
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 January 2021
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.616338
 Mengli Wang1
Mengli Wang1 Lexuan Gao2
Lexuan Gao2 Gengyun Li1
Gengyun Li1 Chengchuan Zhou1
Chengchuan Zhou1 Jinjing Jian1
Jinjing Jian1 Zhen Xing3
Zhen Xing3 Yuguo Wang1
Yuguo Wang1 Wenju Zhang1
Wenju Zhang1 Zhiping Song1
Zhiping Song1 Yonghong Hu4*
Yonghong Hu4* Ji Yang1,4*
Ji Yang1,4*Seed oils are of great economic importance both for human consumption and industrial applications. The nutritional quality and industrial value of seed oils are mostly determined by their fatty acid profiles, especially the relative proportions of unsaturated fatty acids. Tree peony seed oils have recently been recognized as novel edible oils enriched in α-linolenic acid (ALA). However, congeneric species, such as Paeonia ostii and P. ludlowii, showed marked variation in the relative proportions of different unsaturated fatty acids. By comparing the dynamics of fatty acid accumulation and the time-course gene expression patterns between P. ostii and P. ludlowii, we identified genes that were differentially expressed between two species in developing seeds, and showed congruent patterns of variation between expression levels and phenotypes. In addition to the well-known desaturase and acyltransferase genes associated with fatty acid desaturation, among them were some genes that were conservatively co-expressed with the desaturation pathway genes across phylogenetically distant ALA-rich species, including Camelina sativa and Perilla frutescens. Go enrichment analysis revealed that these genes were mainly involved in transcriptional regulation, protein post-translational modification and hormone biosynthesis and response, suggesting that the fatty acid synthesis and desaturation pathway might be subject to multiple levels of regulation.
Plant storage oils are an essential part of the human diet, providing a number of nutrients the body needs including essential fats and vitamins (Damude and Kinney, 2008). Plant oils are also utilized as a major source of calories in our food supply and as feedstocks for non-food uses such as soaps and polymers (Dyer et al., 2008). Due to the concern on the depletion of fossil fuels and the environmental problems caused by the use of fossil fuels, plant oils have recently attracted considerable attention as cleaner, renewable, and sustainable sources for biofuel production (Durrett et al., 2008).
Plant seed oils consist principally of triacylglycerols (TAG’s) and contain both saturated and unsaturated (mono- and polyunsaturated) fatty acids, with the content and fatty acid (FA) composition varying considerably across plants (Voelker and Kinney, 2001). Variations in the ratios of saturated to unsaturated fatty acids (SFA: UFA), and monounsaturated to polyunsaturated fatty acids (MUFA: PUFA) have been widely detected in different plants (Zhang et al., 2015; Ohlrogge et al., 2018). The level of fatty acid unsaturation of oil can directly affect its value and utilization. Polyunsaturated fatty acids, such as linoleic acid (LA), and α-linolenic acid (ALA), are considered to be critical nutrients for human health (Damude and Kinney, 2008), but they are undesirable while being used as biofuel and chemical feedstocks because of their instability in processing, storage and use (Kodali, 2002). The requirements on different optimal fatty acid compositions for different uses have directly led to the growing interest in identifying genetic factors responsible for the variation in seed oil composition to better understand how the relative proportions of fatty acids are regulated in the seed oils derived from different plants.
A considerable amount of research has been conducted to identify genes potentially associated with the differential accumulation of unsaturated fatty acids in seed oils for optimizing the proportions of fatty acids through biotechnology for different industrial and nutrition goals (Rao et al., 2008; Haslam et al., 2013; Chen et al., 2014). Previous studies, however, have mostly focused on the genes encoding for three desaturases, i.e., stearoyl-ACP desaturase (SAD), fatty acid desaturases 2 (FAD2), and fatty acid desaturases 3 (FAD3) (Thambugala et al., 2013; Rajwade et al., 2014, 2016). These three enzymes catalyze stepwise desaturation of fatty acids during oil biosynthesis and determine the relative proportions of three major unsaturated fatty acids, oleic acid (OA), LA and ALA, in seeds. Although a great deal of evidence has shown the conserved roles of SAD, FAD2, and FAD3 across plant species, as well as the positive relations between high levels of fatty acid unsaturation and the upregulation of SAD, FAD2, and FAD3 genes in developing seeds (Ohlrogge and Browse, 1995), it remains elusive how these genes are differentially regulated in different plants so as to collectively control the ratios of SFA: UFA and MUFA: PUFA. Duplicate desaturase genes with altered expression patterns during seed development have also been detected in many plants (Cao et al., 2013; Wang and Liu, 2014; Yurchenko et al., 2014; Han et al., 2017). It is unclear, however, whether the differential transcriptional activities of duplicates are responsible for the differential accumulation of PUFAs in different plants and to what degree the duplicates have diverged within and between species. The question still remains: which genes other than desaturase genes are temporally expressed corresponding to the dynamic synthesis of unsaturated fatty acids in developing seeds, regulating the transcription and catalytic activity of desaturation enzymes and potentially contributing to the variation in unsaturated fatty acids accumulation in different plants. The newly developed methods of genome wide association studies (GWAS) and genome-wide transcription analysis allow for a more efficient discovery of additional genes involved in fatty acid synthesis and desaturation (Li et al., 2013; Branham et al., 2015; Menard et al., 2017; Zhao et al., 2019).
As a traditional ornamental and medicinal plant in China, tree peony has attracted wide attention in the recent past for its relatively high oil content (27%) in seeds and high ratio of unsaturated fatty acids (over 90%) (Li S. et al., 2015). The oil of tree peony has now been recognized as novel edible oil enriched in omega-3 polyunsaturated fatty acid (Han et al., 2016). Comparative studies of oil content and fatty acid composition have revealed significant variations in the relative proportions of three major unsaturated fatty acids among different Paeonia (section Moutan DC) species. The polyunsaturated fatty acid ALA seems to be the dominant component in the oils of the species from the subsection Vaginatae, while the monounsaturated fatty acid OA is predominant in the species of the subsection Delavayanae (Yu et al., 2016). Genes associated with lipid accumulation and polyunsaturated fatty acid synthesis have been cloned and characterized in some tree peony species (Sun et al., 2018; Xiu et al., 2018; Yin et al., 2018). Gene expression patterns associated with differential accumulation of unsaturated fatty acids have also been examined between P. rockii (subsection Vaginatae) and P. lutea (subsection Delavayanae) (Zhang Q. et al., 2018; Zhang et al., 2019). However, their results showed that the predominant unsaturated fatty acid was ALA instead of OA in the oil of P. lutea, while multiple studies have shown that OA was predominated over LA and ALA in P. ludlowii, another species of the subsection Delavayanae (Han et al., 2016; Yu et al., 2016; Xiu et al., 2018). Differences in the ALA, LA, and OA content proportions are in fact indicative of the degree of unsaturation of different seed oils. Comparing the dynamics of fatty acid accumulation and gene expression between congeneric species with similar oil content but varied fatty acid composition can provide valuable clues for identifying genes potentially involved in the regulation of fatty acid desaturation.
In this study, we characterized the fatty acid profiles and the time-course gene expression patterns of P. ostii and P. ludlowii. Based on a comprehensively comparative analysis of homologous genes in sequence divergence, differential expression patterns and their co-expression relationships associated with phenotypic variation, we identified genes encoding desaturases and acyltransferases that were differentially expressed between two species in developing seeds. Network-based comparative transcriptome analysis of the data from the species of Paeonia and two other ALA-rich plants, Camelina sativa and Perilla frutescens, revealed genes potentially involved in regulating ALA biosynthesis and accumulation in plants.
P. ostii was grown in Shanghai Chenshan Botanical Garden, Shanghai (31°4′52″N, 121°10′14 ″E). P. ludlowii was collected from Linzhi, Tibet (29°20′21.9″N, 94°22′35.8″E). Seeds were harvested every 10 days after flowering (DAF), spanning a total of 120 days. Three samples were collected for each species at each developmental stage. Samples used for FA analysis were dried to a constant weight at 60°C, while those used for RNA extraction were soaked in RNAlater solution and stored at −40°C prior to RNA extraction. Young stems and mature leaves were also collected and used for comparative analysis.
Lipid extraction and measurement of FA composition were conducted as previously described (Yu et al., 2016). Dried seeds were ground to fine powder. 0.2 g of the dried powder in a 3 mL mixture of 1:2 chloroform: methanol (v/v) was used to extract lipid. After fatty acid methylation, fatty acid methyl esters were analyzed by GC-MS (GC7890/MS5975, Agilent) equipped with a HP-88 capillary column (60 m, 0.25 mm, 0.2 μm, Agilent), using nonadecanoic acid as the internal standard. Omega-6 and omega-3 desaturation efficiencies (DE’s) were estimated by calculating the proportion of available substrate that was converted to product(s) as described (Menard et al., 2017): omega-6 DE = (18:2 + 18:3)/(18:1 + 18:2 + 18:3) and omega-3 DE = 18:3/(18:2 + 18:3).
Based on the dynamic pattern of oil accumulation, five stages of seed development (0, 40, 50, 60, and 100 DAF) were selected for transcriptome sampling. Total RNA’s were extracted with TIANGEN RNA Prep Plant kit (Tiangen Biotech Co., Ltd.), following the manufacturer’s protocol. The quality and quantity of obtained RNA’s were determined with Nanodrop 2000C spectrophotometry (Thermo scientific) and 2100 Bioanalyzer (Agilent Technologies). RNA samples from five developmental stages of three replicates were pooled for each species and used for de novo transcriptome sequencing and assembly, to obtain comprehensive data on expressed gene sequences. cDNA library construction and Illumina sequencing were conducted at Beijing Genomics Institute (BGI) under the Illumina manufacturer’s instructions (Illumina). In brief, mRNA was isolated from total RNA using Oligo (dT), and subsequently treated with fragmentation buffer. The short-fragmented mRNA was then utilized as template to generate first-strand cDNA. Followed by second strand cDNA synthesis, end reparation, adaptor connection and PCR amplification, the cDNA libraries were sequenced on an Illumina HiSeq 4000. To establish a temporal map of gene expression for each species, RNA’s from the five developmental stages of three replicates were also individually used for cDNA library construction and sequencing. A total of fifteen independent cDNA libraries were generated for each species. The sequence raw data from this study have been submitted to the NCBI Sequence Read Archive (SRA)1 under the BioProject ID PRJNA602603.
