- Research Faculty of Agriculture, Graduate School of Agriculture, Hokkaido University, Sapporo, Japan
Some Asiatic hybrid lily cultivars develop bicolor tepals, which consist of anthocyanin-pigmented upper halves and un-pigmented lower halves. MYB12, a subgroup 6 member of R2R3-MYB that positively regulates anthocyanin biosynthesis, is downregulated in the lower halves. However, MYB12 is usually expressed over entire tepal regions in numerous lily cultivars. Why MYB12 of bicolor cultivars exhibits variable expression spatially in a single tepal remains unclear. Since the lily MYB12 mRNA harbored a binding site for microRNA828 (miR828), the involvement of miR828 in variable spatial accumulation of MYB12 transcripts was evaluated. We analyzed the cleavage of MYB12 mRNA, mature miR828 accumulation, and MYB12 transcript-derived siRNA generation (microRNA-seq). In the bicolor tepals, mature miR828 was more highly accumulated in the lower halves than in the upper halves, and miR828-directed cleavage of MYB12 transcripts was observed predominantly in the lower halves. Moreover, the cleavage triggered the production of secondary siRNA from MYB12 transcripts, and the siRNAs were accumulated predominantly in the lower halves. Consequently, miR828 suppressed MYB12 transcript accumulation in the white region, and the miR828/MYB12 module participated in the development of bicolor patterns in lily flowers. The results present the first example of a microRNA mediating flower color patterns. Finally, we discuss the potential of miR828 creating flower color variations through suppressing the activity of subgroup 6 R2R3-MYB positive regulators in other species.
Introduction
Anthocyanin color patterns are often observed in flowers, including spots, stripes, venation, bud-blush, and bicoloration (two colors in a single petal). The color patterns create major variations in flower appearance and often enhance the ornamental value of flowers. Consequently, the exploration of the underlying mechanisms involved in the development of anthocyanin color patterns could facilitate the breeding of floricultural crops (Nakayama, 2014). Mechanisms of development of anthocyanin color patterns have been reported in some plant species, mainly model plants (Davies et al., 2012; Glover et al., 2013; Albert et al., 2014). The primary mechanism is transcriptional regulation of anthocyanin biosynthesis genes. Subgroup 6 members of R2R3-MYB transcription factors in numerous species and subgroup 5 members of R2R3-MYB transcription factors in orchids predominantly regulate anthocyanin biosynthesis in flowers (the grouping of R2R3-MYBs is according to Stracke et al., 2001), and single plant species often have a couple of R2R3-MYB genes grouped into the subgroups. Each of the genes exhibits spatially and temporally distinct expression profiles and causes restricted pigment deposition, resulting in the development of varied color patterns. For example, petunias have four subgroup 6 R2R3-MYB genes: PhAN2 paints the entire petal region, PhAN4 causes pigmentation in the flower tubes, and DEEP PURPLE and PURPLE HAZE are responsible for venation and bud-blush pigmentation patterns, respectively (Albert et al., 2011). In Phalaenopsis, three subgroup 5 R2R3-MYB genes regulate full-red pigmentation, red spots, and venation patterns, respectively, in petals (Hsu et al., 2015).
Lilies (Lilium spp.) are among the most commercially valuable and globally cultivated floriculture crops (Ministry of Agriculture, Forestry and Fisheries of Japan, 2019). Interspecific hybridization is the principal method of lily breeding. The Lilium species are classified into several sections, and Asiatic hybrid lilies (Lilium spp.) are derived from crosses among species in Sinomartagon and Daurolirion section (Yamagishi and Nakatsuka, 2017; Marasek-Ciolakowska et al., 2018). Since lilies are a major floricultural crop, genetic evaluation of their agricultural traits facilitates the improvement of the character traits of cultivars. However, lilies are not model species, and their genetic evaluation is challenging considering Lilium species have a large genome size (33–36 Gb; Bennett and Leitch, 2011) and long life cycles (3–7 years from sowing to anthesis), and its stable transformation remains challenging.
A high variation in flower color hues is the major selling point for Asiatic hybrid lilies, which accumulate anthocyanins (cyanidin 3-rutinoside, Nørbæk and Kondo, 1999) and carotenoids (Wang and Yamagishi, 2019) in flowers. In addition, a variety of anthocyanin color patterns, such as restricted pigmentation at tepal bases (large spots or blotches) or the upper halves (bicolor), and several types of spots, including raised spots, splatter-type spots, and brush mark spots, are often observed in Asiatic hybrid lily cultivars (Yamagishi, 2013). The underlying mechanisms of such color patterns have been evaluated. Several subgroup 6 members of the R2R3-MYB genes are expressed in lily flowers: MYB12 usually paints entire tepal regions (Yamagishi et al., 2010), MYBSPLATTER (formerly LhMYB12-Latvia, Yamagishi, 2020a) causes splatter-type anthocyanin spots (Yamagishi et al., 2014), MYB19Short and MYB19Long are responsible for raised-spot pigmentation (Yamagishi, 2020b), MYB19Long is indispensable for brush mark pigmentation (Yamagishi, 2020c), and MYB18 is involved in the development of large spots (blotches) at tepal bases (Yamagishi, 2018). Therefore, transcriptional regulation, governed by subgroup 6 members of R2R3-MYB genes, is the primary mechanism of the establishment of a variety of color patterns in lilies.
Some Asiatic hybrid lily cultivars, including Lollypop, develop bicolor tepals where anthocyanin accumulates in the upper halves of tepals but scarcely accumulates at the lower halves of tepals (Figure 1). We have previously compared transcriptome profiles between the upper and lower halves of Lollypop tepals and clarified that the entire anthocyanin biosynthesis pathway is suppressed at the lower halves (Suzuki et al., 2016). Therefore, the bicolor pattern in Lollypop is caused by transcriptional regulation of anthocyanin biosynthesis genes. Putative transcription factors that positively or negatively regulate anthocyanin biosynthesis are surveyed using transcriptome data, among which, certain negatively regulating transcription factors are expressed in Lollypop tepals, including R3-MYB suppressors (Sakai et al., 2019). If such negative regulators are involved in bicolor formation, their expression would be expected to be high in white tepal regions. To date, however, there have been no reports of putative suppressors exhibiting higher expression in the unpigmented regions of flowers (Suzuki et al., 2016; Sakai et al., 2019). Three R2R3-MYB sequences belonging to subgroup 6 are included in the Lollypop transcriptome. Among the three R2R3-MYB sequences, MYB12 is expressed mainly in the tepals, and its expression levels are higher in the upper halves than in the lower halves of the tepals, suggesting that MYB12 is the main transcription factor causing varying anthocyanin biosynthesis activities, and mediating bicolor anthocyanin pigmentation patterns (Suzuki et al., 2016). However, since MYB12 is often responsible for the pigmentation of whole tepal regions in many lilies (Yamagishi et al., 2010, 2012; Yamagishi, 2011), it remains unclear why and how MYB12 expression is suppressed in the lower halves of bicolor tepals.
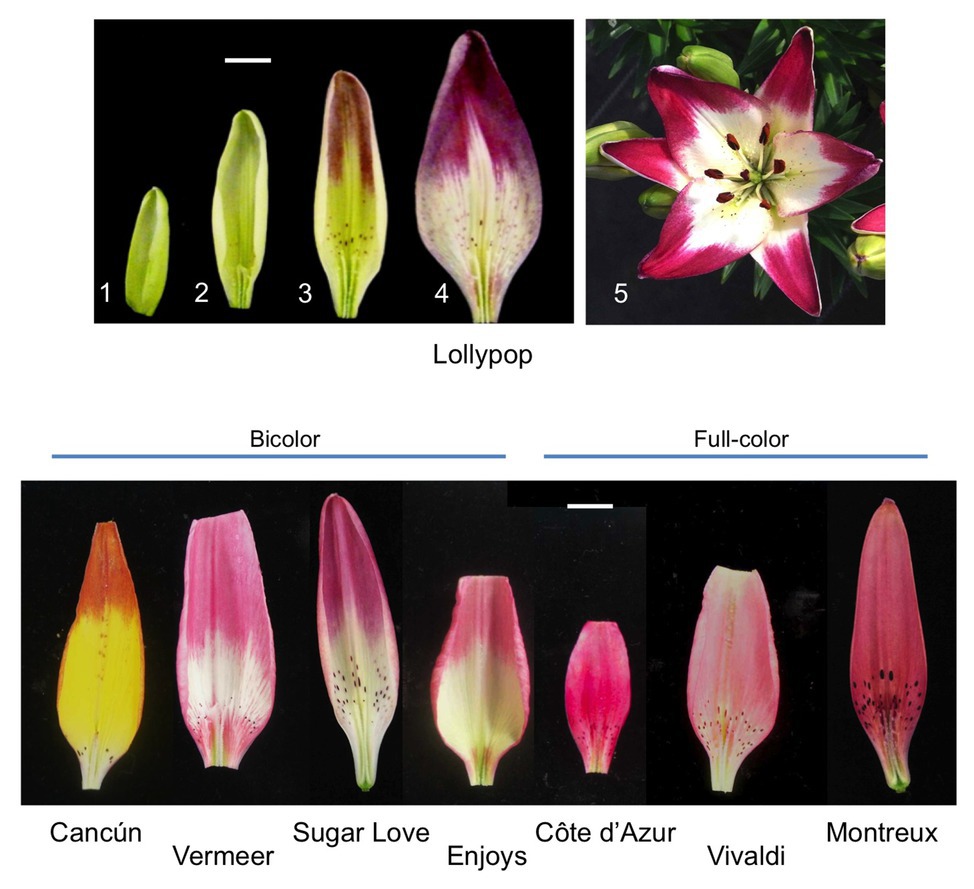
Figure 1. Upper panels: Inner tepals (adaxial surface) of the Lollypop cultivar at floral development stages 1, 2, 3, and 4 and a whole flower of Lollypop at anthesis (stage 5). Lower panel: Inner tepals (adaxial surface) of four bicolor (Cancún, Vermeer, Sugar Love, and Enjoys) and three full-color (Côte d’Azur, Vivaldi, and Montreux) Asiatic hybrid lily cultivars at flower bud development stage 4. The yellow color of Cancún tepals is derived from carotenoids. White bar = 1 cm.
