as well as in the seed cotyledons) resulted in increased amino acid allocation to sinks and co-regulated carbon capture and source-sink allocation. The overall outcome was increased vegetative growth, seed number, and seed protein levels at high N supply (Zhang et al., 2015b). Improvements in seed yield were also achieved when the transgenic plants were exposed to limiting and deficient N levels (Figures 2, 3; Perchlik and Tegeder, 2017). However, factors contributing to the increased NUE varied depending on the N fertilization. Transgenic pea plants exposed to high N soils showed enhanced NUpE, while at low N status NUtE was improved, and at limiting N, both NUpE and NUtE were enhanced. Together, these results support that manipulation of transporters involved in the source-to-sink partitioning of amino acids, and possibly other N metabolites, present a valid strategy to improve NUE. This is further supported by work expressing an S-methyl-methionine transporter (Tan et al., 2010) or a transporter for ureides, an important long-distance N transport form in soybean (Glycine max), in the phloem (Thu et al., 2020). In both cases, plant growth and seed productivity were improved, at least under high N fertilization.
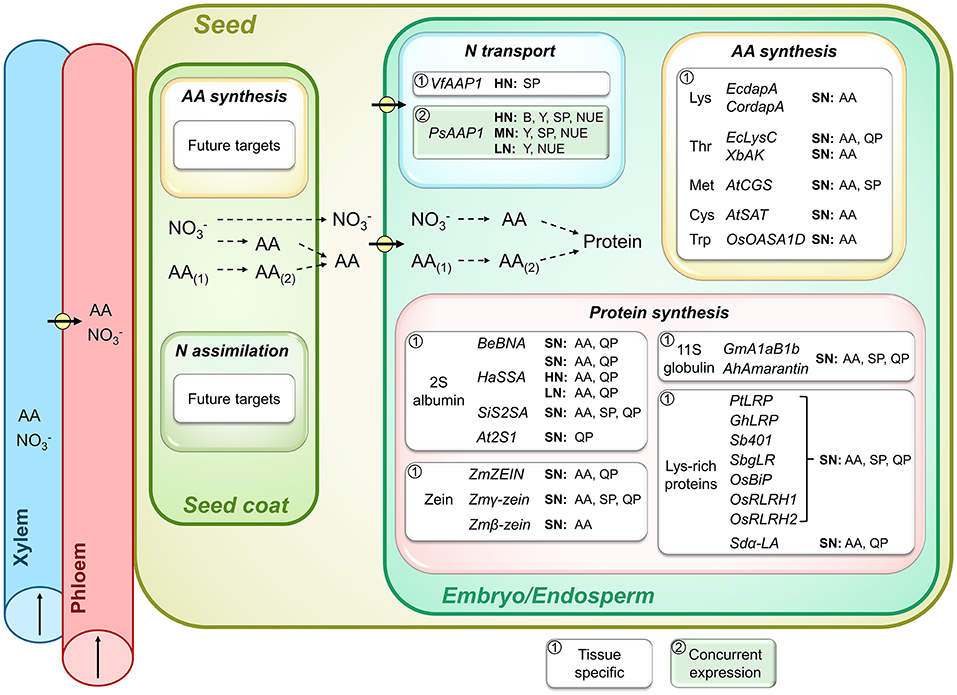
Figure 3. Importance of seed N transport and metabolic processes for seed N accumulation, seed yield, and N use efficiency (NUE). Shown are genes that were successfully overexpressed and resulted in increases in seed levels of specific amino acids (AA), total soluble protein (SP), and specific high-quality proteins (QP), and positively affected shoot biomass (B), seed yield (Y), or NUE. As indicated by a framed white or green background, the transgenic strategies either used (1) seed-specific manipulation or (2) concurrent expression of a particular gene in different source or sink tissues. Plants were grown under high (HN), moderate (MN), sufficient (SN; N supply was not specified), and low N (LN). Pathways of N (re)assimilation, AA synthesis, and conversion (AA(1) to AA(2)) in seed coat and embryo as well as N import into the embryo (arrows with circle) and subsequent protein synthesis are shown. Arrows in xylem and phloem indicate the direction of translocation. Positive results were achieved for embryo expression of genes encoding for AA transporters (VfAAP1, Rolletschek et al., 2005; Weigelt et al., 2008; PsAAP1, Perchlik and Tegeder, 2017), and genes related to synthesis of sulfur-rich (AtCGS, Hanafy et al., 2013; Song et al., 2013; Cohen et al., 2014, 2016; AtSAT, Tabe et al., 2010) and other essential AA (EcdapA, Karachi et al., 1994; Zhu and Galili, 2004; Angelovici et al., 2009; CordapA, Falco et al., 1995; Huang et al., 2005; Frizzi et al., 2007; EcLysC, Karachi et al., 1993, 1994; XbAK, Qi et al., 2011; OsOASA1D, Kita et al., 2010) as well as seed storage protein synthesis (BeBNA, Altenbach and Simpson, 1989; Altenbach et al., 1992; Pickardt et al., 1995; Saalbach et al., 1995; Demidov et al., 2003; HaSSA, Molvig et al., 1997; Tabe and Droux, 2002; Hagan et al., 2003; Chiaiese et al., 2004; SiS2SA, Lee et al., 2003, 2005; At2S1, De Clercq et al., 1990; ZmZEIN, Dinkins et al., 2001; Zmγ-zein, Li et al., 2005; Zmβ-zein. Guo et al., 2020c; GmA1aB1b, Takaiwa et al., 1995; Katsube et al., 1999; Momma et al., 1999; AhAmarantin, Rascón-Cruz et al., 2004; PtLRP, Liu et al., 2016; GhLRP, Yue et al., 2014; Sb401, Yu et al., 2004; SbgLR, Wang et al., 2013b; OsBiP, Yasuda et al., 2009; Kawakatsu et al., 2010; OsRLRH1 and OsRLRH2, Wong et al., 2015; Sdα-LA, Bicar et al., 2008). Names of genes that were overexpressed are provided, however in contrast to Figures 1, 2, the transgenic plant species are not indicated as the respective genes have often been overexpressed in multiple species. See the manuscript text or listed references for more detailed information.
Numerous studies support the fact that the development of vegetative and reproductive sinks is source-limited and depends on the amount of N (and carbon) allocated to sinks (see above; Koch et al., 2003; Chiaiese et al., 2004; Pélissier and Tegeder, 2007; Schmidt et al., 2007; Tan et al., 2010; Zhang et al., 2010, 2015b; Santiago and Tegeder, 2016, 2017; Lu et al., 2020; Thu et al., 2020). On the other hand, seed filling and quality seem to be influenced by seed-localized transport and metabolic processes (Borisjuk et al., 2002; Rosche et al., 2002; Rolletschek et al., 2005; Weigelt et al., 2008; Sanders et al., 2009; Hunt et al., 2010; Ladwig et al., 2012; Pandurangan et al., 2012; c.f. Zhang et al., 2007; Tegeder and Rentsch, 2010; Galili et al., 2016), which may especially be critical when N is limited and individual seeds need to compete for the phloem-derived N.
Seed N transporters are thought to regulate N flux toward, and uptake into, the embryo by controlling the rate of allocation, amount, and composition of amino acids (Tegeder, 2012; Dinkeloo et al., 2018; Tegeder and Hammes, 2018). Amino acid transporters are especially needed at the seed coat–embryo interface where symplasmic connections are lacking (Lanfermeijer et al., 1989; Zhang et al., 2007; Ladwig et al., 2012; Tegeder et al., 2013; Besnard et al., 2018; Tegeder and Hammes, 2018). Following efflux from the seed coat into the apoplasm, the amino acids may either be taken up into the embryo/cotyledons via epidermal cells located opposite to the seed coat or they are imported into the storage parenchyma cells from the seed apoplasm (Tegeder et al., 2000, 2007; Offler et al., 2003; Zhang et al., 2007; Tegeder, 2014). In pea and Vicia narbonensis, N uptake into the embryo could be increased when overexpressing an amino acid transporter (VfAAP1) in the storage parenchyma cells of the cotyledons (Figure 3; Rolletschek et al., 2005; Weigelt et al., 2008). This led to changes in seed metabolism and increased storage protein accumulation under standard N nutrition, including in the field. Further analysis of the transgenic V. narbonensis plants revealed that they take up more N for allocation to seeds and over an extended time period, and that carbon fixation and allocation to seeds are improved as well (Götz et al., 2007). Other studies showed that overexpression of the endogenous PsAAP1 in the epidermal transfer cells of pea cotyledons also resulted in more seed protein when plants were grown at high N fertilization and positively affects carbon metabolism (Zhang et al., 2015b). In those plants, PsAAP1 was concurrently overexpressed in the phloem, triggering enhanced amino acid partitioning to sinks and increased seed number and yield under high N. When exposed to a 50% reduction in N fertilization, the transgenic PsAAP1 pea plants still outperformed wild-type control plants with respect to seed yield and NUE, and seed protein levels were unchanged compared to wild-type (Figure 3; Perchlik and Tegeder, 2017). Together, this supports the fact that the higher amounts of phloem amino acids in the transgenic pea plants accommodate both the increased number of seeds as well as improved seed loading of amino acids. It also demonstrates that even under strongly reduced N supply, the simultaneous upregulation of phloem and embryo loading is a successful strategy for avoiding a dilemma generally observed in crop plants (e.g., in soybean, rice, and maize), which is an increase in seed yield at the cost of seed protein levels or vice versa (Simmonds, 1995; Wilcox and Guodong, 1997; Triboi et al., 2006; Rotundo et al., 2009; Gambín and Borrás, 2010). Even under severe N deficiency (80% less N), seed yield was still higher in the PsAAP1 lines compared to the non-transgenic plants, but at the cost of protein amounts in individual pea seeds (Perchlik and Tegeder, 2017). Nevertheless, total protein yields per plant were always higher in the PsAAP1 overexpressors, independent of how much N was supplied. It is important to point out that the experiments with PsAAP1 overexpressors were done with non-nodulated pea plants that solely relied on soil N. It still needs to be determined, if nodulated PsAAP1 plants, that additionally fix atmospheric N, also outperform wild-type. Together, the above findings support the fact that seed protein levels can be controlled through amino acid transporter functions in the embryo and that their increased activity enables the individual seeds to compete for the phloem N, including under limited N conditions (Figure 3; Rolletschek et al., 2005; Götz et al., 2007; Weigelt et al., 2008; Zhang et al., 2015b; Perchlik and Tegeder, 2017).
While generally a broad spectrum of amino acids is found in the phloem and delivered to seeds, asparagine, glutamine, aspartate, glutamate, alanine, and/or homoserine often dominate, dependent on the plant species, intrinsic factors, and environmental cues (Urquhart and Joy, 1982; Fisher and Macnicol, 1986; Winter et al., 1992; Lohaus et al., 1995; Gattolin et al., 2008). To provide a balanced pool of amino acids and accommodate seed function, amino acid catabolism, N re-assimilation, and amino acid synthesis takes place, either in the seed coat or developing embryo (Figure 3; Rainbird et al., 1984; Ranocha et al., 2001; Lee et al., 2008; Galili et al., 2016; Amir et al., 2018; Tegeder and Masclaux-Daubresse, 2018; Wang et al., 2018c). Amino acids that seem to be primarily produced in the seed coat and released into the seed apoplast are alanine, glutamine, threonine, serine, and valine (Fisher and Macnicol, 1986; Murray, 1987; Rochat and Boutin, 1991; Gallardo et al., 2007). In addition, the sulfur-containing, non-protein amino acid S-methylmethionine is converted to methionine in the seed coat (Ranocha et al., 2001; Gallardo et al., 2007; Lee et al., 2008). Why amino acid metabolism occurs in the seed coat is uncertain, but the conversion of amino acids in the maternal tissue may be important for creating steep concentration gradients for specific amino acids to facilitate their fast movement toward the embryo or to accommodate amino acid transporters involved in embryo uptake of the respective substrate(s). We are not aware of studies examining N metabolism in the seed coat with consequences for seed yield, NUE, or seed protein accumulation, with the exception of work done by Pandurangan et al. (2012). They observed that during soybean seed development asparagine synthetase (ASN) levels in the seed coat decrease while those of asparaginase (ASPGB), involved in asparagine catabolism, increase. Further, ASPGB1a levels and activity were higher in seed coats of a soybean cultivar associated with high seed protein concentrations, suggesting enhanced N flux toward protein synthesis compared to a low-protein cultivar. Overall, the work suggests that seed coat asparagine metabolism may control N partitioning to developing embryos and storage protein accumulation, and could be an important trait for the genetic improvement of seed proteins, at least in soybean.
