
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 January 2021
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.596065
Beige and Chediak Higashi (BEACH) domain proteins mediate membrane-dependent processes in eukaryotic cells. The plant BEACH domain protein SPIRRIG in A. thaliana (AtSPI) was shown to display a similar molecular behavior as its yeast and animal homologs, along with a range of cell morphological defects. In addition, AtSPI was shown to interact with the P-body component DCP1, to differentially effect RNA levels and to be involved in the regulation of RNA stability in the context of salt stress responses. To determine, whether the dual function of SPI in apparently unrelated molecular pathways and traits is evolutionary conserved, we analyzed three Aaspi alleles in Arabis alpina. We show that the molecular behavior of the SPI protein and the role in cell morphogenesis and salt stress response are similar in the two species, though we observed distinct deviations in the phenotypic spectrum.
The mechanisms of cell growth and morphogenesis are in the focus of plant science for decades. Due to fully sequenced model organisms like Arabidopsis thaliana and excellent model systems for cell differentiation like root hairs and trichomes (Mathur and Hülskamp, 2002; Bögre et al., 2008), genetic and environmental factors regulating growth, size and shape of plant organs are better understood nowadays. Morphogenesis is thought to be controlled predominantly by endoreduplication and thereby cell size control, cytoskeletal dynamics, vesicle transport and small GTPase signaling (Guimil and Dunand, 2007).
A group of mutants with distorted cell shapes was identified in EMS screens in Arabidopsis thaliana (Koornneef et al., 1983, 1987; Hülskamp et al., 1994), which show altered trichome morphology, irregular shapes of leaf pavement cells, epidermal hypocotyl cells and epidermal root cells. The corresponding distorted genes were found to encode proteins of the actin regulating ARP2/3 or SCAR/WAVE complex (Mathur et al., 2003; Schwab et al., 2003; Basu et al., 2005), with one exception: the BEACH domain protein SPIRRIG (Saedler et al., 2009).
BEACH domain proteins are named after their corresponding mutations in mice and humans, the beige and Chediak Higashi (BEACH) syndromes (De Lozanne, 2003). They are known facilitators of membrane dynamics (De Lozanne, 2003) and regulators of endosomal sorting processes (Cullinane et al., 2013). In agreement with these essential functions, the BEACH domain protein family is well conserved in mammals, plants and yeast, often exhibiting several members within one species (Saedler et al., 2009).
In Arabidopsis, only the BEACH domain protein SPIRRIG has been characterized so far (Saedler et al., 2009). In the N-terminus, SPI carries tandemly repeated Armadillo domains, each approximately 42 amino acids long. Armadillo domains share a conserved three-dimensional structure of three α helices, which fold together and interact to form a surface for protein-protein interactions (Coates, 2003). The Armadillo repeats are followed by a Concanavalin A (ConA)-like lectin domain, which is thought to be involved in oligosaccharide binding and to mediate membrane fusion events (Burgess et al., 2009). In the C-terminus, SPI exhibits a Pleckstrin-Homology (PH) domain, followed by the name giving BEACH domain. PH domains were found to interact with BEACH domains to form a large groove, possibly serving as a ligand-binding site (Jogl et al., 2002). The BEACH domain of SPI is followed by several WD40 repeats, which can mediate protein-protein interactions.
The spirrig mutants in A. thaliana share all cell morphogenesis defects observed in other distorted mutants including lower complexity of epidermal pavement cells, weakly distorted and curled trichomes, disconnected and out-curled hypocotyl cells in dark grown seedlings as well as defects in vacuolar integrity (Saedler et al., 2009). AtSPI seems to act in two very different pathways. First, it was shown that AtSPI physically interacts with the ATPase Suppressor of K+-Transport Growth Defect 1 (AtSKD1) and LYST Interacting Protein 5 (AtLIP5; Steffens et al., 2017) suggesting that it functions in the context of the endosomal sorting complex required for transport (ESCRT) machinery. Second, AtSPI localizes to and facilitates the formation of processing bodies (P-bodies). In addition, Atspi mutations lead to pleiotropic transcriptional changes and the stabilization of several mRNAs and their recruitment to P-bodies under salt stress conditions. Also this pathway was suggested to be evolutionary conserved because the AtSPI protein interacts with DECAPPING PROTEIN 1 (DCP1) from other species (Steffens et al., 2015).
The unusual finding that the Arabidopsis thaliana SPI gene has a dual role in unrelated pathways and traits raises the question, whether this is specific to one species or evolutionary conserved. We therefore studied this question in another distantly related Brassicaceae species that enables us to use genetic approaches and to unambiguously identify the true SPI ortholog by sequence similarity and synteny. We compared the developmental/physiological and molecular function between A. thaliana and Arabis alpina. A. alpina is suitable for laboratory use, since it is diploid, self-fertile, has a small and mostly sequenced genome and can be transformed with Agrobacterium tumefaciens (Wang et al., 2009). A. alpina was chosen for this work, since it is sufficiently closely related to A. thaliana to clearly identify orthologous genes, but also distant enough (26–40 million years; Koch et al., 2006; Beilstein et al., 2010) to expect phenotypic and molecular variations. The SPI proteins of A. thaliana and A. alpina are highly conserved, especially in the C-terminus. The sequence identity reaches 94% for the whole protein and 97% for the PH-BEACH-WD40 (PBW) domain. Would Aaspi mutants show the same phenotypic spectrum as Atspi mutants? Would the molecular behavior be comparable? Using three Aaspi mutants identified in an exhaustive EMS screen for trichome mutants in A. alpina (Chopra et al., 2019) we reveal a similar molecular behavior and phenotypes, though the phenotypes show distinct differences.
Arabis alpina EMS spirrig mutants spi-1, spi-2, and spi-3 were confirmed by sequencing of the SPIRRIG (Aa_G228370) transcripts obtained from PCR (N-terminus: 5′-CGTGTTTAGAGAGAGAAAG-3′, 5′-TATGAACAACAGCAA GGTGA-3′; Internal fragment: 5′-GCAACACGAACAGAACA TT-3′, 5′-CATCGTTCAAGCTTTTTGC-3′; C-terminus: 5′-AA TCCAAAGGGTCTGAAG-3′, 5′-ACAGATGGGAGCTATACA AT-3′; GATC/Eurofins, Ebersberg). Seeds were grown on soil, or surface-sterilized and grown on full MS medium (Murashige and Skoog, 1962). The seeds were stratified for 5 days. Subsequently, plants were grown under long day conditions at 21 ± 1°C and 100 ± 20 μmol/m2s light intensity. To analyze plant growth in the dark, seeds were removed from light conditions after 4 h, wrapped light-tight and vertically grown for 7 days. For germination assays, seeds were grown on MS plates and MS plates supplemented with 125 mM NaCl. Over the course of 20 days, seeds were checked for breakthrough of either root or cotyledons daily under a stereo microscope. For primary root growth assays, seeds were planted on MS plates, grown vertically, and transferred to MS plates and MS plates supplemented with different concentrations of NaCl after 7 days. Growth was then documented for 4 or 8 days. Seedling growth under transpiring conditions was carried out on soil under standard conditions, plants were watered by dipping every 3–4 days with water until day 20, followed by water or water supplemented with 125 mM NaCl until day 30. For transcript measurements, two to three 5–7 day old seedlings were transferred from plates to liquid 1/2 MS or liquid 1/2 MS with 125 mM NaCl for 4 h under constant shaking and subsequently frozen with liquid nitrogen. For short term salt stress of leaves, samples were transferred to 1/2 MS with 125 mM NaCl for 60–80 min.
Fluorescein diacetate was used as a negative stain to identify vacuole patterns in root hairs. Plants were treated with a 0.05% w/v solution of FDA (Sigma) in ddH2O for 15–20 min and analyzed under the fluorescence microscope.
Agarose imprints were used to visualize the cell shapes of epidermal pavement cells. A 3% w/v agarose solution was applied to young cotyledons, peeled off after hardening and analyzed under the light microscope.
Stereo microscopy was carried out with the Leica MZ 16F stereo microscope and the LAS AF software. Light and fluorescence microscopy was carried out with the Leica DMRB microscope equipped with a Sony Alpha 6000 camera or with the Leica DMRE microscope equipped with the Leica DFC 7000 T camera using the LAS X software. Confocal laser scanning microscopy was carried out with the Leica DM5500 and DM6000 CS Microscopes and documented with the TCS-SPE and TCS-SP8 imaging systems, respectively (Leica Microsystems, Heidelberg, Germany).
Total RNA was isolated (TRI Reagent, Ambion by Life Technologies) and treated with DNaseI (Thermo Fisher Scientific). Integrity of RNA was confirmed on a bleach gel (Aranda et al., 2012) and the concentration was quantified spectrophotometrically. One microgram of total RNA was reverse-transcribed (SuperScriptIII, Invitrogen).
qPCRs were carried out in a QuantStudio 5 System (ABI/Life Technologies) equipped with a 96 well block. The qPCRs were performed using plates (96 well, 0.2 ml) and cover foil (Opti-Seal Optical Disposable Adhesive, BIOplastics) and SYBR Green reagent (Thermo Fisher Scientific). All results represent the average of three biological and three technical replicates. Analysis was carried out with the QuantStudio TM Design and Analysis Software version 1.4.1 and Excel 2007. Outliers were removed using a two-sided Grubbs test at a significance level of 0.05. Efficiency of primers was calculated in cDNA dilution series of 1:10, 1:20, 1:40, 1:80, 1:160, and 1:320. Higher dilutions were not possible due to the concentration of RNA/cDNA. Primers for reference genes were described before (Wang et al., 2009; Stephan et al., 2019). Primers for genes of interest were accepted with an efficiency of 80–120% and a correlation between −1 and −0.99 (Supplementary Table S1). Normalization against one reference gene was carried out using the ΔΔCt method. Normalization against two reference genes was carried out using normalization factors, according to the geNorm manual (Vandesompele et al., 2002). Standard deviations between biological replicates were calculated over the means of the single replicates, rather than the raw data.
