- 1Graduate School of Biotechnology, Kyung Hee University, Yongin, South Korea
- 2Department of Life Science, College of Life Science and Natural Resources, Dong-A University, Busan, South Korea
The fine-tuning of inorganic phosphate (Pi) for enhanced use efficiency has long been a challenging subject in agriculture, particularly in regard to rice as a major crop plant. Among ribonucleases (RNases), the RNase T2 family is broadly distributed across kingdoms, but little has been known on its substrate specificity compared to RNase A and RNase T1 families. Class I and class II of the RNase T2 family are defined as the S-like RNase (RNS) family and have showed the connection to Pi recycling in Arabidopsis. In this study, we first carried out a phylogenetic analysis of eight rice and five Arabidopsis RNS genes and identified mono-specific class I and dicot-specific class I RNS genes, suggesting the possibility of functional diversity between class I RNS family members in monocot and dicot species through evolution. We then compared the in silico expression patterns of all RNS genes in rice and Arabidopsis under normal and Pi-deficient conditions and further confirmed the expression patterns of rice RNS genes via qRT-PCR analysis. Subsequently, we found that most of the OsRNS genes were differentially regulated under Pi-deficient treatment. Association of Pi recycling by RNase activity in rice was confirmed by measuring total RNA concentration and ribonuclease activity of shoot and root samples under Pi-sufficient or Pi-deficient treatment during 21 days. The total RNA concentrations were decreased by < 60% in shoots and < 80% in roots under Pi starvation, respectively, while ribonuclease activity increased correspondingly. We further elucidate the signaling pathway of Pi starvation through upregulation of the OsRNS genes. The 2-kb promoter region of all OsRNS genes with inducible expression patterns under Pi deficiency contains a high frequency of P1BS cis-acting regulatory element (CRE) known as the OsPHR2 binding site, suggesting that the OsRNS family is likely to be controlled by OsPHR2. Finally, the dynamic transcriptional regulation of OsRNS genes by overexpression of OsPHR2, ospho2 mutant, and overexpression of OsPT1 lines involved in Pi signaling pathway suggests the molecular basis of OsRNS family in Pi recycling via RNA decay under Pi starvation.
Introduction
Inorganic phosphate (Pi) is a crucial component of major organic molecules such as RNA, DNA, and ATP in all organisms. In plants, Pi (H2PO4– or HPO4–2) is mainly absorbed by plant roots in the soil. Pi starvation promotes changes in root system architecture and reduces crop productivity. To overcome Pi starvation in plants, plants enhance the activity of RNases, phosphatases, and Pi transporters that promote Pi acquisition by roots (Vance et al., 2003; Plaxton and Tran, 2011; Zhu et al., 2019). Ribonucleases are ubiquitous components of cells that catalyze the cleavage of RNA and act on either single-stranded, double-stranded, or DNA–RNA hybrid substrates. Ribonucleases, which hydrolyze RNA to 3′ mononucleotides via 2′,3′ cyclic nucleotides, have been classified into three functional groups: the RNase A, RNase T1, and RNase T2 families. The RNase A family consists of proteins with a molecular mass between 13 and 14 kDa and either an alkaline (7–8) or weakly acidic (6.5–7) pH preference; moreover, they constitute a group of homologous proteins that have been isolated from many vertebrates, but not from invertebrates (Beintema et al., 1997; Beintema and Kleineidam, 1998). In turn, proteins in the RNase T1 family have a molecular mass of ∼12 kDa and pH optima between 7 and 8, and are found in fungi and bacteria. Finally, the RNase T2 family includes RNases with an average molecular mass around 25 kDa that were originally categorized as having acid RNase activity (Irie, 1999; Deshpande and Shankar, 2002). In addition, RNase T2 enzymes are transferase-type endoribonucleases that produce oligonucleotides and/or mononucleotides with a terminal 3′ phosphate via a 2′,3′ cyclic phosphate intermediate. The RNase T2 family exists in virtually all eukaryotes and seems to play important roles in a variety of biological processes.
The classification of the RNase T2 family in plants into three classes (I–III) is based on phylogenetic analyses (Igic and Kohn, 2001; MacIntosh et al., 2010; Ramanauskas and Igic, 2017). Some of plant class III RNases are involved in self-incompatibility, but in the class I and II RNases, none of the genes have been involved in self-incompatibility. Plant class I RNases are involved in abiotic stress responses such as salt stress and phosphate starvation stresses (Bariola et al., 1999; MacIntosh et al., 2010; Zheng et al., 2014); plant class II RNases participate in phosphate starvation (Bariola et al., 1999) and senescence (Taylor et al., 1993), and also has a housekeeping role in recycling ribosomal RNA (rRNA) (Hillwig et al., 2011; MacIntosh and Bassham, 2011; Floyd et al., 2015, 2017; Morriss et al., 2017); and besides self-incompatibility, some class III RNases are involved in stress such as phosphate starvation without being involved in self-incompatibility (Rojas et al., 2013, 2015, 2018). Thus, we defined plant class III RNases as S-RNases, and plant class I and II RNases as S-like RNases.
Several RNase genes have been identified in dicots, including Arabidopsis, Nicotiana benthamiana (tobacco), and Solanum lycopersicum (tomato) (Taylor et al., 1993; Bariola et al., 1994; Hillwig et al., 2011). Recent studies reported an association between class I RNSs and senescence and environmental signals (Bariola et al., 1999; MacIntosh et al., 2010; MacIntosh and Bassham, 2011), including inorganic phosphate (Pi) starvation, in Arabidopsis and Nicotiana (Rojas et al., 2013); in contrast, the class II RNS genes are constitutively expressed and likely perform housekeeping functions.
In cultivated S. lycopersicum (tomato) cells, the RNase LE and RNase LX genes are induced under Pi starvation (Kaletta et al., 1998). Two Arabidopsis RNase T2 genes, RNS1 and RNS2, are also expressed under Pi starvation (Taylor et al., 1993; Bariola et al., 1994, 1999). RNS2 and RNase LX are upregulated during senescence (Taylor et al., 1993; Lers et al., 1998). Although several studies reported the genome-wide identification of S-like RNase family genes in dicot plants, the global understanding of this family remains limited in monocot plants, including rice. Transcriptome analysis is a very simple and powerful tool that can be used to obtain functional clues for genes with uncharacterized features. Several studies have revealed important components of the sensing and signaling networks involved in Pi starvation responses in rice. OsPHR2 (Rice Phosphate Starvation Response 2), which is an AtPHR1-like gene, has been reported as one of the central regulators of Pi signaling pathway because it regulates the expression of Osa-miR399 and OsPHT1s by binding to the P1BS elements in the promoters of Pi-starvation-induced (PSI) genes (Zhou et al., 2008; Liu et al., 2010; Li et al., 2015). P1BS is also present in the promoters of many key PSI genes (Li et al., 2015; Ruan et al., 2015; Gho et al., 2018). Osa-miR399 represses the expression of OsPHO2, a ubiquitin-conjugating E2 enzyme, which triggers the degradation of several OsPHT1 family genes under normal conditions (Liu et al., 2010; Wu and Wang, 2011; Sun et al., 2012; Cao et al., 2014). Thus, OsPHR2, miR399, OsPHO2, and OsPTs are significant components of the Pi-signaling network. In this study, we carried out a comparative phylogenetic analysis using five Arabidopsis and eight rice RNS genes. Based on a sequence-similarity analysis and domain organization, we assigned two subgroups to this family. We also found that the class I RNS genes in rice, unlike those in Arabidopsis, a representative dicotyledonous plant, were spatially regulated in response to Pi starvation and various environmental stresses. In addition, the amount of total RNA was decreased by 60% in shoots and 80% in roots, respectively, under Pi starvation; concomitantly, RNase activity was enhanced under Pi starvation. Finally, a detailed data analysis of RNS family genes in rice associated with phosphate use efficiency is presented and the functional significance and further applications of these findings are discussed.
Materials and Methods
Plant Materials
Rice (Oryza sativa L. cv. Dongjin) seeds were germinated on Murashige Skoog medium under controlled conditions of 28°C day/25°C night temperatures, 8-h light/16-h dark cycle, and 78% relative humidity after sterilization with 50% (w/v) commercial bleach for 30 min with gentle shaking. For anatomical expression analysis, roots, leaf sheaths, leaf blades, panicles before heading, flowers at the heading stage, and seeds at 10 and 15 days after pollination were harvested to extract total RNAs. Four biological replicates were prepared and analyzed independently. For differential Pi-starvation-inducible expression analysis in shoots and roots, we used rice cv. Dongjin seedlings grown for 21 days in Pi-sufficient (0.320 mM Pi) or Pi-deficient (0 mM Pi) Yoshida solution (Cock et al., 1976) after germination for 10 days in MS media. The pH of the culture solution was adjusted to pH 5.5 using 6 M NaOH every 1 day, and total Yoshida solution was replaced every 3 days. In all hydroponic experiments, seedlings were directly grown in each of the culture solutions (8 L) with an 8-h light (28°C)/16-h dark (22°C) photoperiod.
RNA Extraction, RNA Concentration, Semi-Quantitative RT–PCR, and Real-Time PCR
The anatomical tissues, phosphate stress-treated samples, and roots and shoots of OsPHR2-OX, OsPT1-OX, and ospho2 were frozen in liquid nitrogen and ground with a Tissue Lyzer II (Qiagen; Hilden, Germany). RNAs were extracted with the RNAiso Plus Kit according to the manufacturer’s protocol (Takara Bio; Kyoto, Japan). Complementary DNA (cDNA) was synthesized. To compare RNA concentration in roots and shoots between phosphate-deficient plants and phosphate-sufficient plants, we used 40 mg for the shoot samples and 60 mg for the root samples to isolate RNA using the RNAiso Plus Kit and melted them into 50 μl of distilled water (DW). This experiment was repeated three times using the same amount of independent samples and was measured by UV spectroscopy (Thermo Scientific NanoDrop 2000). To determine the tissue-preferential expression patterns, phosphate-deficient expression, and the expression patterns in the above three genetic resources via real-time PCR, we used OsUbi5 as the reference gene (Jain et al., 2006). We used cycling conditions of 95°C for 15 s, 57°C for 30 s, and 72°C for 60 s. This experiment was repeated three times by using independent biological replicates. Relative transcript levels and fold changes were calculated by the 2ΔCt and 2ΔΔCt methods (Schmittgen and Livak, 2008), respectively.
Preparation of Phosphate Signaling Mutants
To confirm the expression analysis of the OsRNS family gene regulated by the PSI signaling gene, we used rice phosphate 2 (ospho2) T3 mutants, rice phosphate transporter1 overexpressing T5 line (OsPT1-OX), and rice phosphate starvation response 2 overexpressing T2 line (OsPHR2-OX). The ospho2 T-DNA insertion mutant lines, PFG_1C-06032, were obtained from RiceGE1. The OsPT1-OX line, which expresses high-affinity phosphate transporters in rice, was obtained from Dong A University, South Korea (Seo et al., 2008). The overexpression of OsPHR2 was generated. The 1326 bp of the coding region of OsPHR2 including the Kozak sequence was amplified from the rice (O. sativa L. cv. Dongjin) cDNA using the primers listed in Supplementary Table S1. The PCR products were directly cloned into the pGA3438 vector by Infusion Cloning (Infusion HD Cloning Kit, Clontech, 639644, California, United States) (Kim et al., 2009). The expression vectors were transferred to Agrobacterium tumefaciens strain LBA4404 by transformation and co-cultivation in rice callus (O. sativa L. cv. Dongjin), as described previously (Moon et al., 2019). Three mutants used in this study were further confirmed by qRT-PCR with rice ubiquitin 5 (OsUbi5, LOC_Os01g22490) as the reference gene. The primers used in these analyses are summarized in Supplementary Table S1.