After quality filtering, de novo transcriptome assembly was performed using Trinity (Grabherr et al., 2011). The software TGICL (Pertea et al., 2003) was then used to cluster the assembled transcripts to generate a single non-redundant unigene set for each species.
The BLASTx program was used to align the assembled unigenes against the NCBI non-redundant protein (Nr) database2, the Swiss-Prot protein sequence database3 and the Arabidopsis protein database at the Arabidopsis Information Resource (TAIR4) with an E-value threshold of 10–10. Functional annotation by Gene Ontology (GO5) terms were obtained using the Blast2GO program (Conesa et al., 2005). In addition, we obtained metabolic pathway annotations for each hit by searching against the Kyoto Encyclopedia of Genes and Genomes (KEGG6) pathway database (Ogata et al., 1999) using the BLASTx program with an E-value cut off of 10–10.
The clean reads generated from 15 independent cDNA libraries were separately aligned to the de novo assembled transcriptome for each species, respectively. The program RSEM (Li and Dewey, 2011) was used for estimating the expression levels of each unigene, normalizing into Fragments Per Kilobase per Million reads mapped (FPKM). The NOISeq R package was used to screen genes differentially expressed at different stages of seed development (Tarazona et al., 2011). Hierarchical clustering of genes was performed using the hclust function in R v.3.5.3 (Ihaka and Gentleman, 1996). The expression stoichiometries of oil accumulation-related genes were calculated with the methods used in previous studies (Troncoso-Ponce et al., 2011). The coefficient of variation (CV) was calculated to evaluate the expression consistence among growth stages and species.
To validate the reliability of the gene expression data obtained by RNA-Seq, the expression levels of genes of interest were measured by real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) with the LightCycler 96 system (Roche) using SYBR Green PCR Master Mix (TAKARA). Three independent biological replicates and three technical replicates were performed for each reaction. Based on previous studies (Li S.S. et al., 2015; Liu et al., 2015) and the coefficient of variation of expression level, a total of 5 candidate reference genes (ubiquitin, PUF1639, RPS9, Unigene15510_F/Unigene12802_D, and CL5272.Contig1_F/Unigene12410_D) were evaluated for the normalization of RT-qPCR. Three widely-used programs, GeNorm3.5 (Vandesompele et al., 2002), BestKeeper (Pfaffl et al., 2004), and NormFinder (Andersen et al., 2004), were used to rank the stability of the 5 selected reference genes. CL5272.Contig1_F/Unigene12410_D were consistently ranked as the most stably expressed in different tissues, at different developmental phases, and between two species, and were thus used as the reference genes. Gene-specific primers (Supplementary Table 1) were designed using Primer Premier 5.0 (Lalitha, 2000) based on assembled sequences. Relative expression levels of target genes were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
To explore the divergence and phylogenetic relationships of the genes of interest, the full-length coding sequences of interested genes were cloned and sequenced. RT-PCR was conducted to clone and validate the sequences of genes with complete open reading frames (ORFs) assembled. Primers (Supplementary Table 1) were designed based on the predicted coding sequences and alignments against the GenBank database. The first-strand cDNA’s were synthesized using the PrimeScript RT (Perfect Real Time) Kit (Takara Bio). Purified PCR products were cloned into pMDTM 18-T vector (Takara Bio) and sequenced. For partial coding unigenes, the RACE (Rapid Amplification of cDNA Ends) method was implemented using the SMARTer® RACE Amplification Kit (Takara Bio). Gene-specific primers for the 5′- and 3′-RACE reactions were designed based on conserved regions identified from the multiple sequence alignments of different genes. Following cloning and sequencing the overlapping 5′ and 3′-RACE fragments, pairs of “end-to-end” primer combinations were designed and used to amplify the full-length cDNA. The amplified products were then purified and cloned into pMDTM 18-T vector for sequencing. Sequence identities were confirmed by BLAST searches against the GenBank database. The gene sequences obtained in this study have been submitted to the GenBank database with the accession codes: MH748779-MH748792. Phylogenetic analyses were carried out using the neighbor-joining method implemented in MEGA (Tamura et al., 2007).
To identify genes potentially associated with the varied accumulation of unsaturated fatty acids in different species, network-based comparative analyses were carried out based on time course gene expression data. TransDecoder7 was used to predict the coding sequences (CDS) and the peptide sequences from the assembled transcripts. Cd-hit was used to cluster contigs sharing highly identical CDS (with identity of over 90% and coverage of over 80%) (Li and Godzik, 2006). Expression values of the members included in a cluster were summed up, and the longest transcript was retained as the representative sequence. The Reciprocal Best Hits (RBH) procedure was applied to identify one to one orthologous pairs between P. ostii and P. ludlowii, which allowed for a detailed cross-species comparison (Movahedi et al., 2012). The NOISeq R package was used to screen interspecifically differentially expressed genes (Tarazona et al., 2011). Gene co-expression networks were constructed using the WGCNA R software package (Langfelder and Horvath, 2008). The matrices of pairwise Pearson correlation coefficients between all pairs of transcripts were generated and transformed into adjacency matrices using the formula: adjacency value = |(1 + correlation)/2| β. β represents the soft-thresholding power for the correlation matrix, which gives greater weight to strongest correlations while maintaining the gene-gene connectivity. The resulting adjacency matrices was then converted to topological overlap (TO) matrices via TOM similarity algorithm, and genes were hierarchically clustered based on TO similarity. The dynamic tree-cutting algorithm was used to cut the hierarchal clustering dendrogram, and modules were defined as branches from the tree cutting. The modules containing genes encoding core fatty acid synthesis enzymes and showing similar expression patterns were selected and merged to generate the oil biosynthesis module. The robustness of the merged module was validated using 5,000 permutation tests (P value < 10–3; Zhan et al., 2015). The hyper-geometric test was performed for validating the selected gene set associated with oil biosynthesis (P value < 10–6). The program Cytoscape was used to visualize the network (Shannon et al., 2003).
To confirm the potential roles of predicted genes in promoting polyunsaturated fatty acid synthesis and accumulation, the expression data of seed development of two other ALA-rich plants, C. sativa and P. frutescens, were integrated into analysis (Kagale et al., 2016; Kim et al., 2016). The PhyloMCL algorithm was used to sort the predicted peptide sequences of different species into groups of putative orthologs on the basis of sequence similarity with default parameters (Zhou et al., 2020). Gene co-expression networks were constructed and analyzed using the aforementioned methods.
Paeonia ostii and P. ludlowii exhibited a similar trend of change in oil content, though the total oil contents in mature seeds were a bit different between species (Supplementary Figure 1). At the early stage of seed development (0∼40 DAF), the oil content remained constant and low in both species. A critical transition occurred at around 40 DAF, with a sharp increase in oil content that peaked at 90 DAF and dropped slightly afterward (Figure 1A). Developmental stage-specific changes were detected not only in oil contents but also in oil compositions. Fatty acid profiling revealed five predominant components of the tree peony seed oil, i.e., stearic acid (SA), palmitic acid (PA), OA, LA, and ALA. The profiles of FA composition did not show significant difference between species at the early stages of seed development. Interspecific changes in FA composition emerged along with the massive accumulation of oil starting from 40 DAF to seed maturation. The major difference lay in the relative proportions of mono- and polyunsaturated fatty acids in the oils of different species. ALA was the most abundant fatty acid in the oil of P. ostii followed by LA and OA, whereas in the oil of P. ludlowii, OA was the predominant fatty acid (Figure 1A). Compared with that of P. ludlowii, the oil of P. ostii had a higher degree of fatty acid unsaturation. Interestingly, this pattern was reversed in stem and leaf tissues. Comparing the fatty acid profiles of mature seeds with those derived from stem and leaf tissues of P. ostii and P. ludlowii revealed that P. ludlowii contained a remarkably higher proportion of PUFA (LA and ALA) than P. ostii in stems and leaves (Figure 1B). The results also showed that long chain SFA’s including arachidic acid (C20:0) and behenic acid (C22:0) were relatively abundant in stem tissues, leading to a higher ratio of SFA:UFA in stems. By contrast, the high concentration of OA in mature seeds increased the ratios of MUFA:PUFA in seed oils, especially in P. ludlowii. Calculating the product-substrate ratio revealed a relatively lower omega-6 desaturation efficiency in P. ludlowii than in P. ostii, but not for omega-3 desaturation reaction (Supplementary Figure 2).
