- 1USDA-ARS-SHRS, National Clonal Germplasm Repository, Miami, FL, United States
- 2Singapore Botanic Gardens, National Parks Board, Singapore, Singapore
- 3Institute of Environment, Florida International University, Miami, FL, United States
One of the two major clades of the endemic American Amaryllidaceae subfam. Amaryllidoideae constitutes the tetraploid-derived (n = 23) Andean-centered tribes, most of which have 46 chromosomes. Despite progress in resolving phylogenetic relationships of the group with plastid and nrDNA, certain subclades were poorly resolved or weakly supported in those previous studies. Sequence capture using anchored hybrid enrichment was employed across 95 species of the clade along with five outgroups and generated sequences of 524 nuclear genes and a partial plastome. Maximum likelihood phylogenetic analyses were conducted on concatenated supermatrices, and coalescent-based species tree analyses were run on the gene trees, followed by hybridization network, age diversification and biogeographic analyses. The four tribes Clinantheae, Eucharideae, Eustephieae, and Hymenocallideae (sister to Clinantheae) are resolved in all analyses with > 90 and mostly 100% support, as are almost all genera within them. Nuclear gene supermatrix and species tree results were largely in concordance; however, some instances of cytonuclear discordance were evident. Hybridization network analysis identified significant reticulation in Clinanthus, Hymenocallis, Stenomesson and the subclade of Eucharideae comprising Eucharis, Caliphruria, and Urceolina. Our data support a previous treatment of the latter as a single genus, Urceolina, with the addition of Eucrosia dodsonii. Biogeographic analysis and penalized likelihood age estimation suggests an origin in the Cauca, Desert and Puna Neotropical bioprovinces for the complex in the mid-Oligocene, with more dispersals than vicariances in its history, but no extinctions. Hymenocallis represents the only instance of long-distance vicariance from the tropical Andean origin of its tribe Hymenocallideae. The absence of extinctions correlates with the lack of diversification rate shifts within the clade. The Eucharideae experienced a sudden lineage radiation ca. 10 Mya. We tie much of the divergences in the Andean-centered lineages to the rise of the Andes, and suggest that the Amotape—Huancabamba Zone functioned as both a corridor (dispersal) and a barrier to migration (vicariance). Several taxonomic changes are made. This is the largest DNA sequence data set to be applied within Amaryllidaceae to date.
Introduction
Amaryllidaceae J.St.-Hil. is a family of herbaceous monocots with a cosmopolitan distribution containing approximately 90 genera and 1,700–1,800 species (Meerow and Snijman, 1998) that originated in Africa (Meerow et al., 1999). The family comprises three subfamilies (APG, 2003, 2009): Allidoideae Herb. (=Alliaceae J.G. Agardh, the economically-important onion family, 13 genera and 795 spp., the majority in Allium L.), South African Agapanthoideae Endl. (former Agapanthaceae F. Voight, one genus of ca. nine spp.), and Amaryllidoideae Burnett (Amaryllidaceae s.s., ca. 75 genera and ca. 900 spp.). The latter is the subject of this study and has a broad distribution, with a center of diversity in the southern hemisphere tropics. To this subfamily belong many of the world’s most celebrated ornamental bulbs, including daffodils (Narcissus L.), snowdrops (Galanthus L.) and “amaryllis” (Hippeastrum Herb.). Its species are characterized by 6-parted flowers with undifferentiated tepals and inferior ovaries, borne in pseudoumbels atop a leafless stalk known as a scape.
Three tribes of Amaryllidaceae subfam. Amaryllidoideae are almost entirely restricted to Africa (Amaryllideae Dumort., Cyrtantheae W. Aiton, and Haemantheae Hutch.) and one to Australasia (Calostemmateae D.Müll.-Doblies & U.Müll-Doblies) (Meerow and Snijman, 1998; Meerow et al., 1999). These tribes represent the first branches in published phylogenies of the subfamily (Meerow et al., 1999; Ito et al., 1999; Meerow, 2010; Rønsted et al., 2012). Perhaps the most unexpected resolution in early plastid DNA phylogenies of the family (Meerow et al., 1999; Ito et al., 1999), was the sister relationship of the endemic American clade of subfam. Amaryllidoideae to the Eurasian clade of the subfamily (tribes Galantheae Salisb., Lycorideae Traub, Narcisseae Endl., and Pancratieae Salisb.; Meerow et al., 2006). The dispersal of the Old World genus Crinum L. (tribe Amaryllideae) into the Americas is considered a separate event (Meerow et al., 2003; Kwembeya et al., 2007). This Eurasian/American clade relationship has led some to declare it as evidence of a boreotropical (Wolfe, 1975; Tiffney, 1985a,b; Tiffney and Manchester, 2001) origin for the American clade (Christenhusz and Chase, 2013), despite cladistic patterns that are inconsistent with expectations of the boreotropics hypothesis, an absence of fossils (Meerow et al., 2000), and a lack of biogeographic analyses.
The relationships of the endemic American genera were inferred using the internal transcribed spacer regions of nuclear ribosomal DNA (Meerow et al., 2000). These major relationships have also been supported by plastid data (Meerow et al., 1999, 2000; Meerow and Snijman, 2005; Meerow, 2010). The endemic American genera of the family were resolved as two major clades (Meerow et al., 1999, 2000). The “hippeastroid” clade are diploid (x = 11) or dysploid (x = 6, 8, 9), and primarily the extra-Andean element of the family, comprising the Brazilian endemic tribe Griffinieae Ravenna (Cearanthes Ravenna, Griffinia Ker-Gawl. and Worsleya Traub) (Campos-Rocha et al., 2019a,b), and the tribe Hippeastreae Herb. ex Sweet. (García et al., 2019). Several genera within the hippeastroid clade were recovered as polyphyletic (Rhodophiala C. Presl., Zephyranthes Herb.) with ITS (Meerow et al., 2000) and the possibility of reticulate evolution (i.e., early hybridization) in these lineages was hypothesized (Meerow, 2010; Meerow et al., 2000), and later confirmed with further analysis of plastome and multiple nuclear gene sequences (García et al., 2014, 2017). Hippeastreae constitutes two main clades, the subtribe Hippeastrinae Walp and the Chilean endemic subtribe Traubiinae D. & U. Müll.-Doblies (García et al., 2014, 2017, 2019). In contrast to the Hippeastrinae, the Traubiinae exhibits a mostly tree-like pattern of evolution (García et al., 2017). García et al. (2019) presented a new classification scheme for Hippeastreae that reflects its reticulate phylogeny.
The second major clade in the Americas constitutes the tetraploid-derived (n = 23) Andean-centered tribes (Figure 1), most of which have 46 chromosomes (Meerow, 1987c). This somatic number may have arisen by duplication or fission of one chromosome in an ancestor with 2n = 22, considered ancestral in the subfamily (Meerow and Snijman, 1998), followed by genome doubling, which restored secondary balance (Satô, 1938; Bose, 1962). Many genera of the clade have a staminal corona or cup formed by the proximal connation of the filaments (Meerow and Snijman, 1998; Waters et al., 2013). The Andean clade is characterized by three consistent indels in nrDNA, two in the ITS1 and one in the ITS2 regions (Meerow et al., 2000).
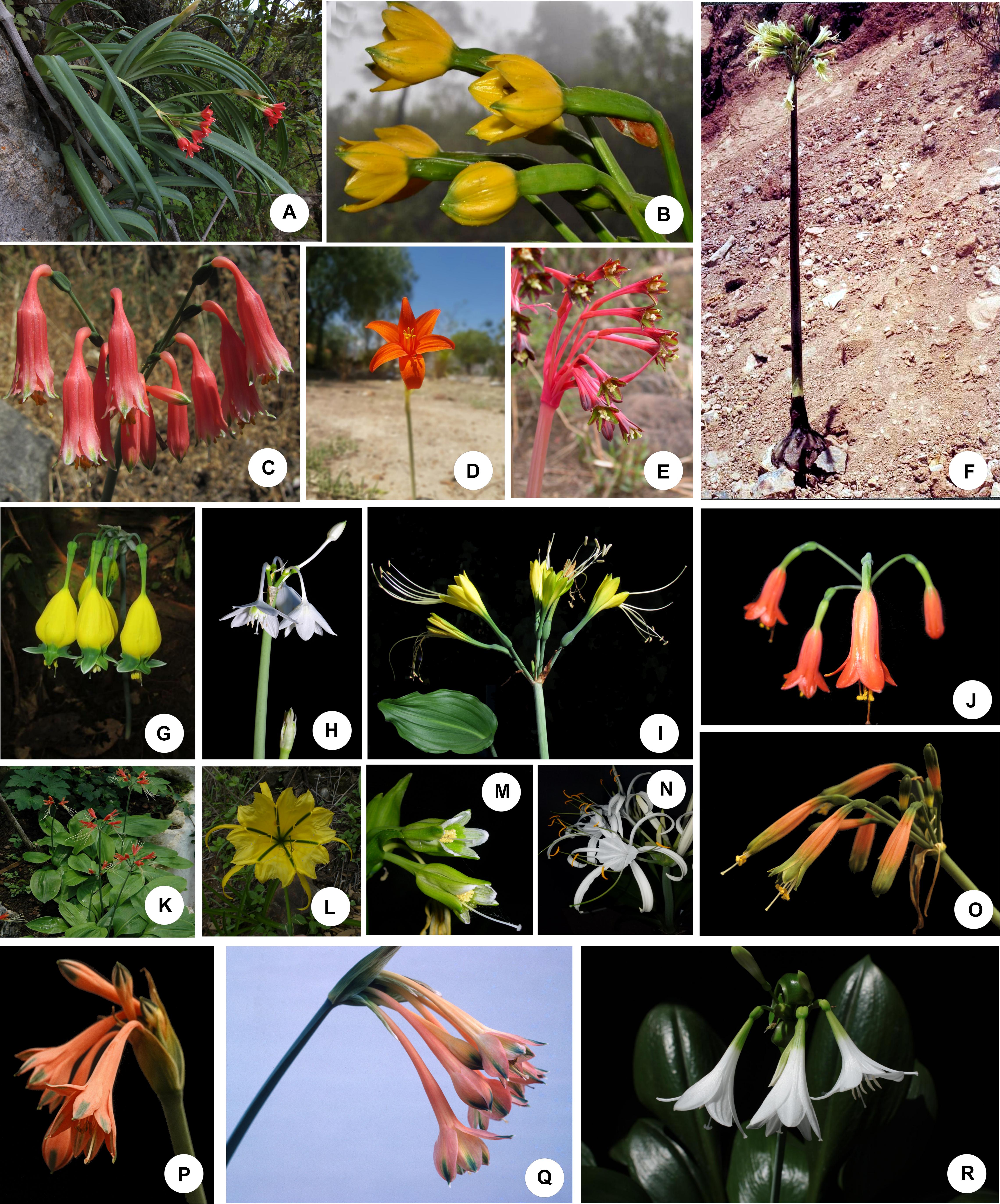
Figure 1. Representative species of the Andean tetraploid clade of the American Amaryllidaceae. (A–C,P,Q) Tribe Clinantheae. (D,E) Tribe Eustephieae. (F–K,O,R) Tribe Eucharideae. (L–N) Tribe Hymenocallideae. (A) Paramongaia milagroantha (Leiva & Meerow) Meerow. (B) Pamianthe ecollis Silverst., Meerow & Sánchez-Taborda. (C). Clinanthus campodensis (Ravenna) Meerow. (D) Pyrolirion tubiflorum L’Hér.) M.Roem. (E) Hieronymiella latifolia (R.E.Fr.) Di Fulvio & Hunz. (F) Rauhia albescens Meerow & Sagást. (G) Urceolina pendula Herb. (H) Eucharis cyaneosperma Meerow. (I) Eucrosia dodsonii Meerow & Dehgan. (J) Stenomesson ecuadorense Meerow, Oleas & L.Jost. (K) Eucrosia bicolor Ker Gawl. (L) Leptochiton helianthus (Ravenna) Gereau & Meerow. (M) Ismene parviflora Meerow & A. Cano. (N) Hymenocallis speciosa (L.f. ex Salisb.) Salisb. (O) Phaedranassa ventricosa Baker. (P) Clinanthus incarnatus (Kunth) Meerow. (Q) Clinanthus variegatus (Ruiz & Pav.) Meerow. (R) Caliphruria subedentata Baker. Photo credits: (A,C,L) Segundo Leiva. (B) Jhon A. Sánchez-Taborda. (D,I–K,N–R) Alan Meerow. (E) John Wood. (F) Abundio Sagástegui. (G,H) Günter Gerlach. (M) Asuncíon Cano. Photos used by permission.
Currently, there are 18 recognized genera in the clade, and ca. 200 spp. (Meerow and Snijman, 1998). Tribe Eustephieae Hutch., which is most diverse from southern Peru through Bolivia and Argentina, is the first branch of this clade, and lacks the indel in ITS2. The tribe Hymenocallideae (D. & U.Müll.-Doblies) Meerow (Meerow et al., 2002), which extends from the central Andes north to the southeastern United States (Figures 1L–N), and its exclusively Andean sister tribe Clinantheae Meerow (Clinanthus Herb., Figures 1C,P,Q; Paramongaia Velarde, Figure 1A; Pamianthe Stapf, Figure 1B) were recognized. Clinanthus was segregated from a polyphyletic Stenomesson Herb., which is now reserved for species of the genus with pseudopetiolate leaves (Meerow et al., 2000). A pseudopetiolate-leafed clade, containing elements of both Eucharideae Hutch. and Stenomesseae Traub (tribe Eucharideae, Figures 1F–K,O,R), was also resolved.
Despite this progress, several relationships within the Andean tetraploid clade were unresolved or weakly supported in these previous studies. Sequence capture using anchored hybrid enrichment, also known as Hyb-Seq (Cronn et al., 2012; Lemmon et al., 2012; Lemmon and Lemmon, 2013; Weitemier et al., 2014) has been informative at various taxonomic levels in plants (Fragoso-Martínez et al., 2017; Wanke et al., 2017; Carter et al., 2019; Granados Mendoza et al., 2020; Larridon et al., 2020; Murphy et al., 2020). The methodology yields sequences of hundreds of single or low copy nuclear loci.
In this paper, we present the results of such a phylogenomic approach across the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae. We also analyze a partial plastome alignment of the clade, extracted and assembled from the sequence capture raw data, and from these two data matrices infer potential reticulation events, biogeographic patterns, and dates of divergence. We sought to test the following hypotheses: (1) phylogenomic data support the previous tribal and generic classification of the tetraploid Andean clade inferred from ITS and several plastid loci, (2) reticulation with each tribe is limited to interspecific gene flow at the generic level, (3) geological events in the Andean region were primary factors in the generic diversification of the clade, and (4) polyploidy and resulting paralogy are not necessary impediments to accurate phylogenomic inference. To date, this is the largest DNA sequence data set applied to Amaryllidaceae.
Materials and Methods
Sampling
We used genomic DNA from 100 species for the sequence capture, 95 from the Andean clade, with Pancratium zeylanicum L. (Eurasian clade) as outgroup, but also including four species of the hippeastroid clade (Supplementary Table 1). Every genus in the Andean clade was sampled, except for Mathieua Klotzch, presumed extinct and likely synonymous with Stenomesson (Meerow, 1987d). The most heavily sampled genus was Hymenocallis Salisb., with 42 samples. Approximate percent sampling of recognized spp. of each genus in our study is as follows: Caliphruria, 75%; Chlidanthus Herb., 25%; Clinanthus, 36%; Eucharis, 39%; Eucrosia, 71%; Eustephia Cav., 25%; Hieronymiella Pax, 17%; Hymenocallis Salisb., 59%; Ismene Salisb., 25%; Leptochiton Sealy, 50%; Pamianthe, 66%; Paramongaia, 100%; Phaedranassa, 44%; Plagiolirion Baker, 100%; Pyrolirion Herb., 50%; Rauhia Traub, 20%; Stenomesson Herb., 35%; Urceolina Rchb., 25%.
Sequencing
We identified putative orthologous single-copy nuclear (SCN) loci from two floral transcriptomes (Griffinia liboniana C. Morren and Rauhia staminosa Ravenna) previously obtained through Beijing Genomics Institute (Shenzhen, China) using MarkerMiner 1.0 (Chamala et al., 2015) with a minimum sequence length of 300 bp and the SCN gene reference set of Oryza sativa L. curated by De Smet et al. (2013). MarkerMiner identified 751 SCN orthogroups which were reduced to 735 loci by excluding the orthogroups containing apparent paralogs, and sequence alignments were provided to Rapid Genomics (Gainesville, FL, United States) for probe design. A total of 45,054 probes of 120 bp were designed at 3× coverage across predicted exons of longer than 120 bp. DNA was extracted from 20 to 100 mg of dried or fresh leaf tissue using FastDNA Kit (MP Biomedicals, Irvine, CA, United States) and provided to Rapid Genomics with whom we contracted for sequence capture and initial bioinformatic analysis. DNA libraries were made by random mechanical shearing of our genomic DNA to an average size of 400 bp followed by an end-repair reaction, ligation of an adenine residue to the 3′-end of the blunt-end fragments to allow the ligation of barcoded adapters, and PCR-amplification of the library. SureSelect probes (Agilent Technologies, Santa Clara, CA, United States) were used for solution-based target enrichment of a pool of 16 libraries following the SureSelectxt Target Enrichment System for Illumina Paired-End Multiplexed Sequencing Library protocol. Captured libraries were sequenced using Illumina HiSeq 3000 (Illumina, San Diego, CA, United States) to generate paired-end 100-bp reads.
Plastome Assembly
Adapter sequences and base calls with a quality score of lower than 36 were trimmed from raw reads using Trimmomatic 0.36 (Bolger et al., 2014), and duplicate reads were removed with Dedupe software in the BBMap package v36.92 (Bushnell, 2014). Quality-filtered reads of Clinanthus variegatus (Ruiz & Pav.) Meerow were aligned against the plastid genome of Hippeastrum cipoanum (Ravenna) Meerow (García et al., 2017) using BWA 0.7.12-r1039 with the MEM algorithm (Li and Durbin, 2009) and other default parameters. The resulting alignment BAM file was imported, visually inspected, and manually corrected in Geneious 9.1.7 (Kearse et al., 2012), and the consensus sequence was extracted from the alignment. Quality-filtered reads of the other 99 species were initially aligned against the C. variegatus reference sequence using BWA, and a consensus sequence of each species was extracted in Geneious as described above. Then, the sequence reads of each species were aligned against the consensus sequence of its own species in the second round to fill gaps resulting from mismatches between the C. variegatus reference sequence and reads of other species. The mapping step was repeated until no more additional reads were aligned to the reference. Because the coverage of the small single copy region (SSC) of the plastomes was very low for most of the samples, our final matrices only include the large single copy region (LSC) and one of the inverted repeat regions (IRb). We manually trimmed difficult to align (mostly AT-rich) non-coding portions.
Bioinformatics of Nuclear DNA
Rapid Genomics used a pipeline described in Breinholt et al. (2018) with a few modifications to process the capture data for phylogenetic analyses. Briefly, adaptors and bases with a Phred score (Ewing and Green, 1998; Ewing et al., 1998) below 20 were removed from raw Illumina reads using Trim Galore! ver. 0.4.01 allowing a minimum read size of 30 bp. Filtered reads of each taxon highly similar to probe regions were selected and assembled de novo for each exon with iterative baited assembly (IBA) using the Python script IBA.py (Breinholt et al., 2018) with a minimum of 5× kmer coverage. To estimate copy numbers and orthology of captured sequences, assembled sequences were aligned with MAFFT 7.x (Katoh and Standley, 2013), trimmed to the probe region using extract_ probe_ region.py (Breinholt et al., 2018), and mapped to the Phalaenopsis equestris (Schauer) Rchb.f. genome using BLASTN (Camacho et al., 2009). The blast results were filtered with the scripts s_ hit_ checker.py and ortholog_filter.py (Breinholt et al., 2018), and those sequences that mapped to the same single location on the P. equestris genome were considered orthologs. The orthologous sequences including flanking regions were aligned with MAFFT, and loci that were represented by at least 70% of the sampled taxa were included in the final dataset. Difficult to align columns in flanking regions in the alignments were removed using the scripts alignment_DE_trim.py and flank_dropper.py (Breinholt et al., 2018).