Multiple Sequence Alignment and Phylogenetic Analysis
To perform a phylogenetic tree analysis of RNS family genes in two monocot plants (rice and Zea mays) and four dicot plants (Arabidopsis, Brassica rapa, S. lycopersicum, and Medicago truncatula), we collected RNS family protein sequences from six plant species using the Interactive Phylogenetics Module tool based on rice RNS locus ID from the PLAZA 4.0 database2 (Van Bel et al., 2018). The multiple alignment of the amino acid sequences was carried out using the ClustalX program version 2.0.11 (Higgins et al., 1996). Phylogenetic analysis was performed using MEGA 7 under neighbor-joining tree method (Tamura et al., 2011). In addition, to perform a phylogenomic analysis of RNS genes in rice and Arabidopsis thaliana, we collected eight family members from the Rice Genome Annotation Project using locus IDs (RGAP3) and five Arabidopsis family members from a previous report (MacIntosh et al., 2010). The multiple alignment of the amino acid sequences was carried out using the ClustalX program version 2.0.11 (Higgins et al., 1996). Phylogenetic analysis was performed using MEGA 6 and the following parameters: neighbor-joining tree method, complete deletion, and bootstrap with 1000 replicates (Tamura et al., 2011).
Meta-Analysis of Tissue Expression Profiles
The integration of transcriptomes into a phylogenetic context can direct experimental strategies for further functional analysis (Jung et al., 2010). Therefore, we used meta-analysis of the expression profiles of RNS genes in six tissues/organs based on data from 983 Affymetrix arrays downloaded from the NCBI gene expression omnibus (GEO4) (Cao et al., 2012). We then uploaded the log2 normalized intensity data in tab-delimited text format into Multiple Experiment Viewer (MEV5) and illustrated these data using heat maps. In addition, we analyzed the meta-expression patterns of RNS genes in six tissues/organs of Arabidopsis using the Arabidopsis Affymetrix microarray data series GSE5630, GSE5633, GSE5631, GSE5632, GSE5634, GSM943445, and GSM943446. Similar to the rice data analysis, we generated meta-expression data. For comparative gene expression analysis, we used data from six tissue/organ types for both rice and Arabidopsis. As a result, the meta-expression data of all RNS genes were analyzed. The log2 intensity ranges of microarray data were differently shown between rice and Arabidopsis by a range from 5 to 15 and a range from 5 to 13, respectively, due to normalization. Yellow-colored boxes indicate a high level of expression and blue ones denote a low level of expression. Integrated meta-expression data were used to determine functional conservancy in terms of anatomy between rice and Arabidopsis RNS ortholog pairs.
Global Identification of Rice RNS Genes Stimulated Under Phosphate Starvation Using Public RNA-Sequencing Data
To identify the role of RNS genes under phosphate starvation, we used the RNA-seq data from rice and microarray data from Arabidopsis obtained under phosphate-starvation conditions in a recent report (Woo et al., 2012; Secco et al., 2013; Gho et al., 2018). The fragments per kilobase of transcript per million mapped reads (FPKM) values from whole samples used in this study were downloadable from the National Center for Biotechnology Information6 under accession number SRA097415. Arabidopsis microarray data (GSE34004) were downloaded from the NCBI GEO4. Subsequently, we selected the generation of fold changes between the phosphate-sufficient condition and phosphate starvation for 21 days + 1 h (21 d + 1 h) in shoots and roots in rice and for 10 days in shoots and roots of Arabidopsis. Moreover, we confirmed additional fold changes to compare Pi recovery conditions (+1 h) after phosphate starvation for 21 days with phosphate starvation in rice and Pi recovery conditions for 3 days after phosphate starvation for 10 days with phosphate starvation in Arabidopsis (Supplementary Figure S2). We used average normalized three FPKM values and three intensity values of RNS genes from RNA-seq and microarray data. Heat map images were produced using the MEV software (Chu et al., 2008).
RNase Activity
RNase activity was analyzed by agarose gel electrophoresis. Total RNA was prepared from 21-day-old rice shoot tissues using the purelink plant RNA reagent kit (Invitrogen). Crude protein extracts were prepared from 31-day-old rice plants under diverse experimental conditions (10 days of solid MSO followed by 21 days of P-sufficient or -starvation condition in a hydroponic culture system with shoot and root tissues). After each sample was ground with liquid nitrogen, the samples were vigorously mixed with 10 mM Tris–HCl buffer (pH 7.4) in 4°C. The mixed samples were centrifuged for 10 min, at 12,000 r/min, and 4°C, and the supernatant was calibrated with Quanti-iT Protein Assay (Thermo scientific). For each lane, 1 μg of total RNA was incubated in 25 mM Tris–HCl buffer (pH 7.4) containing 25 mM KCl and 5 mM MgCl2 in the presence of 0.5 and 0.1 μg of total protein from different samples at 30°C for 5 min. After incubation, the RNA samples were loaded onto 1.2% agarose gels containing 1 × MOPS and 2.2 M formaldehyde. This experiment was repeated three times using independent biological replicates.
Assessment of the Integrity of RNA Samples Using Electropherograms
An integrity assessment analysis of RNA samples was performed by a company (Macrogen, Inc., South Korea) using an Agilent 2100 Bioanalyzer and RNA Bioanalyzer Pico 6000 chip (Agilent Technologies, Inc., Santa Clara, CA, United States).
Measurements of Total Pi Content in Plants
Samples were ground in liquid nitrogen after fresh-weight measurement (about 50–70 mg) and then homogenized in 200 μl of 10% perchloric acid (PCA). After adding 1.8 ml of 5% PCA, the homogenate was mixed and placed on ice for 30 min. After centrifugation at 10,000 × g for 10 min at 4°C, the supernatant was used to measure Pi using the molybdate-blue method: 0.4% ammonium molybdate melted in 0.5 M H2SO4 (solution A) was mixed with 10% ascorbic acid (solution B) (A:B = 6:1). Two milliliters of this solution was added to 1 ml of the sample solution and incubated in a water bath at 40°C for 20 min. The absorbance was measured at 820 nm after cooling on ice for 5 min, and the Pi content was calculated from the absorbance value per fresh weight. This experiment was repeated three times by using independent biological replicates. We conducted inorganic Pi measurement according to Yang et al. (2014).
Results
Identification of Rice S-Like RNase Family (RNS) Genes and Comparative Phylogenetic Analysis With Arabidopsis RNS Genes
MacIntosh et al. (2010) performed a phylogenetic analysis of the RNS family that included eight rice and five Arabidopsis RNS genes present in Figures 1A,B. Among them, OsRNS1, OsRNS3, OsRNS4, OsRNS5, and OsRNS8 are class I RNS genes, while OsRNS2 and OsRNS6 are class II RNS genes (MacIntosh et al., 2010). However, in contrast with Arabidopsis, the role of the individual family members was not well characterized in rice. To overcome this limitation, we attempted to assign biological functions to the RNS genes in rice according to tissues/organs or various stresses, including Pi starvation. Although, in the previous study, plant RNSs were already clustered based on phylogenetic analyses (MacIntosh et al., 2010; Ramanauskas and Igic, 2017), we have redrawn the phylogenetic tree using two monocot [rice (eight genes) and Z. mays (five genes)] and four dicot [Arabidopsis (five genes), B. rapa (eight genes), S. lycopersicum (six genes), and M. truncatula (seven genes)] RNSs to identify the evolutionary difference between class I and class II RNS in monocot and dicot plants. As a result, there was no significant difference in class II RNS family between rice and Arabidopsis as described in the previous report (MacIntosh et al., 2010), but in class I RNS family, it has been identified that it was divided into three groups (class I monocot-common, class I monocot-specific, and class I dicot-specific RNS subgroups) in a little more detail (Supplementary Figure S1). These results have already been mentioned. The diversification of class I RNS is due to a process characterized by differential maintenance and expansion in different lineages by gene classification, i.e., rapid gene replication and inactivation that occurs differentially between lineages (Zhang et al., 2000; MacIntosh et al., 2010).
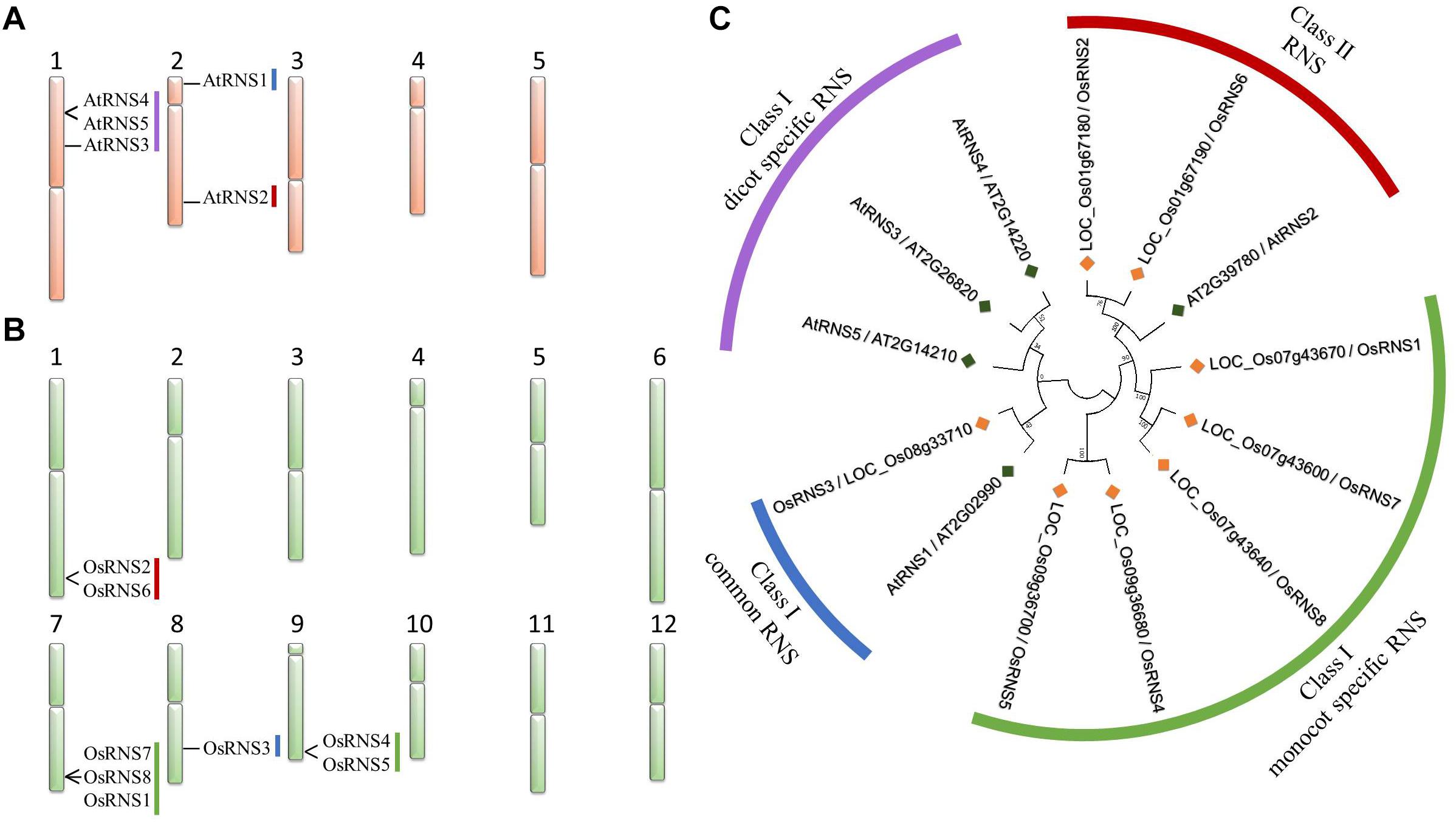
Figure 1. Chromosome map and phylogenetic tree of rice and Arabidopsis RNS family genes. The chromosome map of Arabidopsis RNS family genes (A) and rice RNS family genes (B). Phylogenetic tree of the RNS family comprising eight rice (Oryza sativa) and five Arabidopsis RNS proteins (C). The phylogenetic tree was built using the neighbor-joining method with a bootstrap value of 1000 using MEGA 7.
