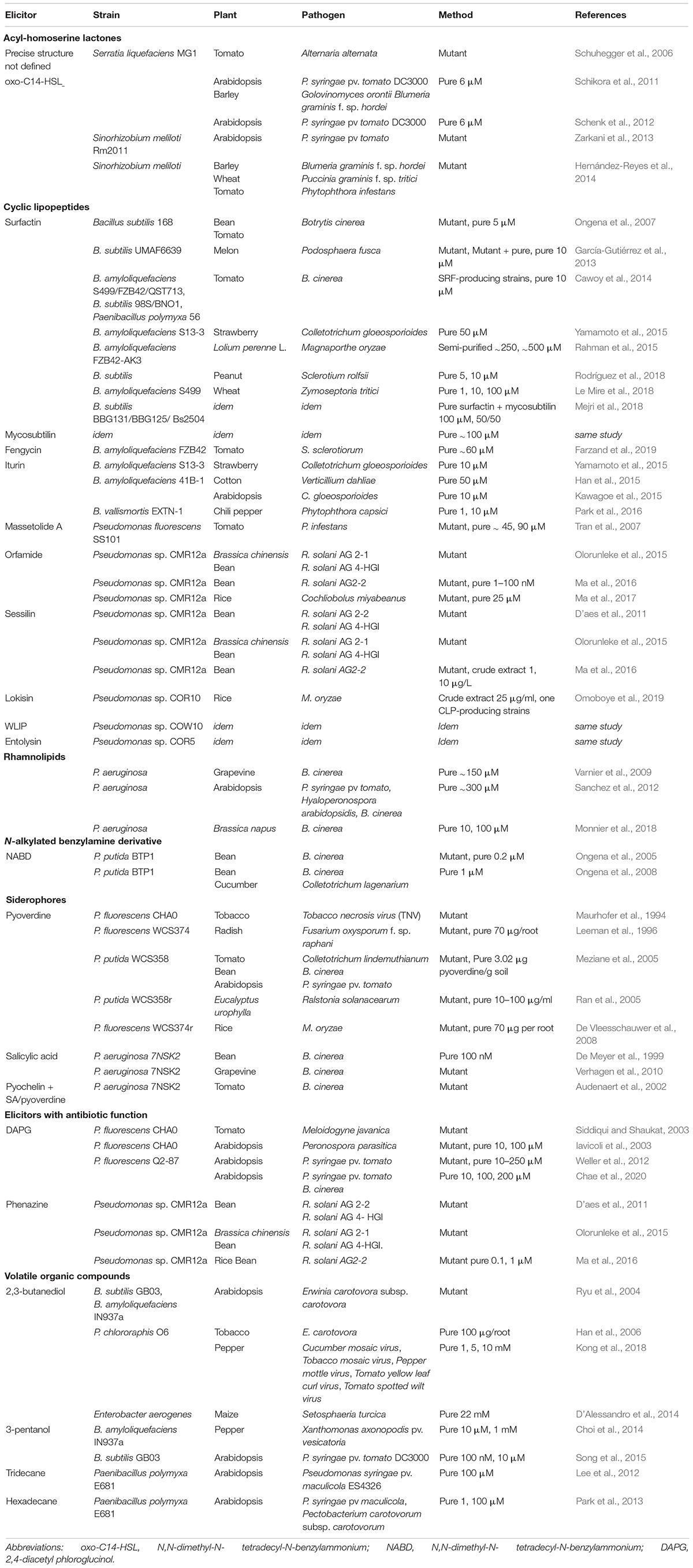
- Microbial Processes and Interactions Laboratory, Terra Teaching and Research Center, Gembloux Agro-Bio Tech, University of Liège, Gembloux, Belgium
The molecular basis of plant immunity triggered by microbial pathogens is being well-characterized as a complex sequential process leading to the activation of defense responses at the infection site, but which may also be systemically expressed in all organs, a phenomenon also known as systemic acquired resistance (SAR). Some plant-associated and beneficial bacteria are also able to stimulate their host to mount defenses against pathogen ingress via the phenotypically similar, induced systemic resistance phenomenon. Induced systemic resistance resembles SAR considering its mechanistic principle as it successively involves recognition at the plant cell surface, stimulation of early cellular immune-related events, systemic signaling via a fine-tuned hormonal cross-talk and activation of defense mechanisms. It thus represents an indirect but efficient mechanism by which beneficial bacteria with biocontrol potential improve the capacity of plants to restrict pathogen invasion. However, according to our current vision, induced systemic resistance is specific considering some molecular aspects underpinning these different steps. Here we overview the chemical diversity of compounds that have been identified as induced systemic resistance elicitors and thereby illustrating the diversity of plants species that are responsive as well as the range of pathogens that can be controlled via this phenomenon. We also point out the need for further investigations allowing better understanding how these elicitors are sensed by the host and the diversity and nature of the stimulated defense mechanisms.
Introduction
Some bacteria isolated from the root microbiome have been selected for their remarkable beneficial effect provided to their host plant and are referred to as plant growth-promoting rhizobacteria (PGPR) (Backer et al., 2018; Singh et al., 2019). These PGPR favor plant growth notably by facilitating nutrient availability and modulating the host’s hormonal balance but also display plant protective activity toward pathogen ingress. This biocontrol potential relies on several traits including the ability to efficiently compete for space and nutrients with pathogens, a strong direct antagonistic activity based on secretion of low-size antimicrobials or hydrolytic enzymes and the capacity to stimulate induced systemic resistance (ISR) (Pieterse et al., 2014; Köhl et al., 2019). ISR is a systemically expressed resistance state that renders the host less susceptible to subsequent infection, and it is of great interest from an agronomical perspective because effective against a broad spectrum of microbial pathogens, nematodes, and insects (Pieterse et al., 2014; Grady et al., 2016; Rashid and Chung, 2017; Mhatre et al., 2018). Phenotypically, ISR resembles the systemic acquired resistance (SAR) mounted upon pathogen perception, which culminates from a complex immune-related process. At the front line, molecular features of invading microbes are detected with high specificity and sensitivity in the apoplast by a range of plasma membrane-anchored immune sensors referred to as pattern recognition receptors (PRR). These PRRs bind precise structural motifs (epitopes) of larger (macro)molecules retaining important functions for microbial fitness and thus widely conserved across species and termed microbe-associated molecular patterns (MAMPs). Some of the best described MAMPs from bacterial pathogens are flg22, a 22 amino acids portion of the flagellin protein, elf18 which is the epitope of the elongation factor EF-Tu, surface-exposed lipopolysaccharides (LipidA), and peptidoglycan (sugar backbone) (Schellenberger et al., 2019). The binding of MAMP by PRR initiates within minutes early immune-related events in responsive cells such as ion fluxes, oxidative burst, and phosphorylation cascade. Furthermore, defense mechanisms sensu stricto such as cell wall reinforcement and production of antimicrobial enzymes and secondary metabolites, referred to as phytoalexins, are stimulated (Piasecka et al., 2015). This PRR-mediated immune response [pattern triggered immunity (PTI)] is robust as it can detect MAMPs at nanomolar concentrations leading to fast and strong defensive responses (high transcriptional activation) but associated with growth-defense tradeoffs that reduce plant fitness (Huot et al., 2014). However, adapted pathogens use protein effectors injected into host cells to dampen PTI and subvert this first line of immune reaction. In turn, plants have evolved intracellular sensors called R (resistance) proteins, which interfere with these effectors leading to the effector-triggered type of immunity (ETI) which may, like PTI, lead to SAR (Kanyuka and Rudd, 2019; Schellenberger et al., 2019).
