- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
- 2Apple Engineering and Technology Research Center of Shaanxi Province, Northwest A&F University, Yangling, China
- 3Shaanxi Key Laboratory Bio-resources, College of Bioscience and Engineering, Shaanxi University of Technology, Hanzhong, China
Anthocyanins are responsible for the red pigmentation in the peel of apple (Malus × domestica Borkh.) fruit. Relatively few studies have investigated anthocyanins at the posttranscriptional level. MicroRNAs play an important role in plant growth and development by regulating gene expression at the posttranscriptional level. In this study, mdm-miR828 showed a relatively low expression level during the rapid fruit coloration period. However, the mdm-miR828 expression level increased in the late fruit coloration stage. Overexpression of mdm-miR828 inhibited anthocyanin synthesis in apple and Arabidopsis. Dual-luciferase and yeast one-hybrid assays showed that MdMYB1 is capable of binding to the promoter of mdm-MIR828b to promote its expression. The results indicate that mdm-miR828 is involved in a feedback regulatory mechanism associated with anthocyanin accumulation in apple. In addition, mdm-miR828 is involved in the inhibition of anthocyanin accumulation in response to high temperature.
Introduction
Apple (Malus × domestica Borkh.) is an economically important fruit crop that is cultivated worldwide. The peel color is an important phenotypic trait of apple fruit, which largely determines the commodity value of the fruit. Anthocyanins, a diverse group of secondary metabolites in plants is responsible for the red coloration in apple peel (Takos et al., 2006). In addition, anthocyanins may be nutritionally and medically beneficial to human health (Chaves-Silva et al., 2018). The mechanism of anthocyanin synthesis has been elucidated in many plant species, for example, petunia (Petunia hybrida; de Vetten et al., 1997; Quattrocchio et al., 1999; Spelt et al., 2000; Albert et al., 2011, 2014), Arabidopsis thaliana (Borevitz et al., 2000; Gonzalez et al., 2008; Petroni and Tonelli, 2011), grape (Vitis vinifera; Kobayashi et al., 2004; Hichri et al., 2010; Matus et al., 2010), nectarine (Prunus persica; Ravaglia et al., 2013), and strawberry (Fragaria × ananassa; Aharoni et al., 2001; Lin-Wang et al., 2010; Medina-Puche et al., 2014). Anthocyanins are synthesized via the flavonoid metabolism pathway (Kim et al., 2003). Many enzymes participate in this pathway, such as chalcone synthase (CHS), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and flavonoid 3-O-glycosyltransferase (UFGT; Holton and Cornish, 1995; Shirley et al., 1995; Kim et al., 2003). These enzymes are coregulated by the MYB-bHLH-WD40 (MBW) transcription factor complex (Goff et al., 1992; Baudry et al., 2004; Lloyd et al., 2017). Three alleles MdMYB10, MdMYB1, and MdMYBA that regulate anthocyanin synthesis in apple have been identified. These transcription factors directly bind to the promoter of anthocyanin biosynthesis genes to promote expression of the latter (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007). For example, a cold-induced bHLH transcription factor, MdbHLH3, directly binds to the promoter of the structural gene MdUFGT and that of MdDFR to activate their expression and also binds to the promoter of the regulatory gene MdMYB1 to promote its expression (Xie et al., 2012).
MicroRNA (miRNA) is a type of endogenous noncoding single-stranded small RNA in eukaryotes that predominantly guides posttranscriptional gene-silencing activities (Cuperus et al., 2011). The miRNA genes are first transcribed into primary transcripts in the nucleus and then processed into miRNA precursors (pre-miRNAs/MIRNA; Song et al., 2010). Subsequently, these pre-miRNAs are processed to form mature miRNAs (Voinnet, 2009). In addition to miRNAs, small interfering RNAs (siRNAs) are synthesized in plants (Rajagopalan et al., 2006), of which one is trans-acting small interfering RNA (ta-siRNA). The miRNA cleaves Trans-Acting SiRNA (TAS) loci transcripts, and then with the aid of RNA-dependent RNA polymerase 6 (RDR6) and suppressor of gene silencing 3 (SGS3), double-stranded RNA is synthesized and is cleaved by the DCL4 enzyme to produce ta-siRNAs. Its mechanism of action is similar to that of miRNA, that is, cleavage of the target transcript to interfere with gene expression (Allen et al., 2005).
MiRNAs are involved in the regulation of anthocyanin synthesis in plants (Liu et al., 2016; Sun et al., 2017). In Arabidopsis, SPL9, the target gene of miR156, competes with a bHLH transcription factor to bind to the MYB75 (PAP1) transcription factor, thereby disrupting the structure of the MBW protein complex and, ultimately, negatively regulating anthocyanin synthesis (Gou et al., 2011). Arabidopsis overexpressing miR778 accumulates higher amounts of anthocyanins than the wild type (WT) under phosphorus deficiency (Wang et al., 2015). The miR858 induced by elongated hypocotyl5 (HY5) targets the anthocyanin inhibitor MYBL2 to promote anthocyanin accumulation in Arabidopsis (Wang et al., 2016b). The miR828 was the first miRNA identified to negatively regulate anthocyanin accumulation. In Arabidopsis, miR828 targets the positive regulator of anthocyanin synthesis MYB113 and cleaves TAS4 to produce TAS4-siR81(−), whereas siR81(−) targets the anthocyanin-related transcription factors PAP1, PAP2, and MYB113 (Rajagopalan et al., 2006). Xia et al. (2012) conducted sequencing of sRNAs from the leaf, root, flower, and fruit of “Golden Delicious” apple and observed that mdm-miR828 cleaves MdTAS4 to derive mdm-siR81(−), and it was predicted that mdm-siR81(−) might target MdbHLH3. RNA gel blot analysis showed that mdm-miR828 predominantly accumulates in flowers. However, “Golden Delicious” is an apple cultivar with yellow fruit peel. It is unclear whether mdm-miR828 and mdm-siR81(−) are involved in the coloration of fruit with red peel, and the relationship between mdm-miR828 and MdMYB1 is uncertain.
In the present study, we investigated the expression pattern of mdm-miR828 during fruit coloration in the red-peeled apple “Starkrimson Delicious.” Overexpression of mdm-miR828 inhibited anthocyanin accumulation in the fruit peel, and heterologous overexpression of mdm-miR828 inhibited anthocyanin accumulation in Arabidopsis. MdMYB1 induced expression of mdm-miR828, which indicated that a feedback regulatory mechanism controlled the anthocyanin content in the peel. In addition, mdm-miR828 may be involved in the inhibition of anthocyanin accumulation in response to high temperature.
Materials and Methods
Plant Materials
Plants of apple “Starkrimson Delicious” were grown at the Apple Experimental Farm of Northwest A&F University, Shaanxi Province, China. Fruit were sampled at 110, 116, 122, 128, 134, 140, and 146 days after blooming. Nine fruit were selected at each time point with three fruit considered as one biological replicate. The peel was excised with a fruit peeler, immediately frozen with liquid nitrogen, and stored at −80°C until use. Transient transformation and high-temperature treatment were applied to fruit that were wrapped in a paper bag (Hongtai, Shanxi, China) at 45 days after blooming and harvested at 120 days after blooming.