Six rice and four Arabidopsis RNS genes were assigned to class I, and two rice and one Arabidopsis RNS gene were assigned to class II (Figure 1C). We found evolutionary conserved RNS genes between rice and Arabidopsis, such as LOC_Os08g33710/OsRNS3, which is an ortholog of A. thaliana AT2G02990/AtRNS1 in class I RNS genes, whereas LOC_Os01g67180/OsRNS2 and LOC_Os01g67190/OsRNS6 are orthologs of AT2G39780/AtRNS2 in class II RNS genes (Supplementary Table S2). We hypothesized that rice and Arabidopsis RNS genes that are clustered in the same subgroup have similar biological functions. Conversely, AT1G26820/AtRNS3, AT1G14220/AtRNS4, and AT1G14210/AtRNS5 formed a class I dicot-specific RNS subgroup, whereas three rice RNS genes (OsRNS1, OsRNS7, and OsRNS8) on chromosome 7 and two rice RNS genes (OsRNS4 and OsRNS5) on chromosome 9 formed a monocot-specific subgroup in class I RNS genes. In addition, three rice RNS genes on chromosome 7 and two genes on chromosome 9, together with OsRNS2, OsRNS6, AtRNS4, and AtRNS5, were tandemly duplicated and more likely functionally to be redundant. The estimation of the functional roles of RNS genes belonging to class I monocot-specific and dicot-specific RNS subgroups requires additional data, such as expression profiles and their functional studies.
Functional Assignment of Rice and Arabidopsis RNS Genes Using a Meta-Expression Analysis in Six Tissues/Organs
Although we inferred structural similarity among rice and Arabidopsis RNS genes clustered in the same subgroup using a phylogenetic analysis, additional data are required to determine the conservation of their biological function. Therefore, we performed a meta-expression analysis based on a large collection of microarray data for all RNS genes in six tissues/organs (Figure 2A). Among the Arabidopsis class I RNS genes, AtRNS1 showed the highest expression in seeds and moderate expression in flowers, with AtRNS3 exhibiting similar expression patterns. AtRNS4 and AtRNS5 showed root-preferred expression patterns, but the level of expression of AtRNS5 was higher than that of AtRNS4. Among the rice class I RNS genes, OsRNS4 and OsRNS5 showed shoot-preferred expression patterns; moreover, OsRNS5 exhibited moderate expression, while OsRNS4 was expressed at a low level, in flowers. OsRNS1 exhibited a root-preferred expression pattern, and OsRNS3 was expressed at high level in shoots/leaves and flowers, and at moderate level in seeds, roots, and anthers. OsRNS7 and OsRNS8 exhibited a very low level of expression, and its expression might have been inhibited during evolution. AtRNS4 and AtRNS5 of A. thaliana and OsRNS4 and OsRNS5 of rice showed high amino acid similarity due to tandem redundancy and showed similar anatomical expression patterns, suggesting that they may have been cloned evolutionarily to enhance robustness of gene function. OsRNS3 was clustered with AtRNS1, but the expression patterns of the two genes were dissimilar, suggesting functional divergence between them. The expression profiles of RNS genes belonging to monocot-specific and dicot-specific subgroups suggest that OsRNS1 is a functional ortholog of AtRNS4 and AtRNS5, because all of these genes had root-preferred expression patterns in the class I RNS family. In addition, the expression patterns of OsRNS4 and OsRNS5 informed that the class I RNS subgroup in rice plays unique roles in leaves or shoots compared with their Arabidopsis counterparts. Conversely, AtRNS1 and AtRNS3 might have a unique function in seed development compared with their counterparts in rice. In the class II RNS subgroup, OsRNS2 might be a functional ortholog of AtRNS2 because both genes showed ubiquitous expression patterns that were suggestive of a housekeeping role. In rice, this subgroup includes an additional member, OsRNS6, the expression pattern of which exhibited lower level of expression than both OsRNS2 and AtRNS2. Based on the comparative anatomical expression analysis of RNS family genes between rice and Arabidopsis, we estimated that AtRNS2 and OsRNS2 have a conserved function as housekeeping genes in the class II RNS subgroup, whereas OsRNS1, AtRNS4, and AtRNS5 have a conserved function in root development in the class I RNS subgroup.
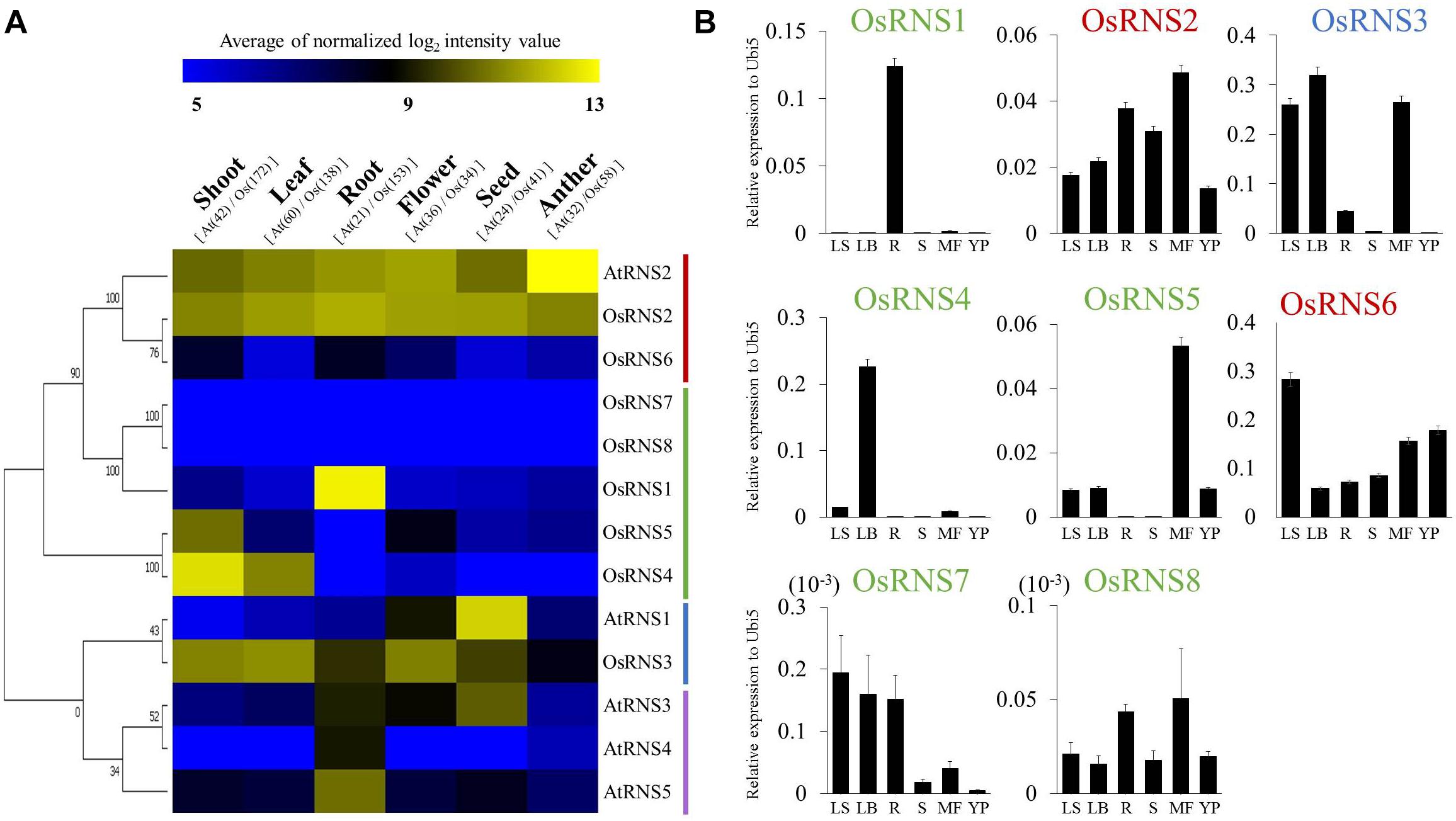
Figure 2. Anatomical meta-expression analysis of data from Affymetrix array platforms, and validation of the meta-expression patterns of eight RNS genes in various tissues/organs via qRT–PCR. (A) Affymetrix expression data from six tissues/organs were used to create the heat map with the MeV software. Blue, low levels of expression; yellow, high levels of expression. (B) qPCR was performed in six tissues/organs (LS, leaf sheath; LB, leaf blade; R, root; S, developing seeds; MF, mature flowers; and YP, young panicle). X-axis, tissues/organs used for qPCR analysis; Y-axis, relative expression level compared with that of rice ubiquitin 5 (OsUbi5, LOC_Os01g22490). Four biological replicates were prepared and analyzed independently. The numbers on the heat map in (A) represent the number of Arabidopsis (At) and rice (Os) samples for each organ. In (B), green letters indicate class I monocot-specific RNS; blue letters indicate class I common RNS; and red letters indicate class II RNS.
Validation of the Anatomical Meta-Expression Patterns of Rice RNS Genes Using qRT-PCR Analysis
To confirm the meta-anatomical expression patterns observed for the rice RNS genes, we carried out a qRT-PCR analysis using leaf sheaths, leaf blades, seedling roots, developing seeds, mature flowers, and young panicles. The three class I RNS genes (OsRNS1, OsRNS4, and OsRNS5) showed a tissue-dependent expression pattern: OsRNS1 had a root-preferred expression pattern; OsRNS4 was significantly expressed in leaf blades; and OsRNS5 showed a mature flower-preferred expression pattern. Interestingly, OsRNS3 was highly expressed in three above-ground tissues. OsRNS2, which was suggested to be a housekeeping gene, was ubiquitously expressed in all tested tissues/organs. OsRNS6 also showed expression in all tested tissues, with the highest expression in leaf sheaths. The expression level of the remaining genes was very weak. In general, qRT-PCR data were well matched with the meta-expression data, indicating that meta-expression information based on a large collection of reference data is highly reliable and effective in estimating gene function (Figure 2B).
A Close Association Between Rice RNS Genes and Phosphate Starvation Was Revealed by Both Meta-Expression and qRT–PCR Analyses
To determine the biological significance of rice RNS genes, we analyzed the diverse meta-expression data under various stress conditions. We found that rice RNS family genes showed significant differential expression under Pi starvation conditions (Woo et al., 2012; Secco et al., 2013; Gho et al., 2018). Supplementary Figure S1 shows the differential expression of these genes in the roots and shoots under Pi starvation compared with the normal condition. In class I RNS family, five rice RNS genes exhibited a > 2-fold upregulation in shoots or roots under Pi starvation. In contrast, AtRNS1 alone showed a > 2-fold upregulation in shoots and roots under Pi starvation in class I RNS family. This result indicates that the rice RNS genes are more closely associated with Pi starvation than are Arabidopsis RNS genes. To confirm the expression patterns observed for the rice RNS genes under Pi starvation, we carried out a qRT-PCR analysis using shoots and roots that were incubated for 21 days under Pi starvation after germination for 10 days. To evaluate the quality of the samples under Pi starvation that were used for RT-PCR analysis, we first checked the expression patterns of two marker genes: rice SULFOQUINOVOSYLDIACYLGLYCEROL 2 (OsSQD2, LOC_Os01g04920) and rice PHOSPHATE TRANSPORTER 6 (OsPT6). We found that OsSQD2 was significantly induced in both roots and shoots under Pi starvation, whereas OsPT6 was only significantly induced in roots (Figure 3 and Supplementary Figure S3). Subsequently, we determined that five RNS genes (OsRNS1, OsRNS3, OsRNS4, OsRNS7, and OsRNS8) in the class I subgroup and two RNS genes (OsRNS2 and OsRNS6) in the class II subgroup were significantly induced under Pi starvation. OsRNS1 showed a root-preferred expression pattern, OsRNS4 showed a shoot-preferred expression pattern, and OsRNS3 was highly expressed in shoots and moderately expressed in roots. Although OsRNS7 and OsRNS8 were expressed at extremely low level, OsRNS7 exhibited PSI repression in the shoot and OsRNS8 exhibited Pi-starvation-inducible positive expression pattern in the root. Between OsRNS2 and OsRNS6, which have been shown to be ubiquitously expressed in most tissues, OsRNS2 was upregulated under Pi starvation in both roots and shoots, while OsRNS6 was upregulated under Pi starvation only in roots (Figure 3). Conversely, OsRNS5 was repressed in shoots under Pi starvation, as assessed in an RNA-seq analysis (Secco et al., 2013), with confirmation of its expression patterns in qRT-PCR analysis. Taken together, these results suggest that six (OsRNS1, OsRNS2, OsRNS3, OsRNS4, OsRNS5, and OsRNS6) out of eight RNS genes in rice are strongly associated with Pi starvation.
