
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 November 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.575591
The Brassica coenospeceis have treasure troves of genes that could be beneficial if introgressed into cultivated Brassicas to combat the current conditions of climate change. Introducing genetic variability through plant speciation with polyploidization is well documented, where ploidy augmentation of inter-generic allohexaploids using somatic hybridization has significantly contributed to genetic base broadening. Sinapis alba is a member of the Brassicaceae family that possesses valuable genes, including genes conferring resistance to Sclerotinia sclerotiorum, Alternaria brassicae, pod shattering, heat, and drought stress. This work aimed to synthesize stable allohexaploid (AABBSS) Brassica while incorporating the yellow-seed trait and resistance to S. sclerotiorum stem rot. The two fertile and stable allohexaploids were developed by polyethylene glycol mediated protoplast fusions between Brassica juncea (AABB) and S. alba (SS) and named as JS1 and JS2. These symmetric hybrids (2n = 60) were validated using morphological and molecular cytology techniques and were found to be stable over consecutive generations. The complete chromosome constitution of the three genomes was determined through genomic in situ hybridization of mitotic cells probed with S. alba genomic DNA labeled with fluorescein isothiocyanate. These two allohexaploids showed 24 hybridization signals demonstrating the presence of complete diploid chromosomes from S. alba and 36 chromosomes from B. juncea. The meiotic pollen mother cell showed 30 bivalent sets of all the 60 chromosomes and none of univalent or trivalent observed during meiosis. Moreover, the backcross progeny 1 plant revealed 12 hybridization signals out of a total of 48 chromosome counts. Proper pairing and separation were recorded at the meiotic metaphase and anaphase, which proved the stability of the allohexaploid and their backcross progeny. When screening, the allohexaploid (JS2) of B. juncea and S. alba displayed a high degree of resistance to S. sclerotiorum rot along with a half-yellow and half-brown (mosaic) seed coat color, while the B. juncea and S. alba allohexaplopid1 (JS1) displayed a yellow seed coat color with the same degree of resistance to Sclerotinia rot.
The Brassica coenospecies (Habard, 1976) are rich reservoirs for genes that confer resistance or tolerance to many biotic and abiotic diseases, such as Alternaria blight, Sclerotinia stem rot, insect pests, pod shattering, high temperature, and drought (Sharma et al., 2002; Li et al., 2009). At the moment, resistance gene sources for these diseases are not available within cultivated germplasms because of the established bottleneck through the continuous selection for yield traits. However, enormous genetic variations do exist in important agronomic traits, including disease resistance within the Brassicaceae family (Warwick, 1993). Camelina sativa, B. desnottesii, Capsella bursa-pastoris, Diplotaxis catholica, D. erucoides, D. tenuisiliqua, and S. alba are well documented for their resistance to Alternaria brassicae (Brun et al., 1988; Conn et al., 1988; Sharma et al., 2002), where S. alba, Erucastrum cardaminoides, E. abyssinium, B. fruticulosa, D. tenuisiliqua show resistance to Sclerotinia sclerotiorum (Li et al., 2009; Garg et al., 2010; Rana et al., 2017, 2019; Atri et al., 2019). Out of these, S. alba was found to be the most promising due to its resistance to major biotic and abiotic diseases that affect rapeseed and mustard, such as Alternaria blight (Kolte, 1985; Brun et al., 1987; Ripley et al., 1992; Sharma and Singh, 1992; Hansen and Earle, 1995, 1997; Sharma et al., 2002), Sclerotinia stem rot (Kolte, 1985; Li et al., 2009), blackleg, beet cyst nematode (Lelivelt et al., 1993), flea beetles (Bodnaryk and Lamb, 1991) heat, drought (Downey et al., 1975; Primard et al., 1988; Downey and Röbbelen, 1989; Brown et al., 1997; Kumari et al., 2018), and pod shattering (Downey, 1987; Chandler et al., 2005; Wang et al., 2007). Along with disease resistance properties, S. alba also carries quality traits, such as the yellow seed color, where yellow seed varieties are superior due to their 2–5% higher oil content relative to black seed varieties of the same genetic background (Daun and DeClercq, 1988), in addition to the high-quality and transparent oil without pigment accumulation (Slominski et al., 1994; Tang et al., 1997; Meng et al., 1998; Li et al., 2009, 2012). Therefore, breeding for yellow seed varieties is desirable by edible oil industries. Although S. alba is cultivated as condiment mustard in Canada with oil content less than B. juncea’s cultivated germplasms, the introgression of genes governing the yellow seed color and resistance to diseases into the cultivated Brassica background could help develop resistant varieties.
The allohexaploid Brassica has the potential to generate new genetic variations and introduce inter-subgenomic regions to mitigate the impact of changing climatic conditions (Gupta et al., 2016; Wei et al., 2016). The third subgenome taken from a close or wild relative could benefit oilseed crops by adding disease resistance (Chen et al., 2011) and heat tolerance genes (Kumari et al., 2018). However, the development of interspecific allohexaploids was attempted by earlier researchers by crossing of allotetraploids and diploid species followed by ovary culture, where they were treated with chromosomes doubling agents (Meng et al., 1998; Rahman, 2001; Pradhan et al., 2010; Gupta et al., 2016; Mwathi et al., 2017, 2020), but their stability was still dubious (Mwathi et al., 2019). Mason et al. (2012) used unreduced gametes by crossing all three allotetraploids of U’s triangle species, but they only obtained a single near-allohexaploid. The inter-generic fertile and stable allohexaploid development was stalled owing to pre and post-hybridization barriers between the genera in conventional breeding programs with subsequent irregular meiosis. Therefore, somatic hybridization is the best alternative approach to assemble the inter-generic genomes within a cell through allohexaploids development. The protoplast fusion of somatic cells of B. napus + S. alba (Primard et al., 1988; Lelivelt et al., 1993; Wang et al., 2005a) and B. juncea + S. alba (Gaikwad et al., 1996) were attempted earlier, but none of the stable somatic hybrids were recovered due to meiotic abnormalities or multivalent formation. Also, the back-cross progenies were derived from the somatic hybrids of B. napus and S. alba to expand their fertility (Wang et al., 2005b). Whereas in our previous attempt, we have developed the first stable and fertile somatic hybrids of B. juncea and S. alba (Australian accession) that possess brown seeds along with resistance to Alternaria blight and heat tolerance (Kumari et al., 2018). Of these three, two somatic hybrids have initiated flowering when day temperature rose above 30°C and efficiently set seeds up to 40°C day temperature.
In the present study, we have taken an attempt for Polyethylene glycol (PEG) mediated protoplast fusion between B. juncea cv. RLM-198 and an Indian accession of S. alba (Gene Bank Accession No. DRMR-2183) procured from the ICAR-Directorate of Rapeseed-Mustard Research, Bharatpur (India) to the genome of two different genera into a single genome by developing the fertile inter-generic allohexaploid (AABBSS) Brassica with a yellow-colored seed coat trait along with resistance to Sclerotinia stem rot. The two allohexaploids reported in the present manuscript have variabilities from earlier published hybrids for reduced plant height, early flowering, yellow, and mottle seed coat colors, and resistance to stem rot. The two allohexaploids developed during the present study acquired the mitochondria from the B. juncea while the two out of three somatic hybrids published earlier carried recombinant mitochondria of their parents. The genomic constitution by genomic in situ hybridization (GISH), genetic stability, gametic fertility, stable resistance against S. sclerotiorum, and the yellow seed color were investigated in subsequent self-generations of allohexaploids with their first back cross progeny. Furthermore, we studied the thickness of the palisade (pigment deposition) layer in both yellow and mottle seeded allohexaploids with their parents to uncover the histological variations inside of the yellow and mottle seed development.
For the development of stable inter-generic allohexaploid Brassica, we have used B. juncea cv. RLM-198 as the cultivated germplasm with Indian S. alba (Gene Bank Accession No. DRMR-2183) as a donor for the yellow seed color and resistance to S. sclerotiorum. The B. juncea cv. RLM-198 was selected as a fusion parent due to its excellent regeneration potential. The three experiments included in this study were conducted using the same fusion parents.
The protoplast of hypocotyl cells of B. juncea cv. RLM-198 and mesophyll cells of S. alba were isolated using the protocol of Kirti and Chopra (1990), followed by protoplast fusion, regeneration, and acclimatization according to Kirti et al. (1995). A total of five plants (clones) from each event were transferred to pots filled with soil:solarite (1:1) under net house conditions at the Indian Agriculture Research Institute (IARI), New Delhi, India and maintained till harvesting.
Young leaves were used for the extraction and purification of genomic DNA as reported by Kirti et al. (1995), while the NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, United States) was used for DNA quantification. The mitochondrial genomic constitution was determined using polymerase chain reaction (PCR) primers specific to orf 108 gene conferring male sterility, present in S. alba (Kumar et al., 2012). In primer combination (F1/R1), F1 is determined for forward and designed from the upstream region of orf108 while R1 for a reverse primer from the coding region of atp1. In another primer combination MF/R1, the forward primer MF was designed from the DNA sequence of M. arvensis (Naresh et al., 2016). To determine the chloroplast genome, we tested nine plastid specific simple sequence repeats (SSR) markers, where three were given polymorphic amplicons between the parents and used to identify the chloroplast genome of somatic hybrids (Flannery et al., 2006). The PCR reaction and conditions were as described by Kumari et al. (2018), where the PCR products were resolved on a 3% agarose gel and visualized by ethidium bromide staining in a gel electrophoresis unit.
The fusion parents were planted with both allohexaploids in the net house at the IARI, New Delhi, India during the crop season (2015–2016). The morphological traits were recorded and compared to the parents, where the phenotypic data of five plants (clones) of both the allohexaploids of B. juncea and S. alba (JS1 and JS2) were collected at maturity during April–May, 2016. Plant height (cm), stem collar diameter, flower characteristics, main shoot length, number of siliquae on the main shoot, number of seeds per silique, and seed weight of 1,000 seeds. The same traits were recorded in the 16 lines of the first backcross progeny that was derived from both allohexaploids (JS1 and JS2) and their subsequent selfing generations across two seasons (crop season 2016–2017 and off-season 2017). The anthers were stained with 1% (w/v) aceto-carmine solution, while the pollen mother cells (PMCs) were teased with needles. Pollen grain viability was determined as the ratio of the number of stained pollen grains to the total number of pollen grains, where each microscopic field comprised 400 pollen grains that helped calculate the pollen grain viability percentage in each microscopic field.
