- 1Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao, China
- 2Key Laboratory of Cell Activities and Stress Adaptations, Ministry of Education, School of Life Sciences, Lanzhou University, Lanzhou, China
RNA splicing is an essential post-transcriptional regulation in plant mitochondria and chloroplasts. As the mechanism of RNA splicing remains obscure, identification and functional elucidation of new splicing factors are necessary. Through a characterization of two maize mutants, we cloned Empty pericarp 24 (Emp24) and Empty pericarp 25 (Emp25). Both Emp24 and Emp25 encode mitochondrion-targeted P-type PPR proteins. EMP24 is required for the splicing of nad4 introns 1 and 3, which was reported (Ren Z. et al., 2019), and EMP25 functions in the splicing of nad5 introns 1, 2, and 3. Absence of either Nad4 or Nad5 proteins blocks the assembly of mitochondrial complex I, resulting in the formation of a sub-sized complex I of similar size in both mutants. Mass spectrometry identification revealed that the subcomplexes in both mutants lack an identical set of proteins of complex I. These results indicate that EMP24 and EMP25 function in the splicing of nad4 and nad5 introns, respectively, and are essential to maize kernel development. The identification of the subcomplexes provides genetic and molecular insights into the modular complex I assembly pathway in maize.
Introduction
Mitochondria are the main energy powerhouse of the cell, which perform oxidative phosphorylation through the electron transport chain (ETC) (Knoop, 2013). The cytochrome pathway of ETC is composed of four protein complexes, I, II, III, and IV. Complex I (NADH dehydrogenase) is the inception point of ETC, which functions to remove two electrons from NADH and transfers to ubiquinone (UQ). Complex II (succinate dehydrogenase) is a parallel electron transport pathway to complex I and delivers additional electrons to the quinone pool (Q). Complex III (cytochrome c reductase) transfers electrons from ubiquinol to cytochrome c, which is a water-soluble electron carrier. Complex IV (cytochrome c oxidase) removes electrons from cytochrome c to oxygen (O2) for water production (Sweetlove et al., 2007; Sazanov, 2015). In the ETC reactions, the protons are transported from the matrix to the intermembrane space, while complex V (ATP-synthase) pumps protons back to the matrix for adenosine triphosphate (ATP) synthesis, which keeps the balance of electron charge between the matrix and the intermembrane space (Zhou et al., 2015). The subunits of these complexes are encoded by both the mitochondrial and nuclear genome; thus, coordination between the two genetic systems is important (Poyton and McEwen, 1996).
As the descendant of α-proteobacteria endosymbiont, mitochondrion in higher plants retains its own genome, which encodes approximately 40 identified proteins (variable in different species) and numerous open-reading frames (ORFs) with unknown origin and function (Unseld et al., 1997; Adams et al., 2002; Notsu et al., 2002; Clifton et al., 2004). The expression of mitochondrial genes is regulated at the post-transcriptional level by nucleus-encoded factors through RNA splicing, cleavage, stabilization, and editing (Binder and Brennicke, 2003; Liere et al., 2011; Hammani and Giege, 2014). These processes are critical for mitochondrial functions as perturbations of these events usually result in defects in growth and development (Meyer et al., 2009; Liu et al., 2013), hormone response (Jiang et al., 2014; Sechet et al., 2015), and cytoplasmic male sterility (Hu et al., 2012; Tang et al., 2014).
Among the post-transcriptional regulation processes, RNA splicing of group II introns in higher plant mitochondria relies on the splicing factors due to the loss of self-splicing capacity of these introns (Bonen, 2008). Different from the splicing facilitated by specific maturase (Mat) that is encoded within the intron itself in bacterial and yeast mitochondria (Bonen and Vogel, 2001), the plant mitochondrial genes contain only one MatR in the nad1 intron 4 and four MATs (nMAT) are encoded by the nucleus in Arabidopsis (Keren et al., 2009, 2012; Cohen et al., 2014). These maturases are required for the splicing of a large number of introns (Keren et al., 2009, 2012; Cohen et al., 2014; Sultan et al., 2016). Additionally, nucleus-encoded factors of different RNA-binding protein families have been identified to function in mitochondrial intron splicing. These include pentatricopeptide repeat (PPR) proteins, plant organellar RNA recognition (PORR) domain protein, mitochondrial transcription termination factor (mTERF), mitochondrial CAF-like splicing factor (mCSF), RNA DEAD-box helicases, and regulator of chromosome condensation (RCC1) protein (Brown et al., 2014).
The PPR protein family is remarkably large, >400 members in land plants. Its members are composed of up to 30 degenerate repeats of a 35-amino-acid motif arrayed in tandem (Small and Peeters, 2000) and are categorized into two major subfamilies according to the motif type. The P-subfamily consists of a canonical 35-residue motif (P-type), while the PLS-subfamily is composed of short (S), long (L), and P-type motifs often with additional C-terminal domains (E, E +, and DYW) (Lurin et al., 2004). Most PPR proteins are targeted to organelles to function in post-transcriptional regulation (Small and Peeters, 2000). However, owing to its large number, functions of many PPR proteins remain to be identified. In this study, we cloned Empty pericarp 24 (Emp24) and Empty pericarp 25 (Emp25) in mutants of maize seed development. EMP24 is the same as EMP602, which is required for the splicing of nad4 introns 1 and 3 (Ren Z. et al., 2019), and EMP25 is required for the splicing of nad5 introns. Lack of either Nad4 or Nad5 blocks the assembly of complex I holoenzyme but allows the formation of a sub-sized complex I containing N and Q modules in the matrix arm and proximal (PP) module modules in the membrane arm, which is similar to the complex I assembly in Arabidopsis (Ligas et al., 2019). Analysis of the subcomplex reveals that the absence of either Nad4 or Nad5 leads to the formation of a subcomplex that misses an identical set of subunits. Based on the proposed modular pathway of mitochondrial complex I assembly in Arabidopsis (Ligas et al., 2019), our results provide genetic evidence to the modular complex I assembly pathway in monocots.
Materials and Methods
Plant Materials
The emp24 and emp25 alleles were obtained from the UniformMu population, which was generated by introgressing Mu active line into the W22 inbred genetic background (McCarty et al., 2005). All plants were cultivated in the field under natural conditions.
Light Microscopy of Cytological Sections
WT and mutant sibling kernels were harvested from emp25 self-pollinated heterozygous ears at different days after pollination (DAP), respectively. The sections’ preparation, staining, and observation were performed as previously described (Shen et al., 2013).
Gene Cloning and Linkage Analysis
The Mu inserted flanking sequences were isolated by the Mu-seq high-throughput sequencing approach (Liu et al., 2016). emp24 and emp25 were 2 of 14 independent mutants in the 14 × 12 grid. Linkage analysis was performed as previously described (Sun et al., 2015), using EMP24-R1 and TIR8 as primers (62°C annealing temperature running 8 cycles followed by 56°C annealing temperature running 32 cycles). The same PCR condition was used in emp25 linkage analysis with primers EMP25-R1 and TIR8. The primers are listed in Supplementary Table S1.
Subcellular Localization of EMP24 and EMP25
The full length of Emp24 or the 669-bp (start from ATG) fragment of Emp25 was ligated into pENTR/D-TOPO (Invitrogen, United States) and then introduced into binary vector pBI221 (CaMV 35S promoter) and pGWB5 (CaMV 35S promoter) via Gateway technology, respectively. The mitochondria-localized protein ATPase was fused with RFP in the C-terminal in the 326-RPF vector (CaMV 35S promoter). EMP24-GFP and ATPase-RFP were co-transiently expressed in Arabidopsis thaliana leaf protoplast by Agrobacterium tumefaciens (EHA105) (Yoo et al., 2007). EMP25-GFP was transiently expressed in Nicotiana tabacum leaves by A. tumefaciens (EHA105) infiltration (van Herpen et al., 2010). The fluorescence signals were detected by a ZEISS LSM 700 confocal microscope. Mitochondria were labeled by the MitoTracker Red (Life Technologies, United States).
RNA Extraction, RT-PCR, Quantitative RT-PCR, and cRT-PCR
Total RNA was extracted with the Trizol reagent (Invitrogen, United States) and treated with RNase-free DNase I (NEB, United States) to eliminate g-DNA contamination and then reverse transcribed using random hexamers as primers. RT-PCR analysis of full-length transcript level difference in emp25 with sibling WT in 34 protein-coded maize mitochondrial genes was performed with ZmActin (ZmActin-F1/R1) as normalization. The mitochondrial gene primers were used as previously described (Liu et al., 2013). In the RT-PCR of nad5 splicing analysis, four pairs of primers (nad5-F1/R1, nad5-F2/R2, nad5-F3/R3, and nad5-F4/R4) were designed to test four introns splicing, respectively. PCR conditions were 95°C for 30 s, 56°C for 30 s, and 72°C for 120 s, with a total of 30 cycles. ZmActin (Zmactin-F1/R1) was used as normalization. Primers for qRT-PCR analysis were designed according to Colas des Francs-Small et al. (2014). qRT-PCR was performed as described above.
Primers for quantitative RT-PCR (qRT-PCR) analysis were designed according to Colas des Francs-Small et al. (2014). cDNAs were obtained from three biological repeats, respectively. qRT-PCR was performed using SYBR Green on a LightCycler® 96 (Roche). PCR conditions are 95°C for 10 s and 60°C for 10 s, with a total of 40 cycles, following 65°C for 5 s and 95°C for 5 s for melt curve. qRT-PCR was performed as described above. Circular RT-PCR (cRT-PCR) analysis was performed as previously described (Zhang et al., 2019). All primers are listed in Supplementary Table S1.
