
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Plant Sci. , 23 December 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.597481
This article is part of the Research Topic Nitrogen Use Efficiency and Sustainable Nitrogen Management in Crop Plants View all 23 articles
Feeding the world population increasing from current 7.7 to 9.7 billion by 2050 is a big challenge (United Nations, 2019). This is further serious in developing countries where degradation of soil health, increasing fertilizers cost and reducing cultivable lands are the major constraints (St Clair and Lynch, 2010). Presently, 119.41 million tons of nitrogen (N) fertilizers are applied worldwide in crops to achieve desirable yield (FAO, 2018). Plant N uptake, transport, utilization/assimilation and remobilization are controlled by a complex network of genes involved in these biological processes. Significant research advancements have been made in nitrogen use efficiency (NUE) in plants like Arabidopsis, rice, maize and wheat (Li et al., 2017), and physiological and molecular mechanisms underlying N pathways have been elucidated in plants (Kant et al., 2011). Although, many studies have been undertaken in different N regimes and candidate genes have been identified for increasing NUE but success in achieving N-use efficient genotypes is limited due to its complex genetics and genotype by environment interaction (Baligar et al., 2001). Interestingly, a considerable number of transgenic plants with increased NUE have been developed in cereals (Li et al., 2020). Notably, progress in CRISPR/Cas9 [clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9] genome editing combined with base-editing technology provides a great opportunity for enhancing NUE in plants (Khatodia et al., 2016; Li et al., 2018).
Potato (Solanum tuberosum L.) is the fourth most important food crop of the world after rice, wheat and maize. Potato is an N fertilizer intensive crop that requires 180–240 kg N/ha fertilizers to produce high tuber yield (35–45 t/ha); of total applied N, plants acquire only 40–50% and remaining N is lost in environment (Trehan and Singh, 2013). Owing to the adverse impacts and high production cost caused by excess N fertilizers application, improving NUE of plant is an environmental-friendly approach to achieve sustainable crop yield (Fageria et al., 2008). This opinion article highlights prospects for improving NUE in potato based on the lessons learnt from the transgenics to the CRISPR/Cas9 genome editing research in plants.
Transgenic technology has been applied in plants to create genetically modified organism (GMO) by overexpression or knockout/silencing of genes. Genes have been transferred within or across the species to introduce new or enhance/alter endogenous gene expression (Good et al., 2007). Whereas, gene silencing (RNAi) process inhibits gene expression or translation by disrupting targeted mRNA (Liang et al., 2014). N metabolism pathways genes such as nitrate or ammonium transporters, assimilation genes and transcription factors (TFs) have been manipulated for improving NUE in cereals (Li et al., 2020). Generally, overexpression of genes driven by the constitutive (Ubiquitin and CaMV35S) or tissue-specific (e.g., OsNAR2.1) promoters has been deployed to develop N-use efficient transgenic plants (Chen J. et al., 2016). Hu et al. (2015) have demonstrated overexpression of OsNRT1.1B allele of indica rice into japonica to increase NUE. RNAi technology has been deployed to knockdown the NAC-like TF OsNAP to improve NUE in rice (Liang et al., 2014).