The root of the subsequent selfing generation, shoot apex, and anthers of the five regenerated plants (clones) of each allohexaploid were used in the mitotic and meiotic chromosome smear preparations. In terms of the backcross progeny, three plants of each line were selected for the mitotic and meiotic preparations, where the root tips of germinating seeds or shoot leaf primordia in the mitotic studies were prefixed in 2 mM solution of 8-hydroxyquinoline for 3 h, kept in 4°C for 2 h, fixed in Carnoy’s solution I (Carnoy, 1887) with a ratio of ethyl alcohol to acetic acid of 3:1 for 72 h, transferred to 70% (v/v)ethanol, and finally stored at 4°C. The root and shoot apex were pretreated with 2% cellulase (w/v) and pectolyase (w/v) mixture for 1 h, then compressed with a 2% (w/v) solution of aceto-orcein. Similar to the mitotic studies, the floral buds were fixed in Carnoy’s solution II with a ratio of ethyl alcohol to chloroform to acetic acid of 6:3:1 for 48 h in the meiotic studies and transferred to 70% (v/v) ethyl alcohol at 4°C. The anthers were teased into a drop of 2% (w/v) acetic acid carmine solution on microscope glass slides to release the PMC, which were observed at the metaphase-I, and anaphase-I stages. Probe preparation and in situ hybridizations were performed as described by Kumari et al. (2020b). Post-hybridization washing was conducted at 42°C for 5 min in a 2x SSC water bath. Next, the slides were incubated in 4x SSC followed by 8x SSC, prepared in 0.2% Tween-20 (EIA grade, BioRad, United States) for 10 min, cooled down to room temperature in 2x SSC for 5 min, and then put through a dehydration series of 70, 95, and 100% alcohol. The chromosomes were stained with 2 mM 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, United States) and mounted with a Vecta shield medium (Sigma-Aldrich, United States), which was visualized under a fluorescent microscope (ImagerZ2 AX10, ZEISS, Germany).
The plants were covered with a cloth bag to develop self-seeds that were used to grow the subsequent generation of allohexaploids. Simultaneously, to develop backcross progeny from allohexaploids, all the 10 clones of both hybrids were used as female parent and B. juncea cv. RLM-198 and B. juncea var. IJ-31 (high yielding variety) as the male parent. Each clone of allohexaploid was used for backcrossing with both B. juncea varieties. A total of about 30–40 flower buds for each cross were emasculated that were ready to bloom the next day and the remaining small buds were removed from the emasculated buds to avoid selfing and unwanted seed development. The fresh pollens were collected from newly opened flowers of both B. juncea varieties (RLM-198 and IJ-31) in the morning and pollinate the emasculated buds. The pollinated buds were covered with paper bags to avoid foreign pollen contamination. The paper bags were removed after 5–6 days and harvested all the crosses after maturity. A total of 20 back-crosses were developed and recovered approximately 22–29 siliquae from each cross. The silique developed 3–5 seeds and about 67–84 seeds harvested from each backcross. The seeds harvested from each backcross were treated as a single line in further experiments.
The physiologically matured 10 siliquae of subsequent selfing generation of each allohexaploid and their parents were fixed in formalin-acetic acid alcohol (FAA) and stored in 70% ethanol until use. Sample dehydration and clearing were performed through the alcohol xylene series, which were kept in infiltration tubes containing xylene and paraffin wax with a low melting point of 50°C under sunlight. This caused the wax to dissolve in the xylene, which was placed in an oven at 67°C for the wax to penetrate the cells and remove any traces of xylene. Next, embedding the siliquae was performed in the melting paraffin wax at 60°C, which was poured into the paper boat and infiltrated the siliqua that was placed immediately into a paper boat. Then, the boat was transferred into cold water for faster solidification of the paraffin. The solid wax cake was cut into small blocks containing a single object, where these blocks were mounted onto the block holder of the microtome to be cut into sections of 15 μ thickness and placed on clean slides with Meyer’s adhesive (Johansen, 1940). These sections were spread on the slides, dried, and stained in Delafield’s Hematoxylin, then mounted in Canada balsam, which were observed under the microscope for histological examination.
The virulent culture of S. sclerotiorum was procured from Indian Type Culture Collection (ITCC), IARI, Pusa, New Delhi, India (accession: ITCC-3292). The fungal mycelium was inoculated into the potato dextrose agar medium in the Petri plates, where the S. sclerotiorum culture was maintained for further experiments in the cultured media at 22°C ± 1°C under alternating 12 h light and dark conditions. The developed sclerotia were harvested 12–15 days after inoculation and maintained at 4°C. The allohexaploids (JS1 and JS2) and their parents were screened with S. sclerotiorum during 2015–2016 to identify the resistant and susceptible plants. A total of 16 lines of BC1 generation of the allohexaploids, expressing high pollen grain fertility, good yield characteristics, and vigorous growth, were selected for the screening of stem rot disease along with both allohexaploids and their parents during the rabi season in 2017–2018 (season-I) at the IARI agriculture farm, Pusa Campus, New Delhi, India and rescreened during the offseason in 2018 (season-II, June to September) at the IARI Regional Station, Katrain, Kullu, Himachal Pradesh, India. The plants of BC1 line, both allohexaploids, and their parents were sown in a randomized block design with three replications. Five plants from each replication were selected for pathogen inoculation after the initiation of flowering, where the selected plants were inoculated with 6 mm discs of actively growing virulent pathogens on agar plates, at just above the first node on a hand scratched point, and tied with parafilm (Garg et al., 2010). The disease progress was monitored regularly, and the final stem lesion sizes were recorded after 21 days of inoculation. The plants were categorized according to lesion size in terms of immune (no sign of disease), highly resistant (0–2.0 cm), resistant (2.1–4.0 cm), moderately resistant (4.1–6.0 cm), tolerant (6.1–8.0 cm), susceptible (8.1–10.0 cm), and highly susceptible (more than 10.0 cm).
To the development of the allohexaploids for introgression Sclerotinia stem rot resistance along with yellow seed coat color, the PEG mediated somatic hybridization was done between B. juncea cv. RLM-198 and S. alba. Hybridity was evident from morphological features, chromosomal constitutions by Genomic in situ hybridizations (GISH), and organelle constitutions determined through molecular analysis. Disease resistance was confirmed by screening using virulent isolate of S. sclerotiorum of the two allohexaploids and their first backcrossed progeny and parental checks in field conditions.
Morphologically distinct protoplasts were used for protoplast fusion, i.e., hypocotyl protoplasts of dark-grown seedlings of B. juncea were colorless, while the mesophyll protoplasts of S. alba were visibly green, therefore, the hetero-fusion products were easily differentiated. Protoplasts of both parents were kept unfused as a control. The hybrid fusion products were detected using the green chloroplasts that were grouped at the center 2 days after fusion (Figure 1A). Cell wall synthesis began 56 h after fusion when the protoplasts lost their spherical shape (Figure 1B). Cell division was initiated 72 h after fusion with consecutive cell divisions observed up to 7 days after fusion. The division also began after 72 h in the protoplast of B. juncea RLM-198, which was kept unfused. However, in the S. alba control, neither the protoplast division nor the cell wall formation was observed. The division rate was inconsistent in the three experiments and ranged between 20 and 50%. The fusion product developed in the form of micro-colonies 20 days after fusion. A total of 954 microcalli were obtained from the three experiments, where a total of 346 calli were transferred into the regeneration medium that was standardized for B. juncea cv. RLM-198 regeneration and the pH was maintained at 5.8 after the addition of plant hormones and obtain 56 shoots on the 5th week of protoplast culture (Figure 1C). All 56 shoots were treated as 56 independent events. The majority of these shoots readily developed roots on the half-strength Murashige and Skoog medium. A total of 54 plantlets showed morphological resemblance to B. juncea more than S. alba, while only two plantlets named JS1 and JS2 after transplantation in the net house showed an intermediate morphology between their parents, which were transferred into pots and grown under the net house (Figure 1D). During the early growth stages, we found that JS1 and JS2 showed more resemblance to S. alba, but after a few weeks, their morphology became an intermediate of the two parents in terms of leaf shape, leaf size, leaf texture, stem, size, stem shape, presence of prominent trichomes on leaves and stem, the emergence of flower buds, flower shape and size, anther, siliquae shape and size, and beak shape.