Following amino acid uptake into the embryo, conversion and transamination reactions also take place in the filial tissue to produce the amounts and spectrum of amino acids required for embryo growth and metabolism, and to produce seed storage proteins (Weber et al., 2005; Tegeder and Rentsch, 2010; Amir et al., 2018). Essential amino acids including aliphatic, aromatic, basic and hydroxyl-group, and sulfur-containing amino acids are critical for human nutrition, but tend to be present in low amounts in many crop species. To achieve higher seed nutritional quality, many studies have therefore targeted genes involved in synthesis of the essential amino acids, often by using seed-specific promoters (Figure 3; Karachi et al., 1993, 1994; Lee et al., 2001; Zhu and Galili, 2004; Kita et al., 2010; Qi et al., 2011; Song et al., 2013; Cohen et al., 2014, 2016; c.f. Galili and Amir, 2013; Wang et al., 2017; Amir et al., 2018). For example, recent work showed that overexpression of cystathionine γ-synthase genes (AtCGS) involved in regulating carbon flow toward methionine synthesis (Amir et al., 2002) resulted in improved levels of free methionine in legume and Arabidopsis seeds (Hanafy et al., 2013; Song et al., 2013; Cohen et al., 2014) and an increased usage of methionine and other amino acids for protein synthesis in tobacco and Arabidopsis seeds (Matityahu et al., 2013; Cohen et al., 2014, 2016). Further, free cysteine levels could be improved in seeds of narrow leaf lupin (Lupinus angustifolius) by manipulating a serine acetyltransferase (AtSAT) (Tabe et al., 2010), and lysine content was increased by expressing feedback insensitive dihydrodipicolinate synthases (DHDPS) in Arabidopsis (E. coli EcdapA, Zhu and Galili, 2004; Angelovici et al., 2009), tobacco (EcdapA, Karachi et al., 1994), canola and soybean (Corynebacterium glutamicum CordapA, Falco et al., 1995), rice (Zmdhps, Lee et al., 2001), and maize seeds (CordapA, Huang et al., 2005; Frizzi et al., 2007). Other studies overexpressed aspartate kinases in soybean (EcAK, Karachi et al., 1993, 1994; Xenorhabdus bovienii XbAK, Qi et al., 2011) or an anthranilate synthase subunit in rice (OsOASA1D, Kita et al., 2010) and successfully enhanced free threonine and tryptophan levels, respectively. Most approaches for altering amino acid synthesis in seeds not only resulted in an increase of the targeted amino acid, but also positively affected levels of other free amino acids (Karachi et al., 1994; Zhu and Galili, 2004; Huang et al., 2005; Qi et al., 2011; Hanafy et al., 2013; Song et al., 2013; Cohen et al., 2014, 2016; Galili et al., 2016; Wang et al., 2017, 2018c). However, unfortunately only a few studies analyzed total soluble protein levels and/or reported increases in seed proteins (Matityahu et al., 2013; Song et al., 2013; Cohen et al., 2014, 2016) or found changes in the amounts of high-quality proteins, such as the methionine-rich globulins and albumins (Figure 3; Karachi et al., 1993, 1994; Angelovici et al., 2009; Cohen et al., 2014, 2016). Seemingly, information on if or how changes in seed amino acid synthesis can affect seed yield and NUE is also lacking.
Seeds of many crop species contain relatively few amounts of high-quality proteins, such as the sulfur-rich albumins and legumin-like globulins. Much research has been done to enhance seed N use and increase the amount and quality of proteins by seed-specific overexpression of storage protein genes (Figure 3; c.f. Ufaz and Galili, 2008; Galili and Amir, 2013; Häkkinen et al., 2018). For example, 2S albumin genes (BNA, SA, or 2SA) have been successfully expressed in seeds of rice (Sesamum indicum SiS2SA, Lee et al., 2003, 2005; Helianthus annuus HaSSA, Hagan et al., 2003), tobacco (Bertholletia excelsa BeBNA, Altenbach and Simpson, 1989; Saalbach et al., 1995; At2S1, De Clercq et al., 1990), canola (At2S1, De Clercq et al., 1990; BeBNA, Altenbach et al., 1992), Vicia narbonensis (BeBNA, Pickardt et al., 1995; Saalbach et al., 1995; Demidov et al., 2003), Cicer arietinum (chickpea; HaSSA, Chiaiese et al., 2004), narrow leaf lupin (HaSSA, Molvig et al., 1997; Tabe and Droux, 2002), and Arabidopsis (At2S1, De Clercq et al., 1990). Further methionine-rich zein proteins from maize could be produced in soybean seeds (ZmZEIN, Dinkins et al., 2001; Zmγ-zein, Li et al., 2005; Zmβ-zein, Guo et al., 2020c), and 11S globulins from soybean or amaranth (Amaranthus hypochondriacus) were expressed in rice (GmA1aB1b, Katsube et al., 1999; Momma et al., 1999) and tobacco (GmA1aB1b, Takaiwa et al., 1995), and maize (AhAmarantin, Rascón-Cruz et al., 2004), respectively. Besides the successes in increasing the methionine/sulfur-rich proteins, only a few studies reported additional positive effects on total seed proteins (Figure 3; Momma et al., 1999; Lee et al., 2003; Rascón-Cruz et al., 2004; Li et al., 2005). If and how varying N fertilization influences seed quality and protein levels, or seed protein yields of transgenic plants expressing storage protein genes has generally not been addressed, with the exception of work by Chiaiese et al. (2004). The authors showed that chickpea plants expressing a sunflower 2S albumin gene (HaSSA) achieved higher seed albumin levels under high and low N supply, while seed yield was not changed at high N but decreased at low N fertilization. However, since sulfur was sequestered in the sulfur-rich protein, the negative effects were mainly attributed to sulfur rather than N deficiency (Chiaiese et al., 2004).
Seed quality is also influenced by the lysine content of proteins, and so-called lysine-rich proteins (LRP) have been successfully increased in rice (Liu et al., 2016) and maize (Solanum berthaultii Sb401, Yu et al., 2004; Psophocarpus tetragonolobus PtLRP, Liu et al., 2016; SbgLR, Wang et al., 2013b; Gossypium hirsutum GhLRP, Yue et al., 2014; Figure 3). Other proteins with high lysine content have also been used to improve the protein quality of seeds, including a binding protein (OsBiP, Yasuda et al., 2009; Kawakatsu et al., 2010) and lysine-rich histone proteins (OsRLRH1 and OsRLRH2, Wong et al., 2015) in rice, and a porcine α-lactalbumin gene in maize (Sus domesticus Sdα-LA, Bicar et al., 2008). In addition, the high-lysine lines displayed increased total seed protein levels, while seed yield was not analyzed (Figure 3). The increases in both specific and total seed proteins support enhanced seed N supply, probably due to increased sink strength (MacKown et al., 1992; Rolletschek et al., 2005; Götz et al., 2007) and elevated N assimilate availability for protein synthesis, which is reinforced by higher seed amino acid levels observed in numerous studies (Figure 3; Momma et al., 1999; Rascón-Cruz et al., 2004; Yasuda et al., 2009; Kawakatsu et al., 2010; Liu et al., 2016).
Improvements in quality proteins have generally been observed at sufficient or high N fertilization, and it is not clear if the transgenic plants maintain their advantage at low N supply, and if and how total seed protein and yield would be affected under non-optimal N conditions. Since seed protein concentrations generally depend on N assimilate availability (Rolletschek et al., 2005; Weigelt et al., 2008; Rotundo et al., 2009; Pandurangan et al., 2012; Santiago and Tegeder, 2016), it also remains to be evaluated if increasing amino acid source-to-sink allocation (c.f. Zhang et al., 2015b; Perchlik and Tegeder, 2017), in addition to upregulation of seed amino acid (see above) or protein synthesis, can further improve seed protein yields and quality, even under low N status, and leads to improved NUE.
The movement of N from the soil to final sink cells involves numerous transport and metabolic processes in source and sink, including (i) N uptake and partitioning within cells and over short and long distances, and (ii) N (re)assimilation, amino acid synthesis and catabolism, and protein synthesis. Since any of these processes could potentially present a bottleneck for the efficient use of N for biomass and seed production, associated key genes have been targets for improving plant performance and NUE, but, as reviewed above, with varying success (Figures 1–3). The inconsistent outcomes might, in part, be caused by unwanted pleiotropic effects due to the use of constitutive promoters, an imbalance in N metabolite pools triggering negative feedback inhibition of N uptake or metabolism, substrate limitations for amino acid synthesis, or post-transcriptional and post-translational regulation (Thomsen et al., 2014; Takabayashi et al., 2016; Osanai et al., 2017; Sweetlove et al., 2017; Gao et al., 2019b; Fernie et al., 2020). To circumvent such detrimental effects and to improve NUE, future research may consider the selection or design of “super promoters” for genetic manipulation that allow tissue or cell-specific, developmentally regulated and enhanced gene expression. Such promoters could, for example, be used for the overexpression of amino acid transporters or synthesis genes to drive N uptake from the soil throughout the vegetative and reproductive phases, or to accelerate source N remobilization and leaf export during leaf senescence. Other research studies could consider a “gene pyramiding approach” (Halpin, 2005; Li et al., 2010; Naqvi et al., 2010; McAllister et al., 2012; Lee et al., 2020a). While selective metabolism and transporter genes have been successful targets to improve NUE in some plant species, at least under high N nutrition (see above), simultaneous manipulation of several genes may be required for a broader approach to improve NUE at high and low N status. For example, improvements in N assimilation or amino acid synthesis may only be successful if metabolic and transport processes are coordinated through concurrent manipulation of two or more genes (c.f., Lee et al., 2020a). Further, one gene could be simultaneously manipulated in root and shoot tissues, or source and sink (c.f., Zhang et al., 2015a; Lee et al., 2020a). For instance, to improve N flux from source to sink and seed N supply, leaf N export and phloem loading, as well as seed transport processes including embryo loading, will need to happen at a high rate and throughout sink development (c.f., Perchlik and Tegeder, 2017; Tegeder and Hammes, 2018). Approaches to improve crop productivity and NUE should also take into account regulatory aspects of N assimilation and transport, as well as the sensing and signaling mechanisms involved (Wang et al., 2018a; Chen et al., 2020; Feng et al., 2020; Han et al., 2020; Vidal et al., 2020). Consideration should further be given to the tight interaction between carbon, N, and sulfur metabolism and their transport processes (Zhang et al., 2015b; Tegeder and Masclaux-Daubresse, 2018; Kopriva et al., 2019; Fernie et al., 2020).
NUE is a complex polygenic trait making its genetic dissection and engineering challenging. In fact, despite the notable progress that has been made in understanding plant NUE, successful agronomic applications of engineered crop plants with altered N transport and metabolism or other physiological functions have not yet been achieved. To better grasp the genetic basis of NUE for genetic manipulation, future strategies might take greater advantage of the available natural variation for NUE between and within crop species (Hawkesford and Griffiths, 2019; Swarbreck et al., 2019). For example, using genetic association studies or genomic selection, in conjunction with high-throughput phenotyping, large crop populations, or different cultivars could be screened to discover further genes associated with improved NUE (Bhat et al., 2016; van Bueren and Struik, 2017; Nguyen and Kant, 2018; York, 2019; Sinha et al., 2020). Alternatively, or in addition, comparative transcriptome and proteome studies will allow for identification of fundamental NUE genes (Simons et al., 2014; Li et al., 2020; Zhang et al., 2020b). Importantly, to develop improved crop cultivars for a range of growing regions, such analyses will have to be done with plants exposed to a variety of locations, latitudes, soil conditions, and varying N regimes and agricultural systems. These should include systems that do not solely rely on mineral N fertilizer but also consider organic matter for plant N nutrition, like organic farming or conservation agriculture, and the need of amino acid transporters for N uptake. Further, to improve NUE for a sustainable agriculture, future climate and environmental changes will need to be addressed (Nguyen et al., 2017; Kanter et al., 2020). For example, exposure of crop plants to drought stress will not only require adjustments with respect to N uptake, partitioning, and metabolic processes, but also other physiological as well as morphological changes. As it would be extremely challenging to conduct field experiments for conventional breeding, marker-assisted modern breeding, genomic selection, and genetic engineering under numerous scenarios, the use of crop modeling that integrates genetics, environmental factors, and management practices will be a critical tool for making progress in NUE (Zhang et al., 2015a; Messina et al., 2020; Peng et al., 2020). Indeed, interdisciplinary strategies and multiscale collaborations are essential to achieve a more robust and holistic understanding of NUE that can lead to global successes for crop production and a more sustainable agroecological system (c.f., Cassman and Grassini, 2020).
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This work was funded by the Agriculture and Food Research Initiative (AFRI) competitive award 2017-67013- 26158 from the United State Department of Agriculture, National Institute of Food and Agriculture, the United Soybean Board (Grant No. 1820-152-0136/1920-152-0136), and the U.S. National Science Foundation (IOS-1932661).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alam, M. K., Bell, R. W., Haque, M. E., Islam, M. A., and Kader, M. A. (2020). Soil nitrogen storage and availability to crops are increased by conservation agriculture practices in rice–based cropping systems in the Eastern Gangetic Plains. Field Crops Res. 250, 107764. doi: 10.1016/j.fcr.2020.107764
Altenbach, S. B., Kuo, C. C., Staraci, L. C., Pearson, K. W., Wainwright, C., Georgescu, A., et al. (1992). Accumulation of a Brazil nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol. Biol. 18, 235–245. doi: 10.1007/BF00034952
Altenbach, S. B., and Simpson, R. B. (1989). Manipulation of methionine-rich protein genes in plant seeds. Trends Biotechnol. 8, 156–160. doi: 10.1016/0167-7799(90)90162-Q
Ameziane, R., Bernhard, K., and Lightfoot, D. (2000). Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 221, 47–57. doi: 10.1023/A:1004794000267
Amir, R., Galili, G., and Cohen, H. (2018). The metabolic roles of free amino acids during seed development. Plant Sci. 275, 11–18. doi: 10.1016/j.plantsci.2018.06.011
Amir, R., Hacham, Y., and Galili, G. (2002). Cystathionine γ-synthase and threonine synthase operate in concert to regulate carbon flow towards methionine in plants. Trends Plant Sci. 7, 153–156. doi: 10.1016/S1360-1385(02)02227-6
Anas, M., Liao, F., Verma, K. K., Sarwar, M. A., Mahmood, A., Chen, Z. L., et al. (2020). Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol. Res. 53, 1–20. doi: 10.1186/s40659-020-00312-4
Andrews, M., Morton, J. D., Lieffering, M., and Bisset, L. (1992). The partitioning of nitrate assimilation between root and shoot of a range of temperate cereals and pasture grasses. Ann. Bot. 70, 271–276. doi: 10.1093/oxfordjournals.aob.a088469
Angelovici, R., Fait, A., Zhu, X., Szymanski, J., Feldmesser, E., Fernie, A. R., et al. (2009). Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol. 151, 2058–2072. doi: 10.1104/pp.109.145631
Atkins, C. A., and Beevers, L. (1990). “Synthesis, transport and utilization of translocated solutes of nitrogen,” in Nitrogen in Higher Plants, ed. Y. P. Abrol (Somerset: Research Studies Press), 223–295.