Determination of Anthocyanin Content
The total anthocyanin content of the fruit was determined using the method described by Xie et al. (2012). A sample (0.5 g) of peel was extracted in 5 ml of 1% (vol/vol) HCl-methanol and incubated in the dark at 4°C for 24 h. After centrifugation at 13,000 × g for 5 min, the absorbance of the upper liquid was measured at 530, 620, and 650 nm. The anthocyanin content was calculated using the formula OD = (A530 − A620) − 0.1(A650 − A620). One unit of anthocyanin content was expressed as a change of 0.1 OD (unit × 103 g−1 fresh weight). Measurement of cyanidin 3-galactoside content was performed as described previously (Liu et al., 2013). A high-performance liquid chromatograph (Waters, Milford, MA, United States) was used for all analyses. Cyanidin 3-galactoside standard (Sigma Chemical, St Louis, MO, United States) was used. Separation of cyanidin 3-galactoside was accomplished on a C18 column (5-μm internal diameter, 250 × 4.6 mm; Waters). The anthocyanin content of Arabidopsis was determined using the method of Wang et al. (2016b).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA was extracted using TRIzol RNA Plant Plus Reagent (Tiangen, Beijing, China) and then treated with DNase I (TaKaRa, Dalian, China) to remove genomic DNA contamination. The RNA was reverse-transcribed with the PrimeScript™ RT Reagent Kit (TaKaRa, Dalian, China). The reverse transcription of mdm-miR828 and mdm-siR81(−) used specific stem-loop primers (Supplementary Table S1; Chen et al., 2005), and that of the internal reference gene MdU6 used gene-specific primers (Supplementary Table S1). Quantitative real-time polymerase chain reaction (RT-qPCR) analysis was conducted using SYBR® Premix Ex Taq™ II (TaKaRa, Dalian, China) on an ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Waltham, MA, United States). Poly (A) polymerase (PAP) RT-qPCR was implemented using the miRcute Plus miRNA First-Strand cDNA Kit (Tiangen, Beijing, China) and miRcute Plus miRNA qPCR Kit (Tiangen, Beijing, China). For normalization, 5S ribosomal RNA was used. MdActin was used to normalize the coding genes. The 2−ΔΔCT method was used to calculate the relative expression level. Quantitative primer sequences used are listed in Supplementary Table S1.
RNA Ligase-Mediated 5'-RACE
To verify the cleavage of MdTAS4 by mdm-miR828 and that of MdbHLH3 by mdm-siR81(−), RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) experiments was performed using the SMARTer® RACE 5'/3' Kit (TaKaRa, Dalian, China). First, total RNA was reverse-transcribed using SMARTScribe™ reverse transcriptase, and the 5' adaptor was attached. Next, 5' universal primers, 3' gene-specific primers, and the cDNA were used for PCR amplification. Gel extraction, in-fusion cloning, and sequencing were then performed.
Vector Construction and Genetic Transformation
The coding sequence (CDS) of MdbHLH3 was cloned from the cDNA derived from “Starkrimson Delicious” fruit peel. The sequence of Md-pri-miR828b (comprising the Md-MIR828b sequence and approximately 100-bp upstream and downstream) was amplified from DNA extracted from “Starkrimson Delicious” peel. The sequences were inserted into the pCAMBIA2301 vector to form 35S:MdbHLH3 and 35S:mdm-miR828. The primer sequences used are listed in Supplementary Table S1. The constructed vector was transformed into Agrobacterium tumefaciens strain GV3101.
Agrobacterium-mediated transformation followed the methods of Yang et al. (2019) and Tian et al. (2015). Agrobacterium cells harboring the overexpression vector were grown, collected, and resuspended in Agrobacterium infiltration buffer (200 mM acetosyringone, 10 mM MES, and 10 mM MgCl2) to OD = 1.2. The suspension was incubated at room temperature for 4 h. The sampled bagged covered apples were infiltrated with Agrobacterium carrying 35S:MdbHLH3, 35S:mdm-miR828, or the pCAMBIA2301 empty vector and then placed at room temperature in the dark for 24 h and transferred to an incubator maintained at 22°C illuminated with white light (200 μmol m−2 s−1) for 3 days.
The 35S:mdm-miR828 vector was used to transform A. thaliana using the floral-dip method (Clough and Bent, 1998). T3 generation of 35S:miR828 transformed Arabidopsis was used for the experiment.
Transient Dual-Luciferase Assay
A transient dual-luciferase assay was performed using tobacco (Nicotiana benthamiana) leaves. The mdm-MIR828b promoter was amplified and cloned into the pGreenII 0800-LUC vector. The CDS of MdMYB1 was amplified and cloned into the pGreenII 62-SK vector. The constructs were transformed into Agrobacterium strain GV3101 carrying the pSoup vector. Agrobacterium cells harboring the different vectors were cultured, collected, and resuspended in infiltration buffer (200 mM acetosyringone, 10 mM MES, and 10 mM MgCl2). After incubation for 2 h, tobacco leaves were injected with the cell suspension. After initial incubation for 24 h in the dark, the leaves were incubated under light (200 μmol m−2 s−1) with a 16 h/8 h (light/dark) photoperiod at 25/22°C (light/dark) for 3 days. Leaf discs were punched from tobacco leaves of similar size, and a microplate reader (Infinite M200 PRO, Tecan, Switzerland) was used to determine the firefly luciferase and Renilla luciferase activities using the Dual-Glo® Luciferase Assay System (Promega, Madison, WI, United States) in accordance with the manufacturer’s instructions.
Yeast One-Hybrid Assay
The CDS of MdMYB1 was inserted into the pGADT7 vector to generate the recombinant vector MdMYB1-AD, and the mdm-MIR828b promoter fragment was inserted into the pHIS2.1 vector to generate the recombinant vector proMIR828b-HIS. Yeast (Saccharomyces cerevisiae) strain Y187 cells carrying the proMIR828b-HIS vector were cultured in −T−H screening medium supplemented with different concentrations of 3-amino-1,2,4-triazole (3-AT). The MdMYB1-AD and proMIR828b-HIS vectors were cotransformed into yeast Y187 cells, which were then cultured in −T−H−L selective medium containing an optimal concentration of 3-AT.
High-Temperature Treatment
Bagged covered fruit of “Starkrimson Delicious” apple were harvested at 120 days after blooming. The harvested fruit were placed in an incubator at 35°C under white light (200 μmol m−2 s−1). The control was placed in an incubator at 23°C and under white light (200 μmol m−2 s−1). The fruit were sampled after 12, 24, and 48 h for analysis of anthocyanin content and gene expression. Three biological replicates, consisting of three fruit per replicate, were sampled at each time point.
Results
Anthocyanin Content and mdm-miR828 Expression During Apple Coloration
We determined the total anthocyanin content in the peel of “Starkrimson Delicious” fruit without covering at 110, 116, 122, 128, 134, and 146 days after blooming. No change in anthocyanin content was observed from 110 to 116 days, whereas anthocyanins accumulated rapidly in the peel from 116 to 134 days (the rapid coloration period). The anthocyanin content stabilized from 134 to 146 days (the late fruit coloration stage; Figures 1A,B). The content of cyanidin 3-galactoside, which accounted for more than 94% of the total anthocyanin content, showed a similar trend (Supplementary Figure S1).