Due to the conserved nature of MAMPs, the plant’s receptor equipment would enable detection not only of pathogenic bacteria, but also of beneficials such as rhizobial symbiots and root-associated epiphytic PGPR (Zipfel and Oldroyd, 2017). These beneficial microbes have thus to evade (by hiding or changing their MAMPs) or suppress (by secreting effectors that interfere with immune responses and signaling) PTI in order to establish a mutualistic relationship with their host (Stringlis et al., 2018; Yu et al., 2019). This has been recently illustrated with the Pseudomonas simiae strain WCS417 first detected as a pathogen via its flg22 epitope, but which then strongly attenuates the host immune response presumably via T3SS-mediated delivery of effectors (Berendsen et al., 2015; Stringlis et al., 2019). Subversion of host immune reaction allows PGPR to sustainably colonize the rhizoplane and establish threshold populations necessary for providing beneficial functions including ISR triggering via the secretion of other elicitors not related to MAMPs.
Here we provide an updated overview of those PGPR determinants responsible for ISR elicitation that are in most cases small-size compounds secreted by the colonizing bacteria even if some proteins isolated from Brevibacillus laterosporus, B. amyloliquefaciens, and Saccharothrix yanglingensis have also been recently proposed as bacterial triggers (Table 1) (Wang et al., 2015, 2016; Zhang et al., 2018). We highlight their chemical diversity and structure- and dose-dependent activity but only refer to compounds that have been tested in pure (proven) form at biologically relevant concentrations and/or via specifically suppressed mutants and for which the ability to stimulate systemic resistance was clearly demonstrated to be independent from direct antimicrobial effect.
PGPR Metabolites Identified as Elicitors of Plant Systemic Resistance
Acyl-Homoserine Lactones
Many Gram-negative bacteria produce N-acyl-homoserine lactones (AHLs) as quorum-sensing molecules involved in cell-to-cell communication in order to monitor their behavior according to population density. Some AHLs are not only the mean of communication between bacterial cells, but also may be used as signal in inter-kingdom interaction and act as plant growth promoting compounds and/or as immunity elicitors (reviewed in Schikora et al., 2016). A first study showed, by using an AHL-suppressed mutant of Serratia liquefaciens MG1, that induced resistance in tomato toward Alternaria alternata by this strain is AHL-dependent (Schuhegger et al., 2006). Several works performed with purified molecules further revealed that AHL bioactivity is structure-dependent. AHLs with short length acyl chains have been mostly demonstrated to promote plant growth, whereas AHLs with longer fatty acid (C12, C14) are better described as elicitors of resistance (Schenk et al., 2012; Zarkani et al., 2013; Schikora et al., 2016). The long-chain N-3-oxo-tetradecanoyl-L-homoserine lactone (oxo-C14-HSL; Figure 1-1) has been amply demonstrated for immunity elicitation in many pathosystems. Upon treatment at 6 μM, oxo-C14-HSL successfully induced systemic resistance against biotrophs (Blumeria graminis f. sp. hordei, Puccinia hordei, Golovinomyces orontii) and hemibiotroph (P. syringae DC3000) in barley and Arabidopsis, but not against the necrotrophs Botrytis cinerea and Plectosphaerella cucumerina BMM (Schikora et al., 2011; Schenk et al., 2012; Wehner et al., 2019). ISR-eliciting activity of oxo-C14-HSL was also observed in wheat and tomato against Puccinia graminis f. sp. tritici and Phytophthora infestans, respectively (Hernández-Reyes et al., 2014). Collectively, these data indicate that the plant defensive response triggered by the same oxo-C14-HSL molecule may not be efficient in enhancing resistance against phytopathogens with necrotrophic lifestyle.
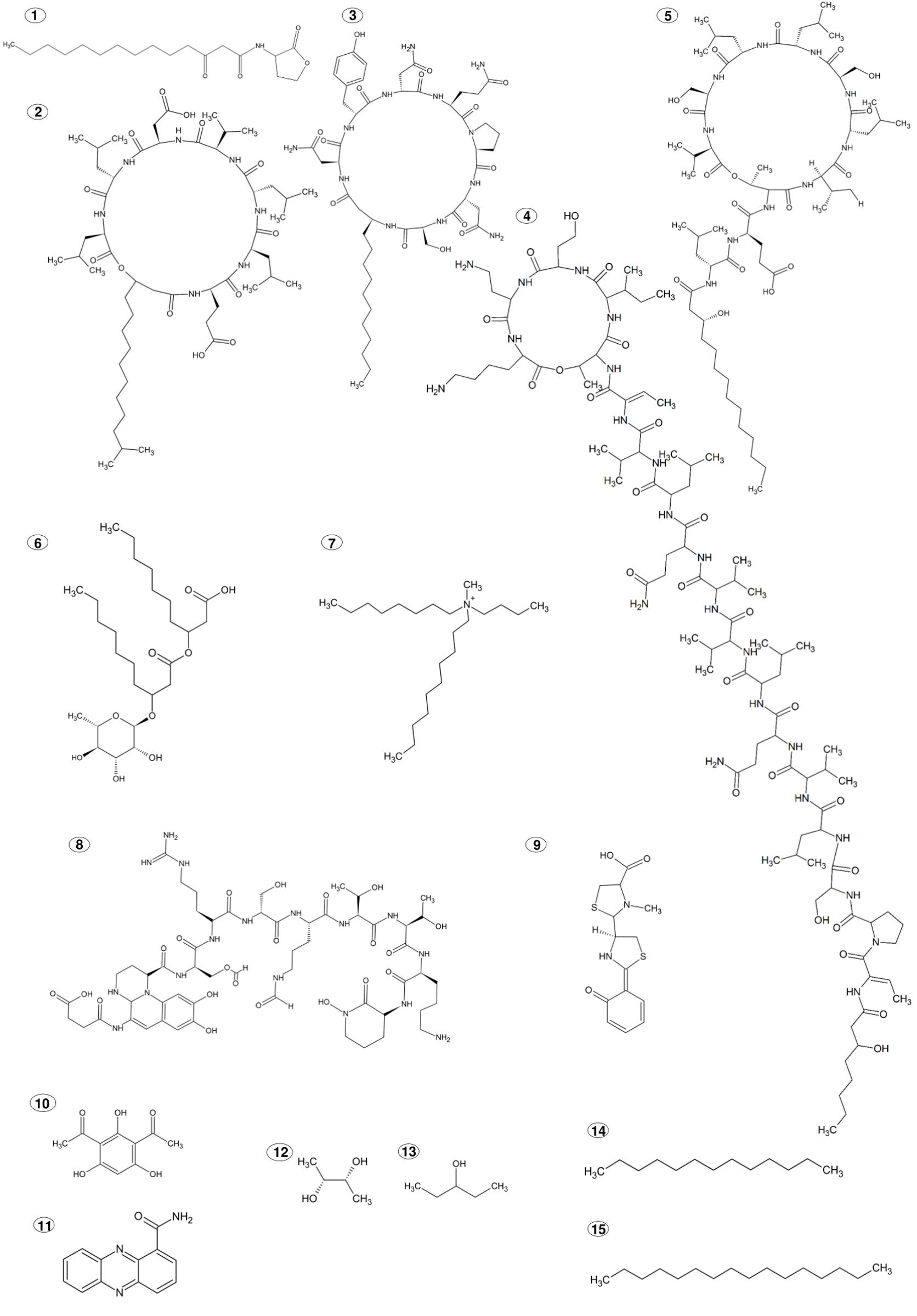
Figure 1. Chemical structures of PGPR produced ISR elicitors. (1) N-3-oxo-tetradecanoyl-L-homoserine lactone; (2) Surfactin; (3) Iturin; (4) Sessilin; (5) Orfamide; (6) Rhamnolipid, L-rhamnosyl-3-hydroxydecanoyl-3-hydroxydecanoic acid; (7) N,N-dimethyl-N- tetradecyl-N-benzylammonium; (8) Pyoverdine; (9) Pyochelin; (10) 2,4-diacetyl phloroglucinol; (11) phenazine-1-carboxamide; (12) 2,3-butanediol; (13) 3-pentanol; (14) Tridecane; (15) Hexadecane.