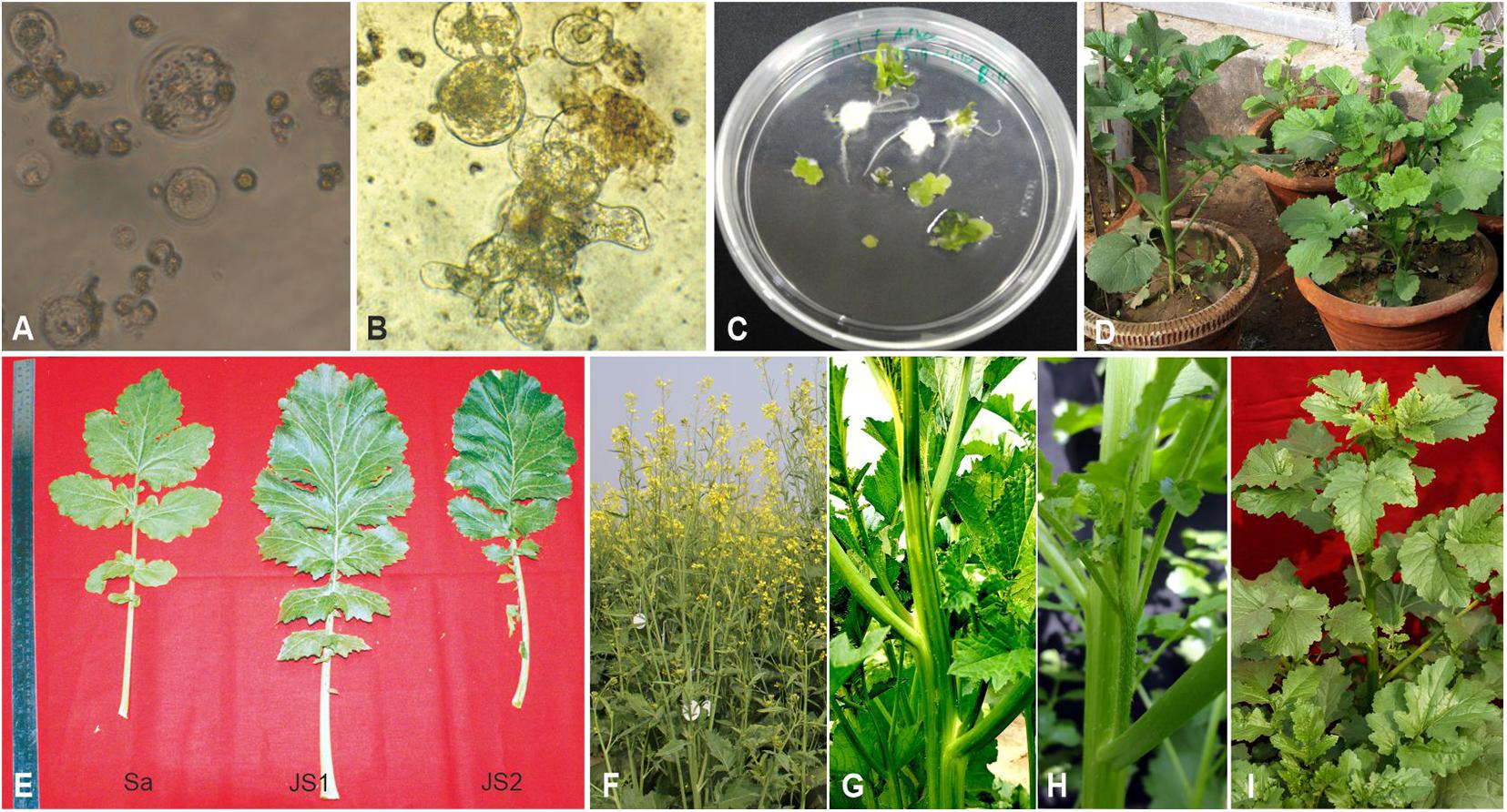
Figure 1. Protoplast fusion between B. juncea and S. alba. (A) Protoplast fusion product after 48 h of fusion. (B) Subsequent cell division and cell wall formation. (C) Initiation of shoot regeneration from calli after 35 days of fusion. (D) Allohexaploids established in pots. (E) Morphological variation in leaf. (F) Plant morphology of B. juncea cv. RLM-198. (G) Stem morphology of JS1 showing angular stem with prominent trichomes. (H) Stem morphology of JS2 showing prominent trichomes. (I) Plant morphology of S. alba.
The stem of B. juncea was smooth, light green, and without trichomes (Figure 1F). The stems of JS1 and JS2 were angular, branched, and covered with dense trichomes (Figures 1G,H). Moreover, the stems of S. alba also found rough, angular with dense trichomes (Figure 1I). The leaf and flower bud margins were similar to S. alba with JS1 and JS2 attaining an average height of 216 and 184 cm, respectively. The delayed flowering was recorded in the allohexaploids relative to B. juncea, where the leaves were considerably larger, thicker, more cuticular, deeper green in color, had more irregular pinnately lobed serrate margins, and was more covered with dense trichomes on both surfaces (Figure 1E). The inflorescence of JS1 showed an S. alba like condense pattern, while JS2 had an intermediate pattern of the parents. JS1 and JS2 flower sizes were larger and intermediate relative to the parents’ flowers, respectively. The stigma was larger than that of the parents with hairy projections on the style, while the stamens of allohexaploids were more prominent than in the parents (Table 1). The pollen grain viability in both allohexaploids was between 91 and 93% (Figure 4A) with female fertility remaining high as revealed by their seed setting. The siliquae of the allohexaploids were found to be intermediate in size relative to their parents with an increased number of seeds (3–5 seeds/silique) compared to S. alba. These allohexaploids produced viable seeds when selfing and backcrossing with B. juncea, which were found to be intermediate in size relative to the parents. However, the weight of 1,000 JS1 and JS2 seeds was higher and less than that of B. juncea and S. alba, respectively (Table 1). Allohexaploid JS1 produced yellow seeds (Figure 4B) that were similar to S. alba, while JS2 interestingly produced half yellow half brown (mosaic) colored seeds (Figure 4C). To the best of our knowledge, this was the first instance of obtaining mosaic and yellow seeds from somatic hybridization of S. alba and B. juncea, where the seeds colors were stable over the generations in both allohexaploids.
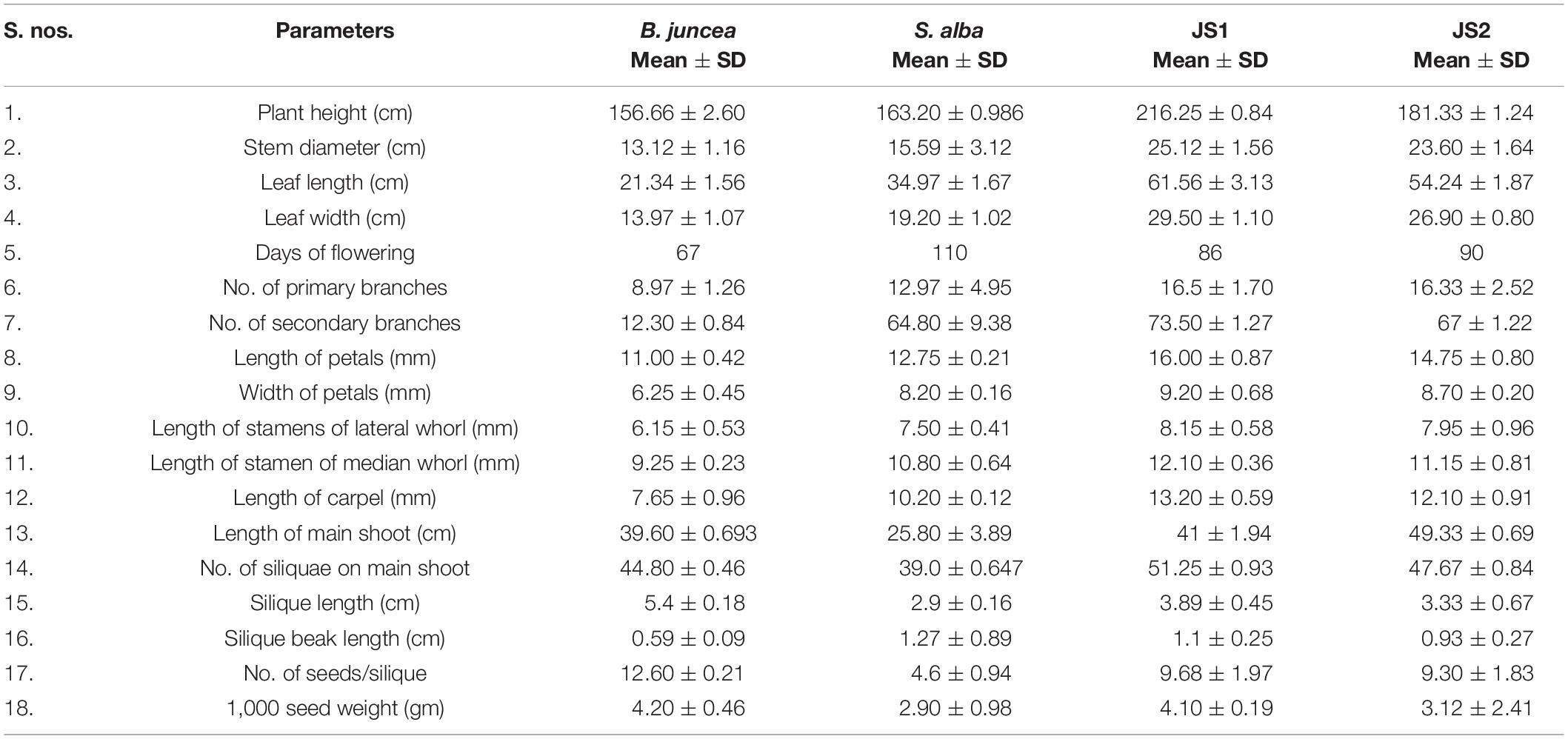
Table 1. Comparative morphological characterization of B. juncea cv. RLM-198, S. alba, JS1, and JS2.
Out of the nine tested chloroplast genome-specific SSR markers, only three markers showed polymorphism between the parents that were used to determine the chloroplast constitutions of the allohexaploids. JS1 and JS2 provided amplicons specific to B. juncea and confirmed the acquisitions of chloroplast from the B. juncea in both allohexaploids (Figure 2A). Similarly, the mitochondrial constitution of both allohexaploids was also found as similar to B. juncea, which was determined through PCR using primers that corresponded to orf108 upstream of the atpA gene. The primer combination of F1/R1 gave amplicon ∼530 bp in S. alba while absent in B. juncea and both allohexaploids. Another primer combination MF/R1 produced polymorphic amplicons between the parents for 1 kb and ∼680 bp in S. alba and B. juncea, respectively. However, both allohexaploids produced similar amplicons of B. juncea (Figure 2B and Table 2).