Editing Analysis of nad4 and nad5 mRNA in WT and emp24/emp25
Total RNA was extracted with the Trizol reagent (Invitrogen, United States) and treated with RNase-free DNase I (NEB, United States) to eliminate g-DNA contamination and then reverse transcribed using random hexamers as primers. The full length of nad4 and nad5 was amplified by using primers nad4-F1/R1 and nad5-F1/R1, respectively. An RNA editing analysis was conducted from these samples by directly sequencing nad4 and nad5, as described in Liu et al. (2013).
Mitochondrial Complex I Activity and Western Blotting Assay
Crude and intact mitochondria were isolated from about 10 ml of maize kernel (removed pericarp) at 13 DAP. Blue native PAGE and in-gel complex I activity assay were performed as previously described (Meyer et al., 2009). Protein concentration was determined using the Bradford assay (Bio-Rad). Western blotting was performed as previously described (Sun et al., 2015).
Results
Molecular Cloning of Emp24 and Emp25
The high-throughput Mu-seq analysis was employed to isolate the causal gene of emp24 and emp25 (McCarty et al., 2013). After all germinal insertions are distributed into samples in the grid (14 × 12) based on barcodes, the phenotype of emp24-1 was found to co-segregate with the Mu insertion at +45 bp start from ATG of GRMZM2G464510 (Supplementary Figure S1A). Linkage analysis by using GRMZM2G464510 gene-specific primer with Mu primer TIR8 showed a tight linkage in 224 individual plants in an F2 progeny of emp24-1 mutant (Supplementary Figure S1B). To confirm that the emp24-1 phenotype is due to mutation of GRMZM2G464510, we isolated two alleles with Mu inserted in the coding region of GRMZM2G464510 from Maize Genetics Cooperation Stock Center, UFMu08492 and UFMu01097. The Mu transposon insertion position of UFMu08492 (+ 45 bp start from ATG) is identical to the one in emp24-1, and the UFMu01097 with Mu inserted at + 483 bp (start from ATG) was named emp24-2. Self-pollinated heterozygotes for emp24-2 segregated emp kernels in a recessive manner, and crosses between emp24-1 and emp24-2 produced emp kernels (Supplementary Figure S2A). These results confirmed that GRMZM2G464510, which has been identified as Emp602 encodes a P-type PPR protein that is specific for the splicing of mitochondrial nad4 introns 1 and 3 (Ren Z. et al., 2019), is the causal gene for the emp24 phenotype.
For emp25-1, the Mu-Seq analysis identified a Mu insertion at +98 bp start from ATG of GRMZM2G312954 (Supplementary Figure S1C). Linkage analysis in 147 members of emp25-1/ + selfed progeny by using TIR8 and GRMZM2G312954 gene specific primers found a tight linkage between the Mu insertion and the emp phenotype (Supplementary Figure S1D). A second allele was identified in UFMu-02672 (+88 bp start from ATG) from the Maize Stock Center, referred to as emp25-2. Crossed ears between emp25-1/+ and emp25-2/+ exhibited emp kernels (Supplementary Figure S2B). These results show that GRMZM2G312954 is the causal gene for the emp25 phenotypes, referred to as Emp25 hereafter.
Loss of Function in Emp25 Impairs Embryo and Endosperm Development
The emp25 mutant showed a 3:1 segregation ratio of wild type (WT) and empty pericarp kernels in self-pollinated heterozygous plants, indicating that the mutation is monogenetic and recessive. The mutant kernels of emp25 could be clearly identified at 11 DAP (Figure 1A), and at maturity, the pericarp collapsed and wrinkled, giving rise to an emp phenotype (Figures 1B,C).
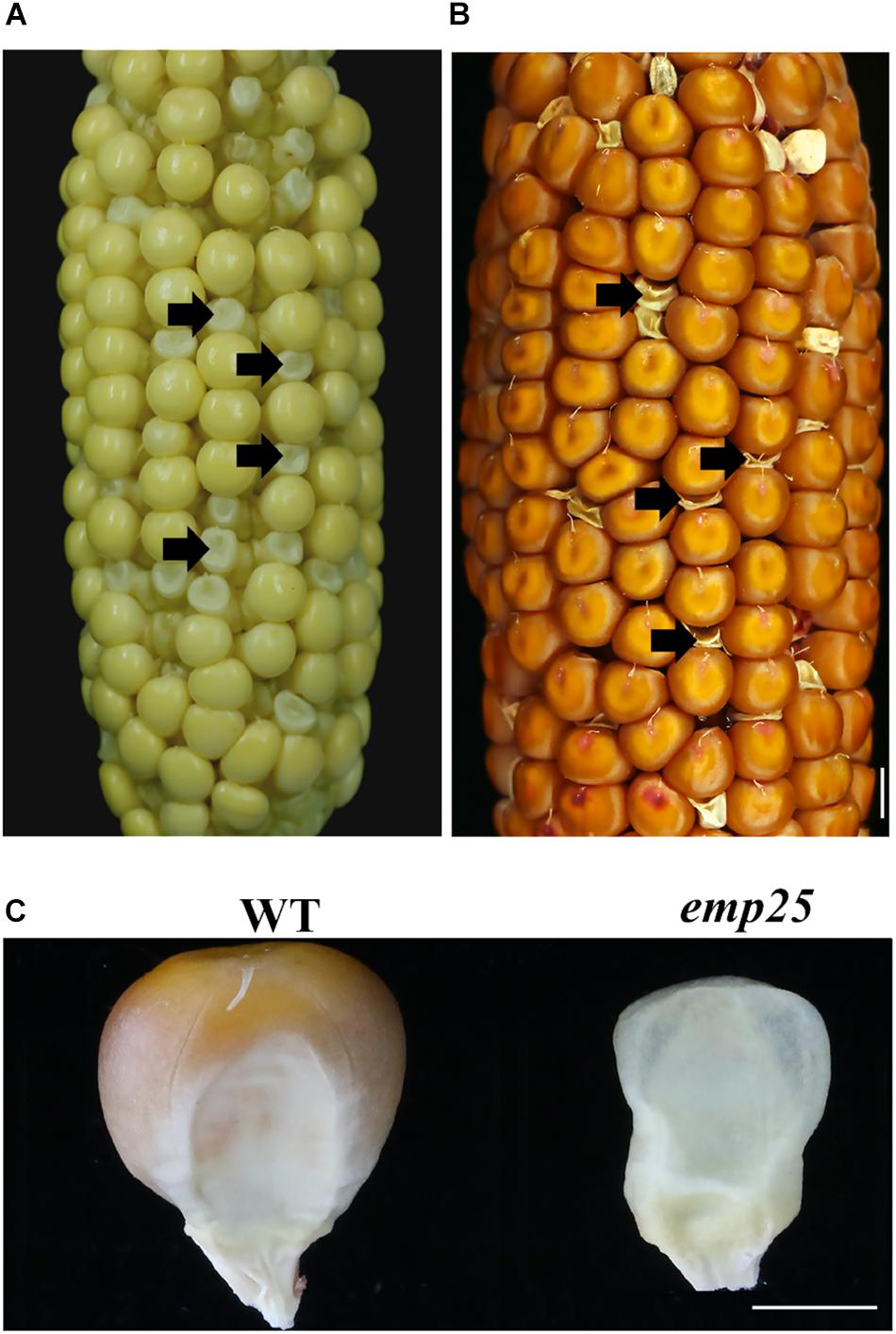
Figure 1. Phenotype of emp25. (A,B) The ear segregates the emp25 kernels at 11 days after polination (DAP) (A) and maturity (B). Arrows indicate the mutants. Scale bar = 0.5 cm. (C) Germinal side view of the WT and emp25 kernel. Scale bar = 0.2 cm.
The embryo development of maize is divided into transition, coleoptilar, and late embryogenesis stage (Fontanet and Vicient, 2008). The endosperm development in maize is classified into coenocytic, cellularization, differentiation, and maturation stages (Olsen, 2001). To underpin the developmental arrest, the emp25 and the WT sibling kernels in the same ear were analyzed at different DAP by paraffin section. At 10 DAP, the WT kernels differentiated clear coleoptile, shoot apical meristem, and suspensor, but the emp25 mutant just formed an embryo proper attached to the suspensor (Figures 2A,B). At 14 DAP, the WT embryo reached late embryogenesis stage with primary leaves, but the emp25 mutant embryo reached the transition stage (Figures 2C,D) and remained at that stage after, suggesting that the embryo development of emp24 was arrested at the transition stage. Both the WT and the emp25 mutant endosperm finished the cellularization and reached the differentiation stage at 14 DAP, but the smaller size of mutant endosperm compared with WT endosperm and the hollow between endosperm and pericarp in mutant suggested that endosperm development was seriously affected in the emp25 (Figures 2B,D).