Cyclic Lipopeptides
Rhizobacterial cyclic lipopeptides (CLPs) are multifunctional secondary metabolites involved in developmental processes such as motility and biofilm formation and in biocontrol primarily based on their antimicrobial activity (Ongena and Jacques, 2008; Geudens and Martins, 2018). However, some CLPs secreted by beneficial bacilli and pseudomonads have emerged as an important category of plant immunity elicitors as well. Surfactin is a heptapeptide occurring as a mix of naturally co-produced homologs varying in the length of the fatty acid chain (Figure 1-2). This CLP is among the Bacillus compounds best described as trigger of systemic resistance. When applied as pure compound on roots, in micromolar amounts (5–10 μM), surfactin demonstrated to induce ISR in bean, tomato, tobacco, against B. cinerea, in melon against Podosphaera fusca, and peanut, against Sclerotium rolfsii (Ongena et al., 2007; García-Gutiérrez et al., 2013; Cawoy et al., 2014; Rodríguez et al., 2018). The structure of this CLP may strongly impact its elicitor activity as observed on tobacco cells. Linear and/or methylated derivatives are much less efficient and only homologs with long C14 and C15 acyl chains are active at inducing early immune-related events, unlike C12 and C13 (Jourdan et al., 2009; Henry et al., 2011). Despite its high bioactivity on dicot plant roots, surfactin shows low competence in mounting immunity when applied on dicots leaves or monocots roots (Rahman et al., 2015; Yamamoto et al., 2015; Mejri et al., 2018). Induction of systemic resistance and/or plant defenses by other CLPs produced by biocontrol bacilli has also been occasionally reported. In tomato and grapevine, fengycin triggered ISR against B. cinerea and Plasmopara viticola (Farzand et al., 2019; Li et al., 2019), while CLPs from the iturin group (Figure 1-3) were shown to have a similar role in strawberry (Yamamoto et al., 2015), cotton (Han et al., 2015), grapevine (Farace et al., 2015) and Arabidopsis (Wu et al., 2018). Additionally illustrating the dependence of CLP-mediated ISR on the plant organ, iturin showed bioactivity on roots at concentration of 50 μM, whereas it was active already at 1–10 μM when applied on leaves (Han et al., 2015; Kawagoe et al., 2015; Yamamoto et al., 2015; Park et al., 2016). The activation of defense genes in Arabidopsis by iturin foliar treatment was also dependent on the molecule’s structure, i.e., cyclization and/or length of the b-hydroxy fatty acid chain (Kawagoe et al., 2015).
Several studies have also reported the involvement of Pseudomonas sp. CLPs in plant resistance stimulation on various pathosystems. Massetolide A was the first reported for its resistance-inducing activity in tomato against P. infestans (Tran et al., 2007). Recent works conducted with Pseudomonas sp. strain CMR12a revealed that two other types of CLPs, sessilin and orfamide (Figures 1-4,5, respectively) are involved in ISR-triggering albeit differently according to the pathosystem tested. Works combining the use of biosynthesis mutants and CLP extracts showed that both sessilin and orfamide are determinants of ISR in bean against Rhizoctonia solani, but that a balanced production is needed for optimal effect. These two compounds are indeed active at precise range of concentrations from 0.001 to 0.1 μM for pure orfamide, and 0.001 and 0.01 mg/L for sessilin as crude extract (Olorunleke et al., 2015; Ma et al., 2016). In monocots, such as rice, Pseudomonas CLP structure is crucial in determining elicitor effect on a given pathosystem. For example, orfamide applied at 25 μM was identified as elicitor inducing resistance to Cochliobolus miyabeanus, but it is not active against Magnaporthe oryzae (Ma et al., 2017). Nevertheless, CLPs such as WLIP, lokisin, and entolysin were recently described to successfully induce resistance toward this last pathogen, confirming elicitor specificity for certain pathosystems (Omoboye et al., 2019).
Rhamnolipids
Rhamnolipids (RLs) are amphiphilic glycolipids produced by various species including pathogenic isolates, but also some plant beneficial Pseudomonas and Burkholderia species (Perneel et al., 2008; Abdel-Mawgoud et al., 2010). They are essential for bacterial surface motility and biofilm development (Vatsa et al., 2010; Chrzanowski et al., 2012). Mono- and di-RLs (Figure 1-6) tested as pure compounds have been shown to elicit plant defense responses and to induce resistance against various pathogens in grapevine, Arabidopsis, and Brassica napus using a wide range of concentrations from approximately 10 up to 300 μM (Varnier et al., 2009; Sanchez et al., 2012; Monnier et al., 2018, 2020).
N-Alkylated Benzylamine Derivative
Although its function for bacterial life is not clear, an N-alkylated benzylamine derivative (NABD; Figure 1-7) produced by P. putida BTP1, was identified as elicitor (Ongena et al., 2005). Treatment of bean and cucumber roots with pure NABD at low micromolar concentration elicited similar protective effect compared to living cells. On the other hand, in tomato, the pure elicitor induced a lower protective effect than observed with the producing strain, suggesting the synthesis of an additional ISR determinant in that case (Ongena et al., 2008).
Siderophores
To ensure their growth in iron-limited environments, microorganisms have evolved powerful Fe3+-acquisition systems based on the secretion of high-affinity iron-chelating molecules termed siderophores. PGPR siderophores are also known to antagonize pathogen populations by decreasing iron amounts in soil, resulting in competition for this essential element (Kramer et al., 2020). However, some siderophores also act as plant immunity elicitors and pyoverdines (also referred to as pseudobactines; Figure 1-8) produced by various fluorescent pseudomonads were, in the 90’s, among the first PGPR metabolites proposed as ISR elicitors. Their key role in systemic resistance induction was notably shown for P. fluorescens CHA0 on tobacco infected by Tobacco necrosis virus, and in the protection provided by P. fluorescens WCS374 to radish against Fusarium wilt (Maurhofer et al., 1994; Leeman et al., 1996). These chromopeptides were further described as inducers of resistance in various plant species such as bean, tomato, Arabidopsis, tobacco, eucalyptus and rice against a range of microbial pathogens (Meziane et al., 2005; Ran et al., 2005; De Vleesschauwer et al., 2008). In addition to pyoverdine, P. aeruginosa 7NSK2 also forms the chelating agent pyochelin (Figure 1-9) and its precursor salicylic acid (SA). When tested on pathosystem grapevine/B. cinerea, SA produced by P. aeruginosa 7NSK2 was crucial for mounting the plant immunity (De Meyer et al., 1999; Verhagen et al., 2010). However, for ISR stimulated in tomato by the same strain, an important role for SA could not be excluded, but probably combined with the action of other metabolites including pyochelin which may somehow also retain some eliciting activity (Audenaert et al., 2002).