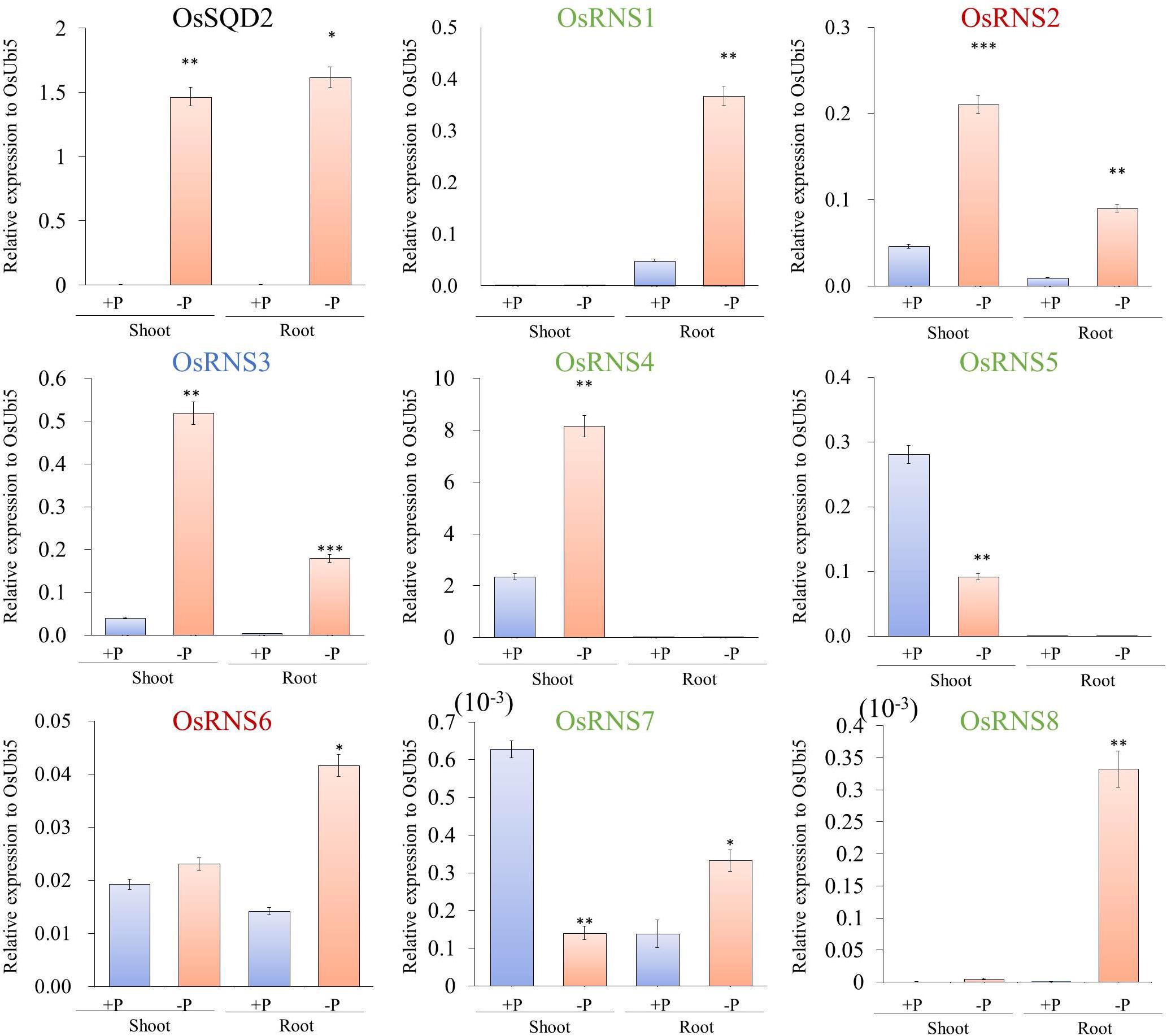
Figure 3. Expression analysis of eight RNS rice genes under phosphate starvation [+P (0.320 mM), –P (0 mM)] via qRT-PCR. X-axis, tissues and conditions used for the qPCR analysis; Y-axis, relative expression level compared with that of OsUbi5. Values are means ± SE (n = 4), and the asterisk indicates that the values of the phosphate starvation differ significantly (P < 0.05) compared with the phosphate-sufficient condition. ***P < 0.001, **P < 0.01, *P < 0.05, based on a t-test. Green letters indicate class I monocot-specific RNS; blue letter indicates class I common RNS; and red letters indicate class II RNS.
Phosphate Deficiency Leads to RNA Degradation Mediated by RNS Genes
To further test the relationship between Pi starvation and the function of RNS genes in rice, we measured the total RNA concentration using a spectrophotometer (Thermo Scientific NanoDrop 2000), ribonuclease activity by RNA gel electrophoresis assay, and the RNA integrity number (RIN) using a Bioanalyzer Pico 6000 chip (Agilent Technologies, Inc., Santa Clara, CA, United States) in samples from shoots and roots that were grown under phosphate-sufficient or -deficient conditions for 21 days. We found that the samples obtained from shoots and roots grown under Pi starvation exhibited a decrease of > 60–80% in the total RNA concentration compared with the normal condition, which was accompanied by increased ribonuclease activity in both shoots and roots under Pi starvation (Figures 4A,B).
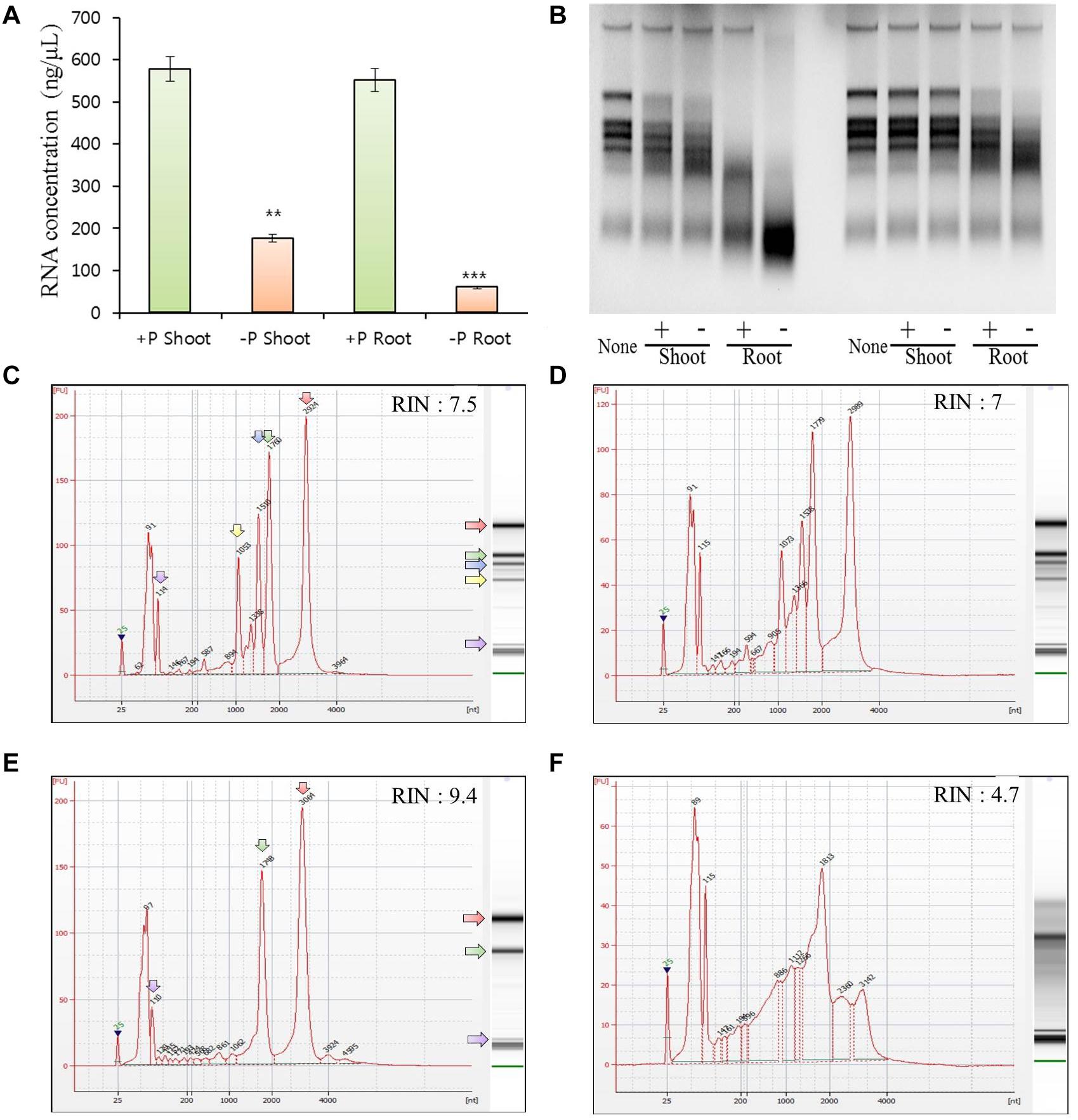
Figure 4. Tissue preferential RNA yield, RNase activity, and RNA integrity assessment of 21 days after germination rice grown under phosphate-sufficient (0.32 mM) or phosphate-starvation (0 mM) conditions. RNA yield of the samples from shoots and roots under phosphate-sufficient or -deficient conditions for 21 days using NanoDrop 2000 (A). Four biological replicates were prepared and analyzed independently. Statistical significance was analyzed using Student’s t-test (∗∗∗P < 0.001; ∗∗P < 0.01). RNase activity was analyzed by agarose gel electrophoresis: 1 μg of total RNA from 21-day-old rice shoot tissues was incubated with 0.5 μg (left) and 0.1 μg (right) of protein from crude extracts from shoot and root samples grown in phosphate-sufficient or -deficient conditions for 21 days. The incubated total RNAs were equally measured (1 μg) and loaded (B). Although we conducted experiments using three biological replicates, we presented only one dataset. Electropherograms of RNA samples from shoots (C,D) and roots (E,F) grown under phosphate-sufficient (C,E) or -deficient (D,F) conditions for 21 days. A standard size ladder in nucleotides (nt) is shown in the X-axis at the bottom of each graph. FU, fluorescent units at the Y-axis. The electrophoresis column generated by the program is located on the right side of each sample graph. The purple arrow indicates the peak of small RNAs. The positions of the 18S and 25S ribosomal RNAs are indicated by a green and red arrow, respectively. Compared with the root RNAs, the leaf RNAs exhibited additional fragments corresponding to the 23S and 16S chloroplastic rRNA fragments, respectively. The positions of the 16S and 23S ribosomal RNAs are indicated by a yellow and blue arrow, respectively.
