- Shanghai Key Lab of Protected Horticultural Technology, Horticultural Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai, China
Powdery mildew (PM), caused by Podosphaera xanthii (Px), is one of the most devastating fungal diseases of melon worldwide. The use of resistant cultivars is considered to be the best and most effective approach to control this disease. In this study, an F2 segregating population derived from a cross between a resistant (wm-6) and a susceptible cultivar (12D-1) of melon was used to map major powdery mildew resistance genes using bulked segregant analysis (BSA), in combination with next-generation sequencing (NGS). A novel quantitative trait locus (QTL) named qCmPMR-12 for resistance to PM on chromosome 12 was identified, which ranged from 22.0 Mb to 22.9 Mb. RNA-Seq analysis indicated that the MELO3C002434 gene encoding an ankyrin repeat-containing protein was considered to be the most likely candidate gene that was associated with resistance to PM. Moreover, 15 polymorphic SNPs around the target area were successfully converted to Kompetitive Allele-Specific PCR (KASP) markers (P < 0.0001). The novel QTL and candidate gene identified from this study provide insights into the genetic mechanism of PM resistance in melon, and the tightly linked KASP markers developed in this research can be used for marker-assisted selection (MAS) to improve powdery mildew resistance in melon breeding programs.
Introduction
Melon (Cucumis melo L.), which belongs to the Cucurbitaceae family, is an important horticultural and economic crop worldwide (Garcia-Mas et al., 2012). In view of its unique biological characteristics, such as a sweet fruit, unique aroma and rich nutritional value, melon is highly favored by consumers. Powdery mildew (PM) is a fungal disease that occurs commonly on leaves, petioles, and stems of most cucurbit crops in both field and greenhouse conditions (Perez-Garcia et al., 2009). This disease can result in a decrease in weight-based productivity and a reduction in fruit quality (Candido et al., 2014), thereby causing severe economic losses in many areas of the world (Romero et al., 2008).
Melon PM is often caused by Podosphaera xanthii (Px) and Golovinomyces cichoracearum (Gc) (Křístková et al., 2009; Li et al., 2017). These two fungi can be distinguished by observation of the pathogens’ cleistothecia, conidia germination, microscopic morphology, and host identification (Liang et al., 2010; Liu et al., 2010). Both pathogens exist either as an asexual stage (conidia) or sexual stage (Cleistothecia); the morphological characteristics of the asexual stage are similar for the two pathogens, and the morphological distinction between them is based on whether fibrous bodies occur on the spore (Px spores possess fibrous bodies, whereas these are absent from those of Gc) (Wang et al., 2013). In China, Px is generally considered to be the main causal agent of PM in melon (Cheng et al., 2011; Zhang et al., 2013). Recent reports have shown that melon PM is caused by Px in Shanghai (Gu et al., 2010; Li et al., 2015). The five phases of the Px life cycle are separately germinating conidia, the formation of a primary germ tube, hyphae, conidiophores, and colonization (Wang et al., 2013). After a spore lands on the leaf cuticle, the interaction is initiated by the formation of a primary germ tube and is followed by the formation and maturation of an appressiorial germ tube; the next step involves haustorium formation within an attacked epidermal cell and fungal development proceeds via the formation of secondary hyphae and haustoria and terminates with sporulation (Wolter et al., 1993). Px can coexist with the diseased plants in the soil as hyphae, conidia and cleistothecia, or it can overwinter as hyphae on greenhouse plants, and when the temperature rises the subsequent year, the conidia are distributed by air or water to infect the plants (Zhang et al., 2007).
Currently, the main method applied to control powdery mildew is chemical prevention by the application of chemical fungicides, which is often time-consuming and labor-intensive (Chen, 2014). Furthermore, chemical control not only leads to the appearance of resistance and to mutations in PM, which therefore means that this treatment becomes ineffective, but it also increases the input costs and has a negative impact on the environment (McGrath, 2001; Hollomon et al., 2002). Therefore, breeding for disease resistance is a safe alternative or a complement to the chemical control of this disease (Perchepied et al., 2005). Marker-assisted selection (MAS) is a powerful genomic tool that assists phenotypic selection for the development of disease-resistant cultivars and can help breeders incorporate and pyramid resistance genes into breeding material, thereby reducing disease severity (Ribaut et al., 2002; Chen, 2013; Zhu et al., 2019). At present, MAS has been extensively applied to search for the molecular markers that are linked to a specific trait during the development of disease-resistant cultivars (Teixeira et al., 2008). To date, a variety of molecular markers have been developed, such as RFLPs, RAPDs, SSRs, InDels and SNPs, to detect allelic variation within different samples at the DNA level (Wang et al., 2015).
Bulked segregant analysis (BSA) is an important technique used to map quantitative trait loci (QTLs) and identify DNA markers. Compared with the traditional QTL mapping method, which is time-consuming and involves screening polymorphic markers and genotyping, BSA provides a convenient and rapid method with which to identify resistance genes by generating two DNA bulks with a contrasting target trait (Michelmore et al., 1991; Abe et al., 2012; Nie et al., 2015). Recently, due to the release of sequenced genomes and the significant reduction in the costs of next-generation sequencing (NGS), whole-genome resequencing has been coupled with BSA to map the genes of interest that are associated with a given phenotype. The combined application of BSA with NGS (BSA-Seq) has accelerated the identification of tightly linked markers for gene identification and QTL mapping (Zou et al., 2016). To date, BSA-Seq has been successfully used in mapping the traits of early flowering, flesh thickness and downy mildew resistance in cucumber (Lu et al., 2014; Xu et al., 2015; Win et al., 2017), cold tolerance and blast resistance in rice (Yang et al., 2013; Zheng et al., 2016), cotyledon color, and a high-sucrose and low-oil seed phenotype in soybean (Dobbels et al., 2017; Song et al., 2017).
At present, the availability of sequence information has facilitated the identification and development of single nucleotide polymorphism (SNP) markers, which have largely replaced simple sequence repeats (SSRs) as markers in crop species (Semagn et al., 2014). Because of the low assay cost, high genomic abundance, ease of documentation, locus specificity, co-dominant inheritance, the potential for high-throughput analysis, and relatively low genotyping error rates, the use of SNPs has emerged as a powerful approach for many genetic applications in areas such as germplasm characterization, quality control (QC) analysis, linkage mapping, linkage-based and linkage disequilibrium-based QTL mapping, allele mining, marker-assisted backcrossing (MABC), genomic selection (GS), and MAS (Rafalski, 2002; Schlotterer, 2004; Semagn et al., 2014). Kompetitive Allele-Specific PCR (KASP) is a high-throughput SNP genotyping platform. Due to its low cost and genotyping error rates, and its high reliability and reproducibility, KASP has evolved to become a global benchmark technology and has been widely used for genetic mapping and trait-specific marker development (He et al., 2014; Ertiro et al., 2015; Rasheed et al., 2016; Tan et al., 2017).
To date, several genes and QTLs that confer resistance to powdery mildew have been identified in melon, such as the genes of Pm-w from WMR 29 (Pitrat, 1991), Pm-x from PI 414723 and Pm-y from VA 435 (Périn et al., 2002), and Pm-1 from the AF125Pm–1 Cantalupensis Charentais-type breeding line (Teixeira et al., 2008), and the QTLs of PmV.1 and PmXII.1 from PI 124112 (Perchepied et al., 2005), Pm-R from TGR-1551 (Yustelisbona et al., 2011) and BPm12.1 from MR-1 (Li et al., 2017). In previous studies, many of these genes and QTLs have been found on chromosomes 2, 4, 5, and 12 (Pitrat, 1991; Périn et al., 2002; Fukino et al., 2008; Zhang et al., 2013; Li et al., 2017). Differing views exist concerning the genetic basis of PM resistance in melon. Some studies have indicated that PM resistance in melon is controlled by a single dominant gene (Epinat et al., 1992; Zhang et al., 2008; Wang et al., 2016; Li et al., 2017), whereas other research has reported that it is controlled by a recessive gene (McCreight and Coffey, 2011), by two dominant genes (Clements, 2014), or by one dominant and one recessive gene (Sun et al., 2010; Yuste-Lisbona et al., 2010). Moreover, it is also reported that resistance to PM in melon is controlled by different sets of QTL (Perchepied et al., 2005).
The melon cultivars wm-6 and 12D-1 are both high-generation inbred lines developed by our group and our previous study has shown that the melon cultivar wm-6 is highly resistant to PM, whereas 12D-1 is highly susceptible (Data not shown). In this study, we obtained an F2 population from a cross between wm-6 (female) and 12D-1 (male). A major QTL that confers PM resistance on chromosome 12 was identified by BSA-Seq analysis, and a most likely candidate gene was predicted from RNA-Seq data in wm-6 melon. In addition, 15 suitable KASP markers were developed by the KASP SNP genotyping method. This will facilitate the cloning and functional validation of the candidate resistance gene and the linked markers will further provide a useful tool for MAS in melon breeding programs.
Materials and Methods
Plant Materials, Growth Conditions and Inoculation With Powdery Mildew Fungus
Two inbred lines, wm-6 (P1, resistant to PM) and 12D-1 (P2, susceptible to PM), were used as parental lines to generate F1 and F2 populations for the QTL mapping of PM resistance in melon (Cucumis melo L.). The P1, P2, F1 and F2 individuals were all placed in a culture room at a temperature of 25/20°C (day/night) with a photoperiod of 14 h light and relative humidity of 50–75%. All the seeds used in this study were provided by the Shanghai Academy of Agricultural Sciences.