The recently discovered CRISPR/Cas9 system has revolutionized the plant research. CRISPR/Cas9 (type-II, originates from Streptococcus pyogenes) is an adaptive immunity found in bacteria or archaea to combat with invading nucleic acids (Khatodia et al., 2016). The unprecedented advances in CRISPR/Cas9 facilitate an easy, versatile and robust technology to accelerate genomics-assisted crop improvement. CRISPR/Cas9 has been successfully deployed to edit N transporter gene to enhance NUE by introgression of NRT1.1B-indica allele into japonica rice (Li et al., 2018). Until now, CRISPR/Cas9 has been mostly applied to mutate negative regulators, instead of overexpression of positive regulators. The gene BT2, a member of the Bric-a-Brac/Tramtrack/Broad gene family, suppresses nitrate uptake and NUE; and overexpression of BT2 decreased NUE in rice under low nitrate by decreasing expression of NRT2.1 and NRT2.4 genes (Araus et al., 2016). Further, symbiotic N fixation (SNF)-associated genes have also been inactivated by CRISPR/Cas9 in Lotus japonicus (Wang et al., 2016), and thus progress in genome editing would accelerate SNF research in legumes and non-legumes. Recently, cytosine- and adenine- base editors (CBEs/ABEs) called base-editing, based on CRISPR/Cas9, have emerged as a newer technology for precise modification of nucleotides [C to T (or G to A), and A to G (or T to C)] for gain or loss of gene functions in eukaryotes (Li et al., 2018; Mishra et al., 2020). The base-editing has been demonstrated in rice for nitrate transporter gene OsNRT1.1B to improve NUE (Lu and Zhu, 2017; Zong et al., 2018). Collectively, the successful examples of a few N-use efficient plants (transgenics and genome-edited) are summarized in Supplementary Table 1.
A number of N transporter genes such as low-affinity nitrate transporter NRT1.1b (Fan et al., 2015; Hu et al., 2015), high-affinity nitrate transporters NRT2.1 (Chen J. et al., 2016; Chen et al., 2017), NAR2.1 and NRT2.3a (Chen et al., 2020), NRT2.3 (Fu et al., 2015), and NRT2.3b (Fan et al., 2016), peptide transporter PTR9 (Fang et al., 2013), ammonium transporter AMT1;1 (Ranathunge et al., 2014), and quantitative trait loci qNGR9, synonymous with gene DEP1 (Sun et al., 2014) have shown to enhance NUE in rice. Similarly, TFs such as MADS25 (Yu et al., 2015) and NAC2-5A (He et al., 2015) have also been found effective in developing N use efficient rice and wheat, respectively. The roles of microRNA miR166 targeting Dof TF RDD1 have also been confirmed to promote ammonium uptake and transport in rice (Iwamoto and Tagiri, 2016). Recently, functions of several genes have been elucidated in plants for high NUE such as nitrate transporter OsNPF4.5 (Wang et al., 2020), NAC42-activated nitrate transporter (Tang et al., 2019) and nitrate reductase gene OsNR2 (Gao et al., 2019). Collectively, genetic engineering in N transporters have been proven successful to increase plant growth, root architecture, N uptake and transport and total N content, and thus improved NUE of plants (Supplementary Table 1).
Several genes have been engineered to enhance N utilization efficiency in plants. For example NIN-LIKE PROTEIN 7 (NLP7) (Yu et al., 2016), asparagine synthetase ASN1 (Lam et al., 2003), autophagy-related gene ATG7-1 (Wada et al., 2015) and glutamine synthetase GS1;2 (Brauer et al., 2011) have improved NUE in Arabidopsis/rice. The functions of TFs such as bZIP AtTGA4 (Zhong et al., 2015), HY5 (Chen X. B. et al., 2016), NAC-like NAP (Liang et al., 2014), Dof1 ZmDof1 (Yanagisawa et al., 2004; Kurai et al., 2011), NAC1-type NAC-S (Zhao et al., 2015) and Nuclear Factor Y NFYA-B1 (Qu et al., 2015) have been demonstrated in development of N-use efficient plants of Arabidopsis/rice/wheat. Importantly, barley AlaAT (alanine aminotransferase) has most successful in increasing NUE in rice (Shrawat et al., 2008), canola (Good et al., 2007) and sugarcane (Snyman et al., 2015). The miR166 targeting Dof TF RDD1 enhances transport of nutrients including ammonium and sucrose, N uptake and content, and grain yield under low N in rice (Iwamoto and Tagiri, 2016) (Supplementary Table 1).