Sequences were taken from the Genomic resources for Arabis alpina website1. In silico sequence analysis was carried out with CLC DNA Workbench version 5.6.1. Conserved domains were determined using CD-Search (Marchler-Bauer and Bryant, 2004) and/or PROSITE (Hulo et al., 2007). Phylogenetic trees were created using the Neighbor Joining method with the Unipro UGENE software (version 1.16.0) and subsequently modified for clarity using Adobe Illustrator CS4 version 14.0.0.
All plant measurements were carried out using ImageJ (Fabrice Cordelieres, Institut Curie, Orsay, France). Data was subsequently analyzed with Microsoft Excel 2007. Cell complexity was calculated using the formula .
Statistical analysis was carried out with OriginPro 8.5 0G SR0, Microsoft Excel 2007 or R Studio. Significance levels were tested as indicated in the results section.
The coding sequences of AaSPI-PBW (Aa_G228370), AaDCP1 (Aa_G368370), AaSKD1 (Aa_G151760), AaLIP5 (Aa_G86640), AaVPS60.1 (Aa_G137100), AaVPS60.2 (Aa_G304080) were amplified from cDNA of pep1 (Supplementary Table S1). Gateway donor or destination vectors containing AtSPI-PBW (AT1G03060), AtDCP1 (AT1G08370), AtDCP2 (AT5G13570), AtDCP5 (AT1G26110), AtSKD1 (AT2G27600), AtLIP5 (AT4G26750), AtVPS60.1 (AT3G10640), AtVPS60.2 (AT5G04850), AtTTG1 (AT5G24520), AtGL3 (AT5G41315), and AtMYC1 (AT4G00480) have been described before (Zimmermann et al., 2004; Shahriari et al., 2010; Zhao et al., 2012; Steffens et al., 2014, 2015; Tian et al., 2015). Gateway vectors pENSG:YFP/CFP, pEXSG:YFP/CFP (Feys et al., 2005) and pAMARENA (M. Jakoby, NCBI:txid905036) were used for localization, pSYN/pSYC (Jakoby et al., 2006) and pCL112/113 (provided by J. F. Uhrig) were used for BiFC, pACT/pAS (Clonetech) were used for yeast two-hybrid analyses.
Yeast two-hybrid assays were carried out as described previously (Gietz et al., 1995). The selection on synthetic dropout interaction media lacking leucine (-L), tryptophan (-W), and histidine (-H), was performed with 3-aminotriazole (3AT) concentrations up to 30 mM. Bimolecular Fluorescence Complementation (BiFC) analysis was done in Arabidopsis leaves by biolistic transformation as described before (Mathur et al., 2003). Comparability of BiFC samples was guaranteed by fixed laser intensities, gain and line accumulation. The raw data was enhanced for clarity using Adobe Photoshop CS4 version 14.0.0 and ImageJ (Fabrice Cordelieres, Institut Curie, Orsay, France). Co-localization of AaSPI-PBW with ESCRT and P-body components was carried out in leaves of 3 week old Arabis seedlings by biolistic transformation (Mathur et al., 2003).
Phylogenetic analysis has shown that BEACH domain proteins can be clustered into four groups (A–D), each containing plant as well as animal genes (Saedler et al., 2009). The Arabidopsis SPI gene belongs to group A, which includes animal genes that were described to be involved in membrane trafficking (De Lozanne, 2003). In our phylogenetic analysis, we found the SPI gene well conserved in all plant species analyzed (Supplementary Figure S1). In Arabis alpina, the orthologous SPI gene was identified by sequence homology and the synteny of homologous genes on the chromosome (Chopra et al., 2019). Three spi mutants were identified in an EMS mutagenesis screen for trichome phenotypes (Chopra et al., 2019) in the perpetual flowering 1 (pep1) mutant, in the pajares (paj) ecotype background (Wang et al., 2009). This line harbors a mutation in the floral repressor PEP1, which causes the plants to flower without vernalization (Albani et al., 2012). Therefore, working in the pep1 background greatly facilitates the genetic analysis of processes not related to flowering. All three alleles display early STOP codons (Figure 1A). To verify that the three mutant lines are indeed three spi alleles we performed complementation tests. F1 plants from pairwise crosses between the three mutants showed a severe trichome phenotype indicating the mutations in the SPI gene are causative for the observed trichome phenotype (Supplementary Figure S2).
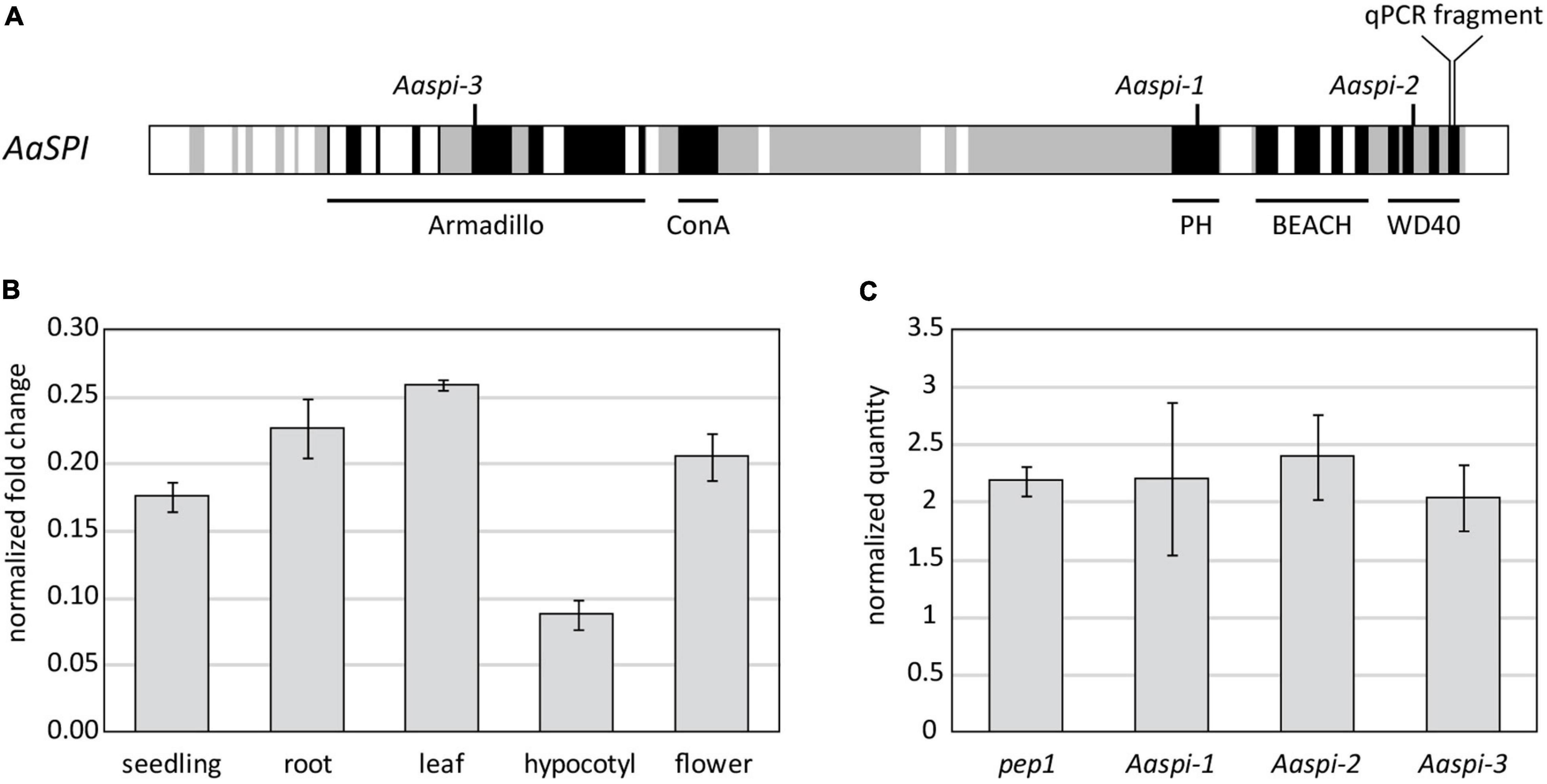
Figure 1. Molecular characterization of the SPIRRIG gene in Arabis alpina. (A) Schematic presentation of AaSPIRRIG. Exons are shown in gray, functional domains are depicted in black. The positions of early stop codons within the spirrig alleles are indicated above. (B) The expression levels of AaSPI were determined in five tissues by qPCR. Normalization was carried out using AaRAN3 (Wang et al., 2009). (C) The expression levels of AaSPI were determined in the above ground parts of seedlings of pep1, Aaspi-1, Aaspi-2, and Aaspi-3 by qPCR. Normalization was carried out using AaTUA5 and AaCAC (Stephan et al., 2019).
The expression profile of AaSPI was determined in whole seedlings, roots, mature leaves, hypocotyls and flowers (Figure 1B). The transcript is expressed throughout the whole plant, similar as observed in Arabidopsis thaliana (Saedler et al., 2009). The expression is lower in the hypocotyl, compared to other tissues of the plant. The expression levels in the three mutant alleles are similar to that in pep1, which is used as the wild-type reference for AaSPI (Figure 1C). This shows that the aberrant transcripts are not recognized and degraded by non-sense mediated decay and that the truncation of the protein is the cause for the mutant phenotype.