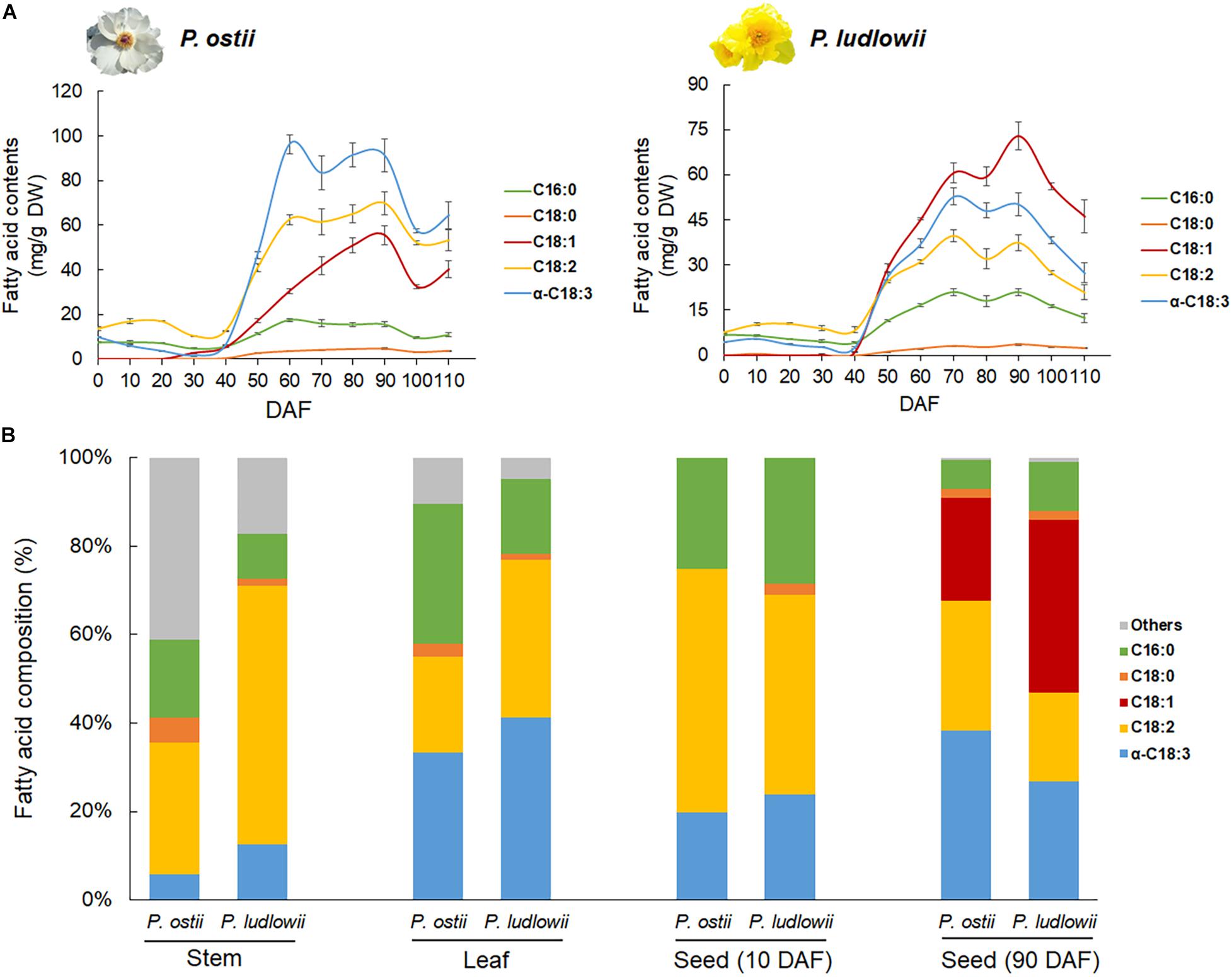
Figure 1. Fatty acid content and composition of the seed oils of P. ostii and P. ludlowii. (A) Accumulation dynamics of fatty acids during seed development, showing the similar trends of fatty acid accumulation but different fatty acid proportions between two species; (B) Relative proportions of various fatty acids in different tissues, showing inter-tissue variations in the fatty acid composition. The long chain SFA’s including arachidic acid (C20:0) and behenic acid (C22:0) were grouped into a single category (Others). Quoted values are means ± SE (N = 3). DAF: days after flowering.
De novo transcriptome assembly using 73.71 million and 73.63 million clean reads generated 54,655 and 57,665 unigenes in P. ostii and P. ludlowii, respectively, with an average length of 911 and 868 nt, N50 length of 1,447 and 1,328 nt, and overall annotation rate of 63.59 and 59.31% (Supplementary Table 2). The analogous distribution patterns of categorized annotations suggested that the expressed genes of two species were involved in similar functional categories and metabolic pathways (Supplementary Figure 3). The results of BLAST searches against the Arabidopsis Lipid Gene Database8 revealed 819 and 759 unigenes in P. ostii and P. ludlowii respectively, to be associated with lipid metabolism and signaling.
A total of 30 independent cDNA libraries were generated and sequenced for evaluating temporal gene expression patterns during seed development in P. ostii and P. ludlowii. Over 20 million clean reads were generated for each library. The clean reads were then aligned respectively, to P. ostii and P. ludlowii reference transcriptomes assembled. More than 90% of the reads were successfully aligned to the reference transcriptomes, with the least mapped sample showing a ratio of 86.11% (Supplementary Table 3).
A total of 8,089 and 14,218 unigenes identified in P. ostii and P. ludlowii, respectively, were differentially expressed at different seed development stages. Hierarchical clustering of differentially expressed genes (DEGs) generated ten clusters of DEG with similar expression dynamics in each species (Supplementary Figure 4). The unigenes included in each cluster were listed in Supplementary Tables 4, 5. Of them, the unigenes included in the cluster1, 4, and 5 of P. ostii and those from the cluster1, 4, and 7 of P. ludlowii showed bell-shaped expression patterns in agreement with the seed oil accumulation dynamics in both species. Functional enrichment analyses indicated that these genes were majorly involved in starch and sucrose metabolism, ALA metabolism, LA metabolism, biosynthesis of secondary metabolites, etc., (Supplementary Table 6).
The key biological processes involved in oil synthesis have been well studied in plants: FA’s are exclusively synthesized in plastids and then the activated FA’s are exported to endoplasmic reticulum (ER) for modification and TAG assembly (Bates et al., 2013). Genes encoding the core enzymes involved in FA synthesis, modification and subsequent incorporation into TAG’s have been identified, which are conserved among plants (Troncoso-Ponce et al., 2011). It is thus possible to detect the potential rate-limiting steps and enzymes associated with interspecific variation in oil components by calculating and comparing the transcriptional abundance of genes encoding for the core enzymes based on seed-specific genome-wide expression data. The results showed that the genes involved in de novo FA synthesis in plastids displayed similar stoichiometries between species (Figure 2), and maintained strong correlations throughout seed development with r2 values ranging from 0.74 to 0.96, with an average inter-species expression stoichiometry CV value of 0.37. In contrast to the globally conserved expression stoichiometry of genes involved in de novo FA synthesis, genes associated with FA modification and TAG assembly underwent divergence in transcriptional abundance. Of them, the genes encoding FAD2, FAD3, DGAT (acyl-CoA: diacylglycerol acyltransferase), and PDAT (phospholipid: diacylglycerol acyltransferase) exhibited distinct stoichiometries in different species, with the CV values of stoichiometry of 0.49, 0.69, 0.65, and 0.41, respectively. The correlations of expression stoichiometries of various genes involved in TAG assembly were not statistically significant during oil-accumulation stages (Figure 2C).
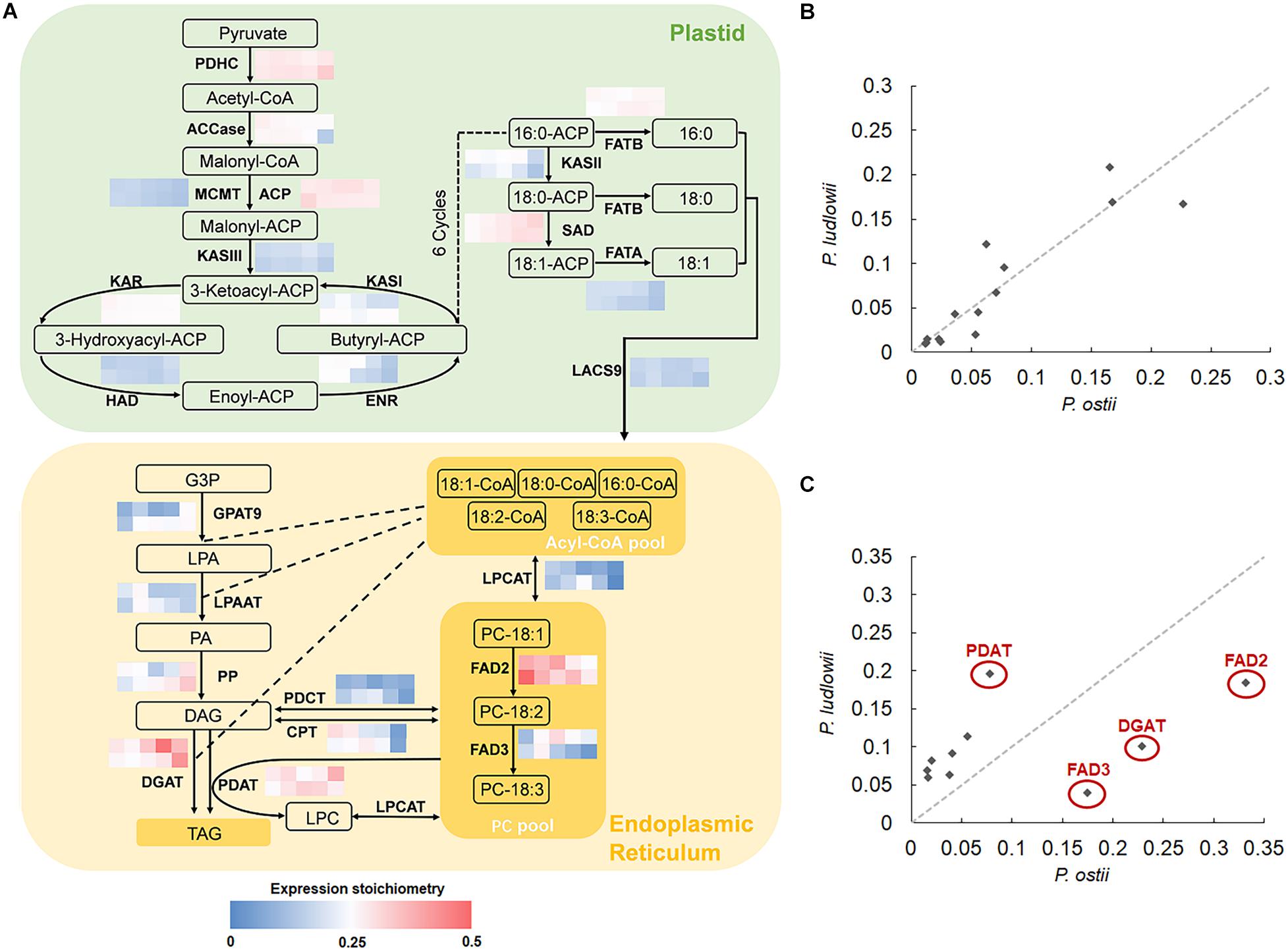
Figure 2. Cross-species comparison of expression stoichiometry of genes involved in fatty acid synthesis (FAS) and triacylglycerol (TAG) assembly. (A) Relative transcript abundance of genes encoding the core enzymes (indicated in bold) involved in lipid synthesis, with the grid showing the ratios of transcripts of each gene to the sum of FAS and TAG assembly transcripts at different developmental stages (0, 40, 50, 60, and 100 DAF from left to right) in P. ostii (upper) and P. ludlowii (lower), respectively; (B) Correlations of the expression stoichiometries of the 14 enzymes involved in de novo fatty acid synthesis at 50 DAF between P. ostii and P. ludlowii; (C) Correlations of the expression stoichiometries of the 10 enzymes involved in TAG assembly at 50 DAF between two species. Abbreviations are as follows: PDHC, plastidial pyruvate dehydrogenase complex; ACC, acetyl-CoA carboxylase; CoA, coenzyme A; ACP, acyl carrier protein; MCMT, malonyl-CoA ACP transferase; KAS, ketoacyl-ACP synthase; KAR, 3-ketoacyl-ACP reductase; HAD, 3-hydroxyacyl-ACP dehydratase; ENR, 2-enoyl-ACP reductase; FATA, acyl-ACP thioesterase A; FATB, acyl-ACP thioesterase B; SAD, stearoyl-ACP desaturases; LACS, long-chain acyl-CoA synthetase; FAD2, fatty acid desaturase 2; FAD3, fatty acid desaturase 3; GPAT9, glycerol 3-phosphate acyltransferase 9; LPAAT, lysophosphatidic acid acyltransferase; LPCAT, lysophosphatidylcholine acyltransferase; DGAT, acyl-CoA: diacylglycerol acyltransferase; PDAT, phospholipid: diacylglycerol acyltransferase; CPT, CDP-choline: DAG cholinephosphotransferase; PDCT, phosphatidylcholine:diacylglycerol cholinephosphotransferase; G3P, glycerol-3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; DAG, diacylglycerol; TAG, triacylglycerol.