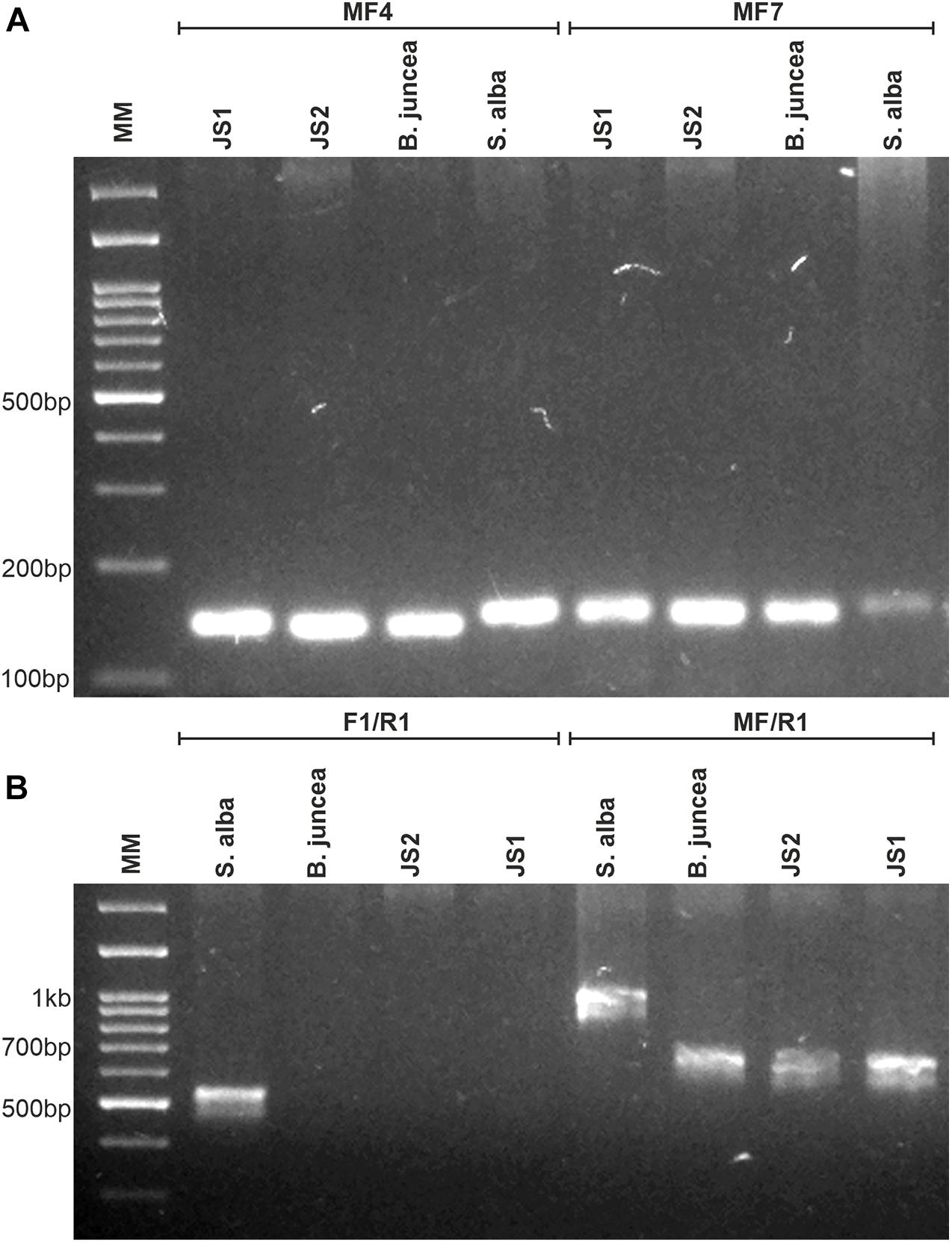
Figure 2. Molecular analysis for determination of organelles constitution of allohexaploids and their parents (B. juncea and S. alba). (A) PCR amplification patterns of MF4 and MF7 markers specific for chloroplast genome, showing inheritance of B. juncea chloroplast in both allohexaploids. (B) PCR amplification obtained in both allohexaploids and their parents using orf-108 primers specific for mitochondria, the F1/R1 and MF/R1 primer combinations showing B. juncea type mitochondria in allohexaploids (JS1 and JS2).
The mitotic chromosome count of JS1 and JS2 revealed that both allohexaploids carried 60 chromosomes reflecting a cumulative chromosome count of B. juncea (2n = 36) and S. alba (2n = 24) (Figures 3A,B). The S. alba chromosome constitution was detected by 24 strong hybridization signals using genomic in situ hybridization (GISH) with a fluorescein isothiocyanate labeled genome-specific probe of S. alba (Figure 3A). This chromosomal stability was also been observed in subsequent generations, where a total of 30 bivalents were noted at meiosis-I in the PMCs of JS1 (Figure 3C) and maintained a high degree of fertility. The BC1 progeny of hybrids using the B. juncea backcrossing was found to be highly fertile in terms of the male and female gametes. This fertility was maintained with a haploid set (n = 12, S) of the S. alba chromosomes along with a diploid set of B. juncea chromosomes (2n = 36, AABB), a total 48 chromosomes were evident from mitosis of first backcross (BC1) generation (Figure 3D). The normal 24 bivalents were observed at diakinesis of meiosis-I in both allohexaploid (JS1 and JS2) (Figures 3E,F). All 48 chromosomes were observed at the equator as normal bivalents at metaphase-I and no laggard chromosome evident (Figure 3G) and equal distribution toward the poles at anaphase-I was observed (Figure 3H).
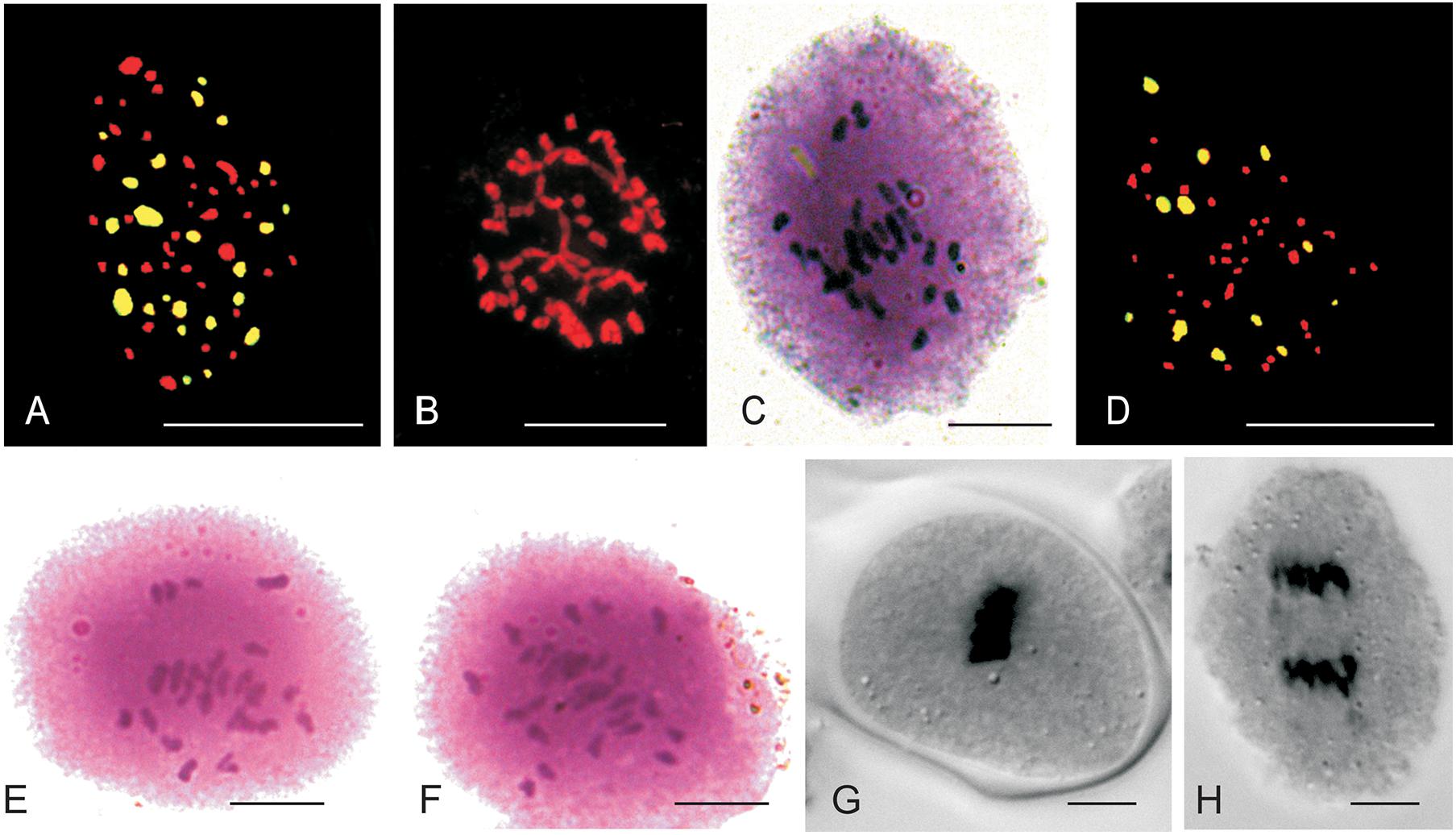
Figure 3. Mitotic and Meiotic studies of allohexaploids and BC1 progeny. (A) 24 FITC labeled S. alba (green) chromosomes and 36 B. juncea chromosomes (red) of JS1. (B) Somatic chromosomes of JS2 (2n = 60). (C) Thirty bivalent observe in PMCs of JS1. (D) 12 FITC labeled S. alba chromosomes in BC1 of JS2 (AABBS = 48) with green color and 36 chromosomes of B. juncea in red color. (E,F) 24 bivalents seen in JS1 and JS2 at diakinesis of meiosis-I, respectively. (G) Meiotic metaphase of BC1 (JS1). (H) Meiotic anaphase-I of BC1 (JS1) (Scale bar 10 μm).
The histological study of the seed coat revealed that the pigment distribution was mainly observed in the palisade layer, where the highest pigmentation was observed in B. juncea (Figure 4D), moreover S. alba and yellow seeded allohexaploid (JS1) showed very low pigmentations in their palisade layer (Figures 4E,F). The mottled seed allohexaploid (JS2) exhibited nearly B. juncea like pigment deposition in the palisade layer (Figure 4G).