Atkins, C. A., Pate, J. S., and Sharkey, P. J. (1975). Asparagine metabolism—key to the nitrogen nutrition of developing legume seeds. Plant Physiol. 56, 807–812. doi: 10.1104/pp.56.6.807
Avila-Ospina, L., Moison, M., Yoshimoto, K., and Masclaux-Daubresse, C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65, 3799–3811. doi: 10.1093/jxb/eru039
Bailey, K. J., and Leegood, R. C. (2016). Nitrogen recycling from the xylem in rice leaves: dependence upon metabolism and associated changes in xylem hydraulics. J. Exp. Bot. 67, 2901–2911. doi: 10.1093/jxb/erw132
Bao, A., Liang, Z., Zhao, Z., and Cai, H. (2015). Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Internat. J. Mol. Sci. 16, 9037–9063. doi: 10.3390/ijms16059037
Bao, A., Zhao, Z., Ding, G., Shi, L., Xu, F., and Cai, H. (2014). Accumulated expression level of cytosolic glutamine synthetase 1 gene (OsGS1;1 or OsGS1;2) alter plant development and the carbon-nitrogen metabolic status in rice. PLoS ONE 9:e95581. doi: 10.1371/journal.pone.0095581
Bárzana, G., Aroca, R., Bienert, G. P., Chaumont, F., and Ruiz-Lozano, J. M. (2014). New insights into the regulation of aquaporins by the arbuscular mycorrhizal symbiosis in maize plants under drought stress and possible implications for plant performance. Mol. Plant Microbe Interact. 27, 349–363. doi: 10.1094/MPMI-09-13-0268-R
Beatty, P. H., Carroll, R. T., Shrawat, A. K., Guevara, D., and Good, A. G. (2013). Physiological analysis of nitrogen-efficient rice overexpressing alanine aminotransferase under different N regimes. Botany 91, 866–883. doi: 10.1139/cjb-2013-0171
Beevers, L., and Hageman, R. H. (1969). Nitrate reduction in higher plants. Annu. Rev. Plant Physiol. 20, 495–522. doi: 10.1146/annurev.pp.20.060169.002431
Besnard, J., Pratelli, R., Zhao, C., Sonawala, U., Collakova, E., Pilot, G., et al. (2016). UMAMIT14 is an amino acid exporter involved in phloem unloading in Arabidopsis roots. J. Exp. Bot. 67, 6385–6397. doi: 10.1093/jxb/erw412
Besnard, J., Zhao, C., Avice, J. C., Vitha, S., Hyodo, A., Pilot, G., et al. (2018). Arabidopsis UMAMIT24 and 25 are amino acid exporters involved in seed loading. J. Exp. Bot. 69, 5221–5232. doi: 10.1093/jxb/ery302
Bhat, J. A., Ali, S., Salgotra, R. K., Mir, Z. A., Dutta, S., Jadon, V., et al. (2016). Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 7:221. doi: 10.3389/fgene.2016.00221
Bicar, E. H., Woodman-Clikeman, W., Sangtong, V., Peterson, J. M., Yang, S. S., Lee, M., et al. (2008). Trangenic maize endosperm containing a milk protein has improved amino acid balance. Transgenic Res. 17, 59–71. doi: 10.1007/s11248-007-9081-3
Bittsánszky, A., Pilinszky, K., Gyulai, G., and Komives, T. (2015). Overcoming ammonium toxicity. Plant Sci. 231, 184–190. doi: 10.1016/j.plantsci.2014.12.005
Borisjuk, L., Wang, T. L., Rolletschek, H., Wobus, U., and Weber, H. (2002). A pea seed mutant affected in the differentiation of the embryonic epidermis is impaired in embryo growth and seed maturation. Development 129, 1595–1607.
Bouchnak, I., Brugière, S., Moyet, L., Le Gall, S., Salvi, D., Kuntz, M., et al. (2019). Unraveling hidden components of the chloroplast envelope proteome: opportunities and limits of better MS sensitivity. Mol. Cell. Proteom. 18, 1285–1306. doi: 10.1074/mcp.RA118.000988
Brauer, E. K., Rochon, A., Bi, Y. M., Bozzo, G. G., Rothstein, S. J., and Shelp, B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant 141, 361–372. doi: 10.1111/j.1399-3054.2011.01443.x
Britto, D. T., and Kronzucker, H. J. (2002). toxicity in higher plants: a critical review. J. Plant Physiol. 159, 567–584. doi: 10.1078/0176-1617-0774
Cai, H., Zhou, Y., Xiao, J., Li, X., Zhang, Q., and Lian, X. (2009). Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 28, 527–537. doi: 10.1007/s00299-008-0665-z
Cañas, R. A., Yesbergenova-Cuny, Z., Belanger, L., Rouster, J., Brulé, L., Gilard, F., et al. (2020). NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 9:130. doi: 10.3390/plants9020130
Cassman, K. G., and Grassini, P. (2020). A global perspective on sustainable intensification research. Nat. Sustain. 3, 262–268. doi: 10.1038/s41893-020-0507-8
Cerezo, M., Tillard, P., Filleur, S., Munos, S., Daniel-Vedele, F., and Gojon, A. (2001). Major alterations of the regulation of root uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127, 262–271. doi: 10.1104/pp.127.1.262
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Chapin, F. S., Moilanen, L., and Kielland, K. (1993). Preferential use of organic nitrogen for growth by a non-mycorrhizal arctic sedge. Nature 361, 150–153. doi: 10.1038/361150a0
Chardon, F., Barthélémy, J., Daniel-Vedele, F., and Masclaux-Daubresse, C. (2010). Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J. Exp. Bot. 61, 2293–2302. doi: 10.1093/jxb/erq059
Chardon, F., Noël, V., and Masclaux-Daubresse, C. (2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J. Exp. Bot. 63, 3401–3412. doi: 10.1093/jxb/err353
Chen, J., Fan, X., Qian, K., Zhang, Y., Song, M., Liu, Y., et al. (2017). pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15, 1273–1283. doi: 10.1111/pbi.12714
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Chen, K. E., Chen, H. Y., Tseng, C. S., and Tsay, Y. F. (2020). Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat. Plants 6, 1126–1135. doi: 10.1038/s41477-020-00758-0
Chen, Q., Soulay, F., Saudemont, B., Elmayan, T., Marmagne, A., and Masclaux-Daubresse, C. (2019). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60, 343–352. doi: 10.1093/pcp/pcy214
Chiaiese, P., Ohkama-Ohtsu, N., Molvig, L., Godfree, R., Dove, H., Hocart, C., et al. (2004). Sulphur and nitrogen nutrition influence the response of chickpea seeds to an added, transgenic sink for organic sulphur. J. Exp. Bot. 55, 1889–1901. doi: 10.1093/jxb/erh198
Chichkova, S., Arellano, J., Vance, C. P., and Hernández, G. (2001). Transgenic tobacco plants that overexpress alfalfa NADH-glutamate synthase have higher carbon and nitrogen content. J. Exp. Bot. 52, 2079–2087. doi: 10.1093/jexbot/52.364.2079
Cliquet, J. B., Deleens, E., and Mariotti, A. (1990). C and N mobilization from stalk and leaves during kernel filling by 13C and 15N tracing in Zea mays L. Plant Physiol. 94, 1547–1553. doi: 10.1104/pp.94.4.1547
Cohen, H., Israeli, H., Matityahu, I., and Amir, R. (2014). Seed-specific expression of a feedback-insensitive form of CYSTATHIONINE-γ-SYNTHASE in Arabidopsis stimulates metabolic and transcriptomic responses associated with desiccation stress. Plant Physiol. 166, 1575–1592. doi: 10.1104/pp.114.246058
Cohen, H., Pajak, A., Pandurangan, S., and Amir, R. (2016). Higher endogenous methionine in transgenic Arabidopsis seeds affects the composition of storage proteins and lipids. Amino Acids 48, 1413–1422. doi: 10.1007/s00726-016-2193-4
Cooper, M., Tang, T., Gho, C., Hart, T., Hammer, G., and Messina, C. (2020). Integrating genetic gain and gap analysis to predict improvements in crop productivity. Crop Sci. 60, 582–604. doi: 10.1002/csc2.20109
Coque, M., and Gallais, A. (2006). Genomic regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor. Appl. Genet. 112, 1205–1220. doi: 10.1007/s00122-006-0222-5
Crawford, N. M. (1995). Nitrate: nutrient and signal for plant growth. Plant Cell 7:859. doi: 10.2307/3870042
Crawford, N. M., and Glass, A. D. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
Cren, M., and Hirel, B. (1999). Glutamine synthetase in higher plants regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 40, 1187–1193. doi: 10.1093/oxfordjournals.pcp.a029506
Crété, P., Caboche, M., and Meyer, C. (1997). Nitrite reductase expression is regulated at the post-transcriptional level by the nitrogen source in Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant J. 11, 625–634. doi: 10.1046/j.1365-313X.1997.11040625.x
De Clercq, A., Vandewiele, M., Van Damme, J., Guerche, P., Van Montagu, M., Vandekerckhove, J., et al. (1990). Stable accumulation of modified 2S albumin seed storage proteins with higher methionine contents in transgenic plants. Plant Physiol. 94, 970–979. doi: 10.1104/pp.94.3.970
Demidov, D., Hosrtmann, C., Meixner, M., Pickardt, T., Saalbach, I., Galili, G., et al. (2003). Additive effects of the feed-back insensitive bacterial aspartate kinase and the Brazil nut S2 albumin on the methionine content of transgenic narbon bean (Vicia narbonensis L.). Mol. Breeding 11, 187–201. doi: 10.1023/A:1022814506153
Diaz, C., Lemaître, T., Christ, A., Azzopardi, M., Kato, Y., Sato, F., et al. (2008). Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 147, 1437–1449. doi: 10.1104/pp.108.119040
Dickson, R. E., Vogelmann, T. C., and Larson, P. R. (1985). Glutamine transfer from xylem to phloem and translocation to developing leaves of Populus deltoides. Plant Physiol. 77, 412–417. doi: 10.1104/pp.77.2.412
Dinkeloo, K., Boyd, S., and Pilot, G. (2018). Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin. Cell Dev. Biol. 74, 105–113. doi: 10.1016/j.semcdb.2017.07.010
Dinkins, R. D., Reddy, R., Curtis, A., Meurer, C. A., Yan, B., Trick, H., et al. (2001). Increased sulfur amino acids in soybean plants overexpressing the maize 15 kD zein protein. In Vitro Cell Dev. Pl. 37, 742–747. doi: 10.1007/s11627-001-0123-x
Djennane, S., Chauvin, J. E., and Meyer, C. (2002). Glasshouse behaviour of eight transgenic potato clones with a modified nitrate reductase expression under two fertilization regimes. J. Exp. Bot. 53, 1037–1045. doi: 10.1093/jexbot/53.371.1037
Du, C., Lin, J., Yang, Y., Liu, H., Li, C., Zhou, Y., et al. (2014). Molecular cloning, characterization and function analysis of a GDH gene from Sclerotinia sclerotiorum in rice. Mol. Biol. Rep. 41, 3683–3693. doi: 10.1007/s11033-014-3233-3
Du, C. Q., Lin, J. Z., Dong, L. A., Liu, C., Tang, D. Y., Yan, L., et al. (2019). Overexpression of an NADP(H)-dependent glutamate dehydrogenase gene, TrGDH, from Trichurus improves nitrogen assimilation, growth status and grain weight per plant in rice. Breed. Sci. 69, 429–438. doi: 10.1270/jsbbs.19014
Dzuibany, C., Haupt, S., Fock, H., Biehler, K., Migge, A., and Becker, T. W. (1998). Regulation of nitrate reductase transcript levels by glutamine accumulating in the leaves of a ferredoxin-dependent glutamate synthase-deficient gluS mutant of Arabidopsis thaliana, and by glutamine provided via the roots. Planta 206, 515–522. doi: 10.1007/s004250050428
Egami, T., Wakayama, M., Aoki, N., Sasaki, H., Kisaka, H., Miwa, T., et al. (2012). The effects of introduction of a fungal glutamate dehydrogenase gene (gdhA) on the photosynthetic rates, biomass, carbon and nitrogen contents in transgenic potato. Plant Biotechnol. 29, 57–64. doi: 10.5511/plantbiotechnology.12.0127a
Eisenhut, M., Hocken, N., and Weber, A. P. (2015). Plastidial metabolite transporters integrate photorespiration with carbon, nitrogen, and sulfur metabolism. Cell Calcium 58, 98–104. doi: 10.1016/j.ceca.2014.10.007
Enggrob, K. L., Jakobsen, C. M., Pedersen, I. F., and Rasmussen, J. (2019). Newly depolymerized large organic N contributes directly to amino acid uptake in young maize plants. New Phytol. 224, 689–699. doi: 10.1111/nph.16070
Epstein, E., and Bloom, A. J. (2005). Mineral Nutrition of Plants: Principles and Perspectives, 2nd edn. Sunderland: Sinauer Assoc. Inc.