Elicitors With Antibiotic Function
To ensure fitness in the competitive rhizosphere niche, PGPR produce a wide range of secondary metabolites best identified for their antimicrobial function such as non-ribosomal peptides, polyketides, bacteriocins, terpenes, phenazines, quinolones, or rhamnolipids (Raaijmakers and Mazzola, 2012; Zhao and Kuipers, 2016; Tracanna et al., 2017). Interestingly, some of these antibiotics were also shown to act as signal for ISR stimulation at similar concentrations, making them promising tools for biocontrol with dual action on pathogen populations (Kenawy et al., 2019). One such antibiotic 2,4-diacetyl phloroglucinol (Figure 1-10), formed by P. fluorescens, is triggering resistance in Arabidopsis against various pathogens, such as Peronospora parasitica, P. syringae pv. tomato, and B. cinerea, upon application at relatively high 10–100 μM concentrations (Iavicoli et al., 2003; Weller et al., 2012; Chae et al., 2020). 2,4-diacetyl phloroglucinol can also induce resistance against nematodes as shown with P. protegens CHA0 for the reduction of infection caused by Meloidogyne javanica on tomato roots (Siddiqui and Shaukat, 2003). Besides, other Pseudomonas antibiotics of the phenazine-type were also reported to induce resistance. Notably phenazine-1-carboxamide (Figure 1-11) in rice toward M. oryzae at 0.1–1 μM, in bean toward R. solani, and pyocyanin produced by P. aeruginosa 7NSK2 in the pathosystem tomato/B. cinerea (Audenaert et al., 2002; D’aes et al., 2011; Ma et al., 2016).
Volatile Organic Compounds
Most of the well-characterized PGPR elicitors are soluble compounds, but some volatile organic compounds (VOCs) were as well-reported to induce systemic resistance in the host plant, showing that these metabolites can also act as infochemicals involved in inter-kingdom communication (Kai et al., 2016). The most studied VOC immunity elicitor is 2,3-butanediol (2,3-BD; Figure 1-12) produced from glucose in the central metabolism (Yang et al., 2013). Its bioactivity was first assessed on Arabidopsis by application on roots, where B. subtilis GB03 induced resistance against Erwinia carotovora subsp. carotovora, while mutants deprived in 2,3-BD or its precursor acetoin production were inactive (Ryu et al., 2004). Bioactivity, when treated on roots in relatively high mM concentrations, was demonstrated as well in the pathosystems maize/Setosphaeria turcica, tobacco/Erwinia carotovora, and in pepper against multiple viruses (Table 1), where the immunity eliciting ability was structure-dependent. Namely, among the three forms, 2R,3R-BD (R), 2S,3S-BD (S) and 2R,3S-BD (meso), the S form is the least active (Han et al., 2006; D’Alessandro et al., 2014; Kong et al., 2018). However, on pepper against Xanthomonas axonopodis pv. vesicatoria, another VOC 3-pentanol (10 μM, 1 mM; Figure 1-13) showed higher activity than 2,3-BD (Choi et al., 2014). Beside 3-pentanol, long-chain VOCs tridecane and hexadecane (Figures 1-14,15, respectively) showed bioactivity as well at μM (100) concentration (Lee et al., 2012; Park et al., 2013).
Molecular Basis of Plant Immunization: PGPR vs. MAMP Elicitors
The potential of PGPR to induce plant immunity mainly relies on the secretion of a range of structurally diverse low-molecular weight metabolites. However, the molecular mechanisms driving recognition of these elicitors at the plant cell surface are poorly understood. By contrast with MAMPs from pathogens, there is no indication so far for specific PRRs involved in the perception of PGPR elicitors. They activate immune responses only at relatively high μM concentrations compared to MAMPs, suggesting that they are not sensed via high-affinity receptors. CLPs are known to promptly insert into biological membranes causing defects, pore formation and cell lysis in a range of (micro)organisms (Balleza et al., 2019). Furthermore, immune response triggered by the Bacillus CLP surfactin was fully conserved in protease-treated tobacco cells, and successive applications do not lead to some refractory state due to the saturation of high-affinity binding sites (as receptors). Based on these and the fact that surfactin readily interact with sphingolipid-enriched microdomains in the plasma membrane, it was suggested that this CLP is perceived by plant cells via a lipid-mediated process. This is supported by the strong structure-dependent activity showing that only long fatty acid chain homologs are active at triggering early immune-related events in tobacco cells because they should display stronger interaction with lipid bilayers (Jourdan et al., 2009; Henry et al., 2011). Rhamnolipids also readily fit into plant lipid-based bilayer models. Therefore, it was proposed that by inserting into plasma membranes, these compounds provoke subtle changes in lipid dynamics that could be related to plant defense induction (Davis et al., 2010; Monnier et al., 2018; Schellenberger et al., 2019). As other alkyl-chain containing elicitors, AHLs could also be perceived by plant cells via a receptor-independent but lipid-mediated process, by analogy with data recently obtained on mammalian cells (Schikora et al., 2016; Song et al., 2018). It may explain why a given compound does not act at the same level on different targets, considering that the lipid composition in the plasma membrane differs according to the plant species/organs.
The way PGPR elicitors are perceived at the plasma membrane level is not clear, but there is still a clear convergence between PGPR-triggered immunity and pathogen-induced PTI in the early steps of downstream signaling. The best studied PGPR elicitors induce similar early immune events as observed upon pathogen perception, such as oxidative burst, ion fluxes, and phosphorylation cascade (Jourdan et al., 2009; Schikora et al., 2011; Cho et al., 2013; Cawoy et al., 2014; Farace et al., 2015; Rahman et al., 2015). Detection of MAMPs from pathogens leads to a fast and strong defensive response, but also to a costly reduction of the plant growth and fitness (Huot et al., 2014). On the other hand, PGPR and/or their elicitors usually prime the host to stimulate defense mechanisms but only after pathogen challenge as observed upon treatment with surfactin, AHLs, or with bacteria producing NADB or pyoverdines (De Vleesschauwer et al., 2008; Mariutto et al., 2011; Cawoy et al., 2014; Debois et al., 2015; Schikora et al., 2016). Again by contrast with PTI, PGPR elicitor-priming is not associated with major transcriptional reprogramming until the pathogen is detected and does not involve fitness costs but still prepares the plant for mounting a robust defense (Martinez-Medina et al., 2016; Mauch-Mani et al., 2017). Globally, the defense mechanisms restricting pathogen ingress that are stimulated by beneficial bacteria or their elicitors resemble those observed upon pathogen MAMP perception. It notably means up-regulation of genes involved in the synthesis of antimicrobial enzymes/proteins or metabolites and in hormone signaling. Higher expression of genes such are pathogenesis- related, lipoxygenase, plant defensin factor, and phenylalanine ammonia lyase, is often reported in elicitor pretreated plants after pathogen infection (Ongena et al., 2007; García-Gutiérrez et al., 2013; Zarkani et al., 2013; Song et al., 2015; Park et al., 2016; Yi et al., 2016; Kong et al., 2018; Song et al., 2019). Besides, PGPR elicitors also trigger stomatal closure and enhance cell wall reinforcement which serves as a structural barrier to pathogen invasion via callose deposition or accumulation of phenolic compounds and lignin (De Vleesschauwer et al., 2006; García-Gutiérrez et al., 2013; Schenk et al., 2014; Rodríguez et al., 2018).
Discussion
From an agronomic perspective, ISR triggered by PGPR is interesting since the phenomenon is considered to provide long-lasting and broad-spectrum protection without causing growth cost and is not a priori conducive for development of resistance in pathogens (Köhl et al., 2019). However, the success of PGPR as stimulators of plant defenses is so far rather limited due to a range of factors. These include our global lack of knowledge about the nature and mode of action of their elicitors. Indeed, ISR determinants were identified mainly from a limited number of species (Pseudomonas spp. and Bacillus spp.), leaving elicitors from many other PGPR to be discovered. Moreover, in the last decades, tremendous advances have been done on understanding the basics of MAMP perception during PTI, but the mechanistic of PGPR elicitor recognition at the plant plasma membrane level and the molecular events underlying PGPR-induced priming remain largely obscure. As the lipid phase is suspected to act as docking platform for some of these elicitors, experimental biophysics and in silico dynamic modeling using appropriate biomimetic vesicles represent interesting approaches to get further insights into the physico-chemical basis of the interactions (Deleu et al., 2014; Balleza et al., 2019; Nishimura and Matsumori, 2020). It would help to explain why some compounds are only efficient on specific plants/tissues according to the nature and proportions of lipids in their domain-structured plasma membranes (Gronnier et al., 2018). Also, the variety of pathosystems tested so far is still limited and additional research on agriculturally important crops and pathogens is needed in order to better appreciate their potential at a larger market scale.