Based on assembled transcripts, the complete coding sequences of FAD2 and FAD3 were cloned and sequenced in both P. ostii and P. ludlowii. The expression levels of all desaturase and terminal acyltransferase genes throughout seed development were validated by RT-qPCR. The results revealed the existence of distinct transcripts for all of the genes examined. Sequence comparison and phylogenetic analysis showed that P. ostii and P. ludlowii had at least two FAD2 and three FAD3 paralogous genes, designated respectively, as Po/PlFAD2-1, Po/PlFAD2-2, Po/PlFAD3-1, Po/PlFAD3-2, and Po/PlFAD3-3. The tree peony FAD2 paralogous genes formed two separate groups with those from other plants in the phylogenetic tree, suggesting that they were derived from a duplication event happened in the common ancestor of different plants (Supplementary Figure 5). Instead, the FAD3 paralogs pairs from P. ostii and P. ludlowii clustered together in the phylogenetic tree, forming an independent clade with three paralogous branches (Supplementary Figure 6A). Two lineage-specific duplication events might have taken place subsequently prior to the divergence of tree peony species to produce the three FAD3 paralogous genes. All tree peony FAD3 genes contained three histidine rich motifs. The FAD3-1 branch was distinct from the other two branches for lacking of the isoleucine to valine substitution in the third histidine rich motif (Supplementary Figure 6B).
A significant correlation was found between the results of RT-qPCR and RNA-Seq, as evaluated by linear regression analysis (r2 = 0.91; p < 0.0001; Supplementary Figure 7). Differential expressions of paralogous genes were detected in both FAD2 and FAD3 genes. As shown in Figure 3A, FAD2-2 was highly expressed in leaves and young seeds, and was down-regulated at seed maturing stages. In contrast, FAD2-1 was exclusively expressed in seeds. It was up regulated during seed maturation, and showed significantly higher expression levels in P. ostii than in P. ludlowii. Similarly, FAD3-1 was mainly expressed in leaves and young seeds, while FAD3-2 and FAD3-3 were mainly expressed in maturing seeds. The expressions of FAD3-2 and FAD3-3 were evidently increased in the oil accumulating stages. The relative expression level of FAD3-3 was much higher than FAD3-2 in both species, and the total level of expression of FAD3-2 and FAD3-3 in P. ostii was significantly higher than that in P. ludlowii.
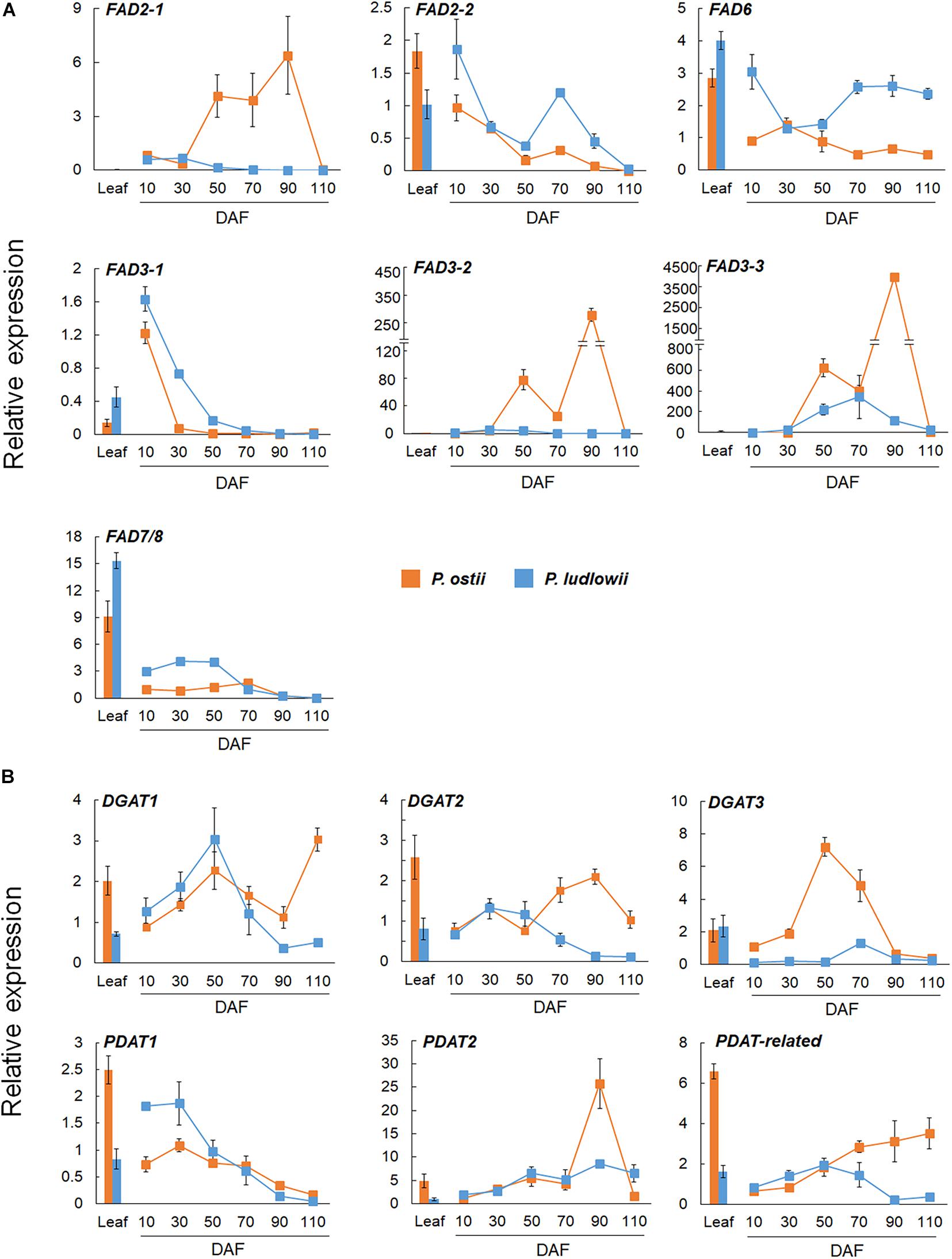
Figure 3. RT-qPCR validation of distinctly expressed desaturase and acyltransferase genes in P. ostii and P. ludlowii. (A) Differential expression patterns of the duplicates of FAD2 and FAD3 in different tissues and at different stages of seed development; (B) Differential expression patterns of analogous DGAT and paralogous PDAT genes in different tissues and at different stages of seed development. Relative expression values, normalized to the selected reference gene, were shown as 2–ΔΔCT relative to 10 DAF of P. ostii. Error bars represent the SE of three biological replicates with three technical replicates each.