Falco, S. C., Guida, T., Locke, M., Mauvais, J., Sanders, C., Ward, R. T., et al. (1995). Transgenic canola and soybean seeds with increased lysine. Nat. Biotechnol. 13, 577–582. doi: 10.1038/nbt0695-577
Fan, T., Yang, W., Zeng, X., Xu, X., Xu, Y., Fan, X., et al. (2020). A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 11:588. doi: 10.3389/fpls.2020.00588
Fan, X., Gordon-Weeks, R., Shen, Q., and Miller, A. J. (2006). Glutamine transport and feedback regulation of nitrate reductase activity in barley roots leads to changes in cytosolic nitrate pools. J. Exp. Bot. 57, 1333–1340. doi: 10.1093/jxb/erj110
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., and Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68, 2463–2475. doi: 10.1093/jxb/erx011
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016b). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. P. Natl. Acad. Sci. U.S.A. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Fan, X. R., Feng, H. M., Tan, Y. W., Xu, Y. L., Miao, Q. S., and Xu, G. H. (2016a). A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58, 590–599. doi: 10.1111/jipb.12382
Fang, Z., Bai, G., Huang, W., Wang, Z., Wang, X., and Zhang, M. (2017). The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front. Plant Sci. 8:1338. doi: 10.3389/fpls.2017.01338
Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S., Rentsch, D., et al. (2013). Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11, 446–458. doi: 10.1111/pbi.12031
Feller, U., and Fischer, A. (1994). Nitrogen metabolism in senescing leaves. CRC Crit. Rev. Plant Sci. 13, 241–273. doi: 10.1080/07352689409701916
Feng, H., Fan, X., Miller, A. J., and Xu, G. (2020). Plant nitrogen uptake and assimilation: regulation of cellular pH homeostasis. J. Exp. Bot. 71, 4380–4392. doi: 10.1093/jxb/eraa150
Feng, H., Li, B., Zhi, Y., Chen, J., Li, R., Xia, X., et al. (2017). Overexpression of the nitrate transporter, OsNRT2.3b, improves rice phosphorus uptake and translocation. Plant Cell Rep. 36, 1287–1296. doi: 10.1007/s00299-017-2153-9
Fernie, A. R., Bachem, C. W., Helariutta, Y., Neuhaus, H. E., Prat, S., Ruan, Y. L., et al. (2020). Synchronization of developmental, molecular and metabolic aspects of source–sink interactions. Nat. Plants 6, 55–66. doi: 10.1038/s41477-020-0590-x
Ferreira, S., Moreira, E., Amorim, I., Santos, C., and Melo, P. (2019). Arabidopsis thaliana mutants devoid of chloroplast glutamine synthetase (GS2) have non-lethal phenotype under photorespiratory conditions. Plant Physiol. Biochem. 144, 365–374. doi: 10.1016/j.plaphy.2019.10.009
Fichtner, F., Dissanayake, I. M., Lacombe, B., and Barbier, F. (2020). Sugar and nitrate sensing: a multi-billion-year story. Trends Plant Sci. in press. doi: 10.1016/j.tplants.2020.11.006
Fisher, D. B., and Macnicol, P. K. (1986). Amino acid composition along the transport pathway during grain filling in wheat. Plant Physiol. 82, 1019–1023. doi: 10.1104/pp.82.4.1019
Fontaine, J. X., Tercé-Laforgue, T., Armengaud, P., Clément, G., Renou, J. P., Pelletier, S., et al. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24, 4044–4065. doi: 10.1105/tpc.112.103689
Fowler, D., Coyle, M., Skiba, U., Sutton, M. A., Cape, J. N., Reis, S., et al. (2013). The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 368:20130164. doi: 10.1098/rstb.2013.0164
Frizzi, A., Huang, S., Gilbertson, L. A., Armstrong, T. A., Leuthy, M. H., and Malvar, T. M. (2007). Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol. J. 6, 13–21. doi: 10.1111/j.1467-7652.2007.00290.x
Fuentes, S. I., Allen, D. J., Ortiz-Lopez, A., and Hernández, G. (2001). Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 52, 1071–1081. doi: 10.1093/jexbot/52.358.1071
Fujiki, Y., Teshima, H., Kashiwao, S., Kawano-Kawada, M., Ohsumi, Y., Kakinuma, Y., et al. (2017). Functional identification of AtAVT3, a family of vacuolar amino acid transporters, in Arabidopsis. FEBS Lett. 591, 5–15. doi: 10.1002/1873-3468.12507
Galili, G., and Amir, R. (2013). Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol. J. 11, 211–222. doi: 10.1111/pbi.12025
Galili, G., Amir, R., and Fernie, A. R. (2016)., The regulation of essential amino acid synthesis and accumulation in plants. Annu. Rev. Plant Biol. 67, 153–178. doi: 10.1146/annurev-arplant-043015-112213
Gallardo, K., Firnhaber, C., Zuber, H., Héricher, D., Belghazi, M., Henry, C., et al. (2007). A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol. Cell Proteom. 6, 2165–2179. doi: 10.1074/mcp.M700171-MCP200
Gambín, B. L., and Borrás, L. (2010). Resource distribution and the trade-off between seed number and seed weight: a comparison across crop species. Ann. Appl. Biol. 156, 91–102. doi: 10.1111/j.1744-7348.2009.00367.x
Ganeteg, U., Ahmad, I., Jämtgård, S., Aguetoni-Cambui, C., Inselsbacher, E., Svennerstam, H., et al. (2017). Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant Cell Environ. 40, 413–423. doi: 10.1111/pce.12881
Gao, Y., de Bang, T. C., and Schjoerring, J. K. (2019b). Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 17, 1209–1221. doi: 10.1111/pbi.13046
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019a). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10:5207. doi: 10.1038/s41467-019-13110-8
Garnett, T., Plett, D., Heuer, S., and Okamoto, M. (2015). Genetic approaches to enhancing nitrogen-use efficiency (NUE) in cereals: challenges and future directions. Funct. Plant Biol. 42, 921–941. doi: 10.1071/FP15025
Gattolin, S., Newbury, H. J., Bale, J. S., Tseng, H. M., Barrett, D. A., and Pritchard, J. (2008). A diurnal component to the variation in sieve tube amino acid content in wheat. Plant Phys. 147, 912–921. doi: 10.1104/pp.108.116079
Gaufichon, L., Marmagne, A., Belcram, K., Yoneyama, T., Sakakibara, Y., Hase, T., et al. (2017). ASN1-encoded asparagine synthetase in floral organs contributes to nitrogen filling in Arabidopsis seeds. Plant J. 91, 371–393. doi: 10.1111/tpj.13567
Gaufichon, L., Masclaux-Daubresse, C., Tcherkez, G., Reisdorf-Cren, M., Sakakibara, Y., Hase, T., et al. (2013). Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant Cell Environ. 36, 328–342. doi: 10.1111/j.1365-3040.2012.02576.x
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M., and Birnbaum, K. D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U.S.A. 105, 803–808. doi: 10.1073/pnas.0709559105
Glass, A. D., Britto, D. T., Kaiser, B. N., Kinghorn, J. R., Kronzucker, H. J., Kumar, A., et al. (2002). The regulation of nitrate and ammonium transport systems in plants. J. Exp. Bot. 53, 855–864. doi: 10.1093/jexbot/53.370.855
Good, A. G., Johnson, S. J., De Pauw, M., Carroll, R. T., Savidov, N., Vidmar, J., et al. (2007). Engineering nitrogen use efficiency with alanine aminotransferase. Botany 85, 252–262. doi: 10.1139/B07-019
Good, A. G., Shrawat, A. K., and Muench, D. G. (2004). Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605. doi: 10.1016/j.tplants.2004.10.008
Götz, K. P., Staroske, N., Radchuk, R., Neil Emery, R. J., Wutzke, K. D., Herzog, H., et al. (2007). Uptake and allocation of carbon and nitrogen in Vicia narbonensis plants with increased seed sink strength achieved by seed-specific expression of an amino acid permease. J. Exp. Bot. 58, 3183–3195. doi: 10.1093/jxb/erm164
Grallath, S., Weimar, T., Meyer, A., Gumy, C., Suter-Grotemeyer, M., Neuhaus, J. M., et al. (2005). The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol. 137, 117–126. doi: 10.1104/pp.104.055079
Gramma, V., Kontbay, K., and Wahl, V. (2020). Crops for the future: on the way to reduce nitrogen pollution. Am. J. Bot. 107, 1211–1213. doi: 10.1002/ajb2.1527
Griffiths, M., and York, L. M. (2020). Targeting root ion uptake kinetics to increase plant productivity and nutrient use efficiency. Plant Physiol. 182, 1854–1868. doi: 10.1104/pp.19.01496
Grzechowiak, M., Sliwiak, J., Jaskolski, M., and Ruszkowski, M. (2020). Structural studies of glutamate dehydrogenase (isoform 1) from Arabidopsis thaliana, an important enzyme at the branch-point between carbon and nitrogen metabolism. Front. Plant Sci. 11:754. doi: 10.3389/fpls.2020.00754
Guo, C., Liu, X., Chen, L., Cai, Y., Yao, W., Yuan, S., et al. (2020c). Elevated methionine content in soybean seed by overexpressing maize β-zein protein. Oil Crop Sci. 5, 11–16. doi: 10.1016/j.ocsci.2020.03.004
Guo, N., Gu, M., Hu, J., Qu, H., and Xu, G. (2020a). Rice OsLHT1 Functions in Leaf-to-Panicle Nitrogen Allocation for Grain Yield and Quality. Front. Plant Sci. 11:1150. doi: 10.3389/fpls.2020.01150
Guo, N., Hu, J., Yan, M., Qu, H., Luo, L., Tegeder, M., et al. (2020b). Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 103, 395–411. doi: 10.1111/tpj.14742
Guo, S., Kaldenhoff, R., Uehlein, N., Sattelmacher, B., and Brueck, H. (2007). Relationship between water and nitrogen uptake in nitrate-and ammonium-supplied Phaseolus vulgaris L. plants. J. Plant Nutr. Soil Sci. 170, 73–80. doi: 10.1002/jpln.200625073
Habash, D. Z., Massiah, A. J., Rong, H. L., Wallsgrove, R. M., and Leigh, R. A. (2001). The role of cytosolic glutamine synthetase in wheat. Ann. App. Biol. 138, 83–89. doi: 10.1111/j.1744-7348.2001.tb00087.x
Hagan, N. D., Upadhyaya, N., Tabe, L. M., and Higgins, T. J. V. (2003). The redistribution of protein sulfur in transgenic rice expressing a gene for a foreign, sulfur-rich protein. Plant J. 34, 1–11. doi: 10.1046/j.1365-313X.2003.01699.x
Häkkinen, S. T., Nuutila, A. M., and Ritala, A. (2018). “Modifying seeds to produce proteins,” in Proteins in Food Processing, ed. R. Y. Yada. (Cambridge: Woodhead Publishing), 413–441. doi: 10.1016/B978-0-08-100722-8.00017-6
Halpin, C. (2005). Gene stacking in transgenic plants – the challenge for 21st century plant biotechnology. Plant Biotechnol. J. 3, 141–155. doi: 10.1111/j.1467-7652.2004.00113.x
Han, M., Okamoto, M., Beatty, P. H., Rothstein, S. J., and Good, A. G. (2015). The genetics of nitrogen use efficiency in crop plants. Annu. Rev. Genet. 49, 269–289. doi: 10.1146/annurev-genet-112414-055037
Han, X., Wu, K., Fu, X., and Liu, Q. (2020). Improving coordination of plant growth and nitrogen metabolism for sustainable agriculture. aBIOTECH 1, 255–275. doi: 10.1007/s42994-020-00027-w
Hanafy, M. S., Rahman, S. M., Nakamoto, Y., Fujiwara, T., Naito, S., Wakasa, K., et al. (2013). Differential response of methionine metabolism in two grain legumes, soybean and azuki bean, expressing a mutated form of Arabidopsis cystathionine γ-synthase. J. Plant Physiol. 170, 338–345. doi: 10.1016/j.jplph.2012.10.018
Hatfield, J. L., and Walthall, C. L. (2015). Meeting global food needs: realizing the potential via genetics × environment × management interactions. Agron. J. 107, 1215–1226. doi: 10.2134/agronj15.0076
Havé, M., Marmagne, A., Chardon, F., and Masclaux-Daubresse, C. (2017). Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529. doi: 10.1093/jxb/erw365
Hawkesford, M. J., and Griffiths, S. (2019). Exploiting genetic variation in nitrogen use efficiency for cereal crop improvement. Curr. Opin. Plant Biol. 49, 35–42. doi: 10.1016/j.pbi.2019.05.003
Hawkesford, M. J., and Riche, A. B. (2020). Impacts of G x E x M on nitrogen use efficiency in wheat and future prospects. Front. Plant Sci. 11:1157. doi: 10.3389/fpls.2020.01157
Hirner, A., Ladwig, F., Stransky, H., Okumoto, S., Keinath, M., Harms, A., et al. (2006). Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18, 1931–1946. doi: 10.1105/tpc.106.041012
Hoyos, M. E., Palmieri, L., Wertin, T., Arrigoni, R., Polacco, J. C., and Palmieri, F. (2003). Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 33, 1027–1035. doi: 10.1046/j.1365-313X.2003.01685.x
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–878. doi: 10.1038/ng.3337
Hu, M., Zhao, X., Liu, Q., Hong, X., Zhang, W., Zhang, Y., et al. (2018). Transgenic expression of plastidic glutamine synthetase increases nitrogen uptake and yield in wheat. Plant Biotechnol. J. 16, 1858–1867. doi: 10.1111/pbi.12921
Huang, N.-C., Liu, K.-H., Lo, H.-J., and Tsay, Y.-F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Huang, S., Kruger, D. E., Frizzi, A., D'Ordine, R. L., Florida, C. A., Adams, W. R., et al. (2005). High-lysine corn produced by the combination of enhanced lysine biosynthesis and reduced zein accumulation. Plant Biotechnol. J. 3, 555–569. doi: 10.1111/j.1467-7652.2005.00146.x
Hunt, E., Gattolin, S., Newbury, H. J., Bale, J. S., Tseng, H. M., Barrett, D. A., et al. (2010). A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 61, 55–64. doi: 10.1093/jxb/erp274