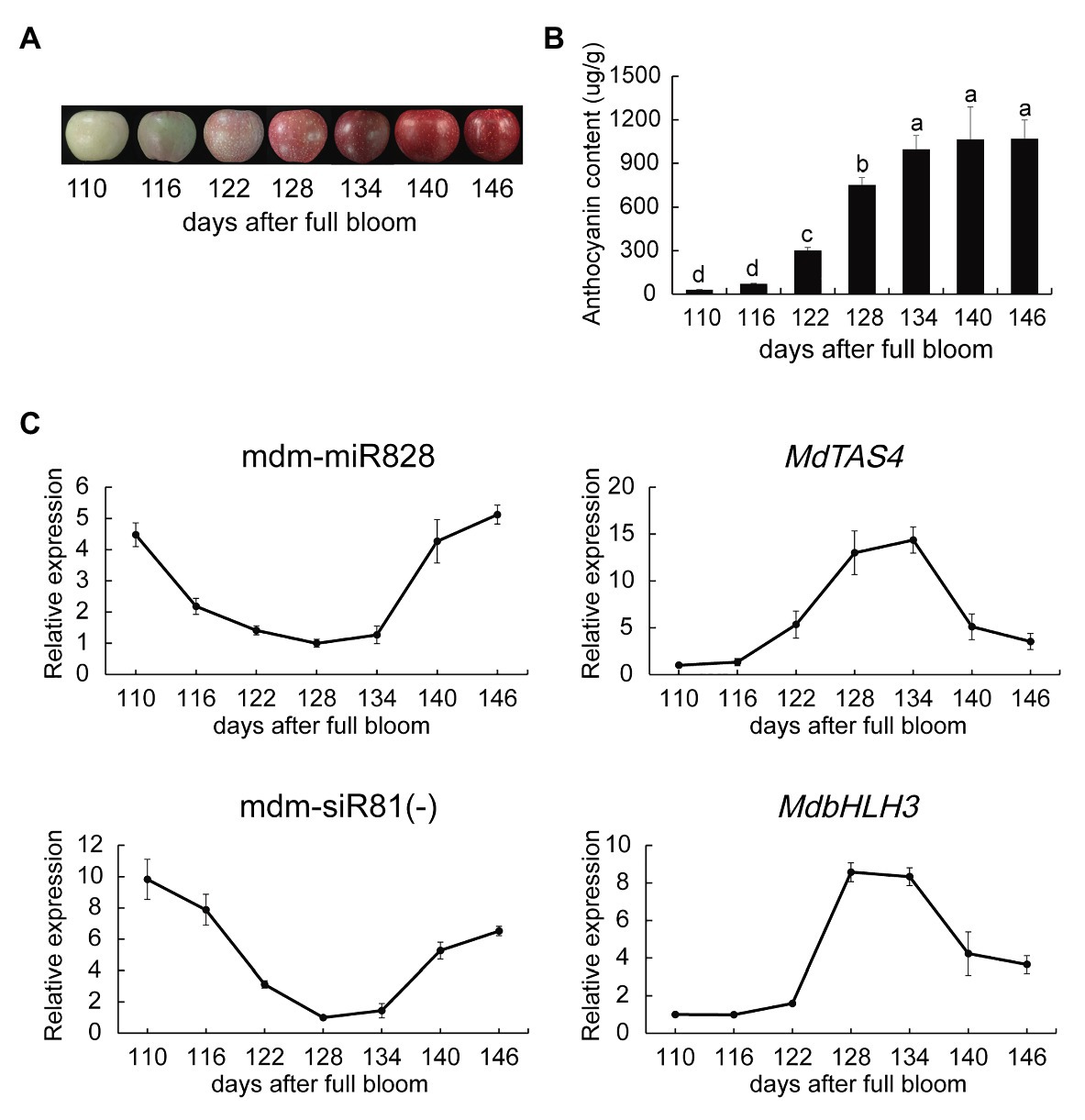
Figure 1. Anthocyanin content and mdm-miR828 expression during coloration of apple “Starkrimson Delicious” fruit. (A) Change in peel color during coloration. (B) Peel anthocyanin content during coloration. (C) Relative expression levels of mdm-miR828, mdm-siR81(−), and anthocyanin biosynthesis-related genes. The expression levels of mdm-miR828 and mdm-siR81(−) were detected by the stem-loop method. MdU6 was used as an internal reference. Error bars represent the standard deviation of three biological replicates. Different letters above the bars indicate a significant difference (p < 0.05; one-way ANOVA and LSD test).
We used the stem-loop method to detect the expression of mdm-miR828 in the peel. Expression of mdm-miR828 decreased rapidly from 110 to 116 days and showed a relatively low expression level during the period of rapid anthocyanin synthesis (116–134 days). However, mdm-miR828 expression increased from 134 to 146 days (Figure 1C). The PAP RT-qPCR method yielded similar results (Supplementary Figure S2). In addition, we measured the expression levels of MdTAS4, mdm-siR81(−), and MdbHLH3 (Figure 1C). The expression level of MdTAS4 increased from 116 to 134 days and thereafter decreased. That of mdm-siR81(−) declined rapidly from 116 to 128 days, remained relatively low from 128 to 134 days, and increased after 134 days. The expression level of MdbHLH3 increased sharply from 122 to 128 days and thereafter decreased from 134 to 146 days. The expression level of anthocyanin structural genes (MdCHS, MdDFR, MdANS, and MdUFGT) increased from 116 to 134 days and then decreased to varying degrees (Supplementary Figure S2).
Overexpression of mdm-miR828 Inhibits Anthocyanin Accumulation in Apples
To confirm that mdm-miR828 is involved in anthocyanin biosynthesis in the peel, we generated the construct 35S:mdm-miR828 and injected the fruit peel of “Starkrimson Delicious” with Agrobacterium carrying this construct. Transient overexpression of mdm-miR828 inhibited anthocyanin accumulation around the injection site, whereas no reduction in anthocyanin content was observed after injection of Agrobacterium containing the empty vector. Thus, mdm-miR828 was indicated to negatively regulate anthocyanin accumulation in apple. In addition, overexpression of MdbHLH3 significantly promoted accumulation of anthocyanins in the peel (Figures 2A,B).
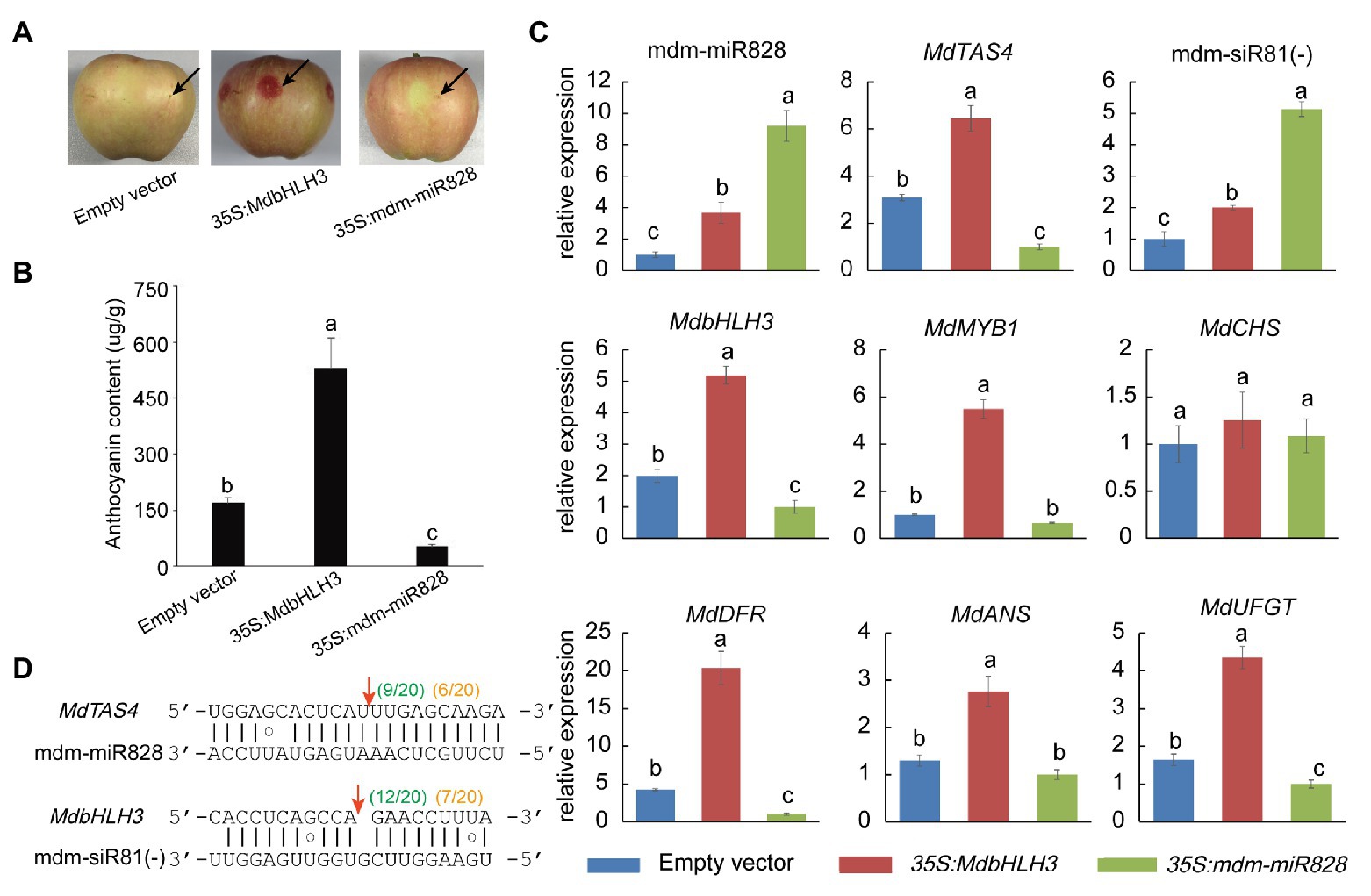
Figure 2. Transient expression of mdm-miR828 in apple “Starkrimson Delicious” fruit. (A) Overexpression of mdm-miR828 inhibits anthocyanin accumulation in the peel. The black arrows represent the injection sites. (B) Anthocyanin content in the peel of fruit overexpressing mdm-miR828 or MdbHLH3. (C) Overexpression of mdm-miR828 and MdbHLH3 affects the expression level of anthocyanin biosynthesis-related genes in the peel. (D) The binding positions of mdm-miR828 and mdm-siR81(−) in the target genes MdTAS4 and MdbHLH3. Green numbers and orange numbers in parentheses indicate the proportion of 5'-RACE clones at the sites where the target gene is cleaved by mdm-miR828 and mdm-siR81(−) in mdm-miR828-overexpressing peel and mixed peels (harvested at 134, 140, and 146 days after blooming), respectively. Error bars represent the standard deviation of three biological replicates. Different letters above the bars indicate a significant difference (p < 0.05, one-way ANOVA and LSD test).