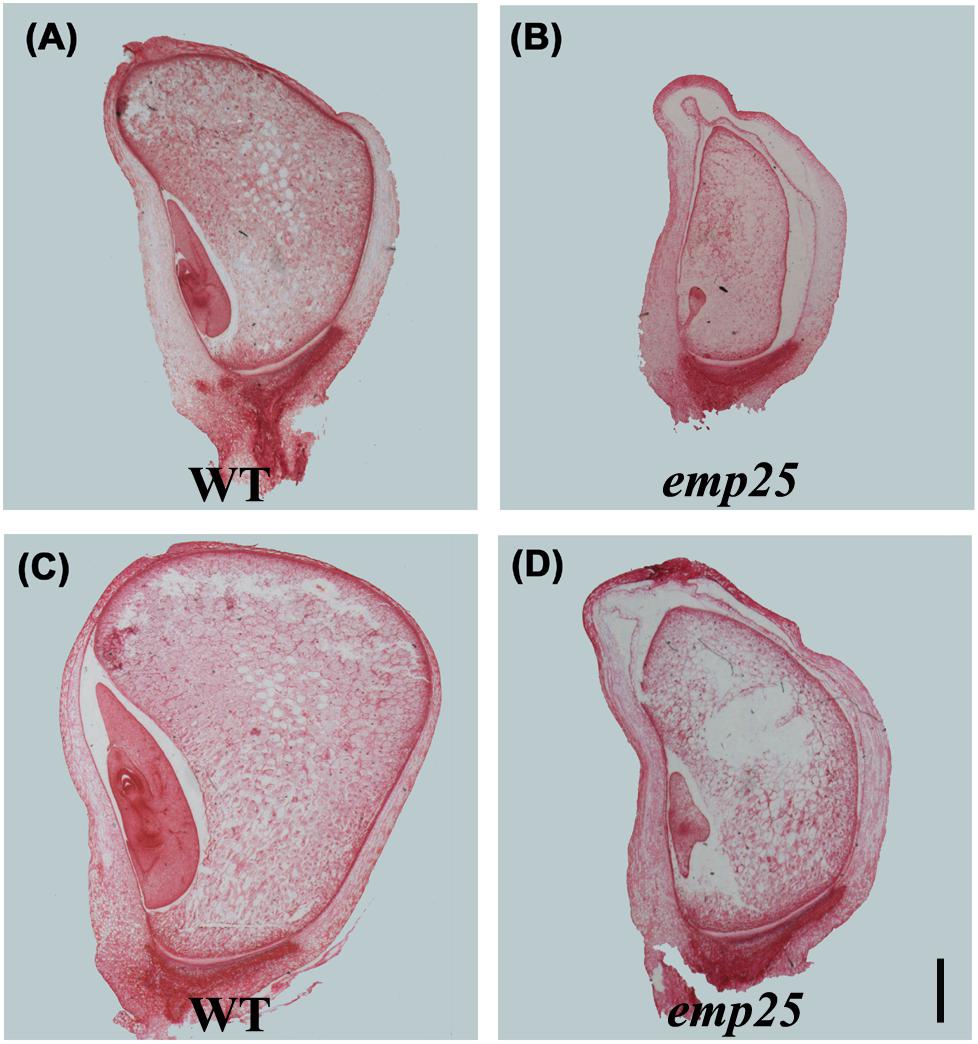
Figure 2. Embryo and endosperm development of emp25. Paraffin sections of the emp25 mutant kernels and the corresponding WT siblings at 10 DAP (A,B) and 14 DAP (C,D). Scale bar = 1 mm.
EMP25 Is Localized in Mitochondria
The Emp25 gene contains five introns (Supplementary Figure S1C), also encoding a P-type PPR protein with 20 PPR motifs (Figure 3A). EMP25 shares a 92% identity with Sb02g380100 in Sorghum bicolor, 70% identity with Os05g19390 in Oryza sativa, and 41% identify with At1g19290 (TANG2, Colas des Francs-Small et al., 2014) in A. thaliana. We tested the expression of Emp25 in the mutant alleles by RT-PCR. No WT transcript was detected in two alleles (Supplementary Figure S3A). Also, the transcript level of Emp25 in main tissues during maize growth and development was analyzed by qRT-PCR. Expression was relatively higher in shoot, pollen, leaf, and roots than in bract and tassel. During kernel development, Emp25 is expressed at a higher level in the early developing kernels at 9 DAP than at later stages (Supplementary Figure S3B). These data indicate that Emp25 is a ubiquitously expressed gene, rather than a seed-specific gene.
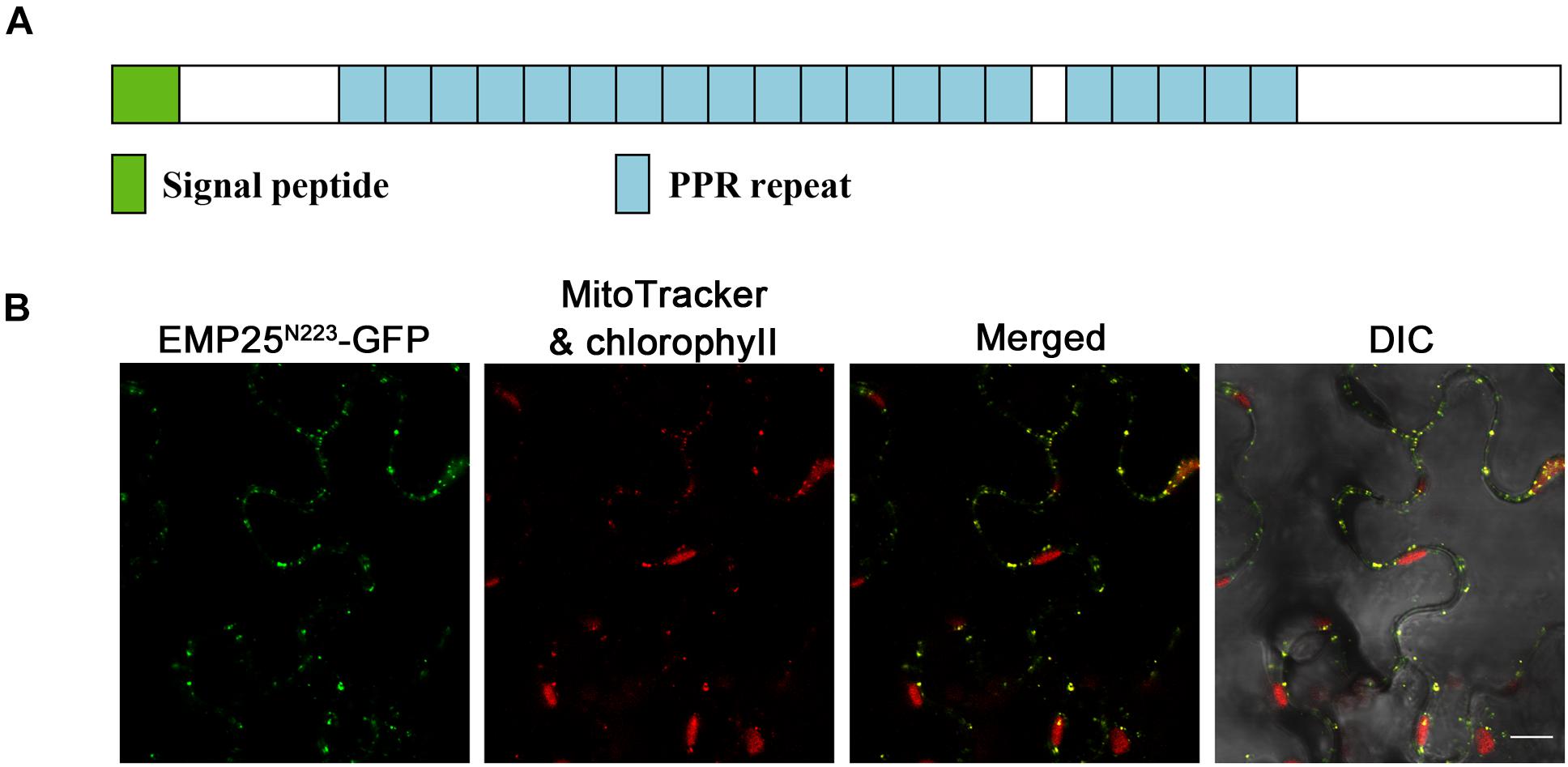
Figure 3. EMP25 is a mitochondrion-localized P-type PPR protein. (A) Schematic diagram of the EMP25 protein. (B) Subcellular localization of EMP25. The N-terminus (223 amino acids) of EMP25 was fused with the green fluorescence protein (GFP). The EMP25N223aa–GFP fusion was transiently expressed in tobacco leaf epidermal cells and the signal was detected by confocal fluorescent microscopy. Mitochondria were detected with MitoTracker. DIC, differential interference contrast. Scale bar = 10 μm.
Because full EMP25-GFP fusion did not produce detectable signals, we fused the N-terminal 223 amino acids of EMP25 with GFP (GFP at the C-terminal). The fusion protein was transiently expressed in tobacco leaf epidermal cells and the MitoTracker red was used as the mitochondrial marker. The results showed that the GFP signals in small dots were completely merged with the MitoTracker red signals (Figure 3B), indicating that EMP25 is localized in mitochondria.
The emp25 Mutant Is Deficient in the Splicing of nad5 Introns
Mitochondrial localized P-type PPR proteins have been shown to function in intron splicing (Barkan and Small, 2014). To uncover the functions of EMP25, we first examined the transcript levels of the 34 mitochondrial protein-coding genes in kernels at 13 DAP (pericarp removed) (Clifton et al., 2004). The primers were designed to amplify nearly full-length transcripts and the RNA templates were treated with DNase and normalized against ZmActin (GRMZM2G126010) (Li et al., 2014). Among the 34 genes, no significant difference in transcript levels was detected in most genes between WT and mutants. However, for emp25, the nad5 transcript is missing (Figure 4A). Such results were consistent with the mRNA accumulation analysis of these 34 genes by qRT-PCR (Figure 4B).