The three Aaspi alleles had been identified by their weak distorted trichome phenotype (Chopra et al., 2019; Figure 2). In Arabidopsis, it was shown that spi mutant trichomes exhibit a reduced stalk length and width and a reduced average branch length (Saedler et al., 2009). In addition, we found that Arabidopsis Atspi mutants have a decreased number of branches (Supplementary Table S2). To compare the phenotypic spectrum of the Aaspi mutants with that in Atspi mutants, we studied the trichome phenotype in more detail. We found that almost all branches were curled or distorted in Aaspi mutants (Figure 2). Stalk length was significantly reduced in all three Aaspi alleles. Also, trichome branching was clearly reduced in the three Aaspi mutants (Figure 2). Stalk width and branch length varied in the three alleles such that some of the Aaspi alleles showed a difference to pep1 and others not (Table 1). We therefore considered these two aspects of the distorted phenotype as not affected by the Aaspi mutations.
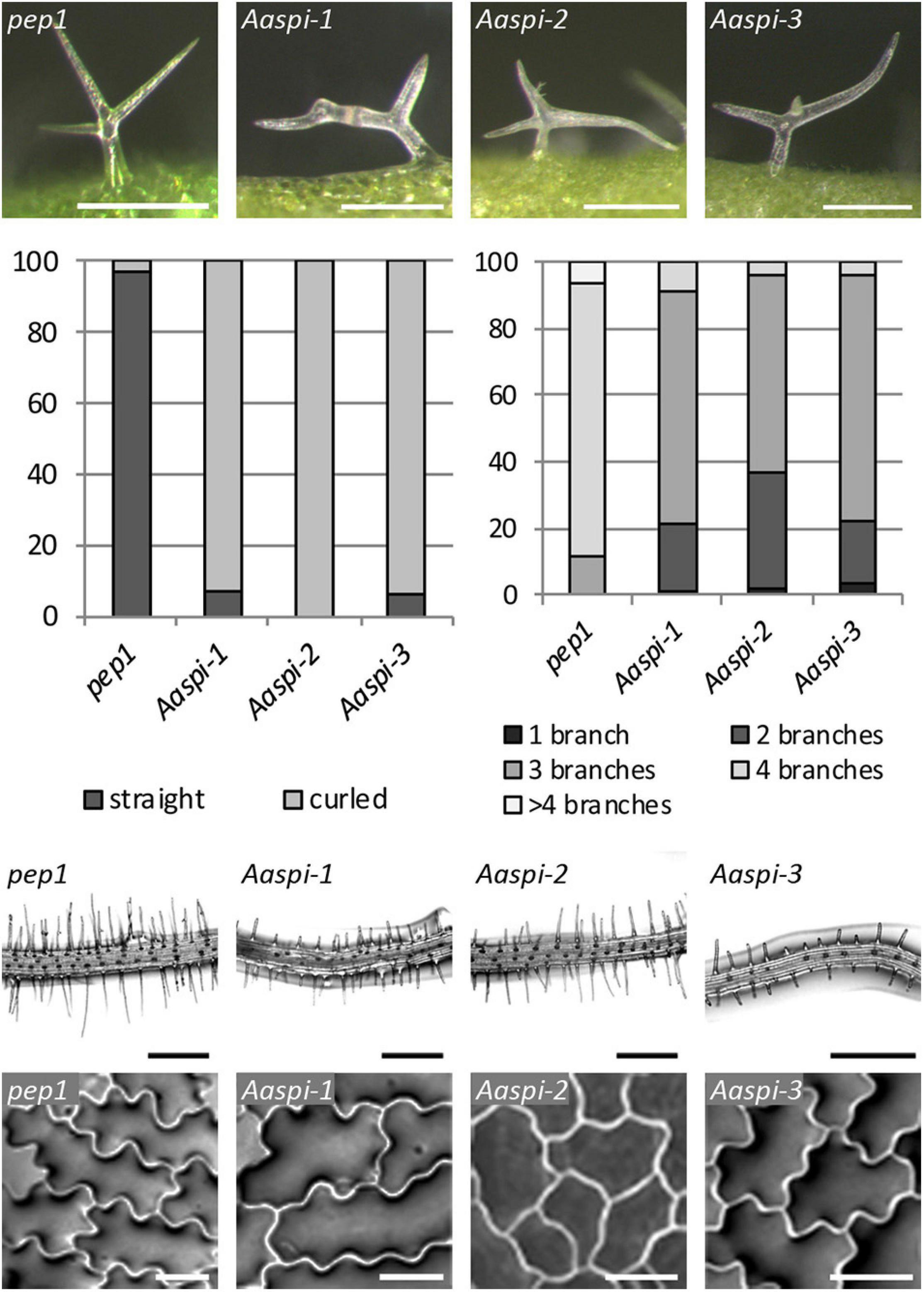
Figure 2. The Aaspi distorted phenotype of trichomes, roots and epidermal pavement cells. First row: Microscopic pictures of distorted trichomes. Second row, left: Ratio of straight and curled trichomes in pep1 and Aaspi mutants. Second row, right: Number of trichome branches in pep1 and Aaspi mutants. Graph shows the relative abundance in% in more than 100 trichomes per line. Third row: Microscopic images of root hair phenotypes to illustrate the shorter root hairs in the Aaspi mutants. Scale bare: 300 μm. Bottom row: Microscopic images of epidermal pavement cells to illustrate the lower complexity of epidermal pavement cells on cotyledons. Scale bar: 50 μm.
Arabidopsis spi mutants are characterized by morphological changes in other cell types as well. In Atspi mutants root hairs are shorter as compared to wild-type (Saedler et al., 2009). We analyzed Aaspi mutant root hairs about 1 mm below the border between hypocotyl and the root of 7 days old seedlings. We found that Aaspi mutants also have significantly shorter root hairs than pep1 (Figure 2 and Table 1).
In Arabidopsis, Atspi epidermal pavement cells show less lobing and, as a result, less complexity than wild-type cells (Saedler et al., 2009). To assess whether this phenotype is also found in Aaspi mutants, we inspected the pavement cells on cotyledons. We show that Aaspi mutant epidermal cells are less lobed than pep1 cells (Figure 2). To quantify the difference in the lobing of cells we determined the mean complexity as done before (Dewitte and Murray, 2003) and found a significant reduction in all three Aaspi alleles (Table 1).
In Arabidopsis, young spi seedlings show an almost twofold reduction of the hypocotyl length (Saedler et al., 2009). Our analysis of Aaspi seedlings revealed no decrease of hypocotyl length as compared to pep1 (Table 2). If at all, hypocotyls of Aaspi seedlings were longer as observed in Aaspi-2 and Aaspi-3. We also never observed individual hypocotyl cells tearing out of the epidermal cell surface when grown in the dark. These observations correlate with our finding that AaSPI expression is relatively lower in the hypocotyl as compared to Arabidopsis, suggesting that AaSPI is less important for hypocotyl growth.
In Arabidopsis root hairs vacuoles were found to be fragmented, giving the initial hint toward a function of AtSPI in membrane trafficking/organization similar as reported for BEACH domain proteins in other species (Saedler et al., 2009). Toward this end we analyzed Fluorescein Diacetate (FDA) stained root hairs of pep1 and Aaspi mutants. Our analysis revealed a low frequency of root hairs with fragmented vacuoles in the range of 5% in both pep1 and all Aaspi mutants. This indicates that Aaspi mutants are not affected in vacuolar integrity of root hairs (Figure 3 and Table 3). This finding raised the question, whether the AaSPI protein interacts with ESCRT proteins, similar as reported for AtSPI (Steffens et al., 2017). In a first step, we studied the intercellular localization by transiently transforming Arabis alpina epidermal cells with YFP tagged proteins. In these experiments, YFP-AaSPI localized to the cytoplasm (Figure 4). Also, the Arabis ESCRT components CFP-AaSKD1 and CFP-AaLIP5 were found in the cytoplasm. CFP-AaVPS60.1 and CFP-AaVPS60.2 were found in the cytoplasm and in dot-like structures (Figure 4). In a second step, we studied the interaction of the AaSPI-PBW fragment with the four ESCRT proteins from A. alpina. In addition, we tested the interaction of AaSPI-PBW with ESCRT proteins from A. thaliana to assess the interspecies interactions (Table 4). In these experiments we found interactions between AaSPI-PBW and all Arabidopsis and Arabis ESCRT proteins, similar as reported for Arabidopsis SPI-PBW. To study the interaction behavior in Arabis alpina cells, we performed Bimolecular Fluorescence Complementation (BiFC) experiments. BiFC interaction was found between AaSPI and all ESCRT proteins from Arabis and Arabidopsis (Figures 5, 6). Positive and negative BiFC controls are shown in Supplementary Figures S5–S7. It would be desirable to further clarify the interaction behavior of AaSPI, however, also pull-down assays failed in our hands. Assays with bacterially expressed proteins did not work since we could not recover sufficient amounts of soluble proteins.