Reduction of Paralogs
Because the assembled loci contained considerable numbers of paralogs (Supplementary Figure 1), we used the tree-based pipeline described by Yang and Smith (2014) to generate sets of one-to-one orthologs and prune extraneous paralogs. First, we inferred a phylogenetic tree for each locus using FastTree2 (Price et al., 2010), or for the CDS-only data sets, RAxML 8.2.12 (Stamatakis, 2014). The scripts used to implement the Yang and Smith (2014) pipeline were downloaded from https://bitbucket.org/yangya/phylogenomic_dataset_construction/src/master/. Pruning then proceeded as follows: we used trim_tips.py to remove terminal tips with an absolute branch length of >0.5 substitutions/site. We then masked mono- and para- phyletic tips with the same sample name using mask_tips_by_taxonID_transcripts.py, thus removing alleles and extremely shallow (within individual) paralogs. Using cut_long_internal_branches.py, we cut deep paralogs based on an internal branch length of >0.5 substitutions/site. For final ortholog pruning, we used prune_paralogs_MO.py, retaining trees with at least 10 tips remaining. These pruned trees and sub-trees were used to write new fasta files with the write_fasta_files_from_trees.py script. The new locus files were re-aligned using MAFFT (Katoh and Standley, 2013). To compare this method with a paralog-agnostic approach, we also analyzed a supermatrix with RAxML version 8.2.12 (Stamatakis, 2014) using consensus sequences generated from all of the paralogs.
Several supermatrices were used for analysis, differing by the percentage of missing data. For each data set, the supermatrix was constructed using the fasta_merge.py script from HybPiper (Johnson et al., 2016). Supermatrices of coding regions only were also analyzed. Compilation and editing of all alignments was conducted with Geneious Prime 2020.0.52.
Data Analyses
Supermatrices
RAxML version 8.2.12 (Stamatakis, 2014) was used to conduct partitioned maximum likelihood (ML) best tree and thorough bootstrap analyses on the CIPRES Science Gateway (Miller et al., 2010), with an estimate of 25 per site rate categories, with joint branch length optimization, and the GTRCAT model. Three hundred bootstrap replicates were run, and a 50% majority rule bootstrap consensus tree was constructed using the SumTrees component of the package DendroPy v. 4.4.0 (Sukumaran and Holder, 2010). We manually created a “tanglegram” from the best ML 70% taxon coverage nuclear supermatrix and the plastome tree to highlight any cytonuclear discordance.
Species Tree Estimation
For each locus, a (ML) gene tree was inferred using RAxML under either the GTRCAT (for data sets with >50 samples) or GTRGAMMA (for data sets with <50 samples) models, with 100 rapid bootstrap replicates. Nodes with less than 30% bootstrap support were collapsed using SumTrees (Sukumaran and Holder, 2010). These collapsed trees were used to infer coalescent-based species trees with ASTRAL-III (Zhang et al., 2018). Node support was calculated using both local posterior probability (LPP, normalized quartet scores, representing gene tree concordance at each node) and bootstrap (100 replicates).
Species Network Analysis
SplitsTree v. 4.15.1 (Huson, 1998; Huson and Bryant, 2006) was used to infer reticulate evolution within the Andean tetraploid clade. Three separate analyses were conducted: Clinantheae, Eucharideae and the genus Hymenocallis. Pyrolirion albicans Herb. (Eustephieae) was included as outgroup in analyses of Clinantheae and Eucharideae; for Hymenocallis, Leptochiton quitoensis (Herb.) Sealy was designated as outgroup. The concatenated supermatrix of 70 nuclear gene alignments with 99% taxon coverage across our original 100 samples as analyzed (120 kbp). Paramongaia viridiflora (Ruiz & Pav.) Meerow was dropped from the Clinantheae alignment because only eight genes (of 70) were successfully recovered from this herbarium sample. We used the NeighborNet distance transformation with uncorrected P distance to draw equal-angle hybridization networks (Albrecht et al., 2012) with 300 bootstrap iterations, and also conducted phi tests for recombination (Bruen et al., 2006).
Dating and Diversification Rate Analyses
We estimated age divergence within the Andean clade using penalized likelihood estimation on the best 90% taxon coverage supermatrix tree. This was conducted with the chronos function in APE v.5.3 (Paradis and Schliep, 2019) in R v. 4.0 (R Core Team, 2017), with two different models, “relaxed” and “correlated” (Kim and Sanderson, 2008; Paradis, 2013). Two calibrations points were placed on the tree. We estimated lower and upper bounds of 30–33 Mya for the stem node of the American clade, based on an inferred estimate from the dated tree in Santos-Gally et al. (2012) for the most recent common ancestor (MRCA) of the Eurasian and American clades of Amaryllidaceae subfam. Amaryllidoideae, and 10–12 Mya for the stem node of Rauhia Traub, based on fossil-supported estimated age of seasonally dry inter-Andean forests in the central Andes (Caetano et al., 2008; Quintana et al., 2017a). Rauhia is endemic to the Marañon dry woodlands of northern Peru (Meerow and Nakamura, 2019). The smoothing parameter (λ) was chosen using the cross-validation method in the chronopl function (testing λ = 0 and 0–5), selecting the value of λ that minimized the cross-validation statistic (Sanderson, 2002). The resulting trees were visualized using the densiTree function in phangorn 2.4.0 (Schliep, 2011), and the time calibrated tree representing the central tendency of these analyses was selected. Because the cross-validation method (Sanderson, 2002) failed to detect a single preferred value for the smoothing parameter (λ), we ran the analysis with 21 values of λ, from 10–5 to 1015. The resulting trees were visualized using the densiTree function in phangorn 2.4.0 (Schliep, 2011), and because several had equally likelihood scores, the time calibrated tree representing the central tendency of these analyses was selected. Confidence intervals on node ages are not provided in this study since the penalized likelihood approach used does not address model or calibration placement and timing uncertainty. A geologic timescale based on the strat2012 dataset was added to the tree using PHYLOCH v 1.5-3 (Heibl, 2008). The best tree was visualized using FigTree v. 1.44 (Rambaut, 2019).
To determine whether there have been any diversification rate shifts in the Andean tetraploid clade, we performed a speciation-extinction analysis on the relaxed dated tree produced by APE with BAMM v 2.5.0 (Rabosky et al., 2013, 2014a; Rabosky, 2014) and the R program BAMMtools (Rabosky et al., 2014b). We ran 50 M Markov Chain Monte Carlo (MCMC) generations with four chains, using priors set by BAMMtools from our tree. We estimated the amount of missing data for each clade (usually = genus) in the tree, and set the expected number of rate shifts to one. Convergence of the runs and the number of sampled rate shifts were tested using effective sample sizes (ESS) with R package coda v. 0.19-3 (Plummer et al., 2006).
Biogeographic Analysis
We used BioGeoBears (Matzke, 2012, 2013, 2014) as implemented through RASP v. 4.2 (Yu et al., 2015) with R v.4.0 to conduct biogeographic analyses. The input tree was the best 90% taxon coverage supermatrix tree. Outgroups were stripped from the tree as recommended by the RASP authors. We first implemented the six model test to determine the most appropriate model for our data by the weighted AICc statistic, subsequently implementing the model with the highest score. The only consequence of using a single tree was that the S-DIVA method could neither be evaluated nor used. Fifteen singular areas were designated in the analyses derived from the Neotropical bioregions and provinces of Morrone (2014) and Antonelli et al. (2018)with the exception of the Nearctic Realm/Eastern North America Bioregion (Ricketts et al., 1999): A = Napo Province, B = Cauca Province, C = Puna Province, D = Desert Province, E = Páramo Province, F = Yungas Province, G = Ecuadorian Province, H = Eastern North America Bioregion (Nearctic Realm), I = Western Ecuador Province, J = Ucayali Province, K = Chocó-Darién Province, L = Prepuna Province, M = Mesoamerica Bioregion, N = Guianan Lowlands Province, and O = West Indies Bioregion. We limited areas at any node to three, and mapped only the most likely on the tree. We also similarly ran an analysis on the best ML tree found by RAxML for the partial plastome supermatrix.
Results
Sequence Alignments
The complete sequence supermatrix included all loci that survived the paralog pruning, and was 730,500 bp in length, comprising 524 genes (Supplementary Table 2). We also created a supermatrix of all genes that included sequences from 70% (260 genes, 277,173 bp), 90% (137 genes, 201,059 bp), and 99% (70 genes, 120,000 bp) of the taxa, respectively.
The plastome supermatrix was 122,549 bp in length. The LSC consisted of 94,938 bp; the IR was 27,611 bp. Genera of the tribe Eucharideae exhibited substantial re-organization and pseudogenization of the genes of the ndh family.
Supermatrix ML Analyses
All of the supermatrix analyses resolved a well-supported monophyletic Andean tetraploid clade (Figure 2) consisting of the four tribal subclades: Clinantheae, Eucharideae, Eustephieae, and Hymenocallideae, all with 100% bootstrap (BP) with a single exception in Clinantheae. The genus Pamianthe in the analyses with all genes (both coding + flanking regions) is sister to Clinantheae/Hymenocallideae (Figure 2,100% BP), but with coding regions alone (Supplementary Figure 2), it is the first branch of Clinantheae (94% BP). Trees from the 70 and 90% taxa coverage supermatrices (Supplementary Figures 3, 4) placed Pamianthe as the first branch in Clinantheae with 94 and 91% BP, respectively.
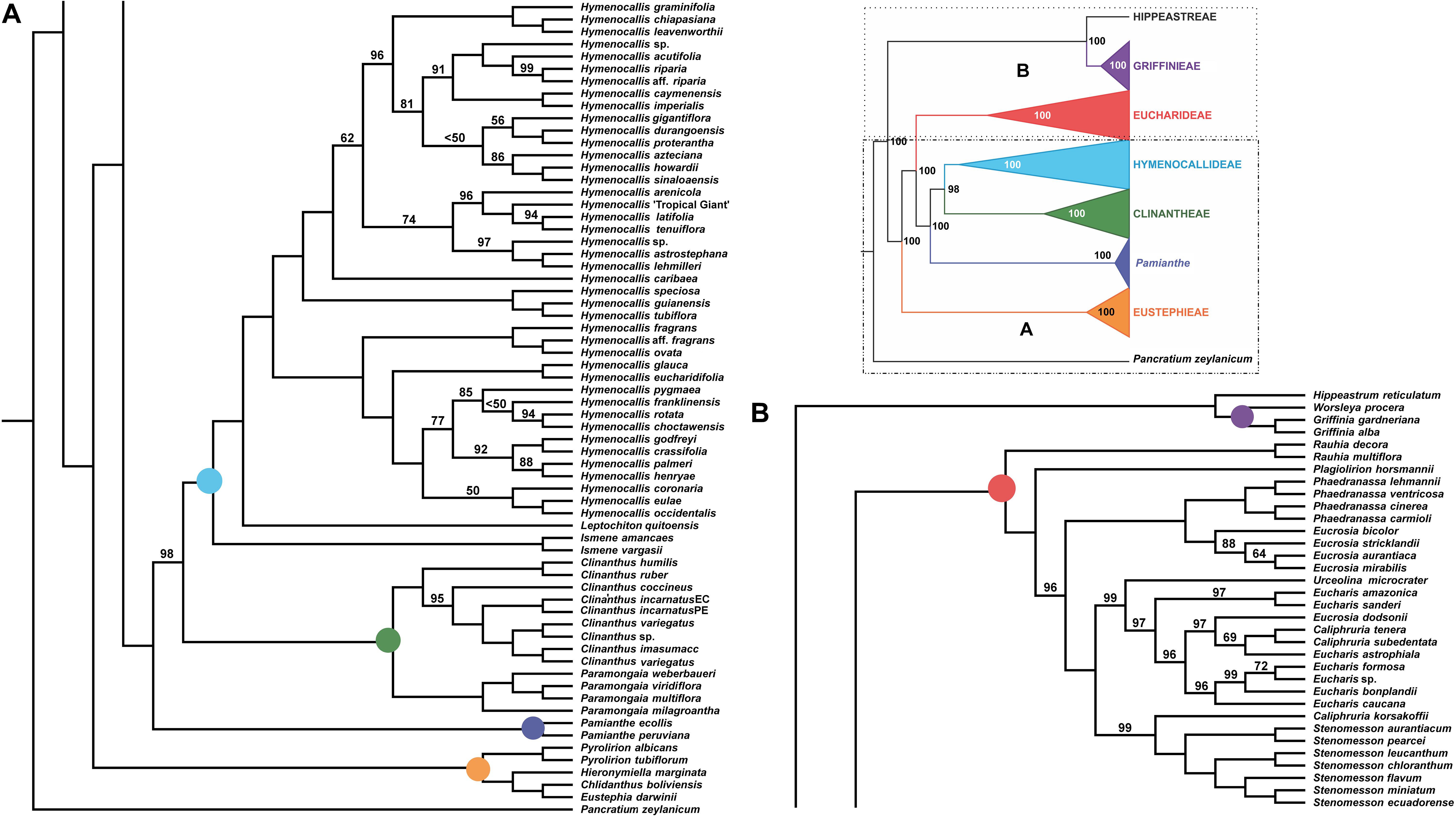
Figure 2. Best tree from maximum likelihood analysis of partitioned 524 nuclear gene supermatrix of the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae, with bootstrap percentages <100 shown above branches (all unmarked branches have 100% BP). The tree is divided into two sections (A,B) for readability; refer to the colored inset (a cartoon of the tree collapsed to the tribal level) as a guide. Colored circles at the nodes in the full tree conform to clade colors in the inset.
The tribe Eustephieae was sister to rest of the Andean clade, within which Pyrolirion Herb. was the first branch, followed by Hieronymiella Pax, then a sister relationship between Chlidanthus Herb. and Eustephia Cav. This topology occurred in all of our trees, as well as the plastome analysis, with 100% BP at each node. Clinantheae (less Pamianthe) and Hymenocallideae were sister tribes with 98% BP. Together with Pamianthe, they formed the sister clade to Eucharideae. The Paramongaia subclade of Clinantheae was supported with 100% BP by all supermatrices. Clinanthus formed two subclades. The larger of the two represented Clinanthus incarnatus (Kunth) Meerow, C. variegatus and their variants. Clinanthus coccineus (Ruiz & Pav.) Meerow was the first branch. C. incarnatus was monophyletic and distinct from C. variegatus.
Within Hymenocallideae, Ismene was the first branch, followed by Leptochiton, and then Hymenocallis. The latter had a consistent topology of two subclades, both of which consisted of a small, pseudopetiolate-leafed, forest understory subclade sister to a larger, more diverse group (Figure 2). One pseudopetiolate group was sister to a clade containing all other West Indian and most of the Mesoamerican species in our sampling. Some of the more terminal subclades were either poorly or not supported (Figure 2). The West Indian Hymenocallis caribaea (L.) Herb. was the first branch with 100 BP (Figure 2) in the complete supermatrix and in the 90% coverage (Supplementary Figure 4), but formed an unsupported sister to Hymenocallis gigantiflora Meerow3 in the 70% coverage best tree (Supplementary Figure 3). The second pseudopetiolate subclade is sister to a clade that united two southern Mexican pseudopetiolate species, Hymenocallis eucharidifolia and Hymenocallis glauca M. Roem., with a monophyletic southeastern United States subclade (Figure 2). A small clade of mostly West Indian species resolved with 94 BP (Figures 2, 3), uniting Hymenocallis arenicola, Hymenocallis latifolia, Hymenocallis tenuiflora and a cultivar of Caribbean origin, H. ‘Tropical Giant.’
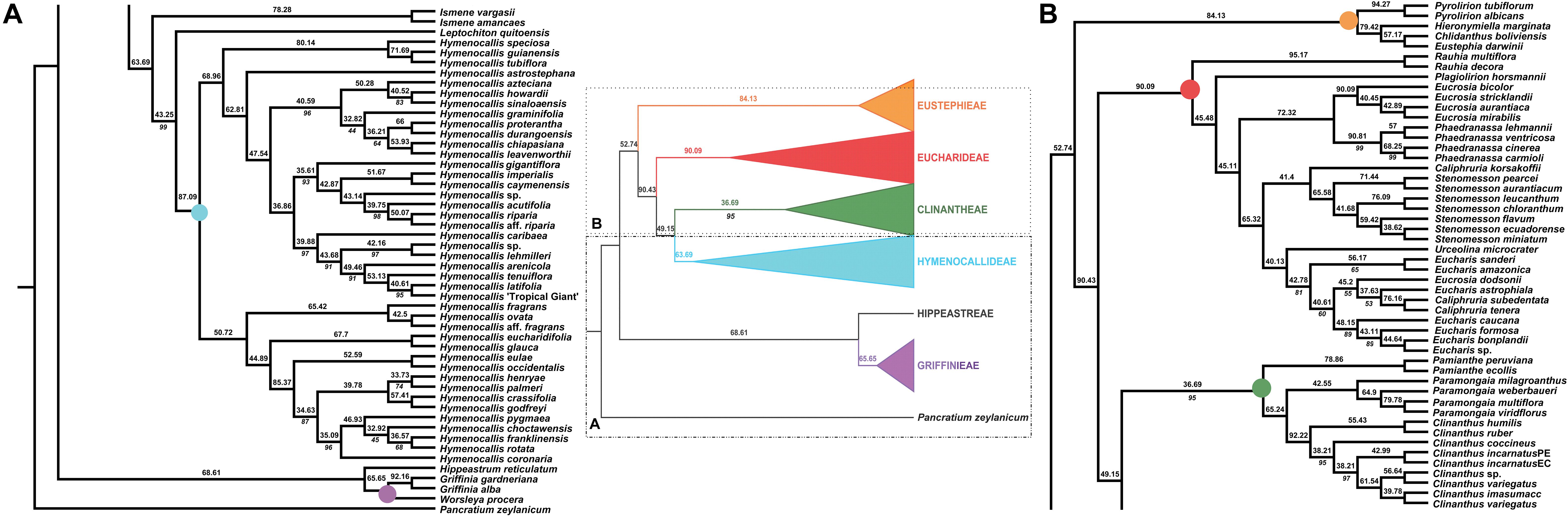
Figure 3. Local posterior probability (LPP) coalescent species tree from ASTRAL III analysis of gene trees of 526 nuclear genes. LPP scores appear above the branches; bootstrap percentages <100 shown below branches (all unmarked branches have 100% BP). The tree is divided into two sections (A,B) for readability; refer to the colored inset (a cartoon of the tree collapsed to the tribal level) as a guide. Colored circles at the nodes in the full tree conform to clade colors in the inset.
In Eucharideae, Rauhia was the first branch, followed by the monotypic Colombian endemic Plagiolirion (Figure 2). Next, Eucrosia (less Eucrosia dodsonii Meerow & Dehgan) and Phaedranassa formed sister clades (100% BP). Stenomesson was monophyletic and sister to Caliphruria korsakoffii (Traub) Meerow, the only Peruvian species of that genus. The remaining clade of Eucharideae united the Colombian Caliphruria, Eucharis, and Urceolina, with the inclusion of E. dodsonii. Urceolina was the first branch, though not consistently in all of the supermatrix analyses (Figure 2 and Supplementary Figures 2–4). Eucharis was not monophyletic. An unexpected and consistent subclade united Eucharis astrophiala (Ravenna) Ravenna, the Colombian Caliphruria spp., and E. dodsonii (97% BP).
A ML tree derived from a slightly different whole gene supermatrix (five samples missing) using ambiguity codes to account for all paralogs (Supplementary Figure 5) showed very strong congruence with trees from our paralog-pruned alignments (Figure 2 and Supplementary Figures 2–4).
Species Tree Analyses
The ASTRAL-III species tree analyses (Figure 3) were largely congruent with the ML trees. Pamianthe was resolved in all species trees as the first branch within Clinantheae. The topology of all of the other major subclades was the same. LPP scores ranged from 33 to 94, but BP was high at the majority of nodes (Figure 3). Little change occurred with increased percentage coverage across taxa (Supplementary Figures 6, 7). There occurred some re-alignment within the two main subclades of Hymenocallis, mostly at nodes with low bootstrap support in the full data set (Figure 3) and some erosion of BP overall. The sister status of Caliphruria korsakoffii to Stenomesson was resolved but not supported by the 70% coverage species tree bootstrap (Supplementary Figure 6). In the 90% species tree, it was sister to the Eucharis/Urceolina/Caliphruria/E. dodsonii clade with weak (63%) BP (Supplementary Figure 7).
Plastome Analysis
The ML analysis of the partial plastome was congruent at the tribal clade level with the nuclear genome, but resolved yet a third resolution for Pamianthe as sister to Eucharideae with 100% BP (Figure 4). The relationships of the Eustephieae clade were identical to the nuclear trees (Figures 2, 3). Paramongaia milagroantha (S. Leiva & Meerow) Meerow was sister to Clinanthus (100%), and C. incarnatus did not resolve as monophyletic (Figure 4). Hymenocallideae had the greatest number of unsupported nodes. The southern Caribbean/Guianan clade of Hymenocallis (Hymenocallis guianensis, Hymenocallis speciosa, and Hymenocallis tubiflora) became the first branch in the genus (Figure 4) instead of sister to the Caribbean/Mexican subclade as with nuclear data (Figures 2, 3). In the other subclade of Hymenocallis, Hymenocallis astrostephana T. M. Howard/H. glauca became the first branch (Figure 4) with no support, followed by the West Indian pseudopetiolate clade. A SE U.S. clade received 100% support (Figure 4), but had poor internal resolution. It was sister to a group of mostly Mexican spp. that resolved within the other main subclade with nuclear genes (Figures 2, 3). Hymenocallis caribaea was a well-supported sister sp. to H. ‘Tropical Giant’ (100% BP), which joined with H. arenicola and H. gigantiflora in a well-supported subclade (98% BP). H. latifolia and H. tenuiflora (100% BP) were also the first branch in a clade of mixed Mexican and West Indian taxa.