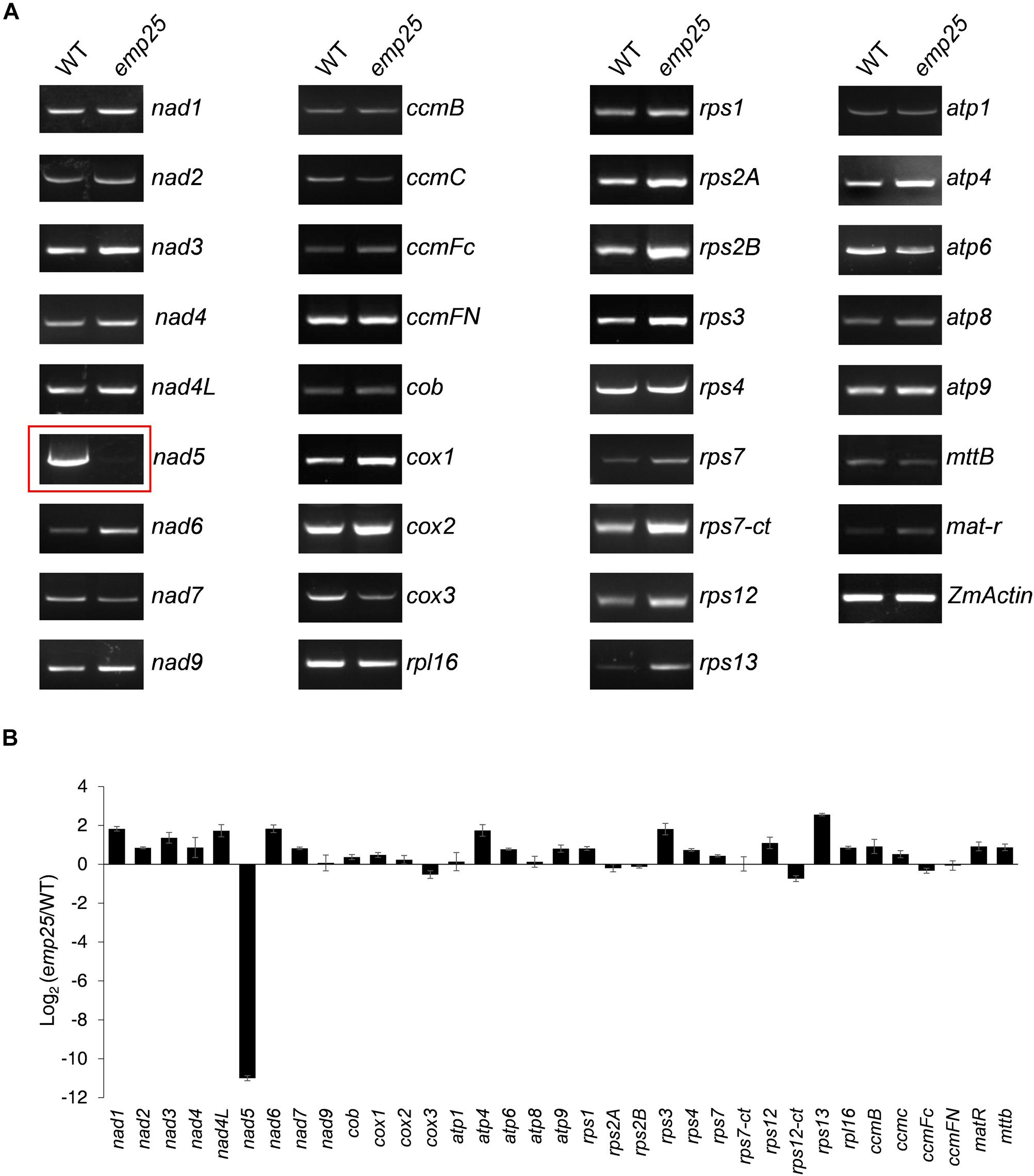
Figure 4. Analysis of mitochondrial gene transcripts in the emp25 kernels. (A) RT-PCR analysis of the 34 mitochondrial protein coding genes in emp25 with corresponding WT siblings. (B) qRT-PCR analysis of the mitochondrial transcripts in WT and emp25. Values represent the mean and standard deviation of three biological replicates. RNA was extracted from 13 DAP kernels without pericarp and was normalized against ZmActin.
The maize mitochondrial nad5 contains four introns, two cis- and two trans-introns (Figure 5A). To test whether the absence of mature transcripts is due to defects in intron splicing, we analyzed the splicing efficiency of each intron by RT-PCR with specific primers anchored across each intron. In emp25, the splicing of nad5 intron 1 (cis) was drastically decreased, and the splicing of nad5 trans-introns 2 and 3 was abolished. For unspliced cis-introns, accumulation of a larger size fragment was detected, which is the unspliced intron as confirmed by sequencing. For trans-intron, such a large fragment was not detected, suggesting that no spliced transcript is produced in the mutants. To independently verify the splicing defects in the emp25, we determined the splicing efficiency of the mitochondrial 22 introns in maize by qRT-PCR analysis in two alleles. The result showed that the splicing efficiency of nad5 introns 1, 2, and 3 was decreased 16-, 2, 000-, and 128-fold in the emp25 mutant when compared to the WT (Figure 5B). Furthermore, in order to confirm that EMP25 is specific in intron splicing of nad5, we detected the editing sites and 5′/3′ maturation of nad5 in emp25. No editing sites were changed (Supplementary Figure S4), and 5′/3′ maturation of nad5 is normal in emp25 (Supplementary Figure S5). These results confirmed that the loss of function in EMP25 affects the splicing of nad5 introns 1, 2, and 3 in mitochondria.
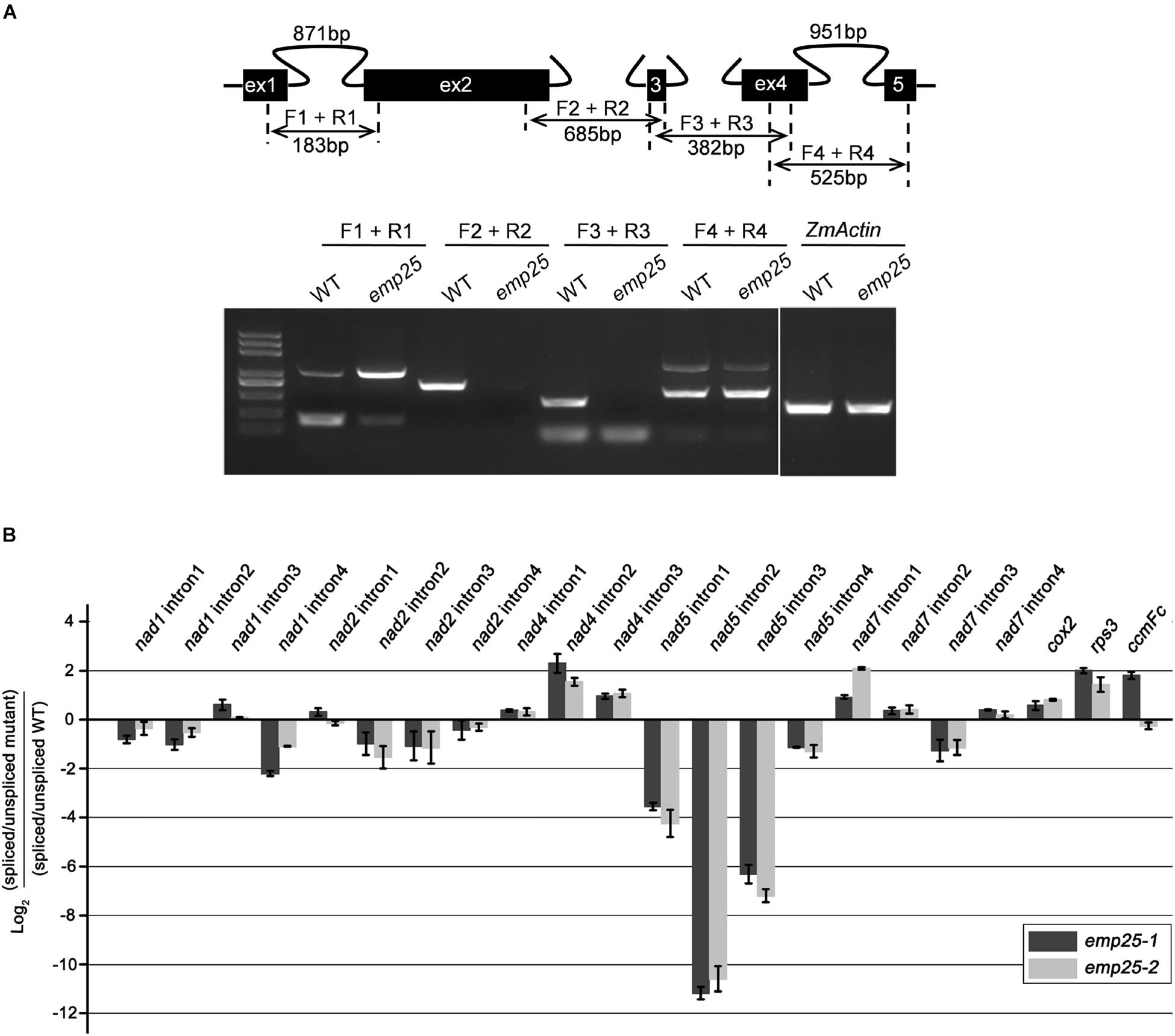
Figure 5. EMP25 is required for the splicing of nad5 introns. (A) Schematic diagram of the maize mitochondrial nad5 gene and RT-PCR analysis of intron splicing efficiency in the WT and emp25. Introns 1 and 4 are cis-splicing introns. Introns 2 and 3 are trans-splicing introns. The expected amplification products using different primer pairs are indicated. The arrows indicate the unspliced fragments of introns 1 and 4 (cis-splicing). (B) Quantitative RT-PCR (qRT-PCR) analysis of the 22 group II intorns in maize mitochondrial genes in two alleles of emp25. Values represent the mean and standard deviation of three biological replicates.