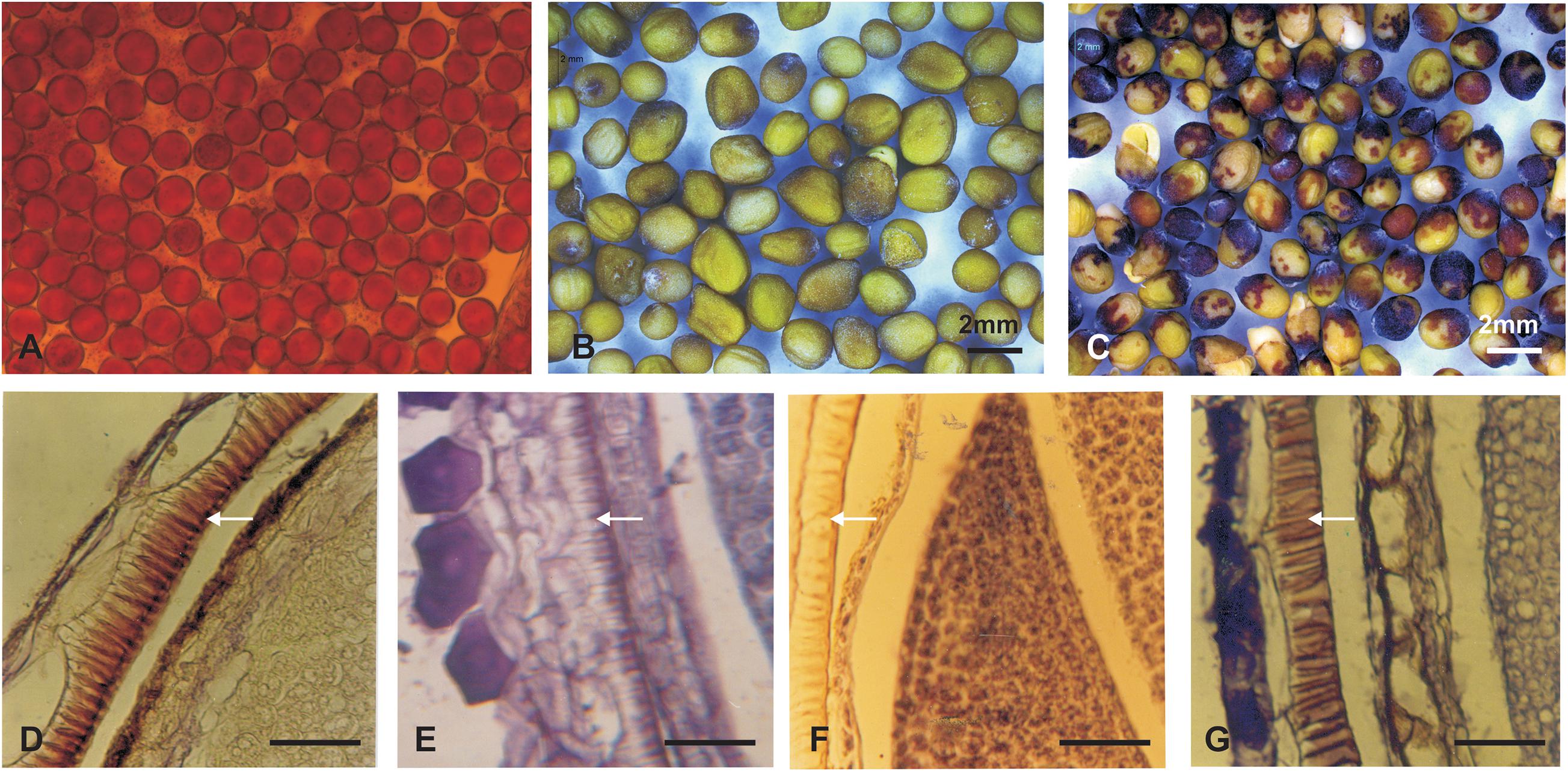
Figure 4. Pollen viability and seed coat colors (upper panel). (A) Pollen grain viability test of JS2. (B) Yellow color seeds of JS1. (C) Yellow–brown color seeds of allohexaploid JS2. (D–G) Mature silique histology showing depositions in palisade layer of the seed coat (lower panel). (D) B. juncea. (E) S. alba. (F) Allohexaploid JS1. (G) Allohexaploid JS2 (Scale bar 25 μm).
The allohexaploids (JS1 and JS2) screened for stem rot resistance during 2015–2016 showed a complete immune response against the pathogen as it was unable to grow on the stem surface. However, B. juncea was found highly susceptible to the disease as the complete stem was rotted by the pathogen, while S. alba showed very limited growth on the stem surface, but it did not develop any characteristic disease symptom. In terms of the BC1 generation of both allohexaploids, a total of 16 lines were selected for disease screening experiments based on their pollen grain fertility (> 90%), good yield, and vigor along with both the allohexaploids (JS1 and JS2) and their parents (Figure 5). The stem rot pathogen produced typical disease symptoms on the susceptible parent B. juncea cv. RLM-198 with lesion sizes between 22.4–24.9 cm and 23.1–26.6 cm during season-I and season-II, respectively. The complete collar area of the stem was rotten (Figure 6A) and produced sclerotia within it at advanced disease stages (Figure 6B). The pathogen grew on the stem surface of S. alba without developing any characteristic rotten disease symptoms (Figure 6C). However, lesion sizes were recorded between 1.0–1.2 cm and 1.0–1.1 cm during both experimental seasons on the upper epidermis only. The stems of both hybrids grew vigorously and did not show any sign of disease at the inoculation point till harvesting (Figures 6D,E). The results of season-I showed that 13 lines of BC1 were of the highly resistant category as the disease lesion size was recorded between 0.6 and 1.1 cm (mean = 0.8 cm) (Figure 6F). Another 3 BC1 lines were found to be resistant to the disease as their lesion sizes ranged between 2.1 and 2.8 cm (mean = 2.3 cm). As expected, it was found that the BC1 generation showed highly resistant responses to the stem rot pathogen compared to the susceptible B. juncea parent. In season-II, the 14 and 2 lines of BC1 progeny were found highly resistant to the disease with mean lesion sizes of 0.9 and 2.2 cm, respectively. These results indicated that BC1 generation was consistently resistant to the disease and had very little variation in mean lesion sizes in both years (Figure 7).
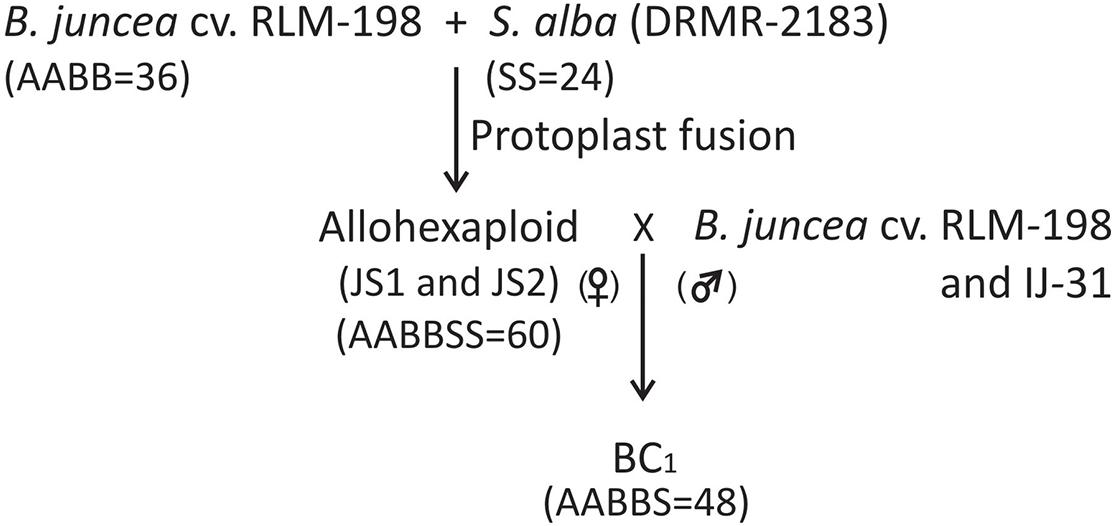
Figure 5. Flow chart diagram showing the development of allohexaploids and their first backcross progeny (BC1).
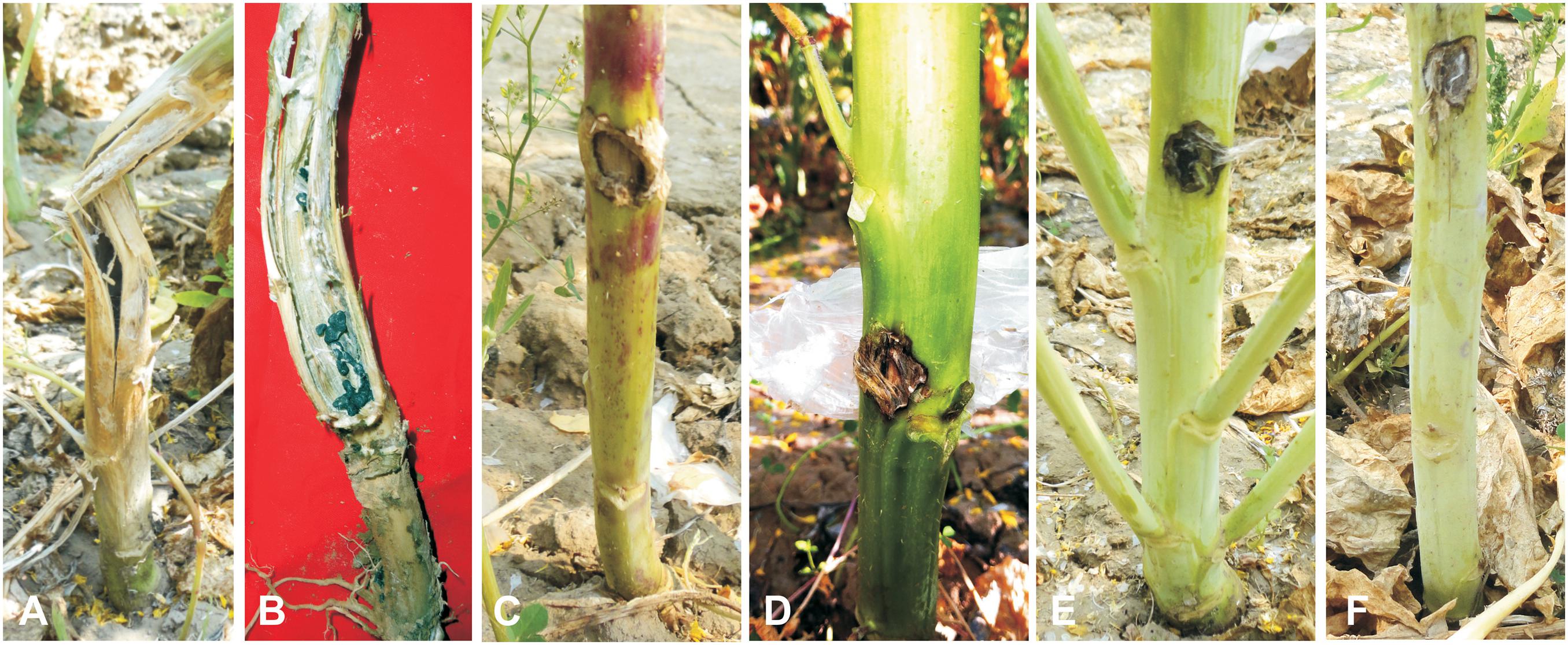
Figure 6. Screening against Sclerotinia sclerotiorum. (A,B) Sclerotinia rot symptoms on B. juncea stems as a susceptible check with sclerotia formation. (C) Inoculated plant of S. alba as a resistant check. (D) Allohexaploid JS1. (E) Allohexaploid JS2. (F) BC1 progeny of Allohexaploid JS1.
