- 1State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 2School of Biological Science, Jining Medical University, Rizhao, China
Nitrogen (N) is one of the most essential macronutrients for plant growth and development. Nitrate (NO3–), the major form of N that plants uptake from the soil, acts as an important signaling molecule in addition to its nutritional function. Over the past decade, significant progress has been made in identifying new components involved in NO3– regulation and starting to unravel the NO3– regulatory network. Great reviews have been made recently by scientists on the key regulators in NO3– signaling, NO3– effects on plant development, and its crosstalk with phosphorus (P), potassium (K), hormones, and calcium signaling. However, several novel aspects of NO3– regulation have not been previously reviewed in detail. Here, we mainly focused on the recent advances of post-transcriptional regulation and non-coding RNA (ncRNAs) in NO3– signaling, and NO3– regulation on leaf senescence and the circadian clock. It will help us to extend the general picture of NO3– regulation and provide a basis for further exploration of NO3– regulatory network.
Introduction
Nitrogen (N) is a macronutrient required by many important biological processes in plants and also a limiting factor for crop production in the agricultural system. Most crops such as maize and wheat absorb nitrate (NO3–) as the main N forms. NO3– uptake, transport, and assimilation are the principal processes for NO3– utilization and have long been studied. Besides its nutritional role, NO3– can also serve as a signaling molecule that modulates plant growth and development. During the last decade, great progress has been made on NO3– regulation in root system architecture, shoot growth, seed dormancy, flowering time, as well as crosstalk with other signals (hormone, calcium) and nutrients (P and K) (Footitt et al., 2011, 2013; Bouguyon et al., 2015, 2016; Riveras et al., 2015; Yan et al., 2016; Yuan et al., 2016; Li et al., 2017; Lin and Tsay, 2017; Gaudinier et al., 2018; Gras et al., 2018; Du et al., 2019; Hu et al., 2019; Medici et al., 2019). Some crucial components involved in NO3– signaling have been identified, such as ANR1, NRT1.1, NLP6/7, TCP20, LBD37/38/39, TGA1/4, SPL9, bZIP1, NAC4, NRG2, and NIGT/HRS1 (Zhang and Forde, 1998; Remans et al., 2006; Ho et al., 2009; Rubin et al., 2009; Krouk et al., 2010; Konishi and Yanagisawa, 2013; Marchive et al., 2013; Alvarez et al., 2014; Guan et al., 2014; Para et al., 2014; Vidal et al., 2014; Xu et al., 2016; Cao et al., 2017; Kiba et al., 2018; Maeda et al., 2018; Sun et al., 2018a, b; Zhao et al., 2018). These progress have been summarized in several excellent review papers (Gutiérrez, 2012; O’Brien et al., 2016; Mu and Luo, 2019; Jia and von Wirén, 2020; Liu et al., 2020; Luo et al., 2020; Vidal et al., 2020). However, the research on nitrate regulation at post-transcriptional levels is still lacking. Using nitrogen use efficiency (NUE) genes to breed high NUE cultivars is essential to sustain crop productivity. Several genes regulating NUE have been characterized in crops, such as OsNRT1.1B, OsNRT2.1, OsNRT2.3b, OsBT1/2, OsGRF4, OsDEP1, OsGCR1, OsGPA1, OsNAC42, ZmNAC7, TaNFYA, TaDOF1 (Sun et al., 2014; Chakraborty et al., 2015, 2019; Hu et al., 2015; Qu et al., 2015; Araus et al., 2016; Chen M. et al., 2016; Fan et al., 2017; Peña et al., 2017; Li et al., 2018; Liang et al., 2018; Sharma et al., 2018; Tang et al., 2019; Zhang et al., 2019; Pathak et al., 2020), which have also been well reviewed (Wang et al., 2018a; Zuluaga and Sonnante, 2019; Vidal et al., 2020).
Following the application of new technologies like multi-omics, post-transcriptional regulators of NO3– including the mRNA splicing factors CPSF30-L, FIP1, and non-coding RNAs (ncRNAs) have been characterized as new players in NO3– signaling (Zhao et al., 2011; Li et al., 2017; Wang et al., 2018b; Liu et al., 2019). In addition, novel functions and mechanisms of NO3– on plant development have been revealed. Scientists found that NRT1.5 and NLA (Nitrogen Limitation Adaptation) could repress N deficiency-induced leaf senescence (Meng et al., 2016; Liu et al., 2017). Regarding the effects of N on circadian clock, the center clock genes CCA1, LHY, and TOC1 have been found to play important roles (Gutiérrez et al., 2008; Yuan et al., 2016; Teng et al., 2017). In this review, we mainly focus on these advances in NO3– regulation. It will provide more insights to better understand the complexity and mechanism of the NO3– regulation and lay a fundamental base for further deciphering the NO3– regulatory network and improving NUE in agriculture.
Post-transcriptional Regulation of Nitrate
The importance of post-transcriptional modifications of genes associating with a wide range of plant responses has been already described in plants (Kim et al., 2008; Kwon et al., 2009; Berr et al., 2010; Nelissen et al., 2010). However, the role of post-transcriptional modifications in N regulation is still poorly understood. The factor IWS1 has been shown to be involved in several aspects of transcription, like transcription elongation, recruitment of chromatin-remodeling factors, mRNA processing and export, and post-transcriptional histone modification (Krogan et al., 2002; Yoh et al., 2007, 2008; Zhang et al., 2008). Lepetit’s group found that IWS1 represses NRT2.1 transcription in response to high N supply, which is associated with an IWS1-dependent accumulation of histone H3 lysine 27 trimethylation (H3K27me3) (Widiez et al., 2011). The result suggests that IWS1 represses gene expression by specifically promoting H3K27me3 methylation under high N conditions. This is the first evidence demonstrating that post-transcriptional chromatin modifications regulate N acquisition. More recently, Coruzzi’s group reported that histone methyltransferase SET DOMAIN GROUP8 (SDG8) mediates genome-wide changes of H3K36me3 after NO3– treatments, resulting in the altered expression of its target genes, RNA processing, and physiological responses (Li et al., 2020). More epigenetic marks, such as other histone modifications (e.g., histone acetylation), chromatin landscape, and DNA methylation need to be investigated in the future to gain a comprehensive understanding of chromatin regulation in NO3– signaling.
RNA processing is important for the post-transcriptional regulation of gene expression, and is a rate-limiting step in the expression of proteins. The CPSF30 gene encodes the cleavage and polyadenylation specificity factor, which forms a larger protein (CPSF30-L) with 65 kDa and a smaller protein (CPSF30-S) with 28 kDa. Both proteins possess three characteristic C3H zinc finger motifs and act as an RNA-binding protein and an endonuclease (Addepalli and Hunt, 2007). The CPSF30-L contains an additional YT512-B Homology (YTH) domain along with the three zinc finger motifs (Li et al., 2017). By using forward genetics, the CPSF30-L gene was cloned and found that it plays a crucial role in NO3– signaling, transport, and assimilation in Arabidopsis (Li et al., 2017). Molecular and genetic assays revealed that CPSF30-L works upstream and regulates the expression of NRT1.1, while it functions independently of NLP7 in NO3– signaling. FIP1, another RNA-binding protein, is also a core component of a pre-mRNA processing complex (Forbes et al., 2006). Further research showed that FIP1 interacts with CPSF30-L and regulates NO3– signaling and assimilation (Wang et al., 2018b). Both CPSF30-L and FIP1 modulate the expression of NO3– regulatory genes CIPK8 and CIPK23 and influence the alternative polyadenylation (APA) of the 3′-UTR of NRT1.1 mRNA (Li et al., 2017; Wang et al., 2018b). These results indicate that CPSF30-L and FIP1 may modulate NO3– signaling through influencing the APA processing of NRT1.1.
