
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 February 2021
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.619417
This article is part of the Research Topic Vernalization and Flowering Time: Celebrating 20 Years of FLC View all 15 articles
 Ayasha Akter1,2
Ayasha Akter1,2 Etsuko Itabashi3
Etsuko Itabashi3 Tomohiro Kakizaki3
Tomohiro Kakizaki3 Keiichi Okazaki4
Keiichi Okazaki4 Elizabeth S. Dennis5,6
Elizabeth S. Dennis5,6 Ryo Fujimoto1*
Ryo Fujimoto1*The genus Brassica includes oil crops, vegetables, condiments, fodder crops, and ornamental plants. Brassica species underwent a whole genome triplication event after speciation between ancestral species of Brassica and closely related genera including Arabidopsis thaliana. Diploid species such as Brassica rapa and Brassica oleracea have three copies of genes orthologous to each A. thaliana gene, although deletion in one or two of the three homologs has occurred in some genes. The floral transition is one of the crucial events in a plant’s life history, and time of flowering is an important agricultural trait. There is a variation in flowering time within species of the genus Brassica, and this variation is largely dependent on a difference in vernalization requirements. In Brassica, like in A. thaliana, the key gene of vernalization is FLOWERING LOCUS C (FLC). In Brassica species, the vernalization response including the repression of FLC expression by cold treatment and the enrichment of the repressive histone modification tri-methylated histone H3 lysine 27 (H3K27me3) at the FLC locus is similar to A. thaliana. B. rapa and B. oleracea each have four paralogs of FLC, and the allotetraploid species, Brassica napus, has nine paralogs. The increased number of paralogs makes the role of FLC in vernalization more complicated; in a single plant, paralogs vary in the expression level of FLC before and after vernalization. There is also variation in FLC expression levels between accessions. In this review, we focus on the regulatory circuits of the vernalization response of FLC expression in the genus Brassica.
Brassica is an economically important genus including many agricultural crops such as Chinese cabbage and turnip (Brassica rapa L.), cabbage, broccoli, cauliflower, kale, and kohlrabi (Brassica oleracea L.), and oilseed or rapeseed and rutabaga (Brassica napus L.). Three diploid species, B. rapa, Brassica nigra L., and B. oleracea are denoted as the A, B, and C genomes, respectively. Allotetraploid species, Brassica juncea L. (AABB), Brassica carinata L. (BBCC), and B. napus (AACC) contain two diploid genomes, and this genomic relationship is known as the “Triangle of U” (Nagaharu, 1935). Arabidopsis thaliana L. is a close relative species to the genus Brassica, and both are contained in the Brassicaceae.
The genus Brassica needs vernalization for induction of flowering except for early flowering accessions, which have lost the genes responsible for a vernalization requirement. The length of cold treatment required for a vernalization response varies between accessions within a species; for example, the natural variation in flowering time in B. napus in response to vernalization is characterized into three groups: winter type (high vernalization requirement), semiwinter type (intermediate vernalization requirement), and spring type (low or no vernalization requirement) (Raman et al., 2016).
The molecular mechanism of vernalization is well studied in A. thaliana, and genes involved in vernalization networks have been characterized (Berry and Dean, 2015; Whittaker and Dean, 2017). No one doubts that FLOWERING LOCUS C (FLC) is a key gene in vernalization. FLC encodes a MADS-box transcription factor and acts as a floral repressor (Michaels and Amasino, 1999; Sheldon et al., 1999). FLC is expressed before cold exposure, and FRIGIDA (FRI) is involved in activation of FLC expression. FLC expression is repressed during cold exposure through epigenetic regulation (Groszmann et al., 2011; Berry and Dean, 2015; Whittaker and Dean, 2017). Upon return to warm conditions, silencing of FLC is maintained (Berry and Dean, 2015; Whittaker and Dean, 2017).
In the genus Brassica, FLC is also a key gene in the vernalization process, and the mechanism of vernalization in the genus Brassica has much in common with that in A. thaliana (Itabashi et al., 2018). However, molecular mechanisms specific to Brassica have also been identified. In this review, we describe the research findings on vernalization in the genus Brassica.
The tribe Brassiceae has undergone a whole genome triplication after speciation, and the whole genome sequence of B. rapa confirmed the triplication of the B. rapa genome relative to A. thaliana (Wang et al., 2011). The total number of genes in B. rapa is much less than three times the gene number in A. thaliana due to gene loss after the whole genome triplication in B. rapa (Wang et al., 2011). In the genus Brassica, three subgenomes, the least fractioned subgenome (LF) and two more fractionated subgenomes (MF1 and MF2), are recognized. In B. rapa, the LF subgenome retains 70% of A. thaliana orthologs, while 46 and 36% of A. thaliana orthologs are retained in MF1 and MF2 subgenomes, respectively (Wang et al., 2011).
B. rapa has multiple copies of orthologs for each A. thaliana gene. For example, there are two FRI paralogs (BrFRIa, BrFRIb) and four FLC paralogs (BrFLC1, BrFLC2, BrFLC3, BrFLC5) (Table 1; Schranz et al., 2002; Wang et al., 2017; Shea et al., 2018). BrFLC1, BrFLC2, and BrFLC3 are located on LF, MF2, and MF1 subgenomes, respectively, indicating that whole genome triplication state is preserved. However, it is not clear how BrFLC5 was generated (Wang et al., 2011). In B. oleracea, there are two FRI and four FLC paralogs (Table 1), and BoFLC5 is a pseudogene (Schranz et al., 2002; Irwin et al., 2012; Itabashi et al., 2019). The allotetraploid species, B. napus, has four FRI and nine FLC paralogs (Table 1; Chalhoub et al., 2014; Shea et al., 2018; Yi et al., 2018; Schiessl et al., 2019). The relationship of orthologs in FRI and FLC among three species is shown in Table 1.