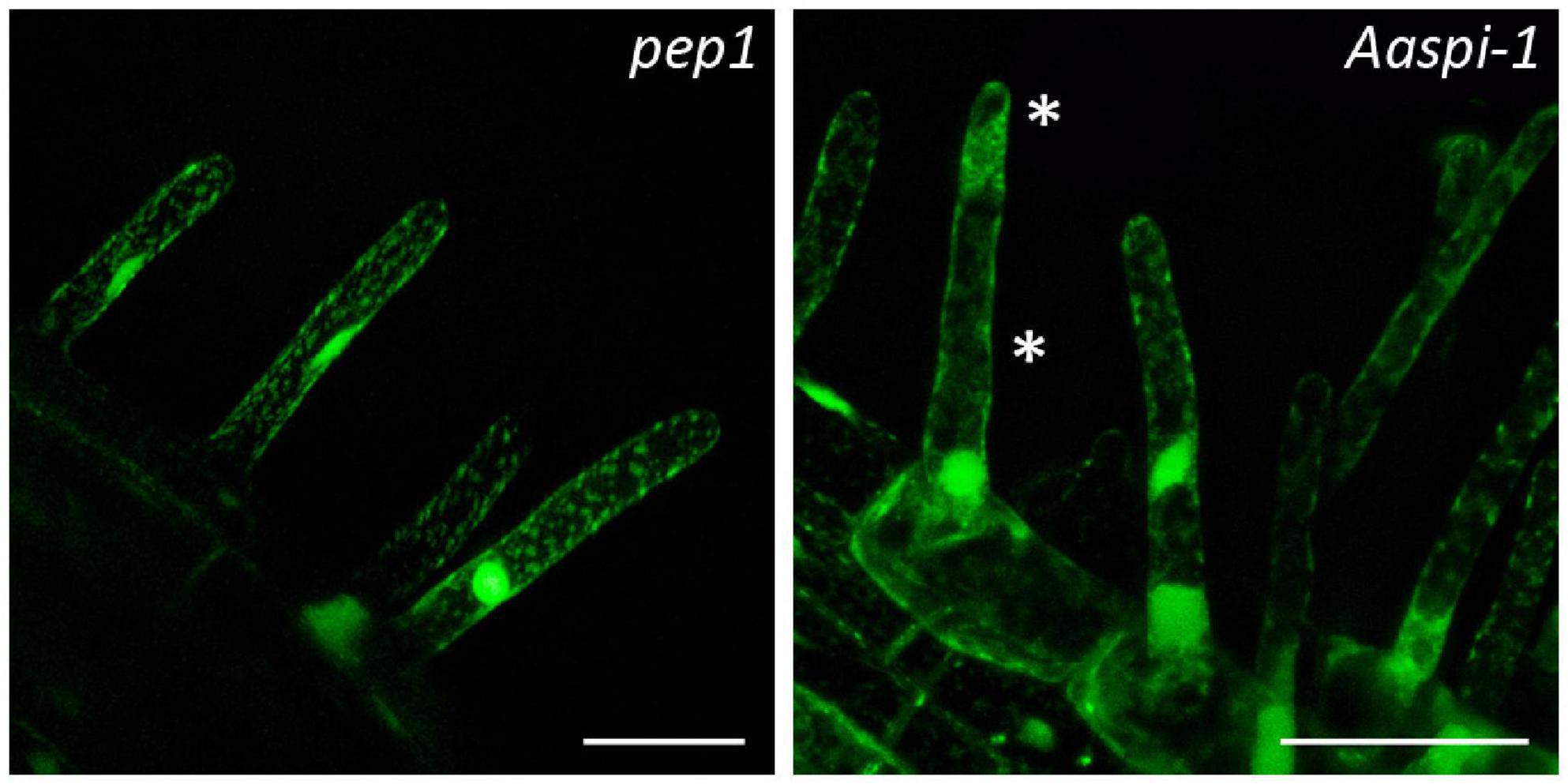
Figure 3. Fragmented root hair vacuoles in pep1 and Aaspi-1 mutants. Left: Representative picture of an intact vacuole in pep1. Right: Representative picture of a fragmented vacuole in Aaspi-1. Asterisks indicate fragmented vacuoles. Root hairs were stained with FDA and integrity of vacuoles was analyzed in > 100 root hairs per line. Scale bars: 50 μm.
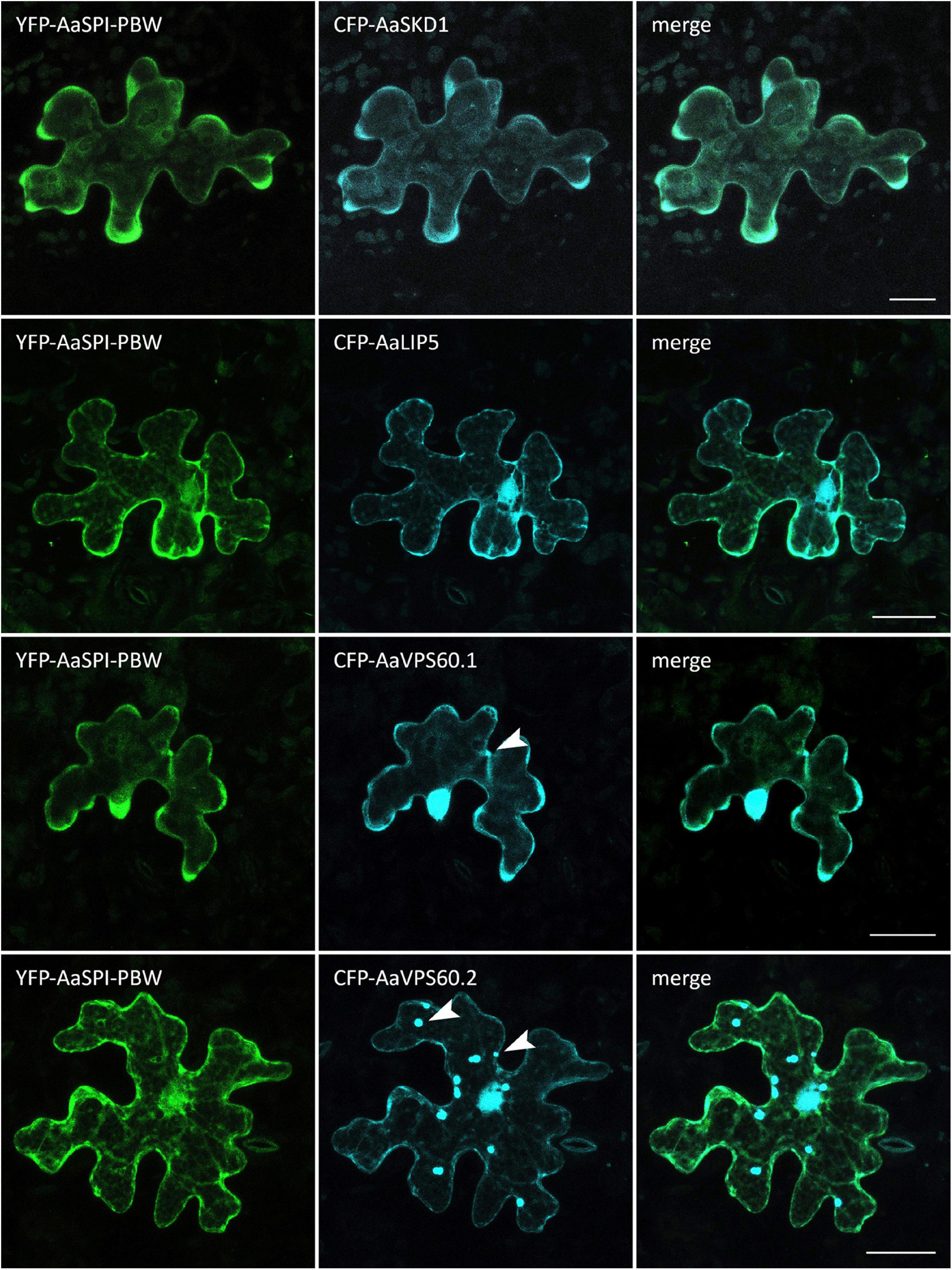
Figure 4. Intracellular localization of CFP-AaESCRT components and YFP-AaSPI-PBW. Transiently expressed CFP tagged AaESCRT proteins and YFP-AaSPI-PBW in Arabis alpina epidermal pavement cells. Arrow heads indicate dot-like structures. Scales bars: 25 μm.
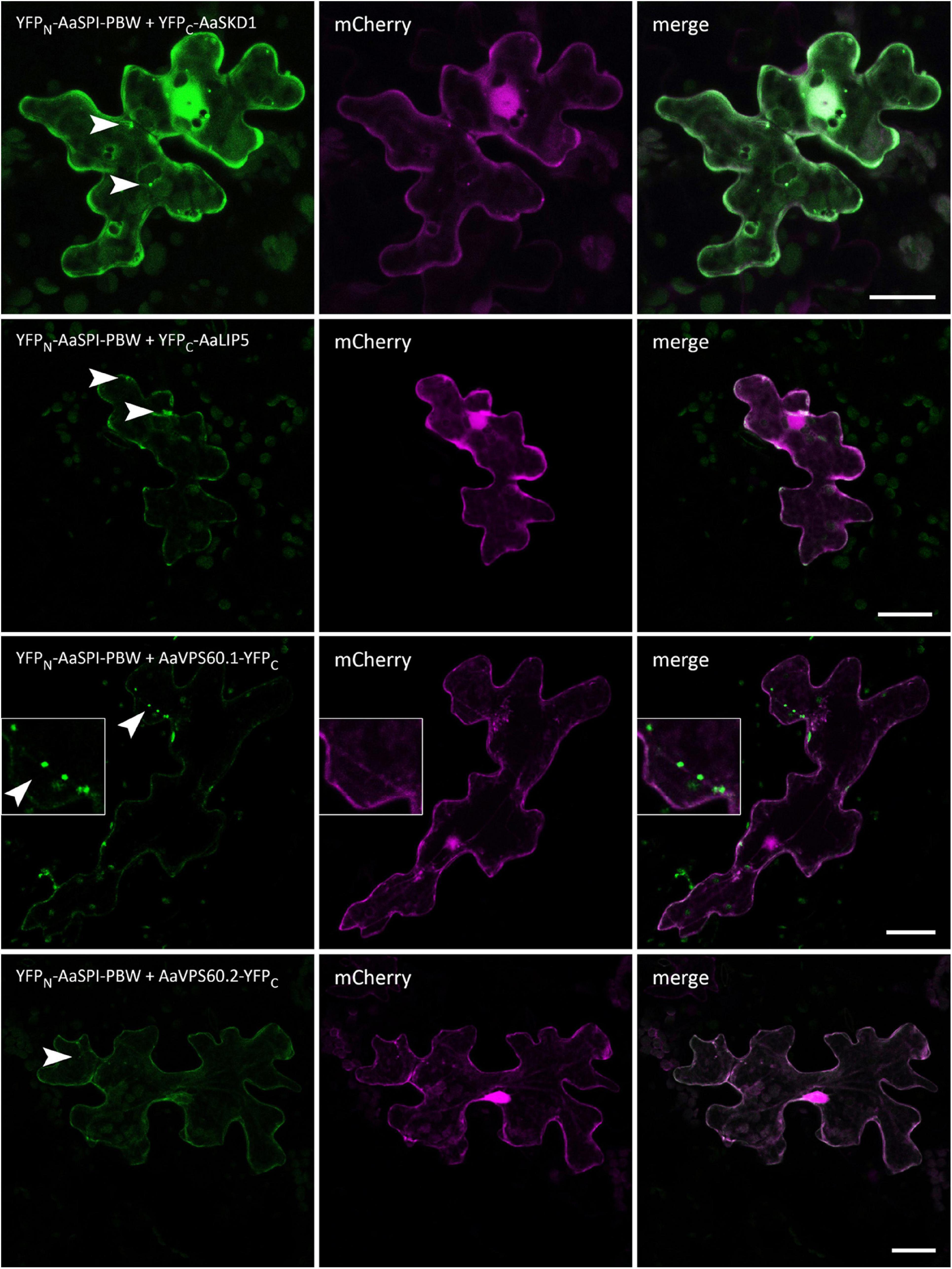
Figure 5. BiFC interactions of AaSPI-PBW with ESCRT components AaSKD1, AaLIP5, AaVPS60.1, and AaVPS60.2. Left—BiFC, middle—transformation control, right—overlay of BiFC and transformation control. Arrow heads indicate the sites of interaction. Scales bars: 25 μm.