RNA can be chemically modified with many different reactions, among which methylation is one of the most well studied modification. N6-methyladenosine (m6A) is a pivotal internal mRNA modification, which plays crucial roles in plant growth and development by regulating gene expression at the post-transcriptional level. Recently, it has been reported that the EVOLUTIONARILY CONSERVED C-TERMINAL REGION2 (ECT2) acts as an m6A reader depending on the presence of the YTH domain (Wei et al., 2018; Yue et al., 2019). As CPSF30-L contains an YTH domain, whether it also serves as the m6A reader to regulate NO3– signaling needs further investigation.
There are over 100 types of RNA modifications (Roundtree et al., 2017; Bailey-Serres et al., 2020), yet our knowledge about their occurrence and functions are still limited. One thing is certain though: post-transcriptional regulation of gene expression is much more intricate than previously thought. Unraveling the functions and underlying mechanisms of RNA modifications are essential to understand the roles of NO3– regulatory genes and will provide valuable insights into post-transcriptional regulation of NO3–.
Nitrate and ncRNAs
Whole-genome sequencing analyses have identified thousands of ncRNAs playing vital roles in numerous biological processes in plants (Laporte et al., 2007; Rymarquis et al., 2008). Several microRNAs (miRNAs) and a long ncRNA (lncRNA) have been reported to function in NO3– regulation in Arabidopsis and crops (Miin-Feng et al., 2006; Vidal et al., 2010; Liu et al., 2019; Table 1). In Arabidopsis, miR167 targets two auxin responsive factors ARF6 and ARF8. The miR167/ARF8 module regulates lateral root growth in response to NO3– by controlling a group of NO3–-responsive genes (Miin-Feng et al., 2006; Gifford et al., 2008). The miR393 and AFB3 can respond to NO3– by integrating internal organic N signals, external NO3– availability and root auxin sensitivity to control root architecture (Vidal et al., 2010). Upon N starvation, the expression of miR160, miR164, miR167, miR780, miR826, miR842, miR846, and miR5090 is induced, whereas the expression of miR169, miR171, miR395, miR397, miR398, miR399, miR408, miR827, and miR857 is repressed (Xu et al., 2011; Liang et al., 2012; He et al., 2014; Li et al., 2016). Among them, miR160, miR167, and miR171 can be responsible for the development of root systems under N-starvation conditions (Liang et al., 2012). Moreover, miR826 and miR5090 suppress the expression of ALKENYL HYDROXALKYL PRODUCING 2 (AOP2), which is involved in the modification of the glucosinolate side chain. Overexpression of miR826 and miR5090 results in less glucosinolate and enhanced tolerance to N starvation (He et al., 2014). Overexpression of miR169 reduces N uptake and total N content by decreasing the expression of NRT1.1 and NRT2.1 and thus promotes N starvation-induced early leaf senescence (Zhao et al., 2011; Figure 1A). In addition, the miR5640 post-transcriptionally regulates the expression of its target AtPPC3 in response to NO3– (Vidal et al., 2013). The N-responsive miRNA miR156 works upstream of SPL9, a SBP-box transcription factor (TF) identified to control the genes involved in the NO3– primary response (Krouk et al., 2010), implying that the miR156/SPL9 module functions in NO3– regulation. MiRNAs have also been found to be responsive to N limitation in crops (Xu et al., 2011; Trevisan et al., 2012). MiR169 is up-regulated in maize but down-regulated in rice under N starvation condition (Xu et al., 2011; Li et al., 2016). Two novel putative miR169 species miRC10 and miRC68 are reported to play major roles in the adaptation to NO3– limitation in maize seedlings (Zhao et al., 2013). Furthermore, OsmiR444a plays multiple roles including NO3–-dependent root growth, NO3– accumulation and phosphate-starvation responses in rice (Yan et al., 2014). Chengcai Chu’s group reported that OsmiR3979 functions as a key regulator to optimize root growth in response to NO3– (Li et al., 2016). Moreover, OsmiR159a.1 can be strongly repressed by ammonium, which is opposite to its response to NO3–. It targets LOC_Os06g40330 and LOC_Os01g59660, encoding MYB family TFs, and cleaves them at the predicted cleavage sites (Jeong et al., 2011; Li et al., 2016). Those results suggest that miRNAs play critical roles in NO3– regulation by modulating the expression of their target genes.
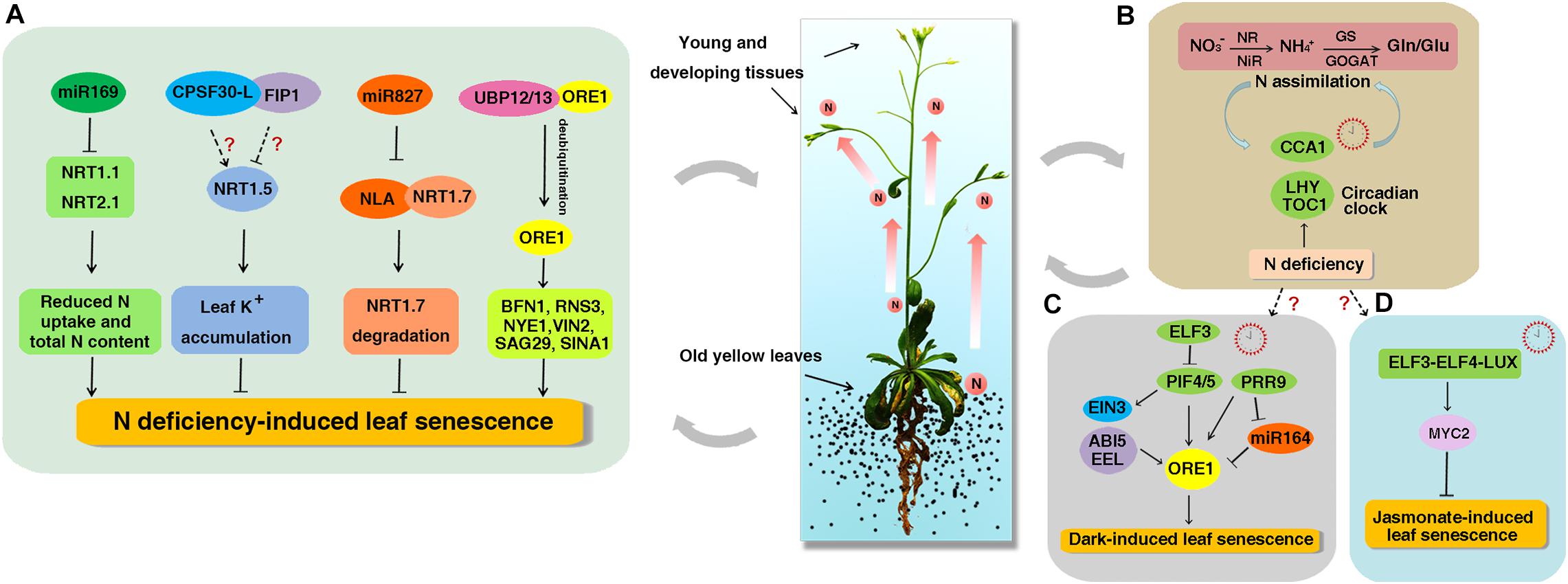
Figure 1. The working model of NO3− regulation on leaf senescence and circadian clock. Leaf senescence accompany with N reallocation to meet the demand of N in young and developing tissues. (A) The miR169 reduces the N uptake and total N content by decreasing the expression of NRT1.1 and NRT2.1 and promotes the early senescence induced by N starvation. NRT1.5 increases foliar K+ levels to suppress NO3− starvation-induced leaf senescence. CPSF30-L interacts with FIP1 and both can regulate the expression of NRT1.5, but if both genes participate in NO3− starvation-induced leaf senescence remains unknown. As a target of miR827, NLA directly interacts with NRT1.7 and promotes its degradation to repress leaf senescence. UBP12 and UBP13 interact with ORE1 and promote its deubiquitination to prevent ORE1 from degradation. The increased ORE1 levels activate the expression of downstream SAGs such as BFN1, RNS3, NYE1, VNI2, SAG29, and SINA1 to promote N deficiency-induced leaf senescence. (B) A feedback loop between N assimilation and circadian clock. Organic N influences the CCA1 phase and CCA1 in turn regulates the expression of organic N-regulated genes. N deficiency condition can increase the amplitudes of CCA1, LHY, and TOC1 transcripts. (C) ELF3 inhibits the expression of PIF4/PIF5 to repress dark-induced leaf senescence. PIF4 and PIF5 promote dark-induced senescence by directly activating the expression of ORE1 and indirectly activating the expression of EIN3, ABI5, and EEL. PRR9 can activate the expression of ORE1 directly or suppress the expression of miR164 to promote dark-induced leaf senescence. (D) The circadian evening complex (ELF3-ELF4-LUX) directly binds to the promoter of MYC2 to repress jasmonate-induced leaf senescence. However, if circadian clock is involved in N deficiency-induced senescence remains unknown. The solid and dotted arrows represent known and unknown functions, respectively.