Multiple copies of orthologs may lead to subfunctionalization in the genus Brassica. In A. thaliana, loss of function of the single-copy AtFRI is the main cause of natural variation in flowering time (Berry and Dean, 2015; Whittaker and Dean, 2017). However, there are few reports of FRI being a major contributor of flowering time variation in the genus Brassica. Since both FRIa and FRIb act as activators of FLC in the genus Brassica (Irwin et al., 2012; Takada et al., 2019), they complement each other so that there is little chance of simultaneous loss of FRI function in nature. In contrast, there are reports that loss of FLC function results in variation in flowering time in the genus Brassica.
To identify the key genes involved in vernalization, quantitative trait locus (QTL) analyses have been performed. QTLs affecting flowering time have been identified in different populations in B. rapa, B. oleracea, and B. napus (Shea et al., 2018). At the beginning of QTL analyses in the genus Brassica, a population derived from early and late flowering parental accessions was used. Some QTLs overlapped with the region covering the FLC gene, and some indicated that loss of FLC function results in early flowering (Kole et al., 2001; Schranz et al., 2002; Lou et al., 2007; Okazaki et al., 2007; Li et al., 2009; Zhao et al., 2010); loss of FLC2 function has been detected in early flowering accessions of B. rapa and B. oleracea (Table 2; Okazaki et al., 2007; Li et al., 2009; Wu et al., 2012). In cauliflower (B. oleracea), there is an association between BoFLC2 allelic variation and floral induction within populations of inbred lines (Ridge et al., 2015).
Loss of FLC function causes markedly early flowering and loss or reduction in the vernalization requirement. However, there is a variation in vernalization requirement among accessions not including early flowering accessions. QTL analysis using a population derived from parental accessions showing different vernalization requirements but both having a vernalization requirement also identified the colocalization of QTLs and the FLC gene. Using an F2 population derived from a cross between two parental accessions of Chinese cabbage (B. rapa) both having a vernalization requirement, QTLs for flowering time colocalized with BrFLC1 and BrFLC5 (Kakizaki et al., 2011). The repression rate of BrFLC1 and BrFLC5 following cold treatment in the later flowering time accession was lower than in the earlier flowering time accession (Table 2; Kakizaki et al., 2011), suggesting that this different rate of repression of FLC expression may be involved in the flowering time difference. It was considered that BrFLC5 is a pseudogene because of a mutation in the splicing donor site (G to A). However, a functional BrFLC5 allele has been identified in some accessions and shown to be a weak regulator of flowering time (Xi et al., 2018). We confirmed that the later flowering time accession used in Kakizaki et al. (2011) had a functional FLC5 allele in spite of sequence basis (G allele), suggesting that colocalization of QTL and BrFLC5 is due to the difference of BrFLC5 function between parental accessions. QTL analysis was also performed using 194 Chinese cabbage accessions including 40 spring, 37 summer, and 117 autumn types, of which 177 accessions showed a vernalization requirement. The order of increasing vernalization requirement is spring, autumn, and summer accessions. B. rapa Vernalization Insensitive 3.1 (BrVIN3.1) and BrFLC1 have been identified as sources of variation in bolting time (Su et al., 2018). In A. thaliana, VIN3 is involved in FLC silencing during cold treatment and is induced by cold treatment (Kim and Sung, 2013). Five haplotypes of BrVIN3.1 with sequence variation in the promoter regions have been identified. Three haplotypes of BrFLC1 with two main groups and one minor group had sequence polymorphism in non-coding regions (Su et al., 2018). These sequence polymorphisms in non-coding regions in BrVIN3.1 or BrFLC1 may result in variation in the vernalization requirement in B. rapa. In case of the difference in vernalization requirement of B. oleracea, a QTL overlapping BoFLC2 has been identified. There are cis polymorphisms in BoFLC2 that influence the expression dynamics in response to cold treatment especially the reactivation rate on return to warm conditions in purple sprouting broccoli (Table 2; Irwin et al., 2016). In the European winter oilseed rape (B. napus), BnaFLC.A02 (BrFLC2 ortholog) has been detected as candidate gene giving rise to the QTL. Higher expression levels of BnaFLC.A02 before cold treatment, lower repression rate following cold treatment, and higher reactivation rate on return to warm conditions of BnaFLC.A02 were found in the later flowering time accession. The amino acid sequence of BnaFLC.A02 was identical between the earlier and later flowering time accessions. In contrast, there were sequence polymorphisms in non-coding intronic regions between them, which could result in a difference in FLC expression and modified vernalization requirement (Table 2; Tudor et al., 2020).
FLC1, FLC2, and FLC3 act as floral repressors in both B. rapa and B. oleracea (Kim et al., 2007; Itabashi et al., 2019; Takada et al., 2019). It has not been shown whether amino acid sequence differences among functional FLC paralogs could lead to differences in the function as floral repressor or variation in vernalization requirement. There is a variation in expression levels among FLC paralogs before and following vernalization, and this is also shown in the repression rate of their expression following vernalization (Itabashi et al., 2019; Schiessl et al., 2019; Takada et al., 2019). These results suggest that variation in vernalization requirement is due to the difference in transcriptional repression rate of FLC between accessions, and this may be due to sequence polymorphisms in the non-coding regions. Furthermore, the fact that a change in one FLC paralog affects vernalization requirement suggests that the FLC paralogs may be functioning additively.
Transposable elements (TEs) are DNA sequences that can change their location in the genome, and they can play a role in plant genome evolution (Fujimoto et al., 2012; Hosaka and Kakutani, 2018). In B. rapa, TE DNA is methylated, and TEs are epigenetically silenced (Fujimoto et al., 2008b; Sasaki et al., 2011; Takahashi et al.,2018a,b). The variation in the genome structure of orthologous loci between B. rapa and B. oleracea is due to TE insertions; the B. oleracea genome has more TE insertions than the B. rapa genome in orthologous genomic regions, which results in a larger genome size in B. oleracea than in B. rapa (Fujimoto et al., 2006a; Liu et al., 2014). TE insertions can cause loss of protein function or gene expression or generate a new expression pattern (Miura et al., 2001; Fujimoto et al., 2006b, 2008a, 2011; Saze and Kakutani, 2007; Naito et al., 2009; Hosaka and Kakutani, 2018).
