- 1Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China
- 2College of Life Sciences, University of Chinese Academy of Science, Beijing, China
- 3Guangdong Provincial Key Laboratory of Digital Botanical Garden, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China
- 4School of Integrative Plant Science, Section of Plant Biology and the L.H. Bailey Hortorium, Cornell University, Ithaca, NY, United States
- 5Center of Conservation Biology/Economic Botany/Plant Ecology, Core Botanical Gardens, Chinese Academy of Sciences, Guangzhou, China
The asymmetric flower, lacking any plane of symmetry, is rare among angiosperms. Canna indica L. has conspicuously asymmetric flowers resulting from the presence of a half-fertile stamen, while the other androecial members develop as petaloid staminodes or abort early during development. The molecular basis of the asymmetric distribution of fertility and petaloidy in the androecial whorls remains unknown. Ontogenetic studies have shown that Canna flowers are borne on monochasial (cincinnus) partial florescences within a racemose inflorescence, with floral asymmetry likely corresponding to the inflorescence architecture. Given the hypothesized role of CYC/TB1 genes in establishing floral symmetry in response to the influence of the underlying inflorescence architecture, the spatiotemporal expression patterns of three Canna CYC/TB1 homologs (CiTBL1a, CiTBL1b-1, and CiTBL1b-2) were analyzed during inflorescence and floral development using RNA in situ hybridization and qRT-PCR. In the young inflorescence, both CiTBL1a and CiTBL1b-1 were found to be expressed in the bracts and at the base of the lateral florescence branches, whereas transcripts of CiTBL1b-2 were mainly detected in flower primordia and inflorescence primordia. During early flower development, expression of CiTBL1a and CiTBL1b-1 were both restricted to the developing sepals and petals. In later flower development, expression of CiTBL1a was reduced to a very low level while CiTBL1b-1 was detected with extremely high expression levels in the petaloid androecial structures including the petaloid staminodes, the labellum, and the petaloid appendage of the fertile stamen. In contrast, expression of CiTBL1b-2 was strongest in the fertile stamen throughout flower development, from early initiation of the stamen primordium to maturity of the ½ anther. Heterologous overexpression of CiTBL genes in Arabidopsis led to dwarf plants with smaller petals and fewer stamens, and altered the symmetry of mature flowers. These data provide evidence for the involvement of CYC/TB1 homologs in the development of the asymmetric Cannaceae flower.
Introduction
Floral symmetry, one of the major features defining the shape of a flower, has been a special focus of botanists for many years. According to the number of symmetry planes, three main types of floral symmetry are recognized: actinomorphy (with several symmetry planes; i.e., polysymmetry or radial symmetry), zygomorphy (with one symmetry plane; i.e., monosymmetry or bilateral symmetry), and asymmetry (no symmetry plane; Endress, 1999; Citerne et al., 2010; Hileman, 2014). Actinomorphy is considered as the ancestral state of flowering plants and it appears in many clades, especially the early angiosperm lineages (Sauquet et al., 2017). Zygomorphic flowers evolved relatively late in the fossil record (Turonian, Upper Cretaceous, ca. 90 mya), and zygomorphy is considered a derived trait relative to actinomorphy (Gandolfo et al., 1998). Evolution of floral zygomorphy is thought to facilitate plant–pollinator interactions by enhancing pollination specificity and success (Neal et al., 1998). Zygomorphy is dominant in some species-rich lineages such as Orchidaceae, Fabaceae, and Lamiales (Endress, 2012). Floral asymmetry, like floral zygomorphy, appears in different forms that are not all homologous and can involve different floral organs and organ whorls. Compared with actinomorphic or zygomorphic flowers, asymmetric flowers are much less common across the angiosperms, and the genetic and molecular mechanisms underlying asymmetry have not been studied in detail as they have for zygomorphy (Endress, 2012).
Cannaceae (Zingiberales) is one of the few families that possesses truly asymmetric flowers. Ten species were recognized in Cannaceae, all of them placed in the single genus Canna (Maas-van de Kamer and Maas, 2008; Prince, 2010). The asymmetry of the Canna flower is mostly associated with an asymmetric distribution and elaboration of petaloidy and organ abortion across the six members of the androecium. Canna flowers consist of two androecial whorls, each with three members (Figure 1A). In the outer whorl, one androecial member develops into a mature petaloid staminode (i.e., infertile stamen) whereas the other two primordia typically abort shortly after initiation (Kirchoff, 1983; Miao et al., 2014). The inner whorl contains two petaloid staminodes plus the fertile stamen, which is in fact a “half-fertile stamen” comprising an anther with a single theca and a petaloid appendage (Kirchoff, 1983; Almeida et al., 2013; Fu et al., 2014; Miao et al., 2014; Tian et al., 2016, 2018). Of the two inner whorl staminodes, one is curved and referred to as the “labellum”. Both the half-fertile stamen and the presence of the labellum contribute to the asymmetry of the androecium. The style is also laminar (Morioka et al., 2015), making the gynoecium appear asymmetric as well. The presence of asymmetry across several floral whorls makes Canna a good taxon to study the genetic mechanisms involved in the development and elaboration of floral asymmetry.
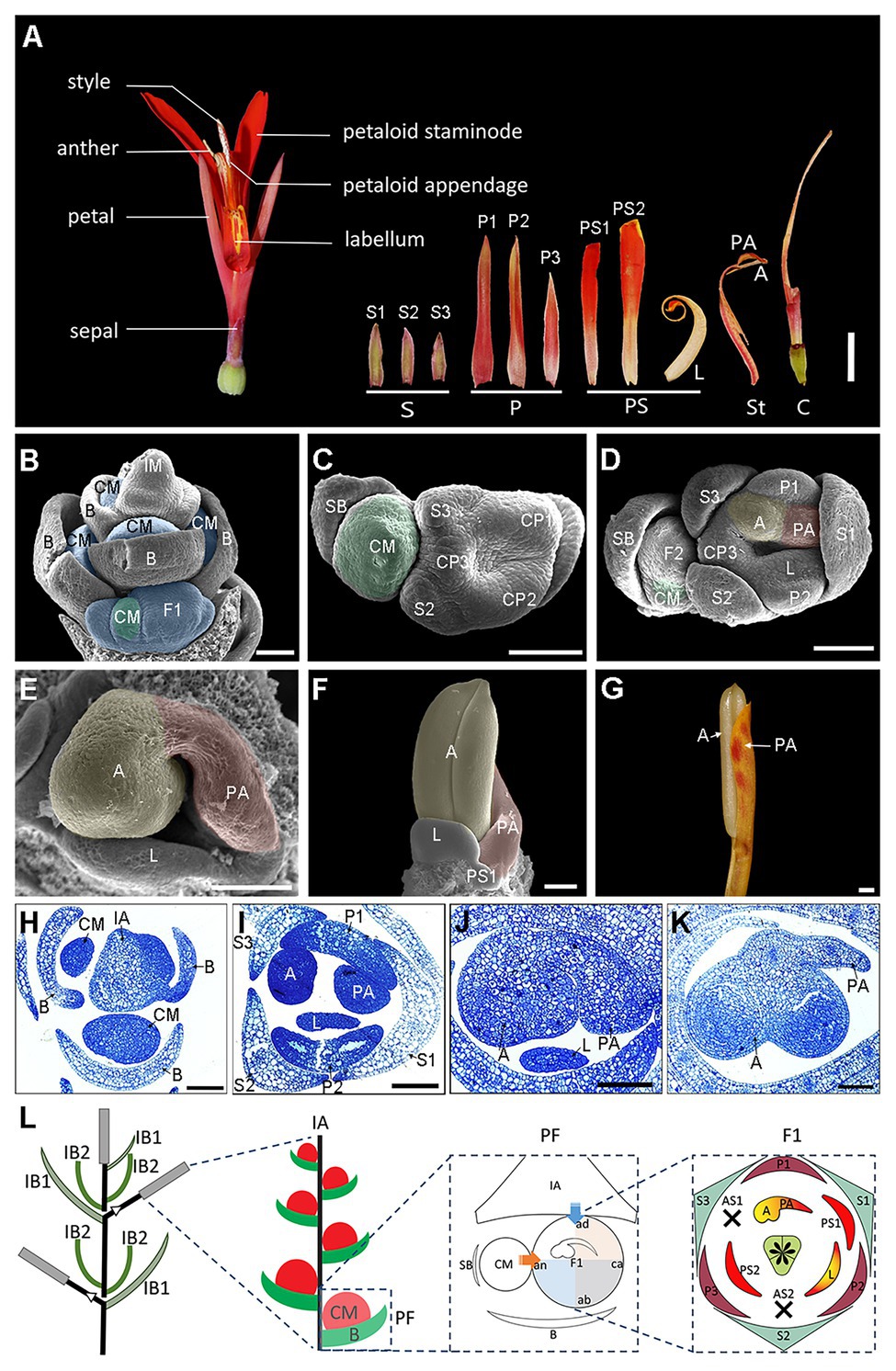
Figure 1. Flower anatomy and ontogeny of Canna indica. (A) Flower dissection showing the floral structure. (B–F) Flower organogenesis observed under a scanning electronic microscopy. (B) Lateral view of a young inflorescence with developing cincinni meristems subtended by the primary bracts. (C) Top view of a cincinni. Three sepal primordia and three petal-stamen common primordia have already formed in the primary flower. (D) Further development of the cincinni. CP1 separates into the first petal and the adaxial stamen, and the later one develops into an anther primordium in the left side and a petaloid appendage primordium in the right side. CP2 separates into the second petal and the labellum primordium. The second floral meristem has formed, and the cincinnus meristem moves to the abaxial side of the secondary flower. (E) Close-up of the asymmetric pattern of the adaxial stamen in the primary flower. The anther primordium is growing quickly while the petaloid appendage slows down its growing. (F) Front view of a fully developed anther. The petaloid appendage continues to develop in a petaloid way together with other petaloid staminodes (PS2 is not shown in this figure). (G) A fertile stamen before mature. (H–K) Transverse semithin sections stained with toluidine blue representing the same developmental stages with (B,D,E,F), respectively. (L) Illustrated inflorescence and floral diagram. The arrows and shades in blue and orange represent the presumed impact on the primary flower by the main florescence axis and the cincinnus meristem, respectively. A, anther; ab, abaxial; ad, adaxial; an, anodic; AS, aborted staminode; B, primary bract; C, carpel; ca, cathodic; CM, cincinni meristem; CP, petal-stamen common primordium; FM, flower meristem; IA, florescence axis; IB1, the bract enclosing the primary rachis of the inflorescence; IB2, the bract which are placed at the base of the new rachis at each branching point, enclosing the new rachis. IM, inflorescence meristem; L, labellum; P, petal; PA, petaloid appendage; PF, partial florescence; PS, petaloid staminode; S, sepal; SB, secondary bract. Blue shades in (B) indicate partial florescences enclosed by the primary bracts. Green shades in (B–D) indicate cincinnus meristems. Yellow and red shades in (D–F) indicate the anther primordium and the petaloid appendage primordium, respectively. Bars: (A) = 1 cm; (B–K) = 100 μm.