Plant tissues have three types of rRNAs: chloroplastic (5S, 16S, and 23S rRNAs), cytosolic (5S, 18S, and 25S rRNAs), and mitochondrial (12S and 16S rRNAs). Compared with the root RNAs, the leaf RNAs exhibited additional fragments corresponding to the 23S and 16S chloroplastic rRNA fragments. Thus, they had lower RIN values than those of root samples (Babu and Gassmann, 2016). Our result also indicates that the RIN of shoot RNA was lower than that of root RNA (Figures 4C–F). RNAs from rice shoots grown under long-term Pi starvation exhibited a slightly lower RIN value compared with those obtained from plants grown under Pi-sufficient conditions; moreover, RNAs from rice roots grown under long-term Pi starvation exhibited markedly decreased RIN values compared with those obtained under Pi-sufficient conditions. We also found that the 25S rRNA was severely degraded after long-term Pi starvation in rice roots. Our results suggest that rice root RNAs are more sensitive to degradation in the presence of Pi starvation than are those from rice shoots (Figures 4C–F). Taken together, our findings indicate that the increase in ribonuclease activity triggered by the upregulation of OsRNS genes under Pi starvation might contribute to RNA degradation.
Cis-Acting Regulatory Element (CRE) Analysis of RNS Family Gene Promoters via in silico Analysis
To identify consensus cis-acting regulatory elements (CREs) for PSI expression in eight RNS genes, we first extracted 2-kb sequences upstream of the ATG for these eight genes and analyzed the promoter sequences in the PLANTPAN 2.0 database. The in silico analysis of CREs revealed that 31 CREs were conserved among the eight genes; among them, we selected two consensus CREs in the promoters of the eight RNS genes as candidate CREs involved in Pi-starvation-inducible expression: P1BS/GNATATNC and WBOXNTERF3/TGACY (Supplementary Figure S4). The P1BS, which is an AtPHR1 binding site, is also present in the promoters of many crucial PSI genes (Li et al., 2015; Ruan et al., 2015; Gho et al., 2018). WBOXNTERF3 is one of the WRKY binding sequences, and previous studies have shown that OsWRKY74, AtWRKY45, and AtWRKY75 are involved in regulating PSI responses in rice and Arabidopsis (Devaiah et al., 2007; Dai et al., 2016). It has also been reported that WBOXNTERF3 exists in the promoter area of high-affinity phosphate transporters (PHT1 group) in rice and wheat.
Spatial Regulation of the RNS Family Genes in PSI Signaling
To determine whether OsRNS genes are involved in the PSI signaling pathway via the degradation of RNAs involved in the recycling phosphate sources, we examined their expression patterns in the rice phosphate starvation response 2 overexpressing line (OsPHR2-OX), rice phosphate 2 (ospho2) mutant, and rice phosphate transporter1 overexpressing line (OsPT1-OX) and compared them with those of the wild-type control. To prove the above hypothesis, we acquired an ospho2 mutant that contains a T-DNA insertion in the 5’UTR of OsPHO2 from RiceGE1 and OsPT1-OX as a high-affinity phosphate transporter from DongA University (Seo et al., 2008; Sun et al., 2012). Moreover, we generated OsPHR2-OX transgenic plant and reaffirmed the results that overexpression of OsPHR2 caused the excessive accumulation of Pi. However, we identified that the RNA concentration increased unlike expectation (Figure 5). The loss of function of OsPHO2, a Tos17 transposon insertion mutant line, led to excessive accumulation of Pi and presented leaf tip necrosis (Zhou et al., 2008; Hu et al., 2011; Cao et al., 2014). ospho2 mutant by the T-DNA insertion also revealed the excessive accumulation of Pi (Figures 5A,B) and leaf tip necrosis in mature plant (Supplementary Figure S5). Furthermore, RNA concentration was significantly decreased with degraded RNA integrity in the ospho2 mutant (Figure 5). The function of OsPT1 was reported in Pi starvation response and OsPT1-OX accumulated almost twice as much phosphate (Figure 5). We confirmed the overexpression of OsPHR2 in OsPHR2-OX and of OsPT1 in OsPT1-OX, and the knockout of OsPHO2 in the ospho2 mutant by qRT-PCR analysis and morphology (Supplementary Figures S5–S7). Subsequently, we examined the expression patterns of the eight RNS genes that exhibited differential expression patterns in PSI signaling gene mutants (Figure 6). We found that the expression of OsRNS1 alone was significantly altered in the roots of OsPHR2-OX, ospho2, and OsPT1-OX. The expression of OsRNS4 and OsRNS5, which had shoot-preferred expression, was enhanced in the shoots of OsPHR2-OX and ospho2 compared with the wild type, but not in OsPT1-OX. OsRNS3 was upregulated in the tissues of both OsPHR2-OX and ospho2. Although OsRNS7 was upregulated in OsPHR2-OX and OsPT1-OX, its level of expression was very low. There was no significant difference in expression of OsRNS8 with very low level of expression. OsRNS2 and OsRNS6 are class II RNS genes with ubiquitous expression. Although the extents were very slight, the latter was positively regulated, while the former was negatively regulated only in the shoots of OsPHR2-OX, ospho2, and OsPT1-OX lines. The induction of the OsRNS1, OsRNS3, OsRNS4, and OsRNS5 class I RNS genes by Pi starvation can be modulated through the OsPHR2- and OsPHO2-mediated PSI signaling pathway (Figure 6). Overall, we propose that the regulation of the rice RNS family of genes might be strongly associated with phosphate recycling and scavenging via RNA degradation under Pi starvation through the OsPHR2- and OsPHO2-mediated PSI signaling pathway (Figure 7).
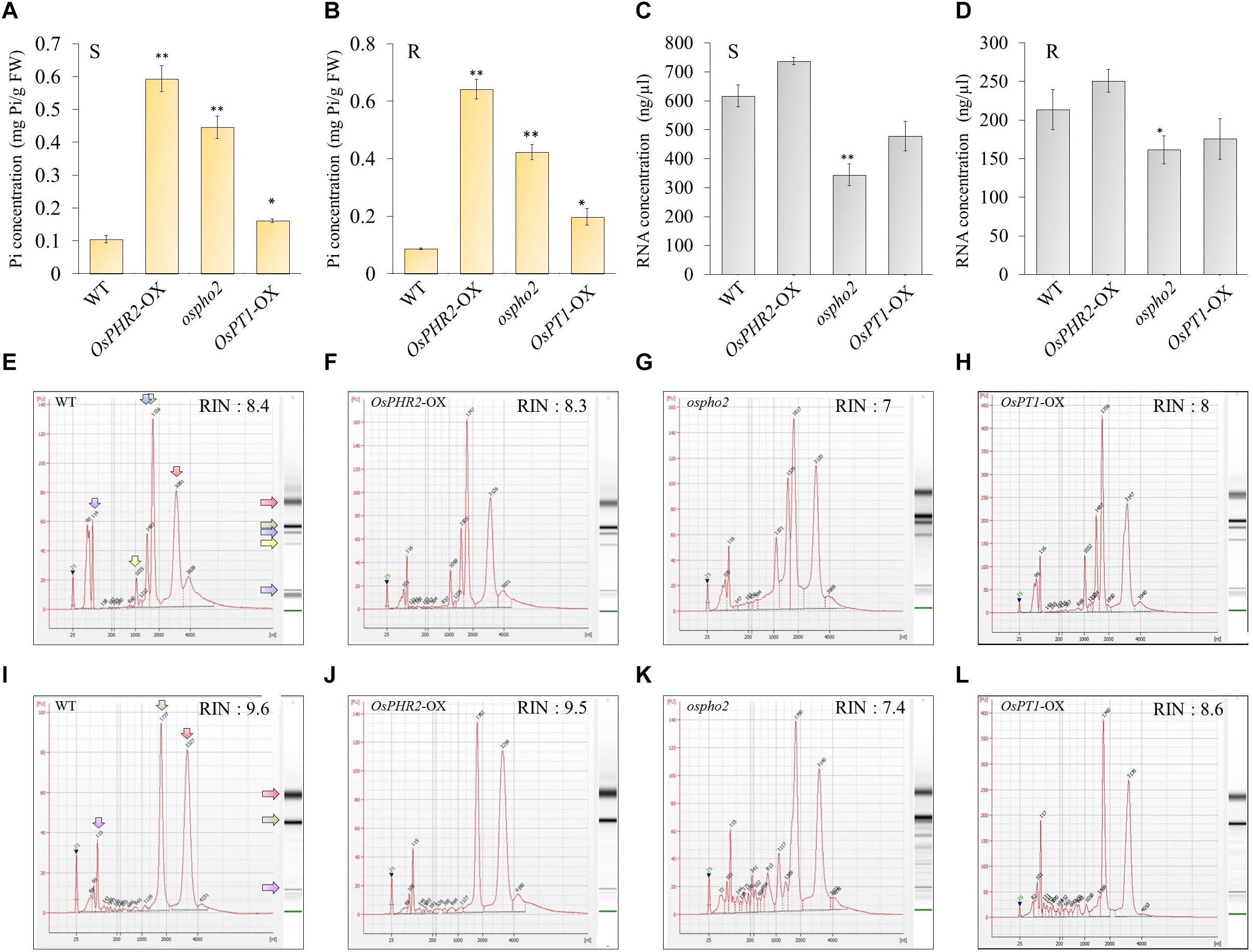
Figure 5. Pi content, RNA concentration, and RIN values of the bioanalyzer data from the roots and shoots of Phosphate Starvation Response 2 (OsPHR2-OX)-overexpressing, Phosphate 2 (ospho2) knockout, and phosphate transporter 1 (OsPT1-OX)-overexpressing lines in the phosphate-starvation-induced (PSI) pathway. Pi content of OsPHR2-OX, ospho2, and OsPT1-OX in the shoots (A) and roots (B) of 10-day seedlings. Determination of the RNA yield of OsPHR2-OX, ospho2, and OsPT1-OX in the shoots (C) and roots (D) of 10-day seedlings using NanoDrop 2000. Values are means ± SE (n = 3), and the asterisk indicates that the values of the mutants or Ox plants differ significantly (P < 0.05) compared with their corresponding wild types. Statistical significance was analyzed using Student’s t-test (∗∗P < 0.01; ∗P < 0.05). Electropherograms of RNA samples from WT (E), OsPHR2-OX (F), ospho2 (G), and OsPT1-OX (H) in shoots and WT (I), OsPHR2-OX (J), ospho2 (K), and OsPT1-OX (L) in roots. A standard size ladder in nucleotides (nt) is shown in the X-axis at the bottom of each graph. FU, fluorescent units at the Y-axis. The electrophoresis column generated by the program is located on the right side of each sample graph. The positions of the 18S and 25S ribosomal RNAs are indicated by a green and red arrow, respectively. The positions of the 16S and 23S ribosomal RNAs are indicated by a yellow and blue arrow, respectively. Although we conducted experiments using three biological replicates with similar results, we presented only one dataset.