The PM fungus (P. xanthii) used in this study was isolated from leaves of diseased Cucurbitaceae plants according to the method of Nie et al. (2015). The plants were grown on the experimental farm of the Shanghai Academy of Agricultural Sciences, and the PM fungus was maintained by infection of susceptible melon cultivar plants. When the three true seedling leaves of melon plants were fully expanded, the fungus was collected and suspended in sterile distilled water containing 0.01% Tween 20 and was then used to inoculate plants at a concentration of 1 × 106 as previously described (Zhang et al., 2011).
Disease Evaluation for Resistance to Powdery Mildew
Phenotyping for powdery mildew resistance of melon was performed according to Zhang et al. (2013) with some modifications at 12-d post-inoculation (dpi), and each infected leaf was analyzed individually. Briefly, the disease grade of powdery mildew was categorized on a scale of 0–5 as follows: Class 0, no infection; Class 1, infection of less than 30% of the leaf; Class 3, infection of less than 70% of the leaf; Class 5, infection of approximately the entire leaf and coverage with heavy sporulation. The disease severity index (DSI) was calculated from the disease-rating scale using the following formula: DSI = 100 × Σ[(disease grade × number of plants in that grade)/(Total number of plants × maximum disease grade)]. For the inheritance study, lines with a DSI ≤ 20 were considered resistant and lines with a DSI > 20 were considered susceptible. For the F2 population, the same protocol was followed to identify resistant and susceptible plants. The phenotype of the two parental lines was analyzed at least three independent experiments with >15 seedlings examined in each experiment. The DSI of the two parental lines and the F1 hybrid plants was separately measured three times for 20 seedlings in each measurement. The DSI of F2 generation was calculated based on the phenotype of 193 F2 plants. On the basis of the DSI scores, plants with a DSI of 0–1 were categorized as resistant, and those with a DSI of 3–5, as susceptible.
RNA Extraction, Library Construction and Sequencing
For RNA-Seq analysis, the two parental lines wm-6 and 12D-1 were separately treated with water (mock) or PM fungus. After 3 days, the leaves of wm-6 and 12D-1 plants (named wm-6K, wm-6P, 12D-1K, 12D-1P, respectively) were harvested, immediately frozen in liquid nitrogen and stored at −80°C before RNA extraction.
Total RNA was extracted using the Trizol reagent kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and analyzed using RNase-free agarose gel electrophoresis. Following total RNA extraction, eukaryotic mRNA was enriched by Oligo(dT) beads, whereas prokaryotic mRNA was enriched by removing rRNA with the Ribo-ZeroTM Magnetic Kit (Epicentre, Madison, WI, United States). The enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTPs and buffer. The cDNA fragments were purified with a QiaQuick PCR extraction kit (Qiagen, Venlo, The Netherlands), and following end repair and the addition of poly(A), was ligated to Illumina sequencing adapters. The ligation products were selected according to size by agarose gel electrophoresis, amplified by PCR, and sequenced using Illumina HiSeq2500 by the Gene Denovo Biotechnology Company (Guangzhou, China).
Genomic DNA Extraction, Library Construction for Bulked Segregant Analysis and Whole-Genome Resequencing
Young leaves from the two parental lines, and from the F1 and the F2 populations were collected, and total genomic DNA was extracted using the CTAB method (Doyle, 1991). For bulked segregant analysis, four DNA pools were constructed, consisting of two parent bulks and two F2 segregating bulks. The parent bulks were separately constructed from the female parent (wm-6) and male parent (12D-1), and the two F2 segregating bulks were separately constructed by mixing an equal amount of DNA extracted from 25 extremely resistant (R-bulk) and 25 susceptible (S-bulk) F2 plants. After the four sequencing libraries were prepared according to the standard protocol of Illumina, they were sequenced on an Illumina HisSeq platform (Illumina, San Diego, CA, United States). Short reads obtained from the four bulks were aligned against the melon reference genome sequence to obtain the consensus sequence using BWA software. SNP calling was performed with GATK tools. The heterozygous alleles in both parents were filtered out during the process. The raw sequence reads are deposited in the NCBI Sequence Read Archive (SRA; Accession number: PRJNA655764).
Genetic Mapping
In this study, four methods (SNP-ratio, ED4, G value and LOD) were used to map QTLs that underlay resistance to PM. The SNP-ratio (resistant alleles/sensitive alleles) of the R-bulk and S-bulk were calculated as described by Soyk et al. (2017) and the SNP-ratio of the R-bulk was then divided by the SNP-ratio of the S-bulk and plotted across the genomic regions that showed ratio peaks, which indicate the possible existence of the QTLs. The read depth for each allele at segregating allelic SNPs in 500-kb sliding windows was summed using a 100-kb step increment. The Euclidean distance of each SNP (ED-SNP) was calculated as described by Hill et al. (2013) and the ED was the sum of 100 ED-SNP values within a window of 100 consecutive SNPs. The ED4 was calculated by raising ED to the fourth power. The G value averaged across neighboring SNPs was calculated according to Magwene et al. (2011). The LOD (logarithm of the odds) score was calculated as described by Zhang et al. (2019).
Development and Analysis of KASP Markers
The polymorphic SNPs identified around the target regions that associated with powdery mildew resistance were converted into KASP markers using PolyMarker software1. For each SNP, two allele-specific forward primers and one common reverse primer based on the flanking sequences around the variant position (SNP) were designed using Primer 3 software. The polymorphic SNP primers were converted to KASP markers to test their ability to differentiate the polymorphism by genotyping the two parents, and the KASP markers were then verified with the entire F2 population. Each KASP reaction was carried out using a 3-μL reaction mixture consisting of 1.48 μL KASP 2 × reaction mix, 50 ng DNA template, 0.17 μM Hex forward primer, 0.17 μM FAM forward primer and 0.42 μM universal reverse primer. The cycling conditions were as follows: 94°C for 15 min followed by 10 touchdown cycles at 95°C for 20 s and 65°C for 60 s (dropping 0.8°C per cycle); after the final annealing temperature of 56°C was reached, 26 cycles were performed at 94°C for 20 s and at 57°C for 60 s. Thermocycling and fluorescence readings were performed on a Hydrocycler and PHERAstar of LGC SNPline platform. Genotyping data were viewed as a cluster plot by SNPviewer software supported from LGC Genomics2. The significance of the correlation coefficients between phenotype and genotype was determined with t-tests. Linkage groups were constructed using JoinMap 4.1.
Results
Evaluation of Resistance to Powdery Mildew in wm-6 × 12D-1
The artificial inoculation results showed that the parental line wm-6 was highly resistant to P. xanthii (DSI = 8.0), whereas the other parental line 12D-1 (DSI = 94.0) was susceptible to the fungus (Figure 1, Table 1 and Supplementary Table 1). The DSI of the F1 plants was 68.0, which meant that the F1 generation was susceptible to PM fungus (Table 1 and Supplementary Table 1). The evaluation of infection by PM indicated that 58 F2 plants showed PM resistance and 135 showed PM susceptibility, with a 1:3 segregation between resistant and susceptible individuals (χ2 = 2.63, P = 0.11) (Table 1). This indicated that resistance to P. xanthii in wm-6 was conferred by a single recessive gene.
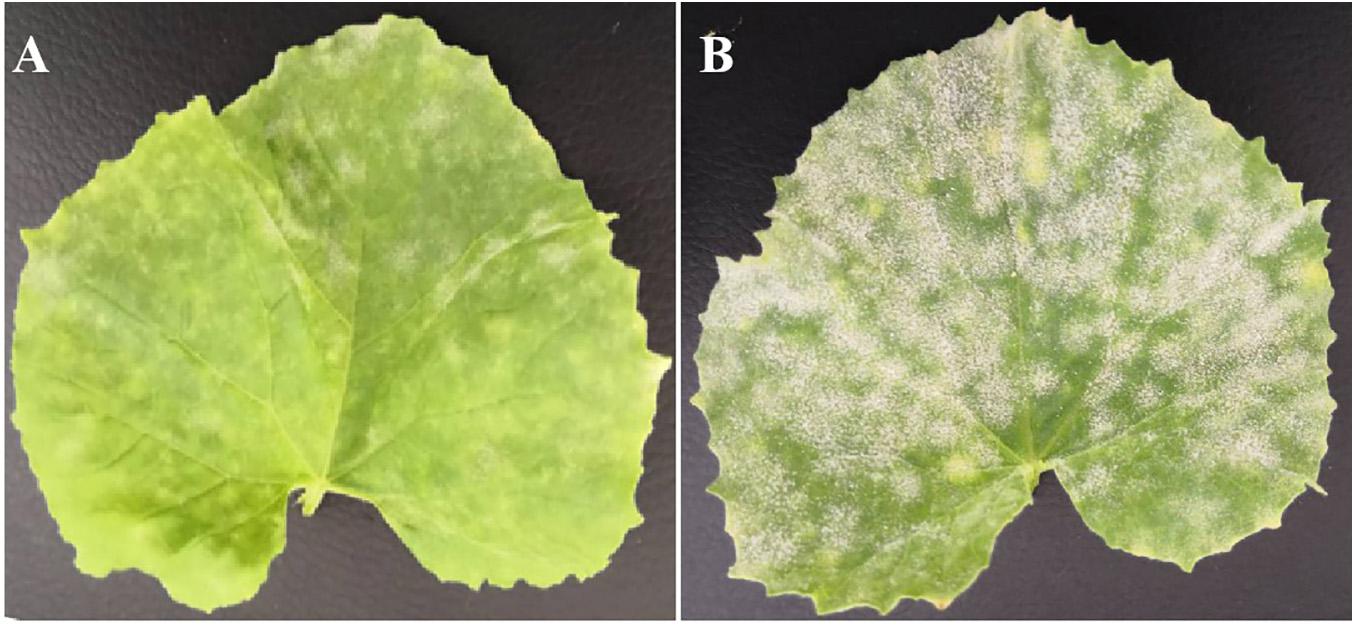
Figure 1. Phenotype of resistant parent wm-6 and susceptible parent 12D-1, 12 days after inoculation with P. xanthii. (A) The phenotype of the resistant parent wm-6, 12 days after inoculation with P. xanthii. (B) The phenotype of the resistant parent 12D-1, 12 days after inoculation with P. xanthii. The experiments were repeated at least three times independently, with similar results.