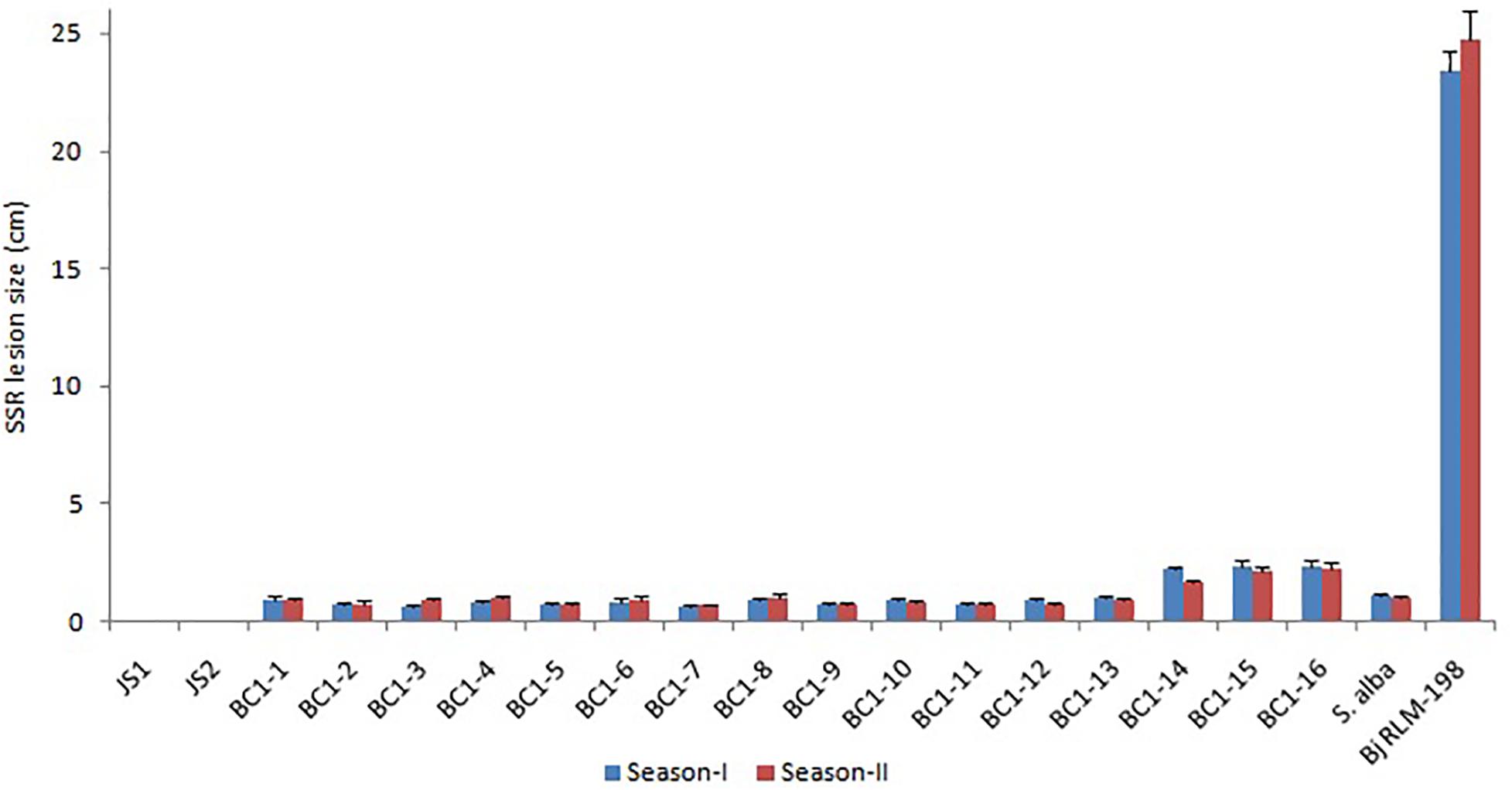
Figure 7. The Sclerotinia stem rot screening results of allohexaploids and their BC1 generation with parental checks during two crop seasons (Bar-StDv).
Inter-generic allohexaploid Brassicas are a potential source for gene introgression conferring resistance against major biotic and abiotic stresses of cultivated Brassica (Primard et al., 1988; Lelivelt et al., 1993; Hansen and Earle, 1997; Wang et al., 2005a, 2006; Kumari et al., 2018). The somatic hybridization is an approach to overcome problems associated with inter-generic incompatibility barriers and assists the inter-generic hybridization (Kirti et al., 1995).
There were two major impediments in successful introgression of the disease resistance from coenospecies to crop Brassicas by somatic hybridization i.e., poor regeneration frequency of fusion products and meiotic abnormalities in allohexaploids. A very low regeneration frequency (0.1 and 0.18–0.81%) was recorded after the protoplast fusion of B. napus and B. oleracea with S. alba by Primard et al. (1988) and Hansen and Earle (1997), respectively. Although, we have successfully overcome the limited regeneration by standardization of the regeneration medium and recorded a 5.9% regeneration frequency during our earlier study (Kumari et al., 2018). Further, significant improvement was observed with a 16.2% regeneration frequency from the protoplast fusion of S. alba and B. juncea in present experiments. The high regeneration frequency confirmed the advantage of standardization of pH after adding hormones into the regeneration medium. The lower male and female fertility were another constrain to move toward inter-generic allohexaploid development through somatic hybridization (Kirti et al., 1992, 1995). The multivalent formations were frequently observed at the meiosis of inter-generic allohexaploids, resulting in low male and female fertility (Primard et al., 1988; Wang et al., 2005b, 2006). Besides this, the male sterility has been recorded even after the predominant pairing at meiosis in allohexaploids of B. juncea + S. alba and resultant seed setting failed during self-pollination. Whereas, satisfactory seed setting was recorded only after backcrossing with B. juncea pollens and using allohexaploids as a female parent (Gaikwad et al., 1996). Nonetheless, we have observed a high male and female fertility after normal pairing and formation of 30 bivalents at diakinesis in three allohexaploids of B. juncea + S. alba (Kumari et al., 2018). We have also obtained high degree male and female fertility in the present two allohexaploids developed by using the Indian accession of S. alba (DRMR-2183) instead of the Australian accession which was used in our previous study. Gaikwad et al. (1996) confirmed that the allohexaploids of B. juncea and S. alba acquired chloroplast constitution from the S. alba. However, in the present study both allohexaploids (JS1 and JS2) acquired their chloroplasts from B. juncea, and the same chloroplast constitution was observed in our earlier three stable hybrids (Kumari et al., 2018). Thus, there might be a possibility that S. alba chloroplast causing male sterility in allohexaploids even after predominant pairing during meiosis. We have also found B. juncea type mitochondria in both allohexaploid (JS1 and JS2). Whereas, recombinant mitochondrial constitutions have been evident in our previous two allohexaploids (H1 and H3) out of three while H2 acquired their mitochondria from B. juncea (Kumari et al., 2018). The recombinant mitochondrial constitutions of their parents were reported frequently in inter-generic somatic hybrids (Kirti et al., 1992, Kirti et al., 1995; Lelivelt et al., 1993).
The inter-generic allohexaploids developed through somatic hybridization were often reported to possess intermediate morphology of parents presumably due to the presence of cumulative chromosome numbers of their parents (Kirti et al., 1992; Gaikwad et al., 1996; Wang et al., 2005a, 2006). Similarly, we have found intermediate morphology at some growth stage during our previous study like time to flower, while for other traits the allohexaploids were superior to their parents such as the presence of apparent trichomes on stems and leaves, vigorous plant growth, and branching patterns. Nonetheless, the seed color of all three allohexaploids (H1, H2, and H3) was brown similar to B. juncea parent (Kumari et al., 2018). However, in the present study, the intermediate phenotypes were observed such as seed color in JS2 (Half yellow and half brown), flowering initiation time, siliquae length, siliquae beak length, number of seeds per silique, and seed weight in both allohexaploids JS1 and JS2. Whereas other traits resembled their resistant donor parent (S. alba) such as the presence of trichomes on stems and leaves, yellow seed color of JS1. The plant height in both the allohexaploids was found more than their parents but much smaller than the earlier brown seeded (H1 and H2) allohexaploids which were also reported tolerant to high temperature (up to 40°C) during seed set.