Long non-coding RNAs are a class of ncRNAs with a length longer than 200 nucleotides and poor protein-coding potential (Pang et al., 2006; Ponting et al., 2009). A multitude of lncRNAs have been identified by using next-generation sequencing during the last several years, but only a few have been characterized (Xin et al., 2011; Liu et al., 2012; Meng et al., 2012). It has been reported that lncRNAs play crucial roles in flowering, phosphate starvation, polar auxin transport, photomorphogenesis, mRNA alternative splicing, and plant immune response (Francozorrilla et al., 2007; Amor et al., 2009; Swiezewski et al., 2009; Heo and Sung, 2011; Bardou et al., 2014; Seo et al., 2017). Several papers described changes of lncRNA responding to N treatments or N deficiency in Populus and maize (Vidal et al., 2013; Alvarez et al., 2014; Chen M. et al., 2016; Lv et al., 2016). However, the role of lncRNAs in N regulation remains largely unclear. Our group found six NO3–-induced lncRNAs using RNA-seq technology and validated by qRT-PCR. Further investigation revealed that the TCONS_00005120 (T5120), showing the highest induction by NO3–, could regulate NO3– response and assimilation. Molecular and genetic assays showed that NLP7 directly binds to the promoter of T5120 to regulate its expression. In addition, the expression of T5120 was modulated by NRT1.1 (Liu et al., 2019). This is the first attempt to investigate the role of lncRNAs in NO3– signaling and provides a fundamental base for discovering more lncRNAs functioning in NO3– regulation.
Nitrate and Leaf Senescence
Leaf senescence is a developmental process accompanying resource rearrangement (Havé et al., 2017). It occurs in a coordinated manner, starting with the inhibition of leaf expansion, and then the induction of metabolic changes followed by degradation of organic substances and nutrient remobilization (Soltabayeva et al., 2018). Some environmental stresses can induce early leaf senescence, among which N deficiency tends to promote leaf senescence to meet the demand of N in young and developing tissues (Aguera et al., 2010).
Although it is previously known that N deficiency can accelerate leaf senescence (Schildhauer et al., 2008; Gregersen et al., 2013), the underlying mechanisms are still poorly understood. It has been found that NRT1.5 functions in transporting NO3– from the roots to the shoots (Lin et al., 2008) and the expression of NRT1.5 is strongly induced by leaf senescence in Arabidopsis (Van Der Graaff et al., 2006). Jiming Gong’s lab has recently reported that NRT1.5 suppresses the leaf senescence induced by NO3– (but not nitrogen) starvation (Meng et al., 2016) and this suppression is independent of the NO3– transport function of NRT1.5. Further analyses showed that foliar K+ levels decreased in nrt1.5 when exposed to NO3– starvation and adding K+ could restore the early leaf senescence phenotype of nrt1.5 plants, suggesting that NRT1.5 may increase foliar K+ levels to suppress NO3– starvation-induced leaf senescence (Meng et al., 2016; Figure 1A). Lately, it was found that the expression of NRT1.5 could be positively modulated by CPSF30-L (Li et al., 2017) and suppressed by FIP1 (Wang et al., 2018b). Therefore, it would be interesting to investigate if both genes participate in NO3– starvation-induced leaf senescence (Figure 1A).
Nitrogen limitation adaptation encodes a RING E3 ubiquitin ligase. A previous study showed that the nla mutant could not accumulate anthocyanin and instead exhibited early senescence phenotype under N deficiency condition (Peng et al., 2008). Recently, Wenxue Li’s group confirmed this result and found that the content of 15N in the young leaves of nla mutant is significantly higher than that of wild-type (WT), indicating that NLA decreases translocation of N from the old leaves to the young leaves (Liu et al., 2017). It has been reported that NRT1.7 works in transporting NO3– from the old to young leaves as less 15NO3 spotted onto old leaves is remobilized to young leaves in nrt1.7 mutant compared with WT (Fan et al., 2009). Further investigation demonstrated that NLA directly interacts with NRT1.7 and promotes NRT1.7 degradation in vivo to repress early senescence. The NLA gene is a target of miRNA827. N limitation strongly repressed the NLA protein abundances in WT, while the loss of function of miRNA827 abolished the repression of NLA protein and down-regulated the NRT1.7 protein abundance under N limitation condition, indicating that N deficiency-induced repression of NLA is dependent on miRNA827 (Liu et al., 2017; Figure 1A).
An extensive reprogramming of gene expression has been found to be involved in leaf senescence to meet the complex biochemical and structural changes. Up to now, about 100 TFs that belong to about 20 different families (in particular, bZIP, WRKY, NAC, C2H2 zinc finger, MYB, and AP2-EREBP) have been described to play important roles in leaf senescence (Guo et al., 2004; Buchanan-Wollaston et al., 2005; Ay et al., 2014). ORESARA1 (ORE1, also known as NAC92) is a key factor that promotes age-dependent leaf senescence by regulating the expression of hundreds of senescence-associated genes (SAGs). These SAGs function mainly in the breakdown of nucleic acids and proteins, and the transport of sugar in Arabidopsis (Balazadeh et al., 2010a; Matallana-Ramirez et al., 2013). Nam-Hai Chua’s group showed that ORE1 interacts with NLA and UBP12/13 (ubiquitin-specific protease 12/13) to regulate leaf senescence (Park et al., 2014, 2018, 2019). Under N sufficient condition, ORE1 is polyubiquitinated by NLA and its E2 conjugase PHO2 (UBC24), and then degraded by 26S proteasomes leading to delayed leaf senescence (Park et al., 2018). But when plants were exposed to N-deficient conditions, UBP12/UBP13 antagonize the action of NLA to maintain ORE1 homeostasis by promoting the deubiquitination of ORE1 (Park et al., 2019). The elevated ORE1 levels activate the expression of downstream SAGs such as BFN1, RNS3, NYE1, VNI2, SAG29, and SINA1 resulting in early leaf senescence (Park et al., 2019; Figure 1A). Those results provide us the post-translational regulation of N deficiency-induced leaf senescence. In addition, miR164 regulates age-induced leaf senescence by targeting ORE1 mRNA cleavage (Kim et al., 2009; Balazadeh et al., 2010b). But miR164 does not regulate the expression of ORE1 at transcriptional levels under N-starvation condition (Liang et al., 2015; Park et al., 2018).