DGAT’s were a group of enzymes that play critical roles in TAG synthesis and accumulation. Previous studies have shown that DGAT’s were encoded by multiple genes that evolved independently but became functionally convergent during evolution (Liu et al., 2012). In this study, three analogous DGAT transcripts were identified in each species, Po/PlDGAT1, Po/PlDGAT2, and Po/PlDGAT3, which were homologous to Arabidopsis At2g19450, At3g51520, and At1g48300, respectively. Among them, DGAT3 was mainly expressed during the oil-accumulating development stage, and showed a much higher expression level in P. ostii than in P. ludlowii. DGAT1 and DGAT2 were expressed in both leaves and seeds with a lower expression level compared to DGAT3. As shown in Figure 3B, the expression levels of DGAT1 and DGAT2 in P. ostii were lower than those in P. ludlowii during the early stages of seed development, but the situation reversed at the later stages. PDAT has been proposed to be involved in TAG biosynthesis via an acyl-CoA independent pathway, enhancing oil accumulation (Bates et al., 2013). The PDAT transcripts homologous to Arabidopsis PDAT1 (At5g13640), PDAT2 (At3g44830), and the PDAT-related gene (At4g19860) respectively, were identified in both P. ostii and P. ludlowii. The expression level of PoPDAT2 in mature seeds was much higher than that of PlPDAT2. PDAT1 was clearly down-regulated with progressing seed development, whereas the PDAT-related gene was continuously up-regulated in P. ostii but not in P. ludlowii.
The gene co-expression network was constructed using WGCNA. A total of 23 co-expression modules were generated for P. ostii, consisting of 21,842 genes. Highly correlated modules were merged to generate the oil biosynthesis module, which contained 3,745 genes. Among the 3,745 co-expressed genes, 687 genes were differentially expressed between P. ostii and P. ludlowii, suggesting their probable involvement in the regulation of FA desaturation and promoted ALA accumulation in P. ostii.
To validate the potential regulatory roles of genes co-expressed with the oil biosynthesis pathway genes in P. ostii, the gene co-expression networks of two ALA-rich species C. sativa and P. frutescens were also constructed based on the gene expression profiles during seed development. The co-expression networks comprised 21,294 genes in C. sativa, and 24,754 in P. frutescens. The oil biosynthesis modules of C. sativa and P. frutescens consisted of 4,454 and 2,329 genes, respectively. Comparing the co-expressed gene sets with that of P. ostii revealed 265 orthogroups that were shared by three ALA-rich species. A total of 362 genes co-expressed in P. ostii were also co-expressed in other two species. Of them, 91 were differentially expressed between P. ostii and P. ludlowii (Supplementary Table 7). Go enrichment analysis revealed that these genes were majorly involved in transcriptional regulation (such as genes encoding the oil store regulating genes WRI1, the basic helix-loop-helix proteins bHLH113, and the ethylene responsive factor ERF72), protein modification (such as the protein kinase genes and the ubiquitin-conjugating enzyme gene UBC5), signal transduction (such as JAZ1 involved in the jasmonic acid mediated signaling pathway), and metabolic processes (Supplementary Table 8). A few genes were found to be conservatively co-expressed with FAD2 and FAD3 across three ALA-rich species (Figure 4 and Supplementary Figure 8). Apart from some genes previously known to regulate desaturase genes, such as FUSCA3 (FUS3) and LEAFY COTYLEDON1-like (L1L), these genes included genes encoding the flavin-containing monooxygenase family protein (YUC10), the auxin-responsive GH3 family protein (DFL1), gibberellin-regulated family protein (GASA6), and genes encoding the proteins from the major facilitator superfamily. Few genes were detected to be conservatively co-expressed with DGAT3 and PDAT2 in three ALA-high species.
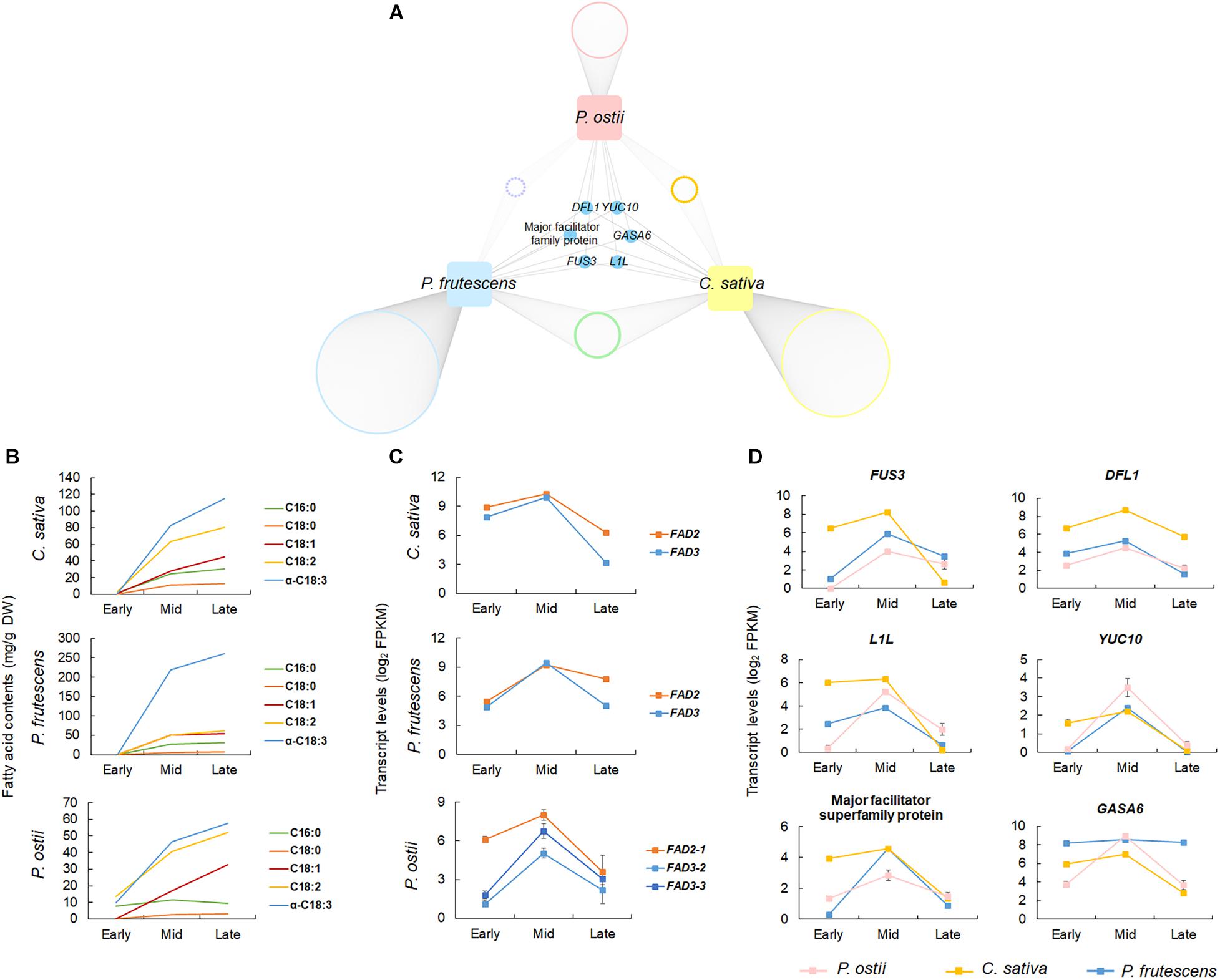
Figure 4. Comparative co-expression network analyses across P. ostii, C. sativa, and P. frutescens. (A) Network showing genes conservatively co-expressed with FAD2 and FAD3 in the three ALA-rich species; (B) Accumulation dynamics of fatty acids during seed development, showing the similar trends of fatty acid accumulation between three species; (C) Expression patterns of FAD2 and FAD3 at different stages of seed development in the three species; (D) Expression patterns of the conservatively co-expressed genes. Error bars indicate the SE of the mean.
Given the indispensability of PUFA for human health and the distinct properties required for industrial uses, it is of great necessity to decipher key regulators and possible mechanisms underlying variations in seed oil fatty acid composition (Dyer et al., 2008). P. ostii and P. ludlowii are congeneric species but have diverged in FA accumulation patterns, with ALA and OA being the most abundant fatty acids in P. ostii and P. ludlowii seed oils, respectively. The higher percentage of polyunsaturated fatty acid indicated a greater degree of fatty acid unsaturation in the P. ostii seed oil. However, within stems and leaves, the levels of LA and ALA in P. ostii were lower than in P. ludlowii. It was also found that the abundance of OA remained very low in stems and leaves, which could hardly be detected in both species. These results not only suggested that the differences in seed oil composition between P. ostii and P. ludlowii were unlikely to be plastic changes induced by environmental variation since previous studies have shown increased proportions of unsaturated fatty acids in plants at lower temperatures/higher altitudes (Linder, 2000; Zhang et al., 2015; Guerin et al., 2020), but also indicated that the reactions of fatty acid desaturation were controlled by distinct regulatory mechanisms in different organs.
The relative proportions of unsaturated FA’s in seed oils were largely determined by the stepwise desaturation processes in the oil biosynthesis pathway. The expression levels of individual FAD’s and their specificity and efficiency in catalyzing fatty acid desaturation play a key role in controlling the component ratios and the unsaturation level of seed oils. The FAD2 and FAD3 genes of tree peony have experienced gene duplication events during evolution. The varied expression patterns of the FAD2 and FAD3 duplicates in different organs clearly showed subfunctionaliztion of FAD2 and FAD3 paralogs following duplication. The significantly higher expressions of FAD2-1, FAD3-2, and FAD3-3 in P. ostii during seed maturing period were consistent with the high proportion of PUFA’s in the P. ostii seed oil. Instead, the downregulation of FAD2-1 in the maturing seeds of P. ludlowii probably induced reduction in omega-6 desaturation efficiency, decreasing the conversion rate of OA to LA and consequently leading to the higher accumulation of OA in P. ludlowii.