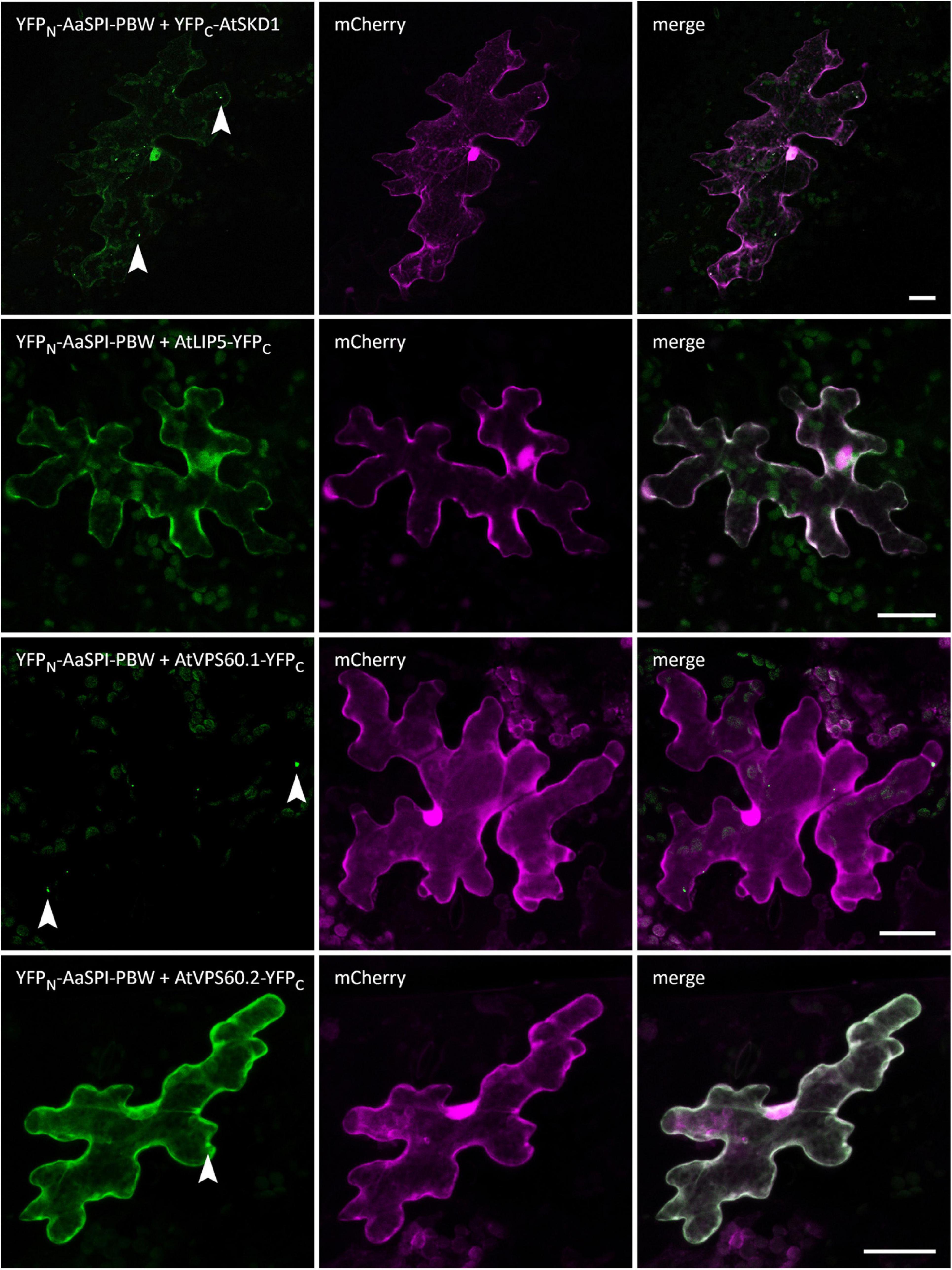
Figure 6. BiFC interactions of AaSPI-PBW with ESCRT components AtSKD1, AtLIP5, AtVPS60.1, and AtVPS60.2. Left—BiFC, middle—transformation control, right—overlay of BiFC and transformation control. Arrow heads indicate sites of interaction. Scale bars: 25 μm.
The PBW domain of AtSPI had been shown to interact with DCP1 homologs from mammals and yeast (Steffens et al., 2015). To determine, whether this behavior is conserved in A. alpina, we studied the interaction of AaSPI with DCP1 in yeast two-hybrid and BiFC assays. We found interactions with the AaDCP1 and AtDCP1 proteins (Table 5 and Figure 7).
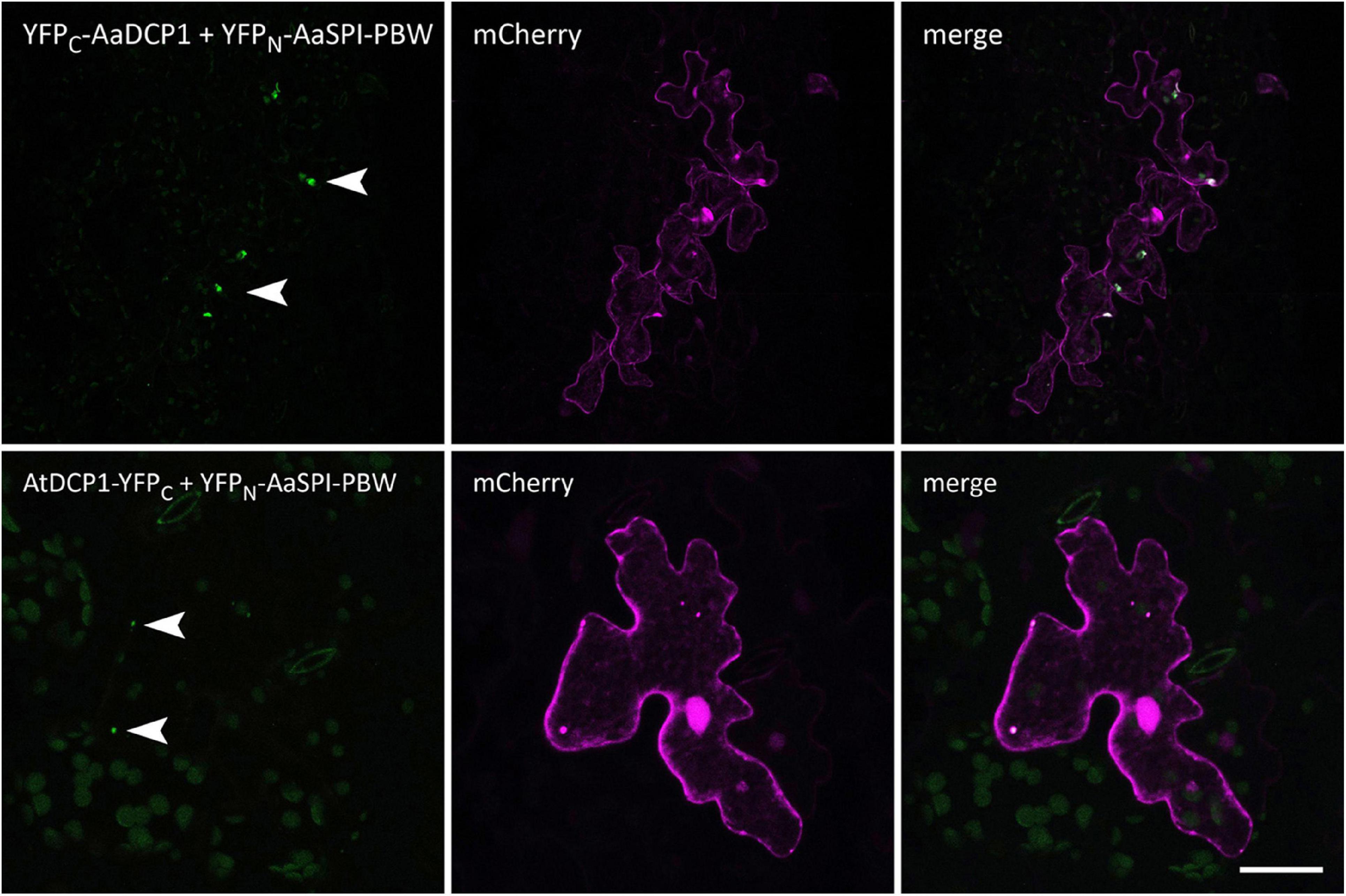
Figure 7. BiFC interactions of AaSPI-PBW with the P-body component DCP1 from A. thaliana and A. alpina. Left—BiFC, middle—transformation control, right—overlay of BiFC and transformation control. Arrow heads indicate sites of interaction. Scale bars: 25 μm.