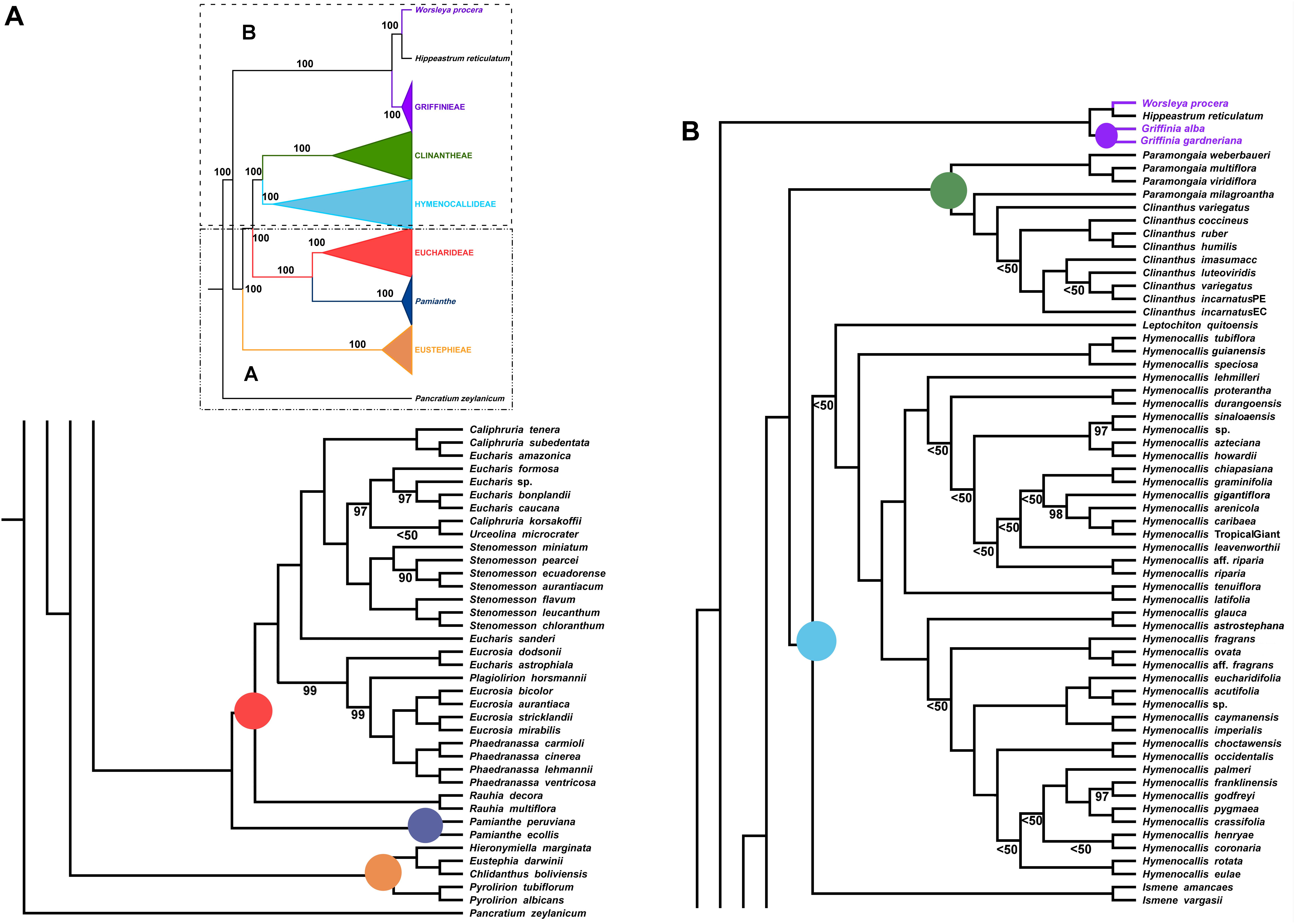
Figure 4. Best tree from maximum likelihood analysis of partial plastome sequences across the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae, with bootstrap percentages <100 shown above branches (all unmarked branches have 100% BP). The tree is divided into two sections (A,B) for readability; refer to the colored inset (a cartoon of the tree collapsed to the tribal level) as a guide. Colored circles at the nodes in the full tree conform to clade colors in the inset.
In the Eucharideae, a monophyletic Stenomesson was embedded within Eucharis, Caliphruria, and Urceolina (Figure 4), and E. sanderi Baker was the first branch. C. korsakoffii was sister to Urceolina microcrater Kraenzl. without support. E. astrophiala was sister to E. dodsonii, together in turn resolved as sister to a Plagiolirion/Eucrosia/Phaedranassa clade. Rauhia was the first branch in the tribe after Pamianthe. We also analyzed only the coding and tRNA regions of the plastome and obtained the same topology as Figure 4 with variably increased or decreased BP at various nodes (not shown).
Cytonuclear Discordance
The congruence between supermatrix and species tree topologies of our nuclear genes (Figures 2, 3 and Supplementary Figures 2–4, 6, 7) suggested that there was no reticulation between major lineages (tribes) of the Andean tetraploid clade, with the possible exception of Pamianthe (see section “Discussion”). Our plastome data (Figure 4) resolved the same very well supported tribal clades as the nuclear data. However, the tanglegram (Figure 5) clearly illustrated cytonuclear discordance within the genera Clinanthus and Hymenocallis, and in the tribe Eucharideae, particularly among the genera Caliphruria, Eucharis, and Urceolina. None was observed for sister genera Eucrosia and Phaedranassa. There was also a disagreement on the position of Worsleya procera relative to the other species from the hippeastroid clade (Figure 5). The plastome tree resolved this monotypic genus as sister to Hippeastrum reticulatum, while the nuclear trees placed it within the Griffinieae.
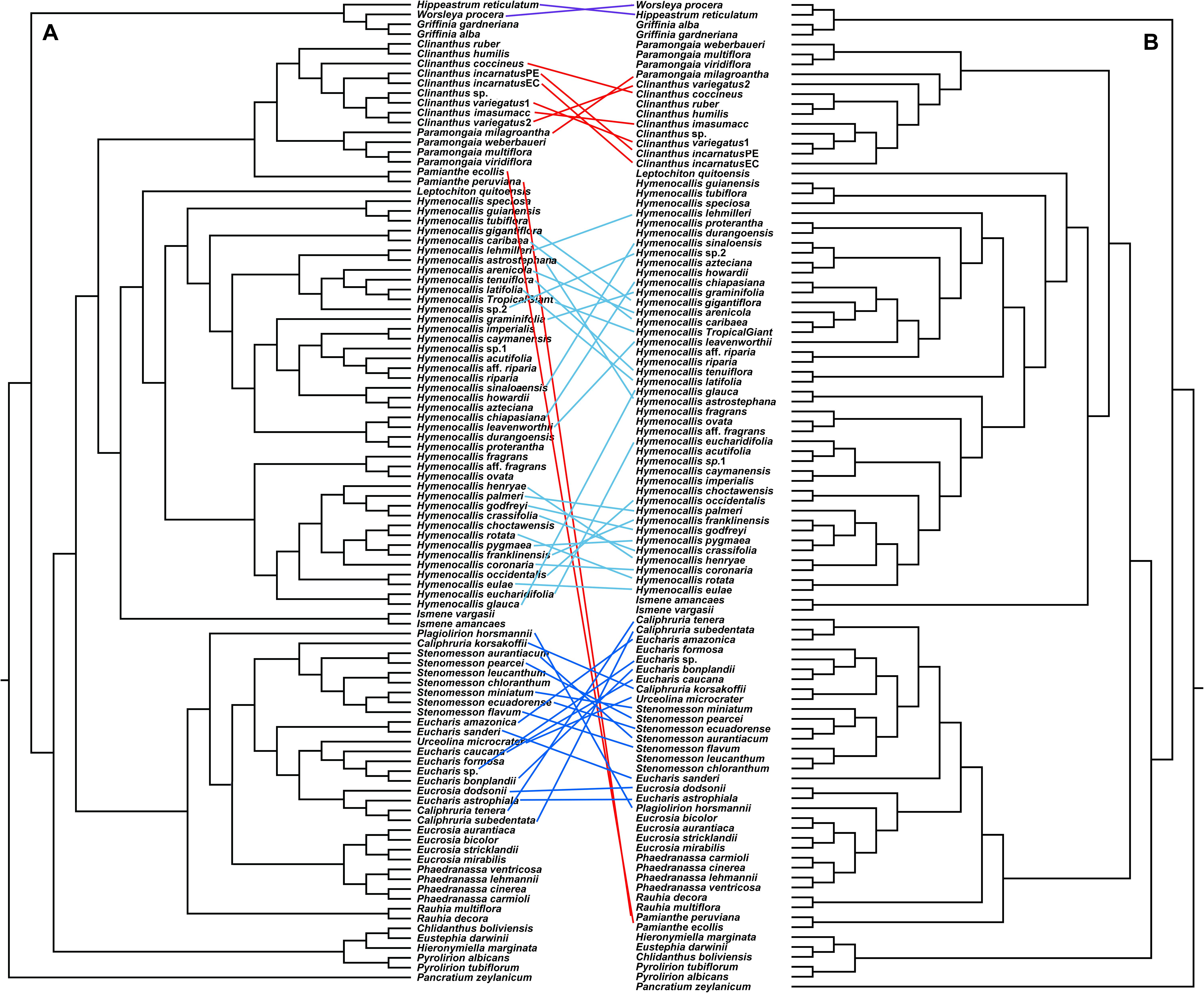
Figure 5. “Tanglegram” created from the best ML 70% taxon coverage nuclear supermatrix (Supplementary Figure 3) and the plastome trees (Figure 4) graphically showing cytonuclear discordance between the two partitions. A = nuclear tree, B = plastome tree.
Species Network Analysis
Hybridization networks of Clinantheae, Eucharideae, and Hymenocallis supported varying degrees of putative reticulation within each clade, and the phi test detected significant recombinant signal in all three supermatrices at p = 0.0. All branches in the networks were strongly supported by the bootstrap. Clinantheae (Figure 6) had the least amount of putative hybridization, and none between genera. There was some minor reticulation evident between Pamianthe ecollis Silverst., Meerow & Sánchez-Taborda, and P. peruviana and between Paramongaia multiflora Meerow and Paramongaia weberbaueri Velarde. Recombinant signal was highest among C. variegatus, Clinanthus imasumacc (Vargas) Meerow and related species, but not between them and C. incarnatus, the latter exhibiting genetic exchange between Peruvian and Ecuadorian collections. Minor reticulation was also apparent between Clinanthus humilis (Herb.) Meerow and Clinanthus ruber (Herb.) Meerow & A. Cano.
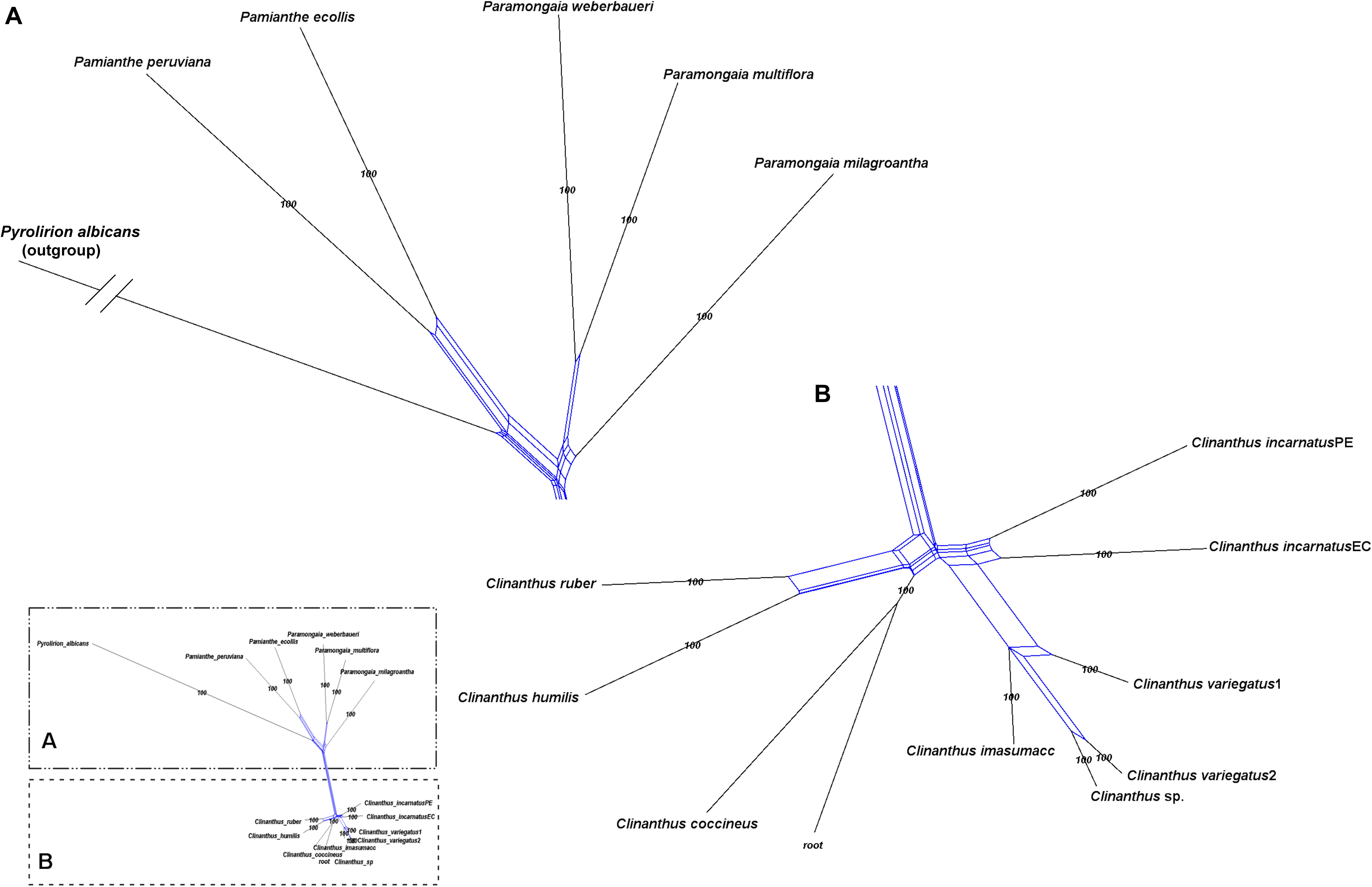
Figure 6. Equal angle 360° hybridization network generated by SplitsTree v. 4.15.1 based on the 99% taxon coverage supermatrix (70 nuclear genes) across the tribe Clinantheae. Numbers above branches are percentages from 300 bootstrap iterations. The entire network is shown in the inset, with subclades (A) and (B) magnified for clarity.
Within the Eucharideae, there was only minor reticulation between sister species of Eucrosia and Phaedranassa (Figure 7), respectively, with considerably more evident among species of Stenomesson, and possibly in the early diversification of Stenomesson from the Caliphruria/Eucharis/Urceolina subclade (i.e., position of C. korsakoffii). Recombinant signal suggested that the monotypic Plagiolirion horsmanii Baker may have arisen via reticulation between the branch leading to Eucrosia and Phaedranassa and the ancestral lineage of Rauhia. However, the reticulate origins of Eucharis, Colombian Caliphruria, Urceolina, and E. dodsonii were strongly evident, as was the misdiagnosis of the latter as a species of Eucrosia.
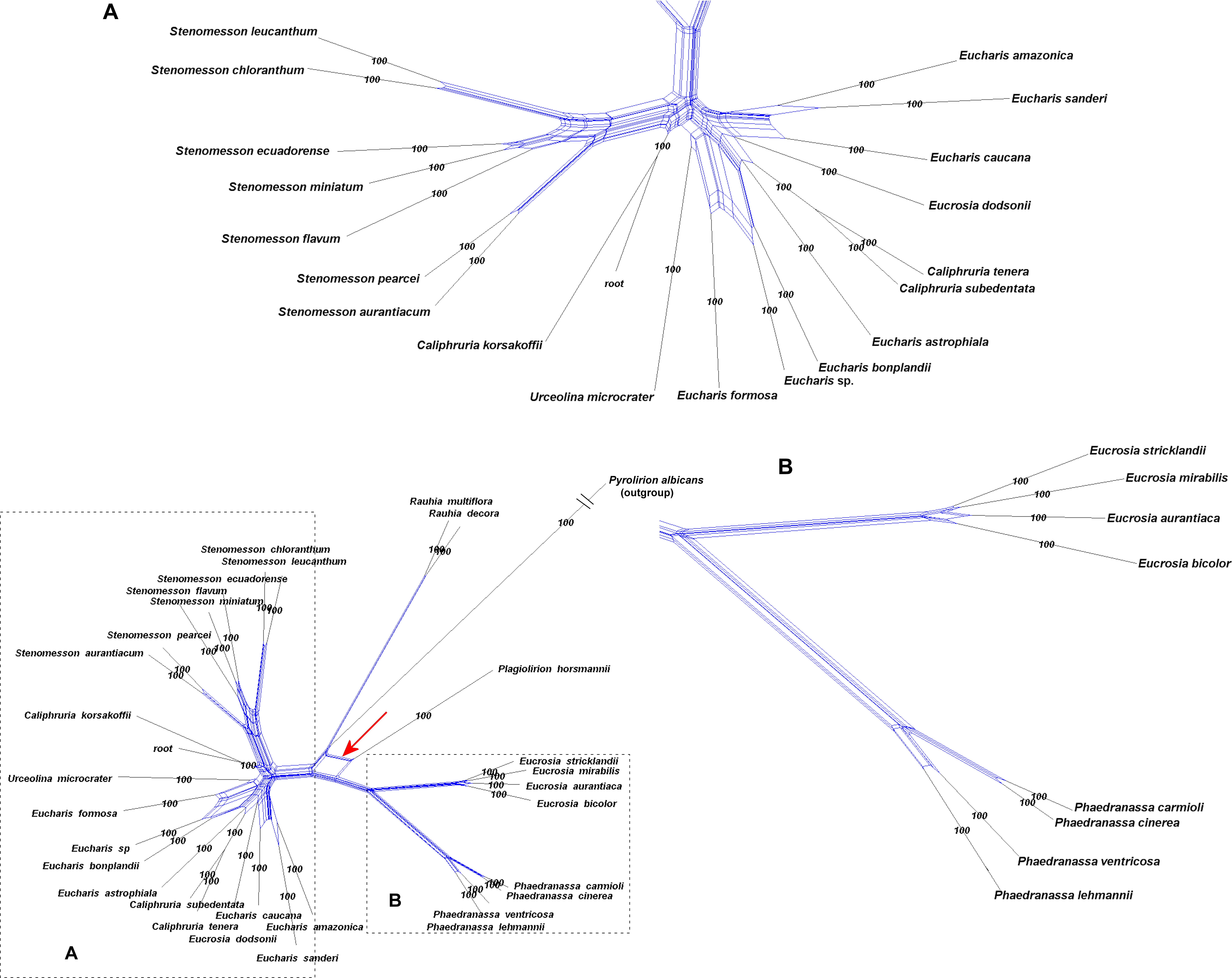
Figure 7. Equal angle 360° hybridization network generated by SplitsTree v. 4.15.1 based on the 99% taxon coverage supermatrix (70 nuclear genes) across the tribe Eucharideae, with percentages from 300 bootstrap iterations. The entire network is shown in the inset, with subclades (A) and (B) magnified for clarity. Red arrow indicates possible reticulate origin of the monotypic Plagiolirion.
Hymenocallis exhibited the greatest degree of interspecific reticulation in our network analyses (Figure 8). Some minor putative recombination signal occurred in both of the small pseudopetiolate subclades. But within both the larger SE U.S. subclade and the Mesoamerican/West Indian subclade, strong interspecific reticulate patterns were evident.