The expression of anthocyanin synthesis-related genes was further analyzed (Figure 2C). The expression levels of MdbHLH3, MdbMYB1, MdDFR, MdANS, and MdUFGT increased to varying degrees compared with those of the control (empty vector) in the peel of fruit overexpressing MdbHLH3. In addition, mdm-miR828, MdTAS4, and mdm-siR81(−) expression levels were higher than those of the control. In the peel of fruit overexpressing mdm-miR828, both mdm-miR828 and mdm-siR81(−) were up-regulated compared with those of the control (empty vector), whereas the expression levels of MdTAS4, MdbHLH3, MdDFR, MdANS, and MdUFGT decreased compared with those of the control.
The RLM-RACE procedure was applied using the peel of fruit overexpressing mdm-miR828 to determine the cleavage site of mdm-miR828 on the MdTAS4 transcript and that of mdm-siR81(−) on the MdbHLH3 transcript. Some predicted 5' free ends of MdTAS4 and MdbHLH3 (between the 10th and 11th nucleotides of the miRNA binding site to the target gene) were detected (Figure 2D). In addition, we mixed samples of peels (collected at 134, 140, and 146 days) during the coloration period and performed the RACE experiment. The cleavage sites of mdm-miR828 and mdm-siR81(−) on the target genes MdTAS4 and MdbHLH3 were also detected.
Overexpression of mdm-miR828 in Arabidopsis Inhibits Anthocyanin Accumulation
To confirm the function of mdm-miR828 in anthocyanin biosynthesis, we generated mdm-miR828-overexpressing Arabidopsis transformants. Sucrose can induce the accumulation of anthocyanins in Arabidopsis seedlings (Teng et al., 2005). Therefore, seeds of WT and mdm-miR828-overexpression lines were cultured on half-strength Murashige and Skoog solid medium supplemented with 2% sucrose. The anthocyanin content was determined after culture for 4 days. The anthocyanin content in the mdm-miR828-overexpression lines was significantly lower than that of the WT. This result indicated that mdm-miR828 negatively regulated anthocyanin synthesis in Arabidopsis (Figures 3A,B).
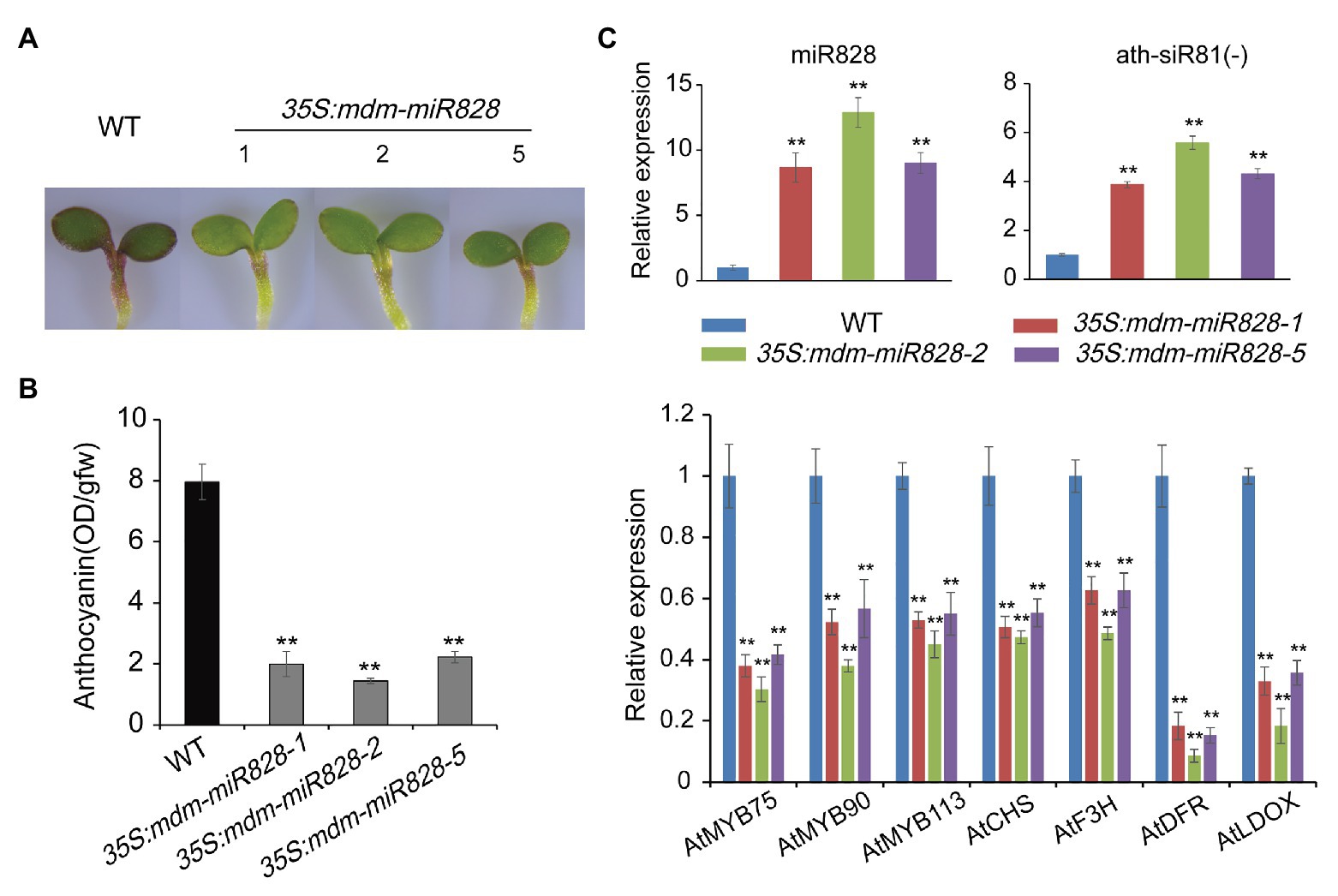
Figure 3. Overexpression of mdm-miR828 inhibits anthocyanin accumulation in Arabidopsis seedlings. (A) Phenotypes of mdm-miR828-overexpressing and wild-type Arabidopsis grown on half-strength Murashige and Skoog medium supplemented with 2% sucrose for 4 days. 35S:mdm-miR828-1, 35S:mdm-miR828-2, and 35S:mdm-miR828-5 represent three transgenic lines. (B) Anthocyanin content in mdm-miR828-overexpressing and wild-type Arabidopsis. (C) Relative expression levels assessed by qRT-PCR analysis showed that mdm-miR828 overexpression affects expression of anthocyanin-related genes in Arabidopsis. Error bars represent the standard deviation of three biological replicates. **p < 0.01 (Student t-test).