MicroRNAs are non-coding RNA species that play key roles in plant development and stress responses. MicroRNAs usually suppress the activity of target genes post-transcriptionally through cleavage of the transcripts or translational attenuation. Many transcription factor genes, including MYB family genes, are targets of microRNAs. MicroRNA production starts from primary microRNA (pri-miRNA), which is cleaved in the nucleus into smaller precursor microRNA (pre-miRNA) with a hairpin structure. These pre-miRNAs are then exported to the cytoplasm and cleaved by DICER-LIKE 1 (DCL1) to form 20–24 nucleotide-long mature microRNAs. Subsequently, the mature microRNAs are loaded into the ARGONAUTE (AGO) effector protein, a part of RNA-induced silencing complexes (RISCs). Afterward, the mature microRNAs guide the RISCs to target mRNA(s) through sequence complementarity and eventually mediate gene silencing (Zhang, 2015; D’Ario et al., 2017). In addition, some 22 nucleotides-long microRNAs are capable of initiating secondary small interfering RNAs (siRNAs). The RNA fragments downstream (or upstream in rare cases) of the cleaved site are converted into a double-stranded RNA molecule by RNA-dependent RNA polymerase 6, which is processed by the DCL4 enzyme into approximately 21 nucleotide-long siRNAs, which are called phased siRNAs (phasiRNAs). Some phasiRNAs can further regulate the expression of other target genes in trans and are called trans-acting siRNAs (tasiRNAs; Shivaprasad et al., 2012; Fei et al., 2013; Deng et al., 2018).
Among the microRNAs identified in a wide range of plant species, miR828, miR858, and miR159 are capable of targeting R2R3-MYB genes (Li and Lu, 2014; Jia et al., 2015; Sharma et al., 2016). Among them, miR828 often suppresses the expression of R2R3-MYB genes belonging to subgroup 15 and subgroup 4, which negatively regulate anthocyanin accumulation (Xia et al., 2012; Zhu et al., 2012). Therefore, anthocyanin pigment amounts increase in fruits or tubers with an increase in miR828 accumulation (Bonar et al., 2018; Tirumalai et al., 2019). However, in Arabidopsis, miR828 directly targets AtMYB113 and indirectly targets AtMYB75 (AtPAP1) and AtMYB113 through TAS4-siR81(-), one of the tasiRNAs derived from Trans-Acting SiRNA Gene 4 (TAS4) transcripts (Rajagopalan et al., 2006). Since AtMYB75 and AtMYB113 are subgroup 6 members of R2R3-MYB that positively regulate anthocyanin biosynthesis, miR828 and TAS4 suppress anthocyanin biosynthesis and are involved in feedback regulation of anthocyanin accumulation in vegetative tissues under stress conditions (Luo et al., 2012).
An miR828 target sequence was found in the nucleotide sequence of lily MYB12 (shown in the results section); however, sequences potentially recognized by miR159, miR858, or TAS4-siR81(-) were not detected. In addition, partial sequences of pri-MIR828 were included in the Lollypop transcriptome (Suzuki et al., 2016). Therefore, the potential of miR828 to suppress MYB12 expression predominantly in the white tepal region was investigated in the present study. We demonstrated that lily miR828 was highly accumulated in the white tepal region and cleaved MYB12 mRNA. MYB12 transcript-derived secondary siRNA molecules investigated using microRNA-seq were also highly accumulated in the white region. Consequently, we conclude that the miR828/MYB12 module has a key role in the development of bicolor tepals in lilies.
Materials and Methods
Plant Materials
Five Asiatic hybrid lily bicolor cultivars, Lollypop, Cancún, Vermeer, Sugar Love, and Enjoys, and three full-color cultivars, Côte d’Azur, Vivaldi, and Montreux (2n = 2x = 24), were examined in the present study (Figure 1). The lily plants were grown in a greenhouse (unheated and natural photoperiod) in the experimental farm of Hokkaido University, Sapporo, Japan. The flower bud developmental stages were defined according to Nakatsuka et al. (2003), i.e., no obvious anthocyanin pigmentation in stage 1, emergence of anthocyanin color only on raised spots in stage 2, beginning of anthocyanin coloration of adaxial tepal surface in stage 3, full pigmentation of adaxial tepal surface in stage 4 (1 day before anthesis), and flower blossoming in stage 5 (Figure 1). Tobacco plants (Nicotiana tabacum “Xanthi NC”) were grown in a growth chamber at 20°C under a 16-h light-8-h dark photoperiod.
Extraction of RNA
Low molecular weight (LMW) RNA was isolated from lily organs using a High Pure miRNA Isolation Kit (Roche Diagnostics K. K., Tokyo, Japan). Total RNA was extracted from lily tepals and tobacco leaves using a NucleoSpin® RNA kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany).
Sequencing
The full-length cDNA sequence of pri-MIR828 was obtained by rapid amplification of cDNA ends (RACE)-PCR (Lai et al., 2012) using gene-specific primers (Supplementary Table S1) designed using the partial pri-MIR828 sequence (Suzuki et al., 2016) and total RNA isolated from Lollypop tepals. The amplified fragments were cloned into the pGEM-T Easy Vector (Promega, Tokyo, Japan) and sequenced [DNA DataBank of Japan (DDBJ) accession numbers LC569960 and LC569961]. The nucleotide sequences were aligned using the default parameters in Clustal W in the Kyoto University Bioinformatics Center website.1
RNA ligase mediated (RLM)-RACE PCR was carried out to validate the targets of miR828. RNA adapter and total RNA isolated from lily tepals or tobacco leaves were ligated using RNA ligase (Takara, Otsu, Japan). The ligated RNA was used for cDNA synthesis using the reverse primer of MYB12 and PrimeScript II reverse transcriptase (Takara). Nested PCR was performed using the two sets of primers. Sequences of the RNA adapter and primers are listed in Supplementary Table S1. The amplified fragments of the appropriate size were cloned into the pMD20-T vector (Takara) and sequenced.
Agrobacterium-Mediated Transient Assay (Agroinfiltration)
The putative pre-MIR828 sequence (141 bp), which included the guide and passenger strands, and made a hairpin structure, was PCR-amplified using primers containing XbaI or SacI recognition sites (Supplementary Table S1). Subsequently, the MIR828 fragment was inserted downstream of the constitutive cauliflower mosaic virus 35S promoter (35S-p) using the restriction sites. Other plasmid constructs were derived from Yamagishi (2016, 2018).
Agrobacterium tumefaciens (EHA105) harboring the 35S-p::MIR828 construct and harboring the 35S-p::MYB12 construct were separately or simultaneously infiltrated into tobacco leaves using a syringe (Suzuki et al., 2015). In another experiment, A. tumefaciens harboring 35S-p::MIR828, 35S-p::MYB12, 35S-p::LhbHLH2, or Lilium hybrid dihydroflavonol 4-reductase (LhDFR)-promoter::intron-containing ß-glucuronidase (iGUS) constructs were infiltrated into several combinations. Six or three days after infiltration, leaf segments were harvested and gene expression levels were evaluated by quantitative reverse transcription PCR (qRT-PCR).
Expression Analysis
Mature miR828 was transcribed into cDNA by reverse transcribing LMW RNA using the stem-loop pulsed RT protocol and, then, end-point PCR was performed to amplify 60-nt fragments that included the mature miR828 sequences (Varkonyi-Gasic et al., 2007) using primers listed in Supplementary Table S1. To confirm the quality of LMW RNA, we amplified U6 small nuclear RNA by reverse transcribing LMW RNA with the U6br primer, followed by PCR amplification with the primers U6af and U6br (Supplementary Table S1). For the analysis of mRNA accumulation, cDNA was synthesized from total RNA using the ReverTraAce® qPCR RT Master Mix with gDNA Remover (Toyobo, Tokyo, Japan).
Quantitative reverse transcription PCR of mature miR828, pri-MIR828, anthocyanin biosynthesis genes [chalcone synthase a (CHSa), CHSb, flavanone 3-hydroxylase (F3H), DFR, and anthocyanidin synthase (ANS)], and other protein-coding sequences were conducted using the THUNDERBIRD® SYBR® qPCR Mix (Toyobo) and PCR primers listed in Supplementary Table S1. Signals were monitored using the CFX Connect Real-Time System (Bio-Rad, Tokyo, Japan). The amount of lily U6, lily ACTIN, tobacco ubiquitin (UBQ), or Renilla luciferase (Rluc) on the LhDFR-promoter::iGUS construct (agroinfiltration) in each sample was used to normalize the amount of each target mRNA, using the formula 2−∆Ct, where ∆Ct = Ct(target gene) − Ct(reference gene). Five or three biological replicates were used to calculate the mean values and standard error. The statistical differences were analyzed using Tukey’s honestly significant difference (Tukey’s HSD) test using R v3.3.12 or the t-test.