The nad5 gene in maize contains a 22-bp exon 3 (22 bp) adjacent to two trans-splicing introns 2 and 3 (Clifton et al., 2004; Figure 6A). It was reported that the trans-splicing of nad5 intron 3 must occur before the trans-splicing of intron 2 or it will generate a variety of mis-spliced products of intron 2, which also results in the accumulation of unspliced fragments of intron 2 (Elina and Brown, 2010; Brown et al., 2014; Colas des Francs-Small et al., 2014). To determine whether the accumulation of unspliced intron 2 is due to the dysfunctional EMP25 or mis-splicing, we quantitated the splicing intermediates of the nad5 transcripts with primers as previously described (Figure 6A; Colas des Francs-Small et al., 2014). The accumulation levels of both ex 2–3 and ex 2–4 decreased in emp25, and the result of ex2-in3, which indicates the accumulation level of mis-spliced fragments, also decreased in emp25 (Figure 6B). These results, coupled with low splicing efficiency of introns 2 and 3 of nad5 (Figure 5B), confirm the true splicing defects of nad5 in emp25.
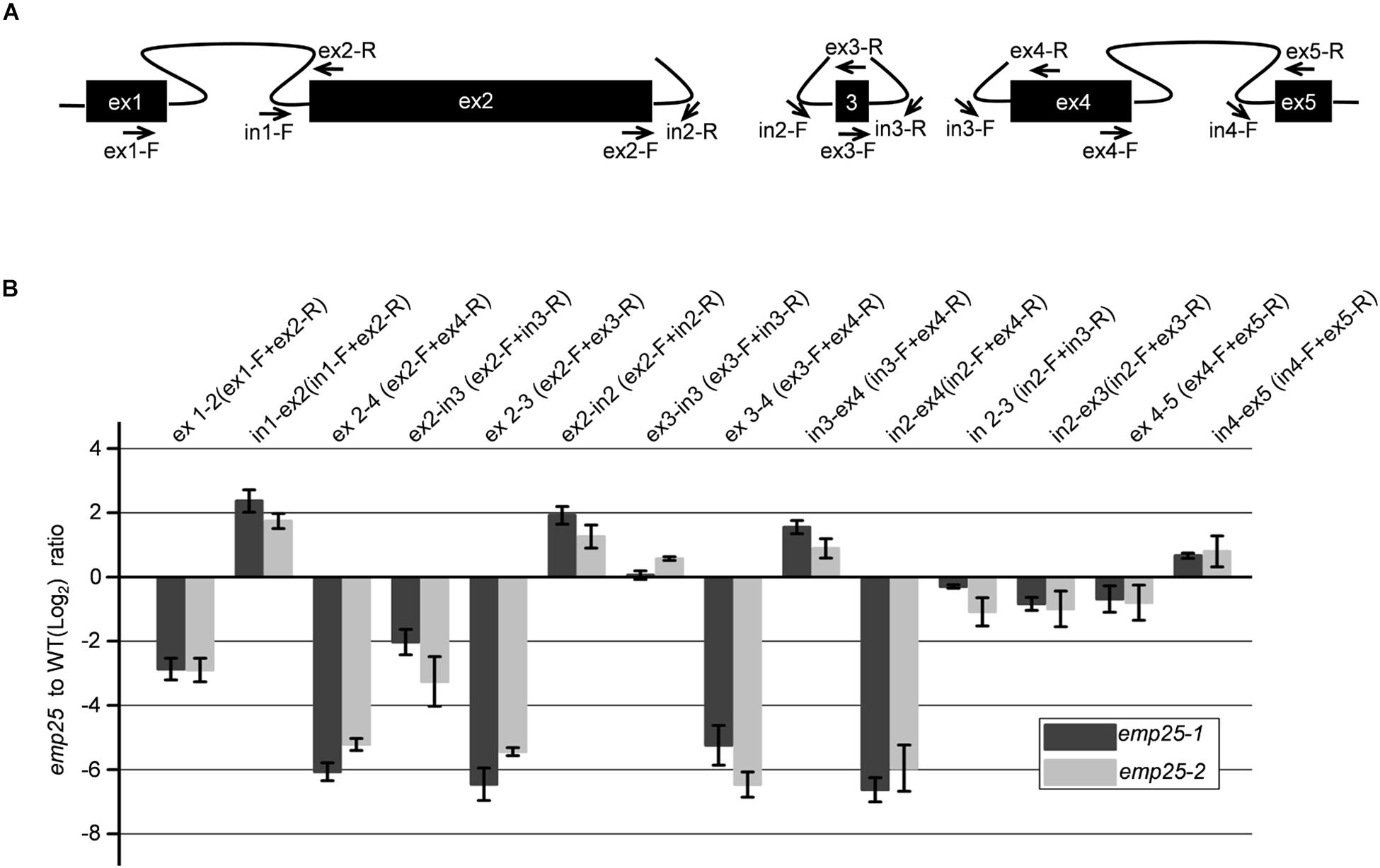
Figure 6. The splicing of nad5 introns 1, 2, and 3 is impaired in emp25. (A) Diagram of the splicing events of nad5 transcript and positions of the primers used. (B) qRT-PCR analysis of the mature and unspliced transcripts of individual nad5 exons in emp25 two alleles. Values represent the mean and standard deviation of three biological replicates.
Loss of Function in EMP24 and EMP25 Affects Mitochondrial Complex I Assembly and Activity
Nad4 and Nad5 are the subunits of complex I in the mitochondrial electron transfer chain (Colas des Francs-Small et al., 2014; Subrahmanian et al., 2016). The complete deficiency in nad4 intron splicing in emp24 and nad5 intron splicing in emp25 causes absence of Nad4 and Nad5 proteins. These mutants provide a good genetic model to test the role of each subunit in the assembly and function of the complex. By using blue native polyacrylamide gel electrophoresis (BN-PAGE) and in-gel assay of the NADH dehydrogenase activity, we analyzed the abundance and activity of the complexes in emp24 and emp25. In the Coomassie blue staining, emp24 and emp25 showed a similar profile of the complexes. Compared with the WT, complex I (CI) and supercomplex I + III (CI + III) were absent in both mutants, and complex III (CIII) and V (CV) were slightly increased (Figures 7A,B). Instead, two subcomplexes smaller than CI appeared in the mutants. Subsequent in-gel NADH dehydrogenase activity staining showed that the two smaller complexes have the activity, indicating that they are CI subcomplexes, hence designated as CI′ and CI″ (Figures 7A,B). Such results of emp24 were consistent with the reports in emp602 (Ren Z. et al., 2019). We also surveyed the protein abundance of the representative subunit of the mitochondrial complexes by Western blot analysis in emp25. Compared to the WT, Nad9 (CI) was slightly increased, whereas Cytc1 (CIII) and ATPase-A (CV) were dramatically increased in emp25 (Figure 7C). This result is consistent with the BN-PAGE analysis. In addition, the assembly of CIII, CIV, and CV was complete using representative antibody blot after transferring membrane from BN-gel (Supplementary Figure S6A) and respiratory rate was dramatically decreased in emp25 (Supplementary Figure S6B). These results indicated that absence of Nad4 or Nad5 protein as a result of loss-of-function mutation in EMP24 or EMP25 impairs the assembly and activity of mitochondrial complex I.
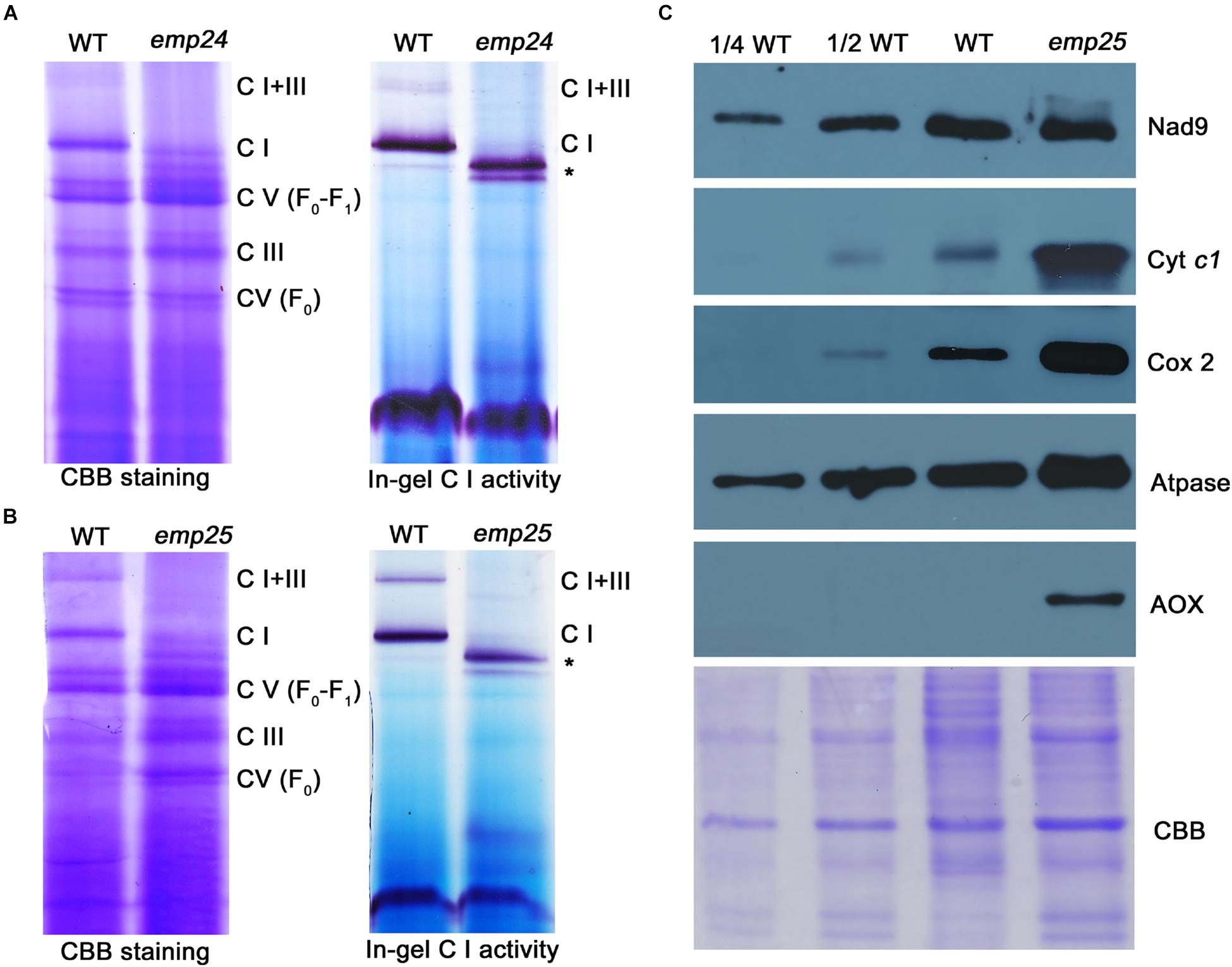
Figure 7. Defect in the complex assembly and activity in emp24 and emp25. (A,B) The blue native polyacrylamide gel electropheresis (BN-PAGE) gel was stained with Coomasssie Brilliant Blue (CBB). In-gel NADH dehydrogenase activity assay of complex I used dihydrolipoamide dehydrogenase (DLDH) as a loading control. C I: Complex I; C III: Complex III; C V: Complex V; *: sub-complex I detected in mutant. (C) Crude mitochondrial extracts from endosperm and embryo of emp25 with sibling WT kernels were used for Western blot analysis with antibodies against Nad9, Cyt c1, Atpase, Cox2, and AOX. CBB staining was used as a loading control.