Furthermore, PGPR elicitors in most instances are active at micromolar doses but only a few studies suggest that quantities produced by bacteria in planta are sufficient to locally reach such threshold around the roots (D’Alessandro et al., 2014; Debois et al., 2015). Determining the amounts of elicitors produced by PGPR under natural conditions is still important, but not an easy task. It would require optimal extraction from rhizosphere samples and the most-advanced MS-based metabolomics allowing high sensitivity for their detection and quantification. Also, environmental factors may affect ISR efficiency (Williams et al., 2018), but their impact on the production of elicitors by PGPR remains poorly appreciated. Low temperature, acidic pH or poor oxygen availability are among the rhizosphere specific abiotic parameters that affect bacterial physiology and which may also modulate the production of secondary metabolites including elicitors as reported for Bacillus lipopeptides (Fahim et al., 2012; Pertot et al., 2013; Zhao and Kuipers, 2016). Biotic factors such as interactions with other microbial species of the soil microbiome or chemical cross-talk with the host plant, may also influence elicitor production under natural conditions (Debois et al., 2015; Wu et al., 2015; Venturi and Keel, 2016; Andrić et al., 2020). A better evaluation of the impact of all these factors deserve further investigation and is necessary to better anticipate inconsistencies in PGPR efficacy observed upon application under field conditions.
Integrating all this missing information should also lead to more rational determine the range of pathosystems, and environmental conditions in which PGPR-based bioproducts would be the most efficient. An alternative is to make from these bacterial immunogenic compounds microbial-derived products for the phytosanitary market provided that they can be produced and formulated in cost-effective industrial processes. This is feasible for some metabolites with high production rate in bioreactors, such as Bacillus lipopeptides (Zanotto et al., 2019; Brück et al., 2020), but the dose and structure dependent activity should guide a rational selection of the strain, optimization of culture conditions and extensive testing in field experiments in comparison with chemical products.
Author Contributions
Both authors conceived the idea, designed the outlines of the review, and wrote the manuscript.
Funding
Research in the laboratory is supported by the EU Interreg FWVL V portfolio SmartBiocontrol and by the EOS project ID30650620 from the FWO/F.R.S.-FNRS (Fonds National de la Recherche Scientifique). MO is senior research associate at the F.R.S.-F.N.R.S.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge S. Andrić, F. Boubsi, A. Anckaert, G. Gilliard, and T. Mayer for critically reading the manuscript and their valuable suggestions and discussions.
References
Abdel-Mawgoud, A. M., Lepine, F., and Deziel, E. (2010). Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86, 1323–1336. doi: 10.1007/s00253-010-2498-2
Andrić, S., Meyer, T., and Ongena, M. (2020). Bacillus responses to plant-associated fungal and bacterial communities. Front. Microbiol. 11:1350. doi: 10.3389/fmicb.2020.01350
Audenaert, K., Pattery, T., Cornelis, P., and Höfte, M. (2002). Induction of Systemic Resistance to Botrytis cinerea in Tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Interact. 15, 1147–1156. doi: 10.1094/MPMI.2002.15.11.1147
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Balleza, D., Alessandrini, A., and Beltrán García, M. J. (2019). Role of lipid composition, physicochemical interactions, and membrane mechanics in the molecular actions of microbial cyclic lipopeptides. J. Memb. Biol. 252, 131–157. doi: 10.1007/s00232-019-00067-4
Berendsen, R. L., van Verk, M. C., Stringlis, I. A., Zamioudis, C., Tommassen, J., Pieterse, C. M. J., et al. (2015). Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358. WCS374 and WCS417. BMC Genom. 16:539. doi: 10.1186/s12864-015-1632-z
Brück, H. L., Coutte, F., Dhulster, P., Gofflot, S., Jacques, P., and Delvigne, F. (2020). Growth dynamics of bacterial populations in a two-compartment biofilm bioreactor designed for continuous surfactin Biosynthesis. Microorganisms 8:679. doi: 10.3390/microorganisms8050679
Cawoy, H., Mariutto, M., Henry, G., Fisher, C., Vasilyeva, N., Thonart, P., et al. (2014). Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant Microbe Interact. 27, 87–100. doi: 10.1094/MPMI-09-13-0262-R
Chae, D. H., Kim, D. R., Cheong, M. S., Lee, Y. B., and Kwak, Y. S. (2020). Investigating the induced systemic resistance mechanism of 2,4-diacetylphloroglucinol (Dapg) using dapg hydrolase-transgenic arabidopsis. Plant Pathol. J. 36, 255–266. doi: 10.5423/PPJ.OA.02.2020.0031
Cho, S. M., Kim, Y. H., Anderson, A. J., and Kim, Y. C. (2013). Nitric oxide and hydrogen peroxide production are involved in systemic drought tolerance induced by 2R,3R-butanediol in Arabidopsis thaliana. Plant Pathol. J. 29, 427–434. doi: 10.5423/PPJ.OA.07.2013.0069
Choi, H. K., Song, G. C., Yi, H. S., and Ryu, C. M. (2014). Field evaluation of the bacterial volatile derivative 3-pentanol in priming for induced resistance in pepper. J. Chem. Ecol. 40, 882–892. doi: 10.1007/s10886-014-0488-z
Chrzanowski, Ł, Ławniczak, Ł, and Czaczyk, K. (2012). Why do microorganisms produce rhamnolipids? World J. Microbiol. Biotechnol. 28, 401–419. doi: 10.1007/s11274-011-0854-8
D’aes, J., Hua, G. K. H., De Maeyer, K., Pannecoucque, J., Forrez, I., Ongena, M., et al. (2011). Biological control of rhizoctonia root rot on bean by phenazine and cyclic lipopeptide-producing Pseudomonas CMR12a. Phytopathology 101, 996–1004. doi: 10.1094/PHYTO-11-10-0315
D’Alessandro, M., Erb, M., Ton, J., Brandenburg, A., Karlen, D., Zopfi, J., et al. (2014). Volatiles produced by soil-borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant Cell and Environment 37, 813–826. doi: 10.1111/pce.12220
Davis, B. M., Jensen, R., Williams, P., and O’Shea, P. (2010). The interaction of N-acylhomoserine lactone quorum sensing signaling molecules with biological membranes: implications for inter-kingdom signaling. PLoS One 5:e0013522. doi: 10.1371/journal.pone.0013522
De Meyer, G., Capieau, K., Audenaert, K., Buchala, A., Métraux, J. P., and Höfte, M. (1999). Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe Interact. 12, 450–458. doi: 10.1094/MPMI.1999.12.5.450
De Vleesschauwer, D., Cornelis, P., and Höfte, M. (2006). Redox-active pyocyanin secreted by Pseudomonas aeruginosa 7NSK2 triggers systemic resistance to Magnaporthe grisea but enhances Rhizoctonia solani susceptibility in rice. Mol. Plant Microbe Interact. 19, 1406–1419. doi: 10.1094/MPMI-19-1406
De Vleesschauwer, D., Djavaheri, M., Bakker, P. A. H. M., and Höfte, M. (2008). Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol. 148, 1996–2012. doi: 10.1104/pp.108.127878