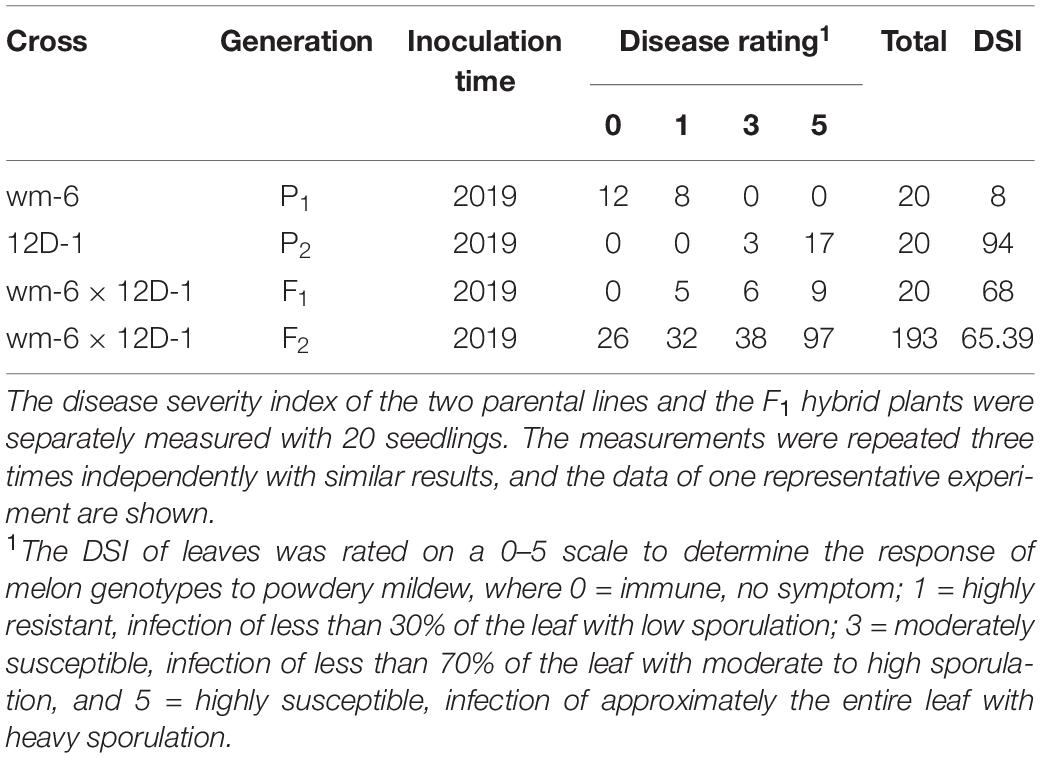
Table 1. Disease evaluation of powdery mildew in wm-6, 12D-1, F1 and F2 plants at 12 days post inoculation with P. xanthii.
Sequencing Data Analysis of Four DNA Bulks
BSA-Seq analysis was performed with the DNAs of four libraries (wm-6, 12D-1, R-bulk and S-bulk) using the Illumina HiSeq 2500 platform. In total, 60,401,327 and 57,755,318 clean reads for wm-6 and 12D-1, respectively, and 92,327,580 and 101,809,441 short reads for the R-bulk and S-bulk libraries, respectively, were generated. The GC content ranged from 36.76% to 37.1% and the Q20 and Q30 of each pool were over 98% and 93%, respectively. Over 95% of the reads were mapped to the melon reference genome, and the coverage rates were 95.78% in wm-6, 95.85% in 12D-1, 97.02% in R-bulk, and 97.05% in S-bulk, approximately resulting in a 23 × coverage depth for parental bulks and at least 35 × coverage for two F2 progeny bulks (Table 2). These results indicated that the quantity and quality of the data were sufficient for further analysis.
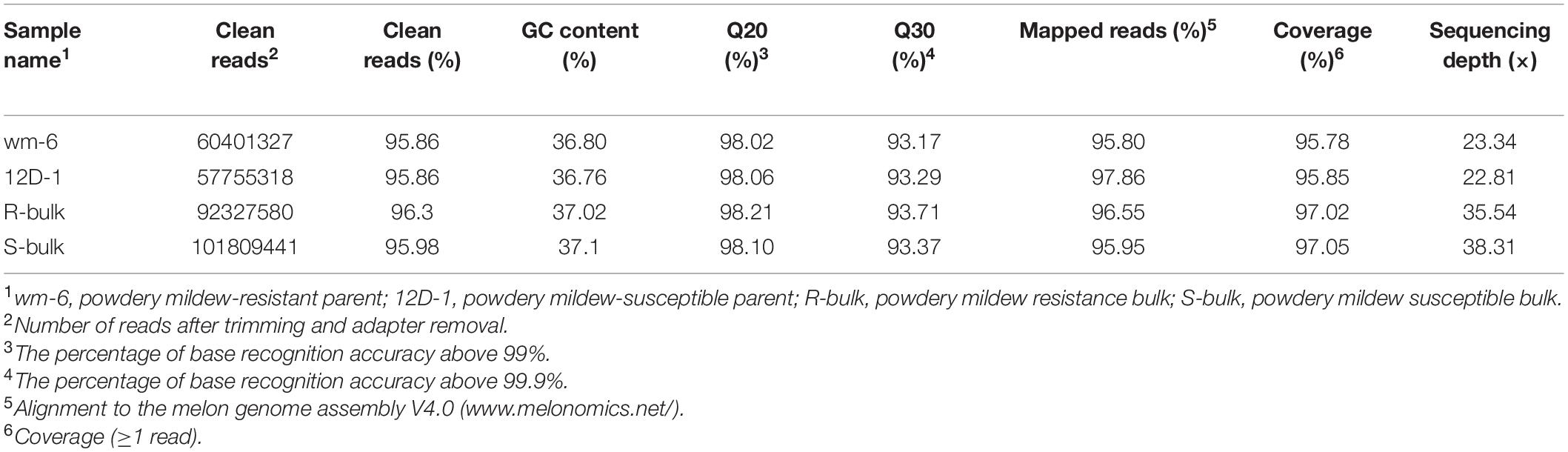
Table 2. Summary of sequencing data and the data aligned to the melon reference genome for the parental lines and the resistant and susceptible pools by BSA-Seq.
QTL Mapping of PM Resistance
After alignment to the reference genome of melon, 2,624,079 SNPs were identified between the parental lines wm-6 and 12D-1, and the distribution of these SNPs on each chromosome is listed in Supplementary Table 2. The resistance-related candidate regions were identified by analyzing the resistant pool and the susceptible pool. Using four statistic methods, a 0.9-Mb region spanning 22.0–22.9 Mb on chromosome 12 was defined as the target region associated with PM resistance (Figures 2A–D), and the significant QTL was designated qCmPMR-12. Within the candidate region, a total of 4,033 SNPs showed polymorphisms, 115 of which were located within the 3′ UTR, 78 in the 5′ UTR, 547 in the downstream region, 397 in the exonic region, 2,243 in the intergenic region, 1,005 in the intronic region, and 735 in the upstream region (Table 3). In addition, the target region included 182 non-synonymous SNV, 3 stopgain, 1 stoploss and 211 synonymous SNV polymorphisms (Table 4). Moreover, the candidate region contained 476 small InDels, 29 of which were located within the 3′ UTR, 23 in the 5′ UTR, 199 in the downstream region, 28 in the exonic region, 892 in the intergenic region, 387 in the intronic region, and 298 in the upstream region (Table 3). The InDels included 9 frameshift deletions, 6 frameshift insertions, 8 non-frameshift deletions and 5 non-frameshift insertions (Table 4).