In potato, several studies have reported on application of soil and agronomic practices for N management (review by Trehan and Singh, 2013), but very limited on genomics uses to enhance NUE. Hence, knowledge about genes and regulatory elements such as TFs and microRNAs (miRNAs) are important to improve NUE. Moreover, the underlying molecular and physiological mechanisms and genetic factors remain elusive in potato for root system architecture, carbon-nitrogen economy and N metabolism (uptake, transport, utilization and remobilization). Recently, we have reviewed application of integrated genomics, physiology and breeding approaches for improving NUE (Tiwari et al., 2018) and traits phenotyping under aeroponic in potato (Tiwari et al., 2020d). Further, recent studies provide information about the genes and miRNAs associated with N stress in potato (Tiwari et al., 2020a,b,c; Zhang et al., 2020). Potato is highly amenable to tissue culture and therefore transgenics protocols are well established. Also, CRISPR/Cas9 tool has been applied in potato such as creation of homozygous mutants, knockdown of steroidal glycoalkaloids, caroteinoid biosynthesis and phosphate transport (review by Nadakuduti et al., 2018; Dangol et al., 2019).
We summarize here potential candidate genes for improving NUE in potato based on the recent research (Tiwari et al., 2020a,b,c; Zhang et al., 2020). Our studies indicate that in potato roots, high-affinity nitrate transporters are the key candidate genes for manipulation in N uptake and transport. Moreover, genes like ferric chelate reductase, protein phosphatase 2 C, glutaredoxin, GDSL esterase/lipase, cytochrome P450 hydroxylase and TFs also appear important in roots. In stolons, nitrate transporter, urea active transporter and sodium/proline symporter facilitate N transport. We have also elucidated miRNAs (up-regulated: miR156/157 and miR482, and down-regulated: miR397 and miR398) in roots under N stress. Further, glutaredoxin gene family has been found the most prominent candidate gene under N stress in shoots. Another study shows effect of overexpression of glutaredoxin gene OsGRX6 on signaling and N status in rice (El-Kereamy et al., 2015). We have identified tartrate-resistant acid phosphatase, glycerophosphodiester phosphodiesterase and TFs (Myb and WRKY), and miRNAs (up-regulated: miR156 and miR319, and down-regulated: miR398 and miR5303) in shoots under N stress. Indeed, stolon formation is a critical stage of tuber formation in potato. Hence, carbohydrate metabolism genes like glucose-6-phosphate/phosphate translocator 2 and glucose-1-phosphate adenylyltransferase, and amino acid synthesis genes such as 2-oxoglutaratedependent dioxygenase, malate synthase and branched-chainamino-acid aminotransferase play crucial roles in potato tuberization. Likewise, inhibitors (cysteine protease and metallocarboxypeptidase), storage protein (patatin), TFs (heat stress, BTB/POZ and LOB domains, F-box), dehydration-responsive protein RD22 and hydroxyproline-rich glycoprotein are essentially involved in potato tuberization. Recently, Zhang et al. (2020) have observed key roles of nitrate transporters (StNRT2. 4, StNRT2. 5, and StNRT2. 7), glutamate dehydrogenase, glutamine synthetase and carbonic anhydrase in N metabolism in potato. Thus, like other plants, gene manipulation of N transporters in roots and assimilatory genes of carbohydrate and amino acids metabolism in shoots/stolons, and TFs (Myb and WRKY) could be manipulated by constitutive or tissue-specific promoters. Further, gene knockdown could be applied via RNAi (miR156, miR397, miR398, miR319, and miR482) or others targeting N pathways genes for improving NUE in potato (Figure 1). Moreover, CRISPR/Cas9 has been deployed in potato for multiple genes like Acetolactate synthase 1 (Butler et al., 2015) and granule-bound starch synthase (GBSS) (Andersson et al., 2017). Overall, candidate genes, TFs and miRNAs could be attempted for genetic manipulation to increase NUE in potato via transgenic or CRISPR/Cas9 or base-editing technologies.