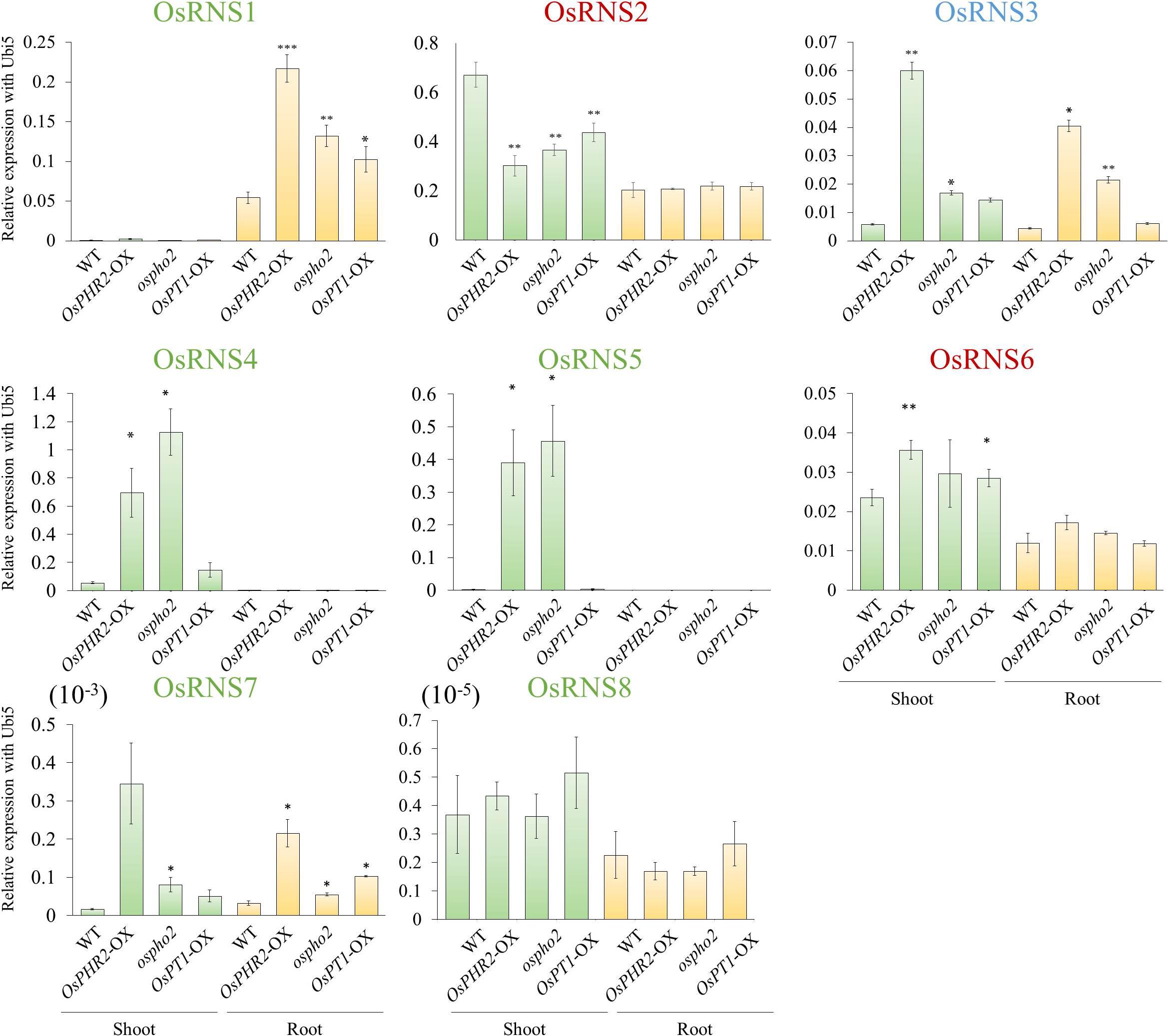
Figure 6. Expression analysis of eight rice RNS genes in the phosphate-starvation-induced (PSI) pathway using OsPHR2-OX, ospho2, and OsPT1-OX via qRT-PCR. X-axis, PSI mutant and tissues used for qPCR analysis; Y-axis, relative expression level compared with OsUbi5. Values are means ± SE (n = 3), and the asterisk indicates that the values of the mutants or Ox plants differ significantly (P < 0.05) compared with their corresponding wild types. ∗∗P < 0.01, ∗P < 0.05, based on a t-test.
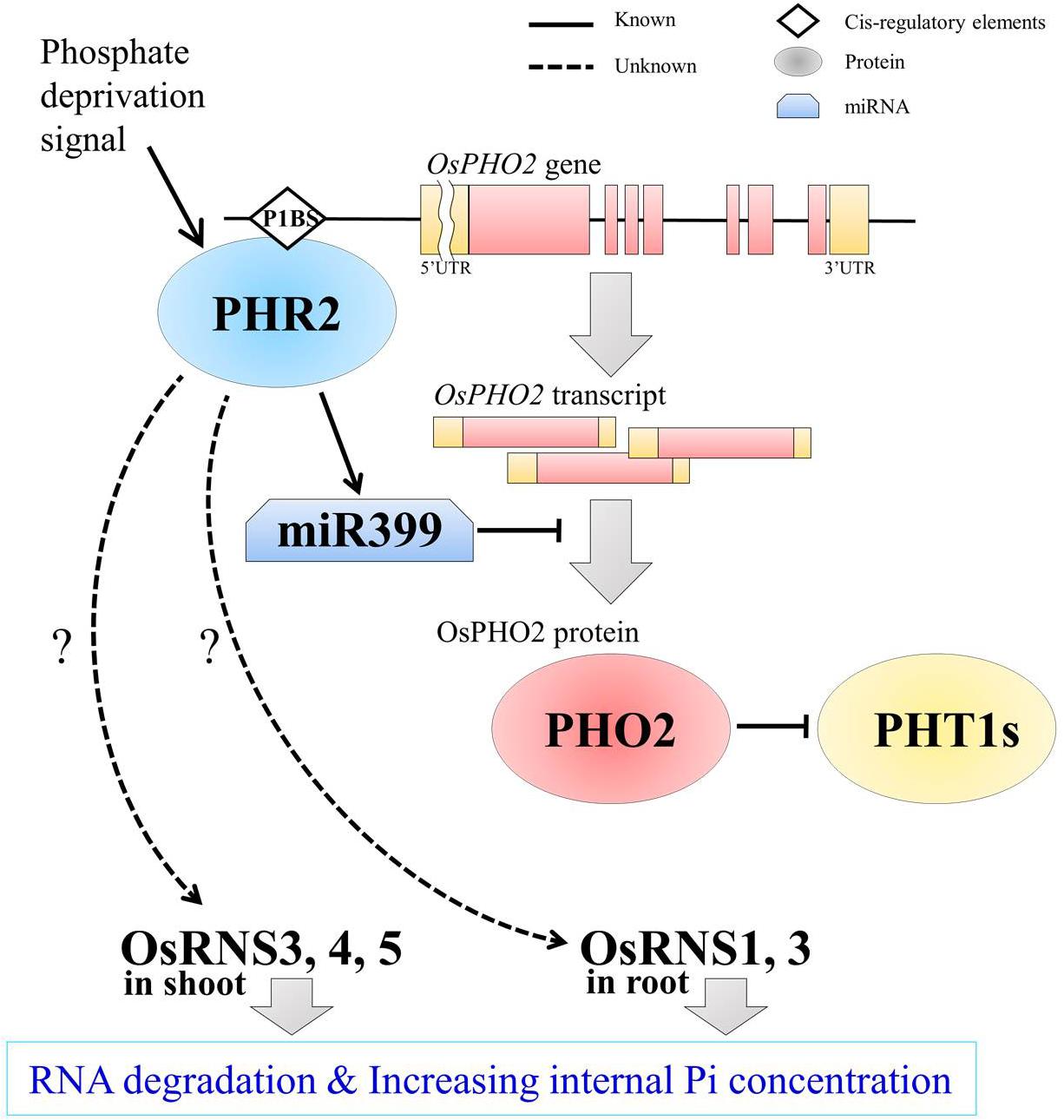
Figure 7. A schematic representation of OsRNSs regulation in the PHR2- and PHO2-mediated PSI signaling pathway for increasing internal Pi concentration in rice. PHR2, as a central regulator of the Pi signaling pathway, increases miR399 levels, thereby inhibiting the E2 ligase, PHO2, at the post-transcriptional level. Meanwhile, the direct binding of PHR2 on P1BS cis-regulatory element in the promoter region of PHO2 was reported for the possibility of transcriptional regulation of PHO2 by PHR2. After the PHO2 protein was generated, it ubiquitinates the phosphate transporters, PHT1s, for maintaining phosphate homeostasis. Our results suggest that OsRNS1, 3, 4, and 5 are negatively regulated by OsPHO2 and that the RNSs are positively regulated by OsPHR2 through expression profiling analysis using the mutants. However, it is questionable that Pi contents were enhanced in OsPHR2 overexpression lines together with the increased amount of the total RNAs including rRNA, the substrate of RNS for rRNA catabolism. The shoot preferential OsRNS3, OsRNS4, and OsRNS5 and the root preferential OsRNS1 and OsRNS3 seem to be spatially separated and regulated. Finally, it is expected that the accumulation of OsRNSs in each tissue will promote RNA degradation and increase the internal Pi concentration. Proteins are represented by circle; cis-regulatory elements, rhombus; mRNA, distorted square; known pathway, solid line; unknown but expected pathway, dotted line.
Discussion
Arabidopsis comprises four class I RNS genes, whereas rice contains six class I RNS genes. Among them, with the exception of a class I common RNS gene (conserved between rice and Arabidopsis), four genes were in the class I monocot-specific RNS subgroup and three genes were in the class I dicot-specific RNS subgroup. In the class I common RNS gene, AtRNS1 is expressed in most tissues but exhibits stronger expression in seeds and flower, whereas OsRNS3, which is the ortholog of AtRNS1, is expressed in most tissues, but has a low level of expression in roots and seeds (Bariola et al., 1994). In rice, OsRNS1 exhibited root-preferred expression, OsRNS4 had leaf blade-preferred expression, and OsRNS5, the tandemly duplicated form of OsRNS4, had flower-preferred expression, while AtRNS4 in the Arabidopsis class I dicot-specific RNS subgroup showed root-specific expression, and the remaining genes did not show specific expression patterns according to the type of tissues/organs. These results suggest that class I RNS genes in rice might be more precisely regulated according to tissues/organs than those of Arabidopsis. Meanwhile, combination of phylogenetic and gene expression analyses suggested that class II RNS genes in rice carry out a housekeeping function in plant species (MacIntosh et al., 2010; Hillwig et al., 2011). AtRNS2 is a unique Arabidopsis class II RNS gene that is expressed in all tissues and senescence and Pi starvation at high levels and is localized at an intracellular compartment (Hillwig et al., 2011; Floyd et al., 2017). In rice, the OsRNS2 and OsRNS6 class II RNS genes are expressed in all tissues and are positively associated with Pi deficiency, similar to AtRNS2 (Figures 2, 3).
Although previous reports elucidated the functional roles of RNS genes in tomato, Arabidopsis, and humans associated with senescence, autophagy, tumor suppression, and antifungal activity, respectively (Lers et al., 1998; Acquati et al., 2011; Floyd et al., 2015; Huang et al., 2015; Islam et al., 2019; Singh et al., 2020), the functional roles of RNS genes in rice have been poorly discovered. In our study, tissue-preferential regulation in response to Pi deficiency is demonstrated. We found that most of class I monocot-specific RNS genes in rice were differentially regulated in response to Pi deficiency according to the type of tissues/organs. OsRNS1 exhibited root-preferred upregulation under Pi starvation, and OsRNS4 showed shoot-preferred upregulation under Pi starvation. AtRNS1 and AtRNS2 are induced by Pi starvation in shoots and roots, while AtRNS3, AtRNS4, and AtRNS5 are not (Taylor et al., 1993; Bariola et al., 1994). It was also reported that the extracellular RNase LX and intracellular RNase Le from S. lycopersicum are induced in tomato suspension cells under Pi starvation (Köck et al., 1998). Furthermore, we found that two class I monocot-specific RNS genes, OsRNS4 and OsRNS5, were upregulated by salt stress and drought stress in addition to responses to Pi deficiency through publicly available transcriptome data analysis (Supplementary Figure S8). It was also reported that OsRNS4 is involved in abscisic acid (ABA)-mediated responses as well as salinity treatment (Zheng et al., 2014).