Jahn, T. P., Møller, A. L., Zeuthen, T., Holm, L. M., Klærke, D. A., Mohsin, B., et al. (2004). Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett. 574, 31–36. doi: 10.1016/j.febslet.2004.08.004
Jensen, E. S., Carlsson, G., and Hauggaard-Nielsen, H. (2020). Intercropping of grain legumes and cereals improves the use of soil N resources and reduces the requirement for synthetic fertilizer N: A global-scale analysis. Agron. Sustain. Dev. 40:5. doi: 10.1007/s13593-020-0607-x
Ji, Y., Huang, W., Wu, B., Fang, Z., and Wang, X. (2020). The amino acid transporter AAP1 mediates growth and grain yield by regulating neutral amino acid uptake and reallocation in Oryza sativa. J. Exp. Bot. 71, 4763–4777. doi: 10.1093/jxb/eraa256
Jones, D. L., Healey, J. R., Willett, V. B., Farrar, J. F., and Hodge, A. (2005a). Dissolved organic nitrogen uptake by plants—an important N uptake pathway?. Soil Biol. Biochem. 37, 413–423. doi: 10.1016/j.soilbio.2004.08.008
Jones, D. L., Shannon, D., Junvee-Fortune, T., and Farrar, J. F. (2005b). Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biol. Biochem. 37, 179–181. doi: 10.1016/j.soilbio.2004.07.021
Ju, X., Liu, X., Zhang, F., and Roelcke, M. (2004). Nitrogen fertilization, soil nitrate accumulation, and policy recommendations in several agricultural regions of China. AMBIO 33, 300–305. doi: 10.1579/0044-7447-33.6.300
Kanter, D. R., Winiwarter, W., Bodirsky, B. L., Bouwman, L., Boyer, E., Buckle, S., et al. (2020). A framework for nitrogen futures in the shared socioeconomic pathways. Glob. Environ. Change 61:102029. doi: 10.1016/j.gloenvcha.2019.102029
Karachi, H., Shaul, O., and Galili, G. (1993). Seed-specific expression of a bacterial desensitized aspartate kinase increases the production of seed threonine and methionine in transgenic tobacco. Plant J. 3, 721–727. doi: 10.1111/j.1365-313X.1993.00721.x
Karachi, H., Shaul, O., and Galili, G. (1994). Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc. Natl. Acad. Sci. U.S.A. 91, 2577–2581. doi: 10.1073/pnas.91.7.2577
Katsube, T., Kurisaka, N., Ogawa, M., Maruyama, N., Ohtsuka, R., Utsumi, S., et al. (1999). Accumulation of soybean glycinin and its assembly with the glutelins in rice. Plant Phys. 120, 1063–1073. doi: 10.1104/pp.120.4.1063
Kawakatsu, T., Wang, S., Wakasa, Y., and Takaiwa, F. (2010). Increased lysine content in rice grains by over-accumulation of BiP in the endosperm. Biosci. Biotechnol. Biochem. 74, 2529–2531. doi: 10.1271/bbb.100619
Kendall, A. C., Wallsgrove, R. M., Hall, N. P., Turner, J. C., and Lea, P. J. (1986). Carbon and nitrogen metabolism in barley (Hordeum vulgare L.) mutants lacking ferredoxin-dependent glutamate synthase. Planta 168, 316–323. doi: 10.1007/BF00392355
Kita, Y., Nakamoto, Y., Takahashi, M., Kitamura, K., Wakasa, K., and Ishimoto, M. (2010). Manipulation of amino acid composition in soybean seeds by the combination of deregulated tryptophan biosynthesis and storage protein deficiency. Plant Cell Rep. 29, 87–95. doi: 10.1007/s00299-009-0800-5
Knoblauch, M., Knoblauch, J., Mullendore, D. L., Savage, J. A., Babst, B. A., Beecher, S. D., et al. (2016). Testing the Münch hypothesis of long distance phloem transport in plants. Elife 5:e15341. doi: 10.7554/eLife.15341
Koch, W., Kwart, M., Laubner, M., Heineke, D., Stransky, H., Frommer, W. B., et al. (2003). Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 33, 211–220. doi: 10.1046/j.1365-313X.2003.01618.x
Komarova, N. Y., Thor, K., Gubler, A., Meier, S., Dietrich, D., Weichert, A., et al. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148, 856–869. doi: 10.1104/pp.108.123844
Kopittke, P. M., Menzies, N. W., Wang, P., McKenna, B. A., and Lombi, E. (2019). Soil and the intensification of agriculture for global food security. Environ. Int. 132:105078. doi: 10.1016/j.envint.2019.105078
Kopriva, S., Malagoli, M., and Takahashi, H. (2019). Sulfur nutrition: Impacts on plant development, metabolism, and stress responses. J. Exp. Bot. 70, 4069–4073. doi: 10.1093/jxb/erz319
Krapp, A. (2015). Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr. Opin. Plant Biol. 25, 115–122. doi: 10.1016/j.pbi.2015.05.010
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A. S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Kumar, A., Kaiser, B. N., Siddiqi, M. Y., and Glass, A. D. (2006). Functional characterisation of OsAMT1.1 overexpression lines of rice, Oryza sativa. Funct. Plant Biol. 33, 339–346. doi: 10.1071/FP05268
Kusano, M., Tabuchi, M., Fukushima, A., Funayama, K., Diaz, C., Kobayashi, M., et al. (2011). Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J. 66, 456–466. doi: 10.1111/j.1365-313X.2011.04506.x
Ladwig, F., Stahl, M., Ludewig, U., Hirner, A. A., Hammes, U. Z., Stadler, R., et al. (2012). Siliques are Red1 from Arabidopsis acts as a bidirectional amino acid transporter that is crucial for the amino acid homeostasis of siliques. Plant Physiol. 158, 1643–1655. doi: 10.1104/pp.111.192583
Lam, H. M., Coschigano, K., Schultz, C., Melo-Oliveira, R., Tjaden, G., Oliveira, I., et al. (1995). Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7, 887–898. doi: 10.1105/tpc.7.7.887
Lam, H. M., Coschigano, K. T., Oliveira, I. C., Melo-Oliveira, R., and Coruzzi, G. M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Lancien, M., Martin, M., Hsieh, M. H., Leustek, T., Goodman, H., and Coruzzi, G. M. (2002). Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 29, 347–358. doi: 10.1046/j.1365-313X.2002.01218.x
Lanfermeijer, F., Koerselman-Kooij, J., Kollöffel, C., and Borstlap, A. (1989). Release of amino acids from cotyledons of developing seeds of pea (Pisum sativum L.). J. Plant Physiol. 134, 592–597. doi: 10.1016/S0176-1617(89)80153-1
Lea, P. J., and Azevedo, R. A. (2007). Nitrogen use efficiency. 2. Amino acid metabolism. Ann. Appl. Biol. 151, 269–275. doi: 10.1111/j.1744-7348.2007.00200.x
Lea, P. J., and Ireland, R. J. (1999). “Nitrogen metabolism in higher plants”, In Plant Amino Acids: Biochemistry and Biotechnology, ed. B. K. Singh (New York, NY: CRC Press), 1–47.
Lea, P. J. and Miflin, B. J. (2011). “Nitrogen assimilation and its relevance to crop improvement,” in Nitrogen Metabolism in Plants in the Post-Genomic Era. Annual Plant Reviews, Vol. 42 (Oxford: Wiley-Blackwell), 1–40. doi: 10.1002/9781119312994.apr0448
Lea, P. J., Sodek, L., Parry, M. A., Shewry, P. R., and Halford, N. G. (2007). Asparagine in plants. Ann. App. Biol. 150, 1–26. doi: 10.1111/j.1744-7348.2006.00104.x
Lee, M., Huang, T., Toro-Ramos, T., Fraga, M., Last, R. L., and Jander, G. (2008). Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J. 54, 310–320. doi: 10.1111/j.1365-313X.2008.03419.x
Lee, S., Marmagne, A., Park, J., Fabien, C., Yim, Y., Kim, S. J., et al. (2020a). Concurrent activation of OsAMT1; 2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 103, 7–20. doi: 10.1111/tpj.14794
Lee, S., Park, J., Lee, J., Shin, D., Marmagne, A., Lim, P. O., et al. (2020b). OsASN1 overexpression in rice increases grain protein content and yield under nitrogen-limiting conditions. Plant Cell Physiol. 61, 1309–1320. doi: 10.1093/pcp/pcaa060
Lee, S. I., Kim, H. U., Lee, Y., Suh, S., Lim, Y. P., Lee, H., et al. (2001). Constitutive and seed-specific expression of a maize lysine-feedback insensitive. Mol. Breeding 8, 75–84. doi: 10.1023/A:1011977219926
Lee, T. T. T., Chung, M. C., Kao, Y. W., Wang, C. S., Chen, L. J., and Tzen, J. T. C. (2005). Specific expression of a sesame storage protein in transgenic rice bran. J. Cereal Sci. 41, 23–29. doi: 10.1016/j.jcs.2004.08.014
Lee, T. T. T., Wang, M. M., Hou, R. C., Chen, L. J., Su, R. C., Wang, C. S., et al. (2003). Enhanced methionine and cysteine levels in transgenic rice seeds by the accumulation of sesame 2S albumin. Biosci. Biotechnol. Biochem. 67, 1699–1705. doi: 10.1271/bbb.67.1699
Lee, Y.-H., Foster, J., Chen, J., Voll, L., Weber, A., and Tegeder, M. (2007). AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 50, 305–316. doi: 10.1111/j.1365-313X.2007.03045.x
Lehmann, S., Gumy, C., Blatter, E., Boeffel, S., Fricke, W., and Rentsch, D. (2011). In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 62, 787–796. doi: 10.1093/jxb/erq320
Lemaire, G., and Ciampitti, I. (2020). Crop mass and N status as prerequisite covariables for unraveling nitrogen use efficiency across genotype-by-environment-by-management scenarios: a review. Plants 9:1309. doi: 10.3390/plants9101309
Li, M., Xu, J., Gao, Z., Tian, H., Gao, Y., and Kariman, K. (2020). Genetically modified crops are superior in their nitrogen use efficiency-A meta-analysis of three major cereals. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-65684-9
Li, P., Chen, F., Cai, H., Liu, J., Pan, Q., Liu, Z., et al. (2015). A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 66, 3175–3188. doi: 10.1093/jxb/erv127
Li, T., Liao, K., Xu, X., Gao, Y., Wang, Z., Zhu, X., et al. (2017). Wheat Ammonium Transporter (AMT) Gene family: diversity and possible role in host–pathogen interaction with stem rust. Front. Plant Sci. 8:1637. doi: 10.3389/fpls.2017.01637
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., and Glass, A. D. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Li, Y., Chang, S. X., Tian, L., and Zhang, Q. (2018). Conservation agriculture practices increase soil microbial biomass carbon and nitrogen in agricultural soils: A global meta-analysis. Soil Biol. Biochem. 121, 50–58. doi: 10.1016/j.soilbio.2018.02.024
Li, Z., Meyer, S., Essig, J. S., Liu, Y., Schapaugh, M. A., Muthukrishnan, S., et al. (2005). High-level expression of maize γ-zein protein in transgenic soybean (Glycine max). Mol. Breeding 16, 11–20. doi: 10.1007/s11032-004-7658-6
Li, Z., Moon, B. P., Xing, A., Liu, Z. B., McCardell, R. P., Damude, H. G., et al. (2010). Stacking multiple transgenes at a selected genomic site via repeated recombinase-mediated DNA cassette exchanges. Plant Phys. 154, 622–631. doi: 10.1104/pp.110.160093
Lightfoot, D. A., Mungur, R., Ameziane, R., Nolte, S., Long, L., Bernhard, K., et al. (2007). Improved drought tolerance of transgenic Zea mays plants that express the glutamate dehydrogenase gene (gdhA) of E. coli. Euphytica 156, 103–116. doi: 10.1007/s10681-007-9357-y
Liu, G., Ji, Y., Bhuiyan, N. H., Pilot, G., Selvaraj, G., Zou, J., et al. (2010). Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22, 3845–3863. doi: 10.1105/tpc.110.079392
Liu, Q., Chen, X., Wu, K., and Fu, X. (2015). Nitrogen signaling and use efficiency in plants: what's new?. Curr. Opin. Plant Biol. 27, 192–198. doi: 10.1016/j.pbi.2015.08.002
Liu, X., Zhang, C., Liu, Q., Yuan, D., Pan, G., Sun, S. S. M., et al. (2016). Development of high-lysine rice via endosperm-specific expression of a foreign LYSINE RICH PROTEIN gene. BMC Plant Biol. 16, 147–160. doi: 10.1186/s12870-016-0837-x
Liu, Y., Ahn, J.-E., Datta, S., Salzman, R. A., Moon, J., Huyghues-Despointes, B., et al. (2005). Arabidopsis vegetative storage protein is an insect acid phosphatase. Plant Physiol. 139, 1545–1556. doi: 10.1104/pp.105.066837
Liu, S., Wang, D., Mei, Y., Xia, T., Xu, W., Zhang, Y., et al. (2020). Overexpression of GmAAP6a enhances tolerance to low nitrogen and improves seed nitrogen status by optimizing amino acid partitioning in soybean. Plant Biotechnol. J. 18, 1749–1762. doi: 10.1111/pbi.13338
Lohaus, G., Winter, H., Riens, B., and Heldt, H. W. (1995). Further studies of the phloem loading process in leaves of barley and spinach. The comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Plant Biol. 108, 270–275. doi: 10.1111/j.1438-8677.1995.tb00860.x
Loqué, D., Ludewig, U., Yuan, L., and von Wirén, N. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. doi: 10.1104/pp.104.051268
Loqué, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Lothier, J., Gaufichon, L., Sormani, R., Lemaître, T., Azzopardi, M., Morin, H., et al. (2011). The cytosolic glutamine synthetase GLN1; 2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J. Exp. Bot. 62, 1375–1390. doi: 10.1093/jxb/erq299
Lu, K., Wu, B., Wang, J., Zhu, W., Nie, H., Qian, J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 16, 1710–1722. doi: 10.1111/pbi.12907
Lu, M. Z., Snyder, R., Grant, J., and Tegeder, M. (2020). Manipulation of sucrose phloem and embryo loading affects pea leaf metabolism, carbon and nitrogen partitioning to sinks as well as seed storage pools. Plant J. 101, 217–236. doi: 10.1111/tpj.14533
Luo, L., Zhang, Y., and Xu, G. (2020). How does nitrogen shape plant architecture?. J. Exp. Bot. 71, 4415–4427. doi: 10.1093/jxb/eraa187
MacKown, C. T., Van Sanford, D. A., and Zhang, N. (1992). Wheat vegetative nitrogen compositional changes in response to reduced reproductive sink strength. Plant Phys. 99, 1469–1474. doi: 10.1104/pp.99.4.1469