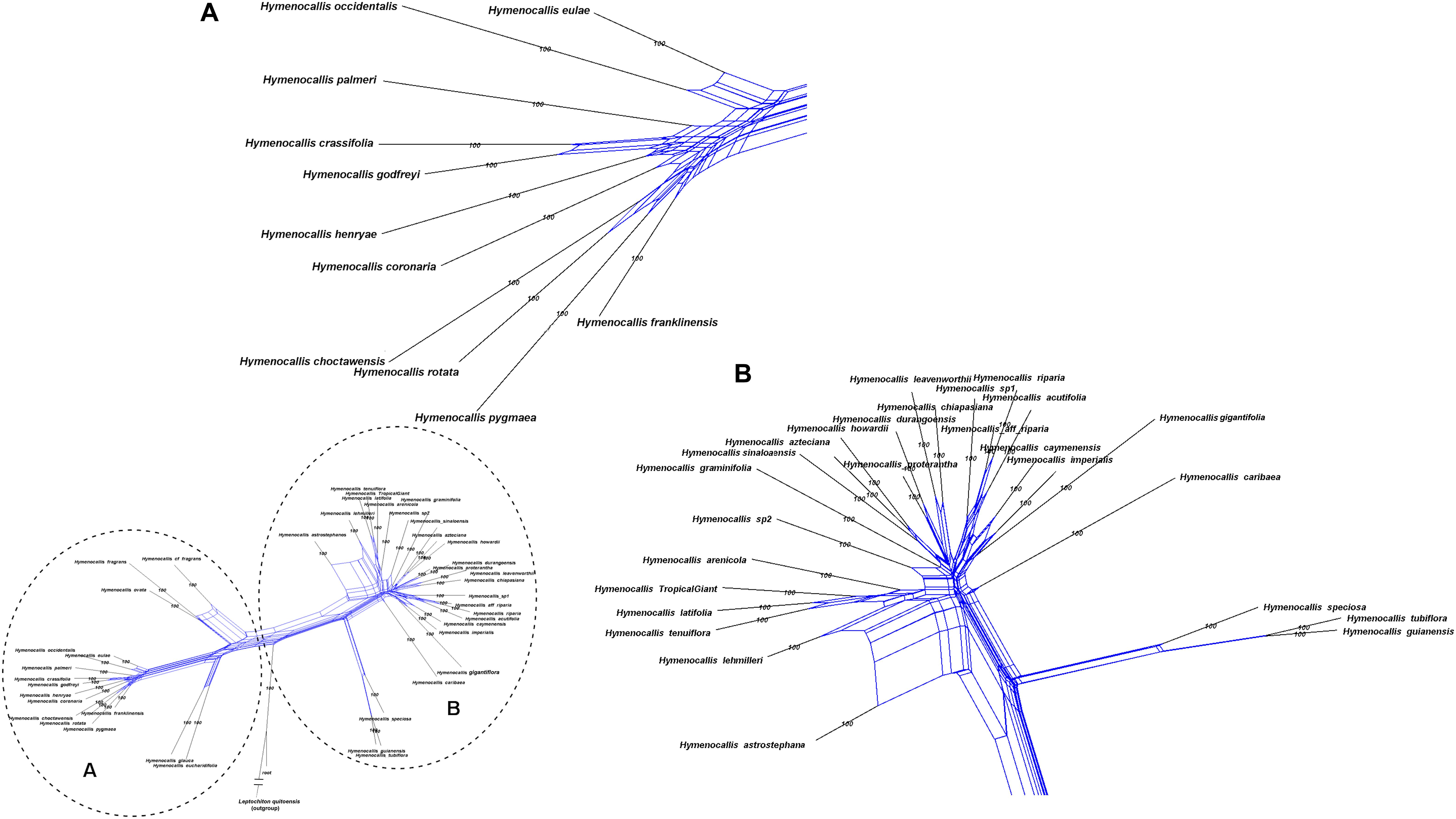
Figure 8. Equal angle 360° hybridization network generated by SplitsTree v. 4.15.1 based on the 99% taxon coverage supermatrix (70 nuclear genes) across the genus Hymenocallis, with percentages from 300 bootstrap iterations. The entire network is shown in the inset, with subclades (A) and (B) magnified for clarity.
Dating and Diversification Rate Analyses
The relaxed model was much less sensitive to the value of λ than the correlated, and several trees had equally high penalized likelihood scores. We therefore used the tree calibrated under the relaxed model and λ = 1 for further analyses (Supplementary Figure 8). The resulting tree (Figure 9), dated the crown node of the Andean clade at 30.9 Mya (stem = 32.1 Mya), thus an early Oligocene origin, with the first branch (Eustephieae) crowning at around 24 Mya in the late Oligocene (Table 1). The MRCA of Clinantheae and Hymenocallideae was dated at 28 Mya, and both tribes’ crown ages are ca. 26 Mya. Ismene and Leptochiton diverged from Hymenocallis by the late Oligocene (ca. 26 and 24.5 Mya, respectively). The crown age of Hymenocallis was ca. 21 Mya, and the two main subclades were contemporaneous at almost 18 Mya. Pamianthe diverged from Clinanthus and Paramongaia ca. 27 Mya, with a Miocene crown age of ca. 18 Mya. Paramongaia and Clinanthus split in the late Oligocene (Figure 10), but the former is older than Clinanthus by ca. 8 MY. The very recent divergence between P. multiflora and P. viridiflora (ca. 130 Ky) may be somewhat artefactual as only eight genes were successfully captured from the latter, which was extracted from an herbarium sample. While the stem node of Eucharideae appeared as late mid-Oligocene (ca. 28 Mya), its generic radiation was a Miocene phenomenon, occurring rapidly between the mid-Miocene through the Pliocene (Figure 9 and Table 1). There was very sudden radiation of lineages between 10 and 11 Mya (Figure 9).
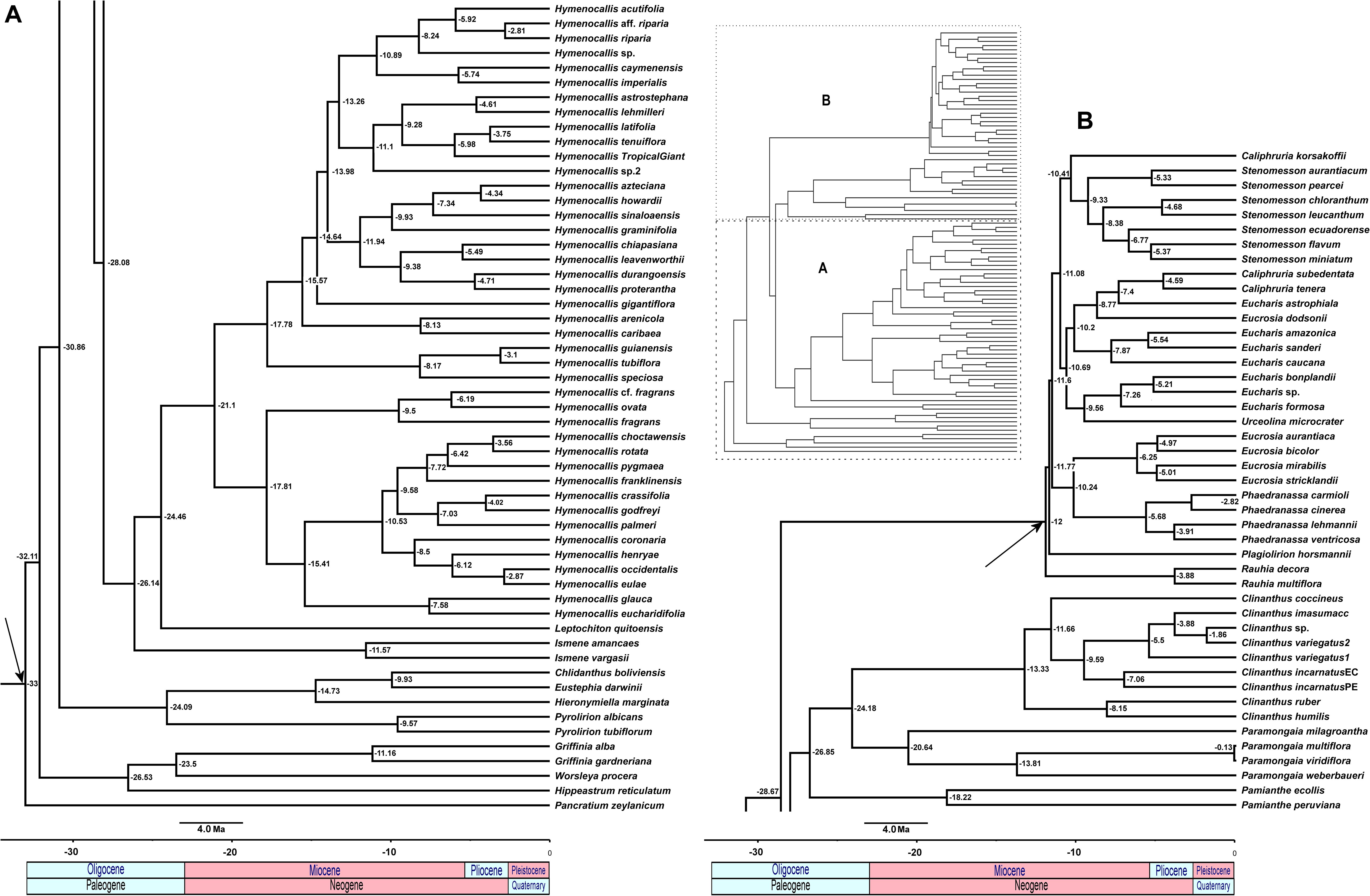
Figure 9. Relaxed penalized likelihood chronogram of the Andean tetraploid clade using the chronos function of APE with the best 90% taxon coverage supermatrix tree found by RAxML. Arrows indicate calibration nodes used in the analysis. Numbers along branches are millions of years before present. The entire tree is shown in the inset, with subclades (A) and (B) magnified for clarity.
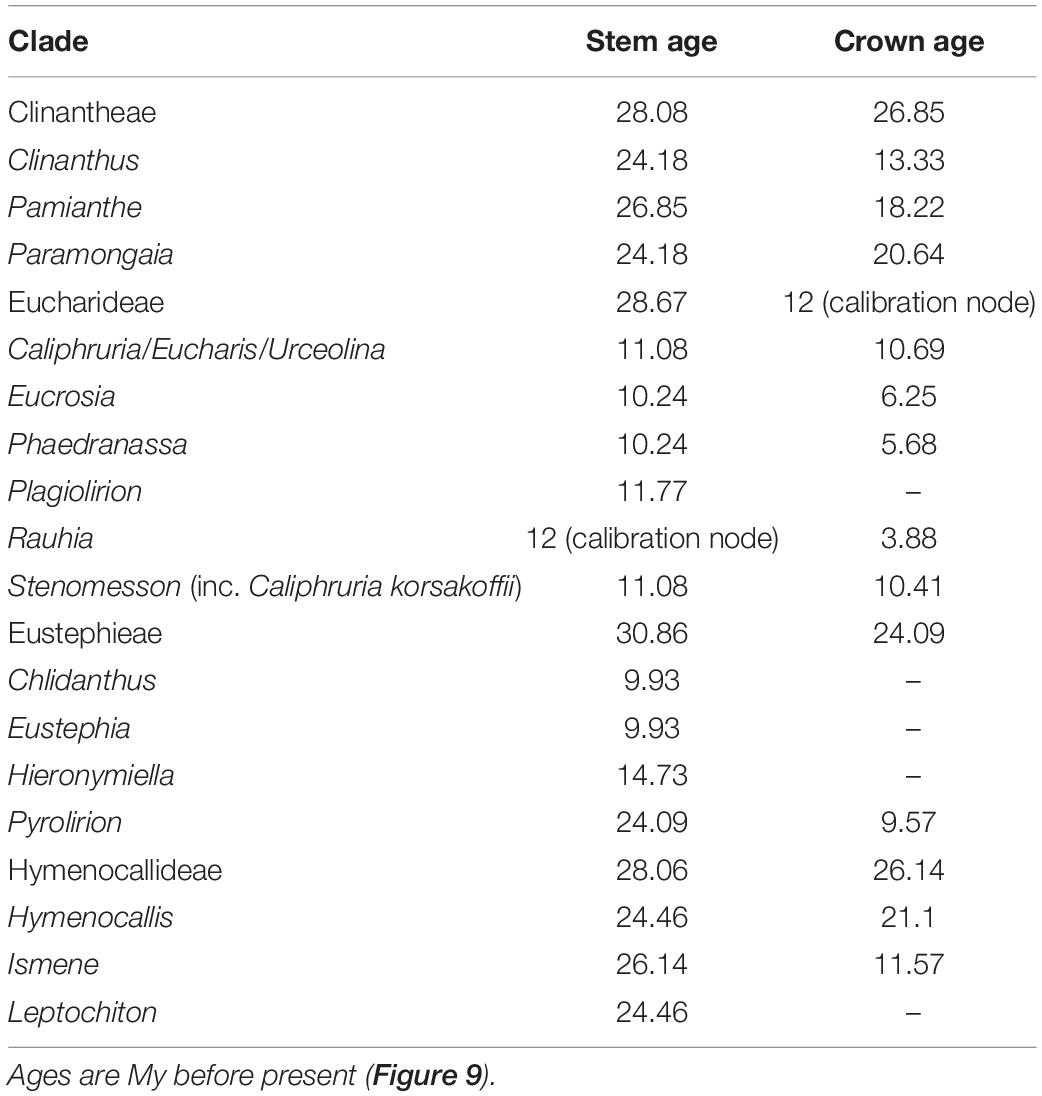
Table 1. Estimated ages of major clades of the Andean tetraploid clade using penalized likelihood with the R package ape on the on the best 90% taxon coverage supermatrix tree found by maximum likelihood with RAxML.
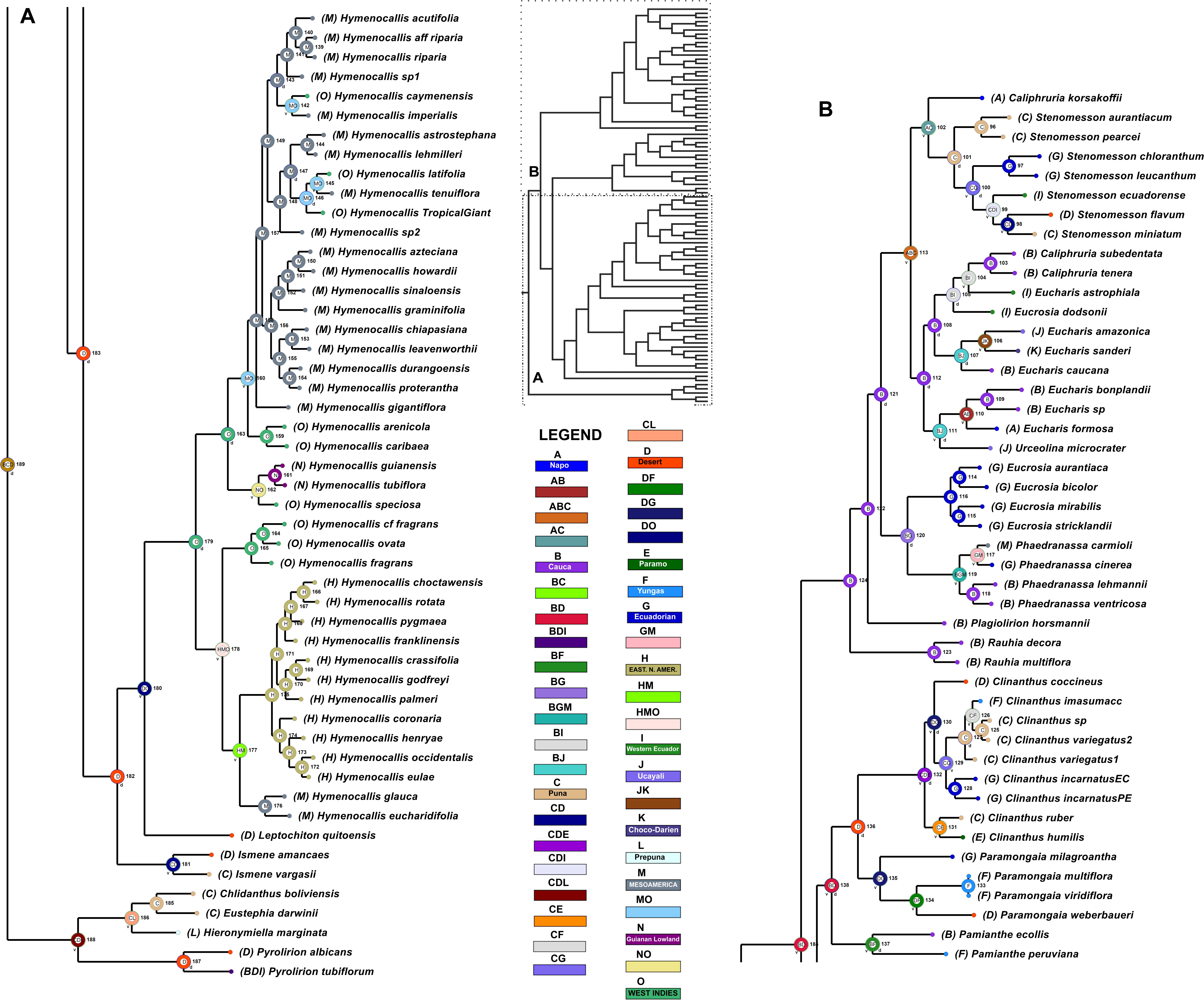
Figure 10. Area cladogram generated by Divalike+J analysis with BioGeoBears through RASP 4.2, using the best tree found by maximum likelihood of the 90% taxon coverage supermatrix with RAxML. Areas at any node were limited to three, and only the most likely was mapped onto the tree. Lower case “v” and “d” adjacent to a node = vicariance or dispersal, respectively. The tree is divided into two sections (A,B) for readability; refer to the inset as a guide.
Both ESS scores of our BAMM run were >3000, indicating good convergence across the MCMC generations and number of rate shifts sampled. Post run analyses with BAMMtools all supported the null model of no rate shifts, thus the absence of rate-shift heterogeneity in the Andean tetraploid clade.
Biogeographic Analysis
The DIVAlike model had the highest weighted AICc score of the six models tested with BioGeoBears, and probability values supported the +J add-on (founder effect speciation); thus that model was applied to our tree (Figure 10). Globally, the model supported 36 dispersal events, 32 vicariance events and no extinction (Supplementary Table 3). The largest number of dispersals originated from the Desert Province (10.17), followed by Cauca Province (9.17). Puna Province was the destination for the largest number of dispersals (4.0) The Mesoamerica Bioregion had the largest number of speciation events within an area (18), followed by the Eastern North America bioregion (10), no doubt due to the relatively large number of samples of Hymenocallis endemic to both areas. Cauca Province was third, with nine.
The ancestral area (node 189, p = 0.0565, Figure 10) encompassed the Cauca, Desert and Puna Provinces (BCD). Two events took place from this node; dispersal into the Prepuna Province (L) at the crown node of Eustephieae (188, p = 0.3340), from which representation within the Cauca Province was eliminated, and vicariance between the Desert (D) and Puna (C)/Prepuna (L) Provinces. At node 187 (the crown node of Pyrolirion, p = 0.7468) two dispersals take place into Cauca and Western Ecuador Provinces. Vicariance between the Puna (C) and Prepuna (L) Provinces occurs at node 186 (p = 0.9847). At node 184 (p = 0.2534), vicariance between the Cauca (B) and Desert (D) Provinces separates the stem node of Hymenocallideae and Clinantheae (183) and the crown node of Eucharideae (124), respectively. From node 183 (p = 0.1040), a return dispersal to Cauca Province took place to the crown node of Clinantheae (138, p = 0.9030), while the crown node of Hymenocallideae (182, p = 0.7938) remains optimized in the Desert Province (D), followed by a range expansion to Puna Province (C) at the crown node of Ismene (181, p = 0.9696), after which vicariance separates these two provinces between the two species of Ismene in our sampling. From this same node, a second, presumably long distance, dispersal to the West Indies Bioregion (O) occurs at the stem node of Hymenocallis (180, p = 0.8065), with subsequent vicariance between the West Indies (O) at the crown node of Hymenocallis (179, p = 0.2944) and the divergence of L. quitoensis. From the crown of Hymenocallis, dispersal to the Eastern North America (H) and Mesoamerican (M) Bioregions occurs (178, p = 0.3234), with subsequent vicariance between the West Indies (165, p = 1.0000), and both the Mesoamerica and Eastern North American Bioregions (177, p = 0.9455) in one of the two subclades of Hymenocallis, followed by vicariance between the two latter bioregions (nodes 175 and 176, p = 1.0000 for both). The lack of either vicariance or dispersal at nodes 166–175 suggests that the entry of the genus into the SE U.S. was a solitary event.
From the West Indian crown node of the second subclade of Hymenocallis (163, p = 0.8308), two dispersals expanded the range to the Guianan Lowlands Province (162, p = 0.9499) and the Mesoamerican Bioregion (160, p = 0.9277), followed by vicariance between the two areas. The remaining events in Hymenocallis were (1) secondary dispersals to the West Indies Bioregion from Mesoamerica (143, 147; p = 0.9030, 0.3412), followed by vicariance (142, 145; p = 0.9052, 0.8709.