HY5 is a key regulator of photomorphogenesis (Lee et al., 2007). About 30% of the senescence-related regulatory genes are predicted as putative targets of the HY5, such as AP2-EREBP, NAC, and WRKY (Breeze et al., 2011; Ay et al., 2014), suggesting that HY5 may play an important role in the regulatory network of leaf senescence. Interestingly, HY5 has been found to activate the expression of NIA1, NIA2, and NRT2.1, but inhibit the expression of NRT1.1 (Jonassen et al., 2009a, b; Chen X. et al., 2016), implying that HY5 may be involved in NO3– regulation. Recently, the light-signaling protein FAR-RED ELONGATED HYPOCOTYL3 (FHY3) was reported to negatively regulate salicylic acid (SA) biosynthesis and light-mediated leaf senescence (Tian et al., 2020). Both the fhy3 mutant and WRKY28-overexpressing plants exhibited early leaf senescence under high (Red/Far-Red) R: FR light conditions. FHY3 could directly bind to the promoter of WRKY28 to repress its expression. These results indicate that the FHY3–WRKY28 module prevents leaf senescence under high R: FR light conditions (Tian et al., 2020). It was reported previously that FHY3 and HY5 physically interact with each other and co-regulate the expression of CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1), a multifunctional E3 ubiquitin ligase, in response to photomorphogenic UV-B (Huang et al., 2012). Further investigation is needed to determine if FHY3, COP1, and HY5 function in N starvation-induced leaf senescence.
Although N starvation induces senescence in plants, it has also been reported that senescence is reversible in some cases after the resupply of N (Schildhauer et al., 2008). Mueller-Roeber’s group further performed transcriptome analysis to investigate the N resupply induced reversal of senescence in Arabidopsis. Their results showed that the senescence program was tuned by the N status, indicating that plants undergoing senescence retain the capacity to sense and respond to the N availability (Balazadeh et al., 2014). Crops such as maize (Girardin et al., 1985), rice (Wang et al., 2012), or barley (Schildhauer et al., 2008), have also been found to have the ability to stop or even reverse the senescence induced by N deficiency when sufficient N is resupplied. However, the underlying mechanism still remains unclear. It is nevertheless evolutionarily important to have a regulatory system capable of monitoring and integrating such environmental stimuli.
Nitrate and Circadian Clock
The circadian clock is one of the most central endogenous factors that allow organisms to synchronize internal biological activities with the external environment. There is increasing evidence that circadian clock regulates N assimilation (Tucker et al., 2004; Cookson et al., 2005; Gutiérrez et al., 2008; Teng et al., 2017). It has been shown that organic N such as Glu or Gln can serve as signals to regulate the expression of N-related genes in plants (Oliveira and Coruzzi, 1999). Coruzzi’s group performed a transcriptome analysis on plants treated with organic N (Gutiérrez et al., 2008). By using systems biology, they found that the center clock gene CCA1 could regulate the expression of organic N-regulated genes. Further investigation revealed that CCA1 could directly bind to the promoters of the glutamine synthetase gene (GLN1.3) and the glutamate dehydrogenase gene (GDH1) to regulate their expression. Moreover, the phase response curve analysis exhibited that organic N could influence the CCA1 phase. These data indicate that organic N may regulate circadian rhythms through CCA1-mediated mechanism (Gutiérrez et al., 2008). Another group found that low N conditions increased the amplitudes of CCA1, LHY, and TOC1 transcripts throughout the circadian cycle, whereas high N conditions decreased the amplitudes of these genes in WT plants (Yuan et al., 2016). Further study showed that CCA1 influence nitrate reductase activity by binding to the CCA1-binding site in the promoters of NIA1 and NIA2 in vitro and activating the NIA1 expression while repressing NIA2 expression (Teng et al., 2017). These results provide basic insight for the link between N and circadian clock (Figure 1B).
It has been reported that TCP20 binds to the promoters of NIA1, NRT1.1, NRT2.1 and acts in systemic NO3– signaling that directs NO3– foraging in Arabidopsis roots (Guan et al., 2014). Furthermore, TCP20 can physically interact with NLP6/7 and ARF8, and their interactions are required for activating the expression of the G2/M cell-cycle marker gene CYCB1;1 to regulate lateral root initiation and growth (Guan et al., 2017; Fan et al., 2019a, b). Interestingly, TCP20 has also been found to be a circadian clock factor that can interact with LIGHT-REGULATED WD1 (LWD1). Both TCP20 and LWD1 can bind to the promoter of CCA1 to activate its expression (Wu et al., 2016). These results imply that TCP20 may be involved in connecting the N signaling with circadian clock.
During last several years, scientists have demonstrated the roles of the circadian clock genes in the regulation of leaf senescence in plants (Sakuraba et al., 2014; Song et al., 2014; Kim et al., 2018; Zhang et al., 2018). PIF4/PIF5 promote dark-induced leaf senescence by directly activating the expression of ORE1 and indirectly activating the expression of ETHYLENE INSENSITIVE 3 (EIN3) in ethylene signaling and bZIP factors ABA INSENSITIVE 5 (ABI5) and ENHANCED EM LEVEL (EEL) in ABA signaling (Sakuraba et al., 2014; Song et al., 2014). ELF3 inhibits leaf senescence by repressing the activity of PIF4/PIF5 (Sakuraba et al., 2014; Figure 1C). The transcript levels of ORE1 and its repressor miR164 show circadian rhythmic patterns (Kim et al., 2018). PRR9 can activate the expression of ORE1 directly or suppress the expression of miR164 to promote dark-induced leaf senescence (Kim et al., 2018; Figure 1C). In addition, elf4 and lux show obviously early leaf senescence than WT in darkness (Sakuraba et al., 2014; Kim et al., 2018). The circadian evening complex (ELF3-ELF4-LUX) can repress jasmonate-induced leaf senescence by directly binding to the promoter of MYC2, a key activator of jasmonate-induced leaf senescence (Zhang et al., 2018; Figure 1D). Ubiquitin-specific protease genes UBP12 and UBP13 are regulated by circadian clock and they in turn regulate circadian clock and photoperiodic flowering through GI and CO, which extends our understanding of deubiquitination in circadian clock and photoperiodic flowering regulation at posttranslational level (Cui et al., 2013). These results demonstrate a strong interplay between the circadian clock and leaf senescence. However, if circadian clock is involved in N deficiency-induced senescence remains unknown.
Conclusions and Perspectives
During last several years, research on NO3– regulation has expanded to multiple regulatory levels such as post-transcriptional modulations and also to some new developmental processes such as leaf senescence and circadian clock. The progress on these fields has started to shed light on better understanding the complex NO3– regulatory network. However, we still lack an integrated view on how NO3– regulates these processes and the full picture of NO3– regulatory network is far from complete.