Small RNA Sequencing
Low molecular weight RNA isolated from the lower and upper halves of Lollypop tepals at stage 4 was processed into sequencing libraries using adapted Illumina protocols and sequenced by Hokkaido System Science Co., Ltd. (Sapporo, Japan) using an Illumina HiSeq sequencer (Illumina Inc., CA, USA). FASTQ file generation, adapter removal, and paired-read merging were performed using Cutadapt (Martin, 2011) and Fastq-join.3 The obtained sequences were plotted (allowing 100% homologous matching) onto the sense and antisense orientations of the MYB12 mRNA sequence using Bowtie (Langmead et al., 2009).
Results
Feature of Lily MicroRNA828
Lily miR828 was characterized in the present study to investigate the underlying mechanism of suppression of the expression of MYB12 at the lower halves of Lollypop tepals. First, the primary transcripts of MIR828 were defined using 5'- and 3'-RACE PCR. Two sequences (636 and 696 bp) were detected: guide and passenger strands appeared at the 5'-terminal region with identical sequences. However, sequences downstream of the region were slightly different (Supplementary Figure S1). As lily cultivars are genetically heterozygous, the two sequences should be allelic. RNA structure4 predicted that pre-MIR828 formed a hairpin loop structure. The two sequences exhibited similar structures (the structure of pri-MIR828-2 is presented in Supplementary Figure S2). One mismatched base pair and one bulged base were observed in the duplex of guide and passenger strands.
Target genes of miR828 in lily were predicted at the “psRNATarget: A Plant Small RNA Target Analysis Server” website5 (Dai et al., 2018), mainly using the Lollypop tepal transcriptome data as a target file. Fifteen sequences were detected under an expectation value lower than 4 (Supplementary Table S2). Out of the 45 R2R3-MYB genes expressed in Lollypop tepals (Suzuki et al., 2016; Yamagishi, 2020b), five were predicted as potential targets, including the all subgroup 6 members (MYB12, MYB15like, MYB16, and MYB19S).
Lily MicroRNA828 Cleaves MYB12 mRNA in Tobacco Leaves
The putative pre-MIR828 sequence was cloned into a pBI121 vector under the control of the 35S promoter to perform transient expression assays in N. tabacum to verify the biological activity of Lily miR828. To that end, a construct harboring 35S-p::MYB12 was infiltrated alone or together with the 35S-p::MIR828 construct, and the transcript abundance of MYB12 and NtDFR was monitored 6 days after infiltration (Figure 2A). NtDFR is a tobacco endogenous gene, whose expression is stimulated by lily MYB12 (Yamagishi, 2020a). Co-infiltration of the 35S-p::MIR828 construct reduced the expression levels of MYB12 and, in turn, suppressed NtDFR expression. RLM-RACE PCR revealed that MYB12 mRNA was cleaved in tobacco leaves at the nucleotide corresponding to the site between the 10th and 11th nucleotides of miR828 (Figure 2B and Supplementary Figure S3A).
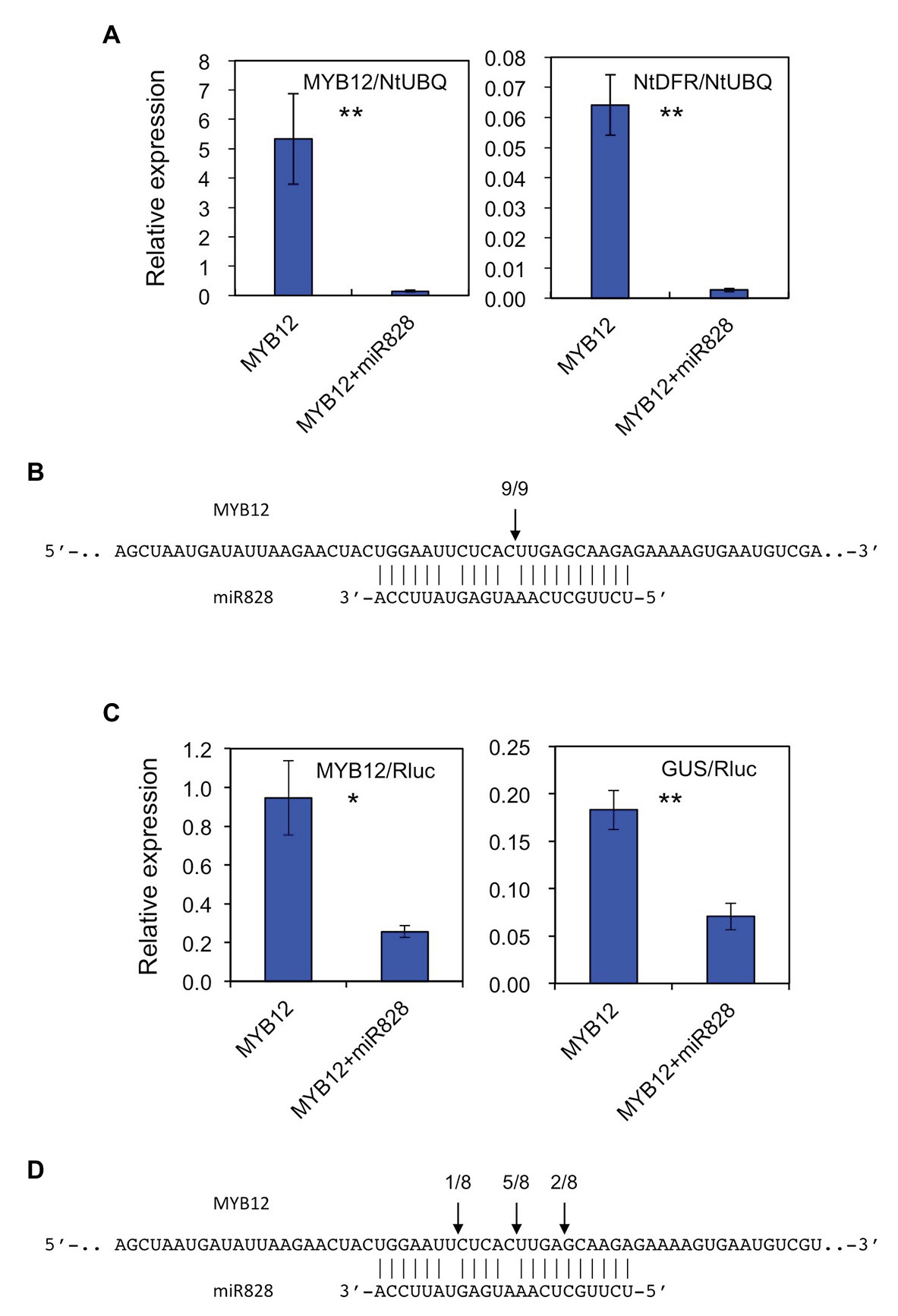
Figure 2. (A) Relative expression levels of MYB12 and NtDFR in tobacco (Nicotiana tabacum) leaves (five plants) infiltrated with Agrobacterium harboring 35S-p::MYB12 or Agrobacterium with 35S-p::pre-MIR828 plus Agrobacterium carrying 35S-p::MYB12. The levels of expression were analyzed 6 days after infiltration. The tobacco UBIQUTIN (NtUBQ) gene was used to normalize the expression of target genes. (B) The cleavage site (arrow) in MYB12 mRNA predicted using RNA ligase mediated (RLM)-rapid amplification of cDNA ends (RACE) PCR (RLM-RACE PCR). (C) Relative expression levels of MYB12 and GUS in tobacco leaves (three plants). A construct harboring 35S-p::MYB12 or a construct containing 35S-p::pre-MIR828 plus a construct containing 35S-p::MYB12 were introduced in tobacco leaves using Agrobacterium, together with 35S-p::LhbHLH2 and lily DFR-p::iGUS constructs. Expression levels were examined 3 days after infiltration. The NOS-p-driven Rluc gene in the DFR-p::iGUS construct was used to normalize the expression levels of target genes. (D) The cleavage sites (arrows) in MYB12 mRNA predicted by RLM-RACE PCR. In (A,C) the vertical bars indicate the standard errors of the means of five or three plants. * and ** Indicate significant differences at the 5 and 1% significance levels, respectively (t-test). In (B,D) segments of the MYB12 (upper) and mature miR828 (lower) sequences are shown. Numerals at the arrows indicate the number of reads with 5' terminal ends at arrowed sites/total number of reads.
Similarly, the constructs harboring 35S-p::MYB12, 35S-p::bHLH2 or lily DFR promoter-driven iGUS were infiltrated into tobacco leaves with or without the 35S-p::MIR828 construct (Figure 2C). Three days after infiltration, while expression of MYB12 activated the lily DFR promoter in tobacco leaves, co-infiltration of the 35S-p::MIR828 construct downregulated MYB12 expression and suppressed the promoter activity of lily DFR. The main cleavage site in MYB12 mRNA was between the 10th and 11th nucleotide from the 5' end of miR828 (Figure 2D and Supplementary Figure S3B). The results illustrated in Figure 2 suggest that MYB12 mRNA is the target of miR828 and is cleaved in the presence of mature miR828, which was processed from pre-MIR828 in tobacco.