Debois, D., Fernandez, O., Franzil, L., Jourdan, E., de Brogniez, A., Willems, L., et al. (2015). Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin. Environ. Microbiol. Rep. 7, 570–582. doi: 10.1111/1758-2229.12286
Deleu, M., Crowet, J., Nasir, M. N., and Lins, L. (2014). Complementary biophysical tools to investigate lipid speci fi city in the interaction between bioactive molecules and the plasma membrane: a review. Biochim. Biophys. Acta Biomemb. 1838, 3171–3190. doi: 10.1016/j.bbamem.2014.08.023
Fahim, S., Dimitrov, K., Gancel, F., Vauchel, P., Jacques, P., and Nikov, I. (2012). Bioresource Technology Impact of energy supply and oxygen transfer on selective lipopeptide production by Bacillus subtilis BBG21. Bioresour. Technol. 126, 1–6. doi: 10.1016/j.biortech.2012.09.019
Farace, G., Fernandez, O., Jacquens, L., Coutte, F., Krier, F., Jacques, P., et al. (2015). Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 16, 177–187. doi: 10.1111/mpp.12170
Farzand, A., Moosa, A., Zubair, M., Khan, A. R., Massawe, V. C., Tahir, H. A. S., et al. (2019). Suppression of Sclerotinia sclerotiorum by the induction of systemic resistance and regulation of antioxidant pathways in tomato using fengycin produced by Bacillus amyloliquefaciens FZB42. Biomolecules 9:613. doi: 10.3390/biom9100613
García-Gutiérrez, L., Zeriouh, H., Romero, D., Cubero, J., de Vicente, A., and Pérez-García, A. (2013). The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 6, 264–274. doi: 10.1111/1751-7915.12028
Geudens, N., and Martins, J. C. (2018). Cyclic lipodepsipeptides from Pseudomonas spp. - biological swiss-army knives. Front. Microbiol. 9:1867. doi: 10.3389/fmicb.2018.01867
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Factories 15:203. doi: 10.1186/s12934-016-0603-7
Gronnier, J., Gerbeau-Pissot, P., Germain, V., Mongrand, S., and Simon-Plas, F. (2018). Divide and rule: plant plasma membrane organization. Trends Plant Sci. 23, 899–917. doi: 10.1016/j.tplants.2018.07.007
Han, Q., Wu, F., Wang, X., Qi, H., Shi, L., Ren, A., et al. (2015). The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ. Microbiol. 17, 1166–1188. doi: 10.1111/1462-2920.12538
Han, S. H., Lee, S. J., Moon, J. H., Park, K. H., Yang, K. Y., Cho, B. H., et al. (2006). GacS-dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol. Plant Microbe Interact. 2006 19, 924–930. doi: 10.1094/MPMI-19-0924
Henry, G., Deleu, M., Jourdan, E., Thonart, P., and Ongena, M. (2011). The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell. Microbiol. 13, 1824–1837. doi: 10.1111/j.1462-5822.2011.01664.x
Hernández-Reyes, C., Schenk, S. T., Neumann, C., Kogel, K. H., and Schikora, A. (2014). N-acyl-homoserine lactones-producing bacteria protect plants against plant and human pathogens. Microb. Biotechnol. 7, 580–588. doi: 10.1111/1751-7915.12177
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Iavicoli, A., Boutet, E., Buchala, A., and Métraux, J.-P. (2003). Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 16, 851–858. doi: 10.1094/MPMI.2003.16.10.851
Jourdan, E., Henry, G., Duby, F., Dommes, J., Barthélemy, J. P., Thonart, P., et al. (2009). Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol. Plant Microbe Interact. 22, 456–468. doi: 10.1094/MPMI-22-4-0456
Kai, M., Effmert, U., and Piechulla, B. (2016). Bacterial-plant-interactions: approaches to unravel the biological function of bacterial volatiles in the rhizosphere. Front. Microbiol. 7:108. doi: 10.3389/fmicb.2016.00108
Kanyuka, K., and Rudd, J. J. (2019). Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8. doi: 10.1016/j.pbi.2019.02.005
Kawagoe, Y., Shiraishi, S., Kondo, H., Yamamoto, S., Aoki, Y., and Suzuki, S. (2015). Cyclic lipopeptide iturin A structure-dependently induces defense response in Arabidopsis plants by activating SA and JA signaling pathways. Biochem. Biophys. Res. Commun. 460, 1015–1020. doi: 10.1016/j.bbrc.2015.03.143
Kenawy, A., Dailin, D. J., Abo-Zaid, G. A., Abd Malek, R., Ambehabati, K. K., Zakaria, K. H., et al. (2019). “Biosynthesis of antibiotics by PGPR and their roles in biocontrol of plant diseases,” in Plant Growth Promoting Rhizobacteria for Sustainable Stress Management, ed. R. Z. Sayyed (Singapore: Springer), 1–36. doi: 10.1007/978-981-13-6986-5_1
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kong, H. G., Shin, T. S., Kim, T. H., and Ryu, C. M. (2018). Stereoisomers of the bacterial volatile compound 2,3-butanediol differently elicit systemic defense responses of pepper against multiple viruses in the field. Front. Plant Sci. 9:90. doi: 10.3389/fpls.2018.00090
Kramer, J., Özkaya, Ö, and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Le Mire, G., Siah, A., Brisset, M.-N., Gaucher, M., Deleu, M., and Jijakli, M. (2018). Surfactin protects wheat against Zymoseptoria tritici and activates both salicylic acid- and jasmonic acid-dependent defense responses. Agriculture 8:11. doi: 10.3390/agriculture8010011
Lee, B., Farag, M. A., Park, H. B., Kloepper, J. W., Lee, S. H., and Ryu, C. M. (2012). Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS One 7:e48744. doi: 10.1371/journal.pone.0048744
Leeman, M., Den Ouden, F. M., Van Pelt, J. A., Dirkx, F. P. M., Steijl, H., Bakker, P. A. H. M., et al. (1996). Iron availability affects induction of systemic resistance to Fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86, 149–155. doi: 10.1094/Phyto-86-149
Li, Y., Héloir, M. C., Zhang, X., Geissler, M., Trouvelot, S., Jacquens, L., et al. (2019). Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 20, 1037–1050. doi: 10.1111/mpp.12809
Ma, Z., Hoang Hua, G. K. H., Ongena, M., and Höfte, M. (2016). Role of phenazines and cyclic lipopeptides produced by Pseudomonas sp. CMR12a in induced systemic resistance on rice and bean. Environ. Microbiol. Rep. 8, 896–904. doi: 10.1111/1758-2229.12454
Ma, Z., Ongena, M., and Höfte, M. (2017). The cyclic lipopeptide orfamide induces systemic resistance in rice to Cochliobolus miyabeanus but not to Magnaporthe oryzae. Plant Cell Rep. 36, 1731–1746. doi: 10.1007/s00299-017-2187-z
Mariutto, M., Duby, F., Adam, A., Bureau, C., Fauconnier, M. L., Ongena, M., et al. (2011). The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 11:29. doi: 10.1186/1471-2229-11-29
Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C. M. J., Pozo, M. J., et al. (2016). Recognizing plant defense priming. Trends Plant Sci. 21, 818–822. doi: 10.1016/j.tplants.2016.08.005
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: an adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 485–512. doi: 10.1146/annurev-arplant-042916
Maurhofer, M., Hase, C., Meuwly, P., Metraux, J. P., and Defago, G. (1994). Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology 84, 139–146. doi: 10.1094/Phyto-84-139