CYCLOIDEA/TEOSINTE BRANCHED1 (CYC/TB1)-like genes, a group of class II TCP transcription factors, are implicated in the evolution of key morphological traits (Cubas et al., 1999a; Howarth and Donoghue, 2006; Martin-Trillo and Cubas, 2010). CYC was first described in the zygomorphic flower of Antirrhinum majus and is now widely accepted as the main floral symmetry gene. Research in Antirrhinum revealed that CYC can respond to a common dorsoventral prepattern of the flowers in the lateral position of a raceme (Clark and Coen, 2002). The CYC gene is expressed in the dorsal region of the zygomorphic floral meristem and functions in stamen abortion and the regulation of petal growth by affecting the rate of cell division (Luo et al., 1996; Gaudin et al., 2000). Numerous studies across the angiosperms revealed that CYC homologs are expressed in the dorsal part of the lateral floral meristem in many of the major clades containing zygomorphic flowers (Martin-Trillo and Cubas, 2010; Hileman, 2014; Spencer and Kim, 2018). However, expression of a CYC/TB1-like gene (at least as one copy) was found on the ventral side of Commelina (Commelinales; Preston and Hileman, 2012), Heliconia, and Costus (Zingiberales; Bartlett and Specht, 2011) flowers, suggesting there may be a divergent mechanism specifying floral symmetry in the commelinid monocots.
Canna flowers are typically situated in an inflorescence that is considered a sympodial thyrse, with monochasial (cymose) terminal units or “partial florescences” subtended by the primary bracts of the racemose main florescence. Our previous observations found that when the partial florescence transformed into a single flower or a dichasial cyme, the flower of that modified florescence develops as zygomorphic due to changes in the degree of stamen petaloidy (Yu et al., in review). Those aberrant zygomorphic flowers suggest that the asymmetry of the normal Canna flower is strongly correlated with inflorescence structure (Yu et al., in review). It is proposed that CYC/TB1-like genes may respond to the asymmetric inflorescence system, resulting in their asymmetric expression in the floral primordium and leading to the asymmetric form of the mature flower.
In this research, we chose C. indica as a system to study the developmental mechanisms of floral asymmetry from both morphogenic and genetic aspects. Morphological and anatomical patterns were observed throughout the establishment of floral asymmetry via SEM and histological sectioning following the ontogeny of individual flowers and their inflorescences. CYC/TB1 homologous genes in C. indica were identified and RNA in situ hybridization and qRT-PCR were carried out to investigate the spatiotemporal expression patterns of gene expression during inflorescence and floral development. Finally, we observed the Arabidopsis overexpression lines for three Canna-specific CYC/TB1 genes (CiTBL1a, CiTBL1b-1, and CiTBL1b-2) to characterize the possible functions of these genes during plant development. Altogether, our work in C. indica is the first attempt to propose a mechanism for the origins of floral asymmetry in this lineage, and results will provide a foundation for studying the genetic basis controlling floral asymmetry across flowering plants.
Materials and Methods
Plant Materials and Growth Conditions
All the plant materials of C. indica used in this study were collected from the Ginger Garden and Aquatic Plants Section of South China Botanical Garden, Guangzhou, China.
Arabidopsis thaliana seeds were sterilized and germinated on MS medium (pH 5.8). The seedlings were transplanted to pots containing the mixture of vermiculite and Pindstrup substrate (Pindstrup, Ryomgaad, Denmark; 1:1), and grown to flowering stage in a growth chamber at 22°C with 16-h light/8-h dark cycles. All Arabidopsis transgenic lines were in the Columbia background, which was also used as the wild-type control.
Scanning Electron Microscopy (SEM)
Inflorescence and floral tissues at different developmental stages fixed with FAA were dissected as necessary to reveal internal floral organs, and then dehydrated in an alcohol series. Specimens were critical point dried with CO2, mounted on stubs, sputter-coated with gold, and observed under a JSM-6360LV scanning electron microscope (JEOL, Tokyo, Japan). Micrographs were adjusted for brightness, contrast, and color balance using Adobe Photoshop CC (Adobe, San Jose, CA, USA).
Histology
Inflorescence and floral tissues were dissected and stored in 2% glutaraldehyde in 0.1 M phosphate buffer at pH 7.2–7.4 overnight at 4°C. The specimens were washed six times in phosphate-buffered saline (PBS) and postfixed in 1% w/v osmium tetroxide for 4 h, and then washed six times in PBS for 2 h. Samples were dehydrated in a graded ethanol series, embedded in Spurr resin, and sectioned at 2 μm with glass knives on a LKB-11800 microtome. Sections were stained with toluidine blue and observed under a Leica DFC550 Microscopy (Wetzlar, Germany).
Gene Retrieval, Protein Sequence Alignment, and Phylogenetic Analysis
The sequences of CYC/TB1 homologous gene were retrieved from the C. indica inflorescence transcriptome we generated before (Tian et al., 2016) by BLAST similarity searches. Sequence information for the four CiTBL genes is available in GenBank under accession numbers MW176099 to MW176102.
To affirm they are in the CYC/TB1 subclade of TCP family, phylogenetic analysis was carried out using MEGA version 6.06 by the full length of protein sequences from GenBank according to Luo et al. (1996, 1999), Doebley et al. (1997), Takeda et al. (2003), Yuan et al. (2009), Martin-Trillo and Cubas (2010), Lin et al. (2016), and Chai et al. (2017). Distance matrices used the PAM-Dayhoff model of amino acid substitution; the phylogenetic tree was constructed with the maximum-likelihood method, and bootstrap analyses used 1,000 resampling replicates. The Zingiberales TBL ML tree was re-conducted according to Bartlett and Specht (2011). Additional sequences of C. glauca were retrieved from the unpublished transcriptome, and sequences of Musa acuminate subsp. malaccensis were retrieved from The Banana Genome Hub (D’Hont et al., 2012).1
Putative protein sequence alignment of CiTBLs and Antirrhinum CYC was conducted by Clustal X2 with default settings.