A TE insertion has been identified in some alleles of FLC in the genus Brassica. Long TE insertions in the first intron of BrFLC2 and BrFLC3 were detected in the extremely late flowering time accession, Tsukena No. 2 (Kitamoto et al., 2014). In A. thaliana, the Landsberg erecta (Ler) accession has a TE in the first intron, and this insertion results in a low level of FLC expression (Gazzani et al., 2003). In contrast, insertion of a TE in the first intron of BrFLC2 or BrFLC3 in Tsukena No. 2 does not affect the FLC expression per se. However, weak repression of BrFLC2 and BrFLC3 following cold exposure has been detected, suggesting that the TE insertion disrupts the vernalization response (Table 2; Kitamoto et al., 2014). A similar phenomenon has been observed in radish (Raphanus sativus); a 1,627-bp insertion in the first intron of RsFLC2 did not affect the RsFLC2 expression level before cold treatment but weakened its repression rate during vernalization, resulting in a late-bolting phenotype (Wang et al., 2018). These results suggest that a TE insertion might disrupt a cis element of the vernalization response in the first intron or inhibit the recruitment of the POLYCOMB REPRESSIVE COMPLEX 2 (PRC2), which plays a role in epigenetic silencing of FLC during vernalization, to the FLC locus. In addition to the TE insertion, an insertion/deletion in the second intron of BoFLC1 may alter gene expression and is associated with variation in flowering time of cabbage varieties (Abuyusuf et al., 2019).
In B. napus, there was TE insertion in the promoter region of some FLC paralogs, and this insertion affects the vernalization requirement. A 621-bp TE insertion in the upstream region of BnaFLC.A10 (BrFLC1 ortholog) was associated with lower rate of BnaFLC.A10 repression during cold treatment (Table 2; Hou et al., 2012). This 621-bp TE insertion is observed in 73% of winter-type accessions (449 of 619 accessions including Tapidor and Darmor-bzh) and associated with high vernalization requirement (Yin et al., 2020). A 4,422-bp TE insertion is observed in BnaFLC.A10 of the semiwinter type (Zhongshuang 11, Ningyou 7), and its expression level before and following cold treatment in semiwinter type was lower than in winter types (Table 2). About 57% of semiwinter accessions have this 4,422-bp TE insertion (Yin et al., 2020). A TE insertion results in loss of FLC function in spring-type accessions; a 5,625-bp TE insertion in the first exon of BnaFLC.A10 (in about 55% of spring-type accessions) and 810-bp TE insertion in the exon 7 of BnaFLC.A02 (BrFLC2 ortholog) (about 7%) cause loss of function as does a 2,833-bp insertion in the first intron of BnaFLC.A02 (about 18%) (Table 2; Chen et al., 2018; Yin et al., 2020).
These data indicate that TE insertions play an important role in the determination of flowering time and vernalization requirement in the genus Brassica.
Histone modification is an epigenetic modification that plays a role in the regulation of gene expression (Pfluger and Wagner, 2007; Richards, 2011; Fujimoto et al., 2012). Genome-wide histone modification states have been examined by chromatin immunoprecipitation sequencing (ChIP-seq) in many plant species. In B. rapa, di-methylation of histone H3 lysine 9 (H3K9me2) and trimethylation of histone H3 lysine 27 (H3K27me3) have been mapped genome wide (Takahashi et al., 2018a; Akter et al., 2019; Payá-Milans et al., 2019). In B. rapa, H3K9me2 was overrepresented in TEs, and there were low number of genes having H3K9me2 marks (Takahashi et al., 2018a). About 30% of genes had H3K27me3 marks, and H3K27me3 marks were associated with low level of gene expression and high level of tissue-specific gene expression (Akter et al., 2019; Payá-Milans et al., 2019). H3K27me3 modification is catalyzed by the PRC2, and in B. rapa, a mutant of the CURLY LEAF (CLF) gene, which is the enzymatic core of PRC2, showed decreased H3K27me3 level (Payá-Milans et al., 2019), suggesting that PRC2 plays a role in H3K27me3 in B. rapa as in other plant species.
In A. thaliana, repression of FLC expression during vernalization is associated with switching of chromatin states from enrichment of active histone marks (H3K4me3, H3K36me3) to repressive histone marks (H3K27me3) at the FLC locus (Berry and Dean, 2015). During prolonged cold treatment, there is accumulation of H3K27me3 within the nucleation region, which consists of a DNA region associated with one or two nucleosomes and harboring the first exon and the start of first intron. H3K27me3 spreads over the entire region of FLC when temperatures are warm (Xi et al., 2020). A cis DNA element in the first intron of FLC [termed the RY element (CATGCA)] that is essential for FLC repression during vernalization and the maintenance of FLC silencing when temperatures are warm has been identified (Qüesta et al., 2016; Yuan et al., 2016). In B. rapa, H3K4me3 accumulation was found in the nucleation region of all four BrFLC paralogs before exposure to cold (Kawanabe et al., 2016). During vernalization, accumulation of H3K27me3 within the nucleation region of all four BrFLC paralogs with decreased expression was observed (Akter et al., 2020; Figure 1). After return to warm conditions, H3K27me3 accumulation spreads across the four BrFLC paralogs maintaining silencing (Kawanabe et al., 2016; Akter et al., 2019; Figure 1). The position of the nucleation region is conserved between A. thaliana and B. rapa, but there is less conservation of first intron sequences (Figure 1). Two RY elements are conserved in four BrFLC paralogs except in BrFLC1 where there is a mutation in one of the two RY elements. Like in A. thaliana, PRC2 could be recruited to the nucleation regions of the four BrFLC paralogs (Akter et al., 2019).