Mejri, S., Siah, A., Coutte, F., Magnin-Robert, M., Randoux, B., Tisserant, B., et al. (2018). Biocontrol of the wheat pathogen Zymoseptoria tritici using cyclic lipopeptides from Bacillus subtilis. Environ. Sci. Pollut. Res. 25, 29822–29833. doi: 10.1007/s11356-017-9241-9
Meziane, H., Van Der Sluis, I., Va Loon, L. C., Höfte, M., and Bakker, P. A. (2005). Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6, 177–185. doi: 10.1111/j.1364-3703.2005.00276
Mhatre, P. H., Karthik, C., Kadirvelu, K., Divya, K. L., Venkatasalam, E. P., Srinivasan, S., et al. (2018). Plant growth promoting rhizobacteria (PGPR): a potential alternative tool for nematodes bio-control. Biocatal. Agricult. Biotechnol. 17, 119–128. doi: 10.1016/j.bcab.2018.11.009
Monnier, N., Cordier, M., Dahi, A., Santoni, V., Guenin, S., Clement, C., et al. (2020). Semi-purified rhamnolipid mixes protect Brassica napus against Leptosphaeria maculans early infections. Phytopathology 110, 834–842. doi: 10.1094/phyto-07-19-0275-r
Monnier, N., Furlan, A., Botcazon, C., Dahi, A., Mongelard, G., Cordelier, S., et al. (2018). Rhamnolipids from Pseudomonas aeruginosa are elicitors triggering Brassica napus protection against Botrytis cinerea without physiological disorders. Front. Plant Sci. 9:1170. doi: 10.3389/fpls.2018.01170
Nishimura, S., and Matsumori, N. (2020). Chemical diversity and mode of action of natural products targeting lipids in the eukaryotic cell membrane. Nat. Prod. Rep. 37, 677–702. doi: 10.1039/c9np00059c
Olorunleke, F. E., Hua, G. K. H., Kieu, N. P., Ma, Z., and Höfte, M. (2015). Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ. Microbiol. Rep. 7, 774–781. doi: 10.1111/1758-2229.12310
Omoboye, O. O., Oni, F. E., Batool, H., Yimer, H. Z., De Mot, R., and Höfte, M. (2019). Pseudomonas cyclic lipopeptides suppress the rice blast fungus Magnaporthe oryzae by induced resistance and direct antagonism. Front. Plant Sci. 10:901. doi: 10.3389/fpls.2019.00901
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends in Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Ongena, M., Jourdan, E., Adam, A., Schäfer, M., Budzikiewicz, H., and Thonart, P. (2008). Amino acids, iron, and growth rate as key factors influencing production of the Pseudomonas putida BTP1 benzylamine derivative involved in systemic resistance induction in different plants. Microb. Ecol. 55, 280–292. doi: 10.1007/s00248-007-9275-5
Ongena, M., Jourdan, E., Schäfer, M., Kech, C., Budzikiewicz, H., Luxen, A., et al. (2005). Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 18, 562–569. doi: 10.1094/MPMI-18-0562
Park, H. B., Lee, B., Kloepper, J. W., and Ryu, C. M. (2013). One shot-two pathogens blocked: exposure of Arabidopsis to hexadecane, a long chain volatile organic compound, confers induced resistance against both Pectobacterium carotovorum and Pseudomonas syringae. Plant Signal. Behav. 8:e24619. doi: 10.4161/psb.24619
Park, K., Park, Y. S., Ahamed, J., Dutta, S., Ryu, H., Lee, S. H., et al. (2016). Elicitation of induced systemic resistance of chili pepper by iturin a analogs derived from Bacillus vallismortis EXTN-1. Can. J. Plant Sci. 96, 564–570. doi: 10.1139/cjps-2015-0199
Perneel, M., D’Hondt, L., De Maeyer, K., Adiobo, A., Rabaey, K., and Hofte, M. (2008). Phenazines and biosurfactants interact in the biological control of soil-borne diseases caused by Pythium spp. Environ. Microbiol. 10, 778–788. doi: 10.1111/j.1462-2920.2007.01501.x
Pertot, I., Puopolo, G., Hosni, T., Pedrotti, L., Jourdan, E., and Ongena, M. (2013). Limited impact of abiotic stress on surfactin production in planta and on disease resistance induced by Bacillus amyloliquefaciens S499 in tomato and bean. FEMS Microbiol. Ecol. 86, 505–519. doi: 10.1111/1574-6941.12177
Piasecka, A., Jedrzejczak-Rey, N., and Bednarek, P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. doi: 10.1111/nph.13325
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced Systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Rahman, A., Uddin, W., and Wenner, N. G. (2015). Induced systemic resistance responses in perennial ryegrass against Magnaporthe oryzae elicited by semi-purified surfactin lipopeptides and live cells of Bacillus amyloliquefaciens. Mol. Plant Pathol. 16, 546–558. doi: 10.1111/mpp.12209
Ran, L. X., Li, Z. N., Wu, G. J., Van Loon, L. C., and Bakker, P. A. H. M. (2005). Induction of systemic resistance against bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. Eur. J. Plant Pathol. 113, 59–70. doi: 10.1007/s10658-005-0623-3
Rashid, M. H. O., and Chung, Y. R. (2017). Induction of systemic resistance against insect herbivores in plants by beneficial soil microbes. Front. Plant Sci. 8:1816. doi: 10.3389/fpls.2017.01816
Rodríguez, J., Tonelli, M. L., Figueredo, M. S., Ibáñez, F., and Fabra, A. (2018). The lipopeptide surfactin triggers induced systemic resistance and priming state responses in Arachis hypogaea L. Eur. J. Plant Pathol. 152, 845–851. doi: 10.1007/s10658-018-1524-6
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Paré, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Sanchez, L., Courteaux, B., Hubert, J., Kauffmann, S., Renault, J. H., Clément, C., et al. (2012). Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiology 160, 1630–1641. doi: 10.1104/pp.112.201913
Schellenberger, R., Touchard, M., Clément, C., Baillieul, F., Cordelier, S., Crouzet, J., et al. (2019). Apoplastic invasion patterns triggering plant immunity: plasma membrane sensing at the frontline. Mol. Plant Pathol. 20, 1602–1616. doi: 10.1111/mpp.12857
Schenk, S. T., Hernández-Reyes, C., Samans, B., Stein, E., Neumann, C., Schikora, M., et al. (2014). N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26, 2708–2723. doi: 10.1105/tpc.114.126763
Schenk, S. T., Stein, E., Kogel, K. H., and Schikora, A. (2012). Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal. Behav 7, 178–181. doi: 10.4161/psb.18789
Schikora, A., Schenk, S. T., and Hartmann, A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol. 90, 605–612. doi: 10.1007/s11103-016-0457-8
Schikora, A., Schenk, S. T., Stein, E., Molitor, A., Zuccaro, A., and Kogel, K. H. (2011). N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 157, 1407–1418. doi: 10.1104/pp.111.180604
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., Vogg, G., et al. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
Siddiqui, I. A., and Shaukat, S. S. (2003). Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite, 2,4-diacetylpholoroglucinol. Soil Biol Biochem. 35, 1615–1623. doi: 10.1016/j.soilbio.2003.08.006
Singh, M., Singh, D., Gupta, A., Pandey, K. D., Singh, P. K., and Kumar, A. (2019). “Plant growth promoting rhizobacteria: application in biofertilizers and bio-control of phytopathogens,” in PGPR Amelioration in Sustainable Agriculture, eds A. K. Singh, A. Kumar, and P. K. Singh (Cambridge: Woodhead Publishing, Inc), 2019.