Expression of siR81(−) in the mdm-miR828-overexpression Arabidopsis seedlings was increased compared with that of the WT. The expression levels of anthocyanin biosynthesis-related regulatory genes (AtMYB75, AtMYB90, and AtMYB113) and structural genes (AtCHS, AtDFR, AtF3H, and AtLDOX) were reduced compared with those of the WT. These results suggested that mdm-miR828 and ath-miR828 performed similar functions and negatively regulated anthocyanin synthesis in Arabidopsis (Figure 3C).
MdMYB1 Promotes Expression of mdm-miR828
An autoregulatory feedback loop operates in Arabidopsis, and up-regulated expression of PAP1/MYB75 can activate miR828 expression (Hsieh et al., 2009; Luo et al., 2012). As a consequence, miR828 generates a greater amount of siR81(−) by directing the cleavage of TAS4 transcripts, thereby enhancing the cleavage of the target genes PAP1/MYB75, PAP2/MYB90, and MYB113 (Hsieh et al., 2009; Luo et al., 2012). In the present experiment, the expression levels of mdm-miR828 and mdm-siR81(−) increased in the late stage of apple coloration (134–146 days; Figure 1C), whereas MdMYB1 maintained a high level of expression in the late stage of coloration (Figure 4A). We speculated that MdMYB1 directly regulates expression of mdm-miR828. To test this hypothesis, we used a dual-luciferase assay to assess the correlation between MdMYB1 and the mdm-MIR828 promoter. First, we detected the expression levels of mdm-MIR828a (or pre–mdm-miR828a) and mdm-MIR828b. Expression of mdm-MIR828a was essentially undetectable, whereas the expression level of mdm-MIR828b was increased in the late fruit coloration stage (134–146 days; Supplementary Figure S3). Therefore, we cloned the 1,514-bp sequence of the mdm-MIR828b promoter and ligated it into the pGreenII 0800-LUC vector to form the fusion vector pMIR828b:LUC (Figure 4B). We then used Agrobacterium carrying pMIR828b:LUC and those harboring 35S:MdMYB1 to cotransform N. benthamiana leaves. Compared with the empty plasmid (pGreenII 62-SK) control, transient overexpression of MdMYB1 significantly up-regulated LUC activity driven by the mdm-MIR828b promoter (Figure 4C). This result indicated that MdMYB1 promoted the expression of mdm-miR828. Using the PlantCARE database, we detected multiple potential MYB transcription factor recognition sites in the 645-bp segment upstream of the mdm-MIR828b promoter (Figure 4D). A yeast one-hybrid assay showed that MdMYB1 is able to bind to this segment in the mdm-MIR828b promoter (Figure 4E).
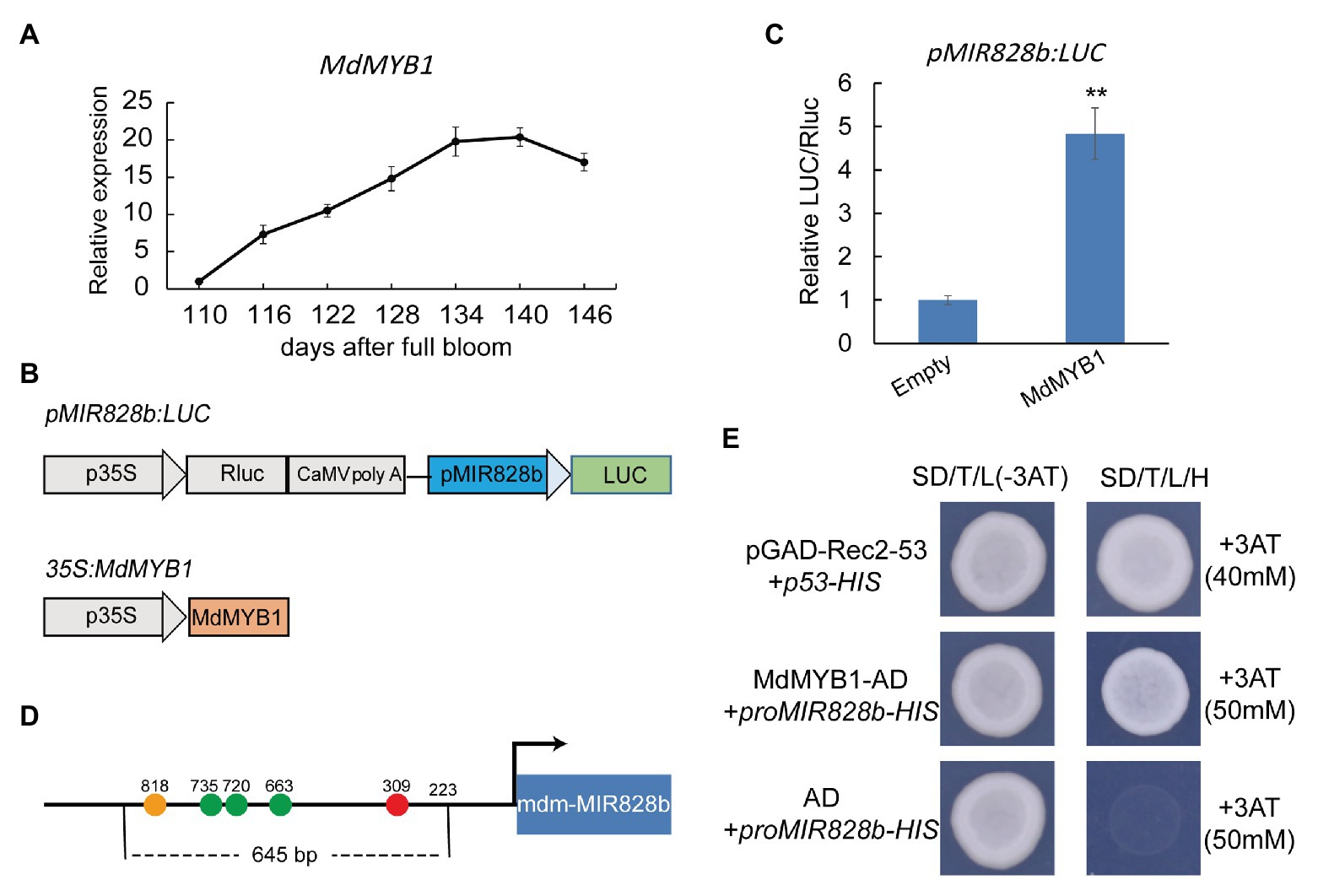
Figure 4. MdMYB1 promotes mdm-MIR828b expression by directly binding to its promoter. (A) Expression pattern of MdMYB1 during the coloration period of apple “Starkrimson Delicious” fruit. (B) Schematic representation of the construct used for the dual-luciferase assay. (C) Relative LUC/REN ratio. REN, Renilla luciferase activity; LUC, firefly luciferase activity. Error bars represent the standard deviation of three biological replicates. **p < 0.01 (Student t-test). (D) Schematic diagram of the mdm-MIR828b promoter structure. Circles represent the potential MYB transcription factor recognition sites: orange (TAACCA), green (CAACCA), and red (TAACTG). The number represents the distance (bp) from the transcription start site. (E) Yeast one-hybrid assay for binding of the mdm-MIR828b promoter and MdMYB1. pGAD-Rec2-53 and p53-HIS were used as positive controls.
mdm-miR828 Participates in the Process of High-Temperature Inhibition of Anthocyanin Accumulation
High temperature can inhibit anthocyanin accumulation in apple (Fang et al., 2019). As a potential negative regulator of anthocyanin accumulation, it is not clear whether mdm-miR828 is involved in the inhibition of anthocyanin accumulation in response to high temperature. We placed previously bagged covered “Starkrimson Delicious” fruit in an incubator maintained at either 35 or 23°C (the control). Under continuous light for 24 h, apple anthocyanin accumulation was reduced at 35°C compared with that of the control group at 23°C, and the difference was more obvious at 48 h (Figures 5A,B). The expression level of mdm-miR828 was elevated under high temperature compared with that of the control. The expression level of mdm-siR81(−) also increased in response to high temperature, whereas the expression levels of the anthocyanin synthesis-related genes MdTAS4, MdbHLH3, MdMYB1, MdCHS, MdDFR, MdANS, and MdUFGT were significantly reduced under high temperature compared with those of the control (Figure 5C). These results suggested that mdm-miR828 may be involved in the process of high-temperature inhibition of anthocyanin accumulation.