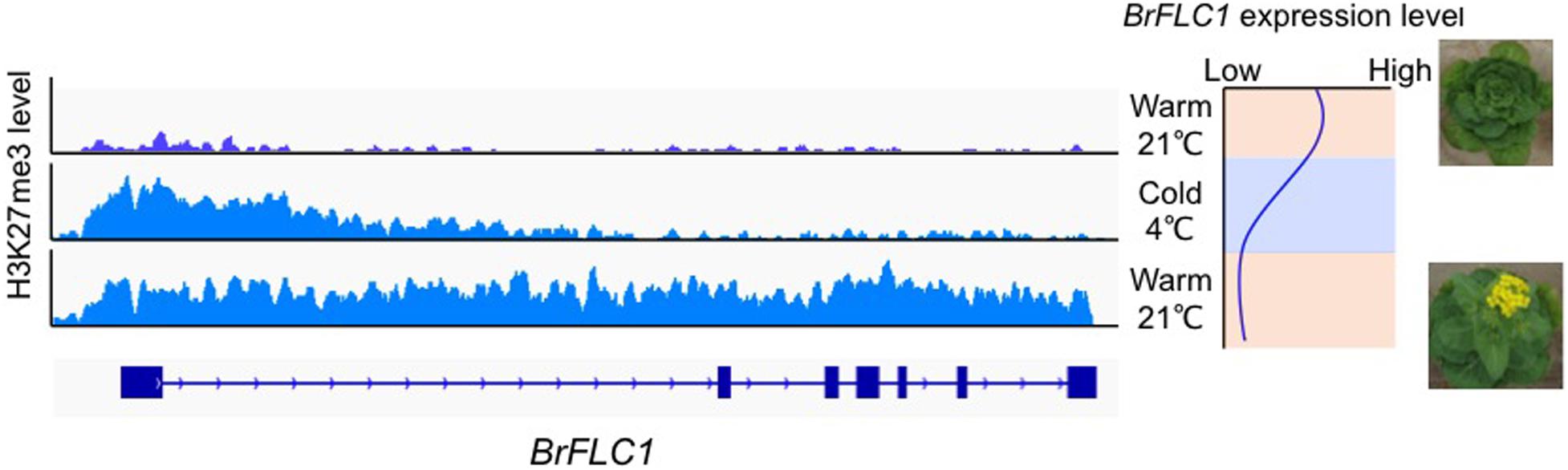
Figure 1. H3K27me3 pattern during vernalization in BrFLC1. Left panel shows the pattern of accumulation of H3K27me3 marks during vernalization. Right panel shows the expression pattern of BrFLC1 during vernalization. BrFLC2, BrFLC3, and BrFLC5 showed similar H3K27me3 pattern to BrFLC1 (Akter et al., 2019).
The genome-wide level of H3K27me3 in B. rapa was compared between non-vernalized, 4 weeks vernalized, and sample upon return to warm temperature after 4 weeks vernalization. Between the non-vernalized and 4 weeks vernalized samples, 6,814 genes showed a difference in H3K27me3 level, and 99% of genes showed decreased H3K27me3 following vernalization. Between the non-vernalized and the sample upon return to warm temperature after 4 weeks vernalization, 189 genes showed a difference in H3K27me3 level, and 85% of genes showed increased H3K27me3 following vernalization. In A. thaliana, 380 genes showed a difference in H3K27me3 level between non-vernalized and sample upon return to warm temperature following 4 weeks vernalization, and only the FLC gene showed increased H3K27me3 levels following vernalization (Akter et al., 2019). A trend toward increased levels of H3K27me3 following vernalization in A. thaliana has been reported (Xi et al., 2020). A limited number of genes such as B. rapa MADS AFFECTING FLOWERING (BrMAF) genes, which are related to FLC, showed a similar H3K27me3 accumulation pattern; H3K27me3 accumulated in a specific region during vernalization and then spread upon returning to warm temperatures after vernalization (Akter et al., 2019). At the transcriptional level, some genes showed a similar expression pattern to FLC during cold treatment in A. thaliana (Xi et al., 2020). Some genes showed a change in H3K27me3 accumulation following vernalization, but only a limited number of genes showed an H3K27me3 modification pattern similar to that of FLC, where vernalization leads to H3K27me3 accumulation in the specific regions (nucleation region) and then spread throughout the gene when temperature is returned (Akter et al., 2019).
In A. thaliana, three cold-induced non-coding RNAs (COOLAIR, COLD ASSISTED INTRONIC NON-CODING RNA (COLDAIR), and COLDWRAP) have been identified. COOLAIR is transcribed in the antisense direction, and COLDAIR and COLDWRAP are transcribed in the sense direction relative to FLC messenger RNA (mRNA) transcription. COOLAIR transcripts start from downstream of the poly-A site to the promoter region of FLC (Figure 2; Swiezewski et al., 2009). COLDAIR is derived from the first intron and COLDWRAP from the promoter region (starts at 225 bp upstream from transcription start site) of FLC (Figure 2; Heo and Sung, 2011; Kim and Sung, 2017). These non-coding RNAs are involved in FLC silencing and maintenance of the repressed condition (Swiezewski et al., 2009; Heo and Sung, 2011; Kim and Sung, 2017; Whittaker and Dean, 2017). COOLAIR-like transcripts have been identified in five species of Brassicaceae, Arabidopsis lyrata, Arabis alpina, Capsella rubella, Eutrema salsugineum, and B. rapa. Although there is low conservation of COOLAIR-like sequences between species, secondary structures show similarity, suggesting that the functional role, the regulation of FLC expression during vernalization, may be conserved in Brassicaceae species (Hawkes et al., 2016).