Quantitative Real-Time RT-PCR
Total RNA was extracted from young flowers of three different stages (1.0, 1.5, and 2.0 cm in length) and seven different organs (petal, petaloid staminode, labellum, anther, petaloid appendage, carpel, and leaf) using RNAprep Pure Kit (Tiangen, Beijing, China). All the floral organs dissected are half the length of the mature organ to make sure they are in rapid growth. About 3 μg total RNA was reversely transcribed to the first strand of the cDNA in a 20-μl volume using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China) with oligo (dT) primers. qRT-PCR was performed using specific primer pairs (Supplementary Table S1) under the manual of LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). Each qPCR reaction (total volume of 20 μl) contains 10 μl of Top Green qPCR SuperMix (TransGen, Beijing, China), 0.2 μl of each primer, 1 μl of 1:5 diluted cDNA, and 8.2 μl ddH2O. Thermal cycling consisted of a hold at 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and 60°C for 30 s. The temperature was then gradually raised by 0.5°C every 10 s for performing melting curve analysis. Three biological replicates were performed with three technical replicates for each sample. The ΔΔCt method was employed with CiActin and CiPP2A (protein phosphatase 2A) as endogenous controls (Livak and Schmittgen, 2001).
RNA in situ Hybridization
To generate specific digoxigenin-labeled riboprobes, nonconservative region of CiTBL1a (291 nt), CiTBL1b-1 (256 nt), and CiTBL1b-2 (244 nt) were amplified with primers listed in Supplementary Table S1 and cloned into pGEM-T vector (Promega Fitchburg, WI, USA) following manufacturer instructions. In vitro transcription was performed using SP6/T7 DIG RNA Labeling Kit (Roche, Basel, Switzerland). Inflorescence and floral tissues were fixed, sectioned, and hybridized to the probes as described previously (Tian et al., 2016, 2018). The hybridized sections were visualized under brightfield illumination with a Leica DFC550 Microscope (Wetzlar, Germany).
Construction of CiTBL Transgenic Plants in Arabidopsis
The ORF of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 were amplified using the primer pairs listed in Supplementary Table S1. The overexpression construct of CiTBL allele, 35S::CiTBL1a, 35S::CiTBL1b-1, and 35S::CiTBL1b-2 was engineered to pCAMBIA1302 using One Step Cloning Kit (Vazyme, Nanjing, China). The sequence confirmed vectors were then transformed into Agrobacterium strain GV3101. The Arabidopsis floral dip transformation protocol was followed by Clough and Bent (1988). The floral dip procedure was repeated once again after a week for obtaining more transformed seeds. Two to five putative transgenic A. thaliana T1 seedlings were selected for each construction by antibiotic hygromycin and PCR confirmed (2 T1 lines for 35S, CiTBL1a, 5 T1 lines for 35S::CiTBL1b-1, and 3 T1 lines for 35S::CiTBL1b-2). Several representative T1 plants with obvious phenotypes were selfed into T2 plants to document the phenotypic effect of each construct. The mRNA expression level of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 in flowers of at least five independent T2 transgenic plants was verified by semi-quantitative RT-PCR. AtActin was chosen as a reference gene and amplified with 30 cycles using primers AtActinF/R (Supplementary Table S1). For each allele construction, 15–20 independent T2 transgenic individuals of overexpressing CiTBL1a, CiTBL1b-1, and CiTBL1b-2, together with T1 plants, were summarized for the phenotypic effects on plant form and floral morphology. The images of petal epidermal cells were taken under a Leica DVM6 Digital Microscope (Wetzlar, Germany). To measure the cell size of the adaxial epidermal petal cells, the middle regions of the petals were zoomed in and the cell numbers in a 100 × 100 μm2 square were counted.
Results
The Establishment of Floral Asymmetry in Canna
The inflorescence of C. indica is a terminal, sympodially branched thyrse, comprising repeated units of florescences. Each florescence is monochasial cyme/cincinnus (Figures 1B,H,L). Bracts (B) on the main florescence axis (IA) subtend a lateral cincinnus meristem (CM), which will ultimately give rise to one or two flowers (Figures 1B,H). In Canna, the secondary bract that subtends a terminal flower is present on the left (anodic, an) side of the primary flower in a polar view. This is the side of the flower that lies in the direction of the rise of the phyllotactic helix which is typically sinistrose, or left-handed, in Canna. The cathodic side (ca) is the side that lies opposite the rise of the phyllotactic helix. As the secondary flower (F2) often ceases development when it is still quite small, only the development of the primary flower (F1) is described for this study. The terms adaxial, abaxial, anodic, and cathodic all refer to the primary flower of a florescence unit that is considered a cincinnus.
In Canna, floral organs of each whorl develop in a clockwise sequence (Kirchoff, 1983; Almeida et al., 2013; Miao et al., 2014). Three sepal primordia initiate sequentially at the three angles of the floral apex. The individual sepal (S1) opposite to the cincinnus meristem arises first and it is larger than the other two at early stages (Figure 1C), with the sepals all becoming almost equal in size at maturity (Figure 1A). Three common stamen-petal primordia (part of the ring primordium) arise after sepal initiation (Figure 1C). The shape of floral apex is approximately zygomorphic, and the symmetry plane passes through the secondary bract and the cincinnus meristem, suggesting the monochasial partial inflorescence structure may affect the shape of the flower at a very early stage. As the common primordia widen, separation occurs between the petals (toward the outside) and inner androecial members (to the inside). The three petals are basally connate and unequal at maturity, with the lateral one remaining smaller than the other two (Figure 1A). The adaxial inner stamen primordium develops to two features – an anther (A) and a petaloid appendage (PA). To better elucidate the asymmetric development of the fertile stamen, we divided the continuous process into three main stages using several landmark events. In stage 1, the anther primordium and the petaloid appendage primordium are equal in size at initiation (Figures 1D,I). In stage 2, the petaloid appendage primordium slows in growth while the anther primordium develops relatively rapidly as a radial structure (Figures 1E,J). The anther is fully formed in stage 3, while the petaloid appendage primordium, which is accompanied by the labellum and other petaloid staminode primordia, continues to develop with a dorsiventrality to become a petaloid structure (Figures 1F,G,K).
In a Canna flower, the half-fertile stamen forms on the adaxial-anodic portion of the floral apex and is adjacent to both the florescence axis and the next older flower in a cincinnus (Figure 1L). The petaloid structures including the inner petaloid staminode, the labellum, and the petaloid appendage are either distal to the florescence axis or the cincinnus meristem, indicating that the fate of the meristem tissue in the androecial whorls is somewhat linked with the flower’s position within the inflorescence (Figure 1L).
CYC/TB1 Homologs in C. indica
Sequences of four putative CYC/TB1 homologs, CiTBL1a (Unigene32176_All), CiTBL1b-1 (Unigene10453_All), CiTBL1b-2 (Unigene5663_All), and CiTBL2 (CL8453.Contig2), were obtained from an existing transcriptome database (Tian et al., 2016). Phylogenetic analysis of protein sequences placed all four CiTBL genes into the CYC/TB1 clade of Class II TCP transcription factors. In this clade, the four CiTBL genes were clustered with Antirrhinum CYC and DICH, maize TB1, and Arabidopsis TCP1, BRC1, BRC2, demonstrating their homology with CYC/TB1-like genes (Figure 2A).