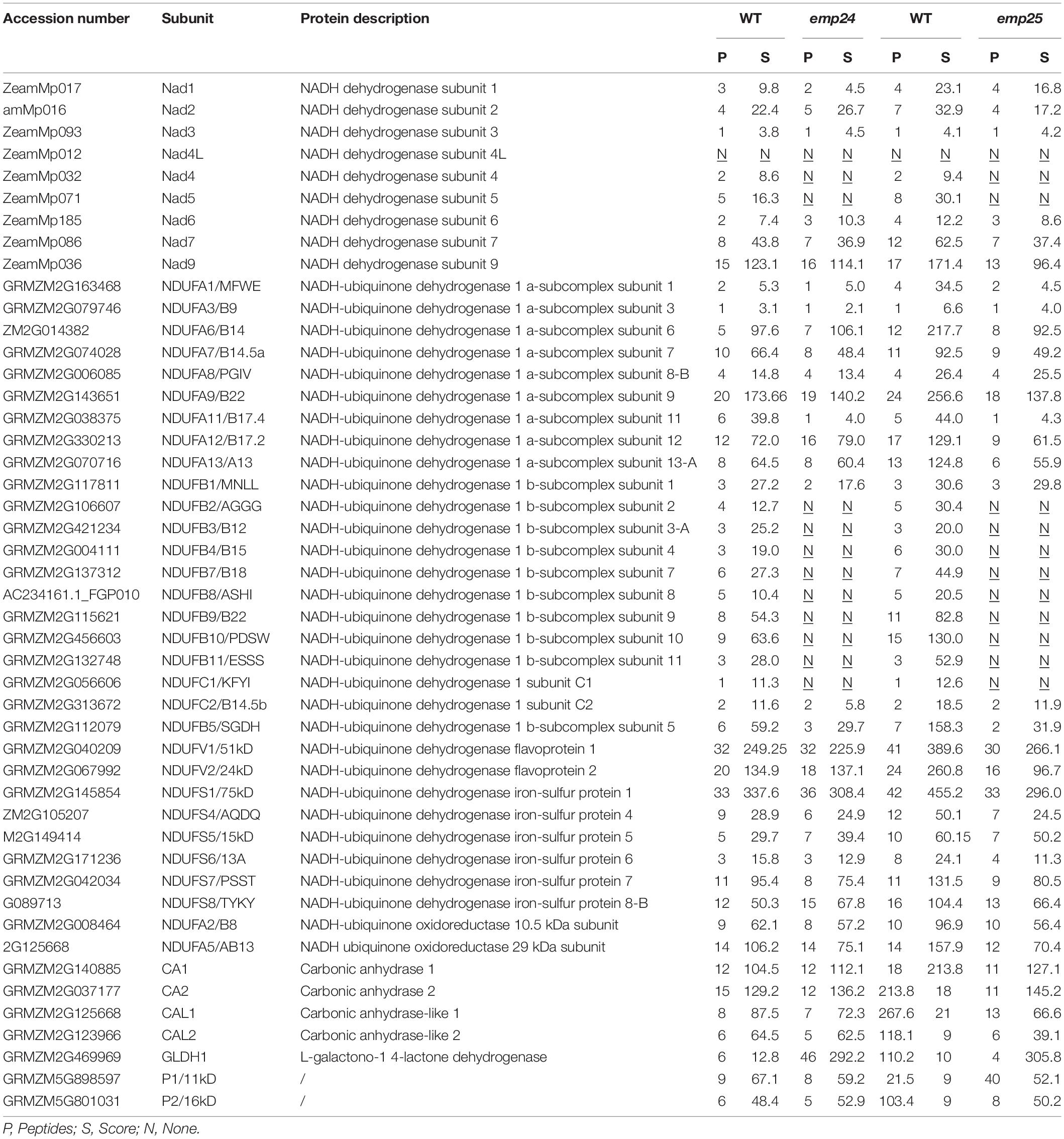
The formation of subcomplexes in the emp24 and emp25 mutants provided an opportunity to investigate the role of Nad4 and Nad5 in complex I assembly. We collected the CI′ and CI″ band from the BN-PAGE in two mutants, respectively, and subjected it to mass spectrometry analysis. The CI in the WT was analyzed as a reference. As shown in Table 1, 46 proteins were identified in the CI complex in the WT, but the emp24 and emp25 mutants showed identical deficiency in 11 proteins, i.e., Nad4, Nad5, NADH-ubiquinone dehydrogenase 1 beta subcomplex subunit 2, 3-A, 4, 7, 8, 9, 10, and 11 (also named NDUFB2, 3, 4, 7, 8, 9, 10, and 11), and NADH-ubiquinone dehydrogenase 1 subunit C1 (NDUFC1/KFYI) (Liu et al., 2019). These results showed that the lack of either Nad4 or Nad5 blocks the assembly of complex I and its activity, resulting in the formation of subcomplexes. Surprisingly, absence of Nad4 or Nad5 leads to the formation of a subcomplex lacking an identical set of proteins.
Discussion
EMP24 and EMP25 Function in the Splicing of nad4 and nad5 Introns, Respectively, in Mitochondria
This study has cloned Emp24 and Emp25. The correct cloning of the genes is confirmed by molecular and genetic analysis of independent alleles (Supplementary Figures S1, S2). Emp24 has been identified as Emp602 that encodes a mitochondrion-localized P-type PPR protein, which functions in the splicing of nad4 introns 1 and 3 (Ren Z. et al., 2019). Emp25 encodes a P-type PPR protein targeted to mitochondria that is required for the splicing of nad5 introns 1, 2, and 3 (Figure 3). EMP25 shares a 41% identity with the Arabidopsis TANG2; the function of these two proteins is either distinct or diverged. Loss of function of TANG2 affects the splicing of introns 1, 2, and 2–3, whereas TANG2 is required mainly for the splicing of nad5 intron 3 in Arabidopsis, and the reduced splicing efficiency of nad5 introns 1 and 2 could be indirectly influenced by intron 3 splicing defects (Colas des Francs-Small et al., 2014). Although mutation of Emp25 also exhibited defects in the splicing of nad5 introns 1, 2, and 2–3, the splicing of intron 2 shows more serious defects compared with intron 3 (Figures 5, 6). It is possible that TANG2 and EMP25 are derived from an ancestor protein that is originally recruited for the splicing of nad5 intron 3. During evolution, however, mutations in the maize nad5 introns 1 and 2 promoted the recruitment of EMP25 for efficient splicing, whereas in Arabidopsis, either different mutations occurred or other proteins were recruited for the splicing of nad5 introns 1 and 2. Thus, the true ortholog of EMP25 cannot be identified in Arabidopsis, reflecting that both the introns and the PPR proteins co-evolve fast to maintain efficient splicing.