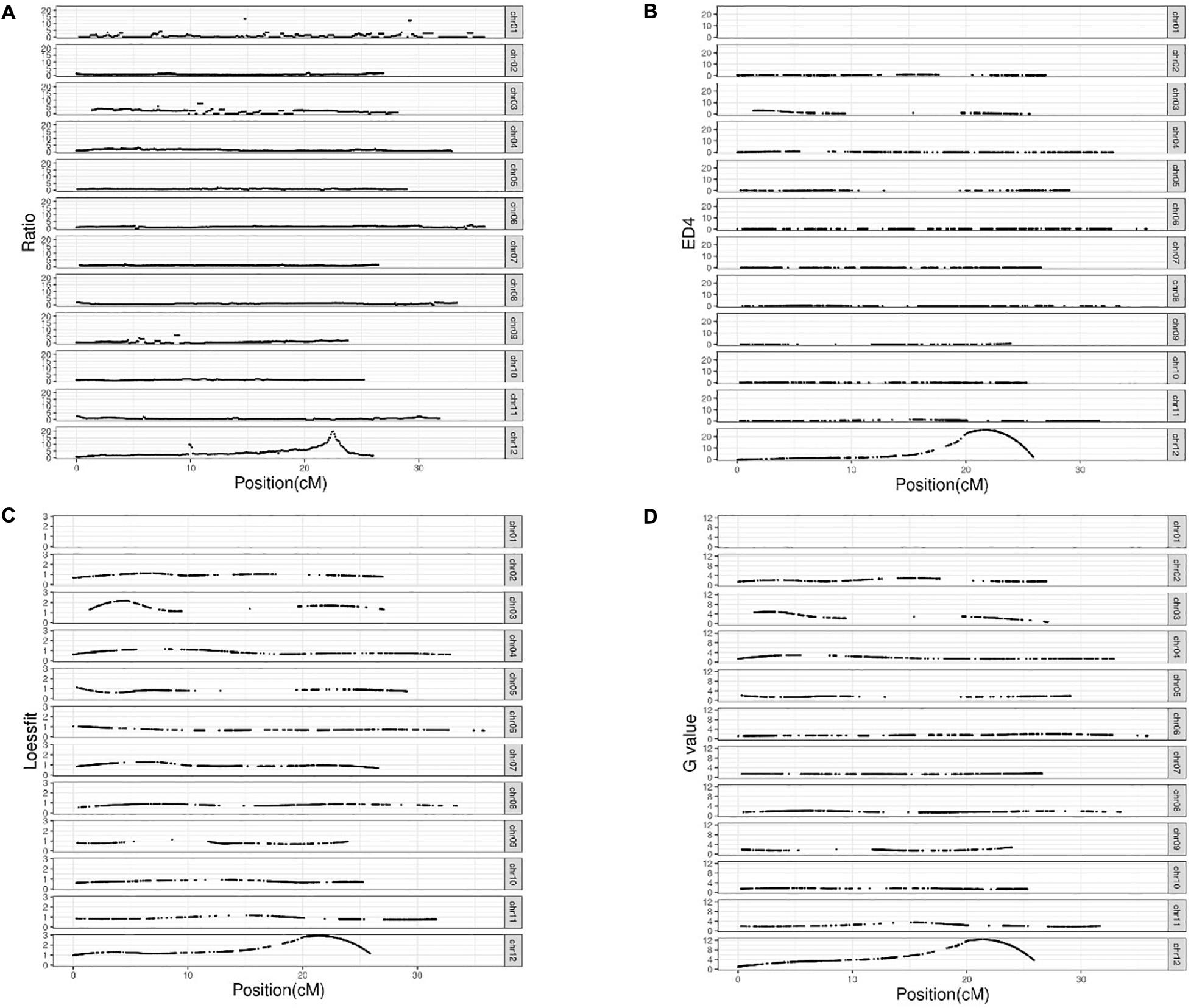
Figure 2. BSA-Seq results. Analysis of the QTL location by (A) the SNP-Ratio method; (B) the ED4 method; (C) the LOD method; (D) the G-value method.
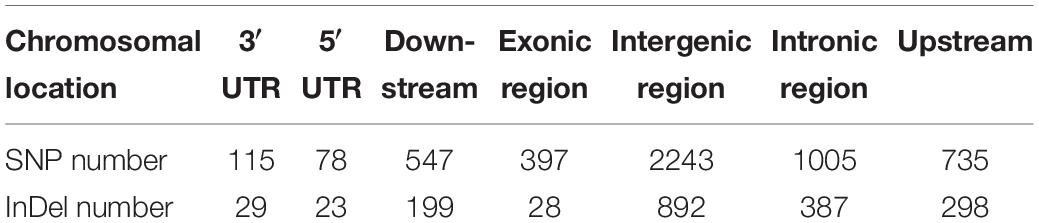
Table 3. The chromosomal locations and numbers of SNPs and InDels in the target QTL region associated with powdery mildew resistance between the melon parental lines wm-6 and 12D-1.
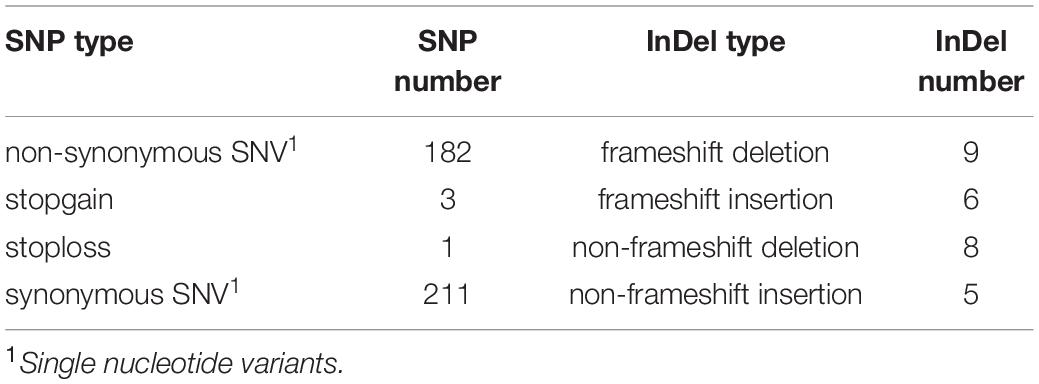
Table 4. The types and numbers of SNPs and InDels in the target QTL region associated with powdery mildew resistance between the melon parental lines wm-6 and 12D-1.
According to the melon gene annotation database3, the target region contains approximately 126 annotated genes, and the specific information for these genes is listed in Supplementary Table 3.
RNA-Seq and Candidate Gene Analysis
To elucidate the changes in gene expression of resistant and susceptible melon lines after inoculation with P. xanthii, we performed an RNA-Seq analysis at 3 dpi. The dataset submitted to NCBI include the raw reads of the assembled transcriptome sequences from control and pathogen-treated melon plants. All transcriptome raw reads have been deposited in NCBI SRA4 under the accession number PRJNA670091.
Analysis of the RNA-Seq results for the 126 annotated genes obtained by BSA-Seq indicated that the transcript levels of the two genes MELO3C002434 and MELO3C002477 (| log2FC| > 1) were clearly up-regulated in the parental lines after inoculation with P. xanthii5. The two genes respectively encode an ankyrin repeat-containing (ANK) protein and a homeobox-leucine zipper protein (Supplementary Table 3). Further analysis showed that only the expression level of MELO3C002434 was significantly higher in the resistant line wm-6 than in the susceptible line 12D-1 following PM infection. Notably, BSA-Seq showed that this gene contained 10 SNPs and 1 InDel (Table 5). Previous studies have shown that ANK proteins play important roles in regulating immune responses against various pathogens (Cao et al., 1997; Ryals et al., 1997; Yan et al., 2002; Li et al., 2013). Therefore, we speculated that MELO3C002434 may confer PM resistance, or at least be a PM resistance-related gene in melon.
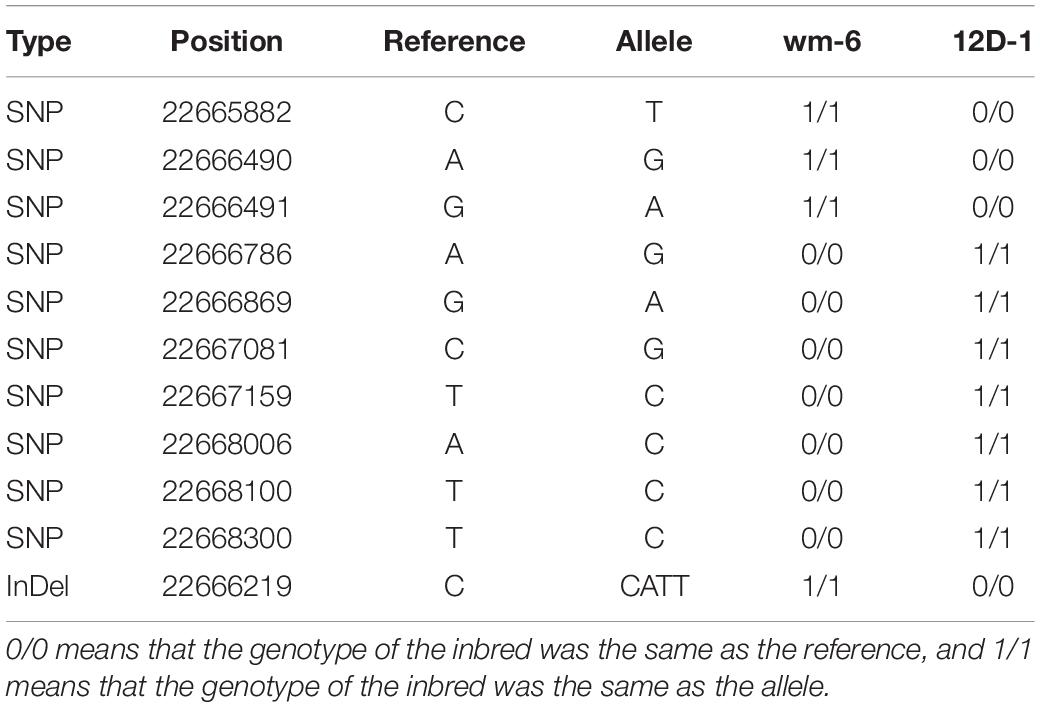
Table 5. Analysis of SNPs and InDels present within the MELO3C002434 gene in two parental melon lines.