The last step in TAG biosynthesis was catalyzed by two types of enzymes: DGAT and PDAT. DGAT catalyzed the acyl-CoA-dependent biosynthesis of TAG, which has been proposed to be a bottleneck for oil accumulation in some plant species (Cahoon et al., 2007). Different types of DGAT’s with similar catalytic properties and no sequence homology have been identified (Liu et al., 2012; Bates et al., 2013). Gene expression profiling revealed that the tree peony DGAT genes homologous to Arabidopsis DGAT1, DGAT2 and DGAT3, respectively, were differentially expressed at different stages of seed development and between two peony species. The expression profiles of different genes shared a common feature that the expression levels of each gene in P. ostii were significantly higher than in P. ludlowii from mid to late stages of seed development (50-110 DAF). DGAT3 showed the most significant difference between species. Previous studies have shown that DGAT3 was distinct from DGAT1 and DGAT2 for its cytoplasmic localization due to lack of transmembrane domains. Although the soluble DGAT3 has been reported to participate in the cytosolic pathway of TAG synthesis, its specific role in TAG synthesis remains unclear. As demonstrated in peanut, soybean and Arabidopsis (Saha et al., 2006; Hernandez et al., 2012; Turchetto-Zolet et al., 2016), the putative DGAT3 homologs in peony species were ubiquitously expressed, with higher expression levels in maturing seeds. The main difference lay in the most abundant accumulation of the DGAT3-related transcripts occurring at the mid but not the late stages of seed development. The correlations between the expression levels of DGAT3 and the distinct fatty acid profiles observed in different peony species were yet to be clarified. Unlike DGAT’s, PDAT catalyzed the acyl-CoA-independent formation of TAG in plants. Previous studies on ALA-rich species, such as Linum usitatissimum and Perilla frutescens, have suggested the activity of PDAT to preferentially transfer ALA into TAG (Pan et al., 2013; Kim et al., 2016). Thus, compared to DGAT, PDAT might contribute more to TAG synthesis in seeds that were high in polyunsaturated fatty acids. The increased expression of peony PDAT2 and the PDAT-related gene in P. ostii seemed to support this issue, though functional and expression divergence of PDAT paralogs had been detected in this study and in other plants (Pan et al., 2013, 2015; Zang et al., 2019).
The congruent patterns of variation at the phenotypic and molecular levels suggested that the relative transcriptional abundances of the enzymes involved in FA desaturation and TAG assembly were correlated with the differential accumulation of unsaturated FA’s in different peony species. The higher expression of FAD2-1, FAD3-2, and FAD3-3 possibly promoted sequential FA desaturation in P. ostii, pushing the reaction toward the formation of PUFA. The functional relationships between the raised expression of DGAT2, DGAT3, and PDAT2 and the preferential accumulation of ALA in P. ostii remain to be elucidated. Previous studies have shown the ALA-preference of a few DGAT and PDAT enzymes in other plants (Pan et al., 2013; Kim et al., 2016; Xu et al., 2018). Experimental evaluation of the selectivity of DGAT’s and PDAT’s in peony species will be helpful to clarify whether the enhanced expression of DGAT2, DGAT3, and PDAT2 have contributed to the enrichment of ALA in P. ostii seed oil by preferentially transferring ALA into TAG. It was evident that the alterations in the relative proportions of the component fatty acids in seed oils and the developmental stage-specific changes in fatty-acid concentrations were not dependent solely on the activity of one single gene encoding enzyme, but on the synergistic effects of all pathway genes. Not only was the mRNA level of each gene encoding enzyme under the transcriptional control during seed development, but there could be some regulators participated in the spatial-temporal co-regulation of different genes and the coordination of upstream and downstream reaction modules. Deciphering the components that coordinately regulated the expression of multiple lipid biosynthetic genes and enzyme activity was essential for dissecting the regulatory mechanisms underlying variations in FA content and composition in seed oils.
Genes with coordinated expression were usually assumed to work cooperatively in related pathways, sharing similar function and regulation, especially when the co-expression relationships were conserved across species (Su et al., 2011; Medema and Osbourn, 2016). Gene co-expression analysis has been increasingly used to prioritize genes that might underlie a phenotype. In this study, we screened co-expressed gene pairs conserved across evolutionarily distant species and identified genes that were conservatively co-expressed with genes known to be involved in FA synthesis and desaturation. The results clearly showed that some of the co-expressed genes were well known as key regulators of seed oil generation, such as the transcription factor WRI1, which was identified to govern the flux of carbon through glycolysis and fatty acid synthesis by regulating genes associated with fatty acid biosynthesis in plastids (Cernac and Benning, 2004; Baud et al., 2007). The higher expression level of WRI1 of P. ostii than that of P. ludlowii was also in accordance with the slightly higher oil content of P. ostii.
In addition to WRI1, a dozen other transcription factors of different families were also tightly co-expressed with the oil biosynthesis pathway genes, including the basic helix-loop-helix proteins (bHLH113), the ethylene responsive factors (ERF72), the MYB transcription factors (MYB61, MYB86) and the NAC transcription factors (NAC100), the B3 protein family (HSL1). Most of them or their homologs have been found to be associated with diverse aspects of lipid metabolism, though very few of them have been shown to modify fatty acid proportions. For instance, bHLH113 has been shown to be up-regulated at different stages of mesocarp development to produce higher oil yield in oil palm and to be co-expressed with major lipid genes in Hiptage benghalensis (Wong et al., 2017; Tian et al., 2019). The role of hormones in regulating seed development and storage compound accumulation has been widely recognized (Yeap et al., 2017; Grimberg et al., 2018). Mutations in hormone-related transcription factors could also induce changes in the fatty acid profiles of seed oil, showing varied SFA: UFA (Hughes et al., 2008) and MUFA:PUFA ratios (Geilen et al., 2017). The functional relationships of the NAC and MYB transcription factors identified in this study to the high-level accumulation of ALA in seed oils remained largely unknown, without empirical evidences showing how they were connected. Nevertheless, such connections were supported by the genome-wide association study of A. thaliana for identifying determinants of variation in seed oil composition, which detected a group of MYB genes (MYB10, MYB64, MYB67, and MYB103) linked to the SNP’s associated with variation in fatty acid proportions (Branham et al., 2015).
Some genes involved in protein post-translational modification, such as protein phosphorylation (Protein kinase superfamily protein) and ubiquitination (UBC5), were also tightly co-expressed with fatty acid biosynthesis genes. Previous studies have shown that a serine/threonine/tyrosine protein kinase participated in the regulation of the content and composition of the Arabidopsis seed oil by mediating phosphorylation of oil body proteins (Ramachandiran et al., 2018). It has also been revealed that the seed-expressed casein kinase I enhanced the bHLH-mediated transactivation of the FAD2 gene promoter via phosphorylation of the bHLH transcription factor (Kim et al., 2010), while the ubiquitin proteasome pathway modulated FAD3 protein amounts in response to temperature (O’Quin et al., 2010). The results of this study were thus consistent with previous studies that suggested that post-translational regulatory mechanisms played an important role in modulating FAD2 and FAD3 protein abundance and stability (Martz et al., 2006; Bourassa, 2008).
Network-based comparative transcriptome analysis also revealed genes with diverse functions to be conservatively co-expressed with FAD2 and FAD3 across different species. Some of them (FUS3 and L1L) were previously known to participate in the regulation of FAD3 via (bZIP67:L1L:NF-YC2) transcriptional complex, whose induction was triggered by FUS3 (Mendes et al., 2013). Though bZIP67 was only found to be co-expressed with FAD2 and FAD3 in C. sativa, another group A bZIP member, AREB3, which was closed related to bZIP67 was found to be co-expressed with FAD2 and FAD3 in P. frutescens and P. ostii, and was differentially expressed between P. ostii and P. ludlowii, previous research has also identified that AREB3 was able to enhance FAD3 expression in the presence of L1L and NF-YC2 (Mendes et al., 2013). In P. ostii, NF-YC2 gene was also found to be co-expressed with FAD3 and FAD2 while in P. frutescens and C. sativa, some other NF-YC family members were found to exhibit similar expression patterns with FAD2 and FAD3, suggesting the potential role of other NF-YC family members in regulating desaturase genes. Apart from them, other co-expressed genes have not yet been reported to date to play roles in regulating fatty acid proportions in plants. Of them, YUC10 (Jia et al., 2014), DFL1 (Nemhauser et al., 2006) and GASA6 (Qu et al., 2016) were associated with hormone biosynthesis or response. The function of the major facilitator superfamily proteins remained unclear in plants, though it has been shown that the major facilitator superfamily domain-containing protein was an omega-3 fatty acid transporter in human (Zhang W. et al., 2018; Zhou et al., 2019). However, their relationships with the variation of fatty acid composition were not only demonstrated by the conserved co-expression patterns in different plants shown in this study, but were also supported by early genome-wide association studies (Menard et al., 2017; Zhao et al., 2019). The roles of these genes in regulating fatty acid composition remained to be functionally characterized in future research.
Fatty acid synthesis and desaturation involved a coordinated series of enzymatic reactions. Lineage-specific duplication and subsequent expression alterations of desaturase genes obviously contributed to the phenotypic divergence in the relative proportions of unsaturated fatty acids in the seed oils of peony species. However, the variation in fatty acid proportions was not entirely dependent on desaturase genes. A few genes associated with transcriptional and translational control (such as NAC100, MYB61, and UBC5) and hormonal regulation (such as YUC10, DFL1, and GASA6) seemed to be also involved to regulate the desaturase activity and coordinate various enzymatic steps. These genes were highly correlated with phenotypic variation, and were tightly co-expressed with desaturase genes, providing new targets for future functional characterization and bioengineering to extend our understanding of the regulatory mechanisms underpinning variations in fatty acid composition and unsaturation levels, and to help improve the composition of nutritional and industrial oils.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MW performed the wet lab work and data analysis and drafted the manuscript. GL, CZ, and JJ helped to set up experiments. ZX helped the material collection. YW, WZ, and ZS participated in the data analysis. LG, YH, and JY conceived the idea, participated in the design of the study, and finalized the manuscript. All of the authors read and approved the final manuscript.