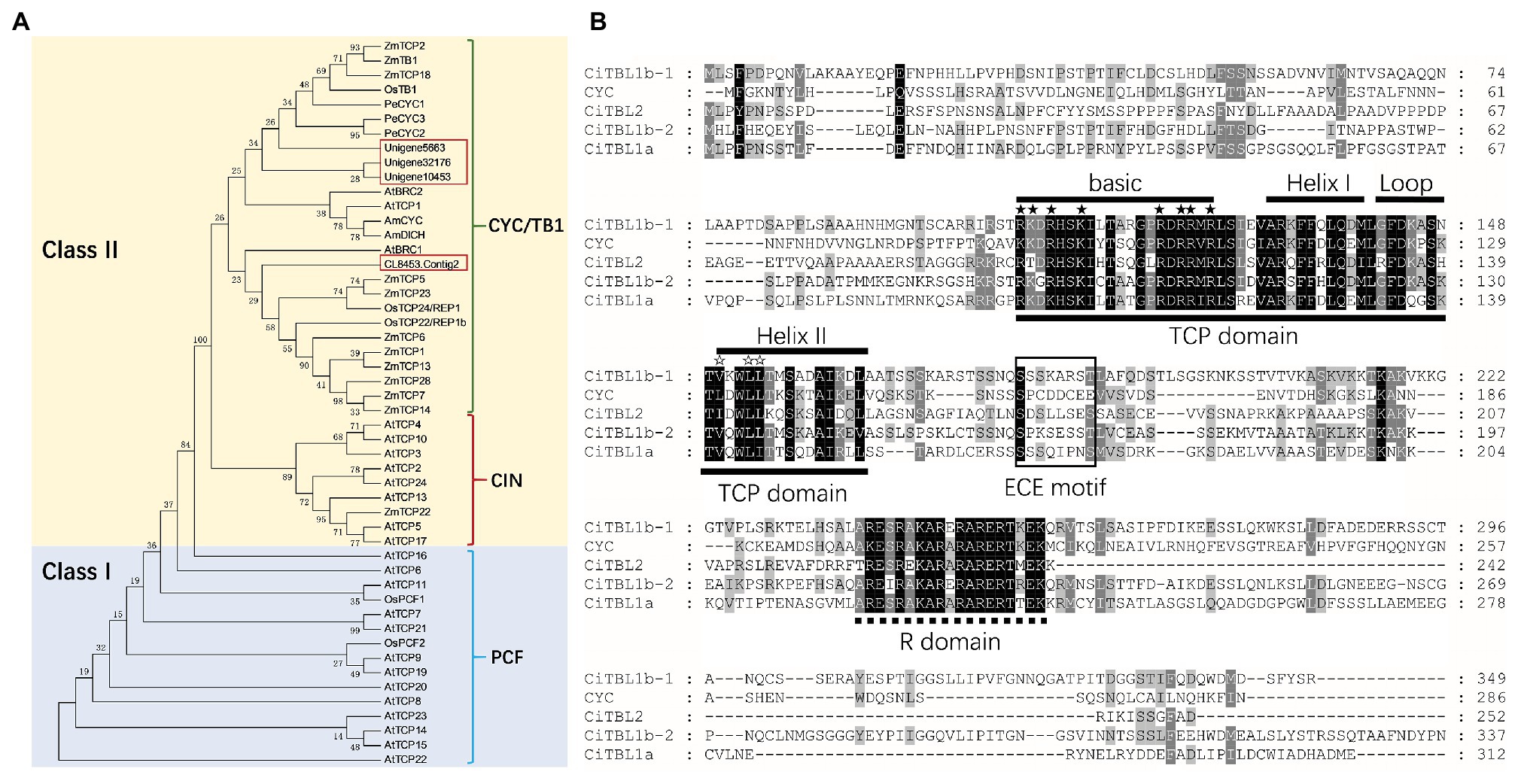
Figure 2. Phylogenetic analysis and protein sequences analysis of CiTBLs. (A) Unrooted phylogram of protein ML analysis with 1,000 bootstraps. All of the AtTCPs and AtBRCs are from Arabidopsis; AmCYC and AmDICH, Antirrhinum; OsTCPs, OsPCFs, and OsTB1, rice; ZmTCPs and ZmTB1, maize; PeCYCs, Phalaenopsis equestris. (B) Multiple alignment of putative protein sequences of CiTBL1a, CiTBL1b-1, CiTBL1b-2, and CYC. TCP domain is underlined with the solid line and R domain is underlined with the dotted line. ★ Indicates the conserved basic residues, and ☆ indicates the LXXLL motif.
To better understand the evolutionary relationships of Zingiberales CYC/TB1-like genes, we combined the recovered Canna sequences with nucleotide sequences derived from a previous publication (Bartlett and Specht, 2011) and an unpublished transcriptome database for C. indica and C. glauca. The results of the phylogenetic analyses of this combined dataset generally conform to previous phylogenetic hypotheses for Zingiberales TB1 homologs, recovering three major clades (ZinTBL1a, ZinTBL1b, and ZinTBL2). In this study, the ZinTBL1b clade was further divided into two subclades: ZinTBL1b-1 and ZinTBL1b-2 (Supplementary Figure S1). Four TBL copies were found in both C. indica and C. glauca belonging to ZinTBL1a, ZinTBL1b-1, ZinTBL1b-2, and ZinTBL2 clades, respectively, suggesting that the gene duplication event leading to these four clades occurred before the divergence of modern species of Cannaceae.
The full-length ORFs of CiTBL1a, CITBL1b-1, CiTBL1b-2, and CiTBL2 are 933, 1,047, 1,131, and 756 bp and encode putative proteins of 311, 349, 337, and 252 amino acids, respectively. Each protein contains a conserved 59-amino acid TCP domain, a 19-amino acid R domain, and a 7-amino acid ECE motif (Figure 2B). Sequence alignment shows the putative proteins of these four CiTBL genes share 34.05, 35.53, 35.96, and 30.87% similarity with Antirrhinum CYC protein, respectively. As to the TCP domain and R domain, CiTBL1a, CiTBL1b-1, CiTBL1b-2, and CiTBL2 share 84.62, 84.62, 83.88, and 79.49% amino acid sequence similarity with Antirrhinum CYC. The ECE motifs and the non-conserved intervening regions of CiTBLs and AmCYC are clearly divergent.
Spatiotemporal Expression Patterns of CiTBL1a, CiTBL1b-1, and CiTBL1b-2
Based on previous RNA-Seq results (Tian et al., 2016), CiTBL2 is minimally expressed in both floral primordia (FP) and differentiated flowers (DF) when compared with the other three CiTBL genes (Supplementary Figure S2). Thus, we focus our study on CiTBL1a, CiTBL1b-1, and CiTBL1b-2. Expression of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 were detected by RNA in situ hybridization (Figure 3) and by qRT-PCR (Figure 4) to observe their overall expression pattern during development and investigate their potential role in establishing asymmetric development of the Canna flower.
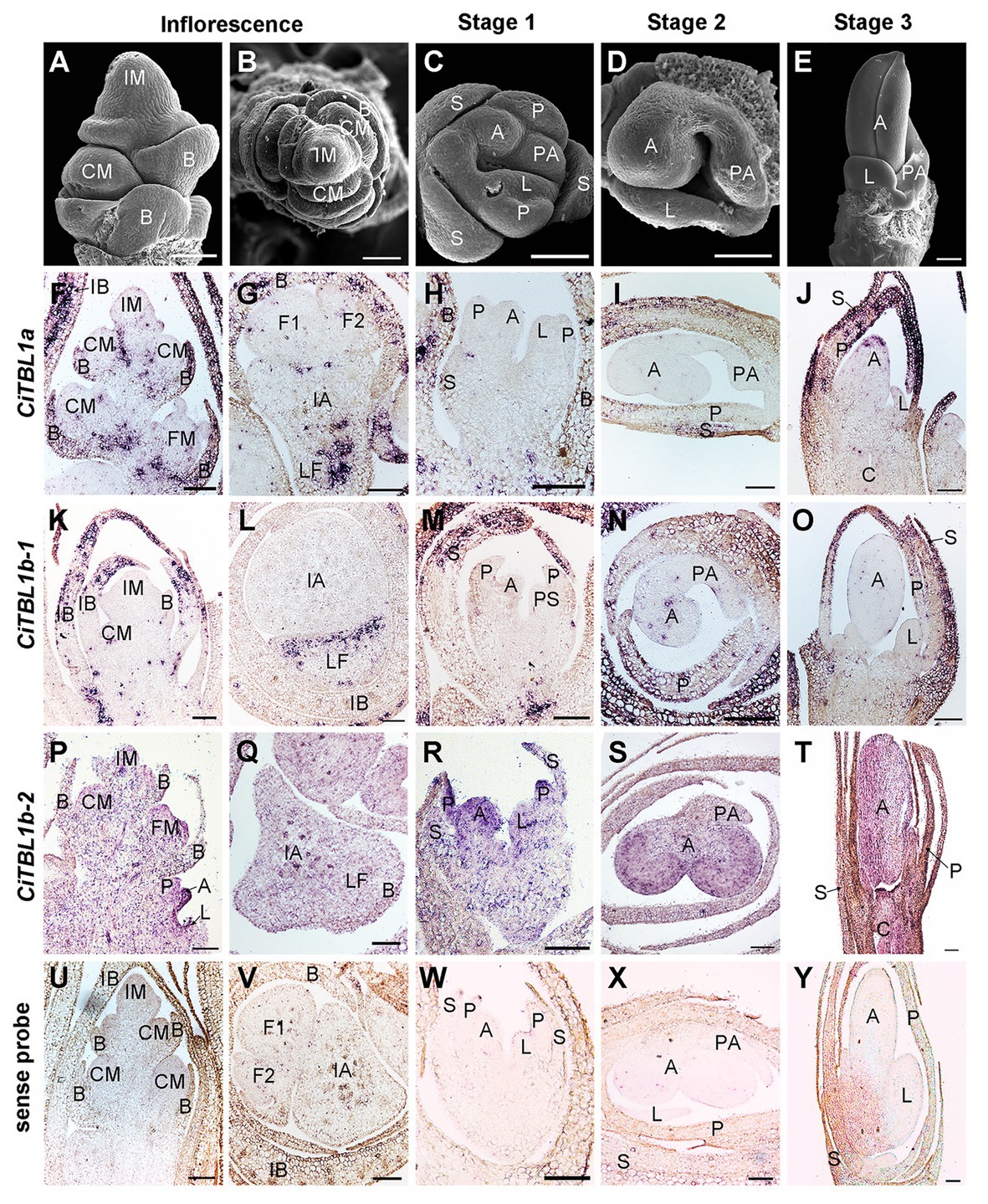
Figure 3. Expression patterns of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 during inflorescence and early flower developmental stages. (A–E) SEM microphotographs of successive developmental stages. (F–T) RNA in situ hybridizations with antisense probes of (F–J) CiTBL1a, (K–O) CiTBL1b-1, and (P–T) CiTBL1b-2. (U–Y) Sections hybridized with CiTBL1b-1 sense probe as negative controls. (G,I,L,Q,S,V,X) were transverse sections, and all the others were longitudinal sections. IB, inflorescence bract; LF, lateral florescence. Bars = 100 μm.