Mandal, S., Shurin, J. B., Efroymson, R. A., and Mathews, T. J. (2018). Heterogeneity in nitrogen sources enhances productivity and nutrient use efficiency in algal polycultures. Environ. Sci. Technol. 52, 3769–3776. doi: 10.1021/acs.est.7b05318
Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. doi: 10.1105/tpc.106.042689
Martinez-Feria, R. A., Castellano, M. J., Dietzel, R. N., Helmers, M. J., Liebman, M., Huber, I., et al. (2018). Linking crop-and soil-based approaches to evaluate system nitrogen-use efficiency and tradeoffs. Agric. Ecosyst. Environ. 256, 131–143. doi: 10.1016/j.agee.2018.01.002
Masclaux, C., Valadier, M. H., Brugière, N., Morot-Gaudry, J. F., and Hirel, B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. doi: 10.1007/s004250000310
Masclaux-Daubresse, C., and Chardon, F. (2011). Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 62, 2131–2142. doi: 10.1093/jxb/erq405
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Masclaux-Daubresse, C., Reisdorf-Cren, M., Pageau, K., Lelandais, M., Grandjean, O., Kronenberger, J., et al. (2006). Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 140, 444–456. doi: 10.1104/pp.105.071910
Matityahu, I., Godo, I., Hacham, Y., and Amir, R. (2013). Tobacco seeds expressing feedback-insensitive cystathionine gamma-synthase exhibit elevated content of methionine and altered primary metabolic profile. BMC Plant Biol. 13:206. doi: 10.1186/1471-2229-13-206
Matson, P. A., Parton, W. J., Power, A. G., and Swift, M. J. (1997). Agricultural intensification and ecosystem properties. Science 277, 504–509. doi: 10.1126/science.277.5325.504
Mauceri, A., Bassolino, L., Lupini, A., Badeck, F., Rizza, F., Schiavi, M., et al. (2020). Genetic variation in eggplant for Nitrogen Use Efficiency under contrasting NO3-supply. J. Integr. Plant Biol. 62, 487–508. doi: 10.1111/jipb.12823
McAllister, C. H., Beatty, P. H., and Good, A. G. (2012). Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 10, 1011–1025. doi: 10.1111/j.1467-7652.2012.00700.x
McAllister, C. H., and Good, A. G. (2015). Alanine aminotransferase variants conferring diverse NUE phenotypes in Arabidopsis thaliana. PLoS ONE 10:e0121830. doi: 10.1371/journal.pone.0121830
Meister, R., Rajani, M. S., Ruzicka, D., and Schachtman, D. P. (2014). Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 19, 779–788. doi: 10.1016/j.tplants.2014.08.005
Melo-Oliveira, R., Oliveira, I. C., and Coruzzi, G. M. (1996). Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc. Natl. Acad. Sci. U.S.A. 93, 4718–4723. doi: 10.1073/pnas.93.10.4718
Messina, C. D., Cooper, M., Reynolds, M. P., and Hammer, G. L. (2020). Crop science: A foundation for advancing predictive agriculture. Crop Sci. 60, 544–546. doi: 10.1002/csc2.20116
Miflin, B. J., and Habash, D. Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987. doi: 10.1093/jexbot/53.370.979
Miflin, B. J., and Lea, P. J. (1977). Amino acid metabolism. Annu. Rev. Plant Physiol. 28, 299–329. doi: 10.1146/annurev.pp.28.060177.001503
Migge, A., Bork, C., Hell, R., and Becker, T. W. (2000a). Negative regulation of nitrate reductase gene expression by glutamine or asparagine accumulating in leaves of sulfur-deprived tobacco. Planta 211, 587–595. doi: 10.1007/s004250000322
Migge, A., Carrayol, E., Hirel, B., and Becker, T. W. (2000b). Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210, 252–260. doi: 10.1007/PL00008132
Millard, P. (1988). The accumulation and storage of nitrogen by herbaceous plants. Plant Cell Environ. 11, 1–8. doi: 10.1111/j.1365-3040.1988.tb01769.x
Miller, A. E., Bowman, W. D., and Suding, K. N. (2007). Plant uptake of inorganic and organic nitrogen: neighbor identity matters. Ecology 88, 1832–1840. doi: 10.1890/06-0946.1
Miller, A. J., Fan, X., Shen, Q., and Smith, S. J. (2008). Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J. Exp. Bot. 59, 111–119. doi: 10.1093/jxb/erm208
Milne, R. J., Grof, C. P., and Patrick, J. W. (2018). Mechanisms of phloem unloading: shaped by cellular pathways, their conductances and sink function. Curr. Opin. Plant Biol. 43, 8–15. doi: 10.1016/j.pbi.2017.11.003
Miyashita, Y., Dolferus, R., Ismond, K. P., and Good, A. G. (2007). Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 49, 1108–1121. doi: 10.1111/j.1365-313X.2006.03023.x
Moison, M., Marmagne, A., Dinant, S., Soulay, F., Azzopardi, M., Lothier, J., et al. (2018). Three cytosolic glutamine synthetase isoforms located in different order veins work together for N remobilization and seed filling in arabidopsis. J. Exp. Bot. 69, 4379–4393. doi: 10.1093/jxb/ery217
Moll, R., Kamprath, E., and Jackson, W. (1982). Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron. J. 74, 562–564. doi: 10.2134/agronj1982.00021962007400030037x
Molvig, L., Tabe, L. M., Eggum, B. O., Moore, A. E., Craig, S., Spencer, D., et al. (1997). Enhanced methionine levels and increased nutritive value of seeds of transgenic lupids (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 8393–8398. doi: 10.1073/pnas.94.16.8393
Momma, K., Hashimoto, W., Ozawa, S., Kawai, S., Katsube, T., Takaiwa, F., et al. (1999). Quality and safety evaluation of genetically engineered rice with soybean glycinin: Analyses of the grain composition and digestibility of glycinin in transgenic rice. Biosci. Biotechnol. Biochem. 63, 314–318. doi: 10.1271/bbb.63.314
Moran-Zuloaga, D., Dippold, M., Glaser, B., and Kuzyakov, Y. (2015). Organic nitrogen uptake by plants: reevaluation by position-specific labeling of amino acids. Biogeochemistry 125, 359–374. doi: 10.1007/s10533-015-0130-3
Mueller, N. D., West, P. C., Gerber, J. S., MacDonald, G. K., Polasky, S., and Foley, J. A. (2014). A tradeoff frontier for global nitrogen use and cereal production. Environ. Res. Lett. 9:P054002. doi: 10.1088/1748-9326/9/5/054002
Müller, B., Fastner, A., Karmann, J., Mansch, V., Hoffmann, T., Schwab, W., et al. (2015). Amino acid export in developing Arabidopsis seeds depends on UmamiT facilitators. Curr. Biol. 25, 3126–3131. doi: 10.1016/j.cub.2015.10.038
Murray, D. R. (1987). Nutritive role of seedcoats in developing legume seeds. Am. J. Bot. 74, 1122–1137. doi: 10.1002/j.1537-2197.1987.tb08724.x
Naqvi, S., Farre, G., Sanahuja, G., Capell, T., Zhu, C., and Christou, P. (2010). When more is better: multigene engineering in plants. Trends Plant Sci. 15, 48–56. doi: 10.1016/j.tplants.2009.09.010
Näsholm, T., Kielland, K., and Ganeteg, U. (2009). Uptake of organic nitrogen by plants. New Phytol. 182, 31–48. doi: 10.1111/j.1469-8137.2008.02751.x
Nguyen, G. N., Joshi, S., and Kant, S. (2017). “Water availability and nitrogen use in plants: effects, interaction, and underlying molecular mechanisms,“ in Plant Macronutrient Use Efficiency, ed. M. A. Hossain, T. Kamiya, D. J. Burritt, L. P. Tran, T. Fujiwara (San Diego, CA: Academic Press), 233–243. doi: 10.1016/B978-0-12-811308-0.00013-2
Nguyen, G. N., and Kant, S. (2018). Improving nitrogen use efficiency in plants: effective phenotyping in conjunction with agronomic and genetic approaches. Funct. Plant Biol. 45, 606–619. doi: 10.1071/FP17266
Nunes-Nesi, A., Fernie, A. R., and Stitt, M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996. doi: 10.1093/mp/ssq049
Offler, C. E., McCurdy, D. W., Patrick, J. W., and Talbot, M. J. (2003). Transfer cells: cells specialized for a special purpose. Annu. Rev. Plant Biol. 54, 431–454. doi: 10.1146/annurev.arplant.54.031902.134812
Okumoto, S., Koch, W., Tegeder, M., Fischer, W. N., Biehl, A., Leister, D., et al. (2004). Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J. Exp. Bot. 55, 2155–2168. doi: 10.1093/jxb/erh233
Oliveira, I. C., Brears, T., Knight, T. J., Clark, A., and Coruzzi, G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1170–1180. doi: 10.1104/pp.020013
Osanai, T., Kuwahara, A., Otsuki, H., Saito, K., and Yokota Hirai, M. (2017). ACR11 is an activator of plastid-type glutamine synthetase GS2 in Arabidopsis thaliana. Plant Cell Physiol. 58, 650–657. doi: 10.1093/pcp/pcx033
Palmieri, L., Todd, C. D., Arrigoni, R., Hoyos, M. E., Santoro, A., Polacco, J. C., et al. (2006). Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim. Biophys. Acta (BBA)-Bioenergetics 1757, 1277–1283. doi: 10.1016/j.bbabio.2006.03.025
Pandurangan, S., Pajak, A., Molnar, S. J., Cober, E. R., Dhaubhadel, S., Hernández-Sebastiá, C., et al. (2012). Relationship between asparagine metabolism and protein concentration in soybean seed. J. Exp. Bot. 63, 3173–3184. doi: 10.1093/jxb/ers039
Pate, J. S., Sharkey, P. J., and Lewis, O. A. M. (1975). Xylem to phloem transfer of solutes in fruiting shoots of legumes, studied by a phloem bleeding technique. Planta 122, 11–26. doi: 10.1007/BF00385400
Patrick, J. W. (1997). Phloem unloading: sieve element unloading and post-sieve element transport. Annu. Rev. Plant Biol. 48, 191–222. doi: 10.1146/annurev.arplant.48.1.191
Patrick, J. W. (2013). “Fundamentals of phloem transport physiology,” in Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions, ed. G. A. Thompson and A. J. E. van Bel (Oxford: Wiley-Blackwell), 30–59. doi: 10.1002/9781118382806.ch3
Patrick, J. W., and Offler, C. E. (2001). Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 52, 551–564. doi: 10.1093/jexbot/52.356.551
Paungfoo-Lonhienne, C., Lonhienne, T. G. A., Rentsch, D., Robinson, N., Christie, M., Webb, R. I., et al. (2008). Plants can use protein as a nitrogen source without assistance from other organisms. Proc. Natl. Acad. Sci. U.S.A. 105, 4524–4529. doi: 10.1073/pnas.0712078105
Pélissier, H., and Tegeder, M. (2007). PvUPS1 plays a role in source-sink transport of allantoin in French bean (Phaseolus vulgaris). Funct. Plant Biol. 18, 282–291. doi: 10.1071/FP06277
Peña, P. A., Quach, T., Sato, S., Ge, Z., Nersesian, N., Dweikat, I. M., et al. (2017). Molecular and phenotypic characterization of transgenic wheat and sorghum events expressing the barley alanine aminotransferase. Planta 246, 1097–1107. doi: 10.1007/s00425-017-2753-1
Peng, B., Guan, K., Tang, J., Ainsworth, E. A., Asseng, S., Bernacchi, C. J., et al. (2020). Towards a multiscale crop modelling framework for climate change adaptation assessment. Nat. Plants 6, 338–348. doi: 10.1038/s41477-020-0625-3
Peng, B., Kong, H., Li, Y., Wang, L., Zhong, M., Sun, L., et al. (2014). OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5, 1–12. doi: 10.1038/ncomms5847
Perchlik, M., Foster, J., and Tegeder, M. (2014). Different and overlapping functions of Arabidopsis LHT6 and AAP1 transporters in root amino acid uptake. J. Exp. Bot. 65, 5193–5204. doi: 10.1093/jxb/eru278
Perchlik, M., and Tegeder, M. (2017). Improving plant nitrogen use efficiency through alteration of amino acid transport processes. Plant Physiol. 175, 235–247. doi: 10.1104/pp.17.00608
Perchlik, M., and Tegeder, M. (2018). Leaf amino acid supply affects photosynthetic and plant nitrogen use efficiency under nitrogen stress. Plant Physiol. 178, 174–188. doi: 10.1104/pp.18.00597
Pickardt, T., Saalbach, I., Waddell, D., Meixner, M., Muntz, K., and Schieder, O. (1995). Seed specific expression of the 2S albumin gene from Brazil net (Bertholletia excelsa) in transgenic Vicia narbonensis. Mol. Breeding 1, 295–301. doi: 10.1007/BF02277429
Plett, D. C., Ranathunge, K., Melino, V. J., Kuya, N., Uga, Y., and Kronzucker, H. J. (2020). The intersection of nitrogen nutrition and water use in plants: new paths toward improved crop productivity. J. Exp. Bot. 71, 4452–4468. doi: 10.1093/jxb/eraa049
Poorter, H., and Evans, J. R. (1998). Photosynthetic nitrogen-use efficiency of species that differ inherently in specific leaf area. Oecologia 116, 26–37. doi: 10.1007/s004420050560
Potel, F., Valadier, M. H., Ferrario-Méry, S., Grandjean, O., Morin, H., Gaufichon, L., et al. (2009). Assimilation of excess ammonium into amino acids and nitrogen translocation in Arabidopsis thaliana–roles of glutamate synthases and carbamoylphosphate synthetase in leaves. FEBS J. 276, 4061–4076. doi: 10.1111/j.1742-4658.2009.07114.x
Purnell, M. P., Skopelitis, D. S., Roubelakis-Angelakis, K. A., and Botella, J. R. (2005). Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222, 167–180. doi: 10.1007/s00425-005-1510-z
Qi, Q., Huang, J., Crowley, J., Ruschke, L., Goldman, B. S., Wen, L., et al. (2011). Metabolically engineered soybean seed with enhanced threonine levels: biochemical characterization and seed-specific expression of lysine-insensitive variants of aspartate kinases from the enteric bacterium Xenohabdus bovienii. Plant Biotechnol. J. 9, 193–204. doi: 10.1111/j.1467-7652.2010.00545.x