This study was supported by the Chenshan Key Scientific Research Projects of Shanghai Municipal Administration of Landscaping and City Appearance (F132419) and the grant from Science and Technology Commission of Shanghai Municipality (14DZ2260400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Ning Jiang (Fudan University) and Luoyan Zhang (Shandong Normal University) for the helpful advice on data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.616338/full#supplementary-material
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Bates, P. D., Stymne, S., Ohlrogge, J., Browse, J., and Farmer, E. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi: 10.1016/j.pbi.2013.02.015
Baud, S., Mendoza, M. S., To, A., Harscoët, E., Lepiniec, L., and Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Bourassa, L. (2008). Post-translational regulation of plant fatty acid desaturases as expressed in Saccharomyces cerevisiae. New Orleans, LA: University of New Orleans. [master’s thesis].
Branham, S. E., Wright, S. J., Reba, A., and Linder, C. R. (2015). Genome-Wide association study of Arabidopsis thaliana identifies determinants of natural variation in seed oil composition. J. Hered. 107, 248–256. doi: 10.1093/jhered/esv100
Cahoon, E. B., Shockey, J. M., Dietrich, C. R., Gidda, S. K., Mullen, R. T., and Dyer, J. M. (2007). Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol. 10, 236–244. doi: 10.1016/j.pbi.2007.04.005
Cao, S., Zhou, X., Wood, C., Green, A. G., Singh, S. P., Liu, L., et al. (2013). A large and functionally diverse family of Fad2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13:5. doi: 10.1186/1471-2229-13-15
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Chen, Y., Meesapyodsuk, D., and Qiu, X. (2014). Transgenic production of omega-3 very long chain polyunsaturated fatty acids in plants: accomplishment and challenge. Biocatal. Agric. Biotechnol. 3, 38–43. doi: 10.1016/j.bcab.2013.08.007
Conesa, A., Gotz, S., Garciagomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Damude, H. G., and Kinney, A. J. (2008). Enhancing plant seed oils for human nutrition. Plant Physiol. 147, 962–968. doi: 10.1104/pp.108.121681
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Dyer, J. M., Stymne, S., Green, A. G., and Carlsson, A. S. (2008). High−value oils from plants. Plant J. 54, 640–655. doi: 10.1111/j.1365-313X.2008.03430.x
Geilen, K., Heilmann, M., Hillmer, S., and Böhmer, M. (2017). WRKY43 regulates polyunsaturated fatty acid content and seed germination under unfavourable growth conditions. Sci. Rep. 7:14235. doi: 10.1038/s41598-017-14695-14690
Grabherr, M., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grimberg, Å, Lager, I., Street, N. R., Robinson, K. M., Marttila, S., Mähler, N., et al. (2018). Storage lipid accumulation is controlled by photoperiodic signal acting via regulators of growth cessation and dormancy in hybrid aspen. New Phytol. 219, 619–630. doi: 10.1111/nph.15197
Guerin, C., Serret, J., Montúfar, R., Vaissayre, V., Bastos-Siqueira, A., Durand-Gasselin, T., et al. (2020). Palm seed and fruit lipid composition: phylogenetic and ecological perspectives. Ann. Bot. 125, 157–172. doi: 10.1093/aob/mcz175
Han, J., Liu, Z., Li, X., Li, J., and Hu, Y. (2016). Diversity in seed oil content and fatty acid composition in three tree peony species with potential as sources of omega-3 fatty acids. J. Hortic. Sci. Bi. 91, 175–179. doi: 10.1080/14620316.2015.1133538
Han, Y., Xu, G., Du, H., Hu, J., Liu, Z., Li, H., et al. (2017). Natural variations in stearoyl-acp desaturase genes affect the conversion of stearic to oleic acid in maize kernel. Theor. Appl. Genet. 130, 151–161. doi: 10.1007/s00122-016-2800-2805
Haslam, R. P., Ruiz-Lopez, N., Eastmond, P., Moloney, M., Sayanova, O., and Napier, J. A. (2013). The modification of plant oil composition via metabolic engineering—better nutrition by design. Plant Biotechnol. J. 11, 157–168. doi: 10.1111/pbi.12012
Hernandez, M. L., Whitehead, L. F., He, Z., Gazda, V., Gilday, A. D., Kozhevnikova, E., et al. (2012). A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 160, 215–225. doi: 10.1104/pp.112.201541
Hughes, R., Spielman, M., Schruff, M. C., Larson, T. R., Graham, I. A., and Scott, R. J. (2008). Yield assessment of integument−led seed growth following targeted repair of auxin response factor 2. Plant Biotechnol. J. 6, 758–769. doi: 10.1111/j.1467-7652.2008.00359.x
Ihaka, R., and Gentleman, R. (1996). R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314. doi: 10.1080/10618600.1996.10474713
Jia, H., Suzuki, M., and Mccarty, D. R. (2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 3, 135–145. doi: 10.1002/wdev.126
Kagale, S., Nixon, J., Khedikar, Y., Pasha, A., Provart, N. J., Clarke, W. E., et al. (2016). The developmental transcriptome atlas of the biofuel crop Camelina sativa. Plant J. 88:879. doi: 10.1111/tpj.13302
Kim, H. U., Lee, K. R., Shim, D., Lee, J. H., Chen, G. Q., and Hwang, S. (2016). Transcriptome analysis and identification of genes associated with ω-3 fatty acid biosynthesis in Perilla frutescens (L.) var. frutescens. BMC Genomics 17:474. doi: 10.1186/s12864-016-2805-2800
Kim, M. J., Go, Y. S., Lee, S. B., Kim, Y. S., Shin, J. S., Min, M. K., et al. (2010). Seed-expressed casein kinase I acts as a positive regulator of the SeFAD2 promoter via phosphorylation of the SebHLH transcription factor. Plant Mol. Biol. Report. 73, 425–437. doi: 10.1007/s11103-010-9630-9637
Kodali, D. R. (2002). High performance ester lubricants from natural oils. Ind. Lubr. Tribol. 54, 165–170. doi: 10.1108/00368790210431718
Lalitha, S. (2000). Primer premier 5. Biotech. Softw. Internet. Rep. 1, 270–272. doi: 10.1089/152791600459894
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Peng, Z., Yang, X., Wang, W., Fu, J., Wang, J., et al. (2013). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–72. doi: 10.1038/ng.2484
Li, S. S., Wang, L. S., Shu, Q. Y., Wu, J., Chen, L. G., Shao, S., et al. (2015). Fatty acid composition of developing tree peony (Paeonia section Moutan DC.) seeds and transcriptome analysis during seed development. BMC Genomics 16:208. doi: 10.1186/s12864-015-1429-1420
Li, S., Yuan, R., Chen, L., Wang, L., Hao, X., Wang, L., et al. (2015). Systematic qualitative and quantitative assessment of fatty acids in the seeds of 60 tree peony (Paeonia section Moutan DC.) cultivars by GC-MS. Food Chem. 173, 133–140. doi: 10.1016/j.foodchem.2014.10.017
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658. doi: 10.1093/bioinformatics/btl158
Linder, C. R. (2000). Adaptive evolution of seed oils in plants: accounting for the biogeographic distribution of saturated and unsaturated fatty acids in seed oils. Am. Nat. 156, 442–458. doi: 10.1086/303399
Liu, H., Gao, L., and Hu, Y. (2015). Reference genes discovery and selection for quantitative real-time PCR in tree peony seed and petal tissue of different development stages. J. Agric. Biotechnol. 23, 1639–1648. doi: 10.3969/j.issn.1674-7968.2015.12.014
Liu, Q., Siloto, R. M., Lehner, R., Stone, S. J., and Weselake, R. J. (2012). Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 51:350. doi: 10.1016/j.plipres.2012.06.001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martz, F., Kiviniemi, S., Palva, T. E., and Sutinen, M.-L. (2006). Contribution of omega-3 fatty acid desaturase and 3-ketoacyl-ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J. Exp. Bot. 57, 897–909. doi: 10.1093/jxb/erj075
Medema, M. H., and Osbourn, A. (2016). Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat. Prod. Rep. 33, 951–962. doi: 10.1039/C6NP00035E
Menard, G., Martin, M. J., Bryant, F., Munoz-Azcarate, O., Kelly, A. A., Hassani-Pak, K., et al. (2017). Genome wide analysis of fatty acid desaturation and its response to temperature. Plant Physiol. 173, 1594–1605. doi: 10.1104/pp.16.01907
Mendes, A., Kelly, A. A., van Erp, H., Shaw, E., Powers, S. J., Kurup, S., et al. (2013). bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25, 3104–3116. doi: 10.1105/tpc.113.116343
Movahedi, S., Van Bel, M., Heyndrickx, K. S., and Vandepoele, K. (2012). Comparative co−expression analysis in plant biology. Plant Cell Environ. 35, 1787–1798. doi: 10.1111/j.1365-3040.2012.02517.x
Nemhauser, J. L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475. doi: 10.1016/j.cell.2006.05.050
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/28.1.27
Ohlrogge, J., and Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. doi: 10.1105/tpc.7.7.957
Ohlrogge, J., Thrower, N., Mhaske, V., Stymne, S., Baxter, M., Yang, W., et al. (2018). PlantFAdb: a resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships. Plant J. 96, 1299–1308. doi: 10.1111/tpj.14102
O’Quin, J. B., Bourassa, L., Zhang, D., Shockey, J. M., Gidda, S. K., Fosnot, S., et al. (2010). Temperature-sensitive post-translational regulation of plant omega-3 fatty-acid desaturases is mediated by the endoplasmic reticulum-associated degradation pathway. J. Biol. Chem. 285, 21781–21796. doi: 10.1074/jbc.M110.135236