KASP Marker Development and Physical Map Construction
KASP assays were designed for SNPs across the 20.0–23.9 Mb region on chromosome 12 and were tested on the F2 population to determine which markers showed the highest association with PM resistance. In this region, thirty-seven chromosome-specific SNPs selected for conversion to KASP markers were used to screen the parents and bulks to confirm their polymorphisms, and 15 out of 37 markers successfully distinguished the parents and bulks (Figure 3), indicating that these KASP markers were suitable for use in MAS to improve the level of powdery mildew resistance in melon breeding. Specific information and the sequences of the polymorphic KASP markers are separately listed in Supplementary Tables 4, 5. The close-up view of QTL and KASP markers linked to the target regions is shown in Figure 4.
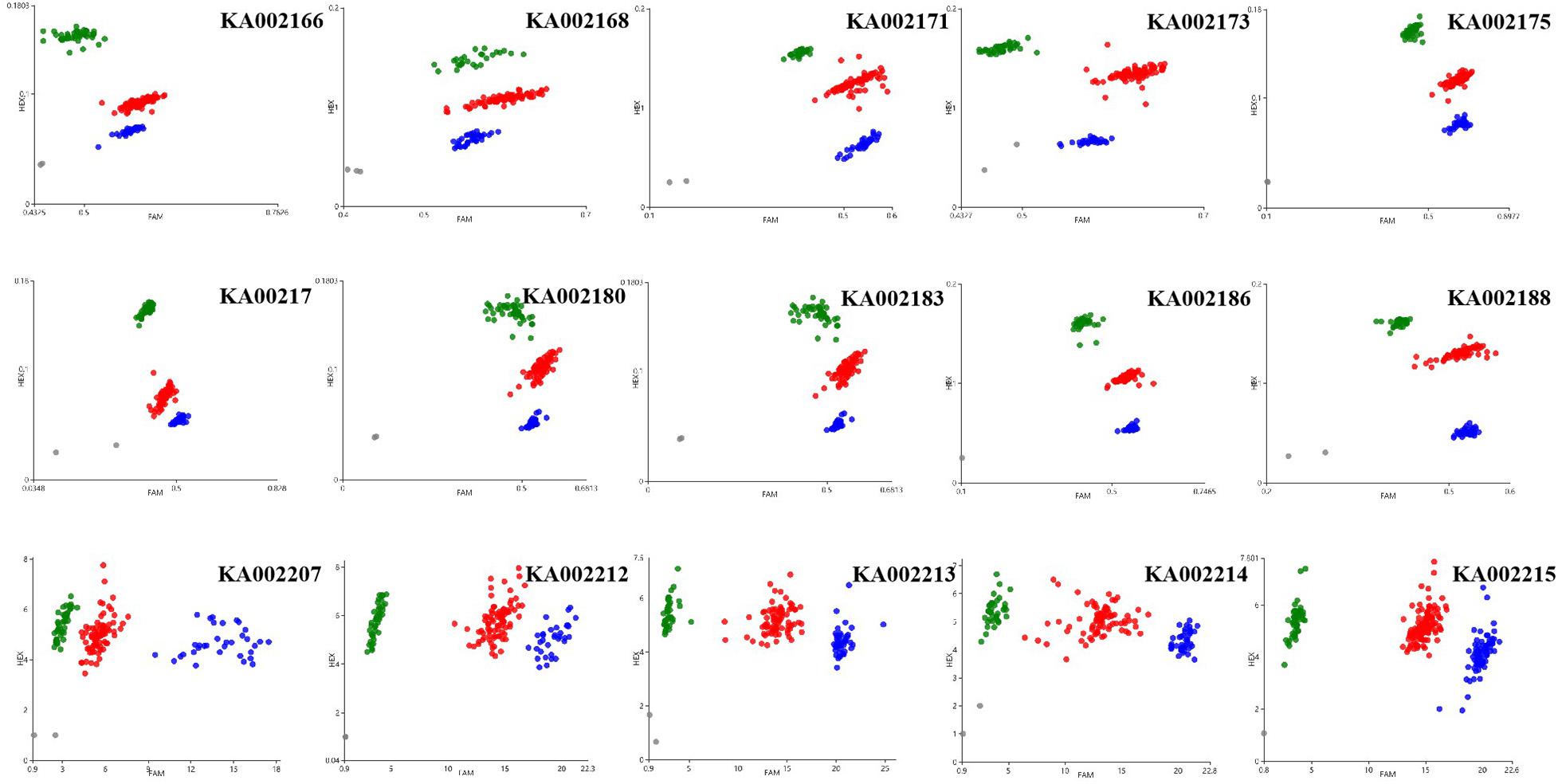
Figure 3. Results of genotyping using the Kompetitive Allele Specific PCR (KASP) assay. Scatter plots for selected KASP assays showing clustering of varieties on the X- (FAM) and Y- (HEX) axes. The green and blue dots represent the homozygous F2 lines and the red dot represents heterozygous F2 lines from the mapping population of the wm-6 × 12D-1 cross. The gray dots represent the NTC (non-template control).
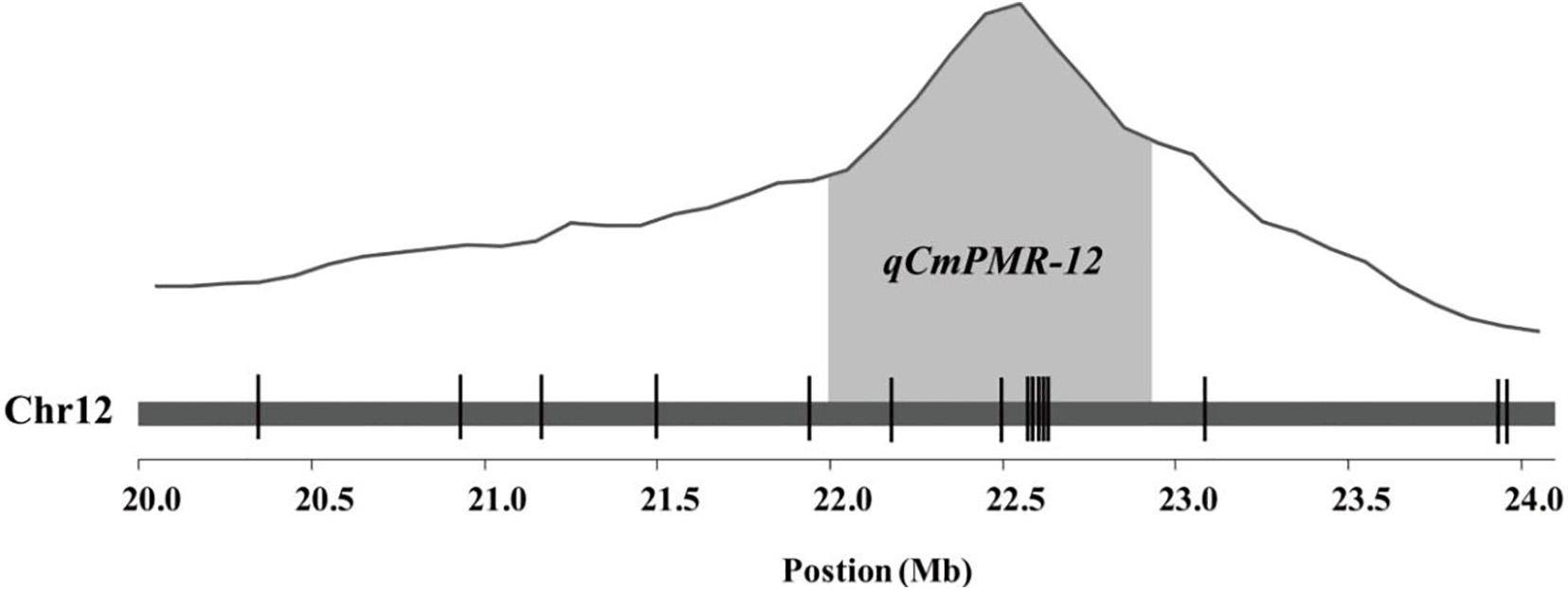
Figure 4. Quantitative trait locus (QTL) analysis of resistance to powdery mildew and the distribution of KASP markers linked to the target region. The gray areas represent the QTL region. The positions of KASP markers are shown by vertical lines.
Using the 15 KASP markers, the genetic physical map and linkage map were constructed by JoinMap 4.1 (Figures 5A,B). As shown in the linkage map, the peak of the QTL was located in a 0.6-cM interval spanned by KASP markers KA002213 and KA002215 (Figure 5B).