From the crown node of Clinantheae (138, p = 0.1060) in the Cauca (B) and Desert Provinces (D), dispersal to the Yungas Province took place at the crown node of Pamianthe (137, p = 0.9642), while vicariance restricted the range of the stem node of Clinanthus and Paramongaia (136, p = 0.0621) to the Desert Province (D). Vicariance subsequently isolated P. ecollis in Cauca Province and P. peruviana in Yungas Province. The crown node of Paramongaia (135, p = 0.4363) resolved within the Desert (D) and Ecuadorian (G) Provinces, from where it dispersed into Yungas Province (134, p = 0.9680) while vicariance separated P. milagroantha from the other spp. The crown node of Clinanthus (132, p = 0.0956) was optimized within the Puna (C), Desert (D) and Páramo (E) Provinces. The genus dispersed to the Ecuadorian Province and vicariance separated the crown nodes of the two subclades into a Desert and Ecuadorian group (DG, 130, p = 0.3268) and a Puna and Páramo (CE) group (131, p = 0.9250). Vicariance split the latter small subclade of C. ruber and C. humilis between the two provinces. There are a number of additional species with affinity to this group that were not sampled. The larger clade of Clinanthus extended its range into the Ecuadorian (G) Province (130, p = 0.3268), and C. coccineus was isolated by vicariance in the Desert Province. The group then dispersed into Puna Province (129, p = 0.8733), followed by vicariance between the Puna and Ecuadorian Provinces (127, p = 0.8077; 128, p = 1.0000), and a terminal range extension by C. imasumacc into Yungas Province.
The crown node of Eucharideae (124, p = 0.9666) was assigned to Cauca Province, and no subsequent events occurred until node 121 (p = 0.1035) when three dispersals extended the range at node 120 (p = 0.2882) to the Ecuadorian Province and to Napo (A) and Puna (C) Provinces at node 113 (p = 0.2329). At node 120, range extension into Mesoamerica occurs at the crown node of Phaedranassa (119, p = 0.5591), with subsequent vicariance between Cauca Province (118, p = 1.0000), and the Ecuadorian Province/Mesoamerica Bioregion (117, p = 0.9663). It should be noted that the majority of Phaedranassa species are endemic to the Ecuadorian Province, of which only one is represented here. The Eucrosia clade remains within the Ecuadorian Province (114–116, p = 1.0000). Vicariance occurs at node 113 (p = 0.2329) between Cauca (112, p = 0.3327) and Napo/Puna Provinces (102, p = 0.6748). From node 112, the crown node of the “Eucharis-Urceolina” clade, dispersal takes place to Ucayali Province (111, p = 0.6193) with a mosaic of subsequent dispersals and vicariant events taking place between that province, Western Ecuador (I), Cauca (B), Napo (A), and Chocó-Darién (K) Provinces (nodes 103-111, see Supplementary Table 3 for probabilities) among the species of Eucharis, Caliphruria, and Urceolina (and E. dodsonii) represented in this subclade. The second subclade (102), the crown node of Stenomesson (including Caliphruria korsakoffii), was marked by dispersal and vicariance events involving Desert (Stenomesson flavum Herb., Ecuadorian [Stenomesson chloranthum Meerow & van der Werff, Stenomesson leucanthum (Ravenna) Meerow & van der Werff], Puna (Stenomesson aurantiacum Herb., Stenomesson miniatum, Stenomesson pearcei Baker), and Western Ecuador (Stenomesson ecuadorense) Provinces; see Figure 10 and Supplementary Table 3 for details.
For the plastome tree, the BAYAREAlike model had the best weighted AICc score with BioGeoBears, and probability value supported the +J option, thus that model was applied to the tree (Supplementary Figure 9). The number of dispersal events = 67, vicariance = 33, and there was a single extinction. This extinction event occurred at node 107, the stem node of Stenomesson, with the elimination of putative species in the Cauca Province at the crown node of the genus. Probabilities for each event were considerably higher for the plastome tree (Supplementary Table 4) compared to the nuclear supermatrix tree (Supplementary Table 3), with much fewer compound distributions at ancestral nodes. Most notably, Western Ecuador Province, rather than Puna Province was designated as one of three ancestral areas at node 189 in the plastome tree. The crown node of Hymenocallis is situated with greatest likelihood within the Mesoamerica Bioregion (Supplementary Figure 9), though both the Guianan Lowland Province and West Indies Bioregion are also included (with lower likelihood) if all potential areas are shown. See Supplementary Table 4 for details.
Discussion
Overview
Both nuclear and plastome genomics leave no doubt that the recognition of four monophyletic tribes, Clinantheae, Eucharideae, Eustephieae, and Hymenocallideae, within the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae is a highly supported, and stable evolutionary classification for the group (Meerow et al., 2000; Meerow, 2010; Meerow and Nakamura, 2019). Only the position of Pamianthe retains ambiguity (Figures 2–4), which in the nuclear phylogenomic data is eliminated by the coalescent species tree (Figure 3) and supermatrices with less missing data (Supplementary Figures 3, 4). Plastome data support a third possible resolution for this enigmatic genus of three epiphytic or lithophytic species (Meerow et al., 2019), as sister to Eucharideae (Figure 4). This is discussed below under “Clinantheae.” Chase et al. (2009) recognized both tribes Eucharideae and Stenomesseae (which we consider synonymous), but not Clinantheae. As they did not list component genera for the tribes of subfam. Amaryllidoideae, it is impossible to determine how they circumscribed these tribes. While there is only minor incongruence between the supermatrix (Figure 2 and Supplementary Figures 2–4) and the coalescent species trees (Figure 3 and Supplementary Figures 6, 7), strong cytonuclear incongruence is evident at the intra-tribal and generic levels (Figure 5).
Our diversification analysis robustly found no rate shifts anywhere in the Andean tetraploid clade, and the nuclear gene biogeographic analysis did not detect any extinction events. A high number of extinction events can be a trigger for diversification rate shifts (Brocklehurst et al., 2015), and in that sense, these two analyses mutually support the relative absence of both in the history of the Andean Amaryllidaceae, but may indicate that an Oligocene origin is an over-estimation of the age of the clade.
Cytonuclear Discordance
Cytonuclear discordance is the phenomenon wherein phylogenies inferred from nuclear genes differ markedly from trees constructed using organellar genes (Rieseberg and Soltis, 1991; Folk et al., 2017; Morales-Briones et al., 2018; Lee-Yaw et al., 2019). Whole or partial plastomes have increasingly been used for phylogenetic analysis, and when contrasted with various large nuclear gene sets, cytonuclear discordance is frequently encountered (Huang et al., 2014; Bruun-Lund et al., 2017; Folk et al., 2017; Sloan et al., 2017; Morales-Briones et al., 2018). Cytonuclear discordance may arise via (1) incomplete lineage sorting (ILS) of ancestral polymorphisms, such that phylogenetic relationships from organellar markers do not capture a true evolutionary history of the taxa under study (Maddison and Knowles, 2006; Joly et al., 2009; Stolzer et al., 2012; Solís-Lemus and Ané, 2016; Knowles et al., 2018), (2) selection operating within organellar genomes independent of speciation (Sloan et al., 2017; Lee-Yaw et al., 2019), and (3) hybridization and/or chloroplast transfer, upon which selection may also play a role (García et al., 2017; Sloan et al., 2017; Bastide et al., 2018; Degnan, 2018; Folk et al., 2018; Morales-Briones et al., 2018). Hippeastreae subtribe Hippeastrinae exhibited a great deal of cytonuclear discordance, but also showed combined effects of ancient reticulation and incomplete linkage sorting (García et al., 2014, 2017). While there was initial evidence from nrDNA ITS sequences of early reticulation in Hippeastreae (Meerow et al., 2000; Meerow, 2010; García et al., 2014, 2017), there was none in the Andean tetraploid clade (Meerow, 2010), although the clade had lower sampling taxonomically and genetically than our current study.
The hybridization network (Figure 7) suggests ancestral reticulation within the subclade of Eucharideae (Figure 7) containing Eucharis, Caliphruria (except C. korsakoffii), Urceolina, and the surprising inclusion of E. dodsonii, which has consequences for the taxonomy of the subclade (discussed below under “Eucharideae”). The only other putative reticulate origin at the generic level is the monotypic Plagiolirion (Figure 7). We are inclined to weight the agreement of supermatrix and coalescent analytical approaches to our nuclear data as evidence that ILS has been rare in the evolution of the Andean tetraploid clade as has been argued with other plant taxa (Stephens et al., 2015; Folk et al., 2017) as well as theoretically (Bayzid and Warnow, 2013; Roch and Warnow, 2015). The two exceptions might be the genus Pamianthe, which resolved in three different places among our three main trees (Figures 2–4), and perhaps the apparent reticulate origin of the Colombian endemic monotypic Plagiolirion (Figure 8). However, the supermatrices with less missing data (Supplementary Figures 2, 3) are in agreement with the species tree (Figure 3) that Pamianthe is the first branch of the tribe Clinantheae. We thus favor reticulation over ILS as the primary cause of observed cytonuclear discordance (Figure 5), with Pamianthe the most likely instance of ILS, and perhaps Plagiolirion as well.
The resolution of Worsleya as sister to H. reticulatum in our plastome tree could be evidence that Worsleya represents a hybrid of Griffinia and Hippeastrum, and was previously observed but unreported (N. García, pers.comm.). Though outside of the scope of this paper, this discordance needs to be further analyzed with greater sampling of Hippeastreae and Griffinieae.
We now discuss each of the four resolved monophyletic tribes of the Andean tetraploid clade, placing our phylogenetic and biogeographic inferences into a clade-based context.
Eustephieae
The first branch of the Andean tetraploid clade constitutes the tribe Eustephieae, which consists of four genera, Chlidanthus, Eustephia, Hieronymiella, and Pyrolirion. The staminal corona is absent (Pyrolirion), reduced to basal connation of the subulate filaments (Chlidanthus), present, but modified (Hieronymiella) or reduced to basal connation, but with acute appendages distally on both sides of the primarily filiform filaments (Eustephia). The uniflorus Pyrolirion has the broadest distribution of the four, occurring from Ecuador to Bolivia, on the western slopes and inter-Andean valleys, predominantly within the Prepuna and Puna Provinces, but also the Peruvian coastal fog desert lomas (Desert Province). The style is branched apically in this genus. Due to uniflory, fused spathe bracts, and perigone shape, it was believed to be allied with Zephyranthes Herb. until recently (Meerow, 2010). Pyroliron albicans bears superficial resemblance in floral morphology to the uniflorus Hippeastrum Herb. subg. Tocantinia (Ravenna) Nic. García (García et al., 2019). Huaylla et al. (in press) lists eight species for the genus, but this may not reflect synonym in the absence of a thorough revision (Cowley, 1989).
The Eustephieae represents the southernmost element of the Andean tetraploid clade, outside of the reported occurrence of C. humilis in the Andes of northern Chile (Ravenna, 1974a). It is the least well understood of the Andean tetraploid clade tribes, and lacks one of three indel regions of nrDNA ITS that are apomorphic for the rest of the Andean clade (Meerow et al., 2000).
Eustephia, with perhaps four species, is restricted to southern Andean Peru (Puna Bioprovince). The majority of Hieronymiella spp. are found in the seasonally arid Prepuna Province (Cabrera, 1994) of northern Argentina (Arroyo-Leuenberger and Leuenberger, 2004), with extensions into montane central and southern Bolivia (Lara and Huaylla, 2015; Lara, 2018). There are six to eight species in the genus (Arroyo-Leuenberger and Leuenberger, 2004; Lara and Huaylla, 2015; Lara, 2018), which show triploid and aneuploid derivations of the ancestral 2n = 46 chromosomes (Di Fulvio, 1973). It is most diverse genus of the tribe in terms of floral morphology (Arroyo-Leuenberger and Leuenberger, 2004). Chlidanthus is found only in the lomas of Central and southern Peru (Desert Province) and Andean Bolivia (Lara and Céspedes, 2019, Puna Province). All of the 3–4 species have long-tubed yellow flowers.
In our reconstruction, a gap of ca. 6 Mya separates the stem and crown nodes of Eustephieae, suggesting a period of stasis until the other three genera diverged from Pyrolirion (Figure 9). The tribe may have originated in the Desert, Prepuna, or Puna Provinces comprising Central and/or southern Peru and perhaps Bolivia (Figure 10). The accuracy of biogeographic scenarios in Eustephieae is compromised by limited sampling of all of the genera with presence in more than one biogeographic area. Nonetheless, it is likely that diversification of the tribe was promoted by the uplift of the eastern cordillera of the central Andes starting in the Miocene (Garzione et al., 2008; Antonelli et al., 2009; Luebert and Weigend, 2014), which might account for the predominance of vicariant events in the internal nodes of the Eustephieae phylogeny (Figure 10). That uplift in Andean Peru may have proceeded from the south to the north (Picard et al., 2008) probably influenced the diversification timing of the tribe relative to the other groups in the Andean clade.
Clinantheae
According to our results, the Clinantheae may have diverged from its sister clade Hymenocallideae in the Oligocene (Figure 9), with a crown origin in the Cauca and Desert Bioprovinces (Figure 10). The first branch, Pamianthe, is difficult to parse biogeographically because the exact locality of P. peruviana in Peru is unknown, and we have no sequence data for the Ecuadorian endemic P. parviflora Meerow (Meerow, 1984). Meerow and Nakamura (2019) and Meerow et al. (2019)proposed that the genus, with its broad range from Colombia to Bolivia may indeed represent a relictual epiphytic and lithophytic lineage that colonized cloud forests of the western Andes that have been in retreat since at least the Miocene and have also suffered large-scale and largely undocumented destruction (Dillon et al., 1995; Mutke et al., 2017). Both Clinanthus and its sister genus Paramongaia originated in the Desert Province, and diversified in the Ecuadorian, Paramo, Puna, and Yungas Provinces, largely within Peru, both dispersing into Bolivia, but only Clinanthus into southern Peru and Ecuador, although not until the Pliocene (Figure 9). Clinanthus is most diverse in northern and central Peru. The northern Peruvian (Ecuadorian Province) spp. of Paramongaia [P. milagroantha and P. mirabilis (Ravenna) Meerow], the latter not available for the sequence capture, but sister to P. milagroantha with ITS (Meerow and Nakamura, 2019), were isolated from the rest of the genus in the early Miocene.
Clinanthus separated into two sister subclades discussed by Esquerre and Meerow (2020) that diverged about 13 Mya in our analysis. The larger of the two conforms to Stenomesson subg. Fulgituba Ravenna as conceived by Ravenna (1974b) in his polyphyletic concept of that genus. The species of this subclade typically have lorate leaves, generally >1 cm. wide, long funnelform-tubular flowers, the tubes long relative to the limb, and various patterns of green in the relatively short limb. They occur in a diversity of habitats (Esquerre and Meerow, 2020). The second subclade typically has narrowly lorate to linear leaves <1 cm wide, and usually no green on the tepals. They appear younger than then the first group (Figure 9), and include the alpine species C. humilis, which occurs at over 4,000 m elevation and maintains its scape inside the bulb until seed capsule maturity (Meerow, 1987b). The genus as a whole may number two dozen or more species (León et al., 2013; Meerow and Cano, 2019; Esquerre and Meerow, 2020), many of which are known from single localities.
Hymenocallideae
The Hymenocallideae is the one lineage of the Andean tetraploid clade that has extended its range the furthest into the West Indies, Mesoamerica and Eastern North America Bioregions. The complex taxonomic history of the tribe is reviewed by Meerow et al. (2002). It is a tenable although untested hypothesis whether long distance dispersal was facilitated by the curious fleshy seeds with photosynthetic integuments and starch storing embryos adapted for hydrochory (Whitehead and Brown, 1940; Flint and Moreland, 1943), relative to the flattened, winged, and presumably wind dispersed seeds of Clinantheae, most Eucharideae, and Eustephieae.
In our reconstruction, the tribe diverged from Clinantheae in the Oligocene, about 28 Mya (Figure 9), perhaps in the Desert Province of northern Peru, dispersing to Puna Province first, ca. 2 My later, the stem age of Ismene, which is the first branch of the tribe. Incomplete sampling might bias the area optimization of Ismene, other species of which range to Ecuador to the north, and southern Peru and Bolivia to the south (Meerow and Cano, 2019), but they remain primarily in the higher elevation Puna Bioprovince. Leptochiton diverged 2 My after Ismene. The genus is endemic to the Desert Province, specifically the low elevation Tumbes-Piura dry forest of NE Peru and SE Ecuador (Leal-Pinedo and Linares-Palomino, 2005; Linares-Palomino, 2006; Loaiza, 2013), with a very short season of active growth. Snoad (1952) reported 2n = 24 chromosomes for L. quitoensis; we counted 2n = 34 (unpubl. data). These would be the lowest reported somatic chromosome numbers in the entire Andean clade, and may actually represent karyotype reduction, which might have had some adaptive value to the Tumbes-Piura biome, considered the most xeric of the seasonally dry tropical forest types in South America (Loaiza, 2013). Meerow et al. (2002) considered Leptochiton the least derived genus of the Hymenocallideae because of its numerous ovules per locule and the presence of phytomelanin in the seed testa, but neither nuclear phylogenomics nor plastomics support this (Figures 2–4).
The ancestor of Hymenocallis managed to disperse to the West Indies Bioregion sometime between 21–24 Mya (Figures 8, 9), certainly the greatest instance of long distance dispersal in the Andean clade. Each of the two small, pseudopetiolate clades that resolve as first branches in their respective clades of Hymenocallis (Figures 2, 3) were established by the start of the upper Miocene (Figure 9). There is no evidence from the hybridization network (Figure 8) that there was reticulation between the two large subclades of the genus, though our partial plastome tree (Figure 4) resolves the H. guianensis/speciosa/tubiflora clade as the first branch in the genus, instead of sister to the subclade that contains the Eastern North American clade, and brings additional Mesoamerican species into this subclade.
The nuclear tree suggests that Hymenocallis entered Mesoamerica twice from the West Indies, establishing a small beachhead in the Mexican southern states of two species retaining pseudopetiolate leaves, H. eucharidifolia Baker in wet forests and H. glauca in drier habitats, before dispersing, presumably just once, to Eastern North America (Figure 10), either a second instance of long distance dispersal, or a gap in our sampling. All of the other Mesoamerican species that we sampled resolved in the second subclade (Figures 3, 4), which dispersed from the West Indies Province (Figure 10). There were either returns to the West Indies in the later history of this clade, or the mosaic was the consequence of reticulation (Figure 8). The alternative position for the H. guianensis/speciosa/tubiflora subclade in the BioGeoBears analysis of the best plastome ML tree (Supplementary Figure 9) places the crown node of the genus in the Mesoamerica Province, (Supplementary Figure 9), the product of a single dispersal. This may be a more realistic scenario of how Hymenocallis made its entry into Mesoamerica, given the degree of reticulation revealed by nuclear genes (Figure 8).
With the exception of Bush et al. (2010), previous molecular phylogenetic studies of Hymenocallis included no more than five species (Meerow et al., 2000, 2002). Bush et al. (2010) utilized a combination of 23 ISSR presence/absence and 10 qualitative and quantitative morphological characters to examine relationships among the 15 endemic species of the genus from the Eastern North American Bioregion recognized by Smith (Smith and Garland, 2003); one additional was recently described (Smith and Garland, 2009). Our data (Figures 2, 3) agreed with some of the conclusions reached by Bush et al. (2010) with combined ISSR and morphological data, e.g., the sister relationship of H. henryae Traub and Hymenocallis palmeri S. Watson, the inclusion of Hymenocallis eulae Shinners as a synonym of Hymenocallis occidentalis (Leconte) Kunth (Smith and Garland, 2003), placement of Hymenocallis choctawensis Traub, Hymenocallis franklinensis G.Lom.Sm., L.C.Anderson & Flory, and Hymenocallis pygmaea Traub in the same subclade, but was completely at odds with their resolution of Hymenocallis crassifolia Herb., Hymenocallis godfreyi G.Lom.Sm. & Darst, and Hymenocallis rotata (Ker Gawl.) Herb. While the Bush et al. (2010) tree was congruent with the Traub (1962) system of “alliances” (Caroliniana and Henryae) for the endemic Eastern North American taxa, our cladistic resolution of the clade was not as straight forward. Traub (1962, 1980) recognized four additional alliances for the rest of the genus. Our data indicate that all four are para- or polyphyletic.
In addition to the unusual seed morphology, the seeds of Hymenocallis can be polyembryonic (Traub, 1966; Borys et al., 2005). The genus also exhibits high instances of polyploidy and aneuploidy (Flory and Schmidhauser, 1957; Flory, 1976) from the ancestral 2n = 46 (Flory, 1976), which may be further evidence of interspecific hybridization. These biological characteristics of the genus when coupled with the ability to reproduce vegetatively, may confound a complete understanding of the systematics of the genus.
Eucharideae
This monophyletic tribe represents the pseudopetiolate genera of the Andean tetraploid clade. This character, which is broadly homoplasious throughout the family (Meerow and Snijman, 1998), and evolved at least twice in Hymenocallis, became fixed in this group, along with the loss of ndhF from the plastome (Meerow, 2010), and pseudogenization of other members of the ndh family (Supplementary Table 5). The short branch lengths of the stem lineages suggest a rapid mid-Miocene radiation (Figure 9), perhaps a consequence of the Andean orogeny (Gentry, 1982) and/or environmental heterogeneity and spatial dynamics (Mutke et al., 2014).