Interestingly, the OsRNS4 and OsRNS5 shared high amino acid similarity including the two CAS domains, which are conserved in a “monocot inactive cluster” (MacIntosh et al., 2010; Ramanauskas and Igic, 2017). However, their expression by tissue and response under Pi deficiency had different patterns (Figure 3). It is inferred that tandemly duplicated OsRNS4 and OsRNS5 acquire functional diversity by regulation of promoters with different regulatory elements. Such cases have already been reported. The tandemly duplicated PRX genes in rice, Os1-CysPrxA and Os1-CysPrxB, also have different expression patterns. Although tandemly duplicated PRX genes in rice, Os1-CysPrxA and Os1-CysPrxB, might be functionally redundant due to high amino acid sequence similarity, their anatomical expression patterns are quite different (Gho et al., 2017). It has been reported that their promoters contain unique tissue-preferred CREs: the promoter of Os1-CysPrxA has CREs for endosperm preferred expression, while that of Os1-CysPrxB has embryo or root-preferred CREs. Thus, OsRNS4 and OsRNS5 are likely to have lost their ability to cleave RNA, and their tight regulation and fine-grained regulatory mechanisms indicate the potential for a role in phosphate deficiency signaling. In addition, our results also suggest that tissue-specific control of the RNS family triggered by gene duplication events in rice exhibits an increase in functional diversity during evolution compared to Arabidopsis.
In addition to the Pi deficiency signal, recent studies have shown that RNA metabolism, including RNA disruption, RNA stability, and RNA processing, plays an important role in responding to abiotic stresses including heat, salt, and nutrient deficiencies (Huang et al., 2015; Kawa and Testerink, 2017; Matsui et al., 2019; Diaz-Baena et al., 2020). Here, we performed various analyses of RNA content, RNase activity, and RNA stability that suggest that RNS induced under Pi deficiency makes Pi acquisition by RNA degradation (Figures 3–5). Our results could also reveal that RNase activity and RNA degradation were enhanced in both Pi-deficient shoots and roots. In addition, it was confirmed that the 25S rRNA fragment was significantly reduced, and the peaks and bands of rRNAs in total RNA were degraded in both the Pi-deficient shoots and roots, resulting in an increase in small RNA fragments (Figures 4D,F). Based on these results, the possibility of providing Pi through rRNA catabolism in OsRNSs induced by Pi deficiency was suggested. Meanwhile, several reports have shown that RNS genes play an important role in nitrogen deficiency in various plant species through rRNA catabolism. In nitrogen-deficient wheat, TaRNS2 has been reported to increase RNA degradation and is involved in the catabolism of nucleotides for growth (Melino et al., 2018) and Rny1, which is an RNase T2 enzyme from Saccharomyces cerevisiae, plays a major role in autophagy-dependent RNA degradation under nitrogen-starvation conditions (Huang et al., 2015). Furthermore, recently, AtRNS1 was reported to be involved in the generation of tRNA-derived fragments (tRFs) under Pi-deficient conditions (Megel et al., 2019). Unfortunately, our data did not analyze the involvement of rice RNS family in the generation of tRFs, but it is necessary to elucidate the association through subsequent studies.
It has been reported that overexpression of OsPHR2, loss of function of OsPHO2, and overexpression of OsPT1 have excessively accumulated phosphoric acid in plants (Zhou et al., 2008; Cao et al., 2014). Therefore, it was questioned whether the cause of this excessive accumulation of Pi was affected by OsRNSs, so we tried to clarify this through RNA content, Pi content, and RNA degradation analyses between each of the lines and wild type. Although it was confirmed that the Pi content was increased in all three lines, the ospho2 mutant alone exhibited a significantly decreased RNA concentration and degraded rRNA (Figures 5C,D). Moreover, we confirmed that OsRNS1, 3, 4, and 5 were strongly enhanced by the ospho2 mutant and OsPHR2-OX, indicating that OsRNS1, 3, 4, and 5 might be involved in the OsPHR2- and OsPHO2-mediated PSI signaling pathway. However, it is questionable that Pi contents were enhanced in OsPHR2 overexpression lines together with increased amount of the total RNAs including rRNA, the substrate of RNS for rRNA catabolism. These results suggest that there are unknown mechanisms for increasing the Pi content or increase of the RNA anabolism in OsPHR2 overexpression lines.
In summary, we have shown that Pi starvation leads to the induction of OsRNS genes encoding ribonucleases and stimulates RNA degradation for Pi acquired in rice by OsRNS family genes. In addition, tissue-preferential expression of the OsRNS genes under phosphate deficiency plays an important role in RNA remodeling and Pi recycling, and this mechanism is regulated by OsPHR2 and OsPHO2. The role of the OsRNS genes in the PSI signaling process has been explained to some extent through the expression analysis of the OsRNS gene family in the OsPHR2-OX, OsPT1-OX, and ospho2 mutant lines, but the RNS family affects various stress signals and complex plant homeostasis. Thus, the various features of this family in rice as a model crop plant require further studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Y-SG and K-HJ conceived, designed, and supervised the experiments, the data collection, and analyses. Y-SG, HC, and K-HJ performed the experiments, analyzed the data, and wrote the draft of the manuscript. Y-SG and HC performed the molecular analysis. MS, SM, and D-HK provided the reagents, materials, and advice. Y-SG, HC, and K-HJ wrote the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01325901 to K-HJ), the Rural Development Administration (PJ01492703 to K-HJ), and the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (No. 2018043004 to K-HJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Gynheung An and Professor Jong-Seong Jeon for valuable comments and providing T-DNA insertional mutants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.585561/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree of Class I and Class II RNS family proteins in two monocot and four dicot plants obtained by the neighbor-joining method. The comparison of two monocots (Rice and Z. mays) and four dicots (Arabidopsis, B. rapa, Solanum lycopersicum, and Medicago truncatula) RNSs by the phylogenetic tree revealed two classes (i.e., Class I and Class II) of RNS family in plant species and in Class I RNS family, we found three subgroups: Class I dicot specific RNS (purple vertical line), Class I common RNS (blue), and Class I monocot specific RNS (yellow green). Class II RNS was marked as red vertical line. The phylogenetic tree was built using MEGA 7 under the neighbor-joining method.
Supplementary Figure 2 | Transcriptome analysis of eight rice RNS genes and five Arabidopsis RNS genes under phosphate starvation based on public RNA-seq data. Heat map are presented graphically differential expression under log2-(Pi-deficient conditions)/+(Pi-sufficient condition) comparison, or log2-(Pi-resupply condition)/- comparison (left). Red, upregulation of gene expression; green, downregulation of gene expression. Average normalized three FPKM values and three intensity value of RNS genes from RNA-seq and microarray data (right). Blue, lowest level of gene expression; yellow, highest level of expression.
Supplementary Figure 3 | Expression analysis of the marker gene (OsPT6) with root-preferred expression under phosphate station. X-axis, tissues and conditions used for qPCR analysis; Y-axis, relative expression level compared with OsUbi5. Values are means ± SE (n = 4) and asterisk indicates that the values of the phosphate starvation differ significantly (P < 0.05) compared with phosphate sufficient condition. ∗∗∗P-value < 0.001, ∗∗P-value < 0.01, ∗P-value < 0.05, based on a t-test.
Supplementary Figure 4 | Analysis of cis-acting regulatory elements in RNS-family gene promoters (A), detailed CRE information (B).
Supplementary Figure 5 | Schematic position of T-DNA insertional regions on the OsPHO2 genomic structure, expression analysis, and phenotype of the knockout of Phosphate 2 (pho2). Schematic position of T-DNA insertional region on the OsPHO2 genomic structure and genotyping result using a T-DNA primer (TubR3) and gene-specific primers (F/R) in ospho2 mutant (A). The combination of F and TubR3 primers revealed the T-DNA insertion and that of F and R primers indicates knockout mutation in OsPHO2 gene. Confirmation for expression level of OsPHO2 genes in ospho2 mutant (B). Values are means ± SE (n = 3) and asterisk indicates that the OsPHO2 expression values of ospho2 mutants differ significantly (P < 0.05) compared with the expression values of WT. Y-axis, relative expression level compared to OsUbi5. ∗∗∗P-value < 0.001, ∗∗P-value < 0.01, ∗P-value < 0.05, based on a t-test. Image of the T5 plants at 10 days after germination being grown in MS media (C). Image of the T5 plants at 90 days after germination being grown in the field condition (D).
Supplementary Figure 6 | Expression analysis and phenotype of the overexpression of Phosphate Starvation Response 2 (OXPHR2). Confirmation of expression level of OsPHR2-OX (A). Values are means ± SE (n = 3) and asterisk indicates that the OsPHR2 expression values of OsPHR2-OX differ significantly (P < 0.05) compared with the expression values of WT. Y-axis, relative expression level compared to OsUbi5. ∗∗∗P-value < 0.001, ∗∗P-value < 0.01, ∗P-value < 0.05, based on a t-test. Image of the T3 plants at 10 days after germination being grown in MS media (B). Image of the T2 plants at 90 days after germination being grown in the field condition (C).
Supplementary Figure 7 | Expression analysis and phenotype of the overexpression of phosphate transporter 1 (OXPT1) lines. Confirmation of expression level of OsPT1-OX (A). Values are means ± SE (n = 3) and asterisk indicates that the OsPT1 expression values of OsPT1-OX differ significantly (P < 0.05) compared with the expression values of WT. Y-axis, relative expression level compared to OsUbi5. ∗∗∗P-value < 0.001, ∗∗P-value < 0.01, ∗P-value < 0.05, based on a t-test. Image of the T6 plants at 10 days after germination being grown in MS media (B). Image of the T6 plants at 90 days after germination being grown in the field condition (C).
Supplementary Figure 8 | Meta-expression analysis in response to various abiotic stresses based on Affymetrix array platforms. Red, upregulation of gene expression; green, downregulation of gene expression.
Supplementary Table 1 | Summary of the primer sequences used for qPCR analyses in Figures 2, 3, 6 and Supplementary Figures S2, S5.
Supplementary Table 2 | Detailed information of the RNS genes in rice and Arabidopsis.
Footnotes
- ^ http://signal.salk.edu/cgi-bin/RiceGE
- ^ https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_dicots/
- ^ http://rice.plantbiology.msu.edu/
- ^ http://www.ncbi.nlm.nih.gov/geo/
- ^ http://www.tm4.org/mev/
- ^ http://www.ncbi.nlm.nih.gov/sra
References
Acquati, F., Monti, L., Lualdi, M., Fabbri, M., Sacco, M. G., Gribaldo, L., et al. (2011). Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells. Oncotarget 2, 477–484. doi: 10.18632/oncotarget.274
Babu, C., and Gassmann, M. (2016). Assessing Integrity of Plant RNA with the Agilent 2100 Bioanalyzer System. (Waldbronn: Agilent technologies), 1–4.
Bariola, P. A., Howard, C. J., Taylor, C. B., Verburg, M. T., Jaglan, V. D., and Green, P. J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6, 673–685. doi: 10.1046/j.1365-313X.1994.6050673.x
Bariola, P. A., Macintosh, G. C., and Green, P. J. (1999). Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol. 119, 331–342. doi: 10.1104/pp.119.1.331
Beintema, J. J., Breukelman, H. J., Carsana, A., and Furiat, A. (1997). Evolution of vertebrate ribonucleases: ribonuclease A superfamily. Ribonucleases 245–269. doi: 10.1016/B978-012588945-2/50009-1
Beintema, J. J., and Kleineidam, R. G. (1998). The ribonuclease A superfamily: general discussion. Cell Mol. Life Sci. 54, 825–832. doi: 10.1007/s000180050211
Cao, P., Jung, K. H., Choi, D., Hwang, D., and Ronald, P. C. (2012). The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice 5:17. doi: 10.1186/1939-8433-5-17
Cao, Y., Yan, Y., Zhang, F., Wang, H.-D., Gu, M., Wu, X.-N., et al. (2014). Fine characterization of OsPHO2 knockout mutants reveals its key role in Pi utilization in rice. J. Plant Physiol. 171, 340–348. doi: 10.1016/j.jplph.2013.07.010
Chu, V. T., Gottardo, R., Raftery, A. E., Bumgarner, R. E., and Yeung, K. Y. (2008). MeV+R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol. 9:R118. doi: 10.1186/gb-2008-9-7-r118
Cock, J., Yoshida, S., and Forno, D. A. (1976). Laboratory Manual For Physiological Studies of Rice. (Los Baños: International Rice Research Institute).