In Arabidopsis, SPI-PBW co-localization to DCP1 is triggered by salt stress, suggesting that this stress treatment causes a re-localization of the AtSPI-PBW protein (Steffens et al., 2015). In a first step we studied the co-localization of AaSPI-PBW and AaDCP1 under normal conditions. AaSPI-PBW alone was evenly distributed in the cytoplasm (Figure 8). When co-expressed with AaDCP1, AaSPI-PBW appeared in the cytoplasm and additionally in AaDCP1-marked P-bodies in the majority of cells (Figure 8). Thus, AaDCP1-PBW is recruited to P-bodies under normal conditions in these experiments. Upon salt stress (up to 300 mM of NaCl) the majority of leaf epidermal cells did not show a response to the treatment. However, we observed an increased number of small P-bodies and additional recruitment of AaSPI-PBW to AaDCP1-marked P-bodies in very few cells after 40 min of salt treatment (Supplementary Figure S3).
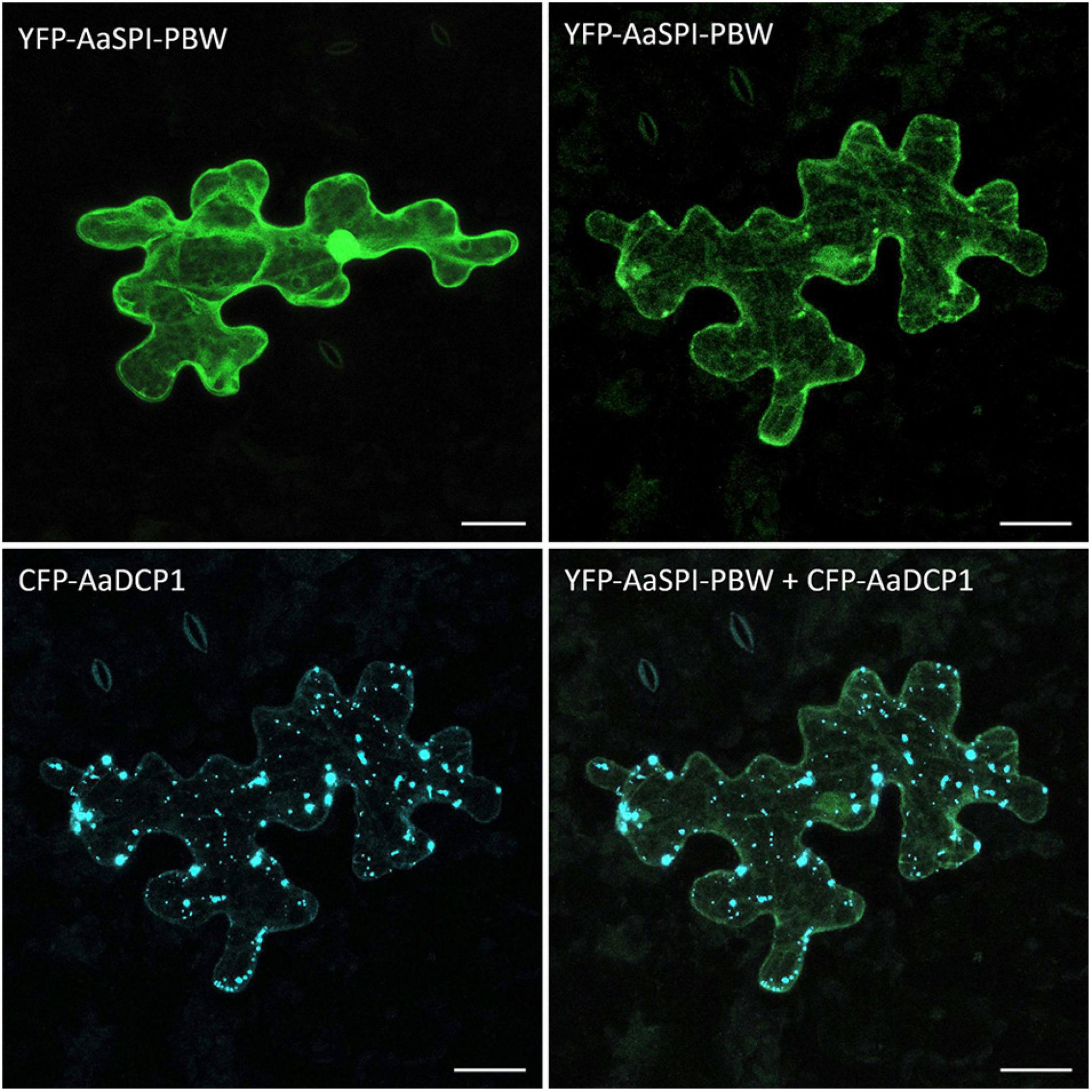
Figure 8. Localization of AaSPI-PBW in pep1 epidermal pavement cells. Transient expression of YFP-AaSPI-PBW (first row), cytosolic (left) and cytosolic with dots (right); and CFP-AaDCP1 (second row), in dots (left) and in dots overlapping the expression of YFP-AaSPI-PBW. Scale bars: 25 μm.
We analyzed the salt response of Aaspi mutants, as SPI was shown to be essential for the salt stress tolerance of Arabidopsis (Steffens et al., 2015). We tested effects of salt on germination, primary root growth and seedling growth, representing different tissues and stages of development.
Germination and primary root growth were significantly inhibited in Aaspi-1 and Aaspi-3 (Figures 9A,C). Strikingly, Aaspi-2 did not show an altered salt response compared to pep1. At a later stage of development and under transpiring conditions, salt hypersensitivity is only visible for Aaspi-1 (Figure 9B). The Aaspi-3 allele is indistinguishable from pep1 at this stage of development, while Aaspi-2 is hyposensitive to salt.
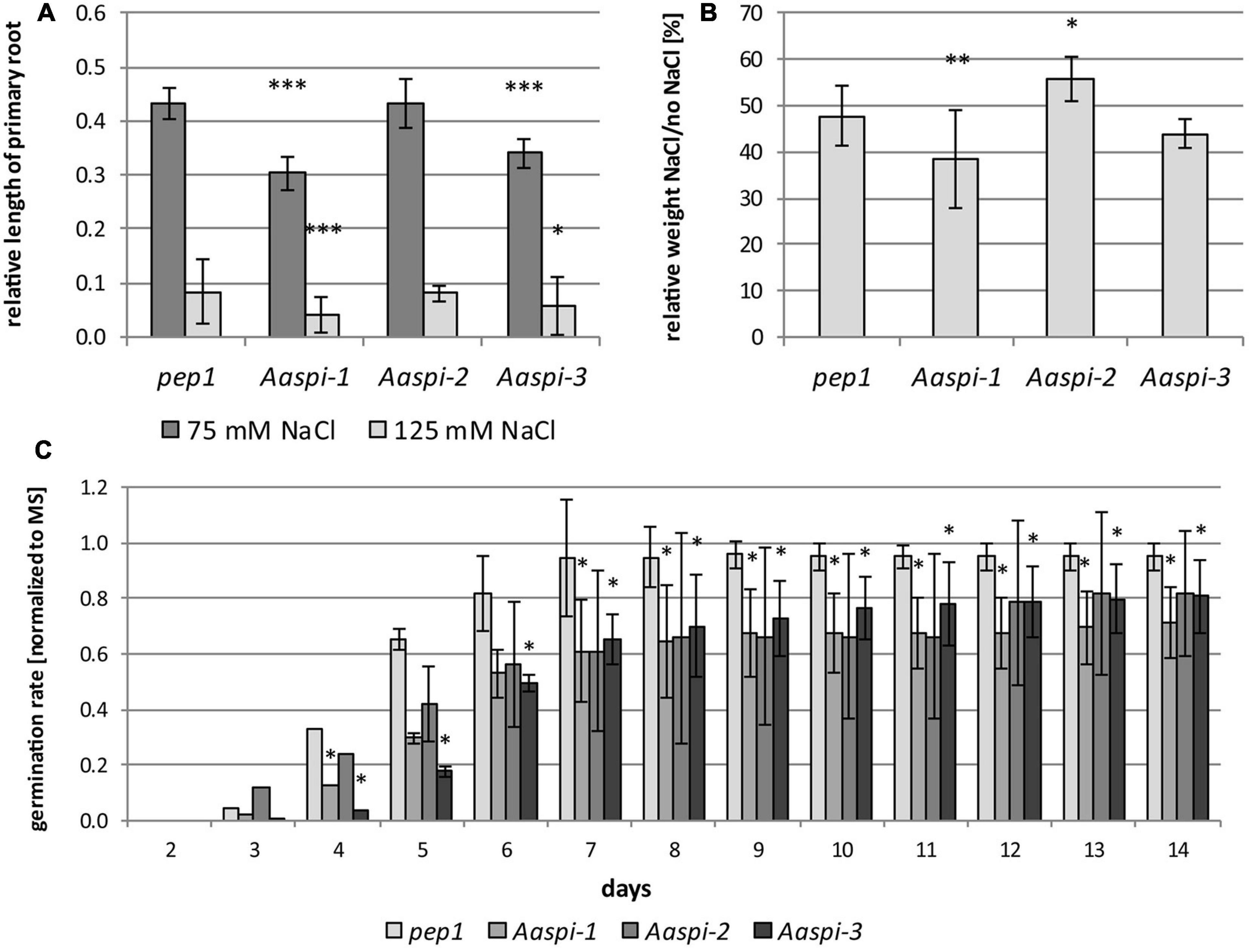
Figure 9. Salt treatments in pep1 and Aaspi alleles. (A) Primary root growth of pep1 and Aaspi on different concentrations of NaCl after 4 days. Primary root growth under salt stress conditions was tested in three independent experiments. The assays were carried out on plates containing 0 mM NaCl, 75 mM NaCl and 125 mM NaCl, respectively. Roots were measured after 4 days of growth. Significance of the data was tested using an ANOVA with p < 0.001 (***), p < 0.01 (**), and p < 0.05 (*). Total N per line per condition 93–128. (B) Seedling growth of Arabis alpina lines on soil under salt stress. Plants were watered every 3–4 days with water for the first 20 days, subsequently plants were watered with either water or water with 125 mM NaCl (N = 27–31 per line and condition). The weight of the seedlings was determined after 30 days. Significance of the data was tested using ANOVA with p < 0.001 (***), p < 0.01 (**), and p < 0.05 (*). (C) Germination of pep1 and spi mutants under salt stress. Germination of pep1 and the three Aaspi mutants was analyzed on MS plates and MS plates supplemented with 125 mM NaCl in three independent sets (N > 115 per line and condition). The relative number of seeds germinated on salt media was normalized against the values from media containing no salt and combined between the sets. Error bars depict standard deviations. Significance was tested using Mann-Whitney-U at a level of *p < 0.10.