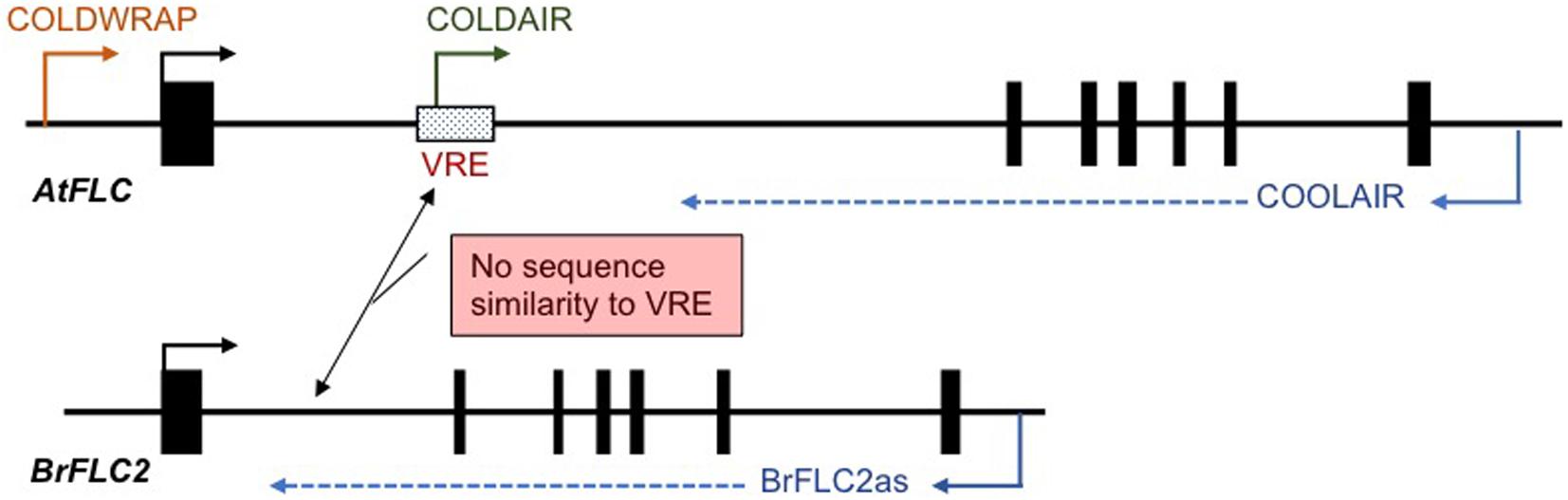
Figure 2. Schematic view of the position of cold-induced non-coding RNAs in A. thaliana and B. rapa. There is no sequence similarity between the putative vernalization response element (VRE) in A. thaliana and the first intron of four BrFLC paralogs. Only BrFLC2 has antisense transcripts, BrFLC2as.
In B. rapa, COOLAIR-like transcripts have been identified only at the BrFLC2 locus and are termed BrFLC2as, while no COOLAIR-like transcript was detected from the other three BrFLC loci (Figure 2; Li et al., 2016; Shea et al., 2019). COOLAIR has two transcripts, proximal site (class I, ∼400 nt) and distal site (class II, ∼750 nt), and BrFLC2as also has two different length transcripts, class I (short, 357, 425, and 510 nt) and class II (long, 665 and 767nt) (Li et al., 2016). BrFLC2as was highly induced by short-term cold treatment, and class II BrFLC2as was at a higher level than class I BrFLC2as (Li et al., 2016; Shea et al., 2019). Similarity of secondary structure and the characteristic of being induced by short-term cold treatment in both BrFLC2as and COOLAIR suggests a functional similarity (Li et al., 2016). However, the other three BrFLC paralogs also show decreased expression during vernalization in spite of no cold-induced COOLAIR-like transcripts being detected (Li et al., 2016; Shea et al., 2019). BrFLC2as may be involved in the silencing of the other three BrFLC paralogs. Overexpression of the BrFLC2as resulted in reduced BrFLC1, BrFLC2, and BrFLC3 expression leading to an early flowering phenotype, suggesting that BrFLC2as may be involved not only in the repression of BrFLC2 but also in the repression of the BrFLC1 and BrFLC3 genes (Li et al., 2016). However, this experiment does not rule out the possibility that repression of the three BrFLC genes is due to posttranscriptional gene silencing because the introduced BrFLC2as sequence includes the exon 1 region of BrFLC2 and may silence all three genes (Li et al., 2016). Furthermore, there is a question whether BrFLC2as is able to function in trans because COOLAIR functions in cis (Csorba et al., 2014). Further study will be needed to elucidate the role of BrFLC2as in the vernalization mechanism in B. rapa.
In A. thaliana, COLDAIR and COLDWRAP play a role in recruitment of PRC2 to the FLC locus (Kim and Sung, 2017). However, neither COLDAIR nor COLDWRAP-like transcripts nor other cold-induced transcripts from the promoter or first intron have been identified in the four FLC paralogs of B. rapa (Li et al., 2016; Shea et al., 2019). However, accumulation of H3K27me3 was confirmed in all four BrFLC paralogs (Akter et al., 2019), leading to the question of how PRC2 is recruited to the BrFLC loci during vernalization.
Cold-induced natural antisense transcripts (NATs) were also identified in two BrMAF genes in B. rapa. In B. rapa, there are five BrMAF genes, and three of these five BrMAF genes were repressed following vernalization (Akter et al., 2019). Accumulation of H3K27me3 during vernalization also has been shown in these three BrMAF genes (Akter et al., 2019). In addition, MAF1 and MAF2 isolated from Pak-choi (B. rapa var. chinensis), termed BcMAF1 and BcMAF2, respectively, function as floral repressors (Huang et al., 2018, 2019). These cold-inducible NATs derived from BrMAF loci may have a similar function to COOLAIR. In A. thaliana, MAF4 has an overlapped NAT termed MAS. MAF4 is induced early in cold treatment, and MAS is coordinately expressed during the cold treatment (Zhao et al., 2018). Repression or silencing of MAF4 expression did not affect MAS expression, while repression of MAS expression reduced MAF4 induction. These results suggest that MAS plays a positive role in MAF4 expression. In B. rapa, coordinate expression of mRNA and the NAT pair following vernalization was observed in some genes but not in BrMAF genes (Shea et al., 2019). This coordinate expression in mRNA and NAT pairs may have a role in vernalization or the cold response.