MicroRNA828 Predominantly Accumulates in the White Tepal Region of Lilies
To further verify the post-transcriptional regulation of MYB12, the accumulation of mature miR828 was evaluated in lily tepals using stem-loop qRT-PCR (Figure 3A). During flower bud developmental stages from 1 to 4, its accumulation level in the lower halves increased with bud growth, while that in the upper halves was low and constant. In each of the developmental stages, accumulation levels were significantly higher in the lower halves than in the upper halves at 1 or 5% levels (t-test). The expression of MYB12 and five anthocyanin biosynthesis gene was mainly detected at stages 3, 4, and 5, and, in contrast to the miR828 accumulation levels, their expression levels were higher in the upper halves than in the lower halves (Figure 4). Sequence-specific cleavage of target RNA provides strong evidence of microRNA involvement in post-transcriptional gene regulation. RLM-RACE PCR revealed that MYB12 mRNA was cleaved in lily tepals predominantly between nucleotides that were annealed to the 10th and 11th nucleotides of miR828 (Figure 3B and Supplementary Figure S3C). Therefore, miR828 potentially contributes to the repression of MYB12 transcript accumulation during flower bud development, resulting in the suppression of anthocyanin biosynthesis in the lower halves of tepals. Pri-MIR828 transcript levels were estimated by qRT-PCR using primers designed at the 3' terminal region. Pri-MIR828 expression levels were higher in the lower halves than in the upper halves and, in contrast to in mature miR828, decreased with increased flower bud development in the lower halves (Figure 3C).
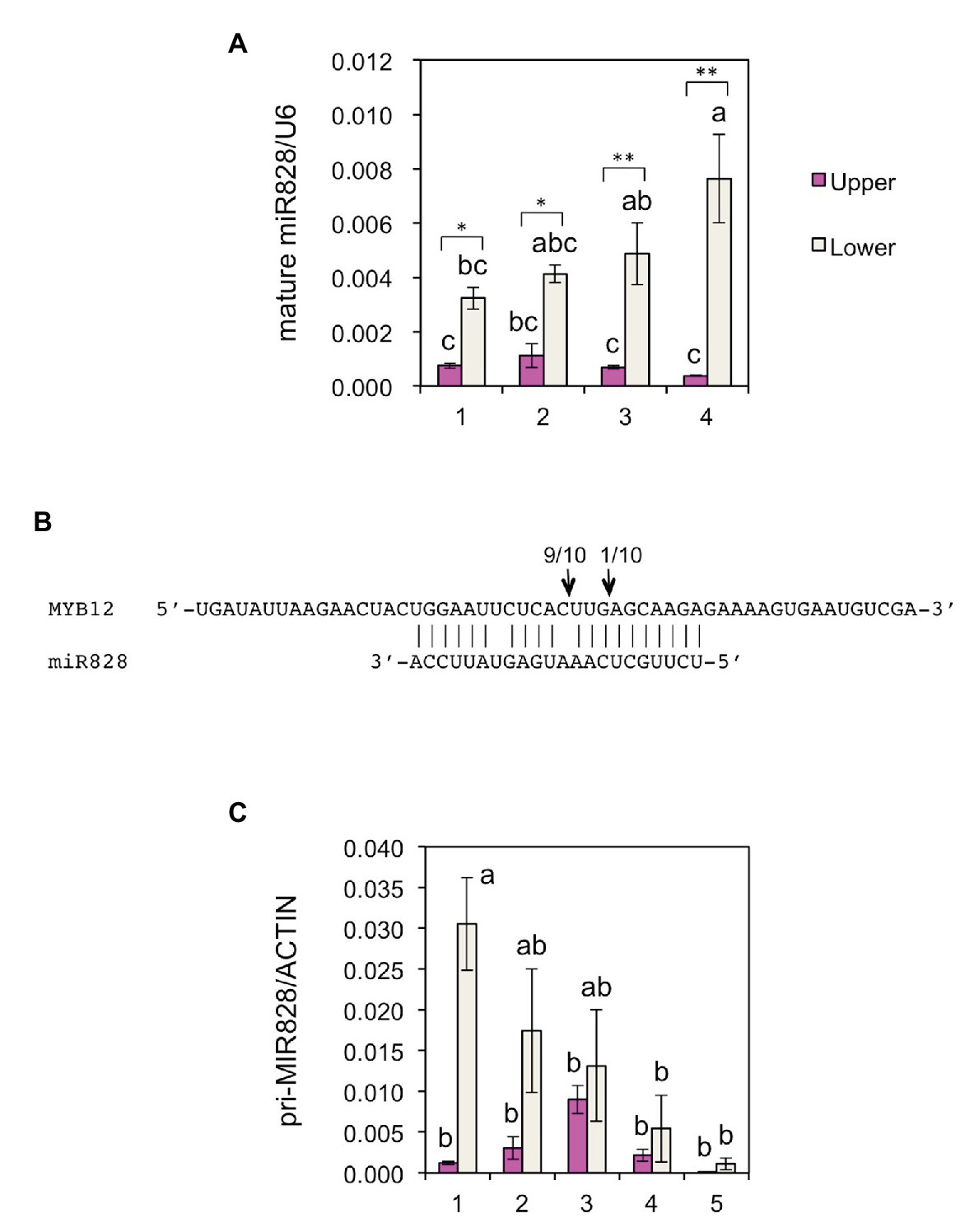
Figure 3. (A) Accumulation of miR828 [stem-loop quantitative reverse transcription PCR (qRT-PCR)] in upper and lower halves of Lollypop tepals at flower bud developmental stages 1, 2, 3, and 4. U6 was used to normalize the accumulation. Vertical bars show the standard errors (SE) of the means of three replicates. The same letters above the columns indicate that the values are not statistically significant (p < 0.05) based on Tukey’s honest significant difference (HSD) test. * and ** Indicate significant difference at 5 and 1% significance levels, respectively (t-test), between the upper and lower halves in each stage. (B) MiR828 cleavage site (arrows) in MYB12, estimated by RLM-RACE PCR. Numerals at the arrows indicate the number of reads with 5' terminal ends at arrowed sites/total number of reads. (C) Expression of pri-MIR828 in upper and lower halves of Lollypop tepals at flower bud developmental stages 1, 2, 3, 4, and 5. Lily ACTIN was used to normalize the expression of the genes under identical conditions. Vertical bars show the SE of the means of three flowers. The same letters above the columns indicate that the values are not statistically significant (p < 0.05) based on Tukey’s HSD.
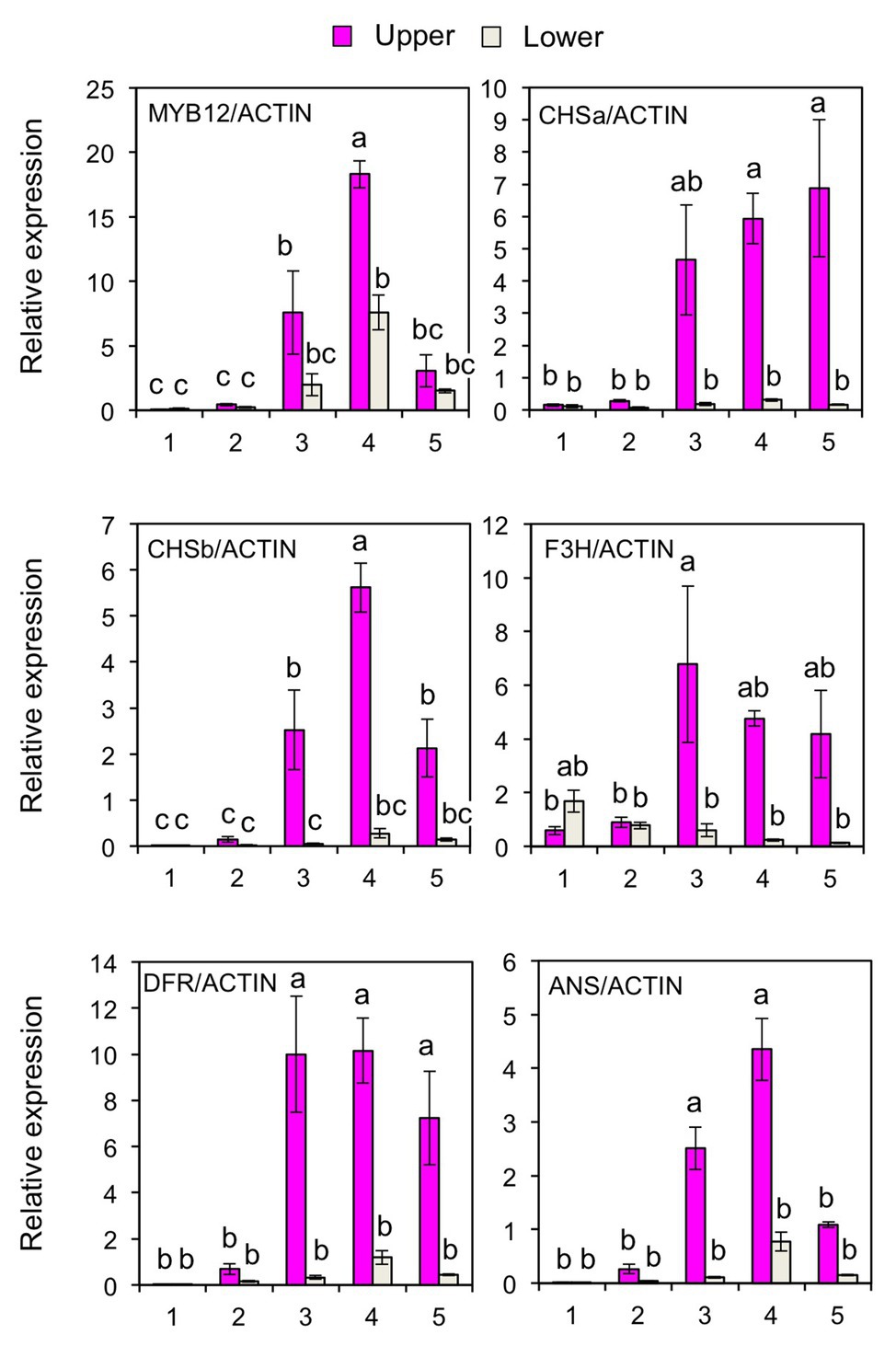
Figure 4. Expression of MYB12, CHSa, CHSb, F3H, DFR, and ANS genes (qRT-PCR) in the upper and lower halves of Lollypop tepals at flower bud development stages 1, 2, 3, 4, and 5. Lily ACTIN was used to normalize the expression of genes under identical conditions. Vertical bars represent the standard errors of the means of three flowers. The same letters above columns indicate that the values are not significantly different (p < 0.05) based on Tukey’s HSD test.