On the transcript level, we identified four genes which are differentially regulated under salt stress in Aaspi mutants (Figure 10). These genes were chosen following an RNAseq analysis in A. thaliana (Steffens et al., 2015). Two of which showed a similar salt response of all Aaspi alleles (Aa_G78660/AT3G07250 and Aa_NA/AT1G71520), while the other two reflected the phenotypic differences between the alleles (Aa_G499040/AT5G59310 and Aa_NA/AT3G44860). AT3G07250 is the nuclear transport factor family protein AtNTF2. This protein has a function in RNA binding and is likely to function as a ribonucleoprotein complex (Lamesch et al., 2012). AaNTF2 mRNA levels are strongly increased under salt stress, with a significantly higher upregulation in all three Aaspi mutants, compared to pep1 (Figure 10). AT1G71520 is an integrase-type DNA-binding superfamily protein with DNA-binding transcription factor activity, which is involved in ethylene-activated signaling, regulation of transcription and response to chitin (Lamesch et al., 2012). Under salt conditions, all Aaspi mutants showed a decreased upregulation compared to pep1 (Figure 10). AT5G59310 encodes the lipid transfer protein 4 (AtLTP4). Proteins of this family can bind fatty acids and acylCoA esters. They can transfer phospholipids and function in response to abscisic acid, salt stress and drought (Lamesch et al., 2012). For this gene, Aaspi-2 showed a significantly higher upregulation in salt conditions compared to pep1 and the other two Aaspi alleles (Figure 10), possibly explaining the hyposensitivity to salt in later developmental stages. AT3G44860 encodes FARNESOIC ACID CARBOXYL-O-METHYLTRANSFERASE (AtFAMT). The role of farnesoic acid in plants has not been revealed so far, however, the transcript level of FAMT is increased upon various stresses, indicating that it might have general functions in stress responses (Yang et al., 2006). The qPCR results showed that the increase of transcript was significantly less in Aaspi-1 compared to pep1 and the other two spi alleles (Figure 10), which could explain the strong hypersensitivity of this allele.
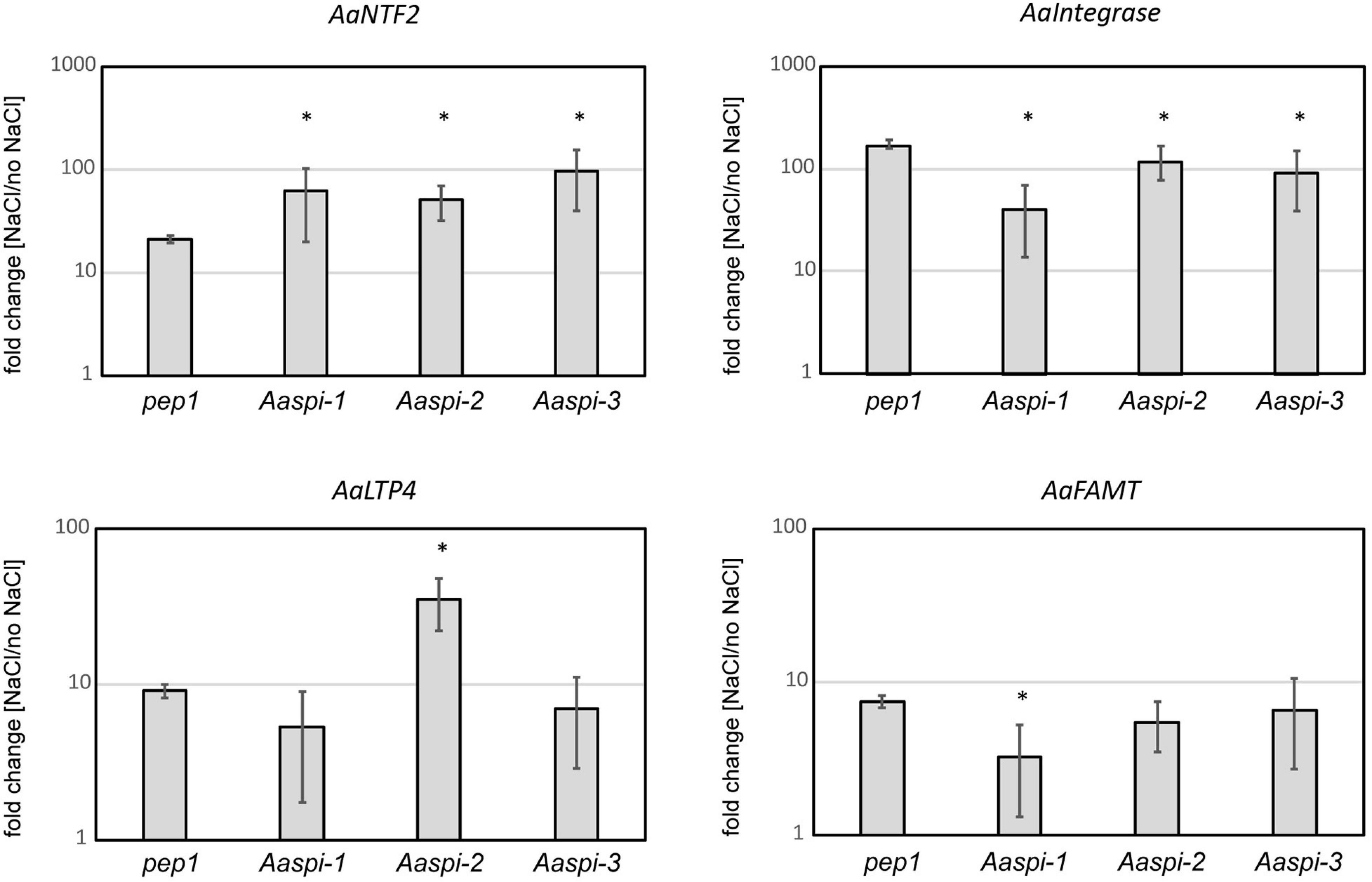
Figure 10. Relative transcript levels in pep1 and Aaspi mutants in response to salt stress. Normalization was carried out against AaCAC and AaTUA5. Error bars depict standard deviation. Significance was determined using a Mann-Whitney-U test at *p < 0.10.
Our phenotypic analysis of Arabis spi mutants revealed that they share most of the morphological phenotypes with Arabidopsis spi mutants. We also found a similar localization and protein-protein interaction behavior of the AtSPI and AaSPI proteins with ESCRT proteins. Taking these observations together, it is conceivable that the regulation of cell morphogenesis and a molecular function in the endosomal pathways is conserved in the two species. In support of this, also spi mutants of the moss Marchantia polymorpha exhibit short rhizoids reminiscent to the root hair phenotype of Aaspi and Atspi (Honkanen et al., 2016).
Interestingly, we also found that Arabis spi alleles show a different response to salt stress than pep1, suggesting that the dual function of SPI in morphogenetic processes and physiological processes is conserved between the two species. The molecular function of AaSPI in salt stress responses and mRNA regulation remains unclear. We found a physical interaction of AaSPI with AtDCP1 and AaDCP1. However, we cannot decide, whether the salt-dependent localization of cytoplasmic SPI protein to P-bodies as observed in Arabidopsis (Steffens et al., 2015) takes place in Arabis as co-expression of DCP1 and SPI is sufficient to trigger a recruitment of SPI to P-bodies. We also found transcriptional changes of several salt-stress response genes when comparing Aaspi alleles and pep1. Whether RNA stability is changed in Aaspi mutants was not tested in this study.