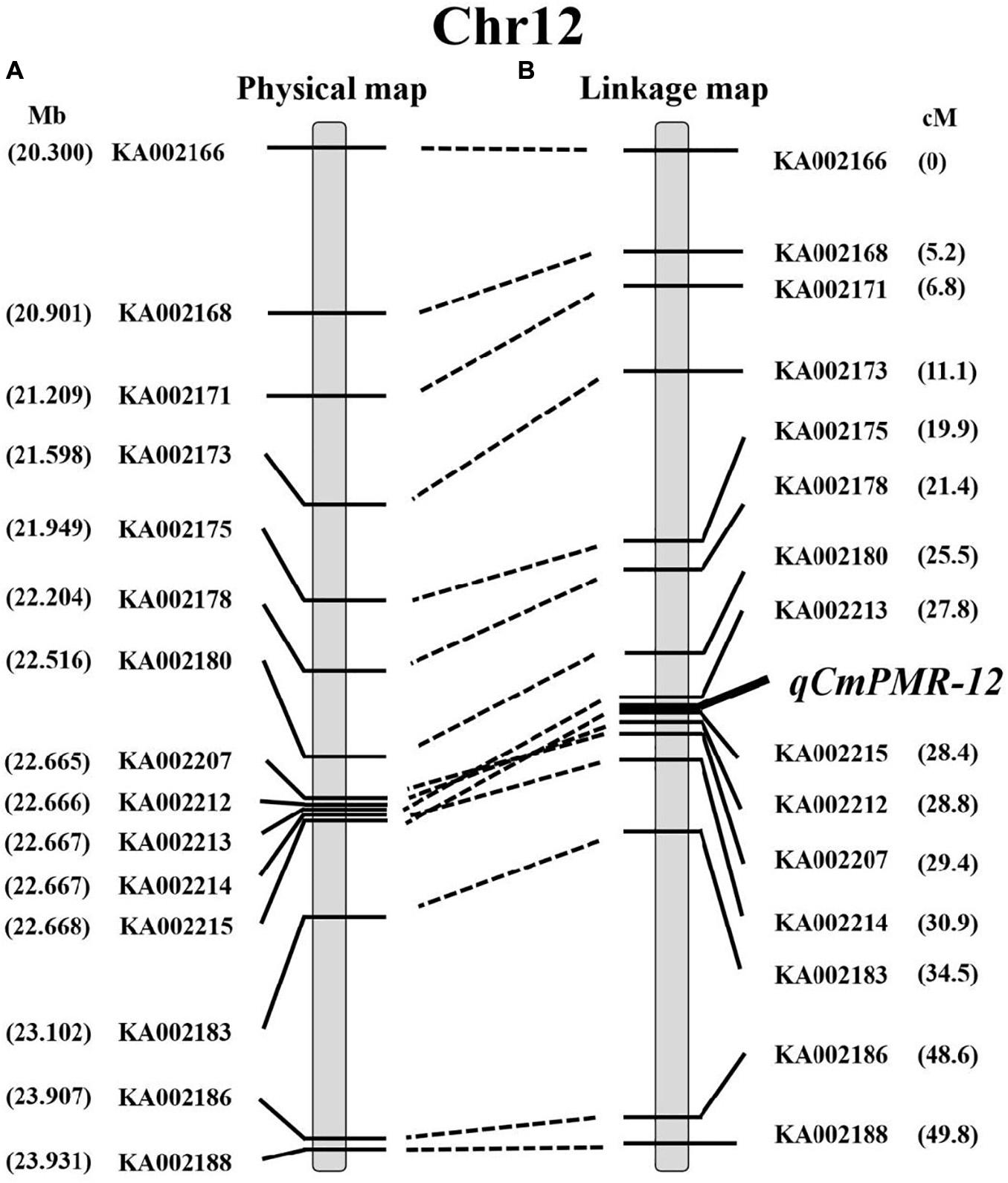
Figure 5. The location of qCmPMR-12 on melon chromosome 12. (A) The physical map around the region of qCmPMR-12; marker names are indicated to the left of the map. (B) The genetic linkage map of melon chromosome 12; map distances are shown on the right. The QTL region was identified by QTL mapping using phenotypic analysis and marker data from F2 populations.
Discussion
The melon cultivar wm-6 is a high-generation inbred line developed by our group, which is highly resistant to powdery mildew (Figure 1A, Table 1 and Supplementary Table 1). In this study, a genomic region ranging from 22.0 Mb to 22.9 Mb on chromosome 12 was identified using BSA-Seq technology. Similarly, Li et al. (2017) detected a major QTL for PM resistance on chromosome 12 and suggested that resistance to P. xanthii in MR-1 was controlled by a single dominant gene. However, in this study, the segregation ratio of the F2 population indicated that the PM resistance in wm-6 was controlled by a single recessive gene, implying a different genetic basis of the PM resistance mechanisms between the melon cultivars wm-6 and MR-1. Other studies have also demonstrated that the genetic basis of resistance to PM differs depending on the tested melon material (Epinat et al., 1992; Zhang et al., 2008; Sun et al., 2010; Yuste-Lisbona et al., 2010; McCreight and Coffey, 2011; Wang et al., 2016).
To identify the PM resistance gene in wm-6, RNA-Seq analysis was performed with the melon lines wm-6 and 12D-1 in parallel. Combination of the results from BSA-Seq and RNA-Seq suggested that the At3g12360-like gene MELO3C002434, which encodes an ANK protein was the most likely candidate gene to confer PM resistance, because only this gene was significantly more highly expressed in the resistant line wm-6 than that in the susceptible line 12D-1 following infection with P. xanthii (see text footnote 5). Previous studies revealed that ANK proteins have critical functions in various biological processes of plant growth and development as well as in response to biotic and abiotic stresses (Albert et al., 1999; Yan et al., 2002; Ha et al., 2004; Hemsley et al., 2005; Garcion et al., 2006; Sakamoto et al., 2008; Shen et al., 2010; Li et al., 2013). It has been reported that the ANK protein NPR1 is important both in the SA-dependent immune response and in SA-independent resistance responses induced by the root-associated bacteria (Cao et al., 1997; Ryals et al., 1997); AKR2 functions in the oxidative metabolism of disease resistance and stress response in Arabidopsis (Yan et al., 2002); a plasma membrane-localized ANK protein, ACD6, is involved in SA-dependent signaling in defense responses and programmed cell death (Lu et al., 2003, 2005), and ectopic expression of the ANK protein OsBIANK1 of rice confers enhanced disease resistance to Botrytis cinerea and Pseudomonas syringae in Arabidopsis (Li et al., 2013). Interestingly, the gene MELO3C002434 contained 10 SNPs and 1 InDel (Table 5) and 5 out of 15 KASP markers that developed in this study were located within this gene (Supplementary Table 5). All of these results further implied that MELO3C002434 is the most likely candidate gene associated with PM resistance in melon wm-6. In addition, the report of Li et al. (2017) suggested MELO3C002434 to be one of the genes related to PM resistance in spite of the different genetic basis of the PM resistance mechanism in melon wm-6 and MR-1. Therefore, further genetic studies and more detailed analyses are required to confirm the role and molecular mechanism of action of MELO3C002434 in the PM defense response.
In this study, 37 SNPs surrounding the candidate region were used to design KASP markers using the PolyMarker website and 15 of these were polymorphic among the bulk and parent populations. To the best of our knowledge, this is the first report that KASP markers have been developed and used in MAS to improve PM resistance in melon breeding, although several markers linked to resistance genes have been reported previously in melon (Ning et al., 2014; Han et al., 2015).
Conclusion
In this study, a major QTL that is associated with PM resistance was identified in a 0.9-Mb interval on chromosome 12 of melon using BSA-Seq technology. Additional RNA-Seq data suggested that an ankyrin repeat-containing gene within this region, MELO3C002434, was implicated to be the most important candidate gene. In addition, 15 suitable KASP markers that were tightly linked to the resistance phenotype were developed for the MAS of melon. These data can be used to improve PM resistance in breeding programs and to facilitate understanding of the molecular mechanisms that underlying PM resistance in melon.
Data Availability Statement
The raw sequence reads produced by BSA-Seq are deposited in the NCBI Sequence Read Archive (SRA; Accession number: PRJNA655764).
Author Contributions
YZ, YC, and HZ designed the research. YC and QD prepared the plant materials. YC performed the experiments, analyzed the data, and wrote the manuscript. YZ, YC, and HJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Shanghai Science and Technology Commission (20392000300), Shanghai Minhang Science and Technology Commission (2019MHC051), Excellent Team of Shanghai Academy of Agricultural Sciences (B-06), and Shanghai Melon and Fruit Industrial Technology System [(2020)1].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.593207/full#supplementary-material
Supplementary Table 1 | Means, standard errors and ranges of DSI of the parents and F1 plants at 12 dpi with P. xanthii in the year of 2019.
Supplementary Table 2 | The number of SNPs distributed on each chromosome of melon detected by BSA-Seq.
Supplementary Table 3 | The positions and annotations of candidate genes in the target region conferring powdery mildew resistance from 22.0 Mb to 22.9 Mb based on the melon reference genome (CM3.6.1).
Supplementary Table 4 | KASP primer sequences used for MAS in melon breeding programs.
Supplementary Table 5 | Genotypes and positions of the 15 KASP markers linked to powdery mildew resistance gene of melon.