Pan, X., Peng, F. Y., and Weselake, R. J. (2015). Genome-wide analysis of PHOSPHOLIPID: DIACYLGLYCEROL ACYLTRANSFERASE (PDAT) genes in plants reveals the eudicot-wide PDAT gene expansion and altered selective pressures acting on the core eudicot PDAT paralogs. Plant Physiol. 167, 887–904. doi: 10.1104/pp.114.253658
Pan, X., Siloto, R. M. P., Wickramarathna, A. D., Mietkiewska, E., and Weselake, R. J. (2013). Identification of a pair of Phospholipid:Diacylglycerol Acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 288:24173. doi: 10.1074/jbc.M113.475699
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Qu, J., Kang, S. G., Hah, C., and Jang, J.-C. (2016). Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 246, 1–10. doi: 10.1016/j.plantsci.2016.01.009
Rajwade, A. V., Joshi, R. S., Kadoo, N. Y., and Gupta, V. S. (2016). Sequence characterization and in silico structure prediction of fatty acid desaturases in linseed varieties with differential fatty acid composition. J. Sci. Food Agric. 96, 4896–4906. doi: 10.1002/jsfa.7775
Rajwade, A. V., Kadoo, N. Y., Borikar, S. P., Harsulkar, A. M., Ghorpade, P. B., and Gupta, V. S. (2014). Differential transcriptional activity of SAD, FAD2 and FAD3 desaturase genes in developing seeds of linseed contributes to varietal variation in α-linolenic acid content. Phytochemistry 98:41. doi: 10.1016/j.phytochem.2013.12.002
Ramachandiran, I., Vijayakumar, A., Ramya, V., and Rajasekharan, R. (2018). Arabidopsis serine/threonine/tyrosine protein kinase phosphorylates oil body proteins that regulate oil content in the seeds. Sci. Rep. 8:1154. doi: 10.1038/s41598-018-19311-19313
Rao, S., Abdelreheem, M., Bhella, R., Mccracken, C., and Hildebrand, D. (2008). Characteristics of high alpha-linolenic acid accumulation in seed oils. Lipids 43, 749–755. doi: 10.1007/s11745-008-3207-3200
Saha, S., Enugutti, B., Rajakumari, S., and Rajasekharan, R. (2006). Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Plant Physiol. 141, 1533–1543. doi: 10.1104/pp.106.082198
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Su, Y., Li, Y., and Ye, P. (2011). Mammalian meiosis is more conserved by sex than by species: conserved co-expression networks of meiotic prophase. Reproduction 142, 675–687. doi: 10.1530/REP-11-0260
Sun, J., Chen, M., Zhu, M., Jiang, Y., Meng, J., Zhao, D., et al. (2018). Cloning, characterization, and expression analysis of three FAD8 genes encoding a fatty acid desaturase from seeds of Paeonia ostii. Molecules 23:929. doi: 10.3390/molecules23040929
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tarazona, S., Garcíaalcalde, F., Dopazo, J., Ferrer, A., and Conesa, A. (2011). Differential expression in RNA-seq: A matter of depth. Genome Res. 21:2213. doi: 10.1101/gr.124321.111
Thambugala, D., Duguid, S., Loewen, E., Rowland, G., Booker, H., You, F. M., et al. (2013). Genetic variation of six desaturase genes in flax and their impact on fatty acid composition. Theor. Appl. Genet. 126, 2627–2641. doi: 10.1007/s00122-013-2161-2162
Tian, B., Lu, T., Xu, Y., Wang, R., and Chen, G. (2019). Identification of genes associated with ricinoleic acid accumulation in Hiptage benghalensis via transcriptome analysis. Biotechnol. Biofuels 12:16. doi: 10.1186/s13068-019-1358-1352
Troncoso-Ponce, M. A., Kilaru, A., Cao, X., Durrett, T. P., Fan, J., Jensen, J. K., et al. (2011). Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68:1014. doi: 10.1111/j.1365-313X.2011.04751.x
Turchetto-Zolet, A. C., Christoff, A. P., Kulcheski, F. R., Loss-Morais, G., Margis, R., and Margis-Pinheiro, M. (2016). Diversity and evolution of plant diacylglycerol acyltransferase (DGATs) unveiled by phylogenetic, gene structure and expression analyses. Genet. Mol. Biol. 39, 524–538. doi: 10.1590/1678-4685-gmb-2016-2024
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–12. doi: 10.1186/gb-2002-3-7-research0034
Voelker, T., and Kinney, A. J. (2001). Variations in the biosynthesis of seed-storage lipids. Annu. Rev. Plant Biol. 52, 335–361. doi: 10.1146/annurev.arplant.52.1.335
Wang, X., and Liu, A. (2014). Expression of genes controlling unsaturated fatty acids biosynthesis and oil deposition in developing seeds of Sacha Inchi (Plukenetia volubilis L.). Lipids 49, 1019–1031. doi: 10.1007/s11745-014-3938-z
Wong, Y. C., Teh, H. F., Mebus, K., Ooi, T. E. K., Kwong, Q. B., Koo, K. L., et al. (2017). Differential gene expression at different stages of mesocarp development in high- and low-yielding oil palm. BMC Genomics 18:470. doi: 10.1186/s12864-017-3855-3857
Xiu, Y., Wu, G., Tang, W., Peng, Z., Bu, X., Chao, L., et al. (2018). Oil biosynthesis and transcriptome profiles in developing endosperm and oil characteristic analyses in Paeonia ostii var. lishizhenii. J. Plant Physiol. 228, 121–133. doi: 10.1016/j.jplph.2018.05.011
Xu, Y., Holic, R., Li, D., Pan, X., Mietkiewska, X., Chen, G., et al. (2018). Substrate preferences of long-chain acyl-CoA synthetase and diacylglycerol acyltransferase contribute to enrichment of flax seed oil with α-linolenic acid. Biochem. J. 475, 1473–1489. doi: 10.1042/BCJ20170910
Yeap, W., Lee, F., Shabari Shan, D. K., Musa, H., Appleton, D. R., and Kulaveerasingam, H. (2017). WRI 1−1, ABI 5, NF−YA 3 and NF−YC 2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 91, 97–113. doi: 10.1111/tpj.13549
Yin, D., Xu, W., Shu, Q., Li, S., Wu, Q., Feng, C., et al. (2018). Fatty acid desaturase 3 (PsFAD3) from Paeonia suffruticosa reveals high α-linolenic acid accumulation. Plant Sci. 274, 212–222. doi: 10.1016/j.plantsci.2018.05.027
Yu, S., Du, S., Yuan, J., and Hu, Y. (2016). Fatty acid profile in the seeds and seed tissues of Paeonia L. species as new oil plant resources. Sci. Rep. 6:26944. doi: 10.1038/srep26944
Yurchenko, O., Park, S., Ilut, D. C., Inmon, J. J., Millhollon, J. C., Liechty, Z. S., et al. (2014). Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 14, 312–312. doi: 10.1186/s12870-014-0312-315
Zang, X., Geng, X., Ma, L., Wang, N., Pei, W., Wu, M., et al. (2019). A genome-wide analysis of the phospholipid: diacylglycerol acyltransferase gene family in Gossypium. BMC Genomics 20:402. doi: 10.1186/s12864-019-5728-5728
Zhan, J., Thakare, D., Ma, C., Lloyd, A., Nixon, N. M., Arakaki, A. M., et al. (2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27, 513–531. doi: 10.1105/tpc.114.135657
Zhang, J., Zhang, S., Zhang, Y., and Kitajima, K. (2015). Effects of phylogeny and climate on seed oil fatty acid composition across 747 plant species in China. Ind. Crops Prod. 63, 1–8. doi: 10.1016/j.indcrop.2014.10.045
Zhang, Q., Yu, R., Sun, D., Rahman, M., Xie, L., Hu, J., et al. (2019). Comparative transcriptome analysis reveals an efficient mechanism of α-linolenic acid in tree peony seeds. Int. J. Mol. Sci. 20:65. doi: 10.3390/ijms20010065
Zhang, Q., Yu, R., Xie, L., Rahman, M. M., Kilaru, A., Niu, L., et al. (2018). Fatty acid and associated gene expression analyses of three tree peony species reveal key genes for α-linolenic acid synthesis in seeds. Front. Plant Sci. 9:106. doi: 10.3389/fpls.2018.00106
Zhang, W., Chen, R., Yang, T., Xu, N., Chen, J., Gao, Y., et al. (2018). Fatty acid transporting proteins: Roles in brain development, aging, and stroke. Prostaglandins Leukotrienes Essent. Fatty Acids 136, 35–45. doi: 10.1016/j.plefa.2017.04.004
Zhao, X., Jiang, H., Feng, L., Qu, Y., Teng, W., Qiu, L., et al. (2019). Genome-wide association and transcriptional studies reveal novel genes for unsaturated fatty acid synthesis in a panel of soybean accessions. BMC Genomics 20:68. doi: 10.1186/s12864-019-5449-z
Zhou, J., Chi, X., Cheng, M., Huang, X., Liu, X., Fan, J., et al. (2019). Zika virus degrades the ω-3 fatty acid transporter Mfsd2a in brain microvascular endothelial cells and impairs lipid homeostasis. Sci. Adv. 5:eaax7142. doi: 10.1126/sciadv.aax7142
Keywords: seed oil, degree of unsaturation, regulatory gene, gene duplication, conserved co-expression
Citation: Wang M, Gao L, Li G, Zhou C, Jian J, Xing Z, Wang Y, Zhang W, Song Z, Hu Y and Yang J (2021) Interspecific Variation in the Unsaturation Level of Seed Oils Were Associated With the Expression Pattern Shifts of Duplicated Desaturase Genes and the Potential Role of Other Regulatory Genes. Front. Plant Sci. 11:616338. doi: 10.3389/fpls.2020.616338
Received: 12 October 2020; Accepted: 22 December 2020;
Published: 13 January 2021.
Edited by:
Aleš Svatoš, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Luisa Hernandez, Institute of Plant Biochemistry and Photosynthesis (IBVF), SpainCopyright © 2021 Wang, Gao, Li, Zhou, Jian, Xing, Wang, Zhang, Song, Hu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Hu, aHV5b25naG9uZ0Bjc25iZ3NoLmNu; Ji Yang, aml5YW5nQGZ1ZGFuLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.