Song, D., Meng, J., Cheng, J., Fan, Z., Chen, P., Ruan, H., et al. (2018). Pseudomonas aeruginosa quorum-sensing metabolite induces host immune cell death through cell surface lipid domain dissolution. Nat. Microbiol. 4, 97–111. doi: 10.1038/s41564-018-0290-8
Song, G. C., Choi, H. K., and Ryu, C. M. (2015). Gaseous 3-pentanol primes plant immunity against a bacterial speck pathogen, Pseudomonas syringae pv. tomato via salicylic acid and jasmonic acid-dependent signaling pathways in arabidopsis. Front. Plant Sci. 6:821. doi: 10.3389/fpls.2015.00821
Song, G. C., Riu, M., and Ryu, C. M. (2019). Beyond the two compartments Petri-dish: Optimising growth promotion and induced resistance in cucumber exposed to gaseous bacterial volatiles in a miniature greenhouse system. Plant Methods 15:9. doi: 10.1186/s13007-019-0395-y
Stringlis, I. A., De Jonge, R., and Pieterse, C. M. J. (2019). The age of coumarins in plant-microbe interactions. Plant Cell Physiol. 60, 1405–1419. doi: 10.1093/pcp/pcz076
Stringlis, I. A., Proietti, S., Hickman, R., Van Verk, M. C., Zamioudis, C., and Pieterse, C. M. J. (2018). Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 93, 166–180. doi: 10.1111/tpj.13741
Tracanna, V., de Jong, A., Medema, M. H., and Kuipers, O. P. (2017). Mining prokaryotes for antimicrobial compounds: from diversity to function. FEMS Microbiol. Rev. 41, 417–429. doi: 10.1093/femsre/fux014
Tran, H., Ficke, A., Asiimwe, T., Höfte, M., and Raaijmakers, J. M. (2007). Role of the cyclic lipopeptide massetolide a in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175, 731–742. doi: 10.1111/j.1469-8137.2007.02138.x
Varnier, A. L., Sanchez, L., Vatsa, P., Boudesocque, L., Garcia-Brugger, A., Rabenoelina, F., et al. (2009). Bacterial rhamnolipids are novel MAMPs conferring resistance to Botrytis cinerea in grapevine. Plant Cell Environ. 32, 178–193. doi: 10.1111/j.1365-3040.2008.01911.x
Vatsa, P., Sanchez, L., Clement, C., Baillieul, F., and Dorey, S. (2010). Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int. J. Mol. Sci. 11, 5095–5108. doi: 10.3390/ijms11125095
Venturi, V., and Keel, C. (2016). Signaling in the Rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Verhagen, B. W. M., Trotel-Aziz, P., Couderchet, M., Höfte, M., and Aziz, A. (2010). Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. J. Exp. Bot. 61, 249–260. doi: 10.1093/jxb/erp295
Wang, H., Yang, X., Guo, L., Zeng, H., and Qiu, D. (2015). PeBL1, a novel protein elicitor from Brevibacillus laterosporus strain A60, activates defense responses and systemic resistance in Nicotiana benthamiana. Appl. Environ. Microbiol. 81, 2706–2716. doi: 10.1128/AEM.03586-14
Wang, N., Liu, M., Guo, L., Yang, X., and Qiu, D. (2016). A novel protein elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 induces systemic resistance in tobacco. Int. J. Biol. Sci. 12, 757–767. doi: 10.7150/ijbs.14333
Wehner, G., Kopahnke, D., Richter, K., Kecke, S., Schikora, A., and Ordon, F. (2019). Priming is a suitable strategy to enhance resistance towards leaf rust in barley. Phytob. J. 3, 46–51. doi: 10.1094/PBIOMES-09-18-0041-R
Weller, D. M., Mavrodi, D. V., Van Pelt, J. A., Pieterse, C. M. J., Van Loon, L. C., and Bakker, P. A. H. M. (2012). Induced systemic resistance in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102, 403–412. doi: 10.1094/PHYTO-08-11-0222
Williams, A., Pétriacq, P., Beerling, D. J., Cotton, T. E. A., and Ton, J. (2018). Impacts of atmospheric CO2 and soil nutritional value on plant responses to rhizosphere colonization by soil bacteria. Front. Plant Sci. 871:1493. doi: 10.3389/fpls.2018.01493
Wu, G., Liu, Y., Xu, Y., Zhang, G., Shen, Q., and Zhang, R. (2018). Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant Microbe Interact. 31, 560–567. doi: 10.1094/MPMI-11-17-0273-R
Wu, K., Fang, Z., Guo, R., Pan, B., Shi, W., Yuan, S., et al. (2015). Pectin enhances bio-control efficacy by inducing colonization and secretion of secondary metabolites by Bacillus amyloliquefaciens SQY 162 in the rhizosphere of tobacco. PLoS One 10:e127418. doi: 10.1371/journal.pone.0127418
Yamamoto, S., Shiraishi, S., and Suzuki, S. (2015). Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 60, 379–386. doi: 10.1111/lam.12382
Yang, T., Rao, Z., Zhang, X., Xu, M., Xu, Z., and Yang, S. T. (2013). Improved Production of 2,3-Butanediol in Bacillus amyloliquefaciens by Over-Expression of Glyceraldehyde-3-Phosphate Dehydrogenase and 2,3-butanediol Dehydrogenase. PLoS One 8:e76149. doi: 10.1371/journal.pone.0076149
Yi, H. S., Ahn, Y. R., Song, G. C., Ghim, S. Y., Lee, S., Lee, G., et al. (2016). Impact of a bacterial volatile 2,3-butanediol on Bacillus subtilis rhizosphere robustness. Front. Microbiol. 7:993. doi: 10.3389/fmicb.2016.00993
Yu, K., Pieterse, C. M. J., Bakker, P. A. H. M., and Berendsen, R. L. (2019). Beneficial microbes going underground of root immunity. Plant Cell Environ. 42, 2860–2870. doi: 10.1111/pce.13632
Zanotto, A. W., Valério, A., de Andrade, C. J., and Pastore, G. M. (2019). New sustainable alternatives to reduce the production costs for surfactin 50 years after the discovery. Appl. Microbiol. Biotechnol. 103, 8647–8656. doi: 10.1007/s00253-019-10123-7
Zarkani, A. A., Stein, E., Röhrich, C. R., Schikora, M., Evguenieva-Hackenberg, E., Degenkolb, T., et al. (2013). Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 14, 17122–17146. doi: 10.3390/ijms140817122
Zhang, Y., Yan, X., Guo, H., Zhao, F., and Huang, L. (2018). A novel protein elicitor BAR11 from Saccharothrix yanglingensis Hhs.015 improves plant resistance to pathogens and interacts with catalases as targets. Front. Microbiol. 9:700. doi: 10.3389/fmicb.2018.00700
Zhao, X., and Kuipers, O. P. O. P. (2016). Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genomics 17:882. doi: 10.1186/s12864-016-3224-y
Keywords: plant immunity, systemic resistance, defense mechanism, molecular patterns, plant growth promoting rhizobacteria, biocontrol
Citation: Pršić J and Ongena M (2020) Elicitors of Plant Immunity Triggered by Beneficial Bacteria. Front. Plant Sci. 11:594530. doi: 10.3389/fpls.2020.594530
Received: 13 August 2020; Accepted: 07 October 2020;
Published: 09 November 2020.
Edited by:
Roeland Lucas Berendsen, Utrecht University, NetherlandsReviewed by:
Pierre Pétriacq, Université de Bordeaux, FranceZhonglin Mou, University of Florida, United States
Copyright © 2020 Pršić and Ongena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jelena Pršić, jelena.prsic@student.uliege.be; Marc Ongena, marc.ongena@uliege.be
 Jelena Pršić
Jelena Pršić