The amounts of the MYB12 mRNA-cleavage products were estimated using PCR: similar to the method of RLM-RACE PCR, RNA adapter was ligated to uncapped RNA and, after cDNA synthesis, PCR was carried out using a forward primer spanning the adapter and MYB12 sequences and an MYB12 gene-specific reverse primer (Figure 5). The cleaved MYB12 transcripts were detected at relatively high levels in the lower halves of tepals but scarcely in the upper halves of tepals, indicating that MYB12 transcripts are predominantly cleaved in the white tepal region.
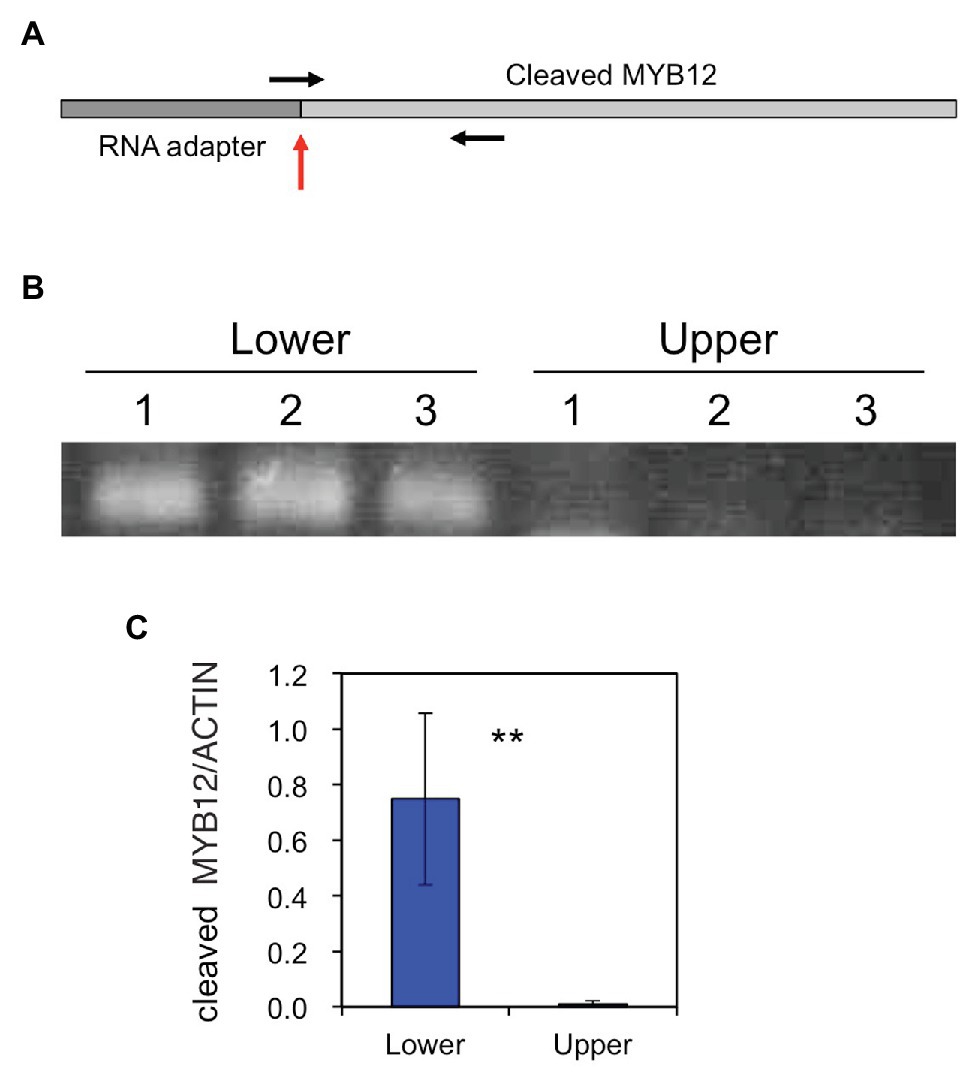
Figure 5. (A) A schematic representation of the miR828-cleaved MYB12 fragment. The vertical red arrow indicates a putative cleavage site and horizontal arrows denote primer positions. (B) PCR products of miR828-cleaved MYB12 fragments that accumulated in the lower and upper halves of Lollypop tepals. 1, 2, and 3 indicate individual flowers collected at stage 4. (C) Relative levels of accumulated miR828-cleaved MYB12 estimated using qRT-PCR. The lily ACTIN gene was used as a reference gene. Vertical bars represent the standard errors of the means of three flowers. **Indicates a significant difference at the 1% significance level (t-test).
To further confirm the functions of miR828 in color pattern development, the accumulation profile of miR828 was compared between bicolor cultivars and full-color cultivars (Figures 1, 6). Its accumulation levels were higher in the lower halves than in the upper halves of tepals in the four bicolor cultivars, while no clear difference was observed between the lower and upper halves of the three full-pink cultivars. Therefore, differences in miR828 levels between the two tepal sections were correlated highly with bicolor development in lily tepals.
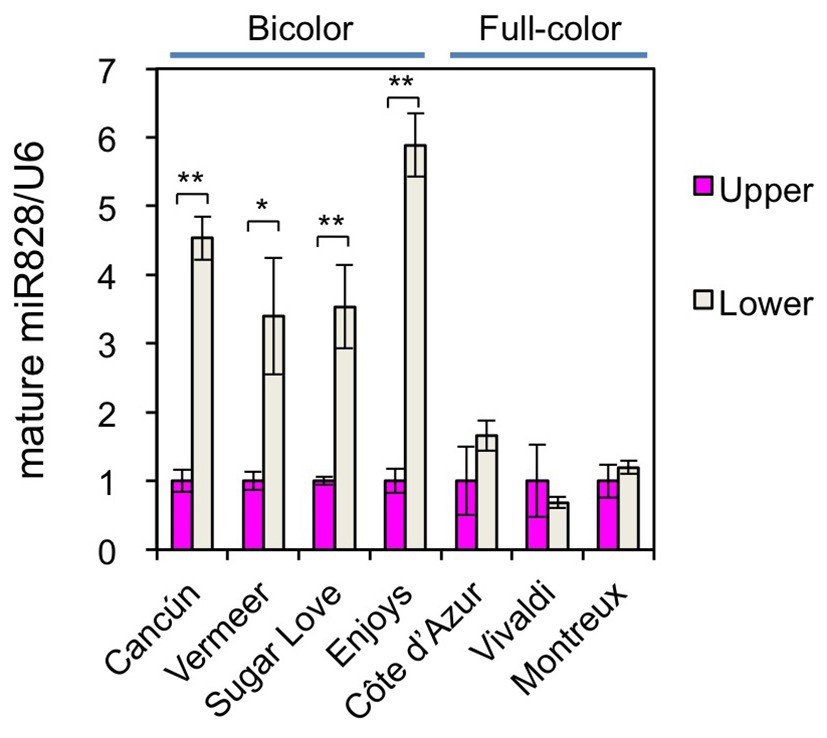
Figure 6. Accumulation of miR828 (stem-loop qRT-PCR) in upper and lower halves of tepals in four bicolor (Cancún, Vermeer, Sugar Love, and Enjoys) and three full-color (Côte d’Azur, Vivaldi, and Montreux) cultivars at flower bud developmental stage 4. U6 was used to normalize the accumulation. Vertical bars represent the standard errors of the means of three replicates. * and **Indicate significant difference at 5 and 1% significance levels, respectively (t-test), between the upper and lower halves.
Mature miR828 accumulation was compared in several Lollypop organs (Figure 7). Floral organs were collected at flower bud developmental stage 4. Its accumulation levels in filaments, ovaries, styles, and leaves were similar to those in the upper halves of tepals, while those in anthers were high, similar to those in the lower halves of tepals. Lilies accumulate high amounts of anthocyanins in anthers (Suzuki et al., 2015). In anthers, anthocyanin biosynthesis gene expression is high during the early stages of flower bud development and low at flower bud developmental stage 4. However, as MYB12 is not expressed (Lai et al., 2012) and other positive regulators are yet to be evaluated in anthers, the significance of a high accumulation of miR828 remains unclear.