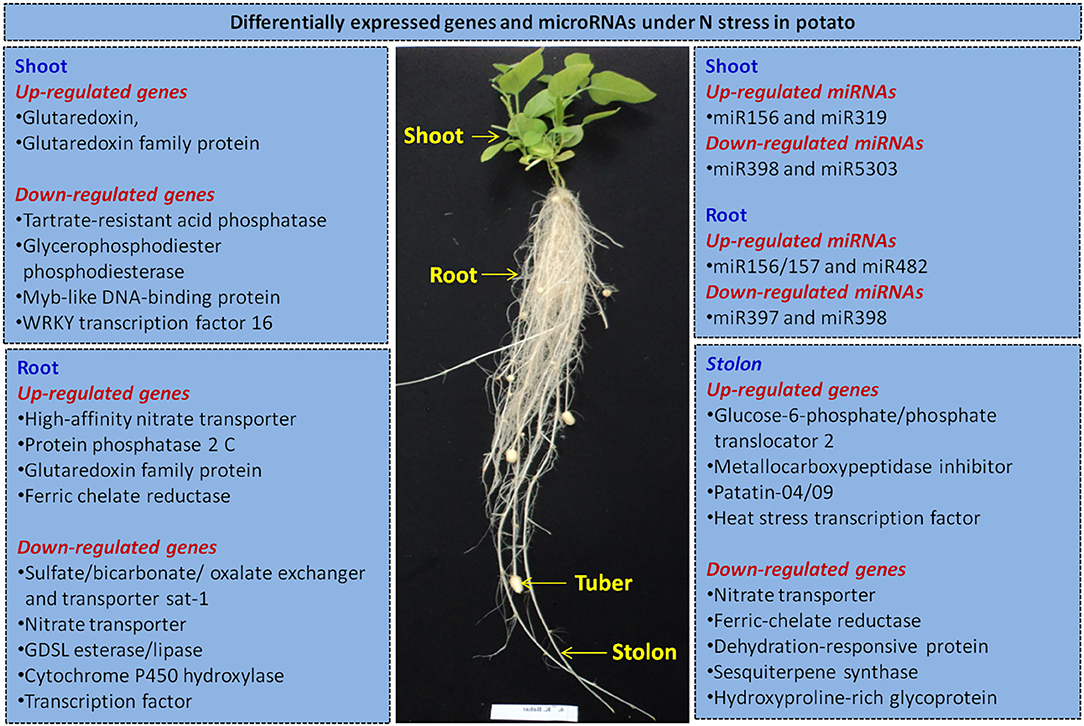
Figure 1. An outline of selected differentially expressed genes identified in potato under N stress based on transcriptome (RNA-seq and small RNA) sequencing of plant grown in aeroponic culture (Tiwari et al., 2020a,b,c). It summarizes the potential candidate genes, transcription factors and microRNAs in different potato tissues (root, shoot/leaf, and stolon) for improving nitrogen use efficiency in potato by gene manipulation via transgenics and/or CRISPR/Cas9 genome editing coupled with based-editing technologies.
Potato is a tetraploid and therefore application of CRISPR/Cas9 is more challenging. Albeit, all four alleles of StGBSS gene in potato have been knocked out and genome-edited mutants have been regenerated (Andersson et al., 2017). Moreover, various transformation methods like Agrobacterium, geminivirus replicon, protoplasts and polyethylene glycol have been suggested for Cas9 application in potato (Butler et al., 2015; Nadakuduti et al., 2018), of which protoplasts is an excellent one (Andersson et al., 2017). Further, suitable sgRNA promoters like Arabidopsis (AtUp) and potato (StU6p), and plant promoters like CAMV35S have been suggested for potato (Belhaj et al., 2013). Nevertheless, selection of target gene, design of guide RNA, efficient CRISPR/Cas9, plant transformation and off-target mutants are the major issues of genome editing in potato.
Plant N metabolism involves a network of genes associated in N uptake, transport, utilization, remobilization and storage processes. NUE is a complex multigenic trait and therefore its improvement becomes difficult particularly in tetraploid potato. However, a substantial success has been achieved through transgenic and little via CRISPR/Cas in plants. CRISPR/Cas9 has been mostly applied to negative regulators of genes, and therefore in future it is expected to discover such additional genes. Here, we have suggested a few candidate genes based on our research findings for improving NUE in potato applying transgenics or CRISPR/Cas9 technologies. Further, strengthening the knowledge on genes, TFs, and microRNAs and elucidating underlying molecular and physiological mechanisms of N pathways are vital for NUE research. Collectively, CRISPR/Cas9 coupled with base-editing strategies represents an invaluable system for precise genome editing. Nonetheless, robust Cas9 array system with multiplexing of targets, transformation and regeneration, phenotypes and people awareness would be challenges in genome editing research.