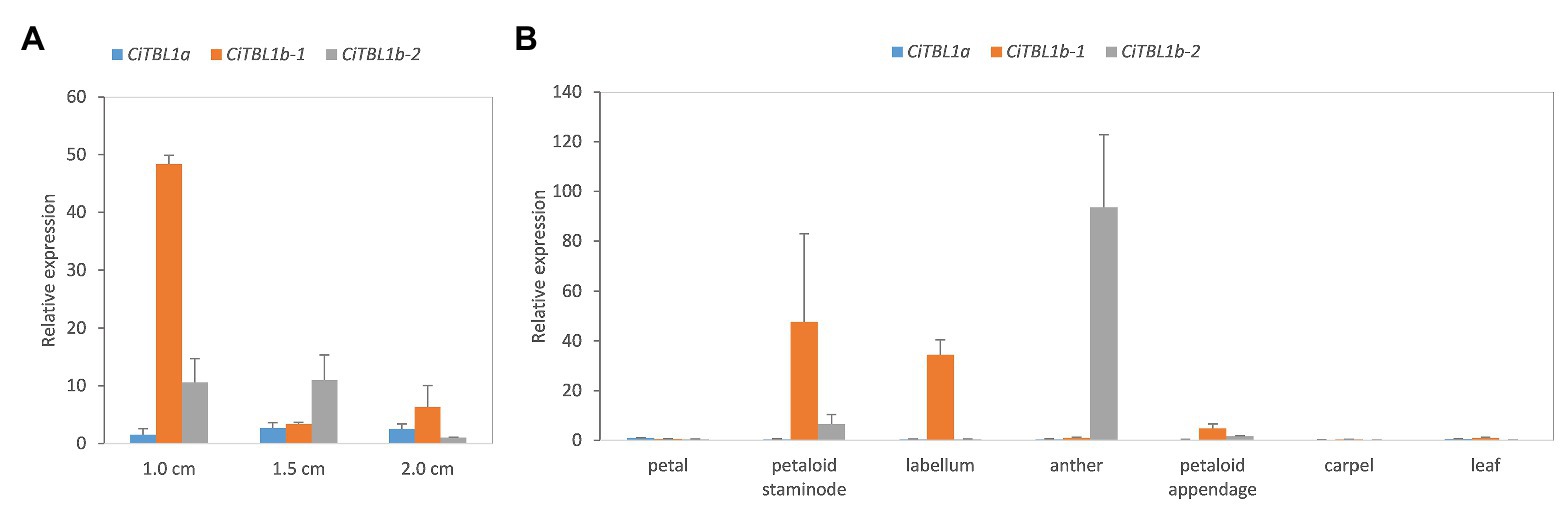
Figure 4. The results of qRT-PCR showing the expression patterns of CiTBL genes in the late floral developmental stages. Relative expression of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 in (A) flower bud of three different lengths and (B) seven different organs. Data were normalized by the relative expression of CiTBL1a in 1.0-cm-long flower bud in (A) and petal in (B). Error bars of gene expression are ±1 SD from three biological replicates.
The results of RNA in situ hybridization show that CiTBL1a and CiTBL1b-1 have similar expression patterns in the young inflorescence (Figures 3A,B,F,G,K,L, 5), with very strong expression in the inflorescence bracts (IB) and the primary bracts (B). The cross-sections of the inflorescence show that CiTBL1a and CiTBL1b-1 are expressed on the adaxial side of the lateral florescence branches (Figures 3G,L, 5). In contrast, CiTBL1b-2 has a broader expression pattern, with transcripts mainly accumulating in the inflorescence meristem (IM), cincinnus meristem (CM), and floral meristem (FM); no adaxial expression was detected in the lateral florescence branches (Figures 3P,Q, 5). After the common primordia differentiates, CiTBL1b-2 is expressed in the petal whorl and inner androecium whorl. After the adaxial stamen primordium differentiates into the anther primordium and the petaloid appendage (Stage 1; Figure 3C), CiTBL1b-2 is expressed in both anther primordium and petaloid appendage, with signal in anther stronger than in petaloid organs (Figures 3R, 5). Expression of CiTBL1a and CiTBL1b-1 are not found in sepal, petal, or stamen primordia during early developmental stages (Figures 3H,M, 5). Later, however, when the anther primordium is growing rapidly (stages 2 and 3), CiTBL1b-2 transcripts are highly enriched in the fertile anther (Figures 3S,T, 5), whereas the transcripts of CiTBL1a and CiTBL1b-1 are mainly observed in developing sepals and petals (Figures 3I,J,N,O, 5).
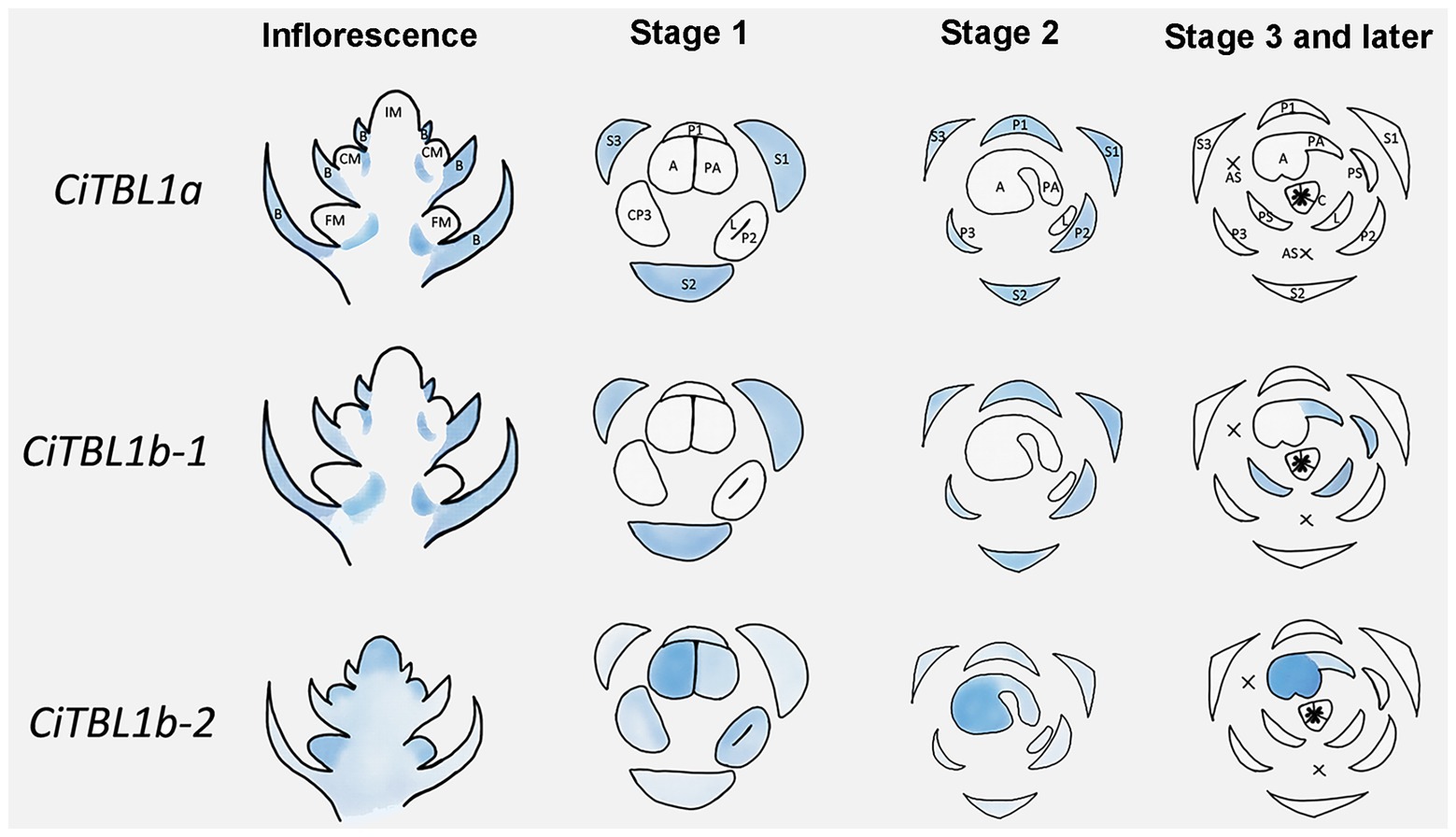
Figure 5. Summary for the expression patterns of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 in the young inflorescence and flowers of different developmental stages. The positions with expression are shaded in blue. The darkness of the blue shades indicates the expression level.