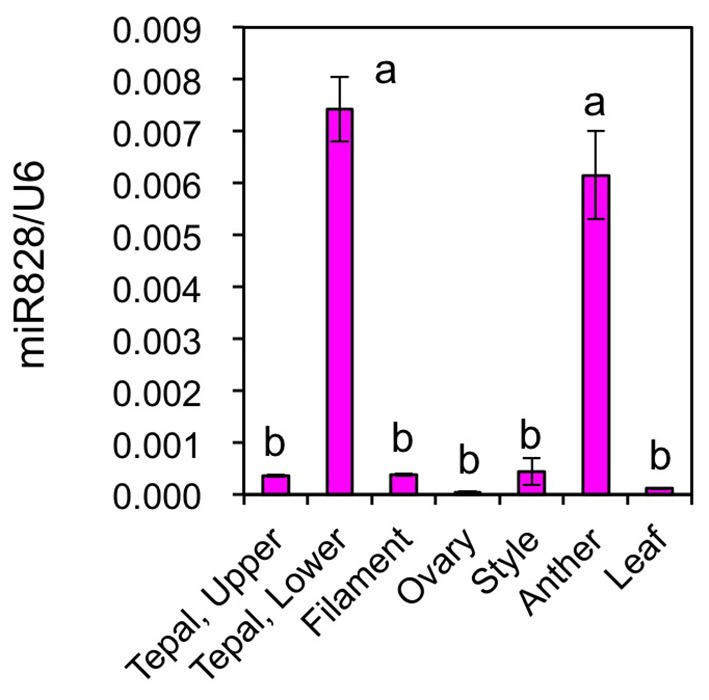
Figure 7. Accumulation of miR828 (stem-loop qRT-PCR) in tepals (upper and lower), filaments, ovaries, styles, anthers, and leaves of the Lollypop cultivar. Floral organs were collected at flower bud development stage 4. The U6 gene was used to normalize accumulation levels. Vertical bars represent the standard errors of the means of three replicates. The same letters above columns indicate that the values are not significantly different (p < 0.05) based on Tukey’s HSD test.
Secondary SiRNAs Initiated by MicroRNA828-Directed Cleavage of MYB12 mRNA
MiR828 was 22 nucleotides long. Since 22 nucleotide-long microRNAs are capable of secondary siRNA generation at their target loci (Shivaprasad et al., 2012; Källman et al., 2013), whether miR828 generated secondary siRNAs from the MYB12 sequence was evaluated using next-generation sequencing. Small RNA was prepared from the lower and upper halves of Lollypop tepals and sequenced using Illumina technology to yield a total of 18,887,232 and 18,997,636 small RNA reads, respectively (Supplementary Table S3). Afterward, the 19–24 nucleotide-long reads were mapped to the MYB12 sequence (Figure 8). Most of the small RNAs with the sense sequences of MYB12 were mapped on the 3' end of MYB12, beginning with the miR828 cleavage site and proceeding in 20–22-nucleotide intervals. The small RNA with the antisense sequence of MYB12 was mapped in 20–22-nucleotide intervals, accounting for the 2-nucleotide offset between the sense and antisense strands. The spacing pattern is consistent with evidence that DCL4 cleaves double-stranded RNA sequentially from the end of the molecule (Yoshikawa et al., 2005) and suggests that miR828-cleavage triggers the production of secondary siRNAs from 3' cleaved transcripts of MYB12. MiR828-mediated phasiRNA production from TAS4 and R2R3-MYB genes has been reported in Arabidopsis, apple, and peach (Rajagopalan et al., 2006; Xia et al., 2012; Zhu et al., 2012). Our results confirmed miR828-mediated secondary siRNA generation in monocots.
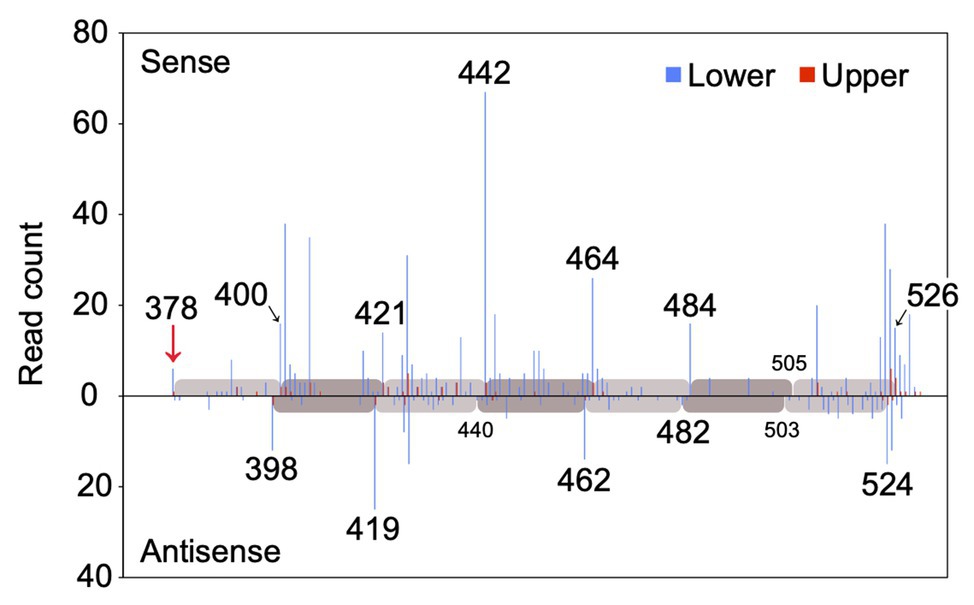
Figure 8. Read count of small RNA (19–24 nucleotides) mapped to the MYB12 mRNA. The number of sequenced small RNAs with 5' residues at each position along the MYB12 mRNA is plotted for the sense and antisense strands. Blue and red bars indicate the read counts of small RNA derived from lower and upper halves of Lollypop tepals (flower bud developmental stage 4), respectively. Numerals in the panel show nucleotide position downstream of the translation initiation site. Only the positions from the 370 to 530 nucleotides are shown. A red arrow indicates miR828-directed cleavage site. Gray and light gray bars represent 20, 21, or 22 nucleotide-intervals.
Out of each of the approximately 19 × 106 small RNA reads, 874 were mapped in the lower halves and 75 in the upper halves (Figure 8 and Supplementary Table S3). The number of the mapped small RNA was more than 10-fold higher in the lower halves than in the upper halves. MYB12 cleavage and subsequent phasiRNA production potentially occurred more frequently in the lower sections of tepals.
Discussion
Bicolor petal patterns, which consist of pigmented and unpigmented regions in a single petal, are often observed in higher plants. Several mechanisms that create bicolor patterns have been elucidated in some species, mainly petunias. One is a post-transcriptional silencing of CHS genes examined in petunias (Koseki et al., 2005) and dahlias (Ohno et al., 2011). In star and picotee petunia lines, siRNA suppresses CHS mRNA accumulation post-transcriptionally in white petal regions, and tandemly arranged CHS sequences are involved in siRNA generation (Morita et al., 2012). This floral color pattern is further modified by the activation of an endogenous virus. Petunia vein clearing caulimovirus (PVCV) proviruses are activated as host plants age, and CHS gene silencing is suppressed by an RNA silencing suppressor of activated PVCV, resulting in anthocyanin biosynthesis in the white regions of star-type petunia petals (Kuriyama et al., 2020). Another mechanism is s space-specific flavonol synthase (FLS) expression. In morn petunia lines, the central regions of flowers are colorless. FLS levels are high, and flavonols accumulate at much higher levels than anthocyanins in the central region (Saito et al., 2006). In Lollypop tepals, the entire anthocyanin biosynthesis pathway was suppressed in the white region, and the key factor that caused the suppression was the MYB12 transcription factor. In addition, most Asiatic hybrid lily cultivars do not accumulate flavonols and flavones in tepals (Lai et al., 2012), and FLS was scarcely expressed in Lollypop tepals (Suzuki et al., 2016). Therefore, in the present study, we explored additional mechanisms.
Lily MYB12 is a target of miR828 because miR828 cleaves MYB12 mRNA in lily tepals, and MIR828 introduced into tobacco leaves cleaved MYB12 mRNA and suppressed MYB12 activity. In addition, mature miR828 and the MYB12-derived secondary siRNA molecules accumulated more abundantly in the white region than in the pigmented region. Therefore, MYB12 expression was significantly lower in the white tepal regions due to targeting by miR828 and potentially by the secondary siRNAs. Other microRNAs that potentially hybridized into MYB12 mRNA were surveyed using the microRNA-seq data, but such a candidate was not found among the small RNAs of 20–24 nucleotides (data not shown). Therefore, miR828 is potentially a solo microRNA suppressing MYB12 expression, and the miR828/MYB12 module is potentially the key mechanism causing discoloration in lily flowers.
Why does mature miR828 predominantly accumulate in the lower halves of tepals? The amounts of pri-MIR828 transcripts were high in the lower halves and, therefore, spatially variable transcription of pri-MIR828 would be one of the primary mechanisms. However, accumulation profiles of mature miR828 and pri-MIR828 transcripts were different: the amounts of mature miR828 increased during flower bud growth, whereas those of pre-MIR828 decreased with flower bud development. Therefore, processing and stability of microRNA were also potentially involved in the predominant accumulation of mature miR828 in the lower halves, and the post-transcriptional regulation of miR828 accumulation unlikely occurs in the upper halves because the amounts do not increase with bud development. MicroRNA abundance is regulated under multiple levels of control, including processing, RISC assembly, and stability (Wang et al., 2019). Different AGO proteins have different effects on the stability of their bound microRNAs, and spatially and temporally distinct AGO5 expression is indispensable for organ-specific function of miR156 (Roussin-Léveillée et al., 2020).