JT conceived the idea and wrote the manuscript. JT, TB, RS, and MK performed research work and improved the manuscript. SK critically read and edited the manuscript. All authors contributed to the article and approved the submitted version.
The work was funded by the Indian Council of Agricultural Research-Central Potato Research Institute, Shimla, Himachal Pradesh, India under the institute Biotechnology research programme and CABin Scheme (ICAR-IASRI, New Delhi).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Competent Authority, Indian Council of Agricultural Research-Central Potato Research Institute, Shimla, Himachal Pradesh, India and CABin Scheme (ICAR-IASRI, New Delhi) for necessary support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.597481/full#supplementary-material
Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell. Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Araus, V., Vidal, E. A., Puelma, T., Alamos, S., Mieulet, D., Guiderdoni, E., et al. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171, 1523–1532. doi: 10.1104/pp.15.01731
Baligar, V. C., Fageria, N. K., and He, Z. L. (2001). Nutrient use efficiency in plants. Commun. Soil Sci. Plant Anal. 32, 921–950. doi: 10.1081/CSS-100104098
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 39–47. doi: 10.1186/1746-4811-9-39
Brauer, E. K., Rochon, A., Bi, Y. M., Bozzo, G. G., Rothstein, S. J., and Shelp, B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant 141, 361–372. doi: 10.1111/j.1399-3054.2011.01443.x
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE 10:e0144591. doi: 10.1371/journal.pone.0144591
Chen, J., Fan, X., Qian, K., Zhang, Y., Song, M., Liu, Y., et al. (2017). pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15, 1273–1283. doi: 10.1111/pbi.12714
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020). Co-overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11:1245. doi: 10.3389/fpls.2020.01245
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Chen, X. B., Yao, Q. F., Gao, X. H., Jiang, C. F., Harberd, N. P., and Fu, X. D. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Dangol, S. D., Barakate, A., Stephens, J., Çaliskan, M. E., and Bakhsh, A. (2019). Genome editing of potato using CRISPR technologies: current development and future prospective. Plant Cell Tissue Organ. Cult. 139, 403–416. doi: 10.1007/s11240-019-01662-y
El-Kereamy, A., Bi, Y. M., Mahmood, K., Ranathunge, K., Yaish, M. W., Nambara, E., et al. (2015). Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front. Plant Sci. 6:934. doi: 10.3389/fpls.2015.00934
Fageria, N. K., Baligar, V. C., and Li, Y. C. (2008). The Role of nutrient efficient plants in improving crop yields in the twenty first century. J. Plant Nutr. 31, 1121–1157. doi: 10.1080/01904160802116068
Fan, X., Feng, H., Tan, Y., Xu, Y., Miao, Q., and Xu, G. (2015). A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58, 590–599. doi: 10.1111/jipb.12382
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., and Yang, M. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U. S. A. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S., Rentsch, D., et al. (2013). Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11, 446–458. doi: 10.1111/pbi.12031
FAO (2018). World Fertilizer Trends and Outlook to 2018. Rome: Food and Agriculture Organization of the United Nations.