Footnotes
- ^ http://polymarker.tgac.ac.uk
- ^ http://www.lgcgenomics.com
- ^ https://www.melonomics.net/
- ^ http://www.ncbi.nlm.nih.gov/sra
- ^ https://submit.ncbi.nlm.nih.gov/subs/sra/SUB8336313/
References
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Albert, S., Despres, B., Guilleminot, J., Bechtold, N., Pelletier, G., Delseny, M., et al. (1999). The EMB 506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J. 17, 169–179. doi: 10.1046/j.1365-313x.1999.00361.x
Candido, V., Campanelli, G., Viggiani, G., Lazzeri, L., and Camele, I. (2014). Melon yield response to the control of powdery mildew by environmentally friendly substances. Entia Horticult. 166, 70–77. doi: 10.1016/j.scienta.2013.12.008
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/s0092-8674(00)81858-9
Chen, X. M. (2013). High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 04, 608–627. doi: 10.4236/ajps.2013.43080
Chen, X. M. (2014). Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 36, 311–326. doi: 10.1080/07060661.2014.924560
Cheng, H., Kong, W., Su, Y., Liu, D., and He, Q. (2011). Identification on powdery mildew and physiological race of melon in china. J. Changjiang Vegetables 18, 1–5. doi: 10.29405/j.bes/411-104725
Clements, J. (2014). Powdery mildew resistance gene of ssr molecular markers linked to the ‘wmr-29’ strains of melon. Northern Horticult. 61, 347–357.
Dobbels, A. A., Michno, J. M., Campbell, B. W., Virdi, K. S., Stec, A. O., Muehlbauer, G. J., et al. (2017). An Induced Chromosomal Translocation in Soybean Disrupts a KASI Ortholog and Is Associated with a High-Sucrose and Low-Oil Seed Phenotype. G3 7, 1215–1223. doi: 10.1534/g3.116.038596
Epinat, C. M., Pitrat, M., and Bertrand, F. (1992). Genetic analysis of resistance of 5 melon lines to powdery mildews. Acta Horticult. 65, 135–144. doi: 10.1007/bf00022575
Ertiro, B. T., Ogugo, V., and Worku, M. (2015). Comparison of Kompetitive Allele Specific PCR (KASP) and genotyping by sequencing (GBS) for quality control analysis in maize. BMC Genomics 16:908. doi: 10.1186/s12864-015-2180-2
Fukino, N., Ohara, T., Monforte, A. J., Sugiyama, M., Sakata, Y., Kunihisa, M., et al. (2008). Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.). Theoret. Appl. Genet. 118, 165–175. doi: 10.1007/s00122-008-0885-1
Garcia-Mas, J., Benjak, A., Sanseverino, W., Bourgeois, M., and Baulcombe, D. C. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. 109, 11872–11877.
Garcion, C., Guilleminot, J., Kroj, T., Parcy, F., Giraudat, J., and Devic, M. (2006). AKRP and EMB506 are two ankyrin repeat proteins essential for plastid differentiation and plant development in Arabidopsis. Plant J. 48, 895–906. doi: 10.1111/j.1365-313x.2006.02922.x
Gu, H. F., Zhang, X., Zhang, W. F., Jin, C. Y., Xu, L., Lu, X. L., et al. (2010). Identification of physiological races of watermelon and melon powdery mildew in shanghai. Acta Agricult. Shanghai 26, 155–158.
Ha, C. M., Jun, J. H., Nam, H. G., and Fletcher, J. C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45, 1361–1370. doi: 10.1093/pcp/pch201
Han, Y., Lv, P., Hou, S., Li, S., Ji, G., Ma, X., et al. (2015). Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in sorghum (Sorghum bicolor L. Moench). PLoS One 10:e0127065. doi: 10.1371/journal.pone.0127065
He, C., Holme, J., and Anthony, J. (2014). SNP genotyping: the KASP assay. Methods Mol. Biol. 1145, 75–86. doi: 10.1007/978-1-4939-0446-4_7
Hemsley, P. A., Kemp, A. C., and Grierson, C. S. (2005). The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17, 2554–2563. doi: 10.1105/tpc.105.031237
Hill, J. T., Demarest, B. L., Bisgrove, B. W., Gorsi, B., Su, Y. C., and Yost, H. J. (2013). Mmappr: Mutation Mapping Analysis Pipeline for Pooled Rna-Seq. Genome Res. 23, 687–697. doi: 10.1101/gr.146936.112
Hollomon, D. W., Wheeler, I. E., Bélanger, R. R., Bushnell, W. R., Dik, A. J., and Carver, T. L. W. (2002). “The Powdery Mildews,” in A Comprehensive Treatise, eds R. R. Bélanger, W. R. Bushnell, A. J. Dik, and T. L. W. Carver (Saint Paul, MN: APS Press), 249–255.
Křístková, E., Lebeda, A., and Sedláková, B. (2009). Species spectra, distribution and host range of cucurbit powdery mildews in the Czech Republic, and in some other European and Middle Eastern countries. Phytoparasitica 37, 337–350. doi: 10.1007/s12600-009-0045-4
Li, B., Zhao, Y., Zhu, Q., Zhang, Z., Fan, C., Amanullah, S., et al. (2017). Mapping of powdery mildew resistance genes in melon (Cucumis melo L.) by bulked segregant analysis. Sci. Horticult. 220, 160–167. doi: 10.1016/j.scienta.2017.04.001
Li, D., Wang, F., Liu, B., Zhang, Y., Huang, L., Zhang, H., et al. (2013). Ectopic Expression of Rice OsBIANK1, Encoding an Ankyrin Repeat-Containing Protein, in Arabidopsis Confers Enhanced Disease Resistance to Botrytis cinerea and Pseudomonas syringae. J. Phytopathol. 161, 27–34. doi: 10.1111/jph.12023
Li, P. F., Zhu, L. L., Yang, X. P., Xu, J. H., Liu, G., Yao, X. F., et al. (2015). Species and physiological race identification of melon powdery mildew fungus in Jiangsu, Zhejiang and Shanghai areas. China Cucurbits Vegetables 28, 16–20.
Liang, Q. L., Xu, B. L., and Yan, H. X. (2010). Hull-less pumpkin powdery mildew and its host range. Mycosystema 29, 636–643.
Liu, L. Z., Chen, Y. Y., Su, Z. H., Zhang, H., and Zhu, W. M. (2010). A sequence-amplified characterized region marker for a single, dominant gene in melon PI 134198 that confers resistance to a unique race of podosphaera xanthii in china. Hortscience 45, 1407–1410. doi: 10.21273/hortsci.45.9.1407
Lu, H., Lin, T., Klein, J., Wang, S., Qi, J., Zhou, Q., et al. (2014). QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127, 1491–1499. doi: 10.1007/s00122-014-2313-z
Lu, H., Liu, Y., and Greenberg, J. T. (2005). Structure-function analysis of the plasma membrane-localized Arabidopsis defense component ACD6. Plant J. 44, 798–809. doi: 10.1111/j.1365-313x.2005.02567.x
Lu, H., Rate, D. N., Song, J. T., and Greenberg, J. T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420. doi: 10.1105/tpc.015412
Magwene, P. M., Willis, J. H., and Kelly, J. K. (2011). Te Statistics of Bulk Segregant Analysis Using Next Generation Sequencing. PLoS Comput. Biol. 7:e1002255. doi: 10.1371/journal.pcbi.1002255
McCreight, J. D., and Coffey, M. D. (2011). Inheritance of resistance in melon PI 313970 to cucurbit powdery mildew incited by podosphaera xanthii race s. HortScience 46, 838–840. doi: 10.21273/hortsci.46.6.838
McGrath, T. M. (2001). Fungicide resistance in cucurbit powdery mildew: experiences and challenges. Plant Dis. 85, 236–245. doi: 10.1094/pdis.2001.85.3.236
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations Proc. Natl. Acad. Sci. U S A 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Nie, J., He, H., Peng, J., Yang, X., Bie, B., Zhao, J., et al. (2015). Identification and fine mapping ofpm5.1: a recessive gene for powdery mildew resistance in cucumber (cucumis sativus L.). Mole. Breed. 35:7.
Ning, X., Wang, X., Gao, X., Zhang, Z., Zhang, L., Yan, W., et al. (2014). Inheritances and location of powdery mildew resistance gene in melon Edisto47. Euphytica 195, 1–9.
Perchepied, L., Bardin, M., Dogimont, C., and Pitrat, M. (2005). Relationship Between Loci Conferring Downy Mildew and Powdery Mildew Resistance in Melon Assessed by Quantitative Trait Loci Mapping. Phytopathology 95, 556–565. doi: 10.1094/phyto-95-0556
Perchepied, L., Marc, B., Catherine, D., and Pitrat, M. (2005). Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology 95, 556–565. doi: 10.1094/phyto-95-0556
Perez-Garcia, A., Romero, D., Fernandez-Ortuno, D., Lopez-Ruiz, F., De Vicente, A., and Tores, J. A. (2009). The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mole. Plant Pathol. 10, 153–160.
Périn, C., Hagen, L., Conto, V. D., Katzir, N., Danin-Poleg, Y., Portnoy, V., et al. (2002). A reference map of Cucumis melo based on two recombinant inbred line populations. Theor. Appl. Genet 104, 1017–1034. doi: 10.1007/s00122-002-0864-x
Pitrat, M. (1991). Linkage groups in Cucumis melo L. Rep. Cucurbit Genetics Cooperative 82, 406–411. doi: 10.1093/oxfordjournals.jhered.a111112
Rafalski, A. (2002). Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5, 94–100. doi: 10.1016/s1369-5266(02)00240-6
Rasheed, A., Wen, W., Gao, F., Zhai, S., Jin, H., Liu, J., et al. (2016). Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theoret. Appl. Genet. 129, 1843–1860. doi: 10.1007/s00122-016-2743-x
Ribaut, J. M., Banziger, M., Betran, J., Jiang, C., Edmeades, G. O., Dreher, K., et al. (2002). “Use of molecular markers in plant breeding: Drought tolerance improvement in tropical maize,” in itor, Quantitative genetics, genomics, and plant breeding, ed. M. S. Kang (Wallingford, UK: CABI Publishing).