Raghuram, N., and Sharma, N. (2019). “Improving Crop Nitrogen Use Efficiency.” In Comprehensive Biotechnology, Vol. 4, ed. M. Moo-Young (Pergamon: Elsevier), 211-220. doi: 10.1016/B978-0-444-64046-8.00222-6
Rainbird, R. M., Thorne, J. H., and Hardy, R. W. (1984). Role of amides, amino acids, and ureides in the nutrition of developing soybean seeds. Plant Physiol. 74, 329–334. doi: 10.1104/pp.74.2.329
Ranathunge, K., El-kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced permeability show superior growth and higher yield under optimal and suboptimal conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Ranocha, P., McNeil, S. D., Ziemak, M. J., Li, C., Tarczynski, M. C., and Hanson, A. D. (2001). The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 25, 575–584. doi: 10.1046/j.1365-313x.2001.00988.x
Rascón-Cruz, Q., Sinagawa-Garca, S., Osuna-Castro, J. A., Bohorova, N., and Paresdes-López, O. (2004). Accumulation, assembly, and digestibility of amarantin expressed in transgenic tropical maize. Theor. Appl. Genet. 108, 335–342. doi: 10.1007/s00122-003-1430-x
Raun, W., and Johnson, G. (1999). Improving nitrogen use efficiency for cereal production. Agron. J. 91, 357–363. doi: 10.2134/agronj1999.00021962009100030001x
Rawat, S. R., Silim, S. N., Kronzucker, H. J., Siddiqi, M. Y., and Glass, A. D. (1999). AtAMT1 gene expression and uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J. 19, 143–152. doi: 10.1046/j.1365-313X.1999.00505.x
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U.S.A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Renné, P., Dreßen, U., Hebbeker, U., Hille, D., Flügge, U. I., Westhoff, P., et al. (2003). The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35, 316–331. doi: 10.1046/j.1365-313X.2003.01806.x
Robinson, S. A., Slade, A. P., Fox, G. G., Phillips, R., Ratcliffe, R. G., and Stewart, G. R. (1991). The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 95, 509–516. doi: 10.1104/pp.95.2.509
Rochat, C., and Boutin, J. P. (1991). Metabolism of phloem-borne amino acids in maternal tissues (Pisum sativum L.). J. Exp. Bot. 42, 207–214. doi: 10.1093/jxb/42.2.207
Rochat, C., and Boutin, J. P. (1992). Temporary storage compounds and sucrose-starch metabolism in seed coats during pea seed development (Pisum sativum). Physiol. Plantarum 85, 567–572. doi: 10.1111/j.1399-3054.1992.tb04756.x
Rolland, N., Curien, G., Finazzi, G., Kuntz, M., Maréchal, E., Matringe, M., et al. (2012). The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu. Rev. Genet. 46, 233–264. doi: 10.1146/annurev-genet-110410-132544
Rolletschek, H., Hosein, F., Miranda, M., Heim, U., Götz, K. P., Schlereth, A., et al. (2005). Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 137, 1236–1249. doi: 10.1104/pp.104.056523
Rosche, E., Blackmore, D., Tegeder, M., Richardson, T., Schroeder, H., Higgins, T. J. V., et al. (2002). Seed-specific expression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 30, 165–175. doi: 10.1046/j.1365-313X.2002.01282.x
Rotundo, J. L., Borrás, L., Westgate, M. E., and Orf, J. H. (2009). Relationship between assimilate supply per seed during seed filling and soybean seed composition. Field Crops Res. 112, 90–96. doi: 10.1016/j.fcr.2009.02.004
Saalbach, I., Wadell, D., Pickardt, T., Schieder, O., and Muntz, K. (1995). Stable expression of the sulfur-rich 2S albumin gene in transgenic Vicia narbonensis increases the methionine content of seeds grain legumes. J. Plant Physiol. 145, 674–681. doi: 10.1016/S0176-1617(11)81280-0
Sanders, A., Collier, R., Trethewy, A., Gould, G., Sieker, R., and Tegeder, M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59, 540–552. doi: 10.1111/j.1365-313X.2009.03890.x
Santiago, J. P., and Tegeder, M. (2016). Connecting source with sink: the role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 171, 508–521. doi: 10.1104/pp.16.00244
Santiago, J. P., and Tegeder, M. (2017). Implications of nitrogen phloem loading for carbon metabolism and transport during Arabidopsis development. J. Integr. Plant Biol. 59, 409–421. doi: 10.1111/jipb.12533
Schmidt, R., Stransky, H., and Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226, 805–803. doi: 10.1007/s00425-007-0527-x
Schobert, C., and Komor, E. (1990). Transfer of amino acids and nitrate from the roots into the xylem of Ricinus communis seedlings. Planta 181, 85–90. doi: 10.1007/BF00202328
Schoenbeck, M. A., Temple, S. J., Trepp, G. B., Blumenthal, J. M., Samac, D. A., Gantt, J. S., et al. (2000). Decreased NADH glutamate synthase activity in nodules and flowers of alfalfa (Medicago sativa L.) transformed with an antisense glutamate synthase transgene. J. Exp. Bot. 51, 29–39. doi: 10.1093/jxb/51.342.29
Schulze-Siebert, D., Heineke, D., Scharf, H., and Schultz, G. (1984). Pyruvate-derived amino acids in spinach chloroplasts: synthesis and regulation during photosynthetic carbon metabolism. Plant Physiol. 76, 465–471. doi: 10.1104/pp.76.2.465
Seiffert, B., Zhou, Z., Wallbraun, M., Lohaus, G., and Möllers, C. (2004). Expression of a bacterial asparagine synthetase gene in oilseed rape (Brassica napus) and its effect on traits related to nitrogen efficiency. Physiol. Plant. 121, 656–665. doi: 10.1111/j.1399-3054.2004.00361.x
Shrawat, A. K., Carroll, R. T., DePauw, M., Taylor, G. J., and Good, A. G. (2008). Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 6, 722–732. doi: 10.1111/j.1467-7652.2008.00351.x
Sieciechowicz, K. A., Joy, K. W., and Ireland, R. J. (1988). The metabolism of asparagine in plants. Phytochemistry 27, 663–671. doi: 10.1016/0031-9422(88)84071-8
Simmonds, N. W. (1995). The relation between yield and protein in cereal grain. J. Sci. Food. Agric. 67, 309–315. doi: 10.1002/jsfa.2740670306
Simons, M., Saha, R., Guillard, L., Clément, G., Armengaud, P., Cañas, R., et al. (2014). Nitrogen-use efficiency in maize (Zea mays L.): From ‘omics’ studies to metabolic modelling. J. Exp. Bot. 65, 5657–5671. doi: 10.1093/jxb/eru227
Sinha, V. B., Jangam, A. P., and Raghuram, N. (2020). “Biological determinants of crop nitrogen use efficiency and biotechnological avenues for improvement.” In Just Enough Nitrogen, ed. Sutton M. A. (Cham: Springer), 157–171. doi: 10.1007/978-3-030-58065-0_11
Snowden, C. J., Thomas, B., Baxter, C. J., Smith, J. A. C., and Sweetlove, L. J. (2015). A tonoplast Glu/Asp/GABA exchanger that affects tomato fruit amino acid composition. Plant J. 81, 651–660. doi: 10.1111/tpj.12766
Snyman, S. J., Hajari, E., Watt, M. P., Lu, Y., and Kridl, J. C. (2015). Improved nitrogen use efficiency in transgenic sugarcane: phenotypic assessment in a pot trial under low nitrogen conditions. Plant Cell Rep. 34, 667–669. doi: 10.1007/s00299-015-1768-y
Somerville, C. R., and Ogren, W. L. (1980). Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286, 257–259. doi: 10.1038/286257a0
Song, S., Hou, W., Good, I., Wu, C., Yu, Y., Matityahu, I., et al. (2013). Soybean seeds expressing feedback-insensitive cystathionine γ-synthase exhibit a higher content of methionine. J. Exp. Bot. 64, 1917–1926. doi: 10.1093/jxb/ert053
Sonoda, Y., Ikeda, A., Saiki, S., Wirén, N. V., Yamaya, T., and Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1–1;3) in rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Staswick, P. E. (1994). Storage proteins of vegetative plant tissues. Annu. Rev. Plant Biol. 45, 303–322. doi: 10.1146/annurev.pp.45.060194.001511
Streeter, J. (1979). Allantoin and allantoic acid in tissues and stem exudate from filed grown soybean plants. Plant Physiol. 63, 478–480. doi: 10.1104/pp.63.3.478
Suzuki, K., and Ohsumi, Y. (2007). Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581, 2156–2161. doi: 10.1016/j.febslet.2007.01.096
Svennerstam, H., Ganeteg, U., Bellini, C., and Näsholm, T. (2007). Comprehensive screening of Arabidopsis mutants suggests the Lysine Histidine Transporter 1 to be involved in plant uptake of amino acids. Plant Physiol. 143, 1853–1860. doi: 10.1104/pp.106.092205
Svennerstam, H., Ganeteg, U., and Näsholm, T. (2008). Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol. 180, 620–630. doi: 10.1111/j.1469-8137.2008.02589.x
Svennerstam, H., Jämtgård, S., Ahmad, I., Huss-Danell, K., Näsholm, T., and Ganeteg, U. (2011). Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. New Phytol. 191, 459–467. doi: 10.1111/j.1469-8137.2011.03699.x
Swarbreck, S. M., Wang, M., Wang, Y., Kindred, D., Sylvester-Bradley, R., Shi, W., et al. (2019). A roadmap for lowering crop nitrogen requirement. Trends Plant Sci. 24, 892–904. doi: 10.1016/j.tplants.2019.06.006
Sweetlove, L. J., Nielsen, J., and Fernie, A. R. (2017). Engineering central metabolism–a grand challenge for plant biologists. Plant J. 90, 749–763. doi: 10.1111/tpj.13464
Tabe, L. M., and Droux, M. (2002). Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol. 128, 1137–1148. doi: 10.1104/pp.010935
Tabe, L. M., Wirtz, M., Molvig, L., Droux, M., and Hell, R. (2010). Overexpression of serine acetyltransferase produced large increases in O-acetylserine and free cysteine in developing seeds of a grain legume. J. Exp. Bot. 61, 721–733. doi: 10.1093/jxb/erp338
Takabayashi, A., Niwata, A., and Tanaka, A. (2016). Direct interaction with ACR11 is necessary for post-transcriptional control of GLU1-encoded ferredoxin-dependent glutamate synthase in leaves. Sci. Rep. 6, 1–10. doi: 10.1038/srep29668
Takaiwa, F., Katsube, T., Kitagawa, S., Hisago, T., Kito, M., and Utsumi, S. (1995). High level accumulation of soybean glycinin in vacuole-derived protein bodies in the endosperm tissue of transgenic tobacco seed. Plant Sci. 111, 39–49. doi: 10.1016/0168-9452(95)04215-G
Tamura, W., Hidaka, Y., Tabuchi, M., Kojima, S., Hayakawa, T., Sato, T., et al. (2010). Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 39, 1003–1012. doi: 10.1007/s00726-010-0531-5
Tan, Q., Zhang, L., Grant, J., Cooper, P., and Tegeder, M. (2010). Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol. 154, 1886–1896. doi: 10.1104/pp.110.166389
Tan, X., Li, K., Wang, Z., Zhu, K., Tan, X., and Cao, J. (2019). A review of plant vacuoles: formation, located proteins, and functions. Plants 8:327. doi: 10.3390/plants8090327
Tang, D., Peng, Y., Lin, J., Du, C., Yang, Y., Wang, D., et al. (2018). Ectopic expression of fungal EcGDH improves nitrogen assimilation and grain yield in rice. J. Integr. Plant Biol. 60, 85–88. doi: 10.1111/jipb.12519
Taniguchi, M., and Miyake, H. (2012). Redox-shuttling between chloroplast and cytosol: integration of intra-chloroplast and extra-chloroplast metabolism. Curr. Opin. Plant Biol. 15, 252–260. doi: 10.1016/j.pbi.2012.01.014
Tegeder, M. (2012). Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol. 15, 315–321. doi: 10.1016/j.pbi.2012.02.001
Tegeder, M. (2014). Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J. Exp. Bot. 65, 1865–1878. doi: 10.1093/jxb/eru012
Tegeder, M., and Hammes, U. Z. (2018). The way out and in: phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 43, 16–21. doi: 10.1016/j.pbi.2017.12.002
Tegeder, M., and Masclaux-Daubresse, C. (2018). Source and sink mechanisms of nitrogen transport and use. New Phytol. 217, 35–53. doi: 10.1111/nph.14876
Tegeder, M., Offler, C., Frommer, W., and Patrick, J. W. (2000). Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol. 122, 319–325. doi: 10.1104/pp.122.2.319
Tegeder, M., and Rentsch, D. (2010). Uptake and partitioning of amino acids and peptides. Mol. Plant 3, 997–1011. doi: 10.1093/mp/ssq047
Tegeder, M., Ruan, Y. L., and Patrick, J. W. (2013). “Roles of membrane transporters in phloem functions,” in Biochemistry of Phloem, eds G. Thompson, A. Van Bel, (Oxford: Wiley-Blackwell), 63–101. doi: 10.1002/9781118382806.ch4
Tegeder, M., Tan, Q., Grennan, A. K., and Patrick, J. W. (2007). Amino acid transporter expression and localisation studies in pea (Pisum sativum). Funct. Plant Biol. 34, 1019–1028. doi: 10.1071/FP07107
Tegeder, M., and Ward, J. M. (2012). Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3:21. doi: 10.3389/fpls.2012.00021
Tercé-Laforgue, T., Bedu, M., Dargel-Grafin, C., Dubois, F., Gibon, Y., Restivo, F. M., et al. (2013). Resolving the role of plant glutamate dehydrogenase: II. Physiological characterization of plants overexpressing the two enzyme subunits individually or simultaneously. Plant Cell Physiol. 54, 1635–1647. doi: 10.1093/pcp/pct108
Thomsen, H. C., Eriksson, D., Møller, I. S., and Schjoerring, J. K. (2014). Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency?. Trends Plant Sci. 19, 656–663. doi: 10.1016/j.tplants.2014.06.002