Fu, Y., Yi, H., Bao, J., and Gong, J. (2015). LeNRT2.3 functions in nitrate acquisition and long-distance transport in tomato. FEBS Lett. 589, 1072–1079. doi: 10.1016/j.febslet.2015.03.016
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10:5207. doi: 10.1038/s41467-019-13110-8
Good, A. G., Johnson, S. J., De Pauw, M., Carroll, R. T., and Savidov, N. (2007). Engineering nitrogen use efficiency with alanine aminotransferase. Can. J. Bot. 85, 252–262. doi: 10.1139/B07-019
He, X., Qu, B., Li, W., Zhao, X., Teng, W., Ma, W., et al. (2015). The nitrate inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169, 1991–2005. doi: 10.1104/pp.15.00568
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838 doi: 10.1038/ng.3337
Iwamoto, M., and Tagiri, A. (2016). MicroRNA-targeted transcription factor gene RDD1 promotes nutrient ion uptake and accumulation in rice. Plant J. 85, 466–477 doi: 10.1111/tpj.13117
Kant, S., Bi, Y. M., and Rothstein, S. J. (2011). Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62, 1499–1509. doi: 10.1093/jxb/erq297
Khatodia, S., Bhatotia, K., Passricha, N., Khurana, S. M., and Tuteja, N. (2016). The CRISPR/Cas genome-editing tool: application in improvement of crops. Front. Plant Sci. 7:506. doi: 10.3389/fpls.2016.00506
Kurai, T., Wakayama, M., Abiko, T., Yanagisawa, S., Aoki, N., and Ohsugi, R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9, 826–837 doi: 10.1111/j.1467-7652.2011.00592.x
Lam, H. M., Wong, P., Chan, H. K., Yam, K. M., Chen, L., Chow, C. M., et al. (2003). Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 132, 926–935. doi: 10.1104/pp.103.020123
Li, H., Hu, B., and Chu, C. (2017). Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, M., Xu, J., Gao, Z., Tian, H., Gao, Y., and Kariman, K. (2020). Genetically modified crops are superior in their nitrogen use efficiency- a meta-analysis of three major cereals. Sci. Rep. 10:8568. doi: 10.1038/s41598-020-65684-9
Li, X., Wang, Y., Liu, Y., Yang, B., Wang, X., Wei, J., et al. (2018). Base editing with a Cpf1–cytidine deaminase fusion. Nat. Biotechnol. 36, 324–327. doi: 10.1038/nbt.4102
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U. S. A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Lu, Y., and Zhu, J. K. (2017). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 10, 523–525. doi: 10.1016/j.molp.2016.11.013
Mishra, R., Joshi, R. K., and Zhao, K. (2020). Base editing in crops: current advances, limitations and future implications. Plant Biotechnol. J. 18, 20–31. doi: 10.1111/pbi.13225
Nadakuduti, S. S., Buell, C. R., Voytas, D. F., Starker, C. G., and Douches, D. S. (2018). Genome editing for crop improvement-applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front. Plant Sci. 9:1607. doi: 10.3389/fpls.2018.01607
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A wheat CCAAT boxbinding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y. M., and Rothstein, S. J. (2014). AMT1;1 transgenic rice plants with enhanced anitha permeability show superior growth and higher yield under optimal and suboptimal anitha conditions. J. Exp. Bot. 65, 965–979. doi: 10.1093/jxb/ert458
Shrawat, A. K., Carroll, R. T., DePauw, M., Taylor, G. J., and Good, A. G. (2008). Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 6, 722–732. doi: 10.1111/j.1467-7652.2008.00351.x
Snyman, S. J., Hajari, E., Watt, M. P., Lu, Y., and Kridl, J. C. (2015). Improved nitrogen use efficiency in transgenic sugarcane: phenotypic assessment in a pot trial under low nitrogen conditions. Plant Cell Rep. 34, 667–669. doi: 10.1007/s00299-015-1768-y
St Clair, S. B., and Lynch, J. P. (2010). The opening of Pandora's box: climate change impacts on soil fertility and crop nutrition in developing countries. Plant Soil 335, 101–115. doi: 10.1007/s11104-010-0328-z
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Tang, W., Ye, J., Yao, X., Zhao, P., Xuan, W., Tian, Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10:5279. doi: 10.1038/s41467-019-13187-1