Whole genome triplication results in multiple copies of FLC paralogs in the genus Brassica. FLC1, FLC2, and FLC3 act as floral repressors in both B. rapa and B. oleracea (Kim et al., 2007; Itabashi et al., 2019; Takada et al., 2019), suggesting that all three FLC paralogs need to be considered to understand the vernalization mechanism. There is a difference in transcriptional level before vernalization among three FLC paralogs in both B. rapa and B. oleracea, and between accessions within species, there are differences in which FLC paralog shows the highest expression (Itabashi et al., 2019; Takada et al., 2019). A similar situation is observed in B. napus, which has nine FLC paralogs (Schiessl et al., 2019). Different expression levels before vernalization between paralogs could be due to sequence polymorphisms in the promoter regions. In addition, sequence polymorphisms in the promoter region could also be involved in the difference in expression levels in each FLC between accessions. The correlation between the total level of FLC expression before vernalization and the vernalization requirement suggests that the level of FLC expression before vernalization is associated with the diversity of vernalization requirement in B. rapa (Takada et al., 2019). We need to identify the region or cis-element controlling FLC expression level to understand the variation in vernalization requirement between accessions.
There is also a variation in the rate of repression of FLC expression among paralogs during vernalization with a difference between accessions as to which FLC paralog shows a low repression rate. A low repression rate of FLC genes is an important factor in generating a vernalization requirement, and in this review, some examples are shown such as a TE insertion in the first intron or a TE insertion in the promoter region, which affect this requirement (Gazzani et al., 2003; Hou et al., 2012; Kitamoto et al., 2014; Yin et al., 2020). There is a need to clarify the complexity that arises from multiple FLC paralogs. Furthermore, non-coding RNAs, which can recruit PRC2 to FLC loci, have not been identified, and a vernalization responsive element (VRE) in the genus Brassica has not been identified. Identification of a VRE may clarify the association between VRE sequence polymorphism and variation in repression rate following vernalization between paralogs or between accessions within species.
TK, KO, ED, and RF conceptualized the manuscript. AA, EI, ED, and RF wrote the manuscript. TK, KO, ED, and RF edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Grant-in-Aid for Scientific Research (B) (15H04433 and 18H02173) from the Japan Society for the Promotion of Science (JSPS) and Fund for the Promotion of International Joint Research (16KK0171) from JSPS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abuyusuf, M., Nath, U. K., Kim, H. T., Islam, M. R., Park, J. I., and Nou, I. S. (2019). Molecular markers based on sequence variation in BoFLC1.C9 for characterizing early-and late-flowering cabbage genotypes. BMC Genet. 20:42. doi: 10.1186/s12863-019-0740-1
Akter, A., Miyazaki, J., Shea, D. J., Nishida, N., Takada, S., Miyaji, N., et al. (2020). Gene expression analysis in response to vernalization in Chinese cabbage (Brassica rapa L). Hort. J. 89, 268–277. doi: 10.2503/hortj.UTD-150
Akter, A., Takahashi, S., Deng, W., Shea, D. J., Itabashi, E., Shimizu, M., et al. (2019). The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Res. 26, 433–443. doi: 10.1093/dnares/dsz021
Berry, S., and Dean, C. (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 83, 133–148. doi: 10.1111/tpj.12869
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, L., Dong, F., Cai, J., Xin, Q., Fang, C., Liu, L., et al. (2018). A 2.833-kb insertion in BnFLC.A2 and its homeologous exchange with BnFLC.C2 during breeding selection generated early-flowering rapeseed. Mol. Plant 11, 222–225. doi: 10.1016/j.molp.2017.09.020
Csorba, T., Questa, J. I., Sun, Q., and Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 111, 16160–16165. doi: 10.1073/pnas.1419030111
Fujimoto, R., Kinoshita, Y., Kawabe, A., Kinoshita, T., Takashima, K., Nordborg, M., et al. (2008a). Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 4:e1000048. doi: 10.1371/journal.pgen.1000048
Fujimoto, R., Okazaki, K., Fukai, E., Kusaba, M., and Nishio, T. (2006a). Comparison of the genome structure of the self-incompatibility (S) locus in interspecific pairs of S haplotypes. Genetics 173, 1157–1167. doi: 10.1534/genetics.104.037267
Fujimoto, R., Sasaki, T., Inoue, H., and Nishio, T. (2008b). Hypomethylation and transcriptional reactivation of retrotransposon-like sequences in ddm1 transgenic plants of Brassica rapa. Plant Mol. Biol. 66, 463–473. doi: 10.1007/s11103-007-9285-1
Fujimoto, R., Sasaki, T., Ishikawa, R., Osabe, K., Kawanabe, T., and Dennis, E. S. (2012). Molecular mechanisms of epigenetic variation in plants. Int. J. Mol. Sci. 13, 9900–9922. doi: 10.3390/ijms13089900
Fujimoto, R., Sasaki, T., Kudoh, H., Taylor, J. M., Kakutani, T., and Dennis, E. S. (2011). Epigenetic variation in the FWA gene within the genus Arabidopsis. Plant J. 66, 831–843. doi: 10.1111/j.1365-313X.2011.04549.x
Fujimoto, R., Sugimura, T., Fukai, E., and Nishio, T. (2006b). Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol. Biol. 61, 577–587. doi: 10.1007/s11103-006-0032-9
Gazzani, S., Gendall, A. R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132, 1107–1114. doi: 10.1104/pp.103.021212
Groszmann, M., Greaves, I. K., Albert, N., Fujimoto, R., Helliwell, C. A., Dennis, E. S., et al. (2011). Epigenetics in plants-vernalisation and hybrid vigour. Biochim. Biophys. Acta 1809, 427–437. doi: 10.1016/j.bbagrm.2011.03.006
Hawkes, E. J., Hennelly, S. P., Novikova, I. V., Irwin, J. A., Dean, C., and Sanbonmatsu, K. Y. (2016). COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 16, 3087–3096. doi: 10.1016/j.celrep.2016.08.045