Thornton, B. (2004). Inhibition of nitrate influx by glutamine in Lolium perenne depends upon the contribution of the HATS to the total influx. J. Exp. Bot. 55, 761–769. doi: 10.1093/jxb/erh066
Thu, S. W., Lu, M. Z., Carter, A. M., Collier, R., Gandin, A., Sitton, C. C., et al. (2020). Role of ureides in source-to-sink transport of photoassimilates in non-fixing soybean. J. Exp. Bot. 71, 4495–4511. doi: 10.1093/jxb/eraa146
Toka, I., Planchais, S., Cabassa, C., Justin, A. M., De Vos, D., Richard, L., et al. (2010). Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol. 152, 1851–1862. doi: 10.1104/pp.109.152371
Triboi, E., Martre, P., Girousse, C., Ravel, C., and Triboi-Blondel, A. M. (2006). Unravelling environmental and genetic relationships between grain yield and nitrogen concentration for wheat. Eur. J. Agron. 25, 108–118. doi: 10.1016/j.eja.2006.04.004
Tsay, Y. F., Chiu, C. C., Tsai, C. B., Ho, C. H., and Hsu, P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581, 2290–2300. doi: 10.1016/j.febslet.2007.04.047
Turgeon, R., and Wolf, S. (2009). Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60, 207–221. doi: 10.1146/annurev.arplant.043008.092045
Ufaz, S., and Galili, G. (2008). Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 147, 954–961. doi: 10.1104/pp.108.118091
Urquhart, A. A., and Joy, K. W. (1982). Transport, metabolism, and redistribution of xylem-bourne amino acids in developing pea shoots. Plant Physiol. 69, 1226–1232. doi: 10.1104/pp.69.5.1226
Urriola, J., and Rathore, K. S. (2015). Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res. 24, 397–407. doi: 10.1007/s11248-014-9852-6
van Bel, A. J. (1990). Xylem-phloem exchange via the rays: the undervalued route of transport. J. Exp. Bot. 41, 631–644. doi: 10.1093/jxb/41.6.631
van Bel, A. J. E. (1984). Quantification of the xylem-to-phloem transfer of amino acids by use of inulin [14C] carboxylic acid as xylem transport marker. Plant Sci. Lett. 35, 81–85. doi: 10.1016/0304-4211(84)90162-7
van Bueren, E. T. L., and Struik, P. C. (2017). Diverse concepts of breeding for nitrogen use efficiency. A review. Agron. Sustain. Dev. 37:50. doi: 10.1007/s13593-017-0457-3
Vidal, E. A., Alvarez, J. M., Araus, V., Riveras, E., Brooks, M., Krouk, G., et al. (2020). Nitrate 2020: Thirty years from transport to signaling networks. Plant Cell 32, 2094–2119. doi: 10.1105/tpc.19.00748
Vidmar, J. J., Zhuo, D., Siddiqi, M. Y., Schjoerring, J. K., Touraine, B., and Glass, A. D. (2000). Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol. 123, 307–318. doi: 10.1104/pp.123.1.307
Wallsgrove, R. M., Keys, A. J., Lea, P. J., and Miflin, B. J. (1983). Photosynthesis, photorespiration and nitrogen metabolism. Plant Cell Environ. 6, 301–309. doi: 10.1111/1365-3040.ep11612102
Wang, G., Xu, M., Wang, W., and Galili, G. (2017). Fortifying horticultural crops with essential amino acids: a review. Int. J. Mol. Sci. 18, 1306–1316. doi: 10.3390/ijms18061306
Wang, J., Wu, B., Lu, K., Wei, Q., Qian, J., Chen, Y., et al. (2019). The amino acid permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 180, 1031–1045. doi: 10.1104/pp.19.00034
Wang, M., Liu, C., Li, S., Zhu, D., Zhao, Q., and Yu, J. (2013b). Improved nutritive quality and slat resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int. J. Mol. Sci. 14, 9459–9474. doi: 10.3390/ijms14059459
Wang, W., Hu, B., Li, A., and Chu, C. (2020). NRT1.1s in plants: functions beyond nitrate transport. J. Exp. Bot. 71, 4373–4379. doi: 10.1093/jxb/erz554
Wang, W., Hu, B., Yuan, D., Liu, Y., Che, R., Hu, Y., et al. (2018b). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6. 3 confers high yield and early maturation in rice. Plant Cell 30, 638–651. doi: 10.1105/tpc.17.00809
Wang, W., Xu, M., Wang, G., and Galili, G. (2018c). New insights into the metabolism of aspartate-family amino acids in plant seeds. Plant Reprod. 31, 203–211. doi: 10.1007/s00497-018-0322-9
Wang, Y., Fu, B., Pan, L., Chen, L., Fu, X., and Li, K. (2013a). Overexpression of Arabidopsis Dof1, GS1 and GS2 enhanced nitrogen assimilation in transgenic tobacco grown under low-nitrogen conditions. Plant Mol. Biol. Rep. 31, 886–900. doi: 10.1007/s11105-013-0561-8
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018a). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Weber, H., Borisjuk, L., Heim, U., Buchner, P., and Wobus, U. (1995). Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. Plant Cell 7, 1835–1846. doi: 10.1105/tpc.7.11.1835
Weber, H., Borisjuk, L., and Wobus, U. (2005). Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 56, 253–279. doi: 10.1146/annurev.arplant.56.032604.144201
Weber, H., Heim, U., Golombek, S., Borisjuk, L., and Wobus, U. (1998). Assimilate uptake and the regulation of seed development. Seed Sci. Res. 8, 331–346. doi: 10.1017/S0960258500004268
Weigelt, K., Küster, H., Radchuk, R., Müller, M., Weichert, H., Fait, A., et al. (2008). Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 55, 909–926. doi: 10.1111/j.1365-313X.2008.03560.x
Widhalm, J. R., Gutensohn, M., Yoo, H., Adebesin, F., Qian, Y., Guo, L., et al. (2015). Identification of a plastidial phenylalanine exporter that influences flux distribution through the phenylalanine biosynthetic network. Nat. Commun. 6:8142. doi: 10.1038/ncomms9142
Wilcox, J. R., and Guodong, Z. (1997). Relationships between seed yield and seed protein in determinate and indeterminate soybean populations. Crop Sci. 37, 361–364. doi: 10.2135/cropsci1997.0011183X003700020009x
Williams, L. E., and Miller, A. J. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Biol. 52, 659–688. doi: 10.1146/annurev.arplant.52.1.659
Winter, H., Lohaus, G., and Heldt, H. W. (1992). Phloem transport of amino acids in relation to their cytosolic levels in barley leaves. Plant Physiol. 99, 996–1004. doi: 10.1104/pp.99.3.996
Wong, H. W., Liu, Q., and Sun, S. S. M. (2015). Biofortification of rice with lysine using endogenous histones. Plant Mol. Biol. 87, 235–248. doi: 10.1007/s11103-014-0272-z
Xia, L., Lam, S. K., Yan, X., and Chen, D. (2017). How does recycling of livestock manure in agroecosystems affect crop productivity, reactive nitrogen losses, and soil carbon balance?. Environ. Sci. Technol. 51, 7450–7457. doi: 10.1021/acs.est.6b06470
Xie, Y., Liu, Y., Wang, H., Ma, X., Wang, B., Wu, G., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8, 1–11. doi: 10.1038/s41467-017-00404-y
Xu, G., Fan, X., and Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yamaya, T., Obara, M., Nakajima, H., Sasaki, S., Hayakawa, T., and Sato, T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53, 917–925. doi: 10.1093/jexbot/53.370.917
Yang, H., Krebs, M., Stierhof, Y. D., and Ludewig, U. (2014). Characterization of the putative amino acid transporter genes AtCAT2, 3 & 4: the tonoplast localized AtCAT2 regulates soluble leaf amino acids. J. Plant Physiol. 171, 594–601. doi: 10.1016/j.jplph.2013.11.012
Yang, X., Nian, J., Xie, Q., Feng, J., Zhang, F., Jing, H., et al. (2016). Rice ferredoxin-dependent glutamate synthase regulates nitrogen–carbon metabolomes and is genetically differentiated between japonica and indica subspecies. Mol. Plant 9, 1520–1534. doi: 10.1016/j.molp.2016.09.004
Yasuda, H., Hirose, S., Kawakatsu, T., Wakasa, Y., and Takaiwa, F. (2009). Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol. 50, 1532–1543. doi: 10.1093/pcp/pcp098
York, L. M. (2019). Functional phenomics: an emerging field integrating high-throughput phenotyping, physiology, and bioinformatics. J. Exp. Bot. 70, 379–386. doi: 10.1093/jxb/ery379
Yu, J., Peng, P., Zhang, X., Zhao, Q., Zhy, D., Sun, X., et al. (2004). Seed-specific expression of a lysine-rich protein gene sb401 significantly increases both lysine and total protein content in maize seeds. Mol. Breed. 14, 1–7. doi: 10.1023/B:MOLB.0000037990.23718.d6
Yu, J., Zhen, X., Li, X., Li, N., and Xu, F. (2019). Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 10:584. doi: 10.3389/fpls.2019.00584
Yue, J., Li, C., Zhao, Q., Zhu, D., and Yu, J. (2014). Seed-Specific expression of a lysine-rich protein gene GhLRP, from cotton significantly increases the lysine content in maize seeds. Int. J. Mol. Sci. 15, 5350–5365. doi: 10.3390/ijms15045350
Zeng, D. D., Qin, R., Li, M., Alamin, M., Jin, X. L., Liu, Y., et al. (2017). The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genomics 292, 385–395. doi: 10.1007/s00438-016-1275-z
Zhang, J., Wang, Y., Zhao, Y., Zhang, Y., Zhang, J., Ma, H., et al. (2020b). Transcriptome analysis reveals nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS ONE 15:e0240662. doi: 10.1371/journal.pone.0240662
Zhang, L., Garneau, M. G., Majumdar, R., Grant, J., and Tegeder, M. (2015b). Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids. Plant J. 81, 134–146. doi: 10.1111/tpj.12716
Zhang, L., Tan, Q., Lee, R., Trethewy, A., Lee, Y., and Tegeder, M. (2010). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22, 3603–3620. doi: 10.1105/tpc.110.073833
Zhang, W., Zhou, Y., Dibley, K., Tyerman, S., Furbank, R., and Patrick, J. (2007). Review: Nutrient loading of developing seeds. Funct. Plant Biol. 34, 314–331. doi: 10.1071/FP06271
Zhang, X., Davidson, E. A., Mauzerall, D. L., Searchinger, T. D., Dumas, P., and Shen, Y. (2015a). Managing nitrogen for sustainable development. Nature 528, 51–59. doi: 10.1038/nature15743
Zhang, Z., Gao, S., and Chu, C. (2020a). Improvement of nutrient use efficiency in rice: current toolbox and future perspectives. Theor. Appl. Genet. 133, 1365–1384. doi: 10.1007/s00122-019-03527-6
Zhao, L., Liu, F., Crawford, N. M., and Wang, Y. (2018). Molecular regulation of nitrate responses in plants. Int. J. Mol. Sci. 19:2039. doi: 10.3390/ijms19072039
Zhen, X., Li, X., Yu, J., and Xu, F. (2019b). OsATG8c-mediated increased autophagy regulates the yield and nitrogen use efficiency in rice. Int. J. Mol. Sci. 20:4956. doi: 10.3390/ijms20194956
Zhen, X., Xu, F., Zhang, W., Li, N., and Li, X. (2019a). Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS ONE 14:e0223011. doi: 10.1371/journal.pone.0223011
Zhou, X., Lin, J., Zhou, Y., Yang, Y., Liu, H., Zhang, C., et al. (2015b). Overexpressing a fungal CeGDH gene improves nitrogen utilization and growth in rice. Crop Sci. 55, 811–820. doi: 10.2135/cropsci2014.03.0252
Zhou, Y., Liu, H., Zhou, X., Yan, Y., Du, C., Li, Y., et al. (2014). Over-expression of a fungal NADP(H)-dependent glutamate dehydrogenase PcGDH improves nitrogen assimilation and growth quality in rice. Mol. Breeding 34, 335–349. doi: 10.1007/s11032-014-0037-z
Zhou, Y., Zhang, C., Lin, J., Yang, Y., Peng, Y., Tang, D., et al. (2015a). Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta 241, 727–740. doi: 10.1007/s00425-014-2214-z
Zhu, C., Fan, Q., Wang, W., Shen, C., Meng, X., Tang, Y., et al. (2014). Characterization of a glutamine synthetase gene DvGS2 from Dunaliella viridis and biochemical identification of DvGS2-transgenic Arabidopsis thaliana. Gene 536, 407–415. doi: 10.1016/j.gene.2013.11.009
Zhu, C., Zhang, G., Shen, C., Chen, S., Tang, Y., Mei, B., et al. (2015). Expression of bacterial glutamine synthetase gene in Arabidopsis thaliana increases the plant biomass and level of nitrogen utilization. Biologia 70, 1586–1596. doi: 10.1515/biolog-2015-0183
Zhu, S., Vivanco, J. M., and Manter, D. K. (2016). Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 107, 324–333. doi: 10.1016/j.apsoil.2016.07.009
Zhu, X., and Galili, G. (2004). Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues. Plant Physiol. 135, 129–136. doi: 10.1104/pp.103.037168
Zrenner, R., Stitt, M., Sonnewald, U., and Boldt, R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57, 805–836. doi: 10.1146/annurev.arplant.57.032905.105421
Keywords: amino acid partitioning, nitrogen assimilation, crop improvement, nitrogen uptake and transport, nitrogen use efficiency, seed yield and quality, source and sink physiology, sustainable agriculture
Citation: The SV, Snyder R and Tegeder M (2021) Targeting Nitrogen Metabolism and Transport Processes to Improve Plant Nitrogen Use Efficiency. Front. Plant Sci. 11:628366. doi: 10.3389/fpls.2020.628366
Received: 11 November 2020; Accepted: 31 December 2020;
Published: 01 March 2021.
Edited by:
Nandula Raghuram, Guru Gobind Singh Indraprastha University, IndiaReviewed by:
Bertrand Hirel, Institut National de la Recherche Agronomique (INRA), FranceCopyright © 2021 The, Snyder and Tegeder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mechthild Tegeder, dGVnZWRlckB3c3UuZWR1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.