Wang et al. (2005b) have observed inter-genomic association in first backcrossed (BC1) progeny of B. napus + S. alba allohexaploids with the sequidiploid chromosomal constitution (AACCS = 50) and observed 4 trivalent during meiosis. Similarly, we have found a haploid set of S. alba chromosomes (S = 12) in sequidiploid chromosome constitution (AABBS = 48) in the first backcross generation and observed normal pairing between the 48 chromosomes. We have observed 24 bivalents at the meiosis-I of BC1 and no univalent, trivalent and laggard chromosome was seen at meiotic metaphase. However, any precise study regarding autosyndesis in S. alba or allosyndesis within B. juncea and S. alba genomes are not available, some presumption comes from the present study and is supported by Gaikwad et al. (1996). Considering the phylogeny and the cytological literature and BLAST the B. juncea sequences with the draft genome of S. alba both genomes share 78% similar sequences. Therefore the chances for the inter-genomic association are greater, Although, further validation is required to confirm this point of view.
Yellow and half yellow and half brown (mottle) seed color has been recorded for the first time in allohexaploids developed through somatic hybridization and their BC1 generation. So far, the yellow seed color is also not recorded in somatic hybrids. However, the yellow seed color introgression has been observed after four rounds of successive backcrossing (BC4F4) in B. napus + S. alba somatic hybrids in addition to Sclerotinia stem rot resistance (Li et al., 2009). Although, the yellow seed varieties are available within B. juncea but preliminary reports have mentioned that the distinct testa color of S. alba differed from other yellow seeded germplasms available in the crop Brassicas. To the best of our knowledge, the introgression of the yellow seed color was not attempted from S. alba to B. juncea so far. Li et al. (2012) have identified a characteristic S. alba specific 288 bp amplicon responsible for yellow seed color in BC4F4 generation of B. napus + S. alba somatic hybrids. This amplicon sequence was not matched with B. napus genome assembly available in the National Center for Biotechnology Information. Thus, they have suggested that it might be a part of the S. alba-specific gene sequence. We have used this 288 bp sequence DNA sequence in the Basic Local Alignment Search Tool Nucleotide (BLASTN) with the S. alba draft genome assembly (Kumari et al., 2020a) and found three copies of approximately similar sequences in S. alba genome. Apart from this, we have studied the histology of physiologically mature seeds of both allohexaploids (yellow and mottled seeded). The palisade thickening of yellow seed was similar to S. alba, while the mottle seeds revealed that their palisade layer thickening closer to B. juncea which was agreed with the findings of Li et al. (2009) for yellow seeded progenies. However, further validation is required to identify the genes responsible for yellow and mottle seed colors. The subsequent self-generations and backcross progenies of the B. juncea + S. alba allohexaploids were found to be highly stable and resistant to Sclerotinia stem rot disease.
A total of 16 lines from BC1 derived from both allohexaploids with their parent used as checks were screened in the field by stem inoculation and gave an immune response to high degree resistance against a virulent isolate of S. sclerotiorum. A similar resistance response was recorded in BC4F4 progeny of B. napus and S. alba somatic hybrids for Sclerotinia stem rot resistance by in vitro leaf inoculation and found higher resistance than the known resistant rapeseed variety “Zhongshuang 9” (Li et al., 2009). Although, S. sclerotiorum pathogen is attacked on the stem and causing severe yield loss but no significant correlation was noticed between leaf and stem resistance. Taylor et al. (2015) concluded that the resistance in these two different tissues (stem and leaves) might be controlled by different genes. Similar findings were reported in ILs of B. juncea-B. fruticulosa, where highly resistant plants with lesion sizes less than 2.5 cm were recorded (Rana et al., 2017). Atri et al. (2019) identified 13 loci responsible for S. sclerotiorum resistance on seven different B. juncea chromosomes that were used in the set of B. juncea-B. fruticulosa ILs. In our study, the allohexaploids (JS1 and JS2) were found highly resistant to the S. sclerotiorum during three seasons (2015–2016 and 2017–2018 crop).
Thus, the stable allohexaploids of B. juncea + S. alba and their backcross progeny are developed in the present study, can provide the genetic resources for novel agronomically important traits and provides new insights into the genetic inheritance of traits such as the resistance to Sclerotinia stem rot and yellow seed color. Breeding for the introgression of these traits into another Brassica crop could be achieved as these allohexaploids are confirmed their cross ability with diploid and amphidiploids of cultivated Brassicas.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PK designed, conducted lab and field experiments, the data collection, preparation of cytological smears and GISH, and drafted, edited, and finalized the manuscript. KS contributed to seed coat developmental histology, conducted field studies, Sclerotinia sclerotiorum culture maintenance, screening stem rot resistance, the data collection and analysis, and drafted and finalized the manuscript. SK observed and photographed GISH slides, and provided the guidance and lab facilities for cytological studies and genomic in situ hybridization (GISH). DY provided the facility and guidance during phenotyping, and performed the manuscript editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Science and Engineering Research Board, New Delhi, India under FASTTRACK (F. No. SR/LS/068/2010) and the Department of Science and Technology, New Delhi, India under the WOS-A scheme concerning PK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. S. R. Bhat, who is an Emeritus Scientist of the National Institute for Plant Biotechnology, New Delhi, India for his guidance during the somatic hybridization. We acknowledging the kind help of Dr. Rajkumar, the Head of IARI-Regional Station, Katrain, India for providing the facilities to grow off-season crops for disease screenings and generation advancements. We also wish to acknowledge the ICAR-Indian Agriculture Research Institute, New Delhi for providing field facilities for conducting this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.575591/full#supplementary-material
Atri, C., Akhatar, J., Gupta, M., Gupta, N., Goyal, A., Rana, K., et al. (2019). Molecular and genetic analysis of defensive response of Brassica juncea-B. fruticulosa introgression lines to Sclerotinia infection. Sci. Rep. 9:17089. doi: 10.1038/s41598-019-53444-3
Bodnaryk, R. P., and Lamb, R. J. (1991). Mechanisms of resistance to the flea beetle, Phyllotretacruciferae (Goeze), in yellow mustard seedlings, Sinapis alba L. Can. J. Plant Sci. 71, 13–20. doi: 10.4141/cjps91-002
Brown, J., Brown, A. P., Davis, J. B., and Erickson, D. (1997). Inter-generic hybridization between Sinapis alba and Brassica napus. Euphytica 93, 163–168. doi: 10.1023/A1002905816887
Brun, H., Plessis, J., and Renard, M. (1987). Resistance of some Crucifers to Alternaria brassicae (Berk.) Sacc. Proc. 7th Int. Rapeseed Congr. Poznan 1222–1227.
Brun, H., Plessis, J., and Renard, M. (1988). “Resistance of some crucifers to Alternaria brassicae(Berk.) Sacc,” in Proceedings GCIRC 7th International Rapeseed Conference on Plant Breeding and Acclimatization Institute, Warsaw, 1222–1227.
Carnoy, J. B. (1887). Appendice les globule polaires de l’ascaris clavata. La Cell. Recueil Cytol. D’Histol. Gén. 3:276.
Chandler, J., Corbesier, L., Spielmann, P., Dettendorfer, J., Stahl, D., Apel, K., et al. (2005). Modulating flowering time and prevention of pod shatter in oilseed rape. Mol. Breed. 15, 87–94. doi: 10.1007/s11032-004-2735-4
Chen, S., Nelson, M. N., Chevre, A. M., Jenczewski, E., Li, Z., Mason, A. S., et al. (2011). Trigenomic bridges for Brassica improvement. Crit. Rev. Plant Sci. 30, 524–547.
Conn, K. L., Tewari, J. P., and Dahiya, J. S. (1988). Resistance to Alternaria brassicae and phytoalexins-elicitation in rapeseed and other crucifers. Plant Sci. 56, 21–25. doi: 10.1016/0168-9452(88)90180-x
Daun, J. K., and DeClercq, D. R. (1988). Quality of yellow and dark seeds inBrassica campestris canola varieties Candle and Tobin. J. Am. Oil Chem. Soc. 65, 122–126. doi: 10.1007/BF02542562
Downey, R. K., Stringham, G. R., McGregor, D. I., and Steffanson, S. (1975). Breeding rapeseed and mustard crops,” in Oilseed and Pulse crops in Western Canada, ed. J. T. Harapiak (Calgary: Western Cooperative Fertilize Ltd.), 157–183.
Downey, R. K. (1987). “Rapeseed and mustard,” in Principal of Cultivar Development, ed. W. R. Fehr (New York, NY: Macmillan Publishing Campany), 437–486.
Downey, R. K., and Röbbelen, G. (1989). Oilseed Crops of the World: Their Breeding and Utilisation, U.S.A. New York, NY: McGraw-Hill Publishing Company.
Flannery, M. L., Mitchell, F. J. G., Coyne, S., Kavanagh, T. A., Burke, J. I., Salamin, N., et al. (2006). Plastid genome characterization in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 113, 1221–1231. doi: 10.1007/s00122-006-0377-0
Gaikwad, K., Kirti, P. B., Prakash, S., and Chopra, V. L. (1996). Cytological and molecular investigations on somatic hybrids of Sinapis alba and Brassica juncea and their backcross progeny. Plant Breed. 115, 480–483. doi: 10.1111/j.1439-0523.1996.tb00961.x
Garg, H., Atri, C., Sandhu, P. S., Kaur, B., Benton, M., Banga, S. K., et al. (2010). High level of resistance of Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 117, 51–58. doi: 10.1016/j.fcr.2010.01.013
Gupta, M., Atri, C., Agarwal, N., and Banga, S. S. (2016). Development and molecular genetic characterization of a stable Brassica allohexaploid. Theor. Appl. Genet. 129, 2085–2100. doi: 10.1007/s00122-016-2759-2
Habard, D. J. (1976). Cytotaxonomic studies of Brassica and related genera,” in The Biology and Chemistry of the Cruciferae, eds J. G. Vaughan, A. J. Mcleod, and B. M. G. Jones (London: Academic Press), 47–68.