From an ancestral area in the Cauca Bioprovince (Figure 10), Rauhia diverged from the rest of the genera in the mid-Miocene (Figure 9). Rauhia, found only in the seasonally dry forests of the inter-Andean valleys of the Marañon river and its tributary, the Utcubamba, with the exception of one species (Meerow and Nakamura, 2019), is a xerophyte with thick, carnose leaves and universally green (or green and white) flowers. Floral morphology is similar to both Phaedranassa [R. multiflora (Kunth) Ravenna, R. staminosa Ravenna] and Eucrosia (R. decora Ravenna).
The next branch is the monotypic Plagiolirion horsmannii, endemic to the Cauca valley of Colombia, and possibly of reticulate origin (Figure 7). This relict genus, most recently allied with Eucharis (Meerow and Silverstone-Sopkin, 1995), is a clear case of peripheral isolation. It adapted to lower and mid-montane rainforest understory of the Cauca valley, exhibiting some of the characters of Eucharis (thin-textured leaf laminae, ellipsoidal seeds with copious oily endosperm), which we interpret as convergences. Plastome sequences position it as sister to Eucrosia and Phaedranassa (Figure 4).
The sister relationship of Eucrosia and Phaedranassa is ironclad, and their divergence at ca. 10 Mya may have been directly influenced by the rise of the Andes. Phaedranassa is primarily found above 2,000 m in the Ecuadorian Andes, which is not evident by the breadth of our sampling (only a single Ecuadorian species of eight was included), while Eucrosia is markedly a low elevation genus, rarely found above 1,000 m. The leaves of Eucrosia have a smaller length:width ratio than its sister genus, and the perigones are zygomorphic [though not strongly so in Eucrosia stricklandii (Baker) Meerow] vs. the actinomorphy of Phaedranassa. Most importantly, all true Eucrosia spp. have globose nectar glands at the base of the stamens (Meerow, 1987a). Like its sister genus, most of its species diversity is found in Ecuador (Meerow, 1990), but unlike Phaedranassa, which does not occur south of Loja Province, Eucrosia is represented in Peru by one endemic [Eucrosia calendulina Meerow and Sagástegui (1997)] and two species that also occur in Ecuador (Eucrosia bicolor, albeit a distinct variety, and Eucrosia eucrosioides Pax). Only two species, E. bicolor and E. stricklandii, have a conspicuous staminal cup that is deeply cleft dorsally (Meerow, 1987a).
The final and somewhat problematic subclade in the Eucharideae resolved a monophyletic Stenomesson, inclusive of Caliphruria korsakoffii4, the only Peruvian species of that genus. This species was the first branch in Stenomesson (Figures 2, 3), and may represent an isolated eastern relict of early diversification of the subclade (Figure 7), as it is known only from wet forest in the vicinity of Moyobamba, Peru (Meerow, 1989). The two species of Stenomesson that release their pollen in tetrads (Meerow et al., 1986; Meerow and van der Werff, 2004), S. chloranthum Meerow & van der Werff and S. leucanthum, (Ravenna) Meerow & van der Werff, were sister species in every tree generated (Figures 2–4 and Supplementary Figures 2–7). Ravenna (1988) Stenomesson sect. Adnata Ravenna, in which the staminal cup is almost completely fused to the perigone tube, represented in our sampling by S. ecuadorense and S. miniatum, was resolved as monophyletic by the full nuclear gene supermatrix and all of the species trees, but not the 70–90% supermatrix trees nor the plastome supermatrix.
The sister clade to Stenomesson is completely adapted to the shady understory of low to mid-elevation rainforest, and extends the furthest east of any taxa in the tribe (Meerow, 1989). It showed a high degree of reticulation in the hybridization network (Figure 7). In this rainforest understory clade, the Colombian Caliphruria spp. were nested in a paraphyletic Eucharis (Figures 2, 3) that was sister to Urceolina. Eucharis amazonica and E. sanderi, which Meerow (1989) placed in subg. Heterocharis Meerow along with Eucharis moorei (Baker) Meerow and one natural hybrid, and which he considered the least derived species in the genus, constituted the first branch after Urceolina (Figures 2, 3). The most intriguing subclade brought together E. dodsonii, E. astrophiala, and the Colombian Caliphruria spp. (Figures 2, 3). Eucharis astrophiala is the only species of the genus endemic to the lower western Andean slopes of Ecuador, and bears novel morphological characteristics suggestive of peripheral isolation (Meerow, 1989). Like its sister species, E. dodsonii, also endemic to the western Ecuadorean mid-elevation (900–1,500 m) montane rainforest, it has deeply plicate leaves (Meerow and Dehgan, 1985). This small subclade of four species (Figures 2, 3 and Supplementary Figures 3,4) dramatically illustrates the convergent evolutionary machinations of isolation on the tetraploid genomes of the Andean clades (Meerow, 2010). E. dodsonii, misdiagnosed as a Eucrosia because of its long exerted stamens, despite its mesic ecology (Meerow and Dehgan, 1985) and small globose seeds (unknown at the time it was described), is analogous to the divergence of Urceolina from Eucharis, and even bears similar perigonal coloration as the former. Although he was wrong in regard to Mathieua and Plagiolirion (Meerow, 1987d; Meerow and Silverstone-Sopkin, 1995), we recognize the prescience of Traub (1971) in combining Caliphruria (less C. korsakoffii), Eucharis and Urceolina as the latter, and accept this clade as a single genus, Urceolina, now with the addition of E. dodsonii5 (Figures 2–4, 7).
Did the Amotape—Huancabamba Zone (AHZ) Function as a Barrier in the Early Diversification of Eucharideae?
The northern and central Andes connect in southern Ecuador and northern Peru, and this area does not achieve elevations above 2,145 m (Weigend, 2002). This depression in the Andes was labeled as the Amotope–Huancabamba Zone (AHZ) by Berry (1982), and was discussed in greater detail by Weigend (2002, 2004). It has also been called the Huancabamba deflection, depression or gap (Quintana et al., 2017b). It is the youngest part of the Andean chain (Hoorn et al., 2010).
The AHZ was invaded by marine incursions in the Eocene (see discussion in the next section), and has been considered a dispersal barrier for lowland taxa from west to east or vice versa (Antonelli et al., 2009). As the Andes rose, these marine incursions ended. Subsequently, the AHZ was considered by some authors to have become a more recent barrier for upland plant taxa in a north/south (and reverse) direction on the basis of biogeographic study (Ayers, 1999; Beck and Richter, 2008; Antonelli et al., 2009; Cosacov Martinez et al., 2009; Antonelli and Sanmartín, 2011; Quintana et al., 2017b), despite previous contrary conclusions (Berry, 1982; Weigend, 2002). Quintana et al. (2017b) concluded that the AHZ functioned as a migration corridor for taxa both in latitudinal and longitudinal directions, but not as a barrier. Mutke et al. (2014) also strongly rejected the barrier hypothesis, and point out the high degree of plant diversity in the zone.
We believe that the distributions of the first branches in the Eucharideae clade suggest that the AHZ may have functioned both as an east-west and north-south corridor, as well as a north-south barrier to migration. The first branch of the tribe is the xerophytic genus Rauhia, the five species of which are found only at the southern limits of the AHZ (Meerow and Nakamura, 2019), generally at 500–800 m elevation. The next branch, Plagiolirion, is endemic to Colombia, but may be of recombinant origin (Figure 7), or perhaps another instance of ILS. It is impossible to say if it represents a long distance dispersal event (Figure 10), or an element of a once more widespread lineage of rainforest understory taxa. The next subclade, sister genera Eucrosia and Phaedranassa, are lowland and highland elements, respectively, the diversity of which is concentrated in Ecuador (Meerow, 1990). Phaedranassa is not known south of Loja Province in Ecuador at the northern limits of the AHZ (Meerow, 1990), while Eucrosia has managed to migrate west of the Andes to the Pacific coastal plain (E. bicolor), and south into Peru [E. bicolor var. plowmanii Meerow, E. eucrosioides (Meerow, 1987a) and the endemic E. calendulina (Meerow and Sagástegui, 1997)].
Additional Biogeographical and Historical Inferences and Caveats
In calibrated age diversification studies of woody plant lineages from seasonally dry Andean tropical forests, splits between clades dating from the Miocene to Pliocene have been observed with strong geographic structure (Pennington et al., 2004, 2010; Särkinen et al., 2012; Lavin et al., 2018), attributed to speciation via enforced geographic isolation (Pennington et al., 2009). This certainly seems to be the case in many of the genera of the Andean tetraploid clade, in which closely related species are found in isolated inter-Andean valleys, e.g., Rauhia (Meerow and Nakamura, 2019), Eucrosia (Meerow, 1987a) and Phaedranassa (Oleas et al., 2012, 2013, 2016) over relatively short distances.
Though the exact timing and rate of Andean uplift remains controversial (Garzione et al., 2008; Picard et al., 2008; Antonelli et al., 2009; Luebert and Weigend, 2014; Mutke et al., 2014), there is greater consensus on the ways in which the vast and heterogeneous orogenic chain has directly affected plant diversification (Simpson, 1975, 1983; Antonelli and Sanmartín, 2011; Hoorn et al., 2013; Luebert and Weigend, 2014). The main classes of effects are (1) creation of new high elevation habitats, (2) providing barriers that result in vicariance (Figure 10), (3) providing a bi-directional north-south migration corridor, and (4) generating new environments for colonization outside of the Andes. All of these seem to have contributed to the diversification of the Andean clade of Amaryllidaceae. Phaedranassa is a good example of the first. The second and third are significantly broad effects on the entire clade (Figure 10). Lastly, the divergence of Hymenocallis from its Andean roots might be an example of the fourth.
From the Eocene to the Middle Miocene, there appear to have been incursions from the Pacific that flooded a low elevation area between the Northern and Central Andes in the vicinity of southern Ecuador/northern Peru, known as the “Western Andean Portal” (WAP) or “‘Guayaquil Gap”’ (Hoorn, 1993; Hoorn et al., 1995; Steinmann et al., 1999; Santos et al., 2008; Antonelli et al., 2009). The WAP has been cited as an important barrier that split lineages of various biota (Molau, 1988; Sparre and Andersson, 1991; Hofreiter and Rodríguez, 2006; Antonelli et al., 2009; Meerow et al., 2015). We believe that this may have been a contributing factor for the relative dearth of species of both Clinanthus and Stenomesson north of Peru.
The flooding of western Amazonia caused by the uplift of the Eastern Cordillera of the Central Andes in the early Miocene and subsequent Caribbean marine incursion from the north (Lundberg et al., 1998; Wesselingh et al., 2002; Wesselingh, 2006; Wesselingh and Salo, 2006; Antonelli et al., 2009; Meerow et al., 2009, 2014; Roncal et al., 2013), created an enormous system of long-lived lakes and wetlands (“Lake Pebas” or the “Pebas Sea”) that provided a significant barrier to dispersal between the eastern Amazonian and Guianan region and the Andes lasting until the Late Miocene. We suggest that this event isolated Hymenocallis from its ancestors in the Andean region.
Age of these deeper internal nodes in our penalized likelihood age divergence tree (Figure 9) should be approached with caution in the absence of fossil calibration, especially since in Bayesian age diversification analyses, it is the deeper branches in phylogenetic trees that typically show the broadest 95% highest prior densities (e.g., Meerow et al., 2009, 2015; Salas-Leiva et al., 2013; Calonje et al., 2019). Consequently, considering that the deeper calibration is based on a previous chronogram from analysis of a few plastid regions, and only four American species (Santos-Gally et al., 2012), an Oligocene origin for the clade might be an overestimation of its age. However, the other calibration point is based on a solid basis of studies with tree fossil support of a minimum age for the type of inter-Andean dry forests in which the genus Rauhia is endemic (Caetano et al., 2008; Pennington et al., 2010; Luebert and Weigend, 2014; Quintana et al., 2017a). Consequently, the node ages above the stem of Rauhia should have greater veracity.
A final caveat is one that needs to be at least acknowledged. Before Europeans arrived on South American shores, successive human cultures inhabited the Andean region, and the final one, the Inca, though relatively short-lived, ruled an empire that stretched from Ecuador to southern Peru (Burger, 1995). The amaryllis subfamily of Amaryllidaceae produce beautiful flowers, and its members contain many novel alkaloids with biological properties. Ceremonial drinking vessels (keros) from the Incan culture bear iconic floral depictions of the species C. humilis, S. miniatum and perhaps a Pyrolirion sp. (Vargas, 1962). A festival celebrating the June gregarious flowering of Ismene amancaes Herb. on the lomas near Lima that lasted until the middle of the 19th century may have predated the arrival of Europeans (Stewart, 1832). Remains of the plant have been found in Peruvian archeological sites (Browman, 1978). S. pearcei, which has generally been thought to occur only in the mountains above Cusco (Macbride, 1936), was recently found in five populations in the Kañaris District, Lambayeque Department between 2,600 and 3,000 m (B. Esquerre, pers. comm), perhaps an anthropogenically facilitated disjunction. Northern Peru is where the greatest diversity of Stenomesson diversity can be found (Meerow, 1987b; Meerow and van der Werff, 2004). The possible influence of humans on the modern range of species in the Andean clade, though difficult to document, should not be discounted.
Conclusion
The Andean tetraploid clade of the Amaryllidaceae subfam. Amaryllidoideae has been characterized as a highly canalized complex suffused with convergent patterns of divergence within the four tribal lineages (Meerow, 2010). This is not merely a matter of taxonomic judgment, but reflects the evolutionary forces at work since its polyploid origin. An association between polyploidy and both morphological and ecological divergence has long been noted (Ohno, 1970; Levin, 1983), but in the genomic age, our understanding of the degree to which polyploidy fuels evolutionary change in plants has become more explicit (Adams and Wendel, 2005; Soltis et al., 2015). Polyploids may undergo fast and sweeping genomic changes (Lynch and Conery, 2000; Seoighe and Gehring, 2004). Evidence has accumulated showing that duplicate genes either diversify in function or undergo subfunctionalization (Lynch and Force, 2000; Blanc and Wolfe, 2004). Polyploidy also effects gene expression, including gene silencing (Kashkush et al., 2002; Adams et al., 2003; He et al., 2003).
Of the external selective forces operating on the polyploid genome of the Andean clade, the most obvious is the rise of the Andes itself, an association that has been noted repeatedly (Meerow and Dehgan, 1985; Meerow, 1987c,d, 2010) and is obvious from our biogeographic analyses (Figure 10 and Supplementary Figure 9). Meerow (2010) characterized the Andean tetraploid clade’s polyploid genome as grist for the mill of the Andean orogeny. Northern Peru in particular, comprising in part the Neotropical bioprovinces Cauca, Desert and Ecuadorian (Morrone, 2014), with its complex of microhabitats, has been a hotspot of diversification in the Andean clade (Meerow and van der Werff, 2004; Meerow and Cano, 2019; Meerow and Nakamura, 2019; Esquerre and Meerow, 2020). The distributions of Rauhia in Peru and Phaedranassa in Ecuador (Meerow, 1990; Oleas et al., 2012, 2013, 2016) fit the allopatric models of dispersal or vicariance (Figure 10 and Supplementary Figure 9) followed by isolation described for Andean complexes of Buddleja L. (Norman, 2000), Calceolaria L. (Molau, 1988), Fuchsia L. (Berry, 1982), and many other taxa (Ayers, 1999; Beck and Richter, 2008; Antonelli et al., 2009; Pennington et al., 2010; Anthelme et al., 2014; Luebert and Weigend, 2014; Bacon et al., 2018).
Compared to the tribe Hippeastreae (Meerow, 2010; García et al., 2014), reticulation has not played a significant role in the generic radiation of the Andean tetraploid clade, except in Eucharideae, where we must concede that Eucharis and Colombian Caliphruria cannot be logically separated from Urceolina (Traub, 1971). However, there is evidence of large scale interspecific hybridization within Hymenocallis in the SE U.S. and Mexico, and to a lesser extent in Clinanthus. We only see ILS as a significant factor in the genus Pamianthe, and perhaps Plagiolirion as well.
In the context of the four hypotheses we tested, (1) phylogenomic data support the previous tribal and generic classification of the tetraploid Andean clade inferred from ITS and several plastid loci, (2) reticulation was primarily limited to interspecific gene flow at the generic level, with one notable exception in the Eucharideae, (3) the Andean orogeny was the primary factor in the generic diversification of the clade and (4) our resolute phylogeny suggest that polyploidy and resulting paralogy do not necessarily impede accurate phylogenomic inference.
It may be tempting for some taxonomic pundits to declare that there has been superfluous recognition of genera within the Andean clade, and suggest further mergers beyond our acceptance of Traub (1971) concept of Urceolina. We believe that would be an error of judgment that obscures the phylogenetic complexity of this Andean-centered polyploid radiation, could result in loss of information, confusion, and would create further nomenclatural chaos in a family already rife with it (García et al., 2019).
Data Availability Statement
Illumina HiSeq reads (fastq files) and associated information from the sequence capture are deposited in the Sequence Read Archive (SRA) database of The National Center for Biotechnology Information as Bioproject PRJNA635412. Alignments, analysis data files and trees are deposited in DRYAD Project: doi: 10.5061/dryad.573n5tb4j.
Author Contributions
AM and KN designed the study. KN conducted all lab work at USDA prior to the contracted services from Rapid genomics, and assembled the plastome data. AM and EG conducted all analyses. AM, EG, and KN wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the many individuals and institutions who provided leaf material for extraction of DNA, or bulbs from their collections, and the curators of FLAS, FTG, and NA for housing voucher specimens. Fieldwork in South America was supported by USDA, and National Science Foundation Grants BSR-8401208, DEB-968787 and 0129179 to AM. Antonio Campos-Rocha, Asuncíon Cano, Norton Cuba, Julie Dutilh, Boris Esquerre, Segundo Leiva, Blanca Léon, Nicolás García, Nora Oleas, and Agostina Sassone at various times stimulated useful discussions on Amaryllidaceae. We are also grateful to the reviewers of the manuscript, all of whom helped improve it. AM dedicates this paper to the memory of Timothy C. Plowman, who first encouraged him to work on the systematics of the family.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.582422/full#supplementary-material
Supplementary Text 1 | Taxonomic changes.
Supplementary Figure 1 | Heat map of paralog presence across the unpruned sequence capture loci from 95 species of Andean Amaryllidaceae and 5 outgroups. White = absent, black = one copy, yellow-to-red = 2 to 11 copies (paralogs).
Supplementary Figure 2 | Best tree from maximum likelihood analysis of partitioned 524 nuclear genes, coding-only supermatrix of the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae, with bootstrap percentages < 100 shown above branches (all unmarked branches have 100% BP). Refer to Supplementary Table 1 for corrected taxon names.
Supplementary Figure 3 | Best tree from maximum likelihood analysis of partitioned 260 nuclear gene supermatrix (only genes amplified across 70% of the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae), with bootstrap percentages < 100 shown above branches (all unmarked branches have 100% BP). Refer to Supplementary Table 1 for corrected taxon names.
Supplementary Figure 4 | Best tree from maximum likelihood analysis of partitioned 137 nuclear gene supermatrix (only genes amplified across 90% of the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae), with bootstrap percentages < 100 shown above branches (all unmarked branches have 100% BP).
Supplementary Figure 5 | Maximum likelihood tree with bootstrap using an earlier complete supermatrix of the Andean tetraploid clade of Amaryllidaceae in which consensus sequences of all paralogs with ambiguity codes were used.
Supplementary Figure 6 | Local posterior probability (LPP) coalescent species tree from ASTRAL III analysis of gene trees from 260 nuclear genes with 70% taxon coverage. LPP scores appear above the branches; bootstrap percentages < 100 shown below branches (all unmarked branches have 100% BP). Refer to Supplementary Table 1 for corrected taxon names.
Supplementary Figure 7 | Local posterior probability (LPP) coalescent species tree from ASTRAL III analysis of gene trees from 70 nuclear genes with 90% taxon coverage. LPP scores appear above the branches; bootstrap percentages < 100 shown below branches (all unmarked branches have 100% BP). Refer to Supplementary Table 1 for corrected taxon names.
Supplementary Figure 8 | Results of the densiTree comparison of relaxed and correlated penalized likelihood age-calibration of the best maximum likelihood tree found by RAxML using the 90% taxon coverage supermatrix and a range of values for λ (0–5).
Supplementary Figure 9 | Area cladogram generated by Bayarealike +J analysis with BioGeoBears through RASP 4.2, using the best maximum likelihood tree found for the partial plastome supermatrix. Areas at any node were limited to three, and only the most likely was mapped onto the tree.
Supplementary Table 1 | Samples used in the phylogenomic analyses of the Andean tetraploid clade of Amaryllidaceae subfam. Amaryllidoideae, and accession numbers for fastq files from the sequence capture.
Supplementary Table 2 | Nuclear gene alignments generated from anchored hybrid sequence capture of 100 taxa of the of Amaryllidaceae subfam. Amaryllidoideae.