MYB12 has been found to be the primary transcription factor regulating bicoloration (Suzuki et al., 2016), and we experimentally confirmed the predominant cleavage of MYB12 mRNA in the lower halves of tepals. However, compared with the upper halves, the levels of accumulated MYB12 transcript in tepals were observed to be reduced by 40% in the lower halves, in which the expression of biosynthesis genes was markedly suppressed (Figure 4; Suzuki et al., 2016). We suspect that the observed differences in suppression levels could be attributable to the fact that miR828 not only cleaves the transcripts but also arrests MYB12 translation in lily cells. In addition to the cleavage of target mRNA, translational attenuation of target genes is involved in microRNA-mediated gene regulation (Dalmay, 2013), and it is often reported that suppression of the accumulation of target gene transcripts is weaker than that of downstream genes regulated by the target genes (Gandikota et al., 2007; Wang et al., 2016; Gattolin et al., 2018).
Sequences of mature miR828 are highly conserved in many plants, including lilies, with an exception in Arabidopsis, in which one nucleotide is different. Major targets of miR828 are R2R3-MYB transcription factor genes and TAS4: MdWEREWOLF (WER, subgroup 15 member of R2R3-MYB), MdC1 (R2R3-MYB), and MdTAS4 in apple (Xia et al., 2012), AtMYB113 (subgroup 6 member of R2R3-MYB), AtMYB82 (ungrouped R2R3-MYB), and AtTAS4 in Arabidopsis (Rajagopalan et al., 2006), VvMYB114 (WER like MYB having EAR suppressor motif) and VvWER in Vitis vinifera (Tirumalai et al., 2019), PpMYB (subgroup 15 member of R2R3-MYB), two PpMYB (ungrouped MYB), and PpTAS4 in peach (Zhu et al., 2012), and R2R3-MYB and StTAS4 in potato (Bonar et al., 2018). R3 repeat sequences in R2R3-MYB genes are highly conserved, and the target site of miR828 exists near the end of the R3 repeat. Therefore, miR828 often targets several MYB genes (Xia et al., 2012; Zhu et al., 2012; Li and Lu, 2014). In Lollypop, 15 sequences were predicted to be targets, although whether they were cleaved by miR828 has not yet been evaluated, and five of the 15 sequences were R2R3-MYB (Supplementary Table S2). MYB15-like exhibited homology with LrMYB15 in Lilium regale, which regulates bud blush anthocyanin pigmentation (Yamagishi, 2016), while, in Asiatic hybrid lilies, its expression levels are not high and its function is unknown (Yamagishi, 2018). MYB16 is also expressed weakly in tepals, and its function is unknown (Yamagishi, 2018). Zm1-like Myb-related protein is a subgroup 2 member of R2R3-MYB (Suzuki et al., 2016). Although its function in lily is unknown, subgroup 2 members are not involved in pigmentation in other species. MYB19S is predominantly expressed in stage 2 flower buds and is involved in the pigmentation at raised spots developed at the basal parts of tepals (Yamagishi, 2020b). Therefore, the R2R3-MYB genes, other than MYB12, are unlikely to be involved in bicoloration.
The involvement of miR828 in pigment accumulation has been reported. MiR828 highly accumulates in pigmented grape fruits and potato tubers and inhibits the expression of grape MYB114 and potato R2R3-MYB, which act as suppressors of anthocyanin biosynthesis (Bonar et al., 2018; Tirumalai et al., 2019). In lilies, miR828 accumulates in the unpigmented regions of tepals and suppresses the positive regulation of anthocyanin biosynthesis. Therefore, the phenomenon in lilies is distinct from those observed in grapes and potatoes.
Subgroup 6 members of R2R3-MYB in numerous species and subgroup 5 members of that in Poaceae and orchids are major positive regulators of anthocyanin biosynthesis. To determine whether miR828 targets these positive regulator genes, putative miR828 target sites found in the genes were compared (Supplementary Figure S4). Base-pairing between miR828 and the putative target sites showed that the number of mismatched nucleotides was three or fewer in R2R3-MYB genes of oil palm, onion, Arabidopsis, snapdragon, and Salvia. In Arabidopsis, AtMYB113 is the direct target of miR828 (Luo et al., 2012), and AtPAP1 and AtMYB113 expression is suppressed in miR828-overexpressed Arabidopsis plants (Yang et al., 2013), indicating that miR828 regulates anthocyanin accumulation in Arabidopsis through the suppression of the positive regulators. However, as miR828 predominantly accumulates in siliques (Rajagopalan et al., 2006), miR828 is unlikely to be involved in flower coloration in Arabidopsis. Deep sequencing analysis revealed that SmMYB36, a subgroup 6 R2R3-MYB in Salvia, is a target of miR828 (Li and Lu, 2014), although the function of SmMYB36 has not been clarified yet. Numerous mismatched nucleotides have been observed in the sequences of other eudicot species, including petunia, grape, and apple, suggesting that miR828 does not target such positive regulators directly in these species. Consequently, the bicolor formation mediated by the modules of miR828 and subgroup 6 members of R2R3-MYB, observed in lily flowers, should be rare in eudicots. Conversely, in addition to the lily MYBs, two of the three subgroup 6 members of R2R3-MYB (EgVIR in oil palm and AcMYB1 in onion) in monocots are putative targets of miR828, although the number of subgroup 6 members of R2R3-MYB genes isolated and characterized in monocots is limited. Therefore, further investigation of miR828-mediated pattern formation should provide novel insights in monocots. Subgroup 5 members of the R2R3-MYBs are unlikely targets of miR828.
Several mechanisms of the development of anthocyanin color patterns have been clarified in flowers. The major mechanisms are spatially distinct expression of R2R3-MYB positive regulators in single petals (Fattorini and Glover, 2020), post-transcriptional gene silencing of CHS (Morita et al., 2012), and the insertion of transposable elements into anthocyanin biosynthesis genes (Momose et al., 2013; Uchiyama et al., 2013). MicroRNA-mediated pattern formation, presented in this study, is likely the novel mechanism of development of the attractive color patterns.
Data Availability Statement
These deep sequencing files have been deposited in the DDBJ Sequence Read Archive under the accession number DRA010576.
Author Contributions
MY designed the research, conducted the experiments, and wrote the manuscript. MS and MY analyzed the data. Both the authors have read and approved the submission of the manuscript.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (no. 19H02945) from the Japan Society for the Promotion of Science and the Akiyama Life Science Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.590791/full#supplementary-material
Footnotes
1. https://www.genome.jp/tools-bin/clustalw
3. https://expressionanalysis.github.io/ea-utils/
4. https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html
References
Albert, N. W., Davies, K. M., and Schwinn, K. E. (2014). Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal. Behav. 9:e29526. doi: 10.4161/psb.29526
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K. E., Jameson, P. E., and Davies, K. M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Bennett, M. D., and Leitch, I. J. (2011). Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann. Bot. 107, 467–590. doi: 10.1093/aob/mcq258
Bonar, N., Liney, M., Zhang, R., Austin, C., Dessoly, J., Davidson, D., et al. (2018). Potato miR828 is associated with purple tuber skin and flesh color. Front. Plant Sci. 9:1742. doi: 10.3389/fpls.2018.01742
D’Ario, M., Griffiths-Jones, S., and Kim, M. (2017). Small RNAs: big impact on plant development. Trends Plant Sci. 22, 1056–1068. doi: 10.1016/j.tplants.2017.09.009
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Dalmay, T. (2013). Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 54, 29–38. doi: 10.1042/bse0540029
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
Deng, P., Muhammad, S., Cao, M., and Wu, L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 16, 965–975. doi: 10.1111/pbi.12882
Fattorini, R., and Glover, B. J. (2020). Molecular mechanisms of pollination biology. Annu. Rev. Plant Biol. 71, 487–515. doi: 10.1146/annurev-arplant-081519-040003
Fei, Q., Xia, R., and Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in post-transcriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Gandikota, M., Birkenbihl, R. P., Höhmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313X.2006.02983.x
Gattolin, S., Cirilli, M., Pacheco, I., Ciacciulli, A., Linge, C. D. S., Mauroux, J. B., et al. (2018). Deletion of the miR172 target site in a TOE-type gene is a strong candidate variant for dominant double-flower trait in Rosaceae. Plant J. 96, 358–371. doi: 10.1111/tpj.14036
Glover, B. J., Walker, R. H., Moyroud, E., and Brockington, S. F. (2013). How to spot a flower. New Phytol. 197, 687–689. doi: 10.1111/nph.12112