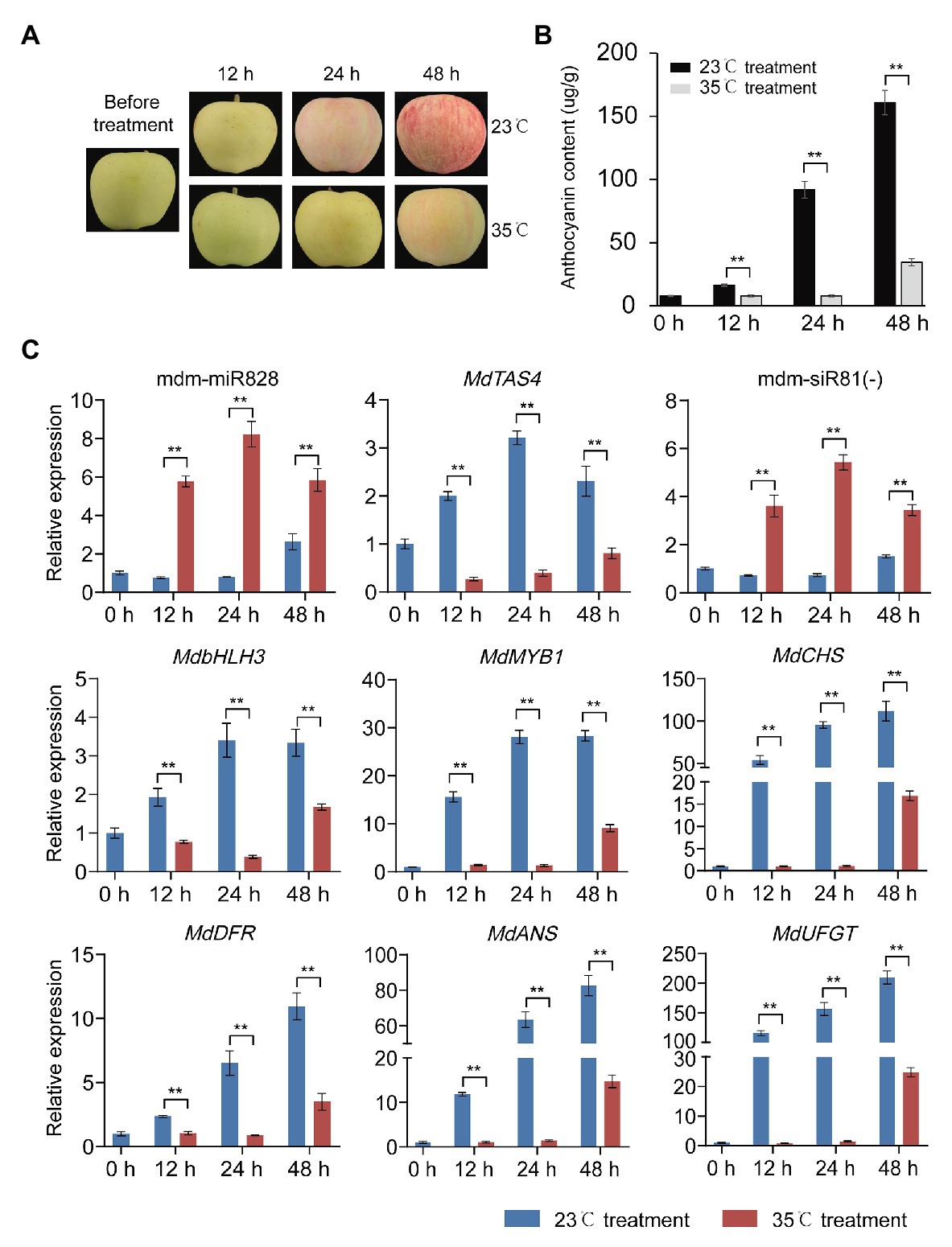
Figure 5. High temperature inhibits anthocyanin accumulation in apple peel. (A) Change in color of “Starkrimson Delicious” peel under high temperature. (B) Anthocyanin content in the peel under high temperature. (C) Relative expression level of mdm-miR828, mdm-siR81(−), and anthocyanin-related genes (MdTAS4, MdbHLH3, MdbMYB1, MdCHS, MdDFR, MdANS, and MdUFGT) under high temperature. Error bars represent the standard deviation of three biological replicates. **p < 0.01 (Student t-test).
Discussion
Anthocyanins are an important subclass of flavonoids that not only provide rich pigmentation to flowers and fruits, but also have anticancer, antioxidation, antiaging, and other beneficial properties for human health (Venancio et al., 2017). In addition, anthocyanins are involved in resistance to diverse biotic and abiotic stresses (Guo et al., 2008; Kovinich et al., 2015). The synthesis of anthocyanins in plants is strictly regulated by sophisticated regulatory networks. Previous studies of anthocyanin synthesis have predominantly focused on regulation at the transcriptional level, whereas research at the posttranscriptional level is relatively rare. In plants, miRNAs play a vital regulatory role at the posttranscriptional level largely through cleavage of target genes. Therefore, identification of miRNAs associated with fruit coloration is important for improvement of fruit quality.
In plants, the MBW complex, composed of an MYB transcription factor, bHLH transcription factor, and WD40 protein, is highly conserved and coordinately regulates structural genes of the anthocyanin synthesis pathway (Gonzalez et al., 2008). A deletion mutation of TT8 (a member of the bHLH transcription factor family) in Arabidopsis causes a severe decrease in the expression level of DFR and BAN genes, which causes a change in the seed coat color to yellow (Nesi et al., 2000). In apples, the transcription factor MdbHLH3 induced by low temperature can interact with MdMYB1 and promotes the expression of MdDFR and MdUFGT structural anthocyanin biosynthesis genes. In addition, MdbHLH3 may bind to the promoter of the regulatory gene MdMYB1 to promote its expression (Xie et al., 2012). In the present study, during the period of rapid synthesis of anthocyanins, MdbHLH3 was significantly increased, and overexpression of MdbHLH3 in the fruit promoted anthocyanin accumulation. These findings are consistent with previous results (Xie et al., 2012) and reveal that MdbHLH3 is essential for anthocyanin synthesis. The TAS4 gene is conserved in many dicotyledonous plants. Luo et al. (2012) compared TAS4 sequences from A. thaliana, Theobroma cacao, Euphorbia esula, Actinidia chinensis, V. vinifera, and Mimulus guttatus and observed that the binding site of miR828 is conserved. In the present study, RACE proved that mdm-miR828 directs cleavage of the MdTAS4 transcript between the 10th and 11th nucleotides, which indicated that mdm-miR828-mediated cleavage of MdTAS4 transcripts is conserved in apple. In Arabidopsis, ath-siR81(−) can target the anthocyanin-related transcription factors PAP1/MYB75, PAP2/MYB90, and MYB113. However, there is no sequence complement between mdm-siR81(−) and the anthocyanin-related transcription factor MdMYB1 in apples. Interestingly, mdm-siR81(−) is complementary to MdbHLH3 sequence, whereas mdm-siR81(−) and MdbHLH3 show an inverse expression profile to each other. In addition, the RACE experiment demonstrated that mdm-siR81(−)-directed cleavage of MdbHLH3 transcripts occurs in the peel of apple fruit.