Supplementary Table 3 | Event tally for DIVALIKE+J biogeographic analysis of the best 90% taxon coverage supermatrix tree found by maximum likelihood. Node numbers refer to Figure 10.
Supplementary Table 4 | Event tally for BAYAREALIKE +J biogeographic analysis of the partial plastome supermatrix best ML tree. Node numbers refer to Supplementary Figure 10.
Supplementary Table 5 | Pseudogenization of ndh genes in plastomes of Eucharideae.
Footnotes
- ^ https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- ^ https://www.geneious.com
- ^ Hymenocallis gigantiflora Meerow, nom. nov. See Supplementary Text 1 for full details.
- ^ Stenomesson korsakoffii (Traub) Meerow, comb. nov. See Supplementary Text 1 for full details.
- ^ Urceolina dodsonii (Meerow & Dehgan) Meerow, comb. nov. See Supplementary Text 1 for full details.
References
Adams, K. L., Cronn, R., Percifield, R., and Wendel, J. F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. U.S.A. 100, 4649–4654. doi: 10.1073/pnas.0630618100
Adams, K. L., and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141. doi: 10.1016/j.pbi.2005.01.001
Albrecht, B., Scornavacca, C., Cenci, A., and Huson, D. H. (2012). Fast computation of minimum hybridization networks. Bioinformatics 28, 191–197. doi: 10.1093/bioinformatics/btr618
Anthelme, F., Jacobsen, D., Macek, P., Meneses, R. I., Moret, P., Beck, S., et al. (2014). Biodiversity patterns and continental insularity in the tropical high Andes. AAAR 46, 811–828. doi: 10.1657/1938-4246-46.4.811
Antonelli, A., Nylander, J. A., Persson, C., and Sanmartin, I. (2009). Tracing the impact of the Andean uplift on Neotropical plant evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 9749–9754. doi: 10.1073/pnas.0811421106
Antonelli, A., and Sanmartín, I. (2011). Why are there so many plant species in the Neotropics? Taxon 60, 403–414. doi: 10.1002/tax.602010
Antonelli, A., Zizka, A., Carvalho, F. A., Scharn, R., Bacon, C. D., Silvestro, D., et al. (2018). Amazonia is the primary source of Neotropical biodiversity. Proc. Nat. Acad. Sci. U.S.A. 115, 6034–6039. doi: 10.1073/pnas.1713819115
APG (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 141, 399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x
APG (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
Arroyo-Leuenberger, S. C., and Leuenberger, B. E. (2004). Hieronymiella (Amaryllidaceae), a little known genus from Argentina and Bolivia. Herbertia 58, 23–45.
Ayers, T. (1999). Biogeography of Lysipomia (Campanulaceae), a high elevation endemic: an illustration of species richness at the Huancabamba Depression. Peru. Arnaldoa 6, 13–27.
Bacon, C. D., Velásquez-Puentes, F. J., Hinojosa, L. F., Schwartz, T., Oxelman, B., Pfeil, B., et al. (2018). Evolutionary persistence in Gunnera and the contribution of southern plant groups to the tropical Andes biodiversity hotspot. PeerJ 6:e4388. doi: 10.7717/peerj.4388
Bastide, P., Solís-Lemus, C., Kriebel, R., William Sparks, K., and Ané, C. (2018). Phylogenetic comparative methods on phylogenetic networks with reticulations. Syst. Biol. 67, 800–820. doi: 10.1093/sysbio/syy033
Bayzid, M. S., and Warnow, T. (2013). Naive binning improves phylogenomic analyses. Bioinformatics 29, 2277–2284. doi: 10.1093/bioinformatics/btt394
Beck, E., and Richter, M. (2008). Ecological aspects of a biodiversity hotspot in the Andes of southern Ecuador. Biodivers. Ecol. Ser. 2, 195–217.
Berry, P. E. (1982). The systematics and evolution of Fuchsia sect. Fuchsia (Onagraceae). Ann. Missouri Bot. Gard. 69, 1–198. doi: 10.2307/2398789
Blanc, G., and Wolfe, K. H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691. doi: 10.1105/tpc.021410
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borys, M., Leszczynska-Borys, H., Galvan, J., and Naturales, A.-R. (2005). Hymenocallis Salisb germination variants and seedling yield. Acta Horticult. 673, 191–198. doi: 10.17660/ActaHortic.2005.673.22
Bose, S. (1962). Cytotaxonomy of Amaryllidaceae. Nelumbo 4, 27–38. doi: 10.20324/nelumbo/v4/1962/76460
Breinholt, J. W., Earl, C., Lemmon, A. R., Lemmon, E. M., Xiao, L., and Kawahara, A. Y. (2018). Resolving relationships among the megadiverse butterflies and moths with a novel pipeline for anchored phylogenomics. Syst. Biol. 67, 78–93. doi: 10.1093/sysbio/syx048
Brocklehurst, N., Ruta, M., Müller, J., and Fröbisch, J. (2015). Elevated extinction rates as a trigger for diversification rate shifts: early amniotes as a case study. Sci. Rep. 5, 1–10.
Bruen, T. C., Philippe, H., and Bryant, D. (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681. doi: 10.1534/genetics.105.048975
Bruun-Lund, S., Clement, W. L., Kjellberg, F., and Rønsted, N. (2017). First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Mol. Phylogenet. Evol. 109, 93–104. doi: 10.1016/j.ympev.2016.12.031
Bush, C. M., Rollins, D., and Smith, G. L. (2010). The phylogeny of the Southeastern United States Hymenocallis (Amaryllidaceae) based on ISSR fingerprinting and morphological data. Castanea 75, 368–380. doi: 10.2179/09-012.1
Bushnell, B. (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner. Available online at: https://www.osti.gov/biblio/1241166 (accessed October 22, 2020).
Caetano, S., Prado, D., Pennington, R., Beck, S., Oliveira-Filho, A., Spichiger, R., et al. (2008). The history of seasonally dry tropical forests in eastern South America: inferences from the genetic structure of the tree Astronium urundeuva (Anacardiaceae). Mol. Ecol. 17, 3147–3159. doi: 10.1111/j.1365-294X.2008.03817.x
Calonje, M., Meerow, A. W., Griffith, M. P., Salas-Leiva, D., Vovides, A. P., Coiro, M., et al. (2019). A time-calibrated species tree phylogeny of the New World cycad genus Zamia L.(Zamiaceae. Cycadales). Int. J. Plant Sci. 180, 286–314. doi: 10.1086/702642
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Campos-Rocha, A., Meerow, A. W., and Lima, D. A. V. (2019a). The rediscovery of Griffinia alba (Amaryllidaceae), a poorly known and endangered species. Brittonia 71, 134–143. doi: 10.1007/s12228-018-9561-1
Campos-Rocha, A., Meerow, A. W., Lopes, E. F. M., Semir, J., Mayer, J. L. S., and Dutilh, J. H. A. (2019b). New and reassessed species of Griffinia (Amaryllidaceae) from the Brazilian Atlantic Forest. Syst. Bot. 44, 310–318. doi: 10.1600/036364419X15562052252199
Carter, K. A., Liston, A., Bassil, N. V., Alice, L. A., Bushakra, J. M., Sutherland, B. L., et al. (2019). Target capture sequencing unravels Rubus evolution. Front. Plant Sci. 10:1615. doi: 10.3389/fpls.2019.01615
Chamala, S., García, N., Godden, G. T., Krishnakumar, V., Jordon-Thaden, I. E., De Smet, R., et al. (2015). MarkerMiner 1.0: a new application for phylogenetic marker development using angiosperm transcriptomes. Appl. Plant Sci. 3:1400115. doi: 10.3732/apps.1400115
Chase, M. W., Reveal, J. L., and Fay, M. F. (2009). A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot. J. Linn. Soc. 161, 132–136. doi: 10.1111/j.1095-8339.2009.00999.x
Christenhusz, M. J. M., and Chase, M. W. (2013). Biogeographical patterns of plants in the Neotropics - dispersal rather than plate tectonics is most explanatory. Bot. J. Linn. Soc. 171, 277–286. doi: 10.1111/j.1095-8339.2012.01301.x
Cosacov Martinez, A., Sersic, A. N., Sosa, V., De-Nova, J. A., Nylinder, S., and Cocucci, A. A. (2009). New insights into the phylogenetic relationships, character evolution, and phytogeographic patterns of Calceolaria (Calceolariaceae). Amer. J. Bot. 96, 2240–2255. doi: 10.3732/ajb.0900165
Cronn, R., Knaus, B. J., Liston, A., Maughan, P. J., Parks, M., Syring, J. V., et al. (2012). Targeted enrichment strategies for next-generation plant biology. Amer. J. Bot. 99, 291–311. doi: 10.3732/ajb.1100356
De Smet, R., Adams, K. L., Vandepoele, K., Montagu, M. C. E., Van Maere, S., and Van de Peer, Y. (2013). Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. U.S.A. 110, 2898–2903. doi: 10.1073/pnas.1300127110
Degnan, J. H. (2018). Modeling hybridization under the network multispecies coalescent. Syst. Biol. 67, 786–799. doi: 10.1093/sysbio/syy040
Di Fulvio, T. (1973). Contribution to the karyological knowlege of Amaryllidaceae. I. Chromosome study in Hieronymiella and other related genera. Kurtziana 7, 117–131.
Dillon, M. O., Sagástegui Alva, A., Sánchez Vega, I., Llatas Quiroz, S., and Hensold, N. (1995). “Floristic inventory and biogeographic analysis of montane forests in northwestern Peru,” in Proceedings of the Neotropical Montane Forest Biodiversity and Conservation Symposium (1993), eds S. P. Churchill, H. Balslev, E. Forero, and J. L. Luteyn (Bronx: NYBG Press), 251–269.
Esquerre, B., and Meerow, A. W. (2020). A new species of Clinanthus (Amaryllidaceae: Amaryllidoideae: Clinantheae) from Cordillera de los Tarros, Northwest Peru, and notes on related species. PhytoTaxa 438:9. doi: 10.11646/phytotaxa.438.1.7
Ewing, B., and Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194. doi: 10.1101/gr.8.3.186
Ewing, B., Hillier, L., Wendl, M. C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185. doi: 10.1101/gr.8.3.175
Flint, L. H., and Moreland, C. F. (1943). Note on photosynthetic activity in seeds of the spider lily. Amer. J. Bot. 30, 315–317. doi: 10.2307/2437461
Flory, W., and Schmidhauser, T. (1957). Mitotic chromosome numbers in Hymenocallis with a consideration of factors possibly affecting numbers and karyotypes. Genetics 42, 369–370.
Flory, W. S. (1976). Distribution, chromosome numbers and types of various species and taxa of Hymenocallis. Nucleus 19, 204–227.
Folk, R. A., Mandel, J. R., and Freudenstein, J. V. (2017). Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Syst. Biol. 66, 320–337. doi: 10.1093/sysbio/syw083
Folk, R. A., Soltis, P. S., Soltis, D. E., and Guralnick, R. (2018). New prospects in the detection and comparative analysis of hybridization in the tree of life. Amer. J. Bot. 105, 364–375. doi: 10.1002/ajb2.1018
Fragoso-Martínez, I., Salazar, G. A., Martínez-Gordillo, M., Magallón, S., Sánchez-Reyes, L., Lemmon, E. M., et al. (2017). A pilot study applying the plant Anchored Hybrid Enrichment method to New World sages (Salvia subgenus Calosphace; Lamiaceae). Mol. Phylogenet. Evol. 117, 124–134. doi: 10.1016/j.ympev.2017.02.006
García, N., Folk, R. A., Meerow, A. W., Chamala, S., Gitzendanner, M. A., Oliveira, R. S. D., et al. (2017). Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae). Mol. Phylogenet. Evol. 111, 231–247. doi: 10.1016/j.ympev.2017.04.003
García, N., Meerow, A. W., Arroyo-Leuenberger, S., Oliveira, R. S., Dutilh, J. H., Soltis, P. S., et al. (2019). Generic classification of Amaryllidaceae tribe Hippeastreae. Taxon 68, 481–498. doi: 10.1002/tax.12062
García, N., Meerow, A. W., Soltis, D. E., and Soltis, P. S. (2014). Testing deep reticulate evolution in Amaryllidaceae tribe Hippeastreae (Asparagales) with ITS and chloroplast sequence data. Syst. Bot. 39, 75–89. doi: 10.1600/036364414x678099
Garzione, C. N., Hoke, G. D., Libarkin, J. C., Withers, S., MacFadden, B., Eiler, J., et al. (2008). Rise of the Andes. Science 320, 1304–1307. doi: 10.1126/science.1148615
Gentry, A. H. (1982). Neotropical floristic diversity: phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Ann. Missouri Bot. Gard. 69, 557–593. doi: 10.2307/2399084
Granados Mendoza, C., Jost, M., Hágsater, E., Magallón, S., van den Berg, C., Lemmon, E. M., et al. (2020). Target nuclear and off-target plastid hybrid enrichment data inform a range of evolutionary depths in the orchid genus Epidendrum. Front. Plant Sci. 10:1761. doi: 10.3389/fpls.2019.01761
He, P., Friebe, B., Gill, B., and Zhou, J.-M. (2003). Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol. Biol. 52, 401–414. doi: 10.1023/A:1023965400532
Heibl, C. (2008). PHYLOCH: R Language Tree Plotting Tools and Interfaces to Diverse Phylogenetic Software Packages. Available online at: http://www.christophheibl.de/Rpackages.html (accessed October 22, 2020).
Hofreiter, A., and Rodríguez, R. (2006). The Alstroemeriaceae in Peru and neighbouring areas. Rev. Peru. Biol. 13, 5–69. doi: 10.15381/rpb.v13i1.1765
Hoorn, C. (1993). Marine incursions and the influence of Andean tectonics on the Miocene depositional history of northwestern Amazonia: results of a palynostratigraphic study. Palaeogeogr. Palaeocl. Paleoecol. 105, 267–309. doi: 10.1016/0031-0182(93)90087-Y
Hoorn, C., Guerrero, J., Sarmiento, G. A., and Lorente, M. A. (1995). Andean tectonics as a cause of changing drainage patterns in Miocene northern South America. Geology 23, 237–240. doi: 10.1130/0091-76131995023<0237:ATAACF<2.3.CO;2
Hoorn, C., Mosbrugger, V., Mulch, A., and Antonelli, A. (2013). Biodiversity from mountain building. Nat. Geosci. 6:154. doi: 10.1038/ngeo1742
Hoorn, C., Wesselingh, F. P., TerSteege, H., Bermudez, M. A., Mora, A., Sevink, J., et al. (2010). Amazonia through time: andean uplift, climate change, landscape evolution, and biodiversity. Science 330, 927–931. doi: 10.1126/science.1194585
Huang, D. I., Hefer, C. A., Kolosova, N., Douglas, C. J., and Cronk, Q. C. B. (2014). Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 204, 693–703. doi: 10.1111/nph.12956
Huaylla, H., Llalla, O., Torras-Claveria, L., and Bastida, J. (in press). Alkaloid profile in Pyrolirion albicans Herb.(Amaryllidaceae), a Peruvian endemic species. South Afr. J. Bot. doi: 10.1016/j.sajb.2020.03.016
Huson, D. H. (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73. doi: 10.1093/bioinformatics/14.1.68
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Ito, M., Kawamoto, A., Kita, Y., Yukawa, T., and Kurita, S. (1999). Phylogenetic relationships of amaryllidaceae based on matK sequence data. J. Plant Res. 112, 207–216. doi: 10.1007/PL00013874
Johnson, M. G., Gardner, E. M., Liu, Y., Medina, R., Goffinet, B., Shaw, A. J., Zerega, N. J., et al. (2016). HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4:1600016. doi: 10.3732/apps.1600016
Joly, S., McLenachan, P. A., and Lockhart, P. J. (2009). A statistical approach for distinguishing hybridization and incomplete lineage sorting. Amer. Nat. 174, E54–E70. doi: 10.1086/600082
Kashkush, K., Feldman, M., and Levy, A. A. (2002). Gene loss, silencing, and activation in a newly synthesized wheat allotetraploid. Genetics 160, 1651–1659.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, J., and Sanderson, M. J. (2008). Penalized likelihood phylogenetic inference: bridging the parsimony-likelihood gap. Syst. Biol. 57, 665–674. doi: 10.1080/10635150802422274
Knowles, L. L., Huang, H., Sukumaran, J., and Smith, S. A. (2018). A matter of phylogenetic scale: distinguishing incomplete lineage sorting from lateral gene transfer as the cause of gene tree discord in recent versus deep diversification histories. Amer. J. Bot. 105, 376–384. doi: 10.1002/ajb2.1064
Kwembeya, E. G., Bjora, C. S., Stedje, B., and Nordal, I. (2007). Phylogenetic relationships in the genus Crinum (Amaryllidaceae) with emphasis on tropical African species: evidence from trnL-F and nuclear ITS DNA sequence data. Taxon 56, 801–810. doi: 10.2307/25065862
Lara, R. F., and Céspedes, Á (2019). Una especie nueva de Chlidanthus Herbert (Amaryllidaceæ), de Bolivia. Fontqueria 57, 27–32.
Lara, R. F., and Huaylla, H. (2015). El género Hieronymiella Pax (Amaryllidaceæ) en Bolivia. Fontqueria 56, 383–392.
Larridon, I., Villaverde, T., Zuntini, A. R., Pokorny, L., Brewer, G. E., Epitawalage, N., et al. (2020). Tackling rapid radiations with targeted sequencing. Front. Plant Sci. 10:1655. doi: 10.3389/fpls.2019.01655
Lavin, M., Pennington, R. T., Hughes, C. E., Lewis, G. P., Delgado-Salinas, A., Duno de Stefano, R., et al. (2018). DNA sequence variation among conspecific accessions of the legume Coursetia caribaea reveals geographically localized clades here ranked as species. Syst. Bot. 43, 664–675. doi: 10.1600/036364418X697382
Leal-Pinedo, J. M., and Linares-Palomino, R. (2005). Los bosques secos de la reserva de biosfera del noroeste (Perú): diversidad arbórea y estado de conservación. Caldasia 27, 195–211.
Lee-Yaw, J. A., Grassa, C. J., Joly, S., Andrew, R. L., and Rieseberg, L. H. (2019). An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 221, 515–526. doi: 10.1111/nph.15386
Lemmon, A. R., Emme, S. A., and Lemmon, E. M. (2012). Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61, 727–744. doi: 10.1093/sysbio/sys049
Lemmon, E. M., and Lemmon, A. R. (2013). High-throughput genomic data in systematics and phylogenetics. Annu. Rev. Ecol. Evol. Syst. 44, 99–121.
León, B., Sagástegui, A., Sánchez, I., Zapata, M., and Meerow, A. (2013). Amaryllidaceae endémicas del Perú. Rev. Peruana Biol. 13, 690–697.
Levin, D. (1983). Polyploidy and novelty in flowering plants. Amer. Nat. 122, 1–25. doi: 10.1086/284115
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Linares-Palomino, R. (2006). “Phytogeography and floristics of seasonally dry tropical forests in Peru in Neotropical Savannas and Seasonally Dry Forests,” in Neotropical savannas and Seasonally Dry Forests: Plant Diversity, Biogeography, and Conservation, eds R. T. Pennington and J. A. Ratter (Boca Raton: CRC press), 257–279.
Loaiza, C. R. (2013). The Tumbesian center of endemism: biogeography, diversity and conservation. Biogeogr. Bull. SEBA 6, 4–10.
Luebert, F., and Weigend, M. (2014). Phylogenetic insights into Andean plant diversification. Front. Ecol. Evol. 2:27. doi: 10.3389/fevo.2014.00027
Lundberg, J. G., Marshall, L. G., Guerrero, J., Horton, B., Malabarba, M. C. S. L., and Wesselingh, F. (1998). “The stage for Neotropical fish diversification, a history of tropical South American rivers,” in Phylogeny and Classification of Neotropical Fishes, eds L. R. Malabarba, R. E. Reis, R. P. Vari, Z. M. Lucena, and C. A. S. Lucena (Porto Alegre: Edipucrs), 13–48.
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473.
Macbride, J. (1936). Amaryllidaceae Lind. in J.F.Macbride, ed., Flora of Peru. Publ. Field Mus. Nat. Hist., Bot. Ser 13, 631–690.
Maddison, W. P., and Knowles, L. L. (2006). Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55, 21–30. doi: 10.1080/10635150500354928
Matzke, N. J. (2012). Founder-event speciation in BioGeoBEARS package dramatically improves likelihoods and alters parameter inference in Dispersal–Extinction–Cladogenesis (DEC) analyses. Front. Biogeogr. 4:210.
Matzke, N. J. (2013). BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R package, version 0.2.1. Available online at: http://CRAN.R-project.org/package=BioGeoBEARS (accessed July 27, 2013).