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., and Chen, H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Jia, X., Shen, J., Liu, H., Li, F., Ding, N., Gao, C., et al. (2015). Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293. doi: 10.1007/s00425-015-2305-5
Källman, T., Chen, J., Gyllenstrand, N., and Lagercrantz, U. (2013). A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162, 741–754. doi: 10.1104/pp.113.214643
Koseki, M., Goto, K., Masuta, C., and Kanazawa, A. (2005). The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 46, 1879–1883. doi: 10.1093/pcp/pci192
Kuriyama, K., Tabara, M., Moriyama, H., Kanazawa, A., Koiwa, H., Takahashi, H., et al. (2020). Disturbance of floral colour pattern by activation of an endogenous pararetrovirus, petunia vein clearing virus, in aged petunia plants. Plant J. 103, 497–511. doi: 10.1111/tpj.14728
Lai, Y., Shimoyamada, Y., Nakayama, M., and Yamagishi, M. (2012). Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Lilium spp.). Plant Sci. 193–194, 136–147. doi: 10.1016/j.plantsci.2012.05.013
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Li, C., and Lu, S. (2014). Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15:277. doi: 10.1186/1471-2164-15-277
Luo, Q., Mittal, A., Jia, F., and Rock, C. D. (2012). An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80, 117–129. doi: 10.1007/s11103-011-9778-9
Marasek-Ciolakowska, A., Nishikawa, T., Shea, D. J., and Okazaki, K. (2018). Breeding of lilies and tulips—interspecific hybridization and genetic background. Breed. Sci. 68, 35–52. doi: 10.1270/jsbbs.17097
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Ministry of Agriculture, Forestry and Fisheries of Japan (2019). The 93rd statistical yearbook of Ministry of Agriculture, Forestry and Fisheries. Available at: https://www.maff.go.jp/e/data/stat/93th/index.html#14 (Accessed July 22, 2020)
Momose, M., Nakayama, M., Itoh, Y., Umemoto, N., Toguri, T., and Ozeki, Y. (2013). An active hAT transposable element causing bud mutation of carnation by insertion into the flavonoid 3'-hydroxylase gene. Mol. Gen. Genomics. 288, 175–184. doi: 10.1007/s00438-013-0742-z
Morita, Y., Saito, R., Ban, Y., Tanikawa, N., Kuchitsu, K., Ando, T., et al. (2012). Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 70, 739–749. doi: 10.1111/j.1365-313X.2012.04908.x
Nakatsuka, A., Izumi, Y., and Yamagishi, M. (2003). Spatial and temporal expression of chalcone synthase and dihydroflavonol 4-reductase genes in the Asiatic hybrid lily. Plant Sci. 165, 759–767. doi: 10.1016/S0168-9452(03)00254-1
Nakayama, M. (2014). A research strategy to understand the mechanisms that govern flower color pattern formation. Jpn. Agric. Res. Q. 48, 271–277. doi: 10.6090/jarq.48.271
Nørbæk, R., and Kondo, T. (1999). Anthocyanins from flowers of Lilium (Liliaceae). Phytochemistry 50, 1181–1184. doi: 10.1016/S0031-9422(98)00661-X
Ohno, S., Hosokawa, M., Kojima, M., Kitamura, Y., Hoshino, A., Tatsuzawa, F., et al. (2011). Simultaneous post-transcriptional gene silencing of two different chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta 234, 945–958. doi: 10.1007/s00425-011-1456-2
Rajagopalan, R., Vaucheret, H., Trejo, J., and Barte, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Roussin-Léveillée, C., Silva-Martins, G., and Moffett, P. (2020). ARGONAUTE5 represses age-dependent induction of flowering through physical and functional interaction with miR156 in Arabidopsis. Plant Cell Physiol. 61, 957–966. doi: 10.1093/pcp/pcaa022
Saito, R., Fukuta, N., Ohmiya, A., Itoh, Y., Ozeki, Y., Kuchitsu, K., et al. (2006). Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in Petunia. Plant Sci. 170, 828–834. doi: 10.1016/j.plantsci.2005.12.003
Sakai, M., Yamagishi, M., and Matsuyama, K. (2019). Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep. 38, 609–622. doi: 10.1007/s00299-019-02391-4
Sharma, D., Tiwari, M., Pandey, A., Bhatia, C., Sharma, A., and Trivedi, P. K. (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 171, 944–959. doi: 10.1104/pp.15.01831
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Suzuki, K., Suzuki, T., Nakatsuka, T., Dohra, H., Yamagishi, M., Matsuyama, K., et al. (2016). RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genomics 17:611. doi: 10.1186/s12864-016-2995-5
Suzuki, K., Tasaki, K., and Yamagishi, M. (2015). Two distinct spontaneous mutations involved in white flower development in Lilium speciosum. Mol. Breed. 35:193. doi: 10.1007/s11032-015-0389-z
Tirumalai, V., Swetha, C., Nair, A., Pandit, A., and Shivaprasad, P. V. (2019). miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 70, 4775–4791. doi: 10.1093/jxb/erz264
Uchiyama, T., Hiura, S., Ebinuma, I., Senda, M., Mikami, T., Martin, C., et al. (2013). A pair of transposons coordinately suppresses gene expression, independent of pathways mediated by siRNA in Antirrhinum. New Phytol. 197, 431–440. doi: 10.1111/nph.12041
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., and Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi: 10.1186/1746-4811-3-12
Wang, J., Mei, J., and Ren, G. (2019). Plant microRNAs: biogenesis, homeostasis, and degradation. Front. Plant Sci. 10:360. doi: 10.3389/fpls.2019.00360
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Wang, X., and Yamagishi, M. (2019). Mechanisms suppressing carotenoid accumulation in flowers differ depending on the hybrid groups of lilies (Lilium spp.). Sci. Hortic. 243, 159–168. doi: 10.1016/j.scienta.2018.08.025
Xia, R., Zhu, H., An, Y. Q., Beers, E. P., and Liu, Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13:R47. doi: 10.1186/gb-2012-13-6-r47
Yamagishi, M. (2011). Oriental hybrid lily Sorbonne homologue of LhMYB12 regulates anthocyanin biosyntheses in flower tepals and tepal spots. Mol. Breed. 28, 381–389. doi: 10.1007/s11032-010-9490-5
Yamagishi, M. (2013). How genes paint lily flowers: regulation of colouration and pigmentation patterning. Sci. Hortic. 163, 27–36. doi: 10.1016/j.scienta.2013.07.024
Yamagishi, M. (2016). A novel R2R3-MYB transcription factor regulates light-mediated floral and vegetative anthocyanin pigmentation patterns in Lilium regale. Mol. Breed. 36:3. doi: 10.1007/s11032-015-0426-y
Yamagishi, M. (2018). Involvement of a LhMYB18 transcription factor in large anthocyanin spot formation on the flower tepals of the Asiatic hybrid lily (Lilium spp.) cultivar ‘Grand Cru’. Mol. Breed. 38:60. doi: 10.1007/s11032-018-0806-1
Yamagishi, M. (2020a). White with partially pink flower color in Lilium cernuum var. album is caused by transcriptional regulation of anthocyanin biosynthesis genes. Sci. Hortic. 260:108880. doi: 10.1016/j.scienta.2019.108880
Yamagishi, M. (2020b). Isolation and identification of MYB transcription factors (MYB19Long and MYB19Short) involved in raised spot anthocyanin pigmentation in lilies (Lilium spp.). J. Plant Physiol. 250:153164. doi: 10.1016/j.jplph.2020.153164
Yamagishi, M. (2020c). MYB19LONG is involved in brush-mark pattern development in Asiatic hybrid lily (Lilium spp.) flowers. Sci. Hortic. 272:109570. doi: 10.1016/j.scienta.2020.109570
Yamagishi, M., and Nakatsuka, T. (2017). LhMYB12, regulating tepal anthocyanin pigmentation in Asiatic hybrid lilies, is derived from Lilium dauricum and L. bulbiferum. Hortic. J. 86, 528–533. doi: 10.2503/hortj.OKD-057
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., and Masuda, K. (2010). Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 51, 463–474. doi: 10.1093/pcp/pcq011
Yamagishi, M., Toda, S., and Tasaki, K. (2014). The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 201, 1009–1020. doi: 10.1111/nph.12572
Yamagishi, M., Yoshida, Y., and Nakayama, M. (2012). The transcription factor LhMYB12 determines anthocyanin pigmentation in the tepals of Asiatic hybrid lilies (Lilium spp.) and regulates pigment quantity. Mol. Breed. 30, 913–925. doi: 10.1007/s11032-011-9675-6
Yang, F., Cai, J., Yang, Y., and Liu, Z. (2013). Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tissue Organ Cult. 115, 159–167. doi: 10.1007/s11240-013-0349-4
Yoshikawa, M., Peragine, A., Park, M. Y., and Poethig, R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. doi: 10.1101/gad.1352605
Zhang, B. (2015). MicroRNAs: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. doi: 10.1093/jxb/erv013
Keywords: anthocyanin, color pattern, bicoloration, phased-siRNA, post-transcriptional gene regulation, subgroup 6 R2R3-MYB
Citation: Yamagishi M and Sakai M (2020) The MicroRNA828/MYB12 Module Mediates Bicolor Pattern Development in Asiatic Hybrid Lily (Lilium spp.) Flowers. Front. Plant Sci. 11:590791. doi: 10.3389/fpls.2020.590791
Edited by:
Prem Bhalla, The University of Melbourne, AustraliaReviewed by:
Kevin Davies, The New Zealand Institute for Plant and Food Research Ltd, New ZealandMasayoshi Nakayama, National Agricultural Research Organisation, Uganda
Copyright © 2020 Yamagishi and Sakai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masumi Yamagishi, eWFtYWdpc2lAcmVzLmFnci5ob2t1ZGFpLmFjLmpw
 Masumi Yamagishi
Masumi Yamagishi Moeko Sakai
Moeko Sakai