Post-transcriptional regulation of NO3– has attracted more attention recently and gained further insights into NO3– regulation. The C3H-type zinc finger motif-containing protein CPSF30-L has been characterized to function in NO3– signaling (Li et al., 2017). The C3H-type zinc finger protein family contains 68 members and whether other members of this family participate in NO3– regulation needs to be investigated. Although ncRNAs have been found to be involved in the regulation of multiple biological processes, the underlying mechanisms still remain elusive, especially in NO3– signaling. The leaf senescence is finely tuned by N status. However, our understanding of the mechanisms that N availability affects leaf senescence is fragmentary and more efforts are still needed. One interesting avenue for future research is to screen mutants without early senescence symptoms under N-deficient conditions to identify the key components. In addition, systems biology may be used to globally reveal novel regulatory genes involved in N deficiency-induced leaf senescence. The core circadian genes CCA1, LHY, TOC1 are the main factors that connect circadian clock and NO3– regulation reported so far. Further studies are needed to find out more genes involved in circadian clock and NO3– regulation and explore their intrinsic mechanisms. The application of translatomics, structural biology and other new technologies will surely promote the clarification of the mechanisms that NO3– regulates plant development. Notably, the major part of the results therein resumed have been obtained in Arabidopsis, which although being an excellent model for studying NO3– signaling and providing useful fundamental knowledge for the scientific community, is quite different from crops (especially monocots), for which specific and targeted research projects are required. With the identification of more novel NO3– regulators and decryption of NO3– regulatory networks, as well as combined with scientific and rational fertilization management, we will effectively improve NUE and reduce the environmental pollution caused by loss of N fertilizers.
Author Contributions
HF and YW wrote the manuscript. SXQ, SDQ, and NX provided assistance for further modification of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31970270), the Taishan Scholar Foundation, Funds of Shandong “Double Tops” Program (grant no. SYL2017YSTD01), National Science and Technology Major Projects of China (2016ZX08003005-009) to YW, Natural Science Foundation of Shandong Province (ZR2017BC081), and National Natural Science Foundation of China (31700211) to NX.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
N, nitrogen; NO3–, nitrate; P, phosphorus; K, potassium; NUE, nitrogen use efficiency; WT, wild-type; T5120, TCONS_00005120; AOP2, ALKENYL HYDROXALKYL PRODUCING 2; NLA, nitrogen limitation adaptation; ORE1, ORESARA1; SAGs, senescence-associated genes; UBP12/13, ubiquitin-specific protease 12/13; FHY3, far-red elongated hypocotyl 3; SA, salicylic acid; COP1, constitutive photomorphogenesis1; EIN3, ETHYLENE INSENSITIVE 3; ABI5, ABA INSENSITIVE 5; EEL, ENHANCED EM LEVEL; GLN1.3, glutamine synthetase; GDH1, glutamate dehydrogenase; LWD1, LIGHT-REGULATED WD1; H3K27me3, histone H3 lysine 27 trimethylation; YTH, YT512-B Homology; ECT2, EVOLUTIONARILY CONSERVED C-TERMINAL REGION2; APA, alternative polyadenylation; m6A, N6-methyladenosine; miRNAs, microRNAs; ncRNAs, non-coding RNAs; lncRNAs, long non-coding RNAs.
References
Addepalli, B., and Hunt, A. G. (2007). A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucl. Acids Res. 35, 4453–4463. doi: 10.1093/nar/gkm457
Aguera, E., Cabello, P., and de la Haba, P. (2010). Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol. Plant. 138, 256–267. doi: 10.1111/j.1399-3054.2009.01336.x
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-Lopez, O., Tamayo, K. P., et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13. doi: 10.1111/tpj.12618
Amor, B. B., Wirth, S., Merchan, F., Laporte, P., D’Aubenton-Carafa, Y., Hirsch, J., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19, 57–69. doi: 10.1101/gr.080275.108
Araus, V., Vidal, E. A., Puelma, T., Alamos, S., Mieulet, D., Guiderdoni, E., et al. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171, 1523–1532. doi: 10.1104/pp.15.01731
Ay, N., Raum, U., Balazadeh, S., Seidensticker, T., Fischer, A., Reuter, G., et al. (2014). Regulatory factors of leaf senescence are affected in Arabidopsis overexpressing the histone methyltransferase SUVH2. J. Plant Growth Regul. 33, 119–136. doi: 10.1007/s00344-013-9384-y
Bailey-Serres, J., Zhai, J., and Seki, M. (2020). The dynamic kaleidoscope of RNA biology in plants. Plant Physiol. 182, 1–9. doi: 10.1104/pp.19.01558
Balazadeh, S., Schildhauer, J., Araújo, W. L., Munné-Bosch, S., Fernie, A. R., Proost, S., et al. (2014). Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomics consequences. J. Exp. Bot. 65, 3975–3992. doi: 10.1093/jxb/eru119
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana-Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010a). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Balazadeh, S., Wu, A., and Mueller-Roeber, B. (2010b). Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 5, 733–735. doi: 10.4161/psb.5.6.11694
Bardou, F., Ariel, F., Simpson, C., Romero-Barrios, N., Laporte, P., Balzergue, S., et al. (2014). Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30, 166–176. doi: 10.1016/j.devcel.2014.06.017
Bardou, F., Ariel, F., Simpson, C., Romero-Barrios, N., Laporte, P., Balzergue, S., et al. (2014). Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30, 166–176. doi: 10.1016/j.devcel.2014.06.017
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T., Shen, W. H., et al. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414. doi: 10.1104/pp.110.161497
Bouguyon, E., Brun, F., Meynard, D., Kubeš, M., Pervent, M., Leran, S., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1:15015. doi: 10.1038/NPLANTS.2015.15
Bouguyon, E., Perrine-Walker, F., Pervent, M., Rochette, J., Cuesta, C., Benkova, E., et al. (2016). Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol. 172, 1237–1248. doi: 10.1104/pp.16.01047
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P. O., Nam, H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585. doi: 10.1111/j.1365-313X.2005.02399.x
Cao, H., Qi, S., Sun, M., Li, Z., Yang, Y., Crawford, N. M., et al. (2017). Overexpression of the maize ZmNLP6 and ZmNLP8 can complement the Arabidopsis nitrate regulatory mutant nlp7 by restoring nitrate signaling and assimilation. Front. Plant Sci. 8:1703. doi: 10.3389/fpls.2017.01703
Chakraborty, N., Kanyuka, K., Jaiswal, D. K., Kumar, A., Arora, V., Malik, A., et al. (2019). GCR1 and GPA1 coupling regulates nitrate, cell wall, immunity and light responses in Arabidopsis. Sci. Rep. 9:5838. doi: 10.1038/s41598-019-42084-2
Chakraborty, N., Sharma, P., Kanyuka, K., Pathak, R. R., Choudhury, D., Hooley, R., et al. (2015). G-protein α-subunit (GPA1) regulates stress, nitrate and phosphate response, flavonoid biosynthesis, fruit/seed development and substantially shares GCR1 regulation in A. thaliana. Plant Mol. Biol. 89, 559–576. doi: 10.1007/s11103-015-0374-2