It is also not clear, whether AaSPI plays a role in salt stress responses. A salt stress phenotype is seen in Aaspi-1 and Aaspi-3 but not in Aaspi-2. One way to interpret this is to conclude that AaSPI is not involved in salt stress response because not all alleles show the phenotype. In this scenario, background mutations in Aaspi-1 and Aaspi-3 would cause the salt stress phenotype. Alternatively, it is possible that the different behavior of the three alleles is due to the type of mutations that they carry. While in Aaspi-1 and Aaspi-3 alleles premature stop codons lead to a loss of the PH-BEACH domain, the stop codon in Aaspi-2 leads to a truncation of only two of the four WD40 repeats in the C-terminus. Thus, it is possible that the protein region between the stop codons is relevant for the salt stress response. In this scenario it would be possible to assign protein domains that would be responsible for salt stress responses but not for cell morphogenesis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
LS, MJ, and MH designed this study and wrote the manuscript. LS performed the research and analyzed the data. MH directed the project. All authors contributed to the article and approved the submitted version.
This work was supported by the Deutsche Forschungsgemeinsc- haft, Priority Program Sonderforschungsprogramm 635 (MH) and an International Max Planck Research School fellowship (LS). Part of the study was conducted in the course of a doctoral thesis (LS) and has been previously published on the Cologne University Publication Server.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Divykriti Chopra and Maria Albani for sharing their expertise on Arabis alpina. We also thank Jessica Pietsch for critically reading the manuscript and Sabine Lohmer for excellent technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.596065/full#supplementary-material
Albani, M. C., Castaings, L., Wötzel, S., Mateos, J. L., Wunder, J., Wang, R., et al. (2012). PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 8:1003130. doi: 10.1371/journal.pgen.1003130
Aranda, P. S., LaJoie, D. M., and Jorcyk, C. L. (2012). Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33, 366–369. doi: 10.1002/elps.201100335
Basu, D., Le, J., El-Essal, S. E.-D., Huang, S., Zhang, C., Mallery, E. L., et al. (2005). DISTORTED3/SCAR2 is a putative arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17, 502–524. doi: 10.1105/tpc.104.027987
Beilstein, M. A., Nagalingum, N. S., Clements, M. D., Manchester, S. R., and Mathews, S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 18724–18728. doi: 10.1073/pnas.0909766107
Bögre, L., Magyar, Z., López-Juez, E., Busov, V., Brunner, A., Strauss, S., et al. (2008). New clues to organ size control in plants. Genome Biol. 9:226. doi: 10.1186/gb-2008-9-7-226
Burgess, A., Mornon, J.-P., de Saint-Basile, G., and Callebaut, I. (2009). A concanavalin A-like lectin domain in the CHS1/LYST protein, shared by members of the BEACH family. Bioinformatics 25, 1219–1222. doi: 10.1093/bioinformatics/btp151
Chopra, D., Mapar, M., Stephan, L., Albani, M. C., Deneer, A., Coupland, G., et al. (2019). Genetic and molecular analysis of trichome development in Arabis alpina. Proc. Natl. Acad. Sci. U.S.A. 116, 12078–12083. doi: 10.1073/pnas.1819440116
Coates, J. C. (2003). Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13, 463–471. doi: 10.1016/S0962-8924(03)00167-3
Cullinane, A. R., Schäffer, A. A., and Huizing, M. (2013). The BEACH is hot: a LYST of emerging roles for BEACH-domain containing proteins in human disease. Traffic 14, 749–766. doi: 10.1111/tra.12069
De Lozanne, A. (2003). The role of BEACH proteins in Dictyostelium. Traffic 4, 6–12. doi: 10.1034/j.1600-0854.2003.40102.x
Dewitte, W., and Murray, J. A. H. (2003). The Plant Cell Cycle. Annu. Rev. Plant Biol. 54, 235–264. doi: 10.1146/annurev.arplant.54.031902.134836
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an enhanced disease susceptibility1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. doi: 10.1002/yea.320110408
Guimil, S., and Dunand, C. (2007). Cell growth and differentiation in Arabidopsis epidermal cells. J. Exp. Bot. 58, 3829–3840. doi: 10.1093/jxb/erm253
Honkanen, S., Jones, V. A. S., Morieri, G., Champion, C., Hetherington, A. J., Kelly, S., et al. (2016). The mechanism forming the cell surface of tip-growing rooting cells is conserved among land plants. Curr. Biol. 26, 3238–3244. doi: 10.1016/j.cub.2016.09.062
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., Cuche, B. A., de Castro, E., et al. (2007). The 20 years of PROSITE. Nucl. Acids Res. 36, D245–D249. doi: 10.1093/nar/gkm977
Hülskamp, M., Misàa, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Jakoby, M. J., Weinl, C., Pusch, S., Kuijt, S. J. H., Merkle, T., Dissmeyer, N., et al. (2006). Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 141, 1293–1305. doi: 10.1104/pp.106.081406
Jogl, G., Shen, Y., Gebauer, D., Li, J., Wiegmann, K., Kashkar, H., et al. (2002). Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain. EMBO J. 21, 4785–4795. doi: 10.1093/emboj/cdf502
Koch, M. A., Kiefer, C., Ehrich, D., Vogel, J., Brochmann, C., and Mummenhoff, K. (2006). Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Mol. Ecol. 15, 825–839. doi: 10.1111/j.1365-294X.2005.02848.x
Koornneef, M., Hanhart, C. J., Martinet, E. L., and Van Der Veen, J. (1987). A marker line, that allows the detection of linkage on all Arabidopsis chromosomes. AGRIS 1987, 46-50.
Koornneef, M., van Eden, J., Hanhart, C. J., Stam, P., Braaksma, F. J., and Feenstra, W. J. (1983). Linkage map of Arabidopsis thaliana. J. Hered. 74, 265–272. doi: 10.1093/oxfordjournals.jhered.a109781
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucl. Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucl. Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Mathur, J., and Hülskamp, M. (2002). Microtubules and microfilaments in cell morphogenesis in higher plants. Curr. Biol. 12, R669–R676. doi: 10.1016/S0960-9822(02)01164-8
Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B. P., and Hülskamp, M. (2003). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. doi: 10.1242/dev.00549
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Saedler, R., Jakoby, M., Marin, B., Galiana-Jaime, E., and Hülskamp, M. (2009). The cell morphogenesis gene SPIRRIG in Arabidopsis encodes a WD/BEACH domain protein. Plant J. 59, 612–621. doi: 10.1111/j.1365-313X.2009.03900.x
Schwab, B., Mathur, J., Saedler, R., Schwarz, H., Frey, B., Scheidegger, C., et al. (2003). Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: actin controls the spatial organization of microtubules. Mol. Genet. Genomics 269, 350–360. doi: 10.1007/s00438-003-0843-1
Shahriari, M., Keshavaiah, C., Scheuring, D., Sabovljevic, A., Pimpl, P., Häusler, R. E., et al. (2010). The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64, 71-85. doi: 10.1111/j.1365-313X.2010.04310.x
Steffens, A., Bräutigam, A., Jakoby, M., Hülskamp, M., and Meyer, S. (2015). The beach domain protein SPIRRIG is essential for Arabidopsis salt stress tolerance and functions as a regulator of transcript stabilization and localization. PLoS Biol. 13:e1002188. doi: 10.1371/journal.pbio.1002188
Steffens, A., Jaegle, B., Tresch, A., Hülskamp, M., and Jakoby, M. (2014). Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol. 164, 1879–1892. doi: 10.1104/pp.113.233031
Steffens, A., Jakoby, M., and Hülskamp, M. (2017). Physical, Functional and Genetic Interactions between the BEACH Domain Protein SPIRRIG and LIP5 and SKD1 and Its Role in Endosomal Trafficking to the Vacuole in Arabidopsis. Front. Plant Sci. 8:1969. doi: 10.3389/fpls.2017.01969
Stephan, L., Tilmes, V., and Hülskamp, M. (2019). Selection and validation of reference genes for quantitative Real-Time PCR in Arabis alpina. PLoS One 14:e0211172. doi: 10.1371/journal.pone.0211172
Tian, J., Han, L., Feng, Z., Wang, G., Liu, W., Ma, Y., et al. (2015). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determination by a plant-unique kinesin. Elife 4, e09351. doi: 10.7554/eLife.09351
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 3, research0034.1–research0034.11. doi: 10.1186/gb-2002-3-7-research0034
Wang, R., Farrona, S., Vincent, C., Joecker, A., Schoof, H., Turck, F., et al. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427. doi: 10.1038/nature07988
Yang, Y., Yuan, J. S., Ross, J., Noel, J. P., Pichersky, E., and Chen, F. (2006). An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 448, 123–132. doi: 10.1016/J.ABB.2005.08.006
Zhao, H., Wang, X., Zhu, D., Cui, S., Li, X., Cao, Y., et al. (2012). A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 287, 14109–14121. doi: 10.1074/jbc.M111.280735
Keywords: morphogenesis, membrane trafficking, Arabis alpina, BEACH domain protein, SPIRRIG, salt response
Citation: Stephan L, Jakoby M and Hülskamp M (2021) Evolutionary Comparison of the Developmental/Physiological Phenotype and the Molecular Behavior of SPIRRIG Between Arabidopsis thaliana and Arabis alpina. Front. Plant Sci. 11:596065. doi: 10.3389/fpls.2020.596065
Received: 18 August 2020; Accepted: 03 December 2020;
Published: 07 January 2021.
Edited by:
Jean Molinier, UPR 2357 Institut de Biologie Moléculaire des Plantes (IBMP), FranceReviewed by:
Thomas Potuschak, UPR 2357 Institut de Biologie Moléculaire des Plantes (IBMP), FranceCopyright © 2021 Stephan, Jakoby and Hülskamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hülskamp, bWFydGluLmh1ZWxza2FtcEB1bmkta29lbG4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.