We further examined the spatial-temporal expression of CiTBL genes during late floral developmental stages using qRT-PCR. The results show that the expression level of CiTBL1a is significantly lower in late developmental stages (stage 3 and later), whereas CiTBL1b-1 has the highest expression in flower buds reaching 1.0 cm in length. CiTBL1b-2 is expressed slightly more in 1.0- and 1.5-cm flower buds than it is in 2.0-cm flower buds (Figure 4A). For spatial expression patterns, the transcripts of CiTBL1b-1 are specifically abundant in the petaloid staminode, labellum, and petaloid appendage, whereas CiTBL1b-2 continues to be expressed primarily in the developing anther (Figures 4B, 5).
Functional Analysis of Three CiTBL Genes in Transgenic Arabidopsis Plants
The ORF of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 were ectopically expressed in A. thaliana to elucidate their function during flower morphogenesis.
Constitutive expression of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 led to plants with relatively similar phenotypes. All the transgenic plants were dwarf and exhibited small oval-shaped leaves (Figures 6A,B), indicating that CiTBL genes likely inhibit vegetative growth in Arabidopsis. The flowers, including the individual petals, were slightly smaller in size than those of wild-type (Figures 6C,D). In addition, the induction of ectopic activity of three CiTBL genes in Arabidopsis altered the symmetry of the flower by reducing stamen number. Stamen loss was observed in all transgenic lines. For 35S::CiTBL1a (92 flowers total), 38.1% have 5 stamens as shown in Figure 6C, and 18.5% have more severe phenotypes with only 3–4 developed stamens. For 35S::CiTBL1b-1 (106 flowers total), 25.3% of the flowers have 5 stamens (Figure 6C), whereas 16.9% have only 2–4 stamens. For 35S::CiTBL1b-2 (134 flowers total), 36.6% have 5 stamens (Figure 6C), and 11.1% have 4 stamens or fewer. In addition, growth inhibition of individual stamens is observed in flowers that still maintain six stamens. In WT Arabidopsis, four stamens are long and two are short. In the overexpression lines with six total stamens, either one is long and five are short or two are long and four are short. This phenomenon accounts for 2.8% of flowers in 35S::CiTBL1b-1 and 1.4% of flowers in 35S::CiTBL1b-2. The loss of stamen primordia results in increased space in the floral meristem, and the two petals adjacent to the position of stamen loss appear to fill in the gap (Figure 6C). The overall floral symmetry changes from bilateral (WT) to pronounced zygomorphy with a single plant of symmetry.
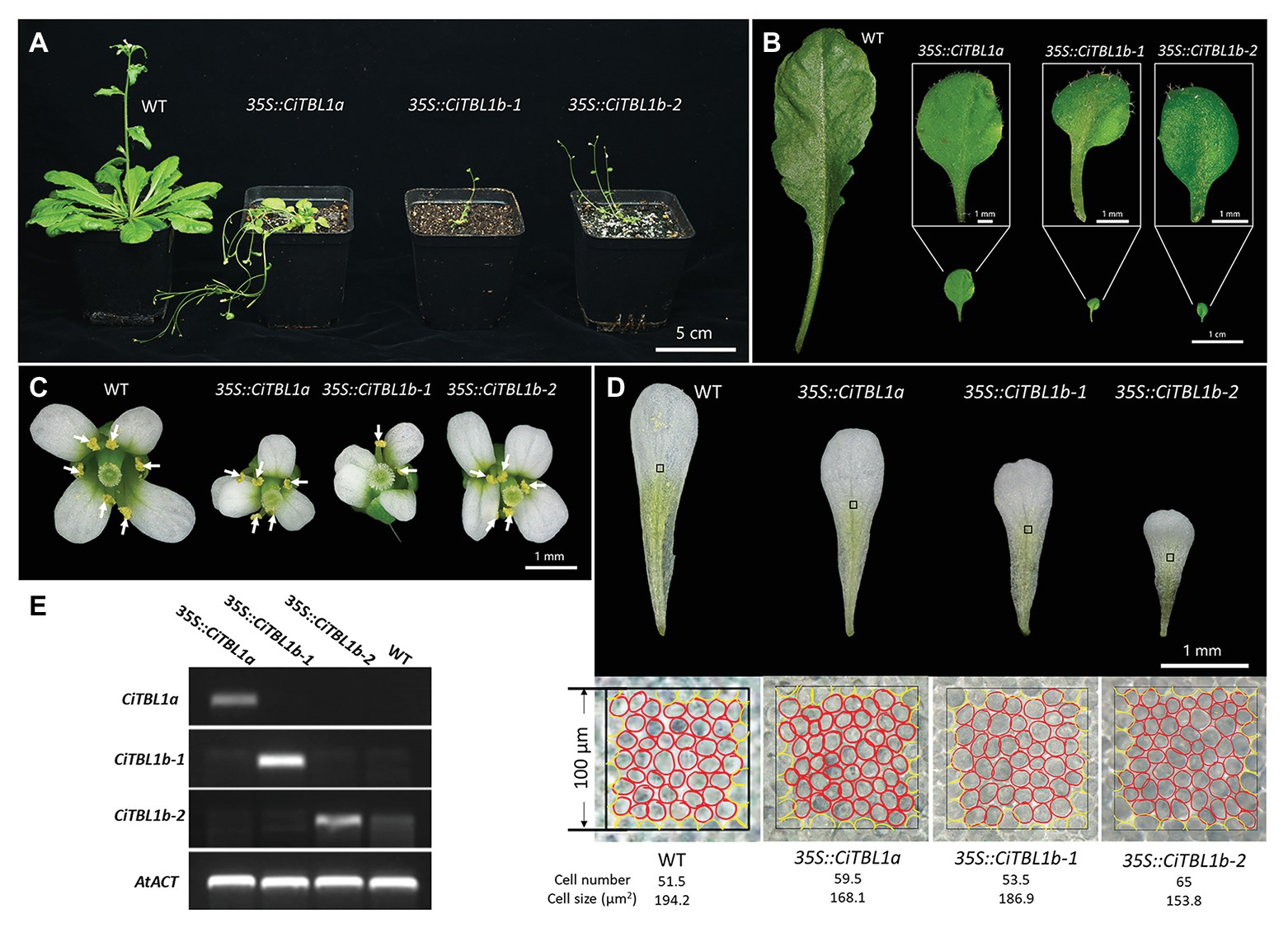
Figure 6. Effects of ectopic expressions of the Canna CiTBL genes in Arabidopsis. Effect of overexpressing CiTBL1a, CiTBL1b-1, and CiTBL1b-2 on Arabidopsis (A) vegetative growth, (B) leaf size and shape, (C) flower symmetry, and (D) petal size. (E) Semi-quantitative RT-PCR results showing strong expression of CiTBL1a, CiTBL1b-1, and CiTBL1b-2 in the flowers of T2 transgenic plants. All the stamens in (C) are indicated with arrows. A red circle in (D) represents one cell, whereas a yellow circle counted for ½ cell.
To determine whether the small petal size of 35S::CiTBL lines was achieved by a decrease in cell proliferation or cell expansion, the cell size of the middle region of adaxial epidermal layer in mature petals was analyzed (Figure 6D). The epidermal cells of wild-type plant petals were about 1.16, 1.04, and 1.26 times bigger than those in 35S::CiTBL1a, 35S::CiTBL1b-1, and 35S::CiTBL1b-2 plants, respectively. The results suggest that the effects of CiTBL genes on petal size is at least partially due to a decrease in cell expansion.
Discussion
CYC/TB1-Like Genes May Be Involved in the Formation of the Novel Organ Petaloid Staminodes in Zingiberales
In flowers of the Zingiberalean ginger families (Costaceae, Zingiberaceae, Cannaceae, and Marantaceae), petaloid staminodes account for the “showy” aspect of the floral display, taking on the role and function typically held by petals. The androecium-derived “petaloid” structures are, however, different from second-whorl petals in their appearance and epidermal cell morphology (Almeida et al., 2015), indicating that mechanisms making a petaloid structure may not be homologous across the whorls. In this research, the genomic presence and expression of endogenous CYC/TB1-like genes was studied to determine their potential role in generating petaloid structures and thereby effecting overall patterns of floral symmetry. Our results indicate CYC/TB1-like genes may play a role in the evolutionary transition from the radial, fertile stamens of the banana families to the laminar, petaloid staminodes that dominate the androecial whorls of the ginger families.