Regulation of anthocyanin synthesis by miRNAs has been observed in many plant species, but the effects of miRNAs on anthocyanins differ. Inhibition of miR156 expression in Arabidopsis results in increased expression of the target gene SPL9 and thus inhibits accumulation of anthocyanins (Gou et al., 2011; Cui et al., 2014). In contrast, overexpression of mdm-miR156a in apple causes decreased expression of the SPL2-like and SPL33 genes and inhibition of anthocyanin accumulation (Yang et al., 2019). Overexpression of miR858 in Arabidopsis targets the anthocyanin inhibitor MYBL2 to promote anthocyanin accumulation (Wang et al., 2016b). However, miR858 in tomato plays a negative role in the biosynthesis of anthocyanins; blockage of miR858 binding increases SlMYB7-like transcript levels and ultimately promotes accumulation of anthocyanins (Jia et al., 2015). In Arabidopsis, siR81(−) derived from TAS4 and miR828 together target the positive regulatory genes PAP1, PAP2, and MYB113 for anthocyanin synthesis (Rajagopalan et al., 2006). Overexpression of miR828 reduces the accumulation of anthocyanins in Arabidopsis (Yang et al., 2013). However, miR828 targets the anthocyanin inhibitor MYB114 in grape to positively regulate anthocyanin accumulation (Tirumalai et al., 2019). In the present study, the mdm-miR828 expression level was relatively low during the rapid fruit coloration period (116 to 134 days), and mdm-siR81(−) expression was also relatively low. In addition, overexpression of mdm-miR828 in apple fruit inhibited anthocyanin synthesis, and overexpression of mdm-miR828 in Arabidopsis inhibited anthocyanin accumulation. These results indicated that mdm-miR828 negatively regulated anthocyanin synthesis in apple.
In the Arabidopsis mutant pap1-D, in which PAP1 is strongly up-regulated, high quantities of anthocyanins accumulate, but the expression levels of ath-miR828 and ath-siR81(−) also increase (Hsieh et al., 2009). Phosphorus deficiency, nitrogen deficiency, and exogenous sugars treatment can increase the expression of the PAP1/MYB75 and PAP2/MYB90 MYB transcription factors, which in turn activates the expression of anthocyanin structural genes and increases anthocyanin synthesis in Arabidopsis. In these plants with high anthocyanin content, the expression levels of ath-miR828 and ath-siR81(−) increased, which induced negative feedback regulation of AtMYB75, AtMYB90, and AtMYB113 expression. These results indicate that a feedback regulatory mechanism in Arabidopsis may control anthocyanin accumulation (Hsieh et al., 2009; Luo et al., 2012). In the current study, the anthocyanin tended to remain stable in the late stage of fruit coloration, whereas MdMYB1 maintained a high level of expression from 134 to 140 days after blooming, and at this stage, the expression level of mdm-miR828 began to increase. The dual-luciferase and yeast one-hybrid assays showed that MdMYB1 is capable of binding to the promoter of mdm-MIR828b to induce its expression. These results indicated that a feedback regulatory mechanism operates in apple. In addition, MdMYB1 and mdm-miR828 expression levels were increased in fruit overexpressing MdbHLH3, which suggested that the feedback pathway was activated.
In addition to sucrose, light (Takos et al., 2006) and low temperature (Xie et al., 2012; Gaiotti et al., 2018) promote anthocyanin synthesis. However, high temperature inhibits biosynthesis of anthocyanins in apple (Wang et al., 2016a). In the present experiment, anthocyanin accumulation under high temperature (35°C) was significantly inhibited compared with that of the control (23°C). The expression levels of mdm-miR828 and mdm-siR81(−) were higher under high temperature compared with those of the control, whereas the expression level of MdbHLH3 was reduced under high temperature compared with that of the control. These findings indicated that mdm-miR828 may be involved in the process of inhibiting anthocyanin accumulation under high temperature.
Based on the present results, we propose a model for mdm-miR828 participation in a feedback regulatory mechanism to balance anthocyanin accumulation in the peel of apple fruit (Figure 6). In this model, mdm-miR828 regulates the expression of MdbHLH3 by cleaving MdTAS4 to derive mdm-siR81(−). During the period of rapid fruit coloration, the expression level of mdm-miR828 is low, and MdbHLH3 and MdMYB1 are expressed in large quantities, which promotes the rapid synthesis of anthocyanins. In the late fruit coloration stage, the high-level expression of MdMYB1, which binds to the promoter of mdm-MIR828b, promotes the expression of mdm-miR828, which in turn inhibits the expression of MdbHLH3 so that anthocyanins do not accumulate indefinitely. In addition, high temperature can also induce the expression of mdm-miR828 and mdm-siR81(−), and then the highly expressed mdm-siR81(−) cleaves MdbHLH3 to prevent the accumulation of anthocyanins.
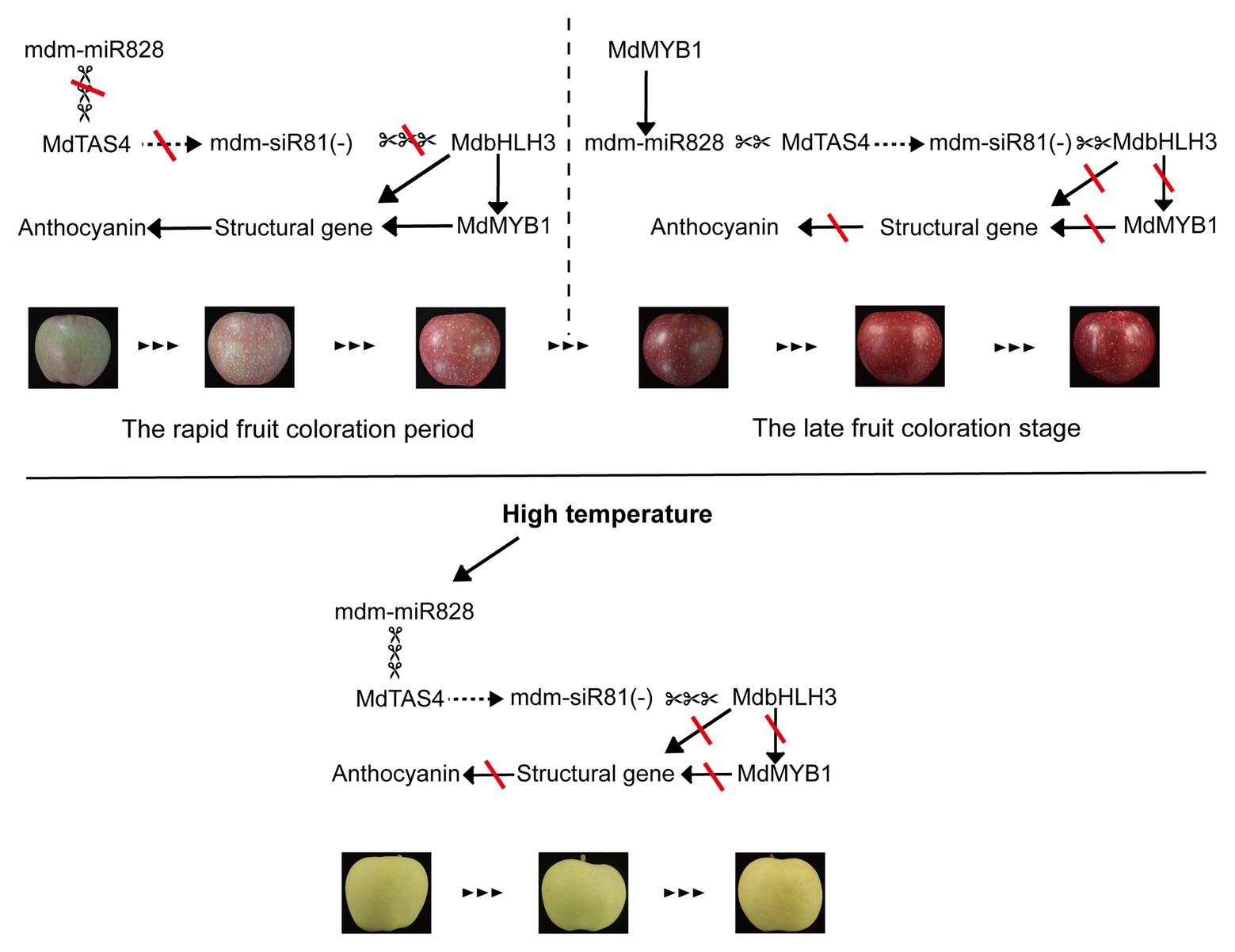
Figure 6. Proposed model for mdm-miR828 participation in a feedback regulation pathway to balance anthocyanin accumulation in the peel of apple fruit. A solid arrow indicates activation, a dashed arrow indicates derivation, scissors indicate cleavage, and red lines indicate closure.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
BZ, H-JY, and Z-YZ conceived the original screening and research plans. BZ, Y-ZY, Z-ZZ, and Y-NL supervised the experiments. BZ, H-JY, and DQ analyzed the data. BZ wrote the article. H-JY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the earmarked fund for the National Natural Science Foundation of China (32072555), and the Modern Agro-industry Technology Research System of China (CARS-27).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.608109/full#supplementary-material
References
Aharoni, A., De Vos, C. H., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K. E., Jameson, P. E., and Davies, K. M. (2011). Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.1105/tpc.12.12.2383
Chaves-Silva, S., Santos, A. L. D., Chalfun-Júnior, A., Zhao, J., Peres, L. E. P., and Benedito, V. A. (2018). Understanding the genetic regulation of anthocyanin biosynthesis in plants ‐ tools for breeding purple varieties of fruits and vegetables. Phytochemistry 153, 11–27. doi: 10.1016/j.phytochem.2018.05.013
Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179. doi: 10.1093/nar/gni178