Tiwari, J. K., Buckseth, T., Devi, S., Varshney, S., Sahu, S., Patil, V. U., et al. (2020a). Physiological and genome-wide RNA-sequencing analyses identify candidate genes in a nitrogen-use efficient potato cv. Kufri Gaurav. Plant Physiol. Biochem. 154, 171–183. doi: 10.1016/j.plaphy.2020.05.041
Tiwari, J. K., Buckseth, T., Zinta, R., Saraswati, A., Singh, R. K., Rawat, S., et al. (2020b). Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 10:1152. doi: 10.1038/s41598-020-58167-4
Tiwari, J. K., Buckseth, T., Zinta, R., Saraswati, A., Singh, R. K., Rawat, S., et al. (2020c). Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato. PLoS ONE 15:e0233076. doi: 10.1371/journal.pone.0233076
Tiwari, J. K., Devi, S., Buckesth, T., Ali, N., Singh, R. K., Zinta, R., et al. (2020d). Precision phenotyping of contrasting potato (Solanum tuberosum L.) varieties in a novel aeroponics system for improving nitrogen use efficiency: in search of key traits and genes. J. Integr. Agric. 19:51–61. doi: 10.1016/S2095-3119(19)62625-0
Tiwari, J. K., Plett, D., Garnett, T., Chakrabarti, S. K., and Singh, R. K. (2018). Integrated genomics, physiology and breeding approaches for improving nitrogen use efficiency in potato: translating knowledge from other crops. Funct. Plant Biol. 45, 587–605. doi: 10.1071/FP17303
Trehan, S. P., and Singh, B. P. (2013). Nutrient efficiency of different crop species and potato varieties-in retrospect and prospect. Potato J. 40, 1–21.
United Nations (2019). Available online at: https://www.un.org (accessed August 6, 2020).
Wada, S., Hayashida, Y., Izumi, M., Kurusu, T., Hanamata, S., Kanno, K., et al. (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73. doi: 10.1104/pp.15.00242
Wang, L., Wang, L., Tan, Q., Fan, Q., Zhu, H., Hong, Z., et al. (2016). Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-Cas9. Front. Plant Sci. 7:1333. doi: 10.3389/fpls.2016.01333
Wang, S., Chen, A., Xie, K., Yang, X., Luo, Z., Chen, J., et al. (2020). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. U. S. A. 117, 16649–16659 doi: 10.1073/pnas.2000926117
Yanagisawa, S., Akiyama, A., Kisaka, H., Uchimiya, H., and Miwa, T. (2004). Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. U. S. A. 101, 7833–7838. doi: 10.1073/pnas.0402267101
Yu, C., Liu, Y., Zhang, A., Su, S., Yan, A., Huang, L., et al. (2015). MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS ONE 10:e0135196. doi: 10.1371/journal.pone.0135196
Yu, L. H., Wu, J., Tang, H., Yuan, Y., Wang, S. M., Wang, Y. P., et al. (2016). Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6:27795. doi: 10.1038/srep27795
Zhang, J., Wang, Y., Zhao, Y., Zhang, Y., Zhang, J., Ma, H., et al. (2020). Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS ONE 15:e0240662. doi: 10.1371/journal.pone.0240662
Zhao, D., Derkx, A. P., Liu, D. C., Buchner, P., and Hawkesford, M. J. (2015). Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 17, 904–913. doi: 10.1111/plb.12296
Zhong, L., Chen, D., Min, D., Li, W., Xu, Z., Zhou, Y., et al. (2015). AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 457, 433–439. doi: 10.1016/j.bbrc.2015.01.009
Keywords: CRISPR-Cas9, nitrogen use efficiency, plant, potato, transgenic
Citation: Tiwari JK, Buckseth T, Singh RK, Kumar M and Kant S (2020) Prospects of Improving Nitrogen Use Efficiency in Potato: Lessons From Transgenics to Genome Editing Strategies in Plants. Front. Plant Sci. 11:597481. doi: 10.3389/fpls.2020.597481
Received: 21 August 2020; Accepted: 10 December 2020;
Published: 23 December 2020.
Edited by:
Laura De Gara, Campus Bio-Medico University, ItalyReviewed by:
Jingguang Chen, Sun Yat-sen University, ChinaCopyright © 2020 Tiwari, Buckseth, Singh, Kumar and Kant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jagesh Kumar Tiwari, amFnZXNodGl3YXJpQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.