P-type PPR proteins have been reported in the splicing of mitochondrial and chloroplast introns. In Arabidopsis, OTP43 functions in the splicing of nad1 intron 1 (de Longevialle et al., 2007), TANG2 functions in the splicing of nad5 intron 3, and OTP439 functions in the splicing of nad5 intron 2 (Colas des Francs-Small et al., 2014). These proteins are all P-type PPR proteins. In maize, P-type PPR proteins EMP10, EMP16, DEK2, and DEK35 have functions in the splicing of mitochondrial introns (Chen et al., 2016; Xiu et al., 2016; Cai et al., 2017; Qi et al., 2017). As some 20 introns exist in the plant mitochondrial genes, EMP24 and EMP25 contribute to the effort of identification of intron splicing factors in mitochondria. The mechanism by which plant organellar group II introns are spliced is not fully understood yet despite the fact that a number of RNA-binding proteins are found to participate in this process. Group II introns can self-splice, but introns in plant organelles have lost the self-splicing activity due to the mutations that accumulated during evolution. It is hypothesized that PPRs and other RNA-binding proteins are recruited to facilitate the formation of such a catalytic active conformation of the intron. Based on this idea, we predicted the EMP25 putative binding sites (as indicated in Supplementary Figure S7) based on the proposed 6, 1′ binding codes of the PPR motif (Barkan et al., 2012; Takenaka et al., 2013; Yagi et al., 2013). Further analysis indicates that these sites are uniquely present in the three introns that require EMP25, but absent in other introns or/and exons in the maize mitochondrial genes. We speculate that these sites may be the cognitive recognition sequences of EMP25. However, as we did not perform the binding assay because of the difficulty in expressing EMP25 recombinant protein, further analysis is necessary.
EMP24 and EMP25 Are Essential for Maize Seed Development
The loss of Emp24 and Emp25 function results in a similar phenotype with arrested embryo and endosperm development in maize (Figure 2). The empty pericarp phenotype highlights the importance of the ETC to seed development. As the powerhouse of the cell, mitochondria provide energy and metabolites to all cellular processes. The mutation in emp24 and emp25 eliminates the synthesis of Nad4 and Nad5, respectively, and blocks the assembly of mitochondrial complex I. As the first and the largest complex of the ETC, complex I transfers electrons onto ubiquinone and pumps protons from the matrix to the inner membrane for proton gradient in ATP synthesis (Brandt, 2006; Hirst, 2013). Deficiency of complex I impairs the function of ETC, disturbs the citric acid (TCA) cycle, and subsequently blocks the energy and intermediate supply, causing arrest of both embryo and endosperm development in the mutant.
Dysfunction in the mitochindrial complexes arresting seed development has been reported in a number of emp/dek mutants (Gutierrez-Marcos et al., 2007; Wang et al., 2017; Yang et al., 2017; Li et al., 2018; Sun et al., 2019). For example, dek43 are deficient in the splicing of nad4 introns and block complex I assembly and activity (Ren R. C. et al., 2019). Mutation in emp21 conditions editing defects in multiple sites of mitochondrial genes and hinders the biogenesis of complexes I and V (Wang et al., 2019). PPR78, which functions in the maturation and stability of mitochondrial nad5, is required for the complex I assembly and seed development (Zhang et al., 2017). The mutation of emp24 and emp25 also disturbs complex I function and subsequently impairs the seed development.
Analysis of the emp24 and emp25 Mutants Provides Genetic Insights Into the Assembly Pathway of Complex I
As important subunits of the mitochondrial complex I, the lack of Nad4 or Nad5 allowed the formation of smaller subcomplex I (CI′) in both the emp24 and emp25 mutants (Figures 7A,B). Analysis of the proteins in CI′ reveals that the subcomplexes in emp24 and emp25 are identical in subunit composition and uniformly lack 11 proteins including Nad4; Nad5; NDUFB2, 3, 4, 7, 8, 9, 10, and 11; and NDUFC1 (Table 1). This result indicates that the absence of Nad4 or Nad5 affects complex I assembly in the same way.
Mitochondrial complex I is an L-shaped holoenzyme with a hydrophobic arm embedded in the cristae membrane and a hydrophilic arm protruding to the matrix. Nad5 is located at the end of the hydrophobic arm, adjacent to Nad4 (Zhu et al., 2016; Liu et al., 2019), and interestingly, all the subunits missing in the emp24 and emp25 mutants are located at the end of the arm, suggesting that the absence of either Nad4 or Nad5 inhibits the assembly of these proteins into complex I.
It is proposed that the assembly of mitochondrial complex I follows a modular pathway in which subunits initially form subcomplexes and then assemble into the holocomplex I (Subrahmanian et al., 2016; Ligas et al., 2019). These subcomplexes are defined as N module (NADH binding), Q module (electrons transfer), and P module (peripheral arm), which is further divided into PP module (proximal section) and PD module (distal section) (Klodmann et al., 2010). The analysis of the complex assembly in emp24 and emp25 provides genetic evidence that Nad4 and Nad5 are the core members of the PD module because deficiency of either protein hinders the assembly. This analysis indicates that nuclear mutants affecting mitochondrial protein expression are valuable genetic materials for the study of mitochondrial complex assembly and other processes, circumventing the inability of creating mitochondrial gene mutants due to cell lethality.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
ZX, LP, YW, and B-CT designed the experiments and analyzed the data. ZX, YW, and LP performed most of the experiments. Other co-authors assisted the experiments and discussed the results. ZX and B-CT wrote the manuscript. All authors contributed to the study conception and design, read, and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31630053, 91735301, and 31700210), the China Postdoctoral Science Foundation Grant (2017M622183), and the Natural Science Foundation of Shandong Province (ZR201702190088).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the protein mass spectrometry assay service that was provided by the proteomics facility of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. We also thank Tsuyoshi Nakagawa (Shimane University, Japan) for pGWB vectors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.608550/full#supplementary-material
Supplementary Figure 1 | Clone and linkage analysis of Emp24 and Emp25.
Supplementary Figure 2 | Allelism test of emp24 and emp25.
Supplementary Figure 3 | Transcript level analysis of Emp25 in mutants and different organs and developing seeds in maize.
Supplementary Figure 4 | The editing profiles of nad5 in WT and emp25-1.
Supplementary Figure 5 | cRT-PCR analysis of the 5′/3′ ends of nad5 transcripts in WT and emp25-1.
Supplementary Figure 6 | The assembly of the mitochondrial CIII, IV, and V and respiratory rate comparison in WT and emp25-1.
Supplementary Figure 7 | Binding sites prediction of EMP25.
Supplementary Figure 8 | Major results of emp24.
Supplementary Figure 9 | The editing profiles of nad4 in WT and emp24-1.
Supplementary Table 1 | Peptides of CI identified details in MS analysis.
Supplementary Table 2 | Primers used in this study.
References
Adams, K. L., Qiu, Y. L., Stoutemyer, M., and Palmer, J. D. (2002). Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 99, 9905–9912. doi: 10.1073/pnas.042694899
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. doi: 10.1371/journal.pgen.1002910
Binder, S., and Brennicke, A. (2003). Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Phil. Trans. R. Soc. Lond. B. 358, 181–189.
Bonen, L. (2008). Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8, 26–34. doi: 10.1016/j.mito.2007.09.005
Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17, 322–331. doi: 10.1016/s0168-9525(01)02324-1
Brandt, U. (2006). Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92. doi: 10.1146/annurev.biochem.75.103004.142539
Brown, G. G., Colas des Francs-Small, C., and Ostersetzer-Biran, O. (2014). Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 5:35. doi: 10.3389/fpls.2014.00035
Cai, M., Shuzhen, L., Sun, F., Sun, Q., Zhao, H., Ren, X., et al. (2017). Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 91, 132–144. doi: 10.1111/tpj.13551
Chen, X., Feng, F., Qi, W., Xu, L., Yao, D., Wang, Q., et al. (2016). Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 Intron 1 and seed development in maize. Mol. Plant. 10, 427–441. doi: 10.1016/j.molp.2016.08.008
Clifton, S. W., Minx, P., Fauron, C. M., Gibson, M., Allen, J. O., Sun, H., et al. (2004). Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136, 3486–3503. doi: 10.1104/pp.104.044602
Cohen, S., Zmudjak, M., Colas des Francs-Small, C., Malik, S., Shaya, F., Keren, I., et al. (2014). nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78, 253–268. doi: 10.1111/tpj.12466
Colas des Francs-Small, C., Falcon de Longevialle, A., Li, Y., Lowe, E., Tanz, S. K., Smith, C., et al. (2014). The Pentatricopeptide repeat proteins TANG2 and organelle transcript processing 439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 165, 1409–1416. doi: 10.1104/pp.114.244616
de Longevialle, A. F., Meyer, E. H., Andres, C., Taylor, N. L., Lurin, C., Millar, A. H., et al. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19, 3256–3265. doi: 10.1105/tpc.107.054841
Elina, H., and Brown, G. G. (2010). Extensive mis-splicing of a bi-partite plant mitochondrial group II intron. Nucleic Acids Res. 38, 996–1008. doi: 10.1093/nar/gkp994
Fontanet, P., and Vicient, C. M. (2008). Maize embryogenesis. Methods Mol. Biol. 427, 17–29. doi: 10.1007/978-1-59745-273-1_2
Cohen, S., Zmudjak, M., Colas des Francs-Small, C., Malik, S., Shaya, F., Keren, I., et al. (2014). nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78, 253–268. doi: 10.1111/tpj.12466
Gutierrez-Marcos, J. F., Dal Pra, M., Giulini, A., Costa, L. M., Gavazzi, G., Cordelier, S., et al. (2007). empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19, 196–210. doi: 10.1105/tpc.105.039594
Hammani, K., and Giege, P. (2014). RNA metabolism in plant mitochondria. Trends Plant Sci. 19, 380–389. doi: 10.1016/j.tplants.2013.12.008
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Jiang, S. C., Mei, C., Wang, X. F., and Zhang, D. P. (2014). A hub for ABA signaling to the nucleus: significance of a cytosolic and nucleus dual-localized PPR protein SOAR1 acting downstream of Mg-chelatase H subunit. Plant Signal. Behav. 9:e972899. doi: 10.4161/15592316.2014.972899
Keren, I., Bezawork-Geleta, A., Kolton, M., Maayan, I., Belausov, E., Levy, M., et al. (2009). AtnMat2, a nucleus-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. doi: 10.1261/rna.1776409
Keren, I., Tal, L., Colas des Francs-Small, C., Araujo, W. L., Shevtsov, S., Shaya, F., et al. (2012). nMAT1, a nucleus-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 71, 413–426.