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Cuperus, J. T., Fahlgren, N., and Carrington, J. C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23, 431–442. doi: 10.1105/tpc.110.082784
de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. doi: 10.1101/gad.11.11.1422
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fang, H., Dong, Y., Yue, X., Chen, X., He, N., Hu, J., et al. (2019). MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control fruit coloration in apple. Plant Cell Physiol. 60, 1055–1066. doi: 10.1093/pcp/pcz023
Gaiotti, F., Pastore, C., Filippetti, I., Lovat, L., Belfiore, N., and Tomasi, D. (2018). Low night temperature at veraison enhances the accumulation of anthocyanins in Corvina grapes (Vitis vinifera L.). Sci. Rep. 8:8719. doi: 10.1038/s41598-018-26921-4
Goff, S. A., Cone, K. C., and Chandler, V. L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. doi: 10.1101/gad.6.5.864
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., and Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Guo, J., Han, W., and Wang, M. -H. (2008). Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: a review. Afr. J. Biotechnol. 7, 4966–4972.
Hichri, I., Heppel, S. C., Pillet, J., Léon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Holton, T. A., and Cornish, E. C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083. doi: 10.2307/3870058
Hsieh, L. C., Lin, S. I., Shih, A. C., Chen, J. W., Lin, W. Y., Tseng, C. Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151, 2120–2132. doi: 10.1104/pp.109.147280
Jia, X., Shen, J., Liu, H., Li, F., Ding, N., Gao, C., et al. (2015). Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293. doi: 10.1007/s00425-015-2305-5
Kim, S. H., Lee, J. R., Hong, S. T., Yoo, Y. K., An, G., and Kim, S. R. (2003). Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 165, 403–413. doi: 10.1016/S0168-9452(03)00201-2
Kobayashi, S., Goto-Yamamoto, N., and Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304:982. doi: 10.1126/science.1095011
Kovinich, N., Kayanja, G., Chanoca, A., Otegui, M. S., and Grotewold, E. (2015). Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 10:e1027850. doi: 10.1080/15592324.2015.1027850
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Liu, Y., Che, F., Wang, L., Meng, R., Zhang, X., and Zhao, Z. (2013). Fruit coloration and anthocyanin biosynthesis after bag removal in non-red and red apples (Malus × domestica Borkh.). Molecules 18, 1549–1563. doi: 10.3390/molecules18021549
Liu, R., Lai, B., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2016). Identification of MicroRNAs and their target genes related to the accumulation of anthocyanins in Litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 7:2059. doi: 10.3389/fpls.2016.02059
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Luo, Q. J., Mittal, A., Jia, F., and Rock, C. D. (2012). An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80, 117–129. doi: 10.1007/s11103-011-9778-9
Matus, J. T., Poupin, M. J., Cañón, P., Bordeu, E., Alcalde, J. A., and Arce-Johnson, P. (2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 72, 607–620. doi: 10.1007/s11103-010-9597-4
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.1105/tpc.11.8.1433
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., Ziosi, V., Hellens, R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13:68. doi: 10.1186/1471-2229-13-68
Shirley, B. W., Kubasek, W. L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F. M., et al. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671. doi: 10.1046/j.1365-313X.1995.08050659.x
Song, L., Axtell, M. J., and Fedoroff, N. V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20, 37–41. doi: 10.1016/j.cub.2009.10.076
Spelt, C., Quattrocchio, F., Mol, J. N., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1632. doi: 10.1105/tpc.12.9.1619
Sun, Y., Qiu, Y., Duan, M., Wang, J., Zhang, X., Wang, H., et al. (2017). Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Mol. Gen. Genomics. 292, 215–229. doi: 10.1007/s00438-016-1268-y
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Tian, J., Peng, Z., Zhang, J., Song, T., Wan, H., Zhang, M., et al. (2015). McMYB10 regulates coloration via activating McF3'H and later structural genes in ever-red leaf crabapple. Plant Biotechnol. J. 13, 948–961. doi: 10.1111/pbi.12331
Tirumalai, V., Swetha, C., Nair, A., Pandit, A., and Shivaprasad, P. V. (2019). miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 70, 4775–4792. doi: 10.1093/jxb/erz264
Venancio, V. P., Cipriano, P. A., Kim, H., Antunes, L. M., Talcott, S. T., and Mertens-Talcott, S. U. (2017). Cocoplum (Chrysobalanus icaco L.) anthocyanins exert anti-inflammatory activity in human colon cancer and non-malignant colon cells. Food Funct. 8, 307–314. doi: 10.1039/C6FO01498D
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016b). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Wang, L., Zeng, J. H., Song, J., Feng, S. J., and Yang, Z. M. (2015). miRNA778 and SUVH6 are involved in phosphate homeostasis in Arabidopsis. Plant Sci. 238, 273–285. doi: 10.1016/j.plantsci.2015.06.020
Wang, N., Zhang, Z., Jiang, S., Xu, H., Wang, Y., Feng, S., et al. (2016a). Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Org. Cult. 127, 217–227. doi: 10.1007/s11240-016-1044-z
Xia, R., Zhu, H., An, Y. Q., Beers, E. P., and Liu, Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13:R47. doi: 10.1186/gb-2012-13-6-r47
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Yang, F., Cai, J., Yang, Y., and Liu, Z. (2013). Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tissue Org. Cult. 115, 159–167. doi: 10.1007/s11240-013-0349-4
Keywords: apple, anthocyanin, mdm-miR828, MdMYB1 , high temperature
Citation: Zhang B, Yang H-J, Yang Y-Z, Zhu Z-Z, Li Y-N, Qu D and Zhao Z-Y (2020) mdm-miR828 Participates in the Feedback Loop to Regulate Anthocyanin Accumulation in Apple Peel. Front. Plant Sci. 11:608109. doi: 10.3389/fpls.2020.608109
Edited by:
Cristina Garcia-Viguera, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Nick Albert, The New Zealand Institute for Plant and Food Research Ltd., New ZealandAntje Feller, University of Tübingen, Germany
Copyright © 2020 Zhang, Yang, Yang, Zhu, Li, Qu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-Yang Zhao, emhhb3p5QG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Bo Zhang
Bo Zhang Hui-Juan Yang
Hui-Juan Yang Ya-Zhou Yang1
,2
Ya-Zhou Yang1
,2
 Ya-Nan Li
Ya-Nan Li Dong Qu
Dong Qu Zheng-Yang Zhao
Zheng-Yang Zhao