Romero, D., Rivera, M. E., Cazorla, F. M., Codina, J. C., Fernandez-Ortuno, D., Tores, J. A., et al. (2008). Comparative histochemical analyses of oxidative burst and cell wall reinforcement in compatible and incompatible melon-powdery mildew (Podosphaera fusca) interactions. J. Plant Physiol. 165, 1895–1905. doi: 10.1016/j.jplph.2008.04.020
Ryals, J., Weymann, K., and Lawton, K. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9, 425–439. doi: 10.1105/tpc.9.3.425
Sakamoto, H., Matsuda, O., and Iba, K. (2008). ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant J. 56, 411–422. doi: 10.1111/j.1365-313X.2008.03614.x
Schlotterer, C. (2004). The evolution of molecular markers-just a matter of fashion? Nat. Rev. Genet 5, 63–69. doi: 10.1038/nrg1249
Semagn, K., Babu, R., Hearne, S., and Olsen, M. (2014). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mole. Breed. 33, 1–14. doi: 10.1007/s11032-013-9917-x
Shen, G., Kuppu, S., Venkataramani, S., Wang, J., Yan, J., Qiu, X., et al. (2010). ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membranebound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell 22, 811–831. doi: 10.1105/tpc.109.065979
Song, J., Li, Z., Liu, Z., Guo, Y., and Qiu, L. (2017). Next-Generation Sequencing from Bulked-Segregant Analysis Accelerates the Simultaneous Identification of Two Qualitative Genes in Soybean. Front. Plant Sci. 8, 919–919. doi: 10.3389/fpls.2017.00919
Soyk, S., Lemmon, Z. H., Oved, M., Fisher, J., Liberatore, K. L., Park, S. J., et al. (2017). Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 169, 1142–1155 e1112.
Sun, X. L., Liu, D., Zhang, H. Q., Huo, N. X., Zhou, R. H., and Jia, J. Z. (2010). Identification and mapping of two new genes conferring resistance to powdery mildew from aegilops tauschii (coss.) schmal. J. Integr. Plant Biol. 48, 1204–1209. doi: 10.1111/j.1744-7909.2006.00328.x
Tan, C., Assanga, S., Zhang, G., Rudd, J. C., Haley, S. D., Xue, Q., et al. (2017). Development and Validation of KASP Markers for Wheat Streak Mosaic Virus Resistance Gene Wsm2. Crop Sci. 57, 340–349. doi: 10.2135/cropsci2016.04.0234
Teixeira, A., Matosobarreto, A. P., and Aranha, L. E. (2008). An AFLP marker linked to the Pm-1 gene that confers resistance to Podosphaera xanthii race 1 in Cucumis melo. Genet. Mol. Biol. 31, 547–550. doi: 10.1590/s1415-47572008000300023
Wang, X. Z., Xu, B. Y., Wang, P., Luan, F. S., Ma, H. Y., and Ma, Y. Y. (2013). Identification of Powdery Mildew Pathogen and Ribosomal DNA-ITS Sequence Analysis on Melon. J. Northeast Agricult. Univ. 10, 18–97.
Wang, Y. H., Wu, D. H., Huang, J. H., Tsao, S. J., Hwu, K. K., and Lo, H. F. (2016). Mapping quantitative trait loci for fruit traits and powdery mildew resistance in melon (cucumis melo). Botanical. Stud. 57:19.
Wang, Y., Ning, Z., Hu, Y., Chen, J., Zhao, R., Chen, H., et al. (2015). Molecular Mapping of Restriction-Site Associated DNA Markers in Allotetraploid Upland Cotton. PLoS One 10:e0124781. doi: 10.1371/journal.pone.0124781
Win, K. T., Vegas, J., Zhang, C., Song, K., and Lee, S. (2017). QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theoret. Appl. Genet. 130, 199–211. doi: 10.1007/s00122-016-2806-z
Wolter, M., Hollricher, K., Salamini, F., and Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. MGG 239, 122–128. doi: 10.1007/bf00281610
Xu, X., Lu, L., Zhu, B., Xu, Q., Qi, X., and Chen, X. (2015). QTL mapping of cucumber fruit flesh thickness by slaf-seq. Scient. Rep. 5:15829.
Yan, J., Wang, J., and Zhang, H. (2002). An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J. 29, 193–202. doi: 10.1046/j.0960-7412.2001.01205.x
Yang, Z., Huang, D., Tang, W., Zheng, Y., Liang, K., Cutler, A. J., et al. (2013). Mapping of Quantitative Trait Loci Underlying Cold Tolerance in Rice Seedlings via High-Throughput Sequencing of Pooled Extremes. PLoS One 8:e68433. doi: 10.1371/journal.pone.0068433
Yustelisbona, F. J., Capel, C., Sarria, E., Torreblanca, R., Gomezguillamon, M. L., Capel, J., et al. (2011). Genetic linkage map of melon (Cucumis melo L.) and localization of a major QTL for powdery mildew resistance. Mole. Breed. 27, 181–192. doi: 10.1007/s11032-010-9421-5
Yuste-Lisbona, F. J., López-Sesé, A. I., and ómez-Guillamón, M. L. G. (2010). Inheritance of resistance to races 1, 2 and 5 of powdery mildew in the melon tgr-1551. Plant Breed. 129, 72–75. doi: 10.1111/j.1439-0523.2009.01655.x
Zhang, C., Ren, Y., and Guo, S. (2013). Application of comparative genomics in developing markers tightly linked to the Pm-2f gene for powdery mildew resistance in melon (Cucumis Melo L.). Euphytica 190, 1–12. doi: 10.12816/0011695
Zhang, H. Y., Su, F., Guo, S. G., Gong, G. Y., Yi, H. P., Wu, M. Z., et al. (2008). Genetic analysis and specific fragments linked to powdery mildew resistant gene Pm-2F in melon. Acta Horticult. Sin. 12, 1773–1780. doi: 10.16420/j.issn.0513-353x.2008.12.009
Zhang, H., Guo, S., Gong, G., Ren, Y., Davis, A. R., and Xu, Y. (2011). Sources of resistance to race 2WF powdery mildew in U.S. watermelon plant introductions. Hortscience 46, 1349–1352. doi: 10.21273/hortsci.46.10.1349
Zhang, H., Wang, X., Pan, Q., Li, P., Liu, Y., Lu, X., et al. (2019). QTG-Seq Accelerates QTL Fine Mapping through QTL Partitioning and Whole-Genome Sequencing of Bulked Segregant Samples. Mol. Plant 12, 426–437. doi: 10.1016/j.molp.2018.12.018
Zhang, S. X., Cui, J., Song, Y. Y., and Liu, S. Q. (2007). Occurrence and control of the melon cultivar. Decid. Fruits 39, 41–42.
Zhang, X. J., Ji, J., Zhai, W. Q., Li, M. H., Wang, H. J., and Yi, H. P. (2013). Identification of physiological races of powdery mildew on Cucumis melo ssp. melo in xinjiang. Xinjiang Agricult. Sci. 50, 1450–1455.
Zheng, W., Wang, Y., Wang, L., Ma, Z., Zhao, J., Wang, P., et al. (2016). Genetic mapping and molecular marker development for Pi65(t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theoret. Appl. Genet. 129, 1035–1044. doi: 10.1007/s00122-016-2681-7
Zhu, H., Zhai, W., Li, X., and Zhu, Y. (2019). Two QTLs controlling clubroot resistance identified from bulked segregant sequencing in pakchoi (brassica campestris ssp. chinensis makino). Scient. Rep. 9:9228.
Keywords: bulked segregant analysis, KASP markers, melon, powdery mildew, resistance gene
Citation: Cao Y, Diao Q, Chen Y, Jin H, Zhang Y and Zhang H (2021) Development of KASP Markers and Identification of a QTL Underlying Powdery Mildew Resistance in Melon (Cucumis melo L.) by Bulked Segregant Analysis and RNA-Seq. Front. Plant Sci. 11:593207. doi: 10.3389/fpls.2020.593207
Received: 10 August 2020; Accepted: 28 October 2020;
Published: 05 February 2021.
Edited by:
Magdalena Arasimowicz-Jelonek, Adam Mickiewicz University, PolandReviewed by:
Christina Cowger, Plant Science Research Unit (USDA-ARS), United StatesZhiyong Liu, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China
Copyright © 2021 Cao, Diao, Chen, Jin, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongping Zhang, enlwMTIzOTQ0QDEyNi5jb20=; Hongmei Zhang, emhhbmdob25nbWVpQHNhYXMuc2guY24=
 Yanyan Cao
Yanyan Cao Qiannan Diao
Qiannan Diao Youyuan Chen
Youyuan Chen Haijun Jin
Haijun Jin Yongping Zhang
Yongping Zhang Hongmei Zhang
Hongmei Zhang