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT 2.1 expression with the OsNAR 2.1 promoter. Plant Biotech. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Chen, M., Wang, C., Bao, H., Chen, H., and Wang, Y. (2016). Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genomics 291, 1663–1680. doi: 10.1007/s00438-016-1210-3
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., and Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Cookson, S. J., Williams, L. E., and Miller, A. J. (2005). Light-dark changes in cytosolic nitrate pools depend on nitrate reductase activity in Arabidopsis leaf cells. Plant Physiol. 138, 1097–1105. doi: 10.1104/pp.105.062349
Cui, X., Lu, F., Li, Y., Xue, Y., Kang, Y., Zhang, S., et al. (2013). Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906. doi: 10.1104/pp.112.213009
Du, X. Q., Wang, F. L., Li, H., Jing, S., Yu, M., Li, J., et al. (2019). The transcription factor MYB59 regulates K+/NO3– translocation in the Arabidopsis response to low K+ stress. Plant Cell 31, 699–714. doi: 10.1105/tpc.18.00674
Fan, H. M., Liu, B. W., Ma, F. F., Sun, X., and Zheng, C. S. (2019a). Proteomic profiling of root system development proteins in chrysanthemum overexpressing the CmTCP20 gene. Plant Sci. 287:110175. doi: 10.1016/j.plantsci.2019.110175
Fan, H. M., Sun, C. H., Wen, L. Z., Liu, B. W., Ren, H., Sun, X., et al. (2019b). CmTCP20 plays a key role in nitrate and auxin signaling-regulated lateral root development in Chrysanthemum. Plant Cell Physiol. 60, 1581–1594. doi: 10.1093/pcp/pcz061
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H., and Tsay, Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7 expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., and Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68, 2463–2475. doi: 10.1093/jxb/erx011
Footitt, S., Douterelo-Soler, I., Clay, H., and Finch-Savage, W. E. (2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. U S A. 108, 20236–20241. doi: 10.1073/pnas.1116325108
Footitt, S., Huang, Z., Clay, H. A., Mead, A., and Finch-Savage, W. E. (2013). Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in 47 winter and summer annual phenotypes. Plant J. 74, 1003–1015. doi: 10.1111/tpj.12186
Forbes, K. P., Addepalli, B., and Hunt, A. G. (2006). An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281, 176–186. doi: 10.1074/jbc
Francozorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubiosomoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gaudinier, A., Rodriguez-Medina, J., Zhang, L., Olson, A., Liseron-Monfils, C., Bågman, A. M., et al. (2018). Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563, 259–264. doi: 10.1038/s41586-018-0656-3
Gifford, M. L., Dean, A., Gutierrez, R. A., Coruzzi, G. M., and Birnbaum, K. D. (2008). Cells pecific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U S A. 105, 803–808. doi: 10.1073/pnas.0709559105
Girardin, P., Tollenaar, M., and Deltour, A. (1985). Effect of temporary N starvation in maize on leaf senescence. Can. J. Plant Sci. 65, 819–829. doi: 10.4141/cjps85-108
Gras, D. E., Vidal, E. A., Undurraga, S. F., Riveras, E., Moreno, S., Dominguez-Figueroa, J., et al. (2018). SMZ/SNZ and gibberellin signaling are required for nitrate-elicited delay of flowering time in Arabidopsis thaliana. J. Exp. Bot. 69, 619–631. doi: 10.1093/jxb/erx423
Gregersen, P. L., Culetic, A., Boschian, L., and Krupinska, K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82, 603–622. doi: 10.1007/s11103-013-0013-8
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitza, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U S A. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U S A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. doi: 10.1111/j.1365-3040.2003.01158.x
Gutiérrez, R. A., Stokes, T. L., Thum, K., Xu, X., Obertello, M., Katari, M. S., et al. (2008). Systems approach identifies an organic nitrogen responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. U S A. 105, 4939–4944. doi: 10.1073/pnas.0800211105
Gutiérrez, R. A. (2012). Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. doi: 10.1126/science.1217620
Havé, M., Marmagne, A., Chardon, F., and Masclaux-Daubresse, C. (2017). Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529. doi: 10.1093/jxb/erw365
He, H., Liang, G., Li, Y., Wang, F., and Yu, D. (2014). Two young microRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol. 164, 853–865. doi: 10.1104/pp.113.228635
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47:834. doi: 10.1038/ng.3337
Huang, X., Ouyang, X., Yang, P., Lau, O. S., Li, G., Li, J., et al. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24, 4590–4606. doi: 10.1105/tpc.112.103994
Jeong, D. H., Park, S., Zhai, J., Gurazada, S. G., De Paoli, E., Meyers, B. C., et al. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23:4185e4207. doi: 10.1105/tpc.111.089045
Jia, Z., and von Wirén, N. (2020). Signaling pathways underlying nitrogen-dependent changes in root system architecture: from model to crop species. J. Exp. Bot. 71, 4393–4404. doi: 10.1093/jxb/eraa033
Jonassen, E. M., Sandsmark, B. A. A., and Lillo, C. (2009a). Unique status of NIA2 in nitrate assimilation. Plant Signal. Behav. 4, 1084–1086. doi: 10.4161/psb.4.11.9795
Jonassen, E. M., Sévin, D. C., and Lillo, C. (2009b). The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in Arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1.1. J. Plant Physiol. 166, 2071–2076. doi: 10.1016/j.jplph.2009.05.010
Kiba, T., Inaba, J., Kudo, T., Ueda, N., Konishi, M., Mitsuda, N., et al. (2018). Repression of nitrogen-starvation responses by members of the Arabidopsis GARP-Type transcription factor NIGT1/HRS1 subfamily. Plant Cell 30, 925–945. doi: 10.1105/tpc.17.00810
Kim, H., Kim, H. J., Vu, Q. T., Jung, S., McClung, C. R., Hong, S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 115, 8448–8453. doi: 10.1073/pnas.1722407115
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Kim, J. M., Kim, To, T., Ishida, J., Morosawa, T., Kawashima, M., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signaling. Nat. Commun. 4:1617. doi: 10.1038/ncomms2621
Krogan, N. J., Kim, M., Ahn, S. H., Zhong, G., Kobor, M. S., Cagney, G., et al. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell Biol. 22, 6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002
Krouk, G., Mirowski, P., LeCun, Y., Shasha, D. E., and Coruzzi, G. M. (2010). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11:R123. doi: 10.1186/gb-2010-11-12-r123
Kwon, C. S., Lee, D., Choi, G., and Chung, W. I. (2009). Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 60, 112–121. doi: 10.1111/j.1365-313X.2009.03938.x
Laporte, P., Merchan, F., Amor, B. B., Wirth, S., and Crespi, M. (2007). Riboregulators in plant development. Biochem. Soc. Trans. 35, 1638–1642. doi: 10.1042/BST0351638
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Li, H., Hu, B., Wang, W., Zhang, Z., Liang, Y., Gao, X., et al. (2016). Identification of microRNAs in rice root in response to nitrate and ammonium. J. Genet. Genomics 43:651e661. doi: 10.1016/j.jgg.2015.12.002
Li, H., Yu, M., Du, X. Q., Wang, Z. F., Wu, W. H., Quintero, F. J., et al. (2017). NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 29, 2016–2026. doi: 10.1105/tpc.16.00972