Klodmann, J., Sunderhaus, S., Nimtz, M., Jansch, L., and Braun, H. P. (2010). Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22, 797–810. doi: 10.1105/tpc.109.073726
Knoop, V. (2013). Plant mitochondrial genome peculiarities evolving in the earliest vascular plant lineages. J. Syst. Evol. 51, 1–12. doi: 10.1111/j.1759-6831.2012.00228.x
Li, X. J., Zhang, Y. F., Hou, M., Sun, F., Shen, Y., Xiu, Z., et al. (2014). Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79, 797–809. doi: 10.1111/tpj.12584
Li, X., Gu, W., Sun, S., Chen, Z., Chen, J., Song, W., et al. (2018). Defective Kernel 39 encodes a PPR protein required for seed development in maize. J. Integr. Plant Biol. 60, 45–64. doi: 10.1111/jipb.12602
Liere, K., Weihe, A., and Borner, T. (2011). The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J. Plant Physiol. 168, 1345–1360. doi: 10.1016/j.jplph.2011.01.005
Ligas, J., Pineau, E., Bock, R., Huynen, M. A., and Meyer, E. H. (2019). The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 97, 447–459. doi: 10.1111/tpj.14133
Liu, B., Yin, G., Whelan, J., Zhang, Z., Xin, X., He, J., et al. (2019). Composition of mitochondrial complex I during the critical node of seed aging in Oryza sativa. J. Plant Physiol. 236, 7–14. doi: 10.1016/j.jplph.2019.02.008
Liu, P., McCarty, D. R., and Koch, K. E. (2016). Transposon mutagenesis and analysis of mutants in UniformMu maize (Zea mays). Curr. Protoc. Plant Biol. 1, 451–465. doi: 10.1002/cppb.20029
Liu, Y. J., Xiu, Z. H., Meeley, R., and Tan, B. C. (2013). Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25, 868–883. doi: 10.1105/tpc.112.106781
Lurin, C., Andres, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyere, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
McCarty, D. R., Latshaw, S., Wu, S., Suzuki, M., Hunter, C. T., Avigne, W. T., et al. (2013). Mu-seq: sequence-based mapping and identification of transposon induced mutations. PLoS One 8:e77172. doi: 10.1371/journal.pone.0077172
McCarty, D. R., Settles, A. M., Suzuki, M., Tan, B. C., Latshaw, S., Porch, T., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44, 52–61. doi: 10.1111/j.1365-313x.2005.02509.x
Meyer, E. H., Tomaz, T., Carroll, A. J., Estavillo, G., Delannoy, E., Tanz, S. K., et al. (2009). Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151, 603–619. doi: 10.1104/pp.109.141770
Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., et al. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268, 434–445. doi: 10.1007/s00438-002-0767-1
Olsen, O. A. (2001). Endosperm development: cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267. doi: 10.1146/annurev.arplant.52.1.233
Poyton, R. O., and McEwen, J. E. (1996). Crosstalk between nucleus and mitochondrial genomes. Annu. Rev. Biochem. 65, 563–607. doi: 10.1146/annurev.bi.65.070196.003023
Qi, W., Yang, Y., Feng, X., Zhang, M., and Song, R. (2017). Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved in nad1 mRNA splicing. Genetics 205, 239–249. doi: 10.1534/genetics.116.196105
Ren, R. C., Wang, L. L., Zhang, L., Zhao, Y. J., Wu, J. W., Wei, Y. M., et al. (2019). DEK43 is a P-type pentatricopeptide repeat (PPR) protein responsible for the cis-splicing of nad4 in maize mitochondria. J. Integr. Plant Biol. 62, 299–313. doi: 10.1111/jipb.12843
Ren, Z., Fan, K., Fang, T., Zhang, J., Yang, L., Wang, J., et al. (2019). Maize empty pericarp602 encodes a P-type PPR protein that is essential for seed development. Plant Cell Physiol. 60, 1734–1746. doi: 10.1093/pcp/pcz083
Sazanov, L. A. (2015). A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388. doi: 10.1038/nrm3997
Sechet, J., Roux, C., Plessis, A., Effroy, D., Frey, A., Perreau, F., et al. (2015). The ABA-deficiency suppressor locus HAS2 encodes the PPR protein LOI1/MEF11 involved in mitochondrial RNA editing. Mol. Plant 8, 644–656. doi: 10.1016/j.molp.2014.12.005
Shen, Y., Li, C., McCarty, D. R., Meeley, R., and Tan, B. C. (2013). Embryo defective12 encodes the plastid initiation factor 3 and is essential for embryogenesis in maize. Plant J. 74, 792–804. doi: 10.1111/tpj.12161
Small, I. D., and Peeters, N. (2000). The PPR motif-a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47.
Subrahmanian, N., Remacle, C., and Hamel, P. P. (2016). Plant mitochondrial Complex I composition and assembly: a review. Biochim. Biophys. Acta 1857, 1001–1014. doi: 10.1016/j.bbabio.2016.01.009
Sultan, L. D., Mileshina, D., Grewe, F., Rolle, K., Abudraham, S., Glodowicz, P., et al. (2016). The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. Plant Cell 28, 2805–2829. doi: 10.1105/tpc.16.00398
Sun, F., Wang, X., Bonnard, G., Shen, Y., Xiu, Z., Li, X., et al. (2015). Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 84, 283–295. doi: 10.1111/tpj.12993
Sun, F., Xiu, Z., Jiang, R., Liu, Y., Zhang, X., Yang, Y. Z., et al. (2019). The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J. Exp. Bot. 70, 963–972. doi: 10.1093/jxb/ery432
Sweetlove, L. J., Fait, A., Nunes-Nesi, A., Williams, T., and Fernie, A. R. (2007). The mitochondrion: an integration point of cellular metabolism and signalling. Crit. Rev. Plant Sci. 26, 17–43. doi: 10.1080/07352680601147919
Takenaka, M., Zehrmann, A., Brennicke, A., and Graichen, K. (2013). Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8:e65343. doi: 10.1371/journal.pone.0065343
Tang, H., Luo, D., Zhou, D., Zhang, Q., Tian, D., Zheng, X., et al. (2014). The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant 7, 1497–1500. doi: 10.1093/mp/ssu047
Unseld, M., Marienfeld, J. R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. doi: 10.1038/ng0197-57
van Herpen, T. W. J. M., Cankar, K., Nogueira, M., Bosch, D., Bouwmeester, H. J., and Beekwilder, J. (2010). Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS One 5:e14222. doi: 10.1371/journal.pone.0014222
Wang, G., Zhong, M., Shuai, B., Song, J., Zhang, J., Han, L., et al. (2017). E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 214, 1563–1578. doi: 10.1111/nph.14507
Wang, Y., Liu, X. Y., Yang, Y. Z., Huang, J., Sun, F., Lin, J., et al. (2019). Empty pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 15:e1008305. doi: 10.1371/journal.pgen.1008305
Xiu, Z., Sun, F., Shen, Y., Zhang, X., Jiang, R., Bonnard, G., et al. (2016). EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J. 85, 507–519. doi: 10.1111/tpj.13122
Yagi, Y., Hayashi, S., Kobayashi, K., Hirayama, T., and Nakamura, T. (2013). Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8:e57286. doi: 10.1371/journal.pone.0057286
Yang, Y. Z., Ding, S., Wang, H. C., Sun, F., Huang, W. L., Song, S., et al. (2017). The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 214, 782–795. doi: 10.1111/nph.14424
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, Y. F., Suzuki, M., Sun, F., and Tan, B. C. (2017). The mitochondrion-targeted pentatricopeptide repeat 78 protein is required for nad5 mature mRNA stability and seed development in maize. Mol. Plant 10, 1321–1333. doi: 10.1016/j.molp.2017.09.009
Zhang, Y. F., Huang, X. Y., Zou, J. Y., Liao, X., Liu, Y. J., Lian, X. T., et al. (2019). Major contribution of transcription initiation to 5’-end formation of mitochondrial steady-state transcripts in maize. RNA Biol. 16, 104–117. doi: 10.1080/15476286.2018.1561604
Zhou, A. N., Rohou, A., Schep, D. G., Bason, J. V., Montgomery, M. G., Walker, J. E., et al. (2015). Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife 4:e10180.
Keywords: EMP24, EMP25, pentatricopeptide repeat protein, mitochondrion, seed development, maize
Citation: Xiu Z, Peng L, Wang Y, Yang H, Sun F, Wang X, Cao S-K, Jiang R, Wang L, Chen B-Y and Tan B-C (2020) Empty Pericarp24 and Empty Pericarp25 Are Required for the Splicing of Mitochondrial Introns, Complex I Assembly, and Seed Development in Maize. Front. Plant Sci. 11:608550. doi: 10.3389/fpls.2020.608550
Received: 21 September 2020; Accepted: 23 November 2020;
Published: 23 December 2020.
Edited by:
Liwen Jiang, The Chinese University of Hong Kong, ChinaReviewed by:
Yihua Wang, Nanjing Agricultural University, ChinaCaiji Gao, South China Normal University, China
Copyright © 2020 Xiu, Peng, Wang, Yang, Sun, Wang, Cao, Jiang, Wang, Chen and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Cai Tan, YmN0YW5Ac2R1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zhihui Xiu1†
Zhihui Xiu1† Ling Peng
Ling Peng Bao-Cai Tan
Bao-Cai Tan