The first line of evidence is based on existing sequencing data. The CYC/TB1-like homolog ZinTBL1b-1 was found exclusively in the four ginger families, which bear flowers with elaborate petaloid staminodes. We did not recover ZinTBL1b-1 in any of the banana families, which lack a predominance of stamen-derived petaloid structures. This indicates that the presence of this particular CYC/TB1-like gene may be important in the formation of the petaloid staminodes that dominate the floral display within the ginger families.
In addition, developmental evidence for the role of CYC/TB1-like genes comes from observations of the ½-fertile stamen growth in Canna sp. In Canna, growth of the ½-fertile stamen is divided into two phases: arrest of anther growth (Figures 1D,E,I,J) and increase in petaloid appendage development (Figures 1F,G,K). We know from previous studies in other lineages that CYC-like genes function in stamen arrest and petal growth by affecting the rate of cell division (Luo et al., 1996; Cubas et al., 1999b; Hileman et al., 2003; Broholm et al., 2008). It is thus possible that CYC/TB1-like genes may suppress stamen fertility while promote the development of a laminar, petaloid appendage in Canna. Although we have not detected differential expression of any CiTBL genes within the Canna ½-fertile stamen, RT-PCR indicates that CiTBL1b-1 is highly expressed late in development when the petaloid appendage is undergoing rapid growth (Figures 1F,G,K, 4A). In addition, CiTBL1b-1 is expressed in all petaloid staminodes, including the labellum and the ½-stamen petaloid appendage (Figure 4B). This is further evidence that CiTBL1b-1 in particular may be responsible for petaloid growth of the androecial members.
Finally, in the core eudicots, loss of function of C-class MADS-box genes is responsible for the transition from stamens to petals (Coen and Meyerowitz, 1991; Causier et al., 2010). Although there is no study that directly indicates the role of CYC/TB1-like genes in the origin of androecial petaloidy, studies have shown that CYC homologs interact with both B-class and C-class genes which are expressed in the androecial whorl. In the Antirrhinum def mutant, cyc expression disappears in the second whorl in the late developmental stage, demonstrating that DEF gene activity is required for later maintenance of CYC in this whorl (Clark and Coen, 2002). In Asteraceae, CYC-like genes also co-operate to regulate stamen and carpel differentiation likely through their interaction with genes controlling cell cycle and floral organ identity (Broholm et al., 2014; Fambrini and Pugliesi, 2016). In Medicago (Fabales), expression of all three MtCYC homologs is significantly upregulated in the AG double mutant which shows abnormality in the dorsal petal (Zhu et al., 2018). The expression pattern of CiTBL1b-2 is similar to that of CiAG-1. Both CiTBL1b-2 and CiAG-1 were expressed slightly higher in the anther primordium than that of petaloid staminodes (Tian et al., 2016). These data indicate that symmetry genes and floral organ identity factors may act together to determine the position and final form of mature floral organs.
Comparison of Expression Patterns of CYC/TB1-Like Genes in Canna and Other Monocot Species
In monocots, expression patterns of CYC/TB1-like genes appear to diverge from those reported for eudicots. For example, eudicot CYC2 genes are typically expressed in the dorsal (adaxial) region of floral meristems, whereas in monocots the CYC/TB1-like genes are expressed on the ventral (abaxial) region in several clades with zygomorphic flowers (Yuan et al., 2009; Bartlett and Specht, 2011; Preston and Hileman, 2012; Lin et al., 2016). In the asymmetric flower of Cannaceae, the “ventral” region is distal to the main florescence axis and to the next order flower of the cincinnus on which it is borne. In this study, CiTBL1b-1 was shown to be expressed ventrally during late stages of floral developmental; i.e., it was expressed in all petaloid staminodes (the 3/4 ventral region of the flower) and was absent from the fertile anther (the dorsal region). The ventral expression patterns of CiTBL1b-1 and some of its monocot homologs suggest that CYC/TB1-like genes may have evolved novel functions within the monocots.
In non-monocot lineages studied to date, CYC-like genes are often expressed in staminodes (Luo et al., 1996; Cubas et al., 1999b; Hileman et al., 2003). However, expression of CYC/TB1 has been documented in fertile stamens in monocots, including rice (REP1, Poacea; Yuan et al., 2009), maize (TB1, Poacea; Hubbard et al., 2002), Tradescantia pallida and Commelina communis (TB1a, Commelinaceae; Preston and Hileman, 2012), Costus spicatus (CsTBL1a and CsTBL2, Costaceae; Bartlett and Specht, 2011), and Alstroemeria aurea (AaTCP1, Alstroemeriaceae; Hoshino et al., 2014). Similar expression patterns have been found in this study; CiTBL1b-2 showed strong expression throughout the development of the single fertile anther. These indicate a potentially distinct mechanism of the CYC/TB1 genes in defining domains of symmetry across monocots in comparison with the well-studied eudicot model systems.
CiTBLs Act as a General Repressor for Both Vegetative and Reproductive Developmental Pathways in Arabidopsis
The ectopic expression of all three CiTBL genes caused reduced vegetative growth in Arabidopsis, consistent with results from previous studies (Costa et al., 2005; Yang et al., 2012; Juntheikki-Palovaara et al., 2014; Liu et al., 2014) and indicating conserved function of CYC/TB1-like genes in vegetative development. However, heterologous overexpression of various CYC/TB1-like genes in Arabidopsis produces differential changes in flower development. Overexpression of Antirrhinum CYC in Arabidopsis produced larger flowers with larger petals (Costa et al., 2005), whereas overexpression of Gerbera CYC2 genes, Primulina CYC1C and CYC1D, and Canna CiTBL genes (this study) all led to smaller flowers (Yang et al., 2012; Juntheikki-Palovaara et al., 2014; Liu et al., 2014), indicating that the function of CYC-like genes on reproductive growth varies. In this study, we also found reduced number of stamens in the transgenic plants, a phenomenon which has not been reported in previous studies. The stamen loss caused modification of floral symmetry in transgenic Arabidopsis plants. Interestingly, a similar phenotype was found in Arabidopsis plants with overexpressed Chrysanthemum CmWUS, indicating a potential interaction between WUSCHEL- and CYC2-like genes in regulating flower development (Yang et al., 2019). Due to the heterologous genetic background of Arabidopsis and Canna, we cannot draw a comprehensive conclusion as to the functions of CiTBL genes during Canna flower development. Genetic knock down or homologous overexpression strategies are needed to investigate the exact functions of CYC/TB1-like genes during the development of the asymmetric flower of Canna.