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Li, Y., Brooks, M., Yeoh-Wang, J., McCoy, R. M., Rock, T. M., Pasquino, A., et al. (2020). SDG8-mediated histone methylation and RNA processing function in the response to nitrate signaling. Plant Physiol. 182, 215–227. doi: 10.1104/pp.19.00682
Li, Z., Wang, R., Gao, Y., Wang, C., Zhao, L., Xu, N., et al. (2017). The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. N. Phytol. 216, 1205–1222. doi: 10.1111/nph.14743
Liang, G., Ai, Q., and Yu, D. (2015). Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci. Rep. 5:11813. doi: 10.1038/srep11813
Liang, G., He, H., and Yu, D. (2012). Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7:e48951. doi: 10.1371/journal.pone.0048951
Liang, Y., Zhao, X., Jones, A. M., and Gao, Y. (2018). G proteins sculp root architecture in response to nitrogen in rice and Arabidopsis. Plant Sci. 274, 129–136. doi: 10.1016/j.plantsci.2018.05.019
Lin, S. H., Kuo, H. F., Canivenc, G., Lin, C. S., Lepetit, M., Hsu, P. K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Lin, Y. L., and Tsay, Y. F. (2017). Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot. 68, 2603–2609. doi: 10.1093/jxb/erx053
Liu, F., Xu, Y., Chang, K., Li, S., Liu, Z., Qi, S., et al. (2019). The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis. N. Phytol. 224, 117–131. doi: 10.1111/nph.16038
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345. doi: 10.1105/tpc.112.102855
Liu, K. H., Diener, A., Lin, Z., Liu, C., and Sheen, J. (2020). Primary nitrate responses mediated by calcium signaling and diverse protein phosphorylation. J. Exp. Bot. 71, 4428–4441. doi: 10.1093/jxb/eraa047
Liu, W., Sun, Q., Wang, K., Du, Q., and Li, W. X. (2017). Nitrogen Limitation Adaptation (NLA) is involved in source-tosink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. N. Phytol. 214, 734–744. doi: 10.1111/nph.14396
Luo, L., Zhang, Y., and Xu, G. (2020). How does nitrogen shape plant architecture? J. Exp. Bot. 71, 4415–4427. doi: 10.1093/jxb/eraa187
Lv, Y., Liang, Z., Ge, M., Qi, W., Zhang, T., Lin, F., et al. (2016). Genomewide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genomics 17:350. doi: 10.1186/s12864-016-2650-1
Maeda, Y., Konishi, M., Kiba, T., Sakuraba, Y., Sawaki, N., Kurai, T., et al. (2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9:1376. doi: 10.1038/s41467-018-03832-6
Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. doi: 10.1038/ncomms2650
Matallana-Ramirez, L. P., Rauf, M., Farage-Barhom, S., Dortay, H., Xue, G. P., Dröge-Laser, W., et al. (2013). NAC transcription factor ORE1 and senescence induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6:14321452. doi: 10.1093/mp/sst012
Medici, A., Szponarski, W., Dangeville, P., Safi, A., Dissanayake, I. M., Saenchai, C., et al. (2019). Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 31, 1171–1184. doi: 10.1105/tpc.18.00656
Meng, S., Peng, J. S., He, Y. N., Zhang, G. B., Yi, H. Y., Fu, Y. L., et al. (2016). Arabidopsis NRT1.5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant 9, 461–470. doi: 10.1016/j.molp.2015.12.015
Meng, Y., Shao, C., Wang, H., and Jin, Y. (2012). Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomics 13:197. doi: 10.1186/1471-2164-13-197
Miin-Feng, W., Qing, T., and Jason, W. R. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
Mu, X., and Luo, J. (2019). Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell Mol. Life Sci. 76, 3753–3764. doi: 10.1007/s00018-019-03164-8
Nelissen, H., De Groeve, S., Fleury, D., Neyt, P., Bruno, L., Bitonti, M. B., et al. (2010). Plant elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. U S A. 107, 1678–1683. doi: 10.1073/pnas.0913559107
O’Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and pesponses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Oliveira, I. C., and Coruzzi, G. M. (1999). Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 121, 301–310. doi: 10.1104/pp.121.1.301
Pang, K. C., Frith, M. C., and Mattick, J. S. (2006). Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22:3. doi: 10.1016/j.tig.2005.10.003
Para, A., Li, Y., Marshall-Colón, A., Varala, K., Francoeur, N. J., Moran, T. M., et al. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 111, 10371–10376. doi: 10.1073/pnas.1404657111
Park, B. S., Yao, T., Seo, J. S., Wong, E. C. C., Mitsuda, N., Huang, C. H., et al. (2018). Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 4, 898–903. doi: 10.1038/s41477-018-0269-8.
Park, B. S., Seo, J. S., and Chua, N. H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26, 454–464. doi: 10.1105/tpc.113.120311
Park, S. H., Jeong, J. S., Seo, J. S., Park, B. S., and Chua, N. H. (2019). Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. N. Phytol. 223, 1447–1460. doi: 10.1111/nph.15879
Pathak, R. R., Jangam, A. P., Malik, A., Sharma, N., Jaiswal, D. K., and Raghuram, N. (2020). Transcriptomic and network analyses reveal distinct nitrate responses in light and dark in rice leaves (Oryza sativa indica var. Panvel1). Sci. Rep. 10:12228. doi: 10.1038/s41598-020-68917-z
Peña, P., Quach, T., Sato, S., Ge, Z., Nersesian, N., Changa, T., et al. (2017). Expression of the Maize DOF1 transcription factor in Wheat and Sorghum. Front. Plant Sci. 8:434. doi: 10.3389/fpls.2017.00434
Peng, M., Hudson, D., Schofield, A., Tsao, R., Yang, R., Gu, H., et al. (2008). Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 59, 2933–2944. doi: 10.1093/jxb/ern148
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A Wheat CCAAT box-binding transcription factor increases the grain yield of Wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U S A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Riveras, E., Alvarez, J. M., Vidal, E. A., Oses, C., Vega, A., and Gutiérrez, R. A. (2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169, 1397–1404. doi: 10.1104/pp.15.00961
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Rubin, G., Tohge, T., Matsuda, F., Saito, K., and Scheible, W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. doi: 10.1105/tpc.109.067041
Rymarquis, L. A., Kastenmayer, J. P., Huttenhofer, A. G., and Green, P. J. (2008). Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 13, 329–334. doi: 10.1016/j.tplants.2008.02.009
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Schildhauer, J., Wiedemuth, K., and Humbeck, K. (2008). Supply of nitrogen can reverse senescence processes and affect expression of genes coding for plastidic glutamine synthetase and lysine-ketoglutarate reductase/saccharopine dehydrogenase. Plant Biol. 1, 76–84. doi: 10.1111/j.1438-8677.2008.00075.x
Seo, J. S., Sun, H. X., Park, B. S., Huang, C. H., Yeh, S. D., Jung, C., et al. (2017). ELF18-INDUCED LONG NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29, 1024–1038. doi: 10.1105/tpc.16.00886
Sharma, N., Sinha, V. B., Gupta, N., Rajpal, S., Kuchi, S., Sitaramam, V., et al. (2018). Phenotyping for nitrogen use efficiency: rice genotypes differ in N-responsive germination, oxygen consumption, seed urease activities, root growth, crop duration, and yield at low N. Front. Plant Sci. 9:1452. doi: 10.3389/fpls.2018.01452
Soltabayeva, A., Srivastava, S., Kurmanbayeva, A., Bekturova, A., Fluhr, R., and Sagi, M. (2018). Early senescence in older leaves of low nitrate-grown Atxdh1 uncovers a role for purine catabolism in N supply. Plant Physiol. 178, 1027–1044. doi: 10.1104/pp.18.00795