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hosaka, A., and Kakutani, T. (2018). Transposable elements, genome evolution and transgenerational epigenetic variation. Curr. Opin. Genet. Dev. 49, 43–48. doi: 10.1016/j.gde.2018.02.012
Hou, J., Long, Y., Raman, H., Zou, X., Wang, J., Dai, S., et al. (2012). A Tourist-like MITE insertion in the upstream region of the BnFLC. A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 12:238. doi: 10.1186/1471-2229-12-238
Huang, F., Liu, T., and Hou, X. (2018). Isolation and functional characterization of a floral repressor, BcMAF1, from Pak-choi (Brassica rapa ssp. chinensis). Front. Plant Sci. 9:290. doi: 10.3389/fpls.2018.00290
Huang, F., Liu, T., Tang, J., Duan, W., and Hou, X. (2019). BcMAF2 activates BcTEM1 and represses flowering in Pak-choi (Brassica rapa ssp. chinensis). Plant Mol. Biol. 100, 19–32. doi: 10.1007/s11103-019-00867-1
Irwin, J. A., Lister, C., Soumpourou, E., Zhang, Y., Howell, E. C., Teakle, G., et al. (2012). Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Biol. 12:21. doi: 10.1186/1471-2229-12-21
Irwin, J. A., Soumpourou, E., Lister, C., Ligthart, J. D., Kennedy, S., and Dean, C. (2016). Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 87, 597–605. doi: 10.1111/tpj.13221
Itabashi, E., Osabe, K., Fujimoto, R., and Kakizaki, T. (2018). Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep. 37, 87–101. doi: 10.1007/s00299-017-2223-z
Itabashi, E., Shea, D. J., Fukino, N., Fujimoto, R., Okazaki, K., Kakizaki, T., et al. (2019). Comparison of cold responses for orthologs of cabbage vernalization-related genes. Hort. J. 4, 462–470. doi: 10.2503/hortj.UTD-059
Kakizaki, T., Kato, T., Fukino, N., Ishida, M., Hatakeyama, K., and Matsumoto, S. (2011). Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breed. Sci. 61, 151–159. doi: 10.1270/jsbbs.61.151
Kawanabe, T., Osabe, K., Itabashi, E., Okazaki, K., Dennis, E. S., and Fujimoto, R. (2016). Development of primer sets that can verify the enrichment of histone modifications, and their application to examining vernalization-mediated chromatin changes in Brassica rapa L. Genes Genet. Syst. 91, 1–10. doi: 10.1266/ggs.15-00058
Kim, D. H., and Sung, S. (2013). Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25, 454–469. doi: 10.1105/tpc.112.104760
Kim, D. H., and Sung, S. (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40, 302–312. doi: 10.1016/j.devcel.2016.12.021
Kim, S. Y., Park, B. S., Kwon, S. J., Kim, J., Lim, M. H., Park, Y. D., et al. (2007). Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L.: ssp. pekinensis). Plant Cell Rep. 26, 327–336. doi: 10.1007/s00299-006-0243-1
Kitamoto, N., Yui, S., Nishikawa, K., Takahata, Y., and Yokoi, S. (2014). A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica 196, 213–223. doi: 10.1007/s10681-013-1025-9
Kole, C., Quijada, P., Michaels, S. D., Amasino, R. M., and Osborn, T. C. (2001). Evidence for homology of flowering-time genes VFR2 from Brassica rapa and FLC from Arabidopsis thaliana. Theor. Appl. Genet. 102, 425–430. doi: 10.1007/s001220051663
Li, F., Kitashiba, H., Inaba, K., and Nishio, T. (2009). A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 16, 311–323. doi: 10.1093/dnares/dsp020
Li, X., Zhang, S., Bai, J., and He, Y. (2016). Tuning growth cycles of Brassica crops via natural antisense transcripts of BrFLC. Plant Biotechnol. J. 14, 905–914. doi: 10.1111/pbi.12443
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Lou, P., Zhao, J., Kim, J. S., Shen, S., Del Carpio, D. P., Song, X., et al. (2007). Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 58, 4005–4016. doi: 10.1093/jxb/erm255
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214. doi: 10.1038/35075612
Nagaharu, U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452.
Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C. N., Richardson, A. O., et al. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134. doi: 10.1038/nature08479
Okazaki, K., Sakamoto, K., Kikuchi, R., Saito, A., Togashi, E., Kuginuki, Y., et al. (2007). Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 114, 595–608. doi: 10.1007/s00122-006-0460-6
Payá-Milans, M., Poza-Viejo, L., Martín-Uriz, P. S., Lara-Astiaso, D., Wilkinson, M. D., and Crevillén, P. (2019). Genome-wide analysis of the H3K27me3 epigenome and transcriptome in Brassica rapa. GigaScience 8:giz147. doi: 10.1093/gigascience/giz147
Pfluger, J., and Wagner, D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. doi: 10.1016/j.pbi.2007.07.013
Qüesta, J., Song, J., Geraldo, N., An, H., and Dean, C. (2016). Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353, 485–488. doi: 10.1126/science.aaf7354
Raman, H., Raman, R., Coombes, N., Song, J., Prangnell, R., Bandaranayake, C., et al. (2016). Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 39, 1228–1239. doi: 10.1111/pce.12644
Richards, E. J. (2011). Natural epigenetic variation in plant species: a view from the field. Curr. Opin. Plant Biol. 14, 204–209. doi: 10.1016/j.pbi.2011.03.009
Ridge, S., Brown, P. H., Hecht, V., Driessen, R. G., and Weller, J. L. (2015). The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. J. Exp. Bot. 66, 125–135. doi: 10.1093/jxb/eru408
Sasaki, T., Fujimoto, R., Kishitani, S., and Nishio, T. (2011). Analysis of target sequences of DDM1s in Brassica rapa by MSAP. Plant Cell Rep. 30, 81–88. doi: 10.1007/s00299-010-0946-1
Saze, H., and Kakutani, T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26, 3641–3652. doi: 10.1038/sj.emboj.7601788
Schiessl, S. V., Quezada-Martinez, D., Tebartz, E., Snowdon, R. J., and Qian, L. (2019). The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci. Rep. 9:14911. doi: 10.1038/s41598-019-51212-x
Schranz, M. E., Quijada, P., Sung, S. B., Lukens, L., Amasino, R., and Osborn, T. C. (2002). Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468.