Conclusion
Asymmetry is a very unusual type of flower form not often included in studies of floral symmetry. Canna bears conspicuous asymmetric flowers due to the presence and distribution of androecial petaloidy within the two androecial whorls. In this study, four CYC/TB1 homologs were isolated and their expression was analyzed to reveal their potential roles in the establishment of asymmetry of the wild-type Canna flower. The results indicate that CiTBL genes are probably involved in asymmetric flower development given their asymmetric expression both early and late during floral development. Notably, ZinTBL1b-1 was expressed in the all androecial petaloid structures, suggesting it may play a role in the morphogenesis of petaloid staminodes. Heterologous overexpression of CiTBLs acts as a general repressor for both vegetative and reproductive developmental pathways in Arabidopsis, and alters floral symmetry by arresting the growth of stamens. Overall, this study provides insights for revealing the developmental mechanisms underlying the formation of asymmetric flowers.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XT and JL conceived and designed the experiments. QY performed the experiments and wrote the manuscript. CL and CS edited the manuscript. JL coordinated all tasks. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science and Technology Infrastructure Program of China (grant number 2015FY210100), Key Research Program of the Chinese Academy of Sciences (grant number KFJ-3W-No1-2), National Natural Science Foundation of China (grant number 31670336), and the UCAS Joint PhD training Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yushi Ye of South China Botanical Garden for help on plant material collection, and Xiaoying Hu and Rufang Deng for technical assistances in SEM and TEM analysis, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.580576/full#supplementary-material
Footnotes
References
Almeida, A. M. R., Brown, A., and Specht, C. D. (2013). Tracking the development of the petaloid fertile stamen in Canna indica: insights into the origin of androecial petaloidy in the Zingiberales. AoB Plants 5:plt009. doi: 10.1093/aobpla/plt009
Almeida, A. M. R., Yockteng, R., and Specht, C. D. (2015). Evolution of petaloidy in the zingiberales: an assessment of the relationship between ultrastructure and gene expression patterns. Dev. Dyn. 244, 1121–1132. doi: 10.1002/dvdy.24280
Bartlett, M. E., and Specht, C. D. (2011). Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot. 98, 227–243. doi: 10.3732/ajb.1000246
Broholm, S. K., Tahtiharju, S., Laitinen, R. A., Albert, V. A., Teeri, T. H., and Elomaa, P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U. S. A. 105, 9117–9122. doi: 10.1073/pnas.0801359105
Broholm, S. K., Teeri, T. H., and Elomaa, P. (2014). Molecular control of inflorescence development in Asteraceae. Adv. Bot. Res. 72, 297–333. doi: 10.1016/b978-0-12-417162-6.00010-9
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Chai, W., Jiang, P., Huang, G., Jiang, H., and Li, X. (2017). Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 23, 779–791. doi: 10.1007/s12298-017-0476-1
Citerne, H., Jabbour, F., Nadot, S., and Damerval, C. (2010). The evolution of floral symmetry. Adv. Bot. Res. 54, 85–137. doi: 10.1016/s0065-2296(10)54003-5
Clark, J. I., and Coen, E. S. (2002). The cycloidea gene can respond to a common dorsoventral prepattern in Antirrhinum. Plant J. 30, 639–648. doi: 10.1046/j.1365-313x.2002.01310.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Costa, M. M., Fox, S., Hanna, A. I., Baxter, C., and Coen, E. (2005). Evolution of regulatory interactions controlling floral asymmetry. Development 132, 5093–5101. doi: 10.1242/dev.02085
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999a). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313x.1999.00444.x
Cubas, P., Vincent, C., and Coen, E. (1999b). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161. doi: 10.1038/43657
D’Hont, A., Denoeud, F., Aury, J. M., Baurens, F. C., Carreel, F., Garsmeur, O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. doi: 10.1038/nature11241
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Endress, P. K. (1999). Symmetry in flowers: diversity and evolution. Int. J. Plant Sci. 160, S3–S23. doi: 10.1086/314211
Endress, P. K. (2012). The immense diversity of floral monosymmetry and asymmetry across angiosperms. Bot. Rev. 78, 345–397. doi: 10.1007/s12229-012-9106-3
Fambrini, M., and Pugliesi, C. (2016). CYCLOIDEA 2 clade genes: key players in the control of floral symmetry, inflorescence architecture, and reproductive organ development. Plant Mol. Biol. Report 35, 20–36. doi: 10.1007/s11105-016-1005-z
Fu, Q., Liu, H., Almeida, A. M., Kuang, Y., Zou, P., and Liao, J. (2014). Molecular basis of floral petaloidy: insights from androecia of Canna indica. AoB Plants 6:plu015. doi: 10.1093/aobpla/plu015
Gandolfo, M. A., Nixon, K. C., and Crepet, W. L. (1998). A new fossil flower from the Turonian of New Jersey: Dressiantha bicarpellata gen. et sp. nov. (Capparales). Am. J. Bot. 85, 964–974. doi: 10.2307/2446363
Gaudin, V., Lunness, P. A., Fobert, P. R., Towers, M., Riou-Khamlichi, C., Murray, J. A., et al. (2000). The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122, 1137–1148. doi: 10.1104/pp.122.4.1137
Hileman, L. C. (2014). Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 369:20130348. doi: 10.1098/rstb.2013.0348
Hileman, L. C., Kramer, E. M., and Baum, D. A. (2003). Differential regulation of symmetry genes and the evolution of floral morphologies. Proc. Natl. Acad. Sci. U. S. A. 100, 12814–12819. doi: 10.1073/pnas.1835725100
Hoshino, Y., Igarashi, T., Ohshima, M., Shinoda, K., Murata, N., Kanno, A., et al. (2014). Characterization of CYCLOIDEA-like genes in controlling floral zygomorphy in the monocotyledon Alstroemeria. Sci. Hortic. 169, 6–13. doi: 10.1016/j.scienta.2014.01.046
Howarth, D. G., and Donoghue, M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U. S. A. 103, 9101–9106. doi: 10.1073/pnas.0602827103
Hubbard, L., McSteen, P., Doebley, J., and Hake, S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162, 1927–1935.
Juntheikki-Palovaara, I., Tahtiharju, S., Lan, T., Broholm, S. K., Rijpkema, A. S., Ruonala, R., et al. (2014). Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 79, 783–796. doi: 10.1111/tpj.12583
Kirchoff, B. K. (1983). Floral organogenesis in five genera of the marantaceae and in Canna (Cannaceae). Am. J. Bot. 70, 508–523. doi: 10.1002/j.1537-2197.1983.tb07878.x
Lin, Y., Chen, Y., Hsiao, Y., Shen, C., Hsu, J., Yeh, C., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67, 5051–5066. doi: 10.1093/jxb/erw273
Liu, B., Pang, H., Yang, X., and Wang, Y. (2014). Functional and evolutionary analyses of Primulina heterotricha CYC1C gene in tobacco and Arabidopsis transformation systems. J. Syst. Evol. 52, 112–123. doi: 10.1111/jse.12067
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., and Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. doi: 10.1016/S0092-8674(00)81523-8
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Maas-van de Kamer, H., and Maas, P. J. M. (2008). The Cannaceae of the world. Blumea. 53, 247–318. doi: 10.3767/000651908X607945
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Miao, M. -Z., Liu, H. -F., Kuang, Y. -F., Zou, P., and Liao, J. -P. (2014). Floral vasculature and ontogeny in Canna indica. Nord. J. Bot. 32, 485–492. doi: 10.1111/j.1756-1051.2013.00311.x
Morioka, K., Yockteng, R., Almeida, A. M., and Specht, C. D. (2015). Loss of YABBY2-like gene expression may underlie the evolution of the laminar style in Canna and contribute to floral morphological diversity in the Zingiberales. Front. Plant Sci. 6:1106. doi: 10.3389/fpls.2015.01106
Neal, P. R., Dafni, A., and Giurfa, M. (1998). Floral symmetry and its role in plant-pollinator systems: terminology, distribution, and hypotheses. Annu. Rev. Ecol. Syst. 29, 345–373. doi: 10.1146/annurev.ecolsys.29.1.345
Preston, J. C., and Hileman, L. C. (2012). Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. Evodevo 3:6. doi: 10.1186/2041-9139-3-6
Prince, L. M. (2010). “Phylogenetic relationships and species delimitation in Canna (Cannaceae)” in Diversity, phylogeny, and evolution in the monocotyledons. eds. O. Seberg, G. Petersen, A. S. Barfod, and J. I. Davis (Denmark: Aarhus University Press), 307–331.
Sauquet, H., von Balthazar, M., Magallon, S., Doyle, J. A., Endress, P. K., Bailes, E. J., et al. (2017). The ancestral flower of angiosperms and its early diversification. Nat. Commun. 8:16047. doi: 10.1038/ncomms16047
Spencer, V., and Kim, M. (2018). Re“CYC”ling molecular regulators in the evolution and development of flower symmetry. Semin. Cell Dev. Biol. 79, 16–26. doi: 10.1016/j.semcdb.2017.08.052
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Tian, X., Yu, Q., Liu, H., and Liao, J. (2016). Temporal-spatial transcriptome analyses provide insights into the development of petaloid androecium in Canna indica. Front. Plant Sci. 7:1194. doi: 10.3389/fpls.2016.01194
Tian, X., Zou, P., Miao, M., Ning, Z., and Liao, J. (2018). RNA-Seq analysis reveals the distinctive adaxial-abaxial polarity in the asymmetric one-theca stamen of Canna indica. Mol. Gen. Genomics 293, 391–400. doi: 10.1007/s00438-017-1392-3
Yang, X., Pang, H., Liu, B., Qiu, Z., Gao, Q., Wei, L., et al. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24, 1834–1847. doi: 10.1105/tpc.112.099457
Yang, Y., Sun, M., Yuan, C., Han, Y., Zheng, T., Cheng, T., et al. (2019). Interactions between WUSCHEL- and CYC2-like transcription factors in regulating the development of reproductive organs in Chrysanthemum morifolium. Int. J. Mol. Sci. 20:1276. doi: 10.3390/ijms20061276
Yuan, Z., Gao, S., Xue, D., Luo, D., Li, L., Ding, S., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149, 235–244. doi: 10.1104/pp.108.128231
Keywords: Canna indica, floral asymmetry, Zingiberales, CYCLOIDEA, TEOSINTE BRANCHED1
Citation: Yu Q, Tian X, Lin C, Specht CD and Liao J (2020) Expression and Function Studies of CYC/TB1-Like Genes in the Asymmetric Flower Canna (Cannaceae, Zingiberales). Front. Plant Sci. 11:580576. doi: 10.3389/fpls.2020.580576
Edited by:
Hongzhi Kong, Chinese Academy of Sciences, ChinaReviewed by:
Yin-Zheng Wang, State Key Laboratory of Systematic and Evolutionary Botany (CAS), ChinaWenheng Zhang, Virginia Commonwealth University, United States
Copyright © 2020 Yu, Tian, Lin, Specht and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingping Liao, bGlhb2pwQHNjYmcuYWMuY24=
 Qianxia Yu
Qianxia Yu Xueyi Tian
Xueyi Tian Canjia Lin1,2,3
Canjia Lin1,2,3 Chelsea D. Specht
Chelsea D. Specht Jingping Liao
Jingping Liao