Song, Y., Yang, C., Gao, S., Zhang, W., Lia, L., and Kuai, B. (2014). Age triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Sun, C. H., Yu, J. Q., Duan, X., Wang, J. H., Zhang, Q. Y., Gu, K. D., et al. (2018a). The MADS transcription factor CmANR1 positively modulates root system development by directly regulating CmPIN2 in chrysanthemum. Hortic. Res. 5:52. doi: 10.1038/s41438-018-0061-y
Sun, C. H., Yu, J. Q., Wen, L. Z., Guo, Y. H., Sun, X., Hao, Y. J., et al. (2018b). Chrysanthemum MADS-box transcription factor CmANR1 modulates lateral root development via homo-/heterodimerization to influence auxin accumulation in Arabidopsis. Plant Sci. 266, 27–36. doi: 10.1016/j.plantsci.2017.09.017
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Tang, W., Ye, J., Yao, X., Zhao, P., Xuan, W., Tian, Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10:5279. doi: 10.1038/s41467-019-13187-1
Teng, Y., Cui, H., Wang, M., and Liu, X. (2017). Nitrate reductase is regulated by CIRCADIAN CLOCK-ASSOCIATED1 in Arabidopsis thaliana. Plant Soil 416, 477–485. doi: 10.1007/s11104-017-3208-y
Tian, T., Ma, L., Liu, Y., Xu, D., Chen, Q., and Li, G. (2020). Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32, 1574–1588. doi: 10.1105/tpc.20.00021
Trevisan, S., Nonis, A., Begheldo, M., Manoli, A., Palme, K., Caporale, G., et al. (2012). Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 35, 1137–1155. doi: 10.1111/j.1365-3040.2011.02478.x
Tucker, D. E., Allen, D. J., and Ort, D. R. (2004). Control of nitrate reductase by circadian and diurnal rhythms in tomato. Planta 219, 277–285. doi: 10.1007/s00425-004-1213-x
Van Der Graaff, E., Schwacke, R., Schneider, A., Desimone, M., Flugge, U., and Kunze, R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. doi: 10.1104/pp.106.079293
Vidal, E. A., Álvarez, J. M., Araus, V., Riveras, E., Brooks, M. D., Krouk, G., et al. (2020). Nitrate in 2020: thirty years from transport to signaling networks. Plant Cell 32, 2094–2119. doi: 10.1105/tpc.19.00748
Vidal, E. A., Álvarez, J. M., and Gutiérrez, R. A. (2014). Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Sig. Behav. 9:e28501. doi: 10.4161/psb.28501
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Vidal, E. A., Moyano, T. C., Krouk, G., Katari, M. S., Tanurdzic, M., Mccombie, W. R., et al. (2013). Integrated RNA-seq and sRNA-seq analysis identifies novel nitrate-responsive genes in Arabidopsis thaliana roots. BMC Genomics 14:701. doi: 10.1186/1471-2164-14-701
Wang, C., Zhang, W., Li, Z., Li, Z., Bi, Y., Crawford, N. M., et al. (2018b). FIP1 plays an important role in nitrate signaling and regulates CIPK8 and CIPK23 expression in Arabidopsis. Front. Plant Sci. 9:593. doi: 10.3389/fpls.2018.00593
Wang, W. H., Köhler, B., Cao, F. Q., Liu, G. W., Gong, Y. Y., Sheng, S., et al. (2012). Rice DUR3 mediates highaffinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 193, 432–444. doi: 10.1111/j.1469-8137.2011.03929.x
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018a). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wei, L. H., Song, P., Wang, Y., Lu, Z., Tang, Q., Yu, Q., et al. (2018). The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30, 968–985. doi: 10.1105/tpc.17.00934
Widiez, T., El-Kafafia, E., Girin, T., Berr, A., Ruffel, S., Krouk, G., et al. (2011). HIGH NITROGEN INSENSITIVE 9 (HNI9)-mediated systemic repression of root NO3– uptake is associated with changes in histone methylation. Proc. Natl. Acad. Sci. U S A. 108, 13329–13334. doi: 10.1073/pnas.1017863108
Wu, J. F., Tsai, H. L., Joanito, I., Wu, Y. C., Chang, C. W., Li, Y. H., et al. (2016). LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7:13181. doi: 10.1038/ncomms13181
Xin, M., Wang, Y., Yao, Y., Song, N., Hu, Z., Qin, D., et al. (2011). Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61. doi: 10.1186/1471-2229-11-61
Xu, N., Wang, R., Zhao, L., Zhang, C., Li, Z., Lei, Z., et al. (2016). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28, 485–504. doi: 10.1105/tpc.15.00567
Xu, Z., Zhong, S., Li, X., Li, W., Rothstein, S. J., Zhang, S., et al. (2011). Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS One 6:e28009. doi: 10.1371/journal.pone.0028009
Yan, D., Easwaran, V., Chau, V., Okamoto, M., Ierullo, M., Kimura, M., et al. (2016). NIN-like protein 8 is a master regulator of nitrate promoted seed germination in Arabidopsis. Nat. Commun. 7:13179. doi: 10.1038/ncomms13179
Yan, Y., Wang, H., Hamera, S., Chen, X., and Fang, R. (2014). miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 78, 44–55. doi: 10.1111/tpj.12446
Yoh, S. M., Cho, H., Pickle, L., Evans, R. M., and Jones, K. A. (2007). The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21, 160–174. doi: 10.1101/gad.1503107
Yoh, S. M., Lucas, J. S., and Jones, K. A. (2008). The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22, 3422–3434. doi: 10.1101/gad.1720008
Yuan, S., Zhang, Z. W., Zheng, C., Zhao, Z. Y., Wang, Y., Feng, L. Y., et al. (2016). Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc. Natl. Acad. Sci. U S A. 113, 7661–7666. doi: 10.1073/pnas.1602004113
Yue, H., Nie, X., Yan, Z., and Weining, S. (2019). N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol. J. 17, 1194–1208. doi: 10.1111/pbi.13149
Zhang, H., and Forde, B. G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. doi: 10.1126/science.279.5349.407
Zhang, J., Fengler, K. A., Van Hemert, J. L., Gupta, R., Mongar, N., Sun, J., et al. (2019). Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biotechnol. J. 17, 2272–2285. doi: 10.1111/pbi.13139
Zhang, L., Fletcher, A. G., Cheung, V., Winston, F., and Stargell, L. A. (2008). Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol. Cell Biol. 28, 1393–1403. doi: 10.1128/MCB.01733-07
Zhang, Y., Wang, Y., Wei, H., Li, N., Tian, W., Chong, K., et al. (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11, 326–337. doi: 10.1016/j.molp.2017.12.017
Zhao, L., Zhang, W., Yang, Y., Li, Z., Li, N., Qi, S., et al. (2018). The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. 8:1487. doi: 10.1038/s41598-018-20038-4
Zhao, M., Ding, H., Zhu, J. K., Zhang, F., and Li, W. X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. N. Phytol. 190, 906–915. doi: 10.1111/j.1469-8137.2011.03647.x
Zhao, Y., Xu, Z., Mo, Q., Zou, C., Li, W., Xu, Y., et al. (2013). Combined small RNA and degradome sequencing reveals novel miRNAs and their targets in response to low nitrate availability in maize. Ann. Bot. 112, 633–642. doi: 10.1093/aob/mct133
Keywords: nitrate regulation, leaf senescence, circadian clock, post-transcriptional regulation, non-coding RNA
Citation: Fan H, Quan S, Qi S, Xu N and Wang Y (2020) Novel Aspects of Nitrate Regulation in Arabidopsis. Front. Plant Sci. 11:574246. doi: 10.3389/fpls.2020.574246
Received: 19 June 2020; Accepted: 18 November 2020;
Published: 10 December 2020.
Edited by:
Nandula Raghuram, Guru Gobind Singh Indraprastha University, IndiaReviewed by:
Silvia Quaggiotti, University of Padua, ItalyAsdrubal Burgos, University of Guadalajara, Mexico
Copyright © 2020 Fan, Quan, Qi, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Wang, d2FuZ3lvbmdAc2RhdS5lZHUuY24=
 Hongmei Fan
Hongmei Fan Shuxuan Quan1
Shuxuan Quan1 Shengdong Qi
Shengdong Qi Na Xu
Na Xu Yong Wang
Yong Wang