Hansen, L. N., and Earle, E. D. (1995). Transfer of resistance to Xanthomonas campestris pv.campestris(L.) by protoplast fusion. Theor. Appl. Genet. 91, 1293–1300. doi: 10.1007/bf00220944
Hansen, L. N., and Earle, E. D. (1997). Somatic hybrids between Brassica oleracea and Sinapis alba L. with resistance to Alternaria brassicae(Berk.) Sacc. Theor. Appl. Genet. 94, 1078–1085. doi: 10.1007/s001220050518
Kirti, P. B., and Chopra, V. L. (1990). Rapid plant regeneration through organogenesis and somatic embryogenesis from cultured protoplasts. Plant Cell Tissue Org. Cult. 20, 65–67. doi: 10.1007/bf00034759
Kirti, P. B., Narasimhulu, S. B., Prakash, S., and Chopra, V. L. (1992). Production and characterization of intergeneric somatic hybrids of Trachystoma ballii and Brassica juncea. Plant Cell Rep. 11, 90–92. doi: 10.1007/BF00235260
Kirti, P. B., Mohapatra, T., Khanna, H., Prakash, S., and Chopra, V. L. (1995). Diplotaxis catholica + Brassica juncea somatic hybrids: molecular and cytogenetic characterization. Plant Cell Rep. 14, 593–597.
Kolte, S. J. (1985). Diseases of Annual Edible Oilseed Crops, Vol-II Rapeseed-Mustard and Sesame Diseases. Boca Raton, FL: CRC Press, 135.
Kumar, P., Naresh, V., Srinivasan, R., and Bhat, S. R. (2012). An evolutionarily conserved mitochondrial orf108 is associated with cytoplasmic male sterility in different alloplasmic lines of Brassica juncea and induces male sterility in transgenic Arabidopsis thaliana. J. Exp. Bot. 63, 2921–2932. doi: 10.1093/jxb/err459
Kumari, P., Bisht, D. S., and Bhat, S. R. (2018). Stable, fertile somatic hybrids between Sinapis alba and Brassica juncea show resistance to Alternaria brassicae and heat stress. Plant Cell Tiss Organ Cult 133, 77–86. doi: 10.1007/s11240-017-1362-9
Kumari, P., Singh, K. P., Bisht, D., and Kumar, S. (2020b). Somatic hybrids of Sinapis alba and Brassica juncea: study of Backcross progenies for morphological variation, chromosome constitution and reaction to Alternaria brassicae. Euphytica 216, 93. doi: 10.1007/s10681-020-02629-3
Kumari, P., Singh, K. P., and Rai, P. K. (2020a). Draft genome of multiple resistance donor plant Sinapis alba: an insight into SSRs, annotations and phylogenetics. PLoS One 15:e0231002. doi: 10.1371/journal.pone.0231002
Lelivelt, C. L. C., Leunissen, E. H. M., Frederiks, H. J., Helsper, J. P. F. G., and Krens, F. A. (1993). Transfer of resistance to the beet cyst nematode (Heterodera schachtii Schm.) from Sinapis alba L. (white mustard) to the Brassica napus L. gene pool by means of sexual and somatic hybridization. Theor. Appl. Genet. 85, 688–696. doi: 10.1007/bf00225006
Li, A. M., Jiang, J., Zhang, Y., Snowdon, R. J., and Liang, G. (2012). Molecular and cytological characterization of introgression lines with yellow seed derived from somatic hybrids between Brassica napus and Sinapis alba. Mol. Breed. 29, 209–219. doi: 10.1007/s11032-010-9540-z
Li, A. M., Wei, C. X., Jiang, J. J., Zhang, Y. T., Snowdon, R. J., and Wang, Y. (2009). Phenotypic variation in the progeny of somatic hybrids between Brassica napus and Sinapis alba. Euphytica 170, 289–296. doi: 10.1007/s10681-009-9979-3
Mason, A. S., Yan, G., Cowling, W. A., and Nelson, M. N. (2012). A new method for producing allohexaploid Brassica through unreduced gametes. Euphytica 186, 277–287. doi: 10.1007/s10681-011-0537-4
Meng, J., Shi, S., Gan, L., Li, Z., and Qu, X. (1998). The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica 103, 329–333. doi: 10.1023/A:1018646223643
Mwathi, M. W., Gupta, M., Atri, C., Banga, S. S., Batley, J., and Mason, A. S. (2017). Segregation for fertility and meiotic stability in novel Brassica Allohexaploids. Theor. Appl. Genet. 130, 767–776. doi: 10.1007/s00122-016-2850-8
Mwathi, M. W., Gupta, M., Martinez, D. Q., Pradhan, A., Batley, J., and Mason, A. S. (2020). Fertile allohexaploid Brassica hybrids obtained from crosses between B. oleracea and B. juncea via ovule rescue and colchicine treatment of cuttings. Plant Cell Tissue Organ Cult. 140, 301–313. doi: 10.1007/s11240-019-01728-x
Mwathi, M. W., Schiessl, S. V., Batley, J., and Mason, A. S. (2019). “Doubled-haploid” allohexaploid Brassica lines lose fertility and viability and accumulate genetic variation due to genomic instability. Chromosoma 128, 521−532. doi: 10.1007/s00412-019-00720-w
Naresh, V., Singh, S. K., Watts, A., Kumar, P., Kumar, V., Rao, K. R. S. S., et al. (2016). Mutations in the mitochondrial orf108 render Moricandia arvensis restorer ineffective in restoring male fertility to Brassica oxyrrhina based cytoplasmic male sterile line of B. juncea. Mol. Breeding 36, 67–75.
Pradhan, A., Plummer, J. A., Nelson, M. N., Cowling, W. A., and Yan, G. (2010). Successful induction of trigenomic hexaploid Brassica from a triploid hybrid of B. napus L. and B. nigra (L.) Koch. Euphytica 176, 87–98. doi: 10.1007/s10681-010-0218-8
Primard, C., Vedel, F., Mathieu, C., Pelletier, G., and Chevre, A. M. (1988). Interspecific somatic hybridization between Brassica napus and Brassica hirta (Sinapis alba L.). Theor. Appl. Genet. 75, 546–552. doi: 10.1007/bf00289119
Rahman, M. H. (2001). Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed. 120, 463–472. doi: 10.1046/j.1439-0523.2001.00640.x
Rana, K., Atri, C., Akhatar, J., Kaur, R., Goyal, A., Singh, M. P., et al. (2019). Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea–Erucastrum cardaminoides introgression lines. Front. Plant Sci. 10:1015. doi: 10.3389/fpls.2019.01015
Rana, K., Atri, C., Gupta, M., Akhatar, J., Prabhjodh, S. S., Kumar, N., et al. (2017). Mapping resistance responses to Sclerotinia infestation in introgression lines of Brassica juncea carrying genomic segments from wild Brassicaceae B. fruticulosa. Sci. Rep. 7:5904. doi: 10.1038/s41598-017-05992-9
Ripley, V., Thorpe, M., Iler, S., Mizier, K., and Beversdorf, W. D. (1992). Isozyme analysis as a tool for introgression of Sinapis alba germplasm into Brassica napus. Theor. Appl. Genet. 84, 403–410. doi: 10.1007/bf00229500
Sharma, G., Kumar, V. D., Haque, A., Bhat, S. R., Prakash, S., and Chopra, V. L. (2002). Brassica coenospecies: a rich reservoir for genetic resistance to leaf spot caused by Alternaria brassicae. Euphytica 125, 411–417. doi: 10.1023/A:1016050631673
Sharma, T. R., and Singh, B. M. (1992). Transfer of resistance to Alternaria brassicae in Brassica juncea through interspecific hybridization among Brassica. J. Genet. Breed. 46, 373–378.
Slominski, B. A., Campbell, L. D., and Guenter, W. (1994). Oligosaccharides in canola meal and their effect on nonstarch polysaccharide digestibility and true metabolizable energy in poultry. Poult. Sci. 73, 156–162. doi: 10.3382/ps.0730156
Tang, Z. L., Li, J. N., Zhang, X. K., Chen, L., and Wang, R. (1997). Genetic variation of yellow-seeded rapeseed lines (Brassica napus L.) from different genetic sources. Plant Breeding 116, 411–414.
Taylor, A., Coventry, E., Jones, J. E., and Clarkson, J. P. (2015). Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in a Brassica napus diversity set. Plant Pathol. 64, 932–940. doi: 10.1111/ppa.12327
Wang, Y. P., Sonntag, K., Rudloff, E., and Chen, J. M. (2005a). Inter-generic somatic hybridization between Brassica napus and Sinapis alba. Integr. J. Plant Biol. 47, 84–91. doi: 10.1111/j.1744-7909.2005.00009.x
Wang, Y. P., Zhao, X. X., Sonntag, K., Wehling, P., and Snowdon, R. J. (2005b). Behaviour of Sinapis alba chromosomes in a Brassica napus background revealed by genomic in-situ hybridization. Chromosome Res. 13, 819–826. doi: 10.1007/s10577-005-1017-2
Wang, Y. P., Sonntag, K., and Rudloff, E. (2006). Production and characterization of somatic hybrids between Brassica napus and Raphanus sativus. Plant Cell Tiss. Organ Cult. 86, 279–283. doi: 10.1007/s11240-006-9118-y
Wang, R., Ripley, V. L., and Rakow, G. (2007). Pod shatter resistance evaluation in cultivars and breeding lines of Brassica napus, B. juncea and Sinapis alba. Plant Breed. 126, 588–595. doi: 10.1111/j.1439-0523.2007.01382.x
Warwick, S. I. (1993). Guide to the Wild Germplasm of Brassica and Allied Crops. Part IV. Wild Species in the Tribe Brassiceae (Cruciferae) as Sources of Agronomic Traits: Technical Bulletin 1993-17E. Ottawa, ON: Agriculture Canada Research Branch, 19.
Keywords: inter-generic somatic hybridization, allohexaploids brassica, genomic in situ hybridization, seed color, Sclerotinia sclerotiorum resistance, Sinapis alba
Citation: Kumari P, Singh KP, Kumar S and Yadava DK (2020) Development of a Yellow-Seeded Stable Allohexaploid Brassica Through Inter-Generic Somatic Hybridization With a High Degree of Fertility and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 11:575591. doi: 10.3389/fpls.2020.575591
Received: 23 June 2020; Accepted: 30 October 2020;
Published: 24 November 2020.
Edited by:
John M. Manners, The University of Queensland, AustraliaReviewed by:
Matthew Nicholas Nelson, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2020 Kumari, Singh, Kumar and Yadava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Preetesh Kumari, cHJlZXRlc2g3OUBnbWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.