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. doi: 10.1093/sysbio/syu056
Meerow, A. (1984). Two new species of pancratioid Amaryllidaceae from Peru and Ecuador. Brittonia 36, 18–25. doi: 10.2307/2806286
Meerow, A. W. (1987a). A Monograph of Eucrosia (Amaryllidaceae). Syst. Bot. 12, 460–492. doi: 10.2307/2418883
Meerow, A. W. (1987c). Chromosome cytology of Eucharis, Caliphruria, and Urceolina (Amaryllidaceae). Amer. J. Bot. 74, 1560–1576. doi: 10.2307/2444050
Meerow, A. W. (1987d). The identities and systematic relationships of Mathieua Klotzsch and Plagiolirion Baker (Amaryllidaceae). Taxon 36, 566–572. doi: 10.2307/1221847
Meerow, A. W. (1989). Systematics of the Amazon Lilies, Eucharis and Caliphruria (Amaryllidaceae). Ann. Missouri Bot. Gard. 76, 136–220. doi: 10.2307/2399347
Meerow, A. W. (1990). “202. Amaryllidaceae,” in Flora of Ecuador 41, eds G. Harling and L. Andersson (Sweden: University of Gothenburg), 52.
Meerow, A. W. (2010). “Convergence or reticulation? Mosaic evolution in the canalized American Amaryllidaceae,” in Diversity, Phylogeny and Evolution in the Monocotyledons, ed. O. Seberg (Aarhus: Aarhus University Press), 145–168.
Meerow, A. W., and Cano, A. (2019). Taxonomic novelties in Amaryllidaceae from the Department of Ancash, Peru, and a new combination in Clinanthus. PhytoKeys 131:115.
Meerow, A. W., and Dehgan, B. (1985). A new species and a new combination in the genus Eucrosia (Amaryllidaceae). Brittonia 37, 47–55. doi: 10.2307/2806243
Meerow, A. W., Dehgan, N. B., and Dehgan, B. (1986). Pollen tetrads in Stenomesson elwesii (Amaryllidaceae). Amer. J. Bot. 73, 1642–1644. doi: 10.2307/2443932
Meerow, A. W., Fay, M. F., Guy, C. L., Li, Q. B., Zaman, F. Q., and Chase, M. W. (1999). Systematics of Amaryllidaceae based on cladistic analysis of plastid rbcL and trnL-F sequence data. Amer. J. Bot. 86, 1325–1345. doi: 10.2307/2656780
Meerow, A. W., Francisco-Ortega, J., Kuhn, D. N., and Schnell, R. J. (2006). Phylogenetic relationships and biogeography within the Eurasian clade of Amaryllidaceae based on plastid ndhF and nrDNA ITS sequences: lineage sorting in a reticulate area? Syst. Bot. 31, 42–60.
Meerow, A. W., Guy, C. L., Li, Q. B., and Clayton, J. R. (2002). Phylogeny of the tribe Hymenocallideae (Amaryllidaceae) based on morphology and molecular characters. Ann. Missouri Bot. Gard. 89, 400–413. doi: 10.2307/3298600
Meerow, A. W., Guy, C. L., Li, Q. B., and Yang, S. L. (2000). Phylogeny of the American Amaryllidaceae based on nrDNA ITS sequences. Syst. Bot. 25, 708–726. doi: 10.2307/2666729
Meerow, A. W., Lehmiller, D. J., and Clayton, J. R. (2003). Phylogeny and biogeography of Crinum L. (Amaryllidaceae) inferred from nuclear and limited plastid non-coding DNA sequences. Bot. J. Linn. Soc. 141, 349–363. doi: 10.1046/j.1095-8339.2003.00142.x
Meerow, A. W., and Nakamura, K. (2019). Two new species of Peruvian Amaryllidaceae, an expanded concept of the genus Paramongaia, and taxonomic notes in Stenomesson. Phytotaxa 416, 184–196.
Meerow, A. W., Noblick, L., Borrone, J. W., Couvreur, T. L., Mauro-Herrera, M., Hahn, W. J., et al. (2009). Phylogenetic analysis of seven WRKY genes across the palm subtribe Attaleinae (Arecaceae) identifies Syagrus as sister group of the coconut. PLoS One 4:e7353. doi: 10.1371/journal.pone.0007353
Meerow, A. W., Noblick, L., Salas-Leiva, D. E., Sanchez, V., Francisco-Ortega, J., Jestrow, B., et al. (2015). Phylogeny and historical biogeography of the cocosoid palms (Arecaceae, Arecoideae, Cocoseae) inferred from sequences of six WRKY gene family loci. Cladistics 31, 509–534. doi: 10.1111/cla.12100
Meerow, A. W., and Sagástegui, A. (1997). A new species of Eucrosia (Amaryllidaceae) from northern Peru. SIDA 17, 761–764.
Meerow, A. W., and Silverstone-Sopkin, P. (1995). The rediscovery of Plagiolirion horsmannii Baker (Amaryllidaceae). Brittonia 47, 426–431. doi: 10.2307/2807573
Meerow, A. W., Silverstone-Sopkin, P. A., Zuluaga-Tróchez, A., and Sánchez-Taborda, J. A. (2019). A remarkable new species of Pamianthe (Amaryllidaceae) from the Department of Cauca. Colombia. Phytokeys 115:73. doi: 10.3897/phytokeys.115.30755
Meerow, A. W., and Snijman, D. (1998). “Amaryllidaceae,” in Flowering Plants: Monocotyledons, ed. K. Kubitzki (Berlin: Springer), 83–110.
Meerow, A. W., and Snijman, D. A. (2005). The never-ending story: multigene approaches to the phylogeny of Amaryllidaceae, and assessing its familial limits. Aliso 22, 353–364. doi: 10.5642/aliso.20062201.29
Meerow, A. W., and van der Werff, H. (2004). Pucara (Amaryllidaceae) reduced to synonymy with Stenomesson on the basis of nuclear and plastid DNA spacer sequences, and a new related species of Stenomesson. Syst. Bot. 29, 511–517. doi: 10.1600/0363644041744400
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proc. Gateway Computing Environm. Workshop (GCE), 14 Nov. 2010 (New Orleans), 1–8.
Molau, U. (1988). Scrophulariaceae I. Calceolarieae. Fl. Neotropica 47. Bronx: New York Botanical Garden.
Morales-Briones, D. F., Liston, A., and Tank, D. C. (2018). Phylogenomic analyses reveal a deep history of hybridization and polyploidy in the Neotropical genus Lachemilla (Rosaceae). New Phytol. 218, 1668–1684. doi: 10.1111/nph.15099
Morrone, J. J. (2014). Biogeographical regionalisation of the Neotropical region. Zootaxa 3782, 1–110. doi: 10.11646/zootaxa.3782.1.1
Murphy, B., Forest, F., Barraclough, T., Rosindell, J., Bellot, S., Cowan, R., et al. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Mol. Phylogen. Evol. 144:106668. doi: 10.1016/j.ympev.2019.106668
Mutke, J., Böhnert, T., and Weigend, M. (2017). Save last cloud forests in western Andes. Nature 541:157. doi: 10.1038/541157e
Mutke, J., Jacobs, R., Meyers, K., Henning, T., and Weigend, M. (2014). Diversity patterns of selected Andean plant groups correspond to topography and habitat dynamics, not orogeny. Front. Genet. 5:351. doi: 10.3389/fgene.2014.00351
Oleas, N. H., Meerow, A. W., and Francisco-Ortega, J. (2012). Population dynamics of the endangered plant. Phaedranassa tunguraguae, from the Tropical Andean Hotspot. J. Hered. 103, 557–569. doi: 10.1093/jhered/ess020
Oleas, N. H., Meerow, A. W., and Francisco-Ortega, J. (2013). Molecular markers and conservation of plant species in the Latin America: the case of Phaedranassa viridiflora (Amaryllidaceae). Bot. Rev. 79, 507–527. doi: 10.1007/s12229-013-9125-8
Oleas, N. H., Meerow, A. W., and Francisco-Ortega, J. (2016). Genetic structure of the threatened Phaedranassa schizantha (Amaryllidaceae). Bot. J. Linn. Soc. 182, 169–179. doi: 10.1111/boj.12444
Paradis, E. (2013). Molecular dating of phylogenies by likelihood methods: a comparison of models and a new information criterion. Mol. Phylogenet. Evol. 67, 436–444. doi: 10.1016/j.ympev.2013.02.008
Paradis, E., and Schliep, K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Pennington, R. T., Lavin, M., and Oliveira-Filho, A. (2009). Woody plant diversity, evolution, and ecology in the tropics: perspectives from seasonally dry tropical forests. Ann. Rev. Ecol., Evol. Syst. 40, 437–457. doi: 10.1146/annurev.ecolsys.110308.120327
Pennington, R. T., Lavin, M., Prado, D. E., Pendry, C. A., Pell, S. K., and Butterworth, C. A. (2004). Historical climate change and speciation: neotropical seasonally dry forest plants show patterns of both Tertiary and Quaternary diversification. Phil. Trans. Roy. Soc. Lond. Ser. B: Biol. Sci. 359, 515–538. doi: 10.1098/rstb.2003.1435
Pennington, R. T., Lavin, M., Särkinen, T., Lewis, G. P., Klitgaard, B. B., and Hughes, C. E. (2010). Contrasting plant diversification histories within the Andean biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 107, 13783–13787. doi: 10.1073/pnas.1001317107
Picard, D., Sempere, T., and Plantard, O. (2008). Direction and timing of uplift propagation in the Peruvian Andes deduced from molecular phylogenetics of highland biotaxa. EPSL 271, 326–336. doi: 10.1016/j.epsl.2008.04.024
Plummer, M., Best, N., Cowles, K., and Vines, K. (2006). CODA: convergence diagnosis and output analysis for MCMC. R News 6, 7–11.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Quintana, C., Girardello, M., Barfod, A. S., and Balslev, H. (2017a). Diversity patterns, environmental drivers and changes in vegetation composition in dry inter-Andean valleys. J. Plant Ecol. 10, 461–475. doi: 10.1093/jpe/rtw036
Quintana, C., Pennington, R. T., Ulloa, C. U., and Balslev, H. (2017b). Biogeographic barriers in the Andes: is the Amotope—Huancabamba Zone a dispersal barrier for dry forest plants. Ann. Missouri Bot. Gard. 102, 542–550. doi: 10.3417/D-17-00003A
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rabosky, D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9:e89543. doi: 10.1371/journal.pone.0089543
Rabosky, D. L., Donnellan, S. C., Grundler, M., and Lovette, I. J. (2014a). Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst. Biol. 63, 610–627. doi: 10.1093/sysbio/syu025
Rabosky, D. L., Grundler, M., Anderson, C., Title, P., Shi, J. J., Brown, J. W., et al. (2014b). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707. doi: 10.1111/2041-210X.12199
Rabosky, D. L., Santini, F., Eastman, J., Smith, S. A., Sidlauskas, B., Chang, J., et al. (2013). Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4:1958. doi: 10.1038/ncomms2958
Rambaut, A. (2019). FigTree Version 1.44. Avaiable at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed October 22, 2020).
Ricketts, T. H., Dinerstein, E., Olson, D. M., Eichbaum, W., Loucks, C. J., Kavanaugh, K., et al. (1999). Terrestrial Ecoregions of North America: A Conservation Assessment. Washington DC: Island Press.
Rieseberg, L. H., and Soltis, D. (1991). Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants 5, 65–84.
Roch, S., and Warnow, T. (2015). On the robustness to gene tree estimation error (or lack thereof) of coalescent-based species tree methods. Syst. Biol. 64, 663–676. doi: 10.1093/sysbio/syv016
Roncal, J., Kahn, F., Millan, B., Couvreur, T. L. P., and Pintaud, J. C. (2013). Cenozoic colonization and diversification patterns of tropical American palms: evidence from Astrocaryum (Arecaceae). Bot. J. Linn. Soc. 171, 120–139. doi: 10.1111/j.1095-8339.2012.01297.x
Rønsted, N., Symonds, M. R., Birkholm, T., Christensen, S. B., Meerow, A. W., Molander, M., et al. (2012). Can phylogeny predict chemical diversity and potential medicinal activity of plants? A case study of Amaryllidaceae. BMC Evol. Biol. 12:182. doi: 10.1186/1471-2148-12-182
Salas-Leiva, D. E., Meerow, A. W., Calonje, M., Griffith, M. P., Francisco-Ortega, J., Nakamura, K., et al. (2013). Phylogeny of the cycads based on multiple single-copy nuclear genes: congruence of concatenated parsimony, likelihood and species tree inference methods. Ann. Bot. 112, 1263–1278.
Sanderson, M. J. (2002). Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109. doi: 10.1093/oxfordjournals.molbev.a003974
Santos, C., Jaramillo, C., Bayona, G., Rueda, M., and Torres, V. (2008). Late Eocene marine incursion in north-western South America. Palaeogeogr. Palaeocl. Paleoecol. 264, 140–146. doi: 10.1016/j.palaeo.2008.04.010
Santos-Gally, R., Vargas, P., and Arroyo, J. (2012). Insights into Neogene Mediterranean biogeography based on phylogenetic relationships of mountain and lowland lineages of Narcissus (Amaryllidaceae). J. Biogeog. 39, 782–798. doi: 10.1111/j.1365-2699.2011.02526.x
Särkinen, T., Pennington, R. T., Lavin, M., Simon, M. F., and Hughes, C. E. (2012). Evolutionary islands in the Andes: persistence and isolation explain high endemism in Andean dry tropical forests. J. Biogeogr. 39, 884–900. doi: 10.1111/j.1365-2699.2011.02644.x
Satô, D. (1938). Karyotype alteration and phylogeny. IV. Karyotypes in Amaryllidaceae with special reference to the SAT-chromosome. Cytologia 9, 203–242.
Schliep, K. P. (2011). Phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Seoighe, C., and Gehring, C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20, 461–464. doi: 10.1016/j.tig.2004.07.008
Simpson, B. B. (1975). Pleistocene changes in the flora of the high tropical Andes. Paleobiology 1, 273–294. doi: 10.1017/S0094837300002530
Simpson, B. B. (1983). An historical phytogeography of the high Andean flora. Rev. Chil. Hist. Nat. 56, 109–122.
Sloan, D. B., Havird, J. C., and Sharbrough, J. (2017). The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 26, 2212–2236. doi: 10.1111/mec.13959
Smith, G. L., and Garland, M. A. (2003). Nomenclature of Hymenocallis taxa (Amaryllidaceae) in southeastern United States. Taxon 52, 805–817. doi: 10.2307/3647354
Smith, G. L., and Garland, M. A. (2009). A new species of Hymenocallis (Amaryllidaceae) from the Apalachicola Forest of the Florida Panhandle. U.S.A. Novon 19, 234–238. doi: 10.3417/2007121
Snoad, B. (1952). Chromosome counts of species and varieties of garden plants. Ann. Rept. John Innes Hortic. Inst. 42:47.
Solís-Lemus, C., and Ané, C. (2016). Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting. PLoS Genet. 12:e1005896. doi: 10.1371/journal.pgen.1005896
Soltis, P. S., Marchant, D. B., Van de Peer, Y., and Soltis, D. E. (2015). Polyploidy and genome evolution in plants. Curr. Opin. Genet. Develop. 35, 119–125. doi: 10.1016/j.gde.2015.11.003
Sparre, B., and Andersson, L. (1991). A taxonomic revision of the Tropaeolaceae. Opera Bot. 108, 1–139.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steinmann, M., Hungerbühler, D., Seward, D., and Winkler, W. (1999). Neogene tectonic evolution and exhumation of the southern Ecuadorian Andes; a combined stratigraphy and fission-track approach. Tectonophysics 307, 255–276. doi: 10.1016/S0040-1951(99)00100-6
Stephens, J. D., Rogers, W. L., Heyduk, K., Cruse-Sanders, J. M., Determann, R. O., Glenn, T. C., et al. (2015). Resolving phylogenetic relationships of the recently radiated carnivorous plant genus Sarracenia using target enrichment. Mol. Phylogenet. Evol. 85, 76–87. doi: 10.1016/j.ympev.2015.01.015
Stewart, C. S. (1832). A Visit to the South Seas, in the US Ship Vincennes, During the Years 1829 and 1830: With Notices of Brazil, Peru, Manilla, the Cape of Good Hope, and St. Helena. London: Fisher, Son, & Jackson.
Stolzer, M., Lai, H., Xu, M. L., Sathaye, D., Vernot, B., and Durand, D. (2012). Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28, I409–I415. doi: 10.1093/bioinformatics/bts386
Sukumaran, J., and Holder, M. T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. doi: 10.1093/bioinformatics/btq228
Tiffney, B. H. (1985a). Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arbor. 66, 73–94.
Tiffney, B. H. (1985b). The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J. Arnold Arbor. 66, 243–273.
Tiffney, B. H., and Manchester, S. R. (2001). The use of geological and paleontological evidence in evaluating plant phylogenetic hypotheses in the Northern Hemisphere Tertiary. Int. J. Plant Sci. 162, S3–S17.
Traub, H. P. (1962). Key to the subgenera, alliances and species of Hymenocallis. PI. Life 18, 55–72.
Vargas, C. (1962). Phytomorphic representations of the ancient Peruvians. Econ. Bot. 16, 106–115. doi: 10.1007/BF02985298
Wanke, S., Mendoza, C. G., Müller, S., Guillén, A. P., Neinhuis, C., Lemmon, A. R., et al. (2017). Recalcitrant deep and shallow nodes in Aristolochia (Aristolochiaceae) illuminated using anchored hybrid enrichment. Mol. Phylogenet. Evol. 117, 111–123. doi: 10.1016/j.ympev.2017.05.014
Waters, M. T., Tiley, A. M. M., Kramer, E. M., Meerow, A. W., Langdale, J. A., and Scotland, R. W. (2013). The corona of the daffodil Narcissus bulbocodium shares stamen-like identity and is distinct from the orthodox floral whorls. Plant J. 74, 615–625. doi: 10.1111/tpj.12150
Weigend, M. (2002). Observations on the biogeography of the Amotope-Huancabamba zone in northern Peru. Bot. Rev. 68, 38–54.
Weigend, M. (2004). Additional observations on the biogeography of the Amotope-Huancabamba zone in Northern Peru: defining the South-Eastern limits. Rev. Peruana Biol. 11, 127–134.
Weitemier, K., Straub, S. C. K., Cronn, R. C., Fishbein, M., Schmickl, R., McDonnell, A., et al. (2014). Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2:1400042. doi: 10.3732/apps.1400042
Wesselingh, F. P. (2006). Miocene long-lived Lake Pebas as a stage of mollusc radiations, with implications for landscape evolution in western Amazonia. Scripta Geol. 133, 1–17.
Wesselingh, F. P., Räsänen, M. E., Irion, G., Vonhof, H. B., Kaandorp, R., Renema, W., et al. (2002). Lake Pebas, a palaeoecological reconstruction of a Miocene, long-lived lake complex in western Amazonia. Cainozoic Res. 1, 35–81.
Wesselingh, F. P., and Salo, J. A. (2006). A Miocene perspective on the evolution of the Amazonian biota. Scripta Geol. 133, 439–458.
Whitehead, M. R., and Brown, C. A. (1940). The seed of the spider lily. Hymenocallis occidentalis. Amer. J. Bot. 27, 199–203. doi: 10.2307/2436880
Wolfe, J. A. (1975). Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Ann. Missouri Bot. Gard. 62, 264–279. doi: 10.2307/2395198
Yang, Y., and Smith, S. A. (2014). Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol. Biol. Evol. 31, 3081–3092. doi: 10.1093/molbev/msu245
Yu, Y., Blair, C., and He, X. (2015). RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 37, 604–606. doi: 10.1093/molbev/msz257
Keywords: anchored hybrid enrichment, Andes, Asparagales, biogeography, geophyte, molecular systematics, monocotyledons, phylogenetics
Citation: Meerow AW, Gardner EM and Nakamura K (2020) Phylogenomics of the Andean Tetraploid Clade of the American Amaryllidaceae (Subfamily Amaryllidoideae): Unlocking a Polyploid Generic Radiation Abetted by Continental Geodynamics. Front. Plant Sci. 11:582422. doi: 10.3389/fpls.2020.582422
Received: 11 July 2020; Accepted: 12 October 2020;
Published: 05 November 2020.
Edited by:
Nina Rønsted, National Tropical Botanical Garden, United StatesReviewed by:
Christine D. Bacon, University of Gothenburg, SwedenJuan José Morrone, National Autonomous University of Mexico, Mexico
Maximilian Weigend, University of Bonn, Germany
Paola de Lima Ferreira, University of Gothenburg, Gothenburg, Sweden in collaboration with reviewer CB.
Copyright © 2020 Meerow, Gardner and Nakamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan W. Meerow, YW1lZXJvd0Bhc3UuZWR1; Z3JpZmZpbmlhQGdtYWlsLmNvbQ==
†Present address: Alan W. Meerow, School of Life Sciences, Arizona State University, Tempe, AZ, United States; Montgomery Botanical Center, Coral Gables, FL, United States
 Alan W. Meerow
Alan W. Meerow Elliot M. Gardner
Elliot M. Gardner Kyoko Nakamura
Kyoko Nakamura