Shea, D. J., Itabashi, E., Takada, S., Fukai, E., Kakizaki, T., Fujimoto, R., et al. (2018). The role of FLOWERING LOCUS C in vernalisation of Brassica: the importance of vernalisation research in the face of climate change. Crop Pasture Sci. 69, 30–39. doi: 10.1071/CP16468
Shea, D. J., Nishida, N., Takada, S., Itabashi, E., Takahashi, S., and Akter, A. (2019). Long noncoding RNAs in Brassica rapa L. following vernalization. Sci. Rep. 9:9302. doi: 10.1038/s41598-019-45650-w
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. doi: 10.1105/tpc.11.3.445
Su, T., Wang, W., Li, P., Zhang, B., Li, P., Xin, X., et al. (2018). A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant 11, 1360–1376. doi: 10.1016/j.molp.2018.08.006
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Takada, S., Akter, A., Itabashi, E., Nishida, N., Shea, D. J., Miyaji, N., et al. (2019). The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 9:13843. doi: 10.1038/s41598-019-50122-2
Takahashi, S., Fukushima, N., Osabe, K., Itabashi, E., Shimizu, M., Miyaji, N., et al. (2018b). Identification of DNA methylated regions by using methylated DNA immunoprecipitation sequencing in Brassica rapa. Crop Pasture Sci. 69, 107–120. doi: 10.1071/CP17394
Takahashi, S., Osabe, K., Fukushima, N., Takuno, S., Miyaji, N., Shimizu, M., et al. (2018a). Genome-wide characterization of DNA methylation, small RNA expression, and histone H3 lysine nine di-methylation in Brassica rapa L. DNA Res. 25, 511–520. doi: 10.1093/dnares/dsy021
Tudor, E. H., Jones, D. M., He, Z., Bancroft, I., Trick, M., Wells, R., et al. (2020). QTL-seq identifies BnaFT.A02 and BnaFLC.A02 as candidates for variation in vernalisation requirement and response in winter oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2466–2481. doi: 10.1111/pbi.13421
Wang, J., Qiu, Y., Cheng, F., Chen, X., Zhang, X., Wang, H., et al. (2017). Genome-wide identification, characterization, and evolutionary analysis of flowering genes in radish (Raphanus sativus L.). BMC Genomics 18:981. doi: 10.1186/s12864-017-4377-z
Wang, Q., Zhang, Y., and Zhang, L. (2018). A naturally occurring insertion in the RsFLC2 gene associated with late-bolting trait in radish (Raphanus sativus L.). Mol. Breed. 38:137. doi: 10.1007/s11032-018-0897-8
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Whittaker, C., and Dean, C. (2017). The FLC Locus: a platform for discoveries in epigenetics and adaptation. Ann. Rev. Cell. Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Wu, J., Wei, K., Cheng, F., Li, S., Wang, Q., Zhao, J., et al. (2012). A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 12:151. doi: 10.1186/1471-2229-12-151
Xi, X., Wei, K., Gao, B., Liu, J., Liang, J., Cheng, F., et al. (2018). BrFLC5: a weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 131, 2107–2116. doi: 10.1007/s00122-018-3139-x
Xi, Y., Park, S. R., Kim, D. H., Kim, E. D., and Sung, S. (2020). Transcriptome and epigenome analyses of vernalization in Arabidopsis thaliana. Plant J. 103, 1490–1502. doi: 10.1111/tpj.14817
Yi, L., Chen, C., Yin, S., Li, H., Li, Z., Wang, B., et al. (2018). Sequence variation and functional analysis of a FRIGIDA orthologue (BnaA3. FRI) in Brassica napus. BMC Plant Biol. 18:32. doi: 10.1186/s12870-018-1253-1
Yin, S., Wan, M., Guo, C., Wang, B., Li, H., Li, G., et al. (2020). Transposon insertions within alleles of BnaFLC. A10 and BnaFLC. A2 are associated with seasonal crop type in rapeseed. J. Exp. Bot. 71, 4729–4741. doi: 10.1093/jxb/eraa237
Yuan, W., Luo, X., Li, Z., Yang, W., Wang, Y., Liu, R., et al. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534. doi: 10.1038/ng.3712
Zhao, J., Kulkarni, V., Liu, N., Del Carpio, D. P., and Bucher, J. (2010). BrFLC2 (FLOWERINGLOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 61, 1817–1825. doi: 10.1093/jxb/erq048
Keywords: FLOWERING LOCUS C, vernalization, epigenetics, non-coding RNA, Brassica
Citation: Akter A, Itabashi E, Kakizaki T, Okazaki K, Dennis ES and Fujimoto R (2021) Genome Triplication Leads to Transcriptional Divergence of FLOWERING LOCUS C Genes During Vernalization in the Genus Brassica. Front. Plant Sci. 11:619417. doi: 10.3389/fpls.2020.619417
Received: 20 October 2020; Accepted: 29 December 2020;
Published: 09 February 2021.
Edited by:
Amy Litt, University of California, Riverside, United StatesReviewed by:
Francisco Madueño, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 Akter, Itabashi, Kakizaki, Okazaki, Dennis and Fujimoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryo Fujimoto, bGVvQHBlb3BsZS5rb2JlLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.