Dai, X., Wang, Y., and Zhang, W.-H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Deshpande, R. A., and Shankar, V. (2002). Ribonucleases from T2 family. Crit. Rev. Microbiol. 28, 79–122. doi: 10.1080/1040-840291046704
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Diaz-Baena, M., Galvez-Valdivieso, G., Delgado-Garcia, E., Pineda, M., and Piedras, P. (2020). Nuclease and ribonuclease activities in response to salt stress: identification of PvRNS3, a T2/S-like ribonuclease induced in common bean radicles by salt stress. Plant Physiol. Biochem. 147, 235–241. doi: 10.1016/j.plaphy.2019.12.016
Floyd, B. E., Morriss, S. C., Macintosh, G. C., and Bassham, D. C. (2015). Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11, 2199–2212. doi: 10.1080/15548627.2015.1106664
Floyd, B. E., Mugume, Y., Morriss, S. C., Macintosh, G. C., and Bassham, D. C. (2017). Localization of RNS2 ribonuclease to the vacuole is required for its role in cellular homeostasis. Planta 245, 779–792. doi: 10.1007/s00425-016-2644-x
Gho, Y.-S., An, G., Park, H.-M., and Jung, K.-H. (2018). A systemic view of phosphate starvation-responsive genes in rice roots to enhance phosphate use efficiency in rice. Plant Biotechnol. Rep. 12, 249–264. doi: 10.1007/s11816-018-0490-y
Gho, Y.-S., Park, S.-A., Kim, S.-R., Chandran, A. K. N., An, G., and Jung, K.-H. (2017). Comparative Expression Analysis of rice and Arabidopsis peroxiredoxin genes suggests conserved or diversified roles between the two species and leads to the identification of tandemly duplicated rice peroxiredoxin genes differentially expressed in seeds. Rice 10:30. doi: 10.1186/s12284-017-0170-5
Higgins, D. G., Thompson, J. D., and Gibson, T. J. (1996). Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402. doi: 10.1016/S0076-6879(96)66024-8
Hillwig, M. S., Contento, A. L., Meyer, A., Ebany, D., Bassham, D. C., and Macintosh, G. C. (2011). RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. U.S.A. 108, 1093–1098. doi: 10.1073/pnas.1009809108
Hu, B., Zhu, C., Li, F., Tang, J., Wang, Y., Lin, A., et al. (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156, 1101–1115. doi: 10.1104/pp.110.170209
Huang, H., Kawamata, T., Horie, T., Tsugawa, H., Nakayama, Y., Ohsumi, Y., et al. (2015). Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 34, 154–168. doi: 10.15252/embj.201489083
Igic, B., and Kohn, J. R. (2001). Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. U.S.A. 98, 13167–13171. doi: 10.1073/pnas.231386798
Irie, M. (1999). Structure-function relationships of acid ribonucleases: lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 81, 77–89. doi: 10.1016/s0163-7258(98)00035-7
Islam, M. S., Proshad, R., Kormoker, T., and Tusher, T. R. (2019). Autophagy-mediated nutrient recycling and regulation in plants: a molecular view. J. Plant Biol. 62, 307–319. doi: 10.1007/s12374-019-0213-0
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Jung, K. H., Cao, P., Seo, Y. S., Dardick, C., and Ronald, P. C. (2010). The Rice Kinase Phylogenomics Database: a guide for systematic analysis of the rice kinase super-family. Trends. Plant Sci. 15, 595–599. doi: 10.1016/j.tplants.2010.08.004
Kaletta, K., Kunze, I., Kunze, G., and Kock, M. (1998). The peptide HDEF as a new retention signal is necessary and sufficient to direct proteins to the endoplasmic reticulum. FEBS Lett. 434, 377–381. doi: 10.1016/S0014-5793(98)01013-8
Kawa, D., and Testerink, C. (2017). Regulation of mRNA decay in plant responses to salt and osmotic stress. Cell. Mol. Life Sci. 74, 1165–1176. doi: 10.1007/s00018-016-2376-x
Kim, S.-R., Lee, D.-Y., Yang, J.-I., Moon, S., and An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73. doi: 10.1007/s12374-008-9008-4
Köck, M., Theierl, K., Stenzel, I., and Glund, K. (1998). Extracellular administration of phosphate-sequestering metabolites induces ribonucleases in cultured tomato cells. Planta 204, 404–407. doi: 10.1007/s004250050273
Lers, A., Khalchitski, A., Lomaniec, E., Burd, S., and Green, P. J. (1998). Senescence-induced RNases in tomato. Plant Mol. Biol. 36, 439–449. doi: 10.1023/a:1005993024161
Li, L. H., Guo, N., Wu, Z. Y., Zhao, J. M., Sun, J. T., Wang, X. T., et al. (2015). P1BS, a conserved motif involved in tolerance to phosphate starvation in soybean. Genet. Mol. Res. 14, 9384–9394. doi: 10.4238/2015.August.14.2
Liu, F., Wang, Z., Ren, H., Shen, C., Li, Y., Ling, H. Q., et al. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. doi: 10.1111/j.1365-313X.2010.04170.x
MacIntosh, G. C., and Bassham, D. C. (2011). The connection between ribophagy, autophagy and ribosomal RNA decay. Autophagy 7, 662–663. doi: 10.4161/auto.7.6.15447
MacIntosh, G. C., Hillwig, M. S., Meyer, A., and Flagel, L. (2010). RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol. Genet. Genomics 283, 381–396. doi: 10.1007/s00438-010-0524-9
Matsui, A., Nakaminami, K., and Seki, M. (2019). Biological Function of Changes in RNA Metabolism in Plant Adaptation to Abiotic Stress. Plant Cell Physiol. 60, 1897–1905. doi: 10.1093/pcp/pcz068
Megel, C., Hummel, G., Lalande, S., Ubrig, E., Cognat, V., Morelle, G., et al. (2019). Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 47, 941–952. doi: 10.1093/nar/gky1156
Melino, V. J., Casartelli, A., George, J., Rupasinghe, T., Roessner, U., Okamoto, M., et al. (2018). RNA catabolites contribute to the nitrogen pool and support growth recovery of wheat. Front. Plant Sci. 9:1539. doi: 10.3389/fpls.2018.01539
Moon, S., Chandran, A. K. N., Kim, Y.-J., Gho, Y., Hong, W.-J., An, G., et al. (2019). Rice RHC encoding a putative cellulase is essential for normal root hair elongation. J. Plant Biol. 62, 82–91. doi: 10.1007/s12374-018-0393-z
Morriss, S. C., Liu, X., Floyd, B. E., Bassham, D. C., and Macintosh, G. C. (2017). Cell growth and homeostasis are disrupted in arabidopsis rns2-2 mutants missing the main vacuolar RNase activity. Ann. Bot. 120, 911–922. doi: 10.1093/aob/mcx099
Plaxton, W. C., and Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Ramanauskas, K., and Igic, B. (2017). The evolutionary history of plant T2/S-type ribonucleases. PeerJ 5:e3790. doi: 10.7717/peerj.3790
Rojas, H., Caspani, C., Escobar, E., Quiroga, R., and Goldraij, A. (2018). NaPi/S X-RNase segregates as a functional S-RNase and is induced under phosphate deficiency in Nicotiana alata. Biol. Plant. 62, 261–268. doi: 10.1007/s10535-018-0783-6
Rojas, H., Floyd, B., Morriss, S. C., Bassham, D., Macintosh, G. C., and Goldraij, A. (2015). NnSR1, a class III non-S-RNase specifically induced in Nicotiana alata under phosphate deficiency, is localized in endoplasmic reticulum compartments. Plant Sci. 236, 250–259. doi: 10.1016/j.plantsci.2015.04.012
Rojas, H. J., Roldan, J. A., and Goldraij, A. (2013). NnSR1, a class III non-S-RNase constitutively expressed in styles, is induced in roots and stems under phosphate deficiency in Nicotiana alata. Ann. Bot. 112, 1351–1360. doi: 10.1093/aob/mct207
Ruan, W., Guo, M., Cai, L., Hu, H., Li, C., Liu, Y., et al. (2015). Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol. Biol. 87, 429–440. doi: 10.1007/s11103-015-0289-y
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Secco, D., Jabnoune, M., Walker, H., Shou, H., Wu, P., Poirier, Y., et al. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25, 4285–4304. doi: 10.1105/tpc.113.117325
Seo, H. M., Jung, Y., Song, S., Kim, Y., Kwon, T., Kim, D. H., et al. (2008). Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnol. Lett. 30, 1833–1838. doi: 10.1007/s10529-008-9757-7
Singh, N. K., Paz, E., Kutsher, Y., Reuveni, M., and Lers, A. (2020). Tomato T2 ribonuclease LE is involved in the response to pathogens. Mol. Plant Pathol. 21, 895–906. doi: 10.1111/mpp.12928
Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X., Ai, P., et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159, 1571–1581. doi: 10.1104/pp.112.196345
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Taylor, C. B., Bariola, P. A., Delcardayre, S. B., Raines, R. T., and Green, P. J. (1993). RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc. Natl. Acad. Sci. U.S.A. 90, 5118–5122. doi: 10.1073/pnas.90.11.5118
Van Bel, M., Diels, T., Vancaester, E., Kreft, L., Botzki, A., Van De Peer, Y., et al. (2018). PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196. doi: 10.1093/nar/gkx1002
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Woo, J., Macpherson, C. R., Liu, J., Wang, H., Kiba, T., Hannah, M. A., et al. (2012). The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12:62. doi: 10.1186/1471-2229-12-62
Wu, P., and Wang, Z. (2011). Molecular mechanisms regulating Pi-signaling and Pi homeostasis under OsPHR2, a central Pi-signaling regulator, in rice. Front. Biol. 6, 242–245. doi: 10.1007/s11515-011-1050-9
Yang, W. T., Baek, D., Yun, D. J., Hwang, W. H., Park, D. S., Nam, M. H., et al. (2014). Overexpression of OsMYB4P, an R2R3-type MYB transcriptional activator, increases phosphate acquisition in rice. Plant Physiol. Biochem. 80, 259–267. doi: 10.1016/j.plaphy.2014.02.024
Zhang, J., Dyer, K. D., and Rosenberg, H. F. (2000). Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. U.S.A. 97, 4701–4706. doi: 10.1073/pnas.080071397
Zheng, J., Wang, Y., He, Y., Zhou, J., Li, Y., Liu, Q., et al. (2014). Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice. Plant Sci. 214, 99–105. doi: 10.1016/j.plantsci.2013.10.003
Zhou, J., Jiao, F., Wu, Z., Li, Y., Wang, X., He, X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Keywords: rice, S-like RNases, phosphate starvation, phosphate recycling, RNA degradation
Citation: Gho Y-S, Choi H, Moon S, Song MY, Park HE, Kim D-H, Ha S-H and Jung K-H (2020) Phosphate-Starvation-Inducible S-Like RNase Genes in Rice Are Involved in Phosphate Source Recycling by RNA Decay. Front. Plant Sci. 11:585561. doi: 10.3389/fpls.2020.585561
Received: 21 July 2020; Accepted: 02 November 2020;
Published: 30 November 2020.
Edited by:
Gustavo C. MacIntosh, Iowa State University, United StatesReviewed by:
Lingyun Cheng, China Agricultural University, ChinaPedro Piedras, University of Córdoba, Spain
Copyright © 2020 Gho, Choi, Moon, Song, Park, Kim, Ha and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ki-Hong Jung, a2hqdW5nMjAxMEBraHUuYWMua3I=
 Yun-Shil Gho
Yun-Shil Gho Heebak Choi
Heebak Choi Sunok Moon1
Sunok Moon1 Doh-Hoon Kim
Doh-Hoon Kim Sun-Hwa Ha
Sun-Hwa Ha Ki-Hong Jung
Ki-Hong Jung